Abstract
Purpose
Although autophagy plays important roles in malignant pathogenesis and drug resistance, there are few clinical agents that disrupt this pathway and the potential therapeutic benefit of autophagy inhibition remains undetermined. We used medicinal chemistry approaches to generate a series of novel agents that inhibit autophagic degradation.
Experimental Design
ROC-325 was selected as a lead compound for further evaluation. Comprehensive in vitro and in vivo studies were conducted to evaluate the selectivity, tolerability, and efficacy of ROC-325 in preclinical models of renal cell carcinoma (RCC) with HCQ serving as a comparator. Markers of autophagy inhibition and cell death were evaluated in tumor specimens.
Results
ROC-325 exhibited superior in vitro anticancer effects than the existing autophagy inhibitor hydroxychloroquine in 12 different cancer cell lines with diverse genetic backgrounds. Focused studies of the mechanism of action and efficacy of ROC-325 in RCC cells showed that drug treatment induced hallmark characteristics of autophagy inhibition including accumulation of autophagosomes with undegraded cargo, lysosomal deacidification, p62 stabilization, and disruption of autophagic flux. Subsequent experiments showed that ROC-325 antagonized RCC growth and survival in an ATG5/7-dependent manner, induced apoptosis, and exhibited favorable selectivity. Oral administration of ROC-325 to mice bearing 786-0 RCC xenografts was well tolerated, significantly more effective at inhibiting tumor progression than HCQ, and inhibited autophagy in vivo.
Conclusions
Our findings demonstrate that ROC-325 has superior preclinical anticancer activity compared to HCQ and support the clinical investigation of its safety and preliminary efficacy in patients with RCC and other autophagy-dependent malignancies.
Keywords: autophagy inhibitor, ROC-325, renal cell carcinoma, hydroxychloroquine, lucanthone
Introduction
Autophagy is an evolutionarily conserved mechanism of lysosomal proteolysis that is used for the turnover of organelles and proteins with long half-lives and also functions to generate alternative sources of metabolic fuel via nutrient recycling under stress conditions (1, 2). Although multiple key studies have demonstrated that autophagy functions as a mechanism of tumor suppression via the elimination of defective pre-malignant cells, overwhelming evidence supports a major role for autophagic degradation in the maintenance of bioenergetic homeostasis under stress conditions including hypoxia and nutrient deprivation (3). Additionally, autophagy has emerged as an important mechanism of resistance to radiation, classical chemotherapy, and targeted anticancer agents due to its ability to augment the survival capacity of malignant cells (4–8). The data supporting roles for autophagy as a mediator of drug resistance and malignant progression provided a logical foundation to devise strategies to impair this process for therapeutic benefit.
Chloroquine (CQ) and hydroxychloroquine (HCQ) have been used for decades to treat malaria, rheumatoid arthritis, and lupus and represent two of a very small group of FDA-approved drugs that disrupt lysosomal function and consequently inhibit autophagy (6). These specific properties of CQ/HCQ spurred numerous preclinical investigations focused on establishing the safety and therapeutic benefit of inhibiting autophagy to increase the efficacy of a diverse range of anticancer agents (9, 10). Based on the positive impact of HCQ in this scenario, we and others initiated a series of phase I and phase I/II trials to investigate the safety and preliminary efficacy of the addition of HCQ to existing anticancer regimens (11–16). Although the addition of HCQ was generally safe and preliminary efficacy was observed in a minority of patients treated with HCQ-based regimens, it was unclear if the maximum tolerated dose (MTD) of HCQ in these studies resulted in complete autophagic inhibition. The results of these initial clinical studies of HCQ-based combination regimens underscored the need for better agents to antagonize lysosomal proteolysis. However, no comprehensive structure activity relationship (SAR) analyses on CQ/HCQ had been conducted to date and it was therefore unclear which specific structural modifications to these agents may result in increased anti-autophagic potency or efficacy.
Lys05 is a novel dimeric form of CQ connected with the spacer N,N-bis(2-aminoethyl)methylamine that was designed to take advantage of the concept of polyvalency and was reported to exhibit superior autophagic inhibition and anticancer activity compared to CQ (17). Polyvalent molecules have the potential to yield nonlinear, multifold potency against their respective targets compared to their corresponding monomers (18, 19). With the goal of generating new autophagy inhibitors with better potency, tolerability, and anticancer efficacy than HCQ, we used logical medicinal chemistry approaches to generate a series of new dimeric compounds containing modified core elements of HCQ, CQ and the anti-schistosomal drug lucanthone (Miracil D) (20). Here we report that ROC-325, a novel compound with structural motifs of both HCQ and lucanthone, inhibits autophagy at significantly lower doses and exhibits significantly superior single agent anticancer activity against a broad range of tumor types compared to HCQ. Focused studies in models of renal cell carcinoma (RCC) demonstrated that ROC-325 treatment led to the deacidification of lysosomes, accumulation of autophagosomes, and disrupted autophagic flux. Targeted knockdown of the essential autophagy genes ATG5 and ATG7 severely blunted the anticancer effects of ROC-325, thus indicating that its autophagy inhibitory properties were a critical component of its mechanism of action. Oral administration of ROC-325 to mice bearing RCC xenografts was well tolerated and yielded dose-dependent inhibition of tumor growth that was significantly more efficacious than a higher dose of HCQ. Analysis of tumor specimens from mice treated with ROC-325 demonstrated in vivo autophagy inhibition, reduced tumor cell proliferation, and apoptosis. Our collective findings establish the foundation for further investigation of ROC-325 as a novel agent for autophagy-dependent malignancies and other disorders where lysosomal activity contributes to disease pathogenesis.
Materials & Methods
Synthesis of ROC-325, other ROC series compounds, and Lys05
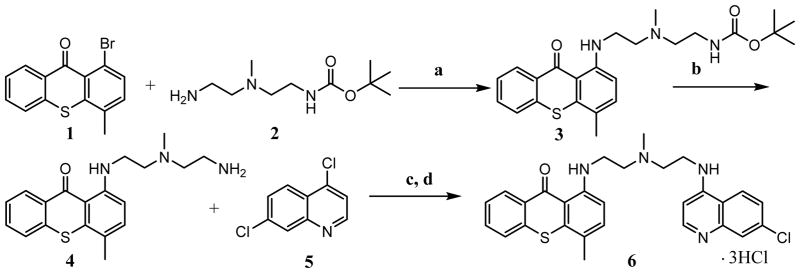
The synthesis of ROC-325 was performed following scheme I.
Scheme I.
a) Pd(II)acetate, BINAP, K3PO4, dioxane, 100 °C, 17 h; b) 4N HCl in dioxane; c) Pd(II)acetate, BINAP, K3PO4, dioxane, 100 °C, 15 h; d) Methanol, 4N HCl in dioxane
The procedures for the synthesis of ROC-325 are representative of the preparation of the derivatives shown in Figure 1A. Compound numbers detailed here correspond to the compound numbers in bold text in the above reaction scheme. A suspension of 1-Bromo-4-methylthiaxanthone (Compound 1, 3.23g, 10.62 mmoles), {2-[(2-Amino-ethyl)-methyl-amine]-ethyl}-carbamic acid tert-butyl ester amine (Compound 2, 1.53g, 7.08 mmoles), Binap (2,2′-bis(diphenylphosphino)-1,1′-binaphthyl) (0.88g, 1.4 mmoles), and K3PO4 (7.5g, 35.0 mmoles) in 70 ml of dioxane was purged with N2 for 25 minutes. The reaction was kept under N2 and Pd(OAc)2 (0.159g, 0.708 mmoles) added. The reaction was heated at 100 °C for 17 hours. The reaction was cooled, dichloromethane (100 ml) added, and the crude mixture filtered and concentrated. Purification by flash silica chromatography eluting with ethyl acetate/hexanes (4:6 to 6:4) provided 1.5 grams of a red solid (Compound 3). 30 ml of 4N HCl was added to Compound 3 (0.53g). The suspension was stirred at room temperature for 1.5 hours and the solid was collected by filtration, washed with dichloromethane (3 x 15 ml) and dried at 50 °C to give 150 mg of a red solid (Compound 4). Next, amine Compound 4 (0.250g, 0.733 mmoles), 4,7-dichloroquinoline Compound 5 (0.329g,1.66mmoles), BINAP (0.091g, 0.146 mmoles), and K3PO4 (0.778g, 3.66 mmoles) were suspended in dry dioxane (25 ml). The mixture was purged with N2 for 25 minutes and then kept under N2. Pd(OAc)2 (0.016g, 0.073 mmoles) was added and the reaction heated for 16 hours at 95 °C. The reaction was cooled and filtered with the aid of dichloromethane (50 ml). The filtrate was concentrated and the crude purified by flash chromatography using ethyl acetate/dichloromethane (1:9) and gave 205 mgs of red foam Compound 6. The tri-HCl salt of Compound 6 was prepared by suspending it in 70 ml of methanol, adding 10 ml of 4N HCl in dioxane, and stirring for 15 hours at room temperature. The precipitate was collected by filtration, washed with methanol, and then ethyl acetate (15/15), and dried at 50 °C under vacuum for 12 hours. The structure of ROC-325 was confirmed by 1H nuclear magnetic resonance (NMR) and high resolution mass spectrometry (HRMS) analyses and these data are included in Figs. S1–2.
Figure 1.
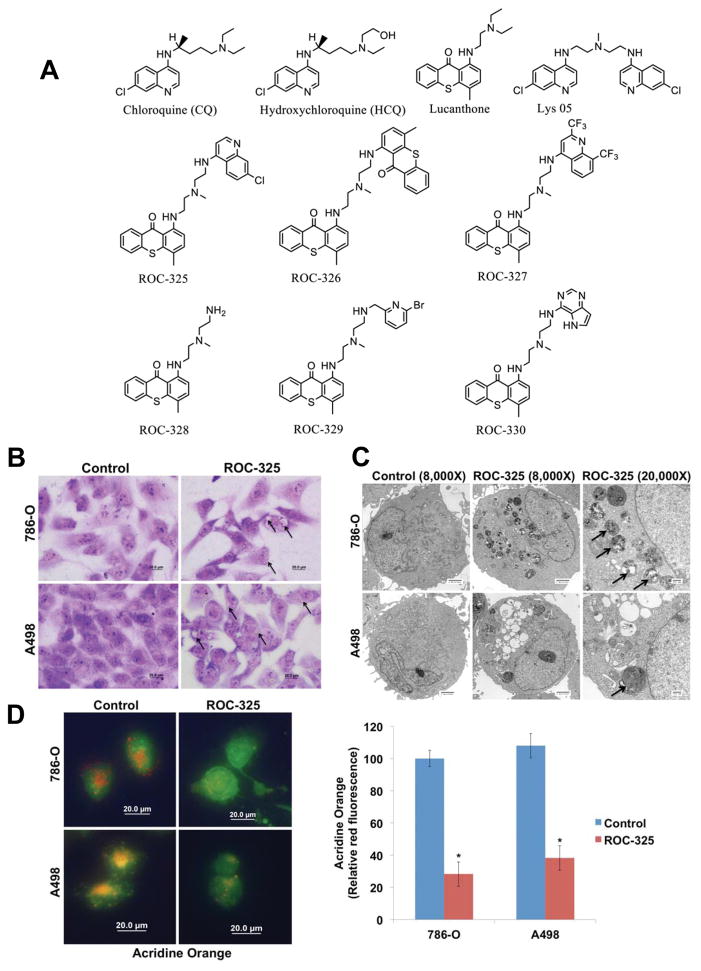
ROC-325 induced vacuolization and lysosome membrane permeability. (A) Chemical structures of ROC-325 and related compounds. (B) ROC-325 induces vacuolization. 786-O and A498 cells were treated for 24 h with 5 μM ROC-325. Cell morphology and vacuolization were visualized by Giemsa staining. Scale bars indicate 20 microns. (C) Electron microscopy demonstrates vacuolization and electron dense particle accumulation. Cells were treated with 5 μM ROC-325 for 24 h. Cells were fixed and prepared for electron microscopy. Arrows indicate vacuoles with undegraded cargo in the cytosol of imaged cells. Scale bars for images with 8000X magnification indicate 2 microns and scale bars for images with 20000X magnification indicate 500 nm. (D) Measurement of lysosome membrane permeability by loss of acridine orange fluorescence. Red acridine orange staining was measured in 786-O and A498 cells by immunofluorescence and quantified using ImageJ software. Mean ± SD, n = 5. *Indicates a significant difference from the controls. P < 0.05.
Cells and cell culture
A498, 786-0, Achn and Caki-2 cells were obtained from ATCC (Manassas, VA). Cells were cultured with medium supplemented with 10% FBS at 37°C with 5% CO2 as previously described (21). Human normal renal proximal tubule epithelial cells (RPTEC) were purchased from Clonetics and cultured in REGM media (REGM BulletKit, Clonetics). Cell lines were authenticated by the source banks using short tandem repeat (STR) DNA profiling techniques.
Lentiviral shRNA gene silencing
Targeted knockdown of ATG5, ATG7, and CTSD was achieved using lentiviral shRNA using commercially available lentiviral shRNAs (Santa Cruz Biotechnology, Cat# sc-41445-V for ATG5, Cat# sc-41447-V for ATG7, Cat# sc-29239-V for CTSD) according to the manufacturer’s instructions.
Chemicals and reagents
Reagents were obtained from the following sources: giemsa stain, acridine orange, bafilomycin A1, propidium iodide, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), hydroxychloroquine, and anti-β tubulin antibody (Sigma, St. Louis, MO), anti-active caspase-3 (Cell Signaling), anti-LC3B, anti-p62, ant-ATG5, and anti-ATG7 (Abcam), anti-cathepsin D (Santa Cruz Biotech), goat anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson Laboratories), sheep anti-mouse-HRP and donkey anti-rabbit-HRP (Amersham). Additional details regarding these reagents were previously described (14).
Measurement of cell proliferation
Cells infected with non-targeted control and ATG5 or ATG7-targeted shRNA were plated in triplicate. Cell numbers were counted daily for 5 consecutive days via automated trypan blue exclusion with the assistance of a Vi-Cell XR system (Beckman-Coulter).
Transmission electron microscopy
Transmission electron microscopy of cells was performed as previously described (22). RCC cells were treated with ROC-325 for 24 h and harvested for imaging. Briefly, sections were cut in an LKB Ultracut microtome (Leica), stained with uranyl acetate and lead citrate, and examined in a JEM 1230 transmission electron microscope (JEOL, USA, Inc.). Images were captured using the AMT Imaging System (Advanced Microscopy Techniques Corp).
Giemsa staining
Cells were plated in chamber slides as previously described and treated with ROC-325 for 24 h (20). Following drug treatment, cells were washed with PBS and fixed in methanol for 5 minutes. Cells were then incubated for 1 h in Giemsa stain diluted 1:20 with deionized water. Cells were rinsed with water and imaged using an Olympus fluorescent microscope with a DP71 camera. Image-Pro Plus software Version 6.2.1 (Media Cybernetics) was used for image acquisition.
Acridine orange staining
Acidic lysosomes were visualized by acridine orange staining as previously described (20). After treatment with ROC-325 for 24 h, cells were stained with 1 μM acridine orange for 15 minutes at 37 °C. Cells were washed with PBS and images were captured using an Olympus fluorescent microscope. Acidic lysosomes appear as orange fluorescent cytoplasmic vesicles. Quantification of 5 random fields of acridine orange intensity and image acquisition were performed using Image-Pro Plus software Version 6.2.1.
Immunocytochemistry
Cells were plated on chamber slides and allowed to attach overnight. Cells were then treated for 24 h with ROC-325. Following drug treatment, cells were fixed with 4% paraformaldehyde, permeabilized using 0.2% triton-X-100, and incubated overnight with anti-LC3B antibody. Alexa Fluor 594 conjugated fluorescent secondary antibody was used to visualize protein localization. DAPI was utilized to stain the nucleus. Images were captured using an Olympus fluorescent microscope with a DP71 camera and a 60X objective. Image-Pro Plus software Version 6.2.1 (MediaCybernetics, Bethesda, MD) was used for image acquisition (20).
Expression microarrays
RCC cells were treated with ROC-325 for 24 h. Total RNAs were isolated using the RNeasy Plus Mini Kit (Qiagen, Germantown, MD) and treated with TURBO DNA-free™ Kit (Applied Biosystems, Foster City, CA). 300 ng of total RNA per sample was amplified and hybridized to GeneChip® Human Gene 1.0 ST arrays (Affymetrix, Inc., Santa Clara, CA) according to the manufacturer’s instructions. Affymetrix CEL files were imported into Partek® Genomics Suite™ 6.4 (Partek Inc., St. Louis, MO) using the default Partek normalization parameters and the robust multi-array average (RMA) analysis adjusted for probe sequence and GC content (GC-RMA). Data normalization was performed across all arrays using quantile normalization (20). Gene ontology enrichments (GO) analyses were performed and heat maps were also generated using Partek software. Microarray data was deposited with NCBI GEO and can be viewed under accession number GSE89766.
Quantitative real time polymerase chain reaction
cDNA from ROC-325 treated cells were used for relative quantification by RT–PCR analyses. First-strand cDNA synthesis was performed from 1 μg RNA in a 20 μl reaction mixture using the high-capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Cathepsin D (CTSD) and GAPDH transcripts were amplified using commercially available TaqMan® Gene expression assays (Applied Biosystems, Foster City, CA). Relative gene expression was calculated with the 2–ΔΔ Ct method using GAPDH as a housekeeping gene (23).
Quantification of drug-induced cytotoxicity
Cell viability was estimated by MTT assay, which detects the conversion of MTT to formazan by the mitochondria of living cells. Cells were seeded into 96-well microculture plates at 10,000 cells per well and allowed to attach for 24 h. Cells were then treated with ROC-325 or HCQ for 72 h. Following drug treatment, MTT was added and formazan absorbance was quantified using a Molecular Devices microplate reader. The estimated cell viability under each experimental condition was calculated by normalizing the respective formazan optical density to the density of control cells. Pro-apoptotic effects following in vitro drug exposure were quantified by propidium iodide (PI) staining and fluorescence-activated cell sorting (FACS) analysis of sub-G0/G1 DNA content as previously described (24) and by measurement of active caspase-3 by flow cytometry using a commercial kit (BD Biosciences).
Immunoblotting
Renal cancer cells were incubated with ROC-325 for 24 h. Cells were harvested and were then lysed as previously described (25). Approximately 50 μg of total cellular protein from each sample were subjected to SDS-PAGE, proteins were transferred to nitrocellulose membranes, and the membranes were blocked with 5% nonfat milk in a Tris-buffered saline solution containing 0.1% Tween-20 for 1 h. The blots were then probed overnight at 4 °C with primary antibodies, washed, and probed with species-specific secondary antibodies coupled to horseradish peroxidase. Immunoreactive material was detected by enhanced chemiluminescence (West Pico, Pierce, Inc.,).
In vivo evaluation of ROC-325 and HCQ
786-O renal cancer cells (5 x 106) were suspended in a mixture of HBSS and Matrigel and subcutaneously implanted into female nude mice (BALB/c background) (26). Tumor-bearing animals from each cell line xenograft were randomized into treatment groups. Mice were treated with vehicle (water), ROC-325 (25, 40, and 50 mg/kg PO), or HCQ (60 mg/kg IP) QDx5 for 6 weeks. Mice were monitored daily and tumor volumes were measured twice weekly. At study completion, tumors from representative animals were excised from each group, formalin-fixed, and paraffin-embedded for immunohistochemical analysis.
Immunohistochemistry
Paraffin-embedded tumor sections were deparaffinized in xylene, exposed to a graded series of alcohol, and rehydrated in PBS (pH 7.5). Heat-induced epitope retrieval on paraffin-embedded sections and probing with specific antibodies was conducted as previously described (14). Positive reactions were visualized using 3,3′-diaminobenzidine (Dako). Images were captured using an Olympus fluorescent microscope with a DP71 camera and a 20X objective. Image-Pro Plus software Version 6.2.1 (MediaCybernetics, Bethesda, MD) was used for image acquisition. ImageJ software was used for quantification of LC3B and p62 levels by densitometric analysis of five random fields containing viable tumor cells as previously described (14). Quantification of cleaved caspase-3 was conducted by counting the number of positive cells in five random fields as previously described (27).
Statistical analyses
Statistical significance of differences observed between paired samples was determined using the Student’s t test. Two-way ANOVA with Tukey post hoc analysis was used to determine the significance of differences between experimental conditions in multi-parameter assays including the in vivo assessment of the effects of ROC-325 and HCQ on RCC xenograft tumors. Differences were considered significant in all experiments at p < 0.05 with two-sided comparisons.
Results
ROC-325 induces lysosomal deacidification and autophagosome formation
We first created a series of novel compounds that contained motifs of HCQ and lucanthone (LUC) with the goal of developing new agents with both improved autophagic inhibition and single agent anticancer activity (Fig. 1A) (6, 20). We evaluated the ability of each of these compounds to reduce the viability of malignant cells and increase the expression of p62, a protein that is known to be specifically turned over by autophagy (28). Based on these initial assays, we identified ROC-325 as a lead agent (Fig. 1A, Supp. Figs. 1–2). Direct comparison of the in vitro anticancer effects of ROC-325 and HCQ in 12 different human cancer cell lines with diverse genetic backgrounds demonstrated that ROC-325 possessed significantly lower IC50s (approximately 10-fold) than HCQ in all models tested (Table 1). We selected RCC as a specific tumor type for further investigation of the pharmacological properties of ROC-325 based on the sensitivity of A498 cells in these preliminary screens, the clinical efficacy we observed in a patient with RCC treated with a combination of HCQ and vorinostat after failing seven lines of prior therapy in our recent investigator-initiated clinical trial, and a new report suggesting that alterations in the autophagy pathway in patients with renal cancer may be relevant for overall survival (14, 29). Giemsa staining of A498 and 786-0 cells demonstrated that ROC-325 induced cytosolic vacuolization (Fig. 1B). Transmission electron microscopy analyses showed that it promoted the accumulation of autophagosomes with undegraded cargo (Fig. 1C). Treatment with ROC-325 also resulted in a significant loss of acridine orange fluorescence, which is consistent with an increase in lysosomal membrane permeability (LMP) and the consequential deacidification of lysosomes (Fig. 1D). These collective findings demonstrated that the chemical refinements of the LUC and HCQ core motifs that we designed to produce ROC-325 resulted in significantly greater anticancer activity while retaining lysosomal disrupting properties.
Table 1.
IC50 values from 72 h MTT assays.
| Cell line | Tumor Type | ROC-325 IC50 (μM) | HCQ IC50 (μM) |
|---|---|---|---|
| A498 | Renal | 4.9 | 52 |
| A549 | Lung | 11 | >75 |
| CFPAC-1 | Pancreas | 4.6 | >75 |
| COLO-205 | Colon | 5.4 | 51 |
| DLD-1 | Colon | 7.4 | >75 |
| IGROV-1 | Ovarian | 11 | >75 |
| MCF-7 | Breast | 8.2 | >75 |
| MiaPaCa-2 | Pancreas | 5.8 | >75 |
| NCI-H69 | Lung | 5.0 | 54 |
| PC-3 | Prostate | 11 | 58 |
| RL | NHL | 8.4 | 35 |
| UACC-62 | Melanoma | 6.0 | >75 |
ROC-325-triggered effects on cathepsin D promote apoptosis, but are not required for autophagy inhibition
We previously showed that CQ/HCQ and LUC induce increased expression of the lysosomal protease cathepsin D (CTSD) (20, 22, 30). The LMP-related effects of HCQ and LUC also promote its subcellular relocalization from lysosomal compartments into the cytosol. Notably, in direct contrast to the majority of lysosomal proteases that require a strict acidic pH environment for their activity, CTSD retains proteolytic function under cytosolic conditions and has been shown to play an active role in apoptosis when relocalized to the cytosol (31–33). Indeed, our earlier work showed that the pro-apoptotic effects of CQ and LUC in malignant cells were significantly impaired when CTSD was targeted using RNAi approaches. We first conducted microarray analyses to assess the global effects of ROC-325 on the gene expression profiles of RCC cells. 786-0 and A498 cells were treated with 5 μM ROC-325 for 24 hours and subjected to Affymetrix microarray quantification of pharmacodynamic changes in gene expression. Heat map analyses identified significant upregulation of genes involved with proteolysis in each cell line (Supp. Fig. 3A). Similar to the autophagy inhibitors CQ/HCQ and LUC, ROC-325 triggered a highly significant increase in CTSD levels, which was confirmed by qRT-PCR (Supp. Fig. 3B). In addition to this specific effect, ROC-325 treatment also stimulated the expression of other genes with important roles in the control of protein turnover and ER stress-induced apoptosis including UBA7, TNFAIP3, DDIT3, PMAIP1, and CASP4 indicating a link between the induction of apoptosis and the disruption of protein homeostasis.
In order to determine if drug-induced expression of CTSD is required for the autophagy inhibitory effects of ROC-325, we used lentiviral shRNA to target CTSD in 786-0 RCC cells (Supp Fig. 4A). These assays showed that antagonizing ROC-325’s ability to trigger increased CTSD levels did not interfere with drug-mediated stabilization of p62, an established marker of autophagy inhibition. However, CTSD knockdown did reduce the pro-apoptotic effects of ROC-325 (Supp. Fig. 4B). These collective findings indicate that CTSD is not essential for ROC-325 to inhibit autophagy and that its increased expression is likely a secondary/downstream pharmacodynamic event that promotes apoptosis.
ROC-325 induces hallmark features of autophagy inhibition and antagonizes autophagic flux
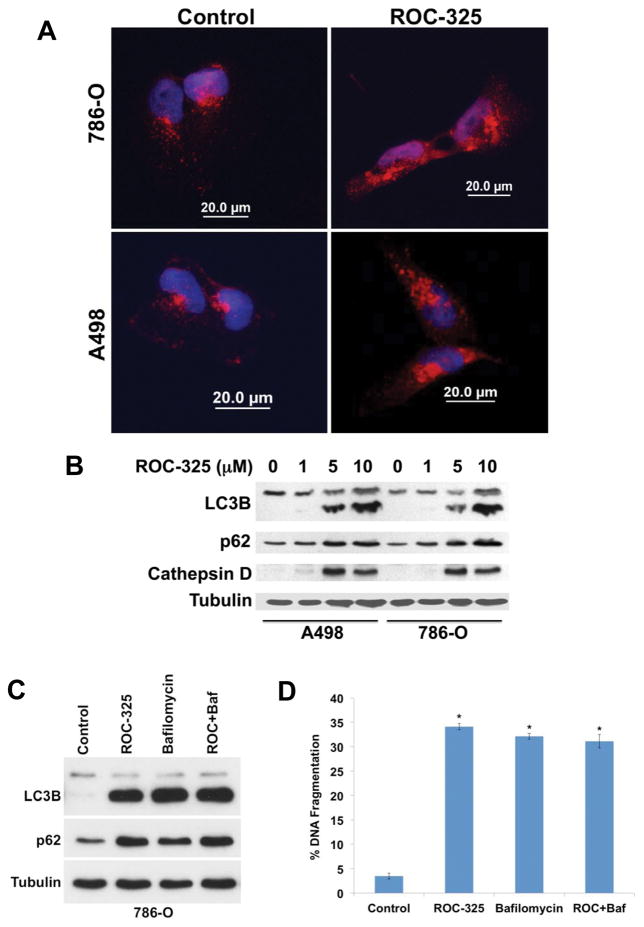
Our morphological and gene expression analyses in RCC cells indicated that ROC-325 treatment yielded pharmacodynamic effects that were consistent with inhibition of autophagy. In order to further investigate the specific effects of ROC-325 on autophagy, we first utilized fluorescent confocal microscopy to quantify the impact of drug treatment on LC3B distribution. Treatment with 5 μM ROC-325 for 24 hours led to the formation of LC3B punctae and a robust increase in LC3B levels in both A498 and 786-0 RCC cells (Fig. 2A). Immunoblotting analyses conducted in both A498 and 786-0 cells demonstrated that ROC-325 promoted a dose-dependent increase in LC3B expression in a manner that correlated with a corresponding increase in the levels of p62 and cathepsin D (Fig. 2B). Washout experiments where cells were treated with ROC-325, the drug was removed, and the effects on p62 expression were followed over time showed that ROC-325 achieved sustained autophagy inhibition for more than 24 h after drug exposure ceased (Supp. Fig. 5). We next used the autophagy inhibitor bafilomycin A1 as a tool to evaluate the effects of ROC-325 on autophagic flux (20). 786-0 cells were treated with ROC-325 alone and in combination with bafilomycin A1 and the levels of LC3B and p62 were quantified by immunoblotting. No significant differences in the levels of either LC3B or p62 were detected when ROC-325 was combined with bafilomycin A1, thus indicating that ROC-325 effectively inhibited autophagic flux (Fig. 2C). Furthermore, the addition of bafilomycin A1 did not significantly augment the ability of ROC-325 to induce apoptosis (Fig. 2D). Our results collectively demonstrate that ROC-325 disrupts autophagic degradation.
Figure 2.
ROC-325 inhibits autophagy. (A) ROC-325 induces LC3B accumulation. RCC cells were treated with 5 μM ROC-325 for 24 h. LC3B accumulation was visualized by immunocytochemistry. (B) ROC-325 increases LC3B, p62, and cathepsin D expression. 786-O and A498 cells were treated with the indicated concentrations of ROC-325 for 24 h. Protein levels were determined by immunoblotting. The lower band on all LC3 blots in the manuscript depicts the lipidated (LC3-II) form of LC3B, which is an established autophagosome marker. (C–D) Bafilomycin A1 does not augment ROC-325-mediated LC3B or p62 accumulation or apoptosis. 786-O cells were treated with 5 μM ROC-325, 100 nM bafilomycin A1, or both agents for 48 h. Protein expression was determined by immunoblotting and apoptosis by PI-FACS analysis. Mean ± SD, n = 3. *Indicates a significant difference from control, P<0.05. There is no significant difference between bafilomycin A1, ROC-325, and ROC-325 + bafilomycin A1 treatments.
The anticancer effects of ROC-325 are severely diminished in cells with genetically impaired autophagy
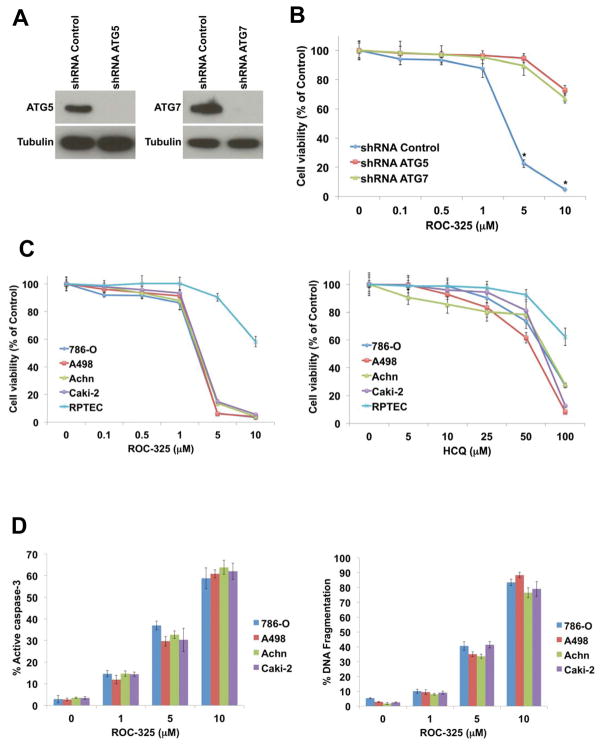
All anticancer drugs have multiple effects. To assess whether autophagic inhibition is a major mechanism of action that contributes to the anticancer properties of ROC-325, we genetically impaired two different genes that have been established to be essential for functional autophagy, ATG5 (34) and ATG7 (35), using lentiviral shRNA approaches in 786-0 RCC cells (Fig. 3A). Cells with targeted knockdown of either gene proliferated more slowly than cells infected with non-targeted control shRNA, indicating that functional autophagy promotes RCC cell proliferation and thereby affirming that autophagy inhibition may be an effective approach to antagonize RCC pathogenesis (Supp. Fig. 6A). Genetic impairment of either ATG5 or ATG7 yielded similar results with respect to the anticancer effects of ROC-325 in that the genetic disruption of autophagy rendered it significantly less effective (Fig. 3B). Supporting experiments with HCQ also showed that this agent was less effective at reducing RCC cell viability when autophagy was genetically impaired (Supp. Fig. 6B). This indicates that autophagy inhibition is a key anticancer mechanism of action of ROC-325 at the concentrations used in this study.
Figure 3.
ROC-325 has selective anticancer effects that are dependent upon autophagy function. (A) The essential autophagy genes ATG5 and ATG7 were knocked down in 786-0 RCC cells using lentiviral shRNA. Knockdown efficiency was assessed by immunoblotting. Tubulin documented equal protein loading. (B) Cells infected with non-targeted control, ATG5- or ATG7-directed lentiviral shRNAs were treated with the indicated concentrations of ROC-325 for 72h. The effects of drug treatment on cell viability were quantified for each experimental condition by MTT assay. Mean ± SD, n = 3. (C) ROC-325 selectively decreases RCC cell line viability more effectively than HCQ. RCC cell lines and normal RPTEC cells were treated with varying concentrations of ROC-325 or HCQ for 72 h. Cell viability was measured using the MTT assay. Mean ± SD, n = 3. (D) ROC-325 stimulates apoptosis. RCC cell lines were treated with the indicated concentrations of ROC-325 for 48 h. Active caspase-3 (left) was measured using a FITC-labeled active caspase-3 antibody followed by flow cytometric analysis. Mean ± SD, n = 3. DNA fragmentation (right) was measured by PI-FACS analysis. Mean ± SD, n = 3.
ROC-325 exhibits therapeutic selectivity and has significantly greater anticancer activity than HCQ
We next treated four different RCC cell lines (786-0, A498, Achn, and Caki-2) with a range of concentrations of ROC-325 and HCQ for 72 hours. The effects of each agent on cell viability were determined by MTT assay. A direct comparison of the activity of ROC-325 and HCQ revealed that ROC-325 was approximately 10-fold more potent than HCQ based on IC50 analyses and exhibited favorable therapeutic selectivity for malignant versus normal renal proximal tubule epithelial cells (RPTEC) treated with ROC-325 (Fig. 3C). Since the reduction in cell viability we observed in RCC cells treated with ROC-325 could have resulted from inhibition of proliferation, cell death, or both, we next quantified the effects of ROC-325 on apoptosis by measuring drug-induced DNA fragmentation and caspase-3 activation. Treatment of RCC cells with ROC-325 for 48 hours led to dose-dependent increases in the percentages of cells with active caspase-3 expression and fragmented DNA (Fig. 3D) in a manner that correlated with the reduction in cell viability. Additionally, treatment of 786-0 cells with ROC-325 in combination with the FDA approved drugs vorinostat, temsirolimus, or sorafenib yielded significantly greater levels of apoptosis than any single agent (Supp. Fig. 7). This suggests that the ability of ROC-325 to trigger apoptosis accounts for a significant portion of its anticancer effects and that it has potential applications as a therapeutic partner to existing anticancer agents.
Oral administration of ROC-325 is well tolerated and antagonizes RCC tumor progression more effectively than HCQ
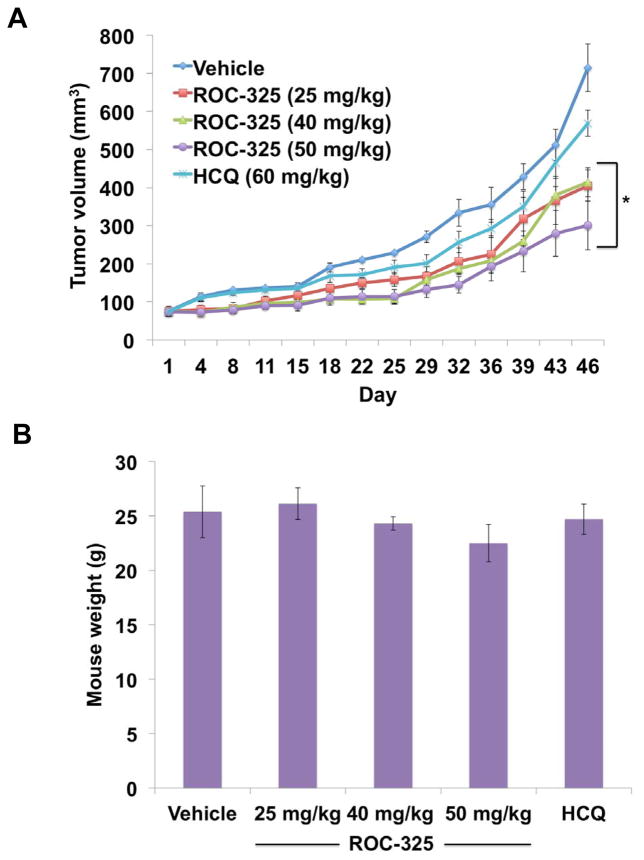
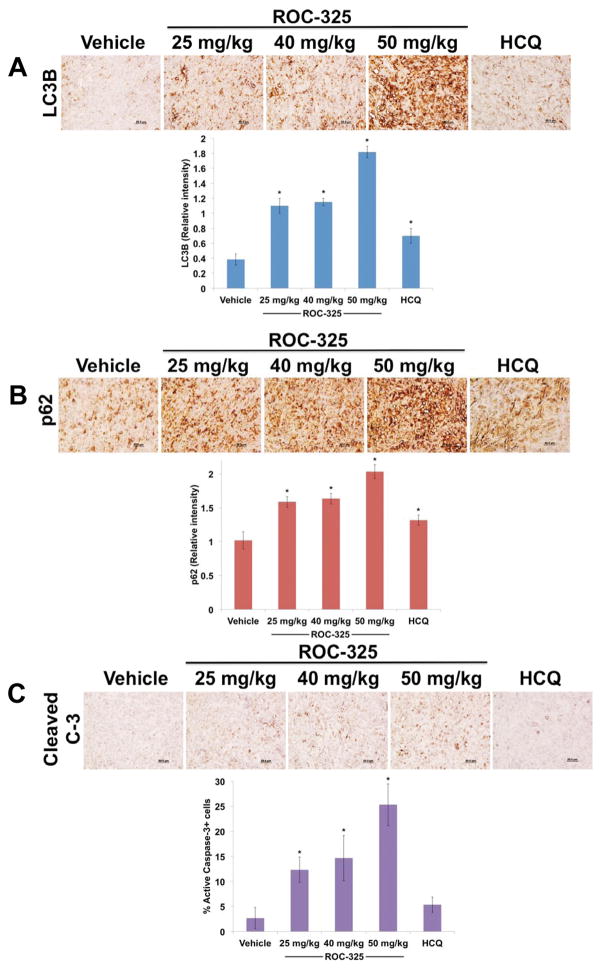
The in vivo anticancer activity of ROC-325 was evaluated by administering vehicle control (water), ROC-325 (25, 40, or 50 mg/kg QD), or HCQ (60 mg/kg QD) to nude mice implanted with 786-0 RCC xenografts. ROC-325 treatment led to significant, dose-dependent inhibition of disease progression in a manner that was superior to HCQ (Fig. 4A). ROC-325 was well tolerated and no notable toxicities were observed other than a very modest, non-significant reduction in mean body weight at the highest dose (Fig. 4B). Immunohistochemical analysis of specimens collected from animals treated with ROC-325, HCQ, or vehicle control demonstrated significant, dose-dependent increases in the autophagic markers LC3B (Fig. 5A) and p62 (Fig. 5B) and increased apoptosis (cleaved caspase-3, Fig. 5C). Our collective data demonstrate that ROC-325 is orally bioavailable, significantly more efficacious as a monotherapy than HCQ, exhibits favorable tolerability, and inhibits autophagy in vivo. These findings support further investigation of the safety and efficacy of ROC-325 as a novel agent for the treatment of autophagy-driven tumors and other disorders where lysosomal activity contributes to disease pathogenesis.
Figure 4.
ROC-325 reduces tumor burden more effectively than HCQ in RCC xenografts. (A) 786-O cells were injected into the flanks of nude mice. Mice were pair-matched and randomized into groups when mean tumor burden reached approximately 100 mm3. Mice were treated with 25, 40, or 50 mg/kg ROC-325 PO and 60 mg/kg HCQ IP QDx5 throughout the course of the study. Tumor volumes were measured twice weekly. Mean ± SEM, n = 5. *Indicates a significant difference in tumor burden compared to vehicle control based on 2-way ANOVA analyses. P < 0.05. (B) ROC-325 is well tolerated in mice. Body weight was determined at the end of the study (Day 46) to quantify drug-induced weight loss. Mean ± SD, n = 5.
Figure 5.
ROC-325 significantly increases LC3B and p62 expression and induces apoptosis in RCC xenografts. (A) Tumors were harvested 6 h following the last dose of each drug was administered. LC3B immunohistochemistry. Tumors were stained with an anti-LC3B antibody and the relative intensity of expression was quantified by densitometry. Mean ± SD, n = 5. (B) p62 immunohistochemistry. Tumors were stained with an anti-p62 antibody and the relative intensity of expression was quantified by densitometry. Mean ± SD, n = 5. (C) Apoptosis was determined by active caspase-3 immunohistochemistry. Tumors were stained with an antibody to cleaved caspase-3. The percentage of positive stained cells was determined manually under 20X magnification. Mean ± SD, n = 5. *Indicates a significant difference from vehicle, P<0.05.
Discussion
The rational ability of autophagic degradation to help fulfill the fundamental need of tumors to maintain energy metabolism to drive their expansion, metastasis, and sustain their survival under therapy- and microenvironment-induced stress gave rise to a new field focused on determining the contributions of autophagy to malignant pathogenesis. More than a decade later, autophagy has been defined as a mechanism that fuels malignant bioenergetics in a manner that contributes to both disease progression and drug resistance in a diverse range of solid and hematological cancers (1, 2). Accordingly, a tremendous number of preclinical investigations showed that genetic or pharmacological inhibition of autophagy diminished drug resistance and augmented the efficacy of a plethora of cytotoxic and targeted anticancer agents and radiation therapy (5, 6). The findings of several of these studies directly established the foundation for multiple early phase clinical trials that investigated the safety and preliminary efficacy of the antimalarial autophagy inhibitor HCQ in combination with several other FDA approved anticancer agents and radiation therapy (11–16). The initial series of HCQ combination clinical trials demonstrated that the addition of HCQ to these specific regimens yielded an acceptable safety profile. Preliminary efficacy was observed in a small subset of patients (such as the RCC patient on our HCQ + vorinostat phase I trial) treated in these clinical studies, but the question of whether the MTD of HCQ in these studies resulted in complete autophagic inhibition remained unanswered.
The modest efficacy that was observed in this first series of trials seeking to deliberately inhibit autophagy as a novel therapeutic strategy likely stemmed from two major issues. First, the lack of validated predictive biomarkers that define genetic features of tumors that render them autophagy-dependent prevented the refinement of the eligibility criteria in a way that would facilitate the targeted enrollment of specific patients that may be more likely to benefit from treatment with autophagy inhibitors. It has been reported in preclinical studies that mutations in RAS confer autophagy addiction and that RAS-driven cancers may be hypersensitive to autophagy inhibition (36, 37). However, this has not been clinically proven to date. It is possible that the importance of RAS in this context may be tumor type specific and that other oncogenes with mechanistic links to the control of autophagy may also be involved in determining autophagy dependence. In order to ultimately position autophagy inhibitors to effectivey benefit patients, it is imperative to define a genetic, epigenetic, metabolic and phenotypic tumor signature that predicts susceptibility to this precision therapeutic approach. Additionally, the anticancer agents whose efficacy is most significantly reduced by therapy-triggered autophagy must be established to identify optimal and rational partners for combination regimens. Advances in both of these areas would dramatically improve the opportunities for this class of drugs to successfully impact cancer outcomes.
A second major underlying cause of the limited efficacy that we and our colleagues observed is likely due to the pharmacological properties of HCQ itself. HCQ has been used for the treatment of malaria, rheumatoid arthritis and systemic lupus for many years. However, it was not designed to be an autophagy inhibitor. Rather, it has been attempted to be repurposed as such due to the lack of rationally designed autophagy inhibitors available for clinical use and the overwhelming interest to quickly translate the excitement of preclinical findings regarding autophagy inhibition into potential therapeutic benefit for patients with cancer. Thus, the limited clinical efficacy of HCQ in this patient population could be due, in part, to the insufficient inhibition of autophagy at doses of HCQ that yield acceptable safety and tolerability.
We sought to develop new agents that exhibited both superior autophagic inhibition and anticancer activity than HCQ. Since no comprehensive SAR studies determining which chemical motifs are essential to disrupt autophagy at the lysosomal level have been conducted to date, we analyzed the structure of HCQ and other reported agents that inhibit autophagy and designed a series of new compounds with various modifications to key chemical motifs in HCQ and these other drugs. Our initial screening assays identified ROC-325 as a lead compound. Notably, the structure of ROC-325 contains elements of both HCQ and LUC, which we previously discovered to inhibit autophagy, but is chemically distinct from Lys05, another new dimeric agent that has been demonstrated to inhibit autophagy (17, 20).
Comprehensive preclinical studies with ROC-325 demonstrated that it induced all of the hallmark features of genetic and pharmacological autophagy inhibition including the formation of LC3B punctae, accumulation of autophagosomes, stabilization of p62 and disruption of autophagic flux. ROC-325 also triggered the expression of the lysosomal protease CTSD, which we previously showed plays a key role in mediating CQ/HCQ and LUC-induced apoptosis in addition to other genes with established roles in controlling protein degradation and endoplasmic reituculum (ER) stress-induced cell death (20, 22, 30). Preliminary studies demonstrated that ROC-325 also appears to augment the anticancer effects of several FDA approved drugs. Although all of these findings are scientifically important and collectively define the mechanism of action of this new drug, the most critical aspect of this study is the evidence demonstrating that ROC-325 is orally bioavailable and inhibits autophagy in vivo while yielding significantly greater single agent efficacy against RCC xenograft tumors than a higher dose of HCQ administered on the same schedule. Taken together, our findings establish ROC-325 as a novel autophagy inhibitor that warrants further investigation to better define its safety and efficacy for the treatment of autophagy-dependent malignancies and other lysosome-centric disorders.
Supplementary Material
Translational Relevance.
Autophagy plays an important role in cancer progression and drug resistance, yet there are currently limited clinically available agents to target this pathway. We used logical medicinal chemistry approaches to develop ROC-325, an orally available novel inhibitor of autophagic degradation. Comprehensive in vitro and in vivo studies in preclinical models of renal cell carcinoma (RCC) demonstrated that ROC-325 induced all of the hallmark features of autophagy inhibition, was well tolerated, and was significantly more effective than hydroxychloroquine. Our findings provide strong rationale for clinical investigation of the safety and efficacy of ROC-325 for RCC and other autophagy-dependent malignancies.
Acknowledgments
Grant support: This work was supported by grants from the Scott Hamilton CARES Initiative (JSC) and the National Cancer Institute (R01CA172443, JSC; R01CA190789, STN; and P30CA023074)
Footnotes
Disclosure of conflicts of interest: JSC and JP are inventors on a pending patent related to ROC-325 and the other novel chemical matter detailed in this manuscript. None of the other authors have any relevant conflicts of interest to declare.
Author contributions:
Conception and design – JSC, JP, and STN
Development of methodology – JSC, JP, STN
Data acquisition, analysis, and interpretation - JSC, CME, WZ, YH, VV, JP, STN
Manuscript preparation – JSC, JP, STN
References
- 1.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–30. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 2011;8:528–39. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 5.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–66. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carew JS, Kelly KR, Nawrocki ST. Autophagy as a target for cancer therapy: new developments. Cancer management and research. 2012;4:357–65. doi: 10.2147/CMAR.S26133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carew JS, Nawrocki ST, Cleveland JL. Modulating autophagy for therapeutic benefit. Autophagy. 2007;3:464–7. doi: 10.4161/auto.4311. [DOI] [PubMed] [Google Scholar]
- 8.Chen S, Zhou L, Zhang Y, Leng Y, Pei XY, Lin H, Jones R, Orlowski RZ, Dai Y, Grant S. Targeting SQSTM1/p62 induces cargo loading failure and converts autophagy to apoptosis via NBK/Bik. Mol Cell Biol. 2014;34:3435–49. doi: 10.1128/MCB.01383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao R, Balusu R, Fiskus W, Mudunuru U, Venkannagari S, Chauhan L, Smith JE, Hembruff SL, Ha K, Atadja P, Bhalla KN. Combination of pan-histone deacetylase inhibitor and autophagy inhibitor exerts superior efficacy against triple-negative human breast cancer cells. Mol Cancer Ther. 2012;11:973–83. doi: 10.1158/1535-7163.MCT-11-0979. [DOI] [PubMed] [Google Scholar]
- 10.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, Schuchter LM, Torigian DA, Panosian JT, Troxel AB, Tan KS, Heitjan DF, DeMichele AM, Vaughn DJ, Redlinger M, Alavi A, Kaiser J, Pontiggia L, Davis LE, O’Dwyer PJ, Amaravadi RK. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1391–402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogl DT, Stadtmauer EA, Tan KS, Heitjan DF, Davis LE, Pontiggia L, Rangwala R, Piao S, Chang YC, Scott EC, Paul TM, Nichols CW, Porter DL, Kaplan J, Mallon G, Bradner JE, Amaravadi RK. Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10:1380–90. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, Tan KS, Heitjan DF, Rodgers G, Gallagher M, Piao S, Troxel AB, Evans TL, DeMichele AM, Nathanson KL, O’Dwyer PJ, Kaiser J, Pontiggia L, Davis LE, Amaravadi RK. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1369–79. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahalingam D, Mita M, Sarantopoulos J, Wood L, Amaravadi RK, Davis LE, Mita AC, Curiel TJ, Espitia CM, Nawrocki ST, Giles FJ, Carew JS. Combined autophagy and HDAC inhibition: a phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy. 2014;10:1403–14. doi: 10.4161/auto.29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, McAfee Q, Fisher J, Troxel AB, Piao S, Heitjan DF, Tan KS, Pontiggia L, O’Dwyer PJ, Davis LE, Amaravadi RK. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10:1359–68. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnard RA, Wittenburg LA, Amaravadi RK, Gustafson DL, Thorburn A, Thamm DH. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy. 2014;10:1415–25. doi: 10.4161/auto.29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McAfee Q, Zhang Z, Samanta A, Levi SM, Ma XH, Piao S, Lynch JP, Uehara T, Sepulveda AR, Davis LE, Winkler JD, Amaravadi RK. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012;109:8253–8. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow LM, Chan TH. Novel classes of dimer antitumour drug candidates. Current pharmaceutical design. 2009;15:659–74. doi: 10.2174/138161209787315576. [DOI] [PubMed] [Google Scholar]
- 19.Hadden MK, Blagg BS. Dimeric approaches to anti-cancer chemotherapeutics. Anti-cancer agents in medicinal chemistry. 2008;8:807–16. doi: 10.2174/187152008785914743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carew JS, Espitia CM, Esquivel JA, 2nd, Mahalingam D, Kelly KR, Reddy G, Giles FJ, Nawrocki ST. Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J Biol Chem. 2011;286:6602–13. doi: 10.1074/jbc.M110.151324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O’Dwyer M, Nawrocki ST, Giles FJ, Carew JS. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 22.Carew JS, Medina EC, Esquivel JA, 2nd, Mahalingam D, Swords R, Kelly K, Zhang H, Huang P, Mita AC, Mita MM, Giles FJ, Nawrocki ST. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J Cell Mol Med. 2010;14:2448–59. doi: 10.1111/j.1582-4934.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahalingam D, Medina EC, Esquivel JA, 2nd, Espitia CM, Smith S, Oberheu K, Swords R, Kelly KR, Mita MM, Mita AC, Carew JS, Giles FJ, Nawrocki ST. Vorinostat enhances the activity of temsirolimus in renal cell carcinoma through suppression of survivin levels. Clin Cancer Res. 2010;16:141–53. doi: 10.1158/1078-0432.CCR-09-1385. [DOI] [PubMed] [Google Scholar]
- 25.Nawrocki ST, Carew JS, Maclean KH, Courage JF, Huang P, Houghton JA, Cleveland JL, Giles FJ, McConkey DJ. Myc regulates aggresome formation, the induction of Noxa, and apoptosis in response to the combination of bortezomib and SAHA. Blood. 2008;112:2917–26. doi: 10.1182/blood-2007-12-130823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75:1322–31. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carew JS, Nawrocki ST, Reddy VK, Bush D, Rehg JE, Goodwin A, Houghton JA, Casero RA, Jr, Marton LJ, Cleveland JL. The novel polyamine analogue CGC-11093 enhances the antimyeloma activity of bortezomib. Cancer Res. 2008;68:4783–90. doi: 10.1158/0008-5472.CAN-07-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lebovitz CB, Robertson AG, Goya R, Jones SJ, Morin RD, Marra MA, Gorski SM. Cross-cancer profiling of molecular alterations within the human autophagy interaction network. Autophagy. 2015 doi: 10.1080/15548627.2015.1067362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–22. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beaujouin M, Baghdiguian S, Glondu-Lassis M, Berchem G, Liaudet-Coopman E. Overexpression of both catalytically active and -inactive cathepsin D by cancer cells enhances apoptosis-dependent chemo-sensitivity. Oncogene. 2006;25:1967–73. doi: 10.1038/sj.onc.1209221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haendeler J, Popp R, Goy C, Tischler V, Zeiher AM, Dimmeler S. Cathepsin D and H2O2 stimulate degradation of thioredoxin-1: implication for endothelial cell apoptosis. J Biol Chem. 2005;280:42945–51. doi: 10.1074/jbc.M506985200. [DOI] [PubMed] [Google Scholar]
- 33.Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, Prebois C, Rochefort H, Vignon F. Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett. 2006;237:167–79. doi: 10.1016/j.canlet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 35.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 36.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, Muller F, Colla S, Nezi L, Genovese G, Deem AK, Kapoor A, Yao W, Brunetto E, Kang Y, Yuan M, Asara JM, Wang YA, Heffernan TP, Kimmelman AC, Wang H, Fleming JB, Cantley LC, DePinho RA, Draetta GF. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–32. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.