Abstract
Background
The metabolism of cancer cells is often reprogrammed by dysregulation of metabolic enzymes. Transketolase-like 1 (TKTL1) is a homodimeric transketolase linking the pentose-phosphate pathway with the glycolytic pathway. It is generally silenced at a transcriptional level in somatic tissues. However, in human cancers its expression is associated with the acquisition of a glycolytic phenotype (the Warburg effect) by cancer cells that contributes to the progression of malignant tumors. In melanoma, defective promoter methylation results in the expression of genes and their products that can affect the tumor cell’s phenotype including the modification of immune and functional characteristics. The present study evaluates the role of TKTL1 as a mediator of disease progression in melanoma associated with a defective methylation phenotype.
Methods
The expression of TKTL1 in metastatic melanoma tumors and cell lines was analysed by qRT-PCR and immunohistochemistry. The promoter methylation status of TKTL1 in melanoma cells was evaluated by quantitative methylation specific PCR. Using qRT-PCR, the effect of a DNA demethylating agent 5-aza-2’-deoxycytidine (5aza) on the expression of TKTL1 was examined. Biochemical and molecular analyses such as glucose consumption, lactate production, invasion, proliferation and cell cycle progression together with ectopic expression and siRNA mediated knockdown were used to investigate the role of TKTL1 in melanoma cells.
Results
Expression of TKTL1 was highly restricted in normal adult tissues and was overexpressed in a subset of metastatic melanoma tumors and derived cell lines. The TKTL1 promoter was activated by hypomethylation and treatment with 5aza induced TKTL1 expression in melanoma cells. Augmented expression of TKTL1 in melanoma cells was associated with a glycolytic phenotype. Loss and gain of function studies revealed that TKTL1 contributed to enhanced invasion of melanoma cells.
Conclusions
Our data provide evidence for an important role of TKTL1 in aerobic glycolysis and tumor promotion in melanoma that may result from defective promoter methylation. This epigenetic change may enable the natural selection of tumor cells with a metabolic phenotype and thereby provide a potential therapeutic target for a subset of melanoma tumors with elevated TKTL1 expression.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2185-5) contains supplementary material, which is available to authorized users.
Keywords: TKTL1, Warburg effect, Aerobic glycolysis, Melanoma, Hypomethylation
Background
Melanoma is a cancer derived from neuroectoderm that is often fatal once it has metastasised [1, 2]. Current treatment approaches include the molecular targeting of oncogenic gene products and immune-based therapies, both of which induce significant responses in many patients with advanced disease [3, 4]. Despite this, many patients develop therapy resistance or do not respond to treatment [5–7]. Since alterations in cellular metabolism may contribute to the malignant phenotype, targeting the metabolic circuitry of melanoma cells may offer a promising additional therapeutic strategy [8, 9].
The reprogramming of cellular metabolism is one of the hallmarks of cancer [10]. Whereas normal cells direct glucose to mitochondrial oxidative phosphorylation to generate ATP when oxygen is abundant, cancer cells generally exhibit greater glucose uptake and lactate secretion, regardless of oxygen availability. This phenomenon is termed “aerobic glycolysis” or the “Warburg effect” [10–12]. This can benefit cancer cells by facilitating increased proliferation, enhanced invasion and resistance to apoptosis, which in turn promote tumor progression and metastasis [13]. These observations have raised the possibility that targeting metabolic pathways that the cancer cell depends upon may be a useful therapeutic strategy. Metabolic profiling of melanoma cells has revealed their dependence upon the Warburg effect as the major source of energy [14]. Moreover, this phenomenon is the basis for 18FDG-PET (Fluorodeoxy glucose positron emission tomography) to image the metabolic activity of cancer in patients [15].
Transketolases are essential and rate-limiting enzymes in the non-oxidative part of the pentose-phosphate pathway [16, 17]. An augmented pentose-phosphate pathway in some tumors enables oxygen-independent glucose conversion to ribose for nucleic acid synthesis, glucose degradation to lactate, and regeneration of redox equivalents [18]. The transketolase family includes Transketolase (TKT), and two Transketolase-like proteins (TKTL1 and TKTL2) [19]. Among them, TKTL1 is the isoform specifically upregulated in different human cancers such as head and neck, lung, breast, stomach, colon, nephroblastoma and endometrial cancer [20–26]. Its overexpression predicts poor patient survival, tumor recurrence and resistance to chemo and radiation therapy in many cancers [23, 27, 28]. TKTL1 expression was demonstrated in invasive tumors which correlated with increased metastasis [29]. Some of the tumor types expressing TKTL1 have responded to the transketolase inhibitor Oxythiamine that effectively blocked cell proliferation [30, 31]. TKTL1 has recently been used as a biomarker in a blood test based on the epitope detection in monocytes (EDIM) technology allowing for the non-invasive detection of neoplasia and tumor recurrence [32]. Although TKTL1 expression in conjunctival melanoma tumors has been reported, the mechanism by which expression becomes unregulated and the functional consequences of aberrant TKTL1 expression in melanoma remains unclear and these warrant further investigation [33].
Methylation defects are commonly observed in melanoma and this can result in the expression of gene products that are otherwise silenced at a transcriptional level in somatic tissues. This can profoundly affect the phenotype of the cancer cell. Among these re-expressed protein are the Cancer Testis or Cancer Germ-line family of antigens (CTAg); so called because they are often immunogenic and can mediate the immune rejection of cancer [34]. They are associated with disease progression and may be associated with poorer clinical outcomes in melanoma and other cancers [35, 36]. Many of these are coded on the X chromosome and their function is poorly understood although the association with stem-like characteristics has been reported [37]. Given that TKTL1 is also an X chromosome coded molecule (Xq28) that, like the CTAg, is generally repressed in somatic tissues, we sought to determine if DNA hypomethylation also induced aberrant expression of TKTL1 in melanoma and to assess its role in promoting the Warburg effect in melanoma cells.
We detected increased expression of TKTL1 in a subset of metastatic melanoma tumors and cell lines and found TKTL1 expression in melanoma tumors was associated with promoter hypomethylation. We demonstrated that the TKTL1 promoter could be activated by treatment with 5-aza-2’-deoxycytidine (5aza) thereby inducing TKTL1 expression in melanoma cells. Elevated TKTL1 expression enhanced the Warburg effect by accelerating glucose utilisation and lactate production and TKTL1 loss and gain of function studies revealed that TKTL1 enhanced invasion in melanoma cells. Taken together, our data suggests that a subset of melanomas with defective methylation rely on TKTL1-dependent aerobic glycolysis and have enhanced tumorigenesis. These may be amenable to inhibition of the Warburg effect by therapies that target TKTL1.
Results
A subset of metastatic melanoma tumors and cell lines express TKTL1
We quantified TKTL1 transcripts by qRT-PCR in a panel of normal human tissues and metastatic melanoma tumors. Figure 1a shows that high expression of TKTL1 mRNA was detected in testis but no other normal human tissues tested including skin and melanocytes. 15 of 38 (40 %) melanoma tumors assessed expressed TKTL1 to varying degrees. We examined TKTL1 by immunohistochemistry using a tissue microarray (TMA) comprising 81 tumors from patients with stage III and IV metastatic melanoma. Four representative tumors with high and diffuse intensity staining of tumor cells for TKTL1 are depicted in Fig. 1b. TKTL1 in tumors varied from homogenous to heterogeneous expression with clusters of TKTL1 positive cells interspersed with TKTL1 negative cells. TKTL1 expression in testis tissue was used as positive control and anti-IgG antibody was used as negative control (Fig. 1b). 31 of 81 (38 %) of metastatic melanoma tumors were scored positive for TKTL1 expression (Fig. 1c). High magnification image shows that TKTL1 localization is predominantly cytosolic and membrane staining is also seen in some melanoma tumor cells. Nuclear staining in some tumors is Melanin (brown) and not TKTL1 (red) (Additional file 1: Figure S1A). We extended our studies to a clinical outcome dataset that subdivided 57 stage IV melanoma samples into high-risk or low-risk melanoma groups based on transcriptome profiling (GSE22153) [38]. We found that tumors expressing high TKTL1 levels were statistically significantly associated with the high-risk group (log-rank p value 0.0277), risk being reduced survival and risk of relapse.
Fig. 1.
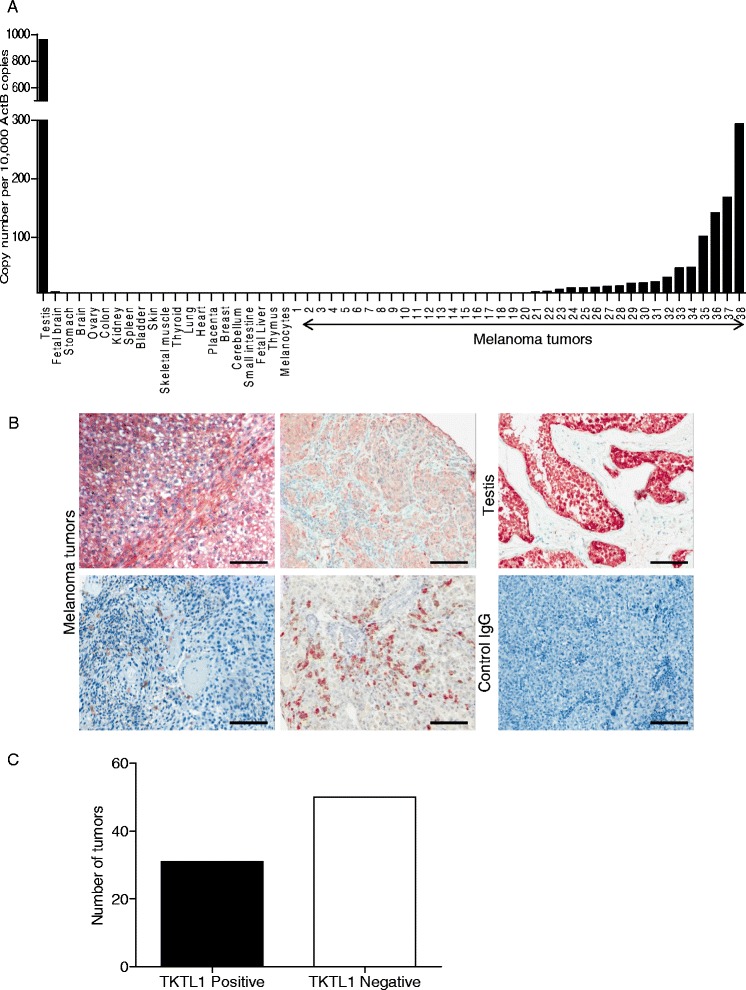
TKTL1 is highly expressed in human testis and melanoma tumors. a qRT-PCR was employed to measure the expression of TKTL1 in a panel of normal human tissues and in 38 metastatic melanoma tumor samples. b TKTL1 immunohistochemical staining in testis as positive control and control IgG staining in tumors as negative control are demonstrated. Representative staining patterns for TKTL1 in metastatic melanoma tumors are shown. Original magnification, 10 ×. c Graph shows number of TKTL1 positive and negative tumors
In a panel of 53 metastatic melanoma cell lines TKTL1 mRNA was detectable in 11 of 53 lines (20 %) by qRT-PCR (Additional file 1: Figure S1B). We selected two cell lines for further functional studies; LM-MEL-59, which showed the highest level of TKTL1 mRNA expression and LM-MEL-44 which lacked TKTL1 expression (Fig. 2a). Immuno-staining confirmed the expression of TKTL1 in LM-MEL-59 and its absence in LM-MEL-44 (Fig. 2b). Both of these cell lines lack activating mutations in the BRAF protein kinase. High magnification image confirms cytosolic TKTL1 localization (Additional file 1: Figure S1C).
Fig. 2.
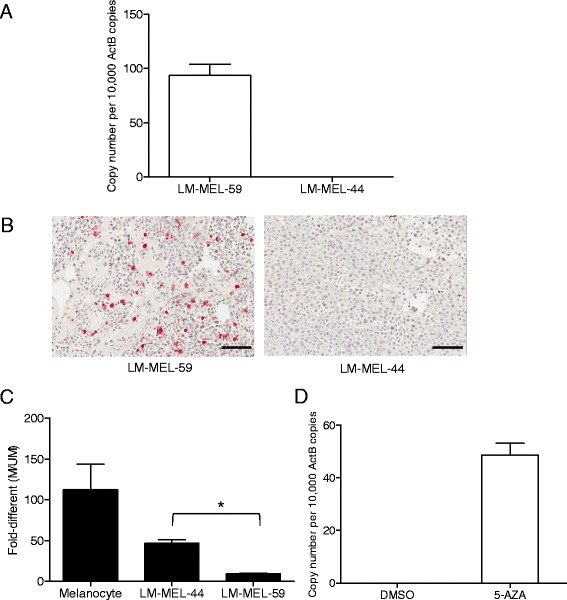
TKTL1 is expressed in metastatic melanoma cell lines and is regulated by promoter hypomethylation. a Using qRT-PCR the expression of TKTL1 in two melanoma cell lines, LM-MEL-59 and LM-MEL-44 was determined. b TKTL1 expression in LM-MEL-59 and LM-MEL-44 was determined by IHC. Original magnification, 20×. c Bisulfite conversion and MS-qPCR was performed to assay the promoter methylation status of TKTL1 in melanocytes and metastatic melanoma cells. Relative levels of methylated and unmethylated products (M/UM) was quantified. d QRT-PCR was employed to detect changes in expression of TKTL1 by 5-azacytidine treatment in LM-MEL-44 cell line. Values are ± SD of three independent experiments in triplicate (*, p < 0.05)
Promoter hypomethylation of the TKTL1 gene is responsible for its activation in melanoma
Expression of TKTL1 has been reported to be controlled by methylation and demethylation of TKTL1 has been previously reported in lung and head and neck cancers [24, 39]. However, little is known about the methylation status of TKTL1 in melanoma. To investigate promoter cytosine-(phosphate)-guanine (CpG) dinucleotide DNA methylation, we quantified the level of TKTL1 promoter methylation using bisulfite modification and MS-qPCR. Melanocytes were found to be hypermethylated which correlated with the lack of TKTL1 expression in those cells (Fig. 2c), while metastatic melanoma cell lines tested showed reduced methylation levels when compared to melanocytes. LM-MEL-44 cell line with no TKTL1 expression showed hypermethylation of the TKTL1 promoter whereas a lesser degree of TKTL1 promoter methylation was detected in LM-MEL-59 with high TKTL1 mRNA expression. Promoter hypermethylation of TKTL1 could be reversed by a DNA demethylating agent 5-aza-2′-deoxycytidine (5aza) in LM-MEL-44 resulting in the expression of TKTL1 in treated LM-MEL-44 cells in contrast to DMSO treated cells (Fig. 2d).
We extended our studies to a publically available clinical melanoma patient dataset to determine whether deregulation of TKTL1 expression was linked to DNA hypomethylation. Using MethPrimer (www.urogene.org/cgi-bin/methprimer/methprimer.cgi), a CpG island prediction program, we identified two CpG sites in the TKTL1 promoter that were highly prone to methylation (data not shown). The primers we designed for MS-qPCR spanned these two CpG sites; cg09892236 (−100 bp of transcription start site (TSS)) and cg23106779 (−250 bp of TSS). We used an in silico approach to assess the methylation status of the promoter region on the TKTL1 gene in 385 metastatic melanoma tissues from the Skin Cutaneous Melanoma (SKCM) dataset of The Cancer Genome Atlas (TCGA) database (www.cbioportal.org) [40, 41] and estimated its association with TKTL1 mRNA expression. We first evaluated DNA methylation of the two CpG sites using the Illumina Infinium Human Methylation 450 k BeadChip array (450 k array). Methylation of the two CpG sites were statistically associated with each other with the melanoma dataset (correlation = 0.68, p-value = 2.2e-16). We next examined the exon and gene expression pattern of TKTL1 in these patient samples in the Illumina Hiseq RNA-Seq Platforms. Spearman correlation coefficients were calculated to test the correlations between DNA methylation and gene expression of TKTL1. We found that methylation status at both the CpG sites was statistically inversely correlated with TKTL1 gene expression in melanoma samples (Table 1 and Additional file 2: Figure S2A&B). Melanoma samples were further sub-divided into two groups, primary (n = 82) and metastatic (n = 303) in order to examine the role of DNA methylation in regulation of TKTL1 gene expression within primary and metastatic melanoma samples. We found positive correlation of TKTL1 expression with methylation in metastatic samples (Table 2). These results confirm that DNA hypomethylation is associated with aberrant activation of TKTL1 expression in metastatic melanoma.
Table 1.
Integrative analysis of methylation and gene expression in Melanoma samples (TCGA data analysis)
| Probe Id (450 k) | Methylation and gene expression (n = 385) | Methylation and exon expression (n = 385) | ||
|---|---|---|---|---|
| Rho | P-value | Rho | P-value | |
| cg09892236 | −0.14 | 0.005 | −0.16 | 0.0024 |
| cg23106779 | −0.11 | 0.04 | −0.13 | 0.01 |
Table 2.
Integrative analysis of methylation and gene expression in primary and metastatic samples (TCGA data analysis)
| Probe Id (450 K) | Methylation and gene expression | Methylation and exon expression | ||||||
|---|---|---|---|---|---|---|---|---|
| Primary (n = 82) | Metastatic (303) | Primary (82) | Metastatic (303) | |||||
| Rho | P-value | Rho | P-value | Rho | P-value | Rho | P-value | |
| cg09892236 | −0.097 | 0.38 | −0.15 | 0.006 | −0.10 | 0.32 | −0.17 | 0.004 |
| cg23106779 | −0.10 | 0.36 | −0.09 | 0.09 | −0.16 | 0.13 | −0.11 | 0.05 |
TKTL1 is co-expressed with Cancer Testis Antigens in melanoma tumors
The epigenetic reactivation of TKTL1 occurs simultaneously with DNA hypomethylation of CTAgs in head and neck cancer [39]. To determine whether TKTL1 expression in melanoma was linked to the expression of CTAgs, we queried the TCGA database comprising of 261 melanoma patient samples. We found a significant tendency toward co-occurrence of many CTAgs with TKTL1 expression in melanoma tumors (Table 3).
Table 3.
Gene expression correlation p value for the co-occurrence of each gene pair across 261 melanoma patient tumors
| Cancer Testis Antigen | Gene symbol | TKTL1 (Adjusted p value) |
|---|---|---|
| Ropporin | ROPN1 | 8.33E-08 |
| Testis-specific serine kinase 6 | TSSK6 | 0.001107 |
| Transcription factor DP family, member 3 | TFDP3 | 0.058881 |
| Sperm autoantigenic protein 17 | SPA17 | 0.061708 |
| Armadillo repeat containing 3 | ARMC3 | 0.061708 |
| Testis expressed 15 | TEX15 | 0.11398 |
| Synovial sarcoma, X breaking point 1 | SSX1 | 0.201966 |
| Centrosomal protein 290 kDa | CEP290 | 0.254303 |
| Synovial sarcoma, X breaking point 5 | SSX5 | 0.280049 |
Altered expression of TKTL1 affects the glycolytic metabolism of melanoma cells
To determine the role of TKTL1 in melanoma, we sought to manipulate TKTL1 levels in melanoma cell lines by siRNA-mediated knockdown and vector-mediated over-expression. Effective suppression of TKTL1 on mRNA and protein levels in LM-MEL-59 was achieved at 10 nM concentration of siRNA (Fig. 3a & b). To confirm that the effects shown by TKTL1 were not off-target effects, we performed microarray [42] and qRT-PCR for the isoforms of TKTL1 namely, TKT and TKTL2. We found that TKT levels were high in both LM-MEL-59 and LM-MEL-44, while TKTL2 levels were low. Additionally, we have used testis tissue as a positive control for the expression of these genes. Upon knockdown with TKTL1 siRNA, TKT levels were affected by one of the siRNAs but only modestly by the other (Additional file 3: Figure S3).
Fig. 3.
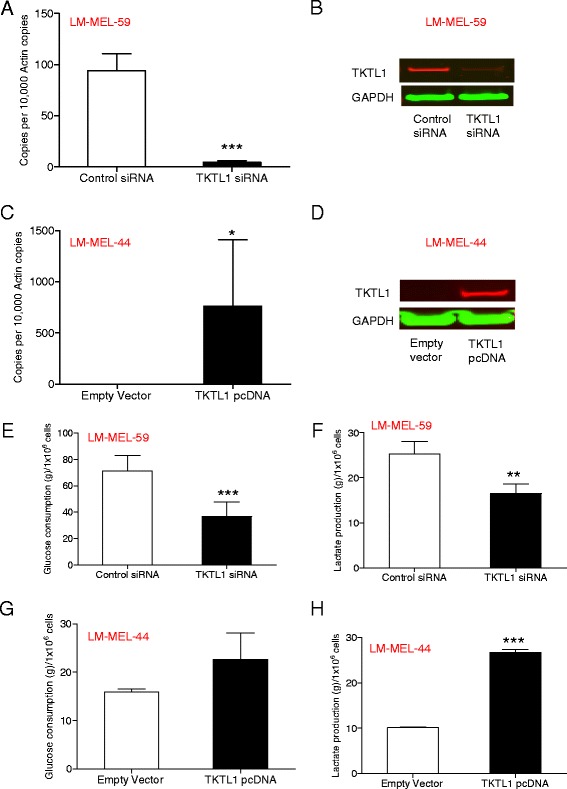
Ectopic overexpression and knockdown of TKTL1 influences the Warburg effect in melanoma cells. a QRT-PCR was employed to evaluate the expression of TKTL1 in LM-MEL-59 after treatment with either TKTL1 or control siRNA for 72 h. b Western Blotting with a mouse monoclonal anti-TKTL1 antibody showed reduction in TKTL1 levels after siRNA treatment in LM-MEL-59 after 72 h. GAPDH was used a loading control. c TKTL1 expression was assessed by qRT-PCR in LM-MEL-44 cells transfected with TKTL1 pcDNA or empty control for 72 h. d Immunoblot of TKTL1 confirmed expression of TKTL1 after transfection of a TKTL1 expression vector in LM-MEL-44. Blots were probed with GAPDH as a control for loading and transfer. Glucose consumption was measured in cell free supernatant of e LM-MEL-59 cell line following treatment with TKTL1 or control siRNA for 72 h and g LM-MEL-44 cell line following over-expression of TKTL1 or empty vector control for 72 h. The production of lactate in culture supernatants was measured in f LM-MEL-59 after knockdown of TKTL1 for 72 h and h LM-MEL-44 after treatment with TKTL1 pcDNA or empty vector control for 72 h. Values are ± SD of three experiments in triplicate (**, p < 0.005, ***, p < 0.0005)
Transfection with a TKTL1 expression plasmid resulted in marked overexpression of TKTL1 in LM-MEL-44, demonstrated by qRT-PCR and immunoblotting (Fig. 3c & d). No additional bands were detected for TKTL1 as evident in Additional file 4: Figure S4A&B. High transfection efficiency was also confirmed with immunofluorescence staining (Additional file 4: Figure S4C). Elevated glucose uptake and lactate production have been detected during malignant transformation. These metabolic parameters of cancer cells are consistent with a pronounced glycolytic state [43, 44]. To investigate whether TKTL1 expression in melanoma was driving the Warburg effect, glucose consumption and lactate production were assayed after TKTL1 interference. Measurements were made on conditioned medium of LM-MEL-59 cells after treatment with TKTL1 or control siRNA for 72 h. Depletion of TKTL1 in these cells caused a substantial decrease in glucose consumption and associated reduction in lactate production (Fig. 3e & f) while ectopic expression of TKTL1 in LM-MEL-44 conferred a more glycolytic phenotype (Fig. 3 g & h). Collectively, these results showed that TKTL1 enhanced aerobic glycolysis in melanoma.
TKTL1 affects proliferation in melanoma cells
TKTL1 plays an important role in tumor proliferation in gastric, colon and hepatocellular carcinomas [19, 26, 45]. We investigated whether TKTL1 plays a similar role in melanoma. TKTL1 knockdown in LM-MEL-59 led to an increase in the percentage of G0-G1 phase cells and a decrease in the percentage of S phase cell population compared to control transfected cells (Fig. 4a, b & e) as demonstrated by cell cycle analysis. Using MTS assay to determine total cell number after TKTL1 knockdown, similar changes were detected. However, effects were only modest after 48 h (Additional file 5: Figure S5A).
Fig. 4.
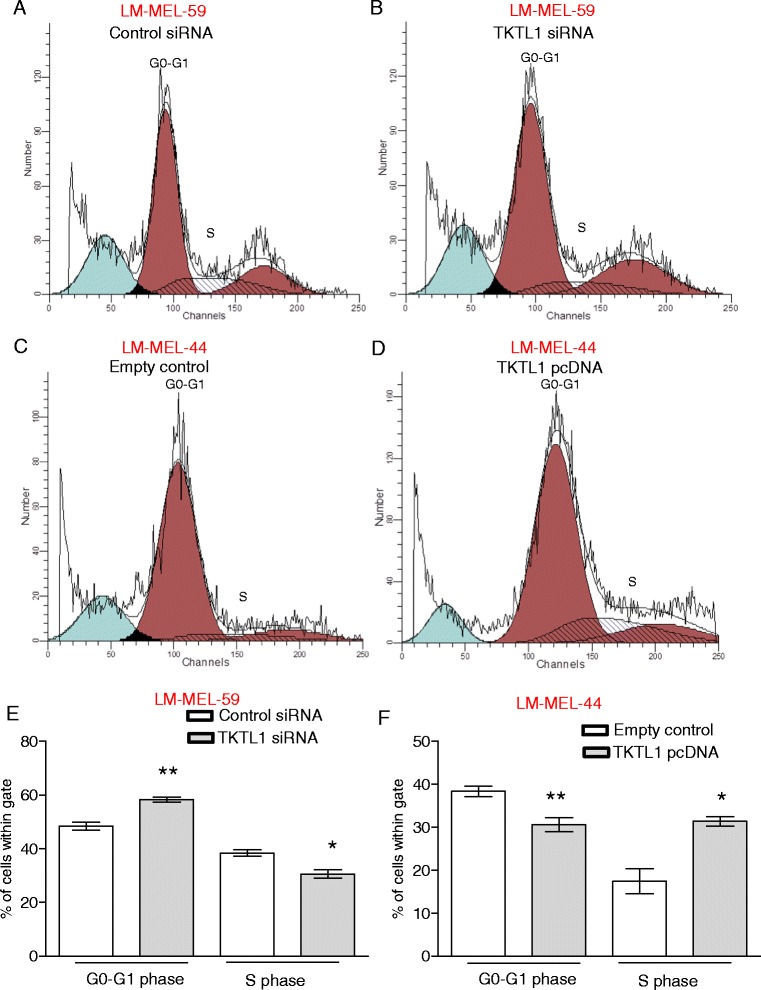
Loss of TKTL1 expression changes cell cycle distribution of melanoma cells. Cell cycle phases were determined by propidium iodide staining of melanoma cells and subsequent flow cytometric analysis. A representative histogram of cell cycle analysis of LM-MEL-59 is shown after a control siRNA treatment and b TKTL1 siRNA treatment. Analysis of percentage of cells in e G0-G1 phase and S phase cell after treatment of LM-MEL-59 with TKTL1 or control siRNA. Values are ± SD of three experiments in triplicate (*, p < 0.05, **, p < 0.005). Histograms depicting distribution of cell cycle phase in LM-MEL-44 after treatment with c empty vector control and d TKTL1 pcDNA. Cell cycle distribution of f GO-G1 and S phase cell population after 48 h of ectopic expression of TKTL1 or empty vector in LM-MEL-44 was performed. Values are ± SD of three experiments in triplicate (*, p < 0.05, **, p < 0.005)
Conversely, TKTL1 overexpression led to a decrease in the percentage of G0-G1 phase cells in LM-MEL-44 and an increase in the percentage of S phase cells (Fig. 4c, d & f). Table 4 depicts the cell cycle phase distributions under the different conditions. Similarly, MTS assay demonstrated slightly enhanced proliferation in TKTL1 overexpressing cells compared to empty vector treated cells, although no significance was achieved (Additional file 5: Figure S5B). Overall, these results indicate that TKTL1 levels may affect melanoma cell proliferation.
Table 4.
Comparison of cell cycle distribution after TKTL1 knockdown and overexpression
| Treatment (LM-MEL-59) | Sub G1 (%) | G0-G1 (%) | S (%) | G2-M (%) |
| Control siRNA | 8.27 | 48.26 | 38.84 | 12.89 |
| TKTL1 siRNA | 7.9 | 58.17 | 30.71 | 11.11 |
| p Value | 0.78 | 0.004 | 0.01 | 0.65 |
| Treatment (LM-MEL-44) | Sub G1 (%) | G0-G1 (%) | S (%) | G2-M (%) |
| Empty vector control | 18.05 | 79.55 | 17.39 | 3.05 |
| TKTL1 pcDNA | 8.89 | 65.55 | 31.42 | 3.05 |
| p Value | 0.02 | 0.001 | 0.01 | 0.99 |
TKTL1 promotes invasion in melanoma cells
Next, we sought to investigate the effect of TKTL1 on cell invasion. TKTL1 depleted LM-MEL-59 cells were significantly less invasive, while TKTL1 overexpressing LM-MEL-44 cells were more invasive than controls in the matrigel-coated Boyden chamber assay (Fig. 5a & c). TKTL1 depleted cells showed 62 % loss and TKTL1 overexpressing cells showed 38 % increase in invasion (Fig. 5b & d). These results demonstrate a substantive effect of TKTL1 levels on melanoma cellular invasion.
Fig. 5.
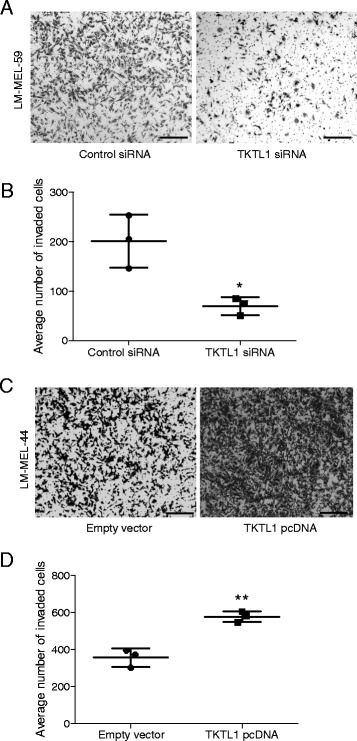
TKTL1 enhances invasive behaviour in melanoma. Melanoma cells were plated out and transfected with either 10nM control siRNA or TKTL1 specific siRNA and subjected to invasion assay. a Representative images of invasion of LM-MEL-59 is shown (scale bar = 100 μm). b The graph shows the total number of invasive cells counted. c Invasiveness of melanoma cells LM-MEL-44 after treatment with empty vector control or TKTL1 pcDNA was tested, images captured (scale bar = 100 μm) and d invasion was quantified as above. Values are mean ± SD of three independent experiments in triplicate (*p < 0.05, **p < 0.005)
Oxythiamine does not inhibit proliferation and invasion in melanoma cells
In tumors expressing high TKTL1, oxythiamine has been used to block proliferation effectively both in vitro and in vivo [46]. We treated both LM-MEL-59 and LM-MEL-44 with the inhibitor in varying dose of 10 and 100 μM (Additional file 6: Figure S6A&B). In both cell lines no reduction in proliferation was detected. To assess if Oxythiamine induced a G1 phase cell cycle arrest, LM-MEL-59 cells treated for 72 h with 100 μM of the inhibitor was subjected to flow cytometric cell cycle analysis. However, Oxythiamine treatment in our hands did not cause a G1 phase arrest in melanoma cells (Additional file 6: Figure S6C). Furthermore, Oxythiamine treatment did not alter invasive potential (Additional file 6: Figure S6D & E).
Discussion
Recent studies have shown that TKTL1 plays an important role in the development and progression of human tumors [20–26, 29, 47, 48]. In this study, we report that TKTL1 expression was upregulated in a significant subset of metastatic melanoma tumors and cell lines. In agreement with other reports, we observed that increased TKTL1 resulted in accelerated glucose uptake and lactate secretion in melanoma cells and this was associated with changes in the proliferation and invasion in melanoma cells. Moreover, we could demonstrate that the TKTL1 promoter is hypomethylated in melanoma cells compared with melanocytes and consequently TKTL1 expression could be induced by incubation of melanoma cells with 5aza. We found a significant association of hypomethylation with TKTL1 gene expression in melanoma patient samples. Additionally, to our knowledge, this is the first study evaluating the effect of TKTL1 expression on metabolic and cellular functions in melanoma.
Abnormal DNA methylation dysregulates gene expression in cancer and this has been associated with changes in phenotype and function. In melanoma X-chromosome coded molecules that can be affected include the CTAg and TKTL1. Simultaneous reactivation of TKTL1 with CTAg such as the melanoma antigen family (MAGE-A) has been reported in lung and head and neck cancers [39]. In melanoma tumors, we have identified that TKTL1 and CTAgs showed significant correlation in expression. Most of these CTAgs have been implicated in tumorigenesis [49–52]. Some of these CTAgs such as Ropporin, TSSK6, SPA17 and TEX15 have been reported to play important roles in sperm motility and knockout mice lacking these genes are often sterile due to profound impairment in sperm motility [53–56]. CTAgs are repressed by epigenetic mechanisms during development and it has been suggested that the coordinated pathologic reversal of developmental epigenetic events often occurs in cancer cells [39]. We speculate that in melanoma aberrant demethylation occurs as a common underlying mechanism that links transcriptional activation of TKTL1 and CTAgs that may cooperate to promote cancer progression by mechanisms such as Warburg effect, motility, proliferation and apoptosis. Aberrant expression of CTAgs has previously been associated with poor outcomes in melanoma and other cancers [34, 36]. It has never been clear why increased expression of these antigens should paradoxically carry a poorer prognosis. This study may provide the clue, that increased antigen expression and TKTL1 expression are both a consequence of a phenotype that results from methylation changes, and it is this phenotype that associates pro-tumorigenic metabolic changes with bystander tumor antigen expression.
Our finding that a subset of metastatic cutaneous melanoma express TKTL1 at the mRNA and protein levels is consistent with the report of Lange et al., showing that conjectival melanoma tumors expressed TKTL1 [33]. We also noted elevated levels of TKTL1 in normal testis tissue, where aerobic glycolysis has been previously detected [57]. Although the Warburg effect appears to be a wasteful form of metabolism compared to the mitochondrial driven oxidative phosphorylation, it is a mechanism for rapid generation of energy [58].
Heterogeneity within LM-MEL-59 for TKTL1 expression is representative of most of the tumors that stained for TKTL1. Ho J et al. reported two distinct populations of cells that rely on either glycolysis or oxidative phosphorylation exist within melanoma tumors and can be metabolically linked such that lactate from the glycolytic portion of the tumor helps fuel ATP production through the oxidative phosphorylation in a process termed metabolic symbiosis [59]. It is conceivable that TKTL1 positive cells may rely solely on the Warburg effect in contrast to the TKTL1 negative cells that may rely on mitochondrial oxidative phosphorylation to support cell growth. Metabolic symbiosis of TKTL1 positive versus negative cells warrants further investigation using purified cell populations.
Recent studies have reported oxidative respiration plays an important role in melanoma metabolism in addition to glycolysis in vitro [14, 59, 60]. Activating mutations in the BRAF protein kinase are the most common genetic alterations in melanoma, found in around 40 % of tumors. Many studies have shown a role for BRAF signalling in the regulation of melanomas metabolism [8, 61, 62]. Melanomas treated with agents that target oncogenic BRAF undergo a reversal of the Warburg effect and become dependent on oxidative phosphorylation [61, 62]. However, both cell lines used to investigate the role of TKTL1 in this study lack the activating mutations in BRAF, so BRAF mutations do not appear to be a prerequisite for TKTL1-driven aerobic glycolysis. This is consistent with TKTL1 being activated through an epigenetic mechanism that is independent of BRAF mutational status. A glycolytic profile of melanoma cell lines has recently been identified by metabolic profiling and the study reported that melanoma cells were inherently more glycolytic than melanocytes and could adapt their metabolic program to environmental challenges and also found that all melanoma cells evaluated maintained functional mitochondria [14]. This does not invalidate a role for the Warburg effect, and suggests that metabolic flexibility may occur, enabling a survival advantage for tumors growing in diverse environments. The mechanisms that tip the balance towards more glycolysis and less respiration in tumors are emerging.
In circumstances where glucose metabolism exceeds the capacity of a cell to assimilate or store glucose-derived carbon, cells with high levels of TKTL1 expression have a growth advantage when compared to those with low levels of TKTL1. A correlation between glucose uptake and proliferation has been demonstrated in several cell lines, for instance in ovarian adenocarcinoma and breast cancer cells [63, 64]. Our finding that TKTL1 expression enhances proliferation is consistent with these findings. The important question is whether this reflects a dependence on TKTL1 as a mechanism for tumor progression or survival. If so, it may be a useful therapeutic target, in those cases where it is overexpressed.
Langbein et al. examined 70 colon cancers and found increased TKTL1 protein levels in invasive tumors compared to non-invasive tumors and healthy tissue [17]. High levels of TKTL1 were also correlated with invasion and poor prognosis in urothelial, laryngeal squamous and colorectal cancer [17, 29]. Our observation that TKTL1 promotes invasiveness in vitro indicates that the metabolic pathways that support tumor progression are likely to be complex. Cells with increased TKTL1 showed increased rate of glycolysis with production of high levels of lactate. Cancer cells in general seem to adapt to the acidic conditions better than surrounding cells and this may promote the degradation of host matrix leading to greater invasiveness and thereby contributing to metastases. Furthermore, the acidic microenvironment can enhance survival of cancer cells by inhibiting anti-cancer immune responses [65, 66]. Thus, the TKTL1-driven metabolic adaptation may promote cancer progression through a variety of mechanisms.
Although our results suggest that TKTL1 could be a therapeutic target in melanoma, treatment with oxythiamine, a thiamine antagonist, was ineffective even though it was previously shown to decrease proliferation in tumors with high TKTL1 expression [46]. Similarly, earlier studies demonstrated that oxythiamine was not effective in most thyroid cancer cells expressing high TKTL1 and in vivo in normal rat tissues that expressed high TKTL1 [30, 67]. Using computer modelling, 38 amino acid deletion was introduced into the normal human TKT gene to obtain a variant analogous to TKTL1. However, this variant lacked catalytic activity exhibited by the TKT protein, and Meshalkina et al. therefore suggest that TKTL1 is distinct from the thiamine diphosphate dependent enzyme TKT [68]. Our data suggests that we cannot rule out the possibility that TKT acts in concert with TKTL1 as we observed that TKT is highly expressed in melanoma cell lines and knockdown of TKTL1 impacts TKT expression, if only modestly. However, given the fact that the overexpression of TKTL1 in LM-MEL-44 showed opposing effects, and that one of the siRNAs is not changing TKT levels significantly while having the same functional effects in LM-MEL-59, TKTL1 is clearly the main player in the observed effects. Recently, more specific inhibitors of transketolases have been developed [69]. Further studies will determine whether these are more effective on TKTL1-expressing cancers.
Conclusion
The functional significance of the increased TKTL1 in melanoma, its association with DNA hypomethylation and the Warburg effect has not previously been reported. Here, we demonstrate that melanoma cells expressing TKTL1 are predominately glycolytic and that TKTL1 expression increases both cellular proliferation and invasion. Melanoma cells exhibit metabolic plasticity and compensatory mechanisms that allow metabolic adaptations have previously made it difficult to exploit metabolic targets for therapy. Here we show that a methylation defect likely leads to TKTL1 over expression. We postulate that this overexpression allows survival within the hypoxic tumor micro-environment and selects for cells that bear it. Consequently their dependence on glycolysis may arise through a mechanism that does not reflect metabolic plasticity. This in turn points to the potential of targeting TKTL1-expressing melanomas particularly if they prove to be dependent on it for their survival. Ultimately the success of such a strategy will depend on this and on the quality of any drugs that are developed. Further investigations will determine the extent of which is useful target for therapeutic intervention in metastatic melanoma.
Methods
Cell culture and archival patient materials
Melanoma cell lines were established from resected melanoma metastases by mechanical dissociation of tissue with subsequent overnight digestion in media containing collagenase IV and DNAse at 37 °C. Established cell lines were Mycoplasma-tested using the MycoAlert test (Lonza Rockland, Inc.) and STR-profiled for identity confirmation.
Following pathological examination post-surgery tumour fragments were formalin-fixed and paraffin-embedded (FFPE) by staff at the Austin Health Tissue Bank. All melanoma tissue was derived from metastases resected from regions distant to the primary tumor; no locoregional metastases or primary tumors were included. Tissue microarray consists of tumor tissue embedded in a block and allows analysis of multiple patient tissue samples on the same slide. The study cohort consisted of 81 patients with stage III and IV metastatic melanoma. All tissue donors provided written informed consent for tissue collection and research, which was covered by protocols approved by the Austin Health Human Research Ethics Committee, Melbourne, Australia (approval number H2012/04446). All cell lines were matched with their donors by HLA-typing. Cells were cultured in RPMI1640 supplemented with 10 % fetal calf serum (FCS) as described previously [42].
Quantitative reverse transcriptase-PCR (qRT-PCR)
Total RNA was extracted using the RNEasy mini kit (Qiagen). We have performed on column DNAase digestion using RNase-Free DNase at room temperature (20–30 °C) for 15 min in accordance to Qiagen RNEasy Kit instructions. Spectrophotometric quantification using Nanodrop confirmed purity of RNA and absence of DNA in our samples. In addition to this to prevent any interference by residual DNA contamination, we have performed qRT-PCR with primers that are intron spanning such that genomic DNA will not be amplified.
RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Following reverse transcription, qRT-PCR was performed using the SensiFAST SYBR Lo-ROX kit (Bioline), and a Vii-A7 thermocycler (Applied Biosystems). The primers for TKTL1 and β-actin (internal control) were synthesized by Sigma Aldrich and the sequences were as follows: TKTL1 primer (forward) 5’-CCA CCT GAT TAC AGA GTT GGT G-3’ and (reverse) 5’-CTC TGT TGT TCG CGT AGC C-3'; β-actin (forward) 5'-CCA ACC GCG AGA AGA TGA-3’ and (reverse) 5'-CCA GAG GCG TAC AGG GAT AG-3’, TKT primer (forward) 5’-TGT GTC CAG TGC AGT AGT GG-3’ and (reverse) 5’-ACA CTT CAT ACC CGC CCT AG-3’; TKTL2 primer (forward) 5’-ACG ACC GGT TCA TCC TCT C-3’ and (reverse) 5’-TCC ACC CAA GCA GCA TAG A-3’.
Immunohistochemistry
Immunohistochemistry was performed on tissue microarrays constructed from formalin-fixed paraffin-embedded (FFPE) metastatic melanomas and melanoma cell lines after citrate buffer (pH 6.0) antigen retrieval for 30 min using the Dako Envision+ kit (Dako). Incubation with 1× protein blocking buffer for one hour at room temperature was followed by overnight incubation at 4 °C with mouse monoclonal anti-TKTL1 antibody (clone JFC12T10; 1:200 dilution) previously described by Langbein et al. [17]. 60 min incubation with secondary anti-mouse antibody HRP (Dako) was performed. 3-amino-9-ethylcarbazole (AEC) was used as chromagen (Vector Laboratories). Slides were counterstained with hematoxylin, images were captured and immunohistochemical reactivity was evaluated by two independent investigators. Patient tumor cores were scored TKTL positive based on cytosolic staining in minimum of 10 % tumor cells. Human testis tissue was used as the positive control for TKTL1 and a negative control, for which the primary antibody was substituted with the same concentration of mouse IgG.
Bisulfite treatment of DNA and quantitative methylation-specific PCR (MS-qPCR)
Genomic DNA was extracted using phenol chloroform (Qiagen), and bisulfite modification of genomic DNA was performed according to the manufacturer’s instructions (Qiagen). Methylation specific and methylation independent primers (Sigma Aldrich) were used to quantify the relative levels of methylated and unmethylated products within PCR amplified samples. The primers are as follows: TKTL1 methylated primer (forward) 5'-GAC GTC TAA AAA ACG AAT AAC GC-3' and (reverse) 5'-AAA GAA CAT TTT GTA TTC GCG C -3'; TKTL1 unmethylated primer (forward) 5'-CAA CAT CTA AAA AAC AAA TAA CAC C-3' and (reverse) 5'-GAA AGA AGA TTT TGT ATT TGT GTG G-3'; β-actin primer (forward) 5'-TGG TGA TGG AGG AGG TTT AGT AAG T -3' and (reverse) 5'-AAC CAA TAA AAC CTA CTC CTC CCT TAA -3'.
Data set analysis
We acquired data form The Cancer Genome Atlas (TCGA) data portal (www.cbioportal.org). We started our analysis with the Level 3 data download (normalized data) of Illumina Infinium Human Methylation 450 k BeadChip array (450 k array) (http://www.illumina.com/products/methylation_450_beadchip_kits.html), and Illumina Hiseq RNA-Seq Platforms with RNA-seq gene and RNA-seq exon expression datasets (http://www.illumina.com/applications/sequencing/rna.html) of melanoma from the TCGA website (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm?mode=ApplyFilter&showMatrix=true&diseaseType=SKCM&tumorNormal=TN&tumorNormal=T&tumorNormal=NT). Methylation measurement of each CpG probe was called β and was calculated as the methylated signal divided by the sum of the methylated and the unmethylated signal. Two methylation probes in accordance with Illumina annotation and UCSC genome browser that overlapped with our targeted region by MS-QPCR were selected. As there were some CpG sites between the two probes that was not covered by 450 k platform, we only investigated the correlation between these two probes in the 450 k platform by Spearman correlation coefficients statistical test (p < 0.05) to estimate methylation level of our targeted region. Subsequently, correlations between the two probes were separately assessed with TKTL1 gene expression and exon expression. All analysis was performed by R statistical programming language (3.1.1).
5-aza-2′-deoxycytidine (5aza) treatment
Melanoma cell line LM-MEL-44 was treated with 0.1 μM of 5aza (Sigma) for 4 days. The medium and the 5aza were refreshed on alternate days. Cells were confluent at time of harvest.
TKTL1 overexpression construct and RNA interference transfection
The plasmid encoding TKTL1 (clone ID 4825931) was obtained from Life Technologies and the TKTL1 gene was subcloned into pcDNA3.1(+) vector (Promega). Cells were transfected using Lipofectamine 2000 (Life Technologies). Empty vector control used was pcDNA3.1(+). For transient siRNA transfection, cells at 30 % confluence were transfected using a control siRNA and two different Silencer select siRNAs targeting TKTL1 (s224894 and s15774) at 10nM final concentration (Ambion) with Lipofectamine RNAiMAX according to the manufacturer’s protocol (Invitrogen).
Immunoblot analysis
Cells were lysed in RIPA buffer (Sigma) and protein concentration quantified using the BCA protein assay (Thermo Scientific). Samples were separated using NuPAGE 4–10 % BisTris gels (Life Technologies) and MES [2-(N-morpholino) ethanesulfonic acid] SDS running buffer (Life Technologies). Transfer to PVDF membrane (Milipore) used a semi-dry transferblot (Bio-Rad Laboratories). Blocking was performed with Odyssey blocking buffer (Millenium Science), followed by incubation with anti-TKTL1 antibody (mouse monoclonal antibody, clone JFC12T10, 1:100) or anti-GAPDH antibody 1:1000 (Cell Signalling) overnight at 4 °C, and IRDye 680/800 secondary goat anti-mouse and anti-rabbit antibodies (Millenium Science) were incubated for one hour at room temperature. Images were acquired using Odyssey infrared imaging system (LI-COR Biosciences).
Immunofluorescence
LM-MEL-44 transfected with TKTL1 overexpression plasmid construct was incubated for 72 h and then fixed with 4 % paraformaldehyde, stained with anti-TKTL1 rabbit polyclonal antibody (ab87187, Abcam) which was applied at 3 μg/mL concentration overnight at 4 °C and with Alexa flour 555 (anti-rabbit) conjugated secondary antibody for 45 min at room temperature (Molecular probes, USA). Cells were counter stained with DAPI for 10 min.
Measurement of extracellular glucose and lactate
Cell-free supernatant of cultured melanoma cells was collected, and glucose and lactate concentrations were determined using colorimetric glucose and lactate assay kits (Biopharma). The assays were performed according to the manufacturer’s instructions and absorbance was measured at 340 nm.
Cytofluorometric analysis
Cell viability and cell cycle analysis was assessed after staining with propidium iodide. TKTL1 was over-expressed or depleted in melanoma cells for 48 h. Cells were stained with 200 μl propidium iodide solution consisting of 50 μg/ml propidium iodide, 0.1 mg/ml RNAse A and 0.05 % Triton-X (all Sigma) in PBS. Cells were incubated overnight at 4 °C and analyzed by flow cytometry using the FL2 channel.
Proliferation assay
MTS colorimetric assay was performed using standard protocols. Twenty thousand cells were seeded per well of a 96 well plate and treated as indicated. Relative cell numbers were measured using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega Corporation).
Invasion assay
Invasion assays were performed with Boyden chamber insert (6.5 mm diameter inserts with 8.0 μm pores) coated with Matrigel (Becton, Dickinson and Company) as described previously [70]. Briefly, melanoma cells were seeded at 40,000 cells per insert. 600 μl of media containing 10 % FCS was added as chemo attractant to the lower compartment. The cells were incubated overnight at 37 °C and subsequently fixed with 4 % paraformaldehyde, stained with 0.1 % Crystal Violet and cells on the upper surface of the insert were removed. Invaded cells on the bottom side were counted from three random fields of view observed with a 10X objective lens. Average of invaded cells across three fields of view was calculated from independent experiments repeated three times.
Oxythiamine treatment
Melanoma cells were treated with varying dose of oxythiamine (Sigma) as indicated for 24, 48 and 72 h. Following treatment cells were subjected to proliferation, invasion and cytofluorometric analyses.
Statistical analysis
All statistical comparisons of data sets were performed using Student’s two-tailed t-test in Prism software version 5.00 (GraphPad Software Inc). Statistical significance was set at p <0.05.
Acknowledgements
We would like to acknowledge the Melanoma Research Alliance (MRA), the Melbourne Melanoma Project (MMP), Ludwig Cancer Research and the Victorian State Government Operational Infrastructure Support Program for partial funding of this project. The establishment of an expression profile in melanoma cell lines was funded by a grant from the Austin Hospital Medical Research Foundation to AB. AJ was supported by the Austin Hospital Medical Research Foundation and Cure Cancer Australia Foundation. We would like to thank Associate Professors Alexander Dobrovic and John Mariadason for helpful comments.
Abbreviations
- 5aza
5-aza-2’-deoxycytidine
- 18FDG-PET
Fluorodeoxy glucose positron emission tomography
- CTAg
Cancer Testis Antigens
- MAGE-A
Melanoma Antigen Family –A
- MS-qPCR
Quantitative Methylation-Specific PCR
- qRT-PCR
Quantitative Reverse Rranscriptase-PCR
- SKCM
Skin Cutaneous Melanoma
- TCGA
The Cancer Genome Atlas
- TKT
Transketolase
- TKTL1
Transketolase-like 1
- TKTL2
Transketolase-like 2
- TMA
Tissue Microarray
- TSS
Transcription Start Site
Additional files
TKTL1 is expressed in a subset of melanoma tumours and melanoma cell lines. (A) Representative staining patterns for TKTL1 in metastatic melanoma tumors are shown. Arrows indicate nuclear staining for Melanin. Original magnification, 200 μm. (B) QRT-PCR for expression level of TKTL1 in a panel of 53 metastatic melanoma cell lines. (C) High magnification staining pattern of TKTL1 in melanoma cell line LM-MEL-59. Arrows indicate nuclear staining for Melanin. Original magnification, 200 μm. (PPTX 1827 kb)
Correlation of TKTL1 gene expression with DNA methylation in melanoma patients. Exon expression data was compared to methylation patterns of TKTL1 and Spearman correlation coefficients were calculated. Methylation status at the CpG site (A) cg09892236 and (B) cg23106779 was inversely correlated with TKTL1 gene expression in melanoma samples. (PPTX 114 kb)
TKT and TKTL2 expression in melanoma cells. (A) TKT, (B) TKTL1 and (C) TKTL2 expression in LM-MEL-59 after treatment with two TKTL1 siRNAs or control siRNA for 72 h. Testis tissue was used as positive control for the expression of TKT, TKTL1 and TKTL2. (PPTX 69 kb)
TKTL1 expression in melanoma cells after gain and loss of function experiments. (A) Western Blotting with the mouse monoclonal anti-TKTL1 antibody showed reduction in TKTL1 levels after siRNA treatment in LM-MEL-59 after 72 h. GAPDH was used a loading control. (B) Western Blotting of TKTL1 confirmed expression of TKTL1 after transfection of a TKTL1 expression vector in LM-MEL-44. Blots were probed with GAPDH as a control for loading and transfer. (C) Immunofluorescence confirmed increase in TKTL1 localization in LM-MEL-44 after transfection with TKTL1 expression vector. In contrast no TKTL1 was detected in LM-MEL-44 transfected with empty control vector. Scale bar =100 μm). (PPTX 590 kb)
TKTL1 expression in melanoma affects proliferation of cells. (A) Measurement of absorbance of LM-MEL-59 cells after TKTL1 siRNA or control siRNA treatment for 48 h by performing MTS assay. (B) Measurement of absorbance of LM-MEL-44 cells after overexpression of TKTL1 or empty vector for 48 h. (PPTX 57 kb)
Oxythiamine treatment in melanoma cells. Measurement of absorbance of (A) LM-MEL-59 or (B) LM-MEL-44 cells after treatment with 10 μM, 100 μM Oxythiamine or control media for 24, 48 and 72 h by performing MTS assay. (C) Cell cycle phases were determined by propidium iodide staining of LM-MEL-59 cells and subsequent flow cytometric analysis of percentage of cells in subG0, G0-G1, S and G2-M phases after treatment of LM-MEL-59 with 100 μM Oxythiamine or control media. Values are ± SD of three experiments. (D) Representative images of invasion of LM-MEL-59 is shown after treatment with 100 μM Oxythiamine (scale bar = 50 μm) and (E) invasion was quantified. (PPTX 183 kb)
Footnotes
Competing interest
The authors declare that they have no competing interest.
Authors’ contributions
AJ and PH carried out the all the molecular biology and functional assays and drafted the manuscript. AC contributed to FACS assays and performed the statistical analysis. PP and MW performed immunostaining and scoring. AJ and MA carried out invasion assays. MD and AC performed bisulfite treatments and MS-qPCR. RM carried out integrative studies of methylation and gene expression with TCGA dataset. AB established melanoma cell lines and provided cell line RNAs. AB and JC conceived and designed the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Aparna Jayachandran, Email: aparna.jayachandran@onjcri.org.au.
Pu-Han Lo, Email: puhan.lo@gmail.com.
Anderly C. Chueh, Email: chueh.a@wehi.edu.au
Prashanth Prithviraj, Email: prashanth.prithviraj@onjcri.org.au.
Ramyar Molania, Email: ramyar.molania@onjcri.org.au.
Mercedes Davalos-Salas, Email: mercedes.davalos@onjcri.org.au.
Matthew Anaka, Email: matt.anaka@gmail.com.
Marzena Walkiewicz, Email: marzena.walkiewicz@onjcri.org.au.
Jonathan Cebon, Email: jonathan.cebon@onjcri.org.au.
Andreas Behren, Phone: +61(3)9496-5837, Email: andreas.behren@onjcri.org.au.
References
- 1.Little EG, Eide MJ. Update on the current state of melanoma incidence. Dermatol Clin. 2012;30(3):355–61. doi: 10.1016/j.det.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355(1):51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 3.Finn L, Markovic SN, Joseph RW. Therapy for metastatic melanoma: the past, present, and future. BMC Med. 2012;10:23. doi: 10.1186/1741-7015-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Girotti MR, Saturno G, Lorigan P, Marais R. No longer an untreatable disease: How targeted and immunotherapies have changed the management of melanoma patients. Mol Oncol. 2014;8(6):1140-58. [DOI] [PMC free article] [PubMed]
- 5.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14(7):455–67. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lovly CM, Shaw AT. Molecular pathways: resistance to kinase inhibitors and implications for therapeutic strategies. Clin. Cancer Res. 2014;20(9):2249–56. doi: 10.1158/1078-0432.CCR-13-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menzies AM, Long GV. Systemic treatment for BRAF-mutant melanoma: where do we go next? Lancet Oncol. 2014;15(9):e371–81. doi: 10.1016/S1470-2045(14)70072-5. [DOI] [PubMed] [Google Scholar]
- 8.Haq R. Metabolic dysregulation in melanoma: cause or consequence? Can. Dis. 2014;4(4):390–1. doi: 10.1158/2159-8290.CD-14-0173. [DOI] [PubMed] [Google Scholar]
- 9.Hersey P, Watts RN, Zhang XD, Hackett J. Metabolic approaches to treatment of melanoma. Clin. Cancer Res. 2009;15(21):6490–4. doi: 10.1158/1078-0432.CCR-09-0251. [DOI] [PubMed] [Google Scholar]
- 10.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 12.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–70. [PubMed] [Google Scholar]
- 13.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23(5):537–48. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, Osterman AL, Smith JW. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J Biol Chem. 2011;286(49):42626–34. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krug B, Crott R, Lonneux M, Baurain JF, Pirson AS, Vander Borght T. Role of PET in the initial staging of cutaneous malignant melanoma: systematic review. Radiology. 2008;249(3):836–44. doi: 10.1148/radiol.2493080240. [DOI] [PubMed] [Google Scholar]
- 16.Furuta E, Okuda H, Kobayashi A, Watabe K. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim Biophys Acta. 2010;1805(2):141–52. doi: 10.1016/j.bbcan.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langbein S, Zerilli M, Zur Hausen A, Staiger W, Rensch-Boschert K, Lukan N, Popa J, Ternullo MP, Steidler A, Weiss C, et al. Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. Br J Cancer. 2006;94(4):578–85. doi: 10.1038/sj.bjc.6602962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittig R, Coy JF. The role of glucose metabolism and glucose-associated signalling in cancer. Perspect Med. Chem. 2008;1:64–82. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, Yang JH, Guo CK, Cai PC. Gene silencing of TKTL1 by RNAi inhibits cell proliferation in human hepatoma cells. Cancer Lett. 2007;253(1):108–14. doi: 10.1016/j.canlet.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Foldi M, Stickeler E, Bau L, Kretz O, Watermann D, Gitsch G, Kayser G, Zur Hausen A, Coy JF. Transketolase protein TKTL1 overexpression: A potential biomarker and therapeutic target in breast cancer. Oncol Rep. 2007;17(4):841–5. [PubMed] [Google Scholar]
- 21.Fritz P, Coy JF, Murdter TE, Ott G, Alscher MD, Friedel G. TKTL-1 expression in lung cancer. Pathol Res Pract. 2012;208(4):203–9. doi: 10.1016/j.prp.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Krockenberger M, Engel JB, Schmidt M, Kohrenhagen N, Hausler SF, Dombrowski Y, Kapp M, Dietl J, Honig A. Expression of transketolase-like 1 protein (TKTL1) in human endometrial cancer. Anticancer Res. 2010;30(5):1653–9. [PubMed] [Google Scholar]
- 23.Schwaab J, Horisberger K, Strobel P, Bohn B, Gencer D, Kahler G, Kienle P, Post S, Wenz F, Hofmann WK, et al. Expression of Transketolase like gene 1 (TKTL1) predicts disease-free survival in patients with locally advanced rectal cancer receiving neoadjuvant chemoradiotherapy. BMC Cancer. 2011;11:363. doi: 10.1186/1471-2407-11-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun W, Liu Y, Glazer CA, Shao C, Bhan S, Demokan S, Zhao M, Rudek MA, Ha PK, Califano JA. TKTL1 is activated by promoter hypomethylation and contributes to head and neck squamous cell carcinoma carcinogenesis through increased aerobic glycolysis and HIF1alpha stabilization. Clin. Cancer Res. 2010;16(3):857–66. doi: 10.1158/1078-0432.CCR-09-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu HT, Allie N, Myer L, Govender D. Anaplastic nephroblastomas express transketolase-like enzyme 1. J Clin Pathol. 2009;62(5):460–3. doi: 10.1136/jcp.2008.063966. [DOI] [PubMed] [Google Scholar]
- 26.Yuan W, Wu S, Guo J, Chen Z, Ge J, Yang P, Hu B, Chen Z. Silencing of TKTL1 by siRNA inhibits proliferation of human gastric cancer cells in vitro and in vivo. Cancer Biol Ther. 2010;9(9):710–6. doi: 10.4161/cbt.9.9.11431. [DOI] [PubMed] [Google Scholar]
- 27.Grimm M, Cetindis M, Lehmann M, Biegner T, Munz A, Teriete P, Kraut W, Reinert S. Association of cancer metabolism-related proteins with oral carcinogenesis - indications for chemoprevention and metabolic sensitizing of oral squamous cell carcinoma? J Transl Med. 2014;12:208. doi: 10.1186/1479-5876-12-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zerilli M, Amato MC, Martorana A, Cabibi D, Coy JF, Cappello F, Pompei G, Russo A, Giordano C, Rodolico V. Increased expression of transketolase-like-1 in papillary thyroid carcinomas smaller than 1.5 cm in diameter is associated with lymph-node metastases. Cancer. 2008;113(5):936–44. doi: 10.1002/cncr.23683. [DOI] [PubMed] [Google Scholar]
- 29.Diaz-Moralli S, Tarrado-Castellarnau M, Alenda C, Castells A, Cascante M. Transketolase-like 1 expression is modulated during colorectal cancer progression and metastasis formation. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0025323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frohlich E, Fink I, Wahl R. Is transketolase like 1 a target for the treatment of differentiated thyroid carcinoma? A study on thyroid cancer cell lines. Investig New Drugs. 2009;27(4):297–303. doi: 10.1007/s10637-008-9174-8. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Zhang X, Ma D, Lee WN, Xiao J, Zhao Y, Go VL, Wang Q, Yen Y, Recker R, et al. Inhibition of transketolase by oxythiamine altered dynamics of protein signals in pancreatic cancer cells. Exp Hematol Oncol. 2013;2:18. doi: 10.1186/2162-3619-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feyen O, Coy JF, Prasad V, Schierl R, Saenger J, Baum RP. EDIM-TKTL1 blood test: a noninvasive method to detect upregulated glucose metabolism in patients with malignancies. Future Oncol. 2012;8(10):1349–59. doi: 10.2217/fon.12.98. [DOI] [PubMed] [Google Scholar]
- 33.Lange CA, Tisch-Rottensteiner J, Bohringer D, Martin G, Schwartzkopff J, Auw-Haedrich C. Enhanced TKTL1 expression in malignant tumors of the ocular adnexa predicts clinical outcome. Ophthalmology. 2012;119(9):1924–9. doi: 10.1016/j.ophtha.2012.03.037. [DOI] [PubMed] [Google Scholar]
- 34.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 35.Barrow C, Browning J, MacGregor D, Davis ID, Sturrock S, Jungbluth AA, Cebon J. Tumor antigen expression in melanoma varies according to antigen and stage. Clin. Cancer Res. 2006;12(3 Pt 1):764–71. doi: 10.1158/1078-0432.CCR-05-1544. [DOI] [PubMed] [Google Scholar]
- 36.Svobodova S, Browning J, MacGregor D, Pollara G, Scolyer RA, Murali R, Thompson JF, Deb S, Azad A, Davis ID, et al. Cancer-testis antigen expression in primary cutaneous melanoma has independent prognostic value comparable to that of Breslow thickness, ulceration and mitotic rate. Eur J Cancer. 2011;47(3):460–9. doi: 10.1016/j.ejca.2010.09.042. [DOI] [PubMed] [Google Scholar]
- 37.Gedye C, Quirk J, Browning J, Svobodova S, John T, Sluka P, Dunbar PR, Corbeil D, Cebon J, Davis ID. Cancer/testis antigens can be immunological targets in clonogenic CD133+ melanoma cells. Cancer Immunol. Immunother. 2009;58(10):1635–46. doi: 10.1007/s00262-009-0672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jonsson G, Busch C, Knappskog S, Geisler J, Miletic H, Ringner M, Lillehaug JR, Borg A, Lonning PE. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin. Cancer Res. 2010;16(13):3356–67. doi: 10.1158/1078-0432.CCR-09-2509. [DOI] [PubMed] [Google Scholar]
- 39.Smith IM, Glazer CA, Mithani SK, Ochs MF, Sun W, Bhan S, Vostrov A, Abdullaev Z, Lobanenkov V, Gray A, et al. Coordinated activation of candidate proto-oncogenes and cancer testes antigens via promoter demethylation in head and neck cancer and lung cancer. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Can. Dis. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behren A, Anaka M, Lo PH, Vella LJ, Davis ID, Catimel J, Cardwell T, Gedye C, Hudson C, Stan R, et al. The Ludwig institute for cancer research Melbourne melanoma cell line panel. Pigment Cell Melanoma Res. 2013;26(4):597–600. doi: 10.1111/pcmr.12097. [DOI] [PubMed] [Google Scholar]
- 43.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18(1):54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19(1):32–7. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, Zur Hausen A, Coy JF, Lochelt M. Transketolase-like protein 1 (TKTL1) is required for rapid cell growth and full viability of human tumor cells. Int. J. Cancer. 2009;124(6):1330–7. doi: 10.1002/ijc.24078. [DOI] [PubMed] [Google Scholar]
- 46.Rais B, Comin B, Puigjaner J, Brandes JL, Creppy E, Saboureau D, Ennamany R, Lee WN, Boros LG, Cascante M. Oxythiamine and dehydroepiandrosterone induce a G1 phase cycle arrest in Ehrlich's tumor cells through inhibition of the pentose cycle. FEBS Lett. 1999;456(1):113–8. doi: 10.1016/S0014-5793(99)00924-2. [DOI] [PubMed] [Google Scholar]
- 47.Coy JF, Dressler D, Wilde J, Schubert P. Mutations in the transketolase-like gene TKTL1: clinical implications for neurodegenerative diseases, diabetes and cancer. Clin Lab. 2005;51(5–6):257–73. [PubMed] [Google Scholar]
- 48.Schmidt M, Voelker HU, Kapp M, Krockenberger M, Dietl J, Kammerer U. Glycolytic phenotype in breast cancer: activation of Akt, up-regulation of GLUT1, TKTL1 and down-regulation of M2PK. J Cancer Res Clin Oncol. 2010;136(2):219–25. doi: 10.1007/s00432-009-0652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiriva-Internati M, Mirandola L, Yu Y, Jenkins MR, Gornati R, Bernardini G, Gioia M, Chiaramonte R, Cannon MJ, Kast WM, et al. Cancer testis antigen, ropporin, is a potential target for multiple myeloma immunotherapy. J Immunother. 2011;34(6):490–9. doi: 10.1097/CJI.0b013e31821ca76f. [DOI] [PubMed] [Google Scholar]
- 50.dos Santos NR, Torensma R, de Vries TJ, Schreurs MW, de Bruijn DR, Kater-Baats E, Ruiter DJ, Adema GJ, van Muijen GN, van Kessel AG. Heterogeneous expression of the SSX cancer/testis antigens in human melanoma lesions and cell lines. Cancer Res. 2000;60(6):1654–62. [PubMed] [Google Scholar]
- 51.Gjerstorff MF, Ditzel HJ. Limited SP17 expression within tumors diminishes its therapeutic potential. Tissue Antigens. 2012;80(6):523–7. doi: 10.1111/tan.12015. [DOI] [PubMed] [Google Scholar]
- 52.Ma Y, Xin Y, Li R, Wang Z, Yue Q, Xiao F, Hao X. TFDP3 was expressed in coordination with E2F1 to inhibit E2F1-mediated apoptosis in prostate cancer. Gene. 2014;537(2):253–9. doi: 10.1016/j.gene.2013.12.051. [DOI] [PubMed] [Google Scholar]
- 53.Chiriva-Internati M, Gagliano N, Donetti E, Costa F, Grizzi F, Franceschini B, Albani E, Levi-Setti PE, Gioia M, Jenkins M, et al. Sperm protein 17 is expressed in the sperm fibrous sheath. J Transl Med. 2009;7:61. doi: 10.1186/1479-5876-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fiedler SE, Dudiki T, Vijayaraghavan S, Carr DW. Loss of R2D2 proteins ROPN1 and ROPN1L causes defects in murine sperm motility, phosphorylation, and fibrous sheath integrity. Biol Reprod. 2013;88(2):41. doi: 10.1095/biolreprod.112.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Sosnik J, Brassard L, Reese M, Spiridonov NA, Bates TC, Johnson GR, Anguita J, Visconti PE, Salicioni AM. Expression and localization of five members of the testis-specific serine kinase (Tssk) family in mouse and human sperm and testis. Mol Hum Reprod. 2011;17(1):42–56. doi: 10.1093/molehr/gaq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang F, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J Cell Biol. 2008;180(4):673–9. doi: 10.1083/jcb.200709057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rolland AD, Lavigne R, Dauly C, Calvel P, Kervarrec C, Freour T, Evrard B, Rioux-Leclercq N, Auger J, Pineau C. Identification of genital tract markers in the human seminal plasma using an integrative genomics approach. Hum Reprod. 2013;28(1):199–209. doi: 10.1093/humrep/des360. [DOI] [PubMed] [Google Scholar]
- 58.Liu L, Duclos G, Sun B, Lee J, Wu A, Kam Y, Sontag ED, Stone HA, Sturm JC, Gatenby RA, et al. Minimization of thermodynamic costs in cancer cell invasion. Proc Natl Acad Sci U S A. 2013;110(5):1686–91. doi: 10.1073/pnas.1221147110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho J, de Moura MB, Lin Y, Vincent G, Thorne S, Duncan LM, Hui-Min L, Kirkwood JM, Becker D, Van Houten B, et al. Importance of glycolysis and oxidative phosphorylation in advanced melanoma. Mol Cancer. 2012;11:76. doi: 10.1186/1476-4598-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.BarbideMoura M, Vincent G, Fayewicz SL, Bateman NW, Hood BL, Sun M, Suhan J, Duensing S, Yin Y, Sander C, et al. Mitochondrial respiration--an important therapeutic target in melanoma. PLoS One. 2012;7(8):e40690. doi: 10.1371/journal.pone.0040690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23(3):302–15. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parmenter TJ, Kleinschmidt M, Kinross KM, Bond ST, Li J, Kaadige MR, Rao A, Sheppard KE, Hugo W, Pupo GM, et al. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Can. Dis. 2014;4(4):423–33. doi: 10.1158/2159-8290.CD-13-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higashi K, Clavo AC, Wahl RL. In vitro assessment of 2-fluoro-2-deoxy-D-glucose, L-methionine and thymidine as agents to monitor the early response of a human adenocarcinoma cell line to radiotherapy. J. Nucl. Med. 1993;34(5):773–9. [PubMed] [Google Scholar]
- 64.Smith TA, Title JC, Eccles SA, McCready VR. 2-Deoxy-D-[1-3H]glucose uptake by MCF7 and T47D breast tumour cells under conditions of active proliferation and serum deprivation. Anticancer Res. 1998;18(3A):1865–9. [PubMed] [Google Scholar]
- 65.Han T, Kang D, Ji D, Wang X, Zhan W, Fu M, Xin HB, Wang JB. How does cancer cell metabolism affect tumor migration and invasion? Cell Adhes Migr. 2013;7(5):395–403. doi: 10.4161/cam.26345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71(22):6921–5. doi: 10.1158/0008-5472.CAN-11-1457. [DOI] [PubMed] [Google Scholar]
- 67.Brin M. Effects of thiamine deficiency and of oxythiamine on rat tissue transketolase. J Nutr. 1962;78:179–83. doi: 10.1093/jn/78.2.179. [DOI] [PubMed] [Google Scholar]
- 68.Meshalkina LE, Drutsa VL, Koroleva ON, Solovjeva ON, Kochetov GA. Is transketolase-like protein, TKTL1, transketolase? Biochim Biophys Acta. 2013;1832(3):387–90. doi: 10.1016/j.bbadis.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 69.Thomas AA, Le Huerou Y, De Meese J, Gunawardana I, Kaplan T, Romoff TT, Gonzales SS, Condroski K, Boyd SA, Ballard J, et al. Synthesis, in vitro and in vivo activity of thiamine antagonist transketolase inhibitors. Bioorg Med Chem Lett. 2008;18(6):2206–10. doi: 10.1016/j.bmcl.2007.11.101. [DOI] [PubMed] [Google Scholar]
- 70.Jayachandran A, Anaka M, Prithviraj P, Hudson C, McKeown SJ, Lo PH, Vella LJ, Goding CR, Cebon J, Behren A. Thrombospondin 1 promotes an aggressive phenotype through epithelial-to-mesenchymal transition in human melanoma. Oncotarget. 2014;5(14):5782–97. doi: 10.18632/oncotarget.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]


