Abstract
During cell division, aberrant DNA structures are detected by regulators called checkpoints that slow division to allow error correction. In addition to checkpoint-induced delay, it is widely assumed, though rarely shown, that merely slowing the cell cycle might allow more time for error detection and correction, thus resulting in a more stable genome. Fidelity by a slowed cell cycle might be independent of checkpoints. Here we tested the hypothesis that a slowed cell cycle stabilizes the genome, independent of checkpoints, in the budding yeast Saccharomyces cerevisiae. We were led to this hypothesis when we identified a gene (ERV14, an ER cargo membrane protein) that when mutated, unexpectedly stabilized the genome, as measured by three different chromosome assays. After extensive studies of pathways rendered dysfunctional in erv14 mutant cells, we are led to the inference that no particular pathway is involved in stabilization, but rather the slowed cell cycle induced by erv14 stabilized the genome. We then demonstrated that, in genetic mutations and chemical treatments unrelated to ERV14, a slowed cell cycle indeed correlates with a more stable genome, even in checkpoint-proficient cells. Data suggest a delay in G2/M may commonly stabilize the genome. We conclude that chromosome errors are more rarely made or are more readily corrected when the cell cycle is slowed (even ∼15 min longer in an ∼100-min cell cycle). And, some chromosome errors may not signal checkpoint-mediated responses, or do not sufficiently signal to allow correction, and their correction benefits from this “time checkpoint.”
Keywords: erv14, delayed cell cycle, chromosome instability, accuracy, speed
CHROMOSOME errors (e.g., DNA breaks, stalled forks, incomplete kinetochore assembly) arise spontaneously due to either the inherent error-prone nature of molecular processes or to exogenous conditions (e.g., radiation). At least two general mechanisms likely minimize error: checkpoints that detect aberrant structures and arrest cell division, and a slower cell cycle that either makes fewer errors or corrects errors in a timely fashion. The first mechanism, checkpoint controls, is well-known and detects different types of errors; the DNA damage checkpoint halts the cell cycle after certain forms of DNA damage, and the spindle assembly checkpoint halts the cell cycle after certain forms of spindle and kinetochore damage (Hartwell and Weinert 1989; Li and Murray 1991). The exact nature and amount of DNA and kinetochore defects are still being evaluated.
The second mechanism of error minimalization involves time. It is generally assumed, and rarely tested, that in WT cells cellular processes have evolved optimally to balance accuracy and speed (Hopfield 1974; Andersson et al. 1986; Mitton-Fry et al. 2004),. That is, a molecular process might be made more error free if the process were slowed to allow elaboration of fidelity mechanisms. We know of only a few instances where it has been demonstrated that indeed a delayed process leads to greater fidelity—one that involves translation and two, of course, that involve checkpoints in cell division (Hopfield 1974; Weinert and Hartwell 1988; Shonn et al. 2000; Johansson et al. 2008) (see Discussion). Nevertheless, it is widely assumed that cells have evolved to sacrifice accuracy for greater speed, especially in cell division. It is nontrivial to test this accuracy and speed dichotomy in cell division and the genome; for example, it is a priori unclear which mutations or chemicals might slow or accelerate the cell cycle, without also affecting chromosome biology by mechanisms other than time. We simply do not know enough about the cell to predict such an only-time-altering condition or mutation.
We entered this area of accuracy and speed of cell division inadvertently while studying genes and DNA sequences that alter genome stability in budding yeast. We were studying a region of a specific chromosome that previously we had shown is associated with high levels of instability (Paek et al. 2009). This region, which we called a fragile site, was immediately next to the ERV14 gene. Unexpectedly, we found that the fragile site sequence does not cause instability (Beyer and Weinert 2016; this study), but rather the expression of the ERV14 gene adjacent to the fragile site paradoxically causes instability (this study). We found that the genome is stabilized by erv14 in the study of three kinds of instability (chromosome loss, allelic recombinants, and unstable chromosomes), in three different chromosome assays, and in cells with spontaneous and induced damage. How might Erv14 cause instability? The Erv14 protein, an ER cargo membrane protein, has roles in endoplasmic reticulum (ER) biology, none of which appear related to chromosome biology. However, erv14 mutant cells do have a plethora of mutant phenotypes. We investigated a myriad of Erv14-dependent pathways (e.g., autophagy, the unfolded protein response, and several others). Ultimately, we inferred that it was no single Erv14-dependent pathway that was responsible for stabilizing the genome, but rather the slowed cell cycle due to the erv14 mutation. We tested this slowed cell cycle hypothesis by examining many genetic mutations and chemical conditions that slowed cell division, and we found that indeed, in general, a slowed cell cycle correlates with a more stable genome. The stabilizing influence of more time appears to act on chromosome error arising from both DNA and kinetochore damage and acts independently of DNA damage and spindle assembly checkpoints (e.g., acts in RAD9 and MAD2 cells, respectively, that have intact checkpoint controls). We conclude that a “time checkpoint” exists that when defective, in faster dividing cells, the genome becomes less stable. Exactly which molecular processes benefit from a slowed cell cycle are unknown.
Materials and Methods
Plasmids
Plasmids were generated using Escherichia coli strain DH5α by standard procedures. The pWL99 plasmid containing the ERV14 gene was constructed by first amplifying ERV14 from WT genomic DNA (gDNA) using primers containing BamHI and EcoRI restriction sequences that were 290 bp upstream of the ERV14 start codon and 348 bp downstream of the ERV14 stop codon, respectively. The ERV14 cassette was then cloned into the pRS406 plasmid using BamHI and EcoRI restriction sites. The pWL100 and pWL101 plasmids containing base sequence mutations altering the ATG start codon were constructed by PCR site-directed mutagenesis of pWL99 that replaced the ERV14 ATG start codon with the SnaBI restriction sequence (pWL100) or the PmlI restriction sequence (pWL101). The two mutations were needed to mutate and efficiently identify ERV14 mutations on both ERV14 alleles on the two copies of ChrVII during strain construction.
Yeast strains
WT ChrVII disome strain (TY200) is MATα HXK2/hxk2::CAN1lys5/LYS5 cyhr/CYHStrp5/TRP5leu1/LEU1 cenVII ade6/ADE6ADE3/ade3, ura3-52. CAN1 on ChrV has been mutated (Carson and Hartwell 1985). The WT and rad9Δ strains were derived from the A364A strain described previously (Weinert and Hartwell 1990; Admire et al. 2006; Paek et al. 2009). All other ChrVII disome mutations were generated from TY200 or TY206 by LiAC/ssDNA/PEG transformation using DNA cassettes amplified by PCR or with plasmids as noted. The mutant rad9Δ erv14atg/atg strain was generated by first digesting pWL100 with AgeI restriction enzyme (cutting the AgeI restriction site 59 bp downstream of the mutant erv14 start codon) and transplacement “pop-in pop-out” (Rothstein 1991) of the cut plasmid into the ChrVII CAN1 homolog of rad9Δ (TY206). Next, pWL101 was similarly transplaced into the ChrVII non-CAN1 homolog of rad9Δ, generating rad9Δ erv14atg/atg (TY660). Verification of both mutant erv14 start codons in rad9Δ was performed by PCR of erv14, followed by restriction digest with SnaBI and PmlI, and gel electrophoresis of the digested PCR products. The single mutant erv14atg/atg (TY661) strain was generated by integrating a pRS406-RAD9 plasmid into the ChrV URA3 locus of rad9Δ erv14atg/atg. Unless otherwise stated, erv14 = erv14atg/atg. The cdc13-F684S strains were generated similarly from the pVL5439 plasmid using transplacement pop-in pop-out (R . Langston and T. Weinert, unpublished data) into ChrV. The rad9Δ ChrVII 403-site deletion strains are specified as follows: Δ403-large deletion ChrVII 400650–406922::KanMX4/NAT; Δ403-small deletion ChrVII 401477–405542 bp::KanMX4/401477–405542 bp:: NAT; Δ403-right deletion ChrVII 403302–405776 bp::HPH/403302–405776 bp::NAT; and Δ403-left deletion 401287–403302 bp::HPH/401287–403302 bp::NAT.
WT ChrV disome strain (TY800) is MATa ade2-1 leu2-3 trp1-1 his3::his3-11,15 can1-100 GAL psi+ 187520–187620 bp::LEU2/187520–187620 bp can1::ADE2/can1::HIS3 541000 bp/541000 bp::CAN1-NAT (P. J. Vinton, R. Langston, and T. Weinert, unpublished data). The WT strain was derived from W303a and was a gift of Angelica Amon.
WTGCR (Gross Chromosomal Rearrangement (GCR)) strain (RDKY6678) is ura3-52 leu2Δ trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072W::HPH (Putnam et al. 2009). erv14GCR was derived from WTGCR and erv14GCR = erv14::KanMX4.
WT397 strain (TWY397a) is MATa ura3his7Ieu2trp1 (Weinert et al. 1994). The erv14397 mutant strain was derived from WT397 by transplacement mutant erv14 for ERV14 (using pWL100 and erv14397 = erv14atg).
Chromosome instability assays
The instability of the ChrVII disome was carried out as described previously (Admire et al. 2006). Briefly, the ChrVII disome is a haploid that contains an extra ChrVII homolog (Figure 1). One homolog contains the CAN1 gene that encodes arginine permease; when intact, the CAN1 gene confers sensitivity to the drug canavanine. The CAN1 gene allows for selection of cells that have an altered ChrVII that can arise by any of several mechanisms that are still being studied (Admire et al. 2006; Paek et al. 2009; Beyer and Weinert 2016). There are three genetic outcomes of instability that we have documented extensively: chromosome loss, allelic recombinants, and an unstable chromosome. The chromosome structures of loss and allelic recombinants are known, and the structures of unstable chromosomes remain speculative, and at least in part include a dicentric. Knowledge of the exact mechanisms of chromosome changes is not central to the hypothesis put forward here that a slowed cell cycle stabilizes the genome, only that a slowed cell cycle stabilizes most or all forms of chromosome change that we can measure.
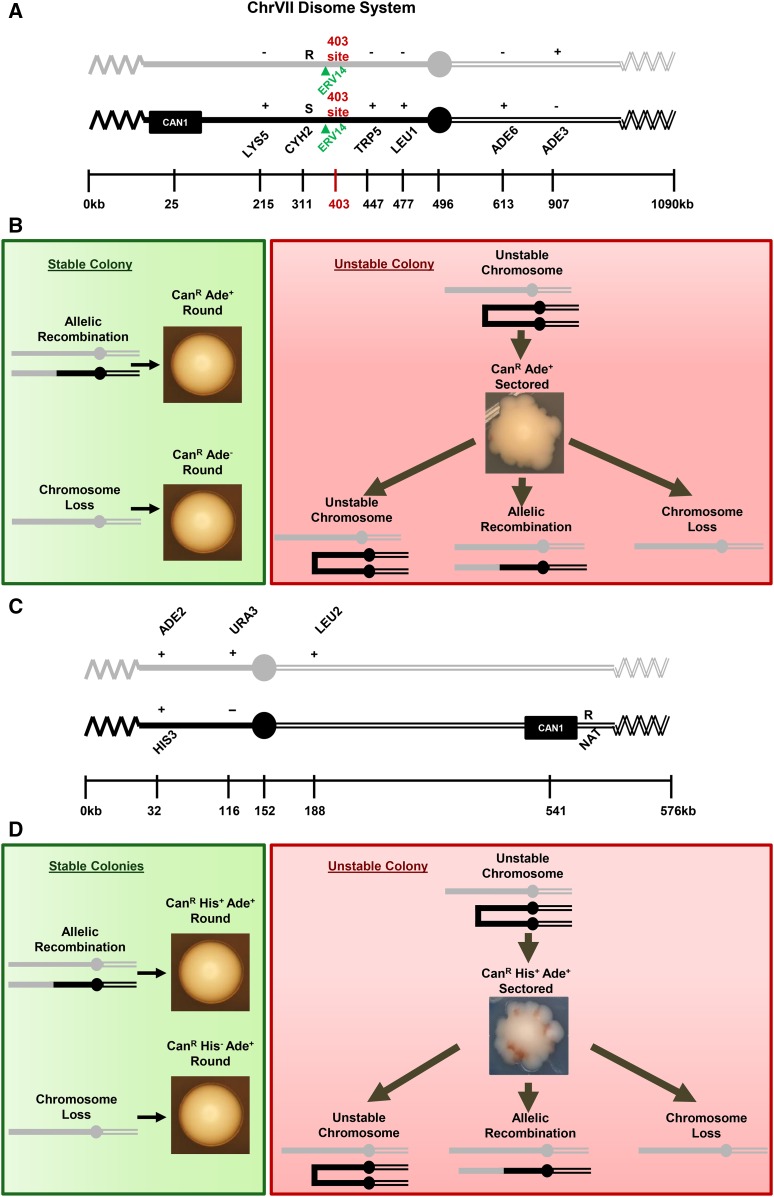
Figure 1.
The ChrVII and ChrV disome systems. (A) Schematic of the ChrVII disome system. Two homologs are shown; the CAN1 homolog is black and the non-CAN1 homolog is gray. The 403-site is shown in red and ERV14 is shown in green due to their relevance in the discovery of slowed cell cycle stabilization. (B) Colony phenotypes. (Green) Allelic recombination and chromosome loss result in stable round colonies. (Red) A cell with an unstable chromosome generates a sectored colony containing three types of cells: cells inheriting the unstable chromosome, cells that have lost the unstable chromosome, and cells that have undergone allelic recombination to stabilize the unstable chromosome. (C) Schematic of the ChrV disome system. Two homologs are shown: the CAN1 homolog is black and the non-CAN1 homolog is gray. (D) Same as in B.
We measure instability using a now-standard assay. Briefly, single cells containing both intact ChrVII homologs were plated on rich media plates (YEPD, 2% dextrose) and grown for 2–3 days at 30° to form colonies; spontaneous chromosome changes occur as cells grow on rich media plates. We typically allowed mutant cells that grow more slowly to grow for a longer time on the rich media plate than their control counterpart (e.g., erv14 and ERV14), to standardize the two strains for number of cell divisions. We typically measure instability on colonies that have 1 × 106–2 × 106 cells. We had done this previously for slower-growing mutant rrm3 cells compared to RRM3 cells (Admire et al. 2006). This being said, we note that we have not detected substantial differences in frequencies of events (less than two-fold) of colonies of different sizes. In any case, we grow colonies to similar sizes. To identify cells with chromosome changes, the colonies were suspended in double-distilled water (ddH2O), cells counted with a hemocytometer, and plated on complete media to determine viability or on media containing canavanine (60 μg/ml) lacking either arginine and serine, or arginine, serine, and adenine to determine instability events. Percentage of viability was calculated by counting the number of cells that had formed a microcolony vs. the number of cells that had not (microcolony with typically >50 cells = viable cell; microcolony with ≤6 cells = inviable cell), unless otherwise noted. We then determined the frequencies of the three types of chromosome events: loss, allelic recombinants, and unstable chromosomes, using a combination of colony morphology and genetics. Chromosome loss generates CanR Ade− cells (Figure 1) that form round colonies on canavanine media with adenine. Allelic recombinants generate CanR Ade+ cells that form round colonies most easily detected on canavanine media without adenine. Unstable chromosomes generate CanR Ade+ cells that form sectored colonies most easily detected on canavanine media without adenine. We infer the first cell to form a CanR Ade+ sectored colony was unstable because the colony contains cells of multiple different phenotypes (Admire et al. 2006). For each mutant strain tested, all three forms of instability were determined from at least six colonies, representing six biological replicas, from each of two genetically generated isolates, unless otherwise noted. We report the median frequency and quartile deviations, and statistical significance was calculated using the Kruskal–Wallis method (Kruskal and Wallis 1952).
FACS analysis
To determine the distribution of cells in the cell cycle, cells were grown to log phase overnight in YEPD liquid media, ∼5 × 106 cells were pelleted in an Eppendorf tube, washed in 1 ml ddH2O, then suspended in 400 μl ddH2O and sonicated to break up cell clumps using Fisher Scientific Sonic Dismembrator Model 500; microtip; output = 10%, 10 half second pulses. Then, 950 μl of 100% ethanol was added (final concentration = 70%) to fix the cells at room temperature for ≤1 hr. Cells were pelleted and supernatant was removed and washed in 800 μl of 50 mM sodium citrate (pH 7.2). Cells were pelleted again, supernatant was removed, and cells were resuspended in 500 μl of 1 μg/ml RNase A Solution, incubated at 37° for 1–2 hr, followed by addition of 50 μl of 20 mg/ml proteinase K and incubation at 50° for an additional 1–2 hr. Cells were then sonicated as before and 500 μl of Sytox Green solution was added. FACS analysis was performed using the Attune Acoustic Focusing Cytometer. FACS analysis was done in duplicate for all strains tested, and the average of two biological replicas is reported, and one representative profile is shown.
Daughter cell vs. mother cell instability assay
We sought to determine whether a slowed cell cycle in G1 in daughter cells vs. mother cells would stabilize the genome of the daughter cell. Single rad9Δ::ura3adh4Δ::KanMX4 cells were plated to rich media plates containing G418 (Geneticin) to select against loss of the ChrVII CAN1 homolog (KanMx4 was inserted at the ADH4 locus 15 kb from the left end of ChrVII CAN1 homolog) and allowed to grow ∼40 hr into colonies. Approximately 70 colonies were put into 3 ml PBS in a 15-ml Falcon tube and vortexed, and then approximately 50 ml from the 15 mL Falcon tube was aliquoted and put into a 1.6 ml Eppendorf tube and 1 mL PBS was added to the Eppedorf tube. Cells were then sonicated to break up cell clumps using Fisher Scientific Sonic Dismembrator Model 500; microtip; output = 10%, 10 − half second pulses. A total of 13 μl of WGA-Alexa Fluor 647 dye was then added and gently mixed by vortexing. The tube was then covered with aluminum foil and allowed to incubate at room temperature for 60–90 min, gently mixing/vortexing every 15 min, to allow staining of bud scars. Cells were then washed twice with PBS and transferred to a 5-ml BD Falcon tube with 5 ml PBS. Unstained cells, representing unscarred daughter cells, were used to calibrate and set sorting gates on the BD FACSCanto II. Mother and daughter cells were then sorted into two separate Falcon tubes. Mother and daughter cells were then plated to selective media to determine chromosome instability, demarcating the initial time point (to). The remaining cells were then pelleted and resuspended in rich liquid media to allow them to cycle. Three hours later, cultures that at t = 0 were mother and daughter cells were again plated to selective media to determine chromosome instability (to + 3 hr). Cell budding morphology was also determined at the to and to + 3 hr time points using light microscopy to ensure cells were cycling.
Slowed cell cycle and single cell cycle experiments using temperature-sensitive mutant cdc13 strains
We measured instability in cdc13 and cdc13erv14 cells dividing continuously at 30°, a semipermissive temperature, and in a more elaborate protocol in which cells were limited for Cdc13 function during a single cell cycle. Limiting Cdc13 function by either protocol induced instability (R. Langston and T. Weinert, unpublished data). Test for instability at the semipermissive temperature is done as with CDC+ strains; cells are plated to rich media plates and allowed to form colonies at 30°, then plated to selective media for CanR colonies at 25°, the permissive temperature for cdc13ts. For the more elaborate single cell cycle experiment, cells were initially grown on minimal media plates at the 25° permissive temperature for ∼72 hr. A flask with 90 ml YEPD was inoculated with cells grown on the minimal media plate, and they were grown overnight at 25° to mid-log phase (∼5 × 106 cells/ml). A total of 10 ml of 2.0 M hydroxyurea (HU) was added to a final concentration of 0.2 M to synchronize cells at the G1–S transition, and agitated for 1.5 hr at 25° to effect arrest. Each culture was divided into two 50-ml Falcon tubes (one for 25° Control incubation and one for 37°), each pelleted and washed twice by gently pipetting with 50 ml 25° YEPD, resuspended in 50 ml 25° YPD by gentle pipetting, and another 50 ml YEPD was added and gently mixed to ensure homogenous resuspension of cells. One culture was then put at 25° as a control (permissive temperature) and the other at 37° (restrictive temperature), and aliquots of 1.5 ml were taken at given time points (3 or 4 hr post-temperature shift). Cells from each aliquot were pelleted, resuspended in 1 ml ddH2O, sonicated, shifted to 25° to allow resumption of cell division, and simultaneously plated for instability.
Determining the cell doubling time
We determined the time of cell division for cells as they grow on solid media agar plates, the condition under which they undergo instability in our assays. Cells were grown overnight to log phase in liquid minimal media to select for cells with intact ChrVIIs. Aliquots of 1 ml were then placed into 1.6-ml Eppendorf tubes and sonicated to disrupt clumps of cells. Cells were plated onto YEPD rich agar media plates to allow detection of growth of single cells. Cells were plated in quadrants on agar media to allow comparison of growth of different strains on the same plate. As a control, one quadrant contained rad9Δ cells. Plates were marked to allow tracking microscopically of the same set of cells over time. Approximately 50 cells of each strain were then counted and tracked under a light microscope with a 40× objective. Cell counts were taken at five time points ∼1.5 hr apart, and the doubling time was calculated between consecutive time points. The median doubling time was determined for each strain tested and normalized to the rad9Δ strain present on the same plate. All doubling time assays were performed in duplicate for each strain. The data for the “instability frequency vs. doubling time” plots use the median doubling time of each strain. The doubling times were calculated using the math formula attained from Roth (2006) Doubling Time Computing, available from: http://www.doubling-time.com/compute.php. For chemically treated cells, we grew cells overnight on agar plates containing the drug to acclimate the cells to the drug, then washed the cells off the plate, sonicated them, replated on agar media containing the drug, and doubling time was determined as described above.
DNA damage sensitivity assay
To determine the sensitivity of cells to DNA damage, we first grew cells overnight to log phase in 6 ml YEPD liquid at 30°. Cells were then pelleted and resuspended in 75 μl fresh YEPD. Three 50-ml flasks were prepared with 10 ml of YEPD, HU-YEPD (0.3 M HU), and methyl methanesulfonate (MMS)-YEPD (0.005% MMS) for the ERV14+ and erv14 strains of interest. An ∼1.5-ml aliquot of culture was taken from each flask at 2-, 4-, or 6-hr time points, pelleted and washed twice with YEPD, and then resuspended in 500 μl YEPD, sonicated, plated on YEPD agar plates, and viability determined microscopically after growth for ∼18 hr at 25°. The percentage of viability was calculated by counting the number of cells that had formed microcolonies vs. the number of cells that had not formed a microcolony (microcolony with ≥50 cells = viable cell; microcolony with ≤16 cells = inviable cell).
L-canavanine exclusion assay 1: Screening for chromosome loss, ChrV disome system
We carried out two experiments to rule out that ERV14 status was affecting drug uptake, thus potentially confounding our assay of instability. In one assay, we determined the frequency of loss by a screen, not a selection, using mutant cells with a very high frequency of loss (median of 210 × 10−4) of in the ChrV system (Table 1; ChrV rad17). We grew individual mutant cells on YEPD agar plates at 30° for ∼48 hr to create individual colonies that contained cells that had lost a chromosome. We then pooled 50 colonies, and plated ∼600 cells onto synthetic complete (SC) media plates (five times) to assay each for loss. The cells were incubated for ∼2 days at 30°, and resulting colonies were replica plated on media that identify loss (lacking leucine and histidine: SC −leu −his). Of the 600 colonies screened, cells that had incurred loss would form a colony that when replica plated would not grow on −leu −his plates. Plates were incubated overnight at 30° after replica plating and scored for chromosome loss frequencies.
Table 1. Mutant DDR and spindle checkpoint strains.
| Genotype | Unstable Chr. (×10−5) | Allelic rec. (×10−5) | Chr. loss (×10−4) | ||||
|---|---|---|---|---|---|---|---|
| Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | ||
| ChrVII disome mutants | ChrVII RAD+ (wild type) | 6.0 (3.5, 7.6) | 1 | 6.8 (3.5, 10) | 1 | 3.7 (1.1, 5.2) | 1 |
| erv14 | 1.7 (1.3, 2.2) | 3.5* | 1.1 (0.6, 2.2) | 6.2* | 0.8 (0.6, 3.3) | 4.6 | |
| rad9∆ | 57 (40, 71) | 1 | 6.0 (2.8, 8.2) | 1 | 22 (16, 39) | 1 | |
| rad9∆ erv14 | 7.2 (2.5, 16) | 7.9* | 2.2 (0.83, 4.2) | 2.7* | 7.2 (3.6, 15) | 3.1* | |
| rad17∆a | 560 (410, 660) | 1 | 28 (14, 34) | 1 | 39 (34, 70) | 1 | |
| rad17∆ erv14 | 140 (110, 160) | 4.0* | 5.4 (5.4, 11) | 5.2 | 24 (11, 27) | 1.6 | |
| tel1∆a | 49 (44, 54) | 1 | 72 (59, 82) | 1 | 15 (12, 24) | 1 | |
| tel1∆ erv14 | 31 (29, 36) | 1.6* | 5.6 (3.7, 7.6) | 13* | 8.3 (7.3, 12) | 1.8 | |
| rad51∆ | 280 (240, 320) | 1 | 12 (9.2, 17) | 1 | 27 (2.2, 68) | 1 | |
| rad51∆ erv14 | 110 (98, 140) | 2.6* | 2.3 (0.0, 3.0) | 5.3* | 37 (31, 49) | 0.72 | |
| mad2∆ | 16 (15, 32) | 1 | 14 (11, 18) | 1 | 54 (32, 81) | 1 | |
| mad2∆ erv14 | 13 (10, 16) | 1.3 | 2.2 (1.7, 3.1) | 6.5* | 13 (9.8, 21) | 4.2* | |
| ChrV disome mutants | ChrV RAD+ (wild type) | 13 (9.8, 16) | 1 | 21 (17, 35) | 1 | 0.52 (0, 1.5) | 1 |
| erv14 | 14 (10, 20) | 0.93 | 2.1 (1.0, 2.3) | 10* | 2.6 (1.0, 5.7) | 0.20* | |
| rad9∆ | 300 (240, 340) | 1 | 22 (13, 31) | 1 | 170 (130, 210) | 1 | |
| rad9∆ erv14 | 210 (180, 230) | 1.4* | 3.1 (2.1, 4.2) | 7.1* | 68 (53, 96) | 2.5* | |
| rad17∆a | 1800 (1600, 1900) | 1 | 210 (150, 240) | 1 | 210 (150, 300) | 1 | |
| rad17∆ erv14 | 900 (710, 1000) | 2.0* | 45 (23, 74) | 4.7* | 80 (45, 100) | 2.6* | |
| mad2∆a | 26 (18, 27) | 1 | 17 (12, 24) | 1 | 34 (33, 42) | 1 | |
| mad2∆ erv14 | 27 (17, 31) | 0.96 | 5.2 (4.2, 6.6) | 3.3* | 8.4 (5.5, 19) | 4.0* | |
Instability frequencies and genome fold stabilization of erv14− strains normalized to their ERV14+ counterparts. Cells with a light gray background indicate genome fold stabilization increase; cells with a white background indicate decreased fold stabilization (<1.0 = increased instability) or no change in stabilization (=1.0). Statistically significant appears in boldface type. Kruskal–Wallis test, * P < 0.01. Rec., recombinant; stabil., stabilization for all tables.
Sample sizes: rad17∆ N = 6 one isolate; tel1∆ N = 6 one isolate; mad2∆ N = 3 three isolates; ChrV rad17∆ N = 3 three isolates; ChrV mad2∆ N = 3 two isolates, rad9∆ N = 3 one isolate; rad9∆ ERV14x2 N = 3 one isolate.
L-canavanine exclusion assay 2: Selecting for chromosome loss using cycloheximide in the ChrVII disome system
The second test of whether ERV14 status was affecting drug uptake utilized the drug cycloheximide instead of canavanine. The CYHS allele is dominant to the cyhR allele, thus the original cell (CYHS/cyhR) is cycloheximide sensitive, while a cell that loses the CYHS allele, by chromosome loss for example, is cycloheximide resistant. To determine loss by selecting for cycloheximide resistance, cells with the initial disome were grown on YEPD agar plates at 30° for ∼48 hr to create individual colonies that had undergone instability. Six colonies were pooled together in ddH2O (Table S2). A total of 104 cells were plated to SC +cycloheximide (5 μg/ml cycloheximide) plates (two times) and grown for ∼48 hr at 30° (cells that grow into colonies on SC +cycloheximide media plates indicate loss of the ChrVII CAN1 homolog, see Figure 1A). SC +cycloheximide plates were replica plated to SC and SC −ade plates. Plates were incubated overnight at 30° and scored for chromosome loss frequencies.
Data availability
Strains are available on request. Supplemental Material, Table S7 contains genotypes of all strains used.
Results
The ChrVII and ChrV disome instability systems
The yeast model systems that allowed us to identify and characterize erv14 are shown in Figure 1. We used both a well-studied ChrVII disome, and a newly developed ChrV disome (details to be published elsewhere). The two disomes behave very similarly in terms of genome instability. The principle of both disomes is that chromosome error is rare, necessitating a selection for cells that harbor a changed chromosome. The CAN1 gene in both systems provides a positive selection for such a change; cells that retain the CAN1 gene (CanS cells) are killed by the drug canavanine, and cells that undergo a chromosome change to lose CAN1 (CanR cells) survive the drug. Both disomes generate three different types of chromosome changes, including unstable chromosomes, chromosome loss, and allelic recombination (Figure 1, B and D). We have reported previously that errors in DNA repair checkpoints cause chromosome instability, e.g., Admire et al. (2006). We typically do not know where errors occur on the chromosome, except for certain instances where error occurs in the telomere (Beyer and Weinert 2016; R. Langston and T. Weinert, unpublished data; a telomere biology mutation (cdc13) was also used in this study). (File S1, File S2, File S3).
To measure genome instability, individual ChrVII or ChrV disome cells bearing intact chromosomes are grown on rich media agar plates to allow chromosome changes to occur (changes are typically spontaneous in this study, except when we use a cdc13 conditional telomere mutation). Chromosome changes are rare (10−3 to 10−6, depending on type of event and mutant background), and we thus detect changes by selecting for loss of CAN1 to form CanR cells that form CanR colonies. We then determine the type of chromosome change by a combination of CanR colony shape and genetic phenotypes. Chromosome loss is detected as round colonies on canavanine-selection plates and prove to be CanR Ade− for ChrVII loss and CanR His− for ChrV loss. Allelic recombinants are detected as round colonies on canavanine-selection plates lacking adenine (ChrVII) and histidine (ChrV). The selection media allows CanR Ade+ and CanR His+ cells to form colonies, which prove to be allelic recombinants (see Materials and Methods). Finally, unstable chromosomes are generated in a third event that is also detected in both disome systems. Unstable chromosomes are detected as sectored colonies (Figure 1, B and D) on canavanine-selection plates that are also lacking adenine (ChrVII system) or histidine (ChrV system). Unstable chromosomes may be dicentrics. They form in rich media just prior to selection and rearrange as the first CanR cell divides on selective media, forming sectored colonies containing cells with multiple different genotypes. In contrast, round CanR colonies (from loss or allelic recombinants) contain progeny with predominantly one genotype.
The initial molecular events leading to the three rearrangements are in general unknown. It is likely that a replication error of some type underlies all three events, or alternatively some defect in chromosome segregation presumably due to a defect in the kinetochore (Paek et al. 2009; Kaochar et al. 2010; Beyer and Weinert 2016). We use the term “instability” to include all three instability outcomes.
Investigation of a fragile site implicates erv14 in genome stability
The circuitous path of discovery that a slowed cell cycle stabilizes the genome began with the investigation of a region on ChrVII that we previously found was associated with chromosomal changes. We found that unstable chromosomes that form sectored colonies, in particular, are frequently resolved to stability in a 4-kb region located 403 kb from the left telomere of ChrVII (Admire et al. 2006; Figure 1A). This 4-kb region contains inverted repeats (of sigma and delta sequences, ends of retrotransposons) that we showed fuse and form a dicentric. This 4-kb site also has four other sigma or delta sequences, as well as fragments of the mitochondrial genome; such fragments arise as the consequence of DNA break and repair, consistent with the instability of this region, as discussed in Admire et al. (2006).
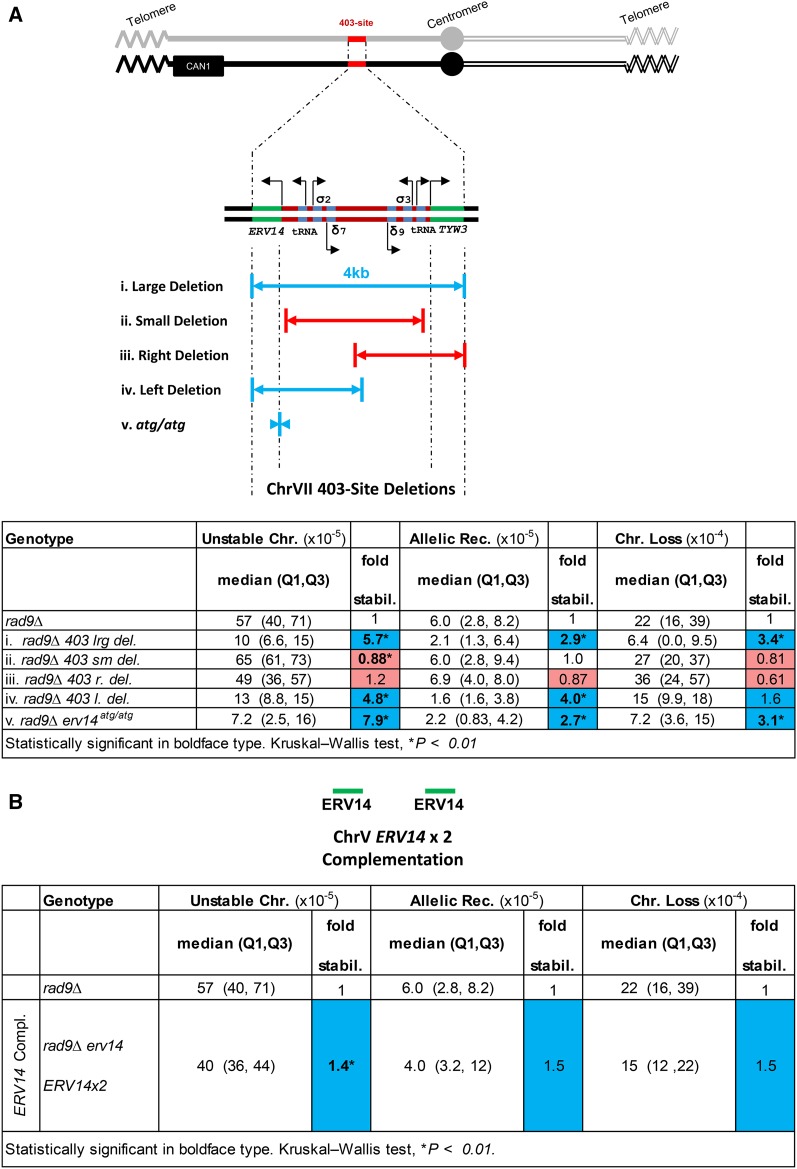
In the current study we initially posited and tested whether the inverted repeats in the 403-site initiate instability and form unstable chromosomes. We deleted specific portions of the 403-site and measured instability. (We modified DNA sequences on both ChrVII homologs as events could potentially initiate on either homolog.) We performed these studies of the 403-site in rad9Δ mutant cells because of its relatively high frequency of instability compared to WT cells, giving us more sensitivity to detect stability differences as different portions of the 403-site were deleted.
When we deleted sequences in the 403-site, including sequences that disrupted the adjacent genes (ERV14 and TWY3), the genome was stabilized for all three events (large deletion, Figure 2A, i). However, when we made smaller deletions that included only the inverted repeats and transfer RNA (tRNA) genes, leaving ERV14 and TWY3 intact, much to our surprise we found frequencies of instabilities were not altered (Figure 2A, ii). We subsequently generated deletions centered on the left and right of the 403-site, and found that only deletions that disrupted ERV14 stabilized the genome (Figure 2A, i, iv, and v), while those that disrupted TWY3 had no effect (Figure 2A, iii). To ask whether the Erv14 protein or ERV14 DNA sequence per se was important in genome stability, we generated mutations totaling only seven bases that altered the ERV14 ATG start codons to non-ATG codons, and on both ERV14 copies on the two ChrVII homologs (the mutation termed erv14atg/atg; Figure 2A, v). We found that the erv14atg/atg mutations also suppressed instability of rad9Δ, and to a similar degree as the large 403-site deletion (Figure 2A, i). We then asked whether expression of ERV14 per se causes instability. We inserted ERV14 elsewhere in the genome, and found that this ERV14 insertion complemented the erv14 phenotype; cells became unstable again (though unstable chromosome frequency was not quite restored to the level of rad9Δ, possibly due to high variance in rad9Δ; Figure 2B and Table 2). Extra copies of ERV14 (four copies vs. two copies) did not increase instability further (Table 2). We conclude that most of the genome stability caused by an ERV14 loss-of-function mutation is due to loss of the Erv14 protein (Figure 2). (In this study erv14 = erv14atg/atg mutation, unless otherwise noted.) (In our subsequent study of erv14 genome stabilization below, we continue using the erv14atg/atg mutation instead of a null mutation with an insertion of foreign DNA, because the erv14atg/atg mutant cells exhibit a lower variance in genome stabilization frequencies than ERV14 null cells, and because the base sequence changes may minimize any potential “neighboring gene effect” on the genome (Wang et al. 2011; Baryshnikova and Andrews 2012). We also determined the effect of the start codon mutation on the ERV14 gene (417 bp) by inspecting the sequence; the next out-of-frame ATG sequence is at 119 bp followed by a TAA stop codon sequence at 125 bp, and the next in-frame ATG sequence is at 351 bp. We infer that if an Erv14atg/atg protein is generated, it will be a loss-of-function protein.
Figure 2.
The 403-site deletions and ERV14 complementation. (A) Explanations of the various 403-site deletions. Deletions/mutations are on both ChrVII homologs. Various rad9Δ 403-site deletions normalized to rad9Δ. (i) Large deletion of the 403-site, includes deletions of ERV14 and TYW3. (ii) Small deletion of the 403-site, ERV14 and TYW3 are left intact. (iii) Right deletion of the 403-site, ERV14 is intact and TYW3 is deleted. (iv) Left deletion of the 403-site, ERV14 is deleted and TYW3 is intact. (v) atg/atg deletion, only ERV14 start codons mutated. (B) ERV14 complementation of rad9Δ erv14. The rad9Δ erv14 ERV14x2 strain is normalized to rad9Δ. Blue and red squares correspond to genome fold stabilization increase or genome fold stabilization decrease (<1.0 = increased instability), respectively.
Table 2. ERV14 complementation and copy number.
| Genotype | Unstable Chr. (×10−5) | Allelic rec. (×10−5) | Chr. loss (×10−4) | |||
|---|---|---|---|---|---|---|
| Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | |
| rad9∆ | 57 (40, 71) | 1 | 6.0 (2.8, 8.2) | 1 | 22 (16, 39) | 1 |
| rad9∆ erv14 ERV14x2 | 40 (36, 44) | 1.4* | 4.0 (3.2, 12) | 1.5 | 15 (12, 22) | 1.5 |
| rad9∆ ERV14x2a | 51 (43, 54) | 1.1 | 7.2 (6.4, 8.6) | 0.83 | 21 (19, 24) | 1.0 |
Complementation section: instability frequencies and genome fold stabilization of ERV14 complementation of rad9Δ erv14 normalized to rad9Δ erv14; ERV14 copy number section: instability frequencies and genome fold stabilization of rad9Δ with extra copies of ERV14, normalized to rad9Δ. Cells with a light gray background indicate genome fold stabilization increase; cells with a white background indicate decreased fold stabilization (<1.0 = increased instability) or no change in stabilization (=1.0). Statistically significant appears in boldface type. Kruskal–Wallis test, * P < 0.01.
Sample sizes: rad17∆ N = 6 one isolate; tel1∆ N = 6 one isolate; mad2∆ N = 3 three isolates; ChrV rad17∆ N = 3 three. isolates; ChrV mad2∆ N = 3 two isolates, rad9∆ N = 3 one isolate; rad9∆ ERV14x2 N = 3 one isolate.
Since our initial studies of erv14 were in a mutant rad9Δ, we reasoned that stabilization of the chromosome could be due to an effect of Erv14 on a Rad9-specific function. We therefore asked whether erv14 suppresses instability arising in other DNA damage response (DDR) mutant strains. We found that an erv14 mutation, to statistical significance with varying degrees, indeed stabilized the genomes of other DDR mutant strains (rad17Δ, rad51Δ, and tel1Δ; Table 1). Furthermore, erv14 stabilizes the genome in both checkpoint-deficient and checkpoint-proficient cells (e.g., rad9Δ and rad51Δ, respectively) and stabilized the genome of an otherwise wild-type cell. We conclude that erv14 genome stabilization is not rad9Δ specific, stabilizes multiple forms of ChrVII instability (e.g., all three types of events), and stabilizes the genome in checkpoint-deficient and proficient cells. Below we will return to show that erv14, and other conditions that slow the cell cycle, also stabilize the genome in a conditional mutant cdc13 that induces DNA damage and instability in the telomere.
An erv14 mutation stabilizes chromosomes made unstable by defects in spindle assembly repair pathways
Next, we tested whether an erv14 mutation suppresses defects in the spindle checkpoint pathway. We measured instability in cells defective in the spindle assembly checkpoint. mad2Δ mutant cells exhibit instability in the ChrVII disome, and we found that erv14 significantly suppresses instability of chromosome loss and allelic recombinants, indicating that defects in the spindle assembly checkpoint are ameliorated by erv14 (Table 1).
An erv14 mutation marginally decreases DNA damage sensitivity
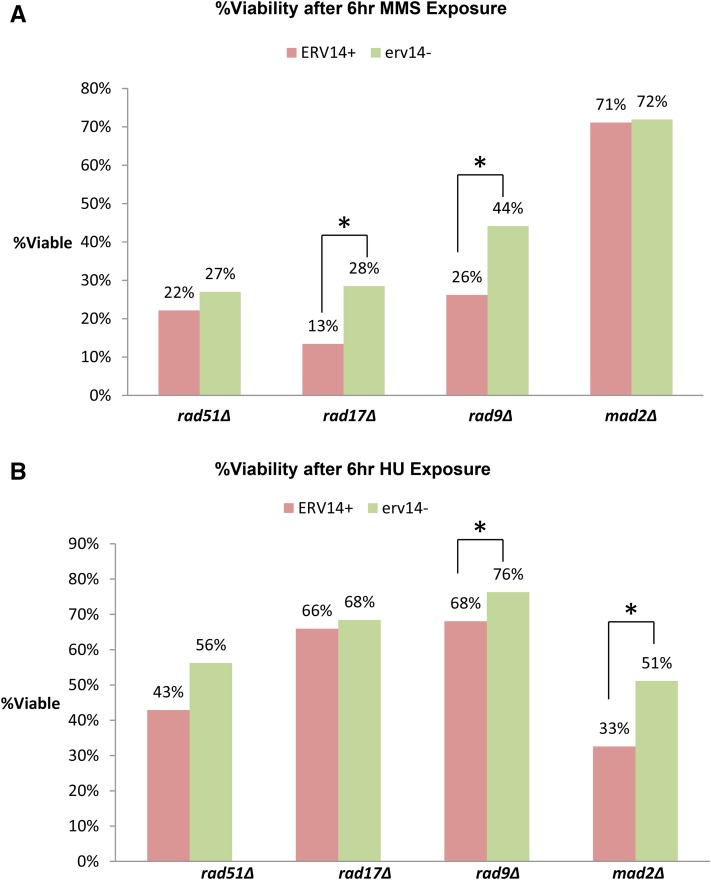
We next determined whether DNA damage sensitivity per se is suppressed by erv14. We tested whether erv14 suppressed DNA damage sensitivity caused by MMS, an alkylating agent that methylates DNA bases and causes DNA replication errors, and HU, a dNTP pool depleting drug. We found that there was statistically significant increased resistance to MMS DNA damage in rad17Δ erv14 and rad9Δ erv14 cells compared to their ERV14+ counterparts (Figure 3A), and increased HU DNA damage resistance in rad9Δ erv14 and mad2Δ erv14 compared to rad9Δ and mad2Δ, respectively (Figure 3B). We conclude erv14 increases DNA damage resistance and enhances DNA repair.
Figure 3.
DNA damage sensitivity assays. (A) Percentage of viability after 6-hr exposure to 0.005% MMS liquid rich media for rad51Δ, rad17Δrad9, rad9Δ, and mad2Δ vs. erv14− counterparts. (B) Percentage of viability after 6-hr exposure to 0.15 M HU liquid rich media for rad51Δ, rad17Δrad9Δ, rad9Δ, and mad2Δ vs. erv14− counterparts. *Statistically significant P ≤ 0.05 using Kruskal–Wallis test.
An erv14 mutation does not alter frequency of point mutations
Finally, we tested whether erv14 affects point-mutation frequency, a measure of errors by DNA polymerase and correction by repair mechanisms. We measured the mutation frequency of CAN1 in a WT397 haploid cell (Weinert et al. 1994), and did not find a statistically significant effect by erv14 on mutations in CAN1 (Figure S1 and Table S1B). We conclude that erv14 may not affect base-pair-change error frequency and repair.
An erv14 mutation stabilizes other chromosomes
We next determined whether stabilization by erv14 is indeed genomewide, or whether stability is conferred only to ChrVII. To determine whether instability is affected genomewide, we used two additional assays of instability, both of which measure events on ChrV. First, we used a ChrV disome that we developed to determine whether findings of ChrVII instability generalize to other chromosomes (to be reported elsewhere). We generated erv14 mutations in a variety of mutant pathways and found that the erv14 mutation indeed renders these wild-type and mutant strains more stable in one or more instability types tested (Table 1).
We then tested whether the erv14 mutation stabilizes the genome in the extensively used GCR assay of ChrV developed by Kolodner and colleagues (Chen and Kolodner 1999; Putnam et al. 2009; Putnam and Kolodner 2010). In this assay, the URA3 and CAN1 genes are placed ∼15 kb from the telomere in a haploid cell; loss of CAN1 and URA3 is selected by CanR FOAR and arises from loss of DNA that can extend to the first essential gene ∼20 kb from the telomere. We found that an erv14 mutation in this system also resulted in a more stable genome (figure 1 in Putnam et al. 2009; Table S1A). We have not further analyzed the CanR FOAR products to determine whether a specific subtype of genome instability is suppressed by an erv14 mutation. Because erv14 increases the stability of ChrVII and ChrV, and in disomes and in a haploid, we conclude that erv14 has a general stabilizing effect on the genome.
Preliminary summary: erv14 stabilizes the genome
From these results we conclude that erv14 increases stability in cells defective for both DNA or spindle damage, and acts genomewide. Before discussing mechanism further, we comment briefly on two aspects of our study. First, we are unable to interpret why erv14 suppresses some but not all events in various mutations; e.g., why erv14 suppresses allelic recombination in tel1Δ but less so in rad17Δ. We do not yet understand the mechanistic difference sufficiently between the three events to interpret our results of suppression. Second, it is curious but apparently without biological meaning that the ERV14 gene is present on ChrVII in a region that is highly unstable.
How might Erv14 cause instability, or how does the erv14 mutation stabilize the genome?:
Thus far we have shown that the erv14 mutation stabilizes the genome of WT cells and also in cells defective in a variety of chromosome pathways. So how does erv14 stabilize the genome, and against different types of error? Erv14’s known major role is as an ER cargo membrane protein (Herzig et al. 2012), with no known direct connection to nuclear biology. Erv14 is a protein whose function is to transport membrane and secretory proteins from the ER to the Golgi and is known to transport at least 30 proteins (Herzig et al. 2012). Given the large number of Erv14 cargo proteins, it is not surprising that an erv14 deletion is pleiotropic, inducing what is called “ER stress” due to the trapping of secretory and membrane proteins in the ER. ER stress itself can “induce” multiple responses and “inhibit” multiple processes, ranging from the unfolded protein response (UPR) (an ER stress response), ER-associated nuclear protein degradation (n-ERAD) (degradation of misfolded proteins), nucleophagy (nuclear membrane autophagy), apoptosis, sphingolipid synthesis, ER chaperone protein folding, and ER-to-Golgi protein transport, to the potential of causing defects in chromatin organization (we infer chromatin organization based on the fact that Erv14 may transport inner nuclear membrane (INM) proteins, though there is no direct evidence of INM protein transport).
We systematically examined each of these ERV14-dependent, ER-linked responses to determine whether any explained the effect of erv14 stabilizing mutant genomes. We took either of two approaches to examine whether a particular ERV14-associated response or process was stabilizing the genome. Do mutations in other genes stabilize the rad9 genome, like erv14? If so, a gene X rad9 double mutation, like an erv14rad9 double mutation, would be more stable (Table 3). Alternatively, it could be that the rad9erv14 cell requires a gene Y to stabilize the genome; if so, a rad9erv14 gene y triple mutation would redestabilize the genome to rad9 levels (Table 4). For example, if erv14 is inducing the UPR pathway that somehow stabilizes the rad9Δ genome, then disrupting the UPR pathway in a UPR stabilized rad9Δ erv14 genome should increase instability.
Table 3. ER stress, sphingolipid, and chromatin anchoring mutations in rad9∆ background.
| Genotype | Unstable Chr. (×10−5) | Allelic rec. (×10−5) | Chr. loss (×10−4) | |||
|---|---|---|---|---|---|---|
| Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | |
| rad9∆ | 57 (40, 71) | 1 | 6.0 (2.8, 8.2) | 1 | 22 (16, 39) | 1 |
| rad9∆ erv25∆ | 26 (23, 37) | 2.3* | 4.2 (3.1, 5.4) | 1.5 | 12 (7.5, 23) | 1.8* |
| rad9∆ scj1∆ | 28 (22, 36) | 2.0* | 3.7 (2.9, 5.3) | 1.6* | 8.5 (7.1, 11) | 2.6* |
| rad9∆ isc1∆ | 34 (24, 39) | 1.7* | 4.0 (2.0, 5.0) | 1.5* | 14 (11, 27) | 1.6* |
| rad9∆ scs7∆ | 77 (51, 154) | 0.74 | 8.6 (5.9, 11) | 0.70 | 66 (33, 120) | 0.33* |
| rad9∆ sur2∆ | 45 (39, 53) | 1.3 | 4.5 (3.0, 6.2) | 1.3 | 15 (11, 27) | 1.5 |
| rad9∆ esc1∆ | 96 (78, 120) | 0.59* | 5.3 (2.9, 7.7) | 1.1 | 34 (24, 46) | 0.65 |
| rad9∆ esc1∆ yku80∆ | 75 (61, 100) | 0.76* | 4.1 (3.1, 5.7) | 1.5 | 29 (15, 63) | 0.76 |
Instability frequencies and genome fold stabilization in mutant protein folding/transport, sphingolipid, and chromatin anchoring in rad9Δ background normalized to rad9Δ. Cells with a light gray background indicate genome fold stabilization increase; cells with a white background indicate decreased fold stabilization (<1.0 = increased instability) or no change in stabilization (=1.0). Statistically significant appears in boldface type. Kruskal–Wallis test, * P < 0.01.
Table 4. UPR, apoptosis, n-ERAD, and nucleophagy mutations in rad9∆ erv14 background.
| Genotype | Unstable Chr. (×10−5) | Allelic rec. (×10−5) | Chr. loss (×10−4) | |||
|---|---|---|---|---|---|---|
| Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | |
| rad9∆ erv14 | 7.2 (2.5, 16) | 1 | 2.2 (0.83, 4.2) | 1 | 7.2 (3.6, 15) | 1 |
| rad9∆ erv14 ire1∆a | 14 (11, 18) | 0.51 | 1.9 (1.6, 2.1) | 1.2 | 7.2 (6.3, 8.0) | 1.0 |
| rad9∆ erv14 hac1∆a,b | 10 (9.0, 15) | 0.72 | 2.5 (1.5, 3.5) | 0.88 | 7.5 (5.0, 11) | 0.92 |
| rad9∆ erv14 stm1∆ | 2.2 (1.1, 4.4) | 3.3* | 1.6 (1.1, 3.5) | 1.4 | 15 (0.0, 24) | 0.48 |
| rad9∆ erv14 kex1∆a | 13 (10, 14) | 0.55 | 1.6 (1.0, 2.1) | 1.5 | 8.0 (5.7, 33) | 0.90 |
| rad9∆ erv14 doa10∆ | 8.9 (6.9, 13) | 0.81 | 1.7 (1.1, 3.1) | 1.3 | 17 (6.4, 35) | 0.42 |
| rad9∆ erv14 nvj1∆a | 20 (13, 24) | 0.36 | 1.9 (1.6, 2.2) | 1.2 | 12 (5.9, 15) | 0.60 |
Instability frequencies and genome fold stabilization in mutant UPR, apoptosis, n-ERAD, and nucleophagy in rad9Δ erv14 background normalized to rad9Δ erv14. Cells with a light gray background indicate genome fold stabilization increase; cells with a white background indicate decreased fold stabilization (<1.0 = increased instability) or no change in stabilization (=1.0). Statistically significant appears in boldface type. Kruskal–Wallis test, * P < 0.01.
Sample sizes: rad9∆ erv14 ire1∆ N = 2 two isolates; rad9∆ erv14 hac1∆ N = 3 three isolates, rad9∆ erv14 kex1∆ N = 3 three isolates, rad9∆ erv14 nvj1∆ N = 3 two isolates.
Viability not tested.
We therefore undertook an extensive analysis of seven pathways (Figure S2), using the ChrVII disome, and tested 14 genes. Of the Erv14 and ER-related responses and processes tested, most mutations did not identify pathways (neither gene X nor gene Y) that are regulated by Erv14 to regulate genome stability. In Figure S2, explanatory text, we provide a brief rational for each pathway tested.
We also ruled out that a defect in drug uptake in an erv14 mutant strain might lead to the appearance of genome stabilization (Table S2 and explanatory text). Additionally there is an ERV14 paralog called ERV15 with 63% sequence identity to ERV14. We asked whether we might achieve an even larger genome stabilization with an erv14erv15 double mutation. We found that a mutation in ERV15 destabilizes the genome, as a single mutation or in combination with erv14; see Table S3 and explanatory text. Erv15 must transport different cargos than Erv14, and thus have a different effect on cell physiology.
Though we did not identify any particular single ER-related pathway that stabilized the genome, we did find that three loss-of-function mutations did restore chromosome stability to rad9Δ (though not as effectively as erv14): mutations in sphingolipid processing (isc1Δ), chaperone protein folding (scj1Δ), and protein transport (erv25Δ) (Table 3). All three ER-linked mutations reside in different pathways, which did not indicate a unique mechanism by which erv14 might stabilize the genome (e.g., how Erv14 might destabilize the genome). However, all three did share another phenotype, and one in common with erv14 mutant strains; a slowed cell cycle (based on qualitative analysis of colony size growth for the same length of time). This key observation sparked our investigation into the role of a slower cell cycle and genome stabilization.
Slowing the cell cycle by different methods stabilizes checkpoint-deficient and checkpoint-proficient genomes:
Delaying the cell cycle seems a priori a simple matter, but many chemicals and genetic mutations that delay the cell cycle may also alter other aspects of cell physiology that can adversely affect genome stability. With this in mind, we screened a variety of different chemicals and mutations to determine whether any slowed the cell cycle and stabilized the genome. For those chemicals or mutations that stabilize the genome we also asked whether stabilization occurs in checkpoint proficient (WT) and checkpoint-deficient (rad9Δ) cells (Table 5 and Table 6, respectively).
Table 5. WT instabilities using different chemical growth media.
| Genotype | Unstable Chr. (×10−5) | Allelic rec. (×10−5) | Chr. loss (×10−4) | ||||
|---|---|---|---|---|---|---|---|
| Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | ||
| ChrVII RAD+ (wild type) | 6.0 (3.5, 7.6) | 1 | 6.8 (3.5, 10) | 1 | 3.7 (1.1, 5.2) | 1 | |
| Cycloheximide | WT 0.04 μg/ml cyc. | 19 (14, 27) | 0.32* | 6.3 (3.9, 7.1) | 1.1 | 4.2 (1.6, 5.2) | 0.88 |
| WT 0.06 μg/ml cyc. | 22 (15, 29) | 0.27* | 10 (5.8, 17) | 0.68 | 4.7 (4.2, 6.7) | 0.79 | |
| D-glucosamine | WT 2% Dex, 0.2% D-gluc | 7.6 (7.4, 7.9) | 0.8 | 4.3 (2.5, 5.7) | 1.6 | 0.52 (0, 1.1) | 7.1* |
| WT 2% Dex, 0.7% D-gluc | 7.6 (5.2, 10) | 0.79 | 2.9 (1.4, 4.2) | 2.3 | 1.6 (1.1, 2.2) | 2.3 | |
| WT 2% Dex, 1.0% D-gluca | 4.2 (3.9, 8.0) | 1.4 | 4.8 (2.8, 5.6) | 1.4 | 6.4 (0.0, 7.0) | 0.58 | |
| WT 2% Dex, 1.4% D-gluc | 8.6 (7.7, 11) | 0.70* | 4.8 (1.6, 7.5) | 1.4 | 5.1 (3.9, 6.9) | 0.73 | |
| WT 2% Dex, 1.8% D-gluc | 8.0 (4.2, 9.5) | 0.75 | 5.6 (4.7, 12) | 1.2 | 2.1 (1.3, 3.6) | 1.8 | |
| WT 2% Dex, 2.0% D-gluc | 9.1 (7.4, 12) | 0.66 | 3.5 (2.4, 4.5) | 1.9 | 3.6 (0.30, 7.3) | 1.0 | |
| Myriocin | WT 200 ng/ml myriocin | 6.2 (3.8, 8.9) | 0.97 | 6.8 (5.6, 9.9) | 1.0 | 2.1 (1.3, 2.9) | 1.8 |
| WT 400 ng/ml myriocin | 6.1 (2.5, 9.3) | 0.98 | 4.6 (3.4, 6.5) | 1.5 | 1.1 (1.1, 1.1) | 3.4 | |
| WT 600 ng/ml myriocin | 5.6 (4.7, 8.0) | 1.1 | 6.5 (3.8, 11) | 1.0 | 0.0 (0.0, 1.0)* | N/A | |
| YEPG | WT 2% glycerol | 3.2 (1.7, 8.9) | 1.9 | 4.9 (3.1, 40) | 1.4 | 61 (2.1, 420) | 0.06 |
Instability frequencies and genome fold stabilization of WT cells grown on media plates containing D-glucosamine, myriocin, YEPG (glycerol), and cycloheximide normalized to WT cells grown on YEPD. Cells with a light gray background indicate genome fold stabilization increase; cells with a white background indicate decreased fold stabilization (<1.0 = increased instability) or no change in stabilization (=1.0). Statistically significant appears in boldface type. Kruskal–Wallis test, * P < 0.01. N/A, not applicable.
Sample size N = 5.
Table 6. rad9Δ instabilities using different chemical growth media.
| Genotype | Unstable Chr. (×10−5) | Allelic rec. (×10−5) | Chr. loss (×10−4) | ||||
|---|---|---|---|---|---|---|---|
| Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | ||
| rad9Δ | 57 (40, 71) | 1 | 6.0 (2.8, 8.2) | 1 | 22 (16, 39) | 1 | |
| Cycloheximide | rad9Δ 0.04 μg/ml cyc. | 78 (67, 99) | 0.73 | 8.4 (4.6, 12) | 0.71 | 47 (28, 57) | 0.47 |
| rad9Δ 0.06 μg/ml cyc. | 86 (67, 97) | 0.66 | 6.4 (3.1, 12) | 0.94 | 36 (19, 53) | 0.61 | |
| D-glucosamine | rad9Δ 2% Dex, 0.2% D-gluc | 62 (54, 90) | 0.92 | 2.2 (1.1, 4.8) | 2.7 | 6.9 (6.0, 8.2) | 3.2* |
| rad9Δ 2% Dex, 0.7% D-gluc | 72 (43, 130) | 0.79 | 9.0 (8.1, 9.3) | 0.67 | 11 (7.1, 16) | 2.0 | |
| rad9Δ 2% Dex, 1.0% D-gluc | 33 (23, 42) | 1.7 | 1.1 (0.28, 4.3) | 5.5* | 30 (14, 51) | 0.73 | |
| rad9Δ 2% Dex, 1.8% D-gluc | 27 (19, 30) | 2.1* | 3.3 (2.2, 5.9) | 1.8 | 14 (8.8, 19) | 1.6 | |
| Myriocin | rad9Δ 800 ng/ml myriocin | 59 (52, 71) | 0.97 | 7.0 (4.7, 10) | 0.86 | 10 (8.7, 16) | 2.2* |
| rad9Δ 1000 ng/ml myriocin | 43 (36, 50) | 1.3 | 2.2 (1.4, 3.0) | 2.7* | 5.8 (4.9, 8.1) | 3.8* | |
| NaCl | rad9Δ 0.125 M NaCla | 33 (27, 36) | 1.7 | 2.5 (2.0, 7.5) | 2.4 | 21 (12, 27) | 1.00 |
| rad9Δ 0.25 M NaCla | 44 (34, 55) | 1.3 | 13 (11, 17) | 0.46* | 92 (60, 105) | 0.24 | |
| rad9Δ 0.5 M NaCla | 47 (35, 54) | 1.2 | 12 (10, 14) | 0.50* | 76 (67, 130) | 0.30* | |
| rad9Δ 1.0 M NaCla | 120 (94, 180) | 0.48* | 9.0 (5.5, 23) | 0.67 | 4900 (3900, 6300) | 0.0045* | |
| YEPG | rad9Δ 2% glycerolb | 29 (15, 31) | 2.0 | 2.5 (1.4, 6.1) | 2.4 | 710 (350, 810) | 0.031* |
Instability frequencies and genome fold stabilization of rad9Δ grown on media plates containing various concentrations of cycloheximide (protein synthesis inhibitor), D-glucosamine (glucose competitor), myriocin (sphingolipid inhibitor), NaCl (osmotic stress inducer), and YEPG (glycerol, respiration energy source) normalized to rad9Δ grown on YEPD media plates. Cells with a light gray background indicate genome fold stabilization increase; cells with a white background indicate decreased fold stabilization (<1.0 = increased instability) or no change in stabilization (=1.0). Statistically significant appears in boldface type. Kruskal–Wallis test, * P < 0.01.
Viability not tested.
Sample size: N = 5.
Chemicals that slow the cell cycle
We tested the following five chemicals: cycloheximide, which inhibits protein synthesis that is of course required for all aspects of cell function, including cell growth and cell division; NaCl, which is known to slow cell division by inducing osmotic stress, when budding yeast cells under osmotic stress slow or stop their cell cycle to respond to the stress to survive (Wuytswinkel et al. 2000); glycerol, which induces a diauxic shift in yeast and poorer generation of ATP by respiration, slowing cell division (budding yeast can grow using nonfermentable carbon sources, but do so more slowly; e.g., Hasselbring 1913; Roberts and Hudson 2006; Levy et al. 2007); D-glucosamine which is a D-glucose analog that is poorly metabolized by budding yeast, thus slowing growth; and myriocin, which is a drug that inhibits sphingolipid biosynthesis necessary in creating more membrane material during division. We found that only D-glucosamine and myriocin stabilized both the WT and rad9Δ genomes (Table 5 and Table 6, respectively). We describe a slowed cell cycle and genome stabilization by using these two chemicals next (see Table S6 for effect of chemicals on doubling time).
Genome stabilization by D-glucosamine and myriocin
D-glucosamine can be used in conjunction with D-glucose to delay the cell cycle. Budding yeast optimally grows in environments containing fermentable carbon sources, such as D-glucose, and D-glucosamine competes with D-glucose during the initial hexokinase step of glycolysis (McGoldrick and Wheals 1989). While keeping the dextrose concentration constant (2%), we varied amounts of D-glucosamine. At a concentration of 1.8% D-glucosamine, we found that unstable chromosomes are suppressed about twofold in rad9Δ (allelic recombinants and chromosome loss did not significantly change) (Table 6). At a concentration of 0.2% D-glucosamine, chromosome loss is stabilized in WT Rad+ (unstable chromosomes and allelic recombinants did not significantly change).
Myriocin is a drug that inhibits sphingolipid biosynthesis. More membrane lipids are synthesized as cells divide. We examined myriocin because we have found that isc1Δ modestly suppressed instability in rad9Δ, and Isc1 is a sphingolipid processing protein. At a concentration 1000 ng/ml of myriocin, rad9Δ allelic recombinants and chromosome loss were significantly suppressed 2.7-fold and 3.8-fold, respectively, and unstable chromosomes did not significantly change (Table 6). At a concentration of 600 ng/ml, chromosome loss was significantly stabilized in WT RAD+ cells (unstable chromosomes and allelic recombinants did not significantly change; Table 5).
Other mutations that slowed the cell cycle and stabilized the genome
We next investigated the correlation between cell doubling time and genome stabilization by analyzing other mutations reported to slow cell division. We searched the PROPHECY database, a database containing growth curves of budding yeast gene deletion library strains, and selected YME1 (mitochondrial protein quality control), HAP3 (regulator of respiratory gene expression), HTD2 (mitochondrial dehydratase required for respiratory growth and normal mitochondrial morphology), and SAM37 (involved in sorting mitochondrial membrane proteins). Only mutations in YME1 significantly and globally stabilized a rad9Δ mutant genome (Table 7). We conclude that, as with our screen of chemicals that slow cell division, not all mutations that slow cell division stabilize the genome. We suggest some mutations with slower cell division may also compromise chromosome biology, obscuring any genome stabilization that may occur by a slower cell cycle.
Table 7. Slowed cell cycle mutations in rad9Δ background.
| Genotype | Unstable Chr. (×10−5) | Allelic rec. (×10−5) | Chr. loss (×10−4) | |||
|---|---|---|---|---|---|---|
| Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | |
| rad9Δ | 57 (40, 71) | 1 | 6.0 (2.8, 8.2) | 1 | 22 (16, 39) | 1 |
| rad9Δ hap3Δ | 41 (36, 53) | 1.4* | 5.9 (3.0, 6.0) | 1.0 | 26 (3.5, 7.1) | 0.85* |
| rad9Δ htd2Δ | 53 (46, 74) | 1.1 | 5.5 (2.7, 12) | 1.1 | 5.5 (3.6, 12) | 4.0* |
| rad9Δ sam37Δ | 77 (48, 100) | 0.7 | 13 (5.8, 28) | 0.46* | 20 (9.4, 28) | 1.1 |
| rad9Δ yme1Δ | 36 (29, 40) | 1.6* | 1.5 (0.0, 3.5) | 4.0* | 0.58 (0.0, 1.3) | 38* |
Instability frequencies and genome fold stabilization for genetically induced slowed cell cycle stabilization in rad9Δ background normalized to rad9Δ. Cells with a light gray background indicate genome fold stabilization increase; cells with a white background indicate decreased fold stabilization (<1.0 = increased instability) or no change in stabilization (=1.0). Statistically significant appears in boldface type. Kruskal–Wallis test, * P < 0.01.
Genome stabilization in spindle checkpoint deficient cells (mad2Δ) and telomere-defective cells (cdc13) by D-glucosamine, myriocin, as well as erv14
Given that D-glucosamine and myriocin modestly stabilize the genome, we next asked whether these drugs stabilize the genome in two other strains, mad2Δ and cdc13. As described above, mutant mad2 strains have a high frequency of chromosome loss, ∼14-fold higher than WT (Table 1), and loss is suppressed (∼4-fold) by an erv14 mutation (Table 1). We found treating mad2Δ cells with D-glucosamine or myriocin significantly stabilized chromosome loss (Table 8).
Table 8. Genetically induced and chemically induced slowed cell cycle in mad2∆.
| Genotype | Unstable Chr. (×10−5) | Allelic rec. (×10−5) | Chr. loss (×10−4) | |||
|---|---|---|---|---|---|---|
| Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | |
| mad2∆ | 16 (15, 32) | 1 | 14 (11, 18) | 1 | 54 (32, 81) | 1 |
| mad2∆ erv14 | 13 (10, 16) | 1.3 | 2.2 (1.7, 3.1) | 6.5* | 13 (9.8, 21) | 4.2* |
| mad2∆ 200 ng/ml myriocin | 19 (16, 23) | 0.84 | 6.5 (5.2, 14) | 2.2 | 21 (15, 24) | 2.6* |
| mad2∆ 400 ng/ml myriocin | 24 (17, 26) | 0.67 | 5.4 (3.3, 6.4) | 2.6 | 12 (8.7, 15) | 4.5* |
| mad2∆ 2% Dex, 0.5% D-gluc | 11 (8.4, 15) | 1.5* | 4.2 (2.9, 4.8) | 3.3 | 11 (7.7, 16) | 4.9* |
| mad2∆ 2% Dex, 1.0% D-gluc | 11 (5.3, 13) | 1.5* | 4.7 (2.4, 6.6) | 3.0 | 15 (12, 26) | 3.6* |
Genetic section: instability frequencies and genome fold stabilization of mad2Δ erv14 normalized to mad2Δ; myriocin section: instability frequencies and genome fold stabilization of mad2Δ grown on myriocin YEPD plates normalized to mad2Δ grown on YEPD; D-glucosamine section: instability frequencies and genome fold stabilization of mad2Δ grown on D-glucosamine YEPD plates normalized to mad2Δ grown on YEPD. Cells with a light gray background indicate genome fold stabilization increase; cells with a white background indicate decreased fold stabilization (<1.0 = increased instability) or no change in stabilization (=1.0). Statistically significant appears in boldface type. Kruskal–Wallis test, * P < 0.01.
We also explored instability arising from telomeres due to a defect in Cdc13, a single strand DNA-binding protein found at telomere G-tails (Gao et al. 2007; de Lange 2009). Cdc13 provides an essential role in telomere maintenance and protection and is thought to predominantly function at the telomere (Mitton-Fry et al. 2004). In our study, we use the cdc13-F684S mutation, which is compromised for DNA binding and is temperature sensitive (Paschini et al. 2012; unless otherwise noted, cdc13 = cdc13-F684S). Elsewhere we will report extensive studies of instability arising in cdc13 mutant cells (R. Langston and T. Weinert, unpublished data). We use cdc13 in thus study of slowed cell cycle stabilization because with a cdc13 defect, we know where the damage arises (the telomere), and we can induce chromosome instability in one cell cycle using the temperature-sensitive mutant cdc13 (see Table S4 and explanatory text). We can restrict a Cdc13 defect to a single cell cycle, so we might define when a longer delay stabilizes the genome. Here we find that a cdc13 defect leads to an 8-fold higher frequency of unstable chromosomes, and importantly cdc13 mutant cells are stabilized 8-fold by erv14 (Table 9). We also performed a single-cell-cycle instability experiment with cdc13 and cdc13erv14, and found a 4-fold stabilization by erv14 (Table S4 and explanatory text). We also found treating cdc13 cells with D-glucosamine or myriocin significantly suppressed all forms of instability, though a myriocin concentration of 400 ng/ml increased allelic recombinants (Table 9). Therefore, defects in both a spindle checkpoint control, and in a telomere binding protein, cause instability that is suppressed by erv14 and by two drugs.
Table 9. Genetically induced and chemically induced slowed cell cycle in cdc13 (-F684s).
| Genotype | Unstable Chr. (×10−5) | Allelic rec. (×10−5) | Chr. loss (×10−4) | ||||
|---|---|---|---|---|---|---|---|
| Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | Median (Q1, Q3) | Fold stabil. | ||
| cdc13 30° | 49 (37, 69) | 1 | 7.3 (4.0, 22) | 1 | 4.8 (3.1, 12) | 1 | |
| ChrVII RAD+ (wild type) | 6.0 (3.5, 7.6) | 8.2* | 6.8 (3.5, 10) | 1.07 | 3.7 (1.1, 5.2) | 1.3 | |
| Genetically slowed cell cycle | cdc13 erv14 30° | 6.2 (3.7, 8.7) | 7.9* | 2.2 (1.1, 4.5) | 3.3* | 2.2 (1.1, 4.2) | 2.20 |
| D-glucosamine | cdc13 2% Dex, 0.7% D-gluc 30° | 23 (21, 37) | 2.10 | 0.0 (0.0, 0.88)* | N/A | 0.59 (0.0, 2.3) | 8.1* |
| cdc13 2%Dex, 1.4% D-gluc 30° | 13 (0.0, 19) | 3.8* | 1.3 (0.0, 3.1) | 5.6* | 4.0 (1.6, 6.3) | 1.20 | |
| Myriocin | cdc13 400 ng/ml Myr. 30° | 18 (12, 24) | 2.7* | 13 (10, 27) | 0.56* | 2.7 (2.2, 4.7) | 1.80 |
| cdc13 800 ng/ml Myr. 30° | 13 (11, 15) | 3.8* | 7.6 (5.9, 8.4) | 0.96 | 3.2 (2.4, 3.3) | 1.50 | |
Instability frequencies and genome fold stabilization for genetically induced and chemically induced slowed cell cycle stabilization in cdc13 normalized to cdc13 grown on YEPD media plates. Instability frequencies and genome fold stabilization of cdc13 erv14 in the temperature shift single cell cycle experiment normalized to cdc13. Experiment was performed in triplicate. Cells with a light gray background indicate genome fold stabilization increase; cells with a white background indicate decreased fold stabilization (<1.0 = increased instability) or no change in stabilization (=1.0). Statistically significant appears in boldface type. Kruskal–Wallis test, * P < 0.01.
Quantification of doubling time and cell cycle delay suggests G2/M delay stabilizes the genome
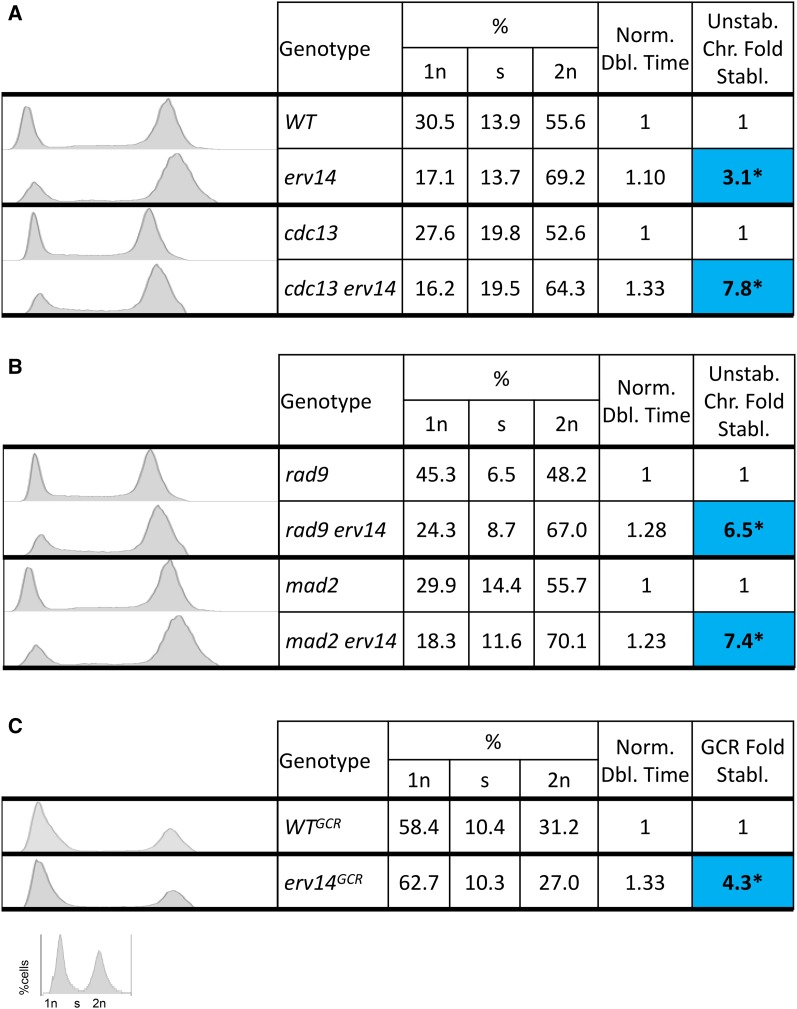
We next examined quantitatively the relationship between a slowed cell cycle and genome stabilization. First, using cells grown on solid media, we quantified the doubling time of five mutant strains and plotted the stabilized effect of the mutation (e.g., yme1Δ) on the rad9Δ genome (Figure S3A); we find a general linear correlation with R2 values of 0.64 for unstable chromosomes and 0.69 for allelic recombinants. Then we analyzed nuclear DNA content and morphology of the same five mutant strains and saw a general increase in the G2/M fraction of the slowed cell cycle rad9Δ mutant strains compared to rad9Δ (Figure S3, B and C).
Next, we performed a similar analysis of cell doubling time, DNA content by FACS analysis and nuclear morphology, for WT, cdc13Δ, and mad2Δ and their erv14 counterparts (Figure 4, A and B and Figure S4). Again, we found an increase in cell doubling time, an increase in the G2/M fraction, and increased stability by erv14. Note that we observe a strong correlation between cell cycle delay and genome stabilization in both checkpoint-deficient cells (rad9Δ and mad2Δ) and in checkpoint-proficient cells (cdc13 and WT cells).
Figure 4.
DNA content FACS analysis and nuclear profiling. (A) DNA content FACS analysis of checkpoint-proficient, WT and cdc13 cells with their erv14−counterparts. Also shown are corresponding doubling times and genome fold stabilization. (B) DNA content FACS analysis of checkpoint-deficient cells rad9Δ and mad2Δ with their erv14− counterparts. Also shown are corresponding doubling times and genome fold stabilization. (C) DNA content FACS analysis of WTGCR with its erv14− counterpart. Also shown are corresponding doubling times and genome fold stabilization. *Statistically significant P < 0.01 using Kruskal–Wallis test.
A longer G1 phase in daughter cells compared to mother cells does not stabilize the genome
It is known that budding yeast daughter cells have a longer G1 cell phase than their progenitor mother cells (di Talia et al. 2009). We therefore designed an experiment to measure, in one cell cycle, whether daughters are more stable than mothers (see Materials and Methods). We isolated mother cells from daughter cells and allowed the cells to progress through one cell cycle, and then determined the frequency of instability. We did not detect any significant difference in relative instability between normal G1-cycling mother cells and longer G1-cycling daughter cells (Table S5). These data suggest that the length of the G1 phase, to the extent it is longer in daughter vs. mother cells, has a minimal impact on genome stabilization. We note that we can detect instability, and its suppression by erv14, in one cell cycle in cdc13-defective cells (above).
Genome stabilization by mih1Δ that delays cells by 15 min in G2/M
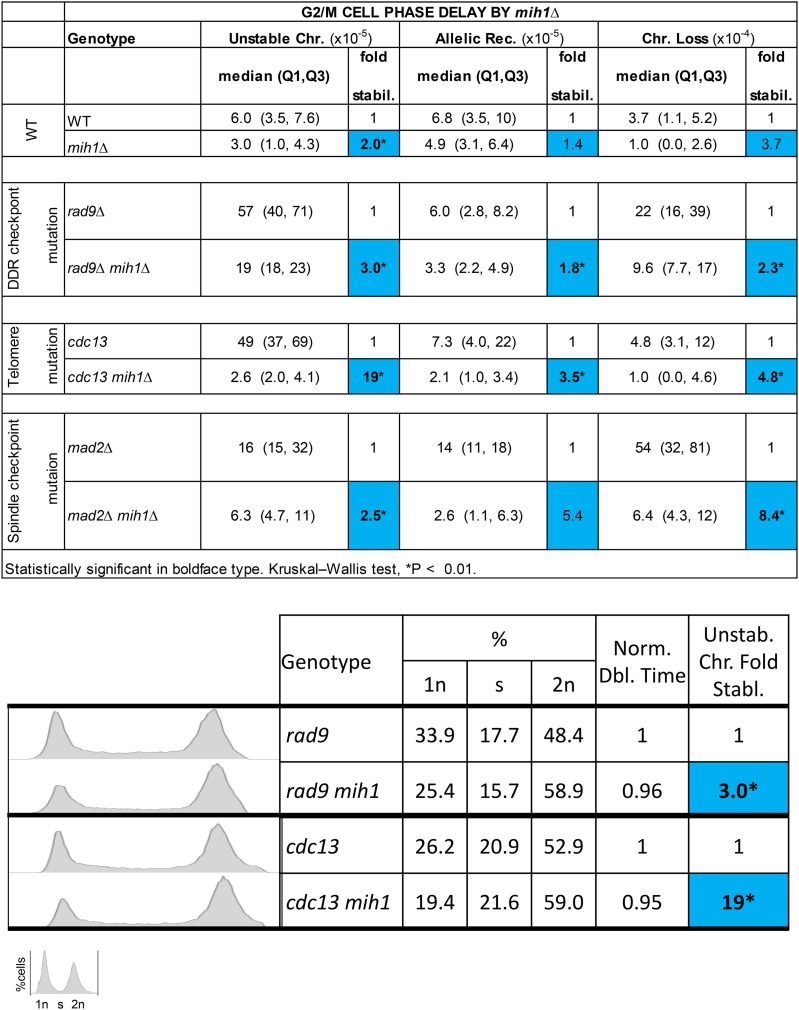
During the course of this work, a colleague (A. Rudner) suggested we test genome stabilization in mih1 mutant cells that are known to slow the cell cycle in G2/M by ∼15 min (Rudner et al. 2000). Mih1 is a phosphatase that regulates the CDK1/CDC28 encoded protein kinase by dephosphorylation. Deletion of MIH1 induces a delay in the G2/M region of the cell cycle due to a defect in Cdc28 dephosphorylation (Russell et al. 1989; Rudner et al. 2000). We generated mih1Δ, rad9Δ mih1Δ, mad2Δ mih1Δ, and cdc13mih1Δ mutant strains to determine whether their genomes are more stable than their MIH1+ counterparts. We found that an mih1 null mutation indeed stabilized their genomes: rad9Δ 3.0-fold stabilization of unstable chromosomes, mad2Δ 8.4-fold stabilization of chromosome loss, cdc13 and WT ∼19-fold and 2-fold stabilization of unstable chromosomes, respectively (Figure 5). We quantified doubling times of mih1− and MIH1+ cells and investigated whether the G2/M phase was also delayed in mih1− cells of our ChrVII disome system (Figure 5). The doubling time for mih1− cells and MIH1+ cells remains about the same; perhaps mih1 daughter cells delay less at START due to growth in the previous G2/M. By FACS, we do detect an extended G2/M and an abbreviated G1 phase of the cell cycle in mih1 null mutations compared to MIH1 cells. Based on the doubling time, FACS analysis, and nuclear profiling results, and the previous genetically induced and chemically induced delayed cell cycle genome stabilization results, we infer that a slowed cell cycle stabilizes the genome, and often by specifically delaying the G2/M phase of the cell cycle (Figure 5 and Figure S5).
Figure 5.
G2/M phase delay by mih1Δ. Instability table of checkpoint-proficient and -deficient cells and their mih1Δ counterparts. DNA content FACS analysis of checkpoint-deficient and -proficient rad9Δ and cdc13 cells with their mih1− counterparts. Also shown are corresponding doubling times and genome fold stabilization. Blue and red squares correspond to genome fold stabilization increase or genome fold stabilization decrease (<1.0 = increased instability), respectively. *Statistically significant P < 0.01 using Kruskal–Wallis test.
Discussion
We began this study asking how deleting a 4-kb locus on a chromosome rendered that chromosome more stable. We found that a gene located within the 4-kb locus, ERV14, encodes a protein that promotes genome instability, or stated another way; loss of Erv14 function stabilizes the genome. Specifically, a start codon ATG mutation in ERV14 stabilized the rad9Δ genome, and expressing the Erv14 protein from another chromosome complements erv14 (destabilizes the genome). The stabilizing influence of erv14 extends to both checkpoint-deficient and -proficient cells, spans the three events detected (unstable chromosomes, allelic recombination, and loss), extends to two other chromosome systems (both using ChrV) and thus is genome-wide, and is not due to any Erv14-specific function tested. Rather, we infer that genome stability arises from a slowed cell cycle, and likely a longer G2/M phase of the cell cycle. Certain other mutations (e.g., yme1Δ) and chemical conditions (e.g., D-glucosamine) also delay the cell cycle and more modestly stabilize the genome. We most directly induce a G2/M phase delay by deleting the G2/M phase transitioning phosphatase MIH1; an MIH1 mutation stabilizes the genome, and in checkpoint-proficient and -deficient cells, and extends their G2/M phases by as little as 15 min (Rudner et al. 2000). From the wealth of our evidence, we conclude that a slower cell cycle, and most likely an extended delay in G2/M, stabilizes the genome.
The nature of accuracy and speed
To our knowledge, there are two types of studies that previously provided correlation between a time delay and accuracy. One type of study is of protein translation in E. coli (Hopfield 1974; Andersson et al. 1986; Johansson et al. 2008). Protein translation involves an initial charged tRNA binding step in the A-site of the ribosome, during which there is a selection/proofreading step (accuracy step). A mutation in the ribosome increases the time the tRNA ternary complex interfaces with the ribosome and mRNA, thus increasing translation accuracy, but also delaying and slowing translation in the process. Is it possible that Erv14 and Mih1, for example, decrease genome stability by accelerating some analogous molecular step as in the translation example? While it may be possible that Erv14 and Mih1 accelerate some specific molecular process in chromosome biology that compromises genome stability, we think it is more likely that Erv14 and Mih1 act more globally to accelerate the cell cycle, decreasing the time spent in the G2/M phase, and thus destabilizing the genome.
The slowed cell cycle model is more aligned with the Murray laboratory findings that showed meiotic budding yeast mutant mad2Δ strains benefitted from a delayed cell cycle with respect to proper disjunction (Shonn et al. 2000). Shonn et al. (2000) found that in a mad2Δ mutant strain, nondisjunction occurred more frequently as chromosomes mis-segregated. They predicted that inducing a metaphase delay in a mad2Δ mutant strain would restore proper disjunction by allowing more time for spindles to reorient themselves correctly. Their prediction proved to be correct; inducing a metaphase delay in mad2Δ mutant strain restored disjunction back to WT levels (Shonn et al. 2000). We see a similar phenomenon in our mutant mad2Δ strain during mitosis; delaying the cell cycle in mad2Δ by using erv14, mih1Δ, sphingolipid inhibition, or glucose competition reduced chromosome loss during mitosis by approximately fourfold, eightfold, fourfold, and fivefold, respectively (Table 8). Of course, the initial study by Weinert and Hartwell (1988) also showed that slowing the cell cycle in G2/M in a rad9 mutant cell increased radiation resistance (as did other studies: Al-Khodairy and Carr 1992; Walworth et al. 1993).
What molecular events are made less error prone by a cell cycle delay?
Currently we do not know what molecular processes are made more accurate by the modest cell cycle delays. DNA replication per se may not be affected; ERV14+ and erv14 cells have similar point mutation frequencies, though our data are not extensive in this regard (Figure S1 and Table S1B). Certainly with mad2Δ, we can speculate that increasing the time to assemble a proper spindle is a key event made more error-free by a delay (Figure 6A). That a cell cycle delay increases accuracy in DDR mutations (e.g., rad51Δ), as well as in DNA damage checkpoint mutations (rad9Δ, rad17Δ; Figure 6C), and in cells defective for Cdc13 acting at telomeres, suggests defects in multiple molecular events may be suppressed by a delay. In addition, some of those molecular errors that benefit from a slower cell cycle must not themselves signal the checkpoint system very effectively, for the slow cell cycle stabilization occurs in checkpoint-proficient cells as well, most convincingly in conditional telomere protein mutations (cdc13; Figure 6B).
Figure 6.
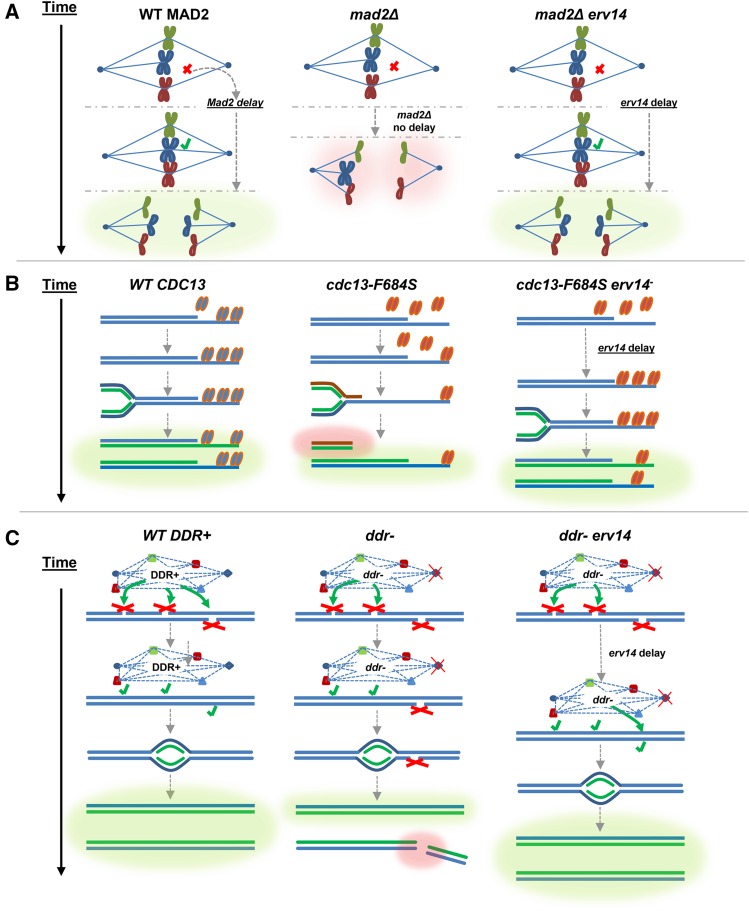
Slowed cell cycle stabilization models of mutant spindle checkpoint, telomere biology, and DDR cells. Green glow indicates WT/WT-like outcome; red glow indicates aberrant/catastrophic outcome. (A, left) WT MAD2. Mad2 activates spindle checkpoint cell cycle delay, spindle attaches, chromosomes segregate properly. (Center) mad2Δ. Defective spindle checkpoint, no cell cycle delay, chromosomes mis-segregate. (Right) mad2Δ erv14. In a defective spindle checkpoint (mad2Δ) cell, the erv14-induced cell cycle delay creates time for spindle attachment; chromosomes segregate properly. (B, left) WT CDC13 Cdc13 protects telomere during replication, and chromosomes replicate properly. (Center) cdc13. Defective Cdc13 is compromised for telomere binding, allowing degradation of exposed chromosome end by exonucleases, resulting in shorter chromosomes with no telomere end protection, resulting in further degradation and instability. (Right) cdc13 erv14. Mutant Cdc13 is compromised for telomere binding, and the erv14-induced cell cycle delay creates time for more mutant Cdc13 to bind to telomere, resulting in chromosomes replicating properly. (C, left) WT DDR+. DDR is fully functional, so a cell cycle delay allows DNA lesions to be recognized and repaired efficiently, and thus chromosomes replicate properly. (Center) In a ddr− cell some DNA lesions escape detection or suffer incomplete repair, resulting in DNA lesions/ssDNA gaps, which persist during DNA replication, generating shorter and damaged chromosomes. (Right) In a ddr− erv14 cell, the partially functioning DDR is permitted more time to repair DNA lesions during the erv14-induced cell cycle delay.
The slowed cell cycle model
In conclusion, we infer that constitutively delaying the cell cycle, in particular in G2/M, potentially provides a time checkpoint. More time allows a more optimal error detection and/or correction. Our genetic analyses show that proteins that normally accelerate the cell cycle destabilize the genome, and one would therefore posit that other loss-of-function mutations may also accelerate the cell cycle and increase instability. Such mutations that reduce the time checkpoint, accelerating the cell cycle, may result in both a selective advantage in shorter cell division time, and in a higher error frequency that creates greater genetic diversity enabling cancer cell evolution. Whether certain cancer cells exhibit unstable genomes because their cell cycles are faster remains speculative (Wakonig-Vaartaja and Hughes 1965; Duesberg et al. 1998; Lengauer et al. 1998).
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.197590/-/DC1.
Acknowledgments
We thank Lisa Shanks, Tracey Beyer, and Rachel Langston for frequent discussions of this work; Christopher D. Putnam and Richard D. Kolodner for supplying us with the RDKY6678 GCR strain; and Angelika Amon for supplying us with the 14479 strain used in developing our ChrV disome system. T.W. was funded by National Institutes of Health R01-GM076186-05, and P.J.V. was funded in part by T32-GM08659.
Footnotes
Communicating editor: J. A. Nickoloff
Literature Cited
- Admire A., Shanks L., Danzl N., Wang M., Weier U., et al. , 2006. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev. 20(2): 159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khodairy F., Carr A. M., 1992. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1: 1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D. I., van Verseveld H. W., Stouthamer A. H., Kurland C. G., 1986. Hicrobiology suboptimal growth with hyper-accurate ribosomes. Arch. Microbiol. 144: 96–101. [DOI] [PubMed] [Google Scholar]
- Baryshnikova A., Andrews B., 2012. Neighboring-gene effect : a genetic uncertainty principle. nature publishing group 9 (4). Nature Methods 9: 341, 343. [DOI] [PubMed] [Google Scholar]
- Beyer T., Weinert T. A., 2016. Ontogeny of unstable chromosomes generated by telomere error in budding yeast. PLoS Genet. 12: e1006345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M. J., Hartwell L. H., 1985. CDC17: An essential gene that prevents telomere elongation in yeast. Cell 42: 249–257. [DOI] [PubMed] [Google Scholar]
- Chen C., Kolodner R. D., 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23(1): 81–85. [DOI] [PubMed] [Google Scholar]
- de Lange T., 2009. How Telomeres Solve the End-Protection Problem. Science 326(5955): 948–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Talia S., Wang H., Skotheim J. M., Rosebrock A. P., Futcher B., et al. , 2009. Daughter-specific transcription factors regulate cell size control in budding yeast. PLoS Biol. 7: (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P., Rausch C., Rasnick D., Hehlmann R., 1998. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc. Natl. Acad. Sci. USA 95(23): 13692–13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Cervantes R. B., Mandell E. K., Otero J. H., Lundblad V., 2007. RPA-like proteins mediate yeast telomere function.Nat. Struct. Mol. Biol. 14: 208–14. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A., 1989. Checkpoints : controls that ensure the order of cell cycle events. Science 246(5930): 629–634. [DOI] [PubMed] [Google Scholar]
- Hasselbring H., 1913. Metabolism of fungi. Bot. Gaz. 56(6): 504–513. [Google Scholar]
- Herzig Y., Sharpe H. J., Elbaz Y., Munro S., Schuldiner M., 2012. A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by erv14. PLoS Biol. 10(5): e1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield J. J., 1974. Biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. USA 71(10): 4135–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Lovmar M., Ehrenberg M., 2008. Rate and accuracy of bacterial protein synthesis revisited. Curr. Opin. Microbiol. 11(2): 141–147. [DOI] [PubMed] [Google Scholar]
- Kaochar S., Shanks L., Weinert T. A., 2010. Checkpoint genes and Exo1 regulate nearby inverted repeat fusions that form dicentric chromosomes in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 107(50): 21605–21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal W. H. and W. A. Wallis, Use of Ranks in One-Criterion Variance Analysis. Journal of the American Statistical Association 47(260): 583–621.
- Lengauer C., Kinzler K. W., Vogelstein B., 1998. Genetic instabilities in human cancers. Nature 396: 643–649. [DOI] [PubMed] [Google Scholar]
- Levy S., Ihmels J., Carmi M., Weinberger A., Friedlander G., et al. , 2007. Strategy of transcription regulation in the budding yeast. PLoS One 2(2): e250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Murray A. W., 1991. Feedback control of mitosis in budding yeast. Cell 66(3): 519–531. [DOI] [PubMed] [Google Scholar]
- McGoldrick E. M., Wheals A. E., 1989. Controlling the growth rate of Saccharomyces cerevisiae cells using the glucose analogue D-glucosamine. J. Gen. Microbiol. 135(9): 2407–2411. [DOI] [PubMed] [Google Scholar]
- Mitton-Fry R. M., Anderson E. M., Theobald D. L., Glustrom L. W., Wuttke D. S., 2004. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 338(2): 241–255. [DOI] [PubMed] [Google Scholar]
- Paek A. L., S., Kaochar H., Jones A., Elezaby L., Shanks, et al. , 2009. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev. 23(24): 2861–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschini M., T. B., Toro J. W., Lubin B., Braunstein-Ballew D. K., Morris, et al. , 2012. A naturally thermolabile activity compromises genetic analysis of telomere function in Saccharomyces cerevisiae. Genetics 191: 79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Kolodner R. D., 2010. Determination of gross chromosomal rearrangement rates. Cold Spring Harb. Protoc. 2010(9): pdb.prot5492. 10.1101/pdb.prot5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Hayes T. K., Kolodner R. D., 2009. Specific pathways prevent duplication-mediated genome rearrangements. Nature 460(7258): 984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts G. G., Hudson A. P., 2006. Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol. Genet. Genomics 276: 170–186. [DOI] [PubMed] [Google Scholar]
- Roth V., 2006. Doubling Time Computing. Available at: http://www.doubling-time.com/compute.php.
- Rothstein R., 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods in Enzymol. 194: 281–301. [DOI] [PubMed] [Google Scholar]
- Rudner A. D., Hardwick K. G., Murray A. W., 2000. Cdc28 activates exit from mitosis in budding yeast. J. Cell. Biol. 149(7): 1361–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P., Moreno S., Reed S. I., 1989. Conservation of mitotic controls in fission and budding yeasts. Cell 57: 295–303. [DOI] [PubMed] [Google Scholar]
- Shonn M. A., McCarroll R., Murray A. W., 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289(5477): 300–303. [DOI] [PubMed] [Google Scholar]
- Wakonig-Vaartaja R., Hughes D. T., 1965. Chromosomal anomalies in dysplasia, carcinoma-in-situ, and carcinoma of cervix uteri. Lancet 2: 756–759. [DOI] [PubMed] [Google Scholar]
- Walworth N., Davey S., Beach D., 1993. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature 363: 368–371. [DOI] [PubMed] [Google Scholar]
- Wang G., Lercher M. J., Hurst L. D., 2011. Transcriptional coupling of neighboring genes and gene expression noise: evidence that gene orientation and noncoding transcripts are modulators of noise. Genome Biol. Evol. 3: 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H., 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241: 317–322. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H., 1990. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in Cell Cycle Arrest after DNA Damage. Mol. Cell. Biol. 10: 6554–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Kiset G. L., Hartwell L. H., 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8: 652–665. [DOI] [PubMed] [Google Scholar]
- Wuytswinkel O. V., Reiser V., Siderius M., Kelders M. C., Ammerer G., et al. , 2000. Response of Saccharomyces cerevisiae to severe osmotic stress: evidence for a novel activation mechanism of the HOG MAP kinase pathway. Mol. Microbiol. 37: 382–397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available on request. Supplemental Material, Table S7 contains genotypes of all strains used.