Abstract
Most of the risk loci identified from genome-wide association (GWA) studies do not provide direct information on the biological basis of a disease or on the underlying mechanisms. Recent expression quantitative trait locus (eQTL) association studies have provided information on genetic factors associated with gene expression variation. These eQTLs might contribute to phenotype diversity and disease susceptibility, but interpretation is handicapped by low reproducibility of the expression results. To address this issue, we have generated a set of consensus eQTLs by integrating publicly available data for specific human populations and cell types. Overall, we find over 4000 genes that are involved in high-confidence eQTL relationships. To elucidate the role that eQTLs play in human common diseases, we matched the high-confidence eQTLs to a set of 335 disease risk loci identified from the Wellcome Trust Case Control Consortium GWA study and follow-up studies for 7 human complex trait diseases—bipolar disorder (BD), coronary artery disease (CAD), Crohn's disease (CD), hypertension (HT), rheumatoid arthritis (RA), type 1 diabetes (T1D), and type 2 diabetes (T2D). The results show that the data are consistent with ∼50% of these disease loci arising from an underlying expression change mechanism.
Introduction
A main challenge in interpreting personal genomes is to identify the causal variants underlying human complex traits and their functional consequences. In the past decade, genome-wide association (GWA) studies have successfully identified thousands of genetic variants associated with numerous human complex traits, including diseases. So far, the GWA studies (GWASs) catalog of the National Human Genome Research Institute lists ∼19,200 single-nucleotide polymorphisms (SNPs) associated with one or more complex traits, gathered from ∼2070 GWA studies (www.genome.gov/gwastudies/ [May 2016]). Each of these disease-associated loci must harbor some underlying mechanism whereby the presence of a causal variant alters some molecular-level process and in turn that perturbation affects higher level processes and pathways. Generally, there is little direct evidence on how these variants affect molecular-level processes. A number of different mechanisms may be involved, including altered protein folding, half-life, and function through missense SNPs (Sunyaev et al., 2000; Wang and Moult, 2001), SNPs that affect splicing (Wang and Cooper, 2007), and SNPs affecting RNA expression level (Nicolae et al., 2010). One major source of difficulty in identifying the mechanism is that genetic variants in a locus found to be associated with disease (the markers) are a small part of a larger set, all in linkage disequilibrium (LD) with each other, and any one of these might be causal.
GWA studies have also been used to discover expression quantitative trait loci (eQTLs) by finding correlations between transcript expression levels and the presence of genetic variants (Jansen and Nap, 2001). The emergence of high-throughput technologies, particularly transcription microarrays and RNA sequencing, provides an efficient way to simultaneously measure the expression levels of thousands of genes. Microarray technology has also been used for large-scale genotyping, and comparison of these two types of data then allows eQTL mapping in a large number of individuals (Lappalainen et al., 2013; Liang et al., 2013; Montgomery et al., 2010). Initially, data derived from Epstein–Barr virus-transformed immortalized lymphoblastoid cell lines (LCLs) were used for population-wide eQTL analysis in humans (Dixon et al., 2007; Duan et al., 2008; Stranger et al., 2007). Recently, a number of studies have performed eQTL mapping on various human tissues, such as brain (Gibbs et al., 2010; Myers et al., 2007), liver (Greenawalt et al., 2011; Innocenti et al., 2011; Schadt et al., 2008), adipose (Emilsson et al., 2008; Greenawalt et al., 2011; Nica et al., 2011), fibroblasts (Dimas et al., 2009), and skin (Ding et al., 2010; Grundberg et al., 2012; Nica et al., 2011). Thousands of cis- and trans-regulatory eQTLs have now been discovered in a variety of human tissues and populations.
A complication in relating eQTLs to disease GWASs is the apparent unreliability of individual eQTL studies, arising from a variety of issues in statistical analysis as well as experimental factors. So far, most eQTLs have not been reproducible in multiple studies, even within studies conducted on the same cell types in the same population (Dixon et al., 2007; Göring et al., 2007; Myers et al., 2007; Stranger et al., 2007; Veyrieras et al., 2008). To address this issue, we have integrated human genome-wide eQTL data from 16 publicly available studies to identify higher confidence eQTL relationships on the basis of consensus, both generally and within several specific cell types.
A number of studies have used eQTL association results and disease GWAS findings to improve the functional interpretation of disease-associated loci (Chu et al., 2011; Ertekin-Taner, 2011; Gibson et al., 2015; Heid et al., 2010; Hrdlickova et al., 2011; Hsu et al., 2010; Lango Allen et al., 2010; Li et al., 2015; Moffatt et al., 2007; Peters et al., 2016; Repnik and Potočnik, 2016; Richards et al., 2012; Schaub et al., 2012; Speliotes et al., 2010; Wu et al., 2012). Several studies have shown that SNPs associated with human traits and chemotherapeutic drug susceptibility are in general enriched for eQTLs (Cookson et al., 2009; Gamazon et al., 2010; Nicolae et al., 2010). Most studies have used eQTL data from the most accessible cell type, LCL, and it is not clear how good a proxy these are for human cells and tissues relevant to nonimmune-related disease, such as psychiatric traits or cancers (Choy et al., 2008; Nicolae et al., 2010). Some studies have used eQTL results from tissues partially appropriate to the disease of interest when linking to disease-associated SNPs (Ding et al., 2010; Fransen et al., 2010; Innocenti et al., 2011; Kang et al., 2012a, 2012b; Liu et al., 2011; Maranville et al., 2011; Schadt et al., 2008; Zhong et al., 2010). For example, Ding et al. (2010) reported an eQTL study of human skin that aimed to elucidate the role of regulation of gene expression in psoriasis. Richards et al. (2012) assigned eQTL status to schizophrenia susceptibility alleles based on eQTL data derived from the adult human brain.
In principle, it is possible to find which disease-associated loci harbor an underlying expression mechanism by comparing the set of markers from a disease GWAS with the set of markers from an eQTL study: if the cause of disease risk is a change in expression discovered in an eQTL, the two sets of markers should overlap or be in LD. The comparison is complicated by the sparse sampling of the full SNP set provided by microarrays. When full genotyping information is available for sampled SNPs, imputation methods (Howie et al., 2009) may be used to obtain estimated association p values for many SNPs not directly measured, potentially addressing this issue. Often these data are not readily available, and an alternative approach is required. We made use of one set of disease GWAS data with complete genotype information to investigate the properties of full marker distributions and, on that basis, devised a method that can be applied to cases where only microarray marker SNP information is available.
In this study, we sought to identify which loci associated with complex trait disease may harbor an underlying expression mechanism, making use of a set of consensus eQTLs. To this end, we examined each of a set of disease-associated loci to ascertain whether any known eQTL relationship may have produced the disease association data.
Materials and Methods
Data sources
All eQTL association data in this study were collected from 16 publicly available studies that had been performed on various human tissues and populations, listed in Table 1.
Table 1.
Expression Quantitative Trait Locus Data for the 16 Selected Genome-Wide Expression Quantitative Trait Locus Association Studies
| Study ID | Samples (size) | Cell type | eQTL associations | exSNPs | exGenes | References |
|---|---|---|---|---|---|---|
| HA | HapMap CEU—Caucasians (30) | LCL | 3858 | 3686 | 239 | Stranger et al. (2007) |
| HapMap CHB (45) | LCL | 4066 | 3780 | 253 | ||
| HapMap JPT (45) | LCL | 5254 | 5061 | 274 | ||
| HapMap YRI–Africans (30) | LCL | 3524 | 3283 | 306 | ||
| BR | Caucasians (193) | Brain Cortex | 624 | 545 | 209 | Myers et al. (2007) |
| AS | Childhood asthma (206) | LCL | 21116 | 12121 | 2632 | Dixon et al. (2007) |
| LV | Caucasian liver donors (427) | Liver cell | 4362 | 2527 | 3824 | Schadt et al. (2008) |
| HA2 | 30 HapMap CEU—Caucasians (30) | LCL | 4453 | 3699 | 722 | Duan et al. (2008) |
| 30 HapMap YRI (30) | LCL | 5027 | 4086 | 1659 | ||
| 3C | Caucasians (75) | LCL | 554 | 544 | 436 | Dimas et al. (2009) |
| Caucasians (75) | Fibroblast | 522 | 508 | 424 | ||
| Caucasians (75) | T cell | 546 | 540 | 429 | ||
| MO | German (1490) | Monocyte | 37694 | 29948 | 2752 | Zeller et al. (2010) |
| HRC | HapMap CEU–Caucasians (60) | LCL | 8908 | 3896 | 930 | Montgomery et al. (2010) |
| HRY | HapMap YRI–Africans (69) | LCL | 799 | 779 | 786 | Pickrell et al. (2010) |
| BR2 | Caucasians (150) | Cerebellum | 5243 | 4399 | 317 | Gibbs et al. (2010) |
| Caucasians (150) | Frontal cortex | 5512 | 5198 | 329 | ||
| Caucasians (150) | Temporal cortex | 5335 | 4059 | 385 | ||
| Caucasians (150) | Pons | 3411 | 3284 | 275 | ||
| SKN | Healthy skin individuals (57) | Skin | 5410 | 4782 | 222 | Ding et al. (2010) |
| LV2 | Liver donors (266) | Liver cell | 1170 | 1161 | 1170 | Innocenti et al. (2011) |
| IM | British (288) | Monocyte | 33740 | 28956 | 6063 | Fairfax et al. (2012) |
| British (288) | B cell | 22453 | 20333 | 5449 | ||
| MuTHER | Caucasian female twins (∼160) | LCL | 211977 | 149684 | 3945 | Grundberg et al. (2012) |
| Caucasian female twins (∼160) | Skin | 103537 | 82933 | 2495 | ||
| Caucasian female twins (∼160) | Adipose | 138885 | 109689 | 3136 | ||
| MRC | MRCA (405) and MRCE (950) | LCL | 176848 | 109763 | 1251 | Liang et al. (2013) |
| E-GEUV | 1000 Genomes—EUR (373) | LCL | 390813 | 281446 | 3048 | Lappalainen et al. (2013) |
| 1000 Genomes—YRI–Africans (89) | LCL | 19314 | 16932 | 472 |
eQTL, expression quantitative trait locus; LCL, lymphoblastoid cell line.
Data preparation
To compare exSNP-exGene association pairs between these studies, all transcript names, probe IDs, and alias gene names were converted to current unique Entrez gene IDs and gene names (NCBI build 37.2). Ambiguities in alias gene names were resolved using chromosome location information. Transcript clusters (TCs) identified in the HA2 study were converted to Entrez gene IDs by mapping the region of each TC to gene ranges on human genome assembly hg19. Retired and discontinued SNP IDs were filtered out and all SNP IDs were converted to the current dbSNP IDs (dbSNP build 134). Retired or unmappable gene names were also eliminated from the study. Any SNP with multiple chromosome coordinates on NCBI reference assembly 37.2 (dbSNP b134) was removed.
Linkage disequilibrium
LD information between pairs of SNPs was acquired from the HapMap project phase III (release 27) (The International HapMap 3 Consortium, 2010) or derived from the 1000 Genomes Project (phase1 release) (The 1000 Genomes Project Consortium, 2010) for several ethnic populations (CEU, YRI, CHB, and JPT for HapMap; EUR and YRI for YRI). For 1000 Genomes LD data, the r2 values for pairs of SNPs with minor allele frequencies (MAFs) >5% and located within 200,000 bp of each other were calculated using PLINK (v. 1.07) (Purcell et al., 2007). Spearmen correlation between LD values from the HapMap project and 1000 Genomes is 0.89.
Where both HapMap and 1000 Genomes provided LD values for an SNP pair, the HapMap value was used. Where possible, appropriate population LD data were used for each dataset. HA_CEU, HA2_CEU, HRC, AS, BR, LV, 3C, BR, and BR2 datasets are from Caucasian (CEU) populations and HA_YRI, HA2_YRI, and HRY datasets are from Yoruba (YRI) populations. HA_CHB and HA_JPT datasets are for Chinese (CHB) and Japanese (JPT) populations, respectively. No clear ethnic identity is available for the MO and LV2 sets. For the LV2 dataset, individuals are mostly from the mixture of Caucasian and African populations and so an intersection LD set for CEU and YRI populations was used. For the MO study, we generated an intersection of LD sets among all four populations, CEU, CHB, JPT, and YRI.
Hierarchical clustering
The distance between each pair of datasets was defined as (1-f), where f is the fraction of common exGenes between the two sets. The hclust module in R was used.
High-confidence eQTL data
Figure 1 summarizes the procedure used to identify high-confidence eQTLs based on consensus within the included 16 independent human genome-wide eQTL studies. For disease analysis, a high-confidence eQTL relationship is defined as one that is identified in at least two studies of these 16. The number of high-confidence eQTL relationships so defined varies with the LD criterion used. For the disease analysis, the most conservative LD level (r2 > 0.8) was used, providing a total of 4252 unique genes with an expression level associated with the presence of at least one high-confidence eQTL SNP.
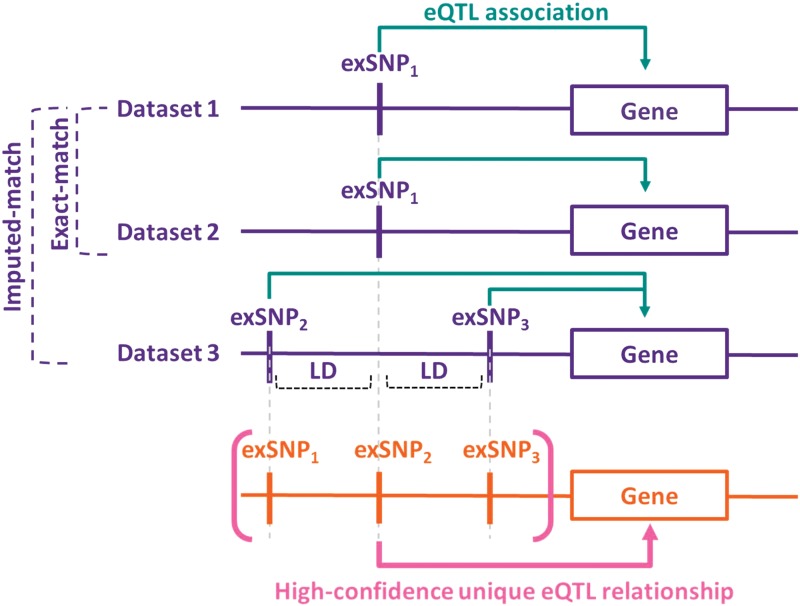
FIG. 1.
Identification of high-confidence unique eQTL relationships. A high-confidence eQTL relationship is defined as one found in two or more datasets. This figure illustrates the two ways, exact-match or imputed-match, used to determine consensus associations. Exact-match: In Dataset 1, the presence of an exSNP1 is associated with altered expression of the gene. Dataset 2 contains the exact same SNP-gene association, sufficient to classify the association as high confidence. Imputed-match: Dataset 3 has an association between two other SNPs, exSNP2 and exSNP3, and the expression level of the same gene. These SNPs are both in LD with exSNP1, so are considered to represent the same underlying relationship. eQTL, expression quantitative trait locus; LD, linkage disequilibrium; SNP, single-nucleotide polymorphism.
GWA studies of human common diseases
Loci significantly associated with disease susceptibility for seven specific human common diseases (bipolar disorder [BD], coronary artery disease [CAD], Crohn's disease [CD], hypertension [HT], rheumatoid arthritis [RA], type 1 diabetes [T1D], and type 2 diabetes [T2D]) were collected from the Wellcome Trust Case Control Consortium (WTCCC1) GWA study (The Wellcome Trust Case Control Consortium, 2007) and from other related meta-analyses and follow-up studies in the GWAS catalog (www.genome.gov/gwastudies/ [March 2013]).
CentiMorgan distance calculation
The genetic map data of all human chromosomes, calculated from HapMap II data with LDhat (ldhat.sourceforge.net/instructions.shtml), were acquired from NCBI FTP (ftp.ncbi.nlm.nih.gov/hapmap/recombination/2011-01_phaseII_B37/). Where necessary, the centiMorgan (cM) coordinates of disease-associated marker SNPs and expression-associated eQTLs were interpolated from those of the closest SNPs with defined cM values based on chromosomal distance.
Comparison of disease and eQTL markers
The procedure used to estimate whether or not the detected disease and eQTL markers in a locus arise from the same underlying causal variant is illustrated in Figure 2. We used complete genotype data for the WTCCC1 study of seven complex trait diseases (The Wellcome Trust Case Control Consortium, 2007) to examine the relationship between disease association p value distributions and eQTL markers. Complete microarray genotype data were downloaded for the WTCCC1 study. The probabilities of each genotype for the SNPs in each disease locus not represented on the microarray were imputed using IMPUTE2 (Howie et al., 2009) and the disease association p value of each SNP was then calculated using SNPTEST (Ferreira and Marchini, 2011). Imputed disease association p value distributions were compared with marker SNPs for high-confidence eQTL relationships derived from the 16 eQTL studies (AllCell_AllPop).
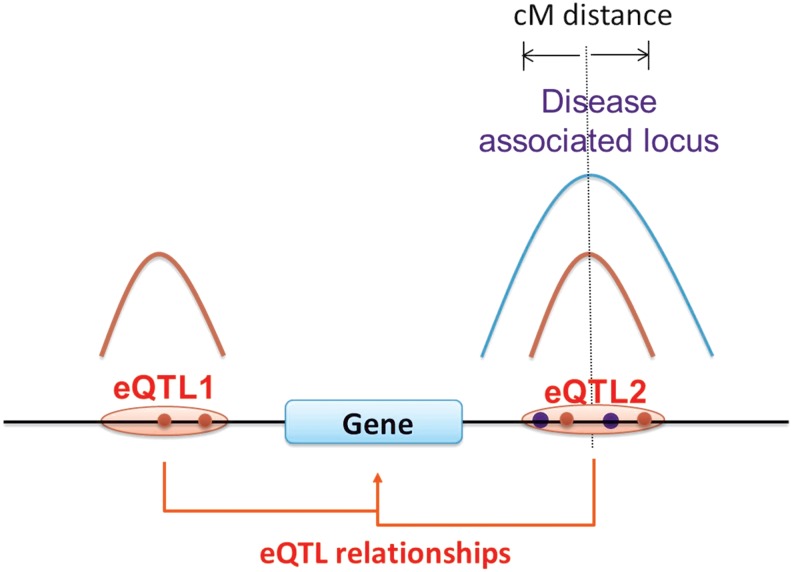
FIG. 2.
Model for identifying those disease-associated loci with a probable underlying expression mechanism. In this hypothetical case, a causal variant, at the position of the vertical dotted line, is related to disease susceptibility as a result of altering the expression level of the nearby gene. Because of LD, the presence of the causal variant will usually result in one or more nearby SNPs also being associated with disease risk, and the blue curve represents the expected p value distribution of these. Sparse sampling with a microarray and noise factors result in only one or a few of these associations being detected (blue dots). Since the causal variant affects expression, the same SNPs will be associated with expression level of the gene, with a colocated expected p value distribution, represented by the red curve, and again because of noise and other factors, only some markers will be identified (red dots). In this example, there is another eQTL in this region (eQTL1) where SNPs are associated with the expression level of the same gene, but unrelated to disease susceptibility, and so its eQTL p value distribution does not overlap with that for disease association.
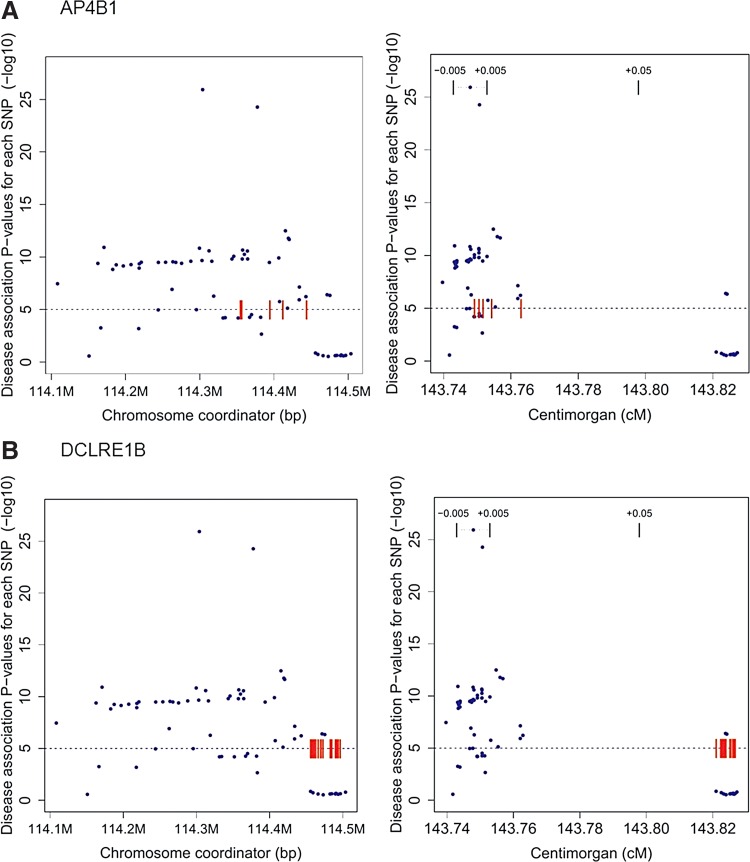
Figure 3 shows Manhattan plots of these data for one region, where SNPs are significantly associated with the risk of T1D in the WTCCC1 study, and that also contains eQTL associations. The left-hand plots show the distribution of disease association p values and the location of the expression marker SNPs as a function of the chromosome coordinate in base-pair units. In plots of this type, it is often not possible to determine whether or not the disease and expression signals share a causal variant. The right-hand plot shows the same data as a function of crossover event probability measured in cM. The cM scale provides a clear distinction between a situation where the underlying causal variant for the disease and expression signals is the same (AP4B1, Fig. 3A) and where they are different (DCLRE1B, Fig. 3B). In (A), significant p value disease-associated SNPs overlap with the eQTL marker SNPs. In (B), there is a 0.06 cM separation between the eQTL marker SNPs, the closest significant disease-associated SNP.
FIG. 3.
Manhattan plots for a locus associated with type 1 diabetes in the WTCCC1 data. These plots show the relationship between disease association p value for all SNPs in the region (blue points) and the location of high-confidence expression-associated SNPs (red dashes). There are two separate high-confidence eQTL relationships in this region, each involving a different gene. The horizontal dotted line indicates the significance threshold for disease p values (1E-05). The left plots show the p value distribution of disease and expression SNPs as a function of chromosome coordinates and the right plots show the same data as a function of genetic map position, in cM. (A) Disease associations and high-confidence eQTL SNPs associated with the expression level of AP4B1 (adaptor-related protein complex 4, beta 1 subunit). In chromosome coordinates (left), the disease markers appear widely spread and there is no clear distinction between these and eQTL markers. On the cM scale (right), it is clear that the disease marker SNPs and eQTL SNPs occupy the same narrow range in the crossover coordinate. (B) High-confidence eQTL SNPs associated with DCLRE1B (DNA cross-link repair 1B) in the same locus. In chromosome coordinates (left), it is unclear whether these markers overlap with the disease markers or not. On the cM scale (right), there is clear separation between expression and disease markers, reflecting low linkage disequilibrium between the two sets of markers so that it is unlikely the same causal variant generates both signals. Together, these plots show that the data are consistent with a disease susceptibility causal variant affecting the expression of AP4B1 and inconsistent with an expression effect on DCLRE1B.
For the other 16 of the 21 WTCCC1 loci that contain at least one high-confidence eQTL relationship, 14 have eQTL markers for at least one gene that overlap with the disease marker SNPs (data not shown). There are often multiple genes in a locus, so the Manhattan plots show a wide variety of situations, but consistently, where there is overlap, the shortest distance between a disease marker and an eQTL marker is <0.05 cM, and in no case without overlap is there a distance <0.05. On that basis, we adopted three thresholds for confidence that the disease and expression signals arise from a common underlying variant: when there is an exact match between a disease marker and an expression marker (i.e., these are the same SNP), when the closest disease and expression markers are with 0.005 cM, and when the two closest markers are within 0.05 cM.
Results
Genome-wide eQTL data
Table 1 summarizes the 16 publicly available genome-wide eQTL studies used, categorized into 29 datasets by tissue and population. The majority of studies were performed on LCLs, and 10 datasets are from that source. Most studies used a combination of genotyping microarrays and transcription microarrays. Three studies, HRC (Montgomery et al., 2010), HRY (Pickrell et al., 2010), and E-GEUV (Lappalainen et al., 2013), all on LCLs, used RNA sequencing technology rather than the older microarray technology to determine expression levels. One study, E-GEUV, used the 1000 Genomes Project populations (EUR and YRI) and so was able to include the genotypes of all SNPs down to about a frequency of 1% instead of the limited number represented on a genotyping microarray.
We define exSNPs as those SNPs that correlate with change of expression of one or more genes. The corresponding genes are referred to as exGenes, and an eQTL association represents the relationships between one exSNP and its associated exGene. After processing the raw data from the 16 studies, there are totally 796,908 unique eQTL associations covering 15,170 unique exGenes and 548,344 unique exSNPs. The number of eQTL associations varies widely across studies (522–390,813). Variation in population sample size is probably the biggest factor in this spread (sample sizes range from 30 to 1490). The expression level of most exGenes is associated with the presence of multiple exSNPs, primarily as a result of LD, and in most cases, only a single variant is likely actually causative of a change in expression.
As is common practice, we consider cis-eQTL associations to be those where the exSNPs are located within 1 Mb of either the 5′ or 3′ end of the associated exGene. eQTL associations between an exGene and an exSNP located more than 1 Mb distance away from the gene region are referred to as trans-eQTL associations. Supplementary Figure S1 shows the proportion of cis- and trans-eQTLs in each dataset. Most datasets have a much higher fraction (>60%) of cis-eQTLs. The predominance of cis-eQTLs is largely a consequence of the increased statistical power obtained by limiting the genome window in which associations are examined, thereby greatly reducing the size of multitesting correction needed in assessing the significance of an association. Supplementary Figure S2 shows the distribution of distances between exSNP-exGene pairs. The density falls off rapidly with distance, and 85% of cis-regulatory exSNPs are within 200 kb of the corresponding exGene. cis-eQTLs are approximately symmetrically distributed both upstream and downstream of the corresponding exGene, as well as within the gene. About 25% of cis-regulatory exSNPs fall within a gene region and were assigned a distance of zero. Although LD broadens this distribution, it is still apparent that the majority of SNPs involved in cis-eQTL relationships are located in the vicinity of the affected gene, including the 5′ and 3′ untranslated regions, and neighboring upstream and downstream regions. Because of LD, it is difficult to determine the exact location of the underlying causal variants that directly affect gene expression.
LD relationships between eQTLs
We presume that the underlying mechanistic origin of a cis-eQTL relationship is that a particular SNP or other variant falls on a functional element such as a transcription factor binding site, a microRNA binding site, or splice site where the change leads to nonsense-mediated decay. Then, an association study will reveal a statistical relationship between the presence of that causal SNP and the level of expression of the gene. In principle, it might be possible to identify which of the set of such SNPs is causal from the strength of the correlation between its presence and the level of gene expression. In practice, LD is often close to 1 for a number of neighboring SNPs, and the data are usually noisy, so it is not possible to make such a determination. Furthermore, because of low sampling of SNPs using typical microarray genotyping technology, it is unlikely the causal SNP will itself be assayed. In spite of these limitations, it is usually possible to group exSNPs into LD blocks and approximately identify the number of unique causal relationships—each block will usually represent one relationship.
Table 2 shows the number of total eQTL associations and the corresponding number of unique exSNPs and unique exGenes in each dataset. It also shows the number of unique eQTL relationships, each of which represent a set of LD-related exSNPs associated with the same exGene, at three LD thresholds, r2 > 0.8, 0.5, and 0.3. In this study, each eQTL relationship likely represents one mechanistic relationship between the presence of a causal variant and the expression level of the gene. The proportion of exGenes with a single eQTL relationship ranges from 54% to 100% using an LD threshold of 0.8 to 72–100% at a threshold of 0.3.
Table 2.
Expression Quantitative Trait Locus Associations and Unique Expression Quantitative Trait Locus Relationships for Each Dataset
| Dataset | Unique eQTL associations | Unique exGenes | Unique exSNPs | Unique eQTL relationships (r2 ≥ 0.8) | Unique eQTL relationships (r2 ≥ 0.5) | Unique eQTL relationships (r2 ≥ 0.3) |
|---|---|---|---|---|---|---|
| HRC | 4362 | 930 | 3896 | 1453 | 1116 | 1038 |
| HA_CEU | 3787 | 239 | 3686 | 451 | 286 | 252 |
| HA2_CEU | 4163 | 722 | 3699 | 1273 | 1166 | 1141 |
| EGEUV_EUR | 390696 | 3048 | 281446 | 135826 | 103879 | 88142 |
| HRY | 794 | 786 | 779 | 794 | 792 | 790 |
| HA_YRI | 3419 | 306 | 3283 | 619 | 372 | 336 |
| HA2_YRI | 5027 | 1659 | 4086 | 3007 | 2835 | 2813 |
| EGEUV_YRI | 19314 | 472 | 16932 | 9349 | 6887 | 5709 |
| HA_CHB | 3930 | 253 | 3780 | 453 | 293 | 265 |
| HA_JPT | 5165 | 274 | 5061 | 481 | 317 | 290 |
| AS | 14348 | 2632 | 12121 | 6596 | 4178 | 3328 |
| MRC | 119958 | 1251 | 109763 | 17019 | 10894 | 8959 |
| 3CL | 554 | 436 | 544 | 531 | 494 | 469 |
| 3CF | 522 | 424 | 508 | 501 | 462 | 443 |
| 3CT | 546 | 429 | 540 | 525 | 475 | 462 |
| MuTHER_Fat | 128181 | 3136 | 109689 | 19704 | 9056 | 5367 |
| MuTHER_LCL | 189983 | 3945 | 149684 | 28861 | 12913 | 7379 |
| MuTHER_Skin | 96412 | 2495 | 82933 | 14236 | 6471 | 3883 |
| SKN | 4916 | 222 | 4782 | 384 | 243 | 227 |
| MO | 37580 | 2752 | 29948 | 29690 | 23130 | 17598 |
| IM_MO | 31914 | 6063 | 28956 | 27794 | 23695 | 19929 |
| IM_B | 21674 | 5449 | 20333 | 19244 | 16665 | 14361 |
| LV | 4171 | 3824 | 2527 | 4145 | 4126 | 4117 |
| LV2 | 1170 | 1170 | 1161 | 1170 | 1170 | 1170 |
| BR | 624 | 209 | 545 | 358 | 323 | 315 |
| BR2_Cer | 5241 | 317 | 4399 | 572 | 374 | 344 |
| BR2_FC | 5429 | 329 | 5198 | 625 | 381 | 347 |
| BR2_TC | 5280 | 385 | 4059 | 681 | 441 | 409 |
| BR2_P | 3389 | 275 | 3284 | 475 | 312 | 285 |
| Total | 808512 | 15170 | 578094 | 249346 | 177435 | 142019 |
Pair-wise comparisons show low agreement between eQTL datasets
To investigate how often the same eQTL relationships are found in different studies, we compared the eQTL associations between each pair of datasets and identified the common exGenes and exSNPs that are associated with these. Supplementary Table S1 summarizes the level of agreement among the 16 different eQTL datasets. In general, the agreement of most (92%) pair-wise comparisons between datasets is low, with only 4–49% of exGenes shared between datasets. There is also a low level of agreement between exSNP-exGene relationships.
Some differences between eQTL studies presumably arise from different biology as a function of cell type and population. However, the fractions of common exGenes for studies on the same population and cell type are also often low. For example, the fractions of common exGenes among studies performed in LCLs for Caucasian populations (HRC, HA_CEU, HA2_CEU, and EGEUV_EUR) are usually not high (8–27%), with one exception at 57%. With a couple of exceptions, studies on the same cell line, but different populations, also have agreements of 7–35%. Agreement for studies in different cell types from the same population tends to be a little higher, but is still low. The MuTHER study (Grundberg et al., 2012; Nica et al., 2011) used adipose, LCL, and skin in a Caucasian population. In this study, levels of agreements are high (54–60%).
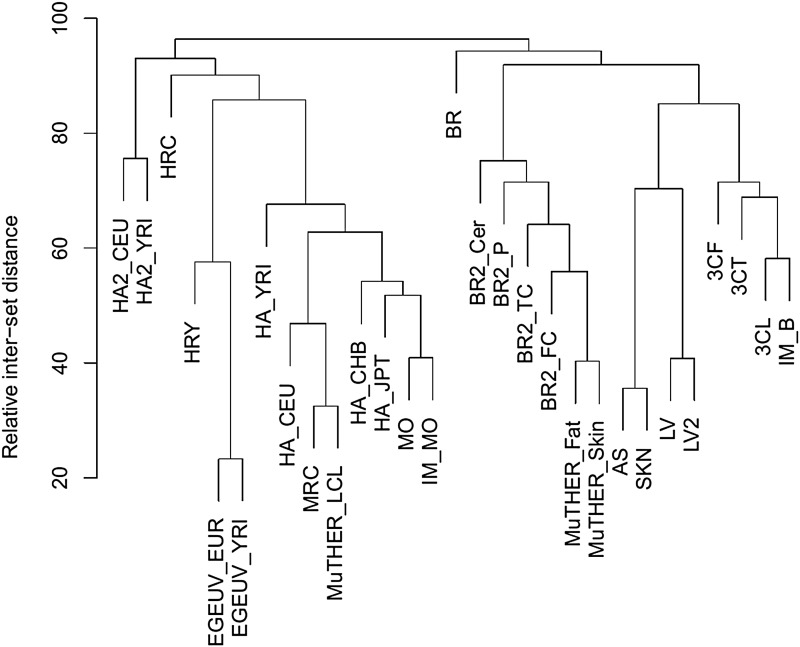
Figure 4 shows a hierarchical clustering comparison of the datasets based on the fraction of common exGenes. Two factors dominate the tree topology—cell type and specific study. Most of the datasets that used LCLs are grouped in one major branch, and studies that used monocytes or liver are also grouped. Datasets from the same study are usually grouped together, for example, the 3C study, the BR2 study, and the MuTHER study (excepting MuTHER_LCL, which is in the LCL group).
FIG. 4.
Hierarchical clustering of the fraction of common exGenes between pairs of eQTL datasets. Distance scale is based on the percentage of common exGenes between pairs of datasets.
High-confidence eQTLs
Given the high level of variability between the studies apparently due to nonbiological causes, it is desirable to identify the more reliable eQTL relationships. For this purpose, we compiled eQTL relationships that have been observed in at least two studies, for studies within the same population, studies on the same cell types, and across all studies, independent of population and cell type. In all, there are 13 subsets (Supplementary Table S2).
A high-confidence eQTL relationship is defined as one for which supporting eQTLs are found in more than one study within an integrated set. We identified high-confidence unique eQTL relationships within the eight integrated sets that contain more than one study. The number of studies in which a particular eQTL is found provides an approximate confidence score. Supplementary Table S3 shows the number of unique eQTL relationships and high-confidence unique eQTL relationships in each integrated set at various LD levels. For the largest integrated set, AllCell_AllPop, including all the data, at the lowest LD threshold (r2 > 0.3), the 133,658 unique eQTL relationships result in 5928 high-confidence unique eQTL relationships involving a total of 4252 exGenes (HC-exGenes). In general, most exGenes (77%) contain only one high-confidence unique eQTL relationship in each integrated set at the lowest LD level (r2 > 0.3) (data not shown).
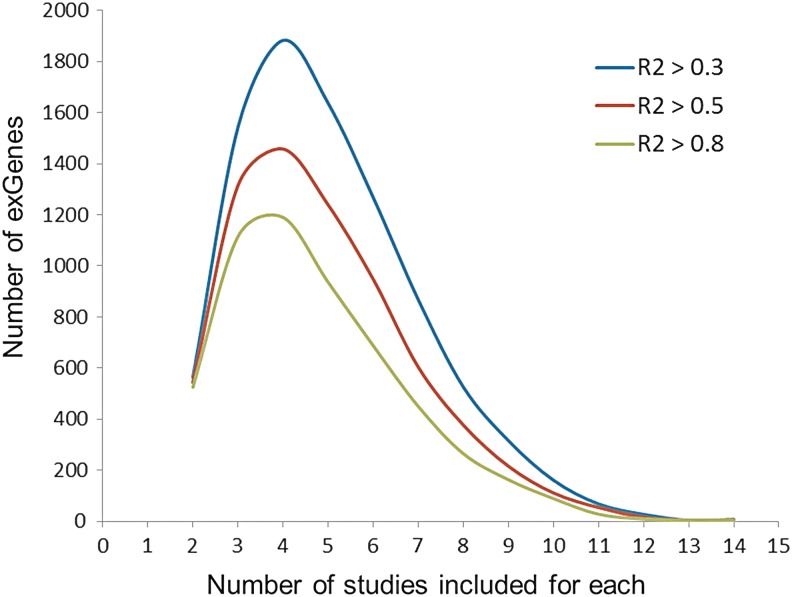
Figure 5 shows the distribution of the number of studies in which each high-confidence exGene is identified, at various LD levels, for the AllCell_AllPop integrated set. Most HC-exGenes appear in more than the minimum of two studies, with four the most common.
FIG. 5.
Number of HC-exGenes with support from 1, 2, 3, … studies at various LD thresholds (R2) in the AllCell_AllPop integrated set.
As an estimate of the relative quality of eQTL datasets, we calculated the fraction of HC-exGenes in each dataset of the LCL_CEU integrated set (Supplementary Fig. S3). This quality measure varies widely. The lowest fraction of HC-exGenes is for the HA2 dataset (6.5%). The MRC dataset has the highest fraction (84%).
Tissue and population dependence of eQTL relationships
We made use of the data for different tissue types included in the 16 eQTL studies to perform limited testing on the extent to which eQTLs are conserved across tissue types. As noted earlier, only a fraction of eQTLs are found in multiple studies even when the same tissue and population have been used, so simply looking at the fraction eQTLs common to studies in different tissues is not an adequate approach. To address this, we restricted the comparisons to situations where there are pairs of studies that share a tissue type, providing a reference level of agreement, and that also have data on other tissues.
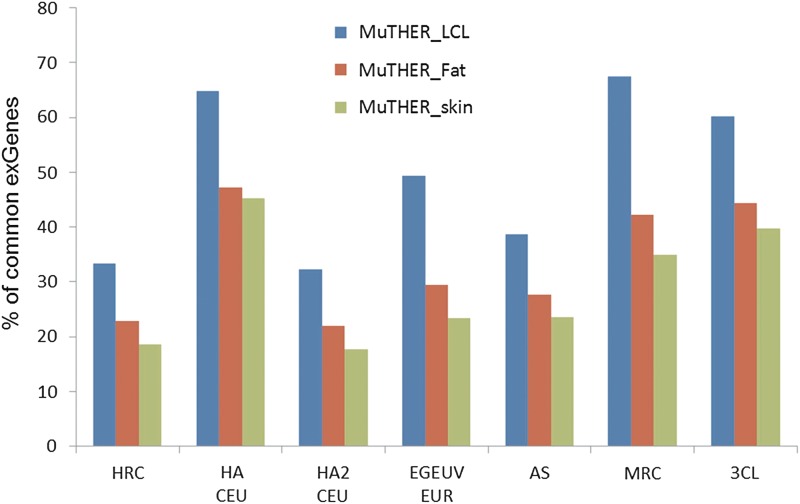
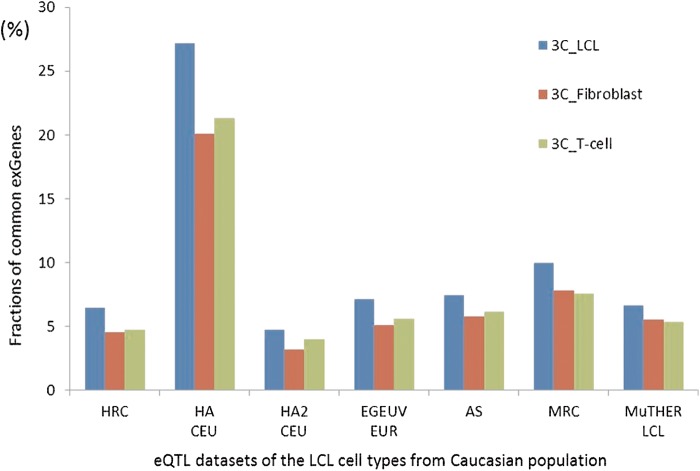
Two studies, each on LCLs and two other tissues, can each be used for this purpose: MuTHER with LCL, fat, and skin (Grundberg et al., 2012; Nica et al., 2011) and 3C with LCL, fibroblast, and T cell (Dimas et al., 2009). Both studies are in Caucasian populations and so can be compared with the other LCL studies on that population. Figure 6 shows the fraction of common exGenes between each of three MuTHER tissues and seven other studies conducted with LCLs. The fraction of exGenes common to pairs of datasets varies widely, from 33% to 68%, reflecting the differing experimental and other factors discussed earlier. However, in all seven comparisons, the fraction of common exGenes is higher between LCL-LCL dataset pairs than for LCL with other tissue comparisons, indicating a level of tissue specificity. For the LCL-fat comparisons, the common exGene fraction is between 27% and 39% lower than for LCL-LCL, and for LCL-T-cell comparisons, it is 29–50% lower. Similar levels of tissue conservation were found within the 3C study. Figure 7 shows similar comparisons between the seven reference LCL sets and the LCL, fibroblast, and T-cell data for the 3C study. In this study, the differences between cell types appear generally rather small: 16–32% fewer for LCL with fibroblast comparisons, and 15–27% less for LCL with T-cell comparisons.
FIG. 6.
Comparisons of fractions of common exGenes between pairs of eQTL datasets of the same cell type and pairs with different cell types for the MuTHER study. The blue bar shows the fractions of common exGenes between various LCL datasets and the MuTHER_LCL dataset. The red and green bars show the fractions of common exGenes between the other LCL datasets and the MuTHER_Fat and MuTHER_skin datasets, respectively. LCL, lymphoblastoid cell line.
FIG. 7.
Comparisons of fractions of common exGenes between pairs of eQTL datasets of the same cell type and pairs with different cell types for the 3C study. The blue bars show the fractions of common exGenes between the LCL datasets and the 3C_LCL dataset. The red and green bars show the fractions of common exGenes between the LCL datasets and the 3C_Fibroblast and 3C_T-cell datasets, respectively. In both sets of comparisons, there is evidence of limited tissue specificity.
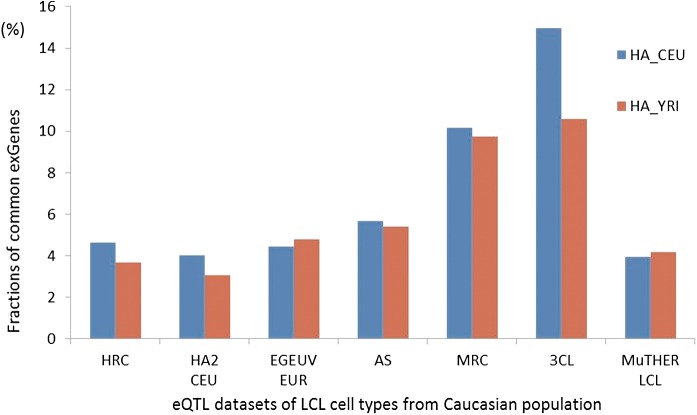
A similar analysis can be made for the population dependence of eQTLs, comparing data from LCLs across Caucasian and African populations, using the HA study. Figure 8 shows the fraction of common exGenes between those datasets and those in other studies on Caucasian populations. Differences within and across population fractions are usually small, with the exception of the 3C comparison, where the fraction of common exGenes is about 25% smaller across the populations than within Caucasians.
FIG. 8.
Comparisons of the fraction of common exGenes between datasets in the same population versus datasets from different populations. The blue bars show the fractions of common exGenes between various Caucasian datasets in the HA_CEU dataset. The red bars are the fractions of common exGenes between the other Caucasian datasets and the HA_YRI dataset. The results indicate low population dependence of eQTLs.
These are very limited comparisons, but suggest that generally the level of conservation of eQTLs across tissues is fairly high and that between populations is also high, allowing extrapolation between tissue types, although at the expense of some false positives.
Comparison of eQTL and disease GWAS data
To investigate the role expression regulation plays in disease susceptibility, we compared results from disease GWA studies and those from eQTL GWA studies. For each identified disease risk locus in a set of common diseases, we estimated whether there is an eQTL consistent with an underlying expression mechanism driving altered disease risk. Analogously with the eQTL analysis, we assume that in each disease risk locus, an underlying causal/mechanism variant affects disease risk. Because of LD, it usually results in a set of SNPs (marker SNPs), including the causal one if that is an SNP, occurring at a different frequency in disease populations than in control populations and so being detectable in GWA studies. If the disease causal variant affects the expression level of a gene, there should also be a set of overlapping marker SNPs discovered in eQTL studies. Thus, comparison of the location of disease markers and of nearby eQTL markers in a locus provides a means of estimating whether a known eQTL relationship provides a possible basis for the disease mechanism. For this purpose, we used the AllCell_AllPop high-confidence eQTLs derived from the full set of 16 studies, with the most conservative LD level (r2 > 0.8), a total of 18,615 unique high-confidence eQTL relationships involving 4252 unique genes. The procedure for comparing disease and eQTL markers is described in the Materials and Methods section. There are 21 disease risk-associated loci reported in the seminal WTCCC1 GWA study of seven diseases (The Wellcome Trust Case Control Consortium, 2007) and a further 316 risk loci from meta-analyses and subsequent studies, extracted from the GWAS catalog (www.genome.gov/gwastudies/), were included.
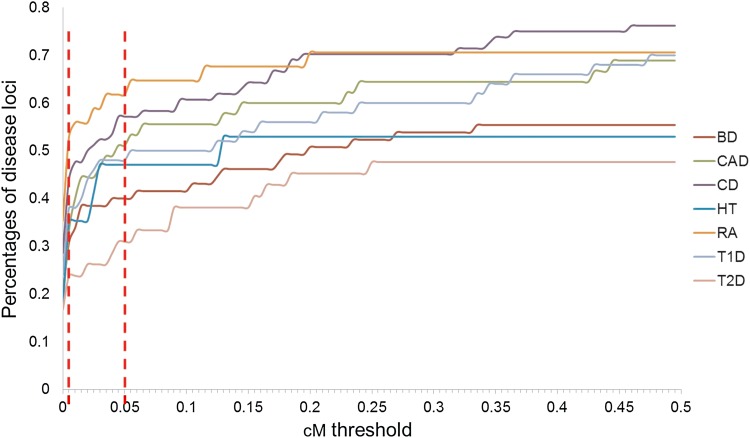
For each disease-associated locus in each set, we collected all disease marker SNPs and all neighboring marker SNPs involved in high-confidence eQTLs within 200 kb of any disease marker. The cM distance between each disease marker and each eQTL marker SNP was estimated using the Caucasian HapMap genetic map (a distance of 1 cM between locations corresponds to a recombination frequency of 1% per generation and provides the measure of genetic linkage). Figure 9 shows the percentage of loci for each disease type where disease markers match high-confidence eQTL markers as a function of cM threshold. The number of loci included rises steeply at low cM values, but less steeply above 0.005 cM. The steep slope at low values is likely a consequence of different tag SNPs used on the microarray chips for disease and expression association studies—often the exact disease marker SNP is not present on the expression chip, but there is one very close in cM space. Above 0.05 cM, the curves begin to plateau, but some extra loci do accumulate as the distance increases. Coverage converges at between 45% and 73% of loci, depending on the disease.
FIG. 9.
Percentage of disease loci with possible expression mechanisms as a function of the cM distance between the closest disease and expression marker SNPs. The AllCell_AllPop eQTL set was used. Two vertical dotted lines indicate the cM thresholds, 0.005 and 0.05. The maximum threshold used in this study is 0.05 cM.
Matches between disease and eQTL markers were collected for three thresholds, cM distances of zero, <0.005, and <0.05, based on the analysis described in the Materials and Methods section. Table 3 shows the number of disease loci that meet these criteria. With the 0 cM threshold, 15–32% of the disease risk loci for each disease have putative expression mechanisms and that increases to 23–52% at a threshold of 0.005 cM and 29–61% at a 0.05 cM threshold. There is considerable variation in the fraction of putative expression loci across the seven diseases, with T2D having the lowest values (31% at the 0.05 threshold) and RA and CD having the highest (62% and 57%, respectively, at the 0.05 threshold). Supplementary Table S4 shows all candidate expression loci for the seven diseases at a cM threshold 0.05 and the eQTL-associated genes for each locus. Each of these genes is a candidate for involvement in disease mechanism based on the eQTL data.
Table 3.
Number of Disease Risk Loci with Possible Underlying Expression Mechanisms in Seven Common Diseases
| Disease set | BD | CAD | CD | HT | RA | T1D | T2D |
|---|---|---|---|---|---|---|---|
| All loci included | 65 | 45 | 84 | 17 | 34 | 50 | 42 |
| 0 cM | 13 | 8 | 24 | 3 | 12 | 12 | 7 |
| 0.005 cM | 20 | 15 | 37 | 6 | 18 | 19 | 10 |
| 0.05 cM | 26 | 23 | 48 | 8 | 21 | 24 | 13 |
Data at three thresholds of agreement between disease and expression markers are included where at least one disease and expression SNP are identical (0 cM), where a disease and expression marker are <0.005 cM apart, and where the markers are <0.05 cM apart.
BD, bipolar disorder; CAD, coronary artery disease; CD, Crohn's disease; HT, hypertension; RA, rheumatoid arthritis; SNP, single-nucleotide polymorphism; T1D, type 1 diabetes; T2D, type 2 diabetes.
To place these results in the context of previous studies, we defined three categories of eQTL-associated disease candidate genes. Genes in category A are those where expression change has already been related to the relevant disease. Those in category B are cases where the eQTL candidate gene has already been proposed as disease involved, usually from a GWA study, but an expression mechanism has not previously been suggested. The genes in category C are those that have not previously been proposed as disease relevant. (Genes in the strong LD immune protein region on chromosome 6 are not included because of ambiguous candidate gene assignments.) Table 4 shows the number of loci with genes in each category for each disease. Only 15 disease candidate genes have a previously proposed expression mechanism. There are 94 genes in category B—previously disease-associated genes where we have now identified a putative expression mechanism. False positives are most likely to be in category C, but we do expect that a substantial fraction of these new disease candidate genes will turn out to be correct. As illustrated below, in some cases, the new candidates are supported by circumstantial evidence of biological relevance.
Table 4.
Number of Genes in Each Category for Each Disease
| Category | BD | CAD | CD | HT | RA | T1D | T2D |
|---|---|---|---|---|---|---|---|
| A | 1 | 4 | 4 | 0 | 1 | 2 | 3 |
| B | 21 | 12 | 38 | 3 | 12 | 9 | 4 |
| C | 25 | 23 | 67 | 8 | 26 | 35 | 14 |
Category A genes are those where an expression mechanism has previously been suggested and the new analysis supports that finding. Category B genes are those where the disease candidate gene has previously been suggested and we have now identified a putative expression mechanism. Category C genes are those where the expression-related candidate genes have not previously been suggested as disease relevant.
Examples of disease-associated eQTL relationships
GALNT4 and hypertension
A marker SNP, rs2681472, on chromosome region 12q21.3 is significantly associated with HT in European origin and East Asian populations (Cho et al., 2012; Hong et al., 2010) and these GWA studies have proposed the ATP2B1 gene (ATPase, Ca++ transporting, plasma membrane 1) as a nearby candidate gene for involvement in HT. A recent study has shown that ATP2B1 is involved in calcium homeostasis related to essential HT (Hirawa et al., 2013). From our eQTL analysis, we found no eQTL SNPs in this region associated with ATP2B1 expression. However, two studies (Grundberg et al., 2012; Zeller et al., 2010) included in the integrated set have several SNPs that are within 0.005 cM of the disease marker and that are significantly associated with the expression level of another nearby gene, GALNT4 (polypeptide N-acetylgalactosaminyltransferase 4). Although there is no GWA study showing an association between GALNT4 and HT, one recent GWA study suggested that GALNT4 plays a causal role in susceptibility to atherosclerosis related to high blood pressure (Erbilgin et al., 2013). N-acetylgalactosaminyltransferase 4 is thought to be involved in endothelial–platelet interactions by O-glycosylating the threonine residues of the P-selectin glycoprotein ligand (PSGL-1) (Erbilgin et al., 2013). Thus, the underlying mechanism in the 12q21.3 region associated with HT likely involves altered expression of GALNT4.
GSDMB, ORMDL3, and immune-related diseases
Several marker SNPs, including rs2872507, rs2305480, and rs2290400, in chromosome region of 17q12 have been identified as associated with the risk of several diseases, especially immune-related ones, such as CD (Barrett et al., 2008; Franke et al., 2010; Repnik and Potočnik, 2016), RA (Okada et al., 2014; Stahl et al., 2010), asthma (Bønnelykke et al., 2013; Moffatt et al., 2007), and T1D (Barrett et al., 2009). Different studies have proposed different disease-relevant candidate genes for this locus. For CD, GSMDL, ZPBP2, ORMDL3, and IKZF3 were reported. In contrast, only IKZF3 was reported as a candidate for RA and only ORMDL3 for asthma and T1D. Based on the eQTL analysis, six genes, GSDMA, GSDMAB, KRT222, ORMDL3, PGAP3, and ZPBP2, are found to have an eQTL association with these marker SNPs. Three of these eQTL genes, KRT222 (Montgomery et al., 2010), ZPBP2 (Grundberg et al., 2012), and PGAP3 (Grundberg et al., 2012), were discovered in only a single eQTL study. Two genes, GSDMB and ORMDL3, are in high-confidence eQTL relationships at the highest LD threshold (r2 > 0.8). Thus, the eQTL analysis suggests that these two genes are likely to be involved in susceptibility to these immune-related diseases. In support of this conclusion, previous studies have shown that changes in the binding of an insulator protein, CTCF, and related chromatin remodeling on this autoimmune associated locus may lead to altered cis-regulation of these two genes (Verlaan et al., 2009).
Discussion
There have now been a number of high-throughput studies for finding eQTLs in human populations and tissues, providing a wealth of data about the relationship between genetic variation and the level of gene expression. At present, although reproducibility between studies is low (Dixon et al., 2007; Göring et al., 2007; Myers et al., 2007; Stranger et al., 2007; Veyrieras et al., 2008), we were interested in obtaining a conservative, but relatively reliable, set of eQTLs for use in identifying those human complex disease loci where a genetic variant affecting expression of a gene may be contributing to disease susceptibility. To this end, we compared the results of 16 independent eQTL studies to find those variant/expression relationships that have been observed more than once. Across the 16 studies considered, more than 15,000 different genes have been reported as involved in an eQTL relationship, usually with a nearby (cis) variant. The number of human genes that are expressed at a high enough level for eQTL associations to be detected is probably not much larger than this, so at face value, almost every human gene has its expression affected by at least one variant. This remarkable observation may be misleading; however, only a little over a quarter of these genes have so far been found to be involved in the same eQTL more than once across the included studies. Most commonly, each gene is found to be involved in a single eQTL relationship.
In addition to differences in expression behavior across cell types and populations, discussed later, there are several possible reasons for low consistency between studies. First, a variety of genotyping arrays, with different tag SNPs and different probes have been used. Second, early studies relied on RNA microarrays to estimate transcript levels. Only three studies used more recent RNA-Seq technology. Third, the analysis procedures and statistical models used in each study vary (e.g., linear regression models, Spearman rank correlation). In addition, there are other possible confounders arising in the experimental procedures, for example, the history of a cell culture and culture conditions, and differences in experimental protocols. Despite these issues, there is evidence that a substantial proportion of the cis-eQTL findings are reproducible (Greenawalt et al., 2011; Innocenti et al., 2011). Innocenti et al. estimated 49–67% cis-eQTL reproducibility between several datasets conducted in the liver, which is consistent with the comparison between LV and LV2 (59%) in our datasets. A recent Genotype-Tissue Expression (GTEx) pilot study (Ardlie et al., 2015) also found a considerable fraction (68%) of previously reported (Westra et al., 2013) exGenes in blood.
For the purpose of relating expression-related SNPs to disease, we include only those eQTLs observed in at least two independent studies. The assumption that such consensus eQTLs are more reliable than those only observed once requires statistical independence of each study. Each of the studies was performed by different investigators, and in general, different genotyping and transcription profiling technologies were used. Additionally, a third factor affecting reliability, the statistical analysis technique used, varies across studies.
It has long been appreciated that expression mechanisms may play a major role in complex trait disease, and some studies have already provided data to support this idea (Cookson et al., 2009; Nicolae et al., 2010). Up to now, it has not been possible to determine how generally this is the case or which disease-associated loci may harbor expression-related mechanisms. In this study, by combining current eQTL data and disease GWAS data, we have been able to address these questions on a relatively large scale. We find that (conservatively) approaching 50% of disease loci have a high-confidence eQTL relationship consistent with an underlying expression mechanism. With the criteria used, the fraction of loci with putative expression mechanisms ranges from 30% to 60%, depending on the disease. We have illustrated that these data are useful for better identifying disease-relevant genes in particular loci. Each proposed expression mechanism defines possible follow-up experiments.
eQTL relationships may vary depending on cell type and also cell state—whether an immune system cell is active, for example. In complex trait disease, it is often difficult to know which cell type is implicated in each disease locus, and even if this is clear, expression data for that cell in that state are unlikely to be available. Typically, it has been assumed that these differences are secondary, and most disease/expression studies have used eQTLs from LCLs (Cookson et al., 2009; Nicolae et al., 2010). One study across multiple tissue types has suggested that the degree of tissue dependence is large (69–80%) (Dimas et al., 2009).
The inclusion of studies with data derived from different tissue types allowed us to estimate the extent to which eQTLs are conserved. The data are limited, and the presence of large amounts of noise also restricts analysis, but nevertheless, the available comparisons suggest a substantial number, larger than 50%, of at least partially tissue-independent eQTLs. In support of this, a recent study found more than 50% of all detected eQTLs to be common to nine tissues (adipose, tibial artery, heart, lung, muscle, tibial nerve, skin, thyroid, and whole blood) (Ardlie et al., 2015). However, it should be also noted that although a study may be tissue specific, the tissue will often include a range of cell types. For instance, in eQTL studies of brain tissue (Gibbs et al., 2010; Myers et al., 2007), various types of cells are included, such as blood cells, subtypes of neuronal cells, and different glial cells, so it is difficult to distinguish the eQTL relationships for each specific cell type.
A key step in identifying which disease loci have a potential underlying expression-related mechanism is comparing markers from eQTL studies with those from disease GWASs. Other studies, for example (Ardlie et al., 2015), have used a single LD r2 threshold as the criterion for deciding whether an eQTL relationship provides a possible mechanism underlying a disease GWAS association (Ardlie et al., 2015). In this study, we show that the cM distance between disease marker SNPs and eQTL SNPs provides a better measure and have used three confidence thresholds. For 79 loci, there is exact agreement between at least one disease marker and one eQTL marker. In a further 125 of loci, the two markers are very close in LD space, less than 0.005 cM. The remaining 51%, out at a separation of 0.05 cM, are still within a conservative threshold.
The presence of an eQTL mechanism in a disease locus does not necessarily mean that it is the dominant mechanism contributing to disease susceptibility. There are several possible molecular-level mechanisms, including high- and low-impact missense, and auxiliary splicing that contribute to common complex trait diseases. In previous work (Pal and Moult, 2015), we have also shown that a significant fraction of the disease loci have a potential high-impact missense SNP disease mechanism. Expression effects for the eQTL data analyzed here are usually relatively small, with a median value of 2.2-fold change in the level of expression. In contrast, high-impact missense variants typically change in vivo activity of a protein by 5- to 10-fold, sometimes more (Yampolsky and Stoltzfus, 2005). Where both mechanisms are present in a locus, a high-impact mechanism does not necessarily dominate. In the model we have previously proposed (Pal et al., 2015), in the same locus, a gene with a high-impact mechanism may be weakly coupled to the disease phenotype, whereas a gene with a lower impact mechanism is tightly coupled and so dominates.
Many more eQTLs in various tissues and populations will be identified in the near future. The NIH GTEx project has recently published impressive pilot study results (Ardlie et al., 2015; Consortium et al., 2013) and has a goal of determining eQTLs in 20,000 tissue samples from 900 donors from predominantly healthy humans. These data, together with results from other studies, will ultimately provide a comprehensive view of the relationship between genetic variants and altered expression and splicing, as well as the role of these mechanisms in disease.
Conclusions
In this study, we identified those loci associated with complex trait disease that may harbor an underlying expression mechanism (Yu, 2014). Our study shows that the data are consistent with ∼50% of these disease loci arising from an underlying expression change mechanism. In many cases, the results provide a proposed expression mechanism for genes previously suggested as disease relevant, but with no known mechanism, and in others, new disease-relevant genes are identified.
Supplementary Material
Abbreviations Used
- BD
bipolar disorder
- CAD
coronary artery disease
- CD
Crohn's disease
- cM
centiMorgan
- eQTLs
expression quantitative trait loci
- GTEx
Genotype-Tissue Expression
- GWA
genome-wide association
- GWASs
GWA studies
- HT
hypertension
- LCL
lymphoblastoid cell line
- LD
linkage disequilibrium
- RA
rheumatoid arthritis
- SNP
single-nucleotide polymorphism
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- TC
transcript cluster
- WTCC1
Wellcome Trust Case Control Consortium
Acknowledgments
This work was supported, in part, by NIH grants R01GM102810 and R01GM104436. Additional detailed methods and results of this research study can be found in C.-H.Y.'s doctoral dissertation (University of Maryland, http://hdl.handle.net/1903/16079).
Data Availability
The results of this study are accessible through a web database, ExSNP (www.exsnp.org), which includes the original eQTLs, high-confidence eQTLs, cell type-dependent eQTLs, population-dependent eQTLs, and disease-associated eQTLs. The website also incorporates a genome browser that allows visualization of the relative positions of eQTL SNPs and their associated genes, as well as other neighboring genes, and the relationship with functional elements and disease.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Ardlie KG, Deluca DS, Segre AV, et al. (2015). The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, et al. (2009). Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 41, 703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, et al. (2008). Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet 40, 955–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bønnelykke K, Matheson MC, Pers TH, et al. (2013). Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat Genet 45, 902–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Chen C-H, Hu C, et al. (2012). Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in East Asians. Nat Genet 44, 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E, Yelensky R, Bonakdar S, et al. (2008). Genetic analysis of human traits in vitro: Drug response and gene expression in lymphoblastoid cell lines. PLoS Genet 4, e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X, Pan C-M, Zhao S-X, et al. (2011). A genome-wide association study identifies two new risk loci for Graves' disease. Nat Genet 43, 897–901 [DOI] [PubMed] [Google Scholar]
- Consortium TG, Lonsdale J, Thomas J, et al. (2013). The Genotype-Tissue Expression (GTEx) project. Nat Genet 45, 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, and Lathrop M. (2009). Mapping complex disease traits with global gene expression. Nat Rev Genet 10, 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimas ASA, Deutsch S, Stranger BEB, et al. (2009). Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325, 1246–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Gudjonsson JE, Liang L, et al. (2010). Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet 87, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, et al. (2007). A genome-wide association study of global gene expression. Nat Genet 39, 1202–1207 [DOI] [PubMed] [Google Scholar]
- Duan S, Huang RS, Zhang W, et al. (2008). Genetic architecture of transcript-level variation in humans. Am J Hum Genet 82, 1101–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, et al. (2008). Genetics of gene expression and its effect on disease. Nature 452, 423–428 [DOI] [PubMed] [Google Scholar]
- Erbilgin A, Civelek M, Romanoski CE, et al. (2013). Identification of CAD candidate genes in GWAS loci and their expression in vascular cells. J Lipid Res 54, 1894–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertekin-Taner N. (2011). Gene expression endophenotypes: A novel approach for gene discovery in Alzheimer's disease. Mol Neurodegener 6, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfax BP, Makino S, Radhakrishnan J, et al. (2012). Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet 44, 502–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira T, and Marchini J. (2011). Modeling interactions with known risk loci—A Bayesian model averaging approach. Ann Hum Genet 75, 1–9 [DOI] [PubMed] [Google Scholar]
- Franke A, McGovern DPB, Barrett JC, et al. (2010). Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet 42, 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen K, Visschedijk MC, van Sommeren S, et al. (2010). Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn's disease. Hum Mol Genet 19, 3482–3488 [DOI] [PubMed] [Google Scholar]
- Gamazon ER, Huang RS, Cox NJ, and Dolan ME. (2010). Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc Natl Acad Sci U S A 107, 9287–9292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, et al. (2010). Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet 6, e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G, Powell JE, and Marigorta UM. (2015). Expression quantitative trait locus analysis for translational medicine. Genome Med 7, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring HHH, Curran JE, Johnson MP, et al. (2007). Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet 39, 1208–1216 [DOI] [PubMed] [Google Scholar]
- Greenawalt DM, Dobrin R, Chudin E, et al. (2011). A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res 21, 1008–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E, Small KS, Hedman ÅK, et al. (2012). Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 44, 1084–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid IM, Jackson AU, Randall JC, et al. (2010). Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet 42, 949–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirawa N, Fujiwara A, and Umemura S. (2013). ATP2B1 and blood pressure: From associations to pathophysiology. Curr Opin Nephrol Hypertens 22, 177–184 [DOI] [PubMed] [Google Scholar]
- Hong K-W, Jin H-S, Lim J-E, et al. (2010). Non-synonymous single-nucleotide polymorphisms associated with blood pressure and hypertension. J Hum Hypertens 24, 763–774 [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, and Marchini J. (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlickova B, Westra H-J, Franke L, and Wijmenga C. (2011). Celiac disease: Moving from genetic associations to causal variants. Clin Genet 80, 203–313 [DOI] [PubMed] [Google Scholar]
- Hsu Y-H, Zillikens MC, Wilson SG, et al. (2010). An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet 6, e1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti F, Cooper GM, Stanaway IB, et al. (2011). Identification, replication, and functional fine-mapping of expression quantitative trait Loci in primary human liver tissue. PLoS Genet 7, e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, and Nap JP. (2001). Genetical genomics: The added value from segregation. Trends Genet 17, 388–391 [DOI] [PubMed] [Google Scholar]
- Kang HP, Morgan A, Chen R, Schadt EE, and Butte AJ. (2012a). Coanalysis of GWAS with eQTLs reveals disease-tissue associations. AMIA Jt Summits Transl Sci Proc 2012, 35–41 [PMC free article] [PubMed] [Google Scholar]
- Kang HP, Yang X, Chen R, et al. (2012b). Integration of disease-specific single nucleotide polymorphisms, expression quantitative trait loci and coexpression networks reveal novel candidate genes for type 2 diabetes. Diabetologia 55, 2205–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, et al. (2010). Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedländer MR, et al. (2013). Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pouladi N, Achour I, et al. (2015). eQTL networks unveil enriched mRNA master integrators downstream of complex disease-associated SNPs. J Biomed Inform 58, 226–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Morar N, Dixon AL, et al. (2013). A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res 23, 716–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, and Boerwinkle E. (2011). dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat 32, 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranville JC, Luca F, Richards AL, et al. (2011). Interactions between glucocorticoid treatment and cis-regulatory polymorphisms contribute to cellular response phenotypes. PLoS Genet 7, e1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Kabesch M, Liang L, et al. (2007). Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–473 [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, et al. (2010). Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464, 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, et al. (2007). A survey of genetic human cortical gene expression. Nat Genet 39, 1494–1499 [DOI] [PubMed] [Google Scholar]
- Nica AC, Parts L, Glass D, et al. (2011). The architecture of gene regulatory variation across multiple human tissues: The MuTHER study. PLoS Genet 7, e1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, and Cox NJ. (2010). Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from GWAS. PLoS Genet 6, e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Wu D, Trynka G, et al. (2014). Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal LR, and Moult J. (2015). Genetic basis of common human disease: Insight into the role of missense SNPs from genome-wide association studies. J Mol Biol 427, 2271–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal LR, Yu C-H, Mount SM, and Moult J. (2015). Insights from GWAS: Emerging landscape of mechanisms underlying complex trait disease. BMC Genomics 16, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Lyons PA, Lee JC, et al. (2016). Insight into genotype-phenotype associations through eQTL mapping in multiple cell types in health and immune-mediated disease. PLoS Genet 12, e1005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Marioni JC, Pai AA, et al. (2010). Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464, 768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repnik K, and Potočnik U. (2016). eQTL analysis links inflammatory bowel disease associated 1q21 locus to ECM1 gene. J Appl Genet 57, 1–10 [DOI] [PubMed] [Google Scholar]
- Richards AL, Jones L, Moskvina V, et al. (2012). Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry 17, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, et al. (2008). Mapping the genetic architecture of gene expression in human liver. PLoS Biol 6, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, and Snyder M. (2012). Linking disease associations with regulatory information in the human genome. Genome Res 22, 1748–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, et al. (2010). Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42, 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Raychaudhuri S, Remmers EF, et al. (2010). Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet 42, 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, et al. (2007). Population genomics of human gene expression. Nat Genet 39, 1217–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, and Bork P. (2000). Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet 16, 198–200 [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. (2010). A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap 3 Consortium. (2010). Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium. (2007). Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan DJ, Ge B, Grundberg E, et al. (2009). Targeted screening of cis-regulatory variation in human haplotypes. Genome Res 19, 118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras J-B, Kudaravalli S, Kim SY, et al. (2008). High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet 4, e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-S, and Cooper TA. (2007). Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8, 749–761 [DOI] [PubMed] [Google Scholar]
- Wang Z, and Moult J. (2001). SNPs, protein structure, and disease. Hum Mutat 17, 263–270 [DOI] [PubMed] [Google Scholar]
- Westra H-J, Peters MJ, Esko T, et al. (2013). Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45, 1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Miao X, Huang L, et al. (2012). Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet 44, 62–66 [DOI] [PubMed] [Google Scholar]
- Yampolsky LY, and Stoltzfus A. (2005). The exchangeability of amino acids in proteins. Genetics 170, 1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C-H. (2014). Analysis of consensus genome-wide expression-QTLS and their relationships to human complex trait diseases. (Doctoral dissertation, University of Maryland; ) http://hdl.handle.net/1903/16079 [Google Scholar]
- Zeller T, Wild P, Szymczak S, et al. (2010). Genetics and beyond—The transcriptome of human monocytes and disease susceptibility. PLoS One 5, e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Beaulaurier J, Lum PY, et al. (2010). Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet 6, e1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The results of this study are accessible through a web database, ExSNP (www.exsnp.org), which includes the original eQTLs, high-confidence eQTLs, cell type-dependent eQTLs, population-dependent eQTLs, and disease-associated eQTLs. The website also incorporates a genome browser that allows visualization of the relative positions of eQTL SNPs and their associated genes, as well as other neighboring genes, and the relationship with functional elements and disease.