Abstract
Alu elements are one of the most successful families of transposons in the human genome. A portion of Alu elements is transcribed by RNA Pol III, whereas the remaining ones are part of Pol II transcripts. Because Alu elements are highly repetitive, it has been difficult to identify the Pol III–transcribed elements and quantify their expression levels. In this study, we generated high-resolution, long-genomic-span RAMPAGE data in 155 biosamples all with matching RNA-seq data and built an atlas of 17,249 Pol III–transcribed Alu elements. We further performed an integrative analysis on the ChIP-seq data of 10 histone marks and hundreds of transcription factors, whole-genome bisulfite sequencing data, ChIA-PET data, and functional data in several biosamples, and our results revealed that although the human-specific Alu elements are transcriptionally repressed, the older, expressed Alu elements may be exapted by the human host to function as cell-type–specific enhancers for their nearby protein-coding genes.
Transposable elements and other repeats contribute to roughly half of the human genome (International Human Genome Sequencing Consortium 2001). Among them, Alu elements represent one of the most successful families, with 1.2 million copies totaling ∼11% of the human genome (International Human Genome Sequencing Consortium 2001). They belong to the primate-specific short interspersed nuclear element (SINE) family of retrotransposons; comparison of primate genomes revealed that the rapid expansion of Alu elements during primate evolution has contributed to wide-ranging genetic diversity in humans, including genetic defects owing to disruption of coding regions and splicing events (Deininger and Batzer 1999; Kazazian 2004; Ade et al. 2013). Although more active during the earlier stage of primate evolution, Alu elements continue to be transcribed (Conti et al. 2015) and inserted into modern human genomes (Konkel et al. 2015), potentially making a profound impact on human biology.
A typical Alu element is ∼280 nucleotides (nt) long and is thought to have evolved from the head-to-tail fusion of two distinct 7SL RNA-derived monomers (left and right arms) (Kriegs et al. 2007). The left arm has bipartite promoter elements (A-box and B-box) (Paolella et al. 1983), bound by the RNA polymerase (Pol) III transcription factor TFIIIC, which in turn initiates Pol III transcription of the Alu element (Orioli et al. 2012). Besides modifying the human genome via retrotransposition, Alu RNAs can also regulate mRNA transcription (Mariner et al. 2008), protein translation (Hasler and Strub 2006), and microRNA biogenesis (Gu et al. 2009; for review, see Deininger 2011; Chen and Yang 2017).
To systematically evaluate the impact of Alu RNAs on gene regulation and cellular function, it is important to quantify the transcription levels of individual Alu elements across diverse cell and tissue types. Previous attempts have been made using RNA-seq (Conti et al. 2015), CAGE (Faulkner et al. 2009; Fort et al. 2014; Li et al. 2018), or ChIP-seq of Pol III factors (Barski et al. 2010; Moqtaderi et al. 2010; Oler et al. 2010); however, it is challenging to assign the short sequencing reads produced by these assays among the highly repetitive Alu elements (Goerner-Potvin and Bourque 2018). Moreover, 58% of the annotated Alu elements in the human genome are located in the introns or 3′ untranslated regions of Pol II–transcribed genes, and it is challenging to distinguish the primary Alu transcripts resulting from Pol III transcription as opposed to the bystander RNAs from Pol II transcription of the host genes.
RAMPAGE is a 5′-complete cDNA sequencing assay that captures the transcription start site (TSS) at single-nucleotide resolution and provides transcript connectivity via paired-end sequencing (Batut et al. 2013). These features allowed it to identify expressed transposons in flies (Batut et al. 2013) and to hold promise for delineating repetitive Alu RNAs. In this study, we developed a new computational pipeline for accurately quantifying the expression levels of individual Alu elements using RAMPAGE data and characterized genomic and epigenetic signatures associated with their transcription profile and potential function as cell-type–specific enhancers.
Results
Identification of primary Alu transcripts using RAMPAGE data
RAMPAGE uses several strategies to enrich for TSSs (Batut and Gingeras 2013): terminator digestion to remove 5′-monophosphorylated RNAs, ribo-zero to remove rRNAs, cap trapping to enrich for 5′-capped RNAs, and template switching to enrich for 5′-triphosphorylated and 5′-capped RNAs. Like other Pol III–transcribed RNAs, Alu RNAs have a 5′-triphosphorylated end but lack the 5′-cap structure (Reddy 1988; Burke et al. 2016). Although the cap trapping assay enriches for RNAs with a 5′-cap, the assay still captures uncapped 5′-triphosphorylated transcripts (Takahashi et al. 2012). Thus, Alu RNAs are enriched by RAMPAGE, although not as enriched as Pol II–transcribed RNAs with 5′-capped ends. Furthermore, with single-nucleotide resolution for identifying TSS and connectivity between the TSS and the downstream transcript, RAMPAGE holds great promise for annotating repetitive Alu elements.
As part of the ENCODE Project, we produced RAMPAGE and RNA-seq data in 155 biosamples with high quality and reproducibility (Supplemental Figs. S1, S2A; Supplemental Table S1; Supplemental Material). To use RAMPAGE peaks to identify individual primary Alu RNAs transcribed by Pol III, we must overcome two challenges. First, a full-size Alu element contains two repeated arms both evolved from the 7SL RNA (Supplemental Fig. S2B), and the 1.2 million copies of Alu elements in the human genome have highly similar sequences; thus, it is challenging to assign short sequencing reads to the bodies of individual Alu elements. Instead, we relied on the downstream unique sequences captured by RAMPAGE: Pol III transcription initiates at the 5′-end of each Alu using the internal promoter elements A-box and B-box located in the left arm (Supplemental Fig. S2B) and terminates downstream from the Alu body because the body lacks internal termination elements (Erwin et al. 2014). Furthermore, paired-end RAMPAGE reads link the downstream sequences to the TSSs of the corresponding Alu elements, so we designed our computational pipeline to capture this information (Fig. 1A; Supplemental Methods). Second, the Pol III–transcribed Alu RNAs are generally expressed at low levels in somatic cells (Paulson and Schmid 1986; Conti et al. 2015), and these weak signals can be contaminated by the typically much stronger Pol II transcription signals even when they are not near a TSS. We developed two measurements—entropy (E) and effective length (L) to filter out false-positive RAMPAGE peaks (Fig. 1A; Supplemental Methods). For GENCODE-annotated genes, TSS-overlapping RAMPAGE peaks have much higher entropies than the other peaks, and an entropy cutoff of 2.5 clearly separated the two populations of peaks (Supplemental Fig. S2C); thus, we applied the same entropy cutoff to RAMPAGE peaks that annotated Alu elements. As primary Alu transcripts are generally unspliced (Conti et al. 2015) and the fragment sizes of RAMPAGE libraries are <1 kb (Batut and Gingeras 2013), we used an effective length cutoff of 1000 nt to further filter out RAMPAGE peaks with improperly long genomic spans (Supplemental Fig. S2D).
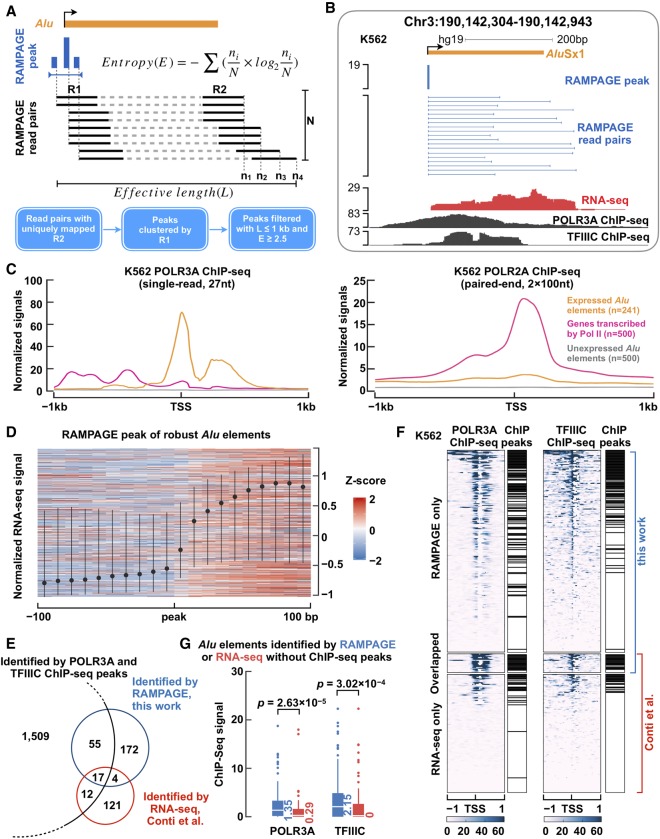
Figure 1.
Genome-wide identification of expressed Alu elements using RAMPAGE data. (A) A computational pipeline to call RAMPAGE peaks and annotate expressed Alu elements. Paired-end RAMPAGE reads (R1 and R2, black; N, read pairs in total) with uniquely mapping R2 reads were first clustered to call peaks at the 5′-end of R1, and the resulting peaks (blue bars) were further filtered with entropy (E) and effective length (L) to annotate expressed Alu elements (orange bar) (for more details, see Supplemental Methods). (B) An expressed AluSx1 element in an intergenic region. RAMPAGE peak marks the TSS of the AluSx1 element precisely, with RAMPAGE read pairs linking the TSS to downstream positions. RNA-seq and ChIP-seq data sets of POLR3A and TFIIIC further confirm the expression of this AluSx1 element. (C) POLR3A (left) and POLR2A (right) ChIP-seq signals are shown with respect to the TSS of expressed Alu elements (orange line), protein-coding and lncRNA genes transcribed by Pol II (pink line), and unexpressed Alu elements (gray line). The signals were averaged over all genes in each set. (D) RNA-seq signal in the ±100-bp window (in 10-nt bins) centered on the TSSs of robustly expressed Alu elements identified by RAMPAGE (identified in three or more biosamples). To avoid overlap with the TSSs of Pol II–transcribed genes, only intronic and intergenic Alu elements were included. Each row of the heatmap corresponds to one such Alu element in one biosample with 10 or more nonzero RNA-seq signal bins, and the dot and bars correspond to the median and first and third quartiles of all Alu elements. (E) Only 84 of the 1593 Alu elements that overlap POLR3A and TFIIIC ChIP-seq peaks (Moqtaderi et al. 2010) are expressed according to RAMPAGE or RNA-seq data in K562 cells. On the other hand, 297 expressed Alu elements (by RAMPAGE or RNA-seq; four by both) do not overlap POLR3A and TFIIIC peaks. (F) Heatmap of normalized read densities for POLR3A and TFIIIC ChIP-seq data around the TSSs of expressed Alu elements identified by RAMPAGE, RNA-seq (Conti et al. 2015), or both techniques in K562 cells. ChIP-seq peaks (Moqtaderi et al. 2010) are labeled on the right of the corresponding heatmaps. (G) Among the 219 expressed Alu elements (by RAMPAGE or RNA-seq) that do not overlap POLR3A or TFIIIC peaks, the Alu elements identified by RAMPAGE show significantly higher POLR3A and TFIIIC signals than the Alu elements defined by RNA-seq. Wilcoxon rank-sum test P-values are shown.
Thus, our computational pipeline took advantage of the two unique features of RAMPAGE technique (TSS identification and connection to the downstream transcripts) and the features of primary Alu elements (unspliced and transcribed past its body). This pipeline allowed us to comprehensively identify Pol III–transcribed Alu elements using only RAMPAGE data. Figure 1B shows an example intergenic AluSx1 element, which is transcribed by Pol III from its 5′-end (bound by its main subunits POLR3A and TFIIIC) to downstream (RNA-seq signal). One RAMPAGE peak with 19 reads precisely annotates the TSS of this AluSx1 element with the RAMPAGE read pairs further depicting its transcriptional profile (Fig. 1B). Supplemental Figure S2E shows four more examples with unfiltered RAMPAGE reads, two intergenic and two intronic.
We performed a series of computational tests and confirmed that the expressed Alu elements identified by our RAMPAGE pipeline resulted from Pol III transcription (Fig. 1C,D; Supplemental Figs. S3A–D, S4; Supplemental Material). We also compared the expressed Alu elements identified by our RAMPAGE pipeline with the expressed Alu elements using RNA-seq and ChIP-seq data (Moqtaderi et al. 2010; Conti et al. 2015), and our method showed higher sensitivity and specificity than the earlier approaches (Fig. 1E–G; Supplemental Fig. S3E,F; Supplemental Material).
Transcribed Alu elements show high tissue specificity
We applied our pipeline to the RAMPAGE data in 155 biosamples (derived from 27 cell lines, 16 primary cell types, and 45 tissues, with some cell and tissue types having multiple biosamples as detailed in Supplemental Table S1) and identified 17,249 Alu elements that were expressed in at least one biosample (Fig. 2A; Supplemental Table S2), which accounted for 1.44% of the 1,194,734 annotated Alu elements in the human genome. This result is consistent with a previous RNA-seq study reporting that only a limited number of Alu elements were transcribed into primary transcripts (Conti et al. 2015). Roughly twice as many expressed Alu elements were detected in tissues than in cell lines or primary cells (median N = 181, 91, 82, respectively) (Supplemental Fig. S5A, left), reflecting the heterogeneous cell type compositions in the tissue samples. The expression levels of the expressed Alu elements were slightly higher in primary cells than in cell lines and tissues (median = 0.76, 0.73, and 0.71 RPM, respectively) (Supplemental Fig. S5A, right). Most of the expressed Alu elements (10,622 of 17,249; 62%) were transcribed in a single biosample (Fig. 2B), and the Alu elements expressed in multiple biosamples had higher expression levels than those expressed in a single biosample (Supplemental Fig. S5B). We defined Alu elements expressed in three or more biosamples as robustly transcribed Alu elements henceforth.
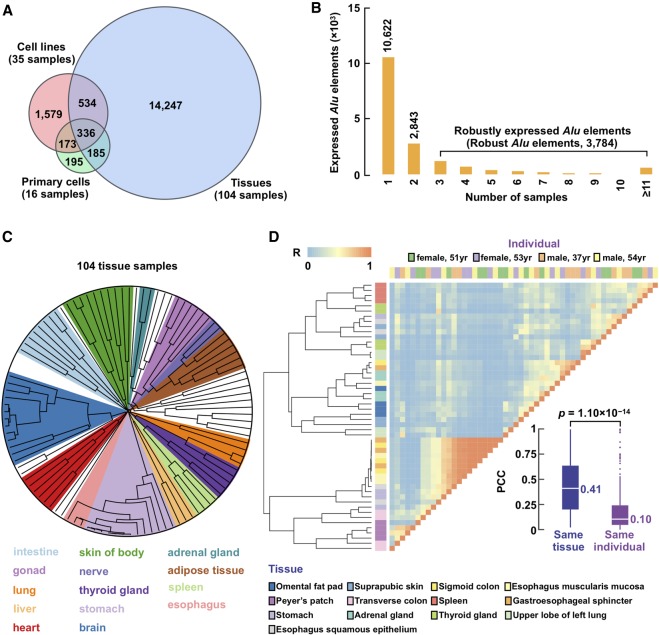
Figure 2.
Expressed Alu elements show high tissue specificity. (A) Venn diagram showing expressed Alu elements defined using RAMPAGE data in cell lines (red), primary cells (green), and tissues (blue). (B) Histogram showing counts of Alu elements expressed in different numbers of biosamples. Note that Alu elements identified in three or more biosamples were defined as robustly expressed Alu elements (robust Alu elements). (C) Dendrogram resulting from agglomerative hierarchical clustering of tissue biosamples based on their Alu expression. Each leaf of the tree represents one RAMPAGE tissue sample, and subtrees dominated by one tissue type are highlighted. (D) Correlation matrix of expressed Alu elements across libraries belonging to 13 tissues of four individuals. Note that correlations between biosamples in the same tissue but from different individuals are significantly higher than correlations between biosamples from the same individual but different tissues. Wilcoxon rank-sum test P-values are shown.
Retrotransposons are generally expressed with high tissue specificity (Faulkner et al. 2009). We asked whether transcribed Alu elements showed tissue specificity. We first clustered the 104 tissue biosamples by their expression profiles of Alu elements, and biologically related biosamples clearly grouped together (Fig. 2C; Supplemental Fig. S5C; Supplemental Table S3). We then perform a detailed analysis on a subset of 52 RAMPAGE data sets from 13 tissues of four individuals. Because most of the expressed Alu elements were transcribed in only one biosample, the overall correlations between biosamples were low, yet similar tissues still clustered together (Fig. 2D). For example, gastroesophageal sphincter, esophagus muscularis mucosa, and sigmoid colon, which are all components of the digestive system, clustered together with high correlations. Furthermore, biosamples from the same tissue but different individuals had substantially higher correlations than biosamples from different tissues of the same individual (median Pearson correlation coefficient 0.41 vs. 0.10; P-value = 1.10 × 10−14) (Fig. 2D). We also observed distinct expression patterns for Alu elements in specific tissue types (e.g., testis) (Supplemental Fig. S5D,E) or associated with specific factors (e.g., DICER1) (Supplemental Material). Together, these results indicate that the expression profile of Alu elements reflects the regulatory programs of the specific tissue type.
Expression of primary Alu transcripts relies more on the genomic context than on primary sequences
We asked whether expressed Alu elements tended to be younger and had stronger regulatory sequence motifs than unexpressed elements. Alu elements are classified into three subfamilies with decreasing evolutionary ages, AluJ, AluS, and AluY, and it has been proposed that AluY elements, being the youngest and least degenerated in sequences, might represent the most retrotranspositionally active subfamily (Bennett et al. 2008). Indeed, AluY is the only known subfamily currently active in retrotransposition in the human genome (Konkel et al. 2015).
In contrast to expectation, there was a slight depletion of expressed AluS and AluY elements compared with the oldest subfamily, AluJ (Supplemental Fig. S6A). Furthermore, we did not observe any difference in evolutionary divergence between expressed and unexpressed Alu elements within each subfamily (Supplemental Fig. S6B). To investigate this problem in greater depth, we classified expressed Alu elements into five groups, B0–B4, depending on whether they were in four other primate genomes (chimpanzee, gorilla, orangutan, or macaque), with B4 defining the human-specific Alu elements (N = 103), i.e., the Alu elements that did not exist in the other four primate genomes, and B0–B3 were collectively designated nonhuman-specific (Fig. 3A; Supplemental Fig. S6C; Supplemental Table S4; Supplemental Methods). As expected, the majority of human-specific Alu elements (88%) were in the AluY class (Fig. 3B); nevertheless, these human-specific AluY elements (N = 91) had significantly lower sequence divergence than the remaining AluY elements (median = 1.6 vs. 6.7; P-value = 2.42 × 10−50) (Fig. 3C), whereas the differences were much less apparent for the AluS and AluJ subfamilies (Fig. 3C). Indeed, the human-specific Alu elements were expressed at significantly lower levels than nonhuman-specific Alu elements (P-value = 0.04) (Supplemental Fig. S6D, left), and our results become more significant when we contrasted human-specific AluY elements against nonhuman-specific AluY elements (P-value = 1.83 × 10−4) (Fig. 3D, left) or human-specific AluS and AluJ elements (P-value = 1.15 × 10−3) (Fig. 3D, right). To test whether our mapping strategy might be biased against younger elements, we modified our pipeline by permitting multiple-mapping reads. The results still showed that human-specific Alu elements showed lower expression levels than nonhuman-specific ones (P-value = 0.02) (Supplemental Fig. S6E, left), especially in the AluY subfamily (P-value = 3.54 × 10−5) (Supplemental Fig. S6E, right). These results suggest that compared with other expressed Alu elements, human-specific AluY elements are relatively repressed in human cells. Furthermore, our B0–B4 classification can better capture the potential activities of Alu elements than the subfamily classification, as we did not detect a significant difference between AluY and AluS/J subfamilies (Supplemental Fig. S6D, right).
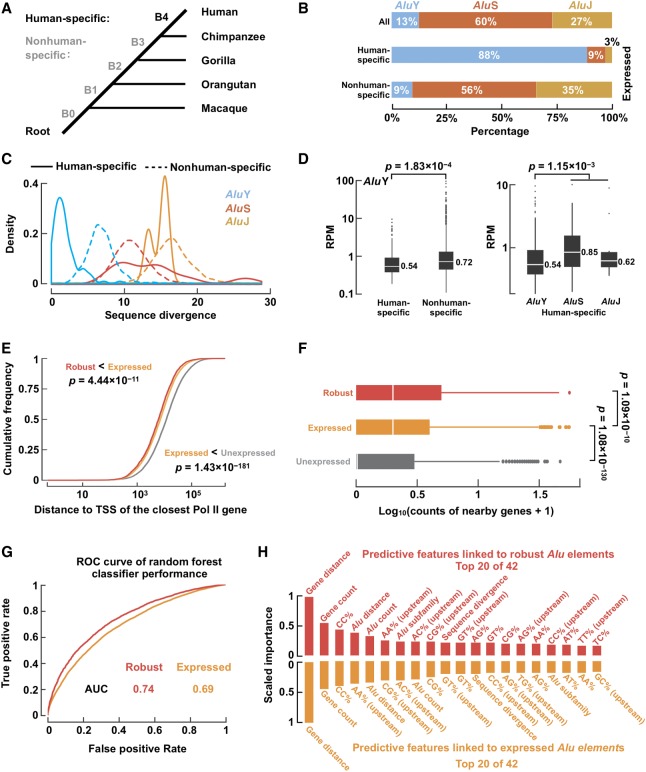
Figure 3.
Expressed Alu elements show distinct genomic context and sequence features. (A) The primate phylogeny. B4 denotes the human-specific branch, and B0–B3 denote nonhuman-specific branches. (B) The AluY subfamily accounted for a large proportion of human-specific Alu elements. (C) Sequence divergence distributions of human-specific (solid lines) and nonhuman-specific (dashed lines) Alu elements in each Alu subfamily. Note that human-specific AluY elements show lower sequence divergence than nonhuman-specific AluY elements. (D) Human-specific AluY elements showed lower expression levels (measured by RPM) than nonhuman-specific AluY elements (left) and human-specific AluS/J elements (right). Wilcoxon rank-sum test P-values are shown. (E) Robustly expressed (red) and expressed (orange) Alu elements are closer to the TSS of Pol II–transcribed genes than are unexpressed (gray) Alu elements. Wilcoxon rank-sum test P-values are shown. (F) Robustly expressed (red) and expressed (orange) Alu elements are more likely to be located in gene-rich regions than are unexpressed (gray) Alu elements. Wilcoxon rank-sum test P-values are shown. (G) Receiver operating characteristic (ROC) curve of random forest classifiers for distinguishing robustly expressed or expressed against unexpressed Alu elements using genomic context and sequence features. (AUC) Area under the curve. (H) The top 20 most important features of the random forest classifiers for distinguishing robustly expressed (red bars) or expressed (orange bars) Alu elements against unexpressed Alu elements, ordered by feature importance.
To understand whether expressed and unexpressed Alu elements were regulated differently, we performed de novo motif finding on these two sets of elements separately (Supplemental Methods). We identified highly significant A-box and B-box motifs, but they were nearly identical between the two sets of elements (Supplemental Fig. S6F,G). Thus, we concluded that the primary sequence of an Alu element is not a strong determinant of its transcriptional activity.
Oler et al. (2010) reported that for Pol III–transcribed genes, especially tRNA genes, genomic context was an important factor for determining whether they could be efficiently transcribed and that expressed tRNA genes generally resided near the TSSs of Pol II–transcribed genes. We tested whether expressed Alu elements also showed such characteristics. Indeed, 65% of robustly expressed, 60% of expressed, and only 45% of unexpressed Alu elements reside within 10 kb of the TSSs of Pol II–transcribed genes, and the differences among the three groups are highly significant (Fig. 3E). Furthermore, there are significantly more genes around robustly expressed and expressed than around unexpressed Alu elements (Fig. 3F). Robustly expressed Alu elements tend to have more nearby Alu elements (regardless of their expression; within 10 kb) than unexpressed Alu elements do (Supplemental Fig. S6H), likely because the former congregate around genes. When we performed this analysis on a per-biosample basis, the difference was significant in 39 of the biosamples (Wilcoxon rank-sum test FDR-adjusted P-value < 0.05). These results are consistent with our results that expressed Alu elements are enriched in the introns of Pol II–transcribed genes (Supplemental Material).
To discern the relative importance of the various aforementioned sequence and genomic context features in determining the transcription status of an Alu element, we trained random forest classifiers using these features (in total 42 features) (Supplemental Methods) to discriminate expressed from unexpressed Alu elements. We could distinguish robustly expressed or expressed Alu elements from unexpressed Alu elements at the area under the receiver operating characteristic (ROC) curve (AUC) of 0.74 and 0.69, respectively (AUC = 0.5 for random performance) (Fig. 3G). The top two most important features for the random forest classifiers were the distance to nearby genes (gene distance) and the number of nearby genes (gene count) (Fig. 3H), consistent with our analysis described above concluding that the genomic context, but less of the primary sequence, of an Alu element determines whether it is expressed.
Alu expression corresponds to open chromatin and active histone modifications
As described above, most Alu elements were expressed in only one biosample. Figure 4A shows the overlaps among Alu elements that were expressed in the three cell lines K562, GM12878, and PC-3; nine elements were expressed in all three cell lines, whereas 416 elements were expressed in only one cell line. To explore possible mechanisms for this cell-type–specific expression, we examined chromatin accessibility (DNase-seq), ChIP-seq of the histone variant H2AZ1 (previously known as H2AZ) and 10 histone modifications, and whole-genome bisulfite sequencing of DNA methylation in these three cell lines (DNA-methylation data were not available in PC-3).
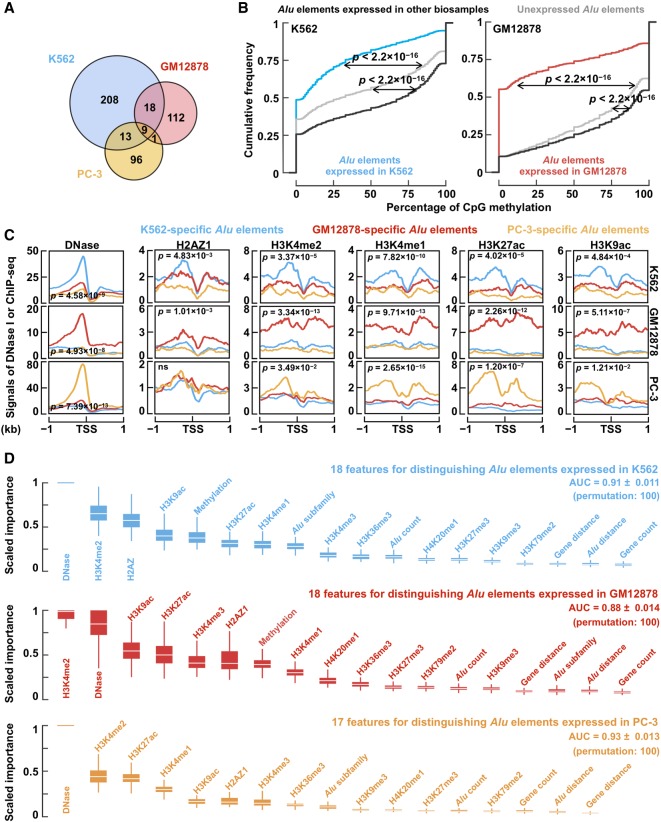
Figure 4.
Cell-type–specific Alu expression corresponds to cell-type–specific histone modifications. (A) Cell-type–specific Alu expression in K562 (blue), GM12878 (red), and PC-3 (yellow) cells. (B) DNA methylation profile of expressed and unexpressed Alu elements. Wilcoxon rank-sum test P-values are shown. (C) Average signal of DNase-seq as well as ChIP-seq signals of H2AZ1, H3K4me2, H3K4me1, H3K27ac, and H3K9ac per cell type (rows) in the ±1-kb regions centered on the TSSs of Alu elements specifically expressed in each of the three cell types. Student's t-test P-values are shown for comparing the average signals in the corresponding cell type against the other two cell types (e.g., DNase signals in K562 cells for K562-specific Alu elements against DNase signals in K562 cells for GM12878-specific and PC-3-specific Alu elements). (D) Features in random forest models for distinguishing cell-type–specific Alu expression in K562 (blue), GM12878 (red), and PC-3 (yellow) cells against 150 randomly sampled Alu elements expressed in other cell types. The features are ordered by their importance.
Low DNA methylation has been associated with active transcription of Alu elements (Jordà et al. 2017). Indeed, the levels of DNA methylation at expressed Alu elements in K562 or GM12878 cells were significantly lower than the levels at unexpressed Alu elements (Fig. 4B). However, it is intriguing that the levels at Alu elements expressed in other samples were even higher than the levels at Alu elements unexpressed in any of the 155 biosamples (Fig. 4B), suggesting that DNA methylation is a mechanism that actively represses Alu elements in a cell-type–specific manner.
We further observed that the levels of DNase, H2AZ1, and four histone marks (H3K4me2, H3K4me1, H3K27ac, and H3K9ac) were higher at the Alu elements specifically expressed in each cell line than at the Alu elements specifically expressed in the other two cell lines (Fig. 4C). These epigenetic marks are typically enriched at enhancers and promoters (Heintzman et al. 2007; Calo and Wysocka 2013). The differences were weaker or insignificant for the other six histone marks (H3K4me3, H3K79me2, H3K36me3, H4K20me1, H3K9me3, and H3K27me3) (Supplemental Fig. S7), which are enriched at transcribed promoters, gene bodies transcribed by Pol II, or repressed promoters, but not at enhancers (Barski et al. 2007). Thus, our results suggest that expressed Alu elements show open chromatin and epigenetic signatures of active enhancers.
To quantify the relative importance of these epigenetic signals in predicting cell-type–specific expression of Alu elements, we constructed random forest classifiers using these signals as features (Supplemental Methods), supplemented by the five best-performing genetic features identified in the previous section: in total, 18 features for K562 and GM12878 and 17 features for PC-3 (sans DNA methylation). These classifiers achieved highly accurate predictions (AUC = 0.91, 0.88, and 0.93 for K562, GM12878, and PC-3, respectively). The relative importance of the features is shown in Figure 4D, with enhancer features ranked at the top, further supporting our hypothesis that expressed Alu elements have enhancer-like chromatin signatures.
Expressed Alu elements may act as cell-type–specific enhancers for nearby genes
As shown above, Alu elements are expressed in a tissue-specific manner, and the expressed Alu elements show epigenetic signatures consistent with active regulatory elements in the corresponding cell types; furthermore, expressed Alu elements tend to be near the TSSs of Pol II genes. Thus, we asked whether expressed Alu elements might function as enhancers for their neighboring Pol II genes in a cell-type–specific manner.
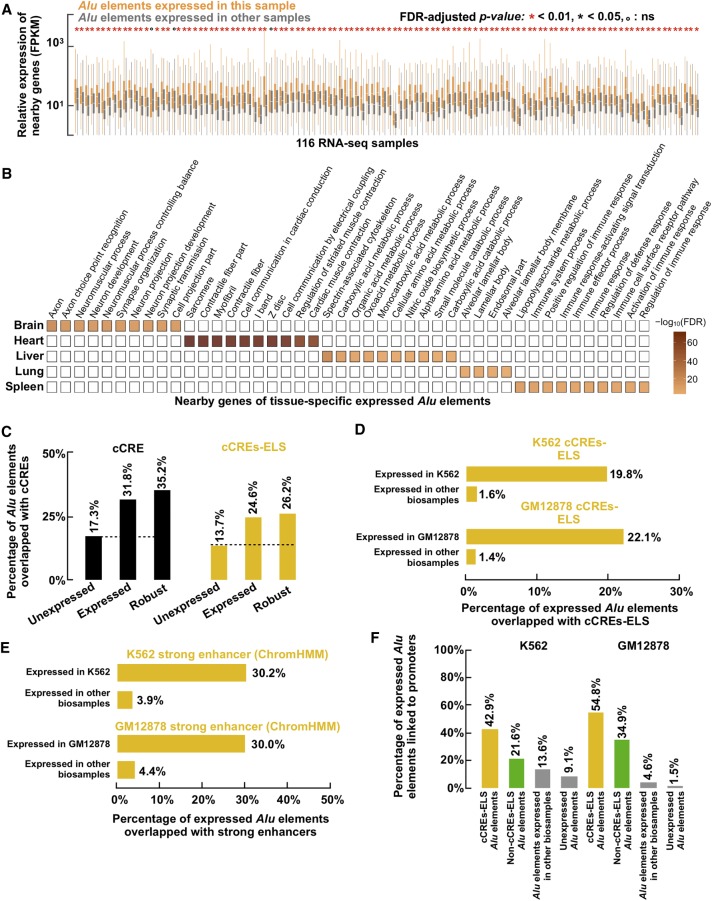
Across the 116 biosamples with at least 50 expressed Alu elements each, the Pol II genes near (TSS located at ≤10 kb) the expressed Alu elements in a biosample were significantly more highly expressed than were the genes near the Alu elements not expressed in that biosample but were expressed in another biosample with RAMPAGE data (median fold-change across 116 biosamples = 2.01) (Fig. 5A). The conclusion remained the same when only the nearest gene was used (Supplemental Fig. S8A). Accordingly, the genes near expressed Alu elements showed significantly higher DNase signals at their TSSs than genes near Alu elements unexpressed in that biosample but expressed in other biosamples (median fold-change across 38 biosamples also with DNase-seq data = 1.76) (Supplemental Fig. S8B). To understand the potential functions of these nearby genes, we performed Gene Ontology (GO) analysis on the genes near expressed Alu elements in five tissues by combining the RAMPAGE data in the biosamples that belonged to each of these tissues (Supplemental Table S3). Our analysis revealed GO terms highly specific to each tissue: axon and neuronal projection for the brain, cardiac muscle for the heart, metabolic processes for the liver, alveolar lamellar body for the lung, and immune responses for the spleen (Fig. 5B; Supplemental Table S5). We further performed motif enrichment analysis on the Alu elements expressed in each of the five tissues and detected some master transcription factors known to be involved in the biological processes that largely define the identities of the respective tissues (Supplemental Fig. S8C; Supplemental Table S6; Supplemental Material). These results are compatible with the hypothesis that expressed Alu elements may contribute to the transcriptional regulation of neighboring genes that have essential functions for the specific tissues.
Figure 5.
Specifically expressed Alu elements may function as cell-type–specific enhancers. (A) Genes within 10 kb of Alu elements expressed in a biosample tend to be more highly expressed in the same biosample than genes within 10 kb of Alu elements expressed in other biosamples. Only biosamples with more than 50 expressed Alu elements were included in this analysis. Wilcoxon rank-sum test FDR-adjusted P-values are reported. (B) Gene Ontology (GO) terms enriched in genes near tissue-specific expressed Alu elements are shown with color intensities corresponding to their FDR values. (C) Compared with unexpressed Alu elements annotated in the human genome (dashed line), expressed and robustly expressed Alu elements showed significant enrichments in cCREs (left; P-value < 2.2 × 10−16, chi-squared test) and cCREs-ELS (right; P-value < 2.2 × 10−16). (D) Alu elements expressed in K562 or GM12878 cells were enriched in K562-specific (top; P-value = 8.05 × 10−96, chi-squared test) or GM12878-specific (bottom; P-value = 4.27 × 10−81) cCREs-ELS, respectively. (E) K562 (top) and GM12878 (bottom) cCREs-ELS Alu elements were enriched in the strong enhancer chromatin state annotated by ChromHMM in the respective cell types (K562, P-value = 1.89 × 10−67; GM12878, P-value = 2.69 × 10−32, chi-squared tests). (F) cCREs-ELS Alu elements expressed in K562 or GM12878 cells were more frequently linked to nearby promoters in the corresponding biosample than non-cCREs-ELS Alu elements expressed in the corresponding biosample (P-values ≤ 7.17 × 10−2, chi-squared test), Alu elements expressed in other biosamples (P-values ≤ 3.40 × 10−7), and unexpressed Alu elements (P-values ≤ 1.61 × 10−15), with links measured by Pol II ChIA-PET interactions.
Next, we investigated whether expressed Alu elements were enriched in annotated cis-regulatory elements, especially enhancers (Supplemental Table S7). We first considered the ENCODE Registry of 1.31 M candidate cis-regulatory elements (cCREs), which were defined using DNase-seq and H3K4me4, H3K27ac, and CTCF ChIP-seq data in hundreds of human biosamples and contained 125,798 and 90,692 cCREs predicted to be active in K562 or GM12878 cells, respectively. Compared with unexpressed Alu elements, expressed and robustly expressed Alu elements showed 1.84- and 2.03-fold enrichment for cCREs, and similar levels of enrichment were observed when the subset of cCREs with enhancer-like signatures (cCREs-ELS) was considered (P-values < 2.2 × 10−16) (Fig. 5C). When comparing Alu elements expressed in K562 or GM12878 cells with Alu elements expressed in other biosamples, we observed 12.3- and 15.7-fold enrichment for cCREs-ELS annotated in the corresponding cell types (P-values ≤ 4.27 × 10−81) (Fig. 5D). We next considered the strong enhancers in K562 and GM12878 defined by ChromHMM with a large panel of histone modifications (Ernst et al. 2011) and observed a 7.74- and 6.82-fold enrichment for Alu elements expressed in K562 or GM12878 cells, respectively, versus Alu elements expressed in other biosamples (chi-squared test P-value = 1.89 × 10−67 and 2.69 × 10−32 for K562 and GM12878, respectively) (Fig. 5E).
To assess the likelihood that expressed Alu elements might act as enhancers for their nearby genes, we further examined whether Alu elements expressed in K562 or GM12878 formed chromatin interactions with their neighboring genes using Pol II ChIA-PET data in these two cell types. Compared with Alu elements expressed in other biosamples or unexpressed Alu elements, there is a significant enrichment for expressed Alu elements in these two cell types to form chromatin interactions with neighboring protein-coding genes, especially those expressed Alu elements that were also cCREs-ELS in the corresponding cell type (Fig. 5F). Moderate enrichment of POLR2A ChIP-seq signals was also observed at these expressed Alu elements in the corresponding cell types where they were also cCREs-ELS (Supplemental Fig. S8D). Taken together, these results suggest that expressed Alu elements might act as enhancers to regulate neighboring genes that function specifically in the corresponding cell types.
Specific binding of TFs associated with cell-type–specific expression of Alu elements
Having observed that expressed Alu elements had characteristic chromatin features of active enhancers and were enriched in chromatin interactions with their nearby genes, which were also expressed and functioned specifically in the corresponding tissues, we asked whether these expressed Alu elements were bound by transcription factors in the corresponding cell types. The ENCODE Consortium had performed ChIP-seq experiments on a large number of TFs, including 275 TFs in K562 cells and 141 TFs in GM12878 cells. Thus, we tested whether there was a significant enrichment of TF binding in Alu elements expressed in each of these two cell types compared with Alu elements expressed in other biosamples.
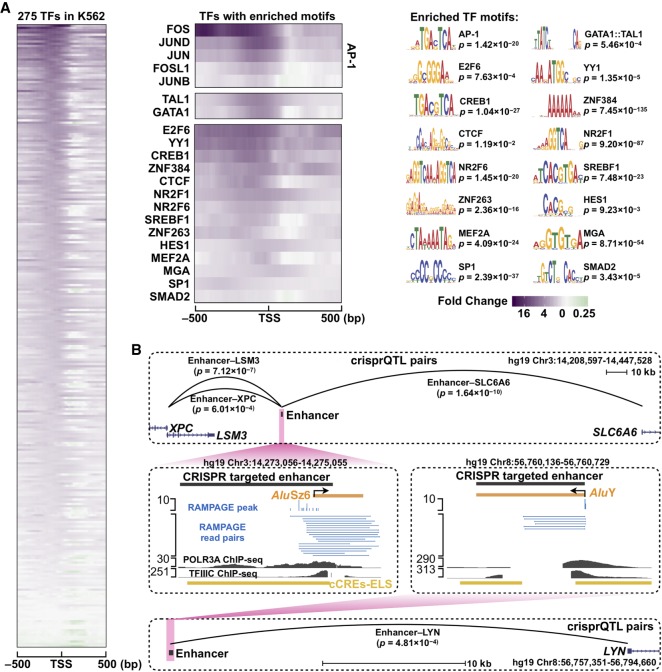
We observed significant enrichments at the upstream regions of expressed Alu elements for most of the TFs with ChIP-seq data (260 TFs in K562 cells [Fig. 6A, left]; 130 TFs in GM12878 cells [Supplemental Fig. S9A, left]). However, only a small subset of these TFs also showed enrichment for their motifs at expressed Alu elements (21 TFs in K562 cells [Fig. 6A, middle]; 16 TFs in GM12878 cells [Supplemental Fig. S9A, right]; Supplemental Table S6). We reasoned that the TFs with enriched motifs bind specifically to the expressed Alu elements, whereas the other TFs may cobind via protein–protein interactions or bind nonspecifically facilitated by the favorable chromatin conditions; thus, we focused our subsequent analysis on the subset of TFs with enriched motifs. Previous studies provided evidence that some of these TFs could enhance the transcription of Alu elements (Ullu and Weiner 1985; Chesnokov and Schmid 1996; Conti et al. 2015). YY1, SP1, and the MEF2 family of TFs were shown to bind Alu elements during developmental processes (Oei et al. 2004) and macrophage responses to tuberculosis infection (Bouttier et al. 2016). AP-1 (heterodimer of FOS and JUN) binds to Pol III genes (Raha et al. 2010; Ahuja and Kumar 2017) and recruits EP300 to increase histone acetylation and stabilize TFIIIC at their core promoters (Mertens and Roeder 2008; Ahuja and Kumar 2017), promoting Pol III transcription at these genes. Thus, AP-1 and EP300 may also activate Alu transcription. Indeed, we detected enriched ChIP-seq signals for FOS and EP300 at the Alu elements specifically expressed in K562 or GM12878 cells (Supplemental Fig. S9B). We stratified expressed Alu elements into three sets—with high, medium, and low FOS ChIP-seq signals, and the EP300 levels scaled positively with FOS occupancy (Supplemental Fig. S9C), providing evidence that AP-1 might recruit EP300 to regulate cell-type–specific expression of Alu elements. In addition to activating Pol III transcription at these Alu loci, these TFs may activate Pol II transcription at nearby genes.
Figure 6.
Enrichment of transcription factor binding at expressed Alu elements reveals the potential to function as cell-type–specific enhancers. (A) The left heatmap shows the fold changes of TF ChIP-seq signals in the ±500-bp window centered on the TSSs of Alu elements specifically expressed in K562 cells compared with Alu elements expressed in other biosamples. Each row is a TF, and all 275 TFs with ChIP-seq data in K562 are included in the heatmap and sorted by their fold changes. The top left heatmaps indicate the TFs with enriched signals and motifs, with the motif logos shown at top right. (B) crisprQTL data (Gasperini et al. 2019) showed that two Alu elements expressed in K562 functioned as enhancers with significant regulatory effects on the expression of nearby genes.
When we compared the TFs that showed enrichments of both ChIP-seq signals and sequence motifs at expressed Alu elements in K562 versus GM12878 (Fig. 6A vs. Supplemental Fig. S9A), some TFs (e.g., GATA1::TAL1 in K562 cells and PAX5 in GM12878 cells) were enriched in one cell type but not in the other, and such TFs were ranked as top master transcription factors associated with cell proliferation and lineage commitment (Supplemental Material), suggesting these cell-type–specific Alu elements can attract the binding of master TFs, which in turn activate lineage-specific transcriptional programs.
Functional data support enhancer activities at expressed Alu elements
We further looked for self-transcribing active regulatory region sequencing (STARR-seq) and massively parallel reporter assay (Sharpr-MPRA) data supporting that expressed Alu elements may function as active enhancers. Barakat et al. (2018) performed STARR-seq on the regions bound by NANOG, POU5F1, H3K4me1, or H3K27ac, identified by ChIP-seq, in primed and naive embryonic stem cells (ESCs). The Alu elements expressed in ESCs (Supplemental Methods) had significantly more STARR-seq reads than did Alu elements expressed in other biosamples (Wilcoxon rank-sum test P-value = 2.42 × 10−3 in primed ESCs and 1.19 × 10−4 in naive ESCs) and unexpressed Alu elements (P-value = 2.17 × 10−4 in primed ESCs and 1.12 × 10−6 in naive ESCs) (Supplemental Fig. S10A, top). When we overlapped Alu elements expressed in ESCs with active enhancers defined by Barakat et al. (reads per plasmid [RPP] ≥138), we observed 4.1- and 4.6-fold enrichment over Alu elements expressed in other biosamples (chi-squared test P-value = 3.00 × 10−7 and 1.11 × 10−7 in primed and naive ESCs, respectively) and 5.9- and 6.6-folds of enrichment over unexpressed Alu elements (P-value = 1.76 × 10−14 and 6.17 × 10−19 in primed and naive ESCs, respectively) (Supplemental Fig. S10A, bottom). These functional data support that expressed Alu elements may function as enhancers in ESCs.
In another study, Ernst et al. (2016) developed the Sharpr-MPRA technique to test more than 15,000 regions (4.6 million nt in total), which were annotated as enhancers by ChromHMM, at 5-nt resolution in K562 cells. Five Alu elements expressed in K562 were among the regions tested by them, and four showed positive Sharpr-MPRA activity scores, indicating that they could activate the transcriptional activity of the reporter. Three of these Alu elements had strong activity scores near their TSSs (one or more averaged over the ±50-bp window centered on the TSS, defined as primarily activating with false-discovery rate ≤5%) (Ernst et al. 2016), and two are shown in Supplemental Figure S10B (top). Compared with Alu elements expressed in other biosamples and unexpressed Alu elements, Alu elements expressed in K562 cells showed significantly stronger Sharpr-MPRA activity scores at their promoter region (P-value = 1.98 × 10−31 and 2.03 × 10−81) (Supplemental Fig. S10B, bottom). Together, these results suggest that some expressed Alu elements have enhancer activities, especially at the regions near their TSSs.
To further evaluate how expressed Alu elements may function as enhancers to regulate the expression of nearby genes, we analyzed the recent crisprQTL data (Gasperini et al. 2019), which yielded 664 enhancer–gene pairs by introducing random combinations of CRISPR/Cas9-mediated perturbations into 5920 predicted enhancers and measuring their effects by single-cell transcriptome profiling in K562 cells. Fifteen of Alu elements expressed in K562 were located in the enhancer regions they surveyed, and two Alu elements showed significant regulatory effects on the expression of neighboring genes (Fig. 6B).
Discussion
We produced RAMPAGE data in 155 biosamples as part of the ENCODE Project and built an atlas of expressed Alu elements using this large collection of data. We identified 17,249 Alu elements that were expressed in at least one of the 155 biosamples, a mere 1.44% of the 1.2 million Alu elements annotated in the human genome, and 61.6% of these 17,249 Alu elements were expressed in only one biosample. These results indicate that Pol III–transcribed Alu expression is overall very low and highly cell-type specific. Contrary to the expectation that the youngest Alu elements—those AluY elements that only exist in the human genome, which are the least divergent from the consensus Alu sequence—should be most expressed, these youngest Alu elements are significantly less expressed than older Alu elements. With the caveat that our results might be influenced by the read-mapping strategy in our pipeline (although we did test allowing multiple-mapping reads and still obtained the same conclusion), these results suggest that humans are highly effective in suppressing young Alu elements, which may still possess the capability of retrotransposition. One active mechanism for repressing Alu expression is DNA methylation (Kochanek et al. 1993, 1995; Bakshi et al. 2016), and accordingly, we found that the Alu elements that were not expressed in a particular biosample (but were expressed in other biosamples) had significantly higher DNA methylation levels in that biosample than did Alu elements that were not expressed in any of the 155 biosamples that we surveyed.
Are the few expressed Alu elements escapees of the active repressive mechanism in the host human cells, or alternatively, can they possibly serve some biological functions? We performed a series of analyses, and our results suggest the latter: Expressed Alu elements may, in some instances, function as cell-type–specific enhancers for nearby protein-coding genes. Expressed Alu elements are significantly more likely to be intronic and exonic than intergenic with respect to genomic Alu elements (2.17% vs. 0.43%, P-value < 2.2 × 10−16). We compared the genetic and epigenetic features at expressed and unexpressed Alu elements and found that distance to Pol II genes, chromatin accessibility, and active histone marks characteristic of active enhancers were predictive of cell-type–specific Alu expression. Furthermore, biosamples in related tissues clustered together by their Alu expression profiles, and the protein-coding genes near expressed Alu elements tended to be expressed and function toward the tissue specificity of the corresponding biosamples. We also observed that expressed Alu elements were significantly enriched in enhancers defined using epigenetic signals and that ChIA-PET data further supported the chromatin interaction between expressed Alu elements and their neighboring genes in the matching cell types. Expressed Alu elements are significantly bound by many transcription factors that work with RNA Pol II, and the binding is cell-type specific. Finally, we found some functional data (STARR-seq, Sharpr-MPRA, and crisprQTL) in ESCs and K562 cells, indicating that some Alu elements expressed in these cell types functioned as enhancers to regulate expression of nearby genes. Because Alu elements are highly repetitive, there have been few studies on individual elements. The atlas of expressed Alu elements that resulted from our study will stimulate more studies targeting individual elements.
A recent study showed that Alu elements could explain a significant amount of disease heritability, especially blood traits (Hormozdiari et al. 2018). Another study analyzed nucleosome occupancy, histone modification, and sequence motif features at Alu elements genome-wide and concluded that Alu elements showed characteristics of enhancers (Su et al. 2014). These studies did not investigate whether these potentially functional Alu elements were expressed in the corresponding cell type and consequently did not distinguish Alu elements transcribed by Pol III versus by Pol II. Here, we focused on the Alu elements transcribed by Pol III and further concluded that the Alu elements transcribed in a biosample by Pol III are more likely to be enhancers than are Alu elements not transcribed by Pol III in the corresponding biosample. Does Pol III play a role in the enhancer functions of these Alu elements that they transcribe? Policarpi et al. (2017) showed that in cortical neurons a subset of SINEs recruited Pol III transcription in a stimulus-dependent manner. They performed in-depth experiments on one such enhancer-like SINE near the Fos gene in the mouse and showed that its Pol III transcript interacted with Pol II at the Fos promoter and was required for the functions of cortical neurons. Thus, we propose that the expressed Alu elements that we identified in this study could act in a similar manner to enhance the transcription of neighboring protein-coding genes. This model is further supported by ChIP-seq data that showed Pol II occupancy at Pol III loci, although that study did not investigate Alu elements (Barski et al. 2010). On the other hand, the moderate enrichment of Pol II ChIP-seq signals at expressed Alu elements in the specific cell types (Supplemental Fig. S8D) suggests that Pol II may facilitate Pol III in its transcription of Alu elements by modifying the local chromatin structure as proposed for other Pol III–transcribed genes (Barski et al. 2010; Raha et al. 2010).
In summary, we have identified 17,249 Pol III–transcribed Alu elements that were expressed in at least one of 155 biosamples. Our integrative analyses showed that although the approximately 100 human-specific Alu elements are actively repressed in transcription, the older expressed Alu elements may have been exapted by the human host to function as enhancers for their neighboring protein-coding genes.
Methods
A computational pipeline for annotating primary Alu transcripts using RAMPAGE data
We developed a new computational pipeline by combining the unique features of the RAMPAGE assay and primary Alu elements to precisely annotated the TSSs of Pol III transcribed Alu elements.
Characterization of Pol III–transcribed Alu elements
Tissue specificity, evolutionary conservation, genomic context, and sequence features were systematically characterized and compared between expressed and unexpressed Alu elements. DNase-seq, ChIP-seq of the histone variant H2AZ1 and 10 histone modifications, and whole-genome bisulfite sequencing of DNA methylation were used to profile the cell-specific expression of Pol III–transcribed Alu elements. Random forest classifiers were implemented to train models for predicting the transcriptional states of Alu elements.
Functional analysis of expressed Alu elements as active enhancers
Expressed Alu elements were overlapped with cCREs and ENCODE ChromHMM annotations (Ernst et al. 2011). STARR-seq (Barakat et al. 2018) and Sharpr-MPRA (Ernst et al. 2016) data were analyzed to assess the enhancer activity of expressed Alu elements. Pol II ChIA-PET data (Li et al. 2012; Tang et al. 2015) and crisprQTL data (Gasperini et al. 2019) were used to evaluate the regulatory impacts of expressed Alu elements on the expression of nearby genes.
Software availability
The source code of our Alu identification pipeline is included in the Supplemental Material as Supplemental Code and can also be accessed at the GitHub (https://github.com/kepbod/rampage_alu).
Supplementary Material
Acknowledgments
We thank members of the Weng laboratory for helpful comments on this manuscript. This project was funded by National Institutes of Health (NIH) grant U24-HG009446 to Z.W. and NIH grant U54-HG004557 to T.R.G.
Author contributions: X.-O.Z. and Z.W. conceived and designed the project. T.R.G. led the production of all RAMPAGE and RNA-seq data. Z.W. supervised the project. X.-O.Z. performed the bioinformatics analysis. X.-O.Z. and Z.W. analyzed the data and wrote the paper with contributions from T.R.G.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.249789.119.
References
- Ade C, Roy-Engel AM, Deininger PL. 2013. Alu elements: an intrinsic source of human genome instability. Curr Opin Virol 3: 639–645. 10.1016/j.coviro.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja R, Kumar V. 2017. Stimulation of Pol III-dependent 5S rRNA and U6 snRNA gene expression by AP-1 transcription factors. FEBS J 284: 2066–2077. 10.1111/febs.14104 [DOI] [PubMed] [Google Scholar]
- Bakshi A, Herke SW, Batzer MA, Kim J. 2016. DNA methylation variation of human-specific Alu repeats. Epigenetics 11: 163–173. 10.1080/15592294.2015.1130518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat TS, Halbritter F, Zhang M, Rendeiro AF, Perenthaler E, Bock C, Chambers I. 2018. Functional dissection of the enhancer repertoire in human embryonic stem cells. Cell Stem Cell 23: 276–288.e8. 10.1016/j.stem.2018.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. 2010. Pol II and its associated epigenetic marks are present at Pol III–transcribed noncoding RNA genes. Nat Struct Mol Biol 17: 629–634. 10.1038/nsmb.1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut P, Gingeras TR. 2013. RAMPAGE: promoter activity profiling by paired-end sequencing of 5′-complete cDNAs. Curr Protoc Mol Biol 104: 25B.11.1–25B.11.16. 10.1002/0471142727.mb25b11s104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut P, Dobin A, Plessy C, Carninci P, Gingeras TR. 2013. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res 23: 169–180. 10.1101/gr.139618.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. 2008. Active Alu retrotransposons in the human genome. Genome Res 18: 1875–1883. 10.1101/gr.081737.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouttier M, Laperriere D, Memari B, Mangiapane J, Fiore A, Mitchell E, Verway M, Behr MA, Sladek R, Barreiro LB, et al. 2016. Alu repeats as transcriptional regulatory platforms in macrophage responses to M. tuberculosis infection. Nucleic Acids Res 44: 10571–10587. 10.1093/nar/gkw782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JM, Kincaid RP, Nottingham RM, Lambowitz AM, Sullivan CS. 2016. DUSP11 activity on triphosphorylated transcripts promotes Argonaute association with noncanonical viral microRNAs and regulates steady-state levels of cellular noncoding RNAs. Genes Dev 30: 2076–2092. 10.1101/gad.282616.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49: 825–837. 10.1016/j.molcel.2013.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Yang L. 2017. ALUternative regulation for gene expression. Trends Cell Biol 27: 480–490. 10.1016/j.tcb.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Chesnokov I, Schmid CW. 1996. Flanking sequences of an Alu source stimulate transcription in vitro by interacting with sequence-specific transcription factors. J Mol Evol 42: 30–36. 10.1007/BF00163208 [DOI] [PubMed] [Google Scholar]
- Conti A, Carnevali D, Bollati V, Fustinoni S, Pellegrini M, Dieci G. 2015. Identification of RNA polymerase III-transcribed Alu loci by computational screening of RNA-Seq data. Nucleic Acids Res 43: 817–835. 10.1093/nar/gku1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. 2011. Alu elements: know the SINEs. Genome Biol 12: 236 10.1186/gb-2011-12-12-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. 1999. Alu repeats and human disease. Mol Genet Metab 67: 183–193. 10.1006/mgme.1999.2864 [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Melnikov A, Zhang X, Wang L, Rogov P, Mikkelsen TS, Kellis M. 2016. Genome-scale high-resolution mapping of activating and repressive nucleotides in regulatory regions. Nat Biotechnol 34: 1180–1190. 10.1038/nbt.3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin JA, Marchetto MC, Gage FH. 2014. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci 15: 497–506. 10.1038/nrn3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571. 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, et al. 2014. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet 46: 558–566. 10.1038/ng.2965 [DOI] [PubMed] [Google Scholar]
- Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS, et al. 2019. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176: 377–390.e19. 10.1016/j.cell.2018.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerner-Potvin P, Bourque G. 2018. Computational tools to unmask transposable elements. Nat Rev Genet 19: 688–704. 10.1038/s41576-018-0050-x [DOI] [PubMed] [Google Scholar]
- Gu TJ, Yi X, Zhao XW, Zhao Y, Yin JQ. 2009. Alu-directed transcriptional regulation of some novel miRNAs. BMC Genomics 10: 563 10.1186/1471-2164-10-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler J, Strub K. 2006. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res 34: 2374–2385. 10.1093/nar/gkl246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Hormozdiari FI, van de Geijn B, Nasser J, Weissbrod O, Gazal S, Ju CJ-T, O'Connor L, Hujoel MLA, Engreitz J, Hormozdiari F, et al. 2018. Functional disease architectures reveal unique biological role of transposable elements. bioRxiv 10.1101/482281 [DOI] [PMC free article] [PubMed]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Jordà M, Díez-Villanueva A, Mallona I, Martín B, Lois S, Barrera V, Esteller M, Vavouri T, Peinado MA. 2017. The epigenetic landscape of Alu repeats delineates the structural and functional genomic architecture of colon cancer cells. Genome Res 27: 118–132. 10.1101/gr.207522.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH. 2004. Mobile elements: drivers of genome evolution. Science 303: 1626–1632. 10.1126/science.1089670 [DOI] [PubMed] [Google Scholar]
- Kochanek S, Renz D, Doerfler W. 1993. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J 12: 1141–1151. 10.1002/j.1460-2075.1993.tb05755.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanek S, Renz D, Doerfler W. 1995. Transcriptional silencing of human Alu sequences and inhibition of protein binding in the box B regulatory elements by 5′-CG-3′ methylation. FEBS Lett 360: 115–120. 10.1016/0014-5793(95)00068-K [DOI] [PubMed] [Google Scholar]
- Konkel MK, Walker JA, Hotard AB, Ranck MC, Fontenot CC, Storer J, Stewart C, Marth GT, Genomes C, Batzer MA. 2015. Sequence analysis and characterization of active human Alu subfamilies based on the 1000 Genomes Pilot Project. Genome Biol Evol 7: 2608–2622. 10.1093/gbe/evv167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. 2007. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet 23: 158–161. 10.1016/j.tig.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98. 10.1016/j.cell.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lenhard B, Luscombe NM. 2018. Integrated analysis sheds light on evolutionary trajectories of young transcription start sites in the human genome. Genome Res 28: 676–688. 10.1101/gr.231449.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29: 499–509. 10.1016/j.molcel.2007.12.013 [DOI] [PubMed] [Google Scholar]
- Mertens C, Roeder RG. 2008. Different functional modes of p300 in activation of RNA polymerase III transcription from chromatin templates. Mol Cell Biol 28: 5764–5776. 10.1128/MCB.01262-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. 2010. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 17: 635–640. 10.1038/nsmb.1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oei SL, Babich VS, Kazakov VI, Usmanova NM, Kropotov AV, Tomilin NV. 2004. Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters. Genomics 83: 873–882. 10.1016/j.ygeno.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. 2010. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17: 620–628. 10.1038/nsmb.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. 2012. RNA polymerase III transcription control elements: themes and variations. Gene 493: 185–194. 10.1016/j.gene.2011.06.015 [DOI] [PubMed] [Google Scholar]
- Paolella G, Lucero MA, Murphy MH, Baralle FE. 1983. The Alu family repeat promoter has a tRNA-like bipartite structure. EMBO J 2: 691–696. 10.1002/j.1460-2075.1983.tb01486.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson KE, Schmid CW. 1986. Transcriptional inactivity of Alu repeats in HeLa cells. Nucleic Acids Res 14: 6145–6158. 10.1093/nar/14.15.6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policarpi C, Crepaldi L, Brookes E, Nitarska J, French SM, Coatti A, Riccio A. 2017. Enhancer SINEs link Pol III to Pol II transcription in neurons. Cell Rep 21: 2879–2894. 10.1016/j.celrep.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. 2010. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci 107: 3639–3644. 10.1073/pnas.0911315106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R. 1988. Compilation of small RNA sequences. Nucleic Acids Res 16(Suppl): r71–r85. 10.1093/nar/16.suppl.r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Han D, Boyd-Kirkup J, Yu X, Han JD. 2014. Evolution of Alu elements toward enhancers. Cell Rep 7: 376–385. 10.1016/j.celrep.2014.03.011 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Lassmann T, Murata M, Carninci P. 2012. 5′ end–centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat Protoc 7: 542–561. 10.1038/nprot.2012.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. 2015. CTCF-Mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163: 1611–1627. 10.1016/j.cell.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Weiner AM. 1985. Upstream sequences modulate the internal promoter of the human 7SL RNA gene. Nature 318: 371–374. 10.1038/318371a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.