Dai et al. show that the loss of ARL13, a small GTPase already known for its role in ciliary protein import, also impairs the export of membrane-associated proteins from Chlamydomonas cilia via the BBSome pathway. Thus, ARL13 and BBSome deficiencies lead to overlapping defects in ciliary membrane composition.
Abstract
The GTPase Arl13b participates in ciliary protein transport, but its contribution to intraflagellar transport (IFT), the main motor-based protein shuttle of cilia, remains largely unknown. Chlamydomonas arl13 mutant cilia were characterized by both abnormal reduction and accumulation of select membrane-associated proteins. With respect to the latter, a similar set of proteins including phospholipase D (PLD) also accumulated in BBSome-deficient cilia. IFT and BBSome traffic were apparently normal in arl13. However, transport of PLD, which in control cells moves by BBSome-dependent IFT, was impaired in arl13, causing PLD to accumulate in cilia. ARL13 only rarely and transiently traveled by IFT, indicating that it is not a co-migrating adapter securing PLD to IFT trains. In conclusion, the loss of Chlamydomonas ARL13 impedes BBSome-dependent protein transport, resulting in overlapping biochemical defects in arl13 and bbs mutant cilia.
Introduction
Arl13b, a conserved ARF-like GTPase, is one of several ARL and RAB-like GTPases that regulates ciliary protein traffic (Fisher et al., 2020; Humbert et al., 2012; Yan and Shen, 2021). Mutations in Arl13b interfere with ciliary assembly and signaling in model organisms and, in humans, cause Joubert syndrome (JS), an autosomal recessive ciliopathy (Cantagrel et al., 2008; Joubert et al., 1969; Parisi, 2009; Sun et al., 2004). The role of Arl13b’s GTP cycle in cilia remains elusive as Arl13b spontaneously exchanges guanine nucleotides in vitro and possesses only weak GTPase activity due to the lack of a critical conserved glutamine residue (Ivanova et al., 2017). In vitro studies showed that Chlamydomonas ARL13 functions as a guanine nucleotide exchange factor (GEF) for the small GTPase ARL3, mutations of which also cause Joubert syndrome in humans (Alkanderi et al., 2018; Gotthardt et al., 2015). Since Arl13b is a resident ciliary protein, it will encounter and activate ARL3 once it enters the cilium (Caspary et al., 2007; Cevik et al., 2010; Hori et al., 2008). Then, ARL3-GTP will bind to the carrier proteins PDE6δ and UNC119, inducing the release of their lipidated protein cargoes into the cilium (Gotthardt et al., 2015; Ivanova et al., 2017; Miertzschke et al., 2014; Zhang et al., 2016). Thus, Arl13b is part of a pathway that mediates the transport of membrane and membrane-associated proteins through the ciliary gate by regulating their release from permeable carrier proteins into the cilium.
However, the proposed role of Arl13b in ciliary protein import fails to satisfactorily explain other features of Arl13b deficiency such as defects in Hedgehog (Hh) signaling, a pathway that regulates cell fate determination, growth, and survival (Caspary et al., 2007). In Arl13b mutant cells, the distribution of Hh signaling proteins is affected (Bangs and Anderson, 2017; Larkins et al., 2011). The GPCR smoothened (Smo), for example, accumulates in the cilia of control cells only after activation of the Hh pathway by a ligand or agonist (Corbit et al., 2005). In Arl13b mutant cilia, however, Smo atypically accumulates even without pathway activation (Caspary et al., 2007). A similar defect in the distribution of Smo is also characteristic for defects in retrograde IFT, which removes proteins from cilia (Firestone et al., 2012; Kim et al., 2009; Ocbina et al., 2009). Removal of Smo and other GPCRs from cilia further requires the BBSome, an octameric cargo adapter that travels on IFT trains and mediates contact between the trains and a select number of signaling proteins (Blacque et al., 2004; Desai et al., 2020; Nachury et al., 2007; Shinde et al., 2020). Thus, mutations in Arl13b, the BBSome, or retrograde IFT all interfere with protein export from cilia. How Arl13b participates in ciliary protein export by the IFT/BBS pathway remains to be explored.
To address this question, we used Chlamydomonas, which facilitates an unbiased biochemical analysis of isolated cilia and in vivo imaging of ciliary protein transport (Engel et al., 2009; Lechtreck, 2016; Lechtreck et al., 2009; Liu and Lechtreck, 2018; Pazour et al., 2005). Chlamydomonas possesses only a single ARL13 gene whereas the gene is duplicated in most metazoans, i.e., Arl13a and Arl13b (Kahn et al., 2008; Schlacht et al., 2013; Vargova et al., 2021). Chlamydomonas ARL13 (527 residues) and human Arl13b (428 residues) are the reciprocal best hits with a score of 2e−56 in protein BLAST and possess 44% sequence identity in the N-terminal G-loop, indicating that the two are orthologs, whereas Arl13a deviates considerably. In cilia of a Chlamydomonas arl13 mutant, the abundant triacylglycerol lipase flagellar associated protein 12 (FAP12) and a few other proteins were severely reduced, whereas a set of lipidated proteins including phospholipase D (PLD) were abnormally accumulated; similar biochemical defects were also observed in the cilia of an arl3 mutant. We previously showed that PLD can enter cilia by diffusion and attaches in a BBSome-dependent manner to IFT trains in cilia, leading to its removal from cilia (Lechtreck et al., 2009; Liu and Lechtreck, 2018). In arl13, the composition and traffic of IFT and the BBSome was apparently normal, but IFT of PLD was not observed. Tagged ARL13 itself only rarely moves by IFT indicating that it is not an adapter linking PLD to the IFT/BBSome carriers. We propose that the loss of Chlamydomonas ARL13 disables cargo transport by BBSome carriers. The hedgehog signaling defects associated with ARL13b mutations in mammals could be explained by similar defects in BBSome-dependent ciliary protein export.
Results
Loss of ARL13 cilia causes abnormal reduction and accumulation of membrane-associated proteins in Chlamydomonas cilia
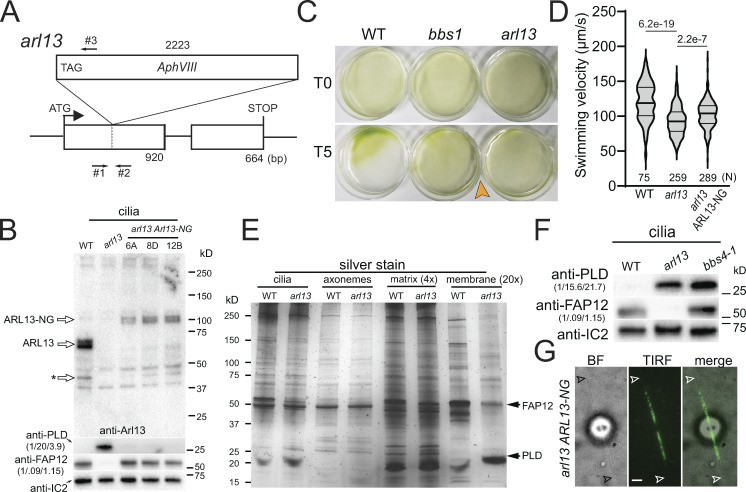
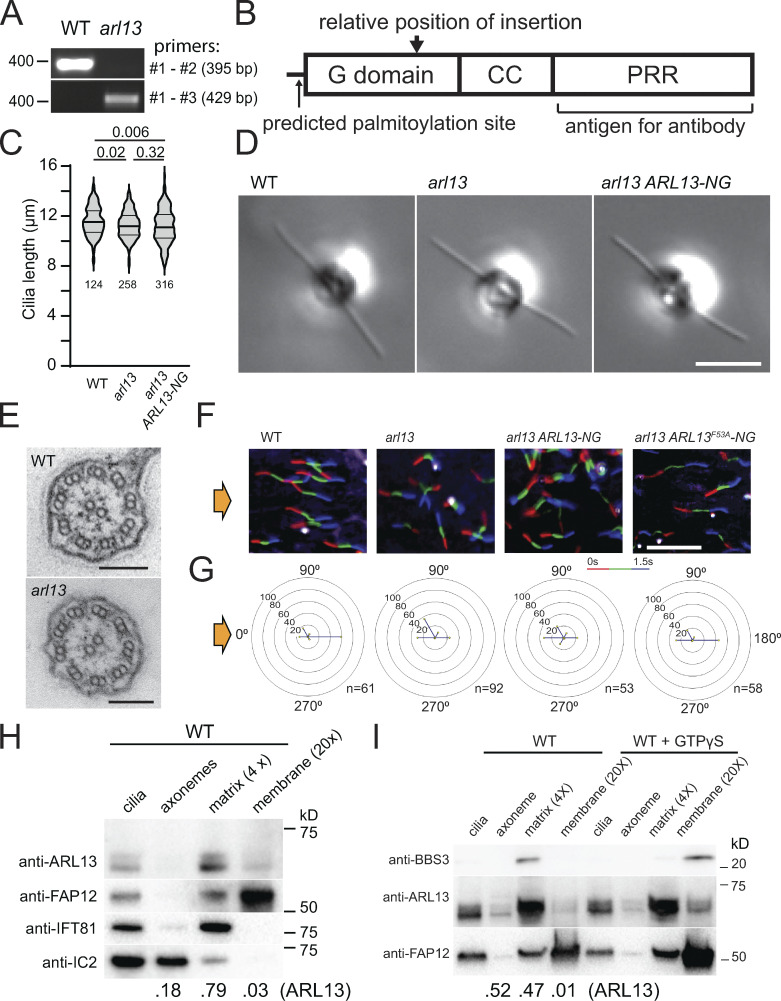
From the CLiP library, we obtained Chlamydomonas strain LMJ.RY0402.181356, which carries an insertion in the first exon of the ARL13 gene (Fig. 1 A; Li et al., 2019). The insertion was confirmed by PCR and is predicted to introduce a premature stop codon after 474 bp, which, if residual expression should occur, will lead to a severely truncated and likely non-functional ARL13 lacking parts of its P-loop GTPase domain and most of the α6 helix, which participates in the binding of ARL3 (Fig. 1 A; and Fig. S1, A and B; Gotthardt et al., 2015). A polyclonal antibody raised against the polypeptide encoded by the second exon of ARL13 detected a double band at ∼60 kD in control cilia, which is close to the predicted size of ∼55 kD for Chlamydomonas ARL13 (Fig. 1 B and Fig. S1 B). The immunoreactive bands were absent in LMJ.RY0402.181356 cilia, and we refer to this strain as arl13 (Fig. 1 B). To remove possible second-site mutations, we out-crossed arl13 to the wild-type strain g1 and used arl13 mutant progeny from the first and second outcross for subsequent experiments. Cilia of the arl13 mutant were of normal length and apparently normal ultrastructure as assessed by standard transmission electron microscopy (Fig. S1, C–E). The mutant cells swam somewhat slower and typically failed to properly phototax, a complex behavior that is modulated, for example, by the circadian rhythm and the nutritional state of the cells (Fig. 1, C and D; and Fig. S1, F and G; Bruce, 1972; Wakabayashi et al., 2011; Witman, 1993).
Figure 1.
Chlamydomonas arl13 shows defects in swimming, phototaxis, and ciliary composition. (A) Schematic presentation of ARL13 genomic DNA and the insertion in arl13. The start codon of ARL13 and the positions of the primers used to track the insertion are indicated (arrows #1–3). (B) Western blot of isolated cilia from control (g1, WT), arl13, and three different arl13 ARL13-NG strains, probed with antibodies against ARL13, PLD, FAP12 and, as a loading control, IC2. The star marks a potential ARL13 fragment. The quantification of the band strengths normalized for those of IC2 are shown in brackets and is based on one experiment. See Fig. S2, A and B, for average data based on several biological replicates. (C) Population phototaxis assay of control (g1, WT), bbs1, and arl13. The direction of light and time of exposure in minutes are indicated. (D) Violin plot of the swimming velocities of control (g1, WT), arl13, and arl13 ARL13-NG cells. N, the number of cells analyzed. The P values of a two-tailed t test are indicated. (E) Silver stained SDS-PAGE of isolated cilia and ciliary subfractions from Triton X-114 phase partitioning of the control (g1, WT) and arl13 strains. (F) Western blot comparing isolated cilia from the control (g1, WT), arl13, and bbs4-1 strains, probed with antibodies against PLD, FAP12, and, as a loading control, IC2. The quantification of the band strengths normalized for those of IC2 is shown in brackets and is based on one experiment. (G) Bright-field (BF), TIRF, and merged image showing the distribution of ARL13-NG. Arrowheads mark the ciliary tips. Bar = 2 µm. Source data are available for this figure: SourceData F1.
Figure S1.
ARL13 is a ciliary matrix protein. (A) PCR products from WT or arl13 genomic DNA. The expected product size and the position of the nearest DNA size marker are indicated. The positions of the primers are marked in Fig. 1 A. (B) Schematic presentation of ARL13 protein showing its G domain, coiled coil (CC) domain, and proline rich region (PRR). The predicted site of palmitoylation (C12), the position corresponding to the site of the insertion in arl13, and the region used for antibody production are marked. (C) Violin plot of the ciliary lengths of control (g1, WT), arl13, and arl13 ARL13-NG cells. The number of cilia analyzed and the P values of a two-tailed t test are indicated. (D) DIC image of live control (g1, WT), arl13, and arl13 ARL13-NG cells. Bar = 10 µm. (E) TEM images of control (g1, WT) and arl13 cilia in cross section. Bar = 100 nm. (F and G) Single cell motion analysis of the control (g1, WT), arl13, and arl13 ARL13-NG and arl13 ARL13F53A-NG strains. The direction of light is indicated (yellow arrows). (F) Composite micrographs showing the tracks of individual cells. Each of the three frames taken during a 1.5 s period was assigned a different color in the order red (first image), green, blue (last image) to visualize the swimming direction. Bar = 100 µm. (G) Radial histograms showing the percentage of cells swimming in a particular direction (six bins of 60° each). (H) Western blot of isolated control cilia and ciliary fractions probed with antibodies against ARL13, the membrane protein FAP12, the matrix protein IFT81, and the axonemal IC2. The numbers at the bottom indicate the distribution of ARL13 based on the band intensities of the axonemal, matrix, and membrane fractions. Note that some IFT81 is present in the axonemal fraction and some FAP12 in the matrix fraction indicating that the extraction with Triton X-114 and the phase separation were incomplete. (I) Western blot of isolated ciliary samples and ciliary fractions from wild-type cells probed with antibodies against BBS3, ARL13, and FAP12. Triton X-114 phase partitioning was carried out with and without GTPγS as indicated. The numbers at the bottom indicate the distribution of ARL13 based on the band intensities. Source data are available for this figure: SourceData FS1.
In mammals, an association of Arl13b with both the axoneme and the ciliary membrane were reported (Larkins et al., 2011; Revenkova et al., 2018). After fractionation of isolated Chlamydomonas cilia using Triton X-114 phase partitioning and centrifugation, most ARL13 (∼65%; n = 2) were found together with the IFT-B protein IFT81 in the soluble aqueous phase representing the ciliary matrix, while smaller amounts were present in the axonemal (∼35%) and membrane (∼2%) fractions (Fig. S1 H). Similar to metazoan Arl13b, Chlamydomonas ARL13 is predicted to be palmitoylated on cysteine12, a modification that allows certain small GTPases to be associated with the membrane when in the GTP-bound state (Fig. S1 B; Cevik et al., 2010; Jin et al., 2010; Li et al., 2010; Mariani et al., 2016; Ren et al., 2008). The addition of a non-hydrolyzable GTP analog GTP-γ-S during phase partitioning moved the GTPase ARL6/BBS3 from the matrix to the membrane fraction, whereas most ARL13 remained in the matrix fraction (Fig. S1 I). The fractionation experiments indicate that Chlamydomonas ARL13, similar to IFT and BBSome proteins, is a mostly soluble ciliary matrix protein.
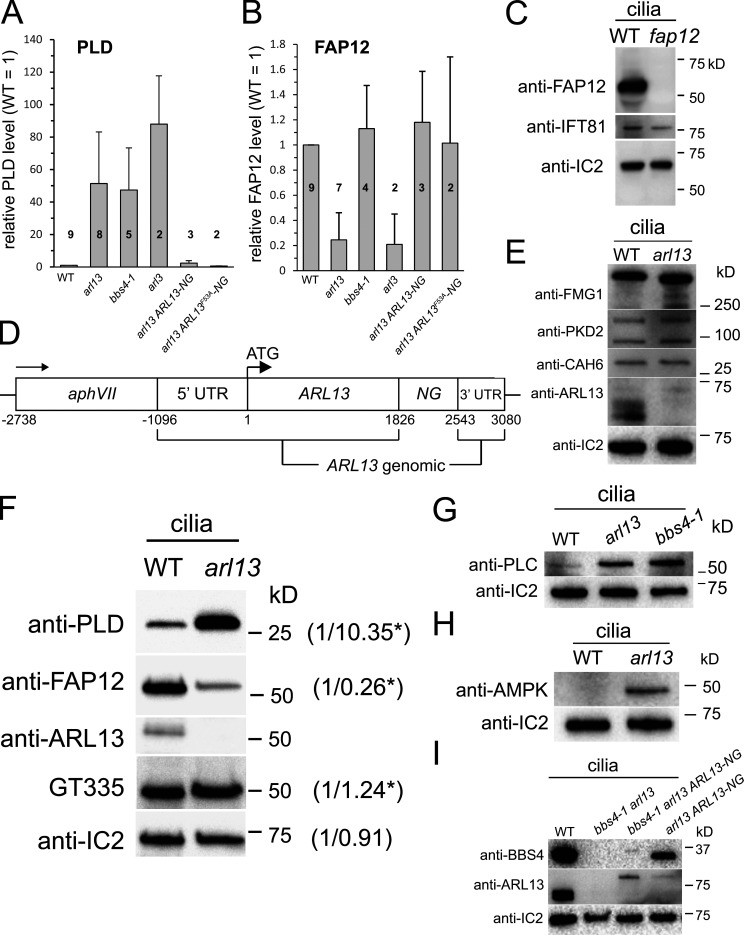
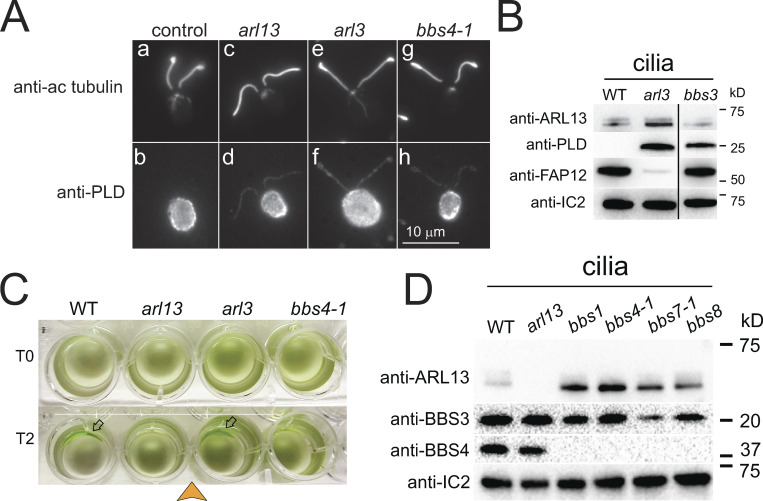
ARL13B is commonly implicated in ciliary protein import in metazoans (Cevik et al., 2010; Gotthardt et al., 2015; Humbert et al., 2012). To investigate how ARL13 deficiency impacts ciliary protein composition in Chlamydomonas, isolated wild-type and arl13 cilia were fractionated as above and analyzed by SDS-PAGE and silver staining (Fig. 1 E). While the axonemal and matrix fractions showed only minor differences between the wild-type and mutant samples, pronounced differences were apparent in the two membrane samples, and most prominently a band of ∼50 kD was significantly reduced in arl13, and a band of ∼25 kD was abnormally accumulated (Fig. 1 E). Mass spectrometry (MS) of the excised bands identified the smaller protein as phospholipase D (PLD, ∼25 kD), a negative regulator of phototaxis (Liu and Lechtreck, 2018), and the larger as flagellar associated protein 12 (FAP12, ∼50 kD), an ciliary triacyl glycerol lipase of unknown function. FAP12 is an abundant ciliary membrane protein and its reduction in arl13 cilia is visible on silver-stained gels of whole cilia samples (Fig. 1 E, lane 1 and 2). Western blotting using a previously characterized antibody against PLD and a novel antibody against FAP12 confirmed both the strong reduction of FAP12 (25% of control, SD 21%, n = 7 independent cilia isolates) and the striking increase of PLD (51×, SD 32×, n = 8 cilia isolates) in arl13 cilia (Fig. 1, B and F; and Fig. S2, A–C; Lechtreck et al., 2013). Accumulation of PLD in the ciliary membrane is also a hallmark of Chlamydomonas bbs mutants, and previously, we linked PLD accumulation in cilia to the loss of phototaxis in bbs mutants (Lechtreck et al., 2009; Liu et al., 2021). The amount of FAP12, however, is normal in bbs4-1 cilia (Fig. 1 F).
Figure S2.
arl13 cilia accumulate AMPK and PLC. (A and B) Bar graphs showing the relative levels of PLD (A) and FAP12 (B) in the strains indicated based on Western blot quantifications. The number of biological repeats, i.e., cilia preparations, is indicated; error bars show the standard deviation. (C) Western blot characterizing the anti-FAP12 antibody using isolated cilia of control (WT, g1) and a fap12 insertional mutant (CLiP strain LMJ.RY0402.206664), which carries an insertion in the fourth exon deleting four bases. Anti-IC2 and anti-IFT81 were used as loading controls. The fap12 mutant will be characterized elsewhere. (D) Schematic presentation of the ARL13-NG expression vector. (E) Western blot of isolated cilia from control (g1, WT) and arl13 strains probed with antibodies against FMG1, PKD2, CAH6, ARL13, and IC2, as a loading control. The analysis is based on several membranes with equal loading; the lanes stained with anti-ARL13 and anti-IC2 were also used for the Western blot shown in Fig. 2 A. (F) Western blot of isolated cilia from control (g1, WT) and arl13 strains probed with GT335 to detect polyglutamylated protein, anti-ARL13, anti-FAP12, anti-PLD and anti-IC2, as a loading control. The numbers indicate band intensities (WT = 1), the stars indicate that the values were adjusted for loading based on the IC2 signal. One of two biological replicates is shown. (G) Western blot of isolated cilia from control (g1, WT), arl13 and bbs4-1 strains probed with antibodies against PLC and IC2, as a loading control. Antibodies to PLC were characterized by Awasthi et al. (2012). (H) Western blot of isolated cilia from control (g1, WT) and arl13 probed with antibodies against AMPK, and IC2, as a loading control. (I) Western blot of isolated cilia from control (g1, WT), bbs4-1 arl13, bbs4-1 arl13 ARL13-NG, and arl13 ARL13-NG strains probed with antibodies against BBS4, ARL13, and IC2, as a loading control. Source data are available for this figure: SourceData FS2.
Since our antibody against ARL13 failed in immunofluorescence experiments (not shown), we expressed Chlamydomonas ARL13 fused at its C-terminus to mNeonGreen (ARL13-NG) under the control of the native ARL13 flanking sequences in the arl13 mutant (Fig. S2 D). In most cells, ARL13-NG was concentrated in the proximal half of the cilia and tapered off toward the tip (Fig. 1 G). This supports previous data from C. elegans, which also possesses only one copy of ARL13, and mammals showing that Arl13b preferentially localized to the proximal inversin compartment (Cevik et al., 2010; Cevik et al., 2013; Warburton-Pitt et al., 2014; Zhang et al., 2016). Expression of ARL13-NG restored near wild-type levels of FAP12 and PLD in mutant cilia, confirming that these biochemical defects are caused by the loss of ARL13 (Fig. 1 B; and Fig. S2, A and B). The arl13 ARL13-NG strain swam faster than arl13, but phototaxis remained inconsistent (Fig. 1 D; and Fig. S1, F and G). ARL13-NG was expressed at levels below that of endogenous ARL13, which could explain the incomplete rescue (Fig. 1 B).
Chlamydomonas arl13 and bbs mutants accumulate a similar set of proteins in the cilia
Western blotting and mass spectrometry were used to further explore how the loss of ARL13 affects the biochemical composition of Chlamydomonas cilia. Western blotting showed that the major ciliary glycoprotein FMG-1, the TRP-channel PKD2, and the dual-lipidated membrane-associated protein carbonic anhydrase 6 (CAH6) were present at near wild-type levels in arl13 cilia (Fig. S2 E; Bloodgood et al., 2019; Huang et al., 2007; Witman et al., 1972; Yu et al., 2020). In mammalian cells and C. elegans, Arl13b knock-down reduces the level of tubulin polyglutamylation in cilia (He et al., 2018; Larkins et al., 2011; Warburton-Pitt et al., 2014). In Chlamydomonas, however, Western blots using anti-GT335 detected near normal levels of polyglutamylated tubulin in arl13 cilia (Fig. S2 F; Wolff et al., 1992). Potentially, these differences could be related to differences in cilia age: primary cilia of differentiated cells are typically long-lasting whereas Chlamydomonas disassembles its cilia day-to-day during cell division. These differences in cilia biology could also explain why ARL13b deficiency causes axonemal defects in C. elegans and mammalian cilia whereas those of the Chlamydomonas mutant were apparently normal (Fig. S1 E; Caspary et al., 2007; Cevik et al., 2010; Li et al., 2010; Warburton-Pitt et al., 2014).
Mass spectrometry (MS) of ciliary membrane fractions from three biological replicates of each wild type and arl13 confirmed the accumulation of PLD in arl13 and showed increased amounts of the AMP-regulated serine/threonine kinase AMPK, the calcium/calmodulin-dependent protein kinase CDPKK1, and the catalytic domain of the phosphatidylinositol-specific phospholipase C (PLC) in arl13 cilia (Table 1). PLD, AMPK, and CDPKK1 were previously shown to be also enriched in Chlamydomonas bbs cilia (Lechtreck et al., 2009; and Lechtreck, unpublished observations). Western blotting confirmed the accumulation of AMPK in arl13 cilia and the accumulation of PLC in arl13 and bbs4-1 cilia (Fig. S2, G and H). The four accumulated proteins lack transmembrane domains but are predicted to be dual- or triple-lipidated on N-terminal MGC-/MGACC-/MGNVFSC-motifs by CSS-Palm 4.0 and the NMT MYR predictor with high scores (Table 1). MS analyses also confirmed the reduction of FAP12 in arl13 cilia and indicated a reduction in FAP138, an uncharacterized coiled-coil protein. Further, FAP124/UBA1, a ubiquitin-activating enzyme E1, and the protein tyrosine-phosphatase PTP1 were abundant in all three control samples but not detected in the arl13 samples (Table 1). FAP12, FAP138, and PTP1 carry N-terminal MGC- or MG-motifs, respectively, suggesting that they might be (dual)-lipidated. In summary, the lack of ARL13 results in the abnormal loss or reduction and accumulation of mostly membrane-associated proteins suggesting a dual role of ARL13 in ciliary import and export of such proteins. Although the biochemical defects in arl13 cilia are broader than those resulting from BBSome deficiency, arl13 and bbs mutants accumulate the same set of proteins in their cilia, indicating an overlapping role of ARL13 and the BBSome in ciliary protein export.
Table 1.
Abundance of select ciliary membrane proteins in control and arl13 samples
| WT | arl13 | |||||||
|---|---|---|---|---|---|---|---|---|
| Coverage (%) | Total peptides | Unique peptides | Coverage (%) | Total peptides | Unique peptides | |||
| Protein (ID #) | N-terminus | Exp.1/Exp.2/Exp.3 (average ±) | Exp.1/Exp.2/Exp.3 (average ±) | Exp.1/Exp.2/Exp.3 (average ±) | Exp.1/Exp.2/Exp.3 (average ±) | Exp.1/Exp.2/Exp.3 (average ±) | Exp.1/Exp.2/Exp.3 (average ±) | |
| Accumulated | PLD | mgcasskeev | 0/0/26.9 (9.0 ± 15.54) | 0/0/7 (2.3 ± 4.04) | 0/0/4 (1.33 ± 2.31) | 52.9/78.9/57.4 (63.1 ± 13.90) | 11/40/35 (28.7 ± 15.50) | 10/18/12 (13.3 ± 4.16) |
| AMPK | mgaccsqpse | 0/0/7.5 (2.5 ± 4.3) | 0/0/5 (1.7 ± 2.9) | 0/0/3 (1 ± 1.7) | 32.5/42.1/45.0 (39.8 ± 6.6) | 16/34/49 (33 ± 16.5) | 14/17/16 (15.7 ± 1.5) | |
| PLC | mgnvfscfet | 0/0/0 (0) | 0/0/0 (0) | 0/0/0 (0) | 30.5/45.3/30.9 (35.6 ± 8.4) | 22/40/41 (34.3 ± 10.7) | 16/18/14 (16 ± 2) | |
| CDPKK1 | mgcvgskeda | 0/0/0 (0) | 0/0/0 (0) | 0/0/0 (0) | 13/15.3/20.1 (16.1 ± 3.7) | 8/24/28 (20 ± 10.6) | 8/11/13 (10.7 ± 2.5) | |
| Reduced | FAP12 | mgcgasvmnr | 78.2/80.6/72.0 (76.9 ± 4.4) | 154/384/462 (333.3 ± 160.1) | 53/52/47 (50.7 ± 3.2) | 47.8/77.6/71.6 (65.7 ± 15.8) | 37/132/289 (127.3 ± 152.7) | 27/41/44 (37.3 ± 9.1) |
| FAP138 | mgcgasvmnr | 35.2/41.4/28.1 (34.9 ± 6.6) | 10/21/13 (14.7 ± 5.7) | 7/9/6 (7.3 ± 1.5) | 13.6/0/9.9 (7.8 ± 7.02) | 4/0/4 (2.3 ± 2.7) | 3/0/2 (1.7 ± 1.5) | |
| FAP124 | mskrtadgst | 24.4/25.9/33.1 (27.8 ± 4.6) | 21/49/56 (42 ± 18.5) | 21/22/27 (23.3 ± 3.2) | 0/0/0 (0) | 0/0/0 (0) | 0/0/0 (0) | |
| PTP1 | mgsgastqvq | 15.9/10.2/7.1 (11.1 ± 4.5) | 12/10/11 (11 ± 1) | 9/6/4 (6.3 ± 2.5) | 0/0/0 (0) | 0/0/0 (0) | 0/0/0 (0) | |
Summary of mass spectrometry analyses of isolated matrix fractions. Only proteins, which were enriched or reduced in all three replicates are shown. Peptide coverage, total peptides identified, number of unique peptides identified, and the corresponding averages across three experiments are listed. PLD, phospholipase D (Cre13.g591900); AMPK, AMP-regulated kinase (Cre16.g657350); PLC, phosphatidylinositol phospholipase C (Cre06.g270200); CDPKK1: calcium/calmodulin-dependent protein kinase kinase (Cre10.g428650); FAP12: flagellar associated protein 12 (Cre09.g390615); FAP138: flagellar associated protein 138 (Cre14.g632350); FAP124: flagellar associated protein 124 (Cre09.g386400); PTP1: protein tyrosine phosphatase 1 (Cre07.g325724).
ARL13 is required for PLD transport by the BBSome/IFT pathway
The established role of ARL13B in ciliary protein import could explain the observed loss and reduction of ciliary proteins in Chlamydomonas. Therefore, we focused on the question how ARL13 loss causes an accumulation of PLD in Chlamydomonas cilia. In wild-type cells, small amounts of PLD cycle via the BBSome/IFT machinery through cilia with the BBSome functioning as an adapter to mediate the association of PLD to IFT trains (Liu and Lechtreck, 2018). In bbs mutants, PLD continues to enter the cilia by diffusion and because its BBSome-dependent export is impaired, accumulates over time (Lechtreck et al., 2013). Because BBSome traffic depends on IFT, PLD also accumulates in hypomorphic retrograde IFT mutants or when IFT is switched off using a conditional mutant (Lechtreck et al., 2013).
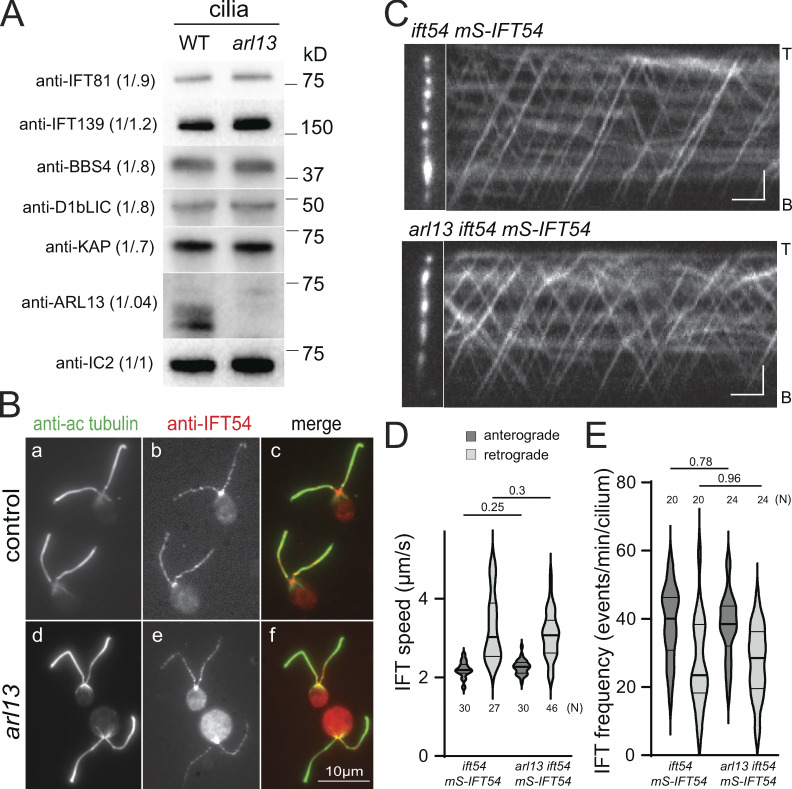
To explore which step of the PLD export pathway is disturbed in arl13, we first analyzed IFT. Western blot analysis of arl13 cilia showed normal or near normal levels of the IFT-A protein IFT139, the IFT-B protein IFT81, the retrograde motor subunit D1bLIC, and anterograde motor subunit KAP (Fig. 2 A). Immunofluorescence showed a similar distribution of IFT54 in control and arl13 (Fig. 2 B). An accumulation of IFT proteins at the ciliary tip, as reported for Arl13b-knockout RPE1 cells, was not observed (Nozaki et al., 2017). To image IFT in vivo, we expressed IFT54 tagged with mScarlet (i.e., mS-IFT54) in an arl13 ift54 double mutant. Total internal reflection fluorescence (TIRF) microscopy revealed that both the speed and frequency of anterograde and retrograde IFT were normal in arl13 ift54 mS-IFT54 cells (Fig. 2, C–E). We concluded that IFT is unaffected in arl13.
Figure 2.
IFT is normal in arl13 mutant cilia. (A) Western blot of isolated control (g1, WT) and arl13 cilia probed with antibodies against IFT81, IFT139, BBS4, D1BLIC, KAP, ARL13, and, as a loading control, IC2. The quantification of the band strengths normalized for those of IC2 are shown in brackets. The analysis is based on several membranes with equal loading; the lanes stained with anti-ARL13 and anti-IC2 were also used for the Western blot shown in Fig. S2 E. (B) Immunofluorescence staining of control (g1; a–c) and arl13 (d–f) cells with antibodies to acetylated tubulin (a and d) and IFT54 (b and e). Merged images are shown in c and f. (C) TIRF images and corresponding kymograms showing IFT in ift54 mS-IFT54 and arl13 ift54 mS-IFT54 cilia. Anterograde IFT results in trajectories from the bottom left to the top right whereas top left to bottom right trajectories result from retrograde IFT. The ciliary tips (T) and bases (B) are indicated. Bars = 2 µm and 2 s. (D) Violin plot of anterograde and retrograde velocities of mS-IFT54 in ift54 mS-IFT54 and arl13 ift54 mS-IFT54 cilia. The P values of a two-tailed t test are indicated. N, the number of IFT trains analyzed. (E) Violin plot of the anterograde and retrograde IFT frequencies of mS-IFT54 in ift54 mS-IFT54 and arl13 ift54 mS-IFT54 cilia. The P-values of a two-tailed t test are indicated. N, the number of cilia analyzed. Source data are available for this figure: SourceData F2.
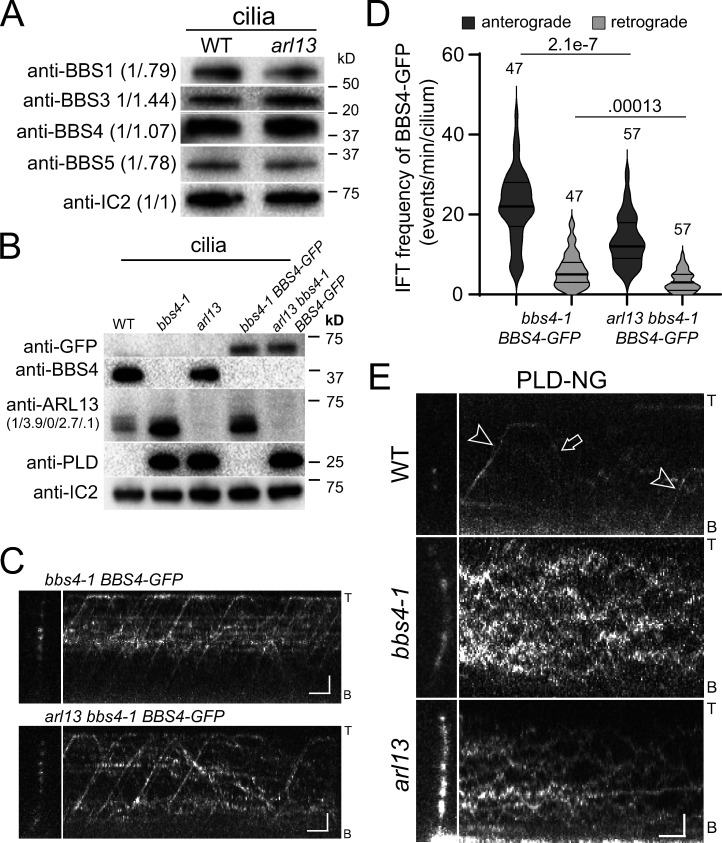
To assess the presence of BBSomes, control and arl13 cilia were probed with antibodies against the BBSome subunits BBS1, BBS4, and BBS5 and the small GTPase ARL6/BBS3. The latter regulates BBSome recruitment to membranes and stabilizes its open configuration for cargo binding (Jin et al., 2010; Liu et al., 2021; Singh et al., 2020; Yang et al., 2020). All four proteins were present in near normal amounts in arl13 cilia (Fig. 3 A). To image BBSome traffic, we generated an arl13 bbs4 BBS4-GFP strain by mating (Fig. 3 B). As previously described, the expression of BBS4-GFP was below that of endogenous BBS4 in controls (Lechtreck et al., 2009). Cilia of the parental bbs4-1 BBS4-GFP strain had normal amounts of PLD whereas ARL13 remained elevated (Fig. 3 B). In arl13 cilia, BBS4-GFP moved processively by anterograde and retrograde IFT albeit at somewhat lower frequencies compared to controls (22.2 vs. 13.5 and 6.2 vs. 3.3 for anterograde and retrograde transport, respectively; Fig. 3, C–D). In most cell types, only intact BBSomes enter cilia and move by IFT (Hsu et al., 2021; Lechtreck et al., 2013; Wei et al., 2012). We conclude that BBSome assembly and traffic are mostly normal in arl13 but for the reduction in frequency observed for BBS4-GFP.
Figure 3.
PLD fails to move by IFT carriers in arl13 cilia. (A) Western blot of isolated cilia from control (g1, WT) and arl13 probed with antibodies against BBS1, BBS3, BBS4, BBS5, and IC2, as a loading control. The quantification of the band intensities normalized for those of IC2 is shown in brackets and is based on one experiment. (B) Western blot of isolated cilia from WT, bbs4-1, arl13, bbs4-1 BBS4-GFP, and arl13 bbs4-1 BBS4-GFP strains probed with antibodies against GFP, BBS4, ARL13, PLD, and IC2 as a loading control. The quantification of the anti-ARL13 band strengths normalized for those of IC2 are shown in brackets and is based on one experiment. Note that ARL13 remained elevated in cilia of the bbs4-1 BBS4-GFP strain. (C) TIRF images and corresponding kymograms of bbs4-1 BBS4-GFP and arl13 bbs4-1 BBS4-GFP cilia. The ciliary tips (T) and bases (B) are indicated. Bars = 2 µm and 2 s. (D) Violin plot of the anterograde and retrograde IFT frequencies of BBS4-GFP in bbs4-1 BBS4-GFP and arl13 bbs4-1 BBS4-GFP cilia. The number of cilia analyzed and the P values of a two-tailed t test are indicated. (E) TIRF images and corresponding kymograms of PLD-NG in control (g1, WT), bbs4-1 and arl13 cilia. Note anterograde (arrowheads) and retrograde (arrow) IFT of PLD-NG in control cilia. The ciliary tips (T) and bases (B) are indicated. Bars = 2 µm and 2 s. Source data are available for this figure: SourceData F3.
For in vivo analysis of PLD in cilia, we expressed an NG-tagged version of PLD (PLD-NG) in control, bbs4-1 and arl13 (Fig. 3 E and Fig. S2 D). In wild-type cilia, which contain only small amounts of PLD-NG, transport by IFT was regularly observed (Fig. 3 E). In bbs4-1 cilia, IFT of PLD was impaired and PLD moved by diffusion as previously described (Liu and Lechtreck, 2018). Similarly, PLD-NG moved by diffusion in arl13 cilia, and IFT of PLD-NG was not observed (Fig. 3 E). We consider it unlikely that the reduced IFT frequency of BBS4-GFP observed in a transgenic strain that was used to monitor BBSome traffic in vivo (Fig. 3 D) explains the complete absence of PLD transport by IFT in arl13. In conclusion, loss of ARL13 incapacitates PLD transport by apparently normal IFT/BBSome carriers.
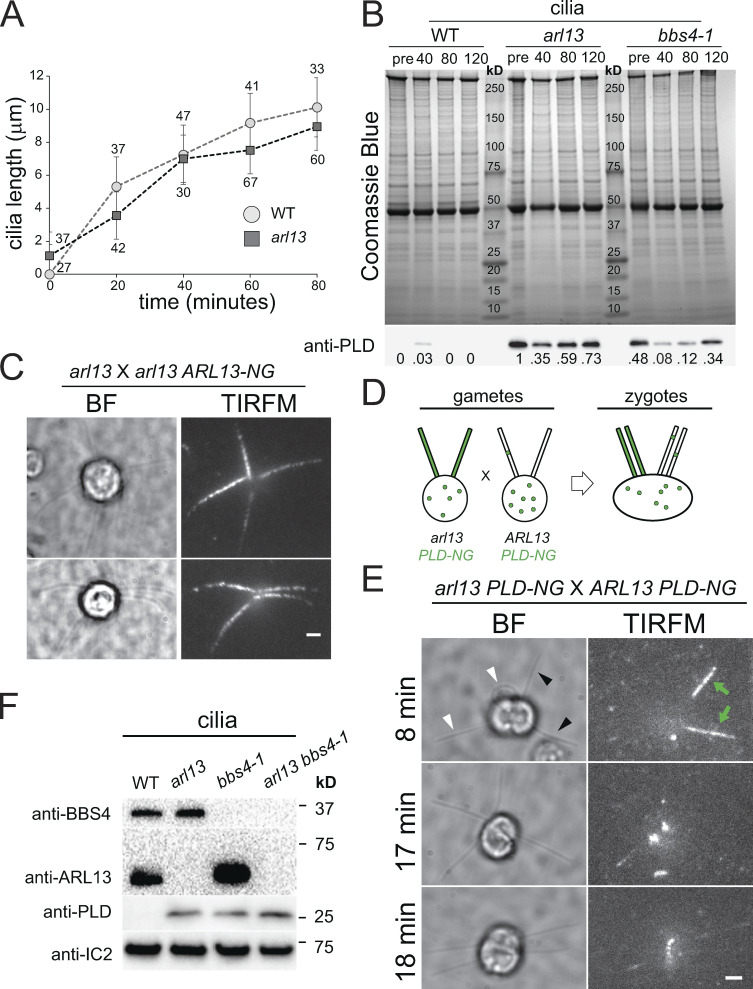
To analyze the dynamics of PLD accumulation in arl13 cilia, we used cilia regeneration and dikaryon rescue experiments (Fig. S3). The arl13 mutant regrew cilia with similar kinetics as wild type (Fig. S3 A). In newly assembled bbs mutant cilia, PLD accumulates slowly over the course of several hours (Fig. S3 B; Lechtreck et al., 2013). Similarly, PLD accumulated over time in newly formed arl13 cilia and, at 2 h after the onset of ciliary regeneration, its level was still below that observed in arl13 cilia prior to deciliation (Fig. S3 B). We previously showed that PLD is rapidly (∼10 min) removed from bbs4-derived cilia when intact BBSomes are re-introduced by mating of the bbs4 gametes to wild-type gametes; the latter will provide BBS4 to the shared cytoplasm of the zygotes (Lechtreck et al., 2013; Yu et al., 2020). To test if ARL13 will enter full-length cilia lacking it, i.e., arl13 cilia, we mated arl13 and arl13 ARL13-NG. Tagged ARL13 was observed in all four cilia of the resulting zygotes analyzed between 10 and 20 min after mixing of the gametes, revealing that ARL13-NG quickly enters arl13-derived mutant cilia (n = 7 zygotes, Fig. S3 C). In similar dikaryon rescue experiments using arl13 PLD-NG × ARL13 PLD-NG zygotes, early stage zygotes had two PLD-NG positive cilia derived from the arl13 mutant parent and two cilia with very low levels of PLD-NG derived from the control strain (Fig. S3 E, 8 min). Low levels of PLD-NG were observed in all four cilia of five of nine such zygotes analyzed within ∼20 min after mixing of the gametes, and in 9 of 12 zygotes analyzed in the 20–40 min time window (Fig. S3 E). As not all gametes fuse immediately after mixing of the gametes (Hunnicutt and Snell, 1991), the data indicate that PLD is rapidly removed from arl13-derived cilia when ARL13 is reintroduced. In cilia of an arl13 bbs4-1 double mutant, PLD levels were comparable to those of the respective single mutants, suggesting that ARL13 and the BBSome act on PLD transport through the same pathway (Fig. S3 F). To summarize, the ciliary levels and transport kinetics of PLD are affected similarly in arl13 and bbs mutants and during the repair of zygotic mutant cilia.
Figure S3.
PLD accumulates slowly in arl13 cilia. (A) Cilia regeneration kinetics after deciliation by a pH shock for control (WT; circles) and arl13 (squares). One cilium was measured per cell, and the number of cells analyzed is indicated. (B) Coomassie Blue stained gel (top) and Western blot of a replicate gel using anti-PLD (bottom) of control (g1, WT), arl13 and bbs4-1 cilia harvested at full-length (pre; no deciliation) and during regeneration at 40, 80, and 120 min after deciliation by a pH shock. PLD accumulated over time in regenerating arl13 and bbs4-1 cilia. Note transient presence of PLD in control cilia during early regeneration. Numbers indicate the anti-PLD signal strength normalized for the Coomassie Blue stained samples. One of two biological replicates is shown with the other experiment omitting the 40 min-samples and the bbs4-1 strain. (C) Bright field (BF) and TIRF images of two arl13 × arl13 ARL-NG zygotes. The images were recorded within 20 min after mixing of the gametes indicating rapid entry of ARL13-NG in arl13-derived cilia. Bar = 2 µm. (D) Schematic presentation of the dikaryon rescue experiment using arl13 PLD-NG and control PLD-NG gametes. After fusion, ARL13 provided by wild-type parent is available for entry into arl13-derived cilia and its effect on the distribution of PLD-NG can be analyzed by TIRFM. (E) Gallery of bright field (BF) and TIRF images of arl13 PLD-NG × PLD-NG zygotes. The time passed since mixing of the gametes is indicated. In the top row, green arrows and black arrowheads indicate the PLD-NG positive cilia; white arrowheads mark cilia derived from the PLD-NG parent. Bar = 2 µm. (F) Western blot of isolated cilia from control (g1, WT), arl13, bbs4-1 and arl13 bbs4-1 double mutant probed with antibodies against BBS4, ARL13, PLD, and IC2, as a loading control. Source data are available for this figure: SourceData F3.
ARL13 is not an adapter for IFT of PLD
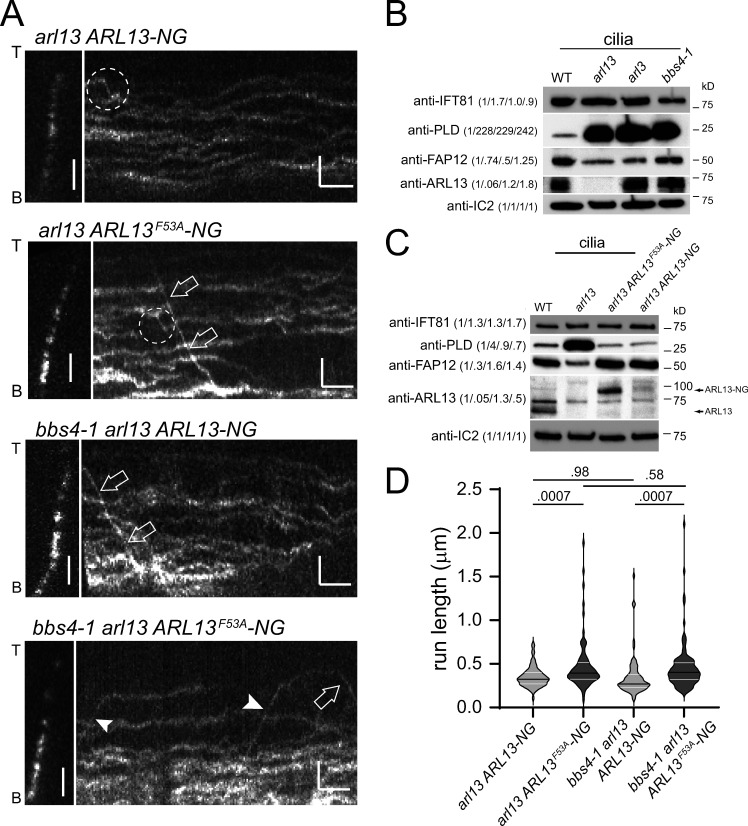
Previously, IFT or IFT-like transport of tagged Arl13b were reported, and ARL13b binds certain IFT-B proteins in vitro (Cevik et al., 2010; Nozaki et al., 2017; Williams et al., 2014). As both bbs and arl13 mutants lack PLD transport, we wondered if Chlamydomonas ARL13 functions similar to the BBSome, which is a co-migrating adapter linking PLD to IFT. If correct, one would expect that ARL13 moves by IFT in Chlamydomonas cilia. However, ARL13-NG, while restoring low levels of PLD in the arl13 cilia, predominately moved by slow diffusion, and processive movements by IFT were exceedingly rare and well below those observed for tagged PLD (∼1 transport/minute/cilium (Fig. 4 A and Fig. S4 A, Table 2; Liu and Lechtreck, 2018). Thus, Chlamydomonas ARL13-NG is not a co-adapter that travels together with BBSomes on IFT trains to mediate PLD transport.
Figure 4.
IFT of Chlamydomonas ARL13 is rare. (A) Gallery of still images and corresponding kymograms showing ciliary movements of ARL13-NG and ARL13F53A-NG in arl13 and bbs4-1 arl13 cilia. Arrowheads mark anterograde and open arrows retrograde IFT of ARL13; the dashed circles mark putative IFT events. The ciliary tips (T) and bases (B) are indicated. Bars = 2 µm and 2 s. (B) Western blot of control (g1, WT), arl13, arl3, and bbs4-1 cilia probed with antibodies against IFT81, PLD, FAP12, ARL13, and IC2. The quantification of the band strengths normalized for those of IC2 is shown in brackets and is based on one experiment. (C) Western blot of control (g1, WT), arl13, arl13 ARL13F53A-NG, and arl13 ARL13-NG cilia probed with antibodies against IFT81, PLD, FAP12, ARL13, and IC2 as a loading control. The quantification of the band strengths normalized for those of IC2 is shown in brackets and is based on one experiment. (D) Violin plot of the run lengths, i.e., distance transported processively by IFT, of ARL13-NG, and ARL13F53A-NG in the arl13 and the bbs4-1 arl13 strains. Transport of ARL13F53A-NG was more processive than that of ARL13-NG in arl13 and arl13 bbs4-1 mutants. The P values of a two-tailed t test are indicated; see Table 2 for additional information. Source data are available for this figure: SourceData F4.
Figure S4.
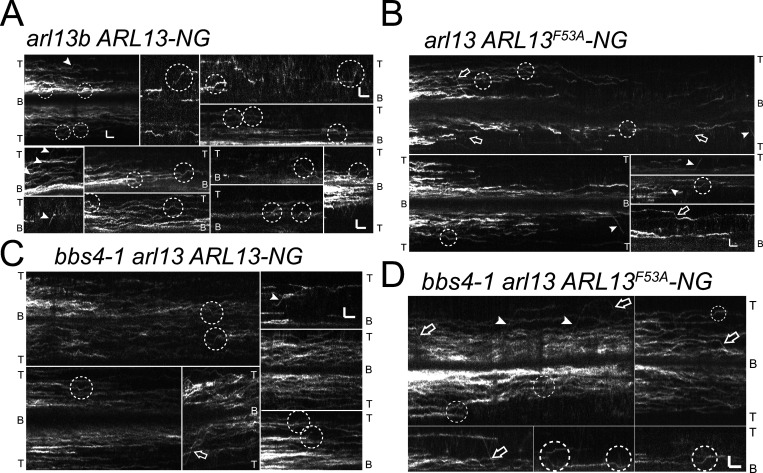
IFT of ARL13-NG. (A–D) Compilation of kymograms showing the behavior of ARL13-NG (A and C) and ARL13F53A-NG (B and D) in arl13 (A and B) and bbs4-1 arl13 (C and D) cilia. IFT events are in marked by arrowheads (anterograde) and arrows (retrograde). The dashed circles indicate putative IFT events and/or IFT-like movements. The tips (T) and bases (B) of the cilia are indicated. Bars = 2 s and 2 µm.
Table 2.
ARL13-NG transport by IFT
| Strain | Cilia (#) | Time (s) | IFT events (a/r) | Velocity (a/r) | Run length (s) (SD) | Frequency (events/min/cilium) | Runs >1 s (a/r) | Frequency >1 s (events/min/cilium) |
|---|---|---|---|---|---|---|---|---|
| arl13 ARL13-NG | 109 | 2,428 | 72 (47/25) | 1.55/1.69 | 0.65 (SD 0.22) | 0.016 | 6 (5/1) | 0.0014 |
| arl13 ARL13 F53A -NG | 107 | 2,091 | 77 (48/29) | 1.46/1.8 | 0.89 (SD 0.55) | 0.02 | 18 (15/3) | 0.0048 |
| bbs4-1 arl13 ARL13-NG | 79 | 1,849 | 75 (31/44) | 1.47/1.88 | 0.65 (SD 0.43) | 0.031 | 8 (5/3) | 0.0032 |
| bbs4-1 arl13 ARL13 F53A -NG | 63 | 1,444 | 102 (58/44) | 1.46/1.55 | 0.85 (SD 0.51) | 0.067 | 24 (14/10) | 0.016 |
The table lists the number of cilia analyzed for each strain, the imaging time, the number of IFT events, their velocity, average run lengths, and frequency, and the number and frequency of long-distance (>1 s) transports. a, anterograde transports; r, retrograde transports.
Chlamydomonas arl3 cilia recapitulate the biochemical defects of arl13 cilia
In vitro, Chlamydomonas ARL13 functions as a GEF for ARL3 increasing its rate of GTP hydrolysis ∼70-fold (ElMaghloob et al., 2021; Fujisawa et al., 2021; Gotthardt et al., 2015; Pandey et al., 2020). Mutations affecting the GEF activity of human ARL13B result in Joubert syndrome, suggesting that the ARL13/ARL3 relationship is relevant in vivo (Ivanova et al., 2017). To test if ARL13-dependent export of PLD from Chlamydomonas cilia involves ARL3, we obtained CLiP strain LMJ.RY0402.182282, which carries an insertion in the fourth exon of Cre04.g218250 encoding Chlamydomonas ARL3. Similar to arl13, PLD was strikingly accumulated in isolated arl3 mutant cilia, whereas FAP12 was reduced (Fig. 4 B). Immunofluorescence showed that PLD is distributed along the length of arl3 cilia, similar to observations in arl13 and bbs4-1 mutants (Fig. S5 A). ARL13 was present in arl3 cilia suggesting that ARL13 alone is unable to mediate PLD export from cilia in the absence of ARL3 (Fig. 4 B and Fig. S5 B). Despite the accumulation of PLD, which we previously characterized as a negative regulator of Chlamydomonas phototaxis, arl3 displayed phototactic behavior (Fig. S5 C; Liu and Lechtreck, 2018). Possibly, the lack of ARL3 blunts PLD’s inhibitory effect on phototaxis but a satisfactory explanation for this observation is currently missing. Chlamydomonas arl13 and arl3 mutants similarly affect ciliary levels of PLD and FAP12 supporting the notion that they form a functional pair in vivo.
Figure S5.
PLD accumulates along the length of arl13, arl3, and bbs4-1 cilia. (A) Immunofluorescence of methanol (−20°C) treated control (a/b), arl13 (c/d), arl3 (e/f), and bbs4-1 (g/h) cells stained with anti-acetylated tubulin (a, c, e, and g) and anti-PLD (b, d, f, and h). In the cell body, most PLD is located below the plasma membrane. Bar = 10 µm. (B) Western blot of isolated cilia from control (g1, WT), arl3 and bbs3 probed with antibodies against ARL13, PLD, FAP12, and IC2, as a loading control. The black line indicates that an unrelated lane was cropped out; see source file for the uncropped blot. (C) Population phototaxis assay of control (g1, WT), arl13, arl3, and bbs4-1. The direction of light (arrowhead) and time of exposure in minutes are indicated. Arrows, accumulated cells. (D) Western blot of isolated cilia from control (g1, WT), arl13, and bbs mutants probed with antibodies against ARL13, BBS3, BBS4, and, as a loading control, IC2. Source data are available for this figure: SourceData FS5.
To further test whether ARL13 acts as a GEF through ARL3 to mediate BBSome-dependent export of PLD, we expressed ARL13F53A-NG, which lacks most (∼95%) of its GEF activity for ARL3 in the arl13 mutant (Gotthardt et al., 2015). ARL13F53A-NG rescued both the reduction of FAP12 and the accumulation of PLD in arl13 cilia (Fig. 4 C). Phototactic behavior was partially restored in the arl13 ARL13F53A-NG strain, which could be related to the higher expression of ARL13F53A-NG in comparison to that of ARL13-NG in the arl13 ARL13-NG rescue strain (Fig. S5 C and Fig. 4 C). The most parsimonious interpretation is that Chlamydomonas ARL13 is not acting via its GEF activity through ARL3 to prevent PLD accumulation in cilia. In related observations, mice expressing Arl13bR79Q/R79Q, a derivative with impaired GEF-activity for Arl3, were shown to develop normally (Suciu et al., 2021). Thus, two widely used models for ciliopathies do not recapitulate the grave effects of ARL13b mutations with diminished GEF activity on humans. Possibly, other cellular GEFs compensate for the loss of ARL13.
IFT of ARL13F53A-NG was more processive and therefore easier to detect (Fig. 4, A and D; Fig. S4; Table 2; and Video 1). While the reason for the more stable association of ARL13F53A-NG with IFT is unknown, it is possible that ARL13’s GEF activity regulates the duration of its transport. Since the arl13 mutant has a defect in BBSome-dependent cargo transport, we wondered if IFT of ARL13 is BBSome-dependent, putatively explaining why ARL13 accumulates in bbs mutant cilia (Fig. 3 B). Thus, we expressed tagged ARL13 and ARL13F53A-NG in the bbs4-1 arl13 double mutant (Fig. 4, A and D). The overall behavior of tagged ARL13/ARL13F53A-NG in the bbs4-1 cilia was similar to that observed in wild-type controls indicating that the high levels of PLD present in bbs mutant cilia do not alter ARL13-NG mobility in a striking manner (Fig. 4 A). IFT of ARL13-NG and the more processive ARL13F53A-NG occurred with similarly low frequencies in arl13 single and bbs4-1 arl13 double mutants (Fig. 4, A and D; Fig. S2 I; and Table 2). Thus, the observed transport of ARL13-NG is not BBSome-dependent, indicating ARL13-IFT rather than ARL13-BBSome interactions.
Video 1.
In vivo imaging of ARL13F53A-NG. Movie obtained by TIRF microscopy (left) and corresponding kymogram (right) showing ARL13F53A-NG in cilia of the arl13 mutant. IFT events are highlighted by exclamation marks in the corresponding frames of the movie and in the kymogram. A central part of the movie (∼50 s, see timer) was removed as during this period no IFT events were observed. The movie was recorded at 10 fps and the timer counts seconds. The movie is related to Fig. 4 and Fig. S4.
IFT and BBSome deficiency affects the amount and phosphorylation state of ciliary ARL13
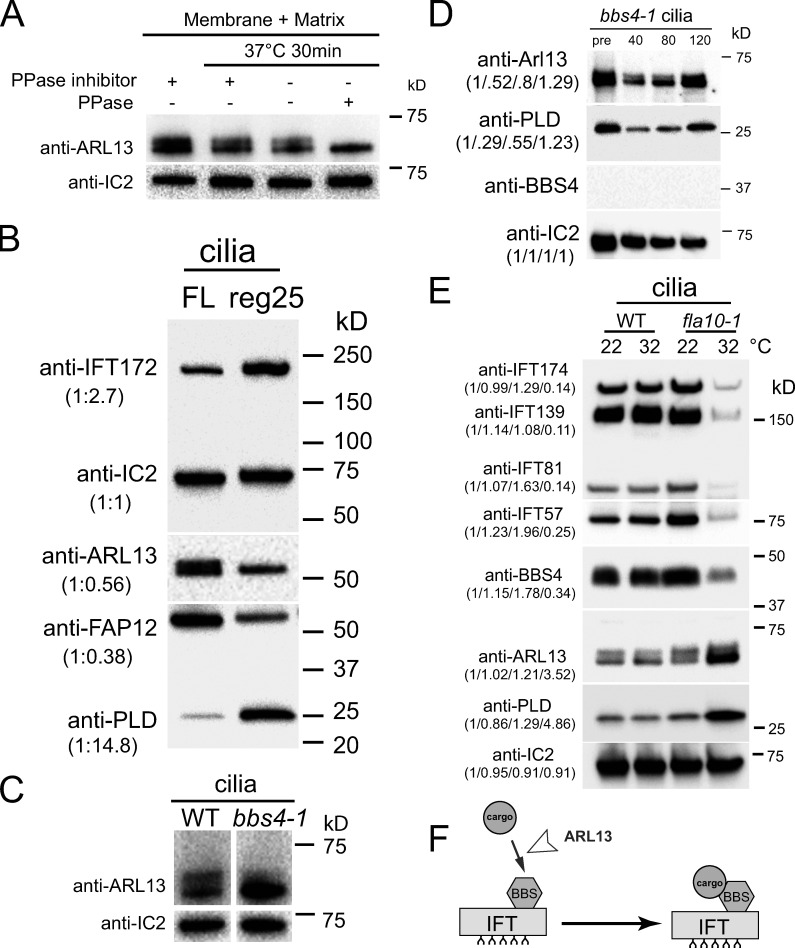
On most Western blots of isolated cilia, endogenous and tagged ARL13 ran as double bands with the faster migrating band typically being somewhat more dominant (Fig. 5 A; see also Figs. 1 B, 2 A, 3 B, and Fig. S5 B). Mammalian Arl13b is phosphorylated by casein kinase 2 in vitro (Ivanova et al., 2017). To test if Chlamydomonas ARL13 from isolated cilia is phosphorylated, the samples were treated with calf intestinal phosphatase prior to SDS-PAGE (Fig. 5 A). This treatment eliminated the upper ARL13 band revealing that it represents phospho-ARL13. Phospho-ARL13 was largely absent from early regenerating control cilia (N = 3 biological replicates; Fig. 5 B). In comparison to full-length cilia, such short growing cilia had reduced levels of FAP12 whereas those of PLD were increased. These correlations could mean that non-phosphorylated ARL13 is imported into cilia, and that phospho-ARL13 is critical for the transport of these lipidated proteins in and out of cilia.
Figure 5.
ARL13 accumulates in BBSome-deficient cilia. (A) Western blot of isolated control (g1, WT) cilia treated with phosphatase inhibitor or protein phosphatase (PPase) as indicated and probed with antibodies against ARL13 and, as a loading control, IC2. (B) Western blot comparing full-length and regenerating control cilia; the latter were harvested ∼25 min after the onset of cilia regrowth. Note relative accumulation of IFT172 and PLD and the reduction of FAP12 and phospho-ARL13 in short growing cilia. The quantification of the band strengths normalized for those of IC2 is shown in brackets and is based on one experiment. (C) Western blot of control (g1, WT) and bbs4-1 cilia probed with antibodies against ARL13 and, as a loading control, IC2. (D) Western blot of pre-deciliation (pre) and regenerating bbs4-1 cilia (time in minutes after cilia amputation is indicated) probed with antibodies against ARL13, PLD, BBS4 and, as a loading control, IC2. Note the accumulation of ARL13 over time. The quantification of the band strengths normalized for those of IC2 is shown in brackets and is based on one experiment. This figure is also part of Fig. S3 B. See Fig. S5, B and D for analyses of other bbs mutants. (E) Western blot analysis of cilia from control (g1, WT) and fla10-1 maintained at the permissive temperature of 22°C or the restrictive temperature of 32°C for 2.5 h prior to cilia isolation. Three replica blots were probed with antibodies to IFT proteins, BBS4, ARL13, PLD, and as a loading control, IC2. Quantifications of the band is shown in brackets and is based on one experiment. (F) Model proposing that ARL13 is required for BBSome-PLD interaction. Source data are available for this figure: SourceData F5.
In BBSome-deficient cilia, but not in the arl6/bbs3 strain, the overall amount of endogenous ARL13 was elevated, whereas the relative amount of phospho-ARL13 was diminished (Fig. 5 C and Fig. S5 D). The analysis of regenerating bbs4-1 cilia indicated that ARL13 accumulates slowly over time as the cilia mature (Fig. 5 D).
To further test the role of the IFT in regulating ciliary ARL13 levels, we employed the conditional kinesin-2 mutant fla10-1, which allows us to switch off IFT by shifting cells from 22°C to the non-permissive temperature of 32°C (Kozminski et al., 1995; Vashishtha et al., 1996). After 2.5 h at the restrictive temperature, the amount of several IFT proteins in fla10-1 cilia was diminished, whereas non-phospho ARL13 and (as previously reported) PLD, were accumulated (Fig. 5 E). While the role of ARL13 phosphorylation is unknown, the data show that both active IFT and the BBSome are required to prevent ARL13 from accumulating in Chlamydomonas cilia. To summarize, the IFT/BBSome system regulates the ciliary levels of ARL13 and in turn, ARL13 is required for PLD transport by IFT/BBSome carriers (Fig. 5 F).
Discussion
Chlamydomonas ARL13 is required for PLD transport by IFT/BBS carriers
ARL13 and the BBSome function in ciliary protein transport, especially that of membrane proteins (Cevik et al., 2010; Nachury et al., 2007). The BBSome has been mostly implicated in protein export from cilia (Lechtreck et al., 2009; Nachury, 2018; Wingfield et al., 2018). In contrast, ARL13, together with ARL3 and carrier proteins such as UNC119 is thought to function mostly in ciliary protein import (Gotthardt et al., 2015). Here, we show that Chlamydomonas bbs and arl13 mutants accumulate a similar set of membrane-associated proteins in cilia. This includes PLD, which can enter cilia by diffusion and, in control cells, migrates in a BBSome-dependent manner by IFT ensuring its export from cilia (Liu and Lechtreck, 2018). Despite the presence of apparently normal IFT/BBSomes carriers, IFT of PLD was abolished in arl13 causing PLD to accumulate in cilia. We conclude that Chlamydomonas ARL13, in addition to its contribution to ciliary protein import, also participates in ciliary protein export via the IFT/BBSome pathway.
A role of ARL13 in regulating BBSome interactions is supported by data from other models. In C. elegans, which employs two distinct anterograde IFT motors, the BBSome mediates the integrity of the IFT trains (Ou et al., 2005). Similar to bbs mutants, IFT-A and IFT-B particles move with distinct velocities in C. elegans arl-13 and arl-3 mutants suggesting that they are required to maintain proper BBSome-IFT contacts (Cevik et al., 2010; Li et al., 2010). The concept of ARL13 regulating BBSome-cargo interactions could also explain the abnormal distribution of the Hedgehog proteins Smo and GPR161 in mammalian arl13 mutants as these and other GPCRs are removed from cilia by the IFT/BBS pathway (Bangs and Anderson, 2017; Desai et al., 2020; Eguether et al., 2014; Larkins et al., 2011; Nozaki et al., 2017; Shinde et al., 2020).
FAP12 and some other ciliary proteins are reduced or missing in Chlamydomonas arl13 (and arl3) cilia but are unaffected in those of the bbs mutants. These proteins could enter cilia using the previously described ARL13/ARL3/UNC119 import system (Gotthardt et al., 2015). An UNC119 ortholog is encoded in the Chlamydomonas genome (Blast score 9e−52 with human UNC119 over 96% of the sequence). Thus, Chlamydomonas ARL13 has a dual role in ciliary protein traffic participating in both ciliary protein entry and IFT/BBSome-dependent export. This causes a broader biochemical defect of arl13 mutant cilia in comparison to those of bbs mutants. If this holds true for mammalian Arl13b, it could explain the phenotypical overlap between Bardet–Biedl syndrome and Arl13b-related Joubert syndrome, with the latter being the more severe ciliopathy (Lee and Gleeson, 2011; Novarino et al., 2011; Waters and Beales, 2011).
How could ARL13 support BBSome-dependent cargo traffic?
In arl13, cargo transport could be impaired due to defects in the integrity of the BBSome carriers, the transportability of PLD, or other conditions required for PLD transport. BBSome composition and traffic were mostly normal in arl13, indicating that ARL13 is neither a BBSome assembly factor nor is it needed for IFT of the BBSomes. In mammalian systems, cargo binding by the BBSome requires activation of its GPCR cargoes by ligand binding and the resulting downstream signaling events (Wingfield et al., 2018; Ye et al., 2018). Further, ubiquitination of GPCRs earmarks them for IFT/BBSome-dependent export from cilia (Desai et al., 2020; Shinde et al., 2020). Increased levels of ubiquitinated proteins are also present in Chlamydomonas bbs4 cilia (Shinde et al., 2020). Similar to cargoes of the BBSome in mammalian bbs cilia, one would expect ubiquitinated forms of PLD to accumulate in Chlamydomonas bbs cilia, which, however, was not apparent on Western blots using anti-PLD (Desai et al., 2020; Shinde et al., 2020). Also, the N-terminal 20 residues of PLD, encompassing the putative dual lipidation motif and a sole lysine, are sufficient for its transport by BBSome-dependent IFT (Liu and Lechtreck, 2018). We therefore consider it unlikely that ARL13 mediates ubiquitination or another covalent modification of PLD as a prerequisite for its attachment to IFT/BBSome carriers. Tagged Chlamydomonas ARL13 is occasionally and transiently moved by IFT but the frequency was well below that of IFT of tagged PLD. Further, IFT of ARL13 was not BBSome-dependent, putatively indicating that ARL13 interacts with the IFT core. Thus, a role of ARL13 as a co-migrating adapter fastening PLD to IFT/BBSome carriers is excluded. However, the ARL13/IFT encounters described here, many more of which could have escaped detection due to their briefness, could stimulate the BBSome to bind its cargoes. Of note, the small GTPase RABL2 binds the IFT-B complex and thereby seems to also regulate BBSome-dependent protein export from cilia (Duan et al., 2021; Kanie et al., 2017; Nishijima et al., 2017). Thus, the binding of small GTPases to IFT-B, which carries the BBSome, might modulate BBSome cargo binding. Finally, ARL13 could generate conditions, e.g., indirectly via its role in protein import or as a GEF or GTPase to ensure that PLD can be picked up by the BBSome; then, ARL13 loss would lead to a ciliary environment impeding BBSome-dependent export of PLD.
ARL13, ARL3, and ARL6/BBS3 functionally overlap in regulating BBSome-dependent cargo transport
PLD also accumulates in a Chlamydomonas mutant that lacks arl6/bbs3 in its cilia; however, this mutant is not null for ARL6/BBS3 (Liu et al., 2021). In agreement with a more limited role of ARL6/BBS3 in BBSome-dependent protein export, the amount of FAP12 in mutant cilia was normal (Fig. S5 B). ARL6/BBS3 binds to BBS1, recruits the BBSome to membranes, and stabilizes the BBSome in an open configuration for loading (Jin et al., 2010; Mourao et al., 2014; Singh et al., 2020; Yang et al., 2020). Despite this clear evidence for direct BBSome-ARL6/BBS3 interaction, travel of tagged Chlamydomonas ARL6/BBS3 by processive IFT was not observed, and BBSomes enter and move by IFT in the arl6/bbs3-mutant cilia (Liu et al., 2021). This recapitulates our observations on ARL13 suggesting that BBS3 might also transiently interact with the BBSomes to ensure PLD export. It should be noted that IFT of ARL6/BBS3 was observed in C. elegans and mammalian cilia (Fan et al., 2004; Williams et al., 2014). Further, PLD accumulates in Chlamydomonas arl3 cilia supporting the idea that ARL13 and ARL3 form a functional pair during the regulation of BBSome-dependent export of PLD as they apparently do in protein import (Gotthardt et al., 2015). Thus, ARL13, ARL3, and ARL6/BBS3 functionally overlap in mediating PLD transport by the IFT/BBS system. In a hypothetical scenario, these three ARLs could initiate BBSome-cargo interactions, e.g., by activating the BBSome or by making membrane-bound cargoes available for BBSome pick-up.
IFT and the BBSome regulate ciliary ARL13 levels
ARL13 accumulated in cilia of several bbs mutants or when IFT was switched off, suggesting that maintaining its level in cilia requires both the BBSome and moving IFT trains. Similarly, PLD also accumulates in cilia of bbs mutants and when IFT is switched off because it requires moving IFT/BBS carriers to exit cilia (Lechtreck et al., 2009; Liu and Lechtreck, 2018). This mechanism seems unlikely to explain the accumulation of ARL13 in bbs cilia since IFT of tagged ARL13 was rare and not BBSome-dependent. It is also unlikely that the accumulation of ARL13 is caused by the elevated levels of PLD as the amounts of ARL13 were normal or only marginally elevated in the bbs3 and arl3 mutants, respectively, which both accumulate PLD in cilia. As a possible solution for this conundrum, we speculate that the BBSome and IFT could regulate ciliary ARL13 levels independently of each other. In detail, influx of ARL13 into cilia could be increased in bbs mutants, whereas the loss of IFT could prevent ARL13 export from cilia, both causing ARL13 to accumulate over time albeit by distinct mechanisms. The BBSome has been repeatedly implicated in the regulation of ciliary protein import. Neuronal cilia of mammalian Bbs−/− mutants, for example, lack certain GPCRs possibly indicating that these are imported in a BBSome-dependent manner (Berbari et al., 2008; Loktev and Jackson, 2013; Nachury et al., 2007). In a variation of entry control, we recently showed that the Chlamydomonas BBSome restricts the entry of tagged carbonic anhydrase 6 (CAH6, a predicted dual lipidated protein like PLD and FAP12) into one of the two cilia of a given cell (i.e., the cilium attached to the younger basal body; Yu et al., 2020). Thus, the BBSome, which is present at the ciliary base, could—directly or indirectly—limit ARL13 entry into cilia, and low-frequency BBSome-independent IFT of ARL13 could be sufficient to balance ARL13 influx; loss of IFT or the BBSome would cause ARL13 levels to rise (Ansley et al., 2003). This model could also explain why low-level expression of tagged BBS4 in the bbs4 mutant was sufficient to restore normal ciliary PLD levels whereas ARL13 remained elevated (Fig. 3 B): a reduced number of BBSomes could still be enough to export small amounts of PLD via IFT but insufficient to limit ARL13 influx into cilia. Regardless of the molecular mechanism of ARL13 homeostasis in cilia, our data indicate the presence of a hypothetical feedback loop or compensation mechanism, in which IFT and the BBSome regulate the amount of ARL13 in cilia and, if IFT or BBSome numbers decrease, more ARL13 will enter cilia to upregulate PLD transport by IFT/BBSome carriers.
Materials and methods
Strains, culture conditions, and genotyping
The Chlamydomonas strains used in this study are listed in Table 3. The arl13 (LMJ.RY0402.181356) mutant and the arl3 (LMJ.RY0402.182282) mutant are available from the Chlamydomonas Library Project (CLiP; https://www.chlamylibrary.org/allMutants; Li et al., 2019). Cells were maintained in modified M medium at ∼24°C with a light/dark cycle of 14:10 h (https://www.chlamycollection.org/methods/media-recipes/minimal-or-m-medium-and-derivatives-sager-granick/). Large cultures (1–4 liters) used to isolate cilia were aerated with air supplemented with 0.5% CO2.
Table 3.
List of strains used in this study
| Name | Genotype | Reference or source |
|---|---|---|
| g1 (wild type) | nit1, agg1, mt + | (Pazour et al., 1995) |
| CC-5325 | cw15, mt − | CLiP (Li et al., 2019) |
| arl13 CLiP | cw15, arl13, mt − | CLiP (Li et al., 2019) |
| arl3 CLiP | cw15, arl3, mt − | CLiP (Li et al., 2019) |
| fap12 | cw15, fap12, mt − | CLiP (Li et al., 2019) |
| CC-4371 (bbs1-1) | bbs1 | (Lechtreck et al., 2009) |
| CC-4377 (bbs4-1) | nit1, agg1, bbs4, mt + | (Lechtreck et al., 2009; Pazour et al., 1995) |
| CC-4381 (bbs7-1) | nit1, agg1, bbs7, mt + | (Lechtreck et al., 2009; Pazour et al., 1995) |
| bbs8 | nit1, agg1, bbs8, mt + | (Lechtreck et al., 2013) |
| arl13 ARL13-NG | arl13 ARL13-NG-TG | This study |
| CC-5860 (ift54 IFT54-mS) | nit1, agg1, ift54, mt-, mS-IFT54-TG | (Wingfield et al., 2017) |
| PLD-NG | nit1, nit2, mt + , PLD-NG-TG | (Liu and Lechtreck, 2018) |
| bbs4-1 BBS4-GFP | nit1, agg1, bbs4, mt + , BBS4-GFP-TG | (Lechtreck et al., 2009) |
| arl13 ift54 mS-IFT54 | arl13, ift54, mS-IFT54-TG | This study |
| arl13 PLD-NG | arl13, PLD-NG-TG | This study |
| arl13 ARL13 F53A -NG | arl13 ARL13 F53A -NG-TG | This study |
| arl13 bbs4-1 | arl13, bbs4-1 | This study |
| arl13 bbs4-1 BBS4-GFP | arl13, bbs4, BBS4-GFP-TG | This study |
| arl13 bbs4-1 ARL13-NG | arl13, bbs4, ARL13-NG-TG | This study |
| arl13 bbs4-1 ARL13 F53A -NG | arl13, bbs4, ARL13F53A-NG-TG | This study |
The CC numbers refer to the Chlamydomonas stock collection number. TG, transgene.
PCR using isolated genomic DNA was used to verify and track the insertion in the mutant and derived progeny. Primers 1–3 (see Table 4) were used to track the arl13 insertional allele, primers 4 and 5 were used to amplify a part of the gβ gene to verify DNA quality, and primers 6–9 were used to determine the mating type (Zamora et al., 2004).
Table 4.
List of primers used in this study
| Primer no. | Sequence (5′ to 3′) |
|---|---|
| 1 | 5′-CGCTTCGAGGAGTCCAAAAT-3′ |
| 2 | 5′-CAGCAAAAGGAGGAGGAGGA-3′ |
| 3 | 5′-TGTCGCTGAAAGTGGAGGTC-3′ |
| 4 | 5′-CAAGCTGAAGAACAACCTGGTG-3′ |
| 5 | 5′-CTTGCTGGTGATGTTGAACTCG-3′ |
| 6 | 5′-TCCAACGCATAGCCATCAAC-3′ |
| 7 | 5′-TGTTTGCTAGGGGTGCAATG-3′ |
| 8 | 5′-ACCGGTGTTTACCGTCGAGT-3′ |
| 9 | 5′-CCTTTCTGTAGGGCCACCTG-3′ |
| 10 | 5′-GCGTCTAGACTCCAGCGCCAGCTCGAGAG-3′ |
| 11 | 5′-CGCCTCGAGTGCGTCATTGGGGGCTCCC-3′ |
| 12 | 5′-GCGAAGCTTCTCCAGCGCCAGCTCGAGAG-3′ |
| 13 | 5′-CGCGAATTCCTACTTGTACAGCTCGTCCATGCC-3′ |
| 14 | 5′-GCGGAATTCTCGGTGCCACACCTGAACGC-3′ |
| 15 | 5′-CGCGAATTCGTTCCGATAGTTTTCCCTGCC-3′ |
| 16 | 5′-AAGGATTTCAGAATTCAGACCTCTGGAGGCGCATCC-3′ |
| 17 | 5′-TAGAGGATCCGAATTCTATGCGTCATTGGGGGCTC-3′ |
| 18 | 5′-ATTGGATCCAATGGGTTGCGGAGCGAGCGTCATG-3′ |
| 19 | 5′-GACAAGCTTTTACGCTGCCG GCGGCGGCGG-3′ |
Transgenic strain and mating progeny generation
To clone ARL13 for tagging with NG, ARL13 was amplified from control genomic DNA using primers 10 and 11 omitting the stop codon and cloned upstream of the NG gene. The 5′-UTR region of ARL13-NG was amplified using primers 12 and 13 and inserted upstream of ARL13. Then, the cassette was transferred to pKL3, a plasmid which contains the aph7″ gene conferring resistance to hygromycin. Finally, the 3′-UTR of ARL13 was amplified using primers 14 and 15 and inserted downstream of the NG gene (Table 4). The resulting plasmid pKL3-ARL13-NG was linearized with NdeI and transformed into arl13 by electroporation (NEPA21 Super Electroporator Type II). Transformants were selected on TAP plates containing 10 µg/ml hygromycin (Bio Basic). Transformants expressing ARL13-NG were identified by TIRF microscopy. To express ARL13F53A-NG, a DNA fragment encoding the corresponding mutation (TTC to GCC) was synthesized (Genewiz) and used to replace the corresponding control fragment in the pKL3-ARL13-NG plasmid by restriction digestion with HpaI and AvrII.
To express mS-IFT54, PLD-NG, and BBS4-GFP in the arl13 mutants, we mated arl13 to ift54 mS-IFT54, PLD-NG, and bbs4-1 BBS4-GFP, respectively. The arl13 bbs4-1 double mutant was generated by mating of the corresponding arl13 and bbs4-1 single mutants. The mating procedure has been described previously (Liu et al., 2020). In short, cells were grown in M medium to a density of 2 × 106 cells/ml, transferred to M-N medium (M medium without nitrogen; https://www.chlamycollection.org/%20methods/media-recipes/minimal-or-m-medium-and-derivatives-%20sager-granick/), and aerated overnight in constant light. The next morning, the plus and minus cells were mixed, incubated for 4–6 h in light, transferred to M medium 4% agar or 1.8% Phytogel (Sigma-Aldrich) plates, air-dried, and incubated in constant light overnight. Then, the plates were incubated in the dark for ≥10 d, transferred to −20°C for 2 d, defrosted, dried, and incubated in light for ∼10 d to allow the zygotes to germinate. The resulting colonies were streaked for single cells, and progeny of the desired genotype were identified by PCR, TIRF imaging, and Western blotting. The same procedure was used for dikaryon rescue experiment using the arl13 PLD-NG and PLD-NG strains and the zygotes were analyzed using TIRFM starting immediately after mixing of the gametes.
Isolation of cilia
To isolate cilia and ciliary subfractions, we followed the procedures described by Witman (1986) and Lechtreck et al. (2009). In brief, cells were concentrated and washed in 10 mM HEPES, pH 7.4, resuspended in HEPES-Magnesium-Sucrose (HMS; 10 mM HEPES, 5 mM MgSO4, and 4% sucrose), and deciliated by adding dibucaine to a final concentration of 4.17 mM (Sigma-Aldrich) and vigorous pipetting. Immediately after deciliation, HMS supplemented with 0.7 mM EGTA was added, and the cell bodies were separated by centrifugation (1,150 g, 3 min, 4°C; Sorvall Legend XTR; Thermo Fisher Scientific). The remaining cell bodies were removed by underlaying the supernatant with a 25% sucrose in HMS cushion and centrifugation (1,700 g, 4°C, 10 min). Then, the cilia were sedimented by centrifugation (31,000 g, 4°C, 15 min; Avanti JXN-26; Beckman Coulter) and resuspended in HEPES-Magnesium-EGTA-Potassium (HMEK; 30 mM HEPES, 5 mM MgSO4, 0.5 mM EGTA, and 25 mM KCl) with 1% protease inhibitor cocktail (P9599; Sigma-Aldrich) and, in some preparations, a phosphatase inhibitor cocktail (NaF, β-glycerophosphate, Na3VO4). To obtain axoneme, matrix, and membrane fractions, Triton X-114 was added to a final concentration of 1% on ice. The axonemes were removed through centrifugation (31,000 g, 4°C, 15 min) and resuspend in HMEK with 1% protease inhibitor cocktail. The supernatant was incubated at 32°C for 3–5 min to induce phase separation, and the Triton X-114 micelles were harvested by centrifugation (3,300 g, RT, 5 min). The phase separation was repeated once by adding Triton X-114 (1–2% final concentration) to the aqueous phase (resulting in the matrix fraction) and HMEK to the detergent phase. Proteins in the detergent phase were isolated by methanol-chloroform precipitation resulting in the membrane fraction.
Protein analysis and antibodies
For silver staining and Western blotting, SDS sample buffer was added to protein samples, and the samples were incubated at 95°C for ∼10 min. Proteins were separated by SDS-PAGE using precast gels (Bio-Rad TGX; 4–15%). After silver-staining (Silver Stain Plus Kit; Bio-Rad Laboratories), bands of interest were excised from the gel and subjected to mass spectroscopy, using an Orbitrap Elite system. For Western blotting, proteins were transferred to PVDF membrane (Millipore), blocked in 4% milk in TBS-T and incubated overnight at 4°C with agitation in the primary antibodies. The primary antibodies and the dilutions used in this study are listed in Table 5. After washing, the membranes were incubated for 60–90 min at RT with agitation in the secondary antibodies (i.e., anti-mouse and anti-rabbit IgGs conjugated to horseradish peroxidase; Invitrogen 31432/AB_228302 and 31460/AB_228341, respectively). For visualization, membranes were incubated in chemiluminescence substrate (SuperSignal West Pico PLUS or Atto; Thermo Fisher Scientific), and the images were captured using a Bio-Rad ChemiDoc MP imaging system and the Image Lab software. To quantify band intensities, we used Bio-Rad Image lab or the ROI Manager in ImageJ/Fiji. CSS-Palm (biocuckoo.org) and NMT—The MYR predictor (imp.ac.at) were used to predict putative myristoylation and palmitoylation sites of ARL13 (Ren et al., 2008).
Table 5.
List of primary antibodies used in this study. rb, rabbit; mo, mouse, rt, rat; WB, Western blotting; IF, immunofluorescence
| Name | Host | Dilution WB | Dilution if | Reference or source |
|---|---|---|---|---|
| Anti-ARL13 | rb | 1:25–250 | This study | |
| Anti-IFT81 | mo | 1:1,000 | (Cole et al., 1998) | |
| Anti-IFT139 | mo | 1:100 | (Cole et al., 1998) | |
| Anti-IFT54 | rb | 1:100 | (Wingfield et al., 2017) | |
| Anti-BBS1 | rb | 1:500 | (Xue et al., 2020) | |
| Anti-BBS3 | rb | 1:2,000 | (Xue et al., 2020) | |
| Anti-BBS4 | rb | 1:500 | (Lechtreck et al., 2009) | |
| Anti-D1BLIC | rb | 1:2,000 | (Hou et al., 2004) | |
| Anti-KAP | rt | 1:2,000 | Invitrogen, Carlsbad, CA | |
| Anti-FMG1 | mo | 1:500 | (Bloodgood and Salomonsky, 1994) | |
| Anti-PKD2 | rb | 1:3,000 | (Huang et al., 2007) | |
| Anti-CAH6 | rb | 1:500 | (Yu et al., 2020) | |
| Anti-FAP12 | rb | 1:1,000–2,500 | This study | |
| Anti-PLD | rb | 1:1,000 | 1:80 | (Lechtreck et al., 2013) |
| Anti-AMPK | rb | 1:1,000 | (Liu and Lechtreck, 2018) | |
| Anti-PLC | rb | 1:3,000 | (Awasthi et al., 2012) | |
| Anti-IC2 | mo | 1:1,000 | (King and Witman, 1990) | |
| Anti-ac tubulin 6-11B-1 | mo | 1:1,000 | 1:800 | (LeDizet and Piperno, 1987); Sigma 32-2,700 |
| Anti-GT335 | mo | 1:3,000 | (Wolff et al., 1992); AdipoGen AG-20B-0020-C100 |
To generate the anti-ARL13 antibody, the second exon of ARL13 was cloned into the EcoRI site in the pMAL-cRI vector (downstream of the Maltose Binding Protein sequence) using primers 16 and 17. The fusion protein was expressed in E. coli, purified using amylose resin following the instructions of the manufacturer (New England Biolabs), and sent to Pocono Rabbit Farm and Laboratory for polyclonal antibody generation in rabbits. Sera containing the anti-ARL13 antibody were affinity-purified using the MBP-ARL13 fusion protein.
Antibodies against FAP12 were generated as follows: a 1.45 kb fragment of the FAP12 cDNA was amplified by PCR from a Chlamydomonas gametic λ phage cDNA library using primers 18 and 19 (Table 4), digested with BamH1 and HindIII, and cloned into cut pTrcHisXa, a modified version of pTrcHis-TOPO plasmid (Invitrogen; Life Technologies) with an inserted factor Xa protease cleavage site to facilitate the removal of hexa-histidine tag. His6-FAP12 was expressed in E. coli TOP10 cells, purified using HisPut Cobalt Resin (Catalog #89965; Pierce; Thermo Fisher Scientific, Inc.,) following the instructions of the manufacturer, and used to raise a rabbit polyclonal antibody (Open Biosystems; Thermo Fisher Scientific, Inc.,). The serum was immunopurified using His6-FAP12 immobilized on nitrocellulose.
Swimming velocity
To measure the swimming velocity, cells were resuspended in fresh M medium, placed in a chambered plastic slide (14-377-259; Fisherbrand), and observed using an inverted light microscope (TMS; Nikon). Images were recorded using a MU500 camera (Amscope) and the associated Topview software at a fixed exposure time of 1 s. The length of the swimming trajectories was analyzed in ImageJ (National Institutes of Health) and converted into μm/s. Excel was used for statistical analysis and violin plots were prepared using GraphPad Prism.
In vivo microscopy and electron microscopy
For in vivo imaging, 10 μl cell suspension was placed on a 24 × 60–mm No. 1.5 coverslip and allowed to settle for ∼1–3 min. A ring of petroleum jelly or vacuum grease was added around the droplet and a 22 × 22–mm No. 1.5 coverslip with 5-μl drop of 10 mM HEPES, pH 7.4, and 5 mM EGTA was inverted onto the larger cover glass to form a sealed observation chamber. For TIRF imaging, a Nikon Eclipse Ti-U inverted microscope equipped with a 60×/1.49 numerical aperture (NA) TIRF objective and a through-the-objective TIRF illumination system was used. Excitation light was provided a 40-mW, 488-nm diode laser (Spectraphysics), filtered by a Nikon GFP/mCherry TIRF filter cube (Lechtreck, 2013), and images were recorded at 10 frames/s using an EMCCD camera (Andor iXon ×3 DU897) and the Elements software package (Nikon). Specimens were imaged at room temperature (24°C). ImageJ was used to analyze the videos and to extract still images, image averages, and kymograms. Photoshop (Adobe) was used to adjust brightness and contrast, and the figures were assembled in Illustrator (Adobe).
For electron microscopy, cells were fixed in glutaraldehyde and processed as previously described (Wilkerson et al., 1995). Images were collected using a JEOL JEM1011 electron microscope and processed as described above.
Immunofluorescence
For indirect immunofluorescence, cells were sedimented, resuspended in HMEK, allowed to settle onto polyethyleneimine (0.2%) coated multiwell slides for 1–2 min, and submerged into −20°C methanol for 8 min. The slides were air dried, blocked (1% BSA in PBS-T), washed with PBS, incubated with the primary antibodies in blocking buffer overnight, washed, stained with secondary antibodies (1:800 AlexaFluor anti-rb-565 and anti-mo-488; Invitrogen A11036/AB_10563566 and A11029/AB_2534088, respectively), washed in PBS-T, submerged briefly in 80% ethanol, air dried, and mounted in ProlongGold (Invitrogen). For widefield epifluorescence microscopy, images were taken using a 60 × 1.49 objective Eclipse Ti-U microscope (Nikon) equipped with a Lumen200 light source (PRIOR) and filters for FITC and TexasRed. Images were documented as described above.
Phototaxis assay
Phototaxis assays were performed as previously described (Liu and Lechtreck, 2018). In brief, population phototaxis assays were performed using cells (∼107 cells per milliliter) harvested during the first half of the light phase, washed in fresh M medium, and placed in a Petri dish or 24-well cell culture plate. Images were taken prior and after illumination for 2–10 min with bright light from one side. Single cell phototaxis assays were performed by placing 20-µL cell suspension in a chambered plastic slide (14-377-259; Fisherbrand). Cells were observed for 10–15 s using nonphototactic red light illumination and an inverted microscope (Eclipse 55i; Nikon). Then, the slides were illuminated from one side with actinic light, three images were taken in series at 2 fps using an MU500 camera (Amscope), and the swimming tracks were analyzed and combined in ImageJ.
Online supplemental material
Fig. S1 characterizes the arl13 mutant and analyzes the solubility of ciliary ARL13. Fig. S2 analyzes the biochemical composition of arl13 mutant cilia by Western blotting. Fig. S3 studies the kinetics of PLD accumulation in regenerating cilia and PLD removal from zygotic arl13 cilia. Fig. S4 provides a gallery of kymograms showing the behavior of ARL13-NG and ARL13F53A-NG in arl13 and arl13 bbs4-1 cilia. Fig. S5 characterizes behavior and ciliary biochemical composition of a novel arl3 mutant and additional bbs mutants. Video 1 shows diffusion and IFT of ARL13F53A-NG in arl13 cilia.
Supplementary Material
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S5.
Acknowledgments
We thank Dr. Suneel Kateriya (Department of Biochemistry, University of Delhi, New Delhi, India) for the generous gift of anti-PLC antibodies and Drs. Aaron Harris and Jenna Wingfield (both University of Georgia, Athens, GA) for TEM analysis of arl13. We thank Drs. Tamara Caspary, Richard Kahn (both Emory University, Atlanta, GA), and Marek Eliáš (University of Ostrava, Ostrava, Czech Republic) for advice on ARL13 gene models and discussion of the manuscript.
This study was supported by grants by the National Institutes of Health (R01GM110413 to K. Lechtreck) and by the National Natural Science Foundation of China (32070698 to Z.-C. Fan and 32100541 to B. Xue). B. Mekonnen was supported by a University of Georgia (UGA) CURO Research Assistantship for undergraduate researchers. We acknowledge expert assistance by UGA’s Electron Microscopy and the Proteomics and Mass Spectrometry core facilities; the latter is funded in part by the National Institutes of Health (S10RR028859). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: J. Dai, G. Zhang, R.A. Alkhofash, B. Mekonnen, S. Saravanan, and K. Lechtreck: investigation, visualization, validation, formal analysis. J. Dai, P. Liu and K. Lechtreck: Conceptualization. G. Zhang, B. Xue, Z.-C. Fan, E. Betleja, D. Cole and P. Liu: resources. Z.-C. Fan and K. Lechtreck: funding acquisition. J. Dai and K. Lechtreck: writing—original draft. J. Dai, B. Xue, P. Liu and K. Lechtreck: writing—review, and editing. J. Dai, P. Liu, D. Cole and K. Lechtreck: supervision.
References
- Alkanderi, S., Molinari E., Shaheen R., Elmaghloob Y., Stephen L.A., Sammut V., Ramsbottom S.A., Srivastava S., Cairns G., Edwards N., et al. 2018. ARL3 mutations cause Joubert syndrome by disrupting ciliary protein composition. Am. J. Hum. Genet. 103:612–620. 10.1016/j.ajhg.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansley, S.J., Badano J.L., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., et al. 2003. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 425:628–633. 10.1038/nature02030 [DOI] [PubMed] [Google Scholar]
- Awasthi, M., Batra J., and Kateriya S.. 2012. Disulphide bridges of phospholipase C of Chlamydomonas reinhardtii modulates lipid interaction and dimer stability. PLoS One. 7:e39258. 10.1371/journal.pone.0039258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs, F., and Anderson K.V.. 2017. Primary cilia and mammalian hedgehog signaling. Cold Spring Harbor Perspect. Biol. 9:a028175. 10.1101/cshperspect.a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari, N.F., Lewis J.S., Bishop G.A., Askwith C.C., and Mykytyn K.. 2008. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. USA. 105:4242–4246. 10.1073/pnas.0711027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque, O.E., Reardon M.J., Li C., McCarthy J., Mahjoub M.R., Ansley S.J., Badano J.L., Mah A.K., Beales P.L., Davidson W.S., et al. 2004. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Gene Dev. 18:1630–1642. 10.1101/gad.1194004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood, R.A., and Salomonsky N.L.. 1994. The transmembrane signaling pathway involved in directed movements of Chlamydomonas flagellar membrane glycoproteins involves the dephosphorylation of a 60-kD phosphoprotein that binds to the major flagellar membrane glycoprotein. J. Cell Biol. 127:803–811. 10.1083/jcb.127.3.803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood, R.A., Tetreault J., and Sloboda R.D.. 2019. The Chlamydomonas flagellar membrane glycoprotein FMG-1B is necessary for expression of force at the flagellar surface. J. Cell Biol. 132:jcs233429. 10.1242/jcs.233429 [DOI] [PubMed] [Google Scholar]
- Bruce, V.G. 1972. Mutants of the biological clock in Chlamydomonas reinhardi. Genetics. 70:537–548. 10.1093/genetics/70.4.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel, V., Silhavy J.L., Bielas S.L., Swistun D., Marsh S.E., Bertrand J.Y., Audollent S., Attie-Bitach T., Holden K.R., Dobyns W.B., et al. 2008. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am. J. Hum. Genet. 83:170–179. 10.1016/j.ajhg.2008.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary, T., Larkins C.E., and Anderson K.V.. 2007. The graded response to sonic hedgehog depends on cilia architecture. Dev. Cell. 12:767–778. 10.1016/j.devcel.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Cevik, S., Hori Y., Kaplan O.I., Kida K., Toivenon T., Foley-Fisher C., Cottell D., Katada T., Kontani K., and Blacque O.E.. 2010. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J. Cell Biol. 188:953–969. 10.1083/jcb.200908133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik, S., Sanders A.A., Van Wijk E., Boldt K., Clarke L., van Reeuwijk J., Hori Y., Horn N., Hetterschijt L., Wdowicz A., et al. 2013. Active transport and diffusion barriers restrict Joubert syndrome-associated ARL13B/ARL-13 to an Inv-like ciliary membrane subdomain. PLoS Genet. 9:e1003977. 10.1371/journal.pgen.1003977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., and Rosenbaum J.L.. 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141:993–1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit, K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., and Reiter J.F.. 2005. Vertebrate smoothened functions at the primary cilium. Nature. 437:1018–1021. 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Desai, P.B., Stuck M.W., Lv B., and Pazour G.J.. 2020. Ubiquitin links smoothened to intraflagellar transport to regulate hedgehog signaling. J. Cell Biol. 219:e201912104. 10.1083/jcb.201912104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, S., Li H., Zhang Y., Yang S., Chen Y., Qiu B., Huang C., Wang J., Li J., Zhu X., and Yan X.. 2021. Rabl2 GTP hydrolysis licenses BBSome-mediated export to fine-tune ciliary signaling. EMBO J. 40:e105499. 10.15252/embj.2020105499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether, T., San Agustin J.T., Keady B.T., Jonassen J.A., Liang Y., Francis R., Tobita K., Johnson C.A., Abdelhamed Z.A., Lo C.W., and Pazour G.J.. 2014. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell. 31:279–290. 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElMaghloob, Y., Sot B., McIlwraith M.J., Garcia E., Yelland T., and Ismail S.. 2021. ARL3 activation requires the co-GEF BART and effector-mediated turnover. Elife. 10:e64624. 10.7554/eLife.64624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel, B.D., Lechtreck K.F., Sakai T., Ikebe M., Witman G.B., and Marshall W.F.. 2009. Total internal reflection fluorescence (TIRF) microscopy of Chlamydomonas flagella. Methods Cell Biol. 93:157–177. 10.1016/S0091-679X(08)93009-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y., Esmail M.A., Ansley S.J., Blacque O.E., Boroevich K., Ross A.J., Moore S.J., Badano J.L., May-Simera H., Compton D.S., et al. 2004. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat Genet. 36:989–993. 10.1038/ng1414 [DOI] [PubMed] [Google Scholar]
- Firestone, A.J., Weinger J.S., Maldonado M., Barlan K., Langston L.D., O'Donnell M., Gelfand V.I., Kapoor T.M., and Chen J.K.. 2012. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 484:125–129. 10.1038/nature10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, S., Kuna D., Caspary T., Kahn R.A., and Sztul E.. 2020. ARF family GTPases with links to cilia. Am. J. Physiol. Cell Physiol. 319:C404–C418. 10.1152/ajpcell.00188.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa, S., Qiu H., Nozaki S., Chiba S., Katoh Y., and Nakayama K.. 2021. ARL3 and ARL13B GTPases participate in distinct steps of INPP5E targeting to the ciliary membrane. Biol. Open. 10:bio058843. 10.1242/bio.058843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt, K., Lokaj M., Koerner C., Falk N., Gießl A., and Wittinghofer A.. 2015. A G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins. Elife. 4:e11859. 10.7554/eLife.11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K., Ma X., Xu T., Li Y., Hodge A., Zhang Q., Torline J., Huang Y., Zhao J., Ling K., and Hu J.. 2018. Axoneme polyglutamylation regulated by Joubert syndrome protein ARL13B controls ciliary targeting of signaling molecules. Nat. Commun. 9:3310. 10.1038/s41467-018-05867-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, Y., Kobayashi T., Kikko Y., Kontani K., and Katada T.. 2008. Domain architecture of the atypical Arf-family GTPase Arl13b involved in cilia formation. Biochem. Biophys. Res. Commun. 373:119–124. 10.1016/j.bbrc.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Hou, Y., Pazour G.J., and Witman G.B.. 2004. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell. 15:4382–4394. 10.1091/mbc.e04-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, Y., Seo S., and Sheffield V.C.. 2021. Photoreceptor cilia, in contrast to primary cilia, grant entry to a partially assembled BBSome. Hum. Mol. Genet. 30:87–102. 10.1093/hmg/ddaa284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K., Diener D.R., Mitchell A., Pazour G.J., Witman G.B., and Rosenbaum J.L.. 2007. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J. Cell Biol. 179:501–514. 10.1083/jcb.200704069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert, M.C., Weihbrecht K., Searby C.C., Li Y., Pope R.M., Sheffield V.C., and Seo S.. 2012. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl. Acad. Sci. USA. 109:19691–19696. 10.1073/pnas.1210916109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunnicutt, G.R., and Snell W.J.. 1991. Rapid and slow mechanisms for loss of cell adhesiveness during fertilization in Chlamydomonas. Dev. Biol. 147:216–224. 10.1016/s0012-1606(05)80019-3 [DOI] [PubMed] [Google Scholar]
- Ivanova, A.A., Caspary T., Seyfried N.T., Duong D.M., West A.B., Liu Z., and Kahn R.A.. 2017. Biochemical characterization of purified mammalian ARL13B protein indicates that it is an atypical GTPase and ARL3 guanine nucleotide exchange factor (GEF). J. Biol. Chem. 292:11091–11108. 10.1074/jbc.M117.784025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., White S.R., Shida T., Schulz S., Aguiar M., Gygi S.P., Bazan J.F., and Nachury M.V.. 2010. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 141:1208–1219. 10.1016/j.cell.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert, M., Eisenring J.J., Robb J.P., and Andermann F.. 1969. Familial agenesis of the cerebellar vermis. A syndrome of episodic hyperpnea, abnormal eye movements, ataxia, and retardation. Neurology. 19:813–825. 10.1212/wnl.19.9.813 [DOI] [PubMed] [Google Scholar]
- Kahn, R.A., Bruford E., Inoue H., Logsdon J.M. Jr., Nie Z., Premont R.T., Randazzo P.A., Satake M., Theibert A.B., Zapp M.L., and Cassel D.. 2008. Consensus nomenclature for the human ArfGAP domain-containing proteins. J. Cell Biol. 182:1039–1044. 10.1083/jcb.200806041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanie, T., Abbott K.L., Mooney N.A., Plowey E.D., Demeter J., and Jackson P.K.. 2017. The CEP19-RABL2 GTPase complex binds IFT-B to initiate intraflagellar transport at the ciliary base. Dev. Cell. 42:22–36.e12. 10.1016/j.devcel.2017.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kato M., and Beachy P.A.. 2009. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. USA. 106:21666–21671. 10.1073/pnas.0912180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.M., and Witman G.B.. 1990. Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J. Biol. Chem. 265:19807–19811. 10.1016/s0021-9258(17)45444-5 [DOI] [PubMed] [Google Scholar]
- Kozminski, K.G., Beech P.L., and Rosenbaum J.L.. 1995. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131:1517–1527. 10.1083/jcb.131.6.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins, C.E., Aviles G.D., East M.P., Kahn R.A., and Caspary T.. 2011. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol. Biol. Cell. 22:4694–4703. 10.1091/mbc.E10-12-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.F. 2013. In vivo imaging of IFT in Chlamydomonas flagella. Methods Enzymol. 524:265–284. 10.1016/B978-0-12-397945-2.00015-9 [DOI] [PubMed] [Google Scholar]
- Lechtreck, K.F. 2016. Methods for studying movement of molecules within cilia. Methods Mol. Biol. 1454:83–96. 10.1007/978-1-4939-3789-9_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.F., Brown J.M., Sampaio J.L., Craft J.M., Shevchenko A., Evans J.E., and Witman G.B.. 2013. Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J. Cell Biol. 201:249–261. 10.1083/jcb.201207139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.F., Johnson E.C., Sakai T., Cochran D., Ballif B.A., Rush J., Pazour G.J., Ikebe M., and Witman G.B.. 2009. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187:1117–1132. 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet, M., and Piperno G.. 1987. Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc. Natl. Acad. Sci. USA. 84:5720–5724. 10.1073/pnas.84.16.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.E., and Gleeson J.G.. 2011. Cilia in the nervous system: Linking cilia function and neurodevelopmental disorders. Curr. Opin. Neurol. 24:98–105. 10.1097/WCO.0b013e3283444d05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Patena W., Fauser F., Jinkerson R.E., Saroussi S., Meyer M.T., Ivanova N., Robertson J.M., Yue R., Zhang R., et al. 2019. A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat. Genet. 51:627–635. 10.1038/s41588-019-0370-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Wei Q., Zhang Y., Ling K., and Hu J.. 2010. The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J. Cell Biol. 189:1039–1051. 10.1083/jcb.200912001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., and Lechtreck K.F.. 2018. The Bardet-Biedl syndrome protein complex is an adapter expanding the cargo range of intraflagellar transport trains for ciliary export. Proc. Natl. Acad. Sci. USA. 115:E934–E943. 10.1073/pnas.1713226115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., Lou X., Wingfield J.L., Lin J., Nicastro D., and Lechtreck K.. 2020. Chlamydomonas PKD2 organizes mastigonemes, hair-like glycoprotein polymers on cilia. J. Cell Biol. 219:e202001122. 10.1083/jcb.202001122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.X., Xue B., Sun W.Y., Wingfield J.L., Sun J., Wu M., Lechtreck K.F., Wu Z., and Fan Z.C.. 2021. Bardet-Biedl Syndrome 3 protein promotes ciliary exit of the signaling protein phospholipase D via the BBSome. Elife. 10:e59119. 10.7554/eLife.59119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loktev, A.V., and Jackson P.K.. 2013. Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep. 5:1316–1329. 10.1016/j.celrep.2013.11.011 [DOI] [PubMed] [Google Scholar]
- Mariani, L.E., Bijlsma M.F., Ivanova A.A., Suciu S.K., Kahn R.A., and Caspary T.. 2016. Arl13b regulates Shh signaling from both inside and outside the cilium. Mol. Biol. Cell. 27:3780–3790. 10.1091/mbc.E16-03-0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miertzschke, M., Koerner C., Spoerner M., and Wittinghofer A.. 2014. Structural insights into the small G-protein Arl13B and implications for Joubert syndrome. Biochem. J. 457:301–311. 10.1042/BJ20131097 [DOI] [PubMed] [Google Scholar]
- Mourao, A., Nager A.R., Nachury M.V., and Lorentzen E.. 2014. Structural basis for membrane targeting of the BBSome by ARL6. Nat. Struct. Mol. Biol. 21:1035–1041. 10.1038/nsmb.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury, M.V. 2018. The molecular machines that traffic signaling receptors into and out of cilia. Curr. Opin. Cell Biol. 51:124–131. 10.1016/j.ceb.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury, M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., and Jackson P.K.. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213. 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Nishijima, Y., Hagiya Y., Kubo T., Takei R., Katoh Y., and Nakayama K.. 2017. RABL2 interacts with the intraflagellar transport-B complex and CEP19 and participates in ciliary assembly. Mol. Biol. Cell. 28:1652–1666. 10.1091/mbc.E17-01-0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino, G., Akizu N., and Gleeson J.G.. 2011. Modeling human disease in humans: The ciliopathies. Cell. 147:70–79. 10.1016/j.cell.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki, S., Katoh Y., Terada M., Michisaka S., Funabashi T., Takahashi S., Kontani K., and Nakayama K.. 2017. Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J. Cell Sci. 130:563–576. 10.1242/jcs.197004 [DOI] [PubMed] [Google Scholar]
- Ocbina, P.J., Tuson M., and Anderson K.V.. 2009. Primary cilia are not required for normal canonical Wnt signaling in the mouse embryo. PLoS One. 4:e6839. 10.1371/journal.pone.0006839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, G., Blacque O.E., Snow J.J., Leroux M.R., and Scholey J.M.. 2005. Functional coordination of intraflagellar transport motors. Nature. 436:583–587. 10.1038/nature03818 [DOI] [PubMed] [Google Scholar]
- Pandey, M., Huang Y., Lim T.K., Lin Q., and He C.Y.. 2020. Flagellar targeting of an arginine kinase requires a conserved lipidated protein intraflagellar transport (LIFT) pathway in Trypanosoma brucei. J. Biol. Chem. 295:11326–11336. 10.1074/jbc.RA120.014287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M.A. 2009. Clinical and molecular features of Joubert syndrome and related disorders. Am. J. Med. Genet. C. Semin. Med. Genet. 151C:326–340. 10.1002/ajmg.c.30229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Agrin N., Leszyk J., and Witman G.B.. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113. 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Sineshchekov O.A., and Witman G.B.. 1995. Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J. Cell Biol. 131:427–440. 10.1083/jcb.131.2.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, J., Wen L., Gao X., Jin C., Xue Y., and Yao X.. 2008. CSS-palm 2.0: An updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21:639–644. 10.1093/protein/gzn039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova, E., Liu Q., Gusella G.L., and Iomini C.. 2018. The Joubert syndrome protein ARL13B binds tubulin to maintain uniform distribution of proteins along the ciliary membrane. J. Cell Sci. 131:jcs212324. 10.1242/jcs.212324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacht, A., Mowbrey K., Elias M., Kahn R.A., and Dacks J.B.. 2013. Ancient complexity, opisthokont plasticity, and discovery of the 11th subfamily of Arf GAP proteins. Traffic. 14:636–649. 10.1111/tra.12063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde, S.R., Nager A.R., and Nachury M.V.. 2020. Ubiquitin chains earmark GPCRs for BBSome-mediated removal from cilia. J. Cell Biol. 219:e202003020. 10.1083/jcb.202003020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.K., Gui M., Koh F., Yip M.C., and Brown A.. 2020. Structure and activation mechanism of the BBSome membrane protein trafficking complex. Elife. 9:e53322. 10.7554/eLife.53322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suciu, S.K., Long A.B., and Caspary T.. 2021. Smoothened and ARL13B are critical in mouse for superior cerebellar peduncle targeting. Genetics. 218:iyab084. 10.1093/genetics/iyab084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z., Amsterdam A., Pazour G.J., Cole D.G., Miller M.S., and Hopkins N.. 2004. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 131:4085–4093. 10.1242/dev.01240 [DOI] [PubMed] [Google Scholar]
- Vargova, R., Wideman J.G., Derelle R., Klimes V., Kahn R.A., Dacks J.B., and Elias M.. 2021. A eukaryote-wide perspective on the diversity and evolution of the ARF GTPase protein family. Genome Biol. Evol. 13:evab157. 10.1093/gbe/evab157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishtha, M., Walther Z., and Hall J.L.. 1996. The kinesin-homologous protein encoded by the Chlamydomonas FLA10 gene is associated with basal bodies and centrioles. J. Cell Science. 109:541–549. 10.1242/jcs.109.3.541 [DOI] [PubMed] [Google Scholar]
- Wakabayashi, K., Misawa Y., Mochiji S., and Kamiya R.. 2011. Reduction-oxidation poise regulates the sign of phototaxis in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 108:11280–11284. 10.1073/pnas.1100592108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton-Pitt, S.R., Silva M., Nguyen K.C., Hall D.H., and Barr M.M.. 2014. The nphp-2 and arl-13 genetic modules interact to regulate ciliogenesis and ciliary microtubule patterning in C. elegans. PLoS Genet. 10:e1004866. 10.1371/journal.pgen.1004866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, A.M., and Beales P.L.. 2011. Ciliopathies: An expanding disease spectrum. Pediatr. Nephrol. 26:1039–1056. 10.1007/s00467-010-1731-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Q., Zhang Y., Li Y., Zhang Q., Ling K., and Hu J.. 2012. The BBSome controls IFT assembly and turnaround in cilia. Nat. Cell Biol. 14:950–957. 10.1038/ncb2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson, C.G., King S.M., Koutoulis A., Pazour G.J., and Witman G.B.. 1995. The 78, 000 M(r) intermediate chain of Chlamydomonas outer arm dynein isa WD-repeat protein required for arm assembly. J. Cell Biol. 129:169–178. 10.1083/jcb.129.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C.L., McIntyre J.C., Norris S.R., Jenkins P.M., Zhang L., Pei Q., Verhey K., and Martens J.R.. 2014. Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat. Commun. 5:5813. 10.1038/ncomms6813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield, J.L., Lechtreck K.F., and Lorentzen E.. 2018. Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem. 62:753–763. 10.1042/EBC20180030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield, J.L., Mengoni I., Bomberger H., Jiang Y.Y., Walsh J.D., Brown J.M., Picariello T., Cochran D.A., Zhu B., Pan J., et al. 2017. IFT trains in different stages of assembly queue at the ciliary base for consecutive release into the cilium. Elife. 6:e26609. 10.7554/eLife.26609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G.B. 1986. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134:280–290. 10.1016/0076-6879(86)34096-5 [DOI] [PubMed] [Google Scholar]
- Witman, G.B. 1993. Chlamydomonas phototaxis. Trends Cell Biol. 3:403–408. 10.1016/0962-8924(93)90091-e [DOI] [PubMed] [Google Scholar]
- Witman, G.B., Carlson K., Berliner J., and Rosenbaum J.L.. 1972. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54:507–539. 10.1083/jcb.54.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, A., de Nechaud B., Chillet D., Mazarguil H., Desbruyeres E., Audebert S., Edde B., Gros F., and Denoulet P.. 1992. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59:425–432 [PubMed] [Google Scholar]
- Xue, B., Liu Y.X., Dong B., Wingfield J.L., Wu M., Sun J., Lechtreck K.F., and Fan Z.C.. 2020. Intraflagellar transport protein RABL5/IFT22 recruits the BBSome to the basal body through the GTPase ARL6/BBS3. Proc. Natl. Acad. Sci. USA. 117:2496–2505. 10.1073/pnas.1901665117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X., and Shen Y.. 2021. Rab-like small GTPases in the regulation of ciliary Bardet-Biedl syndrome (BBS) complex transport. FEBS J. 10.1111/febs.16232 [DOI] [PubMed] [Google Scholar]
- Yang, S., Bahl K., Chou H.T., Woodsmith J., Stelzl U., Walz T., and Nachury M.V.. 2020. Near-atomic structures of the BBSome reveal the basis for BBSome activation and binding to GPCR cargoes. Elife. 9:e55954. 10.7554/eLife.55954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, F., Nager A.R., and Nachury M.V.. 2018. BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J. Cell Biol. 217:1847–1868. 10.1083/jcb.201709041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, K., Liu P., Venkatachalam D., Hopkinson B.M., and Lechtreck K.F.. 2020. The BBSome restricts entry of tagged carbonic anhydrase 6 into the cis-flagellum of Chlamydomonas reinhardtii. PLoS One. 15:e0240887. 10.1371/journal.pone.0240887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora, I., Feldman J.L., and Marshall W.F.. 2004. PCR-based assay for mating type and diploidy in Chlamydomonas. Biotechniques. 37:534–536. 10.2144/04374BM01 [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Li Y., Zhang Y., Torres V.E., Harris P.C., Ling K., and Hu J.. 2016. GTP-binding of ARL-3 is activated by ARL-13 as a GEF and stabilized by UNC-119. Sci. Rep. 6:24534. 10.1038/srep24534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S5.