Abstract
Objectives
Myofibroblasts are key effector cells in the extracellular matrix remodeling of systemic sclerosis-associated interstitial lung disease (SSc-ILD), however the diversity of fibroblast populations present in the healthy and SSc-ILD lung is unknown, and has prevented the specific study of the myofibroblast transcriptome. We sought to identify and define the transcriptomes of myofibroblasts and other mesenchymal cell populations in human healthy and SSc-ILD lungs to understand how alterations in fibroblast phenotypes lead to SSc-ILD fibrosis.
Methods
We performed droplet-based single-cell RNA-sequencing with integrated canonical correlation analysis of 13 explanted lung tissue specimens (56,196 cells) from 4 healthy control and 4 SSc-ILD patients, with findings confirmed by cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) in additional samples.
Results
Examination of gene expression in mesenchymal cells identified two major, SPINT2hiand MFAP5hi, and one minor, WIF1hi, fibroblast populations in the healthy control lung. Combined analysis of control and SSc-ILD mesenchymal cells identified SPINT2hi, MFAP5hi, few WIF1hi fibroblasts, and a new large myofibroblast population with evidence of actively proliferating myofibroblasts. We compared differential gene expression between all SSc-ILD and control mesenchymal cell populations, as well as amongst the fibroblast subpopulations, showing that myofibroblasts undergo the greatest phenotypic changes in SSc-ILD and strongly upregulate expression of collagens and other profibrotic genes.
Conclusions
Our results demonstrate previously unrecognized fibroblast heterogeneity in SSc-ILD and healthy lungs, and define multimodal transcriptome-phenotypes associated with these populations. Our data indicate that myofibroblast differentiation and proliferation are key pathologic mechanisms driving fibrosis in SSc-ILD.
Keywords: Systemic sclerosis, fibroblast, pulmonary fibrosis
INTRODUCTION
Systemic sclerosis (SSc) is an autoimmune disorder with diverse clinical manifestations including fibrosis of the skin and visceral organs, as well as vasculopathy. With limited effective treatments available, SSc continues to result in substantial morbidity and mortality. Pulmonary complications, including interstitial lung disease (ILD) and pulmonary arterial hypertension (PAH), remain the leading cause of disease-related mortality in SSc.[1] While the current paradigm for disease pathogenesis suggests multiple processes, including activation of the innate and adaptive immune system, small vessel vasculopathy, and aberrant TGF-β signaling inducing fibroblast dysfunction, the precise pathophysiology remains uncertain.[2–4]
In fibrotic ILD, myofibroblasts play a pivotal role in aberrant extracellular matrix remodeling due to their dual features: having the collagen-synthesizing capacity of fibroblasts and the contractile capacity of smooth muscle.[5] With enhanced contractility, myofibroblasts also induce progressive tissue stiffness, creating a perpetuating profibrotic stimulus.[6, 7] Myofibroblasts are currently believed to derive from multiple sources, including resident mesenchymal progenitors, pericytes, bone marrow derived fibrocytes, resident fibroblasts, and mesenchymal transition of endothelial cells and epithelial cells.[8–14] Studies evaluating the origin and behavior of myofibroblasts have traditionally defined these cells as fibroblasts expressing ACTA2 (α-smooth muscle actin), lacking specificity as various other cell types including pericytes, smooth muscle cells, and myoepithelial cells also express it.
To better examine changes occurring in the lungs of patients with SSc-ILD, we utilized droplet-based single-cell RNA-sequencing (scRNA-seq) and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq),[15] a method for simultaneously measuring cell surface proteins and mRNA transcripts at the single-cell level, for multimodal analysis of lung tissues from patients with SSc-ILD and healthy controls. We identified all the major cell populations in healthy and SSc-ILD lungs, including myofibroblasts and multiple previously unrecognized fibroblast subpopulations, confirmed with epitope expression by CITE-seq, allowing for detailed analysis of the transcriptome-phenotype of each unique fibroblast population.
METHODS
ScRNA-seq library preparation was performed using the 10X Genomics Chromium System per the manufacturer’s protocol. Libraries were sequenced using an Illumina NextSeq-500 through the University of Pittsburgh Genomics Core Sequencing Facility. Data analysis was performed with the R package Seurat v 2.3.4 and R v 3.5. [16, 17]. To minimize batch effects in combining multiple samples for integrated analysis, an individual object was created for each sample, then aligned for canonical correlation analysis using Seurat’s RunMultiCCA function.[18] Differential gene expression analysis for SSc-ILD versus control cells for each cluster was performed using the Wilcoxon rank sum and MAST statistical tests.[19] A Bonferroni correction was made to correct for multiple comparisons of Wilcoxon p-values. Changes in mean proportion of cells composed of each cell type was compared using a non-parametric Kruskal-Wallis test with Dunn’s multiple comparison test for the overall cell types, and a Mann-Whitney test for the fibroblast subpopulations. P-values less than 0.05 were considered to be statistically significant. Patients and the public were not involved in the design of this study.
Other comprehensive experimental methods and specific materials are detailed in supplementary methods (Supplementary File 1).
RESULTS
Study population
We analyzed 13 lung tissue specimens by scRNA-seq from explanted tissues obtained from four patients with SSc-ILD at the time of lung transplant, and four healthy controls (organ donors with lungs unable to be transplanted). Separate upper and lower lobe samples were included for each SSc-ILD patient (8 SSc-ILD samples), and from one control, with only one sample available for all other controls (5 control samples). Explanted tissue from two additional SSc-ILD samples and one additional healthy control was used for confirmation of scRNA-seq findings at the transcriptome and epitope level. We reviewed the pathology of adjacent lung tissue and clinical information for all samples (Table 1, Supplemental Figure 1). Seven of the SSc-ILD samples showed usual interstitial pneumonia (UIP) on histology, with varied amounts of lymphoid aggregates, increased chronic inflammation, and myointimal thickening of the pulmonary arteries. One upper lobe SSc-ILD sample showed nonspecific interstitial pneumonia (NSIP) with acute lung injury. While NSIP is the most common histopathologic pattern of ILD occurring in SSc overall, [20] the predominance of UIP within these samples may reflect the end stage lung disease seen more commonly in patients requiring transplant, and is consistent with a prior microarray study of SSc-ILD which observed UIP in all explant samples. [21]
Table 1.
Characteristics of Patient Samples
| Sample | Sex | Age | Tissue Type | Number of Cells | Pathology | mPAP (mmHg) | PVR (Woods units) | Immunosuppression |
|---|---|---|---|---|---|---|---|---|
| Control 1 | Male | 55 | Control | 3,498 | Mild increase in neutrophils with few mucus plugs | |||
| Control 2 | Female | 57 | Control | 4,353 | No abnormal tissue | |||
| Control 3 | Male | 18 | Control | 3,354 | Mild increase in muscularity of small airways | |||
| Control 4 | Female | 23 | Control upper lobe | 6,167 | No abnormal tissue | |||
| Control 5 | Female | 23 | Control lower lobe | 4,396 | No abnormal tissue | |||
| SSC 1 | Female | 67 | SSc upper lobe | 4,382 | UIP with minimal acute lung injury | 24 | 3.3 | mycophenolate rituximab |
| SSC 2 | Female | 67 | SSc lower lobe | 4,026 | UIP with minimal acute lung injury | 24 | 3.3 | mycophenolate rituximab |
| SSC 3 | Male | 43 | SSc upper lobe | 4,075 | UIP with minimal acute lung injury | 36 | 2 | cyclophosphamide |
| SSC 4 | Male | 43 | SSc lower lobe | 3,297 | UIP with minimal acute lung injury | 36 | 2 | cyclophosphamide |
| SSC 5 | Male | 53 | SSc upper lobe | 5,297 | NSIP with acute and organizing lung injury | 40 | 6.47 | none |
| SSC 6 | Male | 53 | SSc lower lobe | 4,962 | UIP with prominent lymphoid aggregates | 40 | 6.47 | none |
| SSC 7 | Male | 64 | SSc upper lobe | 4,385 | UIP with prominent lymphoid aggregates | 59 | 15 | mycophenolate prednisone |
| SSC 8 | Male | 64 | SSc lower lobe | 4,004 | UIP with prominent lymphoid aggregates | 59 | 15 | mycophenolate prednisone |
Demographics, the number of cells analyzed after filtering in the scRNA-seq analysis, pathological review of adjacent tissue, and clinical characteristics of the patient samples included. Mean pulmonary artery pressure (mPAP) and pulmonary vascular resistance (PVR) measurements are from the last right heart catheterization preceding lung transplantation. Immunosuppression listed includes the medications received in the 90 days preceding lung transplantation.
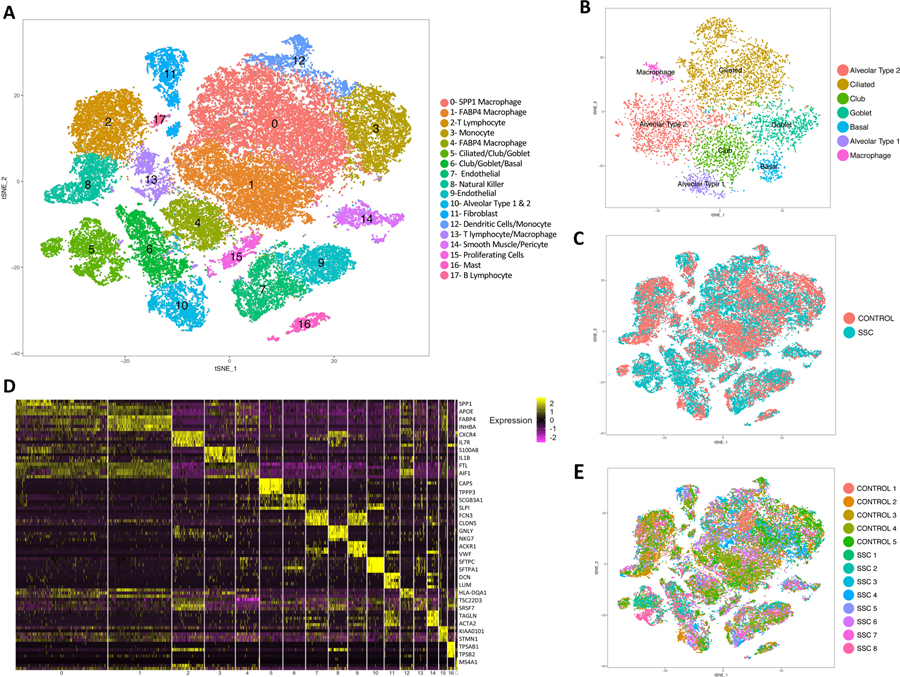
Analysis of SSc-ILD and control transcriptomes reveals multiple cell populations
In total, 56,196 cells were analyzed, with 21,768 cells from healthy controls and 34,428 cells from SSc-ILD patients. Clusters were labeled by cell type using expression of previously described markers (Figure 1, Supplementary Figure 2). Macrophages and monocytes were identified in seven separate clusters, which divided into three primary subgroups: the first expressing SPP1, CCL2, and MERTK (SPP1hi), the second expressing FABP4, INHBA, and SERPING1 (FABP4hi), and the third a population of monocytes expressing FCN1, IL1B, and IL1R2 (FCN1hi) (Supplementary Figure 3A). Proliferating cells from multiple cell types, predicted to be in G2/S phase by cell phase analysis, clustered separately by their unique high expression of genes associated with active cell proliferation (Supplementary Figure 3B, 4A, 4C). Proliferating macrophages of the FABP4hi phenotype were the predominant proliferating cell population, however a higher proportion of proliferating SPP1hi macrophages appeared in SSc-ILD samples compared to healthy controls (Supplementary Figure 3C). Cluster 13, containing control and SSc-ILD macrophages and lymphocytes with low gene expression, likely represented damaged and dying cells.
Figure 1.
Single-cell RNA-sequencing analysis of 5 human healthy control and 8 SSc-ILD lung tissue samples. (A) Visualization of clustering by t-SNE plot of all 13 combined healthy control and SSc-ILD samples, identified by cell type. (B) Reclustering of the original clusters (clusters 5,6, and 10 in Figure 1A) containing multiple bronchial and alveolar epithelial cell types demonstrating separation into individual epithelial cell types. (C) t-SNE plot of cells colored according to disease status, all clusters contained cells from both SSc and control samples (D) Heat map of scaled gene expression data for the top 5 differentially expressed genes identifying each cluster, with selected genes listed. (E) t-SNE plot of cells colored according to sample of origin, demonstrating all clusters contain cells from all samples. t-SNE, t-distributed stochastic neighbor embedding; SSc-ILD, systemic sclerosis-associated interstitial lung disease.
Changes in cell populations present
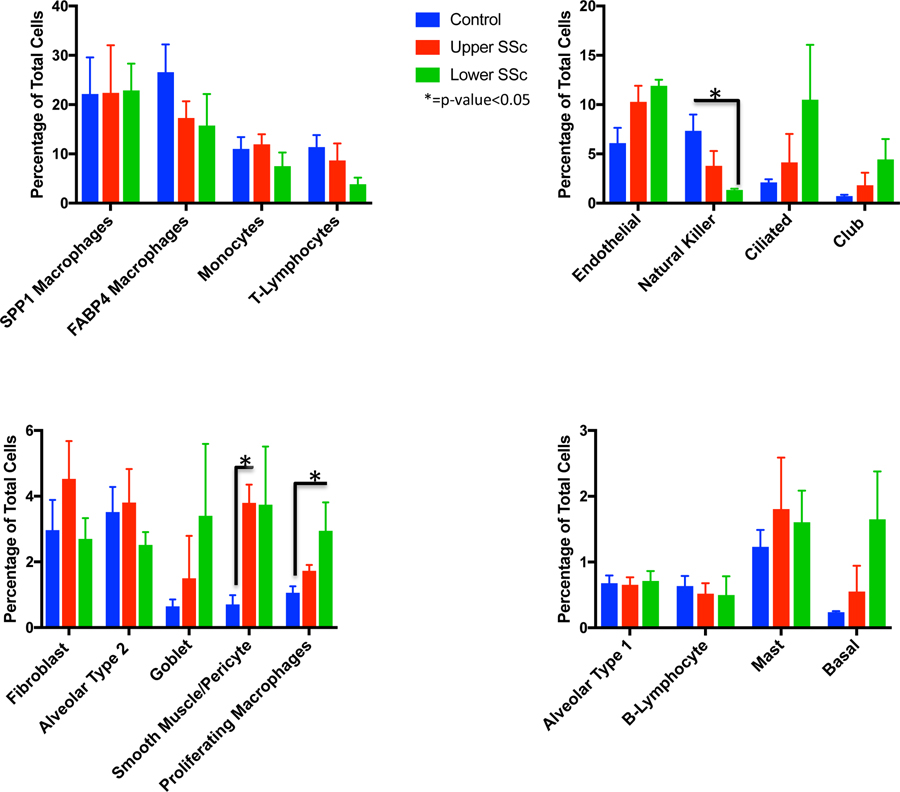
Analyzing the proportion of total cells present in each population by sample and disease status revealed several significant changes between normal lungs and SSc-ILD (Figure 2). The populations of smooth muscle cells and pericytes increased significantly in SSc-ILD upper lobes compared to healthy control lungs (p=0.0209), with endothelial cells also showing a trend toward increased numbers in SSc-ILD lungs. Total macrophages and monocytes fell from 60.84% of cells in controls to 53.33% of cells in upper lobe SSc and 49.09% of cells in lower lobe SSc. Proliferating macrophages increased from only 1.06% of cells in controls to 1.73% in SSc-ILD upper lobes and 2.95% in SSc-ILD lower lobes (p-value 0.0111). The proportion of natural killer cells decreased significantly from 7.34% of cells in controls to 3.80% in SSc-ILD upper lobes and 1.34% in SSc-ILD lower lobes (p-value 0.0125). No consistent changes were seen in the proportion of fibroblasts, alveolar type 1, or alveolar type 2 cells, while ciliated, club, and basal cells all trended toward increased numbers in a graded fashion, i.e., more cells in SSc-ILD upper lobes than controls, and more cells in SSc-ILD lower lobes than SSc-ILD upper lobes, likely reflecting the mucociliary and basal epithelial cells lining honeycomb cysts.[22, 23]
Figure 2.
Mean percentage of total cells comprised of each cell type, comparing control, upper lobe SSc-ILD, and lower lobe SSc-ILD samples. Bars indicate the mean percentage of total cells with error bars indicating the standard error of the mean. *=p-value<0.05. SSc-ILD, systemic sclerosis-associated interstitial lung disease.
Smooth muscle cells and pericytes
Fibroblasts, smooth muscle cells, and pericytes, as well as fibroblasts from the proliferating cell cluster (Supplementary Figure 4), were combined and reclustered to allow clearer identification of these and any rare mesenchymal cell subpopulations (Figure 3A, Supplementary Figure 5). Fibroblasts composed four clusters, as marked robustly by the expression of LUM, PDGFRA, and FBLN1, with fibroblast subpopulations as detailed below. Smooth muscle cells highly expressed ACTA2, DES, MYH11, and PLN (Figure 3E, Supplementary Figure 5). Differential gene expression between SSc-ILD and control lungs was analyzed for all mesenchymal populations (Supplementary File 2).
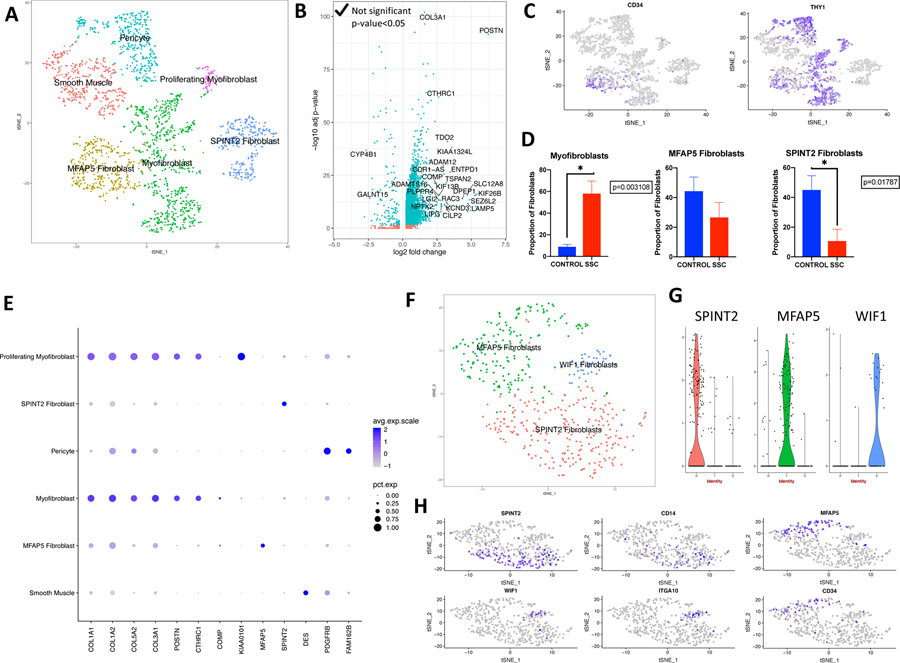
Figure 3.
Single-cell RNA-sequencing analysis of human healthy control and SSc-ILD mesenchymal cell populations. (A) t-SNE plot of combined fibroblast, smooth muscle/pericyte, and proliferating fibroblast cells as identified by cell type and fibroblast subpopulations. (B) Volcano plot of differentially expressed genes (log2 fold change>0.5, adjusted p-value <0.05) from the comparison of all SSc fibroblasts to all control fibroblasts. Results showed that 461 genes were up-regulated and 115 genes were down-regulated by greater than twofold. (C) Gene expression of CD34, demonstrating high CD34 expression in MFAP5 fibroblasts, and THY1, demonstrating high THY1 expression in myofibroblasts, MFAP5hi fibroblasts, and pericytes. (D) Mean proportion of total fibroblasts that each fibroblast subpopulation comprises in SSc-ILD and control lungs, calculated by individual sample. (E) Expression of selected collagen and cell type specific genes by mesenchymal population. Dot size corresponds to the percentage of cells in a cluster expressing the gene, and dot color corresponds to the average expression level for the gene in the cluster. (F) t-SNE plot of healthy control fibroblasts only (G) Violin plots of gene expression of SPINT 2, MFAP5, and WIF1 by control fibroblast cluster. (H) Gene expression plots demonstrating high expression of SPINT2 and CD14 in SPINT2hi fibroblasts, MFAP5 and CD34 in MFAP5hi fibroblasts, and WIF1 and ITGA10 in WIF1hi fibroblasts. Dot color corresponds to the level of gene expression in each cell. d t-SNE, t-distributed stochastic neighbor embedding; SSc-ILD, systemic sclerosis-associated interstitial lung disease.
A distinct pericyte population, markedly expanded in SSc-ILD (p-value 0.0295), was identified by its expression of the known markers RGS5, PDGFRB, MCAM, CSPG4 (NG2), and NES (Figure 3E, Supplementary Figures 5,6).[24, 25] Although none of these markers were exclusive to pericytes, this cluster was the only population expressing all of these genes. The identified pericyte population showed enhanced expression of FAM162B, CHN1, IGFBP2, and HIGD1B compared to other mesenchymal cells, with FAM162B the most specific identifier of this population. Pericytes did not separate into the previously described sub classification of type-1 (NES-/CSPG4+) and type-2 (NES+/CSPG4+).[26, 27]
Fibroblast subpopulations in controls
To guide our understanding of fibroblast heterogeneity in the combined analysis of control and SSc-ILD lungs, we separately analyzed only the fibroblasts from healthy control lungs. Two major and one minor subpopulations of fibroblasts emerged, with all groups containing cells from each control sample (Figure 3F–H). The first major population was defined by expression of SPINT2, CD14, LMCD1, FGFR4, and FIGF (SPINT2 hi fibroblasts). A second major population was defined by expression of MFAP5, CD34, THY1, SLPI, and PLA2G2A (MFAP5 hi fibroblasts). A distinct minor population was distinguished by expression of WIF1 and ITGA10 (WIF1 hi fibroblasts).
Fibroblast subpopulations in SSc-ILD
Examining fibroblast populations in the combined analysis of control and SSc-ILD mesenchymal cells, we again identified distinct populations of SPINT2 hi and MFAP5 hi fibroblasts, with each containing both control and SSc-ILD fibroblasts. An additional large population of fibroblasts, containing primarily SSc-ILD cells, expressed the highest level of ACTA2 (3.04-fold increase compared to other fibroblast populations), consistent with this population representing the contractile myofibroblasts. This population did not express the smooth muscle specific markers MYH11 and DES, and had the highest expression of several collagen genes amongst the mesenchymal cells (Figure 3E). The myofibroblasts exhibited high expression of THY1 and low expression of CD34. A group of proliferating myofibroblasts clustered separately based on co-expression of cell proliferation genes and myofibroblast genes. The total myofibroblasts increased from 10.66% of fibroblasts in controls to 62.75% of fibroblasts in SSc-ILD (Figure 3D). This may overestimate the proportion of control myofibroblasts, however, as the WIF1hi population was counted within this group, and no myofibroblast population was identified when analyzing the control fibroblasts alone. The minor WIF1hi population consisted almost entirely of healthy control fibroblasts. Intersample variability in the presence of and marker gene expression for each fibroblast population is detailed in Supplemental Figures 6 and 7.
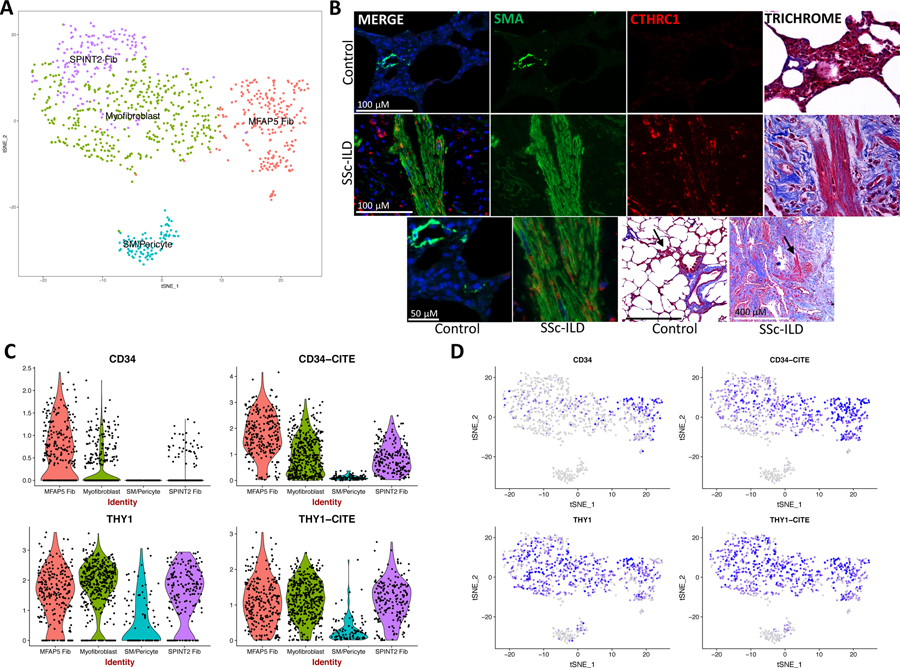
We identified the presence of the myofibroblasts, SPINT2hi fibroblasts, and MFAP5hi fibroblasts by their transcriptome signature and CITE-seq as well in separate SSc-ILD and healthy control samples using the surface markers CD34 and CD90 (THY1) (Figure 4A, C-D). Although increased CD34 mRNA distinguished MFAP5hi fibroblasts, this difference was reduced on examining surface protein expression (Figure 4C–D). Thus, neither of these proteins distinguished the myofibroblast population well. To confirm the population putatively identified as myofibroblasts we stained SSc-ILD and control lungs for α-smooth muscle actin and CTHRC1, a gene we found highly and selectively up-regulated in the myofibroblasts (Figure 4B).
Figure 4.
CITE-seq of four additional SSc-ILD (SSc 9–12) and one additional healthy control lung (Control 6) and immunofluorescence staining of SSc-ILD and control lung. (A) t-SNE plot of combined fibroblast, smooth muscle, and pericyte cells, identified by subpopulation/cluster. (B) Serial sections of control and SSc-ILD lung with immunofluorescence with DAPI nuclear staining and trichrome staining. SMA and CTHRC1 coexpress in areas of disorganized myofibroblasts, with SMA+/CTHRC1- cells staining smooth muscle. Trichrome staining demonstrates excessive collagen deposition (blue) in SSc-ILD lungs. (C) Violin plots of gene expression of CD34 and THY1 and protein expression of CD34 (labeled as CD34-CITE) and CD90/THY1 (labeled as THY1-CITE) as detected by oligonucleotide-labeled antibodies. D) Gene and protein expression of CD34 and THY1 (CD90). Dot color corresponds to the level of gene expression in each cell. t-SNE, t-distributed stochastic neighbor embedding; SSc-ILD, systemic sclerosis-associated interstitial lung disease; DAPI, 4’,6-diamidino-2-phenylindole; SMA, α-smooth muscle actin; CTHRC1, collagen triple helix repeat containing 1.
Differential gene expression in SSc-ILD is driven by myofibroblasts
Comparing differential gene expression of all fibroblasts from SSc-ILD to controls, POSTN, CORIN, KIF26B, FNDC1, SEZ6L2, and LAMP5 were the top up-regulated genes (Figure 3B). Many of the non-collagen up-regulated genes were discretely present in the SSc-ILD myofibroblasts, including POSTN, KIAA1324L, COMP, TDO2, ADAM12, MXRA5, ALDH1A3, and LRRC17, supporting the hypothesis that myofibroblasts undergo the greatest phenotypic changes in SSc-ILD.
Differential expression of myofibroblasts
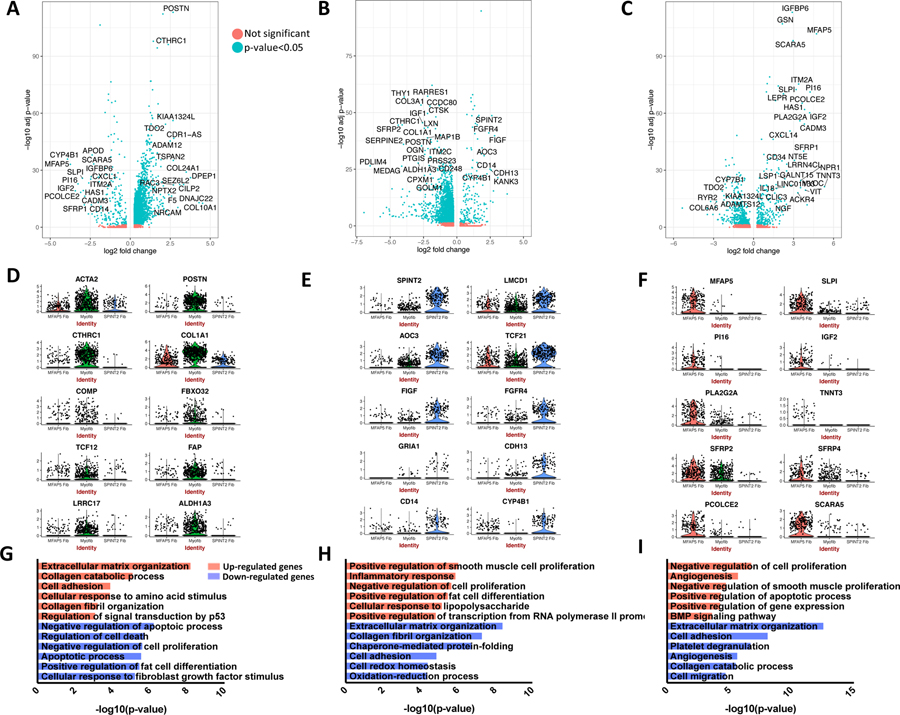
To evaluate the synthetic properties and functions of the three major fibroblast populations, differentially expressed genes (adjusted p-value <0.05, absolute log2 fold change>0.5) distinguishing each cluster from the other two were identified and evaluated for enriched gene ontology (GO) biological processes (Supplementary File 3). In myofibroblasts, 237 genes were up-regulated by greater than twofold, including COL10A1 (22.93 fold), DPEP1 (13.61 fold), TSPAN2 (6.45 fold), POSTN (6.39 fold), and CTHRC1 (5.18 fold) (Figure 5A, 5D). Fifteen collagen genes, numerous other genes essential to collagen synthesis, multiple metalloendopeptidases, and the post-translational modification inducing E3 ubiquitin ligase FBXO32 were among the up-regulated genes. Enriched processes for the up-regulated genes reflect the increased collagen and extracellular matrix synthesis by the myofibroblasts (Figure 5G). Enriched processes amongst the down-regulated genes reflect reduced response to normal regulatory processes including decreased regulation of cell proliferation and cell death, consistent with the pathologic expansion of this subgroup in SSc-ILD.
Figure 5.
Differential gene expression comparing each major fibroblast cluster (myofibroblast, SPINT2hi fibroblast, MFAP5hi fibroblast) to the other two. A-C Volcano plots include all differentially expressed genes with absolute value log2 fold change>0.5, and are colored by adjusted p-value <0.05 or not. D-F Violin plots demonstrate gene expression of selected distinguishing genes for each fibroblast subpopulation. G-I Enriched GO biological processes for the differentially expressed up-regulated and down-regulated genes for each comparison of one major fibroblast cluster to the other two. Functional enrichment analysis was performed using DAVID with all differentially expressed genes with adjusted p-value <0.05 and absolute value log2 fold change>0.5 included. (A), (D), and (G) display results of myofibroblast to SPINT2 hi fibroblast and MFAP5 hi fibroblast comparison. (B), (E), and (H) display results of SPINT 2 hi fibroblast to myofibroblast and MFAP5 hi fibroblast comparison. (C), (F), and (I) display results of MFAP5 hi fibroblast to myofibroblast and SPINT2 hi fibroblast comparison. DAVID, Database for Annotation, Visualization, and Integrated Discovery.
Differential expression of SPINT2 hi fibroblasts
Within the SPINT2 hi fibroblasts, GRIA1 (13.74 fold), KANK3 (8.44 fold), FIGF (7.13 fold), SPINT2 (6.16 fold), FGFR4 (5.27 fold), and TCF21 (2.97 fold) were among 98 genes with greater than twofold increased expression (Figure 5B, 5E). While this subgroup exhibited substantially less expression of the abundant collagen genes COL12A1, COL1A1, and COL1A2, other collagens including COL13A1 (2.63 fold) and COL6A6 (1.60 fold) showed increased expression within this subgroup.
Differential expression of MFAP5 hi fibroblasts
In the MFAP5 hi fibroblasts, TNNT3 (39.89 fold), MFAP5 (26.34 fold), PI16 (18.87 fold), IGF2 (18.49 fold), and ACKR3 (16.16 fold) were among 114 genes with greater than twofold increased expression (Figure 5C, 5F). Three members of the Wnt-related secreted frizzled-related protein (SFRP) family: SFRP1 (13.45 fold), SFRP4 (3.34 fold), and SFRP2 (2.52 fold) were up-regulated, though none were exclusive to this fibroblast subset.
DISCUSSION
There is presently a gap in knowledge regarding the heterogeneity of fibroblast populations in the human lung, and their detailed phenotypic changes in SSc-ILD. In this study we provide a comprehensive view of fibroblasts and other mesenchymal cell populations present in both SSc-ILD and healthy control lungs, and identify new markers of myofibroblasts and other lung fibroblast populations. While fibroblasts with high expression of ACTA2 formed a distinct subtype, other fibroblast subgroups expressed low level ACTA2 in both normal and diseased tissues, precluding its use as a unique marker of myofibroblasts. Given the high synthetic capacity of the myofibroblasts, excessive levels of type I collagen transcription, dramatic expansion, and evidence of active proliferation in SSc-ILD, our results support the current disease paradigm that myofibroblasts are the key profibrotic effecter cell.
Though transcriptome data alone cannot conclusively identify myofibroblast origin, our data supports the model that, in SSc-ILD, myofibroblasts first differentiate from other lung mesenchymal populations, then proliferate. Control lungs demonstrated a paucity of myofibroblasts when examined alone, with a striking expansion of myofibroblasts appearing in SSc-ILD samples, including a subpopulation of actively proliferating myofibroblasts. While myofibroblasts likely differentiate from multiple sources in disease, we hypothesize MFAP5 hi fibroblasts may act as progenitors in SSc-ILD. The MFAP5 hi fibroblasts clustered geographically closest to the myofibroblasts, reflecting their more similar transcriptome, expressed greater collagen than the SPINT2 hi fibroblasts, and had elevated expression of multiple Wnt regulators, consistent with overexpression of SFRP genes previously reported in both idiopathic pulmonary fibrosis lungs and SSc skin.[28–30]
Pericytes are believed to contribute to fibrosis through their transformation into myofibroblasts, as well as their direct production of collagen.[11, 31, 32] While the SSc-ILD myofibroblasts expressed significantly more collagen than the other mesenchymal cell populations (Figure 3E), the SSc-ILD pericytes expressed COL1A2 and COL3A1 at levels similar to the SPINT2 hi and MFAP5 hi fibroblasts, whereas healthy control pericytes expressed much less collagen. In vitro and murine studies have demonstrated TGF-β1 induces transformation of pericytes to myofibroblasts. [13, 33] Although our data provide no direct evidence of pericyte to myofibroblast transformation, the marked expansion of pericytes in SSc-ILD samples is consistent with the possibility that pericytes play an important role in SSc-ILD. As all of the SSc-ILD patients in our study also had WHO Group 3 pulmonary hypertension due to chronic lung disease, the pericyte expansion may also play a role in this complication.
Comparing our scRNA-seq data with recently published analyses of murine lung mesenchymal populations [34, 35], increased expression of COL13A1 and TCF21 amongst the SPINT2 hi fibroblasts was analogous to the description of a subgroup of COL13A1 matrix fibroblasts in mice.[34] ITGA8, NPNT, LBH, and MFAP4, amongst others, were also conserved across species between these groups of fibroblasts, although none were exclusive to the SPINT2hi group, and most were expressed to a lesser degree by the myofibroblasts as well. However, none of the fibroblast subpopulations we observed were consistent with previously described lipofibroblasts, characterized by the lipid-droplet trafficking protein perilipin 2 (also known as ADRP, or adipose differentiation-related protein), as this was similarly expressed in all human fibroblasts.[8, 36] Other proposed lipofibroblast markers including LIPA, LPL, and FABP5 also did not differentiate any specific fibroblast population, suggesting the lipofibroblast designation is not as phenotypically relevant in human lung, or possibly that these cells were lost during scRNA-seq processing.[37, 38] Recent studies have reported varying results as to whether lipid-droplet containing cells are present in the human lung.[8, 39] No analogous human population corresponded to the murine COL14A1 matrix fibroblasts, and the newly proposed markers of murine myofibroblasts, such as Hhip, Mustn1, and Grem2, did not distinguish the human myofibroblast population. In comparison to the SFRP2/DPP1 fibroblasts recently identified in human skin,[40] both the MFAP5hi lung fibroblasts and myofibroblasts expressed SFRP2 and DPP4, with the MFAP5hi fibroblasts expressing higher PCOLCE2 and CD55 compared to the other fibroblasts. Unlike in the skin, WIF1hi lung fibroblasts did not express SFRP2 or NKD2, and other markers of dermal fibroblasts did not differentiate pulmonary fibroblast subpopulations.
Although all samples were from end-stage disease and predominantly demonstrated UIP on histology, we examined both typically less advanced upper lobes and more fibrotic lower lobes in order to capture tissues reflective of a spectrum of the disease course. Case series identifying NSIP as the predominant histopathology in SSc utilized surgical lung biopsies,[20] and may only reflect the distribution of disease patterns in early disease, rather than at transplant or death. For example, a 2001 case series including pathology from autopsy and biopsy noted a UIP pattern in 44% of cases,[41] and a past report of gene expression in SSc-ILD explants observed UIP in all samples.[21] Studying explant tissue is valuable as these patients all progressed to end stage disease, and thus there is the most critical need for improved understanding of their disease pathogenesis in order to develop new therapeutic options. Comparing our data to a previous microarray analysis of non-end stage NSIP SSc-ILD tissue obtained by surgical lung biopsy, amongst the top 40 differentially expressed genes by microarray, COMP, POSTN, FKBP11, COL3A1, COL1A1, and TDO2 were all distinctly expressed by the SSc-ILD myofibroblasts in our scRNA-seq analysis, thus strongly supporting the generalizability of our findings to patients with NSIP and earlier disease.
Our study was limited by its relatively small cohort of patient and control samples. Because patients with SSc-ILD now rarely undergo surgical lung biopsy, explanted lungs are the only consistent source of tissue for new investigative analyses. As many transplant centers continue to avoid transplanting patients with SSc due to their coincident esophageal disease, the availability of tissue is limited to select centers and precludes acquiring large numbers of samples. We were unable to perform age and sex matching due to reliance on explanted tissue, and thus average age and sex of control (35.2 years, 40% male) and SSc-ILD lung subjects (56.75 years, 75% male) differed. The canonical correlation analysis methodology aligned cell types well despite such potential bias.[18] Additionally, due to a difference in the reagent chemistry and digestion protocol used in processing these samples, we did not combine the two SSc-ILD and one control sample used for CITE-seq with the other thirteen samples, and instead chose to utilize these samples as a validation cohort. Analytic methods for scRNA-seq data are rapidly advancing and may in the near future allow for improved normalization and integrated analysis of multiple samples, despite interindividual variation and batch effects, in order to create larger combined datasets from multiple investigators. Our study was also limited by the inability to complete immunohistological verifications of all fibroblast subpopulations at this time, due to the absence of reliable antibodies for immunohistochemistry of the relevant markers.
In summary, our analysis harnesses the distinct capacity of scRNA-seq and CITE-seq to discern new fibroblast heterogeneity in human SSc-ILD and healthy control lungs, providing new insights into these pathogenic cells at an unprecedented multimodal level. The expression signature of mRNAs and select surfaces proteins (or transcriptome map) now available for the pathogenic myofibroblasts add considerably to our knowledge of this key effector cell in fibrotic lung diseases and provides new insights to their functional importance.
Supplementary Material
KEY MESSAGES.
What is already known about this subject?
Systemic sclerosis-associated interstitial lung disease (SSc-ILD) is a devastating complication of SSc, with high morbidity and mortality, and limited effective treatments.
In SSc-ILD, myofibroblasts are the key fibrotic effector cell due to their excessive extracellular matrix production and acquired contractile phenotype.
What does this study add?
For the first time, we identify previously unrecognized fibroblast heterogeneity in SSc-ILD and healthy human lung and the transcriptome-phenotype of these and other mesenchymal cell populations.
How might this impact on clinical practice or future developments?
These results provide new insights into the transcriptome of pathogenic myofibroblasts, supporting the future development of new targeted therapies directing at these key cells.
Acknowledgements
The authors would like to acknowledge the University of Pittsburgh Medical Center lung transplantation team for procurement of the lungs, the Center for Organ Research and Education (CORE), and the organ donors and their families for the generous donation of tissues used in the study.
Funding
Research reported in this publication was supported by the National Institutes of Health National Institute of Arthritis and Musculoskeletal and Skin Diseases under award number 2P50AR060780 (RL) and the National Heart, Lung, and Blood Institute under award numbers R01HL123766 (RL) and 2T32HL007563–31 (EV). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing Interests
RL has received consulting fees from PRISM Biolab, Merck, Bristol Myers Squibb, Biocon, Formation, Genentech/Roche, UCB, and Sanofi and grant support from Elpidera, Kiniksa, and Regeneron, outside the submitted work.
Data Sharing
Raw counts in sparse matrix format for single-cell RNA-seq data are available at GEO (GSE 128169). Sample identification key for uploaded data is detailed in Supplementary file 1.
REFERENCES
- 1.Steen VD, Medsger TA. Changes in causes of death in systemic sclerosis, 1972–2002. Ann Rheum Dis 2007. July; 66(7):940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dowson C, Simpson N, Duffy L, O’Reilly S. Innate Immunity in Systemic Sclerosis. Curr Rheumatol Rep 2017. January; 19(1):2. [DOI] [PubMed] [Google Scholar]
- 3.Lafyatis R Transforming growth factor beta--at the centre of systemic sclerosis. Nat Rev Rheumatol 2014. December; 10(12):706–719. [DOI] [PubMed] [Google Scholar]
- 4.van den Hoogen F, Khanna D, Fransen J, Johnson SR, Baron M, Tyndall A, et al. 2013 classification criteria for systemic sclerosis: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann Rheum Dis 2013. November; 72(11):1747–1755. [DOI] [PubMed] [Google Scholar]
- 5.Bagnato G, Harari S. Cellular interactions in the pathogenesis of interstitial lung diseases. Eur Respir Rev 2015. March; 24(135):102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinz B The extracellular matrix and transforming growth factor-beta1: Tale of a strained relationship. Matrix Biol 2015. September; 47:54–65. [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, Tager AM, et al. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J Cell Biol 2010. August 23; 190(4):693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Agha E, Moiseenko A, Kheirollahi V, De Langhe S, Crnkovic S, Kwapiszewska G, et al. Two-Way Conversion between Lipogenic and Myogenic Fibroblastic Phenotypes Marks the Progression and Resolution of Lung Fibrosis. Cell Stem Cell. 2017. February 2; 20(2):261–273 e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez IE, Eickelberg O. New cellular and molecular mechanisms of lung injury and fibrosis in idiopathic pulmonary fibrosis. Lancet 2012. August 18; 380(9842):680–688. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto N, Phan SH, Imaizumi K, Matsuo M, Nakashima H, Kawabe T, et al. Endothelial-mesenchymal transition in bleomycin-induced pulmonary fibrosis. Am J Respir Cell Mol Biol 2010. August; 43(2):161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung C, Linn G, Chow YH, Kobayashi A, Mittelsteadt K, Altemeier WA, et al. Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 2013. October 1; 188(7):820–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phan SH. Genesis of the myofibroblast in lung injury and fibrosis. Proc Am Thorac Soc 2012. July; 9(3):148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rock JR, Barkauskas CE, Cronce MJ, Xue Y, Harris JR, Liang J, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A 2011. December 27; 108(52):E1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh SM, Worrell JC, Fabre A, Hinz B, Kane R, Keane MP. Novel Differences in Gene Expression and Functional Capabilities of Myofibroblast Populations in Idiopathic Pulmonary Fibrosis. Am J Physiol Lung Cell Mol Physiol 2018. August 9. [DOI] [PubMed] [Google Scholar]
- 15.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017. September; 14(9):865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015. May 21; 161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015. May; 33(5):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018. June; 36(5):411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 2015. December 10; 16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouros D, Wells AU, Nicholson AG, Colby TV, Polychronopoulos V, Pantelidis P, et al. Histopathologic subsets of fibrosing alveolitis in patients with systemic sclerosis and their relationship to outcome. Am J Respir Crit Care Med 2002. June 15; 165(12):1581–1586. [DOI] [PubMed] [Google Scholar]
- 21.Hsu E, Shi H, Jordan RM, Lyons-Weiler J, Pilewski JM, Feghali-Bostwick CA. Lung tissues in patients with systemic sclerosis have gene expression patterns unique to pulmonary fibrosis and pulmonary hypertension. Arthritis Rheum 2011. March; 63(3):783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans CM, Fingerlin TE, Schwarz MI, Lynch D, Kurche J, Warg L, et al. Idiopathic Pulmonary Fibrosis: A Genetic Disease That Involves Mucociliary Dysfunction of the Peripheral Airways. Physiol Rev 2016. October; 96(4):1567–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seibold MA, Smith RW, Urbanek C, Groshong SD, Cosgrove GP, Brown KK, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS One. 2013; 8(3):e58658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008. September 11; 3(3):301–313. [DOI] [PubMed] [Google Scholar]
- 25.Rowley JE, Johnson JR. Pericytes in chronic lung disease. Int Arch Allergy Immunol 2014; 164(3):178–188. [DOI] [PubMed] [Google Scholar]
- 26.Birbrair A, Zhang T, Files DC, Mannava S, Smith T, Wang ZM, et al. Type-1 pericytes accumulate after tissue injury and produce collagen in an organ-dependent manner. Stem Cell Res Ther 2014. November 6; 5(6):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birbrair A, Zhang T, Wang ZM, Messi ML, Enikolopov GN, Mintz A, et al. Skeletal muscle pericyte subtypes differ in their differentiation potential. Stem Cell Res 2013. January; 10(1):67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frost J, Estivill X, Ramsay M, Tikly M. Dysregulation of the Wnt signaling pathway in South African patients with diffuse systemic sclerosis. Clin Rheumatol 2018. September 20. [DOI] [PubMed] [Google Scholar]
- 29.Gardner H, Shearstone JR, Bandaru R, Crowell T, Lynes M, Trojanowska M, et al. Gene profiling of scleroderma skin reveals robust signatures of disease that are imperfectly reflected in the transcript profiles of explanted fibroblasts. Arthritis Rheum 2006. June; 54(6):1961–1973. [DOI] [PubMed] [Google Scholar]
- 30.Yang IV, Burch LH, Steele MP, Savov JD, Hollingsworth JW, McElvania-Tekippe E, et al. Gene expression profiling of familial and sporadic interstitial pneumonia. Am J Respir Crit Care Med 2007. January 1; 175(1):45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang FC, Chou YH, Chen YT, Lin SL. Novel insights into pericyte-myofibroblast transition and therapeutic targets in renal fibrosis. J Formos Med Assoc 2012. November; 111(11):589–598. [DOI] [PubMed] [Google Scholar]
- 32.Popescu FC, Busuioc CJ, Mogosanu GD, Pop OT, Parvanescu H, Lascar I, et al. Pericytes and myofibroblasts reaction in experimental thermal third degree skin burns. Rom J Morphol Embryol 2011; 52(3 Suppl):1011–1017. [PubMed] [Google Scholar]
- 33.Sun W, Tang H, Gao L, Sun X, Liu J, Wang W, et al. Mechanisms of pulmonary fibrosis induced by core fucosylation in pericytes. Int J Biochem Cell Biol 2017. July; 88:44–54. [DOI] [PubMed] [Google Scholar]
- 34.Xie T, Wang Y, Deng N, Huang G, Taghavifar F, Geng Y, et al. Single-Cell Deconvolution of Fibroblast Heterogeneity in Mouse Pulmonary Fibrosis. Cell Rep 2018. March 27; 22(13):3625–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct Mesenchymal Lineages and Niches Promote Epithelial Self-Renewal and Myofibrogenesis in the Lung. Cell. 2017. September 7; 170(6):1134–1148 e1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz CJ, Torres E, Londos C, Torday JS. Role of adipocyte differentiation-related protein in surfactant phospholipid synthesis by type II cells. Am J Physiol Lung Cell Mol Physiol 2002. August; 283(2):L288–296. [DOI] [PubMed] [Google Scholar]
- 37.Imamura M, Inoguchi T, Ikuyama S, Taniguchi S, Kobayashi K, Nakashima N, et al. ADRP stimulates lipid accumulation and lipid droplet formation in murine fibroblasts. Am J Physiol Endocrinol Metab 2002. October; 283(4):E775–783. [DOI] [PubMed] [Google Scholar]
- 38.Li A, Ma S, Smith SM, Lee MK, Fischer A, Borok Z, et al. Mesodermal ALK5 controls lung myofibroblast versus lipofibroblast cell fate. BMC Biol 2016. March 16; 14:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tahedl D, Wirkes A, Tschanz SA, Ochs M, Muhlfeld C. How common is the lipid body-containing interstitial cell in the mammalian lung? Am J Physiol Lung Cell Mol Physiol 2014. September 1; 307(5):L386–394. [DOI] [PubMed] [Google Scholar]
- 40.Tabib T, Morse C, Wang T, Chen W, Lafyatis R. SFRP2/DPP4 and FMO1/LSP1 Define Major Fibroblast Populations in Human Skin. J Invest Dermatol 2018. April; 138(4):802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujita J, Yoshinouchi T, Ohtsuki Y, Tokuda M, Yang Y, Yamadori I, et al. Non-specific interstitial pneumonia as pulmonary involvement of systemic sclerosis. Ann Rheum Dis 2001. March; 60(3):281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.