Graphical abstract
Keywords: HIV-1, Nef, ABCA1, Molecular modeling, Molecular dynamics, Virtual screening
Abstract
HIV-1 infection impairs cellular cholesterol efflux by downmodulating the cholesterol transporter ABCA1, leading to metabolic co-morbidities like cardio-vascular disease. The main mechanism of this effect is impairment by the HIV-1 protein Nef of the ABCA1 interaction with the endoplasmic reticulum chaperone calnexin, which leads to a block in ABCA1 maturation followed by its degradation. However, ABCA1 is also downmodulated by Nef delivered with the extracellular vesicles, suggesting involvement of a direct Nef:ABCA1 interaction at the plasma membrane. Here, we present an optimized model of the Nef:ABCA1 interaction, which identifies interaction sites and provides an opportunity to perform a virtual screening for potential inhibitors. Interestingly, the predicted sites on Nef involved in the ABCA1 interaction overlap with those involved in the interaction with calnexin. The compounds previously shown to block Nef:calnexin interaction were among the top ranking ligands in docking simulations with ABCA1-interacting sites on Nef, suggesting the possibility that both interactions can be inhibited by the same chemical compounds. This study identifies a series of compounds for potential development as inhibitors of Nef-mediated co-morbidities of HIV infection.
1. Introduction
HIV-1 protein Nef is a major viral pathogenic factor, as Nef-deficient virus causes only a mild disease in animals and people [1], [2]. This multifunctional protein stimulates HIV-1 replication by counteracting activity of the innate anti-viral proteins SERINC3 and 5 [3], [4] and impairs the anti-HIV immune responses by downregulating CD4 and MHC I on the immune cells [5]. Nef also contributes to HIV-associated co-morbidities, such as cardio-vascular disease and neurological impairment, which persist even after suppression of the virus with anti-retroviral treatment (ART) [6]. The reason for this phenomenon is likely a continuous release of Nef, as a component of extracellular vesicles (exNef), from HIV-infected cells located in the viral reservoirs [7], [8], [9], [10], [11], [12], [13].
Although the detailed mechanism of HIV-associated co-morbidities’ pathogenesis has not been fully understood, an important component of these pathologies is impairment of cholesterol metabolism [7], [14], [15]. Nef is a key viral factor responsible for cholesterol metabolism impairment via downmodulation of the cellular cholesterol transporter ABCA1 [16]. ABCA1 is a transmembrane protein, which, together with ABCG1, mediates cholesterol efflux to HDL [17]. Nef-mediated downregulation of ABCA1 involves impaired delivery to the plasma membrane and increased degradation of the newly synthesized ABCA1, as well as increased endocytosis and impaired recycling of the plasma membrane ABCA1 [18], [19].
Nef downregulates CD4, MHC I and several other cell surface molecules important for immune responses by binding to the cytoplasmic tails of these molecules and targeting them to degradation pathways [20]. Our studies demonstrated that Nef interacts with ABCA1 via a specific region in the ABCA1, the C-terminal cytoplasmic region [18], [21]. However, deletion of this region, while preventing Nef:ABCA1 interaction, did not prevent ABCA1 downmodulation in Nef-transfected cells [18]. Later studies identified an alternative mechanism: Nef can bind to the endoplasmic reticulum chaperone calnexin and disrupt its interaction with ABCA1, thus impairing ABCA1 maturation and leading to its eventual degradation [22], [23], [24].
To screen for potential inhibitors of the Nef:calnexin interaction, we created a molecular model of this interaction using global docking based on the models of Nef and calnexin built by us previously [22], [23]. The produced model showed interaction sites in both Nef and calnexin and was utilized to perform virtual screening, which gave a number of potential inhibitors predicted to target these sites [23]. The sites identified in Nef were 1-MGGKWSKRS-9 (site I), 75-RPQVPLRPMTYKAALD-90 (site II), and 121-TQGY-124 (site III). An extended site II appeared to be a major site interacting with calnexin [22]. Virtual screening using this interaction model produced a number of potential inhibitors.
These inhibitors were tested for functional activity pointing at ZINC03953858 (NSC13987) as the most potent inhibitor of the Nef:calnexin interaction [23]. NSC13987 is a highly lipophilic compound due to the presence of benzanthrone and anthraquinone moieties. The anthraquinone and secondary amine were identified as critical for the biological activity. Thus, to improve solubility and functional activity, our initial ligand refinement efforts were aimed at preserving these structural elements and focused on iterative alterations to the benzanthrone ring, leading to the development of AMS-55 [22].
The mechanism described above explains the ABCA1 downmodulation in Nef-transfected and HIV-infected cells. Recent studies demonstrated that ABCA1 downmodulation and cholesterol efflux impairment are also observed when cells are treated with exNef [15], [25]. This activity is particularly important in the ART era, as HIV suppression dramatically decreases the number of HIV-infected cells, leaving circulating exNef as the main potential source of this pathogenic factor. Given that exNef delivers Nef to the plasma membrane of the target cells, where functional ABCA1 is localized, we surmised that in this case, increased endocytosis and impaired recycling of the plasma membrane ABCA1 may be the primary event in ABCA1 downmodulation, and direct Nef:ABCA1 interaction may play the key role in exNef-mediated pathogenesis.
To initiate the development of inhibitors of Nef:ABCA1 interaction, we relied on the computational strategy, including molecular modeling, virtual screening, docking and molecular dynamics. Such approach has been previously used to discover a number of inhibitors for potential therapeutic applications [26], [27], [28], [29], [30]. These results identified candidate molecules for future development efforts.
2. Methods
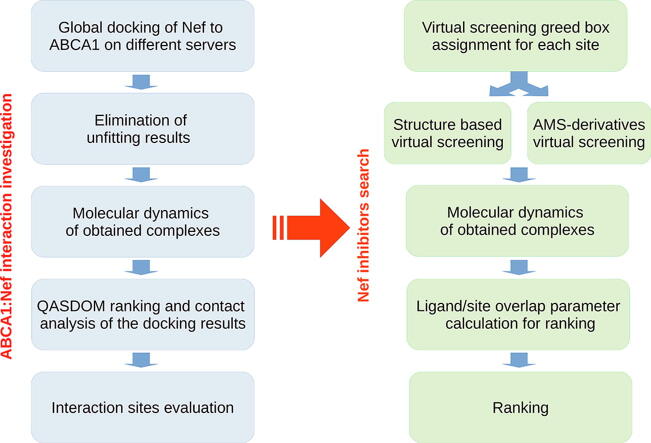
2.1. Study workflow: This study was composed of the following steps.
-
1.
Global docking was performed, using several servers, of the Nef structure built by us previously [22], [23] to ABCA1 structure modeled as described in the Structure modeling section below. This resulted in 64 Nef:ABCA1 complexes.
-
2.
The docking results were superposed with the ABCA1 structure embedded in a membrane. All Nef molecules overlapping with the membrane were removed from the dataset. This led to 21 complexes of Nef:ABCA1. Out of these 21 complexes, those in which Nef did not interact with the membrane were excluded, as we are interested in the interaction at the membrane where both ABCA1 and Nef are targeted [31], [32]. The resulting 8 complexes were used for further tests.
-
3.
The 8 complexes of Nef:ABCA1 from the global docking were processed in MD simulations for 50 ns.
-
4.
The details of the Nef:ABCA1 interaction were obtained by QASDOM metaserver [33] via analysis of the dataset of the 8 Nef:ABCA1 complexes after MD from the previous stage.
-
5.
The Nef:ABCA1 interaction sites I-IV in the Nef structure were identified from the data obtained at the previous stage.
-
6.
For each of the 4 sites in the Nef structure, a grid box for virtual screening was assigned (Supplementary Table S1).
-
7.
A structure-based virtual screening using the 4 grid boxes from the previous step was conducted using program Vina, of the following compounds. (1) Freely available small molecule compounds from the NCI Plated 2007 dataset, and (2) Derivatives of the compounds previously found to inhibit Nef:calnexin interaction (AMS compounds) [22], [23]. AMS derivatives were docked to each of the four interaction sites of Nef, resulting in 4 different complexes for each of the AMS analogs.
-
8.
Each of the complexes obtained by virtual screening, with the best Vina ranking, was processed by MD simulation for 30 ns to test if the ligand remains in docking pocket or moves away.
-
9.
Overlap parameter was calculated as a number of atomic contacts between Nef interaction site and ligand normalized to the number of site residues for each of the interaction sites.
-
10.
Ligands were ranked according to the overlap parameter.
The concise version of the above workflow is shown in the graphical abstract.
Structure modeling: ABCA1 modeling was based on the PDB:5XJY structure [34] (uniprot entry O95477, ABCA1_HUMAN). A model was prepared containing the transmembrane and cytoplasmic parts of ABCA1, where interaction with Nef was expected [18], [21] and a fragment of the lipid membrane. For this purpose, regions 49–623 and 1371–1635 belonging to the extracellular part of the transmembrane domain have been locked by GGG tripeptide sequences. The N- and C-termini of these regions in the initial structure were separated from each other by approximately 7.5 Å, therefore replacing them with short flexible loops should not disrupt the transmembrane domain structure. The resulted structure was inserted into the bilayer membrane made of DDPC lipids with the CHARMM-GUI membrane builder server [35] in accordance with uniprot data on the ABCA1 transmembrane regions. Then it was relaxed in water solution with 0.15 NaCl with a molecular dynamics (MD) run for 100 ns using Gromacs software [36].
Nef modeling was performed for the target sequence P03407 (HIV-1 group M subtype B isolate ARV2/BRU) and the model was built by us using several servers for protein structure prediction: Phyre2 [37], iTasser [38], RaptorX [39], M4T [40], SwissModel [41]. The final structure was built using server QA-RecombineIt [30], as described in [20].
Modeling of interaction interfaces: Prediction of Nef:ABCA1 interaction sites was carried out by modeling the complete interfaces of protein–protein interaction. Modeling was performed by global docking on different docking servers using the approach developed by us [31] and QASDOM metaserver (http://qasdom.eimb.ru) [32]. The Cluspro [33], HEX [34], SwarmDock [35] and Zdock [36] modeling servers were used. From the entire set of Nef:ABCA1 docking models, biologically incorrect complexes (i.e., those where Nef had no contact with the membrane or overlapped the membrane region) were excluded. For the remaining structures, MD simulations of the system that included Nef and the membrane were carried out for 50 ns to obtain the relaxed and energetically favorable complexes. Interaction sites in ABCA1 and Nef in these complexes were selected according to the QASDOM criteria as linear clusters (stretches of sequence) with the number of residue interactions greater than the dataset median, and where the number of atom–atom interactions was greater than the median for the dataset.
Molecular dynamics simulations: All the molecular dynamics simulations were performed with Gromacs software package [36] and CHARMM36 force field. All initial structures were inserted in DPPC bilayer membrane with CCHARMM-GUI server [35] and energy was minimized consecutively with the steepest descent and conjugated gradients algorithms. Then they were equilibrated in water with the NaCl concentration of 150 mM during six consecutive MD runs under position restraints for 1,875 ps according to the usual equilibration protocol for the membrane calculations or 200 ps NVT and NPT equilibration for the protein–ligand systems. Simulations were carried out using the particle mesh Ewald technique with repeating boundary conditions and 1.2 nm cut-offs in a Verlet cutoff-scheme, using the LINCS constraint algorithm with a 2-fsec time step. Vdw type was cutoff and vdw-modifier was force-switch. For temperature coupling, the V-rescale algorithm was used and a constant temperature of 300 K was maintained.
Virtual screening for inhibitors: Docking-based (structure-based) virtual screening was performed with locally installed software package AutoDock VINA 1.1.2 (http://vina.scripps.edu/) [42]. The three-dimensional Grid box for virtual screening was set using AutoDock tool 1.5.6 [43]. The Grid box was positioned to include active binding sites and all essential residues. The box vectors varied from 12 to 30 A. Detailed information on grid boxes for each interaction site is listed in Supplementary Table S1. Eight cpu cores were used for each virtual screening procedure. As a database of low molecular weight compounds, the publicly accessible ZINC database (http://zinc.docking.org/) [44] was used. For compatibility with our previous work [23], we used the NCI Plated 2007 dataset of 139,735 compounds (http://zinc.docking.org/catalogs/ncip). We also docked 26 AMS-series compounds derived from AMS-55 [22].
Ligand preparation: All the ligand structures and their modeling parameters were obtained from Automatic Topology builder [45] by semi-empirical quantum mechanics calculations according to their SMILES strings or PDB structures from ZINC database.
3. Results
Global docking of Nef to ABCA1 provided a large dataset of complex structures, where substantial part was biologically irrelevant, i.e. the structures that contradicted the accumulated experimental data describing these proteins. For example, there have been complexes in which Nef interacted with the transmembrane regions of ABCA1, rather than with the cytoplasmic domains [21]. To eliminate such models, we have superimposed the docking results on ABCA1 structure with the membrane and removed all Nef structures that were embedded in the membrane or did not contact it, since it is known that Nef interacts with the membrane [31].
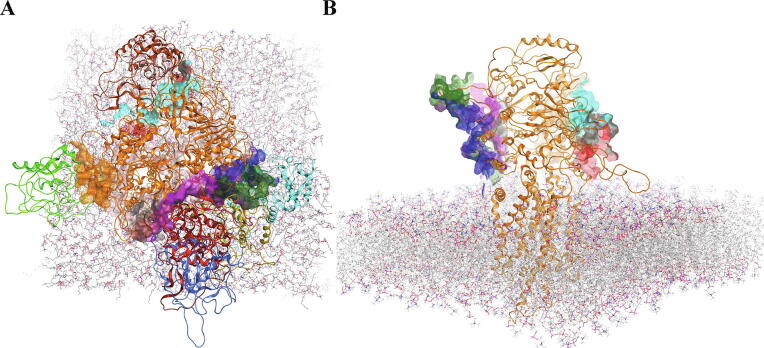
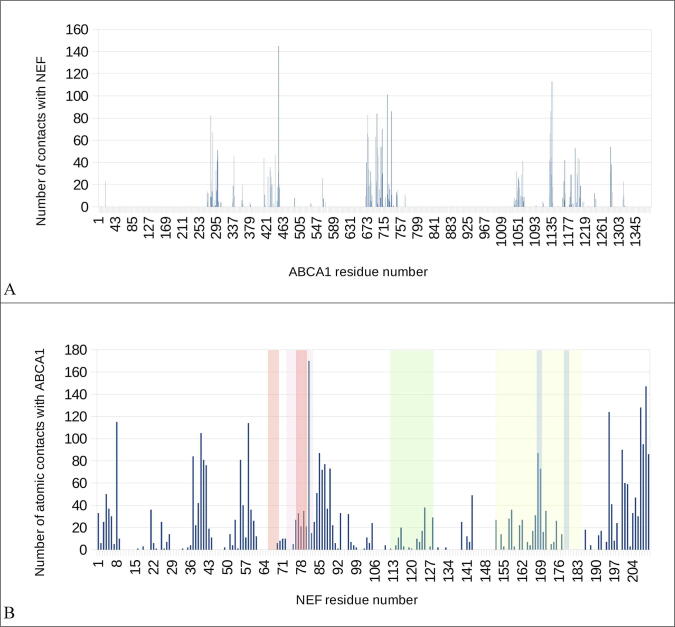
Thus, we have obtained 8 docking models (out of 64 models provided by global docking) of Nef:ABCA1 complexes that included the membrane. Of note, only cytoplasmic domains of ABCA1 were considered for these models. Global protein–protein docking gives only a rough approximation of interprotein interaction since rigid protein structure models are mostly used in that kind of docking. To rectify such models, molecular dynamics simulations of the resulting complexes with the membrane have been performed for 50 ns to obtain relaxed equilibrium structures. Final structures viewed from the top (panel A) or from the side (panel B) are shown in Fig. 1, and side views for each of the 8 docking models are shown in Supplementary Figs. S1-S8. The TM-score calculated for 8 ABCA1 structures from these models was 0.572, indicating a close structural similarity [46], [47]. For the Nef protein, the TM-score of 8 Nef structures was 0.649. The structures are very similar as illustrated in the Supplementary Fig. S9, which shows superimposed Nef structures after 50 ns of MD. Supplementary Table S2 lists the Nef amino acids involved in the interaction with ABCA1 in each model. Although some sites were present in several models, there were significant differences between the models (Table S2). To organize the data, the complex structures were analyzed by QASDOM server [33] to identify the Nef:ABCA1 interaction sites, showing that in 4 out of 8 docking models Nef was interacting with the same region of ABCA1 (Fig. 2A). Targeting the Nef sites was chosen as a preferred approach in developing inhibitory compounds, so the same analysis was performed for Nef. Compact regions displaying strong interaction with ABCA1 are clearly seen (Fig. 2B).
Fig. 1.
Results of 50 ns molecular dynamics simulations of the best 8 docking models of Nef:ABCA1 interaction. ABCA1 is colored orange, interaction regions are shown as surfaces of different colors. A – view from above the membrane, B – lateral view. Only the cytoplasmic part of ABCA1 was modeled (on top of the membrane in panel B). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Number of atom–atom contacts between ABCA1 and Nef after 50 ns of MD, overall data for 8 complex models. A - ABCA1 residues, B - Nef residues. Color shading shows residues involved in Nef interactions with other cellular proteins. MHC-I and AP-1 interaction sites 66–69 and 76–79 are shown with red; PTE1 interaction site 112–128 is colored green; binding region 152–184 to ATP6V1H is colored yellow; amino acids 168-LL-169 and 178-ED-179, necessary for CD4 internalization are colored light blue; the SH3 binding site 73-PVRPQVPLRP-82 is colored violet. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To put these regions in the context of previously identified interactions between Nef and other cellular proteins, these interaction sites are overlayed on Fig. 2B. We have identified four potential sites in Nef for inhibition of Nef:ABCA1 interaction: 1-MGGKWSKRSMGG-12 (Site I), 75-RPQVPLRPM-83 and 121-TQGY-124 (Site II), 84-TYKAALDI-91 (Site III), 201-ELHPEYYKDC-210 (Site IV) (Fig. 3). These sites, together with previously identified sites of Nef interaction with other cellular proteins, are shown in Supplementary Fig. S10. Nef regions 37-VSRDLE-42 and 55-TNADC-59 that showed strong interactions in the MD simulations have been excluded from the final data set because they are located in highly disordered parts of long loop areas of the protein and display high levels of RMSF (Fig. S11), indicating the extreme flexibility. Therefore, virtual screening for these areas would be unreliable. Two of the sites (Sites II and III) are contiguous and could be treated as one larger site (75-RPQVPLRPMYKAALD-90). However, the 75-RPQVPLRPMT-84 is a non-structured region, i.e. a loop stretched along the protein surface (Fig. 3), with length of about 25 A. As the mean length of the ligand is 15 A, we decided to break this long site into two separate sites (Site II and Site III) with the size consistent with ligand length. We also noticed in our previous studies that Autodock Vina does not work properly with larger grid boxes, leading to artifacts: usually all the docked molecules in a large box were placed in the same small area. This was avoided with grid boxes of 20 A or smaller (Table S1). Yet another reason to break the entire docking area into several smaller fragments is that, because of flexibility, the location of the docking site I relative to sites II and III varies considerably during MD simulations and therefore we did not observe a fixed docking pocket in this part of the protein even if the sites were treated as a single docking interface.
Fig. 3.
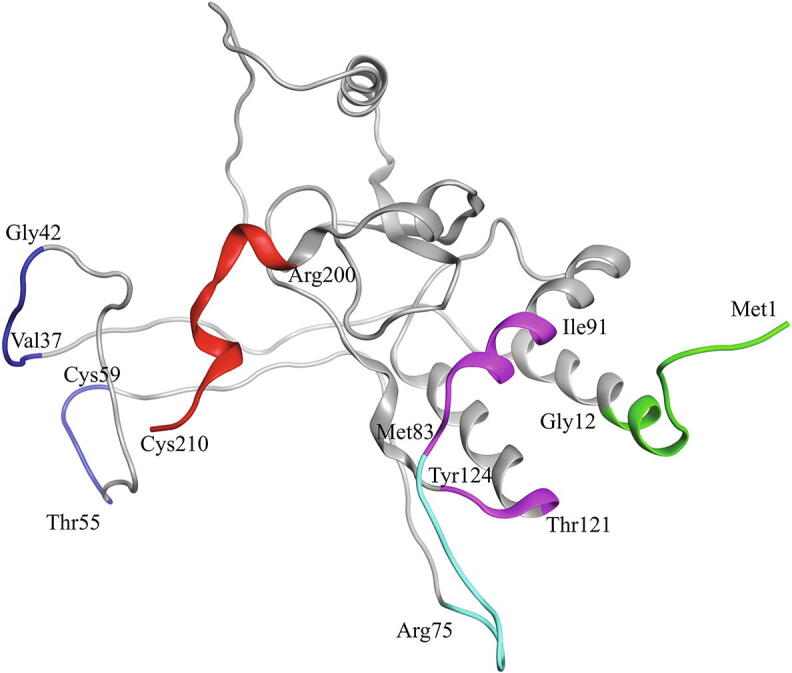
Potential ABCA1 interaction sites in Nef identified using the MD simulations. Site I is colored green, Site II is colored magenta, Site III is colored cyan and Site IV is colored red. Flexible (after MD simulations) sites 37-VSRDLE-42 and 55-TNADC-59 are shown in dark blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To search for potential inhibitors of the Nef:ABCA1 interaction, the following virtual screening runs have been carried out. (1) Virtual screening of Nef sites I-IV by docking program Vina [42] using ZINC NCI Plated 2007 dataset, which was used in our virtual screen for inhibitors of the Nef-calnexin interaction [23]. Ten putative ligands have been identified for each of the four sites and prioritized according to the Vina ranking (Table S3). (2) Since Sites I-III of the Nef:ABCA1 interface overlapped with the Nef sites previously modelled by to interact with calnexin [22], [23], we have included in the current screening the compounds ZINC03953858 (NSC13987) and AMS-55, which were predicted and experimentally confirmed in the above studies to bind to these sites and block Nef:calnexin interaction. We also docked a series of AMS-55-derived compounds, modified to improve potential effectiveness (see Discussion) and solubility (Table S4). In addition, 3 compounds provided by Dr. Smithgall (FC-8052 [48], FC-7976 [48], and DFP-4AP [49]), which have been shown to bind Nef with high affinity and potently inhibit HIV replication [48], were docked to each of these four sites (Table S5). In total, 144 putative ligands have been obtained (40 compounds from the ZINC screening and 26 additional compounds each docked to four sites). To improve the accuracy of docking of these compounds, a 30 ns MD simulations of each Nef:ligand complex in water with 0.15 M NaCl and pH of 7.0 was performed. Ten best putative ligands are listed in Table 1. Tables S3 and S5 provide the full dataset of the predicted putative ligands. The tables list the predicted inhibitory effectiveness parameters and ligand ranking for each of the compounds obtained from ZINC screening and the AMS-analogs. Tables S6–S9 show the detailed data of the compounds interactions with each of the 4 Nef sites for each of the 144 compounds, including docking affinity and description of the interaction between the compound and Nef after MD simulation.
Table 1.
Top 10 putative ligands for Nef interaction sites in the Nef:ABCA1 complexes. Ligands were selected after 30 ns MD simulations by the highest value of the overlap parameter from the full dataset of the predicted ligands.
| Ligand name | Image | Overlap parameter of each interaction site after MD simulations of Nef – ligand complexes. I-IV – Nef sites. | Total overlap parameter of interaction sites with the ligand |
|---|---|---|---|
| AMS-55 |  |
I 2.0 | 3.87 |
| II 0.77 | |||
| III 1.0 | |||
| IV 0.1 | |||
| FC-8052 |  |
I 1.25 | 3.84 |
| II 0.77 | |||
| III 1.22 | |||
| IV 0.60 | |||
| AMS-188 | 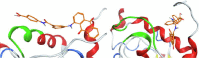 |
I 1.58 | 3.76 |
| II 0.85 | |||
| III 1.33 | |||
| IV 0.0 | |||
| AMS-123 | 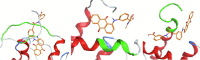 |
I 1.67 | 3.6 |
| II 1.15 | |||
| III 0.78 | |||
| IV 0.0 | |||
| AMS-191 | 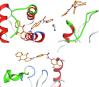 |
I 1.08 | 3.24 |
| II 1.38 | |||
| III 0.78 | |||
| IV 0.0 | |||
| AMS-201 | 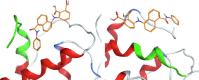 |
I 1.0 | 3.07 |
| II 0.54 | |||
| III 1.33 | |||
| IV 0.2 | |||
| ZINC03953858 (NSC13987) |  |
I 0.75 | 1.70 |
| II 0.62 | |||
| III 0.33 | |||
| IV 0.0 | |||
| ZINC05218086 |  |
I 0.25 | 1.38 |
| II 0.31 | |||
| III 0.22 | |||
| IV 0.60 | |||
| ZINC04522761 |  |
I 0.5 | 1.29 |
| II 0.23 | |||
| III 0.56 | |||
| IV 0.0 | |||
| ZINC03875800 |  |
I 0.58 | 1.26 |
| II 0.46 | |||
| III 0.22 | |||
| IV 0.0 |
The resulting structures of the Nef:ligand complexes were analyzed with QASDOM metaserver [33]. The overlap parameter has been calculated as a number of all Nef residues interacting with the putative ligand (compound) divided by the number of residues located in the site, for every site in every structure. Total overlap parameter is the sum of overlap parameters for four Nef sites, it characterizes the overall effectiveness of a compound as a putative ligand for all sites. This shows relative predicted effectiveness of inhibitors for the whole Nef:ABCA1 interaction in general. After MD simulations, the ligand either stayed at the initial docked position, or shifted to another site, or alternatively lost interaction with the protein (in the last case the ligand was excluded from the final data). As a result, the ligand could appear as docked to a single site or more than one site. In the detailed table of Nef interactions with ligands (Table 1) it can be seen that the cases where the ligand was interacting with two sites simultaneously or has shifted to the neighboring interaction site were quite frequent. For the formal analysis of all these cases for each of the putative ligands, overlap parameters for each of the four docking sites have been calculated. Notably, a compound can appear more than once in the screening results for different sites if it was identified by the Vina rating system for more than one site. For such ligands, there is more than one MD result available, as was seen for the AMS-55-derived ligands, resulting in an increase in overlap parameter for such compounds.
Hence, in every Nef:ligand complex structure after MD, overlap parameters have been calculated for each site and then summarized across all the MD systems (Tables S3, S5). In the tables, the total overlap parameters are given. The sum of these overlap parameter values for each site for a ligand can be treated as an indicator of its effectiveness in putative blocking of the interactions between ABCA1 and Nef. The ligands from the ZINC screening that appeared more than once subsequently gained an increase in rating. The cumulative ranking of the compounds across all 4 sites is shown in Table 1 and Tables S6-S9. These tables show that AMS-55-derived ligands previously found to block Nef:calnexin interaction [22] demonstrated stronger interaction with all the predicted ABCA1 binding sites in Nef, compared to the newly identified ZINC compounds.
The most effective ligands in our MD simulations were AMS-55 and its derivatives, AMS-11, AMS-123, AMS-188, AMS-191, and AMS-201 (Tables 1 and S5). To prove the stable binding of the ligands to Nef, we calculated the root mean square deviation (RMSD), root mean square fluctuation (RMSF), solvent accessible surface area (SASA), and Hbond binding energies of the AMS-55 ligand after MD simulation. Results presented in Supplementary Fig. S11 show values consistent with stable binding of the ligand to Nef.
4. Discussion
In this study, we built a computational model of the interaction between Nef and ABCA1. This model refines our previously published model [21], which focused on ABCA1, and identified the sites in Nef responsible for the interaction. We believe that, given the central role that ABCA1 plays in cholesterol metabolism [50], targeting the sites in Nef is a better approach for development of compounds that block the exNef-mediated downregulation of cell surface ABCA1. Interestingly, the Nef sites identified in this study to mediate interaction with ABCA1 overlapped with the sites involved in the interaction with calnexin [22], [23]. Nef is a relatively small protein (210 amino acids), but plays a major role in HIV pathogenesis by interacting with several cellular proteins (reviewed in [51]). Therefore, overlapping of some interaction sites may be expected.
We have performed docking and virtual screening, and used molecular dynamics to evaluate potential inhibitors of the Nef:ABCA1 interaction. In addition to the new compounds obtained from the virtual screening of the ZINC library, we docked to the Nef interaction sites a number of AMS compounds derived from the compound ZINC03953858 (NSC13987), which was identified in the previous screen and shown to block Nef-calnexin interaction [22], [23]. The NSC13987 compound also appeared in the two ZINC screening results in this study, for Nef sites I and III. It was not the only compound appearing in screening results for more than one site, but it was the best of all 40 compounds from the ZINC screening dataset according to its total overlap parameter. In addition, we docked to Nef sites 3 previously characterized Nef inhibitors, FC-8052 [48], FC-7976 [48], and DFP-4AP [49]. The MD approach provided an opportunity to fine-tune the ranking of the compounds according to their potential inhibitory activity. Unexpectedly, AMS-55 and some of its derivatives turned out to be among the top ranked compounds among the potential inhibitors of Nef:ABCA1 interaction. This appears to be a result of similarity in the interaction areas of Nef with ABCA1 and calnexin predicted by docking. This result suggests that these compounds may potentially inhibit both arms of the Nef effects on ABCA1: the effect on ABCA1 maturation in the endoplasmic reticulum via prevention of Nef-mediated disruption of the ABCA1:calnexin interaction, and the effect on the plasma membrane ABCA1 via prevention of the Nef interaction with ABCA1.
NSC13987 had a total overlap parameter of 2.67 (Table S5) and showed good rating for 3 out of 4 ABCA1 binding sites in Nef. It also came up in the ZINC screening results for 3 sites out of 4 (Table S3). There, because of a different screening (docking) method, it had a lower total overlap parameter. NSC13987 is a highly lipophilic compound due to the presence of benzanthrone and anthraquinone moieties, therefore, a number of derivatives were included in our virtual screening. We envisioned that simplifying the NSC13987 structure by replacing the polycyclic, hydrophobic components with relatively hydrophilic moieties would improve the bioavailability and drug likeliness of the resultant compounds. Chemically, NSC13987 is composed of three components: secondary amine, benzanthrone, and anthraquinone rings. The AMS-55 derivative has naphthylamide moiety replacing benzanthrone. This modification improved the solubility and activity of the compound [22]. We hypothesized that the placement of polar moieties on the phenyl ring in AMS-55 would improve the binding affinity to the identified ABCA1 interaction sites in Nef. Thus, we included in the test analogues with phenolic (AMS-191, AMS-192) and carboxylic acid (AMS-123) moieties on the phenyl ring. The benzene analogue (AMS-194) was designed to test the importance of naphthyl ring in AMS-55.
The redox-sensitive, toxic anthraquinone group was removed in some compounds subjected for docking. We proposed quinone, quinoxaline, and phenazine analogues that offered similar hydrogen bond donor–acceptor properties as anthraquinone. We replaced the anthraquinone in AMS-55 with phthalic acid, morpholine, piperazine and piperazinone in analogues AMS-201 to AMS-204. Analogues AMS-205 and AMS-206 with acyclic secondary amine and carboxylic acid moieties were developed to examine the impact of conformational freedom on biological activity. We also proposed the replacement of carboxamide with sulfonamide in analogues AMS-207 to AMS-209. Analogues AMS-201 to AMS-209 feature at least one site for ionization at physiological pH. Consequently, these compounds were predicted to display improved aqueous solubility as compared to AMS-55 (Table S4).
The most effective ligands in our MD simulations were AMS-55 and its derivatives, AMS-11, AMS-123, AMS-188, AMS-191, and AMS-201 (Tables 1 and S5). These compounds form stable interactions with the interaction sites I-IV, however, the overall interaction with site IV was weaker and less specific than it was with sites I-III. This result was expected as these compounds were constructed to block Nef:calnexin interaction sites, overlapping with the Nef:calnexin interaction sites I–III. In contrast with these ligands, the newly identified ZINC compounds were potentially more effective in blocking individual Nef sites. When these compounds appeared in more than one complex with Nef, the interaction was weaker and the blocked area smaller than that of the majority of the AMS-55-derived ligands.
The finding that one of the 3 previously characterized Nef inhibitors, the hydroxypyrazole compound FC-8052, exhibited impressive activity in binding to the predicted Nef sites involved in the interaction with ABCA1 was unexpected, as this compound was initially used as a negative control. The binding sites of this compound on HIV Nef have not been identified, but it was found to inhibit Nef dimerization [48]. The site responsible for Nef dimerization (Arg105-Asp123 [52]) overlaps with part of Site II (Thr121-Tyr124) of the Nef:ABCA1 interaction interface (Figs. 2 and S10). However, our computational model predicts a strong interaction between FC-8052 and Nef at two other sites: site III (aa 83–91) and site IV (aa 201–210 at the C-end). Unique H-bond donor/acceptor characteristics in FC-8052 may account for its potent binding to Nef. Given that hydroxypyrazole compounds were shown to have potent anti-HIV activity [48], it will be important to verify the ability of FC-8052 to suppress ABCA1 downmodulation by exNef. If this activity is confirmed, this compound would be a good candidate for further development, as it may inhibit both active HIV infection and co-morbidities associated with active and suppressed infection. Given the overlap between the polyproline Nef site (73-PVRPQVPLRP-82), which is responsible for Nef interactions with MHC-I, AP-1 and SH3 domain of Src-family kinase (SFK) [53], [54], with ABCA1 binding Site III, it is possible that other Nef inhibitors targeting these sites, such as Nef:SFK interaction inhibitors [55], [56], may be effective in blocking the Nef:ABCA1 interaction.
The mechanism by which exNef induces ABCA1 internalization remains unknown. We have previously noticed that endogenously expressed Nef reduces ABCA1 exposure on the cell surface [19] and that exNef selectively reduces ABCA1 in the lipid rafts [15]. These activities may be mediated by the known ability of Nef to connect certain plasma membrane proteins to the cell’s endocytosis machinery, directing them to the lysosomes for degradation [57]. Indeed, increased lysosomal degradation of ABCA1 was observed in Nef-expressing cells, and lysosome degradation inhibitor chloroquine reversed Nef-mediated depletion of ABCA1 [19].
In summary, a model of Nef:ABCA1 interaction described in this report allowed identification of potential inhibitory compounds that are predicted to inhibit the inactivating effect that Nef has on the activity of this critical regulator of cholesterol metabolism. The next step is to perform the in vitro testing of the high ranking candidates. Since some of them (e.g. AMS-55) have been shown to reverse the effects on ABCA1 of endogenously expressed Nef [22], it appears likely that they will be effective against exNef. Further optimization based on the created model will accelerate the development efforts. Given the established role of the pathogenic effect of Nef on ABCA1 function and cholesterol metabolism in pathogenesis of HIV-associated co-morbidities [7], development of these inhibitors is expected to advance therapeutic options in treatments of these diseases.
CRediT authorship contribution statement
Alexei A. Adzhubei: Methodology, Software, Validation. Amol Kulkarni: Conceptualization. Anna P. Tolstova: Investigation. Anastasia A. Anashkina: Investigation. Dmitri Sviridov: . Alexander A. Makarov: Supervision. Michael I. Bukrinsky: Supervision, Writing - original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to Dr. Thomas Smithgall for providing FC-8052, FC-7976 and DFP-4AP compounds. The study was supported by the Russian Foundation for Basic Research grant 17–54–30021, NIH grants R01NS102163, R01HL131473, R01HL158305 and P30AI117970.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.06.050.
Contributor Information
Alexei A. Adzhubei, Email: alexei.adzhubei@eimb.ru.
Michael I. Bukrinsky, Email: mbukrins@gwu.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Kestier H.W., Ringler D.J., Mori K., Panicali D.L., Sehgal P.K., Daniel M.D. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65(4):651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 2.Zaunders J., Dyer W.B., Churchill M. The Sydney Blood Bank Cohort: implications for viral fitness as a cause of elite control. Curr Opin HIV AIDS. 2011;6(3):151–156. doi: 10.1097/COH.0b013e3283454d5b. [DOI] [PubMed] [Google Scholar]
- 3.Usami Y., Wu Y., Göttlinger H.G. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526(7572):218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosa A., Chande A., Ziglio S., De Sanctis V., Bertorelli R., Goh S.L. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature. 2015;526(7572):212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piguet V., Schwartz O., Gall S., Trono D. The downregulation of CD4 and MHC-I by primate lentiviruses: a paradigm for the modulation of cell surface receptors. Immunol Rev. 1999;168(1):51–63. doi: 10.1111/j.1600-065x.1999.tb01282.x. [DOI] [PubMed] [Google Scholar]
- 6.Zicari S., Sessa L., Cotugno N., Ruggiero A., Morrocchi E., Concato C. Immune activation, inflammation, and non-AIDS co-morbidities in HIV-infected patients under long-term ART. Viruses. 2019;11(3):200. doi: 10.3390/v11030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sviridov D., Mukhamedova N., Makarov A.A., Adzhubei A., Bukrinsky M. Comorbidities of HIV infection: role of Nef-induced impairment of cholesterol metabolism and lipid raft functionality. AIDS. 2020;34:1–13. doi: 10.1097/QAD.0000000000002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raymond AD, Lang MJ, Chu J, Campbell-Sims T, Khan M, Bond VC, Pollard RB, Asmuth DM, Powell MD. Plasma-derived HIV Nef+ exosomes persist in ACTG384 study participants despite successful virological suppression. bioRxiv. 2019:708719.
- 9.Kodidela S., Gerth K., Haque S., Gong Y., Ismael S., Singh A. Extracellular vesicles: a possible link between HIV and Alzheimer's disease-like pathology in HIV subjects? Cells. 2019;8(9):968. doi: 10.3390/cells8090968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferdin J., Goričar K., Dolžan V., Plemenitaš A., Martin J.N., Peterlin B.M. Viral protein Nef is detected in plasma of half of HIV-infected adults with undetectable plasma HIV RNA. PLoS One. 2018;13(1):e0191613. doi: 10.1371/journal.pone.0191613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNamara R.P., Costantini L.M., Myers T.A., Schouest B., Maness N.J., Griffith J.D. Nef secretion into extracellular vesicles or exosomes is conserved across Human and Simian Immunodeficiency Viruses. MBio. 2018;9(1) doi: 10.1128/mBio.02344-17. e02344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sami Saribas A, Cicalese S, Ahooyi TM, Khalili K, Amini S, Sariyer IK. HIV-1 Nef is released in extracellular vesicles derived from astrocytes: evidence for Nef-mediated neurotoxicity. Cell Death Dis. 2017;8:e2542. [DOI] [PMC free article] [PubMed]
- 13.Khan M.B., Lang M.J., Huang M.B., Raymond A., Bond V.C., Shiramizu B. Nef exosomes isolated from the plasma of individuals with HIV-associated dementia (HAD) can induce Abeta1-42 secretion in SH-SY5Y neural cells. J Neurovirol. 2016;22:179–190. doi: 10.1007/s13365-015-0383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ditiatkovski M., Mukhamedova N., Dragoljevic D., Hoang A., Low H., Pushkarsky T. Modification of lipid rafts by extracellular vesicles carrying HIV-1 protein Nef induces redistribution of APP and Tau causing neuronal dysfunction. J Biol Chem. 2020;295:13377–13392. doi: 10.1074/jbc.RA120.014642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukhamedova N., Hoang A., Dragoljevic D., Dubrovsky L., Pushkarsky T., Low H. Exosomes containing HIV protein Nef reorganize lipid rafts potentiating inflammatory response in bystander cells. PLoS Pathog. 2019;15(7):e1007907. doi: 10.1371/journal.ppat.1007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mujawar Z., Rose H., Morrow M.P., Pushkarsky T., Dubrovsky L., Mukhamedova N. Human immunodeficiency virus impairs reverse cholesterol transport from macrophages. PLoS Biol. 2006;4(11):e365. doi: 10.1371/journal.pbio.0040365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yvan-Charvet L., Wang N., Tall A.R. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol. 2010;30(2):139–143. doi: 10.1161/ATVBAHA.108.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mujawar Z., Tamehiro N., Grant A., Sviridov D., Bukrinsky M., Fitzgerald M.L. Mutation of the ATP cassette binding transporter A1 (ABCA1) C-terminus disrupts HIV-1 Nef binding but does not block the Nef enhancement of ABCA1 protein degradation. Biochemistry. 2010;49(38):8338–8349. doi: 10.1021/bi100466q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui H.L., Grant A., Mukhamedova N., Pushkarsky T., Jennelle L., Dubrovsky L. HIV-1 Nef mobilizes lipid rafts in macrophages through a pathway that competes with ABCA1-dependent cholesterol efflux. J Lipid Res. 2012;53(4):696–708. doi: 10.1194/jlr.M023119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buffalo C.Z., Iwamoto Y., Hurley J.H., Ren X., Pierson T.C. How HIV Nef Proteins Hijack membrane traffic to promote infection. J Virol. 2019;93(24) doi: 10.1128/JVI.01322-19. e01322-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob D., Hunegnaw R., Sabyrzyanova T.A., Pushkarsky T., Chekhov V.O., Adzhubei A.A. The ABCA1 domain responsible for interaction with HIV-1 Nef is conformational and not linear. Biochem Biophys Res Commun. 2014;444(1):19–23. doi: 10.1016/j.bbrc.2013.12.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adzhubei AA, Anashkina AA, Tkachev YV, Kravatsky YV, Pushkarsky T, Kulkarni A, Makarov AA, Bukrinsky MI. Modelling interaction between HIV-1 Nef and calnexin. AIDS. 2018;32:2103-11. [DOI] [PMC free article] [PubMed]
- 23.Hunegnaw R., Vassylyeva M., Dubrovsky L., Pushkarsky T., Sviridov D., Anashkina A.A. Interaction between HIV-1 Nef and calnexin: from modeling to small molecule inhibitors reversing HIV-induced lipid accumulation. Arterioscler Thromb Vasc Biol. 2016;36(9):1758–1771. doi: 10.1161/ATVBAHA.116.307997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jennelle L., Hunegnaw R., Dubrovsky L., Pushkarsky T., Fitzgerald M.L., Sviridov D. HIV-1 protein Nef inhibits activity of ATP-binding cassette transporter A1 by targeting endoplasmic reticulum chaperone calnexin. J Biol Chem. 2014;289(42):28870–28884. doi: 10.1074/jbc.M114.583591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubrovsky L., Ward A., Choi S.H., Pushkarsky T., Brichacek B., Vanpouille C. Inhibition of HIV replication by apolipoprotein A-I binding protein targeting the lipid rafts. mBio. 2020;11:e02956–e3019. doi: 10.1128/mBio.02956-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mollica A., Zengin G., Durdagi S., Ekhteiari Salmas R., Macedonio G., Stefanucci A. Combinatorial peptide library screening for discovery of diverse alpha-glucosidase inhibitors using molecular dynamics simulations and binary QSAR models. J Biomol Struct Dyn. 2019;37:726–740. doi: 10.1080/07391102.2018.1439403. [DOI] [PubMed] [Google Scholar]
- 27.Poli G., Dimmito M.P., Mollica A., Zengin G., Benyhe S., Zador F. Discovery of novel micro-opioid receptor inverse agonist from a combinatorial library of tetrapeptides through structure-based virtual screening. Molecules. 2019;24:3872. doi: 10.3390/molecules24213872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma A., Thelma B.K. Pharmacophore modeling and virtual screening in search of novel Bruton's tyrosine kinase inhibitors. J Mol Model. 2019;25:179. doi: 10.1007/s00894-019-4047-y. [DOI] [PubMed] [Google Scholar]
- 29.Gentile D., Patamia V., Scala A., Sciortino M.T., Piperno A., Rescifina A. Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar Drugs. 2020;18(4):225. doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chahal V., Nirwan S., Pathak M., Kakkar R. Identification of potent human carbonic anhydrase IX inhibitors: a combination of pharmacophore modeling, 3D-QSAR, virtual screening and molecular dynamics simulations. J Biomol Struct Dyn. 2020;1–16 doi: 10.1080/07391102.2020.1860132. [DOI] [PubMed] [Google Scholar]
- 31.Gerlach H., Laumann V., Martens S., Becker C.F.W., Goody R.S., Geyer M. HIV-1 Nef membrane association depends on charge, curvature, composition and sequence. Nat Chem Biol. 2010;6(1):46–53. doi: 10.1038/nchembio.268. [DOI] [PubMed] [Google Scholar]
- 32.Klappe K., Hummel I., Hoekstra D., Kok J.W. Lipid dependence of ABC transporter localization and function. Chem Phys Lipids. 2009;161(2):57–64. doi: 10.1016/j.chemphyslip.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Anashkina AA, Kravatsky Y, Kuznetsov E, Makarov AA, Adzhubei AA. Meta-server for automatic analysis, scoring and ranking of docking models. Bioinformatics. 2018;34:297-9. [DOI] [PubMed]
- 34.Qian H., Zhao X., Cao P., Lei J., Yan N., Gong X. Structure of the human lipid exporter ABCA1. Cell. 2017;169(7):1228–1239. doi: 10.1016/j.cell.2017.05.020. e10. [DOI] [PubMed] [Google Scholar]
- 35.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 36.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B. High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 37.Kelley L.A., Sternberg M.J.E. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 38.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Källberg M., Wang H., Wang S., Peng J., Wang Z., Lu H. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7(8):1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rykunov D., Steinberger E., Madrid-Aliste C.J., Fiser A. Improved scoring function for comparative modeling using the M4T method. J Struct Funct Genomics. 2009;10(1):95–99. doi: 10.1007/s10969-008-9044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195-201. [DOI] [PubMed]
- 42.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanner M.F. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 44.Sterling T., Irwin J.J. ZINC 15–ligand discovery for everyone. J Chem Inf Model. 2015;55(11):2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koziara K.B., Stroet M., Malde A.K., Mark A.E. Testing and validation of the Automated Topology Builder (ATB) version 2.0: prediction of hydration free enthalpies. J Comput Aided Mol Des. 2014;28(3):221–233. doi: 10.1007/s10822-014-9713-7. [DOI] [PubMed] [Google Scholar]
- 46.Dong R., Pan S., Peng Z., Zhang Y., Yang J. mTM-align: a server for fast protein structure database search and multiple protein structure alignment. Nucl Acids Res. 2018;46:W380–W386. doi: 10.1093/nar/gky430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26:889-95. [DOI] [PMC free article] [PubMed]
- 48.Shi H., Tice C.M., Emert-Sedlak L., Chen L.i., Li W.F., Carlsen M. Tight-binding hydroxypyrazole HIV-1 Nef inhibitors suppress viral replication in donor mononuclear cells and reverse Nef-Mediated MHC-I downregulation. ACS Infect Dis. 2020;6(2):302–312. doi: 10.1021/acsinfecdis.9b00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emert-Sedlak L., Kodama T., Lerner E.C., Dai W., Foster C., Day B.W. Chemical library screens targeting an HIV-1 accessory factor/host cell kinase complex identify novel antiretroviral compounds. ACS Chem Biol. 2009;4(11):939–947. doi: 10.1021/cb900195c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang N., Tall A.R. Regulation and mechanisms of ATP-binding cassette transporter A1-mediated cellular cholesterol efflux. Arterioscler Thromb Vasc Biol. 2003;23(7):1178–1184. doi: 10.1161/01.ATV.0000075912.83860.26. [DOI] [PubMed] [Google Scholar]
- 51.Saxena R., Vekariya U., Tripathi R. HIV-1 Nef and host proteome analysis: current perspective. Life Sci. 2019;219:322–328. doi: 10.1016/j.lfs.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 52.Poe J.A., Smithgall T.E. HIV-1 Nef dimerization is required for Nef-mediated receptor downregulation and viral replication. J Mol Biol. 2009;394(2):329–342. doi: 10.1016/j.jmb.2009.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuo L.S., Baugh L.L., Denial S.J., Watkins R.L., Liu M., Garcia J. Overlapping effector interfaces define the multiple functions of the HIV-1 Nef polyproline helix. Retrovirology. 2012;9(1):47. doi: 10.1186/1742-4690-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pene-Dumitrescu T., Shu S.T., Wales T.E., Alvarado J.J., Shi H., Narute P. HIV-1 Nef interaction influences the ATP-binding site of the Src-family kinase. Hck. BMC Chem Biol. 2012;12(1):1. doi: 10.1186/1472-6769-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Betzi S., Restouin A., Opi S., Arold S.T., Parrot I., Guerlesquin F. Protein protein interaction inhibition (2P2I) combining high throughput and virtual screening: application to the HIV-1 Nef protein. Proc Natl Acad Sci USA. 2007;104(49):19256–19261. doi: 10.1073/pnas.0707130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dikeakos J.D., Atkins K.M., Thomas L., Emert-Sedlak L., Byeon I.-J., Jung J. Small molecule inhibition of HIV-1-induced MHC-I down-regulation identifies a temporally regulated switch in Nef action. Mol Biol Cell. 2010;21(19):3279–3292. doi: 10.1091/mbc.E10-05-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.daSilva L.L.P., Sougrat R., Burgos P.V., Janvier K., Mattera R., Bonifacino J.S. Human immunodeficiency virus type 1 Nef protein targets CD4 to the multivesicular body pathway. J Virol. 2009;83(13):6578–6590. doi: 10.1128/JVI.00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.