Abstract
Cellular differentiation, mating, and filamentous growth are regulated in many fungi by environmental and nutritional signals. For example, in response to nitrogen limitation, diploid cells of the yeast Saccharomyces cerevisiae undergo a dimorphic transition to filamentous growth referred to as pseudohyphal differentiation. Yeast filamentous growth is regulated, in part, by two conserved signal transduction cascades: a mitogen-activated protein kinase cascade and a G-protein regulated cyclic AMP signaling pathway. Related signaling cascades play an analogous role in regulating mating and virulence in the plant fungal pathogen Ustilago maydis and the human fungal pathogens Cryptococcus neoformans and Candida albicans. We review here studies on the signaling cascades that regulate development of these and other fungi. This analysis illustrates both how the model yeast S. cerevisiae can serve as a paradigm for signaling in other organisms and also how studies in other fungi provide insights into conserved signaling pathways that operate in many divergent organisms.
Fungi are eukaryotic organisms that diverged from a common ancestor with multicellular eukaryotic animals some 800 to 1,000 million years ago. Despite this evolutionary divergence, fungi are more closely related to animals than to plants, algae, bacteria, or archea and thus share important features with mammalian cells. This is perhaps most apparent in the signaling cascades that regulate cell function. The yeast Saccharomyces cerevisiae expresses at least three members of the G protein-coupled family of serpentine receptors, which are in turn coupled to a heterotrimeric G protein and a G protein alpha subunit homolog. Mating in yeast cells is regulated by a mitogen-activated protein (MAP) kinase cascade that is highly conserved with MAP kinase cascades in mammalian cells. Finally, the signal transduction components that are targeted by the immunosuppressive drugs cyclosporin A, FK506, and rapamycin are remarkably conserved from yeasts to humans. Thus, studies on signal transduction in S. cerevisiae and other genetically tractable fungi promise to reveal common conserved mechanisms of signal transduction.
Recent studies have revealed that the model yeast S. cerevisiae undergoes a dimorphic transition to filamentous growth in response to nutritional signals in the environment, particularly nitrogen limitation. Filamentous growth occurs in both haploid and diploid cells in different environments and may play novel roles in the life cycle of this organism. At least two conserved signal transduction cascades that regulate filamentous growth have been defined, and remarkably related signaling pathways also operate during differentiation of other fungi, including pathogens of both plants and animals. These recent findings suggest that S. cerevisiae is also an excellent model system with great potential to provide insights into signaling in other fungi. We review here studies on the signal transduction cascades that regulate yeast filamentous growth and the related signaling pathways that operate in the fission yeast Schizosaccharomyces pombe, in human fungal pathogens (Candida albicans and Cryptococcus neoformans), in plant fungal pathogens (Ustilago maydis, Magnaporthe grisea, and Cryphonectria parasitica), and in model filamentous fungi (Aspergillus nidulans and Neurospora crassa). Taken together, these studies reveal a high degree of conservation between divergent organisms and illustrate conserved basic principles in the molecular determinants of life. Several other excellent reviews on yeast signal transduction have been published recently and also cover some additional topics in fungal development (5, 8, 10, 17, 18, 30, 139, 140, 177, 178, 263, 276, 289, 290).
FILAMENTOUS GROWTH IN SACCHAROMYCES CEREVISIAE
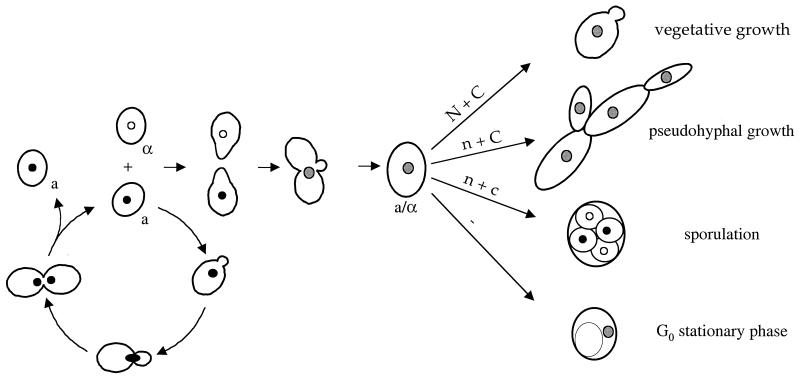
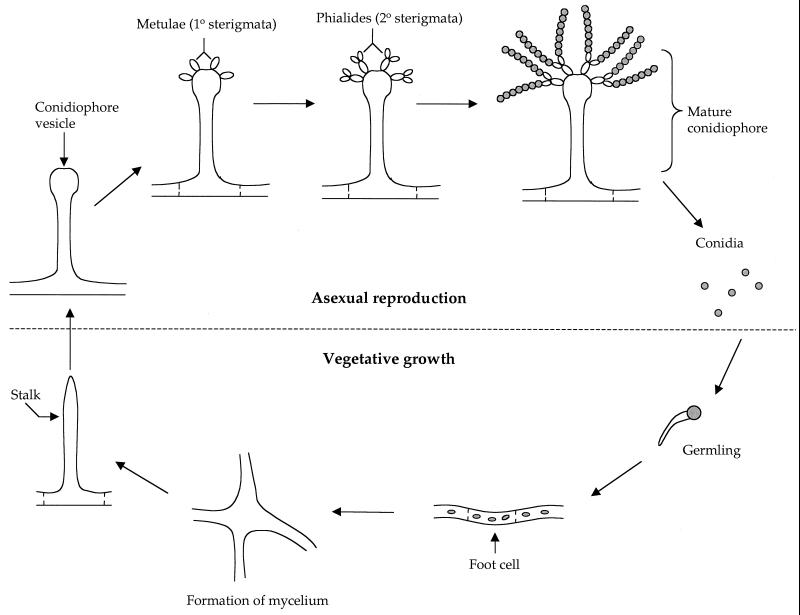
In response to nitrogen limitation and abundant fermentable carbon source, diploid cells of S. cerevisiae undergo dimorphic transition to a filamentous growth form referred to as pseudohyphal differentiation (93) (Fig. 1 and 2). Filamentous growth represents a dramatic change in the normal pattern of cell growth in which the cells become elongated, switch to a unipolar budding pattern, remain physically attached to each other, and invade the growth substrate (Fig. 2). This alternative growth form may enable this nonmotile species to forage for nutrients under adverse conditions.
FIG. 1.
Life cycle of S. cerevisiae. Haploid yeast cells mate on rich medium. Diploid yeast cells adopt alternative fates depending on the availability of nutrients. N + C, abundant nitrogen and fermentable carbon source; n + C, limiting nitrogen and abundent fermentable carbon source; n + c, limiting nitrogen and nonfermentable carbon source; −, no nutrients.
FIG. 2.
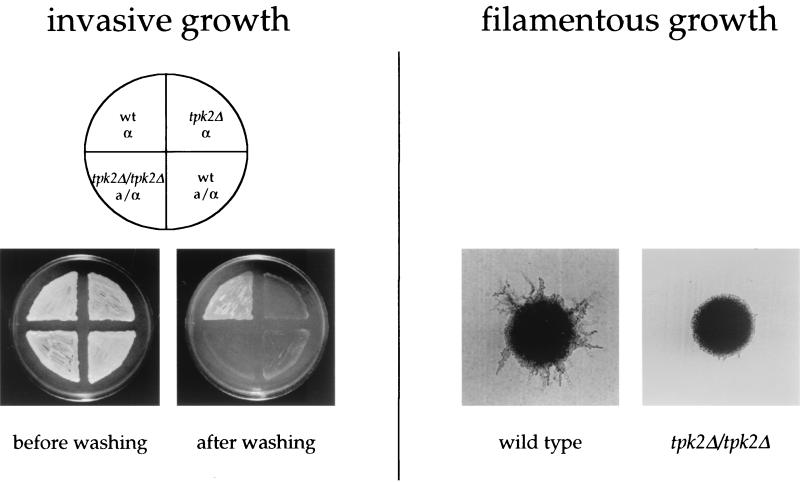
Filamentous and invasive growth of S. cerevisiae. On the left, invasive growth of haploid and diploid wild-type (wt) and tpk2 mutant yeast colonies is shown. On the right, the typical morphology of yeast pseudohyphal colonies is depicted, and a tpk2/tpk2 mutant with a filamentous growth defect is shown for comparison.
Conserved MAP Kinase Cascade
At least two conserved signaling pathways regulate yeast filamentous growth (Fig. 3). The first cascade involves components of the MAP kinase pathway that is also required for mating in haploid yeast cells in response to pheromones (56, 161, 180). The components of this MAP kinase cascade required for filamentous growth include the Ste20, Ste11, Ste7, and Kss1 kinases and the Ste12 transcription factor. In addition, another transcription factor, Tec1, forms a heterodimer with Ste12 that regulates expression of Tec1 itself and additional targets, such as the cell surface flocculin Flo11 required for agar invasion and filamentation (90, 165, 176). The pheromones, pheromone receptors, and subunits of the pheromone-activated heterotrimeric G protein are dispensable for filamentous growth and are not expressed in diploid cells (161).
FIG. 3.
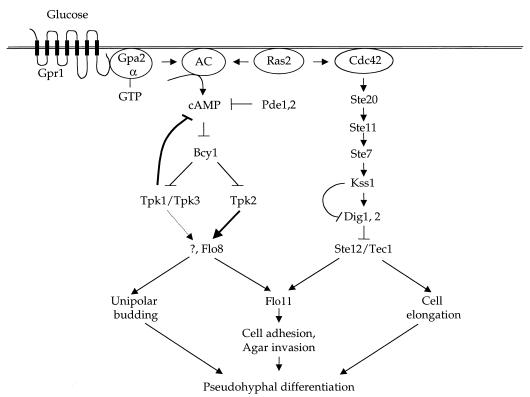
Signal transduction cascades regulating pseudohyphal differentiation in S. cerevisae. Two parallel signaling cascades regulating filamentous growth are depicted: a nutrient-sensing cAMP-PKA pathway, and the MAP kinase cascade. AC, adenylylcyclase.
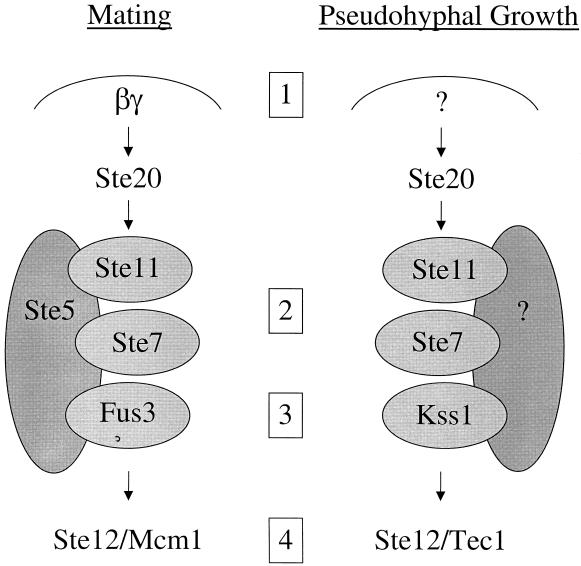
Thus, S. cerevisiae is able to use common components and yet couple them into two different pathways that sense two different environmental signals and give rise to two completely different developmental fates: mating in haploid cells in response to pheromone, and filamentous growth in diploid cells in response to nitrogen limitation and other environmental signals (Fig. 1). Signaling specificity is achieved by at least four different specializations in this signaling pathway (Fig. 4). First, the MAP kinase cascade is activated by the βγ subunits of the pheromone-activated heterotrimeric G protein during mating of haploid cells. During filamentous growth, the MAP kinase pathway is activated by a different mechanism involving the Cdc42, Ras2, and 14-3-3 proteins Bmh1 and Bmh2 (201, 203, 236). Second, the components of the MAP kinase cascade are tethered together by the scaffold protein Ste5 during mating, whereas Ste5 is not required during filamentous growth, and another protein may serve this scaffolding function. It has been suggested that the Spa2 protein, which physically interacts with many of the MAP kinase cascade components, might function as the scaffold during filamentous growth (239). The third level of specialization is at the level of the MAP kinase itself. In S. cerevisiae, the Fus3 and Kss1 kinases have diverged, so that Fus3 is specialized to regulate mating and actually inhibits invasive growth, whereas Kss1 is specialized to regulate invasive and filamentous growth (56, 180). In addition, Kss1 has both positive and negative regulatory roles during filamentous growth (21, 22, 176, 178), in part by relieving repression of Ste12 by the Dig1 and Dig2 proteins (55). Finally, during mating Ste12 interacts with the Mcm1 protein to activate transcription of genes containing pheromone response elements, whereas in diploid cells Ste12 forms a heterodimer with Tec1 that activates transcription of genes with filamentation response elements (176). In this way, one common protein, Ste12, can yield two different patterns of appropriate transcriptional responses in haploid and diploid cells.
FIG. 4.
Signaling specificity during mating and pseudohyphal growth in S. cerevisiae. Two related MAP kinase signaling cascades allow haploid and diploid yeast cells to respond to two different environmental signs, pheromone and nitrogen limitation, and give rise to two completely different developmental fates, mating in haploid cells and filamentous growth in diploid cells. Signaling specificity is achieved by at least four mechanisms. First, pheromone activates a G protein whose βγ subunits activate the MAP kinase cascade, and these components are not expressed in diploid yeast cells and do not regulate filamentous growth. Second, the Ste5 scaffold tethers the components during mating but does not play a role in diploid filamentous growth. Third, the MAP kinase has diverged and specialized: Fus3 to regulate mating and Kss1 to regulate filamentous growth. Finally, Ste12 homodimers or heterodimers with Mcm1 activate pheromone response element-regulated genes in mating or with Tec1 to regulate filamentation response element-driven genes during diploid filamentous growth.
Nutrient-Sensing cAMP Pathway
A second signaling pathway functions in parallel with the MAP kinase pathway to regulate pseudohyphal differentiation (Fig. 3). This second pathway is a nutrient-sensing pathway and involves a novel G protein-coupled receptor, Gprl, the G proteins Gpa2 and Ras2, adenylyl cyclase, cyclic AMP (cAMP), and cAMP-dependent protein kinase (12, 143, 171, 172, 223, 238, 276, 314). In S. cerevisiae, three genes encode the catalytic subunits of cAMP-dependent protein kinase, which play redundant roles in vegetative growth but specialized roles in filamentous growth. The Tpk2 catalytic subunit positively regulates filamentous growth by regulating the transcription factor Flo8, which in turn regulates Flo11 expression (223). Flo11 is a glycosyl-phosphatidylinositol (GPI)-linked cell surface protein that is required for pseudohyphal and haploid invasive growth. In particular, Flo11 plays a role in mother-daughter cell adhesion, which is required for the integrity of pseudohyphal filaments (150, 165). Flo8 was previously shown to be required for pseudohyphal differentiation, and the common lab strain S288C harbors a naturally occurring flo8 mutation that prevents filamentous differentiation (162). Tpk2 also inhibits a transcriptional repressor, Sfl1, which also regulates Flo11 expression (238). The Tpk1 and Tpk3 catalytic subunits play a negative role in regulating filamentous growth, possibly by a feedback loop that inhibits cAMP production (212).
Yeast cells express only two heterotrimeric Gα protein subunits: Gpa1, which plays a well-established role in mating, and Gpa2, which was discovered in 1988 by low-stringency hybridization with a mammalian Gα subunit but whose physiological function was unknown for many years (207). Gpa2 was subsequently discovered to be required for yeast filamentous growth, but it signals in a pathway distinct from the MAP kinase cascade (143, 171). Filamentous growth of gpa2 mutant strains is restored by exogenous cAMP, suggesting that Gpa2 regulates a cAMP signaling pathway regulating filamentous growth. In fact, earlier studies had suggested that Gpa2 might play a role in regulating cAMP production in response to extracellular glucose (207).
The Gpr1 G protein-coupled receptor was subsequently identified by a two-hybrid screen with the Gα protein Gpa2 (314, 322). This is one of only a few examples in which integral membrane proteins have been studied in the two-hybrid system; in this case, the C-terminal soluble tail of Gpr1 was found to interact with the coupled Gα protein Gpa2. The Gpr1 receptor was found to play a role important for vegetative growth, and gpr1 mutations are nearly synthetically lethal with ras2 mutations (314). Recent evidence suggests that the ligand of the Gpr1 receptor may be glucose and other structurally related fermentable sugars. When yeast cells are starved for glucose, readdition of glucose triggers a rapid and transient increase in intracellular cAMP levels (reviewed in reference 276). Most interestingly, both the Gpr1 receptor and the Gα protein Gpa2 are required for cAMP production in response to glucose readdition (54, 137, 172, 321). The Gpr1 receptor is coupled to the heterotrimeric G protein α subunit Gpa2 and is also required for pseudohyphal differentiation and plays a role in nutrient sensing (12, 172, 223, 271). Early studies suggested Gpa2 might stimulate cAMP production by adenylyl cyclase. Consistent with this, cAMP stimulates pseudohyphal differentiation and suppresses the filamentation defects of both gpr1 and gpa2 mutant strains (143, 171, 172, 223). Dominant activated Gpa2 mutations also suppress the pseudohyphal defect of mutant strains lacking the Gpr1 receptor, supporting the hypothesis that Gpa2 signals downstream of Gpr1.
Interestingly, several observations suggest that the Gpr1-Gpa2 receptor system plays a dual role in sensing both fermentable carbon sources and limiting nitrogen source. First, expression of the GPR1 receptor gene is dramatically induced by nitrogen starvation (314). Second, cAMP or dominant active alleles of Gpa2 signal filamentous growth even when nitrogen levels are increased to levels that repress differentiation of wild-type strains (171). Third, Gpa2 has been shown to bind to and inhibit the meiotic regulatory kinase Ime2 specifically under nitrogen-limiting conditions (74). These findings suggest that the sensitivity of this carbon-sensing mechanism is enhanced under nitrogen starvation conditions and that Gpa2 may receive input from other sources that sense nitrogen levels.
Interestingly, a recent report has suggested that the yeast phospholipase C homolog is in a physical complex with the Gpr1 receptor and is required for association of the Gα subunit Gpa2 with the receptor and for pseudohyphal differentiation (12). Plc1 cleaves PIP2 to produce inositol 1,4,5-triphosphate (IP3) and diacylglycerol (DAG) (83). Recent studies have revealed two inositol kinases, Ipk1 and Ipk2, which phosphorylate IP3 and its products (245, 246, 318). IP3 is phosphorylated to IP4 and IP5 by the dual-specificity kinase Ipk2, which regulates transcription (219), and the Ipk1 kinase then phosphorylates IP5 to produce IP6, which regulates mRNA export from the nucleus (318). Some studies have suggested that glucose regulates a pathway involving Ras and Plc1 in phosphatidylinositol metabolism and regulation of the plasma membrane H+-ATPase (32, 53, 124). Further studies will be required to eludicate the role of Plc1 in yeast filamentous growth. In summary, a second signaling pathway comprising the Gpr1 receptor, the Gpa2 Gα protein, and cAMP regulates pseudohyphal growth in parallel to and independently of the MAP kinase pathway (Figure 3).
The target of cAMP in S. cerevisiae is the cAMP-dependent protein kinase PKA. Yeast and mammalian PKA are similar, and both enzymes consist of a regulatory and a catalytic subunit. In yeast cells, the PKA regulatory subunit is encoded by a single gene, BCY1, and three catalytic subunits are encoded by the TPK1, TPK2 and TPK3 genes (39, 279, 280). In both S. cerevisiae and mammals, PKA in resting cells is an inactive tetramer composed of two regulatory subunits bound to two catalytic subunits. In response to external signals that increase intracellular cAMP levels, cAMP binds to the regulatory subunit and triggers conformational changes that release the active catalytic subunits. The yeast cAMP-dependent protein kinase is required for vegetative growth. Triple mutants lacking the Tpk1, Tpk2, and Tpk3 catalytic subunits are inviable, whereas mutant strains expressing any one of the three Tpk subunits are viable. These findings led to the model that the three PKA catalytic subunits are largely redundant for function.
Recent studies reveal that cAMP-dependent protein kinase plays a central role in regulating yeast pseudohyphal differentiation (223, 238). First, mutations of the PKA regulatory subunit Bcy1 dramatically enhance filamentous growth (223). Second, the PKA catalytic subunits play distinct roles in regulating filamentous growth: the Tpk2 subunit activates filamentous growth (Fig. 2), whereas the Tpk1 and Tpk3 subunits primarily inhibit filamentous growth (223, 238). The unique activating function of the Tpk2 subunit is linked to structural differences in the catalytic region of the kinase and not to differences in gene regulation or the unique amino-terminal region of the protein (223). Genetic epistasis experiments support a model in which Tpk2 functions downstream of the Gpr1 receptor and the Gα protein Gpa2 (172, 223). Importantly, activation of PKA by mutation of the Bcy1 regulatory subunit restores pseudohyphal growth in mutants lacking elements of the MAP kinase pathway, including ste12, tec1, and ste12 tec1 mutants (223, 243). Thus, the MAP kinase and PKA pathways independently regulate filamentous growth. Further analysis reveals that the PKA pathway regulates the switch to unipolar budding and invasion, whereas the MAP kinase pathway is required for cell elongation and invasion (Fig. 3).
Recent studies have defined a role for the PKA pathway in activating pseudohyphal growth via transcriptional regulation of the cell surface flocculin Flo11 by the Flo8 transcription factor (223, 243). The transcriptional repressor Sfl1 was also found to interact with the Tpk2 catalytic subunit in a two-hybrid screen (238). Tpk2 appears to enhance Flo11 expression by inhibiting the repression activity of Sfl1. How PKA regulates either Flo8 or Sfl1 is not yet understood in molecular detail. The FLO11 gene promoter is extremely large, ∼3,000 bp, and is regulated by a complex set of transcription factors that includes Ste12/Tec1, Flo8, Sfl1, Msn1/Mss10/Phd2, and Mss11 (87, 243). Taken together, these findings reveal an intimate role for the cAMP-dependent kinase in the regulation of yeast dimorphism.
Two other proteins that appear to regulate the Gpr1-Gpa2-cAMP signaling pathway under certain physiological conditions have recently been identified. One is the low-affinity cAMP phosphodiesterase Pde1 (175). Yeast cells express two different cAMP phosphodiesterases: a high-affinity form called Pde2, and a low-affinity form called Pde1. Interestingly, pde1 mutations dramatically enhance production of cAMP in response to glucose readdition, whereas pde2 mutations have little effect (175). This is in contrast to pseudohyphal differentiation, where pde2 mutations enhance filamentous growth (143, 171, 223). The effects of pde1 mutations on filamentous growth are not yet known. The Pde1 enzyme has a single PKA consensus phosphorylation site and is a phosphoprotein in vivo, and phosphorylation in crude extracts leads to modest increases in enzyme activity (175). These findings suggest that Pde1 could be part of the feedback loop that limits cAMP excursions in response to glucose signaling. A second novel regulator of the pathway is a regulator of G protein signaling (RGS) protein homolog, Rgs2, which binds to the Gpa2-GTP complex and stimulates GTP hydrolysis (288). rgs2 mutations enhance cAMP production in response to glucose, whereas overexpression of Rgs2 attenuates this response. In this regard, Rgs2 functions analogously to the RGS protein Sst2, which stimulates GTP hydrolysis of the Gpa1-GTP complex to attenuate the pheromone response in haploid yeast cells. A role for Rgs2 in pseudohyphal differentiation has not yet been established.
In summary, studies on pseudohyphal differentiation have resulted in the elucidation of several novel signaling features of the PKA pathway. First is the identification of a novel G protein-coupled receptor, Gpr1, which may be the first known G protein-coupled receptor whose ligand is a nutrient. Interestingly, Gpr1 is conserved in C. albicans and S. pombe, and the coupled G protein is known to be present in many different fungi. Second, the Gα protein Gpa2 has sequence identity with other Gα subunits of heterotrimeric G proteins, and it appears to be coupled to a classic G protein-coupled receptor. However, no associated βγ subunits have been identified. A number of candidate β and γ subunits have been mutated, but none conferred phenotypes related to pseudohyphal growth (171). We propose that Gpa2 may function either as a solo Gα subunit or in complex with other proteins such as Plc1 and Ras2. In either case, this represents a new paradigm for signaling from a G protein-coupled receptor via a very unusual type of G protein. Finally, a very interesting finding is the specialized role of the PKA catalytic subunits in regulating filamentous growth. The three catalytic subunits of PKA were thought to be functionally redundant because the essential vegetative function can be satisfied by expression of any one of the three. In contrast, Tpk2 plays a specialized role to activate filamentous growth, whereas Tpk1 and Tpk3 play an inhibitory role. The positive function of Tpk2 involves regulation of the Flo8 and Sfl1 transcription factors. The negative role of Tpk1 and Tpk3 may involve the known role of PKA in a feedback loop that inhibits cAMP production (212). Studies to localize the catalytic and regulatory subunits of PKA under different nutritional conditions have just begun (98) and may provide insights into the unique and specialized roles of Tpk2 compared to Tpk1 and Tpk3.
Cross Talk between MAP Kinase and cAMP Pathways
Recent studies have revealed several examples of cross talk between the MAP kinase and cAMP signaling pathways in the regulation of filamentous growth. First, the small G protein Ras2 plays a dual signaling role, activating both the MAP kinase signaling pathway and adenylyl cyclase (171, 201–203). Second, both the MAP kinase and the PKA signaling pathways converge to regulate the large, complex promoter of the FLO11 gene (223, 243), which encodes a cell surface protein required for pseudohyphal growth (150, 165). Third, cAMP inhibits the expression of MAP kinase-regulated reporter genes (171), and high levels of cAMP or PKA activity enhance filamentous growth but give rise to filaments comprised of round rather than elongated cells (223). Because the MAP kinase pathway regulates cell elongation, these observations suggest that the PKA pathway may antagonize signaling by the MAP kinase pathway at some level. Recent findings have suggested that during haploid invasive growth, PKA may also positively regulate signaling by the MAP kinase cascade at a level downstream of the Ste20 kinase (202). The Ste12 transcription factor has several consensus PKA phosphorylation sites, and overexpression of Ste12 can in some cases suppress pseudohyphal defects, whereas the constitutive active Ste11–4 mutant does not (170), suggesting that Ste12 could represent a target of the PKA pathway that is both positively and negatively regulated depending on cell type.
Haploid Invasive Growth
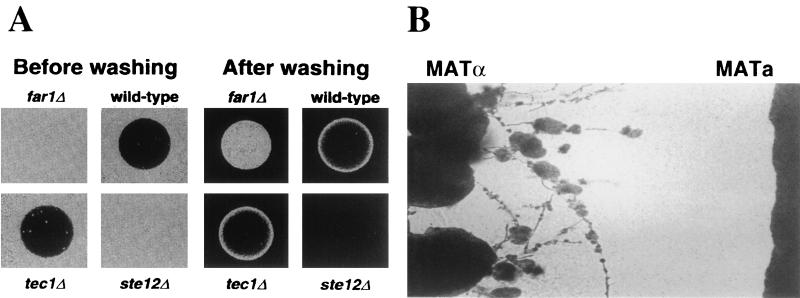
Pseudohyphal growth occurs in diploid cells in response to the presence of an abundant fermentable carbon source and limiting nitrogen source. A related morphological process, termed haploid invasive growth, has also been described, which shares some features with diploid pseudohyphal growth (237) (see Fig. 2). During diploid filamentous growth, some cells invade the agar to produce chains of cells that cannot be removed by vigorous washing. Although haploid strains do not undergo pseudohyphal differentiation under standard conditions, haploid strains derived from the Σ1278b background can invade the agar when grown on rich medium for an extended period of time. Many of the same signaling components that regulate diploid filamentous growth are also required for haploid invasive growth, including several components of the MAP kinase pathway (Ste20, Ste11, Ste7, Ste12, and Tec1) (237). The two related MAP kinases play opposing roles, and fus3 mutants are hyperinvasive whereas kss1 mutants have a defect in agar invasion. Components of the PKA pathway also regulate filamentous growth, including Gpr1, Gpa2, Ras2, Tpk2, Flo8, and Flo11 (165, 172, 202, 223). Taken together, these findings suggest that haploid invasive growth shares many features with diploid filamentous growth.
In contrast to diploid filamentous growth, which occurs on nitrogen-limiting medium, haploid invasive growth normally occurs on rich growth media, including YPD (yeast extract-peptone-dextrose) medium. It has been suggested that nutritional limitation might occur beneath colonies and stimulate haploid invasive growth, even on rich medium. When yeast cells begin to exhaust nutrients in rich medium, cellular proteins and amino acids are catabolized to produce nitrogen, resulting in the production of short-chain alcohols derived from amino acids, which are called fusel oils. Recent studies reveal that several short-chain alcohols, including isoamyl alcohol and butanol, dramatically stimulate pseudohyphal differentiation of haploid yeast strains (72, 168). How these alcohols are sensed is not yet known, but these or other metabolic products could regulate differentiation under certain culture conditions.
Other recent findings reveal that mating pheromones regulate invasive and filamentous growth of haploid S. cerevisiae strains (see Fig. 11). In the wild, most strains of S. cerevisiae are diploid, and the haploid state of the life cycle essentially represents gametes that are short-lived in nature. An elaborate pattern of axial budding has evolved in haploid yeast cells and is thought to promote more rapid mating and diploidization following meiosis. Thus, the main activity of haploid yeast strains would appear to be locating a mating partner. Recent studies using genome arrays reveal that pheromone induces several genes that are known to be induced during filamentous growth, including a hydrolytic enzyme encoded by the PGU1 gene (179, 235).
FIG. 11.
Pheromones stimulate filamentous growth of S. cerevisiae and C. neoformans. (A) Haploid MATa cells of S. cerevisiae are stimulated to invade the agar by pheromone. The disk containing synthetic pheromone was placed on the surface of the lawn of cells, and following growth, the surface of the agar was washed. (B) MATα cells of C. neoformans are stimulated to haploid fruit by confrontation with MATa cells (293). (Panel A was reprinted from reference 235 with permission of the authors and publisher; © 2000, American Association for the Advancement of Science.)
Remarkably, low concentrations of mating pheromones were found to increase agar-invasive growth (235) (see also Fig. 11). These observations suggest that haploid invasive growth may be a mechanism by which yeast cells can locate mating partners at a distance, and yeast cells are known to be responsive to gradients of mating pheromone. During invasive growth, the haploid cells become elongated, switch from an axial pattern of budding to bipolar and unipolar budding, and invade the agar (235, 237). Thus, while the role of filamentous growth in diploids may be to forage for nutrients, the role in haploid cells may be to forage for mating partners. Haploid invasive growth can occur to some extent in the absence of a mating partner. This may be attributable to a basal level of signaling in the absence of ligand. Alternatively, low concentrations of pheromone may result from cells that have switched mating type in the culture or in which repression of the silent mating type cassettes is inefficient, as is known to be the case in older yeast cells. The finding that the pheromone receptors and coupled G protein are not required for standard haploid invasive growth indicates that pheromones may not normally be involved. Pheromone-induced invasive growth requires Ste12 but is independent of Tec1 (235), whereas haploid invasive growth in the absence of pheromone requires both Ste12 and Tec1 (237). Interestingly, filamentous differentiation of haploid S. cerevisiae cells that occurs in response to mating pheromones is analogous to the recent discovery that filamentation and sporulation (haploid fruiting) of MATα cells of the human fungal pathogen C. neoformans are dramatically induced by factors secreted by MATa mating partner cells (293) (see Fig. 11). Taken together, these studies illustrate that mating and filamentous growth are linked, which is perhaps most apparent in the basidiomycetes in which mating results in a filamentous dikaryon.
These observations on the activation of filamentous growth by pheromone may be related to the previous finding that filamentation reporter genes are inappropriately activated by basal signaling of the pheromone response pathway in fus3 mutant haploid cells (177). This example of cross talk requires the pheromone-activated Gβ subunit Ste4, occurs via misactivation of Kss1 on the Ste5 scaffold, and can be further enhanced by mating pheromone.
Another example of cross talk between the MAP kinase and cAMP signaling pathways occurs in haploid cells responding to pheromone. Normally, when glucose is readded to yeast cells that have been starved for glucose, cAMP levels increase (reviewed in reference 276). In contrast, in haploid cells that have first been exposed to mating pheromone, readdition of glucose fails to stimulate cAMP production (13). The pheromone receptor Ste2 and the β subunit of the heterotrimeric G protein (Ste4) are required for inhibition of cAMP production by pheromone, but further-downstream components of the pheromone signaling pathway are not. Expression of a constitutively activated Ras2 mutant (Val-19) restored cAMP production in the presence of pheromone, and pheromone failed to inhibit the modest increase in cAMP in response to glucose that still occurs in a gpa2 mutant strain (225). Taken together, these findings support a model in which pheromone-stimulated release of the βγ subunit of the heterotrimeric G protein inhibits activation of adenylyl cyclase by the Gα subunit Gpa2. Although Gpa2 does not appear to have its own dedicated βγ subunit, this cross talk between the pheromone-regulated G protein and Gpa2 could serve to inhibit signaling via the PKA pathway under certain conditions, such as high levels of pheromone, to promote mating and inhibit filamentous growth. This pathway can only operate in haploid cells, and not during diploid filamentous growth, because the pheromones, pheromone receptors, and βγ subunits are only expressed in haploid cells.
Other Signaling Pathways
Other proteins that regulate yeast filamentous growth have been identified that do not appear to be components of either the MAP kinase or the cAMP signaling pathway. These include the ammonium transporter Mep2, which is required for filamentous differentiation in response to ammonium-limiting conditions (169), and the transcription factors Phd1, Sok2, and Ash1 (43, 92, 224, 296). How limiting nitrogen sources are sensed during pseudohyphal differentiation is not understood in detail. Map 2 ammonium permease is required for filamentous differentiation and plays a unique role not shared with the related Mep1 and Mep3 ammonium permeases (169, 170). mep/mep2 mutant strains fail to differentiate but have no growth defect on medium limiting for ammonium, leading to the hypothesis that the Mep2 protein may function to both transport and sense ammonium ions (169). The Gln3 transcriptional activator and the Gln3 repressor Ure2 that regulate the nitrogen catabolite response (NCR) are also required for pseudohyphal growth. Both gln3 mutants unable to induce the NCR response and ure2 mutants in which the NCR response is constitutive fail to differentiate, suggesting that the ability both to induce and to repress the NCR genes involved in nitrogen source utilization is required for filamentous differentiation (169). The GATA family transcription factor Ash1 represses HO endonuclease expression in haploid daughter cells, restricting mating type switching to haploid mother cells. Ash1 is also required for pseudohyphal differentiation and is localized to daughter cell nuclei in filament cells (43), where it may function to regulate Flo11 expression (224). The transcription factor Sok2 was recently found to regulate a transcription factor cascade involving Phd1, Swi5, and Ash1, which in turn regulates expression of proteins and enzymes (Flo11, Egt2, and Cts1) involved in mother-daughter cell adhesion and separation (224). Several additional proteins also appear to inhibit filamentous growth, including the Elm1 protein kinase homolog and the B subunit of protein phosphatase 2A (Cdc55) (28). Recent studies suggest that these proteins regulate Cdc28 via the Hsl1-Hsl7-Swe1 regulatory cascade and that the Cdc28-cyclin complexes regulate filamentous growth (76).
In addition, several recent studies indicate that both the G1 cyclins Cln1, Cln2, and Cln3 and the Clb2 mitotic cyclin may play roles in filamentous growth (6, 167). cln1 and cln2 mutations inhibit filamentous growth, whereas cln3 mutations enhance filamentation. Mutations in the F-box component of the Skp1-Cdc53-F box protein (SCF) ubiquitin ligase complex, Grr1, dramatically enhance the stability of Cln1 and Cln2 and enhance filamentous growth. Epistasis analysis suggests that Cdc28-Cln1/2 may act at an early step in the MAP kinase cascade (167). In addition, the Cdc28-Cln1/2 complex can phosphorylate Ste20, and cln1 and cln2 mutations are synthetically lethal with mutations in the CLA4 gene, which encodes an Ste20 homolog (220, 307). These findings imply that the MAP kinase cascade may be regulated at the level of Ste20 by Cdc28 in complex with G1 cyclins. Further studies will be required to understand these additional regulatory elements in molecular detail.
MATING IN FISSION YEAST SCHIZOSACCHAROMYCES POMBE
Like budding yeast, the fission yeast S. pombe is homothallic and has a defined sexual cycle involving mating between haploid cells of opposite mating types; in S. pombe, the mating partners are called h+ and h− cells (reviewed in reference 315). In contrast to S. cerevisiae, mating in S. pombe is regulated by both pheromones and nutrients. Nitrogen limitation induces many of the components of the pheromone response pathway and is required for mating (reviewed in references 78 and 211). Also in contrast to S. cerevisiae, the diploid state of S. pombe is relatively unstable, and meiosis and sporulation usually occur immediately following cell fusion during mating. The difference in life style between these two yeasts is thought to have arisen because S. cerevisiae has evolved primarily to be diploid in the wild, whereas S. pombe occurs predominantly as a haploid in nature. As a consequence, the diploid yeast S. cerevisiae responds to nutrient limitation by undergoing either pseudohyphal differentiation or meiosis and sporulation, whereas when the haploid yeast S. pombe encounters adverse nutritional conditions, it must first mate and can then undergo meiosis and sporulate.
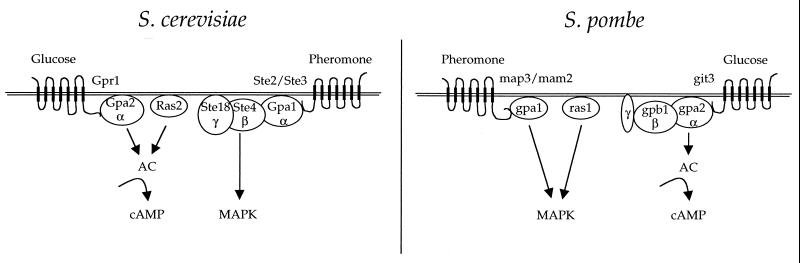
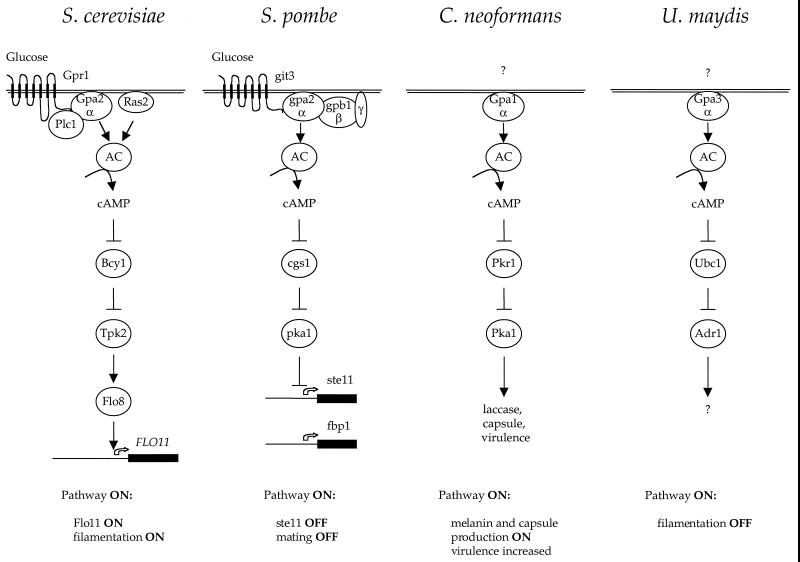
This difference in life cycle has led to an interesting difference in the signaling pathways regulating mating in the two yeasts. In S. cerevisiae, mating is primarily regulated by a linear signaling pathway composed of the pheromone-activated MAP kinase cascade. In S. pombe, two signaling pathways coordinately regulate mating (Fig. 5). One is a pheromone-activated MAP kinase signaling pathway similar to the one that operates during mating in S. cerevisiae. The second is a nutrient-regulated, G protein-cAMP-PKA signaling pathway quite similar to the nutrient-sensing Gpr1-Gpa2-PKA pathway that regulates pseudohyphal differentiation in S. cerevisiae. Thus, in one organism (S. cerevisiae), these two signaling pathways have evolved cell type specificity, so that one functions in response to pheromone during mating and both function to detect nutrients in diploid cells for filamentous growth (Fig. 3 and 6). In contrast, in the other organism (S. pombe), these two pathways function coordinately in the same cell type to regulate mating under adverse nutrient conditions and in response to pheromone (Fig. 5).
FIG. 5.
Signaling pathways regulating mating of S. pombe. Similar to S. cerevisiae diploid pseudohyphal growth, mating in S. pombe is regulated by two parallel signaling pathways. One responds to pheromones and involves a conserved MAP kinase (MAPK) signaling pathway. The second is a nutrient-sensing pathway that, in the presence of abundant nutrients, stimulates cAMP production to repress ste11 expression and inhibit mating. CAP, cyclase-associated protein.
FIG. 6.
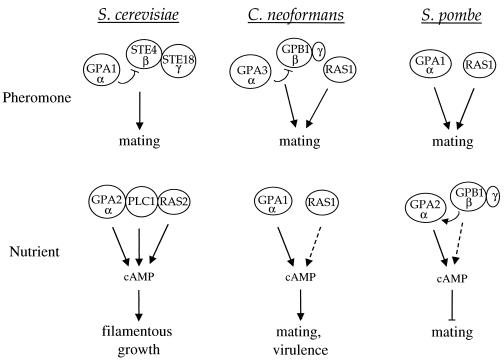
Role of G proteins in pheromone response in budding and fission yeasts and the human fungal pathogen C. neoformans. The G protein subunits regulating pheromone and nutrient responses in these organisms are depicted. Dashed arrows depict weaker signaling.
Nutrient-Sensing G Protein-cAMP-PKA Pathway
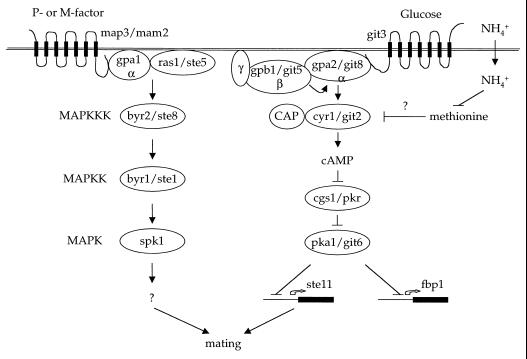
A conserved nutrient-sensing signaling pathway has been defined that regulates mating and gene repression in S. pombe (Fig. 5). The components of this pathway were identified by two different approaches. In the first, Yamamoto and colleagues defined a role for cAMP in repressing mating and then set out to identify components of the cAMP signaling pathway (195, 315). In the second, Hoffman and Winston discovered that glucose dramatically repressed expression of the fbp1 gene, encoding fructose-1, 6-bisphosphatase, and by using lacZ reporter fusions isolated mutations in 10 complementation groups that block repression by glucose to identify the git genes (glucose-insensitive transcription) (38, 109, 110).
By these two different approaches, the elements of a conserved signaling pathway that senses nutrients and regulates cAMP production were defined. This pathway is remarkably similar to the Gpr1-Gpa2-cAMP-PKA pathway discussed above that regulates pseudohyphal differentiation in S. cerevisiae. As in budding yeast, glucose stimulates cAMP production in S. pombe, and classic studies revealed that either glucose limitation or nitrogen limitation results in a reduction in cAMP levels in S. pombe (195). cAMP levels decline rapidly in response to glucose starvation, whereas cAMP levels decline more gradually in response to nitrogen limitation, suggesting that there are different levels of control for carbon compared to nitrogen source sensing. S. pombe expresses git3, which is a homolog of the S. cerevisiae Gpr1 receptor (298). Interestingly, git3 lacks the asparagine-rich third intracellular loop found in Gpr1, indicating that this unusual region does not play an obligate role in signaling. The git3 G protein-coupled receptor homolog is required for glucose-induced repression of the fbp1 gene, and git3 mutants mate on nutrient-rich medium (38, 298). Genetic studies support a model in which the git3 receptor is coupled to the Gα subunit gpa2 (118, 213, 298). gpa2 is most homologous to the nutrient-sensing Gpa2 protein of S. cerevisiae that is required for pseudohyphal growth (143, 171). In contrast to wild-type cells, gpa2 mutant cells mate under nutrient-rich conditions, indicating a defect in nutrient sensing.
Most interestingly, the single Gβ subunit that has been identified in S. pombe is required for adenylyl cyclase activation in response to glucose and repression of the fbp1 gene by glucose (151). Moreover, the phenotype of gpb1 mutant cells is identical to that of gpa2 mutant strains, namely, mating under nutrient-rich conditions. These findings suggest that gpb1/git5 is coupled to the gpa2 Gα protein and not to the pheromone-activated gpa1 Gα protein, in contrast to an earlier report (130) (Fig. 7). gpb1/git5 is not required for basal cAMP production, whereas gpa2 is, suggesting some Gβ-independent action of gpa2 on adenylyl cyclase.
FIG. 7.
Role of G proteins in filamentous growth and mating in budding and fission yeasts. In both yeasts, two Gα proteins regulate cellular responses to pheromone and nutrients. In both, one functions as a heterotrimeric G protein with a βγ partner, and the other signals in conjunction with Ras. Interestingly, the heterotrimeric G protein is coupled to pheromone receptors in S. cerevisiae but to the nutrient sensor in S. pombe. Importantly, the βγ subunits appear to function with only one of the two Gα subunits in both organisms. MAPK, MAP kinase; AC, adenylylcyclase.
Additional git mutations that render transcription of the fbp1 gene insensitive to glucose are suppressed by cAMP and encode functions upstream of adenylyl cyclase (cyr1/git2), including git1, git7, and git10. These additional components may represent the cyclase-associated protein, the Gγ subunit, hexokinase, or Plc1, all of which are known or would be predicted to regulate glucose signaling to adenylyl cyclase in other systems.
Several other components of the cAMP signaling pathway in S. pombe have also been identified. They include adenylyl cyclase itself, cyr1/git2. Quite interestingly, Hoffman and Winston discovered examples of intragenic complementation between different alleles of adenylyl cyclase (109). This unusual genetic behavior usually indicates that a protein has two functional domains. Consistent with this notion, alleles of one class of git2 mutations had no defect in cAMP production in vitro, whereas the other class failed to make cAMP in extracts. These findings suggest that adenylyl cyclase may exist as a dimer in which different domains of the protein have unique functions. In addition to adenylyl cyclase, both the regulatory (cgs1) and the catalytic subunits (pka1) of cAMP-dependent protein kinase have been identified (69, 182). PKA represses the transcription of both the ste11 gene (266), a key regulator of many mating-specific genes, and the fbp1 gene.
To summarize (see also Fig. 15), when glucose levels are high, cAMP is produced, PKA is active, and mating of S. pombe is repressed. When glucose or nitrogen levels fall, cAMP production is reduced, PKA is less active, and ste11 is induced. An important point is that because the role of PKA is to mediate repression of ste11, cAMP has a negative effect on mating in S. pombe. This is in contrast to the positive role that cAMP plays in promoting filamentous growth in S. cerevisiae. Because the pathways can be wired to have either positive or negative regulatory effects, one can essentially achieve either outcome with one common input to the signaling cascade.
FIG. 15.
Role of cAMP in fungal morphogenesis. The nutrient-sensing receptor-G protein-cAMP-PKA signaling pathways are depicted. Two related receptors, Gpr1 and git3, play a conserved role in glucose sensing in S. cerevisiae and S. pombe; homologous receptors in C. neoformans and U. maydis remain to be identified. The receptors are coupled to a highly conserved Gα protein (variously called Gpa2, gpa2, Gpa1, and Gpa3), and the C-terminal domain that is known to interact with receptor has >80% sequence identify. The Gα protein is coupled to adenylyl cyclase, and cAMP regulates PKA. Targets of PKA largely remain to be identified but will likely include a variety of transcription factors. Note that cAMP functions to activate or inhibit development depending on the particular organism. AC, adenylyl cyclase.
By analogy with S. cerevisiae, the most likely ligand for the git3 receptor is glucose. An important unsolved question, then, is the role of nitrogen limitation in mating in S. pombe. Most interestingly, mating and meiosis occur in response to either nitrogen or glucose limitation. This suggests that nitrogen signaling also impinges on the git3-gpa2 signaling pathway or that overlapping signaling pathways sense glucose and nitrogen sources. One very intriguing recent report is that the addition of methionine can stimulate mating of S. pombe on nutrient-rich medium, which normally represses mating (249). One possible model is that methionine biosynthesis is stimulated and represses cAMP production in response to nitrogen source limitation. cAMP levels were found to decline to lower levels in cells cultured with methionine. However, methionine also restored ste11 and fbp1 expression in cells exposed to high levels of exogenous cAMP, suggesting that the effects of methionine may occur downstream or independently of cAMP production (249). We found no effect of methionine, cysteine, S-adenosylmethionine, S-adenosylhomocysteine, or biosynthetic intermediates of either amino acid on pseudohyphal growth in S. cerevisiae (M. E. Cardenas and J. Heitman, unpublished results), suggesting that this aspect of nitrogen sensing may differ in the two yeasts.
Pheromone-Regulated MAP Kinase Pathway
The components of a pheromone-regulated MAP kinase signaling pathway that coordinately regulates mating in S. pombe have also been identified (Fig. 5, 6, and 7). The P- and M-pheromones have been identified and synthesized, and the pheromone receptors (map3 and mam2) have also been identified, sequenced, and mutated (reviewed in reference 315). These components are required for mating in S. pombe and serve to regulate the production of conjugation tubes and early cell fusion steps. As discussed below, these components also function later and are required for meiosis of the diploid cell. The pheromone receptors are coupled to what appears to be the α subunit of a heterotrimeric G protein, gpa1 (215). Importantly, gpa1 plays an active role in mating (Fig. 6 and 7). gpa1 mutants are sterile, and dominant active alleles trigger conjugation tube formation in both mating types. The active signaling role played by the Gα subunit gpa1 is in contrast to budding yeast, in which the Gβγ subunit signals mating and the Gα subunit couples the Gβγ subunit to the receptor, inhibits signaling by Gβγ in the absence of pheromone, and also functions in adaptation. Only one Gβ subunit has been identified, and its function is clearly coupled to the nutrient sensor and not to the pheromone receptor. This is similar to the situation in budding yeast, in which the Gβ subunit Ste4 is coupled to only one of the two Gα subunits, in that case to the pheromone-regulated Gpa1 protein (Fig. 6). These observations suggest that, in contrast to mammals, in which Gβγ subunits are thought to interact with and regulate multiple Gα subunits, in fungi Gβγ subunits are coupled to only one of the two Gα subunits. This implies either that the other Gα subunit, gpa1 in S. pombe and Gpa2 in S. cerevisiae, functions with a nontraditional type of subunit, or that these Gα proteins function as solo Gα subunits, similar to other small GTP-binding proteins such as Ras.
Most interestingly, the ras1 homolog that has been identified in S. pombe functions in activating that MAP kinase pathway during mating and does not appear to play a prominent role in regulating cAMP production (85, 86, 206). In S. cerevisiae, Ras1 and Ras2 both regulate cAMP production by adenylyl cyclase, and Ras2 also regulates activation of the MAP kinase pathway during filamentous growth (202, 203). ras1 mutants of S. pombe have a mating defect, and dominant active ras1 alleles render cells hypersensitive to pheromone. The exchange factor that activates Ras1 to the GTP-bound form is called ste6, and ste6 expression is markedly induced by nitrogen starvation, suggesting that ras1 may be activated by regulated expression of its GDP-GTP exchange factor (115).
These studies illustrate a very interesting parallel with the signaling pathway regulating S. cerevisiae filamentous growth (Fig. 6 and 7). In that case, the nutrient-sensing G protein-coupled receptor (GPCR) Gpr1 is coupled to Gpa2, and Gpa2 in conjunction with Ras2 regulates cAMP production by adenylyl cyclase. There are no known Gβ8 subunits for Gpa2. In S. pombe, it is instead the pheromone receptors that are coupled to a Gα subunit, gpa1, with no apparent Gβ8 subunits, and again there is a similar role for signaling by both the Gα subunit gpa1 and the ras1 protein. We propose that these represent two examples of a novel class of GPCR-G protein system which lacks a classic heterotrimeric G protein. In these cases, the Gα subunit may be physically and functionally coupled to either or both Ras and phospholipase C and would thus represent very novel types of heterodimeric or heterotrimeric (but not αβγ) G proteins. Clearly, further study of these very interesting signaling modules is of significant interest.
The components of a conserved MAP kinase module have also been found to regulate mating in S. pombe. These include homologs of the MAP kinase kinase kinase (byr2/ste8), the MAP kinase kinase (byr1/ste1), and a single MAP kinase (spk1). Interestingly, several of these components can function when heterologously expressed in S. cerevisiae mutant strains, suggesting that the pathway has been functionally conserved during the divergence of budding and fission yeasts (210).
Pheromones Required for Meiosis
Interestingly, mating pheromone signaling is also required for meiosis and sporulation of diploid S. pombe cells. This is in stark contrast to S. cerevisiae, in which, following cell fusion, the genes encoding pheromone receptors and pheromones are repressed and no longer expressed in the diploid zygote (263). This repression does not occur following fusion of haploid S. pombe cells, and continued pheromone signaling is required for meiosis and sporulation. Thus, diploid mutants lacking either the M-factor or the P-factor receptor undergo meiosis normally, but diploid mutants lacking both receptors are unable to undergo meiosis (272). Synthetic pheromones have also been employed to demonstrate regulation of meiosis-regulating genes by pheromone signaling (305).
These findings further underscore the intimate link that exists between mating, meiosis, and sporulation in fission yeast, which is similar to the life cycle of basidiomycetes, such as U. maydis and C. neoformans, in which pheromones regulate cell fusion and the heterokaryon then undergoes a dimorphic transition to filamentous growth. In the basidiomycetes, continued pheromone signaling is required for filament formation and may serve to signal clamp cell fusion during nuclear division and migration.
SIGNAL TRANSDUCTION IN CANDIDA ALBICANS
C. albicans is the most common human fungal pathogen and causes a wide range of superficial mucosal diseases as well as life-threatening systemic infections in immunocompromised patients (216). This fungus is dimorphic and can either grow as a budding yeast (blastospores) or switch to a filamentous form (either hyphae or pseudohyphae) under a variety of environmental conditions (216). The ability to undergo these reversible dimorphic switches is essential for the virulence of this pathogen (164). C. albicans is a diploid organism for which the sexual cycle has only very recently been discovered (117, 183). The characterization of a filamentous state in the baker's yeast S. cerevisiae has made it possible to use this as a model to compare regulation of filamentation in C. albicans and S. cerevisiae (93).
MAP Kinase Signaling Pathway
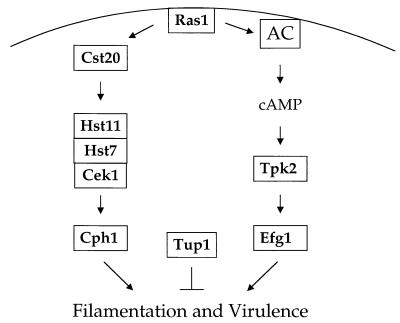
Multiple pathways have been shown to regulate the morphogenic transition between budding and filamentous growth in C. albicans (Fig. 8). A transcription factor, Cph1, which is homologous to the Ste12 transcription factor that regulates mating and pseudohyphal growth in S. cerevisiae, has been identified. (160, 184). In S. cerevisiae, Ste12 is regulated by a conserved MAP kinase cascade. A related MAP kinase cascade has been identified in C. albicans. The cascade consists of the kinases Cst20 (homologous to the p21-activated kinase [PAK] kinase Ste20), Hst7 (homologous to the MAP kinase kinase Ste7), and Cek1 (homologous to the Fus3 and Kss1 MAP kinases) (51, 132, 154, 299). Null mutations in any of the genes in the MAP kinase cascade (Cst20, Hst7, or Cek1) or the transcription factor Cph1 confer a hyphal defect on solid medium in response to many inducing conditions; however, all of these mutants filament normally in response to serum (63, 132, 154). Interestingly, although a cek1/cek1 MAP kinase mutant strain forms morphologically normal filaments in response to serum, it has a minor growth defect on serum-containing medium (63). The cek1/cek1 mutant strain also has a virulence defect that may be attributible to this growth defect (63, 100). This indicates that the Cek1 MAP kinase may function in more than one pathway or that deletion of the gene causes aberrant cross talk between distinct MAP kinase cascades, similar to the altered signaling that occurs in a fus3 mutant of S. cerevisiae. The other elements of the pathway have small but varied effects on virulence. cst20/cst20 mutant strains have a modest virulence defect in a mouse model of systemic candidiasis (154). However, hst7/hst7 and cph1/cph1 mutant strains are able to cause lethal infection in mice at rates comparable to wild-type strains (154, 164).
FIG. 8.
Signaling cascades regulating virulence of C. albicans. Two parallel signaling cascades involving a MAP kinase and a cAMP-PKA signaling pathway regulate filamentation and virulence of this human fungal pathogen. AC, adenylycyclase.
In addition to these components, a MAP kinase phosphatase, Cpp1, has been identified which regulates filamentous growth in C. albicans (62). Disruption of both alleles of the CPP1 gene derepresses hyphal production and results in a hyperfilamentous phenotype. This hyperfilamentation is suppressed by deletion of the MAP kinase Cek1 (62). cpp1/cpp1 mutant strains are also reduced for virulence in both systemic and localized models of candidiasis (62, 99).
EFG1 Transcription Factor
In addition to the transcription factor Cph1 and the MAP kinase module that regulates it, a second transcription factor, Efg1, that regulates filamentous growth in C. albicans has been identified (Fig. 8) (265). Efg1 is a conserved basic helix-loop-helix-type protein that is homologous to the Phd1 transcription factor of S. cerevisiae. Similar to Phd1, heterologous overexpression of Efg1 can induce pseudohyphal growth in S. cerevisiae (265). Overexpression of Efg1 can also induce filamentous growth in C. albicans (265). An efg1/efg1 mutant strain has a moderate but not complete defect in hyphal growth in response to many environmental conditions (164). efg1/efg1 mutant strains also show an aberrant morphology in the presence of serum, forming exclusively pseudohyphae instead of the true hyphae that are produced by wild-type strains (164).
An efg1/efg1 cph1/cph1 double mutant strain has a much more severe defect in filamentous growth and does not filament under almost any conditions tested, including the presence of serum (164). In addition, while the efg1/efg1 mutant strain has a minor reduction in virulence and the cph1/cph1 mutant has little or no defect, an efg1/efg1 cph1/cph1 double mutant strain is essentially avirulent (164). Thus, Cph1 and Efg1 define elements of two separate pathways that together are essential for both filamentation and virulence in C. albicans.
cAMP-PKA Pathway
A PKA-dependent pathway has been shown to regulate filamentous growth of C. albicans under some conditions (Fig. 8). An increase in cAMP levels accompanies the yeast to hyphal transition, and inhibition of the cAMP phosphodiesterase induces this transition (244). Moreover, two cell-permeating PKA inhibitors, myristoylated protein kinase inhibitor (myrPKI) (14–24)amide and the small-molecule PKA inhibitor H-89, both block hyphal growth induced by N-acetylglucosamine, but not in response to serum (42).
More recently, a PKA gene, TPK2, was identified and shown to be required for hyphal differentiation in response to a number of conditions, including the presence of serum (260). Hyphal formation in the tpk2/tpk2 mutant strain was not completely eliminated, however, and the defect was less apparent during growth at 37°C (260). Overexpression of the Tpk2 catalytic subunit also induced hyphal growth under normally noninducing conditions both in liquid culture and on solid medium (260). This is consistent with a model in which PKA acts positively in one of two or more pathways regulating dimorphism.
The phenotypes of a tpk2/tpk2 mutant strain were suppressed by overexpression of either the MAP kinase Cek1 or the transcription factor Efg1 (260). On the other hand, overexpression of Tpk2 could suppress the hyphal defect of a cek1/cek1 mutant strain but not that of an efg1/efg1 mutant strain (260). Efg1 has a single PKA consensus phosphorylation site, which may be required for its transcriptional activation activity (260). These findings suggest that the Efg1 transcription factor may lie downstream of cAMP and the PKA homolog Tpk2 in one of two major pathways regulating hyphal morphogenesis in C. albicans. Interestingly, while Efg1 plays a prominent role in regulating filamentation in C. albicans, mutation of the related protein Phd1 confers only a minor defect in S. cerevisiae filamentation (92, 164, 296).
Function of Ras
In S. cerevisiae, the Ras2 protein regulates pseudohyphal growth and functions upstream of both the MAP kinase and cAMP-PKA pathways and, in conjunction with Ras1, is essential for viability. A single Ras homolog, Ras1, has been identified in C. albicans which is not essential for survival (81). ras1/ras1 mutant strains have a severe defect in hyphal growth in response to serum and other conditions (81). In addition, while a dominant negative Ras1 mutation (Ras1A16) caused a defect in filamentation, a dominant active Ras1 mutation (Ras1V13) enhanced the formation of hyphae (81). ras1/ras1 mutant strains exhibit a filamentation defect similar to that of efg1/efg1 cph1/cph1 mutant strains, and evidence from S. cerevisiae indicates that Ras2 lies upstream of both the cAMP-PKA and MAP kinase pathways regulating pseudohyphal growth (171, 201, 223, 236). These findings are consistent with a model in which Ras1 lies upstream of the related pathways in C. albicans and regulates hyphal morphogenesis. However, these studies indicate that additional pathways may also control filamentation in C. albicans, because even the ras1/ras1 and efg1/efg1 cph1/cph1 mutant strains are able to form hyphae under some conditions (81, 234).
Repression of Filamentation
Another signaling protein that regulates morphogenesis of C. albicans is Tup1, a homolog of the general transcriptional repressor in S. cerevisiae (33) (Fig. 8). S. cerevisiae tup1/tup1 mutant strains have multiple phenotypes, including increased flocculence, temperature-sensitive growth, and inability to sporulate. Heterologous expression of the C. albicans Tup1 protein can suppress these mutant phenotypes. S. cerevisiae tup1/tup1 mutant strains are also defective in pseudohypal growth, but this is complicated by the fact that Tup1 is required for maintenance of the diploid state in S. cerevisiae (33). In contrast, the tup1/tup1 deletion in C. albicans results in a constitutive hyphal growth phenotype under all conditions tested (33). Therefore, although Tup1 does seem to act as a general transcriptional repressor in C. albicans as it does in S. cerevisiae, it seems to have a unique function in repressing the induction of the hyphal phase in this fungus. In addition, although the Tup1 protein has been shown to be epistatic to the transcription factor Cph1, recent genetic evidence indicates that it does not lie downstream of either Cph1 or Efg1 (34, 252), but rather in an independent pathway that regulates filamentation. Furthermore, a cph1/cph1 efg1/efg1 tup1/tup1 triple mutant retains the ability to filament in response to environmental signals, providing further evidence that additional pathways regulating filamentous growth exist (34). In addition, the hyperfilamentous tup1/tup1 mutant strain has a virulence defect, which is consistent with the finding that a cpp1/cpp1 mutant strain also exhibits enhanced hyphal formation and impaired virulence. These findings support a model in which both the yeast form and the filamentous form are required to maintain a stable infection.
Downstream Effectors of Efg1
Although there seems to be a clear link between dimorphism and virulence in C. albicans, it is difficult to unambiguously establish this because it is possible that the signaling elements also regulate other cellular processes. One of the ways to dissect the relative contributions of these various components to the virulence of the organism is to identify the downstream effectors of these signaling molecules. One of the genes recently shown to be regulated by Efg1 is HWP1, which encodes a protein of unknown function that is homologous to GPI-anchored proteins (252). An hwp1/hwp1 mutant strain is reduced for hyphal growth under a variety of different environmental conditions (252). Expression of Hwp1 is reduced in an efg1/efg1 mutant and increased in a tup1/tup1 mutant (252). Thus, Efg1 and Tup1 regulate at least some of the same target genes and may be acting coordinately to modulate dimorphism in C. albicans. Ece1, which is expressed during cell elongation, is also not expressed in an efg1/efg1 mutant (252). However, Ece1 is not regulated by Tup1, and an ece1/ece1 mutation does not affect filamentous growth despite being regulated by Efg1 (252).
Efg1 has also been shown to regulate other morphogenic events, including chlamydospore production and phenotypic switching (259, 261). Because Efg1 is a complex signaling molecule that regulates a variety of morphogenic processes, it cannot yet be determined to what degree each of these functions of Efg1 contributes to virulence. Recent studies of single and multiple mutant strains lacking Tup1, Cph1, and Efg1 reveal that each makes a unique contribution to the regulation of genes involved in filamentous growth, suggesting that multiple pathways rather than a single central regulatory pathway are responsible (34). Further analysis of the downstream effectors of each of these transcription factors will be required to determine the relative contribution of the processes controlled by these proteins to the overall virulence of the organism.
Filamentation in Response to Serum
It has long been known that C. albicans filaments in response to a variety of environmental conditions, including serum. However, recent evidence has disputed the notion that the active portion of serum that causes filamentation is albumin. Feng and colleagues have shown that the active molecule in serum is actually a serum filtrate <1 kDa in size (81). Because this molecule could not be purified, they speculated that it may be multiple small molecules that act coordinately to induce this response. Several small molecules that regulate filamentous growth of S. cerevisiae have recently been identified, including glucose and other carbohydrates, nitrogen sources, short-chain alcohols, and mating pheromone (168, 169, 172, 235).
Sexual Cycle in C. albicans!
Although C. albicans is diploid and was thought to be an asexual organism, several lines of evidence suggested that a cryptic sexual cycle could exist. First, it was shown some time ago that DNA exchange could be forced between strains of C. albicans following mild UV irradiation and that these events involved recombination rather than merely the formation of heterokaryons (268). Furthermore, more recently, Graser and colleagues used population genetics to show that the population structure of C. albicans suggests that some form of recombination is occurring within this species (97). Several homologs of genes regulating mating in S. cerevisiae have been identified in C. albicans. In addition to a G protein α subunit homolog and the MAP kinase cascade components, a homolog of the Ste6 protein which transports a-factor pheromone in S. cerevisiae has been identified in C. albicans (230).
Recently, regions of the C. albicans genome resembling the MATa and MATα mating type loci in S. cerevisiae have been discovered and named the MTLα and MTLa loci (MTL stands for mating type-like) (116). The two chromosomes are only 48% identical in this region but greater than 99% identical in the rest of the genome, findings that are consistent with other MAT loci that have been identified (116). In addition, this locus contains homologs of the a1, α1, and α2 genes that regulate mating type in S. cerevisiae, and these genes are arranged in the same manner as in S. cerevisiae (116). In S. cerevisiae, the diploid state is maintained by the action of the a1/α2 protein complex, which represses the expression of haploid-specific genes. Evidence suggests that the a1/α2 homolog functions similarly in C. albicans. The heterotrimeric G protein α subunit homolog gene CAG1 has a consensus binding site for the a1/α2 complex. Moreover, expression of the CAG1 gene in either S. cerevisiae or C. albicans has been shown to be regulated by the a1 and α2 proteins (116).
Recent studies by two independent groups have demonstrated that mating can occur in C. albicans. Magee and Magee engineered MTLa/MTLa and MTLα/MTLα homozygous strains by inducing loss and reduplication of chromosome V by selection for growth on sorbose (183). With appropriately marked auxotrophic strains, they observed the production of prototrophic tetraploid strains following coincubation. Independently, Hull et al. engineered strains with mutations in the a1 or α1 and α2 genes of the mating type-like loci (117). In this case, mating was observed following coinfection of mice with marked strains and again gave rise to prototrophic tetraploid progeny. In these later studies, no mating has thus far been detected in vitro, suggesting that strain differences may contribute to the ability to detect mating in vitro. These studies overturn the long-held dogma that C. albicans is asexual and open the door to further genetic analysis in this important human fungal pathogen. An important next step will be to determine whether these tetraploid products of mating can be induced to undergo meiosis and sporulate and to establish where in vivo mating occurs and whether there is an impact on virulence.
Conclusions and Perspectives
Proper regulation of dimorphism in C. albicans appears to be important for the virulence of this human pathogen. Dimorphism is regulated by two major pathways, a conserved MAP kinase module culminating in the transcription factor Cph1, and the cAMP-PKA pathway that regulates the transcription factor Efg1p. Dimorphism is also regulated by the transcriptional repressor Tup1. Disruption of signaling by deletion of Efg1 and Cph1 causes nearly complete abrogation of hyphal growth. However, because the filamentation observed in a cph1/cph1 efg1/efg1 tup1/tup1 triple mutant is still environmentally regulated, additional signaling elements may be important in the regulation of dimorphism. Finally, although C. albicans has long been considered an asexual diploid, recent evidence reveals that mating does occur. By comparison with the signaling processes in similar fungi, many of the same elements required for dimorphism may also be important for this newly discovered sexual cycle.
SIGNAL TRANSDUCTION IN CRYPTOCOCCUS NEOFORMANS
C. neoformans is a basidiomycetous fungus and a significant human pathogen with worldwide distribution. Its importance as an opportunistic pathogen has increased in the past two decades, largely as a result of AIDS, cancer chemotherapy, and immunosuppression for organ transplants (193). Infections begin in the lung following inhalation of small, dessicated yeast cells or spores, both of which are small enough to fit into the alveoli of the lung (209, 267). The organism then spreads via the blood to other organs, especially the central nervous system (193). Survival in the central nervous system is enhanced, possibly by the presence of abundant neurotransmitters that can be scavenged as the diphenolic precursors to synthesize the virulence factor melanin. Cryptococcal infection is now the leading cause of fungal meningoencephalitis, which is uniformly fatal if untreated (40).
Basic and Molecular Biology
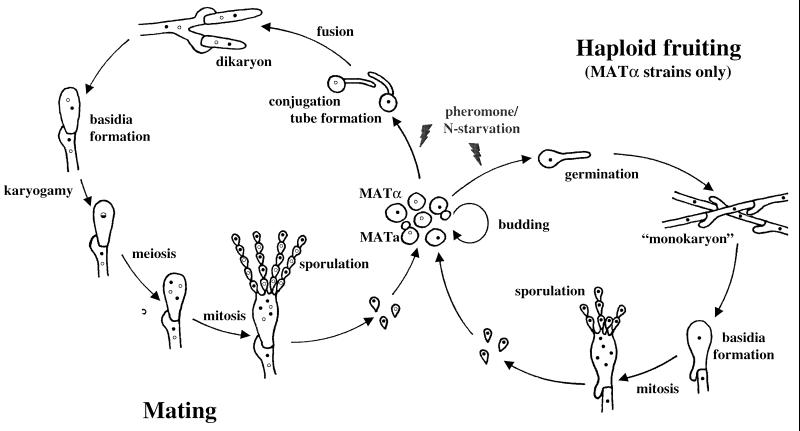
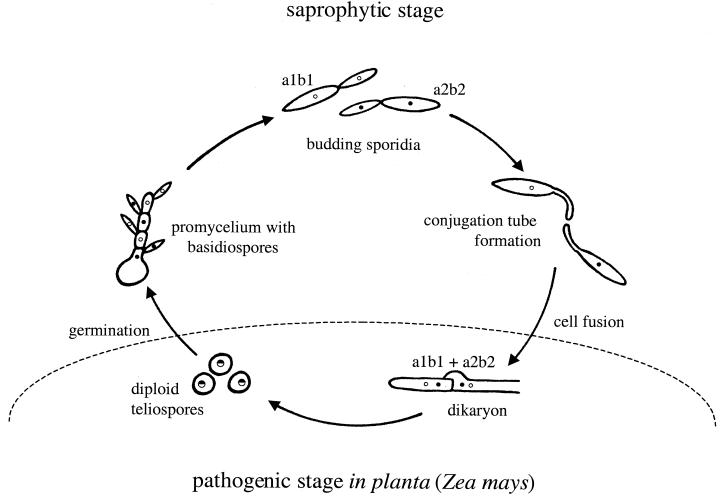
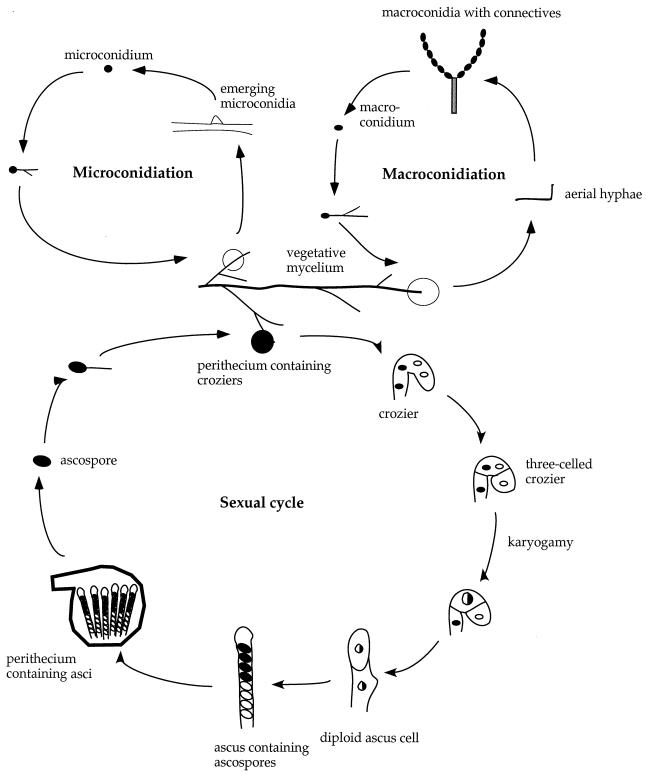
C. neoformans exists predominantly as a haploid yeast. It is heterothallic and has a bipolar mating system, with both MATa and MATα mating types (8, 144, 145). Two signals regulate mating, nutrient limitation and pheromones. Opposite mating types form conjugation tubes and fuse to produce the filamentous heterokaryon, which is also known in the teleomorphic designation as Filobasidiella neoformans (Fig. 9). Following cell fusion, dikaryotic filaments with fused clamp connections are formed. The tips of these filaments differentiate to form terminal basidia, where nuclear fusion (karyogamy) occurs. Meiosis and sporulation ensue, and four long chains of basidiospores are produced that can subsequently germinate into yeast cells.
FIG. 9.
Life cycle of C. neoformans. Mating begins with conjugation tube formation, leading to cell fusion, followed by formation of dikaryotic filaments with fused clamp connections. Terminal basidia form from the tips of mating filaments, where nuclear fusion (karyogamy) occurs. Meiosis and sporulation follow, and long chains of basidiospores are produced, which subsequently germinate into yeast cells. Filamentation independent of a mating partner (haploid fruiting) is observed in MATα strains in response to nitrogen limitation and dessication.
MATα cells can also filament and sporulate in response to nitrogen starvation in the absence of MATa cells (80, 301). This process, termed haploid or monokaryotic fruiting, has been observed in MATα strains in response to severe nitrogen and water deprivation (301). MATα strains undergo robust haploid fruiting in response to factors secreted by MATa cells (293), likely MFa pheromone (see Fig. 11).
C. neoformans strains are classified into five serotypes (A, B, C, D, and AD) and three varieties: C. neoformans var. grubii (A), C. neoformans var. neoformans (D), and C. neoformans var. gattii (B and C) (84, 146). C. neoformans var. gattii is thought to be limited in nature to regions surrounding certain eucalyptus trees, and most infections with this variety occur in tropical and subtropical regions. C. neoformans var. neoformans is not geographically limited and is associated with pigeon nests and soil contaminated with bird excreta. Recent reports also suggest that serotype A strains are associated with certain types of trees in South America (E. Castaneda, personal communication). Of the subtypes, C. neoformans var. grubii serotype A is the most common cause of human disease. Interestingly, mating type is linked to both prevalence in the environment and virulence. In a survey of natural and clinical isolates, the MATα mating type was 40-fold more abundant in environmental isolates and 30-fold more abundant in clinical isolates than its MATa counterpart (147). Furthermore, in a direct comparison of congenic strains, MATα strains were more virulent than MATa strains in a murine model of systemic cryptococcal infection (148).
C. neoformans is an excellent model organism for the study of biological processes in pathogenic fungi. First, a congenic pair of serotype D C. neoformans isolates, differing only at the mating type locus, were created by consecutive backcrossing (105, 148). Auxotrophs have been derived from these strains, facilitating classical genetic analysis. Furthermore, two transformation techniques have been described. Biolistic transformation has been used successfully in both serotype A and D strains (64, 282). With this technique, several genes have been disrupted by homologous recombination at rates of 2 to 25% (7, 9, 59, 166, 217, 293, 320). Electroporation and a positive-negative selection method have been used to transform serotype D strains (77), and several genes have been disrupted by this approach (44, 45, 247). Both the ADE2 and URA5 genes have been used commonly as selectable markers for transformation. Additionally, positive selection systems using resistance to hygromycin B or nourseothricin have been described that allow genes to be disrupted in wild-type or unmarked strains (57, 189).
The pathophysiologic consequences of gene disruption can be studied in animal models that mimic various stages of human infection. These models include the rabbit intracisternal model of cryptococcal meningitis (227), the murine intravenous model (149), and the rat and murine inhalation models (96).
Signaling Pathways Regulating Differentiation and Pathogenicity
Several different signaling pathways regulating morphogenesis and virulence have been investigated in C. neoformans, including a cAMP-PKA pathway, a conserved MAP kinase pathway, and a calcineurin-regulated pathway (Fig. 10) (290).
FIG. 10.
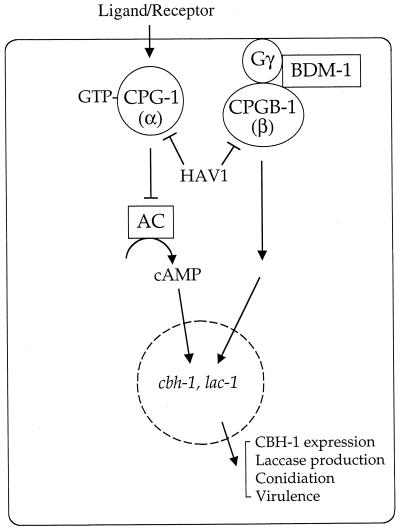
Signal transduction pathways in C. neoformans. Lightly shaded proteins have been identified in C. neoformans. Darkly shaded proteins are mating type specific. Two of the 24 proteins depicted here have not yet been identified in C. neoformans but are hypothesized from characterization of their roles in other fungi. These are the γ subunit of the pheromone-regulated G protein and the Gpr1 receptor homolog, which are depicted with question marks and dashed symbols. CAM, calmodulin.
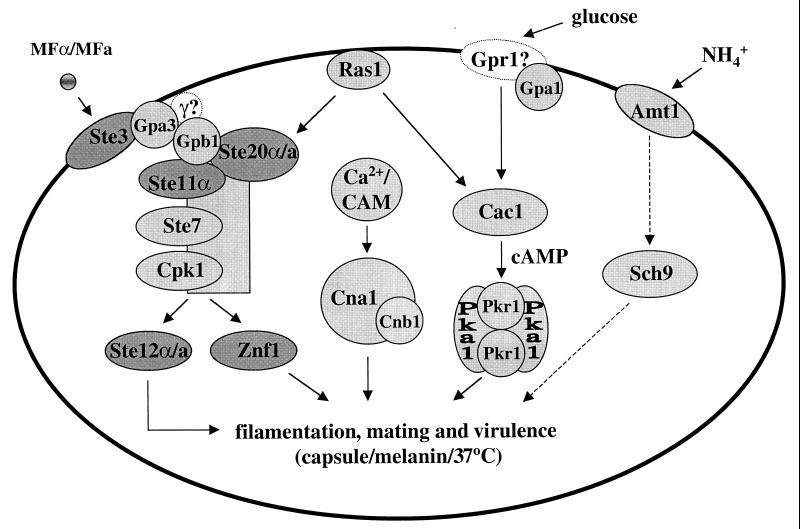
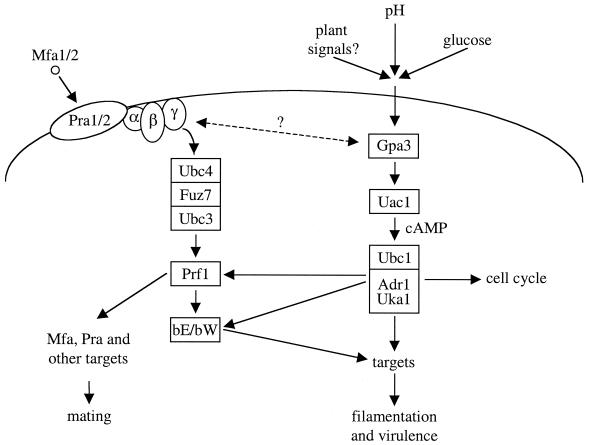
Elements of a cAMP-PKA pathway have been identified, including the Gα protein GPA1, and this pathway has been shown to be important for morphogenesis and virulence (9). Additionally, a member of the RAS family of small GTP-binding proteins, RAS1, has been identified (7). GPA1 is a component of a signaling cascade that transduces changes in extracellular concentrations of nutrients via the cAMP pathway (Fig. 6, 10, and 15). RAS1 has been implicated in control of both the cAMP pathway and a MAP kinase cascade and may mediate cross talk between MAP kinase and cAMP-dependent signaling pathways. A heterotrimeric G protein β subunit, GPB1, has also been identified and functions in response to and activates a pheromone MAP kinase cascade during mating; Gpb1 does not function in the nutrient-regulated Gpa1 pathway (Fig. 10 and 15) (293).
The finding that MATα cells are innately more virulent than MATa cells has motivated molecular analysis of the MATα mating type locus. One candidate downstream target of the MAP kinase cascade, the STE12α homolog, is present only in MATα cells and may play a role in the increased virulence of this mating type in serotype D strains (47, 300, 320). Finally, the role of calcineurin in both serotypes A and D has been investigated, as well as the potential for calcineurin inhibitors to serve as antifungal compounds (60, 61, 68, 217).
cAMP-Mediated Signaling Cascades
A cAMP-dependent pathway regulates several important cellular processes in C. neoformans, including capsule production, melanin formation, mating, and virulence (Fig. 10 and 15). GPA1, a G protein α subunit, was cloned (283), and a gpa1 mutant strain was created by biolistic transformation and homologous recombination using the ADE2 gene as a selectable marker (9). Interestingly, this mutant strain failed to induce two known virulence factors, melanin and capsule, and was also defective in mating. Additionally, the gpa1 mutant strain was avirulent in the rabbit model of cryptococcal meningitis. Exogenous cAMP suppresses the gpa1 mutant phenotypes in vitro, suggesting that a cAMP cascade regulates mating and melanin and capsule production.
Interestingly, while the GPA1 protein serves as a common signaling element for these diverse environmental signals, there is little cross-induction of cellular responses. For example, capsule is induced in response to iron limitation but not in response to glucose or nitrogen starvation. Importantly, the cell surface or intracellular receptors that initiate these signaling cascades have not yet been identified, and these as well as other unidentified signaling components likely direct the appropriate cellular response to nutrient starvation. The GPA1 homolog in S. cerevisiae is the Gpa2 Gα protein, which senses nutrients and regulates pseudohyphal differentiation by an analogous signaling cascade (143, 171, 223) (see Fig. 6 and 15). Gpa2 physically interacts with the G protein-coupled receptor Gpr1 that is also involved in sensing nutrients during pseudohyphal growth (172, 314). By analogy with these findings in S. cerevisiae, a Gpr1 receptor homolog may act upstream of the nutrient-sensing Gα protein GPA1 in C. neoformans.
Pheromone-Activated MAP Kinase Cascades
Elements of a C. neoformans MAP kinase cascade have been identified that function in a pheromone response pathway during mating (198, 293) (Fig. 10). In this organism, GPB1, the β subunit of a heterotrimeric G protein, has been shown to play an active role in mediating responses to pheromones in the early steps of mating via a MAP kinase cascade. Thus, the role of GPB1 in C. neoformans is analogous to the role of the Ste4-Ste18 βγ subunits in activating mating in S. cerevisiae (293) (Fig. 6). gpb1 mutants are sterile and exhibit a severe defect in a cell fusion assay. In contrast, GPB1 gene overexpression stimulates conjugation tube formation. Furthermore, overexpression of the conserved MAP kinase homolog CPK1 suppresses the mating defect of gpb1 mutant strains, whereas cAMP does not (293). This is in contrast to gpa1 mutants, in which the mating defect is suppressed by exogenous cAMP but not by overexpression of the CPK1 gene. These findings support a model in which the GPA1 Gα and GPB1 Gβ subunits are components of two different signaling pathways, one that senses nutrients and signals via a cAMP cascade, and another that senses mating pheromones and signals via a MAP kinase cascade.
Monokaryotic (haploid) fruiting in C. neoformans is also regulated by GPB1 via a MAP kinase cascade (293). gpb1 mutant strains are defective in haploid fruiting. This defect is suppressed by overexpression of the presumptive MAP kinase target STE12α on galactose filament agar.
The finding that the pheromone-regulated GPB1 Gβ subunit regulates haploid fruiting came as a complete surprise, because it had been thought that haploid fruiting in C. neoformans was quite analogous to pseudohyphal differentiation in S. cerevisiae cells (301). In S. cerevisiae, filamentous differentiation occurs in diploid cells, which do not express pheromones, pheromone receptors, or the coupled G protein, and these components do not regulate pseudohyphal differentiation. The finding that the pheromone-regulated Gβ protein GPB1 is required for haploid fruiting in C. neoformans caused us to reevaluate the signals that normally trigger haploid fruiting. Remarkably, we discovered (293) that confrontation of MATα cells with MATa cells dramatically enhanced haploid fruiting, and robust filamentation and basidium and basidiospore production was now observed in 2 to 4 days, as opposed to the modest filamentation that occurs in 2 to 4 weeks with MATα cells alone (Fig. 11). It was shown that this is haploid fruiting and not mating because the filaments produced were monokaryotic, linked by unfused clamp connections, and basidia were produced with short chains of basidiospores that were all MATα mating type. In addition, the effects of MATa cells on fruiting of adjacent MATα cells could occur through a dialysis membrane, suggesting that a small factor such as MFa pheromone activates fruiting, explaining why a component of the pheromone-sensing pathway might be involved.
These observations suggest that the normal function of haploid fruiting may be during the early steps in mating, to allow MATα cells to forage for MATa mating partners. Foraging could occur via filamentation or dispersal of aerial spores by wind currents. Pheromone-regulated haploid fruiting is quite distinct from S. cerevisiae pseudohyphal differentiation, which occurs in diploid cells, is not pheromone regulated, and likely allows cells to forage for nutrients under adverse conditions. Interestingly, Charlie Boone's and Mike Snyder's laboratories have recently discovered that low doses of mating pheromone stimulate haploid S. cerevisiae cells to undergo filamentous growth involving cell elongation, unipolar budding, and agar invasion (235; S. Erdman and M. Snyder, personal communication). This form of yeast filamentous growth may enable haploid yeast cells to forage for mating partners. In summary, pheromones and mating partner cells stimulate filamentous growth in divergent ascomycetous and basidiomycetous yeasts, suggesting that filamentous growth is intimately linked with the sexual cycle (Fig. 11). In diploid yeast cells and C. neoformans MATα cells alone, these signaling cascades may have been coopted to forage for nutrients or to produce infections basidiospores in response to adverse nutritional conditions.
Ras Signaling and Cross Talk between Signaling Cascades
A Ras homolog has recently been identified in C. neoformans (7). Previous studies in S. cerevisiae have shown that two Ras proteins, Ras1 and Ras2, sense changes in the extracellular environment and regulate cAMP synthesis and cell cycle progression (122, 129, 281). The yeast Ras2 protein also regulates MAP kinase signaling and regulates filamentous and invasive growth via both MAP kinase and cAMP pathways (171, 202, 203).
A gene encoding a C. neoformans Ras homolog, RAS1, was disrupted in a serotype A strain (M001) by homologous recombination using the ADE2 gene as a selectable marker (7). The resulting mutant strain was viable but unable to grow at 37°C, defective in mating and agar adherence, and avirulent in a rabbit model of cryptococcal meningitis. The ras1 mutant strain showed no defect in melanin or capsule production, two inducible cAMP-regulated phenotypes (9). A dominant active RAS1 mutant allele, RAS1Q67L, induced haploid fruiting in the serotype A strain H99 (7, 320). Haploid fruiting could not be induced, however, when the RAS1Q67L mutant allele was introduced into several isogenic ste12α mutant strains (320). Interestingly, exogenous cAMP only partially suppressed the ras1 mating defect and fully suppressed the agar adherence defect. The mating defect was fully suppressed in ras1 strains overexpressing GPB1, the gene encoding a G protein β subunit that regulates a pheromone response MAP kinase pathway. Conversely, introduction of the dominant active RAS1Q67L allele into a gpb1 mutant strain did not restore mating competence. These results suggest that RAS1 functions upstream of GPB1 in the mating process. Taken together, these data suggest the potential for RAS-mediated cross talk between MAP kinase and cAMP signaling pathways (Fig. 10). Finally, it is significant that the high-temperature growth defect of the ras1 mutant strain was not restored by exogenous cAMP, overexpression of MAP kinase signaling elements, or both. This suggests the presence of another, as yet unidentified, RAS-specific signaling cascade in C. neoformans.
Mating Type Locus and Virulence
As previously discussed, MATα strains are more prevalent in nature and in clinical specimens. They are also capable of haploid fruiting, while MATa strains are not. This has led to the hypothesis that infectious spores resulting from haploid fruiting may account for the increased prevalence of MATα in clinical isolates (301). Spores are the correct size to fit into the alveoli of the lung, the initial site of infection. However, even in a murine model in which the cells are injected intravenously, bypassing the lung entirely, MATα strains are inherently more virulent than MATα strains (148).
This linkage of both virulence and monokaryotic fruiting to the MATα locus is unique among basidiomycetes and has motivated molecular analysis of the MATα mating type locus. An approximately 40-kb region of the MATα locus was identified and found to contain a gene encoding a small peptide pheromone homolog (198). Subsequently, an additional, distinct region of the MATα locus was found to encode the Ste12α homolog (300). In S. cerevisiae, Ste12 is a transcriptional activator and the target of a MAP kinase cascade including the proteins Ste20, Ste11, and Ste7. Furthermore, the growth conditions that lead to pseudohyphae are very similar to the conditions that result in haploid fruiting in C. neoformans (301). Overexpression of the C. neoformans STE12α homolog stimulates haploid fruiting and production of the known virulence factor melanin by inducing the expression of CNLAC1, which encodes diphenol oxidase (247, 300, 306). This finding led to the hypothesis that STE12α, through activation of laccase, was a molecular link between mating type and virulence in serotype D strains of C. neoformans (300).
The STE12α gene has been disrupted in both serotype A and D strains of C. neoformans (47, 320). In both serotypes, Ste12α plays only a very modest role in mating but is absolutely required for haploid fruiting in response to nitrogen starvation.
The role of STE12α in regulating virulence has been further characterized at the molecular level in the congenic serotype D laboratory strain JEC21 (47). Promoters of the known virulence-associated gene CNLAC1 and the capsule-associated genes CAP59, CAP60, and CAP64 were individually fused with the β-glucoronidase gene (GUS) in a URA5 plasmid. These reporter constructs were then transformed into ste12α mutant and wild-type strains. All four constructs produced significantly lower GUS activity in the ste12α mutant strain than in the wild type. However, on visual inspection, no decrease in capsule or melanin production could be appreciated. Conversely, a strain in which STE12α was placed under the control of the GAL7 promoter was subsequently transformed with the GUS reporter constructs. When transformants were grown in galactose to stimulate overexpression of STE12α, GUS activity increased. This increase was most dramatic with the CAP64 promoter. However, GUS activity also increased in the two control constructs, one in which the ADE2 promoter was fused to the GUS gene and the other in a vector with a promoterless GUS gene.
Virulence studies in animal models have been conducted in both serotype A and D strains, with differing results (47, 320). Mice were infected via injection in the lateral tail vein with the wild-type serotype D MATα strain JEC21 (B-4500), a protrophic first-generation progeny of a mutant ste12α strain, and a protrophic first-generation offspring of an ste12α mutant strain reconstituted with the wild-type STE12α gene (47). The ste12α mutant strain produced significantly lower mortality compared to the wild type; virulence was nearly restored with the reconstituted strain compared to the wild type. Similar numbers of CFU were observed in the brains of mice infected with the wild type and the ste12α mutant strain, suggesting no difference in growth rate (47). Furthermore, the in vivo capsule size was smaller in the ste12α mutant strain, ranging from 0.5 to 3.0 μm, compared to the wild type and reconstituted strains, with capsule sizes of 1 to 7 μm. Lastly, histopathologic comparisons of the ste12α mutant strain and wild-type strain show similar disruption of mouse neural parenchyma, with large, multilocular lesions containing yeast cells and foamy macrophages. However, more inflammatory cells were present in samples obtained from the ste12α mutant, suggesting that inoculation of the mutant strain provoked a stronger immune response than inoculation with the wild type.
The STE12α gene was similarly isolated from a pathogenic serotype A isolate of C. neoformans (H99) and disrupted by homologous recombination (320). ste12α mutant strains showed only modest defects in mating. Importantly, ste12α mutant strains were completely defective in monokaryotic fruiting. However, ste12α mutant strains were fully virulent in both a rabbit model of cryptococcal meningitis and the murine systemic tail vein injection model (320). STE12α thus does not appear to play a primary role in either mating or virulence in serotype A, but rather is specialized to regulate filamentous growth during haploid fruiting (320). In summary, Ste12α may not be required for virulence of pathogenic serotype A clinical isolates, but may be required for virulence in the congenic laboratory serotype D strain JEC21, either because strain JEC21 is attenuated and separated by 12 backcrosses from a clinical isolate or as a result of significant divergence of serotype A and D strains (105, 148).
Calcineurin Regulates Growth and Virulence
Calcineurin is a Ca2+-calmodulin-activated phosphatase with catalytic and regulatory subunits (106). Calcineurin has been identified in several fungi, including S. cerevisiae, in which it regulates cation homeostasis, cell wall biosynthesis, and recovery from pheromone growth arrest. In C. neoformans, the calcineurin A gene has been identified and disrupted by homologous recombination in both serotype A and D strains (61, 217). Calcineurin was found to be essential for growth at 37°C and virulence in both the rabbit model of cryptococcal meningitis and a murine inhalation model. Serotype A mutants are hypersensitive to Na+ and Li+ compared to the isogenic wild type, suggesting a role for calcineurin analogous to that in S. cerevisiae, in which calcineurin regulates expression of the Na+ and Li+ efflux pump Pmr2 (61). In serotype D, however, calcineurin does not appear to regulate cation homeostasis (61).
Recently, it has been suggested that inhibitors of calcineurin A may have potential as antifungal compounds. Cyclosporin A (CsA) and FK506 are toxic to pathogenic fungi, including C. neoformans (61, 131, 218, 284). CsA binds to cyclophilin A, and FK506 binds to the immunophilin FKBP12. The target of both of these complexes is calcineurin. While the in vivo immunosuppressive activity of CsA and FK506 exacerbates cryptococcal infections in a rabbit model of C. neoformans meningitis, nonimmunosuppressive analogs of both CsA and FK506 that retain antifungal activity have been identified (61, 218). The CsA and FK506 analogs are toxic at 37°C but not at 24°C, consistent with the temperature-dependent growth of calcineurin mutant strains. Furthermore, mutations on the cyclophilin A-CsA binding surface of calcineurin (Val344Arg and Val344Lys) confer resistance to CsA analogs, and two mutant strains completely lacking the cyclophilin A protein are resistant to CsA analogs. Interestingly, one of the nonimmunosuppressive analogs has a γ-hydroxy substitution on the leucine 4 residue on the effector surface of CsA that appears to prevent binding to mammalian calcineurin but increases affinity for calcineurin relative to CsA-cyclophilin A in C. neoformans (61).
The in vitro anticryptococcal activity of FK506 and its nonimmunosuppressive analog has also been evaluated in combination with other known antifungal agents (bafilomycin, capsofungin, and fluconazole) (68). In these studies, FK506 and its analog, L-685,818, exhibited marked synergistic effects with all of these agents. Interestingly, the mechanism of action of these drug combinations appears to be independent of calcineurin inhibition but requiring FKBP12 (FK506 plus bafilomycin); dependent upon FKBP12-dependent inhibition of calcineurin by FK506 (FK506 plus caspofungin); and independent of FKBP12 and calcineurin but potentially involving the known ability of FK506 to inhibit multidrug resistance pumps known to export azoles from fungal cells (FK506 plus fluconazole). These synergistic combinations have not yet been tested in animal models of cryptococcal infection but suggest that signaling pathways can be targeted by novel antifungal agents.
LIFE CYCLE OF USTILAGO MAYDIS
The phytopathogenic hemibasidiomycete U. maydis is the causal agent of corn smut disease. The life cycle of U. maydis is characterized by a dimorphic switch between a haploid yeast-like cell form, known as sporidia, and an infectious filamentous dikaryon (17, 19) (Fig. 12).
FIG. 12.
Life cycle of U. maydis. The life cycle of U. maydis is characterized by a dimorphic switch from a saprophytic yeast form to a pathogenic filamentous growth form. The maintenance of the pathogenic stage is dependent on the presence of the plant host Zea mays.
Sporidia, the asexual form of U. maydis, have a slightly elongated cell shape and divide by budding, growing as saprophytes on refuse and manure. Infectious dikaryotic hyphae are formed following fusion of two sporidia with different alleles at the mating type loci a and b. In response to a mating partner, cells develop filament-like protrusions called conjugation tubes (Fig. 12). Cell fusion occurs at the tips of the conjugation tubes, and a dikaryotic filamentous mycelium is formed. The resulting hyphae are pathogenic on maize and are able to infect meristematic tissue in any above-ground part of the plant. Hyphal proliferation in plant tissues induces neoplastic growth and gall formation. Fungal cells inside these tumor-like structures round up and karyogamy occurs, giving rise to uninucleate diploid teliospores. When the tumors burst, millions of black spores are released and can be spread by wind and rain. The resulting soot-like appearance gave this fungus its common name of smut fungus. In response to proper environmental conditions, meiosis occurs and the teliospores germinate to form a promycelium consisting of four haploid cells, including the original teliospore. Finally, basidiospores bud off the promycelium and germinate to produce sporidia once again.
Pheromone Response Pathway
Sexual development in U. maydis is regulated by the a and b mating type loci (reviewed in references 41 and 141). Recognition of the compatible mating partner, conjugation tube formation, and fusion of the two sporidia are largely regulated by the biallelic a mating type locus. Both a1 and a2 contain two tightly linked genes (mfa and pra) that encode a secreted pheromone and a G protein-coupled pheromone receptor. In addition to regulating the initial events during mating, the a locus also contributes to the maintenance of filamentous growth of U. maydis in planta (29, 262).
The establishment of the infectious dikaryotic filaments and the pathogenicity of this stage are both regulated by the b locus. This multiallelic locus encodes two divergently transcribed homeodomain proteins termed bE and bW. Fused sporidia that are heterozygous for the b locus form an active bEx/bWy heterodimer, generating an active transcription factor that regulates filamentous growth and pathogenicity. There is a functional linkage between the a and b mating type loci. The activation of the receptor by pheromone binding induces several genes, including the a- and b-specific genes. Following cell fusion, the active bE/bW heterodimer represses a gene transcription.
Pheromone binding to its cognate receptor is thought to activate a MAP kinase signal transduction pathway similar to the well-characterized pheromone response pathway in S. cerevisiae. Homologs for several components of this pathway have recently been identified in U. maydis (Fig. 13).
FIG. 13.
Signaling networks in U. maydis. Mating, filamentation, and virulence in the smut fungus U. maydis are regulated by a pheromone response pathway (left) and the cAMP-dependent signaling pathway acting through PKA (Ubc1, Adr1, and Uka1) (right).
Four α subunits for heterotrimeric G proteins (Gpa1 to Gpa4) have been identified in U. maydis by a PCR-based approach (231). Each of the four gpa genes was disrupted, and only gpa3 mutant strains exhibited a mating defect. Additional experiments also revealed a role for gpa3 in filament formation and pathogenicity. Furthermore, strains deleted for gpa3 fail to induce specific genes (like mfa and the b genes) in response to pheromone, whereas a constitutively active gpa3 mutant allele induced expression of these genes 50-fold. These experiments reveal that gpa3 plays a central role in pheromone response and sexual development of U. maydis, and it was proposed that this might represent the α subunit of the G protein coupled to the pheromone receptor. However, a constitutive active form of Gpa3 does not allow pheromone-independent mating, and gpa3 mutants are elongated, similar to uac1 mutants lacking adenylyl cyclase. Budding yeast growth is restored in gpa3 mutants by cAMP, suggesting that gpa3 functions upstream of adenylyl cyclase (142).
Quite interestingly, Gpa3 is closely related to Gpa2 from S. cerevisiae, gpa2 from S. pombe, and GPA1 from C. neoformans, which are all involved in a nutrient-regulated cAMP-dependent signaling pathway (Fig. 14) (9, 118, 171). Gpa3 appears to be involved in a similar pathway in U. maydis. The Gα subunit in the pheromone response pathway remains to be identified. In C. neoformans, the pheromone signal is transmitted by the G protein βγ complex (293). If this is also the case in U. maydis, then disruption of the α subunit responsible for the pheromone response will not inhibit signal transmission but activate it instead, increasing mating of the mutant strain. Alternatively, the gpa1 to gpa4 genes could have redundant functions, which could explain why the single mutant strains do not show a mating defect. A third, less likely possibility would be a dual role for Gpa3 in both the pheromone-activated MAP kinase and the cAMP-dependent signaling pathways.
FIG. 14.
Evolutionary tree showing the relationship between fungal Gα proteins. The branch on the left includes a family of Gα proteins that function in nutrient sensing, some of which are contained within the box. (Adapted from Figure 2 of Loubradou et al. [174] with permission of the authors and publisher; © 1999, Genetics Society of America.) Um, U. maydis; Sc, S. cerevisiae; Ca, C. albicans; An, A. nidulans; Cp, C. parasitica; Mg, M. grisea; Nc, N. crassa; Cn, C. neoformans; Sp, S. pombe; Cc, Coprinus congregatus; Uh, Ustilago hordei; Pa, Podospora anserina; Kl, Kluyveromyces lactis; Pc, Pneumocystis carinii..
Several putative components of a conserved MAP kinase module have been identified in U. maydis (Fig. 13). Ubc4, a MAP kinase kinase kinase (MAPKKK) homolog, and Ubc3, a MAP kinase homolog, were identified in a screen for suppressors of the filamentous uac1 adenylyl cyclase mutation (187). ubc3 was also identified by a degenerate PCR approach and termed kpp2 (204). A similar approach identified Fuz7, a MAP kinase kinase (MAPKK) homolog (20). The ubc3 and fuz7 genes have been characterized in more detail.
The analysis of fuz7 deletion strains showed that this MAPKK homolog is involved in a-dependent processes as well as a-independent processes (20). fuz7-deleted strains fail to respond to mating pheromone and produce no conjugation tubes. In addition, fuz7 mutant strains are unable to induce conjugation tubes in a mating partner, likely due to reduced production or secretion of pheromone. Interestingly, the induction of mfa transcription by pheromone does not appear to be affected (231). Filament formation in mating assays is reduced in a dosage-dependent manner. In addition, no hyphae are formed if diploid strains homozygous for a fuz7 deletion and bearing different a and b alleles are shifted from rich to nitrogen starvation medium, whereas this dimorphic transition is readily observed in wild-type diploid strains. Both in planta tumor formation and teliospore formation are impaired by the fuz7 mutation. Taken together, these results provide evidence that Fuz7 might act in two different signal transduction pathways: an a-dependent pathway that is activated by pheromone, and an a-independent pathway that may be stimulated by plant signals (20). fuz7 deletion strains expressing the constitutive active allele of gpa3 are indistinguishable from wild-type strains bearing this allele, further suggesting that Gpa3 is not directly involved in the pheromone response pathway (231).
Strains deleted for the MAP kinase homolog ubc3 (kpp2) have several phenotypes in common with fuz7 mutant strains. Both ubc3 (kpp2) and fuz7 mutants fail to produce conjugation tubes in response to pheromone, exhibit defects in pheromone production, and mate poorly (188, 204). In a solopathogenic background, ubc3 deletion leads to defects in filamentation and tumor formation. These findings indicate a role for Ubc3 in virulence of U. maydis, although the effect is less dramatic compared to fuz7-deleted strains. One explanation for this finding might be, by analogy to budding yeast, that a second MAP kinase is present that partially compensates for loss of ubc3. The reduced virulence can be partially overcome by overexpressing an active bE/bW heterodimer, indicating that Ubc3 is probably a component of the pheromone response pathway and is needed for virulence in the plant host. Further evidence that Ubc3 is part of the MAP kinase cascade is the reduced basal-level expression of mfa1 and the loss of induction by the pheromone signal, which are not observed in fuz7 mutants. Whether Fuz7 and Ubc3 are components of the same MAP kinase module remains to be determined.
A MAPKKK homolog, Ubc4, has also been identified but still remains to be characterized in further detail. ubc3 and ubc4 were identified in the same screen for suppressors of adenylyl cyclase mutants, and the two have indistinguishable phenotypes in this genetic background (187). Therefore, it seems likely that Ubc4 is also a component of a MAP kinase module that mediates pheromone response.
In budding yeast, the MAP kinase module activates a downstream transcription factor in response to pheromone. This is also true in U. maydis, in which a transcription factor of the HMG-box class, termed Prf1, has been identified (103). Interestingly, the Prf1 protein contains several phosphorylation sites for both MAP kinases and cAMP-dependent PKA, and Prf1 is regulated posttranscriptionally by both pathways (104). prf1 mutant strains are completely sterile and nonpathogenic. The same phenotypes are exhibited but are less dramatic in strains expressing a prf1 allele in which the MAP kinase phosphorylation sites have been removed by site-directed mutagenesis (204). These mutant strains retain the ability to respond to environmental signals at a reduced level, probably due to activation by the cAMP-dependent signal transduction pathway.
Prf1 activates transcription by binding to a promoter element termed the pheromone response element, which is also found in the promoter of the prf1 gene (286). Beyond this autoregulation, the main function of Prf1 is proper regulation of the mating type genes within the a and b loci during mating and virulence. Overexpression of an active bE/bW heterodimer restores virulence of prf1 deletion strains (103). In addition to transcriptional regulation in response to pheromone, prf1 is also regulated by nutritional signals (carbon source). A 40-bp upstream activating sequence (UAS) was found to be important for induction of prf1 by carbon source, and this UAS also mediates repression by increased cAMP levels (104). A novel protein that binds to this UAS, called Ncp1, has been identified. Ncp1 is not required for prf1 regulation by nutrients, likely because redundant factors play an overlapping role. The PKA pathway plays a similar role in S. cerevisiae in sensing fermentable carbon sources via the glucose receptor Gpr1 and the Gα protein Gpa2. Because Gpa3 of U. maydis plays a similar function in the PKA pathway, it may also interact with a glucose receptor homologous to Gpr1 and activate the cAMP-dependent pathway in response to various fermentable carbon sources.
cAMP-Dependent Signaling
In addition to pheromone, other signals are also important for mating in U. maydis (Fig. 13 and 15). The first evidence that a cAMP-dependent signaling pathway plays a vital role in differentiation of U. maydis came from the observation that deletion mutants lacking adenylyl cyclase (uac1) exhibit constitutive filamentous growth (23, 94). This phenotype is suppressed by exogenous cAMP, which restores a yeast-like budding pattern. Interestingly, although gpa3 mutant strains do not produce filaments, they do produce highly elongated cells. This phenotype is suppressed by exogenous cAMP, providing further evidence that Gpa3 is a component of the cAMP-dependent signaling pathway (142). In addition, when either uac1 mutants or wild-type strains are exposed to exogenous cAMP, they exhibit a multiple budding pattern with lateral buds and clustering of cells (94, 142). This observation implies that the cAMP signaling pathway might be important for both sexual differentiation and proper progression through the cell cycle. Haploid uac1 deletion strains exhibit hyphal growth but are not pathogenic, indicating that hyphal growth alone is not sufficient for full virulence (23). Fully compatible crosses of uac1 deletion strains are unable to grow in planta and are not pathogenic. Strains deleted for adenylyl cyclase show reduced levels of basal pheromone expression. The transmission of the pheromone signal is blocked in uac1 deletion mutants, which fail to induce pheromone expression in the presence of an activating pheromone. The constitutive activation of Uac1 increases transcription of pheromone-inducible genes about 50-fold (see above). These findings provide clear evidence for cross talk between the MAP kinase and cAMP-dependent signaling pathways (142).
Several components of the MAP kinase signaling pathway were identified as suppressors of uac1-deleted strains (94, 187). Another gene identified by this approach was ubc1, which encodes the regulatory subunit of cAMP-dependent PKA. Deleting the regulatory subunit of PKA results in a constitutively active protein kinase mimicking high intracellular levels of cAMP. Strains deleted for ubc1 show a multiple budding phenotype similar to cells treated with high levels of cAMP. In addition, these strains are unable to form filaments in response to changes in O2 and CO2 levels, like wild-type cells. Filament formation was greatly reduced in compatible crosses homozygous for the ubc1 mutation in vitro and in planta (94). Although phytohormone production and initial pathogenicity symptoms like chlorosis were present, no tumors formed. Diploid strains homozygous for a ubc1 deletion are also reduced for virulence, indicating that postfusion events necessary for pathogenicity are blocked (75, 95). The basal expression level of pheromone-inducible genes in ubc1-deleted strains was increased to levels similar to those in cells treated with exogenous cAMP or expressing a constitutively activated adenylyl cyclase (142). Strains deleted for both uac1 and ubc1 were almost indistinguishable from ubc1 single mutants, showing a slightly elongated cell shape in addition to the multiple budding phenotype. Moreover, no difference between these two mutants was observed in virulence studies, indicating that activation of PKA (via a ubc1 mutation) suppresses the loss of adenylyl cyclase (uac1) (94).
In addition to the regulatory subunit, the genes encoding two catalytic subunits of PKA were identified and named adr1 and uka1 (75). adr1 mutants resemble uac1 adenylyl cyclase mutant strains and undergo filamentous growth as haploid strains and are impaired for virulence in planta in crosses or as diploid strains. In contrast, uka1 mutations deleting the second catalytic subunit resulted in no obvious phenotype. adr1 uka1 double mutants show slightly reduced filament formation in comparison to an adr1 single mutant. There are also preliminary data for the existence of a third catalytic subunit of PKA in U. maydis, which may explain why these mutants are viable. Alternatively, PKA may be dispensable for vegetative growth in U. maydis. Taken together, these findings suggest that Uka1 and Adr1 may have partially overlapping functions but Adr1 is the major catalytic subunit of PKA.
Conclusions and Perspectives
The pheromone response pathway and the cAMP-dependent signal transduction pathway are crucial for the sexual development and virulence of U. maydis (Fig. 13). Many of the components of these pathways have been identified, revealing remarkable conservation in the signaling cascades regulating development, mating, and virulence of the model ascomycetous yeasts S. cerevisiae and S. pombe and the pathogenic basidiomycetes C. neoformans and U. maydis. As depicted in Fig. 15, the conserved nutrient signaling pathway utilizes a conserved G protein-coupled receptor, Gα protein (Gpa2, gpa2, GPA1, Gpa3), cAMP, and PKA. Interestingly, how these pathways are wired differs; for example, cAMP activates filamentous growth in S. cerevisiae and positively regulates virulence in C. neoformans, whereas cAMP plays a negative role and inhibits mating in S. pombe and inhibits filamentous growth in U. maydis. PKA is activated by cAMP, but can then play both positive and negative roles. By activating or inhibiting transcriptional activators and repressors (such as Flo8 and Sfl1), one common pathway can promote or inhibit development in different organisms.
Several interesting questions remain. Some data provide evidence for cross talk between the MAP kinase and cAMP pathways, but the level at which this occurs remains to be resolved. One type of cross talk might be through the transcription factor Prf1, which has phosphorylation sites for both MAP kinase and PKA. Furthermore, proper regulation of PKA activity seems to be critical for the development and virulence of U. maydis. High PKA activity in the fungus is necessary for colonization of the host plant, whereas lower PKA activity favors gall formation and sporulation. Additional studies will be required to dissect processes activated by different levels of intracellular PKA activities, including the identification of targets of PKA. Probably the most interesting question still to be answered is which signals are provided by the host plant that enable the progression of U. maydis through its sexual life cycle within the plant. Recently Ruiz-Herrera et al. (242) succeeded in developing conditions that support the complete life cycle of U. maydis, enabling growth of the fungus on embryonic plant cell cultures. This should allow large-scale screens to identify biologically active substances or fungal mutants that fail to respond to plant signals.
RICE BLAST FUNGUS MAGNAPORTHE GRISEA
M. grisea causes a devasting fungal disease of rice plants known as rice blast. The fungus can also cause a similar disease in over 50 grasses, including economically important crops such as barley, wheat, and millet. Detailed studies of the pathogenesis of this fungus have established it as one of the model organisms among plant-pathogenic fungi.
M. grisea is a heterothallic ascomycete, and it has two mating types, MAT1-1 and MAT1-2. In addition, M. grisea, like Neurospora crassa, produces nuclei that function as either male or female gametes. Thus, the sexual cycle involves fusion of a male and a female gamete, which must be of opposite mating types. Under suitable environmental conditions, sexual reproduction initiates with the fusion of strains of opposite mating types and concludes with the production of mature perithecia with numerous eight-spored asci. M. grisea mating type genes have been identified by genomic subtraction (126). Pairs of nearly isogenic strains were created by switching the MAT idiomorphs and used to demonstrate that the mating type locus controls mating identity and responses. By using these strains, pheromone precursor genes of M. grisea have been successfully isolated (253). Gene deletion experiments suggest that M. grisea pheromones play an important role in the initial step of sexual development (W.-C. Shen, personal communication). To further elucidate molecular signaling during sexual differentiation, other components involved in the mating response pathway need to be identified.
Appressorium Development
The infection cycle of M. grisea initiates when an asexual spore lands on the host surface. Hydration of the conidium triggers the release of spore tip mucilage, which attaches the conidium tightly to the hydrophobic leaf surface (101). Once attached, the spore germinates and then forms a short germ tube which differentiates at the tip to produce the specialized infectious structure, the appressorium (112). The cell wall of the appressorium then thickens, and melanin is produced (113). Newly synthesized melanin is layered around the appressorial cell wall, and the increased accumulation of glycerol inside the appressorium results in the build-up of enormous internal turgor pressure (67). This increased pressure allows the penetration peg to break through the cuticle and into the underlying epidermal cell layers (111, 114). Infectious hyphae ramify through the epidermal cells and differentiate into bulbous, branched secondary hyphae, which continue to spread intercellularly and intracellularly. Finally, ellipsoidal lesions become visible on the infected tissues, and the fungus then produces aerial conidiophores that sporulate to resume another round of the disease cycle.
The appressorium is a unique fungal structure that is used by a diverse group of fungi to penetrate host surfaces (66, 79). Appressorium development is a complex morphogenetic process, and multiple external signals, physical and chemical, appear to be involved (66, 133, 158, 308). In M. grisea, surface hydrophobicity and hardness, adhesion, and other environmental factors have all been reported to regulate appressorium formation (121, 159, 309, 310). Lee and Dean demonstrated that induction of appressorium formation correlates with the hydrophobicity of the contact surface (159). Artificial substrates, like wax paper, polystyrene, glass coverslips, and gel bond (hydrophobic side), mimic the hydrophobicity of the rice plant surface and support appressorium formation.
MPG1 encodes a fungal hydrophobin that is abundantly expressed during appressorium formation, symptom development, and conidiation (270) (Fig. 16). mpg1 deletion mutants are severely impaired in their ability to cause disease on susceptible rice cultivars due to an inability to produce appressoria. The evidence that purified fungal hydrophobins from Schizophyllum commune can self-assemble at any hydrophilic-hydrophobic interphase suggest that MPG1 could play a role in attachment, infection site preparation, or topological signaling (270).
FIG. 16.
Signaling pathways regulating virulence of the rice blast fungus M. grisea. Appressorium formation and virulence of the rice blast fungus M. grisea involve both conserved MAP kinase signaling cascades and the cAMP-PKA signaling pathway.
A novel transmembrane protein, PTH11, has been identified in a mutagenesis screen (71). pth11 mutant strains are nonpathogenic due to a failure in appressorium maturation. A low level of appressorium formation in pth11 mutant strains suggests that PTH11 is not required for appressorium morphogenesis but is involved in surface recognition. Functional characterization of PTH11 in multiple M. grisea strains indicates that PTH11 can activate or repress appressorium differentiation in response to different contact surfaces. Exogenous activation of cellular signaling suppressed pth11 mutant phenotypes. Taken together, these findings suggest that PTH11 may function as an upstream effector of appressorium differentiation in response to surface cues (71).
cAMP Signaling Pathway
How external inductive signals are perceived by M. grisea is not yet clear, but it is clear that a cAMP-dependent signal transduction pathway is involved in appressorium formation (159) (Fig. 16). Exogenous cAMP induces M. grisea germinating conidia or vegetative mycelia to produce appressoria on noninductive surfaces (159). Following this initial finding, the components involved in the cAMP signaling pathway were identified and characterized in detail.
The gene encoding the catalytic subunit of the cAMP-dependent protein kinase (CPKA) was independently identified by three groups (194, 269, 313). Mitchell and Dean reported that disruption of the CPKA gene resulted in mutants defective in appressorium formation on onion epidermal tissue or Gelbond, an artificial hydrophobic surface. cpkA mutants also failed to cause lesions on a susceptible rice cultivar. The failure of appressorium formation in cpkA mutants was not rescued by exogenous cAMP. cpkA mutants also exhibited no defects in growth or asexual or sexual development.
Xu et al. subsequently conducted thorough, side-by-side comparisons of different cpkA mutants from three different sources that further clarified the cpkA mutant phenotype (313). A wild-type level of appressoria was observed in all of the mutants, but appressorium development was delayed and the appressoria produced by the cpkA mutants were smaller than those produced by wild-type strains. In pathogenicity assays, all of the cpkA mutants were still capable of causing disease symptoms on wounded hosts but not on healthy plants. Penetration assays on onion epiderminal strips further confirmed that cpkA mutants are defective in penetration. These results suggest that cAMP signaling plays a role in appressorial penetration. Because cpkA mutants are delayed in appressorium formation, it has been postulated that cAMP responsiveness in the cpkA mutants might also be delayed. Contrary to the original report, Xu et al. found that all cpkA mutants were induced to form appressoria on hydrophilic surfaces after 24 h of incubation by the addition of cAMP (313). These results indicate that another PKA catalytic subunit may be present and function in surface sensing in rice blast fungus (313).
In addition to CPKA, other genes involved in cAMP signaling have also been cloned and characterized (Fig. 16). The gene encoding adenylyl cyclase (MAC1) was cloned by a degenerate PCR approach (50). M. grisea adenylyl cyclase mutants have pleiotropic phenotypes. They fail to form appressoria on a hydrophobic surface of Gelbond film, onion skin, or rice leaves. As a consequence, mac1 mutants are nonpathogenic in susceptible plants. The mac1 mutant strains also fail to produce wild-type spreading lesions when artificially injected into the host. The mac1 mutation significantly reduces vegetative growth and conidiation and confers a sterile phenotype in which no perithecia are produced. Conidial germination is also delayed in mac1 mutants. Appressorium formation and normal conidial germination can be restored in the presence of exogenous cAMP derivatives, such as monobutyryl-cAMP (50).
Studies of the bypass suppressors of the mac1 (sum) mutation revealed that sum mutations completely restore growth, sexual and asexual morphogenesis, and appressorium formation on nonhydrophobic surfaces (1). One sum mutation has been shown to alter the regulatory subunit of PKA. A single amino acid change in the first cAMP-binding domain in the sum mutant suppresses some but not all of the defects conferred by the mac1 adenylyl cyclase mutation. This result suggests that the mutant regulatory subunit retains some activity or that an additional regulatory subunit gene exists.
Gα Protein MAGB Required for Morphogenesis
Heterotrimeric guanine nucleotide-binding proteins (G proteins) are involved in regulating a variety of cellular functions in eukaryotic cells. They serve as critical transducers between activated cell surface receptors and intracellular effectors (91, 255). In M. grisea, three G protein α subunit genes, MAGA, MAGB, and MAGC, have been cloned and characterized (163). Sequence comparison revealed that MAGA, -B, and -C share sequence identity with other fungal G protein α subunits (Fig. 14). Characterization of disruption mutants revealed that MAGA has no role in the life cycle other than the production of mature asci. Interestingly, MAGA is quite similar to the Gpa2 and Gpa1 nutrient sensors of S. cerevisiae and C. neoformans (Fig. 14). MAGC appears to be involved in the production of asexual and sexual propagules.
Disruption of MAGB significantly reduced vegetative growth, conidiation, and, most importantly, appressorium formation (163). The appressorium formation defect of magB mutant strains can be restored by the addition of cAMP or 1,16-hexadecanediol, but the relationship between MAGB and adenylyl cyclase and how 1,16-hexadecanediol is perceived through different pathways to regulate appressorium development in M. grisea need to be further examined. In Fig. 16, MAGB is depicted activating adenylyl cyclase. We note that MAGB is quite divergent from the Gα proteins that regulate nutrient sensing in S. cerevisiae and S. pombe (Fig. 14). Thus, MAGB is likely coupled to a different type of receptor and likely responds to different signals to regulate development. One additional interesting phenotype of magB mutant strains is the inability to form perithecia, suggesting that MAGB has dual functions in sexual development and pathogenesis.
Beckerman et al. reported that S. cerevisiae α-factor pheromone blocked appressorium formation in a mating type-specific manner. Furthermore, culture filtrates of a MAT1-1 strain contained an activity that inhibited appressorium formation of mating type MAT1-2 strains. These findings suggest that activation of the pheromone response pathway may interfere with the signals leading to appressorium formation (24). Following this work, Shen et al. reported the isolation of pheromone precursor genes in M. grisea. Interestingly, a gene structurally similar to the S. cerevisiae α-factor precursor gene was isolated from the MAT1-2 strain, whereas a lipopeptide pheromone precursor gene similar to S. cerevisiae a-factor was isolated from the MAT1-1 strain (253). Because MAT1-2 strains were inhibited by S. cerevisiae α-factor, an unmodified peptide, it was surprising that MAT1-2 cells actually respond to a lipid-modified peptide produced by MAT1-1 cells. This result makes it unlikely that S. cerevisiae α-factor and the M. grisea MAT1-1 pheromone act on the same receptor. In fact, new segregation analysis of the progeny from a cross indicate that the α-factor sensitivity-resistance trait segregates independently from the mating type locus (253). In order to resolve this contradiction, components involved in the pheromone response pathway of M. grisea need to be further characterized.
Multiple MAP Kinases Regulate Appressorium Formation and Function
In addition to the G protein α subunit and components involved in the cAMP signaling pathway, MAP kinase genes have also been isolated through degenerate PCR screens and found to be required for appressorium development and function in M. grisea (311, 312) (Fig. 16). M. grisea pathogenicity MAP kinase 1 protein (PMK1) has significant similarity with S. cerevisiae Fus3 and Kss1 and functionally complements the mating defect of an S. cerevisiae fus3 kss1 double mutant strain. More importantly, PMK1 is essential for pathogenicity and is also required for invasive growth and viability in the host. The virulence defect of pmk1 mutant strains is attributable to a failure to complete the late stages of appressorium morphogenesis. pmk1 mutant strains produce more germ tube hooking and swelling structures on a hydrophilic surface in response to cAMP, suggesting that PMK1 functions in parallel with a cAMP-dependent signal (311).
In the same degenerate PCR screen, another MAP kinase gene, MPS1, was isolated (312). Mps1 is 64% identical and 85% similar to the S. cerevisiae Slt2 MAP kinase and complements the autolysis defect of slt2 null mutants at 37°C. M. grisea mps1 mutants have defects in cell wall integrity similar to yeast slt2 mutants. In addition, mps1 mutant strains are female sterile (as noted above, M. grisea mating involves fusion of gametes of opposite sex and mating type) and have reduced asexual sporulation. Moreover, the appressoria produced by mps1 mutants fail to penetrate plant cell surfaces, and thus mps1 mutants are nonpathogenic. Interestingly, mps1 mutants do not penetrate but still induce the plant defense response (312).
To penetrate the plant cuticle, M. grisea generates turgor pressures in excess of 8.0 MPa inside the appressorium (114). Biochemical characterization of the contents extracted from the appressoria indicates that glycerol accumulates inside the appressoria and causes the turgor to increase (67). Osm1, a homolog of the S. cerevisiae Hog1 MAP kinase, appears to have a role in controlling turgor generation inside the appressoria, because osm1 mutants are sensitive to osmotic stress (73). However, osm1 mutants still accumulate glycerol, generate turgor, and cause disease.
Conclusions and Perspectives
Studies of pathogenesis in the rice blast fungus M. grisea have revealed that heterotrimeric GTP-binding proteins, MAP kinases, and cAMP signaling elements all play essential roles in pathogenic morphogenesis. A model of the signaling components regulating pathogenesis and other aspects of the responses is shown in Fig. 16. Genetic experiments indicate that these elements are involved in multiple pathways. For example, the MAP kinase Mps1 plays roles in both appressorium formation and sexual development. The interesting questions that remain are how the cell perceives the environmental signals and specifies the signaling responses. To address these issues, upstream components such as receptor(s) and downstream targets of these pathways need to be identified and characterized.
SIGNAL TRANSDUCTION IN CRYPHONECTRIA PARASITICA
C. parasitica is an ascomycetous fungus that is responsible for chestnut blight, which destroyed the American chestnut (Castanea dentana) that once dominated the eastern United States. Regulatory mechanisms mediated by GTP-binding proteins, the inositol trisphosphate-Ca2+-calmodulin-calcineurin pathway, and the general amino acid control response have been all implicated as important regulators of growth, development, and virulence in C. parasitica. The double-stranded RNA viruses HAV1 and CHV1-713, which attenuate virulence, have also provided a unique tool with which to examine virulence mechanisms in this organism (214). G proteins involved in signal transduction have been identified (49, 88, 127, 128, 292). Figure 17 depicts a model of C. parasitica G protein-mediated signal transduction involved in gene expression, asexual reproduction, and virulence.
FIG. 17.
Signaling pathways regulating virulence of the chestnut blight fungus C. parasitica; current model of the G protein signaling pathways that affect virulence and hypovirulence as a result of signal perturbation by HAV in C. parasitica. The Gα protein CPG-1 negatively regulates a putative adenylyl cyclase (AC), whereas the Gβ protein CPGB-1 and BDM-1 may function via a different pathway to regulate gene expression. HAV is hypothesized to function on G proteins or upstream targets.
Distinct Functions of G Protein α Subunits CPG-1 and CPG-2
C. parasitica G proteins regulate growth and virulence in C. parasitica. Two G protein α subunit homologs, CPG-1 and CPG-2, have thus far been identified in C. parasitica. CPG-1 has the highest level of amino acid identity with the Gα subunits GNA-1 from Neurospora crassa (285), FadA from Aspergillus nidulans (319), and MAGB from the rice blast fungus M. grisea (163) (Fig. 14). All of these fungal Gα subunits are homologous to the mammalian Giα family. The Gα protein CPG-2 is homologous to the Gα proteins MAGA of M. grisea (163) and MOD-D of the filamentous ascomycete Podospora anserina (174) and the nutrient-sensing Gα subunits (Gpa2, gpa2, and GPA1) from budding and fission yeasts and C. neoformans (Fig. 14).
Functionally, CPG-1 plays a major role in regulating gene expression in C. parasitica. Modulation of CPG-1 by either suppression of CPG-1 expression or cpg-1 gene disruption results in pleiotropic phenotypes, including defects in pigment production, asexual reproduction, female fertility, and virulence. In addition, the expression of several genes, including that for laccase (lac-1), is reduced (49, 88). In contrast, mutation of the second Gα subunit gene cpg-2 causes only minor effects on growth and asexual reproduction (88). This is similar to the finding that mutation of the MAGA Gα protein confers no readily identifiable phenotype in M. grisea. Given that the S. cerevisiae Gα protein Gpa2 languished in obscurity for over 10 years following its initial discovery and was then found to be a central component of a nutrient-sensing pathway regulating pseudohyphal differentiation, it seems likely that equally important physiological functions remain to be discovered for the highly related Gα proteins MAGA of M. grisea and CPG-2 of C. parasitica.
The Gi family of mammalian Gα proteins negatively regulate adenylyl cyclase and function to reduce intracellular cAMP production. The C. parasitica Gα protein CPG-1 has a similar role. In mutant strains in which the expression of CPG-1 was suppressed to a nondetectable level by a mechanism called sense suppression (49), cAMP levels were increased four to fivefold. Interestingly, cAMP levels were also increased three to fivefold in strains infected with the virus HAV1 (48). The current model is that activation of CPG-1 by an as yet unidentified ligand-receptor complex inhibits adenylyl cyclase, lowers intracellular cAMP levels, and impairs activation of a cAMP-dependent protein kinase homolog that regulates downstream targets.
CPGB-1 and BDM-1 Positively Regulate Virulence
The importance of G protein function in C. parasitica has been further illustrated by the characterization of two additional components of the G protein complex, the Gβ subunit CPGB-1 and a mammalian phosducin homolog BDM-1, which has been proposed to be functionally associated with the Gβ subunit CPGB-1 (Fig. 17) (128).
Both the Gβ subunit CPGB-1 and BDM-1 exhibit a positive regulatory role similar to that of the Gα subunit CPG-1. However, CPGB-1 and BDM-1 also play a negative regulatory role in fungal vegetative growth on synthetic medium. CPGB-1 is the only Gβ subunit homolog identified from C. parasitica, and cpgb-1-specific mutant strains exhibit defects in pigmentation, asexual reproduction, female fertility, and virulence (127). Because of the similar role exhibited by both the Gα subunit CPG-1 and Gβ subunit CPGB-1, these two components may be functionally coupled to each other. However, no physical interaction has yet been demonstrated. CPG-1 protein levels are reduced in cpgb-1 mutant strains, suggesting that the Gα subunit may be unstable in the absence of its Gβ partner (88).
The newly identified protein BDM-1 has provided further insight into G protein signaling in C. parasitica. bdm-1 mutant strains exhibit a phenotype strikingly similar to that of cpgb-1 mutant strains with respect to asexual reproduction, pigmentation, and virulence as well as increased vegetative growth on synthetic medium (128). The BDM-1 protein shows amino acid identity in several key regions with the mammalian protein phosducin, which sequesters the transducin Gβγ subunits Tβγ and modulates the function of the transducin Tα subunit in the retina. In contrast to mammalian phosducin, however, BDM-1 exhibits a positive regulatory function and is proposed to destabilize the Gα subunit CPG-1 and assist the function of the Gβ subunit CPGB-1 (128). Two additional phosducin homologs, Plp1 and Plp2, have recently been identified in S. cerevisiae (82). In this case, biochemical studies confirm that both Plp1 and Plp2 can interact with βγ subunits, but genetic studies reveal no readily discernible phenotype with respect to G protein signaling for either protein (82). That three phosducin homologs have thus far been identified in microorganisms further emphasizes the conservation in evolution between mammals and fungi.
Virulence Attenuation
Disrupting or interfering with signal transduction modulates fungal virulence. For example, cAMP-dependent and MAP kinase-mediated signaling pathways are both important for formation of the infectious appressorium in the rice blast fungus M. grisea (for a review, see reference 102). Similarly, the signal transduction components CPG-1, CPGB-1, and BDM-1 regulate the ability of C. parasitica to infect chestnut trees. One established link between the G protein α subunit CPG-1 and virulence is that expression of the putative virulence factor cellobiohydrolase (encoded by the cbh-1 gene) requires CPG-1 (292). G proteins are not the only regulatory mechanisms governing virulence in C. parasitica. Regulatory pathways, including the IP3-Ca2+-calmodulin-calcineurin pathway and the general amino acid control pathway that is activated in response to amino acid starvation, are also important in regulating pigment production and virulence (152, 291).
Infection of C. parasitica by the virus HAV1 causes a broad range of phenotypic changes, many of which mimic phenotypes of mutant strains with defects in G protein signaling. This suggests that the virus may interfere with signal transduction by G proteins (214). Indeed, in a study using a differential display technique to examine mRNA transcripts in C. parasitica, 65% of the changes in mRNA observed in virus-infected cells were also observed in mutant strains in which CPG-1 expression was suppressed (48). Furthermore, cAMP levels were three- to fourfold higher in strains containing the virus, similar to cpg-1 mutant strains. Because phenotypic changes associated with viral infection are more severe than those caused by the cpg-1 mutation, signal transduction mediated by pathways other than G proteins likely also plays an important role.
ASEXUAL DEVELOPMENT OF ASPERGILLUS NIDULANS
The signal transduction cascades regulating asexual sporulation in the ascomycetous filamentous fungi A. nidulans and N. crassa have been well studied. These fungi are amenable to genetic analysis and serve as model organisms for the study of eukaryotic multicellular development. We note that some of the signaling cascades elucidated thus far, especially in A. nidulans, appear somewhat divergent from those discussed up to this point. They may represent unique specializations that have occurred during the development and evolution of the filamentous fungi. On the other hand, given the prominent role of filamentation in the life cycles of S. cerevisiae, C. neoformans, C. albicans, and U. maydis, it seems likely that conserved regulatory cascades will emerge in A. nidulans and N. crassa as their genomes are sequenced and analyzed, as has already become apparent with the PKA pathway in N. crassa.
A. nidulans is homothallic (self-fertile), and sexual conjugation can occur when spores of genetically distinct, vegetatively compatible homokaryotic strains are mixed (278). A heterokaryon is formed when hyphae of these strains anastomose, followed by nuclear division and movement. Heterokaryons of this fungus contain both haploid and diploid nuclei and, following asexual development, can give rise to both haploid and diploid conidia.
Asexual Sporulation Cycle
The asexual sporulation cycle of A. nidulans can be divided into two phases: a growth phase, in which cells become competent to respond to sporulation-inducing signals, and an asexual reproduction phase, including initiation of the sporulation pathway and formation of spore-bearing structures (Fig. 18). This fungus grows by forming an ordered network of filaments or hyphae that form a mycelium. Each hypha is a linear array of vegetative cells that are haploid, multinucleate, and separated by perforated septa to allow nuclear migration. The hyphae grow by apical extension and multiply by branching, giving rise to a radially symmetrical colony that expands at a constant rate. One of the functions of hyphae is to absorb nutrients from the growth medium, facilitated by the secretion of degradative enzymes. After a fixed period of time following vegetative growth, some hyphal cells within the center of the mycelium produce aerial branches that initiate asexual sporulation. This involves the formation of multicellular structures called conidiophores that bear chains of spores called conidia (reviewed in reference 5). Development requires a shift from highly polarized hyphal growth to growth by budding and transition from a multinucleate to a uninucleate state. These changes are directed by an interplay between cell cycle regulators and the developmental pathway (317). Conidia germinate when they contact a nutrient-rich medium, giving rise to hyphae and completing the cycle.
FIG. 18.
Asexual sporulation cycle of A. nidulans. Asexual sporulation of A. nidulans involves two phases: a vegetative growth phase, in which spore germlings give rise to hyphae and the mycelium, and an asexual reproductive phase, in which specialized hyphal cells called foot cells give rise to aerial stalks that produce spore-bearing structures called conidiophores. Stalks produce a vesicle that buds to produce branching cells called sterigmata. Further mitotic divisions of secondary sterigmata called phialides give rise to chains of conidia which germinate on an appropriate medium to complete the cycle.
Physiological Requirements for Asexual Sporulation
A number of physiological criteria regulate A. nidulans asexual development. The initiation of conidiation is contingent upon cells having undergone a defined period of growth (20 h) to reach developmental competence (15). Developmental competence is an intrinsically programmed process that is independent of external changes in growth medium; continuous transfer to fresh growth medium and growth of cells at low densities have no effect on the timing of conidiation (226). In fact, while sporulation in many microorganisms occurs in response to adverse environmental conditions, the signals that induce A. nidulans differentiation are not well defined. Under normal growth conditions, the only environmental requirement is exposure to air (15, 200). This requirement for air does not appear to involve O2 or CO2 levels, but rather results from changes in the cell surface induced by the abrupt formation of an air-water interface.
Although sporulation largely occurs in a medium-independent manner, it can be induced in hyphae grown in submerged culture by nutrient (carbon or nitrogen)-limiting conditions (256). However, nutritional stress must play a secondary role in induction of sporulation, because nutrient limitation reduces conidiation of surface-grown colonies and continuous replacement of growth medium beneath a surface-grown colony does not repress conidiation (226). A fraction of hyphal cells may be nutrient limited and thereby induced to develop. Conidiation in confluent cultures of A. nidulans depends upon inoculum density, and higher initial cell densities promote sporulation (37). However, it is not known whether this promotes sporulation by depleting nutrients.
Conidiation is also dependent upon exposure of hyphae to red light. Light sensing is regulated by the velvet gene (veA), which is proposed to be a negative regulator of development because a veA+ strain does not conidiate as well in the dark as a veA1 mutant does, even when the colony is exposed to air (196). The relief of repression by red light exposure can be partially reversed by exposure to far-red light, similar to responses mediated by phytochrome, the red light photoreceptor in higher plants, suggesting that a phytochrome-like molecule may regulate development of A. nidulans.
Genetic Requirements for Asexual Sporulation
Induction of morphogenesis involves temporal and spatial regulation of several hundred genes, many of which function in conidiophore assembly or conidiospore differentiation (31, 186, 277, 303, 323). The sequential expression of bristle (brlA), abacus (abaA), and wet-white conidia (wetA) establishes a central regulatory pathway (brlA → abaA → wetA) required for the transition from vegetative growth to asexual reproduction (2, 185). brlA encodes a C2H2 zinc finger DNA-binding protein that functions as a primary transcriptional activator of sporulation-specific gene expression (2, 4, 46). brlA activity is required at the time of conidiophore vesicle formation. brlA null mutants form bristle-like aerial stalks due to a block in transition from polar growth of the conidiophore stalk to swelling of the conidiophore vesicle (52). brlA mutants fail to differentiate vesicles, metulae, phialides, or spores, and genes in the central regulatory pathway, such as abaA and wetA, are not expressed (31). The activity of this central regulatory pathway is required for the normal expression of most conidiation-specific genes and is influenced by the products of other genes, including stuA and medA, which act as developmental modifiers (36, 52, 192).
The activation of the central pathway is dependent upon at least six early regulatory genes, fluG, flbA, flbB, flbC, flbD, and flbE, which were identified as recessive mutations that result in a predominantly “fluffy” colony morphology, impaired conidiation, and reduced brlA expression (3, 155, 157, 302, 303). Dominant mutations that confer a fluffy phenotype have also been isolated and characterized (303, 304); among these, mutations in the fadA gene, which encodes a Gα protein (Fig. 14), have shed light on the influence of vegetative growth on sporulation (319).
The role of the fluG and flbA genes in the initiation of conidiation has been studied in detail (Fig. 19). The aconidial phenotype of fluG mutants is partially rescued by growth on solid minimal medium, suggesting that sporulation can occur in response to nutrient limitation in the absence of FluG (3). fluG mutants can also be induced to conidiate when grown in proximity to either a wild-type strain or strains with mutations in different fluffy genes. Rescue of the aconidial phenotype of fluG mutants also occurs when two juxtaposed strains are separated by a dialysis membrane with a pore size of 6 to 8 kDa, suggesting that fluG is required for the biosynthesis of a small, diffusible, extracellular molecule that initiates asexual development (155). Overexpression of fluG can overcome a developmental block normally imposed when hyphae are grown in submerged culture and results in the production of complex conidiophores (156). FluG-induced sporulation requires the activities of other early-acting developmental regulators, including flbA, flbB, flbC, flbD, and flbE. Although the isolation of the extracellular factor and the exact function of fluG remain elusive, the C-terminal region of FluG has significant similarity to prokaryotic glutamine synthetases and is required to induce sporulation (155; C. A. D'Souza, B. N. Lee, and T. H. Adams, submitted for publication). FluG does not synthesize glutamine but may have a related enzymatic activity.
FIG. 19.
Model for the signal transduction pathways regulating asexual sporulation in A. nidulans. Initiation of asexual sporulation in A. nidulans is governed by two parallel antagonistic pathways. Initial mycelial growth is driven by activation of the Gα protein FadA to its GTP-bound form. This pathway represses sporulation until the fungus gains developmental competence. FluG is necessary for the synthesis of an extracellular factor (ECF) that initiates the asexual sporulation pathway by inducing the activities of FlbA, FlbB, FlbC, FlbD, and FlbE, resulting in expression of the brlA, abaA, and wetA genes. Additionally, stimulation of FlbA causes inactivation of FadA, releasing the inhibition of sporulation. The progression of sporulation is also influenced by brlA-dependent induction of the activities of cell cycle kinases NimXcdc2 and NimA, promoting morphogenesis from the multinucleate, filamentous form to a uninucleate, budding form.
To gain further insight into the role of FluG in development, a genetic screen for dominant suppressors of fluG (dsg) was undertaken to identify components acting downstream of FluG in the asexual sporulation pathway (D'Souza et al., submitted). This screen identified an interesting mutation in the dsgA gene. Conidiophores of dsgA1 mutants grown on solid medium are abnormal and possess long branched sterigmata that produce conidia and occasionally bear secondary conidiophores. dsgA1 mutants also produce conidiophores when grown in liquid cultures, indicating that the requirement for air to induce sporulation has been bypassed. DsgA could be the receptor for the FluG-dependent extracellular factor (Fig. 19).
Among other early regulatory genes required for initiation of conidiation, flbA is the best candidate to be part of the signal transduction pathway regulated by the extracellular factor (Fig. 19). FlbA has a C-terminal RGS domain that is shared by a family of proteins found in evolutionarily diverse organisms (70, 107). RGS proteins negatively regulate G protein-mediated signaling pathways by activating the intrinsic GTPase activity of heterotrimeric G protein α subunits (27, 297). Colonies of flbA mutants are described as fluffy autolytic because undifferentiated masses of hyphae autolyse as the colonies mature (157). In contrast, overexpression of flbA in vegetative cells inhibits hyphal growth and induces conidiophore development in liquid culture (156, 157). The ability of flbA overexpression to modulate growth and activate sporulation is dependent on the activities of other genes, including fluG (155, 156). Because fluG-dependent activation of development requires flbA, flbA may regulate a signal transduction pathway that responds to the FluG signal.
The dependence of fluG- and flbA-induced sporulation on the activities of flbB, flbC, flbD, and flbE indicates that they function downstream of fluG and flbA. flbB, flbC, flbD, and flbE mutants still produce the extracellular conidiation factor, rescue the fluG aconidial mutant phenotype (155), and conidiate following prolonged growth. Additionally, genetic analysis revealed that fluG and flbA are both epistatic to flbB, flbC, flbD, and flbE (303). flbB, flbC, and flbD are predicted to encode DNA-binding proteins and may regulate transcriptional activation of developmental regulators such as brlA in response to fluG- and flbA-generated signals (5). FlbE shows no similarity with other proteins.
Antagonistic Mycelial Proliferation Pathway
In addition to the sporulation pathway described above, a parallel pathway involving a heterotrimeric G protein α subunit encoded by the fadA gene (fluffy autolytic dominant) regulates mycelium proliferation and antagonizes conidiophore development (319) (Fig. 19). Dominant active fadA mutations cause uncoordinated growth, autolysis, and inhibition of sporulation, similar to the flbA mutant phenotype, indicating that activation of the mycelial growth pathway inhibits sporulation. The flbA mutant phenotype can also be suppressed by either dominant negative fadAG203R or fadAΔ mutations. These observations suggest that the primary role of the RGS domain protein FlbA in sporulation is to prevent activation of FadA by stimulating GTPase activity. FlbA, following activation by FluG, may promote asexual sporulation by inactivating FadA-dependent growth signaling (108, 319). However, overexpression of flbA activates asexual development in both wild-type and fadAΔ strains, suggesting that FlbA may have additional targets. Dominant or recessive loss-of-function fadA mutations suppress the requirement for flbA but not fluG in sporulation, indicating that FluG has a FadA-independent, development-specific role in addition to its proposed role in repressing growth through FlbA. Initiation of development may require both functions of FluG. dsgA1, isolated as a dominant suppressor of fluG, also bypasses the flbA requirement for sporulation and suppresses inhibition imposed by dominant activating fadA mutations, suggesting that DsgA may function downstream of FadA (D'Souza et al., submitted). Alternatively, suppression of flbA and fadA by dsgA1 could result from hyperactivation of the asexual developmental pathway.
Cross talk occurs between the two parallel antagonistic pathways signaling mycelial growth and sporulation, and asexual development is directed by the dominant pathway (Fig. 19). Growth signal transduction mediated by FadA is thought to involve its activation by GDP-to-GTP exchange, presumably initiated by the binding of a growth factor ligand to a putative G protein-coupled receptor (208). In addition to the Gα protein FadA, mycelial proliferation is also in part regulated by the Gβγ subunit, because deletion of the sfaD gene encoding the Gβ subunit retards growth (240). The sfaDΔ mutation also causes sporulation under normally repressive conditions and suppresses all defects of null mutations in flbA, suggesting that Gβ may also be a target of FlbA. The FadA-independent activity of FlbA may involve interaction with Gβγ subunits, based on the observation that a dominant negative fadAG203R mutation causes sporulation in submerged culture but a fadAΔ mutation does not (319). The sfaDΔ mutation does not suppress mycelial proliferation caused by dominant active fadA mutations, indicating that constitutively active FadA promotes growth independently of SfaD. Although the data suggest that FadA and SfaD are part of the same G protein, a definitive interaction between these proteins has not yet been demonstrated.
The asexual development pathway does not initiate until the fungus progresses through a defined period of growth (20 h postgermination). The fluG gene plays a pivotal role in the switch from vegetative growth to initiation of sporulation (3, 155, 156). In the model shown in Fig. 19, FluG positively regulates the production and secretion of an extracellular factor(s) that binds to a cell surface receptor and induces the asexual sporulation pathway depending on the occurrence of two events: first, activation of FlbA by FluG to inhibit FadA-mediated growth signaling (108, 319), and second, activation by FluG of sporulation-specific functions dependent on the products of the flbB, flbC, flbD, flbE, and brlA genes. DsgA is the most likely link between FluG and the two downstream events. DsgA may encode the receptor for the extracellular conidiation factor or a signal transduction component such as a conidiation pathway-specific G protein (D'Souza et al., submitted).
PKA Signaling Pathway
Given the involvement of a Gα subunit (FadA) in the regulation of sporulation in A. nidulans, it will not be surprising to find a cAMP-dependent protein kinase involved in a G protein-mediated signaling pathway. Indeed, the pkaC gene, encoding the catalytic subunit of PKA, has been isolated from A. nidulans, but its involvement in asexual sporulation remains to be investigated (N. P. Keller, personal communication). The possible involvement of a cAMP-dependent protein kinase, PkaC, in sporulation has been studied in the related fungus Aspergillus niger. Overexpression of pkaC resulted in alterations in growth and development, but transformants varied in phenotype ranging from dense to no sporulation (26). A Ras homolog may regulate cAMP signaling and morphogenesis in A. nidulans (257). Differentiation from vegetative growth to asexual sporulation may be regulated by different concentrations of Ras, and progression of asexual sporulation is dependent upon an initially high level of active Ras followed by a gradual decrease (257).
Perspectives
The precise mechanisms by which extracellular signals are transduced to activate asexual development in A. nidulans still remain to be elucidated. Many components of the signal transduction cascade regulating asexual sporulation of A. nidulans have been isolated by complementation of the aconidial phenotype of developmental mutants. Surprisingly, few kinases and no receptors specific for asexual sporulation have thus far been identified. These components may be either redundant in function or essential for viability. The proposal that dsgA encodes the receptor for the FluG-dependent extracellular factor remains to be verified by isolation and characterization of the dsgA gene. Recent sequencing of the A. nidulans genome is expected to provide an invaluable resource for identifying additional components of the asexual sporulation pathway that may have been missed by conventional genetic screens for aconidial mutants. The identification of a Ste12-like transcription factor, SteA, required for sexual development suggests that a MAP kinase cascade may be involved in sexual morphogenesis (287). While MAP kinase cascades have not been implicated in asexual sporulation of A. nidulans, identification of the Ras and FadA G proteins suggests that a cAMP-dependent signaling cascade may be involved.
SIGNAL TRANSDUCTION IN NEUROSPORA CRASSA
Life Cycle
The ascomycete N. crassa has defined asexual and sexual cycles (Fig. 20). Under nutrient-rich conditions, N. crassa proliferates by the extension and branching of multinucleate vegetative hyphal cells to form a multicellular mycelium. During the vegetative cycle, N. crassa produces two types of vegetative spores, macroconidia and microconidia (reviewed in references 65 and 264). Macroconidia are multinucleate and differentiate from specific organs called conidiophores that arise from aerial hyphae. Microconidia differentiate from vegetative mycelia, are smaller and uninucleate, and germinate poorly.
FIG. 20.
Life cycle of N. crassa. In response to environmental conditions, the vegetative mycelium produces conidia (macroconidia and microconidia) and protoperithecia. Macroconidia arise from conidiophores that differentiate from aerial hyphae, and microconidia arise directly from vegetative hyphae. Once a protoperithecium is fertilized with a male organ, sexual development ensues and ascospores are produced.
N. crassa is heterothallic and has two mating types, A and a. The sexual cycle initiates in response to nitrogen starvation (191). Either A or a cells of vegetative hyphae produce fruiting-body precursors called protoperithecia. Specialized hyphae called trichogynes grow directionally from the protoperithecium toward an opposite mating type cell such as a conidium or a hyphal fragment, which functions as a male gamete. Once a nucleus from a male gamete enters a protoperithecium, the protoperithecium becomes a perithecium. The male and female nuclei divide as a dikaryon in ascogenous hyphae. Dikaryotic cells at the crest of hook-shaped structures, croziers, undergo premeiotic DNA synthesis and nuclear fusion (karyogamy) to produce the only diploid cells in the life cycle of N. crassa. Meiosis and one round of mitosis immediately follow to produce eight-spored asci. The mature ascospores are ejected and can be induced to germinate by heat shock or exposure to ethers, producing cells that reenter the vegetative phase of the life cycle.
In response to nutrient deprivation, desiccation, and light, N. crassa initiates asexual conidiation (65, 264). The major determinants regulating the onset of conidiation are the availability of carbon source (181, 233) and light (153). In response to light, N. crassa cultures produce conidia faster and in greater numbers compared to cultures grown in complete darkness. In addition to these environmental factors, the endogenous clock of the organism affects the conidiation process: conidiation occurs during the subjective morning. Clock-controlled genes expressed during the subjective morning have been found to encode proteins expressed during conidiation (25). Therefore, to complete fungal development in response to various extracellular and intracellular stimuli, signal transduction pathways sensing these stimuli must function coordinately.
cAMP Signaling Pathway
In N. crassa, two Gα subunit genes, gna-1 and gna-2, have been identified (285) (Fig. 14 and 21). The deduced amino acid sequence of GNA1 has homology with members of the Gαi family; for example, it is 55.2% identical to rat Gi2. GNA1 also possesses consensus sequences for addition of myristic acid and labeling by pertussis toxin, which are diagnostic of the Gi family (285). GNA2 is most similar to S. pombe Gpa1 (47.7% identity), which is involved in pheromone sensing.
FIG. 21.
Model for signal transduction pathways in N. crassa. A cAMP signaling pathway consisting of G protein α subunits (GNA1 and GNA2), adenylyl cyclase (CR1), and a PKA regulatory subunit (MCB) is involved in conidiation, morphogenesis, mating, and stress tolerance. Ras-, MAP kinase (MAPK)-, and PKA-related kinase-mediated signaling pathways also regulate conidiation. How signaling is coordinated between these pathways remains to be elucidated.
Disruption of the gna-1 gene causes pleiotropic defects in asexual and sexual development (119, 120, 316). During vegetative growth, Δgna-1 strains exhibit slower apical extension of hyphae and slower growth, and hyperbranching of the mycelium occurs on solid minimal medium (119). Addition of NaCl, KCl, or sorbitol exacerbates the growth defect of Δgna-1 mutants on solid medium. Furthermore, Δgna-1 strains have defects in forming aerial hyphae and macroconidia. In addition to defects in morphogenesis, Δgna-1 mutants show a higher carotenoid content than wild-type strains (316). Carotenoids have been shown to provide resistance to oxidative stress through quenching of free radicals (197, 248). Furthermore, resistance to oxidative stress is often linked to heat tolerance. Consistent with this, the Δgna-1 strain is more resistant to heat and oxidative stress by hydrogen peroxide (316). During sexual development, Δgna-1 mutants differentiate small and infertile protoperithecia and produce fewer protoperithecia than wild-type strains (119).
Strains expressing activated alleles of the gna-1 gene (R178C and Q204L) exhibit opposite phenotypes with respect to defects observed in Δgna-1 strains (316). In contrast to Δgna-1 strains, gna-1R178C and gna-1Q204L mutant strains produce longer, more abundant aerial hyphae than the wild type. In spite of the presence of more aerial hyphae, the activated-allele-containing strains produce almost the same amount of conidia as the wild type. Although Δgna-1 strains produce fewer aerial hyphae, they also make the same amount of conidia as the wild type. Both activated gna1 alleles complement the sensitivity of Δgna-1 mutant strains to NaCl, KCl, and sorbitol (316). gna-1R178C and gna-1Q204L mutant strains produce less carotenoids than the wild type and are more sensitive to heat and hydrogen peroxide. In sexual development, the Q204L and R178C strains are female fertile but produce fewer perithecia than the wild type (316).
Some of the phenotypes observed in the Δgna-1, gna-1R178C, and gna-1Q204L strains are consistent with their effects on cAMP production. As described below, adenylyl cyclase-defective cr-1 mutants exhibit morphological aberrations, defective conidium formation, and thermotolerance. Moreover, previous studies suggest that aerial hyphal development is positively regulated by cAMP (205, 273, 316). Carotenoid accumulation has been shown to be negatively correlated with cAMP levels (138). In accord with these findings, cAMP levels in the Δgna-1 mutants are lower than those in the wild type, and levels of cAMP in the gna-1R178C and gna-1Q204L mutant strains are higher (120, 316). In vitro experiments have demonstrated that GNA1 positively regulates adenylyl cyclase activity, possibly via direct interactions with adenylyl cyclase (120). Interestingly, a mutation in the gna-1 locus affects both phosphodiesterase and adenylyl cyclase activities (120).
Deletion of gna-2 or introduction of an activated allele (gna-2R179C or gna-2Q205L) does not result in major abnormalities during vegetative growth or sexual development (16). However, combination of the Δgna-2 and Δgna-1 mutations exacerbates Δgna-1 mutant phenotypes (16, 120). Δgna-1 Δgna-2 double mutants show a slower rate of hyphal apical extension than Δgna-1 strains on hyperosmotic solid medium containing sorbitol, KCl, or NaCl (16). Adenylyl cyclase and phosphodiesterase activities in the Δgna-1 Δgna-2 strains are lower than those in the Δgna-1 single mutant strains (120). During the sexual cycle, Δgna-1 Δgna-2 mutant strains produce fewer protoperithecia, which fail to differentiate ascospores (16). These data indicate that GNA1 and GNA2 have overlapping roles in both the asexual and sexual cycles.
Adenylyl Cyclase
Two mutations of N. crassa that cause defects in cAMP metabolism have been isolated (241): one is frost (fr) (250, 251), and the other is crisp-1 (cr-1) (274). Originally, these mutations were characterized as morphological, causing a colonial morphology due to hyperbranching of hyphae (89, 228). Both mutants exhibit reduced adenylyl cyclase activity and low endogenous levels of cAMP (274). The abnormal morphology of fr mutants is not corrected by exogenous cAMP or phosphodiesterase inhibitors, indicating that the morphology of fr mutants is not caused by low endogenous cAMP levels (241). Very recently, the fr gene was isolated and found to encode a homolog of the S. cerevisiae Cdc1 protein (258), which regulates Mn2+ homeostasis in S. cerevisiae (221, 222). The fr gene product may also be involved in Mn2+ homeostasis and regulation of hyphal morphology (258).
cr-1 mutant strains exhibit colonial growth with few or no aerial hyphae and undergo a characteristic uniform, dense, and premature conidiogenesis. Even in liquid culture, germ tubes emerging from conidia differentiate early into branched conidiophores (123). Another characteristic of the cr-1 mutant is an inability to use glycerol and other carbon sources (275). Exogenous cAMP or dibutyryl-cAMP stimulates the rate of mycelial elongation and formation of aerial hyphae in cr-1 mutants (273, 274). These observations suggest that a defect in cAMP production is the primary cause of the cr-1 phenotype. Consistent with this, the cr-1 gene was found to encode adenylyl cyclase (134, 135). As observed in S. cerevisiae, adenylyl cyclase-deficient mutant strains exhibit increased thermotolerance (58, 254). In summary, in N. crassa adenylyl cyclase is involved in hyphal tip growth, formation of aerial hyphae, conidiation, and thermotolerance.
Regulatory Subunit of cAMP-Dependent Protein Kinase
N. crassa mcb mutant strains lacking the PKA regulatory subunit exhibit temperature-sensitive morphological abnormalities (35). mcb mutant strains also show a complete loss of growth polarity, which is restricted to the apical cell of hyphal filaments when strains are cultured at 37°C (35). The isotropic mutant hyphae are distended at the restrictive temperature, resulting in an increase in hyphal length as well as hyphal diameter. This phenotype is not suppressed by osmotic stabilization, indicating that cell wall integrity is not compromised in the mcb mutant at 37°C.
Cytological studies on the mcb mutants revealed that these strains have defects in cytoskeleton organization (35). In wild-type hyphae, actin patches are concentrated at the hyphal tip and associated with the cell cortex. When mcb mutant hyphae are incubated at 37°C, actin patches are uniformly distributed over the surface of hyphae but are still concentrated at sites of septation. Calcofluor white staining revealed septal abnormalities in mcb mutants at 37°C. Some septa were immediately adjacent to each other, and most septa were asymmetric. Cytoplasmic microtubule tracks in wild-type cells are located in regions of hyphae and extend from distal to apical regions of hyphae. In the mcb mutant, the hyphae themselves are shorter and organized in overlapping and concentrated networks at 37°C.
The predicted amino acid sequence of the MCB protein has similarity to regulatory subunits of cAMP-dependent protein kinase (PKA), including S. cerevisiae Bcy1 (62%) and S. pombe Cgs1 (57%) (35). Thus, it is most likely that N. crassa PKA regulates hyphal growth polarity, possibly through organization of the cytoskeleton. As mentioned above, components of the cAMP-mediated signaling pathway, including GNA1, GNA2, and adenylyl cyclase (CR1), are involved in morphology and conidiation. Morphogenesis depends on polarized hyphal growth, which is governed by hyphal extension. Conidiation requires polarized growth of hyphae. Therefore, growth polarity and cytoskeletal organization regulated by PKA must be important for conidiation as well as morphology.
Surprisingly, the cytological defects observed in mcb mutant strains with constitutive PKA activity were suppressed by adenylyl cyclase (cr-1) mutations, and it was concluded that the cr-1 mutation is epistatic to the mcb mutation (35). This is surprising because a mutant lacking the regulatory subunit of PKA should have constitutive PKA activity that is no longer regulated by cAMP. Possible explanations are that there is a second PKA regulatory subunit in N. crassa, that there is a second target of cAMP in N. crassa, or that the mcb mutation is a hypomorphic allele that could retain residual regulation by cAMP. Further studies will be required to resolve this issue.
Ras Homologs NC-ras and NC-ras2
Two ras homologs, NC-ras and smco7/NC-ras2, have been identified in N. crassa (11, 125). NC-ras was isolated from an N. crassa cDNA library using the S. cerevisiae RAS2 gene as the probe (11). NC-ras2 was isolated from an N. crassa genomic library using the mammalian v-H-ras gene as the probe (125). Although the roles of the N. crassa ras genes are largely unknown, the NC-ras2 gene has been deleted in N. crassa (125). The NC-ras2 gene is not essential for growth, but mutation confers slow growth and an abnormal morphology during vegetative culture on solid medium. The growth rate of the mutants is about one-tenth that of wild-type cells. The NC-ras2 mutants produce more branching hyphae at peripheral regions of the colony. In addition, the apical compartment in these mutants is shorter and thinner than in the wild type. Because growth rate and morphology depend on hyphal growth, which is limited to the apex of the hyphae in filamentous fungi, the NC-RAS2 protein appears to regulate apical growth of hyphae. Moreover, the NC-ras2 mutants produce fewer aerial hyphae and conidia than the wild type, suggesting that NC-RAS2 protein also controls formation of aerial hyphae and conidia (125).
The morphological characteristics of the NC-ras2 mutant are very similar to those of a morphological mutation, smco7 (semicolonial-7), which had been mapped on the same linkage group as NC-ras2 (89, 199). In fact, the smco7 mutation is an allele of NC-ras2 (125).
MAPKKK homolog and cAMP-Dependent Protein Kinase-Related Kinase
To isolate N. crassa mutations affecting the regulation of the clock-controlled gene 1 (ccg-1) (173), the nrc-1 and nrc-2 mutants were isolated by insertional mutagenesis (136). The ccg-1 gene is one of the earliest genes expressed during conidiation, and its expression is regulated by extracellular glucose levels, circadian rhythm, and light (14, 173, 190). ccg-1 is induced greater than 500-fold in response to glucose limitation (190, 295) and regulated 5- to 10-fold by the circadian clock, with the highest level of expression in the morning, when conidiation occurs (173). ccg-1 expression is also induced by blue light (14). Because expression of the ccg-1 gene is regulated by three distinct stimuli which induce conidiation, it was anticipated that mutants defective in conidiation could be isolated with ccg-1 as a reporter gene. In fact, in both the nrc-1 and nrc-2 mutants, ccg-1 is constitutively expressed and conidiation is defective (136).
Although exposure to air and glucose limitation are necessary for conidiation, the nrc-1 mutant can produce conidiophores in glucose-rich solid and liquid medium. Even in constant darkness, the nrc-1 mutant can differentiate and produce conidia more efficiently than the wild-type strain. Therefore, in the nrc-1 mutant, conidiation occurs constitutively and the asexual developmental program is no longer regulated by glucose and light levels, suggesting that the nrc-1 gene is necessary to repress asexual development. In the sexual cycle, the nrc-1 mutant exhibits a female sterile phenotype and fails to produce protoperithecia because of constitutive entry into the conidiation program and failure to produce vegetative hyphae from which protoperithecia arise. The nrc-1 mutant also shows an ascospore-lethal phenotype when crossed with a wild-type strain as the male partner. Thus, the nrc-1 gene has roles in ascospore development and asexual development.
The deduced amino acid sequence of the nrc-1 gene reveals homology to the S. cerevisiae STE11 and S. pombe byr2 gene products, both of which are MAPKKKs (136, 232, 294). The carboxy-terminal region containing the protein kinase catalytic domain is the most highly conserved. The Ste11 and byr2 kinases are necessary for pheromone-induced sexual differentiation in budding and fission yeasts. Moreover, the S. cerevisiae MAP kinase signaling pathways in which Ste11 functions are also involved in haploid invasive growth and diploid pseudohyphal growth (see section on S. cerevisiae). Interestingly, inactivation of the nrc-1 gene results in enhanced invasive growth in N. crassa (136).
On the other hand, a mutation in the nrc-2 gene results in a complete loss of vegetative hyphae (136). The nrc-2 mutant strain produces thin and meandering hyphae that closely resemble wild-type aerial hyphae. Although some of the mutant hyphae generate chains of proconidia, these conidia do not mature fully and the conidia remain attached together (136). During sexual development, the nrc-2 mutant fails to produce protoperithecia, resulting in female sterility. This may be attributable to derepression of conidial differentiation (136).
The predicted amino acid sequence of NRC2 has the highest level of amino acid sequence identity with putative serine/threonine protein kinases encoded by the KIN82 and YNR047w genes in S. cerevisiae and the kad5+ gene in S. pombe (136). These kinases are closely related to cAMP-dependent protein kinases, but their cellular functions are unknown. Although both nrc-2 and nrc-1 mutant strains were isolated as mutants that fail to repress ccg-1 expression and are unable to regulate conidiation, it is unclear whether the NRC2 kinase acts in the same signaling cascade as the NRC1 MAPKKK.
Conclusions
In summary, a cAMP signaling pathway regulates morphology, conidiation, mating, and responses to heat shock and oxidative stress in N. crassa. The mcb mutant phenotypes suggest that regulation of morphology and conidiation may involve integrity of cellular growth polarity and cytoskeletal organization. The similarity in phenotypes between mutations affecting adenylyl cyclase (cr-1), the MAPKKK (nrc-1), and the PKA homolog (nrc-2) suggests that they coordinately regulate development, possibly in two parallel signaling pathways (MAP kinase and cAMP-PKA), as is the case in S. cerevisiae, S. pombe, C. albicans, U. maydis, and C. neoformans. Further studies are required to establish the relationships among these signaling pathways. In addition to the mutants discussed here, more than 100 mutants of N. crassa exhibiting abnormal morphology have been isolated (229). N. crassa is genetically tractable, and the genome sequencing project for this organism is in progress. Thus, N. crassa is an excellent model organism for the study of signaling cascades regulating morphogenesis and differentiation in filamentous fungi.
FINAL OVERVIEW
Studies in different fungi, including both ascomycetes and basidiomycetes, have converged to define two broadly conserved signal transduction cascades that regulate fungal development and virulence. One is a MAP kinase cascade that mediates responses to pheromone. The second is a nutrient-sensing GPCR, Gα protein-cAMP-PKA cascade. These pathways function coordinately to regulate mating, filamentation, and virulence. How these pathways are organized differs in detail, likely as organisms have evolved to live predominantly as haploid or diploid, to mate under nutrient-rich or -poor conditions, or to respond to host signals as they infect plants or animals. What is most striking is how the general mechanisms of signal sensing are conserved. For example, a highly conserved Gα protein (Gpa2, gpa2, GPA1, and Gpa3) functions in a nutrient-sensing pathway to regulate cAMP production during pseudohyphal growth in S. cerevisiae, mating in S. pombe, mating and virulence in the human fungal pathogen C. neoformans, and mating and virulence of the maize pathogen U. maydis, respectively. These studies illustrate how flexible regulatory networks can be adopted by divergent organisms as they adapt to unique environmental challenges. These signaling cascades have much to teach us about conserved principles of signal transduction that are likely also applicable to more complex multicellular eukaryotes such as vertebrates and mammals. These signaling cascades also represent excellent targets for novel antifungal drugs.
ACKNOWLEDGMENTS
The first eight authors made equal contributions in the preparation of this review.
We thank Olen Yoder and Marian Carlson for the invitation to prepare this review and Tina Jeffries for patient assistance with manuscript preparation.
Our studies are supported by R01 grants AI39115, AI41937, and AI42159 from NIAID and program project P01 award AI44975 to the Duke University Mycology Research Unit from NIAID. Joseph Heitman is a Burroughs Wellcome Scholar in Molecular Pathogenic Mycology and an associate investigator of the Howard Hughes Medical Institute.
ADDENDUM IN PROOF
A Gα subunit closely related to the nutrient-sensing Gα proteins Gpa2 from S. cerevisiae and gpa2 from S. pombe has recently been identified from A. nidulans and named GNA-3 (A. M. Kays, P. S. Rowley, R. A. Baasiri, and K. A. Borkovich, Mol. Cell. Biol. 20:7693–7705, 2000). Both the intracellular levels of cAMP and the protein levels of adenylyl cyclase were found to be reduced in gna-3 mutant strains, leading to the proposal that this Gα subunit might function by regulating the stability, rather than the activity, of adenylyl cyclase. Further studies will be required to test if GNA-3 plays a role in glucose sensing similar to that of the related Gα subunits in budding and fission yeast.
A recent elegant study has examined in detail the cellular functions of the flocculin proteins Flo11, Flo10, Flo1, and Fig2 in S. cerevisiae, revealing that these proteins can substitute for each other if they are appropriately expressed (B. Guo, C. A. Styles, Q. Feng, and G. R. Fink, Proc. Natl. Acad. Sci. USA 97:12158–12163, 2000). In addition, the cell surface flocculin Flo11 that is required for pseudohyphal differentiation was found to be expressed in filamentous diploid cells at the distal tips but not in nonfilamentous vegetative diploid cells. These experiments suggest that additional levels of regulation of Flo11 expression are involved in differentiation into the filamentous state in S. cerevisiae.
A report on the regulation of haploid filamentous growth by mating pheromone in S. cerevisiae is now in press (S. Erdman and M. Snyder, Genetics, in press). Finally, an excellent review on signal transduction in S. pombe has recently appeared (J. Davey, Yeast 14:1529–1566, 1998).
REFERENCES
- 1.Adachi K, Hamer J E. Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. Plant Cell. 1998;10:1361–1374. doi: 10.1105/tpc.10.8.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams T H, Boylan M T, Timberlake W E. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell. 1988;54:353–362. doi: 10.1016/0092-8674(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 3.Adams T H, Hide W A, Yager L N, Lee B N. Isolation of a gene required for programmed initiation of development by Aspergillus nidulans. Mol Cell Biol. 1992;12:3827–3833. doi: 10.1128/mcb.12.9.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams T H, Timberlake W E. Developmental repression of growth and gene expression in Aspergillus. Proc Natl Acad Sci USA. 1990;87:5405–5409. doi: 10.1073/pnas.87.14.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams T H, Wieser J K, Yu J H. Asexual sporulation in Aspergillus nidulans. Microbiol Mol Biol Rev. 1998;62:35–54. doi: 10.1128/mmbr.62.1.35-54.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn S H, Acurio A, Kron S J. Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol Biol Cell. 1999;10:3301–3316. doi: 10.1091/mbc.10.10.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alspaugh J A, Cavallo L M, Perfect J R, Heitman J. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol. 2000;36:352–365. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- 8.Alspaugh J A, Davidson R C, Heitman J. Morphogenesis of Cryptococcus neoformans. In: Ernst J F, Schmidt A, editors. Dimorphism in human pathogenic and apathogenic yeasts. Vol. 5. Basel, Switzerland: Karger; 2000. pp. 217–238. [DOI] [PubMed] [Google Scholar]
- 9.Alspaugh J A, Perfect J R, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alspaugh J A, Perfect J R, Heitman J. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet Biol. 1998;25:1–14. doi: 10.1006/fgbi.1998.1079. [DOI] [PubMed] [Google Scholar]
- 11.Altschuler D L, Muro A, Schijman A, Almonacid F B, Torres H N. Neurospora crassa cDNA clones coding for a new member of the ras protein family. FEBS Lett. 1990;273:103–106. doi: 10.1016/0014-5793(90)81061-r. [DOI] [PubMed] [Google Scholar]
- 12.Ansari K, Martin S, Farkasovsky M, Ehbrecht I M, Kuntzel H. Phospholipase C binds to the receptor-like GPR1 protein and controls pseudohyphal differentiation in Saccharomyces cerevisiae. J Biol Chem. 1999;274:30052–30058. doi: 10.1074/jbc.274.42.30052. [DOI] [PubMed] [Google Scholar]
- 13.Arkinstall S J, Papasavvas S G, Payton M A. Yeast α-mating factor receptor-linked G protein signal transduction suppresses Ras-dependent activity. FEBS Lett. 1991;284:123–128. doi: 10.1016/0014-5793(91)80777-z. [DOI] [PubMed] [Google Scholar]
- 14.Arpaia G, Loros J J, Dunlap J C, Morelli G, Macino G. Light induction of the clock-controlled gene ccg-1 is not transduced through the circadian clock in Neurospora crassa. Mol Gen Genet. 1995;247:157–163. doi: 10.1007/BF00705645. [DOI] [PubMed] [Google Scholar]
- 15.Axelrod D E, Gealt M, Pastushok M. Gene control of developmental competence in Aspergillus nidulans. Dev Biol. 1973;34:9–15. doi: 10.1016/0012-1606(73)90335-7. [DOI] [PubMed] [Google Scholar]
- 16.Baasiri R A, Lu X, Rowley P S, Turner G E, Borkovich K A. Overlapping functions for two G protein alpha subunits in Neurospora crassa. Genetics. 1997;147:137–145. doi: 10.1093/genetics/147.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banuett F. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- 18.Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banuett F. Ustilago maydis, the delightful blight. Trends Genet. 1992;8:174–180. doi: 10.1016/0168-9525(92)90220-x. [DOI] [PubMed] [Google Scholar]
- 20.Banuett F, Herskowitz I. Identification of Fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 1994;8:1367–1378. doi: 10.1101/gad.8.12.1367. [DOI] [PubMed] [Google Scholar]
- 21.Bardwell L, Cook J G, Zhu-Shimoni J X, Voora D, Thorner J. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc Natl Acad Sci USA. 1998;95:15400–15405. doi: 10.1073/pnas.95.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bardwell L, Cook J G, Voora D, Baggott D M, Martinez A R, Thorner J. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 1998;12:2887–2898. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett K J, Gold S E, Kronstad J W. Identification and complementation of a mutation to constitutive filamentous growth in Ustilago maydis. Mol Plant-Microbe Interact. 1993;6:274–283. doi: 10.1094/mpmi-6-274. [DOI] [PubMed] [Google Scholar]
- 24.Beckerman J L, Naider F, Ebbole D J. Inhibition of pathogenicity of the rice blast fungus by Saccharomyces cerevisiae α-factor. Science. 1997;276:1116–1119. doi: 10.1126/science.276.5315.1116. [DOI] [PubMed] [Google Scholar]
- 25.Bell-Pedersen D, Dunlap J C, Loros J J. Distinct cis-acting elements mediate clock, light, and developmental regulation of the Neurospora crassa eas (ccg-2) gene. Mol Cell Biol. 1996;16:513–521. doi: 10.1128/mcb.16.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bencina M, Panneman H, Ruijter G J, Legisa M, Visser J. Characterization and overexpression of the Aspergillus niger gene encoding the cAMP-dependent protein kinase catalytic subunit. Microbiology. 1997;143:1211–1220. doi: 10.1099/00221287-143-4-1211. [DOI] [PubMed] [Google Scholar]
- 27.Berman D M, Kozasa T, Gilman A G. The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J Biol Chem. 1996;271:27209–27212. doi: 10.1074/jbc.271.44.27209. [DOI] [PubMed] [Google Scholar]
- 28.Blacketer M J, Koehler C M, Coats S G, Myers A M, Madaule P. Regulation of dimorphism in Saccharomyces cerevisiae: involvement of the novel protein kinase homolog Elm1p and protein phosphatase 2A. Mol Cell Biol. 1993;13:5567–5581. doi: 10.1128/mcb.13.9.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bölker M, Urban M, Kahmann R. The a mating type locus of U. maydis specifies cell signaling components. Cell. 1992;68:441–450. doi: 10.1016/0092-8674(92)90182-c. [DOI] [PubMed] [Google Scholar]
- 30.Borges-Walmsley M I, Walmsley A R. cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 2000;8:133–141. doi: 10.1016/s0966-842x(00)01698-x. [DOI] [PubMed] [Google Scholar]
- 31.Boylan M T, Mirabito P M, Willett C E, Zimmerman C R, Timberlake W E. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol Cell Biol. 1987;7:3113–3118. doi: 10.1128/mcb.7.9.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandao R L, Magalhaes-Rocha N M d, Alijo R, Ramos J, Thevelein J M. Possible involvement of a phosphatidylinositol-type signaling pathway in glucose-induced activation of plasma membrane H+- ATPase and cellular proton extrusion in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1994;1223:117–124. doi: 10.1016/0167-4889(94)90080-9. [DOI] [PubMed] [Google Scholar]
- 33.Braun B R, Johnson A D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 34.Braun B R, Johnson A D. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics. 2000;155:57–67. doi: 10.1093/genetics/155.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruno K S, Aramayo R, Minke P F, Metzenberg R L, Plamann M. Loss of growth polarity and mislocalization of septa in a Neurospora mutant altered in the regulatory subunit of cAMP-dependent protein kinase. EMBO J. 1996;15:5772–5782. [PMC free article] [PubMed] [Google Scholar]
- 36.Busby T M, Miller K Y, Miller B L. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics. 1996;143:155–163. doi: 10.1093/genetics/143.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bussink H J, Osmani S A. A cyclin-dependent kinase family member (PHOA) is required to link developmental fate to environmental conditions in Aspergillus nidulans. EMBO J. 1998;17:3990–4003. doi: 10.1093/emboj/17.14.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byrne S M, Hoffman C S. Six git genes encode a glucose-induced adenylate cyclase activation pathway in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1993;105:1095–1100. doi: 10.1242/jcs.105.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cannon J F, Tatchell K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1987;7:2653–2663. doi: 10.1128/mcb.7.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, D.C.: ASM Press; 1998. [Google Scholar]
- 41.Casselton L A, Olesnicky N S. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castilla R, Passeron S, Cantore M L. N-Acetyl-d-glucosamine induces germination in Candida albicans through a mechanism sensitive to inhibitors of cAMP-dependent protein kinase. Cell Signal. 1998;10:713–719. doi: 10.1016/s0898-6568(98)00015-1. [DOI] [PubMed] [Google Scholar]
- 43.Chandarlapaty S, Errede B. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:2884–2891. doi: 10.1128/mcb.18.5.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang Y C, Kwon-Chung K J. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect Immun. 1998;66:2230–2236. doi: 10.1128/iai.66.5.2230-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang Y C, Penoyer L A, Kwon-Chung K J. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect Immun. 1996;64:1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang Y C, Timberlake W E. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics. 1993;133:29–38. doi: 10.1093/genetics/133.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang Y C, Wickes B L, Miller G F, Penoyer L A, Kwon-Chung K J. Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J Exp Med. 2000;191:871–882. doi: 10.1084/jem.191.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen B, Gao S, Choi G H, Nuss D L. Extensive alteration of fungal gene transcript accumulation and elevation of G-protein-regulated cAMP levels by a virulence-attenuating hypovirus. Proc Natl Acad Sci USA. 1996;93:7996–8000. doi: 10.1073/pnas.93.15.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi G H, Chen B, Nuss D L. Virus-mediated or transgenic suppression of a G-protein α subunit and attenuation of fungal virulence. Proc Natl Acad Sci USA. 1995;92:305–309. doi: 10.1073/pnas.92.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi W, Dean R A. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell. 1997;9:1973–1983. doi: 10.1105/tpc.9.11.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark K L, Feldmann P J, Dignard D, Larocque R, Brown A J, Lee M G, Thomas D Y, Whiteway M. Constitutive activation of the Saccharomyces cerevisiae mating response pathway by a MAP kinase kinase from Candida albicans. Mol Gen Genet. 1995;249:609–621. doi: 10.1007/BF00418030. [DOI] [PubMed] [Google Scholar]
- 52.Clutterbuck A J. A mutational analysis of conidial development in Aspergillus nidulans. Genetics. 1969;63:317–327. doi: 10.1093/genetics/63.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coccetti P, Tisi R, Martegani E, Teixeira L S, Brandao R L, Castro I, Thevelein J M. The PLC1 encoded phospholipase C in the yeast Saccharomyces cerevisiae is essential for glucose-induced phosphatidylinositol turnover and activation of plasma membrane H+-ATPase. Biochim Biophys Acta. 1998;1405:147–154. doi: 10.1016/s0167-4889(98)00099-8. [DOI] [PubMed] [Google Scholar]
- 54.Colombo S, Ma P, Cauwenberg L, Winderickx J, Crauwels M, Teunissen A, Nauwelaers D, de Winde J H, Gorwa M, Colavizza D, Thevelein J M. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:3326–3341. doi: 10.1093/emboj/17.12.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cook J G, Bardwell L, Kron S J, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 56.Cook J G, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 57.Cox G M, Toffaletti D L, Perfect J R. Dominant selection system for use in Cryptococcus neoformans. J Med Vet Mycol. 1996;34:385–391. [PubMed] [Google Scholar]
- 58.Cruz A K, Terenzi H F, Jorge J A, Terenzi H F. Cyclic AMP-dependent, constitutive thermotolerance in the adenylate cyclase-deficient cr-1 (crisp) mutant of Neurospora crassa. Curr Genet. 1988;13:451–454. doi: 10.1007/BF00365668. [DOI] [PubMed] [Google Scholar]
- 59.Cruz M C, Cavallo L M, Görlach J M, Cox G, Perfect J R, Cardenas M E, Heitman J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol. 1999;19:4101–4112. doi: 10.1128/mcb.19.6.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cruz M C, Poeta M D, Wang P, Wenger R, Zenke G, Quesniaux V F J, Movva N R, Perfect J R, Cardenas M E, Heitman J. Immunosuppressive and nonimmunosuppressive cyclosporin analogs are toxic to the opportunistic fungal pathogen Cryptococcus neoformans via cyclophilin-dependent inhibition of calcineurin. Antimicrob Agents Chemother. 2000;44:143–149. doi: 10.1128/aac.44.1.143-149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cruz M C, Sia R A L, Olson M, Cox G M, Heitman J. Comparison of the roles of calcineurin in physiology and virulence in serotype D and serotype A strains of Cryptococcus neoformans. Infect Immun. 2000;68:982–985. doi: 10.1128/iai.68.2.982-985.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Csank C, Makris C, Meloche S, Schröppel K, Röllinghoff M, Dignard D, Thomas D Y, Whiteway M. Derepressed hyphal growth and reduced virulence in a VH1 family-related protein phosphatase mutant of the human pathogen Candida albicans. Mol Biol Cell. 1997;8:2539–2551. doi: 10.1091/mbc.8.12.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Csank C, Schröppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas D Y, Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davidson R C, Cruz M C, Sia R A L, Allen B, Alspaugh J A, Heitman J. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet Biol. 2000;29:38–48. doi: 10.1006/fgbi.1999.1180. [DOI] [PubMed] [Google Scholar]
- 65.Davis R H, Serres F J D. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;71A:79–143. [Google Scholar]
- 66.Dean R A. Signal pathways and appressorium morphogenesis. Annu Rev Phytopathol. 1997;35:211–234. doi: 10.1146/annurev.phyto.35.1.211. [DOI] [PubMed] [Google Scholar]
- 67.De Jong J C, McCormack B J, Smirnoff N, Talbot N T. Glycerol generates turgor in rice blast. Nature. 1997;389:244–245. [Google Scholar]
- 68.Del Poeta M, Cruz M C, Cardenas M E, Perfect J R, Heitman J. Synergistic antifungal activities of bafilomycin A(1), fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans. Antimicrob Agents Chemother. 2000;44:739–746. doi: 10.1128/aac.44.3.739-746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Voti J, Seydoux G, Beach D, McLeod M. Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. EMBO J. 1991;10:3759. doi: 10.1002/j.1460-2075.1991.tb04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Vries L, Farquhar M G. RGS proteins: more than just GAPs for heterotrimeric G proteins. Trends Cell Biol. 1999;9:138–144. doi: 10.1016/s0962-8924(99)01515-9. [DOI] [PubMed] [Google Scholar]
- 71.DeZwaan T M, Carroll A M, Valent B, Sweigard J A. Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell. 1999;11:2013–2030. doi: 10.1105/tpc.11.10.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dickinson J R. “Fusel” alcohols induce hyphal-like extensions and pseudohyphal formation in yeasts. Microbiology. 1996;142:1391–1397. doi: 10.1099/13500872-142-6-1391. [DOI] [PubMed] [Google Scholar]
- 73.Dixon K P, Xu J R, Smirnoff N, Talbot N J. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell. 1999;11:2045–2058. doi: 10.1105/tpc.11.10.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Donzeau M, Bandlow W. The yeast trimeric guanine nucleotide-binding protein α subunit, Gpa2p, controls the meiosis-specific kinase Ime2p activity in response to nutrients. Mol Cell Biol. 1999;19:6110–6119. doi: 10.1128/mcb.19.9.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dürrenberger F, Wong K, Kronstad J W. Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc Natl Acad Sci USA. 1998;95:5684–5689. doi: 10.1073/pnas.95.10.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edgington N P, Blacketer M J, Bierwagen T A, Myers A M. Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol Cell Biol. 1999;19:1369–1380. doi: 10.1128/mcb.19.2.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edman J C, Kwon-Chung K J. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990;10:4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Egel R, Nielsen O, Weilguny D. Sexual differentiation in fission yeast. Trends Genet. 1990;6:369–373. doi: 10.1016/0168-9525(90)90279-f. [DOI] [PubMed] [Google Scholar]
- 79.Emmett R W, Parbery D G. Appressoria. Annu Rev Phytopathol. 1975;13:147–167. [Google Scholar]
- 80.Erke K H. Light microscopy of basidia, basidiospores, and nuclei in spores and hyphae of Filobasidiella neoformans (Cryptococcus neoformans) J Bacteriol. 1976;128:445–455. doi: 10.1128/jb.128.1.445-455.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feng Q, Summers E, Guo B, Fink G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol. 1999;181:6339–6346. doi: 10.1128/jb.181.20.6339-6346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Flanary P L, DiBello P R, Estrada P, Dohlman H G. Functional analysis of Plp1 and Plp2, two homologues of phosducin in yeast. J Biol Chem. 2000;275:18462–18469. doi: 10.1074/jbc.M002163200. [DOI] [PubMed] [Google Scholar]
- 83.Flick J S, Thorner J. Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5861–5876. doi: 10.1128/mcb.13.9.5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Franzot S P, Salkin I F, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999;37:838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fukui Y, Kaziro Y. Molecular cloning and sequence analysis of a ras gene from Schizosaccharomyces pombe. EMBO J. 1985;4:687–691. doi: 10.1002/j.1460-2075.1985.tb03684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fukui Y, Kozasa T, Kaziro Y, Takeda T, Yamamoto M. Role of ras homolog in the life cycle of Schizosaccharomyces pombe. Cell. 1986;44:329–336. doi: 10.1016/0092-8674(86)90767-1. [DOI] [PubMed] [Google Scholar]
- 87.Gagiano M, van Dyk D, Bauer F F, Lambrechts M G, Pretorius I S. Msn1p/Mss10p, Mss11p and Muc1p/Flo11p are part of a signal transduction pathway downstream of Mep2p regulating invasive growth and pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Microbiol. 1999;31:103–116. doi: 10.1046/j.1365-2958.1999.01151.x. [DOI] [PubMed] [Google Scholar]
- 88.Gao S, Nuss D L. Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc Natl Acad Sci USA. 1996;93:14122–14127. doi: 10.1073/pnas.93.24.14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garnjobst L, Tatum E L. A survey of new morphological mutants in Neurospora crassa. Genetics. 1967;57:579–604. doi: 10.1093/genetics/57.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gavrias V, Andrianopoulos A, Gimeno C J, Timberlake W W. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 91.Gilman A G. G-proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 92.Gimeno C J, Fink G R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 94.Gold S, Duncan G, Barrett K, Kronstad J. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 1994;8:2805–2816. doi: 10.1101/gad.8.23.2805. [DOI] [PubMed] [Google Scholar]
- 95.Gold S E, Brogdon S M, Mayorga M E, Kronstad J W. The Ustilago maydis regulatory subunit of a cAMP-dependent protein kinase is required for gall formation in maize. Plant Cell. 1997;9:1585–1594. doi: 10.1105/tpc.9.9.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goldman D, Lee S C, Casadevall A. Pathogenesis of pulmonary Cryptococcus neoformans infection in the rat. Infect Immun. 1994;62:4755–4761. doi: 10.1128/iai.62.11.4755-4761.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Graser Y, Volovsek M, Arrington J, Schonian G, Presber W, Mitchell T G, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Griffioen G, Anghileri P, Imre E, Baroni M D, Ruis H. Nutritional control of nucleocytoplasmic localization of cAMP-dependent protein kinase catalytic and regulatory subunits in Saccharomyces cerevisiae. J Biol Chem. 2000;275:1449–1456. doi: 10.1074/jbc.275.2.1449. [DOI] [PubMed] [Google Scholar]
- 99.Guhad F A, Csank C, Jensen H E, Thomas D Y, Whiteway M, Hau J. Reduced pathogenicity of a Candida albicans MAP kinase phosphatase (CPP1) mutant in the murine mastitis model. APMIS. 1998;106:1049–1055. doi: 10.1111/j.1699-0463.1998.tb00257.x. [DOI] [PubMed] [Google Scholar]
- 100.Guhad F A, Jensen H E, Aalbaek B, Csank C, Mohamed O, Harcus D, Thomas D Y, Whiteway M, Hau J. Mitogen-activated protein kinase-defective Candida albicans is avirulent in a novel model of localized murine candidiasis. FEMS Microbiol Lett. 1998;166:135–139. doi: 10.1111/j.1574-6968.1998.tb13194.x. [DOI] [PubMed] [Google Scholar]
- 101.Hamer J E, Howard R J, Chumley F G, Valent B. A mechanism for surface attachment in spores of a plant pathogenic fungi. Science. 1988;239:288–290. doi: 10.1126/science.239.4837.288. [DOI] [PubMed] [Google Scholar]
- 102.Hamer J E, Talbot N J. Infection-related development in the rice blast fungus Magnaporthe grisea. Curr Opin Microbiol. 1998;1:693–697. doi: 10.1016/s1369-5274(98)80117-3. [DOI] [PubMed] [Google Scholar]
- 103.Hartmann H A, Kahmann R, Bölker M. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 1996;15:1632–1641. [PMC free article] [PubMed] [Google Scholar]
- 104.Hartmann H A, Krüger J, Lottspeich F, Kahmann R. Environmental signals controlling sexual development of the corn smut fungus Ustilago maydis through the transcriptional regulator Prf1. Plant Cell. 1999;11:1293–1305. doi: 10.1105/tpc.11.7.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heitman J, Allen B, Alspaugh J A, Kwon-Chung K J. On the origins of the congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet Biol. 1999;28:1–5. doi: 10.1006/fgbi.1999.1155. [DOI] [PubMed] [Google Scholar]
- 106.Hemenway C S, Heitman J. Calcineurin: structure, function, and inhibition. Cell Biochem Biophysics. 1999;30:115–151. doi: 10.1007/BF02737887. [DOI] [PubMed] [Google Scholar]
- 107.Hepler J R. Emerging roles for RGS proteins in cell signalling. Trends Pharmacol Sci. 1999;20:376–382. doi: 10.1016/s0165-6147(99)01369-3. [DOI] [PubMed] [Google Scholar]
- 108.Hicks J K, Yu J H, Keller N P, Adams T H. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein-dependent signaling pathway. EMBO J. 1997;16:4916–4923. doi: 10.1093/emboj/16.16.4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoffman C S, Winston F. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 1991;5:561–571. doi: 10.1101/gad.5.4.561. [DOI] [PubMed] [Google Scholar]
- 110.Hoffman C S, Winston F. Isolation and characterization of mutants constitutive for expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics. 1990;124:807–816. doi: 10.1093/genetics/124.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Howard R J. Cell biology of pathogenesis. In: Zeigler R S, Leong S A, Teng P S, editors. Rice blast disease. Wallingford, U.K: CAB International; 1994. pp. 3–22. [Google Scholar]
- 112.Howard R J, Bourett T M, Ferrari M A. Infection by Magnaporthe grisea: an in vitro analysis. In: Mendgen K, Lesemann D E, editors. Electron microscopy of plant pathogens. Berlin, Germany: Springer-Verlag; 1991. pp. 251–264. [Google Scholar]
- 113.Howard R J, Ferrari M A. Role of melanin in appressorium function. Exp Mycol. 1989;13:403–418. [Google Scholar]
- 114.Howard R J, Ferrari M A, Roach D H, Money N P. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc Natl Acad Sci USA. 1991;88:11281–11284. doi: 10.1073/pnas.88.24.11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hughes D A, Yabana N, Yamamoto M. Transcriptional regulation of a Ras nucleotide-exchange factor gene by extracellular signals in fission yeast. J Cell Sci. 1994;107:3635–3642. doi: 10.1242/jcs.107.12.3635. [DOI] [PubMed] [Google Scholar]
- 116.Hull C M, Johnson A D. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- 117.Hull C M, Raisner R M, Johnson A D. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 118.Isshiki T, Mochizuki N, Maeda T, Yamamoto M. Characterization of a fission yeast gene, gpa2, that encodes a Gα subunit involved in the monitoring of nutrition. Genes Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- 119.Ivey F D, Hodge P N, Turner G E, Borkovich K A. The Gαi homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol Biol Cell. 1996;7:1283–1297. doi: 10.1091/mbc.7.8.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ivey F D, Yang Q, Borkovich K A. Positive regulation of adenylyl cyclase activity by a Gαi homolog in Neurospora crassa. Fungal Genet Biol. 1999;26:48–61. doi: 10.1006/fgbi.1998.1101. [DOI] [PubMed] [Google Scholar]
- 121.Jelitto T C, Page H A, Read N D. Role of external signals in regulating the pre-penetration phase of infection by the rice blast fungus, Magnapothe grisea. Planta. 1994;194:471–477. [Google Scholar]
- 122.Jiang Y, Davis C, Broach J R. Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 1998;17:6942–6951. doi: 10.1093/emboj/17.23.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jorge J A, Terenzi H F. An enzymatic alteration secondary to adenylyl cyclase deficiency in the cr-1 (Crisp) mutant of Neurospora crassa. Dev Biol. 1980;74:231–238. doi: 10.1016/0012-1606(80)90065-2. [DOI] [PubMed] [Google Scholar]
- 124.Kaibuchi K, Miyajima A, Arai K-I, Matsumoto K. Possible involvement of RAS-encoded proteins in glucose-induced inositolphospholipid turnover in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83:8172–8176. doi: 10.1073/pnas.83.21.8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kana-uchi A, Yamashiro C T, Tanabe S, Murayama T. A ras homologue of Neurospora crassa regulates morphology. Mol Gen Genet. 1997;254:427–432. doi: 10.1007/s004380050435. [DOI] [PubMed] [Google Scholar]
- 126.Kang S, Chumley F G, Valent B. Isolation of the mating-type genes of the phytopathogenic fungus Magnaporthe grisea using genomic subtraction. Genetics. 1994;138:289–296. doi: 10.1093/genetics/138.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kasahara S, Nuss D L. Targeted disruption of a fungal G-protein β subunit gene results in increased vegetative growth but reduced virulence. Mol Plant-Microbe Interact. 1997;10:984–993. doi: 10.1094/MPMI.1997.10.8.984. [DOI] [PubMed] [Google Scholar]
- 128.Kasahara S, Wang P, Nuss D L. Identification of bdm-1, a gene involved in G protein β-subunit function and α-subunit accumulation. Proc Natl Acad Sci USA. 2000;97:412–417. doi: 10.1073/pnas.97.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- 130.Kim D-U, Park S-K, Chung K-S, Choi M-U, Yoo H-S. The G protein β subunit Gpb1 of Schizosaccharomyces pombe is a negative regulator of sexual development. Mol Gen Genet. 1996;252:20–32. doi: 10.1007/BF02173201. [DOI] [PubMed] [Google Scholar]
- 131.Kirkland T N, Fierer J. Cyclosporin A inhibits Coccidioides immitis in vitro and in vivo. Antimicrob Agents Chemother. 1983;24:921–924. doi: 10.1128/aac.24.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Köhler J R, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signalling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kolattukudy P E, Rogers L M, Li D, Hwang C, Flaishman M A. Surface signaling in pathogenesis. Proc Natl Acad Sci USA. 1995;92:4080–4087. doi: 10.1073/pnas.92.10.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kore-eda S, Murayama T, Uno I. Isolation and characterization of the adenylate cyclase structural gene of Neurospora crassa. Jpn J Genet. 1991;66:317–334. doi: 10.1266/jjg.66.317. [DOI] [PubMed] [Google Scholar]
- 135.Kore-eda S, Murayama T, Uno I. Suppression of the cr-1 mutation in Neurospora crassa. Jpn J Genet. 1991;66:77–83. doi: 10.1266/jjg.66.77. [DOI] [PubMed] [Google Scholar]
- 136.Kothe G O, Free S J. The isolation and characterization of nrc-1 and nrc-2, two genes encoding protein kinases that control growth and development in Neurospora crassa. Genetics. 1998;149:117–130. doi: 10.1093/genetics/149.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kraakman L, Lemaire K, Ma P, Teunissen A W R H, Donaton M C V, Dijck P V, Winderickx J, de Winde J H, Thevelein J M. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- 138.Kritsky M S, Sokolovsky V Y, Belozerskaya T A, Chernysheva E K. Relationship between cyclic AMP level and accumulation of carotenoid pigments in Neurospora crassa. Arch Microbiol. 1982;133:206–208. [Google Scholar]
- 139.Kronstad J, Maria A D, Funnell D, Laidlaw R D, Lee N, de Sá M M, Ramesh M. Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch Microbiol. 1998;170:395–404. doi: 10.1007/s002030050659. [DOI] [PubMed] [Google Scholar]
- 140.Kronstad J W. Virulence and cAMP in smuts, blasts and blights. Trends Plant Sci. 1997;2:193–199. [Google Scholar]
- 141.Kronstad J W, Staben C. Mating type in filamentous fungi. Annu Rev Genet. 1997;31:245–276. doi: 10.1146/annurev.genet.31.1.245. [DOI] [PubMed] [Google Scholar]
- 142.Krüger J, Loubradou G, Regenfelder E, Hartmann A, Kahmann R. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol Gen Genet. 1998;260:193–198. doi: 10.1007/s004380050885. [DOI] [PubMed] [Google Scholar]
- 143.Kübler E, Mösch H U, Rupp S, Lisanti M P. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 144.Kwon-Chung K J. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68:821–833. [PubMed] [Google Scholar]
- 145.Kwon-Chung K J. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- 146.Kwon-Chung K J, Bennett J E. Medical mycology. Malvern, Pa: Lea & Febiger; 1992. pp. 397–446. [Google Scholar]
- 147.Kwon-Chung K J, Bennett J E. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol. 1978;108:337–340. doi: 10.1093/oxfordjournals.aje.a112628. [DOI] [PubMed] [Google Scholar]
- 148.Kwon-Chung K J, Edman J C, Wickes B L. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kwon-Chung K J, Polacheck I, Popkin T J. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J Bacteriol. 1982;150:1414–1421. doi: 10.1128/jb.150.3.1414-1421.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lambrechts M G, Bauer F F, Marmur J, Pretorius I S. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci USA. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Landry S, Pettit M T, Apolinario E, Hoffman C S. The fission yeast git5 gene encodes a Gβ subunit required for glucose-triggered adenylate cyclase activation. Genetics. 2000;154:1463–1471. doi: 10.1093/genetics/154.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Larson T G, Nuss D L. Altered transcriptional response to nutrient availability in hypovirus-infected chestnut blight fungus. EMBO J. 1994;13:5616–5623. doi: 10.1002/j.1460-2075.1994.tb06899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lauter F R, Russo V E A. Blue light induction of conidiation-specific genes in Neurospora crassa. Nucleic Acids Res. 1991;19:6883–6886. doi: 10.1093/nar/19.24.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A R, Brown A J P, Thomas D Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee B N, Adams T H. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 1994;8:641–651. doi: 10.1101/gad.8.6.641. [DOI] [PubMed] [Google Scholar]
- 156.Lee B N, Adams T H. fluG and flbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlAβ activation. EMBO J. 1995;15:299–309. [PMC free article] [PubMed] [Google Scholar]
- 157.Lee B N, Adams T H. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol Microbiol. 1994;14:323–334. doi: 10.1111/j.1365-2958.1994.tb01293.x. [DOI] [PubMed] [Google Scholar]
- 158.Lee Y, Dean R A. Hydrophobicity of contact surface induces appressorium formation in Magnaporthe grisea. FEMS Microbiol Lett. 1994;115:71–76. [Google Scholar]
- 159.Lee Y H, Dean R A. cAMP regulates infection structure formation in the plant pathogenic fungus Magnaporthe grisea. Plant Cell. 1993;5:693–700. doi: 10.1105/tpc.5.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu H, Köhler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 161.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 162.Liu H, Styles C A, Fink G R. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu S, Dean R A. G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol Plant-Microbe Interact. 1997;10:1075–1086. doi: 10.1094/MPMI.1997.10.9.1075. [DOI] [PubMed] [Google Scholar]
- 164.Lo H-J, Köhler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 165.Lo W-S, Dranginis A M. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lodge J K, Jackson-Machelski E, Toffaletti D L, Perfect J R, Gordon J I. Targeted gene replacement demonstrates that myristoyl-CoA:protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans. Proc Natl Acad Sci USA. 1994;91:12008–12012. doi: 10.1073/pnas.91.25.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Loeb J D J, Kerentseva T A, Pan T, Sepulveda-Becerra M, Liu H. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics. 1999;153:1535–1546. doi: 10.1093/genetics/153.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lorenz M C, Cutler N S, Heitman J. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lorenz M C, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lorenz M C, Heitman J. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics. 1998;150:1443–1457. doi: 10.1093/genetics/150.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lorenz M C, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lorenz M C, Pan X, Harashima T, Cardenas M E, Xue Y, Hirsch J P, Heitman J. The G protein-coupled receptor GPR1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Loros J J, Denome S A, Dunlap J C. Molecular cloning of genes under control of the circadian clock in Neurospora. Science. 1989;243:385–388. doi: 10.1126/science.2563175. [DOI] [PubMed] [Google Scholar]
- 174.Loubradou G, Begueret J, Turcq B. MOD-D, a Gα subunit of the fungus Podospora anserina, is involved in both regulation of development and vegetative incompatibility. Genetics. 1999;152:519–528. doi: 10.1093/genetics/152.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ma P, Wera S, Dijck P V, Thevelein J M. The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling. Mol Biol Cell. 1999;10:91–104. doi: 10.1091/mbc.10.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Madhani H D, Fink G R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 177.Madhani H D, Fink G R. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- 178.Madhani H D, Fink G R. The riddle of MAP kinase signaling specificity. Trends Genet. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- 179.Madhani H D, Galitski T, Lander E S, Fink G R. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc Natl Acad Sci USA. 1999;96:12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 181.Madi L, McBride S K, Bailey L A, Ebbole D J. rco-3, a gene involved in glucose transport and conidiation in Neurospora crassa. Genetics. 1997;146:499–508. doi: 10.1093/genetics/146.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Maeda T, Watanabe Y, Kunitomo H, Yamamoto M. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J Biol Chem. 1994;269:9632–9637. [PubMed] [Google Scholar]
- 183.Magee B B, Magee P T. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 184.Malathi K, Ganesan K, Datta A. Identification of a putative transcription factor in Candida albicans that can complement the mating defect of Saccharomyces cerevisiae ste12 mutants. J Biol Chem. 1994;269:22945–22951. [PubMed] [Google Scholar]
- 185.Marshall M A, Timberlake W E. Aspergillus nidulans wetA activates spore-specific gene expression. Mol Cell Biol. 1991;11:55–62. doi: 10.1128/mcb.11.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Martinelli S D, Clutterbuck A J. A quantitative survey of conidiation mutants in Aspergillus nidulans. J Gen Microbiol. 1971;69:261–268. doi: 10.1099/00221287-69-2-261. [DOI] [PubMed] [Google Scholar]
- 187.Mayorga M E, Gold S E. Characterization and molecular genetic complementation of mutants affecting dimorphism in the fungus Ustilago maydis. Fungal Genet Biol. 1998;24:364–376. doi: 10.1006/fgbi.1998.1078. [DOI] [PubMed] [Google Scholar]
- 188.Mayorga M E, Gold S E. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol Microbiol. 1999;34:485–497. doi: 10.1046/j.1365-2958.1999.01610.x. [DOI] [PubMed] [Google Scholar]
- 189.McDade, H. C., and G. M. Cox. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol., in press. [DOI] [PubMed]
- 190.McNally M T, Free S J. Isolation and characterization of a Neurospora glucose-repressible gene. Curr Genet. 1988;14:545–551. doi: 10.1007/BF00434079. [DOI] [PubMed] [Google Scholar]
- 191.Metzenberg R L, Glass N L. Mating type and mating strategies in Neurospora. Bioessays. 1990;12:53–59. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- 192.Miller K Y, Wu J, Miller B L. stuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992;6:1770–1782. doi: 10.1101/gad.6.9.1770. [DOI] [PubMed] [Google Scholar]
- 193.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mitchell T K, Dean R A. The cAMP-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. Plant Cell. 1995;7:1869–1878. doi: 10.1105/tpc.7.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mochizuki N, Yamamoto M. Reduction in the intracellular cAMP level triggers initiation of sexual development in fission yeast. Mol Gen Genet. 1992;233:17–24. doi: 10.1007/BF00587556. [DOI] [PubMed] [Google Scholar]
- 196.Mooney J L, Yager L N. Light is required for conidiation in Aspergillus nidulans. Genes Dev. 1990;4:1473–1482. doi: 10.1101/gad.4.9.1473. [DOI] [PubMed] [Google Scholar]
- 197.Moore M M, Breedveld M W, Autor A P. The role of carotenoids in preventing oxidative damage in the pigmented yeast, Rhodotorula mucilaginosa. Arch Biochem Biophys. 1989;270:419–431. doi: 10.1016/0003-9861(89)90524-9. [DOI] [PubMed] [Google Scholar]
- 198.Moore T D E, Edman J C. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol Cell Biol. 1993;13:1962–1970. doi: 10.1128/mcb.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Morgan M P, Garnjobst L, Tatum E L. Linkage relations of new morphological mutants in linkage group V of Neurospora crassa. Genetics. 1967;57:605–612. doi: 10.1093/genetics/57.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Morton A G. The induction of sporulation in mold fungi. Proc R Soc Lond Biol Sci. 1961;153:548–569. [Google Scholar]
- 201.Mösch H-U, Fink G R. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mösch H-U, Kubler E, Krappmann S, Fink G R, Braus G H. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:1325–1335. doi: 10.1091/mbc.10.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mösch H-U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Müller P, Aichinger C, Feldbrügge M, Kahmann R. The MAP kinase Kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol Microbiol. 1999;34:1007–1017. doi: 10.1046/j.1365-2958.1999.01661.x. [DOI] [PubMed] [Google Scholar]
- 205.Murayama T, Fujisawa Y, Okano Y. A suppressor mutation which suppresses adenylyl cyclase mutations in Neurospora crassa. Exp Mycol. 1995;19:320–323. doi: 10.1006/emyc.1995.1039. [DOI] [PubMed] [Google Scholar]
- 206.Nadin-Davis S A, Nasim A. A gene which encodes a predicted protein kinase can restore some functions of the ras gene in fission yeast. EMBO J. 1988;7:985–993. doi: 10.1002/j.1460-2075.1988.tb02905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nakafuku M, Obara T, Kaibuchi K, Miyajima I, Miyajima A, Itoh H, Nakamura S, Arai K-I, Matsumoto K, Kaziro Y. Isolation of a second yeast Saccharomyces cerevisiae gene (GPA2) coding for guanine nucleotide-binding regulatory protein: Studies on its structure and possible functions. Proc Natl Acad Sci USA. 1988;85:1374–1378. doi: 10.1073/pnas.85.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Neer E J. Heterotrimeric G proteins: organizers of transmembrane signals. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 209.Neilson J B, Fromtling R A, Bulmer G S. Cryptococcus neoformans: size range of infectious particles from aerosolized soil. Infect Immum. 1977;17:634–638. doi: 10.1128/iai.17.3.634-638.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Neiman A M, Stevenson B J, Xu H P, Sprague G F, Herskowitz I, Wigler M, Marcus S. Functional homology of protein kinase required for sexual differentiation in Schizosaccharomyces pombe and Saccharomyces cerevisiae suggests a conserved signal transduction module in eukaryotic organisms. Mol Biol Cell. 1993;4:107–120. doi: 10.1091/mbc.4.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nielsen O. Signal transduction during mating and meiosis in S. pombe. Trends Cell Biol. 1993;3:60–65. doi: 10.1016/0962-8924(93)90162-t. [DOI] [PubMed] [Google Scholar]
- 212.Nikawa J, Cameron S, Toda T, Ferguson K M, Wigler M. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1987;1:931–937. doi: 10.1101/gad.1.9.931. [DOI] [PubMed] [Google Scholar]
- 213.Nocero M, Isshiki T, Yamamoto M, Hoffman C S. Glucose repression of fbp1 transcription in Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein α subunit encoded by gpa2 (git8) Genetics. 1994;138:39–45. doi: 10.1093/genetics/138.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Nuss D L. Using hypoviruses to probe and perturb signal transduction processes underlying fungal pathogenesis. Plant Cell. 1996;8:1845–1853. doi: 10.1105/tpc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Obara T, Nakafuku M, Yamamoto M, Kaziro Y. Isolation and characterization of a gene encoding a G-protein alpha subunit from Schizosaccharomyces pombe: involvement in mating and sporulation pathways. Proc Nat Acad Sci USA. 1991;88:5877–5881. doi: 10.1073/pnas.88.13.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Odds F C. Candida and candidosis: a review and bibliography. 2nd ed. London, U.K: Bailliere Tindall; 1988. [Google Scholar]
- 217.Odom A, Muir S, Lim E, Toffaletti D L, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Odom A, Poeta M D, Perfect J, Heitman J. The immunosuppressant FK506 and its nonimmunosuppressive analog L-685,818 are toxic to Cryptococcus neoformans by inhibition of a common target protein. Antimicrob Agents Chemother. 1997;41:156–161. doi: 10.1128/aac.41.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Odom A R, Stahlberg A, Wente S R, York J D. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 220.Oehlen L J, Cross F R. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J Biol Chem. 1998;273:25089–25097. doi: 10.1074/jbc.273.39.25089. [DOI] [PubMed] [Google Scholar]
- 221.Paidhungat M, Garrett S. Cdc1 and the vacuole coordinately regulate Mn2+ homeostasis in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:1787–1798. doi: 10.1093/genetics/148.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Paidhungat M, Garrett S. Cdc1 is required for growth and Mn2+ regulation in Saccharomyces cerevisiae. Genetics. 1998;148:1777–1786. doi: 10.1093/genetics/148.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pan X, Heitman J. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol Cell Biol. 2000;20:8364–8372. doi: 10.1128/mcb.20.22.8364-8372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Papasavvas S, Arkinstall S, Reid J, Payton M. Yeast α-mating factor receptor and G-protein-linked adenylyl cyclase inhibition requires RAS2 and GPA2 activities. Biochem Biophys Res Commun. 1992;184:1378–1385. doi: 10.1016/s0006-291x(05)80035-x. [DOI] [PubMed] [Google Scholar]
- 226.Pastushok M, Axelrod D E. Effect of glucose, ammonium and media maintenance on the time of conidiophore initiation by surface colonies of Aspergillus nidulans. J Gen Microbiol. 1976;94:221–224. doi: 10.1099/00221287-94-1-221. [DOI] [PubMed] [Google Scholar]
- 227.Perfect J R, Lang S D R, Durack D T. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol. 1980;101:177–194. [PMC free article] [PubMed] [Google Scholar]
- 228.Perkins D D. New markers and multiple point linkage data in Neurospora. Genetics. 1959;44:1185–1208. doi: 10.1093/genetics/44.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Perkins D D, Radford A, Newmeyer D, Bjorkman M. Chromosomal loci of Neurospora crassa. Microbiol Rev. 1982;46:426–570. doi: 10.1128/mr.46.4.426-570.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Raymond M, Dignard D, Alarco A M, Mainville N, Magee B B, Thomas D Y. A Ste6p/P-glycoprotein homologue from the asexual yeast Candida albicans transports the a-factor mating pheromone in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:587–598. doi: 10.1046/j.1365-2958.1998.00704.x. [DOI] [PubMed] [Google Scholar]
- 231.Regenfelder E, Spellig T, Hartmann A, Lauenstein S, Bölker M, Kahmann R. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 1997;16:1934–1942. doi: 10.1093/emboj/16.8.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Rhodes N, Connell L, Errede B. STE11 is a protein kinase required for cell-type-specific transcription and signal transduction in yeast. Genes Dev. 1990;4:1862–1874. doi: 10.1101/gad.4.11.1862. [DOI] [PubMed] [Google Scholar]
- 233.Ricci M, Krappmann D, Russo V E A. Nitrogen and carbon starvation regulate conidia and protoperithecia formation of Neurospora crassa grown on solid media. Fungal Genet Newsl. 1991;38:87–88. [Google Scholar]
- 234.Riggle P J, Andrutis K A, Chen X, Tzipori S R, Kumamoto C A. Invasive lesions containing filamentous forms produced by a Candida albicans mutant that is defective in filamentous growth in culture. Infect Immun. 1999;67:3649–3652. doi: 10.1128/iai.67.7.3649-3652.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y D, Dai H, Walker W L, Hughes T R, Tyers M, Boone C, Friend S H. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 236.Roberts R, Mösch H-U, Fink G R. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- 237.Roberts R L, Fink G R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 238.Robertson L S, Fink G R. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Roemer T, Vallier L, Sheu Y, Snyder M. The Spa2-related protein, Sph1p, is important for polarized growth in yeast. J Cell Sci. 1998;111:479–494. doi: 10.1242/jcs.111.4.479. [DOI] [PubMed] [Google Scholar]
- 240.Rosen S, Yu J H, Adams T H. The Aspergillus nidulans sfaD gene encodes a G protein β subunit that is required for normal growth and repression of sporulation. EMBO J. 1999;18:5592–5600. doi: 10.1093/emboj/18.20.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Rosenberg G, Pall M L. Properties of two cyclic nucleotide-deficient mutants of Neurospora crassa. J Bacteriol. 1979;137:1140–1144. doi: 10.1128/jb.137.3.1140-1144.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ruiz-Herrera J, Leon-Ramirez C, Cabrera-Ponce J L, Martinez-Espinoza A D, Herrera-Estrella L. Completion of the sexual cycle and demonstration of genetic recombination in Ustilago maydis in vitro. Mol Gen Genet. 1999;262:468–472. doi: 10.1007/s004380051107. [DOI] [PubMed] [Google Scholar]
- 243.Rupp S, Summers E, Lo H, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converage on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sabie F T, Gadd G M. Effect of nucleosides and nucleotides and the relationship between cellular adenosine 3′:5′-cyclic monophosphate (cyclic AMP) and germ tube formation in Candida albicans. Mycopathologia. 1992;119:147–156. doi: 10.1007/BF00448812. [DOI] [PubMed] [Google Scholar]
- 245.Saiardi A, Caffrey J J, Snyder S H, Shears S B. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 2000;468:28–32. doi: 10.1016/s0014-5793(00)01194-7. [DOI] [PubMed] [Google Scholar]
- 246.Saiardi A, Erdjument-Bromage H, Snowman A M, Tempst P, Snyder S H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 247.Salas S D, Bennett J E, Kwon-Chung K J, Perfect J R, Williamson P R. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Schroeder W A, Johnson E A. Singlet oxygen and peroxyl radicals regulate carotenoid biosynthesis in Phaffia rhodozyma. J Biol Chem. 1995;270:18374–18379. doi: 10.1074/jbc.270.31.18374. [DOI] [PubMed] [Google Scholar]
- 249.Schweingruber A-M, Hilti N, Edenharter E, Schweingruber M E. Methionine induces sexual development in the fission yeast Schizosaccharomyces pombe via an ste11-dependent signalling pathway. J Bacteriol. 1998;180:6338–6341. doi: 10.1128/jb.180.23.6338-6341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Scott W A. Adenosine 3′:5′-cyclic monophosphate deficiency in Neurospora crassa. Proc Natl Acad Sci USA. 1976;73:2995–2999. doi: 10.1073/pnas.73.9.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Scott W A, Solomon B. Adenosine 3′,5′-cyclic monophosphate and morphology in Neurospora crassa: drug-induced alterations. J Bacteriol. 1975;122:454–463. doi: 10.1128/jb.122.2.454-463.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Sharkey L L, McNemar M D, Saporito-Irwin S M, Sypherd P S, Fonzi W A. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J Bacteriol. 1999;181:5273–5279. doi: 10.1128/jb.181.17.5273-5279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Shen W, Bobrowicz P, Ebbole D J. Isolation of pheromone precursor genes of Magnaporthe grisea. Fungal Genet Biol. 1999;27:253–263. doi: 10.1006/fgbi.1999.1151. [DOI] [PubMed] [Google Scholar]
- 254.Shin D, Matsumoto K, Iida H, Uno I, Ishikawa T. Heat shock response of Saccharomyces cerevisiae mutants altered in cyclic AMP-dependent protein phosphorylation. Mol Cell Biol. 1987;7:244–250. doi: 10.1128/mcb.7.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Simon M I, Strathmann M P, Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 256.Skromne I, Sanchez O, Aguirre J. Starvation stress modulates the expression of the Aspergillus nidulans brlA regulatory gene. Microbiology. 1995;141:21–28. doi: 10.1099/00221287-141-1-21. [DOI] [PubMed] [Google Scholar]
- 257.Som T, Kolaparthi V S R. Developmental decisions in Aspergillus nidulans are modulated by Ras activity. Mol Cell Biol. 1994;14:5333–5348. doi: 10.1128/mcb.14.8.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Sone T, Griffiths A J F. The frost gene of Neurospora crassa is a homolog of yeast cdc1 and affects hyphal branching via manganese homeostasis. Fungal Genet Biol. 1999;28:227–237. doi: 10.1006/fgbi.1999.1169. [DOI] [PubMed] [Google Scholar]
- 259.Sonneborn A, Bockmuhl D P, Ernst J F. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect Immun. 1999;67:5514–5517. doi: 10.1128/iai.67.10.5514-5517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sonneborn A, Bockmuhl D P, Gerads M, Kurpanek K, Sanglard D, Ernst J F. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol Microbiol. 2000;35:386–396. doi: 10.1046/j.1365-2958.2000.01705.x. [DOI] [PubMed] [Google Scholar]
- 261.Sonneborn A, Tebarth B, Ernst J F. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect Immun. 1999;67:4655–4660. doi: 10.1128/iai.67.9.4655-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Spellig T, Bölker M, Lottspeich F, Frank R W, Kahmann R. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 1994;13:1620–1627. doi: 10.1002/j.1460-2075.1994.tb06425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Sprague G F, Jr, Thorner J W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- 264.Springer M L. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. Bioessays. 1993;15:365–374. doi: 10.1002/bies.950150602. [DOI] [PubMed] [Google Scholar]
- 265.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 1991;5:1990–1999. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- 267.Sukroongreung S, Kitiniyom K, Nilakul C, Tantimavanich S. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med Mycol. 1998;36:419–424. [PubMed] [Google Scholar]
- 268.Suzuki T, Rogers A L, Magee P T. Inter- and intra-species crosses between Candida albicians and Candida guilliermondii. Yeast. 1986;2:53–58. doi: 10.1002/yea.320020104. [DOI] [PubMed] [Google Scholar]
- 269.Sweigard J A, Carroll A M, Farrall L, Chumley F G, Valent B. Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol Plant-Microbe Interact. 1998;11:404–412. doi: 10.1094/MPMI.1998.11.5.404. [DOI] [PubMed] [Google Scholar]
- 270.Talbot N J, Ebbole D J, Hamer J E. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Tamaki H, Miwa T, Shinozaki M, Saito M, Yun C, Yamamoto K, Kumagai H. GPR1 regulates filamentous growth through FLO11 in yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2000;267:164–168. doi: 10.1006/bbrc.1999.1914. [DOI] [PubMed] [Google Scholar]
- 272.Tanaka K, Davey J, Imai Y, Yamamoto M. Schizosaccharomyces pombe map3+ encodes the putative M-factor receptor. Mol Cell Biol. 1993;13:80–88. doi: 10.1128/mcb.13.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Terenzi H F, Flawia M M, Tellez-inon M T, Torres H N. Control of Neurospora crassa morphology by cyclic adenosine 3′,5′-monophosphate and dibutyryl cyclic adenosine 3′,5′-monophosphate. J Bacteriol. 1976;126:91–99. doi: 10.1128/jb.126.1.91-99.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Terenzi H F, Flawia M M, Torres H N. A Neurospora crassa morphological mutant showing reduced adenylate cyclase activity. Biochem Biophys Res Commun. 1974;58:990–996. doi: 10.1016/s0006-291x(74)80241-x. [DOI] [PubMed] [Google Scholar]
- 275.Terenzi H F, Jorge J A, Roselino J E, Migliorini R H. Adenylyl cyclase deficient cr-1 (crisp) mutant of Neurospora crassa: cyclic AMP-dependent nutritional deficiencies. Arch Microbiol. 1979;123:251–258. doi: 10.1007/BF00406658. [DOI] [PubMed] [Google Scholar]
- 276.Thevelein J M, de Winde J H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 277.Timberlake W E. Developmental gene regulation in Aspergillus nidulans. Dev Biol. 1980;78:497–510. doi: 10.1016/0012-1606(80)90349-8. [DOI] [PubMed] [Google Scholar]
- 278.Timberlake W E. Molecular genetics of Aspergillus development. Annu Rev Genet. 1990;24:5–36. doi: 10.1146/annurev.ge.24.120190.000253. [DOI] [PubMed] [Google Scholar]
- 279.Toda T, Cameron S, Sass P, Zoller M, Scott J D, McMullen B, Hurwitz M, Krebs E G, Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 281.Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- 282.Toffaletti D L, Rude T H, Johnston S A, Durack D T, Perfect J R. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Tolkacheva T, McNamara P, Piekarz E, Courchesne W. Cloning of a Cryptococcus neoformans gene, GPA1, encoding a G-protein α-subunit homolog. Infect Immun. 1994;62:2849–2856. doi: 10.1128/iai.62.7.2849-2856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Tropschug M, Barthelmess I B, Neupert W. Sensitivity to cyclosporin A is mediated by cyclophilin in Neurospora crassa and Saccharomyces cerevisiae. Nature. 1989;342:953–955. doi: 10.1038/342953a0. [DOI] [PubMed] [Google Scholar]
- 285.Turner G E, Borkovich K A. Identification of a G protein α subunit from Neurospora crassa that is a member of the Gi family. J Biol Chem. 1993;268:14805–14811. [PubMed] [Google Scholar]
- 286.Urban M, Kahmann R, Bölker M. Identification of the pheromone response element in Ustilago maydis. Mol Gen Genet. 1996;251:31–37. doi: 10.1007/BF02174341. [DOI] [PubMed] [Google Scholar]
- 287.Vallim M A, Miller K Y, Miller B L. Aspergillus SteA (sterile 12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol Microbiol. 2000;36:290–313. doi: 10.1046/j.1365-2958.2000.01874.x. [DOI] [PubMed] [Google Scholar]
- 288.Versele M, de Winde J H, Thevelein J M. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 1999;18:5577–5591. doi: 10.1093/emboj/18.20.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Vivier M A, Lambrechts M G, Pretorius I S. Coregulation of starch degradation and dimorphism in the yeast Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1997;32:405–435. doi: 10.3109/10409239709082675. [DOI] [PubMed] [Google Scholar]
- 290.Wang P, Heitman J. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr Opin Microbiol. 1999;2:358–362. doi: 10.1016/S1369-5274(99)80063-0. [DOI] [PubMed] [Google Scholar]
- 291.Wang P, Larson T G, Chen C H, Pawlyk D M, Clark J A, Nuss D L. Cloning and characterization of a general amino acid control transcriptional activator from the chestnut blight fungus Cryphonectria parasitica. Fungal Genet Biol. 1998;23:81–94. doi: 10.1006/fgbi.1997.1023. [DOI] [PubMed] [Google Scholar]
- 292.Wang P, Nuss D L. Induction of a Cryphonectria parasitica cellobiohydrolase I gene is suppressed by hypovirus infection and regulated by a GTP-binding-protein-linked signaling pathway involved in fungal pathogenesis. Proc Natl Acad Sci USA. 1995;92:11529–11533. doi: 10.1073/pnas.92.25.11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wang P, Perfect J R, Heitman J. The G-protein β subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol Cell Biol. 2000;20:352–362. doi: 10.1128/mcb.20.1.352-362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Wang Y, Xu H P, Riggs M, Rodgers L, Wigler M. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol Cell Biol. 1991;11:3554–3563. doi: 10.1128/mcb.11.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wang Z, Deak M, Free S J. A cis-acting region required for the regulated expression of grg-1, a Neurospora glucose-repressible gene: two regulatory sites (CRE and NRS) are required to repress grg-1 expression. J Mol Biol. 1994;237:65–74. doi: 10.1006/jmbi.1994.1209. [DOI] [PubMed] [Google Scholar]
- 296.Ward M P, Gimeno C J, Fink G R, Garrett S. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol Cell Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Watson N, Linder M E, Druey K M, Kehrl J H, Blumer K J. RGS family members: GTPase-activating proteins for heterotrimeric G-protein α-subunits. Nature. 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- 298.Welton R M, Hoffman C S. Glucose monitoring in fission yeast via the gpa2 Gα, the git5 Gβ and the git3 putative glucose receptor. Genetics. 2000;156:513–521. doi: 10.1093/genetics/156.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Whiteway M, Dignard D, Thomas D Y. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interefere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc Natl Acad Sci USA. 1992;89:9410–9414. doi: 10.1073/pnas.89.20.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Wickes B L, Edman U, Edman J C. The Cryptococcus neoformans STE12α gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol. 1997;26:951–960. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]
- 301.Wickes B L, Mayorga M E, Edman U, Edman J C. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc Natl Acad Sci USA. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Wieser J, Adams T H. fibD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes Dev. 1995;9:491–502. doi: 10.1101/gad.9.4.491. [DOI] [PubMed] [Google Scholar]
- 303.Wieser J, Lee B N, Fondon J W, Adams T H. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr Genet. 1994;27:62–69. doi: 10.1007/BF00326580. [DOI] [PubMed] [Google Scholar]
- 304.Wieser J, Yu J H, Adams T H. Dominant mutations affecting both sporulation and sterigmatocystin biosynthesis in Aspergillus nidulans. Curr Genet. 1997;32:218–224. doi: 10.1007/s002940050269. [DOI] [PubMed] [Google Scholar]
- 305.Willer M, Hoffmann L, Styrkarsdottir U, Egel R, Davey J, Nielsen O. Two-step activation of meiosis by the mat1 locus in Schizosaccharomyces pombe. Mol Cell Biol. 1995;15:4964–4970. doi: 10.1128/mcb.15.9.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Williamson P R. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wu C, Leeuw T, Leberer E, Thomas D Y, Whiteway M. Cell cycle- and Cln2p-Cdc28p-dependent phosphorylation of the yeast Ste20p protein kinase. J Biol Chem. 1998;273:28107–28115. doi: 10.1074/jbc.273.43.28107. [DOI] [PubMed] [Google Scholar]
- 308.Wynn W K. Appressorium formation over stomates by the bean rust fungus: response to a surface contact stimulus. Phytopathology. 1976;66:136–146. [Google Scholar]
- 309.Xiao J, Ohshima A, Kamakura T, Ishiyama T, Yamaguchi I. Extracellular glycoprotein(s) associated with cellular differentiation in Magnaporthe grisea. Mol Plant-Microbe Interact. 1994;5:639–644. [Google Scholar]
- 310.Xiao J-Z, Watanabe T, Kamakura T, Ohshima A, Yamaguchi I. Studies on cellular differentiation of Magnaporthe grisea: physicochemical aspects of substratum surfaces in relation to appressorium formation. Physiol Mol Plant Pathol. 1994;44:227–236. [Google Scholar]
- 311.Xu J-R, Hamer J E. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 312.Xu J R, Staiger C J, Hamer J E. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc Natl Acad Sci USA. 1998;95:12713–12718. doi: 10.1073/pnas.95.21.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Xu J R, Urban M, Sweigard J A, Hamer J E. The CPKA gene of Magnaporthe grisea is essential for appressorial penetration. Mol Plant-Microbe Interact. 1997;10:187–194. [Google Scholar]
- 314.Xue Y, Batlle M, Hirsch J P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Yamamoto M, Imai Y, Watanabe Y. Mating and sporulation in Schizosaccharomyces pombe. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 1037–1106. [Google Scholar]
- 316.Yang Q, Borkovich K A. Mutational activation of a Gαi causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa. Genetics. 1999;151:107–117. doi: 10.1093/genetics/151.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Ye X S, Lee S L, Wolkow T D, McGuire S L, Hamer J E, Wood G C, Osmani S A. Interaction between developmental and cell cycle regulators is required for morphogenesis in Aspergillus nidulans. EMBO J. 1999;18:6994–7001. doi: 10.1093/emboj/18.24.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.York J D, Odom A R, Murphy R, Ives E B, Wente S R. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 319.Yu J-H, Wieser J, Adams T H. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 1996;15:5184–5190. [PMC free article] [PubMed] [Google Scholar]
- 320.Yue C, Cavallo L M, Alspaugh J A, Wang P, Cox G M, Perfect J R, Heitman J. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Yun C, Tamaki H, Nakayama R, Yamamoto K, Kumagai H. Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;252:29–33. doi: 10.1006/bbrc.1998.9600. [DOI] [PubMed] [Google Scholar]
- 322.Yun C-W, Tamaki H, Nakayama R, Yamamoto K, Kumagai H. G-protein-coupled receptor from yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1997;240:287–292. doi: 10.1006/bbrc.1997.7649. [DOI] [PubMed] [Google Scholar]
- 323.Zimmermann C R, Orr W C, Leclerc R F, Barnard E C, Timberlake W E. Molecular cloning and selection of genes regulated in Aspergillus development. Cell. 1980;21:709–715. doi: 10.1016/0092-8674(80)90434-1. [DOI] [PubMed] [Google Scholar]