Abstract
DNA barcoding is a fast-developing technique to identify species by using short and standard DNA sequences. Universal selection of DNA barcodes in ferns remains unresolved. In this study, five plastid regions (rbcL, matK, trnH-psbA, trnL-F and rps4-trnS) and eight nuclear regions (ITS, pgiC, gapC, LEAFY, ITS2, IBR3_2, DET1, and SQD1_1) were screened and evaluated in the fern genus Adiantum from China and neighboring areas. Due to low primer universality (matK) and/or the existence of multiple copies (ITS), the commonly used barcodes matK and ITS were not appropriate for Adiantum. The PCR amplification rate was extremely low in all nuclear genes except for IBR3_2. rbcL had the highest PCR amplification rate (94.33%) and sequencing success rate (90.78%), while trnH-psbA had the highest species identification rate (75%). With the consideration of discriminatory power, cost-efficiency and effort, the two-barcode combination of rbcL+ trnH-psbA seems to be the best choice for barcoding Adiantum, and perhaps basal polypod ferns in general. The nuclear IBR3_2 showed 100% PCR amplification success rate in Adiantum, however, it seemed that only diploid species could acquire clean sequences without cloning. With cloning, IBR3_2 can successfully distinguish cryptic species and hybrid species from their related species. Because hybridization and allopolyploidy are common in ferns, we argue for including a selected group of nuclear loci as barcodes, especially via the next-generation sequencing, as it is much more efficient to obtain single-copy nuclear loci without the cloning procedure.
Introduction
DNA barcording is a method to achieve accurate and rapid species identification by using short and standard DNA regions [1]. To find a locus that is universal, readily sequenced and has sufficiently high sequence divergence at the species-level, Chase et al. [2] assessed rbcL and nuclear ITS, and found both markers performed well in identifying plants. Kress et al. [3] initially proposed rbcL as a DNA barcode for plants because of its high universality. A global plant DNA barcode system was evaluated by comparing amplification universality and sequence divergence levels for nine putative barcode loci [4], and they recommended the combination of rbcL+ trnH-psbA as a two-locus global land plant barcode. Hollingsworth et al. [5] evaluated seven candidate plastid regions (rpoC1, rpoB, rbcL, matK, trnH-psbA, atpF-atpH, and psbK-psbI) in three divergent plants groups. Their results revealed that no single locus had high levels of universality and resolvability in these groups [5], and they proposed various three-locus combinations involving rpoC1, rbcL, matK and trnH-psbA to identify these groups. CBOL plant working group [6] recommended a two-locus combination of rbcL+ matK as the land plant barcode. The second internal transcribed spacer (ITS2) was proposed as a universal DNA barcode [7], and then the China Plant BOL Group further argued that ITS/ITS2 should be incorporated as a core barcode for seed plants [8]. In the past five years, DNA barcoding is fast evolving to include genome skimming [9].
DNA barcoding studies on ferns (monilophytes) and lycophytes are relatively few in comparison with those on seed plants, even though DNA barcoding may be of great value on the identification of their gametophytes, a free-living and featureless generation in the life cycle. Ebihara et al. [10] tested the utility of rbcL and trnH-psbA using 733 taxa, and demonstrated that these two barcodes were effective to identify the Japanese pteridophyte flora. de Groot et al. [11] evaluated the discriminatory power of rbcL and trnL-F, and suggested rbcL + trnL-F can be used as a two-locus barcode to identify NW-European fern species. Li et al. [12] assembled sequences of rbcL, matK (designed specific primers for each of the major clades), and trnH-psbA from 74 species of 37 families, and trnL-F from 32 species of 19 families in major fern lineages. They suggested that matK + rbcL can provide a two-locus barcode with strong resolving power in ferns, and the study favored trnL-F over trnH-psbA as a potential back-up locus if the universal primers of matK failed. Schneider & Schuettpelz [13] used rbcL sequence to determine the identity of a sterile gametophyte of unknown origin, and successfully identified it as Osmunda regalis. Li et al. [14] developed a procedure “Tissue-direct PCR”, which can make the identification of diminutive and characterless stages of ferns (gametophytes and young sporophytes) easy and rapid when it was combined with plant barcodes. Pryer et al. [15] used rbcL, atpA, and trnG-R sequences to identify a cultivated plant marketed as Cheilanthes wrightii in the horticultural trade but the plant was actually C. distans.
At present DNA barcoding in ferns has relied solely on plastid loci, which are uniparentally inherited [10–13,15]. Ferns are characterized by frequent hybridizations among closely related species [16] as well as polyploidy [17–18], and have relatively frequent apomictic lineages [19]. DNA barcoding was thought to likely show low levels of discrimination success rate in taxa having high rate of hybridization and polyploidy [20]. Combining biparentally inherited nuclear barcodes with uniparentally inherited plastid barcodes may be useful for species discrimination in allopolyploid species and those of hybrid origin.
The Chinese Adiantum is herein recognized as a good model to evaluate the candidate barcodes because most species in the region have been clearly defined, while a few problematic species can be used to test the effectiveness of selected barcodes. Adiantum consists of about 150~200 species, of which most species are distributed in the tropical to subtropical regions, with the greatest diversity in the Neotropics [21–24]. Ching [25] treated Adiantum as an early diverged and unique genus, and recognized the monotypic family Adiantaceae. Smith et al. [26] included Adiantum in Pteridaceae and Tryon et al. [24] recognized the group as a subfamily Adiantoideae in Pteridaceae. Molecular evidence supported the placement of Adiantum in Pteridaceae [27–29]. There are some eurychoric species showing high morphological divergences due to divergent habitats, while there are also high morphological similarities among some species, especially in series Venusta [23,30]. Lin [23] divided Chinese Adiantum into seven series: Reniformia, Gravesiana, Caudata, Pedata, Flabellulata, Venusta, and Venericapilliformia, and recognized 31 species, five varieties, and four forms in China. Lin et al. [30] recognized three additional species, A. meishanianum F. S. Hsu ex Yea C. Liu & W. L. Chiou and A. formosanum Tagawa from Taiwan, and A. subpedatum Ching from Zhejiang province of eastern China, with a total of 34 species and three varieties of Adiantum, of which 16 are endemic to China.
The DNA barcoding approach has been greatly advocated since the concept was proposed [1], and it has been shown to be an important tool for species identifications, and a supplement to traditional morphology-based taxonomy [31–32]. Combining DNA sequences with existing morphological characters may facilitate species identification and classification [33–36].
The objectives of this study are to (1) screen and evaluate potential plastid and nuclear barcodes in Adiantum; (2) combine the barcodes with morphological characters for assessing species delimitations in several species groups of Adiantum; and (3) discuss general guidelines for barcoding fern species.
Materials and Methods
Taxon Sampling
A total of 154 samples representing 33 species of Adiantum were collected in this study. The taxon names were mainly based on the recent treatments [30,37]. Six samples of A. menglianense [38] and two samples of A. ailaoshanense were also included in this study. At least two individuals were sampled from different populations of each species except for the stenochoric species A. fengianum, A. mariesii and A. lianxianense, which were only sampled from one population. More individuals (3~20) of eurychoric species were sampled to represent their distributional range. All taxa included in this study, together with voucher information, were listed in S1 and S2 Tables (Supporting information).
DNA extraction, PCR amplification and sequencing
Total DNAs were extracted from silica-gel dried-leaf material and herbarium specimens using the CTAB procedure [39] and Dneasy (QIAGEN) extraction kits. Polymerase chain reaction (PCR) amplifications were performed in a 20 μL reaction mixture containing 1×Taq buffer [50 mM (NH4) 2SO4; 75 mM Tris–HCl (pH 8.3); 50 mM KCl; 0.001% gelatin]; 2.5 mM MgCl2, 0.4 mM of dNTPs, 0.5 μM of each primer, 1.0 U of Taq DNA Polymerase (TaKaRa Biotechnology Co. Ltd., Dalian, China), and 1 μL of genomic DNA (25–30 ng). Purified PCR products were sequenced in both directions with the PCR primers on an ABI 3730xl DNA Sequencer (Applied Biosystems, Foster City, USA).
Five candidate plastid regions (rbcL, matK, trnH-psbA, trnL-F and rps4-trnS) and eight candidate nuclear regions (ITS, pgiC, gapC, LEAFY, ITS2, IBR3_2, DET1, and SQD1_1) [40–48] were screened and evaluated in Adiantum. The primer information and thermocycling conditions used in this study were listed in S3 Table.
A subset of samples were sequenced via direct sequencing without cloning using the amplification primers of IBR3_2, but this resulted in chromatograms with multiple peaks for most species. We then cloned IBR3_2 from all samples, using the ZeroBack Fast Ligation Kit (Tiangen, Beijing) and following standard protocols for cloning, colony selection, and post-cloning re-amplification with pZeroBack/Blunt Vector primers. At least six and up to 20 colonies were picked for sequencing for each individual. Some taxa ultimately yielded fewer than six sequences despite multiple cloning attempts because of technical difficulties in generating sequences.
Data analysis
Sequences were aligned with Clustal X [49] and then manually adjusted in Geneious 4.8.2 (Biomatters Ltd., NZ). The genetic pairwise distance for each marker was calculated using MEGA 5.2 based on pairwise deletion and the P-distance model [50]. Intra- and inter-specific genetic divergences of the four candidate DNA regions were analyzed by Wilcoxon signed-rank tests [51], and “barcoding gap” was estimated by comparing the intra- and inter-specific divergences of each candidate locus using taxonDNA [52]. The neighbor-joining (NJ) trees were constructed based on single markers and combinations of two/three/four markers in MEGA 5.2, with pairwise deletion based on the P-distance model, and used to evaluate whether individual samples of a species clustered in species-specific monophyletic clades. Robustness of inference was assessed by running 5, 000 bootstrap replicates [53].
Results
Screening the DNA barcodes for Adiantum
For the plastid DNA barcodes, matK showed the lowest PCR amplification success rate (33.33%), while rbcL, trnH-psbA, trnL-F, and rps4-trnS showed much higher PCR amplification and sequencing success rates. PCR amplification success rate of the four DNA regions was 94.33% (rbcL), 93.20% (trnH-psbA), 85.82% (trnL-F), and 94.20% (rps4-trnS), and sequencing success rate of the four markers was 90.78%, 84.16%, 77.01%, and 83.57%, for rbcL, trnH-psbA, trnL-F, and rps4-trnS, respectively (Table 1). Thus, these four barcodes were used for subsequent analyses in this study.
Table 1. Summary of genetic variability and sequence characteristics of the candidate barcodes and their main combinations in this study.
| rbcL | trnH-psbA | trnL-F | rps4-trnS | R+S | R+P | R+F | P+S | P+F | F+S | R+P+S | R+P+F | R+F+S | P+F+S | R+P+F+S | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. taxa | 30 | 28 | 30 | 30 | 29 | 28 | 29 | 27 | 27 | 29 | 27 | 27 | 27 | 25 | 24 |
| No. sequences | 133 | 122 | 116 | 126 | 110 | 113 | 108 | 104 | 93 | 99 | 95 | 89 | 92 | 82 | 77 |
| Aligned length (bp) | 1143 | 520 | 826 | 792 | 1935 | 1663 | 1969 | 1312 | 1346 | 1618 | 2455 | 2489 | 2761 | 2183 | 3281 |
| Average intra-distance (%) | 0.06 | 0.05 | 0.16 | 0.10 | 0.06 | 0.06 | 0.09 | 0.10 | 0.13 | 0.14 | 0.07 | 0.09 | 0.10 | 0.13 | 0.10 |
| Average inter-distance (%) | 5.74 | 7.38 | 15.76 | 10.69 | 7.62 | 6.22 | 9.38 | 9.62 | 12.68 | 13.04 | 7.70 | 9.16 | 9.85 | 12.14 | 9.72 |
| Variable sites (%) | 23.10 | 28.85 | 53.87 | 42.55 | 30.85 | 24.23 | 35.65 | 36.51 | 43.09 | 47.28 | 29.61 | 33.19 | 36.98 | 40.95 | 34.75 |
| Informative sites (%) | 21.96 | 27.88 | 52.30 | 40.40 | 29.20 | 23.03 | 34.33 | 34.68 | 41.60 | 45.80 | 27.94 | 31.90 | 35.46 | 38.75 | 32.64 |
| PCR success (%) | 94.33 | 93.20 | 85.82 | 94.20 | N/a | N/a | N/a | N/a | N/a | N/a | N/a | N/a | N/a | N/a | N/a |
| Sequencing success (%) | 90.78 | 84.16 | 77.01 | 83.57 | N/a | N/a | N/a | N/a | N/a | N/a | N/a | N/a | N/a | N/a | N/a |
| Identification success (%) | 73.33 | 75 | 66.67 | 77.33 | 79.31 | 78.57 | 79.31 | 81.48 | 77.78 | 79.31 | 77.78 | 77.78 | 92.59 | 92.00 | 92.00 |
| No. unidentified species† | 2 | 1 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
† the number of unidentified species except for four species groups.
For the eight nuclear DNA barcodes, the PCR amplification rates in seven candidate nuclear regions (ITS, ITS2, pgiC, gapC, LEAFY, DET1, and SQD1_1) were low (<50%) or did not obtain clean sequences by direct sequencing without cloning. IBR3_2 had 100% PCR amplification success (S1 Fig), but only 23 sequences of 13 species were obtained by direct sequencing, and additional 244 sequences representing 53 individuals of 32 species (including a recently published species A. ailaoshanense) were obtained by sequencing the clones (S2 Table). IBR3_2 had sufficient discriminatory power in series Caudata, and was used to identify hybrid individuals in Adiantum in the present study.
Variation of barcoding markers
The newly acquired DNA sequences have been deposited in GenBank and their accession numbers were provided in S1 and S2 Tables. Fourteen sequences (rbcL: 5; trnH-psbA: 1; trnL-F: 4; rps4-trnS: 4) were downloaded from GenBank. The data analyses included 133 rbcL sequences, 122 trnH-psbA sequences, 126 rps4-trnS sequences, and 116 trnL-F sequences.
Homopolymer (e.g., poly-A/G/C) within trnH-psbA was detected in taxa in ser. Reniformia, ser. Gravesiana, ser. Caudata, and ser. Venusta. The mononucleotide repeats near the end of sequences led to unclean reverse sequences in some cases. So only forward sequences of psbA in some individuals were used for the analyses. There are some poly structures in trnL-F and rps4-trnS, so the sequences of the “f” end of trnL-F and those of the trnS-F end of rps4-trnS of some samples were difficult to obtain. The forward reads alone were added to the analyses in these samples that the mononucleotide repeats exists.
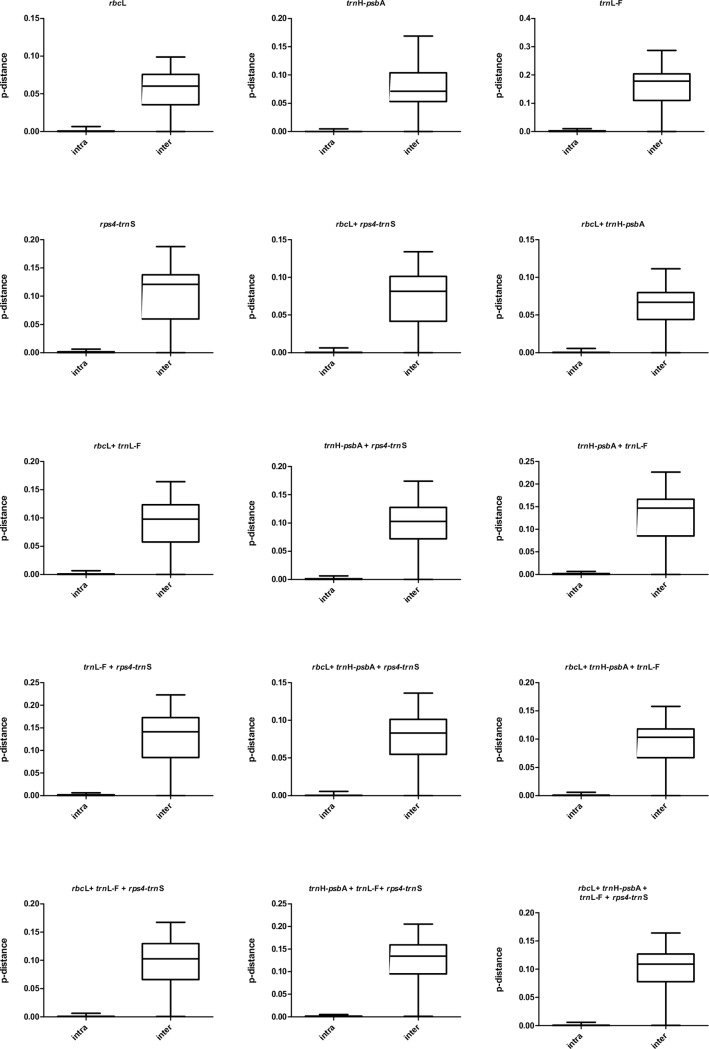
The trnL-F marker had the highest mean intraspecific divergence followed by rps4-trnS, rbcL, and trnH-psbA, and had the highest mean interspecific divergences followed by rps4-trnS, trnH-psbA, and rbcL (Tables 1 and 2). The interspecific and intraspecific genetic divergences of the four DNA regions were analyzed with the Wilcoxon signed-rank tests [51]. At the intraspecific level, genetic divergences exhibited no significant difference among the four barcodes (Table 3). The barcoding gap was detected for the four markers, which was indicative of high sequence variation among species for the four barcodes (Fig 1).
Table 2. Wilcoxon signed-rank tests of interspecific divergence among DNA markers.
| W+ | W- | Relative ranks | N-value | P-value | Result | |
|---|---|---|---|---|---|---|
| W+ | W- | |||||
| trnH-psbA | rbcL | 44905 | 7745 | 325 | ≤0.001 | trnH-psbA > rbcL |
| trnL-F | rbcL | 61424 | 1 | 351 | ≤0.001 | trnL-F > rbcL |
| rps4-trnS | rbcL | 70933 | 320 | 378 | ≤0.001 | rps4-trnS > rbcL |
| trnL-F | trnH-psbA | 44832 | 18 | 300 | ≤0.001 | trnL-F > trnH-psbA |
| rps4-trnS | trnH-psbA | 52279 | 371 | 325 | ≤0.001 | rps4-trnS > trnH-psbA |
| rps4-trnS | trnL-F | 14 | 61411 | 351 | ≤0.001 | rps4-trnS<trnL-F |
Table 3. Wilcoxon signed-rank tests of intraspecific divergences among DNA markers.
| W+ | W- | Relative ranks | N-value | P-value | Result | |
|---|---|---|---|---|---|---|
| W+ | W- | |||||
| trnH-psbA | rbcL | 39 | 66 | 24 | 0.397 | trnH-psbA = rbcL |
| trnL-F | rbcL | 147 | 24 | 24 | 0.007 | trnL-F = rbcL |
| rps4-trnS | rbcL | 90 | 46 | 26 | 0.255 | rps4-trnS = rbcL |
| trnL-F | trnH-psbA | 98 | 7 | 22 | 0.004 | trnL-F = trnH-psbA |
| rps4-trnS | trnH-psbA | 85 | 20 | 24 | 0.041 | rps4-trnS = trnH-psbA |
| rps4-trnS | trnL-F | 40 | 150 | 24 | 0.027 | rps4-trnS = trnL-F |
Fig 1. The distributions of divergences for four markers (rbcL, trnH-psbA, trnL-F, and rps4-trnS).
Applicability for species discrimination
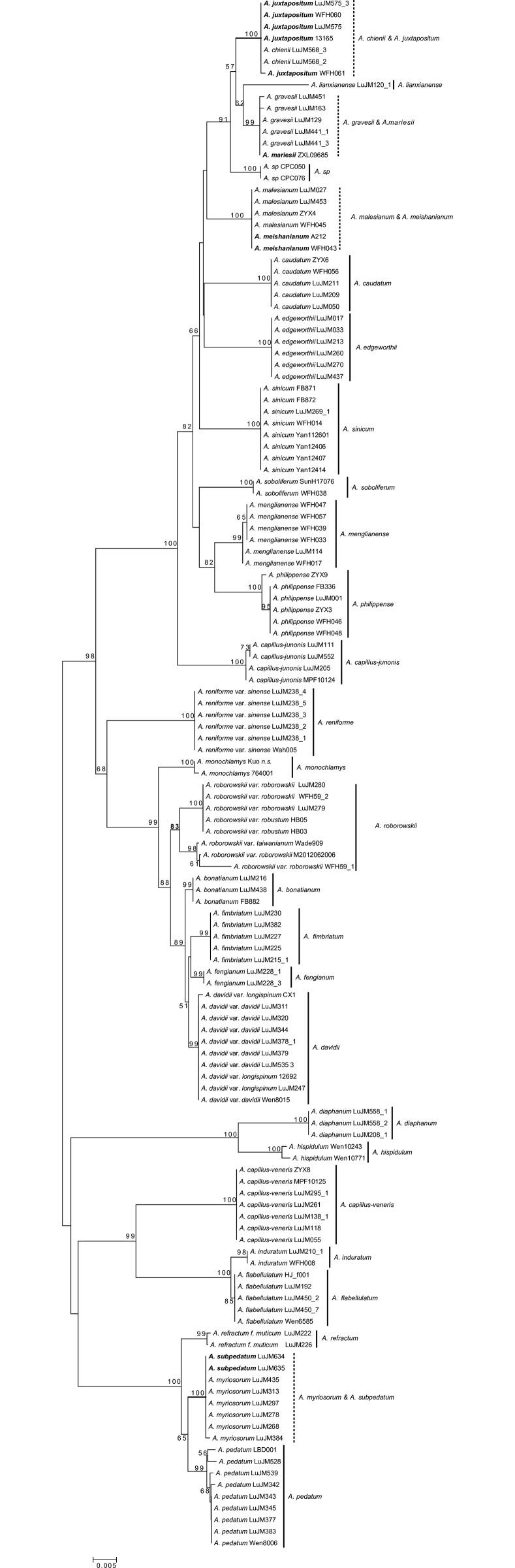
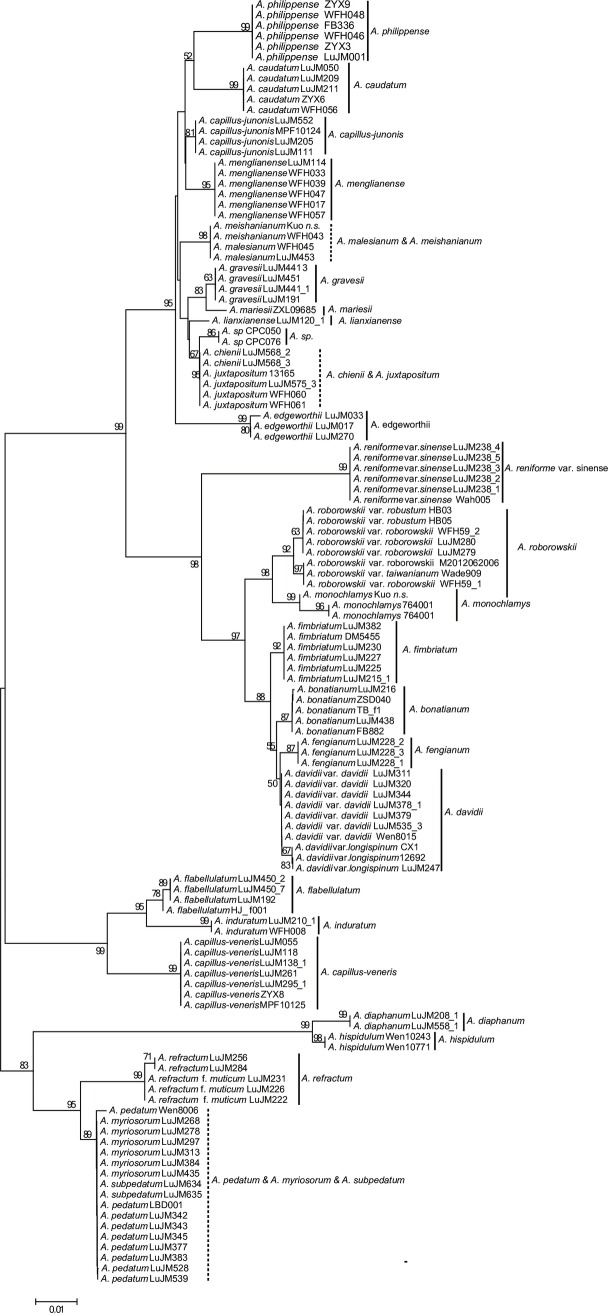
A tree-based method (NJ) was used for the species identification of Adiantum. Based on the single barcode, trnH-psbA showed the highest species discrimination power among the four DNA regions at 75%, followed by rps4-trnS (73.33%) and rbcL (73.33%), and trnL-F (65.52%) (Table 1).
Four groups of species including A. subpedatum and A. myriosorum, A. formosanum and A. refractum, A. juxtapositum and A. chienii, A. meishanianum and A. malesianum were not identified by the present data. Adiantum formosanum (Kuo430) strongly clustered with A. refractum. Samples of A. juxtapositum (LuJM575, WFH060, WFH 061, CSH 13165) and A. chienii (LuJM568) grouped together with a high BP value. The samples of two Vietnamese individuals of “A. juxtapositum” (CP050 and CP076) did not cluster with the Chinese A. juxtapositum clade (Figs 2–5). Two samples of A. erythrochlamys Diels (HB03 and HB05 from Hunan) strongly clustered with A. roborowskii var. roborowskii in the NJ trees (Figs 2–5).
Fig 2. The NJ tree based on the single barcode rbcL using the p-distance model (dotted vertical line: unidentified group).
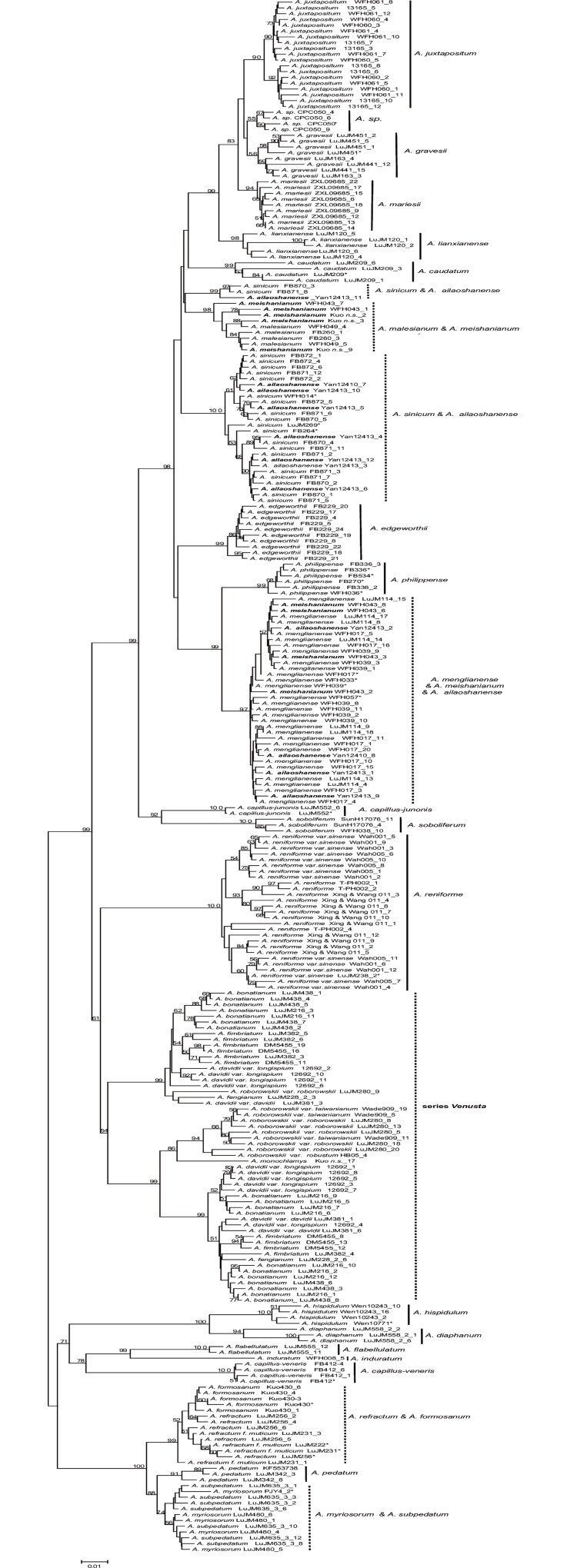
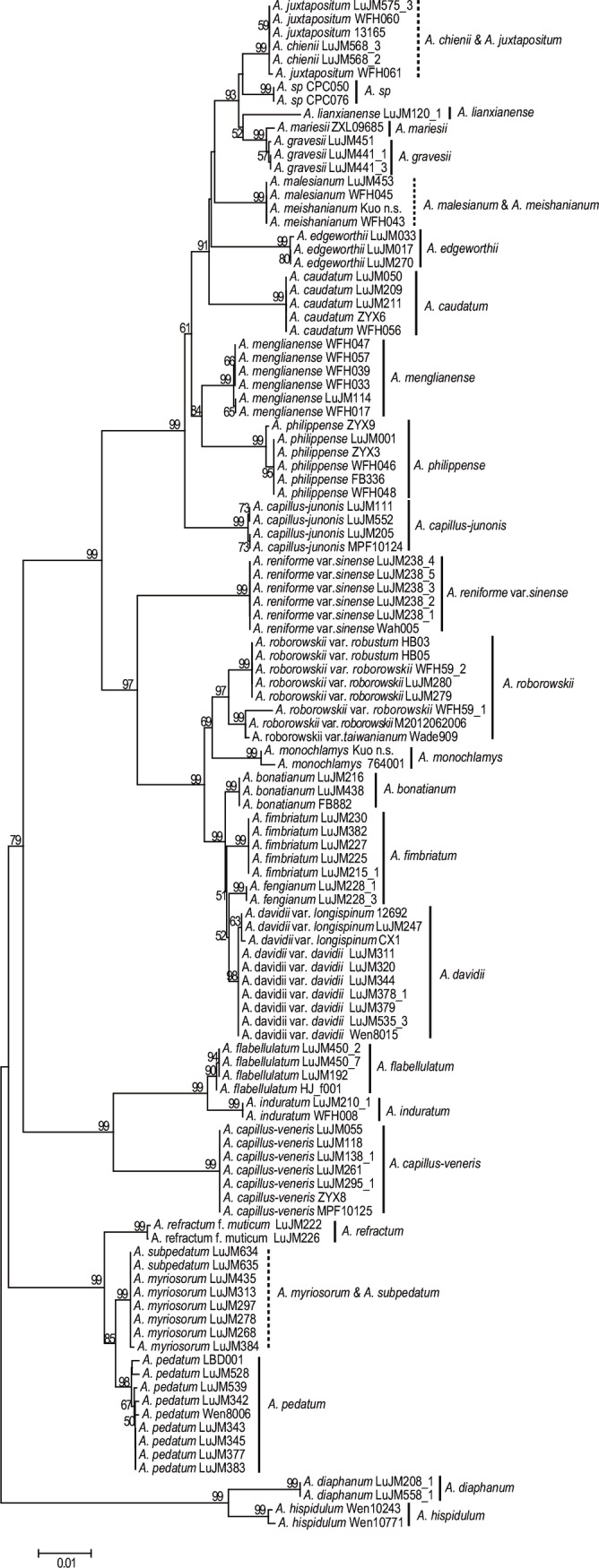
Fig 5. The NJ tree based on the IBR3_2 marker using the p-distance model (dotted vertical line: unidentified group).
Twenty-one of the 28 species were each supported to be monophyletic in the trnH-psbA tree (Fig 3). Three species—A. davidii, A. pedatum and A. myriosorum were not identified successfully in the NJ tree. However, A. myriosorum can be distinguished from A. pedatum by a 9-bp insertion “TTGAAAAGA” in the trnH-psbA sequences, and these two species thus can be successfully identified by trnH-psbA. Adiantum davidii var. longispinum fell into a monophyletic group in the A. davidi clade, even though the A. davidi clade was only weakly supported, and failed to be identified (BS<50). With the lack of sampling of the close relative A. formosanum, A. refractum was successfully identified in the trnH-psbA tree.
Fig 3. The NJ tree based on the single barcode trnH-psbA using the p-distance model (dotted vertical line: unidentified group).
For rbcL and rps4-trnS, 22 of the 30 species each formed a clade with high bootstrap values (Fig 3 and S2 Fig). Except for the four unidentified groups described above, A. gravesii and A. mariesii failed to be identified in the rbcL and rps4-trnS trees. Adiantum refractum was successfully identified in the rbcL tree, and A. myriosorum was successfully identified in rps4-trnS tree, with the absence of A. formosanum and A. subpedatum in the individual datasets.
Based on the trnL-F sequences, 20 of the 30 species were each strongly supported as a monophyletic group (S3 Fig). Five groups of species including A. subpedatum and A. myriosorum, A. formosanum and A. refractum, A. juxtapositum and A. chienii, A. meishanianum and A. malesianum, A. gravesii and A. mariesii were not identified in the trnL-F tree (S3 Fig). Adiantum mariesii clustered within the A. gravesii clade, although the two species differed in two nucleotide positions. Adiantum davidii var. longispinum fell into a monophyletic group in the A. davidi clade, although the support value was moderate (BS = 67).
The combination of DNA barcodes can slightly improve the ability of species identification (Table 1).
IBR3_2 was successful in identifying the parental species of the presumed hybrid taxa. The maternal parent of A. meishanianum was shown to be A. malesianum, and the maternal parent of the hybrid species A. ailaoshanense was identified to be A. sinicum in the present analysis (Fig 5). However the interspecific relationship in series Venusta was more complicated than other series in Adiantum because of the high frequent polyploidy (e.g., hexaploid of A. bonatianum, and octaploid of A. davidii).
Discussion
Evaluation of the potential chloroplast barcodes for Adiantum
An ideal DNA barcode should be routinely retrievable with a single primer pair with little requirement for manual editing of sequence traces, can provide maximal discrimination among species [6,54], and exhibit a “barcode gap” between intraspecific and interspecific divergences [51].
Although matK is one of the most variable coding regions within cpDNA [55–56], it is often difficult to be amplified in ferns because of the loss of the flanking trnK exons [57]. Taxon specific primers need to be designed for ferns [12,57]. Even though Li et al. [12] endorsed matK as a barcode for ferns, matK showed low universality in Adiantum using the same primers FWPtmatKF1 and FWPtmatK rAGK designed by Li et al. [12] based on Cheilanthes of the same family (Pteridaceae), and primers FWPtmatK fEDR and FWPtmatK rAGK designed by Kuo et al. [57]. Our study illustrated the need for further matK primer development in ferns to ensure efficient PCR and sequencing, and at present this marker seems to be a difficult barcode locus for ferns.
Relatively well-defined gaps between intraspecific and interspecific divergences of the four selected barcodes (rbcL, trnH-psbA, trnL-F, and rps4-trnS) were shown in this study (Fig 1). rbcL is widely used for molecular phylogenetic inferences and has been proposed as a DNA barcode in ferns as well [10–11,13–14]. Our study also showed that rbcL provided the highest level of universality in PCR and sequencing with the primer pair 1F and 1379R, and the second highest species discriminatory power (73.33%).
trnH-psbA has been widely used as a plant barcode [3–4,58]. Previous studies suggested that the variation in the relatively short trnH-psbA region was enough to differentiate species [10,59]. The species discrimination of trnH-psbA was 75% in our study; and only Adiantum davidii could not be distinguished (BS<50), except that the four species groups (eight species) cannot be differentiated by all four barcode markers. trnH-psbA was ranked the second for PCR amplification success in Adiantum, however, its sequencing success rate was lower because of the existence of poly-A/G/C repeats [10,59–60]. trnH-psbA can be used as an effective barcode in Adiantum with its short length and relatively high variability, and may be used to identify specimens or traditional medicinal materials.
Reported as the most variable locus in ferns [12,61], trnL-F is shown here to have the highest sequence divergence, even at the intraspecific level in Adiantum. The amplification of trnL-F was, however, relatively difficult. PCR amplification success rate was 82.8% when total DNAs were extracted using the CTAB procedure. Higher-quality DNA can improve this rate, and it can reach about 90% when Dneasy extraction kits were used to extract the total DNAs. The sequencing of this marker was also relatively difficult (sequencing success rate at 77.01%) because of the universal presence of mononucleotide repeats in the intergenic spacer. The presence of mononucleotide repeat in trnL-F affected sequence quality, and reduced the universality of this marker.
With the rates of sequence divergence just lower than trnL-F, rps4-trnS ranked the third for PCR amplification rate, but the lowest sequencing success rate. Only a single forward sequence was obtained from 26% samples because of the presence of the mononucleotide repeat structure in trnL-F IGS and rps4-trnS IGS.
Five species groups including A. subpedatum and A. myriosorum, A. formosanum and A. refractum, A. juxtapositum and A. chienii, A. meishanianum and A. malesianum, A. gravesii and A. mariesii cannot be identified in the trnL-F tree because the species pairs had identical trnL-F sequences. Adiantum refractum was successfully identified in the rbcL tree, and A. myriosorum was successfully identified in rps4-trnS tree, wih the absence of A. formosanum and A. subpedatum in the individual datasets. In fact, the identified species were the same by rbcL, trnL-F,and rps4-trnS, with all five groups of species not identified while trnH-psbA could identify A. mariesii from A. gravesii.
Combination of DNA barcodes slightly improved the ability for species identifications. All combinations except for the three combinations discussed below identified all species except for the four species groups mentioned above. Adiantum davidii fell into two respective clades (A. davidii var. davidii and A. davidii var. longispimum in the trnH-psbA+rps4-trnS NJ tree (S7 Fig). The combination of rbcL +rps4-trnS failed to separate A. gravesii and A. mariesii in the NJ tree (S4 Fig). The three barcode combinations of rbcL+ trnH-psbA +rps4-trnS also failed to separate A. gravesii and A. mariesii in the NJ tree (S10 Fig), although the two species can be distinguished by a 19-bp insertion and a 2-bp inversion in trnH-psbA of A. gravesii. Considering the discriminatory power, cost-efficiency and effort, the two-barcode combination of rbcL+ trnH-psbA (Fig 4) seems to be the best choice for barcoding Adiantum. Furthermore, trnL-F had the highest variation and can be used as a barcoding marker at the intraspecific level.
Fig 4. The NJ tree based on the single barcode rbcL+trnH-psbA using the p-distance model (dotted vertical line: unidentified group).
Need for nuclear barcodes for ferns
Chase et al. [2] suggested that multiple, low-copy nuclear markers with sufficient genetic variability and PCR-reliability need to be developed, which may permit researchers to detect hybrids. Four nuclear regions, the nuclear ribosomal internal transcribed spacer (ITS) region [62–63], the introns of the transcription factor LEAFY (LFY) [42,64–66], the cytosolic phosphoglucose isomerase gene pgiC [40,43,45,63,66–68], the two regions of the plastidicl glyceraldehydes-3-phosphate dehydrogenase (gapCp) -gapCpSh (gapCp “short”) [41,44,63,68–71], and gapCpLg (gapCp “long”) [71] were used to study the evolution of closely related fern species in recent years.
Rothfels et al. [70] presented 20 novel single-copy nuclear regions across ten distinct protein-coding genes: ApPEFP_C, cryptochrome 2 gene, cryptochrome 4 gene, DET1, gapCpSh, IBR3, pgiC, SQD1, TPLATE, and transducin gene which were readily amplified and sequenced from 15 diploid Polypodiales species. The results showed that IBR3_2, DET1, and SQD1_1 were amplified well, and clean sequences were obtained via direct Sanger sequencing from most taxa of Polypodiales examined.
Of the eight nuclear candidate markers (ITS, ITS2, pgiC, gapC, LEAFY, IBR3_2, DET1, and SQD1_1) screened in this study, ITS exhibited high PCR amplification success, but it did not produce a single band for PCR amplification due to additive banding and incomplete concerted evolution. ITS is thus difficult to be used as a barcode in ferns. The PCR amplification rates in six candidate nuclear regions (ITS2, pgiC, gapCp, LEAFY, DET1 and SQD1_1) were extremely low and/or clean sequences were not generated via direct sequencing without cloning.
Ishikawa et al. [40] developed primers to amplify the pgiC gene in ferns. The PCR products of primers 14F/16R containing two introns are moderate in size (534–1,000 bp) and the pgiC gene is possibly of value for phylogenetic reconstruction at the specific and generic levels. The primers 14F/15R and 15F/16R were developed and applied to study mating systems and other population genetic traits [40]. The pgiC gene was also used to detect the origins of polyploids and hybrids in Dryopteris [43,45,68]. But relatively universal primers of the pgiC gene are lacking [67].
Schneider et al. [63] employed three nuclear regions—ITS, gapCp, and pgiC, to explore patterns of reticulate evolution in Asplenium. The results showed that all three nuclear markers amplified well and several copies were recovered by cloning PCR products in Asplenium. Rothfels et al. [70] designed one novel primer pair for pgiC situated in exons 14 and 16, to amplify introns 14, 15, and exon 15 (about 600–700 bp); however, samples of Adiantum failed to be amplified and directly sequenced in their study.
We amplified pgiC using the primers of 14F/16R and 15F/16R, and obtained relatively weak bands in Adiantum when the 14F/16R primers were used. However, the sequences of pgiC are only about 300 bp in Adiantum, and showed multiple peaks in the sequencing signals. The primer pair 15F/16R failed to amplify taxa of Adiantum.
Schuettpelz et al. [41] designed primers to amplify part of the nuclear gapCp gene that encodes the glyceraldehyde-3-phosphate dehydrogenase. Their survey across ferns demonstrated that these primers are nearly universal for ferns, and holds considerable potential for addressing species-level questions across the tree of life in ferns. Rothfels et al. [70] designed specific primers for a region covering introns 8~10 of gapCpSh, which overlaps with the gapCp region amplified with the primers of Schuettpelz et al. [41], and ranges from 450 to 590 bp. In general gapCpSh amplified and sequenced well in fern taxa, however, they did not obtain clean sequences for Adiantum (only a partial 283 bp sequence of A. pedatum was obtained) and a few other genera (cloning not attempted). We amplified gapCp using the primers of Schuettpelz et al. [41], and obtained two or three bands in Adiantum.
IBR3_2 had a high PCR amplification rate even though it is difficult to obtain sequences via direct sequencing. It seems that clean sequences may be obtained from autoploid species whereas allopolyploid or hybrid ones failed. IBR3_2 successfully distinguished two presumed hybrid species—A. meishanianum from its maternal parent A. malesianum [72], and A. ailaoshanense [73] from its maternal parent A. sinicum by the degenerate base. The hybrid A. meishanianum [72] and its maternal A. malesianum had the same plastid sequences. Nevertheless, they can be distinguished in morphology and the IBR3_2 sequences. Therefore, IBR3_2 has the potential to be further explored as a nuclear barcode locus in some fern groups such as Adiantum and its close relatives.
Taxonomic implications of the barcoding results in the context of morphology
DNA barcoding is generally successful for species identification in Adiantum, and the result is nearly congruent with morphology-based taxonomy except for a few species. Ultimately the systematics community relies on morphology for species delimitation [74–75]. Four species groups—A. subpedatum and A. myriosorum, A. formosanum and A. refractum, A. juxtapositum and A. chienii, A. meishanianum and A. malesianum cannot be identified by the plastid barcodes. We herein discuss these species groups in light of a morphological framework.
Adiantum subpedatum was recorded only from the Longtang Mountain in Zhejiang province [76], and was thought to be a possible depauperate form of A. myriosorum [30]. The primary differences between A. subpedatum and A. myriosorum lie in plant size and sori number [30]. The height of A. subpedatum is about 24–28 cm, whereas A. myriosorum is taller, about 40–60 cm. Adiantum subpedatum was described to have 1–2 sori per pinnule [76] while the latter has 4–6 sori. However, we noted that there are a few plants with 2–3 sori in some populations of A. myriosorum, while most samples from the two populations of the Longtang Mountain (type locality) of A. subpedatum have 3 sori per pinnule. Plant size and sori number may vary with the habitats. Adiantum subpedatum and A. myriosorum fell into the same clade in nuclear IBR3-2 tree (Fig 5). Based on the DNA barcoding results and our morphological observations, A. subpedatum is perhaps best treated as a synonym of A. myriosorum.
Adiantum formosanum, endemic to Taiwan, cannot be distinguished from A. refractum, because the two species have identical sequences of trnL-F, rps4-trnS (rbcL and trnH-psbA sequences are not available). Both species are epilithic plants, and have 2–4 sori on the thin papery and fan-shaped pinnules. The primary differences between A. formosanum and A. refractum are plant height and pinnule size. Although A. formosanum differ with A. refractum in a few nucleotides, it still fell into A. refractum clade in nuclear IBR3-2 tree (Fig 5). Based on the similar DNA sequences and the minor morphological differences, the species status of A. formosanum needs to be reevaluated and it may be treated as a synonym of A. refractum.
Lu et al. [77] proposed that A. juxtapositum might be a synonym of A. chienii based on chloroplast sequences and field observations. Samples of the Chinese A. juxtapositum and A. chienii clustered together in all NJ trees with a high BP value (Figs 2–4 and S2, S3 and S13 Figs). However, two samples of “A. juxtapositum” from Vietnam (CPC050 and CPC076 A. sp.) [78] did not cluster with the Chinese A. juxtapositum. The veins on lower surface of the samples from Vietnam are more visible than those of the Chinese samples. The fronds of the former are rather leathery while the latter are sub-leathery. The Chinese A. juxtapositum thus may be best treated as a synonym of A. chienii while the samples from Vietnam probably represent a new species.
Adiantum meishanianum was validated in 2009 [79]. A cryptic species related to A. philippense was suggested as its paternal species, and A. malesianum as its maternal parent [72]. The presumed hybrid species A. meishanianum and its maternal A. malesianum had identical plastid sequences. They can be distinguished by morphology (barely hirsute on rachis and lamina in A. meishanianum vs. densely hirsute on rachis and lamina in A. malesianum; pinnules with articulated stalks in A. meishanianum vs. pinnules nearly without stalks in A. malesianum). Adiantum meishanianum can also be distinguished from the maternal parent A. malesianum using IBR3_2.
Wang et al. [80] used the primers of Rothfels et al. [70] to amplify a region in the third exon of CRY2 and they detected a new hybrid species, A. × ailaoshanense, which probably originated from A. sinicum x A. menglianense [73]. However, the amplification and sequencing of CRY2 were not ideal in other series of Adiantum in our present study. Our IBR3_2 data also support the parents of the hybrid species A. ailaoshanense (Yan 12413 and Yan 12410) to be A. sinicum and A. menglianense (Fig 5).
Mao et al. [37] placed A. erythrochlamys as a synonym of A. roborowskii var. robustum. The diagnostic characters between A. roborowskii and A. erythrochlamys are the number of false indusia (one vs. two) and the shape of pinnule margin (entire or undulate-crenate vs. bluntly serrate) [23]. The different states of the two characters can be observed in one population (WFH59) in Longnan, Gansu province, China. The samples with two false indusia and entire or undulate-crenate pinnule margins clustered with A. roborowskii var. taiwanianum whereas another sample was nested into another subclade in the A. roborowskii var. roborowskii clade. Our DNA barcoding results and morphology thus support the treatment of Mao et al. [37].
Adiantum menglianense was published in 1992 [38], and was included in the Flora Yunnanica treatment [81]. Adiantum menglianense formed a highly-supported clade, and was sister to the closely related species A. philippense with strong support (BP≥82) in all trees (Figs 2, 4 and 5 and S2–S13 Figs) except for the trnH-psbA tree (the closely related species of A. menglianense is A. capillus-junonis in Fig 3). The margin of pinnules is deeply lobed in A. menglianense (vs. subentire in A. philippense); the false indusia are short and straight in A. menglianense (vs. long and cupped in A. philippense); and pinnules are palmate and thin in A. menglianense (vs. semilunar and thick in A. philippense). Adiantum menglianense was described to have 6–10 sori per pinnule [38] while A. philippense has 2–6 sori [30]. Zhang et al. [72] pointed out that a cryptic species under A. philippense is the paternal parent of A. meishanianum. The present analyses confirmed the paternal species of A. meishanianum as A. menglianense.
Adiantum gravesii and A. mariesii failed to be identified except by trnH-psbA and IBR3_2. Adiantum gravesii can be distinguished from A. mariesii using morphological characters with the former being a bigger plant with reniform or lunate indusia vs. the latter a smaller plant with circular indusia.
Conclusions and Outlook
Due to low primer universality of matK and the existence of multiple copies of ITS, these two commonly used barcodes were not appropriate for Adiantum. With the consideration of discriminatory power, cost-efficiency and effort, the two-barcode combination of rbcL+ trnH-psbA seems to be the best choice for barcoding Adiantum, and perhaps basal polypod ferns in general. Coupling DNA barcoding with morphology provides important insights into species delimitations for several taxa in our case study. Overall DNA barcoding provides additional DNA diagnostic characters to discriminate the individuals lacking diagnostic features because of rapid diversification, morphological stasis, and phenotypic variations [82]. Because hybridizations and allopolyploidy are common in ferns, we argue for including a selected group of nuclear loci as barcodes, especially via the next-generation sequencing, as it is more efficient and economical to obtain single-copy nuclear loci without the cloning procedure [9, 83–84]. With the drastic decrease in cost with the next-generation sequencing, fern barcoding can also effectively incorporate whole plastome data as organelle barcodes (e.g., [85]).
Supporting Information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We are grateful to Drs. Yue-Hong Yan, Li-Yaung Kuo, and Mr Xing-Xing Mao, Ms Ai-Hua Wang for sample collection, Ms Yi-Ping Wang for her assistance with the experiments, Drs. Li-Yaung Kuo, Lian-Ming Gao and Xue-Jun Ge for their helpful comments on the earlier versions of the manuscript. We also thank two reviewers for their constructive comments.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The study was supported by The National Key Basic Research Program of China (Grant No. 2014CB954100), the Project of Knowledge Innovation Program of the Chinese Academy of Sciences (Grant No. KSCX2-EW-J-24), and the Applied Fundmental Research Foundation of Yunnan Province (Grants No. 2014GA003, 2014FB168), and the Research Fund for the Large-scale Scientific Facilities of the Chinese Academy of Sciences (Grant No. 2009-LSF-GBOWS-01), the National Natural Science Foundation of China (Grant No. 31070199).
References
- 1.Hebert PD, Cywinska A, Ball SL. Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci. 2003; 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haidar N, et al. Land plants and DNA barcodes: short-term and long-term goals. Phil Trans R Soc Lond B Biol Sci. 2005; 360: 1889–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. P Natl Acad Sci USA. 2005; 102: 8369–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE. 2007; 2: e508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollingsworth ML, Clark AA, Forrest LL, Richardson J, Pennington RT, Long DG, et al. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour. 2009; 9: 439–457. 10.1111/j.1755-0998.2008.02439.x [DOI] [PubMed] [Google Scholar]
- 6.CBOL Plant Working Group. A DNA barcode for land plants. P Natl Acad Sci USA. 2009; 106: 12794–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao H, Song JY, Liu C, Luo K, Han JP, Li Y, et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE. 2010; 5: e13102 10.1371/journal.pone.0013102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. P Natl Acad Sci USA. 2011; 108: 19641–19646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollingsworth PM, Li DZ, van der Bank M, Twyford AD. Telling plant species apart with DNA: from barcodes to genomes. Phil Trans R Soc B. 2016; 371: 20150338 10.1098/rstb.2015.0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebihara A, Nitta JH, Ito M. Molecular species identification with rich floristic sampling: DNA barcoding the pteridophyte flora of Japan. PLoS ONE. 2010; 5: e15136 10.1371/journal.pone.0015136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Groot GA, During HJ, Maas JW, Schneider H, Vogel JC, Erkens RHJ. Use of rbcL and trnL-F as a two-locus DNA barcode for identification of NW-European ferns: an ecological perspective. PLoS ONE. 2011; 6: e16371 10.1371/journal.pone.0016371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li FW, Kuo LY, Rothfels CJ, Ebihara A, Chiou WL, Windham MD, et al. rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS ONE. 2011; 6: e26597 10.1371/journal.pone.0026597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider H, Schuettpelz E. Identifying fern gametophytes using DNA sequences. Mol Ecol Notes. 2006; 6: 989–991. [Google Scholar]
- 14.Li FW, Kuo LY, Huang YM, Chiou WL, Wang CN. Tissue–direct PCR, a rapid and extraction–free method for barcoding of ferns. Mol Ecol Resour. 2010; 10: 92–95. 10.1111/j.1755-0998.2009.02745.x [DOI] [PubMed] [Google Scholar]
- 15.Pryer KM, Schuettpelz E, Huiet L, Grusz AL, Rothfels CJ, Avent T, et al. DNA barcoding exposes a case of mistaken identity in the fern horticultural trade. Mol Ecol Resour. 2010; 10: 979–985. 10.1111/j.1755-0998.2010.02858.x [DOI] [PubMed] [Google Scholar]
- 16.Barrington DS, Haufler CH, Werth CR. Hybridization, reticulation, and species concepts in the ferns. Am Fern J. 1989; 79: 55–64. [Google Scholar]
- 17.Manton I. Problems of cytology and evolution in the Pteridophyta. Cambridge: Cambridge University Press; 1950. [Google Scholar]
- 18.Lǒve A, Lǒve D, Pichi-Sermolli REG. Cytotaxonomical atlas of the Pteridophyta Vaduz: Federal Republic of Germany, J. Cramer; 1977. [Google Scholar]
- 19.Lovis JD. Evolutionary patterns and processes in ferns. Adv Bot Res. 1977; 4: 229–415. [Google Scholar]
- 20.Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS ONE. 2011; 6: e19254 10.1371/journal.pone.0019254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ching RC. On the genus Adiantum L. of China with notes on some related species from neighbouring regions. Acta Phytotax Sin. 1957; 6: 301–354. [Google Scholar]
- 22.Lin YX. New taxa of Adiantum L. in China. Acta Phytotax Sin. 1980; 18: 101–105. [Google Scholar]
- 23.Lin YX. Adiantaceae In: Flora Reipublicae Popularis Sinicae Tomus Editorial Committee, editors. Flora Reipublicae Popularis Sinicae. Vol. 3 (1). Beijing: Science Press; 1990. pp.173–216. [Google Scholar]
- 24.Tryon R, Tryon A, Kramer K. Pteridaceae In: Kubitzki K, editor. The families and genera of vascular plants. Vol. 1 Berlin: Springer-Verlag; 1990. pp. 230–256. [Google Scholar]
- 25.Ching RC. On natural classification of the family "Polypodiaceae". Sunyatsenia. 1940; 5: 201–268. [Google Scholar]
- 26.Smith AR, Pryer KM, Schuettpelz E, Korall P, Schneider H, Wolf PG. A classification for extant ferns. Taxon. 2006; 55: 705–731. [Google Scholar]
- 27.Schuettpelz E, Korall P, Pryer KM. Plastid atpA data provide improved support for deep relationships among ferns. Taxon. 2006; 55: 897–906. [Google Scholar]
- 28.Schuettpelz E, Schneider H, Huiet L, Windham MD, Pryer KM. A molecular phylogeny of the fern family Pteridaceae: assessing overall relationships and the affinities of previously unsampled genera. Mol Phylogenet Evol. 2007; 44: 1172–1185. [DOI] [PubMed] [Google Scholar]
- 29.Schuettpelz E, Pryer KM. Fern phylogeny inferred from 400 leptosporangiate species and three plastid genes. Taxon. 2007; 56: 1037–1050. [Google Scholar]
- 30.Lin YX, Prado J, Gilbert MG. Adiantum L In: Wu CY, Raven PH, Hong DY, editors. Flora of China. Vol. 2–3 Beijing: Science Press; St Louis: Missouri Botanical Garden Press; 2013. pp. 238–250. [Google Scholar]
- 31.Hebert PD, Gregory TR. The promise of DNA barcoding for taxonomy. Syst Biol. 2005; 54: 852–859. [DOI] [PubMed] [Google Scholar]
- 32.Packer L, Gibbs J, Sheffield C, Hanner R. DNA barcoding and the mediocrity of morphology. Mol Ecol Resour. 2009; 9: 42–50. 10.1111/j.1755-0998.2009.02631.x [DOI] [PubMed] [Google Scholar]
- 33.Desalle R. Species discovery versus species identification in DNA barcoding efforts: response to Rubinoff. Conserv Biol. 2006; 20: 1545–1547. [DOI] [PubMed] [Google Scholar]
- 34.Smith MA, Fisher BL, Hebert PD. DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar. Phil Trans R Soc Lond B Biol Sci. 2005; 360: 1825–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Will KW, Mishler BD, Wheeler QD. The perils of DNA barcoding and the need for integrative taxonomy. Syst Biol. 2005; 54: 844–851. [DOI] [PubMed] [Google Scholar]
- 36.Hajibabaei M, Singer GA, Hebert PD, Hickey DA. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 2007;23: 167–172. [DOI] [PubMed] [Google Scholar]
- 37.Mao XX, Liu X, Zhang GM. Taxonomic revision of Adiantum ser. Venusta Ching (Pteridaceae) from Pan-Himalayas. Plant Diversity and Resources. 2014; 36:453–467. [Google Scholar]
- 38.Qian YY. A new species of Adiantum from Yunnan. Acta Botanica Austro Sinica. 1992; 8: 37–38. [Google Scholar]
- 39.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987; 19: 11–15. [Google Scholar]
- 40.Ishikawa H, Watano Y, Kano K, Ito M, Kurita S. Development of primer sets for PCR amplification of the PgiC gene in ferns. J Plant Res. 2002; 115: 65–70. [DOI] [PubMed] [Google Scholar]
- 41.Schuettpelz E, Grusz AL, Windham MD, Pryer KM. The Utility of nuclear gapCp in resolving polyploid fern origins. Syst Bot. 2008; 33: 621–629. [Google Scholar]
- 42.Shepherd LD, Perrie LR, Brownsey PJ. Low-copy nuclear DNA sequences reveal a predominance of allopolyploids in a New Zealand Asplenium fern complex. Mol Phylogenet Evol. 2008; 49: 240–248. 10.1016/j.ympev.2008.06.015 [DOI] [PubMed] [Google Scholar]
- 43.Chang HM, Chiou WL, Wang JC. Molecular evidence for genetic heterogeneity and the hybrid origin of Acrorumohra subreflxipinna from taiwan. Am Fern J. 2009; 99: 61–77. [Google Scholar]
- 44.Nitta JH, Ebihara A, Ito M. Reticulate evolution in the Crepidomanes minutumspecies complex (Hymenophyllaceae). Am J Bot. 2011; 98: 1782–1800. 10.3732/ajb.1000484 [DOI] [PubMed] [Google Scholar]
- 45.Juslén A, Väre H, Wikström N. Relationships and evolutionary origins of polyploid Dryopteris (Dryopteridaceae) from Europe inferred using nuclear pgiC and plastid trnL-F sequence data. Taxon. 2011; 60: 1284–1294. [Google Scholar]
- 46.Dyer RJ, Savolainen V, Schneider H. Apomixis and reticulate evolution in the Asplenium monanthes fern complex. Ann Bot-London. 2012; 110: 1515–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaruwattanaphan T, Matsumoto S, Watano Y. Reconstructing hybrid speciation events in the Pteris cretica group (Pteridaceae) in Japan and adjacent regions. Syst Bot. 2013; 38: 15–27. [Google Scholar]
- 48.Rothfels CJ, Larsson A, Li FW, Sigel EM, Huiet L, Burge DO, et al. Transcriptome-mining for single-copy nuclear markers in ferns. PLoS ONE. 2013; 8: e76957 10.1371/journal.pone.0076957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequences alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer CP, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 2005; 3: e422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meier R, Kwong S, Vaidya G, Ng PKL. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol. 2006; 55: 715–728. [DOI] [PubMed] [Google Scholar]
- 53.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985; 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 54.Kress WJ, Garcia-Robledo C, Uriarte M, Erickson DL. DNA barcodes for ecology, evolution, and conservation. Trends Ecol Evol. 2015; 30: 25–35. 10.1016/j.tree.2014.10.008 [DOI] [PubMed] [Google Scholar]
- 55.Neuhaus H, Link G. The chloroplast tRNALys (UUU) gene from mustard (Sinapis alba) contains a class II intron potentially coding for a maturase-related polypeptide. Curr Genet. 1987; 11: 251–257. [DOI] [PubMed] [Google Scholar]
- 56.Olmstead RG, Palmer JD. Chloroplast DNA systematics: a review of methods and data analysis. Am J Bot. 1994; 81: 1205–1224. [Google Scholar]
- 57.Kuo LY, Li FW, Chiou WL, Wang CN. First insights into fern matK phylogeny. Mol Phylogenet Evol. 2011; 59: 556–566. 10.1016/j.ympev.2011.03.010 [DOI] [PubMed] [Google Scholar]
- 58.Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, et al. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE. 2008; 3: e2802 10.1371/journal.pone.0002802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma XY, Xie CX, Liu C, Song JY, Yao H, Luo K, et al. Species identification of medicinal pteridophytes by a DNA barcode marker, the chloroplast psbA-trnH intergenic region. Biological Pharm Bull. 2010; 33: 1919–1924. [DOI] [PubMed] [Google Scholar]
- 60.Burgess KS, Fazekas AJ, Kesanakurti PR, Graham SW, Husband BC, Newmaster SG, et al. Discriminating plant species in a local temperate flora using the rbcL+ matK DNA barcode. Methods Ecol Evol. 2011; 2: 333–340. [Google Scholar]
- 61.Chen CW, Huang YM, Kuo LY, Nguyen QD, Luu HT, Callado JR, et al. TrnL-F is a powerful marker for DNA identification of field vittarioid gametophytes (Pteridaceae). Ann Bot-London. 2013; 111: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reid JD, Plunkett GM, Peters GA. Phylogenetic relationships in the heterosporous fern genus Azolla (Azollaceae) based on DNA sequence data from three noncoding regions. Int J Plant Sci. 2006;167: 529–538. [Google Scholar]
- 63.Schneider H, Navarro-Gomez A, Russell SJ, Ansell S, Grundmann M, Vogel J. Exploring the utility of three nuclear regions to reconstruct reticulate evolution in the fern genus Asplenium. J Syst Evol. 2013; 51: 142–153. [Google Scholar]
- 64.Adjie B, Masuyama S, Ishikawa H, Watano Y. Independent origins of tetraploid cryptic species in the fern Ceratopteris thalictroides. J Plant Res. 2007; 120: 129–138. [DOI] [PubMed] [Google Scholar]
- 65.Chen CW, Kuo LY, Wang CN, Chiou WL. Development of a PCR primer set for intron 1 of the low-copy gene LEAFY in Davalliaceae. Am J Bot. 2012; 99: e223–e225. 10.3732/ajb.1100498 [DOI] [PubMed] [Google Scholar]
- 66.Wang L, Schneider H, Wu ZQ, He LJ, Zhang XC, Xiang QP. Indehiscent sporangia enable the accumulation of local fern diversity at the Qinghai-Tibetan Plateau. BMC Evol Biol. 2012; 12: 158 10.1186/1471-2148-12-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chao YS, Dong SY, Chiang YC, Liu HY, Chiou WL. Extreme multiple reticulate origins of the Pteris cadieri complex (Pteridaceae). Int J Mol Sci. 2012; 13: 4523–4544. 10.3390/ijms13044523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sessa EB, Zimmer EA, Givnish TJ. Unraveling reticulate evolution in North American Dryopteris (Dryopteridaceae). BMC Evol Biol. 2012;12: 104 10.1186/1471-2148-12-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ebihara A, Ishikawa H, Matsumoto S, Lin S J, Iwatsuki K, Takamiya N, et al. Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the Vandenboschia radicans complex (Hymenophyllaceae) in Japan and adjacent areas. Am J Bot. 2005; 92: 1535–1547. 10.3732/ajb.92.9.1535 [DOI] [PubMed] [Google Scholar]
- 70.Rothfels CJ, Schuettpelz E. Accelerated rate of molecular evolution for vittarioid ferns is strong and not driven by selection. Syst Biol. 2013; 63: 31–54. 10.1093/sysbio/syt058 [DOI] [PubMed] [Google Scholar]
- 71.Zhang RS, Liu T, Wu W, Li YQ, Chao LF, Huang LS, et al. Molecular evidence for natural hybridization in the mangrove fern genus Acrostichum. BMC Plant Biol. 2013; 13: 74 10.1186/1471-2229-13-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang WY, Kuo LY, Li FW, Wang CN, Chiou WL. The hybrid origin of Adiantum meishanianum (Pteridaceae): A rare and endemic species in Taiwan. Syst Bot. 2014; 39: 1034–1041. [Google Scholar]
- 73.Wang Y, Shang H, Zhou XL, Zhao GH, Dai XL, Yan YH. Adiantum × ailaoshanense (Pteridaceae), a new natural hybrid from Yunnan, China. Phytotaxa. 2015; 236: 266–272. [Google Scholar]
- 74.Wen J, Ickert-Bond S, Appelhans MS, Dorr LJ, Funk VA. Collections-based systematics: opportunities and outlook for 2050. J Syst Evol. 2015; 53: 477–488. [Google Scholar]
- 75.Spooner DM. Species delimitations in plants: lessons learned from potato taxonomy by a practicing taxonomist. J Syst Evol. 2016; 54: 191–203. [Google Scholar]
- 76.Ching RC, Zhang CF. New ferns of ZheJiang province. Bull Bot Res. 1983; 3: 2–3. [Google Scholar]
- 77.Lu JM, Wen J, Lutz S, Wang YP, Li DZ. Phylogenetic relationships of Chinese Adiantum based on five plastid markers. J Plant Res. 2012; 125: 237–249. 10.1007/s10265-011-0441-y [DOI] [PubMed] [Google Scholar]
- 78.Van The P, Averyanov L, Loc PK. Notes on Adiantum juxtapositum (Adiantaceae) and Abrodictyum pluma (Hymenophyllaceae) for the fern flora of Vietnam. Taiwania. 2013; 58: 151–155. [Google Scholar]
- 79.Liu YCh, Huang YM, Chiou WL. Validation of the name Adiantum meishanianum (Pteridaceae), a species endemic to Taiwan. Novon. 2009; 19: 59–61. [Google Scholar]
- 80.Wang Y, Shang H, Gu YF, Wei HJ, Zhao GH, Dai XL, et al. A new cryptic hybrid species of Adiantum L. (Pteridaceae) identified by nuclear and chloroplast DNA sequences. Chinese Sci Bull. 2015; 60: 922–932. [Google Scholar]
- 81.Zhang GF. Adiantaceae In: Zhu WM, editor. Flora Yunnanica. Vol. 20 Beijing: Science Press; 2006. pp. 308–323. [Google Scholar]
- 82.Xiang JY, Wen J, Peng H. Evolution of the eastern Asian-North American biogeographic disjunctions in ferns and lycophytes. J Syst Evol. 2015; 53: 2–32. [Google Scholar]
- 83.Bybee SM, Bracken-Grissom H, Haynes BD, Hermansen RA, Byers RL, Clement MJ, et al. Targeted amplicon sequencing (TAS): A scalable next-gen approach to multilocus, multitaxa phylogenetics. Genome Biol Evol. 2011; 3: 1312–1323. 10.1093/gbe/evr106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coissac E, Hollingsworth PM, Lavergne S, Taberlet P. From barcodes to genomes: extending the concept of DNA barcoding. Mol Ecol. 2016; 25: 1423–1428. 10.1111/mec.13549 [DOI] [PubMed] [Google Scholar]
- 85.Yang JB, Tang M, Li HT, Zhang ZR, Li DZ. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol Biol. 2013;13: 84 10.1186/1471-2148-13-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.