Abstract
Acinetobacter baumannii frequently causes nosocomial infections and outbreaks. Whole-genome sequencing (WGS) is a promising technique for strain typing and outbreak investigations. We compared the performance of conventional methods with WGS for strain typing clinical Acinetobacter isolates and analyzing a carbapenem-resistant A. baumannii (CRAB) outbreak. We performed two band-based typing techniques (pulsed-field gel electrophoresis and repetitive extragenic palindromic-PCR), multilocus sequence type (MLST) analysis, and WGS on 148 Acinetobacter calcoaceticus-A. baumannii complex bloodstream isolates collected from a single hospital from 2005 to 2012. Phylogenetic trees inferred from core-genome single nucleotide polymorphisms (SNPs) confirmed three Acinetobacter species within this collection. Four major A. baumannii clonal lineages (as defined by MLST) circulated during the study, three of which are globally distributed and one of which is novel. WGS indicated that a threshold of 2,500 core SNPs accurately distinguished A. baumannii isolates from different clonal lineages. The band-based techniques performed poorly in assigning isolates to clonal lineages and exhibited little agreement with sequence-based techniques. After applying WGS to a CRAB outbreak that occurred during the study, we identified a threshold of 2.5 core SNPs that distinguished nonoutbreak from outbreak strains. WGS was more discriminatory than the band-based techniques and was used to construct a more accurate transmission map that resolved many of the plausible transmission routes suggested by epidemiologic links. Our study demonstrates that WGS is superior to conventional techniques for A. baumannii strain typing and outbreak analysis. These findings support the incorporation of WGS into health care infection prevention efforts.
INTRODUCTION
An important role of clinical microbiology is to identify relationships between bacterial isolates. At a broad level, phenotypic and genotypic tests are used to categorize bacterial isolates into the same or different species. Within a bacterial species, techniques are used to group isolates into clonal lineages, which are groups of closely related bacteria that share a recent common ancestor but have spread regionally or globally. At a more local level, infection control practitioners must determine whether a group of isolates constitutes a hospital outbreak by ascertaining whether the isolates have a degree of similarity consistent with a common source within the hospital. Once isolates belonging to a hospital outbreak have been identified, similarities and differences between these isolates can be exploited to generate a transmission map to aid in finding the source of the outbreak and in disrupting ongoing pathways of transmission. For some groups of bacteria, such as Acinetobacter, discernment at each level has medically important consequences and therefore must be accomplished by hospital-associated clinical microbiology laboratories.
Within the Acinetobacter genus, the Acinetobacter calcoaceticus-Acinetobacter baumannii (ACB) complex encompasses the phenotypically related pathogens A. baumannii, A. pittii (formerly genomospecies 3), and A. nosocomialis (formerly genomospecies 13TU), and one species, A. calcoaceticus, which is not known to cause human disease (1). Of these, A. baumannii is the most common cause of infection. This bacterium is frequently isolated from critically ill hospitalized patients and often causes outbreaks (2–4). Infections with A. baumannii have been associated with high attributable mortality and increased length of hospital stay, with multidrug resistance often being a predictor of poor clinical outcomes (5, 6). However, the frequencies of A. nosocomialis and A. pittii as human pathogens are increasingly recognized (7, 8). Several studies have shown that these different species within the ACB complex exhibit unique epidemiologic niches, drug resistance patterns, and virulence characteristics within the nosocomial environment (9, 10). In addition, recent studies have demonstrated that patients infected with non-baumannii ACB complex bacteria have fewer comorbidities and improved clinical outcomes than patients with A. baumannii infections (7, 11–13). Thus, determining the species within the ACB complex that is responsible for an infection has important medical implications. Unfortunately, commercially available platforms and phenotypic identification methods used in clinical microbiology laboratories are unable to differentiate related species within the ACB complex, and, as such, infections are often reported clinically as “A. baumannii complex” or simply grouped together as “A. baumannii” (14). Molecular techniques, such as rpoB gene sequence analysis, are necessary to accurately identify ACB complex isolates to the species level (15, 16).
In recent years, multidrug-resistant (MDR) strains of A. baumannii have become common, and substantial effort has been devoted to defining the epidemiology of these strains. Initial studies attributed the global spread of MDR A. baumannii to three major clonal lineages identified by amplified fragment length polymorphism (AFLP) analysis that are referred to as international clones (ICs)-I, -II, and -III (17, 18). More recently, multilocus sequence typing (MLST) has become the gold standard for investigating the population structure of A. baumannii (19) and has linked sequence types (STs), such as ST1, ST2, and ST3, with IC-I, -II, and -III, respectively (20). MLST analyses have identified at least six additional clonal lineages with geographically broad distributions (17–19, 21, 22). The study of the global epidemiology and population structure of A. baumannii remains an area of active interest.
A. baumannii is a frequent cause of intrahospital outbreaks, and the ability to distinguish clinical A. baumannii isolates as genetically unique is of particular importance for outbreak analysis. The goal is to distinguish outbreak isolates from nonoutbreak isolates and to use strain typing information to define routes of transmission within an outbreak. Numerous different bacterial strain typing techniques have been described for this purpose. Older band-based fingerprint techniques, such as pulsed-field gel electrophoresis (PFGE), AFLP analysis, and repetitive extragenic palindromic-PCR (Rep-PCR), rely on indirect measures of bacterial genetic composition (22–26). Of these, PFGE arguably remains the gold standard for A. baumannii outbreak investigations (19). More recently, partial direct genetic sequencing techniques have been developed, such as MLST and blaoxa-51-like sequencing (20, 27, 28). Although band-based techniques are often highly discriminatory, direct sequence-based methods can provide greater genetic resolution and are more reproducible and portable (29–31).
For years, high-throughput typing of multiple A. baumannii isolates for short-term outbreak investigations or long-term regional or global surveillance was limited to these band-based or partial sequence-based techniques (32). The decreasing cost, complexity, and turnaround time of whole-genome sequencing (WGS) may soon allow the application of this technology to routine bacterial strain typing in clinical microbiology laboratories (33, 34). A number of recent studies have used WGS to characterize the genetic relatedness of clinical bacterial isolates for various purposes. Some studies have used WGS for bacterial species identification and investigations of the population structure and global spread of bacteria (31, 35, 36). Other studies have analyzed hospital outbreaks in greater detail and have used WGS to distinguish between possible transmission scenarios, suggest alternative transmission links, and identify previously unrecognized colonized patients (37–39). Although WGS may be the most powerful and adaptable tool for bacterial species identification, epidemiologic surveillance, and outbreak analysis, the application of WGS to these endeavors has yet to be fully explored. Careful characterization of the advantages of WGS is necessary to determine whether they outweigh the costs of commercially available next-generation sequencing instruments or services. Also, the criteria for discrimination between outbreak and nonoutbreak isolates or between clonal lineages have yet to be clearly defined for WGS. In contrast, the criteria for these purposes have been well described for interpreting PFGE, Rep-PCR, and MLST results (20, 40–42).
In this study, we compared the performance of WGS to that of two band-based typing techniques (PFGE and Rep-PCR) and to a sequence-based typing technique (MLST) to characterize the genetic relatedness and epidemiology of a large collection of ACB complex clinical bloodstream isolates collected over 7 years at our institution. We compared WGS to conventional typing at the level of Acinetobacter species identification, assignment to clonal lineages, inclusion within an intrahospital outbreak, and outbreak transmission mapping. Our results indicate that WGS provides useful information for each of these purposes and may eventually obviate multiple different techniques within clinical microbiology laboratories.
MATERIALS AND METHODS
Ethics, consent, and permissions.
This study was approved by the institutional review board of Northwestern University, Chicago, IL, with a waiver of informed consent. Informed consent was not obtained because this was a retrospective study, and data collection occurred after patients left the hospital and/or died from their illness. No additional specimens were collected beyond those in routine clinical care, and no diagnostic or treatment decisions were affected by our study.
Bacterial isolates and identification.
One hundred fifty-four bloodstream isolates (designated ABBL isolates) collected between January 2005 and November 2012 from hospitalized patients at Northwestern Memorial Hospital (NMH), a 900-bed tertiary-care academic medical center in Chicago, IL, were analyzed in this study. These isolates were previously reported (13). In addition, 13 blood and respiratory isolates collected during the investigation of a carbapenem-resistant A. baumannii (CRAB) intensive care unit (ICU) outbreak from June 2013 to December 2013 (designated ABOB isolates) were included. Phenotypic identification of isolates to the ACB complex level was performed using the Vitek 2 system (bioMérieux, Marcy l'Etoile, France). Antimicrobial susceptibility testing was performed using a combination of the Vitek 2, disk diffusion test, and Etest (bioMérieux) and was interpreted in accordance with the Clinical and Laboratory Standards Institute guidelines (43). For the purposes of this study, all isolates were identified to the species level using the partial rpoB gene sequence. Zone 1 rpoB sequences were identified from the assembled whole-genome sequences using an in-house in silico PCR script (44). Briefly, assembly contigs were screened to identify sequences flanked by the virtual primers Ac696F (TAYCGYAAAGAYTTGAAAGAAG) and Ac1093R (CMACACCYTTGTTMCCRTGA), as described by La Scola et al. (45). In silico amplified sequences were identified by BLAST alignment against the NCBI nucleotide database (46).
Pulsed-field gel electrophoresis.
During the period of collection of these strains, the NMH clinical microbiology laboratory performed PFGE surveillance on all CRAB isolates in our hospital, as previously described (25). Banding patterns were interpreted and strain types assigned according to the criteria of Tenover et al. (40). Isolates with indistinguishable, closely related, and possibly related PFGE patterns were considered the same type, as they are often analyzed together during outbreak investigations.
DNA extraction.
Bacterial isolates were streaked from −80°C frozen stocks, inoculated in Luria-Bertani (LB) broth, and grown with shaking overnight at 37°C. Genomic DNA was extracted from the cultures using either the Qiagen EZ1 Advanced robotic workstation (Valencia, CA) or the Promega Maxwell 16 instrument (Madison, WI), according to each manufacturer's instructions. Genomic DNA extracts were either kept on ice for immediate use or were stored at −20°C for future use.
Repetitive extragenic palindromic-PCR.
For Rep-PCR analyses, approximately 300 ng of genomic DNA was used in the PCRs, along with the REP1+REP2 oligonucleotide primer set (47). The reaction mixtures contained 100 pmol each primer and 25 μl of AccuStart II PCR Supermix (Quanta Biosciences, Gaithersburg, MD), which includes 2× reaction buffer, 3 mM MgCl2, 0.3 mM each deoxynucleoside triphosphate (dNTP), and AccuStart II Taq DNA polymerase. Sterile distilled water was added to a final volume of 50 μl. The PCR conditions were as follows: initial denaturation of 94° for 3 min, 30 cycles of 94°C for 1 min, 45°C for 1 min, and 65°C for 8 min, and a final elongation of 65°C for 16 min. Samples (20 μl) of each PCR end product were analyzed on a 1% agarose gel with ethidium bromide added. Rep-PCR patterns were visualized with UV transillumination using the AlphaImager system (ProteinSimple Biosciences, Santa Clara, CA). Gel images were uploaded into the BioNumerics version 7.0 program (Applied Maths, Austin, TX) to perform clustering analysis. Briefly, after normalization of inter- and intragel variation using molecular weight standards, individual bands were manually chosen, and a clustering dendrogram was created using the unweighted pair group method using average linkages (UPGMA). The Dice statistic was used, and band tolerance was set to 1%. Isolates clustering together at >90% similarity based on the UPGMA dendrogram were considered to be the same type (23, 48, 49).
Whole-genome sequencing and assembly.
Genomic DNA libraries were prepared and indexed using the Nextera XT kit (Illumina, San Diego, CA). DNA library concentrations were quantified using the Qubit 2.0 fluorometer (Life Technologies, Grand Island, NY). Equal amounts (200 ng) of each library were pooled and run on either the Illumina HiSeq 2000 system with 100-bp paired-end reads or on the Illumina MiSeq system with 250-bp paired-end reads. Sequencing was performed by staff at the University of Maryland Institute for Genome Sciences, Baltimore, MD, who were blinded to all clinical data. Raw sequence reads were then assembled de novo using Ray version 1.7.0 (50). Six bloodstream isolates with low-quality assemblies, defined as a sequence size of assembled contigs of ≥500 bp totaling >4.5 Mb, were excluded from further analysis. The assembly statistics are displayed in Table 1.
TABLE 1.
Date of isolation, sequencing, and assembly statistics for new Acinetobacter bloodstream and outbreak isolates
| Isolate | Culture date (mo/day/yr) | Avg read length (bp) | No. of raw reads | Predicted coverage (fold) | No. of raw reads after downsampling | No. of contigs >500 bp | Total size of contigs >500 bp (bp) | Contig N50 (bp) | SRA accession no. | GenBank WGS accession no.a |
|---|---|---|---|---|---|---|---|---|---|---|
| ABBL001 | 1/30/2005 | 101 | 5,751,032 | 145 | 3,168,318 | 79 | 3,871,836 | 122,252 | SRR2558732 | LLCI00000000 |
| ABBL003 | 2/25/2005 | 101 | 5,878,804 | 148 | 3,168,318 | 72 | 3,959,187 | 114,915 | SRR2558733 | LLCJ00000000 |
| ABBL004 | 5/17/2005 | 101 | 4,935,656 | 125 | 3,168,318 | 76 | 3,857,255 | 129,801 | SRR2558836 | LLCK00000000 |
| ABBL005 | 6/21/2005 | 101 | 5,456,552 | 138 | 3,168,318 | 66 | 4,105,919 | 146,012 | SRR2558848 | LLCL00000000 |
| ABBL006 | 7/6/2005 | 101 | 5,276,928 | 133 | 3,168,318 | 79 | 4,070,748 | 92,563 | SRR2558860 | LLCM00000000 |
| ABBL007 | 8/1/2005 | 234 | 4,672,638 | 273 | 1,367,522 | 770 | 3,762,155 | 8,012 | SRR2558871 | LLCN00000000 |
| ABBL008 | 8/11/2005 | 101 | 6,100,156 | 154 | 3,168,318 | 89 | 4,066,927 | 139,396 | SRR2558883 | LLCO00000000 |
| ABBL010 | 8/15/2005 | 101 | 5,917,234 | 149 | 3,168,318 | 62 | 4,050,651 | 126,884 | SRR2558907 | LLCQ00000000 |
| ABBL011 | 9/4/2005 | 101 | 5,431,532 | 137 | 3,168,318 | 71 | 4,006,992 | 118,605 | SRR2558919 | LLCR00000000 |
| ABBL012 | 9/23/2005 | 101 | 6,780,124 | 171 | 3,168,318 | 87 | 3,927,097 | 110,206 | SRR2558734 | LLCS00000000 |
| ABBL013 | 9/19/2005 | 101 | 4,344,706 | 110 | 3,168,318 | 138 | 4,010,638 | 85,500 | SRR2558776 | LLCT00000000 |
| ABBL014 | 9/25/2005 | 101 | 6,420,868 | 162 | 3,168,318 | 193 | 3,857,237 | 52,131 | SRR2558787 | LLCU00000000 |
| ABBL015 | 10/23/2005 | 101 | 6,262,008 | 158 | 3,168,318 | 68 | 4,191,244 | 186,354 | SRR2558808 | LLCV00000000 |
| ABBL016 | 11/7/2005 | 101 | 6,536,664 | 165 | 3,168,318 | 35 | 3,880,175 | 212,743 | SRR2558820 | LLCW00000000 |
| ABBL017 | 11/16/2005 | 101 | 5,637,164 | 142 | 3,168,318 | 92 | 4,028,200 | 106,423 | SRR2558831 | LLCX00000000 |
| ABBL018 | 12/9/2005 | 101 | 4,978,776 | 126 | 3,168,318 | 125 | 4,318,316 | 105,657 | SRR2558832 | LLCY00000000 |
| ABBL019 | 12/13/2005 | 101 | 5,935,086 | 150 | 3,168,318 | 24 | 3,888,930 | 343,426 | SRR2558833 | LLCZ00000000 |
| ABBL020 | 1/17/2006 | 101 | 7,809,576 | 197 | 3,168,318 | 83 | 3,806,309 | 108,333 | SRR2558834 | LLDA00000000 |
| ABBL021 | 1/31/2006 | 101 | 7,992,930 | 202 | 3,168,318 | 64 | 3,952,727 | 132,629 | SRR2558835 | LLDB00000000 |
| ABBL022 | 2/22/2006 | 101 | 6,912,504 | 175 | 3,168,318 | 83 | 3,994,985 | 105,023 | SRR2558837 | LLDC00000000 |
| ABBL023 | 3/21/2006 | 232 | 7,041,594 | 408 | 1,382.290 | 579 | 3,982,278 | 12,335 | SRR2558838 | LLDD00000000 |
| ABBL024 | 4/4/2006 | 101 | 4,864,248 | 123 | 3,168,318 | 91 | 3,892,382 | 107,246 | SRR2558839 | LLDE00000000 |
| ABBL025 | 4/24/2006 | 101 | 5,678,228 | 143 | 3,168,318 | 71 | 4,070,327 | 171,086 | SRR2558840 | LLDF00000000 |
| ABBL026 | 5/1/2006 | 217 | 10,727,026 | 581 | 1,478,060 | 602 | 3,906,420 | 10,017 | SRR2558841 | LLDG00000000 |
| ABBL027 | 5/13/2006 | 251 | 2,226,438 | 140 | 1,274,900 | 84 | 4,051,543 | 115,039 | SRR2558842 | LLDH00000000 |
| ABBL028 | 5/25/2006 | 239 | 585,626 | 35 | NAb | 909 | 3,667,117 | 6,740 | SRR2558843 | LLDI00000000 |
| ABBL029 | 6/5/2006 | 101 | 5,887,830 | 149 | 3,168,318 | 90 | 3,922,574 | 87,612 | SRR2558844 | LLDJ00000000 |
| ABBL030 | 7/4/2006 | 251 | 2,237,610 | 140 | 1,274,900 | 93 | 4,022,189 | 84,636 | SRR2558846 | LLDK00000000 |
| ABBL031 | 8/9/2006 | 151 | 2,697,432 | 102 | 2,119,206 | 82 | 4,143,212 | 122,907 | SRR2558847 | LLDL00000000 |
| ABBL032 | 8/31/2006 | 101 | 7,469,508 | 189 | 3,168,318 | 111 | 3,917,819 | 66,618 | SRR2558849 | LLDM00000000 |
| ABBL033 | 11/6/2006 | 101 | 7,242,078 | 183 | 3,168,318 | 119 | 3,800,726 | 75,277 | SRR2558850 | LLDN00000000 |
| ABBL034 | 1/9/2007 | 101 | 6,643,438 | 168 | 3,168,318 | 89 | 4,041,141 | 111,495 | SRR2558851 | LLDO00000000 |
| ABBL035 | 1/23/2007 | 232 | 3,782,874 | 219 | 1,379,310 | 966 | 3,811,475 | 6,705 | SRR2558852 | LLDP00000000 |
| ABBL036 | 6/8/2007 | 101 | 14,194,932 | 358 | 3,168,318 | 500 | 3,816,808 | 15,165 | SRR2558853 | LLDQ00000000 |
| ABBL037 | 6/20/2007 | 101 | 8,609,660 | 217 | 3,168,318 | 75 | 4,179,466 | 109,239 | SRR2558854 | LLDR00000000 |
| ABBL038 | 7/7/2007 | 101 | 10,957,150 | 277 | 3,168,318 | 86 | 4,045,591 | 110,028 | SRR2558855 | LLDS00000000 |
| ABBL039 | 8/2/2007 | 101 | 8,767,486 | 221 | 3,168,318 | 118 | 4,005,659 | 59,483 | SRR2558856 | LLDT00000000 |
| ABBL040 | 8/12/2007 | 101 | 11,439,386 | 289 | 3,168,318 | 439 | 3,889,680 | 16,992 | SRR2558857 | LLDU00000000 |
| ABBL041 | 8/12/2007 | 101 | 5,683,398 | 144 | 3,168,318 | 105 | 4,014,785 | 85,413 | SRR2558858 | LLDV00000000 |
| ABBL042 | 8/13/2007 | 251 | 2,148,934 | 135 | 1,274,900 | 104 | 4,023,065 | 95,503 | SRR2558861 | LLDW00000000 |
| ABBL043 | 8/28/2007 | 251 | 2,304,972 | 145 | 1,274,900 | 113 | 4,193,893 | 79,829 | SRR2558862 | LLDX00000000 |
| ABBL044 | 9/20/2007 | 251 | 2,114,926 | 133 | 1,274,900 | 107 | 4,108,999 | 64,973 | SRR2558863 | LLDY00000000 |
| ABBL045 | 9/20/2007 | 101 | 8,425,048 | 213 | 3,168,318 | 103 | 4,052,607 | 101,492 | SRR2558864 | LLDZ00000000 |
| ABBL046 | 10/14/2007 | 101 | 11,383,666 | 287 | 3,168,318 | 117 | 3,917,523 | 70,298 | SRR2558865 | LLEA00000000 |
| ABBL047 | 10/28/2007 | 101 | 6,941,066 | 175 | 3,168,318 | 57 | 4,100,149 | 154,707 | SRR2558866 | LLEB00000000 |
| ABBL048 | 11/27/2007 | 251 | 2,144,992 | 135 | 1,274,900 | 75 | 4,161,657 | 120,446 | SRR2558867 | LLEC00000000 |
| ABBL049 | 12/12/2007 | 251 | 2,480,548 | 156 | 1,274,900 | 88 | 4,142,974 | 116,905 | SRR2558868 | LLED00000000 |
| ABBL050 | 1/22/2008 | 101 | 13,089,350 | 331 | 3,168,318 | 292 | 4,173,128 | 27,050 | SRR2558869 | LLEE00000000 |
| ABBL051 | 1/31/2008 | 101 | 13,714,298 | 346 | 3,168,318 | 858 | 3,801,250 | 7,151 | SRR2558870 | LLEF00000000 |
| ABBL052 | 2/24/2008 | 101 | 8,137,460 | 205 | 3,168,318 | 106 | 4,109,803 | 75,223 | SRR2558873 | LLEG00000000 |
| ABBL053 | 3/17/2008 | 101 | 7,715,994 | 195 | 3,168,318 | 88 | 4,085,835 | 90,003 | SRR2558874 | LLEH00000000 |
| ABBL054 | 3/22/2008 | 101 | 11,043,836 | 279 | 3,168,318 | 108 | 3,435,547 | 70,480 | SRR2558875 | LLEZ00000000 |
| ABBL055 | 4/5/2008 | 101 | 9,541,470 | 241 | 3,168,318 | 110 | 4,102,482 | 60,720 | SRR2558876 | LLFA00000000 |
| ABBL056 | 7/11/2008 | 101 | 7,449,168 | 188 | 3,168,318 | 84 | 4,012,566 | 80,470 | SRR2558877 | LLFB00000000 |
| ABBL057 | 7/22/2008 | 101 | 7,511,856 | 190 | 3,168,318 | 78 | 4,016,266 | 236,259 | SRR2558878 | LLFC00000000 |
| ABBL058 | 7/25/2008 | 101 | 9,584,202 | 242 | 3,168,318 | 117 | 4,010,913 | 80,750 | SRR2558879 | LLFD00000000 |
| ABBL059 | 8/7/2008 | 101 | 5,781,700 | 146 | 3,168,318 | 226 | 3,989,506 | 37,323 | SRR2558880 | LLFE00000000 |
| ABBL060 | 9/15/2008 | 101 | 20,003,004 | 505 | 3,168,318 | 198 | 4,033,351 | 37,283 | SRR2558881 | LLFF00000000 |
| ABBL061 | 9/29/2008 | 101 | 5,134,402 | 130 | 3,168,318 | 93 | 3,960,869 | 101,526 | SRR2558882 | LLFG00000000 |
| ABBL062 | 10/1/2008 | 101 | 5,970,402 | 151 | 3,168,318 | 72 | 3,873,658 | 103,774 | SRR2558884 | LLFH00000000 |
| ABBL063 | 10/23/2008 | 101 | 10,606,350 | 268 | 3,168,318 | 263 | 3,680,335 | 27,301 | SRR2558885 | LLFI00000000 |
| ABBL064 | 12/8/2008 | 101 | 12,685,308 | 320 | 3,168,318 | 479 | 3,871,975 | 15404 | SRR2558886 | LLFJ00000000 |
| ABBL065 | 12/9/2008 | 101 | 8,501,444 | 215 | 3,168,318 | 75 | 4,039,674 | 154,921 | SRR2558887 | LLFK00000000 |
| ABBL066 | 12/11/2008 | 101 | 8,860,706 | 224 | 3,168,318 | 72 | 4,319,668 | 106,135 | SRR2558889 | LLFL00000000 |
| ABBL067 | 12/21/2008 | 101 | 4,584,514 | 116 | 3,168,318 | 78 | 4,032,722 | 114,946 | SRR2558890 | LLFM00000000 |
| ABBL067a | 5/12/2005 | 101 | 12,508,224 | 316 | 3,168,318 | 320 | 3,992,826 | 22,794 | SRR2558891 | LLFN00000000 |
| ABBL067b | 9/14/2005 | 101 | 13,860,118 | 350 | 3,168,318 | 357 | 4,059,209 | 22,441 | SRR2558892 | LLFO00000000 |
| ABBL067c | 12/25/2005 | 101 | 12,256,156 | 309 | 3,168,318 | 396 | 4,013,666 | 19,274 | SRR2558893 | LLFP00000000 |
| ABBL067e | 12/25/2005 | 101 | 13,185,332 | 333 | 3,168,318 | 370 | 3,999,590 | 21,308 | SRR2558894 | LLFQ00000000 |
| ABBL067f | 1/11/2006 | 101 | 12,775,890 | 323 | 3,168,318 | 444 | 3,969,780 | 16,568 | SRR2558896 | LLFR00000000 |
| ABBL067g | 1/22/2006 | 101 | 12,501,658 | 316 | 3,168,318 | 431 | 3,971,931 | 16,858 | SRR2558897 | LLFS00000000 |
| ABBL067h | 4/3/2006 | 101 | 12,791,622 | 323 | 3,168,318 | 404 | 4,002,856 | 19,517 | SRR2558898 | LLFT00000000 |
| ABBL067i | 4/23/2006 | 101 | 6,390,358 | 161 | 3,168,318 | 418 | 3,982,233 | 17,399 | SRR2558900 | LLFU00000000 |
| ABBL067j | 5/20/2007 | 101 | 10,486,564 | 265 | 3,168,318 | 136 | 4,088,444 | 52,671 | SRR2558901 | LLFV00000000 |
| ABBL067k | 12/12/2007 | 101 | 6,091,926 | 154 | 3,168,318 | 98 | 4,002,082 | 82,155 | SRR2558902 | LLFW00000000 |
| ABBL067l | 1/16/2008 | 101 | 8,391,138 | 212 | 3,168,318 | 102 | 4,001,051 | 86,209 | SRR2558903 | LLFX00000000 |
| ABBL068 | 1/14/2009 | 101 | 6,696,118 | 169 | 3,168,318 | 215 | 4,134,094 | 35,378 | SRR2558904 | LLFY00000000 |
| ABBL069 | 2/2/2009 | 101 | 32,813,908 | 829 | 3,168,318 | 319 | 4,091,029 | 25,495 | SRR2558905 | LLFZ00000000 |
| ABBL070 | 2/5/2009 | 101 | 26,881,038 | 679 | 3,168,318 | 265 | 4,020,734 | 31,563 | SRR2558906 | LLGA00000000 |
| ABBL071 | 4/5/2009 | 101 | 6,291,266 | 159 | 3,168,318 | 116 | 4,093,661 | 63,583 | SRR2558908 | LLGB00000000 |
| ABBL072 | 4/9/2009 | 101 | 23,436,746 | 592 | 3,168,318 | 198 | 4,324,713 | 45,835 | SRR2558909 | LLGC00000000 |
| ABBL073 | 4/22/2009 | 101 | 16,950,024 | 428 | 3,168,318 | 131 | 4,092,205 | 78,381 | SRR2558910 | LLGD00000000 |
| ABBL074 | 5/4/2009 | 101 | 7,723,174 | 195 | 3,168,318 | 211 | 3,975,531 | 36,237 | SRR2558911 | LLGE00000000 |
| ABBL075 | 5/12/2009 | 101 | 26,829,806 | 677 | 3,168,318 | 288 | 4,159,435 | 30,922 | SRR2558912 | LLGF00000000 |
| ABBL076 | 5/25/2009 | 101 | 13,587,060 | 343 | 3,168,318 | 386 | 3,868,391 | 19,568 | SRR2558913 | LLGG00000000 |
| ABBL077 | 5/26/2009 | 101 | 35,514,828 | 897 | 3,168,318 | 305 | 3,957,932 | 27,593 | SRR2558914 | LLGH00000000 |
| ABBL078 | 6/15/2009 | 101 | 16,773,812 | 424 | 3,168,318 | 297 | 3,973,781 | 24,307 | SRR2558916 | LLGI00000000 |
| ABBL079 | 7/3/2009 | 101 | 28,887,508 | 729 | 3,168,318 | 342 | 4,265,921 | 22,807 | SRR2558917 | LLGJ00000000 |
| ABBL080 | 8/7/2009 | 101 | 26,434,436 | 667 | 3,168,318 | 428 | 3,976,763 | 19,694 | SRR2558918 | LLGK00000000 |
| ABBL082 | 9/1/2009 | 101 | 25,406,536 | 642 | 3,168,318 | 292 | 3,852,334 | 25,470 | SRR2558920 | LLGL00000000 |
| ABBL083 | 8/30/2009 | 101 | 30,989,798 | 782 | 3,168,318 | 301 | 3,940,840 | 25,555 | SRR2558921 | LLGM00000000 |
| ABBL085 | 9/11/2009 | 101 | 27,206,694 | 687 | 3,168,318 | 241 | 3,813,467 | 29,772 | SRR2558922 | LLGN00000000 |
| ABBL086 | 9/10/2009 | 101 | 16,590,444 | 419 | 3,168,318 | 348 | 4,058,485 | 22,997 | SRR2558923 | LLGO00000000 |
| ABBL088 | 9/13/2009 | 101 | 17,105,376 | 432 | 3,168,318 | 330 | 4,021,290 | 25,565 | SRR2558924 | LLGP00000000 |
| ABBL089 | 10/6/2009 | 101 | 34,503,130 | 871 | 3,168,318 | 327 | 3,997,927 | 26,072 | SRR2558925 | LLGQ00000000 |
| ABBL090 | 10/9/2009 | 101 | 38,910,236 | 982 | 3,168,318 | 310 | 3,817,936 | 24,138 | SRR2558926 | LLGR00000000 |
| ABBL091 | 11/7/2009 | 101 | 28,234,296 | 713 | 3,168,318 | 436 | 4,115,100 | 19,684 | SRR2558927 | LLGS00000000 |
| ABBL092 | 11/13/2009 | 101 | 30,855,824 | 779 | 3,168,318 | 399 | 3,883,553 | 19,303 | SRR2558928 | LLGT00000000 |
| ABBL093 | 11/12/2009 | 101 | 19,490,714 | 492 | 3,168,318 | 309 | 3,874,412 | 25,329 | SRR2558929 | LLGU00000000 |
| ABBL094 | 12/14/2009 | 101 | 18,463,954 | 466 | 3,168,318 | 369 | 3,903,234 | 19,927 | SRR2558737 | LLGV00000000 |
| ABBL095 | 12/19/2009 | 101 | 12,628,086 | 319 | 3,168,318 | 218 | 3,885,321 | 30,203 | SRR2558739 | LLGW00000000 |
| ABBL096 | 1/20/2010 | 101 | 37,281,074 | 941 | 3,168,318 | 306 | 3,898,693 | 23,502 | SRR2558766 | LLGX00000000 |
| ABBL097 | 2/19/2010 | 101 | 29,085,722 | 734 | 3,168,318 | 261 | 3,905,810 | 28,228 | SRR2558767 | LLGY00000000 |
| ABBL098 | 3/6/2010 | 101 | 25,439,548 | 642 | 3,168,318 | 269 | 3,853,967 | 27,320 | SRR2558768 | LLGZ00000000 |
| ABBL099 | 3/6/2010 | 101 | 17,538,744 | 443 | 3,168,318 | 231 | 3,840,161 | 27,302 | SRR2558769 | JPDG00000000 |
| ABBL100 | 4/9/2010 | 101 | 27,451,932 | 693 | 3,168,318 | 241 | 3,939,568 | 30,882 | SRR2558772 | LLHA00000000 |
| ABBL101 | 5/3/2010 | 101 | 19,403,912 | 490 | 3,168,318 | 382 | 3,931,279 | 20,041 | SRR2558773 | LLHB00000000 |
| ABBL102 | 6/4/2010 | 101 | 14,401,494 | 364 | 3,168,318 | 231 | 3,776,367 | 31,608 | SRR2558774 | LLHC00000000 |
| ABBL103 | 7/29/2010 | 101 | 12,252,832 | 309 | 3,168,318 | 207 | 3,987,056 | 37,009 | SRR2558775 | LLHD00000000 |
| ABBL105 | 9/6/2010 | 101 | 37,753,256 | 953 | 3,168,318 | 376 | 4,118,335 | 21,631 | SRR2558777 | LLHE00000000 |
| ABBL106 | 9/6/2010 | 101 | 40,287,706 | 1,017 | 3,168,318 | 370 | 4,124,964 | 21,780 | SRR2558778 | LLHF00000000 |
| ABBL107 | 9/6/2010 | 101 | 22,537,832 | 569 | 3,168,318 | 292 | 3,979,777 | 27,143 | SRR2558779 | LLHG00000000 |
| ABBL109 | 9/6/2010 | 101 | 22,392,882 | 565 | 3,168,318 | 279 | 3,954,675 | 29,282 | SRR2558780 | LLHH00000000 |
| ABBL110 | 9/27/2010 | 101 | 15,009,192 | 379 | 3,168,318 | 250 | 3,694,263 | 30,739 | SRR2558781 | LLHI00000000 |
| ABBL111 | 11/16/2010 | 101 | 28,853,656 | 729 | 3,168,318 | 419 | 4,108,846 | 24,331 | SRR2558782 | LLHJ00000000 |
| ABBL113 | 12/15/2010 | 101 | 14,977,074 | 378 | 3,168,318 | 308 | 4,019,479 | 26,483 | SRR2558783 | LLHK00000000 |
| ABBL114 | 1/20/2011 | 101 | 39,589,430 | 1,000 | 3,168,318 | 337 | 4,137,977 | 25,732 | SRR2558784 | LLHL00000000 |
| ABBL115 | 2/21/2011 | 101 | 32,786,960 | 828 | 3,168,318 | 487 | 3,942,056 | 15,490 | SRR2558785 | LLHM00000000 |
| ABBL116 | 2/26/2011 | 101 | 12,053,200 | 304 | 3,168,318 | 377 | 4,044,273 | 23,431 | SRR2558786 | LLHN00000000 |
| ABBL117 | 3/24/2011 | 101 | 16,212,654 | 409 | 3,168,318 | 550 | 4,115,897 | 14,029 | SRR2558788 | LLHO00000000 |
| ABBL118 | 4/5/2011 | 101 | 28,120,660 | 710 | 3,168,318 | 419 | 3,938,213 | 18,512 | SRR2558790 | LLHP00000000 |
| ABBL120 | 6/16/2011 | 101 | 13,437,746 | 339 | 3,168,318 | 536 | 3,979,007 | 14,187 | SRR2558791 | LLHQ00000000 |
| ABBL121 | 7/25/2011 | 101 | 6,002,822 | 152 | 3,168,318 | 466 | 4,039,842 | 17,305 | SRR2558792 | LLHR00000000 |
| ABBL122 | 7/30/2011 | 101 | 40,444,704 | 1,021 | 3,168,318 | 306 | 3,922,223 | 23,597 | SRR2558797 | LLHS00000000 |
| ABBL123 | 8/23/2011 | 101 | 14,220,382 | 359 | 3,168,318 | 596 | 3,795,535 | 12,218 | SRR2558798 | LLHT00000000 |
| ABBL124 | 9/3/2011 | 101 | 4,054,356 | 102 | 3,168,318 | 197 | 4,013,619 | 34,239 | SRR2558799 | LLHU00000000 |
| ABBL125 | 11/8/2011 | 101 | 20,151,532 | 509 | 3,168,318 | 456 | 3,780,263 | 15,263 | SRR2558800 | LLHV00000000 |
| ABBL126 | 11/8/2011 | 101 | 20,027,030 | 506 | 3,168,318 | 349 | 3,821,336 | 20,964 | SRR2558805 | LLHW00000000 |
| ABBL127 | 11/13/2011 | 101 | 9,442,450 | 238 | 3,168,318 | 356 | 4,152,329 | 22,917 | SRR2558807 | LLHX00000000 |
| ABBL128 | 11/23/2011 | 101 | 20,775,318 | 525 | 3,168,318 | 458 | 3,950,643 | 19,092 | SRR2558809 | LLHY00000000 |
| ABBL129 | 1/5/2012 | 101 | 5,353,678 | 135 | 3,168,318 | 186 | 4,027,647 | 38,732 | SRR2558810 | LLHZ00000000 |
| ABBL130 | 2/12/2012 | 101 | 22,383,756 | 565 | 3,168,318 | 343 | 3,932,290 | 21,064 | SRR2558811 | LLIA00000000 |
| ABBL131 | 2/14/2012 | 101 | 16,386,078 | 414 | 3,168,318 | 394 | 3,989,817 | 19,923 | SRR2558812 | LLIB00000000 |
| ABBL132 | 3/2/2012 | 101 | 23,009,154 | 581 | 3,168,318 | 383 | 3,983,304 | 19,969 | SRR2558813 | LLIC00000000 |
| ABBL133 | 3/19/2012 | 101 | 14,862,736 | 375 | 3,168,318 | 361 | 3,930,248 | 21,247 | SRR2558814 | LLID00000000 |
| ABBL134 | 3/22/2012 | 101 | 19,728,578 | 498 | 3,168,318 | 369 | 4,054,261 | 23,792 | SRR2558815 | LLIE00000000 |
| ABBL135 | 5/1/2012 | 101 | 24,722,876 | 624 | 3,168,318 | 495 | 3,915,368 | 17,220 | SRR2558816 | LLIF00000000 |
| ABBL137 | 5/14/2012 | 101 | 17,330,758 | 438 | 3,168,318 | 428 | 3,919,186 | 18,907 | SRR2558818 | LLIG00000000 |
| ABBL138 | 5/18/2012 | 101 | 16,430,912 | 415 | 3,168,318 | 435 | 3,882,593 | 17,739 | SRR2558819 | LLIH00000000 |
| ABBL140 | 6/9/2012 | 101 | 49,913,702 | 1,260 | 3,168,318 | 543 | 3,869,396 | 13,065 | SRR2558822 | LLII00000000 |
| ABBL141 | 6/12/2012 | 101 | 13,875,892 | 350 | 3,168,318 | 367 | 4,236,436 | 23,187 | SRR2558824 | LLIJ00000000 |
| ABBL142 | 7/31/2012 | 101 | 11,970,574 | 302 | 3,168,318 | 259 | 3,978,019 | 29,463 | SRR2558825 | LLIK00000000 |
| ABBL143 | 8/31/2012 | 101 | 79,935,976 | 2,018 | 3,168,318 | 478 | 3,908,504 | 16,117 | SRR2558826 | LLIL00000000 |
| ABBL144 | 9/16/2012 | 101 | 14,559,986 | 368 | 3,168,318 | 381 | 3,943,688 | 20,834 | SRR2558827 | LLIM00000000 |
| ABBL147 | 10/10/2012 | 101 | 28,332,702 | 715 | 3,168,318 | 395 | 3,716,396 | 19,347 | SRR2558828 | LLIN00000000 |
| ABBL148 | 10/22/2012 | 101 | 20,550,420 | 519 | 3,168,318 | 423 | 3,549,950 | 15,415 | SRR2558829 | LLIO00000000 |
| ABBL149 | 11/10/2012 | 101 | 30,202,168 | 763 | 3,168,318 | 373 | 3,794,913 | 19,926 | SRR2558830 | LLIP00000000 |
| ABOB01 | 6/20/2013 | 101 | 7,652,372 | 193 | 3,168,318 | 139 | 4,062,889 | 58,695 | SRR2559322 | LLIQ00000000 |
| ABOB02 | 6/29/2013 | 101 | 14,825,292 | 374 | 3,168,318 | 389 | 3,963,019 | 18,994 | SRR2559323 | LLIR00000000 |
| ABOB03 | 7/2/2013 | 101 | 16,796,756 | 424 | 3,168,318 | 370 | 3,977,012 | 21,240 | SRR2559352 | LLIS00000000 |
| ABOB04 | 8/31/2013 | 101 | 19,297,440 | 487 | 3,168,318 | 304 | 3,971,774 | 23,905 | SRR2559353 | LLIT00000000 |
| ABOB04_a | 9/9/2013 | 101 | 16,698,092 | 422 | 3,168,318 | 364 | 3,978,511 | 21,753 | SRR2559354 | LLIU00000000 |
| ABOB04_b | 11/16/2013 | 101 | 15,920,118 | 402 | 3,168,318 | 432 | 4,049,068 | 19,216 | SRR2559355 | LLIV00000000 |
| ABOB06 | 10/25/2013 | 101 | 13,923,144 | 352 | 3,168,318 | 278 | 3,993,533 | 27,005 | SRR2559356 | LLIW00000000 |
| ABOB06_a | 11/21/2013 | 101 | 20,292,790 | 512 | 3,168,318 | 602 | 3,891,945 | 10,555 | SRR2559357 | LLIX00000000 |
| ABOB07 | 10/30/2013 | 101 | 25,414,296 | 642 | 3,168,318 | 1,081 | 3,836,895 | 5,674 | SRR2559358 | LLIY00000000 |
| ABOB08 | 10/31/2013 | 101 | 15,633,066 | 395 | 3,168,318 | 418 | 3,993,223 | 19,275 | SRR2559359 | LLIZ00000000 |
| ABOB09 | 11/8/2013 | 101 | 19,274,076 | 487 | 3,168,318 | 375 | 3,945,737 | 20,698 | SRR2559324 | LLJA00000000 |
| ABOB10 | 11/15/2013 | 101 | 17,320,904 | 437 | 3,168,318 | 460 | 3,948,321 | 17,481 | SRR2559325 | LLJB00000000 |
| ABOB11 | 11/16/2013 | 101 | 20,312,352 | 513 | 3,168,318 | 401 | 3,953,603 | 20,420 | SRR2559326 | LLJC00000000 |
| ABOB12 | 11/18/2013 | 101 | 17,876,054 | 451 | 3,168,318 | 284 | 4,086,064 | 27,116 | SRR2559327 | LLJD00000000 |
| ABOB15 | Unknown | 101 | 15,080,752 | 381 | 3,168,318 | 89 | 4,021,456 | 84,258 | SRR2559328 | LLJE00000000 |
| ABOB16 | Unknown | 101 | 16,396,238 | 414 | 3,168,318 | 324 | 3,852,656 | 22,273 | SRR2559329 | LLJF00000000 |
| ABOBEN | Unknown | 101 | 18,111,886 | 457 | 3,168,318 | 381 | 3,940,575 | 20,023 | SRR2559351 | LLJG00000000 |
WGS, whole-genome sequencing.
NA, not available.
Multilocus sequence typing.
The sequences for the Institut Pasteur MLST genes (20) were extracted from the assembled contigs for all isolates, concatenated, and aligned with MUSCLE (51). A phylogeny was inferred with MEGA version 5.2.2 (52), using the maximum-likelihood method, and exported to FigTree version 1.4.2 for visualization (53). In addition, sequence types were determined for all A. baumannii isolates using the database available on the Institut Pasteur MLST website (http://www.pasteur.fr/mlst).
Whole-genome phylogeny and SNP detection.
Unless otherwise stated, single nucleotide polymorphism (SNP) analyses were based on the core genome, which was defined as sequences found in at least 95% of the ABBL isolates (54, 55). The value of >95% was used to avoid substantial changes in the core-genome definition due to one or two isolates that might have undergone gene deletion or for which assembly errors may have resulted in the omission of genes. The kSNP version 2.1.2 program, which uses k-mers (all possible stretches of k-consecutive nucleotides) from input genomes to identify SNPs, was used for this purpose (56). This program has the advantage of not requiring multiple sequence alignments or comparisons with a reference genome. Thirty-one-base-pair k-mers were used, as suggested by the Kchooser script included with kSNP. SNPs were identified by comparing orthologous k-mers from distinct isolates that were identical except for the central nucleotide (nucleotide 16). The kSNP software has the option of searching for k-mers in either assembled genomic sequences or raw sequencing reads. We chose to use assembled sequences, as the de novo assembly process filters out most nonspecific and low-quality reads. However, de novo assembly of short Illumina reads can also produce errors in some genomes, such as the omission of regions from the final assembled contigs or the collapse of repeat genomic regions with one or more nucleotide differences into a single contig. To correct for these errors, we applied a supplemental bioinformatics approach to the kSNP output. Briefly, the Jellyfish k-mer counting software version 1.1.5 (57) was used to directly extract all possible k-mers from each set of unassembled sequencing reads. A Perl script was then used to query these k-mers against the list of all k-mer outputs by kSNP analysis of assembled contigs. For each isolate, if a particular k-mer was not found in the assembled contigs but was found in five or more sequencing reads, the base at the SNP position of that k-mer was added to the kSNP output matrix for that genome. To avoid miscalling sequencing errors as SNPs, a base was removed from the kSNP output matrix for a genome and replaced with a gap (“-”) if it met the following criteria: the k-mer was found to be mixed in the sequencing reads (i.e., two or more k-mer sequences were identified that differed at the central SNP position only), and the occurrence of the most abundant minority k-mer was at least 10% of the occurrence of the majority k-mer. Likewise, if a core-genome k-mer was missing from a strain, the corresponding gap (“-”) was treated as missing data and did not contribute to the placement of that strain in the phylogenetic tree to avoid potentially false inferences resulting from sequencing errors. When indicated, kSNP results were confirmed by the alignment of raw sequencing reads against a reference sequence using the bwa alignment program version 0.7.6a-r433 (58). SNPs were sometimes examined using a combination of automated variant calling with the programs SAMtools and bcftools (both version 0.1.19-44428cd) (59) and manual examination of regions of interest using the Tablet alignment visualization program version 1.13.05.17 (60). To minimize false-positive SNP calls from raw read alignments to sequence contigs due to ambiguous mapping of reads to repeat regions, SNP calls produced by SAMtools and bcftools from the alignment of reads from the query genome to reference contigs were filtered using SNP calls produced from the alignment of reads from the reference genome back to the reference contigs. Any SNP calls found in both alignments were removed. To visualize the relative SNP density, any remaining high-quality SNPs (quality score, 222) between the query genome and reference genome were plotted using CGView (61). The scripts developed for these and other analyses used in this study can be found at the GitHub website (https://github.com/egonozer/snp_tools).
To determine whether the acquisition of DNA sequences through recombination influenced the phylogenetic relationships we obtained, a subset of 23 arbitrarily selected isolates was chosen for further analysis. Whole-genome alignments were generated by aligning sequencing reads of these 23 isolates to the reference genome of A. baumannii ATCC 17978 (GenBank accession no. CP000521) using the bwa program (58). SNPs were called using SAMtools and bcftools. Indels, positions with variant quality <30, positions covered by <8 reads, or positions with <0.7 of the reads representing either the reference or an alternate base were filtered. A tree was then produced from the alignments using ClonalFrameML (62) to remove potential recombinant regions. A comparison of this tree to one using the kSNP approach described above showed no significant differences in topology (data not shown), demonstrating that SNPs present in recombination regions had a minimal impact on the tree structures we obtained.
To determine pairwise core-genome SNP counts, we developed software based on kSNP to (i) identify a core genome of k-mers from genomic assemblies and sequencing reads, and (ii) count SNPs between pairs of these core-genome k-mer sets. First, the set of core-genome k-mers was identified using the kmer_core.pl software. This software compares k-mers among a set of input genomic DNA sequences and outputs all k-mers with identical 15-nucleotide flanking arms (e.g., identical except perhaps at the central nucleotide) that were present in a specified subset of the genomes. For the purposes of this study, a subset cutoff of k-mers present in ≥95% of the input genomes was chosen (54, 55). The assembled genomes of the A. baumannii ABBL strains were used as input for kmer_core.pl to identify a core k-mer set. A second software package (kmer_compare.pl) was then used to search for these core k-mers in query sequences and perform pairwise comparisons to determine the SNP totals between the genomes. To maximize the sensitivity of identification of core k-mers in the strains sequenced for this study, raw sequencing reads (rather than assembled genomes) were used as input to kmer_compare.pl, and a k-mer was considered present if at least 5 sequencing reads were found to contain the k-mer. For reference strains, the genomic sequence was used to search for core k-mers, and k-mers found at least once were considered present. Conflicting k-mers, defined as two or more k-mers present in a single genome that were identical except at the central base, were filtered out of the k-mer set of each genome. To avoid erroneous filtering of k-mers appearing to be in conflict due to sequencing errors in a few reads, conflicting core k-mers were only removed if the number of reads containing k-mers with the most-abundant minority central base was at least 10% of the number of reads containing k-mers with the majority central base. To generate a jitter plot showing the number of core SNPs between pairs of isolates, Microsoft Excel was used to randomly assign an x axis position to each data point within the boundaries of an appropriate column.
For the ABBL isolates, A. baumannii isolates, and ABOB isolates, FastTree2 (63) was used to estimate maximum-likelihood phylogenetic trees (64) from core SNPs, and trees were then visualized in FigTree version 1.4.2 (53). Whole-genome sequences for representative A. baumannii strains from each of the three major IC lineages (65) were downloaded from the NCBI database and also included in some phylogenetic analyses.
To inform the outbreak transmission map, BEAST 2.3.1 (66) was applied to whole-genome SNP, patient of origin, and date of isolation information of the ABOB isolates to generate a Bayesian phylogenetic reconstruction of the outbreak. A Hasegawa, Kishino, and Yano (HKY) substitution model assuming a constant population size was used for the analysis. This information was combined with epidemiological linkages to estimate transmission patterns.
Outbreak analysis.
From June 2013 through December 2013, a CRAB outbreak occurred at NMH and was centered in two ICUs: ICU A, a cardiothoracic ICU that also houses solid-organ transplant recipients, and ICU B, a medical ICU. As part of its investigation, the NMH Department of Infection Prevention and Control performed PFGE typing and assessed epidemiological links. All CRAB isolates deemed to be part of the outbreak had identical or closely related PFGE types. For the purpose of constructing the transmission map, patient-to-patient spread was considered the most likely mode of transmission, and patients were considered to have a direct epidemiologic link if they overlapped in the same unit for >24 h. Indirect links included overlap in the same unit for <24 h, nonoverlapping stays in the same unit in close proximity, and environmental links, such as exposure to contaminated rooms or equipment. As part of the current study, the first CRAB isolates from 10 patients involved in the outbreak were sequenced using the methods described above. Two additional isolates from patient 4 (ABOB04_a and ABOB04_b) and one additional isolate from patient 6 (ABOB06_a) collected during the outbreak were also sequenced. As controls, an environmental isolate taken from the room of patient 4 (ABOBEN) and an isolate with a nonoutbreak PFGE type obtained from a patient in ICU A during the time of the outbreak (ABOB11) were also sequenced. Finally, two carbapenem-susceptible A. baumannii isolates from patients admitted to different locations in the hospital during the outbreak (ABOB15 and ABOB16) were sequenced and also used as controls.
Core- and accessory-genome analyses of outbreak isolates.
As a first step in defining the accessory genome of the outbreak isolates, a core genome was defined by applying the software program Spine (55) to the assembled sequences of the 116 A. baumannii ABBL bloodstream isolates. Briefly, the set of all pairwise whole-genome alignments was used to identify sequences present in 95% of the 116 ABBL isolates (54, 55), and these sequences were defined as the core genome. The software program AGEnt (55) was then used to perform in silico subtractive hybridization of the core genome from the whole-genome sequences of the CRAB ICU outbreak (ABOB) isolates. In this way, the accessory genome of each ABOB isolate was determined. Using BLAST+ version 2.2.24 (44, 46) and in-house Perl scripts, the accessory-genomic sequences of the ABOB strains were aligned and clustered to identify the set of accessory-genomic elements among these strains. To support the accessory element carriage patterns in each strain and correct for false negatives due to misassembly, sequencing reads from each strain were aligned to the set of accessory element sequences using the bwa alignment program (58). The read coverage at each position in the accessory elements was determined from the alignments, and positions with ≥5 aligning reads were considered to be present in the respective genome. A graphical representation of the accessory element composition of each strain was produced using CGView (61). ABOB accessory genetic elements were annotated using Prokka version 1.9 (67). ABOB accessory genomic elements were compared to the nucleotide sequence of A. baumannii plasmid ABKp1 (NCBI accession no. CP001922.1) using BLAST, and gene similarities between these elements and the plasmid were visualized using EasyFig version 2.1 (68).
Comparison of typing techniques.
The adjusted Wallace coefficient, which compares two sets of partitions from different microbial typing methods, was used to determine the congruence between typing techniques (69). Simpson's index of diversity was used to determine the discriminatory power of each individual typing technique (69, 70). For any given typing system, this index measures the probability that two randomly sampled strains will belong to different types. Calculations were performed with the online tool on the Comparing Partitions website (http://darwin.phyloviz.net/ComparingPartitions/). In addition, a sensitivity analysis was performed to evaluate the performance of conventional typing techniques in comparison to WGS in the context of generating a transmission map for a hospital outbreak. For this analysis, WGS was considered to be the gold standard, and a positive result was defined as one that accurately distinguished two distinct isolates as unique.
Nucleotide sequence accession numbers.
All sequence read sets and assembled sequences are deposited in the National Center for Biotechnology Information (NCBI) database (Table 1).
RESULTS
Determining the bacterial species of isolates within the ACB complex.
Species within the ACB complex differ markedly in the severity of illness they cause and in their resistance to antibiotics (7, 13). Accurate identification of Acinetobacter to the species level is becoming increasingly important in the clinical setting. We therefore attempted to determine the species of 154 clinical ACB complex bloodstream isolates cultured from patients at our institution during the study period. One isolate actually contained a mixed culture and was excluded from further analysis. Five isolates gave poor-quality sequencing assemblies, despite repeated attempts, and were also excluded, leaving 148 isolates for analysis. We identified the species of these 148 isolates by rpoB gene sequence analysis, an accepted method of parsing ACB complex bacteria into species (45, 71). This analysis indicated the following Acinetobacter species distribution: A. baumannii (n = 116), A. pittii (n = 28), A. nosocomialis (n = 3), and A. soli (n = 1). Because A. soli is not a member of the ACB complex, the clinical microbiology laboratory presumably misidentified this isolate. These results confirm previous findings that a substantial number of ACB complex isolates acquired from clinical settings are non-baumannii Acinetobacter species (10).
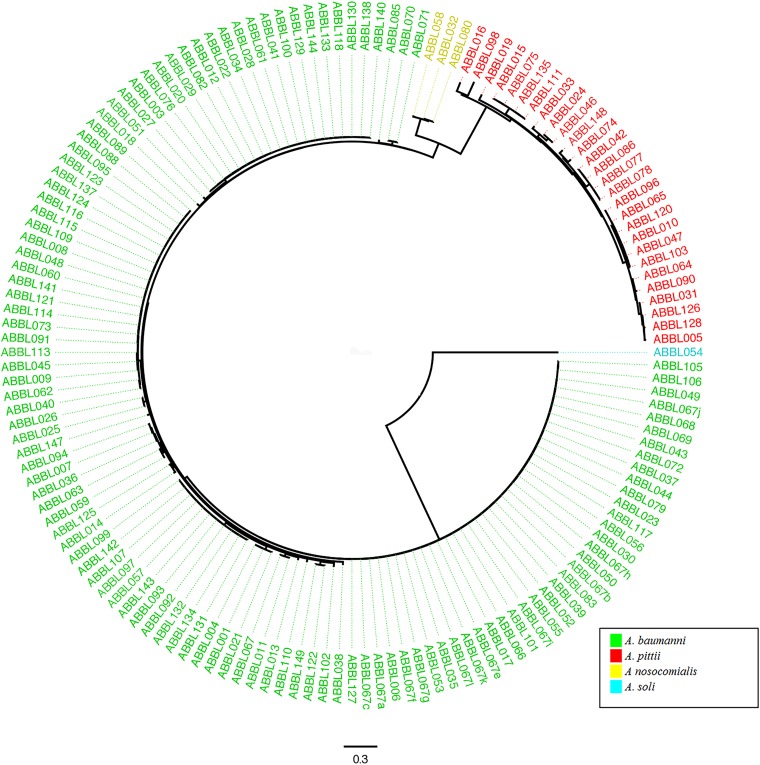
We next examined the utility of using WGS to identify isolates within the ACB complex to the species level. In addition to supplying the sequence of the rpoB gene for each isolate, WGS also allowed the determination of phylogeny via SNP analysis. A phylogenetic tree inferred from core-genome SNPs revealed well-delineated monophyletic lineages corresponding to each species within the ACB complex (Fig. 1). (An alternate version of the tree more clearly demonstrating the species clustering is shown in Fig. S1 in the supplemental material.) Because A. soli is genetically distant from the ACB complex (72, 73), it was used to root the tree. Similar to prior studies, our results showed that A. nosocomialis diverges first from the A. baumannii clade, followed by A. pittii (20). These results confirm that phylogenic analysis based upon SNPs in conserved sequences is capable of distinguishing species within the ACB complex.
FIG 1.
Phylogenetic analysis based on core-genome sequences of all Acinetobacter isolates. A phylogenetic tree for all Acinetobacter isolates was inferred from core SNPs using kSNP version 2. The tree was rooted on isolate ABBL054 (A. soli). The colors correspond to individual ACB complex species, as determined by rpoB gene sequence analysis, and these are indicated in the key. The scale bar indicates branch length, expressed as the number of changes per total number of SNPs.
Next, a band-based typing method was used to examine the collection of ACB complex isolates. Because the NMH clinical microbiology laboratory routinely performed PFGE only on carbapenem-resistant ACB complex isolates (nearly all of which were A. baumannii), we focused on Rep-PCR. This methodology identified 50 unique types among the clinical isolates (Table 2). A dendrogram created from the Rep-PCR fingerprints for all ACB complex isolates demonstrated poor delineation of the isolates into distinct species (see Fig. S2 in the supplemental material). These results demonstrate that Rep-PCR is not a useful genotyping method for determining distant phylogenetic relationships among isolates within the ACB complex.
TABLE 2.
Comparison of typing methods used to characterize CRAB isolatesa
| Typing methodb | No. of types | Simpson's index (95% CI)c | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| PFGE | 40 | 0.892 (0.834–0.950) | 90 | 53 |
| Rep-PCR | 50 | 0.970 (0.950–0.990) | 97 | 57 |
| MLST | 10 | 0.758 (0.706–0.810) | 81 | 100 |
| WGSd | 75 | 0.997 (0.995–1.000) |
In this analysis, sensitivity refers to the ability of the technique to correctly discriminate two unique isolates, and specificity refers to the ability of the technique to correctly group two identical isolates. Sensitivity and specificity were calculated in comparison to WGS as the gold standard.
PFGE, pulsed-field gel electrophoresis; Rep-PCR, repetitive extragenic palindromic-PCR; MLST, multilocus sequence typing; WGS, whole-genome sequencing.
CI, confidence interval.
Isolates were considered the same type by WGS if there were 0 SNPs between them.
Assignment of A. baumannii clinical isolates to ST lineages.
Previous investigations established that certain clonal lineages of A. baumannii have spread widely across and between continents. Some of these lineages have been referred to as international clones (ICs) or European clones (ECs) (17–19, 21). The widespread distribution of these clonal lineages implies that they are highly adapted for persistence and transmission in health care environments. MLST has become the gold standard for assigning A. baumannii isolates to STs that correspond to these lineages. Multilocus sequence types were therefore determined for all A. baumannii strains using the Institut Pasteur database. Twenty-four unique STs were identified among the 116 A. baumannii isolates, including seven new STs, four of which have been assigned: ST496, ST497, ST498, and ST499. However, 90 of 116 (78%) isolates were assigned to one of four STs: ST2, ST79, ST406, and ST499. A phylogeny inferred from the concatenated sequences of the MLST genes for A. baumannii isolates confirmed this clustering (see Fig. S3 in the supplemental material). These results indicated that both previously reported and novel A. baumannii clonal lineages circulated throughout our hospital.
In examining the utility of PFGE and Rep-PCR for identifying clonal lineages of A. baumannii, we found that neither band-based technique accurately grouped strains together in agreement with the MLST data. A dendrogram created using the Rep-PCR results demonstrated only one distinct cluster with >90% similarity (see Fig. S4 in the supplemental material), a cutoff level above which isolates can be considered genetically related (21, 46, 47). This cluster included eight A. baumannii isolates involved in a prolonged multistate outbreak from 2005 to 2006. All of these isolates were ST79, suggesting some concordance between Rep-PCR and MLST in a regional outbreak setting. However, this Rep-PCR cluster also included 11 other isolates that were collected at various times throughout the study and were represented by eight different STs, confirming a lack of agreement with MLST. PFGE likewise showed poor agreement with MLST results (see below).
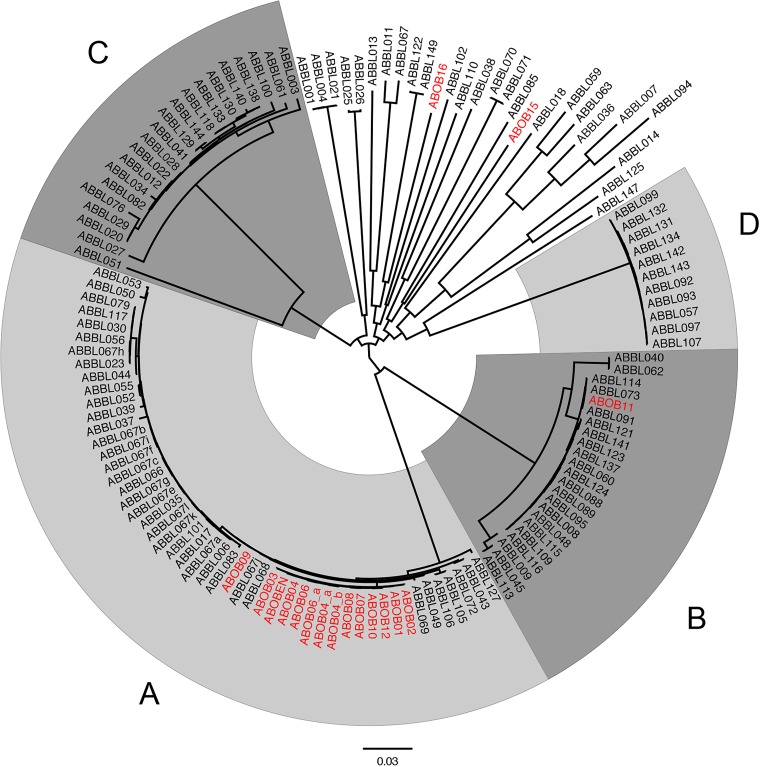
We next examined the ability of WGS to partition A. baumannii isolates into clonal lineages. The core genomes of these isolates were analyzed for the presence of discriminating SNPs, and a phylogenetic tree was created. Four distinct primary clades (labeled A to D) were identified, and also a number of phylogenetically distinct isolates (Fig. 2). Each of the four major clades corresponded to one of the four predominant STs (ST79, ST2, ST406, and ST499 for clades A to D, respectively). Each clade contained isolates collected throughout the 8-year study from patients at differing locations in the hospital, suggesting that they did not represent confined outbreaks within our hospital. To better examine the relationships between the isolates in our hospital and globally disseminated lineages, we added several published strains to our phylogenetic analysis (see Fig. S5 in the supplemental material). Clade B (ST2) strains were closely related to published IC-II strains. Clade C (ST406) is most closely related to IC-I strains, although the tight clustering of the clade C isolates and the phylogenetic distance separating them from the IC-I reference strains suggest that clade C may be a distinct clonal group. Clade A (ST79) isolates do not clearly cluster with any reported IC lineage but are closely related to published strains from the United States and other parts of the world (74–77). Group D (ST499) represents a collection of isolates distinct from known clonal lineages or reported strains. Interestingly, our WGS analysis indicated that IC-III strains did not represent a distinct lineage of strains but rather were phylogenetically diverse. Many of the remaining isolates from our study were interspersed with these published IC-III strains. These findings indicate that WGS accurately assigns A. baumannii isolates to ST lineages and provides additional information regarding the genetic relationships between different lineages.
FIG 2.
Phylogenetic analysis based on core-genome sequences of A. baumannii isolates. A phylogenetic tree for all A. baumannii isolates was inferred from core SNPs using kSNP version 2. Individual taxa and branches of the four major clades are identified by shades of gray and letters (A to D). Isolates collected during the ICU outbreak (those with ABOB) are also shown and colored red. The scale bar indicates branch length, expressed as the number of changes per total number of SNPs.
Use of WGS to define a hospital outbreak of A. baumannii.
A. baumannii frequently causes outbreaks within health care settings, in which a single strain is transferred from one patient to another. Because of the high level of antibiotic resistance associated with some A. baumannii strains, these outbreaks have important consequences for patient care and require the use of considerable resources by infection control personnel. PFGE is routinely used by many hospitals to track nosocomial outbreaks, but several studies indicate that WGS is better suited for this purpose (38, 78, 79). In 2013, a CRAB outbreak occurred in our hospital and was investigated using PFGE and contact tracing. We reexamined this outbreak using WGS to determine whether outbreak isolates and transmissions could be more accurately identified with this technique.
In June 2013, two patients were admitted to ICU B from the same skilled nursing facility within 8 days of each other. Patient 1 grew CRAB (isolate ABOB01) from a respiratory sample 9 days after admission, and patient 2 grew CRAB (isolate ABOB02) with a closely related PFGE type from the blood 10 days after admission. Neither patient was in contact isolation prior to CRAB growth, nor had they overlapped previously with a CRAB patient during their hospital stay. Over the next 5 months, an additional eight patients were identified in ICU A and B who grew CRAB (isolates ABOB03, ABOB04, ABOB06, ABOB07, ABOB08, ABOB09, ABOB10, and ABOB12) with identical or closely related PFGE types to the index cases and were classified as part of the outbreak (see Fig. S6 in the supplemental material). This represented a substantial increase in the monthly number of CRAB cases at the NMH. Three isolates were available from patient ABOB04 (isolates ABOB04, ABOB04_a, and ABOB04_b) and two isolates were from patient ABOB06 (isolates ABOB06 and ABOB06_a). All isolates were susceptible to colistin, and most were susceptible to either doxycycline or minocycline but were otherwise resistant to all antibiotics tested. At the time, epidemiologic contact tracing was performed to estimate a transmission map, which revealed multiple potential infection routes among outbreak patients (Fig. 3A). Interestingly, the PFGE patterns of the two initial isolates ABOB01 and ABOB02 (from patients 1 and 2, respectively) were found to be closely related but not indistinguishable, suggesting that the outbreak may have begun with the introduction of two unique but closely related CRAB isolates into the hospital.
FIG 3.
Possible transmission maps of an ICU CRAB outbreak. Transmission maps were created using patient epidemiologic trace data and PFGE results by the infection control team investigating the outbreak at the time it occurred (A), and additional phylogenetic information inferred from whole-genome sequences (B). (B) Data from whole-genome SNPs were taken into account, as shown in Fig. 5 and Fig. S7 in the supplemental material. Specifically, the most likely patient-to-patient transmissions suggested by BEAST software analysis of WGS data (as shown in Fig. S7 in the supplemental material) were used to resolve ambiguous transmission events in panel A, which in turn yielded the transmission map in panel B. In all maps, nodes represent individual patients, with shaded nodes indicating patients in ICU A and white nodes indicating patients in ICU B. (A) Solid arrows represent direct epidemiological links, while dashed arrows represent indirect or environmental links. (B) Black arrows indicate transmission links supported by WGS data, while red arrows indicate that the transmission is also supported by epidemiologic data. SNF, skilled nursing facility.
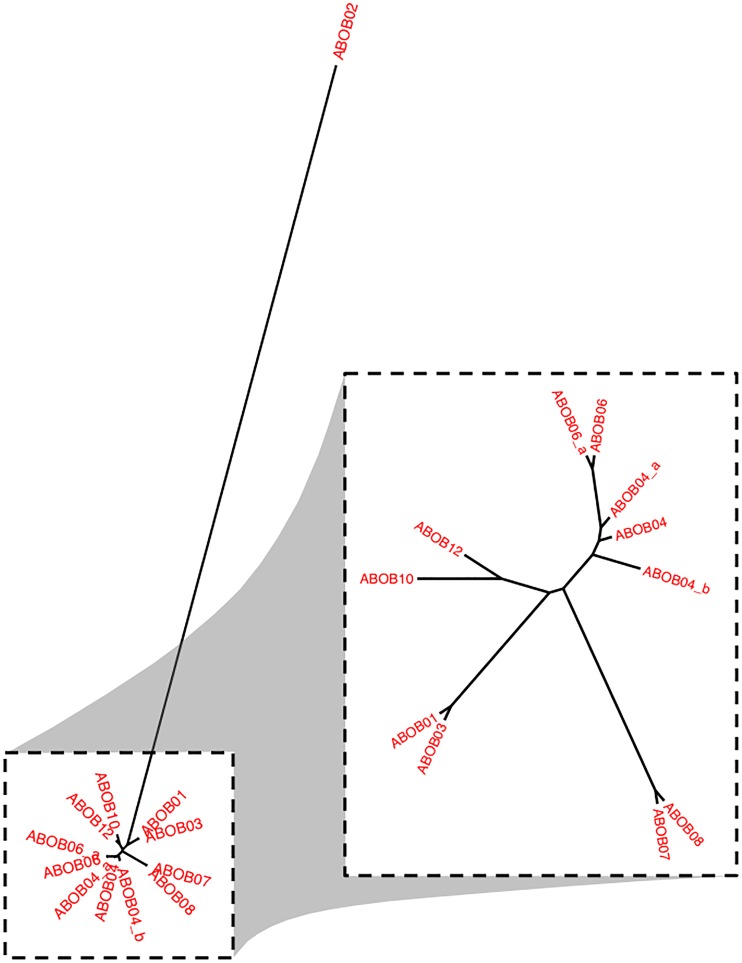
We next applied WGS to investigate this same hospital outbreak. Each outbreak isolate, the control isolates (ABOB11, ABOB15, and ABOB16), and an environmental isolate (ABOBEN) were sequenced, and phylogeny was inferred based on SNPs in the core genome (defined as sequences found in at least 95% of all ABBL isolates) (Fig. 4). As expected, the control isolates were distantly related to the outbreak isolates, which formed a tight cluster. A similar tree including only the clinical outbreak isolates (Fig. 4, first inset) revealed that isolate from patient 9 was genetically distinct, differing by 155 to 157 core SNPs from the other outbreak strains. A closer inspection indicated that these SNPs were not localized to a single small portion of the genome, as would have occurred with a recombination event, but rather were dispersed throughout the chromosome (data not shown). The remaining outbreak isolates differed from each other by 0 to 2 core SNPs. Thus, although patient 9 was infected with a CRAB strain in ICU B during the outbreak, and this CRAB isolate had a PFGE pattern similar to that of the other outbreak isolates, WGS indicated that the patient was not infected with an outbreak strain. ABOB09 was therefore removed from subsequent phylogenetic analyses (Fig. 4, smaller inset).
FIG 4.
Phylogenetic analysis based on core-genome sequences from the ICU CRAB outbreak isolates. A phylogenetic tree for the outbreak and control isolates was inferred from core-genome SNPs using kSNP version 2 and the maximum-likelihood method. Thirteen outbreak isolates (red), three control nonoutbreak isolates (blue), and one environmental outbreak isolate (green) were included. The large inset is the same tree showing only the clinical outbreak (red) isolates and excluding the control and environmental isolates. The small inset excluded isolate ABOB09 to better show the relationships between the remaining isolates. In this analysis, the core genome was defined as sequences present in 95% of the ABBL isolates.
Generation of an intrahospital outbreak transmission map.
The identification of factors allowing an A. baumannii strain to move from patient to patient during a hospital outbreak is necessary to determine the source and mechanisms by which transmission is facilitated in an outbreak. High-resolution genetic information can help distinguish isolates even within the context of a nearly clonal outbreak. We therefore examined whether WGS could discriminate isolates within our ICU outbreak and help generate a more-refined transmission map. To accomplish this, we performed a phylogenetic analysis of whole-genome SNPs in the outbreak isolates (Fig. 5). The core-genome analysis used only SNPs located in the core genome, whereas the whole-genome analysis also included SNPs located in accessory-genome sequences and therefore used more of the information present in the bacterial genomes. In contrast to the core-genome tree, the whole-genome tree placed ABOB02 at an increased distance from the remaining outbreak isolates (Fig. 5). Comparing whole-genome sequences, ABOB02 differed by 98 to 103 SNPs from the remaining outbreak isolates, in contrast to 1 to 2 SNP differences found in the core-genome sequences. At first glance, this difference in the number of whole-genome SNPs suggested that ABOB02 was also not part of the outbreak. However, further examination revealed that most of these SNPs were localized to an ∼14-kb region that is accessory in the ABBL bloodstream isolates but found in each of the outbreak strains (data not shown). Alignment of raw reads from ABOB02 against the ABOB01 sequence and manual examination confirmed these were likely true SNPs and not sequencing errors, with an average of 311 (range, 51 to 696) sequencing reads at each SNP position containing the variant base, and an average of less than one read (range, 0 to 7 reads) containing the reference base (data not shown). The concentration of SNPs in a small region of the genome suggested that this 14-kb accessory region had been acquired by a recent recombination event and therefore did not represent the true genetic history of ABOB02. For this reason, ABOB02 was considered likely to be related to the outbreak. A Bayesian phylogenetic reconstruction of the outbreak (see Fig. S7 in the supplemental material) was generated using the whole-genome sequence SNP information along with isolation dates (see Materials and Methods). The incorporation of these results into the transmission map defined many of the transmission links that were previously ambiguous (Fig. 3B).
FIG 5.
Phylogenetic analysis based on whole-genome sequences from the ICU CRAB outbreak isolates. A phylogenetic tree for 12 outbreak isolates was inferred from SNPs present in the whole genome using kSNP version 2 and the maximum-likelihood method. In the inset, ABOB02 is excluded to better show the genetic distances between the remaining outbreak isolates.
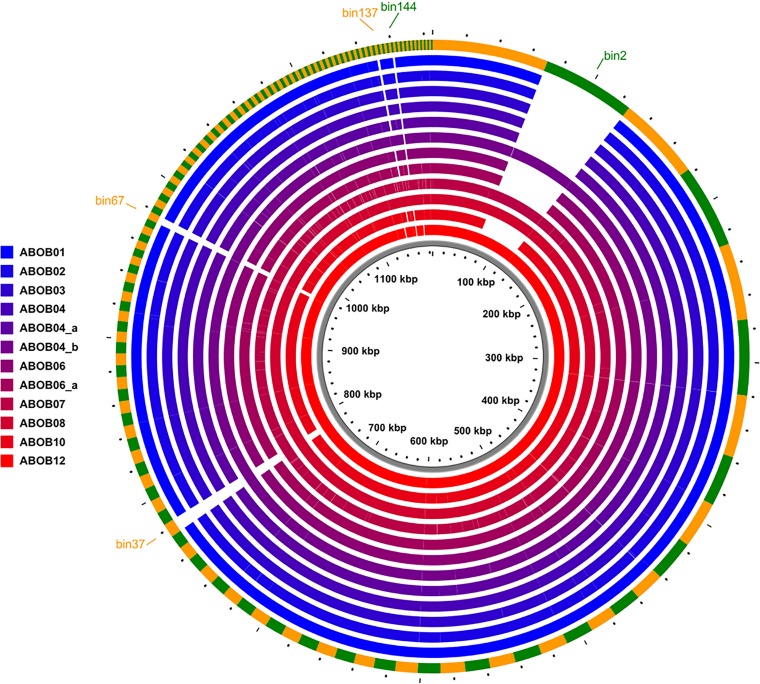
To determine whether there were differences in the accessory genomes of the outbreak isolates beyond SNPs, we looked for accessory sequences that were present in some ABOB isolates but absent from others. We identified four accessory-genome elements (AGEs) present only in isolates ABOB04_b, ABOB07, ABOB08, and ABOB12 and one small AGE present only in isolates ABOB07 and ABOB08 (Fig. 6). These AGEs ranged in size from 1,337 bp to 56,215 bp, totaling 71.8 kb of sequence. These five AGEs are difficult to assemble de novo using short reads, likely due to repeat sequences or ambiguity; however, their presence or absence as a defined set in the outbreak isolates suggested they were in close proximity to or contiguous with each other. This was confirmed by BLAST search, which indicated that together, they were closely homologous with ABKp1, a plasmid found in A. baumannii strain 1656-2 (see Fig. S8 in the supplemental material). Patient 4, who had a prolonged stay in ICU A during the outbreak, harbored isolates both with and without this plasmid. Although plasmids have been shown to be important in the dissemination of antibiotic resistance during outbreaks (80, 81), they are of limited value for defining transmission pathways, because they are frequently gained and lost by closely related circulating strains (74, 82, 83). These results indicated that a plasmid circulated among the outbreak isolates and was the only gross difference in their accessory genomes.
FIG 6.
Accessory-genome differences between ICU CRAB outbreak isolates. All AGEs of >1,000 bp are represented in the circular map, which is organized by size from largest to smallest AGE in a clockwise direction. The outer green and orange ring shows the cumulative collection of AGEs found in at least one of the outbreak isolates. The inner rings indicate the presence or absence of AGEs in each outbreak isolate, colored according to the key. Gaps in color represent AGEs or parts of AGEs that are missing in those isolates. ABOB04, ABOB04_a, and ABOB04_b represent three different isolates from patient 4; ABOB06 and ABOB06_a represent two different isolates from patient 6.
The incorporation of the total information generated by WGS allowed us to refine and remove ambiguities from the previous transmission map based solely on PFGE patterns and epidemiologic links (compare Fig. 3A and B). It also suggested possible transmissions (e.g., from patient 3 to patient 4) that had not been identified by the epidemiological investigation. Such transmission may have occurred indirectly through colonized patients that had not been identified as part of the outbreak. These results suggest that WGS is superior to PFGE for distinguishing A. baumannii outbreak transmission routes, but the quantification of SNPs alone may be misleading, and an examination of both core- and accessory-genome content is beneficial.
SNP thresholds that define isolates as part of a clonal lineage or intrahospital outbreak.
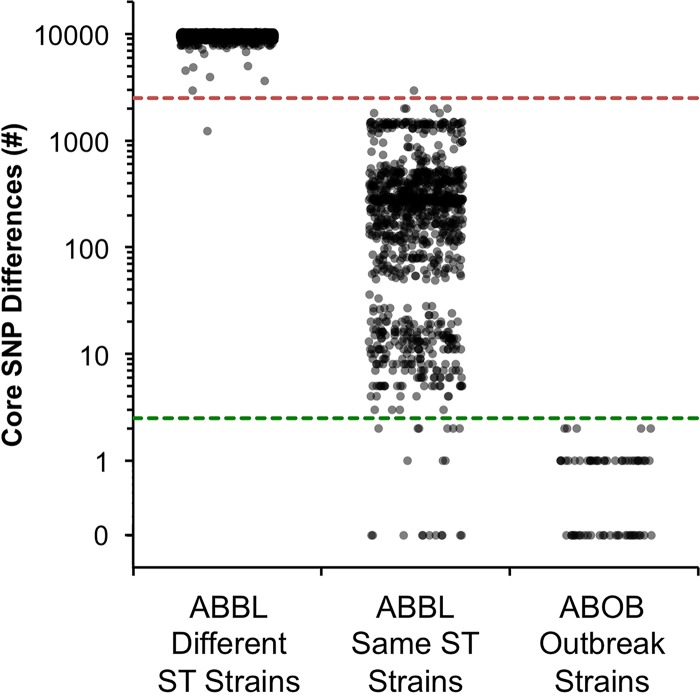
Well-defined criteria are available for interpreting conventional typing techniques for the purpose of parsing clinical isolates into the same clonal lineage or the same hospital outbreak (20, 40–42). Corresponding thresholds of relatedness would be helpful for interpreting WGS results. We hypothesized that pairwise core SNP counts might serve as such thresholds and provide an easy and straightforward method by which infection control practitioners could use WGS data to quickly determine the likelihood of whether any two isolates were part of the same clonal lineage or outbreak. To this end, the total number of core-genome SNPs between any two of the 116 ABBL and 15 ABOB isolates was quantified. These comparisons yielded a minimum of 0 and a maximum of 10,360 core-genome SNPs (median, 9,523 SNPs). We next compared the number of core-genome SNPs between ABBL isolates from different clonal lineages to those from the same clonal lineages (Fig. 7). For the purposes of this analysis, we defined two strains as belonging to the same clonal lineage if they had the same ST. A discontinuity in the distribution of core-genome SNPs was observed between these two groups of isolates. The median number of core-genome SNPs between isolates within the same ST lineage was 261 (interquartile range, 67.5 to 393.5 SNPs) versus 9,613 SNPs (interquartile range, 9,443 to 9,793.75 SNPs) between isolates not within the same ST lineage (Fig. 7). With only two exceptions, a threshold of 2,500 core SNPs distinguished isolates within the same ST lineage from those in different ST lineages (Fig. 7). We next performed a similar analysis on the ABOB outbreak isolates. A maximum of 2 SNPs were found among all pairwise comparisons of the ABOB outbreak isolates. Thus, a threshold of 2.5 SNPs distinguished the vast majority of outbreak isolates from isolates of the same ST lineage that were not part of the outbreak (Fig. 7). Although a number of non-ABOB isolates also differed by <2.5 SNPs, it is possible that some of these represent episodes of unrecognized patient-to-patient transmissions that occurred within our hospital. This analysis needs to be expanded to include other outbreaks at different institutions and of different durations, but these results suggest that core-genome SNP thresholds are useful in preliminarily determining whether an isolate is within or distinct from an ST lineage or intrahospital outbreak.
FIG 7.
Core-genome SNP counts among isolates of the same or different clonal lineage or the same intrahospital outbreak. To generate a jitter plot of core SNP differences, all pairwise core-genome SNP counts were compared among isolates belonging to different clonal lineages (as determined by different STs; column 1, n = 5,026), to the same clonal lineage (the same ST) but not part of a recognized outbreak (column 2, n = 1,079), or to a recognized outbreak (column 3, n = 66). Each dot represents the number of core SNPs between one pair of isolates. The red dashed line indicates a threshold of 2,500 SNPs, and the green dashed line indicates a threshold of 2.5 SNPs.
Comparison of typing methods.
Using the WGS threshold of <2,500 core SNPs, we next compared the four typing techniques (MLST, Rep-PCR, PFGE, and WGS) for their ability to distinguish A. baumannii clonal lineages. Comparisons were restricted to CRAB isolates, because typing results from all methods were available for these isolates. Wallace coefficients, which quantify the congruence between two methods, were calculated for a pairwise comparison of each typing technique (Table 3). Since MLST is currently accepted as the gold standard for the identification of A. baumannii clonal lineages (19), we paid particular attention to how the results of the other techniques compared to MLST. PFGE performed poorly in this regard; two isolates grouped together by PFGE had only a 39% probability of having the same MLST. Rep-PCR performed somewhat better; two isolates grouped together by Rep-PCR had a 65% probability of having the same MLST. As expected based on how the 2,500-core-SNP threshold was derived, two isolates placed in the same clonal lineage by WGS were grouped together by MLST 100% of the time. These results indicate that the band-based techniques PFGE and Rep-PCR differ significantly from MLST in assigning isolates to clonal lineages, but that WGS performed well when a threshold of 2,500 core SNPs was used.
TABLE 3.
Concordance of typing techniques using the adjusted Wallace coefficient for nonoutbreak (ABBL) CRAB isolates
| Typing methoda | Adjusted Wallace coefficient (95% CI)b |
|||
|---|---|---|---|---|
| PFGE | Rep-PCR | MLST | WGSc | |
| PFGE | 0.168 (0.013–0.323) | 0.387 (0.120–0.654) | 0.387 (0.120–0.654) | |
| Rep-PCR | 0.659 (0.603–0.715) | 0.651 (0.415–0.887) | 0.651 (0.415–0.887) | |
| MLST | 0.147 (0.014–0.279) | 0.063 (0–0.131) | 1.0 (1.0–1.0) | |
| WGSc | 0.147 (0.014–0.279) | 0.063 (0–0.131) | 1.0 (1.0–1.0) | |
PFGE, pulsed-field gel electrophoresis; Rep-PCR, repetitive extragenic palindromic-PCR; MLST, multilocus sequence typing; WGS, whole-genome sequencing.
CI, confidence interval.
Isolates were considered the same type by WGS if there were ≤2,500 single nucleotide polymorphisms between them.
We next determined how WGS with the threshold of 2.5 core SNPs compared to PFGE and MLST in correctly identifying A. baumannii isolates as part of an intrahospital outbreak. We compared these techniques using our collection of CRAB isolates, some of which comprised a hospital outbreak (ABOB isolates), but most of which did not (ABBL isolates). Of isolates identified by WGS as having genetic similarity consistent with patient-to-patient transmission (defined as <2.5 core SNPs), 88% had the same PFGE pattern (Table 4). In contrast, of isolates with the same or similar PFGE patterns, only 14% were found to be genetically similar by WGS (<2.5 core SNPs). In other words, PFGE grouped many more isolates together as similar than did WGS, suggesting that WGS has higher discriminatory power. Due to its limited ability to discriminate related strains, MLST showed poor congruence with WGS and PFGE (Table 4).
TABLE 4.
Concordance of typing techniques using the adjusted Wallace coefficient for nonoutbreak (ABBL) CRAB and outbreak (ABOB) CRAB isolates
| Typing methoda | Adjusted Wallace coefficient (95% CI)b |
||
|---|---|---|---|
| PFGE | MLST | WGSc | |
| PFGE | 0.462 (0.228–0.696) | 0.136 (0.087–0.186) | |
| MLST | 0.122 (0.038–0.205) | 0.041 (0.000–0.091) | |
| WGSc | 0.882 (0.841–0.923) | 1.000 (1.000–1.000) | |
PFGE, pulsed-field gel electrophoresis; MLST, multilocus sequence typing; WGS, whole-genome sequencing.
CI, confidence interval.
Isolates were considered the same type by WGS if there were ≤2 single nucleotide polymorphisms between them.
We next directly examined the discriminatory power (i.e., the ability to distinguish any two isolates as distinct) of PFGE, Rep-PCR, MLST, and WGS. The ability to discriminate between two nearly clonal strains is highly important for developing transmission maps during an outbreak. We calculated the Simpson's index, a measure of discriminatory power, of the four typing techniques (Table 2). Overall, both WGS and Rep-PCR were highly discriminatory, with Simpson index values of 0.997 and 0.970, respectively (Table 2). PFGE and MLST were somewhat less discriminatory, with Simpson index values of 0.892 and 0.758, respectively. We also calculated the sensitivity and specificity of the techniques for distinguishing isolates, using WGS as the gold standard. The band-based techniques were highly sensitive for discriminating distinct isolates but suffered from low specificity. In other words, they correctly identified distinct isolates as different but often erroneously distinguished strains that WGS categorized as being the same. MLST was less sensitive in its ability to distinguish isolates but had a specificity of 100%.
DISCUSSION
Hospital-based clinical microbiology laboratories provide a number of services, including the identification of pathogens in clinical specimens and the typing of strains to facilitate regional epidemiological surveillance and hospital outbreak investigation. These tasks require typing techniques with a broad spectrum of discriminatory powers, from distinguishing species of related bacteria to distinguishing individual clones. As a result, different typing techniques have been found to be optimal for these different tasks. In this study, we used conventional band- and sequence-based typing techniques and WGS to characterize the genetic diversity and epidemiology of a large collection of clinical ACB complex isolates and to investigate an ICU CRAB outbreak. Our study adds to the growing body of literature supporting the use of WGS to define Acinetobacter species, characterize the population structure of A. baumannii, clarify the extent of a hospital outbreak, and delineate an outbreak transmission map (84).
Commercially available platforms for the identification of bacteria perform poorly in identifying ACB complex bacteria to the species level, yet distinguishing these species has clinical importance. Compared with non-baumannii ACB complex species, A. baumannii is more frequently multidrug resistant, more often cultured from critically ill patients, and associated with poorer clinical outcomes, likely due to delays in the receipt of appropriate antibiotic therapy (7, 13, 85). A strain typing method that rapidly identifies bacteria within the ACB complex to the species level and identifies resistance genes would lead to a timelier administration of appropriate antibiotic therapy and, presumably, improved patient outcomes. WGS allowed us to assign ACB complex bacterial isolates to an individual species in two ways. First, WGS provided the sequence of the rpoB gene, which by itself can accurately categorize Acinetobacter species (45, 71). Second, phylogenies based on WGS demonstrated distinct clades representing individual Acinetobacter species. The inclusion of the strain types for the relevant Acinetobacter species within these phylogenies would allow for easy assignment of an individual isolate to a species. Although not examined in this study, WGS can also provide ancillary information, such as the presence or absence of virulence factors and antibiotic resistance islands, which are important for developing effective and timely treatment strategies (74, 86). In contrast, the band-based technique Rep-PCR was not able to separate ACB complex isolates into distinct species.
Certain A. baumannii clonal lineages have disseminated widely and are of particular clinical importance. In recent years, MLST has emerged as the gold standard for categorizing A. baumannii isolates into clonal lineages and characterizing their continental or global spread. Indeed, MLST indicated that four major STs (ST2, ST79, ST406, and ST499) were endemic at NMH over the course of our study. A phylogenetic analysis based on WGS also clustered the majority of A. baumannii strains into four groups corresponding to these STs. ST2 (clade B) is part of the IC-II lineage, which is widely prevalent throughout Europe and the United States and is frequently implicated in nosocomial outbreaks (20, 74, 75). ST406 (clade C) was first reported in an epidemiological study of CRAB clinical isolates from Japan (87) and was subsequently identified in Brazil (88). It is closely related to IC-I strains, which are globally disseminated. ST79 (clade A) was first described by Villalón and colleagues (21) during a study of epidemic nosocomial A. baumannii isolates collected over an 11-year period in Spain and has subsequently been described in the United States, Canada, and Argentina (74–77). Although not assigned an IC number, it appears to represent an A. baumannii lineage that has spread globally. We also identified a novel ST, ST499 (clade D), which is closely related to ST123, which was first identified in Las Vegas, NV (75). Many of the remaining isolates from our study were phylogenetically interspersed with strains reported in the literature as belonging to IC-III (see Fig. S5 in the supplemental material). Interestingly, our analysis indicates that many strains previously published as IC-III are in fact quite heterogeneous, with deep branches in the phylogenetic tree. These IC-III strains are more distantly related to each other than IC-I or IC-II strains are to each other and may not represent closely related clonal lineages in the same manner as IC-I and IC-II strains. Consistent with this conclusion, these IC-III strains represented a variety of different STs. Our findings, which are in agreement with the population structure reported in other studies (20, 74, 89), demonstrate that both well-characterized IC lineages and novel clonal lineages of A. baumannii are endemic in NMH, and that WGS accurately identifies these lineages.
In contrast to WGS, the band-based techniques PFGE and Rep-PCR performed poorly in delineating isolates into distinct clonal lineages. In particular, isolates with similar PFGE patterns were of the same ST only 39% of the time. Other studies have also reported poor agreement between PFGE and MLST for A. baumannii (90, 91) and have demonstrated that isolates from a single ST yield markedly different PFGE patterns (21, 89). These disparate results likely reflect the sensitivity of PFGE to accessory-genome changes (such as genomic island acquisition and deletion), which appear to occur relatively frequently and are not limited to isolates with similar evolutionary histories (80). In contrast, MLST reflects SNPs in conserved housekeeping genes and therefore more accurately represents population phylogeny. Although PFGE has improved with standardization, it remains subject to a number of factors that decrease reliability and reproducibility, such as DNA yield and purity, the type of enzyme used, and errors in gel resolution, intergel standardization, and band visualization and identification (25, 26, 49, 92). These findings suggest that PFGE should not be used to determine relationships between distantly related bacterial isolates (93). In our study, Rep-PCR performed better than PFGE but was still poorly congruent with MLST (Table 3). This was somewhat surprising, since Rep-PCR has been used to define globally disseminated A. baumannii clonal lineages, and others have found good agreement between Rep-PCR and MLST when used in this way (22). A possible explanation is that many of the published studies utilized the DiversiLab system, an automated Rep-PCR platform to which we did not have access. DiversiLab uses microfluidic capillary electrophoresis, which allows for high degrees of resolution and reproducibility (94). Our Rep-PCR assays involved individually performed PCRs, traditional gel electrophoresis, and subjectively interpreted banding patterns, which may have resulted in decreased reproducibility, a recognized disadvantage intrinsic to nonautomated band-based techniques (95). Furthermore, it has been shown that Rep-PCR results are affected by the DNA extraction method used (96). We note that these issues are not specific to our laboratory and may make our results more applicable to other clinical microbiology laboratories that do not use the DiversiLab system.
We also investigated the performance of WGS in the context of an intrahospital A. baumannii outbreak. In this setting, analysis of core-genome SNPs via WGS provided greater resolution than conventional PFGE typing and clarified which isolates were part of the outbreak. One isolate (ABOB09) deemed part of the outbreak by epidemiologic tracing and PFGE typing was excluded by WGS. The incorporation of whole-genome SNP information confirmed that the outbreak began with the near-simultaneous introduction into our hospital of two discrete strains from the same skilled nursing facility. Such facilities are known reservoirs for health care-associated bacteria, such as A. baumannii (97, 98). WGS also clarified some of the possible transmission routes suggested by epidemiologic data. In this regard, WGS was superior to PFGE due to its enhanced discriminatory power (Table 2) (0.997 versus 0.892), which has been noted in other studies of bacterial outbreaks (31, 79, 99, 100). In particular, our results are similar to those of Salipante and colleagues (79), who used a different approach to identify SNPs and define outbreak strains but arrived at the same conclusions regarding the deficiencies of PFGE in the context of an A. baumannii outbreak. At the molecular level, this reflects the relatively large number of SNPs that can accumulate in genomes before they are detectable by PFGE (79). In addition, our study found three variants of the same strain within a single patient (ABOB04, ABOB04_a, and ABOB04_b). The existence of multiple strain variants within a single patient, a “cloud of diversity” (101), has been noted by others (82, 102) and complicates the construction of transmission maps. A single patient is capable of spreading different strain variants to multiple other patients, and several strain variants can evolve over time while colonizing or infecting a single patient. These occurrences may confuse transmission events and require that many bacterial isolates from each patient be sequenced if the full complexity of an outbreak is to be captured (101). However, even sequencing a single isolate from most patients allowed us to improve the resolution of our transmission map. Overall, our results agree with prior studies, which have also demonstrated enhanced outbreak analysis with WGS (38, 83, 103). Snitkin and colleagues (38) integrated data from WGS and epidemiologic tracings to more accurately reconstruct transmissions and dissect the progression of a carbapenem-resistant Klebsiella pneumoniae outbreak. Other studies have revealed that WGS is more accurate than conventional techniques in discriminating among alternate transmission scenarios during outbreaks of MDR A. baumannii (78) and carbapenem-resistant Enterobacter cloacae (37). The results of these studies in conjunction with our own reveal how WGS can be a powerful tool in outbreak analysis and can better direct infection prevention efforts, leading to earlier and more successful control of hospital outbreaks.
Before WGS can widely be used by infection control practitioners, user-friendly approaches must be developed that rapidly analyze sequence data to segregate isolates. As stated by Sabat and colleagues (94) in their discussion of the use of WGS for epidemiological surveillance, “…the key challenge will not be to produce the sequence data, but to rapidly compute and interpret the relevant information from large data sets” (94). Easy-to-use criteria to define which isolates are within the same clonal lineage or within the same hospital outbreak are currently available for band-based techniques (23, 40, 48) but not for WGS (104). For this reason, we identified a threshold of 2,500 core SNPs that distinguished isolates of the same clonal lineage from those of different clonal lineages. Using this threshold, WGS was highly congruent with MLST. Similarly, we used a threshold of 2.5 core SNPs to segregate outbreak from nonoutbreak isolates. We acknowledge that several apparently nonoutbreak isolate pairs in our study differed by <2.5 core SNPs (Fig. 7). However, since many of these isolate pairs were collected within 2 weeks of each other, it is possible they were not false positives but rather represent unrecognized patient-to-patient transmissions (data not shown). A threshold of 2.5 core SNPs agrees with other studies of A. baumannii and other bacterial species (39, 79, 105). For example, Salipante and colleagues (79) defined ≤3 SNPs as a threshold for interpreting isolates as clonal, although their value was based on the reproducibility of WGS using technical replicates. To similarly avoid calling sequencing errors as SNPs, we sequenced to a much-higher read number (139 versus ∼50 reads) and filtered low-frequency reads. Our threshold value was also in agreement with sequencing of distinct isolates from the same patient in our ICU outbreak; such isolates had a maximum difference of 1 SNP, demonstrating that A. baumannii accumulated SNPs slowly over the time frame of an intrahospital outbreak. In contrast to our results, some studies have identified greater SNP differences between A. baumannii isolates recovered at different times from the same patient (106, 107). In one study (107), the authors detected SNPs via alignment to a reference genome, followed by SNP filtering based on a “probability score.” Alignment-based methods can result in false-positive SNP calls. Filtering based on multiple parameters with manual verification of SNPs may be more accurate in identifying SNPs (30, 38, 106, 108, 109). Other studies using alignment-based methods with more-stringent filtering have demonstrated far fewer SNPs among similar isolates identified in an outbreak (82). In contrast to reference genome alignment, the kSNP program identifies SNPs by a direct comparison of short oligomers, minimizing false-positive SNP identification resulting from poor alignment. It also filters out high-density SNPs, which are more likely to result from misalignment or misassembly. Whatever method is used, it is important to exclude SNPs acquired through recombination events, as such events can dramatically increase SNP counts and erroneously classify two closely related strains as being genetically divergent (109).
The approach of counting SNPs to categorize A. baumannii isolates does have several limitations. First, it does not take into account the full extent of evolutionary information intrinsic to the locations and types of SNPs present in a collection of isolates. Powerful and sophisticated methodologies are being developed and applied to WGS data to capture this information and more accurately determine transmission maps (110–112). The core-genome SNP thresholds determined in our study should be viewed as rapid and easily obtained first approximations that may require subsequent verification with more sophisticated methodologies. Second, it is likely that core SNP thresholds will vary with the duration of an outbreak. Regional and multistate outbreaks and outbreaks of longer durations will likely require different thresholds. For example, it is anticipated that more SNPs will accumulate in outbreaks lasting a year than in those lasting only a month. Third, our thresholds were based on a single population of isolates collected at one hospital. Studies of additional hospitals and outbreaks are necessary to iteratively refine these values and to enhance their robustness. For this reason, we view the thresholds presented here as starting points for further refinements.
As clinical microbiologists and infection control practitioners consider changing from conventional genotyping techniques to WGS, they must carefully weigh the advantages and disadvantages of both approaches. This study and other recently published studies provide a framework for doing so. We found that WGS was superior to conventional techniques for typing Acinetobacter at the levels of species identification, population structure, and intrahospital outbreak analysis. WGS is also capable of generating reproducible results regardless of the laboratory location or reagents used, and sequencing data can be easily distributed to other epidemiologists, laboratories, and researchers. These benefits of WGS must be balanced against the cost of commercially available next-generation sequencing instruments or services and the current complexity of sequence analysis. Published estimates of per-strain WGS costs approach those for conventional typing techniques (WGS, $35 to 300; PFGE, $150; MLST, $65 to 120; and Rep-PCR, $26 [79, 94, 113]), although these estimates are highly dependent upon how instrument costs are amortized. WGS may allow additional cost savings resulting from earlier control of hospital outbreaks and infection prevention. As sequencing costs continue to fall and resources for fast and accurate sequence interpretation improve, we anticipate that WGS will be regularly incorporated into routine microbiological surveillance and infection control programs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chao Qi and Michael Malczynski from the Northwestern Memorial Hospital Clinical Microbiology Laboratory for assisting with the identification and retrieval of Acinetobacter bloodstream isolates. We also thank Lisa Sadzewicz and Naomi Sengalamay at the University of Maryland Institute for Genome Sciences for assistance with the Illumina sequencing.
M.A.F. participated in the study conception, design, and coordination, carried out the microbiological and molecular studies, performed data analysis, and helped draft the manuscript. E.A.O. participated in the study conception, design, and coordination, performed the bioinformatic analyses, aided in other data analyses, and helped draft the manuscript. A.R.H. participated in the study conception, design, and coordination and helped draft the manuscript. All authors read and approved the final manuscript.
We declare no competing or conflicting interests.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the American Cancer Society, or the Woman's Board of Northwestern Memorial Hospital. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Funding Statement
Funding was provided to Margaret A. Fitzpatrick through the Eleanor Wood Prince Grants Initiative, a project of the Woman's Board of Northwestern Memorial Hospital.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01818-15.
REFERENCES
- 1.Chang HC, Wei YF, Dijkshoorn L, Vaneechoutte M, Tang CT, Chang TC. 2005. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J Clin Microbiol 43:1632–1639. doi: 10.1128/JCM.43.4.1632-1639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 3.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz-Price LS, Zembower T, Penugonda S, Schreckenberger P, Lavin MA, Welbel S, Vais D, Baig M, Mohapatra S, Quinn JP, Weinstein RA. 2010. Clinical outcomes of carbapenem-resistant Acinetobacter baumannii bloodstream infections: study of a 2-state monoclonal outbreak. Infect Control Hosp Epidemiol 31:1057–1062. doi: 10.1086/656247. [DOI] [PubMed] [Google Scholar]
- 5.Falagas ME, Bliziotis IA, Siempos II. 2006. Attributable mortality of Acinetobacter baumannii infections in critically ill patients: a systematic review of matched cohort and case-control studies. Crit Care 10:R48. doi: 10.1186/cc4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheng WH, Liao CH, Lauderdale TL, Ko WC, Chen YS, Liu JW, Lau YJ, Wang LH, Liu KS, Tsai TY, Lin SY, Hsu MS, Hsu LY, Chang SC. 2010. A multicenter study of risk factors and outcome of hospitalized patients with infections due to carbapenem-resistant Acinetobacter baumannii. Int J Infect Dis 14:e764–e769. doi: 10.1016/j.ijid.2010.02.2254. [DOI] [PubMed] [Google Scholar]
- 7.Chuang YC, Sheng WH, Li SY, Lin YC, Wang JT, Chen YC, Chang SC. 2011. Influence of genospecies of Acinetobacter baumannii complex on clinical outcomes of patients with Acinetobacter bacteremia. Clin Infect Dis 52:352–360. doi: 10.1093/cid/ciq154. [DOI] [PubMed] [Google Scholar]
- 8.Karah N, Haldorsen B, Hegstad K, Simonsen GS, Sundsfjord A, Samuelsen Ø, Norwegian Study Group of Acinetobacter . 2011. Species identification and molecular characterization of Acinetobacter spp. blood culture isolates from Norway. J Antimicrob Chemother 66:738–744. doi: 10.1093/jac/dkq521. [DOI] [PubMed] [Google Scholar]
- 9.Park KH, Shin JH, Lee SY, Kim SH, Jang MO, Kang SJ, Jung SI, Chung EK, Ko KS, Jang HC. 2013. The clinical characteristics, carbapenem resistance, and outcome of Acinetobacter bacteremia according to genospecies. PLoS One 8:e65026. doi: 10.1371/journal.pone.0065026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wisplinghoff H, Paulus T, Lugenheim M, Stefanik D, Higgins PG, Edmond MB, Wenzel RP, Seifert H. 2012. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J Infect 64:282–290. doi: 10.1016/j.jinf.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Lee YC, Huang YT, Tan CK, Kuo YW, Liao CH, Lee PI, Hsueh PR. 2011. Acinetobacter baumannii and Acinetobacter genospecies 13TU and 3 bacteraemia: comparison of clinical features, prognostic factors and outcomes. J Antimicrob Chemother 66:1839–1846. doi: 10.1093/jac/dkr200. [DOI] [PubMed] [Google Scholar]
- 12.Lee YT, Kuo SC, Yang SP, Lin YT, Chiang DH, Tseng FC, Chen TL, Fung CP. 2013. Bacteremic nosocomial pneumonia caused by Acinetobacter baumannii and Acinetobacter nosocomialis: a single or two distinct clinical entities? Clin Microbiol Infect 19:640–645. doi: 10.1111/j.1469-0691.2012.03988.x. [DOI] [PubMed] [Google Scholar]
- 13.Fitzpatrick MA, Ozer E, Bolon MK, Hauser AR. 2015. Influence of ACB complex genospecies on clinical outcomes in a U.S. hospital with high rates of multidrug resistance. J Infect 70:144–152. doi: 10.1016/j.jinf.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins PG, Wisplinghoff H, Krut O, Seifert H. 2007. A PCR-based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. Clin Microbiol Infect 13:1199–1201. doi: 10.1111/j.1469-0691.2007.01819.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Ruan Z, Feng Y, Fu Y, Jiang Y, Wang H, Yu Y. 2014. Species distribution of clinical Acinetobacter isolates revealed by different identification techniques. PLoS One 9:e104882. doi: 10.1371/journal.pone.0104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gundi VA, Dijkshoorn L, Burignat S, Raoult D, La Scola B. 2009. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology 155:2333–2341. doi: 10.1099/mic.0.026054-0. [DOI] [PubMed] [Google Scholar]
- 17.Dijkshoorn L, Aucken H, Gerner-Smidt P, Janssen P, Kaufmann ME, Garaizar J, Ursing J, Pitt TL. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J Clin Microbiol 34:1519–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dessel H, Dijkshoorn L, van der Reijden T, Bakker N, Paauw A, van den Broek P, Verhoef J, Brisse S. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res Microbiol 155:105–112. doi: 10.1016/j.resmic.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villalón P, Valdezate S, Medina-Pascual MJ, Rubio V, Vindel A, Saez-Nieto JA. 2011. Clonal diversity of nosocomial epidemic Acinetobacter baumannii strains isolated in Spain. J Clin Microbiol 49:875–882. doi: 10.1128/JCM.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins PG, Dammhayn C, Hackel M, Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother 65:233–238. doi: 10.1093/jac/dkp428. [DOI] [PubMed] [Google Scholar]
- 23.Saeed S, Fakih MG, Riederer K, Shah AR, Khatib R. 2006. Interinstitutional and intrainstitutional transmission of a strain of Acinetobacter baumannii detected by molecular analysis: comparison of pulsed-field gel electrophoresis and repetitive sequence-based polymerase chain reaction. Infect Control Hosp Epidemiol 27:981–983. doi: 10.1086/507286. [DOI] [PubMed] [Google Scholar]
- 24.Koeleman JG, Stoof J, Biesmans DJ, Savelkoul PH, Vandenbroucke-Grauls CM. 1998. Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J Clin Microbiol 36:2522–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol 43:4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundmann HJ, Towner KJ, Dijkshoorn L, Gerner-Smidt P, Maher M, Seifert H, Vaneechoutte M. 1997. Multicenter study using standardized protocols and reagents for evaluation of reproducibility of PCR-based fingerprinting of Acinetobacter spp. J Clin Microbiol 35:3071–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wisplinghoff H, Hippler C, Bartual SG, Haefs C, Stefanik D, Higgins PG, Seifert H. 2008. Molecular epidemiology of clinical Acinetobacter baumannii and Acinetobacter genomic species 13TU isolates using a multilocus sequencing typing scheme. Clin Microbiol Infect 14:708–715. doi: 10.1111/j.1469-0691.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- 28.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol 44:2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foxman B, Zhang L, Koopman JS, Manning SD, Marrs CF. 2005. Choosing an appropriate bacterial typing technique for epidemiologic studies. Epidemiol Perspect Innov 2:10. doi: 10.1186/1742-5573-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roetzer A, Diel R, Kohl TA, Ruckert C, Nubel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rusch-Gerdes S, Supply P, Kalinowski J, Niemann S. 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Belkum A. 2003. High-throughput epidemiologic typing in clinical microbiology. Clin Microbiol Infect 9:86–100. doi: 10.1046/j.1469-0691.2003.00549.x. [DOI] [PubMed] [Google Scholar]
- 33.Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW. 2012. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pallen MJ, Loman NJ, Penn CW. 2010. High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr Opin Microbiol 13:625–631. doi: 10.1016/j.mib.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ, Goldman TD, Feldgarden M, Birren BW, Fofanov Y, Boos J, Wheaton WD, Honisch C, Musser JM. 2010. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci U S A 107:4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mather AE, Reid SW, Maskell DJ, Parkhill J, Fookes MC, Harris SR, Brown DJ, Coia JE, Mulvey MR, Gilmour MW, Petrovska L, de Pinna E, Kuroda M, Akiba M, Izumiya H, Connor TR, Suchard MA, Lemey P, Mellor DJ, Haydon DT, Thomson NR. 2013. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 341:1514–1517. doi: 10.1126/science.1240578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reuter S, Ellington MJ, Cartwright EJ, Koser CU, Török ME, Gouliouris T, Harris SR, Brown NM, Holden MT, Quail M, Parkhill J, Smith GP, Bentley SD, Peacock SJ. 2013. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med 173:1397–1404. doi: 10.1001/jamainternmed.2013.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group, Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O'Connor L, Ip CL, Golubchik T, Batty EM, Finney JM, Wyllie DH, Didelot X, Piazza P, Bowden R, Dingle KE, Harding RM, Crook DW, Wilcox MH, Peto TE, Walker AS. 2013. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higgins PG, Hujer AM, Hujer KM, Bonomo RA, Seifert H. 2012. Interlaboratory reproducibility of DiversiLab rep-PCR typing and clustering of Acinetobacter baumannii isolates. J Med Microbiol 61:137–141. doi: 10.1099/jmm.0.036046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snelling AM, Gerner-Smidt P, Hawkey PM, Heritage J, Parnell P, Porter C, Bodenham AR, Inglis T. 1996. Validation of use of whole-cell repetitive extragenic palindromic sequence-based PCR (REP-PCR) for typing strains belonging to the Acinetobacter calcoaceticus-Acinetobacter baumannii complex and application of the method to the investigation of a hospital outbreak. J Clin Microbiol 34:1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 44.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.La Scola B, Gundi VA, Khamis A, Raoult D. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol 44:827–832. doi: 10.1128/JCM.44.3.827-832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vila J, Marcos MA, Jimenez de Anta MT. 1996. A comparative study of different PCR-based DNA fingerprinting techniques for typing of the Acinetobacter calcoaceticus-A. baumannii complex. J Med Microbiol 44:482–489. doi: 10.1099/00222615-44-6-482. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen JB, Skov MN, Jørgensen RL, Heltberg O, Hansen DS, Schønning K. 2011. Identification of CTX-M15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. Eur J Clin Microbiol Infect Dis 30:773–778. doi: 10.1007/s10096-011-1153-x. [DOI] [PubMed] [Google Scholar]
- 49.Higgins PG, Janssen K, Fresen MM, Wisplinghoff H, Seifert H. 2012. Molecular epidemiology of Acinetobacter baumannii bloodstream isolates obtained in the United States from 1995 to 2004 using rep-PCR and multilocus sequence typing. J Clin Microbiol 50:3493–3500. doi: 10.1128/JCM.01759-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rambaut A, Drummond A. 2014. July 9, 2014. FigTree version 1.4.2. http://tree.bio.ed.ac.uk/software/figtree/.
- 54.Konstantinidis KT, Ramette A, Tiedje JM. 2006. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci 361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozer EA, Allen JP, Hauser AR. 2014. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics 15:737. doi: 10.1186/1471-2164-15-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marçais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 61.Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 62.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 65.Tan SY, Chua SL, Liu Y, Hoiby N, Andersen LP, Givskov M, Song Z, Yang L. 2013. Comparative genomic analysis of rapid evolution of an extreme-drug-resistant Acinetobacter baumannii clone. Genome Biol Evol 5:807–818. doi: 10.1093/gbe/evt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 68.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carriço JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J Clin Microbiol 44:2524–2532. doi: 10.1128/JCM.02536-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee MJ, Jang SJ, Li XM, Park G, Kook JK, Kim MJ, Chang YH, Shin JH, Kim SH, Kim DM, Kang SH, Moon DS. 2014. Comparison of rpoB gene sequencing, 16S rRNA gene sequencing, gyrB multiplex PCR, and the VITEK2 system for identification of Acinetobacter clinical isolates. Diagn Microbiol Infect Dis 78:29–34. doi: 10.1016/j.diagmicrobio.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 72.Kim D, Baik KS, Kim MS, Park SC, Kim SS, Rhee MS, Kwak YS, Seong CN. 2008. Acinetobacter soli sp. nov., isolated from forest soil. J Microbiol 46:396–401. doi: 10.1007/s12275-008-0118-y. [DOI] [PubMed] [Google Scholar]
- 73.Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, Passet V, Vaneechoutte M, Brisse S, Dijkshoorn L. 2011. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol 162:393–404. doi: 10.1016/j.resmic.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 74.Wright MS, Haft DH, Harkins DM, Perez F, Hujer KM, Bajaksouzian S, Benard MF, Jacobs MR, Bonomo RA, Adams MD. 2014. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. mBio 5(1):e00963-13. doi: 10.1128/mBio.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adams-Haduch JM, Onuoha EO, Bogdanovich T, Tian GB, Marschall J, Urban CM, Spellberg BJ, Rhee D, Halstead DC, Pasculle AW, Doi Y. 2011. Molecular epidemiology of carbapenem-nonsusceptible Acinetobacter baumannii in the United States. J Clin Microbiol 49:3849–3854. doi: 10.1128/JCM.00619-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loewen PC, Alsaadi Y, Fernando D, Kumar A. 2014. Genome sequence of an extremely drug-resistant clinical isolate of Acinetobacter baumannii strain AB030. Genome Announc 2(5):e01035-14. doi: 10.1128/genomeA.01035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Traglia G, Vilacoba E, Almuzara M, Diana L, Iriarte A, Centron D, Ramirez MS. 2014. Draft genome sequence of an extensively drug-resistant Acinetobacter baumannii indigo-pigmented strain. Genome Announc 2(6):e01146-14. doi: 10.1128/genomeA.01146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewis T, Loman NJ, Bingle L, Jumaa P, Weinstock GM, Mortiboy D, Pallen MJ. 2010. High-throughput whole-genome sequencing to dissect the epidemiology of Acinetobacter baumannii isolates from a hospital outbreak. J Hosp Infect 75:37–41. doi: 10.1016/j.jhin.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 79.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, Cookson BT. 2015. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol 53:1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valenzuela JK, Thomas L, Partridge SR, van der Reijden T, Dijkshoorn L, Iredell J. 2007. Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J Clin Microbiol 45:453–460. doi: 10.1128/JCM.01971-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adams MD, Chan ER, Molyneaux ND, Bonomo RA. 2010. Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 54:3569–3577. doi: 10.1128/AAC.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eyre DW, Golubchik T, Gordon NC, Bowden R, Piazza P, Batty EM, Ip CL, Wilson DJ, Didelot X, O'Connor L, Lay R, Buck D, Kearns AM, Shaw A, Paul J, Wilcox MH, Donnelly PJ, Peto TE, Walker AS, Crook DW. 2012. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open 2:e001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harris SR, Cartwright EJ, Torok ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rafei R, Kempf M, Eveillard M, Dabboussi F, Hamze M, Joly-Guillou ML. 2014. Current molecular methods in epidemiological typing of Acinetobacter baumannii. Future Microbiol 9:1179–1194. doi: 10.2217/fmb.14.63. [DOI] [PubMed] [Google Scholar]
- 85.Lee HY, Chen CL, Wu SR, Huang CW, Chiu CH. 2014. Risk factors and outcome analysis of Acinetobacter baumannii complex bacteremia in critical patients. Crit Care Med 42:1081–1088. doi: 10.1097/CCM.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 86.Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, Engelthaler DM, Keim P. 2013. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS One 8:e54287. doi: 10.1371/journal.pone.0054287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kouyama Y, Harada S, Ishii Y, Saga T, Yoshizumi A, Tateda K, Yamaguchi K. 2012. Molecular characterization of carbapenem-non-susceptible Acinetobacter spp. in Japan: predominance of multidrug-resistant Acinetobacter baumannii clonal complex 92 and IMP-type metallo-β-lactamase-producing non-baumannii Acinetobacter species. J Infect Chemother 18:522–528. doi: 10.1007/s10156-012-0374-y. [DOI] [PubMed] [Google Scholar]
- 88.Clímaco EC, Oliveira ML, Pitondo-Silva A, Oliveira MG, Medeiros M, Lincopan N, da Costa Darini AL. 2013. Clonal complexes 104, 109 and 113 playing a major role in the dissemination of OXA-carbapenemase-producing Acinetobacter baumannii in Southeast Brazil. Infect Genet Evol 19:127–133. doi: 10.1016/j.meegid.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 89.Villalón P, Valdezate S, Cabezas T, Ortega M, Garrido N, Vindel A, Medina-Pascual MJ, Saez-Nieto JA. 2015. Endemic and epidemic Acinetobacter baumannii clones: a twelve-year study in a tertiary care hospital. BMC Microbiol 15:47. doi: 10.1186/s12866-015-0383-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hamouda A, Evans BA, Towner KJ, Amyes SG. 2010. Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of bla(OXA-51-like) genes. J Clin Microbiol 48:2476–2483. doi: 10.1128/JCM.02431-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cookson BD, Aparicio P, Deplano A, Struelens M, Goering R, Marples R. 1996. Inter-centre comparison of pulsed-field gel electrophoresis for the typing of methicillin-resistant Staphylococcus aureus. J Med Microbiol 44:179–184. doi: 10.1099/00222615-44-3-179. [DOI] [PubMed] [Google Scholar]
- 93.Blanc DS, Francioli P, Hauser PM. 2002. Poor value of pulsed-field gel electrophoresis to investigate long-term scale epidemiology of methicillin-resistant Staphylococcus aureus. Infect Genet Evol 2:145–148. doi: 10.1016/S1567-1348(02)00093-X. [DOI] [PubMed] [Google Scholar]
- 94.Sabat AJ, Budimir A, Nashev D, Sá-Leão R, van Dijl J, Laurent F, Grundmann H, Friedrich AW, ESCMID Study Group of Epidemiological Markers (ESGEM). 2013. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill 18:pii=20380 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20380. [DOI] [PubMed] [Google Scholar]
- 95.Li W, Raoult D, Fournier PE. 2009. Bacterial strain typing in the genomic era. FEMS Microbiol Rev 33:892–916. doi: 10.1111/j.1574-6976.2009.00182.x. [DOI] [PubMed] [Google Scholar]
- 96.Behringer M, Miller WG, Oyarzabal OA. 2011. Typing of Campylobacter jejuni and Campylobacter coli isolated from live broilers and retail broiler meat by flaA-RFLP, MLST, PFGE and REP-PCR. J Microbiol Methods 84:194–201. doi: 10.1016/j.mimet.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 97.Munoz-Price LS. 2009. Long-term acute care hospitals. Clin Infect Dis 49:438–443. doi: 10.1086/600391. [DOI] [PubMed] [Google Scholar]
- 98.Sengstock DM, Thyagarajan R, Apalara J, Mira A, Chopra T, Kaye KS. 2010. Multidrug-resistant Acinetobacter baumannii: an emerging pathogen among older adults in community hospitals and nursing homes. Clin Infect Dis 50:1611–1616. doi: 10.1086/652759. [DOI] [PubMed] [Google Scholar]
- 99.Pendleton S, Hanning I, Biswas D, Ricke SC. 2013. Evaluation of whole-genome sequencing as a genotyping tool for Campylobacter jejuni in comparison with pulsed-field gel electrophoresis and flaA typing. Poult Sci 92:573–580. doi: 10.3382/ps.2012-02695. [DOI] [PubMed] [Google Scholar]
- 100.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. 2014. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paterson GK, Harrison EM, Murray GG, Welch JJ, Warland JH, Holden MT, Morgan FJ, Ba X, Koop G, Harris SR, Maskell DJ, Peacock SJ, Herrtage ME, Parkhill J, Holmes MA. 2015. Capturing the cloud of diversity reveals complexity and heterogeneity of MRSA carriage, infection and transmission. Nat Commun 6:6560. doi: 10.1038/ncomms7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Halachev MR, Chan JZ, Constantinidou CI, Cumley N, Bradley C, Smith-Banks M, Oppenheim B, Pallen MJ. 2014. Genomic epidemiology of a protracted hospital outbreak caused by multidrug-resistant Acinetobacter baumannii in Birmingham, England. Genome Med 6:70. doi: 10.1186/s13073-014-0070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TE. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lindsay JA. 2014. Evolution of Staphylococcus aureus and MRSA during outbreaks. Infect Genet Evol 21:548–553. doi: 10.1016/j.meegid.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 105.Eyre DW, Babakhani F, Griffiths D, Seddon J, Del Ojo Elias C, Gorbach SL, Peto TE, Crook DW, Walker AS. 2014. Whole-genome sequencing demonstrates that fidaxomicin is superior to vancomycin for preventing reinfection and relapse of infection with Clostridium difficile. J Infect Dis 209:1446–1451. doi: 10.1093/infdis/jit598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hornsey M, Loman N, Wareham DW, Ellington MJ, Pallen MJ, Turton JF, Underwood A, Gaulton T, Thomas CP, Doumith M, Livermore DM, Woodford N. 2011. Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J Antimicrob Chemother 66:1499–1503. doi: 10.1093/jac/dkr168. [DOI] [PubMed] [Google Scholar]
- 107.Wen H, Wang K, Liu Y, Tay M, Lauro FM, Huang H, Wu H, Liang H, Ding Y, Givskov M, Chen Y, Yang L. 2014. Population dynamics of an Acinetobacter baumannii clonal complex during colonization of patients. J Clin Microbiol 52:3200–3208. doi: 10.1128/JCM.00921-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJ, Brinkman FS, Brunham RC, Tang P. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 109.Snitkin ES, Zelazny AM, Montero CI, Stock F, Mijares L, NISC Comparative Sequence Program, Murray PR, Segre JA. 2011. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc Natl Acad Sci U S A 108:13758–13763. doi: 10.1073/pnas.1104404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Didelot X. 2013. Genomic analysis to improve the management of outbreaks of bacterial infection. Expert Rev Anti Infect Ther 11:335–337. doi: 10.1586/eri.13.15. [DOI] [PubMed] [Google Scholar]
- 111.Didelot X, Eyre DW, Cule M, Ip CL, Ansari MA, Griffiths D, Vaughan A, O'Connor L, Golubchik T, Batty EM, Piazza P, Wilson DJ, Bowden R, Donnelly PJ, Dingle KE, Wilcox M, Walker AS, Crook DW, Peto TE, Harding RM. 2012. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol 13:R118. doi: 10.1186/gb-2012-13-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Didelot X, Gardy J, Colijn C. 2014. Bayesian inference of infectious disease transmission from whole-genome sequence data. Mol Biol Evol 31:1869–1879. doi: 10.1093/molbev/msu121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sherry NL, Porter JL, Seemann T, Watkins A, Stinear TP, Howden BP. 2013. Outbreak investigation using high-throughput genome sequencing within a diagnostic microbiology laboratory. J Clin Microbiol 51:1396–1401. doi: 10.1128/JCM.03332-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.