Abstract
Androgen receptor (AR) is a major driver of prostate cancer initiation and progression. O-GlcNAc transferase (OGT), the enzyme that catalyzes the covalent addition of UDP-N-acetylglucosamine (UDP-GlcNAc) to serine and threonine residues of proteins, is often highly expressed in prostate cancer with its expression correlated with high Gleason score. In this study, we have identified an AR and OGT coregulated factor, Vpr (HIV-1) binding protein (VPRBP) also known as DDB1 and CUL4 Associated Factor 1 (DCAF1). We show that VPRBP is regulated by the AR at the transcript level, and stabilized by OGT at the protein level. VPRBP knockdown in prostate cancer cells led to a significant decrease in cell proliferation, p53 stabilization, nucleolar fragmentation, and increased p53 recruitment to the chromatin. In human prostate tumor samples, VPRBP protein overexpression correlated with AR amplification, OGT overexpression, a shorter time to postoperative biochemical progression and poor clinical outcome. In clinical transcriptomic data, VPRBP expression was positively correlated with the AR and also with AR activity gene signatures.
Implications:
In conclusion, we have shown that VPRBP/DCAF1 promotes prostate cancer cell proliferation by restraining p53 activation under the influence of the AR and OGT.
Introduction
AR activity plays an important role in the development of localized prostate cancer and also in sustaining treatment-resistant metastatic disease (1). Hence, a comprehensive understanding of AR signaling mechanisms during prostate carcinogenesis is instrumental in developing novel therapies. Studies have shown glycosylation as a key androgen-regulated process in prostate cancer cells (2). AR activation has been shown to enhance flux through hexosamine biosynthetic pathway (HBP) in prostate cancer cell lines (3), which leads to increased bioavailability of UDP-N-acetylglucosamine, a substrate for O-GlcNAcylation as well as N-linked and O-linked glycosylation (4). O-GlcNAcylation, a highly dynamic and often transient posttranslational modification (PTM) is specifically increased in prostate cancer tissues compared with adjacent nonmalignant tissues (5). This PTM is regulated by two enzymes, OGT that catalyzes the covalent addition of UDP-N-acetylglucosamine to serine and threonine residues of cytoplasmic, nuclear and mitochondrial proteins, and O-GlcNAcase (OGA) which removes the O-GlcNAc moiety (6). OGT is considered to be a metabolic rheostat and its expression is elevated in many cancers including prostate cancer, with higher O-GlcNAc levels associated with poor prognosis of patients (7, 8). There is a growing diverse list of proteins which undergo this PTM, including some of the key transcription factors such as c-Myc (9) and p53 (10).
A chromatin immunoprecipitation sequencing (ChIP-seq) study by Itkonen and colleagues has demonstrated that the O-GlcNAc chromatin mark is rapidly diminished by inhibition of OGT activity using a fast-acting inhibitor, OSMI2 (11) in prostate cancer cells. This analysis revealed that the majority of the O-GlcNAc peaks were promoter associated with over 95% overlap with DNase-hypersensitive regions and active chromatin marks. Independent AR ChIP-seq studies from our lab and others have shown that the majority of AR-binding sites are distal intergenic and intronic (12). Genome-wide motif coenrichment, have shown entirely distinct enrichment patterns for O-GlcNAc sites (principally c-Myc and ETS transcription factors), compared with AR sites which show a significant enrichment for Forkhead family transcription factors such as FOXA1 (12). Despite these differences, we know that both AR and OGT contribute to prostate cancer progression. To better understand the interplay between OGT and AR in prostate cancer, we reanalyzed these AR and O-GlcNAc ChIP-seq data focusing on promoter proximal sites and identified a small number of overlapping sites and associated genes. Among these, we focused on VPRBP also known as DCAF1 which has been implicated as a regulator of cell cycle and cell proliferation (13, 14). VPRBP is the substrate recognition component of cullin 4A-ring E3 ubiquitin ligase (CRL4A) complex as well as separate HECT type EDD/UBR5 E3 ligase (15). In this study, we show that VPRBP is a novel AR target as well as an OGT-regulated protein. Knockdown of VPRBP led to a marked reduction in prostate cancer cell proliferation. We go on to show that VPRBP downregulates p53 stability and activity, and that this is in part by maintaining nucleolar integrity. Tissue microarray studies showed a positive correlation of VPRBP expression with AR/OGT expression and an inverse correlation with PSA recurrence-free survival. Furthermore, VPRBP expression in TCGA datasets showed a positive correlation with AR expression and a subset of AR activity gene signatures, and inverse correlation with the p53 pathway. We conclude that VPRBP acts as a novel downstream effector of AR- and OGT-mediated prostate cancer cell proliferation by impairing p53 checkpoint activation.
Materials and Methods
Reagents and consumables
Synthetic androgen, R1881, and dihydrotestosterone were obtained from Sigma-Aldrich. OGT inhibitors, OSMI2 and OSMI3 were kindly provided by Professor Suzanne Walker (Harvard Medical School, Boston, MA). Formaldehyde 16% (F017/3) was purchased from TAAB laboratory. iDeal ChIP-seq Kit for Transcription Factors (C01010170) was obtained from Diagenode. B32B3 (SML1419) and cycloheximide (C4859) were purchased from Sigma-Aldrich. Antibody details are provided in Supplementary Table S1. Lipofectamine RNAiMAX Transfection Reagent (13778075), NE-PER nuclear and cytoplasmic extraction reagents (78835), Click-IT O-GlcNAc Enzymatic Labeling System (C33368), Click-IT Biotin Protein Analysis Detection Kit (33372), and High-Capacity Streptavidin Agarose Resin (20357) were obtained from Thermo Fisher Scientific. Protein A sepharose beads (ab193256) and protein G sepharose beads (ab193259) were from Abcam.
Cell lines
LNCaP and 22Rv1 cells were purchased from ATCC and cultured in RPMI containing 10% FBS and 1% pencillin–streptomycin in a humidified incubator at 37°C and 5% CO2. VCaP cells were obtained from ATCC and cultured in DMEM containing 10% FBS and 1% pencillin–streptomycin in a humidified incubator at 37°C and 5% CO2. Cells were authenticated by ATCC using short tandem repeat (STR) analysis. TP53 CRISPR knockout LNCaP cells were kindly provided by Dr. Peter Nelson (Fred Hutchinson Cancer Research Center, Seattle, WA) and were authenticated by sequencing and karyotyping. All the cells were routinely tested for Mycoplasma contamination using MycoAlert control set (Lonza; LT07–518).
ChIP assays
ChIP assays were carried out as per manufacturer's instructions (details in Supplementary Materials and Methods). ChIP-qPCR primers were obtained from Eurofins genomics and sequences listed in Supplementary Table S2.
Real-time qPCR
The cells were lysed in Qiazol and RNA isolated using Qiagen miRNeasy Mini Kit (catalog no. 217004). Transcriptor First Strand cDNA Synthesis Kit (catalog no. 04897030001, Roche Life Sciences) was used for cDNA preparation. SYBR Green 1 Master (catalog no. 4887352001, Roche Life Science) was used to compare gene expression changes in VPRBP, OGT, CAMKK2, UAP1, COPS3, p53 and p21 by real-time PCR (qPCR) in Roche LightCycler 480 Instrument II. Human large ribosomal protein (RPLPO) was used as the internal control. Primers for qPCR were purchased from either Sigma (KiCqStart predesigned) or from Eurofins genomics. The primer details are provided in Supplementary Tables S3 and S4 respectively.
siRNA transfection
LNCaP cells were seeded on to 6-well plates for siRNA knockdown. Forward transfection was performed the following day using Lipofectamine RNAiMAX Transfection Reagent in OPTI-MEM with 30 pmol of siRNA/well according to the manufacturer's instructions. After overnight incubation, media was changed to RPMI with 10% FBS and antibiotics. Two individual siRNAs were used against each target of interest. The siRNAs used were OGT si1, OGT si2, DCAF1 si1, DCAF1 si2, and Silencer Select Negative Control No.1 siRNA (details are provided in Supplementary Table S5). For studies involving androgen treatment, media were changed to androgen-deprived charcoal-treated media for 3 days prior to stimulation with 1 nmol/L R1881 for 24 hours. The cells were lysed after given number of days post transfection for analysis by Western blot or qPCR.
Western blot analysis
The cells were lysed in RIPA buffer (Sigma) containing protease inhibitor cocktail (Roche) and PhosSTOP (Sigma); protein concentration estimated with Bradford reagent (Bio-Rad) and 30 µg of lysate subjected to electrophoresis using precast 4%–12% NuPage mini-gels (Life Technologies). The resolved proteins were then transferred to polyvinylidene difluoride membrane, blocked with 5% nonfat dry milk, and probed with respective primary antibodies overnight at 4°C. After three washes, the blots were probed with horseradish peroxidase (HRP)-conjugated secondary antibodies and immunoreactivity detected by enhanced chemiluminescence in Syngene G box.
Cycloheximide chase assay
LNCaP cells were transfected with OGT si1 or negative control No.1 siRNA. The cells were treated for 50 µg/mL cycloheximide for indicated time points and lysate collected for Western blot analysis at 72 hours from the start of transfection.
Immunoprecipitation
Briefly, LNCaP cells were lysed in IP lysis buffer (10 mmol/L Tris-Cl pH7.5, 140 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS). Lysates were precleared with protein A sepharose beads for 1 hour at 4°C in a rotator. Immunoprecipitation (IP) was carried out with protein A sepharose beads using 1 mg lysate and 1 µg VPRBP antibody. Lysate was incubated with VPRBP or IgG negative control antibody for 3 hours at 4°C in a rotator followed by overnight incubation with 40 µL of washed protein A sepharose beads. The beads were briefly pelleted and washed thrice in IP wash buffer (10 mmol/L Tris-Cl pH 7.5, 150 mmol/L NaCl, 0.5% Triton X-100). The proteins were eluted by heating in 20 µL Laemmli buffer at 95°C for 5 minutes.
Analysis of VPRBP O-GlcNAcylation
Briefly, cytoplasmic extract was prepared using NE-PER Nuclear and cytoplasmic extraction reagent from LNCaP cells grown with or without 10 µmol/L PUGNaC for 24 hours. Five-hundred micrograms of cytoplasmic extract was labeled according to the Click-iT O-GlcNAc Enzymatic Labelling System protocol, and conjugated with an alkyne-biotin compound as per the Click-iT Protein Analysis Detection Kit protocol as previously described (16). Biotinylated lysates were precipitated using chloroform/methanol, resolubilized in 1% SDS, 50 mmol/L Tris-HCl pH 7.4, and SDS quenched with 1 volume of neutralization buffer (100 mmol/L NaCl, 50 mmol/L Tris-HCl pH 7.4, 5 mmol/L EDTA, 6% NP-40). Lysates were then incubated with high-capacity agarose streptavidin resin with end-to-end rotation at 4°C overnight. Resin was then washed four times in 1 mL low salt buffer (50 mmol/L Tris-HCl pH 7.4, 150 mmol/L NaCl, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate) and once in 1 mL high salt buffer (50 mmol/L Tris-HCl pH 7.4, 500 mmol/L NaCl, 0.2% Triton X-100) Biotinylated proteins were eluted by boiling the resin in 2× Laemmli buffer with dithiothreitol. Western blotting analysis was carried out with anti-VPRBP and anti-G6PD antibodies. Control experiments were carried out in parallel in the absence of the labelling enzyme, GalT1.
Cell counts
Cell counting of siRNA-transfected cells in 12 well plates was performed 4 to 5 days posttransfection. Cells were trypsinized and changes in cell number assessed by cell counting in Countess II Lifetechnologies.
Immunofluorescence
LNCaP cells were seeded on to glass coverslips in a 12-well plate. After reaching 70% to 80% confluence, cells were transfected with scrambled or VPRBP siRNA. After 3 days, cells were fixed with 4% paraformaldehyde in BSA for 10 minutes followed by three washes in PBS for 5 minutes each. The fixed cells were then lysed in 0.1% Triton X-100 in PBS for 10 minutes and blocked in blocking buffer (5% goat serum/1%BSA/0.1% Triton X-100 in PBS) for 1 hour. The cells were then incubated in primary antibodies against VPRBP (1:80) and fibrillarin (1:100) for 2 hours at room temperature followed by Alexa Fluor 594 Goat anti-mouse (Invitrogen, catalog no. A11020) or Alexa Fluor 488 Goat Anti R (Invitrogen, catalog no. A11070) secondary antibodies. Coverslips were mounted on to glass slides using Vectashield with DAPI.
Patients, IHC, and analysis of TCGA and other datasets can be found in Supplementary Materials and Methods.
Statistical analysis
Statistical analyses for studies in LNCaP and VCaP cells were done using either Student t test or one-way ANOVA with Tukey post hoc analysis, as mentioned in the figure legends. For IHC, statistical calculations were performed using JMP 12 software (SAS Institute Inc.). Contingency tables were calculated with the χ2 test. Survival curves were calculated by the Kaplan–Meier method and compared with the log-rank test.
Results
Identification of VPRBP as a novel AR-regulated gene
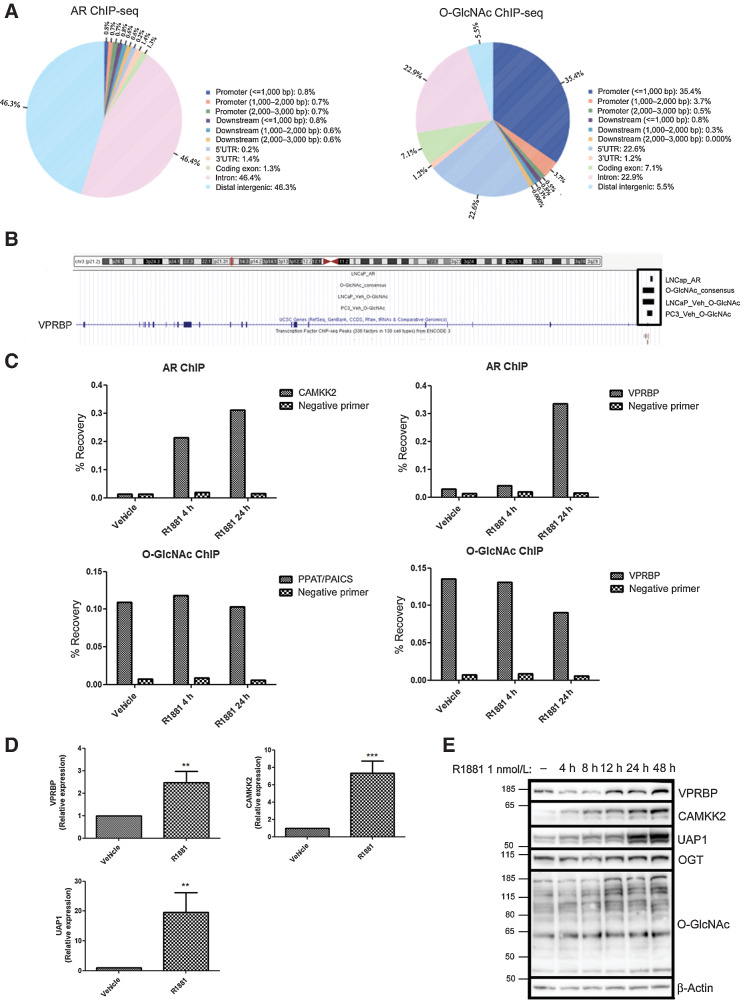
To identify AR and OGT coregulated genes, we reanalyzed published AR (GSE28126; ref. 12) and O-GlcNAc (GSE112667; ref. 3) ChIP-seq data. Comparing peak distribution between AR and O-GlcNAc–binding sites from these two separate studies (Fig. 1A), indicated that, as previously reported the majority of O-GlcNAc–binding sites are promoter proximal, whereas the majority of AR-binding sites are intronic or associated with distal intergenic regions (Fig. 1A). Intersecting LNCaP AR ChIP-seq (R1881 stimulated) binding sites with O-GlcNAc ChIP-seq consensus sites (from LNCaP and PC3) identified only nine overlapping sites, among which a binding site was detected proximal to the VPRBP gene (Fig. 1B). VPRBP is of particular interest because it has been shown to be highly expressed in different tumor tissues (17) and is also known to play a pivotal role in cell-cycle entry and proliferation (13). Moreover, depletion of VPRBP in DU145 prostate cancer cells reduced cell proliferation and number of colony-forming cells (17). However, the roles of VPRBP in mediating androgen response in prostate cancer or its regulation by OGT or O-GlcNAcylation have not been reported so far.
Figure 1.
Identification of VPRBP as an AR and O-GlcNAc co-regulated target. A, Venn diagrams showing the distribution of peaks in relation to genes in LNCaP AR ChIP-seq and O-GlcNAc ChIP-seq (consensus sites from LNCaP and PC3), generated using CEAS tool in galaxy cistrome. B, ChIP-seq enrichment of AR and O-GlcNAc at VPRBP promoter region using UCSC genome browser. “LNCaP_AR” refers to AR ChIP-seq–binding sites in 1 nmol/L R1881-treated LNCaP (GSM696840). “LNCaP_veh_O-GlcNAc” refers to consensus O-GlcNAc ChIP-seq–binding sites (from GSM3076096, GSM3076097 and GSM3076098) in vehicle (DMSO) treated LNCaP. “PC3_veh_O-GlcNAc” refers to consensus O-GlcNAc ChIP-seq–binding sites (from GSM3586373, GSM3586374, GSM3586375) in vehicle (DMSO) treated PC3. C, Percentage recovery of CAMKK2 and VPRBP with AR ChIP; and PPAT/PAICS and VPRBP with O-GlcNAc ChIP in vehicle (0.01% ethanol) and 1 nmol/L R1881 4 -and 24-hour treated LNCaP cells (n = 1). D, mRNA expression of VPRBP, CAMKK2, and UAP1 in LNCaP treated with vehicle or 1 nmol/L R1881 for 24 hours was detected by qRT-PCR. Results are normalized to RPLPO as housekeeping control (n = 3). E, Time dependency of VPRBP expression following exposure to 1 nmol/L R1881 for different time points. LNCaP cells were androgen deprived for 3 days prior to stimulation with vehicle or 1 nmol/L R1881. P values by Student t test. **, P < 0.01; ***, P < 0.001.
To confirm AR and O-GlcNAc enrichment at the VPRBP site, we performed ChIP-qPCR in LNCaP cells stimulated with 1 nmol/L R1881, a synthetic androgen, for 4 and 24 hours following 72 hours of androgen deprivation (12). We confirmed that as expected androgen stimulation resulted in AR enrichment at a CAMKK2-associated site (a known AR target; ref. 12), at both time points (Fig. 1C). In contrast, O-GlcNAc enrichment in the promoter of the PPAT/PAICS gene (a c-Myc–associated site which overlaps with an O-GlcNAc ChIP-seq peak; ref. 18) was not significantly altered in response to androgen treatment (Supplementary Fig. S1A; Fig. 1C). Androgen stimulation resulted in increased binding of AR at the VPRBP promoter region at the 24-hour time point (Fig. 1C; Supplementary Fig. S1A). We also validated an O-GlcNAc site in the promoter of VPRBP by ChIP-qPCR (Fig. 1C), and found that androgen treatment did not significantly affect the signal at this site (Supplementary Fig. S1A).
qPCR analysis revealed that R1881 stimulated binding of AR to VPRBP promoter correlated with a 2.5-fold increase in VPRBP mRNA expression at 24 hours (Fig. 1D), concomitant with significant increases in VPRBP protein levels 24 and 48 hours (Fig. 1E; Supplementary Fig. S1B). OGT and O-GlcNAcylation levels also showed significant increases following R1881 stimulation at 48 hours and 24–48 hours, respectively (Fig. 1E; Supplementary Fig. S1B). CAMKK2 and UAP1 expression levels were used as positive controls (12). Because R1881 is a synthetic androgen, we also tested the effect of endogenous androgen, dihydrotesteosterone (DHT). Similar to R1881, stimulation of LNCaP cells by DHT for 24 hours also increased VPRBP mRNA and protein expression (Supplementary Fig. S1C and S1D). We also found that R1881 and DHT induced upregulation of VPRBP protein expression in VCaP cells following 24-hour treatment (Supplementary Fig. S1E).
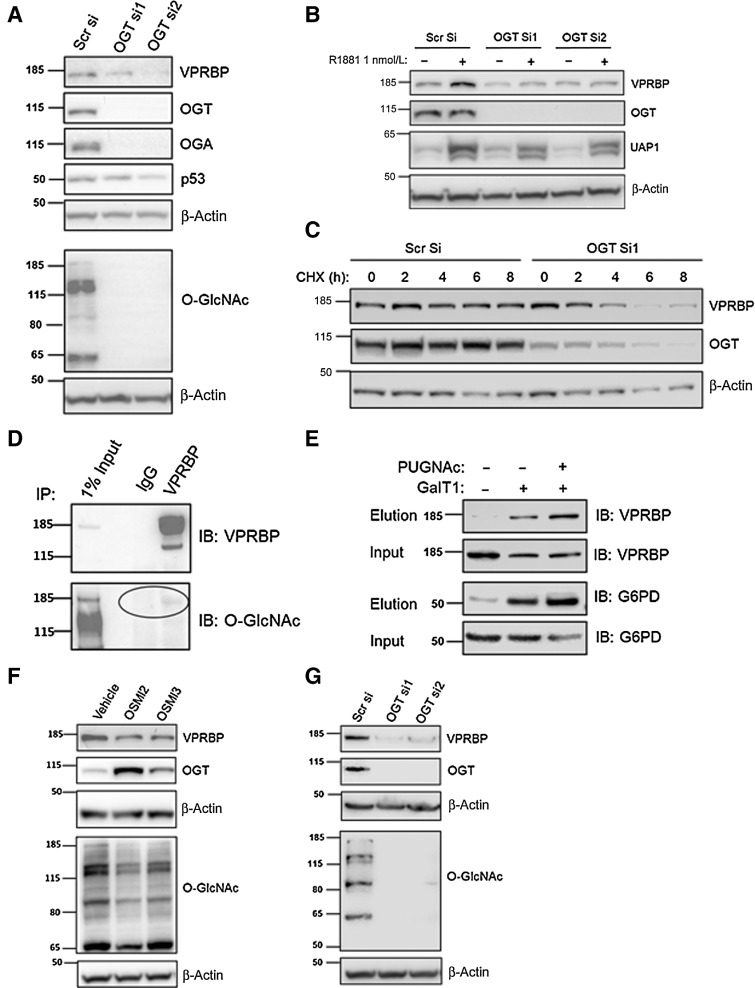
OGT is required for VPRBP stability
To determine whether OGT is required for VPRBP expression, we performed OGT knockdown using siRNA. This revealed that there was no significant change in basal VPRBP mRNA levels in OGT siRNA–transfected cells compared with scrambled control (Supplementary Fig. S2A). Interestingly, OGT knockdown reduced both basal (Fig. 2A) and androgen-induced (Fig. 2B) VPRBP protein expression. OGT siRNA transfection resulted in >90% reduction in basal OGT protein expression and >80% reduction in total O-GlcNAc levels with both siRNAs (Supplementary Fig. S2B). There was approximately 57% reduction in VPRBP protein expression with OGTsiRNA1 and approximately 54% with OGT siRNA2 5 days posttransfection (Supplementary Fig. S2B). Because a reduction in protein levels could either reflect a decrease in translation or an increase in protein degradation, we used a cycloheximide chase experiment (0–8 hours) to assess the impact of OGT knockdown on the half-life of VPRBP. We found that with OGT knockdown VPRBP protein levels significantly reduced from 6-hour time point onwards, whereas in the control siRNA–transfected cells, VPRBP levels did not change significantly in any of the time points tested (Fig. 2C; Supplementary Fig. S2C). This suggests a critical role of OGT in stabilizing VPRBP.
Figure 2.
OGT is required for VPRBP stability. A, LNCaP cells were transiently transfected with OGT siRNAs and cells were harvested 5 days posttransfection to examine protein expression under basal conditions by immunoblot analysis. B, For detection of protein expression under androgen-stimulated conditions, LNCaP cells were transfected with OGT siRNAs or scrambled siRNA (scr si) followed by androgen deprivation for 72 hours prior to 1 nmol/L R1881 stimulation for 24 hours. C, Cycloheximide (50 µg/mL) chase experiment was conducted from 0 to 8 hours to assess the impact of OGT knockdown (72 hours) on the degradation of VPRBP. D, The O-GlcNAcylation of IP VPRBP from LNCaP cells was detected by immunoblotting (IB) with RL2 antibody. E, O-GlcNAcylated VPRBP was detected from LNCaP cytoplasmic extract. Lysates prior to pull down (input) and the captured proteins (elution) were immunoblotted (IB) with an antibody toward VPRBP as well as positive control G6PD. Control experiments in the absence of GalT demonstrated selective labeling of the O-GlcNAcylation on VPRBP and G6PD. F, The effect of OGT inhibitors 40 µmol/L OSMI2 and 10 µmol/L OSMI3 on VPRBP protein levels following 24-hour treatment was detected by immunoblot analysis. G, The effect of OGT knockdown on VPRBP protein expression in 22Rv1 cells by immunoblot analysis.
O-GlcNAcylation has been shown to directly affect stability of proteins like p53 (10), c-Myc (3), and EZH2 (19). To determine whether VPRBP is O-GlcNAcylated, we performed IP with total cell lysate. IP indicates that VPRBP is an O-GlcNAcylated protein (Fig. 2D). To further confirm whether VPRBP is O-GlcNAcylated, we performed Click-IT O-GlcNAc Enzymatic Labeling assay. All O-GlcNAc modified protein from LNCaP cytosolic extract treated in the presence and absence of PUGNAc for 24 hours were enzymatically labeled with azido-N-acetylgalactosamine sugar. Labeled proteins were biotinylated and captured with streptavidin–agarose beads. Subsequent immunoblotting of captured proteins with an antibody against VPRBP clearly showed O-GlcNAcylation of VPRBP (Fig. 2E). G6PD, which was previously shown to be O-GlcNACylated by this method (16), was used as a positive control. PUGNAc-treated samples showed higher levels of O-GlcNACylation of VPRBP (∼2 fold) and G6PD (∼2.7 fold) compared with untreated samples. To further corroborate the role of O-GlcNAcylation in the stability of VPRBP, we treated the cells with inhibitors of OGT activity, OSMI2 and OSMI3 (20). We found that treatment for 24 hours decreased VPRBP expression at the protein level by approximately 73% for 40 µmol/L OSMI2 and approximately 62% for 10 µmol/L OSMI3 (Fig. 2F; Supplementary Fig. S2D). OSMI treatment reduced overall O-GlcNAcylation levels with a compensatory upregulation of OGT expression (Fig. 2F; Supplementary Fig. S2D). There was no significant change in VPRBP expression at the transcript level with OSMI3 treatment, although OSMI2 exhibited approximately 11% reduction of VPRBP transcripts (Supplementary Fig. S2E). We also checked the effect of OGT knockdown on basal levels of VPRBP protein expression in another cell line, 22Rv1, which showed approximately 87% reduction with OGTsiRNA1 and approximately 76% with OGT siRNA2 (Fig. 2G; Supplementary Fig S2F). Overall, these results suggest a possible dual regulation of VPRBP, whereby VPRBP is induced at the mRNA level by AR and is stabilized posttranslationally by OGT activity.
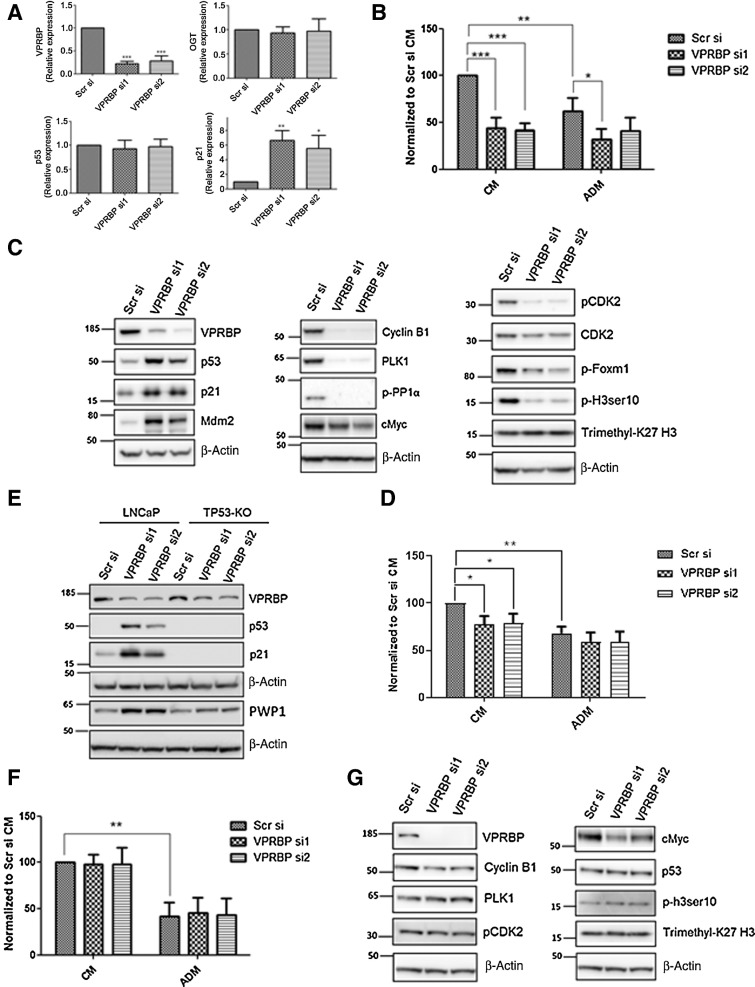
VPRBP downregulation stabilizes p53 and inhibits LNCaP cell proliferation
Having identified VPRBP as an AR and OGT target, we went on to determine the phenotypic effect of VPRBP knockdown in LNCaP cells. Knockdown with VPRBP siRNA resulted in approximately 75% reduction in VPRBP transcripts without any significant change in OGT transcripts (Fig. 3A). A reduction in cell number of approximately 57% was observed with VPRBP siRNAs when cells were grown in complete growth media containing androgens, whereas androgen deprivation on its own resulted in a reduction of approximately 38% in cell numbers (Fig. 3B). Furthermore, chemical inhibition by B32B3, a potent and selective inhibitor of VPRBP kinase activity (17), led to approximately 64% decrease in LNCaP cell proliferation at 5 µmol/L concentration (Supplementary Fig. S3A). B32B2 5µmol/L also decreased histone H2A threonine 120 phosphorylation as previously shown by Kim and colleagues (17) in DU145 cells (Supplementary Fig. S3B). The combination of B32B3 and OGT inhibitors did not show significant decreases in cell numbers compared with B32B3 alone (Supplementary Fig. S3A).
Figure 3.
VPRBP knockdown leads to reduced cell proliferation and p53 stabilization. A, LNCaP cells were transfected with VPRBP siRNA and cells were harvested 3 days posttransfection to detect mRNA expression of VPRBP, OGT, p53, and p21 (n = 4). B, Effect of VPRBP knockdown on LNCaP cell proliferation was assessed by cell counting 5 days posttransfection; the cells were grown in the presence (CM) and absence of androgens (ADM; n = 4). C, Effect of VPRBP knockdown on LNCaP p53, markers of cell cycle and other proteins of interest was assessed by immunoblot analysis of cell lysate prepared 3 days posttransfection. D, Effect of VPRBP knockdown on cell proliferation was assessed by cell counting in TP53 knockout (KO) LNCaP cells grown in the presence (CM) and absence of androgens (ADM; n = 4). E, Effect of VPRBP knockdown on LNCaP versus TP53-KO LNCaP proteins of interest was assessed by immunoblot analysis. F, Effect of VPRBP knockdown on cell proliferation was assessed by cell counting 5 days posttransfection in VCaP cells grown in the presence (CM) and absence of androgens (ADM; n = 3). G, Effect of VPRBP knockdown on VCaP proteins of interest was assessed by immunoblot analysis. Results are expressed as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistical analyses were performed by Student t test for qPCR and one-way ANOVA followed by Tukey post hoc analysis for cell proliferation.
Because VPRBP deletion in T lymphocytes was previously shown to cause p53 stabilization (13), we examined p53 expression following VPRBP knockdown in LNCaP. VPRBP knockdown in LNCaP led to a substantial increase in p53 protein expression and its downstream targets, cyclin-dependent kinase inhibitor p21 (21) and Mdm2 (ref. 22; Mouse double minute 2 homolog; Fig. 3C; Supplementary Fig. S3C). VPRBP knockdown did not affect p53 transcript levels as such, whereas p21 transcript levels were upregulated by approximately 6 fold (Fig. 3A). Although Mdm2 is a negative regulator of p53 at the protein level, there is a complex feedback relationship between Mdm2 and p53 with stabilized p53 transcriptionally activating Mdm2 gene (22). Cyclin-dependent kinase inhibitor, p21 is well known for its role in inducing G1 arrest (21). Reflecting this, we observed a drastic reduction in cell-cycle markers like phospho CDK2 Thr160 (ref. 23; which activates CDK2 complexes and a marker of G1–S), Cyclin B1 (ref. 24; a marker of G2–M phase), polo-like kinase1 (PLK1; ref. 25; a marker of late G2 that promotes mitotic entry), phospho-FoxM1 (ref. 26; a marker of G2–M phase), and phospho-PP1α Thr320 (ref. 27; Fig. 3C; Supplementary Fig. S3C) following VPRBP knockdown. Phospho-histone H3 ser10 (ref. 28; marker of mitotic chromatin condensation) was also significantly reduced following VPRBP knockdown where as another histone mark, Trimethyl-K27 H3 remained unchanged (Fig. 3C; Supplementary Fig. S3C). In summary, VPRBP tightly controls cell proliferation likely by regulating the expression and activity of p53.
To determine whether this was the case we went on to perform VPRBP knockdown in a previously characterized TP53 knockout (TP53-KO) LNCaP cell line (29). As reported early, TP53 KO LNCaP cells exhibited higher growth rates than wild-type (WT) LNCaP in complete growth medium (Supplementary Fig S3D). In TP53 KO cells, we observed approximately 22% reduction in cell numbers with VPRBP knockdown (Fig. 3D) as opposed to approximately 57% in WT LNCaP cells (Fig. 3B). Western blot comparison of TP53 KO and WT cell lysates confirmed the absence of p53 and p21 induction in the knockout cells (Fig. 3E). A recent study by Han and colleagues suggested that VPRBP loss induces p53 activation by impairing ribosome biogenesis by accumulation of ribosome assembly factor PWP1, consequent increase in large ribosomal subunit protein, RPL11, its binding to MDM2 thereby inhibiting p53 ubiquitination and degradation (14). We also observed an increase in PWP1 protein expression with VPRBP knockdown in WT LNCaP suggesting that this may be the case in these cells too (Fig. 3E; Supplementary Fig. S3E). To further assess whether growth effects of VPRBP knockdown require WT p53, we went on to knockdown VPRBP in a p53-mutant cell line, VCaP. VPRBP knockdown in VCaP failed to cause significant changes in cell numbers (Fig. 3F) and cell-cycle markers (Fig. 3G). However, similarly to LNCaP, VPRBP knockdown in VCaP cells reduced c-Myc levels (Fig. 3G). Unlike LNCaP, this downregulation did not lead to significant changes in cell number and this may reflect a difference in the biological processes that are Myc-dependent in the two cell lines or alternatively a weaker correlation between Myc expression and Myc activity in the VCaP cell line. Knocking down VPRBP in another cell line that expresses WT p53, 22Rv1, did result in a significant reduction in cell numbers (Supplementary Fig. S3F), stabilization of PWP1, p53, p21, and a consequent reduction in cell cycle markers like Cyclin B1 and PLK1 (Supplementary Fig. S3G). Together, these results suggest that the growth-inhibitory effects of VPRBP knockdown are highly dependent on p53 stabilization and activation.
Because we previously showed that androgen treatment led to an increase VPRBP expression we went on assess whether R1881 changes p53 expression. We observed a reduction in p53 protein (Supplementary Fig. S4A) and mRNA expression (Supplementary Fig. S4B) in LNCaP cells following R1881 stimulation, and a corresponding decrease in p53 enrichment at p21 (CDKN1A) promoter by ChIP-qPCR (Supplementary Fig. S4C) suggesting that pro-proliferative effects of androgens arise through the suppression of p53 activity in some contexts and that VPRBP may be the mediator of this. Because androgens regulate VPRBP expression and this in turn has an impact on p53, we sought to determine whether stabilizing p53 might have reciprocal suppressive effect on VPRBP levels. To do so, we treated androgen-deprived LNCaP cells with a p53 stabilizer, Nutlin-3a, and this led to a decrease in VPRBP expression (Supplementary Fig. S4F and S4G) although we found no evidence of p53-binding sites in the promoter of VPRBP in a ChIP-seq dataset generated from these treated cells (Supplementary Fig. S4D and S4E). In summary VPRBP seems to be a mediator of reciprocal feedback between the AR and p53 in some contexts.
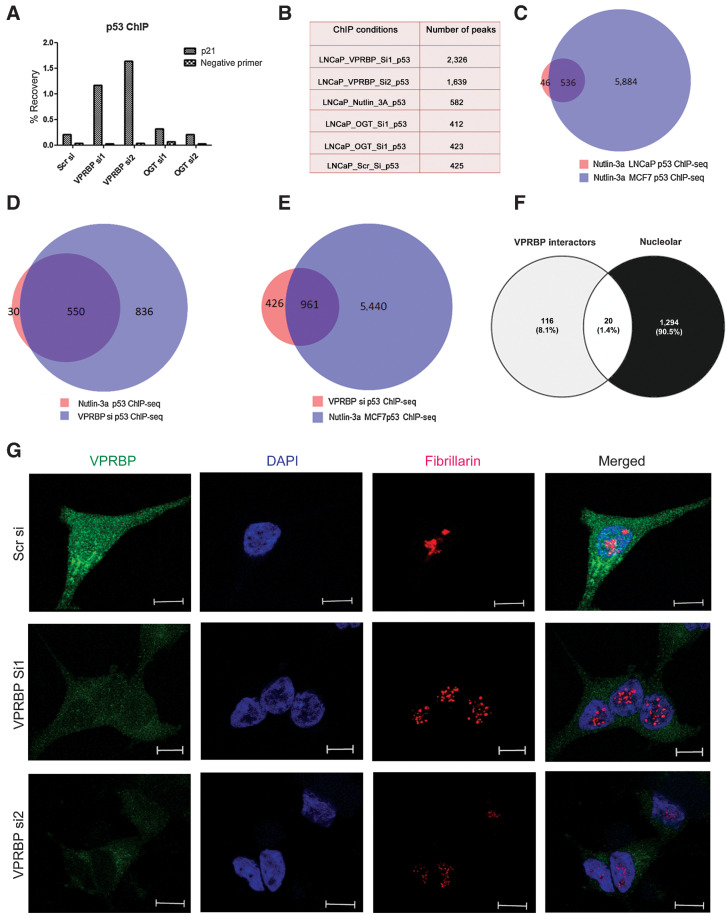
p53 ChIP-seq reveals increased p53 recruitment to the chromatin following VPRBP knockdown
To provide a genome-wide assessment of the impact of VPRBP on p53 activity, we performed p53 ChIP-seq in LNCaP cells following siRNA knockdown of VPRBP, OGT, or treatment with nontargeting siRNAs or nutlin-3a. Canonical p53 target gene p21 (CDKN1A) was used as a positive control to validate ChIP efficiency by ChIP-qPCR (Fig. 4A). This is the first p53 ChIP-seq dataset generated in a prostate cancer cell line. Overall, there were fewer p53 peaks in LNCaP (Fig. 4B) than reported in other cell types (30, 31), suggesting fundamental differences in the accessible chromatin landscape for p53 recruitment in these cells. ChIP-seq in nutlin-3a–treated LNCaP cells returned 582 peaks, the majority of which (>80%) overlapped with nutlin-3a p53 ChIP-seq–binding sites in other cell lines confirming that they were bona fide p53-binding sites (Fig. 4C; Supplementary Fig. S5A). In contrast, VPRBP knockdown led to approximately 4.7-fold increase in the number of p53 genomic-binding sites compared with the scrambled control (Fig. 4B). There were 1,387 consensus p53-binding sites between the two VPRBP knockdown samples (si1 and si2) representing a site overlap between these conditions of approximately 85% (Supplementary Fig. S5B). The vast majority (∼95%) of LNCaP nutlin-3a p53 ChIP-seq sites were present among the consensus p53 sites generated with si1 and si2 (Fig. 4D). However, the much greater number of p53-binding sites upon VPRBP knockdown (Fig. 4D), despite equivalent levels of p53 protein in nutlin-3a and VPRBP knockdown conditions (Supplementary Fig. S5C), suggests that knockdown enhanced chromatin accessibility for p53 recruitment. Importantly, the majority of sites in VPRBP knockdown overlapped with nutlin-3a MCF7 p53 ChIP-seq peaks (Fig. 4E) indicating that VPRBP knockdown was simply permitting a larger proportion of the p53 regulome to be engaged through p53 site occupancy. OGT knockdown gave similar number of peaks as scrambled control (Fig. 4B) and the majority of both OGT si1 and OGTsi2 sites overlapped (Supplementary Fig. S5B).
Figure 4.
VPRBP knockdown increases p53 chromatin recruitment and induces nucleolar stress. A, Bar graph showing percentage recovery of p21 and negative site primer (CCND1) following p53 ChIP in different transfection conditions (n = 1). B, Table showing the number of peaks obtained under different conditions. C, Venn diagram showing the overlap of our nutlin-3a p53 ChIP-seq in LNCaP cells with previously reported nutlin-3a p53 ChIP-seq in MCF7 cells (GSE86164). D, Venn diagram showing the overlap of nutlin-3a p53 ChIP-seq in LNCaP with VPRBPsi p53 ChIP-seq consensus sites. E, Venn diagram showing the overlap of VPRBPsi p53 ChIP-seq consensus sites with previously reported nutlin-3a p53 ChIP-seq in MCF7 cells. F, Venn diagram showing the overlap of VPRBP interactome with nucleolar proteome. G, Representative immunofluorescence images showing VPRBP and fibrillarin staining in scrambled and VPRBP siRNA–transfected LNCaP 3 days posttransfection (scale bar, 10 µm).
Assessing the distribution of p53 peaks revealed that the majority were present in distal intergenic regions (45%–54%), with around 6% binding events in proximal promoters (Supplementary Fig. S5D), resembling the genome-wide site distribution observed in AR ChIP-seq data. The VPRBP si p53 ChIP-seq peaks showed approximately 53% overlap with H2K27Ac peaks and approximately 0.7% overlap with H3K27me3 peaks indicating that the majority of the binding sites are present at sites of transcriptional activity which are not accessible to p53 when VPRBP is expressed (Supplementary Fig. S5E). The genes within 10 kb of p53 sites in the VPRBP knockdown condition were identified (Supplementary Data S2). These included established p53 target genes such as CDKN1A and MDM2. Motif enrichment analysis on the p53 sites in the knockdown condition showed an over-representation of TP53, TP73, and TP63-binding motifs (Supplementary Fig. S5F). Together, these suggest that VPRBP may restrain p53 activity by occluding chromatin and that VPRBP knockdown expands the gene regulatory network affected by increased p53 expression.
VPRBP knockdown induces nucleolar stress in LNCaP cells
Nucleolar stress is known to induce p53 stabilization by perturbing ribosome biogenesis leading to cell-cycle arrest and apoptosis by disrupting Mdm2–p53 interaction (32) and VPRBP knockdown is known to do this too (14). VPRBP has been previously shown to localize in the cytoplasm (33) as well as nucleus (34). A comparison of VPRBP interactome (147 proteins) obtained from BioGRID database (ref. 35; Supplementary Data S3) with nucleolar proteome (1,314 proteins) of human cells derived from the Cell Atlas (36) showed that approximately 14% of VPRBP interactors also show nucleolar localization (Fig. 4F). VPRBP is also reported to be involved in 40S ribosomal subunit biogenesis along with other CRL4 E3 ubiquitin ligase and COP9 signalosome components in a genome wide RNAi screen study (37). Hence, we hypothesised that VPRBP knockdown may enhance p53 stability and diminish ribosome biogenesis by destabilising the nucleolus. Our immunofluorescence studies showed both cytoplasmic and nuclear localization of VPRBP (Fig. 4G) as reported previously (33, 34). Reduction in the number of nucleoli and/or disintegration of nucleolar structures are some of the characteristic features of nucleolar stress (38). We found a marked change in staining pattern of nucleolar protein fibrillarin following VPRBP siRNA knockdown, indicative of nucleolar stress (Fig. 4G). Fibrillarin showed exclusive nucleolar localization with a redistribution to small nucleoplasmic entities in the VPRBP knockdown cells, similar to previously described with actinomycin D treatment (39). To quantify nucleolar fragmentation, we measured the individual nucleolar area based on the fibrillarin staining as previously described (40), which showed that VPRBP knockdown cells have a much smaller nucleolar size compared (∼1.7 µm2) with scrambled si-transfected cells (9.8 µm2; Supplementary Fig S6A). A significant (∼25%) reduction in fibrillarin and 40S ribosomal protein S8 (RPS8) expression, was also observed with VPRBP siRNA2 (Supplementary Fig. S6B and S6C). This suggests that p53 activation may be a downstream consequence of nucleolar stress induced by VPRBP knockdown.
VPRBP expression correlates with AR expression in clinical samples and is prognostic
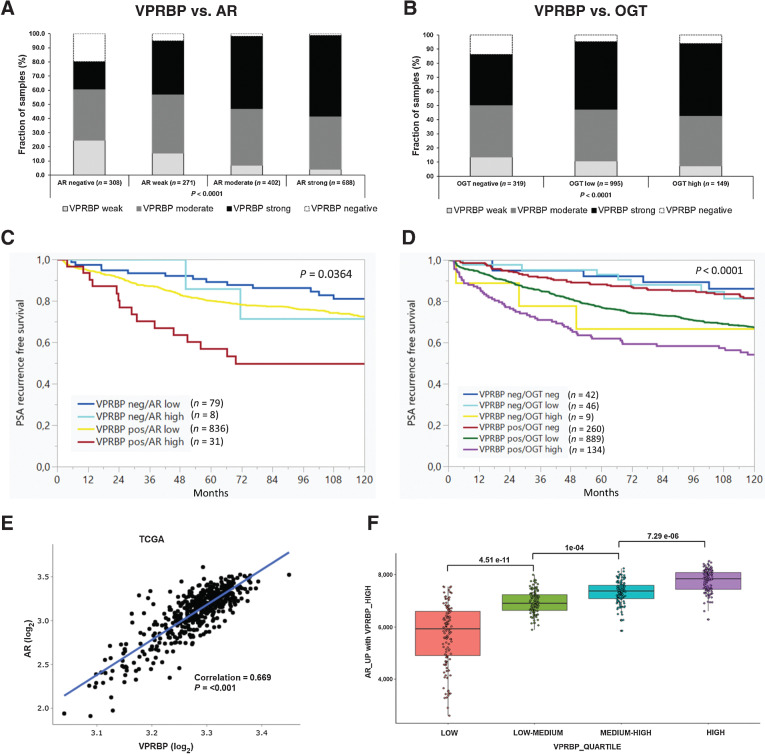
To further understand the prognostic potential of VPRBP in prostate cancer, we analyzed its expression in tissue microarrays (TMA) by IHC. The representative images of negative, low, intermediate, and high staining of VPRBP are shown in Supplementary Fig. S6D. We assessed its correlation with prostate cancer pathology, and with the AR and OGT. In assessing VPRBP expression in the TMA, we observed that 46% of all tumors stained strongly for VPRBP expression with a statistically significant increase in protein levels with tumor stage and quantitative Gleason grade (Supplementary Table S6). Although there were few Gleason score 7 tumors with tertiary Gleason pattern 5 (a powerful predictor of biochemical relapse; ref. 41), all of them were VPRBP-positive. A higher VPRBP staining was observed in cases with positive surgical margins (Supplementary Table S6). However, there were no significant differences in VPRBP expression between patients with no regional lymph node metastasis (N0) and patients with metastasis (N+). Significant correlations were also not seen between VPRBP expression and preoperative PSA levels (Supplementary Table S6). To determine whether VPRBP expression associated with poor prognosis disease, we evaluated its expression versus postoperative biochemical (PSA) recurrence-free survival. We observed a statistically significant reduction in PSA recurrence-free survival in patients with any VPRBP expression compared with those who were negative (Supplementary Fig. S6E). We went on to assess the coexpression of VPRBP with AR or OGT in the TMA. For both, we observed a statistically significant positive correlation in staining which suggests that the expression and activity of these proteins may be relevant in subsets of patients (Fig. 5A and B). Furthermore, VPRBP-positive/AR-high groups exhibited a statistically significant reduction in PSA recurrence-free survival when compared with VPRBP-negative/AR-low groups (P = 0.0019); and so did VPRBP-positive/OGT-high groups compared with VPRBP-negative/OGT-negative (P = 0.0013) (Fig. 5C and D).
Figure 5.
VPRBP protein expression correlates with AR amplification, OGT overexpression, and poor prognosis. Bar graphs showing positive correlation of VPRBP expression with AR (A) and OGT expression (B) by IHC in TMA sections. C, PSA recurrence-free survival curves in patients expressing low or high levels of AR where VPRBP expression was present or absent and survival curves (D) in patients expressing no, low or high levels of OGT where VPRBP expression was present or absent. E, Scatter plot comparing AR mRNA expression to VRPBP mRNA expression in TCGA PanCancer Atlas prostate dataset. F, Box plot showing association between VPRBP expression quartiles and the 139 upregulated AR genes from heatmap. The P-values in C and D indicate the overall significance.
We next sought to determine whether VPRBP transcript expression in clinical samples is associated with activity gene signatures reflecting AR activity. First, we compared the VPRBP mRNA expression in the TCGA datasets and found a positive correlation with AR mRNA expression (Fig. 5E). There have been multiple studies reporting AR activity gene signatures. To identify the subset of AR activity gene signatures that are highly expressed in VPRBP-high tumors, we combined AR activity gene signatures from Dorothea AR regulon, KEGG prostate cancer pathway and an additional 8 gene sets from the Molecular Signatures Database (MSigDB; refs. 42–44) and clustered them on VPRBP-high and -low expression quartiles as a heatmap (Combined AR gene list in Supplementary Data S4; Supplementary Fig. S7). Combined, these studies have attributed the expression of 467 genes to AR activity. Because it is not clear which may be most relevant in a given patient group, we assessed all of them relative to VPRBP quartile expression in the TCGA Prostate Cancer dataset (Supplementary Fig. S7). 139 of these 467 genes positively correlated with VPRBP expression and 92 negatively correlated with VPRBP expression (Supplementary Data S5). The association between VPRBP expression quartiles and the 139 genes is illustrated in a box plot and is statistically significant between the quartiles Fig. 5F. Importantly, the 139 genes most significantly associated with AR responsive pathways but did not include some canonical AR targets such KLK2 and KLK3. To further refine this association, we generated coexpression tables for the TCGA dataset ranking all genes that were coexpressed with VPRBP according to a Spearman rank correlation coefficient. We also did this for two other datasets, SU2C/PCF Dream Team, Cell 2015 and MSKCC, Cancer Cell, 2010 from cBioPortal. Forty-eight genes among the 139 genes identified were significantly coexpressed with VPRBP (Spearman rank coefficient threshold >0.4) in all the three studies (Supplementary Data S5). This represents putative VPRBP-dependent AR regulome that will form the basis for future mechanistic studies on the interplay between VPRBP and AR activity.
VPRBP expression inversely correlates with p53 activity signatures
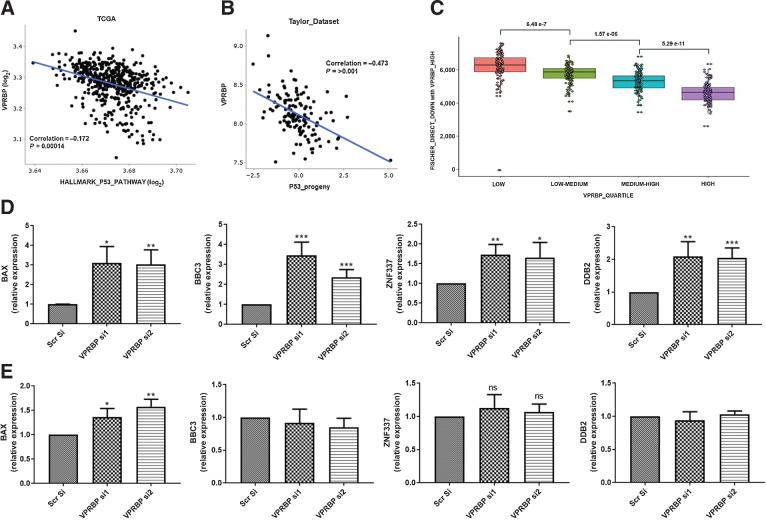
Because our in vitro studies also suggested that a reduction in VPRBP expression enhances p53 activity as measured by recruitment of p53 to chromatin and the expression of p53 target genes, we looked at the p53 activity signatures. We found an inverse correlation of VPRBP expression with HALLMARK_P53_pathway (45) (Fig. 6A). We also evaluated VPRBP expression versus a second gene signature reflecting p53 activity derived from perturbation experiments (progeny; ref. 46) in the Taylor datasets. This also showed a statistically significant inverse correlation (Fig. 6B). To further understand the relationship between VPRBP expression and p53 gene regulation we used a third measure of p53 activity, genes defined as direct p53 targets based both on perturbation experiments and the presence of p53-binding sites within their promoters as determined by ChIP-seq (47). Heatmap was generated to cluster gene lists on VPRBP-high/low quartiles in TCGA (Supplementary Fig. S8) and the p53 direct genes downregulated/upregulated in VPRBP high quartile were extracted. 127 p53 direct bound genes were downregulated and 83 upregulated in VPRBP-high groups (Supplementary Data S6). The statistically significant association between VPRBP expression quartiles and the 127 downregulated genes is illustrated in Fig. 6C. Among these 33, downregulated genes overlapped with gene list from VPRBP si p53 ChIP-seq (supplementary file 6). p53, acting as a transcription factor, can have pleiotropic effects on a wide range of biological processes including metabolic pathways, DNA damage response, cell-cycle regulation and other important contributors to the regulation of cell viability and proliferation. Importantly, the 33 genes identified were significantly enriched for a subset of these processes, specifically cytochrome C release, G2 cell-cycle arrest, DNA damage response, mitochondrial permeability and apoptotic signaling pathways by Enrichr (Supplementary Data S6). We validated some of these genes in VPRBP knockdown LNCaP samples and TP53-KO samples. VPRBP knockdown in WT LNCaP showed significant upregulation of BBC3, ZNF337, and DDB2 transcripts (Fig. 6D) without significant changes in TP53-KO LNCaP (Fig. 6E). BAX showed approximately 3.3-fold increase with VPRBP knockdown in LNCaP (Fig. 6D) versus approximately 1.4-fold increase in TP53-KO LNCaP (Fig. 6E). Further characterization of these genes may be crucial to uncover VPRBP-regulated biology in prostate cancer. Together these studies underscore the significance of VPRBP in promoting prostate cancer growth and progression.
Figure 6.
VPRBP expression inversely correlates with p53 activity signatures. A, Scatter plot comparing GSEA Hallmark “P53 Pathway” to VRPBP mRNA expression in TCGA PanCancer Atlas prostate dataset. B, Scatter plots showing VPRBP expression in Taylor data sets versus p53 progeny scores. C, Box plot showing association between VPRBP expression quartiles and the 127 down-regulated Fischer p53 direct bound genes from heat map. D, Effect of VPRBP knockdown on p53 target genes in LNCaPs by qPCR (n = 3). E, Effect of VPRBP knockdown on p53 target genes in TP53-KO LNCaPs by qPCR (n = 3). Results are expressed as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student t test.
Discussion
Our previous studies attributed the impact of O-GlcNAc/OGT on prostate cancer cells to effects on c-Myc stability (3), and based on ChIP-seq data, to the over-representation of c-Myc at O-GlcNAc–binding sites in the genome (11). Although the majority of O-GlcNAc peaks in the genome are promoter proximal and associated with histone marks indicative of active transcription (11), the majority of AR sites are distal intergenic or intronic. The main challenge of using antibody against O-GlcNAc moiety in ChIP studies is that we do not know the nature of the proteins getting O-GlcNAcylated. Although there are some phosphor-protein–specific antibodies that are available for use in ChIP-seq, there are no O-GlcNAc-protein specific antibodies. However, we observed that a small number of AR-binding sites overlapped with O-GlcNAc sites on chromatin and hypothesized that these might help us to identify factors that were coregulated by the AR and OGT. The potential biological significance of these sites is underscored by the fact that in our case they have led us to uncover VPRBP as a novel AR and OGT target.
VPRBP was initially identified as a protein targeted by HIV-1 viral protein R (Vpr) to initiate host cell response leading to cell-cycle arrest at G2–M by hijacking the CUL4A E3 ubiquitin ligase machinery (33, 48). This implies that VPRBP can in some settings support cell-cycle progression and indeed we know this to be the case from previous studies (49). We show that VPRBP is transcribed in response to androgen treatment (Fig. 1D) and that its protein stability is dependent on OGT (Fig. 2A–G). We went on to show that knockdown of VPRBP by siRNA led to significant decrease in LNCaP proliferation accompanied by stabilization of tumor suppressor p53 (Fig. 3C). Guo and colleagues (13) demonstrated similar stabilization of p53 in T cells following VPRBP deletion suggesting its requirement in Mdm2-mediated p53 polyubiquitination (13). They further show that for T-cell proliferation to occur, VPRBP promotes cell-cycle entry by restraining p53 activation while a VPRBP-dependent, p53-independent programme possibly involving c-Myc dictates cell growth in naïve T cells after T-cell receptor (TCR) activation (13). One can draw some similarities between T cells progressing from quiescence to proliferation and prostate cancer cells during androgen stimulation. During TCR activation, cells undergo drastic metabolic changes with increased glucose and glutamine uptake, and a concomitant increase in O-GlcNAcylation (50). Comparably, androgen stimulation of prostate cancer cells increased glucose uptake and anabolic synthesis of glutamine (12). Androgen-stimulated cells also displayed higher HBP pathway enzymes and protein O-GlcNAcylation levels (Fig. 1E). Interestingly, VPRBP is upregulated upon TCR activation in T cells as well as with AR activation in prostate cancer cells. Collectively, our data suggest that the similar dependencies on both VPRBP and OGT for cell proliferation exist for both prostate cancer cells and T cells.
Other than stabilization, the regulation of p53 transcriptional activity by VPRBP has been previously described by Kim and colleagues (51) to occur at the chromatin level. In this study, they showed that VPRBP is recruited to target promoters by p53 to attenuate p53-dependent transcription by selectively binding to the unacetylated histone H3 tails in the absence of any stress stimuli rendering it inaccessible to Histone acetyl transferases. They also showed that VPRBP knockdown led to activation of p53 target genes, so did its phosphorylation at ser-895 by DNA-activated protein kinase (DNA-PK). A follow up study by the same group further identified a novel intrinsic kinase activity of this protein towards histone H2A on threonine 120 which favors its localization to tumor suppressor genes and chromatin silencing (17). In our study, we have shown that VPRBP knockdown significantly enhances the recruitment of p53 to chromatin as assessed by a significant increase in genome-wide p53-binding sites. Collectively, these suggest that VPRBP is a multistage inhibitor of p53 activation, impacting on both chromatin binding and p53 stability and expression. This impact may however be most profound in cells expressing WT p53 because a mutant p53 cell line, VCaP, did not show significant reductions in p53 levels with VPRBP knockdown (Fig. 3G). Moreover, a p53 knockout LNCaP line failed to achieve similar extent of decrease in cell numbers with VPRBP knockdown compared with WT LNCaP (Fig. 3D) We also tested the feedback effects of p53 activation on VPRBP expression and have found that stabilizing p53 pharmacologically with nutlin-3a diminishes VPRBP stability (Supplementary Fig. S4F). We believe this is predominantly a posttranslational/protein turnover effect because there are no significant changes in transcript levels (Supplementary Fig. S4G) and no evidence of p53 binding to the VPRBP promoter (Supplementary Fig. S4E). All the above studies suggest a reciprocal relation between p53 and VPRBP in prostate cancer cells.
We have also shown previously that inhibiting guanine nucleotide biosynthesis disrupts nucleolar function leading to p53 stabilization and c-Myc downregulation (18). In that study, we reported that inhibiting IMPDH2 with a drug, mycophenolic acid, led to p53 stabilization by depleting cellular GTP levels, promoting degradation of nucleolar proteins such as GNL3 and thereby inducing nucleolar stress (18). Interestingly mycophenolic acid was developed and used initially to restrict T- and B-cell proliferation for the purposes of enhancing graft take in patients undergoing renal transplant surgery (52). This further reinforces the idea that there are significant commonalities in the biological processes that support immune activation mediated T-cell proliferation and prostate cancer cell proliferation. Because VPRBP was previously shown to sustain 40S ribosome subunit biogenesis by supporting nucleolar integrity (37), we were led to test how targeting VPRBP might affect this compartment. Imaging of nucleolar marker, fibrillarin revealed marked changes in nucleolar staining indicative of nucleolar stress (Fig. 4G; Supplementary Fig. S6A). A recent study by Han and colleagues revealed a critical role for VPRBP in rRNA processing and ribosome biogenesis by regulating a previously unknown substrate, the ribosome assembly factor PWP1 (14). Interestingly, VPRBP loss leads to accumulation of free ribosomal protein L11 (RPL11), resulting in L11–MDM2 association and p53 activation. Together, these suggest that p53 activation may be a downstream consequence of nucleolar stress induced by VPRBP knockdown.
By examining the VPRBP protein expression in tissue from a highly annotated prostate cancer patient cohort, we established that expression increases significantly with stage and grade and furthermore correlates positively with high expression of the AR and OGT. We attempted to identify candidates by stratifying human prostate cancer cases according to VPRBP expression quartiles and then focusing on known p53 target genes that are most significantly inversely correlated with VPRBP expression in the uppermost and lowermost quartiles. That gave us 33 p53 direct bound genes which were downregulated in VPRBP high expression quartile and overlapped with our VPRBPsi-p53 ChIP-seq gene list. We validated few of those which were include important mediators of the interplay between cell-cycle control and activation of the DNA damage response checkpoints in cells. Future studies will also need to further dissect the functional impact of VPRBP in a range of other mutational backgrounds include RB loss, PTEN loss, and p53 point mutation. In conclusion, VPRBP represents the first AR and OGT coregulated protein to promote prostate cancer cell proliferation by limiting p53 activation and as such may be an early determinant of prostate cancer progression. On the basis of our studies and previous studies on T-cell proliferation and activation, we believe that it works hand-in-glove with c-Myc to support proliferation. It would be relevant to include VPRBP in patient stratification for treatment optimization in men with prostate cancer. The implication is that VPRBP-high expressing tumors may be p53 WT but p53 inactive due to the impact of VPRBP in occluding chromatin from p53 recruitment and the impact on p53 protein levels. In other words, VPRBP expression is a suppressor of p53 activity. As a result, we are suggesting that some patients with localized prostate cancer, in which there is known to be low burden of somatic p53 mutations (typically around 10%), will be functionally p53-low due to VPRBP expression and consequently more treatment resistant and more likely to progress postsurgery/postradiotherapy. These are the patients that may therefore benefit most from neoadjuvant therapy at the time of surgery with chemotherapy or small molecules.
Supplementary Material
Acknowledgments
This research was supported by the Norwegian Research Council (230559). I.G. Mills is also supported by the Prostate Cancer UK/ Movember Centre of Excellence (CEO13_2–004) and the John Black Charitable Foundation. Authors would like to acknowledge Genomic Core Technology Units, Queens University Belfast for the assistance with ChIP-seq studies. We sincerely thank Dr. Michael Nyquist and Prof. Peter Nelson for providing us TP53 knockout LNCaP cells.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
N. Poulose: Conceptualization, formal analysis, investigation, writing–original draft, writing–review and editing. N. Forsythe: Formal analysis, visualization, writing–review and editing. A. Polonski: Formal analysis, investigation, writing–review and editing. G. Gregg: Methodology. S. Maguire: Formal analysis. M. Fuchs: Formal analysis. S. Minner: Formal analysis, writing–review and editing. G. Sauter: Supervision. S.S. McDade: Formal analysis, supervision, methodology, writing–review and editing. I.G. Mills: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Feng Q, He B. Androgen receptor signaling in the development of castration-resistant prostate cancer. Front Oncol 2019;9:858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Munkley J. Glycosylation is a global target for androgen control in prostate cancer cells. Endocr Relat Cancer 2017;24:R49–64. [DOI] [PubMed] [Google Scholar]
- 3. Itkonen HM, Minner S, Guldvik IJ, Sandmann MJ, Tsourlakis MC, Berge V, et al. O-GlcNAc transferase integrates metabolic pathways to regulate the stability of c-MYC in human prostate cancer cells. Cancer Res 2013;73:5277–87. [DOI] [PubMed] [Google Scholar]
- 4. Akella NM, Ciraku L, Reginato MJ. Fueling the fire: emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol 2019;17:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gu Y, Gao J, Han C, Zhang X, Liu H, Ma L, et al. O-GlcNAcylation is increased in prostate cancer tissues and enhances malignancy of prostate cancer cells. Mol Med Rep 2014;10:897–904. [DOI] [PubMed] [Google Scholar]
- 6. Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol 2015;208:869–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fardini Y, Dehennaut V, Lefebvre T, Issad T. O-GlcNAcylation: a new cancer hallmark? Front Endocrinol 2013;4:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kamigaito T, Okaneya T, Kawakubo M, Shimojo H, Nishizawa O, Nakayama J. Overexpression of O-GlcNAc by prostate cancer cells is significantly associated with poor prognosis of patients. Prostate Cancer Prostatic Dis 2014;17:18–22. [DOI] [PubMed] [Google Scholar]
- 9. Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol 2017;18:452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, et al. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol 2006;8:1074–83. [DOI] [PubMed] [Google Scholar]
- 11. Itkonen HM, Urbanucci A, Martin SE, Khan A, Mathelier A, Thiede B, et al. High OGT activity is essential for MYC-driven proliferation of prostate cancer cells. Theranostics 2019;9:2183–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J 2011;30:2719–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo Z, Kong Q, Liu C, Zhang S, Zou L, Yan F, et al. DCAF1 controls T-cell function via p53-dependent and -independent mechanisms. Nat Commun 2016;7:10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han XR, Sasaki N, Jackson SC, Wang P, Li Z, Smith MD, et al. CRL4(DCAF1/VprBP) E3 ubiquitin ligase controls ribosome biogenesis, cell proliferation, and development. Sci Adv 2020;6:eabd6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schabla NM, Mondal K, Swanson PC. DCAF1 (VprBP): emerging physiological roles for a unique dual-service E3 ubiquitin ligase substrate receptor. J Mol Cell Biol 2019;11:725–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rao X, Duan X, Mao W, Li X, Li Z, Li Q, et al. O-GlcNAcylation of G6PD promotes the pentose phosphate pathway and tumor growth. Nat Commun 2015;6:8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim K, Kim JM, Kim JS, Choi J, Lee YS, Neamati N, et al. VprBP has intrinsic kinase activity targeting histone H2A and represses gene transcription. Mol Cell 2013;52:459–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barfeld SJ, Fazli L, Persson M, Marjavaara L, Urbanucci A, Kaukoniemi KM, et al. Myc-dependent purine biosynthesis affects nucleolar stress and therapy response in prostate cancer. Oncotarget 2015;6:12587–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lo PW, Shie JJ, Chen CH, Wu CY, Hsu TL, Wong CH. O-GlcNAcylation regulates the stability and enzymatic activity of the histone methyltransferase EZH2. Proc Natl Acad Sci U S A 2018;115:7302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin SES, Tan ZW, Itkonen HM, Duveau DY, Paulo JA, Janetzko J, et al. Structure-based evolution of low nanomolar O-GlcNAc transferase inhibitors. J Am Chem Soc 2018;140:13542–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. [DOI] [PubMed] [Google Scholar]
- 22. Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J 1993;12:461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J 1992;11:3995–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindqvist A, van Zon W, Karlsson Rosenthal C, Wolthuis RM. Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol 2007;5:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gheghiani L, Loew D, Lombard B, Mansfeld J, Gavet O. PLK1 activation in late G2 sets up commitment to mitosis. Cell Rep 2017;19:2060–73. [DOI] [PubMed] [Google Scholar]
- 26. Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene 2009;28:4295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berndt N, Dohadwala M, Liu CW. Constitutively active protein phosphatase 1alpha causes Rb-dependent G1 arrest in human cancer cells. Curr Biol 1997;7:375–86. [DOI] [PubMed] [Google Scholar]
- 28. Van Hooser A, Goodrich DW, Allis CD, Brinkley BR, Mancini MA. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J Cell Sci 1998;111:3497–506. [DOI] [PubMed] [Google Scholar]
- 29. Nyquist MD, Corella A, Coleman I, De Sarkar N, Kaipainen A, Ha G, et al. Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress. Cell Rep 2020;31:107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andrysik Z, Galbraith MD, Guarnieri AL, Zaccara S, Sullivan KD, Pandey A, et al. Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome Res 2017;27:1645–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sammons MA, Zhu J, Drake AM, Berger SL. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res 2015;25:179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. James A, Wang Y, Raje H, Rosby R, DiMario P. Nucleolar stress with and without p53. Nucleus 2014;5:402–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang S, Feng Y, Narayan O, Zhao LJ. Cytoplasmic retention of HIV-1 regulatory protein Vpr by protein-protein interaction with a novel human cytoplasmic protein VprBP. Gene 2001;263:131–40. [DOI] [PubMed] [Google Scholar]
- 34. Belzile JP, Abrahamyan LG, Gerard FC, Rougeau N, Cohen EA. Formation of mobile chromatin-associated nuclear foci containing HIV-1 Vpr and VPRBP is critical for the induction of G2 cell cycle arrest. PLoS Pathog 2010;6:e1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006;34:D535–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, et al. A subcellular map of the human proteome. Science 2017;356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 37. Badertscher L, Wild T, Montellese C, Alexander LT, Bammert L, Sarazova M, et al. Genome-wide RNAi screening identifies protein modules required for 40S subunit synthesis in human cells. Cell Rep 2015;13:2879–91. [DOI] [PubMed] [Google Scholar]
- 38. Yang K, Yang J, Yi J. Nucleolar stress: hallmarks, sensing mechanism and diseases. Cell Stress 2018;2:125–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Y, Hu Y, Che L, Jia J, Chen M. Nucleolar localization of Small G protein RhoA is associated with active RNA synthesis in human carcinoma HEp-2 cells. Oncol Lett 2016;11:3605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu B, Lee KK, Zhang L, Gerton JL. Stimulation of mTORC1 with L-leucine rescues defects associated with Roberts syndrome. PLoS Genet 2013;9:e1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hattab EM, Koch MO, Eble JN, Lin H, Cheng L. Tertiary Gleason pattern 5 is a powerful predictor of biochemical relapse in patients with Gleason score 7 prostatic adenocarcinoma. J Urol 2006;175:1695–9. [DOI] [PubMed] [Google Scholar]
- 42. Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A 2002;99:11890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takayama K, Kaneshiro K, Tsutsumi S, Horie-Inoue K, Ikeda K, Urano T, et al. Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene 2007;26:4453–63. [DOI] [PubMed] [Google Scholar]
- 44. Wang G, Jones SJ, Marra MA, Sadar MD. Identification of genes targeted by the androgen and PKA signaling pathways in prostate cancer cells. Oncogene 2006;25:7311–23. [DOI] [PubMed] [Google Scholar]
- 45. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schubert M, Klinger B, Klunemann M, Sieber A, Uhlitz F, Sauer S, et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat Commun 2018;9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res 2016;44:6070–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Le Rouzic E, Belaidouni N, Estrabaud E, Morel M, Rain JC, Transy C, et al. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 2007;6:182–8. [DOI] [PubMed] [Google Scholar]
- 49. Hrecka K, Gierszewska M, Srivastava S, Kozaczkiewicz L, Swanson SK, Florens L, et al. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc Natl Acad Sci U S A 2007;104:11778–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, van Aalten DM, et al. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat Immunol 2016;17:712–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim K, Heo K, Choi J, Jackson S, Kim H, Xiong Y, et al. Vpr-binding protein antagonizes p53-mediated transcription via direct interaction with H3 tail. Mol Cell Biol 2012;32:783–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sollinger HW. Mycophenolate mofetil. Kidney Int Suppl 1995;52:S14–7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.