Abstract
Talaromyces is a monophyletic genus containing seven sections. The number of species in Talaromyces grows rapidly due to reliable and complete sequence data contributed from all over the world. In this study agricultural soil samples from Fujiang, Guangdong, Jiangxi, Shandong, Tibet and Zhejiang provinces of China were collected and analyzed for fungal diversity. Based on a polyphasic approach including phylogenetic analysis of partial ITS, BenA, CaM and RPB2 gene sequences, macro- and micro-morphological analyses, six of them could not be assigned to any described species, and one cannot be assigned to any known sections. Morphological characters as well as their phylogenetic relationship with other Talaromyces species are presented for these putative new species. Penicillium resedanum is combined in Talaromyces section Subinflati as T. resedanus.
Keywords: Eurotiales , Penicillium resedanum, polyphasic taxonomy, section Tenues, soil
Introduction
The genus Talaromyces used to accommodate sexual Penicillium species (Benjamin 1955). The generic concept has changed in the last decade due to changes in nomenclatural rules and results of phylogenetic studies. Before 2011, various studies showed that asexual reproducing Penicillium species classified in subgenus Biverticillium and the sexual Talaromyces species form a monophyletic clade distinct from Penicilliumsensu stricto (LoBuglio et al. 1993; Berbee et al. 1995; Ogawa et al. 1997; Ogawa and Sugiyama 2000; Wang and Zhuang 2007; Houbraken and Samson 2011). In 2011, following the concepts of nomenclatural priority and single name nomenclature, Samson et al. (2011) transferred the majority of accepted Penicillium subgenus Biverticillium species to Talaromyces. A monograph of Talaromyces was provided based on a polyphasic species concept with seven sections Bacillispori, Helici, Islandici, Purpurei, Subinflati, Talaromyces and Trachyspermi (Yilmaz et al. 2014). This sectional classification was further supported by a four gene phylogeny (Chen et al. 2016). The number of species in Talaromyces grows rapidly due to reliable and complete sequence data contributed from all over the world (Visagie et al. 2015; Crous et al. 2016, 2017, 2018; Yilmaz et al. 2016a, b; Guevara-Suarez et al. 2017; Peterson and Jurjević 2017; Barbosa et al. 2018; Varriale et al. 2018; Rodríguez-Andrade et al. 2019; Rajeshkumar et al. 2019; Guevara-Suarez et al. 2020). It is noteworthy that in China many new species were discovered with another 19 new species reported (Chen et al. 2016; Wang QM et al. 2016; Wang XC et al. 2016, 2017; Jiang et al. 2018; Su and Niu 2018).
Talaromyces species are commonly distributed in a wide range of substrates, mostly in soil. Their main interest to food mycologists lies in their production of heat resistant ascospores and association with spoilage of pasteurized fruit juices and fruit-based products; the most commonly isolated heat resistant species include T. bacilisporus, T. helicus, T. macrosporus, T. stipitatus and T. trachyspermus (Dijksterhuis 2007; Pitt and Hocking 2009; Yilmaz et al. 2014). In addition, T. flavus, T. funiculosus, T. pinophilus, T. purpurogenus, T. rugulosus and T. wortmannii have been found quite frequently in food, including fruit, nuts and cereals (Pitt and Hocking 2009). Talaromyces islandicus can cause the yellowing of stored rice and has been reported from e.g. flour, peanuts, pecans, soybeans and maize (Saito et al. 1971; Sakai et al. 2005; Oh et al. 2008; Pitt and Hocking 2009).This species produces unique mycotoxins such as cyclochlorotine, islanditoxin, erythroskyrine and luteoskyrin, which are carcinogenic liver and kidney toxins (Uraguchi et al. 1961, 1972; Uraguchi 1962; Ueno and Ishikawa 1969; Bouhet et al. 1976; Stark et al. 1978). Other mycotoxins produced by Talaromyces members include rugulosin and skyrin (by members of sectionIslandici), botryodiploidin (T. coalescens and T. stipitatus), rubratoxin (T. purpurogenus), rugulovasine (T. purpurogenus and T. wortmannii) and secalonic acid D & F (T. dendriticus, T. flavovirens, T. funiculosus, T. minioluteus, T. pseodostromaticus, T. siamensis and T. stipitatus) (Yilmaz et al. 2014).
Talaromyces contains several species that are reported to cause infections in humans. Talaromyces marneffei has been exclusively associated with acquired immunodeficiency syndrome (AIDS) caused by human immunodeficiency virus (HIV) infections (Supparatpinyo et al. 1994; Limper et al. 2017). Other species like T. indigoticus, T. helicus, T. piceus, T. purpurogenus, T. radicus, T. rugulosus and T. verruculosus have been reported in superficial or disseminated, fatal infections (Horré et al. 2001; Santos et al. 2006; de Vos et al. 2009; Weisenborn et al. 2010; Tomlinson et al. 2011; de Hoog et al. 2014). Recently, four new members of Talaromyces were reported from clinical sources, and more studies are needed to complete the distribution and the relevance of these new fungi in human and animal disease (Guevara-Suarez et al. 2017).
On the other hand, species in Talaromyces are good producers of anticancer, antibacterial and antifungal compounds (Bladt et al. 2013; Zhai et al. 2016; Nicoletti et al. 2018), antiproliferative and antioxidative compounds (Kumari et al. 2018), enzymes (Isbelia et al. 1999; Narikawa et al. 2000; Pol et al. 2012; Maeda et al. 2013; Antonopoulou et al. 2018; Lian et al. 2018; Xu et al. 2018) and natural colourants (Frisvad et al. 2013; Zaccarim et al. 2018). Several species also proved to be effective biocontrol agents against soil-borne pathogens. Talaromyces flavus suppresses Verticillium wilt of tomato, eggplant and potato (Dutta et al. 1981; Marois et al. 1984; Fahima and Henis 1995), parasitizes Sclerotinia sclerotiorum, Rhizoctonia solani and Sclerotium rolfsii (Boosalis 1956; McLaren et al. 1986), degrades cell walls of Pythium ultimum and Fusarium equisetii (Inglis and Kawchuk 2002), shows antagonistic activities against Cylindrocarpon destructans, Fusarium oxysporum, Rhizoctonia solani, Sclerotinia nivalis, Botrytis cinerea, Phytophthora capsici and increases the dehiscence ratios of ginseng seed (Kim et al. 2017). Talaromyces pinophilus shows antagonistic activity and mycoparasitic behavior on Rhizoctonia solani and Botrytis cinerea (Alagesaboopathi 1994; Abdel-Rahim and Abo-Elyousr 2018) and shows plant growth-promoting effects on Waito-C rice (Khalmuratova et al. 2015).
In this study, we collected agricultural soil samples from Fujiang, Guangdong, Jiangxi, Shandong, Tibet and Zhejiang provinces in China. After isolation and identification, six Talaromyces species could not be assigned to any known species. A polyphasic approach including phylogenetic analysis of partial ITS, β-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) gene sequences and macro- and micro-morphological data were used to delimitate the new species and section in this genus.
Materials and methods
Isolates
Soil samples were collected from six provinces from China as mentioned above. A general dilution-plate method was used to isolate fungi, bacteria and actinomycetes. As for fungi, Potato Dextrose Agar (PDA, Guangdong huankai microbiological technology co., LTD) and Rose Bengal Medium (RBM, Beijing luqiao technology co., Ltd) with antibiotics (tetracycline hydrochloride and chloramphenicol with the final concentration of 100 mg/ml) were used. Obtained strains were purified and sub-cultured on malt extract agar (MEA, Guangdong huankai microbiological technology co., Ltd). Reference strains used in this study were obtained from the China General Microbiological Culture Collection Center (CGMCC), Beijing, China, the CBS culture collection and the working collection of the Applied and Industrial Mycology department (DTO), both housed at the Westerdijk Fungal Biodiversity Institute (Utrecht, the Netherlands). An overview of strains is listed in Table 1. For other strains used in the phylogenetic analyses, readers are referred to Chen et al. 2016; Crous et al. 2016, 2017, 2018; Wang QM et al. 2016; Wang XC et al. 2016, 2017; Yilmaz et al. 2016a, b; Guevara-Suarez et al. 2017; Peterson and Jurjević 2017; Barbosa et al. 2018; Jiang et al. 2018; Su and Niu 2018; Varriale et al. 2018; Rajeshkumar et al. 2019; Rodríguez-Andrade et al. 2019; Guevara-Suarez et al. 2020.
Table 1.
Talaromyces strains used in this study.
| Section | Species name | Strain no. | Substrate and origin | GenBank accession nr. | |||
|---|---|---|---|---|---|---|---|
| ITS | BenA | CaM | RPB2 | ||||
| Talaromyces | Talaromyces brevis | CBS 141833T= DTO 349-E7 | Soil, Beijing, China | MN864269 | MN863338 | MN863315 | MN863328 |
| DTO 307-C1 | Soil, Zonguldak, Turkey | MN864270 | MN863339 | MN863316 | MN863329 | ||
| CBS 118436 = DTO 004-D8 | Soil, Maroc | MN864271 | MN863340 | MN863317 | MN863330 | ||
| Talaromyces rufus | CBS 141834 T= DTO 349-D7 = CGMCC 3.13203 | Soil, Yunnan, China | MN864272 | MN863341 | MN863318 | MN863331 | |
| DTO 274-C5 | Soil, Korea | MN864273 | MN863342 | MN863319 | n.a. | ||
| Talaromyces aspriconidius | CBS 141835 T= DTO 340-F8 | Soil, Yunnan, China | MN864274 | MN863343 | MN863320 | MN863332 | |
| Tenues | Talaromyces tenuis | CBS 141840 T = DTO 340-G9 | Soil, Guizhou, China | MN864275 | MN863344 | MN863321 | MN863333 |
| Trachyspermi | Talaromyces albisclerotius | CBS 141839 T = DTO 340-G5 | Soil, Guizhou, China | MN864276 | MN863345 | MN863322 | MN863334 |
| Subinflati | Talaromyces guizhouensis | CBS 141837 T = DTO 340-G8 | Soil, Guizhou, China | MN864277 | MN863346 | MN863323 | MN863335 |
| DTO 054-C8 | Soil from rainforest, Langkawi, Malaysia | MN864278 | MN863347 | MN863324 | MN863336 | ||
| DTO 054-A7 | Soil from rainforest, Langkawi, Malaysia | MN864279 | MN863348 | MN863325 | MN863337 | ||
| Talaromyces resedanus | CBS 181.71T = DTO 376-A7 = ATCC 22356 = FRR 578 = IMI 062877 = NRRL 578 | Soil, A1 horizon of Podzol, Victoria, Seychelles | MN864280 | MN863349 | MN863326 | n.a. | |
| CBS 184.90 = DTO 376-A8 = UPSC 2879 | Soil in greenhouse, Sweden | MN864281 | MN863350 | MN863327 | n.a. | ||
DNA extraction, PCR amplification and sequencing
Strains were grown for 1 wk on MEA prior to DNA extraction. DNA was extracted using the Ultraclean TM Microbial DNA isolation Kit (MoBio, Solana Beach, U.S.A.) and stored at -20 °C. The ITS, BenA, CaM, and RPB2 genes were amplified and sequenced using methods and primers previously described (Houbraken and Samson 2011; Yilmaz et al. 2014).
Phylogenetic analysis
For sectional classification in Talaromyces, a four-gene phylogeny combining ITS, BenA, CaM and RPB2 sequences was used. Prior to combining the datasets, single gene alignments were generated using MAFFT v. 7 (Katoh et al. 2019), and then trimmed at both ends. Aligned datasets were subsequently concatenated using Mesquite v 3.6 (Maddison and Maddison 2018). For each section, single gene phylogenies were generated to determine the phylogenetic relationship among species. The most suitable substitution model was determined using FindModel (Posada and Crandall 1998). Bayesian analyses were performed with MrBayes v. 3.2 (Ronquist et al. 2012). The sample frequency was set to 100 and the first 25% of trees were removed as burn-in. Maximum likelihood analyses including 1000 bootstrap replicates were run using RAxML (Kozlov et al. 2019). Trichocoma paradoxa (CBS 788.83T) was used as an outgroup in the Talaromyces phylogeny. Sequences of T. trachyspermus (CBS 373.48T), T. dendriticus (CBS 660.80T) and T. purpurogenus (CBS 286.36T) were used as outgroups in Talaromyces sections Subinflati, Talaromyces and Trachyspermi, respectively. The resulting trees were visualized with FigTree v1.4.2 and edited in Adobe Illustrator CS5. Bayesian inference (BI) posterior probabilities (pp) values and bootstrap (bs) values are labelled on nodes. Values less than 0.95 pp and 75% bs are not shown. Branches with posterior probability values of 1 and bootstrap values higher than 95% are thickened. Newly obtained sequences were deposited in GenBank.
Morphological analysis
Macroscopic characters were studied on Czapek yeast autolysate agar (CYA), CYA supplemented with 5% NaCl (CYAS), yeast extract sucrose agar (YES), creatine sucrose agar (CREA), dichloran 18% glycerol agar (DG18), oatmeal agar (OA) and malt extract agar (MEA; Oxoid malt) (Samson et al. 2010). Isolates were inoculated at three points on 90 mm Petri dishes and incubated for 7 d at 25 °C in darkness. Additional CYA plates were incubated at 30 and 37 °C, and an additional MEA plate was incubated at 30 °C. After 7 d of incubation, colony diameters were recorded. The colony texture, degree of sporulation, obverse and reverse colony colors, the production of soluble pigments and exudates were noted. Acid production on CREA is indicated by a change in the pH sensitive bromocresol purple dye, from a purple to yellow color in media surrounding colonies. For ascoma production, OA, MEA and CYA plates were incubated for up to four wks. Color codes used in description refer to Rayner (1970).
Microscope preparations were made from 1 wk-old colonies grown on MEA. Production of ascomata, asci and ascospores was determined on 2–3 wk-old colonies on OA. Size of ascospores and conidia were measured without ornamentation. Lactic acid (60%) was used as mounting fluid and 96% ethanol was applied to remove the excess of conidia. A Zeiss Stereo Discovery V20 dissecting microscope and Zeiss AX10 Imager A2 light microscope equipped with Nikon DS-Ri2 cameras and software NIS-Elements D v4.50 were used to capture digital images.
Results
Phylogeny
The individual ITS, BenA, CaM and RPB2 datasets consist of 653, 591, 782 and 802 characters, respectively, and were combined to study the relationship within Talaromyces. The most optimal model for each dataset is listed in Table 2. Eight well-supported lineages are present in the multigene phylogenic analysis (Fig. 1). Seven lineages agree with sectional classification by Yilmaz et al. 2014 and one lineage, represented by a new species described here (Talaromyces tenuis), could not be assigned to any known section. This lineage is sister to sections Talaromyces and Helici but cannot be assigned into any of them. Based on its phylogenetic and morphological peculiarity (see description below), the lineage is described as a new section named Tenues. Furthermore, five new species are distributed over three sections, T. brevis, T. rufus and T. aspriconidius in section Talaromyces; T. albisclerotius in section Trachyspermi and T. guizhouensis in section Subinflati.
Table 2.
Sequence data sets and models used in phylogeny.
| Section | Sequence data sets | |||||||
|---|---|---|---|---|---|---|---|---|
| ITS (bp) | Substitution model | BenA (bp) | Substitution model | CaM (bp) | Substitution model | RPB2 (bp) | Substitution model | |
| Overview Talaromyces | 653 | GTR+G | 591 | K2P+G | 782 | GTR+G | 802 | GTR+G |
| section Subinflati | 751 | GTR+G | 458 | K2P+G | 520 | GTR+G | 893 | K2P+G |
| section Talaromyces | 539 | GTR+G | 398 | HKY+G | 528 | GTR+G | 838 | GTR+G |
| section Trachyspermi | 505 | GTR+G | 422 | GTR+G | 556 | GTR+G | 852 | GTR+G |
Figure 1.
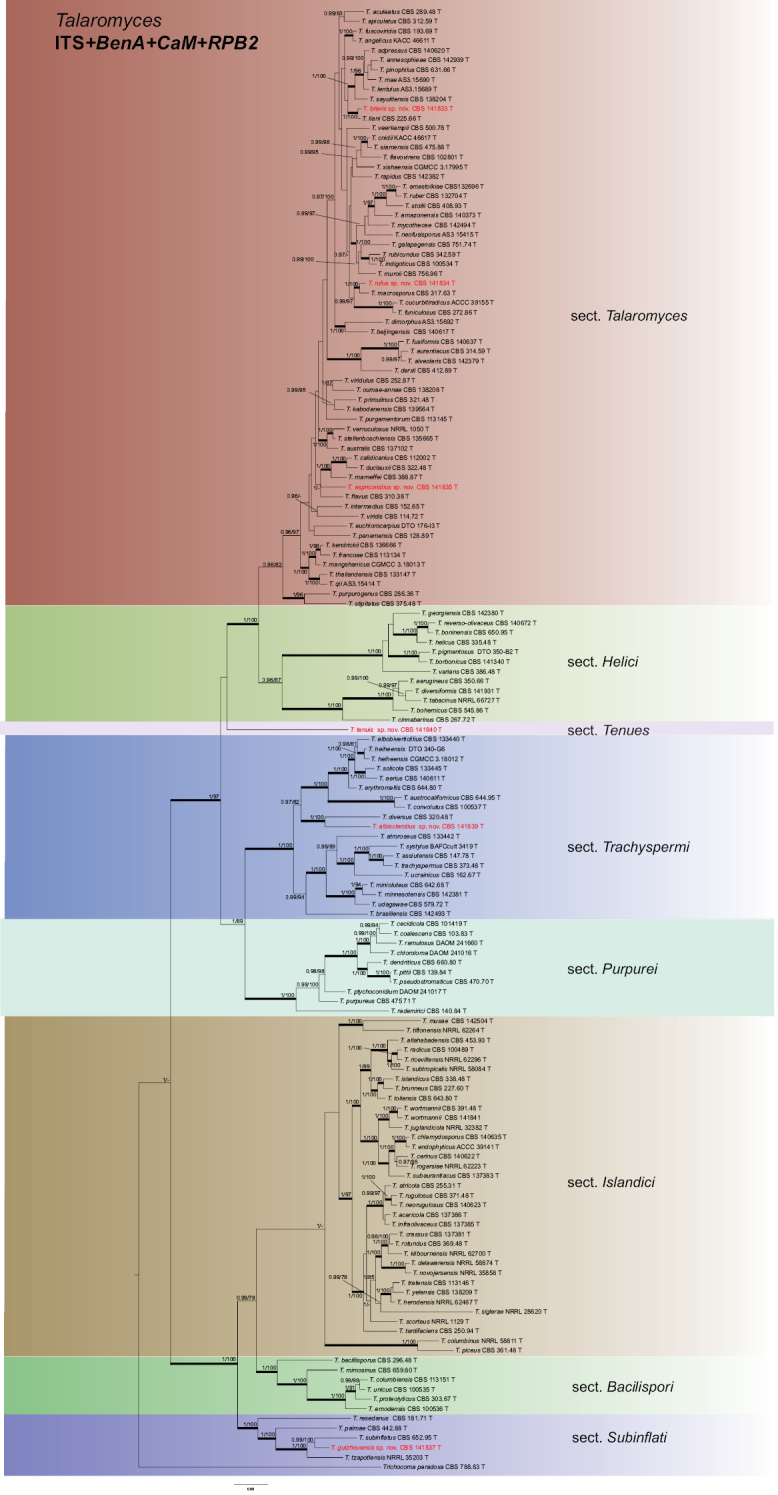
Concatenated phylogeny of the ITS, BenA, CaM and RPB2 gene regions of species from Talaromyces. Branches with values more than 1 pp and 95% bs are thickened, supports lower than those values are indicated with a dash (-). Trichocoma paradoxa (CBS 788.83T) was chosen as outgroup. T: ex-type.
In section Talaromyces, T. rufus and T. aspriconidius can be separated via each single gene phylogram. Talaromyces rufus is close to T. macrosporus based on BenA, CaM and RPB2 phylograms and forms a separate lineage in ITS phylogram. Talaromyces aspriconidius is close to T. primulinus based on RPB2 phylogram, but clusters with T. flavus based on BenA phylogram, and forms a separate lineage in CaM and ITS phylograms. Talaromyces brevis is closely related to T. liani, it can be differentiated via BenA, CaM and RPB2 phylograms, but not via ITS phylogram (Fig. 2; Suppl. materials: 1–3).
Figure 2.
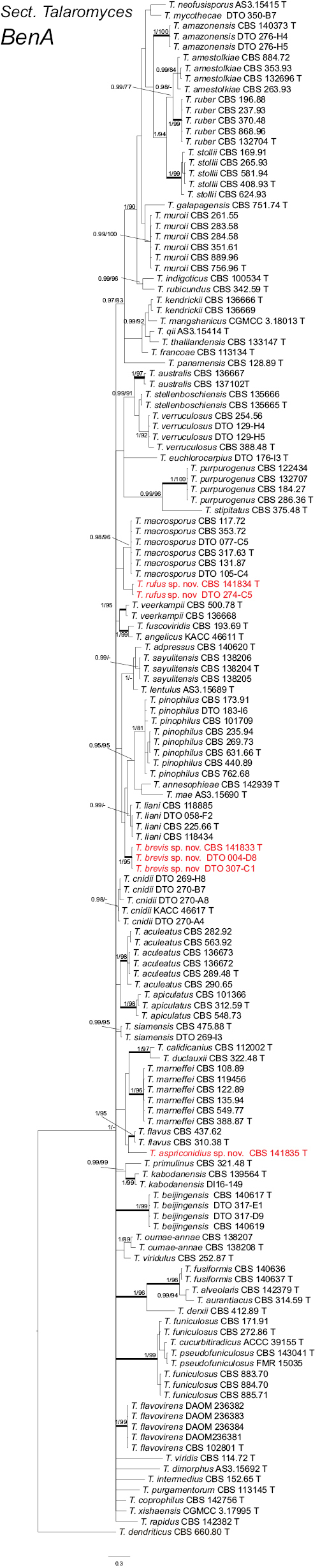
Phylogeny of BenA for species classified in Talaromyces section Talaromyces. Branches with values more than 1 pp and 95% bs are thickened, supports lower than those values are indicated with a dash (-). Talaromyces dendriticus (CBS 633.80T) was chosen as outgroup. T: ex-type.
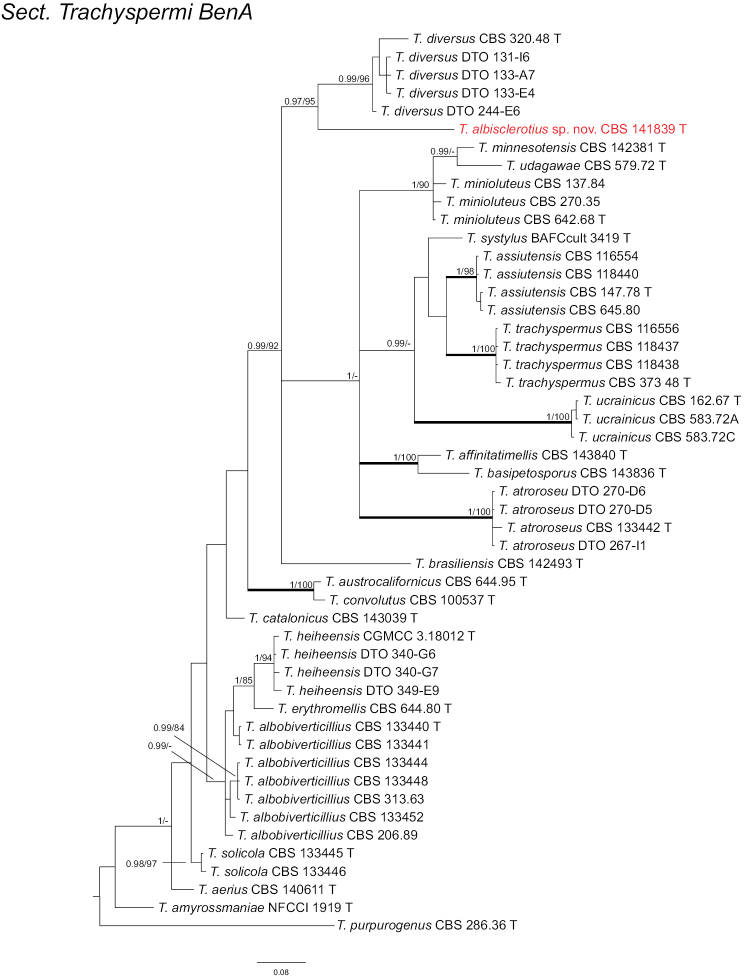
In section Trachyspermi, T. albisclerotius can be well-separated in four single phylograms; this species clusters with T. diversus in BenA, CaM and RPB2 phylograms, and forms a separate lineage in ITS phylogram (Fig. 3; Suppl. materials: 4–6).
Figure 3.
Phylogeny of BenA for species classified in Talaromyces section Trachyspermi. Branches with values more than 1 pp and 95% bs are thickened, supports lower than those values are indicated with a dash (-). Talaromyces purpurogenus (CBS 286.36T) was chosen as outgroup. T: ex-type.
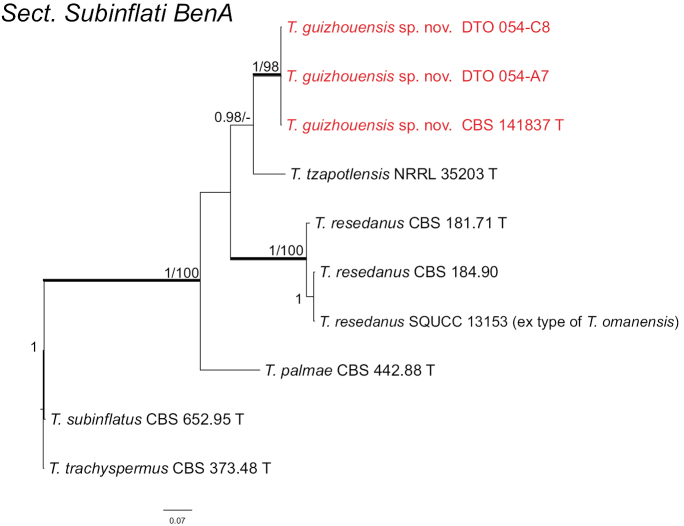
Talaromyces guizhouensis is assigned in section Subinflati and P. resedanum also belongs to this section according to our multigene analysis. With the newly described T. tzapotlensis and T. omanensis the total number of taxa belonging to section Subinflati increased from two to five since it was established in 2014. Talaromyces omanensis shares same ITS, BenA and CaM sequences with T. resedanus CBS 184.90. Talaromyces guizhouensis is close to T. tzapotlensis and T. subinflatus in each single gene phylogram (Fig. 4, Suppl. materials: 7–9).
Figure 4.
Phylogeny of BenA for species classified in Talaromyces section Subinflati. Branches with values more than 1 pp and 95% bs are thickened, supports lower than those values are indicated with a dash (-). Talaromyces trachyspermus (CBS 373.48T) was chosen as outgroup. T: ex-type.
Identification
The five new species T. albisclerotius, T. aspriconidius, T. guizhouensis, T. rufus, T. tenuis can be identified by ITS, BenA, CaM and RPB2 sequences. Talaromyces brevis cannot be separated from T. liani (strains CBS 225.66T, CBS 118885, CBS 118434 and DTO 058-F2) by its ITS sequence, but it can be differentiated from T. liani by BenA (97.3% similarity, 366/376 bp), CaM (99.5% similarity, 463/465 bp) and RPB2 (99% similarity, 838/846 bp).
Taxonomy
Talaromyces section Tenues
B.D. Sun, A.J. Chen, Houbraken & Samson sect. nov.
6770C3CD-40B6-57FB-A11D-00B30DAF2128
833138
Typus.
Talaromyces tenuis B.D. Sun, A.J. Chen, Houbraken & Samson
Description.
Conidiophores monoverticillate or biverticillate, with hyaline, thin stipes, colonies grow restrictedly on CYA, YES, DG18, slightly faster on MEA and OA, no growth on CYAS and CREA at 25 °C and CYA incubated at 37 °C.
Phylogenetic analysis places Talaromyces section Tenues sister to sections Talaromyces and Helici (Fig. 1); however, statistical support for this relationship is lacking. Using a nine-gene sequence data set, Houbraken et al. (2020) confidently shows that Talaromyces sp. CBS 141840 (= T. tenuis, the sole representative of the section) is sister to sect. Purpurei and Trachyspermi. section Trachyspermi species produce abundant red pigments (Yilmaz et al. 2014), while Talaromyces tenuis does not. section Purpurei species generally grow rapidly on CYA and MEA, and usually produce synnemata after two to three weeks of incubation (Yilmaz et al. 2014).
Etymology.
Named after the type species of the section, Talaromyces tenuis.
Talaromyces tenuis
B.D. Sun, A.J. Chen, Houbraken & Samson sp. nov.
84420C8A-B025-53A1-808E-5777DBFB18A0
833136
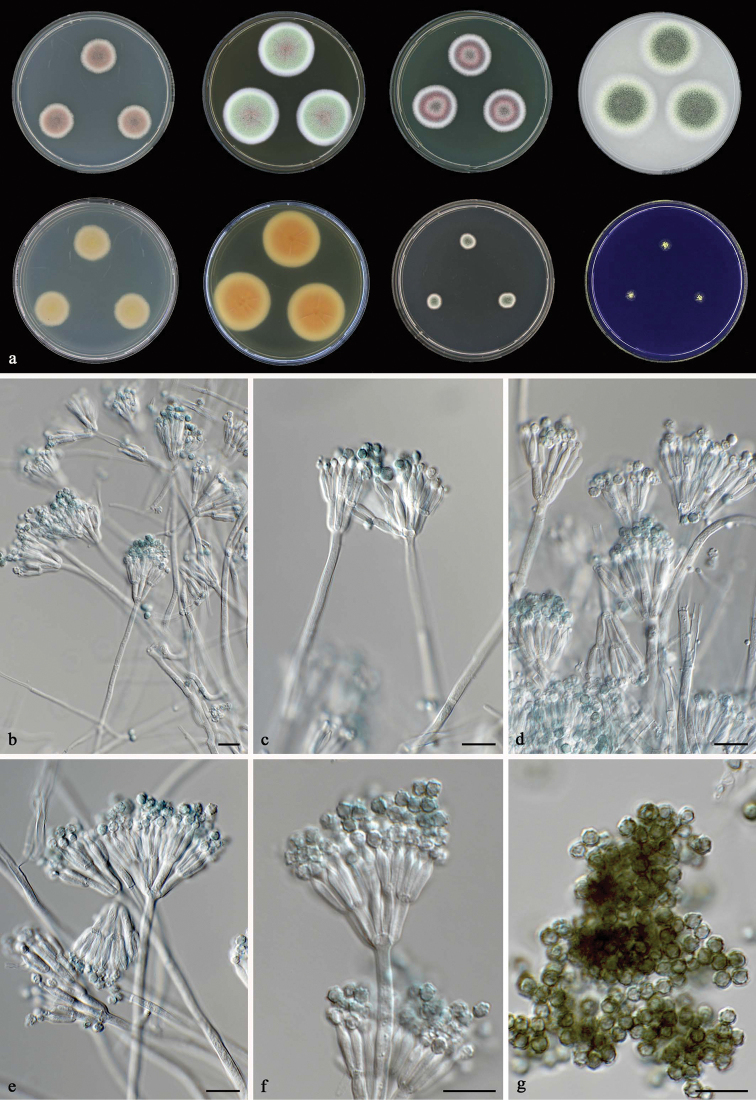
Figure 5.
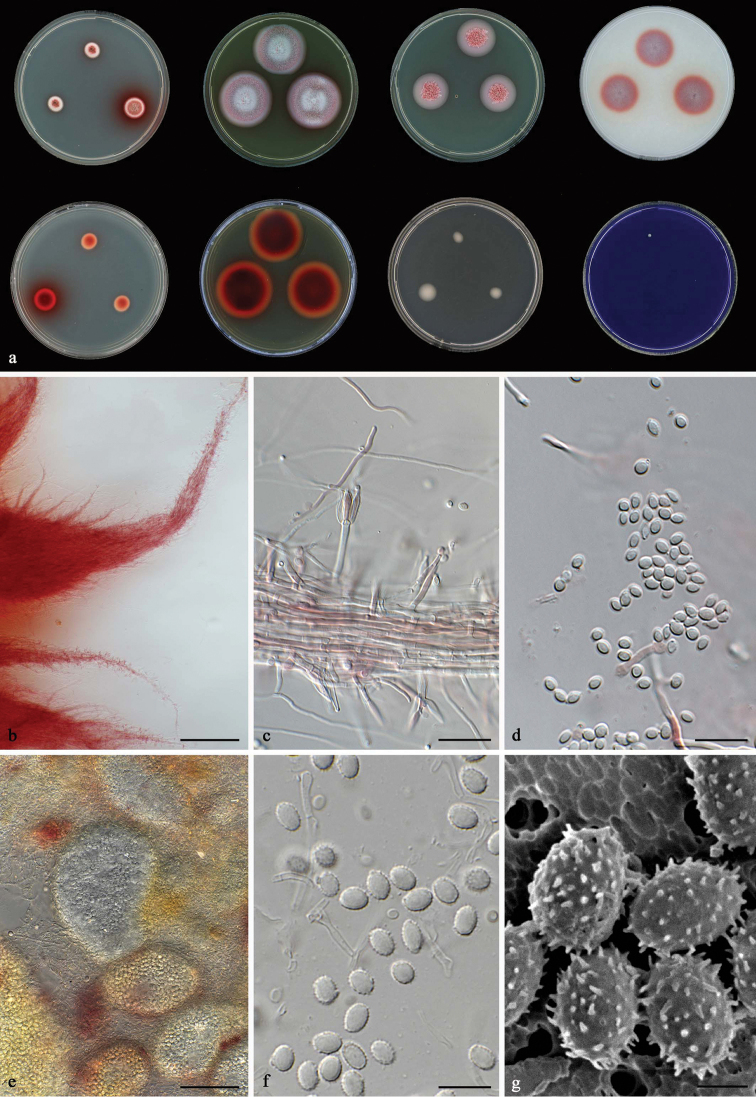
Talaromyces tenuis CBS 141840Ta colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG 18 and CREAb–g conidiophores and conidia. Scale bars: 10 μm (b–g).
Typus.
China, Guizhou, soil, 2014, isolated by X.Z. Jiang, Holotype CBS H-22838, culture ex-holotype CBS 141840 = DTO 340-G9.
ITS barcode.
MN864275. Alternative identification markers: BenA = MN863344, CaM = MN863321, RPB2 = MN863333.
Diagnosis.
Talaromyces tenuis produces hyaline, thin conidiophores, yellow mycelium on MEA and OA, and grows very restrictedly on CYA, YES and DG18.
In.
Talaromyces section Tenues
Colony diam, 7 d (mm).
CYA 7–8; CYA 30 °C 5–8; CYA 37 °C No growth; MEA 18–20; MEA 30 °C 10–11; OA 12–14; YES 9–10; CREA No growth; CYAS No growth; DG18 2–3.
Colony characters.
CYA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse white. MEA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) or ochreous (44); texture floccose; sporulation sparse; conidia en masse white or greyish yellow-green (68); soluble pigments absent; exudates absent; reverse ochreous (44) to umber (9). YES 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse buff (45). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse white. OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium pale luteous (11); texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse buff (45).
Micromorphology.
Conidiophores monoverticillate or biverticillate, stipes smooth, 80–150 × 2–3 μm; metulae 2–3, divergent, 8–9 × 2–2.5 μm; phialides 2–3, acerose, 8–9.5 × 2–2.5 μm; conidia smooth, globose to subglobose, 2–3 × 2–2.5 μm. Ascomata not observed.
Note.
Talaromyces tenuis is phylogenetically distinct and is basal to species belonging to sections Talaromyces and Helici (Fig. 1). In a nine-gene phylogeny, it is sister to sections Purpurei and Trachyspermi (Houbraken et al. 2020). This species is characterized by hyaline, thin conidiophores, and grows very restrictedly on CYA, YES and DG18; colonies on MEA and OA have prominent yellow mycelia.
Etymology.
Latin, tenuis, refers to its thin conidiophores.
Talaromyces albisclerotius
B.D. Sun, A.J. Chen, Houbraken & Samson sp. nov.
6F3414A0-09C1-57DE-B5A0-A1BC48EF98A4
833135
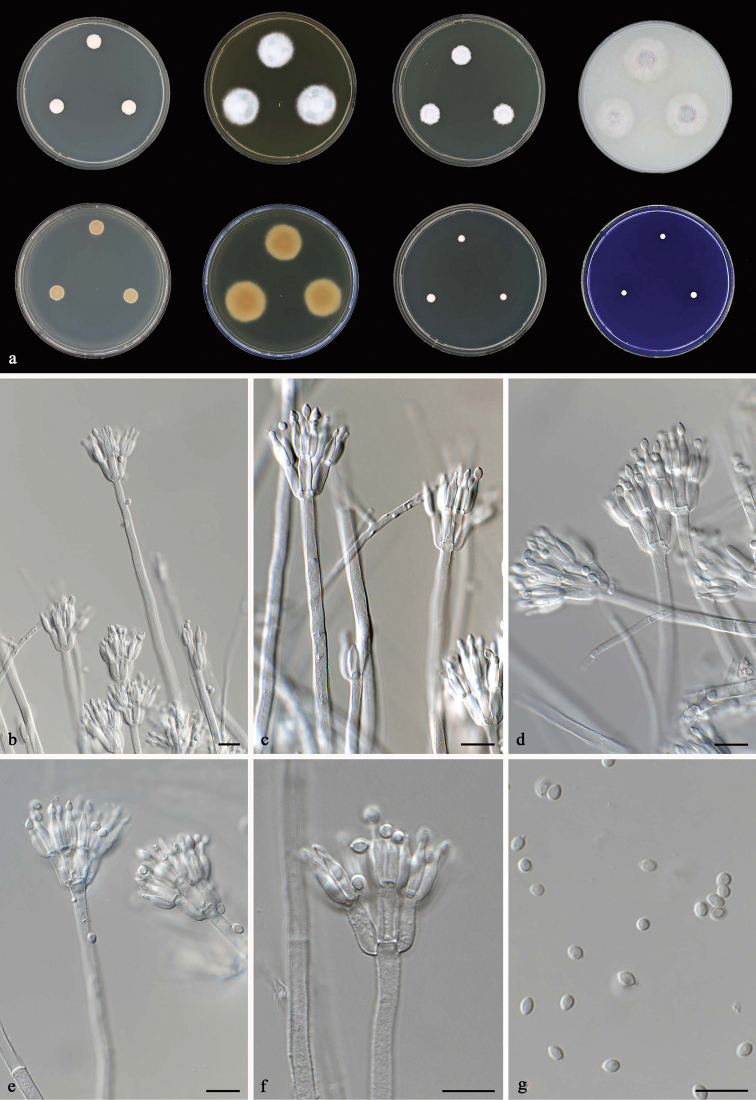
Figure 6.
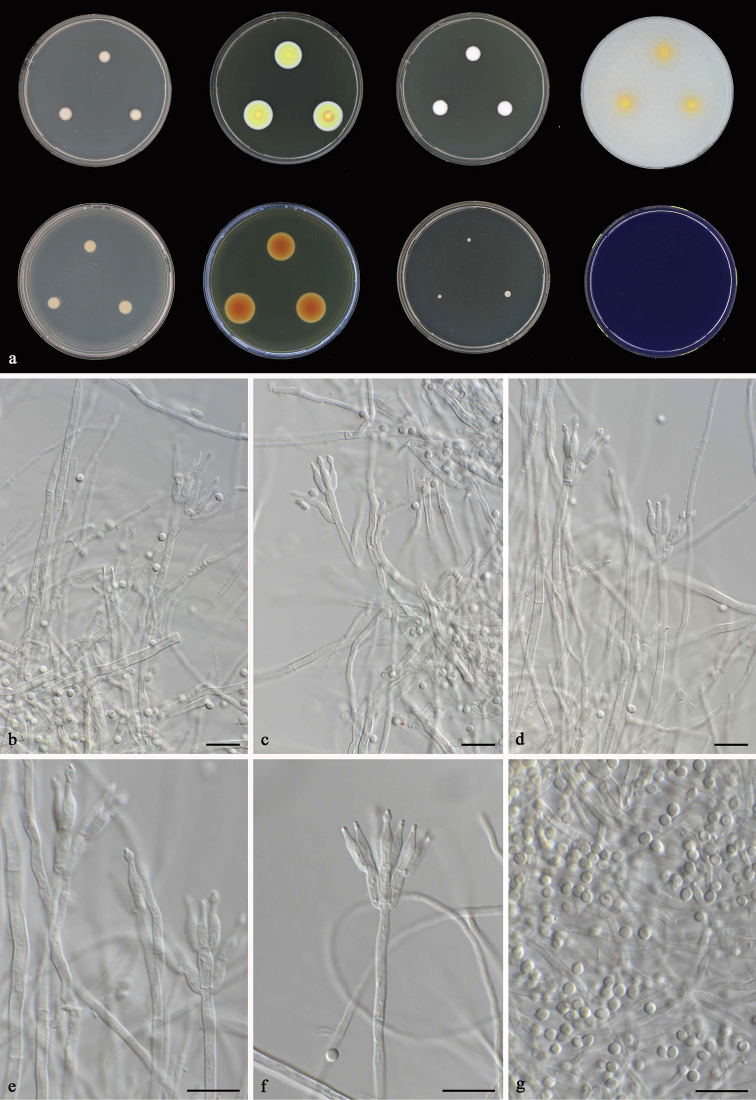
Talaromyces albisclerotius CBS 141839Ta colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG 18 and CREAb sclerotia on OA after two weeks c–g conidiophores and conidia. Scale bars: 1000 μm (b), 10 μm (c–g).
Typus.
China, Xinjiang, soil, 2002, isolated by L. Cai, Holotype CBS H-22837, culture ex-holotype CBS 141839 = DTO 340-G5.
ITS barcode.
MN864276. Alternative identification markers: BenA = MN863345, CaM = MN863322, RPB2 = MN863334.
Diagnosis.
Talaromyces albisclerotius produces white sclerotia on OA, grows restrictedly on CYA, YES, DG18 and OA and does not grow on CYAS.
In.
Talaromyces section Trachyspermi
Colony diam, 7 d (mm).
CYA 5–8; CYA 30 °C 3–4; CYA 37 °C No growth; MEA 19–20; MEA 30 °C 8–9; OA 13–14; YES 6–7; CREA No growth; CYAS No growth; DG18 5–6.
Colony characters.
CYA 25 °C, 7 d: Colonies moderately deep, slight sulcate; margins entire; mycelium white; texture floccose; sporulation dense; conidia en masse greyish yellow-green (68); soluble pigments absent; exudates absent; reverse buff (45). MEA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white and primrose (66); texture floccose; sporulation dense; conidia en masse pistachio green (92); soluble pigments absent; exudates absent; reverse ochreous (44). YES 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse pistachio green (92); soluble pigments absent; exudates absent; reverse buff (45). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse greyish yellow-green (68); soluble pigments absent; exudates absent; reverse sulphur yellow (15). OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and primrose (66); texture velvety; sporulation dense; conidia en masse yellow green (71); soluble pigments absent; exudates clear droplets; reverse greyish yellow-green (68). CREA 25 °C, 7 d: No growth.
Micromorphology.
Conidiophores biverticillate, with a minor proportion having subterminal branches; stipes smooth, 70–130 × 3–4 μm, extra branches 10–20 μm; metulae 3–5, divergent, 8.5–11 × 4–4.5 μm; phialides 4–6, acerose, 9–11 × 3–5 μm; conidia smooth, subglobose to fusiform, 2–4.5× 3–4 μm. Ascomata not observed, white sclerotia present on OA after 1 wk.
Notes.
Talaromyces albisclerotius is characterized by the production of white sclerotia on OA after 1 wk incubation; these sclerotia remain sterile and no ascospores are observed after prolonged incubation up to eight wk. Talaromyces assiutensis and T. trachyspermus could produce white ascomata, but their ascomata mature after weeks and release ascospores (Yilmaz et al. 2014). Phylogenetically, T. albisclerotius clusters with T. diversus and T. brasiliensis, but T. diversus grows faster on MEA, and T. brasiliensis produces rough conidia (Yilmaz et al. 2014; Barbosa et al. 2018).
Etymology.
Latin, albisclerotius, refers to its white sclerotia produced on OA.
Talaromyces aspriconidius
B.D. Sun, A.J. Chen, Houbraken & Samson sp. nov
BAA53332-CBC6-523E-8EF3-808AE135CCC9
833134
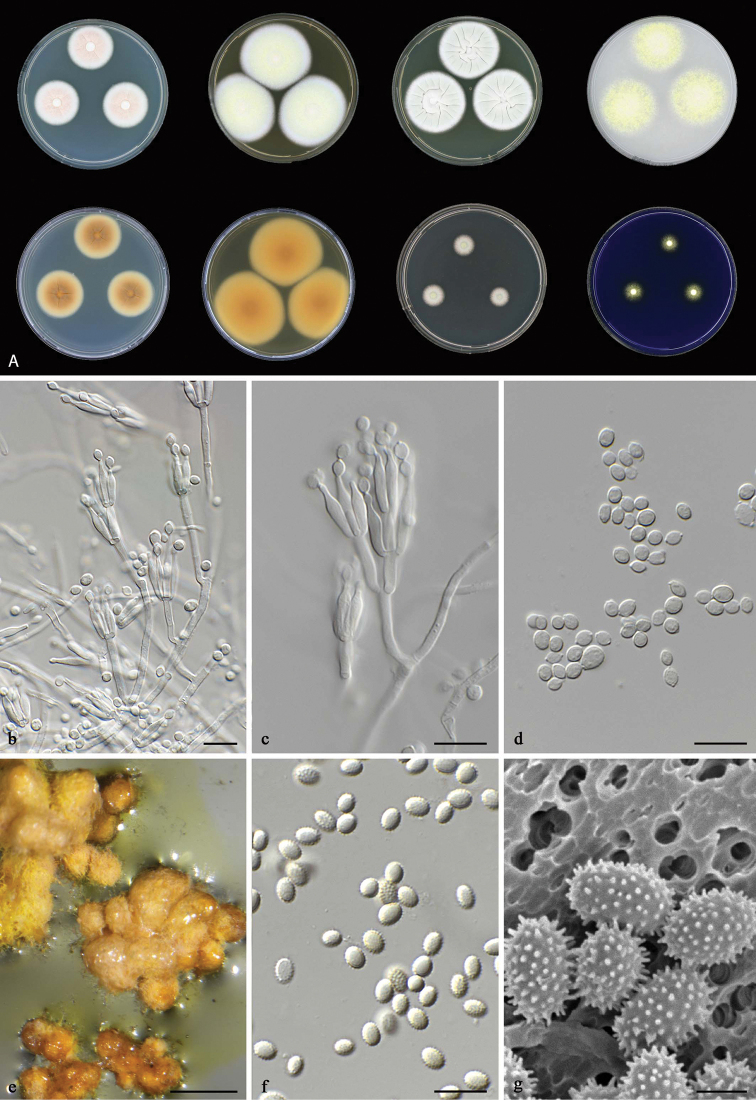
Figure 7.
Talaromyces aspriconidius CBS 141835Ta colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG 18 and CREAb–g conidiophores and conidia. Scale bars: 10 μm (b–g).
Typus.
China, Yunnan, soil, 2008, isolated by L. Cai, Holotype CBS H-22833, culture ex-holotype CBS 141835 = DTO 340-F8.
ITS barcode.
MN864274. Alternative identification markers: BenA = MN863343, CaM = MN863320, RPB2 = MN863332.
Diagnosis.
Talaromyces aspriconidius produces strikingly roughened, globose conidia, grows moderately on CYA and CYA at 30 °C, reaches 22–23 mm and 25–26 mm after 7 d.
In.
Talaromyces section Talaromyces
Colony diam, 7 d (mm).
CYA 22–23; CYA 30 °C 25–26; CYA 37 °C 22–23; MEA 36–37; MEA 30 °C 44–45; OA 38–42; YES 28–29; CREA 7–8; CYAS No growth; DG18 10–11.
Colony characters.
CYA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and peach (4); texture floccose; sporulation moderately dense; conidia en masse greyish yellow-green (68); soluble pigments absent; exudates absent; reverse buff (45). MEA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse leek green (49) and greyish yellow-green (68); soluble pigments absent; exudates brown droplets; reverse saffron (10). YES 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and peach (4); texture floccose; sporulation moderately dense; conidia en masse greyish yellow-green (68); soluble pigments absent; exudates absent; reverse saffron (10). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and primrose (66); texture floccose; sporulation dense; conidia en masse honey (64); soluble pigments absent; exudates absent; reverse greyish yellow-green (68). OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and primrose (66); texture floccose; sporulation dense; conidia en masse yellow-green (71); soluble pigments absent; exudates clear droplets; reverse greyish yellow-green (68). CREA 25 °C, 7 d: Moderate growth, acid production absent.
Micromorphology.
Conidiophores biverticillate, stipes smooth, 150–250 × 3–4 μm, metulae 4–5, divergent, 10–12 × 3–3.5 μm; phialides 4–6, acerose to flask shaped, 8–10.5 × 3–3.5 μm; conidia strikingly roughened, globose, 3–4 μm. Ascomata not observed.
Notes.
Talaromyces aspriconidius is characterized by its strikingly roughened, globose conidia. Talaromyces aculeatus, T. apiculatus, T. diversus, T. solicola and T. verruculosus also produce this kind of conidia. However, T. aspriconidius grows slower than T. aculeatus, T. apiculatus and T. verruculosus, and faster than T. diversus and T. solicola on CYA and CYA at 30 °C (Yilmaz et al. 2014).
Etymology.
Latin, aspriconidius, refers to its strikingly roughened conidia.
Talaromyces brevis
B.D. Sun, A.J. Chen, Houbraken & Samson sp. nov.
D94B7794-43E2-58E9-910F-B836D4CB60DE
833132
Figure 8.
Talaromyces brevis CBS 141833Ta colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG 18 and CREAb–d conidiophores and conidia e ascomata on OA after two weeks f–g ascospores. Scale bars: 10 μm (b–d, f), 1000 μm (e), 2 μm (g).
Typus.
China, Beijing, soil, 2010, isolated by B.D. Sun, Holotype CBS H-22831, culture ex-holotype CBS 141833= DTO 349-E7.
Additional material examined.
Turkey, Zonguldak, soil, 2014, isolated by Rasime Demirel, culture DTO 307-C1. Maroc, soil, 2005, isolated by J. Dijksterhuis, culture CBS 118436 = DTO 004-D8.
ITS barcode.
MN864269. Alternative identification markers: BenA = MN863338, CaM = MN863315, RPB2 = MN863328.
Diagnosis.
Talaromyces brevis produces short conidiophores measuring 15–50 × 3–4 μm, yellow to orange ascomata on OA and spiny ascospores measuring 3.5–4.5 × 3–4 μm.
In.
Talaromyces section Talaromyces
Colony diam, 7 d (mm).
CYA 30–31; CYA 30 °C 28–30; CYA 37 °C 25–26; MEA 50–51; MEA 30 °C 57–60; OA 39–43; YES 42–43; CREA 13–14; CYAS No growth; DG18 13–15.
Colony characters.
CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white and flesh (37); texture floccose; sporulation sparse; conidia en masse white; soluble pigments absent; exudates absent; reverse ochreous (44). MEA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and primrose (66); texture floccose; sporulation sparse; conidia en masse white to greyish yellow-green (68); soluble pigments absent; exudates absent; reverse ochreous (44). YES 25 °C, 7 d: Colonies moderately deep, sulcate; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse ochreous (44). DG18 25 °C, 7 d: Colonies moderately deep, slightly raised at center, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse yellow-green (71); soluble pigments absent; exudates absent; reverse greyish yellow-green (68). OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium primrose (66); texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse primrose (66). Ascomata present. CREA 25 °C, 7 d: Moderate growth, acid production present.
Micromorphology.
Conidiophores monoverticillate and biverticillate; stipes smooth, 15–50 × 3–4 μm; metulae 3–5, divergent, 10–15 × 2.5–3 μm; phialides 4–6, flask-shaped, 9–13 × 2–4 μm; conidia smooth, subglobose to fusiform, 3–4(–5) × 2.5–3.5(–4.5) μm. Ascomata maturing after 2–3 wk of incubation on OA, yellow to orange, globose to subglobose, 400–550 μm; ascospores ellipsoidal, spiny, 3.5–4.5 × 3–4 μm.
Notes.
Talaromyces brevis is morphologically and phylogenetically close to T. liani, but the latter produces larger ascospores measuring 4–6 × 2.5–4 μm and does not produce acid on CREA (except T. liani CBS 118885 produces very weak acid) (Yilmaz et al. 2014).
Etymology.
Latin, brevis, refers to its short conidiophores.
Talaromyces guizhouensis
B.D. Sun, A.J. Chen, Houbraken & Samson sp. nov.
4B355123-B815-57DF-88FF-E4ED77BC6C39
833131
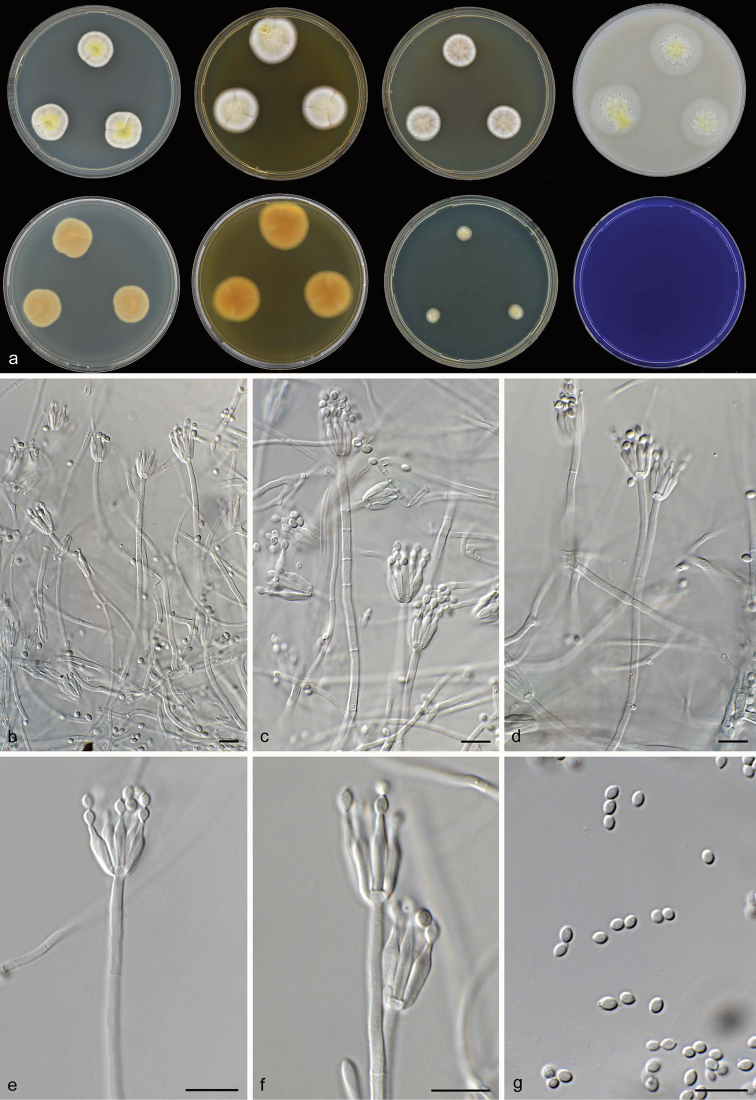
Figure 9.
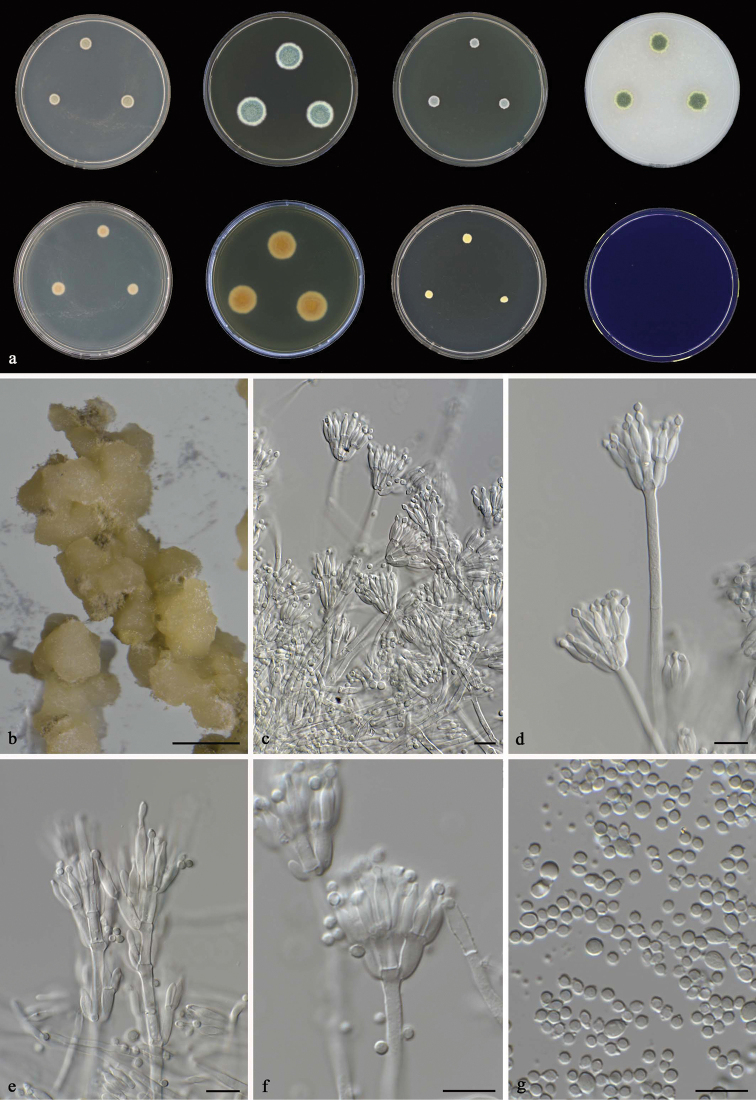
Talaromyces guizhouensis CBS 141837Ta colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG 18 and CREAb–g conidiophores and conidia. Scale bars: 10 μm (b–g).
Typus.
China, Guizhou, soil, 2014, isolated by X.Z. Jiang, Holotype CBS H-22835, culture ex-holotype CBS 141837= DTO 340-G8.
Additional material examined.
Malaysia, Langkawi, soil from rainforest, 2007, isolated by J. Houbraken, culture DTO 054-C8. Malaysia, Langkawi, soil from rainforest, 2007, isolated by J. Houbraken, culture DTO 054-A7.
ITS barcode.
MN864277. Alternative identification markers: BenA = MN863346, CaM = MN863323, RPB2 = MN863335.
Diagnosis.
Talaromyces guizhouensis grows poorly on CREA and DG18, does not produce synnemata as well as ascospores.
In.
Talaromyces section Subinflati
Colony diam, 7 d (mm).
CYA 8–9; CYA 30 °C 10; CYA 37 °C No growth; MEA 24–27; MEA 30 °C 18–19; OA 27–29; YES 12–13; CREA 2–3; CYAS No growth; DG18 4–5.
Colony characters.
CYA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates clear droplets; reverse saffron (10). MEA 25 °C, 7 d: Colonies moderately deep, raised at center, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse pistachio green (92); soluble pigments absent; exudates absent; reverse saffron (10). YES 25 °C, 7 d: Colonies moderately deep, raised at center, plane; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates clear droplets; reverse cream white. DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse cream white. OA 25 °C, 7 d: Colonies moderately deep, raised at center, plane; margins entire; mycelium white; texture floccose; sporulation moderately dense; conidia en masse pistachio green (92); soluble pigments absent; exudates clear droplets; reverse greyish lavender (98) at center, fading into saffron (10). CREA 25 °C, 7 d: Poor growth, acid production absent.
Micromorphology.
Conidiophores biverticillate, stipes smooth to finely rough, 150–300 × 3–4.5 μm, metulae 3–5, divergent, 11–13 × 3–5 μm; phialides 3–5, acerose to flask shaped, 9–10 × 3–3.5 μm; conidia finely rough, subglobose to fusiform, 2.5–4.5 × 2.5–3 μm. Ascomata not observed.
Notes.
section Subinflati previously contained two species namely T. subinflatus and T. palmae. These species do not resemble each other, although both grow poorly on CREA and DG18 (Yilmaz et al. 2014). Talaromyces tzapotlensis was included more recently (Peterson and Jurjević 2017) and we here expand this section with T. guizhouensis and T. resedanus. Like the other species in this section, T. guizhouensis also grows poorly on CREA and DG18. This species is phylogenetically related to T. subinflatus, but the latter grows very restrictedly on common media except MEA (Yilmaz et al. 2014). Talaromyces palmae produces indeterminate synnemata and short stipes (up to 85 μm) (Yilmaz et al. 2014) and these are not observed in T. guizhouensis. Furthermore, T. tzapotlensis grows faster on most media (e.g., 29–30 vs 8–9 mm on CYA; 10–11 vs 4–5 mm on DG18; 20–22 vs 2–3 mm on CREA, all diam. after 7 days (Peterson and Jurjević 2017) and T. resedanus does not grow on CREA and produces smaller conidia measuring 2–3 × 1.5–2 μm.
Etymology.
Latin, guizhouensis, refers to its origin, isolated from Guizhou, China.
Talaromyces resedanus
(McLennan and Ducker) A.J. Chen, Houbraken & Samson comb. nov.
71CF7A9A-5641-5A52-B1CF-4A720AC657EF
302422
Figure 10.
Talaromyces resedanus CBS 181.71Ta colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG 18 and CREAb–g conidiophores and conidia. Scale bars: 10 μm (b–g).
Penicillium resedanum McLennan and Ducker, Aust. J. Bot. 2: 360. 1954. Basionym.
= Talaromyces omanensis Halo, Maharachch., Al-Yahyai and Al-Sadi, Phytotaxa 404: 192. 2019.
Typus.
Australia, Frankston in solo arenoso acido, Holotype IMI 062877, culture ex-holotype CBS 181.71 = DTO 376-A7 = ATCC 22356 = FRR 578 = IMI 062877 = NRRL 578.
Additional material examined.
Sweden, soil in greenhouse, 1989, isolated by O. Constantinescu, culture CBS 184.90 = DTO 376-A8 = UPSC 2879.
ITS barcode.
MN864280. Alternative identification markers: BenA = MN863349, CaM = MN863326, RPB2 = MN969214.
In.
Talaromyces section Subinflati
Colony diam, 7 d (mm).
CYA 19–21; CYA 30 °C 18–20; CYA 37 °C 8–11; MEA 23–25; MEA 30 °C 15–21; OA 26–28; YES 17–19; CREA No growth; CYAS 5–7; DG18 8–9.
Colony characters.
CYA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium sulphur yellow (15) at center, white at edge; texture floccose; sporulation absent; soluble pigments absent; exudates clear droplets; reverse saffron (10). MEA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and sulphur yellow (15); texture floccose; sporulation sparse; conidia en masse white to greyish yellow-green (68); soluble pigments absent; exudates clear droplets; reverse saffron (10). YES 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and buff (45); texture floccose; sporulation sparse; conidia en masse white to greyish yellow-green (68); soluble pigments absent; exudates clear droplets; reverse saffron (10). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse cream white. OA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and sulphur yellow (15); texture floccose; sporulation absent; soluble pigments absent; exudates clear droplets; reverse white. CREA 25 °C, 7 d: No growth.
Micromorphology.
Conidiophores monoverticillate, stipes smooth, 50–150 × 3–4.5 μm, phialides 3–9, flask shaped, 9–12 × 3–3.5 μm; conidia finely rough, ellipsoidal, 2–3 × 1.5–2 μm. Ascomata not observed.
Notes.
Talaromyces resedanus grows restrictedly on DG18 and does not grow on CREA, two features shared with other taxa in section Subinflati. The monoverticillate conidiophores can differentiate T. resedanus from all reported section Subinflati species. Talaromyces aerugineus, T. flavus, T. intermedius, T. rotundus, T. tardifaciens also produce monoverticillate conidiophores, all of them except T. aerugineus can produce ascospores. Talaromyces aerugineus differs from T. resedanus by its shorter conidiophores (10–20 × 2.5–5 μm) and large, globose to ellipsoidal conidia (3–8.5 × 2.5–5 μm).
This species was introduced as Penicillium resedanum (McLennan et al., 1954). Yilmaz et al. (2014) listed it as doubtful species because the ex-type culture CBS 181.71 was not viable at that time. We requested the lyophilized culture of CBS 181.71 and CBS 184.90 deposited in nitrogen, and successfully resurrected them. The concatenated alignment and the single gene phylogenies proved its assignment in section Subinflati. Talaromyces omanensis described by Halo et al. (2019) shares ITS, BenA and CaM (all 100% similarity) sequences with T. resedanus CBS 184.90, have 99.7% (576/578), 98.3% (357/363), 98.8% (487/493) similarity with T. resedanus CBS 181.71T. Its type culture SQUCC 13153 showed good sporulation on CYA and MEA and thus displayed green colony. The monoverticillate conidiophores, size and shape of stipes, phialides and conidia of T. omanensis resemble those of T. resedanus, except that conidiophores of T. omanensis are rough under scanning electron microscope (SEM) (Halo et al. 2019). The photo plate of T. omanensis showed smooth conidiophores under microscope. Based on the molecular and morphological similarity, we considered T. omanensis a synonym of T. resedanus.
Talaromyces rufus
B.D. Sun, A.J. Chen, Houbraken & Samson sp. nov.
A865909C-8672-52AD-BE6E-58C6E86A2122
833133
Figure 11.
Talaromyces rufus CBS 141834Ta colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG 18 and CREAb synnemata on MEA after 1 wk incubation c, d conidiophores and conidia e ascomata f, g ascospores. Scale bars: 200 μm (b), 10 μm (c, d, f), 50 μm (e), 2 μm (g).
Typus.
China, Yunnan, soil, 2009, isolated by T.S. Zhou, Holotype CBS H-22832, culture ex-holotype CBS 141834 = DTO 349-D7 = CGMCC 3.13203.
Additional material examined.
Korea, soil, 2013, isolated by J. Houbraken, culture DTO 274-C5.
ITS barcode.
MN864272. Alternative identification markers: BenA = MN863341, CaM = MN863318, RPB2 = MN863331.
Diagnosis.
This species produces red, determinate synnemata and ellipsoidal, spiny ascospores measuring 5–6 × 4–5 μm.
In.
Talaromyces section Talaromyces
Colony diam, 7 d (mm).
CYA 12–16; CYA 30 °C 18–20; CYA 37 °C 15–16; MEA 37–38; MEA 30 °C 50–51; OA 38–40; YES 26–27; CREA Weak growth; CYAS No growth; DG18 9–13.
Colony characters.
CYA 25 °C, 7 d: Colonies deep, plane; margins entire; mycelium white and scarlet (5); texture floccose; sporulation sparse; conidia en masse greyish yellow-green (68); soluble pigments scarlet (5); exudates absent; reverse scarlet (5). MEA 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white and scarlet (5); texture floccose; sporulation sparse; conidia en masse greyish yellow-green (68); soluble pigments scarlet (5); exudates absent; reverse scarlet (5). YES 25 °C, 7 d: Colonies moderately deep, raised at center, plane; margins entire; mycelium white and scarlet (5); texture floccose; sporulation absent; soluble pigments absent; exudates scarlet (5) droplets; reverse scarlet (5) at center, fading into peach (4). DG18 25 °C, 7 d: Colonies moderately deep, plane; margins entire; mycelium white; texture smooth and sticky; sporulation absent; soluble pigments absent; exudates absent; reverse cream white. OA 25 °C, 7 d: Colonies low, plane; margins entire; mycelium white and scarlet (5); texture floccose; sporulation absent; soluble pigments scarlet (5); exudates absent; reverse scarlet (5). Ascomata present. CREA 25 °C, 7 d: Acid production absent.
Micromorphology.
Conidiophores solitary and monoverticillate; stipes smooth, 5–30 × 2.5–3 μm; phialides1–4, acerose, 10–12 × 3–4 μm; conidia smooth, ellipsoidal to fusiform, 2.5–4.5 × 2–3 μm. Ascomata maturing within 2–3 wk on OA, subglobose to ellipsoildal, 350–600 × 200–350 μm, yellow, ascospores ellipsoidal, spiny, 5–6 × 4–5 μm.
Notes.
Talaromyces rufus is characterized by its red determinate synnemata on all tested media except DG18, CYAS and CREA. According to Yilmaz et al. (2014), twelve Talaromyces species produce determinate or indeterminate synnemata, but T. rufus can be easily distinguished from them by its red synnemata. Phylogenetically, T. rufus is related to T. macrosporus; however, T. macrosporus produces broadly ellipsoidal ascospores and does not produce synnemata.
Etymology.
Latin, rufus, refers to its red synnemata.
Discussion
Previous studies showed that the genus Talaromyces comprises seven sections (Yilmaz et al. 2014). In this study, a comprehensive isolation of soil samples was carried out in China; one new section and six new species were described using a polyphasic approach. Talaromyces section Tenues is newly introduced and contains one species. In our phylogenetic analysis, this section is sister to sections Talaromyces and Helici, though statistical support is lacking. Houbraken et al. (2020) studied the relationships within the Eurotiales and confidently showed that the section is sister to Purpurei and Trachyspermi. section Tenues is morphologically characterized by restricted growth on CYA, YES and DG18, slightly faster growth on MEA and OA and no growth on CREA. Based on these phenotypic characteristics, section Tenues species resemble section Trachyspermi species, but the species in section Trachyspermi are likely to produce abundant red pigments (Yilmaz et al. 2014), while Talaromyces tenuis doesn’t. Talaromyces tenuis also produces thinner conidiophores, and it is interesting to find out whether this character is shared by other species that will be described in this section in the future.
Three of our new species fall into section Talaromyces. This section was first introduced for species that produce yellow ascomata, which can occasionally be white, creamish, pinkish or reddish, and have yellow ascospores (Stolk and Samson 1972). Nowadays, this section is not limited to sexual species, but it still contains the largest number and highest ratio of sexual reproducing species in this genus. Among three new species, T. brevis and T. rufus produce yellow, spiny ascospores, the ascospores of T. brevis are smaller compared to its close relatives T. liani, and T. rufus can be easily distinguished by its red synnemata. Talaromyces aspriconidius is characterized by its strikingly roughened, globose conidia, but ascospores were not observed after long incubation.
Talaromyces albisclerotius is classified in section Trachyspermi. Species in section Trachyspermi show restricted growth on CYA, YES and DG18, grow slightly faster on MEA, and do not, or poorly grow, on CREA. Conidiophores are generally biverticillate and some species produce creamish white or yellow ascomata (Yilmaz et al. 2014). The morphology of T. albisclerotius matches these characters well; however, T. albisclerotius produces white sclerotia and does not produce ascomata and ascospores. Yilmaz et al. (2016c) speculated that Talaromyces species with no known sexual stage may actually be heterothallic. They successfully induced the sexual reproductive structures in T. amestolkiae, which was formerly described as an asexual taxon with black sclerotia. Further study is needed on T. albisclerotius to complete this hypothesis.
Talaromyces guizhouensis and T. resedanus belong to section Subinflati. This section previously contained two morphologically distinct species T. palmae and T. subinflatus. Talaromyces palmae produces short, biverticillate conidiophores and indeterminate synnemata, and T. subinflatus produces longer conidiophores, and grows more restrictedly on all tested media (Yilmaz et al. 2014). Talaromyces tzapotlensis was described by Peterson and Jurjević (2017), it grows well on CREA and DG18. Talaromyces omanensis was considered as a synonym of T. resedanus based on molecular and morphological similarity. The growth rate of T. guizhouensis falls somewhere in between; it is phylogenetically close to T. subinflatus and T. tzapotlensis. Talaromyces resedanus is the only monoverticillate species in this section.
Talaromyces species have a worldwide distribution and are isolated from a wide range of substrates. Soil is their main habitat, but new species were also isolated from indoor air, dust, clinical samples, plants, seed, leaf litter, honey, pollen and stingless bee nests (Sang et al. 2013; Visagie et al. 2014; Chen et al. 2016; Wang QM et al. 2016; Yilmaz et al. 2016a; Guevara-Suarez et al. 2017; Peterson and Jurjević 2017; Barbosa et al. 2018; Crous et al. 2018; Su and Niu 2018; Rodríguez-Andrade et al. 2019). These studies expanded our knowledge on the substrates where Talaromyces species can occur, but on the other hand demonstrated the complicated ecological function of this genus. In this study, one new section and six new species were identified from soil in China. Further research will focus on the Talaromyces diversity from a wide range of substrates.
Supplementary Material
Acknowledgements
This project was supported by the Open Funding Project of the State Key Laboratory of Bioactive Substance and Function of Natural Medicines No. GTZK201903 and Beijing science and technology plan No. Z191100004019025. We acknowledge professor Lei Cai for providing the type culture of Talaromyces aspriconidius and T. albisclerotius.
Citation
Sun B-D, Chen AJ, Houbraken J, Frisvad JC, Wu W-P, Wei H-L, Zhou Y-G, Jiang X-Z, Samson RA (2020) New section and species in Talaromyces. MycoKeys 68: 75–113. https://doi.org/10.3897/mycokeys.68.52092
Funding Statement
Open Funding Project of the State Key Laboratory of Bioactive Substance and Function of Natural Medicines No. GTZK201903 and Beijing science and technology plan No. Z191100004019025
Contributor Information
Xian-Zhi Jiang, Email: jxz@moonbio.com.
Robert A. Samson, Email: r.samson@wi.knaw.nl.
Supplementary materials
Phylogeny of ITS for species classified in Talaromyces section Talaromyces
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of CaM for species classified in Talaromyces section Talaromyces
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of RPB2 for species classified in Talaromyces section Talaromyces
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of ITS for species classified in Talaromyces section Trachyspermi
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces purpurogenus was chosen as outgroup.
Phylogeny of CaM for species classified in Talaromyces section Trachyspermi
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of RPB2 for species classified in Talaromyces section Trachyspermi
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of ITS for species classified in Talaromyces section Subinflati
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of CaM for species classified in Talaromyces section Subinflati
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of RPB2 for species classified in Talaromyces section Subinflati
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
References
- Abdel-Rahim IR, Abo-Elyousr KAM. (2018) Talaromyces pinophilus strain AUN-1 as a novel mycoparasite of Botrytis cinerea, the pathogen of onion scape and umbel blights. Microbiological Research 212–213: 1–9. 10.1016/j.micres.2018.04.004 [DOI] [PubMed]
- Alagesaboopathi C. (1994) Biological control of damping-off disease of cotton seedling. Current Science 66: 865–868. [Google Scholar]
- Antonopoulou I, Iancu L, Jütten P, Piechot A, Rova U, Christakopoulos P. (2018) Screening of novel feruloyl esterases from Talaromyces wortmannii for the development of efficient and sustainable syntheses of feruloyl derivatives. Enzyme and Microbial Technology 120: 124–135. 10.1016/j.enzmictec.2018.08.007 [DOI] [PubMed] [Google Scholar]
- Barbosa RN, Bezerra JDP, Souza-Motta CM, Frisvad JC, Samson RA, Oliveira NT, Houbraken J. (2018) New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie van Leeuwenhoek 111: 1–30. 10.1007/s10482-018-1081-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin CR. (1955) Ascocarps of Aspergillus and Penicillium. Mycologia 47: 669–687. 10.1080/00275514.1955.12024485 [DOI] [Google Scholar]
- Berbee ML, Yoshimura A, Sugiyama J, Taylor JW. (1995) Is Penicillium monophyletic? An evaluation of phylogeny in the family Trichocomaceae from 18S, 5.8S and ITS ribosomal DNA sequence data. Mycologia 87: 210–222. 10.1080/00275514.1995.12026523 [DOI] [Google Scholar]
- Bladt TT, Frisvad JC, Knudsen PB, Larsen TO. (2013) Anticancer and antifungal compounds from Aspergillus, Penicillium and other filamentous fungi. Molecules 18: 11338–11376. 10.3390/molecules180911338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boosalis MG. (1956) Effect of soil temperature and green-manure amendment of unsterilized soil on parasitism of Rhizoctonia solani by Penicillium vermiculatum and Trichoderma sp. Phytopathology 46: 473–478. [Google Scholar]
- Bouhet JC, Van Chuong PP, Toma F, Kirszenbaum M, Fromageot P. (1976) Isolation and characterization of luteoskyrin and rugulosin, two hepatotoxic anthraquinonoids from Penicillium islandicum Sopp and Penicillium rugulosum Thom. Journal of Agricultural and Food Chemistry 24: 964–972. 10.1021/jf60207a028 [DOI] [PubMed] [Google Scholar]
- Chen AJ, Sun BD, Houbraken J, Frisvad JC, Yilmaz N, Zhou YG, Samson RA. (2016) New Talaromyces species from indoor environments in china. Studies in Mycology 84: 119–144. 10.1016/j.simyco.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Hardy GEStJ, Crane C, Barrett S, Cano-Lira JF, Le Roux JJ, Thangavel R, Guarro J, Stchigel AM, Martín MP, Alfredo DS, Barber PA, Barreto RW, Baseia IG, Cano-Canals J, Cheewangkoon R, Ferreira RJ, Gené J, Lechat C, Moreno G, Roets F, Shivas RG, Sousa JO, Tan YP, Wiederhold NP, Abell SE, Accioly T, Albizu JL, Alves JL, Antoniolli ZI, Aplin N, Araújo J, Arzanlou M, Bezerra JDP, Bouchara JP, Carlavilla JR, Castillo A, Castroagudín VL, Ceresini PC, Claridge GF, Coelho G, Coimbra VRM, Costa LA, da Cunha KC, da Silva SS, Daniel R, de Beer ZW, Dueñas M, Edwards J, Enwistle P, Fiuza PO, Fournier J, García D, Gibertoni TB, Giraud S, Guevara-Suarez M, Gusmão LFP, Haituk S, Heykoop M, Hirooka Y, Hofmann TA, Houbraken J, Hughes DP, Kautmanová I, Koppel O, Koukol O, Larsson E, Latha KPD, Lee DH, Lisboa DO, Lisboa WS, López-Villalba Á, Maciel JLN, Manimohan P, Manjón JL, Marincowitz S, Marney TS, Meijer M, Miller AN, Olariaga I, Paiva LM, Piepenbring M, Poveda-Molero JC, Raj KNA, Raja HA, Rougeron A, Salcedo I, Samadi R, Santos TAB, Scarlett K, Seifert KA, Shuttleworth LA, Silva GA, Silva M, Siqueira JPZ, Souza-Motta CM, Stephenson SL, Sutton DA, Tamakeaw N, Telleria MT, Valenzuela-Lopez N, Viljoen A, Visagie CM, Vizzini A, Wartchow F, Wingfield BD, Yurchenko E, Zamora JC, Groenewald JZ. (2016) Fungal Planet description sheets: 469–557. Persoonia 37: 218–403. 10.3767/003158516X694499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Carnegie AJ, Hardy GEStJ, Smith D, Summerell BA, Cano-Lira JF, Guarro J, Houbraken J, Lombard L, Martín MP, Sandoval-Denis M, Alexandrova AV, Barnes CW, Baseia IG, Bezerra JDP, Guarnaccia V, May TW, Hernández-Restrepo M, Stchigel AM, Miller AN, Ordoñez ME, Abreu VP, Accioly T, Agnello C, Agustin CA, Albuquerque CC, Alfredo DS, Alvarado P, Araújo-Magalhães GR, Arauzo S, Atkinson T, Barili A, Barreto RW, Bezerra JL, Cabral TS, Camello RF, Cruz RHSF, Daniëls PP, da Silva BDB, de Almeida DAC, de Carvalho Júnior AA, Decock CA, Delgat L, Denman S, Dimitrov RA, Edwards J, Fedosova AG, Ferreira RJ, Firmino AL, Flores JA, García D, Gené J, Góis JS, Gomes AAM, Gonçalves CM, Gouliamova DE, Groenewald M, Guéorguiev BV, Guevara-Suarez M, Gusmão LFP, Hosaka K, Hubka V, Huhndorf SM, Jadan M, Jurjević Ž, Kraak B, Kučera V, Kumar TKA, Kušan I, Lacerda SR, Lamlertthon S, Lisboa WS, Loizides M, Luangsa-ard JJ, Lysková P, Mac Cormack WP, Macedo DM, Machado AR, Malysheva EF, Marinho P, Matočec N, Meijer M, Mešić A, Mongkolsamrit S, Moreira KA, Morozova OV, Nair KU, Nakamura N, Noisripoom W, Olariaga I, Oliveira RJV, Paiva LM, Pawar P, Pereira OL, Peterson SW, Prieto M, Rodríguez-Andrade E, Rojo De Blas C, Roy M, Santos ES, Sharma R, Silva GA, Souza-Motta CM, Takeuchi-Kaneko Y, Tanaka C, Thakur A, Smith MTh, Tkalčec Z, Valenzuela-Lopez N, Van der Kleij P, Verbeken A, Viana MG, Wang XW, Groenewald JZ. (2017) Fungal Planet description sheets: 625–715. Persoonia 39: 270–467. 10.3767/persoonia.2017.39.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Hardy GEStJ, Gené J, Guarro J, Baseia IG, García D, Gusmão LFP, Souza-Motta CM, Thangavel R, Adamčík S, Barili A, Barnes CW, Bezerra JDP, Bordallo JJ, Cano-Lira JF, de Oliveira RJV, Ercole E, Hubka V, Iturrieta-González I, Kubátová A, Martín MP, Moreau PA, Morte A, Ordoñez ME, Rodríguez A, Stchigel AM, Vizzini A, Abdollahzadeh J, Abreu VP, Adamčíková K, Albuquerque GMR, Alexandrova AV, Álvarez DE, Armstrong-Cho C, Banniza S, Barbosa RN, Bellanger JM, Bezerra JL, Cabral TS, Caboň M, Caicedo E, Cantillo T, Carnegie AJ, Carmo LT, Castañeda-Ruiz RF, Clement CR, Čmoková A, Conceição LB, Cruz RHSF, Damm U, da Silva BDB, da Silva GA, da Silva RMF, de A. Santiago ALCM, de Oliveira LF, de Souza CAF, Déniel F, Dima B, Dong G, Edwards J, Félix CR, Fournier J, Gibertoni TB, Hosaka K, Iturriaga T, Jadan M, Jany JL, Jurjević Ž, Kolařík M, Kušan I, Landell MF, Leite CTR, Lima DX, Loizides M, Luo S, Machado AR, Madrid H, Magalhães OMC, Marinho P, Matočec N, Mešić A, Miller AN, Morozova OV, Neves RP, Nonaka K, Nováková A, Oberlies NH, Oliveira-Filho JRC, Oliveira TGL, Papp V, Pereira OL, Perrone G, Peterson SW, Pham THG, Raja HA, Raudabaugh DB, Řehulka J, Rodríguez-Andrade E, Saba M, Schauflerová A, Shivas RG, Simonini G, Siqueira JPZ, Sousa JO, Stajsic V, Svetasheva T, Tan YP, Tkalčec Z, Ullah S, Valente P, Valenzuela-Lopez N, Abrinbana M, Viana MDA, Wong PTW, Xavier de Lima V, Groenewald JZ. (2018) Fungal Planet description sheets: 716–784. Persoonia 40: 240–393. 10.3767/persoonia.2018.40.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoog GS, Guarro J, Gene J, Figueras MJ. (2014) Atlas of Clinical Fungi. CBS-KNAW Fungal Biodiversity Center, Utrecht, 1126 pp. [Google Scholar]
- de Vos JP, van Garderen E, Hensen H, Tange I, Curfs-Breuker I, Vandevelde B, Meis JF. (2009) Disseminated Penicillium radicum infection in a dog, clinically resembling multicentric malignant lymphoma. Vlaams Diergeneeskundig Tijdschrift 78: 183–188. [Google Scholar]
- Dijksterhuis J. (2007) Heat resistant ascospores. In: Dijksterhuis J, Samson RA. (Eds) Food Mycology.A Multifaceted Approach to Fungi and Food. CRC Press, Boca Raton, 101–117. 10.1201/9781420020984.ch6 [DOI]
- Dutta BK. (1981) Studies on some fungi isolated from the rhizosphere of tomato plants and the consequent prospect for control of Verticillium wilt. Plant Soil 63: 209–216. 10.1007/BF02374599 [DOI] [Google Scholar]
- Fahima T, Henis Y. (1995) Quantitative assessment of the interaction between the antagonistic fungus Talaromyces flavus and the wilt pathogen Verticillium dahliae on eggplant roots. Plant Soil 176: 129–137. 10.1007/BF00017683 [DOI] [Google Scholar]
- Frisvad JC, Yilmaz N, Thrane U, Rasmussen KB, Houbraken J, Samson RA. (2013) Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments. PLoS ONE 8: e84102. 10.1371/journal.pone.0084102 [DOI] [PMC free article] [PubMed]
- Guevara-Suarez M, García D, Cano-Lira JF, Guarro J, Gené J. (2020) Species diversity in Penicillium and Talaromyces from herbivore dung, and the proposal of two new genera of penicillium-like fungi in Aspergillaceae. Fungal Systematics and Evolution 5: 39–75. 10.3114/fuse.2020.05.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Suarez M, Sutton DA, Gené J, García D, Wiederhold N, Guarro J, Cano-Lira JF. (2017) Four new species of Talaromyces from clinical sources. Mycoses 60: 651–662. 10.1111/myc.12640 [DOI] [PubMed] [Google Scholar]
- Halo B, Maharachchikumbura SSN, Al-yahyai RA, Al-nabhani AA, Al-sadi AM. (2019) Talaromyces omanensis sp. nov.: phenotypic and molecular characterization of a novel species isolated from Rhazya stricta in Oman. Phytotaxa 404: 190–202. 10.11646/phytotaxa.404.5.2 [DOI] [Google Scholar]
- Horré R, Gilges S, Breig P, Kupfer B, de-Hoog GS, Hoekstra E, Poonwan N, Schaal KP. (2001) Case report. Fungaemia due to Penicillium piceum, a member of the Penicillium marneffei complex. Mycoses 44: 502–504. 10.1046/j.1439-0507.2001.00710.x [DOI] [PubMed] [Google Scholar]
- Houbraken J, Samson RA. (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51. 10.3114/sim.2011.70.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Kocsubé S, Visagie CM, Yilmaz N, Wang XC, Meijer M, Kraak B, Hubka V, Samson RA, Frisvad JC. (2020) Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Studies in Mycology (accepted). [DOI] [PMC free article] [PubMed]
- Inglis GD, Kawchuk LM. (2002) Comparative degradation of oomycete, ascomycete, and basidiomycete cell walls by mycoparasitic and biocontrol fungi. Canadian Journal of Microbiology 48: 60–70. 10.1139/w01-130 [DOI] [PubMed] [Google Scholar]
- Isbelia R, Louis B, Simard RR, Phillipe T, Hani A. (1999) Characteristics of phosphate solubilization by an isolate of a tropical Penicillium rugulosum and two UV induced mutants. FEMS Microbiology Ecology 28: 291–295. 10.1016/S0168-6496(98)00118-4 [DOI] [Google Scholar]
- Jiang XZ, Yu ZD, Ruan YM, Wang L. (2018) Three new species of Talaromyces sect. Talaromyces discovered from soil in China. Scientific Reports 8: 1–4932. 10.1038/s41598-018-23370-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalmuratova I, Kim H, Nam YJ, Oh Y, Jeong MJ, Choi HR, You YH, Choo YS, Lee IJ, Shin JH, Yoon H, Kim JG. (2015) Diversity and plant growth promoting capacity of endophytic fungi associated with halophytic plants from the west coast of Korea. Mycobiology 43: 373–383. 10.5941/MYCO.2015.43.4.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Shim CK, Kim YK, Hong SJ, Park JH, Han EJ, Kim SC. (2017) Enhancement of seed dehiscence by seed treatment with Talaromyces flavus GG01 and GG04 in Ginseng (Panax ginseng). The Plant Pathology Journal 33: 1–8. 10.5423/PPJ.OA.06.2016.0146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. (2019) RA×ML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 1–3. 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M, Taritla S, Sharma A, Jayabaskaran C. (2018) Antiproliferative and antioxidative bioactive compounds in extracts of marine-derived endophytic fungus Talaromyces purpureogenus Frontiers in Microbiol 9: e1777. 10.3389/fmicb.2018.01777 [DOI] [PMC free article] [PubMed]
- Lian W, Wang W, Tan CP, Wang J, Wang Y. (2018) Immobilized Talaromyces thermophilus lipase as an efficient catalyst for the production of LML-type structured lipids. Bioprocess and Biosystems Engineering 42: 321–329. 10.1007/s00449-018-2036-7 [DOI] [PubMed] [Google Scholar]
- Limper AH, Adenis A, Le T, Harrison TS. (2017) Fungal infections in HIV/AIDS. Lancet Infectious Diseases 17: e334–e343. 10.1016/S1473-3099(17)30303-1 [DOI] [PubMed]
- LoBuglio KF, Pitt JI, Taylor JW. (1993) Phylogenetic analysis of two ribosomal DNA regions indicates multiple independent losses of a sexual Talaromyces state among asexual Penicillium species in subgenus Biverticillium. Mycologia 85: 592–604. 10.2307/3760506 [DOI] [Google Scholar]
- Maddison WP, Maddison DR. (2018) Mesquite: a modular system for evolutionary analysis. Version 3.6 http://www.mesquiteproject.org
- Maeda RN, Barcelos CA, Anna LMMS, Pereira N. (2013) Cellulase production by Penicillium funiculosum and its application in the hydrolysis of sugar cane bagasse for second generation ethanol production by fed batch operation. Journal of Biotechnology 163: 38–44. 10.1016/j.jbiotec.2012.10.014 [DOI] [PubMed] [Google Scholar]
- Marois JJ, Fravel DR, Papavizas GC. (1984) Ability of Talaromyces flavus to occupy the rhizosphere. Soil Biology and Biochemistry 16: 387–390. 10.1016/0038-0717(84)90038-5 [DOI] [Google Scholar]
- McLaren DL, Huang HC, Rimmer SR. (1986) Hyperparasitism of Sclerotinia sclerotiorum by Talaromyces flavus. Canadian Journal of Plant Pathology 8: 43–48. 10.1139/b89-279 [DOI] [Google Scholar]
- Mclennan EI, Tucker SC, Thrower LB. (1954) New soil fungi from Australian heathland: Aspergillus, Penicillium, Spegazzinia. Australian Journal of Botany 2: 355–364. 10.1071/BT9540355 [DOI] [Google Scholar]
- Narikawa T, Shinoyama H, Fujii T. (2000) A β-rutinosidase from Penicillum rugulosum IFO 7242 that is a peculiar flavonoid glycosidase. Bioscience, Biotechnology and Biochemistry 64: 1317–1319. 10.1271/bbb.64.1317 [DOI] [PubMed] [Google Scholar]
- Nicoletti R, Salvatore MM, Andolfi A. (2018) Secondary metabolites of mangrove-associated strains of Talaromyces. Marine Drugs 16: 1–12. 10.3390/md16010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Sugiyama J. (2000) Evolutionary relationships of the cleistothecial genera with Penicillium, Geosmithia, Merimbla and Sarophorum anamorphs as inferred from 18S rDNA sequence divergence. In: Samson RA, Pitt JI. (Eds) Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification.Plenum Press, New York, 149–161.
- Ogawa H, Yoshimura A, Sugiyama J. (1997) Polyphyletic origins of species of the anamorphic genus Geosmithia and the relationships of the cleistothecial genera: Evidence from 18S, 5S and 28S rDNA sequence analyses. Mycologia 89: 756–771. 10.1080/00275514.1997.12026842 [DOI] [Google Scholar]
- Oh JY, Kim EN, Ryoo MI, Kim KD. (2008) Morphological and molecular identification of Penicillium islandicum isolate KU101 from stored rice. Plant Pathology Journal 24: 469–473. 10.5423/PPJ.2008.24.4.469 [DOI] [Google Scholar]
- Peterson SW, Jurjević Ž. (2017) New species of Talaromyces isolated from maize, indoor air, and other substrates. Mycologia 109: 537–556. 10.1080/00275514.2017.1369339 [DOI] [PubMed] [Google Scholar]
- Pitt JI, Hocking AD. (2009) Fungi and Food Spoilage. Springer, New York, 519 pp 10.1007/978-0-387-92207-2 [DOI] [Google Scholar]
- Pol D, Laxman RS, Rao M. (2012) Purification and biochemical characterization of endoglucanase from Penicillium pinophilum MS 20. Indian Journal of Biochemistry and Biophysics 49: 189–194. [PubMed] [Google Scholar]
- Posada D, Crandall KA. (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rajeshkumar KC, Yilmaz N, Marathe SD, Seifert KA. (2019) Morphology and multigene phylogeny of Talaromyces amyrossmaniae, a new synnematous species belonging to the section Trachyspermi from India. MycoKeys 45: 41–56. 10.3897/mycokeys.45.32549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner RW. (1970) A Mycological Colour Chart. Commonwealth Mycological Institute, Kew, 34 pp. [Google Scholar]
- Rodríguez-Andrade E, Stchigel AM, Terrab A, Guarro J, Cano-Lira JF. (2019) Diversity of xerotolerant and xerophilic fungi in honey. IMA Fungus 10: 1–20. 10.1186/s43008-019-0021-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Enomoto M, Tatsuno T. (1971) Yellowed rice toxins. Luteoskyrin and related compounds, chlorine-containing compounds, and citrinin. In: Ciegler A, Kadis S, Ajl SJ. (Eds) Microbial Toxins.Academic Press Inc., New York, 299–308.
- Sakai A, Tanaka H, Konishi Y, Hanazawa R, Ota T, Nakahara Y, Sekiguchi S, Oshida E, Takino M, Ichinoe M, Yoshikawa K, Yoshizawa T, Takatori K. (2005) Mycological examination of domestic unpolished rice and mycotoxin production by isolated Penicillium islandicum. Journal of the Food Hygienic Society of Japan 46: 205–212. 10.3358/shokueishi.46.205 [DOI] [PubMed] [Google Scholar]
- Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. (2010) Food and Indoor Fungi. CBS Laboratory manual. CBS-KNAW Fungal Biodiversity Center, Utrecht, 390 pp. [Google Scholar]
- Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert SW, Peterson SW, Varga J, Frisvad JC. (2011) Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Studies in Mycology 70: 159–183. 10.3114/sim.2011.70.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang H, An TJ, Kim CS, Shin GS, Sung GH, Yu SH. (2013) Two novel Talaromyces species isolated from medicinal crops in Korea. The Journal of Microbiology 51: 704–708. 10.1007/s12275-013-3361-9 [DOI] [PubMed] [Google Scholar]
- Santos PE, Piontelli E, Shea YR, Galluzzo ML, Holland SM, Zelazko ME, Rosenzweig SD. (2006) Penicillium piceum infection: diagnosis and successful treatment in chronic granulomatous disease. Medical Mycology 44: 749–753. 10.1080/13693780600967089 [DOI] [PubMed] [Google Scholar]
- Stark AA, Townsend JM, Wogan GN, Demain AL, Manmade A, Ghosh AC. (1978) Mutagenicity and antibacterial activity of mycotoxins produced by Penicillium islandicum Sopp and Penicillium rugulosum. Journal of Environmental Pathology and Toxicology 2: 313–324. [PubMed] [Google Scholar]
- Stolk AC, Samson RA. (1972) The genus Talaromyces – studies on Talaromyces and related genera II. Studies in Mycology 2: 1–65. 10.2307/3758303 [DOI] [Google Scholar]
- Su L, Niu YC. (2018) Multilocus phylogenetic analysis of Talaromyces species isolated from cucurbit plants in China and description of two new species, T. cucurbitiradicus and T. endophyticus. Mycologia 110: 375–386. 10.1080/00275514.2018.1432221 [DOI] [PubMed] [Google Scholar]
- Supparatpinyo K, Khamwan C, Baosoung V, Nelson KE, Sirisanthana T. (1994) Disseminated Penicillium marneffei infection in southeast Asia. Lancet 344: 110–113. 10.1016/S0140-6736(94)91287-4 [DOI] [PubMed] [Google Scholar]
- Tomlinson JK, Cooley AJ, Zhang S, Johnson ME. (2011) Granulomatous lymphadenitis caused by Talaromyces helicus in a labrador retriever. Veterinary Clinical Pathology 40: 553–557. 10.1111/j.1939-165X.2011.00377.x [DOI] [PubMed] [Google Scholar]
- Ueno Y, Ishikawa I. (1969) Production of luteoskyrin, a hepatotoxic pigment, by Penicillium islandicum Sopp. Applied Microbiology 18: 406–409. 10.1128/AEM.18.3.406-409.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraguchi K. (1962) Malignant hepatoma and so-called carcinogens, with special reference to the toxicity of luteoskyrin in a small dose. Folia Pharmacologica Japonica 58: 1–19.14448886 [Google Scholar]
- Uraguchi K, Uraguchi K, Tatsuno T, Sakaki F, Tsukioka M, Ito H. (1961) Isolation of two toxic agents, luteoskyrin and chlorione-containing peptide, from metabolites of Penicillium islandicum Sopp, with properties thereof. Journal of Experimental Medicine 31: 193–207. [PubMed] [Google Scholar]
- Uraguchi K, Saito M, Noguchi Y, Takahashi K, Enomoto M, Tatsuno T. (1972) Chronic toxicity and carcinogenicity in mice of the purified mycotoxins, luteoskyrin and cyclochlorotine. Food and Cosmetics Toxicology 10: 193–207. 10.1016/S0015-6264(72)80197-4 [DOI] [PubMed] [Google Scholar]
- Varriale S, Houbraken J, Granchi Z, Pepe O, Cerullo G, Ventorino V, Chin-A-Woeng T, Meijer M, Riley R, Grigoriev IV, Henrissat B, de Vries RP, Faraco V. (2018) Talaromyces borbonicus, sp. nov., a novel fungus from biodegraded Arundo donax with potential abilities in lignocellulose conversion. Mycologia 110: 316–324. 10.1080/00275514.2018.1456835 [DOI] [PubMed] [Google Scholar]
- Visagie CM, Yilmaz N, Frisvad JC, Houbraken J, Seifert KA, Samson RA, Jacobs K. (2015) Five new Talaromyces species with ampulliform-like phialides and globose rough walled conidia resembling T. verruculosus. Mycoscience 56: 486–502. 10.1016/j.myc.2015.02.005 [DOI] [Google Scholar]
- Wang L, Zhuang WY. (2007) Phylogenetic analyses of penicillia based on partial calmodulin gene sequences. BioSystems 88: 113–112. 10.1016/j.biosystems.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Wang QM, Zhang YH, Wang B, Wang L. (2016) Talaromyces neofusisporus and T. qii, two new species of section Talaromyces isolated from plant leaves in Tibet, China. Scientific Reports 6: 1–18622. 10.1038/srep18622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XC, Chen K, Qin WT, Zhuang WY. (2017) Talaromyces heiheensis and T. mangshanicus, two new species from China. Mycological Progress 16: 73–81. 10.1007/s11557-016-1251-3 [DOI] [Google Scholar]
- Wang XC, Chen K, Xia YW, Wang L, Li TH, Zhuang WY. (2016) A new species of Talaromyces (Trichocomaceae) from the Xisha Islands, Hainan, China. Phytotaxa 267: 187–200. 10.11646/phytotaxa.267.3.2 [DOI] [Google Scholar]
- Weisenborn JLF, Kirschner R, Cáceres O, Piepenbring M. (2010) Talaromyces indigoticus Takada & Udagawa, the first record from Panama and the American continent. Mycopathologia 170: 203–208. 10.1007/s11046-010-9305-6 [DOI] [PubMed] [Google Scholar]
- Xu Y, Feng X, Jia J, Chen X, Jiang T, Rasool A, Lv B, Qu L, Li C. (2018) A novel β-glucuronidase from Talaromyces pinophilus Li-93 precisely hydrolyzes glycyrrhizin into glycyrrhetinic acid 3-O-mono-β-d-glucuronide. Applied and Environmental Microbiology 84: e00755–18. 10.1128/AEM.00755-18 [DOI] [PMC free article] [PubMed]
- Yilmaz N, Visagie CM, Houbraken J, Frisvad JC, Samson RA. (2014) Polyphasic taxonomy of the genus Talaromyces. Studies in Mycology 78: 175–341. 10.1016/j.simyco.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz N, López-Quintero CA, Vasco-Palacios AM, Frisvad JC, Theelen B, Boekhout T, Samson RA, Houbraken J. (2016a) Four novel Talaromyces species isolated from leaf litter from Colombian Amazon rain forests. Mycological Progress 15: 1041–1056. 10.1007/s11557-016-1227-3 [DOI] [Google Scholar]
- Yilmaz N, Visagie CM, Frisvad JC, Houbraken J, Jacobs K, Samson RA. (2016b) Taxonomic re-evaluation of species in Talaromyces section Islandici, using a polyphasic approach. Persoonia 36: 37–56. 10.3767/003158516X688270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz N, Hagen F, Meis JF, Houbraken J, Samson RA. (2016c) Discovery of a sexual cycle in Talaromyces amestolkiae. Mycologia 108: 70–79. 10.3852/15-014 [DOI] [PubMed] [Google Scholar]
- Zhai MM, Li J, Jiang CX, Shi XP, Di DL, Crews P, Wu QX. (2016) The bioactive secondary metabolites from Talaromyces species. Natural Products and Bioprospecting. 6: 1–24. 10.1007/s13659-015-0081-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogeny of ITS for species classified in Talaromyces section Talaromyces
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of CaM for species classified in Talaromyces section Talaromyces
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of RPB2 for species classified in Talaromyces section Talaromyces
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of ITS for species classified in Talaromyces section Trachyspermi
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces purpurogenus was chosen as outgroup.
Phylogeny of CaM for species classified in Talaromyces section Trachyspermi
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of RPB2 for species classified in Talaromyces section Trachyspermi
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of ITS for species classified in Talaromyces section Subinflati
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of CaM for species classified in Talaromyces section Subinflati
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.
Phylogeny of RPB2 for species classified in Talaromyces section Subinflati
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Bing-Da Sun, Amanda J. Chen, Jos Houbraken, Jens C. Frisvad, Wen-Ping Wu, Hai-Lei Wei, Yu-Guang Zhou, Xian-Zhi Jiang, Robert A. Samson
Data type
phylogenetic
Explanation note
Branches with values more than 1 pp and 95% bs are thickened. Talaromyces dendriticus was chosen as outgroup.