Abstract
Cleavage and polyadenylation is essential for 3′ end processing of almost all eukaryotic mRNAs. Recent studies have shown widespread alternative cleavage and polyadenylation (APA) events leading to mRNA isoforms with different 3′ UTRs and/or coding sequences. Here, we present a compendium of conserved cleavage and polyadenylation sites (PASs) in mammalian genes, based on approximately 1.2 billion 3′ end sequencing reads from more than 360 human, mouse, and rat samples. We show that ∼80% of mammalian mRNA genes contain at least one conserved PAS, and ∼50% have conserved APA events. PAS conservation generally reduces promiscuous 3′ end processing, stabilizing gene expression levels across species. Conservation of APA correlates with gene age, gene expression features, and gene functions. Genes with certain functions, such as cell morphology, cell proliferation, and mRNA metabolism, are particularly enriched with conserved APA events. Whereas tissue-specific genes typically have a low APA rate, brain-specific genes tend to evolve APA. In addition, we show enrichment of mRNA destabilizing motifs in alternative 3′ UTR sequences, leading to substantial differences in mRNA stability between 3′ UTR isoforms. Using conserved PASs, we reveal sequence motifs surrounding APA sites and a preference of adenosine at the cleavage site. Furthermore, we show that mutations of U-rich motifs around the PAS often accompany APA profile differences between species. Analysis of lncRNA PASs indicates a mechanism of PAS fixation through evolution of A-rich motifs. Taken together, our results present a comprehensive view of PAS evolution in mammals, and a phylogenic perspective on APA functions.
Cleavage and polyadenylation is an essential step for 3′ end maturation of almost all mRNAs in eukaryotes (Shi and Manley 2015; Proudfoot 2016). The site of cleavage, also known as poly(A) site (PAS), is defined by surrounding sequence motifs, which vary in different phylogenic groups (Tian and Graber 2012). In mammals, upstream motifs include the UGUA motif, A[A/U]UAAA hexamers or their variants, and U-rich motifs; downstream motifs include U-rich, UGUG, and G-rich motifs (Hu et al. 2005). In contrast, fewer and more degenerate motifs are present in yeasts (Graber et al. 1999; Mata 2013; Schlackow et al. 2013; Liu et al. 2017). PAS motifs function in a concerted fashion to define the strength of the PAS (Cheng et al. 2006), and potent downstream motifs can compensate weak upstream motifs (Nunes et al. 2010). Mutations of PAS motifs have been implicated in human diseases (Higgs et al. 1983; Bennett et al. 2001; Graham et al. 2007; Prasad et al. 2013; Hollerer et al. 2014), highlighting the importance of 3′ end processing for gene expression.
Recent genome-wide studies estimated that 50%–80% of eukaryotic mRNA genes harbor alternative cleavage and polyadenylation (APA) sites, leading to mRNA isoforms (Shepard et al. 2011; Derti et al. 2012; Hoque et al. 2013). APA sites in 3′ UTRs lead to isoforms with different 3′ UTR lengths. Because the 3′ UTR is a hotbed for regulatory elements involved in post-transcriptional control of gene expression, such as miRNA target sites and binding sequences for various RNA-binding proteins (RBPs), APA can modulate aspects of mRNA metabolism, including stability, translation, and localization (Mayr 2016; Tian and Manley 2017). In addition, a sizable fraction of APA sites are located in regions upstream of the last exon, resulting in APA isoforms with different coding sequences (Tian et al. 2007). APA profiles vary in different tissues and cell types (Zhang et al. 2005; Wang et al. 2008; Lianoglou et al. 2013; Sanfilippo et al. 2017). For example, transcripts in brain tend to have long 3′ UTRs, whereas those in testis show the opposite trend (Zhang et al. 2005; Li et al. 2016; Sanfilippo et al. 2017). In addition, APA can be globally regulated in cell proliferation, differentiation, and development, as well as in response to various environmental cues (Tian and Manley 2017). Both sequence motifs and protein factors have been shown to impact APA under different conditions (Zheng and Tian 2014; Xiao et al. 2016; Cannavò et al. 2017).
Recent transcriptomic studies revealed widespread expression of long noncoding RNAs (lncRNAs, >200 nt) (Derrien et al. 2012; Hon et al. 2017). Although lncRNAs are typically expressed at low levels, they are believed to play important roles in the cell, especially for regulatory events in the nucleus (Wu et al. 2017). Some lncRNAs are transcribed from standalone genes with their own promoters, also known as long intergenic ncRNA (lincRNA) genes; some are generated from divergent promoters that also drive the transcription of RNAs in the opposite direction, also known as PROMoter uPstream Transcripts (PROMPTs) or upstream antisense RNAs (uaRNAs); some are generated from enhancer regions, known as eRNAs (Lam et al. 2014; Rothschild and Basu 2017). Although it is generally believed that most, if not all, lncRNAs employ PASs for 3′ end maturation and transcriptional termination, their PAS motifs have not been extensively analyzed.
As 3′ end sequencing efforts advance and method sensitivity improves, an increasing number of PASs have been identified in genomes, posing the question as to what fraction of the PASs are functionally important. Here, we comprehensively map PASs in human, mouse, and rat genomes through 3′ end sequencing of more than 360 samples, totaling approximately 1.2 billion PAS reads. We identify and analyze conserved PASs of different types in mRNA and lncRNA genes. We examine the relevance of PAS conservation to gene expression and impacts of conserved APA on 3′ UTR motifs for post-transcriptional regulation, focusing on mRNA stability. We uncover sequence motifs around different types of APA sites, examine the influence of their mutations on APA changes, and reveal an evolutionary path to PAS fixation.
Results
PAS conservation in human, mouse, and rat genomes
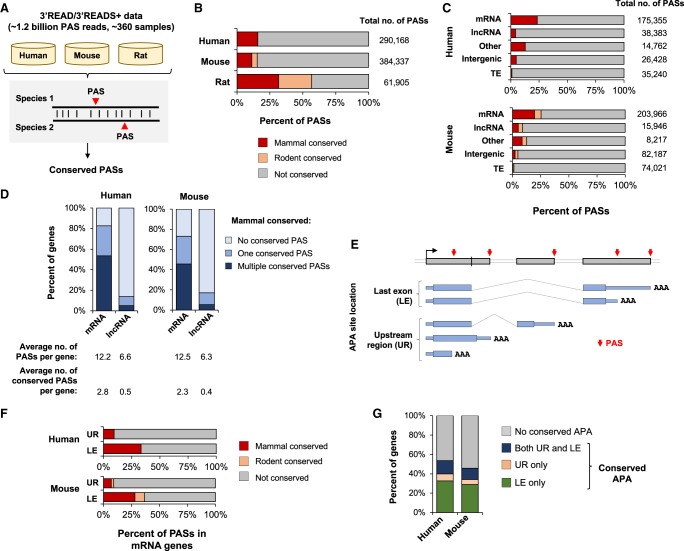
To understand PAS conservation in mammals, we set out to comprehensively map PASs in human, mouse, and rat genomes with 3′ end sequencing methods our laboratory recently developed, i.e., 3′ region extraction and deep sequencing (3′READS) (Hoque et al. 2013) or its updated version 3′READS+ (Zheng et al. 2016). These two methods eliminate the internal priming issue that plagues the 3′ end sequencing methods utilizing oligo(dT) for reverse transcription (Nam et al. 2002). As such, PASs located in A-rich regions can be unequivocally identified (Zheng et al. 2016). We used RNAs from a wide variety of cell and tissue types, totaling 364 samples (Supplemental Table S1). Overall, we obtained approximately 1.2 billion PAS-containing reads (or PAS reads) for the three species (Supplemental Table S1) and identified 290,168, 384,337, and 61,905 PASs in human, mouse, and rat genomes, respectively. The differences in PAS number between species are due mainly to variable numbers of 3′READS/3′READS+ reads used for mapping (Supplemental Table S1). This data set, available through the PolyA_DB database (http://polya-db.org/v3) (Wang et al. 2018), represents the most comprehensive PAS collection for mammals to date.
Using pairwise genome alignments and reciprocal best matches between species (Fig. 1A; Methods), we identified 46,022 mammal-conserved PASs (conserved between human and mouse or rat genomes), accounting for 11.3%–31.6% of the total sites in each of the three species (Fig. 1B). An additional set of 15,887 PASs were found conserved between mouse and rat only, accounting for 4.1% and 25.1% of total sites in mouse and rat, respectively. To determine whether we had sufficient amounts of sequencing reads to identify all conserved PASs, we carried out random sampling of data with different numbers of reads (Supplemental Fig. S1). We found that although human and mouse data were sufficient to cover most conserved PASs in these two species (Supplemental Fig. S1A), rat data were not (Supplemental Fig. S1B,C). Therefore, mammal-conserved PASs were defined mostly by human and mouse data, and rat data were used mainly for comparison with mouse for APA regulation (see below).
Figure 1.
Mapping and statistics of conserved PASs in mammals. (A) Mapping PASs in human, mouse, and rat genomes using 3′READS or 3′READS+ data, and identification of conserved PASs using the reciprocal best-match method. (B) Percentage of PASs conserved in human, mouse, and rat genomes. Two types of conservation are shown, i.e., conserved in mammals (human versus mouse or rat) and conserved in rodents only. Total number of PASs mapped for each species is shown. (C) Percentage of conserved PASs of different gene groups. The number of PASs in each group is indicated. The “other” group contains PASs from overlapping genes (on the same strand) and pseudogenes. (D) Percentage of mRNA or lncRNA genes with conserved PASs. The average number of PASs per gene (with or without conservation in mammals) is indicated. (E) APA sites in different loci of an mRNA gene. The sites are grouped into last exon (LE) or upstream region (UR). (F) Percentage of conserved UR and LE PASs of mRNA genes. (G) Percentage of genes with conserved APA sites in UR, LE, or both; UR APA conservation requires a gene to contain at least one conserved UR PAS, and LE APA conservation requires a gene to contain at least two conserved LE PASs.
Using RefSeq and Ensembl databases (Methods), we classified PASs into genes. To improve 3′ end definition of genes, we used RNA-seq data from the ENCODE project to connect PASs with annotated genes (Methods). On average, 53.1%–60.4% of the PASs of each species were assigned to mRNA, and 0.5%–13.2% to lncRNA genes (Supplemental Fig. S2A). An additional 12.1%–19.3% of PASs were within or near a transposable element (TE) (Supplemental Fig. S2A), and the rest were considered either as intergenic or other (from pseudogenes or overlapping genes) PASs. As expected, mRNA PASs account for most of the PAS reads in each genome (83.5%–88.0%) (Supplemental Fig. S2B), consistent with their higher expression levels than other transcript types.
Based on mammal-conserved PASs, we found that mRNA genes displayed greater PAS conservation than lncRNA genes by 2.8- to 5.6-fold (human and mouse only) (Fig. 1C). Overall, 73.1%–82.7% of the human and mouse mRNA genes contained at least one conserved PAS, and 45.6%–53.5% contained multiple conserved PASs (Fig. 1D). In contrast, 13.7%–17.1% of lncRNAs had at least one conserved PASs, and 5.1%–5.5% had multiple conserved PASs (Fig. 1D). PASs in intergenic regions were much less conserved than genic PASs (Fig. 1C), and TE-associated PASs, which are often species-specific, were least conserved across mammals (Fig. 1C).
Multiple PASs in the last exon (LE) often lead to mRNA isoforms with different 3′ UTR lengths, whereas PASs in an upstream region (UR) of LE could additionally change the coding sequence (Fig. 1E). We found that LE PASs outnumbered UR PASs by 1.4- to 2.6-fold in site (Supplemental Fig. S2A), and 12.5- to 20.2-fold in read number (Supplemental Fig. S2B). In addition, LE PASs were 3.6- to 4.0-fold more likely to be conserved than UR PASs (Fig. 1F). Overall, 40.1%–46.3% of human and mouse mRNA genes had conserved APA events in the last exon (Fig. 1G), and 16.6%–20.8% had conserved upstream region APA (Fig. 1G), with 11.4%–13.6% of genes having conserved events of both types (Fig. 1G). In summary, our comprehensive mapping of conserved PASs indicates that although a large fraction of mammalian PASs vary across species, PASs in mRNA genes, especially those in the last exon, are quite well conserved. Similar patterns were observed with the rat data (Supplemental Fig. S3).
Genes with conserved APA events
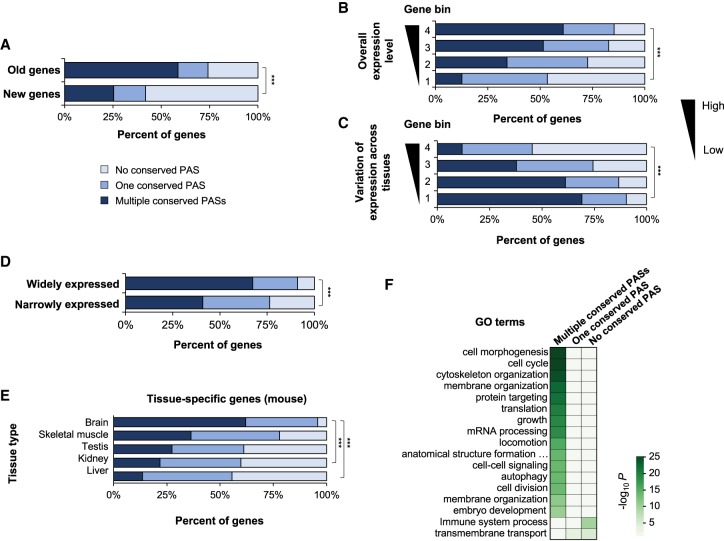
We next asked whether genes with conserved APA had distinct features. In view of our previous finding implicating a correlation between gene age and the frequency of APA sites (Lee et al. 2008), we classified mammalian genes in “old” or “new” groups, based on whether or not a gene had an ortholog in zebrafish (Methods). We found that old genes were about 2.3 times more likely to have conserved APA events than new genes (P = 7.0 × 10−15, χ2 test) (Fig. 2A). A similar trend was observed using more detailed grouping of genes into Eukaryota, Eumetazoa + Opisthokonta, Vertebrata, and Mammalia groups, based on gene classification by Liebeskind et al. (Supplemental Fig. S4A; Liebeskind et al. 2016). Therefore, gene age is an important determinant of APA conservation, suggesting that APA events are generally selected for in evolution.
Figure 2.
Features associated with mRNA genes with conserved APA. (A) Percentage of genes with conserved PASs in old and new genes. Old genes are mammalian genes with orthologs in zebrafish, and new genes are those without. (B) PAS conservation versus gene expression levels using mouse RNA-seq data. Genes were divided into four groups based on expression levels (average transcripts per million [TPM]) across 22 mouse tissues. (C) PAS conservation versus variation of gene expression levels. Genes were divided into four groups based on the coefficient of variation of expression levels across 22 mouse tissues. (D) PAS conservation versus breadth of gene expression (defined by the PaGenBase database). (E) PAS conservation for genes with tissue-specific expression (defined by the PaGenBase database; only tissues with more than 50 specific genes are shown). (F) Gene Ontology (GO) terms enriched for genes with different types of PAS conservation. P-values are shown in a heatmap using the indicated color scheme. Significance of difference between different groups in A–E are indicated: (***) P < 0.001 (χ2 test). Only mouse data are shown in this figure.
We next examined the relationship between APA conservation and gene expression features. Using ENCODE RNA-seq data (GSE36026) from 22 mouse tissues (Mouse ENCODE Consortium et al. 2012), we observed that genes with high expression levels overall (Fig. 2B) or with low variation across tissues (Fig. 2C) tended to have conserved APA events (P < 1.0 × 10−6, χ2 test comparing top and bottom groups) (Fig. 2B,C). These results were corroborated with human gene expression data from the GTEx project (Supplemental Fig. S4B,C; GTEx Consortium 2017). These findings appear in line with an earlier study that indicated that ubiquitously expressed genes are more likely to have APA sites than tissue-restricted genes (Lianoglou et al. 2013). Indeed, using PaGenBase database to define the breath of gene expression (Methods; Pan et al. 2013), we also found that broadly expressed genes had a higher rate of APA conservation than narrowly expressed genes (Fig. 2D). However, we additionally found that genes enriched for some tissues, such as brain, were actually more likely to have conserved APA than genes enriched for some other tissues, such as liver and kidney in both mice (P < 5.0 × 10−73, χ2 test) (Fig. 2E) and humans (P < 5.0 × 10−25, respectively, χ2 test) (Supplemental Fig. S4D), indicating that the tissue type in which a gene is mainly expressed also influences APA conservation.
We found a set of Gene Ontology (GO) terms (biological processes) were significantly enriched for genes with conserved APA, falling largely into three groups, namely, cell morphology (such as “cell morphogenesis” and “cytoskeleton organization”), cell proliferation (such as “cell cycle” and “growth”), and mRNA metabolism (such as “mRNA processing” and “translation”) (Fig. 2F). In contrast, only “immune system process” and “transmembrane transport” were found to be enriched for genes without conserved PASs (Fig. 2F). Transmembrane transport was also enriched for genes with only one conserved PAS with mild significance (Fig. 2F). Because genes involved in immune system process and transmembrane transport tend to evolve rapidly (Sojo et al. 2016), our GO data indicate that evolution of the 3′ end is connected to that of the coding region. Taken together, our result reveals three gene features acting as phylogenetic pressures on APA conservation, i.e., gene age, expression pattern, and biological functions.
Impacts of conserved PASs on 3′ UTR motifs
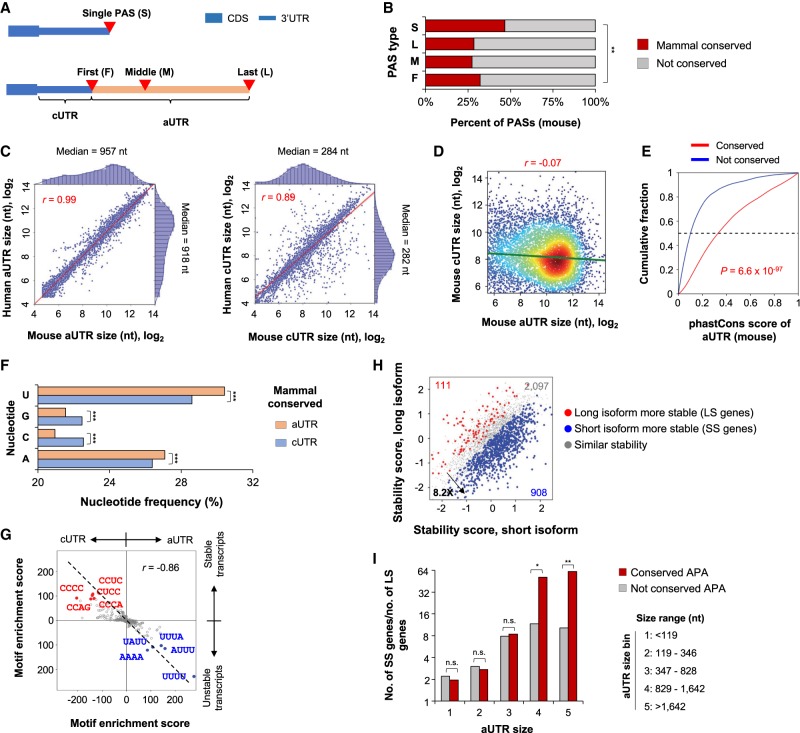
Most APA events of mRNA genes take place in 3′ UTRs located in the last exon (Supplemental Fig. S2A). To further analyze this group of APA, we divided 3′ UTR PASs into four types based on their relative locations: first (F), middle (M), and last (L) PASs when multiple 3′ UTR PASs existed (no M if only two conserved sites), or single PAS (S) when there was only one PAS (Fig. 3A). In mouse mRNA genes, S-type PASs had the highest conservation level than other types (46.7% were mammal conserved) (Fig. 3B), highlighting their functional importance, perhaps for termination of transcription. F-, M-, and L-type PASs displayed similar conservation levels, with ∼30% being conserved across mammals (Fig. 3B). These trends were similar in human and rat genes (Supplemental Fig. S5A).
Figure 3.
Conserved aUTRs. (A) Different types of PASs in the 3′ UTR: (S) single; (F) first; (M) middle; (L) last. As indicated, the region before the first PAS in a 3′ UTR is named common UTR (cUTR), and the region after is alternative UTR (aUTR). (B) Conservation of different types of 3′ UTR PASs: (**) P < 0.01 (χ2 test). (C) Correlation of aUTR length (left) or cUTR length (right) between human and mouse genes. A total of 6706 orthologous genes with conserved 3′ UTR APA are included. 3′ UTRs are divided into aUTRs and cUTRs using the first conserved and last conserved PASs (mammal conserved). Median length value for each species and Pearson correlation coefficient are indicated. (D) Comparison of conserved cUTR size and aUTR size of mouse genes. Pearson correlation coefficient is indicated. (E) Cumulative Distribution Fraction (CDF) curves of phastCons scores of conserved aUTRs versus nonconserved aUTRs. Nonconserved aUTRs are those between the first and last 3′ UTR PASs from genes without any mammal-conserved 3′ UTR PASs. (F) Nucleotide frequencies of conserved aUTRs and cUTRs (mouse sequences): (***) P < 0.001 (χ2 test). (G) Comparison of tetramers enriched in aUTRs versus cUTRs (x-axis) and those enriched for stable versus unstable transcripts in NIH3T3 cells (y-axis). Enrichment score is based on Fisher's exact test (Methods). Pearson correlation coefficient (r) is shown. Top enriched tetramers are indicated. (H) Comparison of mRNA stability between proximal PAS and distal PAS isoforms in NIH3T3 cells. Stability score is based on log2(ratio) of preexisting RNA to newly made RNA (Methods). SS genes (short 3′ UTR isoform significantly more stable than long 3′ UTR isoform) and LS genes (long 3′ UTR isoform significantly more stable than short 3′ UTR isoform) are highlighted. Significance is based on two replicates (Methods). (I) Ratio of gene number (SS genes versus LS genes) versus aUTR size. Genes are divided into five groups based on the aUTR size (range indicated): (*) P < 0.05; (**) P < 0.01; (n.s.) not significant (χ2 test).
The region between the first and last PASs is subject to APA regulation (Fig. 3A). For simplicity, it is named alternative 3′ UTR (aUTR). The median aUTR size between first and last conserved PASs was 957 nt and 918 nt in mouse and human genes, respectively (Fig. 3C). Importantly, the aUTR size was highly correlated between the two species (r = 0.99) (Fig. 3C, left). The region between the stop codon and the first conserved PAS was named common UTR (cUTR), because of its omnipresence in all 3′ UTR isoforms (Fig. 3A). Although the cUTR size was also conserved between human and mouse genes (median = 284 nt and 282 nt in mouse and human genes, respectively; r = 0.89) (Fig. 3C, right), no correlation was discernable between aUTR and cUTR sizes in the same gene (r = −0.07) (Fig. 3D). Using phastCons scores to reflect sequence conservation levels, we found that the aUTR sequences flanked by two conserved PASs were much more conserved than sequences flanked by nonconserved PASs (P = 6.6 × 10−97, Kolmogorov-Smirnov [K-S] test) (Fig. 3E). Similar results were also obtained in mouse versus rat comparisons (Supplemental Fig. S5B–D).
We found that conserved aUTRs tended to have significantly higher uridine (U) and adenosine (A) frequencies than conserved cUTRs (30.4% versus 28.6% for U, and 27.1% versus 26.4% for A; P < 1.0 × 10−140; χ2 test) (Fig. 3F). Consistently, A-rich and U-rich tetramers were highly enriched in conserved aUTRs, with the top five being UUUU, UAAA, AUUU, AAAU, and UUAA (Supplemental Fig. S6A; Supplemental Table S2). In contrast, the conserved cUTRs had higher cytidine (C) and guanosine (G) frequencies (P < 2.1 × 10−8, χ2 test) (Fig. 3F), and the top tetramers were CCCC, GCCC, GGAC, CCAG, and GGCC (Supplemental Fig. S6A; Supplemental Table S2). Because U-rich and C-rich motifs had been implicated in destabilizing and stabilizing mRNAs, respectively (Lee et al. 2010), our result suggested that conserved aUTRs and cUTRs can play opposing roles in mRNA stabilization. To examine this hypothesis, we performed a motif enrichment analysis using the mRNA stability data we recently generated with mouse NIH3T3 cells (Supplemental Fig. S6B; Zheng et al. 2018). Indeed, based on motif enrichment scores (Methods), motifs enriched for aUTRs were also enriched for unstable transcripts, and those for cUTRs were enriched for stable transcripts (Fig. 3G). Consistently, using our isoform-specific mRNA stability data of NIH3T3 cells (Zheng et al. 2018), we found that short 3′ UTR isoforms of a gene were much more likely to be more stable than long isoforms by 8.2-fold (Fig. 3H). Significantly, the longer the aUTR size, the more likely a short isoform was more stable than a long isoform (Fig. 3I). For example, for genes with an aUTR size >1642 nt, the bias in stability was ∼60-fold, whereas those with an aUTR size <119 nt, the bias was 1.9-fold (Fig. 3I). Importantly, this trend was more obvious with conserved APA isoforms than nonconserved ones, especially for genes with long aUTRs (Fig. 3I).
Metazoan 3′ UTRs contain miRNA target sites that exert post-transcriptional controls of gene expression (Bartel 2009). We found that genes with conserved APA tended to have significantly more miRNA target sites overall (P < 6.0 × 10−4, χ2 test) (Supplemental Fig. S6C) and a higher target site density (Supplemental Fig. S6D) compared with genes with only one conserved PAS or no conserved PAS (Supplemental Fig. S6C,D). Conserved aUTR sequences harbored 2.3 times as many miRNA target sites as conserved cUTR sequences (Supplemental Fig. S6C), despite having a lower density by 34.5% (Supplemental Fig. S6D). This result indicates that conserved APA events can have a substantial impact on the presence or absence of miRNA target sites in transcripts. In addition, we found that target sites of highly conserved miRNA families (103 in total) had a greater propensity to be in conserved aUTRs than those of moderately conserved families (114 in total; P = 5.2 × 10−6, K-S test) (Supplemental Fig. S6E), indicating concomitant evolution of aUTRs and miRNAs. Taken together, our data indicate that conserved aUTRs can substantially alter sequence motifs in 3′ UTRs.
Sequence motifs surrounding conserved PASs
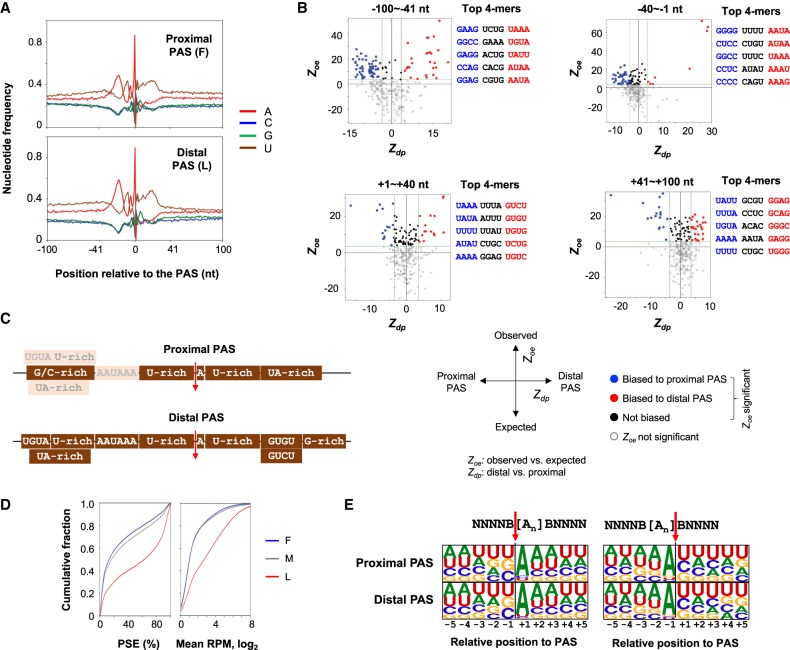
Previous studies indicated that proximal and distal PASs are surrounded with different sequence motifs (Tian et al. 2005). We reasoned that functional motifs could be better defined using conserved proximal and distal PASs, which are under purifying selection. To this end, we compared first and last conserved 3′ UTR PASs—named proximal and distal PASs, respectively—to identify respective motifs. Overall, proximal and distal PASs had similar nucleotide profiles (±100 nt around the PAS) (Fig. 4A). Using an approach similar to the Polyadenylation-Related Oligonucleotide Bidimensional Enrichment (PROBE) method we previously developed (Hu et al. 2005), we generated two values for each tetramer, Zoe for observed occurrence versus expected occurrence, reflecting the significance of enrichment in the PAS region, and Zdp for difference between distal and proximal PASs (Methods; Fig. 4B).
Figure 4.
Motifs around conserved proximal and distal PASs. (A) Nucleotide frequency around (±100 nt) conserved proximal and distal PASs. (B) Zoe versus Zdp for tetramers in four regions around the PAS. The four regions are indicated. Zoe and Zdp greater than 3.6 were used to define significantly biased tetramers. Tetramers with significant Zoe are highlighted, with those biased to distal PASs in red, those biased to proximal PASs in blue, and others in black. The top five tetramers in each category are listed. (C) Summary of B. The darkness of each box reflects its level of enrichment, highlighting the difference between proximal and distal PASs. (D) PSE (left) and mean RPM (right) of first, middle, and last PASs of 3′ UTRs (all mammal conserved). (E) Sequence motifs at the cleavage site, shown as sequence logos. Two scenarios are considered, as illustrated above the logos. (Left) Cleavage site is considered to be immediately after a non-A nucleoside (shown as B) and before an adenosine. (Right) Cleavage site is considered to be immediately after an adenosine and before a B.
We examined four regions around the PAS, namely, −100 to −41 nt, −40 to −1 nt, +1 to +40 nt, and +41 to +100 nt, with the PAS set at position 0 (Fig. 4B; Supplemental Table S3). The motifs enriched for distal PASs compared to proximal PASs included UGUA and UA-rich motifs in the −100 to −41 nt region, AAUAAA (UAAA, AAUA, and AUAA) in the −40 to −1 nt region, UGUG (UGUG and GUGU) and GUCU (GUCU, UCUG, and UGUC) motifs in the +1 to +40 nt region, and G-rich motifs (GGAG, GGGC, GAGG) in the +41 to +100 nt region. In contrast, proximal PASs had G/C-rich motifs enriched in the upstream region of PAS, and U-rich, A-rich, and UA-rich motifs in the downstream region (Fig. 4B; Supplemental Table S3). In addition, both proximal and distal PASs had similar enrichments of U-rich motifs in the −40 to −1 nt region (Fig. 4B). Because the motifs enriched for distal PASs are generally considered to be enhancing elements for PAS usage (Hu et al. 2005; Shi and Manley 2015), this result (Fig. 4C) indicates that distal PASs in general are stronger than proximal PASs. This notion was also supported by expression analysis of APA isoforms. Using the percentage of samples with expression (PSE) and average reads per million across samples to reflect PAS usage levels (Fig. 4D), we found that isoforms using the last conserved PASs were expressed at much higher levels than those using the first conserved PASs (Fig. 4D). In addition, we found that conserved S-type PASs (the sole conserved PAS in 3′ UTR) showed greater similarities in surrounding motifs to distal PASs than to proximal PASs (for correlation coefficients, see Supplemental Fig. S7).
Early biochemical studies and limited PAS surveys indicated the CA motif as the preferred site for 3′ end cleavage (Sheets et al. 1990; Zhao et al. 1999). We thus wanted to examine if this holds with conserved PASs and whether there is a difference between proximal and distal PASs. Because the cleavage site cannot be precisely identified when cleavage takes place next to an adenosine (A residues on genome cannot be distinguished from the poly(A) tail sequence), we considered two scenarios, i.e., the cleavage site is immediately upstream of adenosine residue(s) (scenario 1, Fig. 4E, left) or immediately downstream (scenario 2, Fig. 4E, right). In either case, we found a strong tendency of an adenosine being next to the cleavage site and a mild bias to uridines around the site (Fig. 4E). No enrichment of cytosine could be identified, and no differences could be discerned between proximal and distal PASs (Fig. 4E, top versus bottom). The same result was also obtained without clustering adjacent PASs, eliminating the possibility that merging heterogenous cleavage sites may mask the CA motif (Supplemental Fig. S8). Therefore, our genome-wide analysis using conserved PASs does not support the long-standing notion that the CA motif is preferred for 3′ end cleavage. Instead, cleavage tends to take place next to an adenosine, which is in good agreement with the biochemical study by Chen et al. (1995) and a previous bioinformatic analysis with a much smaller PAS set based on EST sequences from multiple species (Li and Du 2013). In addition, although our sequencing data could not distinguish the two scenarios, we think, based on the biochemical data from Chen et al. (1995), pre-mRNA cleavage immediately upstream of an A residue (scenario 1) is more likely to occur. Therefore, as shown in Figure 4E, we indicate 3′ adenosine as a determining feature for cleavage (Fig. 4E).
PAS conservation impacts robustness of gene expression and APA regulation
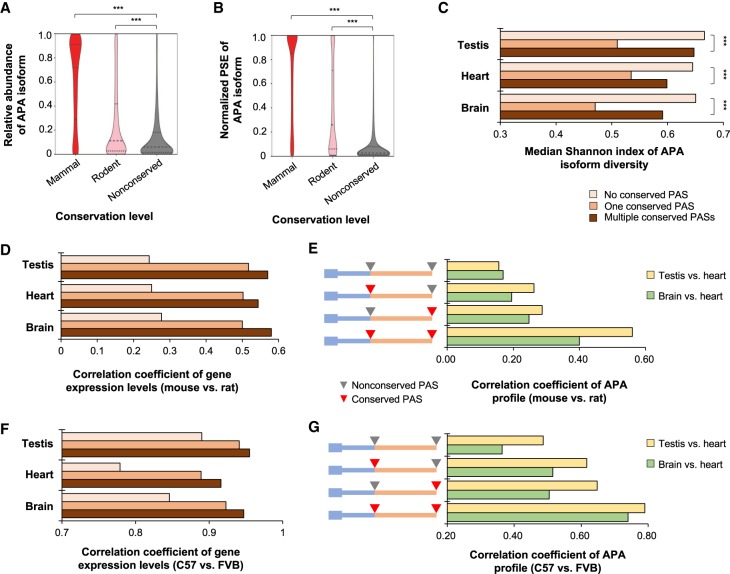
Using relative abundance of APA isoforms (Methods), we found that isoforms with mammal-conserved PASs were generally expressed at higher levels than those with PASs conserved in rodents only or with nonconserved PASs (Fig. 5A). A similar trend was observed using normalized PSE (Methods) to reflect the breadth of transcript expression (Fig. 5B). These results indicated that conserved PASs might function to suppress the usage of nonconserved sites, which might arise from promiscuous 3′ end processing. To further examine this, we used the Shannon index to reflect 3′ end diversity (Methods), and examined APA isoforms in mouse brain, heart, and testis. As expected, genes with multiple conserved PASs had a higher 3′ end diversity than genes with only one conserved PAS (Fig. 5C). However, genes without conserved PASs had significantly higher APA isoform diversity than genes with conserved PASs (P < 5.1 × 10−9 for the three tissues, K-S test) (Fig. 5C).
Figure 5.
PAS conservation impacts gene expression and APA regulation. (A) Relative abundance of APA isoform versus PAS conservation levels. Relative abundance of APA isoform is based on all isoforms of a gene. (B) Isoform expression breadth versus PAS conservation level. Isoform expression breadth is based on PSE normalized to the maximum PSE of all isoforms of the gene. (C) Median Shannon index for APA isoforms of three types of genes, including genes with no conserved PAS, one conserved PAS, and multiple conserved PASs. Data for testis, heart, and brain are shown. For A–C: (***) P < 0.001 (K-S test). (D) Comparison of gene expression levels between mouse and rat for three types of genes. Gene types and tissue types are as in C. (E) Correlation of APA profiles between mouse and rat for four types of genes. As illustrated, the four types of gene are genes with (1) no conserved APA sites, (2) only proximal PAS conserved, (3) only distal PAS conserved, and (4) both proximal and distal PASs conserved. APA profiles are based on testis versus heart or brain versus heart. (F) As in D, except that data comparing two mouse strains, C57BL/6J (C57) and FVB/JN (FVB), are shown. (G) As in E, except that data comparing two mouse strains (C57 versus FVB) are shown.
Because transcripts with different 3′ ends can have different mRNA stability potentials (Geisberg et al. 2014; Zheng et al. 2018), we reasoned that high 3′ end diversity might lead to gene expression variability and, conversely, low 3′ end diversity resulting from PAS conservation might make gene expression levels more stable. To test this hypothesis, we compared matched rat and mouse tissues for consistency of gene expression levels. Indeed, genes with conserved PASs tended to have higher correlation values between corresponding rat and mouse tissues than genes without conserved PASs (Fig. 5D). In addition, genes with multiple conserved PASs were better correlated in expression than genes with only one conserved PAS (Fig. 5D).
We next asked whether conserved APA sites would make APA regulation more consistent across species. To this end, we used the heart sample as a reference and compared its APA profile with that of testis or brain. For each comparison, we used the relative expression difference (RED) score between proximal and distal PASs to reflect APA difference (Δlog2[distal PAS/proximal PAS], brain or testis versus heart) (Methods). As previously reported (Tian and Manley 2017), testis and brain showed global preferences for proximal and distal PAS usages, respectively (Supplemental Fig. S9A). Importantly, the correlation of APA regulation between mouse and rat tissues was much higher for genes with two conserved sites compared to those with one (either proximal or distal) or no conserved PAS (Fig. 5E).
To further explore the relationship between gene expression and APA stability for genes with different APA conservation levels, we extracted RNAs from brain, heart, and testis of two mouse strains, namely, FVB/NJ and C57BL/6J, and subjected them to 3′READS+ analysis. Consistent with the mouse versus rat data, genes with conserved PASs displayed greater correlation of gene expression than those without conserved PASs (Fig. 5F). In addition, APA profiles were much more correlated between the two strains for genes with multiple conserved PASs compared to those with only one or no conserved PASs (Fig. 5G; Supplemental Fig. S9B). Taken together, our data indicate that conservation of PAS leads to low 3′ end diversity, stable gene expression levels, and consistent APA patterns.
Mutations of the U-rich motif lead to APA variations
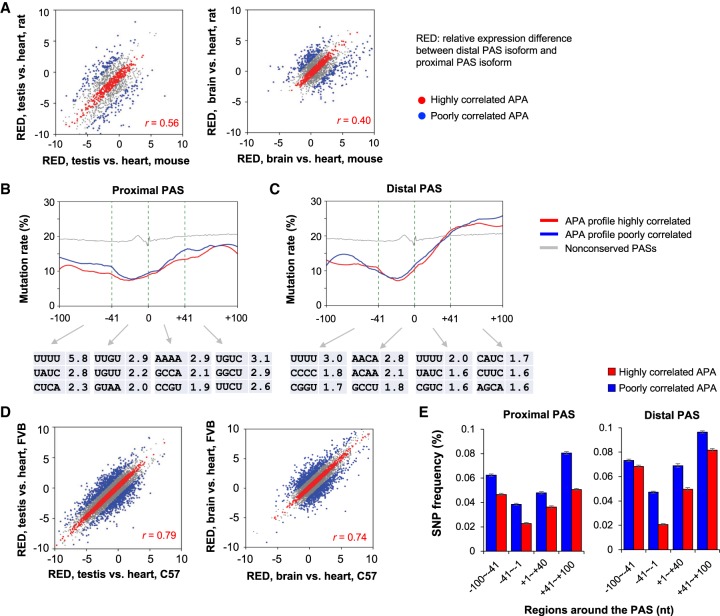
Although APA profiles of conserved sites were generally correlated between mouse and rat, variability was clearly discernable (Fig. 6A). We thus asked how sequence variations contribute to APA changes between the two species. To this end, we first identified conserved APA events that were highly or poorly correlated between mouse and rat, based on testis versus heart and brain versus heart comparisons (Fig. 6A; Supplemental Fig. S9C). We then examined sequence conservation levels around the proximal and distal PASs. As expected, the sequences around the conserved PASs, regardless of proximal or distal, had lower mutation rates than those around nonconserved PASs (Fig. 6B,C). Conservation was particularly high in the −40 to −1 nt region of the PASs, highlighting its importance. In contrast, the downstream region of distal PASs had higher mutation rates than other regions (Fig. 6C), indicating low negative selection pressures after the last PAS of genes. Importantly, the proximal PASs of genes with highly correlated APA profiles showed higher conservation levels than poorly correlated APA profiles, especially in the −100 to −41 nt region (Fig. 6B, blue versus red lines). A similar, albeit less obvious, trend could be discerned with distal PASs (Fig. 6C).
Figure 6.
Sequence variation leads to APA changes. (A) Correlation of APA profiles between mouse and rat. (Left) APA profile based on testis versus heart; (right) APA difference between brain and heart. APA difference was calculated as RED = Δlog2(distal PAS/proximal PAS), testis or brain versus heart. Genes in red have highly correlated APA events (middle 30% of genes based on REDmouse − REDrat), and those in blue have poorly correlated APA events (top and bottom 10% of genes based on REDmouse − REDrat). (B, top) Nucleotide mutation rate (percentage of mutations at each position per gene) around the conserved proximal PAS based on mouse and rat comparison. Red and blue lines are genes with highly correlated and poorly correlated APA profiles from A, respectively. (Bottom) Top three enriched tetramers in mutated sequences of genes with poorly correlated APA events. −log10(P) values (Fisher's exact test) are shown. (C) As in B, except that data for conserved distal PASs are shown. (D) As in A, except that comparisons are based on two mouse strains, C57BL/6J (C57) and FVB/JN (FVB). (E) SNP frequencies around conserved proximal and distal PASs of genes with highly correlated (red) or poorly correlated APA events (blue) between two mouse strains.
We next examined mutation rates of individual tetramers around the poorly correlated PASs versus highly correlated ones. UUUU in the −100 to −41 nt region was found be significantly enriched for the poorly correlated group, for both proximal and distal PASs (Fig. 6B). This result indicates that mutations of U-rich motifs upstream of the PAS contribute to APA variability between species.
To further examine the contribution of sequence variation to APA variability, we calculated single-nucleotide polymorphic (SNP) site frequencies around the PASs of C57BL/6J and FVB/NJ strains (Fig. 6D; Supplemental Fig. S9D; Wong et al. 2012). Consistent with the mouse versus rat analysis, the −40 to +40 regions around both proximal and distal PASs had lower SNP rates (frequency of SNPs in a given region per gene) than flanking regions (Fig. 6E). Importantly, almost in all regions around the PASs, genes with high APA correlation showed lower SNP rates than those with poor APA correlation (Fig. 6E), indicating that sequence variations around the PAS contribute to APA variation between strains. Because of the low frequency of SNP, we were not able to identify specific motifs with statistical significance that were associated with APA variation. Taken together, our data indicate that mutations of sequence motifs around the PAS, especially U-rich motifs, lead to APA changes between species and populations.
Conservation and sequence features of lncRNA PASs
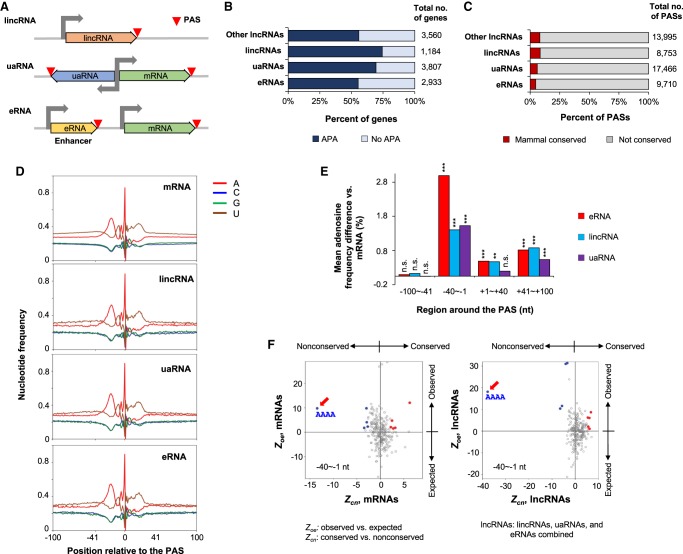
lncRNA PASs in general were less conserved than mRNA PASs (Fig. 1C). Using the lncRNA classification by FANTOM5 project (Hon et al. 2017), we next examined three types of lncRNAs in the human genome, including intergenic lncRNAs with independent promoters (lincRNAs), lncRNAs transcribed from divergent mRNA promoters (uaRNAs), and lncRNAs generated from enhancer regions (eRNAs) (Fig. 7A).
Figure 7.
Conservation analysis of lncRNA PASs. (A) Three types of lncRNAs and their PASs. (B) Frequency of APA in lncRNAs. (C) Conservation rate of lncRNA PASs. (D) Nucleotide frequency around the PASs of different lncRNAs and mRNAs. (E) Difference in adenosine frequency between lncRNAs and mRNAs in four regions around the PAS: (***) P < 0.001; (**) P < 0.01; (n.s.) not significant (binomial test). (F) Comparison of enriched tetramers between conserved and nonconserved PASs in mRNA genes (left) and lncRNA genes (right). Only the −40 to −1 nt region data are shown. “AAAA” motif is highlighted by an arrow.
We found that although all classes of lncRNAs had high frequencies of APA (55.2%–74.4%) (Fig. 7B), only a small fraction of lncRNA PASs were conserved (∼8% for lincRNAs, and ∼5% for uaRNAs and eRNAs) (Fig. 7C). Nucleotide frequency analysis indicated that the PASs of different lncRNA classes were surrounded with similar sequences to those of mRNAs, with an A-rich peak in the upstream −40 to −1 nt region and two U-rich peaks around the PAS (Fig. 7D). However, the surrounding regions of lncRNA PASs appeared to have higher adenosine frequencies, especially in the −40 to −1 nt region (Fig. 7D,E).
We next compared enriched tetramers around the PASs of different types of lncRNAs with those of mRNA PASs. Using Zoe (Methods) to indicate motif enrichment, we found that lncRNAs and mRNAs shared similar PAS motifs (Supplemental Fig. S10). However, one notable exception was AAAA in the −40 to −1 region, which was highly enriched for eRNA and lincRNA PASs (Supplemental Fig. S10, arrows). Because eRNA and lincRNA genes are generally younger than mRNA genes, we thus reasoned that A-rich motifs might be a primitive form of PAS signal, which eventually became stronger motifs when PASs were fixed in evolution. To test this hypothesis, we examined conserved and nonconserved PASs of mRNAs and lncRNAs. Indeed, AAAA in the −40 to −1 nt region was highly enriched for nonconserved PASs compared to conserved ones of both mRNAs and lncRNAs (Fig. 7F, arrows). This comparative analysis also revealed several other motifs that were enriched for conserved mRNA PASs, including upstream UGUA and U-rich motifs, and downstream UGUG and G-rich motifs (Supplemental Fig. S11; Supplemental Table S4). Together, our results indicate that lncRNAs share similar PAS motifs to mRNAs. However, due to the young age of lncRNAs, their PASs tend to use weak A-rich motifs as PAS signals, which becomes stronger motifs, such as AAUAAA, when PASs are fixed in evolution.
Discussion
In this study, we have generated a compendium of conserved PASs in mammals. Our data reveal the extent, rationale, and functional impacts of APA conservation. We show that PAS conservation reduces 3′ end diversity of gene transcripts, stabilizing gene expression and APA profile across species. Sequence analyses using conserved PASs elucidate distinct motifs around proximal and distal APA sites, mutations that influence APA profiles, and an evolutionary path of PAS fixation.
To the best of our knowledge, our effort, based on approximately 1.2 billion PAS reads from roughly 360 samples, represents the most comprehensive PAS collection for mammalian genomes. It is also notable that the sequencing methods we used, 3′READS or 3′READS+, are not affected by the internal priming issue that often leads to false positive and false negative PASs (Nam et al. 2002). As such, we can unequivocally identify many PASs situated in A-rich genomic regions, a large fraction of which are nonconserved, new PASs in lncRNAs.
Rationale of APA in the light of phylogenetics
As the sensitivity of sequencing technologies increases and sequencing data accumulate, the number of PASs in genomes is ever increasing. Given the pervasive nature of transcription and the loose definition of PAS motifs, it is not unreasonable to predict that all genes might display APA under some conditions. It is therefore important to use phylogenetics to filter out cryptic PASs and to understand functional APA events under purifying selection. Our analysis uncovered several gene features that exert evolutionary pressure on APA, including gene age, gene expression patterns, and gene functions, indicating that APA is selected for in evolution for certain groups of genes. Although these features are largely consistent with previous studies (Lee et al. 2008; Lianoglou et al. 2013), using conserved APA events for analysis offers much clarity in understanding the rationale and significance of APA.
The correlation of APA conservation with gene age suggests that genes tend to adopt new PASs in evolution. The enriched GO terms suggest that APA can play a significant role in cell morphology and cell growth. Notably, similar GO terms were also enriched for genes whose expression levels correlated with APA changes in development and differentiation (Ji et al. 2009), suggesting that APA is an integral player of gene expression changes in these processes. APA's role in cell proliferation has been extensively studied (Sandberg et al. 2008), including in the context of cancer (Mayr and Bartel 2009; Xia et al. 2014). In contrast, APA's involvement in cell morphology is so far understudied and requires further investigation in the future.
We found that genes with conserved APA tend to have high expression levels, less variation across tissues, and are broadly expressed. Although these features are largely consistent with the notion that ubiquitously expressed genes tend to have APA, through which they regulate gene expression post-transcriptionally (Lianoglou et al. 2013), we also found that genes with tissue-restricted expression are not necessarily devoid of conserved APA sites. Instead, the type of tissue in which a gene is preferentially expressed has a large role in determining APA evolution. For example, APA is selected for in brain-specific genes, highlighting the importance of having APA isoforms for neural functions. Presumably, APA isoforms can offer diverse subcellular localization potentials in neurons, where control of mRNA localization is widespread (Andreassi and Riccio 2009; Tushev et al. 2018). On the other hand, APA is selected against in some other tissues, such as liver and kidney, suggesting that gene expression in these tissues is controlled more often at the transcriptional level.
We found that aUTRs have a substantial role in mRNA stability through motifs related to mRNA decay. Although largely in line with the finding by Spies et al. (2013), the effect of aUTR in stability control appears stronger in this study. For example, we found that the genes whose short 3′ UTR isoforms are more stable than long 3′ UTR isoforms outnumbered those with the opposite trend by 8.2-fold. In contrast, a 1.5-fold bias was reported by Spies et al. (2013). Whereas both studies used NIH3T3 cells and analyzed a similar number of genes, several factors may contribute to this difference. Our analysis was based on comparison of newly made and preexisting RNA pools (Zheng et al. 2018), whereas transcription shutdown by actinomycin D was used by Spies et al. (2013) for half-life analysis. Our analysis focused on conserved PASs, whereas PAS conservation was not considered in the study by Spies et al. (2013). This latter difference might be important, because we found that nonconserved APA isoforms gave a milder trend in stability difference (Fig. 3I). In addition, we cannot rule out the possibility that the two studies had distinct cell growth conditions, leading to different RNA stability regulations.
PAS motif diversity and evolution
We show that conserved APA sites have distinct sequence motifs depending upon their locations (Fig. 4), echoing our earlier study (Tian et al. 2005). Most of the canonical sequence motifs known to be important for cleavage and polyadenylation are enriched for distal PASs compared to proximal PASs, supporting the notion that proximal sites in general are weaker than distal sites (Proudfoot 2016; Tian and Manley 2017). This arrangement conceivably ensures both regulation of proximal site with a wide controllable range and proper termination of transcription at the distal site. However, U-rich motifs around the PAS is a notable exception. Both proximal and distal PASs tend to be enriched with U-rich motifs (Fig. 4), and mutation analysis based on correlation of APA profiles between matched mouse and rat tissues implies an important role of U-rich motifs in APA control (Fig. 6). Further studies need to delineate how U-rich motif binding RBPs, many of which have been shown to alter APA events (Zheng and Tian 2014; Gruber et al. 2016), contribute to species-specific APA control and gene regulation.
Our comparative analysis of conserved and nonconserved PASs of mRNAs and lncRNAs reveal a path for PAS fixation through evolution of A-rich motifs to stronger motifs. A-rich motifs were previously found to be sufficient to promote cleavage and polyadenylation, as long as they are coupled with strong downstream elements (Nunes et al. 2010). It is thus conceivable that A-rich motifs situated in U-rich regions function as primitive signals for 3′ end processing of young genes. As A-rich motifs become stronger, such as to AAUAAA, the PASs are strengthened. As shown in this study, strengthening of PASs can reduce the 3′ end diversity stemming from cryptic PAS usage and contribute to stabilization of gene expression in evolution.
Methods
3′READS and 3′READS+ data
We collected all the 3′READS and 3′READS+ data our laboratory recently generated, corresponding to a wide variety of cell types and tissues (Supplemental Table S1). Samples with gene perturbations, such as gene knockdown or overexpression, were not used. We additionally generated 3′READS+ for brain, heart, and testis from adult mouse and rat in this study. Briefly, adult male C57BL/6J (C57) and FVB/JN (FVB) mice and rat were anesthetized by CO2 and sacrificed by cervical dislocation. The whole brain was carefully dissected and were free from meninges and cranial nerves. Hearts were rapidly removed from the thoracic cavity by cutting the great vessels, and excess of fatty tissue or blood vessels were removed. Testes were obtained by surgical removal and were cleared from tunica albuginea. All tissue samples were flash frozen in liquid nitrogen. All animal work was conducted according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at Rutgers New Jersey Medical School. Total RNA from cells and tissues was isolated using TRIzol (Invitrogen) or the Qiagen RNeasy kit. RNA samples were checked for integrity by Agilent Bioanalyzer using the RNA 6000 Pico kit (Agilent Technologies). RNA samples with an RNA integrity number (RIN) above 8.0 were used for subsequent processing. 3′READS+ was carried out as previously described (Zheng et al. 2016).
Annotation of PASs in genomes
3′READS and 3′READS+ data were processed to identify PASs using a method previously described (Zheng et al. 2016). Briefly, 3′READS/3′READS+ reads were mapped to the genome using Bowtie 2 (local mode) (Langmead and Salzberg 2012) with a mapping quality score cutoff (MAPQ) ≥10. Reads with two or more nongenomic 5′ Ts after alignment were called PAS-containing reads (PAS reads). PASs within 24 nt from one another were clustered as previously described (Hoque et al. 2013). Only the PASs with at least two reads in at least two samples were used for further analysis. Genome versions used in this study were mm9 (mouse), hg19 (human), and rn5 (rat). Note that the PAS mapping difference between different genome versions is essentially negligible (e.g., ∼0.06% between hg19 and hg38).
PASs mapped by 3′READS/3′READS+ were assigned to genes based on RefSeq (release 83) (Pruitt et al. 2006) and Ensembl databases (release 75 for human, release 67 for mouse, and release 79 for rat) (Aken et al. 2017). Because RefSeq and Ensembl gene annotations often miss PASs at the 3′ end of genes, we used strand-specific, poly(A)+ RNA-seq data sets (Merkin et al. 2012; Mouse ENCODE Consortium et al. 2012; Pervouchine et al. 2015; Mason et al. 2016) to extend the 3′ ends defined by RefSeq and Ensembl. We required a minimum of five reads at each position and allowed gaps <100 nt. We also required that 3′ end extension did not exceed the transcription start site of the downstream gene on the same strand. In total, 5691 million, 1635 million, and 325 million reads were used for 3′ end extensions in human, mouse, and rat genomes, respectively. We annotated genic PASs (both mRNA and lncRNA) by their intron/exon locations based on the representative RefSeq or Ensembl sequences. The sequence with the largest genomic span was used for each gene. When a gene was annotated in both RefSeq and Ensembl databases, RefSeq information was used. For mRNA genes, we further classified PASs into last exon (LE) and upstream region (UR). Most PASs in the last exon are in 3′ UTRs and were further classified into first (F), middle (M), and last (L) PASs. If a gene had only one PAS in the LE, it was named single PAS (S). Human lncRNA annotations were further refined using data from the FANTOM5 database (Hon et al. 2017). lncRNAs were separated into four groups, including intergenic lncRNA (lincRNA), upstream antisense RNA (uaRNA), enhancer RNA (eRNA), and other lncRNAs. We also annotated PASs associated with transposable elements (TEs) using data from the UCSC Genome Bioinformatics Site. PASs located in a TE or close to a TE (within 40 nt from the TE) were categorized as TE-associated PASs. PASs without any annotations were considered as intergenic PASs. PASs in genes overlapping with other genes on the same strand and in pseudogenes were categorized as the “other” group.
Conservation of PASs
We used pairwise genome alignment chain files from the UCSC Genome Bioinformatics Site to obtain syntenic regions between genomes. We used the reciprocal best-match method to identify conserved PASs (Lee et al. 2008). Briefly, two PASs from two species were considered to be orthologous when they were closest reciprocally in the whole genome alignment and were within 24 nt from one another. Two types of conservation were considered. If a PAS was conserved between human (H) and mouse (M) or rat (R) genomes, it was named mammal conserved. If a PAS was conserved between mouse and rat only, it was named rodent conserved.
Gene Ontology (GO) analysis
GO terms associated with genes were obtained from the Gene Ontology Consortium (Ashburner et al. 2000). The Fisher's exact test was used to derive P-values to indicate significance of association between a gene set and a GO term. GO terms associated with more than 1000 genes were considered too generic and were discarded. To remove redundancy, each reported GO term was required to have at least 10% of genes that were not associated with another term with a more significant P-value.
Analysis of gene age, gene expression variation, and tissue-specificity
NCBI HomoloGene database (https://www.ncbi.nlm.nih.gov/homologene) was used to define gene ages. Those with orthologs in zebrafish were considered as “old” genes and those without as “new” genes. More detailed gene age annotations were obtained from Liebeskind et al. (2016). For gene expression variation, we used the ENCODE RNA-seq data set (GSE36026) covering 22 mouse tissues/cell lines (Mouse ENCODE Consortium et al. 2012) and GTEx RNA-seq data set covering 28 tissues (GTEx Consortium 2017). RNA-seq reads were mapped to the mouse (mm9) or human (hg19) genome using STAR (Dobin et al. 2013) with default parameters, and the reads mapped to the coding sequence of each gene were used and normalized to transcripts per million (TPM). The coefficient of variation (CV) of TPM values across samples was calculated for each gene, which reflected its variation of expression. The average TPM value was used to reflect the expression level of a gene. Tissue-specific gene expression information was also obtained from PaGenBase (Pan et al. 2013). The “tissue-specific” and “selective” genes were grouped together as the narrowly expressed gene group, and the “housekeeping” and “repressed” genes were grouped together as the widely expressed gene group. We required that human and mouse orthologs to be in the same group. To define tissue-specificity for human genes using GTEx data, a Z-score based on TPM (minus mean and divided by standard deviation across samples) was calculated for each gene in each tissue. Genes whose largest Z-score greater than 1.6 and TPM ratio greater than 2 between the highest expressing tissue and the second highest were considered as tissue-specific genes.
Analyses of aUTRs and cUTRs
aUTRs and cUTRs were defined in genes with multiple PASs in 3′ UTRs. When there were multiple conserved PASs, the first and last conserved sites were used to define cUTR and aUTR. When there was only one conserved PAS, the nonconserved PAS with most reads was used together with the conserved PAS to define cUTR and aUTR. When there were no conserved PASs, the two PASs with the most reads were used. K-mer frequencies in cUTRs or aUTRs were examined by Biostrings (https://rdrr.io/bioc/Biostrings/). Significance of difference in frequency between cUTRs and aUTRs was calculated using the Fisher's exact test.
miRNA target site analysis in aUTR and cUTR regions
Predicted miRNA target sites (MTS) were downloaded from the TargetScan database (Release 7.1) (Agarwal et al. 2015). A total of 105,509 sites were obtained for 217 miRNA families. miRNA family age was based on annotations in TargetScan. Enrichment in aUTRs was calculated by odds ratio (OR) using the formula OR = (number of MTS of highly conserved miRNAs in conserved regions/number of MTS of highly conserved miRNAs in nonconserved region)/(number of MTS of moderately conserved miRNAs in conserved regions/number of MTS of moderately conserved miRNAs in nonconserved region).
Sequence motif of cleavage site
We considered two scenarios to define the cleavage site motif. Given a cleavage site sequence 5′-NNNNB[An]BNNNN, N is any nucleoside, B is a non-A nucleoside, and An is adenosine of any length. The cleavage site was considered to be before the [An] (scenario 1) or after it (scenario 2). Sequence logos were constructed using seqLogo (https://bioconductor.org/packages/release/bioc/html/seqLogo.html). Note that cleavage can also take place within the [An] region, which cannot be definitively resolved. This is because the poly(A) tail sequence from sequencing reads would align to the region, making it impossible to distinguish genomic template sequence from the poly(A) tail sequence.
Motif analysis of PASs
We define the ±100 nt genomic region surrounding each PAS as the PAS region. K-mers were calculated in four subregions, including −100 to −41 nt, −40 to −1 nt, +1 to +40 nt, and +41 to +100 nt. We used the PROBE method to examine enriched motifs (Hu et al. 2005). Briefly, a Zoe score was calculated for each k-mer, which was based on the difference between the frequency of the k-mer in a given region (observed value) and its expected value generated from randomized sequences of the same region using the first-order Markov chain model. Z-scores to compare distal versus proximal PASs (Zdp) and conserved versus nonconserved PASs (Zcn) were calculated by comparing frequencies between two PAS sets. Z-score greater than 3.6 (Bonferroni-corrected P < 0.05) was used as the cutoff to identify significantly biased tetramers.
Analysis of RNA stability
We used our recently published 3′READS data for newly made RNAs (4sU labeled [4sU]) RNAs and preexisting RNAs (flow-through [Ft]) in NIH3T3 cells (two biological replicates) to examine RNA stability (Zheng et al. 2018). Briefly, a stability score based on log2(Ft/4sU) for each transcript was calculated after adjustment for number of Us in the transcript (Zheng et al. 2018). The 3′ UTRs of genes with top and bottom 10% stability scores were selected for motif enrichment analysis. The top two 3′ UTR isoforms of each gene based on expression were selected for comparison of stability. Signficance of difference in stability between APA isoforms was based on PAS reads in 4sU and Ft samples (P < 0.05, Fisher's exact test) and relative expression difference between 4sU and Ft samples (>5%).
Expression analysis of APA isoforms
To evaluate the expression level of transcripts for a given PAS, we used mean RPM (reads per million) of all 3′READS/3′READS+ samples to reflect overall expression level, and the percentage of samples with expression (PSE) to reflect the breadth of expression. Mean RPM and PSE of each isoform were normalized by summed RPM and maximum PSE of all isoforms of the corresponding gene, respectively, to derive relative abundance and normalized PSE. A PAS was considered expressed in a sample if there were more than two reads in the sample. The mean RPM of each PAS was the averaged RPM value across all the samples in which there were more than two reads. To assess 3′ end diversity, we used the Shannon index with the formula, , where pi was the relative usage of the ith PAS for a given gene with an S number of PASs. RED was calculated as the difference in log2(ratio) of expression levels (RPM) of two PASs (distal and proximal) between two samples.
Mutation analysis of APA regulation
To examine the effect of sequence mutations on APA, we first calculated RED scores using brain versus heart and testis versus heart, and identified highly correlated and poorly correlated APA events using ΔRED = REDmouse − REDrat. The genes with top and bottom 10% ΔRED scores were defined as poorly correlated genes, whereas genes in the middle 30% were considered as highly correlated genes. Genome-wide sequence differences between mouse and rat were extracted from genome alignments obtained from UCSC Genome Bioinformatics Site, and the frequency of nucleotide change for each k-mer in the four flanking regions (−100 to −41 nt, −40 to −1 nt, +1 to +40 nt, and +41 to +100 nt) was calculated. Proximal and distal PASs were analyzed separately. The significance of nucleotide change for each k-mer was calculated using the Fisher's exact test. SNP data for C57BL/6 and FVB/NJ were obtained from the Mouse Genome Project (Wong et al. 2012).
Data access
All sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE111134.
Supplementary Material
Acknowledgments
We thank Lin Yan for technical help, Mainul Hoque and Xiaochuan Liu for contributions at the early stage of this work, and members of the B. Tian laboratory for helpful discussions. This work was funded by the National Institutes of Health grant GM084089 to B.T.
Author contributions: R.W. and B.T. conceived of and designed the work. D.Z. and G.Y. performed the experiments. R.W. analyzed the data. R.W. and B.T. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.237826.118.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4: e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, Billis K, Carvalho-Silva D, Cummins C, Clapham P, et al. 2017. Ensembl 2017. Nucleic Acids Res 45: D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Riccio A. 2009. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol 19: 465–474. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, Brunkow ME, Ramsdell F, O'Briant KC, Zhu Q, Fuleihan RL, Shigeoka AO, Ochs HD, Chance PF. 2001. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA→AAUGAA) leads to the IPEX syndrome. Immunogenetics 53: 435–439. [DOI] [PubMed] [Google Scholar]
- Cannavò E, Koelling N, Harnett D, Garfield D, Casale FP, Ciglar L, Gustafson HE, Viales RR, Marco-Ferreres R, Degner JF. 2017. Genetic variants regulating expression levels and isoform diversity during embryogenesis. Nature 541: 402. [DOI] [PubMed] [Google Scholar]
- Chen F, MacDonald CC, Wilusz J. 1995. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res 23: 2614–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Miura RM, Tian B. 2006. Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics 22: 2320–2325. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. 2014. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 156: 812–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber JH, Cantor CR, Mohr SC, Smith TF. 1999. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci 96: 14055–14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, Plenge RM, Koeuth T, Ortmann WA, Hom G. 2007. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci 104: 6758–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AJ, Schmidt R, Gruber AR, Martin G, Ghosh S, Belmadani M, Keller W, Zavolan M. 2016. A comprehensive analysis of 3′ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res 26: 1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs D, Goodbourn S, Lamb J, Clegg J, Weatherall D, Proudfoot N. 1983. α-Thalassaemia caused by a polyadenylation signal mutation. Nature 306: 398. [DOI] [PubMed] [Google Scholar]
- Hollerer I, Grund K, Hentze MW, Kulozik AE. 2014. mRNA 3′end processing: a tale of the tail reaches the clinic. EMBO Mol Med 6: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, et al. 2017. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. 2013. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods 10: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B. 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11: 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Li W, Rosenfeld MG, Glass CK. 2014. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 39: 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Ji Z, Tian B. 2008. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res 36: 5581–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. 2010. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One 5: e11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XQ, Du D. 2013. RNA polyadenylation sites on the genomes of microorganisms, animals, and plants. PLoS One 8: e79511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Park JY, Zheng D, Hoque M, Yehia G, Tian B. 2016. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol 14: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. 2013. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 27: 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeskind BJ, McWhite CD, Marcotte EM. 2016. Towards consensus gene ages. Genome Biol Evol 8: 1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hoque M, Larochelle M, Lemay JF, Yurko N, Manley JL, Bachand F, Tian B. 2017. Comparative analysis of alternative polyadenylation in S. cerevisiae and S. pombe. Genome Res 27: 1685–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason AS, Fulton JE, Hocking PM, Burt DW. 2016. A new look at the LTR retrotransposon content of the chicken genome. BMC Genomics 17: 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J. 2013. Genome-wide mapping of polyadenylation sites in fission yeast reveals widespread alternative polyadenylation. RNA Biol 10: 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. 2016. Evolution and biological roles of alternative 3′UTRs. Trends Cell Biol 26: 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. 2012. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338: 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse ENCODE Consortium, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, et al. 2012. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen J, Rowley JD, Wang SM. 2002. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci 99: 6152–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes NM, Li W, Tian B, Furger A. 2010. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J 29: 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JB, Hu SC, Shi D, Cai MC, Li YB, Zou Q, Ji ZL. 2013. PaGenBase: a pattern gene database for the global and dynamic understanding of gene function. PLoS One 8: e80747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervouchine DD, Djebali S, Breschi A, Davis CA, Barja PP, Dobin A, Tanzer A, Lagarde J, Zaleski C, See LH, et al. 2015. Enhanced transcriptome maps from multiple mouse tissues reveal evolutionary constraint in gene expression. Nat Commun 6: 5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad MK, Bhalla K, Pan ZH, O'Connell JR, Weder AB, Chakravarti A, Tian B, Chang YP. 2013. A polymorphic 3′UTR element in ATP1B1 regulates alternative polyadenylation and is associated with blood pressure. PLoS One 8: e76290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. 2016. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352: aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. 2006. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild G, Basu U. 2017. Lingering questions about enhancer RNA and enhancer transcription-coupled genomic instability. Trends Genet 33: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilippo P, Wen J, Lai EC. 2017. Landscape and evolution of tissue-specific alternative polyadenylation across Drosophila species. Genome Biol 18: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlackow M, Marguerat S, Proudfoot NJ, Bähler J, Erban R, Gullerova M. 2013. Genome-wide analysis of poly(A) site selection in Schizosaccharomyces pombe. RNA 19: 1617–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP. 1990. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res 18: 5799–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Manley JL. 2015. The end of the message: multiple protein–RNA interactions define the mRNA polyadenylation site. Genes Dev 29: 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojo V, Dessimoz C, Pomiankowski A, Lane N. 2016. Membrane proteins are dramatically less conserved than water-soluble proteins across the tree of life. Mol Biol Evol 33: 2874–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Burge CB, Bartel DP. 2013. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res 23: 2078–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Graber JH. 2012. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA 3: 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Pan Z, Lee JY. 2007. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res 17: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tushev G, Glock C, Heumüller M, Biever A, Jovanovic M, Schuman EM. 2018. Alternative 3′ UTRs modify the localization, regulatory potential, stability, and plasticity of mRNAs in neuronal compartments. Neuron 98: 495–511.e496. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Nambiar R, Zheng D, Tian B. 2018. PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res 46: D315–D319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Bumpstead S, Van Der Weyden L, Reinholdt LG, Wilming LG, Adams DJ, Keane TM. 2012. Sequencing and characterization of the FVB/NJ mouse genome. Genome Biol 13: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Yang L, Chen LL. 2017. The diversity of long noncoding RNAs and their generation. Trends Genet 33: 540–552. [DOI] [PubMed] [Google Scholar]
- Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. 2014. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun 5: 5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao MS, Zhang B, Li YS, Gao Q, Sun W, Chen W. 2016. Global analysis of regulatory divergence in the evolution of mouse alternative polyadenylation. Mol Syst Biol 12: 890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. 2005. Biased alternative polyadenylation in human tissues. Genome Biol 6: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Tian B. 2014. RNA-binding proteins in regulation of alternative cleavage and polyadenylation. Adv Exp Med Biol 825: 97–127. [DOI] [PubMed] [Google Scholar]
- Zheng D, Liu X, Tian B. 2016. 3′READS+, a sensitive and accurate method for 3′ end sequencing of polyadenylated RNA. RNA 22: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Wang R, Ding Q, Wang T, Xie B, Wei L, Zhong Z, Tian B. 2018. Cellular stress alters 3′UTR landscape through alternative polyadenylation and isoform-specific degradation. Nat Commun 9: 2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.