Abstract
MicroRNAs (miRNAs) play roles in various biological processes in plants including growth, development, and disease resistance. Previous studies revealed that some plant miRNAs produce secondary small interfering RNAs (siRNAs) such as phased, secondary siRNAs (phasiRNAs), and they regulate a cascade of gene expression. We performed a genome-wide comparative analysis of miRNAs in Solanaceous species (pepper, tomato, and potato), from an evolutionary perspective. Microsynteny of miRNAs was analysed based on the genomic loci and their flanking genes and most of the well-conserved miRNA genes maintained microsynteny in Solanaceae. We identified target genes of the miRNAs via degradome analysis and found that several miRNAs target many genes encoding nucleotide-binding leucine-rich repeat (NLR) or receptor-like proteins (RLPs), which are known to be major players in defense responses. In addition, disease-resistance-associated miRNAs trigger phasiRNA production in pepper, indicating amplification of the regulation of disease-resistance gene families. Among these, miR-n033a-3p, whose target NLRs have been duplicated in pepper, targets more NLRs belonging to specific subgroup in pepper than those in potato. miRNAs targeting resistance genes might have evolved to regulate numerous targets in Solanaceae, following expansion of target resistance genes. This study provides an insight into evolutionary relationship between miRNAs and their target defense genes in plants.
Keywords: miRNA, degradome, NLR, Solanaceae, plant immunity
1. Introduction
Small RNA-mediated gene silencing is a gene regulatory system that is conserved in eukaryotes.1 Small RNAs are typically 20∼24 nucleotides in length and play important roles in diverse biological processes, such as growth, development, hormone synthesis, and response to biotic/abiotic stresses.2–4 MicroRNAs (miRNAs) are a class of small RNAs that act as gene regulators in plants and animals. In plants, miRNAs are generally 21 nucleotides in length and generated from hairpin-structured RNA precursors, processed by Dicer-like proteins (DCL). These miRNAs associate with Argonaute proteins to form the RNA-induced silencing complex (RISC) and repress target genes via cleavage, which requires near-perfect base-pairing with targets.1
The cleavage of transcripts by miRNAs, which are either 225,6 or 21 nucleotides in length and contain one7 or two target sites,8 can trigger the production of a secondary small RNA cluster. Secondary small interfering RNAs (siRNAs) are derived from the cleaved transcripts, converted into dsRNA by suppressor of gene silencing 3 and RNA-dependent RNA polymerase 6, and processed by DCL4/5 in a phased manner.9,10 Given the nature of their biogenesis, these siRNAs are called phased, secondary siRNA (phasiRNA). PhasiRNA is generally 21 nucleotides in length, and some phasiRNAs are incorporated into the RISC in the same manner as miRNA, leading to silencing of the target mRNA in cis or trans.9,11 Numerous plants, such as rice,12 Arabidopsis,8 alfalfa, and soybean,11 have tens to hundreds of phasiRNA-producing loci (PHAS loci) in their genome. PHAS loci could be protein-coding, or non-coding genes like the trans-acting siRNA-generating loci (TAS).10,13 A well-studied example of the TAS mechanism is the miR390-TAS3-ARF pathway, which is widely conserved in land plants.14 In this pathway, miR390 triggers the production of phasiRNAs from TAS3 transcripts, and these phasiRNAs regulate auxin response factor (ARF) genes in trans.
Various studies have reported that plant miRNAs and phasiRNAs regulate the expression of a range of gene families,9,15–18 including those related to disease resistance. In plants, there are two layers of defense mechanisms. First, pattern recognition receptors (PRRs) recognize specific patterns called pathogen-associated molecular patterns (PAMPs) in pathogens, and induce defense responses (PAMP-triggered immunity, PTI). Second, the pathogens overcoming PTI secrete effectors, which are recognized by resistance proteins (R proteins) that induce a subsequent downstream defense response such as rapid and localized cell death (effector-triggered immunity, ETI).19 Previous studies reported that miRNAs such as those belonging to the miR482/2118 superfamily, miR6019, miR6024, and miR6027 regulate genes encoding nucleotide-binding leucine-rich repeat (NLR) proteins, which belong to a major gene family of R proteins.11,18,20,21 Furthermore, NLR gene family is one of the representative PHAS genes in a broad range of plant families, including Solanaceae, whose trigger is the miR482/2118 superfamily.11,18,20,21
Recently, several studies investigated the evolution of miRNAs across plant lineages via comparative analysis.22,23 These studies revealed that the evolution and divergence of miRNAs are closely associated with genome duplication and evolution of their target genes. Especially, disease-resistance-related miRNAs, such as miR482 superfamily, were studied well in the aspect of co-evolution with target NLRs.18,24,25 Origination of disease-resistance-associated miRNAs is considered to result from tandem duplication of NLRs and these miRNAs have co-evolved with their target NLRs.18 Each plant species experienced divergent evolution of NLRs, therefore, the pool of disease-resistance-associated miRNAs is distinctive in each plant species.18,25
Solanaceae includes many crops that are widely grown in the world, such as tomato (Solanum lycopersicum), potato (Solanum tuberosum), and pepper (Capsicum annuum). Among them, pepper is one of the economically important crops, widely used in food and medicine. Previously, we identified miRNAs and some of their targets through high-throughput sequencing in pepper.15 Another study identified that pepper miRNAs regulate transcription factors, which may play an important role in plant development, via a trans-omics approach.26 In addition, there is a study covering the evolutionary relationship of miR482/2118 superfamily and resistance genes in Solanaceae.24 However, there is no in-depth study that unravels the detailed functions and evolutionary history of pepper miRNAs.
Here, to investigate functions of the pepper miRNAs, we conducted comparative analyses of miRNAs and their targets in pepper, tomato, and potato using degradome analysis. Interestingly, we found that previously identified,15 but not characterized miRNAs (miR-n026 and miR-n033) targeted genes involved in plant defense systems, in addition to several known miRNAs (e.g. miR482). Moreover, these miRNAs were identified as a trigger of phasiRNA biogenesis. Among them, the can-miR-n033 family targets many NLR genes and most of them belong to the subgroup that was highly duplicated in pepper. Performing a comparative analysis based on the estimated divergence times [Capsicum and Solanum: diverged 19 million years ago (mya); potato and tomato: diverged 8 mya],27 we inferred that the miR-n033 family existed in a common ancestor to pepper, tomato, and potato, and the miR-n033a acquired new targets in pepper, a result of NLR gene expansion in this species. This study could advance our understanding of the evolutionary relationship between miRNA and target genes, and target gene regulation by miRNAs and phasiRNAs in plant immunity.
2. Materials and methods
2.1. Exploration of miRNAs in pepper, tomato, and potato
Precursor and mature miRNA sequences from pepper (Capsicum annuum L.), tomato (Solanum lycopersicum L.), and potato (Solanum tuberosum L.) were identified from previous studies and online database (Supplementary Tables S1 and S2).15,17,28–31 In addition, we identified additional miRNAs with their precursor using small RNA-Seq data and almost the same pipeline from our previous study.15 We excluded the expression cut-off (raw reads ≥ 1000) from the previous pipeline to identify additional miRNAs in pepper.15 Some miRNAs with different ID, but having identical sequences among pepper, tomato, and potato were integrated into same ID. Genome sequences and gff files of pepper (v.1.55; http://peppergenome.snu.ac.kr/ (20 April 2018, date last accessed)), tomato (ITAG v.2.3; https://solgenomics.net/ (20 April 2018, date last accessed)), and potato (JGI v.4.03; https://genome.jgi.doe.gov/ (20 April 2018, date last accessed)) were used to locate miRNA loci. Visualization of chromosome location was implemented using in-house Perl scripts. Genomic synteny of miRNAs among Solanaceae was analysed by modified microsynteny-based method.32 In brief, miRNAs between those in pepper and those in tomato or potato were searched for the synteny test using BLASTN (e-value < 1e-05). Five upstream and five downstream flanking proteins of each miRNA gene were retrieved. Synteny of the flanking proteins was analysed by MCScanX with BLAST e-value cut-off < 1e-10.33 Microsynteny was defined as at least 2 out of 10 flanking proteins of a miRNA pair having synteny with that of tomato or potato. Chromosome rearrangement was not considered.
2.2. Degradome sequencing analysis
To identify targets of miRNAs experimentally, pepper (C. annuum CM334), tomato (S. lycopersicum Heinz), and potato (S. tuberosum Phureja) were grown under standard conditions (27 °C/19 °C; 16 h light/8 h dark). Various tissues (leaves, roots, stems, green fruits, red fruits, tubers) were sampled and used for the degradome library. The degradome libraries were constructed and sequenced using an Illumina HiSeq2000.34 From the raw sequencing data, adaptors were removed, and qualified reads that were 15–26 nucleotides in length were obtained. Among these, structural RNAs such as rRNAs, tRNAs, snRNAs, and snoRNAs were removed using BLASTN with default option against Rfam (http://rfam.xfam.org/ (20 April 2018, date last accessed)).35 Subsequently, repeats and transposons were also removed using the repeat database.36 Because UTR sequences have not been identified in pepper, transcripts were defined to coding sequence (CDS) and ±1 kb flanking sequences. Analysis of signal to noise was conducted using the CleaveLand4 pipeline.37 The cut-off for significant targets was a p-value ≤ 0.05 or a score ≤ 4. Significant targets were categorized into five classes according to CleaveLand. Target genes were annotated by a BLASTX search against the NR database and InterPro domains of target genes were analysed using Blast2go.38
2.3. Northern blot analysis
Total RNA was extracted from leaf of pepper, tomato, potato and Nicotiana benthamiana using TRI Reagent (Ambion). A total amount of 20 μg of RNA from leaves of each species was individually separated in a 15% UREA polyacrylamide gel, electrophoretically transferred to Hybond-NX membrane (GE Healthcare), and was chemical cross-linked via 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC).39 For labelling reaction of probes, 2 μl of 10 μM oligo, 2 μl of 10X T4 PNK buffer (Takara), 2.5 μl of [γ-32P] ATP, >7000 Ci/mmole (∼150 μCi/μl), 12.5 μl of dH2O, and 1 μl of T4 polynucleotide kinase (Takara) were added to a 20 μl reaction for 1 h at 37 °C. The labelled probes were further purified from unincorporated labels with PERFORMA Spin Columns (Edge Bio) according to the manufacturers’ instruction. Probe sequences used for northern blot analysis were sequences of the mature miRNAs in pepper (Supplementary Table S1). Hybridization and washing procedures were performed essentially as described.40 The membranes were exposed to a phosphorimager, and signals were analysed using BAS-2500 (Fuji).
2.4. PhasiRNA analysis
The phasiRNA prediction pipeline was written in Python language. Same small RNA-Seq data, which were exploited to identify additional miRNAs in pepper, were used in phasiRNA analysis.15 Small RNA reads were processed to trim adapters, and low-quality reads were filtered and collapsed using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/ (20 April 2018, date last accessed)). Processed reads were aligned to the genomes of pepper, tomato, and potato using Bowtie-1.1.241 (-v 0 -a) and normalized by genome-mapped counts (hit normalize) to prevent overestimating reads that were mapped more than one time to the genome. The criteria to filter the mapped reads were as below. (1) Small RNA reads with abundance less than three were excluded because of computing difficulty. (2) The reads that were mapped to the genome more than 25 times were filtered. After filtering small RNA reads, a P-value was calculated to identify putative PHAS in the genome, filtering with P-value ≤ 0.001, and phasing score was calculated in the expanded PHAS genes (−500 and +1000 bp) as the modified algorithm.42 The filtered PHAS genes, which had phasing scores of 15 or above, were regarded as PHAS genes. PHAS genes were characterized using gene annotation data (v.1.55; https://solgenomics.net/ (20 April 2018, date last accessed)). Furthermore, if they were not associated with non-coding RNAs such as rRNA, tRNA, and snRNA from the Rfam database,35 SILVA (https://www.arb-silva.de/ (20 April 2018, date last accessed)),43 plantRNA database (http://plantrna.ibmp.cnrs.fr/plantrna/ (20 April 2018, date last accessed)),44 and the genome database of each species (https://solgenomics.net/ (20 April 2018, date last accessed)), they were selected for further analysis. PHAS genes were identified with respect to whether miRNAs trigger the production of phasiRNA by degradome analysis via CleaveLand4.37 To predict the target sites of the phasiRNAs, phasiRNAs from the PHAS regions were analysed using CleaveLand4 as the input.37 Phased loci were visualized using the Integrative Genomics Viewer (IGV).45
2.5. Bioinformatic analysis for miR-n026 and miR-n033, and motif analysis targeted by miRNAs
Genome sequences and gff files of pepper, tomato, and potato were used for location of miRNAs. Syntenic relationships between pepper, tomato, and potato genes that flank miRNA sequences were confirmed by MCScanX (BLASTP e-value < 1e-10).33 To examine whether miR-n026 and miR-n033 exist in Solanaceous species, an analysis was conducted using BLASTN with default option against their genome. We referred to evolutionary distances between Solanaceous species from other studies.46,47 The hairpin structures of miR-n026 and miR-n033 were predicted using the RNAfold web server.48 Transposable elements were found as described in a previous study.49 Subgroups of target NLRs in pepper, tomato, and potato were classified according to a previous study.50 The sequences targeted by each miRNA were translated considering the frame and aligned using Clustal Omega.51 The aligned sequences were used to create consensus sequences using WebLogo (http://weblogo.threeplusone.com/ (20 April 2018, date last accessed)).52 Target prediction of miR-n026 and miR-n033 was conducted using psRNAtarget (score ≤ 3.5; http://plantgrn.noble.org/psRNATarget/ (20 April 2018, date last accessed)).53 The phylogenetic tree of NLRs was from our previous study.50
3. Results
3.1. Solanaceae miRNA genes are unevenly distributed with clusters on chromosomes
To perform comprehensive analysis of miRNAs in Solanaceae, the genomic locations of miRNA genes were investigated. Because pre-existing studies identified miRNAs in pepper, tomato, and potato, we used the sequences of miRNAs and their precursors from the literature and databases.15,17,28–31 In the case of pepper, we identified additional miRNAs based on data from our previous study because we had applied strict criteria to identify novel miRNAs, which could cause false negative.15 We collected 145, 123, and 275 mature miRNAs, including those from guide and passenger strand, from the 223, 93, and 393 miRNA genes in pepper, tomato, and potato, respectively (Supplementary Tables S1 and S2). We defined species-specific miRNAs as the miRNAs, whose mature and precursor sequences are not overlapped with those of other species in this study and in miRBase using BLASTN with default option (Supplementary Table S3). These genes were classified into 93, 58, and 126 families, respectively, and only 28 conserved families were shared among the three plants, suggesting the rapid divergence of miRNA genes after divergence of the lineages (Supplementary Fig. S1). Although the number of conserved miRNA families is lower than that of species-specific miRNA families (Supplementary Fig. S1), the number of conserved miRNA genes comprises more than half of the total miRNA genes, indicating that there are more members in each conserved miRNA family than in each species-specific miRNA family (Supplementary Table S3).
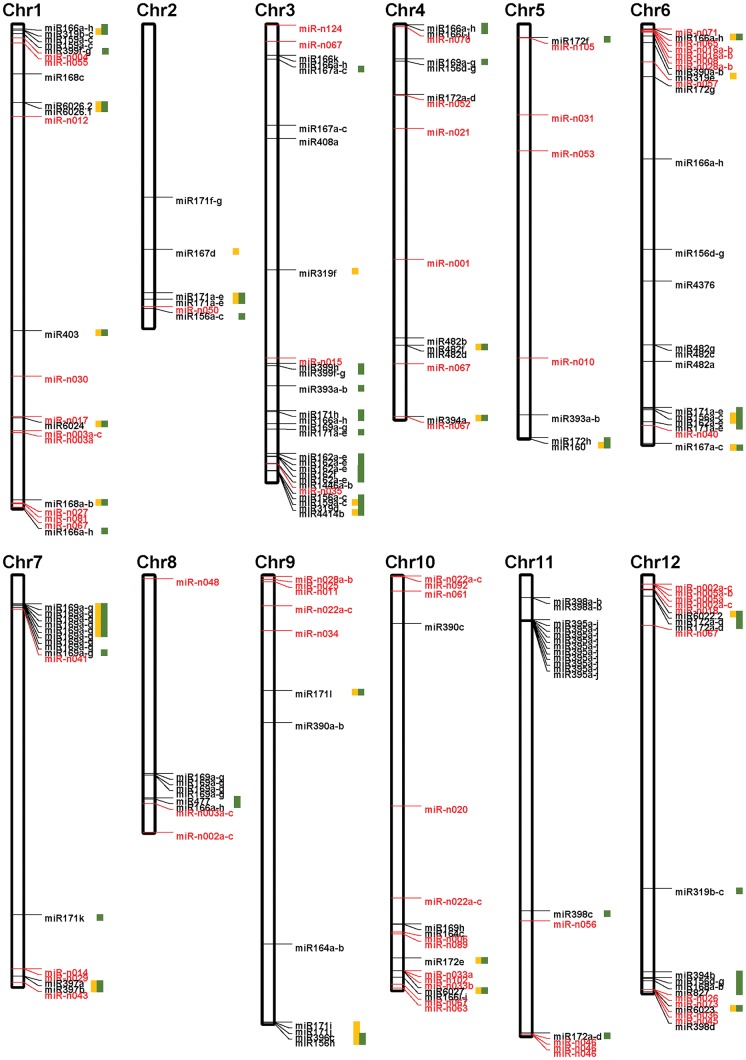
We explored the genomic location of miRNAs in three Solanaceous species, and the 198, 93, and 388 miRNA genes out of 223, 93, and 393 miRNA genes were mapped onto the chromosomes of pepper, tomato, and potato, respectively (Fig. 1, Supplementary Fig. S2, and Table S4). The number of mapped miRNA genes is discordant with the number of miRNA genes because some scaffolds which have miRNA genes were not mapped onto the chromosomes. The genes were not evenly distributed on the 12 chromosomes. We found some clusters of well-conserved miRNA genes in pepper, tomato, and potato. For example, nine miRNA genes producing can-miR169a-g were located on chromosome 7 within 1.9 Mb of each other (Fig. 1). Tomato and potato have similar clusters of miR169 on chromosome 7, although the numbers of miRNAs were different (Supplementary Fig. S2). Therefore, duplication of miR169 on chromosome 7 might have occurred before the divergence of the lineages leading to pepper, tomato, and potato. To perform in-depth analysis, we conducted microsynteny analysis on the miRNA genes and their flanking protein-coding sequences, focusing on pepper. Among 126 conserved mapped miRNA genes in pepper, 77 miRNA genes have at least 2 out of 10 flanking proteins showing synteny with that of tomato or potato, and 51 miRNA genes have more than five flanking proteins with synteny. These results indicate that synteny of many conserved miRNAs in pepper (Capsicum spp.) is stably maintained after the divergence of the lineages to tomato/potato (Solanum spp.; Fig. 1). However, some of the conserved miRNAs do not show synteny in all three species, indicating the evolution of those miRNAs after the divergence of the lineages. For example, can-miR482f and can-miR482d are closely located within a 73 kb region on chromosome 4 (Fig. 1), but only miRNA gene sequence of can-miR482f was similar to those in tomato and potato. Meanwhile, we found some clusters that consist of various miRNAs from different miRNA families. For example, there is a clustered region that contains four miRNA genes (miR-n033a, miR-n102, miR-n033b, and miR6027) on chromosome 10 (Fig. 1). Additionally, there are five miRNA genes (miR-n026, miR-n073, miR6023, miR-n036, and miR-n045) in the clustered region of chromosome 12, but their precursor sequences are not related, and therefore they might have evolved independently. Together, these findings indicate that miRNA genes have undergone dynamic evolution in pepper.
Figure 1.
Genome-wide distribution of miRNA genes in pepper. Black boxes indicate 12 pseudomolecules of pepper. miRNA genes in black and red indicate the location of conserved and pepper-specific miRNA families, respectively. Yellow and green boxes next to miRNAs indicate the miRNA genes and their flanking protein-coding sequences have microsynteny with those of tomato and potato, respectively.
3.2. Degradome analysis revealed that miRNAs are involved in disease-resistance in Solanaceae
To identify targets of miRNAs on a genome-wide level, miRNA-cleaved target libraries from pooled tissues (leaves, roots, stems, green fruits, red fruits, and tubers) of pepper, tomato, and potato were generated (see Materials and methods section). Sequencing of Parallel Analysis of RNA Ends (PARE) libraries, also known as degradome libraries, identifies the remnants of small RNA-directed target cleavage globally by sequencing the 5' ends of uncapped RNAs.54,55 A total of 251,689,162 sequencing reads in pepper, 226,553,118 in tomato, and 371,124,547 in potato were obtained from degradome sequencing (Supplementary Table S5). After removing low-quality or structural RNA reads, 151,815,030, 103,905,693, and 269,954,139 final reads were used for processing and 55.3%, 61.63%, and 73.96% of the reads were mapped to the transcripts of pepper, tomato, and potato, respectively (Supplementary Table S5). The target genes were identified using the CleaveLand4 pipeline.37
A total of 436, 352, and 169 pairs of miRNAs/target genes were identified in pepper, tomato, and potato, respectively. Among these, 99, 72, and 70 miRNAs targeted at least one gene, according to a pre-set standard (e-value ≤ 0.05 or score ≤ 4), respectively (Fig. 2A and Supplementary Table S6). Regarding target genes, 390, 319, and 163 genes were targeted by miRNAs from pepper, tomato, and potato, respectively (Supplementary Table S6). In the case of potato, the total degradome reads are high, but the results might have been underestimated because of the low number of unique reads (Supplementary Table S5). In summary, we found consistent results among targets of conserved miRNAs with the previous study. We confirmed that can-miR396b, can-miR396c, and sly-miR39b targeted domain-rearranged methyltransferase (DRM) gene as validated previously (Supplementary Table S6).15 To explore the targets of miRNAs, we analysed the domains of the target genes (Fig. 2B). We found that a large number of the target genes have the leucine-rich repeat (LRR) and nucleotide-binding (NB) domains shared by APAF-1, R genes, and CED-4 (NB-ARC) in Solanaceae. In particular, pepper had 115 target genes containing the LRR domain; this number is much higher than that in tomato or potato (Fig. 2B). The majority of these NLRs in pepper, tomato, and potato were regulated by conserved miRNAs (miR482 family, miR6024, and miR6027; Supplementary Table S6). Among these, miR6024 and miR6027 were only identified in some Solanaceous species20 but miR482 family exists in many plants18. Although miR6024 and miR6027 are relatively young miRNAs compared with miR482, they also regulate many NLRs in the three species. The second most abundant domain was transcription factors. Specifically, the majority of conserved miRNA targets included transcription factors such as Teosinte Branched 1, Cycloidea, and PCF (TCP), ARF, and MYB.
Figure 2.
Degradome analysis in Solanaceous species. (A) The numbers of miRNAs targeting at least one gene in pepper (Capsicum annuum), tomato (Solanum lycopersicum), and potato (Solanum tuberosum) were indicated in the parenthesis. (B) The top 10 domains of degradome target genes were identified by InterproScan. X and Y axis indicate the number of genes having certain domains and the description of the domain, respectively. NB-ARC: nucleotide-binding domain shared by APAF-1, resistance genes, and CED-4; TCP: teosinte branched1, cycloidea.
We analysed the target genes of species-specific miRNAs in detail (Table 1). In results, we found that many pepper-specific miRNAs had no identified target genes based on degradome analysis. These results are consistent with previous findings that miRNAs have evolved neutrally, so most young miRNAs, which are species-specific, do not have target genes or functions.56,57 On the other hand, some of pepper-specific miRNAs target multiple genes, encoding NLRs, receptor-like proteins (RLPs), accelerated cell death proteins, and F-box proteins. These genes are mostly involved in plant defense mechanisms.58,59 For example, both can-miR-n002a-c and can-miR-n005a-5p target three F-box proteins (Supplementary Table S6). Especially, can-miR-n033a and can-miR-n026 targeted 31 NLRs and 17 RLPs, respectively. In general, only miRNAs which are well-conserved throughout various plant species, such as miR482, have a number of target genes.57 In contrast, most of young miRNAs are weakly expressed and have few target genes.15,57,60,61 However, miR-n026 and miR-n033 were observed to target a number of genes despite their short evolutionary history, indicating their potential role in plant defense. In addition, we found that representative examples of cleaving disease-resistance genes by both conserved miRNAs (miR482, miR6024, and miR6027) and pepper-specific miRNAs (miR-n033a and miR-n026) seems evident in degradome analysis (Supplementary Fig. S3). It supported our hypothesis about the significance of pepper-specific miRNAs in regulation of pepper disease-resistance.
Table 1.
Degradome targets by pepper-specific miRNAs in pepper (Capsicum annuum)
| miRNA | Target description | No. of targets |
|---|---|---|
| miR-n002a-c | F-box/kelch-repeat protein | 3 |
| miR-n003a-c-3p | ACCELERATED CELL DEATH 6-like | 6 |
| miR-n005a-5p | F-box protein CPR30-like | 3 |
| miR-n010 | Receptor-like protein 12-like | 2 |
| miR-n016a-b | Leucine-rich repeat receptor kinase | 2 |
| miR-n022a-c | ATP-citrate synthase beta chain 2 | 2 |
| F-box PP2-B10-like | 2 | |
| miR-n026 | Receptor-like protein 12-like | 17 |
| Acetylajmalan esterase-like | 4 | |
| miR-n033a | Disease-resistance protein, late blight resistance homologa | 31 |
| miR-n033b | Disease-resistance protein, late blight resistance homologb | 4 |
| miR-n056 | Disease-resistance RPP13 | 2 |
and
are different from each other.
3.3. Small RNA clusters and phasiRNAs are also associated with disease-resistance in pepper
Recent studies reported that some miRNAs can produce secondary small RNAs, such as phasiRNA, to regulate a more extensive set of transcripts.11,21,42 Exploiting small RNA libraries from the previous study,15 we performed genome-wide analysis in pepper to identify putative PHAS loci using modified computational algorithms with a stringent threshold, P-value ≤ 0.001.42,62 As a result, we found 31 protein-coding PHAS loci (Table 2). Several gene families were identified as putative PHAS loci, such as NLR, RLP, nuclear transcription factor Y, and transport inhibitor response 1-like (TIR1-like) protein. Most of these protein-coding PHAS genes have been reported in other plant species,11,42,63 indicating a conserved mechanism of phasiRNA biogenesis in various plants. However, another well-conserved phasiRNA gene, TAS, was not identified in our results. Interestingly, most PHAS loci are associated with disease resistance. These include 20 NLRs, 8 RLPs, and 1 TIR1-like protein, which are believed to be involved in ETI and PTI in plants.58
Table 2.
Genes producing phasiRNA cluster by miRNAs in pepper (Capsicum annuum)
| miRNA trigger | Genes producing phasiRNA cluster | No. of genes |
|---|---|---|
| miR482 | Disease-resistance protein, NB-LRRa | 14 |
| miR6027 | Disease-resistance protein, NB-LRRb | 2 |
| miR-n026 | LRR receptor-like protein | 8 |
| miR-n033 | NB-LRR | 4 |
| miR169 | Nuclear transcription factor Y subunit A (NF-YA) | 1 |
| miR171 | GRAS family transcription factor | 1 |
| miR393 | Transport inhibitor response 1-like | 1 |
and
are different from each other.
We identified triggers of phasiRNA biogenesis with degradome analysis using CleaveLand437 (Table 2). Most miRNA triggers of phasiRNA biogenesis in pepper were 22 nucleotides in length, with the exception of can-miR169a-g and can-miR171i, which were 20 and 21 nucleotides in length respectively, consistent with previous studies (Supplementary Table S1).6,11,42 Members of the miR482 family are known as phasiRNA triggers, suggesting amplification of NLR regulation to extensively control disease resistance.11,21 miR6027 also represses NLRs, as in the case of miR482, and also plays a role in phasiRNA biogenesis.20,64 We determined that miR482 and miR6027 initiated phasiRNA biogenesis by cleaving NLRs in pepper (Supplementary Figs S4 and S5). One of the target genes of can-miR482c is coiled-coil NLR (CNL, CA05g06820), and a detailed mapping profile of small RNA and degradome reads demonstrated that the can-miR482c cleavage site in CA05g06820 corresponds to the 21-nucleotide phasing window (Supplementary Fig. S4A). Likewise, other variants of the miR482 family (can-miR482b, d, and f) were also identified as phasiRNA triggers (Supplementary Table S7). can-miR6027-3p targets another CNL (CA04g14510) and induces phasiRNA biogenesis (Supplementary Fig. S5A). However, another miRNA-targeting NLR, miR6024, was not identified as a phasiRNA trigger in pepper. Previous studies revealed that miR6024 acts as a trigger of phasiRNA biogenesis, by targeting the I2 gene in tomato and Rx1 gene in potato.20,65 The length of miR6024 is 22 nucleotides in tomato and potato, but 21 nucleotides in pepper (Supplementary Table S1). Because phasiRNA triggers are typically 22 nucleotides in length,9 can-miR6024-derived phasiRNA biogenesis might not occur in pepper. can-miR393a-b is another conserved miRNA that regulates TIR1-like genes and has been reported as a phasiRNA trigger in soybean, where it is closely linked with the basal defense response.66 It was also identified as a phasiRNA trigger in pepper (Supplementary Table S7); it is therefore likely to be associated with the basal defense response in pepper. We found that pepper-specific miRNAs also induce phasiRNA biogenesis. can-miR-n033 cleaves four NLRs and produces phasiRNA clusters (Supplementary Fig. S6A). In addition, we found that can-miR-n026 triggered phasiRNA production by cleavage of eight RLPs (Table 2; Fig. 3A). Interestingly, all the PHAS loci targeted by pepper-specific miRNAs are associated with disease-resistance.
Figure 3.
can-miR-n026 induces phasiRNA biogenesis by cleaving RLP. (A) Mapping results of small RNA and degradome reads normalized by mapping count to the genome and phasing score data were viewed using IGV. Red arrow indicates the cleavage site by can-miR-n026. Examples of (B) cis-acting phasiRNAs and (C) trans-acting phasiRNAs. Red triangles indicate the cleavage sites by small RNAs. In degradome reads panel, pale red colour indicates reads mapped to Watson strand and blue colour indicates those mapped to Crick strand in the genome.
We exploited degradome data to investigate the function of phasiRNA using CleaveLand4 (P-value ≤ 0.05 or penalty score ≤4).37 We found that a few phasiRNAs from disease-resistance genes act in cis or trans. can-miR482c cleaved a CNL (CA05g06820), producing phasiRNAs that regulated their own NLR in cis or another NLR (CA05g05440) in trans (Supplementary Fig. S4B and C). Additionally, can-miR-n026 cleaved an RLP (CA08g01140), with results similar to that of can-miR482c (Fig. 3B and C). Similar findings were also observed with can-miR-n033a and CA06g02150, except that there was no trans-acting phasiRNA (Supplementary Fig. S6B). These results are consistent with previous observations of cis- or trans-acting of phasiRNAs.11
3.4. Evolution of miRNAs related to defense mechanism in pepper
We found that several miRNAs in pepper targeted many genes-encoding defense-related proteins, such as NLRs and RLPs. In addition, some of the targeted genes produced secondary small RNAs, indicating they might have important roles in plant immunity. Then, we validated expression of the several disease-resistance related miRNAs in leaves of pepper, tomato, potato, and Nicotiana benthamiana via northern blot (Supplementary Fig. S7). We identified that can-miR-n033a-3p and can-miR-n026 were only expressed in pepper and can-miR6027 was expressed in pepper, tomato, and potato. It is consistent with the previous results that can-miR-n033a-3p and can-miR-n026 are pepper-specific miRNAs15 and miR6027 is conserved in tomato and potato.20 However, miR6023 was only expressed in pepper, even though it is also known as a conserved miRNA in tomato and potato (Supplementary Fig. S7).20 The mature sequence for miR6027 is identical across all three species, while miR6023 is polymorphic, which could cause disruption of probe hybridization (Supplementary Table S1). After validating expression of disease-resistance-related miRNAs, we tried to validate the target cleavage by disease-resistance-related miRNAs, especially can-miR-n033a and miR-n026, via 5′ RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) assay. However, we could not clone the target NLRs and RLPs because of their high redundancy. Although we got negative results in 5′ RLM-RACE assay, we got quite significant results in degradome analysis (Supplementary Fig. S3) and there are other studies that validated miRNA-derived cleavage through degradome analysis alone,11,67 therefore, we regarded that disease-resistance-related miRNAs in our study are still reliable and conducted further analyses.
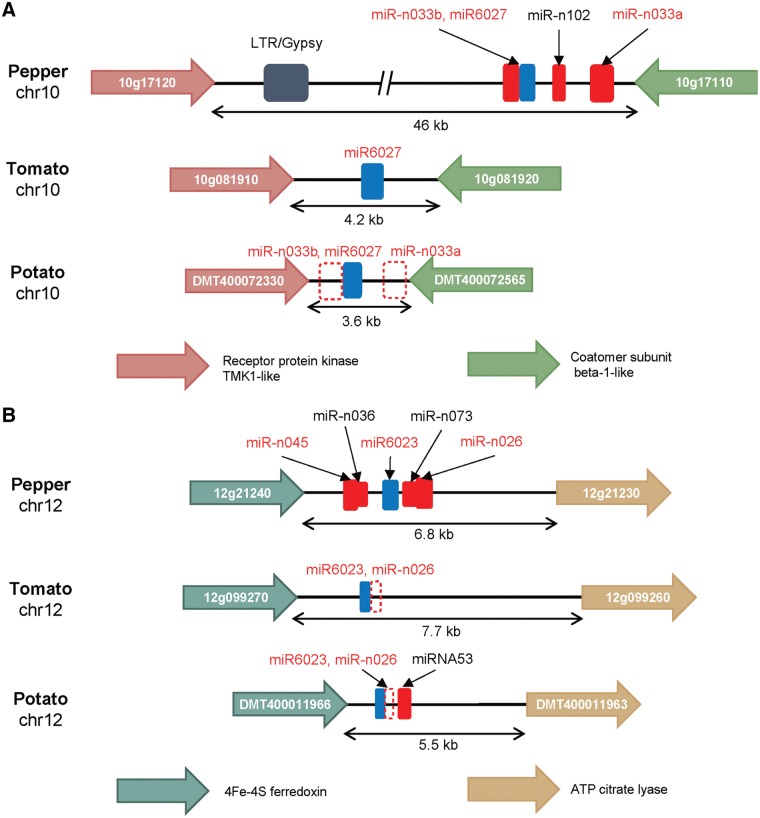
We examined whether genes of miR-n033 and miR-n026 only exist in pepper using BLASTN. We found that homologous regions are located on the intergenic region of chromosome 10 and 12, respectively (Fig. 4). The genes flanking these regions were conserved in tomato and potato. The hairpin sequence of miR6027 was conserved among the three species and can-miR-n033a and can-miR-n033b were located near miR6027 (Fig. 4A). Interestingly, we found candidates of miR-n033a and miR-n033b in potato, which were not annotated previously. Still, there was no similar sequence in tomato (Fig. 4A). In particular, miR-n033b and miR6027 might share the same precursor in pepper and potato, and their precursor sequences are well-conserved (Supplementary Fig. S8A and B). We also confirmed that miR-n033b is expressed in potato using public small RNA sequencing data17,30 (Supplementary Fig. S8E). In the case of miR-n033a, it is also well-conserved in pepper and potato, but it does not exist in tomato (Supplementary Fig. S9). In addition, the length of the intergenic region was ca. 46 kb in pepper, whereas it was 4.2 kb and 3.6 kb in tomato and potato, respectively. The intergenic region was 10 times more expanded and contained the LTR gypsy element in pepper. This may indicate a complex evolutionary history in that region in pepper, tomato, and potato. To identify whether miR-n033 regions were lost in tomato, we examined that miR-n033 regions exist in other Solanaceous species [eggplant (Solanum melongena L.),68Petunia axillaris,27 and Nicotiana benthamiana46] using BLASTN. We found that homologous region of miR-n033a is well-conserved in them, and that of miR-n033b is only conserved in Capsicum and Solanum species (Supplementary Fig. S10). These observations indicate that miR-n033 genes originated in a common ancestor of pepper, tomato, and potato, and then lost in tomato. In the case of miR-n026, the flanking genes were conserved and the length of the intergenic region was not different among pepper, tomato, and potato (Fig. 4B). We found that homologous region of miR-n026 exists in the three species (not annotated previously in tomato and potato), but the precursor sequence of can-miR-n026 is different from those in tomato and potato (Supplementary Fig. S11A–C). miR6023 and miR-n026 might be generated from the same precursor in tomato and potato, but from distinct precursors in pepper (Fig. 4B). This might result from either a pepper-specific translocation of the miR6023 region from the miR-n026 region, or a translocation of the miR6023 region to the miR-n026 region in an ancestor of Solanum species after the divergence of the lineage to pepper. Although hairpin structure of miR-n026 is different between pepper and Solanum species, we confirmed the expression of miR-n026 candidates in tomato and potato using public small RNA libraries.17,30,69,70 Thus, miR-n033 and miR-n026 regions showed the dynamic nature of the evolution, even in syntenic regions.
Figure 4.
Microsynteny analysis of clusters producing miR-n033 and miR-n026, targeting NLRs and RLPs, respectively. Red and blue boxes indicate species-specific and conserved miRNA genes, respectively. Empty boxes with dot line indicate homologous regions of miRNAs that were not annotated. (A) Intergenic regions producing miRNAs targeting NLRs in pepper (Capsicum annuum) and the corresponding regions in tomato (Solanum lycopersicum) and potato (Solanum tuberosum) are depicted. Pink and light green arrows indicate genes encoding receptor protein kinase TMK1-like and Coatomer subunit beta-1-like, respectively. Dark blue box indicates LTR/Gypsy element. miRNAs targeting NLRs were written in red. (B) Intergenic regions producing miRNAs targeting RLPs in pepper and the corresponding regions in tomato and potato are depicted. Khaki and yellow arrows indicate genes encoding 4Fe-4S ferredoxin and ATP citrate lyase, respectively. miRNAs targeting RLPs were written in red.
3.5. Different target affinity of miR-n033 between pepper and potato
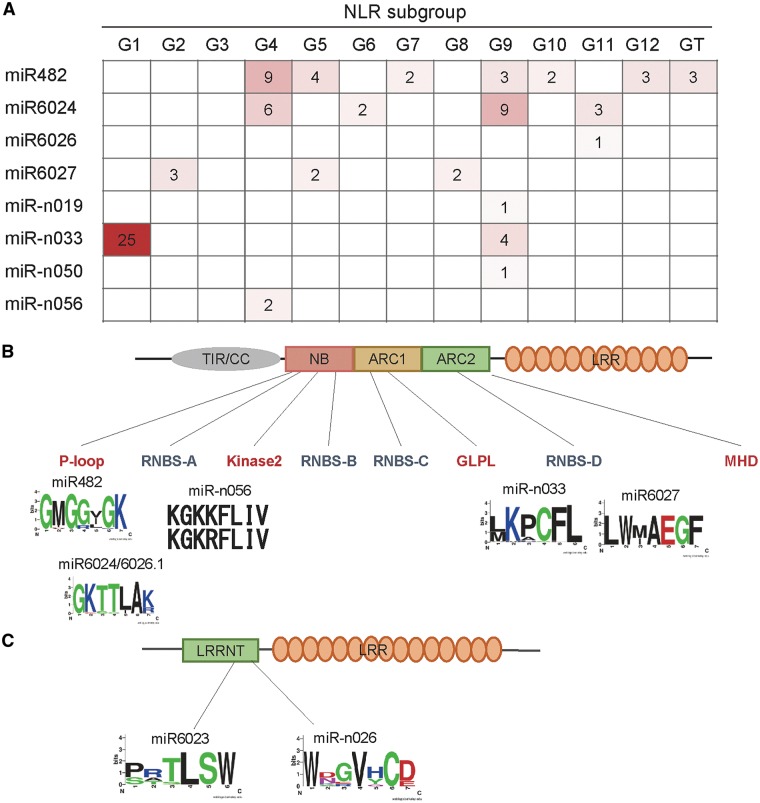
In a previous study, we classified NLRs into 13 subgroups, including 12 CNL types and 1 toll interleukin 1 receptor NLR (TNL) type, based on the phylogeny of NB-ARC domains.50 To perform in-depth analysis of NLR-regulating miRNAs, we analysed subgroups of the target genes. Interestingly, most target genes of can-miR-n033a-3p and can-miR-n033b-3p belong to the CNL-G1 subgroup of CNL (Fig. 5A), which is expanded subgroup since the divergence of pepper from Solanum spp. The duplication events of CNL-G1 peaked at 10 mya.50 Therefore, these miRNAs might have got or reinforced their affinity for CNL-G1 since the divergence of pepper from Solanum spp. This result may indicate that the miR-n033 family has evolved to regulate a number of NLR genes that have recently been duplicated. To investigate how these miRNAs could regulate multiple targets, we identified the region of the NLR targeted by each miRNA (Fig. 5B). The target site of can-miR-n033-3p at the NLR genes was different from that of other NLR-targeting miRNAs with the exception of miR6027. can-miR-n033a-3p and can-miR-n33b-3p targeted nucleotides encoding the resistance nucleotide binding site (RNBS)-D motif of the NB domain, whereas the can-miR482 family and can-miR6024 targeted the P-loop motif (Fig. 5B). We also analysed RLPs targeted by can-miR-n026 using the same approach, but the RLPs could not be classified as subgroups due to sequence variation. Nevertheless, the target regions of miR-n026 and miR6023, leucine-rich repeat N-terminal domain (LRRNT), are conserved (Fig. 5C).
Figure 5.
Group-specific NLR targets in pepper (Capsicum annuum). (A) The number of NLR targets according to their subgroups by miRNAs in pepper were shown as heatmap. NLR subgroups were followed to previous study and subgroups G1–G12 belong to CNL.50 (B) Targeting regions in NLRs by miRNAs in pepper. Domains and motifs of NLRs were marked. Major and minor motifs were written in red and blue, respectively. Representative amino acid sequences encoded by target regions were shown using WebLogo. Some miRNA targets were unclassified because they are thought to target untranslated regions. (C) Targeting regions in RLPs by miRNAs in pepper. Domains of RLPs were marked. Representative amino acid sequences encoded by target regions were shown using WebLogo.
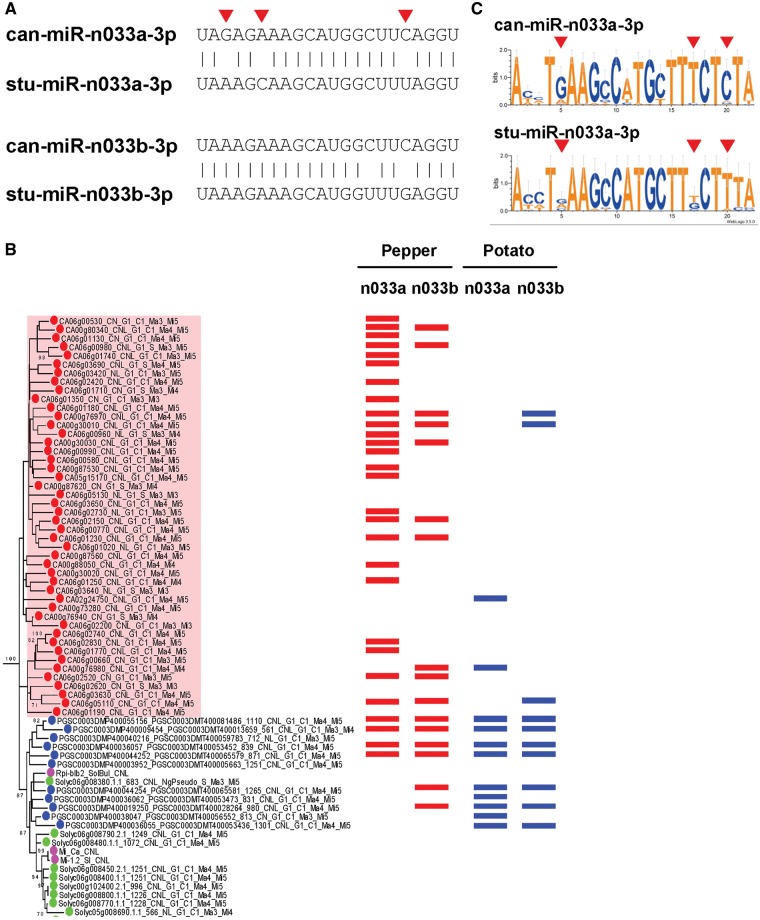
As described above, we found that the homologous region of miR-n026 exists in tomato and potato, and those of miR-n033 family exist in potato. To examine whether difference exists in target affinity of these miRNAs between three species, we extracted candidate sequences of miR-n026 and miR-n033 in tomato and potato, and compared their target NLRs and RLPs in pepper, tomato, and potato. Because of the different depths of the degradome data, we exploited a target prediction program, psRNAtarget, to compare the target genes of miRNAs (score ≤ 3.5).53 Remarkably, can-miR-n033a-3p was predicted to target more NLRs in CNL-G1 group (37, 32%), whereas can-miR-n033b-3p, stu-miR-n033a-3p, and stu-miR-n033b-3p were predicted to target less those (19, 16%; 16, 44% and 12, 33%, respectively; Table 3). It suggests that only can-miR-n033a-3p acquired an ability to target more NLRs which were expanded in pepper, and small portion of expanded NLRs in CNL-G1 group was targeted by can-miR-n033a-3p. We investigated NLRs in CNL-G1 targeted by miR-n033 to answer how can-miR-n033a-3p could get more affinity than other miR-n033s. It might result from the expansion of NLRs in CNL-G1 group of pepper (Fig. 6B).50 The mature miR-n033 sequences are different in these species (Fig. 6A). In addition, the consensus sequences of the CNL-G1 targeted by miR-n033a-3p different between pepper and potato, and these different sequences are corresponded with the sequences which are different between can-miR-n033a-3p and stu-miR-n033a-3p (Fig. 6C). Possible scenario is as described below: (1) miR-n033 genes existed in a common ancestor of pepper, tomato, and potato (Supplementary Fig. S10) and they had a function to regulate CNL-G1; (2) after pepper diverged from the ancestor (19 mya), CNL-G1 in pepper were highly duplicated (Fig. 6B) and it caused diversification of CNL-G1; (3) can-miR-n033a-3p evolved to regulate more CNL-G1 by mutation (Fig. 6A, C and Table 3). Therefore, it might reinforce the affinity of can-miR-n033a-3p for CNL-G1. In the case of stu-miR-n033b-3p, it was predicted to target more NLRs in CNL-G9 group (18) than can-miR-n033a-3p (6), can-miR-n033b-3p (3), and stu-miR-n033a-3p (3). Because, the number of NLRs in CNL-G9 group is slightly different between pepper (73) and potato (55),50 the difference of target affinity might come from mutation of stu-miR-n033b-3p. can-miR-n026 has many predicted target RLPs, but miR-n026 in tomato and potato has few predicted target RLPs (Supplementary Table S8), and their mature sequences are different, similar to the case of miR-n033 (Supplementary Fig. S13). The differences in targeting affinity between miR-n026s might be the result from mutation of miRNAs and/or expansion of target genes.
Table 3.
Comparing target prediction results of miR-n033 in pepper (Capsicum annuum) and potato (Solanum tuberosum)
| miRNA | Species | G1a | G3 | G5 | G6 | G9 | G12 | Ng |
|---|---|---|---|---|---|---|---|---|
| can-miR-n033a-3p | Pepper | 37/116b | 1/37 | 6/73 | 1/17 | 2 | ||
| Potato | 7/36 | 2/17 | 4/55 | |||||
| can-miR-n033b-3p | Pepper | 19/116 | 6/37 | 1/16 | 1/48 | 3/73 | 1/17 | |
| Potato | 12/36 | 4/17 | 8/55 | 1 | ||||
| stu-miR-n033a-3p | Pepper | 4/116 | 1/37 | 3/73 | ||||
| Potato | 16/36 | 1/17 | 1/55 | |||||
| stu-miR-n033b-3p | Pepper | 3/116 | 4/37 | 1/16 | 2/48 | 18/73 | 1/17 | |
| Potato | 12/36 | 3/17 | 2/39 | 9/55 | 1 |
G1 to Ng are classes of NLRs. Subgroups G1–G12 belong to CNL, Ng: non-group.
The number of predicted targets (score cut-off = 3.5)/total number of NLRs belonging to each subgroup.50
Figure 6.
(A) Alignment of mature miR-n033 sequences. Red triangles indicate different sequences between can-miR-n033a-3p and stu-miR-n033a-3p. (B) Detailed phylogenetic tree of CNL-G1 from our previous study (refer Supplementary Fig. S2 from Seo et al.50). Phylogenetic tree of whole CNL-G1 was in Supplementary Fig. S12. Transparent red box indicates duplicated CNL-G1 in pepper. Red boxes indicate the target CNL-G1 of can-miR-n033. Blue boxes indicate the target CNL-G1 of stu-miR-n033. Note, some CNL-G1 NLRs were excluded from the tree, because their NB-domain did not satisfy the criteria to construct a phylogenetic tree. (C) Consensus sequences of CNL-G1 targeted by miR-n033a. Red triangles indicate the sequences, whose complementary sequences in miRNAs are different.
4. Discussion
miRNAs play central roles in the post-transcriptional regulation of gene expression. In this study, we performed a comparative genome-wide analysis of miRNAs and their targets in pepper, tomato, and potato. Many of the miRNAs are conserved and share conserved targets and genomic synteny in plants. In three Solanaceous species, we observed similar patterns. However, some miRNAs, such as miR482d, have evolved in a species-specific manner, even in closely related species.
Degradome analysis provides valuable information to understand the genome-wide targets of miRNAs. In this study, we found consistent results among targets of conserved miRNAs. We confirmed a previous observation that the conserved miR396 targeted DRM, which is found exclusively in pepper and tomato,15 via degradome analysis (Supplementary Table S6). In addition, a number of target genes of miRNAs were classified as being involved in the defense response. Previous studies reported that some conserved miRNAs appear to be involved in defense mechanisms, both PTI and ETI. In Arabidopsis, PAMP induces the accumulation of miR393, which targets F-box auxin receptors, TIR1, and auxin signaling F-box protein 2 and 3.71 Suppression of auxin signalling by miR393 might result in enhanced PTI. In addition, miR160a-targeted ARFs positively regulated callose deposition induced by PAMP like flg22.72 miRNAs play roles in PTI via regulation of hormone signalling, but they also target NLR genes for ETI. For example, miR1507, miR2109, and miR2118 target several hundred NLR genes in Medicago truncatula.11,73 In Solanaceous species—such as tomato, potato, and N. benthamiana—miR482, miR5300, miR6019, and miR6027 were identified to target NLR genes.20,21 In addition, some miRNAs targeting NLRs can trigger the production of phasiRNA to regulate NLRs. For example, three 22-nucleotide miRNAs (miR2118, miR2109, and miR1507) target conserved domains of NLRs and initiate the production of phasiRNAs in legume.11 In N. benthamiana, two miRNAs (nta-miR6019 and nta-miR6020) target the region of the TIR domain in the N gene, and overexpression of these miRNAs attenuate N-mediated resistance to TMV.20
In this study, we found that several miRNAs targeted NLRs and RLPs in pepper. Species-specific miRNAs are considered to have evolved recently,74 and show lower expression levels than those of conserved miRNAs, or are expressed in specific condition.75 In a similar context, many species-specific miRNAs have few target genes, because they evolved neutrally.75 However, can-miR-n033 and can-miR-n026 were each found to target a number of NLRs and RLPs (Table 1), even though they are thought to have evolved recently. In addition, they produce phasiRNAs and regulate the transcripts of NLRs or RLPs in cis or trans manners (Supplementary Fig. S6 and Fig. 3). This indicates that miRNAs and phasiRNAs could regulate a large number of genes involved in defense systems and might have long-term benefits for R gene evolution. For example, NLRs usually induce disease resistance through a hypersensitive cell death response (HR) to infection.76 Several studies exhibited that some NLRs could induce HR in the host without pathogen.77–79 Therefore, if NLRs are not properly regulated, this could result in significant fitness costs in the host plants.80 Consequently, miRNAs regulating NLRs might have evolved to regulate a number of NLRs through the production of phasiRNAs.9,21 It might alleviate the possible problems due to rapid evolution of R genes, such as unexpected HR. Thus, miRNAs and phasiRNAs might facilitate divergent evolution of R genes.
It is known that hundreds of NLRs are present in plant genomes. NLRs might have evolved during an arms race with pathogens. As mentioned above, constant expression of NLRs and overactive immune systems can be harmful to plants with respect to fitness costs.80 For example, a dominant mutant suppressor of npr1-1, constitutive 1, a TNL type gene, shows constitutive activation of defense without pathogen perception, and the resulting phenotype was dwarfed with curly leaves in Arabidopsis.81,82 Therefore, plants should control the expression of NLRs during the absence of pathogens, and fine tuning of NLR expression is important. In this study, we showed that miR-n033 regulates a number of NLR genes in pepper and potato via degradome analysis and target prediction (Table 1). Among them, there are many CNL-G1 NLRs, which belong to a subgroup that is expanded specifically in pepper (Figs 5 and 6),50 whereas many CNL-G9 NLRs are predicted targets of stu-miR-n033b-3p in potato (Table 3). This indicates the divergent evolution of miRNA and their target NLRs in the different species. However, few NLRs in CNL-G2 were found to be targets of miRNAs, although this group was also expanded like CNL-G1 in pepper.50 Because degradome analysis was performed with tissues in normal conditions, more miRNAs targeting NLRs could be identified via miRNA profiling and degradome analysis under pathogen challenge.
We also found that miR-n026 targets RLP genes. There are few studies about miRNA regulation of RLP genes. A previous study reported that the conserved sly-miR6022 and sly-miR6023 cleaved Hcr9, a tomato Cf-9 homolog, which is the target of miR-n026.20 Furthermore, the targeting of other RLPs by sly-miR6022 was verified via degradome analysis in tomato fruit.83 Cf-9 genes confer resistance against Cladosporium fulvum by recognition of pathogen-derived avirulence determinants.84 In that case, necrosis in a host containing Cf-9 was observed to occur during infection by C. fulvum race 5. This suggests the important function of Hcr9 in disease resistance. In another case, an RLP kinase, CaLRR51, was found to play a role in the response of pepper to infection by Ralstonia solanacearum. Moreover, the transient overexpression of CaLRR51 resulted in HR-like cell death responses, indicating the necessity of strict regulation of RLPs.85 We observed in this study that miR-n026 cleaved some Hcr9 genes in pepper (Table 2), and that there are few predicted RLP targets in potato and tomato (Supplementary Table S8). Therefore, RLP-targeting miRNAs in pepper appear to have evolved with their targets to acquire the function of regulating RLPs in order to avoid damage due to unexpected resistance responses. It is possible that other regulatory pathways exist to compensate for the absence of miR-n026 function in tomato and potato.
We regarded that miR-n033 and miR-n026 are pepper-specific miRNAs, but we found homologous sequences of them, and their hairpin structures are also conserved in tomato and potato. However, miR-n033 and miR-n026 were not identified previously in tomato and potato. There are several possibilities that could explain why miR-n033 and miR-n026 were not identified in tomato and potato. First, when we examined their expression in tomato and potato using public small RNA libraries, we merged all libraries from several studies17,30,69,70 for each species. Therefore, expression of miR-n026 and miR-n033 in tomato and potato are too low to be identified as miRNA in the other studies. Also, it might explain why we could not identify miR-n026 and miR-n033 in potato and tomato via northern blot. We extracted total RNA only from leaf in a normal condition, and it is possible that miR-n026 and miR-n033 in tomato and potato were not expressed in that tissue or condition. Second, in the case of miR-n033b and miR-n026, they are generated from the same precursor with miR6027 and miR6023, respectively, so miR-n033b and miR-n026 might be omitted, because the probability that a precursor has two entirely different mature miRNAs is not considered generally. Third, length of the miR-n026 and miR-n033 precursors in tomato and potato is quite long (>200 nt), then it is hard to identify them with general criteria for identification of miRNA. Because of these possibilities, miR-n026 and miR-n033 in tomato and potato require experimental validation for further study.
5. Conclusion
In this study, we systematically investigated the evolutionary patterns of miRNAs and their targets. Our analysis revealed that some miRNAs in pepper mainly targeted genes encoding defense-related proteins. We found that miR-n026 and miR-n033 target a number of NLRs and RLPs in pepper. This study provides insights into evolution of miRNAs and target genes in the plant defense system, which has led to differentiation of miRNA function.
Supplementary Material
Acknowledgements
We thank Dr Barbara Baker from University of California, Berkeley for helpful discussion. We also appreciate the assistance of high-throughput sequencing from the National Instrumentation Center of the Environmental Management College of Agriculture and Life Sciences, Seoul National University. This work was supported by a grant from the Ministry of Science, ICT and Future Planning (MSIP) of the Korean government through the National Research Foundation (NRF-2018R1A2A1A05019892) to D.C. and by a grant from the Next-Generation BioGreen 21 Program (No. PJ01333001), Rural Development Administration, Republic of Korea to C.S.
Availability of data and materials
The dataset supporting the conclusions of this article is available in the Gene Expression Omnibus; GSE102781 (degradome libraries), GSE41654 (public pepper small RNA library), GSE18110, GSE32470, GSE76204 (public tomato small RNA libraries), GSE52599 and GSE32471 (public potato small RNA libraries).
Conflict of interest
None declared.
References
- 1. Axtell M. J., Westholm J. O., Lai E. C.. 2011, Vive la difference: biogenesis and evolution of microRNAs in plants and animals, Genome Biol., 12, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jones-Rhoades M. W., Bartel D. P., Bartel B.. 2006, MicroRNAs and their regulatory roles in plants, Annu. Rev. Plant Biol., 57, 19–53. [DOI] [PubMed] [Google Scholar]
- 3. Sunkar R., Chinnusamy V., Zhu J., Zhu J. K.. 2007, Small RNAs as big players in plant abiotic stress responses and nutrient deprivation, Trends Plant Sci., 12, 301–9. [DOI] [PubMed] [Google Scholar]
- 4. Voinnet O. 2008, Post-transcriptional RNA silencing in plant-microbe interactions: a touch of robustness and versatility, Curr. Opin. Plant Biol., 11, 464–70. [DOI] [PubMed] [Google Scholar]
- 5. Chen H. M., Chen L. T., Patel K., Li Y. H., Baulcombe D. C., Wu S. H.. 2010, 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants, Proc. Natl. Acad. Sci. U.S.A., 107, 15269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cuperus J. T., Carbonell A., Fahlgren N., et al. 2010, Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis, Nat. Struct. Mol. Biol., 17, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Felippes F. F., Marchais A., Sarazin A., Oberlin S., Voinnet O.. 2017, A single miR390 targeting event is sufficient for triggering TAS3-tasiRNA biogenesis in Arabidopsis, Nucleic Acids Res., 45, 5539–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Axtell M. J., Jan C., Rajagopalan R., Bartel D. P.. 2006, A two-hit trigger for siRNA biogenesis in plants, Cell, 127, 565–77. [DOI] [PubMed] [Google Scholar]
- 9. Fei Q., Xia R., Meyers B. C.. 2013, Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks, Plant Cell, 25, 2400–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vazquez F., Vaucheret H., Rajagopalan R., et al. 2004, Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs, Mol. Cell, 16, 69–79. [DOI] [PubMed] [Google Scholar]
- 11. Zhai J., Jeong D. H., De Paoli E., et al. 2011, MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs, Genes Dev., 25, 2540–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song X., Li P., Zhai J., et al. 2012, Roles of DCL4 and DCL3b in rice phased small RNA biogenesis, Plant J., 69, 462–74. [DOI] [PubMed] [Google Scholar]
- 13. Allen E., Xie Z., Gustafson A. M., Carrington J. C.. 2005, microRNA-directed phasing during trans-acting siRNA biogenesis in plants, Cell, 121, 207–21. [DOI] [PubMed] [Google Scholar]
- 14. Xia R., Xu J., Meyers B. C.. 2017, The emergence, evolution, and diversification of the miR390-TAS3-ARF pathway in land plants, Plant Cell, 29, 1232–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hwang D. G., Park J. H., Lim J. Y., et al. 2013, The hot pepper (Capsicum annuum) microRNA transcriptome reveals novel and conserved targets: a foundation for understanding microRNA functional roles in hot pepper, PLoS One, 8, e64238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pantaleo V., Szittya G., Moxon S., et al. 2010, Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis, Plant J., 62, 960–76. [DOI] [PubMed] [Google Scholar]
- 17. Lakhotia N., Joshi G., Bhardwaj A. R., et al. 2014, Identification and characterization of miRNAome in root, stem, leaf and tuber developmental stages of potato (Solanum tuberosum L.) by high-throughput sequencing, BMC Plant Biol., 14, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y., Xia R., Kuang H., Meyers B. C.. 2016, The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them, Mol. Biol. Evol., 33, 2692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dodds P. N., Rathjen J. P.. 2010, Plant immunity: towards an integrated view of plant-pathogen interactions, Nat. Rev. Genet., 11, 539–48. [DOI] [PubMed] [Google Scholar]
- 20. Li F., Pignatta D., Bendix C., et al. 2012, MicroRNA regulation of plant innate immune receptors, Proc. Natl. Acad. Sci. U.S.A., 109, 1790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shivaprasad P. V., Chen H. M., Patel K., Bond D. M., Santos B. A., Baulcombe D. C.. 2012, A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs, Plant Cell, 24, 859–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao M., Meyers B. C., Cai C., Xu W., Ma J.. 2015, Evolutionary patterns and coevolutionary consequences of MIRNA genes and microRNA targets triggered by multiple mechanisms of genomic duplications in soybean, Plant Cell, 27, 546–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abrouk M., Zhang R., Murat F., et al. 2012, Grass microRNA gene paleohistory unveils new insights into gene dosage balance in subgenome partitioning after whole-genome duplication, Plant Cell, 24, 1776–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Vries S., Kloesges T., Rose L. E.. 2015, Evolutionarily dynamic, but robust, targeting of resistance genes by the miR482/2118 gene family in the Solanaceae, Genome Biol. Evol., 7, 3307–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gonzalez V. M., Muller S., Baulcombe D., Puigdomenech P.. 2015, Evolution of NBS-LRR gene copies among Dicot plants and its regulation by members of the miR482/2118 superfamily of miRNAs, Mol. Plant, 8, 329–31. [DOI] [PubMed] [Google Scholar]
- 26. Zhang L., Qin C., Mei J., et al. 2017, Identification of microRNA targets of Capsicum spp. using miRTrans—a trans-omics approach, Front. Plant Sci., 8, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bombarely A., Moser M., Amrad A., et al. 2016, Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida, Nat. Plants, 2, 16074. [DOI] [PubMed] [Google Scholar]
- 28. Fei Z., Joung J. G., Tang X., et al. 2011, Tomato functional genomics database: a comprehensive resource and analysis package for tomato functional genomics, Nucleic Acids Res., 39, D1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kozomara A., Griffiths-Jones S.. 2011, miRBase: integrating microRNA annotation and deep-sequencing data, Nucleic Acids Res., 39, D152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tomato Genome C. 2012, The tomato genome sequence provides insights into fleshy fruit evolution, Nature, 485, 635–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zuo J., Zhu B., Fu D., et al. 2012, Sculpting the maturation, softening and ethylene pathway: the influences of microRNAs on tomato fruits, BMC Genomics, 13, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shen D., Suhrkamp I., Wang Y., et al. 2014, Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes, New Phytol., 204, 577–94. [DOI] [PubMed] [Google Scholar]
- 33. Wang Y., Tang H., Debarry J. D., et al. 2012, MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity, Nucleic Acids Res., 40, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Addo-Quaye C., Eshoo T. W., Bartel D. P., Axtell M. J.. 2008, Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome, Curr. Biol., 18, 758–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burge S. W., Daub J., Eberhardt R., et al. 2013, Rfam 11.0: 10 years of RNA families, Nucleic Acids Res., 41, D226–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J.. 2005, Repbase update, a database of eukaryotic repetitive elements, Cytogenet. Genome Res., 110, 462–7. [DOI] [PubMed] [Google Scholar]
- 37. Brousse C., Liu Q., Beauclair L., Deremetz A., Axtell M. J., Bouche N.. 2014, A non-canonical plant microRNA target site, Nucleic Acids Res., 42, 5270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M.. 2005, Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research, Bioinformatics, 21, 3674–6. [DOI] [PubMed] [Google Scholar]
- 39. Pall G. S., Hamilton A. J.. 2008, Improved northern blot method for enhanced detection of small RNA, Nat. Protocol., 3, 1077–84. [DOI] [PubMed] [Google Scholar]
- 40. Sunkar R., Zhu J. K.. 2004, Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis, Plant Cell, 16, 2001–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langmead B., Trapnell C., Pop M., Salzberg S. L.. 2009, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xia R., Meyers B. C., Liu Z., Beers E. P., Ye S., Liu Z.. 2013, MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA Biogenesis in Eudicots, Plant Cell, 25, 1555–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quast C., Pruesse E., Yilmaz P., et al. 2013, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 41, D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cognat V., Pawlak G., Duchene A. M., et al. 2013, PlantRNA, a database for tRNAs of photosynthetic eukaryotes, Nucleic Acids Res., 41, D273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robinson J. T., Thorvaldsdottir H., Winckler W., et al. 2011, Integrative genomics viewer, Nat. Biotechnol., 29, 24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bombarely A., Rosli H. G., Vrebalov J., Moffett P., Mueller L. A., Martin G. B.. 2012, A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe, 25, 1523–30. [DOI] [PubMed] [Google Scholar]
- 47. Särkinen T., Bohs L., Olmstead R. G., Knapp S.. 2013, A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree, BMC Evol. Biol., 13, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mathews D. H., Disney M. D., Childs J. L., Schroeder S. J., Zuker M., Turner D. H.. 2004, Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc, Natl. Acad. Sci. U.S.A., 101, 7287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim S., Park M., Yeom S. I., et al. 2014, Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species, Nat. Genet., 46, 270–8. [DOI] [PubMed] [Google Scholar]
- 50. Seo E., Kim S., Yeom S. I., Choi D.. 2016, Genome-wide comparative analyses reveal the dynamic evolution of nucleotide-binding leucine-rich repeat gene family among Solanaceae plants, Front. Plant Sci., 7, 1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sievers F., Wilm A., Dineen D., et al. 2014, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega, Mol. Syst. Biol., 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E.. 2004, WebLogo: a sequence logo generator, Genome Res., 14, 1188–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dai X., Zhao P. X.. 2011, psRNATarget: a plant small RNA target analysis server, Nucleic Acids Res., 39, W155–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. German M. A., Luo S., Schroth G., Meyers B. C., Green P. J.. 2009, Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome, Nat. Protocol., 4, 356–62. [DOI] [PubMed] [Google Scholar]
- 55. German M. A., Pillay M., Jeong D. H., et al. 2008, Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends, Nat. Biotechnol., 26, 941–6. [DOI] [PubMed] [Google Scholar]
- 56. Axtell M. J. 2008, Evolution of microRNAs and their targets: are all microRNAs biologically relevant? Biochim. Biophys. Acta 1779, 725–34. [DOI] [PubMed] [Google Scholar]
- 57. Cuperus J. T., Fahlgren N., Carrington J. C.. 2011, Evolution and functional diversification of MIRNA genes, Plant Cell, 23, 431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fei Q., Zhang Y., Xia R., Meyers B. C.. 2016, Small RNAs add zing to the zig-zag-zig model of plant defenses, Mol. Plant Microbe Interact., 29, 165–9. [DOI] [PubMed] [Google Scholar]
- 59. Gou M., Su N., Zheng J., et al. 2009, An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis, Plant J., 60, 757–70. [DOI] [PubMed] [Google Scholar]
- 60. Fahlgren N., Jogdeo S., Kasschau K. D., et al. 2010, MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana, Plant Cell, 22, 1074–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nozawa M., Miura S., Nei M.. 2012, Origins and evolution of microRNA genes in plant species, Genome Biol. Evol., 4, 230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen H. M., Li Y. H., Wu S. H.. 2007, Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A., 104, 3318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Arikit S., Xia R., Kakrana A., et al. 2014, An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes, Plant Cell, 26, 4584–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zheng Y., Wang Y., Wu J., Ding B., Fei Z.. 2015, A dynamic evolutionary and functional landscape of plant phased small interfering RNAs, BMC Biol., 13, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wei C., Kuang H., Li F., Chen J.. 2014, The I2 resistance gene homologues in Solanum have complex evolutionary patterns and are targeted by miRNAs, BMC Genomics, 15, 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wong J., Gao L., Yang Y., et al. 2014, Roles of small RNAs in soybean defense against Phytophthora sojae infection, Plant J., 79, 928–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xia R., Ye S., Liu Z., Meyers B. C., Liu Z.. 2015, Novel and recently evolved microRNA clusters regulate expansive F-BOX gene networks through phased small interfering RNAs in wild diploid strawberry, Plant Physiol., 169, 594–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hirakawa H., Shirasawa K., Miyatake K., et al. 2014, Draft genome sequence of eggplant (Solanum melongena L.): the representative Solanum species indigenous to the old world, DNA Res., 21, 649–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mohorianu I., Schwach F., Jing R., et al. 2011, Profiling of short RNAs during fleshy fruit development reveals stage-specific sRNAome expression patterns, Plant J., 67, 232–46. [DOI] [PubMed] [Google Scholar]
- 70. Wu P., Wu Y., Liu C.-C., et al. 2016, Identification of arbuscular mycorrhiza (AM)-responsive microRNAs in tomato, Front. Plant Sci., 7, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Navarro L., Dunoyer P., Jay F., et al. 2006, A plant miRNA contributes to antibacterial resistance by repressing auxin signaling, Science, 312, 436–9. [DOI] [PubMed] [Google Scholar]
- 72. Li J., Ding J., Zhang W., et al. 2010, Unique evolutionary pattern of numbers of gramineous NBS-LRR genes, Mol. Genet. Genomics, 283, 427–38. [DOI] [PubMed] [Google Scholar]
- 73. Fei Q., Li P., Teng C., Meyers B. C.. 2015, Secondary siRNAs from Medicago NB-LRRs modulated via miRNA-target interactions and their abundances, Plant J., 83, 451–65. [DOI] [PubMed] [Google Scholar]
- 74. Allen E., Xie Z., Gustafson A. M., Sung G. H., Spatafora J. W., Carrington J. C.. 2004, Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana, Nat. Genet., 36, 1282–90. [DOI] [PubMed] [Google Scholar]
- 75. Rajagopalan R., Vaucheret H., Trejo J., Bartel D. P.. 2006, A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana, Genes Dev., 20, 3407–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jones J. D., Dangl J. L.. 2006, The plant immune system, Nature, 444, 323–9. [DOI] [PubMed] [Google Scholar]
- 77. Rairdan G. J., Collier S. M., Sacco M. A., Baldwin T. T., Boettrich T., Moffett P.. 2008, The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling, Plant Cell, 20, 739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Michael Weaver L., Swiderski M. R., Li Y., Jones J. D.. 2006, The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis, Plant J., 47, 829–40. [DOI] [PubMed] [Google Scholar]
- 79. Wang L. C., Ye X. F., Liu H. C., et al. 2016, Both overexpression and suppression of an Oryza sativa NB-LRR-like gene OsLSR result in autoactivation of immune response and thiamine accumulation, Sci. Rep., 6, 24079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tian D., Traw M. B., Chen J. Q., Kreitman M., Bergelson J.. 2003, Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana, Nature, 423, 74–7. [DOI] [PubMed] [Google Scholar]
- 81. Li X., Clarke J. D., Zhang Y., Dong X.. 2001, Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance, Mol. Plant Microbe Interat., 14, 1131–9. [DOI] [PubMed] [Google Scholar]
- 82. Zhang Y., Goritschnig S., Dong X., Li X.. 2003, A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1, Plant Cell, 15, 2636–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Karlova R., van Haarst J. C., Maliepaard C., et al. 2013, Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis, J. Exp. Bot., 64, 1863–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Parniske M., Hammond-Kosack K. E., Golstein C., et al. 1997, Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato, Cell, 91, 821–32. [DOI] [PubMed] [Google Scholar]
- 85. Cheng W., Xiao Z., Cai H., et al. 2017, A novel leucine-rich repeat protein, CaLRR51, acts as a positive regulator in the response of pepper to Ralstonia solanacearum infection, Mol. Plant Pathol., 18, 1089–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset supporting the conclusions of this article is available in the Gene Expression Omnibus; GSE102781 (degradome libraries), GSE41654 (public pepper small RNA library), GSE18110, GSE32470, GSE76204 (public tomato small RNA libraries), GSE52599 and GSE32471 (public potato small RNA libraries).