Abstract
Purpose:
Acral melanoma is the major subtype of melanoma seen in Asian patients with melanoma and is featured by its insidious onset and poor prognosis. The genomic study that elucidates driving mutational events is fundamental to the development of gene-targeted therapy. However, research on genomic profiles of acral melanoma in Asian patients is still sparse.
Experimental Design:
We carried out whole-exome sequencing (WES) on 60 acral melanoma lesions (with 55 primary samples involved), targeted deep sequencing in a validation cohort of 48 cases, RNA sequencing in 37 acral melanoma samples (all from the 60 undergoing WES), and FISH in 233 acral melanoma specimens (54 of the 60 undergoing WES included). All the specimens were derived from Asian populations.
Results:
BRAF, NRAS, and KIT were discerned as significantly mutated genes (SMG) in acral melanoma. The detected COSMIC signature 3 related to DNA damage repair, along with the high genomic instability score, implied corresponding pathogenesis of acral melanoma. Moreover, the copy number gains of EP300 were associated with the response of acral melanoma to targeted therapy of A485 (a p300 inhibitor) and immune checkpoint blockade treatment. In addition, the temporal order in mutational processes of the samples was reconstructed, and copy-number alterations were identified as early mutational events.
Conclusions:
Our study provided a detailed view of genomic instability, potential therapeutic targets, and intratumoral heterogeneity of acral melanoma, which might fuel the development of personalized strategies for treating acral melanoma in Asian populations.
Translational Relevance.
Genomic studies on acral melanoma in Asian populations will provide an important reference for diagnosing and exploring new therapeutic strategies for Asian patients with limited treatment options. Firstly, the detected homologous recombination deficiency in fractional acral melanoma samples reflected by mutational signatures in the present study raised the possibility of targeting DNA repair defects in treating acral melanoma. Moreover, our study showed that copy number gains of EP300 were not only associated with acral melanoma malignancy and the response to targeted therapies embodying A485 (a p300 inhibitor), but could be employed as a potential biomarker for the response to immunotherapy. In addition, the detected genomic differences between acral melanoma lesions arising from different primary sites might refine the targeted therapies with more precision. More importantly, the reconstruction of acral melanoma clonal architecture underpinned the importance of checking clonal status in addition to screening mutations in driver genes in clinical practice.
Introduction
Acral melanoma is the major subtype of melanoma in Asian patients and is featured by its insidious onset and poor prognosis (1, 2). Previously, some seminal genomic studies on European cohorts have been conducted to outline the characteristics of mutations in acral melanoma, which are prominently distinct from those in cutaneous melanoma (3). Specifically, the overall point mutation burdens of acral melanoma are markedly lighter compared with cutaneous melanoma (3). Besides, the majority (51%) of acral melanoma cases are classified into the triple–wild-type (TWT; BRAF-RAS-NF1–wild-type) while the proportion of which in cutaneous melanoma is merely 11% (3). More importantly, acral melanoma is more vulnerable to whole-genome duplication, chromosomal aberrations like aneuploidy, and complex rearrangements (4–6). In parallel, copy-number variants (CNV) intensively favor potential driver genes in acral melanoma that thus exhibit a heavy CNV mutational burden (3, 5). However, Smalley and colleagues have analyzed the genome data and demonstrated that acral nevi are not precursors to the majority of prototypical acral melanomas, indicating that acral melanoma has an evolutionary trajectory different from that of cutaneous melanoma (7).
Nonetheless, most of the genomic studies on acral melanoma were performed among Western populations, whereas those based on Asian patients were sparse. In addition, given that recurrent CNVs favor the driver genes in acral melanoma, as indicated by previous reports, it is of great significance to correlate the CNVs with corresponding clinicopathologic information, so as to evaluate their potential as therapeutic targets. Moreover, a number of metastatic samples were employed in previous analyses, which might hinder the identification of driving mutational processes in early oncogenesis of acral melanoma and necessitates further investigation based on a large cohort of primary lesions. Finally, the spatiotemporal evolutionary patterns of acral melanoma remain uncharted, but advanced analytical approaches of genetic study are yielding new insights into the problem, which could help to comprehend the origins and vulnerabilities of acral melanoma (8). Herein, this study tried to delineate the genomic landscape of acral melanoma in Asian populations by integrating genomic sequencing data with the corresponding clinical information of the patients as well as analyzing the mutational signature, significantly mutated signaling pathways, and intratumoral heterogeneity of acral melanoma, based on primary lesion samples.
Materials and Methods
Patient description
All samples analyzed in the current study were acral melanoma lesions arising from Asian patients collected from the melanoma database of the Department of Dermatology, Xijing Hospital (Fourth Military Medical University, Xi'an, Shaanxi, China). Samples undergoing next-generation sequencing (NGS) were extracted from fresh-frozen tumors and matched adjacent normal tissue or whole blood. Informed written consent was obtained from each subject or each subject's guardian. The research protocol was designed and executed according to the principles of the Declaration of Helsinki and was approved by the ethics review board of Fourth Military Medical University. Samples used for FISH assay were paraffin-embedded. All samples were assessed by melanoma pathologists to confirm the presence of melanoma with greater than 80% tumor content and less than 30% necrosis. The following pathology features were recorded in each patient: Breslow thickness, presence or absence of ulceration, and lymphocytes scores (defined according to previous reports; ref. 9). Patient information and clinicopathologic details of their tumors are shown in Supplementary Table S1.
NGS, variant calling, and copy number analysis
DNA was extracted using Omega Tissue DNA kit (D3396-02) under the manufacturer's protocols. Agilent SureSelect Human All Exon V5/V6 or Agilent SureSelect Kit (1 Kb-499 Kb) were used for the capture of the exome or 77 targeted genes, respectively. Samples were sequenced on Illumina HiSeq PE150. Sequence data were aligned to the GRCh37 assembly using multi-threaded Burrows–Wheeler Aligner (10) and Samblaster 0.1.22 (11) to obtain the comparison result in BAM format. The discovery cohort underwent whole-exome sequencing (WES) to an average depth of 117× (range: 95–163×) and an average coverage ratio of 99.46%. Targeted region sequencing in the validation cohort resulted in an average depth of 1,676× (range: 1,125–2,270×) with a mean coverage rate of 99.53%.
Somatic single-nucleotide variants (SNV) were detected using a dual calling strategy muTect 1.1.4 (12), and insertions/deletions (indel) of 1 to 50 bp in length were called with Strelka v1.0.13 (13). Significantly mutated genes (SMG) were identified using MutSigCV (14) at a threshold of q < 0.2. Genomic copy number assessment was performed with CNVkit (15). GISTIC analysis was performed to determine significant regions of recurrent copy number change (16).
Identification of mutational signatures
The deconstructSigs (https://github.com/raerose01/deconstructSigs) assigning to Catalogue of Somatic Mutations in Cancer (COSMIC) V2 and V3.2 database was employed to identify the mutational signatures (17). Input data for each sample was the deconstructSigs determined signature ‘‘weight’’ of each signature, where only signatures appearing in at least 5% of the samples at a minimum ‘‘weight’’ of 0.2 were used. We further confirmed the curated Signature 3 using SigMA, a computational tool using a likelihood-based measure combined with machine-learning techniques, in order to accurately detect homologous recombination deficiency (HRD)-related signatures even from samples with low mutation counts (18).
Microsatellite instability
Microsatellite instability (MSI) was assessed using two algorithms: mSINGS and MSIsensor. mSINGS was used as previously described by Salipante and colleagues (19). Based on previous validation studies, a fraction of more than 0.2 unstable loci were considered MSI-positive. MSIsensor score was calculated by dividing the number of microsatellite unstable by the total number of microsatellite stable sites detected as previously reported (20). A threshold of 3.5% (of loci called unstable) was recommended for MSIsensor.
Calculation of HRD score
For the determination of HRD scores using WES, BAM files of tumor samples were applied to the Sequenza 2.2.0 followed by the scarHRD R package with a default parameter as previously described (https://github.com/sztup/scarHRD; ref. 21).
Variant calling and clinical annotation of genes associated with homologous recombination repair pathways or PARP inhibitor sensitivity
The variants in the germline exomes of 60 WES samples were called using samtools (22). Germline or somatic variants among 142 protein-coding genes that are either operative in homologous recombination repair (HRR) pathways or unambiguously implicated in PARP inhibitor sensitivity according to Lord and colleagues (23) were extracted from the call set. We searched for the presence of mutations in both the tumor and nonneoplastic tissue from the same individual, which was normally filtered out of the data when looking for somatic mutations. All genotype calls with a genotype quality score <20 were removed, along with sites with a mapping quality score of <30 or sequencing depth <4.
Germline events were considered to be pathogenic if they resulted in a premature stop codon (i.e., nonsense mutation) or a frameshift (excluding BRCA2 p.Lys3326*, which is known to be a low-penetrance allele and is consequently a special case). In addition, missense mutations that were annotated as pathogenic in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) were also considered pathogenic in our analysis. We treated splice-site variants similarly to missense mutations.
In addition, for each of the heterozygous germline variants, we also annotated their allelic imbalance in the corresponding tumor. We assessed the allelic imbalance by tallying reads that supported the reference and the nonreference alleles at the germline site in the corresponding tumor WES data (BAM file). pyLOH 2.2.0 was employed to determine whether the alternate allele was lost in cases of loss of heterogeneity (LOH); cases in which the test yielded P < 0.05 were considered “alternate retained”. For P > 0.05, we defined the LOH status as undetermined (24).
RNA sequencing processing and analysis pipeline
RNA sequencing (RNA-seq) library for acral melanoma samples was generated using NEBNext Ultra™ RNA Library Prep Kit. The library preparations were sequenced on an Illumina Hiseq platform and 150 bp paired-end reads were generated. Index of the reference genome was built using Bowtie v2.2.3 (25) and paired-end clean reads were aligned to the reference genome using HISAT2 v2.0.5 (26). HTSeq was used to count the reads numbers mapped to each gene (27). And then FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) of each gene was calculated based on the length of the gene and the reads count mapped to this gene. Differential expression analysis was performed using the edgeR package (28). GSEA v3.0 was performed using JAVA program and run in preranked mode to identify enriched signatures (29). The gene set enrichment analysis (GSEA) plot, normalized enrichment score, and q values were derived from GSEA output for hallmark signature. The characterization of cell composition was calculated from gene expression of acral melanoma samples via xCell (30).
Statistical analysis
Statistical analyses were performed using SPSS 26.0 software. All statistical analyses were two-sided, and P < 0.05 was considered statistically significant. Corrections for multiple comparisons were performed and the acquired q value was indicated where necessary.
Data availability
The processed data generated in this study are available within the article and its supplementary files. The raw data can only be made available following approval from the ethics review board of the Fourth Military Medical University. Request for access should be directed to the corresponding author.
Additional materials and methods can be found in supplementary materials and methods.
Results
Paired tumor/constitutional WES (average depth: 117×) of acral melanoma samples was performed in a discovery set of 60 acral melanoma samples from Chinese patients, of which 55 (91.7%) provided primary lesions and 5 (8.3%) gave locally recurrent lesions (Supplementary Table S1). Sequentially, targeted deep sequencing (average depth: 1,676×) of 77 candidate genes (Supplementary Table S2) was performed in a validation cohort of 48 specimens (Supplementary Table S1). All the tumors were from primary sites except for one from the locally recurrent site in the validation cohort.
The mutational burden and SMGs
WES identified 11,630 somatic SNVs and 163 somatic small indels. Concerning nonsynonymous events (missense, nonsense, indels, and splice-site), a median of 31 mutations (range: 4–103) across all tumor samples were observed with a median tumor mutational burden (TMB) of 1.09 mutations per megabase (range: 0.18–3.32 per megabase; Supplementary Table S3; Fig. 1A). NRAS, KIT, and BRAF were identified to be SMGs (q < 0.2) in the discovery cohort by MutSigCV (Supplementary Tables S4 and S5). Of these genes, KIT garnered the highest frequency of somatic mutation (n = 10, 16.7%; Fig. 1A). Mutations in KIT were distributed in multiple exons (exon 9, 11, 13, and 17; Fig. 1B). BRAF mutations were mainly V600E substitution (n = 8 of 9) with one infrequent G466V substitution (Fig. 1B). The SNVs of NRAS consisted of variants in Q61R (n = 6), Q61L (n = 2), and Q61K (n = 1; Fig. 1B). Two of four NF1 mutations detected were inactivating mutations (1 nonsense and 1 splice-site indel). Only one sample harbored a concurrent hotspot, i.e., oncogenic Q61L NRAS mutation, and an uncommon G466V BRAF mutation (Supplementary Table S6). The driver genes identified in the discovery cohort also had mutations in parallel loci in the validation cohort (Supplementary Fig. S1A).
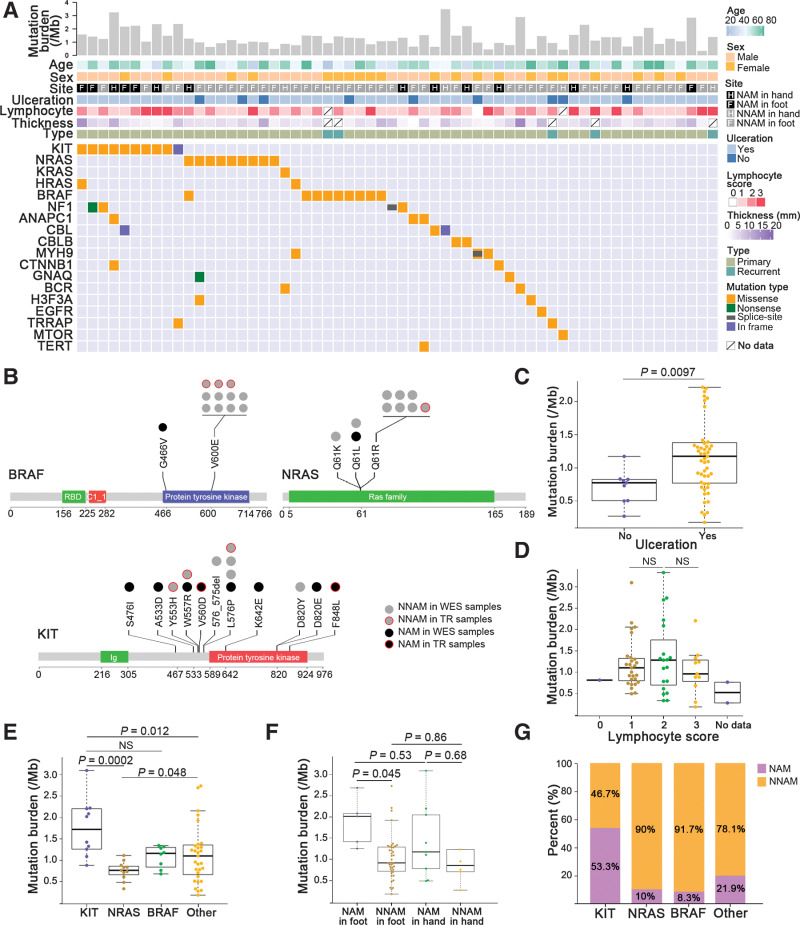
Figure 1.
Somatic variant burden and SMGs. A, From top to bottom: mutation burdens including SNVs and small indels per megabase; age; sex; site of the primary tumor (NAM and NNAM in hand or foot); ulceration; lymphocyte score; thickness; specimen type (primary or recurrent); mutations in driver genes previously described in melanoma and other cancers of the discovery cohort. B, The distribution of mutation sites on the protein coded by BRAF, NRAS, and KIT. Ig, immunoglobulin domain; RBD, Raf-like Ras-binding domain; C1_1, Phorbol esters/diacylglycerol binding domain. C, The difference of mutation burden between ulceration and nonulceration groups. The data are represented as a boxplot, where the middle line is the median, and the bottom and top edges of the box are the first and third quartiles (Mann–Whitney U test). D, The difference of mutation burden between lymphocyte score groups (Kruskal–Wallis test). NS, not significant. E, Mutation burden was significantly different between the genomic subtypes (Kruskal–Wallis test). F, The difference of mutation burden between NAM and NNAM in hand or foot (Kruskal–Wallis test). G, Proportion of NAM and NNAM with BRAF, NRAS, and KIT gene mutations (Fisher exact test).
Some other potentially pathogenic genes in melanoma showed somatic mutations with lower frequencies in our discovery cohort, including CBL, CTNNB1, GNAQ, TRRAP, EGFR, MTOR, and TERT (Supplementary Table S5; Fig. 1A). Another five identified candidate genes, including ANAPC1, MYH9, CBLB, BCR, and H3F3A, have been appreciated as driver mutated genes in other cancers (Supplementary Table S5; Fig. 1A). Notably, a number of latent driver genes for cutaneous melanoma, including TYRP1, TP53, PTEN, DDX3X, RASA2, PPP6C, RAC1, and RB1, carried neither SNVs nor indel mutations in our discovery cohort, implicating that the driving molecular events in acral melanoma differ markedly from those in cutaneous melanoma.
We found that TMB was higher in the ulcerated acral melanoma group (Mann–Whitney U test, P = 0.0097; Fig. 1C). There was no significant difference in TMB between patients with different degrees of lymphocyte infiltration (Kruskal–Wallis test; Fig. 1D). The samples with KIT aberration had higher TMB than those with NRAS mutation (P = 0.0002) or without any mutation of SMGs (Kruskal–Wallis test, P = 0.012; Fig. 1E). We then compared genetic mutational profiles between nail apparatus melanoma (NAM) and nonnail acral melanoma (NNAM) in hand or foot. Significant difference of the SNV burden was observed only between NNAM in foot (n = 40) and NAM in foot (n = 5; Kruskal–Wallis test, P = 0.045; Fig. 1F). Besides, we found significantly differential proportions occupied by the three SMGs between NAM and NNAM based on the combined discovery and validation cohort (25 NAM and 83 NNAM). Most of the mutated BRAF (91.7%) and NRAS (90%) targeted samples of NNAM. NAM samples obtained 53.3% of the total KIT mutations, which was significantly higher than that in BRAF- (P = 0.019), NRAS- (P = 0.041), and other genes- (P = 0.023) mutated subgroups, respectively (Fisher exact test; Fig. 1G).
We leveraged computational workflow that integrates tumor mutation and expression data (DNA sequencing and RNA-seq) by HLA-PRG, NetMHCpan, and pVAC-Seq (31–33), for identification of neoantigens in the discovery cohort (Supplementary Fig. S2A and Supplementary Table S3). Sequentially, in tumors with matching RNA-seq (n = 37), no significant correlation between the neoantigen load of acral melanoma and mutation burden was observed (Pearson correlation, P = 0.19, r = 0.22; Supplementary Fig. S2B). Notably, neoantigens were markedly different among tumors with different lymphocytes scores (Kruskal–Wallis test, P = 0.022; Supplementary Fig. S2C and S2D). However, the neoantigens between NAM and NNAM exhibited no significant difference (Mann–Whitney U test, P = 0.23; Supplementary Fig. S2E).
The mutational signature and DNA damage repair deficiency of acral melanoma
Analysis on the mutation spectrum of the discovery cohort highlighted C > T transitions as dominant somatic base substitutions, which have also been observed in most cancers including cutaneous melanoma (Supplementary Fig. S3). Nevertheless, merely 17.2% to 41.6% of C > T transitions occurred at dipyrimidine sites, which was much lower than that (about 60%) in typical UV-induced mutation signature (ref. 34; Fig. 2A; Supplementary Table S3). The proportions of CC > TT mutations (0.7%, range: 0.5%–1.6%) were also similarly lower than the recognized threshold (5%; Supplementary Table S3). Thus, UV is probably not a cardinal mutagen of acral melanoma in Asian populations.
Figure 2.
Mutational signature contributions and enrichment of DNA repair–related genetic lesion in acral melanoma. A, From top to bottom: TMB; site of the primary tumor (NAM and NNAM in hand or foot); percent of C > T transitions occurring in dipyrimidine sequences; proportions of somatic mutations in the six mutation signatures for each individual; Signature 3–annotated or not by SigMA; HRD score; genetic lesions by type in selected HR-related genes, with mutation type indicated by color-coding according to the key. HomDEL, homozygous deletion. B, Distribution of values of HRD-LOH, telomeric allelic imbalances, large-scale state transitions events, and HRD-sum between Signature 3+ samples (showing contribution of Signature 3 by both deconstructSigs and SigMA) and others (Mann–Whitney U-test). C, Comparison of HRD-sum values in acral melanoma, BRCA1- or BRCA2-deficient TNBC samples and non–BRCA1/2-deficient TNBC samples (Mann–Whitney U test). D, The correlation between the percentage of the genome affected by CNVs [gains of copy number (CN ≥ 3) and copy number loss (CN ≤ 1)] and HRD-sum values in acral melanoma samples (Spearman correlation test). E, The comparisons of Signature 3+ sample proportions between TWT samples and non-TWT samples when annotated (Fisher exact test). NS, not significant.
To determine mutational processes most likely acting on the development of acral melanoma, we identified six single-base substitution mutational signatures in the COSMIC V2 database in the cohort using deconstructSigs (Fig. 2A). Of the identified signatures, Signature 3 represents HRD of double-strand breaks (DSB) and has been curated in mucosal melanoma and TWT melanoma (35, 36). Of the 33 samples identified to harbor Signature 3, 19 showed more than 25% contribution of Signature 3, and 10 samples were simultaneously annotated by Signature 3 in further SigMA validation (Fig. 2A). The age-related Signature 5 that had been discerned in acral melanoma previously was the most prevalent in our discovery cohort (n = 37 of 60; ref. 3). Four other signatures (Signature 16, 9, 12, and 20) presented in our results were not identified in melanoma previously. Signature 16, which has been described exclusively in liver cancers despite being of unknown etiology, was present with more than 25% contribution in 26 acral melanoma specimens (37). Signature 9 that is related to polymerase η with activation-induced deaminase activity during somatic hypermutation and Signature 12 that is also marked as unknown etiology presented with minor contributions (<25%) in most of the annotated samples (17/24 and 13/20, respectively). Finally, only 12 tumors (6/12 having a <25% contribution) were annotated by Signature 20 which is associated with defective DNA mismatch repair. Doublet Base Substitution (DBS) signature analysis established DBS11 (APOBEC mutagenesis) as the most similar signature (Supplementary Fig. S4A and S4B). While small indel–signature analysis identified a total of four signatures, including ID8, ID10, ID12, and ID16 (the latter three are etiology unknown; Supplementary Fig. S5). ID8 was established to be the characteristic of DNA DSB repair by non–homologous recombination–based end-jointing mechanisms, which repairs DSBs compensatively in the absence of effective HRR (23, 38). In addition, deconstructSigs assigning to COSMIC V3.2 also detected some acral melanoma specimens having HRD-related Signature 3 (Supplementary Fig. S6). It deserves noting that a mutational signature that was similar to the UV radiation-related Signature 7 in our cohort was not identified using deconstructSigs assigning to COSMIC V2 or V3.2, though it is reported to dominate most cutaneous melanoma and fractional acral melanoma samples in previous studies (3, 39).
In search of the underlying genetic alteration that contributed to the observed HRD, we examined these 60 cases for any deleterious alterations [e.g., clinically relevant germline or somatic frameshift/nonsense variants, well-characterized pathogenic missense/splice-site germline variants (ClinVar), and/or homozygous deletion] in a catalog of classical genes that are operative HRR pathways or unambiguously implicated in PARP inhibitor (a classical therapeutic approach that target HRD) sensitivity according to Lord and colleagues (23). Besides, we analyzed the LOH (the most common second-hit event) status of the same loci. Concerning alterations of HRD-related genes, 2 patients harbored pathogenetic germline variants of BRCA2 (nonsense with LOH of the wild-type allele and frame-shift insertion, respectively). Two patients carried FANCA (a gene responsible for DNA interstrand cross-link repair) germline mutation, one of which was frame-shift deletion while the other was nonsense with LOH of FANCA. Germline mutations of RECQL4, which promotes DNA end resection in the repair of DNA DSBs (40), were observed in seven tumors. Of the somatic mutations identified, copy number gains of EMSY (a gene that abrogates HRR by causing inactivation of BRCA2; ref. 41) were observed in 14 samples. Additional mutations in ZSWIM7 (germline, frame-shift insertion with LOH), WRN (somatic, homozygous deletions), and MAPK12 (somatic, homozygous deletions) were also detected in our patient cohort (Fig. 2A). Besides, HRD-related mutational events are enriched in Signature 3+ acral melanoma lesions (showing contribution of Signature 3 by both deconstructSigs and SigMA; Mann–Whitney U test, P = 0.026).
We continued to ascertain the HRD by leveraging the corresponding biomarker indices, including HRD-LOH, large-scale state transitions (LST), the number of telomeric allelic imbalances (TAI), and a combination of the three scores (HRD-sum; ref. 21). As a result, in the comparison between Signature 3+ acral melanoma lesions and other lesions, no significant difference was observed in numbers of HRD-LOH events (P = 0.78), TAI (P = 0.49), and LST (P = 0.12), while Signature 3+ lesions tended to have higher HRD-sum values with marginal significance (Mann–Whitney U test, P = 0.09; Fig. 2B). Of note, the distribution of HRD-sum values in acral melanoma lesions was similar to the situation in BRCA1-deficient triple-negative breast cancer (TNBC) samples as reported by de Luca and colleagues (42), and was significantly higher than BRCA2-deficient and non–BRCA1/2-deficient group (Mann–Whitney U test, P < 0.0001; Fig. 2C) and much higher than the value reported in Signature 3–enriched TWT melanoma (35), thus implying that acral melanoma probably harbors features of genomic instability due to HRD. We also found a positive correlation between HRD-sum and the percentage of the genome affected by CNVs [gains of copy number (CN ≥ 3) and copy number loss (CN ≤ 1); Spearman correlation test, r = 0.26, P = 0.046; Fig. 2D]. Furthermore, 19.4% TWT acral melanoma and 12.5% non-TWT acral melanoma samples were identified to be Signature 3+ with no statistical significance between them (Fisher exact test, P = 0.73; Fig. 2E). For the putative mismatch repair deficiency, we sought to identify tumors with mutations in polymerase proofreading and the mismatch repair pathway (POLE, POLD1, MLH1, MSH2, MSH6, and PMS2), but alterations of these genes were not discerned. Next, we measured the somatic microsatellite instability index using a bioinformatics approach with mSINGS and MSIsensor (19, 20). All the 60 acral melanoma samples, however, did not show greatly elevated microsatellite instability levels (Supplementary Fig. S7A and S7B). Therefore, mismatch repair deficiency probably contributes little to acral melanoma arising from Asian populations, at least in the perspective of our samples.
Copy number alterations and signaling pathway activation in acral melanoma
The percentage of the genome affected by CNVs [gains of copy number (CN ≥ 3) and copy number loss (CN ≤ 1)] was variable with a median of 14.2% of the genome affected (range: 0.1–54.7%; Fig. 3A; Supplementary Table S3). A total of 110 recurrent focal CNVs were determined using GISTIC (q < 0.2; Supplementary Fig. S8; Supplementary Table S7). Major gain events comprised KIT, KDR, and PDGFRA (23.3%, 14/60, each) in the same amplicon on chromosome (Chr) 4; TERT (33.3%, 20/60) on Chr 5; CCND1 (35%, 21/60), GAB2 (close to PAK1, 31.7%, 19/60), and YAP1 (13.3%,8/60) on Chr 11; CDK4 (23.3%, 14/60) and MDM2 (16.7%, 10/60) on Chr 12; and EP300 (31.7%, 19/60) on Chr 22, and major loss events comprised CDKN2A (38.3%, 23/60) on Chr 9 and NF1 (11.7%, 7/60) on Chr17. CNV frequency of the genes, which were simultaneously mentioned above and included in targeted deep sequencing, were broadly in line with those in the validation cohort (Supplementary Fig. S1B; Supplementary Table S3). Intriguingly, 57.1% (8/14) of the samples that had copy number gains of CDK4 showed the cooccurrence of MDM2 gains.
Figure 3.
Copy-number variants and signaling pathway alterations in acral melanoma. A, From top to bottom: percent of the genome affected by copy-number aberrations [gains of copy number (CN ≥ 3), normal copy number (CN = 2), and copy number loss (CN ≤ 1)]; age; sex; site of the primary tumor (NAM and NNAM in hand or foot); ulceration; lymphocyte score; Breslow thickness; specimen type (primary or recurrent); CNVs of selected melanoma-associated genes: gain (CN ≥ 3, red), gain and mutation (gain with SNV or indel, dark red), loss (CN ≤ 1, green), loss and mutation (loss with SNV or indel, dark green), and homozygous deletion (CN = 0, black). B, Percentage of tumors with protein-affecting aberrations in candidate driver genes grouped by pathway: substitution/indels (blue), copy number gain (red), and copy number loss (green). C, Frequency of aberrations in pathways as a percentage of tumors in the discovery cohort. D, Kaplan–Meier curves of patients with acral melanoma by CNV and/or SNV in histone modification pathway (with vs. without). HomDEL, homozygous deletion.
We compared the patterns of CNVs between NAM and NNAM in the discovery cohort. The 4q12 focal gains containing the oncogene KIT were detected with a marginal significance (q = 0.061) in NAM, whereas in NNAM, we observed notable copy number gains of TERT, EP300, and CDK4 (Supplementary Fig. S9A). Except for the regions of recurrent copy-number aberrations included in lymphocyte score-low group (lymphocyte score = 0 or 1, n = 28), focal gains of 4q12 containing the oncogene KIT and 11q14.1 containing PAK1 were detected with significance (q < 0.05) in lymphocyte score-high group (lymphocyte score = 2 or 3, n = 30; Supplementary Fig. S9B).
We integrated SNVs and CNVs to identify recurrently targeted pathways and found the three most impacted signaling pathways, including MAPK (81.7%), cell-cycle progression (80%), and histone modification (66.7%; Fig. 3B and C). The patients acquiring CNVs of histone modification pathway tended to have worse overall survival (OS) though with no significance (log-rank, P = 0.099; Fig. 3D).
Genetic aberrations in the EP300 are associated with the responsiveness to p300 inhibitor
Our data showed a higher frequency of EP300 copy number gain in acral melanoma compared with cutaneous melanoma (11.75%) and other cancers (range: 0–10.71%) according to The Cancer Genome Atlas (Supplementary Fig. S10). EP300 encodes the histone acetyltransferase paralogue p300 that manipulates a host of cellular processes (e.g., growth, survival, apoptosis, and DNA repair) and promotes the growth of tumor through its downstream oncogene target MITF (43). To further evaluate the correlation of EP300 pathway aberrations to clinicopathologic features of acral melanoma, we verified the frequency of EP300 and its crucial downstream target MITF affected by copy number gains in acral melanoma by performing the FISH assay in a larger patient cohort (233 paraffin-embedded acral melanoma samples including 54 available tumors from the discovery cohort; Supplementary Table S8). The frequency of copy number gains of EP300 and MITF was 24.5% (n = 57) and 7.3% (n = 17), respectively. In total, 30% (70/233) of the acral melanoma lesion samples carried the copy number gains of the EP300-MITF axis. No significant differences in the median age and sex distribution were observed between patients with different EP300 pathway CNVs. However, we found that the Breslow thickness was markedly increased in subgroups that carried EP300 gains [3.1 (0.75–12.0) vs. 2.7 (0.2–46.0) mm, P = 0.019], MITF gains [4.5 (1.5–46.0) vs. 2.7 (0.2–20.0) mm, P = 0.006], or both [3.8 (0.75–46.0) vs. 2.5 (0.20–20.0) mm, P < 0.001] compared with controls (Table 1). Meanwhile, ulcerations were significantly enriched in tumors with gains of the EP300-MITF axis compared with controls (83.8% vs. 66.9%, P = 0.009). Furthermore, the patients with the copy number gains of EP300 demonstrated a trend toward a higher proportion of stage III/IV (36.8% vs. 24.4%) with borderline significance (P = 0.068). These results indicated that acral melanoma with copy number gains of EP300 pathway tended to be more aggressive.
Table 1.
Correlation of the copy number variation with the EP300-MITF axis to clinicopathologic features in 233 acral melanoma samples.
| EP300 aberration | MITF aberration | Overall aberration | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Clinicopathologic features | Gain | Normal | P a | Gain | Normal | P a | Gainb | Normal | P a |
| Age (year) | |||||||||
| Median (range) | 61 (29–82) | 58 (18–89) | 0.146 | 53 (31–81) | 60 (18–89) | 0.235 | 61 (29–82) | 59 (18–89) | 0.377 |
| Sex n (%) | |||||||||
| Male | 27 (47.4) | 75 (42.6) | 0.529 | 9 (52.9) | 93 (43.1) | 0.429 | 32 (45.7) | 70 (42.9) | 0.696 |
| Female | 30 (52.6) | 101 (57.4) | 8 (47.1) | 123 (56.9) | 38 (54.3) | 93 (57.1) | |||
| Ulceration n (%) | |||||||||
| Yes | 45 (81.8) | 117 (68.8) | 0.062 | 15 (88.2) | 147 (70.7) | 0.121 | 57 (83.8) | 105 (66.9) | 0.009 |
| No | 10 (18.2) | 53 (31.2) | 2 (11.8) | 61 (29.3) | 11 (16.2) | 52 (33.1) | |||
| Missing | 2 | 6 | 0 | 8 | 2 | 6 | |||
| Thickness (mm) | |||||||||
| Median (range) | 3.1 (0.75–12.0) | 2.7 (0.2–46.0) | 0.019 | 4.5 (1.50–46.0) | 2.7 (0.2–20.0) | 0.006 | 3.8 (0.75–46.0) | 2.5 (0.20–20.0) | 0.001 |
| Stagesc, n (%) | |||||||||
| I | 7 (12.3) | 39 (22.2) | 0.135 | 0 (0) | 46 (21.3) | 0.061 | 7 (10.0) | 39 (23.9) | 0.032 |
| II | 29 (50.9) | 94 (53.4) | 13 (76.5) | 110 (50.9) | 39 (55.7) | 84 (51.5) | |||
| III | 16 (28.1) | 28 (15.9) | 4 (23.5) | 40 (18.5) | 19 (27.1) | 25 (15.3) | |||
| IV | 5 (8.8) | 15 (8.5) | 0 (0) | 20 (9.3) | 5 (7.1) | 15 (9.2) | |||
| Advance stagesd, n (%) | 21 (36.8) | 43 (24.4) | 0.068 | 4 (23.5) | 60 (27.8) | 0.706 | 24 (34.3) | 40 (24.5) | 0.127 |
| Total, n (%) | 57 (24.5) | 176 (75.5) | 17 (7.3) | 216 (92.7) | 70 (30.05) | 163 (69.95) | |||
aFor evaluation of age, the two independent sample t tests were used. For evaluation of sex, ulceration, and stages, the χ2 tests were used. For evaluation of thickness, Mann–Whitney U tests were used.
bSamples with copy number gains of either EP300 or MITF or both.
cClinical stages were assessed according to American Joint Committee on Cancer (AJCC) Cancer Staging Manual (seventh edition).
dIII and IV stages.
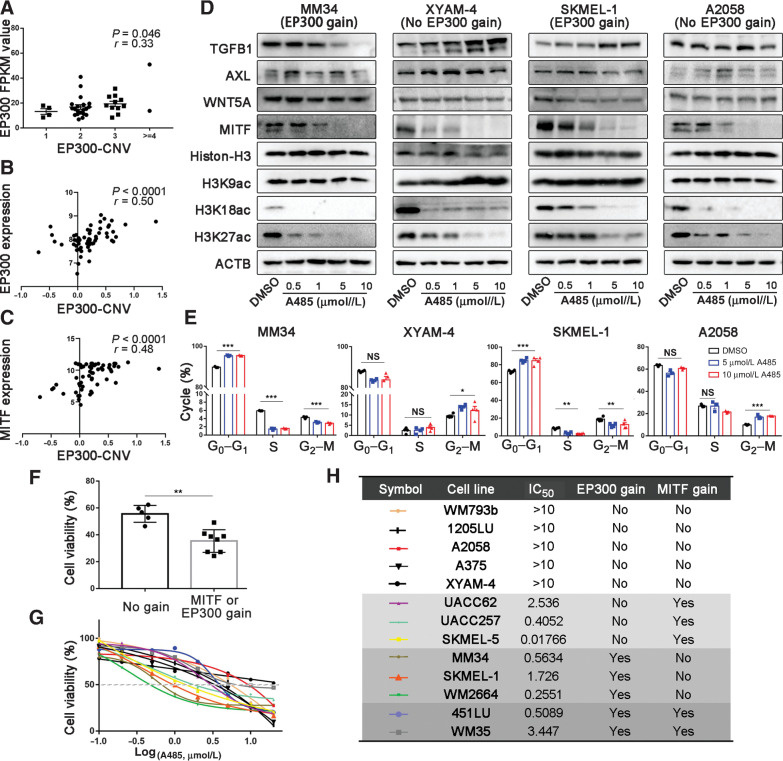
Generally, the gene with a higher copy number could be expressed at a higher level. Indeed, we found a positive correlation between EP300 copy number and p300 expression level in the RNA-seq data of 37 acral melanomas in the discovery cohort (Spearman correlation test, r = 0.33, P = 0.046; Fig. 4A), which was also confirmed by results from Cancer Cell Line Encyclopedia (CCLE) database (Spearman correlation test, r = 0.50, P < 0.0001; Fig. 4B). Moreover, the expression level of MITF was also significantly correlated with the copy number of EP300 (data from CCLE database, Spearman correlation test, r = 0.48, P < 0.0001; Fig. 4C). We observed that A485 (a p300-specific inhibitor) inhibited the acetylation of histone H3K18 and H3K27 and the expression of MITF in a dose-dependent manner in melanoma cell lines MM34, XYAM-4, SKMEL-1, and A2058 (the former two are acral melanoma cell lines; Fig. 4D; ref. 44). However, A485 treatment could only block cell cycle progression of the cell lines with EP300 gain (MM34 and SKMEL-1) instead of those without EP300-MITF axis gain (A2058 and XYAM-4; Fig. 4E; Supplementary Fig. S11), which indicates that the suppressive effect of A485 on the melanoma is associated with EP300-MITF axis aberrations. To assess the efficacy of A485 in suppressing melanoma and further confirm the correlation, we tested the IC50 value of A485 in melanoma cell lines with various EP300 pathway variations. We noted that cell lines containing EP300 gain or MITF gain or gain of both genes were significantly more sensitive to A485 than those without EP300/MITF gains (Fig. 4F–H). Also, copy numbers of both EP300 and MITF were markedly higher in cell lines sensitive to A485 (IC50 value < 10 µmol/L) than those not sensitive (IC50 value > 10 µmol/L; Supplementary Fig. S12). Therefore, the copy number gain of EP300 pathway could be a biomarker of the sensitivity to p300 inhibitor in acral melanoma. Intriguingly, despite that MITF downregulation reportedly switches tumor cells to a more aggressive phenotype (45), Western blotting assays (Fig. 4D) combined with Transwell assays (Supplementary Fig. S13) showed that A485 did not alter the capacity of invasion or migration of the four cell lines in the context of downregulation of MITF expression.
Figure 4.
Melanoma cells with EP300 or MITF copy number gains are sensitive to p300 inhibitor A485. A, The correlation between gene copy number and the mRNA expression of EP300 in our WES samples, n = 60 (Spearman correlation test). B and C, The correlation between EP300 copy number and the mRNA expression of EP300 and MITF by data from CCLE (Spearman correlation test). D, The effect of A485 on the expression of TGFB1, AXL, WNT5A, MITF, total H3 and acetylated H3K9, H3K18, and H3K27 in MM34 (acral melanoma cell line with EP300 gain), XYAM-4 (acral melanoma cell line without EP300 gain), SKMEL-1 (nonacral melanoma cell line with EP300 gain), and A2058 (nonacral melanoma cell line without EP300 gain). E, The effect of A485 on the cell cycle of melanoma cell lines was examined by flow cytometry. F, The comparisons of cell viability after 5 µmol/L A485 treatment between cell lines with MITF/EP300 gains and those without. G and H, Viable cell titer was determined in different melanoma cell lines under the treatment of A485 in concentration gradient. The IC50 values and the state of EP300 or MITF gene copy number variation for these cell lines are shown. * P < 0.05, ** P < 0.01, *** P < 0.001. NS, not significant.
EP300 gain reflects the responsiveness of acral melanoma to anti–PD-1 treatment
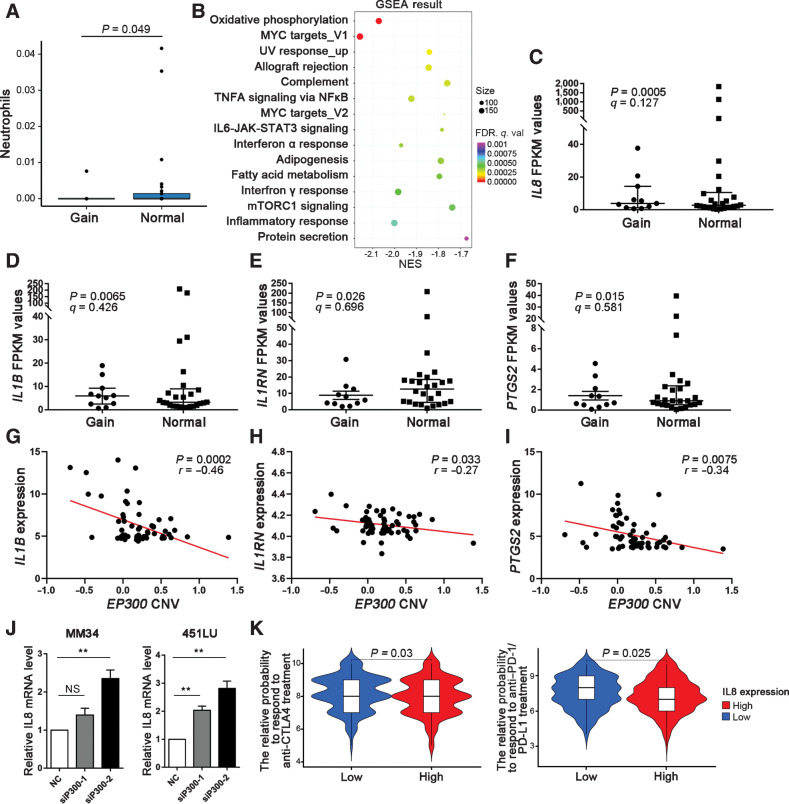
We evaluated intratumoral immune cell compositions in acral melanoma by xCell with the RNA-seq data of 37 acral melanoma samples (30). It turned out that acral melanoma samples without EP300 gains were infiltrated by more neutrophils (Mann–Whitney U test, P = 0.049; Fig. 5A). The GSEA found that genes from several pathways exhibited significantly different expression between EP300-gain and normal samples, among which the enriched inflammatory response pathway indicated the negative correlation between EP300 copy number gain and "smoldering" inflammatory environment that may support the evolution of tumoral immune escape in acral melanoma (q = 0.0005; Fig. 5B). Further analysis showed that EP300-gain samples had decreased expression of proinflammatory genes (IL8, IL1B, IL1RN, and PTGS2; Fig. 5C–F). The expression levels of IL1B, IL1RN, and PTGS2 were negatively correlated with the copy number of EP300 according to CCLE database (Fig. 5G–I). Moreover, the results of quantitative real-time PCR demonstrated that the interference of EP300 by siRNA promoted the expression of IL8 (Fig. 5J).
Figure 5.
Correlation of EP300 copy-number aberrations with patterns of immune infiltration and response to ICB treatment. A, Histograms represent the proportion of neutrophils (from xCell) for different EP300 copy number groups (Mann–Whitney U test). Error bars show the SEM. B, The difference of enriched pathways revealed by GSEA analysis of RNA-seq data between EP300-gain and EP300-normal sample. C–F, Differential expression of IL8, IL1B, IL1RN, and PTGS2 between EP300-gain and EP300-normal tumors. EP300-normal, n = 24; EP300-gain, n = 13. G–I, The correlation between EP300 copy number and the mRNA expression of IL1B, IL1RN, and PTGS2 by data from CCLE (Pearson correlation test). J, The effect of EP300 interference by siRNA on the expression of IL8 in MM34 and A2058 cells. K, The relative probabilities to respond to anti–PD-1/PD-L1 and anti–CTLA-4 treatment between EP300-gain and EP300-normal melanoma groups. NES, normalized enrichment scale; q. val, q value; NS, not significant.
Previous studies have reported that a neutrophil-dominated immune microenvironment hinders the therapeutic efficiency of immune checkpoint blockade (ICB) treatment in tumors (46–49). And it is reported that high systemic and tumor-associated IL8 correlates with the reduced clinical benefit of PD-L1 blockade (50). Hence, we speculated that acral melanoma with downregulated IL8 could be more sensitive to anti–PD-1 treatment. Immunophenoscore (IPS) is a machine learning-based scoring scheme, which could predict patients’ responses to ICB in silico (51). We used two subtypes of IPS values (IPS-PD-1/PD-L1/PD-L2_pos and IPS-CTLA-4_pos) as the surrogates of melanoma patients’ responses to anti–PD-1/PD-L1 and anti–CTLA-4 treatment. In this predictive model, the relative probabilities to respond to anti–PD-1/PD-L1 and anti–CTLA-4 treatment were lower in the IL8-high group (Mann–Whitney U test, P = 0.025 and P = 0.03, respectively; Fig. 5K). The results indicate that patients with EP300 copy number gains and downregulated IL8 expression in acral melanoma might be responsive to ICB treatment.
Temporal ordering of genetic changes in acral melanoma development
Sequencing data from a single biopsy represent a snapshot collected across the time axis of somatic evolution. Although timing estimates of individual events reflect evolutionary periods that differ from one sample to another, they partly define the order in which driver mutations and copy number alterations have occurred in each sample. Assembling those orderings of all the samples in our cohort by PhylogicNDT helped to describe a probabilistic order of somatic mutations acquisition (Fig. 6A). We found that highly enriched copy number events occurred before the formation of the key coding mutation. Specifically, losses in arms 10q, 10p, and 16p were among the earliest events, followed by losses or gains in many chromosomes. Mutations in KIT, BRAF, and NRAS tended to occur at intermediate time points, whereas other rare driver mutations were discerned as later events.
Figure 6.
Characterization of the clonal architecture of driver mutations in acral melanoma. A, Aggregating single-sample ordering reveals preferential ordering diagrams of driver mutations for acral melanoma. B and C, The point mutation (B) and the percentage of the genome affected by CNVs [gains of copy number (CN ≥ 3) and copy number loss (CN ≤ 1; C) along acral melanoma progression (stratified by Breslow thickness). The data are represented as a boxplot, where the middle line is the median, and the lower and upper edges of the box are the first and third quartiles (Mann–Whitney U test). D, The copy-number landscape at each phase of melanoma progression (stratified by Breslow thickness). Significantly different regions affecting copy-number aberrations are highlighted. E, From top to bottom: number of clonal and subclonal mutations identified in 60 acral melanoma cases; sample subtypes; Breslow thickness. F, The correlation between number of subclonal and Breslow thickness (Pearson correlation test). G, Clonality of candidate driver genes in 60 acral melanoma cases studied. Color spectrum: dark blue to light blue corresponds to estimated CCF (Cancer Cell Fraction). Samp, sample.
Sequentially, we stratified the tumors by Breslow thickness that reflects the progression of acral melanoma and found that the SNV and the percentage of the genome affected by CNVs [gains of copy number (CN ≥ 3) and copy number loss (CN ≤ 1)] escalated significantly with the acral melanoma progression (Fig. 6B and C). It is noteworthy that copy-number aberrations preferentially affected certain chromosomal regions in a stereotypic sequential order. Specifically, copy number aberration frequencies in 8q24.22 and 7p11.2 were significantly higher in samples with higher Breslow thickness (q < 0.1; Fig. 6D). Furthermore, a median of 108 subclonal SNV mutations (only missense mutations) were identified in each tumor (range: 32–266). The proportion of subclonal SNV mutations was significantly higher in acral melanoma samples with deeper Breslow thickness (Spearman correlation test, r = 0.34, P = 0.01; Fig. 6E and F), suggesting that in these tumors the SNV subclonal populations were more genetically heterogeneous.
The clonal status of 56 driver SNVs (only missense mutations) was identified by ABSOLUTE in 45 out of 60 acral melanoma specimens in the discovery cohort (Fig. 6G; Supplementary Fig. S14). Twenty-nine out of 45 patients were carriers of clonal mutations (2 of 29 with additional subclones) and the remaining 16 harbored isolated subclonal point mutations. A driver mutation in KIT was clonal in all the tumors (n = 9) garnering this alteration, implicating KIT mutation as an early and critical SNV event in the evolutionary history of acral melanoma. In addition, a group of mutated genes like BRAF, NRAS, and NF1 was identified to comprise both clonal and subclonal mutations, respectively.
Discussion
The current study presented genomic mutational features of acral melanoma and delineated their correlation to clinicopathology. In the jigsaw puzzle of the fitting-together global studies on genomic pathogenesis of acral melanoma, the current study might contribute to some pieces of the “Asian acral melanoma” that otherwise would be lacking. The dominance of primary lesion samples in our patient cohort could substantially reduce the confounding biases that arise from the driver mutations acquired in metastasis and confine the present genomic aberrance to early mutations in acral melanoma evolvement.
The UV-related signatures were not found to be consistently curated or contribute much in the mutational signature analysis using various algorithms based on our cohort. The well-established characteristics of highly frequent C > T or CC > TT mutations at the dipyrimidine sites were not observed in our samples due to the lower proportions than the recognized threshold, either. Hence UV radiation might not be the cardinal reason for the formation of driver mutations in Asian patients with acral melanoma, in contrast with the definite UV footprints in the genome of acral melanoma arising from European cohorts. We surmise the explanation might be that Asians relatively lack sun-seeking behavior and that the skin color of the Asians (mainly Fitzpatrick type III and IV) is more UV-protective compared with Europeans.
Of note, HRD-related mutational signature was observed in acral melanoma samples. Furthermore, mutations in a series of genes (e.g., BRCA2, FANCA) detected in our cohort were probably responsible for genomic HRD borne in corresponding acral melanoma specimens. In addition, HRD-sum value was relatively higher with marginal significance in samples identified to harbor Signature 3 by both deconstructSigs and SigMA when compared with those that do not have. Moreover, the HRD-sum values of the acral melanoma cohort were so high that they approximated to that of BRCA1-mutated TNBC. Therefore, we speculated that the observed footprint of DNA repair deficiency in our samples might result from the separate or synergistic work of mutations or aberrant epigenetic modification of related genes, and baseline endogenous DNA damage overwhelming the originally proficient DNA restorative capacity.
Reliable quantification of HRD in human tumor biopsies is expected to identify patients that are particularly sensitive to platinum or PARP inhibitor–based therapy. A prior randomized phase II trial concluded that the PARP inhibitor, veliparib, could not significantly prolong the OS of patients with metastatic melanoma (52). However, pretreatment verifications of BRCA mutations or HRD were not performed in the trial to screen out potentially sensitive patients, and patients with acral melanoma were not included in the trial. Therefore, the feasibility of using HRD quantification to evaluate the potential of PARP inhibitors in acral melanoma treatment needs further verification.
Our findings may facilitate the selection of therapeutic targets among the enriched CNVs of candidate driver genes in acral melanoma. The EP300-MITF axis was gained at a high rate of 30% in patients with acral melanoma patients. Moreover, melanoma cell lines with copy number gains of the EP300-MITF axis were more sensitive to A485, a p300-targeted inhibitor. As stable genomic lesions, CNVs were superior to mRNA expression levels in predicting the sensitivity to gene-targeted drugs. Therefore, CNVs of the EP300-MITF axis could be targeted and utilized to optimize the screening of patients who may be potentially responsive to p300 inhibitors. Notably, we found that EP300 copy number gains had inherent relation with both decreased expression of IL8 and enhanced neutrophil infiltration as well as levels of inflammatory cytokines. Accordingly, the inclusion of abrogating IL8 function in the treatment strategy for acral melanoma without EP300 copy number gains might enhance the efficacy of immunotherapy to acral melanoma (53).
Intratumoral genetic heterogeneity of acral melanoma was another concern of the current study. We inferred a stereotypical pattern of gradually increased SNVs and CNVs along acral melanoma progression. Single patient trajectories can be aggregated together to build preferential timing models and elucidate the average order of events in a specific cancer type. In this way, our findings implicated that sufficient chromosomal instability is acquired at the initial stages of acral melanoma, which is further supported by a prior study suggesting that gene amplifications occur before the formation of the invasive portion in acral melanoma (54). This is not the case with the known rules of cutaneous melanoma clonal evolution that pathogenic SNVs are often early events while CNVs occur later (54, 55). The findings substantiate both the fundamentality of mechanisms that trigger the genomic instability, especially the occurrence of CNVs, in acral melanoma oncogenesis and the feasibility of employing CNVs as an early diagnostic marker of acral melanoma. Furthermore, the clonal mutations affecting a specific gene suggest that these alterations are possibly acquired at or prior to the most recent selective sweep before sampling. Conversely, the subclonal mutations of driver genes imply the late presence of the variants that probably occur after clonal selection. In this study, KIT mutations were identified to be consistently clonal, and thus might be a critical driver of acral melanoma. Mutations in other common driver genes, such as BRAF and NRAS, could be either clonal or subclonal. Accordingly, a targeted therapy directed at subclonal driver mutations may wane the sensitive clones but conceivably trigger a net growth of neighboring resistant subclones (56). This suggests that the survey of clonal status in addition to screening mutations in driver genes clinically might benefit patients with acral melanoma.
Moreover, our studies, together with previous reports, illustrated the heterogeneity in clinicopathologic phenotypes and the genome among patients with acral melanoma with different primary sites. Wei and colleagues analyzed data of 1,157 patients with acral melanoma and found those with primary lesions in sole were at more advanced stages upon initial diagnosis and had a worse prognosis compared with the patients with primary lesions in palm or nail bed (57). In this study, NAM significantly accumulated SNVs and CNVs of KIT, whereas mutations of BRAF, NRAS, CDK4, EP300, and TERT preferentially targeted NNAM, which indicates differential targeted treatment strategies for the two subtypes of acral melanoma. Furthermore, NAM in the foot harbored higher TMB compared with NNAM in the foot. The genomic differences between different subtypes of acral melanoma primary sites should be explored in a larger patient cohort to propose strategies of more precise targeted therapy.
In aggregate, this WES study provides an overview of the genome aberrations in acral melanoma lesions from Asian patient populations. Acral melanoma mutational signature indicates that DNA damage repair deficiency like HRD might be implicated in acral melanoma development and progression. The study also identifies EP300-MITF axis affected by copy number gains to be clinically targetable. Moreover, the clarification of acral melanoma clonal structure necessitates the exploitation of the clonal status of mutated driver genes in the patients before setting on gene-targeted therapy.
Supplementary Material
Acknowledgments
We thank Prof. Kai Ye, MOE Key Laboratory for Intelligent Networks and Network Security, Faculty of Electronic and Information Engineering, Xi'an Jiaotong University (Xi'an, Shaanxi, China), and Prof. Guanglei Zhuang, State Key Laboratory of Oncogenes and Related genes, Shanghai Center Institute, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University (Shanghai, China), for their kindly help to our study. We thank Prof. Juan Su, Department of Dermatology, Xiangya Hospital, Central South University (Changsha, Hunan, China), for providing us XYAM-4 cell line as a gift. The research leading to these results has received funding from National Natural Science Foundation of China (Nos. 81625020, 81902791, 81903207, and 81702714).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
Q. Shi: Conceptualization, formal analysis, supervision, validation, investigation, writing–review and editing. L. Liu: Data curation, formal analysis, validation, investigation, writing–original draft, writing–review and editing. J. Chen: Formal analysis, validation, writing–original draft, writing–review and editing. W. Zhang: Funding acquisition, writing–original draft, writing–review and editing. W. Guo: Funding acquisition, investigation, writing–review and editing. X. Wang: Data curation, software, formal analysis, validation. H. Wang: Formal analysis, investigation. S. Guo: Resources, methodology. Q. Yue: Formal analysis, investigation. J. Ma: Formal analysis, investigation. Y. Liu: Visualization, methodology. G. Zhu: Resources, funding acquisition, methodology. T. Zhao: Resources, methodology. J. Zhao: Investigation, visualization. Y. Liu: Resources, methodology. T. Gao: Conceptualization, resources, methodology. C. Li: Conceptualization, resources, supervision, funding acquisition, validation, project administration.
References
- 1. Jung HJ, Kweon SS, Lee JB, Lee SC, Yun SJ. A clinicopathologic analysis of 177 acral melanomas in Koreans: relevance of spreading pattern and physical stress. JAMA Dermatol 2013;149:1281–8. [DOI] [PubMed] [Google Scholar]
- 2. Lv J, Dai B, Kong Y, Shen X, Kong J. Acral melanoma in Chinese: a clinicopathological and prognostic study of 142 cases. Sci Rep 2016;6:31432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017;545:175–80. [DOI] [PubMed] [Google Scholar]
- 4. Liang WS, Hendricks W, Kiefer J, Schmidt J, Sekar S, Carpten J, et al. Integrated genomic analyses reveal frequent TERT aberrations in acral melanoma. Genome Res 2017;27:524–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Newell F, Wilmott JS, Johansson PA, Nones K, Addala V, Mukhopadhyay P, et al. Whole-genome sequencing of acral melanoma reveals genomic complexity and diversity. Nat Commun 2020;11:5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in Uveal melanoma. Cancer Cell 2017;32:204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smalley KSM, Teer JK, Chen YA, Wu JY, Yao J, Koomen JM, et al. A mutational survey of acral nevi. JAMA Dermatol 2021;157:831–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Black JRM, McGranahan N. Genetic and non-genetic clonal diversity in cancer evolution. Nat Rev Cancer 2021;21:379–92. [DOI] [PubMed] [Google Scholar]
- 9. Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015;161:1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Faust GG, Hall IM. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 2014;30:2503–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28:1811–7. [DOI] [PubMed] [Google Scholar]
- 14. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016;12:e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gulhan DC, Lee JJ, Melloni GEM, Cortes-Ciriano I, Park PJ. Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nat Genet 2019;51:912–9. [DOI] [PubMed] [Google Scholar]
- 19. Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clin Chem 2014;60:1192–9. [DOI] [PubMed] [Google Scholar]
- 20. Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014;30:1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sztupinszki Z, Diossy M, Krzystanek M, Reiniger L, Csabai I, Favero F, et al. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer 2018;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016;16:110–20. [DOI] [PubMed] [Google Scholar]
- 24. Li Y, Xie X. Deconvolving tumor purity and ploidy by integrating copy number alterations and loss of heterozygosity. Bioinformatics 2014;30:2121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dilthey AT, Gourraud PA, Mentzer AJ, Cereb N, Iqbal Z, McVean G. High-accuracy HLA type inference from whole-genome sequencing data using population reference graphs. PLoS Comput Biol 2016;12:e1005151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: Improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol 2017;199:3360–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, et al. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med 2016;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brash DE. UV signature mutations. Photochem Photobiol 2015;91:15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conway JR, Dietlein F, Taylor-Weiner A, AlDubayan S, Vokes N, Keenan T, et al. Integrated molecular drivers coordinate biological and clinical states in melanoma. Nat Genet 2020;52:1373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Newell F, Kong Y, Wilmott JS, Johansson PA, Ferguson PM, Cui C, et al. Whole-genome landscape of mucosal melanoma reveals diverse drivers and therapeutic targets. Nat Commun 2019;10:3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Letouze E, Shinde J, Renault V, Couchy G, Blanc JF, Tubacher E, et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun 2017;8:1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yeh I, Jorgenson E, Shen L, Xu M, North JP, Shain AH, et al. Targeted genomic profiling of acral melanoma. J Natl Cancer Inst 2019;111:1068–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu H, Shamanna RA, Keijzers G, Anand R, Rasmussen LJ, Cejka P, et al. RECQL4 promotes DNA end resection in repair of DNA double-strand breaks. Cell Rep 2016;16:161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell 2003;115:523–35. [DOI] [PubMed] [Google Scholar]
- 42. de Luca XM, Newell F, Kazakoff SH, Hartel G, McCart Reed AE, Holmes O, et al. Using whole-genome sequencing data to derive the homologous recombination deficiency scores. NPJ Breast Cancer 2020;6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim E, Zucconi BE, Wu M, Nocco SE, Meyers DJ, McGee JS, et al. MITF expression predicts therapeutic vulnerability to p300 inhibition in human melanoma. Cancer Res 2019;79:2649–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lasko L, Jakob C, Edalji R, Qiu W, Montgomery D, Digiammarino E, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017;550:128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Saez-Ayala M, Montenegro MF, Sanchez-Del-Campo L, Fernandez-Perez MP, Chazarra S, Freter R, et al. Directed phenotype switching as an effective antimelanoma strategy. Cancer Cell 2013;24:105–19. [DOI] [PubMed] [Google Scholar]
- 46. McDermott DF, Huseni MA, Atkins MB, Motzer RJ, Rini BI, Escudier B, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 2018;24:749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ugel S, De Sanctis F, Mandruzzato S, Bronte V. Tumor-induced myeloid deviation: when myeloid-derived suppressor cells meet tumor-associated macrophages. J Clin Invest 2015;125:3365–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blank CU, Haanen JB, Ribas A, Schumacher TN. Cancer immunology. The “cancer immunogram”. Science 2016;352:658–60. [DOI] [PubMed] [Google Scholar]
- 50. Yuen KC, Liu LF, Gupta V, Madireddi S, Keerthivasan S, Li C, et al. High systemic and tumor-associated IL-8 correlates with reduced clinical benefit of PD-L1 blockade. Nat Med 2020;26:693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 2017;18:248–62. [DOI] [PubMed] [Google Scholar]
- 52. Middleton MR, Friedlander P, Hamid O, Daud A, Plummer R, Falotico N, et al. Randomized phase II study evaluating veliparib (ABT-888) with temozolomide in patients with metastatic melanoma. Ann Oncol 2015;26:2173–9. [DOI] [PubMed] [Google Scholar]
- 53. Ridker PM, MacFadyen JG, Thuren T, Everett BM, Libby P, Glynn RJ, et al. Effect of interleukin-1beta inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet 2017;390:1833–42. [DOI] [PubMed] [Google Scholar]
- 54. Bastian BC, Kashani-Sabet M, Hamm H, Godfrey T, Moore DH II, Brocker EB, et al. Gene amplifications characterize acral melanoma and permit the detection of occult tumor cells in the surrounding skin. Cancer Res 2000;60:1968–73. [PubMed] [Google Scholar]
- 55. Shain AH, Joseph NM, Yu R, Benhamida J, Liu S, Prow T, et al. Genomic and transcriptomic analysis reveals incremental disruption of key signaling pathways during melanoma evolution. Cancer Cell 2018;34:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613–28. [DOI] [PubMed] [Google Scholar]
- 57. Wei X, Wu D, Li H, Zhang R, Chen Y, Yao H, et al. The clinicopathological and survival profiles comparison across primary sites in acral melanoma. Ann Surg Oncol 2020;27:3478–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed data generated in this study are available within the article and its supplementary files. The raw data can only be made available following approval from the ethics review board of the Fourth Military Medical University. Request for access should be directed to the corresponding author.
Additional materials and methods can be found in supplementary materials and methods.



![Figure 2. Mutational signature contributions and enrichment of DNA repair-related genetic lesion in acral melanoma. A, From top to bottom: TMB; site of the primary tumor (NAM and NNAM in hand or foot); percent of C > T transitions occurring in dipyrimidine sequences; proportions of somatic mutations in the six mutation signatures for each individual; Signature 3–annotated or not by SigMA; HRD score; genetic lesions by type in selected HR-related genes, with mutation type indicated by color-coding according to the key. HomDEL, homozygous deletion. B, Distribution of values of HRD-LOH, telomeric allelic imbalances, large-scale state transitions events, and HRD-sum between Signature 3+ samples (showing contribution of Signature 3 by both deconstructSigs and SigMA) and others (Mann–Whitney U-test). C, Comparison of HRD-sum values in acral melanoma, BRCA1- or BRCA2- deficient TNBC samples and non–BRCA1/2-deficient TNBC samples (Mann–Whitney U test). D, The correlation between the percentage of the genome affected by CNVs [gains of copy number (CN ≥ 3) and copy number loss (CN ≤ 1)] and HRD-sum values in acral melanoma samples (Spearman correlation test). E, The comparisons of Signature 3+ sample proportions between TWT samples and non-TWT samples when annotated (Fisher exact test). NS, not significant.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/afa4/9359751/6b10f67d5fcd/2690fig2.jpg)
![Figure 3. Copy-number alterations and signaling pathway activation in acral melanoma. A, From top to bottom: percent of the genome affected by copy-number aberrations [gains of copy number (CN ≥ 3), normal copy number (CN = 2), and copy number loss (CN ≤ 1)]; age; sex; site of the primary tumor (NAM and NNAM in hand or foot); ulceration; lymphocyte score; Breslow thickness; specimen type (primary or recurrent); CNVs of selected melanoma-associated genes: gain (CN ≥ 3, red), gain and mutation (gain with SNV or indel, dark red), loss (CN ≤ 1, green), loss and mutation (loss with SNV or indel, dark green), and homozygous deletion (CN = 0, black). B, Percentage of tumors with protein-affecting aberrations in candidate driver genes grouped by pathway: substitution/indels (blue), copy number gain (red), and copy number loss (green). C, Frequency of aberrations in pathways as a percentage of tumors in the discovery cohort. D, Kaplan–Meier curves of patients with acral melanoma by CNV and/or SNV in histone modification pathway (with vs. without). HomDEL, homozygous deletion.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/afa4/9359751/e78af5599bc3/2690fig3.jpg)
![Figure 6. Characterization of the clonal architecture of driver genes in acral melanoma. A, Aggregating single-sample ordering reveals preferential ordering diagrams of driver mutations for acral melanoma. The point mutation (B) and the percentage of the genome affected by CNVs [gains of copy number (CN ≥ 3) and copy number loss (CN ≤ 1)] (C) along acral melanoma progression (stratified by Breslow thickness). The data are represented as a boxplot, where the middle line is the median, and the lower and upper edges of the box are the first and third quartiles (Mann–Whitney U test). D, The copy-number landscape at each phase of melanoma progression (stratified by Breslow thickness). Significantly different regions affecting copy-number aberrations are highlighted. E, From top to bottom: number of clonal and subclonal mutations identified in 60 acral melanoma cases; sample subtypes; Breslow thickness. F, The correlation between number of subclonal and Breslow thickness (Pearson correlation test). G, Clonality of candidate driver genes in 60 acral melanoma cases studied. Color spectrum: dark blue to light blue corresponds to estimated CCF. Samp, sample.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/afa4/9359751/502a7dbd7b65/2690fig6.jpg)