Abstract
Temperate coastal marine environments are replete with complex biotic and abiotic interactions that are amplified during spring and summer phytoplankton blooms. During these events, heterotrophic bacterioplankton respond to successional releases of dissolved organic matter as algal cells are lysed. Annual seasonal shifts in the community composition of free-living bacterioplankton follow broadly predictable patterns, but whether similar communities respond each year to bloom disturbance events remains unknown owing to a lack of data sets, employing high-frequency sampling over multiple years. We capture the fine-scale microdiversity of these events with weekly sampling using a high-resolution method to discriminate 16S ribosomal RNA gene amplicons that are >99% identical. Furthermore, we used 2 complete years of data to facilitate identification of recurrent sub-networks of co-varying microbes. We demonstrate that despite inter-annual variation in phytoplankton blooms and despite the dynamism of a coastal–oceanic transition zone, patterns of microdiversity are recurrent during both bloom and non-bloom conditions. Sub-networks of co-occurring microbes identified reveal that correlation structures between community members appear quite stable in a seasonally driven response to oligotrophic and eutrophic conditions.
Introduction
Microbes in coastal marine environments are influenced by multiple biotic and abiotic elements that mediate relationships between community members. Shifts in community structure can be modulated by changes in bottom-up substrate availability induced by seasonal forcing such as solar angle and upwelling and by top–down predator–prey interactions (reviewed in Fuhrman et al., 2015). These dynamics are amplified in temperate coastal marine zones when phytoplankton form massive blooms each spring and summer. Following winter, increases in light and temperature favor algal growth in spring. Upon bloom termination, algae release a complex array of dissolved organic matter. These organic substrates are almost exclusively accessible by heterotrophic bacterioplankton as part of the microbial loop (Azam et al., 1983; Azam and Ammerman, 1984; Ducklow and Carlson, 1992). Many of the bacterioplankton responding to spring bloom events are represented by fast-growing copiotrophs, triggering successions of specialized clades that are able to rapidly incorporate algal-derived dissolved organic matter into their biomass using diverse repertoires of specialized transporters and carbohydrate-active enzymes (Thomas et al., 2011; Koropatkin et al., 2012; Teeling et al., 2012, 2016; Buchan et al., 2014; Cuskin et al., 2015; Xing et al., 2015). In turn, these organisms constitute a new pool of particulate bacterial biomass, which is available to protozoa and other higher trophic levels (Azam et al., 1983). Production that is not catabolized is typically exported to sediments below (Legendre and Le Fèvre, 1989).
Biodiversity studies at long-term sampling stations have revealed broadly recurrent patterns within marine planktonic communities (Steinberg et al., 2001; Gilbert et al., 2012; Chow et al., 2013; Fuhrman et al., 2015). However, many of these studies employ monthly sampling intervals that do not adequately capture community dynamics during productive bloom events, which can elicit rapid shifts in community structure on the order of weeks (Ward et al., 2017), or days (Needham and Fuhrman, 2016). Annually replicated studies employing high-frequency sampling throughout both baseline and bloom periods are required to discern stable forces that initiate, regulate and maintain successional changes in microbial communities, to examine the extent of recurrence across years, and to identify non-random co-occurrences between microorganisms that can be used to formulate hypotheses about ecologically relevant in situ community interactions.
Although previous multi-year studies have identified seasonally repeating patterns of marine microbial diversity based on 16S ribosomal RNA (rRNA) surveys, in many such studies phylogenetic characterization has remained coarsely defined (Fuhrman et al., 2006; Gilbert et al., 2009; Teeling et al., 2016). Microbial clades tend to maintain broadly conserved ecological characteristics (Philippot et al., 2010; Martiny et al., 2015), and in some systems, coarser levels of taxonomy show improved correlations with environmental drivers (Lu et al., 2016). However, it has been shown in marine systems that sub-operational taxonomic unit (OTU) level patterns of microdiversity reveal meaningful variation in response to environmental variables (Eren et al., 2013; Ward et al., 2017) and can display stronger patterns of correlation when used to disentangle highly specific interactions, like those between microbial hosts and their viruses (Needham et al., 2017). High phylogenetic resolution is thus critical to identify relationships and potential interactions between community members as they respond to seasonal fluctuations in environmental variables.
The long-term research site Helgoland Roads (German Bight, North Sea) hosts annual spring and summer phytoplankton blooms. In an effort to improve the resolution of community dynamics during both winter and bloom conditions, we investigated the free-living bacterioplankton community inhabiting Helgoland Roads surface waters using weekly sampling intervals across 3 complete years. In order to improve the resolution of the community diversity, we characterized patterns of microdiversity using Minimum Entropy Decomposition (MED) to cluster 16S rRNA gene amplicons. Using these highly resolved taxonomic units in combination with network analysis, we aimed to identify and characterize recurrent modules of co-varying microbes.
Materials and methods
Sampling site
The long-term Helgoland Roads sampling site is adjacent to Helgoland Island, which lies ~60 km offshore in the southeastern North Sea in the German Bight. This site represents a coastal–oceanic transition zone (Raabe and Wiltshire, 2009) with westerly currents transporting marine water from the English Channel and the rivers Elbe and Weser, providing periodic incursions of freshwater (Wiltshire et al., 2009). Samples were taken at the station 'Kabeltonne' (54° 11' 17.88'' N, 7° 54' 0'' E) from 2010 to 2012, which is located between Helgoland Island and the minor island, Düne, where water depths fluctuate from 7 to 10 m over the tidal cycle. Long-term physicochemical and plankton data have been monitored continuously since 1962 as part of the Helgoland Roads time series and are accessible via the Open Access Library Pangaea (http://www.pangaea.de).
Physicochemical, phytoplankton and bacterioplankton sampling
Physicochemical data collected includes dissolved inorganic nitrogen (NO2−+NO3−+NH4+), silicate (SiO2), phosphate (PO43−), salinity, water temperature, chlorophyll a (chl a) and Secchi disk depth (Supplementary Table S1). Counts of major phytoplankton groups were obtained through microscopic morphological identification (Supplementary Table S2), as described previously (Wiltshire et al., 2009), and include diatoms (pennales and centrales), dinoflagellates, flagellates, coccolithophorids, ciliates, Phaeocystis spp. and Chattonella spp.
Samples of 2010–2012 free-living archaeal and bacterial picoplankton (collectively termed ‘bacterioplankton’) were taken at the station 'Kabeltonne' at bi-weekly to weekly intervals, as described previously (Teeling et al., 2016). In brief, surface seawater samples (ca. −1 m) were pre-filtered at 10 μm, followed by removal of the particle-attached fraction at 3 μm with final collection of the free-living bacterioplankton fraction on 0.2 μm filters. In total, there were 142 free-living samples: 30 in 2010, 68 in 2011 and 44 in 2012. Sampling in 2010 is patchy during winter and late spring, however, 2011 and 2012 are represented by evenly spaced samples. Annual DAPI total cell counts of bacterioplankton abundance were available from a previous study in 2009 (Teeling et al., 2012) and 2010 (Supplementary Table S3).
V4 16S rRNA gene sequencing
DNA was extracted from the free-living 0.2–3 μm fraction and sequenced by the Department of Energy Joint Genome Institute (DOE-JGI, Walnut Creek, CA, USA) using V4 region of the 16S rRNA gene using primers 515 F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′) and Illumina MiSeq 2 × 250 bp chemistry as described previously (Lucas et al., 2015). 16S rRNA gene sequences are available from the DOE-JGI website GOLD database (Project ID: Gp0056779) as part of the community sequencing project COGITO. As a note, the V4 16S rRNA gene primers used in this study do not include primer revisions outlined by Apprill et al., (2015), which better cover the alphaproteobacterial clade SAR11; SAR11 abundances in our data set are thus expected to be significantly under-represented, whereas Gammaproteobacteria might be over-represented (Parada et al., 2016).
Minimum Entropy Decomposition
Raw paired-end reads were merged and quality filtered using Eren et al. (2013) to retain pairs with no mismatches in the overlapping regions, followed by primer trimming with cutadapt (Martin, 2011) with a final average length of 253 bp. Sequences were clustered using Minimum Entropy Decomposition (MED) (Eren et al., 2015). MED applies the principle of oligotyping (Eren et al., 2013), which uses Shannon entropy to iteratively partition amplicons at single nucleotide resolution, often providing more accurate descriptions of closely related but distinct taxa (that is, taxa with >97% sequence identity; Utter et al., 2016). We thus differentiate these MED clusters from conventional OTUs by referring to them as ‘oligotypes’. During MED, we used a minimum substantive abundance (-M) of 100 to filter low-abundant oligotypes with the decomposition of one nucleotide position at a time (-d 1). These settings removed most singletons and doubletons, affecting alpha diversity measures of absolute richness. In total, after merging and quality filtering, 15 014 510 high-quality tags were obtained for MED clustering. A total of 4271 representative nodes were decomposed from the 3-year data set representing 13 259 298 sequences (average 93 375 sequences per sample). The remaining 1 755 212 sequences were identified as outliers by MED owing to filtering by either maximum variation cutoff, as determined intrinsically by MED through mean read length, or by falling below the minimum substantive abundance.
Representative oligotypes were classified against the SILVA v123 database. The following threshold was used to determine a database match: (sequence identity+alignment coverage)/2⩾93%, with the remainder classified as having no relative. In total, 77 of the 4271 OTUs did not contain a database match. Half of these ‘no relatives’ had low score hits to chloroplast and mitochondria. All 404 OTUs classified as chloroplast, mitochondria or ‘no relative’ were removed before downstream analysis (3 867 remaining OTUs). SILVA v123 includes major changes in taxonomy for abundant North Sea clades, particularly for Roseobacter-related lineages including Planktomarina (formerly DC5-80-3 lineage), Amylibacter (formerly NAC11-7 lineage) and Ascidiaceihabitans (formerly Roseobacter OCT lineage). The final abundance matrix includes oligotype ids and SILVA genus-level classifications (Supplementary Table S4). We take proportional read frequencies as a proxy for community relative abundance.
Alpha diversity analysis
Rarefaction (interpolation) and prediction (extrapolation) of Hill number sampling curves for species richness, exponential Shannon diversity and inverse Simpson diversity were performed with the R package iNEXT (v 2.0.9) (Chao et al., 2014; Hsieh et al., 2016). Hill numbers express diversity in units of effective numbers of species: the number of equally abundant species required to give the same value of a diversity measure. The extrapolation end point was set to twice the mean sample size following removal of samples below the 25th percentile in total sequences. Confidence intervals were calculated using the default 50 bootstrap replicates. We calculated Pielou’s J evenness using the equation J=H/log(S) where J=Pielou’s evenness, H=the asymptotic estimated exponential Shannon diversity calculated in iNEXT and S=total number of species.
Oligotype and sample data matrices were managed with the R package phyloseq (v 1.14.0) (McMurdie and Holmes, 2013) and plots were made with ggplot2 (v 2.1.0) (Wickham, 2009). Pairwise sequence identity between closely related oligotypes was calculated in Geneious (v8.0) (Kearse et al., 2012) using percent identical sites. A custom R function called ‘taxplot_grep’ was used to generate the bar plots presented in this paper and is available within the script https://github.com/mc68462/medNS/blob/v0.3/taxplot_functions.R. Using a simple regular expression search of taxonomy, users can explore oligotypes and lineages of interest.
Patterns of oligotype prevalence
We plotted each oligotype in an abundance vs prevalence plot, as in (Barberán et al., 2012), to determine whether oligotypes grouped based on how often they are found across time points. Our measure of prevalence constitutes the percent of samples in which an oligotype appears >500 times, thus filtering out oligotypes present in very low abundance across many samples. The threshold of 500 sequences corresponds to 0.32–1.5% oligotype relative abundance across our 142 samples. We identified oligotypes as being broadly prevalent if found in >75% of samples versus those oligotypes narrowly prevalent in <10% of samples. R code is available at https://github.com/mc68462/medNS/blob/v0.3/oligotype_prevalence.R.
Network analysis
Network construction with SparCC
The aim of our network analysis was to determine whether we could identify recurrent modules of co-varying microbes. Owing to large gaps between sampling time points during winter and late spring (Supplementary Figure S1), we omitted 2010 data from network analysis. Separate unfiltered abundance matrices were extracted for 2011 and 2012 and contained 4250 and 4214 oligotypes, respectively. We performed three filtration steps prior to network construction independent of those described above for our analysis of oligotype prevalence: (1) removal of chloroplast and mitochondria oligotypes, (2) removal of oligotypes present in <10% of samples and (3) removal of oligotypes present only in a single year through an intersection of 2011 and 2012 data sets. In step 1 filtration of chloroplasts and mitochondria, we removed 404 and 391 oligotypes (6.4 and 6.2% of total amplicon abundance) in 2011 and 2012 data sets, respectively. In step 2 filtration of low prevalence oligotypes, we removed an additional 85 and 105 oligotypes (0.1% and 0.1% of total amplicon abundance), respectively; low prevalence oligotypes removed were thus present in very low abundance. In the step 3 intersection of 2011 and 2012 data sets, we removed 97 and 54 total oligotypes (0.5 and 0.2% of total amplicon abundance) respectively; oligotypes unique to a single year were thus also present in very low abundance. We performed step 2 filtration to remove rare oligotypes, which might introduce artifacts in the network inference (Berry and Widder, 2014). We performed step 3 filtration because the downstream identification of consensus network modules required the same set of oligotypes to be used in both 2011 and 2012 networks. A total of 3664 oligotypes were retained for the construction of 2011 and 2012 networks. R code is available at https://github.com/mc68462/medNS/blob/v0.3/pre_processing.R
Following filtration, we constructed two independent networks for 2011 and 2012 using Sparse Correlations for Compositional data (SparCC) Friedman and Alm, 2012). SparCC first performs a log-ratio transformation of relative abundance. With compositional data like 16S rRNA genes, abundances are relative (that is, sum to 1); these proportions are thus not independent and do not capture the true correlations underlying absolute abundances (Pawlowsky-Glahn and Buccianti, 2011; Friedman and Alm, 2012). For example, a large increase in the abundance of one oligotype during a period of low diversity necessarily affects the relative abundance of all other oligotypes resulting in correlations (often negative) that reflect the compositional nature of the data, and not the underlying biological processes (Aitchison, 2003; Friedman and Alm, 2012). In log-ratio transformed data the ratio of the proportions of two oligotypes is independent of which other oligotypes are included in the analysis, and thus maintain ‘subcompositional coherence’. We first implemented SparCC.py, which performs a log-ratio transformation and then estimates compositionality-robust Pearson correlations between all pairs of oligotypes using the average of 10 iterations sampled from the Dirichlet distribution. We then performed 100 bootstraps using MakeBootstraps.py and re-ran SparCC.py with 10 iterations to be used in the calculation of two-sided pseudo-P-values. The resulting networks are comprised of oligotypes represented as vertices and SparCC correlations represented as undirected weighted edges.
Consensus module detection with WGCNA
Following network construction with SparCC, we constructed a topological overlap matrix (TOM) using scripts from the WGCNA R package (Langfelder and Horvath, 2008) as the first step to identify modules of co-varying oligotypes conserved across both 2011 and 2012 SparCC networks (termed consensus modules). As SparCC correlations fall between −1 and 1 and TOM analysis requires values between 0 and 1, we converted the correlations using: 0.5*(cor+1). We performed TOMsimilarity for each network using TOMType=’unsigned’ and then performed a scaling step as in Langfelder and Horvath (2016). In the output TOM matrix, a value of 1 means two nodes are (1) connected and (2) share the same neighbors. A TOM value of 0 means two nodes are unconnected and share no neighbors. We then calculated a consensus TOM from the 2011 and 2012 TOMs using parallel minimum (pmin). If two nodes are highly connected in both years, the pmin TOM value for the node pair will be high. If two nodes are highly connected in only a single year, or if two nodes are unconnected in both years, the pmin TOM value will be low. The consensus TOM was then used as input for average-linkage hierarchical clustering using the hclust function and TOM dissimilarity (1-consensusTOM) to identify clusters as in Langfelder and Horvath (2016). We used cutreeDynamic as part of WGCNA to identify branch boundaries for consensus modules using deepSplit=2, cutHeight=0.995, and minClusterSize=10. It is during this step that oligotypes are assigned to modules if their TOM-based topology is similar in both years; those oligotypes lacking a conserved topology across both 2011 and 2012 SparCC networks are excluded from having membership to a particular module. The large gaps in the 2010 data set spanning 74 and 51 days in winter and late spring, respectively, would have been problematic for this consensus TOM step where inconsistent topology might have arisen from missing data points.
Following module detection, we calculated the module eigengene (ME) (that is, first principal component) for each module using the WGCNA script multiSetMEs, which performs a principal component analysis using the oligotype abundance profiles of each module. We used centered log-ratio (clr) transformed abundance data with a pseudocount of 1e-6 during this step to ensure subcompositional coherence using the R package compositions (van den Boogaart et al., 2014). The calculated MEs can then serve as a representative of each module’s collective abundance profile. We then again performed average-linkage clustering of the MEs using consensusMEDissimilarity and hclust to identify modules sufficiently similar for merging using mergeCloseModules with default cutHeight=0.25. New MEs of merged modules were calculated within this merging step. An overview of our network methods can be found in Supplementary Figure S2. SparCC correlation and P-value matrices are available at https://github.com/mc68462/medNS/tree/v0.3/data alongside WGCNA R code at https://github.com/mc68462/medNS/blob/v0.3/wgcna.R. Network graphs were filtered using igraph and visualized with Gephi (Bastian, Heymann and Jacomy, 2009).
Detection of seasonally structured modules
Using each of the MEs from 2011 and 2012, we performed a redundancy analysis (RDA) to extract variation in the MEs that could be explained by the measured physicochemical variables. Because we performed RDA on the MEs and not the OTU abundance data directly, we refer to this method as an indirect gradient analysis (Legendre and Legendre, 1998; Buttigieg and Ramette, 2014a). Prior to RDA, physicochemical variables were standardized to Z-scores using the R package vegan (Oksanen et al., 2015). Spearman correlations and P-values between MEs and environmental variables were performed with the base R function cor.test from the stats package (R Core Team, 2008). P-values were corrected for multiple testing using the base R function, p.adjust, employing the method of Benjamini and Hochberg (1995). MEs and physicochemical variables measured in this study display seasonal patterns over time. We define season boundaries according to their astronomical definition.
Intersection of 2011 and 2012 module networks
Consensus module detection with WGCNA identifies clusters of oligotypes that are connected and share similar neighbors across both years. To quantify the degree to which correlations between oligotypes are similar within a particular module network in 2011 versus 2012, we performed a graph intersection using the R package igraph (Csardi and Nepusz, 2006). We first extracted the SparCC networks for each consensus module and then performed a filtration step based on the strength of the SparCC correlation between oligotypes. Rather than apply a single arbitrary threshold across all modules, we performed an analysis to determine at which threshold (>abs(r)=0.5) we could trim the edges of each individual module network without losing connectedness with the main graph component (Žure et al., 2017). In other words, we wanted to maximize edge filtration without breaking apart the entire module network. We tested a range of absolute value thresholds from 0 to 1 using igraph. Following the filtration of edge weights, we performed graph intersection to obtain a single network graph per consensus module containing only vertices and edges that are strongly correlated in both 2011 and 2012.
Results
Physicochemical and phytoplankton data
Chl a concentrations reveal a succession of three prominent bloom periods in 2010 with progressively weaker and delayed chl a peaks in 2011 and 2012 (Supplementary Figure S1). Overall diatoms tend to dominate the climax of spring and summer blooms, with the exception of marine raphidophyte Chattonella, which conspicuously replaces diatoms in the spring of 2012. Although the absolute abundance of these major phytoplankton groups is valuable, there are vast differences in cell sizes (see Teeling et al., 2016). Abundance is thus not a reliable indicator of how much each phytoplankton group contributes to the organic substrate pool. We take chl a as a proxy for the eutrophic conditions characteristic of phytoplankton blooms.
Bacterioplankton diversity
Patterns of recurrence in oligotype relative abundance were observed across the three sampled years. Supplementary Figure S3 showcases the oligotype diversity, recurrence and seasonality of major clades. Alphaproteobacteria (A), Gammaproteobacteria (B) and Flavobacteriia (C) comprise 73% of all oligotypes. This value might be higher were it not for the under-representation of SAR11, resulting from primer bias. All three classes increase in relative abundance each spring and summer with markedly higher oligotype diversity within Gammaproteobacteria and Flavobacteriia. Supplementary Figure S3 D-I also includes additional classes that are recurrent, but occur in relatively lower abundances. Acidimicrobiia (D), Betaproteobacteria (E) and Verrucomicrobia (H) are present throughout much of the year. Euryarchaeota (F) and Planctomycetes (I) appear later in the year, whereas Epsilonproteobacteria (G) appear very briefly each year during bloom seasons. Overall, oligotype abundance patterns of all major clades exhibit striking patterns of recurrent oligotype diversity, with a notable delay in the timing of the bacterioplankton response to spring phytoplankton blooms in 2011 and 2012 compared with 2010 within bloom responders Alphaproteobacteria, Gammaproteobacteria and Flavobacteriia.
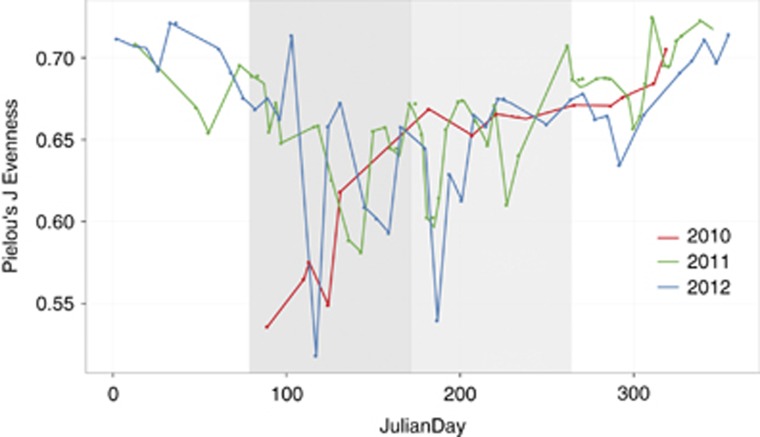
We interpolated and extrapolated alpha diversity using the R package iNEXT (Chao et al., 2014; Hsieh et al., 2016). Asymptotic diversity estimates alongside related statistics are included in Supplementary Table S5. Rarefaction curves calculated from proportional oligotype richness data (q=0) fell between 1249 and 2879 and did not reach saturation (Supplementary Figure S4A). However, exponential Shannon (expShannon) and inverse Simpson (invSimpson) diversity estimates revealed much reduced effective diversity. ExpShannon estimates reported 47–307 effective species and invSimpson estimates reported 6–96 effective species throughout the year (Supplementary Figure S4B). Each spring and summer, invSimpson estimates fall as low as 6 and 14 total species, respectively. Pielou’s J evenness displays this skew in alpha diversity each spring and summer, followed by a return to winter baseline levels of evenness (Figure 1). Overall our analyses indicate that the vast majority of the annual bacterioplankton diversity at Helgoland can be explained by fewer than 100 effective oligotypes, as per the maximum effective diversity calculated with invSimpson. The top 100 most abundant oligotypes comprise 64% of the total 16S rRNA gene amplicon abundance. However, the under-representation of SAR11 in our datasets might have deflated measures of alpha diversity compared with previous work employing the same principle of oligotyping at a coastal sampling site (Eren et al., 2013; Fuhrman et al., 2017). Supplementary Figure S5 displays the relative abundances of the three SAR11 oligotypes we identified (reaching a minimum abundance of 1%) detected with our primers. Our previous study employing SAR11-specific fluorescence in situ hybridization probes (FISH) revealed that SAR11 peaked at ~50% of community abundance shortly prior to spring bloom events; our 16S rRNA gene abundances during this same time period top off at ~8% relative community abundance. Although SAR11 abundance is significantly under-represented in our data set, the pattern over time recapitulates FISH counts from the spring study of Teeling et al., 2016.
Figure 1.
Spring and summer harbor communities of skewed evenness. Pielou’s J metric of evenness is represented on the y axis and Julian days on the x axis. A value of 1.0 represents total community evenness. Spring and summer are denoted by the dark and light gray areas, respectively. Line colors denote year: red=2010, blue=2011, green=2012.
Patterns of oligotype prevalence
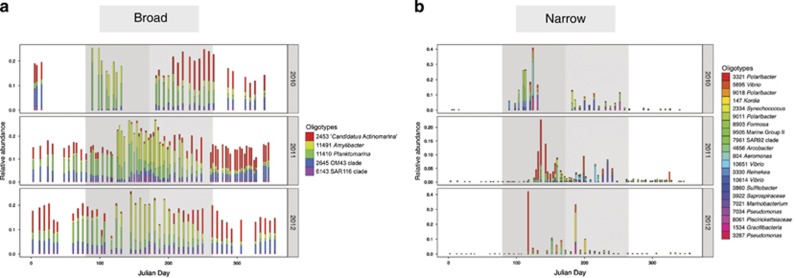
Examining the prevalence of a given oligotype revealed two groups marked by broad and narrow distributions over time (Supplementary Figure S6,Supplementary Table S6). A clear partition is seen for five abundant and broadly prevalent oligotypes present in >75% of samples and include: actinobacterial ‘Candidatus Actinomarina’ oligotype_2453, alphaproteobacterial Amylibacter oligotype_11491, Planktomarina oligotype_11410 and the SAR116 clade oligotype_6143, and the betaproteobacterial OM43 clade oligotype_2645. These five oligotypes occupy ~20% of the total community relative abundance throughout the year (Figure 2a). Planktomarina oligotype_11410 is 100% identical to the formally described and highly abundant Planktomarina temperata RCA23 strain isolated from the southern North Sea (Giebel et al., 2011, 2013), and is known to occupy a large proportion of temperate and polar water masses (Selje et al., 2004). In total we identified 298 oligotypes enriched during spring and summer, which display a narrow prevalence comprising a diverse collection of primarily Gammaproteobacteria and Flavobacteriia (Figure 2b). Narrowly prevalent Gammaproteobacteria include Vibrio oligotypes 5695, 10651 and 10614, SAR92 clade oligotype_7961, Reinekea oligotype_3330 and Aeromonas oligotype_804. Narrowly prevalent Flavobacteriia include Polaribacter oligotypes 3321 and 9018, Kordia oligotype_147 and Formosa oligotype_8903.
Figure 2.
Oligotypes show distinct patterns of prevalence each year. Relative abundance plots show 'broad' (a) and 'narrow' (b) categories of oligotype prevalence patterns. Julian days are represented on the x axis and oligotype relative abundance is represented as a fraction on the y axis. Spring and summer are denoted by the dark and light gray areas, respectively. Broad oligotypes are defined by >75% prevalence and narrow oligotypes are defined by <10% prevalence. All five broad oligotypes identified in Figure S6 are shown in panel A and collectively occupy ~20% of the total community throughout the year. Narrow oligotypes reaching an abundance of 5% in at least one sample are shown in panel B and are enriched in spring and summer. Julian days with no bar indicate the absence of a sample point. Table S6 contains a full list of identified broad and narrow oligotypes.
Oligotype switching
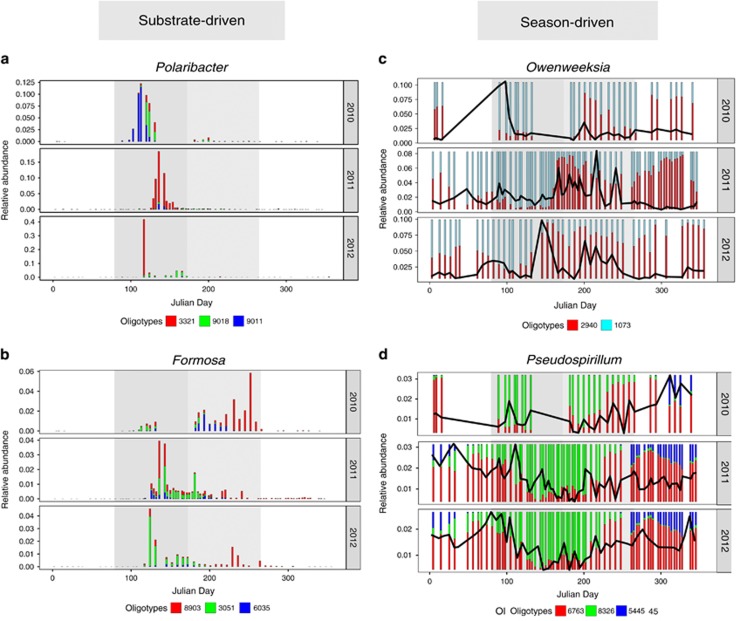
Algal-derived organic biomass dominates spring and summer substrate sources as proxied by chl a. During spring and summer, oligotypes with 99.5% identity within the flavobacterial genera Polaribacter (Figure 3a) and Formosa (Figure 3b) exhibited rapid shifts in dominance, potentially driven by phytoplankton-derived substrate supply. Genera Owenweeksia VIS6 (Flavobacteriia) and Pseudospirillum (Gammaproteobacteria) exhibited seasonally driven changes in oligotype dominance with a more gradual sinusoidal abundance pattern. In late spring/early summer, the dominant Owenweeksia oligotype_1073 switched to oligotype_2940 (Figure 3c), with which it is 98.4% identical, and three oligotypes classified as Pseudospirillum share 97.4% identity but exhibit recurrent seasonal patterns (Figure 3d). These examples display the ability of MED to resolve subtle and ecologically meaningful seasonal shifts in diversity.
Figure 3.
Season and phytoplankton substrate-driven oligotype switching is observed between oligotypes with >97% identity. Relative abundance bar plots show examples of switches in oligotype dominance. Julian days are represented on the x axis and total oligotype relative abundance is represented as a fraction on the y axis. Spring and summer are denoted by the dark and light gray areas, respectively. Flavobacteriia genera Polaribacter (a) and Formosa (b) exhibit potential phytoplankton substrate-driven changes in dominance over short time intervals between oligotypes with 99.5% identity. Genera Owenweeksia, (c, Flavobacteriia) and Pseudospirillum (d, Gammaproteobacteria) exhibit sinusoidal seasonal changes in dominance between oligotypes with >97% identity, visible when relative abundance is scaled to 1 (stacked bars). The total cumulative relative abundance of displayed oligotypes in c and d is represented by the black line (y axis).
Identification of recurrent and seasonally structured module networks
The goal of our network analysis was to identify whether oligotypes could be found within recurrent sub-networks. The TOM-based method we used in WGCNA consensus module detection takes into account correlations and topology to identify clusters of oligotypes that are connected and shared similar neighbors in both 2011 and 2012 SparCC networks. Oligotypes lacking conserved topology would correspondingly lack module membership, however, all 3664 network oligotypes were found within consensus modules. During module detection, we chose to merge similar modules based on the dissimilarity of their MEs (see Materials and Methods). Prior to merging, we identified 11 consensus modules and after merging we obtained a total of seven modules (Supplementary Figure S7). Despite our setting allowing a minimum module size of 10, all detected merged modules contained a large number of oligotypes with between 254 and 628 members (Table 1); pre-merged modules contained between 76 and 622 member oligotypes (data not shown). The vast majority of SparCC correlations between oligotypes within each module were positive; negative correlations tended to be weak (abs(r)<0.5) (Supplementary Figure S8). Given the low values of inverse Simpson diversity in spring and summer (6 and 14, respectively), the use of a compositionality-robust network construction method was particularly important to avoid spurious negative correlations during these periods of low diversity (Weiss et al., 2016).
Table 1. Module network graph statistics.
| Module | Season | Total oligotypes | SparCC cor threshold 2011a | SparCC cor threshold 2012a | Intersecting vertices (%)b | Intersecting edges (%)b |
|---|---|---|---|---|---|---|
| 2 | Autumn | 584 | 0.51 | 0.58 | 72.6 | 44.7 |
| 7 | Winter | 458 | 0.53 | 0.55 | 74.9 | 48.7 |
| 5 | Winter–Spring | 628 | 0.51 | 0.56 | 74.6 | 35.9 |
| 1 | Spring | 622 | 0.61 | 0.61 | 72.7 | 40.2 |
| 4 | Spring–Summer | 538 | 0.62 | 0.57 | 74.1 | 48.7 |
| 3 | Summer | 580 | 0.52 | 0.59 | 53 | 32 |
| 8 | Summer–Autumn | 254 | 0.5 | 0.55 | 67 | 48.2 |
SparCC correlation thresholds were determined according to an analysis of network deconstruction across different thresholds (Supplementary Figure S12).
Intersecting vertices and edges were calculated using igraph (Csardi and Nepusz, 2006).
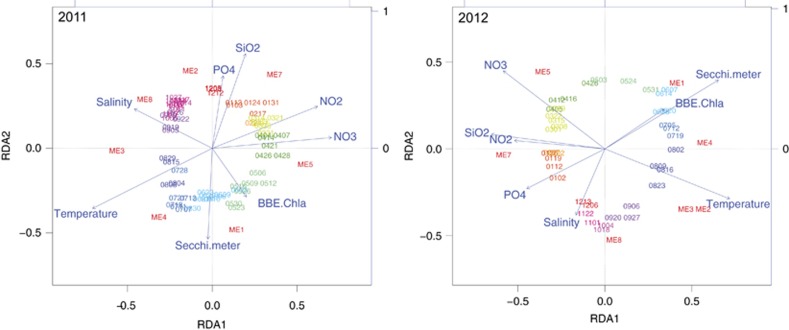
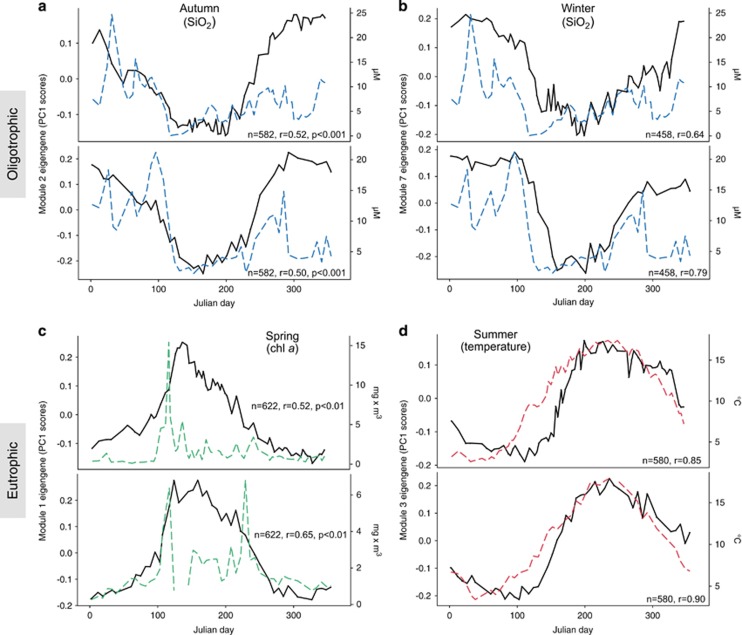
The construction of modules allowed us to identify oligotypes that co-vary. Using MEs of each module, we performed an RDA against physicochemical variables. RDAs revealed a repeating cyclical seasonal pattern of MEs and sample time points across 2011 and 2012 (Figure 4). Modules are differentially driven by oligotrophic and eutrophic conditions (Supplementary Table S7). Positive correlations with inorganic nutrients (PO43−, SiO2, NO2− and NO3−) indicate that heterotrophs are likely subsisting on low concentrations of low molecular weight organic compounds during periods of low primary productivity, whereas positive correlations to chl a indicate a community of heterotrophs that can reach high abundance during periods rich in high molecular weight phytoplankton-derived organic compounds. In Figure 5 we display the MEs of module 2, 7, 1 and 3 against the environmental variable with which they are most strongly and/or significantly correlated. MEs serve as a proxy for the collective abundance of its member oligotypes and for most modules, their pattern closely tracks with seasonal fluctuations of environmental variables. Module diversity (Figure 6) mirrors the seasonal shifts we identified with measures of alpha diversity (Figure 1 and Supplementary Figure S4). We describe each module based on the season in which member oligotypes appear in the highest abundance. Modules of autumn (module 2) and winter (module 7) tend to harbor more diverse and even communities driven by oligotrophic conditions, whereas modules of spring (module 1) and summer (module 3) tend to be driven by eutrophic conditions and harbor fewer oligotypes that reach high relative abundance (Figure 6).
Figure 4.
Module eigengenes (MEs) are structured by seasonally fluctuating environmental variables. Redundancy analysis (RDA) of MEs against measurements of physicochemical variables revealed a repeating cyclical seasonal pattern of MEs and sample time points across 2011 (left panel) and 2012 (right panel). Sample time points are denoted by date (mmdd). MEs are differentially driven by oligotrophic conditions (high inorganic nutrient concentrations: PO43−, SiO2, NO2− and NO3−), eutrophic conditions (high chl a and Secchi disk depth), salinity and/or temperature. Sample points are colored by month: red shades denote winter, green denotes spring, blue denotes summer and purple denotes autumn.
Figure 5.
Module eigengenes (MEs) display recurrent correlations with environmental variables across 2011 and 2012. MEs (black lines) serve as a proxy for the collective abundance of its member oligotypes and are overlaid with their most strongly correlated environmental variable (dashed lines) (a–d) across Julian days represented on the x axis. Inorganic nutrients are denoted by blue dashed lines, chlorophyll a by green dashed lines and temperature by red dashed lines. ME trends are represented on the left y axis and environmental variables on the right y axis. n and r correspond to the total number of module oligotypes and Spearman correlation, respectively. P-values (p) are adjusted employing the method of Benjamini and Hochberg (1995).
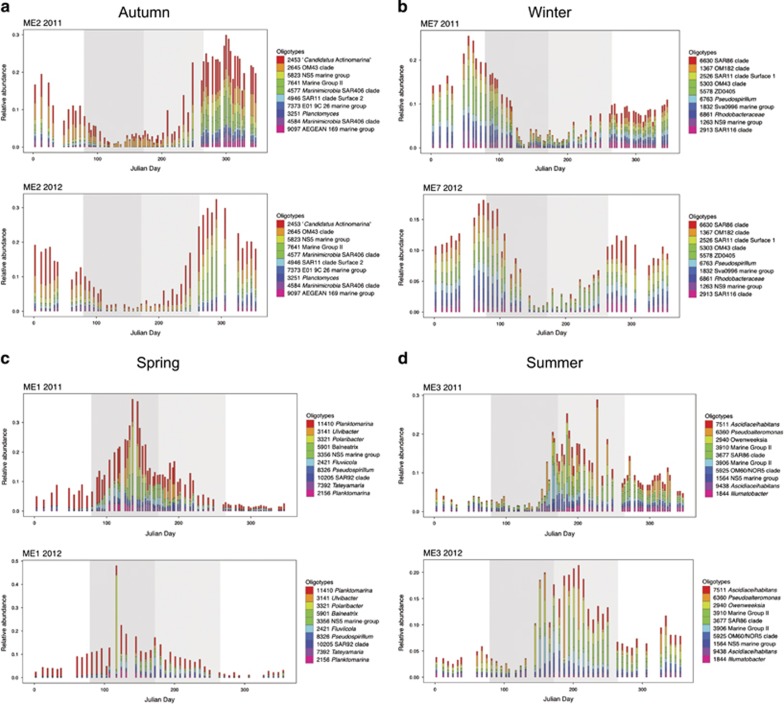
Figure 6.
Module oligotype profiles reveal shifts in diversity patterns depending on the presence of bloom conditions. Relative abundance plots (a–d) show four of the seven identified consensus modules and display the top 10 most abundant module oligotypes. Modules are described according to their season of highest abundance: ME2=‘Autumn’ (a), ME7=‘Winter’ (b), ME1=‘Spring’ (c) and ME3='Summer' (d). Julian days are represented on the x axis and relative abundance is represented as a fraction on the y axis. Spring and summer are denoted by the dark and light gray areas, respectively.
Here we highlight module dynamics characteristic of winter, spring, summer and autumn; Supplementary Figures S9 and S10 contain corresponding plots for all modules. Winter module 7 correlates to oligotrophic conditions marked by high concentrations of inorganic nutrients like SiO2 (Figure 5b) and is represented by equal relative abundances of gammaproteobacterial Oceanospirillales SAR86 clade oligotype_6630 and OM182 clade oligotype_1367, alphaproteobacterial SAR11 clade oligotype_2526 and betaproteobacterial OM43 clade oligotype_5303 (Figure 6b). Spring module 1 is driven by increases in chl a concentrations as active bloom conditions commence (Figure 5c) and is dominated by alphaproteobacterial Planktomarina oligotype_11410, flavobacterial Ulvibacter oligotype_3141 and Polaribacter oligotype_3321, and gammaproteobacterial Balneatrix oligotype_5901 (Figure 6c). The diversity in this spring module is highly skewed by the high relative abundance of Planktomarina and particularly by Polaribacter, which has previously been shown to display copiotrophic growth on algal substrates (Cottrell and Kirchman, 2016). Polaribacter is narrowly prevalent at abundances of 15–40% of the total community (Figure 2b and 3a). Summer module 3 displays a relationship with temperature (Figure 5d), and we begin to observe an increase in community diversity and evenness (Figure 6d). Alphaproteobacterial Ascidiaceihabitans oligotype_7511, flavobacterial Owenweeksia oliogotype_2940 (Figure 3c) and euryarchaeotal Marine Group II (Euryarchaeota) oligotypes 3910 and 3906 comprise the more abundant community members. Although community evenness has partially rebounded since spring, it is within this module that we find the gammaproteobacterial Pseudoalteromonas oligotype_6360 repeatedly reaching 10–20% of the relative community abundance from an abundance near zero (Figure 6d). Prior to module merging (see Materials and Methods), this Pseudoalteromonas oligotype was clustered into module 11 alongside other Gammaproteobacteria (for example, Vibrio and Arcobacter), which display a similar feast-and-famine abundance pattern (Supplementary Figure S11). Autumn module 2 marks a return to oligotrophic conditions characterized by correlations with inorganic nutrients (Figure 5a). Autumn module 2 is dominated by actinobacterial ‘Candidatus Actinomarina’ oligotype_2453 (Figure 6a). Community evenness is slightly skewed by this abundant oligotype, but overall, the community begins a reset to winter baseline conditions of high evenness driven by oligotrophic conditions.
Conservation of module network topology
To evaluate the conservation of module network topology across 2011 and 2012, we extracted the unfiltered SparCC networks for each module and filtered out weak correlations. We employed different correlation thresholds for each module in order to preserve module network integrity (Table 1). The visualization of network graph deconstruction across different correlation thresholds reveals information about how increasing filtration stringency affects graph structures (Supplementary Figure S12). Most modules exhibit a clear threshold where the networks break up into multiple smaller components, as evidenced by the sharp peak in the number of individual graph components appearing between SparCC correlation values of 0.5 and 0.75. P-values for filtered graphs were all ⩽0.001 with the exception of a handful at ⩽0.01. Individual filtered modules from 2011 and 2012 are shown in Supplementary Figure S13. Network graphs make clear the seasonal dynamics of module diversity with even community abundances during autumn and winter and highly skewed abundances spanning spring and summer. Module networks are conserved, even during highly dynamic periods in spring and summer; the same abundant oligotypes are found across both years with similar patterns of topology.
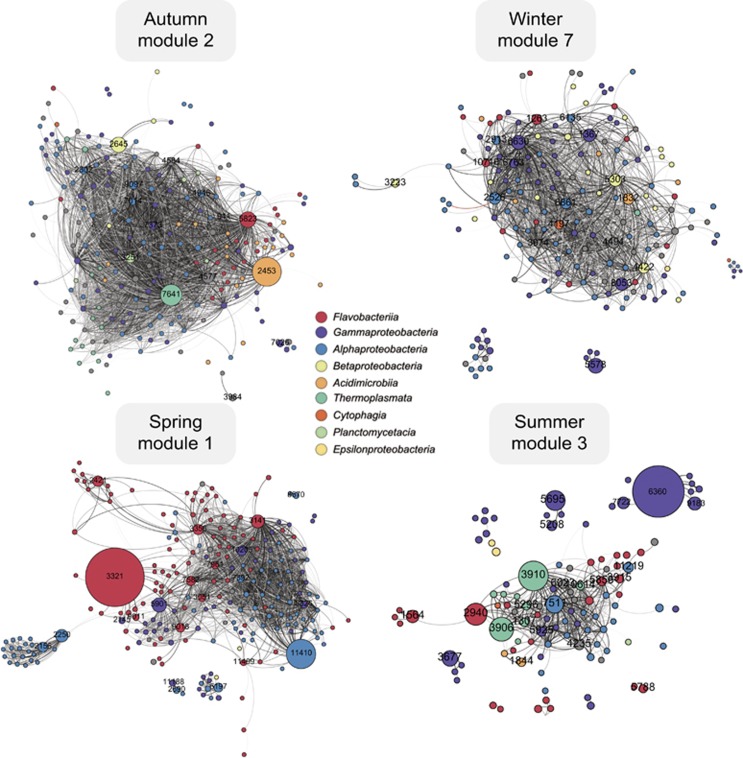
In order to quantify the degree of similarity between 2011 and 2012 filtered module network correlations, we performed a graph intersection, which is the equivalent of retaining the overlapping regions of a Venn diagram. Intersected module networks are depicted in Figure 7 and Supplementary Figure S14, and represent oligotypes and correlations that are conserved in both years. Between 53 and 75% of vertices and between 32 and 49% of edges were shared across 2011 and 2012 (Table 1). Overall, this indicates that module correlation structures are similar across years, sharing on average 70% of vertices and 43% of edges. However, summer module 3 has the lowest overlap in shared network edges and vertices (Supplementary Figure S15), and contains disconnected graph components largely comprised of Gammaproteobacteria oligotypes (Figure 7). Network topology suggests that correlation patterns within the summer module are less conserved across years.
Figure 7.
The intersection of module networks from 2011 and 2012. Individual module networks from 2011 and 2012 were filtered to exclude edges with weak SparCC correlations (Table 1), followed by the removal of vertices which became disconnected from all other vertices (degree=0). Corresponding filtered module networks were then intersected using igraph (Csardi and Nepusz, 2006) to obtain a single graph representative of oligotypes that are strongly correlated across 2011 and 2012 module networks. The percentage of shared vertices and edges is shown in Table 1. Vertex size is represented as the average maximum observed oligotype abundance in 2011 and 2012 and is labeled with an oligotype id if that abundance is >1%. Vertices are colored by class-level taxonomy. Oligotype taxonomic identities can be found in Supplementary Table S4. Edges are colored from light gray to black to denote weak to strong SparCC correlations, respectively. Decreases in community evenness can be seen in bloom compared with non-bloom seasons.
Discussion
Resiliency and microdiversity of microbial communities in the North Sea
We examined the dynamics of free-living bacterioplankton communities at Helgoland Roads over three consecutive years using highly resolved 16S rRNA gene oligotypes. Throughout the course of our study, annual patterns of microdiversity within the free-living bacterioplankton community appeared largely conserved from year to year (for example, Supplementary Figure S3). We used network module eigengenes (MEs) to represent potential guilds of co-varying bacterioplankton, the members of which may exploit similar resources, may interact or may tolerate similar environmental conditions. MEs showed recurrent correlations with seasonally fluctuating environmental parameters across 2011 and 2012 that were broadly defined by oligotrophic and eutrophic conditions (Figure 5). However, as previously found in marine environments (Eren et al., 2013; Ward et al., 2017), we identified seasonal patterns of microdiversity. For example, Cryomorphaceae Owenweeksia-classified oligotypes 1073 and 2940 share 98.4% sequence identity but display seasonality in oligotype dominance (Figure 3c), and they are found within two separate module networks (Figure 6).
Higher frequency shifts in oligotype dominance over short time intervals within Flavobacteriia Polaribacter and Formosa (Figures 3a and b), are more likely owing to subtle shifts in algal-derived substrate that cannot be resolved by measures of chl a. There is evidence that as in the human gut (Koropatkin et al., 2012), polysaccharides can shape marine microbial niches upon their release from algal cells (Xing et al., 2015). Even subtle differences in metabolic pathways and enzyme deployment strategies can differentiate niches of organisms that consume the same algal-derived substrate (Hehemann et al., 2016). Endeavors to evaluate the response of bacterioplankton to specific types of dissolved organic matter molecules suggests that an improved resolution in the characterization of organic substrate is required to uncover the drivers of these more fine-scale shifts in diversity during blooms (Lucas et al., 2016).
In an analysis of prevalence, we identified a distinct grouping of broadly and narrowly abundant oligotypes, indicating the ability of some taxa to exploit resources available throughout much of the year versus those specialized on more ephemeral resource patches. For example, broadly prevalent oligotype_11410 (Figure 2a) is 100% identical to the Planktomarina temperata RCA23 strain (Giebel et al., 2013) known to widely occupy temperate and polar water masses (Selje et al., 2004). Although strains with identical 16S rRNA genes can harbor widely different gene content, oligotype_11410 may correspond to P. temperata RCA23, which was originally isolated near our sampling site in the southern North Sea (Giebel et al., 2011). Genomic analyses suggest that P. temperata RCA23 is a versatile strain with a high proportion of ATP-binding cassette family transporters (Voget et al., 2015) and is capable of aerobic anoxygenic photosynthesis (Giebel et al., 2013). The RCA23 strain thus appears well adapted to nutrient-poor conditions with a relatively streamlined genome compared with other members of the Roseobacter clade, but has also demonstrated high in situ activity across nearly its entire genome during high-nutrient spring bloom conditions in the North Sea (Voget et al., 2015). If this strain does in fact correspond to oligotype_11410, its genomic capabilities would explain the broad prevalence patterns we observe during oligotrophic and eutrophic conditions, even as more copiotrophic Flavobacteriia like the narrowly prevalent Polaribacter oligotypes rise in abundance. Here the partition of niche space likely derives from differential preferences for high molecular weight and low molecular weight compounds; alphaproteobacterial P. temperata might target low molecular weight molecules via ATP-binding cassette transporters, whereas flavobacterial Polaribacter might deploy specialized carbohydrate-active enzymes and transport strategies for the rapid uptake of high molecular weight algal polysaccharides (Xing et al., 2015; Reintjes et al., 2017).
The predictable response of the network modules to environmental variables reveals that each year, resilient autumn and winter bacterioplankton communities re-emerge following dynamic spring and summer bloom disturbance events. Moreover, despite differences in the composition of blooming phytoplankton in 2011 and 2012, there appears in both years a similarly structured secondary bloom within the free-living bacterioplankton community with a marked drop in the conservation of summer module topology across years.
Ecological strategies during spring and summer
Given the patterns of recurrence we have identified across 2011 and 2012, we present a hypothesis regarding the differences in diversity, dynamics and network topology we observed in spring versus summer communities. During winter at Helgoland, system-level productivity is low (for example, low chl a (Supplementary Figure S1) and total cell counts (Supplementary Figure S16)). As viruses and grazers require a critical encounter frequency with their prey to proliferate (Wilcox and Fuhrman, 1994; Cram et al., 2016), Teeling et al., (2016) proposed that spring Helgoland bacterioplankton communities might be subject to less predation following grazer suppression in winter. The KtW (kill-the-winner hypothesis) predicts a balance between bottom-up competition for growth during periods of low productivity and defense against top–down predators during periods of high productivity (Thingstad and Lignell, 1997; Thingstad, 2000; Våge et al., 2013; Thingstad et al., 2014). We hypothesize that spring bacteria responding to the first releases of algal-derived organic matter are competitive for the fresh substrates and capable of rapid growth before grazers and viruses targeting their populations take hold. In contrast, each summer is marked by high productivity (maximal chl a and total cell counts) and thus high potential grazing pressure (KtW). Furthermore, there is evidence from aquatic systems that an increase in predator richness has a positive effect on bacterial richness and evenness (Saleem et al., 2012) stemming primarily from the widespread distribution of resources and resulting niche complementarity (Tilman et al., 1997; Loreau, 1998; Cardinale et al., 2002). The result is an increase in ecosystem biomass and richness (Mulder et al., 2001). We thus hypothesize that the increase in bacterioplankton community diversity and evenness in summer might arise from an increase in grazing diversity and pressure. Any discussion of diversity and evenness must, however, consider the inevitable PCR-bias of the primer set used for sequence retrieval, in our case suppressing SAR11.
Flavobacteriia and Gammaproteobacteria are both specialized in the degradation of algal-derived high molecular weight organic matter, are both enriched during diatom-dominated blooms and both display copiotrophic capabilities (Pernthaler and Amann, 2005; Cottrell and Kirchman, 2016). However, their abundance patterns differ between spring and summer. Flavobacteriia and Gammaproteobacteria in spring of 2011 and 2012 tend to maintain their abundance for several weeks and do not reappear in summer (Figure 6c). Conversely, summer copiotrophic Gammaproteobacteria appear to be governed by a different set of underlying dynamics that result in a pronounced feast-and-famine lifestyle. Gammaproteobacteria have been shown to be opportunistic in nature, harboring the ability to rapidly respond to algal-derived substrates from low abundance (Pernthaler and Amann, 2005). They are also known to be subject to higher viral pressure than their aerobic anoxygenic phototrophic counterparts (for example, Bacteroidetes and alphaproteobacterial Roseobacter and SAR11) (Ferrera et al., 2011). In our data set, Pseudoaltermonas oligotype_6360 reaches high abundance in summer module 3 in both years (Figure 7) and exhibits repeated cycles of rapid growth and decline, much like Psychrobacter oligotype_5208 and Vibrio oligotype_5696 (Supplementary Figure S11), which is also known to be susceptible to lytic phage infection (for example, Kellogg et al., 1995). Our network analysis revealed that these Gammaproteobacteria oligotypes tend to cluster together in a loose connection to the main network graph (Figure 7, Supplementary Figure S13). Although many marine clades are subject to host-phage dynamics that shape diversity abundance over time (Needham et al., 2017), we hypothesize that frequency-dependent phage infections (KtW) (Thingstad and Lignell, 1997; Thingstad, 2000) might contribute to the irregular abundance patterns we have identified in summer Gammaproteobacteria. Such dynamics might explain the destabilization of network conservation across the 2 years investigated here. These shifts in abundance patterns from spring to summer might stem in part from a shift in temperature optima of distinct bacterial taxa as suggested previously (Lucas et al., 2015), but we hypothesize that it might also arise from the shift in the ecological strategies required for proliferation under an increasingly complex and dense web of biochemical activity and top–down pressure.
Conclusion and outlook
We conclude that cycles of deterministic selection pressure driven by stable seasonal forcing has, through the induction of recurrent patters in resource availability, predator–prey dynamics and microbial interactions, allowed for the assemblage of largely stable and resilient free-living microbial communities throughout both non-bloom and bloom conditions. Although we expected the community to return to its stable winter state following bloom events, we did not expect communities with such similar network topologies to emerge during dynamic bloom events. However, we acknowledge that the two years used in network analysis might have been two considerably similar years with regards to the timing of the onset of primary and secondary blooms, which were delayed in 2011 and 2012 compared with 2010. Additional years of data are required in order to capture and predict the more complete range of possible bloom scenarios and community successions that respond to differences in the extent of grazer die-off in winter, bloom timing and phytoplankton composition. Furthermore, the diversity, dynamics and resilience of the particle-attached bacterioplankton fraction are likely very different from that of the free-living fraction measured here. Molecular characterization of particle-attached bacteria and eukaryotic phytoplankton is required to evaluate resilience and identify potential interactions across size fractions and between trophic levels. Permanent, automated systems for global ocean sampling offer a solution for this type of data collection (for example, Davies and Field, 2012; Soltwedel et al., 2013; Davies et al., 2014), which is essential to effectively monitor, describe, model and manage ocean ecosystem diversity. In combination with methods like MED to resolve patterns of microdiversity based on the 16S rRNA gene, these long-term ecological monitoring sites provide opportunities to detect previously undescribed archaeal and bacterial diversity (Buttigieg and Ramette, 2014b).
Nevertheless, in this study we have highlighted the value of multi-year studies in identifying recurrent modular community associations in a dynamic coastal environment. Our use of a sub-compositionally coherent network construction method affords a higher level of confidence in the correlations we measured here between oligotypes, particularly during the low diversity periods of spring and summer, setting forth a foundation for the exploration of ecologically meaningful associations between community members. The modules we present thus serve as targets for further inquiry including genomic characterization and laboratory-based experiments in order to elucidate the mechanisms and potential microbial interactions underlying recurrent patterns of microdiversity.
Acknowledgments
Special thanks to Bernhard Fuchs for assistance in the discussion and revision and to Karen Krüger and T Ben Francis for proofreading. PLB is supported by the European Research Council Advanced Investigator grant ABYSS 294757 to Antje Boetius. AF-G is supported by the European Union’s Horizon 2020 research and innovation program (Blue Growth: Unlocking the potential of Seas and Oceans) under grant agreement no. (634486) (project acronym INMARE). This study was funded by the Max Planck Society. Further support by the Department of Energy Joint Genome Institute (CSP COGITO) and DFG (FOR2406) is acknowledged by HT (TE 813/2-1) and RA (Am 73/9-1).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Aitchison J. (2003) The statistical analysis of compositional data. Blackburn Press: Caldwell, NJ, USA. [Google Scholar]
- Apprill A, McNally S, Parsons R, Weber L. (2015). Minor revision to V4 region SSU rRNA 806 R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75: 29–137. [Google Scholar]
- Azam F, Ammerman JW. (1984) Flows of energy and materials in marine ecosystems. In: Fasham MJR (ed). Springer: Boston, MA, USA. [Google Scholar]
- Azam F, Fenchel T, Field J, Gray J, Meyer-Reil L, Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10: 257–263. [Google Scholar]
- Barberán A, Bates ST, Casamayor EO, Fierer N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6: 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M. (2009), An open source software for exploring and manipulating networks.
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 57: 289–300. [Google Scholar]
- Berry D, Widder S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol 5: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boogaart KG, Tolosana R, Bren M. (2014), compositions: Compositional data analysis.
- Pawlowsky-Glahn V, Buccianti A. (ed. (2011) Compositional data analysis: theory and applications. Wiley: Chichester, West Sussex, UK, pp 400. [Google Scholar]
- Buchan A, LeCleir GR, Gulvik CA, González JM. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12: 686–698. [DOI] [PubMed] [Google Scholar]
- Buttigieg PL, Ramette A. (2014. a). A guide to statistical analysis in microbial ecology: a community-focused, living review of multivariate data analyses. FEMS Microbiol Ecol 90: 543–550. [DOI] [PubMed] [Google Scholar]
- Buttigieg PL, Ramette A. (2014. b). Biogeographic patterns of bacterial microdiversity in Arctic deep-sea sediments (HAUSGARTEN, Fram Strait). Front Microbiol 5: 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale BJ, Palmer MA, Collins SL. (2002). Species diversity enhances ecosystem functioning through interspecific facilitation. Nature 415: 426–429. [DOI] [PubMed] [Google Scholar]
- Chao A, Gotelli NJ, Hsieh TC, Sander EL, Ma KH, Colwell RK et al. (2014). Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol Monogr 84: 45–67. [Google Scholar]
- Chow C-ET, Sachdeva R, Cram JA, Steele JA, Needham DM, Patel A et al. (2013). Temporal variability and coherence of euphotic zone bacterial communities over a decade in the Southern California Bight. ISME J 7: 2259–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell M, Kirchman DL. (2016). Transcriptional control in marine copiotrophic and oligotrophic bacteria with streamlined genomes. Appl Environ Microbiol 82: 6010–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram JA, Parada AE, Fuhrman JA. (2016). Dilution reveals how viral lysis and grazing shape microbial communities. Limnol Oceanogr 61: 889–905. [Google Scholar]
- Csardi G, Nepusz T. (2006). The igraph software package for complex network research. InterJournal, Complex Systems, pp 1695.
- Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA et al. (2015). Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517: 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N, Field D. (2012). Sequencing data: a genomic network to monitor Earth. Nature 481: 45. [DOI] [PubMed] [Google Scholar]
- Davies N, Field D, Amaral-Zettler L, Clark M, Deck J, Drummond A et al. (2014). The founding charter of the genomic observatories network. GigaScience 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducklow HW, Carlson CA. (1992). Oceanic bacterial production. Adv Microb Ecol 12: 113–181. [Google Scholar]
- Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG et al. (2013). Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4: 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Vineis JH, Morrison HG, Sogin ML. (2013). A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS One 8: e66643. [DOI] [PMC free article] [PubMed]
- Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. (2015). Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9: 968–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrera I, Gasol JM, Sebastián M, Hojerová E, Koblížek M. (2011). Comparison of growth rates of aerobic anoxygenic phototrophic bacteria and other bacterioplankton groups in coastal Mediterranean waters. Appl Environ Microbiol 77: 7451–7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Alm EJ. (2012). Inferring correlation networks from genomic survey data. PLoS Comp Biol 8: e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Cram JA, Needham DM. (2015). Marine microbial community dynamics and their ecological interpretation. Nat Rev Microbiol 13: 133–146. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. PNAS 103: 13104–13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel H-A, Kalhoefer D, Gahl-Janssen R, Choo Y-J, Lee K, Cho J-C et al. (2013). Planktomarina temperata gen. nov, sp. nov, belonging to the globally distributed RCA cluster of the marine Roseobacter clade, isolated from the German Wadden Sea. Int J Syst Evol Microbiol 63: 4207–4217. [DOI] [PubMed] [Google Scholar]
- Giebel H-A, Kalhoefer D, Lemke A, Thole S, Gahl-Janssen R, Simon M et al. (2011). Distribution of Roseobacter RCA and SAR11 lineages in the North Sea and characteristics of an abundant RCA isolate. ISME J 5: 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T et al. (2009). The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol 11: 3132–3139. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann J-H, Arevalo P, Datta MS, Yu X, Corzett CH, Henschel A et al. (2016). Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes. Nat Commun 7: 12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TC, Ma KH, Chao A. (2016). iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol 7: 1451–1456. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg CA, Rose JB, Jiang SC, Thurmond JM, Paul JH. (1995). Genetic diversity of related vibriophages isolated from marine environments around Florida and Hawaii, USA. Mar Ecol Prog Ser 120: 89–98. [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. (2012). How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. (2016). Tutorials for the WGCNA package for R: II Consensus network analysis of liver expression data, female and male mice.
- Langfelder P, Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre L, Le Fèvre J. (1989) Hydrodynamic singularities as controls of recycled versus export production in oceans. In: Berger WH, Smetacek VS, Wefer G. (ed). Productivity of the ocean: present and past. Wlley: Chichester, UK, pp 65–83. [Google Scholar]
- Legendre P, Legendre L. (1998) Numerical Ecology. 2nd edn. Elsevier: Amsterdam, The Netherlands. [Google Scholar]
- Loreau M. (1998). Biodiversity and ecosystem functioning: a mechanistic model. PNAS 95: 5632–5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H-P, Yeh Y-C, Sastri AR, Shiah F-K, Gong G-C, Hsieh C. (2016). Evaluating community-environment relationships along fine to broad taxonomic resolutions reveals evolutionary forces underlying community assembly. ISME J 10: 2867–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J, Koester I, Wichels A, Niggemann J, Dittmar T, Callies U et al. (2016). Short-term dynamics of North Sea bacterioplankton-dissolved organic matter coherence on molecular level. Front Microbiol 7: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J, Wichels A, Teeling H, Chafee M, Scharfe M, Gerdts G. (2015). Annual dynamics of North Sea bacterioplankton: seasonal variability superimposes short-term variation. FEMS Microbiol Ecol 91: fiv099. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- Martiny JBH, Jones SE, Lennon JT, Martiny AC. (2015). Microbiomes in light of traits: a phylogenetic perspective. Science 350: aac9323. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder CPH, Uliassi DD, Doak DF. (2001). Physical stress and diversity-productivity relationships: the role of positive interactions. PNAS 98: 6704–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham DM, Fuhrman JA. (2016). Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol 29: 16005. [DOI] [PubMed] [Google Scholar]
- Needham DM, Sachdeva R, Fuhrman JA. (2017). Ecological dynamics and co-occurrence among marine phytoplankton, bacteria and myoviruses shows microdiversity matters. ISME J 11: 1614–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB et al. (2015), vegan: Community ecology package. R package version 2.3-0.
- Parada AE, Needham DM, Fuhrman JA. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18: 1403–1414. [DOI] [PubMed] [Google Scholar]
- Pernthaler J, Amann R. (2005). Fate of heterotrophic microbes in pelagic habitats: focus on populations. Microbiol Mol Biol Rev 69: 440–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Andersson SGE, Battin TJ, Prosser JI, Schimel JP, Whitman WB et al. (2010). The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol 8: 523–529. [DOI] [PubMed] [Google Scholar]
- Raabe T, Wiltshire KH. (2009). Quality control and analyses of the long-term nutrient data from Helgoland Roads, North Sea. J Sea Res 61: 3–16. [Google Scholar]
- R Core Team.. (2008) R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Reintjes G, Arnosti C, Fuchs BM, Amann R. (2017). An alternative polysaccharide uptake mechanism of marine bacteria. ISME J 11: 1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M, Fetzer I, Dormann CF, Harms H, Chatzinotas A. (2012). Predator richness increases the effect of prey diversity on prey yield. Nat Commun 3: 1305. [DOI] [PubMed] [Google Scholar]
- Selje N, Simon M, Brinkhoff T. (2004). A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427: 445–448. [DOI] [PubMed] [Google Scholar]
- Soltwedel T, Schauer U, Boebel O, Nothig E-M, Bracher A, Metfies K et al. (2013). FRAM - FRontiers in Arctic marine Monitoring: Visions for permanent observations in a gateway to the Arctic Ocean. In MTS/IEEE OCEANS - Bergen. IEEE, pp 1–7.
- Steinberg DK, Carlson CA, Bates NR, Johnson RH, Michaels AF, Knap AH. (2001). Overview of the US JGOFS Bermuda Atlantic Time-series Study (BATS): a decade-scale look at ocean biology and biochemistry. Deep Sea Res II 48: 1405–1447. [Google Scholar]
- Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611. [DOI] [PubMed] [Google Scholar]
- Teeling H, Fuchs BM, Bennke CM, Krüger K, Chafee M, Kappelmann L et al. (2016). Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. eLife 5: e11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingstad TF. (2000). Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr 45: 1320–1328. [Google Scholar]
- Thingstad TF, Våge S, Storesund JE, Sandaa R-A, Giske J. (2014). A theoretical analysis of how strain-specific viruses can control microbial species diversity. PNAS 111: 7813–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingstad T, Lignell R. (1997). Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol 13: 19–27. [Google Scholar]
- Thomas F, Hehemann J-H, Rebuffet E, Czjzek M, Michel G. (2011). Environmental and gut Bacteroidetes: The food connection. Front Microbiol 2: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Lehman CL, Thompson KT. (1997). Plant diversity and ecosystem productivity: theoretical considerations. PNAS 94: 1857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utter DR, Mark Welch JL, Borisy GG. (2016). Individuality, stability, and variability of the plaque microbiome. Front Microbiol 7: 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Våge S, Storesund JE, Thingstad TF. (2013). Adding a cost of resistance description extends the ability of virus-host model to explain observed patterns in structure and function of pelagic microbial communities. Environ Microbiol 15: 1842–1852. [DOI] [PubMed] [Google Scholar]
- Voget S, Wemheuer B, Brinkhoff T, Vollmers J, Dietrich S, Giebel H-A et al. (2015). Adaptation of an abundant Roseobacter RCA organism to pelagic systems revealed by genomic and transcriptomic analyses. ISME J 9: 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CS, Yung C-M, Davis KM, Blinebry SK, Williams TC, Johnson ZI et al. (2017). Annual community patterns are driven by seasonal switching between closely related marine bacteria. ISME J 11: 1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y et al. (2016). Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J 10: 1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2009) ggplot2: elegant graphics for data analysis. Springer: New York, NY, USA. [Google Scholar]
- Wilcox RM, Fuhrman JA. (1994). Bacterial viruses in coastal seawater: lytic rather than lysogenic production. Mar Ecol Prog Ser 114: 35–45. [Google Scholar]
- Wiltshire KH, Kraberg A, Bartsch I, Boersma M, Franke H-D, Freund J et al. (2009). Helgoland Roads, North Sea: 45 years of change. Estuaries Coasts 33: 295–310. [Google Scholar]
- Xing P, Hahnke RL, Unfried F, Markert S, Huang S, Barbeyron T et al. (2015). Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J 9: 1410–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žure M, Fernandez-Guerra A, Munn CB, Harder J. (2017). Geographic distribution at subspecies resolution level: closely related Rhodopirellula species in European coastal sediments. ISME J 11: 478–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.