Abstract
Burkholderia pseudomallei is an environmental bacterium that causes melioidosis, a major community-acquired infection in tropical regions. Melioidosis presents with a range of clinical symptoms, is often characterized by a robust inflammatory response, may relapse after treatment, and results in high mortality rates. Lipopolysaccharide (LPS) of B. pseudomallei is a potent immunostimulatory molecule comprised of lipid A, core, and O-polysaccharide (OPS) components. Four B. pseudomallei LPS types have been described based on SDS-PAGE patterns that represent the difference of OPS–type A, type B, type B2 and rough LPS. The majority of B. pseudomallei isolates are type A. We used matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) followed by electrospray ionization quadrupole time-of-flight mass spectrometry (ESI-QqTOF MS) and gas chromatography to characterize the lipid A of B. pseudomallei within LPS type A isolates. We determined that B. pseudomallei lipid A is represented by penta- and tetra-acylated species modified with 4-amino-4-deoxy-arabinose (Ara4N). The MALDI-TOF profiles from 171 clinical B. pseudomallei isolates, including 68 paired primary and relapse isolates and 35 within-host isolates were similar. We did not observe lipid A structural changes when the bacteria were cultured in different growth conditions. Dose-dependent NF-κB activation in HEK cells expressing TLR4 was observed using multiple heat-killed B. pseudomallei isolates and corresponding purified LPS. We demonstrated that TLR4-dependent NF-κB activation induced by heat-killed bacteria or LPS prepared from OPS deficient mutant was significantly greater than those induced by wild type B. pseudomallei. These findings suggest that the structure of B. pseudomallei lipid A is highly conserved in a wide variety of clinical and environmental circumstances but that the presence of OPS may modulate LPS-driven innate immune responses in melioidosis.
Author summary
Melioidosis is a tropical infection caused by Burkholderia pseudomallei. Disease ranges from mild to lethal, and patients who die from melioidosis have greater inflammatory responses than survivors. Identifying variation in bacterial structures that alter the immune response is therefore important. B. pseudomallei expresses lipopolysaccharide, a major stimulator of the immune response. Studies of other bacteria suggest that differences in the lipid A component of lipopolysaccharide result in variable stimulation of the immune system. In this study, we analyzed the chemical structure of lipid A of 171 clinical B. pseudomallei isolates, structural changes caused by different growth conditions, and innate immune responses induced. Surprisingly, we did not observe any variation of lipid A structure or different innate immune activation from different B. pseudomallei isolates. Our data suggest that lipid A structure of B. pseudomallei is highly conserved and that different inflammatory responses in patients may be caused by other factors.
Introduction
Burkholderia pseudomallei is a Gram-negative bacillus and the causative agent of melioidosis. This disease is endemic in Southeast Asia and northern Australia, with a mortality rate up to 40% in northeast Thailand and 20% in Australia [1, 2]. The manifestations of disease vary from indolent chronic infection to acute and fulminant sepsis. The common clinical presentations in melioidosis include severe pneumonia, bacteremia, skin infection, and abscesses in several organs [2]. B. pseudomallei is encapsulated, highly adaptable and able to survive under several extreme conditions in the environment, and clearance of B. pseudomallei in patients is difficult to achieve. The underlying mechanisms that contribute to persistence and death in melioidosis are unclear.
Acute melioidosis is characterized by the up-regulation of pattern recognition receptors (PRRs) and pro-inflammatory cytokine release [3]. Melioidosis patients have increased pro-inflammatory cytokines, such as IL-12, IL-15, IL-18 and IFN-γ and patients who die from melioidosis have higher levels of plasma IL-6 and IL-8 than those who survive [4]. These studies suggest the importance of the innate immune response in the control of infection and pathophysiology of sepsis and mortality in melioidosis.
Host immune cells express several PRRs including membrane bound Toll-like receptors (TLRs) and cytoplasmic receptors that recognize distinct bacterial pathogen-associated molecular patterns (PAMPs) [5]. Recognition of bacterial produces leads to the activation of innate immune response and release of inflammatory cytokines and mediators. Our previous study conducted in 300 healthy Thai donors suggest that lipopolysaccharide (LPS) of B. pseudomallei is a potent immuno-stimulatory molecule in humans [6]. The innate immune response to heat-killed B. pseudomallei is highly correlated with the response to purified LPS. We have also showed that genetic variation in TLR4, which encodes the canonical LPS receptor , TLR4 is associated with susceptibility to melioidosis [7]. These data point to the likely importance of B. pseudomallei LPS in the host-pathogen interaction in melioidosis.
Four B. pseudomallei LPS types have been described based on SDS-PAGE patterns which represent the difference of O-polysaccharides (OPS)–ladder type A, ladder type B, type B2 and rough LPS. Type A and B LPSs are structurally composed of three covalently linked domains of O-polysaccharide, a core-oligosaccharide and a lipid A moiety, while rough LPS lacks OPS. We previously reported that type A is the predominant LPS type expressed by 97% of B. pseudomallei isolates from Thailand and 80% of isolates from Australia [8]. Lipid A, the membrane anchor region of LPS is recognized by the host innate immune system. This process occurs by the presentation of LPS by LPS-binding protein (LBP) in concert with the accessory molecule CD14 to the TLR4/MD-2 complex [9]. This complex is present on a variety of cell types including macrophages and dendritic cells and upon binding of LPS, triggers the production of proinflammatory cytokines by host cells [10].
In several pathogenic Gram-negative bacteria, lipid A variation has been demonstrated both in vivo and in vitro to be associated with the induction of different innate immune responses. In Salmonella enterica serovar Typhimurium, the presence of Ara4N residues on phosphate groups occurs when the bacteria are grown under magnesium-deficient conditions. These modifications decrease the overall negative charge on the surface of bacteria resulting in the lower affinity for cationic antimicrobial peptides (CAMPs) [11]. Temperature-dependent lipid A modifications have been described in Porphyromonas gingivalis, Francisella novicida, and Yersinia pestis [12–14]. Pseudomonas aeruginosa isolates from patients with cystic fibrosis have different lipid A structures that are correlated with disease severity and progression [15, 16].
The first published study of B. pseudomallei lipid A structure in a single B. pseudomallei isolate, strain KHW, proposed a penta-acylated bisphosphorylated disaccharide backbone modified with Ara4N [17]. A second report on B. pseudomallei strains K96243 and 1026b showed the lipid A structure to be a predominantly tetra-acylated with a minor proportion of penta-acylated lipid A [18]. Recently, Norris et al have reported that LPSs from a type B Thai strain 576a and a virulent rough central nervous system-tropic strain MSHR435 from Australia induced higher innate immune responses than the prototype Thai type A strain 1026b. Matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry (MALDI-TOF/TOF MS) analysis demonstrated structural differences in the lipid A of these isolates. They also suggested that the different LPS types may play a role in variable host responses to LPS [19] though a limitation of this study is the minimal number of B. pseudomallei used.
Due to the discrepancy in the findings of these reports, the first aim of this study was to characterize the lipid A structure in a large collection of clinical B. pseudomallei isolates using MALDI-TOF MS followed by electrospray ionization quadrupole- time-of-flight mass spectrometry (ESI-QqTOF MS) and gas chromatography. Because the vast majority of Thai and Australian patients infected with B. pseudomallei type A present with a wide range of clinical symptoms and incur different outcomes, we hypothesized that different innate immune responses to B. pseudomallei type A may be associated with variation in the lipid A structures of clinical isolates. The second aim was to examine the structural change of lipid A under varying laboratory conditions and the third aim was to evaluate the TLR4-dependent immuno-stimulatory effect of different B. pseudomallei type A isolates, as well as the role of OPS and capsule.
Materials and methods
Ethics statement
The study was exempted for ethical review by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (documentary proof of exemption, MUTM-EXMPT 2017–003). All bacterial isolates obtained from human were anonymous.
Bacterial strains and growth conditions
Cultivation of B. pseudomallei was performed in a biosafety level 3 (BSL3) laboratory. Two existing collections of B. pseudomallei isolates were used as follows: (i) 136 isolates from 68 melioidosis patients presenting to Sunpasitthiprasong Hospital, Ubon Ratchathani Northeast Thailand who developed relapsed infection. The relapse was defined as the bacterial isolates from the same patient during distinct episodes, which showed a similar pulsed-field gel electrophoresis banding pattern or multilocus sequence type regarded as originating from the same clone [20]. The first episode of melioidosis was between 1986 and 2004 and relapse was 15 days to 3,757 days after recovery from the primary episode (median = 207.5 days, IQR = 84.5–594.5) [20, 21]; (ii) 35 isolates from 35 individual colonies of 7 clinical specimens (5 colonies for each specimen) from a Thai patient with acute melioidosis who was admitted in 2006 to the same hospital [22]. The clinical specimens were blood, tracheal suction, urine, pus from right leg, pus from left leg, pus from forehead, and wound swab from thigh. Reference or laboratory strains of B. pseudomallei (K96243, 1026b, 153 and 164) were used for comparison. The B. pseudomallei wild type, OPS mutant and capsule mutant strains used during this study are described in S1 Table. Unless stated otherwise, B. pseudomallei was cultured on trypticase soy agar (TSA) and incubated at 37 oC for 16–18 h. All isolates were stored in trypticase soy broth (TSB) with 15% glycerol at -80 oC.
Effect of different culture conditions on B. pseudomallei lipid A structure
The effect of a range of laboratory conditions on lipid A structure was tested for B. pseudomallei strain K96243. The bacteria were cultured on TSA (Oxoid, Hants, UK) for 16–18 h. Approximately 10 colonies were streaked onto agar plates and incubated for 16–18 h under one of the following conditions: (i) TSA (pH 7.4) at 25 oC, (ii) TSA (pH 7.4) at 37 oC, (iii) TSA (pH 7.4) at 42 oC, (iv) TSA (pH 4.5) at 37 oC, (v) TSA (pH 8.5) at 37 oC, (vi) TSA (pH 7.4) plus 2 mM H2O2 (Merck, Darmstadt, Germany) at 37 oC, (vii) TSA (pH 7.4) plus 5 mM H2O2 at 37 oC, (viii) TSA (pH 7.4) plus 50 mM NaNO2 (Sigma-Aldrich, MO, USA) at 37 oC, (ix) TSA (pH 7.4) plus 100 mM NaNO2 at 37 oC, (x) TSA (pH 7.4) plus 1 mM MgCl2 (Fisher Scientific, Leics, UK) at 37 oC, (xi) TSA (pH 7.4) plus 8 μM MgCl2 at 37 oC, (xii) Luria-Bertani agar (LB) (BD, MD, USA) (pH 7.4) at 37 oC, (xiii) Ashdown agar (pH 7.4) at 37 oC, (xiv) blood agar (Oxoid, Hants, UK) (pH 7.4) at 37 oC, (xv) MacConkey agar (Oxoid, Hants, UK) (pH 7.4) at 37 oC, and (xvi) M9 minimal medium agar (Sigma-Aldrich, MO, USA) (pH 7.4) at 37 oC. The bacteria were harvested using a 10 μl loop and further extracted for lipid A.
Lipid A microextraction and MALDI mass spectrometry analysis
Lipid A was extracted from bacteria using a microextraction method as previously described [23] with some modifications. Briefly, the bacteria were cultured on TSA for 16–18 h and harvested as described above. One loop of the bacteria was suspended in 400 μl of 70% isobutyric acid (Sigma-Aldrich, MO, USA) and 1 M ammonium hydroxide (Sigma-Aldrich, MO, USA) at a ratio of 5:3 (v/v). The samples were heated at 100 oC for 1 h followed by incubation on ice for 5 min. After centrifugation at 2000 × g for 15 min, the supernatant was collected, diluted with 1 ml of distilled water and lyophilized. The dried material was washed twice with 1 ml of methanol (Merck, Darmstadt, Germany) by centrifugation at 2000 × g for 15 min. The pellet was extracted for lipid A using 100 μl of a mixture of chloroform:methanol:water in a ratio of 3:1.5:0.25 (v/v/v). The suspension was centrifuged at 2000 × g for 1 min. One microliter of the supernatant was spotted onto a MALDI target plate and allowed to dry in air. The lipid A spots were overlaid with 1 μl of 20 mg/ml norharmane matrix (Sigma-Aldrich, MO, USA). Norharmane was used as a matrix in this study because it has been shown to improve limit of detection and support the characterization of a wide range of lipid A analysis [24]. Each spot was measured in 500 shot steps for a total of 3,000 laser shots using a Bruker Autoflex Speed MALDI-TOF mass spectrometer (Bruker Daltonics, Germany). The instrument was operated in negative ion reflector mode, across the mass range 1500–2000 and calibrated with electrospray ionization (ESI) tuning mix (Agilent Technologies, CA, USA). The mass spectrum was processed for smoothing and baseline subtraction using FlexAnalysis version 3.4 (Bruker Daltonics, Germany). Multiple spectra were displayed in a mass spectrum window for spectra comparison.
Preparation of heat-killed bacteria
To prepare heat-killed B. pseudomallei, bacteria were grown on media including TSA, LB agar, Ashdown agar, blood agar, MacConkey agar and M9 minimal agar at 37 oC in air for 16–18 h, harvested by obtaining two 10 μl loops and suspended in 1 ml of sterile phosphate-buffered saline (PBS) pH 7.4. The bacterial suspension was centrifuged at 10,000 × g for 10 min and washed twice with 1 ml of PBS. The pellet was resuspended in 1 ml PBS. One hundred microliters of bacterial suspension was taken to make a ten-fold serial dilution in PBS. The colony count was performed by spreading 100 μl of each dilution onto TSA plates in triplicate and incubating at 37 oC for 16–18 h. The remaining bacterial suspension was heated at 80 oC for 1 h. One hundred microliters of heat-killed bacteria were plated on TSA in duplicate for a sterility test.
Purification of lipopolysaccharide
Lipopolysaccharide (LPS) was extracted using a modified hot water-phenol method as previously described [25]. Briefly, each of four B. pseudomallei isolates (strains K96243, 1026b, 153 and 164) and B. pseudomallei K96243 derivative with ΔwbiD (mutant defective in OPS synthesis) were cultured on TSA for 50 plates and incubated at 37 oC for 16–18 h. Bacteria were scraped off using a 10 μl loop and suspended in 75 ml of distilled water (DW). The bacterial suspension was mixed with 90% phenol solution at a ratio of 1:1, heated at 80 oC for 5 min with constant mixing, and subsequently cooled down at room temperature. The extract was dialyzed in a dialysis tube with pore size 6–8 kDa (Spectrum, CA, USA) against DW. The dialysate was centrifuged at 8,000 × g for 20 min and the supernatant was lyophilized. The dried material were solubilized in 10 mM Tris-HCl (pH 7.5) buffer containing 1 mM MgCl2, 1 mM CaCl2 at ratio of 30 mg:1 ml and sequentially treated with RNase and DNase at final concentration of 50 μg/ml at 37 oC with agitation for 3 h, and proteinase K (Invitrogen, CA, USA) at final concentration of 50 μg/ml at 60 oC for overnight. LPS was isolated from the supernatant by ultracentrifugation (Beckman Coulter) at 100,000 × g at 4 oC for 3 h. The gel-like pellet was suspended in 10 ml pyrogen-free water and lyophilized. The purified LPS was further analyzed using SDS-PAGE and silver staining as previously described [8]. Protein contamination was examined using Coomassie blue staining [18] and bicinchoninic acid (BCA) assay (Pierce, IL, USA).
Fatty acid analysis
The bacteria were cultured on 10 plates of TSA at 37 oC for 16–18 h, harvested by 10 μl loop and suspended in 18 ml of PBS. Bacteria were inactivated by adding phenol to obtain 1% final concentration, and the bacterial suspension was incubated at 37 oC for overnight. The cell pellet was collected by centrifugation at 12,000 × g for 3 min and wash twice with 20 ml of PBS. The pellet was resuspended in 5 ml PBS. Five hundred microliters of bacteria suspension were plated on TSA for sterility test. The remaining bacterial suspension was lyophilized and collected as a dried bacterial cell.
LPS fatty acids were derivatized to fatty acid methyl esters prior to gas chromatography (GC) analysis as previously described [26]. 20 mg of dried bacterial cells were suspended in 500 μl of DW. Five hundred microliters of 90% hot phenol (80 oC) was added and further incubated at 70 oC for 1 h with constant mixing. The cell suspension was incubated on ice 5 min and centrifuged at 10,000 × g for 10 min. The upper aqueous phase was obtained and washed with 2 ml of diethyl ether (Fisher Scientific, NJ, USA) by centrifugation at 3,000 × g for 5 min. Then, the lower aqueous phase containing LPS was collected and lyophilized. The 200 μg of dried LPS were treated with 200 μl of 2 M anhydrous methanolic HCL (Alltech, KY, USA) at 90 oC for 18 h. Then, the derivatized fatty acid methyl ester was cooled down at room temperature. Two hundred microliters saturated NaCl solution was added. The mixture was subsequently extracted with 400 μl of hexane (Sigma-Aldrich, MO, USA) by mixing using a vortex for 30 sec. The upper layer phase (~400 μl) was analyzed by a gas chromatography-flame ionization detector (GC-FID), HP 5890 series II with a 7673 autoinjector. Bacterial acid methyl ester CP mixture (Matreya, PA, USA) was used as standard control and pentadecanoic acid (Sigma Aldrich, MO, USA) was used as an internal control respectively.
Lipid A hydrolysis and ESI tandem mass spectrometry analysis
Lipid A was liberated from purified LPS by hydrolysis. Purified LPS was dissolved in 10 mM sodium acetate (pH 4.5) with 1% SDS and then heated at 100 oC for 1 h. The sample was lyophilized and then washed with 170 μl of endotoxin free water and 850 μl of acidified ethanol mixture by centrifugation at 5,000 × g for 5 min. The sample was further washed with 1 ml of non-acidified 95% ethanol and followed by 1 ml of 100% ethanol. Finally the sample was lyophilized to yield lipid A. The structural characterization was analyzed on a Waters Synapt G2 QqTOF mass spectrometer (Waters Corporation, Milford, MA, USA) operated in sensitivity mode and negative ion mode. The lipid A was dissolved in a solvent mixture of chloroform and methanol (2:1 v/v). The lipid solution was infused at 3 μl/min flow rate. The source block temperature was set to 150 oC. Tandem MS was carried out using trap collision induced dissection (CID). The instrument standard values (LM resolution 4.7 and HM resolution 15.0) were used for mass selection in the quadrupole. To mitigate the effect of instantaneous signal fluctuations, the data were averaged for 2–3 min.
Cell culture conditions and stimulations
TLR4-transfected human embryonic kidney cells (HEK-BlueTM-hTLR4 cells) were purchased from InvivoGen (San Diego, CA, USA). The HEK-BlueTM-hTLR4 cells are HEK cells that are stably transfected with human TLR4 (hTLR4), myeloid differentiation factor-2, cluster of differentiation-14 (MD-2/CD14) and a secreted embryonic alkaline phosphatase (SEAP) reporter gene. The cells were cultured and maintained at 37 oC with 5% CO2 in complete Dulbecco’s modified Eagle medium (DMEM) (Gibco, NY, USA) which contained 10% heat inactivated fetal bovine serum (FBS) and 1× HEK-Blue selection medium (InvivoGen, San Diego, CA, U.S.A); an antibiotic mixture for maintenance of HEK-Blue hTLR4 cell lines. The cells were seeded at 2.5 × 104 cells/well in a 96-well plate and stimulated for 24 h with heat-killed bacteria at 106 or 107 CFU/ml and purified LPS at 1, 10, 100, 1000, 10,000 ng/ml from B. pseudomallei isolates. Cells stimulated with ultrapure TLR4 ligand Escherichia coli O111:B4 LPS (Sigma-Aldrich, MO, USA) was a positive control. The activation of nuclear transcription factor (NF)-κB in HEK-Blue hTLR4 cells in response to TLR4 agonists was determined by a SEAP reporter assay at a wavelength 620 nm using a microplate reader (TECAN, Grodig, Austria). The results of NF-κB activation in HEK-Blue hTLR4 cells were calculated to account for the difference in weight of wild type LPS and ΔwbiD mutant LPS by a factor 2.5. The mass individually of the lipid A (1948), core (1258), and repeating O-antigen unit (423+ 4250 = 4673) were used to calculated the weight of wild type LPS. The mass individually of the lipid A (1948) and core (1258) were used to calculated the weight of the mutant LPS.
Antimicrobial susceptibility to polymyxin B testing
Susceptibility testing to polymyxin B was performed with B. pseudomallei clinical isolates using a broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [27]. The bacterial isolates used in this experiment were randomly selected from different groups of infections that the lipid A results showed slightly different mass intensity at m/z 1801.6. These included 14 isolates from 7 Thai patients with primary infection (strains: 226a, 855e, 1234a, 1304a, 1620a, 1790a and 2944b) and relapse after recovery from primary infection (strains: 240a, 855h, 1234b, 1304b, 1620d, 1790b and 2944d) and 7 isolates from within-host infection [H3921b(C5), H3921c(C3), H3921d(C2), H3921e(C2), H3921f(C2), H3921g(C3), H3921h(C5)]. The MIC interpretive criteria used for polymyxin B were established in CLSI standard M100-S24 as follows: susceptible ≤ 2 μg/ml, intermediate 4 μg/ml and resistant ≥ 8 μg/ml [28]. Pseudomonas aeruginosa ATCC 27853 was used as a control strain. B. pseudomallei isolates were recovered from the freezer vials at -80 oC by streaking onto Columbia agar and incubating aerobically at 37 oC for 24 h. Bacterial colonies were harvested and suspended in normal saline 0.85%. Bacteria suspension was adjusted at OD600 nm to obtain a concentration of 1 × 108 CFU/ml. Bacteria at a final concentration of 5 × 105 CFU/ml as recommended in CLSI document M07-A9 [29] were used for susceptibility testing of polymyxin B (catalog number P4932; Sigma-Aldrich) at concentrations of 0, 0.25, 0.5, 1, 2, 4, 8, 16, 32, 64, 128, 256, and 512 μg/ml in duplicate. The MIC was read as the lowest drug concentration at which no visible growth was observed following aerobic incubation at 35 ± 2 oC for 16–20 h.
Construction of mutants
Four B. pseudomallei mutants including three OPS mutants and a capsule mutant were constructed from wild types K96243 and 4095A, respectively using a fragment mutagenesis method as described [30] (S1 Table). K96243ΔwbiD was defective in wbiD and did not synthesize the OPS, K96243ΔwbiA and K96243ΔoacA were both defective in acetylation of the OPS. The capsule mutant (4095a ΔwcbB) was defective in wcbB and did not have capsule [30]. Characterization of OPS was examined by SDS-PAGE of proteinase K extracts, silver staining and Western blot using a monoclonal antibody to OPS [8]. Characterization of capsule was examined by latex agglutination and Western blot using a monoclonal antibody to CPS [31].
Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 5; GraphPad Software). One-way ANOVA was used to test the difference among the means of NF-κB activation from different B. pseudomallei isolates. Unpaired t tests were used to test the difference between the mean of results from B. pseudomallei wild type and mutant strains. The data were presented as mean ± standard deviations. Differences were considered statistically significant at a P value < 0.05.
Results
MALDI-TOF MS analysis of B. pseudomallei K96243 lipid A
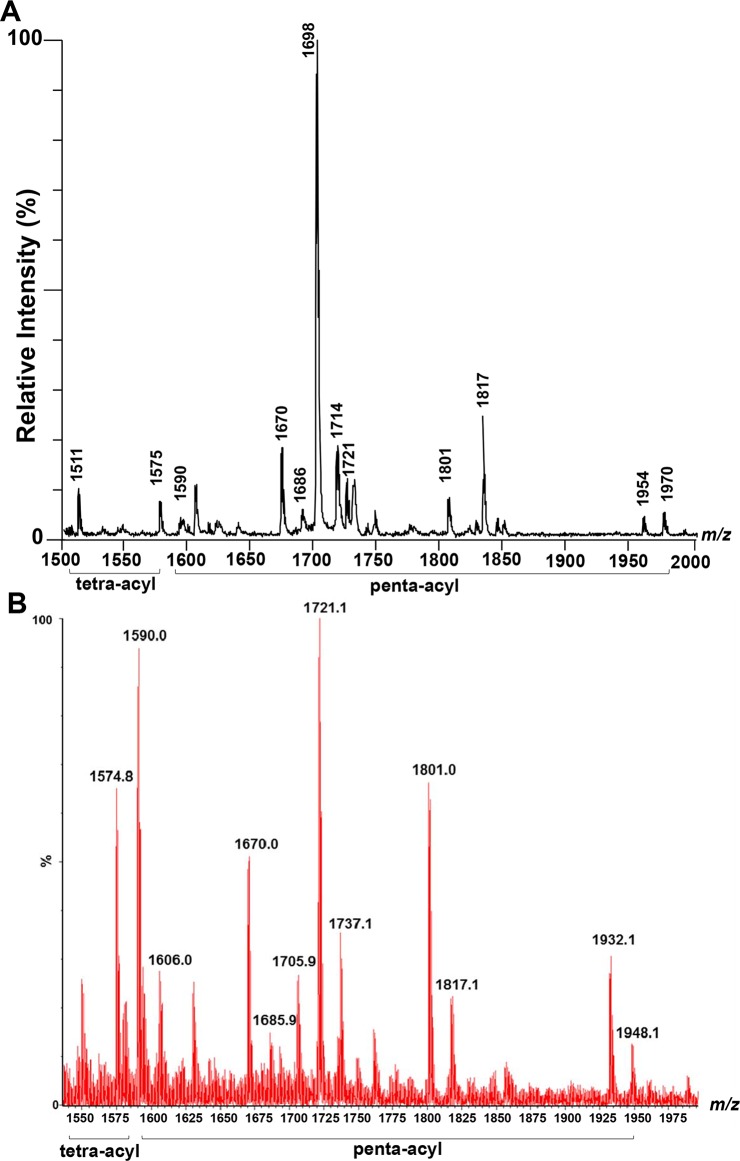
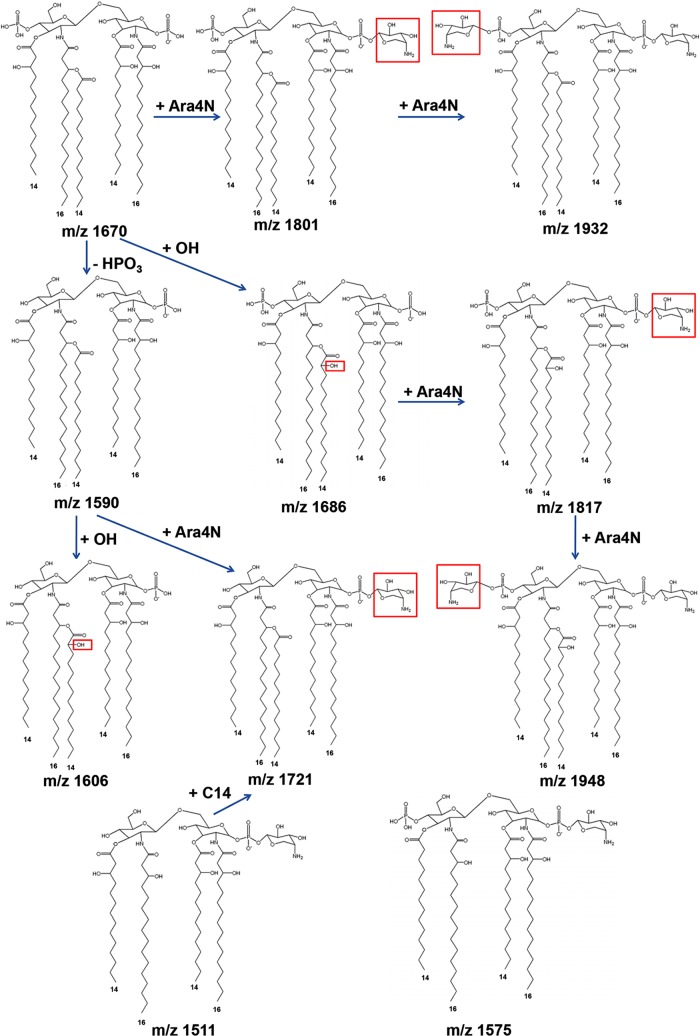
We initially analyzed the lipid A isolated from the reference strain B. pseudomallei K96243. Negative ion mass spectra of K96243 lipid A showed a complex pattern of ions with major ions at m/z 1511, 1575, 1590, 1670, 1686, 1698, 1714, 1721, 1801, 1817, 1954, 1970 (Fig 1A). The predicted structures of fatty acid, phosphate, and carbohydrate substituents on lipid A of K96243 backbone are shown in Fig 2 and Table 1. Based on overall chemical compositions, the data predicted that B. pseudomallei contained penta- and tetra-acylated lipid A species. The ion at m/z 1511 was a representative of tetra-acylated monophosphorylated GlcN disaccharide backbone possessing two C14:0(3-OH) residue in ester linkage and two C16:0(3-OH) residues in amide linkage with one phosphorylated and one 4-amino-4-deoxy-arabinose (Ara4N) attached to the phosphate group. The ion at m/z 1575 was a representative of tetra-acylated bisphosphorylated GlcN disaccharide backbone possessing one C14:0 residue and one C14:0(3-OH) residue in ester linkage and two C16:0(3-OH) residues in amide linkage with two phosphorylated and one Ara4N attached to the phosphate group. The ion at m/z 1590 was a penta-acylated monophosphorylated GlcN disaccharide backbone possessing one C14:0, two C14:0(3-OH) and two C16:0(3-OH) residues with one phosphorylated. The ion at m/z 1670 was a penta-acylated bisphosphorylated GlcN disaccharide backbone possessing one C14:0 residue, two C14:0(3-OH) residues and two C16:0(3-OH) residues with two phosphorylated. The ion at m/z 1686 was a representative of ion at m/z 1670 with the substitution of fatty acid C14:0 for C14:0(2-OH). The ion at m/z 1721 was a representative of ion at m/z 1590 modified with one Ara4N residue attached to the phosphate group (Δm/z + 131). The ion at m/z 1801 was a representative of ion at m/z 1670 modified with an Ara4N residue attached to the phosphate group. The ion at m/z 1817 was a representative of ion at m/z 1686 modified with an Ara4N residue attached to the phosphate group. The ion at m/z 1954 was a representative of ion at m/z 1670 with sodium adduct and modified with two Ara4N residues attached to the phosphate group. The ion at m/z 1970 was a representative of ion at m/z 1686 with sodium adduct and modified with two Ara4N residues attached to the phosphate group. The ions at m/z 1698 and 1714 were detected only by MALDI-TOF and could not be detected for structural identification by ESI-QqTOF.
Fig 1.
Negative ion mass spectrum by MALDI-TOF (A), by ESI (B) of B. pseudomallei K96243 lipid A.
Fig 2. Predicted structure of B. pseudomallei K96243 lipid A.
Table 1. Negative ion MALDI-TOF and ESI-QqTOF MS lipid A species and predicted structures of the fatty acid, phosphate and carbohydrate substituents on the B. pseudomallei K96243 lipid A backbone.
| Lipid A species | Observed ion (m/z) | Theoretical monoisotopic ion (m/z) | Acyl substitution | Predicted structure |
|---|---|---|---|---|
| 1 | 1511.7 | 1511.0 | Tetra | 2 × C14:0(3-OH), 2 × C16:0(3-OH), P, Ara4N |
| 2 | 1574.8 | 1575.0 | Tetra | C14:0, C14:0(3-OH), 2 × C16:0(3-OH), 2 × P, Ara4N |
| 3 | 1590.0 | 1590.1 | Penta | C14:0, 2 × C14:0(3-OH), 2 × C16:0(3-OH), P |
| 4 | 1606.0 | 1606.1 | Penta | C14:0(2-OH), 2 × C14:0(3-OH), 2 × C16:0(3-OH), P |
| 5 | 1670.0 | 1670.1 | Penta | C14:0, 2 × C14:0(3-OH), 2 × C16:0(3-OH), 2 × P |
| 6 | 1685.9 | 1686.1 | Penta | C14:0(2-OH), 2 × C14:0(3-OH), 2 × C16:0(3-OH), 2 × P |
| 7 | 1721.1 | 1721.2 | Penta | C14:0, 2 × C14:0(3-OH), 2 × C16:0(3-OH), P, Ara4N |
| 8 | 1801.0 | 1801.1 | Penta | C14:0, 2 × C14:0(3-OH), 2 × C16:0(3-OH), 2 × P, Ara4N |
| 9 | 1817.1 | 1817.2 | Penta | C14:0(2-OH), 2 ×C14:0(3-OH), 2 × C16:0(3-OH), 2 × P, Ara4N |
| 10 | 1932.1 | 1932.2 | Penta | C14:0, 2 × C14:0(3-OH), 2 × C16:0(3-OH), 2 × P, 2 × Ara4N |
| 11 | 1948.1 | 1948.2 | Penta | C14:0(2-OH), 2 ×C14:0(3-OH), 2 × C16:0(3-OH), 2 × P, 2 × Ara4N |
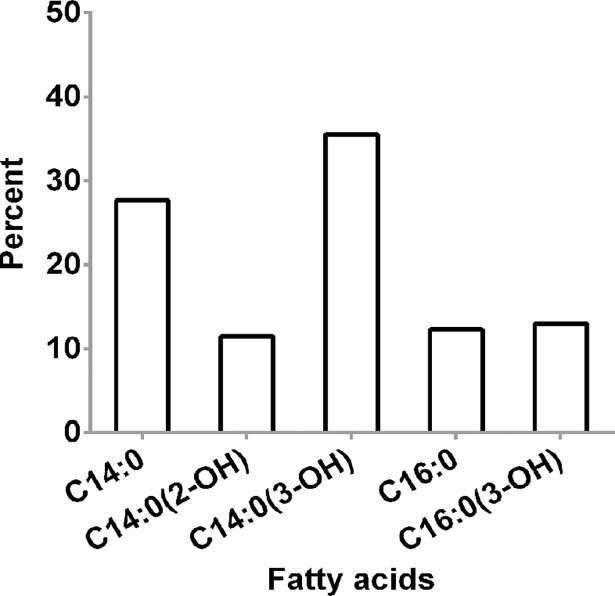
The total fatty acid of B. pseudomallei K96243 lipid A was validated by gas chromatography (GC). The result is presented in Fig 3. Fatty acid composition of B. pseudomallei strain K96243 LPS confirmed the MALDI-TOF MS results for the presence of tetradecanoic acid (C14:0), 2-hydroxytetradecanoic [C14:0(2-OH)], 3-hydroxytetradecanoic acid [C14:0(3-OH)], hexadecanoic acid (C16:0), and 3-hydroxyhexadecanoic acid [C16:0(3-OH)]. Our data suggest that the lipid A species of B. pseudomallei K96243 was predominantly penta-acylated with a combination of C14:0(2-OH), C16:0(3-OH), C14:0(3-OH) or C14:0.
Fig 3. Fatty acid composition of B. pseudomallei K96243 lipid A.
ESI-QqTOF analysis of B. pseudomallei K96243 lipid A
ESI-QqTOF MS analysis with negative ion mode of B. pseudomallei K96243 lipid A showed major ions at m/z 1574.8, 1590.0, 1606.0, 1670.0, 1685.9, 1721.1, 1801.0, 1817.1, 1932.1 and 1948.1 (Fig 1B). ESI results, where ESI-QqTOF gives higher mass accuracy, as compared to MALDI-TOF are shown at Fig 1B. The two MS results are very reproducible. These ions were further characterized by trap collision-induced dissociation (S1 Fig) and the tandem MS results confirm the predicted structures (Fig 2). The ion at m/z 1606.0 was a representative of m/z 1590.0 with the substitution of fatty acid C14:0 for C14:0(2-OH). The ion at m/z 1932.1 was a representative of ion at m/z 1670.0 with two Ara4N residues. The m/z 1698 and 1714 were detected only by MALDI-TOF and could not be detected for structural identification by ESI-QqTOF.
Mass spectra of lipid A from clinical isolates of B. pseudomallei
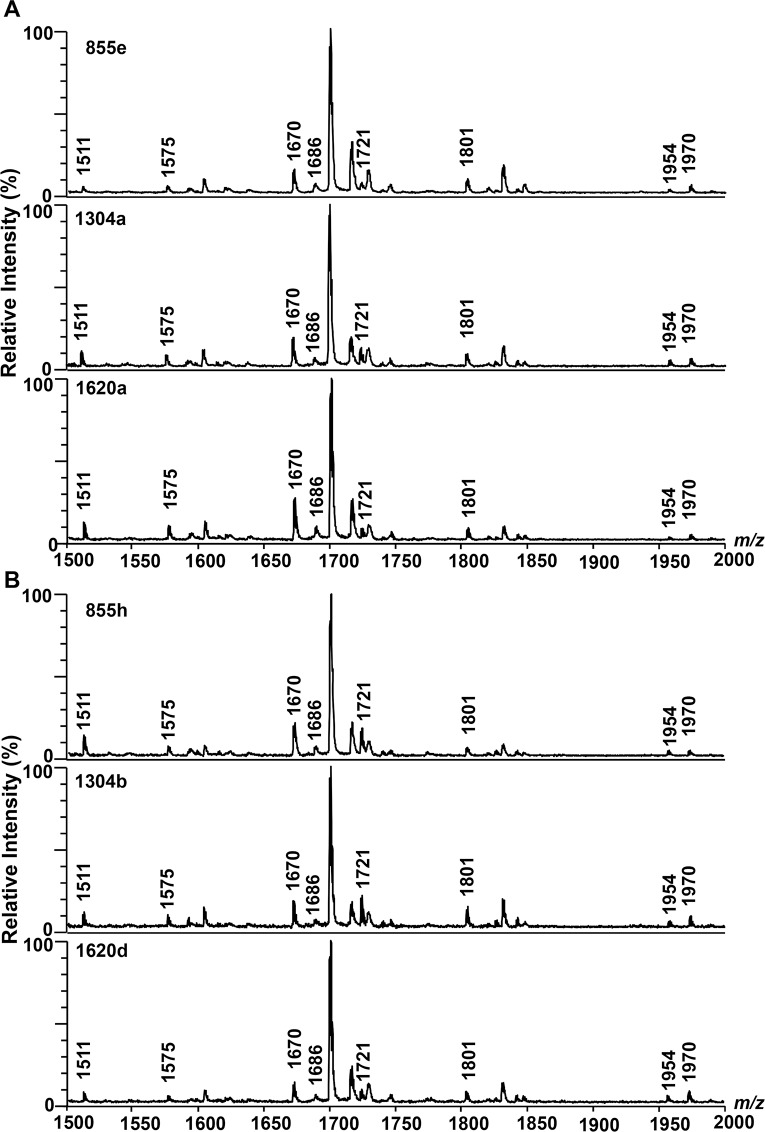
We next investigated the variation of lipid A structure in 136 clinical isolates of B. pseudomallei grown on TSA plates. The mass spectrum of lipid A was determined in two groups of existing Thai isolate collections. The first group represented primary isolates (S2 Fig) and relapse isolates (S3 Fig) from 68 melioidosis patients who had at least one episode of relapse. MALDI-TOF lipid A spectra of representatives of these isolates are shown in Fig 4. Using a negative reflector mode MALDI-TOF MS analysis, we observed similar lipid A spectra in a mass range of 1500–2000 Da in all isolates from both primary and relapse infections. All lipid A species of these isolates contained four main clusters of peaks around m/z of 1575, 1670, 1801 and 1954 corresponding to tetra-acylated modified with one phosphorylated and one Ara4N group, bisphosphorylated penta-acylated, bisphosphorylated penta-acylated modified with only one Ara4N group and bisphosphorylated penta-acylated modified with two Ara4N group, respectively. The second group consisted of 35 B. pseudomallei isolates from a variety of specimen type (blood, tracheal suction, urine, pus from right leg, pus from left leg, pus from forehead, and wound swab from thigh) of a Thai patient with acute melioidosis. The lipid A species of these within-host isolates were not different (S4 Fig), and represented the same four main clusters of peaks at approximately m/z of 1575, 1670, 1801 and 1954. Together, these comprehensive set of data indicate a striking lack of variation in lipid A structure in a large group of clinical isolates cultured from patients with primary melioidosis, as well as in isolates obtained during relapse infection, and from multiple isolates obtained from numerous sites within a single host.
Fig 4. MALDI-TOF lipid A spectra of clinical isolates of B. pseudomallei strains from three Thai patients with primary infection followed by an episode of relapse.
(A) Primary isolates: 855e, 1304a and 1620a. (B) Relapse isolates: 855h, 1304b and 1620d.
Effect of laboratory culture condition on B. pseudomallei K96243 lipid A structure
We considered the possibility that the lipid A structures of B. pseudomallei isolates may be modified during growth on TSA at 37 oC, the conditions used in the initial experiments. To address this, we tested whether the lipid A structure of B. pseudomallei K96243 differed following culture under a range of different laboratory conditions. All of lipid A spectra of B. pseudomallei cultured in different laboratory conditions had the same four main clusters of peaks at approximately m/z of 1575, 1670, 1801 and 1954 (S5 Fig). These data indicate that B. pseudomallei K96243 lipid A molecule was structurally conserved and suggested that the lack of variability in lipid A structure observed in our 136 clinical isolates was not a result of identical growth conditions.
Susceptibility of B. pseudomallei to polymyxin B
Cationic antimicrobial peptides (CAMPs), such as polymyxin B exerts their activity through electrostatic interactions with the lipid A moiety of LPS causing disruption of the outer membrane and cell death. Bacteria can develop resistance to colistin by modifying the structures of the lipid A moiety, hindering colistin binding. Aminoarabinose (AraN4) modification of pathogenic bacteria lipid A has been related to resistance to polymyxin B [32, 33]. B. pseudomallei has been reported to be resistant to polymyxin B [34]. Our lipid A structural characterizations showed that B. pseudomallei lipid A species were modified with Ara4N residues at the terminal phosphate groups that was observed by the ion at m/z 1801, and other associated ions. We postulated that the varying ion intensity of lipid A modified with Ara4N may contribute to variable polymyxin B resistance. To test the susceptibility of B. pseudomallei to polymyxin B, 21 representative isolates were chosen with notable varying ion intensity at m/z 1801. The results demonstrated all 21 isolates were resistant to polymyxin B with a minimum inhibitory concentrations (MICs) for all isolates ≥ 512 μg/ml.
Activation of human TLR4 by B. pseudomallei isolates
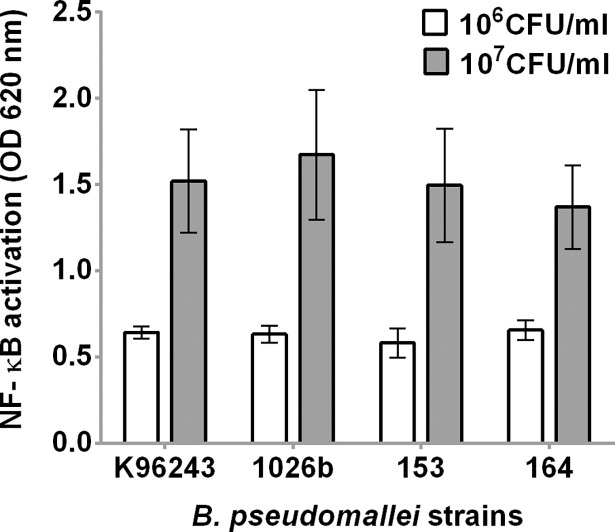
Our previous studies demonstrated that LPS of B. pseudomallei activates the innate immune responses in human monocytes through TLR4 [6, 35]. In this study, we considered whether different B. pseudomallei isolates cultured on the same medium differentially activate TLR4-dependent responses. We prepared heat-killed bacteria from four isolates (K96243, 1026b, 153 and 164) grown on TSA and stimulated HEK-Blue hTLR4 cells, expressing hTLR4-MD2-CD14 (Fig 5). All heat-killed B. pseudomallei isolates at 106 CFU/ml were able to induce NF-κB activation after 24 h at OD levels between 0.49–0.73 (median = 0.63, IQR = 0.58–0.68). At bacterial concentrations of 107 CFU/ml, the NF-κB activation was induced at OD level between 1.02 and 2.17 (median = 1.48, IQR = 1.24–1.72), respectively. We demonstrated no significant difference in TLR4-dependent NF-κB activation in cells stimulated with the four heat-killed bacterial isolates at 106 CFU/ml (P = 0.12) or 107 CFU/ml (P = 0.38).
Fig 5. Activation of NF-κB in HEK-Blue hTLR4 cells by four isolates of B. pseudomallei.
HEK-Blue hTLR4 cells were stimulated with heat-killed B. pseudomallei at 106 or 107 CFU/ml at 37°C in 5% CO2 for 24 h. NF-κB activation was determined in the supernatant by a SEAP reporter assay. Mean ± SD of three independent experiments were illustrated.
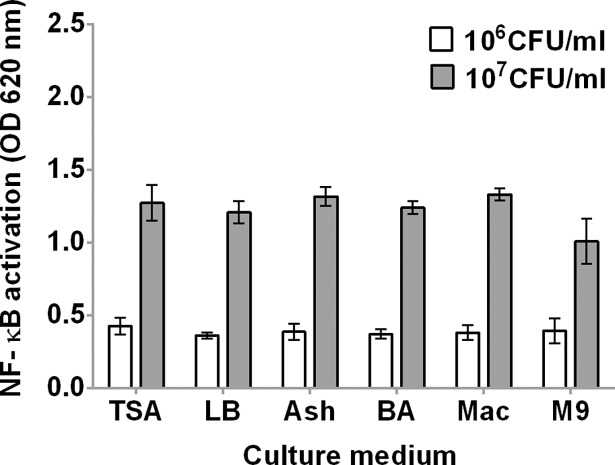
To test whether B. pseudomallei isolates cultured on various media differentially activate TLR4-dependent responses, we prepared heat-killed B. pseudomallei K96243 grown on six different agar media (TSA, LB, Ashdown agar, blood agar, MacConkey agar, M9 minimal medium agar) and stimulated HEK-Blue hTLR4 cells with bacteria at 106 and 107 CFU/ml (Fig 6). We demonstrated no significant difference in NF-κB activation induced by heat-killed B. pseudomallei K96243 on different media at 106 CFU/ml (P = 0.47). However, NF-κB activation induced by 107 CFU/ml of heat-killed B. pseudomallei K96243 cultured on M9 minimal medium agar was significantly lower than that induced by K96243 cultured on other media (P < 0.0001).
Fig 6. Activation of NF-κB in HEK-Blue hTLR4 cells by B. pseudomallei K96243 cultured in different media.
HEK-Blue hTLR4 cells were stimulated with heat-killed B. pseudomallei K96243 at 106 or 107 CFU/ml at 37°C in 5% CO2 for 24 h. NF-κB activation was determined in the supernatant by a SEAP reporter assay with HEK-Blue detection. Mean ± SD of three independent experiment were illustrated. TSA; trypticase soy agar, LB; Luria-Bertani agar, Ash; Ashdown agar, BA; Blood agar, Mac; MacConkey agar, M9; M9 minimal medium agar.
Activation of human TLR4 by LPS of B. pseudomallei isolates
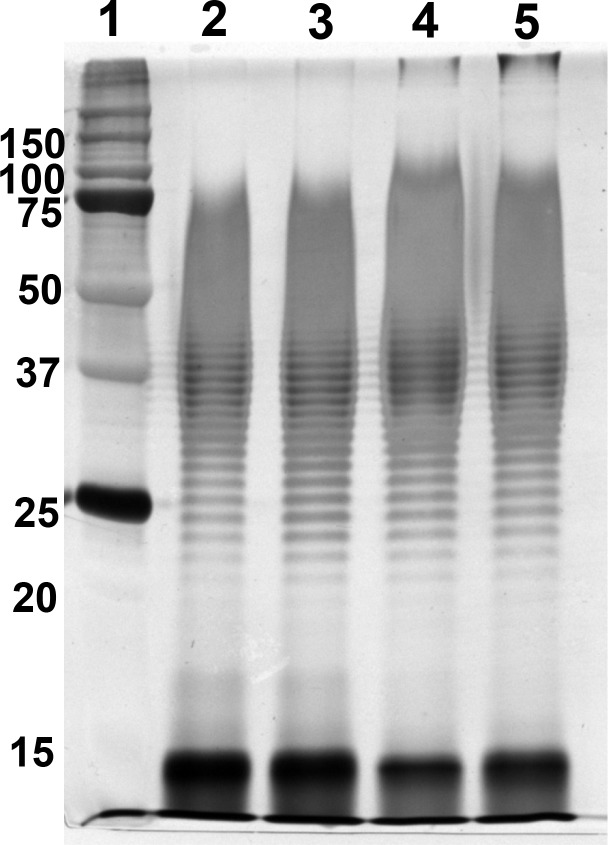
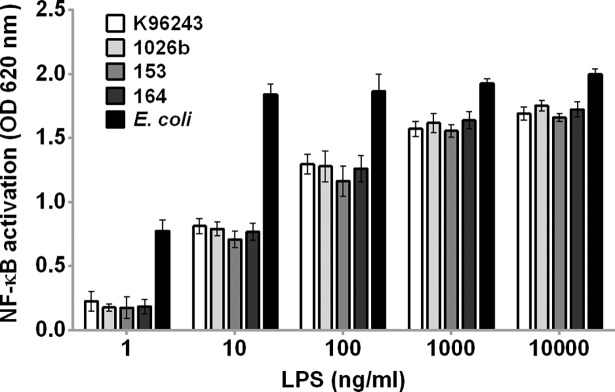
We previously demonstrated that LPS is a main driver of the innate immune response to B. pseudomallei [6]. Therefore, we further investigated whether LPS from different isolates potentially induce variable NF-κB activation. SDS-PAGE and silver stain of purified LPSs from all four isolates showed typical patterns of LPS type A (Fig 7). The Coomassie blue staining and BCA assay results showed no protein contamination. We determined NF-κB activation of HEK-Blue hTLR4 cells at 24 h after stimulation with LPSs from four B. pseudomallei isolates (K96243, 1026b, 153 and 164) at concentrations of 1, 10, 100, 1000, and 10,000 ng/ml. The LPSs of all isolates induced a significant increase of NF-κB activation in HEK-Blue hTLR4 cells above the baseline level in dose-dependent manner (Fig 8). Our results showed no significant difference among different B. pseudomallei isolates in TLR4-dependent NF-κB activation in cells stimulated with LPSs at 1 ng/ml (P = 0.68), 10 ng/ml (P = 0.14), 100 ng/ml (P = 0.33), 1000 ng/ml (P = 0.28) or 10,000 ng/ml (P = 0.29).
Fig 7. Purified LPS from B. pseudomallei isolates K96243, 1026b, 153 and 164.
LPS was purified by a modified hot phenol-water extraction method. Five micrograms of LPS was run by SDS-PAGE electrophoresis followed by silver staining. Lane 1 was a protein marker (number at the left are masses in kilodaltons). Lanes 2–5 were the purified LPS of B. pseudomallei strains K96243, 1026b, 153 and 164, respectively.
Fig 8. Activation of NF-κB in HEK-Blue hTLR4 cells by LPS of four B. pseudomallei clinical isolates.
The HEK-Blue hTLR4 cells were stimulated with LPS from B. pseudomallei K96243, 1026b, 153 and 164 at 1, 10, 100, 1000, and 10,000 ng/ml at 37°C in 5% CO2 for 24 h. The positive control was ultrapure Escherichia coli O111:B4 LPS. NF-κB activation was determined in the supernatant using a SEAP reporter assay. Mean ± SD of two independent experiments were illustrated.
Activation of human TLR4 by heat-killed B. pseudomallei OPS and capsule mutants
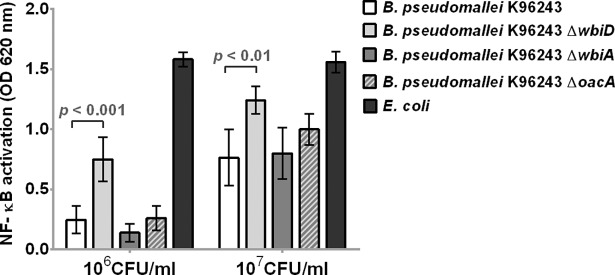
Although lipid A is the ligand for TLR4 [35], we hypothesized that there may be an effect of the OPS component of the LPS on this interaction. To investigate whether the presence of OPS on B. pseudomallei cells was associated with altered TLR4 signaling, we stimulated HEK-BlueTM-hTLR4 cells with heat-killed wild type K96243 and OPS mutant strains containing unmarked deletions of wbiD, wbiA, and oacA, which are involved in B. pseudomallei OPS biosynthesis [30]. We have previously shown that–like wild type B. pseudomallei K96243 –K96243 ΔwbiA and ΔoacA have type A ladder patterns, whereas K96243 ΔwbiD completely lacks OPS [30]. As shown in Fig 9, NF-κB activation was significantly higher in cells stimulated with 106 (P < 0.001) and 107 (P < 0.01) CFU/ml of B. pseudomallei K96243 ΔwbiD compared to those of wild type. We demonstrated no significant difference in TLR4-dependent NF-κB activation in cells stimulated with heat-killed bacteria obtained from B. pseudomallei K96243 ΔwbiA and ΔoacA compared to wild type at 106 CFU/ml or 107 CFU/ml. These data show that the absence of OPS does permit enhanced TLR4 signaling by heat killed B. pseudomallei.
Fig 9. Activation of NF-κB in HEK-Blue hTLR4 cells by heat-killed wild type K96243 and mutant containing unmarked deletions of wbiD, wbiA, and oacA.
HEK-Blue hTLR4 cells were stimulated with heat-killed bacteria at 106 or 107 CFU/ml at 37°C in 5% CO2 for 24 h. The positive control was E. coli ATCC 25922. NF-κB activation was determined in the supernatant by a SEAP reporter assay. Mean ± SD of two independent experiments were illustrated.
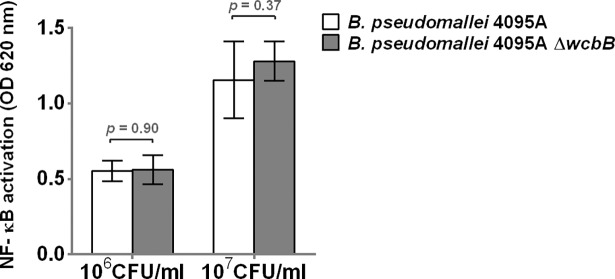
To further test whether the presence of capsule of B. pseudomallei cells interfered with TLR4 signaling, we stimulated HEK-BlueTM-hTLR4 cells with heat-killed wild type strain 4095a and a mutant containing unmarked deletions of wcbB (4095a ΔwcbB), which is involved in B. pseudomallei capsule biosynthesis [30]. Confirmatory testing with latex agglutination, SDS-PAGE, and Western blot of heat-killed bacteria indicated that the 4095a ΔwcbB did not express capsule. The results in Fig 10 demonstrated that NF-κB activation was not different in cells stimulated with 106 (P = 0.90) and 107 (P = 0.37) CFU/ml of B. pseudomallei 4095a ΔwcbB, as compared to those of wild type. Thus, the deletion of the capsule gene does not affect TLR4 signaling by heat killed B. pseudomallei.
Fig 10. Activation of NF-κB in HEK-Blue hTLR4 cells by heat-killed wild type 4095A and mutant containing unmarked deletions of wcbB.
The HEK-Blue hTLR4 cells were stimulated with heat-killed bacteria at 106 or 107 CFU/ml at 37°C in 5% CO2 for 24 h. NF-κB activation was determined in the supernatant by a SEAP reporter assay. Mean ± SD of two independent experiments were illustrated.
Activation of human TLR4 by LPS of B. pseudomallei OPS mutant
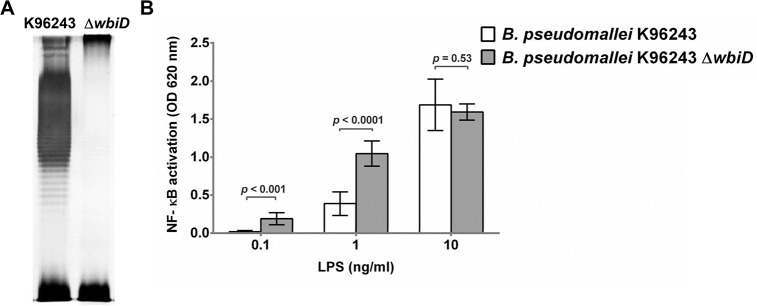
We then tested the impact of the OPS mutant on TLR4-dependent NF-κB activation by B. pseudomallei LPS. LPS was extracted and verified by SDS-PAGE and silver staining confirming the structural difference in LPS between wild type and isogenic mutant. The results in Fig 11A revealed ladder pattern with the presence of OPS for K96243 wild type but not for the K96243 ΔwbiD mutant, indicating the lack of OPS biosynthesis [30]. Stimulation of HEK-BlueTM-hTLR4 cells with different concentrations of LPSs from wild type and ΔwbiD mutant demonstrated that NF-κB activation was significantly higher in cells stimulated with ΔwbiD LPS at 0.1 ng/ml (P < 0.001) and 1 ng/ml, (P < 0.0001) compared with NF-κB activation in cells stimulated with wild type LPS. We observed no difference in NF-kB activation between cells stimulated with 10 ng/ml LPS of wild type and cells stimulated with 10 ng/ml LPS of the ΔwbiD mutant. We noted that the lipid A structure of K96243 ΔwbiD mutant and the wild type were identical (S6 Fig). These data provide additional confirmation that the absence of OPS allows enhanced TLR4 signaling by B. pseudomallei LPS.
Fig 11. Purified LPS from B. pseudomallei strain K96243 and K96243 ΔwbiD.
LPS was purified by a modified hot phenol-water extraction method. Five micrograms of LPS was run by SDS-PAGE electrophoresis followed by silver staining (A). Activation of NF-κB in HEK-Blue hTLR4 cells by LPS at 0.1, 1 and 10 ng/ml of B. pseudomallei K96243 and K96243ΔwbiD (B). NF-κB activation was determined in the supernatant using a SEAP reporter assay. The data were calculated to account for the difference in weight of wild type LPS and ΔwbiD mutant LPS. Mean ± SD of two independent experiments were illustrated.
Discussion
Human melioidosis is characterized by hyperinflammatory responses leading to high mortality rates even in patients who have received appropriate antimicrobial therapy. The initial interaction between B. pseudomallei and innate immune receptors such as the LPS-TLR4 interaction is important in the immunopathogenesis and outcome of infection. In this study, we characterized lipid A structure of B. pseudomallei isolates by MALDI-TOF MS followed by ESI-QqTOF MS and GC and examined the innate immune responses to the different bacterial isolates. Our data revealed the same lipid A profile for B. pseudomallei from a large collection of diverse isolates of LPS type A and demonstrated minimal variability in TLR4 signaling. Despite being cultured in different laboratory-induced conditions, B. pseudomallei K96243 lipid A expressed similar lipid A profiles, as characterized by MALDI-TOF MS. Moreover, we demonstrated that B. pseudomallei OPS interfered with TLR4 activation.
Our data show that the major lipid A species of B. pseudomallei contain a mixture of tetra- and penta-acylated lipid A species that are non-stoichiometrically substituted with Ara4N residues at both phosphate groups. We observed the presence of fatty acid C14:0, C14:0(2-OH), C14:0(3-OH), C16:0, and C16:0(3-OH). In comparison with other bacterial species, the lipid A structure of B. pseudomallei is unique. Previous published data showed that the fatty acid C14:0(2-OH) substituted into lipid A backbone of B. pseudomallei was not present in other closely related Burkholderia species, such as B. thailandensis, B. cepacia and B. mallei [17, 36, 37].
Although genomic analysis of within-host isolates showed substantial divergence from the founder genotype during a short period of acute infection [22], we found a remarkable lack of lipid A structural variation in 35 clinical B. pseudomallei isolates cultured from multiple body sites from a single patient with disseminated melioidosis. The data suggest that the lipid A is essential for B. pseudomallei and the modification during short period of acute infection is not required for bacterial fitness. Lipid A has an important role as it is the anchor for LPS on the outer membrane of B. pseudomallei. We previously showed that the synthesis of lipid A molecules is of vital importance among the various components that are responsible for outer membrane assembly [38].
Surprisingly, our study revealed similar lipid A spectra of 136 clinical isolates obtained from primary and relapse infections of 68 melioidosis patients, thus confirming that all B. pseudomallei isolates in our collection expressed a similar lipid A structure. In comparison to B. pseudomallei, B. mallei also produce both tetra- and penta-acylated lipid A species that are potent stimulators of hTLR-4-dependent cytokine production [36]. Other studies demonstrated that B. cenocepacia strain LMG 12614 expressed only penta-acylated lipid A species and it can induce stronger TNF-α and IL-6 responses than B. multivorans strain LMG 14273 that expressed both tetra- and penta-acylated lipid A [39].
Gram-negative organisms have evolved several LPS modification that benefit these organisms in their interactions with the host innate immune system and hostile environments during infections. The alteration of lipid A structure can promote resistance to antimicrobial peptides and interfere with host recognition by the innate immune system. Alterations are accomplished by many enzymes that modify the lipid A moieties by adding or removing acyl chains and phosphate groups. In some bacteria, lipid A are modified to evade immune recognition and survive within a host by reducing the charge of the bacterial surface via adding sugar onto lipid A molecules. The addition of phosphoethanolamine and aminoarabinose to lipid A molecules can protect bacteria against cationic antimicrobial peptides (CAMPs) [40, 41]. In Salmonella serovar Typhimurium, aminoarabinose modification at both phosphate groups have increased resistance to cationic antimicrobial peptides [11]. Our characterization of lipid A revealed that the lipid A species of all B. pseudomallei isolates were modified with aminoarabinose residues, thus demonstrated the resistance to polymyxin B with MIC value >512 μg/ml.
We observed no alteration of lipid A structure when B. pseudomallei strain K96243 was cultured under standard laboratory-induced conditions. However, we observed a slight decreased TLR4-dependent NF-κB activation after culture using M9 media. A previous study demonstrated that a variety of growth conditions alter the expression of Escherichia coli LPS polysaccharide formation especially in nutrient-depleted conditions [42]. The changes in LPS expression may extend to lipid A synthesis and result less TLR4-dependent NF-κB activation. In contrast, various other Gram-negative bacteria modify their lipid A structure when exposed to the external environment in vitro. For example, temperature-dependent structural changes in lipid A structure have been shown in Porphyromonas gingivalis and Yersinia species. P. gingivalis expresses mainly nonphosphorylated and monophosphorylated tetra-acylated lipid A structure at normal body temperature, whereas the major lipid A produced are monophosphorylated penta-acylated lipid A when the bacteria are grown at 41 oC. The temperature-dependent alteration in P. gingivalis lipid A structure is associated with more potent TLR4-dependent innate immune activation by LPS from bacteria grown at 41 oC than LPS from bacteria cultured at 37 oC [12]. Three pathogenic Yersinia; including Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis synthesize predominantly hexa-acylated lipid A at 21 oC which induces higher levels of cytokine production compared to tetra-acylated lipid A at 37 oC [14]. Additionally, Salmonella enterica serovar Typhimurium undergoes hydroxylation of fatty acid C14:0 to fatty acid C14:0(2-OH) when grown under magnesium-depleted conditions [11].
We also observed that the heat-killed bacteria and LPSs from different B. pseudomallei isolates activated human TLR4 and we demonstrated no significant difference among different B. pseudomallei isolates in TLR4-dependent NF-κB activation. Similarly, LPS from four clinical isolates resulted in similar TLR4-dependent NF-κB activation. These results are concordant with the invariable MALDI-TOF results characterizing B. pseudomallei lipid A. While increased natural diversity or heterogeneity of specific components of LPS, such as lipid A, can produce dramatic changes in host responses, this is not the case for B. pseudomallei.
Our results differ from recent reports. Norris et al. described different lipid A profiles among each of different LPS types in B. pseudomallei and the effect on innate immune activation suggesting that clinical strains with different LPS types may be the cause of clinical outcomes. However, their study used only one strain of B. pseudomallei for each LPS type (type A, B, B2 and rough) [19]. Our study was conducted entirely using LPS type A, which is predominant in Thailand, yet despite analysis of LPS from over 170 clinical isolates, found no differences that could potentially explain clinical variations. Another reason for the discrepancy in results may be due to the different MALDI-TOF matrix used. We used norharmane as a matrix for MALDI-TOF in contrast to the 2,5-dihydroxybenzoic acid used by Norris and colleagues. Norharmane is an optimum matrix for overcoming limitations of lipid A detection [24].
Despite our previous identification of B. pseudomallei lipid A as a TLR4 agonist [35], we also show here that TLR4-dependent NF-κB activation induced by an OPS mutant (rough LPS) is significantly greater than those induced by LPS from wild type B. pseudomallei. However, the response induced by a B. pseudomallei capsule mutant was comparable to those induced by the wild type. This observation is concordant with findings from a previous study showing that rough B. pseudomallei LPS induces higher nitric oxide and TNF-α levels in RAW 264.7 macrophages than smooth LPS [19], and suggests that there may be interference by the OPS on the LBP/CD14-mediated presentation or binding of LPS to the TLR4/MD-2 complex [9].
In conclusion, we have demonstrated that the structural features of B. pseudomallei lipid A are extremely conserved among different clinical B. pseudomallei isolates with LPS type A and result in no variation in TLR4-dependent innate immune activation. Further studies are required to evaluate the lipid A structure of environmental isolates and the role of OPS in the LPS-TLR4 interaction.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank the staff at Sunpasitthiprasong Hospital, Ubon Ratchathani, Mahidol-Oxford Tropical Medicine Research Unit and Department of Microbiology and Immunology, Faculty of Tropical Medicine, Bangkok, Thailand, and Department of Microbial Pathogenesis, University of Maryland, Baltimore, Maryland, USA for their assistance. We thank Onrapak Reamtong for technical assistance and Peeraya Ekchariyawat for useful comments. Authors would like to thank Melinda McFarland and Tim Croley at the FDA, Maryland, USA for access to Waters Synapt G2 mass spectrometer.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Faculty of Tropical Medicine research fund, Mahidol University (www.tm.mahidol.ac.th) to NC. SS was supported by the Royal Golden Jubilee PhD Programme (PHD/0069/2554) (http://rgj.trf.or.th). NC and TEW were supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH/NIAID)(https://www.niaid.nih.gov) under award number U01AI115520. RKE was supported by the National Institute of General Medical Sciences of the NIH (award number R01GM111066). The funders played no role in study design, data collection or interpretation, or the decision to submit the work for publication.
References
- 1.White NJ. Melioidosis. Lancet. 2003;361(9370):1715–22. [DOI] [PubMed] [Google Scholar]
- 2.Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. N Engl J Med. 2012;367(11):1035–44. doi: 10.1056/NEJMra1204699 [DOI] [PubMed] [Google Scholar]
- 3.Wiersinga WJ, Wieland CW, Dessing MC, Chantratita N, Cheng AC, Limmathurotsakul D, et al. Toll-like receptor 2 impairs host defense in gram-negative sepsis caused by Burkholderia pseudomallei (Melioidosis). PLoS Med. 2007;4(7):e248 doi: 10.1371/journal.pmed.0040248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedland JS, Suputtamongkol Y, Remick DG, Chaowagul W, Strieter RM, Kunkel SL, et al. Prolonged elevation of interleukin-8 and interleukin-6 concentrations in plasma and of leukocyte interleukin-8 mRNA levels during septicemic and localized Pseudomonas pseudomallei infection. Infect Immun. 1992;60(6):2402–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbas AK, Lichtman A. H., & Pillai S. Cellular and molecular immunology Philadelphia: Saunders/Elsevier; 2012. [Google Scholar]
- 6.Chantratita N, Tandhavanant S, Myers ND, Seal S, Arayawichanont A, Kliangsa-Ad A, et al. Survey of innate immune responses to Burkholderia pseudomallei in human blood identifies a central role for lipopolysaccharide. PloS one. 2013;8(11):e81617 doi: 10.1371/journal.pone.0081617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West TE, Chierakul W, Chantratita N, Limmathurotsakul D, Wuthiekanun V, Emond MJ, et al. Toll-like receptor 4 region genetic variants are associated with susceptibility to melioidosis. Genes Immun. 2012;13(1):38–46. doi: 10.1038/gene.2011.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anuntagool N, Wuthiekanun V, White NJ, Currie BJ, Sermswan RW, Wongratanacheewin S, et al. Lipopolysaccharide heterogeneity among Burkholderia pseudomallei from different geographic and clinical origins. Am J Trop Med Hyg. 2006;74(3):348–52. [PubMed] [Google Scholar]
- 9.Park BS, Lee JO. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med. 2013;45:e66 doi: 10.1038/emm.2013.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuura M. Structural Modifications of Bacterial Lipopolysaccharide that Facilitate Gram-Negative Bacteria Evasion of Host Innate Immunity. Front Immunol. 2013;4:109 doi: 10.3389/fimmu.2013.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, et al. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276(5310):250–3. [DOI] [PubMed] [Google Scholar]
- 12.Curtis MA, Percival RS, Devine D, Darveau RP, Coats SR, Rangarajan M, et al. Temperature-dependent modulation of Porphyromonas gingivalis lipid A structure and interaction with the innate host defenses. Infect Immun. 2011;79(3):1187–93. doi: 10.1128/IAI.00900-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Powell DA, Shaffer SA, Rasko DA, Pelletier MR, Leszyk JD, et al. LPS remodeling is an evolved survival strategy for bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(22):8716–21. doi: 10.1073/pnas.1202908109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic yersiniae. Mol Microbiol. 2004;52(5):1363–73. doi: 10.1111/j.1365-2958.2004.04059.x [DOI] [PubMed] [Google Scholar]
- 15.Ernst RK, Hajjar AM, Tsai JH, Moskowitz SM, Wilson CB, Miller SI. Pseudomonas aeruginosa lipid A diversity and its recognition by Toll-like receptor 4. J Endotoxin Res. 2003;9(6):395–400. doi: 10.1179/096805103225002764 [DOI] [PubMed] [Google Scholar]
- 16.SenGupta S, Hittle LE, Ernst RK, Uriarte SM, Mitchell TC. A Pseudomonas aeruginosa hepta-acylated lipid A variant associated with cystic fibrosis selectively activates human neutrophils. J Leukoc Biol. 2016;100(5):1047–59. doi: 10.1189/jlb.4VMA0316-101R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novem V, Shui G, Wang D, Bendt AK, Sim SH, Liu Y, et al. Structural and biological diversity of lipopolysaccharides from Burkholderia pseudomallei and Burkholderia thailandensis. Clin Vaccine Immunol: CVI. 2009;16(10):1420–8. doi: 10.1128/CVI.00472-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weehuizen TA, Prior JL, van der Vaart TW, Ngugi SA, Nepogodiev SA, Field RA, et al. Differential Toll-Like Receptor-Signalling of Burkholderia pseudomallei Lipopolysaccharide in Murine and Human Models. PloS one. 2015;10(12):e0145397 doi: 10.1371/journal.pone.0145397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norris MH, Schweizer HP, Tuanyok A. Structural diversity of Burkholderia pseudomallei lipopolysaccharides affects innate immune signaling. PLoS Negl Trop Dis. 2017;11(4):e0005571 doi: 10.1371/journal.pntd.0005571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limmathurotsakul D, Chaowagul W, Chierakul W, Stepniewska K, Maharjan B, Wuthiekanun V, et al. Risk factors for recurrent melioidosis in northeast Thailand. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2006;43(8):979–86. doi: 10.1086/507632 [DOI] [PubMed] [Google Scholar]
- 21.Maharjan B, Chantratita N, Vesaratchavest M, Cheng A, Wuthiekanun V, Chierakul W, et al. Recurrent melioidosis in patients in northeast Thailand is frequently due to reinfection rather than relapse. J Clin Microbiol. 2005;43(12):6032–4. doi: 10.1128/JCM.43.12.6032-6034.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price EP, Hornstra HM, Limmathurotsakul D, Max TL, Sarovich DS, Vogler AJ, et al. Within-host evolution of Burkholderia pseudomallei in four cases of acute melioidosis. PLoS Pathog. 2010;6(1):e1000725 doi: 10.1371/journal.ppat.1000725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Hamidi A, Tirsoaga A, Novikov A, Hussein A, Caroff M. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J Lipid Res. 2005;46(8):1773–8. doi: 10.1194/jlr.D500014-JLR200 [DOI] [PubMed] [Google Scholar]
- 24.Scott AJ, Flinders B, Cappell J, Liang T, Pelc RS, Tran B, et al. Norharmane Matrix Enhances Detection of Endotoxin by MALDI-MS for Simultaneous Profiling of Pathogen, Host, and Vector Systems. Pathog Dis. 2016. doi: 10.1093/femspd/ftw097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry MB, MacLean LL, Schollaardt T, Bryan LE, Ho M. Structural characterization of the lipopolysaccharide O antigens of Burkholderia pseudomallei. Infect Immun. 1995;63(9):3348–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somerville JE Jr., Cassiano L, Bainbridge B, Cunningham MD, Darveau RP. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. J Clin Invest. 1996;97(2):359–65. doi: 10.1172/JCI118423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CLSI. Methods for antimicrobial dilution and disc susceptibility testing of infrequently isolated or fastidious bacteria, 2th edition Approved Standard. A45–A2. Wayne, PA: The Institute; 2010. [Google Scholar]
- 28.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI document M100-S24. Wayne, PA: The Institute; 2014. [Google Scholar]
- 29.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests f or Bacteria That Grow Aerobically; Approved St andard—Ninth Edition CLSI document M07-A9. Wayne, PA: The Institute; 2012. [Google Scholar]
- 30.Wikraiphat C, Saiprom N, Tandhavanant S, Heiss C, Azadi P, Wongsuvan G, et al. Colony morphology variation of Burkholderia pseudomallei is associated with antigenic variation and O-polysaccharide modification. Infect Immun. 2015;83(5):2127–38. doi: 10.1128/IAI.02785-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duval BD, Elrod MG, Gee JE, Chantratita N, Tandhavanant S, Limmathurotsakul D, et al. Evaluation of a latex agglutination assay for the identification of Burkholderia pseudomallei and Burkholderia mallei. Am J Trop Med Hyg. 2014;90(6):1043–6. doi: 10.4269/ajtmh.14-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moskowitz SM, Ernst RK, Miller SI. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol. 2004;186(2):575–9. doi: 10.1128/JB.186.2.575-579.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunn JS. Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res. 2001;7(1):57–62. [PubMed] [Google Scholar]
- 34.Burtnick MN, Woods DE. Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin B resistance. Antimicrob Agents Chemother. 1999;43(11):2648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West TE, Ernst RK, Jansson-Hutson MJ, Skerrett SJ. Activation of Toll-like receptors by Burkholderia pseudomallei. BMC immunology. 2008;9:46 doi: 10.1186/1471-2172-9-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brett PJ, Burtnick MN, Snyder DS, Shannon JG, Azadi P, Gherardini FC. Burkholderia mallei expresses a unique lipopolysaccharide mixture that is a potent activator of human Toll-like receptor 4 complexes. Mol Microbiol. 2007;63(2):379–90. doi: 10.1111/j.1365-2958.2006.05519.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silipo A, Molinaro A, Cescutti P, Bedini E, Rizzo R, Parrilli M, et al. Complete structural characterization of the lipid A fraction of a clinical strain of B. cepacia genomovar I lipopolysaccharide. Glycobiology. 2005;15(5):561–70. doi: 10.1093/glycob/cwi029 [DOI] [PubMed] [Google Scholar]
- 38.Sengyee S, Saiprom N, Paksanont S, Limmathurotsakul D, Wuthiekanun V, Chantratita N. Susceptibility of Clinical Isolates of Burkholderia pseudomallei to a Lipid A Biosynthesis Inhibitor. Am J Trop Med Hyg. 2017;97(1):62–7. doi: 10.4269/ajtmh.16-0858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Soyza A, Ellis CD, Khan CM, Corris PA, Demarco de Hormaeche R. Burkholderia cenocepacia lipopolysaccharide, lipid A, and proinflammatory activity. Am J Respir Crit Care Med. 2004;170(1):70–7. doi: 10.1164/rccm.200304-592OC [DOI] [PubMed] [Google Scholar]
- 40.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem. 2001;276(46):43122–31. doi: 10.1074/jbc.M106961200 [DOI] [PubMed] [Google Scholar]
- 41.Lee H, Hsu FF, Turk J, Groisman EA. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol. 2004;186(13):4124–33. doi: 10.1128/JB.186.13.4124-4133.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson D, Bathgate AJ, Poxton IR. Monoclonal antibodies as probes for detecting lipopolysaccharide expression on Escherichia coli from different growth conditions. Microbiology. 1991;137(12):2741–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.