Abstract
Membrane microdomains that include lipid rafts, are involved in key physiological and pathological processes and participate in the entry of endocellular pathogens. These assemblies, enriched in cholesterol and sphingolipids, form highly dynamic, liquid-ordered phases that can be separated from the bulk membranes thanks to their resistance to solubilization by nonionic detergents. To characterize complexity and dynamics of detergent-resistant membranes of sexual stages of the rodent malaria parasite Plasmodium berghei, here we propose an integrated study of raft components based on proteomics, lipid analysis and bioinformatics. This analysis revealed unexpected heterogeneity and unexplored pathways associated with these specialized assemblies. Protein-protein relationships and protein-lipid co-occurrence were described through multi-component networks. The proposed approach can be widely applied to virtually every cell type in different contexts and perturbations, under physiological and/or pathological conditions.
Membrane microdomains, also referred to as lipid rafts, are specialized assemblies, enriched in cholesterol (CH)1 and sphingolipids, where specific protein-protein or protein-lipid interactions occur (1). The peculiar structural organization of these dynamic platforms also depends on the prevalence of saturated fatty acids in sphingolipids that allow tight CH interactions and the formation of highly packed phases. The liquid ordered nature of CH-rich assemblies renders them resistant to solubilization in nonionic detergents, compared with the liquid-disordered bulk membranes, and allows their enrichment as detergent resistant membranes (DRMs) by sucrose gradient centrifugation.
DRM-rafts, extensively investigated in mammalian cells, have been implicated in key cellular processes such as signal transduction, membrane trafficking or cell polarization (2) and their importance in the pathogenesis of human disorders (3–5) has been elucidated over recent years. Furthermore, a growing number of pathogens have been shown to hijack lipid rafts to enter and survive inside the target cell (6–8).
Given the increasing awareness of the fundamental role of lipid rafts in various cellular functions, these assemblies are being viewed as appealing drug targets. The pharmaceutical potential of raft-associated proteins and lipid/fatty acids are being explored, based on dietary modifications (9), or plant-derived compounds (10).
Raft-like microdomains have been implicated in the control of virulence of numerous parasite protozoa, including Plasmodium falciparum, which causes the most severe human malaria (11, 12).
Malaria is one of the major global health problems with more than 200 million new cases and 438,000 deaths estimated by WHO in 2015. Plasmodium blood stages multiply asexually inside red blood cells (RBCs), causing malaria symptoms, or differentiate into sexual gamete precursors, the male and female gametocytes, responsible for parasite transmission. Circulating gametocytes are arrested in the cell cycle until they are ingested by a female mosquito of the genus Anopheles. Environmental cues, associated with the mosquito midgut, trigger gamete maturation and escape from the host RBC. With a spectacular process named exflagellation, male gametocytes undergo three rounds of DNA replication and axoneme assembly in 15–20 min, leading to the formation of eight motile gametes, competent to fertilize the emerged females, therewith commencing mosquito colonization by the parasite.
Previous studies on Plasmodium asexual blood stages indicated that CH-rich membrane microdomains participate to various biological processes, such as protein sorting/trafficking, host cell invasion, organelle biogenesis and membrane remodeling with a key function in parasite virulence and pathogenesis (13–16). A specific role of these assemblies in sexual stages has not been investigated so far.
Here we propose an integrated method based on label-free quantitative proteomics, lipid analysis and bioinformatics to provide insights on components and buoyancy features of membrane microdomains of Plasmodium sexual stages.
We show here that gametocyte-specific molecules, implicated in either the exit of gametes from the host cell or in gamete fertilization, are recruited into DRMs. This suggests that membrane microdomains may be targeted to block not only parasite invasion and development inside the host erythrocyte but also transmission to the mosquito vector.
EXPERIMENTAL PROCEDURES
Plasmodium Berghei In Vivo Infection and Purification
A high gametocyte producer P. berghei ANKA line (HP) was used for high-yield production of gametocytes, whereas the isogenic nongametocyte producer line (HPE) was used to obtain asexual trophozoites freed from any contamination of sexual stages. Synchronous infections were established in CD1 mice by intravenous injection of purified infective schizonts (17). Blood was collected by heart-puncture under anesthesia and leukocytes were removed using Plasmodipur leukocyte filters (Euro-Diagnostica, Malmoe, Sweden). Schizont- or gametocyte-infected erythrocytes were separated from uninfected cells through Nycodenz density gradient centrifugation (17).
Antibodies
Polyclonal immune sera against the RNA helicase (PBANKA_030680), the conserved protein (PBANKA_0604300) and the deoxyribose-phosphate aldolase (PBANKA_0505800) were obtained in BALB/c mice via intraperitoneal injection of purified recombinant proteins (50 μg in Freund's complete adjuvant) followed by two successive injections at 2-week intervals (25 μg in Freund's incomplete adjuvant). Mice were bled 1 week after the third immunization. The coding fragments used for immunization were cloned in BamHI-NotI sites of PGEX6P-1 and expressed as GST fusions. Peptide sequences and primers used for PCR amplifications are detailed in supplemental Methods. A PbG377 peptide, expressed in pGEX-6P-1 vector detailed in (20) was purified and used to prepare a specific rabbit immune serum for colocalization experiments (1:100 in IFA). Antibodies used for Western blot (WB) and indirect immunofluorescence analysis (IFA) were: α-PbG377 mouse polyclonal (20), 1:300 in IFA, 1:2000 in WB; α-SET rabbit polyclonal (21) 1:200 in IFA, 1:5000 in WB; α-Pb14-3-3 mouse polyclonal (22) 1:200 in IFA, 1:2000 in WB; α-SEP1 rabbit polyclonal (23) 1:5000 in WB; α-alpha-tubulin rabbit monoclonal (Abcam, Cambridge, UK), 1:500 in IFA; α-Flotillin mouse monoclonal (BD Transduction Laboratories, Lexington, KY), 1:1000 in WB; α-Pfhsp70 (kindly provided by Denise Mattei), 1:1000 in WB.
Western Blot Analysis
WB analysis was performed using MINI TRANS-BLOT® Bio-Rad (Hercules, CA) apparatus at constant voltage (100V) for 1h, in transfer buffer (20% methanol, Tris 0,025M, Glycine 0.192 m) onto Protran 0.22 microns membrane (Whatman, Maidstone, UK). Primary and horseradish peroxidase-conjugated secondary antibody were incubated 1h in PBS-Tween (0.05%) 1% nonfat milk and membrane was developed using the ECL system (SuperSignalWest Pico, Thermo Scientific) according to manufacturer's instructions.
Indirect Immunofluorescence Assay
Blood smears were fixed on glass slides for 1h with 4% paraformaldehyde at room temperature (RT), washed in PBS, treated with 0.1% Triton X-100 in PBS (10 min at RT) and washed again in PBS. Blocking was performed overnight in PBS/3% BSA at 4 °C. Slides were then incubated 1h in primary antibody, washed in PBS and incubated 30min in fluorescein- or rodamin-conjugated goat α-mouse or α-rabbit secondary antibodies (1:400 dilution). Cell nuclei were labeled with 4′,6-diamidin-2-fenilindolo (DAPI). The specificity of the immune sera was checked in parallel using pre-immune sera. Subcellular localization of selected proteins was analyzed in P. berghei sexual stages by double IFA. Nonactivated and activated males were recognized using antibodies against the nuclear protein NAP/SET and the alpha-tubulin respectively, whereas nonactivated and activated females were identified using antibodies against PbG377. For each examined antibody, about 100 gametocytes were inspected microscopically and fluorescence pattern was considered representative when detected in 80–90% of the examined cells, in two independent parasite preparations.
DRMs Purification and Fractionation
Nycodenz-purified infected red blood cells (IRBCs) were centrifuged (900 × g,10 min) and RBC membranes selectively ruptured by resuspending cell pellet in 20 volumes of cold lysis buffer (0.15 m NH4Cl, 0.01 m KHCO3, 1 mm EDTA) with the addition of protease mixture inhibitors (Roche, Basel, Switzerland). The suspension was incubated in ice for 10 min. Free parasites were collected by centrifugation, washed three times in PBS to remove hemoglobin and stored at −80 °C for successive analyses (3 × 108-parasite aliquots). Relative protein abundance was evaluated by probing total protein extracts from the stored parasite pellets of mature gametocyte sample A, (MG-A) and asexual trophozoite sample A (T-A), with 14-3-3 and Pfhsp70 specific antibodies (supplemental Fig. S1A).
To isolate DRMs, each parasite pellet (3 × 108 cells) was suspended in 0.75 ml of MES-buffered saline (25 mm MES, pH 6.5, 0.15 m NaCl) containing 1% Triton X-100 (v/v) and homogenized with a potter-elvehjem glass homogenizer. Cell extracts were adjusted to 40% sucrose by the addition of 0.75 ml of 80% sucrose in MES-buffered saline, placed at the bottom of an ultracentrifuge tube (4.5 ml, 13 × 15 mm, Beckman) and overlaid with 1.5 ml of 30% and 1.5 ml of 5% sucrose. Samples were subjected to equilibrium density gradient centrifugation using SW60Ti rotor (Beckman Instruments, Brea, CA) at 45,000 rpm (208,000 × g), for 18 h at 4 °C. 375-μl fractions were then collected from the top of the gradient. To verify linearity of the gradient, sucrose density of the collected fractions was routinely determined through refractive index values (supplemental Fig. S1B). Reproducibility of Flotillin density distribution was always inspected by probing fractions 2–8 with the specific antibody; in our working conditions Flotillin floats to fraction 4 (supplemental Fig. S1B). Proteins in light fractions 2–8 were precipitated, as described in (18), whereas lipids were extracted as described in (19).
To prepare total Triton-insoluble and -soluble parasite extracts, cell pellets (107) were resuspended in 0.1 ml of cold MES-buffered saline supplemented with 1% Triton X-100, homogenized as described above, and centrifuged at 30,000 × g, 30 min. Soluble fractions were collected, while the pellets were washed in cold MES-buffered saline. Insoluble fractions were recovered after centrifugation (30 min at 30,000 × g). and resuspended in 0.1 ml volume of SDS-PAGE loading sample buffer.
Experimental Design and Statistical Rationale
Here we propose a method based on label-free quantitative proteomics to define flotation properties of DRM-associated proteins separated by sucrose gradient centrifugation. DRM analysis was performed on mature gametocytes (sample MG-A) and asexual trophozoites (sample T-A), purified from synchronous P. berghei infections (described above). To limit variability because of the high dynamic nature of these membrane compartments, proteomics and lipid analysis were conducted on a single sample preparation. However, to evaluate the experimental reproducibility, the entire procedure that includes parasite solubilization, DRM separation via equilibrium density sucrose gradient centrifugation, protein and lipid extraction and analysis was performed in three independent experiments. The optimal size of the sample (3 × 108 parasites) was determined experimentally to guarantee efficient cell lysis and avoid sucrose gradient overloading, factors that may affect DRM flotations. Gradient linearity was always verified by measuring refractive index values of the collected fractions.
To obtain protein and lipid constituents of DRMs for proteomics and lipid analysis, fractions 2 to 8, were subjected to chloroform/methanol extraction. Small aliquots of protein samples (about 10%) were probed with antibodies against selected Plasmodium proteins to verify their buoyancy profile in WB analysis (see Fig. 5A and 5B) whereas Flotillin I was used to evaluate reproducibility of sucrose gradient separation (Fig. 5C). Major lipid classes, CH, sphingomyelin (SM), phosphatidylserine (PS), phosphatidylcholine (PC) and phosphatidylethanolamine (PE), were identified by high performance thin layer chromatography (HPTLC), whereas their fatty acid components, i.e. saturated (SFA), mono-unsaturated (MUFA) and poly-unsaturated (PUFA) fatty acids, were identified by Gas Cromatography (GC) analysis.
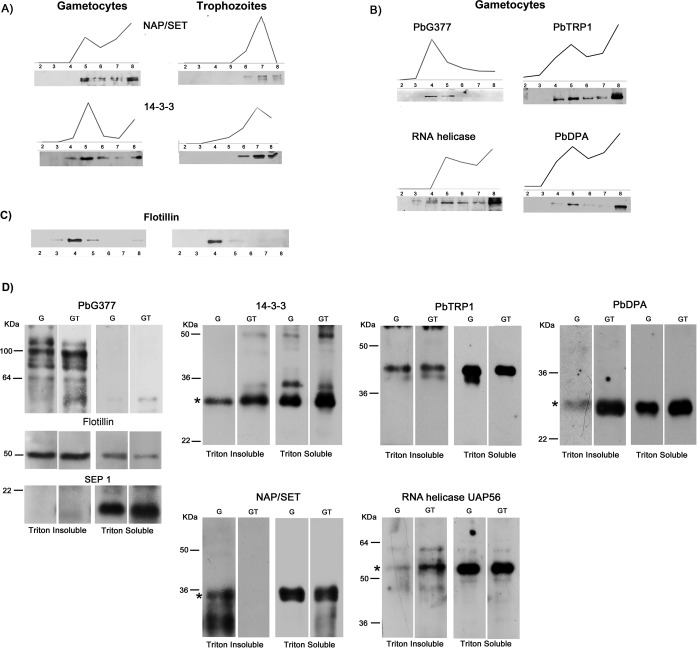
Fig. 5.
DRM partitioning of selected candidates. Floatation profile of selected proteins was determined either by label-free quantitative proteomics (PAP) or by Western blot (WB) analysis. Gradient fractions were probed with specific antibodies raised against: A, the nuclear protein NAP/SET and the nucleo-cytoplasmic protein 14-3-3, partially recruited into DRMs of both sexual and asexual stages; B, the gametocyte-specific protein PbG377, the putative RNA helicase UAP56, the conserved protein of unknown function PbTRP1 and the deoxyribose phosphate aldolase (PbDPA), recruited only in gametocyte DRMs. In all cases, WB profiles are superimposable to PAPs. in our experimental conditions, Flotillin resident into erythrocyte DRMs, floats to fraction 4 (C). To define membrane partioning of the selected proteins, specific antibodies were used in WB analysis of Triton-insoluble and -soluble extracts from nonactivated and 10 min-activated gametocytes (D). To validate the procedure, protein extracts were probed with antibodies against the DRM-resident proteins PbG377 and Flotillin and the integral membrane protein SEP1, not detected in DRM proteome. An asterisk indicates protein bands at the correct molecular mass. Multiple bands, were detected by PbG377-specific antibodies, because of post-translational cleavage.
Proteins detected with a minimum of two unique peptides, in at least two out of three replicates (for search parameter and acceptance criteria see below) were used for further analysis. The abundance of each protein in collected gradient fractions was determined by label-free Top3 method (24). Normalized Top3 values were used to construct protein abundance profiles (PAPs), that describe buoyancy features of each identified protein (see below). Reproducibility of PAPs between replicates was evaluated by the Pearson's correlation coefficient. Biological reproducibility of PAPs was assessed by probing DRM gradient fractions of two independent preparations of mature gametocytes (MG-B and C) and trophozoites (T-B and C) with antibodies against the proteins under study (supplemental Fig. S3).
Protein-related PAPs obtained in the three proteomic replicates were then merged in a single profile of 21 points and submitted to hierarchical average linkage clustering by Cluster 3.0 (2). The resulting dendrogram was manually inspected and clusters of correlated profiles (R ≥ 0.5; p < 0.05) were extracted. A cluster abundance profile was then calculated as the median of the three replicates Top3-medians. Finally, relationships between clusters in the different gradient layers (fractions 4–8) were visualized as stage-specific lipid and protein co-occurrence networks.
Sample Preparation for Proteomic Analysis
Proteins extracted from DRM fractions 2–8 were loaded on SDS-PAGE (home-made 5% stacking-12% resolving bis-tris-PAGE) and run just to allow the protein marker (BenchMark™ Pre-Stained Protein Ladder, Invitrogen, Carlsbad, CA) to enter the resolving gel. After Coomassie Staining (Novex, Colloidal Blue Staining gel, Invitrogen) unresolved bands were excised and in-gel tryptic digestion was performed as already described (16) with slight modifications. Briefly, gel slices were destained by washings in acetonitrile (ACN)/50 mm NH4CO3 (1:1), treated with 10 mm DTT (40 min at 56 °C) and 55 mm iodoacetamide (30 min in the dark at RT) to reduce and alkylate cysteines, shrunken with ACN and rehydrated for 40 min on ice with a solution of 12,5 ng/μl trypsin (Promega, Madison, WI) in 50 mm NH4CO3 and protein digestion was carried out overnight at 37 °C. Trypsin digests were removed and gel pieces were shrunk with 95% ACN, 1% FA. Supernatants were combined with trypsin digests, the volume reduced in speedvac and adjusted to 20 μl with HPLC buffer A (95% ACN, 0.1% FA).
Liquid Chromatography Tandem Mass Spectrometry
Nano-RPLC was performed using a nano-HPLC 3000 Ultimate (Dionex) connected in line to LTQ-XL linear ion trap (Thermo Fisher, Waltham, MA). Tryptic digests were first loaded on a C18 RP-precolumn (300 μm i.d.x5 mm; 5 μm particle size; 100 Å pore size; LC Packings-Dionex), washed by the loading pump at 20 μl/min with buffer A for 5 min and then on an home-made 13 cm × 75 μm- i.d. Silica PicoTip (8 ± 1 μm) column (PicoTip Emitter, NewObjective) packed with Magic C18AQ (5 μm particle size; 200 Å pore size, Michrom Bioresouces Inc.) for chromatographic separations. Peptides were eluted at 0.3 μl/min along a 120-min linear gradient from 20% to 60% of buffer B (95% ACN, 0.1% FA) and electrosprayed directly into the mass spectrometer. Data acquisition was performed in data-dependent Top5. Full-scan MS was set with a maximum injection time of 10 ms and m/z 400–2000 mass range. The five most intense ions were sequentially selected and fragmented in CID mode: maximum injection time of 100 ms; m/z 50–2000 mass range; minimum signal threshold of 100 counts. Wide band activation and dynamic exclusion were enabled.
Mass Spectrometry Data Processing and Analysis
Spectra files were analyzed by Sequest HT search engine with Proteome Discoverer 1.4 (Thermo Fisher) using a homemade database constructed with the Mouse and Rattus Uniprot-Swissprot database (released on June 2012) and PbergheiANKA_9.2 (released on November 2012 on PlasmoDB) and containing also human keratins and decoy database (# sequences: 28632). The carboamidomethylation of cysteines was specified as fixed modification whereas the oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were set as variable modification; only full tryptic peptides were used for identification, mass tolerance was set to 1 Da for precursor ion and 0.4 Da for fragment ions and a maximum of two missed cleavages was allowed. The Percolator tool was used for peptide validation based on the q-value and high confidence was chosen, corresponding to a false discovery rate (FDR) ≤1% on peptide-level. Proteins were identified with a minimum of 2 peptides rank = 1, whereas maximum peptide rank was 2. Score versus Charge State was set as following: Minimal Score for charge state = 1:1.5, Minimal Score for charge state = 2:2, Minimal Score for charge state = 3:2.5, Minimal Score for charge state = 4:3. For peptides and proteins grouping default settings were used. Protein and peptide identification data are reported in supplemental Tables S1 and S2.
Protein Quantification and Analysis
Protein abundance was determined according to the “label-free” Top3 method (24). For a given protein, the average of the three precursor peptides with the highest intensities was calculated (supplemental Tables S3 and S4). Top2 value was reported for proteins identified with only two peptides. Normalized PAPs were then constructed according to PAPij = where Top3i is the Top3 value in the i-th fraction of the j-th protein and Top3jmax is the highest Top3i value found for the j-th protein.
Functional annotation of the identified proteins was extracted from PlasmoDB [http://plasmodb.org/] and manually revised.
Reproducibility Assessment of Proteomic Data
170 proteins were common to the three replicates of gametocyte DRMs, whereas 74 to trophozoite DRMs (supplemental Table S5). To evaluate reproducibility of proteomic data, the Pearson's correlation coefficient of PAP pairs (7 points) was determined. As reported in supplemental Fig. S1C, the percentage of proteins with at least two conserved PAPs (R ≥ 0.7; p < 0.05) is 77.4% for gametocytes and 82.5 for trophozoites. Moreover, 39% and 52.7% of them display PAPs conserved in the three replicates of gametocytes and trophozoites respectively (R ≥ 0.7), 52 and 44.6% in two out of three, whereas 9 and 2.7% have variable profiles (supplemental Table S5).
Lipid Analysis
Lipids were extracted from DRM containing gradient fractions 3–8 using a chloroform/methanol (2:1, v/v) solution according to the procedure described by Folch (19). Extracts were dried under nitrogen atmosphere and used for neutral lipid and phospholipid HPTLC. HPTLC for neutral lipids was done by eluting lipid extract in a solution of hexane/diethylether/acetic acid (70:30:1, v/v). HPTLC for phospholipids was performed by running lipid extract in a chloroform/methanol/acetic acid/formic acid/H2O (35:15:6:2:1, v/v) solution in order to separate SM, PC, and PE. The spots were visualized via copper acid staining (3% w/v). The plate was heated for about 5 min at 180 °C, and the abundance of the different lipid classes in each fraction was estimated in comparison with lipid standards (Sigma-Aldrich Corporation, St. Louis, MO) using a GS-700 imaging densitometer (Bio-Rad). Total lipid amount, calculated by the sum lipid extracted in each gradient fraction, was higher in gametocyte DRM compared with trophozoite stage (176.67 ± 28.32 versus 118.35 ± 4.94 p = 0.02).
Cis and trans fatty acid methyl ester (FAME) used for GC analysis were purchased from Aldrich (Milan, Italy), 6 cis-hexadecenoic acid methyl ester from Lipidox (Lidingö, Sweden), n-hexane from Baker (HPLC grade) (Modena, Italy); all compounds were used without further purification. FAMEs were analyzed by GC (Agilent 6850, Milan) equipped with a 60 m × 0.25 mm × 0.25 μm (50%-cyanopropyl)-methylpolysiloxane column (DB23, Agilent), and a flame ionization detector with the following oven program: temperature started from 165 °C, held for 3 min, followed by an increase of 1 °C/min up to 195 °C, held for 40 min, followed by a second increase of 10 °C/min up to 240 °C, and held for 10min. A constant pressure mode (29 psi) was chosen with helium as carrier gas. Methyl esters were identified by comparison with the retention times of authentic samples. Phospholipid extracts of each DRM fraction were treated with a 0.5 m solution of KOH in methanol (0.5 ml) for 10min at room temperature under inert atmosphere. The reaction mixtures were quenched with brine (0.5 ml) and FAMEs were extracted with n-hexane (3 × 2 ml), the organic phase was dried on anhydrous Na2SO4 and evaporated to dryness. The FAMEs were than dissolved in 10 μl of n -hexane and 1 μl was injected in GC equipment using the split mode (50:1). Under these mild conditions the polyunsatured fatty acids (PUFA) residues present in DRM fractions were quantitatively transformed into their corresponding FAMEs, as determined by comparing the recovery yield with standard references of 18:0 (saturated) and 18:2 (polyunsaturated) fatty acid methyl esters. The efficiency of the extraction-transesterification procedures was estimated as 90 ± 3% based on the theoretical yield expected from the starting PL and using an internal calibration standard (C17:0). Quantitative analysis was performed using the calibration factors of each FAME. Relative abundance of lipids and fatty acids in each gradient fraction was expressed as mean percentage values of two replicates for fatty acids and two out of three replicates for lipids (selecting the closest values).
Generation of Cooccurrence Networks
Protein/lipid cooccurrence networks were constructed for trophozoite and gametocyte DRMs (fractions 4–8). Protein groups obtained by hierarchical clustering represent network nodes, whereas proteins belonging to different clusters but exhibiting correlated PAPs (R ≥ 0.5; p < 0.05) represent network edges. Multi-component networks were visualized using Cytoscape 3.3.0 (http://www.cytoscape.org). Clusters were represented as pie charts proportional to cluster abundances, whereas the abundance of each functional category was calculated as the sum of protein Top3 values in each class. Relative abundances of lipids and fatty acids in each gradient fraction were represented as half ring charts.
RESULTS
A Pipeline for the Generation of a “DRM Barcode”
In this study we propose and validate a method based on label-free quantitative proteomics and lipid analysis to define DRM components (proteins and lipids), and their flotation properties, following enrichment by sucrose gradient centrifugation (Fig. 1). We applied this approach to the analysis of DRMs of sexual gametocytes of the rodent malaria model P. berghei, not investigated so far.
Fig. 1.
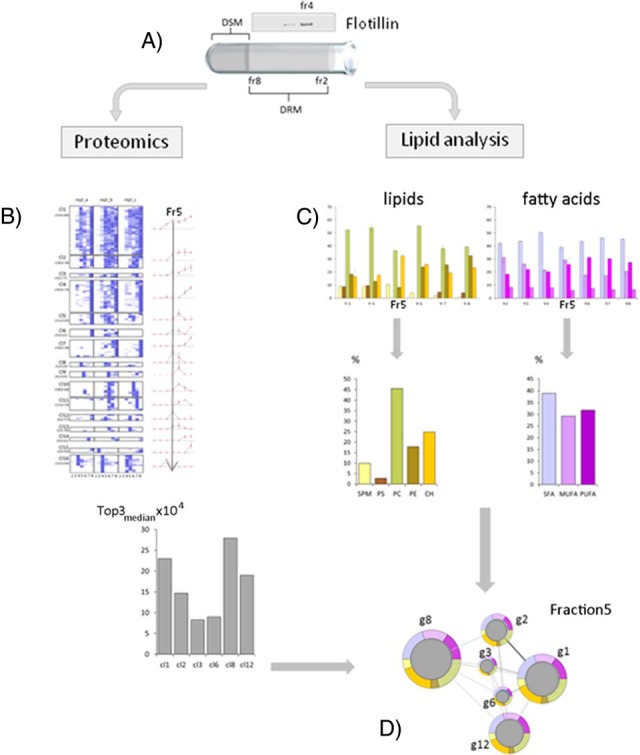
Experimental pipeline for whole-cell DRM analysis. A, Detergent-resistant membranes (DRMs) of Plasmodium-infected erythrocytes were separated from detergent-soluble membranes (DSM) by sucrose gradient centrifugation, exploiting their resistance to solubilization in cold nonionic detergents. The flotation profile of the erythrocyte raft marker Flotillin I was used as gradient control. Light fractions 2–8 were collected and subjected to proteomic or lipid analyses. Proteins detected by LC-MS/MS spectrometry in at least 2 out of 3 biological replicates were quantified (Top3 method) and quantity values in each gradient fraction exploited to generate protein abundance profiles (PAPs). To generate an overall picture of DRM buoyancy features, PAPs were grouped by a hierarchical clustering (an example is shown in Panel B). For each gradient fraction, cluster abundance was calculated as the median of abundance values of each replicate (see Experimental Procedures). Major lipid classes and fatty acid components in DRM fractions were determined by HPTLC and GC, respectively (fraction 5 is shown in Panel C). Relative abundance of sphingomyelin (SM), phosphatidylserine (PS), phosphatidylcholine (PC), phosphatidylethanolamine (PE) and cholesterol (CH), as well as the one of saturated (SFA), monounsaturated (MUFA), and polyunsatured (PUFA) fatty acids was calculated as the percentage of total amount of lipids and fatty acids detected in each gradient fraction. Relationships between protein clusters (circles) in their lipid/fatty acid context (rings), were visualized as fraction-specific multi-component networks. As an example, fraction 5 network is shown in panel D.
To gain insights into conserved and stage-specific functions associated with these membrane assemblies, we compared DRM proteome and lipid components of sexual stages with those obtained from the asexual trophic stage (trophozoite).
IRBCs, collected from peripheral blood of mice were enriched by Nycodenz cushions (17) and hemoglobin was removed by osmotic lysis of the host RBC. Gametocytes or trophozoites were recovered by centrifugation and stored at −80 °C for successive analyses (3 × 108-parasite aliquots).
The thawed parasite pellet was treated with ice-cold Triton X-100 and the resulting cellular extract was subjected to sucrose gradient centrifugation (15). Triton-insoluble floating membranes (DRM-rafts) were collected in light density fractions 2–8, whereas detergent-soluble membranes (DSM) remained in the loading zone (heavy density fractions 9–12). The effectiveness of gradient separation was always verified by probing fractions with antibodies specific for Flotillin I, a raft marker highly enriched in RBCs, floating to fraction 4 in our gradient conditions. Major lipids and proteins, present in light fractions, were determined in three independent replicates.
HPTLC and GC were employed to define and quantify major lipids components, whereas DRM-associated proteins were quantified by label-free mass spectrometry. Normalized quantity values were then exploited to generate abundance profiles characteristic for each identified protein (PAPs). PAPs, obtained from the three experimental replicates, were then submitted to unsupervised hierarchical clustering to identify correlated protein groups. Intercluster relationships and protein-lipid cooccurrence were described by multicomponent networks.
Stage-specific Features of Gametocyte DRM-raft Proteome
Mixed mature male and female gametocyte-IRBCs were isolated from synchronous infections of the high gametocyte producer P. berghei line HP (MG-A sample). Gametocyte purification was performed using a buffer that strongly limits their maturation to gametes (20), even though, triggering of early events of gametogenesis cannot be excluded. MG-A sample was used for proteomics and lipid analyses together with trophozoite-IRBCs prepared from the isogenic, non-gametocyte producer line HPE (T-A sample).
Sucrose gradient centrifugation of gametocyte- or trophozoite-IRBCs, solubilized by cold Triton X-100, were run to determine either protein or lipid/fatty acid components. Proteins were extracted from each of the DRM-containing fractions 2–8 and identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Their relative abundance was calculated as the average of the three highest precursor peptide intensity values detected (24). We detected 257 gametocyte and 103 trophozoite proteins, with a minimum of two unique peptides, in at least two out of three replicates (supplemental Tables S6 and S7).
DRM proteome of trophozoites largely overlaps (88%) the one of sexual stages. It mainly includes proteins involved in basic cellular processes, such as ion transport, protein folding, trafficking and cell redox homeostasis, likely representing a common core of functions associated with parasite DRMs. In both parasite stages, we detected proteins implicated in nuclear activity, translation or endoplasmic reticulum (ER)-specific processes, suggesting that functionally distinct microdomains localize to various membrane compartments inside the cell.
Interestingly, gametocyte DRMs also recruit proteins implicated in processes specific for sexual stages, which naturally occur in mosquito gut. For example, we detected molecules, functionally characterized as involved in male gametogenesis, such as the calcium-dependent protein kinase 4 (CDPK4, PBANKA_061520) with a role in DNA replication and axoneme assembly, an armadillo repeat motif-containing protein (PBANKA_091740), component of the flagellum central apparatus and the actin II, with a role in male gamete development. We also detected components of the osmiophilic bodies (OBs), secretory organelles, implicated in gamete egress from the host erythrocyte, such as the female-specific PbG377 (20) (PBANKA_146300) or the MDV/PEG3 (26) (PBANKA_143220) and GEST (27) (PBANKA_131270), present both in male and female gametocytes.
Buoyancy Characteristics of Plasmodium Blood Stage DRMs
To evaluate flotation features of DRMs, we then generated PAPs using the abundance values of the identified proteins in each gradient fraction. In both sexual and asexual stages, about 80% of proteins display PAPs well superimposable in at least two out of three replicates (correlation values between 1 and 0.7, supplemental Fig. S1C). This conserved set of PAPs represents an internal control of robustness of the adopted method. It is conceivable that the limited PAP variability may be due to intrinsic features of certain microdomains. For this reason, we decided to include all the identified proteins in successive analyses.
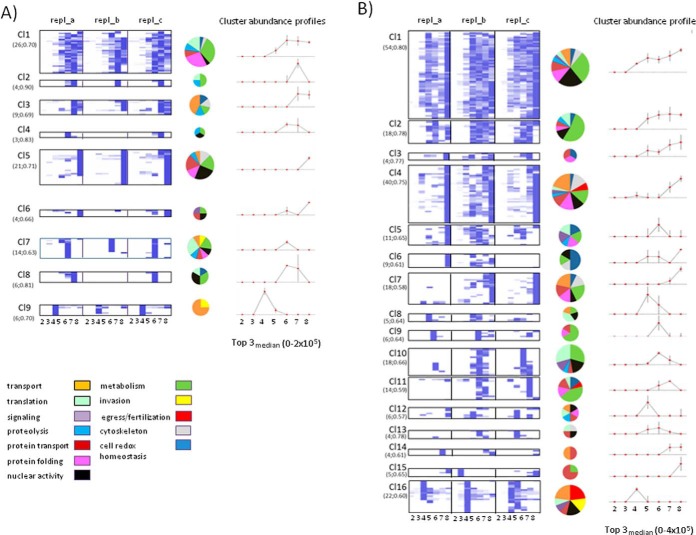
Using a hierarchical clustering method (25), we then analyzed global complexity of stage-related DRMs by grouping proteins based on similarity of their 21-point profiles obtained by merging PAPs of the three replicates. Functional categories were then assigned to each protein group (Fig. 2 and Supplemental Tables S6 and S7).
Fig. 2.
Hierarchical clustering of PAPs. Clustering of PAPs from three replicates of trophozoite (A) and mature gametocyte (B); DRMs are shown as heatmaps. Each cluster includes PAPs with Pearson's correlation coefficient higher than 0.5. The number of proteins and the minimum correlation coefficient in each cluster are reported in parentheses. Cluster abundance profiles were calculated as the median value of the replicate Top3-medians. In the case of trophozoites this value ranges from 0 to 2 × 105, whereas in gametocytes the highest value is 4 × 105. Vertical lines represent the standard error of the mean (S.E.). Results of the functional classification of proteins in each cluster are shown as pie charts the size of which are proportional to cluster dimensions.
Variable PAPs may be suggestive of proteins recruited in membranes with a less-ordered structural organization or proteins weakly binding DRM components in our experimental conditions. For example, four out of five proteins of gametocyte cluster 15 are known components of the ER membrane (Fig. 2 and supplemental Table S6). They float to light fraction 3 in replicate 2, whereas the remaining were confined to the denser fraction 8 in replicates 1 and 3. At variance, PAPs grouped in the neighbor cluster 16, displaying a floating-peak centered at light fraction 4 (where host Flotillin resides), are very well conserved between replicates. This cluster includes parasite aquaglyceroporin and the band 7-related protein, expressed in both asexual and sexual stages, as well as gametocyte-specific factors, such as the female p47 (PBANKA_135970) and the male p48/45 (PBANKA_135960), with a key role in gamete fertilization or the female-specific OB-resident protein PbG377 (20). Notably, the OB components MDV/PEG3 (26) and GEST (27) peak to fraction 4 but also to the denser fractions 7 and/or 8 (cluster 4), suggesting that they are recruited in membrane contexts different from that of PbG377. From a functional point of view, g377 null mutants display only a slight delay in female egress (20), whereas deletion of either mdv1 (26) or gest (27) causes a dramatic impairment in the egress of both male and female gametes that remain trapped inside the host cell.
Interestingly, eight proteins identified both in gametocytes and trophozoites display PAPs highly conserved in both stages (see protein Top list in supplemental Table S5). They include two proteins also identified in DRM proteome of P. falciparum trophozoites (16): the plasmepsin 4, an aspartic protease that localizes in a lysosomal compartment, the food vacuole, with a role in parasite virulence (28) and an ATP-binding cassette (ABC) family member, the multidrug resistance protein 1 (MDR1) (29). We also detected the intra-erythrocytic P. berghei-induced structures 1 (IBIS1) shown to reside in membrane structures exported to the cytoplasm of the host erythrocyte and to the parasitophorous vacuole of liver stages (30). This suggests that in P. berghei, like in P. falciparum (16), DRMs are implicated in the host erythrocyte remodeling.
DRM Environment: Cooccurrence of Proteins and Lipids
Functional analysis indicated that molecules implicated in several and unrelated processes, such as secretion, transport, membrane fusion events or nuclear activities, are compartmentalized in intracellular microdomains with different flotation capacity. Lipid composition of DRMs likely contributes to these distinct chemo-physical properties. To investigate this aspect, lipids were extracted from DRM-containing gradient fractions of both trophozoites and gametocytes and analyzed by HPTLC. As shown in Fig. 3, SM, PC, PE, and PS were the most abundant phospholipids detected, whereas CH was the only neutral lipid revealed by HPTLC. We could quantify lipid content of fractions 3–8 of sexual gametocytes and fractions 4–8 of trophozoites (because of the lower amount of lipids recovered from the asexual parasites).
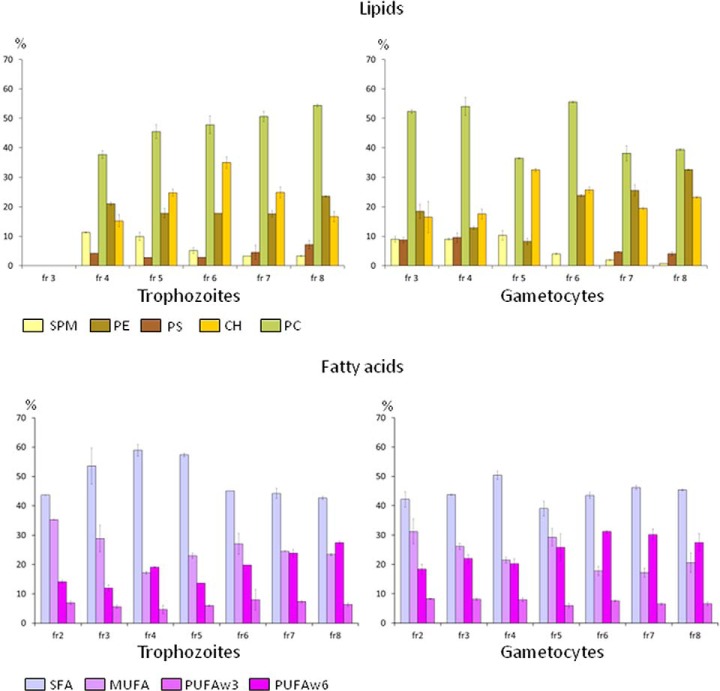
Fig. 3.
Major lipids and fatty acids in DRM containing gradient fractions. Relative abundance of lipids and fatty acid classes in each gradient fraction were calculated by dividing their abundance values by total amount of lipids and fatty acids of the same fraction. Two technical replicates were available for fatty acids and three for lipids. Vertical bars represent the mean standard errors. In the case of lipids the mean values were calculated discarding the abundance value with the highest standard deviation.
PC was the most represented lipid component in each fraction of both asexual and sexual stages. Relative abundance of the SM and CH, lipids known to confer liquid-ordered nature to membrane microdomains, varies along the gradient. In asexual DRMs, the highest concentration of CH was observed in fractions 5 and 6, whereas SM was decreasing from fraction 4 to fraction 8. This reduction of SM level (Fig. 3) is consistent with the acidic profile of DRM fractions, being SM a phospholipid characterized by a high content of saturated fatty acids. We observed, in fact, a marked decrease of saturated fatty acids (SFA) in fractions 6–8, balanced by an increase of PUFA. The lower level of SM accompanied by a higher content in PUFA may justify a decrease in resistance to detergent solubilization of membranes recovered in fractions 7 and 8. At variance, we observed a peak level of SM in fraction 5, and a peak level of CH in fractions 4 and 5 of gametocyte DRMs. Peculiar was also the trend of PS along the gradient: it was slightly reduced in fractions 5 and 6 of trophozoite DRMs, whereas being completely absent in the corresponding DRM fractions of gametocytes, although the total amount of lipids recovered from sexual stages was significantly higher than that of trophozoites (176.67 ± 28.32 versus 118.35 ± 4.94 p = 0.02). Collectively these data indicate that lipid composition of DRMs display differences between asexual and sexual parasites, likely reflecting the remarkable specificity of subcellular structures that characterize these parasite stages as well as the biological processes they are involved in.
Construction of Multicomponent DRM Networks
As seen above, quantitative analysis of DRM-associated molecules revealed an uneven distribution and abundance of both proteins and lipids along the gradient. This suggests the presence of distinct and heterogeneous membrane assemblies.
To investigate the relationships between DRM components, we integrated protein clusters into stage-specific networks (depicted as graphs in supplemental Fig. S2). Network centrality measures (betweenness, closeness and degree), identified few clusters crucial for network architecture, the gametocyte clusters g1, g4, g7, g11, g12 and the trophozoite clusters t1, t5, t7.
To compare relationships between DRM clusters at the different gradient layers, we then derived fraction-specific networks (Fig. 4), which take into account relative abundance of cluster components in each of the collected fractions. Functional categories represented were also reported as pie charts. Relative abundance of the major lipid classes and their fatty acid components were included as half ring pies. Collectively, fraction-specific networks of gametocytes are more populated than the trophozoite ones and nodes much more interconnected, suggesting a higher structural and functional complexity of membrane microdomains of sexual stages.
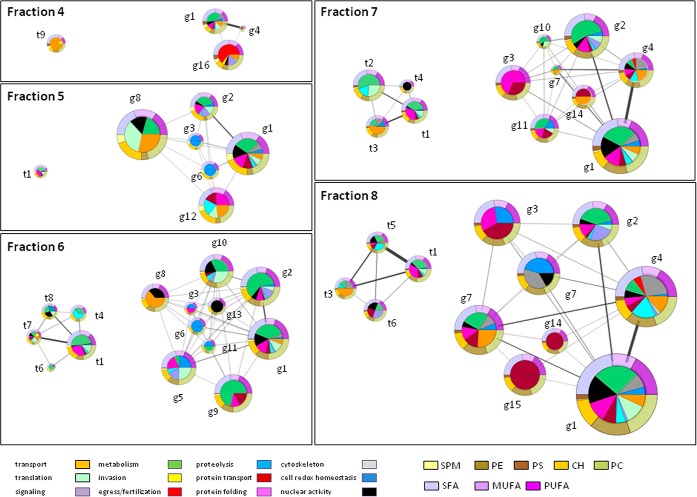
Fig. 4.
Protein/lipid co-occurrence networks. Quantitative data on protein clusters, lipids and fatty acids in DRM fractions were integrated into multi-component networks. The size of nodes (protein clusters) is proportional to their abundance in the considered gradient fraction. Each functional category of the pie charts represents the median of its abundance in the three replicates. The width of edges are proportional to the number of correlated PAPs (correlation coefficient higher than 0.5) between clusters. Each cluster is surrounded by rings that represent the lipid/fatty acid context. Relative abundances of fatty acid (SFA, MUFA and PUFA) and lipid (SM, PS, PC, PE, and CH) classes are shown in the upper and lower half rings respectively.
Dynamics of DRM Components at the Gametocyte-Gamete Transition
Cluster profiles indicate that parasite DRMs are characterized by different floating properties. Some protein groups display a single sharp peak in light gradient fractions, such as the case of g8, g9 or g16, suggesting that they stably reside in membrane microdomains. At difference, molecules floating both to light and heavy fractions, at the DSM boundary, as the case of cluster g1, may be dynamically recruited into DRMs. This cluster occupies a central position in gametocyte network (supplemental Fig. S2) and includes molecules involved in various functions, such as nuclear activity, protein folding/trafficking and proteolysis. It is conceivable that temporal and spatial compartmentalization of these molecules/pathways may be required to guarantee a prompt and effective response to the gametogenesis stimuli.
We then decided to analyze several candidates assigned to cluster 1, to evaluate their expression pattern and subcellular localization at the gametocytes-gametes transition. All selected proteins display similar floating properties, with a peak at the fraction 5 (see supplemental Table S6). This fraction contains the highest relative amount of CH and SM (Fig. 3 and 4), the abundance of which is characteristic of the liquid-ordered membrane microdomains.
Polyclonal rabbit immune sera were available for the nuclear protein NAP/SET (21) and the multifunctional protein 14-3-3 (22), detected both in asexual and sexual stage DRMs. Mouse immune sera were prepared against the conserved Plasmodium protein of unknown function, which contains a tetratricopeptide repeat (TRP) region, known to mediate protein-protein interactions (PBANKA_060430, from now on named PbTRP1), the putative ATP-dependent RNA helicase UAP56 (PBANKA_030680) and the putative deoxyribose-phosphate aldolase (PBANKA_050580, from now on named PbDPA), detected only in gametocyte DRMs.
To further validate the proposed quantitative proteomic approach, we compared PAPs of the selected proteins, with the corresponding antibody signals obtained by probing gradient fractions 2–8 of the same material used for proteomic analysis (gametocyte preparation MG-A). As shown in Fig. 5A and 5B, the intensities of WB signals along the gradient are well superimposable to PAPs. Interestingly, NAP/SET and 14-3-3 detected in both parasite stages, float to the light fractions in gametocytes and to the heavier fractions (7 and 8) in asexual trophozoites (Fig. 5A), suggesting that they are recruited to different membrane contexts. In both stages Flotillin, used as a control, peaks at fraction 4 (Fig. 5C).
Reproducibility of PAPs obtained from gametocyte sample MG-A was confirmed by probing DRM fractions 2 to 8 of two additional mature gametocyte preparations (MC-B and C) with antibodies specific for Flotillin and for the parasite proteins PbG377, NAP/SET and 14-3-3 (supplemental Fig. S3).
When DRM fractions of two additional biological replicates of trophozoites (samples T-B and C) were analyzed by WB, 14-3-3 and NAP/SET were detected in the heavy fraction 8 (supplemental Fig. S3), thus confirming the consistency of buoyancy differences between developmental stages observed for these proteins.
Overall, these results indicate that the proposed approach for membrane microdomain analysis may represent a useful, reproducible and robust tool to characterize DRM dynamics at the whole cell level.
To define membrane partitioning of the selected molecules at the gametocyte/gamete transition, Triton-soluble and -insoluble extracts were prepared from a new sample of nonactivated mature gametocytes and the same cells activated 15 min (see Methods section). After centrifugation DRMs were recovered in the pellet, whereas DSMs remained in the supernatant. The effectiveness of the procedure was verified by probing the extracts with antibodies against the DRM-resident proteins PbG377 and Flotillin as well as the integral membrane protein SEP1, not detected in DRM proteome. As expected (Fig. 5D), both PbG377 and Flotillin were detected in Triton-insoluble extracts, whereas SEP1 was recovered in the soluble fractions of both nonactivated and activated gametocytes. The presence of multiple PbG377-specific bands, likely because of post-translational cleavage, was already described (20).
14-3-3 and PbTRP1 were detected in the soluble and insoluble fractions of both gametocyte and developing gametes, whereas the nuclear protein NAP/SET partitions into DRMs only in nonactivated gametocytes. Notably, PbDPA and RNA helicase-specific signals strongly increase in the Triton-insoluble fraction of activated gametocytes, suggesting that a subset of proteins may be recruited into DRMs during gametogenesis. To further support this observation, we prepared Triton-soluble and -insoluble extracts from two preparations of young gametocytes (YG-A and B), not yet competent to undergo gametogenesis in vitro, and probed them with antibodies specific for the parasite proteins under study. As already observed for mature gametocyte sample (Fig. 5D), in both preparations of young gametocytes (supplemental Fig. S4), PbG377 partitions in Triton-insoluble fraction, NAP/SET and 14-3-3 in soluble and insoluble fractions, whereas SEP1 localizes to Triton-soluble membranes. At difference, PbDPA, RNA-helicase and PbTRP1 were only detected in Triton-soluble extracts, thus supporting the idea that recruitment of these proteins into DRMs may occur in early stages of gametogenesis.
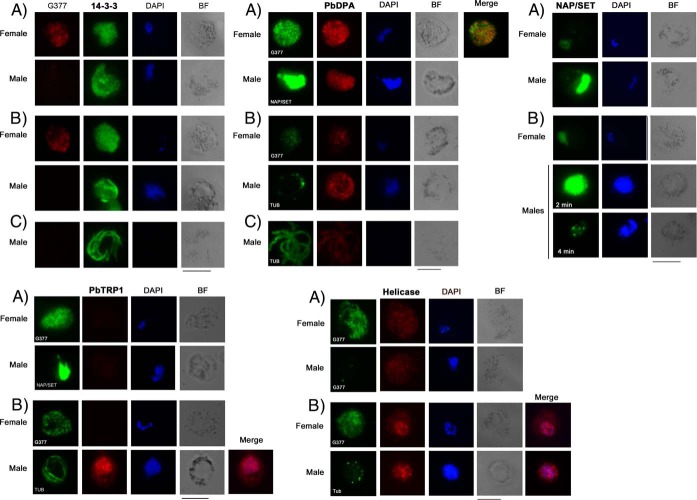
Subcellular localization of these molecules was inspected by IFA on blood smears of nonactivated and activated gametocytes. PbG377 was used as a female-specific marker to distinguish female gametocytes prior and after activation. NAP/SET was used to discriminate male gametocytes in which this nuclear protein is highly abundant (21). Activated male gametocytes and mature male gametes were identified using commercial antibodies against alpha-tubulin, a component of flagella. Representative images are shown in Fig. 6.
Fig. 6.
Immunolocalization of DRM-associated proteins in nonactivated and activated P. berghei gametocytes. To define subcellular localization and dynamics of proteins associated with gametocyte DRMs, antibodies specific for NAP/SET, 14-3-3, PbTRP1, the putative ATP-dependent RNA helicase UAP56 and PbDPA, were used in IFA on fixed nonactivated (A), activated gametocytes (B) and mature male gametes (C). PbG377 and NAP/SET were used as female and male specific markers, respectively. Antibodies against the alpha-tubulin were used to distinguish both the activated male gametocytes and the mature male gametes. Nuclei were stained with DAPI. Activated male gametocytes undergoing DNA replication display bright DAPI fluorescence. PbTRP1 localizes to the nucleus of activated males whereas the RNA helicase UAP56 is detected in the nucleus of both males and females upon activation. NAP/SET is abundant in the nucleus of male gametocytes; whereas the specific fluorescence drops in activated males. 14-3-3 and PbDPA localize to the cytoplasm of nonactivated and activated gametocytes with a dotted pattern. They are also detected in mature male gametes. PbDPA does not colocalizes with the OB-resident PbG377. Bars correspond to 5 μm.
The multifunctional protein 14-3-3 was detected in dot-like structures spread in the cytoplasm of both male and female gametocytes, as well as in activated gametocytes. During male gamete maturation it localizes to the cell periphery, moving then into the forming gametes. These observations are consistent with WB analysis where 14-3-3-specific signal was detected in the insoluble fractions of both gametocytes and developing gametes, (Fig. 5D), suggesting that DRM-associated 14-3-3, likely exerts a role throughout sexual development.
PbTRP1-specific signal was absent in female gametocytes and very faint in males. TRP1 fluorescence greatly increases in the cytoplasm of 2-min activated males and is clearly visible as dotted fluorescence in the nuclei of male gametocytes 10 min upon activation. So far, no functional role has been attributed to PbTRP1, even though its subcellular localization and timing of expression suggests a nuclear-related function. According with PlasmoDB, PbTRP1 contains homology regions with the synaptonemal complex protein 1 (SCP1), and the histone-binding protein N1/N2.
Recruitment of the putative RNA helicase UAP56 into DRMs increases in developing gametes (Fig. 5D) and is accompanied by changes in subcellular localization. In gametocytes, we observed a cytoplasmic punctuate pattern, whereas a few minutes after activation the protein is recruited in the nuclear compartment of both males and females.
As stated above, the nuclear protein NAP/SET (21) is very abundant in male gametocytes and its DRM association occurs only at the gametocyte stage (Fig. 5D). A high protein level is still detected 1–2 min after activation but specific fluorescence drops within 8–10 min, remaining confined in few focal points.
Specific antibodies identify PbDPA to dotted structures in the cytoplasm of both male and female gametocytes. These structures are distinct from the OB population, already characterized in P. berghei for the presence of G377 (20), MDV1 (26) and GEST (27). During gametogenesis, DPA-specific signal decreases in females, whereas it is abundant in activated male gametocytes and in mature gametes.
DISCUSSION
The importance of membrane microdomains enriched in CH and sphingolipids, also referred to as lipid rafts, has been highlighted in recent years (2). However, attempting to capture their complexity at the whole cell level in the physiological as well as in the pathological conditions is challenging because of the dynamic nature of these assemblies. Raft clustering, in fact, rapidly occurs under different stimuli, leading to recruitment and/or stabilization of protein components (31–33). Visualization in living cells of CH-rich microdomains, laterally segregated from the rest of membrane, is limited, whereas biochemical means to assess raft affinity rely on indirect approaches based, for example, on DRM extraction by detergents. This ruling method has been defined as a “working description” of lipid rafts (34). Insolubility in nonionic detergents was, in fact, observed in lipid bilayers, which exist in physical states where lipid packing is tight (35).
Most proteomic approaches identify proteins associated with membrane domains that are visible as a major opaque band in the upper part of the gradient. However, these studies lack information on membrane assemblies, which may populate other density layers.
In this study we developed and validated a method based on label-free quantitative proteomics, lipid analysis and bioinformatics that allows for capturing overall complexity and flotation properties of membrane microdomains at a whole-cell level.
This approach was applied to characterize stage-specific features of DRMs isolated from sexual blood stages of the rodent malaria parasite P. berghei after Triton X-100 treatment and separation by sucrose gradient centrifugation.
Identified proteins were first associated with a characteristic buoyancy profile (PAP) and then grouped based on floating similarities in each replicate. Interestingly, comparative analysis of PAPs showed that 80% of buoyancy profiles were conserved between replicates, indicating that the proposed procedure is robust. A lower reproducibility of buoyancy profiles, observed for a certain number of proteins (about 20% of the DRM proteome), was expected, because of variability inherent to the analysis of these microdomains (36). This variability may be in part explained by intrinsic features of local membrane structures, displaying, for instance, a lower liquid-ordered state, more prone to detergent solubilization or by weak protein-protein and/or protein-lipid interactions.
The lifetime of membrane microdomains depends on their size and factors that may stabilize or destabilize them to cope with cellular processes such as signaling, immune response and cellular trafficking. DRM analysis would be particularly useful in assessing raft potential when able to capture differences in DRM composition triggered by relevant biological events. The ability to highlight buoyancy features of the DRM-associated proteins (PAP analysis) as well as their lipid context, may be valuable to assess proteins going toward a stabilization into or exclusion from functional membrane microdomains after exogenous stimuli or perturbation.
Previous studies (37–39) indicated that membrane microdomains of Plasmodium asexual blood stages have a key role in parasite virulence and pathogenesis as they participate to biological processes, such as protein sorting/trafficking, host cell invasion and membrane remodeling. Here we focused on sexual stage DRMs, not investigated previously. Notably, we identified stage-specific proteins implicated in gamete egress from the host erythrocyte and fertilization, crucial events that occur in mosquito midgut after an infected blood meal. This finding suggests that membrane microdomains may be targeted to block both parasite invasion and transmission.
Further, functions associated with DRMs include molecules residing in the ER, the mitochondrion or secretory organelles. The presence of DRMs in endocellular membrane compartments is not surprising, because evidence is accumulating that suggests a role for membrane microdomains in establishing networks of contact sites (MCS) between organelles, thus ensuring tight coordination of cellular activities, for review see (40). Membranes tethered near one another favor transient or durable exchanges between segregated compartments. Among the others, we detected EMC3, component of the endoplasmic reticulum membrane protein complex (EMC), involved in lipid exchange between ER and mitochondria (41). IEMC3 profile has been assigned to cluster 16 where several DRM-raft resident proteins locate. They include Alba 1 and Alba 4, implicated in translational repression of female gametocytes (42, 43) and the calcium-dependent protein kinase CDPK1, localized to the parasite periphery (44), with multiple key functions in sexual stage development including egress of male gametes and the translational activation of a subset of repressed mRNAs in the ookinete (44).
Stage-specific DRM networks indicated that protein clusters of the highly specialized sexual stages are more interconnected than those of the asexual trophic stages; cluster g1 occupies a central position in the gametocyte network. Floating properties of its components suggest for them a transient association with membrane microdomains during sexual stage development. We showed that nuclear proteins in cluster g1 are dynamically regulated at the gametocyte to gamete transition. Among them, the putative RNA helicase UAP56 moves from the cytoplasm to the nucleus upon gametocyte activation, whereas the conserved protein of unknown function PbTRP1 is detected exclusively in the nuclei of activated males. At difference, the chromatin assembly factor NAP/SET (45, 46), shown to accumulate in the nuclei of male gametocytes (21), is rapidly degraded during male gamete maturation where it is detected only in few focal points.
Recent data support the notion that specific functions segregate in nuclear membrane microdomains (47, 48), even though their organization is still unknown. Cellular nuclei contain high levels of PC and SM, which are partially linked with CH and proteins to form lipid microdomains. These may act as attachment sites for active chromatin during cell proliferation (49) or as a platform for the transcription process (50).
In conclusion, this study reveals unexplored pathways associated with DRMs and highlights unexpected heterogeneity and complexity in membrane microdomains floating at different density layers. The proposed approach could be widely applied to explore structural organization and dynamics of membrane microdomains, using different extraction procedures, in various cellular contexts and perturbations, under physiological or pathological conditions, thus opening new perspectives for the investigation of membrane organization and dynamics.
DATA AVAILABILITY
Raw mass spectrometry data has been deposited in MassIVE (http://massive.ucsd.edu/ProteoSAFe/datasets.jsp) with MassIVE ID MSV000080108.
Supplementary Material
Acknowledgments
We thanks Marco Crescenzi for discussion and critical reading of the manuscript.
Footnotes
Author contributions: E.P. and M.P. designed research; C.R., G.S., C.B., F.G., C.C., A.O., T.P., S.M., and L.P. performed research; F.F., E.P., and M.P. analyzed data; M.P. wrote the paper; F.F. designed and performed proteomic experiments; A.S. and C.F. designed and performed fatty acid analysis; A.D. designed and performed lipid analysis.
* This work was supported by the Italian FLAGSHIP “InterOmics” project (PB.P05) funded by MIUR and coordinated by the CNR and by the project: “Toward multi-stage drugs to fight poverty related and neglected parasitic diseases: synthetic and natural compounds directed against Leishmania, Plasmodium and Schistosoma life stages and assessment of their mechanisms of action”, funded by the italian national research program PRIN-2015, Prot. 20154JRJPP.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CH
- cholesterol
- DAPI
- 4′,6-diamidino-2-phenylindole
- DRM
- detergent resistant membrane
- ER
- endoplasmic reticulum
- FAME
- fatty acid methyl ester
- GC
- gas chromatography
- GST
- glutathione S-transferase
- HPTLC
- high performance thin layer chromatography
- IFA
- immunofluorescence assay
- IRBC
- infected red blood cell
- OB
- osmiophilic body
- PAP
- protein abundance profile
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- PUFA
- polyunsatured fatty acids
- SFA
- saturated fatty acids
- SM
- sphingomyelin
- WB
- Western blot.
REFERENCES
- 1. Simons K., and Gerl M. J. (2010) Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11, 688–699 [DOI] [PubMed] [Google Scholar]
- 2. Simons K., and Sampaio J. L. (2011) Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3 a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mollinedo F., and Gajate C. (2015) Lipid rafts as major platforms for signaling regulation in cancer. Adv. Biol. Regul 57, 130–146 [DOI] [PubMed] [Google Scholar]
- 4. Sonnino S., Aureli M., Mauri L., Ciampa M. G., and Prinetti A. (2015) Membrane lipid domains in the nervous system. Front. Biosci. 20, 280–302 [DOI] [PubMed] [Google Scholar]
- 5. Fridolfsson H. N., and Patel H. H. (2013) Caveolin and caveolae in age associated cardiovascular disease. J. Geriatr. Cardiol. 10, 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldston A. M., Powell R. R., and Temesvari L. A. (2012) Sink or swim: lipid rafts in parasite pathogenesis. Trends Parasitol. 28, 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vieira F. S., Corrêa G., Einicker-Lamas M., and Coutinho-Silva R. (2010) Host-cell lipid rafts: a safe door for micro-organisms? Biol. Cell 102, 391–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hawkes D. J., and Mak J. (2006) Lipid membrane, a novel target for viral and bacterial pathogens. Curr. Drug Targets 7, 1615–1621 [DOI] [PubMed] [Google Scholar]
- 9. Yaqoob P., and Shaikh S. R. (2010) The nutritional and clinical significance of lipid rafts. Curr Opin. Clin. Nutr. Metab. Care 13, 156–166 [DOI] [PubMed] [Google Scholar]
- 10. Verma S. P. (2009) HIV: a raft-targeting approach for prevention and therapy using plant-derived compounds. Curr. Drug Targets 10, 51–59 [DOI] [PubMed] [Google Scholar]
- 11. Murphy S. C., Hiller NL, Harrison T., Lomasney J. W., Mohandas N., and Haldar K. (2006) Lipid rafts and malaria parasite infection of erythrocytes. Mol. Membr. Biol. 23, 81–88 [DOI] [PubMed] [Google Scholar]
- 12. Koshino I., and Takakuwa Y. (2009) Disruption of lipid rafts by lidocaine inhibits erythrocyte invasion by Plasmodium falciparum. Exp. Parasitol. 123, 381–383 [DOI] [PubMed] [Google Scholar]
- 13. Samuel B. U., Mohandas N., Harrison T., McManus H., Rosse W., Reid M., and Haldar K. (2001) The role of cholesterol and glycosylphosphatidylinositol-anchored proteins of erythrocyte rafts in regulating raft protein content and malarial infection. J. Biol. Chem. 276, 29319–29329 [DOI] [PubMed] [Google Scholar]
- 14. Sanders P. R., Gilson P. R., Cantin G. T., Greenbaum D. C., Nebl T., Carucci D. J., McConville M. J., Schofield L., Hodder A. N., Yates J. R. 3rd, and Crabb B. S. (2005) Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J. Biol. Chem. 280, 40169–40176 [DOI] [PubMed] [Google Scholar]
- 15. Di Girolamo F., Raggi C., Birago C., Pizzi E., Lalle M., Picci L., Pace T., Bachi A., de Jong J., Janse C. J., Waters A. P., Sargiacomo M., and Ponzi M. (2008) Plasmodium lipid rafts contain proteins implicated in vesicular trafficking and signalling as well as members of the PIR superfamily, potentially implicated in host immune system interactions. Proteomics 8, 2500–2513 [DOI] [PubMed] [Google Scholar]
- 16. Yam X. Y., Fratini F., Di Girolamo F., Raggi C., Sargiacomo M., Bachi A., Berry L., Fall G., Currà C., Pizzi E., Breton C. B., and Ponzi M. (2013) Proteomic analysis of detergent-resistant membrane microdomains in trophozoite blood stage of the human malaria parasite Plasmodium falciparum. Mol. Cell. Proteomics 12, 3948–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Janse C. J., and Waters A. P. (1995) Plasmodium berghei: The application of cultivation and purification techniques to molecular studies of malaria parasites. Parasitol. Today 11, 138–143 [DOI] [PubMed] [Google Scholar]
- 18. Wessel D., and Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 19. Folch J., Lees M., and Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 20. Olivieri A., Bertuccini L., Deligianni E., Franke-Fayard B., Currà C., Siden-Kiamos I., Hanssen E., Grasso F., Superti F., Pace T., Fratini F., Janse C. J., and Ponzi M. (2015) Distinct properties of the egress-related osmiophilic bodies in male and female gametocytes of the rodent malaria parasite Plasmodium berghei. Cell Microbiol. 17, 355–368 [DOI] [PubMed] [Google Scholar]
- 21. Pace T., Olivieri A., Sanchez M., Albanesi V., Picci L., Siden Kiamos I., Janse C. J., Waters A. P., Pizzi E., and Ponzi M. (2006) Set regulation in asexual and sexual Plasmodium parasites reveals a novel mechanism of stage-specific expression. Mol. Microbiol. 60, 870–882 [DOI] [PubMed] [Google Scholar]
- 22. Lalle M., Currà C., Ciccarone F., Pace T., Cecchetti S., Fantozzi L., Ay B., Breton C. B., and Ponzi M. (2011) Dematin, a component of the erythrocyte membrane skeleton, is internalized by the malaria parasite and associates with Plasmodium 14-3-3. J. Biol. Chem. 286, 1227–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Birago C., Albanesi V., Silvestrini F., Picci L., Pizzi E., Alano P., Pace T., and Ponzi M. (2003) A gene-family encoding small exported proteins is conserved across Plasmodium genus. Mol. Biochem. Parasitol. 126, 209–218 [DOI] [PubMed] [Google Scholar]
- 24. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., and Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 25. de Hoon M. J., Imoto S., Nolan J., and Miyano S. (2004) Open source clustering software. Bioinformatics 20, 1453–1464 [DOI] [PubMed] [Google Scholar]
- 26. Ponzi M., Sidén-Kiamos I., Bertuccini L., Currà C., Kroeze H., Camarda G., Pace T., Franke-Fayard B., Laurentino E. C., Louis C., Waters A. P., Janse C. J., and Alano P. (2009) Egress of Plasmodium berghei gametes from their host erythrocyte is mediated by the MDV-1/PEG3 protein. Cell Microbiol. 11, 1272–1288 [DOI] [PubMed] [Google Scholar]
- 27. Talman A. M., Lacroix C., Marques S. R., Blagborough A. M., Carzaniga R., Menard R., and Sinden R. E. (2011) PbGEST mediates malaria transmission to both mosquito and vertebrate host. Mol. Microbiol. 82, 462–474 [DOI] [PubMed] [Google Scholar]
- 28. Spaccapelo R., Janse C. J., Caterbi S., Franke-Fayard B., Bonilla J. A., Syphard L. M., Di Cristina M., Dottorini T., Savarino A., Cassone A., Bistoni F., Waters A. P., Dame J. B., and Crisanti A. (2010) Plasmepsin 4-deficient Plasmodium berghei are virulence attenuated and induce protective immunity against experimental malaria. Am. J. Pathol. 176, 205–217, January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koenderink J. B., Kavishe R. A., Rijpma S. R., and Russel F. G. (2010) The ABCs of multidrug resistance in malaria. Trends Parasitol. 26, 440–446, September. [DOI] [PubMed] [Google Scholar]
- 30. Ingmundson A., Nahar C., Brinkmann V., Lehmann M. J., and Matuschewski K. (2012) The exported Plasmodium berghei protein IBIS1 delineates membranous structures in infected red blood cells. Mol. Microbiol. 83, 1229–1243, March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schuck S., and Simons K. (2004) Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J. Cell Sci. 117, 5955–5964 [DOI] [PubMed] [Google Scholar]
- 32. Simons K., and Vaz W. L. (2004) Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct 33, 269–295 [DOI] [PubMed] [Google Scholar]
- 33. Meder D., and Simons K. (2006) Lipid rafts, caveolae, and membrane traffic. In Lipid Rafts and Caveolae. From Membrane Biophysics to Cell Biol. (ed. Fielding C. J.) 1.1–1.23 (Wiley-VGH Verlag GmbH & Co., Wienheim, Germany [Google Scholar]
- 34. Brown D. A., and Rose K. J. (1992) Sorting of GPI-anchored proteins to glycolipid enriched membrane subdomains during transport to the apical cell surface. Cell 68, 533–544 [DOI] [PubMed] [Google Scholar]
- 35. London E., and Brown D. A. (2000) Insolubility of lipids in triton X-100, physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts). Biochim. Biophys. Acta 1508, 182–195 [DOI] [PubMed] [Google Scholar]
- 36. Lingwood D., and Simons K. (2007) Detergent resistance as a tool in membrane research. Nat. Protoc. 2, 2159–2165 [DOI] [PubMed] [Google Scholar]
- 37. Koshino I., and Takakuwa Y. (2009) Disruption of lipid rafts by lidocaine inhibits erythrocyte invasion by Plasmodium falciparum. Exp. Parasitol. 123, 381–383 [DOI] [PubMed] [Google Scholar]
- 38. Frankland S., Adisa A., Horrocks P., Taraschi T. F., Schneider T., Elliott S. R., Rogerson S. J., Knuepfer E., Cowman A. F., Newbold C. I., and Tilley L. (2006) Delivery of the malaria virulence protein PfEMP1 to the erythrocyte surface requires cholesterol-rich domains. Eukaryot Cell. 5, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murphy S. C., Hiller N. L., Harrison T., Lomasney J. W., Mohandas N., and Haldar K. (2006) Lipid rafts and malaria parasite infection of erythrocytes. Mol. Membr. Biol. 23, 81–88 [DOI] [PubMed] [Google Scholar]
- 40. Helle S. C. (2013) Organization and function of membrane contact sites. Biochim. Biophys. Acta 1833, 2526–2541 [DOI] [PubMed] [Google Scholar]
- 41. Lahiri S., Chao J. T., Tavassoli S., Wong A. K., Choudhary V., Young B. P., Loewen C. J., and Prinz W. A. (2014) A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLos Biol. 12, e1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mair G. R., Braks J. A., Garver L. S., Wiegant J. C., Hall N., Dirks R. W., Khan S. M., Dimopoulos G., Janse C. J., and Waters A. P. (2006) Regulation of sexual development of Plasmodium by translational repression. Science 313, 667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mair G. R., Lasonder E., Garver L. S., Franke-Fayard B. M., Carret C. K., Wiegant J. C., Dirks R. W., Dimopoulos G., Janse C. J., and Waters A. P. (2010) Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 6, e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sebastian S., Brochet M., Collins M. O., Schwach F., Jones M. L., Goulding D., Rayner J. C., Choudhary J. S., and Billker O. (2012) A Plasmodium calcium-dependent protein kinase controls zygote development and transmission by translationally activating repressed mRNAs. Cell Host Microbe 12, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gill J., Yogavel M., Kumar A., Belrhali H., Jain S. K., Rug M., Brown M., Maier A. G., and Sharma A. (2009) Crystal structure of malaria parasite nucleosome assembly protein: distinct modes of protein localization and histone recognition. J. Biol. Chem. 284, 10076–10087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gill J., Kumar A., Yogavel M., Belrhali H., Jain S. K., Rug M., Brown M., Maier A. G., and Sharma A. (2010) Structure, localization and histone binding properties of nuclear-associated nucleosome assembly protein from Plasmodium falciparum. Malar J. 9, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Graumann K., Irons S. L., Runions J., and Evans D. E. (2007) Retention and mobility of the mammalian lamin B receptor in the plant nuclear envelope. Biol. Cell 99, 553–562 [DOI] [PubMed] [Google Scholar]
- 48. Manfiolli A., Maragno A. L., Baqui M. M., Yokoo S., Teixeira F. R., Oliveira E. B., and Gomes M. D. (2008) FBXO25-associated nuclear domains: a novel subnuclear structure. Mol. Biol. Cell 19, 1848–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Albi E., Lazzarini A., Lazzarini R., Floridi A., Damaskopoulou E., Curcio F., and Cataldi S. (2013) Nuclear lipid microdomain as place of interaction between sphingomyelin and DNA during liver regeneration. Int. J. Mol. Sci. 14, 6529–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cascianelli G., Villani M., Tosti M., Marini F., Bartoccini E., Magni M. V., and Albi E. (2008) Lipid microdomains in cell nucleus. Mol. Biol. Cell 19, 5289–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry data has been deposited in MassIVE (http://massive.ucsd.edu/ProteoSAFe/datasets.jsp) with MassIVE ID MSV000080108.