Abstract
Hepatitis C virus (HCV) is a major global health burden accounting for around 170 million chronic infections worldwide. Although highly potent direct-acting antiviral drugs to treat chronic hepatitis C have been approved recently, owing to their high costs and limited availability and a large number of undiagnosed infections, the burden of disease is expected to rise in the next few years. In addition, HCV is an excellent paradigm for understanding the tight link between a pathogen and host cell pathways, most notably lipid metabolism. HCV extensively remodels intracellular membranes to establish its cytoplasmic replication factory and also usurps components of the intercellular lipid transport system for production of infectious virus particles. Here, we review the molecular mechanisms of viral replicase function, cellular pathways employed during HCV replication factory biogenesis, and viral, as well as cellular, determinants of progeny virus production.
Hepatitis C virus (HCV) remains a major global health burden accounting for around 170 million chronic infections worldwide. Paul et al. review the molecular mechanisms of HCV replicase function, cellular pathways employed during HCV replication factory biogenesis, and viral, as well as cellular, determinants of progeny virus production.
Main Text
Introduction
Infections by the hepatitis C virus (HCV) are characterized by a high rate of chronicity. Despite the mild, often asymptomatic course of acute infection, chronic hepatitis C frequently leads to steatosis, liver cirrhosis, and eventually hepatocellular carcinoma (Yamane et al., 2013). Highly potent direct-acting antiviral drugs (DAAs) have been approved, allowing virus elimination in more than 90% of treated individuals. However, owing to high costs and limited availability in most countries with high HCV prevalence, it is expected that on a global scale the number of HCV-infected individuals will drop only very slowly.
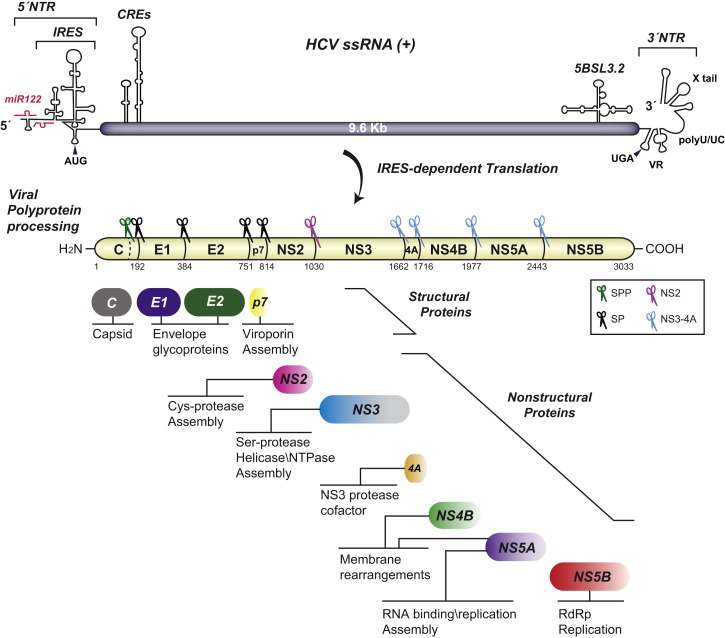
HCV belongs to the Hepacivirus genus of the family Flaviviridae, which also comprises the genera Flavivirus, Pestivirus, and Pegivirus (Simmonds, 2013). HCV enters the cell by receptor-mediated endocytosis involving multiple cell surface molecules (see review by Ding et al., 2014). Upon release into the cytoplasm, the ∼9.6 kb single-stranded RNA genome of positive polarity is directly used for translation at the rough endoplasmic reticulum (ER). The resulting polyprotein precursor has a length of ∼3,000 amino acid residues (aa) and is co- and posttranslationally cleaved by cellular and viral proteases into ten mature products (Figure 1 ); core and envelope glycoproteins E1 and E2 are main constituents of the virus particle, whereas the p7 viroporin and nonstructural protein 2 (NS2) participate in virus assembly. NS3, NS4A, NS4B, NS5A, and NS5B form the replicase complex that is sufficient for viral RNA replication (Lohmann et al., 1999a) occurring via a negative-strand copy. Progeny RNA is either used for translation, thus giving rise to new viral proteins, used for synthesis of new negative strands, or packaged into virus particles that acquire their envelope most likely via budding into the ER lumen. Finally, virions exit the cell via the secretory pathway.
Figure 1.
HCV Genome Organization and Polyprotein Processing
The single-strand (ss) HCV RNA genome is shown on the top. Secondary structures of cis-acting RNA elements (CREs) in the nontranslated regions (NTRs) and the coding region are schematically depicted. Interaction sites with miR-122 in the 5′ NTR that contains an internal ribosome entry site (IRES) are indicated. The polyprotein precursor and cleavage products are shown below. Numbers refer to amino acid positions of the JFH-1 isolate (GenBank accession number AB047639). Scissors indicate proteases responsible for polyprotein cleavage. SP, signal peptidase; SPP, signal peptide peptidase. Functions of cleavage products are indicated below each viral protein. RdRp, RNA-dependent RNA polymerase. VR, variable region in the 3′ NTR.
In this review, we focus on recent insights related to the mechanisms of viral RNA replication, biogenesis of membranous HCV replication factories, and the formation of infectious virus particles. We discuss the involvement of host cell factors in all of these processes, with an emphasis on lipids that play key roles in the HCV replication cycle.
HCV RNA Translation and Regulatory Mechanisms
The HCV genome lacks a 5′-terminal cap and a 3′-terminal poly(A) tract but contains highly structured 5′- and 3′-nontranslated regions (NTRs) flanking a single open reading frame (reviewed in Niepmann, 2013) (Figure 1). The presence of a type III internal ribosomal entry site (IRES) in the 5′ NTR ensures translation initiation by a cap-independent mechanism (Honda et al., 1996). The IRES encompasses most of the 5′ NTR and the following ∼15 nucleotides, which form two domains and a double pseudoknot structure (Niepmann, 2013). Translation of the HCV RNA is stimulated by several cis-acting RNA elements (CREs) residing in the 3′ NTR and by two stem-loop structures located in the core-coding region (Figure 1). In line with current models proposed for other positive-strand RNA viruses, recent studies suggest that the HCV genome can circularize by interactions between motifs in the IRES and stem-loop structures residing in the NS5B coding region (Romero-López et al., 2014). While these in vitro studies argue for direct RNA-RNA interaction, in cells a possible circularization of the HCV genome is likely facilitated by viral and cellular proteins. These include the NFAR proteins and IGF2BP1 (insulin-like growth factor II mRNA-binding protein 1) (Isken et al., 2007, Weinlich et al., 2009). Regardless of the mechanism, circularization of the HCV genome might help to avoid “clashes” between translating ribosomes moving in the 5′ to 3′ direction and the viral replicase complex copying the RNA in the 3′ to 5′ direction. In addition, HCV subverts the highly abundant liver-specific microRNA-122 (miR-122) (Jopling et al., 2005) (Figure 1), which binds to two sites in the 5′ NTR of the HCV RNA genome. This promotes accumulation of viral RNA by several nonexclusive mechanisms, such as stimulation of IRES-mediated translation, enhancement of HCV RNA replication, and protecting viral RNA from degradation (Niepmann, 2013). Thus, in contrast to the usually negative regulation exerted by miRNAs, in case of HCV, miR-122 seems to be a positive regulator.
HCV Replication Machinery and Involved Host Cell Factors
After polyprotein cleavage, the viral replicase complex is constituted (Figure 2 ). It is composed of at least NS3 to NS5B and the genomic RNA template. Although the NS2 protease per se is not essential for RNA replication, cleavage at the NS2/NS3 junction appears to be a rate-limiting step (Madan et al., 2014). Since fully processed NS3 is required for RNA replication, NS2 might indirectly affect replication.
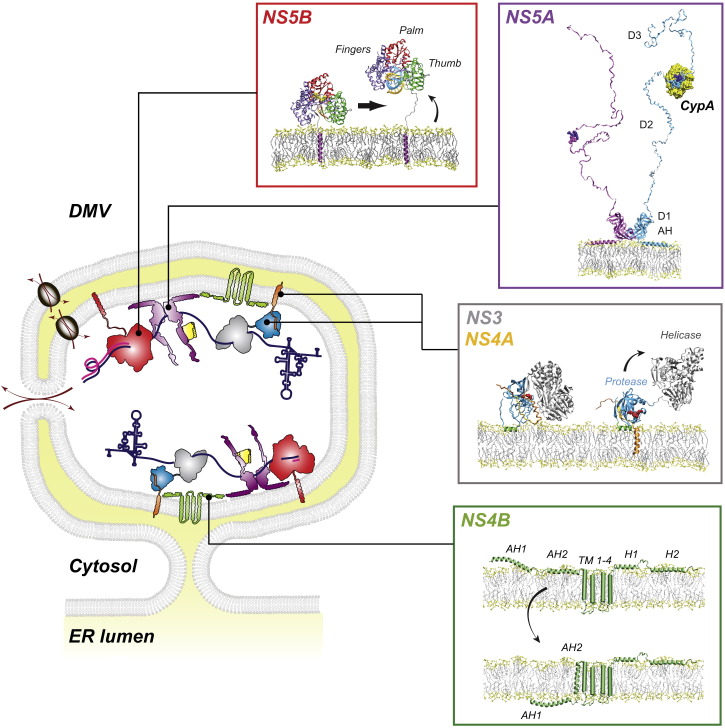
Figure 2.
Model of an HCV-Induced Double-Membrane Vesicle and Hypothetical 3D Structures of Membrane-Associated HCV Proteins
Virus-induced double-membrane vesicles (DMVs) contain HCV nonstructural proteins and RNA and are sites of active RNA replication. The DMV might contain a (transient) opening or a distinct transporter to allow exchange of nucleotides and viral RNA of the DMV interior with the cytoplasm. Note that the viral replicase might also reside on the outer surface of the DMV (not shown). Ribbon diagrams of membrane-associated HCV proteins and assumed conformational changes required for replication are indicated in boxes on the right. NS5B: the structure on the left corresponds to the closed conformation, representing the potential initiation state of the enzyme. The panel on the right shows NS5B in a hypothetical elongation mode. This conformational change would release the RNA binding groove to accommodate a dsRNA replication intermediate (shown in blue and yellow). NS5A: model of a full-length dimer associated to a membrane via the N-terminal amphipathic α helix. Only the clam-like dimer (Tellinghuisen et al., 2005) is shown for simplicity. Domains (D) 2 and 3 are intrinsically unfolded and thought to interact with multiple co-opted host factors, including cyclophilin A (CypA) binding to D2. NS3-4A complex: the presumed membrane orientation of the NS3-4A complex during polyprotein synthesis and prior to self-cleavage at the NS3/4A site is shown on the left. After cleavage, profound structural changes occur, most notably a membrane insertion of the C-terminal tail of NS4A (orange) and a repositioning of the C-terminal NS3-helicase domain (gray) away from the membrane. NS4B: the proposed dual membrane topology is shown. Structures of amphipathic α helices AH2 and H2 have been determined experimentally, while the other structural elements are based on in silico predictions. AH2 potentially traverses the membrane posttranslationally. Structure models are adapted from Bartenschlager et al. (2013) with permission from the publisher and Francois Penin.
NS3 is a bifunctional molecule composed of an N-terminal serine-protease domain, activated by tight interactions with the NS4A cofactor, and a C-terminal helicase domain (Moradpour and Penin, 2013) (Figure 2). While the protease, in complex with NS4A, is responsible for polyprotein cleavage, the role of the helicase domain in viral replication is poorly defined. In vitro it can unwind RNA in an inchworm or ratchet-like manner (Dumont et al., 2006, Gu and Rice, 2010, Appleby et al., 2011), and it was shown that helicase activity is important for RNA replication. In addition, the linker connecting protease and helicase domain appears to be critical for assembly, eventually mediating NS3 interactions with other viral or cellular proteins or by modulating NS3-4A structure (Kohlway et al., 2014b). In fact, the structurally flexible linker might transpose the helicase domain away from the membrane-proximal protease domain by a complex series of conformational changes (Brass et al., 2008) (Figure 2). Why the protease and helicase domains are linked is not known, but accumulating evidence reveals a crosstalk between the two domains (Moradpour and Penin, 2013).
The NS4A protease cofactor is a short transmembrane (TM) protein anchoring NS3 to the ER membrane (Figure 2) and playing a regulatory role for replicase complex function by stimulating both protease and RNA helicase activities of NS3 (Lindenbach et al., 2007). Moreover, formation of NS4A TM homodimers appears to be a prerequisite for efficient HCV replication and virus particle production, suggesting that NS3-4A dimers or oligomers are crucial for the viral life cycle (Kohlway et al., 2014a). Apart from the ER, NS4A seems to target the NS3-4A complex to mitochondrial membranes or defined ER regions closely opposed to mitochondria and designated mitochondria-associated membranes (MAMS) (Horner et al., 2011). This localization plays an important role for blocking the interferon (IFN) response by proteolytic cleavage of the mitochondrial antiviral-signaling protein (MAVS).
NS4B is a poorly characterized protein with a complex TM topology (Figure 2). It can undergo a posttranslational conformational change by flipping its N-terminal amphipathic α helix into the ER lumen (Gouttenoire et al., 2009), which might be regulated by NS5A (Lundin et al., 2006). In addition to its early recognized, central role in inducing membrane alterations (Egger et al., 2002), NS4B was found to be critically involved in RNA replication as well as formation of infectious HCV particles (Jones et al., 2009, Paul et al., 2011). The underlying mechanisms are poorly defined, but NS4B properties are most likely regulated by multiple interactions either with itself by forming homo-oligomers or with other viral proteins, including NS3 and NS5A (Paredes and Blight, 2008, Gouttenoire et al., 2010, Paul et al., 2011).
NS5A is a multifunctional protein associated with intracellular membranes via an N-terminal amphipathic α helix (Figure 2). The protein can form different kinds of homodimers and, eventually, also oligomers (Tellinghuisen et al., 2005, Love et al., 2009, Lambert et al., 2014). NS5A is composed of three domains, separated by low-complexity sequences. Only domain I is structured, whereas domains II and III are intrinsically unfolded. It is widely accepted that NS5A is a RNA-binding phosphoprotein that exists as a basal and a hyperphosphorylated form. The phosphorylation status of NS5A appears to be determined by several kinases, such as casein kinases (CK) I and II, polo-like kinase 1, glycogen synthase kinase 3, protein kinase A, and mitogen-activated protein kinases (MAPKs), but the physiological relevance of these kinases for NS5A phosphorylation remains to be established (Masaki et al., 2014, Cordek et al., 2014). Apart from that, several studies reported a phosphorylation-independent activation of NS5B RNA-dependent RNA polymerase (RdRp) by NS5A, at least in vitro. This activation might be mediated by the RNA-binding ability of NS5A or by direct interaction with NS5B (Shirota et al., 2002, Quezada and Kane, 2013).
No enzymatic activities have been ascribed to NS5A. Its functions are presumably exerted by interactions with various cellular factors, including VAPA (vesicle-associated membrane protein-associated protein A), CypA (cyclophilin A), PI4KIIIα (phosphatidylinositol-4-kinase IIIα), or ApoE (apolipoprotein E), as described in detail below. One of the best-studied factors is CypA, a prolyl-peptidyl isomerase (PPI) that is required for HCV replication (Kaul et al., 2009, Liu et al., 2009). NS5A binds to CypA (Figure 2), eventually altering local folding of domains II and III (Coelmont et al., 2010, Verdegem et al., 2011). CypA appears to increase NS5A RNA-binding capacity, which is reduced by Cyp inhibitors (Foster et al., 2011). Strikingly, the function of NS5A that is necessary for formation of the membranous replication compartment (see below) also seems to require CypA PPI activity (Madan et al., 2014). However, the appealing hypothesis that RNA binding and induction of membrane rearrangements by NS5A are linked remains to be explored.
The key enzyme catalyzing viral RNA replication is the NS5B RdRp (Figure 2). It is composed of an N-terminal catalytic domain, a linker, and a hydrophobic transmembrane domain (TMD) comprising the C-terminal 21 aa, anchoring the protein to intracellular membranes. Although crucial for viral replication in cell culture, the TMD is dispensable for RdRp activity in vitro. 3D crystal structures of NS5Bs from various genotypes revealed a typical right-hand shape with fingers, thumb, and palm subdomains; this feature is shared by most other polymerases (Ago et al., 1999, Bressanelli et al., 1999). A distinctive property of the HCV RdRp is the completely encircled catalytic site, resulting from multiple interactions between the fingers and thumb subdomains, and from structural movements of the linker and a β hairpin of the thumb subdomain that protrudes into the active site (Lesburg et al., 1999). This structure represents the so-called “closed” conformation of the polymerase and is assumed to correspond to the active form of the RdRp responsible for de novo (i.e., primer-independent) initiation of RNA synthesis. This enzyme can bind the single-strand template and priming nucleotides but is too narrow to accommodate double-strand (ds) RNA as formed during RNA synthesis (Simister et al., 2009) (Figure 2).
HCV RNA Synthesis
De novo priming by NS5B requires two nucleotide binding sites in the catalytic pocket of the enzyme to synthesize a dinucleotide (Ferrari et al., 2008). At least in vitro, de novo initiation at the very 3′ end of the template RNA is specifically stimulated by high concentrations of GTP (Lohmann et al., 1999b) that binds to an allosteric site (Bressanelli et al., 2002). Binding of GTP to NS5B appears to play a role in triggering the switch from primer-dinucleotide formation to elongation of RNA synthesis (Harrus et al., 2010, Ranjith-Kumar et al., 2003). This switch requires profound conformational changes in the NS5B structure, including a displacement of the linker and the β-hairpin flap that obstructs the catalytic pocket and blocks the egress of nascent dsRNA (Mosley et al., 2012, Scrima et al., 2012). In this way, the entire enzymatic core is opened up to accommodate dsRNA. In line with this model, the structure of RdRp from the highly replication-competent isolate JFH-1 has an unusually closed active site; accordingly, in vitro, this enzyme shows unprecedented de novo initiation efficiency (Simister et al., 2009).
HCV RNA replication is a multi-step process that is orchestrated by the coordinated action of viral and cellular proteins as well as multiple CREs (Lohmann, 2013) (Figure 1). It is assumed that synthesis of negative-strand RNA initiates at the 3′ end of the viral genome. This reaction appears to be rate-limiting, as positive-strand RNAs are produced in an excess of 5- to 10-fold. Furthermore, it remains unclear how RNA translation and replication are regulated. As described above, circularization of the RNA genome might be one mechanism. Occupation of the 3′ and 5′ NTRs by different viral or cellular proteins or formation of alternative RNA structures are other possibilities. In support of the latter possibility, it was shown that the 5′ NTR of the positive-strand RNA genome and its complementary sequence, i.e., the 3′ NTR of negative-strand RNA, adopt very different secondary structures (Smith et al., 2002). Moreover, RNA sequences in domain II of the IRES are essential for RNA replication. This overlap of signals might be involved in the regulation of a switch from RNA translation to replication.
Structure and Biogenesis of HCV Replication Factories
Like all other positive-strand RNA viruses, HCV extensively remodels intracellular membranes, giving rise to organelle-like membranous structures, commonly referred to as viral replication factories (vRFs) that serve multiple purposes: (i) increasing local concentration of factors required for efficient RNA replication, (ii) spatial coordination of different steps of the viral replication cycle (RNA translation, replication, assembly), and (iii) protecting viral proteins and RNA from antiviral defenses. Membrane rearrangements induced by positive-strand RNA viruses can be assigned to two morphological subclasses: the invaginated vesicle/spherule type and the double-membrane vesicle (DMV) type (Paul and Bartenschlager, 2013). Despite the very distant evolutionary relationship, HCV, picornaviruses, and coronaviruses belong to the DMV type, whereas the more closely HCV-related flaviviruses such as Dengue virus and West Nile virus induce invaginated vesicles within the ER. These morphologies are thought to reflect the subversion of common host cell pathways to establish the membranous vRFs.
Electron microscopy (EM) studies of cells expressing the HCV polyprotein or containing replicating HCV RNAs revealed cytoplasmic vesicular structures embedded into a membranous matrix, designated membranous web (MW) (Egger et al., 2002, Gosert et al., 2003). Follow-up studies with more sophisticated EM methods and using HCV-infected cells identified predominantly DMVs with an average diameter of ∼150 nm that are most likely derived from the endoplasmic reticulum (ER) (Romero-Brey et al., 2012, Ferraris et al., 2013). Several lines of evidence suggest that DMVs are important for HCV replication. First, DMV appearance correlates well with viral RNA replication kinetics upon HCV infection or transfection of subgenomic replicon RNAs independent from the genotype. Second, purified DMVs contain enzymatically active viral replicase, suggesting that they are bona fide HCV vRFs (Paul et al., 2013) (Figure 2). However, the exact topology of the viral replicase with respect to DMV membranes is elusive. The membrane-protected nature of HCV RNA is indicative of DMV-luminal replication sites, which would require transport of metabolites and progeny RNA across the two membranes. Possibly, components of the nuclear pore complex, reported to interact and colocalize with viral proteins in HCV-infected cells, might contribute to this transport (Neufeldt et al., 2013). Alternatively, assuming that DMVs are highly dynamic structures, active replication might occur in the interior of DMVs as long as they are connected to the cytosol (Romero-Brey et al., 2012) but cease once DMV membranes close.
Induction of DMVs does not require viral RNA replication but can be triggered by the sole expression of HCV replicase proteins NS3–5B. NS4B is proposed to be the primary inducer of the MW (Egger et al., 2002). Indeed, mutational analysis has shown that self-interaction of NS4B, when expressed in the context of a NS3–5B polyprotein, is required for DMV induction (Gouttenoire et al., 2010, Paul et al., 2011). Moreover, the C-terminal NS4B domain (Figure 2) has been reported to alter membrane integrity in vitro, underlining intrinsic NS4B membrane activity (Palomares-Jerez et al., 2012). However, recent data suggest that individual expression of NS4B is insufficient to induce DMVs (Romero-Brey et al., 2012). Instead, formation of MW-like structures appears to require a concerted action of all HCV replicase factors (NS3–5B).
Although HCV proteins are the main drivers of membrane remodeling, host factors crucially contribute to vRF formation. For example, CypA that is thought to act on NS5A (Figure 2) appears to contribute to formation of HCV vRFs (Madan et al., 2014). Another example is PSTPIP2 (proline-serine-threonine phosphatase interacting protein 2), which belongs to the BAR (Bin-Amphiphysin-Rvs) domain-containing protein family that act as sensors and/or inducers of positive membrane curvature. PSTPIP2 was recently shown to be required for HCV-induced membrane alterations and, thus, RNA replication (Chao et al., 2012). Both NS4B and NS5A interact with and thereby recruit PSTPIP2 to HCV-remodeled membranes, and depletion of this host factor abrogates DMV formation.
Besides remodeling existing intracellular membranes, HCV induces de novo lipid and membrane biosynthesis via the sterol regulatory element-binding protein (SREBP) pathway (Waris et al., 2007), causing distinct changes in the lipidomic profile of HCV-infected cells (Diamond et al., 2010) (Figure 3 ). Proteolytic cleavage of SREBPs has been observed in HCV-infected as well as core- and NS4B-overexpressing cells (Waris et al., 2007, Park et al., 2009). This leads to elevated levels of lipogenic transcripts, such as fatty acid synthase (FAS) and HMG-CoA (3-hydroxy-3-methylglutaryl coenzyme-A) reductase, the rate-limiting enzyme of the cholesterol biosynthetic mevalonate pathway. In addition, a role of the metabolic intermediate geranylgeranyl phosphate in protein prenylation was shown to be required for viral replication (Ye et al., 2003). HCV also activates lipogenic genes by subversion of an innate immunity pathway. It was found that interaction of the HCV 3′ NTR with DDX3X (DEAD box polypeptide 3 X-linked) activates IKK-α, which in turn facilitates SREBP transcriptional activity (Li et al., 2013). Finally, FAS has recently been described to interact with NS5B and stimulate RdRp activity (Huang et al., 2013).
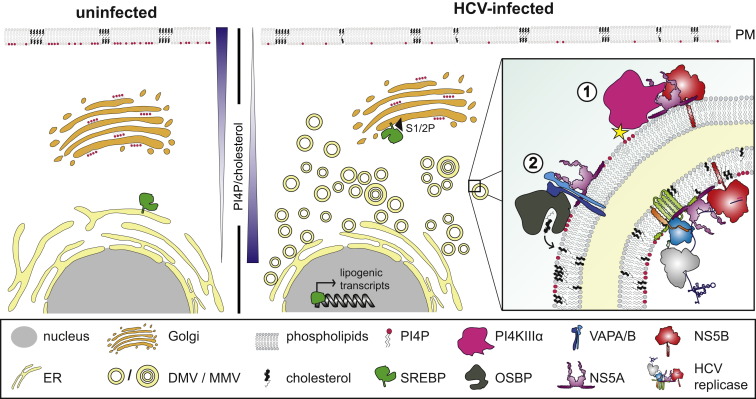
Figure 3.
HCV-Mediated Subversion of Lipid Homeostasis Implicated in MW Biogenesis
HCV induces extensive remodeling of intracellular membranes, most notably double-membrane vesicles (DMVs) and less frequently multimembrane vesicles (MMVs). Right panel: model of HCV-subverted host cell proteins and lipids implicated in MW biogenesis. PI4KIIIα interacts with NS5A and NS5B to induce elevated levels of PI4P (1). Subsequently, oxysterol-binding protein (OSBP) delivers cholesterol to HCV-remodeled membranes. OSBP recruitment is facilitated by locally elevated PI4P levels as well as interactions with VAPs and NS5A (2). For comparison, a naive cell is shown in the left panel.
Another important determinant of vRFs appears to be the local lipid composition. This is best illustrated by the strong dependence of HCV on the lipid kinase PI4KIIIα and its product, phosphatidylinositol-4-phosphate (PI4P). In noninfected cells, PI4P is predominantly found in Golgi membranes and the inner leaflet of the plasma membrane (Figure 3). However, upon HCV infection, presumably via interactions with NS5A and NS5B, subcellular localization of PI4KIIIα is altered, concomitant with an increase in intracellular PI4P levels and a decrease of the PI4P plasma membrane pool (Figure 3) (Reiss et al., 2011, Bianco et al., 2012). PI4KIIIα knockdown impairs HCV replication and causes aggregation of DMVs with significantly reduced diameter (Reiss et al., 2011), which can be phenocopied by pharmacological inhibition of the kinase (Wang et al., 2014). Hence, subversion of PI4KIIIα and locally elevated PI4P levels seem to be dispensable for DMV formation per se but required to modulate lipid content influencing membrane characteristics that are important for DMV functionality. Notably, DMV induction by the distantly related picornaviruses also requires PI4P, arguing for an evolutionarily conserved mechanism (Altan-Bonnet and Balla, 2012).
One pathway linked to PI4P and involving nonvesicular cholesterol transport by oxysterol-binding protein (OSBP) appears to be subverted by HCV for vRF biogenesis (Wang et al., 2014). The N-terminal PI4P-binding pleckstrin-homology domain of OSBP and the C-terminal sterol-binding domain are implicated in cholesterol transport to PI4P-containing HCV-remodeled membranes (Figure 3), consistent with the high cholesterol content of DMVs. This pathway is further stimulated by VAPA and VAPB, which are recruited by NS5A and required for HCV RNA replication (Evans et al., 2004, Gao et al., 2004). Importantly, OSBP interacts with VAPs via its FFAT motif (Mesmin et al., 2013), which is also required to promote cholesterol transport to HCV-remodeled membranes and hence viral replication (Wang et al., 2014). Importantly, distantly related Rhinoviruses subvert this cellular lipid transport system in a strikingly similar way (Roulin et al., 2014), arguing for an evolutionarily conserved mechanism of DMV biogenesis. In case of HCV, reduced DMV diameters were observed both by blocking OSBP-mediated cholesterol transport in HCV-infected cells (Wang et al., 2014) and by depletion of cholesterol from purified DMVs (Paul et al., 2013), arguing that cholesterol is an important structural component of HCV-remodeled membranes. Although DMV morphotypes induced by knockdown of PI4KIIIα or OSBP are very similar, kinase knockdown blocks HCV RNA replication to a greater extent, arguing for additional effector functions of PI4KIIIα and PI4P in viral RNA amplification. In any case, cholesterol enrichment in HCV-induced DMVs could explain the reported association of the viral replicase with detergent-resistant lipid-raft-like assemblies (Shi et al., 2003) that are additionally enriched in sphingolipids. Indeed, HCV infection induces synthesis of specific sphingolipids that enhance NS5B-mediated RNA replication (Hirata et al., 2012). Additionally, HCV has been shown recently to subvert another PI4P effector, namely four-phosphate adaptor protein 2 that, via an analogous mechanism as described above for OSBP, appears to mediate sphingolipid transport to viral replication sites (Khan et al., 2014). These findings highlight the importance of specific lipids in HCV RNA replication, which we are only beginning to understand.
Another cellular pathway eventually exploited by HCV and other positive-strand RNA viruses to build up vRFs is autophagy. One obvious link is the close morphological similarity of autophagosomes and DMV-type vRFs. Moreover, HCV infection induces LC3 lipidation, a key event in autophagosome formation (Dreux et al., 2009), and lipidated LC3 associates with HCV protein-containing membrane fractions (Ferraris et al., 2010). However, conflicting data as to the step of the HCV replication cycle affected by autophagy have been reported. Autophagy was reported to stimulate translation of incoming HCV RNA without affecting RNA replication (Dreux et al., 2009). Others suggest that autophagosomes serve as platforms for HCV RNA synthesis (Sir et al., 2012) or stimulate replication by downregulation of innate immune response (Ke and Chen, 2011). Finally, autophagy might affect virus production (Tanida et al., 2009). Hence, the impact of autophagy on HCV replication might be multifaceted. However, whether the autophagy core machinery plays a role in viral DMV biogenesis remains to be elucidated.
Mechanisms of HCV Particle Production
A remarkable feature of infectious HCV particles is their unusually low buoyant density ranging between <1.06 g/ml in the case of serum-derived particles (André et al., 2002) and ∼1.1 g/ml for cell culture-grown HCV (HCVcc) (Lindenbach et al., 2006). This exceptionally low density of HCV particles is reflected by their lipid composition resembling low-density lipoprotein (LDL) particles (Merz et al., 2011). Thus, HCV virions might exist either as single hybrid particles or as conventional virions decorated with LDL-like particles (Bartenschlager et al., 2011, Lindenbach and Rice, 2013) (Figure 4 ). In any case, HCV particles are pleomorphic, lack discernable surface features, and have a broad size range (40–80 nm diameter) (Gastaminza et al., 2010, Merz et al., 2011, Catanese et al., 2013). For these reasons, even advanced electron tomography techniques failed to define a precise virion structure (Catanese et al., 2013).
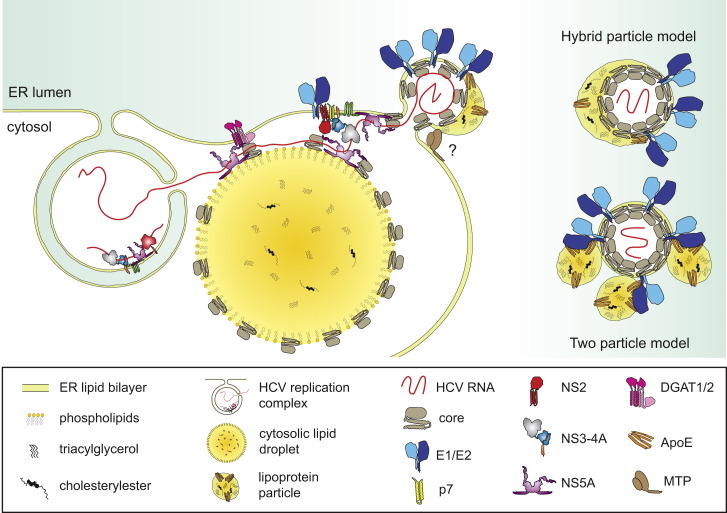
Figure 4.
Model of HCV Particle Production
Left panel: hypothetical model of HCV assembly. Viral progeny RNA is shuttled from replication sites to cytosolic lipid droplets (cLDs), facilitated by NS3 and NS5A. Core protein eventually rerecruited from cLDs to the ER is thought to trigger nucleocapsid formation and budding into the ER lumen. Presumably by tight interaction between NS2 and NS3 protease domains, NS2 brings together structural and nonstructural proteins. Nascent virions incorporate cellular lipoproteins, especially ApoE, presumably required for lipidation of virus particles. This lipidation might occur during budding, according to the hybrid particle model, or during egress via interaction between the virion and VLDL particles, according to the dual-particle model (right panel).
Spatiotemporal control of viral replication and assembly likely requires separation of core protein from the replicase in order to avoid competition at the level of RNA binding. One solution to this problem might be the (temporal) recruitment of core to distinct subcellular compartments. In the case of HCV, core accumulates on the surface of cytosolic lipid droplets (cLDs) (Miyanari et al., 2007). Interestingly, NS5A, which is essential for assembly, also localizes to cLDs (Appel et al., 2008) (Figure 4). Assembly competence of NS5A is mediated by domain III and CK-mediated phosphorylation at a C-terminal serine cluster (Tellinghuisen et al., 2008). Interestingly, CK1-dependent NS5A hyperphosphorylation appears to recruit NS5A to low-density membrane fractions around cLDs, thus promoting interaction with core and nucleocapsid assembly (Masaki et al., 2014). Core recruitment to cLDs is influenced by cellular enzymes involved in LD homeostasis, including cytosolic phospholipase A2 (Menzel et al., 2012) and diacylglycerol acyltransferase-1 (Herker et al., 2010). The latter also binds to and recruits NS5A to cLDs, thus promoting core-NS5A interaction (Camus et al., 2013). Moreover, recruitment of core from cLDs into motile structures, possibly representing assembled HCV particles, has been observed (Counihan et al., 2011). Finally, multiple host factors, such as the RNA binding protein YB-1 (Chatel-Chaix et al., 2013), promoting HCV particle production are recruited to cLDs, underlining their exceptional role as a hub for HCV assembly.
Thus far, it has not been possible to visualize sites of HCV assembly, and therefore detailed knowledge about the individual steps of this process is lacking. Nevertheless, it is generally assumed that nucleocapsid formation and budding are spatially and temporally linked events (Figure 4). One key viral player is NS2, interacting on one hand with a presumed E1/E2/p7 complex and on the other hand most prominently with the NS3-4A complex (Phan et al., 2009, Jirasko et al., 2010, Popescu et al., 2011). In case of the latter, the NS3 linker region and helicase domain are key players (Phan et al., 2009, Kohlway et al., 2014b), but whether helicase activity per se is required for assembly or whether its RNA binding activity is sufficient remains unknown. In addition, homotypic self-interactions of the NS4A TMD influence particle production (Kohlway et al., 2014a). Surprisingly, NS4B and NS5B also affect HCV assembly, but underlying mechanisms are unknown (Jones et al., 2009, Paul et al., 2011, Gouklani et al., 2012). These data illustrate the complexity of HCV assembly since virtually all viral proteins seem to participate in this process. Novel experimental approaches, including trans-complementation assays, as recently established for NS5A (Herod et al., 2014), are needed to resolve the individual steps of HCV virion formation.
HCV particle production is tightly linked to apolipoproteins E, A1, C1, and B. While different degrees of ApoB incorporation into serum-derived versus HCVcc particles have been reported (Bartenschlager et al., 2011), several lines of evidence point toward crucial roles of ApoC1, and especially ApoE, in HCV assembly. First, virus particles are efficiently neutralized by antibodies targeting ApoC1 and ApoE (Meunier et al., 2008, Chang et al., 2007). Second, knockdown of ApoE reduces HCV particle production (Chang et al., 2007). Third, cells depleted from ApoE do not produce infectious HCV particles, and this defect can be restored upon ectopic expression of ApoE (Long et al., 2011, Da Costa et al., 2012). Fourth, ApoE was found to interact with NS5A and viral envelope glycoproteins (Jiang and Luo, 2009, Boyer et al., 2014, Lee et al., 2014). Recent functional data point toward a role of ApoE in a postenvelopment step of HCV particle production (Hueging et al., 2014, Lee et al., 2014), consistent with earlier reported imaging studies (Coller et al., 2012). A drawback of studying HCVcc formation is the inability of most cell lines to produce authentic VLDL particles, which was thought to explain the observed density differences between serum- and cell culture-derived HCV particles. However, this view was challenged by recent studies based on HepG2 cells engineered to allow VLDL secretion; these cells failed to produce ApoB-containing very-low-density HCV lipoviral particles (Jammart et al., 2013).
Formation of VLDL requires the microsomal triglyceride transfer protein (MTP) mediating triacylglycerol incorporation into nascent ER-luminal LDs and lipid loading of ApoB. While some studies observed a decrease in viral titers upon inhibition or knockdown of MTP (Gastaminza et al., 2008, Huang et al., 2007), others could not corroborate these findings (Chang et al., 2007, Jiang and Luo, 2009), perhaps reflecting the use of cell-culture systems differing in their capacity to produce authentic VLDL. HCV virions are thought to be transported along the conventional secretory pathway to the Golgi, where E1 and E2 glycoproteins undergo complex modifications (Vieyres et al., 2014). Moreover, microtubular transport machinery and the endocytic recycling compartment (ERC) are involved in the exit of HCV particles (Coller et al., 2012). Although HCV particle production appears to require components of the ESCRT machinery, a role in membrane scission during budding of HCV particles seems rather unlikely. Instead, ESCRT function might indirectly promote HCV particle release by modulating the ERC (Corless et al., 2010, Tamai et al., 2012).
Conclusions and Future Perspectives
Despite important new insights into structure and function of viral proteins, many details of HCV RNA translation, replication, and assembly, the spatiotemporal coupling of these processes, and the involvement of co-opted host factors remain poorly understood. This is due, at least in part, to the lack of adequate experimental systems. For instance, up to now, in vitro studies of HCV RNA synthesis have been limited to rather simplistic biochemical reactions lacking host cell membranes and proteins or are based on preassembled vRFs isolated from cells and recapitulating only some of the involved reactions. Thus, in vitro systems similar to those developed for picornaviruses are needed to decipher the different steps of the HCV replication cycle. Moreover, deciphering the molecular composition of the membranous HCV vRFs will be instrumental in identifying the plethora of cellular proteins and lipids involved in HCV replication and eventually assembly. Importantly, unraveling of the 3D structures of full-length HCV proteins in their most authentic state, i.e., in association with lipids, will be required to understand their mode of action. Novel approaches including in vitro expression of proteins in the presence of lipids coupled with solid-state NMR or X-ray free-electron laser might help to overcome these hurdles. Another challenge is to decipher the biogenesis of DMVs and their dynamics. Thus far, no intermediates have been detected, and the link to autophagy, if any, remains unclear.
Equally unresolved is the role of LDs in the different steps of virus particle formation and the sites of HCV assembly, which so far could not be visualized. Since biochemical approaches are limited by the rareness of assembly events, imaging techniques such as correlative light electron microscopy offer an attractive alternative. These approaches can be complemented by super-resolution microscopy in combination with novel detection methods, such as metabolic labeling of viral RNA and proteins. Moreover, live-cell imaging has the potential to discover mechanisms orchestrating the spatiotemporal regulation of HCV RNA translation, replication, and packaging. The availability of very powerful molecular virology tools that have been developed for HCV makes it an ideal model system to gain fundamental insights into the relationship between the host cell and a noncytolytic virus.
Author Contributions
D.P. and V.M. contributed equally to drafting the first version of the manuscript and preparing the figures. R.B. edited and commented on the manuscript. All authors edited and approved the final version of the manuscript.
Acknowledgments
We are grateful to Francois Penin for permission to use the models shown in Figure 2. They are reproduced in consent with Nature Publishing Group policy. We apologize to all colleagues whose work could not be cited due to space constraints. Work in the authors’ laboratory is supported by the Deutsche Forschungsgemeinschaft (SFB638, TP5; TRR83, TP13; FOR1202, TP1) and Syspatho (European Union, 7th framework program, 260429).
References
- Ago H., Adachi T., Yoshida A., Yamamoto M., Habuka N., Yatsunami K., Miyano M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure. 1999;7:1417–1426. doi: 10.1016/s0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- Altan-Bonnet N., Balla T. Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem. Sci. 2012;37:293–302. doi: 10.1016/j.tibs.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André P., Komurian-Pradel F., Deforges S., Perret M., Berland J.L., Sodoyer M., Pol S., Bréchot C., Paranhos-Baccalà G., Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 2002;76:6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel N., Zayas M., Miller S., Krijnse-Locker J., Schaller T., Friebe P., Kallis S., Engel U., Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008;4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleby T.C., Anderson R., Fedorova O., Pyle A.M., Wang R., Liu X., Brendza K.M., Somoza J.R. Visualizing ATP-dependent RNA translocation by the NS3 helicase from HCV. J. Mol. Biol. 2011;405:1139–1153. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R., Penin F., Lohmann V., André P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Lohmann V., Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 2013;11:482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- Bianco A., Reghellin V., Donnici L., Fenu S., Alvarez R., Baruffa C., Peri F., Pagani M., Abrignani S., Neddermann P., De Francesco R. Metabolism of phosphatidylinositol 4-kinase IIIα-dependent PI4P Is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog. 2012;8:e1002576. doi: 10.1371/journal.ppat.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer A., Dumans A., Beaumont E., Etienne L., Roingeard P., Meunier J.C. The association of hepatitis C virus glycoproteins with apolipoproteins E and B early in assembly is conserved in lipoviral particles. J. Biol. Chem. 2014;289:18904–18913. doi: 10.1074/jbc.M113.538256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass V., Berke J.M., Montserret R., Blum H.E., Penin F., Moradpour D. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc. Natl. Acad. Sci. USA. 2008;105:14545–14550. doi: 10.1073/pnas.0807298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressanelli S., Tomei L., Roussel A., Incitti I., Vitale R.L., Mathieu M., De Francesco R., Rey F.A. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressanelli S., Tomei L., Rey F.A., De Francesco R. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 2002;76:3482–3492. doi: 10.1128/JVI.76.7.3482-3492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus G., Herker E., Modi A.A., Haas J.T., Ramage H.R., Farese R.V., Jr., Ott M. Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J. Biol. Chem. 2013;288:9915–9923. doi: 10.1074/jbc.M112.434910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanese M.T., Uryu K., Kopp M., Edwards T.J., Andrus L., Rice W.J., Silvestry M., Kuhn R.J., Rice C.M. Ultrastructural analysis of hepatitis C virus particles. Proc. Natl. Acad. Sci. USA. 2013;110:9505–9510. doi: 10.1073/pnas.1307527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.S., Jiang J., Cai Z., Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 2007;81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao T.C., Su W.C., Huang J.Y., Chen Y.C., Jeng K.S., Wang H.D., Lai M.M. Proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), a host membrane-deforming protein, is critical for membranous web formation in hepatitis C virus replication. J. Virol. 2012;86:1739–1749. doi: 10.1128/JVI.06001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L., Germain M.A., Motorina A., Bonneil É., Thibault P., Baril M., Lamarre D. A host YB-1 ribonucleoprotein complex is hijacked by hepatitis C virus for the control of NS3-dependent particle production. J. Virol. 2013;87:11704–11720. doi: 10.1128/JVI.01474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelmont L., Hanoulle X., Chatterji U., Berger C., Snoeck J., Bobardt M., Lim P., Vliegen I., Paeshuyse J., Vuagniaux G. DEB025 (Alisporivir) inhibits hepatitis C virus replication by preventing a cyclophilin A inducedcis-trans isomerisation in domain II of NS5A. PLoS ONE. 2010;5:e13687. doi: 10.1371/journal.pone.0013687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller K.E., Heaton N.S., Berger K.L., Cooper J.D., Saunders J.L., Randall G. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 2012;8:e1002466. doi: 10.1371/journal.ppat.1002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordek D.G., Croom-Perez T.J., Hwang J., Hargittai M.R., Subba-Reddy C.V., Han Q., Lodeiro M.F., Ning G., McCrory T.S., Arnold J.J. Expanding the proteome of an RNA virus by phosphorylation of an intrinsically disordered viral protein. J. Biol. Chem. 2014;289:24397–24416. doi: 10.1074/jbc.M114.589911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless L., Crump C.M., Griffin S.D., Harris M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J. Gen. Virol. 2010;91:362–372. doi: 10.1099/vir.0.017285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counihan N.A., Rawlinson S.M., Lindenbach B.D. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog. 2011;7:e1002302. doi: 10.1371/journal.ppat.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa D., Turek M., Felmlee D.J., Girardi E., Pfeffer S., Long G., Bartenschlager R., Zeisel M.B., Baumert T.F. Reconstitution of the entire hepatitis C virus life cycle in nonhepatic cells. J. Virol. 2012;86:11919–11925. doi: 10.1128/JVI.01066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond D.L., Syder A.J., Jacobs J.M., Sorensen C.M., Walters K.A., Proll S.C., McDermott J.E., Gritsenko M.A., Zhang Q., Zhao R. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., von Schaewen M., Ploss A. The impact of hepatitis C virus entry on viral tropism. Cell Host Microbe. 2014;16:562–568. doi: 10.1016/j.chom.2014.10.009. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M., Gastaminza P., Wieland S.F., Chisari F.V. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. USA. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S., Cheng W., Serebrov V., Beran R.K., Tinoco I., Jr., Pyle A.M., Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D., Wölk B., Gosert R., Bianchi L., Blum H.E., Moradpour D., Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Rice C.M., Goff S.P. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. USA. 2004;101:13038–13043. doi: 10.1073/pnas.0405152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E., He Z., Palermo R.E., Huang H.C. Hepatitis C virus NS5B polymerase exhibits distinct nucleotide requirements for initiation and elongation. J. Biol. Chem. 2008;283:33893–33901. doi: 10.1074/jbc.M803094200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris P., Blanchard E., Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J. Gen. Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- Ferraris P., Beaumont E., Uzbekov R., Brand D., Gaillard J., Blanchard E., Roingeard P. Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection. Cell. Mol. Life Sci. 2013;70:1297–1306. doi: 10.1007/s00018-012-1213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T.L., Gallay P., Stonehouse N.J., Harris M. Cyclophilin A interacts with domain II of hepatitis C virus NS5A and stimulates RNA binding in an isomerase-dependent manner. J. Virol. 2011;85:7460–7464. doi: 10.1128/JVI.00393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Aizaki H., He J.W., Lai M.M. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P., Cheng G., Wieland S., Zhong J., Liao W., Chisari F.V. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 2008;82:2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P., Dryden K.A., Boyd B., Wood M.R., Law M., Yeager M., Chisari F.V. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J. Virol. 2010;84:10999–11009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R., Egger D., Lohmann V., Bartenschlager R., Blum H.E., Bienz K., Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouklani H., Bull R.A., Beyer C., Coulibaly F., Gowans E.J., Drummer H.E., Netter H.J., White P.A., Haqshenas G. Hepatitis C virus nonstructural protein 5B is involved in virus morphogenesis. J. Virol. 2012;86:5080–5088. doi: 10.1128/JVI.07089-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouttenoire J., Castet V., Montserret R., Arora N., Raussens V., Ruysschaert J.M., Diesis E., Blum H.E., Penin F., Moradpour D. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J. Virol. 2009;83:6257–6268. doi: 10.1128/JVI.02663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouttenoire J., Roingeard P., Penin F., Moradpour D. Amphipathic alpha-helix AH2 is a major determinant for the oligomerization of hepatitis C virus nonstructural protein 4B. J. Virol. 2010;84:12529–12537. doi: 10.1128/JVI.01798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M., Rice C.M. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. USA. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrus D., Ahmed-El-Sayed N., Simister P.C., Miller S., Triconnet M., Hagedorn C.H., Mahias K., Rey F.A., Astier-Gin T., Bressanelli S. Further insights into the roles of GTP and the C terminus of the hepatitis C virus polymerase in the initiation of RNA synthesis. J. Biol. Chem. 2010;285:32906–32918. doi: 10.1074/jbc.M110.151316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E., Harris C., Hernandez C., Carpentier A., Kaehlcke K., Rosenberg A.R., Farese R.V., Jr., Ott M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat. Med. 2010;16:1295–1298. doi: 10.1038/nm.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herod M.R., Schregel V., Hinds C., Liu M., McLauchlan J., McCormick C.J. Genetic complementation of hepatitis C virus nonstructural protein functions associated with replication exhibits requirements that differ from those for virion assembly. J. Virol. 2014;88:2748–2762. doi: 10.1128/JVI.03588-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata Y., Ikeda K., Sudoh M., Tokunaga Y., Suzuki A., Weng L., Ohta M., Tobita Y., Okano K., Ozeki K. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog. 2012;8:e1002860. doi: 10.1371/journal.ppat.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M., Ping L.H., Rijnbrand R.C., Amphlett E., Clarke B., Rowlands D., Lemon S.M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- Horner S.M., Liu H.M., Park H.S., Briley J., Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. USA. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Sun F., Owen D.M., Li W., Chen Y., Gale M., Jr., Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. USA. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.T., Tseng C.P., Liao M.H., Lu S.C., Yeh W.Z., Sakamoto N., Chen C.M., Cheng J.C. Hepatitis C virus replication is modulated by the interaction of nonstructural protein NS5B and fatty acid synthase. J. Virol. 2013;87:4994–5004. doi: 10.1128/JVI.02526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueging K., Doepke M., Vieyres G., Bankwitz D., Frentzen A., Doerrbecker J., Gumz F., Haid S., Wölk B., Kaderali L., Pietschmann T. Apolipoprotein E codetermines tissue tropism of hepatitis C virus and is crucial for viral cell-to-cell transmission by contributing to a postenvelopment step of assembly. J. Virol. 2014;88:1433–1446. doi: 10.1128/JVI.01815-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O., Baroth M., Grassmann C.W., Weinlich S., Ostareck D.H., Ostareck-Lederer A., Behrens S.E. Nuclear factors are involved in hepatitis C virus RNA replication. RNA. 2007;13:1675–1692. doi: 10.1261/rna.594207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammart B., Michelet M., Pécheur E.I., Parent R., Bartosch B., Zoulim F., Durantel D. Very-low-density lipoprotein (VLDL)-producing and hepatitis C virus-replicating HepG2 cells secrete no more lipoviroparticles than VLDL-deficient Huh7.5 cells. J. Virol. 2013;87:5065–5080. doi: 10.1128/JVI.01405-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Luo G. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 2009;83:12680–12691. doi: 10.1128/JVI.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirasko V., Montserret R., Lee J.Y., Gouttenoire J., Moradpour D., Penin F., Bartenschlager R. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 2010;6:e1001233. doi: 10.1371/journal.ppat.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.M., Patel A.H., Targett-Adams P., McLauchlan J. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 2009;83:2163–2177. doi: 10.1128/JVI.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kaul A., Stauffer S., Berger C., Pertel T., Schmitt J., Kallis S., Zayas M., Lohmann V., Luban J., Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke P.Y., Chen S.S. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J. Clin. Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I., Katikaneni D.S., Han Q., Sanchez-Felipe L., Hanada K., Ambrose R.L., Mackenzie J.M., Konan K.V. Modulation of hepatitis C virus genome replication by glycosphingolipids and four-phosphate adaptor protein 2. J. Virol. 2014;88:12276–12295. doi: 10.1128/JVI.00970-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlway A., Pirakitikulr N., Barrera F.N., Potapova O., Engelman D.M., Pyle A.M., Lindenbach B.D. Hepatitis C virus RNA replication and virus particle assembly require specific dimerization of the NS4A protein transmembrane domain. J. Virol. 2014;88:628–642. doi: 10.1128/JVI.02052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlway A., Pirakitikulr N., Ding S.C., Yang F., Luo D., Lindenbach B.D., Pyle A.M. The linker region of NS3 plays a critical role in the replication and infectivity of hepatitis C virus. J. Virol. 2014;88:10970–10974. doi: 10.1128/JVI.00745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S.M., Langley D.R., Garnett J.A., Angell R., Hedgethorne K., Meanwell N.A., Matthews S.J. The crystal structure of NS5A domain 1 from genotype 1a reveals new clues to the mechanism of action for dimeric HCV inhibitors. Protein Sci. 2014;23:723–734. doi: 10.1002/pro.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Acosta E.G., Stoeck I.K., Long G., Hiet M.S., Mueller B., Fackler O.T., Kallis S., Bartenschlager R. Apolipoprotein e likely contributes to a maturation step of infectious hepatitis C virus particles and interacts with viral envelope glycoproteins. J. Virol. 2014;88:12422–12437. doi: 10.1128/JVI.01660-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesburg C.A., Cable M.B., Ferrari E., Hong Z., Mannarino A.F., Weber P.C. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 1999;6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- Li Q., Pène V., Krishnamurthy S., Cha H., Liang T.J. Hepatitis C virus infection activates an innate pathway involving IKK-α in lipogenesis and viral assembly. Nat. Med. 2013;19:722–729. doi: 10.1038/nm.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Rice C.M. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 2013;11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Meuleman P., Ploss A., Vanwolleghem T., Syder A.J., McKeating J.A., Lanford R.E., Feinstone S.M., Major M.E., Leroux-Roels G., Rice C.M. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. USA. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B.D., Prágai B.M., Montserret R., Beran R.K., Pyle A.M., Penin F., Rice C.M. The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J. Virol. 2007;81:8905–8918. doi: 10.1128/JVI.00937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Yang F., Robotham J.M., Tang H. Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J. Virol. 2009;83:6554–6565. doi: 10.1128/JVI.02550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V. Hepatitis C virus RNA replication. Curr. Top. Microbiol. Immunol. 2013;369:167–198. doi: 10.1007/978-3-642-27340-7_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V., Körner F., Koch J., Herian U., Theilmann L., Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Lohmann V., Overton H., Bartenschlager R. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 1999;274:10807–10815. doi: 10.1074/jbc.274.16.10807. [DOI] [PubMed] [Google Scholar]
- Long G., Hiet M.S., Windisch M.P., Lee J.Y., Lohmann V., Bartenschlager R. Mouse hepatic cells support assembly of infectious hepatitis C virus particles. Gastroenterology. 2011;141:1057–1066. doi: 10.1053/j.gastro.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Love R.A., Brodsky O., Hickey M.J., Wells P.A., Cronin C.N. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 2009;83:4395–4403. doi: 10.1128/JVI.02352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin M., Lindström H., Grönwall C., Persson M.A. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J. Gen. Virol. 2006;87:3263–3272. doi: 10.1099/vir.0.82211-0. [DOI] [PubMed] [Google Scholar]
- Madan V., Paul D., Lohmann V., Bartenschlager R. Inhibition of HCV replication by cyclophilin antagonists is linked to replication fitness and occurs by inhibition of membranous web formation. Gastroenterology. 2014;146:1361–1372. doi: 10.1053/j.gastro.2014.01.055. e1–e9. [DOI] [PubMed] [Google Scholar]
- Masaki T., Matsunaga S., Takahashi H., Nakashima K., Kimura Y., Ito M., Matsuda M., Murayama A., Kato T., Hirano H. Involvement of hepatitis C virus NS5A hyperphosphorylation mediated by casein kinase I-α in infectious virus production. J. Virol. 2014;88:7541–7555. doi: 10.1128/JVI.03170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel N., Fischl W., Hueging K., Bankwitz D., Frentzen A., Haid S., Gentzsch J., Kaderali L., Bartenschlager R., Pietschmann T. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog. 2012;8:e1002829. doi: 10.1371/journal.ppat.1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz A., Long G., Hiet M.S., Brügger B., Chlanda P., Andre P., Wieland F., Krijnse-Locker J., Bartenschlager R. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 2011;286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- Meunier J.C., Russell R.S., Engle R.E., Faulk K.N., Purcell R.H., Emerson S.U. Apolipoprotein c1 association with hepatitis C virus. J. Virol. 2008;82:9647–9656. doi: 10.1128/JVI.00914-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- Moradpour D., Penin F. Hepatitis C virus proteins: from structure to function. Curr. Top. Microbiol. Immunol. 2013;369:113–142. doi: 10.1007/978-3-642-27340-7_5. [DOI] [PubMed] [Google Scholar]
- Mosley R.T., Edwards T.E., Murakami E., Lam A.M., Grice R.L., Du J., Sofia M.J., Furman P.A., Otto M.J. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J. Virol. 2012;86:6503–6511. doi: 10.1128/JVI.00386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt C.J., Joyce M.A., Levin A., Steenbergen R.H., Pang D., Shields J., Tyrrell D.L., Wozniak R.W. Hepatitis C virus-induced cytoplasmic organelles use the nuclear transport machinery to establish an environment conducive to virus replication. PLoS Pathog. 2013;9:e1003744. doi: 10.1371/journal.ppat.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepmann M. Hepatitis C virus RNA translation. Curr. Top. Microbiol. Immunol. 2013;369:143–166. doi: 10.1007/978-3-642-27340-7_6. [DOI] [PubMed] [Google Scholar]
- Palomares-Jerez M.F., Nemesio H., Villalaín J. Interaction with membranes of the full C-terminal domain of protein NS4B from hepatitis C virus. Biochim. Biophys. Acta. 2012;1818:2536–2549. doi: 10.1016/j.bbamem.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Paredes A.M., Blight K.J. A genetic interaction between hepatitis C virus NS4B and NS3 is important for RNA replication. J. Virol. 2008;82:10671–10683. doi: 10.1128/JVI.00875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.Y., Jun H.J., Wakita T., Cheong J.H., Hwang S.B. Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway. J. Biol. Chem. 2009;284:9237–9246. doi: 10.1074/jbc.M808773200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol. 2013;2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Romero-Brey I., Gouttenoire J., Stoitsova S., Krijnse-Locker J., Moradpour D., Bartenschlager R. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J. Virol. 2011;85:6963–6976. doi: 10.1128/JVI.00502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Hoppe S., Saher G., Krijnse-Locker J., Bartenschlager R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 2013;87:10612–10627. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T., Beran R.K., Peters C., Lorenz I.C., Lindenbach B.D. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 2009;83:8379–8395. doi: 10.1128/JVI.00891-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu C.I., Callens N., Trinel D., Roingeard P., Moradpour D., Descamps V., Duverlie G., Penin F., Héliot L., Rouillé Y., Dubuisson J. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 2011;7:e1001278. doi: 10.1371/journal.ppat.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada E.M., Kane C.M. The Stimulatory Mechanism of Hepatitis C Virus NS5A Protein on the NS5B Catalyzed Replication Reaction In Vitro. Open Biochem J. 2013;7:11–14. doi: 10.2174/1874091X01307010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar C.T., Gutshall L., Sarisky R.T., Kao C.C. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 2003;330:675–685. doi: 10.1016/s0022-2836(03)00613-2. [DOI] [PubMed] [Google Scholar]
- Reiss S., Rebhan I., Backes P., Romero-Brey I., Erfle H., Matula P., Kaderali L., Poenisch M., Blankenburg H., Hiet M.S. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I., Merz A., Chiramel A., Lee J.Y., Chlanda P., Haselman U., Santarella-Mellwig R., Habermann A., Hoppe S., Kallis S. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-López C., Barroso-Deljesus A., García-Sacristán A., Briones C., Berzal-Herranz A. End-to-end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3’X-tail region. Nucleic Acids Res. 2014;42:567–582. doi: 10.1093/nar/gkt841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin P.S., Lötzerich M., Torta F., Tanner L.B., van Kuppeveld F.J.M., Wenk M.R., Greber U.F. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe. 2014;16:677–690. doi: 10.1016/j.chom.2014.10.003. this issue. [DOI] [PubMed] [Google Scholar]
- Scrima N., Caillet-Saguy C., Ventura M., Harrus D., Astier-Gin T., Bressanelli S. Two crucial early steps in RNA synthesis by the hepatitis C virus polymerase involve a dual role of residue 405. J. Virol. 2012;86:7107–7117. doi: 10.1128/JVI.00459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.T., Lee K.J., Aizaki H., Hwang S.B., Lai M.M. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 2003;77:4160–4168. doi: 10.1128/JVI.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirota Y., Luo H., Qin W., Kaneko S., Yamashita T., Kobayashi K., Murakami S. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 2002;277:11149–11155. doi: 10.1074/jbc.M111392200. [DOI] [PubMed] [Google Scholar]
- Simister P., Schmitt M., Geitmann M., Wicht O., Danielson U.H., Klein R., Bressanelli S., Lohmann V. Structural and functional analysis of hepatitis C virus strain JFH1 polymerase. J. Virol. 2009;83:11926–11939. doi: 10.1128/JVI.01008-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. The origin of hepatitis C virus. Curr. Top. Microbiol. Immunol. 2013;369:1–15. doi: 10.1007/978-3-642-27340-7_1. [DOI] [PubMed] [Google Scholar]
- Sir D., Kuo C.F., Tian Y., Liu H.M., Huang E.J., Jung J.U., Machida K., Ou J.H. Replication of hepatitis C virus RNA on autophagosomal membranes. J. Biol. Chem. 2012;287:18036–18043. doi: 10.1074/jbc.M111.320085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.M., Walton C.M., Wu C.H., Wu G.Y. Secondary structure and hybridization accessibility of hepatitis C virus 3′-terminal sequences. J. Virol. 2002;76:9563–9574. doi: 10.1128/JVI.76.19.9563-9574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai K., Shiina M., Tanaka N., Nakano T., Yamamoto A., Kondo Y., Kakazu E., Inoue J., Fukushima K., Sano K. Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology. 2012;422:377–385. doi: 10.1016/j.virol.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Tanida I., Fukasawa M., Ueno T., Kominami E., Wakita T., Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5:937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- Tellinghuisen T.L., Marcotrigiano J., Rice C.M. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature. 2005;435:374–379. doi: 10.1038/nature03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen T.L., Foss K.L., Treadaway J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 2008;4:e1000032. doi: 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdegem D., Badillo A., Wieruszeski J.M., Landrieu I., Leroy A., Bartenschlager R., Penin F., Lippens G., Hanoulle X. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic α-helical propensity and is a substrate of cyclophilin A. J. Biol. Chem. 2011;286:20441–20454. doi: 10.1074/jbc.M110.182436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieyres G., Dubuisson J., Pietschmann T. Incorporation of hepatitis C virus E1 and E2 glycoproteins: the keystones on a peculiar virion. Viruses. 2014;6:1149–1187. doi: 10.3390/v6031149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Perry J.W., Lauring A.S., Neddermann P., De Francesco R., Tai A.W. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146:1373–1385. doi: 10.1053/j.gastro.2014.02.002. e1–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waris G., Felmlee D.J., Negro F., Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J. Virol. 2007;81:8122–8130. doi: 10.1128/JVI.00125-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Weinlich S., Hüttelmaier S., Schierhorn A., Behrens S.E., Ostareck-Lederer A., Ostareck D.H. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3’UTR. RNA. 2009;15:1528–1542. doi: 10.1261/rna.1578409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane D., McGivern D.R., Masaki T., Lemon S.M. Liver injury and disease pathogenesis in chronic hepatitis C. Curr. Top. Microbiol. Immunol. 2013;369:263–288. doi: 10.1007/978-3-642-27340-7_11. [DOI] [PubMed] [Google Scholar]
- Ye J., Wang C., Sumpter R., Jr., Brown M.S., Goldstein J.L., Gale M., Jr. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA. 2003;100:15865–15870. doi: 10.1073/pnas.2237238100. [DOI] [PMC free article] [PubMed] [Google Scholar]