Genetic mapping and coexpression highlights hot spots regulating transcripts linked to diverse biological processes.
Abstract
Variation in gene expression, in addition to sequence polymorphisms, is known to influence developmental, physiological, and metabolic traits in plants. Genetic mapping populations have facilitated identification of expression quantitative trait loci (eQTL), the genetic determinants of variation in gene expression patterns. We used an introgression population developed from the wild desert-adapted Solanum pennellii and domesticated tomato (Solanum lycopersicum) to identify the genetic basis of transcript level variation. We established the effect of each introgression on the transcriptome and identified approximately 7,200 eQTL regulating the steady-state transcript levels of 5,300 genes. Barnes-Hut t-distributed stochastic neighbor embedding clustering identified 42 modules revealing novel associations between transcript level patterns and biological processes. The results showed a complex genetic architecture of global transcript abundance pattern in tomato. Several genetic hot spots regulating a large number of transcript level patterns relating to diverse biological processes such as plant defense and photosynthesis were identified. Important eQTL regulating transcript level patterns were related to leaf number and complexity as well as hypocotyl length. Genes associated with leaf development showed an inverse correlation with photosynthetic gene expression, but eQTL regulating genes associated with leaf development and photosynthesis were dispersed across the genome. This comprehensive eQTL analysis details the influence of these loci on plant phenotypes and will be a valuable community resource for investigations on the genetic effects of eQTL on phenotypic traits in tomato.
The genetic basis of many qualitative and quantitative phenotypic differences in plants has been associated with sequence polymorphisms and the corresponding changes in gene function. However, differences in the levels of steady-state transcripts, without underlying changes in coding sequences, also significantly influence plant phenotypes. Closely related plant species often have little coding sequence divergence; nonetheless, the related species often develop unique physiological, metabolic, and developmental characteristics, indicating that patterns of gene expression are important in species-level phenotypic variation (Kliebenstein, 2009; Koenig et al., 2013). Phenotypic differences attributed to variations in gene expression patterns have been found to influence disease resistance, insect resistance, phosphate sensing, flowering time, circadian rhythm, and plant development (Kroymann et al., 2003; Werner et al., 2005; Clark et al., 2006; Zhang et al., 2006; Svistoonoff et al., 2007; Chen et al., 2010; Hammond et al., 2011).
Global transcript level changes across precise genetic backgrounds have been used to define expression quantitative trait loci (eQTL) by identifying genomic regions responsible for the variation in transcript levels (Jansen and Nap, 2001; Kliebenstein, 2009; Druka et al., 2010; Chitwood and Sinha, 2013). An eQTL is a chromosomal region that drives variation in gene expression patterns (i.e. steady-state transcript abundance) between individuals of a genetic mapping population and can be treated as a heritable quantitative trait (Brem et al., 2002; Kliebenstein, 2009; Cubillos et al., 2012). Depending upon the proximity to the gene being regulated, eQTL can be classified into two groups: cis-eQTL when the physical location of an eQTL coincides with the location of the regulated gene, and trans-eQTL when an eQTL is located at a different position from the gene being regulated (Kliebenstein, 2009). eQTL studies with the model plant Arabidopsis (Arabidopsis thaliana) showed that cis-eQTL have a significant effect on local expression levels, whereas trans-eQTL often have global influences on gene regulation (DeCook et al., 2006; West et al., 2007; Holloway and Li, 2010). Global eQTL studies also identified transacting eQTL hot spots, which contain master regulators controlling the expression of a suite of genes that act in the same biological process or pathway. For example, eQTL hot spots in Arabidopsis colocate with the ERECTA locus, which has been shown to pleiotropically influence many traits, including those regulating morphology (Keurentjes et al., 2007). Similarly, the rice sub1 locus, which regulates submergence tolerance by controlling internode and leaf elongation, controls the activity of ethylene response factors with significant trans effects (Fukao et al., 2006). In addition, the eQTL identified using pathogen-challenged tissues in barley were enriched for genes related to pathogen response (Chen et al., 2010; Druka et al., 2010). Thus, eQTL analyses have the potential to reveal a genome-wide view of the complex genetic architecture of gene expression regulation and the underlying gene regulatory networks and may also identify master transcriptional regulators.
Cultivated tomatoes, along with their wild relatives, harbor broad genetic diversity and large phenotypic variability (Moyle, 2008; Ranjan et al., 2012). Wide interspecific crosses bring together divergent genomes, and hybridization of such diverse genotypes leads to extensive gene expression alterations compared to either parent. Introgression lines (ILs), developed by crosses between wild relatives and the cultivated tomato to bring discrete wild relative genomic segments into the cultivated background, have proved to be a useful genetic resource for genomics and molecular breeding studies. These ILs may vary in the size of the introgressed region that may range from a few genes to more than a thousand genes. ILs developed from the wild desert-adapted species Solanum pennellii and domesticated Solanum lycopersicum cv M82 have proved to be a useful genetic resource (Eshed and Zamir, 1995; Liu and Zamir, 1999). This population has been successfully used to map numerous QTL for metabolites, enzymatic activity, yield, fitness traits, and developmental features, such as leaf shape, size, and complexity (Frary et al., 2000; Holtan and Hake, 2003; Fridman et al., 2004; Chitwood et al., 2013; Muir et al., 2014). Comparative transcriptomics for the two parents enabled identification of transcript abundance variation potentially underlying trait differences between species (Koenig et al., 2013). However, the genetic regulators of these transcriptional differences between the species still need to be elucidated. Therefore, we used a genomics approach in combination with statistical methods to identify the genetic basis of transcript level variation in tomato using the S. pennellii introgression lines.
Here, we report on a comprehensive transcriptome profile of the ILs, a comparison between the transcript abundance patterns of the ILs and the cultivated M82 background (differential gene expression [DE]), as well as a global eQTL analysis to identify patterns of genetic regulation of transcript abundance in the tomato shoot apex. We have identified more than 7,200 cis- and trans-eQTL in total, which regulate the transcript abundance of approximately 5,300 genes in tomato. Additional analyses using Barnes-Hut t-distributed stochastic neighbor embedding (BH-SNE; van der Maaten, 2013) identified 42 modules revealing novel associations between transcript abundance patterns and biological processes. The transcript abundance patterns under strong genetic regulation are related to plant defense, photosynthesis, and plant developmental traits. We also report important eQTL regulating steady-state transcript abundance pattern associated with leaf number, complexity, and hypocotyl length phenotypes.
RESULTS AND DISCUSSION
Transcriptome Profiling and Global eQTL Analysis
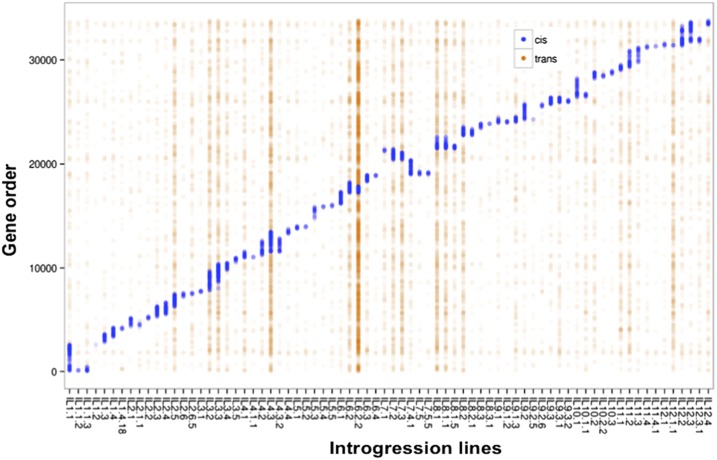
RNA-seq reads obtained from the tomato shoot apex with developing leaves and hypocotyl were used to identify DE genes at the transcript level between each S. pennellii IL and the cultivated M82 (Supplemental Data Set S1). The total number of genes differentially expressed for each IL both in cis (in this population reflecting “local” level regulation either from within a gene itself or other genes in the introgression) and trans, along with the number of genes in the introgression regions, is presented in Figure 1 and Supplemental Table S1. There was a strong correlation between the number of genes in the introgression regions and the number of DE genes in cis (Supplemental Fig. S1A). In contrast, the number of DE genes in trans was poorly correlated with introgression size (Supplemental Fig. S1B). For example, IL12.1.1, despite having one of the smallest introgressions, showed 96% of approximately 500 DE genes regulated in trans (Supplemental Table S1; Supplemental Fig. S2). In contrast, IL1.1 and IL12.3, the ILs with highest number of genes in the introgression regions, showed smaller numbers of total and trans DE genes (Fig. 1; Supplemental Table S1; Supplemental Fig. S2). These examples suggest that specific loci and not the introgression size determine gene regulation in trans. This could, in part, be due to the presence of genes encoding key transcription factors or developmental regulators in the regions with strong influence on transcript expression pattern, as is seen in the ERECTA containing genomic region in Arabidopsis (Keurentjes et al., 2007). A total of 7,943 unique tomato genes were DE between the ILs and cv M82, representing approximately one-third of the approximately 21,000 genes with sufficient sequencing depth to allow DE analysis. There were 2,286 genes, more than one-fourth of unique DE genes between the ILs and cv M82, which showed transgressive expression patterns, that is, those genes were differentially expressed at the transcript level for the IL but not for S. pennellii compared to cv M82 (Supplemental Data Sets S2 and S3). These data suggest that in addition to protein coding differences, transcriptional regulation of less than one-third of all genes accounts for most of the phenotypic and trait differences between the ILs and the cultivated parent.
Figure 1.
Transcriptome profile of the tomato introgression lines. Differentially expressed genes at the transcript level for the ILs compared to cultivated parent M82. The y axis shows all the tomato genes starting from the first gene on chromosome 1 to the last gene on chromosome 12, and the x axis depicts the individual ILs. Genes differentially expressed within the introgression regions (in cis) are shown as blue points and differentially expressed genes in trans (outside) the introgression region are shown as orange points.
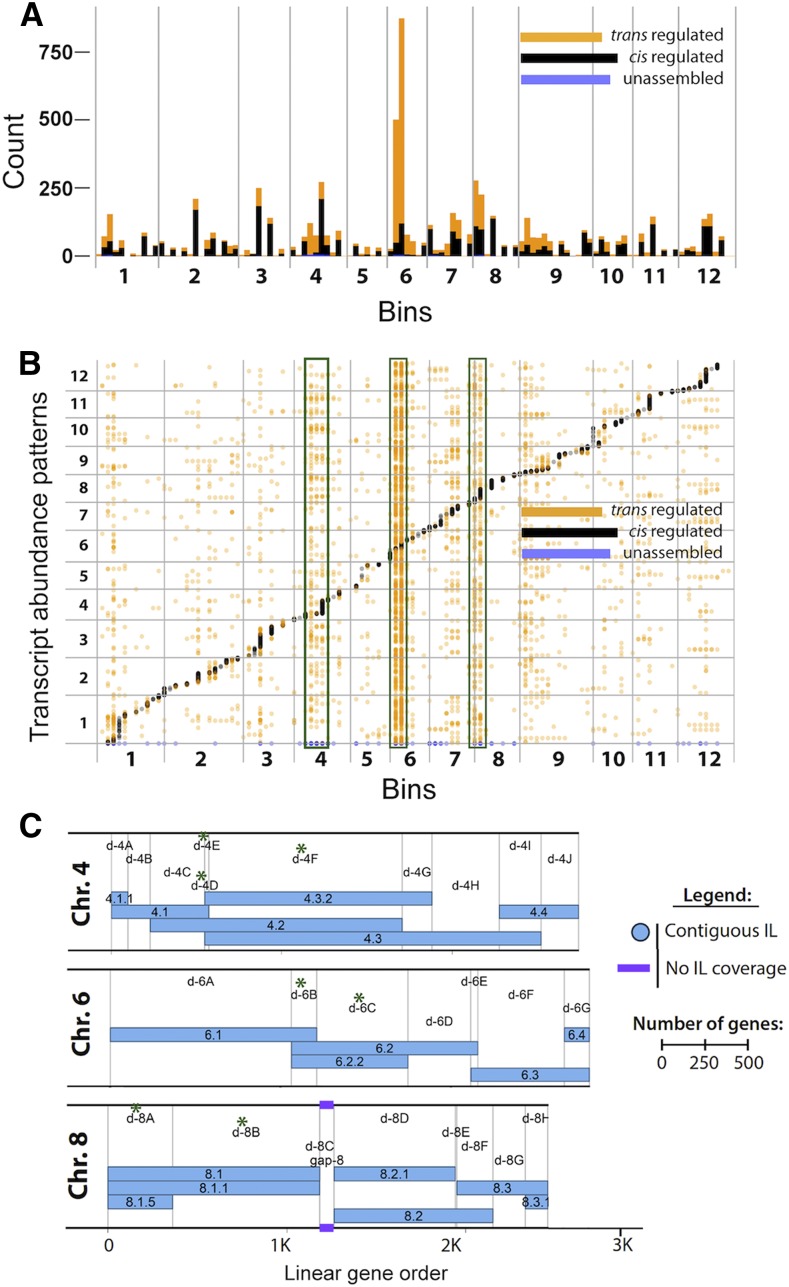
Identifying eQTL localized to subsets of the introgressions, based on overlaps between them, enabled us to narrow down the regions that contain the regulatory loci. This analysis brings us one step closer to identifying potential candidates that influence transcript abundance patterns in tomato. We identified 7,225 significant eQTL (bins) involving 5,289 unique genes across the 74 ILs (Fig. 2; Supplemental Data Set S4). These 7,225 significant eQTL (located in bins) were designated as cis, trans, or chromo0 (unmapped transcripts) as defined in the methods and illustrated in Supplemental Figure S3, and either up or down based on increase or decrease in transcript levels. This correlation resulted in a total of 1,759 cis-up and 1,747 cis-down eQTL, 2,710 transup and 920 transdown eQTL, and 51 chromo0-up and 38 chromo0-down eQTL (Spearman’s rho values; Supplemental Fig. S4; Supplemental Table S2). The majority of genes (>4,000 of 5,289) are under the regulation of a single eQTL (3,134 cis, 1,014 trans, and 19 chromo0; Supplemental Fig. S5). This observation shows the predominance of cis-eQTL for genetic regulation of transcript abundance in the tomato ILs. Similar correlation between transcript level variation and genome-wide sequence divergence within seven Arabidopsis accessions was reported to be due to cis control of a majority of the detected variation (Kliebenstein et al., 2006).
Figure 2.
cis- and trans-eQTL plotted by bin across the 12 chromosomes of S. lycopersicum cv M82. A, Stacked bar graph showing the sum of the number of eQTL mapping to each bin. B, Dotplot showing each eQTL arranged vertically by bin and horizontally by the location of the transcript abundance pattern it regulates. Bins with the largest numbers of trans-eQTL (4D, 4E, 4F, 6B, 6C, 8A, 8B) are highlighted by green boxes. C, Map of chromosomes 4, 6, and 8 showing the overlapping IL regions, which define the bins (modified from Chitwood et al., 2013). Bins with the largest numbers of trans-eQTL are indicated by green asterisks.
The number of genes regulated by eQTL showed large variation across bins. Bins on chromosomes 6, 8, and 4, such as 6B, 6C, 4D, 8A, and 8B, contain predominantly trans-eQTL (Supplemental Data Set S5). In contrast, three bins, 1F, 3I, and 8G, which each contain more than 100 genes, have no significant trans or cis-eQTL and are transcriptionally silent. As expected, bins containing more than 100 significant cis-eQTL are scattered across the genome (Supplemental Data Set S5). The abundance of trans-eQTL on chromosomes 4, 6, and 8 strengthens the idea of trans-eQTL hot spots controlling expression of a large number of transcripts, as reported in other organisms (Brem et al., 2002; Schadt et al., 2003). The resolution in this analysis is at the level of bin, and these significant eQTL likely map to a smaller number of genes within the bins. Functional classification of genes being regulated by these eQTL and phenotypic association with the relevant ILs was undertaken to glean insights into the identity of candidate genes in the bin.
Clustering eQTL Target Genes into Modules Defined by Transcript Abundance Patterns
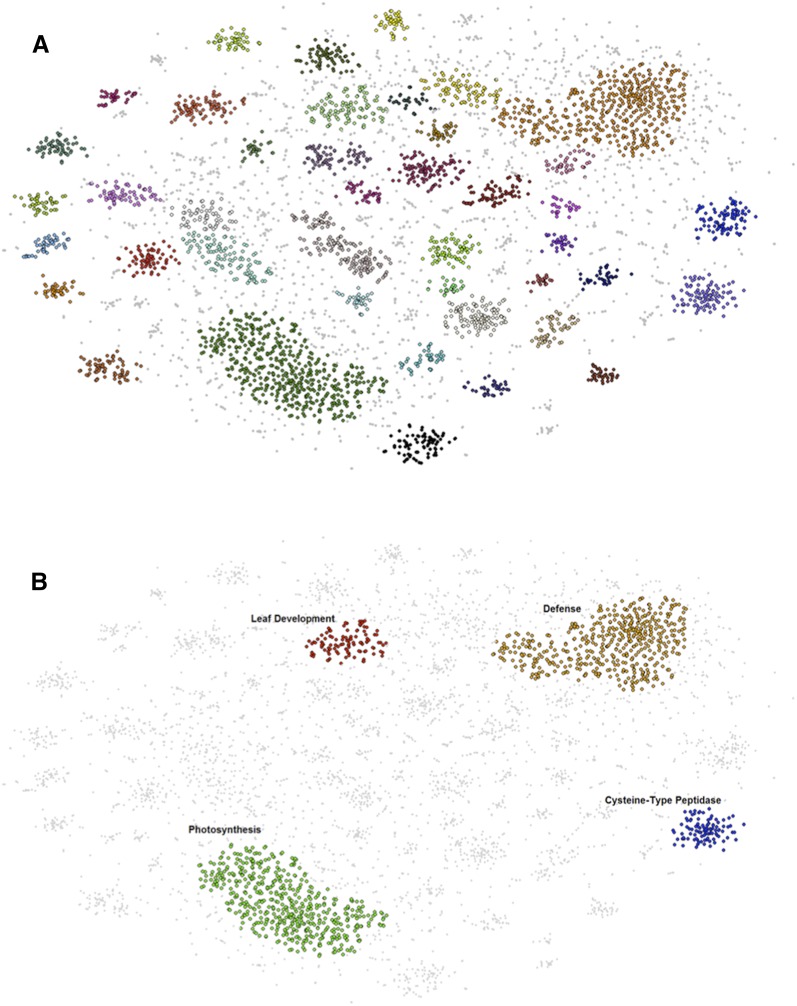
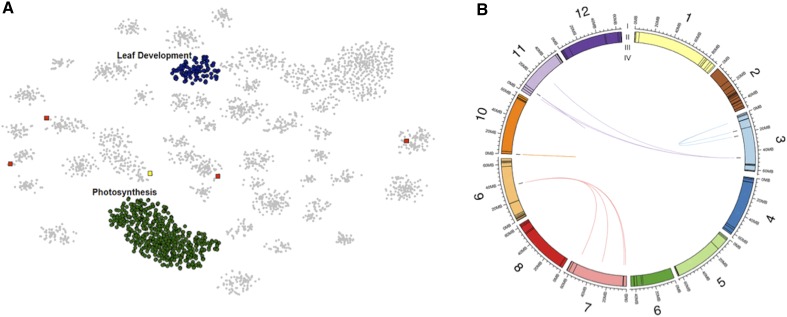
To functionally categorize the eQTL regulated genes, BH-SNE (van der Maaten, 2013) was performed on the 5,289 genes with eQTL to detect novel associations between transcript abundance patterns. This clustering resulted in 42 distinct modules containing 3,592 genes (Fig. 3). Seventeen of these modules had significant Gene Ontology (GO) enrichment (P value < 0.05) with each module consisting of transcript abundance patterns either predominately regulated by cis- or trans-eQTL (Supplemental Table S3). To determine which ILs are important for module regulation, the median transcript abundance value of module genes for each IL was calculated and used to identify ILs with significantly altered module steady-state transcript level.
Figure 3.
BH-SNE 2D mapping of eQTL. A, Forty-two distinct modules identified by DBscan from the mapping generated by BH-SNE analysis. B, The three modules defined as landmark modules: photosynthesis, defense, and Cys-type peptidase activity and the leaf development module’s position within the mapping. Modules are false colored.
Three modules were present in all mappings of the BH-SNE (van der Maaten and Hinton, 2008) determined through iterations of DBscan analysis and GO enrichment and were designated as landmark modules (Fig. 3B; Supplemental Fig. S6; Supplemental Data Set S6; Supplemental Table S3). The largest module had a GO enrichment for photosynthesis and related processes, and significant trans-eQTL scattered widely across the genome with no bin or IL identified as the primary regulating region (Fig. 4B; Supplemental Fig. S6A; Supplemental Data Sets S6 and S7). The second landmark module was enriched for transcript abundance patterns with roles in defense, metabolism, and signaling with the majority of their trans-eQTL mapped to IL6.2 and 6.2.2 (Fig. 4A; Supplemental Fig. S6B; Supplemental Data Sets S6 and S8). The third module, which is enriched for transcript abundance patterns with Cys-type peptidase activity, was predominately composed of genes regulated by cis-eQTL on IL 4.2, 4.3, and 4.3.2 (Bins 4E and 4F; Fig. 4C; Supplemental Fig. S6C; Supplemental Data Sets S6 and S9). A cluster of genes enriched for “peptidase regulation” also emerged from a transcriptome study of leaf development for three tomato species; this cluster was uniquely associated with S. pennellii orthologs at the P5 stage of leaf development, indicating that this species has a unique pattern of gene expression, which involves peptidase regulation (Ichihashi et al., 2014), and may be related to leaf maturation and senescence processes (Díaz-Mendoza et al., 2014).
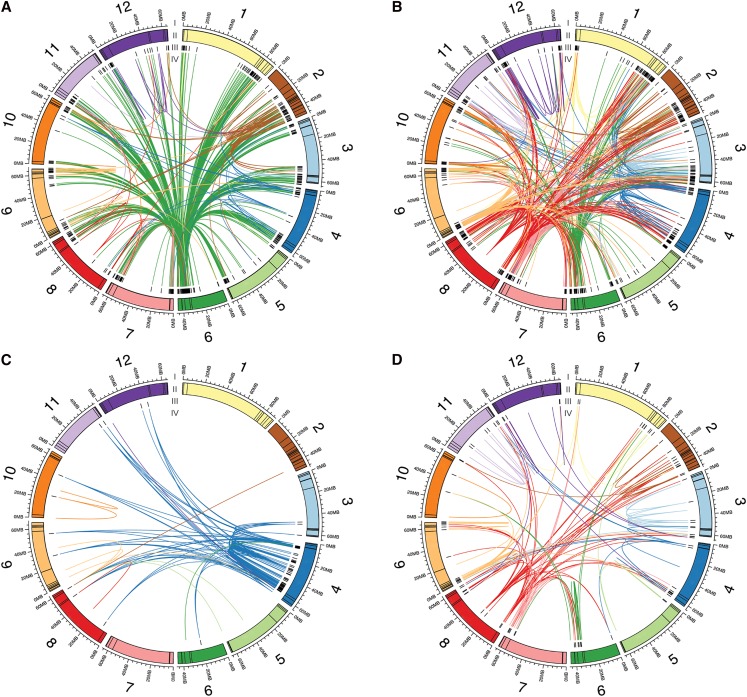
Figure 4.
Connections between eQTL and the genes with correlated transcript level. Each plot includes the genes with eQTL that were clustered together into a module based on transcript level patterns. A, Defense module. B, Photosynthesis module. C, Cys peptidase module. D, Leaf development module. I, The 12 tomato chromosomes in megabases. II, Colored boxes indicate the sizes of each bin. III, Black bars indicate the locations of the genes. IV, Chords connect eQTL to the genes whose transcript level patterns they regulate. Chords are colored by the chromosome location of the eQTL.
Genetic Regulation of Transcriptional Responses Associated with Plant Defense
One of the landmark modules from the clustering analysis was enriched for transcript abundance patterns related to plant defense (Fig. 3B; Supplemental Data Set S8). Therefore, we explored the genetic basis of transcriptional changes associated with plant defense. IL6.2 and IL6.2.2, and associated bins 6B and 6C, in particular, influence of the transcriptional responses of genes associated with plant defense and signaling (Supplemental Data Set S1). The genes showing increased steady-state transcript levels in both ILs compared to cv M82, as well as the genes regulated by the corresponding bins, show enrichment of the GO categories response to stress and stimulus, cell death, defense response, and plant-type hypersensitive response (Supplemental Data Sets S10 and S11). Promoter enrichment analysis for these genes showed enrichment of a W-box promoter motif that is recognized by WRKY transcription factors and influences plant defense response (Supplemental Data Sets S12 and S13; Yu et al., 2001). Both bins, in particular bin 6C, contain genes involved in pathogen, disease, and defense response, such as NBS-LLR resistance genes, WRKY transcription factors, Multidrug resistance genes, Pentatricopeptide repeat-containing genes, Chitinase, and Heat Shock Protein coding genes. This transcriptional response in the ILs is also reflected in the morphology of IL6.2.2; the plants are necrotic and dwarfed (http://tgrc.ucdavis.edu/pennellii_ils.aspx; Sharlach et al., 2013). Previously, a phenotypic study for the chromosome 6 introgression, specifically a 190-kb region in bin 6C, in a pathogen (Xanthomonas perforans)/control experiment was shown to confer hypersensitive response in IL6.2 and 6.2.2 (Sharlach et al., 2013). Taken together, these findings suggest bins 6B and 6C contain master genetic regulators of plant defense response genes, though identification of the causal gene/s that influence many other genes in trans will need further genetic dissection of these bins.
Genetic Regulation of Transcriptional Responses Associated with Leaf Development
Given the striking differences in leaf features between S. pennellii and cv M82 that are manifested in many ILs (Chitwood et al., 2013), the IL population provides an excellent system for determining the extent of genetic regulation of genes controlling leaf development. Previous phenotypic and QTL analyses identified many ILs, such as IL4.3, IL8.1.5, IL8.1.1, and IL8.1, harboring loci regulating leaf and plant developmental traits (Holtan and Hake, 2003; Chitwood et al., 2013; Muir et al., 2014). IL4.3, which harbors loci with the largest contribution to leaf shape and shows larger epidermal cell sizes (Chitwood et al., 2013), exhibited decreased steady-state transcript levels for many genes associated with cell division, such as Cyclin-dependent protein kinase regulator-like protein (CYCA2;3), Cyclin A-like protein (CYCA3;1), and F-box/LRR-repeat protein 2 SKP2A (Supplemental Data Sets S1 and S10). In addition, genes showing differences in transcript levels in IL4.3 were enriched for the promoter motifs MSA (M-specific activators that are involved in M-phase specific transcription) and the E2F binding site (Supplemental Data Set S11). Genes with decreased transcript levels in ILs 8.1.5, 8.1.1, and 8.1, also included genes associated with leaf development and morphology, genes encoding WD-40 repeat family protein LEUNIG, Homeobox-Leu zipper protein PROTODERMAL FACTOR 2, and the transcription factor ULTRAPETALA (Supplemental Data Sets S1 and S10; Abe et al., 2003; Cnops et al., 2004; Carles et al., 2005).
We further investigated the transcript expression dynamics of a set of literature-curated genes related to leaf development (Ichihashi et al., 2014) across the ILs and bins (Supplemental Data Sets S14 and S15). A number of canonical leaf developmental genes such as SHOOT MERISTEMLESS (Solyc02g081120, STM), GROWTH-REGULATING FACTOR 1 (GRF1, Solyc04g077510), ARGONAUTE 10 (AGO10, Solyc12g006790), BELL (BEL1, Solyc08g081400), LEUNIG (Solyc05g026480), and SAWTOOTH 1 (SAW1, Solyc04g079830) were differentially expressed at the transcript level in more than five ILs. At the level of bins, genes involved in leaf development were regulated by eQTL scattered widely across the genome (Fig. 4D). eQTL(bin)-regulation of leaf developmental genes for some of ILs, such as IL 2.1, 4.3, 5.4, 8.1/8.1.1/8.1.5, and 9.1.2 showing strong leaf phenotypes, is summarized in Supplemental Table S4. We then examined the location of literature-curated leaf developmental genes within the identified modules in the BH-SNE mapping (Fig. 3). The highest number of literature-curated leaf developmental genes (108) was located in the photosynthesis module, whereas 19 of these genes were located in the leaf development module (Supplemental Fig. S7B; Supplemental Data Sets S16 and S17), suggesting a relationship between these two modules. Over one-third of the transcript expression patterns in the leaf development module have significant eQTL that map to bins 4D, 8A, and 8B (5.4%, 16.2%, and 15.5%, respectively; Supplemental Data Set S18), suggesting that these bins contain important regulators of leaf development. This enrichment of eQTL for specific bins is also consistent with the strong leaf phenotypes for ILs 4.3, 8.1, 8.1.1, and 8.1.5. Altogether, DE, eQTL, and BH-SNE results indicate that while there is no obvious master regulatory bin for leaf developmental genes, many are under strong genetic regulation by eQTL distributed throughout the genome (Fig. 4D). This observation underscores the highly polygenic regulation of leaf development (Chitwood et al., 2013) as multiple loci, residing in many different chromosomal locations, regulate the expression of key leaf-developmental genes at the transcriptional level.
Genetic Regulation of Transcriptional Responses Associated with Photosynthesis
Since photosynthesis GO terms were enriched for the largest module from the clustering analysis (Fig. 3B) and there was a correlation between photosynthesis and leaf developmental modules (Supplemental Fig. S7B), we examined the genetic regulation of photosynthetic genes by specific ILs and corresponding bins. Genes related to photosynthesis show increased transcript levels across 21 ILs distributed on all chromosomes except chromosome 5 (Supplemental Data Set S10), showing multigenic regulation of photosynthetic traits. Many of these ILs, including 8.1.5, 8.1.1, 8.1, and 4.3, and associated bins showed regulation of genes linked to photosynthesis, chlorophyll biosynthesis, and response to light stimulus (Supplemental Data Sets S10 and S11). This observation indicates that ILs may also differ from each other and from the cultivated M82 background in photosynthetic efficiency. However, no studies, so far, have investigated the photosynthetic phenotype of these ILs.
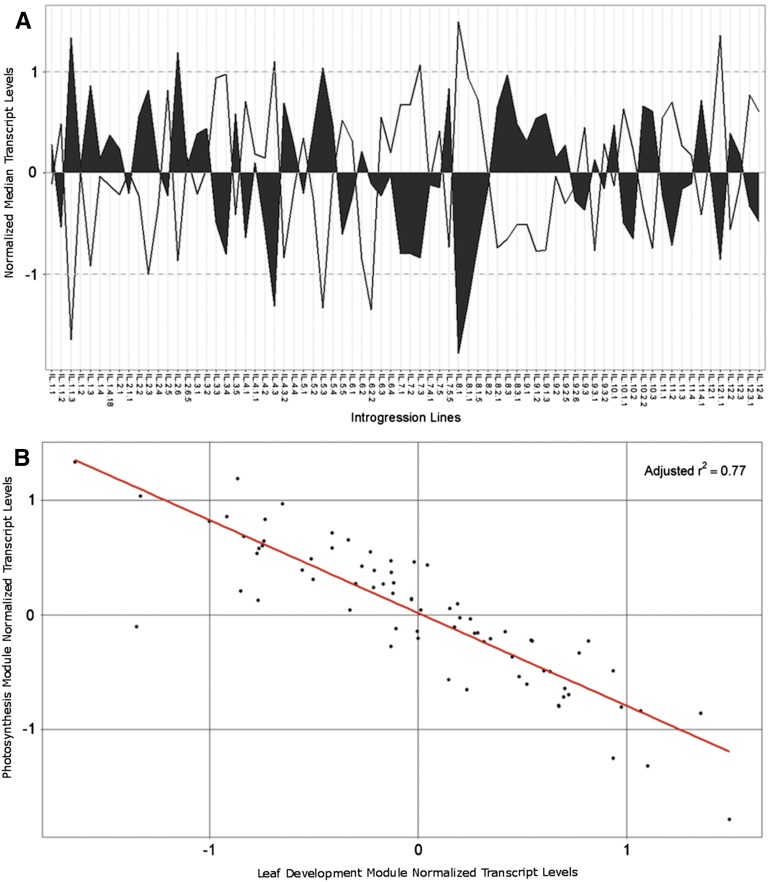
To analyze the relationship between the leaf development and photosynthesis modules, the median transcript abundance value of all genes in each module was compared, resulting in a significant negative correlation (adjusted r2 = 0.77; Fig. 5). This analysis likely reflects the transition from leaf development to leaf maturation captured in our shoot meristem samples. The genes found in the leaf development module may promote developmental processes such as cell division and maintenance or meristematic potential, whereas the leaf development-related genes found in the photosynthesis module may act to suppress this process to allow for maturation of the leaf. The two modules had their most influential eQTL on bins 4D, 8A, and 8B (Supplemental Data Set S6; Supplemental Fig. S7A), suggesting that leaf development and photosynthetic genes not only have transcript levels in opposition but also likely share common regulatory loci. This finding is consistent with the link between leaf development and photosynthesis that we established previously by meta-analysis of developmental and metabolic traits (Chitwood et al., 2013).
Figure 5.
Median transcript level values for leaf development and photosynthesis related modules and expression correlation. A, The median transcript level values of a module for each IL are shown. A consistent negative correlation between photosynthesis and leaf development transcript expression is evident across nearly all 74 ILs. Dashed lines indicate 1 SD from the module mean transcript level. Filled areas represent the median transcript level of the leaf development module, while open areas indicate the photosynthesis module median transcript levels. B, Leaf development median transcript level versus photosynthesis median transcript level values for each IL show a distinct negative correlation with an adjusted R-squared value of 0.77 (calculated by linear regression in R).
Dissection of Identified eQTL to Spatially and Temporally Regulated Development
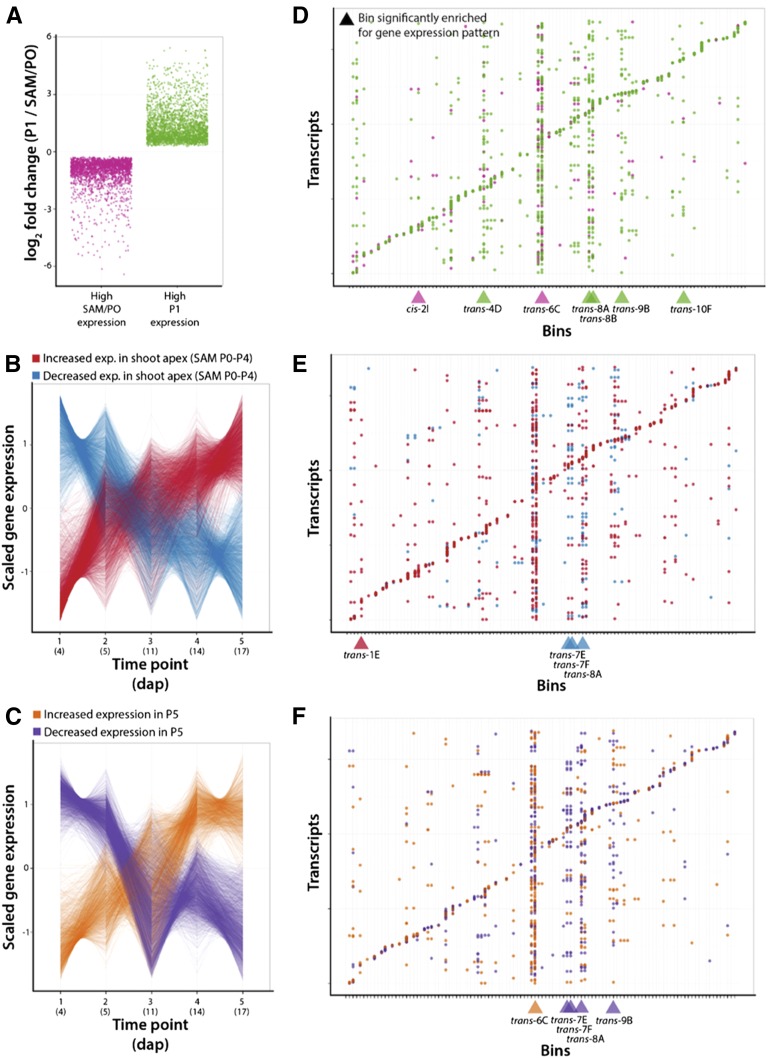
Since the eQTL study used shoot apices that includes the shoot apical meristem (SAM) and developing leaves, we resolved the detected eQTL to specific tissues and temporally regulated development using previous gene expression data. We analyzed transcript abundance in laser microdissected samples representing the SAM + P0 (the incipient leaf) versus the P1 (the first emerged leaf primordium) that represents transcript levels in the meristem (SAM) and the first differentiated leaf (P1; Fig. 6A). We also analyzed hand dissected samples of the SAM + P0-P4 vs. the P5 collected over time (Fig. 6, B and C), representing genes regulated by vegetative phase change (heteroblasty; Chitwood et al., 2015).
Figure 6.
Enriched gene transcript levels that are controlled by specific bins. A, Log fold change values (P1/SAM +P0) for previously identified differentially expressed genes with high transcript levels in the SAM + P0 (magenta) versus P1 (green). B, Scaled transcript level values for previously identified differentially expressed genes with increasing (red) and decreasing (blue) transcript levels over developmental time in the SAM + P0-P4. C, Scaled expression values for previously identified genes with differential levels of transcripts with increasing (orange) and decreasing (purple) transcript levels over developmental time in P5. D, Transcripts (y axis) and bins (x axis) showing the genetic regulation of transcript abundance (eQTL). Colors indicate SAM + P0 (magenta) and P1 (green) transcripts. Bins enriched for genetically regulating genes with specific transcript expression patterns are indicated below with triangles. E, Same as in D, except showing genes with increasing (red) and decreasing (blue) transcript levels over temporal time in the SAM + P0-P4. F, Same as in D, except showing genes with increasing (orange) and decreasing (purple) transcript levels over temporal time in P5. Previously determined transcript abundance patterns are previously published (Chitwood et al., 2015).
Using a bootstrapping approach, we identified bins statistically enriched for genetically regulating genes with previously identified transcript expression patterns (Fig. 6, D and F). Except for one instance (cis-regulated genes with high SAM/P0 expression located in bin 2I), bins enriched for transcript expression patterns represented trans regulation, hinting at predominant regulation of gene expression patterns mediated by transcription factors at the level of transcription. Most SAM/P0 versus P1 enriched bins were enriched for P1 transcript expression (Fig. 6D). We previously showed that genes with high P1 transcript levels are enriched for photosynthetic-related GO terms compared to SAM/P0 genes enriched for transcription, cell division, and epigenetics-related GO terms (Chitwood et al., 2015), suggesting a genetic basis at both a functional and tissue-specific level for genes related to photosynthesis expressed preferentially in the P1 compared to the SAM/P0.
Bins enriched for regulation of genes with temporally dependent steady-state transcript levels were mostly associated with genes with decreasing transcript level over time, for both the SAM + P0-P4 and P5 (Fig. 6, E and F). Interestingly, 3 bins (7E, 7F, and 8A) share enrichment for genes with decreasing transcript levels over time in both the SAM + P0-P4 and P5 (Fig. 6, E and F), suggesting true temporal trans regulation, regardless of tissue, by these loci. Broadly, genes with increasing transcript levels over time are associated with transcription and small RNA GO terms in both the SAM + P0-P4 and P5, whereas decreasing transcript levels over time are associated with translation associated GO terms in the SAM + P0-P4 and photosynthetic activity in the P5 (Supplemental Data Set S19).
Linking Leaf and Hypocotyl Phenotypes to Detected eQTL
To connect detected eQTL with leaf and hypocotyl phenotypes under two different environmental conditions, we correlated transcript abundance with leaf number, leaf complexity (as measured in Chitwood et al., 2014), and hypocotyl length phenotypes of the ILs grown under simulated sun and shade conditions. Significant correlations with transcript abundance patterns were identified for all three phenotypes analyzed under both treatments (Supplemental Table S5). Focusing on a subset of these transcript expression patterns that had associated eQTL enabled us to connect the phenotypes to their regulatory loci (Supplemental Table S5).
Genes negatively correlated with leaf number showed enrichment of leaf development GO terms, whereas positively correlated genes showed enrichment of photosynthesis-related GO terms (Supplemental Fig. S8, A and B; Supplemental Data Set 2 in Chitwood et al., 2014). For the leaf complexity trait, correlations were reversed compared to leaf number (Supplemental Fig. S9, A and B; Supplemental Data Set S20). The transcript levels of these genes associated with leaf number were predominantly regulated by eQTL on chromosomes 7 and 8 (Supplemental Fig. S8, C and D) and those of leaf complexity on chromosomes 4, 7, and 8 (Supplemental Fig. S9, C and D). These results, in combination with DE, eQTL, and BH-SNE, highlight bins on chromosomes 4 and 8 as important genetic regulators of leaf developmental genes.
Five genes were positively correlated with hypocotyl length under simulated shade, and one gene (Solyc10g005120) was negatively correlated with hypocotyl length under both sun and shade (Fig. 7A; Supplemental Data Set S21). eQTL for the positively correlated genes are located on chromosomes 3, 7, and 11, whereas the single cis-eQTL for the negatively correlated gene, Solyc10g005120 (an uncharacterized Flavanone 3-hydroxylase-like gene), was located in bin 10A.1 (Supplemental Fig. S10; Fig. 7B). The transcript is expressed only in IL 10.1, which has the S. pennellii version of the gene and an attenuated shade avoidance response, but is not expressed in IL 10.1.1, which has the M82 version of the gene and a normal shade avoidance response (Supplemental Fig. S11). This indicates that genes in bin 10A, the nonoverlapping regions of 10.1 and 10.1.1, are responsible for the shade avoidance response. Bin 10A includes Solyc10g005120, the one gene negatively correlated with hypocotyl length under both sun and shade.
Figure 7.
eQTL regulation of transcript abundance patterns that correlate with hypocotyl length. A, Forty-two distinct modules identified by DBscan from the eQTL mapping generated by BH-SNE analysis. Modules enriched for genes with leaf development and photosynthesis GO terms are labeled in blue and green, respectively. Genes with transcript levels correlated with hypocotyl length under simulated shade are indicated by squares with positive correlations in red and negative correlations in yellow. B, Genes with transcript levels correlated with hypocotyl length under simulated shade are shown connected to their respective eQTL with chords. I, The 12 tomato chromosomes in megabases. II, Colored boxes indicate the sizes of each bin. III, Black bars indicate the locations of the genes. IV, Chords connect eQTL to the genes whose transcript levels those eQTL regulate. Chords are colored by the chromosome location of the eQTL.
A set of Backcross Inbred Lines (BILs), developed from cv M82 and S. pennellii, provide higher resolution gene mapping with smaller bin sizes (Müller et al., 2016; Fulop et al., 2016). To further explore the role of Solyc10g005120, we used BIL-128, which contains a subregion of bin 10A and has a secondary introgression on chromosome 2 (Supplemental Fig. S10). Influence of the secondary introgression was examined using BIL-033, which shares the introgression on chromosome 2. BIL-128 has an attenuated shade avoidance response, as does 10.1, whereas BIL-033 undergoes a shade avoidance response similar to that of cv M82 (Supplemental Fig. S11). These results rule out the influence of chromosome 2 genes on the attenuated hypocotyl phenotype and confirm the influence of the bin 10A subregion, which includes Solyc10g005120, on the attenuated hypocotyl phenotype (Supplemental Figs. S10 and S11). Solyc10g005120 is an uncharacterized gene, and our observations highlight it as a new candidate regulating shade avoidance responses.
CONCLUSION
In this study, we have investigated the regulation of steady-state transcript levels in the progeny of crosses between cultivated tomato and a wild relative (S. pennellii). A combination of DE, eQTL, and clustering analyses provides a comprehensive picture of genetic regulation of transcript expression patterns in this IL population. Our data show that some biological pathways, such as plant defense, are under the regulation of a limited number of loci with strong effects, whereas loci regulating other pathways, such as photosynthesis and leaf development, are scattered throughout the genome, most likely with weaker individual effects. We correlated transcript levels with leaf and hypocotyl phenotypes and identified the regulatory regions driving these transcript expression patterns. Coupled with comprehensive phenotyping on these ILs, this data set provides a valuable resource to design strategies to achieve a desirable plant phenotype through genetic manipulation of the transcript abundance of key genes or gene modules. Our ability to predict and understand the downstream effects of genes introgressed from wild relatives on gene expression patterns and ultimately phenotypes will be a critical component of crop plant enhancement.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Experimental Design
Plant materials, growth conditions, and experimental design were described in (Chitwood et al., 2013), but are outlined here briefly. Seeds of wild tomato (Solanum pennellii) ILs (Eshed and Zamir, 1995; Liu and Zamir, 1999) and cultivated tomato (Solanum lycopersicum cv M82) were obtained either from Dani Zamir (Hebrew University, Rehovot, Israel) or from the Tomato Genetics Resource Center (University of California, Davis). Seeds were stratified in 50% bleach for 2 min and grown in darkness for 3 d for uniform germination before moving to a growth chambers for 5 d. Six seedlings of each genotype were planted per pot for each replicate. The 76 ILs (and two replicates each of cv M82 and S. pennellii) were divided into four cohorts of 20 randomly assigned genotypes. These cohorts were placed across four temporal replicates in a Latin square design as described in (Chitwood et al., 2013). The seedlings were harvested 5 d after transplanting (13 d of growth in total). Cotyledons and mature leaves >1 cm in total length were excluded, and remaining tissues (including the SAM) above the midpoint of the hypocotyl were pooled, for all individuals in a pot, into 2-mL microcentrifuge tubes and immediately frozen in liquid nitrogen. Two ILs, IL7.4 and IL12.4.1, were not included in the final analysis due to seed contaminations.
Growth Conditions and Quantification of Hypocotyl Length
Seeds 76 ILs (covering the entire genome) along with the parents were sterilized using 70% ethanol, followed by 50% bleach, and finally rinsed with sterile water. This experiment was replicated three times each in 2011 and 2012. Ten to 12 seeds of each IL were sown into Phytatray II (Sigma-Aldrich) containers with 0.5× Murashige and Skoog minimal salt agar. Trays were randomized and seeds germinated in total darkness at room temperature for 48 h. Trays of each IL were randomly assigned to either a sun or shade treatment consisting of 110 μmol PAR with a red to far-red ratio of either 1.5 (simulated sun) or 0.5 (simulated shade) at 22°C with 16-h-light/8-h-dark cycles for 10 d. Three genotypes were excluded from the analyses due to poor germination (IL3.3) or their necrotic dwarf phenotypes (IL6.2, 6.2.2). After 10 d, seedlings were removed from the agar and placed onto transparency sheets containing a moistened kimwipe to prevent dehydration and scanned using an Epson V700 at 8-bit grayscale at 600 dpi. Image analysis was carried out using the software ImageJ (Abramoff et al., 2004).
For hypocotyl length analysis of backcross inbred lines between S. pennellii and S. lycopersicum cv M82, seeds were sterilized in 50% bleach and then rinsed with sterile water. The seeds were then placed in Phytatrays in total dark at room temperature for 72 h and then moved to 16 h light/8 h dark for 4 d. Seedlings were transferred to soil using a randomized design and assigned to either a sun or shade treatment (as described above) for 7 d. Images were taken with an HTC One M8 Dual 4MP camera and hypocotyl lengths measured in ImageJ (Abramoff et al., 2004) using the Simple Neurite Tracer (Longair et al., 2011) plugin.
RNA-Seq Library Preparation and Preprocessing RNA-Seq Data
RNA-seq libraries were prepared and the reads were preprocessed as described in Chitwood et al. (2013) and are outlined here. mRNA isolation and RNA-seq library preparation were performed from 80 samples at a time using a high-throughput RNA-seq protocol (Kumar et al., 2012). The prepared libraries were sequenced in pools of 12 for replicates 1 and 2 (one lane each) and in pools of 80 for replicates 3 and 4 (seven lanes) at the UC Davis Genome Centre Expression Analysis Core using the HiSeq 2000 platform (Illumina). Preprocessing of reads involved removal of low-quality reads (phred score <20), trimming of low-quality bases from the 39 ends of the reads, and removal of adapter contamination using custom Perl scripts. The quality-filtered reads were sorted into individual libraries based on barcodes, and then barcodes were trimmed using custom Perl script.
Read Mapping and Quantification of Transcript Levels
Mapping and normalization were done on the iPLANT Atmosphere cloud server (Goff et al., 2011). S. lycopersicum reads were mapped to 34,727 tomato cDNA sequences predicted from the gene models from the ITAG2.4 genome build (downloadable from ftp://ftp.solgenomics.net/tomato_genome/annotation/ITAG2.4_release/). A pseudo reference list was constructed for S. pennellii using the homologous regions between S. pennellii scaffolds v.1.9 and S. lycopersicum cDNA references above. Using the defined boundaries of ILs, custom R scripts were used to prepare IL-specific references that had the S. pennellii sequences in the introgressed region and S. lycopersicum sequences outside the introgressed region. The reads were mapped using BWA (Li and Durbin, 2009; Roberts and Pachter, 2013) using default parameters except for the following that were changed: bwa aln: -k 1 -l 25 -e 15 -i 10 and bwa samse: -n 0. The bam alignment files were used as inputs for express software to account for reads mapped to multiple locations (Roberts and Pachter, 2013). The estimated read counts obtained for each gene for each sample from express were treated as raw counts for DE analysis. The counts were then filtered in R using the Bioconductor package EdgeR version 2.6.10 (Robinson and Oshlack, 2010) such that only genes that had more than two reads per million in at least three of the samples were kept. Normalization of read counts was performed using the trimmed mean of M-values method (Robinson and Oshlack, 2010), and normalized read counts were used to identify genes that are differentially expressed at the transcript level in each IL compared to cv M82 parent as well as between two parents, S. pennellii and M82. The DE genes for each IL were compared to those between the two parents to identify genes that were differentially expressed for the IL but not for S. pennellii compared to cv M82. Those genes were considered to show transgressive expression pattern at the transcript level for the specific IL, whereas other DE genes were considered to show the transcript expression similar to S. pennellii.
Correlation of Phenotype with Pattern of Steady-State Transcript Levels
Transcript level patterns were correlated with three phenotypes collected from the ILs along with the parents. Normalized estimated read counts with 3 to 4 independent replicates per IL were log2 transformed prior to the analyses. Leaf number and complexity were collected from the ILs as outlined in Chitwood et al. (2014) under both sun and shade treatments. Hypocotyl lengths were measured as detailed above. To test whether the transcript level for a given gene was correlated with a particular phenotype, boostrapping analyses were performed. Transcript levels and phenotype data were randomly permuted (with replacement) using the sample() function against IL and then merged. For each analysis, 1,000 replications were performed and the P values were calculated from the Spearman’s rho value distributions. P values were adjusted for multiple comparisons using the BH correction (Benjamini and Hochberg, 1995). Significant correlations were identified as those with an adjusted P value < 0.05, and the mean rho value (the correlation coefficient) was used to designate the correlation as either positive (positive slope) or negative (negative slope). All analyses were implemented using the statistical software R and custom scripts (R Development Core Team, 2015).
Methods for eQTL Analyses
eQTL mapping analyses were performed to determine whether the transcript level of a gene is correlated with the presence of a specific introgression from S. pennellii into S. lycopersicum cv M82. This correlation was examined at the level of “bin,” with a bin defined as a unique overlapping region between introgressions. Examining eQTL at the bin level enables those eQTL to be mapped to considerably smaller intervals than the ILs themselves (Liu and Zamir, 1999). eQTL mapping analyses were performed on the normalized estimated read counts with 3 to 4 independent replicates per IL, which were log2 transformed prior to the analyses. To test whether the transcript level for a given gene is correlated with the presence of a particular bin, a Spearman’s rank correlation test was used with ties resolved using the midrank method. P values were adjusted for multiple comparisons using the BH correction (Benjamini and Hochberg, 1995). Significant eQTL were identified as those with an adjusted P value < 0.05, and Spearman’s rho (the correlation coefficient) was used to designate the eQTL as up (positive slope) or down (negative slope). Significant eQTL were also designated as cis (defined as local gene regulation within the same bin) if the gene was located on the bin with which it is correlated; trans (distant) if the gene was correlated with a bin that is neither the bin it is on nor a bin that shares an overlapping IL with the correlated bin; or chromo0 if the gene lies in the unassembled part of the genome. When a gene has a designation cis-eQTL, and a secondary correlation was found with a bin that shares an overlapping introgression, this secondary correlation was not designated as an eQTL. When a gene does not have a designated cis-eQTL and a correlation was found with a bin that shares an overlapping introgression, this correlation was designated as a trans-eQTL. All analyses were implemented using the statistical software R and custom scripts (R Development Core Team, 2015).
Methods for eQTL Clustering Analysis
Data Preparation
In preparation for analysis using the Barnes-Hut-SNE algorithm, the data set was log2 transformed. The transcript level for each gene was then normalized across all 74 introgression lines so that the profile had a mean of zero and a sd of one. Normalization of the data allowed for comparison of the relative relationship between each gene expression profile (Bushati et al., 2011).
Barnes-Hut-SNE
t-SNE or t-distributed stochastic neighbor embedding (van der Maaten and Hinton, 2008) is a nonlinear dimensionality reduction method, which faithfully maps objects in high dimensional space (H-space) into low dimensional space (V-space). Crowding is avoided through the long-tailed t-distribution, which forces nonneighbor clusters farther away from each other in V-space than those clusters actually are in H-space (van der Maaten and Hinton, 2008). The exaggerated separation of nonneighboring clusters improves 2D resolution, allowing identification of novel groupings not readily apparent in other clustering methods. However, this method is resource intensive, and with higher dimensionality, the number of genes that can be analyzed is limited. We have used Barnes-Hut-SNE, a newer implementation of t-SNE that greatly increases the speed and number of genes that can be analyzed, for the present analysis (van der Maaten, 2013). Barnes-Hut-SNE accomplishes this efficiency through the use of a Vantage Point tree and a variant of the Barnes-Hut algorithm (van der Maaten, 2013). For clustering, 2D maps were generated using a perplexity of 30 and without the initial PCA step from the Barnes-Hut-SNE R implementation (Rtsne package; Krijthe, 2014). Theta was set to 0.3 based on van der Maaten (2013) to maintain an accurate dimensionality reduction without sacrificing processing speed.
Clustering for Module Selection
The DBscan algorithm (Density Based spatial clustering of applications with noise) was used to select modules from the Barnes-Hut-SNE results (fpc package; Hennig, 2014). This algorithm had the advantage of both selecting modules and removing any genes that fell between modules. The scanning range (epsilon) and minimum seed points (minpts) were selected manually and used to determine if any one point is a member of a cluster based on physical positioning within the mapping relative to neighboring points. A minpts of 25 was used to capture smaller modules on the periphery, and an epsilon of 2.25 was used to avoid the overlapping of internal and closely spaced modules.
Plots
Box plots were generated from normalized transcript abundance values for each module. The ribbon plot was generated from correlated abundance values from leaf development and photosynthesis related modules. These plots were generated using ggplot form the ggplot2 R Package (Wickham, 2009). The median transcript levels of the genes mapped to a module were calculated for each IL and replicated for all modules. Significant ILs were identified as those with a median transcript level >1 sd from the mean of all genes across all ILs in the module.
GO Enrichment Analysis
Differentially expressed genes at the transcript level for individual ILs and genes with significant eQTL were analyzed for enrichment of GO terms at a 0.05 false discovery rate cutoff (goseq Bioconductor package; Young et al., 2010).
Promoter Enrichment Analysis
Promoter enrichment analysis was performed by analyzing the 1,000 bp upstream of the ATG translational start site for genes with significant eQTL using 100 motifs represented in the AGRIS AtTFDB (http://arabidopsis.med.ohio-state.edu/AtTFDB). The Biostrings package was used to analyze the abundance of 100 motifs in groups of genes with significant eQTL compared to motif abundance in promoters of all analyzed genes using a Fisher’s exact test (P < 0.05) with either zero or one mismatch (Ichihashi et al., 2014).
Dissection of eQTL to Different Stages and Time of Development at Shoot Apex
Differentially expressed genes with enriched transcript levels in laser-microdissected SAM/P0 versus P1 samples or hand-dissected samples of the SAM + P0-P4 or P5 sampled over developmental time were obtained from Chitwood et al. (2015). Genes for which a differential expression call could be made (i.e. had enough reads and passed quality filters) were merged with detected eQTL using the merge() function in R (R Development Core Team, 2015). For bootstrapping, cis- and transregulated transcripts were analyzed separately. Merged transcript abundance patterns were randomly permuted (without replacement) using the sample() function against bin identity. For each test, 10,000 permutations were sampled to count the times that a particular transcript expression pattern was assigned to a bin more than the actual count. Resulting frequencies, representing a probability value, were multiple test adjusted using the Benjamini-Hochberg (Benjamini and Hochberg, 1995) method using p.adjust(). Those bins with multiple test adjusted P values < 0.05 were analyzed further using visualizations created with ggplot2 (Wickham, 2009).
Accession Numbers
The quality filtered, barcode-sorted, and trimmed short read data set, which was used to get the normalized read counts and for DE analysis, was deposited to the NCBI Short Read Archive under accessions SRR1013035 to SRR1013343 (Bioproject accession SRP031491).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Number of genes in the introgression region for an IL and the number of differentially expressed genes at the transcript level compared to cv M82.
Supplemental Figure S2. Histograms for differentially expressed genes at the transcript level for the ILs.
Supplemental Figure S3. eQTL and the transcript abundance patterns they regulate.
Supplemental Figure S4. cis- and trans-eQTL.
Supplemental Figure S5. Frequency and distribution of differentially expressed genes at the transcript level for the IL population at the introgression and the bin level.
Supplemental Figure S6. Box plots of the normalized transcript levels for the three landmark modules.
Supplemental Figure S7. Normalized transcript levels of the leaf development module and genes associated with leaf development within the mapping.
Supplemental Figure S8. eQTL regulation of transcript abundance patterns that correlate with leaf number.
Supplemental Figure S9. eQTL regulation of transcript abundance patterns that correlate with leaf complexity.
Supplemental Figure S10.http://www.plantphysiol.org/cgi/content/full/pp.16.00289/DC1 Distributions of introgressions from S. pennellii into S. lycopersicum cv M82.
Supplemental Figure S11.http://www.plantphysiol.org/cgi/content/full/pp.16.00289/DC1 Tomato hypocotyl length under sun and shade treatments.
Supplemental Table S1. Number of DE genes at the transcript level in cis, trans, and the total number of DE genes for the ILs along with number of genes in the introgression region.
Supplemental Table S2. Correlation coefficients (Spearman’s rho) for significant eQTLs.
Supplemental Table S3. GO enrichment and cis or trans regulation of the 42 identified modules.
Supplemental Table S4. Leaf developmental phenotypes of selected ILs and genetic effects of eQTL (bin) on transcript levels of candidate genes.
Supplemental Table S5. Significant correlations between transcript expression patterns and phenotypes.
Supplemental Data Set S1. List of differentially expressed genes at the transcript level.
Supplemental Data Set S2. Transgressive expression of genes at the transcript level among ILs.
Supplemental Data Set S3. Genes with transgressive transcript level.
Supplemental Data Set S4. All genes with significant eQTL.
Supplemental Data Set S5. Number of eQTL and genes per bin.
Supplemental Data Set S6. Number of eQTL and the bin on which those eQTL reside for each of the landmark modules and leaf development module.
Supplemental Data Set S7. Photosynthesis module gene list.
Supplemental Data Set S8. Defense module gene list.
Supplemental Data Set S9. Cysteine-type module gene list.
Supplemental Data Set S10. GO enrichment for DE genes.
Supplemental Data Set S11. GO enrichment for eQTL.
Supplemental Data Set S12. Promoter motif enrichment for DE genes.
Supplemental Data Set S13. Promoter motif enrichment for trans-eQTL.
Supplemental Data Set S14. Differentially expressed leaf developmental genes at the transcript level.
Supplemental Data Set S15. Curated list of leaf developmental genes with eQTLs.
Supplemental Data Set S16. Literature-curated plus list of leaf development genes present in the leaf development modules.
Supplemental Data Set S17. Literature-curated plus list of leaf development genes that are present in all modules.
Supplemental Data Set S18. Leaf development module gene list.
Supplemental Data Set S19. GO enrichment for bins statistically enriched for transcripts expressed spatio-temporally across tissues.
Supplemental Data Set S20. Leaf complexity phenotype of ILs.
Supplemental Data Set S21. Hypocotyl phenotype of ILs.
Supplementary Material
Acknowledgments
We thank Lauren R. Headland, Jason Kao, and Paradee Thammapichai for help in generating plant materials. We also thank Mike Covington for his advice on bioinformatic analyses. We thank the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley (supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303) and computational resources/cyber infrastructure provided by the iPlant Collaborative (www.iplantcollaborative.org), funded by the National Science Foundation (Grant DBI-0735191).
Glossary
- BIL
Backcross Inbred Line
- DE
differential gene expression
- eQTL
expression Quantitative Trait Loci
- GO
Gene Ontology
- IL
introgression line
- SAM
shoot apical meristem
References
- Abe M, Katsumata H, Komeda Y, Takahashi T (2003) Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130: 635–643 [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L (2002) Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Bushati N, Smith J, Briscoe J, Watkins C (2011) An intuitive graphical visualization technique for the interrogation of transcriptome data. Nucleic Acids Res 39: 7380–7389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles CC, Choffnes-Inada D, Reville K, Lertpiriyapong K, Fletcher JC (2005) ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development 132: 897–911 [DOI] [PubMed] [Google Scholar]
- Chen X, Hackett CA, Niks RE, Hedley PE, Booth C, Druka A, Marcel TC, Vels A, Bayer M, Milne I, et al. (2010) An eQTL analysis of partial resistance to Puccinia hordei in barley. PLoS One 5: e8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Kumar R, Headland LR, Ranjan A, Covington MF, Ichihashi Y, Fulop D, Jiménez-Gómez JM, Peng J, Maloof JN, Sinha NR (2013) A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell 25: 2465–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Kumar R, Ranjan A, Pelletier JM, Townsley BT, Ichihashi Y, Martinez CC, Zumstein K, Harada JJ, Maloof JN, Sinha NR (2015) Light-induced indeterminacy alters shade-avoiding tomato leaf morphology. Plant Physiol 169: 2030–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Ranjan A, Kumar R, Ichihashi Y, Zumstein K, Headland LR, Ostria-Gallardo E, Aguilar-Martínez JA, Bush S, Carriedo L, et al. (2014) Resolving distinct genetic regulators of tomato leaf shape within a heteroblastic and ontogenetic context. Plant Cell 26: 3616–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Sinha NR (2013) A census of cells in time: quantitative genetics meets developmental biology. Curr Opin Plant Biol 16: 92–99 [DOI] [PubMed] [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J (2006) A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet 38: 594–597 [DOI] [PubMed] [Google Scholar]
- Cnops G, Jover-Gil S, Peters JL, Neyt P, De Block S, Robles P, Ponce MR, Gerats T, Micol JL, Van Lijsebettens M (2004) The rotunda2 mutants identify a role for the LEUNIG gene in vegetative leaf morphogenesis. J Exp Bot 55: 1529–1539 [DOI] [PubMed] [Google Scholar]
- Cubillos FA, Coustham V, Loudet O (2012) Lessons from eQTL mapping studies: non-coding regions and their role behind natural phenotypic variation in plants. Curr Opin Plant Biol 15: 192–198 [DOI] [PubMed] [Google Scholar]
- DeCook R, Lall S, Nettleton D, Howell SH (2006) Genetic regulation of gene expression during shoot development in Arabidopsis. Genetics 172: 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Mendoza M, Velasco-Arroyo B, González-Melendi P, Martínez M, Díaz I (2014) C1A cysteine protease-cystatin interactions in leaf senescence. J Exp Bot 65: 3825–3833 [DOI] [PubMed] [Google Scholar]
- Druka A, Potokina E, Luo Z, Jiang N, Chen X, Kearsey M, Waugh R (2010) Expression quantitative trait loci analysis in plants. Plant Biotechnol J 8: 10–27 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88 [DOI] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop D, Ranjan A, Ofner I, Covington MF, Chitwood DH, West D, Ichihashi Y, Headland L, Zamir D, Maloof JN, et al. (2016) A new advanced backcross tomato population enables high resolution leaf QTL mapping and gene identification. G3 g3.116.030536, doi/10.1534/g3.116.030536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, Matasci N, Wang L, Hanlon M, Lenards A, et al. (2011) The iPlant Collaborative: Cyberinfrastructure for Plant Biology. Front Plant Sci 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Mayes S, Bowen HC, Graham NS, Hayden RM, Love CG, Spracklen WP, Wang J, Welham SJ, White PJ, et al. (2011) Regulatory hotspots are associated with plant gene expression under varying soil phosphorus supply in Brassica rapa. Plant Physiol 156: 1230–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig C. (2014) FPC: flexible procedures for clustering. R Package Version: 2.1-9
- Holloway B, Li B (2010) Expression QTLs: applications for crop improvement. Mol Breed 26: 381–391 [Google Scholar]
- Holtan HE, Hake S (2003) Quantitative trait locus analysis of leaf dissection in tomato using Lycopersicon pennellii segmental introgression lines. Genetics 165: 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Aguilar-Martínez JA, Farhi M, Chitwood DH, Kumar R, Millon LV, Peng J, Maloof JN, Sinha NR (2014) Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc Natl Acad Sci USA 111: E2616–E2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, Nap JP (2001) Genetical genomics: the added value from segregation. Trends Genet 17: 388–391 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, Terpstra IR, Garcia JM, van den Ackerveken G, Snoek LB, Peeters AJ, Vreugdenhil D, Koornneef M, Jansen RC (2007) Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci USA 104: 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D. (2009) Quantitative genomics: analyzing intraspecific variation using global gene expression polymorphisms or eQTLs. Annu Rev Plant Biol 60: 93–114 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, West MA, van Leeuwen H, Kim K, Doerge RW, Michelmore RW, St Clair DA (2006) Genomic survey of gene expression diversity in Arabidopsis thaliana. Genetics 172: 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D, Jiménez-Gómez JM, Kimura S, Fulop D, Chitwood DH, Headland LR, Kumar R, Covington MF, Devisetty UK, Tat AV, et al. (2013) Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci USA 110: E2655–E2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijthe J. (2014) Rtsne: T-distributed Stochastic Neighbor Embedding using Barnes-Hut implementation. R Package Version: 0.9
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T (2003) Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci USA (Suppl 2) 100: 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Ichihashi Y, Kimura S, Chitwood DH, Headland LR, Peng J, Maloof JN, Sinha NR (2012) A high-throughput method for illumina RNA-Seq library preparation. Front Plant Sci 3: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YS, Zamir D (1999) Second generation L. pennellii introgression lines and the concept of bin mapping. Tomato Genet Coop Rep 49: 26–30 [Google Scholar]
- Longair MH, Baker DA, Armstrong JD (2011) Simple neurite tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27: 2453–2454 [DOI] [PubMed] [Google Scholar]
- Moyle LC. (2008) Ecological and evolutionary genomics in the wild tomatoes (Solanum sect. Lycopersicon). Evolution 62: 2995–3013 [DOI] [PubMed] [Google Scholar]
- Muir CD, Pease JB, Moyle LC (2014) Quantitative genetic analysis indicates natural selection on leaf phenotypes across wild tomato species (Solanum sect. Lycopersicon; Solanaceae). Genetics 198: 1629–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller NA, Wijnen CL, Srinivasan A, Ryngajllo M, Ofner I, Lin T, Ranjan A, West D, Maloof JN, Sinha NR, et al. (2016) Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat Genet 48: 89–93 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Ranjan A, Ichihashi Y, Sinha NR (2012) The tomato genome: implications for plant breeding, genomics and evolution. Genome Biol 13: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pachter L (2013) Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 10: 71–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. (2003) Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302 [DOI] [PubMed] [Google Scholar]
- Sharlach M, Dahlbeck D, Liu L, Chiu J, Jiménez-Gómez JM, Kimura S, Koenig D, Maloof JN, Sinha N, Minsavage GV, et al. (2013) Fine genetic mapping of RXopJ4, a bacterial spot disease resistance locus from Solanum pennellii LA716. Theor Appl Genet 126: 601–609 [DOI] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- van der Maaten L. (2013) Barnes-Hut-SNE. https://arxiv.org/pdf/1301.3342.pdf
- van der Maaten L, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9: 2579–2605 [Google Scholar]
- Werner JD, Borevitz JO, Warthmann N, Trainer GT, Ecker JR, Chory J, Weigel D (2005) Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc Natl Acad Sci USA 102: 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MA, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, St Clair DA (2007) Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009) ggplot2: elegant graphics for data analysis. Springer, New York
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z (2001) Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13: 1527–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Fetch T, Nirmala J, Schmierer D, Brueggeman R, Steffenson B, Kleinhofs A (2006) Rpr1, a gene required for Rpg1-dependent resistance to stem rust in barley. Theor Appl Genet 113: 847–855 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.