Abstract
The incidence of urinary stone disease (USD) has increased four-fold in 50 years. Oxalate, which is degraded exclusively by gut bacteria, is an important constituent in 80% of urinary stones. We quantified the effects of antibiotics and a high fat/high sugar (HFHS) diet on the microbial metabolism of oxalate in the gut. High and low oxalate-degrading mouse models were developed by administering fecal transplants from either the wild mammalian rodent Neotoma albigula or Swiss-Webster mice to Swiss-Webster mice, which produces a microbiota with or without the bacteria necessary for persistent oxalate metabolism, respectively. Antibiotics led to an acute loss of both transplant bacteria and associated oxalate metabolism. Transplant bacteria exhibited some recovery over time but oxalate metabolism did not. In contrast, a HFHS diet led to an acute loss of function coupled with a gradual loss of transplant bacteria, indicative of a shift in overall microbial metabolism. Thus, the effects of oral antibiotics on the microbiome form and function were greater than the effects of diet. Results indicate that both antibiotics and diet strongly influence microbial oxalate metabolism.
Subject terms: Metabolomics, Microbial ecology
Background
The gut microbiome is strongly associated with health and disease, in part because of complex metabolic interactions between host and microbes that contribute to xenobiotic transformations, toxin degradation, immunomodulation, and the production of metabolites that have either pro- or anti-inflammatory effects, among other functions [1–6]. Dysbiosis has been defined as alterations in the microbiome that contribute to disease processes [7]. Dysbiosis can be further separated into two distinct categories. Gain of function dysbiosis is a shift in the microbiome that leads to the emergence of microbial functions that cause disease. Such dysbiosis underpins infectious diseases such as Clostridium difficile infection, cholera, or pneumococcal disease. Loss of function dysbiosis is a shift in the microbiome that leads to the loss of microbial functions that protect health. Loss of function dysbiosis associated with disease is more subtle to detect as there is not an overt growth of a particular disease-causing pathogen or pathogens. However, evidence is accumulating that loss of function dysbiosis is driving the emergence of a number of diseases [8].
Several studies have found significantly lower levels of microbiome diversity associated with disease states, such as Clostridium difficile infection, IBD, diabetes, asthma, colorectal cancer, autism, psoriatic arthritis, Celiac disease, and obesity, implicating loss of function dysbiosis as a contributor to the onset of disease [9–19]. Dysbiosis is often correlated with diet and antibiotic use, which can alter microbial diversity and composition [17, 20–29]. The levels of fats, sugars, and salts in the diet specifically can impact host-microbe interactions in ways that alter inflammation, gut barrier function, endotoxemia, energy balance, intestinal permeability, insulin resistance, and other physiological responses [30–38]. Furthermore, antibiotic use can lead to broad alterations in host-microbe interactions driven by the loss of bacteria [39–44]. Despite the emerging evidence of the association between loss of function dysbiosis and disease, studies that link antibiotic use, diet, or other factors to the loss of specific microbial functions associated with disease are rare [45, 46].
Urinary stone disease (USD) is an ideal condition to examine questions related to loss of function dysbiosis. The incidence of USD has increased four-fold in the last 50 years, which is coincident with the rise of antibiotic use and a shift to a high-fat, high-sugar (HFHS) diet [47–52]. Approximately 80% of stones contain oxalate as a primary constituent [53]. In humans, oxalate comes from two sources. First is the ingestion of oxalate containing plants such as tea, rhubarb, chocolate, nuts, berries, spinach, and potatoes [54–56]. In addition, oxalate is produced endogenously as a terminal metabolite in the liver, primarily from the precursor glyoxylate [57–61]. However, despite the toxicity of oxalate associated with calcium oxalate crystal formation, it is not degraded by mammalian enzymes [62]. Instead, humans and other mammals rely in part on diverse oxalate-degrading bacteria in the gut to minimize the amount of oxalate in circulation [63–69]. The most common pathway for microbial oxalate degradation involves a two-step enzymatic reaction that involves the enzymes oxalyl-CoA decarboxylase and formyl-CoA transferase, along with an oxalate-formate antiporter [70–72]. Oxalobacter formigenes, a bacterium that requires oxalate as a carbon and energy source, is negatively associated with USD, but is also sensitive to many antibiotics [63, 73, 74].
The white-throated woodrat, Neotoma albigula, is a wild rodent that normally consumes a high oxalate diet in the wild and can degrade ~100% of dietary oxalate, even when on a 12% oxalate diet [75]. Studies with this host species have revealed that oxalate metabolism is associated with a diverse and consistent oxalate-degrading microbial network, which increases in relative abundance upon consumption of oxalate [75, 76]. Furthermore, fecal transplants from N. albigula to Sprague-Dawley rats, can induce microbial oxalate metabolism and reduce urinary oxalate excretion more effectively and persistently than microbial transplants containing oxalate-degrading bacteria alone [77, 78].
Given that oxalate is a primary constituent in 80% of stones and is only degraded by gut microbes, the connection between USD and loss of function dysbiosis is more direct compared to other conditions such as diabetes, obesity, inflammatory bowel disorders, or cardiovascular disease where lifestyle and host genetics present additional complex interactions [79–81]. In comparison to subjects with no history of USD, those with an active episode of USD are more likely to have taken oral antibiotics in the last year, have a reduced oxalate-degrading microbial network in the gut, and have a reduced microbe to urinary metabolome co-occurrence network for both the microbiome of the gut and of the urinary tract, which provides a direct mechanism between antibiotic use, loss of function dysbiosis, and the onset of USD [82].
The objective of the current investigation was to examine the effects of antibiotics and a HFHS diet, two factors with known antagonistic interactions with the gut microbiota, on microbial oxalate metabolism directly. Our goal was to determine if these factors cause the loss of microbial oxalate metabolism. Results of the study have implications specifically for the prevention and management of USD along with broader implications for understanding the role of antibiotics and HFHS diets on other dysbiosis-associated diseases.
Methods
Development of mouse models to study loss of function dysbiosis
For the current study, we developed two distinct host-microbe model systems. For both models, Swiss-Webster mice were used as the host to remove any effect of host on the subsequent results. As a negative control, Swiss-Webster mice were given fecal transplants using feces from other Swiss-Webster mice, as discussed below. A diet of 1.5% oxalate is sufficient to induce hyperoxaluria in laboratory rodents such as Swiss-Webster mice, as defined by a 50% increase in urinary oxalate excretion, which is specifically an effect of a lack of microbial oxalate metabolism in the gut [65, 77, 78]. As a positive control and as a baseline for subsequent antibiotic and dietary treatments, Swiss-Webster mice were given fecal transplants from N. albigula. Previous studies show that fecal transplants from N. albigula are sufficient to induce significant and persistent oxalate metabolism [77, 78].
Fecal collection
Feces for fecal transplants were collected from two sources. For the N. albigula feces, 14 individual animals (mixed sex and age) were placed on a 3% oxalate diet in a metabolic chamber at the University of Utah (IACUC #12-12010) to acclimate the microbiome to oxalate metabolism. We have previously determined that 3% oxalate is sufficient to stimulate the oxalate-degrading microbial network in these animals [76]. After 3 days of acclimation, feces were collected from animals within 2 h of defecation, submerged in sterile 15% glycerol, and flushed with CO2 prior to freezing at −80 °C. Feces were then sent to the Cleveland Clinic on dry ice and frozen at −80 °C until use in the diet trials approximately 6 months after receipt (IACUC #2016-1653). For the Swiss-Webster feces, 20 individual animals (Taconic farms, female, six weeks old) were placed on a 0% oxalate diet in cages with a custom-designed insert to separate urine and feces (Fig. S1). Swiss-Webster mouse feces were collected from animals and subjected to the same preservation procedures as the N. albigula feces.
Fecal microbial transplants
During and after fecal transplants, animals were grouped four to a cage, with five cages assigned to each treatment group, for a total of twenty animals per experimental group. To minimize individual variability and eliminate any cage effect in subsequent metrics, samples and data collected from all animals in a cage were considered an individual sample. To develop the two different mouse models, study animals needed to have their native microbiota robustly converted to the study microbiome. The conversion of the gut microbiota was completed in two stages. First, to reduce the native microbiota in animals and maximize the success of fecal transplants, all animals were given neomycin for 5 days, by adding neomycin at 0.5 g/l in their water along with 2 g/l sucralose. Mice were given ad libitum access to water during this time and throughout the experiment. Neomycin is a broad spectrum antibiotic that is poorly absorbed across the gut, and effectively reduces gut microbiota density by up to 90% [83]. After treatment with neomycin, mice were given fecal transplants from either Swiss-Webster mice or from N. albigula. Fecal transplants were performed by first thawing feces, then aseptically mixing 32 g feces per kg body weight directly into powdered mouse chow. Fecal transplants were performed daily for 6 days (Fig. 1). All animals were given a 1.5% oxalate, low fat, low sugar diet (Envigo, Madison, WI) during the antibiotic and fecal transplant periods (composition presented in Table 1). One experimental group was given fecal transplants from Swiss-Webster mice, with four groups given fecal transplants from N. albigula.
Fig. 1.
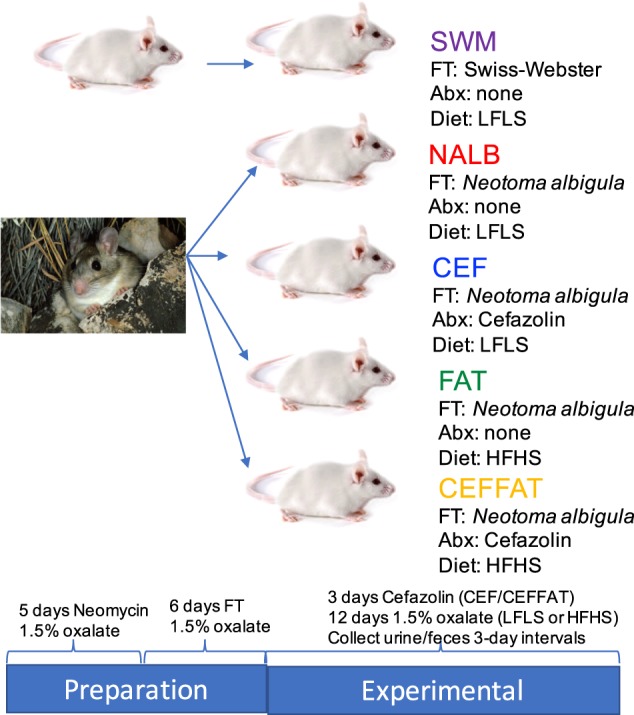
Study design. A population of Swiss-Webster mice (four animals/cage, five cages/group) were initially given Neomycin for 5 days to clear their gut microbiota. Then all mice were given fecal transplants (FT) from either Swiss-Webster mice or Neotoma albigula for 6 days. Some groups were then treated with antibiotics (Abx) and/or were maintained on a low fat/low sugar diet (LFLS) or switched to a high fat/high sugar diet (HFHS) for 12 days. Urine and feces were collected for creatinine, oxalate, and microbial assays at 3-day intervals
Table 1.
Composition of the diets used in the diet trials
| Ingredients | LFLS (g/Kg) | HFHS (g/Kg) |
|---|---|---|
| Casein | 200 | 200 |
| L-cysteine | 3 | 3 |
| Corn starch | 285.5 | NA |
| Maltodextrin | 120 | 120 |
| Sucrose | 100 | 185.4 |
| Anhydrous milkfat | NA | 200 |
| Soybean oil | 70 | 70 |
| Cellulose | 150 | 150 |
| Mineral mix | 42 | 42 |
| Vitamin mix | 12 | 12 |
| Choline bitartrate | 2.5 | 2.5 |
| TBHQ, antioxidant | 0.014 | 0.054 |
| Sodium oxalate | 15 | 15 |
Cefazolin and dietary treatments
To determine the effect of diet and antibiotics on oxalate metabolism, animals were maintained on a 1.5% oxalate diet. Cefazolin (3 day course) was chosen for the antibiotic treatment due to its common use prior to surgical procedures to remove kidney stones [84]. Fat content was added to the HFHS diet in the form of milk fat, while the addition of sugar content was in the form of sucrose for both diets (Table 1, Envigo, Madison, WI). Sucrose was specifically chosen because it has been shown to not alter endogenous oxalate production [58].
Animals receiving fecal transplants from Swiss-Webster mice were maintained on a 1.5% oxalate, LFLS diet with no additional antibiotics (“SWM”) as a negative control. The remaining four treatment groups given fecal transplants from N. albigula and subsequently given the following: (1) A 1.5% oxalate, LFLS diet with no cefazolin (NALB) as a positive control; (2) A 1.5% oxalate, LFLS diet with 3 days of cefazolin administered at 0.125 g/l in the water with 2 g/l sucralose (CEF) starting on the first day after fecal transplants had stopped, to examine the impact of antibiotics; (3) A 1.5% oxalate, HFHS diet with no cefazolin (FAT) to examine the impact of a HFHS diet; and (4) A 1.5% oxalate, HFHS diet with cefazolin for 3 days (CEFFAT) to examine the synergistic effects of antibiotics and a HFHS diet (Fig. 1). All post-transplant diets were maintained for the remainder of the trial (12 days). All urine and feces excreted from the past 24 h were collected from every cage, daily during the diet trial starting on the fourth day of fecal transplants, after the host-microbe models were successfully developed. Samples were pooled by 3-day intervals to longitudinally track changes in urinary creatinine, fecal and urinary oxalate, and the composition of the fecal microbiome. Urine samples were split into two aliquots with one aliquot acidified with 3 M HNO3 for oxalate assays and a second that was unmodified for creatinine assays. Both aliquots were then frozen at −20 °C prior to analysis. Approximately 0.5 g of feces were frozen at −80 °C prior to DNA extraction for microbiome analysis, with the remainder of the samples dried at 45 °C overnight, for oxalate quantification. Body mass, food, water, and oxalate intake, urine output, and dry matter digestibility (DMD) were quantified daily and averaged at 3-day intervals. Oxalate intake was calculated as 1.5% of total daily food intake. The DMD was calculated as 1 (fecal output/food consumed on a dry matter basis), where 1 indicates 100% of dry matter in the diet.
Creatinine and oxalate metrics
Urine oxalate and creatinine were quantified using the enzymatic oxalate assay kit or the colorimetric creatinine assay kit (both from Sigma-Aldrich) following manufacturer’s instructions. Positive controls included a known amount of either oxalate or creatinine and negative controls excluded either oxalate oxidase or creatinine, and were quantified along with each sample. All samples were run in duplicate. Total urinary oxalate and creatinine excretion was quantified for each 3 day period, then divided by 3 to acquire the average daily excretion.
Enzymatic oxalate assays were specifically designed for urinary oxalate and do not require the isolation of oxalate. Preliminary tests of the suitability of the kit for fecal oxalate produced results that often varied by an order of magnitude between duplicates, indicative of the presence of compounds that interfere with the enzymes. Therefore, fecal oxalate excretion was quantified with isolation followed by KMnO4 titration as previously described [75, 77, 78, 85]. Briefly, dried fecal samples were ground to a powder with a sterile mortar and pestle. Oxalate was extracted by submersing 1 g of powder in 6 M H2SO4 at room temperature for 15 min. Following extraction, particulate matter was filtered out, and the remaining solution was brought up to pH 7. Calcium oxalate was then precipitated through the addition of CaCl2, and isolated by centrifugation. Isolated calcium oxalate was re-suspended in de-ionized water and titrated with 0.1 M KMnO4. Titrations were compared to a standard curve. Positive controls included the 1.5% oxalate mouse chow (LFLS & HFHS) and negative controls were the same diets (LFLS & HFHS) without oxalate. Total fecal oxalate excretion was quantified for each 3 day period and then divided by 3 to acquire average daily excretion. Daily oxalate degradation was then quantified as oxalate consumed - (urinary oxalate + fecal oxalate).
DNA extraction and 16S rDNA sequencing
Fecal DNA was extracted from approximately 200 mg of feces using the QIAamp PowerFecal DNA Kit (Qiagen), following manufacturer’s instructions. A total of 125 microbial inventories were generated from all samples across five timepoints, starting on the last 3 days of the fecal transplant. It is at this timepoint that the first differences in gut microbiota composition and oxalate metabolism were expected among groups (Fig. 1). To generate microbial inventories from samples, DNA was sent to Argonne National Laboratory (Chicago, IL) for sequencing of the V4 region of the 16 S rRNA gene, with primers 515 F and 806 R [86]. Samples were multiplexed with 12 bp barcodes for sequencing on a single-lane, MiSeq run with 150 bp paired-end sequencing. Duplicate samples were run for three samples to determine if the sequencing run itself had an effect on the resulting community composition. Additionally, DNA extracted from sterile MilliQ water was sequenced to control for variability due to potential contamination of DNA extraction kits.
Raw sequencing data were demultiplexed and quality controlled with default parameters in QIIME [87]. Operational taxonomic units (OTUs) were assigned using an open-reference strategy with UCLUST [88], with a custom reference database derived from 2011 Greengenes database [89], combined with de novo assigned sequences from previous studies on N. albigula [75–78]. Sequences that did not match the reference database at >97% homology were then assigned de novo. Following taxonomic assignment, OTUs were filtered out if they were assigned as chloroplasts or mitochondria, had fewer than 10 representations across the entire dataset, or were chimeras identified with ChimeraSlayer [90].
Bioinformatic analysis
Unless otherwise noted, bioinformatic analysis was conducted in QIIME. Following the creation of OTU tables, data was normalized with a negative binomial Wald test, using the DESeq2 algorithm [91]. Normalized data were used for alpha and beta diversity analyses. Alpha diversity metrics included Margalef’s species richness, Equitability, Shannon’s Index, and Phylogenetic Diversity. Beta diversity analyses included unweighted and weighted UniFrac analyses, which is a measure of community membership and structure, respectively [92].
To conservatively quantify the bacteria involved in oxalate metabolism, a differential abundance analysis was conducted between the microbial inventories of the NALB group and the SWM group with all timepoints pooled by group, which assumes that the difference in oxalate metabolism between these two groups resulted from the differentially abundant bacteria. Differential abundance analysis is executed as the log fold change of normalized OTU counts, divided by the standard error [91].
To track the oxalate-degrading microbial network in each treatment group, across the diet trial, the co-occurrence of bacteria enriched in the NALB group from the differential abundance analysis, was quantified in each treatment group at each timepoint, using the SparCC algorithm [93]. This algorithm was specifically developed for microbial count data and conducts all pairwise comparisons of defined OTUs. Only strong positive correlations, with a r > 0.6 and p-value = 0 were used in subsequent network analyses. Co-occurrence networks were then visualized in Cytoscape software [94].
Statistical analysis
For food, water, and oxalate intake, urine output, DMD, creatinine and oxalate excretion, and alpha diversity, means were compared with a repeated measures ANOVA comparing treatment group, timepoint, and the interaction between group and timepoint, followed by a Holm’s corrected, Tukey’s honestly significant difference test to determine differences by both group and time point. The slope of body mass change was compared with ANOVA. Prior to ANOVA testing, all data was tested for normality with a Shapiro-Wilk test for normality and sphericity with a Bartlett test for homogeneity of variance, both in R statistical software. For beta diversity, unweighted UniFrac distance matrices were statistically compared with a repeated measures Adonis, with 999 permutations that included treatment Group, Time, and Group*Time as factors. This analysis was followed by post-hoc pairwise comparisons of each group, which were Holm’s corrected for multiple comparisons.
Results
Effect of microbiome, diet, and antibiotics on host and oxalate metrics
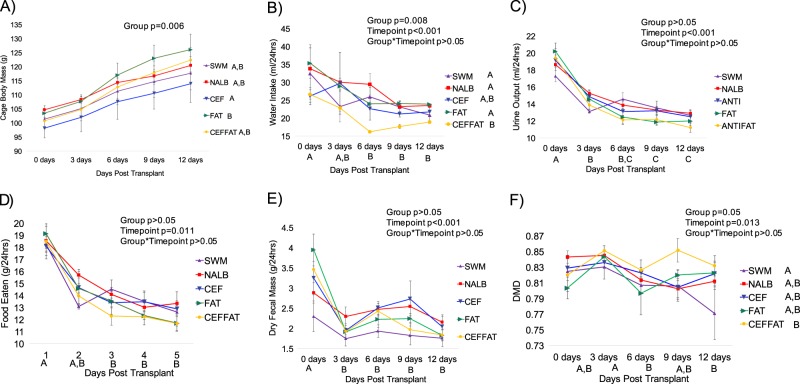
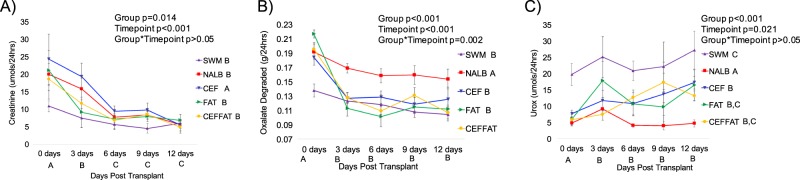
During the fecal transplant period (0 days post transplant), there were significantly higher levels of water intake, urine output, fecal output, urinary creatinine excretion, and food consumed compared to the remaining time points (Figs. 2 and 3). Transplants from N. albigula significantly increased total oxalate metabolism and decreased urinary oxalate excretion at the start of the trial (Fig. 3). Within 3 days, the cefazolin and HFHS diet treatments resulted in an acute and persistent loss of oxalate metabolism, compared to the NALB treatment group (Fig. 3b). The loss of oxalate degradation of these groups led to comparable levels of oxalate metabolism to that observed in the negative control group (SWM). There was no apparent synergistic effect between these treatments. Additionally, both cefazolin and a HFHS diet led to a more moderate, but still significant increase in urinary oxalate excretion, when compared to the NALB group, again with no apparent synergistic effect (Fig. 3c).
Fig. 2.
Effect of treatments on host response. Each timepoint represents the average daily value for the 3-day interval. a Body mass; b Water intake; c Urine output; d Food eaten; e Fecal output; and f DMD. Letters indicate statistically significant differences either by Treatment group (in legend) or by time point (on x-axis) as determined by a repeated measures ANOVA and post-hoc Tukey’s honestly significant difference analysis. Group, timepoint, and interaction ANOVA p-values are presented in the chart. No letters indicate no significant differences or not applicable. For body mass, the slope of body mass change was compared statistically with an ANOVA and therefore differences by timepoint are not applicable
Fig. 3.
Effect of treatments on urinary creatinine and oxalate. Each timepoint represents the average daily value for the 3-day interval. a Urinary creatinine excretion; b Total microbial oxalate metabolism (oxalate consumed minus oxalate excreted); c Urinary oxalate excretion (Urox). Letters indicate statistically significant differences either by Treatment group (in legend) or by time point (on x-axis) as determined by a repeated measures ANOVA and post-hoc Tukey’s honestly significant difference analysis. Group, timepoint, and interaction ANOVA p-values are presented in the chart
Effect of diet and antibiotics on the microbiome
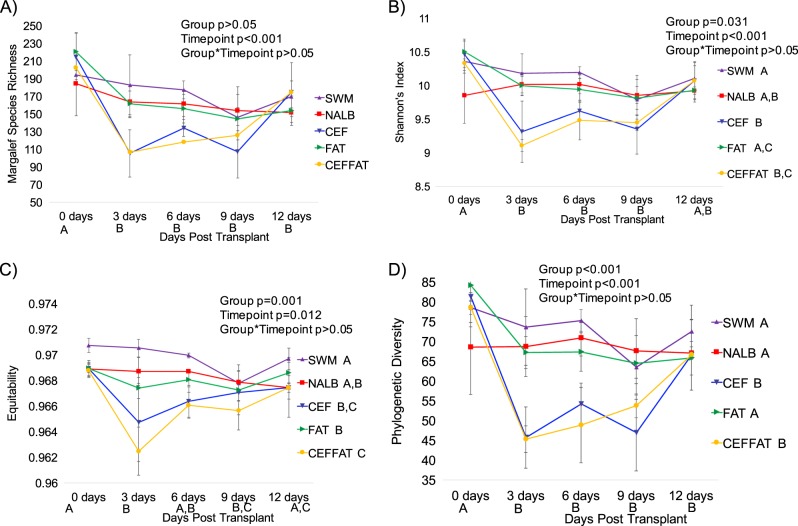
There was a gradual but significant decline in alpha-diversity across all treatment groups, with some recovery apparent depending on the specific metric (Fig. 4). Furthermore, treatment groups that received cefazolin exhibited an acute loss of diversity, followed by a strong recovery to near baseline levels by the end of the diet trial (Fig. 4). This effect was most apparent in the phylogenetic diversity metric, indicative of a taxon-specific response (Fig. 4d).
Fig. 4.
Alpha-diversity by group-time. a Margalef’s species richness; b Shannon’s index; c Equitability; d Phylogenetic diversity. Letters indicate statistically significant differences either by Treatment group (in legend) or by time point (on x-axis) as determined by a repeated measures ANOVA and post-hoc Tukey’s honestly significant difference analysis. No letters indicate no significant differences. Group, timepoint, and interaction ANOVA p-values are presented in the chart
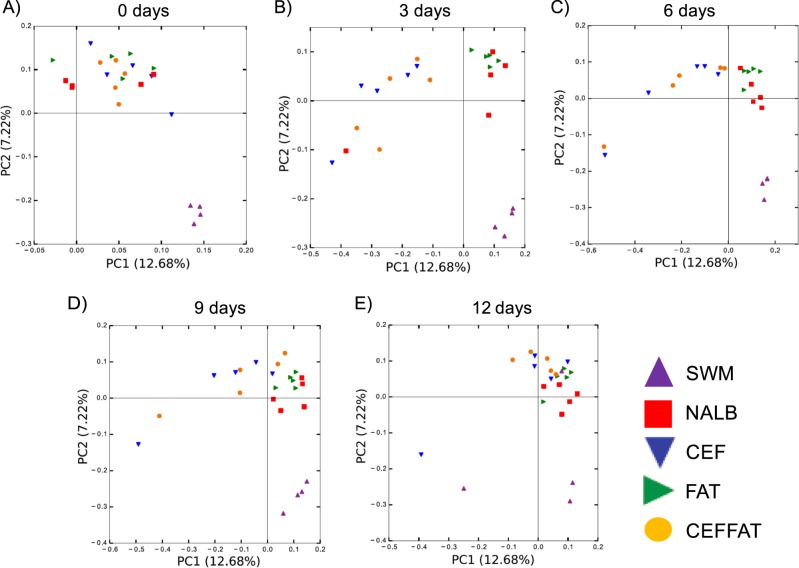
Both cefazolin and a HFHS diet had a significant effect on the composition of the microbiota as assessed by a repeated measures Adonis analysis of the unweighted UniFrac beta diversity (Fig. 5, Table 2). Specifically, all treatment groups were significantly different from each other, except for the CEF and CEFFAT groups. Additionally, cefazolin caused an initial drift in community composition away from all other treatment groups, followed by a return to the post-transplant baseline (Fig. 5, Table 2).
Fig. 5.
Beta-diversity by group-time. PCoA plots are based upon the unweighted UniFrac distance matrices for all groups and timepoints. Beta-diversity plots were created for the entire data set, but points are only shown for each individual timepoint to show how each group changes over time. Plots are labeled with the number of days post transplant: a 0 days; b 3 days; c 6 days; d 9 days; e 12 days. Significant group and group*time differences are indicated in Table 2
Table 2.
Adonis p-values of unweighted UniFrac distance matrices
| Comparison | Adonis p-value | Category |
|---|---|---|
| Group | 0.031a | rmAdonis |
| Time | 0.715 | rmAdonis |
| Group*Time | 0.661 | rmAdonis |
| SWM v NALB | 0.002a | Group × Group |
| SWM v FAT | 0.002a | Group × Group |
| SWM v CEFFAT | 0.002a | Group × Group |
| SWM v CEF | 0.002a | Group × Group |
| NALB v FAT | 0.003a | Group × Group |
| NALB v CEF | 0.002a | Group × Group |
| FAT v CEFFAT | 0.002a | Group × Group |
| FAT v CEF | 0.002a | Group × Group |
| CEF v CEFFAT | 0.847 | Group × Group |
Data matrices were compared by group, time, and group*time using a repeated measures Adonis (rmAdonis), followed by post-hoc pairwise comparisons (Group × Group).
aindicates significant differences between pairwise group comparisons and for each group over the duration of the experiment. p-values were FDR-corrected for multiple comparisons
Effect of diet and antibiotics on the oxalate-degrading microbial network
The oxalate-degrading microbial network is defined as the community of bacteria associated with oxalate metabolism [75–78, 95, 96]. In the current study, we conservatively defined the oxalate-degrading network as the OTUs that were enriched in the NALB group compared to the SWM, as assessed by a significant difference in normalized abundance, with the assumption that the difference in total oxalate metabolism was specifically due to the difference in microbial community composition. Differential abundance analysis resulted in a total of 438 OTUs (Table S1). The taxa identified through this analysis largely overlapped taxa identified in previous studies that used varying analytical techniques in both rodent and human studies (Fig. S2).
To track the oxalate-degrading microbial network across time and treatment group, co-occurrence analyses of bacteria enriched in the NALB group were conducted for each treatment group and time point (Fig. 6). For the NALB group, there was a gradual increase in both the number of bacteria and interactions of the network, indicative of network size and cohesiveness respectively (Figure S3). The increases in number of bacteria and interactions were accompanied by a gradual differentiation in interaction clusters, indicated by the emergence of denser regions within the overall network (Fig. 6a). For the SWM group, there were considerably fewer bacteria and interactions in the defined network compared to the NALB group, which declined over the course of the diet trial. Furthermore, no structural differentiation was apparent in the network (Fig. 6b). Corroborating the alpha- and beta-diversity analyses, the cefazolin-treated groups saw an acute loss of bacteria within the network, followed by a gradual recovery (Figs. 6c, e). Finally, the FAT group exhibited similar levels of bacteria and interactions to the NALB group initially. However, these numbers remained unchanged throughout the diet trial (Fig. 6d, S3).
Fig. 6.
Oxalate-degrading microbial network by group-time. The microbial network was defined as OTUs significantly enriched in the NALB group vs. the SWM group, totaling 438 bacteria (Table S1). Detailed below are the Group, Time point, No. of bacteria present in the network, and No. of Interactions in the network. Also of importance is the level of structural differentiation within each of the networks apparent from denser regions of the network
Discussion
Subjects with an active episode of USD are more likely to have taken oral antibiotics in the previous year and have reduced functional microbial networks in both the microbiome of the gut and the urinary tract compared to subjects with no history of USD [82]. Thus, there is a direct potential mechanism between oral antibiotics, loss of function dysbiosis, and the onset of USD. The current study was designed to further test the potential links between perturbation of the gut microbiota, driven both by antibiotics and diet, and the loss of a specific microbial function associated with the inhibition of USD.
The oral antibiotic cefazolin led to an acute loss of both microbial oxalate metabolism and bacterial diversity (Figs. 3, 4, 6). The loss of diversity was apparent both at the level of the whole community and from the oxalate-degrading microbial network specifically (Figs. 4, 6). Interestingly, the microbial diversity of cefazolin-treated animals recovered back to their post-transplant microbiota composition 9 days after antibiotic use ceased. However, oxalate metabolism had not recovered over this time period, corroborating the incomplete recovery indicated by network analysis (Fig. 6). The loss and incomplete recovery of the gut microbiota due to antibiotic use has been shown previously [22, 23]. It is not known if the function would have fully recovered if the experiment continued or if a second round of fecal transplants would have been required to recover the function.
For the animals receiving the HFHS diet, there was an acute loss of oxalate metabolism, with no significant loss of bacteria either from the whole gut microbiota or the oxalate-degrading network (Figs. 3, 4, 6). However, there was a significant shift in overall gut microbiota composition (Fig. 5, Table 2). These data indicate that a broader shift in microbial metabolism away from oxalate degradation resulted from the switch to a HFHS diet. The lack of a reduction in alpha-diversity from the switch to a HFHS diet is in contrast to our hypothesis and to other studies [24, 97–100]. However, when a HFHS diet is consumed, bacteria in the gut increase the net energy harvest from food, decrease energy expenditure, and exhibit a broad shift in their microbial metabolic phenotype, consistent with the results of the current study [35, 99]. The fact that all animals receiving the N. albigula fecal transplant consistently grouped away from animals receiving the Swiss-Webster transplant and that the microbiota returned to the post-transplant baseline after cefazolin treatment indicate that cefazolin and HFHS diet led to a loss of function dysbiosis in the NALB microbiome rather than a return to the native Swiss-Webster microbiome.
To date, the oxalate-degrading microbial network has been identified through gradual changes to dietary oxalate and association with O. formigenes, in both rodent and clinical studies [75–78, 82, 101]. In the current study, we conservatively identified the oxalate-degrading microbial network by quantifying the OTUs that were significantly enriched in the NALB group compared to the SWM group with the assumption that the difference in oxalate degradation resulted from the difference in community composition between these two groups. In a meta-analysis of studies, a taxonomic core set of bacteria were identified in at least 71% of studies, with diverse taxa apparently opportunistic or dependent on the method of detection (Fig. S2). Of these, the Coprococcus, Lachnospriaceae, Oscillospira, and Ruminococcaceae were associated with the oxalate-degrading microbial network in all studies. Additionally, the Bacteroides, Bifidobacterium, Clostridiales, Lachnospira, Morganella, Oxalobacter, RF39, Rikenellaceae, Roseburia, Ruminococcus, S24-7, and YS2 taxa were found in all but one of the studies. Oxalobacter is a genus of bacteria that requires oxalate as a carbon and energy source. Other taxa, such as the Bifidobacterium or S24-7, have been shown to degrade oxalate in vitro and/or harbor oxalate-degrading genes [68, 102].
We have previously hypothesized that the taxa associated with the oxalate-degrading network relies on the by-products produced by the metabolism of oxalate, CO2 and formate, which can be used subsequently in downstream metabolic pathways such as acetogenesis, methanogenesis, and sulfate-reduction, [75, 76, 103–106]. However, the hypothesis that the by-products of microbial oxalate metabolism are used in downstream metabolic pathways needs to be explicitly tested. Regardless, the data here show that a strong perturbation can effectively and persistently inhibit this functional microbial network.
The incidence of USD and co-morbidities is rapidly increasing. Evidence is accumulating that the increase in incidence is being driven by a loss of bacteria in the gut that normally provide a health benefit. The current study implicates oral antibiotics and a HFHS diet in the loss of a specific function of the gut microbiota, oxalate metabolism, that is associated with USD. We only tracked one specific microbial function associated with a generalized perturbation of the gut microbiota. The results obtained here likely have broad implications for the loss of microbial functions associated with other diseases. In fact, a recent meta-analysis showed many common taxa that were lost between USD and co-morbidities [95]. Thus, our results suggest that treatments for USD and other diseases, particularly centered on antibiotic use and dietary recommendations, need to consider the gut microbiota and the functions that maintain health.
Supplementary information
Acknowledgements
This study was funded with seed funds from Lerner Research Institute and NIH (grant 1F32DK102277-01A1 to Aaron W. Miller) and by the NSF (DEB 1342615 to M. Denise Dearing).
Data availability
Sequence reads are available at the Sequence Read Archive under Accession # SRP179737.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0357-4) contains supplementary material, which is available to authorized users.
References
- 1.Wilson I, Nicholson J. The role of gut microbiota in drug response. Curr Pharm Des. 2009;15:1519–23. doi: 10.2174/138161209788168173. [DOI] [PubMed] [Google Scholar]
- 2.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2010;12:5. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 3.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 5.Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 6.Kohl KD, Weiss, Robert B, Cox J, Dale C, Dearing MD. Gut microbes of mammalian herbivores facillitate intake of plant toxins. Ecol Lett. 2014;17:1238–46. doi: 10.1111/ele.12329. [DOI] [PubMed] [Google Scholar]
- 7.Myers SP. The causes of intestinal dysbiosis: a review. Altern Med Rev. 2004;9:180–97. [PubMed] [Google Scholar]
- 8.Blaser MJ. Missing microbes: how the overuse of antibiotics is fueling our modern plagues. Macmillan; 2014; New York City, NY. [DOI] [PMC free article] [PubMed]
- 9.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile—associated diarrhea. J Infect Dis. 2008;197:435–8. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schippa S, Iebba V, Barbato M, Di Nardo G, Totino V, Checchi MP, et al. A distinctive ‘microbial signature’ in celiac pediatric patients. BMC Microbiol. 2010;10:175. doi: 10.1186/1471-2180-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durbán A, Abellán JJ, Jiménez-Hernández N, Ponce M, Ponce J, Sala T, et al. Assessing gut microbial diversity from feces and rectal mucosa. Microb Ecol. 2011;61:123–33. doi: 10.1007/s00248-010-9738-y. [DOI] [PubMed] [Google Scholar]
- 13.Carroll IM, Ringel‐Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea‐predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:521. doi: 10.1111/j.1365-2982.2012.01891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907–11. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang DW, Park JG, Ilhan ZE, Wallstrom G, LaBaer J, Adams JB, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8:e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 17.Abrahamsson T, Jakobsson H, Andersson AF, Björkstén B, Engstrand L, Jenmalm M. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44:842–50. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 18.Sha S, Liang J, Chen M, Xu B, Liang C, Wei N, et al. Systematic review: faecal microbiota transplantation therapy for digestive and nondigestive disorders in adults and children. Aliment Pharmacol Ther. 2014;39:1003–32. doi: 10.1111/apt.12699. [DOI] [PubMed] [Google Scholar]
- 19.Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67:128–39. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 22.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci. 2011;108:4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 25.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown K, Valenta K, Fisman D, Simor A, Daneman N. Hospital ward antibiotic prescribing and the risks of Clostridium difficile infection. JAMA Intern Med. 2015;175:626–33. doi: 10.1001/jamainternmed.2014.8273. [DOI] [PubMed] [Google Scholar]
- 28.Candon S, Perez-Arroyo A, Marquet C, Valette F, Foray AP, Pelletier B, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One. 2015;10:e0125448. doi: 10.1371/journal.pone.0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vangay P, Ward T, Gerber JS, Knights D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe. 2015;17:553–64. doi: 10.1016/j.chom.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet–induced obesity and diabetes in mice. Diabetes. 2008;57:1470–81. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 31.Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–83. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 32.Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Neyrinck AM, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775–86. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neyrinck AM, Possemiers S, Druart C, Van de Wiele T, De Backer F, Cani PD, et al. Prebiotic effects of wheat arabinoxylan related to the increase in bifidobacteria, Roseburia and Bacteroides/Prevotella in diet-induced obese mice. PLoS One. 2011;6:e20944. doi: 10.1371/journal.pone.0020944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neyrinck AM, Possemiers S, Verstraete W, De Backer F, Cani PD, Delzenne NM. Dietary modulation of clostridial cluster XIVa gut bacteria (Roseburia spp.) by chitin–glucan fiber improves host metabolic alterations induced by high-fat diet in mice. J Nutr Biochem. 2012;23:51–9. doi: 10.1016/j.jnutbio.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Serino M, Luche E, Gres S, Baylac A, Bergé M, Cenac C, et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61:543–53. doi: 10.1136/gutjnl-2011-301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam YY, Ha CW, Campbell CR, Mitchell AJ, Dinudom A, Oscarsson J, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One. 2012;7:e34233. doi: 10.1371/journal.pone.0034233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de La Serre CB, Ellis CL, Lee J, Hartman AL, Rutledge JC, Raybould HE. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol. 2010;299:G440–8. doi: 10.1152/ajpgi.00098.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilck N, Olesen S, Matus M, Balogh A, Dechend R, Alm E, et al. A high-salt diet alters the composition of intestinal microbiota in mice. Hypertension. 2014;64:A321–A321. [Google Scholar]
- 39.Morgun Andrey, Dzutsev Amiran, Dong Xiaoxi, Greer Renee L, Sexton D Joseph, Ravel Jacques, Schuster Martin, Hsiao William, Matzinger Polly, Shulzhenko Natalia. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut. 2015;64(11):1732–1743. doi: 10.1136/gutjnl-2014-308820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank DN, Amand ALS, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, van Zanten SJV. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–41. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–41. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci. 2013;110:9066–71. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scales CD, Smith AC, Hanley JM, Saigal CS, Project UDiA. Prevalence of kidney stones in the United States. Eur Urol. 2012;62:160–5. doi: 10.1016/j.eururo.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy SC, Jacob JT, Varkey JB, Gaynes RP. Antibiotic use in US hospitals: quantification, quality measures and stewardship. Expert Rev Anti Infect Ther. 2015;13:843–54. doi: 10.1586/14787210.2015.1040766. [DOI] [PubMed] [Google Scholar]
- 49.Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis. 2014;14:742–50. doi: 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 50.Aminov RI. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 2010;1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cordain L, Eaton SB, Sebastian A, Mann N, Lindeberg S, Watkins BA, et al. Origins and evolution of the Western diet: health implications for the 21st century. Am J Clin Nutr. 2005;81:341–54. doi: 10.1093/ajcn.81.2.341. [DOI] [PubMed] [Google Scholar]
- 52.De SK, Liu X, Monga M. Changing trends in the American diet and the rising prevalence of kidney stones. Urology. 2014;84:1030–3. doi: 10.1016/j.urology.2014.06.037. [DOI] [PubMed] [Google Scholar]
- 53.Moe OW. Kidney stones: pathophysiology and medical management. lancet. 2006;367:333–44. doi: 10.1016/S0140-6736(06)68071-9. [DOI] [PubMed] [Google Scholar]
- 54.Holmes RP, Goodman HO, Assimos DG. Contribution of dietary oxalate to urinary oxalate excretion. Kidney Int. 2001;59:270–6. doi: 10.1046/j.1523-1755.2001.00488.x. [DOI] [PubMed] [Google Scholar]
- 55.Gasińska A, Gajewska D. Tea and coffee as the main sources of oxalate in diets of patients with kidney oxalate stones. Rocz Panstw Zakl Hig. 2007;58:61–67. [PubMed] [Google Scholar]
- 56.Noonan S, Savage G. Oxalate content of foods and its effect on humans. Asia Pac J Clin Nutr. 1999;8:64–74. [PubMed] [Google Scholar]
- 57.Conyers R, Bais R, Rofe A. The relation of clinical catastrophes, endogenous oxalate production, and urolithiasis. Clin Chem. 1990;36:1717–30. [PubMed] [Google Scholar]
- 58.Ribaya JD, Gershoff SN. Factors affecting endogenous oxalate synthesis and its excretion in feces and urine in rats. J Nutr. 1982;112:2161–9. doi: 10.1093/jn/112.11.2161. [DOI] [PubMed] [Google Scholar]
- 59.Knight J, Wood KD, Lange JN, Assimos DG, Holmes RP. Oxalate formation from glyoxal in erythrocytes. Urology. 2016;88:226. e211–5. doi: 10.1016/j.urology.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lange Jessica N., Wood Kyle D., Knight John, Assimos Dean G., Holmes Ross P. Glyoxal Formation and Its Role in Endogenous Oxalate Synthesis. Advances in Urology. 2012;2012:1–5. doi: 10.1155/2012/819202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holmes RP, Assimos DG. Glyoxylate synthesis, and its modulation and influence on oxalate synthesis. J Urol. 1998;160:1617–24. [PubMed] [Google Scholar]
- 62.Hodgkinson A. Oxalic acid in biology and medicine. Academic Press; London, New York; 1977.
- 63.Allison MJ, Dawson KA, Mayberry WR, Foss JG. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol. 1985;141:1–7. doi: 10.1007/BF00446731. [DOI] [PubMed] [Google Scholar]
- 64.Hatch M, Cornelius J, Allison M, Sidhu H, Peck A, Freel R. Oxalobacter sp. reduces urinary oxalate excretion by promoting enteric oxalate secretion. Kidney Int. 2006;69:691–8. doi: 10.1038/sj.ki.5000162. [DOI] [PubMed] [Google Scholar]
- 65.Hatch M, Gjymishka A, Salido EC, Allison MJ, Freel RW. Enteric oxalate elimination is induced and oxalate is normalized in a mouse model of primary hyperoxaluria following intestinal colonization with Oxalobacter. Am J Physiol-Gastrointest Liver Physiol. 2011;300:G461–9. doi: 10.1152/ajpgi.00434.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller AW, Kohl KD, Dearing MD. The gastrointestinal tract of the white-throated woodrat (Neotoma albigula) harbors distinct consortia of oxalate-degrading bacteria. Appl Environ Microbiol. 2014;80:1595–601. doi: 10.1128/AEM.03742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campieri C, Campieri M, Bertuzzi V, Swennen E, Matteuzzi D, Stefoni S, et al. Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int. 2001;60:1097–105. doi: 10.1046/j.1523-1755.2001.0600031097.x. [DOI] [PubMed] [Google Scholar]
- 68.Turroni S, Bendazzoli C, Dipalo SC, Candela M, Vitali B, Gotti R, et al. Oxalate-degrading activity in Bifidobacterium animalis subsp. lactis: impact of acidic conditions on the transcriptional levels of the oxalyl coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes. Appl Environ Microbiol. 2010;76:5609–20. doi: 10.1128/AEM.00844-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turroni S, Vitali B, Bendazzoli C, Candela M, Gotti R, Federici F, et al. Oxalate consumption by lactobacilli: evaluation of oxalyl‐CoA decarboxylase and formyl‐CoA transferase activity in Lactobacillus acidophilus. J Appl Microbiol. 2007;103:1600–9. doi: 10.1111/j.1365-2672.2007.03388.x. [DOI] [PubMed] [Google Scholar]
- 70.Baetz AL, Allison MJ. Purification and characterization of oxalyl-coenzyme A decarboxylase from Oxalobacter formigenes. J Bacteriol. 1989;171:2605–8. doi: 10.1128/jb.171.5.2605-2608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baetz AL, Allison MJ. Purification and characterization of formyl-coenzyme A transferase from Oxalobacter formigenes. J Bacteriol. 1990;172:3537–40. doi: 10.1128/jb.172.7.3537-3540.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruan ZS, Anantharam V, Crawford IT, Ambudkar SV, Rhee SY, Allison MJ, et al. Identification, purification, and reconstitution of OxlT, the oxalate: formate antiport protein of Oxalobacter formigenes. J Biol Chem. 1992;267:10537–43. [PubMed] [Google Scholar]
- 73.Kaufman DW, Kelly JP, Curhan GC, Anderson TE, Dretler SP, Preminger GM, et al. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol. 2008;19:1197–203. doi: 10.1681/ASN.2007101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lange JN, Wood KD, Wong H, Otto R, Mufarrij PW, Knight J, et al. Sensitivity of human strains of Oxalobacter formigenes to commonly prescribed antibiotics. Urology. 2012;79:1286–9. doi: 10.1016/j.urology.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller AW, Dale C, Dearing MD. Microbiota diversification and crash induced by dietary oxalate in the mammalian herbivore Neotoma albigula. mSphere. 2017;2:e00428–17. [DOI] [PMC free article] [PubMed]
- 76.Miller AW, Oakeson KF, Dale C, Dearing MD. Effect of dietary oxalate on the gut microbiota of the mammalian herbivore Neotoma albigula. Appl Environ Microbiol. 2016;82:2669–75. doi: 10.1128/AEM.00216-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller AW, Dale C, Dearing MD. The induction of oxalate metabolism in vivo is more effective with functional microbial communities than with functional microbial species. mSystems. 2017;2:e00088–17. doi: 10.1128/mSystems.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller Aaron W., Oakeson Kelly F., Dale Colin, Dearing M. Denise. Microbial Community Transplant Results in Increased and Long-Term Oxalate Degradation. Microbial Ecology. 2016;72(2):470–478. doi: 10.1007/s00248-016-0800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Korat AVA, Willett WC, Hu FB. Diet, lifestyle, and genetic risk factors for type 2 diabetes: a review from the Nurses’ Health Study, Nurses’ Health Study 2, and Health Professionals’ Follow-up Study. Curr Nutr Rep. 2014;3:345–54. doi: 10.1007/s13668-014-0103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rankinen T, Sarzynski MA, Ghosh S, Bouchard C. Are there genetic paths common to obesity, cardiovascular disease outcomes, and cardiovascular risk factors? Circ Res. 2015;116:909–22. doi: 10.1161/CIRCRESAHA.116.302888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ng SC, Tang W, Leong RW, Chen M, Ko Y, Studd C et al. Environmental risk factors in inflammatory bowel disease: a population-based case-control study in Asia-Pacific. Gut. 2014;64:1063–71. [DOI] [PubMed]
- 82.Zampini A, Nguyen A, Rose E, Monga M, Miller A. Reduced functional microbial network in patients with urolithiasis. Unpublished.
- 83.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wolf JS, Bennett CJ, Dmochowski RR, Hollenbeck BK, Pearle MS, Schaeffer AJ. Best practice policy statement on urologic surgery antimicrobial prophylaxis. J Urol. 2008;179:1379–90. doi: 10.1016/j.juro.2008.01.068. [DOI] [PubMed] [Google Scholar]
- 85.Miller AW, Oakeson KF, Dale C, Dearing MD. The effect of dietary oxalate on the gut microbiota of the mammalian herbivore Neotoma albigula. Appl Environ Microbiol. 2016;AEM-00216. [DOI] [PMC free article] [PubMed]
- 86.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 89.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lozupone C, Hamady M, Knight R. UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinforma. 2006;7:1. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Batagello CM, Manoj, Miller, AW. Urolithiasis: a case of missing microbes? J Endourol. 2018;32:995–1005. [DOI] [PubMed]
- 96.Miller AW, Dearing D. The metabolic and ecological interactions of oxalate-degrading bacteria in the mammalian gut. Pathogens. 2013;2:636–52. doi: 10.3390/pathogens2040636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–23. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 99.Shen W, Gaskins HR, McIntosh MK. Influence of dietary fat on intestinal microbes, inflammation, barrier function and metabolic outcomes. J Nutr Biochem. 2014;25:270–80. doi: 10.1016/j.jnutbio.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 100.Brinkworth GD, Noakes M, Clifton PM, Bird AR. Comparative effects of very low-carbohydrate, high-fat and high-carbohydrate, low-fat weight-loss diets on bowel habit and faecal short-chain fatty acids and bacterial populations. Br J Nutr. 2009;101:1493–502. doi: 10.1017/S0007114508094658. [DOI] [PubMed] [Google Scholar]
- 101.Miller AWC, David P, Kristina L, Lange, D. Identification of oxalate microbiome in non-stone forming individuals. Unpublished.
- 102.Ormerod KL, Wood DLA, Lachner N, Gellatly SL, Daly JN, Parsons JD, et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome. 2016;4:1–17. doi: 10.1186/s40168-016-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rieu-Lesme F, Morvan B, Collins M, Fonty G, Willems A. A new H2/CO2-using acetogenic bacterium from the rumen: description of Ruminococcus schinkii sp. nov. FEMS Microbiol Lett. 1996;140:281–6. doi: 10.1016/0378-1097(96)00195-4. [DOI] [PubMed] [Google Scholar]
- 104.Robeniol RZG, Serrano VD, Florendo PD, Aquino D, Cruz KJ, Divina CC. Acetogenic and acid utiliizng bacterial content analysis on ruminal fluid of water buffalo (Bubalus bubalis) calves. Int J Agric Technol. 2016;12:2065–71. [Google Scholar]
- 105.Bernalier A, Willems A, Leclerc M, Rochet V, Collins MD. Ruminococcus hydrogenotrophicus sp. nov., a new H2/CO2-utilizing acetogenic bacterium isolated from human feces. Arch Microbiol. 1996;166:176–83. doi: 10.1007/s002030050373. [DOI] [PubMed] [Google Scholar]
- 106.Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol. 2002;52:1615–20. doi: 10.1099/00207713-52-5-1615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence reads are available at the Sequence Read Archive under Accession # SRP179737.