Abstract
BACKGROUND
Forkhead box protein A2 (FOXA2) plays an important in development, cellular metabolism and tumorigenesis. The Cancer Genome Atlas (TCGA) identified a modest frequency of FOXA2 mutations in endometrioid endometrial cancers (EEC). The current study sought to determine the relationship between FOXA2 mutation and clinicopathologic features in EEC and FOXA2 expression.
METHODS
Polymerase chain reaction (PCR) amplification and sequencing were used to identify mutations in 542 EEC. Western blot, quantitative reverse transcriptase PCR (qRT-PCR) and immunohistochemistry (IHC) were used to assess expression. Methylation analysis was performed using combined bisulfite restriction analysis (COBRA) and sequencing. Chi-squared, Fisher's exact, student's t- and log-rank tests were performed.
RESULTS
Fifty-one mutations were identified in 49 tumors (9.4% mutation rate). The majority of mutations were novel, loss of function (LOF) (78.4%) mutations, and most disrupted the DNA-binding domain (58.8%). Six recurrent mutations were identified. Only two tumors had two mutations and there was no evidence for FOXA2 allelic loss. Mutation status was associated with tumor grade and not associated with survival outcomes. Methylation of the FOXA2 promoter region was highly variable. Most tumors expressed FOXA2 at both the mRNA and protein level. In those tumors with mutations, the majority of cases expressed both alleles.
CONCLUSION
FOXA2 is frequently mutated in EEC. The pattern of FOXA2 mutations and expression in tumors suggests complex regulation and a haploinsufficient or dominant-negative tumor suppressor function. In vitro studies may shed light on how mutations in FOXA2 affect FOXA2 pioneer and/or transcription factor functions in EEC.
Introduction
Endometrial cancer (EMCA) is the most common gynecologic malignancy in the United States with over 60,000 new cases per year1. The SEER program and National Center for Health Statistics have shown that the rate of endometrial cancer is rising, particularly in obese and minority populations1,2. There are two distinct pathologic classifications of endometrial cancer (EMCA). The more common, less aggressive type I EMCA or EEC, is associated with younger onset, obesity and earlier stage. Although overall prognosis is favorable1, there are important challenges with identifying early stage EEC patients who would benefit from recurrence risk reduction with adjuvant therapy. Understanding the cellular mechanisms and molecular biology of EEC is central to its prevention and treatment.
Recent genomic discovery efforts of The Cancer Genome Atlas (TCGA) shed light on the molecular complexity of EEC and pointed to novel mutational targets3. Previous work revealed strong associations between EEC and PTEN inactivation, diploid chromosomal content, microsatellite instability, as well as polymerase ε (POLE) mutations3,4. The TCGA reported a modest frequency of Forkhead box A2 gene (FOXA2) mutations in EECs (5.2%). It is noteworthy that among the many cancer types investigated to date, uterine carcinomas and carcinosarcomas have some of the highest rates of FOXA2 mutation (Supplemental Fig.1). FOXA2 is a transcription factor, pioneer factor, and nuclear receptor regulator important in development, cellular metabolism, and tumorigenesis. FOXA2's role in EMCA is not well understood. Work in other malignancies suggests that the effects of FOXA2 abnormalities are highly variable and tissue-specific/context dependent. For example, low FOXA2 expression is associated with increased metastasis in colon cancer, epithelial-to-mesenchymal transition and increased cell migration in lung cancers, and decreased overall survival in gastric cancer5-10. Alternatively, increased expression is associated with esophageal Barrett's metaplasia, dysplasia, and adenocarcinoma11. In the endometrium, murine models suggest that FOXA2 is necessary for deep uterine gland formation12. FOXA2 expression is increased in endometrial complex atypical hyperplasia and associated with hormone receptor regulation in the endometrium13-15. These studies and TCGA results suggest a potential role for FOXA2 in endometrial cancer.
We previously performed whole exome sequencing of eight early stage endometrial cancer patients with poor outcomes. Our analysis (unpublished data) suggested a higher FOXA2 mutation (37.5%, 3 of 8) than reported by TCGA. In the current study we sought to determine the spectrum and frequency of FOXA2 mutations in a large EEC cohort and to determine the relationships between mutation status and molecular and clinicopathologic characteristics.
Methods
Study Population
FOXA2 mutation analysis was completed for 542 EECs (see supplemental extended methods)16-18. Institutional review board (IRB) approval for analyses of these specimen as well as additional fresh frozen EEC samples was approved by The Ohio State University (Columbus, OH) Office of Responsible Research Practices IRB.
Mutation and Methylation Testing
Polymerase chain reaction (PCR) amplification (AmpliTaq Gold DNA Polymerase; Applied Biosystems Inc., Foster City, CA) and Sanger sequencing were used to assess for FOXA2 gene mutations. Both the short (NM_153675) and the long isoform (NM_021784) that differ by 6 amino acids were analyzed, with mutations referencing the the long isoform. Exon 1 and 2 code for residues 1-29 and 30-463, respectively. High quality sequence data for exon 2 were obtained for all cases and for 382 tumors for exon 1. All mutations were confirmed to be somatic by testing the matched normal DNA (blood). Loss of function mutations were defined as frameshift, nonsense, and splice site variants changes. Cloning of PCR products (Topo®TA Cloning Kit; Invitrogen, Carlsbad, CA) was used to confirm cis or trans configuration for two-hit mutants as described in supplemental extended methods.
Promoter methylation in endometrial cancer was analyzed using bisulfite converted DNA. EMBOSS CpGblot identified four CpG islands in the proximal promoter region. Four cell line DNA (A549, HepG2, RL952, AN3CA), primary endometrioid endometrial cancer DNA, and matched blood control DNA were bisulfite converted using EZ DNA Methylation-Gold Kit (Zymo Research). Restriction digests were with: TaqI (T/CGA), BstUI (CG/CG), BsiEI (CGRY/CG), and HpyCH4IV(A/CGT). Digestion products were resolved on polyacrylamide gel with appropriate controls. Cloning and Sanger sequencing was used to confirm and refine COBRA findings.
Expression analysis
FOXA2 transcription was assessed using qRT-PCR assays. RNA prepared from primary tumor samples was DNAse treated (DNAse I; New England Biolabs, Ipswich, MA) and reverse transcribed (Superscript III First-Strand Synthesis System; Invitrogen, Carlsbad, CA). SYBR Green (Thermo Fisher Scientific Inc., Waltham, MA) qRT-PCR assays were performed on using Bio-Rad CFX96 Real-Time System. Each assay was completed three independent times in quadruplicates. High Fidelity Taq DNA polymerase (Phusion High-Fidelity DNA Polymerase; Thermo Fisher Scientific Inc., Waltham, MA) was used for qRT-PCR experiments with primer pair E1-E2 (see supplemental extended methods). Given lack of expression of FOXA2 in endometrial cancer cell lines, HepG2 and A549, two known high FOXA2 expressers without FOXA2 mutations, were used as controls throughout experiments. Sample expression was normalized to HepG2 transcript levels. The average of the means and the variability between independent experiments was calculated and histoplot comparisons created.
Western blot and immunohistochemistry (IHC) were used to assess protein expression. Protein lysates were prepared from cell lines and primary tumors (see supplemental extended methods). Samples were resolved with a 4-20% gradient polyacrylamide gel (Mini-PROTEAN®TGX™, Bio-Rad). Sample loading quantity of protein adjusted based on vinculin protein expression. Membranes were probed using anti-FOXA2 C-terminal specific monoclonal antibody (Abcam EPR4466, detects amino acid 350-450, 1:1000 dilution), anti-FOXA2 N-terminal polyclonal antibody (Millipore 07-633, detects amino acid 7-86, 1:1000 dilution), anti-vinculin polyclonal antibody (Sigma V4139, 1:2000 dilution). Membranes were then incubated with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (GE Healthcare Life Sciences NA934V, Buckinghamshire, UK). ECL (HyGLO™ Quick Spray Chemiluminescent HRP Antibody Detection Reagent, Denville Scientific, Inc.) and FEMTO (SuperSignal® West Femto Masimum Sensitivity Substrate, Thermo Fisher Scientific, Inc.) chemiluminescence were used. IHC was carried out to detect the expression of FOXA2 in mouse endometrium, human endometrium, and both wild-type and mutant endometrial tumors. Anti-FOXA2 monoclonal antibody (Abcam EPR4466) was used for detection of FOXA2. IHC was performed by the Solid Tumor Translational Science Shared Resource team (The Ohio State University) (see supplemental methods).
Statistical analyses
Analyses were based on available demographic and clinicopathologic data as well as publically available mutation and polymorphism data from TCGA3, Ensembl19, cbioportal.org20, and the UCSC Genome Browser (Genome Bioinformatics Group, Santa Cruz, CA). Chi-squared or Fisher's exact tests were used to compare categorical data. T-tests were used for continuous variables. All p-values were two-sided.
Kaplan-Meier estimates were calculated to compare progression-free and overall survival using log-rank method. OS was defined as time from surgical intervention to death from any cause. Patients alive (with or without disease) at time of last follow-up were censored. PFS was characterized as time to recurrence or death from disease. Patients alive without disease at time of last follow-up and patients whose deaths were unrelated to their EC were censored. Perioperative deaths were excluded. Prism 5 (Graphpad Software, Inc., La Jolla, CA) and STATA (STATACorp LP, College Station, TX) were used for statistical calculations.
Results
Frequent frame-shift and stop mutations in FOXA2 in EEC
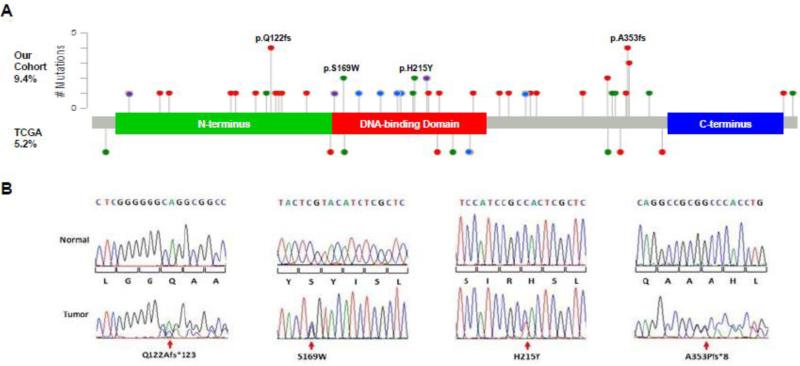
Forty-nine of 542 primary EECs investigated had FOXA2 mutations for a mutation rate of 9.4% (Fig.1A, Supplemental Table 1). Overall coverage of coding sequences was 93.7%. No mutations in the first exon were identified among the 381 cases successfully analyzed. Exon 2 (flanking intron 1 and coding sequence) was analyzed fully in all 542 tumor specimen. Forty-two of 44 different mutations identified were novel. Sequencing the matched normal DNA proved variants were somatic mutations (Fig.1B). The majority of sequence alterations resulted in frameshifts or stops. Fifteen of 51 mutations (29%) were in the N-terminal region. Thirteen of these mutations encode truncated proteins lacking the DNA-binding and C-terminal histone binding domains21. An additional eight tumors carried nonsense or frameshift mutations within the DNA-binding domain. The preponderance of N-terminal and DNA-binding domain mutations is markedly different than the spectrum of mutations reported by TCGA3. Only one of the nonsense mutations identified, p.Q179* (COSM1025156), has been previously reported in endometrial cancer.
Fig. 1. Pattern of FOXA2 somatic mutations in EECs.
(A) Lollipop plot of FOXA2 mutations identified in 542 primary tumors (above protein domain diagram) and as reported by TCGA (below). Red, frameshift mutation; Green, missense mutation; Blue, nonsense mutation; Purple, in-frame deletion. Note: for purposes of comparison, all TCGA mutants were adjusted from previously reported FOXA2 NP_710141 into FOXA2 NP_068556 isoform. (B) Sequence chromatograms for representative strand-slippage, recurrent missense, and novel recurrent hotspot frameshift mutations demonstrating retention of wild-type sequences.
Six recurrent mutations were identified. Two were missense variants (two instances of each mutation). Both p.S169W and p.H215Y are within the DNA-binding domain and are at residues at which a different substitution has been reported in EEC (p.S169L, COSM1025157) or the same mutation (p.H215Y) seen in a lung adenocarcinoma tumor (COSM377059) (Fig.1A, B). There were four recurrent frameshift mutations including three frameshifts in short mononucleotide repeat regions and a deletion of 20 base pairs (Fig.1A, B, Supplemental Table 1). The C6 and G6 insertion/deletions seen, c.363insG, c.363delG, and c.1016dup, are consistent with strand slippage mutations associated with microsatellite instability (MSI)22. The recurrent mutation, c.1056_1075del, was however, seen in both MSI and microsatellite stable (MSS) tumors. No single “hotspot” mutation was identified. However, there were eight frameshifts involving the same 8-20 bases corresponding to amino acids 352-354, including c.1056_1075del, suggesting a potential hotspot region.
Frameshift and nonsense mutations made up 74.5% of the mutations, all of which would result in truncated protein products. A splice-site variant, two in-frame deletions, and missense mutations made up the remaining 25.5% (Supplemental Table 1). The splice-site mutant, p.G30_M52del, is a large deletion of 172 bases including 23 amino acids of exon 2. The two in-frame deletions both map to the DNA-binding domain. The p.F223del (phenylalanine deletion) mutation involves a highly conserved residue. The p.H163_T192del removes 30 amino acids within the DNA-binding domain. The majority of missense mutations are predicted phosphoacceptor alterations or predicted to be damaging or deleterious (Supplemental Table 2).
The large percentage of predicted loss of function mutations suggests FOXA2 is a tumor suppressor gene in the endometrium. However, only two tumors had two mutations, and as such FOXA2 is unlike typical two-hit tumor suppressor genes (Supplemental Table 1). Tumor 1780 had a c.363dup and a c.880_902del. Tumor 1829 had an c.148insA and a c.1056_1075del. Cloning and sequencing confirmed that the mutations in both were in trans (Supplemental Fig.2). None of the tumors with FOXA2 mutations had apparent homozygosity (allelic loss) or revealed loss of the wild-type allele in cDNAs.
FOXA2 Mutations and Mutator Status
Given the preponderance of frameshifts, we predicted that FOXA2 mutations would be more common in tumors with defective DNA repair (MSI cases). The rate of mutations in tumors with MSI was double the rate for tumors with normal mismatch repair, 13.7% vs. 6.2% (Supplemental Fig.3). This difference is consistent with the overall increased rate of mutation in MSI-positive EEC3. However, the pattern of mutations was similar for MSI and MSS cases. In fact, frameshifts were more common in MSS than MSI tumors. Thus, the frameshift mutations seen in MSI tumors are unlikely to be MSI strand slippage mutational noise. FOXA2 mutations were identified in 6.7% of cases with somatic DNA polymerase ε (POLE) exonuclease domain mutations (2/30) (Supplemental Table 1). Both mutations had guanine and cytosine substitutions typical of the POLE mutation signature; tumor 1269 had c.1027C>T and tumor 2112 had c.1101G>T3,4. These mutations were of uncertain functional significance based on prediction algorithms (Supplemental Table 2). Although our data indicate FOXA2 is not particularly vulnerable to mutation in POLE mutant tumors, the mutations identified in the two POLE-mutant tumors may represent mutational noise rather than selected defects.
FOXA2 expression in endometrial cancers and normal endometrium
It has been shown previously that FOXA2 is expressed in the deep glands of the mouse uterus23,24. We confirmed these findings and noted that in normal endometrium, FOXA2 is diffusely expressed in the deep glands with rare expression in individual cells along the surface epithelium (Supplemental Fig.4A-D). In humans, it was reported FOXA2 is expressed at higher levels in complex atypical hyperplasia than normal endometrium13. Using IHC we saw variable FOXA2 expression in EEC, both between tumors, and within tumor samples (Supplemental Fig.4E-I). Quantitative IHC on archived old tissue microarrays was unreliable likely due to epitope stability and/or reduced epitope retrieval for older samples stored in paraffin blocks25. Thus mRNA qRT-PCR and protein western blot assays were utilized for evaluation and comparisons of FOXA2 expression in EEC.
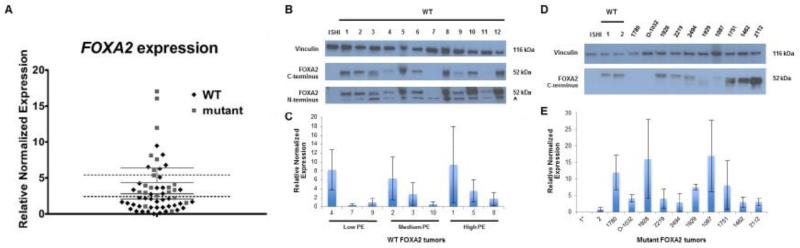
qRT-PCR used to assess expression in primary tumors and cell lines (HepG2, as a positive control26) revealed endometrial cancer cell lines had no or very low FOXA2 transcript levels (Ishikawa, MFE296, HEC1A, AN3CA, RL952, and KLE: data not shown). On the other hand, FOXA2 mRNA expression was high in most primary EECs. Western blot analysis confirmed high level expression of protein in most EECs. One of 12 tumors wild-type for FOXA2 did not express the 52kDa protein at detectable levels (Fig. 2B). qRT-PCR showed that FOXA2 transcripts were low in the non-expressing tumor (0.34 fold HepG2 control). Transcript levels varied among wild-type tumors and did not directly correlate with protein expression (Fig. 2C). These findings suggest complex regulation of FOXA2 protein and transcription.
Fig. 2. Variable FOXA2 expression in EECs.
(A) mRNA levels in primary cancers, normalized to HepG2, shows >15 fold variability in FOXA2 transcripts. Expression was significantly higher in tumors with FOXA2 mutations (N=21, three missense, one in-frame deletion, and 17 stop or frameshift mutations) than in wild-type tumors (N=42), with mean expression levels 8.27 and 3.07, respectively (p ≤ 0.05). Mean relative mRNA expression level for each tumor was based on 3 different independent q-RT PCR experiments performed in quadruplicate. (B) Western blot analysis of FOXA2 using a C-terminal antibody (Abcam EPR4466). Protein levels varied from undetectable in a single tumor (lane 7) to high level expression, comparable to the HepG2 positive control. The Ishikawa cell line (negative control) does not express FOXA2. N-terminal antibody (Millipore 07-633) confirmed variability in protein levels, but because of a nonspecific band at ~36 kDa (arrowhead) it was not possible to reliably test for truncated protein products in mutant samples. (C) mRNA and protein levels are not correlated in FOXA2 wild-type tumors. (D) Western blot analysis of FOXA2 mutant tumor samples shows similar variability in expression of FOXA2 using a C-terminal antibody. Tumors 1780 and 0-1032 each have two mutations, and have no detectable FOXA2. The wild-type 52kDa protein is seen with the C-terminal antibody (stop and frameshift proteins not detectable). Tumors 1929 and 1087 are exceptions in that they have a single mutation (truncating), but have no or very little wild-type FOXA2. Wild-type endometrial tumors and HepG2 served as positive controls, and the Ishikawa cell line served as a negative control. Vinculin loading control. (E) No correlation between type of mutations, transcript levels, and protein expression for tumors with FOXA2 mutations. WT, wild-type; ISHI, Ishikawa cell line; PE, protein expression.
FOXA2 promoter methylation and expression in EEC
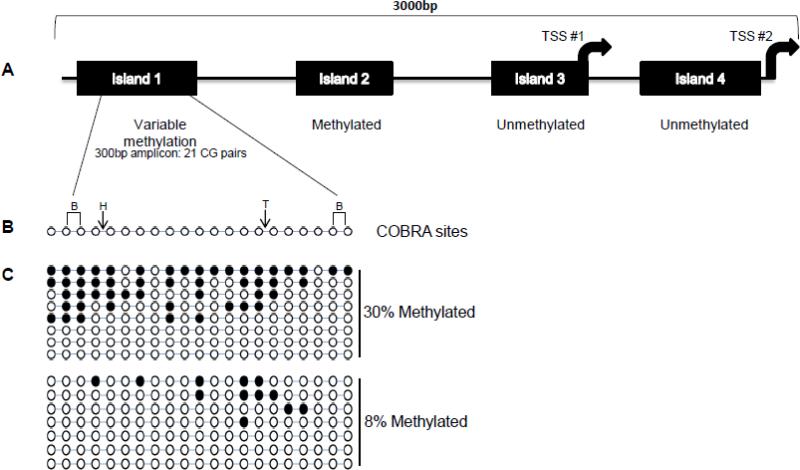
Variable FOXA2 mRNA levels in EEC could reflect multiple epigenetic control mechanisms, including DNA methylation. Methylation of CpG resides upstream of FOXA2 has been shown to correlate with expression in mouse endodermal development of pancreatic beta cells27. We therefore evaluated promoter methylation in four CpG islands in the FOXA2 promoter region (Fig.3A-B). DNA from cell lines that express FOXA2, A549 and HepG2, was compared to the RL952 and AN3CA endometrial cancer cell lines that do not express FOXA2. This survey showed methylation of CpG island 2 in all four cell lines and variable methylation of islands 3 and 4. Island 1 was unmethylated in the cell lines that expressed FOXA2 and methylated in the non-expressing endometrial cancer cell lines. COBRA was performed for 23 primary EECs and 9 matched peripheral white blood normal DNAs. Methylation was variable at Island 1 methylation with 9/23 cases COBRA-positive. Eight of nine matched controls had methylation in Island 1 (89%). Island 2 was methylated in all tumor and normal. Island's 3 and 4 were unmethylated in all samples. Cloning bisulfite converted PCR products showed methylation was highly variable in tumors (Fig.3C). In a tumor classified as unmethylated by COBRA, 0.5% of CpG pairs (1/168 sequenced CGs) were methylated.
Fig. 3. Methylation analysis of the FOXA2 promoter.
(A) The FOXA2 promoter region showing the location of four CpG islands and two transcription start site (TSS). COBRA results for tumors are summarized below each island. (B) Representation of the 21 CpG pairs included in amplicon 1 and the location of cut sites for the restriction enzymes used. B, BstUI; H, HpyCHIV; T, TaqI. (C) Bisulfite sequencing for representative tumors that were predicted by COBRA to be methylated. Black ovals indicate a cytosine (methylated C) and open circles (unmethylated cytosine converted to thymine).
FOXA2 mutants have variable effects on expression
When FOXA2 mutant and wild-type tumors were compared using qRT-PCR, the mean relative normalized expression of FOXA2 showed mutants had a significantly higher expression than wild-type tumor specimens (p=0.014, Fig.2A). cDNA sequencing proved that both the mutant and wild-type alleles were expressed in seven of eight tumors analyzed (data not shown). The single exception was tumor 2494T (c.363delG mutation) that expressed only the wild-type allele. However, a second tumor with the same mutation (2219T) expressed both FOXA2 alleles. The presence of mutant transcripts in cDNAs is consistent with retained heterozgyosity in tumors and absence of nonsense-mediated decay as would be predicted for a two exon gene.
FOXA2 protein levels on Western blot analysis using a C-terminal monoclonal antibody showed variable expression in mutant tumors (Fig.2D). As expected, two-hit mutant tumors had undetectable FOXA2. The missense mutations had high FOXA2 expression. Frameshift and nonsense mutations showed variable expression. The low protein level in tumors with mutations was unexpected given the observed higher transcript levels. Furthermore, similar to wild-type tumors, there was no direct correlation between mRNA expression and FOXA2 protein levels (Fig.2E). Mutant truncated proteins may explain the differences between transcription levels and protein expression. Unfortunately, the available N-terminal gave non-specific bands precluding detection of frameshift and truncated mutant FOXA2 (Fig.2D).
We tested the potential antigenicity of predicted mutant proteins using NetMHC 4.0 (Supplemental Table 3)28. The majority of mutations were predicted to be highly antigenic. Three of eight missense mutations (37.5%) predicted strong binding affinity antigens. Sixteen of 25 frameshift mutations (64%) were predicted to have strongly antigenic mutant proteins. These results suggest that FOXA2 mutants may contribute to immunogenicity and/or alterations in the immune microenvironment.
Relationship between FOXA2 mutant status and demographic and clinicopathologic features
FOXA2 mutation status was significantly associated with lower age at diagnosis and higher tumor grade (Table 1). There were no associations with other clinicopathologic features. The mean age of FOXA2 mutation patients was 67 vs. 63 years for women whose tumors lacked mutations (p=0.03). The association with grade was attributable to increased rate of mutations in grade 2 tumors (59.2% of mutants vs. 32.7% of wild-type) and decreased percent of both grade 1 and 3 tumors (p=0.001). This finding is likely due to the increased rate of FOXA2 mutation in MSI tumors. In both our cohort and the TCGA, MSI is associated with increased rates of grade 2 disease3.
Table 1.
Correlation between mutation status and demographic and clinicopathologic features.
| FOXA2 status | |||
|---|---|---|---|
| Characteristic | WT (N=493) | Mutant (N=49) | p-valuea |
| Age, mean (SD) | 63 (11.7) | 67 (12.3) | 0.03 |
| Grade | |||
| 1 | 253 (51.3%) | 16 (32.7%) | 0.01 |
| 2 | 161 (32.7%) | 29 (59.2%) | |
| 3 | 79 (16%) | 4 (8.2%) | |
| Stage | |||
| I-II | 398 (80.9%) | 45(91.8%) | 0.08 |
| III-IV | 94(19.1%) | 4 (8.2%) | |
| Race | |||
| Caucasian | 432 (87.6%) | 42 (85.7%) | 0.82 |
| African American | 58 (11.8%) | 6 (12.2%) | |
| Other/not reported | 3 (0.6%) | 1 (2%) | |
| BMI, mean (SD) | 34.8 (10.2) | 34.1 (12.5) | 0.62 |
| < 30 (%) | 154 (31.2%) | 17 (34.7%) | |
| ≥ 30 | 282 (57.2%) | 26 (53.1%) | |
| LVSI | |||
| Absent | 317 (64.3%) | 31 (63.3%) | 0.87 |
| Present | 166 (33.7%) | 17 (34.7%) | |
| Recurrence/Progressionb | 74 (15.0%) | 7 (14.3%) | 1.000 |
Significant associations (p < 0.05) were found between mutation status and age (two sided unpaired t-test), and mutation status and grade (two-sided chi-square test).
One FOXA2 wild-type patient was unstaged, no BMI data was available for 63 subjects, and LVSI status was missing for 12 subjects.
Recurrence is defined as time from initial surgical intervention until clinical evidence of disease. WT, wild type; BMI, body mass index; LVSI, lymphovascular space invasion; SD, standard deviation.
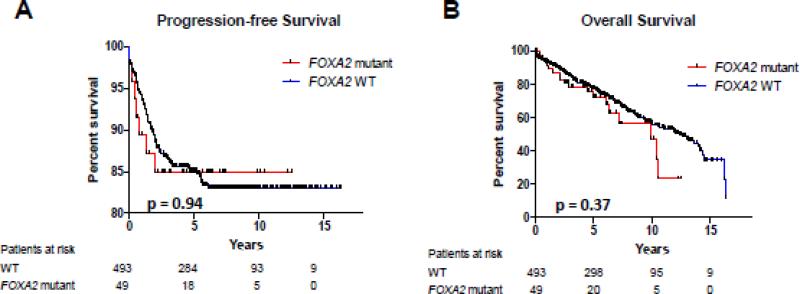
We hypothesized FOXA2 mutation was associated with poor outcomes based on whole exome sequencing of primary early-stage cancers that subsequently recurred (unpublished data). In that small study, 3 of 8 tumors (37.5%) had FOXA2 mutations. In our cohort of 542 cases, there was no association between mutation and recurrence (Table 1) or recurrence-free survival (p=0.80). Kaplan-Meier analysis showed no difference in progression-free survival (PFS) or overall survival (OS) (Fig.4).
Fig. 4. Survival outcomes stratified by FOXA2 mutation status.
Kaplan-Meier curves for (A) progression-free and (B) overall survival. WT, wild-type. P values based on log-rank test.
Discussion
This study presents the first large-scale analysis of FOXA2 mutations in primary EECs, pointing to FOXA2's role as a tumor suppressor gene. In TCGA, FOXA2 mutations were found in just nine EECs 3. Although the overall mutation rate was higher in our study (9.4% vs. 5.2%) it was not statistically significant (Supplemental Table 4). There were a striking number of novel mutations. Unexpectedly, higher rates of frameshift and in-frame deletions were seen at 6.6% vs. 2.3% than previously reported (p=0.04)3. This difference may reflect differences in coverage for GC-rich sequences, such as FOXA2, and showcases the importance of understanding the limitations of different sequencing techniques29,30.
DNA mismatch repair defects are common in EECs3,4,22. FOXA2 mutations could represent unselected mutational noise secondary to mismatch repair defects in endometrial cancer. Frameshift mutations in repeat regions are characteristic MSI associated strand-slippage22,31. Although the majority of FOXA2 mutants were MSI tumors, we saw a similar pattern of frameshift mutations within our MSS tumors. This suggests that FOXA2 mutations are not mutational noise in mismatch repair deficient tumors.
The mutation pattern of FOXA2 overall points to a tumor suppressor gene function in the endometrium. Tumor suppressor genes characteristically have high rates of damaging or loss of function mutations. Considering loss of function mutations, potential function altering in-frame deletions, and missense mutations unanimously damaging amongst all prediction models, 90.2% of mutations (46 of 51) are predicted to significantly change the function of FOXA2. Oncogenic mutation profiles on the other hand favor missense mutations and recurrent mutations, i.e. hotspot mutations. The overwhelming number of predicted loss of function mutations and lack of hotspot mutations strongly suggests FOXA2 as a tumor suppressor gene in EEC. Davoli et al32 predicted that FOXA2 is a uterine tumor suppressor gene based on a computational algorithm that utilized the TCGA mutational profile. Aside from this hypothesis generating paper, we are unaware of any previous studies implicating FOXA2 as a tumor suppressor gene in endometrial cancer. FOXA2 is, however, a tumor suppressor gene in lung, liver, gastric, and pancreatic cancer6,7,9,33-35. Additionally, the rarity of two-hit mutants suggests a tumor suppressor gene with possible dominant negative or haploinsufficient mechanism.
Splice site and frameshift mutations may result in a variety of neomorphic, misfolded or truncated protein products that effect FOXA2 function such as DNA-binding, immunogenicity, autoregulation, and protein degradation. In fact, 64% of frameshift mutations were predicted to create highly antigenic mutant proteins, which may affect the tumor microenvironment and immune regulation. Odom et al described a pattern of auto-regulation of FOXA2 with an upstream promoter site in human hepatocytes using ChIP and high-resolution promoter microarrays, which is a mechanism for a possible dominant-negative effect36. Unfortunately, we were unable to confirm truncated protein products and support our hypothesis. Alternatively, the atypical rare number of two-hit mutants suggests that FOXA2 may join the increasingly recognized group of haploinsufficient tumor suppressor genes in EEC. Heterozygote loss of mouse Foxa2 is associated with age dependent motor deficits, aberrant lipid metabolism and loss of a single allele of FOXA2 results in a phenotypic variant of holoprosencephaly 37-40. In order to further elucidate the functional changes from different FOXA2 mutations, in vitro studies assessing protein stability and self-regulation of FOXA2 expression and defects in DNA-binding and target gene regulation will need to be performed.
Based on mRNA transcription and protein expression assays in our cohort we ascertained that FOXA2 expression in EEC is variable. FOXA2 is highly expressed at the transcript level in both mutant and wild-type FOXA2 EEC and the majority of mutants are expressing the mutant allele. One case had aberrant results, which were likely due to heterogeneity within the sample or the result of alternative regulatory mechanisms. Also, endometrial cancer cell lines appeared to lose expression of FOXA2. As FOXA2 is expressed in the deep glands of the endometrium there could be some environmental pressure of the uterus required for expression. This is supported by previous findings that FOXA2 expression is tissue, gender and developmental stage dependent7,10,24.
CG-rich sequences, like the FOXA2 promoter, can be methylated and with that there is potential for epigenetic regulation. Our survey of methylation included three previously studied promoter islands and one additional island. Island 2 was previously described as being unmethylated in FOXA2 expressing embryonic stem cells, but once differentiated, cells showed methylation in the CpG island and loss of expression of FOXA227. Island 3 and 4 are most proximal to the TSS and methylation of either correlates with higher degrees of FOXA2 expression in NSCLC and melanoma 5. In our survey, we found no evidence of methylation in either tumors or normal blood controls, which may be related to alternate FOXA2 expression regulation. Island 1 was surveyed for the first time in our cohort. The methylated tumors did show a decreased FOXA2 transcript levels, but our survey was underpowered to detect statistically significant differences. Island 1 is more distal to the TSS for both the short and long FOXA2 isoforms, which may make it a more important gene regulator region than previously thought and may further elucidate some of the variability in FOXA2 transcription in wild-type EEC tumors.
We showed that FOXA2 protein expression does not appear to be FOXA2 mutation dependent (Fig.2C). Mutants showed variable expression of wild-type FOXA2 from no to normal levels using a C-terminal specific antibody. Thus, mutant proteins may alter feedback regulation of transcription either through auto-regulation, changes in binding affinity to feedback modulators, or interactions with other expression regulating pathways. The two-hit mutants and p.A353Gfs*11, one of the previously described possible hot-spot mutants, had no or extremely low expression of FOXA2. This suggests an association with protein expression and mutation type but our sample size is too small to draw any definitive conclusions. Unfortunately, exploring expression of truncated proteins using an N-terminal antibody was not possible (Fig.2D). Unstable or misfolded protein products or lack of translation of mutant allele could explain why some mutants are not expressed.
Recent advancements in understanding of other forkhead box proteins’ roles in endometrial tumorigenesis may give insights into the function of FOXA2. FOXA1 and FOXO1 are transcriptional regulators of hormone receptors and low expression levels are reported in endometrial cancer14,15,41,42. In murine models, foxa1 and foxa2 have overlapping gene targets and result in compensatory functional roles attributed to similar DNA-binding motifs43. Interestingly, FOXA2 and FOXA1 mutations were mutually exclusive with the exception of one tumor in the TCGA3. This suggests that loss of either gene may play a role in tumorigenesis and mutations in both genes may have cellular lethality. However, differences in expression of forkhead box genes exist. For example, in endometrial hyperplasia, FOXA1 is downregulated while FOXA2 is upregulated13,14. Therefore, the true function of FOXA2 in the endometrium is unclear.
Although FOXA2 was not found to be prognostic in EEC, it likely plays important roles in the endometrium and endometrial cancer tumorigenesis. The high mutation rate of FOXA2, as well as the large number of loss of function mutants, point to FOXA2 as a potential tumor suppressor gene. In order to understand the impact of FOXA2 mutations in EEC, the role of FOXA2 in the normal endometrium, including pre and post-menopausal, will need to be evaluated. Furthermore, investigation of FOXA2 epigenetic regulation, molecular pathway involvement, cofactors and gene targets in the human endometrium and EEC are warranted.
Supplementary Material
Highlights.
Both MSI and MSS tumors endometrioid endometrial cancers have frequent FOXA2 frameshift mutations
FOXA2 is a haploinsufficient tumor suppressor in endometrial cancer
Most endometrioid tumors express FOXA2
ACKNOWLEDGMENTS
We would like to thank Alexis Chassen for manuscript editing and data curation. We would like to acknowledge Cynthia Timmers and the Ohio State University Solid Tumor Translational Shared Resource and Genomics and Biostatistics shared resources at the Ohio State University Comprehensive Cancer Center. We thank Takeshi Kurita for normal mouse tissues and for his helpful discussions. We are very grateful to all of the patients who contributed specimens to this study and all of the attending physicians and staff at the Washington University School of Medicine Division of Gynecologic Oncology, and The Ohio State University College of Medicine Division of Gynecologic Oncology.
FUNDING
This work was supported by the National Cancer Institute [P30 CA016058], the James Comprehensive Cancer Center Pelotonia Fellowship Program [C.J.W.] and the National Institutes of Health [T32 GM068412 to C.M.R.] and the James CCC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors declare no conflict of interest.
References
- 1.Howlader N, Noone A, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2012. National Cancer Institute; Bethesda, MD: 2015. [Google Scholar]
- 2.Modesitt S, Walker J. Obesity crisis in cancer care: gynecologic cancer prevention, treatment, and survivorship in obese women in the United States. Gynecol Oncol. 2014;133(1):1–3. doi: 10.1016/j.ygyno.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 3.Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billingsley CC, Cohn DE, Mutch DG, Stephens JA, Suarez AA, Goodfellow PJ. Polymerase ε (POLE) mutations in endometrial cancer: clinical outcomes and implications for Lynch syndrome testing. Cancer. 2015;121(3):386–394. doi: 10.1002/cncr.29046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basseres DS, D'Alò F, Yeap BY, et al. Frequent downregulation of the transcription factor Foxa2 in lung cancer through epigenetic silencing. Lung Cancer. 2012;77(1):31–37. doi: 10.1016/j.lungcan.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halmos B, Bassères DS, Monti S, et al. A transcriptional profiling study of CCAAT/enhancer binding protein targets identifies hepatocyte nuclear factor 3 beta as a novel tumor suppressor in lung cancer. Cancer Res. 2004;64(12):4137–4147. doi: 10.1158/0008-5472.CAN-03-4052. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148(1-2):72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70(5):2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Y, Shu G, Yuan X, Jing N, Song J. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res. 2011;21(2):316–326. doi: 10.1038/cr.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu CP, Wang J, Shi B, et al. The transcription factor FOXA2 suppresses gastric tumorigenesis in vitro and in vivo. Dig Dis Sci. 2015;60(1):109–117. doi: 10.1007/s10620-014-3290-4. [DOI] [PubMed] [Google Scholar]
- 11.Wang DH, Tiwari A, Kim ME, et al. Hedgehog signaling regulates FOXA2 in esophageal embryogenesis and Barrett's metaplasia. J Clin Invest. 2014;124(9):3767–3780. doi: 10.1172/JCI66603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong JW, Kwak I, Lee KY, et al. Foxa2 is essential for mouse endometrial gland development and fertility. Biol Reprod. 2010;83(3):396–403. doi: 10.1095/biolreprod.109.083154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villacorte M, Suzuki K, Hirasawa A, et al. β-Catenin signaling regulates Foxa2 expression during endometrial hyperplasia formation. Oncogene. 2013;32(29):3477–3482. doi: 10.1038/onc.2012.376. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Bao W, Qiu M, et al. Forkhead-box A1 suppresses the progression of endometrial cancer via crosstalk with estrogen receptor α. Oncol Rep. 2014;31(3):1225–1234. doi: 10.3892/or.2014.2982. [DOI] [PubMed] [Google Scholar]
- 15.Tangen IL, Krakstad C, Halle MK, et al. Switch in FOXA1 status associates with endometrial cancer progression. PLoS One. 2014;9(5):e98069. doi: 10.1371/journal.pone.0098069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novetsky AP, Zighelboim I, Thompson DM, Powell MA, Mutch DG, Goodfellow PJ. Frequent mutations in the RPL22 gene and its clinical and functional implications. Gynecol Oncol. 2013;128(3):470–474. doi: 10.1016/j.ygyno.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zighelboim I, Goodfellow PJ, Gao F, et al. Microsatellite instability and epigenetic inactivation of MLH1 and outcome of patients with endometrial carcinomas of the endometrioid type. J Clin Oncol. 2007;25(15):2042–2048. doi: 10.1200/JCO.2006.08.2107. [DOI] [PubMed] [Google Scholar]
- 18.Zighelboim I, Schmidt AP, Gao F, et al. ATR mutation in endometrioid endometrial cancer is associated with poor clinical outcomes. J Clin Oncol. 2009;27(19):3091–3096. doi: 10.1200/JCO.2008.19.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham F, Amode MR, Barrell D, et al. Ensembl 2015. Nucleic Acids Res. 2015;43(Database issue):D662–669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karamurzin Y, Rutgers JK. DNA mismatch repair deficiency in endometrial carcinoma. Int J Gynecol Pathol. 2009;28(3):239–255. doi: 10.1097/PGP.0b013e31818d8fe6. [DOI] [PubMed] [Google Scholar]
- 23.Filant J, Zhou H, Spencer TE. Progesterone inhibits uterine gland development in the neonatal mouse uterus. Biol Reprod. 2012;86(5):146, 141–149. doi: 10.1095/biolreprod.111.097089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filant J, Lydon JP, Spencer TE. Integrated chromatin immunoprecipitation sequencing and microarray analysis identifies FOXA2 target genes in the glands of the mouse uterus. FASEB J. 2014;28(1):230–243. doi: 10.1096/fj.13-237446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin F, Chen Z. Standardization of diagnostic immunohistochemistry: literature review and geisinger experience. Arch Pathol Lab Med. 2014;138(12):1564–1577. doi: 10.5858/arpa.2014-0074-RA. [DOI] [PubMed] [Google Scholar]
- 26.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bahar Halpern K, Vana T, Walker MD. Paradoxical role of DNA methylation in activation of FoxA2 gene expression during endoderm development. J Biol Chem. 2014;289(34):23882–23892. doi: 10.1074/jbc.M114.573469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32(4):511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40(10):e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rieber N, Zapatka M, Lasitschka B, et al. Coverage bias and sensitivity of variant calling for four whole-genome sequencing technologies. PLoS One. 2013;8(6):e66621. doi: 10.1371/journal.pone.0066621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aaltonen LA, Peltomäki P, Mecklin JP, et al. Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res. 1994;54(7):1645–1648. [PubMed] [Google Scholar]
- 32.Davoli T, Xu AW, Mengwasser KE, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birnbaum DJ, Adélaïde J, Mamessier E, et al. Genome profiling of pancreatic adenocarcinoma. Genes Chromosomes Cancer. 2011;50(6):456–465. doi: 10.1002/gcc.20870. [DOI] [PubMed] [Google Scholar]
- 34.Pinho AV, Rooman I, Real FX. p53-dependent regulation of growth, epithelial-mesenchymal transition and stemness in normal pancreatic epithelial cells. Cell Cycle. 2011;10(8):1312–1321. doi: 10.4161/cc.10.8.15363. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki A. [Identification of master regulators for induction of hepatocyte differentiation]. Seikagaku. 2012;84(8):675–679. [PubMed] [Google Scholar]
- 36.Odom DT, Dowell RD, Jacobsen ES, et al. Core transcriptional regulatory circuitry in human hepatocytes. Mol Syst Biol. 2006;2:2006.0017. doi: 10.1038/msb4100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaestner KH. The FoxA factors in organogenesis and differentiation. Curr Opin Genet Dev. 2010;20(5):527–532. doi: 10.1016/j.gde.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfrum C, Asilmaz E, Luca E, Friedman JM, Stoffel M. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432(7020):1027–1032. doi: 10.1038/nature03047. [DOI] [PubMed] [Google Scholar]
- 39.Wolfrum C, Howell JJ, Ndungo E, Stoffel M. Foxa2 activity increases plasma high density lipoprotein levels by regulating apolipoprotein M. J Biol Chem. 2008;283(24):16940–16949. doi: 10.1074/jbc.M801930200. [DOI] [PubMed] [Google Scholar]
- 40.Rosenfeld JA, Ballif BC, Martin DM, et al. Clinical characterization of individuals with deletions of genes in holoprosencephaly pathways by aCGH refines the phenotypic spectrum of HPE. Hum Genet. 2010;127(4):421–440. doi: 10.1007/s00439-009-0778-7. [DOI] [PubMed] [Google Scholar]
- 41.Abe Y, Ijichi N, Ikeda K, et al. Forkhead box transcription factor, forkhead box A1, shows negative association with lymph node status in endometrial cancer, and represses cell proliferation and migration of endometrial cancer cells. Cancer Sci. 2012;103(4):806–812. doi: 10.1111/j.1349-7006.2012.02201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korani M, Fallah S, Tehranian A, et al. The evaluation of the FOXO1, KLF9 and YT521 genes expression in human endometrial cancer. Clin Lab. 2013;59(5-6):483–489. doi: 10.7754/clin.lab.2012.120626. [DOI] [PubMed] [Google Scholar]
- 43.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10(4):233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.