Abstract
G protein-coupled receptors (GPCRs) are regulated by various downstream proteins, of which the melanocortin receptor accessory protein 1 (MRAP1) is closely involved in the regulation of melanocortin receptor 2 (MC2R). Assisted by MRAP1, MC2R responds to adrenocorticotropic hormone (ACTH) and stimulates glucocorticoid biogenesis and cortisol secretion. MC2R activation plays an essential role in the hypothalamic-pituitary-adrenal (HPA) axis that regulates stress response, while its dysfunction causes glucocorticoid insufficiency- or cortisol excess-associated disorders. Here, we present a cryo-electron microscopy (cryo-EM) structure of the ACTH-bound MC2R–Gs–MRAP1 complex. Our structure, together with mutagenesis analysis, reveals a unique sharp kink at the extracellular region of MRAP1 and the ‘seat-belt’ effect of MRAP1 on stabilizing ACTH binding and MC2R activation. Mechanisms of ACTH recognition by MC2R and receptor activation are also demonstrated. These findings deepen our understanding of GPCR regulation by accessory proteins and provide valuable insights into the ab initio design of therapeutic agents targeting MC2R.
Subject terms: Cryoelectron microscopy, Cell signalling
Introduction
G protein-coupled receptors (GPCRs) are regulated by various downstream proteins, such as G proteins, kinases, β-arrestins, and accessory proteins, adding to the complexity and fine-tuning of GPCR functional expression and signal transduction.1 The melanocortin receptor accessory protein (MRAP) family contains two members of single transmembrane proteins, MRAP1 and MRAP2, which regulate ACTH receptor formation and energy homeostasis, respectively.2,3 MRAP1 was initially discovered to facilitate the trafficking of melanocortin receptor 2 (MC2R) to the plasma membrane after translation and is required for MC2R activity.4–6 The N-terminus of MRAP1 was further proven to be a prerequisite for the activation of MC2R.4,7,8 MRAP1 was reported to exist as an anti-parallel homodimer with the two copies of MRAP1 inserted into the membrane in ‘N-terminus out’ and ‘C-terminus out’ orientations, respectively, which appears to be the functional conformation of MRAP1 to bind MC2R.9–11 Although the conformation and importance of MRAP1 in regulating energy homeostasis are beginning to be understood, the mechanism by which MRAP1 modulates MC2R activity remains elusive.
MC2R belongs to the melanocortin receptor (MCR) family, which consists of five closely related class A GPCRs, MC1R–MC5R. It was identified in the human adrenal gland and stood out as one of the smallest GPCRs, comprised of only 297 amino acids.12 Unlike its homologs with comparable activity to melanocortin hormones, such as melanocyte-stimulating hormones (MSHs) and ACTH, MC2R exclusively responds to ACTH.13,14 Besides an evolutionally conserved ‘message’ motif (HFRW) shared by all known melanocortin hormones, only ACTH has a unique ‘address’ motif (KKRR), which may explain its selectivity for MC2R (Fig. 1a).15,16 A two-step activation model has been proposed in which both ‘message’ and ‘address’ motifs of ACTH are required for ligand recognition by MC2R.13,17,18 However, the selectivity mechanism of MC2R towards ACTH remains to be clarified.
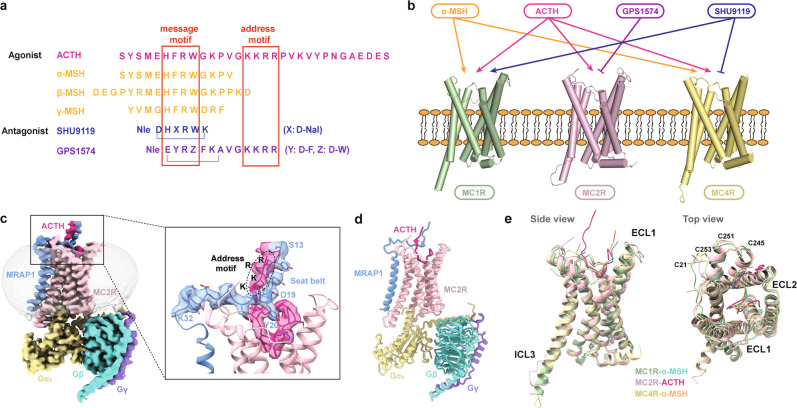
Fig. 1. Ligand selectivity and cryo-EM structure of the ACTH–MC2R–Gs–MRAP1 complex.
a Sequence comparison among peptidic agonists and antagonists of MCRs with the address and message motifs highlighted. b Differential activities of α-MSH, ACTH, GPS1574, and SHU9119 on MC1R, MC2R, and MC4R. c, d Cryo-EM density map with an enlarged presentation of the extracellular region (c) and cartoon representation (d). Polar interactions are indicated by red dashed lines. MC2R, pink; ACTH, deep pink; MRAP1, blue; Gα, yellow; Gβ, turquoise; Gγ, purple. e Structural features of MC2R shown by alignment to MC1R (PDB code: 7F4D) and MC4R (PDB code: 7F53). Left, side view; right, top view.
The polypeptide ACTH, along with MC2R and MRAP1, are critical modulators in the hypothalamic-pituitary-adrenal (HPA) axis and are closely associated with various pathological processes.19 By dictating glucocorticoid production and cortisol release, MC2R signaling regulates multiple physiological functions, including metabolism and immune responses under circadian- and stress-induced conditions, etc.20 Pathologically, inactivating mutations on MC2R or MRAP1 led to familial glucocorticoid deficiency (FGD),21,22 a rare and fatal disease characteristic of isolated cortisol deficiency, with oral hydrocortisone being the only effective treatment.23,24 Conversely, excessive ACTH stimulation causes congenital adrenal hyperplasia (CAH) and Cushing syndrome (CS).25 Blocking ACTH-induced MC2R activation is a potential treatment modality for these disorders.26 Till now, no effective synthetic agonist or antagonist of MC2R has been discovered except the peptide antagonist GPS1574, which may serve as a potential treatment for CS27,28 (Fig. 1a, b). Revealing ligand recognition and activation mechanism of MC2R will facilitate the development of drug candidates for ACTH-MC2R/MRAP1 pathway-relevant diseases.
Recent progress made in the determination of MC1R and MC4R structures bound to agonist or antagonist disclosed their familial conservation and specificity of ligand recognition and functionality.11,29–32 However, the lack of structural information on MC2R hampers our understanding of its activity regulation by MRAP1 and recognition selectivity for ACTH. Here we report the cryo-electron microscopy (cryo-EM) structure of the ACTH-bound MC2R–Gs–MRAP1 complex at a global resolution of 3.3 Å. This structure presents a ‘seat-belt’ model of MRAP1 in MC2R activation and selective features of ACTH recognition by the receptor. Our findings extend the understanding of MC2R signaling modulation and open a new window in drug discovery targeting MC2R.
Results
Structure determination of the ACTH–MC2R–Gs–MRAP1 complex
To facilitate the assembly of the ACTH–MC2R–Gs–MRAP1 complex, we co-expressed MRAP1 and the full-length human MC2R, along with miniGαs, Gβ, and Gγ, in the presence of ACTH in High Five insect cells (Supplementary information, Fig. S1a). By using the cryo-EM analysis, the ACTH–MC2R–Gs–MRAP1 complex was initially determined to the resolution of 3.2 Å (Supplementary information, Fig. S1b–e). Nevertheless, the extracellular region of the complex displayed a strong but discontinuous signal, extending from the intersection of MC2R N-terminus and MRAP1 to the ligand-binding pocket (LBP) of the receptor, indicating the conformational heterogeneity and potential significance of this region. We further performed a focused three-dimensional (3D) classification with a mask containing the area within 3 Å of ACTH and the extracellular part to differentiate conformational states and uncover hidden molecular details in the discontinuous density. The data processing yielded one class with a high-quality extracellular density, and this class was used for the final reconstruction of an integrated structure of the ACTH–MC2R–Gs–MRAP1 complex at 3.3 Å resolution (Supplementary information, Fig. S1d, f, g), including a well-defined ectodomain consisting of the N-terminal loop of MRAP1 and C-terminal portion of ACTH (Fig. 1c; Supplementary information, Fig. S2 and Table S1).
The overall architecture of the MC2R complex
The high-quality density map enables us to precisely position all the components of the complex (Fig. 1d). The agonist-bound MC2R adopts a canonical fold of the seven-transmembrane domain (7-TMD) and exhibits structural features similar to MC1R and MC4R in the active state, including a relatively short ECL2, two disulfide bonds bridging N’-loop and ECL3 (C21NT–C253ECL3 and C245ECL3–C251ECL3) (Fig. 1e). Several notable differences reside in the extracellular and intracellular loops between MC2R and MC1R or MC4R, for instance, the ECL1 of MC2R is more compressed, and its ICL3 is shorter (Fig. 1e). Intriguingly, ICL3 of MC2R is visible, indicative of its more stable conformation relative to its homologs, which is probably attributed to its interaction with MRAP1 (Fig. 1e).
ACTH is embedded in the external cavity of MC2R with a ‘hook’-shaped conformation, forming a total buried interface area of 1816 Å2. The C-terminal fragment of ACTH, carrying the unique address motif compared with other melanocortin hormones, forms an extracellular region together with the N-terminus of MRAP1 (Fig. 1a, c).
The TMD of MRAP1 sits almost parallel to that of MC2R and directly contacts TM5 and TM6 of the receptor, constituting a TM core of the complex consisting of eight helices. Interestingly, residues 20–32 from the N-terminal loop of MRAP1 make an approximately 90-degree sharp turn relative to the TMD of MRAP1, then extend across the extracellular vestibule and orient horizontally to the membrane. In addition, the N-terminal segment 13–19 vertically stretches from the membrane and is closely packed with the C-terminus of ACTH by forming short anti-parallel β-strands. The overall extracellular part of MRAP1 is reminiscent of a seat-belt, anchoring ACTH inside the MC2R ligand-binding cavity (Fig. 1c). Unlike the N-terminus, the C-terminus of MRAP1 at the cytoplasmic side is unstructured, indicating a relatively weak association with the receptor (Fig. 1c; Supplementary information, Fig. S2).
Assembly of MRAP1 with ACTH-bound MC2R
The complex structure provides a template for clarifying the assembly of MRAP with ACTH-bound MC2R and deepening our understanding of the necessity of MRAP1 in MC2R activation. MRAP1 interacts with the ACTH-bound MC2R complex through three major interfaces, including two with MC2R and one with the C-terminus of ACTH (Fig. 2a–d).
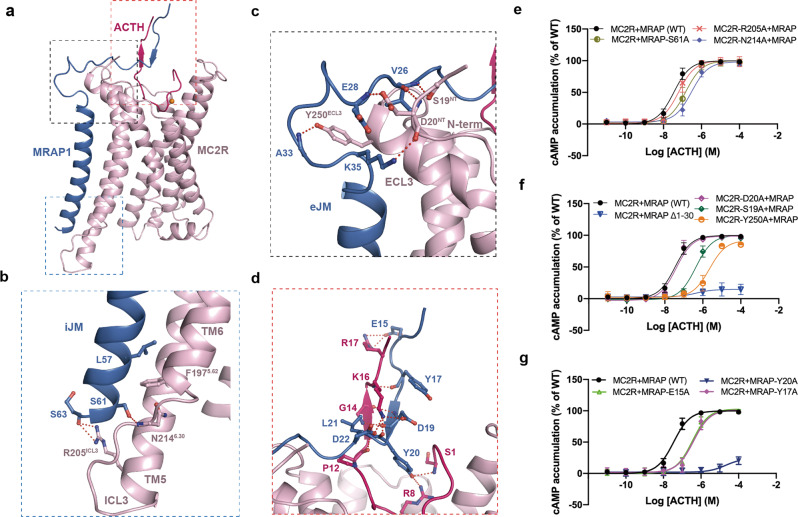
Fig. 2. Molecular mechanism of MRAP1-mediated regulation of ACTH recognition by MC2R.
a Three interfaces formed by MRAP1 contacting MC2R and ACTH. b–d Detailed interactions between MRAP1 iJM motif and MC2R (b), between MRAP1 eJM motif and MC2R (c), and between MRAP1 and ACTH (d), highlighted in the blue, black and red frames in a, respectively. e–g cAMP responses of ACTH-stimulated MC2R–MRAP1 WT or mutants in the interacting interfaces of b–d.
The intracellular juxtamembrane motif (iJM) of MRAP1 and ICL3 of MC2R constitute a cytosolic interface maintained by sidechain–mainchain interaction between S61, S63 of MRAP1 and R205ICL3 and N2146.30 of MC2R, thereby triggering the shift of ICL3 towards MRAP1 (Fig. 2b). Substituting R205ICL3 and N2146.30 with alanine made negligible impact on ACTH activity, indicating an insignificant role of this cytosolic interface in the regulation of MC2R by MRAP1 (Fig. 2e; Supplementary information, Table S2).
More extensive contacts exist at the extracellular interface between the extracellular juxtamembrane motif (eJM) of MRAP1 and the N-terminus as well as ECL3 of the receptor. These interactions stabilize the sharp kink of MRAP1. Notably, Y250ECL3 provides a hinge point for MRAP1 to take a perpendicular turn. S19NT and D20NT of MC2R fall within the distance of the hydrogen bond with the mainchain of MRAP1, probably stabilizing the sharp kink conformation and making MRAP1 stretch to the extracellular entrance of the LBP (Fig. 2c). Truncation of 30 amino acids at the N-terminus of MRAP1 almost abolished ACTH-induced MC2R activation, reflecting an essential role of the extracellular interface compared with the cytosolic interface in the MC2R regulation by MRAP1. Further alanine mutation analysis identified S19NT and Y250ECL3 as two determinants in MC2R responsible for MRAP1 regulation (Fig. 2f; Supplementary information, Table S2).
Besides two major interfaces with MC2R, the N-terminus of MRAP1 forms an additional interface with the C-terminus of ACTH. The interface is primarily stabilized by polar interactions. K16 and R17 in the address motif (KKRR) of ACTH are located at a distance of salt bridges with acidic residues D19 and E15 at the N-terminus of MRAP1, respectively (Fig. 2d). Replacing E15 with alanine notably hampered MC2R activation, indicating the importance of the salt bridge in MC2R activation (Fig. 2g; Supplementary information, Table S2). In addition, Y17 also contributes to ACTH activity, probably by maintaining the extracellular ascending structure (Fig. 2d, g). Intriguingly, hydrogen bonds are established between Y20 of MRAP1 and S1 and R8 of MC2R, resembling the lock of the MRAP1 seat-belt, thus fastening the ligand inside the pocket (Fig. 2d). Mutating Y20 of MRAP1 to alanine completely abrogated ACTH-induced cAMP accumulation, consistent with the previous finding on the significance of Y20 in MC2R activation4 (Fig. 2g; Supplementary information, Table S2). In addition, the map with the discernible density at the extracellular region presenting segments of ACTH, MRAP1, and the receptor stands for a more stable complex conformation, indicating that engagement of MRAP1 with MC2R and ACTH is a prerequisite of MC2R activation.
Both MRAP1 and MRAP2 have been thought to regulate receptor localization to the cell surface and signaling. However, unlike MRAP1, which only regulates ACTH signaling via MC2R, MRAP2 has also been implicated in modulating other GPCR signaling (e.g., ghrelin receptor) appetite and energy homeostasis.33 Sequence alignment reveals an additional ‘LDYL’ motif (L18–D19–Y20–L21) in MRAP1 relative to MRAP2 (Supplementary information, Fig. S3a), which substantially contributes to the association of MRAP1 with ACTH and is involved in ACTH-induced MC2R activation. Adding ‘LDYL’ motif to the wild-type (WT) MRAP2 enabled the MRAP2-LDYL mutant to activate MC2R in synergy with ACTH (Supplementary information, Fig. S3b). These findings provide a rationale for understanding the different effects of the two MRAPs on ACTH-mediated MC2R activation.
Besides MRAPs, receptor activity-modifying proteins (RAMPs) stands out as another type of GPCR accessory protein. RAMPs can be classified into three types: RAMP1, RAMP2, and RAMP3, all of which interact with a broad set of GPCRs and are best studied with class B GPCRs, such as the calcitonin receptor-like receptor (CLR) and calcitonin receptor (CTR).34 RAMPs form heterodimers with CLR/CTR, including calcitonin gene-related peptide receptor (CGRPR, i.e., CLR–RAMP1), adrenomedullin (AM) receptor 1 (AM1R, CLR–RAMP2), AM receptor 2 (AM2R, CLR–RAMP3), amylin receptors AMY1R (CTR–RAMP1), AMY2R (CTR–RAMP2), and AMY3R (CTR–RAMP3), in a conserved assembly mode.35–38 Thus, CGRPR (CLR–RAMP1) was used as a representative heterodimer to compare the GPCR-binding modes between RAMPs and MRAP1. RAMP1 functions similarly to MRAP1, such as facilitating the cell surface expression of CLR and being involved in CGRP recognition.35 MRAP1 and RAMP1 showed a similar N-terminus-out and C-terminus-in organization mode, and no substantial densities were observed at their C-terminal tails, indicating their weak association with G proteins. Intriguingly, TMD of MRAP1 displays a distinct interface with the receptor helices relative to RAMP1 (TM5–TM6 for MC2R–MRAP1 vs TM3–TM4–TM5 for CLR–RAMP1) (Supplementary information, Fig. S4). In addition, unlike MRAP1, which shows extensive interactions with the receptor and ligand peptide, RAMP1 primarily interacts with the extracellular domain of CLR and makes only direct contact with the far C-terminus of CGRP.35
ACTH recognition by MC2R
The residues of ACTH, especially H6, F7, and W9, penetrate deeply into the extracellular core of MC2R (Supplementary information, Fig. S5a). Extensive interactions between ACTH and N-terminus, TM1, TM2, TM3, TM4, and ECL3 are observed inside the LBP of MC2R. Similar to the binding mode of α-MSH in MC1R31 and MC4R,32 these interactions are mainly made by salt bridges, hydrogen bonds, and hydrophobic interactions (Fig. 3a). Mutations of the key residues of MC2R that interact with ACTH, including E28A, Y76A, D103A, D107A, and S169A, diminished cAMP accumulation to different extents (Fig. 3b, c; Supplementary information, Table S2). A calcium ion, linking E5 and F7 from ACTH as well as E80, D103, and D107 from MC2R, strengthens ACTH binding to MC2R (Supplementary information, Fig. S6). This calcium-mediated interaction is believed to be critical for ligand recognition by MCRs.29
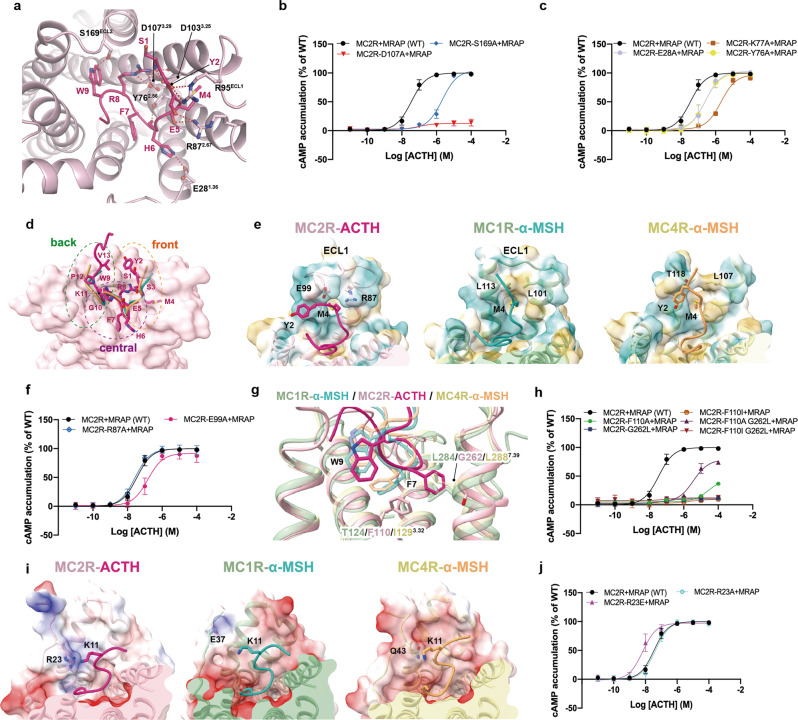
Fig. 3. Ligand recognition by the orthosteric LBP of MC2R.
a Detailed interactions between ACTH and MC2R. Polar interactions are presented by red dashed lines. b, c cAMP signaling of MC2R mutants in the LBP. d Front, central and back regions of MC2R LBP. MC2R surface is colored light pink; ACTH is colored deep pink; and α-MSH in active MC1R and MC4R structures are shown in forest green and orange, respectively. e Structural comparison among the front regions of the LBPs of MC2R, MC1R (PDB code: 7F4D), and MC4R (PDB code: 7F53). LBP surfaces are shown by the hydrophobicity. f cAMP responses of MC2R mutants in the front region. g Rotameric change of ACTH-F7 in the cavity of MC2R central region, compared to α-MSH-F7 in MC1R and MC4R (PDB codes: 7F4D, 7F53). h cAMP signaling of MC2R mutants in the central region. i Structural comparison among the back regions of the LBPs of MC2R, MC1R (PDB code: 7F4D), and MC4R (PDB code: 7F53). LBP surfaces are shown by the electrostatic potential. j cAMP signaling of MC2R mutants in the back region.
Compared to α-MSH bound to MC1R or MC4R, ACTH adopts a more curved conformation (Fig. 3d). Its N-terminus shares an identical sequence with α-MSH, hinting at a similar binding mode for MC2R. However, the structural superposition of ACTH-bound MC2R with α-MSH-bound MC1R (Cα root mean square deviation, RMSD = 0.915 Å) and MC4R (RMSD = 0.868 Å) show that amino acids 1–5, 6–9 (message motif), and 10–13 in both peptides display different binding poses with interacting residues in corresponding regions, designated as the front, central and back regions, respectively (Fig. 3d).
In the front region, MC1R and MC4R provide hydrophobic space to accommodate Y2 and M4 of α-MSH.31,32 Differently, charged residues R87 and E99 in MC2R produce an unfavorable polar environment to engage the hydrophobic M4 (Fig. 3e; Supplementary information, Fig. S2). However, R87A and E99A mutations did not substantially affect ACTH activity (Fig. 3f; Supplementary information, Table S2). Moreover, ECL1 of MC2R prevents the ACTH N-terminal motif from reaching out and pushes Y2 centripetally (Fig. 3e). The unfavorable electrostatic environment and the steric hindrance caused by ECL1 may hamper the binding of α-MSH to MC2R.
In the central region, the space in MC4R accommodating the aromatic group of F7 in the message motif (HFRW) of α-MSH is occupied by F1103.32 in MC2R. The bulkier sidechain of phenylalanine than cognate isoleucine (I1293.32) in MC4R leads to a notable rotamer change of F7 of ACTH to avoid a probable steric clash, thereby alternatively rearranging its sidechain in close proximity to G2627.39. Moreover, compared to the glycine in MC2R, the relatively bulkier L2887.39 in MC4R may sterically clash with F7 of ACTH and impede its rotamer change (Fig. 3g). Single or combined F1103.32A/I and G2627.39L mutations support the necessity of rotamer change of F7 for MC2R activation (Fig. 3h). Additionally, W9 in ACTH bound to MC2R at a position slightly deeper relative to W9 of α-MSH bound to MC1R/MC4R (Fig. 3g).
In the back region, contrary to the extracellular concave surfaces of MC1R/MC4R, which are negatively charged and electrostatically complementary to α-MSH, the bilateral parts of MC2R LBP enrich positively charged residues, thus probably weakening the electrostatic interactions between MC2R and ACTH (Supplementary information, Fig. S5b, c). Specifically, the electrostatic repulsion between R23NT of MC2R and K11 of ACTH pushes the C-terminal portion of the peptide outwards (Fig. 3i) and increases the curvature of the GKPV motif (Fig. 3d). This structural observation is supported by the increased ACTH activity when substituting R23 with the corresponding glutamic acid in MC1R that is compatible to K11 (Fig. 3j; Supplementary information, Table S2). Consequently, the unfavorable environment of MC2R LBP generates steric hindrance and electrostatic repelling effects to prevent short proteolytic products such as α-MSH from binding (Supplementary information, Fig. S5). However, ACTH carrying an elongated C-terminal loop attracted to the MRAP1 seat-belt by electrostatic interactions could be recognized and retained, which may explain the unique selectivity of MC2R for ACTH.
Mechanism of MC2R activation
Structural comparisons between the ACTH-bound MC2R complex and the active and inactive MC4R bound to α-MSH (PDB code: 7F53) and SHU9119 (PDB code: 6W25), respectively, reveal that MC2R is indeed in the active state (Fig. 4a–c). In contrast to the inactive MC4R, the cytoplasmic end of TM6 of MC2R undergoes a pronounced outward displacement to accommodate the C-terminus of α5 helix of Gαs subunit, the hallmark of class A GPCR activation. ACTH-bound MC2R shows a similar overall active conformation compared with α-MSH-bound MC4R. However, several observed conformational differences suggest a receptor-specific activation mode of MC2R relative to MC4R.
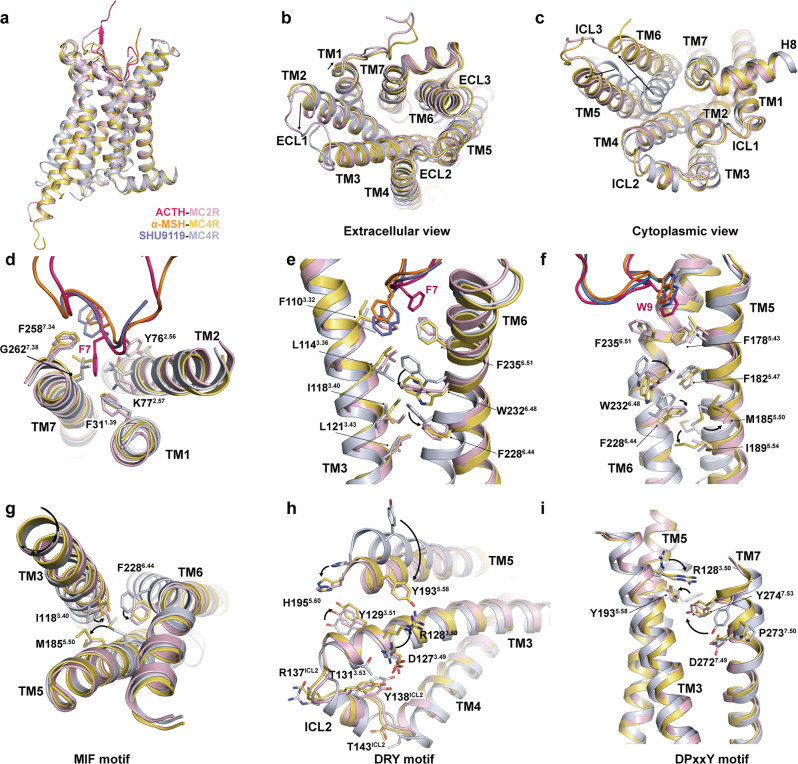
Fig. 4. Activation mechanism of MC2R.
a–c Structural alignment of the active MC2R with active and inactive MC4R (PDB codes: 7F53, 6W25). Side view (a); extracellular view (b); cytoplasmic view (c). d Steric hindrance effect of ACTH-F7 on TM1 and TM2 of MC2R. e–i Conformational changes of MC2R upon activation indicating a conserved activation mechanism across the MCR family. Toggle switch (e, f), MIF motif (g), DRY motif (h), and DPxxY motif (i) are shown.
On the extracellular side, minor differences were observed in TM1 and TM2 between activated MC2R and MC4R (Fig. 4b), which may arise from the steric hindrance between F7 of ACTH and F311.39 and K772.57 of MC2R and the consequent outward movement of TM1 and TM2 (Fig. 4d). At the bottom of the peptide-binding pocket, F7 of α-MSH directly contacts L1333.36 of MC4R. Differently, the benzene ring of F1103.32 of MC2R sterically overlaps with the phenyl group of F7 of α-MSH, substituting its role in engaging the cognate residue L1143.36 of MC2R (Fig. 4e). Although differently triggered, the sidechain of leucine at 3.36 undergoes a similar upward rotamer switch and subsequently leads to downward rotameric switch of W6.48 for MC2R or movement for MC4R, resulting in the rotation of F6.44 and initiating the outward rotation and displacement of the cytoplasmic part of TM6 (Fig. 4f).
These conformational changes further propagate the agonism signal downward through several motifs associated with class A GPCR activation. The M5.50I3.40F6.44 motif, corresponding to the noncanonical PIF motif in MCRs, undergoes active-like conformational changes, concomitant with the large shift of TM5 (Fig. 4c, g). The shift of TM5 relocates Y5.58 and H5.60, establishing polar interactions with R3.50 and Y3.51 in the D3.49R3.50Y3.51 motif, respectively (Fig. 4h). Y5.58 also forms a polar interaction with Y7.53 in the NPxxY motif, further stabilizing the polar interaction network among TM3, TM5, and TM7. The sidechain of Y7.53 is re-orientated and brings TM7 closer to the helical core (Fig. 4i). These significant conformational changes of MC2R are consistent with those in MC1R and MC4R,31 implying a conserved activation signal propagation mechanism across the MCR family.
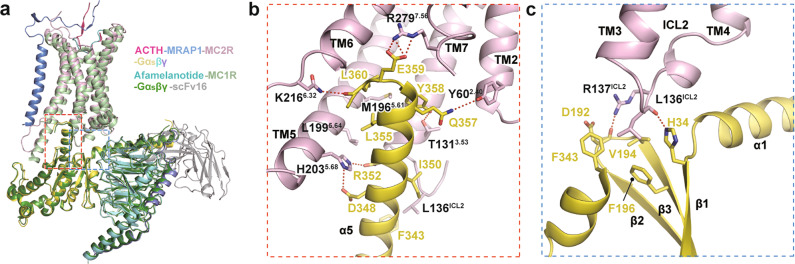
The profound conformational changes of MC2R TMD during activation reform the cytoplasmic interface for Gs protein recruitment. Structural superimposition of the Gs-coupled MC2R with the Nb35-free MC1R–Gs complex (PDB code: 7F4F) shows a largely conserved coupling mode. Notable MC2R conformational differences include a shorter TM5 (Fig. 5a). The major coupling interface is generated by the insertion of helix 5 of Gαs into the intracellular core enclosed by TM2, TM3, TM5, TM6, and TM7. The C-terminal residues of the Gαs subunit, especially the ‘wavy hook’ residues from Q357 to L361, establish extensive contacts with the surrounding TMs of MC2R. Besides, the hydrophobic interactions of F343, I350, and L355 of the Gαs subunit with the nearby aliphatic residues may provide additional stability to the coupling interface (Fig. 5b). ICL2 interacts with the αN–β1 hinge region of the Gαs subunit. L136ICL2 makes hydrophobic contact with V194, F196, and F343. Residues L136ICL2 and R137ICL2 form mainchain–sidechain hydrogen bonds with H34 and D192 of the Gαs subunit (Fig. 5c). Together, our structure provides a template for enhancing the mechanistic understanding of MC2R activation and the MC2R–Gs coupling.
Fig. 5. Intracellular interactions between MC2R and Gs protein.
a Structural alignment of the ACTH–MC2R–Gαβγ–MRAP1 and afamelanotide–MC1R–Gαβγ–scFv16 (PDB code: 7F4F) complexes. b The interface in the cavity at the cytoplasmic region between MC2R and α5 helix of Gα. c The interface between ICL2 of MC2R and α1 helix, β1 sheet, β2–β3 loop of Gαs. Polar interactions are shown as red dashed lines.
Discussion
MC2R, selectively activated by ACTH with the interplay of MRAP1, regulates adrenal steroidogenesis crucial for stress response. In this study, we report the cryo-EM structure of the ACTH-bound MRAP1–MC2R–Gs complex, providing structural evidence for the seat-belt function of MRAP1 to stabilize ACTH binding to MC2R. Both the shape and electrostatic condition of MC2R unfavored the binding of the ACTH N-terminus to the receptor pocket and may lead to ligand binding instability. Alternatively, the two essential elements for MC2R activation, the N-loop of MRAP1 and the address motif of ACTH, are actually bound together to implement ligand recognition.
Earlier data support the theory that MRAP1 adopts a dual topology as a functional form when binding to MC2R.8 Nonetheless, we found a stoichiometry of 1:1 for MRAP1 and MC2R in our complex structure and structurally proved that the N-terminus-out MRAP1 in the monomer state interacts with MC2R and participates in ACTH binding. However, the possibility that the homodimer MRAP1 exists cannot be dismissed. The assembly state of MRAP1 and its relevance to functional regulation have yet to be addressed.
Although the association mode of MRAP1 with ACTH-bound MC2R in a functional state is revealed, the dynamic assembly process of the complex, such as whether MRAP1 captures ACTH prior to its binding to MC2R LBP or restricts the bound ACTH inside the LBP, remains elusive. Interestingly, our focused 3D classification revealed one class with relatively poor but apparent ACTH density inside the LBP of MC2R, in the absence of density for the MRAP1 N-terminus (Supplementary information, Fig. S8a, b). Interestingly, the presence of density for Gs protein in this volume indicates that MC2R is somehow activated, suggesting that MC2R might be bound by ACTH and activated without the seat-belt restraint of MRAP1. However, it is arguable that the MRAP1 seat-belt could be loosened after receptor activation or destroyed during cryo-sample preparation.
We therefore propose that the N-terminal loop of MRAP1 functions as a seat-belt to fasten ACTH in the LBP. Similarly, wrapping of a flexible regulatory loop around the ligand for GPCR activation was observed in glycoprotein hormones (i.e., chorionic gonadotropin (CG) and thyroid-stimulating hormone (TSH)) and their receptors (i.e., luteinizing hormone/choriogonadotropin receptor (LHCGR) and thyroid-stimulating hormone receptor (TSHR)).39,40 The C-terminal seat-belt segment of the β-subunit of CG/TSH encircles the entire α- and β-subunits of the hormone and mediates the hormone binding to the corresponding receptor.39,40 Different from the intramolecular seat-belt for CG/TSH, MRAP1 provides an intermolecular seat-belt loop to activate the ACTH–MC2R complex. Moreover, many accessory proteins in addition to MRAP1 are essential for GPCR activation. For example, single-transmembrane protein RAMP1 allosterically modulates CLR.35 The diversity of GPCR modulation by accessory proteins could provide variable ways to adjust and control GPCR signal transduction vital to many physiological functions.
Taken together, our results offer a molecular explication of selective ligand recognition and activation of MC2R, and the necessity of the MRAP1 N-terminus for this process. It is our hope that the MC2R structure reported herein would support the de novo structure-based design of MC2R ligands and accelerate drug discovery for the treatment of diseases caused by an abnormality in the ACTH-MC2R-MRAP1 pathway.
Materials and methods
Construct preparation
The full-length human MC2R was modified to contain the haemagglutinin (HA) signal peptide followed by an N-terminal 10× His tag and cytochrome b562RIL (BRIL) to enhance the receptor expression; a 15-amino acid polypeptide linker and a LgBiT (Promega) were fused in the C-terminus. The full-length human MRAP1 was modified to include an N-terminal Flag epitope tag. An engineered human Gαs was constructed based on miniGs (PDB code: 5G53)41 with eight mutations (G49D, E50N, L63Y, A249D, S252D, L272D, I372A, and V375I), and two additional dominant-negative mutations (G226A and A366S) to decrease the affinity of nucleotide binding and increase the stability of the heterotrimeric G protein. All these modified cDNAs, as well as the human Gβ fused with HiBiT at its C-terminus, and the human Gγ were cloned into pFastBac vector, respectively.
Complex expression
MC2R, MRAP1, Gαs, Gβ, and Gγ were co-expressed in High Five insect cells (Thermo Fisher) using the baculovirus method as previously described.31 The cells were grown in ESF 921 serum-free medium (Expression Systems) to a density of 2 × 106 cells per mL, and then five baculoviruses at a ratio of 1:1:1:1:1 were added to the cell culture. The cells were harvested 48 h after infection at 27 °C. The cell pellet was collected by centrifugation at 2000× g for 15 min and stored at −80 °C before use.
Complex purification
Cell pellets were thawed in suspension buffer (20 mM HEPES, pH 7.4, 100 mM NaCl, 10 mM MgCl2, and 5 mM CaCl2 supplemented with Protease Inhibitor Cocktail (TargetMol)). The complex formation was initiated by the addition of 25 mU/mL apyrase (Sigma) and 10 μM ACTH (GenScript). The suspension was incubated for 1 h at room temperature (RT). Membranes were collected by centrifugation at 30,000× g for 30 min. Complexes from membranes were solubilized by 0.5% (w/v) lauryl maltose neopentyl glycol (LMNG, Anatrace) supplemented with 0.1% (w/v) cholesteryl hemisuccinate (CHS, Anatrace) for 2 h at 4 °C. The supernatant was then isolated by centrifugation at 65,000× g for 30 min and incubated with M1 anti-Flag affinity resin for 2 h at 4 °C. The resin was collected by centrifugation at 500× g for 10 min and loaded onto a gravity-flow column.
The resin was then washed with 30 column volumes of washing buffer (20 mM HEPES, pH 7.4, 100 mM NaCl, 100 μM TCEP, 10 μM ACTH, 2 mM CaCl2, 0.01% (w/v) LMNG, 0.01% (w/v) GDN, and 0.004% (w/v) CHS) before bound complexes were eluted with the same buffer containing 0.1 mg/mL Flag peptide. The complexes were then concentrated using a 100-kD Amicon Ultra centrifugal filter (Millipore) and loaded onto Superdex 200 10/300 GL column (GE Healthcare) pre-equilibrated with running buffer containing 20 mM HEPES, 100 mM NaCl, 100 μM TCEP, 10 μM ACTH, 2 mM CaCl2, pH 7.4, 0.00075% (w/v) LMNG, 0.00025% (w/v) GDN, and 0.0002% (w/v) CHS. The sample collected from size-exclusion chromatography was analyzed by SDS–PAGE. The monomeric peak fractions were collected and concentrated to 4–6 mg/mL for electron microscopy experiments.
Cryo-EM data acquisition
The purified ACTH–MC2R–Gs–MRAP1 complex was applied to a glow-discharged holey carbon EM grid (Quantifoil, Au 300 R1.2/1.3) in a Vitrobot chamber (FEI Vitrobot Mark IV). The chamber of the Vitrobot was set to 100% humidity at 4 °C. The sample was blotted for 3 s with a blot force of 2 and then plunged into liquid ethane. Cryo-EM data were collected by a Titan Krios G4 (Thermo Fisher) using 300 kV accelerating voltage at the Advanced Center for Electron Microscopy, Shanghai Institute of Material Medica. The micrographs were recorded using a super-resolution counting mode at a pixel size of 0.52 Å using the EPU software. Micrographs were obtained at a dose rate of ~15 e−/(Å2·s) with a defocus ranging from −1.0 to −3.0 μm. A total movie with a total exposure time of 3.33 s for each micrograph was collected and was dose-fractionated to 32 frames, resulting in a total dose of ~50 e−/Å2.
Cryo-EM data processing
All collected 7131 movie stacks were 2 × 2 binned, and motion corrected in Relion 4.0,42 to generate micrographs with a final pixel size of 1.04 Å. The contrast transfer function (CTF) parameters were estimated by CTFFIND4 program.43 Particles picked by Blob picker in Cryosparc 3.2 from micrographs with CTF parameter were subjected to 2D classification to generate input particles to train Topaz in Cryosparc 3.2. A total of 4,347,219 particles were automatically picked by Topaz Extract and screened by several rounds of 2D classification in Cryosparc 3.2.44 1,235,537 selected particles were used to generate ab initio map reference in Cryosparc3.2, and subjected to a cascade of 3D classification and refinement in Relion 4.0.42 After 3D refinement, particle polishing, CTF refinement in Relion 4.0, NU-refinement in Cryosparc 3.2,45 and sharpening in DeepEMhancer,46 the resolution of the initial structure of the ACTH–MC2R–Gs–MRAP1 complex reconstructed by 194,986 particles was 3.2 Å. The resolution was estimated by the gold-standard Fourier shell correlation (FSC, 0.143 criterion). Local resolution distribution was evaluated using Cryosparc 3.2.
To further investigate the complex structure in different states, local 3D classification with a 3D mask surrounding ACTH and the extracellular area was performed using 194,986 particles. This resulted in two states, well-defined and poor-density states, inside the MC2R LBP with high resolution. Further data processing yielded the reconstruction of the integrated and loose structures of the ACTH–MC2R–Gs–MRAP1 complex with the resolution of 3.3 Å for both states, reconstructed by 59,170 and 89,184 particles, respectively.
Model building and refinement
The integrated structure was used to build a model of the ACTH–MC2R–Gs–MRAP1 complex. The α-MSH–MC1R–Gs structure was docked into the EM map in Chimera, and manually adjusted in COOT47 based on the global EM map to build the mainchain of ACTH, MC2R, and Gs. For the MRAP1 structure, AlphaFold248-predicted MRAP1 structure was used to generate a template of MRAP1 transmembrane motif; and N-loop was manually built in COOT. The resulting model was refined against the EM density using real space refinement in PHENIX49 with secondary structure and geometry restraints. The final model after refinement was validated using the PHENIX package. The model statistics are summarized in Supplementary information, Table S1.
cAMP accumulation
WT or mutant MC2R and MRAP constructs were cloned into pcDNA6.0 vector (Invitrogen) for functional studies. HEK293T cells were transiently transfected with the vectors at a ratio of 1:2 using Lipofectamine 2000 transfection reagent (Invitrogen) and incubated at 37 °C in 5% CO2. Twenty-four hours after transfection, the transfected cells were digested with 0.02% (w/v) EDTA, resuspended in stimulation buffer (Hanks’ balanced salt solution (HBSS) supplemented with 5 mM HEPES, 0.5 mM IBMX, and 0.1% (w/v) bovine serum albumin (BSA), pH 7.4) to a density of 6 × 105 cells per mL and added to 384-well white plates (PerkinElmer, 3000 cells per well). cAMP accumulation was measured by a LANCE Ultra cAMP kit (PerkinElmer) according to the manufacturer’s instructions. In brief, transfected cells were incubated for 40 min in a stimulation buffer with different concentrations of ACTH (5 μL) at RT. The reaction was stopped by the addition of lysis buffer containing 5 μL Eu-cAMP tracer and 5 μL ULight-anti-cAMP. Plates were then incubated for 50 min at RT and time-resolved FRET signals were measured at 620 nm and 665 nm, respectively, by an EnVision multilabel plate reader (PerkinElmer). Data were analyzed in GraphPad PRISM 8 and all values were normalized to the WT. All outcomes were repeated at least three times.
To assess the effect of calcium and magnesium ions on cAMP signaling, HEK293T cells were digested with 0.02% (w/v) EDTA and washed three times with calcium/magnesium-free HBSS buffer. Then the cells were resuspended and stimulated with different concentrations of ACTH in Ca2+/Mg2+-free stimulation buffer (aforementioned stimulation buffer supplemented with 1 mM EGTA) or with an additional 1.5 mM CaCl2 or MgCl2 in Ca2+/Mg2+-free stimulation buffer ([Ca2+] = ~0.5 mM).29 The rest of the steps were essentially the same as described above.
Receptor surface expression
Cell surface expression of MC2R, MRAP1, and MRAP2 was determined by flow cytometry 24 h post-transfection in HEK293T (MC2R (mutant) + MRAPs (WT)) or CHO-K1 (MC2R (WT) + MRAPs (mutant)) cells. Briefly, ~2 × 105 cells were blocked with PBS containing 5% BSA (w/v) at RT for 15 min. After that, cells expressing MC2R (mutant) + MRAPs (WT) were incubated with anti-Flag primary antibody (1:300, diluted with PBS containing 5% BSA, Sigma), and those expressing MC2R (WT) + MRAPs (mutant) were incubated with anti-c-Myc antibody (1:200, diluted with PBS containing 5% BSA, Abcam) at RT for 1 h. The cells were then washed three times with PBS containing 1% BSA (w/v) followed by 1 h incubation with anti-mouse Alexa Fluor 488 conjugated secondary antibody (1:1000, diluted with PBS containing 5% BSA, Invitrogen) at 4 °C in the dark. After washing three times, cells were re-suspended in 200 μL PBS containing 1% BSA for detection by Flow Cytometer (BD Biosciences) utilizing laser excitation and emission wavelengths of 488 nm and 530 nm, respectively. For each sample, 10,000 cellular events were collected, and the total fluorescence intensity of the positive expression cell population was calculated. Data were normalized to the WT receptor.
Statistical analysis
All functional study data were analyzed using GraphPad Prism 8.0 (GraphPad Software Inc.) and showed as means ± SEM. from at least three independent experiments in triplicate. The significance was determined with two-sided, one-way ANOVA with Tukey’s test, and *P < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
The cryo-EM data of the ACTH–MC2R–Gs–MRAP1 complex were collected at the Advanced Center for Electron Microscopy, Shanghai Institute of Materia Medica (SIMM). We thank all the staff at the cryo-EM facilities for their technical support. This work was partially supported by the National Natural Science Foundation (32171187 to Y.J., 82121005 to Y.J. and H.E.X., 32130022 to H.E.X., 81872915 to M.-W.W., 81773792 and 81973373 to D.Y.); the Ministry of Science and Technology (China) grants (2018YFA0507002 to H.E.X. and M.-W.W.); the Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 to H.E.X.); Shanghai Municipal Science and Technology Major Project (H.E.X.); the CAS Strategic Priority Research Program (XDB37030103 to H.E.X.).
Author contributions
P.L. designed the expression constructs, purified the MC2R complex protein, prepared the final samples for negative stain and cryo-EM data collection, conducted functional studies, and prepared figures and manuscript draft; W.F. conducted functional studies with the help of A.D., X. Chen, and X. Cai, supervised by M.-W.W. and D.Y.; S.M. participated in the preparation of constructs and expression screening; K.W. and Q.Y. participated in cryo-EM data acquisition; M.-W.W. participated in manuscript preparation; Y.J. and H.E.X. conceived and supervised the project, analyzed the structures, and wrote the manuscript with inputs from all authors.
Data availability
The corresponding coordinates and cryo-EM density map have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with code 8GY7, and in EMDB (http://www.ebi.ac.uk/pdbe/emdb/) with code EMD-34371, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ping Luo, Wenbo Feng.
Contributor Information
Ming-Wei Wang, Email: mwwang@simm.ac.cn.
H. Eric Xu, Email: eric.xu@simm.ac.cn
Yi Jiang, Email: yjiang@lglab.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-022-00751-6.
References
- 1.Cooray SN, Clark AJL. Melanocortin receptors and their accessory proteins. Mol. Cell Endocrinol. 2011;331:215–221. doi: 10.1016/j.mce.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 2.Webb TR, Clark AJL. Minireview: the melanocortin 2 receptor accessory proteins. Mol. Endocrinol. 2010;24:475–484. doi: 10.1210/me.2009-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouault AAJ, Srinivasan DK, Yin TC, Lee AA, Sebag JA. Melanocortin Receptor Accessory Proteins (MRAPs): Functions in the melanocortin system and beyond. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:2462–2467. doi: 10.1016/j.bbadis.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Sebag JA, Hinkle PM. Regions of Melanocortin 2 (MC2) receptor accessory protein necessary for dual topology and MC2 receptor trafficking and signaling. J. Biol. Chem. 2009;284:610–618. doi: 10.1074/jbc.M804413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy S, et al. Mechanisms of melanocortin-2 receptor (MC2R) internalization and recycling in human embryonic kidney (hek) cells: identification of Key Ser/Thr (S/T) amino acids. Mol. Endocrinol. 2011;25:1961–1977. doi: 10.1210/me.2011-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy S, Rached M, Gallo-Payet N. Differential regulation of the human adrenocorticotropin receptor [melanocortin-2 receptor (MC2R)] by human MC2R accessory protein isoforms alpha and beta in isogenic human embryonic kidney 293 cells. Mol. Endocrinol. 2007;21:1656–1669. doi: 10.1210/me.2007-0041. [DOI] [PubMed] [Google Scholar]
- 7.Hinkle PM, Sebag JA. Structure and function of the melanocortin2 receptor accessory protein (MRAP) Mol. Cell Endocrinol. 2009;300:25–31. doi: 10.1016/j.mce.2008.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malik S, Dolan TM, Maben ZJ, Hinkle PM. Adrenocorticotropic Hormone (ACTH) responses require actions of the melanocortin-2 receptor accessory protein on the extracellular surface of the plasma membrane. J. Biol. Chem. 2015;290:27972–27985. doi: 10.1074/jbc.M115.668491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebag JA, Hinkle PM. Melanocortin-2 receptor accessory protein MRAP forms antiparallel homodimers. Proc. Natl. Acad. Sci. USA. 2007;104:20244–20249. doi: 10.1073/pnas.0708916105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maben ZJ, Malik S, Jiang LYH, Hinkle PM. Dual topology of the melanocortin-2 receptor accessory protein is stable. Front. Endocrinol. 2016;7:96. doi: 10.3389/fendo.2016.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyder NA, et al. Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide. Cell Res. 2021;31:1176–1189. doi: 10.1038/s41422-021-00569-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mountjoy KG, Robbins LS, Mortrud MT, Cone RD. The cloning of a family of genes that encode the melanocortin receptors. Science. 1992;257:1248–1251. doi: 10.1126/science.1325670. [DOI] [PubMed] [Google Scholar]
- 13.Schwyzer R. ACTH: a short introductory review. Ann. N. Y. Acad. Sci. 1977;297:3–26. doi: 10.1111/j.1749-6632.1977.tb41843.x. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y, Harmon CM. Molecular signatures of human melanocortin receptors for ligand binding and signaling. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:2436–2447. doi: 10.1016/j.bbadis.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 15.Yang YK, Harmon CM. Molecular determinants of ACTH receptor for ligand selectivity. Mol. Cell Endocrinol. 2020;503:110688. doi: 10.1016/j.mce.2019.110688. [DOI] [PubMed] [Google Scholar]
- 16.Cawley NX, Li Z, Loh YP. 60 YEARS OF POMC: Biosynthesis, trafficking, and secretion of pro-opiomelanocortin-derived peptides. J. Mol. Endocrinol. 2016;56:T77–T97. doi: 10.1530/JME-15-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang L, Angleson JK, Dores RM. Using the human melanocortin-2 receptor as a model for analyzing hormone/receptor interactions between a mammalian MC2 receptor and ACTH(1-24) Gen. Comp. Endocr. 2013;181:203–210. doi: 10.1016/j.ygcen.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Dores RM, Liang L. Analyzing the activation of the melanocortin-2 receptor of tetrapods. Gen. Comp. Endocr. 2014;203:3–9. doi: 10.1016/j.ygcen.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 19.Kim JD, Leyva S, Diano S. Hormonal regulation of the hypothalamic melanocortin system. Front. Physiol. 2014;5:480. doi: 10.3389/fphys.2014.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell G, Lightman S. The human stress response. Nat. Rev. Endocrinol. 2019;15:525–534. doi: 10.1038/s41574-019-0228-0. [DOI] [PubMed] [Google Scholar]
- 21.Chung TT, Chan LF, Metherell LA, Clark AJ. Phenotypic characteristics of familial glucocorticoid deficiency (FGD) type 1 and 2. Clin. Endocrinol. (Oxf) 2010;72:589–594. doi: 10.1111/j.1365-2265.2009.03663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heshmatzad K, Mahdieh N, Rabbani A, Didban A, Rabbani B. The genetic perspective of familial glucocorticoid deficiency: in silico analysis of two novel variants. Int. J. Endocrinol. 2020;2020:2190508. doi: 10.1155/2020/2190508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meimaridou E, et al. Familial glucocorticoid deficiency: new genes and mechanisms. Mol. Cell Endocrinol. 2013;371:195–200. doi: 10.1016/j.mce.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 24.Metherell LA, et al. Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2. Nat. Genet. 2005;37:166–170. doi: 10.1038/ng1501. [DOI] [PubMed] [Google Scholar]
- 25.Chan LF, Metherell LA, Clark AJ. Effects of melanocortins on adrenal gland physiology. Eur. J. Pharmacol. 2011;660:171–180. doi: 10.1016/j.ejphar.2010.11.041. [DOI] [PubMed] [Google Scholar]
- 26.Ghaddhab C, Vuissoz JM, Deladoey J. From bioinactive ACTH to ACTH antagonist: the clinical perspective. Front. Endocrinol. 2017;8:17. doi: 10.3389/fendo.2017.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldenberg AJ, et al. Effect of a melanocortin type 2 receptor (MC2R) antagonist on the corticosterone response to hypoxia and ACTH stimulation in the neonatal rat. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2018;315:R128–R133. doi: 10.1152/ajpregu.00009.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouw E, et al. Development of potent selective competitive-antagonists of the melanocortin type 2 receptor. Mol. Cell Endocrinol. 2014;394:99–104. doi: 10.1016/j.mce.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, et al. Determination of the melanocortin-4 receptor structure identifies Ca2+ as a cofactor for ligand binding. Science. 2020;368:428–433. doi: 10.1126/science.aaz8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Israeli H, et al. Structure reveals the activation mechanism of the MC4 receptor to initiate satiation signaling. Science. 2021;372:808–814. doi: 10.1126/science.abf7958. [DOI] [PubMed] [Google Scholar]
- 31.Ma SS, et al. Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor. Cell Res. 2021;31:1061–1071. doi: 10.1038/s41422-021-00557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang HB, et al. Structural insights into ligand recognition and activation of the melanocortin-4 receptor. Cell Res. 2021;31:1163–1175. doi: 10.1038/s41422-021-00552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berruien NNA, Smith CL. Emerging roles of melanocortin receptor accessory proteins (MRAP and MRAP2) in physiology and pathophysiology. Gene. 2020;757:144949. doi: 10.1016/j.gene.2020.144949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hay DL, Pioszak AA. Receptor activity-modifying proteins (RAMPs): new insights and roles. Annu. Rev. Pharmacol. Toxicol. 2016;56:469–487. doi: 10.1146/annurev-pharmtox-010715-103120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang YL, et al. Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature. 2018;561:492–497. doi: 10.1038/s41586-018-0535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Josephs TM, et al. Structure and dynamics of the CGRP receptor in apo and peptide-bound forms. Science. 2021;372:eabf7258. doi: 10.1126/science.abf7258. [DOI] [PubMed] [Google Scholar]
- 37.Liang YL, et al. Structure and dynamics of adrenomedullin receptors AM1 and AM2 reveal key mechanisms in the control of receptor phenotype by receptor activity-modifying proteins. ACS Pharmacol. Transl. Sci. 2020;3:263–284. doi: 10.1021/acsptsci.9b00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao J, et al. A structural basis for amylin receptor phenotype. Science. 2022;375:eabm9609. doi: 10.1126/science.abm9609. [DOI] [PubMed] [Google Scholar]
- 39.Nunez Miguel R, Sanders J, Furmaniak J, Smith BR. Structure and activation of the TSH receptor transmembrane domain. Auto Immun. Highlights. 2017;8:2. doi: 10.1007/s13317-016-0090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan J, et al. Structures of full-length glycoprotein hormone receptor signalling complexes. Nature. 2021;598:688–692. doi: 10.1038/s41586-021-03924-2. [DOI] [PubMed] [Google Scholar]
- 41.Carpenter B, Nehme R, Warne T, Leslie AG, Tate CG. Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature. 2016;536:104–107. doi: 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimanius D, Dong L, Sharov G, Nakane T, Scheres SHW. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 2021;478:4169–4185. doi: 10.1042/BCJ20210708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Punjani A, Rubinstein JL, Fleet DJ, Brubaker M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 45.Punjani A, Zhang H, Fleet DJ. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods. 2020;17:1214–1221. doi: 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez-Garcia R, et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 2021;4:874. doi: 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The corresponding coordinates and cryo-EM density map have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with code 8GY7, and in EMDB (http://www.ebi.ac.uk/pdbe/emdb/) with code EMD-34371, respectively.