Abstract
The mitochondrial transmembrane (TMEM) protein family has several essential physiological functions. However, its roles in cardiomyocyte proliferation and cardiac regeneration remain unclear. Here, we detected that TMEM11 inhibits cardiomyocyte proliferation and cardiac regeneration in vitro. TMEM11 deletion enhanced cardiomyocyte proliferation and restored heart function after myocardial injury. In contrast, TMEM11-overexpression inhibited neonatal cardiomyocyte proliferation and regeneration in mouse hearts. TMEM11 directly interacted with METTL1 and enhanced m7G methylation of Atf5 mRNA, thereby increasing ATF5 expression. A TMEM11-dependent increase in ATF5 promoted the transcription of Inca1, an inhibitor of cyclin-dependent kinase interacting with cyclin A1, which suppressed cardiomyocyte proliferation. Hence, our findings revealed that TMEM11-mediated m7G methylation is involved in the regulation of cardiomyocyte proliferation, and targeting the TMEM11-METTL1-ATF5-INCA1 axis may serve as a novel therapeutic strategy for promoting cardiac repair and regeneration.
Subject terms: Epigenetics, Cardiovascular diseases
Introduction
Cardiovascular disease is the leading cause of death worldwide, and its occurrence is highly correlated with age. Heart failure is a global challenge primarily caused by myocardial cell death [1–4]. The prevalence of heart failure with preserved ejection fraction, such as hypertensive cardiomyopathy, is increasing and is a major cause of hospitalization for heart failure. Myocardial infarction (MI) leads to a massive loss of functional cardiac cells, which is the basic pathological process that induces heart failure. Previous experimental studies have identified complex signal transduction processes that regulate cardiomyocyte death; however, novel therapeutic interventions are still lacking. As a novel therapeutic avenue, MI can be treated by inducing the proliferation of existing adult cardiomyocytes (ACMs), which produces new cardiomyocytes. Mature cardiomyocytes can reenter the cell cycle through dedifferentiation, proliferation, and redifferentiation to form new cardiomyocytes [5, 6]. Several proteins, such as HIPPO, YAP1, PITX2 and MEIS1, play vital roles in controlling the proliferation of adult mammalian cardiomyocytes [7–11]. The HIPPO signaling pathway is an evolutionarily conserved signaling pathway that restricts development and reproduction mainly by inhibiting the activity of the transcriptional coactivator YAP [10, 12]. Moreover, non-coding RNAs, including miRNAs and lncRNAs, play crucial roles in regulating cardiomyocyte proliferation and cardiac regeneration [13–16]. Although substantial progress has been made in understanding the molecular mechanisms of adult mammalian cardiomyocytes proliferation and cardiac regeneration, an in-depth evaluation of these molecular networks is required.
Mitochondria are highly dynamic organelles involved in various physiological activities, such as cell cycle, development, and morphological change [17]. The mitochondria of cardiomyocytes produce most of their chemical energy in the form of adenosine triphosphate (ATP), which is necessary for cardiac contractility and robust heart function [18]. Although mitochondria play a central role in the onset and progression of heart failure (HF) and other cardiovascular diseases (CVD), there is currently no treatment for mitochondrial dysfunction [19]. The mitochondrial transmembrane protein (TMEM) family has several essential physiological functions, notably autophagy, apoptosis, participation in signal transduction pathways, and the formation of plasma membrane ion channels [20–22]. TMEMs also play critical roles in many pathological processes. For instance, TMEMs can function as tumor suppressors [23, 24], and some TMEMs with predictive endoplasmic reticulum localization can be used for kidney cancer classification [25]. Down-regulation of the TMEM88 gene can inhibit myocardial cell differentiation and promote endothelial cell differentiation [26]. However, the functions of TMEMs in the heart remain largely unknown, and their biological functions in cardiovascular diseases require further exploration.
Transcriptional and post-transcriptional regulation, including methylation, demethylation, and RNA splicing, have significant impact on mRNA turnover, which regulates protein expression diversity during cardiomyocyte proliferation and heart regeneration [27–33]. The N7-methylguanosine (m7G) modification present in the mRNA 5′ cap plays a fundamental role in the regulation of transcription elongation [34], mRNA splicing [35], polyadenylation [36], and translation [37], and therefore it is implicated in nearly all aspects of the mRNA life cycle. m7G is also present in tRNAs and rRNAs, where it is associated with human diseases [38, 39]. A recent study showed that methyltransferase METTL1 mediates m7G modification within mammalian mRNAs, which affects their translation [40]. Additionally, METTL1-dependent N7-methylation of guanosine regulates miRNA structure, biogenesis, and cell migration [41]. These findings indicate that RNA m7G modification is strongly associated with the gene regulatory networks involved in both physiological and pathological processes. However, the cellular and molecular mechanisms underlying dysregulation of m7G modifications in the heart are unknown.
Here, we investigated the previously unrecognized function of TMEM11 in regulating cardiomyocyte proliferation in the postnatal heart and uncovered its underlying mechanism. We demonstrated that TMEM11 suppressed cardiomyocyte cell cycle activity by directly interacting with METTL1 and increasing its RNA m7G-methylation activity, thereby upregulating activating transcription factor 5 (ATF5) expression through hypermethylation of Atf5 mRNA. Further, upregulated ATF5 enhanced the expression of INCA1, an inhibitor of cyclin-dependent kinase (CDK) interacting with cyclin A1, which suppressed cardiomyocyte proliferation. Collectively, our study uncovered a novel mechanism by which TMEM11-dependent m7G modification regulates cardiomyocyte proliferation. Targeting the TMEM11-METTL1-ATF5 axis may be an effective strategy for improving heart regeneration after cardiac injury.
Methods
Primary cardiomyocytes culture and treatment
Cardiomyocytes were isolated from neonatal mice (1–2 days old), and we washed the newborn mouse with 75% alcohol. The hearts were surgically cut out and placed in a petri dish with pre-cooled PBS in a sterile environment. We cleaned the hearts with PBS three times, and we shredded the heart tissue with surgical scissors. The heart tissue was transferred to 10 ml of digestive juice (1.2 mg/ ml trypsin and 0.14 mg/ml collagenase II) and gently stirred in a 37 °C water bath for 6 min. After each digestion, we transferred the supernatant to a centrifuge tube containing serum, which was stored on ice, and we added new digestive juice again until the heart tissue disappeared. Centrifuge at 4 °C at 1000 rpm/min for 10 min, the supernatant was taken and re-suspended with F12/DMEM containing 10% serum. Centrifuge at 4 °C at 1000 rpm/min for 10 min, the supernatant was taken and filtered by 70 um filter. The filtered cardiomyocytes were placed in a 10 cm petri dish. Then the suspended cells were placed in a 5% CO2 humidified incubator at 37 °C for 1.5 h. Fibroblasts were attached to the wall, and the culture medium containing cardiomyocytes was collected into a new centrifuge tube. The supernatant was discarded in a 4 °C centrifuge at 1000 rpm/min for 10 minutes. And added 0.1 mm bromodeoxyuridine. According to the experimental design, the plates were placed in a humidified incubator with 5% CO2 at 37 °C. The next day, the culture medium in the petri dish was replaced, and we continued the culture for another day. On the third day, the cardiomyocytes transfected with adenovirus for 6 h were cultured in a new medium for 24 h.
Generation of TMEM11 knockout (TMEM11 KO) mice and TMEM11 transgenic (TMEM11 Tg) mice
All animal experiments were approved by the Institutional Review Board (IRB) of Qingdao University, and the procedures followed were by the guidelines of Qingdao University institutions. TMEM11 knockout mice were generated using the CRISPR/Cas9 technology to modify Tmem11 gene from Nanjing GemPharmatech company in China. The Tmem11 gene has 3 transcripts. According to the structure of Tmem11 gene, exon2 of Tmem11-201 transcript is recommended as the knockout region. The region contains most of the coding sequence. Knock out the region will result in disruption of protein function. The brief process is as follows: sgRNA was transcribed in vitro. Cas9 and sgRNA were microinjected into the fertilized eggs of C57BL/6JGpt mice. Fertilized eggs were transplanted to obtain positive F0 mice which were confirmed by PCR and sequencing. A stable F1 generation mouse model was obtained by mating positive F0 generation mice with C57BL/6JGpt mice. The following primer sequences were used to amplify the genomic DNA fragments containing the CRISPR/Cas9 target sites and TMEM11 knockout. The forward primer(F1) was 5′-CTCCAATCTGAATACATCCAAAGTCC-3′; the reverse primer(R1) was 5′-GGCATTTCAGAATCTGGGTTAGTACAC-3′; the forward primer(F2) was 5′-TGCTGTCTTACCTGTGAACTTGAGAGA-3′; the reverse primer(R2) is 5′-CTGTTTGTCAGGAGAAATGAGTGAGC-3′.
TMEM11 transgenic mice were generated using the microinjection technology from Nanjing GemPharmatech company in China. The brief process is as follows: In order to construct heart-specific TMEM11 overexpression transgenic mice (TMEM11 Tg), the mouse TMEM11 gene fragment was cloned into a vector (pαMHC-clone26) under the control of the α-myosin heavy chain (MHC) promoter. The constructed transgenic vector was microinjected into the fertilized eggs of C57BL/6JGpt mice. Microinjection was performed according to the standard protocols. Fertilized eggs were transplanted to obtain positive F0 mice which were confirmed by PCR and sequencing. A stable F1 generation mouse model was obtained by mating positive F0 generation mice with C57BL/6JGpt mice. The forward primer(F1) was 5′-GGAAGGAGGCGTCTGGGTC-3′; the reverse primer(R1) was 5′-TCATACTCCACCTGGTACTTGCAG-3′; the forward primer(F2) was 5′-CTAGGCCACAGAATTGAAAGATCT-3′; the reverse primer(R2) is 5′-CTAGGCCACAGAATTGAAAGAT CT-3′.
MeRIP-qPCR
Total RNA was extracted from cardiomyocytes or tissues. RNase MiniElute Kit segmented RNA, we added 50 ul protein A/G magnetic beads to the segmented RNA sample, we added 500 ul 1x IP buffer, we used the pipette to blow and mix, and magnetic beads were suspended again. Place the Eppendorf tube on the magnetic rack until the liquid is clear and discard the supernatant. We added 200 ul 1 x IP buffer to re-suspend protein A/G magnetic beads. Rotate and incubate at room temperature for 30 minutes. Place the Eppendorf tube on the magnetic rack until the liquid is clear and discard the supernatant. Add MeRIP reaction mixture, gently pipette up and down several times, make protein A/G magnetic beads completely re-suspended. Rotate and incubate at 4 °C for 2 hours. Place the Eppendorf tube on the magnetic rack until the liquid is clear and discard the supernatant. Add 100 ul of eluent into the Eppendorf tube, gently pipette up and down several times to make the magnetic beads wholly suspended. Under the condition of 4 °C, continuously shake and incubate for 1 h. Place the Eppendorf tube on a magnetic stand until the liquid is clear. The supernatant containing elution RNA fragments was transferred to a new 1.5 mL Eppendorf tube. After elution, the RNA sample was purified by RNase MiniElute Kit, the RNA sample after MeRIP was obtained. Reverse transcription was performed on the obtained samples, and qPCR was used to detect the expression.
Immunofluorescence
The processed cardiomyocytes or heart tissue sections were fixed with 4% paraformaldehyde for 15 min, washed with PBS three times, and treated with 0.1% Triton X-100 for 15 min, washed with PBS three times, and incubated with 5% BSA solution for 30 min. Specific antibody binding protein signal was used, and the antibodies were as follows: Anti-Ki67 (abcam, Cat: AB16667, 1:200); Anti-Aurora B (abcam, Cat: AB2254, 1:200); Anti-histone H3 (abcam Cat: AB5176, 1:200); Anti-cardiac Troponin T (abcam, Cat: AB8295, 1:200); TMEM11 Rabbit pAb, (Abclonal, A17578, 1:100); Anti-LC3B (abcam,Cat:ab192890,1:1000); Caspase-3 (D3R6Y) Rabbit mAb, (cell signaling technology,Cat:#14220,1:1000). The fluorescence signal was detected by a Leica inverted two-photon laser confocal scanning microscope.
MeRIP-sequencing
RNA was extracted from WT and TMEM11 Tg mice hearts and quantified (using NanoDrop ND-1000). M7G RNA-Seq service was provided by Cloudseq Biotech Inc. (Shanghai, China). Briefly, total RNA was decapped with mRNA decapping enzyme (New England Biolabs), and then subjected to m7G RNA immunoprecipitation with the GenSeqTM m7G RNA IP Kit (GenSeq Inc., China) by following the manufacturer’s instructions. Both the input sample without immunoprecipitation and the m7G IP samples were used for RNA-seq library generation with NEBNext® Ultra II Directional RNA Library Prep Kit (New England Biolabs, Inc., USA). The library quality was evaluated with BioAnalyzer 2100 system (Agilent Technologies, Inc., USA). Library sequencing was performed on an illumina Hiseq instrument with 150 bp paired-end reads. Paired-end reads were harvested from Illumina HiSeq 4000 sequencer, and were quality controlled by Q30. After 3′ adaptor-trimming and low quality reads removing by cutadapt software (v1.9.3). First, clean reads of all libraries were aligned to the reference genome (UCSC MM10) by Hisat2 software (v2.0.4). Methylated sites on RNAs (peaks) were identified by MACS software. Differentially methylated sites were identified by diffReps. These peaks identified by both softwares overlapping with exons of mRNA were figured out and choosed by home-made scripts. GO and Pathway enrichment analysis were performed by the differentially methylated protein coding genes.
Statistical analysis
Data were expressed as the mean ± SD. Statistical analysis was performed using GraphPad Prism 7.0 (GraphPad Software Inc., San Diego, CA). Student’s t-test was used for the comparison of statistical significance between two groups and all tests were determined by unpaired two-sided tests. Comparisons between multiple groups were assessed by one-way ANOVA with Tukey’s multiple comparisons test or two-way ANOVA with Bonferroni’s multiple comparisons test. The experiments were repeated independently at least three times with similar results and p < 0.05 was considered statistically significant.
Results
Inhibition of TMEM11 promotes cardiomyocyte proliferation in vitro
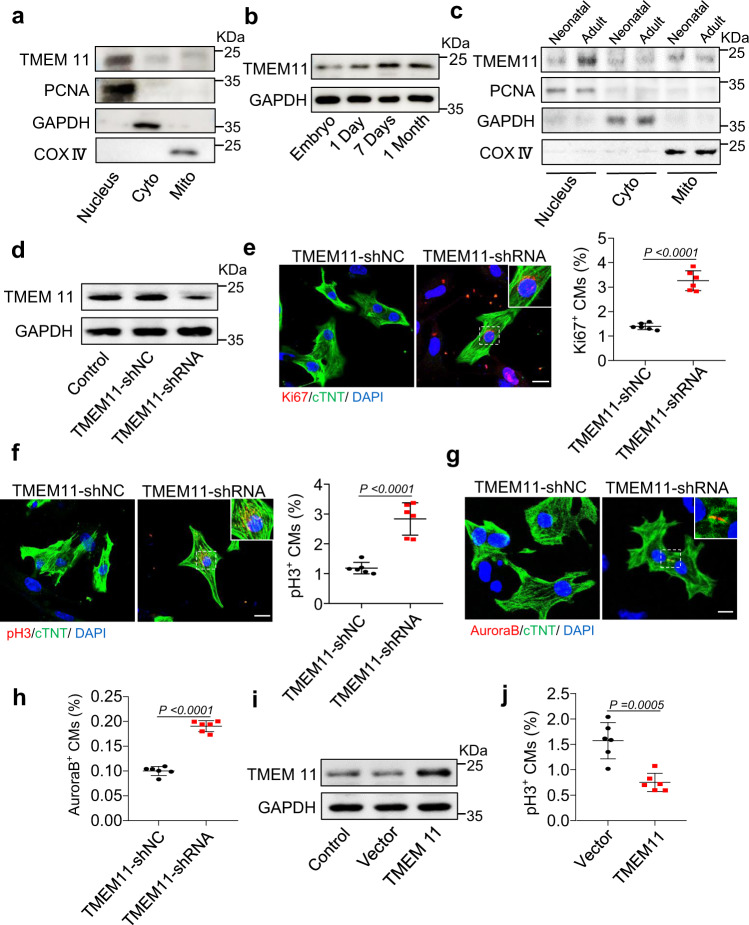
To investigate the potential role of mitochondrial proteins in regulating cardiomyocyte proliferation, we reviewed the mitochondrial proteins previously reported in the literature and analyzed their location and expression in the heart. Interestingly, we observed that the mitochondrial inner membrane protein TMEM11 was distributed in the nucleus, cytoplasm, and mitochondria of cardiomyocytes (Fig. 1a and Supplementary Fig. 1a). The expression pattern of the TMEM11 protein in the postnatal hearts of mice revealed that TMEM11 protein levels were relatively low at birth and increased during adulthood (Fig. 1b), indicating that TMEM11 expression might be associated with the loss of cardiac regenerative capacity in adult mice. Furthermore, increased expression of TMEM11 was mainly observed in the nucleus, whereas no significant increase was detected in the cytoplasm or mitochondria in the adult mouse heart (Fig. 1c). Also, TMEM11 was abundantly expressed in the heart (Supplementary Fig. 1b) and was significantly higher in cardiomyocytes than in fibroblasts (Supplementary Fig. 1c). Thus, we speculated that TMEM11 has a potential regulatory role in cardiomyocyte proliferation and cardiac regeneration.
Fig. 1. TMEM11 participates in the regulation of proliferation in cardiomyocytes.
a Representative western blots showing the expression of TMEM11 in the nucleus, cytoplasm and mitochondria of cardiomyocytes (n = 3 independent experiments). b Representative western blots showing the expression of TMEM11 in hearts at different ages of mice (n = 4 mice). c Representative western blots showing the expression of TMEM11 in the nucleus, cytoplasm and mitochondria of cardiomyocytes (n = 3 mice). d–h Neonatal mice cardiomyocytes were infected with adenovirus harboring TMEM11-shRNA or its control (TMEM11-shNC) for 48 h. d The expression levels of TMEM11 mRNA assessed by immunoblot (n = 3 independent experiments). e Representative confocal images of Ki67-positive cardiomyocytes (Bar = 20 μm) and quantification of Ki67-positive (right) cells (n = 6 independent experiments). f Representative confocal images of pH3-positive cardiomyocytes and quantification of pH3-positive cells in TMEM11-shNC and TMEM11-shRNA infected cardiomyocytes (Bar = 20 μm) (n = 6 independent experiments). Representative confocal images of Aurora-B-positive cardiomyocytes (g) and quantification (h) of Aurora-B-positive cells in TMEM11-shNC and TMEM11-shRNA infected cardiomyocytes (Bar = 20 μm) (n = 6 independent experiments). i, j Neonatal mice cardiomyocytes were infected with adenovirus harboring negative control (Vector) or mouse TMEM11 gene (TMEM11) for 48 h. i Representative western blots showing the expression of TMEM11 (n = 4 independent experiment). j PH3-positive cardiomyocytes were measured in Vector and TMEM11 treated cardiomyocytes (n = 6 independent experiment). Data are presented as the mean ± s.d. Two-sided Student’s t test (e, f, h and j) was performed.
To verify the role of TMEM11 in regulating cardiomyocyte proliferation, we constructed a recombinant adenoviral vector harboring a Tmem11-targeting shRNA (TMEM11-shRNA) to silence TMEM11 expression in neonatal mouse cardiomyocytes (Fig. 1d). Silencing of TMEM11 increased the expression of proliferation markers in cardiomyocytes, as indicated by an increase in Ki67-positive (Fig. 1e), mitosis marker phospho-histone H3 (pH3)-positive (Fig. 1f), and cytokinesis marker Aurora B-positive cells (Fig. 1g, h). Next, we overexpressed TMEM11 in cardiomyocytes (Fig. 1i), and exogenous TMEM11 expression significantly reduced the expression of proliferation markers (Fig. 1j, Supplementary Fig. 1d–f). In addition, we also assessed whether TMEM11 affects mitochondrial functions. Our results demonstrated that knockdown of TMEM11 in cardiomyocytes did not change the mitochondrial membrane potential (Supplementary Fig. 2a, b), which is an important indicator of mitochondrial activity, and TMEM11 had no effect on apoptosis or autophagy (Supplementary Fig. 2c, d). Together, these data suggest that ablation of TMEM11 promotes cardiomyocyte proliferation, whereas its overexpression inhibits cell cycle progression.
TMEM11 deficiency reactivates cardiomyocytes proliferation in the adult hearts
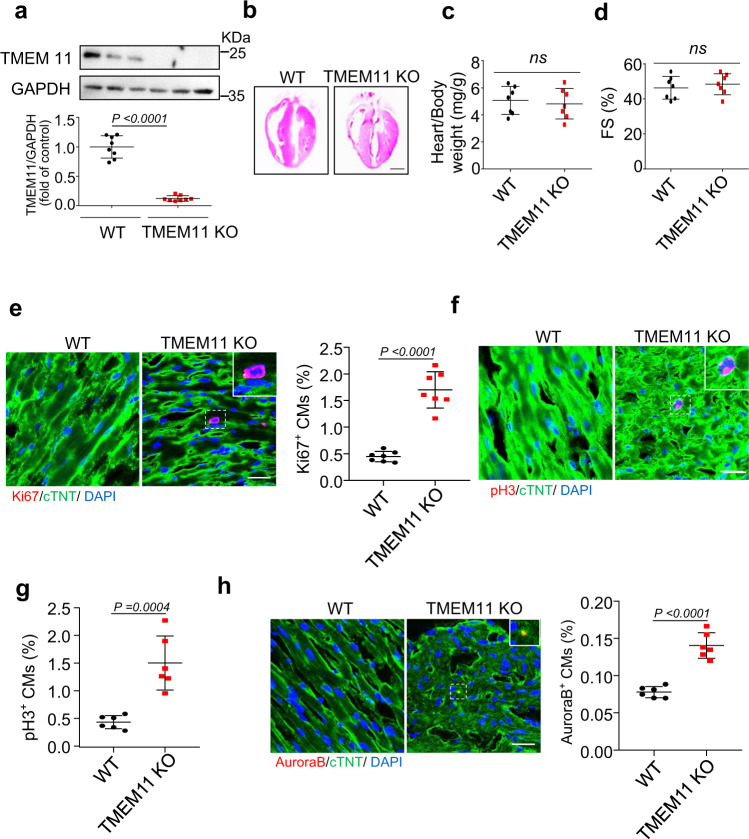
To investigate the effects of TMEM11 knockdown on cardiomyocyte proliferation in vivo, we generated TMEM11 knockout (TMEM11 KO) mice, and the deletion of TMEM11 in mice hearts was confirmed by western blot analysis (Fig. 2a). Cardiac tissue morphology, heart-to-body weight ratio, and heart function were not significantly different between WT and TMEM11 KO mice under physiological conditions (Fig. 2b–d, and Supplementary Fig. 3a, b). We analyzed the cross-sectional area and number of cardiomyocytes in the TMEM11 KO and WT mice. Cardiomyocytes in the TMEM11 KO hearts were smaller in size (Supplementary Fig. 3c, d), and the number of cardiomyocytes was increased in the TMEM11 KO hearts (Supplementary Fig. 3e). In addition, no hypertrophic effects were observed in TMEM11 KO mouse hearts (Supplementary Fig. 3f, g). Further, knockout of TMEM11 increased the number of Ki67-positive cells compared to those in WT mice (Fig. 2e). Likewise, the numbers of pH3-positive (Fig. 2f, g) and Aurora B-positive cells (Fig. 2h) were remarkably higher in Tmem11-deleted adult hearts than in WT mice. Together, these results indicated that the loss of TMEM11 enhanced cardiomyocyte proliferation in vivo.
Fig. 2. TMEM11 mediates cardiomyocytes proliferation in adult hearts.
a Western blot image (top) and statistical data (bottom) showing the myocardial level of TMEM11 in wild type (WT) and TMEM11 KO adult mice hearts (n = 8 mice). b–h TMEM11 silencing increases cardiomyocytes proliferation in adult mice hearts. Representative images of gross morphology and haematoxylin-eosin stained transverse sections of hearts from WT and TMEM11 KO mice (Bar = 2 mm) (b). Heart to body weight ratio (c) and echocardiography analysis of fractional shortening (FS) (d) in WT and TMEM11 KO mice (n = 7 mice). e Representative confocal images of Ki67-positive cardiomyocytes (red, Bar = 25 μm) and quantification of Ki67-positive (right) cells (n = 7 mice per group). cTNT (green) mark cardiomyocytes and DAPI (blue) labels nuclei. f, g Representative confocal images and qualification of pH3-positive cardiomyocytes (red, Bar = 25 μm) in WT and TMEM11 KO hearts. h Aurora-B-positive (red) cardiomyocytes was calculated in WT and TMEM11 KO hearts. cTNT marks cardiomyocytes and DAPI labels nuclei (Bar = 25 μm). (n = 6 mice per group). Data are presented as the mean ± s.d. Two-sided Student’s t test (a, c–e, g and h) was performed.
Knockout of TMEM11 promotes cardiomyocyte proliferation and cardiac regeneration after ischemic injury
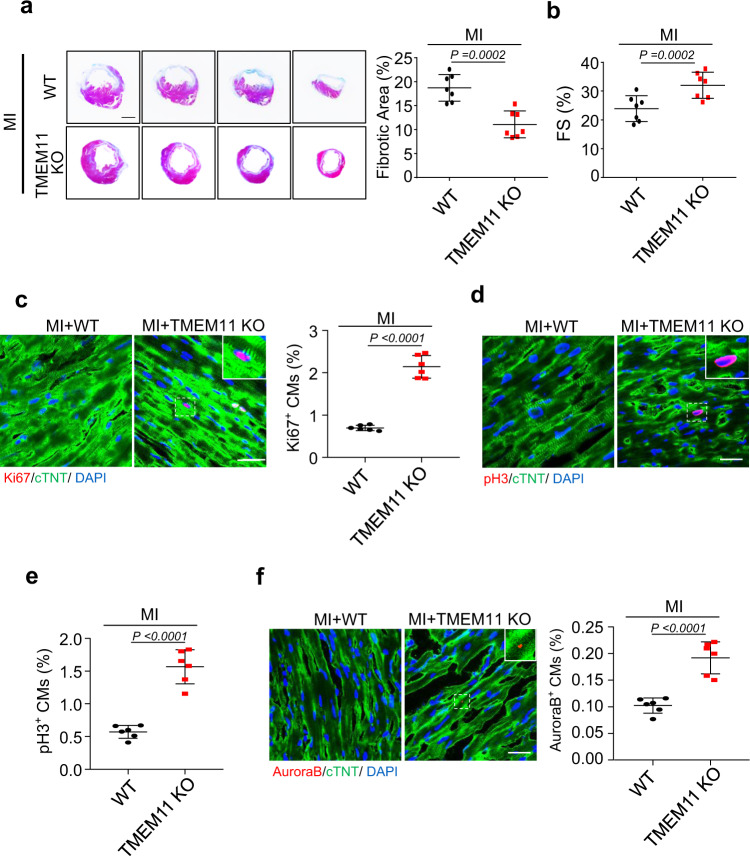
To further validate the impact of TMEM11 silencing on cardiac regeneration in the adult heart, TMEM11 KO and WT mice were subjected to MI induced by coronary artery ligation (Supplementary Fig. 4a). TMEM11 KO significantly reduced scar size compared to that in WT hearts (Fig. 3a), and the left ventricle function was improved in TMEM11 KO mice after MI (Fig. 3b, Supplementary Fig. 4b, c), indicating that TMEM11 silencing mediates the reparative potential of cardiomyocytes after cardiac injury. Meanwhile, the numbers of Ki67-positive, pH3-positive, and Aurora B-positive cells were higher in TMEM11 KO hearts after MI injury (Fig. 3c–f), which suggest that depletion of TMEM11 improved myocardial repair and cardiac function after MI injury by inducing cardiomyocyte proliferation. Furthermore, increased TMEM11 expression (Supplementary Fig. 4d) was observed upon MI injury, and there was no significant change in TMEM11 levels following TAC treatment (Supplementary Fig. 4e). Collectively, these data confirm that suppression of TMEM11 promoted cardiac regeneration after ischemic injury in mice.
Fig. 3. TMEM11 deletion promotes cardiomyocytes proliferation and improves cardiac function after MI injury.
a–f 10-weeks old wild type (WT) and TMEM11 KO mice were subjected to MI and heart samples were collected at 8 weeks post-MI. a Representative images of Masson’s trichrome-stained cross-sections of hearts (Bar = 2 mm) (left panel). The analysis of the percentage of the fibrotic area after MI injury (right panel, n = 7 mice per group). b Echocardiography analysis of left ventricular function (fractional shortening, FS%) after MI (n = 7 mice). c Representative confocal images of Ki67-positive cardiomyocytes (red, Bar = 25 μm) and quantification of Ki67-positive (right) cells (n = 6 mice per group). cTNT (green) marks cardiomyocytes and DAPI (blue) labels nuclei. d, e Representative confocal images and qualification of pH3-positive cardiomyocytes (red, Bar = 25 μm) in WT and TMEM11 KO hearts (n = 6 mice per group). f Aurora-B-positive (red) cardiomyocytes was calculated in WT and TMEM11 KO hearts. cTNT marks cardiomyocytes and DAPI labels nuclei (Bar = 25 μm). (n = 6 mice per group). Data are presented as the mean ± s.d. Two-sided Student’s t test (a–c, e and f) was performed.
TMEM11 overexpression inhibits cardiomyocyte proliferation and cardiac regeneration in neonatal hearts
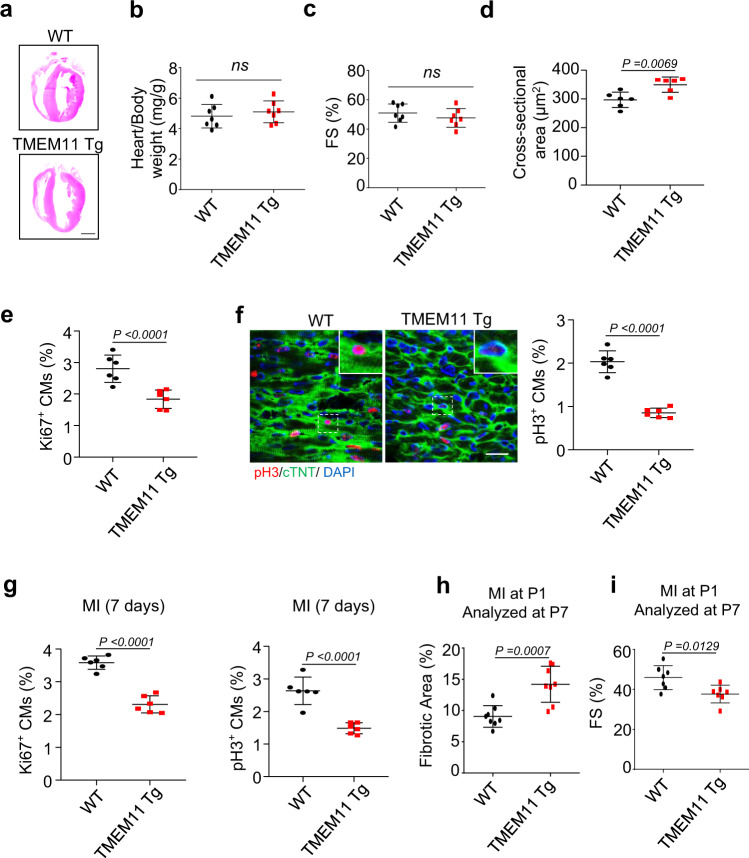
We next explored whether TMEM11 overexpression suppressed cardiomyocyte proliferation and cardiac regeneration in neonatal hearts. We generated cardiac-specific TMEM11 transgenic (TMEM11 Tg) mice (Supplementary Fig. 5a). Under physiological conditions, there were no differences in cardiac morphology, heart-to-body weight ratio, or hypertrophic marker expression and cardiac function between the Tg and WT mice in the adult mice hearts (Fig. 4a–c, Supplementary Fig. 5b–d). However, cardiomyocyte size increased in the hearts of Tg mice compared to that in WT mice (Fig. 4d). Besides, the number of Ki67-positive (Fig. 4e) or pH3-positive cells (Fig. 4f) were significantly reduced in the hearts of 7-day old Tg mice compared with that in WT mice, indicating that TMEM11 overexpression inhibits cardiomyocyte proliferation in mice. To further investigate the effect of TMEM11 overexpression on the regenerative capacity of neonatal hearts, 1-day old TMEM11 Tg mice were subjected to MI, and analyses were carried out on day 7 (P7). The number of Ki67-positive and pH3-positive cells (Fig. 4g, Supplementary Fig. 5e, f) decreased while the fibrotic area increased in TMEM11 Tg mice compared with that in WT mice (Fig. 4h). In addition, the cardiac function in TMEM11 Tg mice was impaired compared to that of WT controls following MI injury (Fig. 4i, Supplementary Fig. 5g). Together, these findings indicate that TMEM11 overexpression hindered cardiac regeneration by inhibiting cardiomyocyte proliferation in postnatal hearts.
Fig. 4. TMEM11 overexpression inhibits cardiomyocytes proliferation and cardiac regeneration in neonatal hearts.
a Representative images of haematoxylin-eosin stained transverse sections of WT and TMEM11 transgenic (TMEM11 Tg) adult mice hearts (Bar = 2 mm). b Heart to body weight ratio in WT and TMEM11 Tg adult mice hearts (n = 7 mice per group). c The left ventricular fractional shortening (FS%) in WT and TMEM11 Tg adult mice hearts (n = 7 mice per group). d Cardiomyocytes surface area was measured in WT and TMEM11 Tg adult mice hearts (n = 6 mice per group). e Quantification of the number of Ki67-positive cardiomyocytes in WT and TMEM11 Tg heart sections at 7 days after birth (n = 6 mice per group). f Representative confocal images and qualification of pH3-positive cardiomyocytes (red, Bar = 25 μm) in WT and TMEM11 Tg heart sections at 7 days after birth (n = 6 mice per group). (g–i) WT and TMEM11 Tg neonatal mice (P1) were subjected to MI. g Quantification of pH3-positive and Ki67-positive cardiomyocytes in heart sections at 7 days after MI (n = 6 mice per group). h Analysis of the percentage of the fibrotic area (n = 8 mice per group). i Echocardiography analysis of fractional shortening (FS, n = 7 mice per group) at P7 post-MI. Data are presented as the mean ± s.d. Two-sided Student’s t test (b–i) was performed.
TMEM11 interacts with METTL1 and regulates its RNA m7G-methylation activity
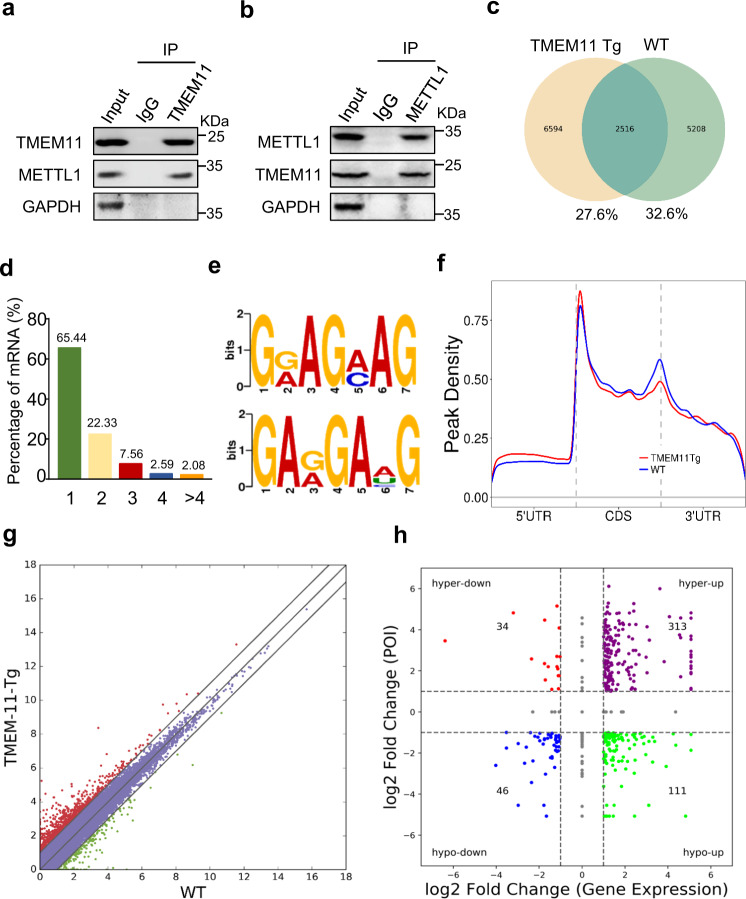
It was our first priority to investigate the proteins that interact with TMEM11 to better understand how TMEM11 regulates cardiomyocyte proliferation. Our experiments involved making protein immunoprecipitations from isolated cardiomyocytes and performing liquid chromatography tandem mass spectrometry (LC-MS/MS) on the samples (Supplementary Table 1). Among the proteins enriched with the anti-TMEM11 antibody, we identified METTL1, an RNA methyltransferase that specifically methylates N7-guanosine (Supplementary Fig. 6a). We further studied METTL1, considering that the function of m7G mRNA methylation in cardiomyocyte proliferation and cardiac regeneration remains unknown. We performed immunoprecipitation followed by western blotting and confirmed the direct interaction of TMEM11 and METTL1 in vivo (Fig. 5a, b). To determine whether TMEM11 regulates METTL1-dependent m7G-methylation in cardiomyocytes, we performed m7G-methylated RNA immunoprecipitation sequencing (MeRIP-seq) in WT and TMEM11 Tg mouse hearts (Fig. 5c and Supplementary Table 2).
Fig. 5. Transcriptome-wide m7G methylome analyses (using m7G-Seq) in TMEM11 Tg mice heart.
Immunoprecipitation assessment using anti-TMEM11 antibody (a) and anti- METTL1 antibody (b) showing the interaction of TMEM11 and METTL1. Representative immunoblots from four independent experiments with similar results. c–f m7G methylated RNA immunoprecipitation and sequencing (MeRIP-seq) was performed in TMEM11 Tg and WT mice hearts. c Numbers of m7G peaks detected in TMEM11 Tg (left circle) and WT (right circle) mice hearts. d Percentage of mRNAs with different numbers of m7G peaks. e Sequence motifs enriched within m7G peaks identified by MeRIP-seq in TMEM11 Tg mice hearts. f Metagene profile showing the distribution of m7G peaks across the length of transcripts composed of three rescaled non-overlapping segments 5′UTR, CDS and 3′UTR in TMEM11 Tg and WT mice hearts. g Scatter plot of differential expression of mRNAs assessed from RNA-seq data. Red dots denote up-regulated genes and green dots denote down-regulated genes. h Correlation between the level of gene expression and changes in m7G level in TMEM11 Tg mice hearts compared to WT.
Among all the detected m7G mRNA transcripts, more than half (65.44%) contained one m7G peak, and 4.67% contained four or more m7G peaks (Fig. 5d). Sequence motif analysis revealed that m7G peaks were highly enriched in two conserved consensus motif sequences of METTL1 (Fig. 5e). In both WT and TMEM11 Tg mice, m7G peaks were predominantly distributed in coding sequences (CDSs), particularly near the start and stop codons. However, m7G peaks had relatively lower density in the area between CDS and 3’ UTR in TMEM11 Tg mice compared to those in WT mice (Fig. 5f and Supplementary Fig. 6b). Gene Ontology analysis showed that genes with upregulated m7G modifications were mainly involved in cellular metabolism, protein modification, and regulation of signal transduction (Supplementary Fig. 6c). Genes with downregulated m7G modifications were mainly involved in the negative regulation of cellular, biosynthetic, and developmental processes (Supplementary Fig. 6d). Next, we performed RNA-seq analysis in TMEM11 Tg and WT mouse hearts to explore the relationship between m7G modifications and gene expression (Fig. 5g) and plotted m7G peak data against RNA-seq gene expression data to correlate the gene expression level with the m7G modification level (Fig. 5h and Supplementary Table 3). We termed the upregulated peaks as hypermethylated m7G peaks, including hyper-down and hyper-up genes. We termed the downregulated peaks as hypomethylated m7G peaks, including hypo-down and hypo-up genes.
TMEM11 promotes METTL1-dependent m7G modification of Atf5 mRNA and enhances its expression
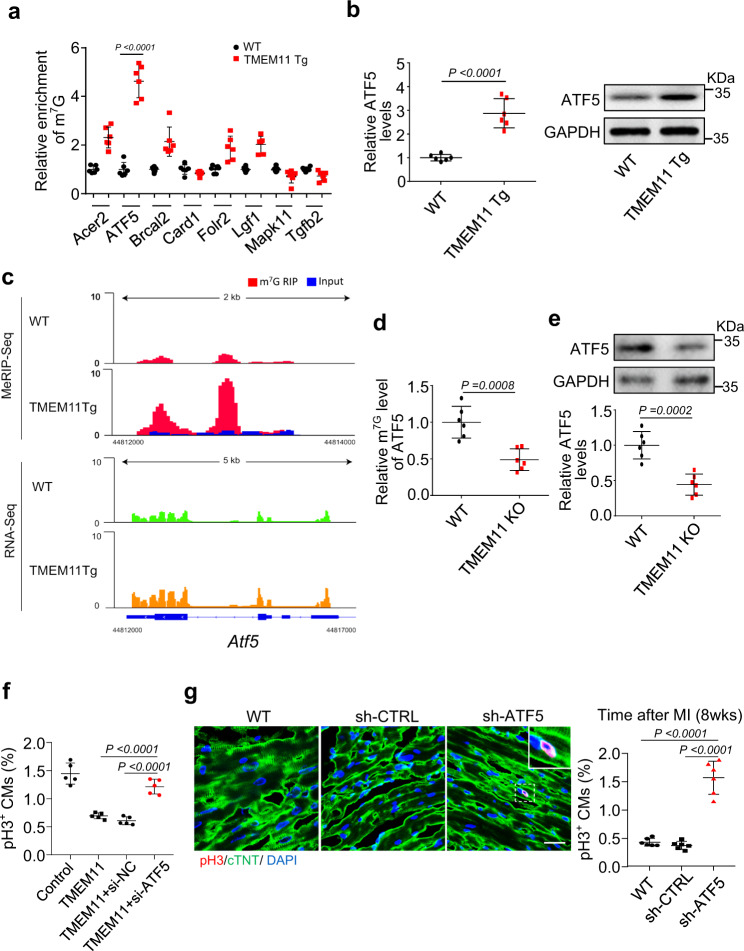
We further investigated the potential downstream targets of METTL1 during cardiomyocyte proliferation in TMEM11 Tg hearts based on the results shown in Fig. 5h. We performed MeRIP-qPCR assays for the differentially methylated genes reported to be associated with proliferation. Our results revealed that the m7G modification of Atf5 mRNA increased most significantly in TMEM11 Tg mice compared to WT mice (Fig. 6a). In addition, the expression of ATF5 significantly increased in TMEM11 Tg hearts (Fig. 6b). Integrative genomics viewer analysis of Atf5 mRNA revealed a remarkable increase in m7G peaks, along with increased expression of Atf5 mRNA, in TMEM11 Tg hearts (Fig. 6c). In contrast, m7G modification in Atf5 mRNA decreased (Fig. 6d), along with decreased expression of Atf5 mRNA and protein, in TMEM11 KO hearts (Fig. 6e). These results prompted us to select ATF5 as a potential target of the TMEM11-METTL1 axis in the regulation of cardiomyocyte proliferation.
Fig. 6. TMEM11 mediates m7G methylation of Atf5 and its expression.
a MeRIP-qPCR validation of m7G modification levels of genes which selected from the results of MeRIP-seq and mRNA-seq data in TMEM11 Tg and WT mice hearts (n = 6 mice per group). b Expression levels of Atf5 mRNA and protein in WT and TMEM11 Tg mice hearts (n = 6 mice per group). c Integrative Genomics Viewer (IGV) tracks showing MeRIP-seq (upper panel) and mRNA-seq (bottom panel) read distribution in Atf5 mRNA from TMEM11 Tg and WT mice hearts. d MeRIP-qPCR analysis in TMEM11 KO and WT mice hearts shows the m7G modification level in Atf5 mRNA (n = 6 mice per group). e Expression levels of ATF5 protein (upper panel) and mRNA (lower panel) in TMEM11 KO and WT mice hearts (n = 6 mice per group). f Neonatal cardiomyocytes were infected with adenovirus harboring TMEM11, and transfected with si-NC or si-ATF5 for 48 h. Qualification of pH3-positive cardiomyocytes was calculated (n = 5 independent experiments). g Knockdown of ATF5 promotes cardiomyocyte proliferation following MI injury. AAV9 loaded with ATF5 shRNA (sh-ATF5) or its control (sh-CTRL) was administered to adult WT mice and subjected to MI, and heart samples were collected at 8 weeks post-MI. Representative confocal images and qualification of pH3-positive cardiomyocytes in heart sections at 8 weeks post-MI (Bar = 25 μm) (n = 6 mice per group). Data are presented as the mean ± s.d. One-way ANOVA (f and g) or two-sided Student’s t test (a, b, d and e) was performed.
We examined whether ATF5 acts as a downstream target of TMEM11 by regulating the proliferation of neonatal cardiomyocytes. TMEM11 overexpression inhibited cardiomyocyte proliferation, which was rescued by transfection with siRNA-ATF5 (Supplementary Figs. 7a, b and 6f). In addition, the knockdown of ATF5 using AAV9 system (sh-ATF5) in adult mice hearts (Supplementary Fig. 7c) promoted cardiomyocytes proliferation, improved cardiac function and reduced infarct size in MI-injured hearts (Fig. 6g and Supplementary Fig. 7d, e), indicating that ATF5 is involved in the regulation of cardiomyocyte proliferation and cardiac regeneration.
We investigated the mechanism by which TMEM11 regulates the expression of ATF5. We found that ATF5 expression was increased in MI-injured hearts (Supplementary Fig. 7f), but did not change significantly after TAC treatment (Supplementary Fig. 7g). The binding of METTL1 to Atf5 mRNA was remarkably decreased in TMEM11 KO hearts compared to WT mice hearts (Supplementary Fig. 7h). Moreover, METTL1 overexpression increased the m7G modification of Atf5 mRNA and elevated ATF5 expression, and these effects were reinforced by concomitant TMEM11 overexpression (Supplementary Fig. 7i). Furthermore, METTL1 knockdown attenuated the m7G modification of Atf5 mRNA and its expression upon TMEM11 overexpression in vivo and in vitro (Supplementary Fig. 8a–c). Together, these results revealed that TMEM11 mediates m7G modification in Atf5 mRNA through METTL1, and that this methylation enhances mRNA stability and translation capacity. As “YAP” is an important regulator of cardiac regeneration, we determine if YAP played a role in the TMEM11-mediated proliferation of cardiomyocytes. In our study, we found that TMEM11 did not affect the expression levels of YAP in cardiomyocytes. (Supplementary Fig. 8d, e). In addition, the interaction between TMEM11 and YAP was not detected by co-immunoprecipitation in cardiomyocytes (Supplementary Fig. 8f). In the results, it was found that YAP is not involved in the pathway of TMEM11-mediated cardiomyocyte proliferation.
TMEM11 regulates INCA1 expression during cardiomyocyte proliferation
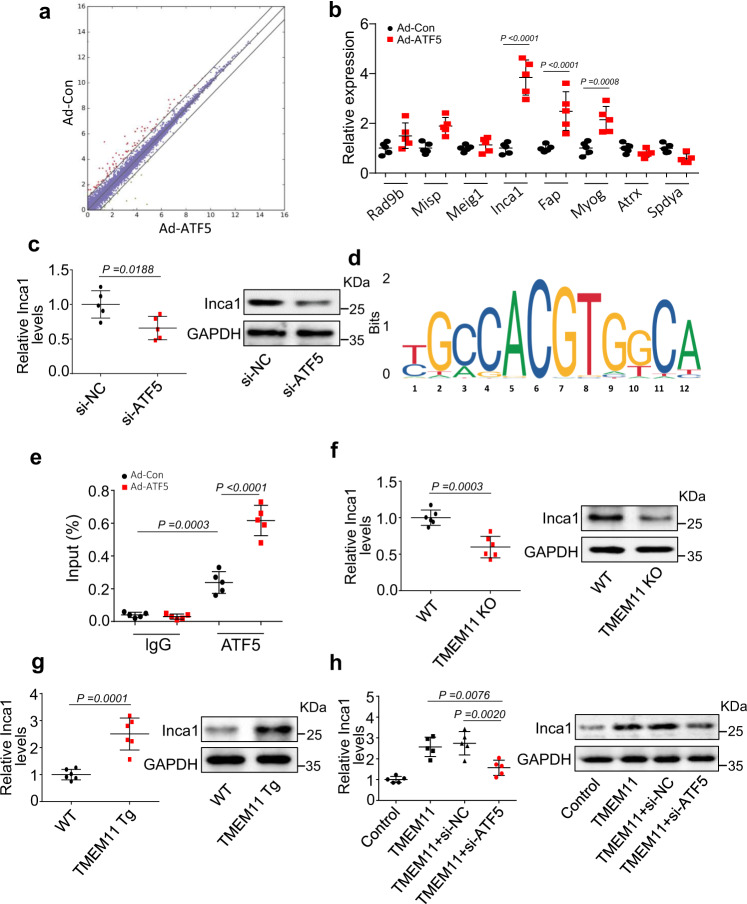
To explore the downstream targets of ATF5, we performed an RNA-seq analysis in ATF5-overexpressing cardiomyocytes (Fig. 7a and Supplementary Table 4). We screened for differentially expressed genes associated with proliferation regulation in the RNA-seq dataset. We validated their expression using qPCR. Among them, only the expression levels of Inca1, Myog, and Fap were significantly increased in ATF5-overexpressing cardiomyocytes compared to those in the control cells (Fig. 7b). Furthermore, changes in the expression levels of these three genes in ATF5-overexpressing cardiomyocytes prompted us to investigate whether TMEM11 also regulates their expression. Only Inca1 expression was decreased in the hearts of TMEM11 KO mice compared to that in WT hearts (Supplementary Fig. 9a), indicating that INCA1 might be a downstream target of the TMEM11-ATF5 pathway. In addition, previous studies have reported that INCA1 inhibits the interaction of CDK2 with cyclin A1, thereby causing a S to G2/M cell cycle arrest and inhibiting cell proliferation [42–44]. In addition, our results showed that knockdown of INCA1 promotes cardiomyocyte proliferation (Supplementary Fig. 9b, c). Therefore, INCA1 was selected for further investigation.
Fig. 7. ATF5 regulates INCA1 expression during cardiomyocyte proliferation.
a mRNA-seq analysis was performed in cardiomyocytes infected with or without adenovirus harboring ATF5 (Ad-ATF5 or Ad-Con). b qPCR validation of the expression levels of genes which were selected from the results of mRNA-seq data in Ad-ATF5 or Ad-Con treated cardiomyocytes (n = 5 independent experiments). c Cardiomyocytes were transfected with ATF5-siRNA (si-ATF5) and its control (si-NC). Inca1 mRNA and protein levels were evaluated by qRT-PCR and western blot (n = 5 independent experiments). d Motif analysis of the ATF5 bound regions. e Isolated neonatal cardiomyocytes were infected with adenovirus harboring ATF5 (Ad-ATF5) or its control (Ad-Con) for 24 h. CHIP-qPCR assay was performed using antibodies against ATF5 or IgG (n = 5 independent experiments). f The mRNA (left panel) and protein (right panel) levels of INCA1 in TMEM11 KO and WT mice hearts (n = 6 mice per group). g The mRNA (left panel) and protein (right panel) levels of INCA1 in TMEM11 Tg and WT mice hearts (n = 6 mice per group). h Neonatal cardiomyocytes were infected with adenovirus harboring TMEM11, and transfected with si-NC or si-ATF5 for 48 h. The mRNA (left panel) and protein (right panel) levels of Inca1 were analyzed (n = 5 independent experiments). Data are presented as the mean ± s.d. One-way ANOVA (h), two-sided Student’s t test (b, c, f and g) or two-way ANOVA (e) was performed.
Next, we demonstrated that ATF5 knockdown reduced Inca1 mRNA and protein expression (Fig. 7c), indicating that the ATF5 transcription factor promoted Inca1 expression. We also observed an ATF5-binding consensus motif in the promoter region of the Inca1 gene (Fig. 7d) and verified ATF5 binding to this sequence using ChIP-qPCR (Fig. 7e). In addition, a marked enrichment of Inca1 promoter region was observed following ATF5 overexpression (Fig. 7e). Consistent with these in vitro findings, Inca1 mRNA and protein expression decreased in TMEM11 KO mouse hearts compared to that in WT hearts (Fig. 7f), whereas Inca1 expression levels dramatically increased in the hearts of TMEM11 Tg mice (Fig. 7g). In cardiomyocytes, TMEM11 increased the levels of Inca1 mRNA and protein, and these effects were attenuated by ATF5 knockdown (Fig. 7h and Supplementary Fig. 9d). Taken together, these results suggested that INCA1 acts as a downstream target of TMEM11/ATF5 pathway, which inhibits postnatal cardiomyocyte proliferation and regeneration.
Discussion
There is currently no information about the molecular mechanisms responsible for the withdrawal of cell cycle and the non-proliferative growth of cardiomyocytes in mammals after birth. In this study, we identified a mitochondrial transmembrane protein, TMEM11, involved in the proliferation of cardiomyocytes, in both postnatal and adult mice. TMEM11 knockout promoted myocardial cell proliferation, reduced MI area, and improved cardiac function in adult mice. Overexpression of TMEM11 inhibited neonatal mouse cardiomyocyte proliferation and cardiac regeneration in mouse hearts. TMEM11 interacted with METTL1 and promoted the m7G methylation of Atf5 mRNA, thereby enhancing its expression and that of its downstream target Inca1 to ultimately suppress cardiomyocyte proliferation. Our study demonstrated that METTL1-dependent m7G RNA hypermethylation is required for cardiomyocyte proliferation; however, substantial expression of TMEM11 after birth intervenes in this event and suppresses cardiomyocyte proliferation.
Several studies have reported that cardiomyocyte proliferation is almost absent in the adult heart, and the death of cardiomyocytes and damage to cardiac function caused by MI are irreparable [45, 46]. Hippo and Wnt signaling pathways are known to regulate cardiomyocyte proliferation. Similarly, inhibition of the transcriptional coactivator Yap restricts cardiac growth and regeneration [12]. In addition, many studies have found that MI causes molecular changes in cardiac cells, resulting in mitochondrial dysfunction, which is the main cause of functional damage in cardiomyocytes [47–49]. A few mitochondrial transcription factors ameliorate mitochondrial defects and HF after MI [50]. Although the functional role of mitochondria in regulating MI is well-documented, the exact function of mitochondrial proteins in myocardial cell proliferation remains largely unclear. Our study focused on the role of the mitochondrial transmembrane protein, TMEM11, in myocardial cell proliferation and MI treatment. Silencing TMEM11 increased cardiomyocyte proliferation, improved cardiac function, and reduced infarct size.
Post birth, the expression of TMEM11 in the heart gradually increased and became abundant in adult mice. TMEM11 inhibited cardiomyocyte proliferation in adult mice. TMEM11 knockdown enhanced myocardial nuclear mitosis and cytokinesis in neonatal mouse cardiomyocytes. In addition, TMEM11 KO mice showed reduced infarct size after MI, restored cardiac function, and increased proliferation of adult mouse cardiomyocytes. Overexpression of TMEM11 in neonatal mouse cardiomyocytes inhibited cardiomyocyte proliferation both in vitro and in vivo. Thus, our data suggests that TMEM11 affects the proliferation of adult cardiomyocytes and that the inhibition of TMEM11 can promote adult cardiomyocyte proliferation and reduce infarct size. There is, however, no current understanding of how TMEM11 suppresses in neonatal hearts or how it is upregulated in young and adult hearts. Further research is needed to determine the mechanisms behind this phenomenon.
TMEM11 is primarily located in the mitochondria of cardiomyocytes. Previous studies have shown that TMEM11 is a molecular switch in the mitochondria that adapts to different physiological needs, affects mitochondrial morphology, and regulates mitochondrial phagocytosis and cell death. Other studies have shown that TMEM11 influences mitochondrial diameter and tubular shape [51, 52]. In this study, we investigated whether TMEM11 affects mitochondrial function. Our results showed that TMEM11 knockdown in cardiomyocytes did not change the mitochondrial membrane potential, which is an important indicator of mitochondrial activity. Furthermore, TMEM11 had no effects on apoptosis and autophagy. However, we cannot completely exclude the possibility that TMEM11 regulates cardiomyocyte proliferation by influencing mitochondrial function and energy metabolism. Further studies are required to dissect the effects of TMEM11 on mitochondrial energy metabolism and cardiomyocyte proliferation.
Hippo signal pathway is an evolutionarily conserved pathway, which is activated by a series of phosphorylation, such as MST1/2, SAV1, LATS1 and LATS2, which jointly phosphorylate YAP, thus regulating the proliferation of cardiac myocytes [53, 54]. We examined whether YAP is required for cardiomyocyte proliferation mediated by TMEM11 in this study, and results showed that TMEM11 had no effect on YAP expression levels. In cardiomyocytes, co-immunoprecipitation failed to detect an interaction between TMEM11 and YAP, indicating that YAP is not involved in TMEM11-mediated pathway. The results of our study demonstrate ATF5 as a target of TMEM11 during regulating the cardiomyocyte proliferation. It was reported that other molecules or pathways including Notch and Wnt/β-catenin also participate in the regulation of development, differentiation and regeneration [55–57]. According to MeRIP-seq and RNA-seq analyses, more than 500 genes were differentially expressed in TMEM11 Tg mouse hearts compared to those in WT hearts; some of these are involved in metabolism, cell division, and growth. These results suggest that other genes may also act as downstream targets of TMEM11 to regulate cardiomyocyte proliferation and warrant further studies.
As a transcription factor, ATF5 targets the mitochondria through its amino-terminal mitochondrial-targeting sequence in the absence of mitochondrial stress. However, during mitochondrial dysfunction, ATF5 cannot be imported into the mitochondria and is transported to the nucleus instead via its nuclear localization signal, inducing the transcription of genes that affect mitochondrial apoptosis inhibition, anti-apoptotic mechanisms, cell growth, and migration [58–63]. In addition, ATF5 has been shown to be a critical regulator of cell proliferation and survival [64–66]. Consistent with these findings, we found that ATF5 regulates cardiomyocyte proliferation. Our results showed that TMEM11 increased ATF5 expression, which promoted the binding of ATF5 to the promoter region of Inca1 and induced the expression of INCA1. Enhanced INCA1 expression inhibited the transition from S to G2/M in the cell cycle, thus preventing cardiomyocytes from entering the cell cycle [65, 67].
The m7G modification exists within tRNAs, rRNAs, and mRNA caps and is implicated in nearly all aspects of the mRNA life cycle. A recent study verified that m7G modification mediated by METTL1 within mammalian mRNA affects mRNA translation [40], miRNA structure, and cell migration [41]. As a matter of fact, its role in cardiac development is unknown, especially when it comes to cardiac proliferation. Our study revealed that transcriptome-wide m7G sequencing analysis in TMEM11-overexpressing hearts revealed that METTL1-induced m7G hypermethylation of Atf5 mRNA was related with enhanced translation of Atf5 mRNA, which was evident from the remarkable increase in Atf5 mRNA and protein levels. Consistent with a previous study [40], our data showed that m7G modification promoted mRNA translation. Here, we investigated the previously unrecognized function of the m7G modification in the regulation of cardiomyocyte proliferation and uncovered the underlying mechanisms of m7G modification-mediated cardiomyocyte proliferation and cardiac regeneration. This study suggests that a novel m7G modification-based therapeutic strategy may be effective for cardiac repair and regeneration.
Supplementary information
Author contributions
KW, PL and YW designed and supervised the experiments. XC, XL, SX, SH, TW, RL, CL, JX and LZ performed experiments. XC, XL, SX, SH and TW analyzed the data. KW, XC, PL and YW wrote the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (82070313, 81770275, 92249303); Guiding Fund of Government’s Science and Technology (YDZX2021004); National special support plan for high-level talents; Chinese Academy of Medical Sciences-Innovation Fund for Medical Sciences (2020-I2M-C&T-B-053); National High Level Hospital Clinical Research Funding (121-BJ-2018-202, BJ-2022-164).
Data availability
All data in this study are provided in the paper and Supplementary Materials.
Competing interests
The authors declare no competing interests.
Ethics approval
Ethical approval was obtained from Ethics Committee of Qingdao University.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xin-Zhe Chen, Xin-Min Li, Shi-Jun Xu, Shen Hu.
Contributor Information
Yun-Hong Wang, Email: wangyunhong123@139.com.
Pei-Feng Li, Email: peifengliqd@163.com.
Kun Wang, Email: Wangk696@qdu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01179-0.
References
- 1.Braunwald E. The war against heart failure: the Lancet lecture. Lancet. 2015;385:812–24. doi: 10.1016/S0140-6736(14)61889-4. [DOI] [PubMed] [Google Scholar]
- 2.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, et al. Executive summary: heart disease and stroke statistics-2014 update a report from the american heart association. Circulation. 2014;129:399–410. doi: 10.1161/01.cir.0000442015.53336.12. [DOI] [PubMed] [Google Scholar]
- 3.Roger VL. Epidemiology of heart failure. Circ Res. 2013;113:646–59. doi: 10.1161/CIRCRESAHA.113.300268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahill TJ, Ashrafian H, Watkins H. Genetic cardiomyopathies causing heart failure. Circ Res. 2013;113:660–75. doi: 10.1161/CIRCRESAHA.113.300282. [DOI] [PubMed] [Google Scholar]
- 5.Wang WE, Li L, Xia X, Fu W, Liao Q, Lan C, et al. Dedifferentiation, proliferation, and redifferentiation of adult mammalian cardiomyocytes after ischemic injury. Circulation. 2017;136:834–U140.. doi: 10.1161/CIRCULATIONAHA.116.024307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Z, von Gise A, Zhou P, Gu F, Ma Q, Jiang J, et al. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ Res. 2014;115:354–63. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian Y, Liu Y, Wang T, Zhou N, Kong J, Chen L, et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci Transl Med. 2015;7:279ra38. doi: 10.1126/scitranslmed.3010841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao G, Kahr PC, Morikawa Y, Zhang M, Rahmani M, Heallen TR, et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature. 2016;534:119–23. doi: 10.1038/nature17959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahmoud AI, Kocabas F, Muralidhar SA, Kimura W, Koura AS, Thet S, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–53.. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Feng J, Song S, Li H, Yang H, Zhou B, et al. gp130 controls cardiomyocyte proliferation and heart regeneration. Circulation. 2020;142:967–82. doi: 10.1161/CIRCULATIONAHA.119.044484. [DOI] [PubMed] [Google Scholar]
- 11.Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Umansky KB, Yifa O, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547:179–84. doi: 10.1038/nature22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, et al. Hippo pathway inhibits wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–61. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W, Feng Y, Liang J, Yu H, Wang C, Wang B, et al. Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration. Nat Commun. 2018;9:700. [DOI] [PMC free article] [PubMed]
- 14.Chen J, Huang Z-P, Seok HY, Ding J, Kataoka M, Zhang Z, et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112:1557–66. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian Y, Liu Y, Wang T, Zhou N, Kong J, Chen L, et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci Transl Med. 2015;7:279ra38. [DOI] [PMC free article] [PubMed]
- 16.Ponnusamy M, Liu F, Zhang Y-H, Li R-B, Zhai M, Liu F, et al. Long noncoding RNA CPR (cardiomyocyte proliferation regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation. 2019;139:2668–84.. doi: 10.1161/CIRCULATIONAHA.118.035832. [DOI] [PubMed] [Google Scholar]
- 17.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Bio. 2007;8:870–9. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 18.Dorn GW, II, Vega RB, Kelly DP. Mitochondrial biogenesis and dynamics in the developing and diseased heart. Gene Dev. 2015;29:1981–91. doi: 10.1101/gad.269894.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian R, Colucci WS, Arany Z, Bachschmid MM, Ballinger SW, Boudina S, et al. Unlocking the secrets of mitochondria in the cardiovascular system path to a cure in heart failure-a report from the 2018 National Heart, Lung, and Blood Institute Workshop. Circulation. 2019;140:1205–16.. doi: 10.1161/CIRCULATIONAHA.119.040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun W, Qiu G, Zou Y, Cai Z, Wang P, Lin X, et al. Knockdown of TMEM45A inhibits the proliferation, migration and invasion of glioma cells. Int J Clin Exp Patho. 2015;8:12657–67. [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu G, Sun W, Zou Y, Cai Z, Wang P, Lin X, et al. RNA interference against TMEM97 inhibits cell proliferation, migration, and invasion in glioma cells. Tumor Biol. 2015;36:8231–8. doi: 10.1007/s13277-015-3552-6. [DOI] [PubMed] [Google Scholar]
- 22.Li B, Huang M-Z, Wang X-Q, Tao B-B, Zhong J, Wang X-H, et al. TMEM140 is associated with the prognosis of glioma by promoting cell viability and invasion. J Hematol Oncol. 2015;8:101. [DOI] [PMC free article] [PubMed]
- 23.Zhou X, Popescu NC, Klein G, Imreh S. The interferon-alpha responsive gene TMEM7 suppresses cell proliferation and is downregulated in human hepatocellular carcinoma. Cancer Genet Cytogen. 2007;177:6–15. doi: 10.1016/j.cancergencyto.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doolan P, Clynes M, Kennedy S, Mehta JP, Germano S, Ehrhardt C, et al. TMEM25, REPS2 and Meis 1: Favourable Prognostic and Predictive Biomarkers for Breast Cancer. Tumor Biol. 2009;30:200–9. doi: 10.1159/000239795. [DOI] [PubMed] [Google Scholar]
- 25.Wrzesinski T, Szelag M, Cieslikowski WA, Ida A, Giles R, Zodro E, et al. Expression of pre-selected TMEMs with predicted ER localization as potential classifiers of ccRCC tumors. Bmc Cancer. 2015;15:518. [DOI] [PMC free article] [PubMed]
- 26.Palpant NJ, Pabon L, Rabinowitz JS, Hadland BK, Stoick-Cooper CL, Paige SL, et al. Transmembrane protein 88: a Wnt regulatory protein that specifies cardiomyocyte development. Development. 2013;140:3799–808. doi: 10.1242/dev.094789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponnusamy M, Li PF, Wang K. Understanding cardiomyocyte proliferation: an insight into cell cycle activity. Cell Mol life Sci. 2017;74:1019–34.. doi: 10.1007/s00018-016-2375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Z, Wang X, Xu Z, Cao Y, Gong R, Yu Y, et al. ALKBH5 regulates cardiomyocyte proliferation and heart regeneration by demethylating the mRNA of YTHDF1. Theranostics. 2021;11:3000–16.. doi: 10.7150/thno.47354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz-Sánchez P, Villalba-Orero M, López-Olañeta MM, Larrasa-Alonso J, Sánchez-Cabo F, Martí-Gómez C, et al. Loss of SRSF3 in cardiomyocytes leads to decapping of contraction-related mRNAs and severe systolic dysfunction. Circ Res. 2019;125:170–83.. doi: 10.1161/CIRCRESAHA.118.314515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathiyalagan P, Adamiak M, Mayourian J, Sassi Y, Liang Y, Agarwal N, et al. FTO-dependent N(6)-methyladenosine regulates cardiac function during remodeling and repair. Circulation. 2019;139:518–32.. doi: 10.1161/CIRCULATIONAHA.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponnusamy M, Liu F, Zhang YH, Li RB, Zhai M, Liu F, et al. Long noncoding RNA CPR (cardiomyocyte proliferation regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation. 2019;139:2668–84.. doi: 10.1161/CIRCULATIONAHA.118.035832. [DOI] [PubMed] [Google Scholar]
- 32.Yang Q, Wu F, Wang F, Cai K, Zhang Y, Sun Q, et al. Impact of DNA methyltransferase inhibitor 5-azacytidine on cardiac development of zebrafish in vivo and cardiomyocyte proliferation, apoptosis, and the homeostasis of gene expression in vitro. J Cell Biochem. 2019;120:17459–71.. doi: 10.1002/jcb.29010. [DOI] [PubMed] [Google Scholar]
- 33.Akerberg AA, Henner A, Stewart S, Stankunas K. Histone demethylases Kdm6ba and Kdm6bb redundantly promote cardiomyocyte proliferation during zebrafish heart ventricle maturation. Dev Biol. 2017;426:84–96. doi: 10.1016/j.ydbio.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem. 2002;277:19639–48. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 35.Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, et al. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol. 2003;23:1368–78. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drummond DR, Armstrong J, Colman A. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985;13:7375–94. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5’-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–7. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 38.Guy MP, Phizicky EM. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014;11:1608–18. doi: 10.1080/15476286.2015.1008360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, Bohnsack MT. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14:1138–52.. doi: 10.1080/15476286.2016.1259781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang LS, Liu C, Ma H, Dai Q, Sun HL, Luo G, et al. Transcriptome-wide Mapping of Internal N(7)-Methylguanosine Methylome in Mammalian mRNA. Mol Cell. 2019;74:1304–16.e8. doi: 10.1016/j.molcel.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandolfini L, Barbieri I, Bannister AJ, Hendrick A, Andrews B, Webster N, et al. METTL1 promotes let-7 MicroRNA processing via m7G methylation. Mol Cell. 2019;74:1278–90.e9. doi: 10.1016/j.molcel.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bäumer N, Tickenbrock L, Tschanter P, Lohmeyer L, Diederichs S, Bäumer S, et al. Inhibitor of cyclin-dependent kinase (CDK) interacting with cyclin A1 (INCA1) regulates proliferation and is repressed by oncogenic signaling. J Biol Chem. 2011;286:28210–22. doi: 10.1074/jbc.M110.203471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li XB, Chen J, Deng MJ, Wang F, Du ZW, Zhang JW. Zinc finger protein HZF1 promotes K562 cell proliferation by interacting with and inhibiting INCA1. Mol Med Rep. 2011;4:1131–7. doi: 10.3892/mmr.2011.564. [DOI] [PubMed] [Google Scholar]
- 44.Zhang F, Bäumer N, Rode M, Ji P, Zhang T, Berdel WE, et al. The inhibitor of growth protein 5 (ING5) depends on INCA1 as a co-factor for its antiproliferative effects. PloS One. 2011;6:e21505. doi: 10.1371/journal.pone.0021505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–75. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 46.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–U186.. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cizkova A, Stranecky V, Mayr JA, Tesarova M, Havlickova V, Paul J, et al. TMEM70 mutations cause isolated ATP synthase deficiency and neonatal mitochondrial encephalocardiomyopathy. Nat Genet. 2008;40:1288–90. doi: 10.1038/ng.246. [DOI] [PubMed] [Google Scholar]
- 48.Mayr JA, Haack TB, Graf E, Zimmermann FA, Wieland T, Haberberger B, et al. Lack of the mitochondrial protein acylglycerol kinase causes sengers syndrome. Am J Hum Genet. 2012;90:314–20. doi: 10.1016/j.ajhg.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai D-F, Chen T, Wanagat J, Laflamme M, Marcinek DJ, Emond MJ, et al. Age-dependent cardiomyopathy in mitochondrial mutator mice is attenuated by overexpression of catalase targeted to mitochondria. Aging Cell. 2010;9:536–44. doi: 10.1111/j.1474-9726.2010.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, et al. Overexpression of mitochondrial transcription factor A ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112:683–90. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- 51.Rival T, Macchi M, Arnaune-Pelloquin L, Poidevin M, Maillet F, Richard F, et al. Inner-membrane proteins PMI/TMEM11 regulate mitochondrial morphogenesis independently of the DRP1/MFN fission/fusion pathways. Embo Rep. 2011;12:223–30. doi: 10.1038/embor.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macchi M, El Fissi N, Tufi R, Bentobji M, Lievens J-C, Martins LM, et al. The Drosophila inner-membrane protein PMI controls crista biogenesis and mitochondrial diameter. J Cell Sci. 2013;126:814–24. doi: 10.1242/jcs.115675. [DOI] [PubMed] [Google Scholar]
- 53.Xiao Y, Leach J, Wang J, Martin JF. Hippo/Yap signaling in cardiac development and regeneration. Curr Treat Options Cardiovasc. Med. 2016;18:38. doi: 10.1007/s11936-016-0461-y. [DOI] [PubMed] [Google Scholar]
- 54.Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:8248–53. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol life Sci. 2009;66:1631–46. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blankesteijn WM. Interventions in WNT signaling to induce cardiomyocyte proliferation: crosstalk with other pathways. Mol Pharmacol. 2020;97:90–101. doi: 10.1124/mol.119.118018. [DOI] [PubMed] [Google Scholar]
- 57.Fan Y, Ho BX, Pang JKS, Pek NMQ, Hor JH, Ng SY, et al. Wnt/β-catenin-mediated signaling re-activates proliferation of matured cardiomyocytes. Stem cell Res. Ther. 2018;9:338. doi: 10.1186/s13287-018-1086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S, Macias-Garcia A, Ulirsch JC, Velazquez J, Butty VL, Levine SS, et al. HRI coordinates translation necessary for protein homeostasis and mitochondrial function in erythropoiesis. Elife. 2019;8:e46976. [DOI] [PMC free article] [PubMed]
- 59.Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–73. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 60.Madarampalli B, Yuan Y, Liu D, Lengel K, Xu Y, Li G, et al. ATF5 connects the pericentriolar materials to the proximal end of the mother centriole. Cell. 2015;162:580–92. doi: 10.1016/j.cell.2015.06.055. [DOI] [PubMed] [Google Scholar]
- 61.Zhang S, Chen J-J. Requirement of activating transcription factor 5 for murine fetal liver erythropoiesis. Br J Haematol. 2020;188:582–5. doi: 10.1111/bjh.16202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fiorese CJ, Schulz AM, Lin Y-F, Rosin N, Pellegrino MW, Haynes CM. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol. 2016;26:2037–43.. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng P, Haynes CM. Mitochondrial dysfunction in cancer: potential roles of ATF5 and the mitochondrial UPR. Semin Cancer Biol. 2017;47:43–9. doi: 10.1016/j.semcancer.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mason JL, Angelastro JM, Ignatova TN, Kukekov VG, Lin G, Greene LA, et al. ATF5 regulates the proliferation and differentiation of oligodendrocytes. Mol Cell Neurosci. 2005;29:372–80. doi: 10.1016/j.mcn.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 65.Liu DX, Qian D, Wang B, Yang JM, Lu Z. p300-Dependent ATF5 acetylation is essential for Egr-1 gene activation and cell proliferation and survival. Mol Cell Biol. 2011;31:3906–16. doi: 10.1128/MCB.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hua ZY, Hansen JN, He M, Dai SK, Choi Y, Fulton MD, et al. PRMT1 promotes neuroblastoma cell survival through ATF5. Oncogenesis. 2020;9:50. [DOI] [PMC free article] [PubMed]
- 67.Baeumer N, Tickenbrock L, Tschanter P, Lohmeyer L, Diederichs S, Baeumer S, et al. Inhibitor of Cyclin-dependent Kinase (CDK) Interacting with Cyclin A1 (INCA1) Regulates Proliferation and Is Repressed by Oncogenic Signaling. J Biol Chem. 2011;286:28210–22. doi: 10.1074/jbc.M110.203471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this study are provided in the paper and Supplementary Materials.