Here, Hizume et al. investigated the mechanisms underlying the proper pausing of replication forks at barriers on chromosomes, which is needed for genome integrity. Using reconstituted replication fork pausing from purified yeast replication proteins, the authors provide new insights into the mechanism of fork pausing and show that the processive properties of the fork against barriers are modulated by the association with regulatory factors.
Keywords: DNA replication, CMG helicase, Polε, replication fork barrier
Abstract
The proper pausing of replication forks at barriers on chromosomes is important for genome integrity. However, the detailed mechanism underlying this process has not been well elucidated. Here, we successfully reconstituted fork-pausing reactions from purified yeast proteins on templates that had binding sites for the LacI, LexA, and/or Fob1 proteins; the forks paused specifically at the protein-bound sites. Moreover, although the replicative helicase Cdc45–Mcm2–7–GINS (CMG) complex alone unwound the protein-bound templates, the unwinding of the LacI-bound site was impeded by the presence of a main leading strand DNA polymerase: polymerase ε (Polε). This suggests that Polε modulates CMG to pause at these sites.
DNA replication machineries assemble at specific sites, called replication origins, and travel on chromosomes to synthesize DNA. However, the replication machinery often faces obstacles, such as unusual secondary and tertiary structures of DNA, damaged DNA, and proteins bound to DNA. Cells manage the overriding of these obstacles by the machineries and maintain replication throughout chromosomes. Although structural hindrance of DNA has been analyzed well in conjunction with DNA polymerases, the manner in which DNA-binding proteins stall replication forks has not been elucidated well. The most analyzed example is the Tus protein, which stalls the replication forks at the termination sites (Ters) on the chromosome of Escherichia coli in an orientation-dependent manner (Khatri et al. 1989; Kobayashi et al. 1989; Hill and Marians 1990; MacAllister et al. 1990; Neylon et al. 2005). The Tus protein not only binds to the specific dsDNA sequence Ter (Hidaka et al. 1989; Hill et al. 1989; Sista et al. 1989) but also has a pocket for the conserved cytosine on the lagging strand template at the edge of the Ter sequence when unwound by the DnaB replicative helicase from the orientation in which the fork stalls (Mulcair et al. 2006). The insertion of a cytosine residue into the pocket locks the binding of Tus to Ters, which explains the orientation-dependent stalling of the replication fork (Mulcair et al. 2006; Pandey et al. 2015). The replication machinery comprises a DNA helicase, DNA polymerases, and other auxiliary factors. It has been demonstrated that a purified helicase stalls at Ters in the presence of Tus (Khatri et al. 1989; Lee et al. 1989).
In eukaryotes, several DNA-binding proteins impede the progression of replication forks in vivo. The yeast Fob1 protein, for example, binds to the replication fork barrier (RFB) at the ribosomal DNA (rDNA) locus (Kobayashi 2003) and stalls the replication fork in an orientation-dependent manner (Kobayashi and Horiuchi 1996; Kobayashi et al. 1998). This stalling requires the action of the fork protection complex, which consists of Tof1 and Csm3 (Mohanty et al. 2009) and works toward the maintenance of the copy number of rDNA (Kobayashi et al. 1998). Conversely, the lac repressor protein LacI binds to the LacO sequence inserted artificially onto chromosomes and stalls replication forks in an orientation-independent manner in vivo (Payne et al. 2006; Sofueva et al. 2011; Beuzer et al. 2014) and in in vitro Xenopus egg extracts (Duxin et al. 2014; Dewar et al. 2015). However, the manner in which these proteins stall replication forks is not understood.
Here, we established an in vitro system that reconstitutes fork pausing at the RFB and LacO sites from purified proteins. This in vitro assay demonstrated that Fob1 and Tof1–Csm3 are necessary and sufficient for fork pausing at the RFB in an orientation-dependent manner. Moreover, the LacI protein was sufficient to stall the replication fork at the LacO site. In contrast, the eukaryotic replicative helicase Cdc45–Mcm2–7–GINS (CMG) overrode the LacI-bound LacO site, and the addition of DNA polymerase ε (Polε; which mainly synthesizes leading strands at the replication forks), appeared to impede the unwinding by the CMG helicase when LacI was bound to the LacO site. Furthermore, the C-terminal noncatalytic domain of Pol2, a catalytic subunit of Polε, alone impeded the unwinding by CMG in a LacI-dependent manner, suggesting that the processive properties of the fork against barriers are modulated by the association with regulatory factors, such as Polε and Tof1–Csm3.
Results and Discussion
In vitro reconstitution of fork pausing at sites of protein binding
To establish an assay system to study the properties of the stalling of the replication fork at replication barriers, we constructed plasmids for use as the template DNA in the replication assay, which contained a replication origin (ARS1) and LacO (Fig. 1A) or SOS-box (Fig. 1B) on both sides of ARS1. LacI monomers bind strongly to LacO sequences (KD = 10−10 M) (Dong et al. 1999). LacI forms homodimers and homotetramers, and each subunit binds stably to DNA; thus, LacI dimers and tetramers exhibit an extremely stable interaction with DNA. The binding of the LexA protein to a DNA fragment containing one highly homologous SOS-box has a KD of 0.2 × 10−9 M (Kitagawa et al. 1985). To test whether these two strong DNA–protein interactions work as a barrier to the replication fork, we performed an in vitro replication assay using these plasmids (Fig. 1C). The in vitro replication assay was performed as summarized in Supplemental Figure S1A according to the method described in Yeeles et al. (2015), and the synthesized DNA was detected in a manner that depended on Cdc6 (Supplemental Fig. S1B), which is an essential factor for the loading of the replicative helicase core—the Mcm2–7 complex—onto origins (Donovan et al. 1997; Tanaka et al. 1997). Using either the LacO plasmid or SOS-box plasmid as a template, we detected two major smeared bands of synthesized DNA (Fig. 1C, lanes 1,9, respectively), which peaked at ∼3 kb and 200–400 base pairs (bp) in alkaline gel electrophoresis. The length of the synthesized DNA peaked at ∼3 kb, which corresponded to half of the plasmid length, suggesting that leading strands initiated at the ARS in each of the two directions and converged at the opposite side of the plasmid. Because we performed this assay without DNA ligase, which is required for Okazaki fragment maturation, the 0.2- to 0.4-kb and 3-kb bands corresponded to the lagging and leading strands, respectively. In the presence of LacI, the 3-kb band disappeared in the reaction using the LacO plasmid (Fig. 1C, lanes 2–4) but not in the reaction using the SOS-box plasmid (Fig. 1C, lanes 10–12). In contrast, in the presence of LexA, the intensity of the 3-kb signal was reduced in the reaction using the SOS-box plasmid (Fig. 1C, lanes 13–15) but not in the reaction using the LacO plasmid (Fig. 1C, lanes 5–7). These results suggest that replication forks reconstituted from purified proteins were blocked by the barrier of the LacI–LacO or LexA–SOS-box interaction, which is consistent with the fork pausing detected in yeast and mammalian cells (Sofueva et al. 2011; Beuzer et al. 2014) or frog extracts (Duxin et al. 2014; Dewar et al. 2015).
Figure 1.
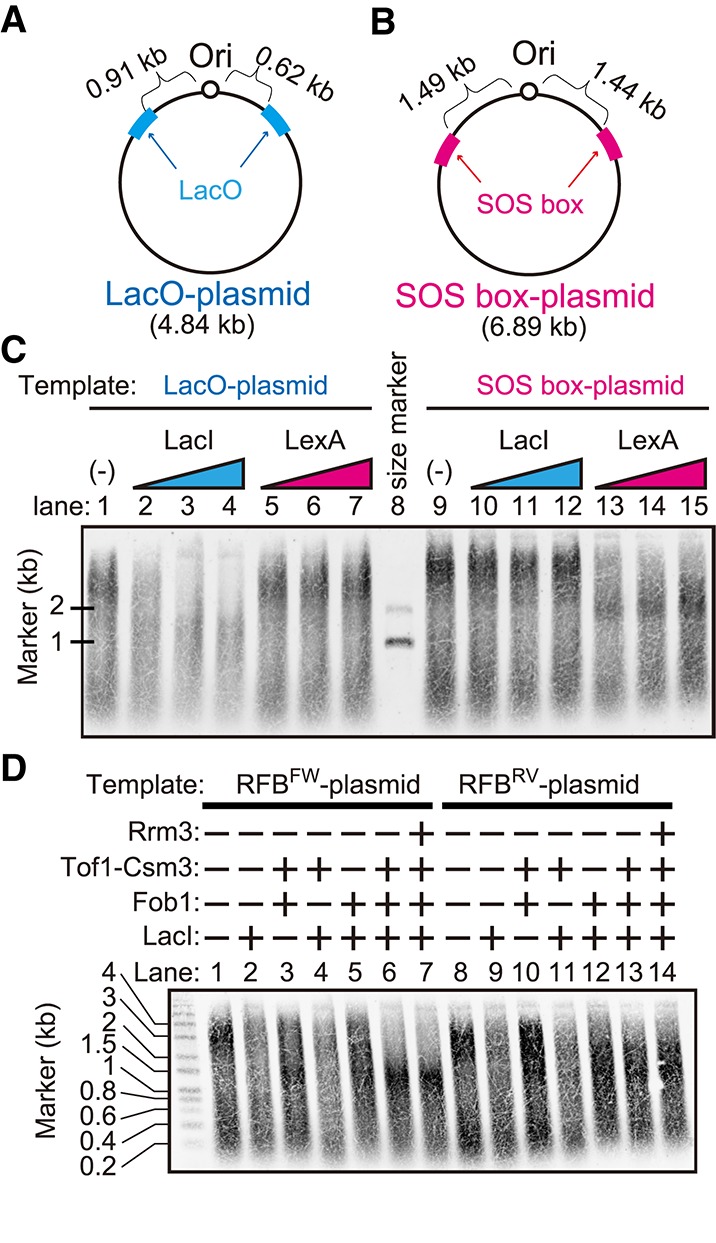
In vitro DNA replication pausing at barriers. (A,B) Plasmids used as templates in the in vitro DNA replication assay. A replication origin (ARS1 from budding yeast) was inserted, and the eight LacO tandem repeats (A) or four SOS-box tandem repeats (B) were inserted at both sides of the origin. (C) In vitro replication assay performed using the plasmids shown in A and B. In the presence of LacI or LexA, the reaction was performed and loaded onto an alkaline agarose gel, followed by blotting onto a membrane. The synthesized DNA incorporating biotin-UTP was detected using IR dye-labeled streptavidin. (D) In vitro replication assay using the plasmids shown in Supplemental Figure S1, A and B. Purified proteins, such as LacI and Fob1 (Supplemental Fig. S1C), were added to the indicated reaction.
Reconstitution of fork pausing at the RFB in a polar manner
Next, we examined the effect of the RFB on fork progression in vitro. Eight LacO and seven RFB copies were inserted on both sides of ARS1, respectively, and the direction of the RFB was constructed in the right or opposite direction against the replication from ARS1 (Supplemental Fig. S2A,B), termed the RFBFW plasmid and the RFBRV plasmid, respectively.
The in vitro replication assay was performed using these plasmids (Fig. 1D), and the measured intensities were plotted (Supplemental Fig. S2C,D). In the presence of LacI (Fig. 1D, lanes 2,9), the intensity of the signal around 3 kb became weaker than those shown in Figure 1D, lanes 1 and 8, suggesting that the synthesis of one of the leading strands was impeded at LacO sites. In the presence of LacI, Fob1, and Tof1–Csm3, both leading strands were blocked at the length of ∼1 kb in the assay using the RFBFW plasmid (Fig. 1D, lane 6). In the assay using the RFBRV plasmid, the length of the synthesized DNA was <3 kb but >1 kb (Fig. 1D, lane 13), suggesting that the pausing of the replication fork was less efficient in the case of the opposite direction of the RFB and that replication fork pausing at the RFB in a polar manner was successfully reproduced in this assay. The comparison between lanes 3 and 10 in Figure 1D clearly indicates fork pausing in an RFB orientation-dependent manner; in the absence of LacI but the presence of Fob1 and Tof1–Csm3 (Fig. 1D, lane 3), the length of the synthesized DNA became shorter than that in the absence of LacI, Fob1, and Tof1–Csm3 in the assay using the RFBFW plasmid (Fig. 1D, lane 1) but behaved similarly in the assay using the RFBRV plasmid (Fig. 1D, cf. lanes 8, 10). The RFB orientation-dependent fork pausing was also observed even in the presence of single RFB (Supplemental Fig. S3). When Fob1 or Tof1–Csm3 was omitted (Fig. 1D, lanes 4,5, respectively), the synthesis of the leading strand was >1 kb, implying that both Fob1 and Tof1–Csm3 are required for replication fork pausing at the RFB. Rrm3, which is a 5′ → 3′ DNA helicase (Ivessa et al. 2002) and another factor that has been reported to be involved in polar fork pausing at the RFB (Mohanty et al. 2006), was added to the assay (Fig. 1D, lanes 7,14), but the length of the synthesized DNA remained unchanged regardless of the presence of Rrm3.
To determine the site of replication fork pausing at each molecule, DNA was purified from the reaction of the condition shown in lane 6 in Figure 1D, digested with the restriction enzyme ScaI (which cut the opposite side of the ARS1) (Supplemental Fig. S2), and visualized by atomic force microscopy (AFM) (Fig. 2A). Replication bubbles with a replicated region in the middle and unreplicated regions at both ends were detected, suggesting that both the LacO and RFB regions block replication forks. Histograms of the measured length of replicated and unreplicated DNA are shown in Figure 2, B and C. The length of the unreplicated DNA was 764 nm ± 245 nm (mean ± SD; n = 44), which was consistent with the expected length of the sequence between the ScaI site and LacO or the RFB (740 nm of 2.2 kb). The length of the replicated DNA was 472 nm ± 287 nm (mean ± SD; n = 48), which was shorter than the expected length of the sequence between the RFB and LacO (540 nm of 1.6 kb), possibly because of shrinking of this ssDNA segment that remained in this region, as indicated by the arrows in Figure 2A. If we added only LacI but not Fob1or added Fob1and Tof1–Csm3 but not LacI, bubbled DNA was not observed by AFM, suggesting that the bubbled DNA was a specific product of fork blocking at both sides of ARS. This analysis using visualization by AFM suggests rigorous pausing of the replication fork at LacI–LacO and RFB sites.
Figure 2.
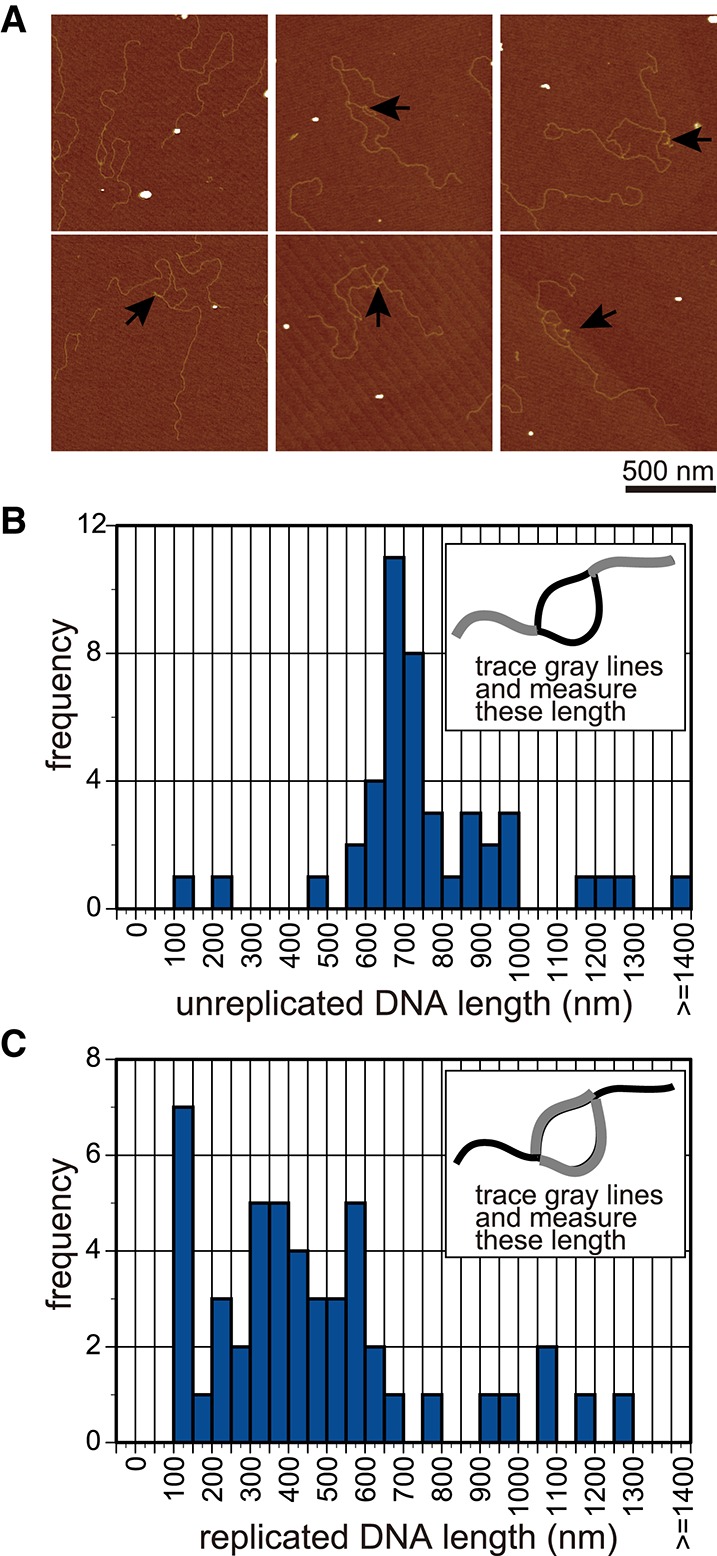
AFM analysis of the replication pausing. (A) AFM images of the DNA that was partially replicated. DNA was purified from the reaction of lane 6 in Figure 1C via treatment with phenol, cleaved with the restriction enzyme ScaI at the opposite side of the origin (Supplemental Fig. S2A,B), and observed by AFM. The ssDNA region, which was unwound and might not have been used for the synthesis of a complementary strand, was observed as the shrinking structure indicated by arrows. (B,C) The lengths of the unreplicated DNA (B) or replicated DNA (C) were measured and are shown in the histogram (n = 44 and n = 48, respectively).
Fork pausing at the RFB has been characterized extensively, and the requirements for this pausing have been identified: Fob1 (Kobayashi and Horiuchi 1996) and Tof1–Csm3 (Mohanty et al. 2006) are required for, and Rrm3 antagonizes, the pausing (Mohanty et al. 2006). However, it has not been known whether these factors are sufficient to explain the orientation-dependent (polar) pausing of the replication forks at the RFB. The results shown here clearly indicate that Fob1 and Tof1–Csm3, but not Rrm3, are sufficient to pause the replication forks at the RFB.
The CMG helicase overcomes barriers
The component at the head of the replication fork would be the CMG helicase. Therefore, it seems reasonable to think that the cause of the replication fork pausing at barriers is “helicase” pausing at these obstacles. In fact, the bacterial replicative helicase DnaB stops the progression at LacI–LacO (Yancey-Wrona and Matson 1992) and Tus–Ter (Khatri et al. 1989; Hiasa and Marians 1992). To test whether the progression of the CMG helicase is blocked by LacI–LacO, we performed a CMG helicase assay using a DNA template containing LacI–LacO.
We mixed the CMG helicase purified from a yeast strain that overexpressed all subunits of the complex (Supplemental Fig. S1C) and showed ATP-dependent helicase activity against a forked DNA substrate (Supplemental Fig. S4A). The forked DNA substrate contained a LacO sequence in the dsDNA region (Fig. 3A). Unexpectedly, CMG unwound the substrate regardless of the presence of LacI.
Figure 3.
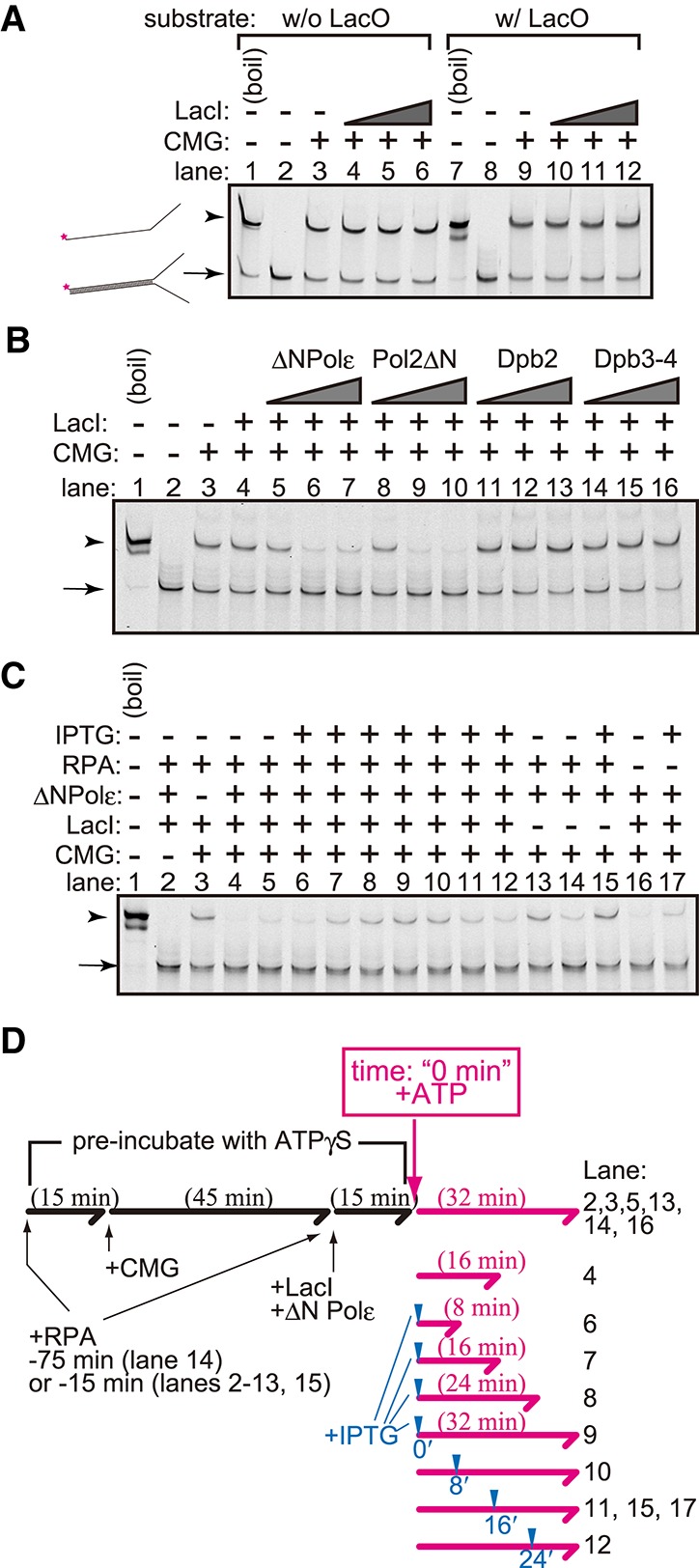
Helicase assay using a purified CMG complex. (A) The CMG helicase assay was performed using forked DNA with (lanes 7–12) or without (lanes 1–6) the LacO sequence as a substrate. CMG (200 fmol) was mixed with 2 fmol of the substrates and ATPγS in the presence (36, 72, and 144 fmol) or absence of LacI. ATP was added, and a helicase reaction was then performed in its presence. The reaction was stopped by adding EDTA and sarkosyl and was used for native PAGE analysis. DNA was detected by the infrared signal label located at the end of the forked substrate. (B) The CMG helicase assay was performed as shown in A, but 6, 18, and 54 fmol of the Polε–pol2ΔN complex (ΔNPolε), pol2ΔN, Dpb2, or the Dpb3–Dpb4 (Dpb3–4) complex was added. (C) The CMG helicase assay was performed as shown in A and B using 144 fmol of LacI, 54 fmol of the Polε–pol2ΔN complex (ΔNPolε), and 275 fmol of RPA. IPTG was added to the reaction at the final concentration of 10 mM at the time point indicated in D. (D) Flowchart of the helicase assay shown in C. The forked DNA substrates were preincubated with proteins in the presence of ATPγS for the indicated period (black lines and characters). The time point of the addition of IPTG and the incubation time after the addition of ATP are indicated by blue arrowheads and the lengths of the magenta arrows, respectively.
To date, covalently bound proteins or proteins with an extremely high affinity for DNA, such as that of avidin for biotinylated DNA, have been shown to block replicative helicases, like a road block (Kaplan 2000; Kaplan and O'Donnell 2002; Shin et al. 2003; Pacek et al. 2006; Fu et al. 2011; Langston and O'Donnell 2017). The E. coli Tus protein may be an example of the latter. It binds to the Ter sequence with extremely high affinity (KD = 3.4 × 10–13 M), thus stalling the helicase. In contrast, LacI binds to DNA with high affinity (KD = 10–10 M), and Fob1 binds to RFB similarly (no measured KD available, but Fob1 at the concentration of 1.4 × 10–9 M made the half of DNA in the reaction mixture be DNA–Fob1 complex) (Kobayashi 2003). Although the E. coli DnaB helicase is impeded by the LacI–LacO barrier (Yancey-Wrona and Matson 1992), the eukaryotic replicative helicase CMG alone unwinds dsDNA even in the presence of the fork barriers (Fig. 3A), probably because of its robust nature.
Requirement of Polε for the pausing of the CMG helicase at the LacI–LacO site
The LacI–LacO interaction blocked replication forks (Fig. 1) but not the CMG helicase (Fig. 3A). We hypothesized that a factor contained in the in vitro replication assay is required for CMG pausing at LacI–LacO and identified Polε as that factor. In the presence of Polε, LacI blocked the unwinding afforded by the CMG helicase (Fig. 3B, lanes 5–7). In this assay, we used a mutant Polε lacking the N-terminal region of the Pol2 subunit because this region contains a domain of DNA polymerase and proofreading exonuclease that degrades the substrate and the product of the helicase assay. In the absence of LacI, the mutant Polε did not affect the unwinding afforded by the CMG helicase (Supplemental Fig. S4B).
Polε consists of four subunits: Pol2, Dpb2, Dpb3, and Dpb4. To specify which subunit was responsible for CMG pausing, each subunit was added to the helicase assay. Dpb2 (Fig. 3B, lanes 11–13) and the Dpb3–Dpb4 complex (Fig. 3B, lanes 14–16) did not affect the unwinding afforded by the CMG helicase in the presence of LacI; in contrast, the addition of Pol2ΔN led to the blockage of CMG helicase-mediated unwinding (Fig. 3B, lanes 8–10). These results suggest that Pol2ΔN is required for the pausing of CMG at the barrier.
To determine whether CMG paused temporarily at the LacI–LacO site or was inactivated irreversibly, we added IPTG into the reaction to release LacI from the substrate DNA (Fig. 3C). We also added RPA, a ssDNA-binding protein, to the reaction because the unwound DNA tends to anneal during the pausing of CMG. In the presence of LacI, unwinding by the CMG helicase was inhibited in a Polε-dependent manner (Fig. 3C, lanes 3,5), but the addition of IPTG restored unwinding by the CMG helicase (Fig. 3C, lanes 10–12). The reaction time after the addition of IPTG was 8, 16, and 24 min in lanes 12, 11, and 10 of Figure 3C, respectively, and the band intensities of these lanes were similar to those of lanes 6, 7, and 8 of Figure 3C, respectively, in which IPTG was added before the addition of ATP, and the reaction times were 8, 16, and 24 min, respectively. This result suggests that CMG paused at the LacI–LacO site while keeping its helicase activity. The mixing of RPA with DNA before the addition of CMG (Fig. 3C, lane 14) led to the inhibition of the helicase activity, suggesting that prebinding of RPA to ssDNA regions blocks CMG loading. This result suggests that when CMG was paused at the LacI–LacO in the reaction shown in lanes 10–12 of Figure 3C, CMG reloading was inhibited by RPA binding to the unwound regions, and CMG paused at LacO sites and restored helicase activity after the addition of IPTG.
In the analysis that was performed using chemical cross-linking with a mass spectrometric readout (Sun et al. 2015), the C-terminal domains of Mcm2 and Mcm6 were cross-linked to the C-terminal half of Pol2. The C-terminal regions of the Mcm2–7 proteins have AAA+ motor domains, which support helicase progression. Thus, we speculate that the C terminus of Pol2 modulates the motor activity of the CMG helicase via an interaction with the motor domain. It is noteworthy that the polymerase domain of Pol2 is dispensable for the modulation of the pausing of the CMG helicase. CMG would be controlled through the direct interaction with the subunit of the leading strand DNA polymerase but not in a polymerase reaction-coupling manner. It is also interesting to note that the interaction between CMG and Polε sometimes enhances the processivity of leading strand synthesis (Schauer and O'Donnell 2017; Yeeles et al. 2017) in the absence of barriers and reduces its processivity in the presence of barriers, as shown here. Although we do not know the precise mechanism underlying this process, the interaction between CMG and Polε seems to tune the replication forks finely.
Pausing of the replication forks at barriers
Our results clearly showed that Polε is important for the modulation of CMG action in the pausing of replication forks. However, the interaction between CMG and Polε cannot account for all of the fork pausing observed in cells. The fork pausing at the RFB was successfully reconstituted in an orientation-dependent manner (Fig. 1D), whereas we did not detect RFB-dependent pausing of the CMG helicase even in the presence of Pol2ΔN or Fob1 and Tof1–Csm3, both of which are required for fork pausing at the RFB (Supplemental Fig. S5). It may be that the fork pausing that occurs at the RFB requires other replication factors or active DNA synthesis.
The activity of the CMG helicase against the DNA fragment containing the SOS-boxes was also tested (Supplemental Fig. S6). Unwinding by the CMG helicase was rarely inhibited by the addition of LexA (Supplemental Fig. S6, lanes 7–9) but was strongly inhibited by the addition of Polε even in the absence of LexA (Supplemental Fig. S6, lanes 10–12). The addition of both LexA and Polε showed the most effective inhibition of the CMG helicase (Supplemental Fig. S6, lanes 13–15), suggesting that Polε promotes CMG pausing at the secondary structure of the SOS-box sequence and that LexA enhances the pausing.
The existence of programmed fork arrest, such as that involving Tus–Ter and Fob1–RFB, showcases the importance of the pausing of the forks for genome integrity. In contrast, the multiple binding sites of LacI have been shown to reduce genome integrity (Sofueva et al. 2011; Beuzer et al. 2014). Thus, the replication forks are thought to be trapped by the sites of protein binding. Our results showed that the replication helicase alone overrides these obstacles, while the replication forks pause in the reconstituted system. Thus, we speculate that replication forks are paused or slowed down at the sites bound by proteins for setting chromosome environments such as chromatin structures with other factors that join these sites.
Materials and methods
Preparation of DNA for the in vitro replication assay
The sequences of the plasmids used in this study are listed in Supplemental Table S1.
Protein purification
Protein purification is described in detail in the Supplemental Material.
In vitro replication assay
The in vitro replication assay was performed as described previously (Yeeles et al. 2015) with some modifications. Details are described in the Supplemental Material.
AFM imaging of the DNA product of the in vitro replication assay
AFM imaging was performed as described previously (Hizume et al. 2017) with some modifications. Preparation of the DNA product of the in vitro replication assay for AFM imaging and detailed AFM imaging are described in the Supplemental Material.
CMG helicase assay
The helicase reaction was performed as described previously (Georgescu et al. 2014) with some modifications. Details are described in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank J.F.X. Diffley for the yeast strain and plasmids that were used to purify Dpb11, CDK, Polα, and Top2; S. Brill for the plasmid expressing RPA; P. Burgers for the plasmids used for the expression of PCNA and RFC; M.E. O'Donnell for the yeast strain used to purify CMG; E. Johansson for the plasmids expressing subunits of Polε; M. Yagura for the plasmids used to purify MCM–Cdt1, DDK, Mcm10, and Polδ; S. Tanaka for the plasmids used to purify Cdc6 and Sld3–Sld7; I. Muramatsu and A. Hosoe for technical assistance and help; and Bio-Nano Electronics Research Center organizers, Toyo University, for allowing us to use the facility in the revising process of this study. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant no. JP15H05975 to H.A.), Japan Society for the Promotion of Science KAKENHI (grant no. JP17H01443 to T.K.), and National Institute of Genetics (NIG)-JOINT (grant no. 85B2018 to K.H.).
Author contributions: K.H., T.K., and H.A. conceived the study. K.H., S.E., and S.M. performed the investigation. K.H. and H.A. wrote the manuscript. H.A. supervised the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.317073.118.
References
- Beuzer P, Quivy JP, Almouzni G. 2014. Establishment of a replication fork barrier following induction of DNA binding in mammalian cells. Cell Cycle 13: 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar JM, Budzowska M, Walter JC. 2015. The mechanism of DNA replication termination in vertebrates. Nature 525: 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Spott S, Zimmermann O, Kisters-Woike B, Muller-Hill B, Barker A. 1999. Dimerisation mutants of lac repressor. I. A monomeric mutant, L251A, that binds lac operator DNA as a dimer. J Mol Biol 290: 653–666. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. 1997. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci 94: 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Dewar JM, Yardimci H, Walter JC. 2014. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell 159: 346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu RE, Langston L, Yao NY, Yurieva O, Zhang D, Finkelstein J, Agarwal T, O'Donnell ME. 2014. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol 21: 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiasa H, Marians KJ. 1992. Differential inhibition of the DNA translocation and DNA unwinding activities of DNA helicases by the Escherichia coli Tus protein. J Biol Chem 267: 11379–11385. [PubMed] [Google Scholar]
- Hidaka M, Kobayashi T, Takenaka S, Takeya H, Horiuchi T. 1989. Purification of a DNA replication terminus (ter) site-binding protein in Escherichia coli and identification of the structural gene. J Biol Chem 264: 21031–21037. [PubMed] [Google Scholar]
- Hill TM, Marians KJ. 1990. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci 87: 2481–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TM, Tecklenburg ML, Pelletier AJ, Kuempel PL. 1989. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc Natl Acad Sci 86: 1593–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizume K, Kominami H, Kobayashi K, Yamada H, Araki H. 2017. Flexible DNA path in the MCM double hexamer loaded on DNA. Biochemistry 56: 2435–2445. [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev 16: 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DL. 2000. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J Mol Biol 301: 285–299. [DOI] [PubMed] [Google Scholar]
- Kaplan DL, O'Donnell M. 2002. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol Cell 10: 647–657. [DOI] [PubMed] [Google Scholar]
- Khatri GS, MacAllister T, Sista PR, Bastia D. 1989. The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell 59: 667–674. [DOI] [PubMed] [Google Scholar]
- Kitagawa Y, Akaboshi E, Shinagawa H, Horii T, Ogawa H, Kato T. 1985. Structural analysis of the umu operon required for inducible mutagenesis in Escherichia coli. Proc Natl Acad Sci 82: 4336–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. 2003. The Replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol 23: 9178–9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1: 465–474. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Hidaka M, Horiuchi T. 1989. Evidence of a ter specific binding protein essential for the termination reaction of DNA replication in Escherichia coli. EMBO J 8: 2435–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12: 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston L, O'Donnell M. 2017. Action of CMG with strand-specific DNA blocks supports an internal unwinding mode for the eukaryotic replicative helicase. Elife 6: e23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EH, Kornberg A, Hidaka M, Kobayashi T, Horiuchi T. 1989. Escherichia coli replication termination protein impedes the action of helicases. Proc Natl Acad Sci 86: 9104–9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAllister T, Khatri GS, Bastia D. 1990. Sequence-specific and polarized replication termination in vitro: complementation of extracts of tus- Escherichia coli by purified Ter protein and analysis of termination intermediates. Proc Natl Acad Sci 87: 2828–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Bairwa NK, Bastia D. 2006. The Tof1p–Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci 103: 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Bairwa NK, Bastia D. 2009. Contrasting roles of checkpoint proteins as recombination modulators at Fob1-Ter complexes with or without fork arrest. Eukaryot Cell 8: 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcair MD, Schaeffer PM, Oakley AJ, Cross HF, Neylon C, Hill TM, Dixon NE. 2006. A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell 125: 1309–1319. [DOI] [PubMed] [Google Scholar]
- Neylon C, Kralicek AV, Hill TM, Dixon NE. 2005. Replication termination in Escherichia coli: structure and antihelicase activity of the Tus–Ter complex. Microbiol Mol Biol Rev 69: 501–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. 2006. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587. [DOI] [PubMed] [Google Scholar]
- Pandey M, Elshenawy MM, Jergic S, Takahashi M, Dixon NE, Hamdan SM, Patel SS. 2015. Two mechanisms coordinate replication termination by the Escherichia coli Tus–Ter complex. Nucleic Acids Res 43: 5924–5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne BT, van Knippenberg IC, Bell H, Filipe SR, Sherratt DJ, McGlynn P. 2006. Replication fork blockage by transcription factor–DNA complexes in Escherichia coli. Nucleic Acids Res 34: 5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer GD, O'Donnell ME. 2017. Quality control mechanisms exclude incorrect polymerases from the eukaryotic replication fork. Proc Natl Acad Sci 114: 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Jiang Y, Grabowski B, Hurwitz J, Kelman Z. 2003. Substrate requirements for duplex DNA translocation by the eukaryal and archaeal minichromosome maintenance helicases. J Biol Chem 278: 49053–49062. [DOI] [PubMed] [Google Scholar]
- Sista PR, Mukherjee S, Patel P, Khatri GS, Bastia D. 1989. A host-encoded DNA-binding protein promotes termination of plasmid replication at a sequence-specific replication terminus. Proc Natl Acad Sci 86: 3026–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Osman F, Lorenz A, Steinacher R, Castagnetti S, Ledesma J, Whitby MC. 2011. Ultrafine anaphase bridges, broken DNA and illegitimate recombination induced by a replication fork barrier. Nucleic Acids Res 39: 6568–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H, O'Donnell ME. 2015. The architecture of a eukaryotic replisome. Nat Struct Mol Biol 22: 976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90: 649–660. [DOI] [PubMed] [Google Scholar]
- Yancey-Wrona JE, Matson SW. 1992. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res 20: 6713–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. 2015. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519: 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Janska A, Early A, Diffley JF. 2017. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell 65: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


