Abstract
The influence of the interaction(s) between the medicinal plant Echinacea purpurea (L.) Moench and its endophytic communities on the production of alkamides is investigated. To mimic the in vivo conditions, we have set up an infection model of axenic in vitro E. purpurea plants inoculated with a pool of bacterial strains isolated from the E. purpurea stems and leaves. Here we show different alkamide levels between control (not-inoculated) and inoculated plants, suggesting that the alkamide biosynthesis may be modulated by the bacterial infection. Then, we have analysed the branched-chain amino acids (BCCA) decarboxylase gene (GenBank Accession #LT593930; the enzymatic source for the amine moiety formation of the alkamides) expression patterns. The expression profile shows a higher expression level in the inoculated E. purpurea tissues than in the control ones. These results suggest that the plant-endophyte interaction can influence plant secondary metabolism affecting the therapeutic properties of E. purpurea.
Introduction
Echinacea purpurea (L.) Moench (Asteraceae) is a medicinal plant with immune-modulatory and anti-inflammatory properties, whose roots and aerial parts are frequently used in Europe and North America for the preparation of therapeutic extracts for common cold1. It is rich in various phytochemicals including caffeic acid derivatives, alkamides and polysaccharides2. The concentrations of these bioactive compounds are species-specific and they may vary due to several factors such as plant material, cultivation techniques, plant tissue treatment, extraction methods and phytosanitary status3. Recently, the attention has been focused on the plant microbiota and its role in the production of secondary metabolites4,5. Many studies aim to investigate the influence of endophytic fungi on the production of plant bioactive molecules6,7 but the interest for the bacterial endophytes is considerably increasing8. Genomics and proteomics approaches have been applied to deepen the understanding of the plant-endophyte interaction9. Differential protein accumulations have been revealed in the proteome of in vitro-grown Zea mays 10 and Chinese hybrid poplar clone 74111 inoculated or not-inoculated with Herbaspirillum seropedicae and Paenibacillus sp., respectively. In particular, the role of the endophytes is investigated related to agricultural aspects (e.g. plant-growth promoting and biocontrol) and very few studies have been conducted on medicinal plants3,12. E. purpurea root extracts are reported to stimulate macrophage TNF-α production but the extracts obtained from in vitro-grown axenic E. purpurea do not induce the same result, supporting the hypothesis that it is originated from the interaction with bacterial endophytes12. Our previous study shows that different compartments of E. purpurea, namely stems and leaves (SL), roots (R) and the rhizosphere (RS), share very few strains13. This finding has been shown to be related to the antagonism existing between strains inhabiting the different compartments14 and to the degree of resistance to antibiotics15. Moreover, the presence of distinct bacterial communities in plant compartments could account for the different bioactive compounds found in the various plant organs. Therefore, E. purpurea represents an interesting and useful in vitro model for plant-bacterial interaction studies on the production of pharmacological relevant secondary metabolites as the alkamides.
Alkamides (alkylamides; fatty acid amides) are lipophilic compounds chemically composed of two moieties, an amine moiety acylated by a fatty acid-derived one16. They are important bioactive compounds dissimilarly distributed in the different compartments of E. purpurea 17. Indeed, a notable difference is reported for the total alkamide content between aerial parts and roots, which is mainly due to a larger presence of non-tetraene alkamides in roots than in other plant organs18. The main Echinacea spp. alkamides are the isomeric dodeca-2E, 4E, 8Z, 10E/Z-tetraenoic acid isobutylamides19. These compounds increase the TNF mRNA expression in macrophages and monocytes binding the cannabinoid CB2 receptor20. Furthermore, the alkylamides decrease mitogens-induced interleukin-2 secretion in Jurkat-T cells21 and show an in vitro inhibitory activity of the 5-lipoxygenase22 and the cyclooxygenase-1 and 2 enzymes23.
Recently, a pyridoxal phosphate-dependent (PLP) decarboxylating enzyme belonging to the Class II tryptophan synthase family that utilizes branched-chain amino acids (BCAA) as substrate has been suggested24. Isotope labelling analyses have revealed the generation of isobutylamine and 2-methylbutylamine (i.e. the amine moiety of the E. purpurea alkamides) from valine and isoleucine, respectively. PLP decarboxylase-like proteins have been identified in the proteome of E. purpurea through in silico analyses and their transcript levels correlated with alkamide accumulation patterns in E. purpurea tissues. Then, a valine decarboxylase (VDC), potentially involved in the generation of the amine moieties of the alkamides, has been identified.
The aim of this work is to check the involvement of the endophytic communities of E. purpurea plants in the regulation of bioactive compound (alkamide) accumulation. To this purpose, we set up an in vitro model system in which axenic E. purpurea plants, in vitro germinated from sterilized seeds, are inoculated with a pool of selected endophytes previously isolated from the aerial compartment of E. purpurea plants cultivated in open field (Fig. 1). We have evaluated the biochemical alkamide profiles estimating different alkamide levels in control (not-inoculated) and inoculated plants. We have also found that the level of VDC gene expression is higher in the inoculated E. purpurea tissues than in the control ones, establishing a close relationship between the endophyte presence and alkamide levels.
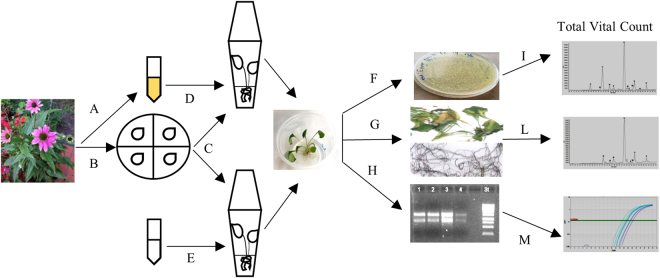
Figure 1.
In vitro model system setting up to study the interaction between Echinacea purpurea plants and their stem/leaves endophytic bacteria. (A) Bacterial endophytes were isolated from the aerial compartment of E. purpurea plants. (B) E. purpurea seeds were provided by the common garden at the “Il Giardino delle Erbe”, Casola Valsenio, Italy and surface-sterilized. (C) Seeds were germinated in De Wit Culture tubes containing 5 ml of Linsmaier & Skoog Medium (LS) including vitamins. After root formation, the seedlings were transferred in Wavin flasks containing 50 ml of LS solid medium, supplemented with 3% sucrose and maintained in a plant growth chamber for a photoperiod of 16 h light a day. (D,E) After about 2 months, five E. purpurea plants were inoculated with 8 × 106 bacterial endophytes isolated from SL compartment of E. purpurea plants (D); five plants were used as control and were inoculated with sterilized saline solution (E). After 45 days, SL and root (R) samples from control and infected plants, were collected separately and sterilized. (F–H) Samples were then, separated in different aliquots (R and SL pooled separately). (I) One aliquot of each tissue was immediately used for the in planta bacterial growth analysis. (L) One aliquot of each tissue was weighed and dried at 60 °C to be used for n-hexane extracts preparation. (M) One aliquot of each tissue was ground to a fine powder in liquid nitrogen and successively used for RNA extraction.
Results
Bacterial endophytes tend to re-colonize the native niche during plant infection
A pool of thirty-seven bacterial strains (Supplementary Table 1), isolated from the SL compartment of E. purpurea plants, was used to inoculate five axenic in vitro 2-months old E. purpurea plants each with 6 or 7 leaflets; five plants of the same age, used as control, were inoculated with sterilized saline solution. The infection experiment was repeated three times. Forty-five days after the infection, plants were analysed for bacterial colonization estimating the total viable count (TVC) as Colony Forming Units (CFU)/g into the host R and SL tissues. Data obtained revealed that the highest CFU/g was detected in the SL compartment (7.06 ± 6.50 log CFU/g), and the lowest one in the roots of infected plants (6.70 ± 5.82 log CFU/g; P < 0.001). This finding could indicate that the endophytes tended to re-colonize the native niche (SL compartment). The absence of bacteria in the control plant tissues and in the washing solutions confirmed the use of an axenic plant model and a successful sterilization procedure, respectively.
Alkamide profiling analysis in different organs of control and infected plants
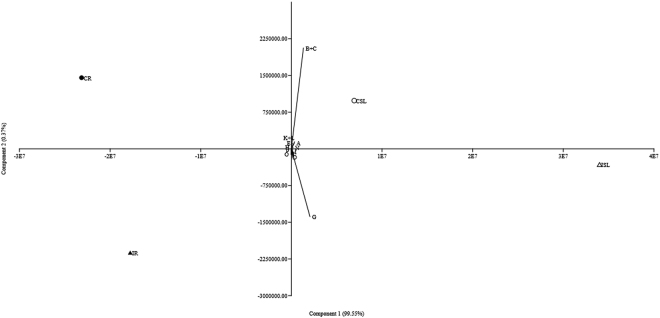
The alkamide profiles of both control and infected R and SL extracts of E. purpurea pooled plants were investigated by means of high performance liquid chromatography (HPLC) coupled to a photo diode array (PDA)/ultraviolet (UV) detector and electrospray ionization tandem mass spectrometry (ESI-MS/MS). The LC-PDA/UV chromatograms of both E. purpurea R and SL extracts showed different alkamide profiles (Supplementary Figs 1 and 2). In particular, the SL extracts appeared to be richer in alkamide content, with twelve identified compounds (1-12, Table 1), respect to the R samples (compounds 5, 7-12, Table 1) even though for the peaks A, B, G, K, and O two isomers can occur and their exact identification was not possible based on MS fragmentation pathway. Some alkamides present in the SL extract (peaks A, B, C, E, F, H, I, and M) were not found in the R one. On the other hand, two R extracts showed the presence of the alkamides 13/14 (peak O) and 15 (peak P), which were absent in the SL ones. In all R and SL samples, the most representative alkamides were a mixture of the two co-eluting isomers dodeca-2E,4E,8Z,10Z-tetraenoic acid isobutylamide (7) and dodeca-2E,4E,8Z,10E tetraenoic acid isobutylamide (8). During the alkamide chromatography of both R and SL extracts, polyacetylene amides (peaks A, B, D, O, and P) elute early, followed by tetraenes (peaks G and K), trienes (peak L), and finally dienes (N). Interestingly, the analysis of the alkamide content in the E. purpurea plant organs also revealed the presence of seven alkamides not previously reported (peaks C, E, F, H, I, J, and M), but ascribable to the alkamide class due to the characteristic absorbance at 260 nm and typical ESI-MS and MS/MS spectra (Supplementary Table 3). A tentative level estimation of the main alkamides was performed on control and infected R and SL extracts by measuring the peaks area and calculating the mean of the replicates (Supplementary Table 4). Data were submitted to Principal Component Analysis (PCA), and the result was reported in Fig. 2: the vectors accounting for the R extracts were differentially oriented than those of the SL ones (F = 10.42; P < 0.001). In particular, the R samples presented C12 diene-diyne alkamides (peak O and P), whilst the SL samples contained C11 diene-diyne alkamides (peak A and B). Both samples showed a prevalence of C12 tetraene alkamides (peak G and K). Furthermore, PCA showed a discrimination among control and infected samples both for R and SL extracts and suggested that such differences were mainly created by the amount of the most abundant alkamide isomers 7 and 8 (peak G). The comparison of means (Supplementary Table 4) revealed that the amount of these alkamide isomers was significantly different for all samples (Tukey HSD P < 0.001). A relative estimation of the alkamide isomers 7 and 8 in the R and SL infected samples was also performed in respect to the levels of the relative control samples showing that the infection resulted in an increase of alkamide levels of about 70% in SL and 87% in R infected samples compared to the controls.
Table 1.
Identification of alkamides detected in roots (R) and stem/leaves (SL) of control and infected E. purpurea plants. Compound numbers are referred to peaks in the chromatograms of Supplementary Figs 1 and 2. Peaks C, E, F, H, I, J, and M remained unidentified.
| Compound | Peaka | Organ | Alkamides |
|---|---|---|---|
| 1 | A | SL | undeca-2E,4Z-diene-8,10-diynoic acid isobutylamide |
| 2 | A | SL | undeca-2Z,4E-diene-8,10-diynoic acid isobutylamide |
| 3 | B | SL | undeca-2E,4Z-diene-8,10-diynoic acid methylbutylamide |
| 4 | B | SL | undeca-2Z,4E-diene-8,10-diynoic acid methylbutylamide |
| 5 | D | R, SL | trideca-2E,7Z-diene-8,10-diynoic acid isobutylamide |
| 6 | D | SL | dodeca-2E,4Z,10E-triene-8-ynoic acid isobutylamide |
| 7 | G | R, SL | dodeca-2E,4E,8Z,10Z-tetraenoic acid isobutylamide |
| 8 | G | R, SL | dodeca-2E,4E,8Z,10E tetraenoic acid isobutylamide |
| 9 | K | R, SL | dodeca-2E,4E,8Z,10Z-tetraenoic acid methylbutylamide |
| 10 | K | R, SL | dodeca-2E,4E,8Z,10E-tetraenoic acid methylbutylamide |
| 11 | L | R, SL | dodeca-2E,4E,8Z-trienoic acid isobutylamide |
| 12 | N | R, SL | dodeca-2E,4E-dienoic acid isobutylamide |
| 13 | O | R | dodeca-2E,4Z-diene-8,10-diynoic acid isobutylbutylamide |
| 14 | O | R | dodeca-2Z,4E-diene-8,10-diynoic acid isobutylbutylamide |
| 15 | P | R | dodeca-2E,4Z-diene-8,10-diynoic acid 2-methylbutylamide |
Figure 2.
Principal Component Analysis of alkamide relative estimations of the four different Echinacea purpurea extracts. Letters on vectors indicate the HPLC peaks accounting for the differentiation of samples (see text for details). CSL, stem/leaves extract from control plants; ISL, stem/leaves extract from infected plants, CR, root extract from control plants; IR: root extract from infected plants.
BCCA decarboxylases genetic expression in different organs of control and infected plants
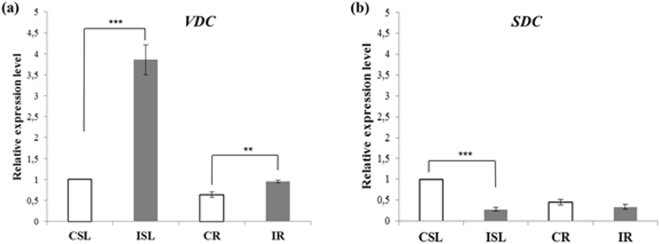
The relative quantification of VDC gene expression in the SL and R tissues of the control and infected E. purpurea plants was calculated in respect to the ubiquitin E2 (UbE2) gene expression in the three biological replicates. The amplification efficiency was optimal as achievable by the R2 and slope values (slopeVDC = –3.113; R2 VDC = 0.904; slopeUbE2 = –3.182; R2 UbE2 = 0.988). As depicted in Fig. 3a, the VDC gene expression levels were higher in the SL samples than the R ones (P < 0.01). The VDC transcription level in the infected SL samples was about 4 times more than in the SL control tissues (P < 0.001). Also, the expression level resulted increased in R infected tissues respect to the relative controls even if at a minor extent (P < 0.01). In parallel, the involvement of an enzyme (serine decarboxylase SDC, GenBank Accession #LT593931.1) not decarboxylating valine or isoleucine (to form isobutylamine or 2-methylbutylamine, the amine moieties of the E. purpurea alkamides) but utilizing the serine as substrate to generate ethanolamine24 was evaluated. The transcription of the SDC gene (slopeSDC = −3.135; R2 SDC = 0.982) appeared down-regulated in the infected SL samples respect to the SL control ones whilst the SDC expression levels were similar between control and infected R samples (Fig. 3b).
Figure 3.
Gene expression of (a) valine decarboxylase (VDC) and (b) serine decarboxylase (SDC) in control and infected samples of Echinacea purpurea plants. Expression levels were normalized to expression in CSL. Data report average from three independent experiments (with 3 technical replicates each). Error bars: standard error of the mean (s.e.m.). Comparison between infected samples and the relative controls was determined by 2-tailed t-test (*P < 0.05, **P < 0.01, ***P < 0.001). CSL, stem/leaves extract from control plants; ISL, stem/leaves extract from infected plants, CR, root extract from control plants; IR: root extract from infected plants.
Discussion
In this work we set up an in vitro system to study the influence of the interaction between bacterial endophytes and E. purpurea plants on the production of plant bioactive compounds.
The infection data revealed the bacterial tendency to reach their niche of origin passing through the R apparatus since a significant number of bacteria was detected also in the R tissues of the infected plants. This was in agreement with the current literature, according to which one of the ways used by endophytes to reach the aerial parts of the host plant was their migration through the xylem vascular system25 once that they have entered the plant roots. Hence, we speculated that all the endophytes could reach the SL compartment. However, we were not able to consider a post-infection time longer than 45 days since it would have induced several plant damages. Therefore, our data confirmed the hypothesis that differences in bacterial strains distribution between R and SL of E. purpurea could be related to the physiological conditions existing in the different plant tissues and organs representing a specific ecological niche, as previously suggested13.
On the basis of data obtained in this work, the R and SL extracts displayed different alkamide profiles, with twelve and ten compounds, respectively identified comparing their HPLC elution orders, ESI-MS/MS, and PDA/UV spectra with data reported in the literature3,26–29. The most abundant alkamides were the 2,4-diene type compounds and the two isomers dodeca-2E,4E,8Z,10Z-tetraenoic acid isobutylamide (7) and dodeca-2E,4E,8Z,10E tetraenoic acid isobutylamide (8) being predominant, according to previous studies performed on in vivo plants30,31. Moreover, seven alkamides (peaks C, E, F, H, I, J, and M) remained not identified, and among these one alkamide (peak F) was present only in the infected SL compartment. To our knowledge, these alkamides were not previously reported in E. purpurea.
The estimated total alkamide level was higher in the SL samples than in R ones, apparently in contrast with the literature data reporting that the E. purpurea roots contain more alkamides than the leaves2. However, the alkamide content in SL samples could be higher than the reported leaves amount since it was the sum of the leaves and the vegetative stems. In fact, in line with the literature32, the R and SL extracts were differentiated by the presence of the C12 and C11 diene-diyne alkamides (mainly reported in the stem), respectively. On the other hand, Qu et al.18 reported that the tetraene alkamides 7 and 8 accounted for the 75% in the aerial part and only for the 9% in the roots of E. purpurea plants cultivated into a field and these estimates resulted discordant with our results (about 50 and 70%, respectively). To this concern, one possible explanation could be that, to the best of our knowledge, this study is the first to estimate the E. purpurea alkamide level in an in vitro model whose experimental conditions could influence plant genetics and/or biochemical synthesis. However, the estimated alkamide level was higher in both infected R and SL, with a relative increase of alkamides 7 and 8 about 87% and 70% respectively, when compared to the controls, suggesting that the alkamide biosynthesis was modulated by the E. purpurea endophyte infection.
Concerning the VDC gene expression profiles, the highest expression level was detected in the infected E. purpurea tissues. By comparing results concerning both the chemical profiling analysis and the VDC gene expression data, it could be observed that the use of the proposed in vitro infection model system allowed us to demonstrate that the infection of E. purpurea axenic plants with their endophytes influenced the alkamide levels. In fact, both alkamide content and VDC transcription level resulted increased after the infection. The transcriptional up-regulation in the infected R samples was lower than the expected one: this result could be explained considering that the increase of the alkamide quantity in the infected R might be due to the transcriptional regulation of other enzymes required for the biosynthesis of the alkamide ammine and fatty acid moieties24. Interestingly, the SDC enzyme did not seem to contribute to the increase of alkamide levels confirming the specificity of the enzymatic source for the amine moiety formation of the alkamides (i.e. the VDC decarboxylase) as reported by Rizhsky et al.24.
The main objective of this work was to develop an in vitro model to study the role of the interaction between E. purpurea and its endophytes in the modulation of the plant secondary metabolism. Bacterial communities differed substantially between E. purpurea organs13 probably exerting a host selectivity able to modulate the community structure as reported for endophytic fungi33. In fact, the E. purpurea endophytes inoculated in axenic plants tended to re-colonize the native niche whose specific properties probably were influenced in turn by the natural endophytes. Chemical profiles and VDC genetic expression resulted quantitatively different between the control and infected plants, in particular in the SL compartment. Therefore, despite the in vivo status during bacterium-host interactions was difficult to mimic in vitro, the infection with E. purpurea SL endophytes modulated the characteristics resulting in axenic conditions, at least to some extent.
Consequently, the whole body of data obtained in this work strongly suggested that the secondary metabolism in E. purpurea was influenced by the plant-endophyte interaction thus possibly contributing to the therapeutic properties of this medicinal plant.
Materials and Methods
Bacterial cultures and plant material
Bacterial endophytes were isolated from the aerial compartment (stem and leaves) of E. purpurea plants grown at the “Il Giardino delle Erbe”, Casola Valsenio, Italy, as previously reported13. Stock cultures were grown at 30 °C on tryptone soy agar (TSA; Bio-Rad, USA) solid medium or tryptone soy broth (TSB, Bio-Rad, USA) liquid medium. E. purpurea seeds were provided by the “Il Giardino delle Erbe”.
Seed sterilization and plating
Seeds were surface sterilized in order to prevent any unwanted fungal or bacterial growth. Seeds were immersed in a 70% (v/v) ethanol for one minute and, subsequently, in a 5% sodium hypochlorite solution for eight minutes. They were then rinsed three times with sterile distilled water, kept overnight at 4 °C in the dark for growth synchronization and then germinated in De Wit Culture tubes (LAB Associates BV, The Netherlands) containing 5 ml of Linsmaier & Skoog Medium (LS) including vitamins (Duchefa Biochemie, The Netherlands) at 24 ± 1 °C in the dark. After root formation, the seedlings were transferred in Wavin flasks (LAB Associates BV, The Netherlands) containing 50 ml of LS solid medium, supplemented with 3% sucrose, for a photoperiod of 16 h light a day for a minimum of two months. In order to validate the sterility of the obtained model system, cultivable endophyte multiplication into host tissues was checked: both shoots and roots were separately collected, washed in saline solution (0,9% NaCl, washing solution), surface sterilized in 1% (v/v) hypochlorite for 8 min and rinsed three times with sterile distilled water. Both samples were homogenized in saline solution and five replications of 100 μl of the homogenates were plated on TSA medium. Bacterial growth was scored after three days of plate incubation at 30 °C.
Plant infection
Inocula of bacterial endophytes, isolated from SL compartment of E. purpurea plants, were incubated for three days at 30 °C in horizontal position and in agitation. The bacterial suspensions were then adjusted to 8 × 108 CFU/ml (OD600 = 1). The optical density (OD) was measured in a biophotometer (Eppendorf, Germany). The pool generated from 100 μl of each diluted 1:10 OD600 suspension cultures was then centrifuged at 4000 rpm for 20 minutes and the pellet suspended in a correspondent volume of 0.9% saline solution. Five 2-months old E. purpurea plants were infected with 100 μl of bacterial suspension culture. Five plants were used as control and were infected with 100 μl of sterilized saline solution. Plants were then incubated in the growth chamber at 24 ± 1 °C. After 45 days, SL and R samples from control and infected plants, were collected separately, firstly washed in saline solution and then sterilized in 1% (v/v) hypochlorite for 8 min. Then, both tissue samples were washed three times with sterile distilled water and separated in different aliquots. R and SL aliquots were weighed and dried at 60 °C to be used for n-hexane extract preparation. R and SL aliquots of fresh material were ground to a fine powder in liquid nitrogen and successively stored at – 80 °C for RNA extraction. Finally, 1.0 g of fresh R and SL tissues were immediately used for the in planta bacterial growth analysis. The experiment was performed in triplicate.
In planta bacterial growth analysis
In order to evaluate endophytes multiplication into host tissues, 1.0 g of each sample was homogenized in saline solution and 100 µl of the homogenate were serially diluted up to 10−7/ml cells. Five replications of each dilution were plated on TSA medium. The washing solution and the distilled water after the last wash were also diluted to check the presence of bacterial cells on the surface of the tissues and the outcome of the sterilization procedure. Bacterial growth was scored after two, three and four days of incubation of the plates at 30 °C.
Sample preparation for HPLC analysis of alkamides
Dried and powdered R and SL of control and infected E. purpurea plants from three independent experiments were pooled and extracted at room temperature with n-hexane (1.0 g of dried drug in 30 ml of solvent for three times, every 24 h) as detailed in Supplementary Table 5. Solutions of each n-hexane residue from R and SL samples were then prepared dissolving the respective n-hexane extract in an opportune volume of methanol and then centrifuging the mixture. Finally, 20 μl of each supernatant solution (2.0 mg/ml) were injected for HPLC-PDA/UV-ESI-MS/MS analysis. The experiment was performed in duplicate.
HPLC-PDA/UV-ESI-MS/MS analyses
Qualitative HPLC-PDA/UV-ESI-MS/MS analyses were performed using a Surveyor LC pump, a Surveyor autosampler, coupled with a Surveyor PDA detector, and a LCQ Advantage ion trap mass spectrometer (ThermoFinnigan) equipped with Xcalibur 3.1 software. Analyses were performed using a 4.6 × 250 mm, 4 µm, Synergi Fusion-RP column (Phenomenex). The eluent was a mixture of methanol (solvent A) and a 0.1% v/v aqueous solution of formic acid (solvent B). A linear gradient of increasing 55% to 85% A was developed within 45 min. The column was successively washed for 15 min with methanol and equilibrated with 55% A for 10 min. Elution was performed at a flow rate of 0.8 ml/min with a splitting system of 2:8 to MS detector (160 ml/min) and PDA detector (640 ml/min), respectively. The volume of the injected methanol solutions was 20 μl. Analyses were performed with an ESI interface in the positive mode. The ionization conditions were optimized and the parameters used were as follows: capillary temperature, 270 °C; capillary voltage, 29.0 V; tube lens offset, 50.0 V; sheath gas flow rate, 60.00 arbitrary units; auxiliary gas flow rate, 3.00 arbitrary units; spray voltage, 4.50 kV; scan range of m/z 150–1200. N2 was used as the sheath and auxiliary gas. PDA data were recorded with 200–600 nm range with preferential channel as the detection wavelength 260 nm.
Quantitative real time PCR (qRT-PCR) analysis
Total RNA from R and SL was extracted from E. purpurea tissues using the RNeasy Micro Kit (Qiagen, USA) and quantified by Qubit® 2.0 fluorimeter. One μg of total RNA for each sample was reverse-transcribed using the Quantitect Reverse Transcription Kit according to the manufacturer’s instructions (Qiagen) including a treatment with 1X gDNA Wipeout Buffer to remove any remaining DNA. The relative abundance of the E. purpurea cDNA for BCAA decarboxylases (VDC, GenBank Accession #LT593930; SDC, GenBank Accession #LT593931.1) was determined on a QUANTSTUDIO 7 FLEX (Applied Biosystems, USA) using the QuantiNova SYBR Green PCR Kit (Qiagen, USA). Ubiquitin E2 genetic expression was used as internal reference to normalize mRNA content. The quantification of the expression was measured by the comparative Ct (2−∆∆Ct) method34. Target gene expression was relative to the control SL cDNA, which has been adopted as calibrator. The experiment was conducted in triplicate. Primer3 software35 was used to design primers specific to SDC template (epa_locus_952_iso_8_len_1801_ver_2) as available in the Medicinal Plant Genomics Resource (http://medicinalplantgenomics.msu.edu). The primer sequences for the VDC gene were reported in Rizhsky et al.24. All primers listed in Supplementary Table 2 were synthesized by Eurofins Genomics (Ebersberg, Germany). Independent RT-PCR products were sequenced to control for primer specificity.
Statistical analyses
Means and standard deviations of the bacterial TVC data of the three biological replicates were estimated and compared by one-way analysis of variance between R and SL samples. To evaluate whether the level estimations of the alkamides (mean peak area values) were useful in reflecting the chemical relationships between R and SL samples (controls and infected ones), a PCA was performed36. One-way analysis of variance followed by Tukey test was used to compare peak area values between control and infected plants and to identify the alkamides mainly responsible of the differences between the samples. Comparison of the qRT-PCR data of the three biological replicates was determined by 2-tailed t-test. P < 0.05 was considered significant (*P < 0.05, **P < 0.01, ***P < 0.001). Error bars are shown as s.e.m. The analyses were performed by using the modules present in the PAST program, version 3.1537.
Data availability
The Authors declare that all the data supporting the findings of this study are available within the manuscript and its Supplementary Material and from the Corresponding Author on request.
Electronic supplementary material
Acknowledgements
We thank Claudio Ciofi and Chiara Natali from the Department of Biology, University of Florence for providing sequencing data. This work was supported by the Ente Cassa di Risparmio di Firenze (project 2016.0936 e 2013.0657).
Author Contributions
V.M., E.R.G., A.M., R.F., P.B. and F.F. conceived and designed the work. V.M. and P.B. conceived and planned the experiments. V.M. and M.D.L. carried out all the experiments. R.V.B.R. and E.M. were involved in data collection. V.M. and A.M. performed the data analysis. V.M., M.D.L., A.M., R.F., L.P. and P.B. contributed to the interpretation of results. V.M. and P.B. wrote the first draft of the manuscript. All authors were involved in critical revision and approval of the final version.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Fabio Firenzuoli and Patrizia Bogani jointly supervised this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17110-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Karsch-Volk M, Barrett B, Linde K. Echinacea for preventing and treating the common cold. Jama. 2015;313:618–619. doi: 10.1001/jama.2014.17145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manayi A, Vazirian M, Saeidnia S. Echinacea purpurea: Pharmacology, phytochemistry and analysis methods. Pharmacognosy reviews. 2015;9:63–72. doi: 10.4103/0973-7847.156353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pellati, F. et al. Chromatographic methods for metabolite profiling of virus- and phytoplasma-infected plants of Echinacea purpurea. Journal of agricultural and food chemistry59 (2011). [DOI] [PubMed]
- 4.Braga RM, Dourado MN, Araujo WL. Microbial interactions: ecology in a molecular perspective. Brazilian journal of microbiology: [publication of the Brazilian Society for Microbiology] 2016;47(Suppl 1):86–98. doi: 10.1016/j.bjm.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanvir, R., Javeed, A. & Bajwa, A. G. Endophyte bioprospecting in South Asian medicinal plants: an attractive resource for biopharmaceuticals. Applied microbiology and biotechnology101 (2017). [DOI] [PubMed]
- 6.Aly AH, Debbab A, Proksch P. Fungal endophytes: unique plant inhabitants with great promises. Applied microbiology and biotechnology. 2011;90:1829–1845. doi: 10.1007/s00253-011-3270-y. [DOI] [PubMed] [Google Scholar]
- 7.Zhai X, et al. The regulatory mechanism of fungal elicitor-induced secondary metabolite biosynthesis in medical plants. Critical reviews in microbiology. 2017;43:238–261. doi: 10.1080/1040841X.2016.1201041. [DOI] [PubMed] [Google Scholar]
- 8.Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A. Metabolic potential of endophytic bacteria. Current opinion in biotechnology. 2014;27:30–37. doi: 10.1016/j.copbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaul S, Sharma T, M KD. “Omics” Tools for Better Understanding the Plant-Endophyte Interactions. Frontiers in plant science. 2016;7:955. doi: 10.3389/fpls.2016.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari CS, et al. Expressed proteins of Herbaspirillum seropedicae in maize (DKB240) roots-bacteria interaction revealed using proteomics. Applied biochemistry and biotechnology. 2014;174:2267–2277. doi: 10.1007/s12010-014-1197-3. [DOI] [PubMed] [Google Scholar]
- 11.Scherling C, Ulrich K, Ewald D, Weckwerth W. A metabolic signature of the beneficial interaction of the endophyte paenibacillus sp. isolate and in vitro-grown poplar plants revealed by metabolomics. Molecular plant-microbe interactions: MPMI. 2009;22:1032–1037. doi: 10.1094/MPMI-22-8-1032. [DOI] [PubMed] [Google Scholar]
- 12.Todd DA, et al. Ethanolic Echinacea purpurea Extracts Contain a Mixture of Cytokine-Suppressive and Cytokine-Inducing Compounds, Including Some That Originate from Endophytic Bacteria. PloS one. 2015;10:e0124276. doi: 10.1371/journal.pone.0124276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiellini C, et al. Endophytic and rhizospheric bacterial communities isolated from the medicinal plants Echinacea purpurea and Echinacea angustifolia. International microbiology: the official journal of the Spanish Society for Microbiology. 2014;17:165–174. doi: 10.2436/20.1501.01.219. [DOI] [PubMed] [Google Scholar]
- 14.Maida I, et al. Antagonistic interactions between endophytic cultivable bacterial communities isolated from the medicinal plant Echinacea purpurea. Environmental microbiology. 2016;18:2357–2365. doi: 10.1111/1462-2920.12911. [DOI] [PubMed] [Google Scholar]
- 15.Mengoni A, et al. Antibiotic resistance differentiates Echinacea purpurea endophytic bacterial communities with respect to plant organs. Research in microbiology. 2014;165:686–694. doi: 10.1016/j.resmic.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Greger H. Alkamides: a critical reconsideration of a multifunctional class of unsaturated fatty acid amides. Phytochem Rev. 2016;15:729–770. doi: 10.1007/s11101-015-9418-0. [DOI] [Google Scholar]
- 17.Barnes J, Anderson LA, Gibbons S, Phillipson JD. Echinacea species (Echinacea angustifolia (DC.) Hell., Echinacea pallida (Nutt.) Nutt., Echinacea purpurea (L.) Moench): a review of their chemistry, pharmacology and clinical properties. The Journal of pharmacy and pharmacology. 2005;57:929–954. doi: 10.1211/0022357056127. [DOI] [PubMed] [Google Scholar]
- 18.Qu L, Chen Y, Wang X, Scalzo R, Davis JM. Patterns of Variation in Alkamides and Cichoric Acid in Roots and Aboveground Parts of Echinacea purpurea (L.) Moench. HortScience: a publication of the American Society for Horticultural Science. 2005;40:1239–1242. [PMC free article] [PubMed] [Google Scholar]
- 19.Woelkart K, Bauer R. The role of alkamides as an active principle of echinacea. Planta medica. 2007;73:615–623. doi: 10.1055/s-2007-981531. [DOI] [PubMed] [Google Scholar]
- 20.Raduner S, et al. Alkylamides from Echinacea are a new class of cannabinomimetics. Cannabinoid type 2 receptor-dependent and -independent immunomodulatory effects. The Journal of biological chemistry. 2006;281:14192–14206. doi: 10.1074/jbc.M601074200. [DOI] [PubMed] [Google Scholar]
- 21.Sasagawa M, Cech NB, Gray DE, Elmer GW, Wenner CA. Echinacea alkylamides inhibit interleukin-2 production by Jurkat T cells. International immunopharmacology. 2006;6:1214–1221. doi: 10.1016/j.intimp.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Muller-Jakic B, et al. In vitro inhibition of cyclooxygenase and 5-lipoxygenase by alkamides from Echinacea and Achillea species. Planta medica. 1994;60:37–40. doi: 10.1055/s-2006-959404. [DOI] [PubMed] [Google Scholar]
- 23.Clifford LJ, Nair MG, Rana J, Dewitt DL. Bioactivity of alkamides isolated from Echinacea purpurea (L.) Moench. Phytomedicine: international journal of phytotherapy and phytopharmacology. 2002;9:249–253. doi: 10.1078/0944-7113-00105. [DOI] [PubMed] [Google Scholar]
- 24.Rizhsky L, et al. Integrating metabolomics and transcriptomics data to discover a biocatalyst that can generate the amine precursors for alkamide biosynthesis. The Plant journal: for cell and molecular biology. 2016;88:775–793. doi: 10.1111/tpj.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardoim PR, et al. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiology and molecular biology reviews: MMBR. 2015;79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cech NB, et al. High performance liquid chromatography/electrospray ionization mass spectrometry for simultaneous analysis of alkamides and caffeic acid derivatives from Echinacea purpurea extracts. Journal of chromatography A. 2006;1103:219–228. doi: 10.1016/j.chroma.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 27.He XG, Lin LZ, Bernart MW, Lian LZ. Analysis of alkamides in roots and achenes of Echinacea purpurea by liquid chromatography electrospray mass spectrometry. Journal of Chromatography A. 1998;815:205–211. doi: 10.1016/S0021-9673(98)00447-6. [DOI] [Google Scholar]
- 28.Hohmann J, et al. Alkamides and a neolignan from Echinacea purpurea roots and the interaction of alkamides with G-protein-coupled cannabinoid receptors. Phytochemistry. 2011;72:1848–1853. doi: 10.1016/j.phytochem.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Spelman K, Wetschler MH, Cech NB. Comparison of alkylamide yield in ethanolic extracts prepared from fresh versus dry Echinacea purpurea utilizing HPLC-ESI-MS. Journal of pharmaceutical and biomedical analysis. 2009;49:1141–1149. doi: 10.1016/j.jpba.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Binns SE, Livesey JF, Arnason JT, Baum BR. Phytochemical variation in echinacea from roots and flowerheads of wild and cultivated populations. Journal of agricultural and food chemistry. 2002;50:3673–3687. doi: 10.1021/jf011439t. [DOI] [PubMed] [Google Scholar]
- 31.Chen CL, Zhang SC, Sung JM. Caffeoyl phenols and alkamides of cultivated Echinacea purpurea and Echinacea atrorubens var. paradoxa. Pharm Biol. 2009;47:835–840. doi: 10.1080/13880200902939291. [DOI] [Google Scholar]
- 32.Perry NB, van Klink JW, Burgess EJ, Parmenter GA. Alkamide levels in Echinacea purpurea: a rapid analytical method revealing differences among roots, rhizomes, stems, leaves and flowers. Planta medica. 1997;63:58–62. doi: 10.1055/s-2006-957605. [DOI] [PubMed] [Google Scholar]
- 33.Heilmann-Clausen J, et al. Citizen science data reveal ecological, historical and evolutionary factors shaping interactions between woody hosts and wood-inhabiting fungi. The New phytologist. 2016;212:1072–1082. doi: 10.1111/nph.14194. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Untergasser A, et al. Primer3–new capabilities and interfaces. Nucleic acids research. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cserhati T. Data evaluation in chromatography by principal component analysis. Biomedical chromatography: BMC. 2010;24:20–28. doi: 10.1002/bmc.1294. [DOI] [PubMed] [Google Scholar]
- 37.Hammer Ø H. D., R. P. PAST : paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Authors declare that all the data supporting the findings of this study are available within the manuscript and its Supplementary Material and from the Corresponding Author on request.