Abstract
Chromatin accessibility states that influence gene expression and other nuclear processes can be altered in disease. The constellation of transcription factors and chromatin regulatory complexes in cells results in characteristic patterns of chromatin accessibility. The study of these patterns in tissues has been limited because existing chromatin accessibility assays are ineffective for archival formalin-fixed, paraffin-embedded (FFPE) tissues. We have developed a method to efficiently extract intact chromatin from archival tissue via enhanced cavitation with a nanodroplet reagent consisting of a lipid shell with a liquid perfluorocarbon core. Inclusion of nanodroplets during the extraction of chromatin from FFPE tissues enhances the recovery of intact accessible and nucleosome-bound chromatin. We show that the addition of nanodroplets to the chromatin accessibility assay formaldehyde-assisted isolation of regulatory elements (FAIRE), does not affect the accessible chromatin signal. Applying the technique to FFPE human tumor xenografts, we identified tumor-relevant regions of accessible chromatin shared with those identified in primary tumors. Further, we deconvoluted non-tumor signal to identify cellular components of the tumor microenvironment. Incorporation of this method of enhanced cavitation into FAIRE offers the potential for extending chromatin accessibility to clinical diagnosis and personalized medicine, while also enabling the exploration of gene regulatory mechanisms in archival samples.
The dynamic regulation of chromatin architecture is a central epigenetic process. DNA wrapped around histone octamers forms nucleosomes, the basic unit of chromatin organization. By restricting access to DNA, nucleosomes present the primary barrier to DNA templated processes including transcription and replication. Nucleosome-depleted regions coincide with active transcription start sites and functional enhancers. The pattern of accessible chromatin is specific to cell states and marks cell identity during normal development and in disease (Patel et al. 2012; Stergachis et al. 2013; Yu et al. 2016; Flavahan et al. 2017; Lewis et al. 2017; Davis and Pattenden 2019). The identification of sites of differential chromatin accessibility also offers diagnostic potential (Patel et al. 2012; Simon et al. 2012, 2014). Consequently, there is considerable interest in the detection of specific chromatin accessibility patterns in both diseased and normal tissues (Stergachis et al. 2013; Corces et al. 2016, 2018; Yu et al. 2016; Cusanovich et al. 2018; Satpathy et al. 2018; Lareau et al. 2019). For instance, chromatin variation has been used to classify previously unidentified cancer subtypes (Corces et al. 2018) and is associated with therapeutic resistance (Goulet et al. 2020). Regulatory element use can define cell identity and developmental trajectory in human leukemias (Corces et al. 2016). Chromatin accessibility patterns enforce lineage-specific transcriptional constraints during development and therefore constitute better early predictors of stem cell fate than gene expression profiles alone (Stergachis et al. 2013; Domcke et al. 2020; Ma et al. 2020). Further, we and others have shown that specific changes in chromatin accessibility can result from treatment with small molecule inhibitors (Hurtado et al. 2011; Patel et al. 2012; Pattenden et al. 2016).
Despite its potential application as a biomarker, studies of chromatin accessibility have focused on fresh tissues or cells. The most frequently used techniques, such as ATAC and DNase I hypersensitivity rely on enzymatic treatments that require intact nuclei. The ability to study chromatin accessibility in archival tissues would enable retrospective studies using patient samples to explore chromatin alterations associated with specific diseases or therapeutic resistance. Formalin fixation followed by paraffin embedding (FFPE) is the standard method to archive diagnostic specimens. Preservation by FFPE is inexpensive and yields tissue samples that can be stored at room temperature for extended periods of time (Bass et al. 2014). FFPE tissues represent a remarkable repository of samples for both biomedical research and diagnostics. Access to archival tissue permits examination of rare diseases that are often associated with clinical information, such as comorbidities, treatment outcomes, and family histories (Donczo and Guttman 2018; Gaffney et al. 2018). Although nucleic acids can be isolated from FFPE tissues using a variety of commercial methods, the extraction of intact chromatin has proven virtually impossible. Harsh treatments needed to reverse the extensive chemical cross-linking degrade the specimen, ultimately preventing successful use of the chromatin in downstream assays. Gentler and uniform sample processing would make FFPE samples amenable to chromatin accessibility assays.
We developed a nanodroplet cavitation enhancement reagent with unique qualities that facilitate robust extraction of high-quality chromatin for chromatin immunoprecipitation (ChIP) in cells (Kasoji et al. 2015; Chiarella et al. 2018). In this study, we asked whether this reagent would enable the detection of chromatin accessibility FFPE cells and tissues. Formaldehyde-assisted isolation of regulatory elements (FAIRE) is a biochemical strategy that depends on stabilized protein–DNA interactions to separate accessible (nucleosome-free chromatin) from histone-bound DNA (Giresi et al. 2007; Giresi and Lieb 2009; Simon et al. 2013). Because the technique depends on formalin cross-linking and fragmented chromatin rather than intact nuclei, it is compatible with fixed samples. The formalin fixation implicit in FFPE processing eliminates the need for additional fixation such that accessible chromatin can be separated directly from the soluble chromatin fraction. Here, we evaluate the inclusion of nanodroplet cavitation enhancement into the FAIRE assay for FFPE tissues to identify chromatin accessibility genome-wide.
Results
Nanodroplets facilitate extraction of high-quality chromatin from cross-linked cells
We had previously showed that nanodroplets can facilitate the extraction of chromatin from cross-linked cells for use in chromatin immunoprecipitation (ChIP) analyzed by the quantitative polymerase chain reaction (qPCR) (Chiarella et al. 2018). Based on this result, we asked whether nanodroplets could similarly enhance the isolation of intact chromatin from FFPE tissues. In contrast to ChIP that depends on the interaction of an antibody-recognized protein with chromatin, FAIRE requires the preservation of histone/DNA interactions. Consequently, FAIRE analyzed by high-throughput sequencing (FAIRE-seq) offers a relevant method to evaluate the preservation of intact (yet fragmented) chromatin.
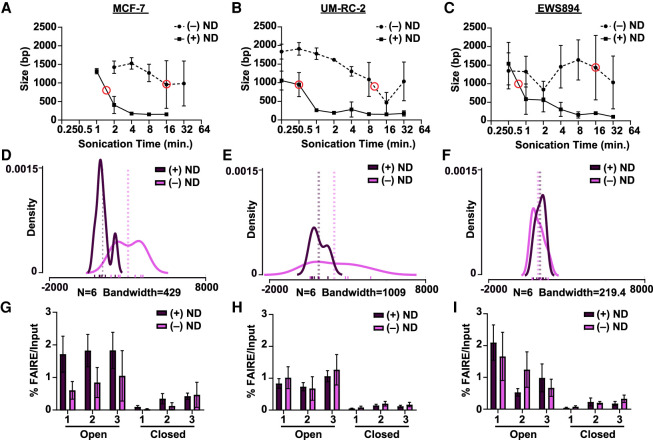
To evaluate the effect of nanodroplets on chromatin fragmentation, luminal breast cancer (MCF-7), clear cell renal cell carcinoma (ccRCC, UM-RC-2), and Ewing sarcoma (EWS894) cells were fixed and embedded into paraffin blocks, and sections were sonicated for various lengths of time with or without nanodroplets. DNA fragment size was then measured (Fig. 1A–C). For all cell lines, inclusion of nanodroplets decreased the sonication time necessary to achieve our target DNA fragment size (750–1000 bp, 10.7-fold for MCF-7, 20.0-fold for UM-RC-2, and 21.3-fold for EWS894). In addition, inclusion of nanodroplets greatly increased the consistency of the fragment sizes for two of the three cell lines (Fig. 1D–F). qPCR quantification (three biological and two technical replicates per cell line) (Fig. 1G–I) showed that inclusion of the nanodroplets did not influence signal enrichment at accessible chromatin (open) relative to inaccessible (closed) (Fig. 1G,H).
Figure 1.
Nanodroplets facilitate extraction of high-quality chromatin from cross-linked cells. (A–C) Peak DNA fragment size (base pairs, y-axis) after sonication time (minutes, x-axis) for MCF-7, UM-RC-2, and EWS894cells (Agilent TapeStation 2200). Data represent the average of three biological replicate samples sonicated with (solid line) or without (dashed line) nanodroplets (ND). Red circles indicate the time point at which peak DNA size reached the target range of 750–1000 bp. (D–F) Density distribution of fragment sizes for FAIRE input DNA with and without nanodroplets from MCF-7, UM-RC-2, and EWS894 cells, respectively, at data points circled (in red) in A, B, and C, respectively. Actual fragment lengths for each of six replicates within each group delineated with rug plot beneath density plots (without nanodroplets, lavender; with nanodroplet, dark purple). Bandwidth for each density plot displayed beneath each graph. Average fragment length within each group per cell line is displayed by dotted vertical lines. (G–I) Percent FAIRE-qPCR signal over input with (dark purple) and without nanodroplets (lavender) from MCF-7, UM-RC-2, and EWS894 cells, respectively, at data points circled (in red) in A, B, C, respectively. (Open) regions of known accessible chromatin; (Closed) regions of known inaccessible chromatin.
Pattern of extracted accessible chromatin signal is unaffected by inclusion of nanodroplets
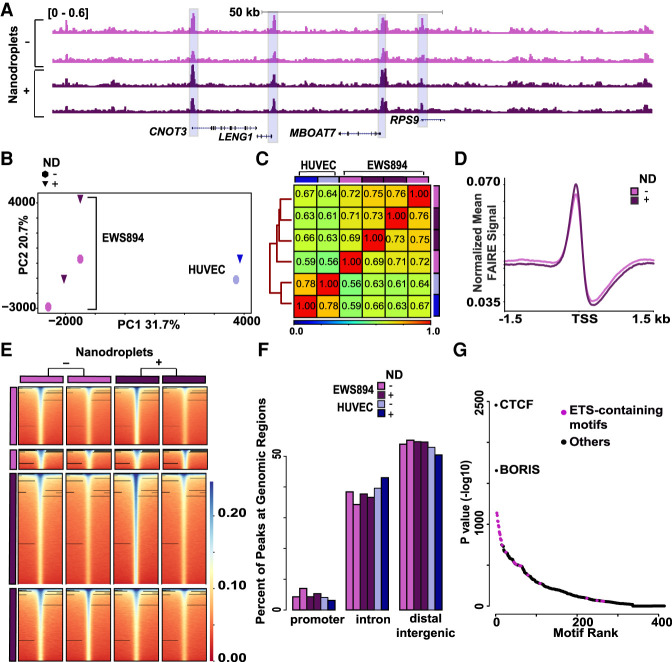
We then asked whether the inclusion of nanodroplets influenced FAIRE signal in FFPE preserved cells genome-wide. FAIRE-seq was performed on chromatin isolated from EWS894 cells and human umbilical vein endothelial cells (HUVEC) with or without nanodroplets. FAIRE signal showed a very similar pattern of enrichment across the conditions and between replicates (Fig. 2A). We then compared FAIRE signal genome-wide. The inclusion of nanodroplets resulted in correlation between FAIRE signals similar to that found between the replicates (no ND vs. ND, r = 0.89; no ND replicates, r = 0.79; ND replicates, r = 0.8) (Supplemental Fig. S1A–C). Principal component analysis on the genome-wide signal matrix showed that inclusion of nanodroplets did not influence clustering along any axes of variation (Fig. 2B; Supplemental Fig. S2). The largest axis of variation (PC1) was associated with cell type. We then calculated the Spearman's correlation coefficients between all replicates and cell types. FAIRE signal clustered by cell type without influence of the inclusion of nanodroplets (Fig. 2C). The signal pattern at transcriptional start sites (TSSs) was unaffected by nanodroplet addition (Fig. 2D). We identified regions of enrichment with and without nanodroplets (n = 2, MACS) (Zhang et al. 2008). FAIRE signal 2 kb around each replicate set of regions of enrichment was similar across all replicates, regardless of whether nanodroplets were included (Fig. 2E). The inclusion of nanodroplets was associated with detection of more regions of signal enrichment. However, signal enrichment was detected at all regions, regardless of sample processing. The distribution of peak locations was not influenced by nanodroplet inclusion (promoters P = 0.47; introns P = 0.55; distal intergenic P = 1) (Fig. 2F; Supplemental Fig. S3). Next, we identified DNA sequence motifs preferentially detected in the FAIRE enriched regions (HOMER) (Heinz et al. 2010) using pooled sequences from all EWS894 replicates. Following CTCF/BORIS, the most highly enriched motifs contained ETS motifs, marked by a central GGAA sequence, to which the critical oncoprotein of Ewing sarcoma, EWSR1-FLI1 (also known as EWS-FLI1), binds (Fig. 2G; Patel et al. 2012). Taken together, these results show that introduction of nanodroplets to FAIRE performed in fixed cells does not influence the identification of accessible chromatin and preserves the ability to detect biologically relevant regions.
Figure 2.
Use of nanodroplets does not influence detection of chromatin accessibility. (A) Normalized sequencing tracks of FAIRE-seq signal without (lavender) or with (dark purple) nanodroplets (EWS894). Signal for each replicate is shown. Regions of enrichment shared across all replicates are highlighted in blue. (B) Top two principal components of mean FAIRE-seq signal in 300-bp windows genome-wide across EWS894 (purple) and HUVEC cell lines (blue) identified by FAIRE with and without nanodroplets. Each symbol represents a FAIRE-seq replicate data set. Circles denote signal identified by FAIRE without nanodroplets, inverted triangles by nanodroplet FAIRE. (C) Spearman's correlation heatmap of genome-wide FAIRE-seq signal (300-bp windows) across replicates in EWS894 and HUVEC cell lines (lavender and sky blue, without nanodroplets; dark purple and blue, with nanodroplets). Clustering distances across samples determined by the nearest point algorithm depicted by tree on the left. (D) Normalized mean EWS894 FAIRE signal by FAIRE without nanodroplets (lavender) and with nanodroplets (dark purple) at all transcription start sites with non-zero signal genome-wide. Signal ±1.5 kb around transcription start sites is shown. (E) Signal around regions of enrichment for each replicate of EWS894 FAIRE signal found by FAIRE with and without nanodroplets. Signal enrichment ±2 kb around peak center shown. All heatmaps sorted by sum of signal in each peak set within which peaks were called. (F) Bar plot of the percentage of peaks at genomic locations called from EWS894 and HUVEC FAIRE in each replicate found without (lavender and sky blue, respectively) and with nanodroplet FAIRE (dark purple and blue, respectively). (G) Enriched motifs identified at union set of peaks identified from pooled replicates of EWS894 FAIRE signal found by FAIRE with and without nanodroplets. Motifs are plotted base on rank (x-axis) and enrichment P-value (y-axis). ETS-containing motifs are shown in purple.
Nanodroplets facilitate the extraction of chromatin from archival tissue
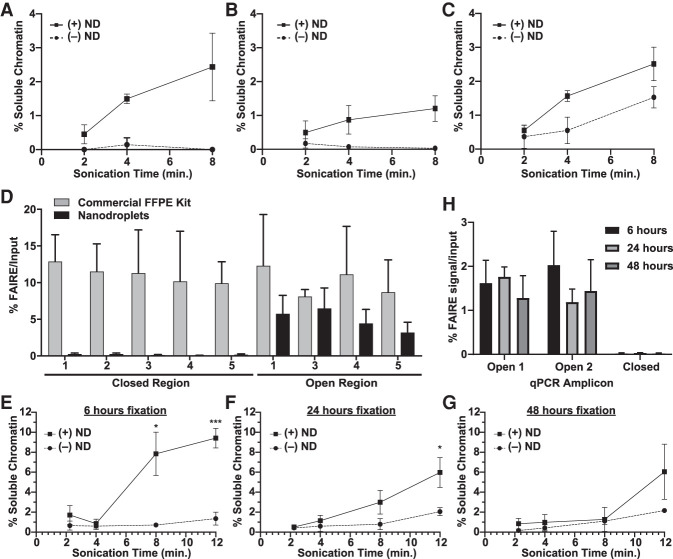
Unlike cultured cells, tissues are characterized by mixtures of cells surrounded by extracellular matrix components. Cross-linking intended to maintain tissue architecture for histology complicates fragmentation. Because nanodroplets greatly enhanced chromatin yield from cross-linked cells, we asked whether this approach would enable the extraction of intact chromatin from FFPE tissue. One 10-µm section of FFPE xenograft tissue (nuclei count) (Supplemental Table S1) in biological triplicate (xenografted tumors from three different mice) was deparaffinized, rehydrated, and then sonicated for different lengths of time in the presence or absence of nanodroplets. Inclusion of nanodroplets significantly increased the fractional yield of soluble chromatin following 4 min of sonication for xenografted UM-RC-2 clear cell renal cell carcinoma (ccRCC) tumors (P = 0.011) (Fig. 3A), EWS894 Ewing sarcoma tumors (P = 0.044) (Fig. 3B), and MDA-MB-231 basal breast tumors (P = 0.008) (Fig. 3C).
Figure 3.
Nanodroplets facilitate the extraction of chromatin from archival tissue. Percent soluble chromatin from one 10-µm section of UM-RC-2 (A), EWS894 (B), and MDA-MB-231 (C) following sonication with (solid) or without (dashed line) for indicated. (D) FAIRE-qPCR was performed on chromatin prepared from EWS894 xenograft tumor tissue with a commercial FFPE chromatin preparation kit (eight 10-µm sections, gray bars) or with nanodroplets (one 10-µm section, black bars). The y-axis indicates the percent FAIRE signal compared to input signal by qPCR. (Open) regions of known accessible chromatin; (Closed) regions of known inaccessible chromatin. (E–H) Xenograft tumor tissue derived from MCF-7 cells was fixed for 6 h (E), 24 h (F), or 48 h (G), and one, 10-μm section of each was deparaffinized, rehydrated, and sonicated with (solid line) or without (dashed line) nanodroplets for indicated time (x-axis, minutes). Percent soluble chromatin (y-axis) was measured by comparing the quantity of DNA in the soluble fraction to that of the insoluble pellet fraction. (H) FAIRE-qPCR was performed on one, 10-µm tissue section sonicated for 8 min with nanodroplets for each fixation time point. The y-axis indicates the percent FAIRE signal compared to input signal by qPCR. (Open) regions of known accessible chromatin; (Closed) regions of known inaccessible chromatin.
Commercially available products currently enable the extraction of chromatin from FFPE tissues for ChIP. However, because of the extraction method, it was not clear whether isolated chromatin would be compatible with FAIRE. Chromatin was prepared from eight 10-µm sections of EWS894 xenograft tissue in biological triplicate according to the manufacturer's instructions. Chromatin from one 10-µm section of the same tissue blocks was also isolated using nanodroplets. FAIRE-qPCR was then performed on regions of chromatin that are known to be accessible (open, FAIRE-positive) or inaccessible (closed, FAIRE-negative) (Fig. 3D). FAIRE signal at accessible regions was not greater than background using chromatin prepared by the kit method. In contrast, chromatin prepared using nanodroplets showed a highly significant enrichment of accessible chromatin. These data indicate that current commercial methods of chromatin extraction are not compatible with FAIRE.
Pathology protocols for tissue fixation and embedding are standardized. However, clinical circumstances may result in variation in fixation times. To evaluate the influence of fixation time on FAIRE, we divided MCF-7 xenografted tumors from three different animals and fixed matched tissue for 6, 24, and 48 h. We then compared soluble chromatin yields from samples (single 10-µm sections) sonicated in the presence or absence of nanodroplets (Fig. 3E–G). As expected, we found that increased fixation time decreased overall soluble chromatin yield for both conditions. However, regardless of fixation time, the presence of nanodroplets greatly increased soluble chromatin yield. We then assessed the quality of the soluble chromatin by FAIRE-qPCR. For all fixation times, including 48 h, soluble chromatin isolated with nanodroplets showed a significant difference between accessible chromatin signal compared to that of background (Fig. 3H).
Nanodroplets enable cell-specific detection of chromatin accessibility in archival tissue
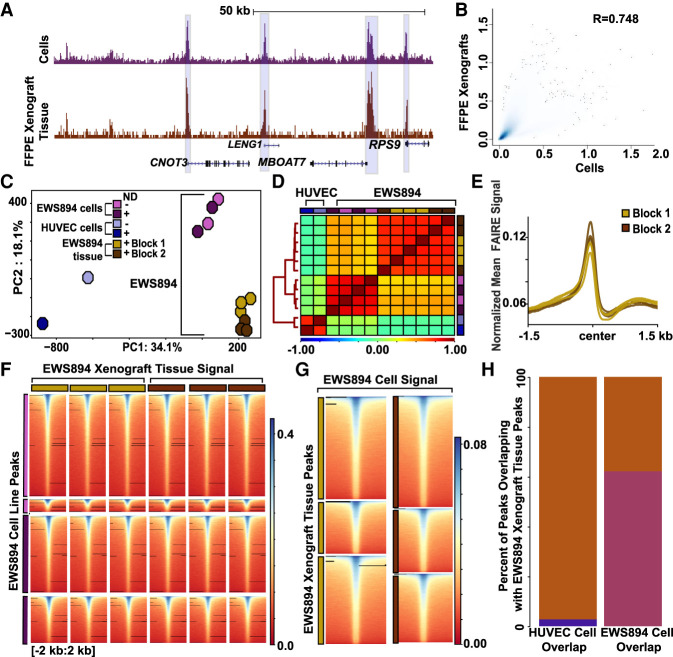
We then compared the genome-wide performance of nanodroplet-assisted FAIRE on archival tissue to that of fixed cells. FAIRE-seq was performed on two biological replicates of EWS894 mouse FFPE xenograft tissue (three technical replicates per xenograft) in the presence of nanodroplets. Tissue and cells showed a similar FAIRE signal (Fig. 4A). Next, we compared genome-wide signal between the replicates of each xenograft block and found high correlation within each block (Spearman's correlation coefficient, r = 0.77–0.83) (Supplemental Fig. S4). We then compared the genome-wide signal from EWS894 cells to the xenografted tissue (Fig. 4B). The genome-wide correlation between the cells and archival tissue was comparable to that found between xenograft block replicates (Spearman's correlation coefficient r = 0.876). We then identified regions of signal enrichment for each replicate of EWS894 xenograft tissue with and without nanodroplets (MACS2). These regions were combined (from Fig. 2) to create one union set. Principle component analysis of the mean FAIRE signal at sites in this set as well as in sites similarly identified in HUVEC cells showed a clear separation by cell type, and this separation was maintained regardless of whether the signal was derived from fixed cells or archival tissue (Fig. 4C). Correlation of signal in the union set of enriched regions clustered by cell type, with EWS894 cell and xenografts more highly correlated with each other than with HUVEC cells (Fig. 4D). FAIRE signal at TSSs was similar across all replicates for each xenograft block (Fig. 4E). We next evaluated xenograft FAIRE signal at regions of signal enrichment detected in cell lines. Regions of enrichment identified in chromatin from cells showed a similar relative signal enrichment in chromatin from xenografted tumors (Fig. 4F). We then explored this relationship in the opposite direction, comparing regions of signal enrichment in xenograft-derived chromatin to that of cell-line-derived chromatin. Again, we found that regions of signal enrichment in archival tissue were also enriched in cells (Fig. 4G). Sixty percent of sites detected in EWS894 cells were also detected in EWS894 xenografts, compared with <10% common peaks between HUVEC cells and EWS894 xenografts (Fig. 4H). These analyses show that chromatin accessibility patterns identified from archival tissues are similar to those from cells in culture. Although not specifically explored here, this similarity is despite likely transcriptional and chromatin differences between cells in culture and grown as xenografts.
Figure 4.
Nanodroplet FAIRE identifies cell-type-specific regions of accessible chromatin from FFPE xenograft tissue. (A) Normalized FAIRE-seq sequencing tracks of EWS894 cell line signal (FAIRE replicates with and without nanodroplets combined) and EWS894 xenograft FFPE tissue (two tissue blocks, three replicates each, combined). Cell line signal depicted in purple, FFPE tissue signal in brown. Regions of enrichment shared across all tracks are highlighted in blue. (B) Correlation of genome-wide signal (300-bp windows), between EWS894 cells (replicates pooled), and EWS894 xenograft tissue (replicates and blocks pooled). Spearman's correlation coefficient is shown. (C) Top two principal components of mean FAIRE-seq signal in the union set of peaks called across all data sets, EWS894 cells (lavender, without nanodroplets; purple, with nanodroplets), EWS894 xenograft tissue (gold, FFPE block 1; brown, FFPE block 2), and HUVEC cell lines (sky blue, without nanodroplets; blue, with nanodroplets). Each symbol represents a FAIRE-seq data set. (D) Spearman's correlation of mean FAIRE-seq signal in union set of peaks called across all replicates in EWS894 (cells and xenografts) and HUVEC (cells). Distances across samples depicted by tree on left, determined by the nearest point algorithm. (E) Normalized mean EWS894 FFPE FAIRE signal in Block 1 (gold) and Block 2 (brown) replicates at all transcription start sites with non-zero signal genome-wide. Enrichment 3 kb around transcription start sites is shown. (F) Heatmap of signal in xenografts tissue replicates 4 kb around center of replicate-specific regions of enrichment identified in EWS894 cell data sets. (G) Mean normalized FAIRE-seq signal in EWS894 cell FAIRE (all replicates pooled), 4 kb around the center of FFPE tissue-replicate-specific regions of enrichment sets from Block 1 (gold) and Block 2 (brown), three replicates each. (H) Bar plot of the percentage of HUVEC cell peaks (all replicates pooled, blue) and EWS894 cell peaks (all replicates pooled, purple) that overlap with EWS894 xenograft peaks called (all replicates pooled, brown).
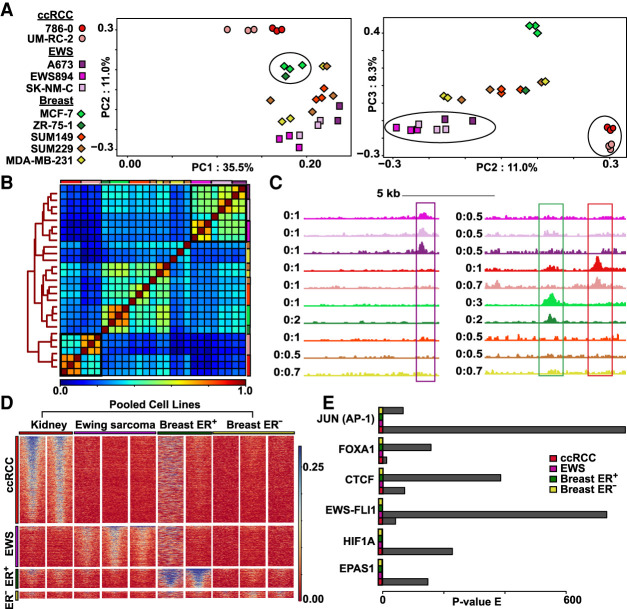
Identification of tumor-type-specific chromatin patterns using nanodroplet FAIRE
Next, we asked whether nanodroplet-assisted FAIRE in xenograft tissues could identify relevant tumor-type-specific regions (TSRs) of chromatin accessibility. Renal cell clear cell carcinoma (ccRCC) (UM-RC-2, 786-0), Ewing sarcoma (EWS894, A673, SK-NM-C), ER-positive luminal breast carcinoma (MCF-7, ZR-75-1), and ER-negative basal breast carcinoma (SUM149, SUM229, MDA-MB-231) cells were xenografted into mice. Xenografts of approximately 1000 mm3 were isolated and fixed for 24 h (unless otherwise indicated) in 10% buffered formalin followed by paraffin embedding to generate FFPE blocks. Nanodroplet-mediated FAIRE was performed on four 10-μm sections. Regions with signal enrichment were identified for each block of each tumor type, and regions were combined to generate a union set. We calculated the mean FAIRE signal for each sample across this set. At these regions, signal segregated by tumor type (PC 1, 2, and 3) (Fig. 5A). Based on Spearman's correlation scores, samples clustered by cell line and then by tumor type (black squares) (Fig. 5B). Breast cancer cell lines clustered primarily based on molecular subtype, with some ER-positive luminal derived tumors separating from ER-negative. The lower correlation between the ER-negative breast cancer xenografts was likely caused by differences in signal-to-noise between the blocks (Supplemental Figs. S5–S8). We observed that the MDA-MB-231 xenografts showed low genome-wide correlation between biological replicate blocks (Supplemental Fig. S9). Comparison of signal between these blocks revealed subgroups of regions with lower correlation (Supplemental Fig. S9A). Signal from these regions derived from reads that mapped to unplaced/unlocalized contigs. Unplaced/unlocalized contigs make up 0.11% of all genome-wide bins. Replicates 1 and 2 had a lower signal-to-noise than replicate 3 resulting in more uniform distribution of reads across the genome, including at noncanonical regions (Supplemental Fig. S9B,C). Other studies that similarly detected high signal at background bins in lower quality samples filtered out these regions (Furey 2012; Berger et al. 2019). We also found a significantly higher distribution of signal in replicate 3 at the union set of FAIRE peaks (Supplemental Fig. S9D,E). Considering this observation, we focused on canonical regions of the genome (Chr 1–22, Chr X, and Chr Y) in subsequent analyses.
Figure 5.
Nanodroplet FAIRE identifies tumor-type-specific chromatin signatures from xenograft FFPE tissue. (A) Top three principal components from mean signal in union set of regions of enrichment called across all xenograft carcinoma tissue blocks. Tumor categories defined by symbols: (circles) ccRCC; (squares) EWS894; (diamonds) breast carcinoma. Number of blocks per cell line: 786-0, n = 3; UM-RC-2, n = 3; A673, n = 2; EWS894, n = 3; SK-NM-C, n = 3; MCF-7, n = 3; ZR-75-1, n = 1; SUM149, n = 3; SUM229, n = 3; MDA-MB-231, n = 3). Xenograft replicates defined by colors. Clustering of ccRCC group, breast ER+ group, and EWS894 group highlighted within PCA by black circles. (B) Spearman's correlation heatmap of mean signal in union set of regions of enrichment called across all xenograft tissue replicates. Xenografts are color coded by cell line outside the heatmap perimeter (top and right); color coding matches legend defined in A. Clustering distance tree across all replicates depicted on the left of the heatmap. Legend of correlation coefficient scores depicted at the bottom of the heatmap. Clusters of tumor-type-specific cell lines defined by tree nodes highlighted with black boxes within the heatmap. (C) Examples of regions of enrichment from within the EWS894, breast ER+, and ccRCC TSRs (tumor-type-specific regions). Replicates for each xenograft pooled into one signal track. Tracks colored by xenograft to match legend defined in A. Signal at depicted signature regions across all cell line tracks highlighted within box (Ewing sarcoma signature region defined by purple box, breast ER+ region by green box, ccRCC region by red box). (D) Heatmap of xenograft tissue FAIRE signal at each TSR. TSRs defined on the left with each region as a row; pooled xenografts grouped by tumor type define columns. Each TSR heatmap is sorted by signal in the first cell line within that tumor type category. (E) Motifs enriched at TSR. P-value of enrichment for each motif within each TSR displayed for six relevant transcription factors.
Next, we identified regions that showed selective enrichment in each tumor type (DESeq2, P < 0.05, log2 fold difference >0.5) (Fig. 5C; Love et al. 2014). FAIRE signal at these regions across the set of xenografted tumor (blocks pooled) supported tumor-type specificity (Fig. 5D). For example, both 786-0 and UM-RC-2, ccRCC xenografts showed greater signal enrichment compared with regions identified in Ewing sarcoma or ER-positive and ER-negative breast cancer xenografts. Similarly, signal was not enriched in the Ewing sarcoma and breast cancer xenografts at those regions detected in ccRCC cells. Variation in FAIRE results were also evident in this comparison. In the absence of replicates, results from the ZR-75-1 analysis were noisier compared to those from the other samples. However, even for ZR-75-1, signal enrichment mostly localized to ER-positive breast cancer TSRs. As mentioned previously, variability in signal-to-noise between blocks likely limited the ability to identify ER-negative breast cancer xenograft-specific sites.
We then asked whether DNA sequence motifs enriched in the TSR showed lineage relevance. TSRs were analyzed using the genome as the background (Heinz et al. 2010). The hypoxia response element was observed at ccRCC xenograft TSRs (Fig. 5E). This association is consistent with the importance of HIF1A (also known as HIF1) and EPAS1 (also known as HIF2A) to ccRCC biology. In contrast, Ewing sarcoma TSRs were marked by the ETS motif, which is bound by the driver oncoprotein for this tumor, EWS-FLI1 (Patel et al. 2012). Motifs identified in breast cancer xenografts TSRs reflected their molecular subtypes (Hurtado et al. 2011; Zhao et al. 2014). Luminal type MCF-7 and ZR-75-1 were enriched for FOXA1 motifs, whereas basal or claudin-low SUM229 and SUM149 were enriched for the JUN portion of the AP-1 DNA binding motif. Collectively, these results show that nanodroplet-assisted FAIRE identifies tumor-relevant chromatin accessibility patterns from archival tissue.
Effect of block age on nanodroplet FAIRE signal detection
We next assessed the impact of block age on chromatin extraction. Among our samples, the youngest tissue blocks at the time of processing were 2 mo of age (Figs. 3, 5), and the oldest tissue blocks were 7 yr of age (EWS894 xenograft tissue) (Fig. 4A–H). One thousand peaks were shared by both the old and young blocks, with 2777 peaks identified only in the old blocks, and 2069 peaks identified only in the young blocks (Supplemental Fig. S10A). However, we found that increased FAIRE signal was evident at all sites (Supplemental Fig. S10B). A quantitative comparison showed that the signals at almost all peaks called were not statistically different between the younger and older blocks (Supplemental Fig. S10C).
Because of the difference in the regions identified between the older and younger blocks, we sought to establish an expectation for variance in the identification of peaks across similar samples. This analysis would offer context when comparing peak sets across experimental variables (e.g., block age). Peaks were repeatedly identified from randomly selected replicates of the older blocks and compared to the remaining replicates. We found that the fraction of shared peaks between the older and younger blocks fell within the range established through our permutation analysis (Supplemental Fig. S10D). This analysis suggests that the differences in shared peaks may not be associated with block age. Differences in peaks identified were attributable to heterogeneity in signal-to-noise among the blocks, because older blocks had more consistency across replicates—Spearman's correlation coefficient in (1) shared: older replicates [0.51,0.60], younger replicates [0.30–0.44]; (2) older only: older replicates [0.36–0.45], younger replicates [0.14–0.25]; (3) younger only: older replicates [0.52–0.58], younger replicates [0.30,0.41] (Supplemental Fig. S10E–G). The signal-to-noise difference is most likely not associated with block age, because many of our other 2-mo-old blocks (786-0, MCF-7) showed a similar range of signal-to-noise to that shown in our older blocks (Supplemental Fig. S10H). All together, these results show that nanodroplet FAIRE-seq can identify regions of chromatin accessibility from FFPE tissue blocks of at least 7 yr of age. In addition, there does not appear to be a substantial difference in FAIRE signal clearly attributable to the age of the block.
Tumor-type-specific chromatin accessibility in xenografts reflects primary tumor chromatin accessibility
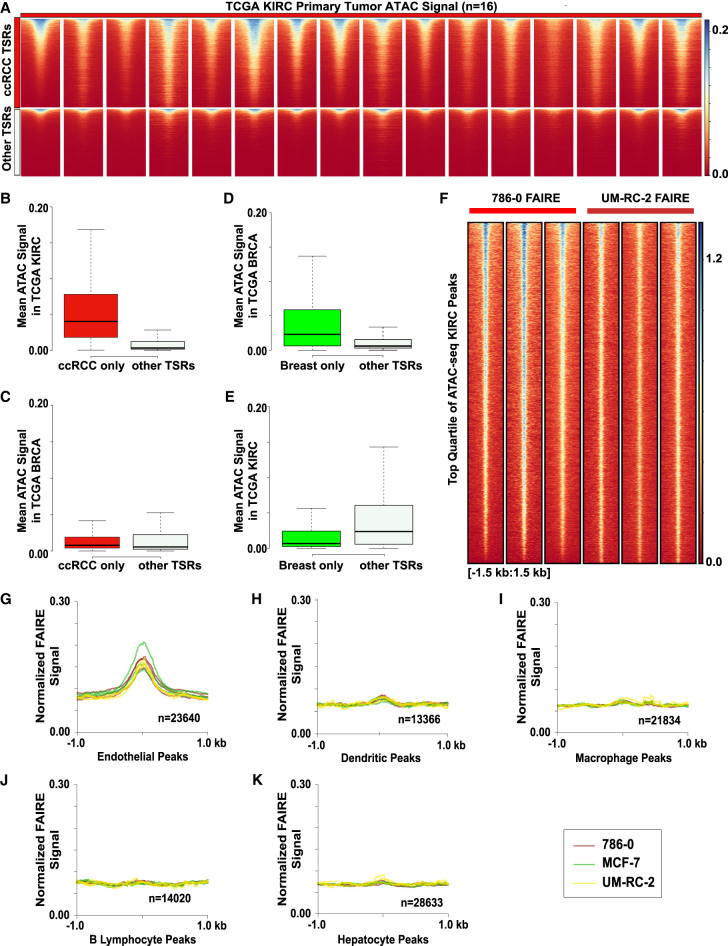
Next, we tested whether chromatin accessibility detected in the xenografted tumors reflected that in primary human tumors. The Cancer Genome Atlas Program applied ATAC-seq in frozen primary human tumors to identified chromatin accessibility patterns (Corces et al. 2018). We compared ATAC-seq results from KIRC (ccRCC, n = 16) and BRCA (invasive breast carcinoma, n = 44) tumors to our FAIRE-seq results from UM-RC-2 and 786-0 ccRCC xenografts. All KIRC tumors analyzed showed increased ATAC signal at the ccRCC cell-line-derived TSRs (Fig. 6A). KIRC ATAC signal enrichment was minimally observed at TSRs identified from the Ewing sarcoma and breast cancer xenografts. To assess the magnitude of these signal differences, we calculated the mean ATAC signal for all KIRC tumors at each ccRCC xenograft TSR. ATAC signal at the TSRs identified in ccRCC xenografts was significantly higher compared to TSRs in the other xenografts (P < 2.2 × 10−16) (Fig. 6B). Breast tumor ATAC signal was not enriched at ccRCC-specific regions (Fig. 6C). We repeated these analyses using the breast cancer cell line xenograft TSRs (ER+ and ER− combined) (Fig. 6D). As observed for ccRCC, BRCA ATAC signal was significantly higher at these TSRs compared with those identified from other xenografts (P < 2.2 × 10−16) (Fig. 6D). KIRC ATAC signal was greater in non-breast cancer xenograft-derived TSRs than at breast cancer TSRs, demonstrating the TSR specificity (Fig. 6E). These findings reveal that despite expected differences between xenografted cultured cells and primary tumors, our strategy identified relevant, tumor-specific accessible chromatin regions.
Figure 6.
Tumor-specific regions identified by nanodroplet FAIRE predict chromatin states in primary tumors and murine chromatin accessibility profiles identify stromal cells. (A) ATAC-seq signal across KIRC primary tumor samples (n = 16, TCGA) at ccRCC TSRs (n = 10,120) and other TSRs (n = 7737). (B) Mean signal across all KIRC samples at each region within the ccRCC TSRs (red) and other TSRs (gray), outliers excluded. (C) Mean signal across all BRCA samples (n = 44) at each region within the ccRCC TSRs (red) and other TSRs (gray), outliers excluded. (D) Mean signal across all BRCA samples at each region within the breast TSRs (n = 3019) and other TSRs (n = 14,838), outliers excluded. (E) Box plot of mean signal across all KIRC samples at each region within the breast TSRs and other TSRs, outliers excluded. (F) Volcano plot of FFPE FAIRE signal in 786-0 and UM-RC-2 replicates 3 kb around the center of the top quartile of TCGA KIRC peaks called. (G–K) Normalized mean xenograft FAIRE signal in 786-0 (brown), MCF-7 (green), and UM-RC-2 (yellow) replicates at cell-type-specific peaks (endothelial, dendritic, macrophage, B lymphocyte, hepatocytes, respectively) as defined from sci-ATAC-seq Mouse Atlas. Signal 2 kb around the peak centers shown.
Regions with disparate chromatin accessibility by ATAC can be detected using FAIRE
Owing to the background signal often present in FAIRE, identification of enriched regions can be compromised. To explore whether FAIRE identifies regions with limited chromatin accessibility signal, we evaluated performance at regions with strong or weak signals as determined by ATAC. Because ATAC is currently not compatible with fixed tissues, we used ATAC-seq from MCF-7 cells grown in culture (Nagarajan et al. 2020). Following down-sampling of aligned MCF-7 ATAC-seq reads to match the read depth from our FAIRE-seq MCF-7 xenograft data, we identified areas of enrichment (MACS2). Regions were classified as “strong” or “weak” based on the Q-value significance (−log10): top quartile (10.11,79.25]; upper (6.84,10.11]; lower (4.68,6.84]; bottom [1.33,4.68]. Because we were comparing across data types (cells vs. tissues) as well as assay types (ATAC vs. FAIRE), we anticipated differences between regions of enrichment. However, despite these differences, we identified FAIRE enrichment at both classes of ATAC peaks (Supplemental Fig. S11A). The top (75th–100th percentile) and upper (50th–75th percentile) quartiles of ATAC peaks significantly overlapped with FAIRE tissue peaks (∼33%–40% and ∼17%–18%, respectively) relative to overlap with shuffled random genomic regions (Supplemental Fig. S11B). Nanodroplet FAIRE also identified ATAC peaks called in the lower and bottom quartiles, with expectedly lower percent overlap. FAIRE signal was detected at peaks from each quartile identified by ATAC (Supplemental Fig. S11C). These analyses showed that nanodroplet facilitated FAIRE from FFPE samples identified regions of both strong and weak signal enrichment; although, as expected, with greater ability to detect regions with stronger signal enrichment.
We then performed a similar analysis comparing primary tumor-derived ATAC signal from TCGA with FAIRE. Regions demonstrating enriched ATAC signal across all KIRC samples (n = 38,426, MACS2 callpeak, default parameters) were separated into quartiles based on the mean signal. We compared the top 75% of ATAC peaks (n = 9607) to FAIRE peaks from each of our ccRCC xenografts to identify regions common to both data sets. Of those peaks identified by FAIRE in the xenografts, 49%–56% of 786-0 peaks and 26%–54% of UM-RC-2 peaks were among those in the top quartile of ATAC peaks. We also evaluated sites at which FAIRE-seq signal was not sufficient to identify ATAC-enriched regions. 786-0 and UM-RC-2 xenograft FAIRE did not identify 23.8% of the high confidence ATAC peaks. However, signal was detected at almost all these regions (Fig. 6F). These analyses show that nanodroplet FAIRE-seq detects regions with strong ATAC-seq signal in primary tumors. Differences in detection may result from technical factors, but also likely reflect the comparison between cell lines and primary tumors.
Identification of chromatin accessibility signals from stromal cells
Intratumoral heterogeneity and the presence of stromal cells can influence chromatin accessibility signals. To assess whether nanodroplet FAIRE could identify the contribution of non-tumor cells, we isolated reads that aligned to the mouse genome from three of our xenograft data sets (786-0, MCF-7, UM-RC-2). Because the tumor cell lines were of human origin, murine sequence must derive from host cells. Mice used in our study (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, Jackson laboratories 005557) do not have lymphocytes but produce defective macrophages and dendritic cells (Ishikawa et al. 2005; Shultz et al. 2005). We compared FAIRE signal with cell-type-specific chromatin accessibility identified in the Mouse sci-ATAC-seq Atlas (Cusanovich et al. 2018), a compendium of chromatin accessibility data from various cell types determined by single-cell ATAC. We evaluated regions of accessibility specific to hepatocytes, endothelial cells, macrophages, dendritic cells, and B cells. Regions that were shared between cell types, that overlapped TSSs, or that were present in <75% of the constituent cells of each cell type were removed. We detected xenograft FAIRE signal at the endothelial-specific regions (Fig. 6G). Some signal enrichment was also observed at dendritic cell-specific regions (Fig. 6H). Enrichment at B lymphocyte, macrophage, and hepatocyte specific regions was not detected (Fig. 6I–K). Because the mice are deficient in B lymphocytes, the absence of B lymphocyte signal was expected. These results show that, despite limited read depth, our technique enables the detection of chromatin accessibility from rare populations of cells in a heterogeneous tissue.
Discussion
The detection of chromatin accessibility from FFPE tissues poses a unique challenge. However, formalin cross-linking of these tissues enables the application of FAIRE to isolate accessible chromatin without inclusion of other processing steps. To overcome the issue of extensive fixation, we found that incorporation of a nanodroplet reagent enhanced cavitation during sonication and enabled extraction of randomly fragmented, high-quality chromatin from fixed tissues in <2 h (Supplemental Methods S1). Nanodroplets decreased sonication time, increased the homogeneity of fragment lengths, and had no observed influence on the detection of accessible chromatin. Nanodroplet-mediated FAIRE-seq identified tumor-type-specific regions of chromatin accessibility from xenografted tumors shared by primary human tumors.
Although nucleic acids are routinely isolated from FFPE tissues, extracting chromatin that preserves the interaction of DNA with associated proteins poses specific challenges. The processing needed to overcome extended cross-linking destroys these interactions and consequently the ability to discriminate between accessible and inaccessible states. DNase I hypersensitivity assays on fixed tissue are compromised by inefficient library preparation for genome-wide analyses (Cooper et al. 2017). The ATAC assay has been considered largely incompatible with cross-linked samples. For instance, ATAC-seq on fixed cells produced short DNA fragments, which reduced library creation efficiency (Buenrostro et al. 2015a; Chen et al. 2016). While this paper was under review, a study was published outlining a technique to repair DNA breaks in ATAC DNA from FFPE tissue (Zhang et al. 2022). Although effective, the method involves a complicated, multistep protocol. A proof-of-concept study was recently published that applied the Universal NicE-seq enzyme-based chromatin accessibility assay to FFPE liver tissue (Chin et al. 2020). To date, however, this method has yet to be tested on a broad range of fixed tissue samples and depends on an enzyme with sequence bias (Ponnaluri et al. 2017; Chin et al. 2020). Several approaches have been developed to extract chromatin from FFPE tissues for use in the ChIP assay (Fanelli et al. 2010, 2011; Amatori et al. 2014; Cejas et al. 2016; Zhong et al. 2019). One of these methods, PAthology Tissue Chromatin Immunoprecipitation (PAT-ChIP) was successfully modified for commercial applications. Although useful for ChIP, we showed that this method did not yield chromatin suitable for chromatin accessibility assays (Fig. 3D).
The ability of nanodroplets to extract intact chromatin from fixed tissue may be linked to their submicron dimension (150–250 nm), which makes them nearly neutrally buoyant and likely to impregnate intracellular spaces created through the process of embedding for enhanced dispersion. Once vaporized, the nanodroplets expand approximately fivefold owing to the liquid to gas volume change. The resulting bubble, which cavitates in the ultrasound field, likely facilitates tissue dispersion, with the additional advantage of enhanced penetration of tissue while in the liquid nanodroplet precursor stage (Pajek et al. 2014). A similar principal has been applied for medical acoustics in which cavitation has been shown to disrupt intravascular blood clots (Kim et al. 2020), as well as endothelial and epithelial barriers (Chen et al. 2013; Fix et al. 2019). Depending on the duration of sonication, the degree of chromatin fragmentation in the presence of nanodroplets can be controlled (Figs. 1, 3). Future studies will examine whether minimally fragmented chromatin from FFPE tissues can be used for enzymatic assays that are currently compatible with sample fixation such as micrococcal nuclease nucleosome positioning assays (Byrum et al. 2011; Zhang and Pugh 2011; Mieczkowski et al. 2016; Chereji et al. 2017). Although the inclusion of nanodroplets enabled the extraction of intact chromatin from archival samples, the signal characteristic of FAIRE can complicate the identification of enriched regions and consequently the detection of accessible chromatin (Simon et al. 2012). Through our comparison of signals based on block age, we noted that factors such as signal-to-noise and overall read coverage can influence the detection of enriched regions. We recommend further qualification of differences in detected peaks through the application of complementary methods (e.g., DESeq2, cSAW) (Lun and Smyth 2016).
There is growing appreciation of cellular heterogeneity and microenvironment as important features of normal and pathological states. As a result, there has been an explosion in the development of single-cell technologies (Schwartzman and Tanay 2015; Wen and Tang 2018). Specifically, epigenetic-based single-cell assays include DNA methylation (Buenrostro et al. 2015b; Cusanovich et al. 2015), chromatin accessibility and nucleosome positioning (Buenrostro et al. 2015b; Cusanovich et al. 2015), ChIP (Rotem et al. 2015; Grosselin et al. 2019; Wang et al. 2019), and assays that examine 3D chromatin architecture such as Hi-C (Nagano et al. 2013). Dissociating individual cells from fixed tissues, however, presents challenges that make these approaches largely unsuitable for archival specimens (Corver and ter Haar 2011; Carrick et al. 2015; Simmons et al. 2015, 2016; Zhang et al. 2017). We found nanodroplet facilitated FAIRE yielded signal that could be deconvoluted to identify microenvironmental contributions. This observation led to a proof-of-concept experiment demonstrating that computational approaches could be applied to detect chromatin accessibility patterns from cell types within archived complex tissues. Overall, we present a strategy to extract chromatin from archival tissues that preserves protein–DNA interactions enabling detection of differential chromatin accessibility. The exploration of chromatin states in archival tissues further expands the range of analytics possible in these tissues.
Methods
Cell culture
Cells were cultured in standard conditions at 37°C with 5% CO2 (Supplemental Methods S1).
FAIRE assay for fixed cells
FAIRE was performed as described (Pattenden et al. 2016; Supplemental Methods S1).
Generation of FFPE xenograft tissue
On the day of injection, 6- to 8-wk-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ female mice originating from The Jackson Laboratory and then bred in-house at UNC were used to establish cell line xenografts. Five million cells from each cell line were collected and resuspended in 250 µL cold phosphate buffered saline (PBS) and 250 µL cold Matrigel (Sigma-Aldrich). Mice were anesthetized (isoflurane) and injected subcutaneously in the right flank (four mice per cell line) with 0.1 mL cell solution. Mice injected with ER+ cell lines (MCF-7, ZR-75-1) were implanted with 0.36 mg estrogen 4 d before cell injection. Estrogen capsules were prepared as described (Ingberg et al. 2012). Tumors were measured twice weekly using a Vernier calipers and body weights were taken once weekly. Following CO2 euthanasia, tumors were sterilely harvested and rinsed with cold PBS. Harvested tumors were immediately divided with half of the tumor flash frozen and stored at −80°C and half fixed for 24 h (unless otherwise indicated) in 10% buffered formalin followed by resuspension in 70% ethanol and storage at 4°C until embedding (up to 24 h). Samples were processed (UNC Animal Histopathology and Laboratory Medicine Core or CGIBD Histology Core) on the standard cycle at 60°C in the Leica ASP300S Tissue processor for ∼8.5 h. After completion of the overnight processing, samples were embedded using the Leica EG1160 Embedding Station. FFPE tissue cassettes were stored at room temperature under nitrogen until needed for sectioning.
FAIRE assay for FFPE tissue
A detailed protocol is included (Supplemental Methods S2). FFPE tissue blocks were sectioned (UNC Translational Pathology Laboratory) to 10-µm thickness, and each section was mounted on a charged glass slide. The top and bottom sections from the first sectioning of each block were stained with Haemotoxylin and Eosin (H&E), visualized on an Aperio ScanScope AT2, and nuclei counted using the Aperio ImageScope software. The slides were deparaffinized and hydrated in xylene, 100%, 85%, and 70% ethanol followed by double deionized water. Tissue sections were scraped from the slide with a razor blade and transferred to a 100-µL glass borosilicate vial (Triangle Biotechnology PL101-BT100) containing 90 µL of Lysis Buffer A (Simon et al. 2012). Just before sonication, 10 µL of nanodroplets (Triangle Biotechnology FF101-5000) (Kasoji et al. 2015) were pipetted directly into the Lysis Buffer A in each sonication tube. Sonication and FAIRE were performed as described for fixed cells. For Figure 3C, chromatin was extracted (Active Motif 53045) from eight 10 µm EWS894 FFPE tissue sections according to the manufacturer's instructions. For the nanodroplet extraction method, chromatin was extracted from one 10-µm tissue section using the protocol described above. Each FFPE FAIRE-seq sample consisted of FAIRE DNA extracted from four adjacent 10-µM tissue sections per tumor (for nuclei count from FFPE tumors, see Supplemental Table S1). Sequencing libraries were prepared (Kapa HyperPrep) according to the manufacturer's instructions.
Sequencing
EWS894 fresh cells and the first round of FFPE tissue FAIRE from EWS894 xenografts were sequenced on the Illumina HiSeq 2500 instrument. Genomic FAIRE samples from fresh HUVEC cells were sequenced on the Illumina NextSeq 500/550. Sequencing of all other genomic FAIRE samples from xenograft tissues were sequenced on the Illumina HiSeq 4000 instrument. All samples were sequenced as single-end 50-bp reads, with a range of sequence depth across data sets of 26,000,000–42,700,000 reads. Sequenced FAIRE-seq data sets have been made available (see Data access).
Genome assembly build
Human-derived sequences were aligned to the hg19 reference genome (GRCh37 genome build). Using hg19, as opposed to the GRCh38 assembly, should not significantly affect our conclusions because our study questions and conclusions are independent of the differences in the genome builds. When aligning our FAIRE reads, we excluded any reads aligning to alternative haplotypes and blacklisted regions of the genome (repetitive, hard to align to regions, like centromeric regions). As a result, any contributions made by using the hg38 assembly in place of hg19 would be filtered out, because major differences between the two assemblies are the incorporation of alternative sequences (alt_sequences) and centromere and mitochondrial genome changes. Use of the hg19 genome also allowed us to remain consistent with tools developed specifically using this construct (Cis-regulatory Element Annotation System). The mm9 reference genome was used for the alignment and annotation of murine data, because the published reference data we used for our deconvolution analysis was also aligned to mm9. As a result, we were unable to align our data to the GRCm38 (mm10) or evaluate whether realigning reads to this construct would significantly affect the conclusions because doing so would make our data incompatible with the cell-type-specific peaks defined in the published single cell ATAC-seq data used as a reference.
FAIRE-seq and ATAC-seq analyses
Following determination of read quality (FastQC/0.11.5), adapter sequences were removed (cutadapt/1.12) (Martin 2011) and reads of poor base quality were removed (fastx_toolkit/0.0.14 [-Q 33 -p 90 -q 20]). Duplicate reads were removed; in the case of ATAC, we associated paired ends, filtering out any read whose matching read from pair had been filtered at earlier steps. We then aligned the remaining reads to the hg19 genome using STAR v. 2.5.2b (Dobin et al. 2013) (Supplemental Methods S1).
Peak calling and overlap
Peaks were called using MACS2 callpeak (MACS2 v. 2.1.2) (Supplemental Methods S1).
Genome coverage tracks
Snapshots of the FAIRE signal distribution at genomic location were exported from the UCSC Genome Browser.
FAIRE composition and correlation analyses
TSS and principal component analyses were performed and Spearman's correlation coefficients were calculated using deepTools (deepTools/3.2.0) (Ramírez et al. 2016) functions (Supplemental Methods S1). An R script was written (R Core Team 2016) to make correlation scatterplots plotting the mean normalized FAIRE signal in genome-wide bins output from multibigWigSummary BED-file (‐‐outRawCounts) (Supplemental Code 1).
Defining FFPE FAIRE tumor-type-specific regions
TSRs were identified using DESeq2 of r/3.3.1 (Supplemental Methods S1).
ATAC-seq signal enrichment analysis in FFPE FAIRE tumor-type-specific regions
The mean ATAC-seq signal in each TSR was calculated using multibigWigSummary BED-file (‐‐outRawCounts) tool of the deepTools/3.2.0 package. An R script was written (R Core Team 2016) to calculate the mean signal within each TSR across all ATAC-seq samples of the TCGA KIRC group (n = 16) and TCGA BRCA group (n = 44) (Supplemental Code 2). Significant differences were then compared in ATAC-seq signal across relevant TSR groups (ccRCC vs. non-ccRCC, breast vs. non-breast). Statistical significance was determined by Mann–Whitney U test in R following tests of normality (shapiro.test, qqplot, ks.test) which defined data sets as nonparametric.
Deconvolution of murine stromal cell chromatin accessibility signal from FAIRE xenograft samples
To identify murine reads, FASTQ files were aligned to the mm9 genome (STAR v. 2.5.2b). Regions of enrichment were identified by MACS2, then deconvoluted into subpopulations using scATAC-seq as a reference (Mouse sci-ATAC-seq Atlas) (Supplemental Methods S1; Cusanovich et al. 2018).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE162443.
Supplementary Material
Acknowledgments
Animal Studies were performed within the University of North Carolina (UNC) Lineberger Animal Studies Core Facility at the University of North Carolina at Chapel Hill. The UNC Lineberger Animal Histopathology Core is supported in part by a National Cancer Institute (NCI) Center Core Support Grant (P30CA016085) to the UNC Lineberger Comprehensive Cancer Center. We thank Nana Feinberg, Bentley Midkiff, and other staff at the Translational Pathology Laboratory (TPL) for expert technical assistance for tissue sectioning, staining, and nuclei counts. The UNC TPL is supported in part by grants from the NCI (5P30CA016080-42), National Institutes of Health (NIH) (U54CA156733), National Institute of Environmental Health Sciences (3P30 EOS010126-17), University Cancer Research Fund, and North Carolina Biotechnology Center (NCBC) (2015-IDG-1007). We thank Carolyn Suitt from the Center for Gastrointestinal Biology and Disease (CGIBD) Histology Core at the University of North Carolina, Chapel Hill (NIH, P30 DK 034987), and Dawud Hilliard from the Animal Histopathology. High-throughput sequencing was performed in the UNC High-Throughput Sequencing Facility. We thank Jacob Kennedy (Amanda Paulovich Laboratory, Fred Hutchinson Cancer Research Center) for advice on deparaffinization and rehydration of tissue sections, and members of the Davis and Pattenden laboratories for helpful advice and comments on the manuscript. This work was supported by funding from the Howard Hughes Medical Institute through the James H. Gilliam Fellowships for Advanced Study (S.S.M.), the National Institute of General Medical Sciences under award T32GM067553 (S.S.M.), NIH/NCI IMAT R33CA206939 (S.G.P., I.J.D., P.A.D.), R21CA232902 (S.G.P., P.A.D.), the North Carolina Translational and Clinical Sciences Institute (UNC Chapel Hill) 550 KR151619 (S.G.P., I.J.D., P.A.D.), and the NCBC 2017-IDG-1005 (S.G.P.).
Author contributions: S.S.M., A.L.Q., M.P.N., I.J.D., and S.G.P. designed research; S.S.M., A.L.Q., M.P.N., O.C.J., M.M.-M., S.A.A., and C.M.S. performed research; B.V., J.K.T., S.K.K., and P.A.D. contributed new reagents or analytical tools; S.S.M., A.L.Q., M.P.N., O.C.J., J.S.P., I.J.D., and S.G.P. analyzed data; and S.S.M., I.J.D., and S.G.P. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275219.121.
Competing interest statement
The authors declare the following competing financial interest(s): S.G.P., I.J.D., P.A.D., and S.K.K. have equity ownership in Triangle Biotechnology, Inc., to which the following technologies used or evaluated in this paper have been licensed from the University of North Carolina at Chapel Hill: nanodroplets (U.S. Patent no. 9,427,410 and U.S. Patent no. 9,982,290). S.G.P. and P.A.D. are inventors on U.S. Patent 9,982,290. P.A.D. is inventor on U.S. Patent no. 9,427,410.
References
- Amatori S, Ballarini M, Faversani A, Belloni E, Fusar F, Bosari S, Pelicci PG, Minucci S, Fanelli M. 2014. PAT-ChIP coupled with laser microdissection allows the study of chromatin in selected cell populations from paraffin-embedded patient samples. Epigenetics Chromatin 7: 18. 10.1186/1756-8935-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BP, Engel KB, Greytak SR, Moore HM. 2014. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: how well do you know your FFPE specimen? Arch Pathol Lab Med 138: 1520–1530. 10.5858/arpa.2013-0691-RA [DOI] [PubMed] [Google Scholar]
- Berger S, Pachkov M, Arnold P, Omidi S, Kelley N, Salatino S, van Nimwegen E. 2019. Crunch: integrated processing and modeling of ChIP-seq data in terms of regulatory motifs. Genome Res 29: 1164–1177. 10.1101/gr.239319.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015a. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21.29.1–21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. 2015b. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523: 486–490. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum S, Mackintosh SG, Edmondson RD, Cheung WL, Taverna SD, Tackett AJ. 2011. Analysis of histone exchange during chromatin purification. J Integr OMICS 1: 61–65. 10.5584/jiomics.v1i1.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrick DM, Mehaffey MG, Sachs MC, Altekruse S, Camalier C, Chuaqui R, Cozen W, Das B, Hernandez BY, Lih CJ, et al. 2015. Robustness of next generation sequencing on older formalin-fixed paraffin-embedded tissue. PLoS One 10: e0127353. 10.1371/journal.pone.0127353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejas P, Li L, O'Neill NK, Duarte M, Rao P, Bowden M, Zhou CW, Mendiola M, Burgos E, Feliu J, et al. 2016. Chromatin immunoprecipitation from fixed clinical tissues reveals tumor-specific enhancer profiles. Nat Med 22: 685–691. 10.1038/nm.4085 [DOI] [PubMed] [Google Scholar]
- Chen CC, Sheeran PS, Wu SY, Olumolade OO, Dayton PA, Konofagou EE. 2013. Targeted drug delivery with focused ultrasound-induced blood-brain barrier opening using acoustically-activated nanodroplets. J Control Release 172: 795–804. 10.1016/j.jconrel.2013.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shen Y, Draper W, Buenrostro JD, Litzenburger U, Cho SW, Satpathy AT, Carter AC, Ghosh RP, East-Seletsky A, et al. 2016. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat Methods 13: 1013–1020. 10.1038/nmeth.4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji RV, Ocampo J, Clark DJ. 2017. MNase-sensitive complexes in yeast: nucleosomes and non-histone barriers. Mol Cell 65: 565–577.e3. 10.1016/j.molcel.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarella AM, Quimby AL, Mehrab-Mohseni M, Velasco B, Kasoji SK, Davis IJ, Dayton PA, Hathaway NA, Pattenden SG. 2018. Cavitation enhancement increases the efficiency and consistency of chromatin fragmentation from fixed cells for downstream quantitative applications. Biochemistry 57: 2756–2761. 10.1021/acs.biochem.8b00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin HG, Sun Z, Vishnu US, Hao P, Cejas P, Spracklin G, Estève PO, Xu SY, Long HW, Pradhan S. 2020. Universal NicE-seq for high-resolution accessible chromatin profiling for formaldehyde-fixed and FFPE tissues. Clin Epigenetics 12: 143. 10.1186/s13148-020-00921-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J, Ding Y, Song J, Zhao K. 2017. Genome-wide mapping of DNase I hypersensitive sites in rare cell populations using single-cell DNase sequencing. Nat Protoc 12: 2342–2354. 10.1038/nprot.2017.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. 2016. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48: 1193–1203. 10.1038/ng.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, Silva TC, Groeneveld C, Wong CK, Cho SW, et al. 2018. The chromatin accessibility landscape of primary human cancers. Science 362: eaav1898. 10.1126/science.aav1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver WE, ter Haar NT. 2011. High-resolution multiparameter DNA flow cytometry for the detection and sorting of tumor and stromal subpopulations from paraffin-embedded tissues. Curr Protoc Cytom Chapter 7: Unit 7.37. 10.1002/0471142956.cy0737s55 [DOI] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J. 2015. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348: 910–914. 10.1126/science.aab1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. 2018. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174: 1309–1324.e18. 10.1016/j.cell.2018.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IJ, Pattenden SG. 2019. Chromatin accessibility as a strategy to detect changes associated with development, disease, and exposure and susceptibility to chemical toxins. In Toxicoepigenetics: core principles and applications (ed. McCullough SD, Dolinoy DC), pp. 85–103. Academic Press, London. 10.1016/b978-0-12-812433-8.00003-4 [DOI] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S, Hill AJ, Daza RM, Cao J, O'Day DR, Pliner HA, Aldinger KA, Pokholok D, Zhang F, Milbank JH, et al. 2020. A human cell atlas of fetal chromatin accessibility. Science 370: eaba7612. 10.1126/science.aba7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donczo B, Guttman A. 2018. Biomedical analysis of formalin-fixed, paraffin-embedded tissue samples: the Holy Grail for molecular diagnostics. J Pharm Biomed Anal 155: 125–134. 10.1016/j.jpba.2018.03.065 [DOI] [PubMed] [Google Scholar]
- Fanelli M, Amatori S, Barozzi I, Soncini M, Dal Zuffo R, Bucci G, Capra M, Quarto M, Dellino GI, Mercurio C, et al. 2010. Pathology tissue-chromatin immunoprecipitation, coupled with high-throughput sequencing, allows the epigenetic profiling of patient samples. Proc Natl Acad Sci 107: 21535–21540. 10.1073/pnas.1007647107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanelli M, Amatori S, Barozzi I, Minucci S. 2011. Chromatin immunoprecipitation and high-throughput sequencing from paraffin-embedded pathology tissue. Nat Protoc 6: 1905–1919. 10.1038/nprot.2011.406 [DOI] [PubMed] [Google Scholar]
- Fix SM, Koppolu BP, Novell A, Hopkins J, Kierski TM, Zaharoff DA, Dayton PA, Papadopoulou V. 2019. Ultrasound-stimulated phase-change contrast agents for transepithelial delivery of macromolecules, toward gastrointestinal drug delivery. Ultrasound Med Biol 45: 1762–1776. 10.1016/j.ultrasmedbio.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, Bernstein BE. 2017. Epigenetic plasticity and the hallmarks of cancer. Science 357: 256–266. 10.1126/science.aal2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey TS. 2012. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet 13: 840–852. 10.1038/nrg3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney EF, Riegman PH, Grizzle WE, Watson PH. 2018. Factors that drive the increasing use of FFPE tissue in basic and translational cancer research. Biotech Histochem 93: 373–386. 10.1080/10520295.2018.1446101 [DOI] [PubMed] [Google Scholar]
- Giresi PG, Lieb JD. 2009. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements). Methods 48: 233–239. 10.1016/j.ymeth.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. 2007. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885. 10.1101/gr.5533506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet DR, Foster JP, Zawistowski JS, Bevill SM, Noël MP, Olivares-Quintero JF, Sciaky N, Singh D, Santos C, Pattenden SG, et al. 2020. Discrete adaptive responses to MEK inhibitor in subpopulations of triple-negative breast cancer. Mol Cancer Res 18: 1685–1698. 10.1158/1541-7786.MCR-19-1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, Dahmani A, Lameiras S, Reyal F, Frenoy O, et al. 2019. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet 51: 1060–1066. 10.1038/s41588-019-0424-9 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. 2011. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–33. 10.1038/ng.730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingberg E, Theodorsson A, Theodorsson E, Strom JO. 2012. Methods for long-term 17β-estradiol administration to mice. Gen Comp Endocrinol 175: 188–193. 10.1016/j.ygcen.2011.11.014 [DOI] [PubMed] [Google Scholar]
- Ishikawa F, Yasukawa M, Lyons B, Yoshida S, Miyamoto T, Yoshimoto G, Watanabe T, Akashi K, Shultz LD, Harada M. 2005. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor γ chainnull mice. Blood 106: 1565–1573. 10.1182/blood-2005-02-0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasoji SK, Pattenden SG, Malc EP, Jayakody CN, Tsuruta JK, Mieczkowski PA, Janzen WP, Dayton PA. 2015. Cavitation enhancing nanodroplets mediate efficient DNA fragmentation in a bench top ultrasonic water bath. PLoS One 10: e0133014. 10.1371/journal.pone.0133014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, DeRuiter RM, Goel L, Xu Z, Jiang X, Dayton PA. 2020. A comparison of sonothrombolysis in aged clots between low-boiling-point phase-change nanodroplets and microbubbles of the same composition. Ultrasound Med Biol 46: 3059–3068. 10.1016/j.ultrasmedbio.2020.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau CA, Duarte FM, Chew JG, Kartha VK, Burkett ZD, Kohlway AS, Pokholok D, Aryee MJ, Steemers FJ, Lebofsky R, et al. 2019. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat Biotechnol 37: 916–924. 10.1038/s41587-019-0147-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L, Crawford GE, Furey TS, Rusyn I. 2017. Genetic and epigenetic determinants of inter-individual variability in responses to toxicants. Curr Opin Toxicol 6: 50–59. 10.1016/j.cotox.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AT, Smyth GK. 2016. csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res 44: e45. 10.1093/nar/gkv1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Zhang B, LaFave LM, Earl AS, Chiang Z, Hu Y, Ding J, Brack A, Kartha VK, Tay T, et al. 2020. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183: 1103–1116.e20. 10.1016/j.cell.2020.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 17: 3. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mieczkowski J, Cook A, Bowman SK, Mueller B, Alver BH, Kundu S, Deaton AM, Urban JA, Larschan E, Park PJ, et al. 2016. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat Commun 7: 11485. 10.1038/ncomms11485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. 2013. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502: 59–64. 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan S, Rao SV, Sutton J, Cheeseman D, Dunn S, Papachristou EK, Prada JG, Couturier DL, Kumar S, Kishore K, et al. 2020. ARID1A influences HDAC1/BRD4 activity, intrinsic proliferative capacity and breast cancer treatment response. Nat Genet 52: 187–197. 10.1038/s41588-019-0541-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajek D, Burgess A, Huang Y, Hynynen K. 2014. High-intensity focused ultrasound sonothrombolysis: the use of perfluorocarbon droplets to achieve clot lysis at reduced acoustic power. Ultrasound Med Biol 40: 2151–2161. 10.1016/j.ultrasmedbio.2014.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Simon JM, Iglesia MD, Wu SB, McFadden AW, Lieb JD, Davis IJ. 2012. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res 22: 259–270. 10.1101/gr.125666.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattenden SG, Simon JM, Wali A, Jayakody CN, Troutman J, McFadden AW, Wooten J, Wood CC, Frye SV, Janzen WP, et al. 2016. High-throughput small molecule screen identifies inhibitors of aberrant chromatin accessibility. Proc Natl Acad Sci 113: 3018–3023. 10.1073/pnas.1521827113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnaluri VKC, Zhang G, Estève PO, Spracklin G, Sian S, Xu SY, Benoukraf T, Pradhan S. 2017. NicE-seq: high resolution open chromatin profiling. Genome Biol 18: 122. 10.1186/s13059-017-1247-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. 2015. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol 33: 1165–1172. 10.1038/nbt.3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, Saligrama N, Buenrostro JD, Wei Y, Wu B, Rubin AJ, Granja JM, Lareau CA, Li R, Qi Y, et al. 2018. Transcript-indexed ATAC-seq for precision immune profiling. Nat Med 24: 580–590. 10.1038/s41591-018-0008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzman O, Tanay A. 2015. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet 16: 716–726. 10.1038/nrg3980 [DOI] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J, et al. 2005. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R γnull mice engrafted with mobilized human hemopoietic stem cells. J Immunol 174: 6477–6489. 10.4049/jimmunol.174.10.6477 [DOI] [PubMed] [Google Scholar]
- Simmons AJ, Banerjee A, McKinley ET, Scurrah CR, Herring CA, Gewin LS, Masuzaki R, Karp SJ, Franklin JL, Gerdes MJ, et al. 2015. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-α-induced apoptosis in vivo. Mol Syst Biol 11: 835. 10.15252/msb.20156282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons AJ, Scurrah CR, McKinley ET, Herring CA, Irish JM, Washington MK, Coffey RJ, Lau KS. 2016. Impaired coordination between signaling pathways is revealed in human colorectal cancer using single-cell mass cytometry of archival tissue blocks. Sci Signal 9: rs11. 10.1126/scisignal.aah4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JM, Giresi PG, Davis IJ, Lieb JD. 2012. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc 7: 256–267. 10.1038/nprot.2011.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JM, Giresi PG, Davis IJ, Lieb JD. 2013. A detailed protocol for formaldehyde-assisted isolation of regulatory elements (FAIRE). Curr Protoc Mol Biol Chapter 21: Unit 21.26. 10.1002/0471142727.mb2126s102 [DOI] [PubMed] [Google Scholar]
- Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M, Ho TH, Kuan PF, Jonasch E, Furey TS, et al. 2014. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res 24: 241–250. 10.1101/gr.158253.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. 2013. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154: 888–903. 10.1016/j.cell.2013.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Xiong H, Ai S, Yu X, Liu Y, Zhang J, He A. 2019. CoBATCH for high-throughput single-cell epigenomic profiling. Mol Cell 76: 206–216.e7. 10.1016/j.molcel.2019.07.015 [DOI] [PubMed] [Google Scholar]
- Wen L, Tang F. 2018. Single cell epigenome sequencing technologies. Mol Aspects Med 59: 62–69. 10.1016/j.mam.2017.09.002 [DOI] [PubMed] [Google Scholar]
- Yu VW, Yusuf RZ, Oki T, Wu J, Saez B, Wang X, Cook C, Baryawno N, Ziller MJ, Lee E, et al. 2016. Epigenetic memory underlies cell-autonomous heterogeneous behavior of hematopoietic stem cells. Cell 167: 1310–1322.e17. 10.1016/j.cell.2016.10.045 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Pugh BF. 2011. High-resolution genome-wide mapping of the primary structure of chromatin. Cell 144: 175–186. 10.1016/j.cell.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Lehmann BD, Shyr Y, Guo Y. 2017. The utilization of formalin fixed-paraffin-embedded specimens in high throughput genomic studies. Int J Genomics 2017: 1926304. 10.1155/2017/1926304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Polavarapu VK, Xing P, Zhao M, Mathot L, Zhao L, Rosen G, Swartling FJ, Sjöblom T, Chen X. 2022. Profiling chromatin accessibility in formalin-fixed paraffin-embedded samples. Genome Res (in press). 10.1101/gr.275269.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Qiao Y, Jonsson P, Wang J, Xu L, Rouhi P, Sinha I, Cao Y, Williams C, Dahlman-Wright K. 2014. Genome-wide profiling of AP-1–regulated transcription provides insights into the invasiveness of triple-negative breast cancer. Cancer Res 74: 3983–3994. 10.1158/0008-5472.CAN-13-3396 [DOI] [PubMed] [Google Scholar]
- Zhong J, Ye Z, Clark CR, Lenz SW, Nguyen JH, Yan H, Robertson KD, Farrugia G, Zhang Z, Ordog T, et al. 2019. Enhanced and controlled chromatin extraction from FFPE tissues and the application to ChIP-seq. BMC Genomics 20: 249. 10.1186/s12864-019-5639-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.