Immunotherapy is among the novel classes of therapy in development for acute myeloid leukemia (AML) and harnesses the specificity of the immune system to eliminate leukemia cells.1 This approach builds upon the success of allogeneic hematopoietic stem cell transplant (allo-SCT) in AML, which can be curative in up to 50% of patients, but often at the cost of high rates of treatment-related toxicities, including graft-vs-host disease (GVHD).2 To avoid this complication, one approach is to refine the potent graft-versus-leukemia (GVL) effect to target discrete leukemia antigens in the form of adoptive cellular therapy (ACT). Importantly, in order to overcome intratumoral heterogeneity, durable clinical efficacy will likely require targeting multiple leukemia antigens simultaneously, an approach that is currently limited by the number of known effective leukemia antigens.3
We previously reported that cathepsin G (CG), an azurophil granule protease, represents a novel leukemia-associated antigen (LAA).4 CG is largely restricted to the myeloid lineage and like proteinase 3 (P3) and neutrophil elastase (NE), the sources of the well-established HLA-A2 (i.e., HLA-A*0201) LAA PR1,5 CG is contained within the primary granules of maturing and mature neutrophils. However, CG is expressed later in myeloid differentiation, resides under a different promoter than NE and P3, and demonstrates a distinct pattern of expression, suggesting that targeting both PR1 and CG could be synergistic.6 In our prior work4, we demonstrated that CG is highly expressed in primary patient AML blasts, AML cell lines, and, critically, in leukemia stem cells (LSCs). We showed that CG is localized outside azurophil granules and is ubiquitinated, favoring antigen presentation. Additionally, we identified an HLA-A2 restricted epitope within the leader sequence of CG, designated as CG1 (amino acid sequence FLLPTGAEA), and showed that targeting CG1 resulted in lysis of AML blasts in vitro. Finally, we detected cytotoxic T-lymphocytes (CTLs) specific for CG1 in the peripheral blood of AML patients after allo-SCT4.
In this study, we further investigate the therapeutic potential of targeting CG, specifically CG1, focusing on the anti-leukemia activity of CG1-specific-CTL in vivo and evaluating their toxicity against normal hematopoietic progenitor cells (HPCs). In addition, we analyze the expression of CG and its presentation by HLA-A2 in primary patient AML blasts through reverse-phase protein array (RPPA) and liquid chromatography (LC)/tandem mass spectrometry (MS/MS).
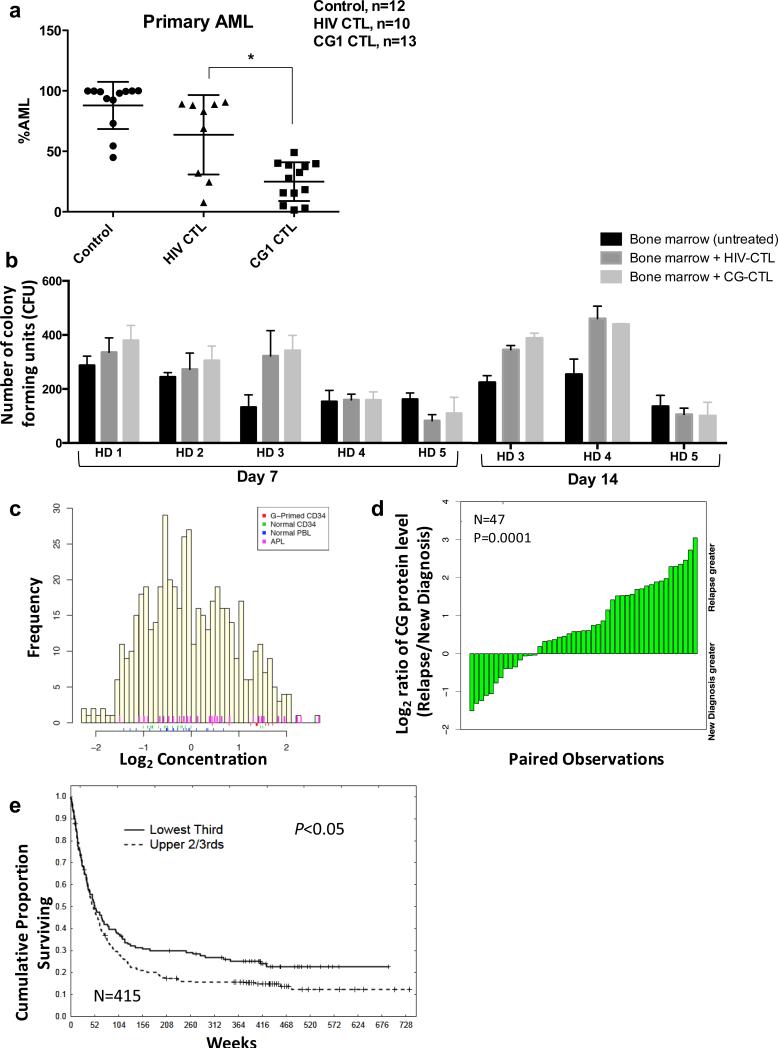
To determine the role of CG as an immunotherapeutic target in vivo, we first studied a patient-derived xenograft (PDX) treatment model of primary AML (UPN#1; Table 1). After confirming leukemia engraftment (~ 3 weeks), NSG mice were treated intravenously with either 0.5 × 106 CG1-CTL or HIV-CTL, or were left untreated. At the time of sacrifice, bone marrow (BM) was harvested and analyzed for leukemia burden. Mice treated with CG1-CTL had a significantly lower AML burden in the BM (24.94 % ± 4.451%; n = 13) compared with HIV-CTL-treated mice (61.46% ± 11.07%; n = 10) and untreated mice (87.9% ± 5.632%; n = 12) (P < 0.01; Figure 1a). The efficacy of targeting CG in vivo using CG1-CTL was also recapitulated in a murine model that incorporated the HLA-A2-transduced U937 myelomonoblastic leukemia cell line (U937-A2) (Supplementary Figures S1-S2).
Table 1.
AML patient samples used in liquid chromatography tandem mass spectrometry (LC-MS/MS). UPN #1 patient blasts were utilized in the patient-derived xenograft (PDX) treatment model.
| Sample | CG1 | Leukemia | Cytogenetic/Molecular Abnormalities | HLA-A typing | Phenotype |
|---|---|---|---|---|---|
| UPN #1 | yes | FAB-M1 | Diploid 46 XX; FLT3-TD; NPM1+; DNMT3A+ | A02:01, A24:02 | CD13/33+; MPO+ |
| UPN #2 | yes | FAB-M1 | Pseudodiploid 46XY; del (5q); CEBPA+ | A02:01, A03:01 | CD33/34/38/117+; HLA-DR+, MPO+ |
| UPN #3 | yes | FAB-M4/M5 | not tested | A02:01 | not tested |
| UPN #4 | no | FAB-M4 | Diploid 46XY; FLT3-TD; NPM1+ | A02:01; A32:01 | CD13/33; MPO+; HLA-DR+ |
| UPN #5 | no | Biphenotypic | Diploid 46XY; t(9;22); BCR-ABL1 | A24:02; A33:01 | CD13/19d/33/34/38+; MPO+; HLA-DR+ |
| UPN #6 | yes | B-ALL | Diploid 46XX; t(9;22); BCR-ABL1 | A02:01; A24:02 | CD10/13/19/33/34/38+; HLA-DR+ |
| UPN #7 | no | B/T-ALL | Hyperdiploid 50XY | A02:01, A03:01 | CD13/19/33/34/38+; HLA-DR+ |
| U937 | no | Monocytic | Complex karyotype (>5 cytogenetic abnormalities) | A03:01; A31:12 | CD13, CD33, CD15, CD11b, CD18 |
| U937-A2 | yes | Monocytic | Complex karyotype (>5 cytogenetic abnormalities) | A03:01; A31:12; A0201 | CD13, CD33, CD15, CD11b, CD18 |
| HL-60 | no | APL | Complex karyotype (>5 cytogenetic abnormalities) | A01:01* | CD13, CD33, CD38, CD117, MPO+ |
| HL-60-A2 | yes | APL | Complex karyotype (>5 cytogenetic abnormalities) | A01:01; A02:01 | CD13, CD33, CD38, CD117, MPO+ |
Abbreviations: UPN, unique patient number; FAB, French-American-British; APL, acute promyelocytic leukemia; MPO, myeloperoxidase.
Wild-type HL-60 cell line is missing a haplotype, hence only expresses one allele at the HLA-A locus.
Figure 1.
Cathepsin G is an effective immunotherapeutic target in vivo and is broadly expressed in AML patients. (a) Irradiated NSG mice were injected intravenously with human primary AML blasts (UPN#1; 7 × 106 blasts) on day 0. After confirming leukemia engraftment (~ 3 weeks), mice were treated with either negative control HIV-CTL (0.5 × 106), CG1-CTL (0.5 × 106) or were left untreated. Mice were sacrificed for all groups when any mouse became moribund or during week 7. The results are expressed as percentage of CD33+/CD3− cells from viable hCD45+/mCD45− population within the bone marrow. Results reflect 4 independent experiments; *P<0.01. (b) Bone marrow from 5 healthy donors (HD 1-5) was cultured alone (untreated) or co-cultured with HIV-enriched T cells (HIV-CTL) or CG1-enriched T cells (CG1-CTL) at a 1:5 ratio for 4 hours in cell media. Cells were then resuspended in methylcellulose semi-solid matrix and co-cultured for 7 days. On day 7, colonies were counted and imaged; for HD 3, 4 and 5, colonies were again counted on day 14. Each group was cultured in triplicate and data represent 4 independent experiments. (c) Reverse-phase protein array (RPPA) was used to quantify protein levels of cathepsin G in blasts from 511 newly diagnosed AML patients (yellow bars) and 21 newly diagnosed APL patients (pink bars). Controls included healthy donor CD34+ progenitor cells (n=21, green bars), healthy donor peripheral blood lymphocytes (n=21, blue bars), and GM-CSF-primed healthy donor CD34+ progenitor cells (n=10, red bars). (d) Cathepsin G levels were assessed for 47 patients at diagnosis and relapse and were compared by paired t-test (P=0.0001). (e) Kaplan-Meier plots showing OS in AML patients (n=415) comparing patients with high CG protein expression by RPPA (upper 2/3) to patients with low CG expression (lowest 1/3). Results are significant by Cox univariate model testing (P= 0.04).
We next sought to confirm that the CG1 peptide is presented on the HLA-A2+ AML cell surface. We used W6/32 antibody to immunoprecipitate (IP) surface HLA Class I molecules and their bound peptides, which were isolated and analyzed via high sensitivity targeted LC-MS/MS. Blasts from seven newly diagnosed acute leukemia patients, including AML (n=4), ALL (n=2), and biphenotypic leukemia (n=1), as well as HL-60 and U937 cell lines and their HLA-A2 transfected counterparts were studied in this analysis. CG1 was identified in the eluted fraction in 3 of 4 patient AML cases (Table 1), including UPN #1 whose disease was used in the PDX model. CG1 was also identified on the surface of blasts from patient UPN #6 with precursor B-ALL, which is in agreement with two studies that confirmed CG expression and validated CG as an immunotherapeutic target in lymphoid malignancies.7, 8 In addition, CG1 was eluted from the surface of both HL-60-A2 and U937-A24 but not from either wild type cell line or from HLA-A2 negative UPN #5 (Supplementary Figure S3). Taken together, these data confirm that CG1 is naturally processed and presented on the surface of HLA*0201-positive leukemic blasts and cell lines, reinforcing its potential as a leukemia-associated antigen.
We previously demonstrated that primary AML blasts and CD34+38− LSCs have higher expression of CG than normal HPC and are preferentially eliminated in in vitro cytotoxicity assays.4 Nevertheless, since CG, like the LAAs NE and P3, is also expressed in healthy myeloid progenitor cells6, we investigated the potential toxicity of CG1-CTL against the formation of typical colonies from healthy donor bone marrow (HDBM) progenitor cells using colony-forming unit (CFU) assays. HDBM (n=5) was cultured in methylcellulose semi-solid matrix either alone or in the presence of HIV-CTL or CG1-CTL (Figure 1b). After 7 days, the mean CFUs were similar among the untreated and the two treatment groups (195.6 ± 66.7 for untreated vs. 234.5 ± 109.5 for HIV-CTL vs. 259.1 ± 118.2 for CG1-CTL). The CFUs for HDs 3-5 were followed for an additional 7 days, and at day 14, the differences among the 3 groups remained non-significant. These data suggest that CG1-CTL, with a range of avidities, do not significantly impair normal hematopoiesis.
Next, given The Cancer Genome Atlas (TCGA) data demonstrating highest CG transcript expression across a diverse set of AML cases (Supplementary Figure S4), we utilized RPPA to study the expression of CG protein in 511 newly diagnosed AML patients, a cohort that has been described previously.9 As shown in Figure 1c, CG was variably expressed across patients and the mean CG protein level was higher in APL and AML patients compared with normal CD34+ HPC (mean Log2 0.47 vs. mean Log2 - 0.01 vs. mean Log2 −0.34, respectively; P = 0.0375 for AML vs. CD34+ HPC). CG expression was higher in AML blasts than in HD CD34+ cells in 230 samples, equal to normal CD34+ cells in 234 samples and less than normal CD34+ cells in 47 samples. We also specifically studied CG levels in the 47 patients from our cohort who experienced relapsed AML and for whom paired samples were available at initial diagnosis and at relapse. Overall, CG level was higher in relapsed disease than at diagnosis (mean Log2 0.46 vs. mean Log2 −0.01, P < 0.001) and in 33 of the 47 (70%) patients with paired samples, CG levels were higher in the relapse sample (Figure 1d). These results confirm that CG is broadly expressed across AML cases and suggest that patients with relapsed disease may be candidates for CG-targeting therapies given their relatively high CG expression.
Although CG has been linked with aggressive behavior in solid tumors,10 the prognostic role of CG in AML patients has not been reported. Analysis of our RPPA data revealed a significant association between CG levels and overall survival (OS) in our patient cohort. In 415 patients for whom survival information was available, patients with CG levels in the lower tertile (as well as those below the median) showed significantly better OS than those whose CG expression ranked in the upper 2/3 (P = 0.04) (Figure 1e). The survival advantage associated with low CG level was most pronounced in the subset of AML patients with intermediate cytogenetics and FLT3 mutations (Supplementary Figure S5), a prevalent subset of patients for whom the optimal treatment of leukemia in first remission is controversial.11
In this report, we validate CG as an immunotherapeutic target in myeloid leukemia. We demonstrate that targeting CG in vivo with CG1-CTL reduces the leukemia burden in NSG mice. In addition, even though CG is expressed in normal hematopoietic cells, similar to other LAAs12, 13, targeting CG does not inhibit normal hematopoiesis. We postulate that CG1-CTL preferentially target leukemia over HPCs, in part, because CG has greater access to the MHC processing and presentation components within leukemic cells, since we previously showed that CG is located outside of azurophil granules and is ubiquitinated preferentially in AML.4
Critically, we also provide direct evidence that the HLA-A2 restricted peptide CG1 is naturally processed presented by primary AML. Lastly, we report that CG is expressed in a large cohort of AML patients and that high CG expression correlates with poor outcomes. We speculate that this association may be due to the potential interaction of CG with oncogenic proteins whose expression levels closely correlate with that of CG (Supplementary Figure S6). For example, there was a correlation between CG and the expression/phosphorylation of members of the Hippo pathway, TAZ and YAP1, which have been shown to play a role in cancer, including AML.14, 15
We have successfully targeted the LAA PR1 using various strategies including vaccination, T-cell based ACT, and a TCR-like antibody.5 We intend to apply similar strategies in the clinic to target CG1, and potentially other CG peptides, ideally in the autologous setting to minimize the risk of GVHD. We recognize that the autologous setting may yield lower affinity CG1-CTL, which could provide a therapeutic advantage clinically in that these CG1-CTL may preferentially lead to killing of high CG1-expressing AML, while sparing normal tissues. However, we also recognize the value of high affinity tumor antigen specific CTL in that they provide potent tumor killing, albeit sometimes at the expense of unwanted toxicity. In conclusion, these data lay the foundation for development of CG-targeting immunotherapies in AML.
Supplementary Material
Acknowledgements
This work is supported by grants from the Leukemia Research Foundation (G.A.), National Institutes of Health National Cancer Institute (G.A.) and Leukemia & Lymphoma Society (G.A. and S.K.)
Footnotes
The authors have no conflicts of interest to declare.
Supplementary information is available at the Leukemia journal website.
References
- 1.Dombret H, Gardin C. An update of current treatments for adult acute myeloid leukemia. Blood. 2016;127(1):53–61. doi: 10.1182/blood-2015-08-604520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornelissen JJ, Blaise D. Hematopoietic stem cell transplantation for patients with AML in first complete remission. Blood. 2016;127(1):62–70. doi: 10.1182/blood-2015-07-604546. [DOI] [PubMed] [Google Scholar]
- 3.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang M, Sukhumalchandra P, Enyenihi AA, St John LS, Hunsucker SA, Mittendorf EA, et al. A novel HLA-A*0201 restricted peptide derived from cathepsin G is an effective immunotherapeutic target in acute myeloid leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(1):247–257. doi: 10.1158/1078-0432.CCR-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sergeeva A, He H, Ruisaard K, St John L, Alatrash G, Clise-Dwyer K, et al. Activity of 8F4, a T-cell receptor-like anti-PR1/HLA-A2 antibody, against primary human AML in vivo. Leukemia. 2016 doi: 10.1038/leu.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garwicz D, Lennartsson A, Jacobsen SE, Gullberg U, Lindmark A. Biosynthetic profiles of neutrophil serine proteases in a human bone marrow-derived cellular myeloid differentiation model. Haematologica. 2005;90(1):38–44. [PubMed] [Google Scholar]
- 7.Gorodkiewicz E, Sienczyk M, Regulska E, Grzywa R, Pietrusewicz E, Lesner A, et al. Surface plasmon resonance imaging biosensor for cathepsin G based on a potent inhibitor: development and applications. Analytical biochemistry. 2012;423(2):218–223. doi: 10.1016/j.ab.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Fujiwara H, Melenhorst JJ, El Ouriaghli F, Kajigaya S, Grube M, Sconocchia G, et al. In vitro induction of myeloid leukemia-specific CD4 and CD8 T cells by CD40 ligand-activated B cells gene modified to express primary granule proteins. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(12):4495–4503. doi: 10.1158/1078-0432.CCR-04-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter BZ, Qiu YH, Zhang N, Coombes KR, Mak DH, Thomas DA, et al. Expression of ARC (apoptosis repressor with caspase recruitment domain), an antiapoptotic protein, is strongly prognostic in AML. Blood. 2011;117(3):780–787. doi: 10.1182/blood-2010-04-280503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson TJ, Nannuru KC, Futakuchi M, Singh RK. Cathepsin G-mediated enhanced TGF-beta signaling promotes angiogenesis via upregulation of VEGF and MCP-1. Cancer letters. 2010;288(2):162–169. doi: 10.1016/j.canlet.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bacher U, Haferlach T, Alpermann T, Kern W, Schnittger S, Haferlach C. Molecular mutations are prognostically relevant in AML with intermediate risk cytogenetics and aberrant karyotype. Leukemia. 2013;27(2):496–500. doi: 10.1038/leu.2012.200. [DOI] [PubMed] [Google Scholar]
- 12.Sergeeva A, Alatrash G, He H, Ruisaard K, Lu S, Wygant J, et al. An anti-PR1/HLA-A2 T-cell receptor-like antibody mediates complement-dependent cytotoxicity against acute myeloid leukemia progenitor cells. Blood. 2011;117(16):4262–4272. doi: 10.1182/blood-2010-07-299248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosen N, Sonoda Y, Oji Y, Kimura T, Minamiguchi H, Tamaki H, et al. Very low frequencies of human normal CD34+ haematopoietic progenitor cells express the Wilms' tumour gene WT1 at levels similar to those in leukaemia cells. British journal of haematology. 2002;116(2):409–420. doi: 10.1046/j.1365-2141.2002.03261.x. [DOI] [PubMed] [Google Scholar]
- 14.Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163(4):811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogawa S, Yokoyama Y, Suzukawa K, Nanmoku T, Kurita N, Seki M, et al. Identification of a fusion gene composed of a Hippo pathway gene MST2 and a common translocation partner ETV6 in a recurrent translocation t(8;12)(q22;p13) in acute myeloid leukemia. Annals of hematology. 2015;94(8):1431–1433. doi: 10.1007/s00277-015-2391-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.