Abstract
RNA-directed DNA methylation (RdDM) is an important de novo DNA methylation pathway in plants. Small interfering RNAs (siRNAs) generated by Dicers from RNA polymerase IV (Pol IV) transcripts are thought to guide sequence-specific DNA methylation. To gain insight into the mechanism of RdDM, we performed whole-genome bisulfite sequencing of a collection of Arabidopsis mutants, including plants deficient in Pol IV (nrpd1) or Dicer (dcl1/2/3/4) activity. Unexpectedly, of the RdDM target loci that required Pol IV and/or Pol V, only 16% were fully dependent on Dicer activity. DNA methylation was partly or completely independent of Dicer activity at the remaining Pol IV- and/or Pol V-dependent loci, despite the loss of 24-nt siRNAs. Instead, DNA methylation levels correlated with the accumulation of Pol IV-dependent 25-50 nt RNAs at most loci in Dicer mutant plants. Our results suggest that RdDM in plants is largely guided by a previously unappreciated class of Dicer-independent non-coding RNAs, and that siRNAs are required to maintain DNA methylation at only a subset of loci.
Keywords: siRNA, non-coding RNA, Pol IV, Dicer-like, RdDM, DNA methylation
Introduction
DNA methylation is an ancient and conserved epigenetic modification that is involved in important biological functions including transposon silencing, genomic imprinting, transcriptional gene regulation during development and environmental responses, and transgenerational epigenetic inheritance1,2,3,4. DNA methylation in plants occurs in three different sequence contexts, CG, CHG and CHH (where H is A, C or T), each maintained by different DNA methyltransferases and pathways. CG methylation is maintained by MET1, the plant homolog of DNMT1, which functions at DNA replication foci and copies methylation patterns from the parental to the daughter strand5,6. CHG methylation is tightly linked to histone methylation and is maintained by a positive feedback loop. The histone methyltransferases KYP, SUVH5 and SUVH6 bind to methylated CHG and catalyze methylation on histone H3 lysine 9 (H3K9me2)7,8,9; the plant-specific DNA methyltransferase CMT3 binds to this mark and promotes CHG methylation10,11. The asymmetric CHH methylation is maintained by two different DNA methyltransferases, CMT2 and DRM2. CMT2 mainly functions at heterochromatic regions and long transposons, and catalyzes ∼70% of total CHH methylation12,13. DRM2 maintains the remaining CHH methylation14, and is a component of the canonical RNA-directed DNA methylation (RdDM) pathway1,2,15. RdDM is also important for de novo DNA methylation in all three sequence contexts2.
Non-coding RNA-mediated transcriptional gene silencing and heterochromatin formation has been observed in many organisms16,17,18. In all cases, small interfering RNAs (siRNAs) were thought to provide the targeting specificity for the enzymes that catalyze repressive epigenetic modifications16. RdDM represents a major RNA-mediated epigenetic silencing pathway in plants19, and has been proposed to proceed through at least two sequential steps: 24-nt siRNA biogenesis and siRNA-guided de novo methylation. The 24-nt siRNAs are generated through the sequential actions of RNA polymerase IV (Pol IV), RNA-dependent RNA polymerase 2 (RDR2) and Dicer-like 3 (DCL3). Current models posit that base-pairing between AGO4-bound siRNAs and the scaffold RNAs produced by RNA polymerase V (Pol V) triggers the recruitment of DRM2 and de novo methylation20.
Maintaining methylation at the RdDM loci sometimes requires repressive histone modifications in addition to pre-existing DNA methylation. It has been shown that an H3K9me2-binding protein, DTF1/SHH1, is required for Pol IV recruitment to a subset of RdDM target loci21,22, whereas two SRA-domain-containing proteins SUVH2/9, which bind to methylated DNA, are required for Pol V occupancy at RdDM loci23,24. Both DTF1/SHH1 and SUVH2/9 interact with putative ATP-dependent chromatin remodeling factors, including CLSY1 and DRD1, which are presumed to facilitate transcription by RNA Pol IV and Pol V, respectively22,24.
Little is known about the characteristics of transcripts generated by Pol IV. The difficulty in detecting Pol IV transcripts could be due to their rapid processing into small RNAs by RNA-dependent RNA polymerases (RDRs) and Dicer-like (DCL) proteins. Indeed, RDR2 co-purifies with RNA Pol IV, and its interaction with Pol IV seems to be important for its activity as RDR2 purified from the nrpd1 mutant, which is deficient in the largest subunit of Pol IV, lacked in vitro polymerase activities25. Arabidopsis contains four DCL proteins. DCL3 is the major Dicer homolog that processes RDR2 products into 24-nt siRNAs26. DCL1 mainly processes miRNAs, whereas DCL2 and DCL4 are responsible for producing 22-nt virus-related siRNAs and 21-nt trans-acting siRNAs (ta-siRNAs), respectively. Functional redundancies among the different DCL proteins have been reported or suggested in many studies27,28,29,30. Despite the role of DCL3 in generating 24-nt siRNAs, the dcl3 mutant does not exhibit a severe decrease in DNA methylation levels at RdDM loci31. It has been thought that other DCL proteins could also process RDR products, compensating for DCL3 when it is absent, and that the 21/22-nt siRNAs, though less efficient, could also be fed into Argonaute (AGO) proteins20. A recent study utilized a dcl2/3/4 mutant background to characterize Pol IV-dependent transcripts and found that they were mostly 100-500 nt in length, had 5′ monophosphates and lacked 3′ polyA tails32.
In addition to Dicer proteins, both animals and plants use AGO proteins for small RNA binding and processing. Arabidopsis contains ten AGO proteins. Among them, AGO4/6/9 were shown to be involved in epigenetic silencing. Although AGO9 mainly functions in reproductive tissues33, AGO4 and AGO6 are more universally expressed and independently affect DNA methylation at RdDM loci34. A recent study has shed light on the molecular mechanism involved in pairing between the AGO-siRNA complex and the scaffold RNA generated by Pol V, and the subsequent recruitment of DNA methyltransferases35. It was found that AGO4 recruits DRM2 dimers, which methylate the strand acting as the template for Pol V transcription35. Further, the base-pairing between the siRNA and scaffold RNA may be important for directing de novo methylation at the last few bases35.
In addition to the canonical RdDM pathway, several studies revealed the existence of a non-canonical RdDM pathway in plants that involves RDR6 for small RNA production19,36. The 21/22-nt small RNAs generated by RDR6 and DCL proteins are believed to be loaded into AGO4/6 and initiate DNA methylation. The methylated DNA can further recruit Pol IV for 24-nt siRNA production and amplification of the silencing signal.
To circumvent possible redundant or independent functions within groups of DCL, AGO and RDR proteins, we took a genetic approach and systematically examined the effect of dcl, ago, rdr and other RdDM mutants on the DNA methylation level of RdDM loci. We observed heterogeneity in the requirement of RdDM components for proper methylation at different target loci. Unexpectedly, although loss of function of all four DCL genes almost completely abolished 21-24 nt small RNAs, it did not lead to significant decreases in DNA methylation at most RdDM target loci, contrary to the prediction of current RdDM models. In fact only about 16% of the RdDM loci fully rely on DCLs for wild-type level methylation. Through RNA sequencing of the dcl quadruple mutant, we identified a new class of Pol IV-dependent small RNAs (“P4 RNAs”) of 25-50 nt, which represent the majority of small RNAs in this mutant. P4 RNAs are produced from RdDM loci, and their abundance positively correlates with DNA methylation levels. Moreover, 68% of 24-nt siRNA reads are substrings of P4 RNA reads. These results demonstrate that DCL proteins and 24-nt siRNAs are not required for DNA methylation at the majority of RdDM loci. We propose that P4 RNAs are transcripts generated by Pol IV and RDRs, and function as trigger RNAs to initiate DNA methylation through RdDM. Further, we suggest that some P4 RNAs are processed by Dicers into 24-nt siRNAs in wild-type plants, which can help maintain DNA methylation at some loci.
Results
Characterization of an atypical RdDM target locus
In our analysis of DNA methylation at the genomic regions targeted by RdDM, we noticed a region with an intriguing requirement for components of the RdDM pathway. The region corresponds to the 5′ UTR of the meiosis recombination gene HEI1037. Using individual locus bisulfite sequencing, we found that DNA methylation at HEI10 was dependent on the largest subunit of Pol IV, NRPD1 (Figure 1A). Small RNA northern blot analysis showed that 24-nt siRNAs accumulated at this locus, and this accumulation was blocked in an nrpd1 mutant (Figure 1B and 1C). The siRNA accumulation was also blocked in a dcl2/3/4 triple mutant (Figure 1B and 1C), which is consistent with the generation of 24-nt siRNAs by DCL326,27. Unexpectedly, DNA methylation at this region was not affected in the dcl2/3/4 mutant (Figure 1A), nor was it affected in dcl1 mutant plants (Supplementary information, Figure S1A). These results strongly suggest that 24-nt siRNAs are not required for DNA methylation at this locus.
Figure 1.
Characterization of DNA methylation and siRNAs at the 5′ UTR of HEI10. (A) The methylation levels in plants of the indicated genotypes, measured by individual locus bisulfite sequencing. (B, C) Small RNA northern blot analysis using probes for the HEI10 locus in wild-type and RdDM mutant plants.
In contrast, we found that DNA methylation at HEI10 requires RDR proteins. Both DNA methylation and 24-nt siRNAs were abolished in an rdr1/2/6 triple mutant (Figure 1A and 1B). Consistent with the role of RDR2 in generating precursors of 24-nt siRNAs, the accumulation of the siRNAs was totally blocked in rdr2 but was only reduced in rdr6 mutant plants (Figure 1C). However, DNA methylation was lost in rdr6 rather than rdr2 mutant plants (Supplementary information, Figure S1B). These results revealed a disconnection between 24-nt siRNA accumulation and DNA methylation. Even though Pol IV, RDR2 and DCL3 are required for siRNA generation at this locus, as the canonical RdDM model predicts, the 24-nt siRNAs are not required for DNA methylation. Instead, it appears that the Pol IV transcript is mainly converted into dsRNA or amplified by RDR6, and RDR6-generated dsRNA or amplified transcript is required for DNA methylation.
DNA methylation was also unaffected in the ago4/6 double mutant or any of the ago1, 2, 3, 5, 7, 8, 9 or 10 single mutants, consistent with the hypothesis that 24-nt siRNAs are not required for DNA methylation at this locus (Figure 1A and Supplementary information, Figure S1C). DNA methylation was also not affected in nrpe1 or nrpb2 mutants (Figure 1A), indicating that Pol V or Pol II are not required at this locus.
In contrast, DNA methylation in all sequence contexts (CG, CHG, CHH) at the HEI10 locus was impaired in met1, cmt3, drm1/2 and ddc (drm1/2/cmt3) methyltransferase mutants (Supplementary information, Figure S1D). These results suggest that DNA methylation in the three sequence contexts is interdependent at this locus.
Genome-wide identification of atyptical RdDM loci
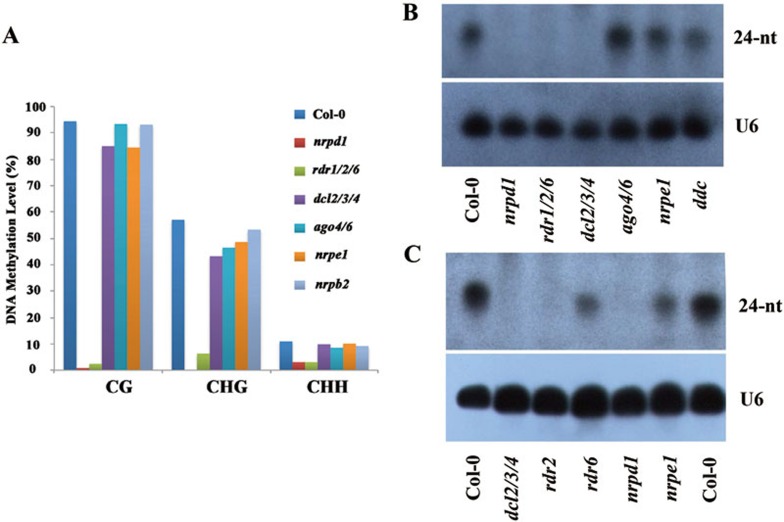
To identify additional genomic loci where DNA methylation is dependent on Pol IV and RDRs but not on DCLs or AGOs, we performed whole-genome bisulfite sequencing of nrpd1 (Pol IV), rdr1/2/6 (RDRs), dcl2/3/4 (Dicers), ago4/6 (AGOs) and nrpe1 (Pol V) mutant plants (Supplementary information, Table S1). We also performed whole-genome bisulfite sequencing of the methyltransferase mutants ddc (drm1/2/cmt3, which maintains non-CG methylation) and met1 (which maintains CG methylation) as controls. All of the plants were in Columbia background and at the same developmental stage (2-week-old seedlings), with two biological replicates for each genotype and each replicate having very high (∼60×) sequence coverage (Supplementary information, Table S1). We identified differentially methylated regions (DMRs) that were hypomethylated (hypo) in the nrpd1 (Pol IV) mutant compared to the wild-type control. We uncovered 2 452 mC hypo-DMRs, 344 mCG hypo-DMRs, 506 mCHG hypo-DMRs and 2 462 mCHH hypo-DMRs (Supplementary information, Table S2). The significantly fewer numbers of mCG and mCHG hypo-DMRs are consistent with the role of Pol IV in maintaining primarily CHH methylation. We focused our subsequent analyses on these Pol IV functional regions.
The DNA methylation levels in rdr1/2/6 at Pol IV functional regions (mC hypo-DMRs) are overall similar to those observed in nrpd1 (Figure 2A). Further, we observed a decrease in DNA methylation levels for the ddc mutant that was comparable to that of nrpd1 and rdr1/2/6 in the CG and CHH contexts. However, the ddc mutant displayed a larger reduction in CHG methylation (Figure 2A), which is consistent with CMT3 being the major CHG maintenance methylation enzyme. In contrast, the DNA methylation levels in dcl2/3/4 and ago4/6 mutants were significantly higher than that in nrpd1 (Supplementary information, Table S3), although the levels were still reduced relative to wild type (Figure 2A and Supplementary information, Table S3). We did not observe reduced methylation levels in the dcl1-100 mutant at the nrpd1 hypo-DMRs (Figure 2A and Supplementary information, Table S3). These results suggest that many of the Pol IV-dependent loci identified in nrpd1 mutant plants do not require siRNAs for proper methylation.
Figure 2.
Profiles of DNA methylation. Boxplots showing the distribution of average DNA methylation levels at (A) hypo-mC-DMRs in nrpd1 (Supplementary information, Table S2), (B) D loci (DCL2/3/4-independent loci), and (C) A loci (AGO4/6-independent loci) (Supplementary information, Table S2). (D) Venn diagram showing the common regions among D loci, A loci and E loci (NRPE1-independent loci; Supplementary information, Table S2). Overlapped regions between D, A, E loci are defined as regions that are overlapped by at least 20% in length (of the shorter DMR), while non-overlapped regions do not physically overlap. Please note that the number discrepancy between DMR number in Supplementary information, Table S2 and the sum of numbers in the venn circles comes from regions that are overlapped by < 20% in length.
We thus characterized the DMR regions where the DNA methylation level in dcl2/3/4 or ago4/6 is significantly higher than in nrpd1 at the Pol IV-dependent loci (please refer to Materials and Methods for detailed description of DMR analyses). For simplicity, we referred to these DMRs identified in dcl2/3/4 as D loci (for higher methylation in dcl2/3/4). The average methylation levels at the 1 395 D loci (Supplementary information, Table S2) are not significantly reduced in dcl1 and dcl2/3/4 plants compared to wild type (Supplementary information, Table S3), whereas DNA methylation levels decrease to a significant and similar extent in rdr1/2/6, nrpe1, ddc and nrpd1 mutants (Figure 2B and Supplementary information, Table S3). Similar to dcl2/3/4, ago4/6 exhibits significantly higher methylation levels compared to nrpd1 mutant (Figure 2B and Supplementary information, Table S3). Using a similar method, we identified 1 157 DMRs (Supplementary information, Table S2) where the methylation levels in the ago4/6 mutant were significantly higher compared to nrpd1 at the Pol IV-dependent loci (Supplementary information, Table S3). These regions were referred to as A loci (for higher methylation in ago4/6). Similar to the D loci, the methylation levels at the A loci were lower in nrpe1, rdr1/2/6 and ddc mutants, as well as nrpd1 mutants, compared to ago4/6 mutants (Figure 2C and Supplementary information, Table S3). Approximately 44% of the A loci were shared by the D loci (referred to as DA loci; Figure 2D). Our findings suggest that DNA methylation at the DA loci is dependent on Pol IV, RDRs, Pol V and the methyltransferases, but is at least partially independent of Dicers and 24-nt siRNAs.
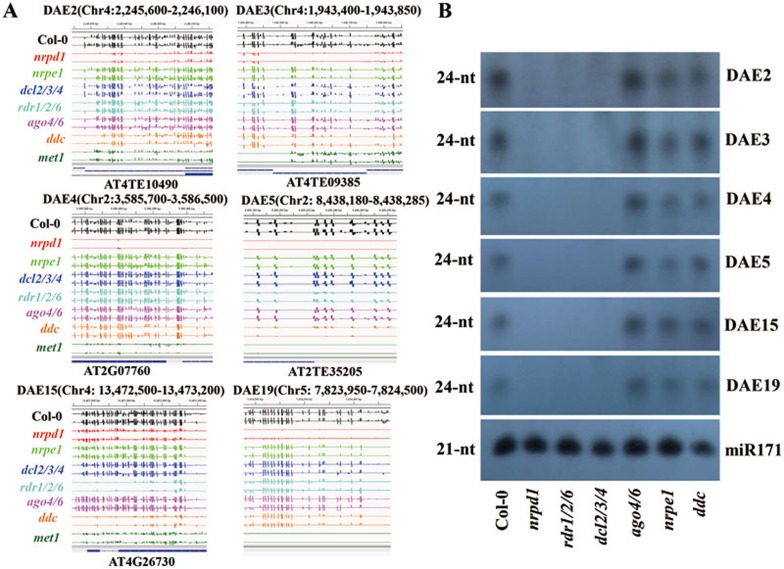
Interestingly, there were 372 loci where DNA methylation was reduced more in nrpd1 (Pol IV) than nrpe1 (Pol V; referred to as E loci, for higher methylation in nrpe1 compared to nrpd1) (Supplementary information, Table S2), and 80 of these were shared with the DA loci (referred to as DAE loci, Figure 2D). The HEI10 locus was within this group (Supplementary information, Figure S2A). We selected 9 additional DAE loci for detailed characterization. Like the DNA methylation at HEI10, DNA methylation at these 9 loci was not reduced relative to wild type in dcl2/3/4 or ago4/6 (Figure 3A and Supplementary information, Figure S2B-S2D). In contrast, DNA methylation was reduced in rdr1/2/6 mutant plants at 5 of the 9 loci (DAE5, DAE15, DAE26, DAE41 and DAE45) but remained unchanged at the remaining 4 loci (DAE2, DAE3, DAE4, and DAE19; Figure 3A and Supplementary information, Figure S2B-S2D). It is possible that DNA methylation at the latter 4 loci is dependent on one of the other three RDRs that are not mutated in the rdr1/2/6 mutant. At all of the 9 loci, 24-nt siRNAs were detected in wild-type, ago4/6, nrpe1 and ddc plants, but were abolished in nrpd1, rdr1/2/6 and dcl2/3/4 mutants (Figure 3B and Supplementary information, Figure S2E), consistent with siRNA generation by the canonical RdDM pathway. The lack of correlation between the accumulation of 24-nt siRNA and DNA methylation in these mutants suggests that 24-nt siRNAs are not necessary for DNA methylation at these loci.
Figure 3.
Characterization of DNA methylation and siRNAs at DAE loci. (A) IGV genome browser screenshots of DNA methylation levels at 6 DAE loci. DNA methylation levels are indicated by the height of vertical bars in each row. (B) Northern blot analysis of 24-nt siRNA levels at the DAE loci.
At 6 of the 9 loci examined (DAE2, DAE3, DAE4, DAE5, DAE15 and DAE45), DNA methylation was reduced to some extent in the ddc mutant relative to wild type (Figure 3A and Supplementary information, Figure S2D). At the other 3 loci (DAE19, DAE26 and DAE41), DNA methylation was not affected in the ddc mutant (Figure 3A and Supplementary information, Figure S2D). Interestingly, MET1, a DNA methyltransferase known for the maintenance of CG methylation3,5, was required for DNA methylation at all of the 9 DAE loci (Figure 3A and Supplementary information, Figure S2D). Thus, MET1 seems to have a critical function in mediating DNA methylation at the DAE loci where 24-nt siRNAs are not required.
Detection of P4 RNAs
DNA methylation at the 9 DAE loci remained unchanged in the nrpe1 (Pol V) mutant (Figure 3A and Supplementary information, Figure S2B-S2D). However, at two DA loci (AT1TE21210 and AT3TE24910) where DNA methylation was impaired in nrpd1 and rdr1/2/6 but not in dcl2/3/4 or ago4/6, the DNA methylation was also abolished in nrpe1 (Supplementary information, Figure S3A). Additionally, DNA methylation at these two DA loci was impaired in ddc but not met1 mutant plants (Supplementary information, Figure S3A). Individual bisulfite sequencing confirmed that the DNA methylation was impaired in nrpd1, nrpe1 and drm1/2 but not in dcl2/3/4 or ago4/6 (Supplementary information, Figure S3B). These results suggest the RdDM components Pol IV, Pol V and DRM1/2, but not 24-nt siRNAs, are involved in methylation at the two DA loci.
Indeed, small RNA northern blotting indicated that 24-nt siRNAs at the two DA loci were eliminated in nrpd1, rdr1/2/6 and dcl2/3/4, and substantially reduced in nrpe1 and ddc mutant plants (Supplementary information, Figure S3C). Interestingly, a smear of RNAs in the size range of ∼25-50 nt appeared at these loci in the dcl2/3/4 mutant (Supplementary information, Figure S3C), and disappeared in the dcl2/3/4/nrpd1 quadruple mutant (Supplementary information, Figure S3D). We refer to these Pol IV-dependent RNAs as “P4 RNAs”. Although less prominent, the RNA smears could also be detected at 6 of the 10 tested DAE loci (including HEI10-DAE1) in dcl2/3/4 mutant plants (Supplementary information, Figure S3E). The DNA methylation levels at the two DA loci were decreased to the nrpd1 mutant level in dcl2/3/4/nrpd1 mutant (Supplementary information, Figure S3B), suggesting that P4 RNAs detected in the dcl2/3/4 mutant are responsible for directing the methylation.
Dicer-independent DNA methylation at RdDM target loci
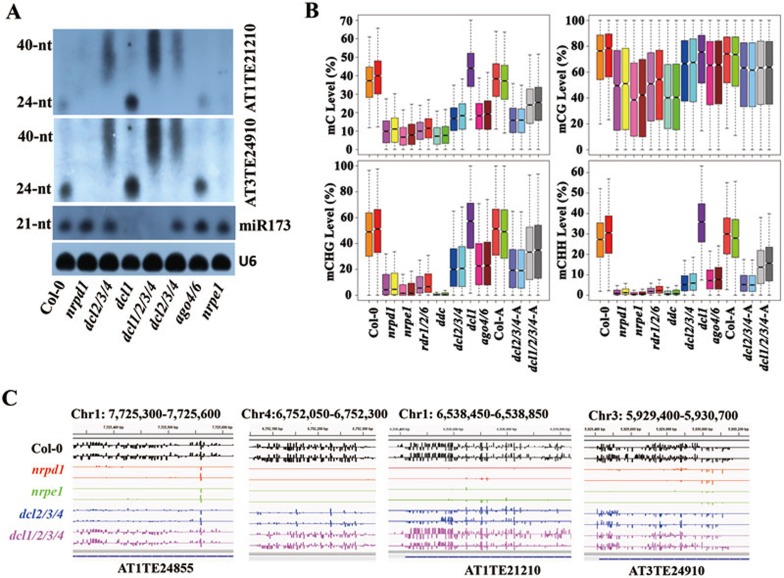
Our results showing that DNA methylation at a subset of RdDM target loci is not reduced in dcl2/3/4 or dcl1 mutant plants suggest that DNA methylation at these loci does not require DCLs. However, it remained possible that a functional redundancy between DCL1 and DCL2, 3 and 4 may have masked the requirement for the DCLs. To further test the requirement for DCLs in RdDM, we constructed dcl1/2/3/4 quadruple mutant plants using CRISPR/Cas938,39 to mutate DCL1 in the dcl2/3/4 background. CRISPR/Cas9 caused different 1-bp insertions at two target sites near the dcl1-100 T-DNA insertion site40 in the DCL1 coding region, resulting in frameshifts (Supplementary information, Figure S4A). The accumulation of miRNA173 was blocked in dcl1/2/3/4 mutant plants but not in dcl2/3/4 mutant plants (Figure 4A). Similar to dcl1-100, the dcl1/2/3/4 mutant exhibited pleiotropic developmental defects, including sterility (Supplementary information, Figure S4B and S4C), indicating that the dcl1-201/202 mutations caused a strong or near complete loss of function of DCL1.
Figure 4.
Levels of siRNAs, P4 (Pol IV-dependent) RNAs and DNA methylation in dcl mutants. (A) Northern blot analysis of miRNA, siRNAs and P4 RNAs at the indicated loci. The dcl1/2/3/4 mutant contained the dcl1-201 allele. (B) Boxplots of DNA methylation levels at nrpd1 hypo-mC-DMRs in the various genotypes. Col-0-A, dcl2/3/4-A and dcl1/2/3/4-A are adult (5-week-old) plants of the Col-0, dcl2/3/4 and dcl1/2/3/4 (left, dcl1-201; right, dcl1-202) genotypes, respectively. Except for the dcl1/2/3/4 mutants, each genotype had two replicates. (C) IGV genome browser screenshots showing the DNA methylation levels at the indicated loci. For dcl1/2/3/4, the upper row had the dcl1-201 allele and the lower one had the dcl1-202 allele.
We selected dcl1/2/3/4 homozygous mutant plants (both the dcl1-201 and dcl1-202 alleles) at the inflorescence stage (5-week-old soil grown) from segregating populations of dcl2/3/4-DCL1+/− for whole-genome bisulfite sequencing, and used dcl2/3/4 and wild-type plants at the same stage as controls. Two biological replicates from each genotype were sequenced at ∼60× coverage, except for dcl1-201/2/3/4 and dcl1-202/2/3/4 (one replicate each; Supplementary information, Table S1). Unexpectedly, the DNA methylation level of dcl1/2/3/4 was not decreased but rather increased compared to that of dcl2/3/4 at nrpd1 hypo-DMRs (Figure 4B and Supplementary information, Table S3). Remarkably, at some of the RdDM target loci where DNA methylation was partially reduced in dcl2/3/4, the methylation in dcl1/2/3/4 was restored to the wild-type level (Figure 4C), suggesting a role for DCL1 in preventing DNA methylation at these loci in the dcl2/3/4 background.
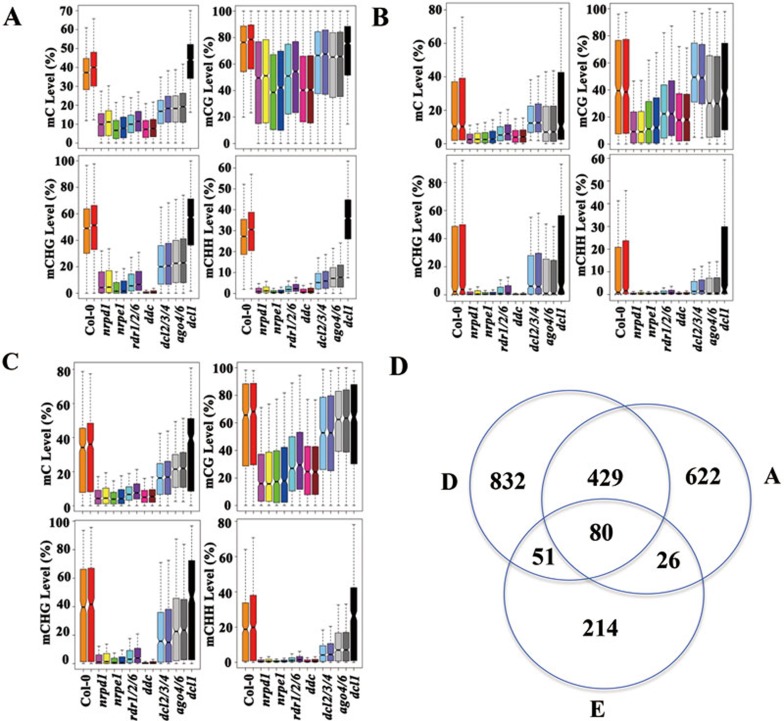
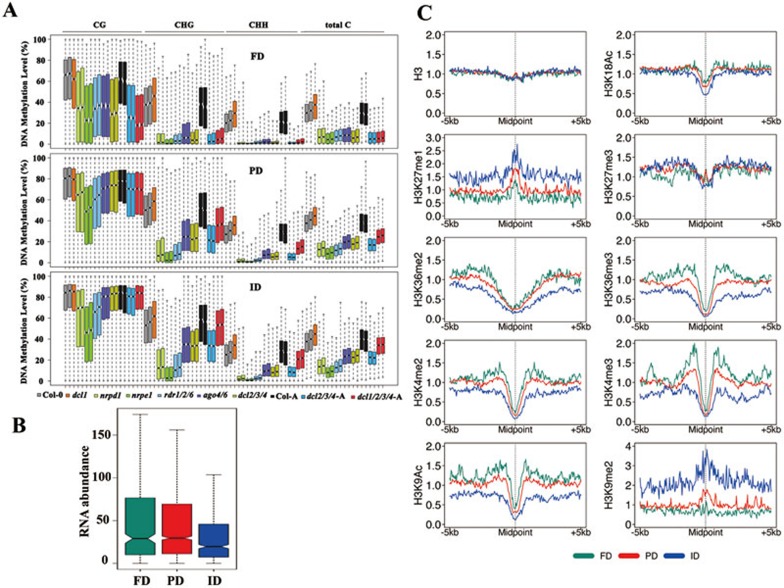
We defined the RdDM target loci (n = 3 995) as the combination of hypo-DMRs identified in the nrpd1 and nrpe1 mutants, and subsequently categorized these RdDM target loci into three groups based on the DNA methylation level in the dcl1/2/3/4 mutant: FD (fully dependent on DCL), PD (partially dependent on DCL) and ID (DCL-independent; Supplementary information, Table S2). Compared to wild type, the DNA methylation levels at the FD loci in dcl1/2/3/4 mutants were reduced to a similar extent as in nrpd1 mutant plants (Figure 5A and Supplementary information, Table S3). Only 619 loci (15.5%) belonged to the FD group, whereas 1 094 of the RdDM target loci (27.4%) belonged to the ID group and did not require DCLs for DNA methylation (Figure 5A and Supplementary information, Table S2). The remaining loci (n = 2 282; 57.1%) were partially dependent on DCLs as their methylation levels in dcl1/2/3/4 are lower than wild type, but higher than nrpd1 and nrpe1 (Figure 5A and Supplementary information, Table S2). The methylation profile of wild type or dcl2/3/4 showed no difference in seedlings and adult plants, indicating comparisons made between different developmental stages are valid (Figure 5A). Except for the FD loci, where dcl1/2/3/4 and dcl2/3/4 had similar methylation levels (Figure 5A), dcl1/2/3/4 showed significantly higher methylation levels than dcl2/3/4 at PD and ID loci (Figure 5A and Supplementary information, Table S3). The dcl1 mutation restored DNA methylation of dcl2/3/4 to wild-type level at ID loci, especially for CG and CHG methylation (Figure 5A). These results demonstrate that DNA methylation at only a small fraction of RdDM target loci is fully dependent on DCLs.
Figure 5.
Epigenetic features of DCL-independent RdDM loci. (A) Boxplots showing the distribution of DNA methylation levels at DCL-fully dependent (FD), DCL-partially dependent (PD), and DCL-independent (ID) RdDM loci. Genotypes of plants are indicated as color code. “-A” indicates adult plants. (B) The abundance of 24-nt siRNAs is significantly lower at ID loci than at PD or FD loci (P-value = 7.701E-05 between FD and ID, P-value = 4.817E-11 between PD and ID, P-value = 0.681 between FD and PD by two-tailed Wilcoxon test). (C) Patterns of histone modifications (specified on the left of each graph) surrounding the RdDM loci. ID, PD and FD loci are indicated by blue, red and green lines, respectively.
We next determined whether the three types of RdDM target loci exhibited distinct epigenetic features. We found that the abundance of 24-nt siRNAs at the ID loci was significantly lower than at PD or FD loci (Figure 5B) in wild-type plants. The abundance of 24-nt siRNAs does not correlate with CHH methylation level at these loci. The average CHH methylation level at FD loci (median ∼21.5%) is significantly lower than that at PD (median ∼28.7%) and ID (median ∼26.1%) loci (Figure 5A and Supplementary information, Table S3). The disconnection between methylation levels and siRNA levels provides further support that factors other than the 24-nt siRNA are triggering DNA methylation at a subset of RdDM loci.
We also examined specific histone modifications associated with the three types of RdDM loci using publicly available data41. We found that the ID and PD loci have significantly higher levels of two heterochromatin marks, H3K9me2 and H3K27me1 than the FD loci (Figure 5C and Supplementary information, Table S3). The enrichment of H3K9me2 at ID and PD loci is associated with significantly higher levels of CHG methylation at these two types of loci than FD loci (Figure 5A and Supplementary information, Table S3). Indeed, the median values of CHG methylation at ID, PD and FD loci are 56%, 51% and 37%, respectively. We also observed that CG methylation levels are significant higher at ID and PD loci than FD loci (Supplementary information, Table S3); the median mCG values at ID, PD and FD loci are 84.8%, 80.6% and 67.3%, respectively. In addition, the FD loci tend to be surrounded by the highest levels of active marks that are usually present in euchromatin, including H3K4me3, H3K4me2, H3K36me2, H3K36me3, H3K9Ac and H3K18Ac42, while the ID loci are surrounded by regions with no preference for those marks (Figure 5C and Supplementary information, Table S3). PD loci are typically found within regions that are intermediate for both heterochromatic and euchromatic marks (Figure 5C and Supplementary information, Table S3). No differences were identified among ID, PD and FD loci in nucleosome density (H3) or a histone modification associated with developmental regulation, H3K27me343 (Figure 5C and Supplementary information, Table S3). These results demonstrate that the ID/PD/FD loci each preferentially associate with different histone modifications and may reside in different local chromatin environments.
Since the Pol IV-interacting protein DTF1/SHH1 binds to H3K9me2 and helps recruit Pol IV to a subset of RdDM loci21,22, we tested the possibility that DTF1/SHH1 preferentially targets ID and/or PD loci. However, we found that the ID loci are less enriched for DTF1-dependent DMRs (Supplementary information, Figure S5), suggesting that multiple factors are involved in recruiting Pol IV to different types of RdDM target loci.
The abundance of P4 RNAs positively correlates with DNA methylation levels at Dicer-independent loci in dcl1/2/3/4 mutant plants
We found that dcl1/2/3/4 mutant plants are deficient in 24-nt siRNA accumulation at the two tested DA loci, AT3TE24910 and AT1TE21210, similar to dcl2/3/4 mutant plants (Figure 4A). Interestingly, P4 RNAs accumulated to a higher level in dcl1/2/3/4 compared to dcl2/3/4 (Figure 4A). This increase in P4 RNAs correlated with increased DNA methylation at the tested loci in dcl1/2/3/4 compared to dcl2/3/4 (Figure 4C). These data suggest that DCL1 suppresses the accumulation of P4 RNAs, and that P4 RNAs may underlie the increased DNA methylation at RdDM target loci in dcl1/2/3/4.
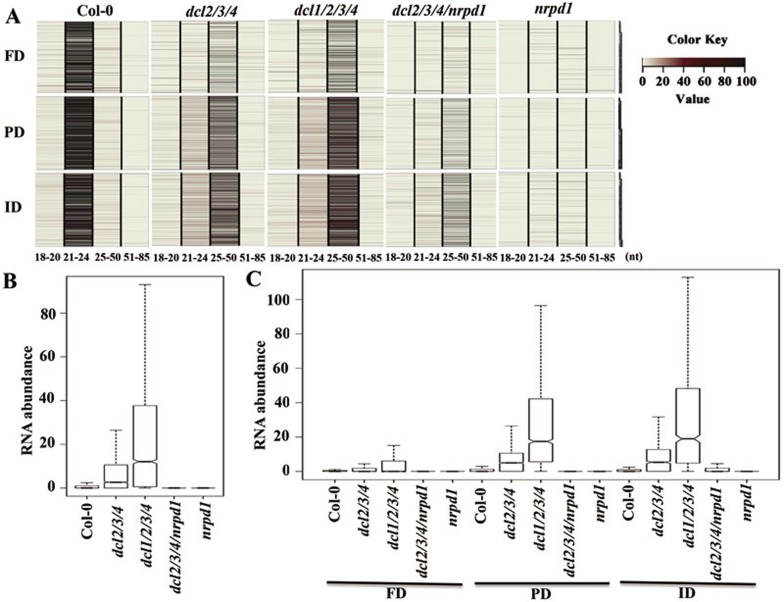
To test whether P4 RNAs are also produced from other RdDM target loci and whether the accumulation of P4 RNAs correlates with the relatively higher DNA methylation levels in dcl1/2/3/4 than dcl2/3/4 mutants, we gel-purified and sequenced 18-85 nt RNAs from wild-type, nrpd1, dcl2/3/4, dcl2/3/4/nrpd1 and dcl1/2/3/4 mutant plants (Supplementary information, Table S1). We found that, on the genome scale, the number of 24-nt siRNA clusters (please refer to Materials and Methods for the definition of siRNA clusters) was reduced to background levels in dcl2/3/4 and dcl1/2/3/4 as well as nrpd1 mutant plants, consistent with the primary role of DCL3 in producing 24-nt siRNAs (Supplementary information, Figure S6A). In contrast, small RNAs in the size range of 25-50 nt account for the majority of reads in the dcl1/2/3/4 mutant (Figure 6A). These results suggest that the accumulation of P4 RNAs is a universal phenomenon across the whole genome.
Figure 6.
RdDM loci accumulate P4 RNAs in dcl2/3/4 and dcl1/2/3/4 mutants. (A) Heatmaps showing the levels of small RNAs of different sizes at the RdDM loci where DNA methylation is fully-dependent (FD), partially-dependent (PD), or independent (ID) of DCLs. (B) Boxplots showing the abundance of 25-50 nt RNAs in WT, dcl2/3/4, dcl1/2/3/4, dcl2/3/4/nrpd1 and nrpd1 mutants in the RdDM loci. (C) Boxplots showing the abundance of 25-50 nt RNAs in WT, dcl2/3/4, dcl1/2/3/4, dcl2/3/4/nrpd1 and nrpd1 mutants at the FD, PD or ID RdDM loci.
Consistent with the northern blot result (Figure 4A), the number of P4 RNA clusters dramatically increased in the dcl2/3/4 mutant plants compared to wild type (Figure 6A and 6B, Supplementary information, Figure S6B and Table S3). As expected, these P4 RNAs depend on RNA Pol IV, as their accumulation was abolished in the dcl2/3/4/nrpd1 mutant (Figure 6A-6C and Supplementary information, Figure S6B). Strikingly, the 25-50 nt P4 RNAs in the dcl1/2/3/4 quadruple mutant not only accumulate to much higher levels than in the dcl2/3/4 triple mutant (Figure 6B and 6C, Supplementary information, Figure S6B and Table S3), but also their distribution pattern along the chromosomes superimposes the pattern of 24-nt siRNA clusters in wild-type plants (Supplementary information, Figure S6C), suggesting that P4 RNAs are primarily produced from RdDM target loci. Indeed, the loci that accumulated P4 RNAs in the dcl1/2/3/4 mutant are enriched for 24-nt siRNA in wild-type (Col-0) plants (Figure 6A, Supplementary information, Figure S7A and S7B).
The increases in P4 RNAs correlated with higher methylation levels at RdDM target loci in dcl1/2/3/4 than in dcl2/3/4 (Figure 4B). We next determined whether the P4 RNAs are preferentially enriched at ID/PD/FD loci. A heatmap of small RNA abundance at the three types of RdDM loci clearly indicates that the ID and PD loci accumulated much more 25-50 nt P4 RNAs than the FD loci in dcl2/3/4 and dcl1/2/3/4 mutants (Figure 6A and Supplementary information, Figure S7B). Abundance of P4 RNAs in the dcl1/2/3/4 mutant at ID and PD loci is significantly higher than in the dcl2/3/4 mutant (Figure 6C, Supplementary information, Figure S7B and Table S3). Thus, the increased DNA methylation level at PD and ID loci observed in dcl1/2/3/4 relative to dcl2/3/4 (Figure 5A) correlates with an increased abundance of P4 RNAs in these regions. These results suggest that the 25-50 nt P4 RNAs may function in directing DNA methylation at PD and ID loci.
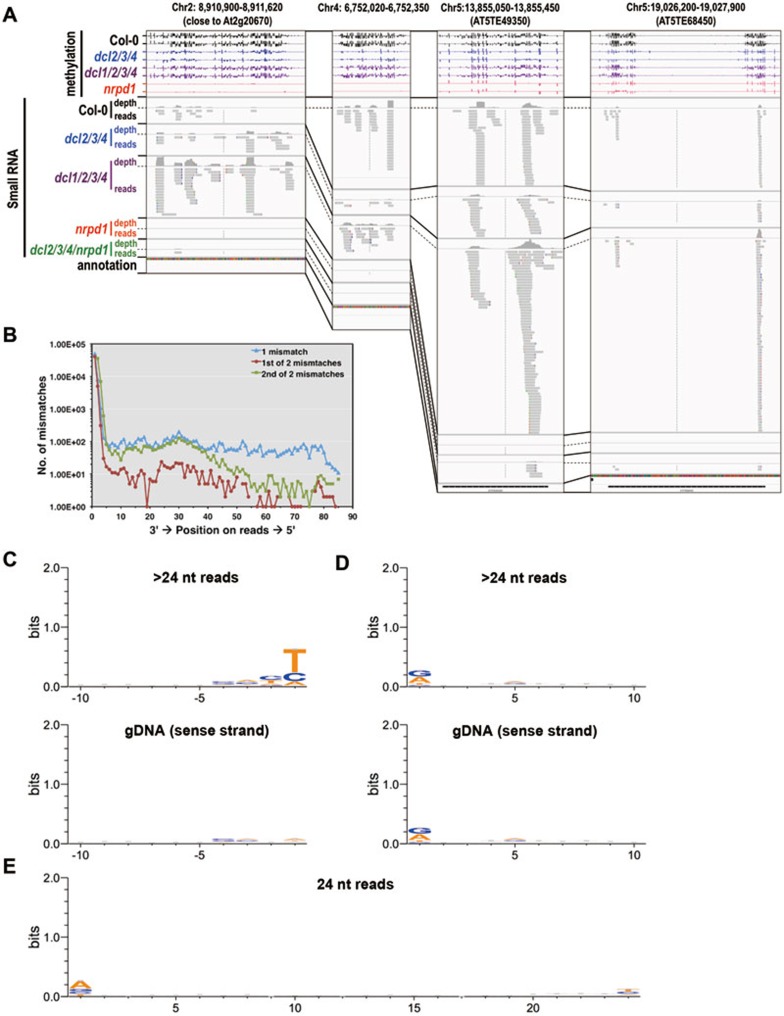
By visual inspection of small RNA reads in Integrated Genomics Viewer (IGV)44,45, we found that P4 RNA reads preferentially mapped to a specific strand, similar to 24-nt siRNA reads, and that P4 RNA and siRNA reads typically map to the same strand (Figure 7A). Approximately 93.6% of the 24-nt siRNAs from WT plants are in the same direction with P4 RNAs in dcl1/2/3/4 mutant plants. At the same locus, P4 RNAs usually accumulate to a higher level in dcl1/2/3/4 than the 24-nt siRNA in wild-type plants (Figure 7A). A survey of our small RNA library indicates that 68% of the 24-nt reads from WT plants are substrings of the 25-85 nt reads that are primarily identified in dcl1/2/3/4 mutants. These analyses strongly suggest that the P4 RNAs are products of RNA Pol IV and precursors of 24-nt siRNAs.
Figure 7.
Characteristics of P4 RNAs. (A) Examples of DCL-independent RdDM loci. The DNA methylation level and small RNA abundance of DCL-independent RdDM loci were visualized in the Integrative Genomics Viewer. The level of DNA methylation is indicated by vertical bars within the range of 100% to −100% (“−” denotes reverse strand). Small RNA abundance is indicated by the sequencing “depth” or the number of “reads” at each locus. Note that the small RNA depth was shown with different data ranges to accommodate variation in small abundance at different loci. Small RNA reads are represented by filled gray boxes with arrows pointing from the 5′ to 3′ end. Colored bars within the gray box indicate location of mismatches. Only uniquely mapped reads were used for this purpose. (B) Number of mismatches at each position of the 25-85 nt P4 RNA reads. The reads were aligned at the 3′ end and the number of mismatches at each position was summed. Note that the data are presented in the direction of 3′ end to 5′ end. (C) Sequence logos of the last 10 nucleotides of 25-85 nt mismatched reads from dcl1/2/3/4 or corresponding genome sense strand sequences that are aligned at the 3′ end. The scale of the y-axis indicates sequence conservation, defined as the difference between the maximum possible entropy and the entropy of the specific position. Thus 2 bits are fully conserved (for example, all A) and 0 bits indicate equal distribution of A/T/C/G. (D) Sequence logos of the first 10 nucleotides of 25-85 nt mismatched reads from dcl1/2/3/4 or corresponding genome sense strand sequences that are aligned at the 5′ end. (E) Sequence logo of 24-nt siRNA sequences from wild-type plants.
An intriguing observation is that P4 RNA reads preferentially have mismatches at the 3′ end of the reads. Mismatched reads account for 22.3% of uniquely mapped > 24-nt reads (structural RNA excluded) in the dcl1/2/3/4 small RNA library. When two mismatches are allowed, many of the uniquely aligned P4 RNA reads from dcl1/2/3/4 contained a significant amount of mismatches at the 3′ end (Figure 7A). Indeed, more than 82.67% of mismatches occur at the last two residues of the reads (Figure 7B). In contrast, the number of mismatches at the 5′ end of the reads are 2-3 orders of magnitude lower, consistent with them being random errors (Figure 7B). Since mismatches predominantly locate to the 3′ end of P4 RNA reads, we reasoned that mismatches may reflect a low selectivity of nucleotide incorporation or lack of proofreading by Pol IV, instead of RDRs. Incorporation of wrong bases at the end of a transcript may result in polymerase stall and release of the transcript. We thus tested whether the template DNA or the RNA product exhibited any sequence features. When we aligned all of the 25-85 nt reads with mismatches by their 3′ ends, a clear preference for pyrimidines, especially thymine, was observed at the last position (Figure 7C), but the mismatched position on the sense strand of the genomic DNA showed no preferences for a particular base (Figure 7C). When we aligned all of the 25-85 nt reads with mismatches by their 5′ ends, a slight preferences for purines was observed at the first position (Figure 7D). The same preference was observed for sense strand sequences, consistent with no skewed mismatches observed at the 5′ ends of P4 RNA reads. Thus, 3′ misincorporation could happen at any type of residues, and pyrimidines, especially thymines, are preferentially inserted. The same level of mismatches was not observed in 24-nt reads in wild-type plants even when mismatches were allowed (Figure 7E), although we did observe preferential presence of adenine at the +1 position, consistent with previous observations46. Although the mechanism and functional significance of the misincorporation in the 3′ end of P4 RNAs remain to be determined, the lack of such misincorporation in 24-nt siRNAs suggests that the 24-nt siRNAs are generated through preferential processing of the 5′ fragments of P4 RNAs by Dicers.
Discussion
RdDM is a form of RNA silencing47,48,49. It is generally believed that a fundamental step in RNA silencing is the Dicer-catalyzed cleavage of dsRNAs into siRNAs, which then guide sequence-specific RNA degradation or epigenetic modifications2,19,20,50,51. In the two decades since RdDM was first discovered, there has been much evidence supporting the important roles of 24-nt siRNAs in this epigenetic pathway2,19,20,27. Nevertheless, as noted recently by Dalakouras and Wassengger52,53, there have been a number of studies involving transgenes where long, non-coding RNAs, but not the siRNAs, were implicated in directing DNA methylation. In many cases, siRNA accumulation at endogenous genomic loci is not sufficient to induce DNA methylation, as in the epiallele fwa-d in Arabidopsis54 or the B-I paramutable allele of maize55. In other cases, a significantly decreased abundance of siRNAs does not reduce DNA methylation levels22.
We systematically characterized the DNA methylome of mutants defective in siRNA biogenesis and RNA-directed DNA methylation. To rule out the possibility that proteins of the same family could function redundantly/independently in the RdDM pathway, we used dcl2/3/4, dcl1/2/3/4, ago4/6 and rdr1/2/6 mutants and analyzed their DNA methylation phenotypes. Although the methylome of rdr1/2/6 largely resembles that of nrpd1, dcl2/3/4 and ago4/6 mutants exhibit significantly higher methylation levels than nrpd1 at > 44% of Pol IV-regulated loci. We also identified over 300 loci whose DNA methylation levels do not rely on Pol V. Our results, particularly those on the dcl1/2/3/4 mutant, demonstrate that DNA methylation at many of the RdDM target loci in Arabidopsis does not correlate with 24-nt siRNAs, and was in fact completely independent of the four DCLs.
Unexpectedly, we found that the dcl1/2/3/4 mutant exhibits significantly higher DNA methylation levels than dcl2/3/4. In fact, the dcl1 mutation restores methylation of the dcl2/3/4 mutant to wild-type levels at over 27% of the RdDM loci. After categorizing RdDM loci into three groups based on their methylation profiles in dcl1/2/3/4 mutant plants, we found that they are associated with very different epigenetic features. The DCL-independent loci are enriched for repressive histone modifications and non-CHH methylation, whereas the DCL-dependent loci are surrounded by active euchromatin marks, and the PD loci represent intermediate states. Thus, it is possible that, due to the active epigenetic state of the surrounding region, RdDM needs more reinforcement by 24-nt siRNAs at FD loci than ID loci. It seems that 24-nt siRNAs are required to maintain DNA methylation at FD loci. Consistent with this hypothesis, we observed a higher abundance of 24-nt siRNAs at FD loci than at ID loci.
The structural features of P4 RNAs strongly suggest that they are products of Pol IV and precursors of 24-nt siRNA. P4 RNAs are mostly 25-35 nt in length and are more abundant at RdDM loci whose methylation is independent of, or partially dependent on, DCL proteins. A previous study that characterized Pol IV- and RDR2-dependent RNAs suggested that the majority of Pol IV-dependent RNAs are 100-500 nt in length32. However, we did not observe > 100 nt Pol IV-dependent transcripts by northern blot analyses (data not shown). Instead, we found that 25-50 nt RNAs are the major class of small RNAs generated from most RdDM loci in DCL mutant plants. Importantly, many of the P4 RNAs overlap with each other. The RNA-seq library preparation procedure in the Li et al. study contained a transcript fragmentation step and accordingly neighboring reads no more than 60 nt apart were joined together computationally32. Since our study did not include a RNA fragmentation step, our sequencing reads should represent the native state of Pol IV-dependent transcripts, and no further assembly of the reads is needed. On the other hand, our results are consistent with the finding of Li et al. that P4 RNAs contain a monophosphate in their 5′ end and no poly(A) tail in the 3′ end32. The T4 RNA ligase used for our RNA-seq library construction requires 5′ monophosphate and 3′ hydroxyl groups of the RNAs.
More than 68% of the 24-nt siRNA reads can be identified as substrings of P4 RNA reads, suggesting that most siRNAs may be generated directly from these P4 RNAs. Visual inspection of P4 RNA reads in the IGV browser indicates that P4 RNAs and 24-nt siRNAs usually have the same 5′ ends but different 3′ ends (Figure 7A). It was previously shown by an in vitro enzymatic assay that DCL4 cut long dsRNAs more efficiently, whereas DCL3 preferred short dsRNA substrates with an A or U in the 5′ phosphorylated end56. Thus, the 25-50 nt P4 RNAs are predicted to be good substrates for DCL3. Mismatches are often observed in the last 2 nucleotides of 25-50 nt reads (Figure 7A and 7B) but not in 24-nt reads (Figure 7E), further suggesting that 24-nt siRNAs are preferentially generated through processing of the 5′ fragments of P4 RNAs. In eukaryotes, TFIIS facilitates Pol II proofreading and elongation57. It is possible that similar factors are missing for Pol IV, and that misincorporation of NTPs facilitates release of the transcript, which is then transcribed by RDR2. Of the 4 464 > 24 nt P4 RNA clusters identified in dcl1/2/3/4 mutant plants, we found that 1 751 overlap with DRM2-dependent (D2) loci and 651 overlap with CMT2-dependent (C2) loci, and 73 overlap with both D2 and C2 loci, consistent with the findings that both D2 and C2 loci produce 24-nt siRNAs13,32. During the preparation of this manuscript, two independent groups reported short, Pol IV-dependent precursors of 24-nt siRNAs58,59, which corroborate our results.
The general requirement for RDRs at Pol IV-dependent loci indicates that the Pol IV transcripts need to be amplified and/or converted to dsRNAs for RdDM. Dalakouras and Wassengger have proposed that long dsRNAs direct DNA methylation, and the siRNAs function in helping to amplify dsRNAs by promoting cleavage of long non-coding RNAs produced by Pol IV and/or Pol V53. The detection of 25-50 nt RNAs that positively correlate with DNA methylation in the dcl1/2/3/4 mutant supports this model. The 25-50 nt non-coding RNAs from Pol IV may be loaded onto AGOs to guide DNA methylation by DRM2 by base-paring with scaffold transcripts produced by Pol V. The requirements for RDRs and AGOs for DNA methylation at some of the DCL-independent RdDM loci (Figure 5A) suggest that the P4 RNAs guiding DNA methylation may be double stranded and one strand may be loaded onto AGOs. Conceivably, non-coding RNAs from Pol IV or even Pol V may also guide DNA methyltransferases such as DRM2 and/or MET1 more directly for sequence-specific DNA methylation. Recently, DNMT1, the MET1 ortholog in mammals, was reported to interact with non-polyadenylated, long non-coding RNAs, and this interaction regulates genomic DNA methylation by interfering with DNMT1 function60. It would be of interest to test whether the plant DNA methyltransferases may interact with and are regulated by non-coding transcripts.
Bond and Baulcombe recently proposed that RdDM can be divided into initiation, establishment and maintenance stages61. We propose that the 25-50 nt P4 RNAs could be responsible for triggering DNA methylation at the initiation stage of RdDM. At the DCL-independent RdDM loci, subsequent establishment and maintenance of DNA methylation does not require 24-nt siRNAs, although some of the initial trigger P4 RNAs are also processed into siRNAs by DCLs. The high abundance of the trigger P4 RNAs at the DCL-independent loci may be sufficient for establishment and maintenance of DNA methylation. The high levels of H3K9me2 and H3K27me1 at these loci may also be important, perhaps by helping to retain the trigger transcripts at the chromatin for efficient initiation, establishment and maintenance of DNA methylation, such that the 24-nt siRNAs are not needed. At the DCL-partially dependent, and maybe even the DCL-fully dependent loci, RdDM may be initiated by 25-50 nt P4 RNAs but establishment and/or maintenance would rely partly or fully on the 24-nt siRNAs produced by DCLs. The 24-nt siRNAs produced at the DCL-independent loci are presumed to be loaded onto AGO4 or AGO6; however, since they are not required for DNA methylation, their functional significance at these loci is unclear. Future studies to determine whether and how the 25-50 nt P4 RNAs may be loaded onto AGOs to initiate RdDM would be of interest.
Materials and Methods
Plant materials and growth conditions
All Arabidopsis plants used in this study were in the Col-0 background. The ago4/6 double mutant, rdr1/2/6, dcl2/3/4 and ddc triple mutants were reported previously20,34,54,62. All other mutants (Supplementary information, Table S5) were either ordered from Arabidopsis Biological Resource Center (http://www.arabidopsis.org) or kindly provided by other scientists (ago1-27 mutant from Vaucheret H; met1-1 from Richards EJ).
Plants that were subjected to DNA methylation and small RNA analyses were 2-week-old seedlings grown on half Murashige Skoog (MS) medium supplemented with 0.7% agar and 1% sucrose, unless indicated otherwise. The growth condition for the seedlings is 16 h of light and 8 h of darkness at 22 °C in growth chambers (Percival, Inc.).
Small RNA extraction and northern blot analyses
Total RNA was extracted from Arabidopsis seedlings (2-week old) or flowers following the standard protocol of TRIzol reagent (Ambion). The small RNA fraction was precipitated using the PEG method. Briefly, an equal volume of PEG 8000 solution (20% PEG 8000, 1 M NaCl) was added to the total RNA. After centrifugation at 16 000× g at 4 °C, an equal volume of isopropanol and 0.1 volume of 3 M NaAC were added to the supernatant. Following another centrifugation at 16 000× g at 4 °C, the small RNA pellet was re-suspended in DEPC-treated water. For each sample, small RNAs from about 100 μg of total RNA were separated on a 17% polyacrylamide gel, which was electrotransferred to a Hybond N+ membrane (GE Lifesciences). Membranes were cross-linked, incubated for 2 h at 80 °C, and hybridized overnight at 38 °C with 32P-labeled DNA probes or oligonucleotides (listed in Supplementary information, Table S4) in PerfectHyb buffer (Sigma). Washed membranes were exposed to X-ray films at −80 °C for 6 days.
Production of dcl1/2/3/4 quadruple mutants
The dcl1-201 and dcl1-202 alleles were generated using CRISPR/Cas9 technology39 in the dcl2/3/4 mutant background27. We designed two sgRNAs targeting the 3′ portion of DCL1, close to the T-DNA insertion site of the dcl1-100 mutant (Supplementary information, Figure S4A)40. All the dcl2/3/4 plants used for transformation were genotyped. Afterwards the T2 plants that show similar developmental defects to dcl1-100 were selected. The two sgRNA target regions were then PCR amplified and sequenced. The resulting dcl1-201 and dcl1-202 alleles contain a “T” insertion at 5 646 bp and a “G” insertion at 5 573 bp of the DCL1 gene, respectively, both of which leads to frameshift and premature stop codons (Supplementary information, Figure S4A).
Individual locus bisulfite sequencing
Genomic DNA was extracted from 2-week-old seedlings using the Qiagen Plant DNeasy Mini Kit. The extracted DNA was then treated with BisulFlash DNA Modification Kit (Epigentek) following the manufacturer's protocol. A 1-μl aliquot of bisulfite converted DNA was used for PCR reaction with primers designed for specific target regions (Supplementary information, Table S4). The PCR products were ligated into the pMD18-T vector (Takara, Japan). The ligation product was transformed into DH5α competent cells and at least 15 single clones were sequenced for each type of transformants. The sequencing results were aligned and analyzed using CyMate (http://www.cymate.org).
Whole-genome bisulfite sequencing and data analysis
Genomic DNA was extracted from either 2-week-old seedlings or adult plants (indicated in the text) and sent to Beijing Genomics Institute (Shenzhen, China) for bisulfite treatment, library construction and high throughput sequencing. Clean reads were generated by trimming off adaptor and low-quality sequences (q < 20) and then were mapped to the Arabidopsis genome (TAIR 10) using BSMAP (Bisulfite Sequence Mapping Program) allowing two mismatches22. DMRs were identified using a published method with slight modifications22. First, the cytosines with a read coverage less than five were filtered out. The DNA methylation level in every 200-bp window with a step size of 50 bp was compared using Fisher's exact test between control and mutant plants (nrpd1 vs wild type; dcl/ago/nrpe1 mutants vs nrpd1). P-values calculated from the tests were then adjusted using the Benjamini-Hochberg method to control for the false discovery rate (FDR). Windows with an adjusted P-value < 0.05 were then tested for the number of DMCs (differentially methylated cytosines), which were defined as cytosines with a P-value < 0.01 in Fisher's exact test. For the hypo-DMRs identified in nrpd1 windows with at least 7 DMCs and 1.5-fold change in DNA methylation levels were combined to generate the final list of DMRs if the gap length between two windows is no more than 100 bp. Regarding the use of replicates, we used the two replicates to identify DMRs separately, and the two lists of DMRs were compared and only DMRs with at least 20% overlap (of the shorter one) were retained as the final list of DMRs.
For the D/A/E loci, windows with at least 5 DMCs and 1.5-fold higher DNA methylation levels (dcl/ago/nrpe1 vs nrpd1) were combined to generate the list of loci. Overlapped regions between D, A, E loci are defined as regions that are overlapped by at least 20% in length (of the shorter DMR), while non-overlapped regions do not physically overlap. Please note that the number discrepancy between DMR number and the sum of numbers in the Venn circles comes from regions that are overlapped by < 20% in length.
For the DCL-independent (ID), -partially dependent (PD) and -fully dependent (FD) loci, we first defined RdDM loci by combining the hypo-DMRs identified in either nrpd1 or nrpe1. Then the average DNA methylation level at each RdDM locus was determined and the difference between dcl1/2/3/4 and wild-type plants (dcl1/2/3/4-WT) was calculated. Each locus was classified based on the methylation level of WT (A), dcl1/2/3/4 (B) and lower one of nrpd1 and nrpe1 (denoted as C). If B > A – 0.25 × (A-C), the locus was classified as ID. If B < A – 0.75 × (A-C), it was classified as FD. The rest were PD.
Genome-wide small RNA sequencing and analysis
Total RNA samples were prepared from 2-week-old seedlings following the standard protocol of TRIzol reagent (Sigma). Total RNA was then separated on denaturing polyacrylamide gels and < 100-nt fractions were cut out and purified for standard small RNA library preparation and sequencing at BGI (Shenzhen, China). After sequencing, adapter sequences were trimmed and clean reads with sizes ranging from 18 to 85 nt were mapped to the Arabidopsis genome (TAIR10) using Bowtie with parameter “-v 0 −k 10”63. Reads that were mapped to annotated structure RNAs including tRNAs, rRNAs, snRNAs and snoRNAs were excluded. Read counts were normalized to Reads Per Ten million (RPTM) based on total mapped reads. The “hits-normalized-abundance” (HNA) values were calculated by dividing the normalized abundance (in RPTM) for each small RNA hit, where a hit is defined as simply the number of loci at which a given sequence perfectly matches the genome64 and they were used for boxplot generation. The boxplots were generated in R using function “boxplot” with parameter “range = 1.5, outline = F, notch = T”. To generate the sRNA heat map, the percentage of HNA value in different size ranges was calculated for each RdDM locus.
The generation of sRNA clusters was modified from65: reads with > 24-nt or 24-nt were merged using merge from BEDTools66 to form “islands”. Islands with fewer than 3 reads were filtered out. Then islands within 100 bp were merged. Finally, those clusters containing 2 or more islands were retained. For cluster with singleton island, it was retained if its length ≥ 100 bp or it contains at least 25 reads.
To characterize Pol IV-dependent > 24-nt RNAs, clean reads were mapped allowing two mismatches. Only uniquely mapped reads that cannot be mapped to annotated structure RNAs were retained for downstream analysis.
Histone features at RdDM loci
The public data used for the analysis was downloaded from Gene Expression Ominbus (Accession No: GSE28398)41. The color-space reads were aligned to TAIR10 genome using Bowtie63 allowing no more than 3 mismatches. Only reads that are uniquely mapped to the genome were retained for the downstream analysis. To generate the relative histone signal distribution in the flanking 5-kb region of the mid-point of DMRs, the whole region was divided into 201 bins with a size of 50 bp and the 101th bin aligning at the middle point of each DMR. The number of reads in each bin was summed.
The relative histone modification signal (y axis) in each bin was defined as: n(Histone) × N(Input)/(N(Histone) × n(Input)) where n are the sum of depth in each bin and N are the number of all mapped reads. Histone and Input represent specific histone modifications and ChIP input, respectively.
Acknowledgments
This work was supported by the Chinese Academy of Sciences and US National Institutes of Health (R01GM070795) to J-K Z.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
DNA methylation levels at the 5' UTR region of HEI10.
DNA methylation and small RNA levels at DAE loci.
DNA methylation and small RNA levels at DA loci.
Introducing dcl1 mutations in the dcl2/3/4 mutant.
Overlap between SHH1/DTF1-dependent loci and DCL-independent (ID), -partially dependent (PD) or -fully dependent (FD) RdDM loci.
The distribution of 24-nt and >24-nt RNAs on the genome.
Size distribution of 18–85 nt RNAs.
Whole genome bisulfite sequencing and RNA sequencing information
Position information of DMRs
Statistic analysis results
List of oligos used in this study
List of mutants used in this study
References
- He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res 2011; 21:442–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 2010; 11:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 2007; 130:851–862. [DOI] [PubMed] [Google Scholar]
- Martienssen RA, Richards EJ. DNA methylation in eukaryotes. Curr Opin Genet Dev 1995; 5:234–242. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, et al. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 2003; 163:1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res 1993; 21:2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 2007; 17:379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Johnson LM, Groth M, et al. Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol Cell 2014; 55:495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 2006; 18:1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 2001; 292:2077–2080. [DOI] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 2012; 151:167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 2014; 21:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013; 153:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 2002; 99 (Suppl 4):16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 2014; 15:394–408. [DOI] [PubMed] [Google Scholar]
- Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 2015; 16:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M. The role of the RNAi machinery in heterochromatin formation. Cell 2005; 122:13–16. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev 2005; 19:1635–1655. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJ. RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 2015; 66:243–267. [DOI] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 2011; 12:483–492. [DOI] [PubMed] [Google Scholar]
- Law JA, Du J, Hale CJ, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 2013; 498:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ma ZY, Zeng L, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci USA 2013; 110:8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Du J, Hale CJ, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 2014; 507:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZW, Shao CR, Zhang CJ, et al. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet 2014; 10:e1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Ream TS, Marasco M, et al. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol Cell 2012; 48:811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2004; 2:E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet 2006; 38:721–725. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 2005; 15:1494–1500. [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 2005; 102:12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 2007; 5:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 2013; 152:352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Vandivier LE, Tu B, et al. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res 2015; 25:235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, et al. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 2010; 22:321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan CG, Zhang H, Tang K, et al. Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation. EMBO J 2015; 34:581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Du J, Hale CJ, et al. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 2014; 157:1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda K, Slotkin RK. Proposed mechanism for the initiation of transposable element silencing by the RDR6-directed DNA methylation pathway. Plant Signal Behav 2013; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Chambon A, et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet 2012; 8:e1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang B, Ding W, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 2013; 23:1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Mao Y, Xu N, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 2014; 111:4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA 2008; 105:8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Sidote DJ, Zhang Y, Kerstetter RA, Michael TP, Lam E. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J 2013; 73:77–90. [DOI] [PubMed] [Google Scholar]
- Qian W, Miki D, Zhang H, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 2012; 336:1445–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 2007; 5:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008; 133:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature 2004; 431:356–363. [DOI] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, van der Winden J, Matzke AJ, Matzke M. RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA 2002; 99 (Suppl 4):16499–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M. RNA-directed DNA methylation. Plant Mol Biol 2000; 43:203–220. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002; 297:1833–1837. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science 2003; 301:798–802. [DOI] [PubMed] [Google Scholar]
- Dalakouras A, Dadami E, Wassenegger M. Viroid-induced DNA methylation in plants. Biomol Concepts 2013; 4:557–565. [DOI] [PubMed] [Google Scholar]
- Dalakouras A, Wassenegger M. Revisiting RNA-directed DNA methylation. RNA Biol 2013; 10:453–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol 2006; 4:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga-Vazquez M, Sidorenko L, Rabanal FA, et al. RNA-mediated trans-communication can establish paramutation at the b1 locus in maize. Proc Natl Acad Sci USA 2010; 107:12986–12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano H, Fukudome A, Hiraguri A, Moriyama H, Fukuhara T. Distinct substrate specificities of Arabidopsis DCL3 and DCL4. Nucleic Acids Res 2014; 42:1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell 1998; 93:627–637. [DOI] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, et al. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 2015; 163:445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Podicheti R, Mishra V, et al. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 2015; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 2013; 503:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond DM, Baulcombe DC. Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc Natl Acad Sci USA 2015; 112:917–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Wu Q, Ito T, et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA 2010; 107:484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Tang K, Qian W, et al. An Rrp6-like protein positively regulates noncoding RNA levels and DNA methylation in Arabidopsis. Mol Cell 2014; 54:418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA 2011; 108:2617–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA methylation levels at the 5' UTR region of HEI10.
DNA methylation and small RNA levels at DAE loci.
DNA methylation and small RNA levels at DA loci.
Introducing dcl1 mutations in the dcl2/3/4 mutant.
Overlap between SHH1/DTF1-dependent loci and DCL-independent (ID), -partially dependent (PD) or -fully dependent (FD) RdDM loci.
The distribution of 24-nt and >24-nt RNAs on the genome.
Size distribution of 18–85 nt RNAs.
Whole genome bisulfite sequencing and RNA sequencing information
Position information of DMRs
Statistic analysis results
List of oligos used in this study
List of mutants used in this study