Abstract
Cdc42p is an essential GTPase that belongs to the Rho/Rac subfamily of Ras-like GTPases. These proteins act as molecular switches by responding to exogenous and/or endogenous signals and relaying those signals to activate downstream components of a biological pathway. The 11 current members ofthe Cdc42p family display between 75 and 100% amino acid identity and are functional as well as structural homologs. Cdc42p transduces signals to the actin cytoskeleton to initiate and maintain polarized gorwth and to mitogen-activated protein morphogenesis. In the budding yeast Saccharomyces cerevisiae, Cdc42p plays an important role in multiple actin-dependent morphogenetic events such as bud emergence, mating-projection formation, and pseudohyphal growth. In mammalian cells, Cdc42p regulates a variety of actin-dependent events and induces the JNK/SAPK protein kinase cascade, which leads to the activation of transcription factors within the nucleus. Cdc42p mediates these processes through interactions with a myriad of downstream effectors, whose number and regulation we are just starting to understand. In addition, Cdc42p has been implicated in a number of human diseases through interactions with its regulators and downstream effectors. While much is known about Cdc42p sturcture and functional interactions, little is known about the mechanism(s) by which it transduces signals within the cell. Future research sould focus on this question as well as on the detailed analysis of the interactions of Cdc42p with its regulators and downstream effectors.
The establishment of cell polarity is an important component of the overall process of cellular morphogenesis, the complex process by which the three-dimensional organization of subcellular constituents, which ultimately determines an organism’s characteristic growth patterns and shape, is generated and maintained. The generation of cell polarity is critical for the control of many cellular and developmental processes such as shape development in early plant and animal embryogenesis, axon migration and neurite outgrowth in early development, the intracellular movement of organelles and proteins in polarized epithelial cells, the stimulated secretion of neurotransmitters, the directed movement of migratory cells, polarized growth within yeast and fungal cells, and the asymmetric partitioning of new cellular constituents during cell division. Establishment of cell polarity involves the generation of cellular asymmetry through the localized temporal and spatial activation of cellular processes and can be divided into several hierarchical and interdependent events. These events include the initial response to endogenous and/or exogenous signals, the determination of an axis of polarization relative to these signals, and the subsequent asymmetric distribution of cellular components along that axis. At the molecular level, cell polarity is best understood in the budding yeast Saccharomyces cerevisiae, but results from studies in fission yeast Schizosaccharomyces pombe, Drosophila, Caenorhabditis elegans, and cultured mammalian cells strongly suggest that the molecular mechanisms controlling cell polarity in S. cerevisiae are highly conserved in other eukaryotes. Due to the abundance of recent reviews on cell polarity and signaling (26, 69, 77, 100, 126, 170, 177, 202, 216, 218, 246, 281, 346, 347, 350, 382, 464, 465, 467, 511, 520, 587) and on the roles of Rho-type GTPases in these processes (46, 61, 120, 141, 171, 186, 192, 225, 241, 288, 290, 325, 326, 337, 344, 419, 441, 473, 477, 478, 544, 550, 552, 572, 578, 638, 644). I will limit this review to a discussion of the Cdc42p GTPase, its identification, its structure and subcellular localization, its function(s) in controlling cell polarity, and its regulators and effectors.
It is becoming increasingly apparent that the Cdc42p GTPase and other Rho-type GTPases play a vital role in regulating the signal transduction pathways that control the generation and maintenance of cell polarity in many, if not all, eukaryotic cell types. The Cdc42p GTPase signaling module consists of regulators of the guanine nucleotide-bound state of Cdc42p, i.e., guanine nucleotide exchange factors (GEFs), guanine nucleotide dissociation inhibitors (GDIs), and GTPase-activating proteins (GAPs), as well as downstream effectors of Cdc42p function (Table 1). The regulators of the guanine nucleotide-bound state of Cdc42p must respond to a variety of exogenous and/or endogenous signals, thereby activating Cdc42p to a GTP-bound state or inactivating it to a GDP-bound state. A myriad of Cdc42p downstream effectors interact with the activated (GTP-bound) form of Cdc42p, thereby inducing a number of downstream events, including rearrangements of the actin cytoskeletal network and protein kinase-dependent induction of transcription, which are increasingly coming into view. Interactions between the 21-kDa Cdc42p GTPase and this host of regulators and effectors must be controlled in a temporal and spatial manner so that Cdc42p can function at different times within the cell cycle and at different places within the cell. Cdc42p function is also regulated by its subcellular localization, which depends on its prenylation state and interactions with its GDI.
TABLE 1.
Cdc42p potential regulators and effectorsa
| Regulator or effector | S. cerevisiae | S. pombe | Drosophila | C. elegans | Mammals |
|---|---|---|---|---|---|
| Regulators | |||||
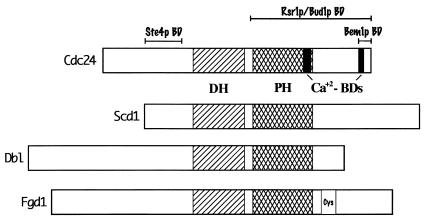
| GEFs | Cdc24 | Scd1 | Drt-GEF? | Unc-73? | Dbl |
| DRho-GEF? | Bcr | ||||
| DRho-GEF2? | Abr | ||||
| Dbs | |||||
| Tiam-1 | |||||
| Ect2 | |||||
| Ost | |||||
| FGD1 | |||||
| Brx | |||||
| GDIs | Rdi1 | ? | ? | ? | Rho-GDI |
| GAPs | Bem3 | ? | RnRac-GAP? | Ce-GAP? | Cdc42GAP/p50rhoGAP |
| Rga1/Dbm1 | Bcr | ||||
| Rga2 | Abr | ||||
| p190GAP | |||||
| n-Chimaerin | |||||
| 3BP-1 | |||||
| Graf | |||||
| RalBP1/RLIP76/RIP1 | |||||
| MgcRacGAP | |||||
| PARG1 | |||||
| myr5 | |||||
| CdGAP | |||||
| Effectors | |||||
| Kinases | Ste20 | Pak1/Shk1 | DPAK | Ce-PAK | PAK1 |
| Cla4 | Pak2 | cMEKKs | PAK2 | ||
| Skm1 | PAK3 | ||||
| ACK-1, ACK-2 | |||||
| MLK3 | |||||
| MEKKs | |||||
| Others | Bni1, Bnr1 | Fus1, Cdc12? | diaphanous | ? | Formins, p140mDia |
| Iqg1/Cyk1 | ? | ? | ? | IQGAPs | |
| Bee1/Las17 | ? | ? | ? | WASPs | |
| Gic1, Gic2 | ? | ? | ? | ? | |
| Zds1, Zds2 | ? | ? | ? | ? | |
| Bem4/Rom7 | ? | ? | ? | ? | |
| ? | ? | ? | ? | pp70-S6 kinase |
Proteins that have in vitro Cdc42p GEF, GAP, or GDI activity or that contain structurally homologous domains are listed. See the text for details on potential in vivo functions and complete references.
CDC42P STRUCTURE AND FUNCTIONAL DOMAINS
Identification of Cdc42
The Cdc42p GTPase was first identified from an S. cerevisiae mutant strain carrying a temperature-sensitive (ts) mutation, cdc42-1ts, that blocked bud formation at 37°C but allowed the cell mass and volume to increase, resulting in greatly enlarged, unbudded cells (2). Although cell division was arrested at 37°C, DNA replication and nuclear division continued into the next cycle, resulting in multinucleate cells as determined by DNA staining with the fluorescent dye 4′,6-diamidino-2-phenylindole (DAPI) and mitotic spindle staining with anti-tubulin antibodies. Fluorescence microscopy with rhodamine-conjugated phalloidin showed that the polarized organization of the actin cytoskeleton (i.e., cortical actin distribution to the regions of new cell growth in the bud and actin cables directed into the enlarging bud) was disrupted, indicating that Cdc42p functioned in the organization of the actin cytoskeleton, which is necessary for polarized cell growth. Chitin and other cell surface materials were deposited uniformly throughout the enlarging cell walls, in contrast to their normal polarized patterns of deposition. Growth of the cdc42-1ts strain at semipermissive temperatures led to a small percentage of cells with elongated buds. Taken together, these observations suggested that Cdc42p controls polarized cell growth during the cell cycle but that isotropic incorporation of new cell wall material was not impaired through the loss of Cdc42p function. Examination of the cdc42 null phenotype in S. cerevisiae and S. pombe indicated that Cdc42p was essential for viability (242, 390).
DNA and predicted amino acid sequence analysis (242) indicated that Cdc42p belongs to the Rho subfamily of the Ras superfamily of GTPases that act as molecular switches in the control of a variety of eukaryotic processes (191, 239, 241, 242, 642) (see below). At about the same time, a ∼25-kDa guanine nucleotide binding protein was purified from bovine brain and human placental membranes (140, 461, 582), and peptide sequences from this protein, termed Gp or G25K, showed a high degree of similarity to S. cerevisiae Cdc42p (242, 461). This protein was shown to be a good in vitro substrate for epidermal growth factor (EGF)-stimulated phosphorylation (209), although the in vivo phosphorylation of Cdc42p has not been reported to date. Subsequent analysis of the predicted amino acid sequence from two independent human cDNA isolates indicated the existence of two highly conserved (95% identical) proteins, the ubiquitously expressed Cdc42Hs (525) and the brain isoform G25K (407). The Cdc42Hs and G25K proteins are identical in both nucleotide and predicted amino acid sequences up to amino acid 163 but diverge from residues 163 to 191, suggesting that these isoforms are differential splicing products of a single gene. Structural and/or functional Cdc42p homologs have subsequently been characterized in the pathogenic yeast Candida albicans (394), S. pombe (390), C. elegans (88, 500), Drosophila (336), chicken (Gallus gallus) cochlea (172), mouse (Mus musculus) liver (172) and brain (367), and dog (Canus familiaris) (GenBank accession no. Z49944), and these homologs are 80 to 95% identical in predicted amino acid sequence (241) (see below) (Fig. 1). S. pombe, Drosophila, and C. elegans Cdc42p, as well as Cdc42Hs and G25K, can complement the cdc42-1ts mutant (88, 390, 407, 507, 525), suggesting that Cdc42p may have conserved functions in these other eukaryotes.
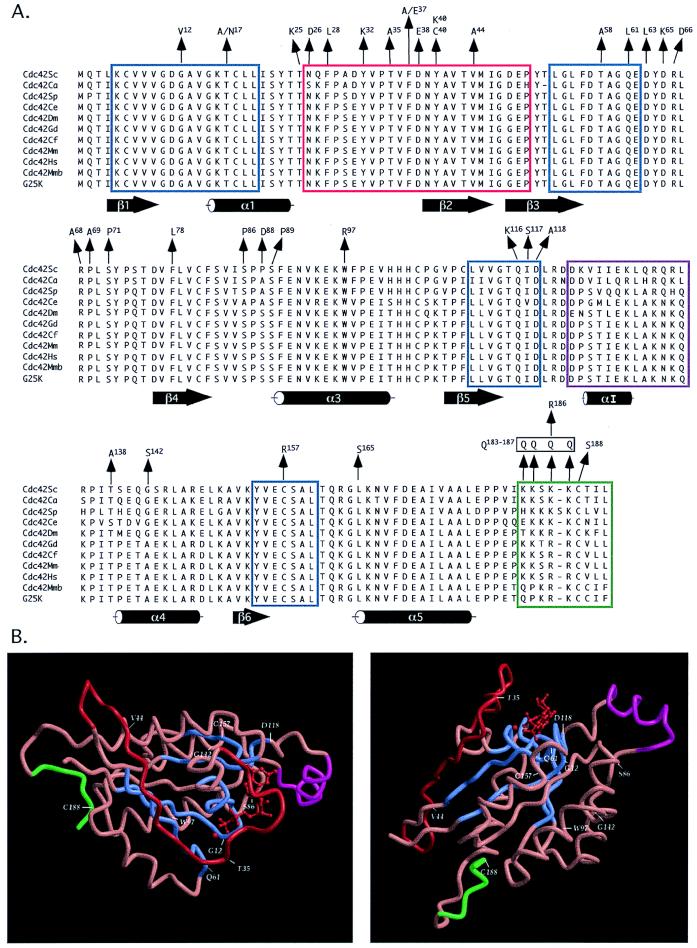
FIG. 1.
Cdc42p mutations and X-ray crystal structure. (A) Comparison of Cdc42p sequences from S. cerevisiae (Sc [242]), C. albicans (Ca [394]), S. pombe (Sp [390]), C. elegans (Ce [88]), D. melanogaster (Dm [336]), chicken (Gd [172]), dog (Cf [GenBank no. Z49944]), mouse (Mm [172]), human (Hs [525]), mouse brain (Mmb [367]), and human brain (G25K [407]). Included are the known Cdc42 mutations mapped onto the primary amino acid sequences and the known functional domains: GTP binding/hydrolysis domains (blue boxes), effector domain (red box), Rho insert domain (purple box), and membrane localization domain (green box). Secondary-structure elements indicated below the sequence are taken from reference 145. (B) Two views of the X-ray crystal structure of Cdc42Hs complexed with GDP (kindly provided by N. Nassar and R. Cerione, Cornell University). Color coding for functional domains is the same as in panel A.
The Cdc42 family of proteins currently has 11 members ranging in size from 190 to 192 amino acids (Fig. 1A). Within this family, there is a very high degree of sequence conservation, ranging from ∼75% amino acid identity between C. albicans Cdc42p and the human brain isoform G25K to 100% identity between the dog, mouse, and human Cdc42p and 100% identity between the mouse brain and human brain (G25K) isoforms. Cdc42 proteins display ∼40% similarity to other Ras-like GTPases, but this similarity is clustered in the four domains implicated in GTP binding and hydrolysis (Fig. 1, blue boxed domains). The most obvious difference between Cdc42p and Ras protein sequences in these domains is in amino acids 115 to 118. Ras proteins contain the diagnostic sequence Asn-Lys-Xaa-Asp (NKXD, where X is any amino acid), while all Cdc42p proteins contain the signature sequence Thr-Gln-Xaa-Asp (TQXD, with X being predominantly an Ile residue). It has been postulated that these differences may account for the ∼10-fold-higher rate of GTP hydrolysis observed with Cdc42p proteins than with Ras proteins (210), but this has not been experimentally tested to date (see “GTPase-activating proteins” below). All Cdc42 proteins contain the C-terminal sequence Cys-Xaa-Xaa-Leu except for the two brain isoforms (Cdc42Mmb and G25K [Fig. 1]), which end in a Phe residue. This conserved domain is necessary for proper membrane anchorage of Cdc42 proteins (see “Prenylation and subcellular localization” below). Much that is known about the functional domains of Cdc42p has been determined by analyzing gain-of-function, loss-of-function, and dominant negative mutations (shown in Fig. 1). A compendium of these mutations, along with their mutant phenotypes, is listed in Table 2.
TABLE 2.
Compendium of cdc42 mutations
| Mutation | Cdc42 testedb | Phenotype | Reference(s)a |
|---|---|---|---|
| G12V | Sc, Sp, Dm, Hs | Dominant activated; GTPase defective; GTP bound | 210, 336, 390, 642 |
| T17N | Sp, Dm, Hs | Dominant negative; apo or GDP bound | 336, 447 |
| T25K | Hs | Wild-type response to Cdc24-stimulated GDP dissociation | 323 |
| N26D | Hs | Wild-type response to Cdc24-stimulated GDP dissociation | 323 |
| F28L | Hs | Rapid nucleotide exchange; transforms NIH 3T3 cells | 327 |
| Y32K | Hs | Loss of Cdc24-stimulated GDP dissociation; reduced binding to Cdc42-GAP, IQGAP1, IQGAP2, and mPAK-3 CRIB domains | 309, 310, 323, 379 |
| T35A | Sc, Sp, Hs | Loss of binding to Cdc24 and CRIB-containing effectors; suppresses G12V and D118A | 116, 447 |
| F37A | Hs | Slight loss of JNK1 activation; disrupts Rac activation; does not affect actin polymerization or filopodium formation | 291 |
| F37E | Hs | Loss of Cdc24-stimulated GDP dissociation | 323 |
| D38E | Hs | Reduced binding to IQGAP1, IQGAP2, and mPAK-3 CRIB domain | 310, 379 |
| Y40C | Hs | Reduced binding to CRIB-containing effectors; does not affect actin polymerization and filopodium formation | 291 |
| Y40K | Hs | Wild-type response to Cdc24-stimulated GDP dissociation | 323 |
| V44A | Sc | Morphogenesis checkpoint; G2/M delay; reduced binding to Cdc24, Cla4, Gic1, Gic2 | 116, 475a |
| T58A | Sc, Sp | ts loss-of-function | 391 |
| Q61L | Sc, Sp, Hs | Dominant activated; GTPase defective; GTP bound | 642 |
| D63L | Hs | Wild-type response to Cdc24-stimulated GDP dissociation | 323 |
| D65K | Hs | Wild-type response to Cdc24-stimulated GDP dissociation | 323 |
| R66D | Hs | Wild-type response to Cdc24-stimulated GDP dissociation | 323 |
| R68A | Hs | Wild-type response to Cdc24-stimulated GDP dissociation | 323 |
| P69A | Hs | Wild-type response to Cdc24-stimulated GDP dissociation | 323 |
| S71P | Sc, Sp | ts loss-of-function | 391 |
| F78L | Hs | Wild-type response to Cdc24-stimulated GDP dissociation | 323 |
| S86P | Sc | Loss of binding to Cdc24; cs dominant negative; suppresses G12V | 116 |
| S88D | Hs | Specifies interaction with p190-GAP | 322 |
| S89P | Sc | cs dominant negative; suppresses G12V | 116 |
| W97R | Sc, Sp | ts (Sc), cs (Sp) loss-of-function | 391 |
| Q116K | Hs | Loss of Cdc24-stimulated GDP dissociation | 323 |
| I117S | Sc | Loss of binding to Cdc24 | 116 |
| D118A | Sc, Sp, Hs | Dominant negative (Sc); activated (Sp); sequesters Cdc24 | 116, 390, 641, 642 |
| T138A | Sc | Loss of binding to Cdc24 | 116 |
| G142S | Sc | Component of cdc42-1ts allele | 391 |
| C157R | Sc | Partial cs dominant negative | 116 |
| L165S | Sc | Loss of binding to Cdc24 | 116 |
| K183-187Q | Sc | Partial delocalization of protein; suppresses G12V | 116 |
| K186R | Sc | ts loss-of-function; higher intrinsic GTPase activity | 391 |
| C188S | Sc, Sp, Hs | Prenylation defective; delocalized protein; intragenic suppressor of G12V, Q61L, D118A mutations | 642, 643 |
This is not a complete set of references for the analysis of these mutations. See Fig. 1 for a map of these mutations on the Cdc42p primary amino acid sequence and the text for additional references.
Sc, S. cerevisiae; Sp, S. pombe; Dm, Drosophila; Hs, H. sapiens.
Cdc42Hs Three-Dimensional Structure
The numerous cdc42 mutations analyzed to date (see below) have greatly aided in defining functional domains within Cdc42p. However, without the information derived from the crystal structure of purified Cdc42, these mutations do little to clarify the global structure of Cdc42 and hence the multiple interactions between Cdc42 and its regulators and effectors. This problem has recently been resolved with the determination of the solution structure of Cdc42Hs by nuclear magnetic resonance (NMR) spectroscopy techniques (145), along with the determination of the X-ray crystal structure of Cdc42Hs bound to GDP (Fig. 1B) (413a). Several of the more interesting and informative mutations have been mapped onto the crystal structure (Fig. 1), and they highlight the potential functional domains of Cdc42p. The four domains implicated in the binding and hydrolysis of GTP are highlighted in blue in Fig. 1. The structure of these domains is similar to those found in the Ras and Rac crystal structures, highlighting the conservation of structure and function between different guanine nucleotide binding proteins.
Clearly, one of the more interesting and functionally important domains is the effector or switch I domain between residues 26 and 50 (highlighted in red in Fig. 1) (see “Effector domain” below). This domain forms an extended β2-strand/loop structure covering a large proportion of one face of the molecule. Based on its extended structure, it is easy to see how different effectors or regulators of Cdc42p could bind to different subdomains of the effector domain, possibly at the same time, as suggested by the analysis of different effector domain mutations. One effector/regulator could be bound to the N-terminal proximal domain around residue 35, which is in close proximity to the bound nucleotide, while another could be bound to the N-terminal distal region around residue 44. In addition, binding of an effector protein to this domain could interfere with the binding of other effector/regulator proteins to this domain, thereby providing a basis for the regulation of the myriad of Cdc42-dependent cellular processes.
The domain that makes Rho-type GTPases unique within the Ras superfamily is the so-called Rho insert domain (highlighted in purple in Fig. 1). This extra ∼13 amino acids is α-helical and has been implicated in Cdc42 interactions with one of its downstream effectors, the IQGAPs (379, 605), as well as its GDIs (605). In studies with a chimeric Cdc42Hs in which the insert domain between residues 120 to 139 was replaced with residues 121 to 127 of Ha-Ras, the resulting Cdc42Hs-ΔL8 protein showed a two- to threefold-reduced affinity for the carboxyl-terminal (97-kDa) half of IQGAP1, which contains the GRD Cdc42 binding domain (379, 605) (see “Cdc42p downstream effectors” below). While this Cdc42Hs-ΔL8 protein did not exhibit altered interactions with Rho-GDI from the perspective of the ability of Rho-GDI to extract Cdc42Hs-ΔL8 from membranes, it had a greatly reduced sensitivity to the Rho-GDI-dependent inhibition of GDP dissociation or GTP hydrolysis (605). These results suggest that the insert domain is mediating some of the effects of the Rho-GDI. Rho-GDI binding to Cdc42Hs requires C-terminal prenylation (307) (see “Mammalian GDIs” below), suggesting that Rho-GDI binds to the C-terminal prenylation domain. The fact that the insert domain is on the other side of the Cdc42 molecule from the C-terminal prenylation domain (Fig. 1B) makes it likely that Rho-GDI binding induces a conformational change in the structure of the insert domain, possibly leading to the insert domain shifting its location to block the guanine nucleotide binding pocket, thereby “locking” the Cdc42 protein in either a GDP- or GTP-bound state and inhibiting GDP dissociation or GTP hydrolysis (see “Mammalian GDIs” below). It is interesting that Ras proteins do not have an insert domain and also do not seem to have physiological interactions with GDI proteins. Recently, the insert domain was shown to play a role in the ability of Cdc42 to transform NIH 3T3 fibroblasts (606). The ΔL8 deletion mutation (see above) could intragenically suppress the transforming ability of the Cdc42F28L mutant protein without affecting its ability to bind GTP, induce Jun N-terminal kinase (JNK) and p21-activated kinase (PAK) activities, or induce filopodium formation (see “Mammals” under “Functional studies” below). These results further highlight the ability of Cdc42p to differentially function in multiple cellular processes through interactions between its different structural domains and downstream effectors (see “Cdc42p downstream effectors” below).
In addition to the effector domain, Cdc24p and other GEFs interact with Cdc42 through other domains, including residues 82 to 100, which encompass the β4-strand–α3-helix region, and residues 140 to 150, which encompass the α4-helix. The α3- and α4-helices lie on the same face of Cdc42p highlighted by the S86, W97, and G142 residues (Fig. 1B, right). The dominant negative S86P mutation lies in the loop region between the β4-strand and the α3-helix and it interferes with interactions between S. cerevisiae Cdc42p and Cdc24p (116). Interestingly, this loop region makes close contacts with residues in the Rho insert domain (see above), and this region undergoes chemical shift changes in NMR spectroscopy studies upon binding of the nonhydrolyzable GTP analog GMPPCP (145), suggesting that it may be an additional switch region (i.e., switch III). The nature of the S86P dominant negative phenotype is unknown, but it is not due to sequestration of Cdc24p as is the mechanism of action of the D118A mutant allele (116). The W97R (α3-helix) and G142S (α4-helix) mutations are ts loss-of-function alleles in S. cerevisiae (391) and although their mechanisms of action are unknown, the W97R mutation leads to a bud site selection defect, implicating Cdc42p in the initial selection of a nonrandom bud emergence site. Taken together, these observations show that the face of Cdc42p defined by the α3- and α4-helices plays a critical role in Cdc42p function. The structure and function of the C-terminal membrane localization domain (highlighted in green in Fig. 1) are discussed below (see “Prenylation and subcellular localization”).
GTP Binding and Hydrolysis Domains
The Cdc42 domains involved in guanine nucleotide binding and GTP hydrolysis (blue boxes in Fig. 1) have been inferred through structural similarities to domains in other GTPases and through the analysis of activated and dominant negative cdc42 mutations. The initial cdc42 mutations were analyzed in S. cerevisiae (642) and were based on the paradigmatic Ras mutations that led to oncogenic transformation. The cdc42G12V and cdc42Q61L mutations are analogous to H-ras mutations that cause a decreased level of intrinsic GTPase activity, thereby shifting the mutant proteins to an “activated” GTP-bound state. In S. cerevisiae, these cdc42 mutations were lethal, resulting in large, multibudded cells with aberrant cortical actin structures localized in multiple buds (642). These phenotypes suggested that the mutant proteins were activated and constitutively interacting with downstream components of the pathway, leading to polarization, albeit incorrectly, of the actin cytoskeleton. The H-rasD119A mutation also leads to an activated phenotype; however, this phenotype was mechanistically due to an increased GDP dissociation rate, which is thought to result in a higher probability of the protein being in a GTP-bound state due to the higher concentration of GTP than of GDP in the cell. The phenotype of the S. cerevisiae cdc42D118A mutant was unexpected and different from the cdc42G12V and cdc42Q61L activated phenotypes. The cdc42D118A mutant phenotype was temperature-dependent dominant lethal and resulted in large, round, unbudded cells that were phenotypically similar to cdc42ts mutants grown at restrictive temperatures. This dominant negative phenotype suggested that the Cdc42D118A mutant protein could bind and sequester the cellular factor(s) necessary for the budding process (see “S. cerevisiae Cdc24p” below).
Expression of equivalent mutant proteins in S. pombe gave different results (390). First, unlike the activated and dominant negative phenotypes seen in S. cerevisiae, the morphological phenotypes of cells overproducing the cdc42G12V, cdc42Q61L, and cdc42D118A mutant gene products were similar to one another. Second, the mutant constructs did not exert a dominant lethal effect in S. pombe cells. Instead, S. pombe cells overproducing these mutant proteins exhibited an abnormal morphology of enlarged, round or misshapen cells with delocalized cortical actin structures, as opposed to the small, round cellular morphology of cdc42 loss-of-function and dominant negative mutants (390, 447). The cdc42D118A mutant phenotype also was temperature dependent in S. pombe, suggesting that the mutant protein loses either a critical interaction or its three-dimensional structure upon shift to higher temperature, leading to its mutant morphology. Interestingly, septum formation was still evident in these mutant cells, even though the presence of an organized actin ring was not, suggesting that septation can occur in the absence of an actin ring. However, this point must be clarified by experiments with either actin mutants or actin polymerization inhibitors such as latrunculin A.
While activated or dominant negative cdc42 mutations have not been analyzed in C. elegans to date, expression of the cdc42G12V allele in Drosophila ovaries led to defects in actin distribution whereas expression of dominant negative cdc42 alleles led to defects in actin organization in imaginal discs and wing hairs (see “Drosophila” under “Functional studies” below), reinforcing a role for Cdc42p in regulating actin function. For a discussion of mutations in mammalian Cdc42p, see “Mammals” under “Functional studies” below.
Effector Domain
The so-called effector domain between residues 26 and 48 of the Ras GTPase is required for downstream effector function (370, 460). The effector or switch I domain between residues 26 and 50 is highly conserved among Cdc42 proteins (Fig. 1A) but diverges among closely related but not functionally homologous Rac GTPases (239). The current paradigm is that GTPases bind to GEFs when in the nucleotide-free or GDP-bound state and bind to GAPs and downstream effectors when in the GTP-bound state. Since the switch I and II (residues 60 to 76) domains are the predominant regions of GTPases that change conformation upon binding different guanine nucleotides, it is likely that multiple factors interact with these regions. Given that Cdc42p interacts with multiple downstream effectors along with regulatory factors such as GEFs and GAPs (see “Cdc42p regulators” and “Cdc42p downstream effectors” below), it is likely that the specificity of interaction will be through either different residues within the Cdc42p effector domain, competition between effectors and regulators, and/or interactions at different times in the cell cycle.
A predominant binding partner for the effector domain is the CRIB (for “Cdc42/Rac interactive binding”) domain (also known as the PBD, GBD, or PAK domain [see Table 3]) found in many Cdc42p downstream effectors, including the PAK family of protein kinases (59). The highest-efficiency binding domain in the CRIB-containing PAK protein was residues 70 to 118, thereby defining the optimal CRIB domain as these 48 amino acid residues (558). In these studies, it was also shown that this domain interacted with Cdc42p at a ∼3- to 10-fold-higher affinity than it interacted with Rac and that it interacted with activated (Q61L) alleles at a 5- to 10-fold-higher affinity than it interacted with the wild type, reinforcing the notion that CRIB-containing interacting proteins function as downstream effectors. A recent study in which NMR spectroscopy was used to probe the interactions between Cdc42Hs and 46 amino acids of the PAK CRIB domain showed that the CRIB binding domain surface on Cdc42Hs was the β2 switch I domain and part of the loop between the α1-helix and β2-strand (185). In addition, nuclear Overhauser effect contacts suggested that the formation of an intermolecular β-sheet was the basis for the Cdc42Hs-CRIB domain interactions. The CRIB domain of the Wiskott-Aldrich syndrome protein (WASP) downstream effector (see “Cdc42p downstream effectors” below) was dissected by a variety of biophysical techniques including fluorescence spectroscopy, surface plasmon resonance, circular dichroism, and NMR spectroscopy (498). The results indicated that a core 26-amino-acid fragment (residues 221 to 257) was necessary for binding to GST-Cdc42, but higher affinity binding was observed with a larger (120-amino-acid) fragment (residues 201 to 321), suggesting that the CRIB domain was necessary but not sufficient for high-level binding. In addition, these studies suggested that the isolated CRIB domain does not exhibit any apparent secondary structure; it is unknown if the CRIB domain would form a secondary structure, possibly β-strands (see above), within the context of the entire protein.
TABLE 3.
Structural and/or functional domains within Cdc42 regulators and effectors
| Domaina | Function | Regulator or effector |
|---|---|---|
| CRIB (also PBD, GBD, PAK) | Binding to Cdc42p effector domain | PAK, ACK, Gek, MIHCK, MLK3, and MEKK4 kinases; Gic1; Gic2; WASPs; N-WASP |
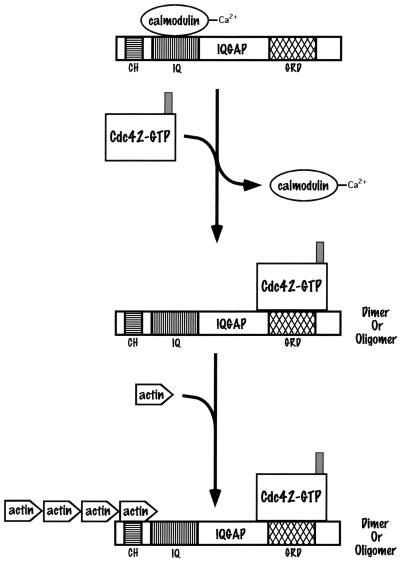
| CH | Binding to actin | Iqg1/Cpk1, IQGAPs |
| DH | GEF catalytic domain | GEFs |
| FH1, FH2 | Binding to profilin, myosin | Bni1, Bnr1, formins |
| GRD | Binding to Cdc42p | Iqg1/Cpk1, IQGAPs |
| IQ (Ile and Gln residues) | Binding to calmodulin | Iqg1/Cpk1, IQGAPs, N-WASP |
| LIM | Protein-protein interactions | Rga1, Rga2 |
| PDZ | Protein-protein interactions | Sif, DRhoGEF2 |
| PH | Membrane localization and protein-protein interactions | GEFs, Bem3, CeGAP, Cla4, Skm1, Pak2, Gek, Bee1, WASPs, N-WASP, Boi1, Boi2 |
| SH3 | Binding to proline-rich domains | Myo3, Myo5, Bem1, Boi1, Boi2, Scd2, DrtGEF |
CRIB, Cdc42/Rac interactive binding; CH, calponin homology; DH, Dbl homology; FH1, FH2, formin homology 1, 2; GRD, GAP-related domain; PH, pleckstrin homology; SH3, src homolog 3.
Mutations that disrupt the interaction between Cdc42p and downstream effectors should define the effector domain and should suppress dominant activated cdc42G12V mutant phenotypes. The T35A allele was thought to be a paradigmatic effector domain mutation in that it could interfere with the ability of S. cerevisiae Cdc42p to interact with the PAK family of protein kinases and could suppress the dominant-activated cdc42G12V mutant but could not complement the loss-of-function cdc42-1ts allele. However, the T35A mutation also suppressed the dominant negative S. cerevisiae cdc42D118A allele and interfered with two-hybrid protein interactions between Cdc42D118Ap and the Cdc24p GEF (116), suggesting that the effector domain may also interact with the Cdc24p GEF. Corroborating this hypothesis are the results obtained with the Y32K and F37E mutations in the Cdc42Hs effector domain, which caused a loss of Cdc24-stimulated GDP dissociation (323), and the Cdc42Hs F28L mutation, which led to rapid nucleotide exchange and transformation of NIH 3T3 cells similar to that seen with the Cdc24p homolog Dbl (327). Interestingly, the T25K, N26D, and Y40K mutations within the Cdc42Hs effector domain did not show a loss of Cdc24-stimulated GDP dissociation (323), which could be a function of the individual mutational changes or could indicate a level of specificity at the individual amino acid residue for interactions with GEFs.
The S. cerevisiae cdc42V44A mutation represents a new class of effector domain mutations in that it could complement the cdc42-1ts allele (475a), suggesting that it did not lead to a nonfunctional protein; it also interfered with interactions with the upstream effector Cdc24p (116). In addition, the cdc42V44A mutant displayed a morphological phenotype of elongated buds with multiple nuclei, which is suggestive of either a delay at the apical/isotropic switch and morphogenesis checkpoint (see “S. cerevisiae” under “Functional studies” below) and/or a defect in cytokinesis (475a). The V44A mutation interfered with two-hybrid protein interactions between Cdc42p and the S. cerevisiae Cla4p PAK-like kinase but not the Ste20p or Skm1p PAK-like kinases and also between Cdc42p and the Gic1p and Gic2p downstream effectors but not the Bnip or Iqg1p scaffold proteins (see “Cdc42p downstream effectors” below). All of these proteins contain CRIB domains, suggesting that the effector domain may differentially interact with multiple CRIB domain-containing effectors. This hypothesis is substantiated by mutations in the Cdc42Hs effector domain that differentially affected interactions with mammalian downstream effectors (291, 379) (see “Cdc42p downstream effectors” below). The Y40C mutation interfered with interactions between Cdc42p and downstream PAKs and other proteins containing CRIB domains, leading to a loss of p65PAK kinase activation in transfected COS fibroblasts, but it did not affect Cdc42p-dependent actin reorganization, as evidenced by normal filopodium and integrin complex formation in Swiss 3T3 cells (291). The D38E mutation interfered with in vitro binding to an mPAK-3 CRIB domain peptide (310) but had no effect on the binding of two other downstream effectors, IQGAP1 and IQGAP2. The Cdc42Hs F37A mutation did not affect interactions with CRIB-containing proteins or actin reorganization (291). Taken together, these results suggest that there are different classes of effector domain mutations that can be distinguished by their morphological phenotypes and protein-protein interactions. These different mutations may define interactions with different effectors or regulators, thereby allowing us to dissect the multiple pathways leading from Cdc42p (see “Cdc42p downstream effectors” below). Given the possibility that the Cdc42p effector domain interacts with CRIB-containing proteins through formation of an intermolecular β-sheet (see above), it is likely that the nature and orientation of amino acid side chains emanating from the β2 strand (Fig. 1) have an influence on this differential binding. It should be noted, however, that not all Cdc42p-interacting proteins contain recognizable CRIB domains, suggesting that there may be multiple mechanisms by which proteins interact with Cdc42p.
GEF Interaction Domains
In addition to the effector/switch I domain mutations that affect interactions between Cdc42 and its GEFs, there are mutations in other domains that affect either binding to GEFs or GEF-induced GDP dissociation. Mutations within the switch II domain, including D63L, D65K, R66D, R68A, P69A, and F78L, did not affect Cdc24-stimulated GDP dissociation, but the Q116K mutation in the Cdc42 signature GTP binding domain did (323). Analysis of chimeric Cdc42Hs-RhoA proteins indicated that residues 82 to 120 and 121 to 155 are necessary for Cdc24 responsiveness (323). Within the first domain, the S86P and S89P mutations in S. cerevisiae Cdc42p led to a cold-sensitive, dominant negative phenotype and, in the case of S86P, led to a loss of interaction with Cdc24p (116). Similar mutations in Drosophila Cdc42p also led to a dominant negative phenotype (see “Drosophila” under “Functional studies” below). Other mutations that led to a loss of interaction with Cdc24p in S. cerevisiae included the I117S mutation within the Cdc42 signature GTP binding domain and the T138A and L165S mutations (116). In addition, the C157R mutation within one of the highly conserved GTP binding domains led to a partially cold-sensitive dominant negative phenotype. The C-terminal membrane localization domain (see “Prenylation and subcellular localization” below) does not seem to be necessary for GEF interactions (323). Interestingly, the S86, S89, and T138 residues all lie on the same face of the Cdc42 protein (Fig. 1B), suggesting that this may be a conserved interactional interface between Cdc42p and its GEFs.
PRENYLATION AND SUBCELLULAR LOCALIZATION
In all organisms examined, Cdc42p is prenylated with a C20 geranylgeranyl isoprene group at a C-terminal Cys residue, and this prenylation is necessary for the membrane attachment of Cdc42p. S. cerevisiae and S. pombe Cdc42p fractionated into both soluble and particulate fractions, suggesting that Cdc42p can exist in two cellular pools (390, 643). S. cerevisiae Cdc42p was found predominantly in the particulate fraction, but a significant soluble pool, up to ∼20% in some instances, could be observed. Given the existence of GDI proteins in S. cerevisiae and mammalian cells that can interact with Cdc42p and extract Cdc42p from membranes (see “Guanine nucleotide dissociation inhibitors” below), the soluble pool of S. cerevisiae Cdc42p is probably either nonprenylated or complexed with the rho-GDI protein Rdi1p (267, 373). The particulate form of Cdc42p could be solubilized by added detergent but not by added NaCl or urea, suggesting that Cdc42p was tightly associated with either a cellular membrane or a cytoskeletal complex. When synchronous cultures were used, the fractionation pattern of S. cerevisiae Cdc42p did not appear to vary through the cell cycle (643). However, recent studies indicating that Cdc42p functions at different places in the cell at different times of the cell cycle (see below) suggest that fractionation patterns may not be a very sensitive measure of Cdc42p localization.
By using Cdc42p-specific antibodies in immunofluorescence and immunoelectron microscopy, S. cerevisiae Cdc42p was localized to the plasma membrane at sites of polarized growth (642, 643). These sites coincided with the sites of cortical actin localization and included invaginations of the plasma membrane at the site of bud emergence, the tips of growing buds, and the tips of mating projections in pheromone-arrested cells (3, 259). This localization pattern was consistent with Cdc42p functioning in controlling polarized cell growth during the mitotic cell cycle and mating. Recently, functional green fluorescent protein (GFP)-Cdc42p fusion proteins have been localized to the mother-bud neck region in S. cerevisiae and the septum area in S. pombe, suggesting that Cdc42p also plays a role in cytokinesis and/or septation in both yeasts (508a). This localization of Cdc42p at different sites of polarized growth during the S. cerevisiae cell cycle, which is mirrored by the localization of the actin cytoskeletal network, suggests that the subcellular localization of Cdc42p is under temporal and spatial control during the cell cycle. It should be noted that Cdc42p localization to sites of polarized growth was not disrupted by incubation with the actin-depolymerizing drug latrunculin-A (21), suggesting that Cdc42p localization occurs independently of actin localization and of the structural integrity of the actin cytoskeleton.
S. cerevisiae Cdc42p contains the C-terminal 183Lys-Lys-Ser-Lys-Lys-Cys-Thr-Ile-Leu sequence which is essential for the membrane localization of Cdc42p. In the Cdc42Hs NMR and crystal structures, this region forms a flexible tether that is separated from the body of Cdc42Hs by two Pro residues at positions 179 and 180 (145) (Fig. 1B), thereby allowing the bulk of Cdc42p to be sterically unhindered by membrane attachment and accessible for binding to other proteins. This region is modified by geranylgeranylation at the Cys188 residue (indicated by an underbar in the above sequence), which is necessary for its anchoring within the plasma membrane (147, 643). This is thought to be followed by proteolytic cleavage of the last three amino acids and carboxyl methylation of the now C-terminal Cys residue, although this has not been experimentally shown with S. cerevisiae Cdc42p (see below). The geranylgeranylation is deemed necessary because the C188S mutation, which renders the protein incapable of being prenylated, can intragenically suppress the dominant lethality associated with the cdc42G12V, cdc42Q61L, and cdc42D118A mutations (642) and because the S. cerevisiae and S. pombe Cdc42C188S mutant proteins, either by themselves or as GFP-Cdc42C188Sp fusion proteins, are nonfunctional, delocalized proteins that fractionate almost exclusively into soluble pools (508a, 642, 643). Whether this prenylation is sufficient for Cdc42p targeting to the sites of polarized growth is not known.
An increase in soluble S. cerevisiae Cdc42p was observed in cdc43-2ts (643) and cdc43-5ts (434) mutant cell extracts, suggesting that membrane localization of Cdc42p depended on Cdc43p-dependent geranylgeranylation. Cdc43p was originally identified by ts mutations that led to cell cycle arrest of large, unbudded cells, a phenotype similar to cdc42ts mutants (2). In addition, cdc43ts cdc42ts double mutants displayed a synthetic lethal phenotype at 23°C, suggesting that these gene products may interact. Another mutation in CDC43, designated cal1-1, was identified by its calcium-dependent phenotype (437); the cal1-1 mutant required 100 mM CaCl2 for growth. The predicted amino acid sequence of the CDC43/CAL1 gene (240, 435) showed significant similarity to the S. cerevisiae DPR1/RAM1 gene (173), which encoded the β subunit of the protein farnesyltransferase (FTase), which modified the C termini of Ras GTPases. Type I protein geranylgeranyltransferase (GGTase I) activity was reconstituted from Escherichia coli cells that overproduced both Cdc43p and Ram2p (377, 539, 540), and S. cerevisiae GGTase I activity was decreased in cdc43ts and ram2 mutants but not ram1 mutants (147, 377, 434), indicating that Cdc43p and Ram2p encoded the β and α subunits, respectively, of the S. cerevisiae GGTase I. The Ram2p α subunit also acts as the α subunit for the S. cerevisiae FTase (214), which may account for the in vitro and in vivo cross-specificity that is observed between FTase and GGTase I activities in S. cerevisiae (66, 565). S. cerevisiae GGTase I is an Mg2+-requiring Zn2+ metalloenzyme (377, 539), but it can also function with Ca2+ as the only divalent cation (377). Added Ca2+ could not rescue the reduced in vitro GGTase I activity from cal1-1 mutant cell extracts (434), but only 20 mM CaCl2 was added, as opposed to the 100 mM CaCl2 needed to rescue the in vivo cal1-1 growth defect (435, 437). The essential targets for S. cerevisiae GGTase I seem to be Cdc42p and Rho1p, because certain cdc43/cal1 alleles can be suppressed by overexpression of one or both of these GTPases (434, 439).
Another localization determinant consists of the four Lys residues that are next to the C-terminal prenylated Cys residue. This polylysine region is not found in most Ras-like GTPases, and its positive charges may be interacting with negatively charged components, either protein or phospholipid, at the membrane site to play a role in enhancing membrane association or specific targeting of Cdc42p. A similar polylysine domain is found in the K-Ras protein and is important for membrane localization; altering the Lys residues to Gln results in delocalized K-Ras protein (196, 197). In addition, the analogous polylysine domain in Rac1 was recently shown to be important for interactions with PAK effector kinases (266). Mutating the four Lys residues to Gln in S. cerevisiae Cdc42p, creating the cdc42K183-187Q mutant protein, led to a partial delocalization of the mutant protein (116), suggesting that this domain played a role in targeting or anchoring Cdc42p to the plasma membrane. The K183-187Q mutation could intragenically suppress the dominant lethal cdc42G12V mutant, and expression of the cdc42K183-187Q mutant gene on a plasmid could complement the cdc42-1ts mutant. The ability of the cdc42K183-187Q mutant gene to complement the cdc42-1ts mutant (in contrast to the nonfunctional cdc42C188S mutant gene, which cannot complement the cdc42-1ts mutant [642]), together with the partial delocalization of the mutant protein, suggested that the K183-187Q mutation had an intermediate effect on Cdc42p function and that the polylysine domain of Cdc42p was necessary but not sufficient for complete plasma membrane localization. Another interesting mutation in this domain, the cdc42K186R mutant allele, exhibited a ts loss-of-function phenotype in S. cerevisiae (391) and displayed a morphological phenotype of elongated buds and multiple nuclei suggestive of either a delay at the morphogenesis checkpoint (see “S. cerevisiae” under “Functional studies” below) and/or a cytokinesis defect at permissive temperatures (106a). The nature of this mutation (Lys to Arg) suggested that these phenotypes were not due to a change in charge or conformation of the protein but more probably were due to altered interactions with another protein. However, recent results indicate that this mutant protein has an increased intrinsic GTPase activity (see “GTPase-activating proteins” below), suggesting that improper negative regulation of this mutant protein may be playing a role in its phenotypes. Therefore, the mechanism by which this mutant protein exerts its effects remains to be fully elucidated.
All known Cdc42 proteins contain the C-terminal sequence Cys-Xaa-Xaa-Leu, with the exception of the mouse and human brain G25K isoforms, which end in a Phe residue instead of Leu (Fig. 1). Mammalian Cdc42p posttranslational modifications have been analyzed biochemically with protein purified from bovine brain cells (22, 612, 613), cultured murine erythroleukemia cells (354), rat and human pancreatic islet cells (274), or rat kidney cells (45), not with recombinant protein, and so it is unclear whether these studies were performed on the Cdc42Hs or G25K isoform. Regardless, it is clear that the membrane-bound form of mammalian Cdc42p is geranylgeranylated at the Cys residue, the last three amino acids are proteolytically removed, and the now C-terminal prenylated Cys residue is carboxyl methylated, resulting in a protein with a S-(all-trans-geranylgeranyl) cysteine methyl ester at its C terminus (612). These modifications are necessary for membrane localization, and, as with S. cerevisiae Cdc42p, mammalian Cdc42p fractionates to both particulate and soluble pools (45, 354).
Carboxyl methylation of soluble Cdc42p from bovine brain (22), rat kidney cells (45), or pancreatic islet cells (274) seems to be GTP stimulated, but methylation of the membrane-bound form is not (613), presumably because the membrane-bound form is already GTP bound. The methyltransferase activity from brain extracts (613) and insulin-secreting cells (318) was membrane bound. Recently, a human myeloid prenylcysteine carboxyl methyltransferase with in vitro activity against Cdc42Hs was shown to localize to the endoplasmic reticulum membrane (114); the S. cerevisiae Ste14p prenylcysteine carboxyl methyltransferase is also found in the endoplasmic reticulum membrane (490), but it has not been shown to have in vitro or in vivo activity against Cdc42p. Interestingly, addition of glucose to pancreatic islet cells extracts stimulated the carboxyl methylation of Cdc42p (274), and inhibition of Cdc42p function by Clostridium difficile toxins A or B resulted in reductions in glucose-stimulated insulin secretion (273), suggesting that Cdc42p may play a role in glucose-stimulated insulin secretion.
Using affinity-purified anti-Cdc42 antibodies, Cdc42p from rabbit liver was shown to associate with a membrane fraction highly enriched in Golgi membranes (137). In pancreatic islet cells (274) and rat kidney cells (45), Cdc42 was predominantly cytosolic, but it was translocated to the particulate pool upon addition of guanosine 5′-(3-O-thio)triphosphate (GTPγS). In NR-6 fibroblasts and rat kidney cells, Cdc42p localized to a perinuclear region that coincided with markers for the Golgi complex including the 110-kDa subunit of the coatomer complex β-COP (137). This localization was rapidly altered to general cytosolic localization upon addition of brefeldin A (BFA), a drug which inhibits vesicle formation at the Golgi membrane by inhibiting the guanine nucleotide exchange activity for the Arf GTPase, suggesting that Cdc42p may play a role in or be subject to intracellular membrane trafficking events. BFA-induced delocalization of Cdc42p was inhibited by AlF4− and by expression of GTPase-defective Arf, while expression of a dominant negative Arf mutant resulted in BFA-independent delocalization of Cdc42p. These results suggest that association of Cdc42p with Golgi membranes is dependent on the guanine nucleotide-bound state of the Arf GTPase. It should be noted that in these experiments, the NR-6 fibroblasts and rat kidney cells did not exhibit polarized growth patterns to a region of their cell periphery. In human HeLa cells transiently transfected with a epitope-tagged Cdc42G12V protein, the epitope-tagged protein localized to focal complexes and to regions of polarized growth within the Cdc42G12V-induced peripheral actin microspikes (PAMs) (see “Mammals” under “Functional studies” below) and colocalized with actin and PAKs within these PAMs (127, 356). In addition, HA-tagged Cdc42G12V protein co-localized with the IQGAP1 downstream effector to cell-cell contact sites of Madin-Darby canine kidney cells (282).
In Drosophila wing disc epithelial cells, Cdc42p localized in a polarized manner to the basal and apical regions (128). In elongating cells, Cdc42p was restricted to the apical and basal membranes, but in nonelongating cells, it was found on lateral membranes as well. This localization pattern was also seen for the actin cytoskeleton, again providing a mechanistic link between Cdc42 and actin rearrangements. C. elegans Cdc42p was shown to fractionate predominantly to a particulate fraction from mixed-stage populations of C. elegans cells and to localize in a polarized manner to both the circumferential and longitudinal boundaries of hypodermal cells during hypodermal cell fusion in embryo elongation, in a pattern similar to that of the C. elegans PAK homolog (87).
In summary, Cdc42 proteins are membrane bound through their posttranslational modifications and are localized to either internal membranes or the plasma membrane at locations where polarized events are occurring. While prenylation is necessary for membrane anchorage, it is not known if it is sufficient for proper targeting. The mechanism by which Cdc42 proteins are targeted to appropriate membranes in regions of polarized cell growth is unknown, but it is likely to be through protein-protein or protein-lipid interactions at the site.
FUNCTIONAL STUDIES
It is clear from the recent explosion of experimental results that Cdc42p functions in a variety of cellular processes in eukaryotic cells. The major functions of Cdc42p seem to be in regulating the rearrangements of the actin cytoskeleton in response to extracellular and intracellular signals as well as in modulating protein kinase cascades that result in the transcriptional activation of genes required for growth control and numerous other cellular processes. Cdc42p also performs other cellular functions, which are independent of actin rearrangements and mitogen-activated protein (MAP) kinase cascades, and so it would be premature to think that the panoply of Cdc42p-dependent processes has been entirely revealed. Although Cdc42p has been implicated in a wide variety of cellular processes, we still have little insight into the mechanisms of action or the conservation of function for Cdc42p within these processes.
Saccharomyces cerevisiae
S. cerevisiae alters its morphology in response to both exogenous and endogenous signals, leading to either bud emergence and enlargement during the mitotic cell cycle, mating-projection (“shmoo”) formation through the mating/pheromone response pathway in response to exogenous mating-factor pheromones, pseudohyphal formation and filamentous growth in response to starvation conditions, or spore formation during meiosis. Cdc42p has been implicated in regulating the first three processes but not in sporulation to date. The mechanisms by which Cdc42p regulates the generation of, and switching between, these different morphogenetic patterns is still unclear, but Cdc42p interactions with the actin cytoskeleton play a critical role in this regulation. The functional connection between Cdc42p and the cortical actin cytoskeleton has recently been reinforced by the observation that Cdc42p can stimulate actin polymerization in permeabilized S. cerevisiae cells (324).
Mitotic cell cycle.
The morphological changes that occur during the S. cerevisiae mitotic cell cycle can be divided into five sequential phases: (i) selection of a nonrandom bud emergence site and the organization of the protein machinery at that bud site, including rearrangement of the cortical actin cytoskeleton; (ii) bud emergence and polarized growth towards and within the emerging bud; (iii) a switch from apical to isotropic bud growth (the “apical-isotropic switch” [see below]); (iv) cytokinesis, septum formation, and cell separation; and (v) isotropic growth of undersized daughter cells after cell separation prior to the initiation of their next cell cycle (reviewed in references 77, 126, and 465) (Fig. 2). A variety of data suggest that Cdc42p can function at multiple stages of the cell cycle. As mentioned above (see “Cdc42p structure and functional domains”), the initial characterization of S. cerevisiae Cdc42p suggested that it plays a role in the actin-dependent generation of cell polarity during the process of bud emergence. Subsequent analysis of ts, dominant activated, and dominant negative cdc42 alleles substantiated this inference and suggested an additional function in the initial selection of the site of bud emergence. Analysis of the cdc42V44A and cdc42K186R mutant alleles, along with the subcellular localization of Cdc42p to the mother-bud neck region in large-budded cells, raises the possibility that Cdc42 functions either within the apical-isotropic switch at a morphogenesis checkpoint (Fig. 2) (see below) and/or in controlling actin-dependent events that occur during cytokinesis and septum formation. In addition, GTP-bound Cdc42p functions with the mitotic cyclin Clb2p-Cdc28p kinase complex to lead to the mitosis-specific phosphorylation of several substrates (see below). A potential model that is consistent with the proposed roles for Cdc42p throughout the cell cycle is presented in Fig. 3; a detailed discussion of individual protein components of the model can be found under the individual protein subsections in “Cdc42p regulators” and “Cdc42p downstream effectors” below.
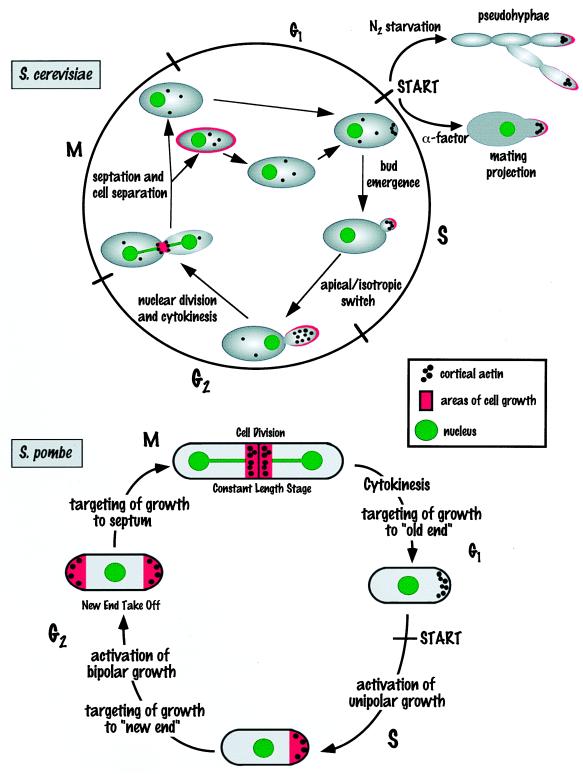
FIG. 2.
Polarized cell growth during the S. cerevisiae and S. pombe cell cycles. See the text for details.
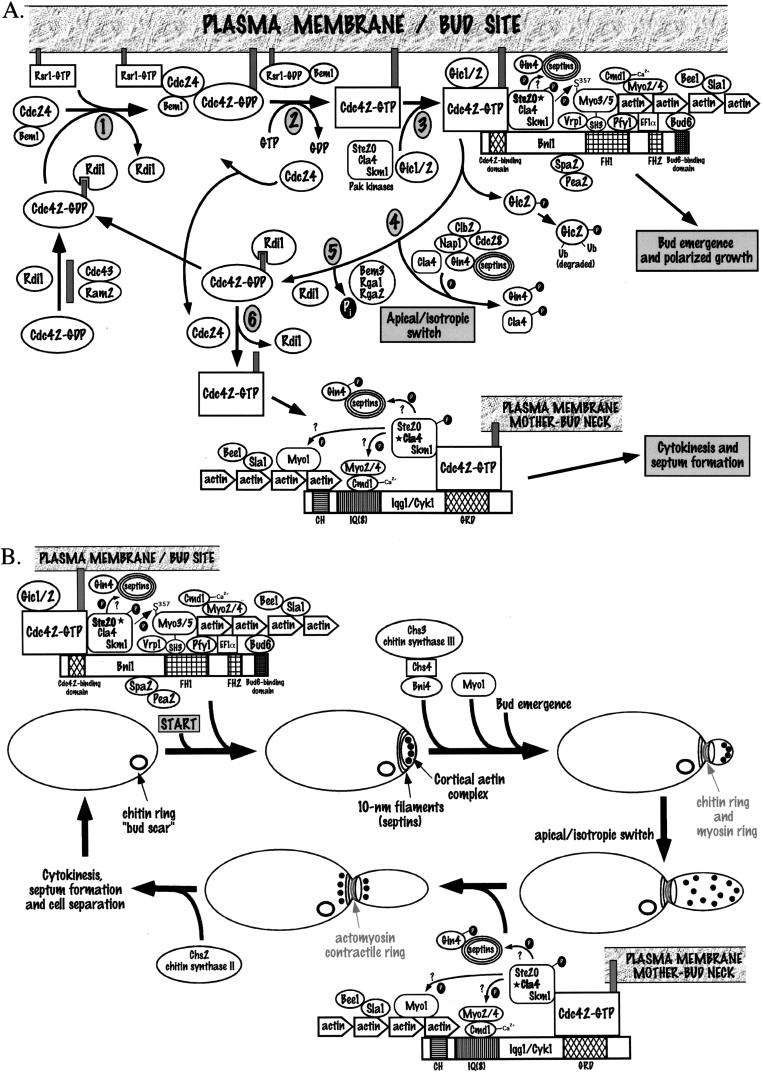
FIG. 3.
Molecular and cytological model for polarized cell growth in S. cerevisiae. (A) Molecular model for the Cdc42-dependent processes during the S. cerevisiae cell cycle. Shaded boxes attached to GTPases are isoprenyl groups. The stars by Ste20 in the bud emergence complex and Cla4 in the cytokinesis complex indicate that they are the likely PAK functioning at this stage of the cell cycle. See the text for details. (B) Cytological model for bud emergence and cytokinesis. Cdc42 complexes are the same as in panel A. See the text for details.
Newly synthesized Cdc42p is geranylgeranylated by the Cdc43p-Ram2p GGTase I and interacts with the Rdi1p rho-GDI within the cytosol (Fig. 3A). Presumably, Cdc42p is in a GDP-bound state at this point, because the Cdc24p GEF is membrane bound. In step 1, the Cdc42p-GDP-Rdi1p complex interacts with a Cdc24p-Bem1p protein complex at the plasma membrane. This interaction and subsequent guanine nucleotide exchange presumably leads to a loss of Cdc42p-Rdi1p binding. The Cdc24p-Bem1p complex could bind to the plasma membrane either through an interaction between Cdc24p and the GTP-bound Rsr1p/Bud1p GTPase, which is already at the plasma membrane at the site of incipient bud emergence, or through the Cdc24p PH domain. It is unclear how, when, or where the Cdc24p guanine nucleotide exchange function is activated, but it is likely that this occurs at the bud site. In step 2, Cdc24p catalyzes the dissociation of GDP from Cdc42p, and GDP is replaced by GTP. As a result of this biochemical exchange reaction, Cdc24p dissociates from both Cdc42p and Bem1p, which can now interact with Rsr1p/Bud1p, which is GDP bound through the action of the Bud2p GAP (not shown in Fig. 3A). Released Cdc24p is free to recycle to the bud site or become available for nucleotide exchange later in the cell cycle (see below).
In step 3, activated GTP-bound Cdc42p interacts with Gic1p and/or Gic2p and/or one of the family of PAK-like kinases (Cla4p, Ste20p, and Skm1p), and this complex binds to the Bni1p scaffold protein (see reference 454 for a review of scaffold proteins). Ste20p is the most likely PAK involved at this step due to its localization to bud tips and its Cln1p–2p/Cdc28p-dependent phosphorylation at this stage of the cell cycle (see below). This interaction brings the PAK-like kinase in close proximity to the Myo3p and/or Myo5p myosin, which can bind to Bni1p through an interaction between their SH3 domains and the FH1 formin domain of Bni1p, leading to the phosphorylation of the S357 residue of Myo3p and/or Myo5p. Bni1p can also bind to a number of other proteins that interact with the actin cytoskeletal network, including the Pfy1p profilin, the EF1α elongation factor, Bud6p, and Spa2p. This complex, along with a number of other actin-binding proteins (not shown in Fig. 3), nucleates the localized assembly of the septin, chitin, and Myo1p rings and the subsequent polymerization of actin at the bud tip, leading to bud emergence and apical bud growth (Fig. 3B). After bud emergence, Gic2p is phosphorylated and ubiquitinated in a Cdc42p-GTP-dependent manner, leading to its degradation (Fig. 3A).
After DNA replication, the apical growth of the bud switches to isotropic growth. This apical-isotropic switch (316) (step 4) depends on activation of the Clb1p–2p/Cdc28p kinase complex, which, along with GTP-bound Cdc42p and Nap1p, is needed for the hyperphosphorylation of Cla4p and Gin4p; it is evidenced by bud shape changes and the distribution of cortical actin and Cdc42p to the sides of enlarging buds. The action of one or more Cdc42p-GAPs (Bem3p, Rga1p, and Rga2p) may also be necessary for the apical-isotropic switch, leading to the conversion of Cdc42p-GTP to a GDP-bound state which can then be extracted from the plasma membrane by the Rdi1p Rho-GDI (Fig. 3A, step 5). Since both GFP-Cdc42p and GFP-Cdc24p are localized to the mother-bud neck region in large-budded cells (508b), in step 6 Cdc42p can be activated to a GTP-bound state by Cdc24p at the mother-bud neck region. Activated GTP-bound Cdc42p can interact again with one or more of the PAK-like kinases, probably Cla4p, whose kinase activity peaks at G2/M, as well as the IQGAP homolog Iqg1p/Cyk1p through its GRD domain. The Iqg1p/Cyk1p IQGAP protein is another scaffold protein that can interact with the Cmd1p calmodulin protein through its IQ domains as well as with actin through its calponin homology (CH) domain. The activated PAK-like kinase might then phosphorylate the Myo1p myosin, which has already formed a ring structure at the mother-bud neck region, the Myo2 and Myo4 myosins, which bind to calmodulin, the septin proteins that comprise the 10-nm filament ring present at the mother-bud neck region, or other as yet undetermined substrates. (Phosphorylation of Myo1p and/or septin proteins may also occur upon assembly in late G1; see “Cla4p” under “Cdc42p downstream effectors”). These interactions lead to the formation of a septin-dependent actomyosin ring at the mother-bud neck region and the subsequent contraction of this ring following anaphase, leading to cytokinesis (Fig. 3B). The localization of cortical actin at the mother-bud neck region, following the contraction of the actomyosin ring and cytokinesis, is a prelude to chitin synthase II-dependent septum formation and eventual cell separation (Fig. 3B).
The means by which cell cycle control is imposed on these morphogenetic events is starting to come into focus (for reviews, see references 41, 316, and 381). The timing of these events is coordinated with the cyclin-dependent kinase (CDK) Cdc28p and the START step of the cell cycle. Formation of the cortical actin patches and formation of the septin, Myo1p, and chitin rings occur around the same time in late G1 following the activation of the G1 cyclins Cln1p-3p–Cdc28p kinase complexes that are necessary for START (Fig. 3B). Bud emergence and apical bud growth occur ∼15 min later. The Cln2p-Cdc28p complex also phosphorylates the Ste20p PAK-like kinase at this stage of the cell cycle, and although this phosphorylation occurs at the time when Ste20p is localized to the emerging bud tips, it does not seem to affect the Ste20p kinase activity (see “Ste20p” below). There also exists a morphogenesis checkpoint that monitors proper actin cytoskeletal structures after bud emergence (313–315, 381, 526). The lack of proper actin structures triggers this morphogenesis checkpoint, causing the Swe1p inhibitory phosphorylation of the mitotic cyclin Clb1p–2p/Cdc28p complexes at the Y19 residue of Cdc28p, leading to G2/M and nuclear division delays. The morphological phenotypes associated with the cdc42V44A effector domain mutant suggest that Cdc42p or Cdc42p-dependent events may also be involved in this checkpoint (475a). Activation of the Clb1p–2p/Cdc28p kinase complex is necessary for the apical-isotropic switch, possibly through the Cdc42p- and septin-dependent phosphorylation of Gin4p and Cla4p (Fig. 3A), and for the promotion of anaphase and nuclear division, while destruction of the mitotic cyclins Clb1p through Clb4p is necessary for cytokinesis and redistribution of cortical actin to the site of septation. While many of the aspects of this proposed cell cycle and morphogenesis model are consistent with published findings (see the sections on individual proteins below), several aspects remain to be experimentally tested. Most notably, the functional consequences of interactions between Cdc42p and Gic1p, Gic2p, and the three PAK-like kinases remain to be fully elucidated and the potential cell cycle-dependent role of the Bni1p, Bnr1p, and Iqg1p/Cyk1p scaffold proteins in brokering Cdc42p-actin interactions is unclear. In addition, the substrates for the Cln1p–3p/Cdc28p and Clb1p–4p/Cdc28p CDK complexes that are necessary for these cell cycle-dependent morphogenetic switches remain to be determined.
Mating pathway.
The S. cerevisiae mating pathway is a classic signal transduction pathway in which an extracellular signal (peptide pheromone) binds to a G-protein coupled transmembrane receptor, thereby activating a MAP kinase cascade that ultimately leads to a number of cellular events, including the transcriptional induction of genes necessary for the mating process, a G1 arrest as a prelude to cell-cell fusion and karyogamy, and the generation of unique morphological structures (mating projections or shmoos) that are the sites of contact for cell-cell fusion and mating (for reviews, see references 26, 297, 350, and 368) (Fig. 4A). The notion that Cdc42p plays a role in the S. cerevisiae mating pathway came from the analysis of cdc42 mutants, the subcellular localization of Cdc42p, and its interactions with the Ste20p protein kinase. The mating efficiencies of loss-of-function cdc42 mutants were reduced, and signaling through the pheromone response pathway was diminished (527, 626), while expression of the dominant activated cdc42G12V mutant allele led to a modest (two- to threefold) increase in signaling as assayed by FUS1-lacZ expression (9), suggesting that Cdc42p plays a role in the activation of the pheromone response MAP kinase cascade (see below). In addition, Cdc42p localized to the tips of mating projections in pheromone-arrested cells (643), suggesting that it plays a direct role in pheromone-induced morphological changes.
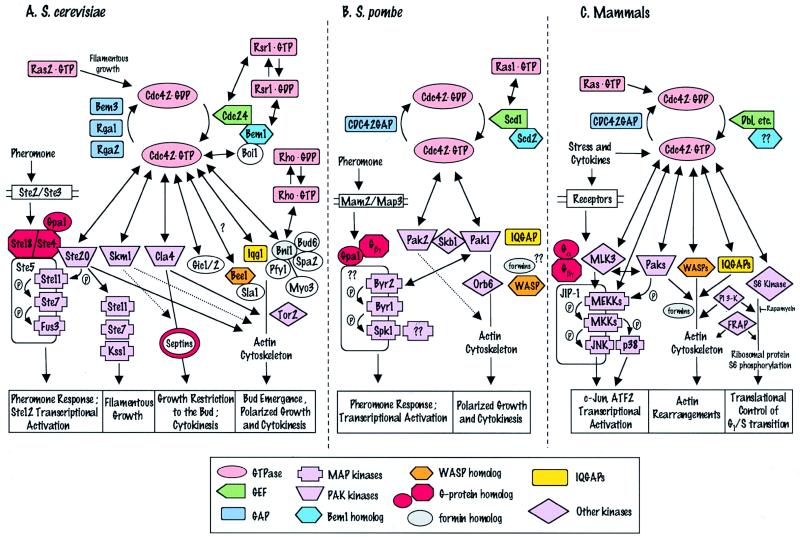
FIG. 4.
Comparison of Cdc42 interactions and dependent processes in S. cerevisiae (A), S. pombe (B), and mammals (C). Color and shape coding is given in the box at the bottom of figure. Two-headed arrows indicate physical interactions. Single-headed arrows indicate pathways; dotted arrows indicate potential involvement in pathways. For simplicity, not all components of the actin cytoskeleton or JNK kinase cascade are shown. See the text for details.
The observation that GTP-Cdc42 can interact with Ste20p, a PAK-like kinase intimately involved in the pheromone response pathway (see “PAK-like kinases” below) suggested that Cdc42p plays a direct role within the pheromone response pathway itself. However, recent results bring this conclusion into question. Most importantly, deletion of the Ste20p CRIB domain and therefore loss of Cdc42 interaction had little effect on either pheromone signaling through Ste20p and the MAP kinase signaling cascade or the generation of properly oriented, morphologically normal mating projections (see “Ste20p” below). Instead, these mutants appeared to have defects in bilateral mating and the cell-cell fusion event preceding zygote formation (298, 456). Also, it was recently shown that the modest reduction of pheromone signaling observed in cdc42-1 and cdc24-1 mutants was partially suppressed by deletion of the Cln1 and Cln2 G1 cyclins or by artificial depletion of the Cln1 protein, resulting in a G1 arrest at START prior to the cdc42 block (432). This result suggested that the effects of the cdc42 mutant on pheromone signaling were indirect, acting by elevating Cdc28-Cln levels at a particular stage of the cell cycle that led to a repression of the pheromone response pathway (430, 584). It seems likely, therefore, that the duties of Cdc42p and Cdc24p (see “Cdc24p-Ste4p interactions” below) in the mating pathway are largely restricted to coupling the G protein and Ste20p to an undetermined event, possibly the polarized deposition of secretory vesicles containing proteins needed for cell-cell adhesion into the mating projections, that occurs prior to cell-cell fusion, and not to induction of the Ste20-Ste11-Ste7-Fus3/Kss1 MAP kinase cascade. Interestingly, a recent report indicates that Spa2p, a protein that binds to the Cdc42p-interacting protein Bni1p at sites of polarized growth (see above and “Cdc42p downstream effectors” below) is needed for the clustering of secretory vesicles at the site of cell-cell fusion during the mating process (162). It remains to be seen if Cdc42p localizes to this region and is involved in the polarization of these secretory vesicles.
Pseudohyphal and invasive growth.
S. cerevisiae cells can alter their morphogenetic patterns in response to starvation conditions, leading to filamentous growth and the generation of pseudohyphae (for reviews, see references 26, 167, 297, 349, and 350). Diploid cells respond to nitrogen starvation by altering their cell cycles, budding patterns, cell shape, and cell separation patterns, resulting in polarized elongated budded cells that resemble fungal hyphae (168, 280). Haploid cells can also be induced to filamentous growth, which is manifested as invasive growth into agar plates (43, 487). A detailed mutational analysis of actin mutants indicated that the actin cytoskeleton plays a critical role in various aspects of pseudohyphal growth (63). The primary signaling route leading to pseudohyphal growth involves the Ras2 GTPase signaling through Cdc42p to several components of the pheromone response MAP kinase cascade, including Ste20p, Ste11p, Ste7p, and Kss1p, thereby activating the Ste12p transcription factor which, together with the Tec1 transcription factor, induces the expression of genes necessary for filamentous growth (Fig. 4A) (168, 330, 348, 351, 405, 487, 488).
The role of Cdc42p in this signaling pathway was deduced by the observations that expression of the dominant negative Cdc42D118A mutant protein inhibited Ras2-dependent filamentous growth and that expression of the activated Cdc42G12V mutant protein induced filamentous growth and transcription from a FG(TyA)::lacZ reporter construct that specifically responds to filamentous growth signals (405). As opposed to effects in the pheromone response pathway (see above), mutations within the Ste20 CRIB domains inhibited filamentous growth (298, 456), indicating that Cdc42p-Ste20p interactions are necessary for this morphological switch. Interestingly, the yeast 14-3-3 proteins Bmh1p and Bmh2p (165, 573) are also necessary for this signaling pathway and interact with Ste20p in vivo (488), although it is not known whether this interaction is mediated through phosphoserine residues in Ste20p as are other 14-3-3 interactions (610). It is interesting that Cln1p–2p/Cdc28p phosphorylation of Ste20p (see “Mitotic cell cycle” above) may also play a role in pseudohyphal and invasive growth in that cells lacking Cln1p and Cln2p fail to undergo pseudohyphal or invasive growth (431). Therefore, in response to nutritional signals, Cdc42p plays a role in activating a MAP kinase cascade that positively regulates the transcriptional induction of key morphogenetic and growth genes. This Cdc42p function is similar to that seen in mammalian cells with the induction of the JNK and p38 MAP kinase cascades leading to c-Jun transcriptional activity (see “Mammals” below).
Schizosaccharomyces pombe
In the rod-shaped fission yeast S. pombe, there are three switches in polarized cell growth patterns during the cell cycle (Fig. 2). First, selective and polar growth is initiated at the beginning of the cell cycle at the “old end” of the cell, which is the end distal to the previous division site (reviewed in reference 425). This growth occurs at the end of the cylindrical cell and can be monitored by staining with the dye Calcofluor and by the presence of cortical actin dots (366, 376). Second, after ∼0.3 of the cell cycle, a switch in polarized growth, referred to as new-end takeoff, occurs from unidirectional at the old end to bidirectional at both ends (Fig. 2). This growth pattern is visualized by the appearance of both Calcofluor staining and cortical actin dots at the new end and depends on the cell attaining a minimal length and completing the S phase. Third, bipolar growth continues until ∼0.75 of the cell cycle, at which time cortical actin reorganizes to the site of septum formation and end growth ceases, resulting in a constant-length stage of the cell cycle. Following cytokinesis and cell separation, polarity must be re-established at the old end as a prelude to unipolar growth in the next cell cycle.
The S. pombe Cdc42p homolog (cdc42+) was isolated from an S. pombe cDNA library by functional complementation of the S. cerevisiae cdc42-1ts mutation (390). The predicted amino acid sequence of S. pombe Cdc42p is 85% identical to those of both S. cerevisiae and human Cdc42p (Fig. 1). Disruption of cdc42+ showed that the gene was essential for growth. The S. pombe cdc42 loss-of-function phenotype was originally determined by generating a null allele in a haploid strain that was complemented by the wild-type allele on a plasmid and then inducing plasmid loss to uncover the loss-of-function phenotype (390). The morphological phenotype consisted of small, round, dense, uninucleate cells, which is strikingly different from that associated with cdc42 loss-of-function alleles in S. cerevisiae (2, 642) (see above). The S. pombe cdc42 null phenotype suggested that macromolecular synthesis continued but incorporation of new cellular material into an enlarging cell was inhibited, hence the small, dense, dead cells. Similar morphologies, as well as reduced mating efficiencies, were observed with the cdc42T17N dominant negative allele (447), suggesting that Cdc42p functions within the mating pathway as well (see below). The uninucleate, 1C phenotype, as assayed by DAPI staining and fluorescence-activated cell sorter analysis, indicated that the mitotic cell cycle was blocked in G1 phase, which is also different from the S. cerevisiae arrest phenotype of multinucleate cells. It is likely that the cell cycle coordination between DNA synthesis and Cdc42 function is more tightly regulated in S. pombe. Taken together, these data are consistent with Cdc42p functioning in the targeting and incorporation of new growth at the old end in G1 phase, possibly by affecting protein secretion or secretory-vesicle fusion to the plasma membrane.
Recent data indicating that S. pombe Cdc42p localizes to the septum region (381a) raises the possibility that Cdc42p plays a direct role in septum formation. In S. pombe, cytokinesis begins in early M phase with the assembly of the actin-based medial ring followed by septum formation and cell separation (reviewed in reference 176). Given that GFP-Cdc42p in S. pombe localizes to the medial area in a ring-like structure in some cells that do not have a visible septum, Cdc42p may play a role in the early steps of medial ring formation prior to septum formation. Interestingly, S. pombe Cdc12p, a homolog of the S. cerevisiae Cdc42-interacting protein Bni1p (139, 232) (see “Cdc42p downstream effectors” below), has also been implicated in medial ring formation (74).
To date, two potential downstream effectors of Cdc42p has been characterized in S. pombe, the Pak1p/Shk1p (364, 447) and Pak2p/Shk2p (515, 616) protein kinases (see “PAK-like kinases” below). Pak1p/Shk1p is a CRIB domain-containing ∼72-kDa serine/threonine protein kinase that belongs to the PAK family of Cdc42-interacting protein kinases. It can autophosphorylate on Ser and Thr residues and binds preferentially to Cdc42p-GTP. The physiological significance of these interactions was supported by the morphological abnormalities associated with the overexpression or absence of Pak1p and the synthetic-overdose phenotypes observed when overexpressing activated or dominant negative cdc42 alleles together with wild-type or kinase-defective pak1 mutants. Deletion of pak1 is lethal, resulting in small, round cells, a morphology reminiscent of cdc42 null mutants. This result indicates that Pak1p provides an essential function in the cell polarity pathway, which is different from its S. cerevisiae homologs Ste20p, Cla4p, and Skm1p. Pak1p and Cdc42p were also required for mating in S. pombe (see “PAK-like kinases” below). Taken together, the data are consistent with Pak1p being a downstream effector of Cdc42p in the cell polarity and mating pathways (Fig. 4B). Pak2p/Shk2p is a nonessential protein with the greatest similarity in predicted amino acid sequence to S. cerevisiae Cla4p and Skm1p (515). Its role as a downstream effector of Cdc42p is unclear (see “PAK-like kinases” below).
Candida albicans
The Candida albicans CDC42 gene was identified by degenerate oligonucleotide PCR and isolated from a C. albicans genomic library by DNA-DNA hybridization to the PCR probe (394). C. albicans Cdc42p is 87.8% identical to S. cerevisiae Cdc42p throughout the entire coding region (Fig. 1A), and DNA-DNA hybridizations suggested that CDC42 is single copy. Analysis of mRNA levels indicated that there is a transient increase in Cdc42p expression in the dimorphic switch to bud emergence, suggesting that C. albicans Cdc42p plays a role in this process. C. albicans homologs of the S. cerevisiae cell polarity proteins Rsr1p/Bud1p (608), Rho1p (272), Cla4p (299), Ste20p and Ste7 (268, 296), Fus3p/Kss1p (110, 595), and Ste12p (329, 352) have also been identified, and many of them have been implicated in hyphal formation and candidiasis. It remains to be seen if Cdc42p also functions in pseudohyphal formation in C. albicans, as it does in S. cerevisiae.
Caenorhabditis elegans
The Caenorhabditis elegans Cdc42 gene seems to be more highly expressed in embryonic cells than in larvae or adults, and when expressed in S. cerevisiae, it could complement the cdc42-1ts allele (88), indicating that it is a functional homolog. The C. elegans Cdc42 protein, expressed as a glutathione S-transferase (GST) fusion, could bind and hydrolyze GTP at rates comparable to the human Cdc42 protein (88). By using anti-Cdc42 antibodies, Cdc42p was shown to fractionate predominantly to a particulate fraction from mixed-stage populations of C. elegans cells and to localize to both the circumferential and longitudinal boundaries of hypodermal cells during hypodermal cell fusion in embryo elongation, in a pattern similar to that of the C. elegans PAK homolog (87). These results suggest that C. elegans Cdc42 plays a role in the actin-dependent process of embryonic-body elongation. Analysis of cdc42 activated or dominant negative mutant alleles in C. elegans has not been reported to date. The unc-73 gene product exhibits structural homologies to the Dbl family of GEFs (see “Mammalian GEFs” below) and has guanine nucleotide exchange activity against Ce-Rac but not Ce-Cdc42 in vitro (536); whether it is a GEF for Cdc42p in vivo is unknown. Unc-73p localized to the nerve ring and ventral nerve cord and was required for neuronal axon guidance; upon injection into cells, it induced actin polymerization at the plasma membrane. Recently, a C. elegans homolog of the mammalian MKK7 protein kinase, which functions in the Cdc42p-JNK MAP kinase signaling cascade (see “Mammals” below), was identified (148). Characterization of this and other components of the JNK signaling cascade should provide valuable insight into Cdc42p functions in C. elegans.
Drosophila
Studies with Drosophila have indicated a role for Cdc42p in a variety of actin-dependent processes (128, 129, 336, 408). These studies have been based on the analysis of the cellular phenotypes associated with overexpression of activated and dominant negative cdc42 alleles. For example, expression of the activated cdc42G12V mutant allele in the nervous system resulted in defects in dendrite and axon outgrowth and in embryo death whereas expression in muscle led to abnormally shaped muscle fibers (336). Expression of the cdc42G12V mutant allele in epithelial cells of the wing imaginal disc led to a high rate of cell death, but expression of a cdc42 dominant negative allele led to changes in the shape of polarized cells, a disruption of apico-basal cell elongation, and a loss of the dense, actin-containing plaques seen on the basal membrane. There were also mild effects on the location of the adherens junctions at the apical face of the cells (128). Expression of the dominant negative cdc42 allele did not affect polarized-protein accumulation in epithelial cells, as evidenced by proper cadherin and yellow-protein localization. In wing epithelial cells that form hairs, expression of the dominant negative cdc42 allele led to a loss or stunting of wing hair formation, which appeared to be a consequence of defects in actin polymerization within the wing hair (128, 129). In ovaries, expression of the activated cdc42G12V allele led to defects in actin distribution, resulting in altered nurse cell structure (i.e., abnormally fused nurse cells) and delocalized ring canals, structures which function as a cytoplasmic conduit between nurse cells and the oocyte but did not affect border cell migration (408). Therefore, Cdc42p seems to regulate actin distribution and function in a number of different cellular processes. In addition, Cdc42p may be involved with various Drosophila MAP kinase and JNK homologs (195, 482, 530) (see below) that regulate a variety of cellular processes including dorsal closure and establishment of cell polarity (for a review, see reference 233).
Mammals
Mammalian Cdc42p has been implicated in a wide variety of in vivo functions including receptor-mediated signal transduction pathways leading to induction of transcription and actin rearrangements, cell cycle progression, and apoptosis. Most of these studies have relied on the phenotypic analysis of ectopic expression of dominant activated and dominant negative cdc42 mutants. In addition, the characterization of mammalian Cdc42p-interacting proteins has implicated Cdc42p in multiple pathways (Fig. 4C), whose regulation is still unclear.
Actin rearrangements.
One of the earliest studies showed that differentiation of human monocytes into macrophages following the addition of phorbol esters led to an increase in the amount of membrane-bound Cdc42p that correlated with an increase in the actin-dependent cell-spreading activity (6). In subsequent studies, microinjection of wild-type or activated Cdc42G12V protein into serum-starved or subconfluent Swiss 3T3 fibroblasts led to the induction of peripheral actin PAMs, actin-containing long peripheral filopodia, and vinculin-containing focal complexes and to a reduction in the number of Rho-dependent actin stress fibers (277, 420). Filopodia are thought to be sensory structures involved in the actin-based polarized cell growth observed in fibroblasts (11) and neuronal cells (229, 254, 531). The Cdc42p-dependent induction of vinculin-containing focal complexes in serum-starved Swiss 3T3 fibroblasts was inhibited by microinjection of the dominant negative RacT17N protein, and the induction of filopodia was seen as a prelude to the formation of Rac-dependent membrane ruffles and lamellipodia and Rho-dependent stress fibers in time-lapse photomicroscopy (277, 420, 476, 479, 480). These results suggest that Cdc42p may function upstream of Rac and Rho in the generation of actin-dependent subcellular structures in these cells (420).
PAM induction was a rapid response that occurred within 5 min after injection and was mimicked by the addition of the mitogenic peptide bradykinin but not by phorbol esters or growth factors, such as lysophosphatidic acid and bombesin, that induced Rho-dependent stress fibers (476) or by platelet-derived growth factor (PDGF), EGF, or insulin, which induced Rac-dependent membrane ruffling (480). Both Cdc42p- and bradykinin-dependent inductions were inhibited by coinjection of either the dominant negative Cdc42T17N protein or the Rho-GDI protein (277), suggesting that the bradykinin effects were mediated by Cdc42p. Induction of PAMs and loss of stress fibers was also observed when the activated Cdc42G12V allele was stably expressed from a tetracycline-repressible promoter in human HtTa-1 cells (127), and this led to an enrichment for actin and the actin-bundling protein plastin, as well as the proline-rich focal adhesion protein VASP and the phosphatidylinositol (PI) 3-kinase p85 regulatory subunit, in the PAMs (127). Additionally, expression of Cdc42G12V in these cells resulted in large multinucleate cells, suggesting a defect in cytokinesis. Expression of Cdc42G12V in the highly motile Bac1 murine macrophage cell line also led to the induction of filopodia, a phenotype that was mimicked by the addition of colony-stimulating factor type 1 (CSF-1) and inhibited by injection of the dominant-negative Cdc42T17N protein (12), suggesting that Cdc42p may mediate CSF-1 actions in this cell type. Recent studies have shown that Cdc42p plays a role in Bac1 cell chemotaxis and response to CSF-1 gradients whereas Rho and Rac are required for cell migration (13). Cdc42p and Rac have also been implicated in the PI 3-kinase-dependent actin rearrangements leading to adenovirus endocytosis (317).
Cdc42p also seems to mediate the actin rearrangements resulting from cell-cell and cell-substratum adhesion. Stably transformed rat basophil leukemia (RBL-2H3) mast cells expressing the dominant negative myc-tagged Cdc42T17N protein exhibited defects in actin cytoskeletal rearrangements, including cell-substratum adhesion, antigen-induced actin plaque assembly, and vinculin localization, along with a 40% inhibition of high-affinity IgE receptor (FcɛRI)-mediated serotonin release and degranulation (183). Comicroinjection of the activated Cdc42G12V allele in Madin-Darby canine kidney cells reversed the cell-cell and cell-substratum adhesion defects observed upon microinjection of the Rho-GDI protein (282), suggesting that Cdc42p plays a role in cell-cell contact, as was also suggested by functional studies with Drosophila (see above). It is likely that the effects of Cdc42p on cell-cell adhesion are mediated through interactions with IQGAP1 (see “IQGAPs as scaffold proteins mediating Cdc42p-actin interactions” below), in that IQGAP1 colocalized with E-cadherin and β-catenin at cell-cell adhesion sites and interacted with both proteins (284). Cdc42p was shown to mediate integrin-dependent adhesion, membrane ruffling, and cell spreading in Rat1 and NIH 3T3 fibroblasts (101, 463), and recently it was shown that disassembly of the fibronectin matrix in human umbilical vein endothelial cells and KD fibroblasts led to the generation of Cdc42p-dependent filopodia and the activation of the Cdc42p downstream effector kinases ACK and p38 (49). In addition, stable expression of Cdc42G12V in murine 2B4 T cells led to a loss of microtubule-organizing center polarization toward antigen-presenting B cells and to a disruption of actin polarization at the junction between the T cells and B cells (541), a region to which Cdc42p also localized (reference 249 and data not shown), suggesting that Cdc42p plays a role in T-cell–B-cell recognition and interaction.
One of the original Cdc42 cDNAs (G25K) was isolated from bovine brain (407), and recently Cdc42/G25K was shown to be expressed in a variety of neuronal tissues within the adult rat brain, including the neocortex, thalamus, cerebellum, and hippocampus (440, 559). Microinjection of wild-type or activated Cdc42G12V into N1E-115 neuroblastoma cells led to the induction of filopodium formation, which was not seen upon microinjection of the dominant negative Cdc42T17N protein (278). Microinjection of the dominant negative Cdc42T17N protein also inhibited neurite outgrowth in these cells and inhibited the acetylcholine-dependent formation of filopodia. Expression of the activated Cdc42Q61L protein in E18/19 cultured cortical rat neuronal cells led to a slight increase in dendrite formation from pyramidal and nonpyramidal neurons, while expression of the dominant negative Cdc42T17N protein resulted in a slight reduction of dendrites and a reduction in cells that underwent the pyramidal-to-nonpyramidal morphological remodelling typically seen during differentiation (559). Also, expression of dominant negative Cdc42T17N led to a reduction of nerve growth factor (NGF)-induced neurite formation in PC12 cells (115). These data suggest that, as with Drosophila Cdc42p, mammalian Cdc42p functions in dendrite and axon outgrowth during neuronal development, probably through rearrangements of the actin cytoskeleton (for reviews, see references 337 and 345).
It is clear from these data that Cdc42p plays an intimate role in controlling actin rearrangements in a number of mammalian cell types. Cdc42p has been shown to induce membrane-dependent actin polymerization in Xenopus extracts (340, 402), in cell extracts of polymorphonuclear leukocytes and Dictyostelium discoideum amoebae (639), in permeabilized S. cerevisiae cells (324), and in neutrophil cell extracts (640), reinforcing the functional connections between Cdc42p and actin rearrangements. However, the intermediary proteins linking Cdc42p and the actin cytoskeleton have yet to be determined. Obvious candidates are the IQGAPs, which can bind both actin and GTP-Cdc42p (see “IQGAPs as scaffold proteins mediating Cdc42p-actin interactions” below), and the PAKs (see “PAK-like kinases” below). For example, expression of a autoinhibitory PAK1 peptide containing amino acids 83 to 149 led to an inhibition of PAK function and a loss of Cdc42-induced PAMs (627).
Another potential Cdc42p effector in the actin pathway is PI 3-kinase and phosphoinositides (for reviews, see references 154 and 372). The p85 subunit of PI 3-kinase from NIH 3T3 and PC12 cell lysates was able to bind to GST-Cdc42Hs-GTPγS as well as to an activated GST-Cdc42Q61L protein but not to the Cdc42T35A effector domain mutant protein (628). These interactions were inhibited by incubation with either the Rho-GAP homology domain of p85 or the Cdc42GAP/p50rhoGAP protein (see “Mammalian GAP” below), suggesting that binding was through interactions between the GAP homology domain and the effector domain. Binding of Cdc42-GTPγS to p85 led to a two- to fourfold increase in PI 3-kinase activity from PC12 cell extracts (628), as well as from rat liver cytosol and COS7 cell lysates (561). This activation was inhibited by a p85-specific peptide antigen and by the PI 3-kinase specific inhibitor wortmannin (561), suggesting that Cdc42p specifically interacts with and activates PI 3-kinase in these cells. Cdc42-induced actin rearrangements and morphological changes observed in T47D mammary epithelial cells were inhibited by the PI 3-kinase inhibitors wortmannin and LY294002, and transfection of these cells with a constitutively activated p110 catalytic subunit of PI 3-kinase led to the same morphological changes and actin rearrangements as activated Cdc42p (256). Cdc42 also displays interactions with PI 4,5-bisphosphate [PI(4,5)P2]. PI(4,5)P2 can bind to pleckstrin homology (PH) domains found in all Cdc42 GEFs (201) (see “Guanine nucleotide exchange factors” below) and can also enhance nucleotide exchange on Cdc42Hs by stimulating GDP release and stabilizing the nucleotide-depleted state in much the same manner as GEFs (632). This effect was specific for PI(4,5)P2, and deletion of the last 7 amino acids of Cdc42Hs, including the prenylation site (Fig. 1), led to a loss of PI(4,5)P2 stimulation without affecting Dbl GEF activity. These data suggested that the PI(4,5)P2 effects were due to specific nucleotide exchange and not to a nonspecific denaturation of the protein due to the high levels of phosphoinositide in the reaction micelles, although this point is in contention (70, 72, 250). PI(4,5)P2 in combination with Cdc42p can also induce actin polymerization in Xenopus extracts (340) and in cell extracts of polymorphonuclear leukocytes and D. discoideum amoebae (639). The role of phosphoinositides in Cdc42p-dependent actin polymerization is speculated to be through either stimulation of guanine nucleotide exchange on Cdc42p or an enhancement of membrane attachment and/or targeting of Cdc42p by disrupting the interactions between Cdc42p and its GDI (94, 340, 639). PI(4,5)P2 also acts directly on the actin cytoskeleton to promote actin assembly through the PI(4,5)P2-dependent dissociation of profilin from actin monomers, inhibition of actin-capping proteins, and inhibition of the actin-severing activities of gelsolin and other actin-severing proteins, thereby leading to an increase in the number of free actin ends available for polymerization (372, 511). Taken together, these data suggest that PI 3-kinase and phosphoinositides play an important physiological role in mediating Cdc42p-dependent actin rearrangements.
Another potential Cdc42 effector in regulating actin rearrangements is the myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK). Coexpression of MRCKα with activated Cdc42G12V resulted in an induction of PAMs, and expression of dominant negative, kinase-defective MRCKα inhibited the effects of Cdc42G12V (311). Another interesting candidate is the myr5 rat unconventional myosin. This protein is capable of binding actin and contains a C-terminal domain, with sequence similarity to Rho-GAP proteins, that is functional as a GAP against Cdc42p in vitro (474). However, recent in vivo and in vitro studies suggest that myr5 may be a specific RhoA-GAP (406). Finally, the CIP4 protein, which was isolated through a two-hybrid interaction with activated Cdc42Q61L and contains domains with similarity to the non-receptor tyrosine kinase FER and to RhoGAP, bound to Cdc42p in vitro and localized to PAMs and membrane ruffles along with Cdc42p (18). Determination of the exact nature of the connections between the Cdc42p signaling module and actin rearrangements should be vigorously pursued in the near future.
Cdc42p and the JNK/SAPK and p38 MAP kinase cascades.
In addition to regulating actin rearrangements, Cdc42p functions to couple cell surface receptors to MAP kinases, thereby transducing extracellular signals to regulate intracellular events, most notably the transcriptional induction of genes essential for a diverse number of cellular processes. These processes include inflammatory and stress responses, mitogenesis, differentiation, cell growth, cell cycle progression, apoptosis, prostaglandin biosynthesis, myocyte hypertrophy, and immunity gene expression (103, 120, 141, 179, 195, 217, 253, 287, 369, 415, 489, 513, 514, 578). The stress response signaling pathway involves the stress-activated protein kinases (SAPKs) (286), which also can phosphorylate Ser63 and Ser73 in the N terminus of the c-Jun subunit of the AP-1 transcription factor (JNKs) (121). These JNK/SAPK protein kinases can be activated by a variety of cell surface receptors and cellular stresses such as heat shock, UV radiation, and changes in osmolarity, by the protein synthesis inhibitors anisomycin and cycloheximide, and by the cytokines interleukin-1 (IL-1) and tumor necrosis factor alpha (TNF-α) (121, 286) and can be inhibited by high cell density (289). The p38/Mpk2 protein kinase, which is another member of this SAPK family and is the mammalian homolog of the S. cerevisiae Hog1p (for “high-osmolarity glycerol”) protein kinase, also regulates the ATF2 and Elk-1 transcriptional activators (150, 194, 300, 493).
In several independent studies, Cdc42p was shown to activate JNK (23, 109, 392, 442, 556) and p38 (23, 625) in vitro kinase activity. Ectopic expression of the wild-type or activated Cdc42Q61L proteins in COS7 fibroblasts led to a 5- to 10-fold increase in the immunoprecipitated JNK in vitro activity against the transcriptional activators c-Jun and ATF2 as substrates but had little or no effect on Ras-dependent MAP kinase (ERK) activity (109, 556) (see below). Similar results were observed in COS1 cells (23), HeLa, NIH 3T3, and Rat1A cells (392), and 293 human kidney fibroblasts (556). This induction also led to an elevation of c-Jun AP-1 transcriptional activity, presumably through a JNK-dependent phosphorylation event (102, 392). These effects were assayed 2 to 3 days posttransfection, and so the timescale for induction of transcription relative to the rapid induction of actin-based filopodia (see above) is unclear.
Expression of activated RacQ61L led to a similar induction of in vitro JNK (109, 556) and p38 (392) kinase activity, as did expression of the Dbl protooncogene product, which has in vitro guanine nucleotide exchange activity against Cdc42p (see “Mammalian GEFs” below). Coexpression of Rho-GDI or p190 Rho-GAP (109) or the CRIB domain of human PAK65 (392) abrogated the Dbl induction but did not affect the Cdc42Q61L induction. In addition, expression of the dominant negative Cdc42T17N protein inhibited the JNK induction by EGF and TNF-α, but not by anisomycin or Cdc42Q61L. Recently, a C-terminal domain of the polycystic kidney disease I (PKD1) gene, which plays a role in autosomal dominant polycystic kidney disease, was shown to induce c-Jun/AP-1 transcriptional activation through the activation of JNK activity, and this induction was inhibited by coexpression of dominant negative Cdc42T17N or RacT17N (17). Expression of dominant negative Cdc42T17N, as well as dominant negative RasT17N, dominant negative Rac1T17N, and dominant negative JNK mutant alleles, in primary cultures of rat hepatocytes inhibited the hyperosmotic-glucose-, TNF-α-, and hepatocyte growth factor-induced stimulation of JNK1 kinase activity, phosphorylation of c-Jun, and up-regulation of DNA synthesis (20). Transient expression of wild-type or activated Cdc42Q61L, as well as wild-type or activated RhoA or Rac, in simian COS7 cells and human T-cell lymphoma Jurkat cells led to a four- to sevenfold induction of NF-κB-dependent transcription from a human immunodeficiency virus (HIV)-luciferase reporter construct (455). This induction was inhibited by coexpression of the IκBα inhibitory subunit, and TNF-α induction of NF-κB-dependent transcription was inhibited by expression of the dominant negative Cdc42T17N protein. Recently, analysis of dominant negative Cdc42T17N protein expression in Rat-2 fibroblasts indicated that Cdc42p mediated the JNK-dependent transformation of these cells by the Fps and Fes nonreceptor tyrosine kinases (320), and studies with αT3-1 pituitary cells indicated that the Cdc42p-JNK pathway mediated gonadotropin-releasing hormone activity (312). The above-mentioned results are consistent with a model in which Cdc42p acts to stimulate JNK/SAPK and p38 kinase activities, leading to translocation of these kinases to the nucleus, where they phosphorylate a number of different transcriptional activators and induce the expression of genes needed for stress responses, mitogenesis, and cell growth.
There are several likely intermediates in the signaling cascades linking Cdc42p to the JNK/SAPK and p38 MAP kinase, including the PAK family of Cdc42p-interacting kinases (see “PAK-like kinases” below), as well as other downstream MAP kinase kinases and MAP kinase kinase kinases (for reviews, see references 141 and 233). Overexpression of the wild-type PAK1 protein kinase stimulated p38 in vitro kinase activity, and overexpression of a dominant negative PAK1K299R mutant protein, as well as the dominant negative Cdc42T17N mutant protein, inhibited the IL-1-dependent induction of p38 activity in HeLa cells (625). Expression of the mPAK-3F91S, G93A, P95A (23) or hPAK1L107F (55) constitutively activated alleles in COS cells led to a modest (mPAK-3F91S, G93A, P95A) to substantial (hPAK1L107F) induction of JNK in vitro protein kinase activity. Transfection of wild-type PAK into 293 human kidney fibroblasts led to induction of the JNK/SAPK and p38 in vitro kinase activities (153), and transfection of activated Cdc42Q61L also led to an induction of the JNK/SAPK kinase activity, which was abrogated in cells cotransfected with a PAK CRIB domain-containing fragment (556). Expression of another CRIB-containing protein kinase, MLK3, whose CRIB domain can interact with Cdc42p in vitro (59), effectively induced JNK activity and enhanced the Cdc42Q61L-dependent induction (555). Of the three known MLKs, MLK2 and MLK3 interact with Cdc42p-GTP and activate the JNK pathway (411). MLK3 and PAK1 coimmunoprecipitated in the presence of activated Cdc42Q61L, and expression of Cdc42Q61L led to an increase in the in vitro MLK3 and PAK1 activity against myelin basic protein. Expression of the dominant negative MLK3K144R mutant allele led to a reduction of the Cdc42Q61L-dependent JNK activation, and expression of a dominant negative SEK mutant allele (see below) reduced MLK3 induction. This complex set of results suggest that Cdc42 can interact with multiple MAP kinase kinase kinases depending on the cell type and that these kinases can also transduce Cdc42p-independent signals to downstream effectors.
The MEKKs (MAP/ERK kinase kinases) function downstream of Cdc42 to regulate the JNK pathway but not the p38 pathway (142, 286, 393, 614). Of the four MEKK proteins (MEKK1 to MEKK4) characterized from COS cells (142), only MEKK4 contains a potential Cdc42p-interacting CRIB domain. However, expression of both MEKK4 and MEKK1 dominant negative kinase-defective mutant proteins, but not MEKK2 or MEKK3, could inhibit Cdc42Q61L-dependent induction of JNK kinase activity. Both MEKK4 and MEKK1 could interact with Cdc42 in in vitro binding assays, although MEKK1 preferentially interacted with GTP-bound Cdc42p whereas MEKK4 could bind to both GDP- and GTP-bound Cdc42p (142). Interestingly, MEKK4 localized to the Golgi in a similar manner to Cdc42p in COS cells (137, 142). Expression of dominant negative kinase-defective MEKK4 and MEKK1 did not affect the PAK-dependent induction of JNK activity in COS cells (142), suggesting that PAKs and MEKKs can function in independent Cdc42-dependent signaling pathways to JNK activation.
The two primary activators of JNK that function downstream of the MEKKs are the MKK4/SEK1 and MKK7 kinases (for a review, see reference 233). MKK7 specifically activates JNK and has homologs in Drosophila (hemipterous), which functions upstream of Drosophila DJNK, and C. elegans (cMKK7), while MKK4/SEK1 can activate both JNK and p38 in vitro. MKK4/SEK1 knockout mice display a specific defect in stress-induced JNK activation and AP-1 transcriptional activation (417, 615), providing elegant in vivo data supporting the role of the JNK signaling pathway in stress-induced AP-1 transcription activation. Taken together, these data suggest that Cdc42p can signal through either PAKs, MLKs, or MEKKs to the MKKs that induce JNK activity and transcriptional activation. Recently, a potential scaffolding protein, JIP-1, was shown to bind to MLKs, MKK7, and JNK but not Cdc42, thereby linking these kinase-signaling components (597). Determination of the mechanisms by which Cdc42p signals to this multitude of protein kinases in vivo should be an area of intensive investigation in the near future.
In addition to functioning within the JNK/SAPK and p38 MAP kinase signaling cascades, Cdc42p has been implicated in the activation of the Ras-dependent ERK MAP kinase pathway. This involvement is consistent with the role of Cdc42p (see below) and Rac (258) in Ras-dependent transformation of NIH 3T3 cells. Expression of wild-type or activated Cdc42G12V alone in 293 kidney cells did not activate ERK2 in vitro kinase activity, but coexpression with wild-type Raf-1 (152) or a constitutively activated Raf BXB mutant (153) resulted in an 11- to 16-fold synergistic stimulation. In addition, expression of the PAK1 CRIB domain inhibited the H-RasG12V stimulation of ERK2 activity. Coexpression of Cdc42Q61L and wild-type Raf-1 also resulted in a synergistic stimulation of Elk1-dependent transcriptional activation in NIH 3T3 cells (152) (Elk1 is a member of the ternary complex factor family of ERK-dependent transcription factors that regulate the c-fos serum response element), providing a physiological correlation between Cdc42p-dependent ERK2 stimulation and ternary complex factor-dependent transcriptional activation. Similar results were seen with ectopic expression of activated Cdc42G12V in NIH 3T3 cells (220) and this activation was not dependent on activation of the JNK/SAPK pathway. This cross talk between the ERK and JNK/SAPK pathways seems to occur through interactions of PAK with the MEK1 MAP kinase kinase because coexpression of wild-type or constitutively activated PAK1 with Raf-1 led to a synergistic stimulation of MEK1 and ERK2 activity and Elk-dependent transcription and because PAK1 could phosphorylate the functionally important MEK1-Ser298 residue in vitro and in NIH 3T3 cells (152). The physiological ramifications of this potential cross talk remain to be determined.
Ras-mediated transformation, cell cycle progression, and apoptosis.
Cdc42p has been implicated in mitogenesis and cell cycle progression, but there are conflicting data concerning its exact role in these processes. Microinjection of activated Cdc42G12V protein into quiescent Swiss 3T3 fibroblasts led to an increase in the bromodeoxyuridine incorporation into DNA, and expression of dominant negative Cdc42T17N blocked serum-induced bromodeoxyuridine incorporation (442), suggesting that Cdc42p is necessary for cell cycle progression in these cells. In contrast, microinjection of wild-type Cdc42p into G1-synchronized NIH 3T3 cells resulted in a dramatic cell cycle arrest at G1/S, whereas microinjection of activated and dominant negative Cdc42 alleles had a similar but less drastic effect (400). Interestingly, this effect was mediated through the p38 MAP kinase pathway but not the JNK/SAPK pathway. Expression of activated Cdc42G12V in Rat1 and NIH 3T3 cells did not seem to lead to increases in low-serum growth or growth to high saturation density (466, 494), which is also in contrast to the cell cycle-stimulatory effects seen in Swiss 3T3 cells (see above). It is likely that these discrepancies are due to different signaling mechanisms in these different cell types, but the nature of these important differences has not been elucidated.
Recently, Cdc42p has been implicated in Ras-dependent transformation of NIH 3T3 cells. Expression of either dominant negative Cdc42T17N or the CRIB domain of the Cdc42p effectors PAK1 or WASP (see “Cdc42p downstream effectors” below) inhibited focus formation and soft-agar growth by H-RasG12V in a dose-dependent manner (445, 466, 494). In addition, stable expression of activated Cdc42G12V in Rat1 fibroblasts led to an increase in anchorage-independent growth in soft agar, and injection of Cdc42G12V-expressing cells into athymic nude mice led to the formation of tumors (466). These data, along with the observation that expression of the Cdc42F28L mutant protein can lead to transformation of NIH 3T3 cells similar to that seen with expression of the Cdc42-GEF dbl oncogene (327), indicate that activation of Cdc42 can lead to malignant transformation and that Cdc42 is a bona fide oncogene.
Recent studies have shed some mechanistic light on the means by which Cdc42 mediates cellular transformation and cell cycle progression. In a detailed comparative study, it was observed that the transformation potential of Dbl GEF family members (see “Cdc42p regulators” below), including Dbl and Dbs, correlated most closely with the transcriptional induction of the cyclin D1 promoter and not with the activation of JNK, p38, SRF, or c-Jun (590). Cyclin D1-Cdk4/6 kinase complexes can phosphorylate the p105 RB retinoblastoma protein, leading to a dissociation of the E2F family of transcription factors that regulate the transcriptional activity of DNA replication genes necessary for the G1-S cell cycle transition (for a review, see reference 522). Ectopic expression of activated Cdc42G12Vp or RacG12V, but not RhoG12V, in NIH 3T3 cells led to an induction of transcription from an E2F-dependent reporter construct, an induction of p105 RB hyperphosphorylation, and a moderate induction of cyclin D1 protein levels (169). The Cdc42/Rac induction of E2F-dependent transcription was reduced in cells expressing the p16ink4, p21cip1, or p27kip1 CDK inhibitors. These data suggest that the effects of Cdc42 on the cell cycle are mediated, at least in part, through cyclin-CDK phosphorylation of RB and subsequent E2F-dependent transcriptional activation. These data are supported by the recent observation that the Rho GTPase plays a role in blocking the Ras-dependent induction of p21cip1, leading to an induction of cell cycle progression (380, 444). In addition, alterations in human capillary endothelial-cell shape and/or actin cytoskeletal structure led to a cyclin D- and p27kip1-dependent block in cell cycle progression (228), suggesting that the integrity of the actin cytoskeleton, as well as growth factors and integrin signaling, can affect progression through the cell cycle. The mechanism by which Cdc42 regulates these effects should be intensively investigated in the near future.
Cdc42p, PAKs, and the JNK/SAPK and p38 protein kinase cascades have also been implicated in stress-activated programmed cell death (apoptosis) (32, 96, 143, 171, 230, 288, 496, 497). Programmed cell death in the immune system is mediated through the coupling of activated Fas receptors to the IL-converting enzyme (ICE) (caspase) protease cascade, which results in proteolytic cleavage of downstream targets, including the PAK2 protein kinase (496, 497, 583), the D4-GDI (409), and components of the JNK pathway (62, 245). Expression of wild-type or activated Cdc42Q61L in Jurkat T lymphocytes resulted in a decreased transfection frequency and induction of apoptotic responses, including characteristic DNA fragmentation and morphological changes (96). Also, expression of activated Cdc42G12V in rat sympathetic (SCG) neurons led to an induction of neuronal apoptosis through activation of the JNK pathway (32). These Cdc42-dependent apoptotic responses were inhibited by coexpression of dominant negative MEKK and MKK/SEK kinases. By contrast, apoptotic responses were induced by expression of a constitutively activated MEKK, indicating that activation of the JNK/SAPK signaling pathway is necessary for apoptotic responses in this cell type, as well as various other cell types (89, 193, 607, 621). These responses were also inhibited by various inhibitors of the ICE/caspase proteases (96; for a review, see reference 105), suggesting that the caspases function downstream of Cdc42p and further implicating PAK2 and D4-GDI in mediating these apoptotic responses. In mink lung epithelial (Mv1Lu) cells, TNF-α-induced JNK activation and apoptosis were dependent on another upstream kinase, ASK1, which displayed sequence similarities to upstream kinases in the S. cerevisiae HOG pathway (230). The Fas receptor has recently been shown to activate ASK1 through interactions with the receptor-associated adapter protein Daxx (75), and expression of constitutively active Gα13 and Gα12 mutants (see “Cdc42p/JNK pathway and ion homeostasis” below) led to ASK1 activation and induction of apoptosis (40). The mechanism by which JNK induces apoptosis is unclear, but an interesting observation is that the JNK1, JNK2, and JNK3 isoforms have potent in vitro kinase activity against the tumor suppressor protein p53 (227) that affects p53 ubiquitination and stability (155, 156); whether this activity depends on Cdc42p has not been determined.
Cdc42p and Nef-dependent HIV replication.
A fascinating connection has recently been made between Cdc42p and Nef-dependent HIV replication and pathogenesis (for reviews, see references 111 and 564). Nef associated with, and activated, a cellular Ser/Thr PAK-like kinase, the Nef-associated kinase (NAK). NAK shares epitopes with PAK but is not one of the three major PAK isoforms (335, 508) (see “PAK-like kinases” below). Activation of NAK appeared to be mediated through Cdc42p (and Rac), in that expression of dominant negative Cdc42T17N reduced NAK activity in transfected COS cells and expression of activated Cdc42G12V enhanced Nef association with and activation of NAK activity (335). This NAK activation also led to serum response element-dependent transcriptional induction that was blocked by Cdc42T17N expression. In addition, Cdc42T17N expression led to a reduction in HIV-1 production in transfected COS cells. Determination of the physiological role of Cdc42p in HIV pathogenesis should be active area of investigation in the near future.
Cdc42p/JNK pathway and ion homeostasis.
Activation of mammalian NHE1, an integral membrane Na+-H+ exchanger subtype involved in regulating intracellular ion homeostasis and pH (421), by the Gα13 GTPase is mediated through the Cdc42-MEKK1-JNK pathway (40, 222, 580, 581). Although it is unclear whether these JNK pathway effects are due to transcriptional activation of the NHE1 gene or induction of NHE1 exchanger activity, expression of dominant negative MEKK1 inhibited the rapid activation of a Gα13/Gαz chimera by the D2-dopamine receptor (222), suggesting that transcriptional regulation is not involved. Recently, the p115 Rho-GEF has been shown to act as a GAP for Gα13 and Gα12 (275), and Gα13 has been shown to stimulate the guanine nucleotide exchange activity of p115 Rho-GEF (207), which can lead to Tec/Bmx nonreceptor tyrosine kinase-dependent induction of serum response factor-dependent transcription (361–363). These results suggest a regulatory connection between these G-protein α subunits and members of the Rho/Rac family of GTPases. Analysis of Gα13 knockout mice indicated that Gα13 functions in thrombin-dependent cell migration, probably in a Cdc42p/actin-dependent manner, and in vascular system development (433). In addition, expression of constitutively active Gα13 and Gα12 in differentiated PC12 cells led to Rho-dependent neurite retraction and cell rounding (255). Another interesting connection between Cdc42p and ion channels was the recent identification of the human homolog (IBP72) of the S. pombe Skb1 protein kinase, which interacts with the S. pombe PAK homolog Shk1/Pak1 (166) (see “S. pombe PAK-like kinases” below) in a two-hybrid protein screen for proteins that interacted with the pICln protein, a putative component of the chloride channel (279). Given the unclear nature of the role of pICln in chloride channel activation, the significance of this interaction remains to be determined.
Cdc42p and host cell responses to bacterial invasion.
Cdc42p also plays a role in host signaling pathways that are activated in response to invasive bacteria (for reviews, see references 125 and 234). These pathways include actin rearrangements seen in Salmonella and Shigella invasion (see below), as well as JNK pathway-dependent induction of proinflammatory cytokines in epithelial cells in response to Neisseria gonorrhoeae invasion (414). Salmonella typhimurium can induce actin rearrangements and macropinocytosis in host cells (149), and expression of the dominant negative Cdc42T17N mutant protein in COS1 fibroblasts inhibited this induction and prevented internalization of the bacteria into the host cells (85). Expression of the activated Cdc42G12V mutant protein did not alter these processes with wild-type Salmonella, but it did allow an invasion-defective Salmonella mutant to invade COS1 and Rat-1 cells. Salmonella invasion also induced JNK kinase activity, and expression of the Cdc42T17N mutant protein prevented this bacterially induced activation. These Salmonella-induced effects were mediated by the Salmonella SopE protein (599), as evidenced by the ability of SopE both to act as a GEF for Cdc42p and Rac1p and to induce cytoskeletal rearrangements and JNK kinase activation (200). Therefore, aspects of Salmonella invasion are mediated through bacterial proteins impinging on host cell signal transduction pathways.
The effects of Cdc42 on actin rearrangements and invasion of HeLa cells by enteropathogenic E. coli (EPEC) were investigated by inhibiting Cdc42 function in three ways (33). Addition of compactin or ToxB led to disruption of normal actin structures in HeLa cells but did not block EPEC-induced formation of actin pedestal structures. Compactin inhibits Cdc42 function by inhibiting the 3-hydroxy-3-methylglutaryl coenzyme A reductase enzyme necessary for isoprene biosynthesis and subsequent isoprenylation, while ToxB, which is the Clostridium difficile toxin B protein, glucosylates and inactivates Cdc42 (247). Expression of the dominant negative Cdc42T17N mutant protein did not affect the formation of actin pedestals, but compactin and ToxB addition did inhibit the ability of EPEC to invade HeLa cells, suggesting that actin pedestal formation is not sufficient for EPEC invasion of HeLa cells. Interestingly, treatment of various mammalian cell lines with reagents that inhibited Rho activity, including Clostridium botulinum exoenzyme C3 transferase, inhibited the ability of Shigella flexneri to invade but did not affect invasion of Salmonella typhimurium (586), suggesting that Shigella flexneri utilizes host Rho protein for invasion. However, generation of the actin tail or “comet” produced by Shigella flexneri for its intracellular movement was dependent on the Cdc42p effector N-WASP (543) (see “Cdc42p downstream effectors” below), suggesting that bacterially induced actin rearrangements can be mediated through multiple Rho-type GTPases. These differences reinforce the notion that different bacteria use different invasion mechanisms (234). The future examination of Cdc42 effects on actin rearrangements and proinflammatory cytokine production in response to bacterial invasion should prove very exciting.
In conclusion, an important question to consider is whether all of the above-mentioned effects of Cdc42 on mammalian cellular processes are mediated through interactions with PAKs and the JNK/SAPK MAP kinase pathways. Clearly, other Cdc42p downstream effectors that function independently of the JNK/SAPK pathway exist in mammalian cells, suggesting that there may be bifurcations in Cdc42p-regulated pathways (Fig. 4C) (see below). Therefore, the exact nature in which Cdc42p impinges on this myriad of mammalian cellular processes remains to be determined.
CDC42P REGULATORS
Guanine Nucleotide Exchange Factors
The activation of G proteins from an inactive GDP-bound state to an active GTP-bound state requires the action of a GEF. While the structures of GEFs for different G-protein families (i.e., Ras, Rho/Rac/Cdc42, Rab, Arf, and heterotrimerics) are quite dissimilar, the mechanism of guanine nucleotide exchange seems to be conserved (for a review, see reference 533). GEFs function by stabilizing the nucleotide-free state of the G protein, through the disruption of both Mg2+ and nucleotide binding sites and subsequent GDP dissociation. However, the means by which GEFs promote these effects can be quite different. GEFs for the Rho/Rac/Cdc42 family of GTPases all contain a Dbl homology (DH) domain (Fig. 5), which is a highly α-helical (331) catalytically active domain, and a PH domain, which functions in membrane localization and has recently been shown to enhance DH-domain-dependent nucleotide exchange (331).
FIG. 5.
Structure of the representative Dbl family member GEFs Cdc24, Scd1, Dbl, and Fgd1. The DH and PH domains are indicated, along with the potential Ca2+ binding domains and Ste4p, Rsr1p/Bud1p, and Bem1p binding domains in Cdc24. See the text for details.
S. cerevisiae Cdc24p.
Cdc24p is believed to be the sole GEF for Cdc42p in S. cerevisiae. CDC24 mutants were among the original cdc mutants isolated and characterized by Hartwell et al. (211–213). The initial analysis of cdc24ts mutants indicated that Cdc24p plays a role in bud emergence, with cdc24ts mutants exhibiting a first cycle arrest as large, round unbudded cells with multiple nuclei. The presence of multiple nuclei in arrested cells was the first evidence that the budding cycle was independent of the DNA synthesis and nuclear division cycles in S. cerevisiae. Interestingly, overexpression of Cdc24p led to similar morphological phenotypes (504, 641), indicating that either a lack or an excess of Cdc24p leads to a loss of cell polarity, presumably through the disruption of multiprotein complexes (see below). Other phenotypes associated with cdc24ts mutants were delocalized chitin deposition throughout the cell instead of the typical chitin ring formation at the mother-bud neck region (528, 529) and defects in bud site selection and localized deposition of mannan (528), localized secretion of acid phosphatase (146), and mating (416, 472) (see below). Delocalized chitin deposition in cdc24 mutants is probably due to the altered assembly of the 10-nm filament septin ring, which is necessary for proper chitin ring formation (119). Disruption of CDC24 led to death (106), indicating that Cdc24p had an essential function in cell growth. The cdc24-3 and cdc24-4 mutant alleles displayed abnormal elongated buds when grown at semipermissive temperatures (528), suggesting that Cdc24p plays a role in the apical-isotropic switch (see “S. cerevisiae” under “Functional studies” above). They also displayed defects in bud site selection to a discrete, nonrandom site in both haploids and diploids (528), suggesting that Cdc24p is involved in the bud site selection process (see below). When cdc24-4ts cells were arrested in S phase with hydroxyurea and then released into media at the restrictive temperature, the predominant arrest phenotype observed consisted of large mother cells with small buds (528), suggesting that Cdc24p also functions after bud emergence to direct growth preferentially into the enlarging bud instead of in the nonenlarging mother cell.
In a screen for S. cerevisiae mutants sensitive to 100 mM Ca2+ in the growth media, 1 of 30 Ca2+-sensitive mutants, csl4, was shown by complementation and genetic linkage tests to be allelic to CDC24 (436, 438). The cls4 mutant arrested in medium containing 100 mM Ca2+ with large, round unbudded cells and had wild-type levels of intracellular Ca2+ and normal rates of uptake of Ca2+, indicating that the Ca2+-sensitive phenotype was not due to an alteration in Ca2+ homeostasis but was more likely to be due to an alteration in a Ca2+-regulated process. Certain cdc24 alleles were also sensitive to high-Na+ growth media and showed synthetic lethality with a vma5 vacuolar ATPase subunit mutant (591), suggesting that Cdc24p may have functions outside its normal role in cell polarity. Taken together, these data indicate that Cdc24p has essential functions in bud site selection, organization of the cell polarity machinery at the bud site, polarized growth into an enlarging bud during the cell cycle, and mating and that calcium may play an important role in Cdc24p function. These functions coincide with Cdc42p functions (see “Functional studies” above), reinforcing the physiological connection between these two proteins.
Cdc24p is a ∼97-kDa protein with no apparent homologs in the S. cerevisiae genome. Cdc24p contains three recognizable functional domains (Fig. 5), including a DH domain that shows a high degree of similarity to the Dbl family of GEFs (residues 283 to 452 [see below]), a PH domain (residues 472 to 681), and two potential Ca2+ binding domains (residues 649 to 658 and 820 to 831) inferred through homology to the α-lactalbumin and S-100a Ca2+ binding domains, respectively (396). It should be noted that one of the Ca2+-sensitive mutations, cls4-2, is within the DH domain, suggesting that Ca2+ plays a role in Cdc24p GEF activity. The observation that overexpression of either S. cerevisiae or human Cdc42p can suppress a cdc24ts mutant (35, 407), along with the similarity in cdc24 and cdc42 mutant phenotypes (see above), suggested that Cdc24p is a GEF for Cdc42p. This possibility was addressed both genetically and biochemically (630, 641). Overexpression of Cdc24p was able to suppress the dominant negative cdc42D118A mutant (641), suggesting that the dominant negative effect was due to the binding and sequestration of Cdc24p away from endogenous Cdc42p. This inference was substantiated by analysis of two-hybrid protein interactions between Cdc24p and Cdc42p (116). A GST-Cdc24p fusion protein stimulated GDP dissociation from a GST-Cdc42p fusion protein in vitro with a half time of ∼5 min and also led to a four- to fivefold increase in [35S]GTPγS binding to GST-Cdc42p (630). Cdc24p did not display these biochemical activities against either Rho1p or Rsr1p/Bud1p (see below). In addition, Cdc24p interacted tightly with a guanine nucleotide-depleted GST-Cdc42p, slightly less so with GST-Cdc42-GDP, and less still with GST-Cdc42-GTPγS (629). These observations are consistent with interactions seen between human Cdc42p and the Dbl GEF and suggest that Cdc24p functions to stabilize the nucleotide-depleted form of Cdc42p as a prelude to guanine nucleotide exchange. These results, along with the observations that Cdc24p is an essential protein with no significant homologs in the S. cerevisiae genome, suggest that Cdc24p is the sole physiological GEF for Cdc42p in S. cerevisiae.
By using Cdc24p-specific antisera, it was shown that Cdc24p fractionated to a particulate pool (383, 397, 465) and seemed to be present at the plasma membrane around the entire periphery of the cell in immunofluorescence microscopy experiments (465). However, recent studies with functional GFP-Cdc24p fusion proteins indicate that Cdc24p localizes to the plasma membrane at the site of incipient bud emergence and to the mother-bud neck region in large-budded cells (508b). The mechanism by which Cdc24p is membrane associated is unknown, but it may be through its PH domain or through interactions with other proteins (see below).
(i) Cdc24p-Rsr1p/Bud1p interactions.
Besides interacting with Cdc42p (see “GEF interaction domains” above), Cdc24p interacts with Rsr1p/Bud1p, Bem1p, and the Ste4p Gβ subunit. Rsr1p was originally identified in a genetic screen for multicopy suppressors of a cdc24-4ts mutant (35). DNA and predicted amino acid sequence analyses indicated that Rsr1p belonged to the Ras subfamily of the Ras superfamily of GTPases. Rsr1p showed 57% identity to the first 120 amino acids of c-Ha-Ras protein and yeast Ras1p and Ras2p; its closest homolog is the Krev-1/rap1a protein, with 56% identity over the entire protein. Deletion of Rsr1p did not result in death, indicating that Rsr1p is not essential for growth, but it did result in a random bud site selection pattern, as occurs with certain cdc24 alleles (see above), suggesting that the role of Rsr1p may be in the selection of the nonrandom site for bud emergence. This possibility was confirmed when rsr1 mutants were identified in a screen for mutants defective in establishing a normal axial budding pattern (80). This screen identified five genes, designated BUD1 through BUD5, of which BUD1 was shown to be allelic to RSR1. Subsequent genetic and biochemical analyses have indicated that Bud2p is a GAP and Bud5p is a GEF for the Rsr1p/Bud1p GTPase (34, 64, 79, 81, 448, 450, 462).
Expression of the rsr1G12V activated allele, but not the rsr1K16N dominant negative allele, suppressed a cdc24 mutant (499), suggesting that Rsr1p/Bud1p must be in an activated state to interact with Cdc24p. This hypothesis was confirmed biochemically by showing interactions between GST-Rsr1p/Bud1p-GTPγS with Cdc24p (449, 629), as well as by the inability of the rsr1/bud1T35A effector domain mutant to suppress a cdc24 mutant (383, 449) or to bind to Cdc24p (449). A truncated protein containing residues 472 to 854 of Cdc24p, which encompasses the PH domain and the potential Ca2+ binding domains (Fig. 5), was capable of interacting with Rsr1p/Bud1p (449); the effects of added Ca2+ on Cdc24p-Rsr1p/Bud1p interactions were not tested. Interestingly, the Bem1p scaffold protein (see below) preferentially bound to GDP-bound Rsr1p/Bud1p (449), suggesting that Rsr1p/Bud1p has differential binding partners depending on its nucleotide-bound state. Binding of Cdc24p to GST-Rsr1p/Bud1p in vitro did not affect the Bud5p-dependent GEF activity on Rsr1p/Bud1p, and binding of GST-Rsr1p/Bud1p to either GDP or GTPγS did not affect the in vitro Cdc24p-dependent GEF activity on Cdc42p (629). However, added Cdc24p did inhibit the intrinsic and GAP-stimulated GTPase activity of Rsr1p/Bud1p, suggesting that binding of Cdc24p to Rsr1p/Bud1p stabilizes the GTP-bound form of Rsr1p/Bud1p. These results suggest that Cdc24p may act to connect the Rsr1p/Bud1p-dependent bud site selection process and the Cdc42p-dependent bud site organization and polarized growth processes.
Rsr1p/Bud1p contains a canonical C-terminal CAAX farnesylation site, which is modified, leading to membrane localization. However, there are conflicting data on the subcellular localization of Rsr1p/Bud1p. One study, using polyclonal anti-Bud1p antibodies, indicated that the protein was distributed entirely to the particulate fraction and was localized around the entire periphery of the cell (383), while another study indicated that ∼10% of a HA-tagged Rsr1p/Bud1p was in a soluble fraction (449). There also are conflicting results on the effects of guanine nucleotide on these fractionation patterns. One study showed no dependence on the guanine nucleotide-bound state of Rsr1p/Bud1p (i.e., presumably GTP-bound Bud1G12Vp and presumably GDP-bound Bud1K16Np both fractionated solely to the particulate fraction) or on the Bud2p GAP or the Bud5p GEF (383). Another study showed Bud1G12Vp in both soluble and particulate pools but Bud1K16Np only in a particulate pool (449), suggesting that GDP-bound Rsr1p/Bud1p was always membrane bound. These discrepancies must be resolved before it is possible to conclude that Rsr1p/Bud1p cycles on and off the membrane as part of its cellular function. Based on the above data, a possible role for Rsr1p/Bud1p is in the targeting and/or anchoring of Cdc24p to the plasma membrane at the site of incipient bud emergence. However, Rsr1p/Bud1p was not necessary for the Cdc24p particulate fractionation pattern (383) (see above) or for localization of Cdc42p to the tips of mating projections (133), suggesting that the membrane localization of Cdc24p and Cdc42p does not solely depend on their interactions with Rsr1p/Bud1p.
(ii) Cdc24p-Bem1p interactions.
Bem1p was identified in three different genetic screens: as a synthetic lethal mutant with msb1 (36), a mutant that was identified as a multicopy suppressor of the cdc24-4ts and cdc42-1ts alleles (35); as a synthetic lethal mutant with a bud5 allele (79); and as a mutant with mating defects that were due to an inability to form mating projections (90, 91). The bem1ts alleles analyzed displayed large, unbudded, multinucleate cells with delocalized chitin and actin reminiscent of cdc24 and cdc42 alleles. Bem1p is not essential but is important for growth, in that Δbem1 cells grew slowly at 23 and 30°C, but were ts and cs for growth (90); however, this phenotype was strain specific in that the Δbem1 mutation was lethal in other strain backgrounds (294). Bem1p fractionated to both particulate and soluble pools and appears to be a phosphoprotein, as evidenced by a protein mobility shift on sodium dodecyl sulfate-polyacrylamide gel electrophoresis upon phosphatase addition (303). Bem1p localized to the site of incipient bud emergence and to the tips of small buds (21, 465), as do Cdc24p and Cdc42p. This localization was not disrupted by incubation with the actin-depolymerizing drug latrunculin-A (21), suggesting that Bem1p localization occurs independently of actin localization.
Bem1p is a ∼62-kDa protein with two N-terminal SH3 domains, and it has a homolog (Scd2p/Ral3p) in S. pombe (73). In GST affinity chromatography and yeast two-hybrid assays, Cdc24p interacted with Bem1p through the C-terminal 35 amino acids of Cdc24p, which contain one of the two potential Ca2+ binding domains (Fig. 5), and the C-terminal 225 amino acids of Bem1p (374, 458, 629); the Bem1p SH3 domains were not necessary for this interaction. The in vitro interaction between GST-Bem1p and Cdc24p was inhibited by adding 2 mM Ca2+, which may explain the Ca2+-sensitive phenotypes of cls4 alleles (see above). As mentioned above, GST-Bem1p interacted in vitro with GDP-bound Rsr1p/Bud1p, but addition of GST-Bem1p did not affect the in vitro, Bud5p-dependent GEF activity toward Rsr1p/Bud1p, the Cdc24p-dependent inhibition of Rsr1p/Bud1p GTPase activity, or the Cdc24p-dependent GEF activity on Cdc42p (629). Therefore, the biochemical consequences of Bem1p interaction remain elusive.
Through its interactions with a number of different proteins, Bem1p is thought to serve as a scaffold protein to bring various proteins into an ∼80S complex (458) during both the mitotic cell cycle and the mating cycle. As mentioned above, Bem1p interacts with Cdc24p, and this interaction was corroborated genetically by the synthetic lethal phenotype of cdc24 bem1 double mutants (458). In coimmunoprecipitation and yeast two-hybrid protein experiments, Bem1p associated independently with the Ste20p protein kinase, Ste5p, and with actin and was necessary for the interaction between Ste20p and actin (303). However, in another study (339), interactions were observed between Bem1p and Ste5p but not between Bem1p and Ste20p. Bem1p-Ste20p interactions were between the second SH3 domain and C-terminal domain of Bem1p and the N-terminal domain, including the CRIB domain, of Ste20p. Interestingly, two bem1 truncation mutants (bem1-s1 and bem1-s2) that were identified by their mating defects (90, 91) were unable to interact with Ste20p but could still interact with Ste5p and actin (303), suggesting that the mating defects were the consequence of loss of Ste20p interactions. Bem1p was also identified in two genetic screens in which overexpressed proteins suppressed dominant-negative ste4 mutants (294) and the α-factor resistance and G1 arrest defect of a fus3-2 mutant (339). In the former screen, Mdg1p was also identified; overexpression of this 41-kDa unique protein could also suppress the bem1-s1 mating defects. In the latter study, it was also shown that Bem1p interacts with Far1p, a Cdc28-cyclin inhibitor, and can stimulate Fus3p protein kinase activity.
Bem1p may link Cdc42p- and Rho-dependent processes. It was observed that bem1 mutants exhibited synthetic lethality with bem2 mutants (458) (Bem2p is a Rho-GAP) and that overexpression of Bem1p suppressed the lethality of a Δrho3 Δrho4 double mutation (375). This suppression was mediated by the second SH3 domain and the C-terminal 35 amino acids of Bem1p (37, 374). The same two domains are essential for Bem1p function in vivo, as evidenced by results with loss-of-function bem1 mutations (374). The second SH3 domain of Bem1p also interacts with two functionally redundant proteins, Boi1p and Boi2p, through their proline-rich domains (37, 374). Boi1p and Boi2p are structurally similar, each containing a N-terminal SH3 domain, a central proline-rich domain, and a C-terminal PH domain. The Δboi1 Δboi2 double mutant displayed bud emergence defects and poor viability, which could be suppressed by overexpression of Rho3p or Rho4p (37, 374). Mutational analysis indicated that the C-terminal PH domain, but not the SH3 domain or the proline-rich region, is necessary and sufficient for Boi1p morphological function (37). In addition, overexpression of Boi1p or Boi2p alone (37, 374) or with Rho3p (37) led to impaired growth and large, unbudded, multinucleate cells, which could be suppressed by overexpression of Cdc42p (37). Interestingly, Boi1p interacted with Cdc42p, but not Rho1p, Rho2p, Rho3p, or Rho4p, in two-hybrid protein assays, and this interaction was through the Boi1p PH domain.
Taken together, these data indicate that Bem1p interacts with numerous proteins that are necessary for the budding cycle and the mating cycle, and although its role in the budding cycle is unclear beyond its ability to interact with multiple components, its role in the mating cycle seems to be in activating the pheromone response kinase cascade through interactions with Ste20p, Ste5p, and kinase components.
(iii) Cdc24p-Ste4p interactions.
Several lines of evidence suggest that Cdc24p functions within the S. cerevisiae mating pathway through interactions with the Ste4p Gβ subunit and Cdc42p and that it functions prior to cell-cell fusion, possibly by affecting the orientation of mating-projection formation in response to localized high concentrations of pheromone (91, 416, 472, 527, 626). The original cdc24ts mutants had reduced mating efficiencies (472), and the cdc24-4ts strain had modest defects in pheromone-induced, Ste4p-dependent transcriptional activation. These defects could be suppressed by overexpression of wild-type or activated CDC42 alleles (527, 626). New cdc24 mutant alleles have been identified in genetic screens for mutants that have either reduced mating efficiencies with mating-enfeebled partners (91) or reduced mating efficiencies with wild-type partners but no effects on the vegetative role of Cdc24p (i.e., wild-type growth, morphology, bud site selection, and actin distribution [416]). The latter cdc24-m1, cdc24-m2, and cdc24-m3 mutants had wild-type phenotypes with respect to pheromone-induced cell cycle arrest, transcriptional activation, mating-projection formation, and actin polarization but had defects in cell-cell fusion, an inability to properly orient to a mating-pheromone gradient, and decreased mating efficiencies with mating-enfeebled partners, suggesting that Cdc24p may be playing a role in mating-projection orientation in response to mating pheromone.
In two-hybrid protein assays, Cdc24p interacted with Ste4p (416, 626), an interaction mediated by the Cdc24p N-terminal domain between residues 170 and 245 (416) (Fig. 5). Interestingly, the cdc24-m1 and cdc24-m3 alleles encode amino acid changes within this domain at residue 189 (S189F and S189P, respectively) and the cdc24-m2 allele encodes a D190G change (416), suggesting that the loss of mating-projection orientation seen in the cdc24-m1, cdc24-m2, and cdc24-m3 mutants may be due to a loss of interaction with Ste4p and not with Cdc42p or Bem1p; this inference was confirmed in two-hybrid protein assays. This N-terminal domain of Cdc24p is separate from the catalytic DH domain and the PH domain. Two-hybrid interactions between Cdc24p and Ste4p required the presence of the Gγ subunit Ste18p, but in vitro GST affinity chromatography experiments indicated that Cdc24p could interact directly with Ste4p in the absence of other yeast proteins (416). Whether interactions between Cdc24p and Ste4p occur in vivo awaits coimmunoprecipitation experiments, but these in vitro and two-hybrid protein data suggest that Cdc24p functions in mating-projection orientation through interactions with the Ste4p Gβ subunit. It is not clear whether Cdc24p is serving as a GEF for Cdc42p in this process or if Cdc24p and Cdc42p are localized to mating projections in the cdc24-m1, cdc24-m2, and cdc24-m3 mutants.
S. pombe GEF.
A potential Cdc24 homolog, named Scd1p, was identified in S. pombe in a genetic screen for mutants with mating defects and round cells (73). Scd1p showed 32% identity to Cdc24p, and this identity was found throughout the coding region, including amino acids 194 to 254 (containing the DH domain) (Fig. 5). Scd1p is the same protein as Ral1p, also identified as a mutant with mating and morphological defects (159). Surprisingly, a scd1 deletion did not lead to cell inviability, as a cdc24 deletion does in S. cerevisiae, but it did result in mating defects and round cells. This result brings into question whether Scd1p is the sole physiological GEF for the essential GTPase Cdc42p; this has not been tested biochemically to date. Overexpression of S. cerevisiae Cdc24p could partially suppress the mating and morphological defects of a scd1 mutant, and although overexpression of S. pombe Cdc42p could not suppress the same defects, it did enhance the suppression by S. cerevisiae Cdc24p. This result is again different from results with S. cerevisiae, in which overexpression of Cdc42p can suppress cdc24ts mutants. In two-hybrid protein assays, Scd1p could interact with Scd2p, a potential Bem1p homolog that was identified in the same genetic screen. In addition, Scd1p could interact with S. pombe Cdc42p, but only when Scd2p or S. pombe Ras1p was overexpressed in the same cells, suggesting that Scd1p may have to be bound to Scd2p or activated by Ras1p in order to interact with Cdc42p. Scd1p also interacted with Ras1p in the presence of overexpressed Scd2p or after the N-terminal 671 amino acids of Scd1p were deleted (Scd1ΔNp); it could also interact with activated mutations of human H-Ras. Interactions between Scd1ΔNp and Scd2p were corroborated by GST affinity chromatography experiments. While interactions between S. pombe Scd1p, Scd2p, and Ras1p are reminiscent of interactions between S. cerevisiae Cdc24p, Bem1p, and Rsr1p/Bud1p (see above), there remain questions about the physiological role of Scd1p in S. pombe as a potential GEF for Cdc42p.
Drosophila and C. elegans GEFs.
Three potential Cdc42 GEFs have been identified in Drosophila to date: Drt-GEF (589), still life (sif) (532), and DRho-GEF2 (29, 190). However, none has been shown to have either in vitro or in vivo GEF activity against Cdc42p. The 658-amino-acid Drt-GEF contains an N-terminal SH3 domain followed by a DH domain and a PH domain. Drt-GEF mRNA is expressed during oogenesis and embryogenesis and seems to be concentrated in the ventral furrow, cephalic furrow, posterior midgut, and anterior midgut involutions, areas which undergo actin-dependent morphological changes during gastrulation. The still life (sif) mutant was identified by reduced locomotor behavior, and the sif gene encodes two differentially spliced products. Both contain two PH domains, a PDZ domain, and a DH domain as well as potential PEST sequences. The sif mRNA was found predominantly in the brain and ventral nerve cord, and anti-Sif antibodies were localized to the neuropils, the location at which neurites form synapses, in both embryonic and adult brains. Overexpression of full-length Sif was not associated with a phenotype, but overexpression of a N-terminal truncation mutant missing the sequences before the first PH domain resulted in defects in axonal extension in Drosophila and induced membrane ruffling in human KB cells. This mutant protein colocalized with actin structures at the ruffles. DRho-GEF2 was identified as a dominant suppressor, Su(Rho1)2B, of DRho1 overexpression (29) and in a screen for maternal effects of zygotic lethal mutations (190). The Su(Rho1)2B mutation could not suppress the DRac1 and Cdc42Dm overexpression phenotypes, suggesting that DRho-GEF2 has Rho1-specific functions. DRho-GEF2 is ∼284 kDa and contains an N-terminal PDZ domain followed by a potential phorbol ester/diacylglyceride binding domain, a DH domain, and a PH domain. Mutants defective in DRho-GEF2 function have abnormalities in ventral-furrow formation and anterior and posterior midgut invaginations that are qualitatively similar to those associated with ectopic expression of a dominant negative Rho1N19 mutant protein and not seen with ectopic expression of dominant negative Cdc42 or Rac1 mutant proteins. This result reinforces the possibility that DRho-GEF2 is an in vivo GEF for Rho1.
As mentioned above (see “C. elegans” under “Functional studies”), the C. elegans unc-73 gene product exhibits structural homologies to the Dbl family of GEFs and has guanine nucleotide exchange activity against CeRac1 but not CeCdc42 in vitro (536); whether it is a GEF for Cdc42p in vivo is unknown. No other potential C. elegans GEFs have been described to date.
Mammalian GEFs.
There are multiple potential Cdc42p-GEFs in mammalian cells, including the Dbl, Dbs, Ost, Bcr, and Abr oncoproteins, the Tiam-1 invasion-inducing protein, the PAK-interacting exchange factor PIX, the FGD1 faciogenital dysplasia protein, and the Brx estrogen receptor binding auxiliary protein (for reviews, see references 44, 71, 441, and 594). The prototypical mammalian GEF is the Dbl oncoprotein, originally identified by malignant transformation of NIH 3T3 cells with transfected DNA from a human B-cell lymphoma (138, 534). Comparison of the predicted amino acid sequence of proto-Dbl with the Cdc24p GEF and the Bcr oncoprotein identified a ∼200-amino-acid domain (residues 498 to 674 in Dbl) with significant sequence similarity (491). Deletion of or mutations in this DH domain (Fig. 5), including the replacement of the highly conserved LLLKELL sequence at amino acids 640 to 646 with the conservative IIIRDII sequence, resulted in loss of Dbl-transforming activity and GDP dissociation activity (206, 491), suggesting that the DH domain is necessary for both activities. Expression of the Dbl DH domain by itself in NIH 3T3 cells did not lead to cellular transformation (634), but fusion of the DH domain to GST resulted in a protein with fully functional exchange activity against Cdc42Hs (206), suggesting that the DH domain is not sufficient for transformation but is sufficient for GEF activity. It should be noted that DH domains have been found in all known or potential Cdc42-GEFs identified to date but that the presence of a DH domain does not determine that a protein will have in vivo Cdc42-GEF activity. Dbl has in vitro GEF activity (i.e., ability to stimulate GDP dissociation from and promote GTP binding to a GTPase) against platelet and recombinant Cdc42Hs (205, 206) as well as RhoA and membrane-bound Rac1 (206, 443, 611), and it is able to bind to the Rho family members Cdc42Hs, RhoA, and Rac1 (206) and murine Cdc42, Rho, RhoC, Rac1, and RhoG (389) in GST affinity chromatography experiments, suggesting that it has a broad in vitro specificity range. Dbl binding to Cdc42 was observed with nucleotide-free Cdc42p and to a lesser extent with GDP-bound Cdc42p but not with GTP-bound Cdc42p (206, 389), suggesting that Dbl may stabilize a nucleotide-free or GDP-bound state of Cdc42p.
All known Dbl family members also have a ∼100-amino-acid PH domain, ubiquitously found C-terminal to the DH domain (Fig. 5). Expression of the Dbl PH domain by itself in NIH 3T3 cells did not lead to cellular transformation but did inhibit the transforming ability of full-length Dbl in a specific manner not seen with expression of the Vav or Cdc24 PH domain (634). Deletion of the Dbl PH domain (residues 703 to 812) or addition of a GST-PH fusion domain had no effect on the DH-dependent GEF activity. However, recent studies suggest that the presence of a PH domain enhances the DH catalytic activity (331). In immunoprecipitation studies with NIH 3T3 cells, usually more than 50% of the Dbl protein was found in Triton X-100-insoluble fractions (178, 634), suggesting that Dbl is localized to the cytoskeletal matrix. Deletion of the Dbl PH domain resulted in a protein with a cytosolic fractionation pattern; addition of the Ras membrane localization signal could restore the particulate fractionation pattern but not the transforming activity of Dbl (634). In addition, a FLAG epitope-tagged Dbl PH domain fractionated to the Triton X-100-insoluble pool. These data suggest that the Dbl PH domain is necessary and sufficient for targeting of Dbl to its proper subcellular location, a function that seems to be conserved in a number of Dbl family members.
The Ost and Dbs Dbl family members have significant amino acid sequence similarity to Dbl both inside and outside of the DH and PH domains (224, 592). Both proteins have in vitro GEF activity against Cdc42Hs and RhoA but not against Rac1. The Bcr and Abr proteins have in vitro GEF activity against Cdc42, RhoA, Rac1, and Rac2 (95); the Bcr protein contains an N-terminal Ser/Thr kinase domain, and both proteins contain C-terminal GAP domains that are functional against Rac1 and Rac2 and to a lesser extent against Cdc42 (see “GTPase-activating proteins” below). While the Tiam-1 protein has in vitro GEF activity against RhoA, Cdc42Hs, and Rac1 (385), in vivo studies strongly suggest that Tiam-1 activity is mediated through Rac1 (131, 189, 223, 386, 535). The PIX GEFs are new members of the Dbl family that were identified as high-affinity binding partners with PAK1 (360). These proteins display in vitro GEF activity against Cdc42 and Rac1, but as with Tiam-1, the in vivo target may be solely Rac1. Interestingly, PIX localized to focal complexes, as did PAK and Cdc42, and it may act in a Cdc42-independent targeting mechanism for targeting PAKs to focal complexes (see “PAK-like kinases” below).
The faciogenital dysplasia protein (FGD1), which is encoded by the genetic locus responsible for Aarskog-Scott syndrome (452, 453), is believed to be a Cdc42-specific GEF in vivo. This inference is based on the observations that (i) an epitope-tagged FGD1 polypeptide containing its DH and PH domains bound specifically to Cdc42, but not to Rho or Rac, in GST affinity chromatography experiments (631); (ii) the same FGD1 polypeptide displayed in vitro GEF activity against Cdc42 but not Rho or Rac (631); (iii) microinjection of FGD1 into Swiss 3T3 fibroblasts led to a stimulation of G1 cell cycle progression, induction of filopodia, and activation of the JNK and p70 S6 kinases in a manner similar to activated alleles of Cdc42p (410, 443, 631); and (iv) FGD1 induction of filopodia was blocked by coexpression of either the dominant negative Cdc42T17N mutant protein (631) or the WASP CRIB domain (410). However, recent evidence suggests that FGD1 may have Cdc42-independent functions in certain signaling pathways (410, 593). Interestingly, a FGD1-related protein, termed frabin, has recently been identified and shown to interact with actin and induce JNK activation and actin-dependent cell shape changes in Swiss 3T3 cells (426); whether frabin has Cdc42 GEF activity was not tested. The Brx estrogen receptor-binding auxiliary protein contains DH and PH domains as well as a diacylglycerol binding domain, and it is believed to function through Cdc42p because expression of the dominant negative Cdc42T17N protein led to a decrease in Brx-dependent induction of estrogen receptor transcriptional activity (495). However, Brx has not been shown to have in vitro or in vivo GEF activity against Cdc42p or other Rho-type GTPases.
There is recent evidence suggesting that these GEFs have the ability to differentially modulate Cdc42-dependent downstream effects. For instance, Dbl was able to stimulate PAK1 activation in COS7 cells to a higher degree than it could stimulate JNK activation, while FGD1 stimulated JNK activation but was unable to stimulate PAK1 activation (635). However, despite this wealth of information on potential mammalian Cdc42p GEFs, the physiologically relevant Cdc42p GEFs and the processes they regulate have not been definitively identified.
GTPase-Activating Proteins
The transition of G proteins from an active GTP-bound state to an inactive GDP-bound state occurs through the intrinsic hydrolysis of GTP to GDP + inorganic phosphate (Pi), a process that can be significantly stimulated by the action of GAPs. Cdc42p displays a ∼10-fold higher intrinsic (GAP-independent) rate of GTP hydrolysis compared to Ras proteins (210), and this rate can be further stimulated by the addition of GTPase-activating proteins. It has been postulated that differences in the GTP binding domain (residues 115 to 118 in Cdc42p) may account for the higher rate of GTP hydrolysis, but this has not been experimentally tested to date. Recently, it was shown that Cdc42p can undergo homodimer formation in vitro and that this homodimer formation can lead to a Cdc42-GTP-stimulated increase in intrinsic GTPase activity (623). In addition, the C-terminal polybasic region of Cdc42p, and specifically the R186 residue, was shown to be necessary for homodimer formation and for this GAP activity (622, 623). Introduction of the K186R mutation into S. cerevisiae Cdc42p leads to an increase in intrinsic GAP activity in vitro, a ts loss-of-function phenotype in vivo, and abnormal cell morphologies at the permissive temperature (622), reinforcing a potential physiological role of the polybasic region in intrinsic GAP activity.
S. cerevisiae Cdc42p GAPs.
Bem3p, Rga1p/Dbm1p, and Rga2p are three potential Cdc42p GAPs identified in S. cerevisiae, but only Bem3p has been shown to have GAP activity against Cdc42p in vitro (630). Bem3p was originally isolated as a multicopy suppressor of bem2ts mutants (36, 630). The 125-kDa Bem3p contains a C-terminal domain (residues 977 to 1140) with significant similarity to Bem2p and other Rho-GAPs (630), and this GAP domain can be subdivided into three subdomains with various levels of sequence similarity (633). Bem3p also contains a PH domain (residues 633 to 739). An E. coli-produced GST-Bem3p GAP domain (residues 751 to 1128) fusion protein had in vitro GAP activity against a GST-Cdc42p fusion protein that was not competed by the GST-Bem2p GAP domain fusion protein (630). The GST-Bem3p GAP domain fusion protein also had in vitro GAP activity against human Cdc42Hs but not against the GTPase-defective Cdc42G12V mutant protein, and it did not affect the binding of GTP to Cdc42Hs (633). Analysis of Bem3p GAP domain deletions indicated that all three GAP homology subdomains were necessary for GAP activity, but analysis of chimeras between Bem2p and Bem3p GAP subdomains indicated that the two N-terminal subdomains were sufficient for Cdc42p binding and GAP activity, albeit at ∼30% of the GST-Bem3p levels (633). Bem3p interacted with Cdc42p, but not with other Rho-type GTPases, in a two-hybrid protein assay; this interaction was enhanced with the GTPase-defective Cdc42Q61L mutant protein (537). A Δbem3 strain was viable with no morphological abnormalities (references 537 and 630 and data not shown).
Rga1p/Dbm1p was identified in two independent genetic screens (84, 537, 538). Rga1p was identified in a genetic screen designed to isolate mutants that could activate the pheromone response pathway in the absence of the Ste4p Gβ subunit, as assayed by FUS1::HIS3 expression (537, 538). Dbm1p was identified as a dominant suppressor of a bem2ts mutant (84). The RGA1/DBM1 gene encodes a predicted ∼113-kDa protein that contains a C-terminal Rho-GAP domain and two N-terminal LIM domains (84, 537), which are thought to bind zinc ions and mediate protein-protein interactions (117, 384). Deletion of RGA1 did not lead to death, indicating that it is not essential for growth, but the Δrga1 mutation led to a shift in the haploid bud site selection pattern from axial to bipolar and increased the expression of a FUS1::lacZ reporter gene (84, 537). Overexpression of Rga1p led to a decrease in FUS1::lacZ expression, which is consistent with Rga1p playing a negative role in the regulation of the pheromone response pathway, as well as a reduction in the restrictive temperature for the cdc42-1ts allele (537) and a reversal of the ability of overexpressed Cdc42p to suppress a cdc24ts mutant (465). The Δrga1 mutation raised the restrictive temperature of a cdc24ts mutant and restored wild-type morphology to a cdc24ts mutant at 30°C (537), which is consistent with Rga1p acting in opposition to Cdc24p as a negative regulator of Cdc42p function. While Δrga1 and Δbem3 single mutants had modest morphological defects, a Δrga1 Δbem3 double mutant displayed an aberrant morphology of elongated cells with enlarged mother-bud neck regions reminiscent of cells delayed in the apical-isotropic switch, suggesting that Cdc42p must be inactivated to induce this switch (84, 537). Interestingly, the double mutant was still viable, suggesting that there may be additional Cdc42-GAPs (see below).
Rga1p interacted with Cdc42p, but not other Rho-type GTPases, in a two-hybrid protein assay; this interaction was enhanced with the GTPase-defective Cdc42Q61L mutant protein (537) and lost with the Cdc42V44A effector domain mutant protein (475a). Rga1p also did not interact with the Ste20p, Ste5p, Ste11p, Ste7p, or Fus3p components of the pheromone response pathway. Taken together, these data suggest that Rga1p is a negative regulator of Cdc42p within the mating pathway and possibly in the budding pathway and that it may be functioning as a GAP, although biochemical characterization of a Rga1p GAP activity has not been reported.
Another potential Cdc42-GAP, Rga2p, was identified through its homology to Rga1p (532a). Rga2p contains a Rho-GAP domain and two LIM domains, and as with Rga1p, overexpression of Rga2p decreased the restrictive temperature of the cdc42-1ts allele and a Δrga2 mutation raised the restrictive temperature of a cdc24ts mutant. However, a physiological role for Rga2p has not been determined.
Drosophila and C. elegans GAPs.
To date, the only potential Cdc42-GAP identified in Drosophila is the RnRac-GAP, the product of the rotund locus (7, 180, 181). Although RnRac-GAP has not been shown to have GAP activity against Cdc42p, overexpression of RnRac-GAP led to defects in actin organization similar to those seen with Drosophila cdc42 mutants (see “Drosophila” under “Functional studies” above).
The only potential Cdc42-GAP identified to date in C. elegans is the Ce-GAP isolated via DNA-DNA hybridization with the GAP domain from the mammalian Bcr GAP (86). However, a GST-Ce-GAP GAP domain fusion protein had in vitro GAP activity against all three Rho-like GTPases in C. elegans (Ce-Rac1, CDC42Ce, and Ce-RhoA), as well as the C. elegans Ras homolog Let-60. Therefore, this Ce-GAP has an even higher degree of in vitro biochemical promiscuity than mammalian GAPs (see below), and the physiological role of this GAP remains to be determined.
Mammalian GAPs.
At least 12 mammalian proteins have in vitro GAP activity against Cdc42p, including CDC42GAP/p50rhoGAP (28, 163, 164, 210, 293, 403), Bcr (124), Abr (95, 215, 548), p190GAP (518), n-chimaerin (8, 276, 357), 3BP-1 (98, 99), Graf (219, 553), RalBP1/RLIP76/RIP1 (65, 244, 451), MgcRacGAP (562), PARG1 (506), myr5 (406, 474), and Cd-GAP (292). However, most of these proteins also have in vitro activity against Rac and Rho proteins (for review, see references 290 and 572), and so the assignment of a subset of these GAPs as specific in vivo Cdc42-GAPs has proven difficult.
In initial in vitro GAP assays, CDC42GAP/p50rhoGAP showed a significant preference for Cdc42p over RhoA or Rac (28, 293, 481). Subsequently, in three different in vitro GAP assays (2-amino-6-mercapto-7-methylpurine ribonucleoside–phosphorylase-coupled assay, [γ-32P]GTP filter binding assay, and tryptophan fluorescence methods), it was determined that CDC42GAP/p50rhoGAP had ∼60-fold-enhanced catalytic efficiency against Cdc42 compared to Bcr, 3BP-1, and p190 (624). However, microinjection of the CDC42GAP/p50rhoGAP GAP domain into Swiss 3T3 cells led to an inhibition of actin-dependent stress fibers, suggesting that it may inhibit RhoA function within these cells (481). Bcr has in vitro GAP activity against Cdc42p and Rac, but microinjection of the Bcr-GAP domain into Swiss 3T3 cells led to inhibition of membrane ruffling, suggesting that Bcr interacts with Rac to regulate ruffling in these cells (481). However, while analysis of bcr-null mutants corroborated the interactions between Bcr and Rac, the data indicated that the cellular functions of Bcr were related to the Rac-dependent neutrophil respiratory burst and not to membrane ruffling (579), suggesting that overexpression of the Bcr-GAP domain may have pleiotropic effects in Swiss 3T3 cells.
The p190GAP has in vitro GAP activity against Cdc42p, Rac, RhoA, and RhoB (518), with a ∼ninefold-increased activity against Rho, and microinjection of p190GAP into Swiss 3T3 cells led to an inhibition of stress fiber formation (481), suggesting that Rho is its physiological substrate. The p190GAP interacted with Ras-GAP through its SH2 domains (56, 226, 518, 519), and the two proteins colocalized to actin structures in EGF-stimulated cells (76), providing a possible mechanistic link between the Ras and Rho signaling pathways. The n-chimaerin GAP has in vitro GAP activity against Rac1 preferentially over Cdc42p, but expression of dominant negative Cdc42T17N in Swiss 3T3 cells inhibited n-chimaerin-induced filopodium formation and expression of dominant negative RacT17N inhibited the n-chimaerin-induced lamellipodium formation (276), suggesting that n-chimaerin can function with both GTPases in vivo. The SH3 domain of Graf interacted with the focal adhesion kinase and localized to cortical actin structures (219), but there are no data to discriminate between Cdc42p or RhoA as its in vivo substrate. Microinjection of the 3BP-1 GAP inhibited Rac-dependent membrane ruffling, suggesting that its in vivo substrate was Rac (99). The myr5 rat unconventional myosin has a C-terminal domain with sequence similarity to Rho-GAP proteins and is functional as a GAP protein against Cdc42p in vitro (474). However, recent in vivo and in vitro studies suggest that myr5 may be a specific RhoA-GAP (406). The PARG1 protein, which was identified by binding the PDZ domain of the PTPL1 protein tyrosine phosphatase, displayed in vitro GAP activity against Cdc42p, Rac, and Rho, but it had greater efficacy against Rho (506). Recently, CdGAP was identified in a yeast two-hybrid protein screen with the Cdc42Y40C effector domain mutant protein as the bait (292). The proline-rich Cd-GAP had in vitro GAP activity against both Cdc42p and Rac1 but not RhoA, and microinjection of Cd-GAP into Swiss 3T3 cells led to the inhibition of Cdc42-dependent, bradykinin-induced filopodium formation as well as Rac-dependent, PDGF-induced membrane ruffles, suggesting that Cd-GAP can down-regulate both Cdc42p and Rac in vivo.
All these GAPs contain a ∼140-amino-acid GAP homology domain, and each contains additional structural motifs, including SH3 and SH2 domains, PH domains, DH domains, and proline-rich SH3 binding domains (290) (Table 3). The recently solved X-ray crystal structures of CDC42GAP/p50rhoGAP complexed with Cdc42Hs or RhoA (484, 485) suggest that CDC42GAP/p50rhoGAP interacts with GTPases predominantly through their switch I and II regions. Interestingly, these structural determinations indicate that CDC42GAP/p50rhoGAP and Ras-GAP have very similar three-dimensional structures while having little amino acid homology. For example, both have their catalytic Arg residues (Arg fingers) in approximately the same position, thereby stabilizing the GTPase in a transition state that promotes GTP hydrolysis (50). These structural and functional similarities suggest that these GAPs may have derived from a common ancestor (31, 483) or may have arisen through convergent evolution (50). Recently, the importance of the conserved Arg305 and Arg306 residues in the catalytic function of CDC42GAP/p50rhoGAP was genetically and biochemically confirmed (309), reinforcing the X-ray crystal structure predictions and supporting a mechanism of action in which the positive charges of the Arg residues stabilize the negative charges that occur upon the interactions between the catalytic Gln61 Cdc42 residue and the GTP β-γ oxygens (309).
Guanine Nucleotide Dissociation Inhibitors
GDIs have a diverse set of in vitro functions including the ability to extract GTPases from membranes and the ability to inhibit guanine nucleotide exchange and GTPase activity, probably by sterically locking a GTPase in either a GDP- or GTP-bound state. The physiological roles for Cdc42-GDIs have not been examined in depth to date, but they will probably be shown to play a key role in regulating Cdc42p function through the cell cycle by altering its subcellular localization.
S. cerevisiae Rdi1p.
Rho-GDI was purified from S. cerevisiae cytosolic fractions by assaying for an activity that inhibited the dissociation of [3H]GDP from bovine rhoA (373). Based on peptide sequences, the Rho-GDI gene, RDI1, was isolated and it encoded a ∼23-kDa polypeptide with 36% identity to human and bovine Rho-GDIs. A GST-Rdi1 fusion protein was active on prenylated yeast Rho1p and mammalian RhoA and Rac1 but not on nonprenylated Rho1p, suggesting that Rdi1p interacts with Rho-like GTPases through the C-terminal membrane localization domain. Disruption of Rdi1p did not lead to death or defects in mating, sporulation, heat shock sensitivity, or budding pattern, suggesting that the protein is not essential for growth or morphogenesis. However, overexpression of Rdi1p, as well as bovine Rho-GDI, resulted in cell death. The morphological phenotypes associated with this cell death have not been reported, but if Rdi1p can extract Cdc42p from cellular membranes as mammalian Rho-GDI can, one would predict that the phenotypes would resemble cdc42 loss-of-function phenotypes. Myc-tagged Rdi1p coimmunoprecipitated with HA-tagged Rho1p and Cdc42p (267), suggesting that Rdi1p could interact with both GTPases within the cell. Rdi1p fractionated exclusively into soluble fractions, and immunofluorescence microscopy with HA-tagged Rdi1p indicated that Rdi1p was present in the cytosol. There was no change in the fractionation pattern of overexpressed HA-Cdc42p in a Δrdi1 mutant compared to the wild type, but overexpression of Rdi1p led to an increase in soluble Cdc42p (267). However, it should be noted that in these experiments, HA-tagged Cdc42p was found predominantly in soluble fractions, which is opposite from the predominant particulate fractionation pattern of endogenous Cdc42p (643). It remains to be seen if Rdi1p displays GDI activity against Cdc42p as exhibited by mammalian Rho-GDI (see below).
Mammalian GDIs.
Three Cdc42-GDI proteins have been identified to date in mammalian cells; they are currently designated RhoGDIα (previously rhoGDI [for a review, see reference 549]), RhoGDIβ (previously LD4, LyGDI, D4, or D4/LyGDI [4, 306, 409, 459, 509]), and RhoGDIγ (also known as RhoGDI-3). The first Cdc42-GDI was purified from bovine brain cytosol by its ability to inhibit the dissociation of labeled GDP from Cdc42Hs (307); it effectively inhibited Dbl-catalyzed GDP dissociation as well. Limited peptide sequence from this 28-kDa purified protein suggested that it was identical to the previously isolated 28-kDa Rho-GDI (160, 569), which was shown to act as a GDI against Cdc42Hs (307) and to cosediment with Cdc42Hs (471). Rho-GDIα mRNA and protein were expressed in the brain, lungs, thymus, spleen, small intestines, and kidney (160, 524), suggesting that Rho-GDIα was a ubiquitous and promiscuous regulator of Rho-type GTPases. This GDI activity required the isoprenylation of Cdc42Hs, in that unprenylated Cdc42Hs isolated from E. coli was not responsive to the GDI (208, 307) and addition of the prenylation inhibitor lovastatin altered the Cdc42Hs sedimentation profile toward a noncomplexed protein (471). Addition of purified bovine brain Rho-GDIα to either human placental membranes or membranes from human epidermoid carcinoma (A431) cells containing Cdc42Hs resulted in a significant dissociation of Cdc42Hs from the particulate pool into the soluble pool, and this dissociation seemed to be insensitive to the Cdc42Hs-bound guanine nucleotide (307), suggesting that the GDI can interact with both GDP-bound and GTP-bound Cdc42Hs. A GST–Rho-GDIα fusion protein could also efficiently extract RhoA and Cdc42Hs, but not Rac1, from rat liver membranes (353). The Rho-GDIα, either native or as a GST fusion protein, also inhibited the intrinsic and GAP-stimulated GTPase activity of Cdc42Hs, and this GTPase inhibitory protein activity also required isoprenylation of Cdc42Hs (208).
The effects of Rho-GDIα in most cell types were in the disruption of actin-dependent structures and processes including cell motility and cellular morphologies in Swiss 3T3 cells (395, 546) and human keratinocytes (305), membrane ruffling in human KB cells (418), and cytoplasmic division in Xenopus embryos (262). In addition, overexpression of Rho-GDIα in C2C12 myoblasts inhibited differentiation into myotubes by affecting the transcription of myogenin and other regulatory factors (547). However, it should be noted that the role of Cdc42Hs–Rho-GDIα interactions in mediating these effects is unknown.
By using N-methylanthraniloyl-GDP (Mant-GDP) and fluorescence spectroscopy (308), it was shown that the saturable binding of Rho-GDIα to Cdc42Hs induced a conformational change resulting in quenching of the Mant-GDP fluorescence and that deleting the last 8 amino acids of Rho-GDIα interfered with this interaction (422) and resulted in a loss of GDI activity (459). As mentioned above (see “Cdc42Hs three-dimensional structure”), it seems likely that this conformational change is within the Cdc42Hs Rho insert domain, thereby leading to a stabilization of the guanine nucleotide-bound form. Recent studies involving Mant-GDP fluorescence with Rho-GDIα deletion mutants and Rho-GDIα NMR structure determinations indicated that the C terminus of Rho-GDIα formed a β-sandwich structure whose open end created a pocket for isoprene binding and that N-terminal residues 23 to 42 were necessary for the GDI inhibitory activity (175). These data suggested that GDI function was mediated through the cooperative action of C-terminal and N-terminal residues.
Rho-GDIβ was identified predominantly in hematopoietic cells, and it had ∼67% identity to Rho-GDIα. The activity of Rho-GDIβ toward Cdc42Hs is ∼10- to 20-fold weaker than that of Rho-GDIα (4, 459), which correlated with a ∼15-fold-weaker interaction between Rho-GDIβ and Cdc42Hs as assayed by Mant-GDP fluorescence (422). Also, Cdc42Hs did not cosediment with Rho-GDIβ isolated from U937 hematopoietic cells (174), suggesting that although Rho-GDIβ may have in vitro GDI activity against Cdc42Hs, it may not be a physiological GDI for Cdc42, a hypothesis corroborated by the lack of significant mutant phenotypes associated with the disruption of both copies of Rho-GDIβ in embryonal stem cells (182).
Human Rho-GDIγ was isolated from a human brain cDNA library by low-stringency DNA-DNA hybridization against Rho-GDIα and Rho-GDIβ cDNA probes (5); a mouse homolog, termed Rho-GDI-3, was identified in a two-hybrid protein screen with RhoB as bait (620). The 52-kDa Rho-GDIγ was ∼50% identical to Rho-GDIα and Rho-GDIβ but contained a highly hydrophobic 30 amino acid N-terminal domain not found in the other Rho-GDIs. Analysis of mRNA levels indicated that Rho-GDIγ was expressed predominantly in the brain and pancreas. In GST affinity chromatography experiments, Rho-GDIγ interacted with RhoA and Cdc42Hs but not with Rac1 or Rac2. Rho-GDIγ displayed in vitro GDI activity against Cdc42Hs, but it was ∼20-fold less potent than Rho-GDIα. The murine Rho-GDI-3 interacted with, and had in vitro GDI activity against, RhoB and RhoG and was expressed in the brain, lungs, kidneys, and testes (620). Determination of the physiological roles for Cdc42-GDIs in the future should provide valuable insights into Cdc42p function and subcellular localization.
CDC42P DOWNSTREAM EFFECTORS
Cdc42p can interact with a myriad of downstream effectors to regulate a diverse set of cellular functions. These effectors preferentially bind to GTP-bound Cdc42p and transduce the Cdc42p-dependent signals downstream to ultimately affect actin rearrangements, induction of transcription, and other cellular processes (see above). It should be noted that the mechanism(s) by which the interactions between Cdc42p and its effectors lead to this signal transduction is still unclear.
PAK-Like Kinases
S. cerevisiae PAK-like kinases.
Ste20p, Cla4p, and Skm1p are the three members of the PAK family of serine/threonine protein kinases found in S. cerevisiae (for a review of PAKs, see reference 516). All three have a highly conserved protein kinase domain in their C termini as well as a CRIB domain in their N termini. Cla4p and Skm1p differ from Ste20p in that they contain a PH domain located N-terminal to the CRIB domain. There are clear instances of overlapping functions for these PAK-like kinases in regulating actin-dependent growth during the cell cycle, the pheromone response pathway, and filamentous growth. However, it remains to be determined whether these overlaps are physiologically relevant or are due to imposed artificial stresses uncovering biochemical redundancies.
(i) Ste20p.
Ste20p was identified in two different genetic screens by its ability, when overexpressed on a multicopy plasmid, to either suppress the sterility caused by overexpression of the dominant negative ste4D62N mutant allele in a Δste4 background (295) (Ste4 encodes the Gβ subunit of the pheromone response pathway heterotrimeric G protein [596]) or induce the transcriptional activation of a FUS1-lacZ fusion protein independent of added pheromone (469). Disruption of the Ste20p kinase domain resulted in cells with defects in mating, pheromone sensitivity, transcriptional induction of mating-specific genes, and induction of mating projections (295, 469), while overexpression of N-terminal truncation mutants in which the CRIB domain and other sequences, but not the kinase domain, were deleted, caused death (298, 469). Epistasis experiments with mutant alleles of other components of the pheromone response pathway indicated that Ste20p functioned at or just below the level of the G protein but above the Ste11p-Ste7p-Fus3p/Kss1p MAP kinase signaling module. Recent data indicates that Ste20p interacts directly with the Ste4p Gβ subunit and that this interaction is enhanced upon pheromone addition (304). A small (14-amino-acid [ANSSLAPLVKLARL]) domain C-terminal to the kinase domain was necessary and sufficient for this interaction, and mutating the underlined Ser and Pro residues led to loss of this interaction. Interestingly, the Cla4p kinase (see below) does not have this highly conserved domain and interacts weakly with Ste4p, suggesting that these interactions play a role in the in vivo specificity of the PAKs.
In addition to its role in the pheromone response pathway, Ste20p was shown to be necessary for the Cdc42p-dependent induction of filamentous growth (405). Interactions between Cdc42p and the CRIB domain of Ste20p were necessary for this induction (298). Recent data also suggested that Ste20p mediates Cdc42p effects on the actin cytoskeleton during the mitotic cell cycle. Overexpression of a GST-Ste20p fusion protein, but not a kinase-inactive mutant protein, could suppress the cdc42-1ts growth and morphological phenotypes, and added GST-Ste20p could reverse the rhodamine-labeled actin polymerization defects observed in permeabilized cdc42-1ts mutant cells (130). There were also defects in rhodamine-labeled actin polymerization observed in permeabilized Δste20 and Δcla4 mutants, which could be reversed by adding GST-Ste20p, suggesting that both Ste20p and Cla4p (see below) can mediate Cdc42p effects on the actin cytoskeleton. Interestingly, the Cln1p–2p/Cdc28p G1 CDK complex can phosphorylate Ste20p at the time that Ste20p is localized to the sites of polarized growth at the bud tips, but this phosphorylation does not seem to affect Ste20p kinase activity (431, 601). However, the Cln2p-Cdc28p phosphorylation of Ste20p does correlate with a repression of the pheromone response signaling pathway (431), suggesting that Ste20p phosphorylation induces a switch between the mitotic cell cycle and mating responses.
Characterization of in vitro and in vivo Ste20p kinase activity indicated that Ste20p is a phosphoprotein that can undergo autophosphorylation that is necessary for its kinase activity and depends on the K649 and T777 residues. Ste20p can phosphorylate the Ste11p kinase in vitro, which is the next downstream kinase in the pheromone response MAP kinase module, on Ser and Thr residues (603). In addition, both Ste20p and Cla4p can phosphorylate S357 of the head domain of the Myo3p myosin I heavy-chain protein in S. cerevisiae (602). This phosphorylation was deemed necessary for Myo3p function because the S357A mutation resulted in a nonphosphorylated, nonfunctional protein and the S357D mutation, which would mimic the phosphorylation state of S357, resulted in a functional protein. The Myo3S357D mutant allele could not suppress the lethality or morphological defects associated with the Δste20 Δcla4 double mutant or Δcla4 single mutant (see below), suggesting that Myo3 was not the sole physiologically important substrate for Ste20p and Cla4p. Phosphorylation of the Dictyostelium myosin I homolog led to actin-stimulated Mg2+-ATPase activity and motor activity (604); it is unknown if Myo3p has similar functions in S. cerevisiae. A myosin I heavy-chain kinase (MIHCK) with sequence similarities to PAKs has been identified in Dictyostelium (302) and Acanthamoeba (57), and the Dictyostelium MIHCK contained a CRIB domain and kinase domain, bound to GTP-Cdc42p and GTP-Rac1p in overlay assays, and autophosphorylated in the presence of GTPγS-Rac1p and GTPγS-Cdc42p (301, 302). In addition, another Ste20-like kinase from Dictyostelium has recently been shown to phosphorylate the Ca2+-dependent actin fragmenting protein severin (132).
By using two-hybrid protein assays, GST affinity chromatography experiments, and immunoprecipitation experiments, it has been shown that Ste20p preferentially interacted with Cdc42p-GTP in vitro and in vivo (298, 456, 527). This interaction was between the effector domain of Cdc42p and the CRIB domain of Ste20p, as evidenced by the loss of binding seen with the Cdc42T35A effector domain mutant protein (456) or the CRIB-deleted Ste20p-Δ335–370 (456) or Ste20p-Δ334–369 (298), by the inability of the Cdc42T35A mutant protein to suppress a ste20K649R dominant negative mutant (456), and by the ability of wild-type Cdc42p and the GTP-bound Cdc42Q61L mutant protein, but not the dominant negative Cdc42D118A mutant protein, to suppress a ste20K649R dominant negative mutant (456). Interestingly, the Cdc42V44A effector domain mutant protein still interacted with Ste20p at comparable levels to wild-type Cdc42p (475a), indicating that Ste20p interacts with a subdomain of the effector domain. Further evidence for an in vivo interaction between Cdc42p and Ste20p came from the examination of a mutant strain containing the Δcla4 and Δste20 mutations along with a cla4-75ts allele (112, 298, 456). This strain is inviable at 37°C but can be complemented by expressing wild-type Ste20p on a plasmid; however, expression of a catalytically inactive Ste20-K649R mutant allele or the CRIB-deleted Ste20p-Δ335–370 mutant allele could not complement the defect (456). Similar results were seen when CRIB-deleted Ste20 mutant alleles were assayed for their ability to suppress a Δcla4 Δste20 double mutant that was viable upon GAL1-CLA4 expression but inviable on glucose media (298). Deletion of STE20 or other downstream components of the pheromone response pathway did not affect the ability of activated Cdc42G12V mutant protein to cause death and abnormal multibudded cells (9, 116), suggesting that Ste20p did not mediate Cdc42G12Vp effects.
Binding of Cdc42p-GTP to Ste20p was not required for Ste20p kinase activity in vitro (298, 456) (an effect of GTP-Cdc42 on Ste20p kinase activity has been reported [527]), and deletion of the N-terminal regulatory domain including the CRIB domain resulted in full and weakly constitutive catalytic activity (456), suggesting that binding of Cdc42p-GTP to the Ste20p N-terminal domain relieved a negative regulatory effect of this domain on the kinase activity. The effects of Cdc42p binding to Ste20p on the mating pathway were examined by analyzing the effects of the CRIB-deleted mutant Ste20p (298, 456). In both studies, it was shown that deletion of the CRIB domain had little effect on the pheromone response pathway as assayed by mating efficiencies, α-factor halo assays, induction of a FUS1::lacZ reporter gene, and generation of properly oriented, morphologically normal mating projections. Interestingly, there were defects in bilateral mating (i.e., mating between cells with the same ste20 mutants) and in mating with far1-c mutants that also display bilateral mating defects. These types of bilateral-mating defects are thought to be the consequence of loss of oriented mating projections toward the opposite mating partner, but this was not shown to be the case with the ste20 mutants (298). Instead, it appeared that these mutants had defects in the cell-cell fusion event preceding zygote formation. Taken together, these results suggest that Cdc42p-Ste20p interactions are not necessary for induction of the pheromone response signaling pathway (see “Mating pathway” under “Functional studies” above). Cdc42p-Ste20p interactions are necessary for the generation of pseudohyphae upon nitrogen starvation (298, 456, 488), and these interactions are mediated through the S. cerevisiae 14-3-3 proteins Bmh1p and Bmh2p as well as components of the pheromone response pathway (see “Pseudohyphal and invasive growth” above).
Wild-type Ste20p was localized to the sites of polarized growth in emerging buds and in mating projections, while the CRIB-deleted Ste20p showed a general cytoplasmic staining (298, 456), indicating that binding to Cdc42p is necessary for proper localization of Ste20p to sites of polarized growth. In summary, loss of Cdc42p binding does not dramatically alter the Ste20p kinase activity or Ste20p function in the pheromone response pathway, which is probably mediated through interactions with the Ste4p Gβ subunit (see above), but does affect Ste20p function in filamentous growth and progression through the cell cycle. The primary role for Cdc42p-Ste20p interactions may be in stabilizing Ste20p in the proper subcellular location so that it can interact with its downstream substrates.
(ii) Cla4p.
Cla4p was originally identified in two genetic screens designed to identify mutants unable to survive in the absence of the two G1 cyclins Cln1p and Cln2p (39, 113). The cla4/erc10 cln1 cln2 triple mutants displayed abnormal morphologies, including elongated buds, wide mother-bud necks, and multinucleate cells, indicative of a delay in either the apical-isotropic bud growth switch and/or nuclear division and/or a cytokinesis defect. Another mutant allele identified in the screen was cla10, which was allelic to CDC12, a member of the septin family of proteins that comprise the 10-nm filaments laid down at the mother-bud neck region in late G1 post-START and are necessary for proper cytokinesis (see below). Cla4p showed significant sequence similarity to Ste20p within its kinase and CRIB domains, with the exception that Cla4p contained a PH domain in its N terminus (112). Deletion of the CRIB domain or the PH domain resulted in a nonfunctional or partially functional protein (38). Deletion of the Cla4p catalytic kinase domain did not result in death but did result in morphogenetic defects similar to the original cla4 mutant (112).
The Cla4p CRIB domain could bind to Cdc42p in [32P]GTP overlay assays, and full-length Cla4p interacted with Cdc42p in the yeast two-hybrid protein assay (112). The Cdc42V44A mutation interfered with Cdc42p-Cla4p interactions in the two-hybrid assay (475a), suggesting that Cla4p interacted with Cdc42p through the effector domain. Cla4p interacted more strongly with Cdc42G12Vp and did not interact with other S. cerevisiae Rho-type GTPases (112), indicating that Cla4p was a bona fide downstream effector of Cdc42. This conclusion was reinforced by the Δcla4 cdc42-1ts and Δcla4 cdc42V44A double-mutant synthetic lethality (112, 475a) and the alteration of Cdc42G12V-dependent abnormal cellular morphologies in a Δcla4 background (116). While Δcla4 and Δste20 single mutants did not die, the Δcla4 Δste20 double mutant was inviable (112), indicating that Cla4p and Ste20p have an overlapping essential function. This essential function was not within the pheromone response pathway or the protein kinase C pathway, as evidenced by the inability of mutations within these pathways to suppress cla4 mutants. Mutations in the HOG (for “high-osmolarity glycerol”) pathway (reviewed in references 26 and 574) exacerbated Δcla4 mutant phenotypes and abolished the ability of high Na+ concentrations to rescue a Δcla4 mutant, suggesting that there may be common functions between these pathways. Interestingly, the morphological defects associated with the cdc42V44A mutant allele could also be remediated by high Na+ concentrations and by overexpression of Cla4p (475a). A cla4-75ts Δste20 double mutant at restrictive temperatures displayed defects in cellular morphologies (enlarged mother-bud necks) and septin ring assembly but not in cortical actin localization (112), suggesting the presence of other downstream effectors that could transduce the Cdc42p signal to the actin cytoskeleton. However, recent data suggests that both Ste20p and Cla4p can mediate Cdc42p effects on the actin cytoskeleton (see the previous section).
Immunoprecipitated Cla4p had in vitro kinase activity against the nonphysiological substrate myelin basic protein (38) as well as the S. cerevisiae Myo3p myosin I protein (602) (see above), although the physiological relevance of this phosphorylation is not known. This kinase activity was reduced three- to fourfold when the Cla4p was isolated from either a cdc42-1 or cdc24-1 mutant background (38), suggesting that functional GTP-bound Cdc42p is needed to activate Cla4p kinase activity. Cla4p kinase activity was cell cycle regulated, with a peak of activity during G2/M. This cyclical kinase activity depended on binding of Cdc42p, because a CRIB-deleted Cla4p did not show this cell cycle control and because expression of activated Cdc42G12Vp, but not dominant negative Cdc42D118Ap, resulted in a ∼2.5-fold increase in kinase activity. Activated Cdc42G12Vp along with the mitotic cyclin Clb2p-Cdc28p kinase complex led to the hyperphosphorylation of Cla4p and subsequent mitosis-specific and septin-specific hyperphosphorylation of the Gin4p protein kinase (14, 67, 333, 560), reinforcing a possible role for Cla4p during mitosis or cytokinesis, although the in vitro Cla4p kinase activity did not seem to be altered by its hyperphosphorylation. It remains to be seen if Cdc42p is needed for the subcellular localization of Cla4p, as it is for Ste20p (see above).
In addition to displaying synthetic lethality with cdc42 mutants, a cla4::LEU2 mutant was synthetic lethal with a cdc12 septin mutant and septin ring localization was defective in cla4 mutant cells (112). The S. cerevisiae mitotic septins, products of the CDC3, CDC10, CDC11, and CDC12 genes, belong to a family of eukaryotic proteins that are involved in regulating cytokinesis and cellular morphogenesis (for reviews, see references 78, 107, and 332). In S. cerevisiae, these proteins are components of a 10-nm filament ring that is set down at the site of bud emergence ∼15 min prior to bud emergence (260) in an actin-independent manner (21) and that persists at the mother-bud neck region through cytokinesis. The function of the septins is unclear, but recently they were shown to be necessary for the localization of various components of the bud site selection machinery (e.g., Bud4p) and the chitin biosynthesis/targeting machinery to the bud emergence site (119, 505), for the mitosis-specific phosphorylation of the Gin4p protein kinase (67, 333), and for the assembly and maintenance of the contractile actomyosin ring needed for cytokinesis (41). Taken together, these data suggest that the primary role of Cla4p is in regulating cytokinesis through interactions with Cdc42p and septins. The detailed analysis of Cdc42p-Cla4p and Cla4p-septin interactions, as well as the identification of physiological Cla4p phosphosubstrates, should greatly enhance our understanding of the function of this key regulatory kinase.
(iii) Skm1p.
Skm1p was identified on chromosome XV through the S. cerevisiae genome-sequencing project (371). It exhibited higher sequence similarity to Cla4p than to Ste20p, including the presence of a PH domain in its N terminus. Skm1p showed weak interactions with Cdc42p in two-hybrid protein assays, but it interacted more strongly with Cdc42G12Vp (475a), indicating that it was a bona fide downstream effector of Cdc42p. As with Ste20p, this interaction was not affected by the Cdc42V44A effector domain mutation, suggesting that Skm1p interacts with another subdomain of the Cdc42p effector domain. Disruption of Skm1p did not cause death (371), indicating that Skm1p does not play an essential role in cell growth. Loss of Skm1p also did not show adverse effects on cellular morphologies, bud site selection, growth on high-osmolarity media, or mating, and did not show a synthetic lethal phenotype with a Δcla4 or Δste20 mutant or mutations in the CDC10 septin, the Rho-GAP BEM2 or CDC42. Expression of Skm1p on a high-copy-number plasmid could not suppress the Δcla4 or Δste20 mutant phenotype; overexpression under an inducible promoter was not tested. However, overexpression of a GST-Skm1p fusion protein led to an abnormal cellular morphology of large, round, multinucleate cells with small, sometimes multiple, buds, suggesting that Skm1p may be functioning in cellular morphogenesis. Deletion analysis indicated that overexpression of the catalytic kinase domain was responsible for this phenotype as well as for an ability to suppress Δste20 mating defects. Overexpression of the Skm1p catalytic domain also led to severe growth defects similar to those seen with N-terminal truncation mutants of Ste20p and Cla4p, reinforcing the notion that the N-terminal domain of these PAK-like kinases containing the CRIB domain plays a negative regulatory function on kinase activity that is relieved by binding of Cdc42p. A detailed genetic analysis of SKM1 is needed to pinpoint its cellular function.
S. pombe PAK-like kinases.
There are two known PAK homologs in S. pombe, Pak1p/Shk1p (364, 447) and Pak2p/Shk2p (515, 616). The pak1+ (447) and shk1+ (364) gene product was isolated in two independent screens by degenerate oligonucleotide PCR based on S. cerevisiae Ste20 sequences; pak2+ was isolated in a cDNA library screen with the pak1 PCR product (447). The pak1+/shk1+ gene encoded a 72-kDa protein with significant amino acid identity to the PAK family, especially within the kinase domain and the N-terminal CRIB domain, and was shown to have in vitro autophosphorylation activity predominantly on Ser residues (447). Analysis of Δpak1/shk1 mutants indicated that Pak1p/Shk1p was essential for cell growth, with mutant cells exhibiting a small, round cellular phenotype reminiscent of cdc42 null mutants (390). Pak1p/Shk1p preferentially interacted with GTP-Cdc42p in GST affinity chromatography experiments and with the Cdc42G12V activated allele in two-hybrid protein assays (447). This interaction was abolished with the T35A effector domain mutation (447) and was not seen with the Cdc42T17N dominant negative allele (364, 447), suggesting that Pak1p/Shk1p is a bona fide downstream effector of Cdc42p. This point was corroborated by the observations that co-overexpression of mutant alleles of cdc42 and pak1 led to lethal growth and morphology defects (447) and that overexpression of wild-type Pak1p or a kinase-defective K415,416R mutant protein (447) or a C-terminal truncation mutant protein that still contained the CRIB domain (364) resulted in cells with abnormal morphologies and delocalized cortical actin structures. Both Pak1p/Shk1p and Cdc42p also functioned in the mating pathway, as evidenced by the reduced mating in the cdc42T17N dominant negative mutant and the pak1K415,416R kinase-defective mutant (364, 447), by the ability of Pak1p/Shk1p to partially suppress the cdc42T17N mating defect (364) and the S. cerevisiae ste20 mating defect (447), and by the ability of a Pak1p/Shk1p N-terminal deletion mutant protein to activate the S. cerevisiae pheromone response pathway (364) and to interfere with two-hybrid protein interactions between the S. pombe Byr1 and Byr2 protein kinases involved in Ras-mediated pheromone response (568).
Pak2p/Shk2p also showed a high degree of sequence similarity to PAK-like kinases and was most similar to S. cerevisiae Cla4p and Skm1p in that Pak2p contained a N-terminal PH domain not found in Pak1p/Shk1p (515, 616). Like Pak1p, Pak2p/Shk2p preferentially interacted with activated Cdc42p in two-hybrid protein assays and GST affinity chromatography experiments, but unlike Pak1p, deletion of pak2/shk2 did not lead to a lethal phenotype and Δpak2/shk2 mutants did not display any morphological or mating defects. However, overexpression of Pak2p/Shk2p led to morphological defects and could suppress the morphological and mating defects associated with Δpak1 mutants, and this suppression required the Pak2p/Shk2p PH and CRIB domains. In addition, co-overexpression of Pak2p/Shk2p and Cdc42p led to cell death and aberrant cellular morphologies. Overexpression of Pak2p/Shk2p in S. cerevisiae could not suppress the mating defects associated with Δste20 mutants or the morphological defects associated with Δcla4 single or Δcla4 ste20 double mutants; effects on Skm1p function were not analyzed. Therefore, it is likely that Pak2p/Shk2p is a downstream effector of Cdc42p in S. pombe, but its function in the polarity pathway is yet unknown.
In a two-hybrid protein screen with Pak1p/Shk1p as bait, a new protein kinase termed Skb1p was identified (166). Skb1p interacted with Pak1p/Shk1p, Pak2p/Shk2p (616), and itself but did not interact with Cdc42p, Scd1p, Scd2p, Ste20p, Ras1p, or mammalian p65PAK. The interaction with Pak1p/Shk1p was through a domain adjacent to the Pak1p/Shk1p CRIB domain and the N-terminal 72 amino acids of Skb1p. Deletion of skb1 did not result in death or mating defects but did result in a slightly slower growth and a mild shortened-cell morphological phenotype that was suppressed by overexpression of Pak1p/Shk1p. Overexpression of Skb1p resulted in hyperelongated cells and, together with overexpression of Pak1p/Shk1p, resulted in suppression of a ras1 mutant morphology. These data are consistent with Skb1p acting as a positive effector of Pak1p/Shk1p function. Recently, a 72-kDa mammalian homolog of Skb1p, termed IBP72, which was 52% similar in predicted amino acid sequence to Skb1p, was shown to interact in vitro and in two-hybrid protein assays with pICln, a protein involved in the regulation of a nucleotide-sensitive chloride current (279). Interestingly, mammalian Cdc42p also has been implicated in ion homeostasis through G-protein coupled Na+-K+ exchange (222) (see “Mammals” under “Functional studies” above).
A new S. pombe Ser/Thr protein kinase, termed Orb6p, was recently implicated in acting downstream of Pak1p/Shk1p in controlling cell polarity (575, 576). Orb6p shows significant amino acid similarity to the mammalian Rho-associated kinase and the myotonic dystrophy kinase DMPK (see below), and orb6ts mutants display defects in polarized cell growth and actin organization. The possibility that Orb6p acts downstream of Pak1p/Shk1p was deduced from the observations that orb6 mutants displayed synthetic lethality with pak1/shk1/orb2ts mutants and that overexpression of Orb6p suppressed the pak1/shk1/orb2ts mutant morphology defects. A physical interaction between Pak1p/Shk1p and Orb6p has not been reported to date.
Drosophila and C. elegans PAK-like kinases.
The Drosophila PAK homolog (DPAK) was identified by low-stringency DNA-DNA hybridization from an embryonic cDNA library (199). The 76-kDa DPAK contained a highly conserved CRIB domain as well as a Ser/Thr kinase domain. A GST fusion to the DPAK N-terminal CRIB domain bound to Drosophila RacA (DRacA) and Cdc42p (Dcdc42) and human Rac1 and Cdc42p (data not shown) in an overlay assay (199). DPAK mRNA and protein were localized ubiquitously throughout embryonic development, with elevated localization in epidermal cells associated with the dorsal vessel and muscle attachment sites as well as the central nervous system. DPAK colocalized with antiphosphotyrosine antibodies to focal adhesions and focal complexes, and with F-actin caps in the syncytial blastoderm and the leading edge of epidermal cells during dorsal closure over the amnioserosa, a process that is inhibited by expression of a dominant negative DRacA transgene (198). Therefore, it is unclear whether DPAK is a physiological Cdc42 effector.
Another potential downstream effector of Drosophila Cdc42p, the Gek protein kinase, was identified in a two-hybrid protein screen for proteins that interacted with the Dcdc42V12 mutant protein (338). Gek also bound to GST-Dcdc42V12 in GST affinity chromatography experiments, and this interaction was abolished by the Dcdc42T35A effector domain mutation and by the GekΔISP mutation within the Gek CRIB domain, suggesting that binding occurs between the Cdc42p effector domain and the Gek CRIB domain. The 1,613-amino-acid Gek protein contained an N-terminal Ser/Thr kinase domain, and immunoprecipitated myc-tagged Gek had histone kinase activity, which was lost when the catalytic Lys residue at position 105 was mutated to Ala. The Gek protein also contained an N-terminal coiled-coil domain, a Cys-rich domain similar to phorbol ester binding domains, a PH domain, and the C-terminal CRIB domain. The Gek kinase domain displayed 63% amino acid identity to the kinase domain of the human myotonic dystrophy protein kinase (DMPK) and 49% identity to the kinase domain of Rho-kinase, although DMPK and Rho kinase do not contain a Cdc42-interacting CRIB domain and are probably not Cdc42p effectors. Generation and characterization of P-element-directed gek deletion mutants suggested that Gek was essential for proper oogenesis and that cortical F-actin assembly around nurse cells required functional Gek. Defects in cortical F-actin assembly seen in Δgek mutants were similar to those seen with expression of activated cdc42G12V mutant allele (408) (see “Drosophila” under “Functional studies” above), suggesting that Gek may be a bona fide Cdc42p downstream effector.
The gene encoding the C. elegans PAK homolog, CePAK, was identified by degenerate PCR, isolated from an embryonic cDNA library, and mapped to chromosome X (87). The 64-kDa CePAK contained a N-terminal CRIB domain and a C-terminal Ser/Thr kinase domain and exhibited ∼52% similarity to DPAK and rat α-PAK. GST-CePAK bound in vitro to GTP-CeRac1 and GTP-Cdc42Ce, but not to the GDP-bound proteins, in filter-binding assays. Immunoprecipitated CePAK displayed weak autophosphorylation activity in the presence of GTP-Cdc42Ce and was found in both soluble and particulate fractions. The levels of CePAK mRNA normalized to actin mRNA levels were highest during embryogenesis and subsequently decreased during larval development. By using anti-CePAK antibodies, the protein was localized to hypodermal cell boundaries during embryonic body elongation and colocalized with Cdc42Ce at these boundaries, suggesting that both proteins function in the actin-dependent elongation of embryonic cell bodies. Recently, CePAK-β-galactosidase and CePAK-GFP fusion proteins were shown to localize to the cell surface of pharyngeal muscle cells as well as motor neurons and distal tip cells (231). Whether CePAK transduces Cdc42p signals in vivo is unknown.
Mammalian PAK-like kinases.
Mammalian PAKs act in response to a variety of intracellular and extracellular signals to mediate a number of different cellular events including growth factor- and stress-induced actin rearrangements and activation of the JNK/SAPK and p38 MAP kinase pathways (see below) (see “Mammals” under “Functional studies” above), Nef- and Nef-associated kinase-dependent HIV-1 replication and pathogenesis (111, 335, 508, 564), thrombin cleavage in platelets (554), cleavage arrest in frog embryos (492), Schwann cell transformation (551), CD28-dependent antigen-specific activation of T cells (248, 249), and T-cell receptor-mediated activation of the nuclear factor of activated T cells transcription factor (609). The original mammalian PAK, designated p65PAK, was identified as a rat brain protein that interacted with [γ-32P]GTP-GST-Cdc42 and [γ-32P]GTP-GST-Rac1, but not with [γ-32P]GTP-GST-RhoA, in an overlay assay (359). This interaction was specific for the GTP-bound form of Cdc42p, suggesting that p65PAK could function as a downstream effector of Cdc42p function. Purified p65PAK displayed Ser/Thr autophosphorylation activity that was stimulated by GTP-bound Cdc42p and Rac1p and had kinase activity against the exogenous myelin basic protein substrate. Currently, there are three major 62- to 68-kDa PAK isoforms in mammalian tissues, designated (in the nomenclature of Sells and Chernoff [561]) PAK1 (previously p65PAK, αPAK, and hPAK-1 [24, 55, 104, 265, 359]), which is found predominantly in brain, muscle and spleen tissue; PAK2 (previously γ-PAK, PAKI, and hPAK65 [235, 265, 492, 554]), which is ubiquitous; and PAK3 (previously PAKβ and mPAK-3 [24, 355]), which is found in brain tissue (for reviews, see references 264 and 516 and references therein). These three isoforms all contain several N-terminal proline-rich domains followed by a CRIB domain that interacts with GTP-bound Cdc42p through its effector domain and a C-terminal Ser/Thr protein kinase domain, whose activity is stimulated by binding of Cdc42p and Rac to the CRIB domain. Although it was believed that the CRIB domain played a role in autoinhibition of PAK activity, recent data suggest that a highly conserved domain C-terminal to the CRIB domain functions in this capacity (151). The PAK N-terminal proline-rich domains can interact with SH3 domains within the Nck adapter protein (47, 161, 263, 334, 468, 517, 542), thereby forming a linkage between growth factor receptors and activation of the PAKs. Interestingly, another Cdc42p effector, the Wiskott-Aldrich syndrome protein (WASP), also interacts with Nck through its SH3 domains (486) (see “Wiskott-Aldrich syndrome proteins mediate actin rearrangements” below).
The subcellular localization of PAKs reinforces the effects of constitutively activated and dominant negative PAK isoforms on actin rearrangements, suggesting that PAKs mediate Cdc42-dependent actin cytoskeletal effects (123, 356, 627) (see “Mammals” under “Functional Studies” above). PAK1 localized to focal complexes when cells were transfected with activated Cdc42G12V (356, 360, 420), suggesting that interactions between GTP-bound Cdc42 and PAK1 were important for this subcellular localization (see below). In unstimulated Swiss 3T3 cells, PAK1 localized to intracellular vesicles, but upon stimulation by PDGF, PAK1 relocalized to regions of actin rearrangements, including membrane ruffles and lamellipodia; a similar localization effect was seen in v-src-transformed 10 T1/2 cells (123). It appeared that PAK1 localization to membrane ruffles preceded the major actin localization to these subcellular regions, suggesting that activated PAK1 functions early in the induction of actin rearrangements, which is corroborated by the observation that constitutively activated PAK1H83L, H86L induced membrane ruffles (123). In PC12 cells, expression of constitutively activated PAK1H83L, H86L containing a membrane-targeting isoprenylation signal led to significant NGF-dependent neurite outgrowth, and endogenous PAK1 fractionated to membrane pools in response to NGF treatment (115), suggesting that membrane localization of PAK1 is important for this actin-dependent process. The question whether the interactions between Cdc42p and PAKs are necessary for proper subcellular localization of the PAK family of kinases is still unanswered. Certain mutations within the CRIB domain of PAK1 can inhibit its binding to Cdc42p without affecting its localization to focal complexes (627), suggesting that Cdc42p-PAK1 interactions are not necessary for PAK1 localization. However, Cdc42p-Ste20p binding is necessary for Ste20p subcellular localization in S. cerevisiae (see “Ste20p” above). These results suggest that different PAKs may have Cdc42p-independent or Cdc42p-dependent mechanisms of localization.
ACK tyrosine kinases.
Two tyrosine protein kinases, enriched in mammalian brain and skeletal muscle tissue, have been identified as specific effectors of Cdc42p. These tyrosine kinases, designated ACK-1 (358) and ACK-2 (617), specifically interact with GTP-bound Cdc42p in vitro and in vivo and contain a CRIB domain along with a tyrosine kinase catalytic domain, an SH3 domain, and a proline-rich domain. The function of ACK-1 is unknown, but incubation of ACK-2-transfected, detached (not adherent) COS7 cells with EGF or bradykinin resulted in an increase in ACK-2 phosphorylation, suggesting that ACKs may link serpentine/G-protein-coupled receptors to Cdc42p signaling pathways.
Bni1p and Bnr1p Formins May Function as Scaffold Proteins
Bni1p (139, 232, 236, 269, 619) is a ∼220-kDa protein that can interact with Rho-type GTPases in S. cerevisiae. It contains four functional domains including a N-terminal Cdc42/Rho interaction domain contained within amino acids 90 to 343; a proline-rich formin homology 1 (FH1) domain (amino acids 1230 to 1330) found in a number of formin family members including the S. pombe genes fus1 (457) and cdc12 (74), Drosophila genes diaphanous (68) and cappuccino (134), the Aspergillus nidulans gene figA/speA (203, 365), and vertebrate formins (342, 566, 585, 600); a formin homology 2 (FH2) domain (amino acids 1516 to 1616); and a C-terminal Bud6p/Aip3p binding domain (within amino acids 1647 to 1953). It should be noted that the only other formin-like protein that has been shown to interact with Rho-type GTPases is murine p140mDia (585), a homolog of Drosophila diaphanous that specifically interacted with GTP-bound RhoA (in vitro interactions with Cdc42p have recently been reported [10]), localized to spreading lamellae and cleavage furrows in Swiss 3T3 cells, and colocalized with RhoA and profilin (see below) in membrane ruffles in HT1080 human fibrosarcoma cells. However, several other formin-like proteins have been implicated in cell polarity processes in their respective organisms.
Bni1p interacted with Cdc42p in two-hybrid protein assays, and this interaction was specific for GTP-bound Cdc42G12Vp and not GDP-bound Cdc42D118Ap (139). This interaction was substantiated by the in vitro binding of an HA-tagged fragment of Bni1p (amino acids 1 to 1214) purified from S. cerevisiae on Sepharose beads to GTPγS-bound, but not GDP-bound or nucleotide-free, Cdc42p. Bni1p was also identified by a two-hybrid protein interaction with the activated Rho1Q68L mutant protein (269). This interaction was between Rho1Q68Lp and amino acids 90 to 489 of Bni1p, and deletion analysis indicated that amino acids 90 to 343 were capable of interacting with Rho1Q68Lp. The two-hybrid interaction was abolished by the Rho1T42A effector domain mutation, suggesting that Bni1p was a downstream effector of Rho1, but a maltose binding protein fusion to amino acid 1 to 524 of Bni1p could bind nonspecifically in vitro to both GDP-bound and GTPγS-bound Rho1p. Bni1p may also interact with Rho3p and Rho4p (unpublished results cited in reference 139), suggesting that Bni1p may be a general effector of Rho-like GTPases in S. cerevisiae.
In two-hybrid protein assays, GST affinity chromatography experiments, and a maltose binding protein tag overlay assay, Bni1p also interacted with the S. cerevisiae Pfy1p profilin, and this interaction occurred through the Bni1p FH1 domain (139, 232). This binding was also substantiated by a loss of interaction with a profilin mutant protein, Pfy1p-3, that had defects in polyproline binding but not actin binding (139). Several C-terminal fragments of the actin-binding protein Bud6p/Aip3p (15), as well as the Act1p actin protein, displayed two-hybrid protein interactions with Bni1p (139). Bni1p-Bud6p two-hybrid protein interactions occurred through the C-terminal ∼300 amino acids of Bni1p, and interactions with actin occurred through the FH1 domain; these interactions may be mediated through Bni1p interactions with profilin (see above). Bni1p can also interact with elongation factor 1α (EF1α), a protein that has actin binding activity, through a domain between the FH1 and FH2 domains, and the binding of Bni1p to EF1α led to a loss of EF1α-actin binding (570). Bni1p also interacts with the SH3 domain of the Myo3p myosin as assayed by two-hybrid protein assays and GST affinity chromatography (48a). Interestingly, the SH3 domains of the Myo3p and Myo5p myosins also bind to the proline-rich protein verprolin (Vrp1p) (16), which has previously been implicated in cell polarity (571). Bni1p displays genetic and physical interactions with Spa2p, a protein of unknown function that localizes to regions of polarized growth, and localization of Bni1p to the tips of enlarging buds requires the presence of Spa2p and the N-terminal Cdc42/Rho interaction domain (157), suggesting that Bni1p is tethered to the plasma membrane through binding to either a Rho-type GTPase or Spa2p or both. Recent results indicate that Spa2p can interact with Bud6p as well as components of several MAP kinase cascades and the Pea2 cell polarity protein (523), suggesting that actin, profilin, verprolin, Myo3 and Myo5 myosins, Bni1p, Spa2p, Bud6p, EF1α, Pea2p, and possibly other actin binding proteins may form a multiprotein complex with Cdc42p during the bud emergence process (Fig. 3).
BNI1 was identified in a screen for mutants that exhibited a randomization of bud site selection in cells exhibiting a bipolar budding pattern (619). A bni1 disruption mutant, in which amino acids 1228 to 1414 containing the FH1 domain were replaced, grew poorly at high temperatures (269); whether this was a true null mutant is unclear, given that the Cdc42/Rho interaction domain would be predicted to still be expressed in this truncated Bni1p. In addition, a transposon insertion mutation in BNI1 led to a defect in filamentous growth (404), suggesting that Bni1p may either mediate actin rearrangements during pseudohyphal growth or be required for the bipolar budding pattern necessary for pseudohyphal formation. Genetic evidence for a role of Bni1p in Rho1p function came from synthetic lethal phenotypes observed between this bni1 disruption mutant and a mutant expressing mammalian RhoA in place of S. cerevisiae Rho1p and between a pkc1 mutant defective in protein kinase C (269), a known downstream effector of Rho1p (251, 423). BNI1 was also shown to be allelic to SHE5, mutants of which are defective in transcriptional expression from the HO endonuclease promoter in mother cells (236); another mutant identified in this screen, she1, was found to be in the MYO4 gene, a class V type minimyosin (188). An HA-epitope-tagged Bni1p localized to the tips of mating projections in pheromone-arrested cells (139), which correlated with the isolation of bni1 mutants in a screen for mutants defective in mating (139). This defect was due to an inability to form pheromone-induced mating projections in bni1 mutants, which was a consequence of a depolarized cortical actin cytoskeleton. This phenotype could not be suppressed by a Bni1 mutant protein lacking its FH1 domain (unpublished results cited in reference 139), reinforcing the role of the FH1 domain in actin interactions.
Overexpression of full-length Bni1p had no phenotypic effect, but overexpression of the Bni1ΔN N-terminal truncation protein, which was missing amino acids 1 to 451 containing the Cdc42/Rho interaction domain, resulted in cell death and a dominant negative phenotype of large, round, unbudded cells with a delocalized cortical actin cytoskeleton and an increased number of cortical actin patches and actin cables (139). This result suggested either that the essential polarization of cortical actin to the site of bud emergence was dependent on Cdc42p binding to Bni1p or that overexpression of the C-terminal portion of Bni1p could lead to the nonproductive sequestration of actin or actin-binding proteins. The striking appearance of cortical actin structures around the periphery of these cells suggested that plasma membrane localization of cortical actin, albeit nonpolarized, may be possible in the absence of Cdc42p binding to Bni1p; it would be very interesting to determine if the Bni1ΔN truncation protein localizes to the plasma membrane and is capable of cross-linking actin in the absence of Cdc42p. The Bni1ΔNp dominant negative phenotype could be suppressed by overexpression of Pfy1p profilin and the two tropomyosins Tpm1p and Tpm2p, suggesting that loss of Cdc42p-Bni1p binding resulted in a Bni1p that can sequester Pfy1p profilin or other actin binding proteins in a nonfunctional manner. Interestingly, overexpression of Pfy1p profilin inhibited the growth of a bni1 disruption mutant (232). Although the cellular morphologies and actin localization patterns associated with this profilin-based inhibition of growth were not reported, this result suggests that the interactions between these two proteins is important for their functions.
A sequence homology search of the S. cerevisiae genome database revealed the presence of a protein, designated Bnr1p, with significant amino acid sequence homology to Bni1p (232). The smaller (1,374-amino-acid) Bnr1p exhibited 19% identity in the Cdc42/Rho interaction domain, 44% identity in the FH1 domain, and 35% identity in the FH2 domain of Bni1p. Bnr1p also interacted with the Pfy1p profilin protein in two-hybrid protein assays, and this interaction also occurred through the Bnr1p FH1 domain. While a bnr1 disruption mutant was viable at all temperatures tested, a bni1 bnr1 double mutant displayed a ts growth defect at 33°C and an arrested phenotype of large, round, unbudded, multinucleate cells with delocalized actin and chitin, highly reminiscent of cdc42 loss-of-function alleles. Interestingly, bud site selection is randomized in haploid bnr1 mutants but not diploids, which is the opposite of the diploid bud site selection defects in a bni1 mutant (619). Also, a bni1 bnr1 double mutant was sensitive to growth on 1 M sorbitol (232), as were pfy1 (187) and certain act1 (424) mutants, suggesting that these proteins function in osmoregulation. In two-hybrid protein assays, Bnr1p interacted only with wild-type and activated (Q70L mutant) Rho4p but not with Rho1p, Rho2p, Rho3p, or Cdc42p, and maltose binding protein-tagged Bnr1p containing the Rho interaction domain of amino acids 63 to 421 bound in vitro to GTPγS-Rho4p but not GDP-Rho4p (232). In addition, Bnr1p and Bni1p interacted with the SH3 domain-containing protein Hof1p, which displayed sequence similarity to the S. pombe Cdc15 protein involved in cytokinesis, and HA-tagged Bnr1p and Hof1p localized to the mother-bud neck region (252). Taken together, these data suggest that Bni1p and possibly Bnr1p can act as scaffold proteins juxtaposing Cdc42p and other Rho-like GTPases with actin and actin binding proteins during bud emergence and possibly cytokinesis, thus serving as a critical link between the Cdc42p-dependent signal transduction machinery and its ultimate target, the cortical actin cytoskeleton (Fig. 3).
IQGAPs as Scaffold Proteins Mediating Cdc42p-Actin Interactions
S. cerevisiae Iqg1p/Cyk1p functions during cytokinesis.
Mammalian IQGAP proteins are potential scaffold proteins that interact with Cdc42p, actin, and calmodulin (see below). In S. cerevisiae, a potential IQGAP termed Iqg1p (135, 446) or Cyk1p (328) was identified in three independent studies. Iqg1p/Cyk1p is a ∼165-kDa protein that contains several, but not all, of the structural motifs found in mammalian IQGAPs, including an N-terminal calponin homology (CH) domain predicted to interact with actin, four or eight IQ domains predicted to interact with calmodulin, a coiled-coil domain that may function in the dimerization of IQGAPs, and a C-terminal GAP homology domain (GRD) predicted to interact with Cdc42p. In two-hybrid protein assays, Iqg1p preferentially interacted with activated (GTP-bound) Cdc42G12Vp, suggesting that it is a downstream effector (446). Iqg1p/Cyk1p coimmunoprecipitated with actin from S. cerevisiae cell lysates (446) and cosedimented with polymerized rabbit skeletal muscle actin; this cosedimentation was dependent on the Iqg1p/Cyk1p CH domain (135). While Iqg1p/Cyk1p has not been shown to interact with calmodulin as do mammalian IQGAPs (see below), calmodulin is delocalized in a Δiqg1 strain (446).
In certain strain backgrounds, deletion of Iqg1p/Cyk1p led to cell death (135, 328), indicating that it is an essential protein, while in other backgrounds, it led to a ts lethal phenotype (446). In promoter turnoff experiments (135, 328), it was apparent that cells depleted of Iqg1p/Cyk1p had defects in various aspects of cytokinesis, including abnormally elongated buds, multiply budded cells with cytoplasmic continuity, and multinucleate cells. Analysis of the Δiqg1ts strain revealed a more heterogeneous population of cellular morphologies at the restrictive temperature, including large, round cells indicative of a G1 block (446). Functional epitope-tagged Iqg1p/Cyk1p was observed at the presumptive site of bud emergence in unbudded cells (446) and in a ring structure at the mother-bud neck region (135, 328, 446), which colocalized with the septin, actin, and Myo1p rings present at the mother-bud neck region (Fig. 3B). It also occasionally appeared as a double-ring structure or as a dot in the middle of the mother-bud neck region (328); this dot colocalized to a Myo1p dot structure that appeared to be the result of the constriction of the actomyosin ring during cytokinesis. This localization was not dependent on the localization of the actin ring, as evidenced by the persistence of the Iqg1p/Cyk1p ring after treatment with the actin-depolymerizing drug latrunculin-A (135), and the organization of cortical actin at sites of polarized growth and at the septum region did not seem to be disrupted in Δiqg1 cells, suggesting that the localizations of Iqg1p/Cyk1p and actin are independent of each other. Cell cycle synchronization experiments suggested that these Iqg1p/Cyk1p ring structures assembled predominantly after the elongation of the mitotic spindle in anaphase and disassembled after anaphase completion (328). Given the localization of both Iqg1p/Cyk1p and Cdc42p, along with the septin, actin, and Myo1 rings, to the mother-bud neck region, it is likely that Iqg1p/Cyk1p functions to mediate Cdc42p interactions with the actomyosin ring during cytokinesis, possibly as a scaffold protein serving to nucleate various essential components of the actomyosin ring (Fig. 3). Interestingly, a Dictyostelium IQGAP has recently been identified, and mutations of this protein cause cytokinesis defects (1), suggesting that IQGAPs have a common function in cytokinesis (see below).
Mammalian IQGAPs mediate Cdc42p-calmodulin-actin interactions.
There are two identified mammalian IQGAPs, designated IQGAP1 and IQGAP2. Human IQGAP1 was originally identified serendipitously in a PCR-based search for matrix metalloproteinase family members (588). IQGAP1 was also identified from COS cell lysates by its ability to interact with GTPγS-GST-Cdc42 on agarose beads (204). This ∼195-kDa protein contains a GRD (residues 997 to 1270) with significant similarity to the catalytic domain of Ras-GAP proteins, as well as a CH domain (residues 48 to 161), which is believed to interact with actin, and four tandemly repeated IQ domain (residues 745 to 865), which are present in a number of proteins that interact with calmodulin. IQGAP1 also contains a single WW domain implicated in protein-protein interactions and six copies of a unique 50- to 60-amino-acid domain with no known matches in the database. The human IQGAP1 gene mapped to chromosome 15p-15q1.1, and RNA blot analysis indicated that IQGAP1 was highly expressed in the placenta, lungs, kidneys, and skeletal muscle but was absent from the brain. A GST-IQGAP1-GRD fusion protein or an in vitro-translated C-terminal IQGAP1 domain polypeptide did not exhibit GAP activity against Ras, Rho, or Cdc42Hs (204, 588), and affinity-purified IQGAP1 did not display GAP activity against Ras, RalA, Cdc42Hs, Rac1, or RhoA (reference 283 and data not shown), suggesting that IQGAPs do not have catalytic GAP activity. Interestingly, expression of the IQGAP1 C-terminal domain peptide in S. cerevisiae resulted in a dominant negative loss-of-polarity phenotype that could be suppressed by overexpression of wild-type S. cerevisiae Cdc42p (204), suggesting that this C-terminal domain could interact with Cdc42p in vivo (see below).
IQGAP1, as well as IQGAP2 (see below), was also identified in rabbit liver fractions (379) and bovine brain cytosol (283) by its ability to preferentially bind to Cdc42Hs-GTPγS on columns. Both proteins could also interact with Rac1 but not RhoA or Ha-Ras. The binding of IQGAP1 p180 and IQGAP2 p175 to Cdc42Hs could not be competed with a functional Cdc42-GAP polypeptide, but it could be competed with the mPAK-3 CRIB domain, suggesting that the IQGAPs bind to Cdc42p through its effector domain. Neither IQGAP1 or IQGAP2 contain a CRIB domain, and their interactions with Cdc42Hs are mediated through the C-terminal GRD (204, 283, 379). IQGAP1 binding to Cdc42Hs seemed to stabilize the GTP-bound state of the protein and inhibited its intrinsic GTPase activity (379), suggesting that it acts as a GTPase inhibitor similar to rho-GDI. Interestingly, when an in vitro MESG/phosphorylase-coupled assay was used to measure γPi release from Cdc42-GTP, it was shown that the IQGAP GRD domain had a ∼10-fold higher affinity toward Cdc42-GTP than did the CRIB domains of PAK1 and WASP (624). IQGAP1 was coimmunoprecipitated with HA-tagged Cdc42G12V protein from COS and Rat1 cells (204) and was also the major protein coimmunoprecipitated with HA-tagged Cdc42Q61L protein from COS7 cells (136), and this association was slightly enhanced after treatment with EGF, indicating that IQGAP1 is a likely physiological Cdc42p effector. Recently, both IQGAP1 and IQGAP2 (see below) were shown to coimmunoprecipitate with Cdc42p from rabbit liver and Chinese hamster ovary (CHO) Golgi membrane-enriched fractions (378), suggesting that IQGAPs may play a role in Cdc42p-dependent membrane trafficking events (see “Prenylation and subcellular localization” above).
IQGAP1 was also identified by nanoelectrospray tandem mass spectrometry from normal (Hs578Bst) and malignant (MCF-7) human breast cell lines by its ability to bind to a calmodulin-Sepharose column in the presence and absence of Ca2+ (243). Although IQGAP1 could bind calmodulin in both the absence and presence of Ca2+ (204, 243), binding was enhanced ∼twofold in the presence of Ca2+ (243). Immunoprecipitation with anti-IQGAP1 antibodies brought down calmodulin from NIH 3T3 cells (204), and immunoprecipitation with anti-calmodulin antibodies brought down IQGAP1 from MCF-7 cells (243). Binding of IQGAP1 to calmodulin occurred through the IQ motifs in the N terminus of IQGAP1 (204). In GST affinity chromatography experiments, it was shown that calmodulin could inhibit the binding of IQGAP1 to Cdc42Hs in a Ca2+- and dose-dependent manner, but this inhibition was not observed when IQGAP1 was already bound to Cdc42Hs (243).
IQGAP1 was also identified from bovine adrenal cytosol by its ability to cosediment with rabbit muscle actin microfilaments (30), and actin from bovine brain cytosol could bind to GST-IQGAP1 columns (158). IQGAP1 copurified with substoichiometric amounts of calmodulin and was found as a dimer in sedimentation equilibrium experiments (30) and as oligomers in gel filtration experiments, with addition of GTPγS-GST-Cdc42Hs enhancing the oligomerization (158). The presence of calmodulin, with or without Ca2+, interfered with the cosedimentation of IQGAP1 with actin microfilaments. IQGAP1 could cross-link F-actin, as evidenced by increases in the viscosity of microfilament networks in the presence of IQGAP1 (30, 158) and by the appearance of actin bundles in negative-stain electron microscopy (30), and this cross-linking was enhanced in the presence of GTPγS-GST-Cdc42Hs (158). In addition, both IQGAP1 and actin were found in an immunoprecipitation complex with HA-tagged Cdc42Hs (136). IQGAP1 colocalized to rhodamine-phalloidin-stained cortical actin in insulin-induced membrane ruffles in KB cells, and this localization was inhibited by prior treatment of the cells with dominant negative Rac1T17N or Cdc42T17N (283). IQGAP1 also localized to lamellipodia and membrane ruffles, but not stress fibers, in monkey kidney, rat kidney, and NIH 3T3 cells (30) and PAE endothelial cells (204) and co-localized with transfected Cdc42Q61Lp in COS7 cells (136). Recently, IQGAP1 was localized to cell-cell junctions in MDCK cells (283) and was shown to colocalize and interact with E-cadherin and β-catenin at sites of cell-cell contact in mouse L fibroblasts (284), suggesting that IQGAP1 may mediate Cdc42p effects on cell-cell adhesion (see “Mammals” under “Functional studies” above). Given the recent localization of IQGAP1 to a perinuclear fraction coincident with Golgi markers (378) (see above), it appears that there are different subcellular pools of IQGAPs that can interface with Cdc42p and actin to mediate multiple cellular events.
IQGAP2 was identified from a mouse brain library by screening with an IQGAP1 probe at low stringency (51). The human IQGAP2 gene mapped to chromosome 5q1.1-1.3, and RNA blot analysis indicated that IQGAP2 was expressed predominantly in the liver and in several hepatoblastoma cell lines. Human IQGAP2 was isolated from a human cDNA library and found to encode a ∼180-kDa protein with 62% identity to IQGAP1 over its entire length and containing all of the structural motifs found in IQGAP1 (see above). As with IQGAP1, IQGAP2 binds calmodulin, as determined by coimmunoprecipitation with HA-tagged IQGAP2, and the IQ domains were necessary for this binding. It preferentially interacted with GTP-bound Cdc42Hs and to a lesser extent with Rac1 in GST affinity chromatography experiments and immunoprecipitations from human liver cell lysates and COS cells, suggesting that it is also a bona fide Cdc42p effector. IQGAP2 did not have GAP activity against Cdc42Hs or other small GTPases, but as with IQGAP1 (see above), it inhibited the intrinsic and Cdc42-GAP-stimulated GTPase activity of Cdc42Hs and Rac1.
IQGAPs do not seem to have catalytic GAP activity against any GTPase, but they do have the characteristics of scaffold proteins capable of interacting with multiple proteins. Taken together, the above data suggest a possible model (Fig. 6) for IQGAP function in which IQGAPs are inactive when bound to Ca2+-calmodulin and a reduction in Ca2+ levels (or other signals) leads to a loss of calmodulin-IQGAP interactions and increased interactions between GTP-bound Cdc42p and IQGAPs. This increased interaction leads to the oligomerization of IQGAP and the formation of a Cdc42p-IQGAP-actin complex, enhancing the ability of IQGAP to cross-link and possibly polymerize actin. Therefore, it seems that IQGAPs in mammalian cells are major downstream effectors linking Cdc42p to the actin cytoskeleton.
FIG. 6.
Molecular model for the generation of an IQGAP-Cdc42p-actin ternary complex. See the text for details.
Wiskott-Aldrich Syndrome Proteins Mediate Actin Rearrangements
S. cerevisiae Bee1p/Las17p.
Another family of potential scaffold proteins linking Cdc42p and the actin cytoskeleton are the WASPs, with the prototypical WASP being encoded by the gene that is defective in Wiskott-Aldrich syndrome patients (see below). Bee1p (321) was identified from the S. cerevisiae genome database by its similarity to the mammalian WASP. Bee1p contains a WASP homology domain (WH1) that is similar to PH domains, as well as a proline-rich domain that binds SH3 domains; it does not seem to contain a CRIB domain for binding Cdc42p as does the mammalian WASP. Deletion of Bee1p resulted in a slow-growth phenotype at room temperature and no growth at high temperatures (321). Morphological characterization of Δbee1 cells indicated defects in bud growth, cytokinesis, and actin organization. In addition, Bee1p localized to cortical actin patches and bound to actin and the actin binding protein Sla1p in immunoprecipitation experiments (321) and to the actin binding protein verprolin in a two-hybrid experiment (413). The Δbee1 cells were also defective in the ability to assemble cortical actin to the buds in an in vitro permeabilized cell system. While Bee1p clearly plays a role in actin cytoskeleton organization, it is unclear if Bee1p interacts with Cdc42p as does its human counterpart (Fig. 3 and 4A).
Mammalian WASPs.
Patients with Wiskott-Aldrich syndrome have multiple immunological defects, including thrombocytopenia with small platelets, eczema, T- and B-lymphocyte defects, and an increased risk of malignancies and autoimmune diseases (for reviews, see references 144, 261, and 427). The severity of these defects has been directly correlated with mutations within the X-linked recessive gene WASP and with defects in cellular actin cytoarchitecture (122, 257, 270, 285, 399, 428, 475, 577, 636, 637). The ∼62-kDa WASP (19, 271, 545) contains several functional domains including an N-terminal WH1 domain (residues 8 to 105), a CRIB domain (residues 238 to 257), a proline-rich domain (residues 312 to 404), two potential actin binding sites with similarity to verprolin and cofilin sequences (residues 430 to 446 and 469 to 489), and an acidic C-terminal region. A human WASP-GST fusion protein containing residues 48 to 321 bound to GTP-Cdc42Hs, but not GTP-bound RhoA or Rac1, in GST affinity chromatography experiments, and this binding depended on the presence of the WASP CRIB domain (545) and the Cdc42 effector domain (291), suggesting that WASP is a Cdc42p-specific effector. Ectopic expression of FLAG-tagged WASP in rat kidney epithelial cells indicated that WASP was present predominantly in cytosolic clusters and colocalized with actin structures excluding stress fibers. Formation of these clusters was inhibited by addition of cytochalasin D and by coexpression of the dominant negative Cdc42T17N mutant protein but not the dominant negative Rho or Rac mutant proteins (545), reinforcing the specific interactions seen between WASPs and Cdc42p. It was reported that the Cdc42 Y40C effector domain mutation abolished interactions with WASP and other CRIB domain-containing proteins but did not affect the generation of actin-dependent morphological structures, suggesting that other downstream effectors mediated Cdc42p-actin interactions (291). However, examination of the data indicates that the Y40C mutation, while interfering with p65PAK kinase activity in immunoprecipitates, reduced but did not abolish the binding of GST-WASP (containing amino acids 201 to 321 of WASP) to [γ-32P]GTP-loaded Cdc42Q61L, Y40C mutant protein in a nitrocellulose overlay assay (291, 388), leaving open the question of the physiological role of WASP-Cdc42 interactions.
WASP was phosphorylated in vivo on Ser and Thr residues (521) and has been shown to interact with the SH3 domains of the Nck (486) and Grb2 (468, 521) adapter proteins and the WIP profilin-binding protein (470). The Grb2p adapter protein mediated interactions between the EGF receptor and WASP in EGF-stimulated A431 cells and in GST affinity chromatography experiments in vitro (521). WASP has also been shown to interact with the SH3 domains of various protein tyrosine kinases (25, 58, 108), and recent data indicates that WASP undergoes tyrosine phosphorylation by the Lyn and Btk tyrosine kinases in RBL-2H3 rat tumor mast cells, which is enhanced in the presence of Cdc42G12Vp (184), and that collagen induces tyrosine phosphorylation of human platelet-associated WASP, leading to its redistribution to the cytoskeleton (429).
The N-WASP homolog was isolated from bovine brain cytosol by its ability to bind to the Grb2 SH3 domain in vitro (387). N-WASP was ∼50% similar to WASP over the entire coding region and also contained an N-terminal WH-1/PH domain, a CRIB domain, a proline-rich domain, and C-terminal actin binding domains and acidic regions. In addition, N-WASP contained a potential IQ domain, suggesting that N-WASP may interact with calmodulin-Ca2+. Analysis of N-WASP mRNA and protein expression indicated that it was expressed predominantly in brain tissue and moderately in heart, lung, and colon tissue. In GST affinity chromatography experiments with N-WASP fusion proteins, it was shown that GST-N-WASP could bind to Grb2 through its proline-rich domain, to phosphatidylinositol-4,5-bisphosphate (PIP2) through its PH domain, to calmodulin in a Ca2+-dependent manner through its IQ domain, to actin through its C-terminal actin binding domain (387), and to GTP-Cdc42 through its CRIB domain (388). Interestingly, the H208D N-WASP CRIB domain mutation abolished interactions with Cdc42p, but the Cdc42-Y40C mutation, which interfered with WASP-Cdc42p interactions (see above), did not affect N-WASP–Cdc42p binding, suggesting that N-WASP may be the physiological effector linking Cdc42p to the actin cytoskeleton. Transient coexpression of N-WASP, but not WASP, with activated Cdc42G12V in COS7 cells led to an increase in elongated actin microspikes not seen in cells transfected with the two proteins separately, and expression of a mutant N-WASP lacking a stretch of four amino acids in the cofilin-like actin binding domain inhibited Cdc42-dependent microspike formation (388). Recently, N-WASP was shown to mediate the assembly of the actin tail that is associated with the intracellular movement of invading Shigella flexneri by interacting with its VirG outer membrane protein (543). Overexpression of N-WASP in COS7 cells led to a loss of thick actin fibers and a relocalization to cortical areas, where N-WASP was localized. A GST–N-WASP fusion protein containing the actin binding domain could depolymerize actin in vitro, as does cofilin (387), and addition of full length N-WASP to actin filaments in vitro had a slight effect on reducing actin viscosity in falling-ball methods, which was significantly enhanced by the addition of GTP-Cdc42p but not GDP-Cdc42p (388). Taken together, these data are consistent with the binding of GTP-Cdc42p to N-WASP leading to an activation of N-WASP actin depolymerizing activity, thereby generating uncapped filament ends that could serve as templates for subsequent actin polymerization events.
Other Effectors
Gic1p and Gic2p.
The S. cerevisiae Gic1p and Gic2p downstream effectors were identified in two independent studies (54, 83). Gic1p was identified by its ability to suppress the bem2-101ts growth and bud site selection phenotypes (83), and Gic1p and Gic2p were identified through a search of the S. cerevisiae genome database for proteins that contained CRIB domains (54). Gic1p and Gic2p were 39% identical and 54% similar in predicted amino acid sequence and were not homologous to other proteins in the database. Deletion of GIC1 or GIC2 did not lead to abnormal phenotypes, but a Δgic1 Δgic2 double mutant did not grow at high temperatures and displayed numerous morphological abnormalities at semipermissive temperatures, including the presence of a large percentage of large, unbudded, multinucleate cells, delocalized chitin deposition, aberrant actin organization, and abnormal mitotic spindles, suggesting a role for Gic1p/Gic2p in cellular morphogenesis, as well as defects in mating-projection formation and reduced mating efficiencies, suggesting a role in the mating pathway.
Cdc42-GTPγS bound to columns containing purified Gic2p, and both Gic1p and Gic2p interacted with Cdc42p in two-hybrid protein assays (54, 83). These interactions were enhanced with the activated Cdc42G12Vp and were between the Gic1/2 CRIB domains and the Cdc42 effector domain, as evidenced by the interactions being abolished by the Cdc42T35Ap effector domain mutation and CRIB domain mutations and/or deletions. Further genetic experiments that solidified the physiological interactions between Gic1p, Gic2p, and Cdc42p included the observations that overexpression of the Gic2p CRIB domain led to a dominant growth arrest that could be suppressed by overexpression of Cdc42p but not Cdc42T35Ap (54); that the Δgic2 mutation exacerbated the cdc42-1 and cdc24-2 ts phenotypes (83); that overexpression of Cdc42p suppressed the Δgic1 Δgic2 ts phenotype (54, 83); that overexpression of the Rga1p GAP exacerbated the Δgic1 Δgic2 ts phenotype (83); that overexpression of Cla4p, but not Ste20p or Skm1p, could partially suppress the Δgic1 Δgic2 double mutant (83); and that Δgic1 Δgic2 Δcla4 triple mutants had a more severe cytokinesis defect than Δcla4 mutants did (83).
The levels of Gic2p cycled through the cell cycle with an accumulation during G1 phase and a peak around the time of septin ring formation ∼15 min prior to bud emergence (54). Cell cycle-dependent regulation of Gic2p levels has recently been shown to be through ubiquitin-dependent degradation shortly after bud emergence (237). This degradation required the SCF (for “Skp1–cullin–F-box”)-Grr1 protein complex and depended on the phosphorylation of Gic2p and the binding of Gic2p to GTP-bound Cdc42p, suggesting that Cdc42p is needed not only for Gic2p function but also for its degradation. Subcellular localization of HA-tagged Gic1p and Gic2p (54) indicated that they were distributed to the site of incipient bud emergence and to the tips of enlarging buds and mating projections in a pattern similar to that seen with Cdc42p (643). However, analysis of GFP-Gic1p localization revealed a more complex picture (83). GFP-Gic1p localized to the site of incipient bud emergence in the mother and daughter progenitor cells in early G1 and to the tips of enlarging buds. It seemed to disappear from the bud tip in medium- to large-budded cells but appeared at sites adjacent to the mother-bud neck regions in haploid cells, from which the bud would emerge in the next cell cycle. In some large-budded cells, GFP-Gic1p appeared in a ring structure at the mother-bud neck region but did not persist there after cytokinesis. Interestingly, there are differing data concerning the role of the CRIB domains in Gic1p and Gic2p function and localization. Gic2p lacking its CRIB domain was unable to complement the Δgic1 Δgic2 double mutant, and HA-Gic2p lacking its CRIB domain was distributed diffusely throughout the cytoplasm (54), suggesting that Gic2p interactions with Cdc42p were necessary for proper function and localization. The localization of GFP-Gic1p, however, was not dramatically altered upon deletion of its CRIB domain, and expression of CRIB-deleted Gic1p could still partially suppress the Δgic1 Δgic2 double mutant (83), suggesting that interactions with Cdc42p are important but not essential for Gic1p function and localization. These analyses of Gic1p and Gic2p function suggest that they play an important role in bud site selection and actin organization through their interactions with Cdc42p.
Zds1p and Zds2p.
S. cerevisiae Zds1p and Zds2p were identified in multiple genetic screens (see below) including a screen for negative regulators of Cdc42p function (42). The 915-amino-acid Zds1p and the 942-amino-acid Zds2p exhibited ∼42% similarity and contained potential coiled-coil domains but did not contain recognizable CRIB domains. Overexpression of Zds1p reduced the restrictive temperatures of the cdc24-10 and cdc42-1 mutants, led to a large proportion of enlarged unbudded cells when expressed in the cdc24-12 mutant, and altered chitin and actin localization in haploid cells (42), reinforcing its potential role as a negative regulator of the cell polarity pathway. Deletion of Zds1p or Zds2p singly had no phenotypic effect (however, see below), but the Δzds1 Δzds2 double mutant showed a reduced growth rate and a high percentage of elongated budded cells with a single nucleus and 2C DNA content, indicating a defect in the apical-isotropic switch and/or a G2/M cell cycle delay, suggesting that Zds1p and Zds2p together are needed for cell cycle progression (42, 618). Subcellular localization of a GST-Zds1p fusion protein by using anti-GST antibodies suggested that Zds1p localized to the sites of incipient bud emergence as well as to tips of enlarging buds and occasionally to the mother-bud neck region, suggesting a colocalization with Cdc42p. However, it should be noted that a direct interaction between Zds1p or Zds2p and Cdc42p has not been reported.
In addition to the above-mentioned phenotypes, Zds1p was identified in a number of other genetic screens (for examples, see personal communications cited in references 42 and 618), and including (i) as a multicopy suppressor of a cdc28-1N CDK mutant, which had defects in promoting entry into mitosis (618); (ii) as a suppressor of an ssd1 hht1 double mutant (HHT1 encodes histone H3); (ii) as a multicopy suppressor of the hsl1 and hsl7 mutants that were isolated by synthetic lethality with a histone H3 hht1 N-terminal deletion mutant (341); and (iii) as a multicopy suppressor (HST1) of the Ca2+ and trifluoperazine sensitivities of an ssd1 mutant (567). Interestingly, a Δswe1 deletion suppressed the hsl1 and hsl7 synthetic lethal mutants and the G2/M delay observed in Δhsl1 and Δzds1 mutants (341, 398). Swe1p is a CDK-inhibitory kinase that phosphorylates Cdc28 on Y19 (48, 501) thereby inhibiting Cdc28p kinase activity and resulting in a G2/M delay from a novel morphogenesis checkpoint monitoring bud emergence (313, 526) (see “S. cerevisiae” under “Functional studies” above). Zds1p was shown to negatively control Swe1p by altering its cell cycle transcriptional regulation; a Δzds1 mutant altered the SWE1 mRNA periodicity away from a peak in G1/S and delayed its repression in G2, suggesting that Zds1p was involved in repressing Swe1p expression in G2/M (398). One possible model, among many, would have Zds1p and Zds2p functioning in combination with Cdc42p in regulating the Swe1p protein kinase at a morphogenesis checkpoint monitoring bud emergence and elongation. The binding of Zds1p to Cdc42p at a key time in the cell cycle, possibly after successful bud emergence or the apical-isotropic switch, could down-regulate Swe1p activity, thereby inducing mitosis and cytokinesis. Clearly, more experiments are needed to elucidate this interesting connection between Cdc42p, Zds1p, and Swe1p.
Bem4p/Rom7p.
S. cerevisiae Bem4p was identified in three separate screens as a multicopy suppressor of the cdc42-1 mutant simultaneously overexpressing the SRO4 gene, as a mutant that required multiple copies of CDC42 to grow, and as a synthetic-lethal mutant with the cdc24-4 mutation (343). The allelic Rom7p was identified as a multicopy suppressor of a dominant negative rho1 mutant (221). In two-hybrid protein studies, Bem4p was shown to interact with Cdc42p, Rho1p, Rho2p, and Rho4p, and it interacted equally well with the Cdc42G12V, Cdc42Q61L, and Cdc42D118A constructs, suggesting that it can interact with both GTP-bound and GDP-bound Cdc42p. However, in vivo interactions between these proteins have not been reported. Deletion of BEM4 led to cell inviability at 37°C with a cellular morphology of large, round, unbudded, multinucleate cells containing delocalized actin, and this ts phenotype could be suppressed by simultaneous overexpression of both Cdc42p and Rho1p. It should be noted, however, that these phenotypes varied in different strain backgrounds. Due to its ability to interact with multiple Rho-type GTPases in S. cerevisiae, the physiological role for Bem4p is elusive, but an interesting observation was that deletion of the Rho-GDI, Rdi1p, led to cell inviability in the presence of a Δbem4 mutation (unpublished results cited in reference 343), suggesting that Bem4p and Rdi1p, while showing no sequence similarity to each other, may have overlapping functions in vivo.
70-kDa S6 kinase.
The mammalian 70-kDa S6 kinase (pp70S6k) is involved in growth control, translation initiation, and progression through the G1/S phase of the cell cycle through its phosphorylation of ribosomal protein S6 and the subsequent translation of 5′ terminal oligopyrimidine tract-containing mRNA (for reviews, see references 92, 238, and 557). The pp70S6k kinase activity is activated by a number of different regulatory signals including growth factors, phorbol esters, and cytokines, and this activation is inhibited by the immunosuppressant drug rapamycin, which functions through its binding to the FK506 binding protein rapamycin-associated protein FRAP (also known as RAFT and TOR), a PI 3-kinase-like protein kinase (52, 53, 502, 503). The pp70S6k kinase activity is also activated by PI 3-kinase (82, 97, 401), and this activation is blocked by the PI 3-kinase inhibitors wortmannin and LY294002. Immunoprecipitated HA-tagged pp70S6k from NIH 3T3 and COS cells had in vitro kinase activity against ribosomal protein S6 as a substrate, and this activity was enhanced by cotransfection with activated GST-Cdc42G12V and GST-Rac1G12V, but not GST-RhoAG12V, and by the Cdc42-GEF Dbl and was inhibited by dominant negative Cdc42 and Rac mutants (93), suggesting that activation of Cdc42p leads to activation of pp70S6k kinase activity. Cotransfection with activated GST-Cdc42G12V and GST-Rac1G12V constructs also increased the phosphorylation of pp70S6k necessary for its kinase activity, and this activation was lost in the Cdc42T35A effector domain mutant and in the Cdc42C189S prenylation mutant, suggesting that proper subcellular localization is necessary for the interaction between Cdc42p and pp70S6k. The growth factor-induced activation of pp70S6k activity seemed to be independent of the activation of the JNK and p38 kinase activities, but the Cdc42p-induced pp70S6k activation was blocked by the addition of rapamycin and wortmannin, suggesting that FRAP and PI 3-kinase function in the activation pathway.
E. coli-produced GST-GTPγS-Cdc42p fusion protein formed an in vitro complex with pp70S6k from NIH 3T3 cell extracts and an in vivo complex in COS cells as assayed by GST affinity chromatography and coimmunoprecipitation of HA-tagged Cdc42p with endogenous pp70S6k (93). This interaction and subsequent activation of pp70S6k activity was not seen with the dominant negative Cdc42T17N mutant protein and was lost in the presence of the T35A mutation, suggesting that pp70S6k is a physiological Cdc42p downstream effector that interacts with the Cdc42p effector domain. Cdc42p bound to hypophosphorylated pp70S6k species and immunoprecipitated complexes did not have kinase activity, suggesting that Cdc42p binds to the inactive pp70S6k. This in vivo complex formation was resistant to addition of wortmannin and rapamycin, suggesting that their actions occur upstream or independent of Cdc42p-pp70S6k binding. Addition of cytochalasin D did not affect Cdc42p-induced activation of pp70S6k activity (reference 93 and data not shown), suggesting that this Cdc42p pathway was independent of actin-associated events. However, recent studies have identified a neural tissue-specific F-actin binding protein termed neurabin that can interact with both F-actin and pp70S6k (60, 412). The physiological ramifications of Cdc42p-pp70S6k interactions are unknown, but they could play a role in Cdc42p-dependent cell cycle progression or growth control (see “Mammals” under “Functional studies” above).
Examination of the S. cerevisiae genome database does not reveal the presence of a recognizable pp70S6k species, but S. cerevisiae does contain two FRAP homologs, named Tor1p and Tor2p (for a review, see reference 557). Both Tor1p and Tor2p are involved in rapamycin-sensitive translation initiation and G1 cell cycle progression (27), but Tor2p is also involved in organization of the actin cytoskeleton (512), and this function occurs through interactions with the Rom2p GEF for the Rho1p and Rho2p GTPases (510). A connection between the S. cerevisiae Tor proteins and Cdc42p has not been described to date.
CDC42 AND HUMAN DISEASE
As detailed in the above sections, the analysis of Cdc42p function in cultured mammalian cells and the characterization of Cdc42p effectors and regulators suggest that Cdc42p functions in a variety of human diseases through modulation of the actin cytoskeleton and JNK-dependent transcriptional induction events (see above for references). First, the observations that (i) Cdc42p is implicated in Ras-dependent cellular transformation, (ii) injection of Cdc42G12V-expressing cells into athymic nude mice led to the formation of tumors, and (iii) expression of the Cdc42F28L mutant protein led to cellular transformation similar to that seen with expression of the dbl oncogene (a Cdc42p GEF) indicate that activation of Cdc42p can lead to malignant transformation and that cdc42 is a bona fide oncogene. It should be noted, however, that the presence of activated Cdc42 alleles in human tumor cells has not been reported to date. Second, the mammalian WAS proteins, encoded by the genetic locus responsible for the Wiskott-Aldrich syndrome immunological disorder, bind specifically to GTP-Cdc42p, but not to GTP-bound RhoA or Rac1, and mediate Cdc42p-actin interactions. Third, the polycystic kidney disease I (PKD1) protein, which plays a role in autosomal dominant polycystic kidney disease, was shown to induce c-Jun/AP-1 transcriptional activation through the activation of the Cdc42-dependent JNK pathway. Fourth, the myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK) interacts with Cdc42p in the regulation of actin rearrangements. Fifth, the faciogenital dysplasia protein FGD1, which is encoded by the genetic locus responsible for the X-linked developmental disorder Aarskog-Scott syndrome, is believed to be a Cdc42-specific GEF in vivo. Sixth, activation of the Nef-associated kinase (NAK) was mediated through Cdc42p, suggesting that Cdc42p plays a role in Nef-dependent HIV replication. Finally, the Salmonella SopE protein acts as a GEF for Cdc42p, leading to the actin rearrangements necessary for Salmonella invasion of host epithelial cells. It is likely that Cdc42p will also play a critical role in other human diseases that involve actin rearrangements or JNK pathway activation, and so a detailed understanding of Cdc42p structure and function could be invaluable in developing therapeutic strategies.
CONCLUSIONS AND FUTURE RESEARCH DIRECTIONS
The experimental results detailed in this review strongly support a model in which Cdc42p interacts with multiple regulators and effectors to activate a variety of cellular processes. It is interesting that Cdc42p has not been implicated as a negative or inhibitory factor in any cellular process; therefore, its roles seem to be positive or stimulatory in nature. The two primary Cdc42-dependent pathways leading to actin rearrangements and transcriptional inductions through protein kinase signaling cascades seem to be conserved in most cell types examined. However, it is dangerous to extrapolate precise Cdc42p functions or mutational phenotypes from one organism to another, given, for instance, the differences in phenotypes seen between analogous cdc42 mutants in S. cerevisiae and S. pombe (see “Functional studies” above). It has been difficult to address whether the downstream effectors that mediate these two pathways are separate and independent or whether there is substantial cross-talk between the physiological pathways. This question should be vigorously addressed in the future.
It is unfortunate that despite all the detailed information we have garnered about Cdc42 functions and interacting proteins, we have little experimental data addressing the specific mechanism(s)-of-action for Cdc42p in these different cellular processes. It seems likely that one of the major roles that Cdc42p plays is in transducing exogenous and/or endogenous signals to downstream effectors by specifically binding and localizing these effectors to the appropriate subcellular locations so that they can interact with further downstream components, in much the same manner that Ras does with Raf. The formation of these multiprotein complexes at discrete locations within the cell in response to different signals could be a primary regulatory mechanism for the specificity of Cdc42p function within different pathways. This hypothesis should be tested in the future through the phenotypic analysis of different Cdc42p effector domain mutations and their interactions with different downstream effectors and through subcellular colocalization studies with epitope-tagged or GFP fusion proteins. The observations that Cdc42p can function at several points in the cell cycle adds an additional layer of complexity to understanding these differential regulatory interactions, but analysis of different effector domain mutations should provide some insight into this aspect of Cdc42p function as well.
So what are the future research directions for deciphering Cdc42p function? The answer to this question will be determined partly by the organism in which experiments are performed. For instance, genetic and biochemical studies in S. cerevisiae and in cultured mammalian cells, and to a lesser extent in S. pombe, have identified a myriad of Cdc42p regulators and effectors, but only recently have experiments designed to test specific protein-protein interactions and multiprotein complex formation been performed. In addition, little is known about the in vivo specificity of assorted GEFs, GAPs, GDIs, or downstream effectors or about the targeting mechanisms for Cdc42p to the plasma membrane at sites of polarized growth in response to different signals or at different times in the cell cycle. Therefore, future experiments with these organisms will probably focus on these issues. Few Cdc42p effectors and regulators have been identified or characterized in Drosophila and C. elegans, and so these proteins must be isolated before detailed mechanistic questions can be addressed. However, the mechanistic studies in yeast and mammalian cells should develop useful paradigms that will allow for more defined questions to be addressed in Drosophila and C. elegans. Interestingly, no bona fide Cdc42p homologs have been identified in fungal systems outside of the unicellular yeast or in plant systems, although multiple Rac homologs have been identified (118, 319, 563, 598). Given the high degree of cellular polarization seen in fungal and plant cell growth patterns, it would be surprising if Cdc42p homologs did not exist and were not involved in these processes. Finally, given the recent determination of the NMR and X-ray crystal structures of Cdc42p and Cdc42p complexed with one of its GAPs, future molecular modeling studies could provide valuable insight into the effects of various loss-of-function, gain-of-function, and effector domain mutations on Cdc42p structure and function and its interactions with downstream effectors and regulators. The much anticipated NMR and/or X-ray crystal structure determinations of Cdc42p complexed with a downstream effector or GEF or GDI should greatly enhance our knowledge of the mechanisms of action of these proteins. All in all, the explosion of research centered on Cdc42p over the past 5 to 10 years has only served to whet our appetite for more details, which will certainly be forthcoming in the very near future.
ACKNOWLEDGMENTS
I thank the many colleagues who have shared their results prior to publication, especially Nicolas Nassar and Rick Cerione for the Cdc42p structure figure (Fig. 1B). I also thank the members of the Johnson laboratory, both past and present, for their many helpful discussions, as well as David Pederson, Gary Ward, and the anonymous reviewers for an insightful and critical examination of this review.
Research in my laboratory has been supported by grants from the National Science Foundation, the American Cancer Society, and the U.S. Department of Agriculture.
REFERENCES
- 1.Adachi H, Takahashi Y, Hasebe T, Shirouzu M, Yokoyama S, Sutoh K. Dictyostelium IQGAP-related protein specifically involved in the completion of cytokinesis. J Cell Biol. 1997;137:891–898. doi: 10.1083/jcb.137.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams A E M, Johnson D I, Longnecker R M, Sloat B F, Pringle J R. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams A E M, Pringle J R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adra C N, Ko J, Leonard D, Wirth L J, Cerione R A, Lim B. Identification of a novel protein with GDP dissociation inhibitor activity for the Ras-like proteins CDC42Hs and Rac1. Genes Chromosomes Cancer. 1993;8:253–261. doi: 10.1002/gcc.2870080408. [DOI] [PubMed] [Google Scholar]
- 5.Adra C N, Manor D, Ko J L, Zhu S, Horiuchi T, Aelst L V, Cerione R A, Lim B. RhoGDIγ: a GDP-dissociation inhibitor for Rho proteins with preferential expression in brain and pancreas. Proc Natl Acad Sci USA. 1997;94:4279–4284. doi: 10.1073/pnas.94.9.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aepfelbacher M, Vauti F, Weber P C, Glomset J A. Spreading of differentiating human monocytes is associated with a major increase in membrane-bound CDC42. Proc Natl Acad Sci USA. 1994;91:4263–4267. doi: 10.1073/pnas.91.10.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agnel M, Roder L, Vola C, Griffin-Shea R. A Drosophila rotund transcript expressed during spermatogenesis and imaginal disc morphogenesis encodes a protein which is similar to human Rac GTPase-activating (racGAP) proteins. Mol Cell Biol. 1992;12:5111–5122. doi: 10.1128/mcb.12.11.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed S, Lee J, Wen L-P, Zhao Z, Ho J, Best A, Kozma R, Lim L. Breakpoint cluster region gene product-related domain of n-chimaerin. Discrimination between Rac-binding and GTPase-activating residues by mutational analysis. J Biol Chem. 1994;269:17642–17648. [PubMed] [Google Scholar]
- 9.Akada R, Kallal L, Johnson D I, Kurjan J. Genetic relationships between the G protein βγ complex, Ste5p, Ste20p, and Cdc42p: investigation of effector roles in the yeast pheromone response pathway. Genetics. 1996;143:103–117. doi: 10.1093/genetics/143.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alberts A S, Bouquin N, Johnston L H, Treisman R. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein β subunits and the yeast response regulator protein Skn7. J Biol Chem. 1998;273:8616–8622. doi: 10.1074/jbc.273.15.8616. [DOI] [PubMed] [Google Scholar]
- 11.Albrecht-Buehler G. Filopodia of spreading 3T3 cells. J Cell Biol. 1976;69:275–286. doi: 10.1083/jcb.69.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen W E, Jones G E, Pollard J W, Ridley A J. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J Cell Sci. 1997;110:707–720. doi: 10.1242/jcs.110.6.707. [DOI] [PubMed] [Google Scholar]
- 13.Allen W E, Zicha D, Ridley A J, Jones G E. A role for Cdc42 in macrophage chemotaxis. J Cell Biol. 1998;141:1147–1157. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altman R, Kellogg D. Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol. 1997;138:119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amberg D C, Zahner J E, Mulholland J W, Pringle J R, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson B L, Boldogh I, Evangelista M, Boone C, Greene L A, Pon L A. The src homology domain 3 (SH3) of a yeast type I myosin, Myo5p, binds to verprolin and is required for targeting to sites of actin polarization. J Cell Biol. 1998;141:1357–1370. doi: 10.1083/jcb.141.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnould T, Kim E, Tsiokas L, Jochimsen F, Gruning W, Chang J D, Walz G. The polycystic kidney disease I gene product mediates protein kinase C α-dependent and c-Jun N-terminal kinase-dependent activation of the transcription factor AP-1. J Biol Chem. 1998;273:6013–6018. doi: 10.1074/jbc.273.11.6013. [DOI] [PubMed] [Google Scholar]
- 18.Aspenström P. A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr Biol. 1997;7:479–487. doi: 10.1016/s0960-9822(06)00219-3. [DOI] [PubMed] [Google Scholar]
- 19.Aspenström P, Lindberg U, Hall A. Two GTPases, Cdc42 and Rac, bind directly to a protein implicated in the immunodeficiency disorder Wiskott-Aldrich syndrome. Curr Biol. 1996;6:70–75. doi: 10.1016/s0960-9822(02)00423-2. [DOI] [PubMed] [Google Scholar]
- 20.Auer K L, Contessa J, Brenz-Verca S, Pirola L, Rusconi S, Cooper G, Abo A, Wymann M P, Davis R J, Birrer M, Dent P. The Ras/Rac1/Cdc42/SEK/JNK/c-Jun cascade is a key pathway by which agonists stimulate DNA synthesis in primary cultures of rat hepatocytes. Mol Biol Cell. 1998;9:561–573. doi: 10.1091/mbc.9.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Backlund P S., Jr GTP-stimulated carboxyl methylation of a soluble form of the GTP-binding protein G25K in brain. J Biol Chem. 1992;267:18432–18439. [PubMed] [Google Scholar]
- 23.Bagrodia S, Derijard B, Davis R J, Cerione R A. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 24.Bagrodia S, Taylor S J, Creasy C L, Chernoff J, Cerione R A. Identification of a mouse p21Cdc42/Rac activated kinase. J Biol Chem. 1995;270:22731–22737. doi: 10.1074/jbc.270.39.22731. [DOI] [PubMed] [Google Scholar]
- 25.Banin S, Truong O, Katz D R, Waterfield M D, Brickell P M, Gout I. Wiskott-Aldrich syndrome protein (WASp) is a binding partner for c-Src family protein-tyrosine kinases. Curr Biol. 1996;6:981–988. doi: 10.1016/s0960-9822(02)00642-5. [DOI] [PubMed] [Google Scholar]
- 26.Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barfod E T, Zheng Y, Kuang W-J, Hart M J, Evans T, Cerione R A, Ashkenazi A. Cloning and expression of a human CDC42 GTPase-activating protein reveals a functional SH3-binding domain. J Biol Chem. 1993;268:26059–26062. [PubMed] [Google Scholar]
- 29.Barrett K, Leptin M, Settleman J. The rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 1997;91:905–915. doi: 10.1016/s0092-8674(00)80482-1. [DOI] [PubMed] [Google Scholar]
- 30.Bashour A M, Fullerton A T, Hart M J, Bloom G S. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links microfilaments. J Cell Biol. 1997;137:1555–1566. doi: 10.1083/jcb.137.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bax B. Domains of rasGAP and rhoGAP are related. Nature. 1998;392:447–448. doi: 10.1038/33040. [DOI] [PubMed] [Google Scholar]
- 32.Bazenet C E, Mota M A, Rubin L L. The small GTP-binding protein Cdc42 is required for nerve growth factor withdrawal-induced neuronal death. Proc Natl Acad Sci USA. 1998;95:3984–3989. doi: 10.1073/pnas.95.7.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Ami G, Ozeri V, Hanski E, Hofmann F, Aktories K, Hahn K M, Bokoch G M, Rosenshine I. Agents that inhibit Rho, Rac, and Cdc42 do not block formation of actin pedestals in HeLa cells infected with enteropathogenic Escherichia coli. Infect Immun. 1998;66:1755–1758. doi: 10.1128/iai.66.4.1755-1758.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bender A. Genetic evidence for the roles of the bud-site-selection genes BUD5 and BUD2 in control of the Rsr1p (Bud1p) GTPase in yeast. Proc Natl Acad Sci USA. 1993;90:9926–9929. doi: 10.1073/pnas.90.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bender A, Pringle J R. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc Natl Acad Sci USA. 1989;86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bender A, Pringle J R. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bender L, Lo H S, Lee H, Kokojan V, Peterson J, Bender A. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in yeast. J Cell Biol. 1996;133:879–894. doi: 10.1083/jcb.133.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benton B K, Tinkelenberg A, Gonzalez I, Cross F R. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol. 1997;17:5067–5076. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benton B K, Tinkelenberg A H, Jean D, Plump S D, Cross F R. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 1993;12:5267–5275. doi: 10.1002/j.1460-2075.1993.tb06222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berestetskaya Y V, Faure M P, Ichijo H, Voyno-Yasenetskaya T A. Regulation of apoptosis by α-subunits of G12 and G13 proteins via apoptosis signal-regulating kinase-1. J Biol Chem. 1998;273:27816–27823. doi: 10.1074/jbc.273.43.27816. [DOI] [PubMed] [Google Scholar]
- 41.Bi E, Maddox P, Lew D J, Salmon E D, McMillan J N, Yeh E, Pringle J R. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bi E, Pringle J R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blacketer M J, Koehler C M, Coats S G, Myers A M, Madaule P. Regulation of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol Cell Biol. 1993;13:5567–5581. doi: 10.1128/mcb.13.9.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 45.Boivin D, Beliveau R. Subcellular distribution and membrane association of Rho-related small GTP-binding proteins in kidney cortex. Am J Physiol. 1995;269:F180–F189. doi: 10.1152/ajprenal.1995.269.2.F180. [DOI] [PubMed] [Google Scholar]
- 46.Boivin D, Bilodeau D, Beliveau R. Regulation of cytoskeletal functions by Rho small GTP-binding proteins in normal and cancer cells. Can J Physiol Pharmacol. 1996;74:801–810. [PubMed] [Google Scholar]
- 47.Bokoch G M, Wang Y, Bohl B P, Sells M A, Quilliam L A, Knaus U G. Interaction of the nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 48.Booher R N, Deshaies R J, Kirschner M W. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34(CDC28) in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48a.Boone, C. Personal communication.
- 49.Bourdoulous S, Orend G, MacKenna D A, Pasqualini R, Ruoslahti E. Fibronectin matrix regulates activation of RHO and CDC42 GTPases and cell cycle progression. J Cell Biol. 1998;143:267–276. doi: 10.1083/jcb.143.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourne H R. The arginine finger strikes again. Nature. 1997;389:673–674. doi: 10.1038/39470. [DOI] [PubMed] [Google Scholar]
- 51.Brill S, Li S, Lyman C W, Church D M, Wasmuth J J, Weissbach L, Bernards A, Snijders A J. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol Cell Biol. 1996;16:4869–4878. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 53.Brown E J, Beal P A, Keith C T, Chen J, Shin T B, Schreiber S L. Control of p70 S6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 54.Brown J L, Jaquenoud M, Gulli M-P, Chant J, Peter M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997;11:2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown J L, Stowers L, Baer M, Trejo J, Coughlin S, Chant J. Human Ste20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr Biol. 1996;6:598–605. doi: 10.1016/s0960-9822(02)00546-8. [DOI] [PubMed] [Google Scholar]
- 56.Bryant S S, Briggs S, Smithgall T E, Martin G A, McCormick F, Chang J-H, Parsons S J, Jove R. Two SH2 domains of p120 Ras GTPase-activating protein bind synergistically to tyrosine phosphorylated p190 Rho GTPase-activating protein. J Biol Chem. 1995;270:17947–17952. doi: 10.1074/jbc.270.30.17947. [DOI] [PubMed] [Google Scholar]
- 57.Brzeska H, Szczepanowska J, Hoey J, Korn E D. The catalytic domain of Acanthamoeba myosin I heavy chain kinase. II. Expression of active catalytic domain and sequence homology to p21-activated kinase (PAK) J Biol Chem. 1996;271:27056–27062. doi: 10.1074/jbc.271.43.27056. [DOI] [PubMed] [Google Scholar]
- 58.Bunnell S C, Henry P A, Kolluri R, Kirchhausen T, Rickles R J, Berg L J. Identification of Itk/Tsk Src homology 3 domain ligands. J Biol Chem. 1996;271:25646–25656. doi: 10.1074/jbc.271.41.25646. [DOI] [PubMed] [Google Scholar]
- 59.Burbelo P D, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 60.Burnett P E, Blackshaw S, Lai M M, Qureshi I A, Burnett A F, Sabatini D M, Snyder S H. Neurabin is a synaptic protein linking p70 S6 kinase and the neuronal cytoskeleton. Proc Natl Acad Sci USA. 1998;95:8351–8356. doi: 10.1073/pnas.95.14.8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cabib E, Drgonová J, Drgon T. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu Rev Biochem. 1998;67:307–333. doi: 10.1146/annurev.biochem.67.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cahill M A, Peter M E, Kischkel F C, Chinnaiyan A M, Dixit V M, Krammer P H, Nordheim A. CD95 (APO-1/Fas) induces activation of SAP kinases downstream of ICE-like proteases. Oncogene. 1996;13:2087–2096. [PubMed] [Google Scholar]
- 63.Cali B M, Doyle T C, Botstein D, Fink G R. Multiple functions for actin during filamentous growth of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1873–1889. doi: 10.1091/mbc.9.7.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Camus C, Boy-Marcotte E, Jacquet M. Two subclasses of guanine exchange factor (GEF) domains revealed by comparison of activities of chimeric genes constructed from CDC25, SDC25, and BUD5 in Saccharomyces cerevisiae. Mol Gen Genet. 1994;245:167–176. doi: 10.1007/BF00283264. [DOI] [PubMed] [Google Scholar]
- 65.Cantor S B, Urano T, Feig L A. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caplin B E, Hettich L A, Marshall M S. Substrate characterization of the Saccharomyces cerevisiae protein farnesyltransferase and type-I protein geranylgeranyltransferase. Biochim Biophy Acta. 1994;1205:39–48. doi: 10.1016/0167-4838(94)90089-2. [DOI] [PubMed] [Google Scholar]
- 67.Carroll C W, Altman R, Schieltz D, Yates III J R, Kellogg D. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castrillon D H, Wasserman S A. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- 69.Cereijido M, Valdes J, Shoshani L, Contreras R G. Role of tight junctions in establishing and maintaining cell polarity. Annu Rev Physiol. 1998;60:161–177. doi: 10.1146/annurev.physiol.60.1.161. [DOI] [PubMed] [Google Scholar]
- 70.Cerione R. A difference of opinion or a matter of perspective? Trends Biochem Sci. 1998;23:100. [Google Scholar]
- 71.Cerione R A, Zheng Y. Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 72.Chabre M, Antonny B, Paris S. PIP2: activator … or terminator of small G proteins? Trends Biochem Sci. 1998;23:98–99. doi: 10.1016/s0968-0004(98)01177-3. [DOI] [PubMed] [Google Scholar]
- 73.Chang E C, Barr M, Wang Y, Jung V, Xu H-P, Wigler M H. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 74.Chang F, Drubin D, Nurse P. Cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang H Y, Nishitoh H, Yang X, Ichijo H, Baltimore D. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science. 1998;281:1860–1863. doi: 10.1126/science.281.5384.1860. [DOI] [PubMed] [Google Scholar]
- 76.Chang J-H, Gill S, Settleman J, Parsons S J. c-Src regulates the simultaneous rearrangement of actin cytoskeleton, p190RhoGAP, and p120RasGAP following epidermal growth factor stimulation. J Cell Biol. 1995;130:355–368. doi: 10.1083/jcb.130.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chant J. Generation of cell polarity in yeast. Curr Opin Cell Biol. 1996;8:557–564. doi: 10.1016/s0955-0674(96)80035-4. [DOI] [PubMed] [Google Scholar]
- 78.Chant J. Septin scaffolds and cleavage planes in Saccharomyces. Cell. 1996;84:187–190. doi: 10.1016/s0092-8674(00)80972-1. [DOI] [PubMed] [Google Scholar]
- 79.Chant J, Corrado K, Pringle J R, Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- 80.Chant J, Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- 81.Chant J, Pringle J R. Bud5p. In: Zerial M, Huber L A, editors. Guidebook to the small GTPases. Oxford, United Kingdom: Oxford University Press; 1995. pp. 193–196. [Google Scholar]
- 82.Cheatham L, Vlahos C J, Cheatham L, Wang L, Blenis J, Kahn C R. Phosphatidylinositol 3-kinase activation is required for insulin stimulation of pp70 S6 kinase, DNA synthesis, and glucose transporter translocation. Mol Cell Biol. 1994;14:4902–4911. doi: 10.1128/mcb.14.7.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen G-C, Kim Y-J, Chan C S M. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997;11:2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen G C, Zheng L, Chan C S. The LIM domain-containing Dbm1 GTPase-activating protein is required for normal cellular morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1376–1390. doi: 10.1128/mcb.16.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen L-M, Hobbie S, Galán J E. Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science. 1996;274:2115–2118. doi: 10.1126/science.274.5295.2115. [DOI] [PubMed] [Google Scholar]
- 86.Chen W, Blanc J, Lim L. Characterization of a promiscuous GTPase-activating protein that has a Bcr-related domain from Caenorhabditis elegans. J Biol Chem. 1994;269:820–823. [PubMed] [Google Scholar]
- 87.Chen W, Chen S, Yap S F, Lim L. The Caenorhabditis elegans p21-activated kinase (CePAK) colocalizes with CeRac1 and CDC42Ce at hypodermal cell boundaries during embryo elongation. J Biol Chem. 1996;271:26362–26368. doi: 10.1074/jbc.271.42.26362. [DOI] [PubMed] [Google Scholar]
- 88.Chen W, Lim H H, Lim L. The CDC42 homologue from Caenorhabditis elegans. J Biol Chem. 1993;268:13280–13285. [PubMed] [Google Scholar]
- 89.Chen Y, Wang X, Templeton D, Davis R J, Tan T. The role of c-Jun N-terminal kinase (JNK) in apoptosis induced by ultraviolet C and gamma radiation. J Biol Chem. 1996;271:31929–31936. doi: 10.1074/jbc.271.50.31929. [DOI] [PubMed] [Google Scholar]
- 90.Chenevert J, Corrado K, Bender A, Pringle J, Herskowitz I. A yeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature. 1992;356:77–79. doi: 10.1038/356077a0. [DOI] [PubMed] [Google Scholar]
- 91.Chenevert J, Valtz N, Herskowitz I. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics. 1994;136:1287–1297. doi: 10.1093/genetics/136.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chou M M, Blenis J. The 70 kD S6 kinase: a kinase with multiple roles in mitogenic signaling. Curr Opin Cell Biol. 1995;7:806–814. doi: 10.1016/0955-0674(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 93.Chou M M, Blenis J. The 70 kDa S6 kinase complexes with and is activated by the Rho family G proteins Cdc42 and Rac1. Cell. 1996;85:573–583. doi: 10.1016/s0092-8674(00)81257-x. [DOI] [PubMed] [Google Scholar]
- 94.Chuang T-H, Bohl B P, Bokoch G M. Biologically active lipids are regulators of RacGDI complexation. J Biol Chem. 1993;268:26206–26211. [PubMed] [Google Scholar]
- 95.Chuang T-H, Xu X, Kaartinen V, Heisterkamp N, Groffen J, Bokoch G M. Abr and Bcr are multifunctional regulators of the Rho GTP-binding protein family. Proc Natl Acad Sci USA. 1995;92:10282–10286. doi: 10.1073/pnas.92.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chuang T H, Hahn K M, Lee J D, Danley D E, Bokoch G M. The small GTPase Cdc42 initiates an apoptotic signaling pathway in Jurkat T lymphocytes. Mol Biol Cell. 1997;8:1687–1698. doi: 10.1091/mbc.8.9.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chung J, Grammer T, Lemon K P, Kaslauskas A, Blenis J. PDGF- and insulin-dependent pp70S6k activation mediated by phosphatidylinositol 3-OH kinase. Nature. 1994;370:71–75. doi: 10.1038/370071a0. [DOI] [PubMed] [Google Scholar]
- 98.Cicchetti P, Mayer B J, Thiel G, Baltimore D. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science. 1992;257:803–806. doi: 10.1126/science.1379745. [DOI] [PubMed] [Google Scholar]
- 99.Cicchetti P, Ridley A J, Zheng Y, Cerione R A, Baltimore D. 3BP-1, an SH3 domain binding protein, has GAP activity for Rac and inhibits growth factor-induced membrane ruffling in fibroblasts. EMBO J. 1995;14:3127–3135. doi: 10.1002/j.1460-2075.1995.tb07315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cid V J, Durán A, Del Rey F, Snyder M P, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Clark E A, King W G, Brugge J S, Symons M, Hynes R O. Integrin-mediated signals regulated by members of the Rho family of GTPases. J Cell Biol. 1998;142:573–586. doi: 10.1083/jcb.142.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clarke N, Arenzana N, Hai T, Minden A, Prywes R. Epidermal growth factor induction of the c-jun promoter by a Rac pathway. Mol Cell Biol. 1998;18:1065–1073. doi: 10.1128/mcb.18.2.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Clerk A, Fuller S J, Michael A, Sugden P H. Stimulation of “stress-regulated” mitogen-activated protein kinases (stress-activated protein kinases/c-Jun N-terminal kinases and p38-mitogen-activated protein kinases) in perfused rat hearts by oxidative and other stresses. J Biol Chem. 1998;273:7228–7234. doi: 10.1074/jbc.273.13.7228. [DOI] [PubMed] [Google Scholar]
- 104.Clerk A, Sugden P H. Activation of p21-activated protein kinase α (αPAK) by hyperosmotic shock in neonatal ventricular myocytes. FEBS Lett. 1997;403:23–25. doi: 10.1016/s0014-5793(97)00020-3. [DOI] [PubMed] [Google Scholar]
- 105.Cohen G M. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Coleman K G, Steensma H Y, Kaback D B, Pringle J R. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation and characterization of the CDC24 gene and adjacent regions of the chromosome. Mol Cell Biol. 1986;6:4516–4525. doi: 10.1128/mcb.6.12.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106a.Collins, C., and D. I. Johnson. Unpublished data.
- 107.Cooper J A, Kiehart D P. Septins may form a ubiquitous family of cytoskeletal filaments. J Cell Biol. 1996;134:1345–1348. doi: 10.1083/jcb.134.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cory G O, MacCarthy-Morrogh L, Banin S, Gout I, Brickell P M, Levinsky R J, Kinnon C, Lovering R C. Evidence that the Wiskott-Aldrich syndrome protein may be involved in lymphoid cell signaling pathways. J Immunol. 1996;157:3791–3795. [PubMed] [Google Scholar]
- 109.Coso O A, Chiariello M, Yu J-C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 110.Csank C, Schroppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas D Y, Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cullen B R. HIV-1: is Nef a PAK animal. Curr Biol. 1996;6:1557–1559. doi: 10.1016/s0960-9822(02)70770-7. [DOI] [PubMed] [Google Scholar]
- 112.Cvrcková F, De Virgilio C, Manser E, Pringle J R, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 113.Cvrcková F, Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dai Q, Choy E, Chiu V, Romano J, Slivka S R, Steitz S A, Michaelis S, Philips M R. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- 115.Daniels R H, Hall P S, Bokoch G M. Membrane targeting of p21-activated kinase 1 (PAK1) induces neurite outgrowth from PC12 cells. EMBO J. 1998;17:754–764. doi: 10.1093/emboj/17.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Davis C R, Richman T R, Deliduka S B, Blaisdell J O, Collins C C, Johnson D I. Analysis of the mechanisms of action of the Saccharomyces cerevisiae dominant lethal cdc42G12V and dominant negative cdc42D118A mutations. J Biol Chem. 1998;273:849–858. doi: 10.1074/jbc.273.2.849. [DOI] [PubMed] [Google Scholar]
- 117.Dawid I B, Breen J J, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 118.Delmer D P, Pear J R, Andrawis A, Stalker D M. Genes encoding small GTP-binding proteins analogous to mammalian rac are preferentially expressed in developing cotton fibers. Mol Gen Genet. 1995;248:43–51. doi: 10.1007/BF02456612. [DOI] [PubMed] [Google Scholar]
- 119.DeMarini D J, Adams A E M, Fares H, DeVirgilio C, Valle G, Chuang J S, Pringle J R. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Denhardt D T. Signal-transducing protein phosphorylation cascades mediated by Ras/Rho proteins in the mammalian cell: the potential for multiplex signalling. Biochem J. 1996;318:729–747. doi: 10.1042/bj3180729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Derijard B, Hibi M, Wu I-H, Barret T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 122.Derry J M J, Ochs H J, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–644. doi: 10.1016/0092-8674(94)90528-2. [DOI] [PubMed] [Google Scholar]
- 123.Dharmawardhane S, Sanders L C, Martin S S, Daniels R H, Bokoch G M. Localization of p21-activated kinase 1 (PAK1) to pinocytic vesicles and cortical actin structures in stimulated cells. J Cell Biol. 1997;138:1265–1278. doi: 10.1083/jcb.138.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Diekmann D, Brill S, Garrett M D, Totty M, Hsuan J, Monfries C, Hall C, Lim L, Hall A. Bcr encodes a GTPase-activating protein for p21rac. Nature. 1991;351:400–402. doi: 10.1038/351400a0. [DOI] [PubMed] [Google Scholar]
- 125.Dramsi S, Cossart P. Intracellular pathogens and the actin cytoskeleton. Annu Rev Cell Dev Biol. 1998;14:137–166. doi: 10.1146/annurev.cellbio.14.1.137. [DOI] [PubMed] [Google Scholar]
- 126.Drubin D G, Nelson W J. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 127.Dutartre H, Davoust J, Gorvel J-P, Chavrier P. Cytokinesis arrest and redistribution of actin-cytoskeleton regulatory components in cells expressing the Rho GTPase CDC42Hs. J Cell Sci. 1996;109:367–377. doi: 10.1242/jcs.109.2.367. [DOI] [PubMed] [Google Scholar]
- 128.Eaton S, Auvinen P, Luo L, Jan Y N, Simons K. CDC42 and Rac1 control different actin-dependent processes in the Drosophila wing disc epithelium. J Cell Biol. 1995;131:151–164. doi: 10.1083/jcb.131.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Eaton S, Wepf R, Simons K. Roles for Rac1 and Cdc42 in planar polarization and hair outgrowth in the wing of Drosophila. J Cell Biol. 1996;135:1277–1289. doi: 10.1083/jcb.135.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Eby J J, Holly S P, vanDrogen F, Grishin A V, Peter M, Drubin D G, Blumer K J. Actin cytoskeleton organization regulated by the PAK family of protein kinases. Curr Biol. 1998;8:967–970. doi: 10.1016/s0960-9822(98)00398-4. [DOI] [PubMed] [Google Scholar]
- 131.Ehler E, vanLeeuwen F, Collard J G, Salinas P C. Expression of Tiam-1 in the developing brain suggests a role for the Tiam-1-Rac signaling pathway in cell migration and neurite outgrowth. Mol Cell Neurosci. 1997;9:1–12. doi: 10.1006/mcne.1997.0602. [DOI] [PubMed] [Google Scholar]
- 132.Eichinger L, Bahler M, Dietz M, Eckerskorn C, Schleicher M. Characterization and cloning of a Dictyostelium Ste20-like protein kinase that phosphorylates the actin-binding protein severin. J Biol Chem. 1998;273:12952–12959. doi: 10.1074/jbc.273.21.12952. [DOI] [PubMed] [Google Scholar]
- 133.Elia L, Marsh L. A role for a protease in morphogenic responses during yeast cell fusion. J Cell Biol. 1998;142:1473–1485. doi: 10.1083/jcb.142.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Emmons S, Phan H, Calley J, Chen W, James B, Manseau L. Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 1995;9:2482–2494. doi: 10.1101/gad.9.20.2482. [DOI] [PubMed] [Google Scholar]
- 135.Epp J A, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- 136.Erickson J W, Cerione R A, Hart M J. Identification of an actin cytoskeletal complex that includes IQGAP and the Cdc42 GTPase. J Biol Chem. 1997;272:24443–24447. doi: 10.1074/jbc.272.39.24443. [DOI] [PubMed] [Google Scholar]
- 137.Erickson J W, Zhang C J, Kahn R A, Evans T, Cerione R A. Mammalian Cdc42 is a brefeldin A-sensitive component of the Golgi apparatus. J Biol Chem. 1996;271:26850–26854. doi: 10.1074/jbc.271.43.26850. [DOI] [PubMed] [Google Scholar]
- 138.Eva A, Vecchio G, Rao C D, Tronick S R, Aaronson S A. The predicted DBL oncogene product defines a distinct class of transforming proteins. Proc Natl Acad Sci USA. 1988;85:2061–2065. doi: 10.1073/pnas.85.7.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Evangelista M, Blundell K, Longtine M S, Chow C J, Adames N, Pringle J R, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 140.Evans T, Brown M L, Fraser E D, Northrup J K. Purification of the major GTP-binding proteins from human placental membranes. J Biol Chem. 1986;261:7052–7059. [PubMed] [Google Scholar]
- 141.Fanger G R, Gerwins P, Widmann C, Jarpe M B, Johnson G L. MEKKs, GCKs, MLKs, PAKs, TAKs, and Tpls: upstream regulators of the c-Jun amino-terminal kinases? Curr Opin Genet Dev. 1997;7:67–74. doi: 10.1016/s0959-437x(97)80111-6. [DOI] [PubMed] [Google Scholar]
- 142.Fanger G R, Johnson N L, Johnson G L. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J. 1997;16:4961–4972. doi: 10.1093/emboj/16.16.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Faure S, Vigneron S, Doree M, Morin N. A member of the Ste20/PAK family of protein kinases is involved in both arrest of xenopus oocytes at G2/prophase of the first meiotic cell cycle and in prevention of apoptosis. EMBO J. 1997;16:5550–5561. doi: 10.1093/emboj/16.18.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Featherstone C. The many faces of WAS protein. Science. 1997;275:27–28. doi: 10.1126/science.275.5296.27. [DOI] [PubMed] [Google Scholar]
- 145.Feltham J L, Dotsch V, Raza S, Manor D, Cerione R A, Sutcliffe M J, Wagner G, Oswald R E. Definition of the switch surface in the solution structure of Cdc42Hs. Biochemistry. 1997;36:8755–8766. doi: 10.1021/bi970694x. [DOI] [PubMed] [Google Scholar]
- 146.Field C, Schekman R. Localized secretion of acid phosphatase reflects the pattern of cell surface growth in Saccharomyces cerevisiae. J Cell Biol. 1980;86:123–128. doi: 10.1083/jcb.86.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Finegold A A, Johnson D I, Farnsworth C C, Gelb M H, Judd S R, Glomset J A, Tamanoi F. Protein geranylgeranyltransferase of Saccharomyces cerevisiae is specific for Cys-Xaa-Xaa-Leu motif proteins and requires the CDC43 gene product, but not the DPR1 gene product. Proc Natl Acad Sci USA. 1991;88:4448–4452. doi: 10.1073/pnas.88.10.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Foltz I N, Gerl R E, Wieler J S, Luckach M, Salmon R A, Schrader J W. Human mitogen-activated protein kinase kinase 7 (MKK7) is a highly conserved c-jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) activated by environmental stresses and physiological stimuli. J Biol Chem. 1998;273:9344–9351. doi: 10.1074/jbc.273.15.9344. [DOI] [PubMed] [Google Scholar]
- 149.Francis C L, Ryan T A, Jones B D, Smith S J, Falkow S. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature. 1993;364:639–642. doi: 10.1038/364639a0. [DOI] [PubMed] [Google Scholar]
- 150.Freshney N W, Rawlinson L, Guesdon F, Jones E, Cowley S, Hsuan J, Saklatvaia J. Interleukin-1 activates a novel protein kinase cascade that results in the phosphorylation of Hsp27. Cell. 1994;78:1039–1049. doi: 10.1016/0092-8674(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 151.Frost J A, Khokhlatchev A, Stippec S, White M A, Cobb M H. Differential effects of PAK1-activating mutations reveal activity-dependent and -independent effects on cytoskeletal regulation. J Biol Chem. 1998;273:28191–28198. doi: 10.1074/jbc.273.43.28191. [DOI] [PubMed] [Google Scholar]
- 152.Frost J A, Steen H, Shapiro P, Lewis T, Ahn N, Shaw P E, Cobb M H. Cross-cascade activation of ERKs and ternary complex factors by Rho family proteins. EMBO J. 1997;16:6426–6438. doi: 10.1093/emboj/16.21.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Frost J A, Xu S, Hutchison M R, Marcus S, Cobb M H. Actions of Rho family small G proteins and p21-activated protein kinases on mitogen-activated protein kinase family members. Mol Cell Biol. 1996;16:3707–3713. doi: 10.1128/mcb.16.7.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fruman D A, Meyers R E, Cantley L C. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 155.Fuchs S Y, Adler V, Buschmann T, Yin Z M, Wu X W, Jones S N, Ronai Z. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 1998;12:2658–2663. doi: 10.1101/gad.12.17.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fuchs S Y, Fried V A, Ronai Z. Stress-activated kinases regulate protein stability. Oncogene. 1998;17:1483–1490. doi: 10.1038/sj.onc.1202184. [DOI] [PubMed] [Google Scholar]
- 157.Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fukata M, Kuroda S, Fujii K, Nakamura T, Shoji I, Matsuura Y, Okawa K, Iwamatsu A, Kikuchi A, Kaibuchi K. Regulation of cross-linking of actin filament by IQGAP1, a target for Cdc42. J Biol Chem. 1997;272:29579–29583. doi: 10.1074/jbc.272.47.29579. [DOI] [PubMed] [Google Scholar]
- 159.Fukui Y, Yamamoto M. Isolation and characterization of Schizosaccharomyces pombe mutants phenotypically similar to ras1−. Mol Gen Genet. 1988;215:26–31. doi: 10.1007/BF00331298. [DOI] [PubMed] [Google Scholar]
- 160.Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H, Araki S, Ueda T, Kikuchi A, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- 161.Galisteo M L, Chernoff J, Su Y-C, Skolnik E Y, Schlessinger J. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 162.Gammie A E, Brizzio V, Rose M D. Distinct morphological phenotypes of cell fusion mutants. Mol Biol Cell. 1998;9:1395–1410. doi: 10.1091/mbc.9.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Garrett M D, Major G N, Totty N, Hall A. Purification and N-terminal sequence of the p21rho GTPase-activating protein, rho GAP. Biochem J. 1991;276:833–836. doi: 10.1042/bj2760833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Garrett M D, Self A J, van Oers C, Hall A. Identification of distinct cytoplasmic targets for ras/R-ras and rho regulatory proteins. J Biol Chem. 1989;264:10–13. [PubMed] [Google Scholar]
- 165.Gelperin D, Weigle J, Nelson K, Roseboom P, Irie K, Matsumoto K, Lemmon S. 14-3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:11539–11543. doi: 10.1073/pnas.92.25.11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gilbreth M, Yang P, Wang D, Frost J, Polverino A, Cobb M H, Marcus S. The highly conserved skb1 gene encodes a protein that interacts with Shk1, a fission yeast Ste20/PAK homolog. Proc Natl Acad Sci USA. 1996;93:13802–13807. doi: 10.1073/pnas.93.24.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gimeno C J, Fink G R. The logic of cell division in the life cycle of yeast. Science. 1992;257:626. doi: 10.1126/science.1496375. [DOI] [PubMed] [Google Scholar]
- 168.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 169.Gjoerup O, Lukas J, Bartek J, Willumsen B M. Rac and Cdc42 are potent stimulators of E2F-dependent transcription capable of promoting retinoblastoma susceptibility gene product hyperphosphorylation. J Biol Chem. 1998;273:18812–18818. doi: 10.1074/jbc.273.30.18812. [DOI] [PubMed] [Google Scholar]
- 170.Glotzer M, Hyman A A. The importance of being polar. Curr Biol. 1995;5:1102–1105. doi: 10.1016/s0960-9822(95)00221-1. [DOI] [PubMed] [Google Scholar]
- 171.Gomez J, Martinez C, Gonzalez A, Rebollo A. Dual role of Ras and Rho proteins: at the cutting edge of life and death. Immunol Cell Biol. 1998;76:125–134. doi: 10.1046/j.1440-1711.1998.00723.x. [DOI] [PubMed] [Google Scholar]
- 172.Gong T W L, Shin J J, Burmeister M, Lomax M I. Complete cDNAs for CDC42 from chicken cochlea and mouse liver. Biochim Biophy Acta Gene Struct Express. 1997;1352:282–292. doi: 10.1016/s0167-4781(97)00027-4. [DOI] [PubMed] [Google Scholar]
- 173.Goodman L E, Perou C M, Fujiyama A, Tamanoi F. Structure and expression of yeast DPR1, a gene essential for the processing and intracellular localization of ras proteins. Yeast. 1988;4:271–281. doi: 10.1002/yea.320040405. [DOI] [PubMed] [Google Scholar]
- 174.Gorvel J P, Chang T C, Boretto J, Azuma T, Chavrier P. Differential properties of D4/LyGDI versus RhoGDI: phosphorylation and rho GTPase selectivity. FEBS Lett. 1998;422:269–273. doi: 10.1016/s0014-5793(98)00020-9. [DOI] [PubMed] [Google Scholar]
- 175.Gosser Y Q, Nomanbhoy T K, Aghazadeh B, Manor D, Combs C, Cerione R A, Rosen M K. C-terminal binding domain of rho GDP-dissociation inhibitor directs N-terminal inhibitory peptide to GTPases. Nature. 1997;387:814–819. doi: 10.1038/42961. [DOI] [PubMed] [Google Scholar]
- 176.Gould K L, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- 177.Govindan B, Novick P. Development of cell polarity in budding yeast. J Exp Zool. 1995;273:401–424. doi: 10.1002/jez.1402730505. [DOI] [PubMed] [Google Scholar]
- 178.Graziani G, Ron D, Eva A, Srivastava S K. The human dbl-proto-oncogene product is a cytoplasmic phosphoprotein which is associated with the cytoskeletal matrix. Oncogene. 1989;4:823–829. [PubMed] [Google Scholar]
- 179.Guan Z H, Buckman S Y, Pentland A P, Templeton D J, Morrison A R. Induction of cyclooxygenase-2 by the activated MEKK1→SEK1/MKK4→p38 mitogen-activated protein kinase pathway. J Biol Chem. 1998;273:12901–12908. doi: 10.1074/jbc.273.21.12901. [DOI] [PubMed] [Google Scholar]
- 180.Guichard A, Bergeret E, Griffin-Shea R. Rotund Rac-GAP. In: Zerial M, Huber L A, editors. Guidebook to the small GTPases. Oxford, United Kingdom: Oxford University Press; 1995. pp. 261–265. [Google Scholar]
- 181.Guichard A, Bergeret E, GriffinShea R. Overexpression of RnRacGAP in Drosophila melanogaster deregulates cytoskeletal organisation in cellularising embryos and induces discrete imaginal phenotypes. Mech Dev. 1997;61:49–62. doi: 10.1016/s0925-4773(96)00619-3. [DOI] [PubMed] [Google Scholar]
- 182.Guillemot J C, Kruskal B A, Adra C N, Zhu S, Ko J L, Burch P, Nocka K, Seetoo K, Simons E, Lim B. Targeted disruption of guanosine diphosphate-dissociation inhibitor for Rho-related proteins, GDID4: normal hematopoietic differentiation but subtle defect in superoxide production by macrophages derived from in vitro embryonal stem cell differentiation. Blood. 1996;88:2722–2731. [PubMed] [Google Scholar]
- 183.Guillemot J C, Montcourrier P, Vivier E, Davoust J, Chavrier P. Selective control of membrane ruffling and actin plaque assembly by the rho GTPases Rac1 and CDC42 in FcɛRI-activated rat basophilic leukemia (RBL-2H3) cells. J Cell Sci. 1997;110:2215–2225. doi: 10.1242/jcs.110.18.2215. [DOI] [PubMed] [Google Scholar]
- 184.Guinamard R, Aspenström P, Fougereau M, Chavrier P, Guillemot J C. Tyrosine phosphorylation of the Wiskott-Aldrich syndrome protein by Lyn and Btk is regulated by CDC42. FEBS Lett. 1998;434:431–436. doi: 10.1016/s0014-5793(98)01016-3. [DOI] [PubMed] [Google Scholar]
- 185.Guo W, Sutcliffe M J, Cerione R A, Oswald R E. Identification of the binding surface on Cdc42Hs for p21-activated kinase. Biochem. 1998;37:14030–14037. doi: 10.1021/bi981352+. [DOI] [PubMed] [Google Scholar]
- 186.Gutkind J S, VitaleCross L. The pathway linking small GTP-binding proteins of the Rho family to cytoskeletal components and novel signaling kinase cascades. Semin Cell Dev Biol. 1996;7:683–690. [Google Scholar]
- 187.Haarer B K, Lillie S H, Adams A E M, Magdolen V, Bandlow W, Brown S S. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Haarer B K, Petzold A, Lillie S H, Brown S S. Identification of MYO4, a second class V myosin gene in yeast. J Cell Sci. 1994;107:1055–1064. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- 189.Habets G G M, Scholtes E H M, Zuydgeest D, van der Kammen R A, Stam J C, Berns A, Collard J G. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 190.Hacker U, Perrimon N. DRhoGEF2 encodes a member of the Dbl family of oncogenes and controls cell shape changes during gastrulation in Drosophila. Genes Dev. 1998;12:274–284. doi: 10.1101/gad.12.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hall A. The cellular functions of small GTP-binding proteins. Science. 1990;249:635–640. doi: 10.1126/science.2116664. [DOI] [PubMed] [Google Scholar]
- 192.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 193.Ham J, Babji C, Whitfield J, Pfarr C M, Lallemand D, Yaniv M, Rubin L L. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 194.Han J, Lee J D, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 195.Han Z Q S, Enslen H, Hu X D, Meng X J, Wu I-H, Barrett T, Davis R J, Ip Y T. A conserved p38 mitogen-activated protein kinase pathway regulates Drosophila immunity gene expression. Mol Cell Biol. 1998;18:3527–3539. doi: 10.1128/mcb.18.6.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hancock J F, Cadwallader K, Paterson H, Marshall C J. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hancock J F, Paterson H, Marshall C J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 198.Harden N, Loh H Y, Chia W, Lim L. A dominant inhibitory version of the small GTP-binding protein Rac disrupts cytoskeletal structures and inhibits developmental cell shape changes in Drosophila. Development. 1995;121:903–914. doi: 10.1242/dev.121.3.903. [DOI] [PubMed] [Google Scholar]
- 199.Harden N, Lee J, Loh H-Y, Ong Y-M, Tan I, Leung T, Manser E, Lim L. A Drosophila homolog of the Rac- and Cdc42-activated serine/threonine kinase PAK is a potential focal adhesion and focal complex protein that colocalizes with dynamic actin structures. Mol Cell Biol. 1996;16:1896–1908. doi: 10.1128/mcb.16.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hardt W-D, Chen L-M, Schuebel K E, Bustelo X R, Galan J E. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 201.Harlan J E, Hajduk P J, Yoon H S, Fesik S W. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature. 1994;371:168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- 202.Harold F M. From morphogens to morphogenesis. Microbiology. 1995;141:2765–2778. doi: 10.1099/13500872-141-11-2765. [DOI] [PubMed] [Google Scholar]
- 203.Harris S D, Hamer L, Sharpless K E, Hamer J E. The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J. 1997;16:3474–3483. doi: 10.1093/emboj/16.12.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hart M J, Callow M G, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996;15:2997–3005. [PMC free article] [PubMed] [Google Scholar]
- 205.Hart M J, Eva A, Evans T, Aaronson S A, Cerione R A. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 206.Hart M J, Eva A, Zangrilli D, Aaronson S A, Evans T, Cerione R A, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- 207.Hart M J, Jiang X, Kozasa T, Roscoe W, Singer W D, Gilman A G, Sternweis P C, Bollag G. Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Gα13. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 208.Hart M J, Maru Y, Leonard D, Witte O N, Evans T, Cerione R A. A GDP dissociation inhibitor that serves as a GTPase inhibitor for the Ras-like protein CDC42Hs. Science. 1992;258:812–815. doi: 10.1126/science.1439791. [DOI] [PubMed] [Google Scholar]
- 209.Hart M J, Polakis P G, Evans T, Cerione R A. The identification and characterization of an epidermal growth factor-stimulated phosphorylation of a specific low molecular weight GTP-binding protein in a reconstituted phospholipid vesicle system. J Biol Chem. 1990;265:5990–6001. [PubMed] [Google Scholar]
- 210.Hart M J, Shinjo K, Hall A, Evans T, Cerione R A. Identification of the human platelet GTPase activating protein for the CDC42Hs protein. J Biol Chem. 1991;266:20840–20848. [PubMed] [Google Scholar]
- 211.Hartwell L H, Culotti J, Pringle J R, Reid B J. Genetic control of the cell division cycle in yeast. A model to account for the order of cell cycle events is deduced from the phenotypes of yeast mutants. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 212.Hartwell L H, Culotti J, Reid B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc Natl Acad Sci USA. 1970;66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hartwell L H, Mortimer R K, Culotti J, Culotti M. Genetic control of the cell division cycle in yeast. V. Genetic analysis of cdc mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.He B, Chen P, Chen S, Vancura K L, Michaelis S, Powers S. RAM2, an essential gene of yeast, and RAM1 encode the two polypeptide components of the farnesyltransferase that prenylates a-factor and Ras proteins. Proc Natl Acad Sci USA. 1991;88:11373–11377. doi: 10.1073/pnas.88.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Heisterkamp N, Kaartinen V, Vansoest S, Bokoch G M, Groffen J. Human ABR encodes a protein with GAP(rac) activity and homology to the DBL nucleotide exchange factor domain. J Biol Chem. 1993;268:16903–16906. [PubMed] [Google Scholar]
- 216.Herskowitz I. Building organs and organisms: elements of morphogenesis exhibited by budding yeast. Cold Spring Harbor Symp Quant Biol. 1997;62:57–63. [PubMed] [Google Scholar]
- 217.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 218.Herskowitz I, Park H-O, Sanders S, Valtz N, Peter M. Programming of cell polarity in budding yeast by endogenous and exogenous signals. Cold Spring Harbor Symp Quant Biol. 1995;60:717–727. doi: 10.1101/sqb.1995.060.01.078. [DOI] [PubMed] [Google Scholar]
- 219.Hildebrand J D, Taylor J M, Parsons J T. An SH3 domain-containing GTPase-activating protein for Rho and Cdc42 associates with focal adhesion kinase. Mol Cell Biol. 1996;16:3169–3178. doi: 10.1128/mcb.16.6.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 221.Hirano H, Tanaka K, Ozaki K, Imamura H, Kohno H, Hihara T, Kameyama T, Hotta K, Arisawa M, Watanabe T, Qadota H, Ohya Y, Takai Y. ROM7/BEM4 encodes a novel protein that interacts with the Rho1p small GTP-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4396–4403. doi: 10.1128/mcb.16.8.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hooley R, Yu C-Y, Symons M, Barber D L. Gα13 stimulates Na+-H+ exchange through distinct Cdc42-dependent and RhoA-dependent pathways. J Biol Chem. 1996;271:6152–6158. doi: 10.1074/jbc.271.11.6152. [DOI] [PubMed] [Google Scholar]
- 223.Hordijk P L, ten Klooster J P, van der Kammen R A, Michiels F, Oomen L C J M, Collard J G. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science. 1997;278:1464–1466. doi: 10.1126/science.278.5342.1464. [DOI] [PubMed] [Google Scholar]
- 224.Horii Y, Beeler J F, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. EMBO J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hotchin N A, Hall A. Regulation of the actin cytoskeleton, integrins and cell growth by the Rho family of small GTPases. Cancer Surv. 1996;27:311–322. [PubMed] [Google Scholar]
- 226.Hu K Q, Settleman J. Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J. 1997;16:473–483. doi: 10.1093/emboj/16.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hu M C T, Qiu W R, Wang Y P. JNK1, JNK2 and JNK3 are p53 N-terminal serine 34 kinases. Oncogene. 1997;15:2277–2287. doi: 10.1038/sj.onc.1201401. [DOI] [PubMed] [Google Scholar]
- 228.Huang S, Chen C S, Ingber D E. Control of cyclin D1, p27Kip1, and cell cycle progression in human capillary endothelial cells by cell shape and cytoskeletal tension. Mol Biol Cell. 1998;9:3179–3193. doi: 10.1091/mbc.9.11.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hynes R O, Lander A D. Contact and adhesive specificities in the associations, migrations, and targeting of cells and axons. Cell. 1992;68:303–322. doi: 10.1016/0092-8674(92)90472-o. [DOI] [PubMed] [Google Scholar]
- 230.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 231.Iino Y, Yamamoto M. Expression pattern of the C. elegans p21-activated protein kinase, CePAK. Biochem Biophys Res Commun. 1998;245:177–184. doi: 10.1006/bbrc.1998.8380. [DOI] [PubMed] [Google Scholar]
- 232.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ip Y T, Davis R J. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 234.Ireton K, Cossart P. Interaction of invasive bacteria with host signaling pathways. Curr Opin Cell Biol. 1998;10:276–283. doi: 10.1016/s0955-0674(98)80151-8. [DOI] [PubMed] [Google Scholar]
- 235.Jakobi R, Chen C J, Tuazon P T, Traugh J A. Molecular cloning and sequencing of the cytostatic G protein-activated protein kinase PAK1. J Biol Chem. 1996;271:6206–6211. doi: 10.1074/jbc.271.11.6206. [DOI] [PubMed] [Google Scholar]
- 236.Jansen R-P, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 237.Jaquenoud M, Gulli M P, Peter K, Peter M. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 1998;17:5360–5373. doi: 10.1093/emboj/17.18.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Jefferies H B J, Thomas G. Ribosomal protein S6 phosphorylation and signal transduction. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 389–409. [Google Scholar]
- 239.Johnson D I. CDC42: a member of the ras superfamily involved in the control of cellular polarity during the Saccharomyces cerevisiae cell cycle. In: Lacal J C, McCormick F, editors. The ras superfamily of GTPases. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 297–312. [Google Scholar]
- 240.Johnson D I, O’Brien J M, Jacobs C W. Isolation and sequence analysis of CDC43, a gene involved in the control of cell polarity in Saccharomyces cerevisiae. Gene. 1991;98:149–150. doi: 10.1016/0378-1119(91)90119-v. [DOI] [PubMed] [Google Scholar]
- 241.Johnson D I, Pringle J R. Cdc42p. In: Zerial M, Huber L A, editors. Guidebook to the small GTPases. Oxford, United Kingdom: Oxford University Press; 1995. pp. 283–287. [Google Scholar]
- 242.Johnson D I, Pringle J R. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Joyal J L, Annan R S, Ho Y D, Huddleston M E, Carr S A, Hart M J, Sacks D B. Calmodulin modulates the interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by nanoelectrospray tandem mass spectrometry. J Biol Chem. 1997;272:15419–15425. doi: 10.1074/jbc.272.24.15419. [DOI] [PubMed] [Google Scholar]
- 244.Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis J H. Bridging Ral GTPases to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 245.Juo P, Kuo C J, Reynolds S E, Konz R F, Raingeaud J, Davis R J, Biemann H-P, Blenis J. Fas activation of the p38 mitogen-activated protein kinase signalling pathway requires ICE/CED-3 family proteases. Mol Cell Biol. 1997;17:24–35. doi: 10.1128/mcb.17.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Jurgens G, Grebe M, Steinmann T. Establishment of cell polarity during early plant development. Curr Opin Cell Biol. 1997;9:849–852. doi: 10.1016/s0955-0674(97)80087-7. [DOI] [PubMed] [Google Scholar]
- 247.Just I, Selzer J, Wilm M, Eichel-Streiber C V, Mann M, Aktories K. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature. 1995;375:500–503. doi: 10.1038/375500a0. [DOI] [PubMed] [Google Scholar]
- 248.Kaga S, Ragg S, Rogers K A, Ochi A. Activation of p21-CDC42/Rac-activated kinases by CD28 signaling: p21-activated kinase (PAK) and MEK kinase 1 (MEKK1) may mediate the interplay between CD3 and CD28 signals. J Immunol. 1998;160:4182–4189. [PubMed] [Google Scholar]
- 249.Kaga S, Ragg S, Rogers K A, Ochi A. Stimulation of CD28 with B7-2 promotes focal adhesion-like cell contacts where Rho family small G proteins accumulate in T cells. J Immunol. 1998;160:24–27. [PubMed] [Google Scholar]
- 250.Kahn R A. PIP2: activator … or terminator of small G proteins? Trends Biochem Sci. 1998;23:99. doi: 10.1016/s0968-0004(98)01177-3. . (Response.) [DOI] [PubMed] [Google Scholar]
- 251.Kamada Y, Qadota H, Python C P, Anraku Y, Ohya Y, Levin D E. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271:9193–9196. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- 252.Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:28341–28345. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- 253.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 254.Kater S B, Rehder V. The sensory role of growth cone filopodia. Curr Opin Neurobiol. 1995;5:68–74. doi: 10.1016/0959-4388(95)80089-1. [DOI] [PubMed] [Google Scholar]
- 255.Katoh H, Aoki J, Yamaguchi Y, Kitano Y, Ichikawa A, Negishi M. Constitutively active Gα12, Gα13, and Gαq induce Rho-dependent neurite retraction through different signaling pathways. J Biol Chem. 1998;273:28700–28707. doi: 10.1074/jbc.273.44.28700. [DOI] [PubMed] [Google Scholar]
- 256.Keely P J, Westwick J K, Whitehead I P, Der C J, Parise L V. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature. 1997;390:632–636. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]
- 257.Kenny D, Cairns L, Remold-O’Donnell E, Peterson J, Rosen F S, Parkman R. Morphological abnormalities in the lymphocytes of patients with the Wiskott-Aldrich syndrome. Blood. 1986;68:1329–1332. [PubMed] [Google Scholar]
- 258.Khosravi-Far R, Solski P A, Clark G J, Kinch M S, Der C J. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Kilmartin J V, Adams A E M. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kim H B, Haarer B K, Pringle J R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kirchhausen T, Rosen F S. Disease mechanism: unravelling Wiskott-Aldrich syndrome. Curr Biol. 1996;6:676–678. doi: 10.1016/s0960-9822(09)00447-3. [DOI] [PubMed] [Google Scholar]
- 262.Kishi K, Sasaki T, Kuroda S, Itoh T, Takai Y. Regulation of cytoplasmic division of Xenopus embryo by rho p21 and its inhibitory GDP/GTP exchange protein (rho GDI) J Cell Biol. 1993;120:1187–1195. doi: 10.1083/jcb.120.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Kitamura Y, Kitamura T, Sakaue H, Maeda T, Ueno H, Nishio S, Ohno S, Osada S, Sakaue M, Ogawa W, Kasuga M. Interaction of Nck-associated protein 1 with activated GTP-binding protein Rac. Biochem J. 1997;322:873–878. doi: 10.1042/bj3220873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Knaus U G, Bokoch G M. The p21Rac/Cdc42-activated kinases (PAKs) Int J Biochem Cell Biol. 1998;30:857–862. doi: 10.1016/s1357-2725(98)00059-4. [DOI] [PubMed] [Google Scholar]
- 265.Knaus U G, Morris S, Dong H-J, Chernoff J, Bokoch G M. Regulation of human leukocyte p21-activated kinases through G protein-coupled receptors. Science. 1995;269:221–223. doi: 10.1126/science.7618083. [DOI] [PubMed] [Google Scholar]
- 266.Knaus U G, Wang Y, Reilly A M, Warnock D, Jackson J H. Structural requirements for PAK activation by Rac GTPases. J Biol Chem. 1998;273:21512–21518. doi: 10.1074/jbc.273.34.21512. [DOI] [PubMed] [Google Scholar]
- 267.Koch G, Tanaka K, Masuda T, Yamochi W, Nonaka H, Takai Y. Association of the Rho family small GTP-binding proteins with Rho GDP dissociation inhibitor (Rho GDI) in Saccharomyces cerevisiae. Oncogene. 1997;15:417–422. doi: 10.1038/sj.onc.1201194. [DOI] [PubMed] [Google Scholar]
- 268.Kohler J R, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, Ohya Y, Takai Y. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- 270.Kolluri R, Shehabeldin A, Peacocke M, Lamhonwah A M, Teichert-Kuliszewska K, Weissman S M, Siminovitch K A. Identification of WASP mutations in patients with Wiskott-Aldrich syndrome and isolated thrombocytopenia reveals allelic heterogeneity at the WAS locus. Hum Mol Genet. 1995;4:1119–1126. doi: 10.1093/hmg/4.7.1119. [DOI] [PubMed] [Google Scholar]
- 271.Kolluri R, Tolias K F, Carpenter C L, Rosen F S, Kirchhausen T. Direct interaction of the Wiskott-Aldrich syndrome protein with the GTPase Cdc42. Proc Natl Acad Sci USA. 1996;93:5615–5618. doi: 10.1073/pnas.93.11.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kondoh O, Tachibana Y, Ohya Y, Arisawa M, Watanabe T. Cloning of the RHO1 gene from Candida albicans and its regulation of β-1,3-glucan synthesis. J Bacteriol. 1997;179:7734–7741. doi: 10.1128/jb.179.24.7734-7741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Kowluru A, Li G D, Rabaglia M E, Segu V B, Hofmann F, Aktories K, Metz S A. Evidence for differential roles of the Rho subfamily of GTP-binding proteins in glucose- and calcium-induced insulin secretion from pancreatic beta cells. Biochem Pharmacol. 1997;54:1097–1108. doi: 10.1016/s0006-2952(97)00314-6. [DOI] [PubMed] [Google Scholar]
- 274.Kowluru A, Seavey S E, Li G, Sorenson R L, Weinhaus A J, Nesher R, Rabaglia M E, Vadakekalam J, Metz S A. Glucose- and GTP-dependent stimulation of the carboxyl methylation of CDC42 in rodent and human pancreatic islets and pure β cells. Evidence for an essential role of GFP-binding proteins in nutrient-induced insulin secretion. J Clin Invest. 1996;98:540–555. doi: 10.1172/JCI118822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kozasa T, Jiang X, Hart M J, Sternweis P M, Singer W D, Gilman A G, Bollag G, Sternweis P C. p115 RhoGEF, a GTPase activating protein for Gα12 and Gα13. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 276.Kozma R, Ahmed S, Best A, Lim L. The GTPase-activating protein n-chimaerin cooperates with Rac1 and Cdc42Hs to induce the formation of lamellipodia and filopodia. Mol Cell Biol. 1996;16:5069–5080. doi: 10.1128/mcb.16.9.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kozma R, Sarner S, Ahmed S, Lim L. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol Cell Biol. 1997;17:1201–1211. doi: 10.1128/mcb.17.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Krapivinsky G, Pu W, Wickman K, Krapivinsky L, Clapham D E. pICln binds to a mammalian homolog of a yeast protein involved in regulation of cell morphology. J Biol Chem. 1998;273:10811–10814. doi: 10.1074/jbc.273.18.10811. [DOI] [PubMed] [Google Scholar]
- 280.Kron S J, Styles C A, Fink G R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Kropf D L. Cytoskeletal control of cell polarity in a plant zygote. Dev Biol. 1994;165:361–371. doi: 10.1006/dbio.1994.1259. [DOI] [PubMed] [Google Scholar]
- 282.Kuroda S, Fukata M, Fujii K, Nakamura T, Izawa I, Kaibuchi K. Regulation of cell-cell adhesion of MDCK cells by Cdc42 and Rac1 small GTPases. Biochem Biophys Res Commun. 1997;240:430–435. doi: 10.1006/bbrc.1997.7675. [DOI] [PubMed] [Google Scholar]
- 283.Kuroda S, Fukata M, Kobayashi K, Nakafuku M, Nomura N, Iwamatsu A, Kaibuchi K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J Biol Chem. 1996;271:23363–23367. doi: 10.1074/jbc.271.38.23363. [DOI] [PubMed] [Google Scholar]
- 284.Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, Shoji I, Matsuura Y, Yonehara S, Kaibuchi K. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin-mediated cell-cell adhesion. Science. 1998;281:832–835. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- 285.Kwan S -P, Hagemann T L, Radtke B E, Blaese R M, Rosen F S. Identification of mutations in the Wiskott-Aldrich syndrome gene and characterization of a polymorphic dinucleotide repeat at DXS6940, adjacent to the disease gene. Proc Natl Acad Sci USA. 1995;92:4706–4710. doi: 10.1073/pnas.92.10.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Kyriakis J, Banerjee P, Nikolakaki E, Dai T, Rubie E, Ahmad M, Avruch J, Woodgett J. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 287.Kyriakis J M, Avruch J. Protein kinase cascades activated by stress and inflammatory cytokines. Bioessays. 1996;18:567–577. doi: 10.1002/bies.950180708. [DOI] [PubMed] [Google Scholar]
- 288.Lacal J C. Regulation of proliferation and apoptosis by Ras and Rho GTPases through specific phospholipid-dependent signaling. FEBS Lett. 1997;410:73–77. doi: 10.1016/s0014-5793(97)00444-4. [DOI] [PubMed] [Google Scholar]
- 289.Lallemand D, Ham J, Garbay S, Bakiri L, Traincard F, Jeannequin O, Pfarr C M, Yaniv M. Stress-activated protein kinases are negatively regulated by cell density. EMBO J. 1998;17:5615–5626. doi: 10.1093/emboj/17.19.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Lamarche N, Hall A. GAPs for rho-related GTPases. Trends Genet. 1994;10:436–440. doi: 10.1016/0168-9525(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 291.Lamarche N, Tapon N, Stowers L, Burbelo P D, Aspenström P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65(PAK) and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 292.Lamarche-Vane N, Hall A. CdGAP, a novel proline-rich GTPase-activating protein for Cdc42 and Rac. J Biol Chem. 1998;273:29172–29177. doi: 10.1074/jbc.273.44.29172. [DOI] [PubMed] [Google Scholar]
- 293.Lancaster C A, Taylor-Harris P M, Self A J, Brill S, van Erp H E, Hall A. Characterization of rhoGAP: A GTPase-activating protein for rho-related small GTPases. J Biol Chem. 1994;269:1137–1142. [PubMed] [Google Scholar]
- 294.Leberer E, Chenevert J, Leeuw T, Harcus D, Herskowitz I, Thomas D Y. Genetic interactions indicate a role for Mdg1p and the SH3 domain protein Bem1p in linking the G-protein mediated yeast pheromone signalling pathway to regulators of cell polarity. Mol Gen Genet. 1996;252:608–621. doi: 10.1007/BF02172407. [DOI] [PubMed] [Google Scholar]
- 295.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A R, Brown A J P, Thomas D Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 298.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall J E, Thomas D Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas D Y. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr Biol. 1997;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- 300.Lee J C, Laydon J T, McDonnell P C, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M J, Heys J R, Landvatter S W, Strickler J E, McLaughlin M M, Siemens I R, Fisher S M, Livi G P, White J R, Adams J L, Young P R. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–745. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 301.Lee S -F, Mahasneh A, de la Roche M, Côté G P. Regulation of the p21-activated kinase-related Dictyostelium myosin I heavy chain kinase by autophosphorylation, acidic phospholipids, and Ca2+-calmodulin. J Biol Chem. 1998;273:27911–27917. doi: 10.1074/jbc.273.43.27911. [DOI] [PubMed] [Google Scholar]
- 302.Lee S F, Egelhoff T T, Mahasneh A, Côté G P. Cloning and characterization of a Dictyostelium myosin I heavy chain kinase activated by Cdc42 and Rac. J Biol Chem. 1996;271:27044–27048. doi: 10.1074/jbc.271.43.27044. [DOI] [PubMed] [Google Scholar]
- 303.Leeuw T, Fourest-Lieuvin A, Wu C, Chenevert J, Clark K, Whiteway M, Thomas D Y, Leberer E. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science. 1995;270:1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- 304.Leeuw T, Wu C L, Schrag J D, Whiteway M, Thomas D Y, Leberer E. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- 305.Leffers H, Nielsen M S, Andersen A H, Honore B, Madsen P, Vandekerckhove J, Celis J E. Identification of two human rho GDP dissociation inhibitor proteins whose overexpression leads to disruption of the actin cytoskeleton. Exp Cell Res. 1993;209:165–174. doi: 10.1006/excr.1993.1298. [DOI] [PubMed] [Google Scholar]
- 306.Lelias J-M, Adra C N, Wulf G M, Guillemot J-C, Khagad M, Caput D, Lim B. cDNA cloning of a human mRNA preferentially expressed in hematopoietic cells and with homology to a GDP-dissociation inhibitor for the rho GTP-binding proteins. Proc Natl Acad Sci USA. 1993;90:1479–1483. doi: 10.1073/pnas.90.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Leonard D, Hart M J, Platko J V, Eva A, Henzel W, Evans T, Cerione R A. The identification and characterization of a GDP-dissociation inhibitor (GDI) for the CDC42Hs protein. J Biol Chem. 1992;267:22860–22868. [PubMed] [Google Scholar]
- 308.Leonard D A, Evans T, Hart M, Cerione R A, Manor D. Investigation of the GTP-binding/GTPase cycle of Cdc42Hs using fluorescence spectroscopy. Biochemistry. 1994;33:12323–12328. doi: 10.1021/bi00206a040. [DOI] [PubMed] [Google Scholar]
- 309.Leonard D A, Lin R, Cerione R A, Manor D. Biochemical studies of the mechanism of action of the Cdc42-GTPase-activating protein. J Biol Chem. 1998;273:16210–16215. doi: 10.1074/jbc.273.26.16210. [DOI] [PubMed] [Google Scholar]
- 310.Leonard D A, Satoskar R S, Wu W J, Bagrodia S, Cerione R A, Manor D. Use of a fluorescence spectroscopic readout to characterize the interactions of Cdc42Hs with its target/effector, mPAK-3. Biochem. 1997;36:1173–1180. doi: 10.1021/bi9622837. [DOI] [PubMed] [Google Scholar]
- 311.Leung T, Chen X-Q, Tan I, Manser E, Lim L. Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol Cell Biol. 1998;18:130–140. doi: 10.1128/mcb.18.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Levi N L, Hanoch T, Benard O, Rozenblat M, Harris D, Reiss N, Naor Z, Seger R. Stimulation of jun N-terminal kinase (JNK) by gonadotropin-releasing hormone in pituitary αT3-1 cell line is mediated by protein kinase C, c-Src, and CDC42. Mol Endocrinol. 1998;12:815–824. doi: 10.1210/mend.12.6.0120. [DOI] [PubMed] [Google Scholar]
- 313.Lew D J, Reed S I. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Lew D J, Reed S I. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 315.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Lew D J, Weinert T, Pringle J R. Cell cycle control in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. 3. Cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 607–695. [Google Scholar]
- 317.Li E, Stupack D, Bokoch G M, Nemerow G R. Adenovirus endocytosis requires actin cytoskeleton reorganization mediated by Rho family GTPases. J Virol. 1998;72:8806–8812. doi: 10.1128/jvi.72.11.8806-8812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Li G, Kowluru A, Metz S A. Characterization of prenyl-cysteine methyl transferase activity in insulin-secreting cells. Biochem J. 1996;316:345–351. doi: 10.1042/bj3160345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Li H, Wu G, Ware D, Davis K R, Yang Z B. Arabidopsis Rho-related GTPases: differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 1998;118:407–417. doi: 10.1104/pp.118.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Li J Z, Smithgall T E. Fibroblast transformation by Fps/Fes tyrosine kinases requires Ras, Rac, and Cdc42 and induces extracellular signal-regulated and c-Jun N-terminal kinase activation. J Biol Chem. 1998;273:13828–13834. doi: 10.1074/jbc.273.22.13828. [DOI] [PubMed] [Google Scholar]
- 321.Li R. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Li R, Zhang B, Zheng Y. Structural determinants required for the interaction between Rho GTPase and the GTPase-activating domain of p190. J Biol Chem. 1997;272:32830–32835. doi: 10.1074/jbc.272.52.32830. [DOI] [PubMed] [Google Scholar]
- 323.Li R, Zheng Y. Residues of the Rho family GTPases Rho and Cdc42 that specify sensitivity to Dbl-like guanine nucleotide exchange factors. J Biol Chem. 1997;272:4671–4679. doi: 10.1074/jbc.272.8.4671. [DOI] [PubMed] [Google Scholar]
- 324.Li R, Zheng Y, Drubin D G. Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J Cell Biol. 1995;128:599–615. doi: 10.1083/jcb.128.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Lim L, Hall C, Monfries C. Regulation of actin cytoskeleton by Rho-family GTPases and their associated proteins. Semin Cell Dev Biol. 1996;7:699–706. [Google Scholar]
- 326.Lim L, Manser E, Leung T, Hall C. Regulation of phosphorylation pathways by p21 GTPases: the p21 Ras-related Rho subfamily and its role in phosphorylation signalling pathways. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 327.Lin R, Bagrodia S, Cerione R, Manor D. A novel Cdc42Hs mutant induces cellular transformation. Curr Biol. 1997;7:794–797. doi: 10.1016/s0960-9822(06)00338-1. [DOI] [PubMed] [Google Scholar]
- 328.Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Liu H, Köhler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 330.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 331.Liu X, Wang H, Eberstadt M, Schnuchel A, Olejniczak E T, Meadows R P, Schkeryantz J M, Janowick D A, Harlan J E, Harris E A S, Staunton D E, Fesik S W. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell. 1998;95:269–277. doi: 10.1016/s0092-8674(00)81757-2. [DOI] [PubMed] [Google Scholar]
- 332.Longtine M S, DeMarini D J, Valencik M L, Al-Awar O S, Fares H, DeVirgilio C, Pringle J R. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- 333.Longtine M S, Fares H, Pringle J R. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Lu W, Katz S, Gupta R, Mayer B J. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr Biol. 1997;7:85–94. doi: 10.1016/s0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- 335.Lu X B, Wu X N, Plemenitas A, Yu H F, Sawai E T, Abo A, Peterlin B M. CDC42 and Rac1 are implicated in the activation of the Nef-associated kinase and replication of HIV-1. Curr Biol. 1996;6:1677–1684. doi: 10.1016/s0960-9822(02)70792-6. [DOI] [PubMed] [Google Scholar]
- 336.Luo L, Liao Y J, Jan L Y, Jan Y N. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 337.Luo L Q, Jan L Y, Jan Y-N. Rho family small GTP-binding proteins in growth cone signalling. Curr Opin Neurobiol. 1997;7:81–86. doi: 10.1016/s0959-4388(97)80124-9. [DOI] [PubMed] [Google Scholar]
- 338.Luo L Q, Lee T, Tsai L, Tang G, Jan L Y, Jan Y N. Genghis Khan (Gek) as a putative effector for Drosophila Cdc42 and regulator of actin polymerization. Proc Natl Acad Sci USA. 1997;94:12963–12968. doi: 10.1073/pnas.94.24.12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Lyons D M, Mahanty S K, Choi K-Y, Manandhar M, Elion E A. The SH3-domain protein Bem1 coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4095–4106. doi: 10.1128/mcb.16.8.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Ma L, Cantley L C, Janmey P A, Kirschner M W. Corequirement of specific phosphoinositides and small GTP-binding protein Cdc42 in inducing actin assembly in Xenopus egg extracts. J Cell Biol. 1998;140:1125–1136. doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Ma X-J, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 342.Maas R L, Jepeal L I, Elfering S L, Holcombe R F, Morton C C, Eddy R L, Byers M G, Shows T B, Leder P. A human gene homologous to the formin gene residing at the murine limb deformity locus: chromosomal location and RFLPs. Am J Hum Genet. 1991;48:687–695. [PMC free article] [PubMed] [Google Scholar]
- 343.Mack D, Nishimura K, Dennehey B K, Arbogast T, Parkinson J, Toh-E A, Pringle J R, Bender A, Matsui Y. Identification of the bud emergence gene BEM4 and its interactions with Rho-type GTPases in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4387–4395. doi: 10.1128/mcb.16.8.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Mackay D J G, Hall A. Rho GTPases. J Biol Chem. 1998;273:20685–20688. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- 345.Mackay D J G, Nobes C D, Hall A. The Rho’s progress: a potential role during neuritogenesis for the Rho family of GTPases. Trends Neurosci. 1995;18:496–501. doi: 10.1016/0166-2236(95)92773-j. [DOI] [PubMed] [Google Scholar]
- 346.Madden K, Costigan C, Snyder M. Cell polarity and morphogenesis in Saccharomyces cerevisiae. Trends Cell Biol. 1992;2:22–29. doi: 10.1016/0962-8924(92)90140-i. [DOI] [PubMed] [Google Scholar]
- 347.Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- 348.Madhani H D, Fink G R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 349.Madhani H D, Fink G R. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- 350.Madhani H D, Fink G R. The riddle of MAP kinase signaling specificity. Trends Genet. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- 351.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 352.Malathi K, Ganesan K, Datta A. Identification of a putative transcription factor in Candida albicans that can complement the mating defect of Saccharomyces cerevisiae ste12 mutants. J Biol Chem. 1994;269:22945–22951. [PubMed] [Google Scholar]
- 353.Malcolm K C, Ross A H, Qiu R-G, Symons M, Exton J H. Activation of rat liver phospholipase D by the small GTP-binding protein RhoA. J Biol Chem. 1994;269:25951–25954. [PubMed] [Google Scholar]
- 354.Maltese W A, Sheridan K M. Isoprenoid modification of G25K (Gp), a low molecular mass GTP-binding protein distinct from p21ras. J Biol Chem. 1990;265:17883–17890. [PubMed] [Google Scholar]
- 355.Manser E, Chong C, Zhao Z S, Leung T, Michael G, Hall C, Lim L. Molecular cloning of a new member of the p21-Cdc42/Rac-activated kinase (PAK) family. J Biol Chem. 1995;270:25070–25078. doi: 10.1074/jbc.270.42.25070. [DOI] [PubMed] [Google Scholar]
- 356.Manser E, Huang H-Y, Loo T-H, Chen X-Q, Dong J-M, Leung T, Lim L. Expression of constitutively active α-PAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Manser E, Leung T, Monfries C, Teo M, Hall C, Lim L. Diversity and versatility of GTPase activating proteins for the p21 rho subfamily of ras G proteins detected by a novel overlay assay. J Biol Chem. 1992;267:16025–16028. [PubMed] [Google Scholar]
- 358.Manser E, Leung T, Salihuddin H, Tan L, Lim L. A non-receptor tyrosine kinase that inhibits the GTPase activity of p21cdc42. Nature. 1993;363:364–367. doi: 10.1038/363364a0. [DOI] [PubMed] [Google Scholar]
- 359.Manser E, Leung T, Salihuddin H, Zhao Z-S, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 360.Manser E, Loo T-H, Koh C-G, Zhao Z-S, Chen X-W, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 361.Mao J, Yuan H, Xie W, Wu D. Guanine nucleotide exchange factor GEF115 specifically mediates activation of Rho and serum response factor by the G protein α subunit Gα13. Proc Natl Acad Sci USA. 1998;95:12973–12976. doi: 10.1073/pnas.95.22.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Mao J H, Xie W, Yuan H D, Simon M I, Mano H, Wu D Q. Tec/Bmx non-receptor tyrosine kinases are involved in regulation of Rho and serum response factor by Gα12/13. EMBO J. 1998;17:5638–5646. doi: 10.1093/emboj/17.19.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Mao J H, Yuan H D, Xie W, Simon M I, Wu D Q. Specific involvement of G proteins in regulation of serum response factor-mediated gene transcription by different receptors. J Biol Chem. 1998;273:27118–27123. doi: 10.1074/jbc.273.42.27118. [DOI] [PubMed] [Google Scholar]
- 364.Marcus S, Polverino A, Chang E, Robbins D, Cobb M H, Wigler M H. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signaling module in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1995;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Marhoul J F, Adams T H. Identification of developmental regulatory genes in Aspergillus nidulans by overexpression. Genetics. 1995;139:537–547. doi: 10.1093/genetics/139.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Marks J, Hagan I M, Hyams J S. Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J Cell Sci Suppl. 1986;5:229–241. doi: 10.1242/jcs.1986.supplement_5.15. [DOI] [PubMed] [Google Scholar]
- 367.Marks P W, Kwiatkowski D J. Genomic organization and chromosomal location of murine Cdc42. Genomics. 1996;38:13–18. doi: 10.1006/geno.1996.0586. [DOI] [PubMed] [Google Scholar]
- 368.Marsh L, Rose M D. The pathway of cell and nuclear fusion during mating in S. cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 827–888. [Google Scholar]
- 369.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 370.Marshall M S. The effector interactions of p21 ras. Trends Biochem Sci. 1993;18:250–254. doi: 10.1016/0968-0004(93)90175-m. [DOI] [PubMed] [Google Scholar]
- 371.Martin H, Mendoza A, Rodriguez-Pachon J M, Molina M, Nombela C. Characterization of SKM1, a Saccharomyces cerevisiae gene encoding a novel Ste20/PAK-like protein kinase. Mol Microbiol. 1997;23:431–444. doi: 10.1046/j.1365-2958.1997.d01-1870.x. [DOI] [PubMed] [Google Scholar]
- 372.Martin T F J. Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu Rev Cell Dev Biol. 1998;14:231–264. doi: 10.1146/annurev.cellbio.14.1.231. [DOI] [PubMed] [Google Scholar]
- 373.Masuda T, Tanaka K, Nonaka H, Yamochi W, Maeda A, Takai Y. Molecular cloning and characterization of yeast rho GDP dissociation inhibitor. J Biol Chem. 1994;269:19713–19718. [PubMed] [Google Scholar]
- 374.Matsui Y, Matsui R, Akada R, Toh-e A. Yeast src homology region 3 domain-binding proteins involved in bud formation. J Cell Biol. 1996;133:865–878. doi: 10.1083/jcb.133.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Matsui Y, Toh-e A. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol Cell Biol. 1992;12:5690–5699. doi: 10.1128/mcb.12.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.May J W, Mitchison J M. Pattern of polar extension of the cell wall in the fission yeast Schizosaccharomyces pombe. Can J Microbiol. 1995;41:273–277. doi: 10.1139/m95-037. [DOI] [PubMed] [Google Scholar]
- 377.Mayer M L, Caplin B E, Marshall M S. CDC43 and RAM2 encode the polypeptide subunits of a yeast type I protein geranylgeranyltransferase. J Biol Chem. 1992;267:20589–20593. [PubMed] [Google Scholar]
- 378.McCallum S J, Erickson J W, Cerione R A. Characterization of the association of the actin-binding protein, IQGAP, and activated Cdc42 with Golgi membranes. J Biol Chem. 1998;273:22537–22544. doi: 10.1074/jbc.273.35.22537. [DOI] [PubMed] [Google Scholar]
- 379.McCallum S J, Wu W J, Cerione R A. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J Biol Chem. 1996;271:21732–21737. doi: 10.1074/jbc.271.36.21732. [DOI] [PubMed] [Google Scholar]
- 380.McCormick F. Signal transduction—why Ras needs Rho. Nature. 1998;394:220–221. doi: 10.1038/28268. [DOI] [PubMed] [Google Scholar]
- 381.McMillan J N, Sia R A L, Lew D J. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381a.Merla, A., and D. I. Johnson. Unpublished results.
- 382.Meyerowitz E M. Genetic control of cell division patterns in developing plants. Cell. 1997;88:299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- 383.Michelitch M, Chant J. A mechanism of Bud1p GTPase action suggested by mutational analysis and immunolocalization. Curr Biol. 1996;6:446–454. doi: 10.1016/s0960-9822(02)00512-2. [DOI] [PubMed] [Google Scholar]
- 384.Michelsen J W, Schmeichel K L, Beckerle M C, Winge D R. The LIM domain defines a specific zinc-binding protein domain. Proc Natl Acad Sci USA. 1993;90:4404–4408. doi: 10.1073/pnas.90.10.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Michiels F, Habets G G, Stam J C, van der Kammen R A, Collard J G. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 386.Michiels F, Stam J C, Hordijk P L, van der Kammen R A, Stalle L R-V, Feltkamp C A, Collard J G. Regulated membrane localization of Tiam1, mediated by the NH2-terminal pleckstrin homology domain, is required for Rac-dependent membrane ruffling and C-Jun NH2-terminal kinase activation. J Cell Biol. 1997;137:387–398. doi: 10.1083/jcb.137.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 388.Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 389.Miki T, Smith C L, Long J E, Eva A, Fleming T P. Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature. 1993;362:462–465. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- 390.Miller P, Johnson D I. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Miller P J, Johnson D I. Characterization of the S. cerevisiae cdc42-1ts allele and new temperature-conditional-lethal cdc42 alleles. Yeast. 1997;13:561–572. doi: 10.1002/(SICI)1097-0061(199705)13:6<561::AID-YEA114>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 392.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signaling pathway and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 393.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 394.Mirbod F, Nakashima S, Kitajima Y, Cannon R D, Nozawa Y. Molecular cloning of a Rho family, CDC42Ca gene from Candida albicans and its mRNA expression changes during morphogenesis. J Med Vet Mycol. 1997;35:173–179. [PubMed] [Google Scholar]
- 395.Miura Y, Kikuchi A, Musha T, Kuroda S, Yaku H, Sasaki T, Takai Y. Regulation of morphology by rho p21 and its inhibitory GDP/GTP exchange protein (rho GDI) in Swiss 3T3 cells. J Biol Chem. 1993;268:510–515. [PubMed] [Google Scholar]
- 396.Miyamoto S, Ohya Y, Ohsumi Y, Anraku Y. Nucleotide sequence of the CLS4 (CDC24) gene of Saccharomyces cerevisiae. Gene. 1987;54:125–132. doi: 10.1016/0378-1119(87)90354-4. [DOI] [PubMed] [Google Scholar]
- 397.Miyamoto S, Ohya Y, Sano Y, Sakaguchi S, Iida H, Anraku Y. A DBL-homologous region of the yeast CLS4/CDC24 gene product is important for Ca2+-modulated bud assembly. Biochem Biophys Res Commun. 1991;181:604–610. doi: 10.1016/0006-291x(91)91233-3. [DOI] [PubMed] [Google Scholar]
- 398.Mizunuma M, Hirata D, Miyahara K, Tsuchiya E, Miyakawa T. Role of calcineurin and Mpk1 in regulating the onset of mitosis in budding yeast. Nature. 1998;392:303–306. doi: 10.1038/32695. [DOI] [PubMed] [Google Scholar]
- 399.Molina I J, Kenney D M, Rosen F S, Remold-O’Donnell E. T cell lines characterize events in the pathogenesis of the Wiskott-Aldrich syndrome. J Exp Med. 1992;176:867–874. doi: 10.1084/jem.176.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Molnar A, Theodoras A M, Zon L I, Kyriakis J M. Cdc42Hs, but not Rac1, inhibits serum-stimulated cell cycle progression at G1/S through a mechanism requiring p38/RK. J Biol Chem. 1997;272:13229–13235. doi: 10.1074/jbc.272.20.13229. [DOI] [PubMed] [Google Scholar]
- 401.Monfar M, Lemon K P, Grammer T C, Cheatham L, Chung J, Vlahos C J, Blenis J. Activation of p70/p85 S6 kinases in interleukin-2-responsive lymphoid cells is mediated by phosphatidylinositol 3-kinase and inhibited by cyclic AMP. Mol Cell Biol. 1995;15:326–337. doi: 10.1128/mcb.15.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Moreau V, Way M. Cdc42 is required for membrane dependent actin polymerization in vitro. FEBS Lett. 1998;427:353–356. doi: 10.1016/s0014-5793(98)00443-8. [DOI] [PubMed] [Google Scholar]
- 403.Morii N, Kawano K, Sekine A, Yamada T, Narumiya S. Purification of GTPase-activating protein specific for the rho gene products. J Biol Chem. 1991;266:7646–7650. [PubMed] [Google Scholar]
- 404.Mösch H-U, Fink G R. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Mösch H-U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Muller R T, Honnert U, Reinhard J, Bahler M. The rat myosin myr 5 is a GTPase-activating protein for Rho in vivo: essential role of arginine 1695. Mol Biol Cell. 1997;8:2039–2053. doi: 10.1091/mbc.8.10.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Munemitsu S, Innis M A, Clark R, McCormick F, Ullrich A, Polakis P. Molecular cloning and expression of a G25K cDNA, the human homolog of the yeast cell cycle gene CDC42. Mol Cell Biol. 1990;10:5977–5982. doi: 10.1128/mcb.10.11.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Murphy A M, Montell D J. Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J Cell Biol. 1996;133:617–630. doi: 10.1083/jcb.133.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Na S, Chuang T H, Turi T G, Hanke J H, Bokoch G M, Danley D E. D4-GDI, a substrate of CPP32 is proteolyzed during Fas-induced apoptosis. J Biol Chem. 1996;271:11209–11213. doi: 10.1074/jbc.271.19.11209. [DOI] [PubMed] [Google Scholar]
- 410.Nagata K, Driessens M, Lamarche N, Gorski J L, Hall A. Activation of G1 progression, JNK mitogen-activated protein kinase, and actin filament assembly by the exchange factor FGD1. J Biol Chem. 1998;273:15453–15457. doi: 10.1074/jbc.273.25.15453. [DOI] [PubMed] [Google Scholar]
- 411.Nagata K-I, Puls A, Futter C, Aspenström P, Schaefer E, Nakata T, Hirokawa N, Hall A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 1998;17:149–158. doi: 10.1093/emboj/17.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Nakanishi H, Obaishi H, Satoh A, Wada M, Mandai K, Satoh K, Nishioka H, Matsuura Y, Mizoguchi A, Takai Y. Neurabin: a novel neural tissue-specific actin filament-binding protein involved in neurite formation. J Cell Biol. 1997;139:951–961. doi: 10.1083/jcb.139.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Naqvi S N, Zahn R, Mitchell D A, Stevenson B J, Munn A L. The WASp homologue Las17p functions with the WIP homologue End5p/verprolin and is essential for endocytosis in yeast. Curr Biol. 1998;8:959–962. doi: 10.1016/s0960-9822(98)70396-3. [DOI] [PubMed] [Google Scholar]
- 413a.Nassar, N., and R. Cerione. Personal communication.
- 414.Naumann M, Rudel T, Wieland B, Bartsch C, Meyer T E. Coordinate activation of activator protein 1 and inflammatory cytokines in response to Neisseria gonorrhoeae epithelial cell contact involves stress response kinases. J Exp Med. 1998;188:1277–1286. doi: 10.1084/jem.188.7.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Nemoto S, Sheng Z L, Lin A N. Opposing effects of Jun kinase and p38 mitogen-activated protein kinases on cardiomyocyte hypertrophy. Mol Cell Biol. 1998;18:3518–3526. doi: 10.1128/mcb.18.6.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Nern A, Arkowitz R A. A GTP-exchange factor required for cell orientation. Nature. 1998;391:195–198. doi: 10.1038/34458. [DOI] [PubMed] [Google Scholar]
- 417.Nishina H, Fischer K D, Radvanyl L, Shahinian A, Hakem R, Ruble E A, Bernstein A, Mak T W, Woodgett J R, Penninger J M. Stress-signaling kinase Sek1 protects thymocytes from apoptosis mediated by CD95 and CD3. Nature. 1997;385:350–353. doi: 10.1038/385350a0. [DOI] [PubMed] [Google Scholar]
- 418.Nishiyama T, Sasaki T, Takaishi K, Kato M, Yaku H, Araki K, Matsuura Y, Takai Y. rac p21 is involved in insulin-induced membrane ruffling and rho p21 is involved in hepatocyte growth factor- and 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced membrane ruffling in KB cells. Mol Cell Biol. 1994;14:2447–2456. doi: 10.1128/mcb.14.4.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Nobes C D, Hall A. Rho, rac and cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem Soc Trans. 1995;23:456–459. doi: 10.1042/bst0230456. [DOI] [PubMed] [Google Scholar]
- 420.Nobes C D, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fiber, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 421.Noël J, Pouysségur J. Hormonal regulation, pharmacology, and membrane sorting of vertebrate Na+/H+ exchanger isoforms. Am J Physiol. 1995;268:C283–C296. doi: 10.1152/ajpcell.1995.268.2.C283. [DOI] [PubMed] [Google Scholar]
- 422.Nomanbhoy T K, Cerione R A. Characterization of the interaction between RhoGDI and Cdc42Hs using fluorescence spectroscopy. J Biol Chem. 1996;271:10004–10009. doi: 10.1074/jbc.271.17.10004. [DOI] [PubMed] [Google Scholar]
- 423.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- 425.Nurse P. Fission yeast morphogenesis—posing the problems. Mol Biol Cell. 1994;5:613–616. doi: 10.1091/mbc.5.6.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Obaishi H, Nakanishi H, Mandai K, Satoh K, Satoh A, Takahashi K, Miyahara M, Nishioka H, Takaishi K, Takai Y. Frabin, a novel FGD1-related actin filament-binding protein capable of changing cell shape and activating c-Jun N-terminal kinase. J Biol Chem. 1998;273:18697–18700. doi: 10.1074/jbc.273.30.18697. [DOI] [PubMed] [Google Scholar]
- 427.Ochs H D. The Wiskott-Aldrich syndrome. Semin Hematol. 1998;35:332–345. [PubMed] [Google Scholar]
- 428.Ochs H D, Slichter S J, Harker L A, Von B W, Clark R A, Wedgwood R J. The Wiskott-Aldrich syndrome: studies of lymphocytes, granulocytes, and platelets. Blood. 1980;55:243–252. [PubMed] [Google Scholar]
- 429.Oda A, Ochs H D, Druker B J, Ozaki K, Watanabe C, Handa M, Miyakawa Y, Ikeda Y. Collagen induces tyrosine phosphorylation of Wiskott-Aldrich syndrome protein in human platelets. Blood. 1998;92:1852–1858. [PubMed] [Google Scholar]
- 430.Oehlen L J W M, Cross F R. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- 431.Oehlen L J W M, Cross F R. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J Biol Chem. 1998;273:25089–25097. doi: 10.1074/jbc.273.39.25089. [DOI] [PubMed] [Google Scholar]
- 432.Oehlen L J W M, Cross F R. The role of Cdc42 in signal transduction and mating of the budding yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:8556–8559. doi: 10.1074/jbc.273.15.8556. [DOI] [PubMed] [Google Scholar]
- 433.Offermanns S, Mancino V, Revel J-P, Simon M I. Vascular system defects and impaired cell chemokinesis as a result of Gα13 deficiency. Science. 1997;275:533–536. doi: 10.1126/science.275.5299.533. [DOI] [PubMed] [Google Scholar]
- 434.Ohya Y, Caplin B E, Qadota H, Tibbetts M F, Anraku Y, Pringle J R, Marshall M S. Mutational analysis of the β-subunit of yeast geranylgeranyl transferase I. Mol Gen Genet. 1996;252:1–10. [PubMed] [Google Scholar]
- 435.Ohya Y, Goebl M, Goodman L E, Petersen-Bjfrn S, Friesen J D, Tamanoi F, Anraku Y. Yeast CAL1 is a structural and functional homologue to the DPR1 (RAM) gene involved in ras processing. J Biol Chem. 1991;266:12356–12360. [PubMed] [Google Scholar]
- 436.Ohya Y, Miyamoto S, Oshumi Y, Anraku Y. Calcium-sensitive cls4 mutant of S. cerevisiae with a defect in bud formation. J Bacteriol. 1986;165:28–33. doi: 10.1128/jb.165.1.28-33.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Ohya Y, Ohsumi Y, Anraku Y. Genetic study of the role of calcium ions in the cell division cycle of Saccharomyces cerevisiae: a calcium-dependent mutant and its trifluoperazine-dependent pseudorevertants. Mol Gen Genet. 1984;193:389–394. doi: 10.1007/BF00382073. [DOI] [PubMed] [Google Scholar]
- 438.Ohya Y, Oshumi Y, Anraku Y. Isolation and characterization of Ca2+-sensitive mutants of Saccharomyces cerevisiae. J Gen Microbiol. 1986;132:979–988. doi: 10.1099/00221287-132-4-979. [DOI] [PubMed] [Google Scholar]
- 439.Ohya Y, Qadota H, Anraku Y, Pringle J R, Botstein D. Suppression of yeast geranylgeranyl transferase I defect by alternative prenylation of two target GTPases, Rho1p and Cdc42p. Mol Biol Cell. 1993;4:1017–1025. doi: 10.1091/mbc.4.10.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Olenik C, Barth H, Just I, Aktories K, Meyer D K. Gene expression of the small GTP-binding proteins RhoA, RhoB, Rac1, and Cdc42 in adult rat brain. Mol Brain Res. 1997;52:263–269. doi: 10.1016/s0169-328x(97)00270-2. [DOI] [PubMed] [Google Scholar]
- 441.Olson M F. Guanine nucleotide exchange factors for the Rho GTPases: a role in human disease? J Mol Med. 1996;74:563–571. doi: 10.1007/s001090050060. [DOI] [PubMed] [Google Scholar]
- 442.Olson M F, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 443.Olson M F, Pasteris N G, Gorski J L, Hall A. Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho GTPases. Curr Biol. 1996;6:1628–1633. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- 444.Olson M F, Paterson H F, Marshall C J. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature. 1998;394:295–299. doi: 10.1038/28425. [DOI] [PubMed] [Google Scholar]
- 445.Osada S, Izawa M, Koyama T, Hirai S, Ohno S. A domain containing the Cdc42/Rac interactive binding (CRIB) region of p65PAK inhibits transcriptional activation and cell transformation mediated by the Ras-Rac pathway. FEBS Lett. 1997;404:227–233. doi: 10.1016/s0014-5793(97)00139-7. [DOI] [PubMed] [Google Scholar]
- 446.Osman M A, Cerione R A. Iqg1p, a yeast homologue of the mammalian IQGAPs, mediates Cdc42p effects on the actin cytoskeleton. J Cell Biol. 1998;142:443–455. doi: 10.1083/jcb.142.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Ottilie S, Miller P J, Johnson D I, Creasy C L, Sells M A, Bagrodia S, Forsburg S L, Chernoff J. Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 1995;14:5908–5919. doi: 10.1002/j.1460-2075.1995.tb00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Park H-O, Chant J S. Bud2p. In: Zerial M, Huber L A, editors. Guidebook to the small GTPases. Oxford, United Kingdom: Oxford University Press; 1995. pp. 200–203. [Google Scholar]
- 449.Park H O, Bi E, Pringle J R, Herskowitz I. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci USA. 1997;94:4463–4468. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Park H O, Chant J, Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365:269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- 451.Park S H, Weinberg R A. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene. 1995;11:2349–2355. [PubMed] [Google Scholar]
- 452.Pasteris N G, Buckler J, Cadle A B, Gorski J L. Genomic organization of the faciogenital dysplasia (FGD1; Aarskog syndrome) gene. Genomics. 1997;43:390–394. doi: 10.1006/geno.1997.4837. [DOI] [PubMed] [Google Scholar]
- 453.Pasteris N G, Cadle A, Logie L J, Porteous M E M, Glover T W, Wilroy R S, et al. Isolation and characterisation of the faciogenital dysplasia (Aarskog-Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell. 1994;79:669–678. doi: 10.1016/0092-8674(94)90552-5. [DOI] [PubMed] [Google Scholar]
- 454.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 455.Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal J C. Activation of the nuclear factor-κ B by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 456.Peter M, Neiman A M, Park H-O, vanLohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 457.Petersen J, Weilguny D, Egel R, Nielsen O. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol Cell Biol. 1995;15:3697–3707. doi: 10.1128/mcb.15.7.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Platko J V, Leonard D A, Adra C N, Shaw R J, Cerione R A, Lim B. A single residue can modify target-binding affinity and activity of the functional domain of the Rho-subfamily GDP dissociation inhibitors. Proc Natl Acad Sci USA. 1995;92:2974–2978. doi: 10.1073/pnas.92.7.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Polakis P, McCormick F. Structural requirements for the interaction of p21ras with GAP, exchange factors, and its biological effector target. J Biol Chem. 1993;268:9157–9160. [PubMed] [Google Scholar]
- 461.Polakis P G, Snyderman R, Evans T. Characterization of G25K, a GTP-binding protein containing a novel putative nucleotide binding domain. Biochem Biophys Res Commun. 1989;160:25–32. doi: 10.1016/0006-291x(89)91615-x. [DOI] [PubMed] [Google Scholar]
- 462.Powers S, Gonzales E, Christensen T, Cubert J, Broek D. Functional cloning of BUD5, a CDC25-related gene from S. cerevisiae that can suppress a dominant-negative RAS2 mutant. Cell. 1991;65:1225–1231. doi: 10.1016/0092-8674(91)90017-s. [DOI] [PubMed] [Google Scholar]
- 463.Price L S, Leng J, Schwartz M A, Bokoch G M. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol Biol Cell. 1998;9:1863–1871. doi: 10.1091/mbc.9.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Priess J R. Establishment of initial asymmetry in early Caenorhabditis elegans embryos. Curr Opin Genet Dev. 1994;4:563–568. doi: 10.1016/0959-437x(94)90073-c. [DOI] [PubMed] [Google Scholar]
- 465.Pringle J R, Bi E, Harkins H A, Zahner J E, De Virgilio C, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harbor Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- 466.Qiu R G, Abo A, McCormick F, Symons M. Cdc42 regulates anchorage-independent growth and is necessary for Ras transformation. Mol Cell Biol. 1997;17:3449–3458. doi: 10.1128/mcb.17.6.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Quatrano R S. Cortical asymmetries direct the establishment of cell polarity and the plane of cell division in the Fucus embryo. Cold Spring Harbor Symp Quant Biol. 1997;62:65–70. [PubMed] [Google Scholar]
- 468.Quilliam L A, Lambert Q T, Mickelson-Young L A, Westwick J K, Sparks A B, Kay B K, Jenkins N A, Gilbert D J, Copeland N G, Der C J. Isolation of a NCK-associated kinase, PRK2, an SH3-binding protein and potential effector of Rho protein signaling. J Biol Chem. 1996;271:28772–28776. doi: 10.1074/jbc.271.46.28772. [DOI] [PubMed] [Google Scholar]
- 469.Ramer S W, Davis R W. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Ramesh N, Anton I M, Hartwig J H, Geha R S. WIP, a protein associated with Wiskott-Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc Natl Acad Sci USA. 1997;94:14671–14676. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Regazzi R, Kikuchi A, Takai Y, Wollheim C B. The small GTP-binding proteins in the cytosol of insulin-secreting cells are complexed to GDP dissociation inhibitor proteins. J Biol Chem. 1992;267:17512–17519. [PubMed] [Google Scholar]
- 472.Reid B J, Hartwell L H. Regulation of mating in the cell cycle of Saccharomyces cerevisiae. J Cell Biol. 1977;75:355–365. doi: 10.1083/jcb.75.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Reif K, Cantrell D A. Networking Rho family GTPases in lymphocytes. Immunity. 1998;8:395–401. doi: 10.1016/s1074-7613(00)80545-2. [DOI] [PubMed] [Google Scholar]
- 474.Reinhard J, Scheel A A, Diekmann D, Hall A, Ruppert C, Bähler M. A novel type of myosin implicated in signalling by rho family GTPases. EMBO J. 1995;14:697–704. doi: 10.1002/j.1460-2075.1995.tb07048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Remold-O’Donnell E, Cooley J, Shcherbina A, Hagemann T L, Kwan S -P, Kenney D M, Rosen F S. Variable expression of WASP in B cell lines of Wiskott-Aldrich syndrome patients. J Immunol. 1997;158:4021–4025. [PubMed] [Google Scholar]
- 475a.Richman, T., and D. I. Johnson. Unpublished results.
- 476.Ridley A F, Hall A. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 477.Ridley A J. Rho: theme and variations. Curr Biol. 1996;6:1256–1264. doi: 10.1016/s0960-9822(02)70711-2. [DOI] [PubMed] [Google Scholar]
- 478.Ridley A J. Signal transduction through the GTP-binding proteins Rac and Rho. J Cell Sci Suppl. 1994;18:127–131. doi: 10.1242/jcs.1994.supplement_18.19. [DOI] [PubMed] [Google Scholar]
- 479.Ridley A J, Hall A. Signal transduction pathways regulating Rho-mediated stress fibre formation: requirement for a tyrosine kinase. EMBO J. 1994;13:2600–2610. doi: 10.1002/j.1460-2075.1994.tb06550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 481.Ridley A J, Self A J, Kasmi F, Paterson H F, Hall A, Marshall C J, Ellis C. rho family GTPase activating proteins p190, bcr, and rhoGAP show distinct specificities in vitro and in vivo. EMBO J. 1993;12:5151–5160. doi: 10.1002/j.1460-2075.1993.tb06210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Riesgo-Escovar J R, Jenni M, Fritz A, Hafen E. The Drosophila Jun-N-terminal kinase is required for cell morphogenesis but not for DJun-dependent cell fate specification in the eye. Genes Dev. 1996;10:2759–2768. doi: 10.1101/gad.10.21.2759. [DOI] [PubMed] [Google Scholar]
- 483.Rittinger K, Taylor W R, Smerdon S J, Gamblin S J. Support for shared ancestry of GAPs. Nature. 1998;392:448–449. doi: 10.1038/33043. [DOI] [PubMed] [Google Scholar]
- 484.Rittinger K, Walker P A, Eccleston J F, Nurmahomed K, Owen D, Laue E, Gamblin S J, Smerdon S J. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 485.Rittinger K, Walker P A, Eccleston J F, Smerdon S J, Gamblin S J. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 486.Rivero-Lezcano O M, Marcilla A, Sameshima J H, Robbins K C. Wiskott-Aldrich syndrome protein physically associated with Nck through Src homology 3 domains. Mol Cell Biol. 1995;15:5725–5731. doi: 10.1128/mcb.15.10.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Roberts R L, Fink G R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 488.Roberts R L, Mosch H U, Fink G R. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- 489.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 490.Romano J D, Schmidt W K, Michaelis S. The Saccharomyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:2231–2247. doi: 10.1091/mbc.9.8.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Ron D, Zannini M, Lewis M, Wickner R B, Hunt L T, Graziani G, Tronick SR, Aaronson S A, Eva A. A region of proto-dbl essential for its transforming activity shows sequence similarity to a yeast cell cycle gene, CDC24, and the human breakpoint cluster gene, bcr. New Biol. 1991;3:372–379. [PubMed] [Google Scholar]
- 492.Rooney R D, Tuazon P T, Meek W E, Carroll J E J, Hagen J J, Gump E L, Monnig C A, Lugo T, Traugh J A. Cleavage arrest of early frog embryos by the G protein-activated protein kinase PAKI. J Biol Chem. 1996;271:21498–21504. doi: 10.1074/jbc.271.35.21498. [DOI] [PubMed] [Google Scholar]
- 493.Rouse J, Cohen P, Trigon S, Morange M, Alonso-Liamazares A, Zamanillo D, Hunt T, Nebreda A R. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 494.Roux P, Gauthier-Rouviere C, Doucet-Brutin S, Fort P. The small GTPases Cdc42Hs, Rac1 and RhoG delineate Raf-independent pathways that cooperate to transform NIH3T3 cells. Curr Biol. 1997;7:629–637. doi: 10.1016/s0960-9822(06)00289-2. [DOI] [PubMed] [Google Scholar]
- 495.Rubino D, Driggers P, Arbit D, Kemp L, Miller B, Coso O, Pagliai K, Gray K, Gutkind S, Segars J. Characterization of Brx, a novel Dbl family member that modulates estrogen receptor action. Oncogene. 1998;16:2513–2526. doi: 10.1038/sj.onc.1201783. [DOI] [PubMed] [Google Scholar]
- 496.Rudel T, Bokoch G M. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–1574. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 497.Rudel T, Zenke F T, Chuang T H, Bokoch G M. p21-activated kinase (PAK) is required for Fas-induced JNK activation in Jurkat cells. J Immunol. 1998;160:7–11. [PubMed] [Google Scholar]
- 498.Rudolph M G, Bayer P, Abo A, Kuhlmann J, Vetter I R, Wittinghofer A. The Cdc42/Rac interactive binding region motif of the Wiskott-Aldrich syndrome protein (WASP) is necessary but not sufficient for tight binding to Cdc42 and structure formation. J Biol Chem. 1998;273:18067–18076. doi: 10.1074/jbc.273.29.18067. [DOI] [PubMed] [Google Scholar]
- 499.Ruggieri R, Bender A, Matsui Y, Powers S, Takai Y, Pringle J R, Matsumoto K. RSR1, a ras-like gene homologous to Krev-1 (smg21A/rap1A): role in the development of cell polarity and interactions with the Ras pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:758–766. doi: 10.1128/mcb.12.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Run J-Q, Steven R, Hung M-S, van Weeghel R, Culotti J G, Way J C. Suppressors of the unc-73 gene of Caenorhabditis elegans. Genetics. 1996;143:225–236. doi: 10.1093/genetics/143.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Russell P, Moreno S, Reed S I. Conservation of mitotic controls in fission and budding yeasts. Cell. 1989;57:295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- 502.Sabatini D M, Erdjument-Bromage H, Lui M, Tempst P, Snyder S H. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 503.Sabers C J, Martin M M, Brunn G J, Williams J M, Dumont F J, Wiederrecht G, Abraham R T. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 504.Sakaguchi S, Miyamoto S, Iida H, Suzuki T, Ohya Y, Anraku Y. Overproduction of Cdc24p (Cls4p), a guanine nucleotide-exchange factor toward Cdc42 GTPase, impairs initiation of budding in Saccharomyces cerevisiae. Protoplasma. 1995;189:142–148. [Google Scholar]
- 505.Sanders S L, Herskowitz I. The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle-dependent manner. J Cell Biol. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Saras J, Franzen P, Aspenström P, Hellman U, Gonez L J, Heldin C-H. A novel GTPase-activating protein for rho interacts with a PDZ domain of the protein-tyrosine phosphatase PTPL1. J Biol Chem. 1997;272:24333–24338. doi: 10.1074/jbc.272.39.24333. [DOI] [PubMed] [Google Scholar]
- 507.Sasamura T, Kobayashi T, Kojima S, Qadota H, Ohya Y, Masai I, Hotta Y. Molecular cloning and characterization of Drosophila genes encoding small GTPases of the rab and rho families. Mol Gen Genet. 1997;254:486–494. doi: 10.1007/s004380050443. [DOI] [PubMed] [Google Scholar]
- 508.Sawai E T, Khan I H, Montbriand P M, Peterlin B M, Cheng-Mayer C, Luciw P A. Activation of PAK by HIV and SIV Nef: importance for AIDS in rhesus macaques. Curr Biol. 1996;6:1519–1527. doi: 10.1016/s0960-9822(96)00757-9. [DOI] [PubMed] [Google Scholar]
- 508a.Sawyer, M., A. Merla, and D. I. Johnson. Unpublished data.
- 508b.Sawyer, M., and D. I. Johnson. Unpublished data.
- 509.Scherle P, Behrens T, Staudt L M. Ly-GDI, a GDP-dissociation inhibitor of the RhoA-binding protein, is expressed preferentially in lymphocytes. Proc Natl Acad Sci USA. 1993;90:7568–7572. doi: 10.1073/pnas.90.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Schmidt A, Bickle M, Beck T, Hall M N. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- 511.Schmidt A, Hall M N. Signaling to the actin cytoskeleton. Annu Rev Cell Dev Biol. 1998;14:305–338. doi: 10.1146/annurev.cellbio.14.1.305. [DOI] [PubMed] [Google Scholar]
- 512.Schmidt A, Kunz J, Hall M N. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc Natl Acad Sci USA. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.See R H, Shi Y. Adenovirus E1B 19,000-molecular-weight protein activates c-Jun N-terminal kinase and C-Jun-mediated transcription. Mol Cell Biol. 1998;18:4012–4022. doi: 10.1128/mcb.18.7.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Seger R, Krebs E G. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 515.Sells M A, Barratt J T, Caviston J, Ottilie S, Leberer E, Chernoff J. Characterization of Pak2p, a pleckstrin homology domain-containing, p21-activated protein kinase from fission yeast. J Biol Chem. 1998;273:18490–18498. doi: 10.1074/jbc.273.29.18490. [DOI] [PubMed] [Google Scholar]
- 516.Sells M A, Chernoff J. Emerging from the PAK: the p21-activated protein kinase family. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- 517.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 518.Settleman J, Albright C F, Foster L C, Weinberg R A. Association between GTPase activators for Rho and Ras families. Nature. 1992;359:153–154. doi: 10.1038/359153a0. [DOI] [PubMed] [Google Scholar]
- 519.Settleman J, Narasimhan V, Foster L C, Weinberg R A. Molecular cloning of cDNAs encoding the GAP-associated protein p190: implications for a signaling pathway from ras to the nucleus. Cell. 1992;69:539–549. doi: 10.1016/0092-8674(92)90454-k. [DOI] [PubMed] [Google Scholar]
- 520.Shapiro L. Protein localization and asymmetry in the bacterial cell. Cell. 1993;73:841–855. doi: 10.1016/0092-8674(93)90266-s. [DOI] [PubMed] [Google Scholar]
- 521.She H Y, Rockow S, Tang J P, Nishimura R, Skolnik E Y, Chen M, Margolis B, Li W. Wiskott-Aldrich syndrome protein is associated with the adapter protein Grb2 and the epidermal growth factor receptor in living cells. Mol Biol Cell. 1997;8:1709–1721. doi: 10.1091/mbc.8.9.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 523.Sheu Y-J, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Shimizu K, Kaibuchi K, Nonaka H, Yamamoto J, Takai Y. Tissue and subcellular distributions of an inhibitory GDP/GTP exchange protein (GDI) for the RHO proteins by use of its specific antibody. Biochem Biophys Res Commun. 1991;175:199–206. doi: 10.1016/s0006-291x(05)81220-3. [DOI] [PubMed] [Google Scholar]
- 525.Shinjo K, Koland J G, Hart M J, Narasimhan V, Johnson D I, Evans T, Cerione R A. Molecular cloning of the gene for the human placental GTP-binding protein Gp (G25K): Identification of this GTP-binding protein as the human homolog of the yeast cell-division-cycle protein CDC42. Proc Natl Acad Sci USA. 1990;87:9853–9857. doi: 10.1073/pnas.87.24.9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Sia R A L, Herald H A, Lew D J. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol Biol Cell. 1996;7:1657–1666. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Simon M N, De Virgilio C, Souza B, Pringle J R, Abo A, Reed S I. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal transduction. Nature. 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- 528.Sloat B F, Adams A, Pringle J R. Roles of the CDC24 gene product in cellular morphogenesis during the S. cerevisiae cell cycle. J Cell Biol. 1981;89:395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Sloat B F, Pringle J R. A mutant of yeast defective in cellular morphogenesis. Science. 1978;200:1171–1173. doi: 10.1126/science.349694. [DOI] [PubMed] [Google Scholar]
- 530.Sluss H K, Han Z, Barrett T, Goberdhan D C I, Wilson C, Davis R J, Ip Y T. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 1996;10:2745–2758. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- 531.Smith S J. Neuronal cytomechanics: the actin-based motility of growth cones. Science. 1988;242:708–714. doi: 10.1126/science.3055292. [DOI] [PubMed] [Google Scholar]
- 532.Sone M, Hoshino M, Suzuki E, Kuroda S, Kaibuchi K, Nakagoshi H, Saigo K, Nabeshima Y-I, Hama C. Still life, a protein in synaptic terminals of Drosophila homologous to GDP-GTP exchangers. Science. 1997;275:543–547. doi: 10.1126/science.275.5299.543. [DOI] [PubMed] [Google Scholar]
- 532a.Sprague, G. Personal communication.
- 533.Sprang S R, Coleman D E. Invasion of the nucleotide snatchers: structural insights into the mechanism of G protein GEFs. Cell. 1998;95:155–158. doi: 10.1016/s0092-8674(00)81746-8. [DOI] [PubMed] [Google Scholar]
- 534.Srivastava S K, Wheelock R H P, Aaronson S A, Eva A. Identification of the protein encoded by the human diffuse B-cell lymphoma (dbl) oncogene. Proc Natl Acad Sci USA. 1986;83:8868–8872. doi: 10.1073/pnas.83.23.8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Stam J C, Sander E E, Michiels F, van Leeuwen F N, Kain H E T, van der Kammen R A, Collard J G. Targeting of Tiam1 to the plasma membrane requires the cooperative function of the N-terminal pleckstrin homology domain and an adjacent protein interaction domain. J Biol Chem. 1997;272:28447–28454. doi: 10.1074/jbc.272.45.28447. [DOI] [PubMed] [Google Scholar]
- 536.Steven R, Kubiseski T J, Zheng H, Kulkarni S, Mancillas J, Morales A R, Hogue C W V, Pawson T, Culotti J. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- 537.Stevenson B J, Ferguson B, De Virgilio C, Bi E, Pringle J R, Ammerer G, Sprague G F. Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev. 1995;9:2949–2963. doi: 10.1101/gad.9.23.2949. [DOI] [PubMed] [Google Scholar]
- 538.Stevenson B J, Rhodes N, Errede B, Sprague G F. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 539.Stirtan W G, Poulter C D. Yeast protein geranylgeranyltransferase type-I: overproduction, purification, and characterization. Arch Biochem Biophys. 1995;321:182–190. doi: 10.1006/abbi.1995.1384. [DOI] [PubMed] [Google Scholar]
- 540.Stirtan W G, Poulter C D. Yeast protein geranylgeranyltransferase type-I: steady-state kinetics and substrate binding. Biochemistry. 1997;36:4552–4557. doi: 10.1021/bi962579c. [DOI] [PubMed] [Google Scholar]
- 541.Stowers L, Yelon D, Berg L J, Chant J. Regulation of the polarization of T cells toward antigen-presenting cells by Ras-related GTPase CDC42. Proc Natl Acad Sci USA. 1995;92:5027–5031. doi: 10.1073/pnas.92.11.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Su Y C, Han J H, Xu S C, Cobb M, Skolnik E Y. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 1997;16:1279–1290. doi: 10.1093/emboj/16.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Suzuki T, Miki H, Takenawa T, Sasakawa C. Neural Wiskott-Aldrich syndrome protein is implicated in the actin-based motility of Shigella flexneri. EMBO J. 1998;17:2767–2776. doi: 10.1093/emboj/17.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Symons M. Rho family GTPases: the cytoskeleton and beyond. Trends Biochem Sci. 1996;21:178–182. [PubMed] [Google Scholar]
- 545.Symons M, Derry J M J, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 546.Takaishi K, Kikuchi A, Kuroda S, Kotani K, Sasaki T, Takai Y. Involvement of rho p21 and its inhibitory GDP/GTP exchange protein (rho GDI) in cell motility. Mol Cell Biol. 1993;13:72–79. doi: 10.1128/mcb.13.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Takano H, Komuro I, Oka T, Shiojima I, Hiroi Y, Mizuno T, Yazaki Y. The Rho family G proteins play a critical role in muscle differentiation. Mol Cell Biol. 1998;18:1580–1589. doi: 10.1128/mcb.18.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Tan E C, Leung T, Manser E, Lim L. The human active breakpoint cluster region-related gene encodes a brain protein with homology to guanine nucleotide exchange proteins and GTPase-activating proteins. J Biol Chem. 1993;268:27291–27298. [PubMed] [Google Scholar]
- 549.Tanaka K, Sasaki T, Kikuchi A, Kaibuchi K, Takai Y. Rho-GDI. In: Zerial M, Huber L A, editors. Guidebook to the small GTPases. Oxford, United Kingdom: Oxford University Press; 1995. pp. 231–234. [Google Scholar]
- 550.Tanaka K, Takai Y. Control of reorganization of the actin cytoskeleton by Rho family small GTP-binding proteins in yeast. Curr Opin Cell Biol. 1998;10:112–116. doi: 10.1016/s0955-0674(98)80093-8. [DOI] [PubMed] [Google Scholar]
- 551.Tang Y, Marwaha S, Rutkowski J L, Tennekoon G I, Phillips P C, Field J. A role for Pak protein kinases in Schwann cell transformation. Proc Natl Acad Sci USA. 1998;95:5139–5144. doi: 10.1073/pnas.95.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Tapon N, Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr Opin Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 553.Taylor J M, Hildebrand J D, Mack C P, Cox M E, Parsons J T. Characterization of Graf, the GTPase-activating protein for Rho associated with focal adhesion kinase. Phosphorylation and possible regulation by mitogen-activated protein kinase. J Biol Chem. 1998;273:8063–8070. doi: 10.1074/jbc.273.14.8063. [DOI] [PubMed] [Google Scholar]
- 554.Teo M, Manser E, Lim L. Identification and molecular cloning of a p21cdc42/rac1-activated serine/threonine kinase that is rapidly activated by thrombin in platelets. J Biol Chem. 1995;270:26690–26697. doi: 10.1074/jbc.270.44.26690. [DOI] [PubMed] [Google Scholar]
- 555.Teramoto H, Coso O A, Miyata H, Igishi T, Miki T, Gutkind J S. Signaling from the small GTP-binding proteins Rac1 and Cdc42 to the c-Jun N-terminal kinase stress-activated protein kinase pathway: A role for mixed lineage kinase 3/protein-tyrosine kinase 1, a novel member of the mixed lineage kinase family. J Biol Chem. 1996;271:27225–27228. doi: 10.1074/jbc.271.44.27225. [DOI] [PubMed] [Google Scholar]
- 556.Teramoto H, Crespo P, Coso O A, Igishi T, Xu N Z, Gutkind J S. The small GTP-binding protein Rho activates c-Jun N-terminal kinases/stress-activated protein kinases in human kidney 293T cells. Evidence for a Pak-independent signaling pathway. J Biol Chem. 1996;271:25731–25734. doi: 10.1074/jbc.271.42.25731. [DOI] [PubMed] [Google Scholar]
- 557.Thomas G, Hall M N. TOR signalling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 558.Thompson G, Owen D, Chalk P A, Lowe P N. Delineation of the Cdc42/Rac-binding domain of p21-activated kinase. Biochemistry. 1998;37:7885–7891. doi: 10.1021/bi980140+. [DOI] [PubMed] [Google Scholar]
- 559.Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 560.Tjandra H, Compton J, Kellogg D. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr Biol. 1998;8:991–1000. doi: 10.1016/s0960-9822(07)00419-8. [DOI] [PubMed] [Google Scholar]
- 561.Tolias K F, Cantley L C, Carpenter C L. Rho family GTPases bind to phosphoinositide kinases. J Biol Chem. 1995;270:17656–17659. doi: 10.1074/jbc.270.30.17656. [DOI] [PubMed] [Google Scholar]
- 562.Toure A, Dorseuil O, Morin L, Timmons P, Jegou B, Reibel L, Gacon G. MgcRacGAP, a new human GTPase-activating protein for Rac and Cdc42 similar to Drosophila rotundRacGAP gene product, is expressed in male germ cells. J Biol Chem. 1998;273:6019–6023. doi: 10.1074/jbc.273.11.6019. [DOI] [PubMed] [Google Scholar]
- 563.Trainin T, Shmuel M, Delmer D P. In vitro prenylation of the small GTPase Rac13 of cotton. Plant Physiol. 1996;112:1491–1497. doi: 10.1104/pp.112.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Trono D, Wang J K. Nef and PAK: Virulence factor and cellular accomplice. Chem Biol. 1997;4:13–15. doi: 10.1016/s1074-5521(97)90232-5. [DOI] [PubMed] [Google Scholar]
- 565.Trueblood C E, Ohya Y, Rine J. Genetic evidence for in vivo cross-specificity of the CaaX-box protein prenyltransferases farnesyltransferase and geranylgeranyltransferase-I in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4260–4275. doi: 10.1128/mcb.13.7.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Trumpp A, Blundell P A, de la Pompa J L, Zeller R. The chicken limb deformity gene encodes nuclear proteins expressed in specific cell types during morphogenesis. Genes Dev. 1992;6:14–28. doi: 10.1101/gad.6.1.14. [DOI] [PubMed] [Google Scholar]
- 567.Tsuchiya E, Matsuzaki G, Kurano K, Fukuchi T, Tsukao A, Miyakawa T. The Saccharomyces cerevisiae SSD1 gene is involved in the tolerance to high concentration of Ca2+ with the participation of HST1/NRC1/BFR1. Gene. 1996;176:35–38. doi: 10.1016/0378-1119(96)00204-1. [DOI] [PubMed] [Google Scholar]
- 568.Tu H, Barr M, Dong D L, Wigler M. Multiple regulatory domains on the Byr2 protein kinase. Mol Cell Biol. 1997;17:5876–5887. doi: 10.1128/mcb.17.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Ueda T, Kikuchi A, Ohga N, Yamamoto J, Takai Y. Purification and characterization from bovine brain cytosol of a novel regulatory protein inhibiting the dissociation of GDP from and the subsequent binding of GTP to rhoB p20, a ras p21-like GTP-binding protein. J Biol Chem. 1990;265:9373–9380. [PubMed] [Google Scholar]
- 570.Umikawa M, Tanaka K, Kamei T, Shimizu K, Imamura H, Sasaki T, Takai Y. Interaction of Rho1p target Bni1p with F-actin-binding elongation factor 1 alpha: implication in Rho1p-regulated reorganization of the actin cytoskeleton in Saccharomyces cerevisiae. Oncogene. 1998;16:2011–2016. doi: 10.1038/sj.onc.1201724. [DOI] [PubMed] [Google Scholar]
- 571.Vaduva G, Martin N C, Hopper A K. Actin-binding verprolin is a polarity development protein required for the morphogenesis and function of the yeast actin cytoskeleton. J Cell Biol. 1997;139:1821–1833. doi: 10.1083/jcb.139.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 573.Van Heusden G P H, Griffiths D J F, Ford J C, Chin-A-Woeng T F C, Schrader P A T, Carr A M, Steensma H Y. The 14-3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur J Biochem. 1995;229:45–53. [PubMed] [Google Scholar]
- 574.Varela J C S, Mager W H. Response of Saccharomyces cerevisiae to changes in external osmolarity. Microbiology. 1996;142:721–731. doi: 10.1099/00221287-142-4-721. [DOI] [PubMed] [Google Scholar]
- 575.Verde F, Mata J, Nurse P. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J Cell Biol. 1995;131:1529–1538. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Verde F, Wiley D J, Nurse P. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc Natl Acad Sci USA. 1998;95:7526–7531. doi: 10.1073/pnas.95.13.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Villa A, Notarangelo L, Macchi P, Mantuano E, Cavagni G, Brugnoni D, Strina D, Patrosso M C, Ramenghi U, Sacco M G, Ugazio A, Vezzoni P. X-linked thrombocytopenia and Wiskott-Aldrich syndrome are allelic diseases with mutations in the WASP gene. Nat Genet. 1995;9:414–417. doi: 10.1038/ng0495-414. [DOI] [PubMed] [Google Scholar]
- 578.Vojtek A B, Cooper J A. Rho family members: activators of MAP kinase cascades. Cell. 1995;82:527–529. doi: 10.1016/0092-8674(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 579.Voncken J W, van Schaick H, Kaartinen V, Deemer K, Coates T, Landing B, Pattengale P, Dorseuil O, Bokoch G M, Groffen J, Heisterkamp N. Increased neutrophil respiratory burst in bcr-null mutants. Cell. 1995;80:719–728. doi: 10.1016/0092-8674(95)90350-x. [DOI] [PubMed] [Google Scholar]
- 580.Voyno-Yasenetskaya T A, Faure M P, Ahn N G, Bourne H R. Gα12 and Gα13 regulate extracellular signal-regulated kinase and c-Jun kinase pathways by different mechanisms in COS-7 cells. J Biol Chem. 1996;271:21081–21087. doi: 10.1074/jbc.271.35.21081. [DOI] [PubMed] [Google Scholar]
- 581.Wadsworth S J, Gebauer G, van Rossum G D V, Dhanasekaran N. Ras-dependent signaling by the GTPase-deficient mutant of Gα12. J Biol Chem. 1997;272:28829–28832. doi: 10.1074/jbc.272.46.28829. [DOI] [PubMed] [Google Scholar]
- 582.Waldo G L, Evans T, Fraser E D, Northup J K, Martin M W, Harden T K. Identification and purification from bovine brain of a guanine-nucleotide-binding protein distinct from Gs, Gi and Go. Biochem J. 1987;246:431–439. doi: 10.1042/bj2460431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Walter B N, Huang Z, Jakobi R, Tuazon P T, Alnemri E S, Litwack G, Traugh J A. Cleavage and activation of p21-activated protein kinase γ-PAK by CPP32 (caspase 3). Effects of autophosphorylation on activity. J Biol Chem. 1998;273:28733–28739. doi: 10.1074/jbc.273.44.28733. [DOI] [PubMed] [Google Scholar]
- 584.Wassmann K, Ammerer G. Overexpression of the G1-cyclin gene CLN2 represses the mating pathway in Saccharomyces cerevisiae at the level of the MEKK Ste11. J Biol Chem. 1997;272:13180–13188. doi: 10.1074/jbc.272.20.13180. [DOI] [PubMed] [Google Scholar]
- 585.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch B M, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Watarai M, Kamata Y, Kozaki S, Sasakawa C. rho, a small GTP-binding protein, is essential for Shigella invasion of epithelial cells. J Exp Med. 1997;185:281–292. doi: 10.1084/jem.185.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587.Way J C, Wang L, Run J Q, Hung M S. Cell polarity and the mechanism of asymmetric cell division. Bioessays. 1994;16:925–931. doi: 10.1002/bies.950161212. [DOI] [PubMed] [Google Scholar]
- 588.Weissbach L, Settleman J, Kalady M F, Snijders A J, Murthy A E, Yan Y-X, Bernards A. Identification of a human RasGAP-related protein containing calmodulin-binding motifs. J Biol Chem. 1994;269:20517–20521. [PubMed] [Google Scholar]
- 589.Werner L A, Manseau L J. A Drosophila gene with predicted rhoGEF, pleckstrin homology and SH3 domains is highly expressed in morphogenic tissues. Gene. 1997;187:107–114. doi: 10.1016/s0378-1119(96)00732-9. [DOI] [PubMed] [Google Scholar]
- 590.Westwick J K, Lee R J, Lambert Q T, Symons M, Pestell R G, Der C J, Whitehead I P. Transforming potential of Dbl family proteins correlates with transcription from the cyclin D1 promoter but not with activation of Jun NH2-terminal kinase, p38/Mpk2, serum response factor, or c-Jun. J Biol Chem. 1998;273:16739–16747. doi: 10.1074/jbc.273.27.16739. [DOI] [PubMed] [Google Scholar]
- 591.White W H, Johnson D I. Characterization of synthetic-lethal mutants reveals a role for the Saccharomyces cerevisiae guanine-nucleotide exchange factor Cdc24p in vacuole function and Na+ tolerance. Genetics. 1997;147:43–55. doi: 10.1093/genetics/147.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Whitehead I, Kirk H, Kay R. Retroviral transduction and oncogenic selection of a cDNA encoding Dbs, a homolog of the Dbl guanine nucleotide exchange factor. Oncogene. 1995;10:713–721. [PubMed] [Google Scholar]
- 593.Whitehead I P, Abe K, Gorski J L, Der C J. CDC42 and FGD1 cause distinct signaling and transforming activities. Mol Cell Biol. 1998;18:4689–4697. doi: 10.1128/mcb.18.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Whitehead I P, Campbell S, Rossman K L, Der C J. Dbl family proteins. Biochim Biophy Acta Rev Cancer. 1997;1332:F1–F23. doi: 10.1016/s0304-419x(96)00040-6. [DOI] [PubMed] [Google Scholar]
- 595.Whiteway M, Dignard D, Thomas D Y. Dominant negative selection of heterologous genes isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc Natl Acad Sci USA. 1992;89:9410–9414. doi: 10.1073/pnas.89.20.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Whiteway M, Hougan L, Dignard D, Thomas D Y, Bell L, Saari G C, Grant F J, O’Hara P, MacKay V L. The STE4 and STE18 genes of yeast encode potential β and γ subunits of the mating factor receptor-coupled G protein. Cell. 1989;56:467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 597.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 598.Winge P, Brembu T, Bones A M. Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol Biol. 1997;35:483–495. doi: 10.1023/a:1005804508902. [DOI] [PubMed] [Google Scholar]
- 599.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 600.Woychik R P, Maas R, Zeller R, Vogt T, Leder P. ‘Formins’: proteins deduced from the alternative transcripts of the limb deformity gene. Nature. 1990;346:850–852. doi: 10.1038/346850a0. [DOI] [PubMed] [Google Scholar]
- 601.Wu C, Leeuw T, Leberer E, Thomas D Y, Whiteway M. Cell cycle- and Cln2p-Cdc28p-dependent phosphorylation of the yeast Ste20p protein kinase. J Biol Chem. 1998;273:28107–28115. doi: 10.1074/jbc.273.43.28107. [DOI] [PubMed] [Google Scholar]
- 602.Wu C, Lytvyn V, Thomas D Y, Leberer E. The phosphorylation site for Ste20-like protein kinases is essential for the function of myosin-I in yeast. J Biol Chem. 1997;272:30623–30626. doi: 10.1074/jbc.272.49.30623. [DOI] [PubMed] [Google Scholar]
- 603.Wu C, Whiteway M, Thomas D Y, Leberer E. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J Biol Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- 604.Wu C L, Lee S F, Furmaniak-Kazmierczak E, Coté G P, Thomas D Y, Leberer E. Activation of myosin-I by members of the Ste20p protein kinase family. J Biol Chem. 1996;271:31787–31790. doi: 10.1074/jbc.271.50.31787. [DOI] [PubMed] [Google Scholar]
- 605.Wu W J, Leonard D A, Cerione R A, Manor D. Interaction between Cdc42Hs and RhoGDI is mediated through the Rho insert region. J Biol Chem. 1997;272:26153–26158. doi: 10.1074/jbc.272.42.26153. [DOI] [PubMed] [Google Scholar]
- 606.Wu W J, Lin R, Cerione R A, Manor D. Transformation activity of Cdc42 requires a region unique to Rho-related proteins. J Biol Chem. 1998;273:16655–16658. doi: 10.1074/jbc.273.27.16655. [DOI] [PubMed] [Google Scholar]
- 607.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 608.Yaar L, Mevarech M, Koltin Y. A Candida albicans RAS-related gene (CaRSR1) is involved in budding, cell morphogenesis and hypha development. Microbiology. 1997;143:3033–3044. doi: 10.1099/00221287-143-9-3033. [DOI] [PubMed] [Google Scholar]
- 609.Yablonski D, Kane L P, Qian D P, Weiss A. A Nck-Pak1 signaling module is required for T-cell receptor-mediated activation of NFAT, but not of JNK. EMBO J. 1998;17:5647–5657. doi: 10.1093/emboj/17.19.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Yaffe M B, Rittinger K, Volinia S, Caron P R, Aitken A, Leffers H, Gamblin SJ, Smerdon S J, Cantley L C. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1998;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 611.Yaku H, Sasaki T, Takai Y. The Dbl oncogene product as a GDP/GTP exchange protein for the Rho family: its properties in comparison with those of Smg GDS. Biochem Biophys Res Commun. 1994;198:811–817. doi: 10.1006/bbrc.1994.1116. [DOI] [PubMed] [Google Scholar]
- 612.Yamane H K, Farnsworth C C, Xie H, Evans T, Howald W N, Gelb M H, Glomset J A, Clarke S, Fung B K-K. Membrane-binding domain of the small G-protein G25K contains an S-(all-trans-geranylgeranyl)cysteine methyl ester at its carboxyl terminus. Proc Natl Acad Sci USA. 1991;88:286–290. doi: 10.1073/pnas.88.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Yamane H K, Fung B K K. The membrane-binding domain of a 23-kDa G-protein is carboxyl methylated. J Biol Chem. 1989;264:20100–20105. [PubMed] [Google Scholar]
- 614.Yan M, Dal T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 615.Yang D, Tournier C, Wysk M, Lu H-T, Xu J, Davis R J, Flavell R A. Targeted disruption of the MKK4 gene causes embryonic death, inhibition of c-Jun NH2-terminal kinase activation and defects in AP-1 transcriptional activity. Proc Natl Acad Sci USA. 1997;94:3004–3009. doi: 10.1073/pnas.94.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Yang P, Kansra S, Pimental R A, Gilbreth M, Marcus S. Cloning and characterization of shk2, a gene encoding a novel p21-activated protein kinase from fission yeast. J Biol Chem. 1998;273:18481–18489. doi: 10.1074/jbc.273.29.18481. [DOI] [PubMed] [Google Scholar]
- 617.Yang W N, Cerione R A. Cloning and characterization of a novel Cdc42-associated tyrosine kinase, ACK-2, from bovine brain. J Biol Chem. 1997;272:24819–24824. doi: 10.1074/jbc.272.40.24819. [DOI] [PubMed] [Google Scholar]
- 618.Yu Y, Jiang Y W, Wellinger R J, Carlson K, Roberts J M, Stillman D J. Mutations in the homologous ZDS1 and ZDS2 genes affect cell cycle progression. Mol Cell Biol. 1996;16:5254–5263. doi: 10.1128/mcb.16.10.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Zahner J E, Harkins H A, Pringle J R. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Zalcman G, Closson V, Camonis J, Honoré N, Rousseau-Merck M F, Tavitian A, Olofsson B. RhoGDI-3 is a new GDP dissociation inhibitor (GDI): identification of a non-cytosolic GDI protein interacting with the small GTP-binding proteins RhoB and RhoG. J Biol Chem. 1996;271:30366–30374. doi: 10.1074/jbc.271.48.30366. [DOI] [PubMed] [Google Scholar]
- 621.Zanke B W, Boudreau K, Rubie E, Winnett E, Tibbles L A, Zon L, Kyriakis J, Fiu F, Woodgett J R. The stress-activated protein kinase pathway mediates cell death following injury induced by cis-platinum, UV irradiation or heat. Curr Biol. 1996;6:606–613. doi: 10.1016/s0960-9822(02)00547-x. [DOI] [PubMed] [Google Scholar]
- 622.Zhang, B., Y. Zhang, C. C. Collins, D. I. Johnson, and Y. Zheng. 1999. A built-in arginine finger triggers the self-stimulatory GTPase-activating activity of Rho family GTPases. 274:2609–2612. [DOI] [PubMed]
- 623.Zhang B, Zheng Y. Negative regulation of Rho family GTPases Cdc42 and Rac2 by homodimer formation. J Biol Chem. 1998;273:25728–25733. doi: 10.1074/jbc.273.40.25728. [DOI] [PubMed] [Google Scholar]
- 624.Zhang B L, Wang Z-X, Zheng Y. Characterization of the interactions between the small GTPase Cdc42 and its GTPase-activating proteins and putative effectors. Comparison of kinetic properties of Cdc42 binding to the Cdc42-interactive domains. J Biol Chem. 1997;272:21999–22007. doi: 10.1074/jbc.272.35.21999. [DOI] [PubMed] [Google Scholar]
- 625.Zhang S, Han J, Sells M A, Chernoff J, Knaus U G, Ulevitch R J, Bokoch G M. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 626.Zhao Z-S, Leung T, Manser E, Lim L. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol Cell Biol. 1995;15:5246–5257. doi: 10.1128/mcb.15.10.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Zhao Z-S, Manser E, Chen X-Q, Chong C, Leung T, Lim L. A conserved negative regulatory region in αPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol. 1998;18:2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.Zheng Y, Bagrodia S, Cerione R A. Activation of phosphoinositide 3-kinase activity by Cdc42Hs binding to p85. J Biol Chem. 1994;269:18727–18730. [PubMed] [Google Scholar]
- 629.Zheng Y, Bender A, Cerione R A. Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J Biol Chem. 1995;270:626–630. doi: 10.1074/jbc.270.2.626. [DOI] [PubMed] [Google Scholar]
- 630.Zheng Y, Cerione R, Bender A. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
- 631.Zheng Y, Fischer D J, Santos M F, Tigyi G, Pasteris N G, Gorski J L, Xu Y. The faciogenital dysplasia gene product FGD1 functions as a Cdc42Hs-specific guanine-nucleotide exchange factor. J Biol Chem. 1996;271:33169–33172. doi: 10.1074/jbc.271.52.33169. [DOI] [PubMed] [Google Scholar]
- 632.Zheng Y, Glaven J A, Wu W J, Cerione R A. Phosphatidylinositol 4,5-bisphosphate provides an alternative to guanine nucleotide exchange factors by stimulating the dissociation of GDP from Cdc42Hs. J Biol Chem. 1996;271:23815–23819. doi: 10.1074/jbc.271.39.23815. [DOI] [PubMed] [Google Scholar]
- 633.Zheng Y, Hart M J, Shinjo K, Evans T, Bender A, Cerione R A. Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. Delineation of a limit Cdc42 GTPase-activating protein domain. J Biol Chem. 1993;268:24629–24634. [PubMed] [Google Scholar]
- 634.Zheng Y, Zangrilli D, Cerione R A, Eva A. The pleckstrin homology domain mediates transformation by oncogenic Dbl through specific intracellular targeting. J Biol Chem. 1996;271:19017–19020. doi: 10.1074/jbc.271.32.19017. [DOI] [PubMed] [Google Scholar]
- 635.Zhou K M, Wang Y, Gorski J L, Nomura N, Collard J, Bokoch G M. Guanine nucleotide exchange factors regulate specificity of downstream signaling from Rac and Cdc42. J Biol Chem. 1998;273:16782–16786. doi: 10.1074/jbc.273.27.16782. [DOI] [PubMed] [Google Scholar]
- 636.Zhu Q, Zhang M, Blaese R M, Derry J M J, Junker A, Francke U, Chen SH, Ochs H D. The Wiskott-Aldrich syndrome and X-linked congenital thrombocytopenia are caused by mutations of the same gene. Blood. 1995;86:3797–3804. [PubMed] [Google Scholar]
- 637.Zhu Q L, Watanabe C, Liu T, Hollenbaugh D, Blaese R M, Kanner S B, Aruffo A, Ochs H D. Wiskott-Aldrich syndrome/X-linked thrombocytopenia: WASP gene mutations, protein expression, and phenotype. Blood. 1997;90:2680–2689. [PubMed] [Google Scholar]
- 638.Zigmond S H. Signal transduction and actin filament organization. Curr Opin Cell Biol. 1996;8:66–73. doi: 10.1016/s0955-0674(96)80050-0. [DOI] [PubMed] [Google Scholar]
- 639.Zigmond S H, Joyce M, Borleis J, Bokoch G M, Devreotes P N. Regulation of actin polymerization in cell-free systems by GTPγS and Cdc42. J Cell Biol. 1997;138:363–374. doi: 10.1083/jcb.138.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Zigmond S H, Joyce M, Yang C, Brown K, Huang M, Pring M. Mechanism of Cdc42-induced actin polymerization in neutrophil extracts. J Cell Biol. 1998;142:1001–1012. doi: 10.1083/jcb.142.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Ziman M, Johnson D I. Genetic evidence for a functional interaction between S. cerevisiae CDC24 and CDC42. Yeast. 1994;10:463–474. doi: 10.1002/yea.320100405. [DOI] [PubMed] [Google Scholar]
- 642.Ziman M, O’Brien J M, Ouellette L A, Church W R, Johnson D I. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol Cell Biol. 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Ziman M, Preuss D, Mulholland J, O’Brien J M, Botstein D, Johnson D I. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol Biol Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Zohn I M, Campbell S L, Khosravi-Far R, Rossman K L, Der C J. Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene. 1998;17:1415–1438. doi: 10.1038/sj.onc.1202181. [DOI] [PubMed] [Google Scholar]