Abstract
Background and Aims
While variation in genome size and chromosome numbers and their consequences are often investigated in plants, the biological relevance of variation in chromosome size remains poorly known. Here, we examine genome and mean chromosome size in the cyperid clade (families Cyperaceae, Juncaceae and Thurniaceae), which is the largest vascular plant lineage with predominantly holocentric chromosomes.
Methods
We measured genome size in 436 species of cyperids using flow cytometry, and augment these data with previously published datasets. We then separately compared genome and mean chromosome sizes (2C/2n) amongst the major lineages of cyperids and analysed how these two genomic traits are associated with various environmental factors using phylogenetically informed methods.
Key Results
We show that cyperids have the smallest mean chromosome sizes recorded in seed plants, with a large divergence between the smallest and largest values. We found that cyperid species with smaller chromosomes have larger geographical distributions and that there is a strong inverse association between mean chromosome size and number across this lineage.
Conclusions
The distinct patterns in genome size and mean chromosome size across the cyperids might be explained by holokinetic drive. The numerous small chromosomes might function to increase genetic diversity in this lineage where crossovers are limited during meiosis.
Keywords: Chromosome number, chromosome size, Cyperaceae, distribution range size, genome size, holocentric chromosomes, holokinetic drive, Juncaceae, Thurniaceae
INTRODUCTION
Genome size, chromosome number and chromosome size are among the most useful traits to characterize an organism’s genome. Considerable attention has been paid to describing and explaining the important variation in genome size and chromosome number in plants (Pellicer and Leitch, 2020). Genome size across flowering plants exhibits an estimated 2400-fold variation (Rice et al., 2015; Hidalgo et al., 2017; Pellicer et al., 2018), whereas variation in chromosome numbers across flowering plants is also pronounced, spanning two orders of magnitude from 2n = 4 to 2n = 640 (Stace, 2000; Carta et al., 2020). While variation in genome size and chromosome number has been characterized in many lineages of plants (e.g. Bennett, 1987; Soltis et al., 2003; Roalson, 2008; Leitch et al., 2010; Rice et al., 2015; Pellicer et al., 2018), differences in chromosome sizes among large groups of species have received relatively little attention in recent decades (for earlier works, see Avdulov, 1931; Hasegawa, 1932; Kostoff, 1939; Levin and Funderburg, 1979).
An important factor in determining the genome and chromosome sizes of a plant is whether its chromosomes are monocentric (i.e. having a ‘localized’ centromere) or holocentric (i.e. having ‘diffuse’ centromeres). In plants with holocentric chromosomes, kinetochores cover a large portion of the poleward surface of chromosomes to which microtubules can attach along their entire length during mitosis (Bureš et al., 2013; Márquez-Corro et al., 2019a). Whereas fission events usually end in chromosome loss in monocentric species (i.e. the majority of plants where the spindle attachment is restricted to a single localized centromere), an advantage of holocentric chromosomes is that fragments resulting from fission are often retained, because their diffuse centromeric activity allows for correct segregation (Melters et al., 2012; Bureš et al., 2013; Márquez-Corro et al., 2019a; Mandrioli and Manicardi, 2020; Lucek et al., 2022). Similarly, chromosomal fusion resulting in dicentric chromosomes can lead to the potentially deleterious consequences of merotelic microtubule attachment in monocentric species, while in holocentric species fused chromosomes segregate regularly (Bureš et al., 2013, and references within). Thus, the size and number of chromosomes can change more readily in holocentric compared with monocentric species. However, the unique architecture of holocentric chromosomes limits the number of crossovers to two per homologous pair, regardless of their size, which is a constraint not present in monocentric chromosomes (Escudero et al., 2012; Bureš et al., 2013). A process that possibly affects plants with holocentric chromosomes is holokinetic drive, which involves the preferential size-dependent inheritance of smaller homologous chromosomes during asymmetric meiosis, facilitating removal of repetitive DNA and/or chromosomal fission (Bureš and Zedek, 2014; Márquez-Corro et al., 2019a). The opposite scenario might also be possible, where larger, fused chromosomes with higher amounts of repetitive DNA are preferentially inherited (Bureš and Zedek, 2014; Veleba et al., 2017).
Variation in both genome size and chromosome number in plants has been shown to be associated with certain environmental conditions. Several studies have shown associations between genome size and soil nutrients (Leitch and Leitch, 2008; Šmarda et al., 2013; Kang et al., 2015; Guignard et al., 2016), climatic variables (Knight and Ackerly, 2002; Knight et al., 2005; Greilhuber and Leitch, 2013; Cacho et al., 2021), distribution range size (e.g. Vinogradov, 2003) and different geographic variables, such as latitude and altitude (Knight et al., 2005; LaBar and Adami, 2020). Attention has also been paid to the association of chromosome number with different environmental variables, but results have not been as conclusive (e.g. Escudero et al., 2012; Carta et al., 2018; Márquez-Corro et al., 2021). Since only a limited number of studies in the previous century focused on the association between chromosome size and environmental growing conditions (see Stebbins, 1966; Levin and Funderburg, 1979), the adaptive role of chromosome size remains largely unknown. Possible explanations for this knowledge gap include limited data associated with genomic traits for the species of interest (making it impossible to estimate chromosome size), the absence of suitable phylogenetic data and large evolutionary distances separating focal species which might obscure signals associated with important biological drivers.
The cyperid clade (families Cyperaceae, Juncaceae and Thurniaceae: Linder and Rudall, 2005; Roalson et al., 2007; Escudero et al., 2016; Baez et al., 2020; Larridon et al., 2021a) is a species-rich lineage sharing a similar grass-like growth form, with species occupying a range of habitats across a nearly global distribution (Goetghebeur, 1998; Larridon et al., 2021b). Variation in chromosome number and genome size, and thus chromosome size, has been shown to be high and to vary within relatively short phylogenetic time scales in this clade (e.g. Roalson, 2008; Márquez-Corro et al., 2019b; Elliott et al., 2022). This large variation coincides with the frequent occurrence of species with holocentric chromosomes.
The few studies focusing on variation in chromosome size with different growing conditions have often made comparisons between tropical and temperate regions, but the pattern of smaller chromosome sizes in plants from tropical regions might have been influenced by the species sampled in these studies (see Avdulov, 1931; Levin and Funderburg, 1979). Tropical regions with less seasonal climatic fluctuations compared with temperate regions might provide more stable growing conditions for plants (Janzen, 1967). Given the direct relationship between chromosome number and recombination rates (‘evolutionary potential’) in cyperids, low chromosome numbers (and, thus, large chromosome size) will be selected for where evolutionary innovations could be maladaptive, such as in the tropics. On the other hand, high chromosome numbers (and, thus, small chromosome size) might be selected for where evolutionary potential is required (Wang et al., 2019), as in the temperate regions.
Here, we capitalize on an international collaborative effort to obtain genome size estimates of species across the phylogeny of cyperids. Our primary goal is to determine whether chromosome size might have an adaptive role in this clade, which we achieve by combining genome size and chromosome number estimates to calculate mean chromosome sizes, followed by comparing these sizes with the different environmental growing conditions associated with each species. We also test the prediction that the mean chromosome size of species in tropical climates is smaller than that of species in temperate climate based on results presented in the small number of previous studies on the topic (see Stebbins, 1966; Levin and Funderburg, 1979). Finally, we hypothesize that plants with small chromosomes will be selected for in temperate regions where high evolutionary potential is needed, whereas large chromosomes will be selected for in the tropics.
MATERIALS AND METHODS
Taxon sampling, nomenclature and phylogenetic reconstruction
We focused our sampling on the cyperids (families Cyperaceae, Juncaceae and Thurniaceae) and referred to the World Checklist of Selected Plant Families (WCSP; Govaerts et al., 2021) to guide nomenclature for the species included in this study. Infraspecific taxa (i.e. subspecies and varieties) were included when possible, as genome size and chromosome number in the cyperids can vary amongst these ranks (Lee et al., 2019). Genome and mean chromosome sizes were compared among the Juncaceae, Thurniaceae and 24 tribes of Cyperaceae, as species numbers were large enough for meaningful comparisons at this phylogenetic scale. Species were not divided into genera for the purpose of this study, as several genera are very small (e.g. Oreojuncus, Patosia and Thurnia; WCVP, 2022) and taxonomy at the generic level is not fully resolved in these three families; for example, Juncus—the largest genus in Juncaceae—is not monophyletic, as indicated by a recent proposal to split it into six separate genera (Drábková and Vlček, 2009; Brožová et al., 2022 ; Elliott et al., in press).
Details of the data utilized in this study are noted in Supplementary data Appendix S1 and Table S1, while the methods used to create the phylogenetic reconstruction are outlined in Elliott et al. (2022). We pruned species from the phylogeny to match those in the genome size data set using R v.4.0.2 (R Core Team, 2020) with the function ‘drop.tip’ in package APE v.5.4-1 (Paradis et al., 2004).
Genome size estimations
Flow cytometric samples were prepared from fresh leaves and processed according to the two-step protocol of Otto et al. (1981) and Galbraith et al. (1983) with concentrations of buffers, dyes and other modifications described in detail by Šmarda et al. (2008). The samples were measured on a CyFlow cytometer (Partec GmbH) using one of the internal standards: Carex acutiformis (2C = 799.93 Mbp), Solanum lycopersicum ‘Stupické polní tyčkové rané’ (2C = 1696.81 Mbp), Pisum sativum ‘Ctirad’ (2C = 7841.27 Mbp; Veselý et al., 2012) and Bellis perennis (2C = 3089.89 Mbp; Šmarda et al., 2014), whose genome sizes were derived from comparisons with the completely sequenced Oryza sativa subsp. japonica ‘Nipponbare’ (International Rice Genome Sequencing Project, 2005); for details see also Temsch et al. (2022; Table 1). Propidium iodide was used as a fluorochrome. Typically, the genome size of each species is represented by one sample measured three times on consecutive days. Genome size estimates of repeated measurements were averaged per sample, and these values were further averaged per each species in cases where two or more individuals were measured for a given species (Supplementary data Appendix S1).
Table 1.
Results from multiple phylogenetic generalized least squares (PGLS) analyses with holoploid genome size (1C: Mbp) as the response variable
| Predictor variable(s) | logLik | Estimate | s.e. | t-value | R 2 adj | AIC | delta AIC | AICw | P |
P
(corrected) |
|---|---|---|---|---|---|---|---|---|---|---|
| Prec_dry_month | –224.93 | 0.03 | 0.01 | 2.42 | 0.01 | 453.87 | 0 | 0.31 | 0.02 | 0.17 |
| EOO | –225.42 | –0.02 | 0.01 | –2.22 | 0.01 | 454.83 | 0.96 | 0.19 | 0.03 | 0.17 |
| Prec_cold_quarter | –225.63 | 0.01 | 0 | 2.11 | 0.01 | 455.27 | 1.40 | 0.15 | 0.04 | 0.17 |
| Prec_ann | –226.31 | 0.09 | 0.05 | 1.77 | 0 | 456.61 | 2.75 | 0.08 | 0.08 | 0.26 |
| SOC | –226.64 | 0.06 | 0.04 | 1.56 | 0 | 457.29 | 3.42 | 0.06 | 0.12 | 0.28 |
| Latitude | –226.72 | 0.03 | 0.02 | 1.51 | 0 | 457.44 | 3.57 | 0.05 | 0.13 | 0.28 |
| Intercept | –227.87 | NA | NA | NA | 0 | 457.73 | 3.87 | 0.04 | NA | NA |
| Diurnal_range | –227.36 | –0.09 | 0.08 | –1.01 | 0 | 458.72 | 4.85 | 0.03 | 0.31 | 0.58 |
| pH | –227.62 | –0.15 | 0.22 | –0.7 | 0 | 459.24 | 5.37 | 0.02 | 0.48 | 0.69 |
| P | –227.85 | –0.01 | 0.06 | –0.15 | 0 | 459.71 | 5.84 | 0.02 | 0.88 | 0.98 |
| Temp_ range |
–227.85 | –0.01 | 0.07 | –0.16 | 0 | 459.71 | 5.84 | 0.02 | 0.88 | 0.98 |
| Niche | –227.86 | 0 | 0.01 | –0.13 | 0 | 459.72 | 5.85 | 0.02 | 0.90 | 0.98 |
| Elevation | –227.46 | 0 | 0 | 0.90 | 0 | 458.92 | 5.05 | 0.02 | 0.37 | 0.60 |
Results are ranked in order of descending AICw values. Analyses were performed using the R package CAPER v.1.0.1 (Orme, 2013) and all variables were log transformed, except for Prec_dry_month, Prec_cold_quart, Niche, Latitude and Elevation, which were transformed by the square root. Lambda was fixed at 0.872. The following abbreviations are used to represent the environmental variables included in these analyses: pH (soil pH); ‘SOC’ (soil organic carbon); ‘P’ (available phosphorus); ‘Diurnal_range’ (mean diurnal air temperature range); ‘Temp_range’ (annual range of air temperature); ‘Prec_ann’ (annual precipitation); ‘Prec_dry_month’ (precipitation of the driest month); ‘Prec_cold_quart’ (precipitation of the coldest quarter); ‘EOO’ (extent of occurrence) and ‘Niche’ (niche size). Corrections for multiple comparisons were implemented using the method of Benjamini and Hochberg (1995), and the alpha value was set at 0.05. Only species with a minimum number of 25 observations were included in the analyses (494 observations retained).
We analysed genome sizes from 594 individual plant specimens representing 436 species of cyperids (Supplementary data Appendix S1). Genome sizes for a further 526 species of cyperids were extracted from published studies (see Supplementary data Appendix S1), while ensuring not to include duplicate records. To allow comparisons with major groupings of plants, we extracted genome size data available from the Plant DNA C-values database (Pellicer and Leitch, 2020) for all seed plant species. Throughout this study, we report holoploid (1C: sensuGreilhuber et al., 2005) genome sizes in megabase pairs (Mbp: 1 pg of 1C-value = 978Mb; Doležel et al., 2003).
Chromosome numbers
The respective chromosome numbers for the species whose genome sizes were included in our study (Supplementary data Appendix S1) were retrieved from the review of Cyperaceae chromosome counts (Roalson, 2008) and from the Chromosome Counts Database (CCDB; Rice et al., 2015). Counts were assessed on a per species basis by considering reliability of sources, pseudoreplication and variation in the data. In general, the median value was selected for each species; however, species were omitted from the chromosome number dataset if the variation in counts was too high (i.e. mid-values differing by >25%), as it was impossible to attribute a count value to a specific genome size because of possible differences in ploidy. For the purposes of this study, the term ‘mean chromosome size’ refers to holoploid genome size multiplied by two (2C) divided by diploid chromosome number (2n).
Genome size, mean chromosomes size and chromosome number statistics
The phylogenetic signal of both genome and mean chromosomes size across the cyperids was calculated with the K statistic (Blomberg et al., 2003). Genome and mean chromosome sizes were compared amongst the major lineages of seed plants (Poales, non-Poales and Lentibulariaceae) and cyperids using the Tukey–Kramer procedure (Tukey, 1949; Dunnett, 1980) with the function TukeyHSD in base R. This function was also used to compare genome and mean chromosome sizes among tribes of Cyperaceae and the families Juncaceae and Thurniaceae. Associations between genome size and diploid chromosome number, as well as mean chromosome size and diploid chromosome number, were evaluated using phylogenetic generalized least square (PGLS) regressions (Freckleton et al., 2002) with the R package CAPER v.1.0.1 (Orme, 2013). Maximum likelihood estimation was used to optimize branch lengths based on the data (lambda = ‘ML’).
Distribution data
We downloaded occurrence records for the species with genome size estimations from the Global Biodiversity Information Facility [(https://www.gbif.org: GBIF Occurrence Downloads doi.org/10.15468/dl.w6gqpk and doi.org/10.15468/dl.yw34a8 (20 October 2020) and doi.org/10.15468/dl.f9cc43 (26 October 2020)] with package RGBIF v.3.5.0 (Chamberlain et al., 2020). For each species, we included ‘PRESERVED_SPECIMEN’, ‘HUMAN_OBSERVATION’ and ‘OBSERVATION’ data as categorized by GBIF.
We removed problematic records flagged by R package COORDINATECLEANER v.2.0-18 (Zizka et al., 2019). The filter we created with this package included records without a latitude/longitude reference system, with zeros in the co-ordinates and the radius around (0/0), with identical latitude/longitude values, outside a latitude/longitude co-ordinate system, with co-ordinates in the oceans and duplicate records. We also filtered occurrence records within 10 km of country capitals, 1 km of geographic centroids of political units, 100 m of biodiversity institutions and 0.5° radius of the GBIF headquarters in Copenhagen.
To further remove any problematic records downloaded from GBIF, we then passed the occurrence data through an additional geographic filter developed by ourselves, which was based on global-scale distribution data as specified as Level-3 Continental and Regional Codes by the International Working Group on Taxonomic Databases for Plant Sciences (TDWG; Brummitt et al., 2001). Level-3 TDWG regions record plant distributions based on geographic units at approximately the level of countries (Brummitt et al., 2001). Codes for each species were downloaded from the WCSP website on 21 January 2021 with a Python (v.3.7.6) script using packages BEAUTIFULSOUP v.4-4.8.2 (Richardson, 2019) and MECHANICALSOUP v.0.12.0 (https://mechanicalsoup.readthedocs.io/en/stable/index.html). For each species, we mapped the cleaned species occurrence records on maps showing the corresponding TDWG regions for that species. We visually inspected the maps and added TDWG regions in cases where they were conspicuously missing, as supported by floras (e.g. Flora of North America Editorial Committee, 1993) and our understanding of the distributions of different species (Supplementary data Appendix S2). We also removed regions that corresponded to naturalized areas or did not seem feasible (Supplementary data Appendix S3). Finally, GBIF occurrence records that were outside of the TDWG regions for each species were removed from the dataset. Computational resources for this and further analyses were supplied on the Czech National Grid Infrastructure (NGI), and all analyses were conducted using R v.4.0.3 with JupyterLab.
Climatic and edaphic data
We extracted 19 bioclimatic variables from the ‘Climatologies at high resolution for the earth’s land surface areas’ database (CHELSA: https://chelsa-climate.org/), which is a high-resolution database (30 arc sec, approx. 1 km) that uses statistical downscaling in its calculations based on observations from 1979 to 2013 (Karger et al., 2017). In addition, two other extended climatic variables were downloaded from CHELSA, the heat sum of growing degree days above 5°C (GDD: CHELSA_gdd_5_1979.2013) and potential evapotranspiration (PET). A temperature threshold of 5°C was chosen for GDD since many cyperid species are capable of growing at relatively low temperatures (Flora of North America Editorial Committee, 1993; WCVP, 2022). In addition, mean annual PET was calculated by applying a mask to the areas corresponding to oceans and by then summing the 12 monthly raster layers.
Soil total nitrogen (N), bulk density of fine earth fraction (Bdod), cation exchange capacity (CEC), organic carbon (SOC) and pH in water (pH) were extracted from the SoilGrids250m v.2.0 database (de Sousa et al., 2020) hosted on the WebDAV server (https://www.isric.org/explore/soilgrids/soilgrids-access). The SoilGrids 2.0 maps have a spatial resolution of 250 m cell size and are created using machine learning methods (de Sousa et al., 2020). For each soil variable, three different soil depths (0–5, 5–15 and 15–30 cm) were extracted and the weighted mean was calculated based on relative soil depths.
Soil phosphorus (P) including labile inorganic P, organic P, occluded P, secondary mineral P, apatite P and total P were downloaded from EARTHDATA (https://daac.ornl.gov/SOILS/guides/Global_Phosphorus_Dist_Map.html), with a resolution of 0.5° (Yang et al., 2013). We then calculated the sum of the labile inorganic P, organic P and secondary mineral P to represent total available P to plants (Yang et al., 2013).
Geographic and environmental area estimations
Species were first classified as either temperate or tropical by overlaying the cleaned co-ordinates downloaded from GBIF for each species on a map showing the TDWG regions where the species is reported to occur. The classification between the temperate and tropical category incorporated broad climatic associations, where temperate species were considered to occur in higher latitudes and altitudes. Species occurring in both temperate and tropical regions were omitted from subsequent analyses (98 omitted species).
For the analyses using continuous environmental variables, 1000 occurrence records were randomly selected with replacement for each species in our genome size dataset, and the corresponding elevations were extracted from the GeoNames geographical database (http://www.geonames.org) with the R package RGBIF (Chamberlain et al., 2020). A mean elevation value was then calculated for each species.
We then calculated two separate metrics of geographic distribution for each species: area of occupancy (AOO) and extent of occurrence (EOO) using functions ‘aoo’ and ‘eoo’ in the R package RED v.1.5.0 (IUCN, 2001; Cardoso, 2017). The AOO was calculated as the area of all known cells (resolution 2 × 2 km) occupied by a species, whereas we calculated EOO as the minimum convex polygon covered by the cleaned GBIF occurrence records for a species (IUCN, 2001). We chose two thresholds of occurrence records (25 and 50) for the calculation of the geographic metrics included in our study after comparing the effects of different minimum threshold values (25, 50, 75 and 100) on the number of species retained for subsequent analyses.
In addition, a niche size metric based on alpha hull volumes (Edelsbrunner and Mücke, 1994; Gardiner et al., 2018) was calculated for each species. We first reprojected the edaphic rasters to match the extent, projection and resolution of the climatic rasters using the ‘projectRaster’ function in R package RASTER v.3.4-5 (Hijmans, 2020). Total P available to plants was omitted from this analysis because of the relatively low resolution of the raster layer compared with the other climatic and edaphic variables (Yang et al., 2013; Karger et al., 2017; de Sousa et al., 2020). Next, we conducted a principal components analysis (PCA) using the ‘rasterPCA’ function in R package RSTOOLBOX v.0.2.6 (Leutner et al., 2017) on the 26 variables. Values corresponding to the cleaned species occurrence data were then extracted from the three PC axes explaining the greatest variance, while removing duplicate co-ordinates from the same grid cell to address spatial clustering (Varela et al., 2014). Finally, alpha hull volumes based on the first three PC axes were calculated for each species with the ‘ashape3d’ (alpha = 1), followed by the ‘volume_ashape3d’ function in the R package ALPHASHAPE3D v.1.3.1 (Lafarge and Pateiro-Lopez, 2020). As with the AOO and EOO metrics, only species with a minimum of 25 cleaned occurrence records were used to calculate alpha hull volumes.
Association of genome and mean chromosome sizes with different predictor variables
Differences were tested between species classified as tropical and temperate with one-factor analyses of variance (ANOVAs) using the ‘aov’ function in base R. The ‘pgls’ function in R package CAPER v.1.0.1 (Orme, 2013) with lambda specified as ‘ML’ was used to incorporate evolutionary relationships among species into the analyses. All genome and mean chromosome size values were log-transformed prior to both analyses.
We assessed collinearity between the 31 predictor variables included in this study (AOO, EOO, niche size, elevation and latitude combined with the 26 climatic and edaphic variables) by calculating Pearson’s correlation coefficients in base R. Variables were considered collinear if their pairwise correlation coefficients were either ≤ –0.70 or ≥0.70, which is a common threshold used in ecological studies (Dormann et al., 2013).
To examine the relationship between the different environmental variables and genome/mean chromosome size, we conducted a series of PGLS regressions. We considered genome and mean chromosome size as the response variables in these analyses based on our reasoning that plants were filtered into their niches based on various environmental variables. These analyses were individually conducted with the selected sub-set of variables that were not collinear, as well as a model including only the intercept. Only those species with a minimum threshold of at least 25 (and 50) cleaned occurrence records per species were included in the analyses. Data were either log- or square-root transformed to meet the assumptions of homoscedasticity and normality of the residuals. To allow for comparisons among models with different predictor variables, a common lambda value was selected by first running each model using the ‘pgls’ function and then calculating the likelihood profiles of branch length transformations using the ‘pgls.profile’ function in R package CAPER v.1.0.1 (Orme, 2013). The mean of the optimal lambdas from each of these preliminary models was then specified as the lambda value for each of the final models, which were then compared with weighted Akaike information criterion (AICw) values. The Benjamini and Hochberg correction (Benjamini and Hochberg, 1995) was applied to P-values to correct for multiple tests for both the genome and mean chromosome size sets of analyses.
RESULTS
Phylogenetic representation
The phylogenetic reconstruction generated for this study is currently the most comprehensive for the cyperid clade, including approx. 30% of all recognized species, as well as species corresponding to all 24 tribes of Cyperaceae, in addition to all genera of Juncaceae and Thurniaceae (Supplementary data Appendix S1; Fig. S1; Table S2).
Genome sizes
Our analyses included genome size estimations for 757 (approx. 13%) recognized cyperid species (Supplementary data Table S3). Although we estimated genome sizes for the majority of tribes of Cyperaceae (18 of 24), as well as both the Juncaceae and Thurniaceae families (Supplementary data Table S3), the sampling of genome size was generally limited in coverage, with only three tribes having genome size estimations for >50% of their species (Bolboschoeneae, Dulichieae and Schoenoplectieae; Supplementary data Table S3).
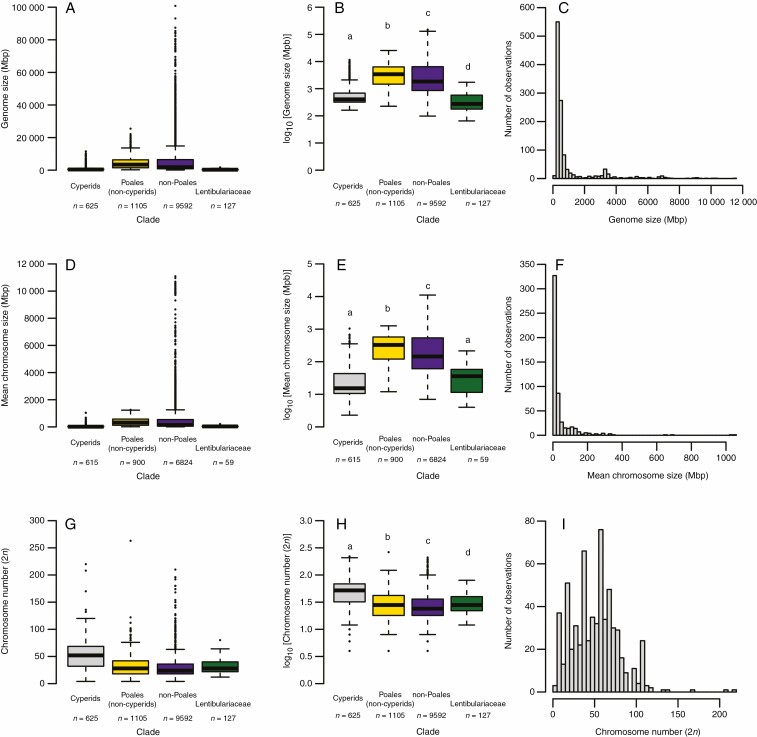
We examined the variation in 1C genome sizes across the cyperid clade and found a 72-fold difference from 161 Mbp in Juncus maritimus Lam. (family Juncaceae) to 11 547 Mbp in Schoenus aureus T.L.Elliott & Muasya (tribe Schoeneae, family Cyperaceae), with a mean of 1020 Mbp (Supplementary data Appendix S1). However, the majority of cyperids have small genome sizes (Fig. 1C). Compared with other major groupings of plants included in our analyses, genome sizes were smaller for the cyperid clade relative to both the non-cyperid Poales and non-Poales (Tukey–Kramer: P <0.001, for both comparisons; Fig. 1A, B). We also compared genome sizes in the cyperids with those of the Lentibulariaceae, which have been reported to have miniature genomes (Veleba et al., 2014; 2020). Our results showed that genome sizes for the cyperid clade were larger than those of the Lentibulariaceae (Tukey–Kramer: P <0.001; Fig. 1A, B).
Fig. 1.
Comparisons between holoploid (1C; A and B) genome sizes, mean chromosome sizes (2C/2n; D and E) and chromosome numbers (2n; G and H) among cyperids (grey), Poales (excluding cyperids: yellow), non-Poales seed plants (violet) and Lentibulariaceae (green). The results from raw data are shown in (A), (D) and (G), whereas (B), (E) and (H) are based on log10-transformed data. Unique letters above the boxplots indicate significant differences among groups, and numbers at the bottom of the boxplots show the sample size (species number) of each group. The distribution of genome sizes, mean chromosome sizes and chromosome numbers within cyperids are shown in (C), (F) and (I), respectively. The boxes cover 50% of the data values ranging between the 25th and 75th percentiles, whereas the whiskers above and below each box represent the interquartile range multiplied by 1.5. The line within each box represents the median, and outlying values are indicated by small circles. The alpha value was set at 0.05.
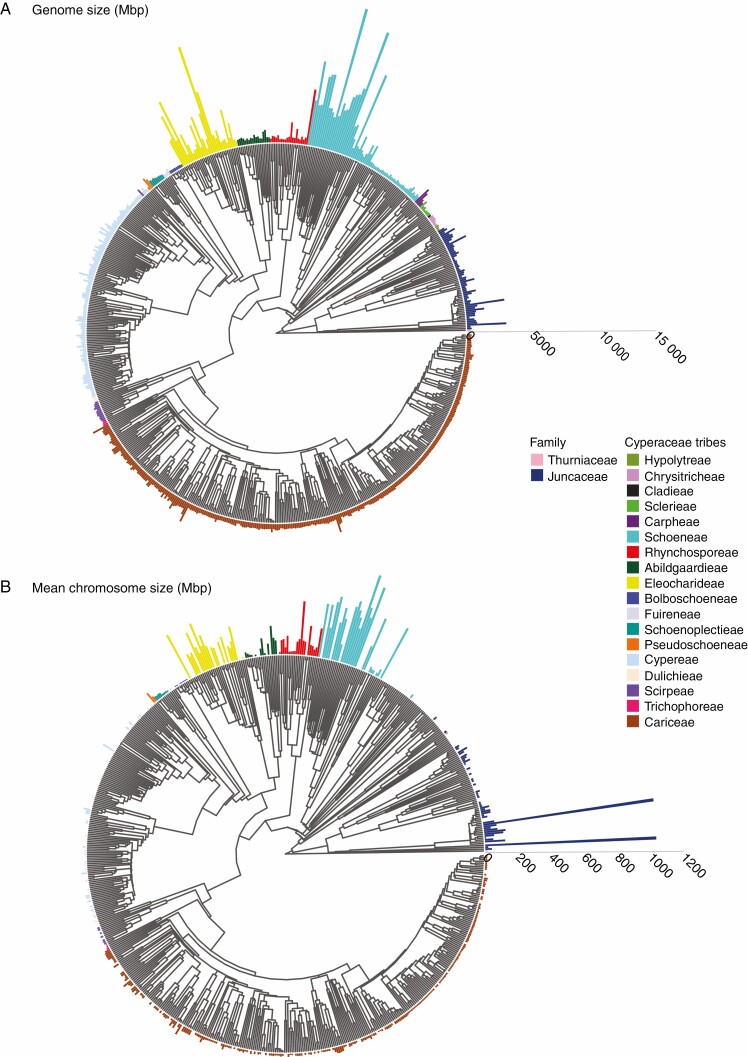
Our analyses revealed differences in genome sizes across the clades of cyperids included in our study. Mean 1C genome size was <500 Mbp for the majority of clades of cyperids; however, mean values >500 Mbp were recorded for tribes Abildgaardieae, Carpheae, Eleocharideae, Pseudoschoeneae, Rhynchosporeae and Schoeneae, as well as the Juncaceae (Fig. 2; Supplementary data Fig. S2). We found that the genome sizes of tribes Eleocharideae and Schoeneae were significantly higher than those of Juncaceae and ten tribes of Cyperaceae (Abildgaardieae, Bolboschoeneae, Chrysitricheae, Cariceae, Cypereae, Fuireneae, Rhynchosporeae, Schoenoplectieae, Scirpeae and Sclerieae: Tukey–Kramer: P <0.05, Fig. 2; Supplementary data Fig. S2). Across the cyperid clade, the phylogenetic signal of genome size was significant but not high (K = 0.254, P = 0.002: Fig. 2A).
Fig. 2.
Holoploid genome (A) and chromosome (B) sizes mapped onto the phylogeny of cyperids. The length of the bars adjacent to the tips of the phylogeny represent the genome (1C: Mbp) and chromosome size (2C/2n: Mbp) of each species, respectively, according to the scale bar on the right side of each phylogeny. Species without chromosome number data do not have bars adjacent to them in (B). Family (Juncaceae and Thurniaceae) and tribal associations (Cyperaceae) are indicated by the colours given in the key. Genome and chromosome sizes were mapped onto the phylogeny with the R package GGTREEEXTRA v.1.0.4 (Xu et al., 2021).
Mean chromosome sizes
Similar to genome size, we examined patterns in mean chromosome sizes across seed plants and within the cyperid clade. Mean chromosome sizes were smaller for the cyperid clade compared with both the non-Poales seed plant and non-cyperid Poales groupings (Tukey–Kramer: P <0.001, for both comparisons; Fig. 1D, E). Furthermore, chromosomes in cyperids were as small as the miniature ones found in Lentibulariaceae (Fig. 1D, E).
Within the cyperid clade, we found a 455-fold difference in mean chromosome sizes across the clade, with sizes ranging from 2.3 Mbp in Cyperus cyperoides (L.) Kuntze to 1042 Mbp in Luzula purpureosplendens Seub. In addition, the distribution of chromosome sizes was strongly right skewed (Fig. 1F). Mean chromosome sizes were higher in Juncaceae than in tribe Cariceae, whereas sizes were also higher in tribe Eleocharideae compared with tribes Cariceae and Cypereae (Fig. 2B; Supplementary data Fig. S3). Moreover, mean chromosome sizes were higher in tribe Schoeneae compared with tribes Abildgaardieae, Bolboschoeneae, Cariceae, Cypereae, Dulicheae, Rhynchosporeae, Schoenoplectieae, Scirpeae and Trichophoreae (Fig. 2B; Supplementary data Fig. S3). Phylogenetic signal across the cyperids for mean chromosome sizes was relatively low (K = 0.178, P = 0.026: Fig. 2B).
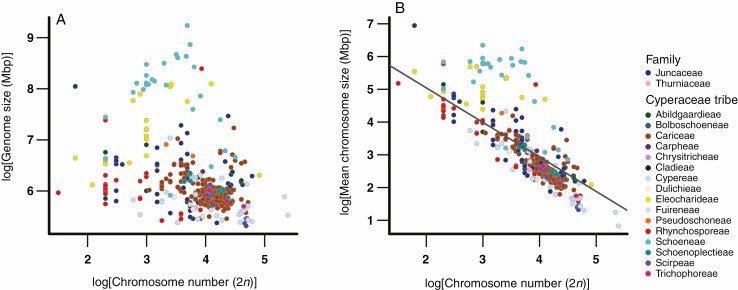
We also compared the three genomic traits (genome size, chromosome size and chromosome number) with each other. Our results indicated that there was not an association between 1C holoploid genome size and chromosome number in the cyperid clade (Fig. 3A; Supplementary data Figs S4, S5 and S6). However, we did find a strong negative association between chromosome number and mean chromosome size (PGLS: estimate = –1.05, R2adj = 0.57, P <0.001: Fig. 3B).
Fig. 3.
The association between diploid chromosome numbers and holoploid genome size (1C; A), as well as mean chromosome size (2C/2n; B) across the cyperids. Values corresponding to the Juncaceae and 16 tribes of Cyperaceae are indicated by the colours shown to the right-hand side of (B). All data were log transformed. Statistical significance (P <0.05) between variables, as determined by phylogenetic generalized least squares (PGLS) performed using the R package CAPER v.1.0.1 (Orme, 2013), is indicated by a solid regression line.
Correlations between environmental variables
Our examination of Pearson’s correlation coefficients between sets of environmental variables revealed collinearity in both the genome and mean chromosome size datasets. Amongst the environmental variables in the genome size dataset, latitude (Lat), soil organic carbon (SOC), annual range of air temperature (Temp_range), annual precipitation (Prec_ann) and precipitation of the driest month (Prec_dry_month) were collinear (r < –0.70 or r >0.70) with at least one other variable when species with a minimum number of 25 observations were included in the analysis (Supplementary data Table S4). These results slightly changed when the minimum threshold of observations increased to 50, as ‘Bdod’ was correlated with more variables compared with ‘SOC’ (Supplementary data Table S5). Pearson’s correlation coefficients between sets of variables included in the mean chromosome size datasets having both 25 and 50 minimum observations revealed similar patterns, with ‘Lat’, ‘Bdod’, ‘Temp_range’, ‘Prec_ann’ and ‘Prec_dry_month’ all being collinear with at least one other variable (Supplementary data Tables S6 and S7). We removed AOO from the genome and mean chromosome size analyses because of its high correlation (r = 0.98) with the number of GBIF observations for each species. Based on these patterns of collinearity, 12 variables were retained for the analyses focusing on the environmental correlates of genome size (25 minimum observations), whereas 11 variables were retained for those analyses including genome size (50 minimum observations), as well as mean chromosome sizes with both 25 and 50 minimum observations (Tables 1 and 2; Supplementary data Tables S8 and S9).
Table 2.
Results from multiple phylogenetic generalized least squares (PGLS) analyses with mean chromosome size (2C/2n) as the response variable
| Predictor variable(s) | logLik | Estimate | s.e. | t-value | R 2 adj | AIC | delta AIC | AICw | P |
P
(corrected) |
|---|---|---|---|---|---|---|---|---|---|---|
| EOO | –241.55 | –0.11 | 0.02 | –4.88 | 0.07 | 487.11 | 0 | 1 | 0 | 0 |
| Niche | –249.36 | –0.05 | 0.02 | –2.73 | 0.02 | 502.73 | 15.62 | 0 | 0.01 | 0.06 |
| Prec_ann | –250.46 | 0.24 | 0.10 | 2.29 | 0.01 | 504.92 | 17.81 | 0 | 0.02 | 0.08 |
| P | –251.84 | 0.03 | 0.02 | 1.56 | 0 | 507.69 | 20.58 | 0 | 0.12 | 0.36 |
| Intercept | –253.07 | NA | NA | NA | 0 | 508.13 | 21.02 | 0 | NA | NA |
| Latitude | –252.07 | –0.18 | 0.13 | –1.41 | 0 | 508.13 | 21.02 | 0 | 0.16 | 0.36 |
| Prec_dry_month | –252.29 | 0.07 | 0.05 | 1.24 | 0 | 508.58 | 21.48 | 0 | 0.22 | 0.36 |
| Diurnal_range | –252.36 | 0.20 | 0.17 | 1.19 | 0 | 508.72 | 21.61 | 0 | 0.24 | 0.36 |
| Bdod | –252.37 | 0.35 | 0.30 | 1.18 | 0 | 508.74 | 21.63 | 0 | 0.24 | 0.36 |
| pH | –252.68 | 0.34 | 0.39 | 0.88 | 0 | 509.36 | 22.25 | 0 | 0.38 | 0.51 |
| Elevation | –252.86 | 0.02 | 0.04 | 0.64 | 0 | 509.72 | 22.61 | 0 | 0.52 | 0.61 |
| Temp_range | –252.89 | –0.09 | 0.15 | –0.59 | 0 | 509.78 | 22.68 | 0 | 0.56 | 0.61 |
Results are ranked in order of ascending AIC values. Analyses were performed using the R package CAPER v.1.0.1 (Orme, 2013) and all variables were log transformed, except for P and Niche, which were transformed by the square root. Lambda was fixed at 0.906. The following abbreviations are used to represent the environmental variables included in these analyses: pH (soil pH); ‘Bdod’ (bulk density of the fine earth fraction); ‘P’ (available phosphorous); ‘Diurnal_range’ (mean diurnal air temperature range); ‘Temp_range’ (annual range of air temperature); ‘Prec_ann’ (annual precipitation); ‘Prec_dry_month’ (precipitation of the driest month); ‘EOO’ (extent of occurrence); and ‘Niche’ (niche size). Corrections for multiple comparisons were implemented using the method of Benjamini and Hochberg (1995), and the alpha value was set at 0.05. Only species with a minimum number of 25 observations were included in the analyses (300 observations retained).
Total N (N) was dropped from both sets of analyses because of its correlation with ‘Lat’ and for not being an appropriate measure of N available to plants, although it has been linked to genome sizes in previous studies (e.g. Kang et al., 2015; Guignard et al., 2017).
Environmental associations
Temperate and tropical species did not differ in genome sizes (P = 0.41; Supplementary data Fig. S7A), and this association did not change when evolutionary relationships were incorporated into the analyses (pgls: P = 0.20). Tropical species had larger mean chromosome sizes compared with temperate species (P < 0.001; Supplementary data Fig. S7B), but this association disappeared after correcting for evolutionary relationships among species (pgls: P = 0.32).
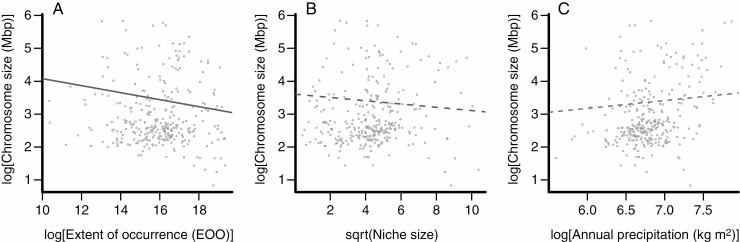
Based on the genome size analysis with a minimum of 25 observations, the environmental variable with the highest AICw value was ‘Prec_dry_month’; however, this variable explained little variation in genome size and was not significant (R2adj = 0.01, estimate = 0.03, P corrected = 0.17, Table 1). The EOO had the highest AICw for the genome size analysis with a minimum of 50 observations, as well as for both analyses including mean chromosome size (Table 2; Supplementary data Tables S8 and S9). While EOO explained little variation in genome size when the minimum threshold of observations was 50 (R2adj = 0.01, estimate = –0.03, P corrected = 0.06; Supplementary data Table S8), this variable explained more variation in the analyses of mean chromosome size for both threshold levels (minimum 25 observations: R2adj = 0.07, estimate = –0.11, P corrected <0.001, Table 2; minimum 50 observations: R2adj =0.08, estimate = –0.12, P corrected <0.001, Supplementary data Table S9). The majority of variables in all other analyses explained little variation in genome or mean chromosome size, with the exception of niche size (‘Niche’: R2adj = 0.02, estimate = –0.05, P corrected = 0.06) and ‘Prec_ann’ (R2adj = 0.02, estimate = 0.24, P corrected = 0.08) in the mean chromosome size analysis with 25 minimum observations (Table 2; Fig. 4), as well as ‘Prec_ann’ (R2adj = 0.02, estimate =0.11, P corrected = 0.08) and ‘Prec_dry_month’ (R2adj = 0.02, estimate = 0.05, P corrected = 0.08) in the mean chromosome size analysis with 50 minimum observations (Supplementary data Table S9).
Fig. 4.
The association between mean chromosome size (2C/2n) and extent of occurrence (A), niche size (B) and annual precipitation (C) in cyperids as determined by phylogenetic generalized least squares (PGLS) performed using the R package CAPER v.1.0.1 (Orme, 2013). These three predictor variables received relatively high AICw and log-likelihood scores, and corrections for multiple comparisons were implemented using the method of Benjamini and Hochberg (1995). Significant relationships are indicated by solid regression lines, whereas marginally significant relationships between variables are indicated by broken lines. All variables were log transformed, except for Niche size, which was square-root transformed.
DISCUSSION
Cyperids have chromosomes that are amongst the smallest in seed plants
Our results demonstrate that mean chromosome sizes in several cyperids are the smallest recorded in seed plants to date. Based on new data generated for this study, the lowest mean chromosome size in cyperids [Bolboschoenus robustus (Pursh) Soják: 1C = 198.0 Mbp; 2n = 107; mean chromosome size (2C/2n) = 3.7 Mbp: Supplementary data Appendix S1, population at Průhonice Botanic Garden] is slightly smaller than that of Genlisea nigrocaulis Steyerm. [1C = 86.0 Mbp; 2n = 40; mean chromosome size (2C/2n) = 4.3 Mbp] and Genlisea pygmaea A.St.-Hil. [1C = 179.0 Mbp; 2n = 80; mean chromosome size (2C/2n) = 4.3 Mbp), which have the smallest values in the Lentibulariaceae – a family reported to have among the smallest genome sizes across plants (Greilhuber et al., 2006; Veleba et al., 2014). Our data show that three species of Cyperus could have even smaller mean chromosome sizes [C. cyperoides (L.) Kuntze, 2.3 Mbp; C. brevifolius (Rottb.) Hassk., 3.7 Mbp; C. esculentus L., 3.5 Mbp], but these values are sensitive to intraspecific differences in cytotypes since the genome size and chromosome count estimations are based on different populations. In addition, a detailed RADseq linkage map and chromosome-level whole-genome sequencing suggest the presence of remarkably small chromosomes in Carex (Escudero et al., 2018; Can et al., 2020; Planta et al., 2022). When compared with other holocentric lineages, there are species with small mean chromosome sizes in the genus Drosera (Veleba et al., 2017), but this pattern in mean chromosome size has yet to be documented in other holocentric clades, such as Cuscuta subgenus Cuscuta (Neumann et al., 2021). Other clades of seed plants could have species with smaller individual chromosomes than those in the cyperids, as our measure of mean chromosome size overlooks chromosome size variation inside the karyotype of a species, with this variation being substantial in bimodal karyotypes (Plačková et al., 2022).
The difference in mean chromosome sizes across cyperids is large compared with other families of seed plants (Supplementary data Figs S8 and S9). This large difference results from a combination of species with many small, fragmented chromosomes in genera such as Carex and Cyperus (e.g. Nishikawa et al., 1984; Roalson et al., 2007; Roalson, 2008; Chung et al., 2012; Lipnerová et al., 2013; Márquez-Corro et al., 2021), and those with fewer, larger chromosomes, for example in Eleocharis (Roalson, 2008; Zedek et al., 2010) and Schoenus (Kaur et al., 2012; Elliott et al., 2022). Within other clades of holocentric plants, this divergent pattern in chromosome sizes among species has also been documented in the Australian Drosera (Veleba et al., 2017).
Relatively small genome sizes in the cyperids
Our comparison with genome sizes available in the Plant DNA C-values database showed that minimum genome sizes in both Cyperaceae and Juncaceae are amongst the smallest compared with 58 other families of seed plants (Supplementary data Figs S10 and S11). These results confirm the relatively small genome sizes in these two families documented by Šmarda et al. (2014), whose dataset was much more restricted in scope. Larger genome sizes in tribes Eleocharideae and Schoeneae of the Cyperaceae contribute to a higher overall mean genome size in cyperids compared with families such as Dipterocarpaceae and Lentibulariaceae (Veleba et al., 2014; Ng et al., 2016), although all major clades in our analyses included species with small genomes (Supplementary data Fig. S2). A relatively low rate of polyploidy across most clades of cyperids (see discussion below), as well as less accumulation or more effective removal of repetitive DNA sequences might contribute to lower genome sizes in this lineage compared with many clades of seed plants, but further research is required to confirm this.
An important factor that might contribute to small genome and mean chromosome sizes of cyperids, as well as the large variation in chromosome sizes is that the genomes of most species in this clade are thought to be composed of holocentric chromosomes. Although earlier several studies thought that the sister families Cyperaceae and Juncaceae (Roalson et al., 2007; Bureš et al., 2013; Escudero et al., 2016), along with Thurniaceae (Zedek et al., 2016), were holocentric, recently Baez et al. (2020) showed that there are monocentric chromosomes in Prionium serratum of the Thurniaceae (the chromosome type of the other three species placed in this family remains unstudied). In addition, monocentric chromosomes are present in some species of Juncus, in the Juncaceae (Guerra et al., 2019; Hofstatter et al., 2022); however, since Juncus is not monophyletic (Brožová et al., 2022; Elliott et al., 2022), it is important to investigate the presence of holocentricity across the family in a phylogenetic context to verify whether this trait is present in some major clades and not others. It is yet to be shown that the entire Cyperaceae family is holocentric, even though it is often assumed so (e.g. Roalson et al., 2007; Escudero et al., 2016; Márquez-Corro et al., 2019b; Zedek et al., 2022) and there is currently no published evidence to the contrary (Krátká et al., 2021), despite a claim by Nijalingappa (1974) of localized centromeres in several species of what is currently circumscribed as Schoenoplectiella (formerly Scirpus; WCVP, 2022). Assuming most species of cyperids are holocentric, a possible explanation for the small genome sizes across this clade is holokinetic drive, which involves the preferential size-dependent inheritance of homologous chromosomes during asymmetric meiosis (Bureš and Zedek, 2014; Veleba et al., 2017; Márquez-Corro et al., 2019a). Aside from holokinetic drive, another possibility is that genome sizes in these holocentric species further decrease when fission occurs, as there might be a loss in some DNA near breakage points before telomeres are formed near the new ends of chromosomes (Roalson et al., 2007). The formation of new telomeres at breakpoints appears to be very fast in holocentric chromosomes (Jankowska et al., 2015).
Strong negative association between chromosome size and number suggests the importance of holokinetic drive
Based on our expectations, holokinetic drive should result in a dichotomy, with species either having many small, fragmented chromosomes or fewer, larger chromosomes with more repetitive DNA sequences. Thus, there should theoretically be an inverse relationship between mean chromosome number and size in holocentric lineages. Our results, showing a pronounced inverse association between mean chromosome size and chromosome number, provide evidence that holokinetic drive is influencing chromosome evolution in cyperids. This association could be obscured, however, by the presence of polyploidy where chromosome number increases while there is a minimal change in chromosome size (Lipnerová et al., 2013). Although polyploidy is reported to be relatively rare in cyperids compared with other seed plant groups, it has been documented in, for example, Carex (Lipnerová et al., 2013), Cyperus (Roalson, 2008), Eleocharis (e.g. Harms, 1968; Bureš, 1998; Zedek et al., 2010; da Silva et al., 2017; Johnen et al., 2020), Rhynchospora (Vanzela et al., 1996; Luceño et al., 1998; Burchardt et al., 2020) and Schoenus (Kaur et al., 2012; Elliott et al., 2022) – of which the last three pertain to tribes with points that lie above the regression line in Fig. 3B.
Whether there is a positive or negative association between genome size and chromosome number in different genera of Cyperaceae has been a source of discussion in recent decades. Nishikawa et al. (1984) first reported a negative association between these two variables in Japanese species of Carex. Focusing on the section Cyperoideae (formerly Carex section Ovales; Global Carex Group et al., 2021), Chung et al. (2012) noted that the association between genome size and chromosome number was weakly negative at deeper phylogenetic scales (i.e. C. subgenus Vignea, excluding species from C. section Cyperoideae) but flat or weakly positive towards the tips of the phylogeny corresponding to C. section Cyperoideae, while variation in genome size in all species was low. A strong negative relationship between genome size and chromosome number in non-polyploid Carex species was observed by Lipnerová et al. (2013). We further explored the relationship between genome size and chromosome number in four cyperid clades that had at least ten observations in our dataset and had not been the subject of previous studies (Juncaceae and tribes Abildgaardieae, Cypereae and Rhynchosporeae). Based on these analyses, there is also a negative association between these two genomic traits in Juncaceae (Supplementary data Table S10). In contrast, the relationship between chromosome number and genome size has been found to be positive in Eleocharis and Schoenus – two genera with higher numbers of polyploid species (Zedek et al., 2010; de Souza et al., 2018; Elliott et al., 2022) – and in the Carex laevigata group, which suggests that it might be due to the proliferation and removal of repetitive DNA sequences (Escudero et al., 2015). At a wider phylogenetic scale, Global Carex Group et al. (2007) noted a negative relationship between chromosome number and genome size in Cyperaceae and Juncaceae combined, albeit with relatively limited sampling (approx. <1% of all species). Our results (see Fig. 3A), based on more comprehensive sampling across the cyperids (approx. 7%: species with both genome size and chromosome number estimates), did not reveal an association between chromosome number and genome size, as the negative and positive relationship characteristic of different genera obscured each other.
The adaptiveness of genome and mean chromosome size in cyperids
Nearly a century ago, various authors noted differences in chromosome sizes between tropical and temperate species. Although an adaptive function was suspected, they did not give explanations for these differences (e.g. Heitz, 1925–1926; Avdulov, 1931). The first speculation explaining this association that we are aware of was proposed by Stebbins (1966); however, he used the term chromosome size interchangeably with DNA content (i.e. genome size) in his interpretations, so his explanations are not strictly related to chromosome size. Whereas Levin and Funderburg (1979) justified that chromosome size is a good estimate of genome size by using data presented in previous studies to calculate a correlation coefficient of 0.90 between the two variables, in our study of cyperids the correlation was only 0.60. Thus, it cannot be assumed that temperate and tropical cyperid species have similar patterns for both genome and chromosome sizes. Once evolutionary relationships were incorporated into our analyses, there was no association between either genome or mean chromosome size and whether cyperids were classified as tropical or temperate. Possible reasons explaining this lack of association in cyperids include few or no differences in selective pressure on recombination rates between temperate and tropical regions, as well as the under-representation of tropical species in our dataset (Supplementary data Figs S7 and S12).
Our results focusing on different continuous environmental variables suggest that mean chromosome size, but not genome size, might have an adaptive function in cyperids. The extent of occurrence (i.e. geographical range size) and niche size were inversely associated with mean chromosome size in our phylogeny-corrected analyses, but these correlations were not recovered for genome size. A negative association between geographical range size and genome size might be expected based on the hypothesis of efficiency of selection, which – although developed for the population scale – could be extended to the global scale if geographical range size is considered a proxy for population size (Brown, 1984; Johnson, 1998; Lynch and Conery, 2003; Lynch, 2007). Based on this hypothesis, deleterious mutations can build up in smaller populations because of genetic drift; however, as population size grows, the relative effect of selection pressure increases, decreasing genome size via the purging of slightly deleterious mutations or unnecessary DNA (Lynch, 2007). A similar association might also be expected between niche and genome size, as a positive correlation between niche and geographical range size has been shown across numerous study systems (Gaston and Spicer, 2001; Slatyer et al., 2013; Cardillo et al., 2019). We did not find support for the hypothesis of efficiency of selection, but we did observe that chromosome size (not genome size) was inversely associated with both geographical range and niche size, which might similarly be associated with an increased adaptive capacity for species with small chromosomes given the unique characteristics of holocentric chromosomes assumed to be present in the majority of cyperids. Since crossover events are limited to a maximum of two per bivalent in holocentrics (Bureš et al., 2013), the breakage of these chromosomes into smaller fragments through chromosomal fission might function to increase genome-wide recombination rates, which could in turn increase the efficacy of selection and facilitate adaptation (Burt, 2000; Stapley et al., 2017). Thus, it is possible that small chromosomes are an adaptive feature that facilitates the survival of these plants in a wider range of growing conditions, leading to larger geographical range and niche sizes. Associations between chromosome size and other environmental variables were not evident in our study, suggesting that it is the combination of growing conditions captured in our geographical range and niche size metrics that these predominantly holocentric plants are adapting to, not individual edaphic or climatic factors.
Future directions
We think that the potential role of mean chromosome size and number in genome-wide recombination rates and how this association might function as an adaptive trait merits further study. As more genome size, chromosome number and phylogenetic sequence data are compiled for the cyperids, we are gaining a better understanding of genome size and chromosome evolution in this lineage, as well as the possible ecological consequences of these traits. More intense sampling in tropical regions and across undersampled lineages of cyperids in the future might reveal additional genome expansion and contraction events, which could help develop further comparative studies focusing on the possibility of holokinetic drive in this lineage.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Appendix S1: identifications, authorities, source of data, genome size and chromosome number estimations for cyperid specimens. Appendix S2: level 3 ‘Botanical countries’, as classified by Biodiversity Information Standards, which were manually added to the list of countries downloaded from the World Checklist of Selected Plant Families database. Appendix S3: level 3 ‘Botanical countries’, as classified by Biodiversity Information Standards, which were manually removed from the list of countries downloaded from the World Checklist of Selected Plant Families database. Figure S1: phylogenetic reconstruction of the cyperids. Figure S2: holoploid genome sizes across Cyperids divided into 17 tribes of Cyperaceae, as well as the Juncaceae family. Figure S3: mean chromosome sizes across cyperids divided into 16 tribes of Cyperaceae, as well as the Juncaceae family. Figure S4: chromosome number across cyperids divided into 16 tribes of Cyperaceae, as well as the Juncaceae family. Figure S5: fold difference in chromosome number across 48 families of seed plants. Figure S6: minimum chromosome number across 48 families of seed plants. Figure S7: comparisons between genome sizes and mean chromosome sizes in temperate vs. tropical cyperid species. Figure S8: fold difference in mean chromosome size across 48 families of seed plants. Figure S9: minimum mean chromosome sizes across 48 families of seed plants. Figure S10: fold difference in genome size across 60 families of seed plants. Figure S11: minimum holoploid genome sizes across 60 families of seed plants. Figure S12: number of species of cyperids plotted per TDWG region. Table S1: changes in species names compared with those presented in the phylogenetic reconstruction of Larridon et al., 2021a Table S2: representation of species in the phylogenetic reconstruction presented here, divided into families and tribes, compared with total species numbers estimated by other sources. Table S3: representation of species with genome size estimations, divided into families and tribes, compared with total species numbers estimated by other sources. Table S4: Pearson’s correlation coefficients >0.70 or less than –0.70 for 31 variables, including holoploid genome size for analyses with a minimum number of 25 observations per species. Table S5: Pearson’s correlation coefficients >0.70 or less than –0.70 for 31 variables, including holoploid genome size for analyses with a minimum number of 50 observations per species. Table S6: Pearson’s correlation coefficients >0.70 or less than –0.70 for 31 variables, including mean chromosome size for analyses with a minimum number of 25 observations per species. Table S7: Pearson’s correlation coefficients >0.70 or less than –0.70 for 31 variables, including mean chromosome size for analyses with a minimum number of 50 observations per species. Table S8: results from multiple phylogenetic generalized least squares (PGLS) analyses with holoploid genome size as the response variable. Table S9: results from multiple PGLS analyses with mean chromosome size as the response variable. Table S10: association between diploid chromosome number and holoploid genome size in three tribes of Cyperaceae and the family Juncaceae.
ACKNOWLEDGEMENTS
The authors thank L. Horová and J. Šmerda for flow cytometric analyses, as well as K. Bauters, J.-A. Viljoen, M.S. González-Elizondo, P. Goetghebeur, G. Mwachala and C.J. Prychid for supplying additional plant samples. Computational resources were supplied by the project ‘e-Infrastruktura CZ’ (e-INFRA CZ ID:90140) supported by the Ministry of Education, Youth and Sports of the Czech Republic. New South African specimens included in this project were collected under CapeNature permit numbers CN35‐28‐4102 and 0028‐AAA008‐ 00233; Province of the Eastern Cape Department of Economic Development, Environmental Affairs and Tourism permit numbers CRO 02/17CR and CRO 03/2017; Eastern Cape Parks & Tourism Agency permit number RA 0256; and South African National Parks permit numbers CRC/2016‐2017/011-2016/V1, CRC/2017‐2018/011-2016/V1 and ELLT/AGR/2019‐2022/011-2016/V2. T.L.E., P.B., A.M.M. and F.Z. conceived the study. Field sampling work was conducted by P.B., A.M.M., G.M., J.J.B., M.L., R.L.B., K.L.W., S.M.B., T.L.E. and Z.H. P.B. managed the flow cytometric analyses and compiled the data. T.L.E., analysed the data and prepared the first draft, while all other authors contributed to subsequent drafts. P.B., F.Z., M.E., P.S. and S.J. provided input for the analyses. The final manuscript was approved by all authors.
Contributor Information
Tammy L Elliott, Department of Botany and Zoology, Faculty of Science, Masaryk University, Kotlářská 2, 611 37 Brno, Czech Republic.
František Zedek, Department of Botany and Zoology, Faculty of Science, Masaryk University, Kotlářská 2, 611 37 Brno, Czech Republic.
Russell L Barrett, National Herbarium of New South Wales, Australian Institute of Botanical Science, Australian Botanic Garden, Locked Bag 6002, Mount Annan, New South Wales 2567, Australia.
Jeremy J Bruhl, Botany and N.C.W. Beadle Herbarium, School of Environmental and Rural Science, University of New England, Armidale, NSW 2351, Australia.
Marcial Escudero, Department of Plant Biology and Ecology, University of Seville, Reina Mercedes 6, 41012 Seville, Spain.
Zdenka Hroudová, Institute of Botany of the Czech Academy of Sciences, 252 43 Průhonice, Czech Republic; National Museum, Department of Botany, Cirkusová 1740, 193 00 Prague 9, Czech Republic.
Simon Joly, Montreal Botanical Garden, 4101, Sherbrooke East, Montreal, QC H1X 2B2, Canada; Institut de recherche en biologie végétale, Département de sciences biologiques, Université de Montréal, 4101, Sherbrooke East, Montreal, QC H1X 2B2, Canada.
Isabel Larridon, Royal Botanic Gardens, Kew, Richmond, Surrey TW9 3AE, UK; Systematic and Evolutionary Botany Lab, Department of Biology, Ghent University, K.L. Ledeganckstraat 35, 9000 Gent, Belgium.
Modesto Luceño, Botany Area, Department of Molecular Biology and Biochemical Engineering, Universidad Pablo de Olavide, ctra. de Utrera km. 1, 41013, Seville, Spain.
José Ignacio Márquez-Corro, Royal Botanic Gardens, Kew, Richmond, Surrey TW9 3AE, UK; Botany Area, Department of Molecular Biology and Biochemical Engineering, Universidad Pablo de Olavide, ctra. de Utrera km. 1, 41013, Seville, Spain.
Santiago Martín-Bravo, Botany Area, Department of Molecular Biology and Biochemical Engineering, Universidad Pablo de Olavide, ctra. de Utrera km. 1, 41013, Seville, Spain.
A Muthama Muasya, Bolus Herbarium, Department of Biological Sciences, University of Cape Town, Private Bag X3, Rondebosch, Cape Town 7701, South Africaand.
Petr Šmarda, Department of Botany and Zoology, Faculty of Science, Masaryk University, Kotlářská 2, 611 37 Brno, Czech Republic.
William Wayt Thomas, New York Botanical Garden, Bronx, NY 10458-5126, USA.
Karen L Wilson, National Herbarium of New South Wales, Australian Institute of Botanical Science, Australian Botanic Garden, Locked Bag 6002, Mount Annan, New South Wales 2567, Australia.
Petr Bureš, Department of Botany and Zoology, Faculty of Science, Masaryk University, Kotlářská 2, 611 37 Brno, Czech Republic.
FUNDING
This work was supported by the Czech Science Foundation (grant GA20-15989S to P.B.).
LITERATURE CITED
- Avdulov NP. 1931. Karyo-systematische untersuchungen der familie Gramineen. Bulletin of Applied Botany, of Genetics and Plant Breeding, Leningrad 44: 1–428. [Google Scholar]
- Baez M, Kuo Y-T, Dias Y, et al. 2020. Analysis of the small chromosomal Prionium serratum (Cyperid) demonstrates the importance of reliable methods to differentiate between mono- and holocentricity. Chromosoma 129: 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological) 57: 28989–28300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Bennett MD. 1987. Variation in genomic form in plants and its ecological implications. New Phytologist 106: 177–200. [Google Scholar]
- Blomberg SP, Garland T, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57: 717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Brown JH. 1984. On the relationship between abundance and distribution of species. The American Naturalist 124: 255–279. doi: 10.1086/284267. [DOI] [Google Scholar]
- Brožová V, Proćków J, Drábková LZ. 2022. Toward finally unraveling the phylogenetic relationships of Juncaceae with respect to another cyperid family, Cyperaceae. Molecular Phylogenetics and Evolution 177: 107588. [DOI] [PubMed] [Google Scholar]
- Brummitt RK, Pando F, Hollis S, Brummitt N. 2001. World geographical scheme for recording plant distributions. Pittsburgh, PA: Hunt Institute for Botanical Documentation, Carnegie-Mellon University. [Google Scholar]
- Burchardt P, Buddenhagen CE, Gaeta ML, Souza MD, Marques A, Vanzela ALL. 2020. Holocentric karyotype evolution in Rhynchospora is marked by intense numerical, structural, and genome size changes. Frontiers in Plant Science 11: 1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureš P. 1998. A high polyploid Eleocharis uniglumis sl (Cyperaceae) from Central and Southeastern Europe. Folia Geobotanica 33: 429–439. doi: 10.1007/bf02803644. [DOI] [Google Scholar]
- Bureš P, Zedek F. 2014. Holokinetic drive: centromere drive in chromosomes without centromeres. Evolution 68: 2412–2420. doi: 10.1111/evo.12437. [DOI] [PubMed] [Google Scholar]
- Bureš P, Zedek F, Marková M. 2013. Holocentric chromosomes. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, eds. Plant genome diversity, Vol. 2. Vienna: Springer, 187–208. [Google Scholar]
- Burt A. 2000. Perspective: sex, recombination, and the efficacy of selection – was Weismann right? Evolution 54: 337–351. doi: 10.1111/j.0014-3820.2000.tb00038.x. [DOI] [PubMed] [Google Scholar]
- Cacho NI, McIntyre PJ, Kliebenstein DJ, Strauss SY. 2021. Genome size evolution is associated with climate seasonality and glucosinolates, but not life history, soil nutrients or range size, across a clade of mustards. Annals of Botany 127: 887–902. doi: 10.1093/aob/mcab028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can M, Wei W, Zi H, et al. 2020. Genome sequence of Kobresia littledalei, the first chromosome-level genome in the family Cyperaceae. Scientific Data 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardillo M, Dinnage R, McAlister W. 2019. The relationship between environmental niche breadth and geographic range size across plant species. Journal of Biogeography 46: 97–109. [Google Scholar]
- Cardoso P. 2017. red – an R package to facilitate species red list assessments according to the IUCN criteria. Biodiversity Data Journal (5): e20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta A, Bedini G, Peruzzi L. 2018. Unscrambling phylogenetic effects and ecological determinants of chromosome number in major angiosperm clades. Scientific Reports 8: 14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta A, Bedini G, Peruzzi L. 2020. A deep dive into the ancestral chromosome number and genome size of flowering plants. New Phytologist 228: 1097–1106. doi: 10.1111/nph.16668. [DOI] [PubMed] [Google Scholar]
- Chamberlain S, Barve V, Mcglinn D, Oldoni D, Desmet P, Geffert L, Ram K. 2020. rgbif: interface to the Global Biodiversity Information Facility API. https://cran.r-project.org/web/packages/rgbif/rgbif.pdf [Google Scholar]
- Chung K, Hipp AL, Roalson EH. 2012. Chromosome number evolves independently of genome size in a clade with nonlocalized centromeres (Carex: Cyperaceae). Evolution 66: 2708–2722. doi: 10.1111/j.1558-5646.2012.01624.x. [DOI] [PubMed] [Google Scholar]
- Doležel J, Bartoš J, Voglmayr H, Greilhuber J. 2003. Nuclear DNA content and genome size of trout and human. Cytometry A 51: 127–128. [DOI] [PubMed] [Google Scholar]
- Dormann CF, Elith J, Bacher S, et al. 2013. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36: 27–46. [Google Scholar]
- Drábková LZ, Vlček C. 2009. DNA variation within Juncaceae: comparison of impact of organelle regions on phylogeny. Plant Systematics and Evolution 278: 169–186. [Google Scholar]
- Dunnett CW. 1980. Pairwise multiple comparisons in the homogeneous variance, unequal sample size case. Journal of the American Statistical Association 75: 789–795. doi: 10.1080/01621459.1980.10477551. [DOI] [Google Scholar]
- Edelsbrunner H, Mücke EP. 1994. Three-dimensional alpha shapes. ACM Transactions on Graphics 13: 43–72. [Google Scholar]
- Elliott TL, Muasya AM, Bureš P. 2022. Complex patterns of ploidy in a holocentric plant clade (Schoenus, Cyperaceae) in the Cape biodiversity hotspot. Annals of Botany. doi: 10.1093/aob/mcac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott TL, Larridon I, Barrett RL, Bruhl JJ, Costa SM, Escudero M, Hipp AL, Jiménez-Mejías P, Kirschner J, Luceño M, Ignacio Márquez-Corro J, Martín-Bravo S, Roalson EH, Semmouri I, Spalink D, Wayt Thomas W, Villaverde T, Wilson KL, Muthama Muasya A. 2022. Addressing inconsistencies in Cyperaceae and Juncaceae taxonomy: Comment on Brožová et al. Molecular Phylogenetics and Evolution. doi: 10.1016/j.ympev.2022.107665. [DOI] [PubMed] [Google Scholar]
- Escudero M, Hipp AL, Hansen TF, Voje KL, Luceño M. 2012. Selection and inertia in the evolution of holocentric chromosomes in sedges (Carex, Cyperaceae). New Phytologist 195: 237–247. doi: 10.1111/j.1469-8137.2012.04137.x. [DOI] [PubMed] [Google Scholar]
- Escudero M, Maguilla E, Loureiro J, Castro M, Castro S, Modesto L. 2015. Genome size stability despite high chromosome number variation in Carex gr. laevigata. American Journal of Botany 102: 233–238. [DOI] [PubMed] [Google Scholar]
- Escudero M, Márquez-Corro JI, Hipp AL. 2016. The phylogenetic origins and evolutionary history of holocentric chromosomes. Systematic Botany 41: 580–585. doi: 10.1600/036364416x692442. [DOI] [Google Scholar]
- Escudero M, Hahn M, Hipp AL. 2018. RAD‐seq linkage mapping and patterns of segregation distortion in sedges: meiosis as a driver of karyotypic evolution in organisms with holocentric chromosomes. Journal of Evolutionary Biology 31: 833–843. doi: 10.1111/jeb.13267. [DOI] [PubMed] [Google Scholar]
- Flora of North America Editorial Committee. 1993. Flora of North America North of Mexico. New York: Oxford University Press. [Google Scholar]
- Freckleton R, Harvey P, Pagel M. 2002. Phylogenetic analysis and comparative data: a test and review of evidence. The American Naturalist 160: 712–726. doi: 10.1086/343873. [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Gardiner JD, Behnsen J, Brassey CA. 2018. Alpha shapes: determining 3D shape complexity across morphologically diverse structures. BMC Evolutionary Biology 18: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston KJ, Spicer JI. 2001. The relationship between range size and niche breadth: a test using five species of Gammarus (Amphipoda). Global Ecology and Biogeography 10: 179–188. doi: 10.1046/j.1466-822x.2001.00225.x. [DOI] [Google Scholar]
- Global Carex Group, Roalson EH, Jiménez-Mejías P, et al. 2021. A framework infrageneric classification of Carex (Cyperaceae) and its organizing principles. Journal of Systematics and Evolution 59: 726–762. [Google Scholar]
- Goetghebeur P. 1998. Cyperaceae. In: Kubitzki K, ed. The families and genera of vascular plants. Flowering plants, Monocotyledons: Alismatanae and Commelinanae (except Gramineae). New York: Springer, 141–190. [Google Scholar]
- Govaerts R, Jiménez-Mejías P, Koopman J, et al. 2021. World checklist of selected plant families. Cyperaceae. Kew: Royal Botanic Gardens, [Google Scholar]
- Greilhuber J, Leitch IJ. 2013. Genome size and the phenotype. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF, eds. Plant genome diversity, Vol. 2. Vienna: Springer, 323–344. [Google Scholar]
- Greilhuber J, Doležel J, Lysák MA, Bennett M. 2005. The origin, evolution and proposed stabilization of the terms ‘Genome Size’ and ‘C-Value’ to describe nuclear DNA contents. Annals of Botany 95: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Borsch T, Müller K, Worberg A, Porembski S, Barthlott W. 2006. Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biology 8: 770–777. [DOI] [PubMed] [Google Scholar]
- Guerra M, Ribeiro T, Felix LP. 2019. Monocentric chromosomes in Juncus (Juncaceae) and implications for the chromosome evolution of the family. Botanical Journal of the Linnean Society 191: 475–483. doi: 10.1093/botlinnean/boz065. [DOI] [Google Scholar]
- Guignard MS, Nichols RA, Knell RJ,, et al. 2016. Genome size and ploidy influence angiosperm species’ biomass under nitrogen and phosphorus limitation. New Phytologist 210: 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guignard M, Leitch A, Acquisti C, et al. 2017. Impacts of nitrogen and phosphorus: from genomes to natural ecosystems and agriculture. Frontiers in Ecology and Evolution 5: 70. [Google Scholar]
- Harms LJ. 1968. Cytotaxonomic studies in Eleocharis subser. Palustres: central United States taxa. American Journal of Botany 55: 966–974. doi: 10.1002/j.1537-2197.1968.tb07456.x. [DOI] [Google Scholar]
- Hasegawa N. 1932. Comparison of chromosome types in Disporum. Cytologia 4: 350–368. [Google Scholar]
- Heitz H. 1925–1926. Der nachweis der chromosomen. Vergleichende Studien über ihre Zahl, Grösse ond Form im Pflanzenreich I. Zeitschrift für Botanik 18: 627–681. [Google Scholar]
- Hidalgo O, Pellicer J, Christenhusz M, Schneider H, Leitch AR, Leitch IJ. 2017. Is there an upper limit to genome size? Trends in Plant Science 22: 567–573. doi: 10.1016/j.tplants.2017.04.005. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ. 2020. Geographic data analysis and modeling [R package raster version 3.4-5]. https://rspatial.org/raster [Google Scholar]
- Hofstatter PG, Thangavel G, Lux T, et al. 2022. Repeat-based holocentromeres influence genome architecture and karyotype evolution. Cell 185: 3153–3168.e18. doi: 10.1016/j.cell.2022.06.045. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. 2005. The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- IUCN. 2001. IUCN Red List categories and criteria: Version 3.1. IUCN Species Survival Commission. Gland, Switzerland and Cambridge, UK: IUCN. [Google Scholar]
- Jankowska M, Fuchs J, Klocke E, et al. 2015. Holokinetic centromeres and efficient telomere healing enable rapid karyotype evolution. Chromosoma 124: 519–528. doi: 10.1007/s00412-015-0524-y. [DOI] [PubMed] [Google Scholar]
- Janzen DH. 1967. Why mountain passes are higher in the tropics? The American Naturalist 101: 233–249. [Google Scholar]
- Johnen L, de Souza TB, Rocha DM, et al. 2020. Allopolyploidy and genomic differentiation in holocentric species of the Eleocharis montana complex (Cyperaceae). Plant Systematics and Evolution 306: 1–17. [Google Scholar]
- Johnson C. 1998. Species extinction and the relationship between distribution and abundance. Nature 394: 272–274. [Google Scholar]
- Kang M, Wang J, Huang H. 2015. Nitrogen limitation as a driver of genome size evolution in a group of karst plants. Scientific Reports 5: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karger DN, Conrad O, Böhner J, et al. 2017. Climatologies at high resolution for the earth’s land surface areas. Scientific Data 4: 170122. doi: 10.1038/sdata.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N, Datson PM, Murray BG. 2012. Genome size and chromosome number in the New Zealand species of Schoenus (Cyperaceae). Botanical Journal of the Linnean Society 169: 555–564. doi: 10.1111/j.1095-8339.2012.01238.x. [DOI] [Google Scholar]
- Knight CA, Ackerly DD. 2002. Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecology Letters 5: 66–76. doi: 10.1046/j.1461-0248.2002.00283.x. [DOI] [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. 2005. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany 95: 177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostoff D. 1939. Evolutionary significance of chromosome size and chromosome number in plants. Current Science 8: 306–310. [Google Scholar]
- Krátká M, Šmerda J, Lojdová K, Bureš P, Zedek F. 2021. Holocentric chromosomes probably do not prevent centromere drive in Cyperaceae. Frontiers in Plant Science 12: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBar T, Adami C. 2020. Genome size and the extinction of small populations. In: Banzhaf W, Cheng BHC, Deb K, et al., eds. Evolution in action: past, present and future. Cham, Switzerland: Springer, 167–183. [Google Scholar]
- Lafarge T, Pateiro-Lopez B. 2020. Implementation of the 3D alpha-shape for the reconstruction of 3D sets from a point cloud. alphashape3d: R package version 1.3.1. https://CRAN.R-project.org/package=alphashape3d
- Larridon I, Spalink D, Jiménez‐Mejías P, et al. 2021a. The evolutionary history of sedges (Cyperaceae) in Madagascar. Journal of Biogeography 48: 917–932. doi: 10.1111/jbi.14048. [DOI] [Google Scholar]
- Larridon I, Zuntini AR, Léveillé-Bourret E, et al. 2021b. A new classification of Cyperaceae (Poales) supported by phylogenomic data. Journal of Systematics and Evolution 59: 852–895. [Google Scholar]
- Lee B, Cho Y, Kim S. 2019. Genome size estimation of 43 Korean Carex. Korean Journal of Plant Taxonomy 49: 334–344. doi: 10.11110/kjpt.2019.49.4.334. [DOI] [Google Scholar]
- Leitch AR, Leitch IJ. 2008. Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483. doi: 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Beaulieu JM, Chase MW, Leitch AR, Fay MF. 2010. Genome size dynamics and evolution in monocots. Journal of Botany 2010: 862516. [Google Scholar]
- Leutner B, Horning N, Schwalb-Willmann J, Hijmans R. 2017. RStoolbox: tools for remote sensing data analysis. R package version 0.1 7. https://CRAN.R-project.org/package=RStoolbox
- Levin DA, Funderburg SW. 1979. Genome size in angiosperms: temperate versus tropical species. The American Naturalist 114: 784–795. doi: 10.1086/283528. [DOI] [Google Scholar]
- Linder HP, Rudall PJ. 2005. Evolutionary history of Poales. Annual Review of Ecology, Evolution, and Systematics 36: 107–124. doi: 10.1146/annurev.ecolsys.36.102403.135635. [DOI] [Google Scholar]
- Lipnerová I, Bureš P, Horová L, Šmarda P. 2013. Evolution of genome size in Carex (Cyperaceae) in relation to chromosome number and genomic base composition. Annals of Botany 111: 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucek K, Augustijnen H, Escudero M. 2022. A holocentric twist to chromosomal speciation? Trends in Ecology & Evolution 37: 655–662. [DOI] [PubMed] [Google Scholar]
- Luceño M, Vanzela ALL, Guerra M. 1998. Cytotaxonomic studies in Brazilian Rhynchospora (Cyperaceae), a genus exhibiting holocentric chromosomes. Canadian Journal of Botany 76: 440–449. [Google Scholar]
- Lynch M. 2007. The frailty of adaptive hypotheses for the origins of organismal complexity. Proceedings of the National Academy of Sciences, USA 104: 8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2003. The origins of genome complexity. Science 302: 1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Mandrioli M, Manicardi GC. 2020. Holocentric chromosomes. PLoS Genetics 16: e1008918. doi: 10.1371/journal.pgen.1008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-Corro JI, Martín-Bravo S, Pedrosa-Harand A, Hipp AL, Luceño M, Escudero M. 2019a. Karyotype evolution in holocentric organisms. In: eLS. Chichester: John Wiley & Sons.doi: 10.1002/9780470015902.a0028758 [DOI] [Google Scholar]
- Márquez-Corro JI, Martín-Bravo S, Spalink D, Luceño M, Escudero M. 2019b. Inferring hypothesis-based transitions in clade-specific models of chromosome number evolution in sedges (Cyperaceae). Molecular Phylogenetics and Evolution 135: 203–209. doi: 10.1016/j.ympev.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Márquez-Corro JI, Martín-Bravo S, Jiménez‐Mejías P, et al. 2021. Macroevolutionary insights into sedges (Carex: Cyperaceae): the effects of rapid chromosome number evolution on lineage diversification. Journal of Systematics and Evolution 59: 776–790. [Google Scholar]
- Melters DP, Paliulis LV, Korf IF, Chan SW. 2012. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Research 20: 579–593. [DOI] [PubMed] [Google Scholar]
- Neumann P, Oliveira L, Čížková J, et al. 2021. Impact of parasitic lifestyle and different types of centromere organization on chromosome and genome evolution in the plant genus Cuscuta. New Phytologist 229: 2365–2377. [DOI] [PubMed] [Google Scholar]
- Ng CH, Lee SL, Tnah LH, Ng KKS, Lee CT, Madon M. 2016. Genome size variation and evolution in Dipterocarpaceae. Plant Ecology & Diversity 9: 437–446. [Google Scholar]
- Nijalingappa B. 1974. Cytological studies in Scirpus (Cyperaceae). Proceedings of the Indian Academy of Sciences 80: 134–138. [Google Scholar]
- Nishikawa K, Furuta Y, Ishitobi K. 1984. Chromosomal evolution in genus Carex as viewed from nuclear DNA content, with special reference to its aneuploidy. The Japanese Journal of Genetics 59: 465–472. doi: 10.1266/jjg.59.465. [DOI] [Google Scholar]
- Orme D. 2013. The CAPER package: comparative analysis of phylogenetics and evolution in R. https://cran.r-project.org/web/packages/caper/index.html.
- Otto F, Oldiges H, Göhde W, Jain V. 1981. Flow cytometric measurement of nuclear DNA content variations as a potential in vivo mutagenicity test. Cytometry 2: 189–191. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pellicer J, Leitch IJ. 2020. The Plant DNA C-values database (release 7.1): an updated online repository of plant genome size data for comparative studies. New Phytologist 226: 301–305. [DOI] [PubMed] [Google Scholar]
- Pellicer J, Hidalgo O, Dodsworth S, Leitch IJ. 2018. Genome size diversity and its impact on the evolution of land plants. Genes 9: 88. doi: 10.3390/genes9020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plačková K, Zedek F, Schubert V, Houben A, Bureš P. 2022. Kinetochore size scales with chromosome size in bimodal karyotypes of Agavoideae. Annals of Botany 130: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta J, Liang Y-Y, Xin H, et al. 2022. Chromosome scale genome assemblies and annotations for Poales species Carex cristatella, Carex scoparia, Juncus effusus and Juncus inflexus. G3 Genes| Genomes| Genetics 12: jkac211. doi: 10.1093/g3journal/jkac211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rice A, Glick L, Abadi S, et al. 2015. The Chromosome Counts Database (CCDB) – a community resource of plant chromosome numbers. New Phytologist 206: 19–26. [DOI] [PubMed] [Google Scholar]
- Richardson L. 2019. Beautiful soup documentation Version 4.4.0. https://beautiful-soup-4.readthedocs.io/en/latest/
- Roalson EH. 2008. A synopsis of chromosome number variation in the Cyperaceae. The Botanical Review 74: 209–393. [Google Scholar]
- Roalson EH, McCubbin AG, Whitkus R. 2007. Chromosome evolution in Cyperales. Aliso 23: 62–71A. [Google Scholar]
- da Silva CRM, de Souza TB, Trevisan R, et al. 2017. Genome differentiation, natural hybridisation and taxonomic relationships among Eleocharis viridans, E. niederleinii and E. ramboana (Cyperaceae). Australian Systematic Botany 30: 183–195. doi: 10.1071/sb17002. [DOI] [Google Scholar]
- Slatyer RA, Hirst M, Sexton JP. 2013. Niche breadth predicts geographical range size: a general ecological pattern. Ecology Letters 16: 1104–1114. doi: 10.1111/ele.12140. [DOI] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L, Foggi B, Rossi G. 2008. Genome size and GC content evolution of Festuca: ancestral expansion and subsequent reduction. Annals of Botany 101: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Hejcman M, Březinová A, et al. 2013. Effect of phosphorus availability on the selection of species with different ploidy levels and genome sizes in a long-term grassland fertilization experiment. New Phytologist 200: 911–921. doi: 10.1111/nph.12399. [DOI] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L, et al. 2014. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proceedings of the National Academy of Sciences, USA 111: E4096–E4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Bennett MD, Leitch IJ. 2003. Evolution of genome size in the angiosperms. American Journal of Botany 90: 1596–1603. doi: 10.3732/ajb.90.11.1596. [DOI] [PubMed] [Google Scholar]
- de Sousa LM, Poggio L, Batjes NH, et al. 2020. SoilGrids 2.0: producing quality-assessed soil information for the globe. Soil 7: 217–240. [Google Scholar]
- de Souza TB, Chaluvadi SR, Johnen L, et al. 2018. Analysis of retrotransposon abundance, diversity and distribution in holocentric Eleocharis (Cyperaceae) genomes. Annals of Botany 122: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stace CA. 2000. Cytology and cytogenetics as a fundamental taxonomic resource for the 20th and 21st centuries. Taxon 49: 451–477. doi: 10.2307/1224344. [DOI] [Google Scholar]
- Stapley J, Feulner PGD, Johnston SE, Santure AW, Smadja CM. 2017. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philosophical Transactions of the Royal Society B: Biological Sciences 372: 20160455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. 1966. Chromosomal variation and evolution: polyploidy and chromosome size and number shed light on evolutionary processes in higher plants. Science 152: 1463–1469. doi: 10.1126/science.152.3728.1463. [DOI] [PubMed] [Google Scholar]
- Temsch EM, Koutecký P, Urfus T, Šmarda P, Doležel J. 2022. Reference standards for flow cytometric estimation of absolute nuclear DNA content in plants. Cytometry A 101: 710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey JW. 1949. Comparing individual means in the analysis of variance. Biometrics 5: 99–114. [PubMed] [Google Scholar]
- Vanzela ALL, Guerra M, Luceño M. 1996. Rhynchospora tenuis Link (Cyperaceae), a species with the lowest number of holocentric chromosomes. Cytobios 88: 219–228. [Google Scholar]
- Varela S, Anderson RP, García-Valdés R, Fernández-González F. 2014. Environmental filters reduce the effects of sampling bias and improve predictions of ecological niche models. Ecography 37: 1084–1091. [Google Scholar]
- Veleba A, Bureš P, Adamec L, Šmarda P, Lipnerová I, Horová L. 2014. Genome size and genomic GC content evolution in the miniature genome‐sized family Lentibulariaceae. New Phytologist 203: 22–28. doi: 10.1111/nph.12790. [DOI] [PubMed] [Google Scholar]
- Veleba A, Šmarda P, Zedek F, Horová L, Šmerda J, Bureš P. 2017. Evolution of genome size and genomic GC content in carnivorous holokinetics (Droseraceae). Annals of Botany 119: 409–416. doi: 10.1093/aob/mcw229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleba A, Zedek F, Horová L, Veselý P, Srba M, Šmarda P, Bureš P. 2020. Is the evolution of carnivory connected with genome size reduction? American Journal of Botany 107: 1253–1259. doi: 10.1002/ajb2.1526 [DOI] [PubMed] [Google Scholar]
- Veselý P, Bureš P, Šmarda P, Pavlíček T. 2012. Genome size and DNA base composition of geophytes: the mirror of phenology and ecology? Annals of Botany 109: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. 2003. Selfish DNA is maladaptive: evidence from the plant Red List. Trends in Genetics 19: 609–614. doi: 10.1016/j.tig.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Wang S, Veller C, Sun F, et al. 2019. Per-nucleus crossover covariation and implications for evolution. Cell 177: 326–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WCVP. 2022. World Checklist of Vascular Plants, version 2.0. Facilitated by the Royal Botanic Gardens, Kew. http://wcvp.science.kew.org/ [Google Scholar]
- Xu S, Dai Z, Guo P, et al. 2021. ggtreeExtra: compact visualization of richly annotated phylogenetic data. Molecular Biology and Evolution 38: 4039–4042. doi: 10.1093/molbev/msab166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Post WM, Thornton PE, Jain A. 2013. The distribution of soil phosphorus for global biogeochemical modeling. Biogeosciences 9: 2525–2537. [Google Scholar]
- Zedek F, Šmerda J, Šmarda P, Bureš P. 2010. Correlated evolution of LTR retrotransposons and genome size in the genus Eleocharis. BMC Plant Biology 10: 265. doi: 10.1186/1471-2229-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F, Veselý P, Horová L, Bureš P. 2016. Flow cytometry may allow microscope-independent detection of holocentric chromosomes in plants. Scientific Reports 6: 27161. doi: 10.1038/srep27161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedek F, Veselý P, Tichý L, et al. 2022. Holocentric plants are more competitive under higher UV‐B doses. New Phytologist 233: 15–21. [DOI] [PubMed] [Google Scholar]
- Zizka A, Silvestro D, Andermann T, et al. 2019. CoordinateCleaner: standardized cleaning of occurrence records from biological collection databases. Methods in Ecology and Evolution 10: 744–751. doi: 10.1111/2041-210x.13152. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.