Abstract
Ewing sarcoma is a fusion oncoprotein–driven primary bone tumor. A subset of patients (∼10%) with Ewing sarcoma are known to harbor germline variants in a growing number of genes involved in DNA damage repair. We recently reported our discovery of a germline mutation in the DNA damage repair protein BARD1 (BRCA1-associated RING domain-1) in a patient with Ewing sarcoma. BARD1 is recruited to the site of DNA double stranded breaks via the PARP protein and plays a critical role in DNA damage response pathways including homologous recombination. We thus questioned the impact of BARD1 loss on Ewing cell sensitivity to DNA damage and the Ewing sarcoma transcriptome. We demonstrate that PSaRC318 cells, a novel patient-derived cell line harboring a pathogenic BARD1 variant, are sensitive to PARP inhibition and by testing the effect of BARD1 depletion in additional Ewing sarcoma cell lines, we confirm that BARD1 loss enhances cell sensitivity to PARP inhibition plus radiation. In addition, RNA-sequencing analysis revealed that loss of BARD1 results in the upregulation of GBP1 (guanylate-binding protein 1), a protein whose expression is associated with variable response to therapy depending on the adult carcinoma subtype examined. Here, we demonstrate that GBP1 contributes to the enhanced sensitivity of BARD1-deficient Ewing cells to DNA damage. Together, our findings demonstrate the impact of loss-of function mutations in DNA damage repair genes, such as BARD1, on Ewing sarcoma treatment response.
Significance:
This work provides preclinical support for the inclusion of pediatric patients with advanced Ewing sarcoma and pathogenic germline variants in BARD1 in future clinical trials testing novel agents inducing DNA damage/targeting DNA damage repair.
Introduction
Ewing sarcoma is a primary bone cancer driven by an aberrant fusion between EWSR1 and a gene encoding an E26 transformation–specific (ETS) transcription factor, most commonly FLI1 (1). Mechanistically, EWS-FLI1 fusions are formed as the result of either reciprocal translocation or chromoplexy events (2). Recent studies have revealed that EWS-FLI1 itself impairs homologous recombination (HR) by sequestering the HR protein BRCA1 (breast cancer gene 1; ref. 3). The disruption of HR by EWS-FLI1 supports the categorization of Ewing sarcoma as a “BRCAness” tumor, phenotypically mimicking loss of BRCA1 expression (4). Clinically, Ewing tumors demonstrate sensitivity to DNA-damaging agents such as doxorubicin (5). Preclinical studies using Ewing sarcoma cell lines and Ewing xenografts have demonstrated tumor sensitivity to compounds that prevent DNA damage repair such as PARP1 inhibitors (PARPi; refs. 6–8).
In addition to the DNA damage repair defects imparted by EWS-FLI1 itself, germline pathogenic variants in DNA damage repair genes, such as APC, BRCA1, FANCC, and RAD51, have been identified in greater than 10% of patients with Ewing sarcoma (9, 10). We recently contributed to this growing body of literature by reporting our discovery of a paternally inherited germline frameshift pathogenic variant in the RING domain of BARD1 (BRCA1-associated RING domain protein 1) in a patient with Ewing sarcoma (11). BARD1, a known tumor suppressor, is an obligatory binding partner of BRCA1. The BRCA1-BARD1 heterodimer functions as an E3 ubiquitin ligase and promotes DNA double-strand break repair by HR (12). The contribution of germline pathogenic variants in DNA damage repair genes, such as BARD1, to the overall sensitivity of Ewing tumors to DNA damage is largely unknown. Patients with pathogenic BRCA1/2 variant hereditary breast and ovarian cancers demonstrate significant response to PARPi/DNA-damaging combinations, a response not seen in patients without these germline variants (13, 14). Thus, we questioned whether the subset of patients with Ewing sarcoma who also harbor germline pathogenic variants in DNA damage repair genes, such as a germline BARD1 mutation, may demonstrate enhanced sensitivity to PARPi/DNA-damaging agent combinations.
Here, we demonstrate that additional hits to tumor DNA damage machinery, such as loss of BARD1 expression, can indeed render Ewing cells more sensitive to DNA-damaging agents, especially when in combination. Using an unbiased RNA-sequencing (RNA-seq) approach, we determined the impact of BARD1 loss on the Ewing cell transcriptome. We found that guanylate-binding protein 1 (GBP1) is significantly upregulated upon BARD1 downregulation and we subsequently demonstrate a role for GBP1 in DNA damage sensitivity in Ewing sarcoma.
Materials and Methods
Reagents
Antibodies were purchased from the following sources: anti-CD99 FITC conjugated (BD Biosciences, catalog no: 555688, concentration 1:20), anti-FLI1 (Abcam, catalog no: 133485, concentration 1:2,000), anti-BARD1 (Bethyl Laboratories, catalog no: A300–263A, 1:2,000), anti-BARD1 (Abcam, catalog no: ab50984, concentration 1:100), anti-GBP1 (Abcam, catalog no: 131255, concentration 1:300 for IHC), anti-GBP1 (Santa Cruz Biotechnology, catalog no: sc-53857, concentration 1:200 for Western blot analysis), anti-phospho (Ser 139)-γH2A.X (Millipore Sigma, catalog no: 05–636, concentration 1:2,500 for immunofluorescence), anti-phospho (Ser 139)-γH2A.X (Invitrogen, catalog no: MA1–2022, concentration 1:1,000 for Western blot analysis), goat anti-mouse IgG AF-488 (Thermo Fisher Scientific, catalog no: A-11001, concentration 1:2,000), tubulin (Cell Signaling Technology, catalog no: 2144S, concentration 1:5,000), vinculin (Cell Signaling Technology, clone E1E9V, catalog no: 13901S, concentration 1:5,000), and anti-rabbit IgG-horseradish peroxidase (HRP) (Promega, catalog no: W401B). Additional specialized reagents include: BMN 673 (talazoparib; Cayman Chemical, catalog no: 19782), MK-4827 tosylate (niraparib; Cayman Chemical, catalog no: 20842), DMSO (MP Biomedicals, catalog no: 196055), doxorubicin (Cayman Chemical, catalog no: 15007), and IFNγ (R&D systems, cat no: 285IF100).
Patient-Derived Relapsed Ewing Sarcoma Cell Line
We have previously described an adolescent patient with relapsed Ewing sarcoma and a paternally inherited, pathogenic germline BARD1 variant (11). This tumor was confirmed as Ewing sarcoma both by EWSR1 FISH and panel sequencing of the tumor. Sequencing revealed a type 3 EWSR1::FLI1 fusion (11). The PSaRC318 patient tumor–derived Ewing sarcoma cell line was generated by our laboratory (studies were approved by the University of Pittsburgh Institutional Review Board, STUDY19030108 and patient written informed consent for sample collection was obtained by the Musculoskeletal Oncology Biobank and Tumor Registry, STUDY20010034; studies were conducted in accordance with U.S. Common Rule ethical guidelines). Briefly, viably preserved tumor biopsy tissue was placed in warm Iscove's modified Dulbecco's medium (IMDM) supplemented with 20% heat-inactivated FBS. The tumor was then physically dissociated using a disposable stainless-steel blade, and individual tumor pieces were embedded in growth factor–reduced Matrigel (Corning). The dissociated tumor was incubated at 37οC and 5% CO2, with media exchanged every 72 hours. Tumor organoids grew over the next 3 weeks and were then harvested. The PSaRC318 Ewing sarcoma cell line was generated by: generating single-cell suspensions, culturing cells on fibronectin-coated (R&D Systems, catalog no: 342000101) plates and serially depleting cultures of fibroblasts. Short tandem repeat (STR) profiling (University of Arizona Genetics Core, Tucson, AZ) was performed on the PSaRC318 cell line to confirm cell authentication prospectively.
Additional Cell Lines and Culture Conditions
A673 and TC71 cells were cultured in RPMI + l-glutamine media supplemented with 10% FBS. CHLA9 and CHLA10 cells were cultured in IMDM + l-glutamine media supplemented with 20% FBS and 1% insulin–transferrin–selenium (ITS, R&D Systems, catalog no: AR013). Early passage stocks of cell lines were obtained in collaboration with the Lawlor laboratory (University of Michigan, Ann Arbor, MI, 2016) and were maintained at 37οC and 5% CO2. All cell lines underwent routine STR profiling for cell authentication (University of Arizona Genetics Core, Tucson, AZ) and regular monitoring for Mycoplasma contamination (MycoAlert PLUS Mycoplasma Detection Kit, Lonza, catalog no:LT07–703).
Radiation
Cells were radiated with X-RAD 320 (Precision X-ray Inc) using Filter 2 (1.5 mm Al + 0.25 mm Cu +0.75 mm Sn) at doses (Gy) noted in individual experiments.
siRNA
BARD1 knockdown in Ewing sarcoma cells was achieved using BARD1 siRNA SMART pool (Dharmacon, catalog no: L-003873–00–0005), which consists of four individual siRNAs targeting BARD1 (catalog nos: J-003873–12–0005, J-003873–11–0005, J-003873–10–0005, and J-003873–09–0005). GBP1 knockdown was achieved by using GBP1 siRNA SMART pool (Dharmacon, catalog no: L-005153–00–0005). ON-TARGET plus nontargeting pool (Dharmacon, catalog no: D-001810–10–2) was used as the control for all siRNA-based experiments. Briefly, 500 μL of optiMEM (Gibco, catalog no: 11058021) was placed per well (6-well plate) along with 3 μL of 20 μmol/L siRNA (reconstituted as per manufacturer's instructions using 5× siRNA buffer, Dharmacon, catalog no: B-002000-UB-100) and 2 μL (A673 and PSaRC318) or 2.5 μL (CHLA10) Liopfectamine RNAiMAX transfection reagent (Life Technologies, catalog no: 13778150). The mixture was incubated with intermittent rocking for 30 minutes prior to the addition of cells.
RT-PCR
RNA isolation was performed using Qiagen RNeasy Plus Mini isolation kit (Qiagen, catalog no: 74134) for >500K cells or Qiagen RNeasy Plus Micro isolation kit (Qiagen, catalog no: 74034) for ≤500K cells. RNA concentration was measured using Nanodrop (Thermo Fisher Scientific). cDNA synthesis was performed on 1 μg of RNA with the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, catalog no: 4374966) and Applied Biosystems Veriti 96-well Thermocycler. qRT-PCR analysis was performed using Taqman probes (Life Technologies, GAPDH Hs02758991_g1, RPLP0 Hs00420895_gH, BARD1 Hs00957655_m1, and GBP1 Hs00977005_m1), Taqman Universal PCR Master Mix (Life Technologies, catalog no: 4304437), and StepOnePlus Real-Time PCR system (Life Technologies).
Immunoblotting
Immunoblot analysis was performed on cell lysates prepared using RIPA or LDS lysis buffer as previously reported (15). After sonication, gel electrophoresis was performed using SDS-PAGE gel electrophoresis. After transfer to nitrocellulose, membranes were blocked with 5% milk in TBST and incubated with primary antibody overnight in 2.5%–5% milk (or BSA for phospho-antibodies)/TBST at 4°C. After washing, membranes were incubated with HRP-conjugated secondary antibody prior to the addition of ECL reagent (Thermo Scientific) and film exposure/processing. ImageJ software (https://imagej.nih.gov) was used to perform densitometry analysis of resulting bands.
Flow Cytometry
Adherent Ewing sarcoma cells were detached from the culture plate using Accutase Cell Detachment Solution (Corning, catalog no: 25–058-Cl). Cells were first stained with Live/Dead Aqua (Life Technologies, #L34957) using a ratio of 1 μL stain per 1 × 106 cells in 1 mL. Cells were then stained for CD99 (FITC, BD Biosciences, catalog no: 555688) using 5 μL of antibody per 1 × 106 cells in 1 mL in a total volume of 100 μL. The percentage of live, FITC-positive cells and the mean fluorescence intensity was determined using a BD FACSAria III or BD FACSAria-II SORP. FlowJo software was used for data analysis and generation of data plots.
Immunofluorescence Staining and Confocal Microscopy
Cells were fixed and immunofluorescently labeled as previously described (16). Slides were mounted using a DAPI-containing mounting solution and cells were imaged using Zeiss LSM 510 and Leica Stellaris 5 confocal microscopes.
IHC
Formalin-fixed, paraffin-embedded (FFPE) Ewing tumor samples underwent deparaffinization, antigen retrieval (buffer pH 8.5–9, catalog no: 950–224), and staining for GBP1 (1:300 dilution, Abcam, catalog no: 131255) via IHC using a RUO DISCOVERY Multimer V2 (v0.00.0083) and Discovery ULTRA Staining Module (Department of Pathology, UPMC ISH Laboratory). Antibody dilution was selected in consultation with a pathologist following stained slide review.
Live-Cell Monitoring and Apoptosis Assays
Cells were seeded into 96-well plates (Corning 3610 or 3596) in 100 μL of Fluorobrite media (Gibco, #A18967–01) containing 5% FBS minimally in triplicate. Because of baseline differences in proliferation between cell types, A673, CHLA10, and PSaRC 318 cells were seeded at a starting cell count of 5,000, 8,000, or 10,000 cells per well, respectively. Cell treatment conditions are described in the individual experiments. For apoptosis assays, IncuCyte Caspase 3/7 green reagent (Essen BioScience, catalog no: 4440) was added to a final dilution of 1:1,000. Phase contrast images of the cells in standard culture conditions were obtained at 3- to 6-hour intervals using an IncuCyte S3 or IncuCyte Zoom (Essen BioScience). Green fluorescence images were additionally captured for apoptosis assays. Experiments were repeated minimally in technical and biologic triplicates.
Statistical Analyses
PRISM software was used to plot individual data points and the SD. Single comparisons were performed through the use of an unpaired, two-tailed Student t test. For the analysis of differences between multiple groups, ANOVA analysis with Tukey multiple comparisons test was utilized.
RNA-Seq
The University of Pittsburgh Health Sciences Sequencing Core at the UPMC Children's Hospital of Pittsburgh measured RNA quantity and quality using Qbit (Thermo Fisher Scientific), and performed mRNA library preparation and RNA-seq using an Illumina platform. Fastq files were prepared for RNA-seq analysis using the programs FastQC version 0.11.5 and Fastp version 0.20.0, and salmon version 0.13.1 was used for quantification of transcript expression. Transcript-level estimates were imported into R, version 4.03, using package tximport, version1.18.0. Differential gene expression analysis was performed with package DESeq2, version 1.30.0. Gene-set enrichment analysis was performed using package fgsea, version 1.16.0, which was also used for figure generation along with package EnhancedVolcano, version 1.8.0.
Data Availability Statement
The RNA-seq data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE 182677. In addition, publicly available data generated by others were used by the authors as follows:
Cancer Cell Line Encyclopedia Analysis
Ewing sarcoma cell lines were queried in the Broad Institute Cancer Cell Line Encyclopedia (CCLE) database (https://portals.broadinstitute.org/ccle). The somatic mutations in each cell line were compared against genes involved in DNA damage repair and classified as pathogenic, likely pathogenic, variant of unknown significance, benign, or likely benign using COSMIC.
PEDS MiONCOseq Data Analysis
PEDS-MiONCOseq is an institutional review board (IRB)-approved, pediatric precision oncology pediatric cohort enrolling since May 2011 as previously described (17). Targeted exome-sequencing data from this cohort of 747 pediatric oncology patients were queried for germline variants in BARD1. Pathogenicity status of variants identified was annotated by searching the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).
St. Jude Cloud PeCan Data Analysis
The St. Jude Cloud PeCan (Pediatric Cancer Knowledgebase; https://www.stjude.cloud, doi:https://doi.org/10.1101/2020.08.24.264614) germline sequencing data from 1,120 patients (18) were queried for germline variants in BARD1 and corresponding pathogenicity status of variants was annotated by searching the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).
Results
The Landscape of Germline BARD1 Variants in Pediatric Oncology
Germline variants in DNA damage repair genes are reported in >10% of patients with Ewing sarcoma (9, 10). Clinically, such germline variants are often first discovered when tumors are sequenced upon relapse, as patients with Ewing sarcoma currently do not routinely undergo germline testing at diagnosis. We have previously reported a patient with Ewing sarcoma who harbors a heterozygous pathogenic germline BARD1 variant (c.176_177AG; p.E59Afs*8) that results in a frameshift that introduces a premature stop codon within the RING domain of BARD1 (ref. 11; Table 1, footnote “a”, index case). The RING domain is the BARD1 interaction site for BRCA1. The premature stop codon eliminates the Ankyrin repeat domain and BRCT (BRCA1 C Terminus) domain, the binding site for PAR (19).
TABLE 1.
The landscape of germline BARD1 variants in a cohort of pediatric oncology patients.
| Cancer type | Location | Effect | Sequence change | Amino acid change | VAF | LOH |
|---|---|---|---|---|---|---|
| Leukemia/Lymphoma | ||||||
| B-cell ALL | chr2:215661812 | Missense | c.188T>C | p.L63S | 44% | NO |
| T-cell ALL | chr2:215645930 | Missense | c.668A>G | p.E223G | 47% | NO |
| B-cell ALL | chr2:215674215 | Missense | c.79G>C | p.E27Q | 44% | NO |
| B-cell ALL | chr2:215674267 | Deletion | p.R5_N9del | 63% | NO | |
| T-cell ALL | chr2:215645322 | Missense | c.1276C>G | p.H426D | 59% | N/A |
| Brain Tumors | ||||||
| Ependymoma | chr2:215645939 | Missense | c.659T>C | p.L220S | 47% | UPD |
| Ependymoma | chr2:215657104 | Missense | c.281A>C | p.D94A | 51% | YES |
| DIPG | chr2:215645939 | Missense | c.659T>C | p.L220S | 45% | N/A |
| Low-grade glioma | chr2:215595215 | Frameshift | c.1921C>T | p.R641fs | 25% | NO |
| Neuroblastoma | ||||||
| Neuroblastoma | chr2:215595169 | Missense | c.1967G>A | p.G656D | 42% | NO |
| Neuroblastoma | chr2:215632365 | Missense | c.1409A>G | p.N470S | 51% | NO |
| Neuroblastoma | chr2:215595181 | Frameshift | c.1935_1954dup | p.E652fs | 31% | NO |
| Neuroblastoma | chr2:215645966 | Stopgain | c.632T>A | p.L211X | 46% | NO |
| Bone sarcomaa | ||||||
| Osteosarcoma | chr2:215645471 | Missense | c.1127C>T | p.S376L | 49% | NO |
| Ewing sarcoma | chr2:215645877 | Missense | c.721T>C | p.S241P | 55% | NO |
| Osteosarcoma | chr2:215645351 | Missense | c.1247T>G | p.L416R | 32% | NO |
| Other | ||||||
| Retinoblastoma | chr2:215645514 | Missense | c.1084T>G | p.C362G | 42% | N/A |
NOTE: Germline sequencing data from PEDS MiONCOseq and St. Jude PeCan datasets was queried for BARD1 variants and the pathogenicity status of the individual variants were determined. Variants highlighted in blue are of unknown significance. Variants highlighted in red and bolded are pathogenic or likely pathogenic.
Abbreviations: ALL, acute lymphoblastic leukemia; LOH, loss of heterozygosity; N/A, data not available; UPD, uniparental disomy; VAF, variant allele frequency.
aIndex case: patient with Ewing sarcoma and a germline pathogenic variant in BARD1: c.176_177AG; p.E59Afs*8
Pathogenic germline BARD1 variants provide a moderate risk for heritable breast cancer and have also been reported in pediatric patients diagnosed with high-risk neuroblastoma (20–22). A study of 4,469 patients with breast cancer in Germany revealed 23 (0.51%) with BARD1 loss-of-function (LOF) variants as compared with 0.1% with LOF variants in controls (22). To better understand the frequency of germline BARD1 mutations across pediatric malignancies, we investigated the landscape of germline BARD1 variants via analysis of two pediatric oncology sequencing databases: PEDS-MiONCOseq (747 patients at the time of analysis) and St. Jude Cloud PeCan (1,120 patients; ref. 18; Table 1). Four pathogenic or likely pathogenic germline BARD1 variants (those which are capable or likely capable of disrupting function/causing disease, denoted in red) were found in patients diagnosed with pediatric cancers including neuroblastoma, glioma, and B-cell acute lymphoblastic leukemia. Our index case of the first report of a patient with Ewing sarcoma and a germline pathogenic variant in BARD1 is listed in the footnote (a) for reference. Fifteen patients with pediatric cancer with variants of uncertain significance (those for which insufficient or conflicting data exists to determine whether they are disease causing, VUS, denoted in blue) were identified, including three patients diagnosed with bone sarcomas. In summary, out of 1,867 patients assessed from two institutions, 15 patients (0.8%) were identified with a BARD1 VUS and 4 patients (0.21%) with a pathogenic/likely pathogenic germline BARD1 variant. Twenty-four benign or likely benign germline BARD1 variants (those unlikely or not the cause of dysfunction/disease) were also noted in this pediatric cancer patient cohort and are included in Supplementary Table S1 for completeness.
PSaRC318: A Patient-Derived Ewing Sarcoma Cell Line Harboring a Germline Frameshift Variant in BARD1
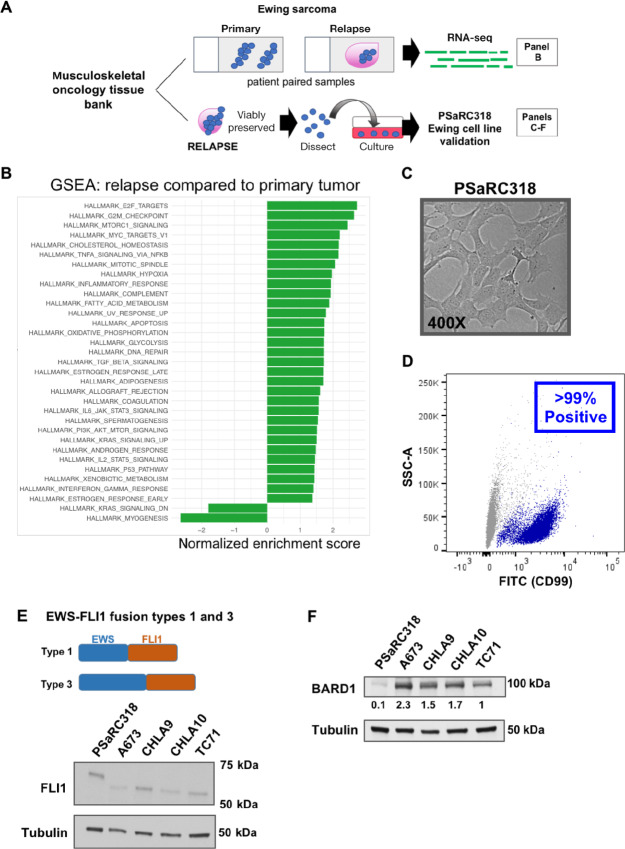
The EWS-FLI1 fusion oncoprotein itself contributes to impaired DNA damage repair, broadly resulting in the inclusion of Ewing sarcoma as one of the “BRCAness” tumors (3, 4). EWS-FLI1 can interact with BARD1 (23). It is unknown whether the presence of a germline pathogenic variant in a DNA damage repair gene such as BARD1 is able to further disrupt DNA damage repair and enhance sensitivity of Ewing tumor cells to DNA-damaging agents. Thus, to address this question, we established and validated a patient-derived Ewing sarcoma cell line (PSaRC318) harboring a germline frameshift pathogenic variant in BARD1 (11). Our workflow of analyzing the relapsed bulk tumor and generating PSaRC318 is summarized in Fig. 1A. RNA-seq analysis was performed on FFPE tumor samples from both the primary and relapsed tumor. Gene-set enrichment analysis (GSEA) was preformed to determine pathways significantly (P < 0.05) up- or downregulated in the relapse as compared with the original primary, pretreatment biopsy specimen (Fig. 1B). In addition to DNA damage–related signatures, we noted a number of hallmark immunoregulatory pathways upregulated upon relapse including, but not limited to, the inflammatory and IFNγ response signatures.
FIGURE 1.
GSEA analysis and validation of a primary cell line from a Ewing tumor with a BARD1 pathogenic variant. A, Schematic overview of tumor samples associated with analyses in B–F. B, Gene-set enrichment analysis (GSEA) of RNA-seq data comparing the lung relapse of the Ewing tumor with a germline BARD1 pathogenic variant to the original primary/pretreatment biopsy. Genesets significantly impacted (P < 0.05) are included. C, Phase contrast image (400×) of the PSaRC318 Ewing tumor cell line. D, Flow cytometry showing presence of surface CD99 expression in the PSaRC318 cell line. E, Schematic detailing the difference between Type 1 and Type 3 EWS-FLI fusions (top) and Western blot analysis with anti-FLI1 antibody of Ewing sarcoma cell lines with type 3 (PSaRC318) versus type 1 (A673, CHLA9, CHLA10, and TC71) EWS-FL1 fusions. F, Western blot demonstrating BARD1 protein expression in the same Ewing sarcoma cell lines as in E. PSaRC318 cells demonstrate significantly (P < 0.05) less BARD1 expression as compared with other Ewing cell lines. Densitometry values below the blot indicate relative expression values. Experiments in D–F were completed minimally in biological triplicate.
Next, viably frozen tumor tissue from a biopsy of the relapsed lung tumor specimen was used to develop a novel Ewing sarcoma cell line, PSaRC318, the morphology of which is shown in Fig. 1C. Flow cytometry confirmed that more than 99% of PSaRC318 cells express surface CD99, a commonly used marker to identify Ewing sarcoma cells (Fig. 1D). Sequencing from the tumor upon relapse revealed a rare Type 3 EWS-FLI1 fusion. Type 3 fusions contain a larger N-terminal portion of EWS (exon 10 breakpoint) as compared with the more common Type 1 EWS-FLI1 fusion (exon 7 breakpoint; see Fig. 1E, schematic). Expression of the Type 3 fusion oncoprotein (with higher molecular weight) was confirmed via anti-FLI1 Western blot analysis of PSaRC318 protein lysate as compared with Ewing cell lines harboring a type 1 EWS-FLI1 fusion (Fig. 1E). In comparison with other Ewing sarcoma cell lines (A673, TC71, CHLA9, and CHLA10), PSaRC318 cells demonstrate significantly less BARD1 expression upon densitometry analysis (P < 0.05; Fig. 1F).
Loss of BARD1 Enhances Ewing Tumor Cell Sensitivity to DNA Damage
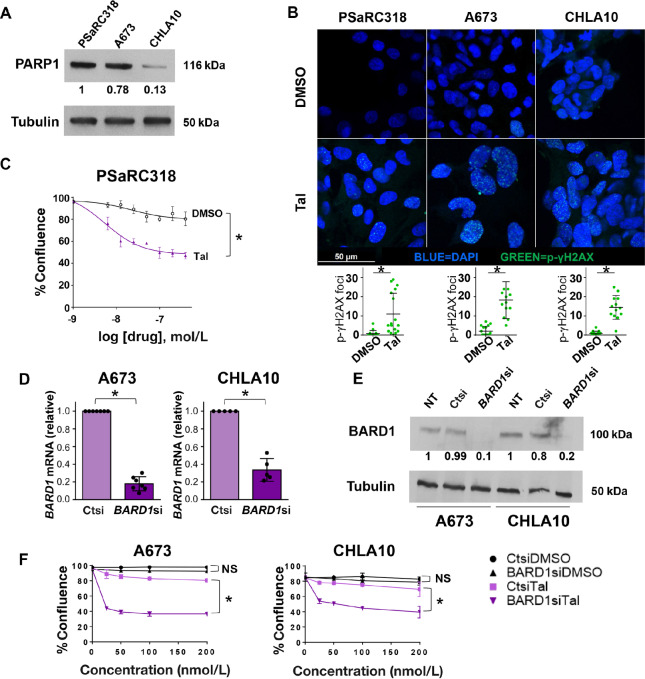
Using the validated PSaRC318 Ewing sarcoma cell line as a tool, we next sought to address the impact of BARD1 loss on Ewing cell sensitivity to DNA damage. Prior in vitro studies have shown that Ewing sarcoma cells demonstrate sensitivity to PARP inhibition (6, 7), although resistance mechanisms do exist (24). PARP inhibitors act by preventing the recruitment of protein complexes key in DNA damage repair to sites of single-strand breaks (25). Given the pathogenic, frameshift variant present in BARD1 in the PSaRC318 cells, we questioned whether PSaRC318 cells would demonstrate sensitivity to PARPi. We confirmed PARP1 expression in PSaRC318, A673, and CHLA10 Ewing sarcoma cells (Fig. 2A). Next, we sought to verify that PARPi treatment–induced DNA damage in our model system. PSaRC318 cells were treated with 100 nmol/L talazoparib (vs. DMSO control) for 24 hours, fixed, and then labeled for phospho-γH2AX (p-γH2AX), a marker of DNA double-strand breaks. Talazoparib treatment did indeed induce p-γH2AX staining (punctate dots)/DNA double-strand breaks in PSaRC318 cells as compared with cells treated with DMSO alone (Fig. 2B). p-γH2AX expression was also determined by Western blot analysis (Supplementary Fig. S1A). To determine the impact of talazoparib treatment on the growth and survival of PSaRC318 cells, live-cell IncuCyte analysis was performed to monitor PSaRC318 cell confluence following treatment with increasing doses of talazoparib versus equivalent volume DMSO (vehicle control). We found that PSaRC318 cell proliferation was significantly (P < 0.01) impaired by talazoparib treatment (Fig. 2C).
FIGURE 2.
Loss of BARD1 enhances Ewing sarcoma cell sensitivity to PARP inhibition. A, Western blot analysis for PARP1 expression in Ewing sarcoma cells. B, p-γH2AX and DAPI immunofluorescence staining of PSaRC318, A673, and CHLA10 cells treated with DMSO or 100 nmol/L talazoparib (Tal). Cells imaged at 630× and p-γH2AX foci were quantified (bottom graphs). C, IncuCyte assay comparing the confluence of PSaRC318 cells treated with DMSO versus 100 nmol/L talazoparib over 1 week. D, qRT-PCR showing BARD1 mRNA expression in A673 or CHLA10 cells treated with control (ctsi) or BARD1 (BARD1si) siRNA. E, Western blot analysis for BARD1 expression in untreated (NT) A673 or CHLA10 cells or cells treated with Ctsi or BARD1si. F, IncuCyte monitoring of cell confluence at increasing concentrations of talazoparib versus DMSO controls in A673 and CHLA10 cells treated with Ctsi versus BARD1si. A673 and CHLA10 cell data is graphed at the 60-hour time point. Normalized expression values from densitometry analyses are included under Western blots. Experiments were completed minimally in biological triplicate. NS, not significant; *, P < 0.01. Error bars, SD.
Next, to more directly determine the impact of BARD1 loss on Ewing cell sensitivity to PARP inhibition, we evaluated the effect of siRNA-mediated knockdown of BARD1 in A673 and CHLA10 Ewing cells. CHLA10 cells, a Ewing sarcoma cell line derived post-chemotherapy/upon disease progression, are known to be relatively less sensitive to PARPi as compared with other Ewing cells. Of note, many pathogenic germline variants in DNA damage repair genes are heterozygous, with some degree of loss of heterozygosity demonstrated within tumors (26, 27). Thus, we specifically chose a pooled BARD1 siRNA-based approach to mimic this scenario by significantly reducing but not eliminating BARD1 expression (see Supplementary Fig. S1B for BARD1 knockdown efficiency using the SMART pool as compared to the four individual siRNAs). Efficient (70%–80%) knockdown of BARD1 was achieved in both A673 and CHLA10 cells as demonstrated by qRT-PCR and Western blot analysis (Fig. 2D and E; P < 0.01). In addition, induction of DNA damage upon talazoparib treatment of A673 and CHLA10 cells was confirmed via p-γH2AX staining (Fig. 2B; Supplementary Fig. S1A for Western blot analysis). Treatment of Ewing cells with talazoparib does not result in loss of BARD1 expression (Supplementary Fig. S1C). Live-cell monitoring of A673 and CHLA10 cells treated with BARD1siRNA revealed enhanced sensitivity to talazoparib as compared with cells treated with control siRNA (Ctsi; Fig. 2F, P < 0.01). Together, these results strongly suggest that loss of BARD1 enhances Ewing cell sensitivity to PARP inhibition.
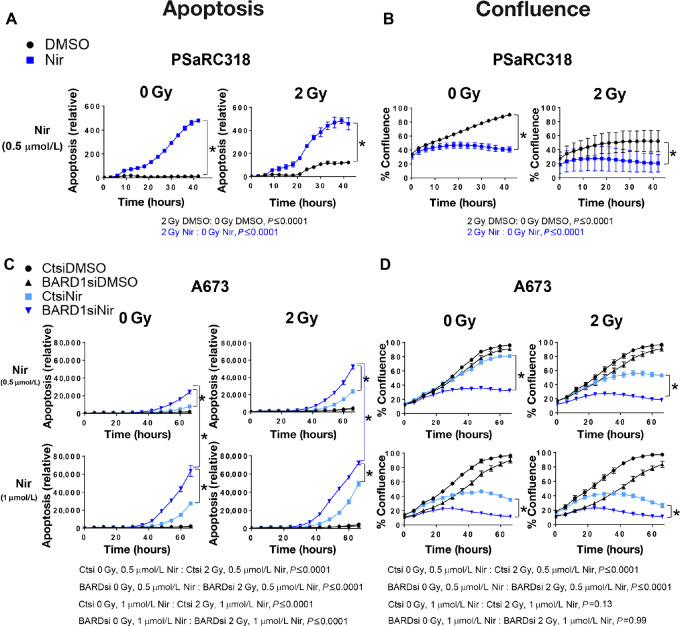
The addition of DNA damage in the setting of PARPi has been shown to enhance apoptosis of Ewing cells (28). We next wanted to determine the impact of reducing BARD1 expression on Ewing cell response to a combination of direct DNA damage (radiation) plus PARPi. We performed this analysis using a second PARPi (niraparib; ref. 29), which is also being tested in clinical trials for patients with Ewing sarcoma (SARC025, NCT02044120). PSaRC318 cells were treated with 0.5 μmol/L niraparib or DMSO control and then treated with or without 2 Gy radiation at 15 hours and monitored via IncuCyte. PSaRC318 cells demonstrate significantly more apoptosis and are less confluence over time (hour 20, 25, 30, etc.) in the setting of niraparib plus radiation as compared with niraparib alone (P < 0.0001; Fig. 3A and B). Of note given the clinical use of radiation in the treatment of Ewing sarcoma, PSaRC318 cells also demonstrate sensitivity to 2 Gy radiation alone (P < 0.0001).
FIGURE 3.
BARD1 loss enhances Ewing sarcoma cell apoptosis in response to niraparib plus radiation. A, Relative apoptosis (caspase 3/7 activity) data from IncuCyte assays showing the effect of 0.5 μmol/L niraparib (Nir) versus DMSO control plus either 0 or 2 Gy radiation on PSaRC318 cells. B, Confluence data from IncuCyte assays showing the effect of 0.5 μmol/L niraparib versus DMSO control plus either 0 or 2 Gy radiation on PSaRC318 cells. C and D, A673 cells were treated with control (Ctsi) or BARD1 (BARD1si) siRNA, niraparib (at doses indicated) versus DMSO control, and either 0 or 2 Gy radiation and monitored via IncuCyte apoptosis assay (C) or confluence assay (D). For these experiments, cells were seeded in the presence of niraparib and radiation was performed at 12–15 hours. Relative apoptosis (caspase 3/7 dye activity) is calculated as green fluorescence in μm2 divided by confluence. Experiments were completed minimally in technical and biological triplicates. *, P < 0.01 as determined by ANOVA analysis with Tukey multiple comparisons test. Error bars, SD.
To more precisely examine the role of BARD1, we compared the response of A673 and CHLA10 cells treated with Ctsi versus BARD1siRNA to niraparib/radiation. A673 cells were treated with 0.5 or 1 μmol/L niraparib and CHLA10 cells were treated with 1 or 1.5 μmol/L niraparib and then monitored over time via IncuCyte analysis. The higher dose of niraparib was used for CHLA10 cells given their relative resistance to PARPi. At approximately 12 hours, cells were treated with 2 Gy radiation. In A673 cells, cells treated with BARDsi demonstrated more apoptosis as compared with Ctsi-treated cells. In addition, significantly more apoptosis was noted over time (hour 40, 50, 60) when comparing BARDsi cells treated with 2 Gy as compared with 0 Gy at both doses of niraparib tested (P ≤ 0.0001, Fig. 3C). BARDsi cells treated with niraparib at either 0.5 or 1 μmol/L doses fail to achieve 50% confluency by 60 hours (Fig. 3D). Multiple statistical comparisons between cell treatment groups are included below the graphs in Fig. 3A–D for reference. Similar results were seen with CHLA10 cells (Supplementary Fig. S2), although as noted, higher doses of niraparib were required to achieve this effect. Together, these data demonstrate that reduction of BARD1 in Ewing sarcoma: (i) enhances sensitivity to both talazoparib and niraparib, (ii) leads to increased apoptosis in the setting of PARPi plus direct DNA damage (radiation).
Enhanced GBP1 Expression is Noted in the BARD1-Deficient Ewing Cell RNA Landscape
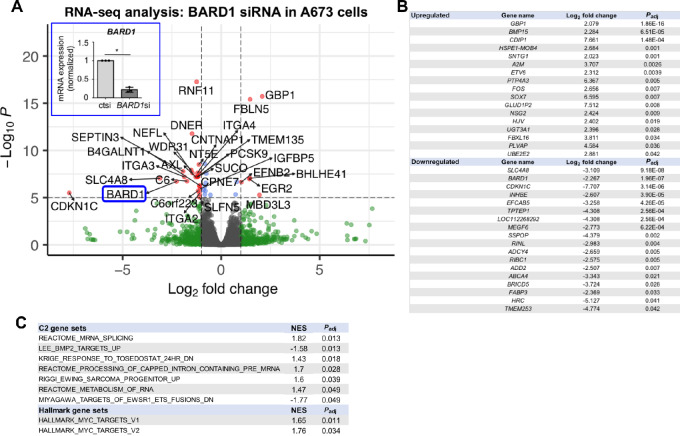
Having determined that Ewing sarcoma cells deficient in BARD1 demonstrate enhanced susceptibility to DNA damage, we next wanted to better understand the changes that occurred in the Ewing sarcoma cell transcriptome upon reducing BARD1 expression. To do this, we utilized an unbiased approach by performing RNA-seq analysis on RNA isolated from A673 Ewing sarcoma cells treated with control or BARD1 siRNA for 72 hours. Knockdown of BARD1 was confirmed via RT-PCR prior to RNA-seq (Fig. 4A, inset, P < 0.05). Analysis of the resultant RNA-seq data comparing BARD1 knockdown to control cells was performed, using cutoffs of 1 for log2 fold change in RNA expression and a P value of <0.05 (5 on the −log10P scale) to generate the volcano plot shown in Fig. 4A. In total, 162 genes were significantly (Padj < 0.05) upregulated and 271 genes were significantly downregulated when comparing BARD siRNA-treated cells to controls (Ctsi). Of these significant (Padj < 0.05) genes, 17 upregulated genes and 18 downregulated genes demonstrated a log2 fold change of 2 or greater (Fig. 4B). Pathway analysis of C2 and Hallmark gene sets was also performed and all significantly (Padj < 0.05) impacted gene sets and the corresponding normalized enrichment scores (NES) are included (Fig. 4C). Interestingly, expression of guanylate-binding protein 1 (GBP1), a protein implicated in the regulation of cellular response to inflammation and chemotherapy responsiveness (30), was the most significantly upregulated gene upon knockdown of BARD1 (Padj = 1.86E-16). GBP1 expression is associated with enhanced motility of lung cancer cells, treatment resistance in ovarian cancer, and better recurrence-free survival in breast cancer. GBP1 functions as a tumor suppressor in colorectal cancer, and in contrast, can protect prostate carcinoma cells from IFNγ-mediated apoptosis (30–34). Given the upregulation of GPB1 in the BARD si-treated cells upon RNA-seq analysis, we sought to determine whether GBP1 is expressed in PSaRC318 cells.
FIGURE 4.
Impact of BARD1 loss on the Ewing sarcoma cell transcriptome. A, Volcano plot of genes up-/downregulated upon loss of BARD1 as compared with A673 Ewing sarcoma cells transfected with Ctsi in three biological replicates. Horizontal dashed lines denote P = 0.05 (5 on −log10 scale). Vertical dashed lines denote a value of 1 on the log2 scale. BARD1 is circled in blue and highlighted as to verify that it is significantly downregulated upon RNA-seq analysis. In addition, the inset image demonstrates RT-PCR analysis of BARD1 expression as a second means by which to validate reduction of BARD1 expression in these RNA samples (×3 biological replicates) as compared with Ctsi-treated cells, *, P < 0.05; error bars, SD. B, List of most significantly (Padj) upregulated and downregulated genes with a log2 fold change of 2 or greater when comparing cells treated with BARD1 siRNA versus Ctsi. C, Pathway analysis (C2 and Hallmark genesets) of BARD1 siRNA-treated cells as compared with Ctsi-treated cells. NES, normalized enrichment score; Padj, adjusted P value.
GBP1 is Expressed and Contributes to DNA Damage Response in PSaRC318 Cells
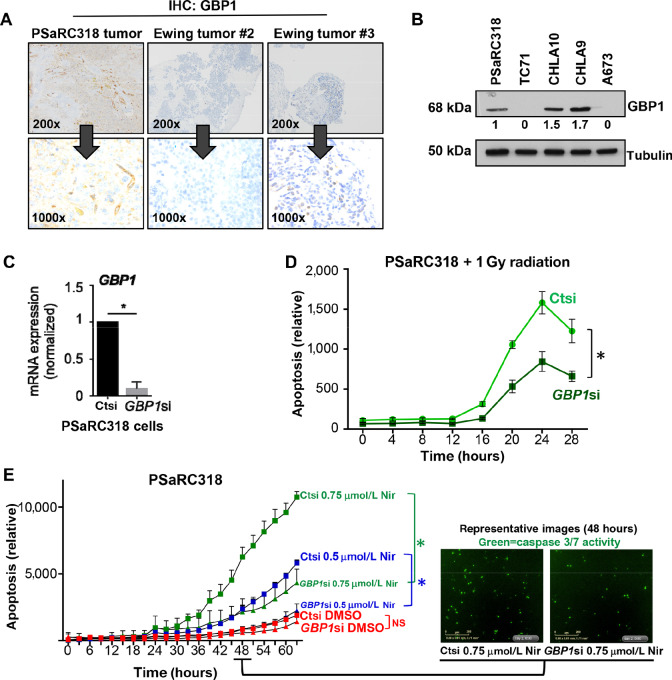
Using IHC to stain for GBP1 expression, a FFPE tissue sample from the original PSaRC318 tumor did indeed demonstrate diffuse cytoplasmic staining for GBP1 (Fig. 5A, left). Additional Ewing tumors were also stained (Fig. 5A, Ewing tumor #2 and #3) for GBP1 and demonstrate no expression (middle) or intercellular heterogeneity in expression (right). To determine whether GBP1 protein is expressed in Ewing sarcoma cell lines, PSaRC318, A673, TC71, CHLA 9, and CHLA10 cell lysates were subjected to Western blot analysis for GBP1. PSaRC318 cells also express GBP1 protein. A673 and TC71 do not express GBP1. CHLA9 and CHLA10 (cell lines from pretreatment and postprogression samples from the same patient) demonstrate GBP1 expression (Fig. 5B). The GBP1 promoter harbors three p53 response elements, an IFN-stimulated response element (ISRE), IFNγ activation site, and a c-Rel site. IFN exposure and NFκB pathway signaling can upregulate GBP1 expression and factors such as VEGF and TGFβ have been reported to downregulate GBP1 expression (30). In cells with no baseline GBP1 protein expression, we demonstrate that expression can be induced upon exposure to IFNγ in both Ctsi- and BARD1 si-treated cells (Supplementary Fig. S3). We conclude that loss of BARD1 expression is only one of many possible factors contributing to GBP1 expression in Ewing sarcoma.
FIGURE 5.
Guanylate-binding protein 1 (GBP1) contributes to Ewing cell sensitivity to DNA damage noted upon loss of BARD1. A, IHC analysis of GPB1 expression in the PSaRC318 patient tumor and in two additional independent Ewing tumors. Images provided at both low (200×) and high (1,000×) power. B, Western blot for GBP1 in Ewing sarcoma cell lines. Numbers under the blot indicate normalized expression as determined by densitometry analysis. C, Normalized GPB1 mRNA expression of PSaRC318 cells treated with control (ct) or GBP1 siRNA (si). *, P < 0.0001. D, PSaRC318 cells were transfected with control or GBP1 siRNA for 72 hours. Cells were then seeded into 96-well plates in quadruplicate, allowed to adhere and then radiated (dose = 1 Gy). Live-cell IncuCyte monitoring of cell confluence and apoptosis (caspase 3/7 activity) was monitored. The graph displays the relative apoptosis (apoptosis/confluence) in control versus GBP1 siRNA-treated cells over time. *, P < 0.05. E, PSaRC318 cells treated with siRNA and seeded as in D and then treated with DMSO, 0.5 μmol/L niraparib (Nir), or 0.75 μmol/L niraparib. The graph displays the relative apoptosis (apoptosis/confluence) over time. Representative IncuCyte images at 48 hours are included (right). NS, not significant; *, P < 0.05. Experiments completed minimally in biological triplicate. Error bars, SD.
Given the varied effects of GBP1 in cancer as detailed earlier, we next sought to determine the functional role(s) of GBP1 expression in PSaRC318 cells. To manipulate GBP1 expression, PSaRC318 cells were transfected with Ctsi (control) or GBP1 siRNA. Figure 5C demonstrates >75% knockdown of GBP1 mRNA expression (P < 0.05). Ctsi- and GBP1 si-treated PSaRC318 cells were then subjected to radiation and monitored for apoptotic activity in real time (IncuCyte). As shown in Fig. 5D, loss of GBP1 expression results in a significant reduction in relative radiation-mediated apoptosis (apoptosis/cell confluence) of PSaRC318 cells (P < 0.05). Nonnormalized confluence data comparing Ctsi and GBP1si cells is included in Supplementary Fig. S4. In addition to reduced relative apoptosis in GBP1 si-treated cells upon radiation treatment, we also noted this effect in GBP1si cells treated with niraparib (Fig. 5E, P < 0.05). GBP1 expression in some adult carcinomas alters cell motility. Using scratch/wound healing assays, we noted no difference in cell motility between Ctsi- and GBP1 si-treated cells (Supplementary Fig. S5). In summary, we find that GBP1 expression is an important factor in the apoptotic response of PSaRC318 cells upon induction of direct DNA damage (radiation) or PARPi.
Discussion
Relapsed and metastatic Ewing sarcoma remains a deadly disease, and to date, personalized medicine approaches have remained elusive, in part due to the rarity of this cancer. While any single germline DNA damage repair gene variant in patients with Ewing sarcoma is rare, as highlighted by our analysis of BARD1 germline variants in the PEDS MiONCOseq and St. Jude PeCan datasets, in aggregate, patients with Ewing sarcoma and pathogenic germline variants in DNA damage repair genes constitute approximately 10%–13% of the Ewing sarcoma patient population (9, 10). The prognostic impact of these pathogenic germline variants on patient outcomes is currently unknown. Therapeutically, it is logical to determine whether Ewing tumors harboring pathogenic germline DNA damage repair gene variants demonstrate enhanced sensitivity to agents which induce DNA damage. The Pediatric MATCH Screening Trial experimental subprotocol H (NCT03155620) enrolls patients with pathogenic ATM, BRCA1, BRCA2, RAD51C, and RAD51D variants to receive the PARP inhibitor olaparib. In this work, we have generated and utilized the novel patient-derived BARD1-mutated Ewing sarcoma cell line PSaRC318 as a model to ask the question: can pathogenic variants in the DNA damage repair gene BARD1 enhance sensitivity to PARPi/DNA damage beyond the vulnerability already imparted by the presence of EWS-FLI1? Indeed, here we show that disrupting BARD1 expression enhances Ewing cell apoptosis in response to PARPi alone and in combination with radiation. On the basis of our current work, we propose the inclusion of patients harboring pathogenic BARD1 variants in similar future clinical trials aimed at testing DNA-damaging agents/combinations in patients with any one of many pathogenic germline variants in DNA damage repair genes such as ATM, BRCA1, RAD51C, etc. and Ewing sarcoma. Better understanding germline DNA damage defects in patients with Ewing sarcoma and discovering novel approaches to treat Ewing tumors with additional impairments in DNA damage repair is an ongoing priority of our work.
Knowing the presence of additional DNA damage repair gene variants is also important when selecting Ewing cell lines to test new agents/effects. Using Cancer Cell Line Encyclopedia (CCLE, Broad Institute) sequencing data, we determined the presence of somatic variants in DNA damage repair genes in the 14 Ewing sarcoma cell lines used in the original analyses of PARP inhibition in Ewing sarcoma cells (6, 7). Ewing sarcoma cell lines such as SKPNDW and ES7 harbor pathogenic somatic variants in FANCM (Fanconi anemia, complementation group M). In addition, the Ewing cell line CADOES1 harbors a pathogenic somatic SLX4 variant (Supplementary Table S2). Both of these genes are involved in DNA damage repair (35, 36) and cells with such additional dysfunctional DNA damage repair may behave differently than Ewing sarcoma cells without such defects.
Through our RNA-seq studies of BARD1 loss, we uncovered a link to GBP1 expression. As noted in the results, GBP1 expression can be regulated by a host of factors and some Ewing cell lines, such as CHLA10, express GBP1 in the absence of BARD1 deficits. We were able to demonstrate that GBP1 expression contributes to the apoptotic response to DNA damage in Ewing sarcoma. On the basis of these findings, future studies to determine the association of Ewing tumor GBP1 expression/expression patterns with patient outcome are warranted.
In addition to GBP1 upregulation, we noted additional genes in our RNA-seq analysis of BARD1 knockdown cells that could also contribute to DNA damage/chemotherapy response, such as CDKN1C. We noted a significant downregulation of cyclin dependent kinase inhibitor 1C (CDKN1C, also known as P57Kip2) upon knockdown of BARD1. CDKN1C expression is strongly induced by EWS-FLI1 (37, 38) and is reported to play a role in chemoresistance. One report has demonstrated that BARD1 can physically interact with EWS-FLI1 (23), and thus it is interesting to speculate how the loss of BARD1, or loss of other BARD1-binding partners such as BRCA1, may impact the transcriptional activity of EWS-FLI1. Our findings could indicate that loss of BARD1 leads to a reduction of EWS-FLI1–induced CDKN1C expression.
Finally, given the significant cross-talk between DNA damage and immunoregulatory pathways, novel DNA-damaging agent/immunotherapy combinations are also worthy of preclinical testing specifically in this unique subset of Ewing tumors. GSEA analysis of our RNA-seq data from the PSaRC318 tumor at relapse demonstrated upregulation of multiple immunoregulatory pathways including TNFα/NFκB, inflammatory response, TGFβ, IL6, IL2, and IFNγ (Fig. 1). Ongoing studies in our laboratory are assessing how concomitantly targeting DNA damage repair and immunoregulatory pathways may be an effective alternative approach for treating Ewing tumors harboring additional germline DNA damage repair defects.
Supplementary Material
Benign germline BARD1 variants.
Additional protein/mRNA expression data (corresponding to Figure 2).
Prevalence of somatic DNA damage repair gene variants in Ewing cell lines included in original studies of PARP inhibition.
BARD1 loss enhances Ewing sarcoma cell apoptosis in response to niraparib plus radiation in CHLA10 Ewing sarcoma cells.
Interferon-gamma-induced GBP1 expression in A673 Ewing sarcoma cells.
PSaRC318 cell confluency data.
Loss of GBP1 expression does not impact PSaRC318 cell migration.
Acknowledgments
The authors would like to thank The Alex's Lemonade Stand Foundation Childhood Cancer Data Laboratory's R-learning workshop, including Dr. Huajin Wang. The authors would like to thank Drs. Steffi Oesterreich and Richard Gorlick for their feedback on this work.
This work was supported by Alex's Lemonade Stand Foundation Million Mile Award (to K.M. Bailey), Hyundai Hope on Wheels Young Investigator Award (to K.M. Bailey), and the NIH: NCI K08CA252178 (to K.M. Bailey) and K12HD052892–13 (to L.M. Maurer, K.M. Bailey, PI: Terence M. Dermody, MD). The Bailey laboratory would like to thank the Morden Foundation for their continued support. The University of Pittsburgh holds a Physician-Scientist Institutional Award from the Burroughs Welcome Fund (to J.D. Daley). Research reported here has been supported by the NIH under their award number T32HD071834 “Research Training Program for Pediatric Subspecialty Fellows” (PI: Terence S. Dermody, MD).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
M.F. Jacobs reports grants from NIH during the conduct of the study. K.A. Janeway reports personal fees from Bayer, Ipsen, Foundation Medicine, and personal fees from Takeda outside the submitted work. P.C. Lucas reports other from Amgen; grants from NSABP Foundation, and grants from NIH outside the submitted work. L. McAllister-Lucas reports other from Amgen and other from Schrodinger Incorporated outside the submitted work. No other disclosures were reported.
Authors’ Contributions
L.M. Maurer: Conceptualization, data curation, formal analysis, writing-original draft, writing-review and editing. J.D. Daley: Conceptualization, data curation, formal analysis, investigation, methodology, writing-review and editing. E. Mukherjee: Data curation, formal analysis, supervision, validation, investigation, methodology, project administration, writing-review and editing. R.E. Venier: Conceptualization, formal analysis, investigation, writing-review and editing. C.M. Julian: Formal analysis, investigation, writing-review and editing. N.G. Bailey: Data curation, software, formal analysis, visualization, methodology, writing-review and editing. M.F. Jacobs: Data curation, formal analysis, investigation, writing-review and editing. C. Kumar-Sinha: Data curation, formal analysis, writing-review and editing. H. Raphael: Investigation, writing-review and editing. N. Periyapatna: Investigation, writing-review and editing. K. Weiss: Resources, writing-review and editing. K.A. Janeway: Investigation, writing-review and editing. R. Mody: Resources, investigation, writing-review and editing. P.C. Lucas: Resources, writing-review and editing. L.M. McAllister-Lucas: Resources, formal analysis, visualization, writing-review and editing. K.M. Bailey: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing-original draft, writing-review and editing.
References
- 1. Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, De Álava E, Kovar H, et al. Ewing sarcoma. Nat Rev Dis Primers 2018;4:5. [DOI] [PubMed] [Google Scholar]
- 2. Anderson ND, de Borja R, Young MD, Fuligni F, Rosic A, Roberts ND, et al. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science 2018;361:eaam8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gorthi A, Romero JC, Loranc E, Cao L, Lawrence LA, Goodale E, et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 2018;555:387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gorthi A, Bishop AJR. Ewing sarcoma fusion oncogene: at the crossroads of transcription and DNA damage response. Mol Cell Oncol 2018;5:e1465014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grier HE, Krailo MD, Tarbell NJ, Link MP, Fryer CJH, Pritchard DJ, et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing's sarcoma and primitive neuroectodermal tumor of bone. N Engl J Med 2003;348:694–701. [DOI] [PubMed] [Google Scholar]
- 6. Brenner JC, Feng FY, Han S, Patel S, Goyal SV, Bou-Maroun LM, et al. PARP-1 inhibition as a targeted strategy to treat Ewing's sarcoma. Cancer Res 2012;72:1608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012;483:570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vormoor B, Curtin NJ. Poly(ADP-ribose) polymerase inhibitors in Ewing sarcoma. Curr Opin Oncol 2014;26:428–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brohl AS, Patidar R, Turner CE, Wen X, Song YK, Wei JS, et al. Frequent inactivating germline mutations in DNA repair genes in patients with Ewing sarcoma. Genet Med 2017;19:955–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parsons DW, Roy A, Yang Y, Wang T, Scollon S, Bergstrom K, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol 2016;2:616–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Venier RE, Maurer LM, Kessler EM, Ranganathan S, Mcgough RL, Weiss KR, et al. A germline BARD1 mutation in a patient with Ewing Sarcoma: implications for familial testing and counseling. Pediatr Blood Cancer 2019;66:e27824. [DOI] [PubMed] [Google Scholar]
- 12. Irminger-Finger I, Jefford CE. Is there more to BARD1 than BRCA1? Nat Rev Cancer 2006;6:382–91. [DOI] [PubMed] [Google Scholar]
- 13. Telli ML, Ford JM. PARP inhibitors in breast cancer. Clin Adv Hematol Oncol 2010;8:629–35. [PubMed] [Google Scholar]
- 14. Chan SL, Mok T. PARP inhibition in BRCA-mutated breast and ovarian cancers. Lancet 2010;376:211–3. [DOI] [PubMed] [Google Scholar]
- 15. Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen Q-R, Yeung C, et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J Natl Cancer Inst 2011;103:962–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bailey KM, Airik M, Krook MA, Pedersen EA, Lawlor ER. Micro-environmental stress induces Src-dependent activation of invadopodia and cell migration in Ewing sarcoma. Neoplasia 2016;18:480–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mody RJ, Wu Y-M, Lonigro RJ, Cao X, Roychowdhury S, Vats P, et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA 2015;314:913–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang J, Walsh MF, Wu G, Edmonson MN, Gruber TA, Easton J, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 2015;373:2336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tarsounas M, Sung P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat Rev Mol Cell Biol 2020;21:284–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet 2013;45:279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Watters AK, Seltzer ES, MacKenzie D, Young M, Muratori J, Hussein R, et al. The effects of genetic and epigenetic alterations of BARD1 on the development of non-breast and non-gynecological cancers. Genes (Basel) 2020;11:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weber-Lassalle N, Borde J, Weber-Lassalle K, Horváth J, Niederacher D, Arnold N, et al. Germline loss-of-function variants in the BARD1 gene are associated with early-onset familial breast cancer but not ovarian cancer. Breast Cancer Res 2019;21:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spahn L, Petermann R, Siligan C, Schmid JA, Aryee DN, Kovar H. Interaction of the EWS NH2 terminus with BARD1 links the Ewing's sarcoma gene to a common tumor suppressor pathway. Cancer Res 2002;62:4583–7. [PubMed] [Google Scholar]
- 24. Kim D-S, Camacho CV, Kraus WL. Alternate therapeutic pathways for PARP inhibitors and potential mechanisms of resistance. Exp Mol Med 2021;53:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rose M, Burgess JT, O'byrne K, Richard DJ, Bolderson E. PARP inhibitors: clinical relevance, mechanisms of action and tumor resistance. Front Cell Dev Biol 2020;8:564601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep 2018;23:239–54.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Riaz N, Blecua P, Lim RS, Shen R, Higginson DS, Weinhold N, et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun 2017;8:857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee H-J, Yoon C, Schmidt B, Park DJ, Zhang AY, Erkizan HV, et al. Combining PARP-1 inhibition and radiation in Ewing sarcoma results in lethal DNA damage. Mol Cancer Ther 2013;12:2591–600. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Antolin AA, Ameratunga M, Banerji U, Clarke PA, Workman P, Al-Lazikani B. The kinase polypharmacology landscape of clinical PARP inhibitors. Sci Rep 2020;10:2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Honkala AT, Tailor D, Malhotra SV. Guanylate-binding protein 1: an emerging target in inflammation and cancer. Front Immunol 2019;10:3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tipton AR, Nyabuto GO, Trendel JA, Mazur TM, Wilson JP, Wadi S, et al. Guanylate-Binding Protein-1 protects ovarian cancer cell lines but not breast cancer cell lines from killing by paclitaxel. Biochem Biophys Res Commun 2016;478:1617–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamakita I, Mimae T, Tsutani Y, Miyata Y, Ito A, Okada M. Guanylate binding protein 1 (GBP-1) promotes cell motility and invasiveness of lung adenocarcinoma. Biochem Biophys Res Commun 2019;518:266–72. [DOI] [PubMed] [Google Scholar]
- 33. Britzen-Laurent N, Lipnik K, Ocker M, Naschberger E, Schellerer VS, Croner RS, et al. GBP-1 acts as a tumor suppressor in colorectal cancer cells. Carcinogenesis 2013;34:153–62. [DOI] [PubMed] [Google Scholar]
- 34. Ascierto ML, Kmieciak M, Idowu MO, Manjili R, Zhao Y, Grimes M, et al. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res Treat 2012;131:871–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 2009;138:78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xue X, Sung P, Zhao X. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev 2015;29:1777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dauphinot L, De Oliveira C, Melot T, Sevenet N, Thomas V, Weissman BE, et al. Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2and c-Myc expression. Oncogene 2001;20:3258–65. [DOI] [PubMed] [Google Scholar]
- 38. Lessnick SL, Dacwag CS, Golub TR. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 2002;1:393–401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Benign germline BARD1 variants.
Additional protein/mRNA expression data (corresponding to Figure 2).
Prevalence of somatic DNA damage repair gene variants in Ewing cell lines included in original studies of PARP inhibition.
BARD1 loss enhances Ewing sarcoma cell apoptosis in response to niraparib plus radiation in CHLA10 Ewing sarcoma cells.
Interferon-gamma-induced GBP1 expression in A673 Ewing sarcoma cells.
PSaRC318 cell confluency data.
Loss of GBP1 expression does not impact PSaRC318 cell migration.
Data Availability Statement
The RNA-seq data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE 182677. In addition, publicly available data generated by others were used by the authors as follows:
Cancer Cell Line Encyclopedia Analysis
Ewing sarcoma cell lines were queried in the Broad Institute Cancer Cell Line Encyclopedia (CCLE) database (https://portals.broadinstitute.org/ccle). The somatic mutations in each cell line were compared against genes involved in DNA damage repair and classified as pathogenic, likely pathogenic, variant of unknown significance, benign, or likely benign using COSMIC.
PEDS MiONCOseq Data Analysis
PEDS-MiONCOseq is an institutional review board (IRB)-approved, pediatric precision oncology pediatric cohort enrolling since May 2011 as previously described (17). Targeted exome-sequencing data from this cohort of 747 pediatric oncology patients were queried for germline variants in BARD1. Pathogenicity status of variants identified was annotated by searching the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).
St. Jude Cloud PeCan Data Analysis
The St. Jude Cloud PeCan (Pediatric Cancer Knowledgebase; https://www.stjude.cloud, doi:https://doi.org/10.1101/2020.08.24.264614) germline sequencing data from 1,120 patients (18) were queried for germline variants in BARD1 and corresponding pathogenicity status of variants was annotated by searching the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).