Abstract
The majority of circular RNAs (circRNAs) spliced from coding genes contain open reading frames (ORFs) and thus, have protein coding potential. However, it remains unknown what regulates the biogenesis of these ORF-containing circRNAs, whether they are actually translated into proteins and what functions they play in specific physiological contexts. Here, we report that a large number of circRNAs are synthesized with increasing abundance when late pachytene spermatocytes develop into round and then elongating spermatids during murine spermatogenesis. For a subset of circRNAs, the back splicing appears to occur mostly at m6A-enriched sites, which are usually located around the start and stop codons in linear mRNAs. Consequently, approximately a half of these male germ cell circRNAs contain large ORFs with m6A-modified start codons in their junctions, features that have been recently shown to be associated with protein-coding potential. Hundreds of peptides encoded by the junction sequences of these circRNAs were detected using liquid chromatography coupled with mass spectrometry, suggesting that these circRNAs can indeed be translated into proteins in both developing (spermatocytes and spermatids) and mature (spermatozoa) male germ cells. The present study discovered not only a novel role of m6A in the biogenesis of coding circRNAs, but also a potential mechanism to ensure stable and long-lasting protein production in the absence of linear mRNAs, i.e., through production of circRNAs containing large ORFs and m6A-modified start codons in junction sequences.
Subject terms: Developmental biology, Molecular biology
Introduction
Spermatogenesis refers to the process through which spermatogonial stem cells differentiate into spermatozoa within the seminiferous epithelium in the testis.1 It consists of three phases: mitotic (proliferation and differentiation of spermatogonia), meiotic (spermatocyte differentiation and division), and haploid (also called spermiogenesis, differentiation of round spermatids into spermatozoa) phases.2 Each of the three phases involves complex regulatory networks that control precise spatiotemporal gene expression.3,4 The haploid phase, spermiogenesis, is unique in that (1) it does not involve proliferation and (2) it displays two prominent regulatory features of gene expression: global shortening of transcripts and delayed translation.5–8 The delayed translation, also called uncoupling of transcription and translation, is due to the fact that transcription ceases upon nuclear condensation, which coincides with spermatid elongation (step 9 in mice), whereas proteins required for the remaining steps (steps 9–16 in mice) of sperm assembly must be translated using mRNAs synthesized prior to transcriptional shutdown.9 Therefore, thousands of transcripts are pre-synthesized in late pachytene spermatocytes and round spermatids (steps 1–7) and then stored in ribonucleoprotein particles (RNPs) until translation in elongating/elongated spermatids when needed. For example, it has been shown that Prm1 mRNAs can be stored for up to 2 weeks before translation.10 The current concept ascribes the prolonged stability of mRNAs to their physical confinement to RNPs, a cytoplasmic sub-compartment known as the RNA processing and storage center.9,11 It is widely believed that these translationally suppressed mRNAs are associated with RNA-binding proteins (RBPs) and small RNAs, which are highly enriched in RNPs and thus, may protect mRNAs from degradation or detrimental precocious translation.12,13 During the last several steps of spermiogenesis, a quick protein turnover is required for efficient sperm assembly in spermiogenesis.14,15 Global shortening of mRNA transcripts during late spermiogenesis has been postulated as a mechanism to ensure efficient translation, because these shorter transcripts have much shorter 3′UTRs, thus containing much fewer binding sites for RBPs and small RNAs.6,7,13 This is opposite to the global transcript lengthening events in brain because longer 3′UTRs in brain mRNAs enable much more complicated posttranscriptional regulation due to more binding sites for RBPs and small RNAs.16 Alternative polyadenylation (APA) has been regarded as the mechanism underlying the production of shortened transcripts in late pachytene spermatocytes and round spermatids.17 However, such an APA factor has not been identified. Recently, we and others have demonstrated that UPF2, a critical factor involved in the nonsense-mediated mRNA decay pathway, can selectively degrade longer transcripts during spermiogenesis,6,7,18 suggesting that selective degradation of longer transcripts, in addition to increased synthesis of shorter transcript, represents an alternative mechanism for achieving global shortening of transcripts in late spermiogenesis. Nevertheless, the increased production of shorter transcripts requires enhanced alternative splicing, which has been linked to a common mRNA modification, called N6-methyladenosine, or m6A.19 It has been shown that elevated levels of m6A on pre-mRNAs leads to more splicing events in cultured cells.20,21 Our recent report demonstrated that longer transcripts tend to contain more m6A compared with shorter ones in pachytene spermatocytes, round, and elongating spermatids.22 Correct splicing for the production of longer transcripts requires significant removal of m6A by ALKBH5, a known m6A eraser in the nucleus of spermatocytes and round spermatids. Inactivation of Alkbh5 leads to higher levels of m6A, causing enhanced splicing and production of numerous shorter transcript isoforms with elevated levels of m6A, which undergo rapid degradation in the cytoplasm of elongating/elongated spermatids.22 Therefore, m6A appears to regulate splicing events in the nucleus and to control mRNA stability in the cytoplasm in pachytene spermatocytes and round and elongating spermatids.22
Clearly, the regulation of gene expression in haploid male germ cell development involves two cellular events: stabilization of mRNAs for delayed translation and enhanced splicing. Both have been linked to the biogenesis and function of a unique class of regulatory RNAs, circular RNAs (circRNAs).23 CircRNAs are RNA circles mostly representing back spliced, exon-containing pre-mRNA fragments circularized by a covalent, 3′,5′-phosphodiester bond.24 These RNA circles used to be regarded as byproducts of erroneous splicing without specific functions. However, this view is no longer valid because thousands of circRNAs have been identified in various tissues of almost all species, and their evolutionary conservation, tissue- and developmental stage-specific expression patterns and highly variable abundance between circRNAs and their corresponding linear forms in the same tissues,25–27 all suggest that circRNAs are purposely synthesized with regulatory functions. Indeed, studies showing circRNA functions have started emerging, e.g., several circRNAs derived from intergenic regions have been found to act as miRNA sponge28–31; a gene knockout (KO) study demonstrated that a circRNA, Cdr1as plays a critical role in regulating brain function.32 Recent studies show that some circRNAs have coding potential and can be translated into proteins in Drosophila, mice and humans.33–38 Despite the progress, little is known regarding factors that affect circRNA biogenesis and their roles in physiological contexts. Here, we report that circRNA levels increase while their corresponding linear forms actively undergo degradation in haploid male germ cells with progression of spermatogenesis. Junction sequences of these circRNAs appear to be enriched for m6A, which is usually located around the start and stop codons in linear mRNAs.22 Consequently, ~50% of these spermiogenesis-enriched circRNAs contain large open reading frames (ORFs) with m6A-modified start codons in their junctions, which can indeed be translated into proteins in both spermatogenic cells and spermatozoa. Our findings suggest that m6A-dependent circRNA accumulation with the progression of spermatogenesis may function to compensate for massive degradation of linear mRNAs in late spermiogenesis so that stable and long-lasting production of proteins in the last several steps of spermiogenesis and in spermatozoa is maintained.
Results
CircRNAs are abundantly expressed in male germ cells during spermatogenesis
To define the entire large RNA transcriptome in purified murine spermatogenic cells, we generated large RNA libraries with and without RNase R treatment and performed RNA-seq with greater depth for two reasons: First, we would like to see whether RNA-seq without RNase R treatment would lead to a higher false positive rate in circRNA annotation. Second, to normalize the circRNA/junction reads, we need to use total linear RNA reads as the internal control. Once treated with RNase R, this normalization method would not be appropriate because the efficiency of RNase R treatment is often affected by many factors (e.g., temperature, digestion time, and enzymatic activities, etc.) and thus, is uncontrollable. To enhance circRNA discovery, we also adopted a recently reported circRNA enrichment method called RNase R treatment followed by polyadenylation and poly(A) + RNA Depletion (RPAD).39 The RPAD method selectively eliminates most of the linear mRNAs, thus enriching circRNAs from total RNA for library construction and deep sequencing (Supplementary information, Fig. S1a).
The overall expression patterns of circRNAs in three spermatogenic cell types (pachytene spermatocytes, round, and elongating spermatids) were similar between the two conventional RNA-seq methods (Fig. 1a, and Supplementary information, Fig. S2). A total of 65,500 circRNAs were annotated from RPAD-seq data (Supplementary information, Table S1; the raw data have been deposited into the CNGB database with accession# CNP0000637). Of importance, ~80% of the circRNAs identified from the regular RNA-seq data could be verified in the RPAD-seq data (Supplementary information, Table S2), suggesting that the false positive rates are similar between both methods. Considering proper normalization method and limited starting materials available, we decided to mainly use circRNAs identified from conventional RNA-seq data (without RNase R treatment) for all the analyses reported below.
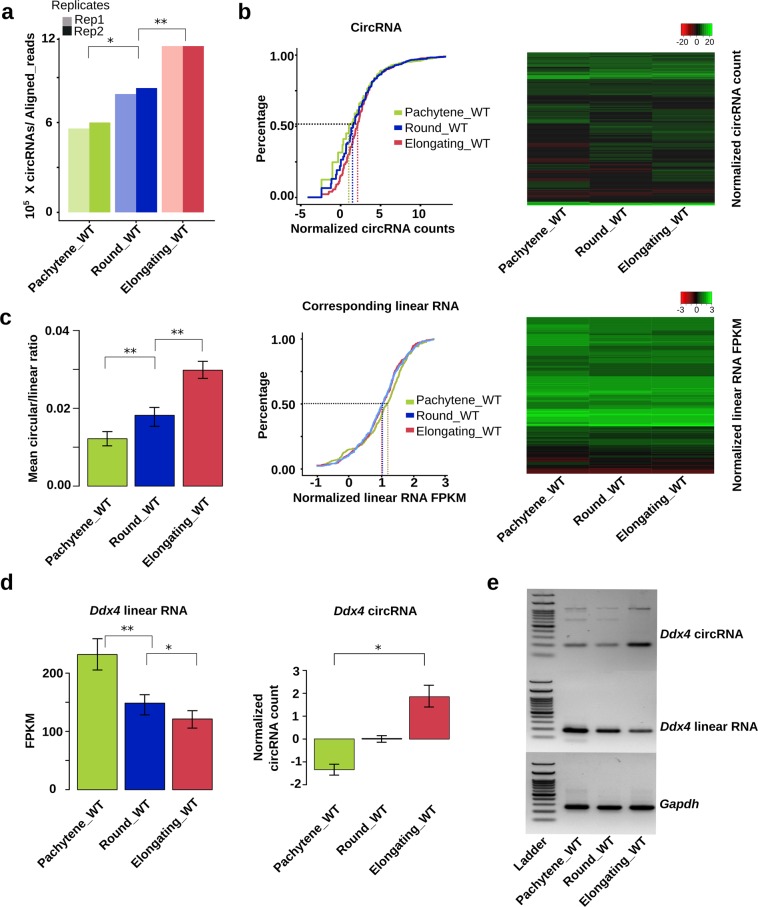
Fig. 1. CircRNA levels increase with progression of spermiogenesis.
a CircRNA expression in wild type (WT) pachytene spermatocytes, round, and elongating spermatids. CircRNA reads were identified from RNA-seq data, followed by normalization against aligned total RNA reads. b CircRNA levels increase while levels of their linear isoforms decrease when WT pachytene spermatocytes develop to round and elongating spermatids. Accumulation curves and heat maps showing normalized circRNA counts and normalized linear RNA levels (FPKM) are used to compare levels of circRNAs and their linear isoforms in WT pachytene spermatocytes, round, and elongating spermatids. c The circular/corresponding linear RNA ratio increases from WT pachytene spermatocytes, to round and elongating spermatids. Normalized counts of circRNAs and their linear isoforms are used to calculate the ratios and to generate the bar graphs. d Ddx4 linear and circRNAs display opposite expression patterns in WT pachytene spermatocytes, round, and elongating spermatids. Normalized circRNA counts and normalized linear RNA levels (FPKM) are compared in WT pachytene spermatocytes, round, and elongating spermatids. e A representative result of semi-quantitative qPCR analyses for levels of Ddx4 circular and linear RNAs in WT pachytene spermatocytes, round, and elongating spermatids. Gapdh was used as a loading control. The Student’s t test was used for statistical analyses. *P < 0.1; **P < 0.05; ***P < 0.001. The data were presented as two biological replicates or means ± SD, biological replicates, n = 2. Sums of two technical repeats in each of the biological replicates were used for analyses.
CircRNA levels increase with progression of spermiogenesis
Accumulating data suggest that the global shortening of mRNA transcripts during spermiogenesis is achieved through both alternative usage of polyadenylation signals and selective degradation of longer 3′UTR transcripts in spermatids.6,17,18 It is also known that circRNA production positively correlates with the splicing activity.26,27,40 Given that progression of spermatogenesis from late meiotic to haploid phases involves enhanced alternative splicing,17,41 we first tested whether circRNA biogenesis also increases from pachytene spermatocytes to round/elongating spermatids. Indeed, we found that both the number (Fig. 1a) and relative abundance (Fig. 1b, upper two panels) of unique circRNAs increased drastically when pachytene spermatocytes developed into round/elongating spermatids (Supplementary information, Table S1). Interestingly, levels of their corresponding linear forms decreased from pachytene spermatocytes to round/elongating spermatids (Fig. 1b, lower two panels). Consistently, the circular/linear ratio increased significantly during the same period of spermatogenesis (Fig. 1c). We experimentally validated this finding by examining levels of both the linear and circular forms of three genes (Ddx4, Spag16, and Trim37), which are all known to be essential for spermatogenesis42 (Fig. 1d, e; Supplementary information, Fig. S3). These data suggest that levels of circRNAs increase while their linear precursors undergo degradation from pachytene spermatocytes and round spermatids to elongating spermatids. Gene ontology (GO) term enrichment analyses revealed that the host genes of these circRNAs were involved in critical events during spermiogenesis, e.g., epigenomic modulations (DNA methylation, histone modifications, and nucleosome organization) and sperm motility (cilium movement, axoneme assembly, and microtubule bundle formation) (Supplementary information, Fig. S4). circRNAs have a much longer half-life than linear RNAs because they are resistant to degradation due to a lack of free ends.43 Given that linear RNAs, especially those with longer 3′UTRs, are actively undergoing degradation in late spermiogenesis,6,13 the increased levels of circRNAs may represent a mechanism through which certain transcripts of particular importance are preserved for protein production at a later time point during spermiogenesis.
CircRNA accumulation correlates with enhanced splicing at m6A sites
Recent data suggest that m6A is involved in the regulation of alternative splicing in both somatic21,44 and male germ22 cells. In our recent report, we have demonstrated that longer mRNA transcripts that are normally expressed in WT pachytene spermatocytes and round spermatids are spliced into shorter transcripts when higher levels of m6A are present in those longer transcripts due to inactivation of ALKBH5, an eraser of m6A.1,22 This is consistent with other data showing that pre-mRNAs with elevated m6A levels appear to bind the spliceosome more tightly, thus leading to enhanced splicing.44 Given that the global shortening of mRNA transcripts during late spermiogenesis (steps 9–16) results from enhanced splicing,17 m6A levels should increase while ALKBH5 levels should decrease if m6A is involved in this process. Indeed, based on our RNA-Seq analyses on purified WT spermatogenic cells, Alkbh5 mRNA levels decreased from pachytene spermatocytes to round/elongating spermatids (Fig. 2a). This pattern is consistent with immunofluorescence staining of adult testes, showing that ALKBH5 protein is abundantly expressed in pachytene spermatocytes and round spermatids, but becomes undetectable in elongating spermatids.22 More interestingly, more circRNAs were identified in Alkbh5 KO testes (~29,000 circRNAs) than in WT control testes (~19,000 circRNAs), supporting the connections among m6A levels, splicing, and circRNA biogenesis (Supplementary information, Table S3). To further confirm that ALKBH5 affects circRNA production, we conducted in vitro Alkbh5 knockdown and minigene reporter assays as reported previously.45,46 Sltm was chosen because both its circular and linear forms were abundantly expressed in the spermatogenic cell types analyzed in our RNA-seq analyses (Supplementary information, Table S2). Using junction PCR, levels of Sltm circRNAs were found to be drastically upregulated, whereas levels of its linear form remained unaffected when Alkbh5 levels were reduced through siRNA-mediated knockdown in NIH 3T3 cells (Supplementary information, Fig. S5a). The data suggest that inhibition of ALKBH5 indeed enhances circRNA production most likely through enhanced splicing, as reported previously.22,47 However, in the minigene reporter assays, no enhanced production of Sltm circRNAs was observed in HEK293 cells (Supplementary information, Fig. S5b). The negative finding is most likely due to the fact that the splicing mechanisms in HEK293 cells are different from those utilized in spermatogenic cells.
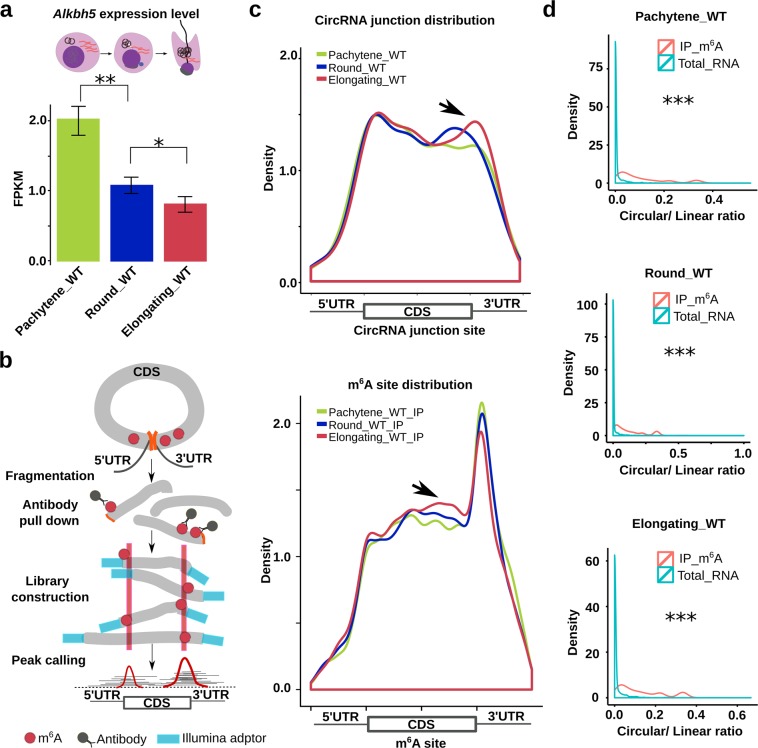
Fig. 2. CircRNA accumulation correlates with enhanced splicing at m6A sites.
a Alkbh5 mRNA levels decrease from pachytene spermatocytes to round and elongating spermatids. Normalized counts of Alkbh5 (FPKM) were used to represent mRNA levels and data are presented as means ± SD, biological replicates, n = 3. b Schematics of circRNA identification from m6A-RIP-seq data. Briefly, RNAs are fragmentized and pulled down using m6A antibodies. Sequencing adapters are added during library construction. After deep sequencing, circRNAs are identified from the RIP-seq data based on junction reads using CIRI. c Density plots showing circRNA junction reads increase around 3′UTRs (upper panel) (biological replicates, n = 2), while this increase coincides with elevated m6A levels nearby (lower panel) (biological replicates, n = 3). Total RNA-seq and m6A-RIP-seq data were used to determine the distribution of circRNA junction reads and m6A sites, respectively. Sequencing was performed in biological triplicates (n = 3). Normalized, relative density is presented on the y-axis, whereas read distribution across 5′UTR, CDS, and 3′UTR is shown on the x-axis. d The circular/linear ratio was much higher in m6A-RIP-seq reads than that in total RNA-Seq reads, suggesting the increased circRNAs during spermiogenesis are enriched with m6A (biological replicates, n = 3). The Student’s t test was used for statistical analyses. *P < 0.1; **P < 0.05; ***P < 0.001.
To further establish the relationship between circRNA production and m6A levels, we conducted m6A RNA immunoprecipitation followed by deep sequencing (m6A-RIP-seq) analyses (Fig. 2b). By mapping the sequence reads to the longest RNA isoform for each gene, we calculated the m6A enrichment score and located m6A sites based on constitutive 100 nt windows, as previously reported.22 From pachytene spermatocytes to round and elongating spermatids, the circRNA junction reads became increasingly enriched near the stop codon (Fig. 2c, upper panel), which correlated well with the increased m6A levels around the same region (Fig. 2c, lower panel), suggesting a link between elevated m6A and circRNA levels. By analyzing circRNA abundance and m6A levels, we found that the more abundant the circRNAs were, the higher m6A levels they contained (Supplementary information, Fig. S6b). Such a positive correlation strongly suggests that circRNAs are spliced from m6A-enriched sites. If this notion is correct, then the m6A immunoprecipitation products should contain abundant circRNAs. Indeed, by annotating circRNAs using the m6A-RIP-seq reads, we observed that the circular/linear ratio increased from pachytene spermatocytes to round and then to elongating spermatids (Supplementary information, Fig. S6a). The circular/linear ratio was significantly higher in m6A-RIP-seq reads than that in total RNA-seq reads, suggesting that the junction regions of circRNAs are significantly enriched for m6A (Fig. 2d). We then divided the junction reads into the head (close to the 5′ end) and tail (close to the 3′ end) fragments, followed by mapping them against their linear mRNAs. Data from this analysis revealed that these m6A-IP-enriched circRNAs contained ORFs of variable lengths, with the majority possessing the start codon and some even with both the start and stop codons (Supplementary information, Fig. S6c). Together, these data imply that circRNA levels increase while the levels of their corresponding linear forms decrease (due to massive degradation) in elongating and elongated spermatids, and that back splicing tends to occur between m6A-enriched sites, which are usually around the start and stop codons of linear mRNAs in spermatogenic cells.22
A subgroup of circRNAs contain ORFs with m6A-modified start codons
Only a few circRNAs derived from intergenic regions have been shown to act as miRNA sponge,31 whereas the majority of the circRNAs spliced from coding genes contain ORFs and thus, have protein-coding potential.33,34 A recent report has demonstrated that some ORF-containing circRNAs can be efficiently translated into proteins using the m6A-modified start codon as the internal ribosome entry site (IRES), which can be recognized by YTHDF3, an m6A reader.35,48 Accumulation of circRNAs from pachytene spermatocytes to round spermatids coincides with the commencement of transcriptional cessation and massive degradation of mRNA transcripts, especially those with longer 3′UTRs, in elongating/elongated spermatids.6,13,22 If these RNA circles can be translated into proteins, then turning linear mRNAs into circRNAs with coding capability would represent an ideal mechanism to bypass the massive mRNA degradation and to maintain the continuous production of certain proteins that are of particular importance for late spermiogenesis. To test this hypothesis, we examined whether these circRNAs contain ORFs and m6A-modified start codons, and whether they are associated with polyribosomes in round and elongating spermatids.
CircRNAs accumulated during late meiotic and early haploid phases of spermatogenesis (pachytene spermatocytes→round spermatids→elongating spermatids) were predominantly derived from exons (Fig. 3a). Interestingly, by mapping the head and tail junction reads to the full-length host linear mRNAs, we observed that the junctions appeared to be enriched in both start and stop codons (arrows pointing to the two peaks in Fig. 3b), suggesting these circRNAs contain potentially full-length or partial ORFs. Furthermore, levels of these ORF-containing circRNAs seemed higher in elongating spermatids than those in pachytene spermatocytes (Fig. 3b). If these circRNAs are translatable, then it means that partial or full-length proteins can be produced in elongating spermatids despite the massive degradation of linear mRNAs. More interestingly, we also observed that circRNAs with higher circular/linear ratios (i.e., circRNAs as the main output of transcription) contained more intact ORFs compared with those with lower circular/linear ratios (i.e., linear RNAs as the main output of transcription) (Fig. 3c). These data suggest that those ORF-containing circRNAs can gradually substitute their linear forms, probably for maintaining continuous protein production even after the degradation of linear mRNAs in elongating and elongated spermatids (steps 9–16).
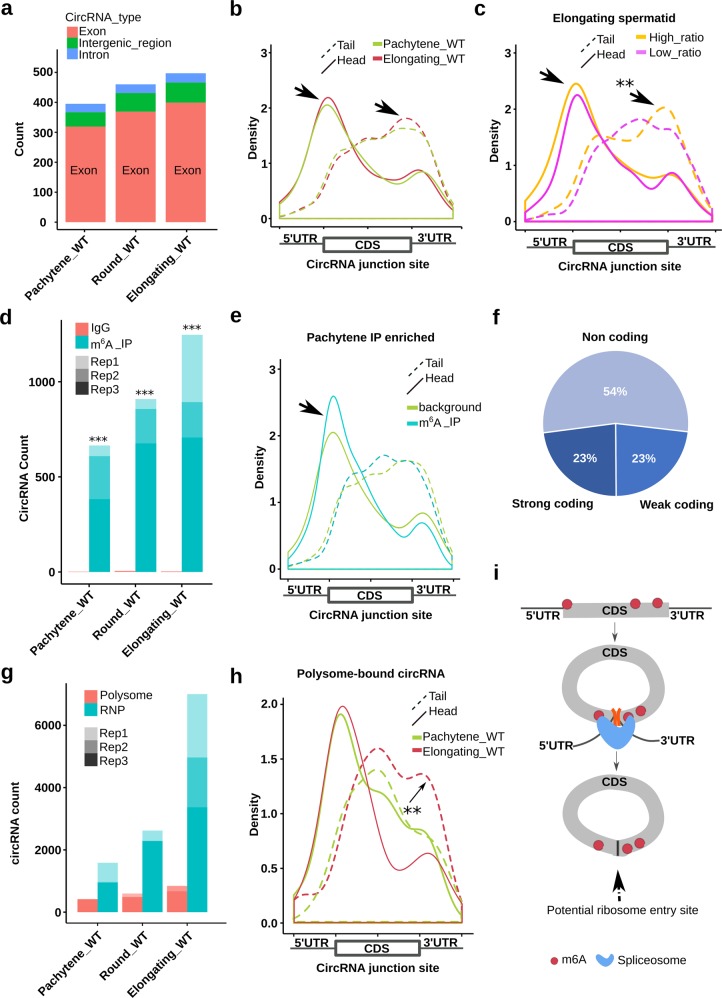
Fig. 3. A subgroup of circRNAs contain ORFs with m6A-modified start codons.
a Exonic circRNAs are the majority in WT pachytene spermatocytes, round, and elongating spermatids. Normalized average circRNA counts derived from exons, introns, and intergenic regions are presented (biological replicates, n = 2). b Density plots showing that circRNAs in elongating spermatids have more coding potential than those in pachytene spermatocytes. The plots were generated by mapping the junction reads containing the 3′ (head) and 5′ (tail) ends of their host mRNAs to the full-length mRNAs. Compared with circRNAs in pachytene spermatocytes, those in elongating spermatids display more junction sequences containing the start and stop codons, suggesting higher coding potential for full-length proteins (biological replicates, n = 2). c Density plots showing that circRNAs with higher circular/linear ratios contain both start and stop codons more often and thus, have higher coding potential. Density plots were generated by mapping junction reads of circRNAs expressed in elongating spermatids against their full-length host mRNAs. The cutoffs for higher and lower circular/linear ratio were >0.2 and <0.1, respectively (biological replicates, n = 2). d CircRNAs that contain m6A were detected in WT pachytene spermatocytes, round and elongating spermatids. Data represent circRNA counts in m6A-RIP-seq data of the three types of spermatogenic cells. m6A-RIP-seq was performed in triplicates and IgG was used as negative controls (biological replicates, n = 3). e Density plots showing that pachytene circRNAs with junction sites containing the start codon tend to have higher levels of m6A. The plots were generated by mapping the head (containing the 5′ ends of their host mRNAs) or tail (containing the 3′ ends of their host mRNAs) junction reads of m6A IP-enriched pachytene circRNAs to the full-length mRNAs. Because IgG could only pull down some junction reads, we used the RNA-seq data (non-IP) as background control (biological replicates, n = 3). f Pie chart showing distribution of circRNAs with or without coding potential in the three types of spermatogenic cells. g Bar graphs showing levels of circRNAs associated with polysomes and RNP in WT pachytene spermatocytes, round and elongating spermatids. Data represent normalized circRNA counts in RNA-seq analyses of RNP-/polysome-associated RNA (biological replicates, n = 3). h Density plots showing that polysome-associated circRNAs tend to contain the start/stop codon more often in elongating spermatids than those in pachytene spermatocytes. Density plots were generated by mapping the head (containing the 5′ ends of their host mRNAs) or tail (containing the 3′ ends of their host mRNAs) junction reads of polysome-associated circRNAs to the full-length mRNAs (biological replicates, n = 3). i A proposed model depicting how m6A near the start and stop codons induces circRNA production and translation. m6A near 5′UTR and 3′UTR tends to recruit spliceosome, leading to back splicing and circRNA production. Junction m6A in the start codon may serve as the IRES for translation initiation. The Student’s t test was used for statistical analyses. *P < 0.1; **P < 0.05; ***P < 0.001.
Next, we looked into whether the junction sites of these circRNAs are enriched for the m6A-modified start codon, which has been shown to serve as the IRES for translational initiation.35,48 m6A-RIP-seq analyses on purified spermatogenic cells revealed that the number of unique circRNAs increased from pachytene spermatocytes to round and elongating spermatids, and peaked in elongating spermatids (Fig. 3d); this pattern is consistent with the increased splicing events and elevated levels of m6A, as discussed earlier (Fig. 2). Further mapping of the head (5′ end) and tail (3′ end) junction reads against the host genes revealed an enrichment of m6A around the start codon (Fig. 3e; Supplementary information, Fig. S7), indicating that these circRNAs indeed contain m6A-modified start codons. Given that the m6A-modified start codon can be recognized by YTHDF3 and acts as the IRES for translational initiation,35,48 this finding suggests that this subgroup of circRNAs may be translatable. To further support this notion, we combined all of the sequencing reads (~300 Gb) from the conventional RNA-seq and assembled the full-length sequences for 68 circRNAs. Interestingly, approximately a half of these circRNAs were predicted to have coding potential (Fig. 3f). Similarly, full-length sequences of 2949 circRNAs were assembled using all of RPAD-seq reads, and 1427 (~48%) were predicted to have coding potential (Supplementary information, Fig. S1b). The majority of these circRNAs were predicted to have coding potential in all three spermatogenic cell types as well as sperm (Supplementary information, Fig. S1c).
Association with polyribosomes is another indication of potential translation.48 Indeed, circRNAs were detected in both RNPs and polysomes (pooled heavier fractions) with increasing levels from pachytene spermatocytes to round/elongating spermatids (Fig. 3g). Levels of RNP-associated circRNAs were always greater than those of circRNAs associated with polysomes, and the highest levels of circRNAs were found in elongating spermatids (Fig. 3g). The distribution patterns are consistent with the fact that transcripts subjected to delayed translation are sequestered into RNPs during spermiogenesis.13 By pair mapping the head and tail junction reads to the host genes, we observed that the circRNAs associated with polysomes appeared to contain intact ORFs in both round and elongating spermatids (Supplementary information, Fig. S8), further supporting the notion that these circRNAs are made for translation. Moreover, polysome-bound circRNAs appeared to contain more intact ORFs in elongating spermatids than those in pachytene spermatocytes, suggesting enhanced coding potential in elongating spermatids (Fig. 3h). Taken together, these data indicate that the circRNAs enriched in late meiotic (spermatocytes) and haploid (spermatids) male germ cells mostly contain both intact ORFs and m6A-modified start codons (Fig. 3i). Given that the host linear transcripts of these circRNAs are undergoing massive degradation in elongating/elongated spermatids, it is likely that these circRNAs are made to maintain a continuous production of proteins, which may be critical for late spermiogenesis and sperm function.
Spermiogenesis-enriched circRNAs shift from RNPs to polyribosomes with progression of spermiogenesis
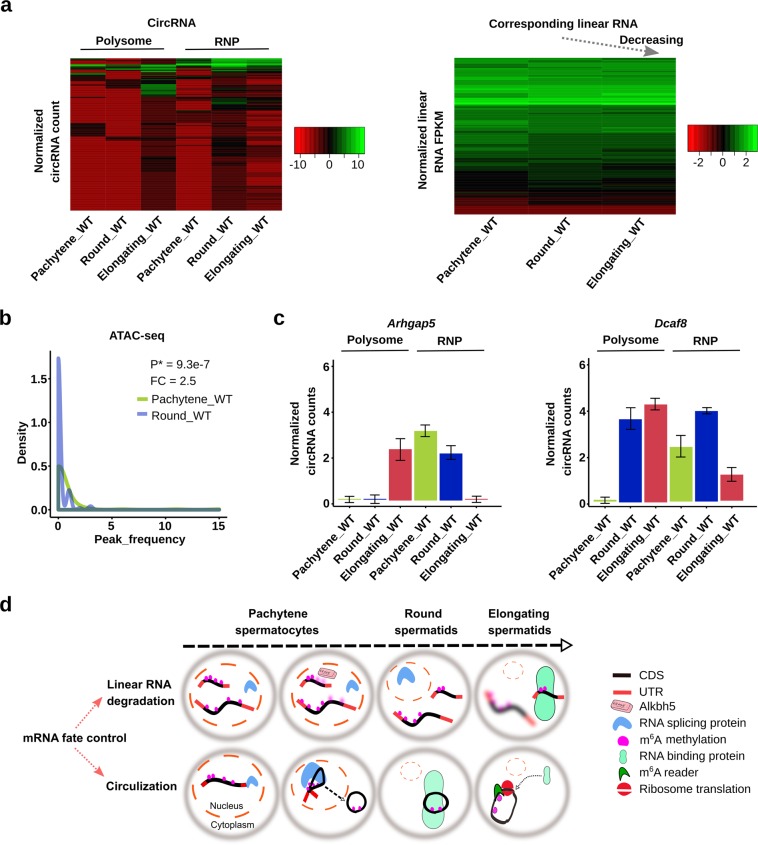
Delayed translation in spermiogenesis allows for continuous supply of proteins in the absence of de novo transcription, which has been shown to be essential for late sperm assembly (steps 9–16 spermatids in mice).9 By analyzing circRNA abundance, we observed that those RNP-enriched circRNAs in round spermatids shifted from RNPs to the polysomes when round spermatids developed into elongating spermatids (Left panel, Fig. 4a and Supplementary information, Table S4), suggesting a transition from a non-translational state in round spermatids to an active translational state in elongating spermatids. Meanwhile, levels of their corresponding linear RNAs continuously decreased due to massive degradation of mRNAs (Fig. 4a, right panel). ATAC-seq relies on transposase accessibility and thus, can be used to detect transcriptional status (open vs. closed chromatin). More transposon peaks indicate higher transcriptional activity/open chromatin. Using published ATAC-seq data on murine spermatogenic cells,49 we mapped the reads representing those regions, from which the circRNAs appear to be shifting from RNPs to polysomes in pachytene spermatocytes and round spermatids (Fig. 4a, left panel), to the genome. We found that regions from which those circRNAs were derived had changed from open to closed chromatin states when pachytene spermatocytes developed into round spermatids (Fig. 4b), suggesting that the circRNAs tend to be produced from loci that are destined to become transcriptionally inactive. Given that transcription remains active in round spermatids, circRNA production from these “pre-silenced” regions strongly suggests that these circRNAs are produced to compensate for the upcoming transcriptional shutdown and massive linear mRNA degradation. Two mRNAs known to display delayed translation (Arhgap5 and Dcaf8) during spermiogenesis were examined and both showed a significant shift of their circular forms from RNPs to polysomes when pachytene spermatocytes developed into round and then elongating spermatids (Fig. 4c). GO term enrichment analyses on those polysome-enriched circRNAs in elongating spermatids revealed that their corresponding host genes were mostly related to flagellar development (e.g., cilium, centriole microtubules, etc.) and cellular structural organization (e.g., cytoskeleton, cell projection, etc.) (Supplementary information, Fig. S9), the most active cellular events in the last several steps of sperm assembly.50,51
Fig. 4. Spermiogenesis-enriched circRNAs shift from RNPs to polyribosomes with progression of spermiogenesis.
a Heat map showing round spermatid RNP-associated circRNAs being shifted and loaded onto polysomes in elongating spermatids (left panels), while levels of their corresponding linear RNAs continuously decrease from pachytene spermatocytes to round and elongating spermatids (right panels). Normalized circRNA counts and normalized linear RNA FPKM represent levels of circRNAs and their corresponding linear transcripts, respectively (biological replicates, n = 3). b Density plots showing that regions from which polysome-enriched circRNAs were derived had changed from open to closed chromatin structures when pachytene spermatocytes developed into round spermatids based on ATAC-seq analyses (biological replicates, n = 3). c Levels of Arhgap5 and Dcaf8, two mRNAs known to display delayed translation, in RNPs and polysomes of pachytene spermatocytes, round, and elongating spermatids. Data represent normalized circRNA counts in RNA-seq analyses of RNP-/polysome-associated RNA (biological replicates, n = 3). d Schematics depicting the role of m6A in mRNA fate control in late meiotic and haploid phases of spermatogenesis. On one hand, higher m6A levels in mRNAs lead to degradation in the cytoplasm of elongating spermatids. Since m6A is mostly located in 3′UTRs, the longer transcripts tend to contain higher levels of m6A because of the longer 3′UTRs and thus, are more prone to degradation compared with shorter transcripts (upper panel). This is a known means to achieve global transcript shortening during late spermiogenesis. On the other hand, the more m6A the transcripts contain, the more they are subjected to splicing, leading to increased circRNA production. Consequently, a large number of circRNAs can be produced from longer transcripts. These RNA circles become associated with RNPs and later loaded onto polysomes for translation (lower panel). This represents an alternative way to protect longer transcripts form massive global degradation events during late spermiogenesis because these proteins remain needed for the last several steps of sperm assembly.
Based on the analyses above, we propose that m6A-directed circRNA biogenesis participates in the regulation of delayed translation during spermiogenesis (Fig. 4d). One way to achieve delayed translation is to lower m6A levels in mRNAs by ALKBH5.22 Since m6A is mostly located in 3′UTRs, the longer transcripts tend to contain more m6A sites because of the longer 3′UTRs and thus, are more prone to degradation compared to shorter ones22 (Fig. 4d). In this way, the overall length of transcripts becomes gradually decreased.13,22 However, the more m6A sites the transcripts contain, the more they are subjected to splicing,20,52 which, in turn, leads to enhanced circRNA production. Consequently, a large number of circRNAs would be produced from those longer transcripts. These RNA circles then become associated with RNPs and are later loaded onto polysomes for translation. This may represent an alternative means to protect the transcripts from massive global degradation events during late spermiogenesis because these proteins are still needed for the last several steps of spermiogenesis.
ALKBH5 and METTL3 affect circRNA biosynthesis through modulating m6A levels
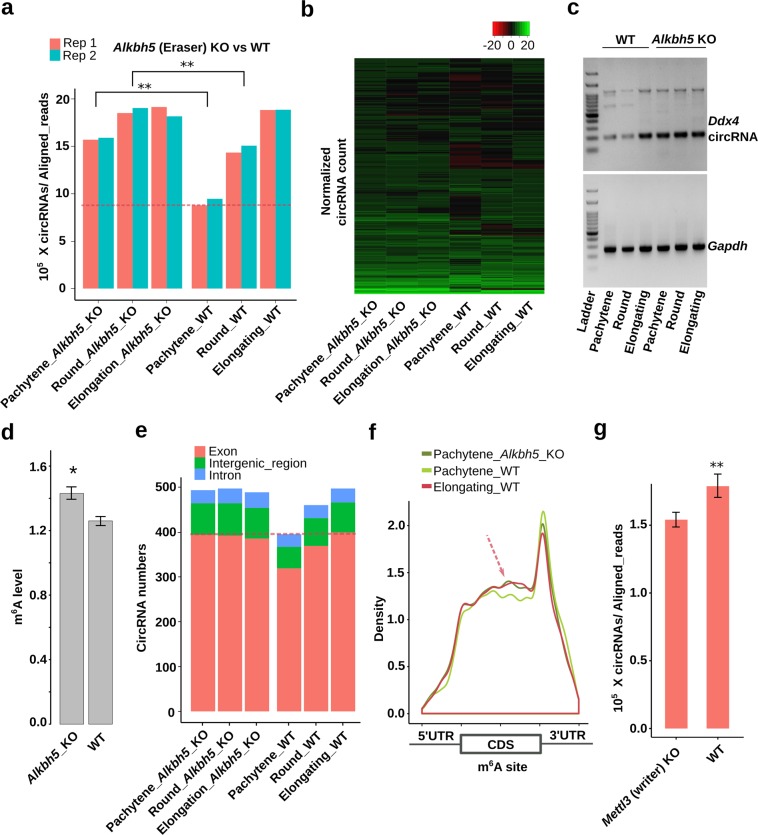
To establish the cause–effect relationship between m6A and circRNAs, we analyzed data from Alkbh5- and Mettl3-null testes.47,53,54 As an eraser of m6A, ALKBH5 has been shown to play a critical role in spermatogenesis.47 Specifically, Alkbh5-null spermatogenic cells display significant higher m6A levels than the wild-type controls.22,47 Since elevated m6A levels correlate with increased splicing,22 which may enhance circRNA biogenesis, we examined the relative abundance of unique circRNAs in Alkbh5-null spermatogenic cells (Fig. 5a). Indeed, instead of a steady increase from spermatocytes to round and elongating spermatids in WT testes, circRNAs levels were uniformly higher among all the three types of Alkbh5-null spermatogenic cells analyzed (Fig. 5a, Supplementary information, Fig. S10a and Table S3). It is noteworthy that the Alkbh5-null pachytene spermatocytes expressed similar levels of circRNAs as compared with wild-type elongating spermatids, which express no or minimal amount of ALKBH5.22 Consistently, quantitative analyses revealed that circRNA levels already peaked in Alkbh5-null pachytene spermatocytes, and the drastically elevated levels persisted in round and elongating spermatids (Fig. 5b). This altered expression pattern was also confirmed by semi-quantitative RT-PCR analyses and RNAseq on Ddx4 circRNAs (Fig. 5c and Supplementary information, Fig. S10b). Consistent with our notion that elevated m6A levels promote circRNA biogenesis, circRNAs in Alkbh5-null spermatogenic cells displayed greater m6A levels than those in the wild-type cells (Fig. 5d). Since enhanced splicing promotes circRNA formation, we then examined levels of circRNAs and their corresponding linear forms. Indeed, we observed significantly upregulated circular/linear ratios in Alkbh5-null spermatogenic cells compared with WT controls (Supplementary information, Fig. S10c). Although circRNA accumulation represented a common trend shared between WT and Alkbh5-null spermatogenic cells, the differences among the three cell types appeared to be much larger in WT cells than those in Alkbh5-null male germ cells (Supplementary information, Fig. S10c), suggesting that elevated m6A levels enhance circRNA production. For example, Gbe1 and Oxr1, both are essential for spermiogenesis,42 were marked with higher levels of m6A on the start and stop codons, and formed circRNAs with the full-length ORF in Alkbh5-null pachytene spermatocytes (Supplementary information, Fig. S10d). The increased circRNA types in Alkbh5-null pachytene spermatocytes were mainly from exons, and the abundance was close to that in WT elongating spermatids (Fig. 5e). Although the m6A distribution pattern in Albkbh5-null pachytene spermatocytes was similar to that in WT elongating spermatids, it was drastically different from that in WT pachytene spermatocytes (Fig. 5f and Supplementary information, Fig. S11). These data indicate a precocious circRNA surge in preparation for delayed translation due to increased m6A levels in Alkbh5-null spermatogenic cells. METTL3 functions as an m6A writer, and a lack of METTL3 has been shown to cause spermatogenic arrest at the meiotic phase and male infertility.53,54 If m6A levels affect circRNA production, then circRNA biogenesis in Metll3-null spermatogenic cells should be downregulated. Indeed, we identified much fewer circRNAs in Mettl3-null testes at postnatal day 12, in which the most advanced spermatogenic cells are pachytene spermatocytes (Fig. 5g and Supplementary information, Table S5). Together, these data further support the notion that m6A affects circRNA production in meiotic and haploid male germ cells.
Fig. 5. ALKBH5 and METTL3 affect circRNA generation through modulating m6A levels.
a CircRNA levels in Alkbh5 KO and WT pachytene spermatocytes, round, and elongating spermatids. CircRNAs were identified from RNA-seq data in two biological replicates (n = 2), followed by normalization against aligned total RNA reads. Note that the circRNA levels in Alkbh5-null pachytene spermatocytes and round spermatids are similar to those in WT or Alkbh5 KO elongating spermatids, and are much higher than those in WT pachytene spermatocytes and round spermatids. b Heatmap showing circRNA expression levels, as represented by normalized circRNA counts in RNA-seq data (biological replicates, n = 2), in Alkbh5 KO and WT pachytene spermatocytes, round, and elongating spermatids. Note that the circRNA expression patterns in all three types of Alkbh5-null spermatogenic cells are similar to those in WT elongating spermatids. c A representative semi-qPCR gel image showing much higher levels of Ddx4 circRNAs in Alkbh5 KO than in WT pachytene spermatocytes, round and elongating spermatids. Gapdh was used as a loading control. Two technical repeats were performed with similar results. d m6A levels in circRNA junction sequences are higher in Alkbh5-null than those in WT spermatogenic cells. The m6A peak-searching was performed around the junction region and m6A levels were determined based on how many times a specific m6A site was detected in three replicates of experiments (1–3). Data are presented as means ± SD, biological replicates, n = 3, *P < 0.05. e Exonic circRNAs are differentially expressed in WT, but not in Alkbh5 KO pachytene spermatocytes, round, and elongating spermatids. Normalized average circRNA counts derived from exons, introns and intergenic regions were calculated based on RNA-seq data in two biological replicates (n = 2). f Density plots showing the distribution of m6A sites in Alkbh5 KO and WT pachytene spermatocytes, as well as WT elongating spermatids. Note that the m6A profile in Alkbh5 KO pachytene circRNAs is similar to that in WT elongating spermatid circRNAs, but very different from that in WT pachytene circRNAs (arrow). m6A sites were determined based on peak searching in the m6A-RIP-seq data in triplicates (biological replicates, n = 3). g Levels of circRNAs in WT and Metll3 KO testes at postnatal day 12, when the most advanced spermatogenic cells are pachytene spermatocytes. Normalized circRNA counts were calculated based on RNA-seq data in duplicates (biological replicates, n = 2) and data are presented as means ± SD. Please note that all the values are log-transformed, and the Student’s t test was used for statistical analyses. *P < 0.1; **P < 0.05; ***P < 0.001.
Sperm carry abundant, evolutionarily conserved circRNAs
Given that circRNAs are much more stable than linear RNAs, we hypothesized that a small amount of circRNAs may be present in spermatozoa. To our surprise, we found that circRNAs were ~ 50-100 times more abundant in spermatozoa than in spermatocytes and spermatids (Fig. 6a and Supplementary information, Fig. S12a). We purified cytoplasmic droplets (CDs), a transient organelle only present in epididymal spermatozoa,52,55 and found that the CDs also contained a large number of circRNAs (Fig. 6a, Supplementary information, Fig. S12b and Table S6). GO analyses revealed that the host genes of these CD circRNAs were enriched in genes involved in sperm mobility and sperm energy metabolism (P < 0.05), whereas the host genes of the whole sperm circRNAs were mostly those involved in chemical modifications of histone and DNA (Fig. 6b and Supplementary information, Fig. S12c). We then analyzed circRNA contents in whole sperm and sperm heads, and the number of unique circRNAs was three times greater in whole sperm than that in sperm heads (Fig. 6c), suggesting that the majority of circRNAs are localized to sperm tail and the connecting piece/neck. We also found that ~30% of the sperm-borne circRNAs could be found in elongating spermatids, suggesting that sperm-borne circRNAs are derived from spermatogenesis (Supplementary information, Fig. S12d).
Fig. 6. Sperm-borne circRNAs are abundant, highly conserved across species and variable with aging and fertility statuses.
a Levels of circRNAs in three types of spermatogenic cells (pachytene spermatocytes, round, and elongating spermatids), whole sperm and cytoplasmic droplets. CircRNA reads were identified from RNA-seq data in triplicates (biological replicates, n = 3 in whole sperm and cytoplasmic droplets), followed by normalization against aligned total RNA reads. b Enriched GO terms for cytoplasmic droplet-enriched and whole sperm-enriched circRNAs. c CircRNA abundance in sperm heads and whole sperm. CircRNA reads were identified from RNA-seq data in biological triplicates (n = 3), followed by normalization against aligned total RNA reads. d Dot plot showing circRNAs are more abundant in young than those in old mouse testes. Normalized FPKM values of circRNAs in old (2-year-old) mouse testes are plotted against those in young (4-month-old) mouse testes (biological replicates, n = 2). The regression line is indicated in blue with gray shading. e Representative semi-qPCR gel images showing levels of the circular and linear forms of two genes (Sept10 and Ttil5) in young and old mouse testes (Technical replicates n = 2, results are similar). f High fertility human sperms contain more circRNAs and less linear RNAs than low fertility human sperms. CircRNA reads were identified from RNA-seq data (biological replicates n = 8 after removing outlier) from whole sperm of healthy donors, followed by normalization against aligned total RNA reads (Upper panel). Normalized counts of circRNAs and their linear isoforms are used to calculate the ratios and to generate the accumulation curve (lower panel). g Venn diagram showing sperm circRNAs are conserved across species. Unique circRNAs in rat, human, and rabbit sperm were identified from RNA-seq data (biological replicates, n = 2–3) of whole sperm, followed by normalization against aligned total RNA reads. h Significantly enriched GO terms in differentially expressed circRNAs between high and low fertility human sperms. GO terms related to nuclear functions are in orange, whereas those involved in sperm motility are in gray. The Student’s t test was used for statistical analyses. *P < 0.1; **P < 0.05; ***P < 0.001.
Since earlier reports have shown that circRNAs accumulate with age in mouse brain,56 we examined circRNA expression in young (4-month old) and aged (2-year old) sperms. To our surprise, we observed the opposite trend, i.e., young sperm contained a much greater number of circRNAs compared with aged sperm (Fig. 6d, e), suggesting that these sperm-borne circRNAs may have positive impact on sperm quality in general. GO term analyses revealed that host genes of these circRNAs are mostly involved in the control of sperm motility (Supplementary information, Fig. S12e). If the sperm-borne circRNAs have regulatory roles, then evolutionary conservation should be expected. To test this, we analyzed sperm-borne circRNAs in four mammalian species using published data.57 Interestingly, we found that 30–50% of circRNAs were conserved among mice, rats, rabbits, and humans (Fig. 6g).
To further explore potential roles of circRNAs in human sperm, we analyzed circRNA expression in high fertility (pregnancy rate in IVF > 25%) and low fertility (pregnancy rate in IVF < 10%) human sperms using RNA-seq. High fertility sperm contained more circRNAs than low fertility sperm (Fig. 6f). Consistently, more linear RNAs were present in low fertility sperm compared to high fertility sperm (Supplementary information, Fig. S12f and Table S7). We extracted the differentially expressed circRNAs and performed GO term analyses on their host genes. Our results showed that the enriched terms were mostly related to regulation of histone modifications (Fig. 6h), suggesting the potential epigenomic differences between these two types of sperms. Overall, these data suggest that sperms contain a large number of circRNAs that may have functions in their post-testicular lives, including regulation of motility and epigenetic modifications during fertilization and early embryonic development. Moreover, these circRNAs may be good biomarkers for predicting epigenomic integrity of human sperm.
CircRNAs can be translated into proteins in both spermatogenic cells and spermatozoa
As described above, several lines of evidence, including (1) large ORFs-containing circRNAs containing m6A-modified start codons at the junction sites and (2) increasingly dynamic associations of circRNAs with polysomes from late pachytene spermatocytes to round and elongating spermatids, all suggest that circRNAs with these characteristics are made for protein production. Given that transcription ceases and massive degradation of linear mRNAs occurs in elongating and elongated spermatids, translation from circRNAs would represent a compensatory mechanism to ensure continuous production of proteins critical for late spermiogenesis. However, direct proof of translation remained lacking. It is very hard to unequivocally demonstrate circRNA translation because both the circRNAs and their homologous linear forms share the same ORFs and thus, one cannot tell the origin of the peptides detected (from circRNAs vs. from their linear forms). Recent reports have shown that the translational machinery, although rarely, can sometimes go over the junction sites, leading to the production of polypeptides encoded by the back-spliced junction sequences, which are unique to the circRNAs.33,35–38 Due to the extremely low abundance, the high-resolution mass spectrometry (LC–MS)-based proteomic approach is required to identify these rare, but unique junction peptides.33,35–37 To directly identify junction peptides, we first generated a customized database containing all peptides possibly encoded by junction sequences of all circRNAs (based on three different coding frames) identified in pachytene spermatocytes, round/elongating spermatids and spermatozoa (Fig. 7a). All peptides identified by LC–MS were then blasted against the database, and hundreds of junction peptides were identified with high confidence (99%) from the three spermatogenic cell types as well as spermatozoa (Fig. 7b and Supplementary information, Table S8). We also blasted these junction peptides against the NCBI protein database and found only 1–2 hits, suggesting the false positive rate is very low (<0.5%). Therefore, the vast majority (99.8%) of the peptides identified were unique to the junction sequences of circRNAs, and this result is consistent with previous reports.33,35–37 More interestingly, a close correlation (R2 = 0.94, P = 0.05) was observed between the counts of junction peptides identified by LC-MS and those of circRNAs annotated using RNA-seq in spermatids and spermatozoa (Fig. 7c), suggesting that these junction peptides are not random protein degradation products, but rather specifically translated from circRNAs. GO term enrichment analyses revealed that the host genes of these coding circRNAs are involved in spermatogenesis in general, and flagellar development, energy metabolism and protein turnover in particular, which all represent the most active cellular events during spermiogenesis35,50,51 (Supplementary information, Fig. S13 and Table S9). CircRNAs derived from two genes, Hook1 and Ranbp9, which are known to be essential for late spermatogenesis,58,59 were most likely translatable because of the specific detection of the polypeptides encoded by their corresponding junction sequences (Fig. 7d). Together, these data strongly suggest that male germ cell circRNAs accumulate in late meiotic and early haploid male germ cells, and function to provide continuous supply of proteins critical for late spermiogenesis and normal sperm functions.
Fig. 7. Detection of circRNA junction peptides using LC-MS.
a Workflow for assembling junction peptide sequence database. RNA-seq data were processed by CIRI2 to map the circRNA junction site to the mouse genome. The circRNA 3′ (pink) and 5′ back splicing site (dark) are joined with 60 bp adjacent sequences (dark green). The predicted, trypsin-digested circRNA junction peptide sequences were collected in the circRNA junction peptide sequence database, which will be used for analyzing LC-MS data. b Number of junction peptides identified by LC–MS in pachytene spermatocytes, round, and elongating spermatids, as well as spermatozoa. Bars represent the total number of junction peptides identified from LC-MS analyses in three replicates (numbers of junction peptides from three biological replicates were combined). c Close correlations between counts of junction peptides identified via LC–MS and those of circRNA junction sequences identified from RNA-seq in pachytene spermatocytes, round, and elongating spermatids, as well as spermatozoa (R2 = 0.94, P = 0.05, counts of three biological replicates were combined for analyses). d Examples of two circRNA junction peptides identified using LC–MS. The junction peptide sequences are fully covered. The relative intensity of each peptide identified is indicated in each vertical line. The y-axis shows the relative ion intensity and x-axis shows the mass to charge ratio (m/z). Peptide fragment ion in the spectrum is labeled in red. Peptide fragment ion missing from the spectrum is labeled in black.
Discussion
Although circRNAs have been discovered for decades,60 it was not until the past several years that these RNA circles started to draw investigators’ attention largely due to the fact that tens of thousands of circRNAs have been identified in almost all species27,61 and their regulatory roles start to emerge.24–27 CircRNAs were identified based on bioinformatic analyses of junction sequences in RNA-seq reads. It was subsequently realized that RNase R-treated total RNAs should be used for library construction to avoid interference of linear RNAs which are dominant in total RNAs. However, recent reports have shown that RNase R treatment cannot remove linear RNAs with modifications, e.g., m6A; thus, even after RNase R treatment, linear RNAs can persist.62 RPAD was developed to remove linear RNAs from total RNAs and thereby enrich circRNAs,39 and its efficiency was proved by our data (~40× enrichment of circRNAs). We used all three methods to identify circRNAs and similar results were obtained, suggesting that conventional RNA-seq with/without RNase R treatment, as long as the sequencing is deep enough, can accurately identify most of the circRNAs. Obviously, RPAD-seq would be much more efficient and economic for circRNA discovery due to significant enrichment of circRNAs.
Given that normalization was performed against total reads, degradation of linear RNAs could increase the relative abundance of circRNAs. However, this is highly unlikely due to several reasons. First, massive linear RNA degradation does not commence until the elongation steps (i.e., in elongating spermatids), whereas levels of circRNA are increased in both pachytene spermatocytes and round spermatids in which the massive linear RNA degradation has not happened yet.13,63–65 Second, it has been well-documented that m6A levels increase with progression of spermatogenesis from pachytene spermatocytes to round/elongating spermatids, coinciding with enhanced splicing to generate shorter 3′UTR transcripts and degradation of longer 3′UTR transcripts.22 In the present study, we discovered that the m6A-guided splicing events positively correlate with elevated levels of circRNAs in Alkbh5-null mice, suggesting that enhanced splicing due to increased m6A levels may increase circRNA biogenesis. Third, circRNAs increase in both total counts and species with progression of spermatogenesis from pachytene spermatocytes to round/elongating spermatids. Based on these data, we, therefore, believe that accumulation of circRNAs mainly resulted from enhanced circRNA biogenesis instead of relative enrichment due to linear RNA degradation; this applies, at least, to those in the progression of spermatogenesis from pachytene spermatocytes to round spermatids. However, the rapid accumulation of circRNAs in elongating spermatids may represent a relative enrichment because of the cessation of transcription and massive degradation of linear RNAs in these cells. Regardless, neither of the scenarios would affect our conclusion that circRNA levels increase with progression of spermiogenesis.
The increased circRNA/linear mRNA ratio suggests that circRNAs become the major RNA output when spermatogenesis progresses from late meiotic phase (late pachytene spermatocytes) to spermiogenesis/haploid phase (round, elongating and elongated spermatids). A similar trend was reported in Drosophila cells when spliceosomes are activated or when approaching transcription termination.66 Thus, switching from linear mRNA production to circRNA biogenesis appears to be a conserved mechanism for post-transcriptional regulation of gene expression. Given their superior stability, circRNAs would be ideal to protect RNAs from degradation. The increased circRNA production and drastic downregulation of their linear forms from pachytene spermatocytes to round and elongating spermatids strongly imply that these RNA circles are made to compensate for the functions of certain linear mRNAs that are destined to be degraded in elongating/elongated spermatids. However, such a compensatory mechanism makes sense only if they can be translated into proteins. Indeed, these spermiogenesis-enriched circRNAs have coding potential based upon the presence of large intact ORFs and m6A-modified start codons at the junction sites. Moreover, these potentially coding circRNAs also display a dynamic shift from RNPs to polysomes when round spermatids develop into elongating spermatids, further supporting their translational potential. Identification of polypeptides encoded by the junction sequences of circRNAs by LC-MS-based deep proteomic analyses provided the ultimate evidence of translation. Out of tens of thousands of circRNAs identified, only three to five hundred junction peptides were detected with high confidence; the low detection rate is consistent with the notion that it is a rare event for the translational machinery to go through the junction sites during translation.33,35–37 A strong correlation between counts of junction peptides identified via LC–MS and those of circRNA junction sequences identified from RNA-seq not only proves that the junction peptides are truly specific to circRNAs, but also provides solid evidence supporting the notion that circRNA translation increases with progression of spermatogenesis from pachytene spermatocytes to round and elongating spermatids. Given that during the transition from round to elongating spermatids transcription ceases and large linear mRNAs undergo massive degradation, circRNAs may serve as the backup version of their degenerating linear forms, thus maintaining stable and long-lasting protein production in elongating/elongated spermatids and in spermatozoa. This notion is supported by the fact that most of those coding circRNAs are synthesized from genes critical for the final several steps of spermiogenesis/sperm assembly and sperm function.
m6A appears to play an important role in the regulation of gene expression in spermatogenesis because KO mice lacking m6A writer (Metll3 and Mettl14),35,53,54 eraser (Alkbh5)22,47 or reader (Ythdc2 and Ythdf2)67,68 genes all display disrupted spermatogenesis and male infertility. This is not surprising given that spermatogenesis is known to have complex gene regulatory networks to ensure precise spatiotemporal gene expression.3,15 Our recent study on Alkbh5 KO mice have revealed that m6A controls correct splicing of longer transcripts in the nuclei and mRNA stability in the cytoplasm in pachytene spermatocytes and round spermatids.22 Significantly, increased circRNA production in Alkbh5-null pachytene spermatocytes and spermatids is consistent with the enhanced, but aberrant splicing of mRNA transcripts. This finding further supports the slicing function of m6A and also the notion that circRNA production results from enhanced splicing.20,21 The preferential enrichment of m6A in the junction sequences of a subgroup of circRNAs indicates that these circles are spliced from m6A-enriched sites in linear mRNAs, which are usually located around the stop and start codons.69–71 Meanwhile, m6A-enriched junction sites may have specific functions, e.g., the m6A-modified start codon can be recognized by the m6A reader YTHDF3 and serves as the IRES.35,48 Our data strongly suggest a direct link between m6A and circRNA biogenesis, and that m6A-guided circRNA production represents an alternative mechanism to ensure stable and more “weather-proof” translation of proteins critical for specific physiological processes.
The presence of abundant circRNAs in spermatozoa is intriguing because it suggests that these sperm-borne circRNAs may play an important role. The high degree of evolutionary conservation in sperm-borne circRNAs further support that these RNA circles are not random carryover products from testicular spermatogenic cells. The facts that cytoplasmic droplet-enriched circRNAs are synthesized from genes involved in energy metabolism and that the sperm head circRNAs are enriched for mRNAs encoding epigenetic regulators strongly suggest that these circRNAs may be able to be translated into proteins either inside sperm or in fertilized eggs. Although mature sperm do not have the typical translational machinery, studies have shown that sperm can translate protein using mitochondrial-type ribosomes during capacitation.72–74 It will be of great significance to test whether these sperm-borne RNA circles can be translated into proteins once released into the ooplasm. These possibilities represent exciting new research directions in the future. Altered circRNA profiles in aged sperm and the differential circRNA contents between high and low fertility human sperms all support the notion that these RNA circles regulate sperm function and may be used as biomarkers for sperm quality. Further investigation would allow these stable RNA circles to be used for diagnostic or even therapeutic purposes in the future.
It is of great interest to note that circRNAs accumulate with aging in brain,56 whereas their levels decrease in aging/aged testes. This opposing trend is worth further investigation. Although several recent reports have shown that circRNAs can be translated into protein,33,35–37 it remains unknown why such a mechanism is needed in physiological contexts. Spermiogenesis represents a perfect model to address this question because transcription and translation are uncoupled during late spermiogenesis.5–8 Despite the massive, global degradation of mRNAs, especially those with longer 3’UTRs, in spermatids,64 some transcripts are still needed for protein production during the remaining several steps of differentiation.5–8 Since physical sequestration into RNPs (e.g., the chromatoid body, nuage, intermitochondrial cements, etc.) is no longer feasible in elongating/elongated spermatids,18 circularization of the minimal ORFs would be an ideal way of preserving those transcripts for delayed translation because of the superior stability of circRNAs. In this way, these circRNAs would represent the continued “fuel” supply for delayed translation when the transcriptional machinery is completely shut down. This mechanism appears to operate not only in late spermiogenesis, but also in post-testicular spermatozoa. It is noteworthy that numerous shorter, non-polysome-associating, non-ORF-containing circRNAs (~50% of all circRNAs identified in spermatogenic cells) also accumulate with progression of spermatogenesis. These circRNAs have no translational potential and their roles need further investigation in the future.
In summary, through comprehensive transcriptomic and epi-transcriptomic analyses, we have discovered not only a novel role of m6A in the biogenesis of circRNAs with coding potential, but also an alternative mechanism to ensure stable and more “weather-proof” translation of proteins critical for specific physiological processes, i.e., through production of circRNAs containing large ORFs and m6A-modified start codons in junction sequences.
Materials and Methods
Animals
Animal use protocol was approved by Institutional Animal Care and Use Committee (IACUC) of the University of Nevada, Reno (Protocol number 00494), and is in accordance with the “Guide for the Care and Use of Experimental Animals” established by National Institutes of Health (NIH) (1996, revised 2011). All mice used in this study were in the C57Bl/6J background and housed under specific pathogen-free conditions in a temperature- and humidity- controlled animal facility at the University of Nevada, Reno. The male Alkbh5 KO mice used in this study were described previously.22,47
Ethical approval and consent to participate
The use of human semen samples was approved by the Institutional Review Board of the Family Planning Research Institute of Guangdong Province, China. All human semen samples used were deidentified. The original Informed Consent Forms are available upon request.
Purification of murine spermatogenic cells
Pachytene spermatocytes and round and elongating/elongated spermatids were purified from adult mouse testes using the STA-PUT method.13
RNP and polysome fractionation
We fractionated the purified murine spermatogenic cells into RNP and polyribosome fractions using a continuous sucrose gradient ultracentrifugation method, as described.13
Murine sperm head and droplets purification
Adult C57Bl/6J mice were euthanized and the epididymides were dissected and transferred into 2.5 ml of a dissection medium (0.15 M KCl solution containing 0.01 M Tris-HCl, pH7.1). The epididymides were further dissected into smaller pieces (5 mm × 5 mm) followed by incubation in a humidity incubator at 37 °C for ~10 min. The spermatozoa-containing supernatants (~2 ml) were collected and subjected to sperm head75 and cytoplasmic droplet55 purification using a discontinuous sucrose density gradient centrifugation method.
Human sperm
Deidentified human semen samples were obtained from the Sperm Bank of Guangdong Province. Donors were healthy adult men (20–30 years of age) whose semen parameters met or exceeded the criteria illustrated in the WHO Laboratory Manual for the Examination and Processing of Human Semen (5th edition). High fertility sperm samples were defined as those with a pregnancy rate >25% (i.e., legally allowed 5 pregnancies in ≤20 IVF attempts on different women with normal reproductive functions), whereas low fertility sperm samples were referred to those with a pregnancy rate <15%. Human semen samples (1 ml per vial) were thawed and washed with the HTF medium for three times by centrifugation at 1000 × g at room temperature. The sperm pellets were then subjected to RNA extraction for RNA-seq, as described below.
RNA extraction
RNA was extracted from cells using the mirVana miRNA Isolation Kit (ThermoFisher, Cat#AM1560), according to the manufacturer’s instructions. Extracted RNA was quantitated using the Qubit RNA High Sensitivity Assay Kit (Invitrogen No. Q32855) and measured on the Qubit 2.0 Fluorometer (Invitrogen).
CircRNA enrichment using RPAD
CircRNAs were enriched from total RNA using the RPAD method as described.39 In brief, 2 μg of whole testis total RNA, 1 μl RNase inhibitor (Thermo Fisher Scientific), 1× RNase R buffer and 20 U RNase R (Epicenter) were incubated for 30 min at 37 °C. The RNA was purified using RNeasy Mini Kit (Qiagen) following the manufacturer's instructions. A 40 μl polyadenylation reaction was prepared with polyA polymerase reaction (NEB) following the manufacturer’s instruction and incubated for 30 min at 37 °C. Oligo-dT Dynabeads (10 μl) from Poly(A)Purist™ MAG Kit (AM1922) were washed three times with the 1× binding buffer provided in the kit and dissolved in 40 μl of 2× binding buffer. The Oligo-dT Dynabeads in 2× binding buffer were added into the poly(A)-tailing reaction mix of RNase R-treated samples and incubated for 5 min at 75 °C followed by 20 min at 25 °C with shaking. The supernatant was collected for RNA isolation.
Semi-quantitative PCR
To validate the bioinformatic data, semi-qPCR analyses were performed as described.76 The sequences for the primers used can be found in Supplementary information, Table S10.
Alkbh5 knockdown assay
NIH 3T3 cells were cultured in a six-well plate (5 × 105 cells/well) in DMEM with high glucose supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin at 37 °C under a humidified atmosphere containing 5% CO2. The cells were transfected with 50 nM or 100 nM small interfering RNA (RiboBio, Guangzhou, China) using Lipofectamine 3000 to knock down Alkbh5. Cells were then harvested 68-72 h post-transfection for RT-PCR analyses. Sequences of primers and oligos used can be found in Supplementary information, Table S10.
Minigene reporter assay
The minigene reporter assay was conducted as described45 with modifications. Briefly, to generate the splicing minigene construct, the genomic region (chr9:70406844-70410668) corresponding to the splicing junction of Sltm and flanking intronic regions was cut out and then inserted into pCITF-KCC2 vector (Cambridge, MA, USA) by USER cloning (NEB cat#M5505S) following the manufacturer’s protocol. HEK293 cells were cultured in a 6-well plate (5 × 105 cells/well) in DMEM with high glucose supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin at 37 °C under a humidified atmosphere containing 5% CO2. The constructs were transiently transfected into HEK293 cells using Lipofectamine® 3000 Transfection Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The cells were then harvested 68–72 h post transfection for RT-PCR analyses. Sequences of primers and oligos used can be found in Supplementary information, Table S10.
m6A RNA immunoprecipitation
Rabbit anti-m6A antibody (ab151230, Abcam) or normal rabbit IgG (10500C, Invitrogen) (6 μg each) was used for m6A RNA immunoprecipitation using procedures described previously.22 Experiments were performed in triplicates.
RNA library construction
Immunoprecipitated RNA (1 ng) and non-immunoprecipitated RNA (300 ng) were constructed into next-generation sequencing libraries (Illumina) using the KAPA Stranded RNA-seq Library Preparation Kit (KK8400) according to the manufacturer’s instructions, as described previously.22
CircRNA junctional peptide database
We generated a customized database containing peptides encoded by RNA sequences spanning back-spliced junctions of all circRNAs identified in this study using the method as reported.35 In brief, CIRI277 was used to identify circRNA junction sites, followed by adding the upstream/downstream 100 bp sequence to the junction. The junction sequences were then translated into three coding frames, and trypsin-digested peptide sequences encoded by the junction sequences were collected into the database, which was used to blast search matching peptides identified by liquid chromatography coupled with LC–MS (see below).
Protein isolation and enzymatic digestion
The sperm samples were lysed a lysis buffer [180 μl H2O + 100 μl 2% Salkosyl + 20 μl 1 M Tris-HCl pH8.0 + 20 μl 1 M KCL + 80 μl 1 M Dithiothreitol (DTT)]. Sperm protein samples were digested and desalted. Protein concentrations were measured using a bicinchoninic acid assay (Thermo-Fisher Scientific, San Jose, CA). Proteins were then reduced and alkylated using DTT and iodoacetamide followed by methanol-chloroform precipitation prior to digestion with endoproteinase Lys-C (Wako, Richmond, VA). After digestion with trypsin (Promega, Madison, WI), the samples were subjected to desalting and concentration using C18 Sep-Pak Cartridges (Waters).
Basic reversed-phase fractionation
Pooled, tandem mass tags labeled peptides were fractionated by basic pH reversed-phase fractionation on an Ultimate 3000 HPLC (Thermo Scientific) using an integrated fraction collector. Elution was performed using a 10 min gradient of 0%–20% solvent B followed by a 50 min gradient of 20%–45% solvent B (Solvent A: 5% Acetonitrile, 10 mM ammonium bicarbonate, pH 8.0. Solvent B: 90% Acetonitrile, 10 mM ammonium bicarbonate, pH 8.0) on a Zorbax 300Extend-C18 column (Agilent) at a flow rate of 0.4 ml/min. A total of 24 fractions were collected at 37 s intervals in a looping fashion for 60 min. Peptide elution was monitored at a wavelength of 220 nm using a Dionex Ultimate 3000 variable wavelength detector (Thermo Scientific). Each fraction was then centrifuged to near dryness and desalted using C18 Sep-Pak Cartridges followed again by centrifugation to near dryness and reconstitution with 20 μl 5% acetonitrile and 0.1% formic acid.
Liquid chromatography coupled with LC–MS
LC-MS was performed on an UltiMate 3000 RSLCnano system (Thermo Scientific, San Jose, CA). The total gradient time was 175 min and the gradient consisted of solvent B from 2%–90% (Solvent A: 0.1% formic acid in water, Solvent B: 0.1% formic acid in acetonitrile) at 50 °C using a digital Pico View nanospray source (New Objectives, Woburn, MA) that was modified with a custom-built column heater and an ABIRD background suppressor (ESI Source Solutions, Woburn, MA). The column was a self-packed Pico-Frit (New Objectives, Woburn, MA) column with a 15 μm tip packed with 40 cm of 1.9 μm ReproSil-Pur 120 C18-AQ (Dr. Maisch GmbH, Germany) at 9000 psi using a nano LC column packing kit (nanoLCMS, Gold River, CA) for the separation.
Mass spectral analysis was performed using an Orbitrap Fusion mass spectrometer (Thermo Scientific, San Jose, CA). The MS precursor selection range is 400–1500 m/z at a resolution of 120,000 and an automatic gain control (AGC) target was set to 2.0 × 105 with a maximum injection time of 100 ms. Quadrupole isolation for MS2 analysis was performed using CID fragmentation in the linear ion trap with a collision energy of 35%. The AGC was set to 4.0 × 103 with a maximum injection time of 150 ms. The instrument was operated in a top speed data-dependent mode with the most intense precursor priority with dynamic exclusion set to an exclusion duration of 60 s with a 10 ppm tolerance. The data were then analyzed using Sequest (Thermo Fisher Scientific, San Jose, CA, version v.27, rev. 11.) and Proteome Discoverer (Thermo Scientific, San Jose, CA. version 2.1).
Bioinformatics
RNA-seq data analysis
Trimmomatic was used to remove adapter sequences and low-quality reads from the sequencing data.78 To identify all the transcripts, we used Tophat2 and Cufflinks to assemble the sequencing reads based on the UCSC MM9 mouse genome.79 The differential expression analysis was performed by Cuffdiff.79 The UTR and alternative splicing analyses were performed using the SpliceR pipeline.80 The sequences without identified coding frames were extracted and subjected to coding potential calculating.81 FPKMs counts are scaled in Cuffdiff analyses via the median of the geometric means of fragment counts across all libraries, as described previously.82
m6A RIP-seq data analysis
Trimmomatic was used to remove adapter sequences and low-quality reads from the sequencing data.78 To reduce bias from potential inaccurate gene structure annotation, we aligned the m6A-seq reads to the assembled gene sequences derived from RNA-Seq cufflink results, using Tophat v2.0.14.79 The longest isoform was used if the gene had multiple isoforms. The peak-calling method was modified from a published work.83 To call m6A peaks, the longest isoform of each gene was scanned using a 100 nt sliding window with 10 nt step. We add one count to all the windows to avoid 0 count in the windows. The peak height threshold was ten counts. For each gene, the read counts in each window were normalized by the median counts of all windows of that gene. A Fisher exact test was used to identify the differential windows between IP and input samples. The window was called positive if the count ≥10 and log2(enrichment score) >0.8, which FDR is close to zero. Overlapping positive windows were merged. The following four numbers were calculated to obtain the enrichment score of each window: (a) read counts of the IP samples in the current window, (b) median read counts of the IP sample in all 100 nt windows on the current mRNA, (c) read counts of the input samples in the current window, and (d) median read counts of the input samples in all 100 nt windows on the current mRNA. The enrichment score of each window was calculated as (a × d)/(b × c).
CircRNA analysis
Reads were aligned to the mouse genome (UCSC mm9 assembly) using the Burrows-Wheeler Aligner (bwa) tool. CIRI277 was used for circRNA annotation (CIRI_v2.0.1.pl –no_strigency -I bwa.sam -O ciriout -F bwaindex/genome.fa). The circRNA counts were normalized in Deseq2, and the circRNA junction ratios were used without normalization. The longest isoforms of the gene were chosen to represent this gene to calculate the circRNA junction locations in genes. The prospective full-length circRNAs and coding potential prediction pipeline are illustrated in Supplementary information, Fig. S14. The clean data are processed by Tophat-Fusion to identify the fusion junction sites. Then the Circseq_cup84 was performed to identify full length circRNAs. The two fusion sites with genomic sequence less than 50 kb in between were concatenated as the back-splicing site reference. Paired-end reads were mapped to the reference and full-length circRNAs were eventually determined by the assembled full-length circRNAs with overlapping contigs. The identified full-length circRNA sequences were sent to Coding Potential Calculator (CPC)81 for coding potential predictions based on the default parameters for support vector machine (SVM) analysis of six features of ORF and BLASTX scores.
Quantification and statistical analysis
Student’s t test was used for statistical analyses. The majority of data followed a lognormal distribution. Student’s t test was also performed on the logarithm data.
Data visualization
For scatterplot (e.g., Fig. 6d), normalized circRNA read counts calculated in DESeq2 were averaged across biological replicates. Scatterplots display normalized values of these averages (ggplot2). The linear regression lines were calculated though ggplot2 geom_smooth.
For m6A location density plot (e.g. Fig. 2c), the peak position of each m6A site was classified into three mutually exclusive mRNA structural regions including 5UTR, CDS, and 3UTR. The relative locations in each gene were normalized by the distance between start (0.25/1) and stop site (0.75/1). The normalized location values were plotted by ggplot2 density function.
For bar plot (e.g. Fig. 1a), the circRNAs were annotated by CIRI2. The total number of the junction reads was summarized by CIRI2, then normalized by total aligned RNA reads. The normalized circular junction counts were plotted by ggplot2 bar plot function.
For heatmap (e.g. Fig. 4a), the counts of the circular/linear RNAs were normalized in DESeq2 across biological replicates. The average normalized counts of the selected circular/linear RNAs in each subject were plotted by gplots heatmap2 function.
For genome browser track (e.g., Supplementary information, Fig. S9d), bed files of m6A IP reads from concordant biological replicates were concatenated and inputted into Integrated Genome Browser.85 Coordinates in mm9 are indicated above genome browser tracks.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and BioProject database (http://www.ncbi.nlm.nih.gov/bioproject/) under accession number (GSE80353, PRJNA420607, GSM747485, GSE81216, PRJNA448275, PRJNA448271). The RPAD-seq datasets are available in the CNGB database (https://db.cngb.org) (Accession# CNP0000637).
Supplementary information
Acknowledgements
This study was supported by grants from the NIH (HD071736 and HD085506 to W.Y.) and the Templeton Foundation (PID: 61174 to W.Y.). RNA-seq was conducted in the Single Cell Genomics Core of the University of Nevada, Reno School of Medicine, which was supported, in part, by the NIH COBRE Grant (P30GM110767 to W.Y.). Bioinformatics and RPAD-seq were, in part, carried out in the BGI Co. Ltd, with the support of a grant from the Science, Technology and Innovation Commission of Shenzhen Municipality (JSGG20170824152728492 to C.T.). The human sperm work was supported by grants from the Natural Science Foundation of Guangdong Province (2015A030313884 and 2018A030313528 to Y.T. and W.Q.), the Science and Technology Projects of Guangzhou (201607010137 and 201804010431 to W.Q. and Y.T.) and the Family Planning Research Institute of Guangdong Province (S2014001 to Y.T.).
Author contributions
W.Y. and C.T. conceived and designed the research. C.T., T.Y., Z.W., Z.X., R.J.W., Y.Z., R.K., H.Z., and D.R.Q. performed experiments on mouse samples. Y.T., X.Z., G.S., W.Z., and W.Q. provided human samples and performed RNA-seq on human samples. C.T., Y.X., N.L., J.W., W.C., and X.W. performed bioinformatics analyses. W.Y. and C.T. wrote the manuscript. All authors reviewed and agreed with the contents of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Chong Tang, Yeming Xie, Tian Yu, Na Liu
Contributor Information
Chong Tang, Email: leochong718@gmail.com.
Wei Yan, Email: wyan@med.unr.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0279-8.
References
- 1.Leblond CP, Clermont Y. Definition of the stages of the cycle of the seminiferous epithelium in the rat. Ann. N.Y. Acad. Sci. 1952;55:548–573. doi: 10.1111/j.1749-6632.1952.tb26576.x. [DOI] [PubMed] [Google Scholar]
- 2.Oakberg EF. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am. J. Anat. 1956;99:507–516. doi: 10.1002/aja.1000990307. [DOI] [PubMed] [Google Scholar]
- 3.Maclean JA, 2nd, Wilkinson MF. Gene regulation in spermatogenesis. Curr. Top. Dev. Biol. 2005;71:131–197. doi: 10.1016/S0070-2153(05)71005-X. [DOI] [PubMed] [Google Scholar]
- 4.Eddy EM. Regulation of gene expression during spermatogenesis. Semin. Cell Dev. Biol. 1998;9:451–457. doi: 10.1006/scdb.1998.0201. [DOI] [PubMed] [Google Scholar]
- 5.Kleene KC. Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenet. Genome Res. 2003;103:217–224. doi: 10.1159/000076807. [DOI] [PubMed] [Google Scholar]
- 6.Bao J, et al. UPF2-dependent nonsense-mediated mrna decay pathway is essential for spermatogenesis by selectively eliminating longer 3'UTR transcripts. PLoS Genet. 2016;12:e1005863. doi: 10.1371/journal.pgen.1005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, et al. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol. 2016;14:6. doi: 10.1186/s12915-016-0229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashiwabara S, Nakanishi T, Kimura M, Baba T. Non-canonical poly(A) polymerase in mammalian gametogenesis. Biochim. Biophys. Acta. 2008;1779:230–238. doi: 10.1016/j.bbagrm.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Steger K. Haploid spermatids exhibit translationally repressed mRNAs. Anat. Embryol. 2001;203:323–334. doi: 10.1007/s004290100176. [DOI] [PubMed] [Google Scholar]
- 10.Braun RE. Temporal translational regulation of the protamine 1 gene during mouse spermatogenesis. Enzyme. 1990;44:120–128. doi: 10.1159/000468752. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen-Chi M, Morello D. RNA-binding proteins, RNA granules, and gametes: is unity strength? Reproduction. 2011;142:803–817. doi: 10.1530/REP-11-0257. [DOI] [PubMed] [Google Scholar]
- 12.Idler RK, Yan W. Control of messenger RNA fate by RNA-binding proteins: an emphasis on mammalian spermatogenesis. J. Androl. 2012;33:309–337. doi: 10.2164/jandrol.111.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. MicroRNAs control mRNA fate by compartmentalization based on 3' UTR length in male germ cells. Genome Biol. 2017;18:105. doi: 10.1186/s13059-017-1243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jha KN, Tripurani SK, Johnson GR. TSSK6 is required for gammaH2AX formation and the histone-to-protamine transition during spermiogenesis. J. Cell Sci. 2017;130:1835–1844. doi: 10.1242/jcs.202721. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka H, Baba T. Gene expression in spermiogenesis. Cell Mol. Life Sci. 2005;62:344–354. doi: 10.1007/s00018-004-4394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3' UTRs in the mammalian brain. Genome Res. 2013;23:812–825. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacDonald CC, McMahon KW. Tissue-specific mechanisms of alternative polyadenylation: testis, brain, and beyond. Wiley Interdiscip. Rev. RNA. 2010;1:494–501. doi: 10.1002/wrna.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanourgakis G, Lesche M, Akpinar M, Dahl A, Jessberger R. Chromatoid body protein TDRD6 supports long 3' UTR triggered nonsense mediated mRNA decay. PLoS Genet. 2016;12:e1005857. doi: 10.1371/journal.pgen.1005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adhikari S, Xiao W, Zhao YL, Yang YG. m(6)A: Signaling for mRNA splicing. RNA Biol. 2016;13:756–759. doi: 10.1080/15476286.2016.1201628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang C, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA. 2018;115:E325–E333. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 25.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starke S, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 28.Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem. Biophys. Res. Commun. 2018;495:2369–2375. doi: 10.1016/j.bbrc.2017.12.050. [DOI] [PubMed] [Google Scholar]
- 29.Weiser-Evans MCM. Smooth muscle differentiation control comes full circle: the circular noncoding RNA, circActa2, functions as a miRNA sponge to fine-tune alpha-SMA expression. Circ. Res. 2017;121:591–593. doi: 10.1161/CIRCRESAHA.117.311722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 31.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 32.Piwecka Monika, Glažar Petar, Hernandez-Miranda Luis R., Memczak Sebastian, Wolf Susanne A., Rybak-Wolf Agnieszka, Filipchyk Andrei, Klironomos Filippos, Cerda Jara Cledi Alicia, Fenske Pascal, Trimbuch Thorsten, Zywitza Vera, Plass Mireya, Schreyer Luisa, Ayoub Salah, Kocks Christine, Kühn Ralf, Rosenmund Christian, Birchmeier Carmen, Rajewsky Nikolaus. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357):eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 33.Pamudurti NR, et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21 e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legnini I, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37 e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Wang Z. Constructing GFP-based reporter to study back splicing and translation of circular RNA. Methods Mol. Biol. 2018;1724:107–118. doi: 10.1007/978-1-4939-7562-4_9. [DOI] [PubMed] [Google Scholar]
- 38.Fan, X., Yang, F. & Wang, Z. Pervasive translation of circular RNAs driven by short IRES-like elements. BioRxiv, 10.1101/473207 (2018). [DOI] [PMC free article] [PubMed]
- 39.Panda AC, et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017;45:e116. doi: 10.1093/nar/gkx297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venables JP. Alternative splicing in the testes. Curr. Opin. Genet. Dev. 2002;12:615–619. doi: 10.1016/S0959-437X(02)00347-7. [DOI] [PubMed] [Google Scholar]
- 42.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat. Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao W, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 45.Bartosovic M, et al. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 2017;45:11356–11370. doi: 10.1093/nar/gkx778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang CX, et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16:e2004880. doi: 10.1371/journal.pbio.2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatomer DC, Wilusz JE. An unchartered journey for ribosomes: circumnavigating circular RNAs to produce proteins. Mol. Cell. 2017;66:1–2. doi: 10.1016/j.molcel.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Maezawa S, Yukawa M, Alavattam KG, Barski A, Namekawa SH. Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res. 2018;46:593–608. doi: 10.1093/nar/gkx1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Donnell L. Mechanisms of spermiogenesis and spermiation and how they are disturbed. Spermatogenesis. 2014;4:e979623. doi: 10.4161/21565562.2014.979623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan W. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol. Cell Endocrinol. 2009;306:24–32. doi: 10.1016/j.mce.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu H, Yuan SQ, Zheng ZH, Yan W. The cytoplasmic droplet may be indicative of sperm motility and normal spermiogenesis. Asian J. Androl. 2013;15:799–805. doi: 10.1038/aja.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu K, et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27:1100–1114. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Z, et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res. 2017;27:1216–1230. doi: 10.1038/cr.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan S, Zheng H, Zheng Z, Yan W. Proteomic analyses reveal a role of cytoplasmic droplets as an energy source during epididymal sperm maturation. PLoS One. 2013;8:e77466. doi: 10.1371/journal.pone.0077466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci. Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuster A, et al. SpermBase: a database for sperm-borne RNA contents. Biol. Reprod. 2016;95:99. doi: 10.1095/biolreprod.116.142190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamauchi Y, Ward MA. Preservation of ejaculated mouse spermatozoa from fertile C57BL/6 and infertile Hook1/Hook1 mice collected from the uteri of mated females. Biol. Reprod. 2007;76:1002–1008. doi: 10.1095/biolreprod.106.059881. [DOI] [PubMed] [Google Scholar]
- 59.Bao J, et al. RAN-binding protein 9 is involved in alternative splicing and is critical for male germ cell development and male fertility. PLoS Genet. 2014;10:e1004825. doi: 10.1371/journal.pgen.1004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Capel B, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-Y. [DOI] [PubMed] [Google Scholar]
- 61.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panda Amaresh C., De Supriyo, Grammatikakis Ioannis, Munk Rachel, Yang Xiaoling, Piao Yulan, Dudekula Dawood B., Abdelmohsen Kotb, Gorospe Myriam. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Research. 2017;45(12):e116–e116. doi: 10.1093/nar/gkx297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang J, Morales CR, Medvedev S, Schultz RM, Hecht NB. In the absence of the mouse DNA/RNA-binding protein MSY2, messenger RNA instability leads to spermatogenic arrest. Biol. Reprod. 2007;76:48–54. doi: 10.1095/biolreprod.106.055095. [DOI] [PubMed] [Google Scholar]
- 64.Gou LT, et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014;24:680–700. doi: 10.1038/cr.2014.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe T, Cheng EC, Zhong M, Lin H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015;25:368–380. doi: 10.1101/gr.180802.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang D, et al. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell. 2017;68:940–954 e943. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wojtas MN, et al. Regulation of m(6)A transcripts by the 3'–>5' RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell. 2017;68:374–387 e312. doi: 10.1016/j.molcel.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 68.Hsu PJ, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu PJ, He C. High-resolution mapping of N (6)-methyladenosine using m(6)A crosslinking immunoprecipitation sequencing (m(6)A-CLIP-Seq. Methods Mol. Biol. 2019;1870:69–79. doi: 10.1007/978-1-4939-8808-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grozhik AV, Linder B, Olarerin-George AO, Jaffrey SR. Mapping m(6)A at individual-nucleotide resolution using crosslinking and immunoprecipitation (miCLIP) Methods Mol. Biol. 2017;1562:55–78. doi: 10.1007/978-1-4939-6807-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Linder B, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gur Y, Breitbart H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006;20:411–416. doi: 10.1101/gad.367606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gur Y, Breitbart H. Protein translation in mammalian sperm. Soc. Reprod. Fertil. Suppl. 2007;65:391–397. [PubMed] [Google Scholar]
- 74.Zhao C, et al. Role of translation by mitochondrial-type ribosomes during sperm capacitation: an analysis based on a proteomic approach. Proteomics. 2009;9:1385–1399. doi: 10.1002/pmic.200800353. [DOI] [PubMed] [Google Scholar]
- 75.Yan W, et al. Birth of mice after intracytoplasmic injection of single purified sperm nuclei and detection of messenger RNAs and MicroRNAs in the sperm nuclei. Biol. Reprod. 2008;78:896–902. doi: 10.1095/biolreprod.107.067033. [DOI] [PubMed] [Google Scholar]
- 76.Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev. Biol. 2007;311:592–602. doi: 10.1016/j.ydbio.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vitting-Seerup K, Porse BT, Sandelin A, Waage J. spliceR: an R package for classification of alternative splicing and prediction of coding potential from RNA-seq data. BMC Bioinform. 2014;15:81. doi: 10.1186/1471-2105-15-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao BS, et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ye CY, et al. Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol. 2017;14:1055–1063. doi: 10.1080/15476286.2016.1245268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nicol JW, Helt GA, Blanchard SG, Jr., Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.