Abstract
Background
Glioblastoma is an aggressive and highly infiltrative brain cancer. Standard surgical resection is guided by enhancement on postcontrast T1-weighted (T1) magnetic resonance imaging (MRI), which is insufficient for delineating surrounding infiltrating tumor.
Objective
To develop imaging biomarkers that delineate areas of tumor infiltration and predict early recurrence in peritumoral tissue. Such markers would enable intensive, yet targeted, surgery and radiotherapy, thereby potentially delaying recurrence and prolonging survival.
Methods
Preoperative multiparametric MRIs (T1, T1-Gad, T2-weighted [T2], T2-fluid-attenuated inversion recovery [FLAIR], diffusion tensor imaging (DTI), and dynamic susceptibility contrast-enhanced [DSC]-MRI) from 31 patients were combined using machine learning methods, thereby creating predictive spatial maps of infiltrated peritumoral tissue. Cross validation was used in the retrospective cohort to achieve generalizable biomarkers. Subsequently, the imaging signatures learned from the retrospective study were used in a replication cohort of 34 new patients. Spatial maps representing likelihood of tumor infiltration and future early recurrence were compared to regions of recurrence on postresection follow-up studies with pathology confirmation.
Results
This technique produced predictions of early recurrence with a mean area under the curve (AUC) of 0.84, sensitivity of 91%, specificity of 93%, and odds ratio estimates of 9.29 (99% CI, 8.95–9.65) for tissue predicted to be heavily infiltrated in the replication study. Regions of tumor recurrence were found to have subtle, yet fairly distinctive multiparametric imaging signatures when analyzed quantitatively by pattern analysis and machine learning.
Conclusion
Visually imperceptible imaging patterns discovered via multiparametric pattern analysis methods were found to estimate the extent of infiltration and location of future tumor recurrence, paving the way for improved targeted treatment.
Keywords: Glioblastoma, Recurrence, Infiltration, Multiparametric, Imaging pattern analysis
Glioblastoma, a grade IV astrocytoma, is the most common and aggressive primary brain tumor with a median survival of 12–15 months after diagnosis.1 Maximal safe surgical resection combined with adjuvant radiotherapy and chemotherapy has been the cornerstone in the management of these high-grade gliomas.2–4 Advanced imaging modalities have improved the precision of radiotherapy through enhanced tumor delineation, whereas conformal radiotherapy and intensity-modulated radiation therapy (IMRT) allow for an increased radiation dose to the tumor bed and surrounding margin while preserving critical brain structures.5,6
Glioblastoma is well known to extend beyond the visible borders of the enhancing tissue on magnetic resonance imaging (MRI),7 which is the primary target of surgical treatment. In particular, stereotactic biopsies have revealed the presence of tumor in regions that were presumed to be either normal or edematous brain on the basis of MRI characteristics.8,9 Inclusion of this peritumoral region within the clinical target volume has been an area of contention when planning postoperative radiotherapy.10,11 This is in large part due to the fact that the risk of radiation toxicity increases with escalating target volume and dose. Nevertheless, dose escalation in the peritumoral region has been shown to confer a survival advantage, despite a higher frequency of white matter abnormalities.12 Meanwhile, mounting evidence in the surgical literature suggests that the aggressive extent of resection, independent of age, is likewise associated with improved survival.3
Thus, the peritumoral region remains a critical problem for both understanding and effective treatment of glioblastoma. An altered imaging characteristic of this region, commonly referred to as peritumoral edema, represents a combination of infiltrating tumor cells13 and vasogenic edema secondary to angiogenic and vascular permeability factors released by adjacent tumor cells,10 where a compromised blood-brain barrier (BBB) allows intravascular proteins and fluid to penetrate into the parenchymal extracellular space. It is difficult to distinguish infiltrating neoplasm from vasogenic edema with standard imaging approaches; therefore, it is important to identify imaging biomarkers that distinguish regions of densely infiltrating glioblastoma from the biophysical response of surrounding brain tissue to inflammatory factors.
Multiparametric MR imaging has been the most informative, noninvasive method for comprehensive characterization of tumor and surrounding brain tissue.14 Various acquisition protocols reflect different properties of tissue; however, no single imaging metric is currently sufficient to delineate areas of nonenhancing tumor infiltration. Pregadolinium and postgadolinium T1-weighted (T1) images contain information about regional angiogenesis and integrity of the BBB in the tumor. T2-weighted (T2) and T2-fluid attenuated inversion recovery (FLAIR) images are helpful for assessing nonenhancing tumor and edema extent15 because they are sensitive to water concentration. Diffusion imaging techniques such as diffusion tensor imaging (DTI) map the diffusion process of water in the brain, affected in part by tumor cell density.16 Dynamic susceptibility contrast-enhanced (DSC) MRI techniques reflect various aspects of perfusion in the brain,17 which provide quantitative measures of regional microvasculature and hemodynamics.18,19 These patterns are variably affected by infiltrating tumor cells, and independently they are not sufficiently specific enough to clearly define areas of tumor infiltration.
In the current study, we test the hypothesis that advanced pattern analysis and machine learning methods applied to multiparametric MRI are able to quantitatively capture subtle and otherwise imperceptible, imaging variations that highlight heterogeneity within edematous peritumoral tissue. Moreover, we test the hypothesis that these methods can provide predictive spatial maps of tumor infiltration and the likeliness of early recurrence. Understanding subtle imaging characteristics of infiltrating glial tumors can lead to more aggressive, yet targeted surgical and radiation treatment strategies that aim to maximize treatment efficacy while maintaining neurological function.
METHODS
Study Setting
Institutional review board approval was obtained for this study. Our research consists of a retrospective cohort and a replication study cohort at the University of Pennsylvania from 2006 to 2012.
Participants
The retrospective cohort included 31 patients who had de novo glioblastoma (World Health Organization grade IV) and were treated at our institution. The inclusion criteria consisted of a preoperative advanced MRI (ie, anatomic, diffusion, and perfusion imaging modalities) and subsequent recurrence. In all subjects, the clinical diagnosis of tumor recurrence was established with histopathologic analysis after repeat resection. The subjects who had a prior tumor, a previous resection, or residual tumor after surgical resection were excluded. Residual tumors were defined as any contrast-enhancing areas identified by the neuroradiologist (M.B., 14 years of experience) on the immediate postoperative MRI. The replication study included an additional 34 patients with de novo glioblastoma who met the aforementioned inclusion and exclusion criteria. All subjects in this analysis received the same treatment, namely gross total resection of enhancing tumor, followed by temozolomide and radiotherapy. The average time to recurrence was 8.04 months (median, 6.5 months).
Quantitative Variables
Preoperative MRI was acquired using a 3 Tesla (3T) scanner (Tim Trio, Siemens Medical Solutions, Erlangen, Germany). T1-weighted (T1), T2-weighted (T2), T2–fluid-attenuated inversion recovery (FLAIR), DTI, and DSC-MRI were captured for all the patients included in this analysis before the surgery (see Figure, Supplemental Digital Content 1). All MRIs were affinely registered intrasubject, smoothed, inhomogeneity corrected, and skull stripped as previously described.20–22 For imaging sequence details, please see Supplemental Digital Content 2.
Tumor Segmentation
To precisely segment the tumor and peritumoral edema, we used state of the art GLISTR software to delineate tumor (enhancing and nonenhancing regions) and edematous regions for which the infiltration maps were analyzed.23 GLISTR software uses an image analysis technique that incorporates probabilistic imaging and biophysical models of tumor growth and appearance to segment brain tumors and their surrounding tissues using 4 MRI modalities (T1, T1-Gad, T2, and T2-FLAIR).
MRI Features
Principal component analysis (PCA) was used to extract all the information conveyed by the DSC-MRI and this information was used to create the model.17 PCA is a standard dimensionality reduction method which was used here to distill the time series of DSC-MRI down to a few parameters that capture the temporal dynamics of blood perfusion (see Figure, Supplemental Digital Content 3).17
The machine-learning model combines information from 4 structural MRI images (T1, T1-Gad, T2, and T2-FLAIR), 4 DTI-derived images (ie, fractional anisotropy [FA], radial diffusivity [RAD], axial diffusivity [AX], and trace [TR]), and perfusion images (PC1-PC5).17 In addition, because relative cerebral blood volume is a commonly used normalized ratio of blood volume, which is calculated based on an estimate of the arterial input function, we used rCBV as an additional feature.
Infiltration Model
A multidimensional pattern classification method called support vector machines (SVMs) was used to create the infiltration tissue pattern classifier. To magnify the heterogeneity within peritumoral edema, 2 extremities were selected to train the model. Near extremity was defined as the area immediately adjacent to the enhancing tumor while the distal edge of the edema was designated as the far extremity (see Figure, Supplemental Digital Content 4). These 2 regions served as reference examples for near-tumor and far-from-tumor tissue based on the expectation that they are likely to have relatively higher and lower infiltration, respectively.8,9 The classifier was trained on the labeled voxels using libSVM24 with a Gaussian kernel function, weighting to balance the classes and parameters C and sigma in a standard SVM model based on a leave-one-subject-out cross-validated grid search on the population.
In our modeling, this voxel-wise map signifies spatial pseudo-probability of infiltration that was named infiltration index. It may be noted that the infiltration index has a value between 0 and 1 representing noninfiltrated and infiltrated tissue, respectively.
Retrospective Cohort Study
Leave-one-subject-out cross-validation was used in the retrospective study. Specifically, to produce the infiltration index map for 1 patient, the data from that patient were put aside, and the classifier was trained using the other 30 patients in the retrospective cohort study, which tested the remaining patient. This process was repeated 31 times, and each time a different patient was left out. This cross-validation process is critical for providing realistic estimates of how well the predictive model is likely to generalize to new patients.
Replication Study
The classifier was trained on 31 patients who participated in the retrospective cohort study. The trained classifier was used to create infiltration maps in 34 new patients who participated in the replication study.
Statistical Methods
The evaluation was performed by comparing the created infiltration map to 2 regions of interest (ROIs): recurring and nonrecurring. Recurring tissue ROIs were manually drawn by experts (H.A., L.M., M.B., R.L.W.) with the intention of selecting a small region on the preoperative MRI from which the recurrence originated (recurrence being estimated from the follow-up MRI). Preoperative mass effect, resection, and inexact registration between preoperative and follow-up recurrence scans made it nearly impossible to precisely delineate on the preoperative images the follow-up recurrence origin. Therefore, the origin of recurrence was approximately placed by the raters in small regions with relatively high certainty. A 3-mm margin around the manually estimated origin-of-recurrence regions was included into the recurrence ROIs, assuming that they were likely to be heavily infiltrated, but also accounting for registration uncertainties in placing the recurrence ROIs. An additional margin between 3 mm and 10 mm was excluded from any labeling as recurrence or nonrecurrence because of its immediate proximity to actual recurrence, but its nonrecurrence status was deemed to render it unsuitable for evaluation, given that tumor recurrence would be considered likely to progress quickly to its surrounding, likely aggressively infiltrated, tissue. Nonrecurring ROIs were defined as all remaining peritumoral edematous regions between 5 and 20 mm around the tumor core (enhancing tumor plus nonenhancing core). The 20-mm maximum distance of evaluation was used to avoid artificially overestimating the predictive value of our analysis because distant edema is naturally unlikely to recur early; therefore, a model predicting this right would have limited added value over common clinical knowledge. Similarly, the 5-mm margin around the tumor was excluded in part to account for the fact that some peritumoral nonenhancing tissue is typically removed in our institution during surgery, and also in part to account for registration uncertainties in mapping recurrence to preoperative scans. The 5-mm margin was also excluded to avoid overly optimistic evaluations of our predictions because the majority of recurrences occur close to the tumor, which is also where the infiltration index is high by construction, thereby rendering the added value of our predictions limited in that region.
To statistically determine the quality of the infiltration index maps, a receiver operating characteristic (ROC) curve was drawn and the area under the curve (AUC) was calculated. To draw the ROC, sensitivity and specificity were calculated using the labeled maps for each subject and the training ROIs identified by the experts. The odds ratio (OR) was calculated to quantify how strongly the estimated preoperative infiltration maps were associated with subsequent recurrence.
RESULTS
Participants
Model Generation Retrospective Cohort Study
The Table shows the results for the 31 retrospective cohort patients, and Figure 1 shows color-coded tumor infiltration index map for a representative patient. Classification results include a mean AUC of 0.80, sensitivity of 93%, specificity of 88%, accuracy of 87% and recurrence odds ratio estimates of 11.17 (99% CI, 10.71–11.64; P < 0.0001) for tissue predicted to be relatively more infiltrated in the retrospective cohort study. All these results were obtained using cross-validation.
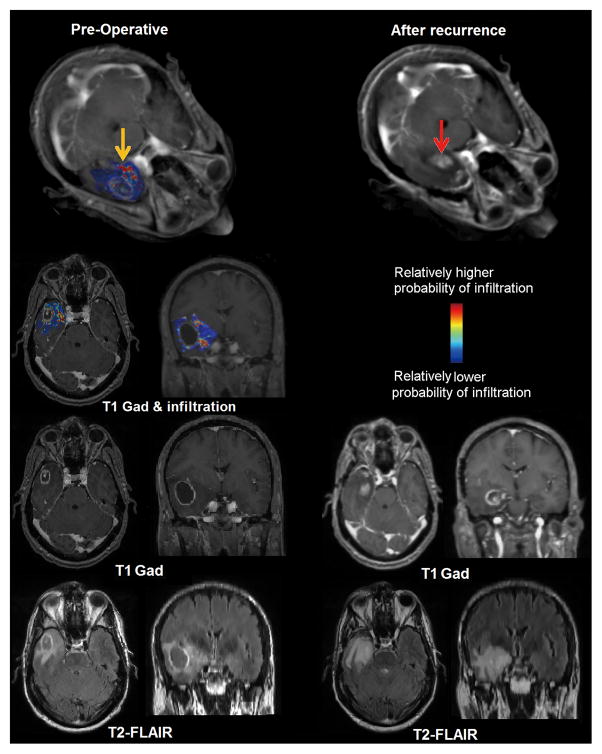
Figure 1. Retrospective study results.
Left panel presents an estimated map for tumor infiltration from preoperative MRI analysis; yellow arrow points to a regions estimated to be relatively more infiltrated. Right panel represents the corresponding MRIs after tumor resection and subsequent recurrence (red arrow) for the same patient. Recurrence occurred in the vicinity of peritumoral tissue originally estimated to be highly infiltrated. The first row represents a three-dimensional rendering of the images. The second, third, and fourth rows show T1-weighted with contrast-fused infiltration map, T1weighted with contrast, and T2-FLAIR respectively.
Replication
After cross-validation, we created a single model using the MRI data of 31 retrospective subjects. This model was utilized to generate the infiltration index maps for 34 new subjects who participated in the replication study. The Table shows the results for this cohort, and Figure 2 shows color-coded tumor infiltration index maps for a representative subject. This method produced predictions with a mean AUC of 0.84, sensitivity of 91%, and specificity of 93%, and recurrence odds ratio estimates of 9.29 (99% CI, 8.95–9.65; P < 0.0001) for tissue predicted to be relatively more infiltrated in the replication study. Figure 3 shows the ROC curve for these subjects.
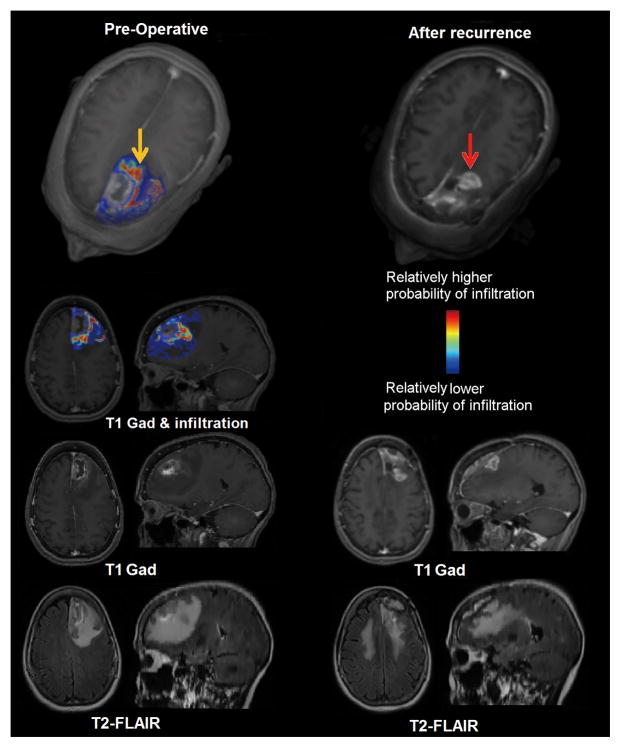
Figure 2. Replication study results.
Left panel presents an estimated map for tumor infiltration from preoperative MRI analysis; yellow arrow points to a region estimated to be relatively more infiltrated. Right panel represents the corresponding MR images after tumor resection and subsequent recurrence (red arrow) for the same patient. Recurrence occurred in the vicinity of peritumoral tissue originally estimated to be highly infiltrated. The first row represents a three-dimensional rendering of the images. The second, third, and fourth rows show T1-weighted with contrast fused infiltration map, T1-weighted with contrast, and T2-FLAIR respectively.
Figure 3. Accuracy Analysis.
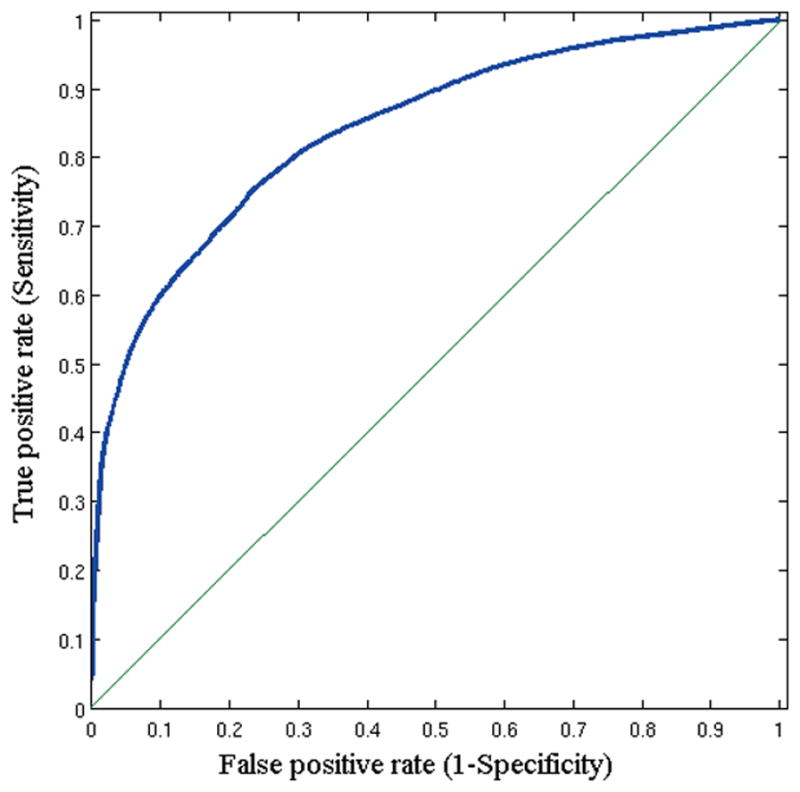
ROC curve for the replication study.
Imaging Features in Recurrence and Nonrecurrence
Five principal components (PCs) captured more than 99% of the variance in the perfusion signal, which quantified all subtleties of these time curves. Each principal component conveys different characteristics of the perfusion signal (see Figure, Supplemental Digital Content 5). MRI signals contained significant discriminating information, which has been organized in Figure 4 into 3 rows in accordance with respective interpretation discussed in the following section (see Figure, Supplemental Digital Content 6). Figure 4 also shows the distribution of our infiltration index in recurring and nonrecurring tissue, indicating markedly greater separation compared to any individual MR signal. The infiltration index is calculated by integrating the top imaging features for both tissue groups to determine how distinctive (separable) the classifier is for each group.
Figure 4. Imaging characteristics of recurrent and nonrecurrent tissue.
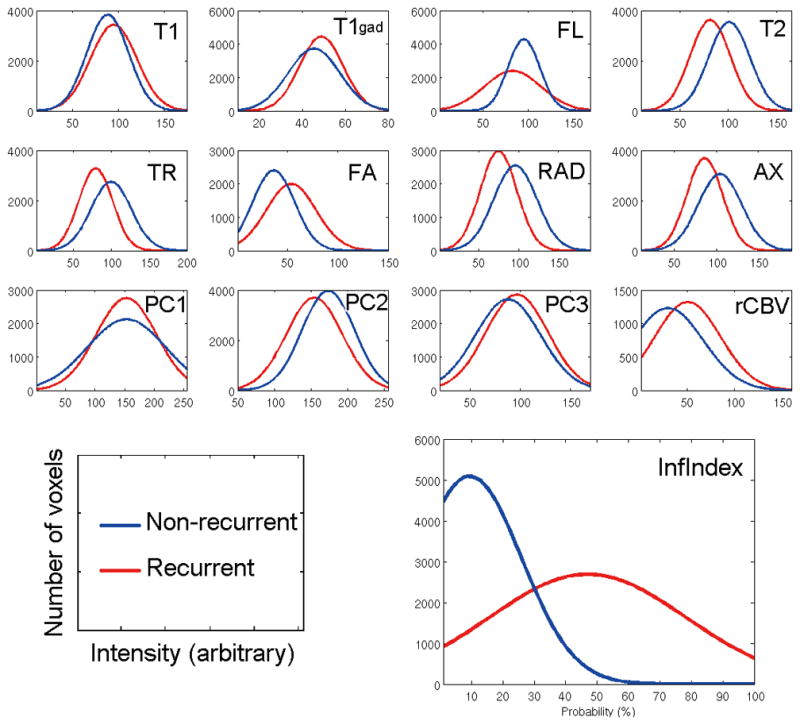
The figures demonstrate the imaging characteristics of the recurrence and nonrecurrence regions within the peritumoral edema on the preoperative MRI. Red represents the probability density function of the recurrence tissues, whereas blue represents the nonrecurrence tissues. T1, T1-weighted (AUC = 0.58); T1gad, T1-weighted contrast-enhanced (AUC,= 0.60); FL, T2–fluid-attenuated inversion recovery (AUC = 0.62); T2, T2-weighted (AUC = 0.76); TR, trace (AUC = 0.73); FA, fractional anisotropy (AUC = 0.73); RAD, radial diffusivity (AUC = 0.74); AX, axial diffusivity (AUC = 0.72); PC1, first principal component (AUC = 0.56); PC2, second principal component (AUC = 0.66); PC3, third principal component (AUC = 0.60); RCBV, relative cerebral blood volume (AUC = 0.72); Inf Index, infiltration index (AUC = 0.89). X-axis shows the intensity in arbitrary unit scaled between 0 and 255 and Y-axis is the number of voxels. The P value of a t-test showed a significant difference between the 2 groups in all modalities (P < .001).
DISCUSSION
Current surgical treatment of glioblastoma is guided largely by enhancement on the postgadolinium T1-weighted MRI, and it is mainly this region that is the focus of resection. Although this practice is known to leave the majority of infiltrating tumor unresected, this is inevitable in the absence of spatially specific knowledge of the infiltration pattern of the tumor. Likewise, the clinical target volume of radiation therapy includes the resection bed plus a variable margin that commonly receives a reduced and largely spatially uniform radiation dose. Although this practice is aimed to address the infiltrative component of glioblastoma, recurrence in this disease is currently certain, and almost always occurs within this peritumoral edematous tissue.11,25 Nonetheless, aggressive use of both treatment modalities has been associated with prolonged survival. Surgical treatment confers a greater survival benefit as the extent of resection increases,26 whereas dose escalation within the peritumoral region similarly leads to longer survival compared to more traditional radiotherapy plans.12 It stands to reason that the efficacy and safety of these therapies could be significantly improved if the spatial extent of the infiltrative component were known and could be specifically targeted.
Key Results and Interpretation
The current study addresses a critical and unresolved need in the field of glioblastoma therapy; specifically, by using advanced analytical methodologies we can quantify subtle, yet important, heterogeneity within the peritumoral region and provide an estimate of the spatial extent and pattern of tumor infiltration. Although we used the current standard-of-care MRI data for our analysis, this information is not apparent when the imaging data are interpreted using conventional means and measurements, but this can be teased out by integrating multiparametric images with an optimized predictive algorithm. These estimated infiltration patterns represent a promising tool for characterizing the spatial pattern of glioblastoma infiltration and pave the way for aggressive, yet targeted, treatment of the infiltrated peritumoral region.
The features used for estimating infiltration fall under 3 categories, each corresponding to a different row in Figure 4. The top row shows conventional imaging features, which suggest that areas of relatively lower water content (ie, lower signal intensity on T2 and FLAIR) have a higher infiltration index. This finding is consistent with the hypothesis that these regions harbor a higher ratio of malignant cells to fluid content. The T1 signal is also higher in those regions, which would be consistent with lower fluid content. Finally, slightly higher T1-Gad implies relatively more compromised BBB in tissue that later recurred, also consistent with infiltrating tumor characteristics. The second row summarizes statistics from features derived from DTI, which reflect lower mean diffusivity and increased fractional anisotropy in regions of higher infiltration. This is expected in areas of higher cellular concentration and is consistent with the literature.27 Radial and axial diffusivity were consistent with overall diffusivity captured by the TR image.
The third set of imaging features relates aspects of tissue vascularization, perfusion, and permeability of blood vessels. Glioblastoma remains the most angiogenic human glioma, therefore exhibiting extensive neovascularization.28 When brain tumors exceed a critical volume, the resultant ischemia triggers the secretion of angiogenic factors. These factors, such as vascular endothelial growth factor (VEGF), promote vascular proliferation leading to the formation and maintenance of tumor vessels.29,30 These new, immature vessels tend to be tortuous and leaky.31 PC2 is inversely related to the magnitude of signal drop in relation to the baseline (see Figure, Supplemental Digital Content 5). Our results indicate a relatively lower PC2 in areas of high infiltration, indicative of a higher degree of BBB compromise and leaky neovasculature. Finally, the relatively higher values of PC3 in these regions were particularly interesting, as it suggested a relative time delay in the contrast agent reaching the highly infiltrated tissue, likely due to higher flow resistance, tortuosity, and other characteristics of tumor vasculature,29,32 which contrast it to healthy brain vasculature known to display extremely low blood flow resistance. In addition to this time delay, the infiltrated tissue was found to have slower and incomplete signal recovery, both indicative of immature, leaky vessels.33 Remarkably, even though each of these MRI signals displayed very subtle differences between recurrence and nonrecurrence, which were not useful for prediction individually, the integration of these complementary signals via machine learning methods provided predictive indices of infiltration and recurrence that can potentially guide personalized treatment.
Limitations
An important limitation of this study is the lack of a histopathological ground truth from the infiltrated, peritumoral areas. Although it is not practical, or sometimes even possible, to finely sample the peritumoral region in routine clinical practice, in our future work we plan to perform targeted biopsies in several of these regions and relate the imaging-based predictions with pathology proven concentration of malignant cells. This model also predicts areas that have a high probability of recurrence, where in fact no recurrence was observed on follow-up imaging. Due to the aggressive and infiltrative nature of glioblastoma, it is quite likely that these areas also harbor a higher concentration of malignant cells that will recur in the future, given enough time. Future work will elaborate how these areas are biologically different.
Finally, it is important to emphasize that the near and far peritumoral tissue ROIs used to train our model does not imply any certainty that there is more infiltration near to as opposed to far from the tumor. These regions should be viewed as part of the algorithmic procedure used to derive the infiltration model, which is then tested against true tumor recurrence. In particular, these regions are used to derive the model that highlights the heterogeneity within peritumoral edema. Although there are certainly other ways of deriving predictive models, our choice was not arbitrary, but it was based on ample literature that generally shows denser malignant cell concentration near the enhancing, solid tumor component.8,9
CONCLUSION
The results of this study demonstrate that new ways of analyzing multiparametric MRI data can elucidate patterns of tumor infiltration within the peritumoral region that predict subsequent tumor recurrence, a task that is virtually impossible based on visual interpretation of standard MRIs. The proposed methodology could offer significant advantages to clinicians treating patients with newly diagnosed glioblastoma. Neurosurgeons could potentially perform supra-total resections when anatomically possible, addressing highly infiltrated peritumoral tissue at higher risk for recurrence. Likewise, radiation oncologists could escalate the radiation dose to high-risk areas. Although our modeling was built specifically for evaluating the infiltrative component of glioblastoma, our methodology is quite general and could be applied to a host of image analysis problems.
Supplementary Material
Table 1. Study results for retrospective (cross-validated) and prospective cohorts;
Statistics are based on positive (recurrence) and negative (non-recurrence) ROIs.
| OR % (CI 99%) | AUC | Accuracy | Sensitivity | Specificity | p value | |
|---|---|---|---|---|---|---|
| Retrospective | 11.17 (10.71–11.64) | 0.80 | 87.11% | 93.55% | 88.06% | < 0.0001 |
| Prospective | 9.29 (8.95–9.65) | 0.84 | 91.25% | 91.18% | 93.48% | < 0.0001 |
OR: Odds ratio; CI: Confidence interval; AUC: Area under the curve.
Footnotes
Disclosures: This work was supported by the National Institutes of Health (NIH) grant R01NS042645. The authors have no personal, financial, or institutional interest in any of the drugs, materials, or devices described in this article.
References
- 1.Yang I, Aghi MK. New advances that enable identification of glioblastoma recurrence. Nature Reviews Clinical Oncology. 2009;6(11):648–657. doi: 10.1038/nrclinonc.2009.150. [DOI] [PubMed] [Google Scholar]
- 2.Brandsma D, Stalpers L, Taal W, Sminia P, van den Bent MJ. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. The lancet oncology. 2008;9(5):453–461. doi: 10.1016/S1470-2045(08)70125-6. [DOI] [PubMed] [Google Scholar]
- 3.Sanai N, Polley M-Y, McDermott MW, Parsa AT, Berger MS. An extent of resection threshold for newly diagnosed glioblastomas: Clinical article. Journal of neurosurgery. 2011;115(1):3–8. doi: 10.3171/2011.2.jns10998. [DOI] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, Van Den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New England Journal of Medicine. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Heesters M, Wijrdeman H, Struikmans H, Witkamp T, Moerland M. Brain tumor delineation based on CT and MR imaging. Implications for radiotherapy treatment planning. Strahlentherapie und Onkologie: Organ der Deutschen Rontgengesellschaft... [et al] 1993;169(12):729–733. [PubMed] [Google Scholar]
- 6.Lorentini S, Amelio D, Giri M, et al. IMRT or 3D-CRT in Glioblastoma? A Dosimetric Criterion for Patient Selection. Technology in cancer research & treatment. 2013;12(5):411–20. doi: 10.7785/tcrt.2012.500341. [DOI] [PubMed] [Google Scholar]
- 7.Konukoglu E, Clatz O, Bondiau P-Y, Delingette H, Ayache N. Extrapolating glioma invasion margin in brain magnetic resonance images: Suggesting new irradiation margins. Medical image analysis. 2010;14(2):111–125. doi: 10.1016/j.media.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Yamahara T, Numa Y, Oishi T, et al. Morphological and flow cytometric analysis of cell infiltration in glioblastoma: a comparison of autopsy brain and neuroimaging. Brain tumor pathology. 2010;27(2):81–87. doi: 10.1007/s10014-010-0275-7. [DOI] [PubMed] [Google Scholar]
- 9.Guo J, Yao C, Chen H, et al. The relationship between Cho/NAA and glioma metabolism: implementation for margin delineation of cerebral gliomas. Acta neurochirurgica. 2012;154(8):1361–1370. doi: 10.1007/s00701-012-1418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang EL, Akyurek S, Avalos T, et al. Evaluation of peritumoral edema in the delineation of radiotherapy clinical target volumes for glioblastoma. International Journal of Radiation Oncology Biology Physics. 2007;68(1):144–150. doi: 10.1016/j.ijrobp.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Oppitz U, Maessen D, Zunterer H, Richter S, Flentje M. 3D-recurrence-patterns of gliobastomas after CT-planned postoperative irradiation. Radiotherapy and oncology. 1999;53(1):53–57. doi: 10.1016/s0167-8140(99)00117-6. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka M, Ino Y, Nakagawa K, Tago M, Todo T. High-dose conformal radiotherapy for supratentorial malignant glioma: a historical comparison. The lancet oncology. 2005;6(12):953–960. doi: 10.1016/S1470-2045(05)70395-8. [DOI] [PubMed] [Google Scholar]
- 13.Barajas RF, Phillips JJ, Parvataneni R, et al. Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR Imaging. Neuro-oncology. 2012;14(7):942–954. doi: 10.1093/neuonc/nos128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galbán CJ, Chenevert TL, Meyer CR, et al. Prospective Analysis of Parametric Response Map–Derived MRI Biomarkers: Identification of Early and Distinct Glioma Response Patterns Not Predicted by Standard Radiographic Assessment. Clinical Cancer Research. 2011;17(14):4751–4760. doi: 10.1158/1078-0432.CCR-10-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurki T, Lundbom N, Valtonen S. Tissue characterisation of intracranial tumours: the value of magnetisation transfer and conventional MRI. Neuroradiology. 1995;37(7):515–521. doi: 10.1007/BF00593707. [DOI] [PubMed] [Google Scholar]
- 16.Lu S, Ahn D, Johnson G, Cha S. Peritumoral diffusion tensor imaging of high-grade gliomas and metastatic brain tumors. American Journal of Neuroradiology. 2003;24(5):937–941. [PMC free article] [PubMed] [Google Scholar]
- 17.Akbari H, Macyszyn L, Da X, et al. Pattern Analysis of Dynamic Susceptibility Contrast-enhanced MR Imaging Demonstrates Peritumoral Tissue Heterogeneity. Radiology. 2014;273(2):502–10. doi: 10.1148/radiol.14132458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wintermark M, Sesay M, Barbier E, et al. Comparative overview of brain perfusion imaging techniques. Journal of neuroradiology. 2005;32(5):294–314. doi: 10.1016/s0150-9861(05)83159-1. [DOI] [PubMed] [Google Scholar]
- 19.Tykocinski ES, Grant RA, Kapoor GS, et al. Use of magnetic perfusion-weighted imaging to determine epidermal growth factor receptor variant III expression in glioblastoma. Neuro-oncology. 2012;14(5):613–623. doi: 10.1093/neuonc/nos073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Medical image analysis. 2001;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 21.Smith SM. Fast robust automated brain extraction. Human brain mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Gooya A, Biros G, Davatzikos C. Deformable registration of glioma images using EM algorithm and diffusion reaction modeling. Medical Imaging, IEEE Transactions on. 2011;30(2):375–390. doi: 10.1109/TMI.2010.2078833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang C-C, Lin C-J. LIBSVM : a library for support vector machines. ACM Transactions on Intelligent Systems and Technology. 2011;2(3):27:21–27:27. [Google Scholar]
- 25.Lee SW, Fraass BA, Marsh LH, et al. Patterns of failure following high-dose 3-D conformal radiotherapy for high-grade astrocytomas: a quantitative dosimetric study. International Journal of Radiation Oncology Biology Physics. 1999;43(1):79–88. doi: 10.1016/s0360-3016(98)00266-1. [DOI] [PubMed] [Google Scholar]
- 26.Senft C, Bink A, Franz K, Vatter H, Gasser T, Seifert V. Intraoperative MRI guidance and extent of resection in glioma surgery: a randomised, controlled trial. The lancet oncology. 2011;12(11):997–1003. doi: 10.1016/S1470-2045(11)70196-6. [DOI] [PubMed] [Google Scholar]
- 27.Gerstner ER, Frosch MP, Batchelor TT. Diffusion magnetic resonance imaging detects pathologically confirmed, nonenhancing tumor progression in a patient with recurrent glioblastoma receiving bevacizumab. Journal of Clinical Oncology. 2010;28(6):e91–e93. doi: 10.1200/JCO.2009.25.0233. [DOI] [PubMed] [Google Scholar]
- 28.Akella NS, Twieg DB, Mikkelsen T, et al. Assessment of brain tumor angiogenesis inhibitors using perfusion magnetic resonance imaging: quality and analysis results of a phase I trial. Journal of Magnetic Resonance Imaging. 2004;20(6):913–922. doi: 10.1002/jmri.20202. [DOI] [PubMed] [Google Scholar]
- 29.Bullitt E, Zeng D, Gerig G, et al. Vessel tortuosity and brain tumor malignancy: a blinded study1. Academic radiology. 2005;12(10):1232–1240. doi: 10.1016/j.acra.2005.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald DM, Choyke PL. Imaging of angiogenesis: from microscope to clinic. Nature medicine. 2003;9(6):713–725. doi: 10.1038/nm0603-713. [DOI] [PubMed] [Google Scholar]
- 31.Thompson G, Mills S, Coope D, O’connor J, Jackson A. Imaging biomarkers of angiogenesis and the microvascular environment in cerebral tumours. British Journal of Radiology. 2011;84(Special Issue 2):S127–S144. doi: 10.1259/bjr/66316279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parikh AH, Smith JK, Ewend MG, Bullitt E. Correlation of MR perfusion imaging and vessel tortuosity parameters in assessment of intracranial neoplasms. Technology in cancer research & treatment. 2004;3(6):585. doi: 10.1177/153303460400300608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldman C, Kim J, Wong W, King V, Brock T, Gillespie G. Epidermal growth factor stimulates vascular endothelial growth factor production by human malignant glioma cells: a model of glioblastoma multiforme pathophysiology. Molecular biology of the cell. 1993;4(1):121. doi: 10.1091/mbc.4.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.