Abstract
Hispanic/Latino patients have a higher incidence of gastric cancer and worse cancer-related outcomes compared to patients of other backgrounds. Whether there is a molecular basis for these disparities is unknown, as very few Hispanic/Latino patients have been included in previous studies. To determine the genomic landscape of gastric cancer in Hispanic/Latino patients, we performed whole-exome sequencing (WES) and RNA sequencing on tumor samples from 57 patients; germline analysis was conducted on 83 patients. The results were compared to data from Asian and White patients published by The Cancer Genome Atlas. Hispanic/Latino patients had a significantly larger proportion of genomically-stable subtype tumors compared to Asian and White patients (65% vs 21% vs 20%, P < 0.001). Transcriptomic analysis identified molecular signatures that were prognostic. Of the 43 Hispanic/Latino patients with diffuse-type cancer, 7 (16%) had germline mutations in CDH1. Mutation carriers were significantly younger than non-carriers (41 vs 50 years, P < 0.05). In silico algorithms predicted 5 variants to be deleterious. For two variants that were predicted to be benign, in vitro modeling demonstrated that these mutations conferred increased migratory capability, suggesting pathogenicity. Hispanic/Latino gastric cancer patients possess unique genomic landscapes, including a high rate of CDH1 germline mutations that may partially explain their aggressive clinical phenotypes. Individualized screening, genetic counseling, and treatment protocols based on patient ethnicity and race may be necessary.
Keywords: Gastric cancer, Hispanic, Latino, cancer disparity, cancer genomics, CDH1, E-cadherin
Introduction
Gastric cancer is the second-deadliest cancer worldwide, causing an estimated 834,000 deaths in 2016(1). Hispanic/Latino patients have different clinicopathologic features than patients of other ethnicities and races. In the United States, Hispanics/Latinos have twice the incidence and mortality from gastric cancer compared to non-Hispanic Whites (2). Hispanic/Latino gastric cancer patients also tend to be diagnosed at a younger age, with more advanced-stage disease, and with a higher proportion of diffuse-type cancers (DGC) (3–5). While environmental exposures and socioeconomic factors likely contribute to the observed clinicopathologic differences, ethnicity/race-associated differences in tumor biology may also be involved. For example, African-American breast cancer patients have higher rates of triple-negative cancers and a higher prevalence of TP53 mutations, as compared with White patients (6,7).
Whether there is a molecular basis for observed outcome differences for gastric cancer patients of different ethnicities/races has been heretofore unanswerable as previous large gastric cancer genomic studies had included very few Hispanic/Latino patients. The TCGA has performed the largest published sequencing study of gastric adenocarcinoma and included only five Hispanic/Latino patients in its 478-patient cohort (8). Other major sequencing efforts of gastric cancer originated in East Asia, including those by Ichikawa et al (207 patients) and Cristescu et al (300 patients); these studies also did not include any Hispanic/Latino patients (9,10). Given the known association between ethnicity/race and tumor biology, the underrepresentation of Hispanic/Latino patients in previously published studies have likely biased our current genomic understanding of gastric cancer (11).
To address this knowledge gap, we performed a large, integrated genomic analysis of samples from 83 Hispanic/Latino gastric cancer patients. Comparative analyses were performed using data from Asian and White patients previously published by The Cancer Genome Atlas (TCGA) (12).
Materials and Methods
Sample acquisition and processing
This study was approved by the University of Texas Southwestern Medical Center Institutional Review Board. All gastric adenocarcinoma patients who were self-reported as being of Hispanic/Latino ancestry were recruited to join the study. All enrolled patients provided written informed consent.
Blood samples were drawn and stored at -80ºC prior to nucleic acid extraction. Tumor and adjacent non-neoplastic gastric tissue were obtained from subjects via endoscopic biopsies or gastric resections. The samples were stabilized immediately in RNAlater (Ambion) for at least 24 hours at 4ºC, then stored in liquid nitrogen until nucleic acid extraction. A second set of adjacent tissue samples from both the tumor and non-neoplastic stomach were also obtained for pathologic examination to confirm the histology, and to provide a microscopic assessment of tumor cellularity and extent of tumor necrosis. These samples were evaluated by a board-certified pathologist with expertise in gastrointestinal malignancies (S.T.G.H.). No samples were excluded on the basis of tumor cellularity. Samples with greater than 10% necrosis were excluded. For some samples, RNA was isolated with mirVana miRNA Isolation Kits (Ambion) and DNA was isolated with QuickGene DNA Tissue Kits (Kurabo). Other samples were processed using the AllPrep DNA/RNA kits (Qiagen). Nucleic acid quality control was ensured with NanoDrop (Thermo Fisher) spectrophotometric quantitation and visualization on an agarose gel.
CDH1 promoter methylation
Tumor and non-tumor DNA were prepared with the EpiTect II DNA Methylation Enzyme Kit (Qiagen). Quantification of methylated DNA was then performed using quantitative-PCR based assay with the EpiTect Methyl II PCR Primer Assay for Human CDH1, CpG Island 105415 (Qiagen) with the RT2 SYBR Green qPCR Mastermix (Qiagen).
Generation of CDH1 mutants
Chinese hamster ovary (CHO) cells (ATCC, CCL-61) were maintained in F-12K medium (Gibco) with 10% fetal bovine serum supplementation. Mycoplasma testing was performed upon receipt of the cells. hE-cadherin-pcDNA3 was a gift from Barry Gumbiner (Addgene plasmid # 45769; http://n2t.net/addgene:45769; RRID:Addgene_45769). Variants were generated using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs). Plasmid transfection was performed with Lipofectamine 3000 (Invitrogen). Selection was performed with G-418 (Sigma). Sanger sequencing was performed to confirm sequences using the following primers (Genewiz):
| Patient ID | PCR Primer F | PCR Primer R | Sanger Seq Primer |
|---|---|---|---|
| P15 | TGTGCCCAGTCGAGAAGTTA | CAGCGTGACTTTGGTGGAAA | TCAGAGCACAAGGAAGTCATC |
| P16 | CCTCTCCCAAGCCTTAGACC | TCAAAGGCTGAGTCACTTGC | ACCTAAATAAAACCCAAGCAGCT |
| P20 | TGTAAAACGGCCAGAGACCT | CATGGCAGTTGGAGCAAAGT | CTGGGAGTGGAGGTCCTTTG |
| P30 | CCCACCATCCCAGTTCTGAT | GCTGTGTGACCTTAGCCAAG | TGTTTCTTCGGAGGAGAGCG |
| P33 | CTGTTGGTTTCGGTGAGCAG | GCCCTCAACCTCCTCTTCTT | TCACCCGGTTCCATCTACCT |
| P50 | GACCAGAGCAAGTTTCACCC | CCTTCCATGACAGACCCCTT | TTTCAGGCCCGCATCTTCAT |
| P71 | AGTCTGGGTGCATTGTCGTA | CTCAAGGGAAGGGAGCTGAA | CTGGGTGCATTGTCGTACCT |
Immunofluorescence
CHO cells were fixed on glass slides with pre-cooled methanol for 15 min at −20ºC and blocked by 1% bovine serum albumin in PBS-T for 1 hour at room temperature. The slides were then incubated with anti-E-cadherin antibody (Abcam, ab76055; 1:1000 dilution) at 4ºC overnight, followed by secondary antibody at room temperature for 1 hour. DAPI was used as a nuclear stain (Vector Laboratories). Images were captured on a Zeiss confocal microscope.
Immunohistochemistry
Antigen retrieval was performed with sodium citrate buffer, followed by incubation with anti-E-cadherin antibody (Abcam, ab76055; 1:1000) at 4ºC overnight. Detection was performed with the ABC kit (Vector Laboratories) and DAB kit (Vector Laboratories) or MOM kit (Vector Laboratories).
Scratch assay
CHO cells transfected with a given plasmid were grown to confluence. A scratch was made and three images were taken of each well. 24 hours later, three more images of each well were taken. The distance between the wound edges was measured using cellSens Dimension software (Olympus). The average of the three images from each time point was used as one biological replicate. Two independent experiments with at least four biological replicates for each genotype were performed.
Statistical analysis
The Mann-Whitney U test was used to compare continuous variables. Categorical variables were presented as counts and proportions and compared with Fisher exact tests. Survival was estimated using the Kaplan-Meier method and compared via the log-rank test. For the epidemiologic studies, data was presented as medians with interquartile ranges and full ranges in box and whisker plots and compared with the Kruskal-Wallis test.
Whole-exome sequencing, RNA sequencing, and bioinformatic analyses
Please see the Supplementary Methods section for details regarding the whole-exome sequencing (WES), RNA sequencing (RNA-seq), and bioinformatic analyses. The data have been deposited with links to BioProject accession number PRJNA611545 in the NCBI BioProject database.
Patient and public involvement statement
Neither patients nor the public were involved in the design, conduct, reporting, or dissemination of our research.
Results
We performed WES and RNA-seq on tissue samples from 57 patients, 55 of whom had not received any treatment. Blood samples were also obtained from 52 of these patients and used as normal controls. For the five patients for whom blood samples were unavailable, we used non-neoplastic gastric tissue as controls. We also performed WES on blood samples from an additional 26 patients (Table 1 and Supplementary Table 1). The mean coverage for WES was 267x for the 57 tumor samples, 209x for the five non-neoplastic gastric samples, and 67x for the 78 blood samples. For RNA-seq, the average number of reads was 96.9 million, with an average mapping rate of 97.6% for the 57 tumors and 5 non-neoplastic gastric samples.
Table 1:
Clinicopathologic characteristics of Hispanic/Latino gastric cancer patients in this study. IQR: interquartile range.
| N (%) | ||
|---|---|---|
| Number of patients | 83 | |
| Sample Sequenced | ||
| Tissue | 57 (69%) | |
| Blood only | 26 (31%) | |
| Age (median, IQR, range) | 53, 45–61, 23–85 | |
| Gender | ||
| Male | 54 (65%) | |
| Female | 29 (35%) | |
| Tumor Location | ||
| Cardia | 24 (29%) | |
| Body | 30 (36%) | |
| Antrum | 24 (29%) | |
| Overlapping | 5 (6%) | |
| Clinical Stage | ||
| Stage I (T1–2N0M0) | 1 (1%) | |
| Stage II and III (T3–4N0M0, TanyN1–3M0) | 46 (55.5%) | |
| Stage IV (TanyNanyM1) | 34 (41%) | |
| Recurrent | 2 (2.5%) | |
| Differentiation | ||
| Well | 1 (1%) | |
| Moderate | 18 (22%) | |
| Moderate/Poor | 1 (1%) | |
| Poor | 57 (69%) | |
| Unknown | 6 (7%) | |
| Lauren Classification | ||
| Diffuse | 43 (52%) | |
| Mixed | 7 (8.5%) | |
| Intestinal | 26 (31%) | |
| Unknown | 7 (8.5%) | |
|
Helicobacter pylori Infection (by histology) | ||
| Yes | 15 (18%) | |
| No | 59 (71%) | |
| Unknown | 9 (11%) | |
Consistent with previous reports, the median age at time of diagnosis for the 83-patient Hispanic/Latino patient cohort was younger than that for the 77 Asian and 172 White patients analyzed by the TCGA (53 years, vs 66 and 66, respectively, P < 0.0001, Supplementary Fig. 1a). To confirm that the Hispanic/Latino cohort’s self-reported ancestry was unique from that of the TCGA Asian and White patients, we compared WES data from each of the three groups to reference data available through the Human Genome Diversity Project (HGDP) (13). Using principal component analysis, we found that the Hispanic/Latino cohort clustered independently from the Asian and White patients in the TCGA groups and were related most closely to the HGDP samples from Central and South America (Fig. 1 and Supplementary Fig. 1b).
Figure 1. Hispanic/Latino (Hs/L) gastric cancer patients are of unique ancestry as compared to Asian and White patients.
Principal component (PC) analysis was performed using Locating Ancestry from SEquence Reads (LASER) comparing Hispanic/Latino patients from this study to Asian and White patients analyzed by The Cancer Genome Atlas (TCGA). The Human Genome Diversity Project (HGDP) was used as the reference.
Gastric Cancers in Hispanic/Latino Patients are Enriched for the Genomically-Stable Subtype
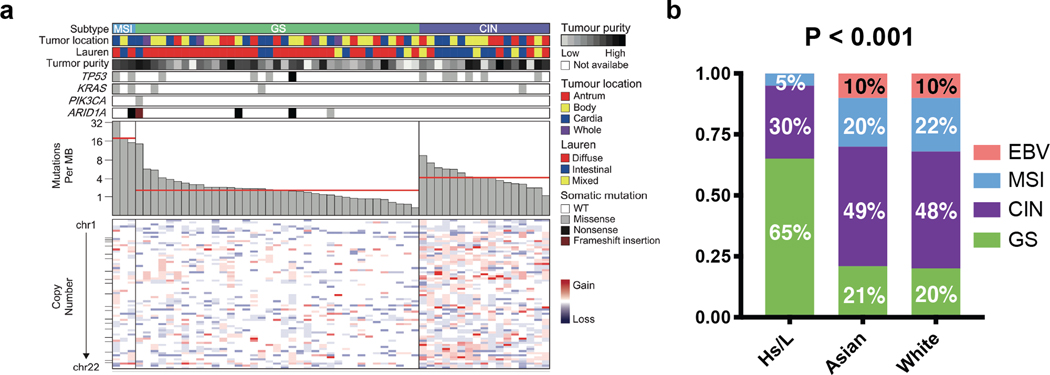
We next classified the 57 Hispanic/Latino gastric cancer samples into one of the four molecular subtypes established by the TCGA (Supplementary Fig. 2a) (12). We did not include African-Americans in this analysis, as there were only four African-American patients in the TCGA cohort. Tumors were first characterized based on Epstein-Barr virus (EBV) infection status, which was determined bioinformatically with PathoScope 2.0 (14). We found no EBV infections, whereas 10% of the TCGA cohort was EBV-positive (12). Next, microsatellite instability (MSI) was assessed bioinformatically using MSISensor, which has previously demonstrated near-perfect concordance with the results of PCR or immunohistochemical analysis (15,16). Three of the 57 samples (5%) had MSIsensor scores of greater than 10, indicating microsatellite instability (Supplementary Fig. 2b). Accordingly, these three samples showed mutation burdens greater than 13 mutations per megabase (Mb), whereas the average mutation burden for the 54 non-MSI samples was 2.5 mutations per Mb.
The remaining samples underwent somatic copy number alteration (SCNA) analysis (10). 17 samples (30%) had high SCNA scores, which placed them into the CIN group, and 37 patients (65%) had low scores and were categorized as genomically stable (GS; Fig. 2a). When compared to the Asian (20%) and White (21%) patients, Hispanic/Latinos had a significantly higher proportion of GS tumors (65%, P < 0.001; Fig. 2b). There were no significant differences between Asian and White patients in the proportions of subtypes. CIN samples showed an average of 3.5 mutations per Mb, while GS tumors had 2.0 mutations per Mb. This is consistent with the TCGA data found on the Broad Firehose, which showed CIN and GS samples as having 3.3 and 1.8 mutations per Mb, respectively (http://firebrowse.org/?cohort=STAD).
Figure 2. Gastric cancer in Hispanic/Latino (Hs/L) patients are predominantly of the genomically stable subtype.
a. Tumors from 57 Hispanic/Latino gastric cancer patients were subtyped and listed by descending mutation burden. Clinical and molecular data are depicted. MB = megabase, WT = wild-type.
b. Molecular classification of samples within each ethnicity/race. P < 0.001.
EBV: Epstein-Barr virus infected, MSI: microsatellite instability, CIN: chromosomal instability, GS: genomically stable.
In the TCGA analysis, the GS subtype was found to be enriched for tumors with diffuse-type histology (12). Accordingly, we found that of the 37 GS patients, 78% had diffuse-type, 16% had intestinal-type, and 6% had mixed-type tumors. In contrast, the CIN cohort was comprised of 23.5% diffuse, 53% intestinal, and 23.5% mixed-type tumors (P < 0.001, Fig. 2a).
Hispanic/Latino Gastric Cancers Recapitulate Key Genomic Features Identified by the TCGA
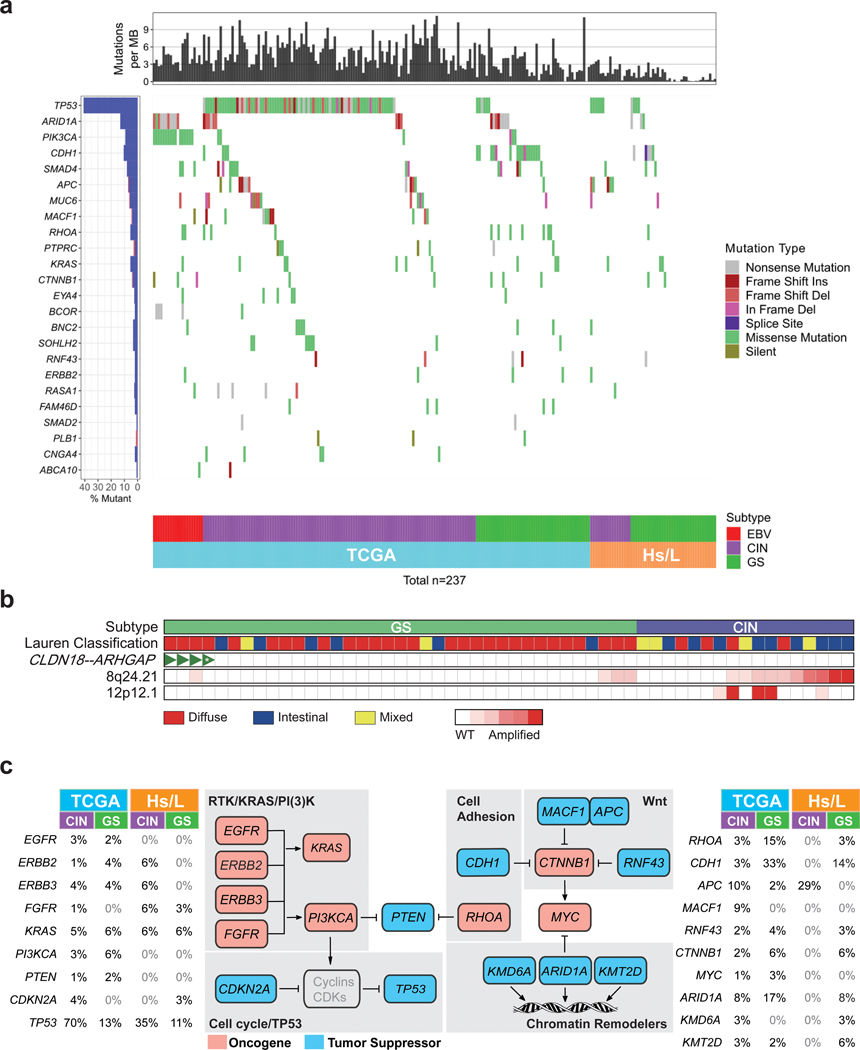
Although the Hispanic/Latino samples were significantly enriched for GS tumors, many defining genomic alterations previously identified by the TCGA were recapitulated in the current cohort. For example, the most common recurrent mutation in Hispanic/Latino gastric cancer samples was TP53, as was the case in the TCGA (Fig 3a.). We also found similar structural variations. The TCGA identified CLDN18-ARHGAP fusions in 15% of their GS-type tumors. These rearrangements lead to dysregulated RHOA signaling and loss of an epithelial phenotype (12,17). Using FusionCatcher and STAR-fusion to evaluate our RNA-seq data, we found that four tumors had this rearrangement (Fig. 3b and Supplementary Table 2) (18,19). All four were GS and diffuse-type. We also observed that 76% of CIN tumors, which were enriched for intestinal-type histology, had amplifications in the 8q24.21 region. This was significantly higher than seen in GS samples, in which only 19% had this copy number abnormality (Fig. 3b; P < 0.001). We confirmed this finding in the TCGA cohort, which similarly showed an enrichment of this structural alteration in CIN samples (12). The 8q24.21 region most notably carries the MYC oncogene, and other groups have noted that amplifications in this region are common in intestinal-type gastric cancers and are associated with worse outcomes in gastric cancer patients (20–22). Finally, we identified five instances of KRAS amplification (12p12.1), all of which occurred in CIN patients, consistent with previous reports (Fig. 3b) (23).
Figure 3. Key genomic features of gastric cancer are identified in Hispanic/Latino (Hs/L) samples.
a. Recurrent somatic mutations identified by the TCGA in non-hypermutated gastric cancer samples from Hispanic/Latino patients.
b. Structural variations seen in Hispanic/Latino gastric cancer samples. CIN: chromosomal instability, GS: genomically stable, WT: wild-type, *: CLDN18-ARHGAP45, all other fusions were CLDN18-ARHGAP26.
c. Comparison of incidence of somatic alterations in select genes involved in RTK/RAS/PI(3)K signaling, cell cycle, cell adhesion, Wnt signaling, and chromatin remodeling, in the TCGA and Hs/L cohorts, stratified by CIN and GS subtypes.
One major difference between the cohorts is the incidence of PIK3CA mutations, which were mainly found in EBV-type tumors that were lacking in the Hispanic/Latino patients. We also found differences in the mutation rates of some key signaling pathway members stratified by molecular subtype (Fig. 3c). We observed that Hispanic/Latino CIN tumors had a lower rate of TP53 mutations (35% vs 70%) but a higher incidence of APC mutations (29% vs 10%). In the Hispanic/Latino patients we also found a lower rate of alterations in RHOA (3% vs 18%, sum of both CIN and GS) and ARID1A (8% vs 25%, sum of both CIN and GS).
Gene Expression Profiling is Prognostic
To further interrogate the RNA-seq dataset, we selected the top 50 most variably expressed genes and performed unbiased consensus clustering to identify patient subgroups. We found five clusters with distinct clinicopathologic profiles (Fig. 4a, Supplementary Table 3). Patients in Clusters 2 and 3 tended to be younger, while Clusters 1 and 5 patients were older. Cluster 3 patients had tumors enriched for diffuse-type and GS tumors. When we compared the survival of each cluster, we found the grouping provided significant prognostic capability (Supplementary Fig. 3a, P < 0.01). Cluster 1 patients had the shortest median survival at 7.7 months, whereas Cluster 4 patients had the longest survival, with median survival not reached. Patients in Clusters 2, 3, and 5 had similar survival that were intermediate to Clusters 1 and 4. When we grouped Clusters 2, 3, and 5 as an intermediate-risk category, its median survival was 19.7 months (Figure 4b, P < 0.001). Importantly, the prognostic value of mRNA clustering was maintained when patients were stratified by molecular subtype or by Lauren classification (Supplementary Fig 3b-e, P < 0.05 for each).
Figure 4. Transcriptomic signatures of gastric cancer from Hispanic/Latino patients are prognostic.
a. Unsupervised consensus clustering based on the top 50 most variably expressed genes. MSI: microsatellite instability, CIN: chromosomal instability, GS: genomically stable.
b. Kaplan-Meier curves comparing overall survival based on clusters. P < 0.001.
c. Normalized enrichment scores from Gene Set Enrichment Analysis (GSEA) comparing Cluster 1 to Clusters 2, 3, 4, and 5. Orange dots denote Hallmark gene sets related to cell cycle, cell growth, and epithelial-mesenchymal transition, all of which had false-discovery rate q-value < 0.01
d. Normalized enrichment scores from GSEA analysis comparing Cluster 4 to Clusters 1, 2, 3, and 5. Red dots denote immune-related Hallmark gene sets, all of which had false-discovery rate q-value < 0.01
When we performed Gene Set Enrichment Analysis (24) to identify pathways that were uniquely overexpressed in Cluster 1 and 4 tumors, we found that the upregulated pathways in Cluster 1 were involved in cell cycle regulation, cell growth, and epithelial-mesenchymal transition (Fig. 4c) while upregulated pathways in Cluster 4 were associated with an activated immune response (Fig. 4d).
Hispanic/Latino Patients with Diffuse-Type Tumors Have Frequent Germline CDH1 Mutations
We analyzed the WES data from either blood or non-neoplastic stomach from 83 patients and identified seven germline CDH1 mutations (Fig. 5a, Table 2). All seven mutations were identified in patients with diffuse-type cancer (DGC; 16%) and were confirmed with Sanger sequencing (Supplementary Fig. 4a, Supplementary Table 4). Two mutations were deletions and five were missense variants. In patients with DGC, the median age of mutation carriers was 41 years (range 36–54 years) while the median age of CDH1 wild-type patients was 50 years (range: 26–76 years; P < 0.05; Supplementary Fig. 4b, Supplementary Table 1).
Figure 5. Hispanic/Latino gastric cancer patients have high rates of germline CDH1 mutations.
a. Seven germline CDH1 mutations were identified in patients with diffuse gastric cancer.
b. Western blot showing E-cadherin expression level upon transfection of plasmids carrying wild-type CDH1, A286G variant, or G1849A variant into Chinese hamster ovary cells.
c. Representative pictures of scratch assays. Distance between the wound edges were measured after 24 hours.
d. Quantification of remaining distance between wound edges, relative to 0h. N ≥ 9 per group, with at least two independent experiments. ** P < 0.01, *** P < 0.001, **** P < 0.0001
Table 2:
Germline CDH1 mutations found in the Hispanic/Latino gastric cancer cohort.
| Patient ID | Age | Mutation | Chromosome position | Amino Acid Changes | Mutation Type | ClinVar | Predicted Effect by SIFT & PolyPhen-2 | gnomAD | gnomAD (Latino) |
|---|---|---|---|---|---|---|---|---|---|
| P15 | 51 | germline | Exon3:c.286A>G | I96V | missense | Benign | Benign | 0.016% | 0.12% |
| P16 | 36 | germline | Exon13:c.1988_2011del, 2012A>C | 663–671del EVGDYKINLKLMDNQN>VGDSNQND | deletion-inframe and missense | N/A | Deleterious | . | . |
| P20 | 37 | germline | Exon12:c.1849G>A | A617T | missense | Benign | Benign | 0.45% | 0.30% |
| P30 | 41 | germline | Exon14:c.2276G>C | G759A | missense | Uncertain | Deleterious | 0.0004% | 0.0029% |
| P33 | 40 | germline | Exon2:c.135delC | H45fs | deletion-frameshift | N/A | N/A | . | . |
| P50 | 41 | germline | Exon6:c.715G>A | G239R | missense | Conflicting | Deleterious | . | . |
| P71 | 54 | germline | Exon16:c.2558C>T | S853L | missense | Uncertain | Deleterious | 0.0016% | 0.009% |
Pathogenic CDH1 germline mutations are known to cause hereditary DGC syndrome. However, none of the germline mutation carriers in our Hispanic/Latino cohort had a family history of gastric cancer or lobular breast cancer, which is another manifestation of the mutations (25). Previous reports have suggested that germline CDH1 alterations contribute to early-onset gastric cancer in patients without family histories of cancer (26). We performed a literature search to estimate the rate of germline CDH1 mutations in gastric cancer patients without family histories of gastric cancer, and identified four studies with relatively large cohorts. These included patients from Italy,(27) Canada,(28) China,(29) and Korea (30). Out of 350 DGC patients, 12 germline mutations in the coding region of CDH1 were identified across 13 patients (3.7%), with 3 patients having deletions, and 10 having missense alterations (Supplementary Table 4). Thus, the prevalence of germline CDH1 mutations in patients without a relevant family history was markedly higher in the Hispanic/Latino cohort than what has been reported in other ethnic/racial groups.
To determine whether the identified CDH1 mutations were pathogenic, we first checked the population frequency of these variants in the Genome Aggregation Database (https://gnomad.broadinstitute.org). All seven variants were found in less than 1% of both the general population and in the Latino cohort, which would be consistent with pathogenicity (Table 2). We next queried the annotations of the five missense mutations in the ClinVar database (31). Two were classified as benign (P15 and P20) while the rest were either of uncertain significance or had conflicting data (P30, P50 and P71). Next, we used SIFT and PolyPhen-2 to predict the variant functionality via a bioinformatic approach. Consistent with ClinVar annotation, P15 and P20 were predicted to be benign, but P30, P50, and P71 were projected to be pathogenic (Table 2). Of the six patients whose tissue samples were available for analysis, we performed immunohistochemistry for E-cadherin, and found that there was either decreased (P15, P16, and P33) or near-complete loss (P20 and P50) of protein expression in five of the six patients, including in P15 and P20, who harbored putatively benign variants (Supplementary Fig. 5).
The variant found in P15, who was a 51-year-old man presenting with locally advanced disease, was a c.286 A>G transition that resulted in an I96V amino acid alteration. Patient 20, who was a 37 year-old woman presenting with metastatic disease, had an c.1849 G>A change that led to an A617T amino acid change. To test the effects of these variants in vitro, we generated plasmids carrying wild-type CDH1 or these two variants and transfected them into Chinese hamster ovary (CHO) cells, which do not express E-cadherin at baseline and has been used extensively by other groups to test CDH1 variant function (32–34). Sanger sequencing confirmed that the mutations were generated correctly. We found that both 286 A>G and 1849 G>A variants generated protein products that were normal in size and cellular localization (Fig. 5b and Supplementary Fig. 6a). There was no difference in protein expression levels.
E-cadherin is involved in cell-cell adhesion and its loss can result in increased cellular migration. We performed scratch assays to test if 286 A>G or 1849 G>A affected the migratory ability of CHO cells. After 24 hours, parental CHO cells had completely covered the scratch. As expected, CHO cells expressing wild-type CDH1 led to significantly reduced cellular migration, with 68% of the wound distance remaining (P < 0.0001). However, 286 A>G expressing cells had only 54% (P < 0.01) and 1849 G>A expressing cells had only 53% (P < 0.001) of their wound distances remaining (Fig. 5c and 5d). Thus, both variants conferred significantly increased migratory capability.
In gastric cancer with both germline and somatic CDH1 mutations, promoter methylation of the other allele is the most common form of second-hit inactivation (35,36). To test the methylation status of the CDH1 promoter, we used paired tumor and non-neoplastic DNA. Of the seven patients carrying germline CDH1 variants, there were six sample pairs available for analysis. We found evidence for strong promoter methylation in one patient, for mild increase in methylation in four patients and no increase of methylation in one patient (Supplementary Fig. 6b).
Discussion
Hispanic/Latino patients experience significant gastric cancer outcome disparities. Whether there is a molecular basis for these differences is unknown, as previous gastric cancer genomic studies included very few Hispanic/Latino patients (9,10,12). To our knowledge, the only study to date that had included a large number of Hispanic/Latino patients was performed by Sahasrabudhe et al (37). However, their analysis of 333 patients from Latin America was limited to targeted sequencing of only five genes involved in DNA repair.
In this study, we have performed a large, integrated analysis of Hispanic/Latino gastric cancer samples and compared our results to those from Asian and White patients’ samples published by the TCGA. We found that Hispanic/Latino gastric cancer patients had a high incidence of germline CDH1 mutations, and that their tumors were enriched for the GS molecular subtype. Our findings indicate that the lack of ethnic and racial diversity in samples analyzed by previous large-scale studies has likely biased our genomic understanding of gastric adenocarcinoma due to the overrepresentation of White and Asian patients. Previous studies in other cancer types also identified genomic differences based on ethnicity and race. Shi et al found that more than 50% of Asian non-small-cell lung adenocarcinoma patients had EGFR mutations, as compared to 20% of White patients (38). In a study of African-American prostate cancer patients, Yamoah et al identified genomic biomarkers related to race that were highly prognostic (39). Thus, having ethnically and racially representative study cohorts will enhance our understanding of fundamental disease biology and ensure that the efficacy of a selected treatment has been tested and confirmed for the patient’s ethnic/racial background (11,40). Improved recruitment of underrepresented patient populations into future clinical and basic scientific studies should be mandatory.
The high rate of germline CDH1 mutations in our Hispanic/Latino DGC cohort is striking. Of the seven mutations we identified, which represented 16% of the DGC patients, two had not been previously reported in ClinVar, three were annotated as uncertain or conflicting, and two were designated as benign. Thus, these variants would likely have been excluded as pathogenic. However, several lines of evidence indicate that these mutations have deleterious effects. First, previous studies have suggested that germline CDH1 mutations may contribute to early-onset DGC (26). The variant carriers in our cohort had a median age of diagnosis of 41 years as compared to DGC patients with wild-type CDH1 who had a median age of 50 at diagnosis. Second, E-cadherin protein expression was decreased or lost in five of the six tumors from CDH1 mutation carriers that were available for analysis. Third, in silico analysis predicted that three of the five missense mutations were pathogenic. Finally, functional modeling of the two missense variants annotated by ClinVar and predicted to be benign by both SIFT and PolyPhen2 demonstrated pathogenic cellular migration phenotype. Our findings speak to the limitations of the currently available tools to predict accurately the pathogenicity of a given variant. When germline CDH1 mutations are identified in patients who have a high pre-test probability of carrying a pathogenic variant, such as in a young DGC patient, more rigorous functional testing should be utilized to determine pathogenicity.
Germline CDH1 mutations are one of the causes hereditary DGC syndrome. Since none of the seven Hispanic/Latino CDH1 variant carriers had a family history of gastric cancer or lobular breast cancer, these mutations are either de novo or exhibited low penetrance. Previous estimates that carriers of pathogenic CDH1 variants have a lifetime risk of up to 80% of developing DGC are likely overestimations as they are based on families that fulfil the International Gastric Cancer Linkage Consortium (IGCLC) guidelines and thus subjected to ascertainment bias (41). Recent studies that examined DGC penetrance in carriers of pathogenic CDH1 variants that do not fulfil IGCLC criteria indicate a lower lifetime gastric cancer risk. Xicola et al found a lifetime risk of 37% in their cohort while Roberts et al estimated risk at 42% for men and 33% for women by age 80 (42,43). These recent reports along with our findings suggest that some germline CDH1 variants require other oncogenic molecular and/or environmental factors to drive DGC formation. This represents an opportunity for precision treatment strategies as we hypothesize that different variants may produce varied biological effects and targets for therapy. Finally, while five of our seven patients would have undergone genetic testing based on IGCLC recommendations to test DGC patients diagnosed before age 50, two did not meet criteria. A recent study by Lowstuter et al found that 65% of CDH1 mutation carriers did not meet IGCLC guidelines for testing (44). This suggests that revisions will be necessary to improve the sensitivity of guidelines for genetic testing to identify germline CDH1 carriers.
Previous analyses of early-onset gastric cancer have identified DGC as being associated with young age (12,45). The high rate of DGC in Hispanic/Latino patients is consistent with the younger age of diagnosis in this cohort. However, the molecular mechanism behind early-onset carcinogenesis in this subgroup is unknown. As discussed above, the high rate of germline CDH1 mutations in the Hispanic/Latino cohort may play a role. Previous studies in non-Hispanic/Latino cohorts showed that germline CDH1 mutations occurs in about 1–3% of non-familial gastric cancer patients (27–30). In addition, the TCGA reported only two CDH1 variants in 295 patients, and these were non-pathogenic (12). Other factors unrelated to Lauren classification and GS subtype clearly affect the etiology of early-onset gastric cancer since three of four very young (<35 years old) patients in our cohort were in the CIN group, with two of them having intestinal-type cancers. This will be an important area for future study, as the incidence of gastric cancer is rising in the United States only amongst young patients and thus will likely disproportionately affect the Hispanic/Latino population and exacerbate gastric cancer outcome disparities (46).
While molecular classification systems proposed by the TCGA and others have provided new paradigms to study gastric cancers, the practical implications of this scheme for patient care remain elusive. Recently, Sohn et al. reported that the TCGA classification may provide both prognostic and predictive value in Korean patients (47). They found that EBV tumors had the best outcomes and GS cancers had the worst. Additionally, Kim et al showed that immunotherapy was effective mainly in EBV or MSI-type tumors, while CIN and GS cancers were generally resistant (48). These findings have significant implications for the Hispanic/Latino cohort that we analyzed since 95% of the patients had either CIN or GS tumors. Using consensus clustering of RNA-seq data, we identified transcriptomic signatures that were prognostic and thus can aid in risk-stratification and treatment planning. Importantly, we found that the signatures were prognostic for subgroups stratified based on both molecular subtypes and by Lauren classification, suggesting of their wide applicability across tumor types. Intriguingly, patients in the low-risk, favorable prognosis group had a gene signature indicative of an activated immune response. Whether these patients will benefit from immune therapy is a possibility that should be tested. Finally, validation in patients of all ancestry will be required to determine if this signature is ethnicity/race-specific.
Future studies of Hispanic/Latino populations will benefit from more refined definitions of ancestry mix. Hispanic and Latino groups encompass a geographically diverse population exhibiting significant genomic heterogeneity, due to the differential admixture of European, Indigenous American, and African populations. Previous studies have shown that ancestry proportions in Hispanic/Latino patients are associated with breast cancer incidence and outcomes (49,50). However, while our study cohort is derived from patients living in North Texas, the country of origin is heterogeneous, as denoted by the relatively broad cluster seen in the ancestry analysis.
In conclusion, while gastric cancer outcome disparities may result from a combination of environmental exposures and socioeconomic factors, inherent tumor biology is also an essential component. Our study analyzing a large cohort of Hispanic/Latino gastric cancer patients is an important step in addressing the outcome disparity that these patients face by providing a genomic context for their disease. We have found that gastric cancers arising in Hispanic/Latino patients exhibit significantly different genomic landscapes than those developing in Asian and White patients. There is an enrichment for GS tumors and a high rate of germline CDH1 mutations. Our findings should be considered in establishing guidelines for screening, genetic counseling, and treatment of Hispanic/Latino gastric cancer patients.
Supplementary Material
Significance.
Gastric cancer in Hispanic/Latino patients has unique genomic profiles that may contribute to the aggressive clinical phenotypes seen in these patients.
Acknowledgements
S.C.W. was a UT Southwestern Disease-Oriented Scholar and an American College of Surgeons Research Faculty Fellow and supported by the Society for Surgery of the Alimentary Tract Health Care Disparity Research Award and the NCI/NIH (K08 CA222611). M.R.P. is a Dedman Family Scholar in Clinical Care. I.N. was supported by the National Center for Advancing Translational Sciences of the NIH, under Award No. UL1TR001105. H.Z. was supported by the NIDDK/NIH (R01DK111588). T.H.H. was supported by the University of Texas Lung Cancer SPORE career development grant (P50CA070907), the American Cancer Society Institutional Research Award (ACS-IRG-02–196), and the UT Southwestern Simmons Comprehensive Cancer Center Support Grant (P30CA142543). We acknowledge the assistance of the University of Texas Southwestern Tissue Resource, a shared resource at the Simmons Comprehensive Cancer Center, which is supported in part by the National Cancer Institute, under award number 5P30CA142543. The authors have no financial disclosures or conflicts of interest. We thank Drs. Herbert J. Zeh, III and Todd A. Aguilera for critical reading of the manuscript.
Footnotes
Conflict of Interest
The authors declare no potential conflicts of interest.
References
- 1.Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Akinyemiju TF, Lami F, Alam T, Alizadeh-Navaei R, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. Jama Oncol. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Fedewa SA, Miller KD, Goding-Sauer A, Pinheiro PS, Martinez-Tyson D, et al. Cancer statistics for Hispanics/Latinos, 2015. Ca Cancer J Clin. 2015;65:457 480. [DOI] [PubMed] [Google Scholar]
- 3.Yao JC, Tseng JF, Worah S, Hess KR, Mansfield PF, Crane CH, et al. Clinicopathologic Behavior of Gastric Adenocarcinoma in Hispanic Patients: Analysis of a Single Institution’s Experience Over 15 Years. J Clin Oncol. 2005;23:3094 3103. [DOI] [PubMed] [Google Scholar]
- 4.Refaie W, Tseng JF, Gay G, Patel-Parekh L, Mansfield PF, Pisters PW, et al. The impact of ethnicity on the presentation and prognosis of patients with gastric adenocarcinoma. Cancer. 2008;113:461 469. [DOI] [PubMed] [Google Scholar]
- 5.Bautista MC, Jiang S-F, Armstrong M, Kakar S, Postlethwaite D, Li D. Significant Racial Disparities Exist in Noncardia Gastric Cancer Outcomes Among Kaiser Permanente’s Patient Population. Digest Dis Sci. 2015;60:984 995. [DOI] [PubMed] [Google Scholar]
- 6.Yuan J, Hu Z, Mahal BA, Zhao SD, Kensler KH, Pi J, et al. Integrated Analysis of Genetic Ancestry and Genomic Alterations across Cancers. Cancer Cell. 2018;34:549 560.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietze EC, Sistrunk C, Miranda-Carboni G, O’Regan R, Seewaldt VL. Triple-negative breast cancer in African-American women: disparities versus biology. Nat Rev Cancer. 2015;15:248 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400 416.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cristescu R, Lee J, Nebozhyn M, Kim K-M, Ting JC, Wong S, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449 456. [DOI] [PubMed] [Google Scholar]
- 10.Ichikawa H, Nagahashi M, Shimada Y, Hanyu T, Ishikawa T, Kameyama H, et al. Actionable gene-based classification toward precision medicine in gastric cancer. Genome Med. 2017;9:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan DS, Mok TS, Rebbeck TR. Cancer Genomics: Diversity and Disparity Across Ethnicity and Geography. J Clin Oncol. 2016;34:91–101. [DOI] [PubMed] [Google Scholar]
- 12.Bass AJ, Thorsson V, Shmulevich I, Reynolds SM, Miller M, Bernard B, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavalli-Sforza LL. The Human Genome Diversity Project: past, present and future. Nat Rev Genet. 2005;6:333 340. [DOI] [PubMed] [Google Scholar]
- 14.Hong C, Manimaran S, Shen Y, Perez-Rogers JF, Byrd AL, Castro-Nallar E, et al. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome. 2014;2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30:1015 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middha S, Zhang L, Nafa K, Jayakumaran G, Wong D, Kim HR, et al. Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data. Jco Precis Oncol. 2017;2017:1 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao F, Kausalya JP, Sia Y, Teo AS, Lee W, Ong AG, et al. Recurrent Fusion Genes in Gastric Cancer: CLDN18-ARHGAP26 Induces Loss of Epithelial Integrity. Cell Reports. 2015;12:272 285. [DOI] [PubMed] [Google Scholar]
- 18.Nicorici D, Satalan M, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, et al. FusionCatcher-a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014;011650. [Google Scholar]
- 19.Haas B, Dobin A, Stransky N, Li B, Yang X, Tickle T, et al. STAR-Fusion: Fast and Accurate Fusion Transcript Detection from RNA-Seq. Biorxiv. 2017;120295. [Google Scholar]
- 20.Jin D-H, Park S-E, Lee J, Kim K-M, Kim S, Kim D-H, et al. Copy Number Gains at 8q24 and 20q11-q13 in Gastric Cancer Are More Common in Intestinal-Type than Diffuse-Type. Plos One. 2015;10:e0137657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burbano R, Assumpção P, Leal M, Calcagno D, Guimarães A, Khayat A, et al. C-MYC locus amplification as metastasis predictor in intestinal-type gastric adenocarcinomas: CGH study in Brazil. Anticancer research. 2006;26:2909 2914. [PubMed] [Google Scholar]
- 22.Wang X, Liu Y, Shao D, Qian Z, Dong Z, Sun Y, et al. Recurrent amplification of MYC and TNFRSF11B in 8q24 is associated with poor survival in patients with gastric cancer. Gastric Cancer. 2016;19:116 127. [DOI] [PubMed] [Google Scholar]
- 23.Wong GS, Zhou J, Liu J, Wu Z, Xu X, Li T, et al. Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat Med. 2018;24:968 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. P Natl Acad Sci Usa. 2005;102:15545 15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansford S, Kaurah P, Li-Chang H, Woo M, Senz J, Pinheiro H, et al. Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. Jama Oncol. 2015;1:23 32. [DOI] [PubMed] [Google Scholar]
- 26.Corso G, Pedrazzani C, Pinheiro H, Fernandes E, Marrelli D, Rinnovati A, et al. E-cadherin genetic screening and clinico-pathologic characteristics of early onset gastric cancer. Eur J Cancer. 2011;47:631 639. [DOI] [PubMed] [Google Scholar]
- 27.Garziera M, Canzonieri V, Cannizzaro R, Geremia S, Caggiari L, Zorzi M, et al. Identification and characterization of CDH1 germline variants in sporadic gastric cancer patients and in individuals at risk of gastric cancer. Plos One. 2013;8:e77035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bacani J, ares, Zwingerman R, di Nicola N, Senz J, Riddell R, et al. CDH1/E-cadherin germline mutations in early-onset gastric cancer. J Med Genet. 2006;43:867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q-H. Novel CDH1germline mutations identified in Chinese gastric cancer patients. World J Gastroentero. 2013;19:909 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho S, Park J, Liu Y, Park Y, Kim J, Yang H, et al. Sporadic Early-Onset Diffuse Gastric Cancers Have High Frequency of Somatic CDH1 Alterations, but Low Frequency of Somatic RHOA Mutations Compared With Late-Onset Cancers. Gastroenterology. 2017;153:536 549.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garziera M, Re V, Geremia S, Seruca R, Figueiredo J, Melo S, et al. A novel CDH1 germline missense mutation in a sporadic gastric cancer patient in north-east of Italy. Clin Exp Med. 2013;13:149 157. [DOI] [PubMed] [Google Scholar]
- 33.Simões-Correia J, Figueiredo J, Lopes R, Stricher F, Oliveira C, Serrano L, et al. E-Cadherin Destabilization Accounts for the Pathogenicity of Missense Mutations in Hereditary Diffuse Gastric Cancer. Plos One. 2012;7:e33783 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira C, Seruca R, Carneiro F. Genetics, pathology, and clinics of familial gastric cancer. Int J Surg Pathol. 2006;14:21 33. [DOI] [PubMed] [Google Scholar]
- 35.Machado J, Oliveira C, Carvalho R, Soares P, Berx G, Caldas C, et al. E-cadherin gene (CDH1) promoter methylation as the second hit in sporadic diffuse gastric carcinoma. Oncogene. 2001;20:1525–8. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira C, Sousa S, Pinheiro H, Karam R, Bordeira–Carriço R, Senz J, et al. Quantification of Epigenetic and Genetic 2nd Hits in CDH1 During Hereditary Diffuse Gastric Cancer Syndrome Progression. Gastroenterology. 2009;136:2137–48. [DOI] [PubMed] [Google Scholar]
- 37.Sahasrabudhe R, Lott P, Bohorquez M, Toal T, Estrada AP, Suarez JJ, et al. Germline Mutations in PALB2, BRCA1, and RAD51C, Which Regulate DNA Recombination Repair, in Patients With Gastric Cancer. Gastroenterology. 2017;152:983 986.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y, Au J, Thongprasert S, nivasan S, Tsai C-M, Khoa M, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9:154 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamoah K, Johnson MH, Choeurng V, Faisal FA, Yousefi K, Haddad Z, et al. Novel Biomarker Signature That May Predict Aggressive Disease in African American Men With Prostate Cancer. J Clin Oncol. 2015;33:2789 2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spratt DE, Chan T, Waldron L, Speers C, Feng FY, Ogunwobi OO, et al. Racial/Ethnic Disparities in Genomic Sequencing. Jama Oncol. 2016;2:1070 1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Post RS, Vogelaar IP, Carneiro F, Guilford P, Huntsman D, Hoogerbrugge N, et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J Med Genet. 2015;52:361 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xicola RM, Li S, Rodriguez N, Reinecke P, Karam R, Speare V, et al. Clinical features and cancer risk in families with pathogenic CDH1 variants irrespective of clinical criteria. J Med Genet. 2019;56:838–43. [DOI] [PubMed] [Google Scholar]
- 43.Roberts ME, Ranola JO, Marshall ML, Susswein LR, Graceffo S, Bohnert K, et al. Comparison of CDH1 Penetrance Estimates in Clinically Ascertained Families vs Families Ascertained for Multiple Gastric Cancers. Jama Oncol. 2019;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowstuter K, Espenschied CR, Sturgeon D, Ricker C, Karam R, LaDuca H, et al. Unexpected CDH1 Mutations Identified on Multigene Panels Pose Clinical Management Challenges. Jco Precis Oncol. 2017;1 12. [DOI] [PubMed] [Google Scholar]
- 45.Kong X, Wang J-L, Chen H-M, Fang J-Y. Comparison of the clinicopathological characteristics of young and Elderly patients with gastric carcinoma: A meta analysis. J Surg Oncol. 2012;106:346 352. [DOI] [PubMed] [Google Scholar]
- 46.Anderson WF, Camargo CM, Fraumeni JF, Correa P, Rosenberg PS, Rabkin CS. Age-specific trends in incidence of noncardia gastric cancer in US adults. Jama. 2010;303:1723 1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sohn B, Hwang J-E, Jang H-J, Lee H-S, Oh S-C, Shim J-J, et al. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin Cancer Res. 2017;23:4441 4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S, Cristescu R, Bass AJ, Kim K-M, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1 14. [DOI] [PubMed] [Google Scholar]
- 49.Fejerman L, Romieu I, John EM, Lazcano-Ponce E, Huntsman S, Beckman KB, et al. European ancestry is positively associated with breast cancer risk in Mexican women. Cancer Epidem Biomar. 2010;19:1074 1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fejerman L, Hu D, Huntsman S, John EM, Stern MC, Haiman CA, et al. Genetic ancestry and risk of mortality among U.S. Latinas with breast cancer. Cancer Res. 2013;73:7243 7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.