Abstract
The origin and evolution of the domestic dog remains a controversial question for the scientific community, with basic aspects such as the place and date of origin, and the number of times dogs were domesticated, open to dispute. Using whole genome sequences from a total of 58 canids (12 gray wolves, 27 primitive dogs from Asia and Africa, and a collection of 19 diverse breeds from across the world), we find that dogs from southern East Asia have significantly higher genetic diversity compared to other populations, and are the most basal group relating to gray wolves, indicating an ancient origin of domestic dogs in southern East Asia 33 000 years ago. Around 15 000 years ago, a subset of ancestral dogs started migrating to the Middle East, Africa and Europe, arriving in Europe at about 10 000 years ago. One of the out of Asia lineages also migrated back to the east, creating a series of admixed populations with the endemic Asian lineages in northern China before migrating to the New World. For the first time, our study unravels an extraordinary journey that the domestic dog has traveled on earth.
Keywords: dog domestication, demography, Chinese indigenous dog, gene flow, artificial selection
Introduction
The domestic dog (Canis lupus familiaris), one of our closest companions in the animal kingdom, has followed us to every continent of the world. As a single species, the domestic dog embodies one of the largest collections of phenotypic diversity for any species living on earth1. Due to their cognitive and behavioral abilities, domestic dogs have been selected to fulfill a wide variety of tasks including hunting, herding and companionship. The genetic and historical basis of these phenotypic changes has intrigued the scientific community, including Darwin2.
The history of dog domestication is often depicted as a two-stage process where primitive dogs were first domesticated from their wild ancestors, the gray wolves, and in the second stage, the primitive forms were further selected to form many dog breeds with specialized abilities and morphology3,4,5. Despite many efforts studying dog evolution, several basic aspects about the origin and evolution of the domestic dog are still in dispute. For example, several different geographical regions have been proposed as the birthplace of domestic dogs, and the date of divergence between wolves and dogs has been estimated between 32 000 years ago and 10 000 years ago6,8,9,10, with relatively weak gene flows found between these two groups since their divergence4,6,7,9. The exact history of dog domestication thus remains to be fully resolved11.
The first comprehensive genetic investigations of the geographical origin of dogs were based on global intraspecific studies of maternally transmitted DNA (mtDNA) in contemporary dogs, which gave a strong indication that dogs originated in the southern part of East Asia7,12. However, several subsequent studies based on diverse genetic markers have given discrepant answers. For example, using mtDNA from ancient dog samples, Thalmann et al. have suggested Europe as the place of origin13. Likewise, using genome-wide genotyping of modern dogs, vonHoldt et al. found high haplotype sharing between Middle Eastern wolves and dogs, proposing the Middle East as the major source of dog diversity14.
Although the datasets and approaches are different in these studies, a common drawback of these single nucleotide polymorphism (SNP) array- and ancient DNA-based studies is a lack of samples from southern East Asia, thus precluding evaluation of the possible scenario that domestic dogs actually originated in this region. In addition, the use of a single locus, especially mtDNA, can skew the conclusion as it is more malleable by stochastic and/or selective forces7,12,13. Thus, the history of dog domestication remains enigmatic and highly controversial11.
Whole genome sequencing provides a powerful holistic approach to understanding the evolutionary history of a species, and is sufficiently robust in mitigating problems such as SNP ascertainment bias or stochastic effects acting on a single marker, which have influenced earlier studies15. In this work, we collected the genome sequences of 58 canids from across the world, including samples from southern and northern parts of East Asia, Africa, Europe, the Middle East, Siberia and the Americas. Population genetic analysis reveals an ancient origin for the domestic dog in southern East Asia about 33 000 years ago. After evolving for several thousand years in East Asia, a subgroup of dogs radiated out of southern East Asia about 15 000 years ago to the Middle East, Africa as well as Europe. One of these out of Asia lineages then migrated back to northern China and made a series of admixtures with endemic East Asian lineages, before traveling to the Americas. Our study, for the first time, reveals the extraordinary journey that the domestic dog has traveled on this planet during the past 33 000 years.
Results
Sample collection and whole genome sequencing
58 canids from around the world were gathered for this study. This collection includes 12 gray wolves from across the Eurasian continent, 11 indigenous dogs from southern East Asia, 12 indigenous dogs from northern East Asia, 4 village dogs from Africa (Nigeria) and a set of 19 diverse dog breeds distributed across the Old World and the Americas.
Chinese indigenous dogs are dogs living in the countryside of China16 (Supplementary information, Data S1 and Figure S1) and were sampled across the geographic range of rural China, including many remote regions in Yunnan and Guizhou in southern China (Supplementary information, Table S1). The breeds include dogs from Central Asia (Afghan Hound) and North Africa (Sloughi), Europe (eight different breeds), the Arctic and Siberia (Greenland dog, Alaska Malamute, Samoyed, Siberian Husky, and East Siberian Laika), the New World (Chihuahua, Mexican and Peruvian naked dog) as well as the Tibetan Plateau (Tibetan Mastiff). These dogs were chosen to cover as many major geographic regions as possible (Figure 1A and Supplementary information, Table S1).
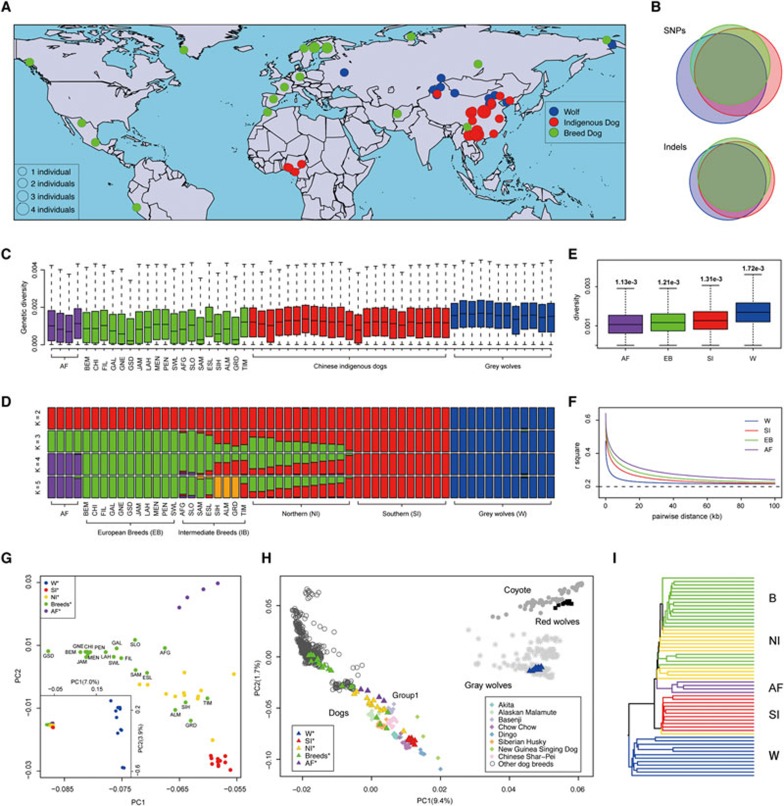
Figure 1.
Population structure and genetic diversity of 58 canids. (A) Geographic locations of the 58 canids sequenced in this study. (B) Amount of of SNPs and small indels called in this study. (C) Genetic diversity for the 58 canids. AF, African village dogs; BEM, Belgian Malinois; CHI, Chihuahua; FIL, Finnish Lapphund; GAL, Galgo; GNE, Gray Norwegian Elkhound; GSD, German Shepherd Dog; JAM, Jamthund; LAH, Lapponian Herder; MEN, Mexican naked (hairless); PEN, Peruvian naked (hairless); SWL, Swedish Lapphund; AFG, Afghan Hound; SLO, Sloughi; SAM, Samoyed; ESL, East Siberian Laika; SIH, Siberian Husky; ALM, Alaska Malamute; GRD, Greenland dogs; TIM, Tibetian Mastiff. (D) Structure analysis of the 58 canids. (E) Genetic diversity of the different groups. AF, African village dogs; EB, European breeds; SI, southern Chinese indigenous dogs; W, wolves. (F) Linkage disequilibrium patterns for the different groups. (G) Principle component analysis of the 58 canids. Inset is for all individuals and the large panel is for dogs only. (H) Principle component plot for a large collection of canids together with our data. (I) A clock-like tree (UPGMA) for all the 58 individuals56.
After DNA extraction, individual genomes were sequenced to an average of 15× coverage (Supplementary information, Table S1). Of the 58 individuals, 4 gray wolves and 6 dogs have been sequenced in a previous study10. DNA sequence analysis was done using the Genome Analysis Toolkit17. After stringent filtering, we identified 20 353 184 SNPs and 3 856 246 small indels (Figure 1B), most of which are shared between groups. For example, 40.3% of the SNPs are shared between wolves, indigenous dogs and dog breeds, reflecting their recent divergence (Figure 1B). Using Sanger sequencing, we verified that the sequencing strategy was highly sensitive (false negative rate around 10%) and the amount of false positives was less than 5% (Supplementary information, Data S2 and Figure S2).
Genetic diversity and population structure
Comparison of the two haploid genomes within each individual yields the genetic diversity θ (4 Nμ) for the 58 individuals. As shown in Figure 1C, genetic diversity shows a decreasing trend from wolves to Chinese indigenous dogs (preserving 78% of the wolf heterozygosity) and subsequently to dog breeds (66% of the wolf heterozygosity), with the African village dogs having a genetic diversity comparable to many dog breeds (69% of the wolf heterozygosity). Among the dog breeds, the levels of variation in genetic diversity are quite dramatic. For example, the East Asian breed Tibetan Mastiff and East Siberian Laika show levels of diversity comparable to the Chinese indigenous dogs, but many of the European dog breeds have considerably reduced genetic diversity. Such dramatic differences in genetic diversity can be influenced both by ancient and recent history of inbreeding.
To explore the genetic relationships among these individuals, we performed a structure analysis using an expectation maximization (EM) algorithm to cluster the individuals into different numbers of groupings18. When partitioning the individuals into two groups, the algorithm separates the dogs from the wolves, with very limited admixture observed (Figure 1D). Further dividing the individuals into three subsets split the dogs into two clusters, with indigenous dogs from southern East Asia representing one subset and the other subset consisting of dog breeds from Europe and South/Central America and the African village dogs. Indigenous dogs from northern China and dog breeds from the Arctic and Central Asia, the Middle East and North Africa show a mixture of these components with varying proportions. This observation implies that there are two divergent groups of dogs: one is East Asian component and the other, non-East Asian component. It is important to emphasize that individuals with mixed constituents identified in the structure analysis are not always due to true admixture events, since populations of intermediate genotypes between these two groups tend to display mixed components (e.g., originated shortly after the split of two clades, Supplementary information, Data S3 and Figure S3). Further partitioning into four and five groups leads to the separation of the African village dogs and the breed dogs from the eastern Arctic regions (i.e., Siberian Husky, Alaska Malamute and the Greenland dog).
Genetic diversity among individuals (Figure 1C) may be heavily influenced by ancient as well as recent history, e.g., breeding programs during the last few thousand years or the past few hundred years. However, combined information from multiple breeds may reveal information about the ancestral populations that gave rise to them, since each breed has experienced separate breeding history. We therefore calculated the genetic diversity (θπ) for the “pure groups” informed by the structure analysis (K = 4, Figure 1D). As shown in Figure 1E, dog breeds, most of which of European origin, carry lower diversity than the Chinese indigenous dogs as a group, but have higher genetic diversity than the African indigenous dogs. This suggests that the ancestral population that gave rise to the European breeds was larger than the ancestral population of the African indigenous dogs. Linkage disequilibrium patterns also show similar trends (Figure 1F).
Principle component and phylogenetic analysis
When projecting the genotypes into a two-dimensional space using a principle component analysis (PCA)19, all dogs cluster together tightly compared with the distribution seen for wolves (Figure 1G, inset). When inspecting the distribution among dogs, we find that dogs spread along three major geographic axes: southern East Asia, Europe and Africa. The northern Chinese indigenous dogs and dog breeds from the Middle East/Arctic regions/Tibet fall between these three extremes (Figure 1G). The observed pattern reflects the overall geographic locations of these groups following a clear East-West gradient, which matches quite well the observation from our structure analysis.
Combining our dataset with data from a previous SNP array study, which included a larger number of samples20, we found that the southern Chinese indigenous dogs together with several East Asian dogs (e.g., Chow Chow, Akita, Chinese Shar-Pei) are closest to wolves (Figure 1H). When the phylogenetic relationships among our 58 samples are inspected, East Asian dogs spread over both sides of the deepest node connecting all dogs, while dogs from other continental areas coalesce into a subclade and then join with East Asian dogs. Thus, East Asian dogs are the most basal lineages connecting to gray wolves (Figure 1I). It is worth pointing out that the genomes of dogs from Oceania (dingoes and New Guinea singing dogs), although being closer to wolves in the PCA plot (Figure 1H), bear strong signals of admixture with gray wolves6, which likely reflects their past history of admixture, before they migrated to Australia and New Guinea (Supplementary information, Data S4 and Figure S4).
Admixture analysis
Using the joint allele frequencies among all populations in our study, we infer the split and admixture history among groups of populations using TreeMix21. If migration tracks are not allowed, then the relationships inferred from the TreeMix analysis (Figure 2A) directly reflect the patterns observed in our previous analyses including the structure (Figure 1D), the phylogenetic (Figure 1I) and the principal component analyses (Figure 1G). Thus, following the divergence between contemporary wolves and domestic dogs, the first partition within dogs is between the southern Chinese indigenous group and all other dogs. This is then followed by branching of the other dogs, largely matching the geographical distance from southern East Asia: first, dogs from Central Asia, northern China, and eastern Arctic, followed by dogs in Africa, the Middle East, and western Arctic, and the final group including all dog breeds in Europe and South/Central America.
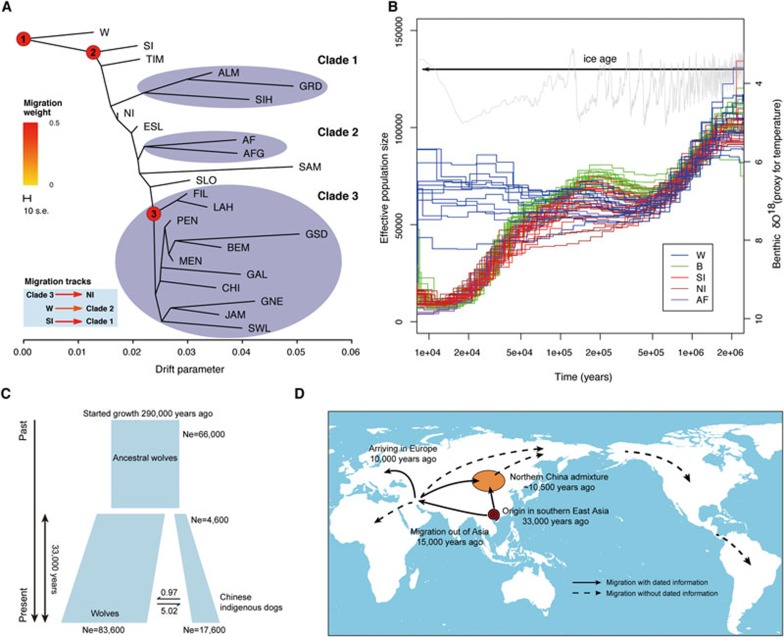
Figure 2.
Demographic and migration histories for the domestic dog. (A) Tree topology inferred from TreeMix when no migratory tracts are allowed. The drift parameter is the amount of genetic drift along each population. Further inferred migratory tracts are shown in the bottom-left corner of the panel. The three important nodes are those that we have provided extensive dating information. (B) The PSMC plot for all the individuals. Gray lines plot the benthic δO18 levels, which are a proxy for global temperature61. The span of the current ice age (Quaternary ice age, 2.58M-now) is shown with an arrow. The x-axis is time plotted in log scale and the y-axis is effective population size. (C) Inferred population demographic history between wolves and southern East Asian indigenous dogs using the joint site frequency spectra. (D) A proposed migratory history for domestic dogs across the world based on the evidence from our study. Solid arrows represent migratory tracts that we have dating information, while dashed arrows indicate those without accurate dating.
If migration tracks are allowed in TreeMix, there is strong statistical support for migrations among a few groups: (1) northern Chinese indigenous dogs show strong admixture from European dogs (Figure 2A and Supplementary information, Data S5, Figure S5, Tables S2 and S3); (2) gene flow from wolves to the African/Middle Eastern dogs (Supplementary information, Figure S5); (3) migratory tracks from the southern Chinese dogs to the eastern Arctic group (i.e., Siberian Husky, Alaska Malamute and the Greenland dog; Supplementary information, Figure S5). When all possible migration events in the history of these samples are examined using the F3/F4 test22, there is again a strong statistical support for all the migration events listed above (Supplementary information, Data S5).
Long-term evolutionary trajectories for wolves and dogs
Using the divergence between the two haploid genomes within individuals, the pairwise sequentially Markovian coalescent (PSMC) model provides a method for investigating the long-term trajectories in population sizes23. To translate demographic history into real-time units, estimation of an accurate mutation rate is very important. Previously, several different mutation rates were used, but they were generally not carefully calibrated (Supplementary information, Data S6)24. Using multiple outgroup species to the dog (e.g., horse and cat), our estimate of the mutation rate for the lineage leading to the domestic dog is 2.2 × 10−9 per site per year (Supplementary information, Data S6 and Table S4), a rate similar to those from several earlier studies25,26. Using this mutation rate, we estimate dates for the population history of dogs and wolves. As shown in Figure 2B, a decrease in the size of the ancestral wolf population started to occur 2 million years ago, reaching a saddle point about 3-400 000 years ago. The ancestral population then increased in size, peaking at around 200 000 years ago. After a subsequent small decline in population size, wolves and dogs started to diverge from each other between 20 000 and 100 000 years ago (see next section for a more precise dating). Although all domestic dogs drastically decreased in population size after the population split, the wolf population experienced a slight growth, possibly as a consequence of the megafauna extinctions (i.e., late Quaternary extinction)27 that provided gray wolves with better food resources due to reduced competition from other predators.
Time of divergence between contemporary wolves and dogs
Treemix and phylogenetic analyses identified southern Chinese indigenous dogs as the most basal population compared to wolves, from which all other dog populations diverged. We therefore used joint allele frequencies between the 12 gray wolves and the 11 southern Chinese indigenous dogs, to infer the demographic history for these two populations with the dadi package28. Similar to the result from the PSMC analysis, the wolf population experienced a very mild population growth (1.26-fold increase) that started around 290 000 years ago (Figure 2C). The time of divergence for the wolf and dog populations is inferred to be around 33 000 years ago, where the domestic dog lineage expanded from a population of 4 600 individuals to about 17 500.
In addition to gauging changes in population size, statistical methods can also estimate the rates of exchange of migrants between two populations. The migration rate (2Nm) from the dog lineage to the wolf lineage is estimated to be 0.97, while the other direction (wolves to dogs) is inferred to be 5.02, showing a clear asymmetry in the migration rates29.
Examination of the sequence divergences between the multiple populations using a Markov chain Monte Carlo (MCMC) approach30,31 (Supplementary information, Data S7, Figures S6-S8, Tables S5 and S6) reveals a similar profile for the history between wolves and dogs, which includes a slight growth in the wolf population and an ancient divergence between wolves and dogs (Supplementary information, Data S7 and Table S5). In summary, multiple levels of genetic information (i.e., both joint site frequencies as well as sequence divergence) support an ancient split between dogs and wolves.
The geographical origins of dogs: a single origin in southern East Asia
In order to identify the most probable geographical origin of dogs, we hypothesized that similar to many organisms, the geographical origin of a species holds the greatest genetic diversity, and the global relationship among multiple populations will, in the absence of strong influence of admixture, follow a serial founder model32,33. In the case of dogs, the wild ancestor, the wolf, has been present along the dog throughout Eurasia, implying that intense dog-wolf admixture could possibly have influenced this pattern.
Despite the concern on the confounding effect of wolf/dog gene flow, the TreeMix analysis, F3/F4 test as well as the demographic analysis suggest that gene flow between dogs and wolves is relatively mild. In Supplementary information, Data S8, we review the evidence for dog/wolf gene flow from our study, as well as from multiple previous studies. The combined evidence shows that the migration rates (2Nm) are mostly around one or less (a maximum of five found in the dadi analysis) and that the admixture proportion is normally around 10%, with a maximum of 16% for the Middle East (Supplementary information, Data S8). Low levels of migration are detected between wolves and dogs across Eurasia when the very sensitive D test is used34,35 (Supplementary information, Data S8). Thus, we conclude that while dog-wolf gene flow has occurred throughout history of the domestic dog, it has been at a moderate level and the level of admixture has been relatively similar across Eurasia (Supplementary information, Data S8). Without the strong influence of admixture32, we may assume that genetic diversity is highest at the place of origin and that the global relationship among the multiple populations follows a serial founder model reflecting their dispersal routes33.
It is tempting to draw conclusions about the origin of dogs from the high genetic diversity observed in the Chinese indigenous dogs. However, comparing breed dogs with indigenous dogs at the individual level is likely misleading since most of the differences in genetic diversity are probably caused by recent bottleneck events rather than their distant origin1. Thus, we combine multiple breeds in each region as a group representing the ancestral haplotype pool giving rise to the contemporary dogs of that region. Our analysis shows that dogs from East Asia have the highest genetic diversity (Figure 1E). This suggests that the ancestral population that gave rise to East Asian dogs was much larger than ancestral populations in other regions (e.g., Europe). The linkage disequilibrium pattern also shows the same trend (Figure 1F). Higher levels of genetic diversity in East Asian dogs are also observed in mtDNA and Y chromosome data7,12,36.
Beside group diversity, in the phylogenetic and TreeMix analyses, the deepest node connecting all dogs separates into two clades, one of which is composed of only East Asian dogs, while the other clade includes both East Asian and non-East Asian dogs (Figures 1I and 2A, and Supplementary information, Figure S5). Dogs from Africa and Europe share a most recent common ancestor, which then coalesces with dogs from East Asia (Figures 1I and 2A). Notably, this basal position of East Asia is robust to the levels of migrations between wolves and dogs (Supplementary information, Data S9, Figure S9, and Table S7). The basal position of East Asian dogs is similar to the pattern observed for Africans within human populations37.
In addition to the observations based on group level diversity and the basal phylogenetic position, the PCA pattern also provides supporting evidence for the southern East Asian origin of dogs. As the amount of genetic drift in basal groups is typically lower due to their larger population sizes, we expect them to display a closer genetic relationship with wolves in the PCA plot (Figure 2A). When we simulate a serial founder model that mimics the history of dog domestication, we can easily generate a pattern that is similar to that shown in Figure 1G (see also Supplementary information, Data S10 and Figure S10). Thus, in our analysis, we find dogs with ancestry in southern East Asia to be closest to wolves, and also a geographical distribution of the populations following a clear east-west gradient, indicating serial founder events. It is important to emphasize that admixture between wolves and dogs is unlikely to have created the observed pattern, given that the dog-wolf admixture rate in East Asia is not higher than that seen in other regions (Supplementary information, Data S8).
Having identified southern East Asia as the likely origin of dogs, we asked whether the domestic dog may have originated in more than one region through separate domestication events. In order to test whether multiple origins are compatible with the observed data, we performed simulations mimicking different scenarios (Supplementary information, Data S11 and Figure S11). Our results show that, if there were multiple origins for dogs from separate wolf populations, the descendant populations would tend to reside in separate clusters in the PCA plot, which is in contrast to what we observe (Figure 1G, inset). Thus, that the domestic dog originated multiple times in different geographical areas is not compatible with the observed genetic patterns found in our genome data.
The out of southern East Asia history for the domestic dog
To study the subsequent global history of the dog, we used an MCMC approach to date several important transitional points among the major clades (Figure 2A). Our analysis supports the split between the southern Chinese indigenous dogs and all other dogs across the world around 15 000 years ago, thus indicating a radiation of dogs out of southern East Asia earlier than the origin of agriculture (Supplementary information, Data S7 and node 2 in Figure 2A and 2D)38. After radiating from southern East Asia, possibly following existing human settlements at the time (Supplementary information, Data S12 and Figure S12), the out of southern East Asia lineage spread to the Middle East/Africa and arrived in Europe by about 10 000 years ago (Supplementary information, Data S7; node 3 in Figure 2A and 2D). Notably, one of the out of southern East Asia lineages migrated back to northern China, meeting endemic Asian lineages that had spread from southern East Asia and yielding a series of admixed populations, including the northern Chinese indigenous dogs and the Arctic dog breeds (Figure 2A and 2D).
Several dog breeds from South and Central America (i.e., Chihuahua, the Mexican and Peruvian naked dog) show no signs of admixture, while the Arctic breeds, Alaska Malamute and the Greenland dog, display extensive admixture from the southern Chinese Indigenous lineage39. Possibly, this reflects that the human colonization of the New World occurred in several waves, in which dogs may have followed in different time periods40 (Figure 2D). Using the patterns of the admixture tracks, we estimate that the time of the admixture for the northern Chinese indigenous dogs was quite ancient (around 10 500 years ago, Supplementary information, Data S13 and Figure S13)40. The relatively recent origin of European dogs (i.e., ∼10 000 years) together with this rather ancient admixture suggests that multiple lineages travelled to the Far East from the Middle East/Europe.
Population structure among wolves
Our structure and principal component analyses do not reveal any population substructure among the gray wolves collected for this study (Figure 1D). The high migratory ability of the gray wolf might allow the populations to remain highly homogenous across the eastern part of Eurasia41. A previous study using wolves from the Middle East (Israel), Europe (Croatia) as well as China found genetic differentiation among these wolf populations6. When these three individuals are overlaid on the large PCA plot, the wolves from western Eurasia do not group together with the wolves we collected from eastern Eurasia, and they are genetically closer to dogs (Supplementary information, Data S14 and Figure S14). Given the fact that Middle Eastern wolves generally have more dog admixture6, the observed difference might not represent true population differentiation among wolves. Nevertheless, it is possible that some wolves have recently diverged from each other8, as there is weak isolation between the wolves from eastern and western Eurasia. Explicit testing for potential admixture between wolves and dogs sampled in our study finds evidence of gene flow between wolves and local dog populations in each region, albeit the magnitude is low (Supplementary information, Table S8). Further study on the genetic and geographic relationships between dogs and wolves is one of the important tasks for the community.
Domestication genes
Our analyses indicate that the Chinese indigenous dogs represent an intermediate form between wolves and breed dogs, and they have not experienced intense artificial selection. Analyses of Chinese indigenous dogs therefore allow us to stratify the domestication process in dogs, and investigate the role of positive selection that occurred specifically during the first stage of domestication. Using a statistical method that explicitly models selective sweeps42, we have identified the top 1% of the genome bearing strong statistical evidence of positive selection in the southern Chinese indigenous dogs. In Table 1, we list the categories of genes that show statistical significance by a gene enrichment-based analysis. Groups of genes showing the strongest evidence of positive selection are those related to metabolism and motility, neurological process and perception as well as sexual reproduction (Table 1 and Supplementary information, Data S15, Tables S9 and S10). Genes that seem to have been positively selected in subsequent evolutionary steps, including dog breed formation, are related to the control of developmental processes and to metabolism (see a full discussion of candidate genes involved in transforming wild wolves to dogs in Supplementary information, Data S15).
Table 1. Gene ontology analysis of genes selected during the first stage of dog domestication.
| GO category | number of genes | P value |
|---|---|---|
| Metabolism and motility | ||
| ATPase activity1 | 19 | 0.0056 |
| ATP binding1 | 55 | 0.0192 |
| Actin binding1 | 17 | 0.0195 |
| Nucleoside triphosphate metabolic process2 | 9 | 0.0208 |
| ATP metabolic process2 | 8 | 0.0211 |
| Phasic smooth muscle contraction2 | 3 | 0.0315 |
| Ribonucleotide metabolic process2 | 9 | 0.0385 |
| Purine nucleoside triphosphate metabolic process2 | 8 | 0.0399 |
| ATPase activity, coupled1 | 14 | 0.0402 |
| mRNA metabolic process2 | 17 | 0.0423 |
| Receptor metabolic process2 | 4 | 0.0448 |
| Drug metabolism3 | 8 | 7.82E-04 |
| Metabolism of xenobiotics by cytochrome P4503 | 7 | 0.0031 |
| ABC transporters3 | 6 | 0.0056 |
| Glutathione metabolism3 | 6 | 0.0075 |
| Retinol metabolism3 | 5 | 0.0452 |
| Neurological process and perception | ||
| Memory2 | 6 | 0.0034 |
| Regulation of sensory perception2 | 3 | 0.0315 |
| Regulation of sensory perception of pain2 | 3 | 0.0315 |
| Learning or memory2 | 8 | 0.0352 |
| Regulation of neurotransmitter levels2 | 6 | 0.0359 |
| Long-term memory2 | 3 | 0.0446 |
| Synaptic transmission2 | 14 | 0.049 |
| Long-term potentiation3 | 6 | 0.0291 |
| Sexual reproduction | ||
| Germ cell development2 | 7 | 0.0481 |
1Molecular function;
2Biological process;
3KEGG pathway
Among the candidates as positively selected genes in the first stage of dog domestication, a class of genes are related to memory and long-term potentiation (LTP), which is widely considered to be the major cellular mechanism underling learning and memory43. For example, GRIA1 (glutamate receptor, ionotropic, AMPA 1) is an important protein that mediates excitatory synaptic transmission in the central nervous system and plays a key role in hippocampal synaptic LTP and long-term depression (LTD). Interestingly, a suite of other genes, including GRIN2A (glutamate receptor, ionotropic, N-methyl D-aspartate 2A), are also found to be heavily involved in LTP and LTD (Table 1). The large physiological and behavioral changes empowered by these genes may have enabled the transformation of gray wolves to domestic dogs, allowing them to flourish in the human environment.
Discussion
Based on genome sequences from a worldwide collection of dogs, especially a large collection of indigenous dogs from southern East Asia, this study provides strong genetic evidence that the domestic dog originated in southern East Asia. The analyses give a coherent picture, where the indigenous dogs in southern East Asia or East Asia in general stand out compared to other populations, with higher genetic diversity as a group, and occupying a basal position next to wolves. Other dog populations show progressive ancestry gradient away from wolves starting from southern East Asia. Notably, these findings corroborate earlier work based on mtDNA and Y-chromosomal DNA7,36. Thus, studies based on comprehensive global samples and diverse types of genetic data (e.g., autosomes, Y chromosome, mtDNA) converge on the same story about the origin of the domestic dog.
The origins of the global domestic dog populations can be traced to two important demographic steps: first, dog and wolf populations started to diverge from each other 33 000 years ago in southern East Asia (matching several previous findings8,10). Subsequently there was a global dispersal of dogs out of southern East Asia around 15 000 years ago. The long persistence of the domestic dog lineage in southern East Asia opens up for interesting scenarios. One possible explanation for the 33 000-year deep divergence between dogs and wolves is that it represents a split among wolf populations, and that South Chinese wolves (ancestors to the dog) were genetically differentiated from the more northern wolves sampled in our study. In this case, the global expansion of dogs out of southern East Asia around 15 000 years ago may correspond with the origins of actual domestic dogs. This scenario is contradicted by the fact that wolves in our study display no apparent genetic substructure (Supplementary information, Data S14). An alternative scenario is that the ancient dog-wolf split actually constitutes the first step in the domestication of wolves and evolution to domestic dogs. It is possible that the ecological niche unique in southern East Asia provided an optimal refuge for both humans and the ancestors of dogs during the last glacial period (110-12k years ago, with a peak between 26 500 and 19 000 years ago)44. The mild population bottleneck in dogs suggests that dog domestication may have been a long process that started from a group of wolves that became loosely associated and scavenged with humans, before experiencing waves of selection for phenotypes that gradually favored stronger bonding with humans (a process called self-domestication)1. That among the candidate genes as positively selected are genes involved in the neurological processes may be a manifestation of this dynamic process (Supplementary information, Data S15). After this long-term nurturing, humans and dogs might have eventually come together with a strong bond for each other. Thus, the history of dogs might involve three major stages: (a) loosely engaged pre-domesticated scavengers, (b) domesticated non-breed dogs with close human-dog interactions, (c) breed formation following intense human selection for diverse sets of phenotypic traits. The study of Chinese indigenous dogs thus provide missing links that connect these three major stages45,46.
The exact time when dogs reached the Middle East is difficult to estimate with our sample since the Middle Eastern dogs (and also African dogs) bear relatively strong signals of introgression from wolves (Figure 2A). However, demographic inferences suggest that dogs had arrived in Europe by about 10 000 years ago (Figure 2D and Supplementary information, Data S7), a short time after the origin of agriculture in the Middle East38. It is notable that the global spread of dogs around 15 000 years ago corresponds well with the generally accepted earliest archaeological evidence of dogs across Eurasia11. As there is little evidence of westward human migrations from southern East Asia around 15 000 years ago, the initial spread of the domestic dog out of Asia may in part have been a self-initiated dispersal driven by environmental factors (e.g., the retreat of the glacial coverage that started about 19 000 years ago). The specific route domestic dogs used to migrate to the Middle East, Africa and Europe remains to be uncovered (Figure 2D and Supplementary information, Data S12). Some of this dispersal might be heavily influenced by humans, as dogs were often part of the civilization package that traveled together as agriculture spread47 (Figure 2D). Further studies using samples from western Eurasia should reveal insights into these early dog migrations6.
Despite the strong patterns presented by the genetic data, archaeological evidence supporting an East Asian origin is missing11. Several important factors further confound current analysis. First, the morphological differences between dogs and gray wolves are not always very clear-cut, especially for specimens from the early phase of dog domestication48. In fact, a recent ancient DNA study has ruled out several ancient dog-like specimens found in Europe13. Second, archaeological studies in the Far East are generally lagging behind those in Europe, with most of the ancient dog-like fossils from before 12 000 years ago being found outside of East Asia11. This could also be due to the unfavorable environmental conditions for preserving fossils in southern East Asia. Nevertheless, it is possible that multiple primitive forms of the dog existed, including in Europe13,49. However, in this case, the genetic pattern presented here shows that those lineages were replaced by dogs that migrated from southern East Asia, and thus made negligible contributions to the modern dog gene pool (Figure 1D).
This study opens many potential avenues for future research (Figure 2D). For example, the history of the American colonization and the scale of wolf-dog admixture in the Middle East and Africa remain largely unexplored, especially given the limited coverage of our African samples50. Analysis of additional samples from other parts of the world (especially the Indian coastal region and northern Eurasia as well as Africa) should allow us to draw a more complete picture of the worldwide migration patterns, and their association with human populations. Comprehensive analyses of ancient canid genomes will provide genetic information from multiple time points for elucidating the initial steps of dog history, and identifying putative population replacements that may have influenced modern day dog's gene pool8.
The study of Chinese indigenous dogs has provided an unprecedented opportunity for illuminating the history of selection during dog domestication. For example, the initial selection on the domestic dog is found be strongly associated with an enrichment of genes affecting behavior and motility. As dogs established stronger bonds with humans, possibly empowered by the origin of modern agriculture in the Middle East and China51, strong selection for genes involved in metabolism and morphology/development emerged (Supplementary information, Data S15). Our study, for the first time, begins to reveal a large and complex landscape upon which a cascade of positive selective sweeps occurred during the domestication of dogs. The domestic dog represents one of the most beautiful genetic sculptures shaped by nature and man.
Materials and Methods
Sample collection and sequencing
Total genomic DNA was extracted from the blood or tissue samples of the animals using the phenol/chloroform method. For each individual, 1-3 μg of DNA was sheared into fragments of 200-800 bp with the Covaris system. DNA fragments were then processed and sequenced using the Illumina HiSeq 2000 platform.
Sequence data pre-processing and variant calling
Raw sequence reads were mapped to the dog reference genome (Canfam3) using the Burrows-Wheeler Aligner (BWA)52. Sequence data were next subjected to a strategic procedure for variant calling using the Genome Analysis Tool Kit (GATK)17. During base and variant recalibration, a list of known SNPs/indels downloaded from the Ensembl database were used as the training set. Small indels were separately called using SAMtools mpileup53.
Genetic diversity, linkage disequilibrium and structure analysis
Beagle was used to impute the missing genotypes and phase of the genotypes into the associated haplotypes54. Genetic diversity for each individual, as well as for several sub-groupings, was calculated using a custom python script. Linkage disequilibrium for the different populations was calculated using the haploview software55. Population structure analysis was done using the EM algorithm implemented in the Frappe package18. Principle component analysis was carried out using the smartPCA program from the Eigensoft package19. Unweighted Pair Group Method with Arithmetic mean (UPGMA) tree was built based on the genetic distances calculated from whole genome data56.
Estimation of mutation rate from between species comparisons
Multiple species alignment data were downloaded from the Ensembl database. We used human as the outgroup and chose a second species (cat, horse or cattle) as the sister species to the dog. For each possible sister species, we did a three species comparison (human, (dog, sister_species)) by extracting information from the multiple species alignments. Branch lengths along the dog lineage were estimated using the baseml package from the PAML package57. Long-term evolutionary rate along the dog lineage was then calculated using the branch length divided by the divergence time between the sister species and the dog.
Population admixture and demographic analysis
Population level admixture analysis was first carried out using the TreeMix program21. The threepop/fourpop module from the TreeMix package was used to perform the F3/F4 test22. PSMC model was used to estimate the population histories from the individual genomes23. Since sequence coverage is an important factor in determining the inferred population sizes, a correction factor was invoked to correct for false negatives in SNP calling (Supplementary information, Data S2).
The joint site frequency spectrum between wolves and the southern Chinese indigenous dogs was used to infer the population history using the dadi package28. Lineage specific substitution matrix was first estimated using the ambiore package58 with the whole genome sequence alignments between the outgroup (dhole) (Supplementary information, Table S1) and the dog genome. A corrected site-frequency spectra (SFS) was then used to perform the demographic inference.
Since the ancestral population of wolves might not have been at equilibrium, we allowed the wolf population to change continuously from an equilibrium population at some time in the past (T1). During the continuous change (i.e., from T1 to now), at some more recent time T2, the dog population split off, and started to change its size continuously from an initial size (S1) to an end size (S2) (Figure 2C).
Bayesian analysis of the species evolutionary history was conducted using both the BPP and G-PhoCS package independently on noncoding sequences extracted from the polymorphism data30,31. Population admixture time was estimated using the HAPMIX program40. We used southern Chinese indigenous dogs and breed dogs as the two source populations for the northern Chinese indigenous dogs. The genetic distances between SNPs were extracted from a previous published genetic map59. The overall admixture time is inferred by maximizing the likelihood combining the likelihood values from all individuals.
Targets of positive selection
The SweepFinder algorithm was used to extract regions of the genome that show the strongest signals of positive selection42. The genome-wide site frequency spectrum is used as the background site frequency distribution before fitting a sweep model to the data. Gene Ontology (GO) analysis was carried using DAVID60.
For detailed Materials and Methods see Supplementary information, Data S16. A separate reference list for Supplementary information is provided at the end of Supplementary information, Data S16.
Accession number
This project has been deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive database. The accession number is SRA307300.
Acknowledgments
This work was supported by grants from the NSFC (91231108, 91531303), the 973 program (2013CB835200 and 2013CB835202), the Breakthrough Project of Strategic Priority Program of the Chinese Academy of Sciences (XDB13000000), the Jane and Aatos Erkko Foundation, the Academy of Finland, and Biocentrum Finland. WZ and H-C Y are supported by the Genome Institute of Singapore. We want to thank Dr Jianjun Liu for his comments on an earlier version of this work. PS is a Royal Swedish Academy of Sciences Research Fellow supported by a grant from the Knut and Alice Wallenberg Foundation. G-D W is supported by the Youth Innovation Promotion Association, Chinese Academy of Sciences.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Nomenclature of non-breed dogs and morphological features of Chinese indigenous dogs
False positive (FP) and false negative (FN) rates for variant calling
Both ancient origin and true admixture are compatible with the mixed components observed in the structure analysis
Admixture in dingoes and New Guinea singing dogs
TreeMix results with our dataset and the F3/F4 test
Mutation rate estimation
BPP/G-PhoCS results
Estimating the amount of gene flow between dogs and wolves
Basal positioning of the East Asian breeds is robust against gene flow between wolves and dogs.
Serial founder models can generate a gradient in distances from the outgroup observed in the PCA plot and are unlikely to be caused by wolf/dog migration
Genetic evidence for a single origin
Two possible migration routes
Dating admixture times using HAPMIX
Population substructure in wolves between the eastern and western parts of Eurasia
Selected genes and gene ontology (GO) analysis
Supplementary materials and methods
The morphological features of Chinese indigenous dogs
FN and FP as a function of sequenced individuals
Simulated scheme to study the mixed components in the structure analysis.
TreeMix analysis using the combined genotype data
Treemix results with different numbers of migration tracks
BPP/G-PhoCS analysis for the first group analysis
BPP/G-PhoCS analysis for the second group analysis
Parameter estimates under a wide variety of demographic simulations
Robustness of the basal positioning of Southern Chinese Indigenous dogs to mild gene flow between dogs and wolves
Serial founder models can create patterns observed in the PCA analysis
Simulation of the independent history and principal component analysis
Two possible migration routes
Estimated admixture time for the Northern Chinese indigenous dogs
PCA plot including the three wolves sampled from an earlier publication
Sample information and sequencing throughput
Significant F3 tests among the samples
Candidate admixed populations revealed by the F4 test.
Mutation rate along the dog lineage
Divergence time estimates using Markov chain Monte Carlo(MCMC) methods
Simulated histories testing BPP/G-PhoCS
The inferred tree topologies under different migration rates
D test results between wolves and dogs
GO categories for genes identified using SweepFinder (extra categories not presented in the main text).
Gene Ontology analysis for genes identified by combining FST and population diversity ratio
References
- Coppinger R, Coppinger L. Dogs: A Startling New Understanding of Canine Origin, Behavior & Evolution. 1st Edition, Scribner, 2001.
- Darwin C. The variation of animals and plants under domestication. O. Judd & company, 1868. [PMC free article] [PubMed]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005; 438:803–819. [DOI] [PubMed] [Google Scholar]
- Wang GD, Xie HB, Peng MS, Irwin D, Zhang YP. Domestication genomics: evidence from animals. Annu Rev Anim Biosci 2014; 2:65–84. [DOI] [PubMed] [Google Scholar]
- Wayne RK, Ostrander EA. Lessons learned from the dog genome. Trends Genet 2007; 23:557–567. [DOI] [PubMed] [Google Scholar]
- Freedman AH, Gronau I, Schweizer RM, et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet 2014; 10:e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang JF, Kluetsch C, Zou XJ, et al. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol 2009; 26:2849–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Ersmark E, Palkopoulou E, Dalen L. Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Curr Biol 2015; 25:1515–1519. [DOI] [PubMed] [Google Scholar]
- Skoglund P, Gotherstrom A, Jakobsson M. Estimation of population divergence times from non-overlapping genomic sequences: examples from dogs and wolves. Mol Biol Evol 2011; 28:1505–1517. [DOI] [PubMed] [Google Scholar]
- Wang GD, Zhai W, Yang HC, et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun 2013; 4:1860. [DOI] [PubMed] [Google Scholar]
- Larson G, Karlsson EK, Perri A, et al. Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proc Natl Acad Sci USA 2012; 109:8878–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science 2002; 298:1610–1613. [DOI] [PubMed] [Google Scholar]
- Thalmann O, Shapiro B, Cui P, et al. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science 2013; 342:871–874. [DOI] [PubMed] [Google Scholar]
- Vonholdt BM, Pollinger JP, Lohmueller KE, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 2010; 464:898–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Hellmann I, Jensen JD, Nielsen R. Population genetic inference from genomic sequence variation. Genome Res 2010; 20:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crapon de Caprona M-D, Savolainen P. Extensive phenotypic diversity among South Chinese dogs. ISRN Evol Biol 2013; 2013:1–8. [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol 2005; 28:289–301. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet 2006; 2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, Pollinger JP, Earl DA, et al. A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Res 2011; 21:1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 2012; 8:e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature 2009; 461:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature 2011; 475:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Bradley DG. How much is that in dog years? The advent of canine population genomics. PLoS Genet 2014; 10:e1004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci USA 2002; 99:803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GE, Matukumalli LK, Sonstegard TS, Shade LL, Van Tassell CP. Genomic divergences among cattle, dog and human estimated from large-scale alignments of genomic sequences. BMC Genomics 2006; 7:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch PL, Barnosky AD. Late quaternary extinctions: State of the debate. Annu Rev Ecol Evol S 2006; 37:215–250. [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet 2009; 5:e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JP. Genetics and the Social Behavior of the Dog: University of Chicago Press 1965.
- Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. Bayesian inference of ancient human demography from individual genome sequences. Nat Genet 2011; 43:1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian species delimitation using multilocus sequence data. Proc Natl Acad Sci USA 2010; 107:9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Reich D. Toward a new history and geography of human genes informed by ancient DNA. Trends Genet 2014; 30:377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA 2005; 102:15942–15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, et al. Ancient admixture in human history. Genetics 2012; 192:1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MA, Malaspinas AS, Durand EY, Slatkin M. Ancient structure in Africa unlikely to explain Neanderthal and non-African genetic similarity. Mol Biol Evol 2012; 29:2987–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding ZL, Oskarsson M, Ardalan A, et al. Origins of domestic dog in southern East Asia is supported by analysis of Y-chromosome DNA. Heredity 2012; 108:507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann RL, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature 1987; 325:31–36. [DOI] [PubMed] [Google Scholar]
- Riehl S, Zeidi M, Conard NJ. Emergence of agriculture in the foothills of the Zagros Mountains of Iran. Science 2013; 341:65–67. [DOI] [PubMed] [Google Scholar]
- van Asch B, Zhang AB, Oskarsson MC, Klutsch CF, Amorim A, Savolainen P. Pre-Columbian origins of Native American dog breeds, with only limited replacement by European dogs, confirmed by mtDNA analysis. Proc Biol Sci 2013; 280:20131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet 2009; 5:e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli K-P, Pollinger J, Godinho R, et al. Genome-wide evidence reveals that African and Eurasian Golden Jackals are distinct species. Curr Biol 2015; 25:2158–2165. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. Genomic scans for selective sweeps using SNP data. Genome Ges 2005; 15:1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature 1993; 361:31–39. [DOI] [PubMed] [Google Scholar]
- Saltzman B. Dynamical Paleoclimatology, Volume 80: Generalized Theory of Global Climate Change Academic Press; 1 Edition 2001.
- Vigne JD, Carrere I, Briois F, Guilaine J. The early process of mammal domestication in the Near East. Curr Anthropol 2011; 52:S255–S271. [Google Scholar]
- Zeder MA. Pathways to Animal Domestication. In: Gepts P, Famula TR, Bettinger RL, et al, eds. Biodiversity in Agriculture: Domestication, Evolution, and Sustainability. Cambridge: Cambridge Univ Press, 2012:227–259.
- Pinhasi R, Fort J, Ammerman AJ. Tracing the origin and spread of agriculture in Europe. PLoS Biol 2005; 3:e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tedford RH. Dogs: Their Fossil Relatives and Evolutionary History. Columbia University Press, 2008.
- Ovodov ND, Crockford SJ, Kuzmin YV, Higham TF, Hodgins GW, van der Plicht J. A 33,000-year-old incipient dog from the Altai Mountains of Siberia: evidence of the earliest domestication disrupted by the Last Glacial Maximum. PLoS One 2011; 6:e22821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Boyko RH, Boyko CM, et al. Complex population structure in African village dogs and its implications for inferring dog domestication history. Proc Natl Acad Sci USA 2009; 106:13903–13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature 2002; 418:700–707. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 2007; 81:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21:263–265. [DOI] [PubMed] [Google Scholar]
- Sokal R, Michener C. A statistical method for evaluating systematic relationships. Univ Kans Scientif Bull 1958; 38:1409–1438. [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 2007; 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Hwang DG, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci USA 2004; 101:13994–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Rui Li Y, Kidd J, et al. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet 2013; 9:e1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- Lisiecki LE, Raymo ME. A Pliocene-Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography 2005; 20:PA1003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nomenclature of non-breed dogs and morphological features of Chinese indigenous dogs
False positive (FP) and false negative (FN) rates for variant calling
Both ancient origin and true admixture are compatible with the mixed components observed in the structure analysis
Admixture in dingoes and New Guinea singing dogs
TreeMix results with our dataset and the F3/F4 test
Mutation rate estimation
BPP/G-PhoCS results
Estimating the amount of gene flow between dogs and wolves
Basal positioning of the East Asian breeds is robust against gene flow between wolves and dogs.
Serial founder models can generate a gradient in distances from the outgroup observed in the PCA plot and are unlikely to be caused by wolf/dog migration
Genetic evidence for a single origin
Two possible migration routes
Dating admixture times using HAPMIX
Population substructure in wolves between the eastern and western parts of Eurasia
Selected genes and gene ontology (GO) analysis
Supplementary materials and methods
The morphological features of Chinese indigenous dogs
FN and FP as a function of sequenced individuals
Simulated scheme to study the mixed components in the structure analysis.
TreeMix analysis using the combined genotype data
Treemix results with different numbers of migration tracks
BPP/G-PhoCS analysis for the first group analysis
BPP/G-PhoCS analysis for the second group analysis
Parameter estimates under a wide variety of demographic simulations
Robustness of the basal positioning of Southern Chinese Indigenous dogs to mild gene flow between dogs and wolves
Serial founder models can create patterns observed in the PCA analysis
Simulation of the independent history and principal component analysis
Two possible migration routes
Estimated admixture time for the Northern Chinese indigenous dogs
PCA plot including the three wolves sampled from an earlier publication
Sample information and sequencing throughput
Significant F3 tests among the samples
Candidate admixed populations revealed by the F4 test.
Mutation rate along the dog lineage
Divergence time estimates using Markov chain Monte Carlo(MCMC) methods
Simulated histories testing BPP/G-PhoCS
The inferred tree topologies under different migration rates
D test results between wolves and dogs
GO categories for genes identified using SweepFinder (extra categories not presented in the main text).
Gene Ontology analysis for genes identified by combining FST and population diversity ratio