Abstract
Rbm38 is a p53 target and a RNA-binding protein known to suppress p53 translation by preventing eukaryotic translation initiation factor 4E (eIF4E) from binding to p53 mRNA. In this study, we show that synthetic peptides corresponding to the binding interface between Rbm38 and eIF4E, including an 8 amino acid peptide (Pep8) derived from Rbm38, are effective in relieving Rbm38-mediated repression of p53. Molecular simulations showed that Ser-6 in Pep8 forms a hydrogen bond with Asp-202 in eIF4E. Substitution of Ser-6 with Lys, but not with Asp, enhanced the ability of Pep8 to inhibit the Rbm38-eIF4E complex. Importantly, Pep8 alone or together with a low dose of doxorubicin potently induced p53 expression and suppressed colony and tumor sphere formation and xenograft tumors in Rbm38- and p53-dependent manners. Together, we conclude that modulating the Rbm38-eIF4E complex may be explored as a therapeutic strategy for cancers that carry wild-type p53.
Keywords: p53, p21, Rbm38, eIF4E, senescence
Introduction
As a focal regulatory component of RNA metabolism, RNA-binding proteins (RBPs) modulate all facets of RNA biogenesis, including RNA surveillance and maturation, subcellular localization and nucleocytoplasmic transport, translation and degradation (1). Given the fact that RBPs control RNA metabolism, defects in the function of RBPs can have far reaching implications, such as neurodegenerative disorders, muscular atrophy, and numerous cancers (1,2). RBPs are also shown to form complex integrated networks, which differentially regulate both oncogenes and tumor suppressors, and thus, can have opposing effects on tumorigenesis (3,4).
The p53 gene is one of the most important and well-studied tumor suppressors. Inactivation of p53 occurs in more than 50% of human cancers and is a hallmark of tumor progression (5,6). Functionally, p53 predominantly acts as a transcription factor that can be activated by multiple stimuli, including DNA damage, hypoxia and oncogene activation (6–8). Activated p53 can stimulate multiple antiproliferative mechanisms by modulating expression of genes involved in cell-cycle arrest, cellular senescence and apoptosis (9). Because of compromised p53 activity in a vast amount of human cancers, regaining wild-type function of p53 is being explored as a therapeutic approach.
RNA-binding motif protein 38 (Rbm38), also known as RNPC1, is a RBP which contains one RNA recognition motif with sequence homology to nucleolin and HuR (10). The Rbm38 gene resides on chromosome 20q13, which has been shown to be frequently amplified in multiple cancers (11–14). Rbm38 functions by positively regulating the mRNA stability of p21, p73, and HuR, while it negatively regulates the mRNA stability of p63 and MDM2 (10,15–19). Furthermore, elevated levels of Rbm38 are correlated with poor prognosis in breast cancer patients (20,21) and is associated with malignant transformation of colorectal adenoma to carcinoma (22,23). Our group discovered that Rbm38 suppresses p53 mRNA translation by directly interacting with eukaryotic translation initiation factor 4E (eIF4E) on p53 mRNA (24). Consistently, Rbm38-deficient mice are prone to premature aging and early onset of multiple tumors in a p53-dependent manner (25). Of interest, phosphorylation of Rbm38 at serine 195 by glycogen synthase kinase-3 (GSK3) inhibits its interaction with eIF4E, switching Rbm38 from a suppressor to an activator of p53 mRNA translation (26). Further, Rbm24, a homologous family member, also interacts with eIF4E leading to decreased p53 translation (27). As a key component of the eIF4F complex, eIF4E binds to the 5’ mRNA cap, which is known to be the rate limiting step in mRNA translation (28,29). Therefore, removing Rbm38 translational inhibition to enhance p53 expression may prove to be an effective therapeutic option for cancers that often carry wild-type p53.
Materials and Methods
Human cell lines
RKO and MCF7 cells were cultured at 37°C in DMEM (Dulbecco’s Modified Eagle’s medium, Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA) in a humidified incubator with 5% CO2. Cell lines RKO (CRL-2577) and MCF7 (HTB-22) were obtained from ATCC between 2007 and 2016 and used below passage 25 or within 2 months after thawing. Cells were tested negative for mycoplasma after thawing and used within two months. Since all cell lines from ATCC have been thoroughly tested and authenticated, we did not authenticate the cell lines used in this study.
Mice
6-week-old athymic nude mice from Charles Rivers were ordered and used to generate xenograft tumors. Mice were maintained in accordance with NIH animal guidelines following protocols approved by UC Davis Institutional Animal Care and Use Committee. Sample size and power analysis was determined by the work done by Hather et al. (30).
Plasmids and cell line generation
Generation of Rbm38-null and p53-null cells lines was achieved by using CRISPR-Cas9, pSpCas9(BB)-2A-Puro vector expressing guide RNA’s ( Rbm38: 5’-TTT TCT CCA TCA GGG CAC GT 3’; p53 guide #1 5’-CCA TTG TTC AAT ATC GTC CG, p53 guide #2 5’-TCC ATT GCT TGG GAC GGC AA ). The cells were selected with puromycin and each individual clone was confirmed by western blot analysis. GST-Rbm38 and His-eIF4E expression plasmids were generated as previously described (26). pTXB1-eIF4E plasmid was generated by amplifying eIF4E using His-eIF4E expression plasmid as template. The amplicon was then cloned into pTXB1 via NdeI and SapI. The primers used to amplify eIF4E were forward primer, 5´- GGT GGT CAT ATG GCG ACT GTC GAA CCG GAA ACC −3´, and reverse primer, 5´- GGT GGT TGC TCT TCC GCA AAC AAC AAA CCT ATT TTT AGT GGT GGA G −3´.
Peptide synthesis and delivery
Peptides were synthesized by GenScript (Piscataway, NJ).
Western blotting
Western blot procedures were as previously described (31). Briefly, membranes were blocked in PBST containing 3% milk for 1 hour at 20°C. Primary antibodies in PBST containing 3% milk were incubated at 4°C rocking overnight. The following morning membranes were washed 3x with PBST followed by the addition of secondary antibody in PBST containing 3% milk at 20°C for 2 hours. Membranes were then washed 3x with PBS. All western blot figures are representative data of at least 2 independent replicates. Band intensities were calculated using ImageJ (32). Antibodies used are: anti-p53 (1C12, Cell Signaling, Danvers, MA), anti-actin (Sigma), anti-GST (B-14, Santa Cruz, Dallas, TX), anti-His (Omni-probe D-8, Santa Cruz), anti-eIF4E (P-2, Santa Cruz), anti-p21 (H164, Santa Cruz), anti-p130 (A-10, Santa Cruz), anti-vinculin (7F9, Santa Cruz), anti-MDM2 (SMP14, Santa Cruz), and anti-HA (Covance, San Diego, CA).
Competitive pull-down assays
TentaGel beads bound to peptides were incubated with cold PBS with 1% BSA for 1 h rotating at 4°C. Beads were washed 5x with PBS and incubated overnight at 4°C with equal amounts of purified eIF4E in PBS containing 0.1% BSA and 0.1% Triton-X100. The following day beads were washed 5X with PBS containing 0.1% Triton-X100 before being eluted with 1x SDS-lysis buffer and subjected to western blot analysis. For GST-Rbm38 competitive pull-down assays pGEX-4T3-Rbm38 plasmid was transformed into BL21 (DE3) competent E. coli. 1-liter culture was grown at 37°C until OD600 = 0.6–0.8 and then induced with a final concentration of 0.1 mM IPTG for 4 hours. Bacteria were spun down and the pellet placed in −80°C overnight. Pellets were lysed, sonicated and centrifuged in 20 mL lysis buffer with 1% Triton-X100, 1 mM DTT and protease inhibitor cocktail. Lysates were then incubated with GST beads rocking at 4°C for 2 hours. Beads were washed 3x with lysis buffer. After brief centrifugation lysates were carefully removed. Beads were then resuspended in lysis buffer with 0.1% Triton-X100 to make a 50% bead slurry. 100 μL bead slurry was incubated in 650 μL lysis buffer, 5 μM peptide (Ctrl, Pep8, Pep8S-D, Pep8S-K) and 5 μM purified eIF4E in a 1.5 mL tube. Samples were rocked overnight, washed 3x with lysis buffer and eluted with 60 μL 1x SDS-loading buffer before western blot analysis.
RNA-ChIP
RKO cells were seeded at 2 × 106 cells per plate in four 10-cm plates. The next day two plates were treated with 1.5 μM Pen-Ctrl and two plates with 1.5 μM Pen-Pep8 for 2 hours. RNA-ChIP was performed as previously described (33).
35S-metabolic labeling
Metabolic labeling of newly synthesized protein was performed as previously described (34). Briefly, RKO cells were treated with Pen-Ctrl or Pen-Pep8 (5 μM) for 18 h. Next, the cells were incubated for 1 h in methionine-free DMEM before being labeled with 100 μCi/mL 35S for 30 min. The amount of 35S incorporation was measured by TCA precipitation and scintillation counting. Equal amounts of 35S-labeled lysates (3.7 × 106 cpm) were incubated with 1.0 μg anti-p53 antibody and Protein G agarose beads (25 μL bed volume). The immunocomplexes were resolved on an 8% SDS-PAGE gel and subjected to autoradiography.
Colony formation
Colony formation assays were performed as previously described (35). Briefly, 1,000 cells were plated in 6-well plates. The following day peptides and/or doxorubicin were added to the media. Following three-day incubation, the media was replaced with fresh complete DMEM. Cells were grown until colonies were visible. Cells were fixed using (7:1) methanol : acetic acid followed by crystal violet staining.
Tumor spheres formation
MCF7 cells were cultured in MammoCult™ (Stemcell, Cambridge, MA) media per manufacturer’s guidelines. 20,000 cells per well were seeded in 6-well ultra-low adherent plates. The following day the cells were treated with 5 μM Pen-Ctrl or Pen-Pep8 for 20 minutes followed by vehicle or 6.25 ng/mL doxorubicin for 7 days. After 7 days, tumor spheres larger than 50 μm were counted.
Xenograft tumor generation
Female 6-week-old athymic nude mice were injected using a 24-guage needle with 1×106 RKO cells in 0.2 mL of serum-free RPMI 1640 mixed with Matrigel at a 1:1 ratio (100 μL of cells mixed with 100 μL of Matrigel (BD Biosciences, San Jose, CA) subcutaneously into the right flank of each nude mouse. Mice were randomly separated into Pen-Ctrl or Pen-Pep8 with or without doxorubicin treated groups. After the xenograft tumors reached a volume larger than 100 mm3, mice were intratumorally injected with 5 μM Pen-Ctrl or Pen-Pep8 (50 μL injection) every other day for 14 days. For the doxorubicin treated groups, mice received two doses (2 mg/kg); one on the first day of peptide treatment and 7 days later. After 14 days, the mice were sacrificed, tumors were excised, weighed and processed for either western blot or H&E staining.
Histological analysis
Xenograft tumors were fixed in 10% (wt/vol) neutral buffered formalin, routinely processed, and embedded in paraffin blocks. H&E staining and IHC were performed on xenograft tissue sections (5 μm) as previously described (35).
Statistical analysis
Experimental values are presented as mean ± SEM. Statistical comparisons between experimental groups were analyzed by a two-tailed Student’s t-test. P values < 0.05 were considered statistically significant. For tumor volume over time multiple t-test with a false discovery rate Q=1% was used for analysis.
Results
Pep8 modulates the Rbm38-eIF4E complex to increase p53 expression
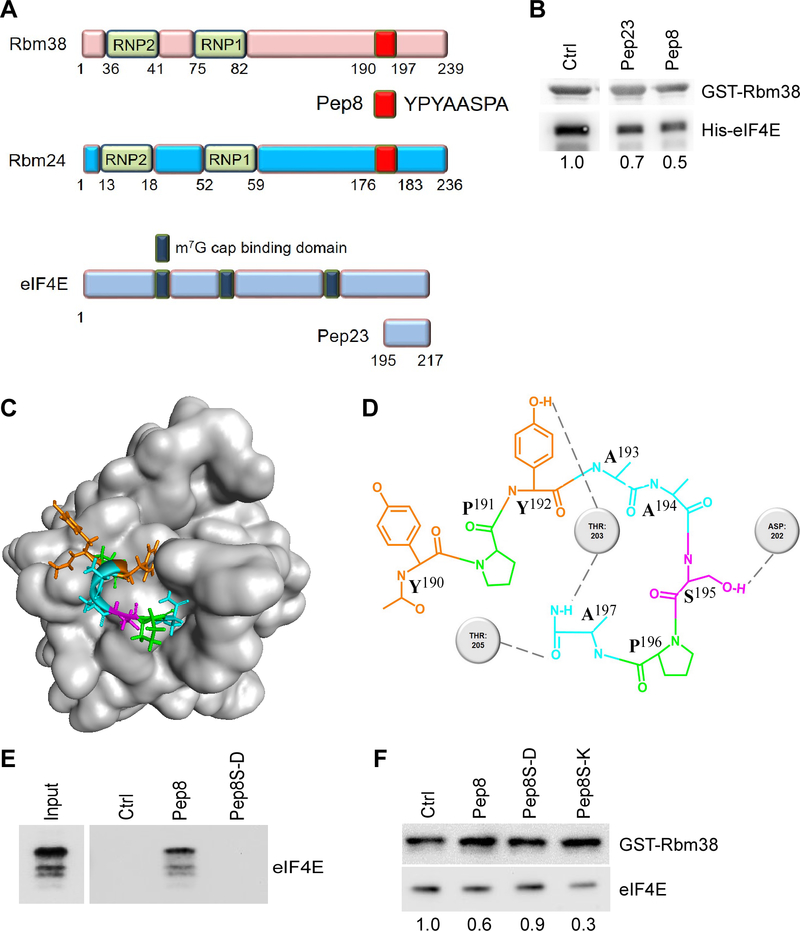
We recently showed that Rbm38 and Rbm24 both directly interact with eIF4E on p53 mRNA and suppress p53 translation (24,27). With this knowledge, we postulated that using a peptide composed of the binding interface from either Rbm38 or eIF4E may impede their binding, thus increase p53 translation. Previously, the binding interfaces between Rbm38/Rbm24 and eIF4E were mapped to their extreme carboxyl termini (Fig. 1A) (26,27). Of interest, the Rbm38/Rbm24 binding interfaces share a common 8 amino acids (Pep8) unique to just Rbm38 and Rbm24 that may be involved in their interaction with eIF4E (Fig. 1A). Based on these findings, several peptides were synthesized, including Pep8 derived from Rbm38/Rbm24 and Pep23 derived from eIF4E (Supplementary Table 1). To test their ability to modulate the interaction between Rbm38 and eIF4E, competitive pull-down assays were performed. We showed that the Rbm38-eIF4E complex was diminished by Pep23 or Pep8 compared to the control peptide (Fig 1B, compare lane 1 with 2–3).
Figure 1.
Pep8 modulates the Rbm38-eIF4E complex. A Visual representation of the peptides used in this study. B Immunoblot for the competitive pull-down assay for GST-Rbm38 and His-eIF4E (2 μg) in the presence of peptide modulators (375 nM). Standard deviation of relative eIF4E band intensities was 0.1 (Pep23) and 0.08 (Pep8). C Visual representation of the docking of Pep8 to eIF4E based on REMDS. D Visual depiction of the key residues involved in the Pep8-eIF4E interaction. E Immunoblot for the pull-down of eIF4E with TentaGel-bound peptides. F Immunoblot for the competitive pull-down assay for GST-Rbm38 and eIF4E (5 μM) with the addition of Pep8 derivatives (5 μM). Standard deviation of relative eIF4E band intensities was 0.07 (Pep8), 0.003 (Pep8S-D) and 0.07 (Pep8S-K). All western blot figures are representative data of at least 2 independent replicates.
Next, replica exchange molecular dynamics simulations (REMDS) were performed and identified a potential binding pocket for Pep8 in eIF4E (Fig. 1C). The model predicted that Tyr-3/Ser-6/Ala-8 in Pep8 form hydrogen bonds with Thr-203/Asp-202/Thr-205 in eIF4E, respectively (Fig. 1D). Since Ser-6 in Pep8 makes a key interaction with Asp-202 in eIF4E, we theorized that Pep8S-D (Ser-6 to Asp-6) would abolish the affinity of Pep8 to eIF4E. To test this, control (11-aa nonspecific peptide), Pep8 and Pep8S-D were conjugated to TentaGel beads and examined for their abilities to pull down purified eIF4E. We showed that only Pep8 was able to pull down eIF4E, suggesting that Ser-6 is necessary for the binding to eIF4E (Fig. 1E). With this insight, we asked whether Pep8S-K (Ser-6 to Lys-6) would enhance the affinity for eIF4E. To that end, competitive pull-down assays were performed and showed that Pep8S-D had no effect on the Rbm38-eIF4E complex, whereas Pep8S-K was even more potent than Pep8 to inhibit the complex formation (Fig. 1F). To establish that Pep8 or Pep8S-K is able to diminish the interaction between Rbm38 and endogenous eIF4E, RKO cells were transfected with HA-tagged Rbm38 and pull-down assays were undertaken in the presence of peptide modulators. We found that both Pep8 and Pep8S-K blocked the interaction between HA-tagged Rbm38 and endogenous eIF4E, whereas both control and Pep8S-D had no effect (Supplementary Fig. 1). Collectively, our data suggest that Pep8 is docked at a pocket in eIF4E where Ser-6 in Pep8 makes a key hydrogen bond with Asp-202 on eIF4E, thereby modulating the Rbm38-eIF4E complex.
Synthetic peptides modulate p53 protein expression
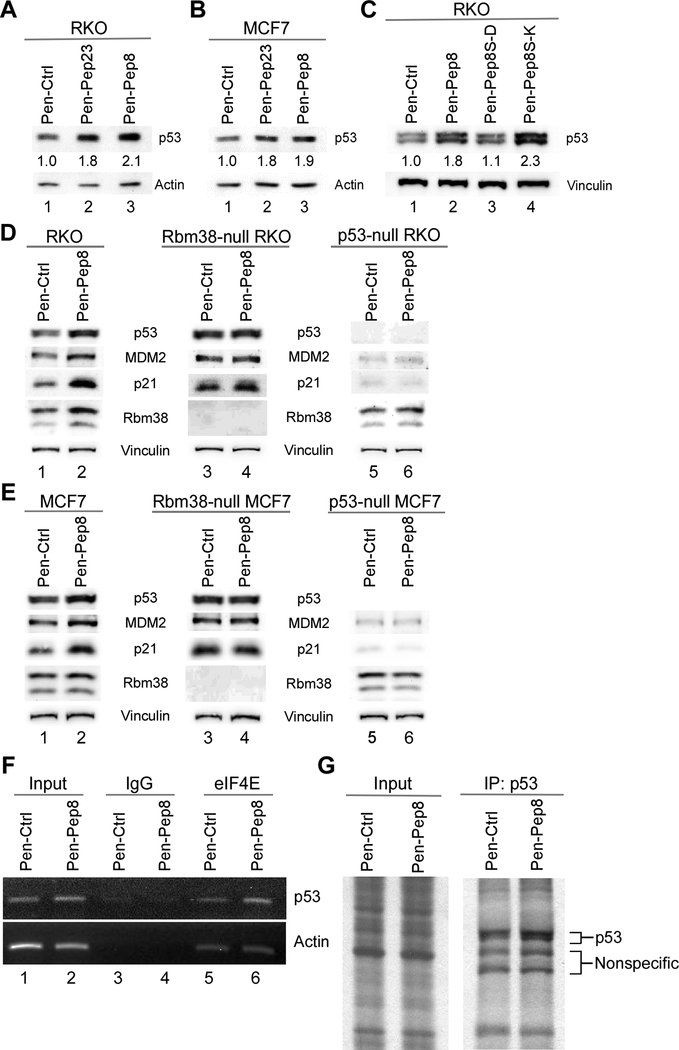
In order for a peptide to interfere with the intracellular Rbm38-eIF4E complex in vivo, the peptide would have to enter the cell. To that end, we applied penetratin (Pen) cell-penetrating peptide (CPP) (36) fused to the amine-termini of the cargo peptides to allow for efficient delivery into cancer cells (Pen-Ctrl, Pen-Pep23 and Pen-Pep8). We found that in both RKO and MCF7 cells, p53 expression was highly induced by Pen-Pep23 and Pen-Pep8 as compared to Pen-Ctrl (Figs. 2A-B compare lane 1 with 2–3). We further demonstrated that p53 was induced in RKO and MCF7 cells by Pep8 in a dose-dependent manner, with 2.5 μM and 5 μM being the most effective (Supplementary Fig. 2A). To exclude the possibility that Pep8 stabilizes p53, the half-life of p53 was examined in cells treated with Pen-Ctrl or Pen-Pep8 followed by cycloheximide. We found that p53 protein half-life was not increased by Pep8 (Supplementary Fig. 2B). Since p53 is highly induced by DNA damage, we determined the level of γ-H2AX, a marker of DNA damage, in cells treated with Pen-Pep8 or doxorubicin as a control. We found that both γ-H2AX and cleavage of PARP, a marker for induction of apoptosis, were highly induced by doxorubicin (Supplementary Fig. 2C). In contrast, Pep8 did not elicit DNA damage, nor did we see evidence of induction of apoptosis (Supplementary Fig. 2C). Since Pep8S-K, but not Pep8S-D, was able to reduce the Rbm38-eIF4E complex, we tested their ability to modulate p53 expression in vivo. We found that Pep8S-K was more potent than Pep8 to increase p53 expression (2.3 vs 1.8 folds of induction) (Fig. 2C), consistent with their ability to moderate the Rbm38-eIF4E complex (Fig. 1F).
Figure 2.
Synthetic peptides modulate p53 protein expression. A Immunoblot for RKO cells treated with 2.5 μM penetratin fusion peptides for 18 h. Standard deviation of relative p53 band intensities was 0.3 (Pep23) and 0.003 (Pep8). B The experiment with MCF7 cells was performed as in A. Standard deviation of relative p53 band intensities was 0.04 (Pep23) and 0.007 (Pep8). C Immunoblot for RKO cells treated with 2.5 μM penetratin-fused peptides (Ctrl, Pep8, Pep8S-D or Pep8S-K) for 18 h. Standard deviation of relative p53 band intensities was 0.15 (Pep8), 0.07 (Pep8S-D) and 0.14 (Pep8S-K). D-E Immunoblots for wild-type, Rbm38- and p53-null RKO and MCF7 cells treated with 5 μM penetratin fusion peptides for 18 h. F RNA-ChIP assay for eIF4E binding to p53 mRNA in RKO cells treated with 1.5 μM Pen-Ctrl or Pen-Pep8 for 2 h. Representative figure from three independent replicates. G 35S-labeling for de novo p53 protein synthesis after treatment with 5 μM Pen-Ctrl or Pen-Pep8 for 18 h. All western blot figures are representative data of at least 2 independent replicates.
Next, we sought to determine whether these peptides increase p53 expression in an Rbm38-dependent manner. To that end, Rbm38- and p53-null RKO and MCF7 cells were generated. As expected, Rbm38 was not detectable in Rbm38-null cells, whereas p53 was not detectable in p53-null cells (Supplementary Figs. 2D-E). Additionally, the level of p53 was increased in Rbm38-null RKO and MCF7 cells, consistent with our previous observations that knockout of Rbm38 leads to increased p53 expression (24). Demonstrating that Pep8 functions through an Rbm38-dependent manner, we showed that Pep8 was capable of increasing p53 expression in wild-type, but not in Rbm38-null RKO or MCF7 cells (Figs. 2D-E, compare lanes 1 and 2 with lanes 3 and 4 respectively, and Supplementary Figs. 2D-E). In addition, MDM2 and p21, both of which are known p53 targets, were upregulated upon treatment with Pen-Pep8 in wild-type, but not in Rbm38- and p53-null cells (Figs. 2D-E, compare lanes 1 and 2, 3 and 4, and 5 and 6, respectively). Further, we demonstrated that both MDM2 and p21 mRNA were increased following Pep8 treatment in wild-type, but not in Rbm38- and p53-null RKO cells (Supplementary Fig. 2F).
Previously, we found that Rbm38 prevents eIF4E from binding to p53 mRNA (24). To determine whether Pep8 modulation of the Rbm38-eIF4E complex formation in vivo leads to increased eIF4E binding to p53 mRNA, RNA-ChIP assays were performed. We found that Pen-Pep8 increased the association of eIF4E with p53 mRNA as compared to Pen-Ctrl (Fig. 2F, compare lanes 5 and 6). To further determine whether Pep8 moderation of the Rbm38-eIF4E complex enhances p53 translation, de novo synthesis of p53 protein was examined in 35S-labeled cells. As shown in figure 2G, the amount of newly synthesized p53 was increased by Pep8 as compared to control. Collectively, these data suggest that Pep8 is capable of increasing p53 expression by modulating p53 mRNA translation.
Pep8 inhibits tumor cell growth in Rbm38- and p53-dependent manners
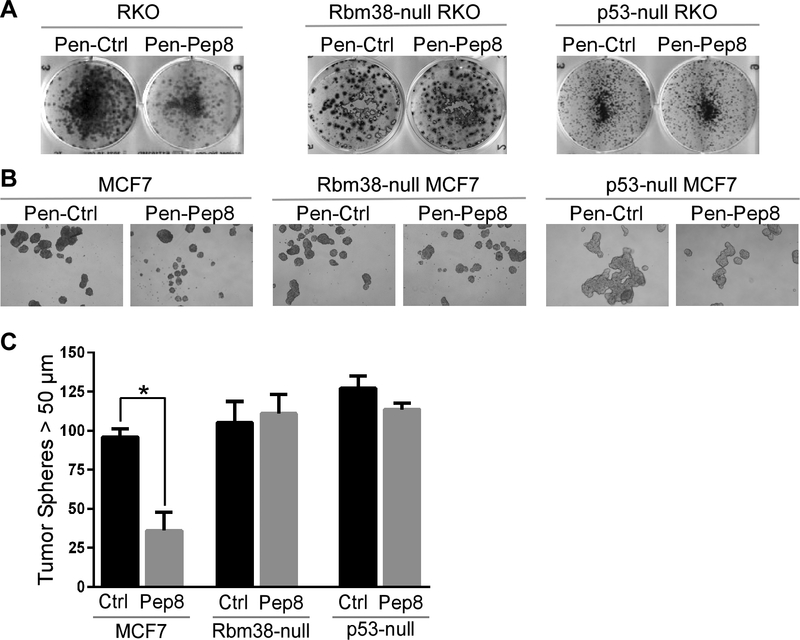
To determine if increased expression of p53 by Pep8 has a physiological response, colony formation assays were performed. We showed that Pep8 caused a substantial decrease in colony formation in wild-type, but not in Rbm38-null RKO and MCF7 cells, suggesting that the effect of Pep8 is dependent on Rbm38 (Fig. 3A and Supplementary Fig. 3, left and middle panels). We also found that Pep8 had no effect on colony formation in p53-null RKO and MCF7 cells, supporting the idea that the effect of Pep8 on colony formation is p53-dependent (Fig. 3A and Supplementary Fig. 2, right panel).
Figure 3.
Pep8 inhibits tumor cell growth in Rbm38- and p53-dependent manners. A Pen-Pep8 (5 μM) treatment decreases colony formation in wild-type, but not Rbm38- or p53-null RKO cells. B Pen-Pep8 (5 μM) treatment reduces MCF7 tumor sphere formation in wild-type, but not in Rbm38- or p53-null MCF7 cells. Tumor spheres (> 50 μm) counted 1 week after peptide treatment. C Quantification of tumor spheres from B. Values represent the mean ± SEM of three independent experiments (* p=0.0013).
Recently, tumor sphere assays were found to accurately represent the potential of a therapeutic compound in vivo (37). Thus, we measured the effect of Pep8 on tumor sphere formation and found that Pen-Pep8 significantly reduced the ability for MCF7 cells to form tumor spheres as compared to Pen-Ctrl (Figs. 3B-C). Additionally, Pep8 had little if any effect on tumor sphere formation in Rbm38- and p53-null MCF7 cells (Figs. 3B-C). Together, these data suggest that Pep-8 suppresses cell growth in Rbm38- and p53-dependent manners.
Pep8 sensitizes tumor cells to doxorubicin
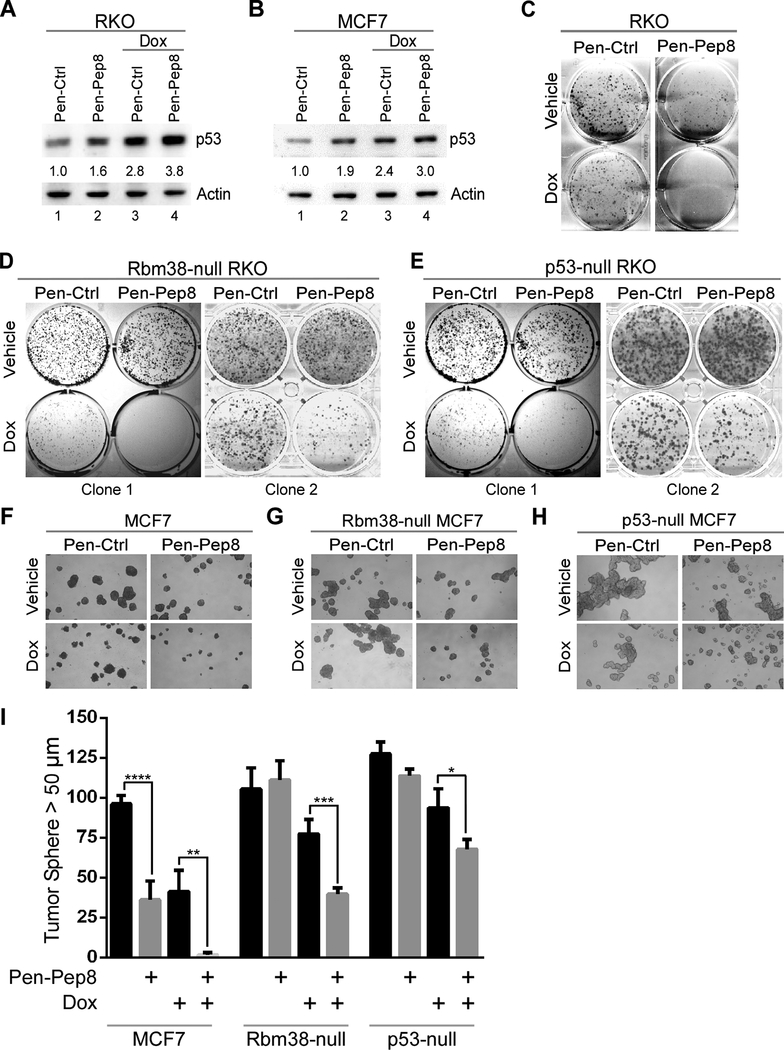
It is widely accepted that DNA damage, including treatment with doxorubicin, induces p53 protein accumulation through multiple post-translational modifications (38). In addition, our group has demonstrated that Rbm38 is capable of decreasing p53 expression even after doxorubicin treatment (24). Thus, we sought to address if Pep8 can cooperate with doxorubicin to increase p53 expression. We used 6.25 ng/mL of doxorubicin, a relatively low dose, as high doses lead to massive p53 induction which would obliterate any changes in p53 by Pep8. To that end, RKO and MCF7 cells were first treated with Pen-Ctrl or Pen-Pep8 for 20 minutes followed by a low dose of doxorubicin (6.25 ng/mL) for 18 hours. Pen-Pep8 increased p53 expression in both RKO and MCF7 cells (Figs. 4A-B; compare lanes 1 and 2), where an additional increase in p53 was observed upon concurrent treatment of Pep8 and doxorubicin in both cell lines (Figs. 4A-B; compare lanes 3 and 4). Furthermore, p53 induction was enhanced by concomitant treatment of either Pen-Pep23 or Pen-Pep8 with etoposide, another topoisomerase II inhibitor (Supplementary Fig. 4).
Figure 4.
Pep8 sensitizes tumor cells to doxorubicin. A Immunoblot for RKO cells treated with peptide alone (Pen-Ctrl or Pen-Pep8, 2.5 μM), or in combination with 6.25 ng/mL doxorubicin for 18 h. Standard deviation of relative p53 band intensities was 0.05 (Pep8), 0.7 (Ctrl + Dox) and 0.8 (Pep8 + Dox). B The experiment with MCF7 cells was performed as in A. Standard deviation of relative p53 band intensities was 0.09 (Pep8), 0.2 (Ctrl + Dox) and 0.5 (Pep8 + Dox). C Colony formation assay for RKO cells treated with 5 μM Pen-Ctrl or Pen-Pep8 with and without 3.125 ng/mL doxorubicin. D-E Colony formation assay for Rbm38- and p53-null RKO cells treated with Pen-Ctrl or Pen-Pep8 with and without 6.25 ng/mL doxorubicin. F-H Tumor sphere assays for wild-type, Rbm38- and p53-null MCF7 cells treated with Pen-Ctrl or Pen-Pep8 (5 μM) with and without 6.25 ng/mL doxorubicin. Tumor spheres (> 50 μm) counted after 7 days of treatment with peptides alone or in combination with doxorubicin. I Quantification of tumor spheres from F-H. Values represent the mean ± SEM of three independent experiments (* p=0.028, ** p=0.0068, *** p=0.0028, **** p=0.0013). All western blot figures are representative data of at least 2 independent replicates.
To determine if Pep8 is capable of sensitizing cancer cells to low dose doxorubicin, colony formation assays were performed. Wild-type RKO cells showed increased sensitivity to doxorubicin upon Pep8 treatment (Fig. 4C). Captivatingly, while Pep8 treatment alone had no effect on Rbm38- or p53-null RKO cells, Pep8 treatment sensitized both Rbm38- and p53-null RKO cells to doxorubicin treatment (Figs. 4D-E). These data suggest that other targets of Pep8 may cooperate with DNA damage to inhibit cell growth independent of Rbm38 and/or p53. Next, tumor sphere assays were performed with MCF7 cells and showed that Pen-Pep8, but not Pen-Ctrl, was able to cooperate with doxorubicin to suppress tumor sphere formation (Fig. 4F). Similar to the observations by colony formation assays, Rbm38- and p53-null MCF7 cells were not sensitive to Pep8 alone, but sensitive to concurrent treatment with Pep8 and doxorubicin (Figs. 4G-I). Collectively, these data suggest that Pep8 together with a low dose of a chemotherapeutic agent has an additive effect on p53 expression and suppression of colony/tumor sphere formation primarily in a p53-dependent manner.
Pep8 increases cellular senescence in Rbm38- and p53-dependent manners
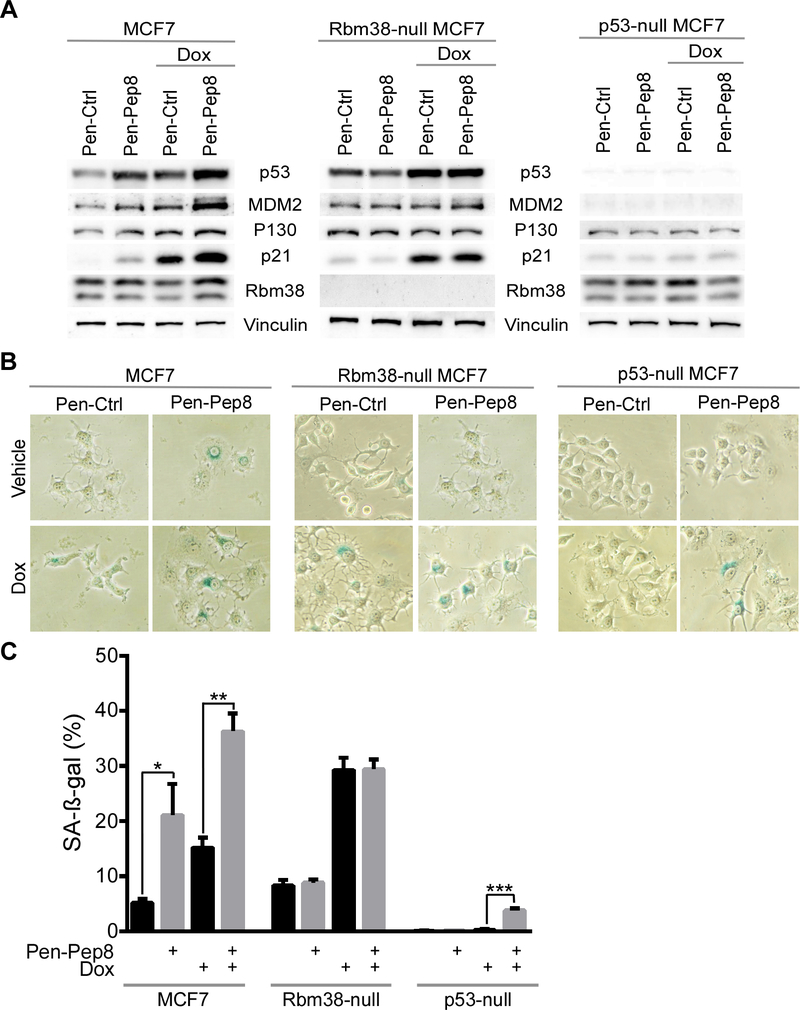
Since Pep8 was shown to increase p21 (Fig. 2D-E), a marker for cellular senescence, we hypothesized that Pep8 may cooperate with doxorubicin to modulate cellular senescence. First, we looked at induction of p21 in cells treated with Pep8 and doxorubicin. We found that in both MCF7 and RKO cells, p21 induction was further enhanced in Rbm38- and p53-dependent manners (Fig. 5A and Supplementary Fig. 5). Similarly, induction of p130, a senescence marker, was enhanced in cells treated with Pep8 and doxorubicin in Rbm38- and p53-dependent manners (Fig. 5A and Supplementary Fig. 5). Next, SA-β-gal staining was performed and showed that cellular senescence was significantly increased by Pen-Pep8 alone or in combination with doxorubicin in wild-type, but not in Rbm38-null MCF7 cells (Figs. 5B-C). Intriguingly, Pen-Pep8 together with doxorubicin led to a small, but significant increase of cellular senescence in p53-null MCF7 cells (Figs. 5B-C). Together, our data suggest that Pep8 alone or in combination with doxorubicin is capable of inducing cellular senescence in cancer cells primarily through induction of p53.
Figure 5.
Pep8 increases cellular senescence in Rbm38- and p53-dependent manners. A Immunoblots for wild-type, Rbm38- and p53-null MCF7 cells treated with 5 μM Pen-Ctrl or Pen-Pep8 with and without 6.25 ng/mL doxorubicin for 18 h. B SA-ß-gal staining of wild-type, Rbm38- and p53-null MCF7 cells after treatment with 5 μM Pen-Ctrl or Pen-Pep8 with and without 6.25 ng/mL doxorubicin for 18 h. C Quantification of SA-ß-gal positive cells as shown in B. Values represent the mean ± SEM of three independent experiments (* p= 0.009, ** p= 0.0006, *** p= 0.0001). All western blot figures are representative data of at least 2 independent replicates.
Pep8 suppresses solid tumor growth in a xenograft model
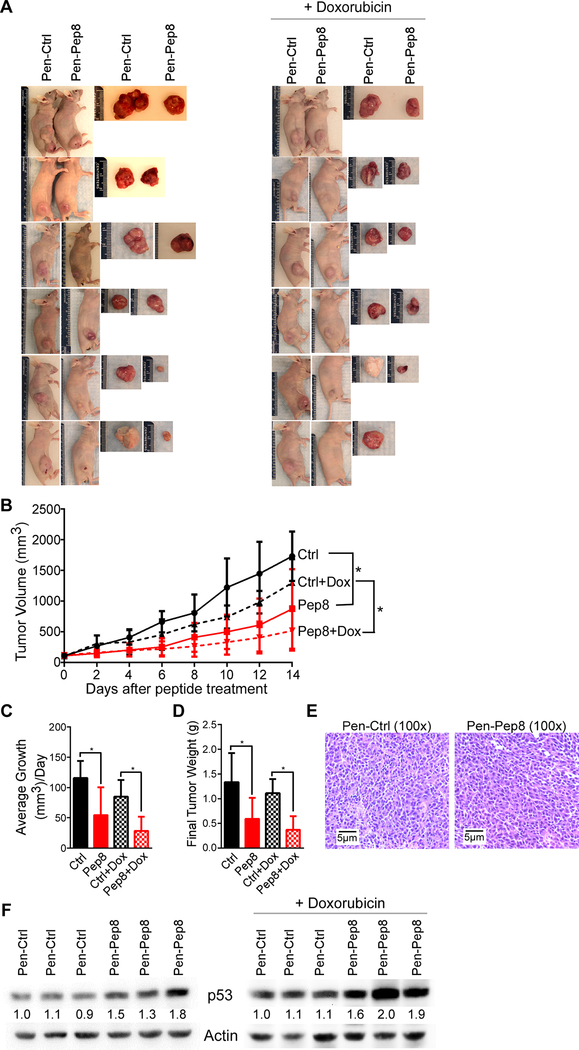
To test the therapeutic potential of Pep8 in vivo, RKO xenograft model was used. Briefly, 6-week-old female nude mice were injected in the right flank with RKO cells. After xenograft tumors reached 100 mm3 in size, Pen-Ctrl or Pen-Pep8 was intratumorally injected every other day for two weeks. We showed that xenograft tumor growth was substantially reduced by Pen-Pep8 alone, or in combination with doxorubicin as compared to Pen-Ctrl treated groups (Fig. 6A). Xenograft volume over time, average tumor volume, and final tumor weight were significantly reduced by Pen-Pep8 or together with doxorubicin compared to that by Pen-Ctrl (Figs. 6B-D).
Figure 6.
Pep8 suppresses solid tumor growth in a xenograft model. A Photographs of RKO xenografts after being excised from animal. RKO cells were injected into the right flank of 6-week-old female nude mice. After tumors reached a size > 100 mm intratumoral injections of Pen-Ctrl or Pen-Pep8 (5 μM) were administered every other day for 14 days. Mice treated with doxorubicin received two doses (2 mg/kg) on the first day of peptide treatment and 7 days later. B Xenograft tumor volume was measured every other day by using the following formula; ½ (Length x Width2). Values represent the mean ± SEM, n=6 per group. * p < 0.05. C Average tumor growth per day for peptide treated tumors with and without doxorubicin. Values represent the mean ± SEM, n=6 per group. * p < 0.05. D Xenograft tumor weight after being excised following treatment. Values represent the mean ± SEM, n=6 per group. * p < 0.05. E Representative H&E staining of xenograft tissue. F Immunoblot of Pen-Ctrl and Pen-Pep8 treated xenograft tumors with and without doxorubicin.
Histological examination showed that all xenograft tumors were indeed human adenocarcinomas (Fig. 6E). Next, we wanted to address if Pen-Pep8 alone or in combination with doxorubicin leads to increased p53 expression in the xenograft tumors. We showed that p53 expression was increased in tumors treated with Pen-Pep8 alone or together with doxorubicin as compared to that in Pen-Ctrl treated tumors (Fig. 6F). Moreover, IHC staining of xenograft tissues for p53 demonstrated that Pen-Pep8 alone, or with doxorubicin was capable of increasing p53 expression in tumor cells (Supplementary Fig. 6). These data are consistent with the results observed in cell lines (Figs. 4A-B), suggesting that Pen-Pep8 has the ability to decrease solid tumor growth by induction of p53 and sensitize tumors to doxorubicin treatment.
Discussion
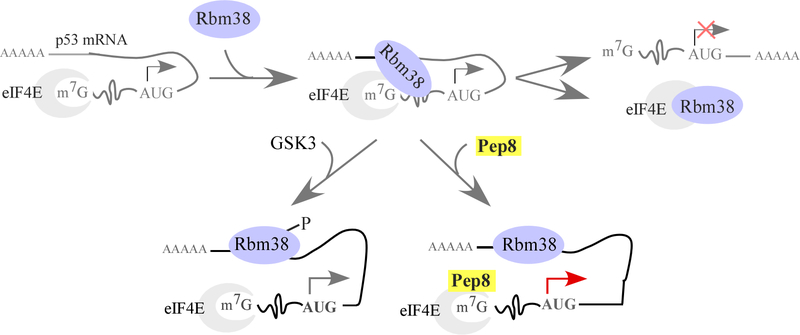
Restoration of wild-type p53 has been explored and found to be an effective strategy to suppress tumor growth in animal models (39–41). Our group previously identified that Rbm38, an RNA-binding protein and a p53 target, can inhibit p53 translation via interaction with eIF4E (24). Interestingly, phosphorylation of Rbm38 at Ser-195 abrogates its ability to interact with eIF4E, converting Rbm38 from a repressor to an activator of p53 translation (Fig. 7) (26). With these data in hand, we aimed to test if peptide modulators of the Rbm38-eIF4E complex could increase p53 translation (Fig. 7) and thus, would have a therapeutic application in treating tumors that harbor wild-type p53. We identified two peptides corresponding to the binding interface between Rbm38/Rbm24 and eIF4E. These peptides were capable of moderating the Rbm38-eIF4E complex, thereby abrogating the translational inhibition of p53 by Rbm38. Significantly, Pep8, an 8 amino acid peptide moderator, is a highly potent inducer of p53 translation, leading to decreased colony/tumor sphere formation and reduced xenograft tumor growth in nude mice. In addition, Pep8 substantially sensitizes cancer cells to doxorubicin. Collectively, these findings support the idea that rationally designed peptide moderators of the Rbm38-eIF4E complex may be explored as a therapeutic agent.
Figure 7.
Working model for how Rbm38 inhibits p53 translation and the mode of action for Pep8 treatment.
Protein-protein interactions (PPIs) are necessary for many cellular functions and can modulate complex biological activities, such as normal cellular homeostasis and pathogenesis of diseases, including cancer (39). The use of peptides to block PPIs comes with certain advantages, such as high potency, target specificity and increased safety (42). Here we tested and showed that rationally designed peptides, especially Pep8, can diminish the Rbm38-eIF4E complex, thereby restoring p53 expression. REMDS simulations predict that Pep8 is docked in a pocket at the carboxyl-terminus of eIF4E, with Ser-6 in Pep8 forming a key hydrogen bond with Asp-202 in eIF4E (Figs. 1C-D). To test this model, we created single point mutants of Pep8 that would either enhance or decrease the ability of Pep8 to form a hydrogen bond with Asp-202 in eIF4E. Indeed, Pep8S-D was found to be inert, likely due to the inability for Asp-6 in Pep8S-D to interact with Asp-202 in eIF4E (Figs. 1E-F, 2C, Supplementary Fig. 1). In contrast, Pep8S-K, in which Lys-6 in Pep8S-K can potentially form a hydrogen bond with Asp-202 in eIF4E, was found to be the most potent inhibitor of the Rbm38-eIF4E complex and inducer of p53 expression (Figs. 1F, 2C, Supplementary Fig. 1). These data suggest that REMDS docking of Pep8 to the pocket in eIF4E is likely correct. Nevertheless, structural analysis is still needed to confirm our modeling in future studies.
Doxorubicin is one of the most effective cancer therapeutic agents and widely used as a first-line agent for a multitude of cancers (43). However, doxorubicin is known to have systemic toxicities, including severe inflammation of multiple organs and especially cardiac damage, resulting in patients unable to finish a chemotherapeutic regimen (44). In this study, we showed that Pep8 cooperates with doxorubicin to increase p53 expression and inhibit colony/tumor sphere formation as well as xenograft tumor growth, indicating that Pep8 significantly sensitizes cancer cells to doxorubicin. These data support that concurrent treatment of Pep8 with a low dose of doxorubicin, and possibly other chemotherapeutic agents, may decrease the systemic toxicities associated with the current dosing of these drugs, which warrants further investigation(s).
Our results showed that Pep8 induces p53 expression, leading to increased cellular senescence and growth suppression in Rbm38- and p53-dependent manners (Figs. 3A-C, 5A-C, Supplementary Fig. 3). However, Pep8 is still capable of inducing growth suppression in Rbm38-null and p53-null RKO and MCF7 cells when combined with doxorubicin (Figs. 4D-E, 4G-I and 5A-C). These data suggest that Pep8 may modulate other targets to sensitize cancer cells to doxorubicin independent of Rbm38/p53. Since Rbm24 can also regulate p53 through the dissociation of eIF4E from p53 mRNA, Pep8 is likely to modulate the Rbm24-eIF4E complex (27). Additionally, Pep8 may compete with Rbm38 for GSK3, a kinase known to phosphorylate Rbm38 at Ser-195 (26). Moreover, since Rbm38 interacts with Ago2 to regulate the stability of multiple transcripts, such as p21, Pep8 may influence growth suppression via other Rbm38/Rbm24 targets (45). However, further investigations are needed.
A major hurdle to design an effective peptide for an intracellular target is how to deliver the peptide into the cell. Moreover, peptide delivery to tumors faces additional challenges. For example, peptides are known to have short circulating plasma half-life, induce systemic cytotoxicities, accumulate in major organs, and lack tissue specificity (42,46,47). In this study, intratumoral injection was used to deliver Pen-Pep8 to xenograft tumors to circumvent these concerns. However, intratumoral injection has its own limitations, such as a diminished accessibility of targeted tumor tissues and decreased dissemination of the peptide throughout the tumor (46,48). Thus, future studies are needed to devise an approach to deliver Pep8 systemically and specifically to tumor cells, such as conjugated antibody delivery and nanoparticle technologies.
Collectively, our findings here demonstrate that Pep8 can modulate the Rbm38-eIF4E complex, thereby increasing p53 expression. While these findings are exciting, other questions are still yet to be addressed. For example, does Pep8 stabilize mutant p53, and is Pep8-induced cell sensitivity to doxorubicin a result of increased p53 expression or another yet to be determined mechanism? Further, does reduced doses of doxorubicin together with Pep8 relieve systemic toxicities but still have therapeutic benefits?
Supplementary Material
Significance.
Disruption of the Rbm38-eIF4E complex via synthetic peptides induces wild-type p53 expression, suppresses tumor growth and progression, and may serve as a novel therapeutic strategy in cancer.
Acknowledgements
CL, JZ, BM, and MC performed the experiments. CL, JZ, BM, and XC designed the experiments and analyzed the data. CL and XC wrote the manuscript. CL, JZ, BM, MC, and XC proofread and edited the manuscript. C. Lucchesi received T32 CA108459. X. Chen received NIH RO1CA076069 and NIH RO1CA195828. B. Ma was funded with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E.
Financial support: RO1 CA076069, RO1 CA195828, HHSN261200800001E, T32 CA108459
Footnotes
The authors declare no conflict of interest.
References
- 1.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease Trends Genet [Internet]. Terry Fox Molecular Oncology Group, and the Bloomfield Center for Research on Aging, Lady Davis Institute for Medical Research, Sir Mortimer B. Davis Jewish General Hospital, Department of Medicine, McGill University, Montreal, Quebec H3T 1E2, Canada; 2008;24:416–25. [DOI] [PubMed] [Google Scholar]
- 2.Kechavarzi B, Janga SC. Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol [Internet]. 2014;15:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim MY, Hur J, Jeong S. Emerging roles of RNA and RNA-binding protein network in cancer cells BMB Rep [Internet]. National Research Lab for RNA Cell Biology, BK21 Graduate Program for RNA Biology and Department of Molecular Biology, Dankook University, Gyeonggi-do 448–701, Republic of Korea; 2009;42:125–30. [DOI] [PubMed] [Google Scholar]
- 4.Wurth L Versatility of RNA-binding proteins in cancer. Comp. Funct. Genomics 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature [Internet]. 2000;408:307–10. [DOI] [PubMed] [Google Scholar]
- 6.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009. [DOI] [PubMed] [Google Scholar]
- 7.Tovy A, Spiro A, McCarthy R, Shipony Z, Aylon Y, Allton K, et al. p53 is essential for DNA methylation homeostasis in naive embryonic stem cells, and its loss promotes clonal heterogeneity Genes Dev [Internet]. 2017/June/14 Department of Molecular Cell Biology, The Weizmann Institute of Science, Rehovot 76100, Israel: Department of Computer Science and Applied Mathematics, The Weizmann Institute of Science, Rehovot 76100, Israel. MD: Anderson Department of Epigenetics and Mol; 2017;31:959–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way Cancer Cell [Internet]. 2009/November/03 Department of Radiation Oncology, School of Medicine, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599–7512, USA: ypzhang@med.unc.edu; 2009;16:369–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript Genes Dev [Internet]. Department of Cell Biology, The University of Alabama at Birmingham, Birmingham, AL 35294, USA; 2006;20:2961–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginestier C, Cervera N, Finetti P, Esteyries S, Esterni B, Adélaïde J, et al. Prognosis and gene expression profiling of 20q13-amplified breast cancers. Clin Cancer Res. 2006; [DOI] [PubMed] [Google Scholar]
- 12.Zheng SL, Xu J, Isaacs SD, Wiley K, Chang BI, Bleecker ER, et al. Evidence for a prostate cancer linkage to chromosome 20 in 159 hereditary prostate cancer families. Hum Genet. 2001; [DOI] [PubMed] [Google Scholar]
- 13.Tanner MM, Grenman S, Koul A, Johannsson O, Meltzer P, Pejovic T, et al. Frequent amplification of chromosomal region 20q12-q13 in ovarian cancer. Clin Cancer Res. 2000; [PubMed] [Google Scholar]
- 14.Korn WM, Yasutake T, Kuo WL, Warren RS, Collins C, Tomita M, et al. Chromosome arm 20q gains and other genomic alterations in colorectal cancer metastatic to liver, as analyzed by comparative genomic hybridization and fluorescence in situ hybridization. Genes Chromosomes Cancer. 1999; [DOI] [PubMed] [Google Scholar]
- 15.Yin T, Cho SJ, Chen X. RNPC1, an RNA-binding protein and a p53 target, regulates macrophage inhibitory cytokine-1 (MIC-1) expression through mRNA stability J Biol Chem. 2013/July/10 Comparative Oncology Laboratory, University of California, Davis, California 95616, USA; 2013;288:23680–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan W, Zhang J, Zhang Y, Jung YS, Chen X. p73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability Mol Cell Biol. 2012/April/18 Comparative Oncology Laboratory, University of California at Davis, Davis, California, USA; 2012;32:2336–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho SJ, Jung YS, Zhang J, Chen X. The RNA-binding protein RNPC1 stabilizes the mRNA encoding the RNA-binding protein HuR and cooperates with HuR to suppress cell proliferation J Biol Chem. 2012/March/01 Center for Comparative Oncology, University of California, Davis, California 95616, USA; 2012;287:14535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu E, Zhang J, Chen X. MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability Oncogene [Internet]. Comparative Oncology Laboratory, Department of Surgical and Radiological Sciences, University of California at Davis, Davis, CA 95616, USA; 2013;32:2169–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Xu E, Chen X. Regulation of Mdm2 mRNA stability by RNA-binding protein RNPC1 Oncotarget. 2013/August/06 Comparative Oncology Laboratory, University of California at Davis, Davis, CA; 2013;4:1121–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006; [DOI] [PubMed] [Google Scholar]
- 21.Jenssen T-K, Kuo WP, Stokke T, Hovig E. Associations between gene expressions in breast cancer and patient survival. Hum Genet. 2002; [DOI] [PubMed] [Google Scholar]
- 22.Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, Van De Wiel MA, et al. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009; [DOI] [PubMed] [Google Scholar]
- 23.Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002; [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Cho SJ, Shu L, Yan W, Guerrero T, Kent M, et al. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas Genes Dev [Internet]. Comparative Cancer Center, Schools of Medicine and Veterinary Medicine, University of California at Davis, USA; 2011;25:1528–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Xu E, Ren C, Yan W, Zhang M, Chen M, et al. Mice deficient in Rbm38, a target of the p53 family, are susceptible to accelerated aging and spontaneous tumors Proc Natl Acad Sci U S A [Internet]. Comparative Oncology Laboratory, and Department of Surgical & Radiological Sciences, School of Veterinary Medicine, University of California, Davis, CA 95616. Department of Pathology & Laboratory Medicine, School of Medicine. Center for Comparative Medici; 2014;111:18637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M, Zhang J, Chen X, Cho SJ, Chen X. Glycogen synthase kinase 3 promotes p53 mRNA translation via phosphorylation of RNPC1 Genes Dev [Internet]. Comparative Oncology Laboratory, University of California at Davis, Davis, California 95616, USA; 2013;27:2246–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Min Enshun Xu, Shakur Mohibi, Michelle de Anda Danielle, Yuqian Jiang, Zhang Jin, Xinbin Chen YZ. Rbm24, a target of p53, is necessary for proper expression of p53 and heart development. Cell Death Differ. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rau M, Ohlmann T, Morley SJ, Pain VM. A reevaluation of the Cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J Biol Chem. 1996; [DOI] [PubMed] [Google Scholar]
- 29.Robichaud N, Sonenberg N. Translational control and the cancer cell response to stress Curr Opin Cell Biol [Internet]. 2017/June/06 Goodman Cancer Research Centre and Department of Biochemistry, McGill University, Montreal, QC, Canada. Goodman Cancer Research Centre and Department of Biochemistry, McGill University, Montreal, QC, Canada: Electronic address: nahum.sonenberg@mcgill.ca.; 2017;45:102–9. [DOI] [PubMed] [Google Scholar]
- 30.Hather G, Liu R, Bandi S, Mettetal J, Manfredi M, Shyu WC, et al. Growth rate analysis and efficient experimental design for tumor xenograft studies Cancer Inf [Internet]. 2015/January/13 Department of Global Statistics, Takeda Pharmaceuticals International Co., Cambridge, MA, USA. Department of Research and Development Systems, Takeda Pharmaceuticals International Co., Cambridge, MA, USA: Department of DMPK, Takeda Pharmaceuticals Interna; 2014;13:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M, Xu E, Zhang J, Chen X. PPM1D phosphatase, a target of p53 and RBM38 RNA-binding protein, inhibits p53 mRNA translation via dephosphorylation of RBM38 Oncogene [Internet]. Comparative Oncology Laboratory, Department of Surgical & Radiological Sciences, School of Veterinary Medicine, University of California, Davis, Davis, CA, USA; 2015;34:5900–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis Nat Methods [Internet]. 2012/June/30 Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany; 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, et al. Immunoprecipitation of mRNA-protein complexes Nat Protoc [Internet]. 2007/April/05 Department of Pharmacology, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania 19104, USA; 2006;1:577–80. [DOI] [PubMed] [Google Scholar]
- 34.Bonifacino JS. Biosynthetic labeling of proteins. Curr Protoc Immunol. 2001; [DOI] [PubMed] [Google Scholar]
- 35.Yang HJ, Zhang J, Yan W, Cho SJ, Lucchesi C, Chen M, et al. Ninjurin 1 has two opposing functions in tumorigenesis in a p53-dependent manner Proc Natl Acad Sci U S A [Internet]. 2017/October/27 Department of Surgical and Radiological Sciences, Schools of Veterinary Medicine and Medicine, University of California, Davis, CA 95616: Department of Surgical and Radiological Sciences, Schools of Veterinary Medicine and Medicine, University of Californ; 2017;114:11500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dom G, Shaw-Jackson C, Matis C, Bouffioux O, Picard JJ, Prochiantz A, et al. Cellular uptake of Antennapedia Penetratin peptides is a two-step process in which phase transfer precedes a tryptophan-dependent translocation Nucleic Acids Res [Internet]. 2003/January/16 Unit of Developmental Genetics, Universite Catholique de Louvain, 73 (boite 82) avenue Mounier, 1200 Brussels, Belgium; 2003;31:556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S, Alexander CM. Tumorsphere assay provides more accurate prediction of in vivo responses to chemotherapeutics Biotechnol Lett [Internet]. 2013/October/26 McArdle Laboratory for Cancer Research, University of Wisconsin-Madison, Madison, WI, 53706, USA, skim@oncology.wisc.edu.; 2014;36:481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage Oncogene [Internet]. Wellcome Trust/Cancer Research Campaign, Institute of Cancer and Developmental Biology, Cambridge University, Tennis Court Road, Cambridge CB2 1QR, UK; 1999;18:7644–55. [DOI] [PubMed] [Google Scholar]
- 39.Pelay-Gimeno M, Glas A, Koch O, Grossmann TN. Structure-Based Design of Inhibitors of Protein-Protein Interactions: Mimicking Peptide Binding Epitopes. Angew. Chemie - Int. Ed. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo Nature [Internet]. 2007/January/26 Center for Cancer Research, Massachusetts Institute of Technology, Cambridge, Massachusetts 02142, USA; 2007;445:661–5. [DOI] [PubMed] [Google Scholar]
- 41.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors Cell [Internet]. 2006/December/22 Cancer Research Institute and Department of Cellular and Molecular Pharmacology, Comprehensive Cancer Center, University of California, San Francisco, San Francisco, CA 94143, USA; 2006;127:1323–34. [DOI] [PubMed] [Google Scholar]
- 42.Fosgerau K, Hoffmann T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015. [DOI] [PubMed] [Google Scholar]
- 43.Verma S, Clemons M. First-line treatment options for patients with HER-2 negative metastatic breast cancer: the impact of modern adjuvant chemotherapy Oncologist [Internet]. Division of Medical Oncology, Sunnybrook and Women’s College Health Sciences Centre, T-Wing, 2nd Floor, TSRCC, Toronto, ON, Canada: sunil.verma@sunnybrook.ca; 2007;12:785–97. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Chen Q, Qi H, Wang C, Wang C, Zhang J, et al. Doxorubicin-Induced Systemic Inflammation Is Driven by Upregulation of Toll-Like Receptor TLR4 and Endotoxin Leakage Cancer Res [Internet]. State Key Laboratory of Pharmaceutical Biotechnology, NJU Advanced Institute for Life Sciences (NAILS), School of life sciences, Nanjing University, Nanjing, China: State Key Laboratory of Quality Research in Chinese Medicine, Institute of Chinese Medical; 2016;76:6631–42. [DOI] [PubMed] [Google Scholar]
- 45.Leveille N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Oude Vrielink J, et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity Nat Commun [Internet]. 2011/October/27 Division of Gene Regulation, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands; 2011;2:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurrikoff K, Gestin M, Langel U. Recent in vivo advances in cell-penetrating peptide-assisted drug delivery Expert Opin Drug Deliv [Internet]. 2015/December/05 a Institute of Technology, University of Tartu, Tartu, Estonia. b Department of Neurochemistry, Stockholm Universitet, Stockholm, Sweden; 2016;13:373–87. [DOI] [PubMed] [Google Scholar]
- 47.Sarko D, Beijer B, Garcia Boy R, Nothelfer EM, Leotta K, Eisenhut M, et al. The pharmacokinetics of cell-penetrating peptides Mol Pharm [Internet]. 2010/September/18 Department of Nuclear Medicine, University Hospital Heidelberg, INF 400, 69120 Heidelberg, Germany; 2010;7:2224–31. [DOI] [PubMed] [Google Scholar]
- 48.Ernsting MJ, Murakami M, Roy A, Li SD. Factors controlling the pharmacokinetics, biodistribution and intratumoral penetration of nanoparticles J Control Release [Internet]. 2013/October/01 Drug Delivery and Formulation, Drug Discovery Program, Ontario Institute for Cancer Research, 101 College Street, Suite 800, Toronto, Ontario M5G 0A3, Canada; Ryerson University, Faculty of Architectural Science and Engineering, Toronto, Ontario M5B 1Z2,; 2013;172:782–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.