Abstract
Background
Recently there has been a growing interest in the potential for host transcriptomic analysis to augment the diagnosis of infectious diseases.
Methods
We compared nasal and blood samples for evaluation of the host transcriptomic response in children with acute respiratory syncytial virus (RSV) infection, symptomatic non-RSV respiratory virus infection, asymptomatic rhinovirus infection, and virus-negative asymptomatic controls. We used nested leave-one-pair-out cross-validation and supervised principal components analysis to define small sets of genes whose expression patterns accurately classified subjects. We validated gene classification scores using an external data set.
Results
Despite lower quality of nasal RNA, the number of genes detected by microarray in each sample type was equivalent. Nasal gene expression signal derived mainly from epithelial cells but also included a variable leukocyte contribution. The number of genes with increased expression in virus-infected children was comparable in nasal and blood samples, while nasal samples also had decreased expression of many genes associated with ciliary function and assembly. Nasal gene expression signatures were as good or better for discriminating between symptomatic, asymptomatic, and uninfected children.
Conclsusions
Our results support the use of nasal samples to augment pathogen-based tests to diagnose viral respiratory infection.
Keywords: diagnosis, human gene expression, microarray, respiratory viruses, viral infection
We compared the human transcriptomic response in nasal and blood samples from children with acute viral respiratory infection and controls. Nasal gene expression signatures were as good or better than blood signatures for discriminating between symptomatic, asymptomatic, and uninfected children.
In recent years, there has been great interest in the potential for host transcriptomic analysis to augment the diagnosis of infectious diseases [1, 2]. One reason is the potential for discriminating between broad classes of infection, such as those caused by viruses versus bacteria [3–6]. This capability would have enormous therapeutic utility in selecting appropriate therapy, especially in situations in which specimens for direct pathogen detection cannot be obtained or do not identify the etiologic agent of infection, or in situations in which it is necessary to distinguish between asymptomatic colonization by a potential pathogen versus significant infection. Most gene expression analyses to date have been directed at acute respiratory infections [7–18] or nonspecific febrile illnesses [4, 19, 20], but other infectious disease applications have included dengue [21], other tropical hemorrhagic fevers [22, 23], and tuberculosis [24–26].
Most previous studies have analyzed the human transcriptomic response in peripheral blood leukocytes (PBLs) [1–26]. However, there may be advantages in analyzing host response at the site of infection (for example, the nose or nasopharynx in acute viral respiratory infection), including the possibility of developing an integrated diagnostic test that simultaneously detects the nucleic acid of pathogens while also analyzing host gene expression [27–29].
To evaluate the potential of using nasal cells for the diagnostic evaluation of host response, we undertook a direct comparison of the host transcriptomic responses in the nose and blood of young children experiencing infection with common respiratory viruses. We compared gene expression in matched blood and nasal samples obtained from children with symptomatic infection to those of asymptomatic children, including some who were virus positive and some who were negative for respiratory viruses. The study was designed to address whether analysis of the nasal transcriptomic response was at least as informative as analysis of the blood response.
METHODS
Subjects and Samples
Subjects were children hospitalized for acute respiratory illness at St. Louis Children’s Hospital with positive results only for a single virus on a multiplex molecular test performed on a nasopharyngeal swab obtained as part of the child’s routine care (BioFire FilmArray Respiratory Panel, Salt Lake City, UT). Other inclusion criteria were age between 3 months and 18 years and presence of a parent or guardian capable of providing informed consent. Exclusion criteria included any underlying medical condition that required regular medical care, receipt of immunosuppressive medications including corticosteroids within the preceding 30 days, and receipt of antibiotics within 7 days. Control subjects were children in the same age range having ambulatory surgery for nonacute conditions. Exclusion criteria were the same as for symptomatic subjects plus fever within the preceding 48 hours. Participating families were called 7 days after enrollment to determine whether any illness had occurred in the control subject since enrollment. (Additional information is provided in Supplementary Methods.) This study was approved by the institutional review boards of Washington University and the University of Rochester, and written informed consent was provided by participants or a parent or guardian.
Nasal samples were collected by a trained coordinator. After obtaining informed consent, the nasopharynx was washed with normal saline, and a nasal swab was obtained using a mid-turbinate flocked swab (Copan Diagnostics, Inc., Murrieta, CA) and immediately placed in RNAprotect (QIAGEN, Valencia, CA). A blood sample was drawn by venipuncture into a Tempus tube (Applied Biosystems, Foster City, CA). Samples from control subjects were obtained at the time of anesthesia induction using the same procedures, except that before the nasal wash an additional nasal swab was obtained using a mid-turbinate flocked swab, placed in universal transport media, and tested for respiratory viruses using the GenMark eSensor respiratory virus panel (GenMark, Carlsbad, CA). Methods used to confirm the identification of rhinoviruses (RVs) and enteroviruses (EVs) are described in Supplementary Methods.
Microarray Sample and Data Processing
Human gene expression data from blood and nasal ribonucleic acid (RNA) samples was generated using Affymetrix Human Clariom-D chips (Affymetrix, Santa Clara, CA). Ribonucleic acid samples were processed using the Affymetrix GeneChip Whole Transcript Pico Reagent Kit recommended by the manufacturer for use with partially degraded RNAs. Microarray data generated in this study are accessible in the GEO database hosted by the National Center for Biotechnology Information of the National Institutes of Health (accession number GSE117827).
Cell-Type Deconvolution
We used the R program “DSA” (digital sorting algorithm) [30] to estimate the relative proportion of hematopoietic and epithelial cells present in nasal samples. For cell markers, we selected 157 genes (112 hematopoietic and 45 laryngeal) from the Tissue-Specific Gene Expression and Regulation (TiGER) database (http://bioinfo.wilmer.jhu.edu/tiger/). The DSA-estimated proportion of epithelial cells in each nasal sample was used to adjust the nasal data sets before selected subsequent analyses as indicated below.
Biological Process Enrichment and Pathway Analysis
Using the 28476 transcript clusters (TCs) with Entrez Gene identifications (IDs), we identified genes with differential mean microarray signal intensity in the 3 comparisons of virus-infected versus negative controls, with separate analyses for nasal and blood samples. To identify enriched biological processes, we used the web gene set enrichment tool “Enrichr” (http://amp.pharm.mssm.edu/Enrichr/) [31] to analyze the genes with significantly increased or decreased expression from the comparisons of the subjects with symptomatic and asymptomatic viral infection and controls. The same genes were further investigated using the Ingenuity Pathway Analysis tool ([IPA] http://analysis.ingenuity.com/pa/) for identification of enriched canonical pathways. Pathways with unadjusted Fisher Exact test –log P values greater than 1.3 (equivalent P < .05) were identified as significant. Both the Enrichr and IPA analyses were carried out using nasal gene signal data that were adjusted by the DSA procedure to account for the variable contribution of inflammatory cells in each sample.
Construction and Cross-Validation of Gene-Based Classification Scores
Classification scores were developed separately for nasal and blood samples. For each, TCs undetectable in all subjects were removed from further consideration, leaving 18523 TCs for nasal and 18435 TCs for blood samples. We sought to develop classification scores based on the smallest number of TCs (genes) capable of distinguishing among the different groups of subjects. Each of the classification scores was developed independently. Selection of component genes was carried out using a modified version of supervised principal components analysis (PCA) [32] described in Supplementary Methods.
External Validation of Gene-Based Classification Scores
We externally validated our gene-based classification scores for respiratory syncytial virus (RSV) versus control (Ctrl) and non-RSV versus Ctrl, for both nasal and blood genes, using the data reported by Do et al [33], that included nasal and blood samples from children with acute RSV and RV infection paired with early recovery samples from the same subjects. Methods used for the validation are described in Supplementary Methods.
Confirmation by Reverse Transcription-Quantitative Polymearse Chain Reaction
Gene expression levels defined by microarray were validated for select genes by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) using predesigned primers/probe reagents labeled with FAM reporter and MGB quencher (IDT, San Jose, CA) run on the Fluidigm Biomark HD system (Fluidigm Corporation, San Francisco, CA) according to the manufacturer’s protocol. Quantitative PCR assays were performed in triplicate. Relative expression levels were normalized to the reference gene ACTB and a calibrator RNA (pool of multiple RNAs of the healthy controls in the study) using the 2−ΔΔCt method [34].
RESULTS
Subjects and Samples
Forty-six subjects were enrolled in the study between March 2016 and January 2017, including 17 with acute respiratory infection (8 with RVs or EVs, 7 with RSV, 1 with adenovirus, and 1 with parainfluenza virus), and 29 ambulatory control subjects, 24 of whom were negative for respiratory viruses and 5 of whom were positive for RV without other viruses. These 5 subjects were considered to have asymptomatic RV (asRV). In accordance with prestudy plans to evaluate approximately 6 subjects in each category, RNA was selected for analysis from 6 of the subjects with RSV (all subjects with RSV were symptomatic), 9 subjects with symptomatic infections with viruses other than RSV, including 6 with RV, 2 with EV, and 1 with adenovirus (non-RSV, or nRSV in figures), all 5 asRV subjects, and 6 virus-negative, asymptomatic Ctrl subjects. Two of the subjects with RSV did not have blood RNA samples available. Thus, in all, we analyzed RNA from 26 nasal swabs and 24 blood samples from 26 subjects. Asymptomatic subjects were older than symptomatic subjects (median ages of 26 months and 8 months), and males were overrepresented among the negative controls. Age and other demographic characteristics of the study subjects are shown in Supplementary Tables S1–S3. Although the RSV group was younger than the other groups, age was not correlated with RNA quality (Supplementary Figure S1) or with gene expression (Supplementary Table S4)
Ribonucleic Acid Characteristics and Quality
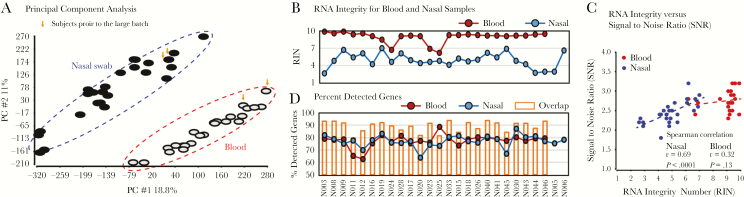
Mean total RNA recovery was 522 ng (range, 77–2413) from the nasal swabs and 4607 ng (range 230–12346) from the blood samples. Results of analysis of RNA quality are shown in Figure 1 and Supplementary Figure S1, which show that RNA metrics were similar in symptomatic and asymptomatic subjects and in patients of different ages. Analysis by PCA of expression patterns for the 28476 coding TCs with Entrez Gene IDs revealed clear separation between nasal and blood samples (Figure 1A). As expected, RNAs from nasal specimens was of moderate quality with RNA integrity number (RIN) ranging from 2.6 to 7.0, whereas RNAs from blood were of high quality with RIN scores of 9 or higher for most samples (Figure 1B). All microarrays for nasal and blood RNA samples passed vendor-recommended key quality-control parameters such as positive versus negative control area under the curve (AUC) >0.8. There was a significant positive correlation between RIN and signal-to-noise ratio for nasal (r = 0.69, P < .0001) but not for blood samples (r = 0.32, P = .13) (Figure 1C). The absence of a significant correlation for the blood samples is probably the result of the relatively narrow range of RINs for the blood samples. The percentage of genes detected in nasal and blood samples was comparable, with an average of 76.3% (range, 67.6%–79.4%) for nasal samples and 79.6% (range, 76.5%–83.1%) for blood samples (Figure 1D).
Figure 1.
Ribonucleic acid (RNA) and microarray quality assessment. (A) Principal component analysis shows that microarray chips from blood and nasal RNAs are separated with greater variability across samples in nasal compared with blood samples. The analysis used quantile-normalized log2 signal data for all genes. Data points from 2 subjects whose samples were analyzed in a separate batch from the other subjects’ samples are marked by arrowheads. (B) Ribonucleic acid integrity numbers (RINs) are higher for blood than for nasal RNAs. (C) Chip signal-to-noise ratio is correlated with RIN for nasal but not for blood RNAs. (D) The percentage of detectable genes from blood and nasal RNAs are generally comparable, substantial, and independent of the RIN. Detectable genes were from among the 28476 well annotated transcript clusters (27823 genes) with an Entrez Gene identification that had signal greater than the mean + 1 standard deviation of the chip-negative control probes. asRV, asymptomatic rhinovirus; Ctrl, control; PC, principal component; nRSV, nonrespiratory syncytial virus; RSV, respiratory syncytial virus.
Cell Marker Gene Expression
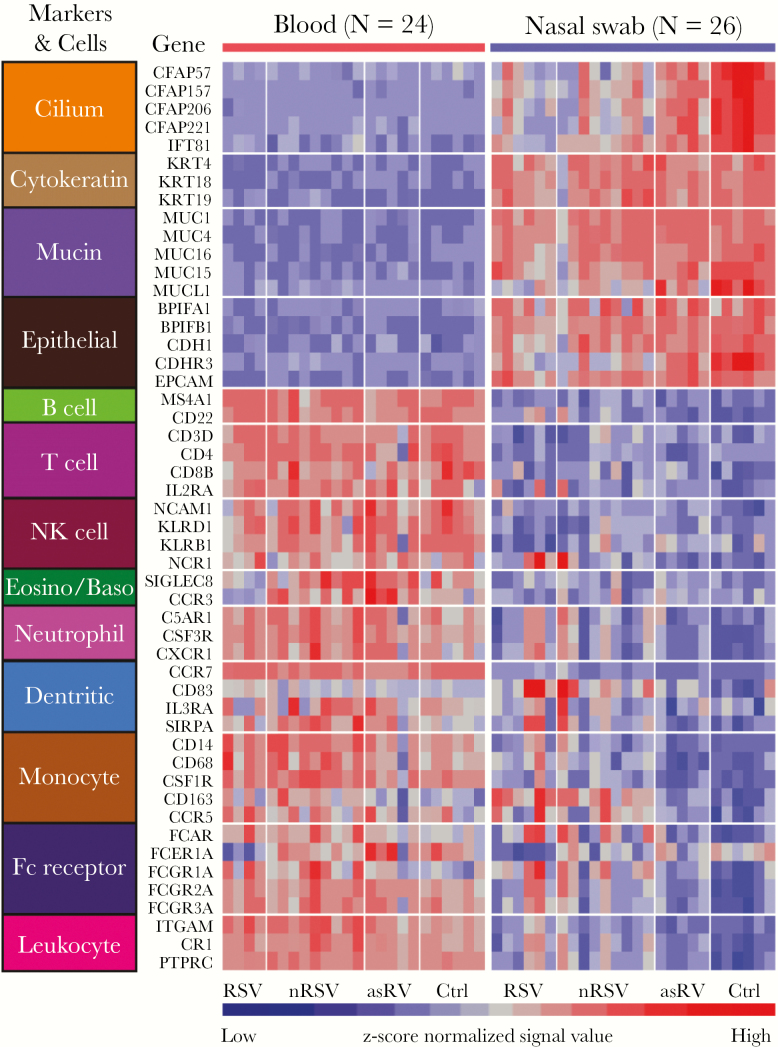
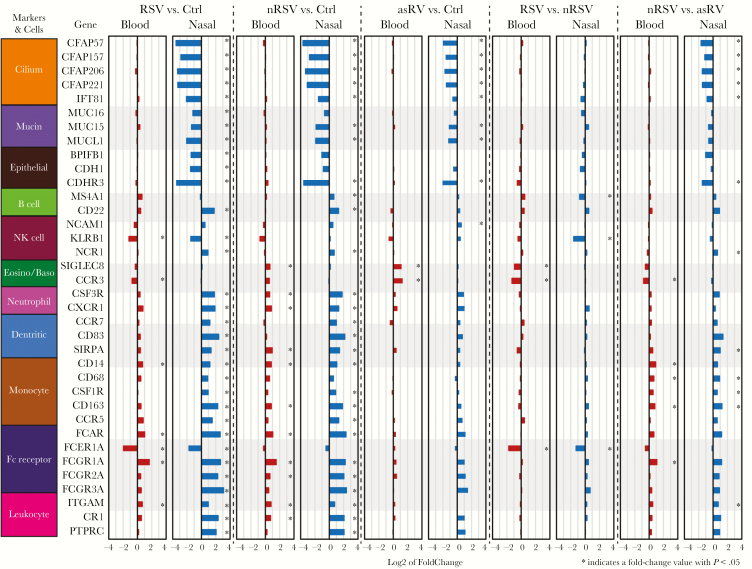
To gain additional understanding of the differences in cellular composition between nasal and blood samples, we analyzed the 2 sample types with regard to the expression of 50 genes that encode putative epithelial and leukocyte markers (Figure 2). As expected, the expression of epithelial genes was higher in nasal samples, and the expression of leukocyte genes was higher in blood samples. Between-group comparisons of these 50 genes in nasal and blood samples revealed that virus-infected subjects had a broad decrease in expression of genes encoding cilia and other epithelial markers in nasal samples and an increased expression of genes encoding neutrophil, monocyte, and dendritic cell markers to Ctrl (Figure 3). Differences in expression were more prominent in nasal than blood samples, consistent with leukocyte recruitment into the nasal mucosa of virus-infected subjects. We extended the cell marker expression analysis to several datasets in the GEO database [12, 33, 35] (and GEO41374 [Ramilo, 2016, unpublished data]) and found that overall expression patterns were consistent with the prior studies in both blood and nasal samples (Supplementary Figure S2).
Figure 2.
Heatmap for the expression profiles of 50 cell marker genes in both blood and nasal samples. Each column displays an individual sample and each row represents a marker gene whose expression value was normalized across these 50 genes by the z-score method. Subject groups are indicated at the bottom. asRV, asymptomatic rhinovirus; Ctrl, control; nRSV, nonrespiratory syncytial virus; RSV, respiratory syncytial virus.
Figure 3.
Fold-change comparison between blood and nasal samples for 36 putative cell marker genes that were significantly different in at least 1 of the comparisons: respiratory syncytial virus (RSV) vs control (Ctrl), nonrespiratory syncytial virus (nRSV) vs Ctrl, and asymptomatic rhinovirus (asRV) vs Ctrl. The length of the bars indicates the log2-fold change in expression for the indicated comparison. Asterisk indicates statistical significance (P < .05).
Deconvolution of Signal From Nasal Samples
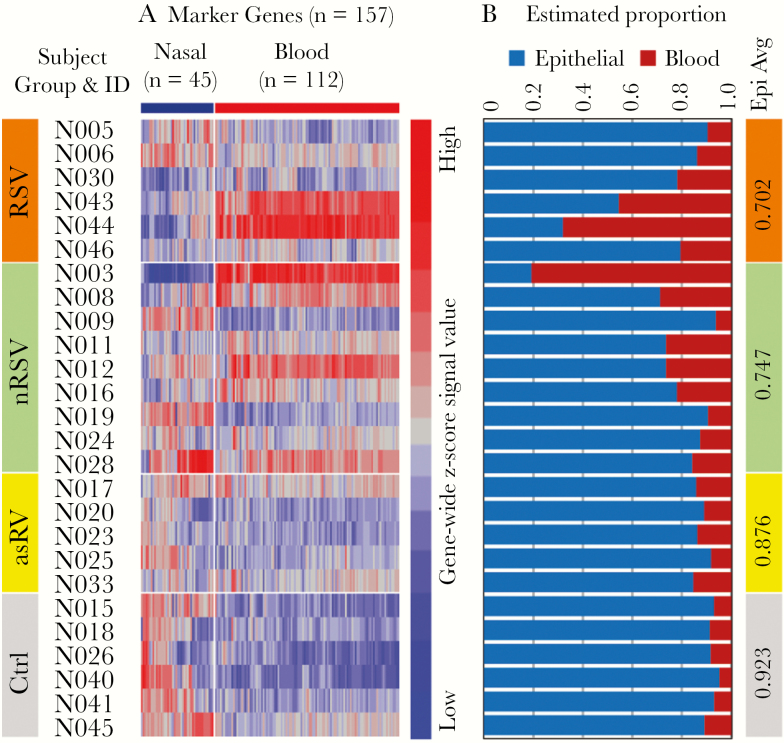
To further understand the implications of the mixed cell population in the nasal samples, we deconvoluted each nasal sample data set into epithelial and hematopoietic components using DSA [30]. As shown in Figure 4, the hematopoietic component showed considerable variability among subjects but was larger in samples from symptomatic subjects (mean, 27.1%; range, 6.2%–80.7%) compared with asymptomatic subjects (mean, 9.9%; range, 4.8%–15.3%; P = .003) and in subjects with asRV (mean, 12.4%; range, 8.1%–15.3%) compared with Ctrl (mean, 7.7%; range, 4.8%–10.8%; P = .03). We also compared the effect of deconvolution on the analysis of gene expression in the 4 subject groups (Supplementary Figure S3). Although application of DSA changed the patterns of gene expression, the distinctions among the 4 groups were similar in magnitude with and without application of DSA.
Figure 4.
Cell type-specific signal deconvolution for nasal gene data set. (A) Expression profiles of 157 marker genes in the nasal samples. (B) The digital sorting algorithm-estimated proportion of signal contributed by epithelial and hematopoietic cells for each of the 26 nasal samples. The numbers in the vertical bar on the right are the mean nasal signal proportion for each of the 4 subject groups. asRV, asymptomatic rhinovirus; Ctrl, control; ID, identification; nRSV, nonrespiratory syncytial virus; RSV, respiratory syncytial virus.
Differentially Expressed Genes
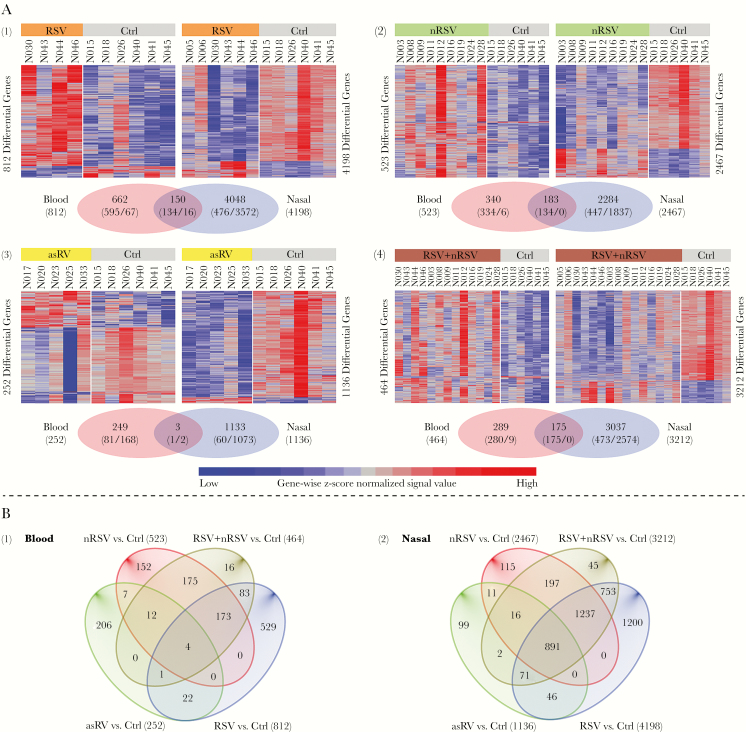
We looked for genes with differential expression in 2-way comparisons between groups of subjects. For initial analysis, comparisons were based on genes with mean normalized signal intensity fold-change of at least 1.5 and uncorrected P < .05 (Figure 5 and Supplementary Figure S4). In these comparisons, the number of genes with differential expression was higher for nasal compared with blood samples and highest for RSV (nasal 4198, blood 812), followed by symptomatic non-RSV (nasal 2467, blood 523), with a smaller but still substantial number in asRV (nasal 1136, blood 252). Of note, most differentially expressed genes in the blood had increased expression, whereas most differentially expressed genes in nasal samples had decreased expression. Overlap of genes with increased expression in nasal and blood samples was only 11.1% for RSV and 19.0% for symptomatic non-RSV.
Figure 5.
Heat maps and Venn diagrams for differentially expressed genes in comparisons between virus-infected groups and negative controls (Ctrl): (1) respiratory syncytial virus (RSV), (2) nonrespiratory syncytial virus (nRSV), (3) asymptomatic rhinovirus (asRV), (4) RSV + nRSV combined. Top panel (A) shows each comparison in matched blood and nasal data sets. These genes were among the fine-filtered 19837 protein-coding transcript clusters ([TCs] 19655 genes). In the heat map, each row represents a TC and each column is an individual study subject. The values used in the heat map were z-score normalized signals from log2 intensity data. Within the Venn diagram, the digits in parenthesis show the number of up- and downregulated genes, displayed before and after the slash, respectively. Bottom panel (B) shows Venn diagram of the 4 comparisons separately in the blood and nasal samples illustrating similarities and differences across different viral groups.
Biological Process and Pathway Analysis
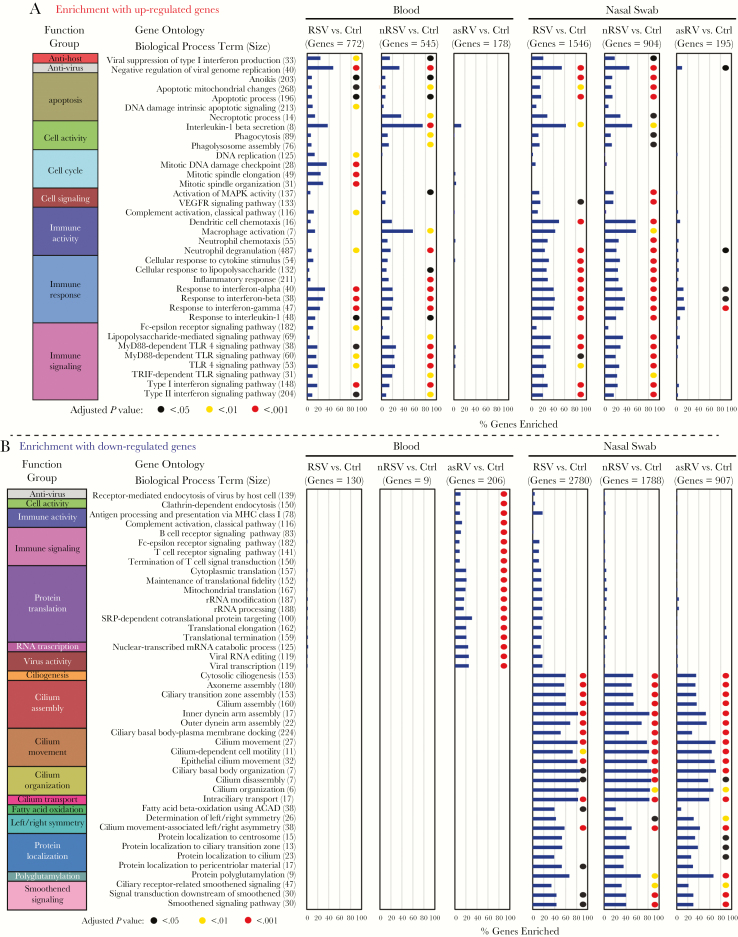
Among the genes with increased expression, similar process terms were enriched in the nasal and blood samples, but the percentage of genes corresponding to most of the processes examined was higher in the nasal samples (Figure 6). Enriched biological processes included antihost, antivirus, apoptosis, cell activity, cell cycle, cell signaling, immune activity, immune response, and immune signaling and were generally similar for RSV and symptomatic non-RSV. An exception was the cell cycle processes, which were enriched only for the subjects with RSV. Enrichment was markedly less for subjects with asRV compared with either of the symptomatic groups. Among the genes with decreased expression, we observed very broad enrichment of cilia-related processes in the nasal samples from the virus-infected groups, including those with asRV. Pathway analysis using IPA revealed that activated pathways were predominantly related to cell signaling, immune activity, and immune signaling (Supplementary Figure S5). Similar pathways were activated in blood and nasal samples, with greater activation in the nasal samples for many of the pathways.
Figure 6.
Gene ontology (GO) enrichment of genes with increased (A) and decreased expression (B). In A, a total of 36 biological process terms were selected for display that included the most highly enriched terms in either or both blood and nasal samples. These terms were arranged in 9 functional groups. The number of genes in each pathway is designated as “size”. The percentage of genes enriched is represented by the horizontal bars, and adjusted P values <.05 are shown by colored circles. In B, a total of 44 biological process terms were selected for display that included the most highly enriched terms in either or both blood and nasal samples. These terms were arranged in 17 functional groups including 10 specific for nasal genes. The percentage of genes enriched is represented by the horizontal bars, and significant enrichments are shown by colored circles. asRV, asymptomatic rhinovirus; Ctrl, control; nRSV, nonrespiratory syncytial virus; RSV, respiratory syncytial virus.
Identification of Gene-Based Classification Scores
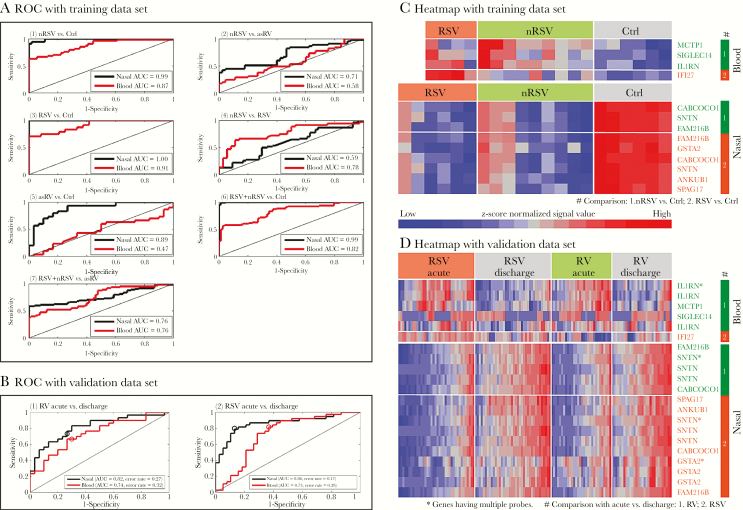
The genes used to derive classification scores for each comparison are shown in Supplementary Table S5 and Supplementary Figure S6. As shown in Figure 7A, we achieved high levels of classification accuracy for most comparisons using a relatively small number of genes, with higher AUCs for nasal compared with blood classifiers for most comparisons. For example, for distinguishing between symptomatic non-RSV and Ctrl, a set of 3 genes assessed in nasal samples achieved a perfect AUC of 1.0, whereas a set of 3 genes assessed in blood had an AUC of 0.94. For distinguishing between symptomatic non-RSV and asRV, a set of 3 genes assessed in nasal samples had an AUC of 0.78, whereas a set of 12 genes assessed in blood had an AUC of 0.71. The only comparison for which blood classifiers outperformed nasal classifiers was RSV versus symptomatic non-RSV.
Figure 7.
Nested cross-validated (CV) receiver operating characteristic (ROC) curves with area under the curve (AUC) for the discriminatory genes based on a modified supervised principal components analysis. A shows ROC curves with corresponding AUCs for gene-based classification scores comprising nasal and blood genes. (1) Nonrespiratory syncytial virus (nRSV) vs control (Ctrl), (2) nRSV vs asymptomatic rhinovirus (asRV), (3) respiratory syncytial virus (RSV) vs Ctrl, (4) nRSV vs RSV, (5) asRV vs Ctrl, (6) RSV + nRSV vs Ctrl, (7) RSV + nRSV vs asRV. B shows ROC curves with corresponding unstratified AUCs and error rates for gene-based classification scores using nasal and blood genes from the present study applied to classify samples from an external dataset (Do et al [33]). (1) RV acute vs discharge, (2) RSV acute vs discharge. Open circles indicate the gene-based classifiers defined by thresholding the mean of 0, yielding the reported error rates. C and D show heat maps displaying the expression of genes used to create the gene-based classification scores the ROC curves displayed in A and B.
External Validation
To assess performance of gene-based classifiers, we applied them to an external data set that included nasal and blood based expression data [33]. Receiver operating characteristic (ROC) curves are shown in Figure 7B, and performance of the component genes are shown in Supplementary Table S6. The classification scores performed well for all 4 tested comparisons (P < .00005 for each) with strong performance of all 10 contributing genes (P < .0011 for each). Figure 7C and D show heatmaps of classifier genes used in the training (Figure 7C) and validation analyses (Figure 7D).
Reverse Transcription-Quantitative Polymearse Chain Reaction Validation
Results of comparing gene expression measured by microarray against that measured by RT-qPCR is shown in Supplementary Figure S7. Signal correlation between RT-qPCR and microarray was high (r = 0.73–0.97, P < .0001) for all genes (Supplementary Figure S7A), and fold-change correlation across the 2 platforms was also high (r = 0.79, P < .0001 for blood and r = 0.95, P < .0001 for nasal samples) (Supplementary Figure S7B). Results of RT-qPCR assays for individual genes are shown in Supplementary Figure S7C (blood) and Supplementary Figure S7D (nasal).
DISCUSSION
This study compared nasal and blood samples for evaluating host response in viral respiratory infections in children. Analysis of nasal samples revealed a rich transcriptomic response that was at least as informative for clinical diagnosis as analysis of the matched blood samples. We were able to identify small sets of genes that distinguished between infected and uninfected children and between those with symptomatic and asymptomatic infection, and nasal gene-based classifiers performed better than blood gene-based classifiers for making these distinctions.
Nasal samples differ in important ways from blood samples that are relevant to diagnostic utility. Most obvious is the different cellular compositions of the 2 locations, with a predominance of epithelial cells of various subtypes in the nose [36] versus leukocytes in the blood. Leukocytes and epithelial cells are both known to participate in the innate immune response to infection [37–39]. However, differences in response are expected because epithelial cells are directly infected during respiratory virus infection, whereas PBLs are responding to a signal emanating from the site of infection, but they are not typically infected themselves. Finally, the microbial environment in the nose is more complex than that of the blood, including a rich resident bacterial microbiota that changes during infection and that can also affect the host transcriptomic response [40].
We found important similarities and differences between the nasal and blood transcriptomes in acute respiratory viral infection. The major difference was that nasal samples had a large number of genes with decreased expression, with strikingly decreased signal from genes involved in cilia structure and assembly. The profile of upregulated genes was generally similar in the 2 sample types, revealing a strong type I interferon response in both locations. Enrichment of biological processes and functional pathways was greater overall in the nasal compared with the blood response, consistent with our finding that diagnostic discrimination was generally superior using nasal samples.
Because of the relatively small number of subjects in our study, it was important to validate our gene classifiers using external data. The only study we found with data appropriate for that purpose used transcriptomic analysis to characterize the pathophysiology of acute RSV and RV in young children [33]. That study differed from our’s in a number of respects, including different ethnic composition of the study populations, comparison of acute versus early recovery samples rather than comparison of infected versus uninfected children in our study, use of nasopharyngeal swabs rather than mid-turbinate (nasal) swabs in our study, and lack of asymptomatic controls. In spite of these differences, our gene-based classification scores performed well at distinguishing between acute and recovery samples from children with acute RSV or RV infection using transcriptomic data from the other study.
Only a few previous studies have analyzed the nasal transcriptome in respiratory virus infection [30, 33, 41, 42]. Proud et al [42] used microarrays to characterize the transcriptomic response of adult volunteers who underwent experimental RV infection. Comparing nasal gene expression of our symptomatic non-RSV patients with the results of that study, the pattern of expression of cell marker genes was similar, as was the increased expression of interferon pathway genes and potential antiviral genes. However, the up-regulation of chemokine genes was less marked in our study compared with that study. Do et al [33] compared nasal and blood transcriptomes in children with acute RSV and RV infection and also found (1) more differentially expressed genes in nasal versus blood samples as well as (2) activation of innate immune response genes in both sample types. Decreased expression of cilia genes in nasal samples was not described. Two other recent studies [43, 44] evaluated expression of specific genes in nasal samples to confirm the presence of respiratory viruses. The genes evaluated in the 2 studies were among the ones that we found that could discriminate between subjects with symptomatic infections and negative controls. Although limited, the group of studies strongly suggest that analysis of nasal gene expression is both practical and informative.
Our study has certain limitations. The number of subjects was small, and this limited our statistical power and our ability to refine, validate, and formally compare genetic classifiers. Nonetheless, rigorous nested leave-one-pair-out cross-validation was used to obtain ROC curves and AUC, and our gene-based classification scores for RSV and symptomatic non-RSV performed well in an independent data set. Future studies must evaluate the impact of underlying conditions and the applicability to viruses other than those included in this study. Finally, gene expression markers should be extended to include markers of bacterial infection.
CONCLUSIONS
In summary, this study provides strong support for the use of nasal samples for assessing the host transcriptomic response to acute viral respiratory illness. This is important because use of nasal samples opens a path to a new generation of diagnostic tests for respiratory infection that can detect pathogens and characterize host response using the same sample and test device, with the practical advantage of not requiring a blood sample. Our studies show a strong and informative host response measurable in nasal samples that can identify individuals with acute respiratory viral infection and can suggest whether the infection is symptomatic. The ability to make these distinctions is at least as good and possibly superior to those made using blood samples. Our future vision includes tests with markers of both viral and bacterial infection and conversion to inexpensive rapid test formats that are currently in development [45, 46]. If realized, this vision would provide the tool needed to limit the overuse of antibiotics for acute respiratory infections, most of which are viral [47].
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Maria Cannella, Sheila Mason, and Richard Buller from the Special Projects Laboratory of the Department of Pediatrics at Washington University for their work processing and testing samples. We also thank Kusal Mihindukulasuriya for assistance in enrolling subjects and Jeanne Holden-Wiltse and Anthony Corbett of the University of Rochester Respiratory Pathogens Research Center Data Management, Informatics and Computational Biology Core for assistance with organization of subject and molecular data. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis.
Disclaimer. This publication is solely the responsibility of the authors and does not necessarily represent the official view of the National Center for Research Resources (NCRR) or the National Institutes of Health (NIH).
Financial support. This work was funded by the National Institute of Allergy and Infectious Diseases, Division of Microbiology and Infectious Diseases, NIH, Department of Health and Human Services, under Contract No. HHSN272201200005C. The Genome Technology Access Center is partially funded by National Cancer Institute Cancer Center Support Grant No. P30 CA91842 to the Siteman Cancer Center and by Institute of Clinical and Translational Sciences/Clinical and Translational Sciences Award Grant No. UL1TR002345 from the NCRR, a component of NIH, and NIH Roadmap for Medical Research.
Potential conflicts of interest. G. A. S. has been a consultant for BioFire Diagnostics (Salt Lake City, UT) and Luminex Corporation (Austin, TX). A. R. F. has received research funding from Merck Sharpe and Dohme, Janssen Pharmaceuticals Inc., Gilead Sciences Inc., Sanofi Pasteur, Pfizer, and Medimmune and has served as an unpaid advisor for Sanofi Pasteur, Pfizer, Novavax, and Gilead Sciences. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Ramilo O, Mejías A. Shifting the paradigm: host gene signatures for diagnosis of infectious diseases. Cell Host Microbe 2009; 6:199–200. [DOI] [PubMed] [Google Scholar]
- 2. Holcomb ZE, Tsalik EL, Woods CW, McClain MT. Host-based peripheral blood gene expression analysis for diagnosis of infectious diseases. J Clin Microbiol 2017; 55:360–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramilo O, Allman W, Chung W, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 2007; 109:2066–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hu X, Yu J, Crosby SD, Storch GA. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc Natl Acad Sci U S A 2013; 110:12792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sweeney TE, Wong HR, Khatri P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci Transl Med 2016; 8:346ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhattacharya S, Rosenberg AF, Peterson DR, et al. Transcriptomic biomarkers to discriminate bacterial from nonbacterial infection in adults hospitalized with respiratory illness. Sci Rep 2017; 7:6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fjaerli HO, Bukholm G, Krog A, Skjaeret C, Holden M, Nakstad B. Whole blood gene expression in infants with respiratory syncytial virus bronchiolitis. BMC Infect Dis 2006; 6:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zaas AK, Chen M, Varkey J, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 2009; 6:207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parnell GP, McLean AS, Booth DR, et al. A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit Care 2012; 16:R157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zaas AK, Burke T, Chen M, et al. A host-based rt-PCR gene expression signature to identify acute respiratory viral infection. Sci Transl Med 2013; 5:203ra126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woods CW, McClain MT, Chen M, et al. A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza H1N1 or H3N2. PLoS One 2013; 8:e52198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mejias A, Dimo B, Suarez NM, et al. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med 2013; 10:e1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bucasas KL, Mian AI, Demmler-Harrison GJ, et al. Global gene expression profiling in infants with acute respiratory syncytial virus broncholitis demonstrates systemic activation of interferon signaling networks. Pediatr Infect Dis J 2013; 32:e68–76. [DOI] [PubMed] [Google Scholar]
- 14. Heinonen S, Jartti T, Garcia C, et al. Rhinovirus detection in symptomatic and asymptomatic children: value of host transcriptome analysis. Am J Respir Crit Care Med 2015; 193:772–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhai Y, Franco LM, Atmar RL, et al. Host transcriptional response to influenza and other acute respiratory viral infections–a prospective cohort study. PLoS Pathog 2015; 11:e1004869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andres-Terre M, McGuire HM, Pouliot Y, et al. Integrated, multi-cohort analysis identifies conserved transcriptional signatures across multiple respiratory viruses. Immunity 2015; 43:1199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsalik EL, Henao R, Nichols M, et al. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci Transl Med 2016; 8:322ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suarez NM, Bunsow E, Falsey AR, Walsh EE, Mejias A, Ramilo O. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J Infect Dis 2015; 212:213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herberg JA, Kaforou M, Wright VJ, et al. Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA 2016; 316:835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mahajan P, Kuppermann N, Mejias A, et al. Association of RNA biosignatures with bacterial infections in febrile infants aged 60 days or younger. JAMA 2016; 316:846–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simmons CP, Popper S, Dolocek C, et al. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis 2007; 195:1097–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caballero IS, Yen JY, Hensley LE, Honko AN, Goff AJ, Connor JH. Lassa and Marburg viruses elicit distinct host transcriptional responses early after infection. BMC Genomics 2014; 15:960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eisfeld AJ, Halfmann PJ, Wendler JP, et al. Multi-platform ‘omics analysis of human ebola virus disease pathogenesis. Cell Host Microbe2017; 22:817–29 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anderson ST, Kaforou M, Brent AJ, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med 2014; 370:1712–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sweeney TE, Braviak L, Tato CM, Khatri P. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir Med 2016; 4:213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singhania A, Verma R, Graham CM, et al. A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nat Commun 2018; 9:2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greninger AL, Chen EC, Sittler T, et al. A metagenomic analysis of pandemic influenza A (2009 H1N1) infection in patients from North America. PLoS One 2010; 5:e13381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Westermann AJ, Gorski SA, Vogel J. Dual RNA-seq of pathogen and host. Nat Rev Microbiol 2012; 10:618–30. [DOI] [PubMed] [Google Scholar]
- 29. Graf EH, Simmon KE, Tardif KD, et al. Unbiased detection of respiratory viruses by use of rna sequencing-based metagenomics: a systematic comparison to a commercial PCR panel. J Clin Microbiol 2016; 54:1000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhong Y, Wan YW, Pang K, Chow LM, Liu Z. Digital sorting of complex tissues for cell type-specific gene expression profiles. BMC Bioinf 2013; 14:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf 2013; 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bair E, Hastie T, Debashis P, Tibshirani R. Prediction by supervised principal components. J Am Stat Assoc 2006; 101:119–37. [Google Scholar]
- 33. Do LA, Pellet J, van Doorn HR, et al. Host transcription profile in nasal epithelium and whole blood of hospitalized children under 2 years of age with respiratory syncytial virus infection. J Infect Dis 2017; 217:134–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–8. [DOI] [PubMed] [Google Scholar]
- 35. Proud D, Sanders SP, Wiehler S. Human rhinovirus infection induces airway epithelial cell production of human beta-defensin 2 both in vitro and in vivo. J Immunol 2004; 172:4637–45. [DOI] [PubMed] [Google Scholar]
- 36. Harkema JR, Carey SA, Wagner JG. The nose revisited: a brief review of the comparative structure, function, and toxicologic pathology of the nasal epithelium. Toxicol Pathol 2006; 34:252–69. [DOI] [PubMed] [Google Scholar]
- 37. Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin Microbiol Rev 2011; 24:210–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hariri BM, Cohen NA. New insights into upper airway innate immunity. Am J Rhinol Allergy 2016; 30:319–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Troy NM, Bosco A. Respiratory viral infections and host responses; insights from genomics. Respir Res 2016; 17:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chu CY, Qiu X, Wang L, et al. The healthy infant nasal transcriptome: a benchmark study. Sci Rep 2016; 6:33994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van den Kieboom CH, Ahout IM, Zomer A, et al. Nasopharyngeal gene expression, a novel approach to study the course of respiratory syncytial virus infection. Eur Respir J 2015; 45:718–25. [DOI] [PubMed] [Google Scholar]
- 42. Proud D, Turner RB, Winther B, et al. Gene expression profiles during in vivo human rhinovirus infection: insights into the host response. Am J Respir Crit Care Med 2008; 178:962–8. [DOI] [PubMed] [Google Scholar]
- 43. Yahya M, Rulli M, Toivonen L, Waris M, Peltola V. Detection of host response to viral respiratory infection by measurement of messenger RNA for MxA, TRIM21, and viperin in nasal swabs. J Infect Dis 2017; 216:1099–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Landry ML, Foxman EF. Antiviral response in the nasopharynx identifies patients with respiratory virus infection. J Infect Dis 2018; 217:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. St John A, Price CP. Existing and emerging technologies for point-of-care testing. Clin Biochem Rev 2014; 35:155–67. [PMC free article] [PubMed] [Google Scholar]
- 46. Kozel TR, Burnham-Marusich AR. Point-of-care testing for infectious diseases: past, present, and future. J Clin Microbiol 2017; 55:2313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fleming-Dutra KE, Hersh AL, Shapiro DJ, et al. Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010–2011. JAMA 2016; 315:1864–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.