A large panel of patient-derived xenograft models of small cell lung cancer reveal that extrachromosomal amplifications of MYC family genes arise after relapse to drive cross-resistance to multiple chemotherapeutic regimens.
Abstract
Small cell lung cancer (SCLC) presents as a highly chemosensitive malignancy but acquires cross-resistance after relapse. This transformation is nearly inevitable in patients but has been difficult to capture in laboratory models. Here, we present a preclinical system that recapitulates acquired cross-resistance, developed from 51 patient-derived xenograft (PDX) models. Each model was tested in vivo against three clinical regimens: cisplatin plus etoposide, olaparib plus temozolomide, and topotecan. These drug-response profiles captured hallmark clinical features of SCLC, such as the emergence of treatment-refractory disease after early relapse. For one patient, serial PDX models revealed that cross-resistance was acquired through MYC amplification on extrachromosomal DNA (ecDNA). Genomic and transcriptional profiles of the full PDX panel revealed that MYC paralog amplifications on ecDNAs were recurrent in relapsed cross-resistant SCLC, and this was corroborated in tumor biopsies from relapsed patients. We conclude that ecDNAs with MYC paralogs are recurrent drivers of cross-resistance in SCLC.
Significance:
SCLC is initially chemosensitive, but acquired cross-resistance renders this disease refractory to further treatment and ultimately fatal. The genomic drivers of this transformation are unknown. We use a population of PDX models to discover that amplifications of MYC paralogs on ecDNA are recurrent drivers of acquired cross-resistance in SCLC.
This article is featured in Selected Articles from This Issue, p. 695
INTRODUCTION
Small cell lung cancer (SCLC) remains one of the most common and deadly human cancers. Each year, it afflicts 30,000 to 35,000 patients in the United States (200,000–250,000 patients worldwide) with a median survival of 7 to 11 months that has not improved significantly over the past 40 years (1–5). In 2012, the United States Congress designated SCLC as a “recalcitrant cancer” due to high incidence, poor prognosis, and stalled progress (6). More than 95% of patients with SCLC present with disseminated disease (7, 8), and even among the rare surgical cases with no detectable metastases, only 40% are cured with resection alone (9). However, untreated SCLC is remarkably sensitive to DNA damage, and for these reasons, chemotherapy is recommended regardless of stage (10). First-line regimens combining etoposide with cisplatin (EP) or carboplatin (EC) yield overall response rates (ORR) of 60% to 70%, rivaling the most effective oncogene-targeted therapies in non–small cell lung cancer (NSCLC; refs. 11–14). A variety of agents can yield similar efficacy, including platinum–irinotecan combinations and cyclophosphamide- or methotrexate-containing regimens (15–18). This broad sensitivity to DNA damage distinguishes untreated SCLC from NSCLC, where responses are far less frequent (ORR, 15%–30%; refs. 19, 20).
For a small minority of patients, concurrent thoracic radiation or immune-checkpoint blockade can extend the initial response to chemotherapy for years or indefinitely (21–25). However, for most patients, SCLC relapses within months as a far less manageable disease. Topotecan and lurbinectedin are approved second-line agents, but treatment responses are infrequent and brief (ORR, 24% and 35%, and duration, 3.3 and 5.3 months, respectively; refs. 26, 27). Other drugs have similar modest efficacy, including temozolomide (TMZ), amrubicin, gemcitabine, bendamustine, vinorelbine, and taxanes (28, 29). In addition to sharing similar efficacy, second-line agents share the same functional biomarker, the chemotherapy-free interval (CTFI), between the conclusion of first-line chemotherapy and progression (29). Tumors with a CTFI of less than 90 days, termed “platinum resistant,” are unlikely to respond to any further chemotherapy (ORR, 14% across regimens). In contrast, “platinum-sensitive” SCLC (CTFI > 90 days) may remain sensitive to first-line chemotherapy and can be rechallenged (ORR, 49%; ref. 30). However, this approach is only transiently effective, as patients with platinum-sensitive SCLC rarely survive more than 7 to 8 months regardless of regimen (30). This common trajectory suggests two categories of SCLC: cancers that remain sensitive to DNA damage and cancers that have acquired broad cross-resistance.
Acquired cross-resistance renders SCLC untreatable and rapidly fatal, as patients rarely receive more than 2 to 3 lines of therapy (31). Despite decades of research, the drivers of this transformation remain unknown. Research has been hindered by a scarcity of tumor samples, as there is no clinical indication to rebiopsy SCLC after relapse. Furthermore, laboratory models that recapitulate clinical cross-resistance are not defined, as genetically engineered mouse models lack clinical histories, and cell lines derived before treatment or after relapse have similar chemosensitivity in vitro (32). Patient-derived xenograft (PDX) models may overcome these barriers. PDXs can be established after relapse from circulating tumor cells (CTC), bypassing the need for rebiopsy (33) and maintaining genomic fidelity to their donor tumors (34). More importantly, PDX models recapitulate the responses of their corresponding patients to DNA-damaging regimens such as first-line EP or EC, and olaparib plus TMZ (OT), a promising experimental regimen for relapsed SCLC (34, 35). This functional fidelity is necessary to capture acquired cross-resistance.
To investigate cross-resistance, we generated comprehensive clinical, molecular, and functional profiles of 51 SCLC PDX models. Each model was treated with three clinical regimens in vivo to quantify cross-resistance, and these values were compared with patient treatment histories to confirm clinical fidelity. A pair of serially derived PDXs from the same patient revealed that a focal amplification of MYC on extrachromosomal DNA (ecDNA) was acquired on treatment to confer cross-resistance. The full PDX panel demonstrated that this was not isolated to one patient, as ecDNA amplifications of MYC, MYCN, and MYCL were recurrent among cross-resistant models derived after relapse. Genomic analysis of tumor biopsies obtained from patients with SCLC showed recurrent focal amplifications of MYC paralogs in the relapsed samples but not in pretreatment samples, consistent with the PDX results. We conclude that ecDNAs harboring MYC paralogs drive acquired cross-resistance in SCLC.
RESULTS
Serial PDX Models Demonstrate Acquired Cross-Resistance and High-Level MYC Expression
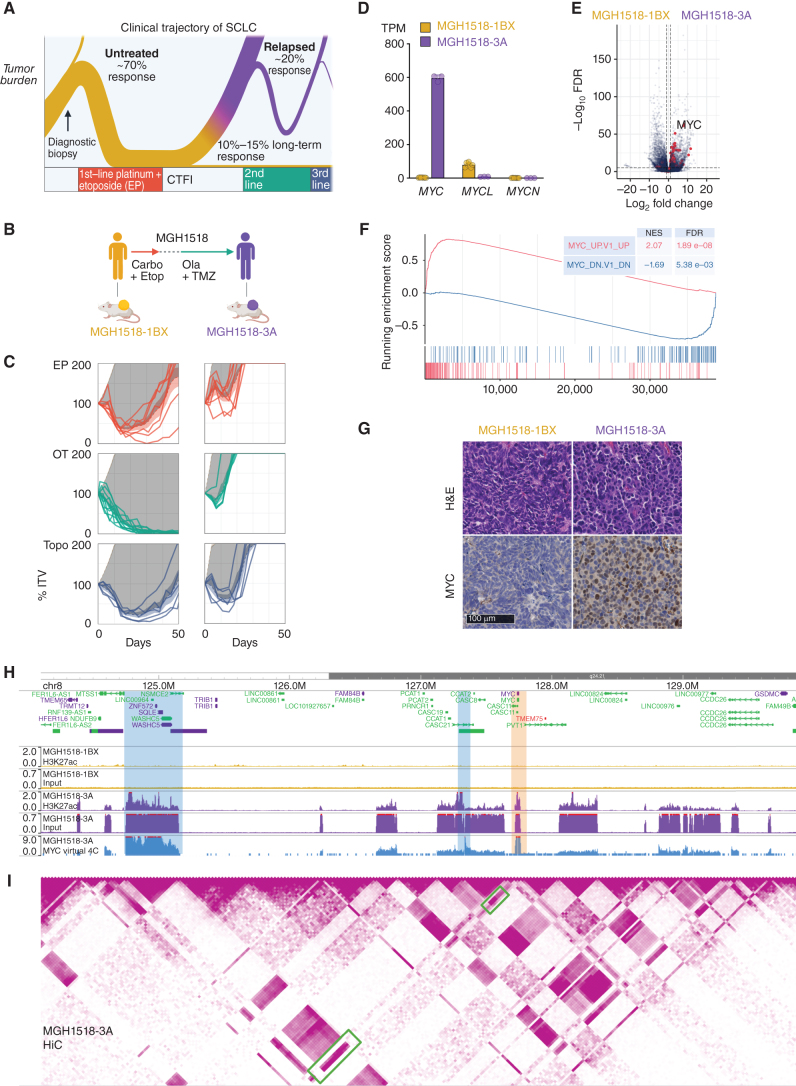
The clinical trajectory of SCLC is biphasic and defined by acquired cross-resistance (Fig. 1A). To discover genetic alterations that underlie this transition, we compared serial PDX models derived from the same patient, MGH1518, before first-line therapy with EC (MGH1518-1BX) and again after a durable response to second-line OT (MGH1518-3A; Fig. 1B; Supplementary Table S1). These models recapitulated the patient's clinical history, with MGH1518-3A acquiring resistance to both EP and OT (Fig. 1C; Supplementary Fig. S1A–S1C). Resistance in this model extended beyond the regimens received by the patient to include topotecan, demonstrating acquired cross-resistance (Fig. 1C). Of note, we used EP in vivo throughout this study to model sensitivity to both first-line EC and EP because the clinical efficacy of these regimens is equivalent (11). We confirmed this experimentally for three PDX models, including the MGH1518 serial pair, with nearly identical responses to EC and EP in vivo (Supplementary Fig. S1D).
Figure 1.
Serial PDX models of SCLC demonstrate cross-resistance and high-level MYC expression acquired after two lines of therapy. A, The biphasic clinical trajectory of SCLC, from broad chemosensitivity to acquired cross-resistance. Ribbon thickness depicts the proportion of patients. Purple = relapse. CTFI = chemotherapy-free interval. B, Clinical treatment history of patient MGH1518. Prior to therapy, PDX model MGH1518-1BX was derived from core biopsy. Patient then received 5 cycles of EC, followed by 65 days off therapy and progression at first restaging. Second-line OT resulted in a durable partial response of 6.8 months. PDX model MGH1518-3A was derived after progression on OT. C, PDX responses to EP, OT, and topotecan (Topo), as described in detail in Supplementary Fig. S1A–S1C for MGH1518-1BX treated with EP (top left). Solid color lines = tumor-volume (TV) curves for treated xenografts starting from initial tumor volume (ITV) of 300–600 mm3. Dashed color lines + color shading = average TV curves ± 95% confidence interval (CI). Tan dashed lines + shading = untreated TV curves ± 95% CI from model growth coefficients calculated from 94 MGH1518-1BX xenografts and 60 MGH1518-3A xenografts. Dark gray shading = difference in the area under treated and untreated average TV curves (ΔAUC). D, PDX MYC paralog transcripts per million (TPM). Circles = replicate xenografts. Error bars = mean and SEM. E, PDX differential gene expression. Red = genes upregulated upon overexpression of MYC per the MSigDB gene set “MYC_UP.V1_UP,” which is derived from ectopic MYC expression in primary breast epithelial cells (36). F, Gene set enrichment plot in MGH1518-3A vs. MGH1518-1BX for genes upregulated and downregulated with MYC overexpression. G, PDX morphology by hematoxylin and eosin stain, and MYC expression by IHC. H and I, PDX Histone H3K27ac ChIP-seq, Hi-C, and virtual 4C viewed in a 6 MB region on chromosome 8q24 containing the MYC locus. H, Normalized input and H3K27ac signals between models. Peak heights = counts per million mapped reads (CPM). Orange shaded region = MYC locus. I, MGH1518-3A Hi-C contact map and degree of interaction with the MYC locus by virtual 4C. Hi-C map color intensity = frequency of interaction between 2 loci. Virtual 4C peak height = relative interaction frequency with the MYC locus. Green boxes and blue-shaded regions = loci with strong interactions with the MYC locus, high H3K27ac peak, and chromatin input indicative of focal amplification. Created with BioRender.com.
The underlying gene-expression changes that might account for cross-resistance were investigated through transcriptome sequencing. This revealed a striking upregulation of the basic helix–loop–helix transcription factor MYC in MGH1518-3A, with nearly absent expression in MGH1518-1BX and only modest expression of its paralog, MYCL (Fig. 1D). Upregulation of MYC was highly impactful, driving a global shift in gene expression (Supplementary Table S2) that was led by changes characteristic of MYC induction (Fig. 1E and F; Supplementary Fig. S2A; ref. 36). This MYC-driven transcriptomic change was reflected in the divergent cellular morphologies of the tumors (Fig. 1G). MGH1518-1BX demonstrated features of classic SCLC, with sheets of fusiform cells with scant cytoplasm, nuclear molding, and inconspicuous nucleoli. In contrast, MGH1518-3A demonstrated the variant morphology originally associated with MYC amplification, with more abundant cytoplasm, round nuclei, and occasional nucleoli (37).
Increased MYC transcription resulted in elevated MYC protein levels and nuclear concentration, as measured by immunoblot and IHC (Fig. 1G; Supplementary Fig. S2B). MYC staining was surprisingly heterogeneous, with nuclear signal nearly absent in some cells and intensely positive in others. To discover epigenetic changes that might account for high but variable MYC induction, we profiled transcriptionally active chromatin through histone H3K27 acetylation chromatin immunoprecipitation and sequencing (H3K27ac ChIP-seq). This revealed a massively increased H3K27ac mark on the MYC locus in MGH1518-3A as compared with MGH1518-1BX, in agreement with their sharp difference in MYC transcription (Fig. 1H). The global shift in gene expression between these models was mirrored by a profoundly altered epigenetic landscape, with >29,000 unique H3K27ac peaks in MGH1518-3A that include MYC targets such as KLF16, whose expression increased 11-fold (Supplementary Fig. S2C and S2D; Supplementary Table S2; ref. 36). These epigenetic and transcriptional changes align with a well-described role for MYC as a global transcriptional amplifier (38).
In addition to the H3K27ac peak at MYC in MGH1518-3A, the input chromatin was also markedly elevated in the surrounding region (Fig. 1H, compared with a control region in Supplementary Fig. S2E). The segmentally increased input signal suggested a focal amplification, but not all of the high-input regions surrounding MYC contained strong H3K27ac peaks, raising the possibility of specific interactions between MYC and distant loci. To further investigate the 3D architecture of this region, we generated high-throughput chromosome conformation capture (Hi-C) maps and performed virtual circular chromosome conformation capture (4C) for the MYC locus (Fig. 1I). This showed strong contact between MYC and two regions, one in the neighboring topologically associating domain (TAD) and the other in a distant TAD, both with high input and H3K27 peaks. These interactions could be explained by structural rearrangements within the amplification to allow MYC to access both local and distant enhancers.
Together, these results suggested a focal amplification of MYC and surrounding enhancers, resulting in high-level but heterogeneous gene expression that radically altered the epigenetic landscape of the tumor. This could be explained by either subclonal amplification or by amplification on circular ecDNA, which is known to drive tumor heterogeneity (39).
High-Level MYC Amplification on ecDNA Was Acquired after the Start of Second-Line Therapy
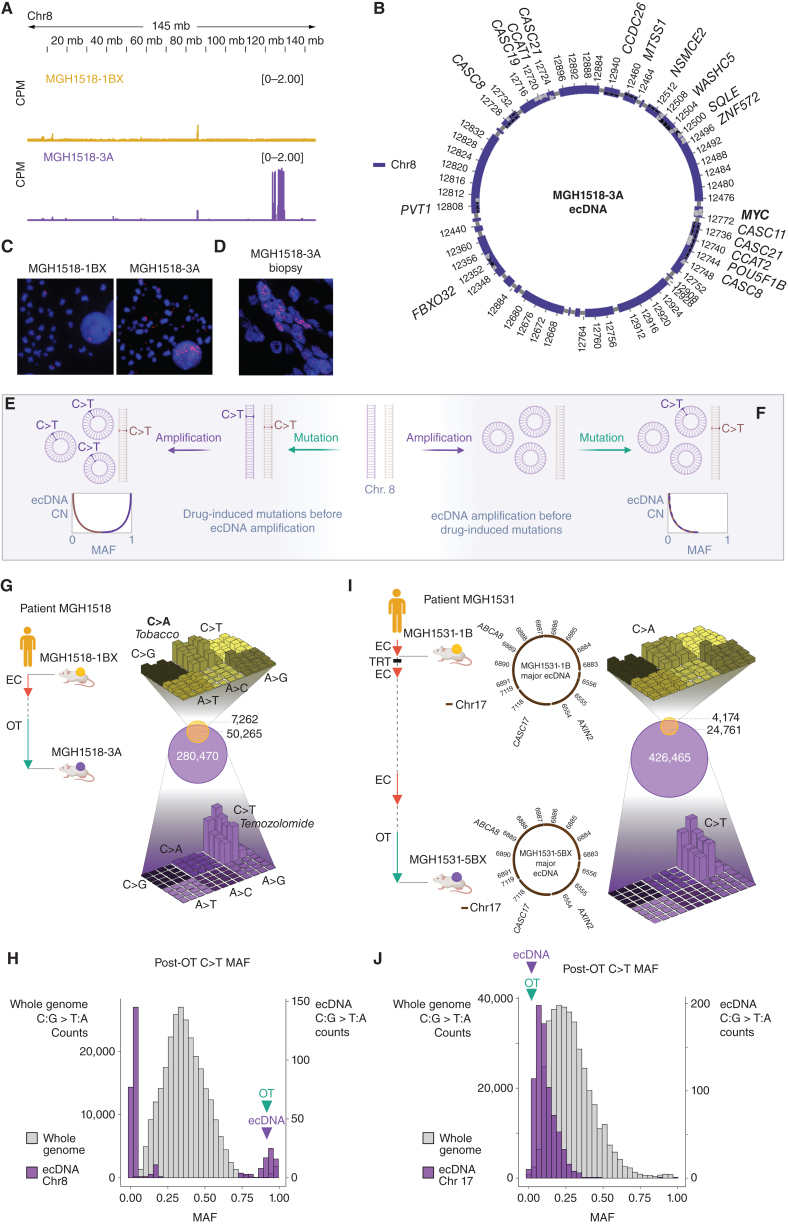
To investigate the focal MYC amplification further, MGH1518-1BX and MGH1518-3A were compared by whole-genome sequencing (WGS). Most copy-number variations were conserved between the models, but MGH1518-3A demonstrated a private 58-copy amplification of 2.09 Mb on chromosome 8 containing the MYC locus (Fig. 2A; Supplementary Fig. S3A). Reconstruction of this amplification with the AmpliconArchitect (AA) computational tool revealed an ecDNA composed of 22 fragments that included MYC in contact with its presumed enhancers, as mapped by H3K27ac ChIP-seq and Hi-C (Fig. 1H and I; Fig. 2B; Supplementary Fig. S3B; refs. 40, 41). The ecDNA was confirmed by MYC fluorescence in-situ hybridization (MYC-FISH) in metaphase cells, which showed multiple dispersed extrachromosomal foci in MGH1518-3A but not MGH1518-1BX (Fig. 2C). Furthermore, MYC-FISH performed on the patient tumor biopsy that gave rise to MGH1518-3A revealed numerous diffuse nuclear foci, demonstrating this amplification was present in the patient's tumor and not an artifact of model derivation (Fig. 2D). High-level focal amplifications of oncogenes and resistance factors on ecDNAs are common across cancer subtypes, including SCLC (42–44). Asymmetric mitotic segregation and positive selection allow ecDNAs to reach extremely high copies per cell, and also generate intratumoral copy-number heterogeneity (39). This may explain the heterogeneity of MYC expression observed in MGH1518-3A cells (Fig. 1G).
Figure 2.
Serial PDX models demonstrate MYC amplification on ecDNA acquired after the start of second-line therapy. A, Copy-number variation on chromosome 8 demonstrated focal amplification of discontinuous segments including MYC in MGH1518-3A but not MGH1518-1BX. Peak heights = counts per million mapped reads (CPM). B, AmpliconArchitect (AA) reconstruction of circular ecDNA containing MYC. C, FISH using probes for MYC in metaphase cells from MGH1518-1BX (left) and MGH1518-3A (right) xenografts following dissociation. MYC foci apart from DAPI-stained chromosomes confirm the presence of ecDNAs in MGH1518-3A but not MGH1518-1BX. D, FISH of probes for MYC in the patient tumor biopsy sample that gave rise to MGH1518-3A. E and F, Model depicting ecDNA amplification either before or after TMZ-induced C>T mutations. E, If TMZ causes a C>T mutation that is amplified on an ecDNA, then the mutant allele fraction (MAF) will increase with ecDNA copy number (CN). Conversely, if TMZ causes a C>T mutation on the unamplified allele, then its MAF will decrease after amplification. F, If ecDNA amplification precedes TMZ, then the C>T MAFs will be inversely proportional to ecDNA CN, regardless of location. G and H, MGH1518 serial model mutation analysis. G, Left: Clinical history of MGH1518 serial models as in Fig. 1B. Right center: Venn diagram compares the shared and unique mutations in serial models to reveal >280,000 new mutations in MGH1518-3A. Right top and bottom: three-dimensional bar plots (also called “Lego plots”) representing mutational signatures in a three-base context for each model (three-base key in Supplementary Fig. S4A). Note that tobacco smoking-induced C>A transversions in MGH1518-1BX (top, yellow) have been overshadowed by TMZ-induced C>T transition mutations in MGH1518-3A (bottom, purple). H, Distribution of C>T MAFs across the whole MGH1518-3A genome (gray, left y-axis) and on ecMYC (purple, right y-axis), in 30 bins ranging from 0 to 1. Bimodal distribution is consistent with ecDNA amplification after the start of TMZ as depicted in E. I and J, MGH1531 serial model mutation analysis. I, Left: Patient MGH1531 received two cycles of EC at a reduced dose with disease stabilization, and PDX MGH1531-1B was derived from a pleural effusion specimen. Patient then responded well to thoracic radiation, further EC, rechallenge with EC, and then OT. After 7.2 months on OT, a brain metastasis was resected and gave rise to MGH1531-5BX. Right: mutation overlap and signatures as in G for MGH1518 models. Center: AA reconstruction of circular ecDNA as in B reveals the same ecDNA in both MGH1531-1B and MGH1531-5BX. J, Distribution of C>T MAFs in MGH1531-5BX, as in H for MGH1518-3A. Absence of a high-MAF peak is consistent with ecDNA amplification before the start of TMZ as depicted in F. Created with BioRender.com.
Formation of circular ecDNAs usually involves DNA damage in the context of tumor suppressor inactivation and frequently occurs through chromothripsis, in which a chromosome is shattered, and the fragments are rearranged or lost (43, 45). SCLC is primed for these events, as the genomic hallmarks of the disease are loss of RB1 and TP53 and the mainstays of therapy are DNA-damaging agents and ionizing radiation (7). We investigated the rearrangements that formed the ecDNA containing MYC (ecMYC) in MGH1518-3A through analysis of germline heterozygous loci (single-nucleotide polymorphisms, or SNPs; Supplementary Fig. S3C). This revealed loss-of-heterozygosity (LOH) around but not within the amplified segments, but it remained unclear whether these deletions occurred on the chromosome that formed ecMYC. To investigate further, the allelic fractions (AF) of the SNPs were compared between MGH1518-1BX and MGH1518-3A (Supplementary Fig. S3D). On the chromosome that formed ecMYC, the SNPs between amplified segments were undetectable in MGH1518-3A, consistent with deletions, whereas the SNPs on the unamplified chromosome remained intact (Supplementary Fig. S3E). Importantly, the retention of an intact copy of chromosome 8 in MGH1518-3A resulted in a bimodal distribution of SNP AFs within the amplified regions, either very low on the unamplified chromosome or very high on ecMYC.
The ecMYC amplification was detected in MGH1518-3A but not in MGH1518-1BX (Fig. 2A and C). This could mean that it was acquired on therapy, but there are other possibilities, such as the presence of ecMYC in a small subclone that was omitted from MGH1518-1BX. We reasoned that the mutant allele fractions (MAF) of therapy-induced mutations could be measured to distinguish these possibilities (Fig. 2E and F). If the ecMYC formed after the therapy-induced mutations, then the MAFs of these preexisting mutations would mirror the SNP AFs (Supplementary Fig. S3E): mutations on amplified sequences would be coamplified, increasing their MAFs, and mutations on the unamplified allele would be diluted by the amplification, decreasing their MAFs to yield a bimodal distribution (Fig. 2E). In contrast, if the ecMYC amplification were preexisting, new therapy-induced mutations would be diluted regardless of location, with uniformly low MAFs (Fig. 2F). Even a modest preexisting amplification would limit subsequent MAFs to less than 0.5.
Patient MGH1518 was treated with OT for more than 6 months, and the comparison of the serial models generated from this patient revealed the emergence of TMZ-induced hypermutation, as is well described for glioblastoma (46). More than 280,000 new mutations were detected in MGH1518-3A, constituting a 5.75-fold increase in tumor mutational burden (TMB; Fig. 2G and ref. 47). Nearly all new mutations were C>T transitions characteristic of TMZ (SBS11 in COSMIC), which became so abundant that they supplanted the tobacco-induced C>A transversion signature (SBS4 in COSMIC; Fig. 2G; Supplementary Fig. S4A; ref. 48). We analyzed the MAFs of C>T mutations to determine the timing of ecMYC amplification relative to the start of OT. The distribution of C>T MAFs across the genome was unimodal with a median of 0.37, reflecting overall near-triploidy (Fig. 2H; Supplementary Fig. S3A). In contrast, the distribution of C>T MAFs within the ecMYC region was bimodal, mirroring the SNP AFs in this region (Fig. 2H; Supplementary Fig. S3D; Supplementary Table S3). The bimodal C>T MAF distribution is consistent with TMZ hypermutation occurring before ecMYC formation (Fig. 2E). The difference between the peaks, with MAFs most frequent below 0.07 and above 0.90, reflects the high copy number of ecMYC.
A second set of serial PDX models provides the counterexample, in which ecDNA amplification precedes TMZ-induced hypermutation (Fig. 2F). After patient MGH1531 received 2 cycles of EC at a reduced dose, MGH1531-1B was derived from an effusion specimen (Fig. 2I; Supplementary Table S1). After 2.5 years of further therapy ending with 7.2 months of OT, MGH1531-5BX was derived from a resected brain metastasis. WGS and AA reconstruction revealed an ecDNA from chromosome 17 containing AXIN and CASC17 (Fig. 2I; Supplementary Fig. S4B and S4C). Unlike the MGH1518 serial models, this ecDNA was detected and highly conserved in both models, derived before and after OT (similarity score, 0.746; P = 7.26e−05). The post-OT model MGH1531-5BX demonstrated extreme TMZ-induced hypermutation, with >400,000 new mutations (15.6-fold TMB over MGH1531-1B) and a clear transition from C>A to C>T predominance (Fig. 2I). In contrast with the ecMYC amplification in MGH1518-3A, the preexisting chromosome 17 ecDNA in MGH1531-5BX demonstrated a C>T MAF distribution that was low and unimodal (Fig. 2J; Supplementary Table S4), as predicted for therapy-induced mutations on preexisting ecDNA amplifications (Fig. 2F).
Finally, a small number of TMZ-induced mutations in MGH1518-3A could be localized directly to either ecMYC or the unamplified chromosome. Ten mutations on chromosome 8 were close enough to rearrangement junctions to be included on the same reads. These mutations could be localized to ecMYC by the presence of fusion sequences on reads that spanned the junctions (Supplementary Fig. S4D and S4E; ref. 49). 8 mutations were detected exclusively on junction reads that contain an ecMYC fusion, and for 7 of 8, the MAF was greater than 0.95 (Supplementary Fig. S4F and Supplementary Table S5). Two mutations were detected exclusively on junction reads that lack a fusion, both with MAF less than 0.05, as expected if they occurred on the unamplified chromosome. The high clonality of TMZ-induced mutations on ecMYC supports a model in which amplification occurred after the start of therapy. We concluded that the SCLC in patient MGH1518 acquired an ecMYC amplification after the start of second-line OT, and this drove the extreme epigenetic and transcriptional changes that accompanied acquired cross-resistance.
High-Level ecMYC Amplification Confers Cross-Resistance to DNA-Damaging Therapies
ecDNAs lack centromeres to mediate spindle attachment during mitosis. Instead, they segregate randomly during cell division to generate a binomial distribution of copy numbers in daughter cells (ref. 50; Supplementary Fig. S5A). This natural heterogeneity provides an opportunity to investigate the effects of copy-number variation on adjacent cells within the same tumor, without further genetic manipulation (Supplementary Fig. S5B). We exploited this powerful tool to address whether ecMYC drives cross-resistance in SCLC.
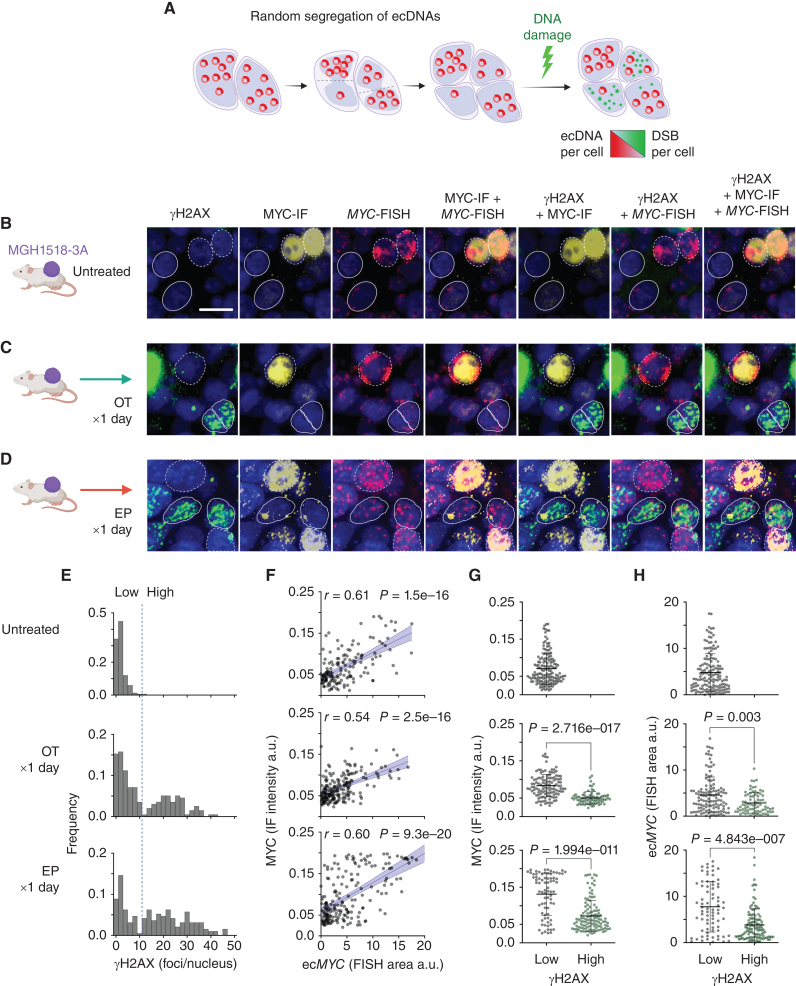
We first asked whether chemotherapy induced less DNA damage in cells that inherited higher ecMYC copy numbers (Fig. 3A; Supplementary Fig. S5C). MGH1518-3A xenografts were treated with one day of EP or OT, resected within 24 hours of the final dose, and compared with untreated controls (Fig. 3B–D). Each resected xenograft was probed for phosphorylated H2AX (γH2AX), MYC protein level, and ecMYC abundance by dual immunofluorescence and MYC-FISH (Supplementary Fig. S5D). Discrete nuclear foci of phosphorylated H2AX (γH2AX) form rapidly at double-stranded breaks, and their levels provide an indirect measure of the generation and repair of these lesions. In interphase nuclei, ecDNAs can be measured readily by FISH, and results correlate strongly with metaphase FISH from the same tumors (39, 51).
Figure 3.
ecMYC protects cells from DNA damage induced by chemotherapy. A, Schema to determine whether ecMYC protects cancer cells from DNA damage. Daughter cells inherit variable numbers of ecDNAs due to random segregation during mitosis. This generates natural copy-number heterogeneity that can be exploited experimentally to measure the effects of ecDNA dosage on therapy-induced DNA damage in individual tumor cells. B–D, MGH1518-3A xenografts (mean 58 copies ecMYC per cell) received either no treatment, a single day of OT, or a single day of EP and then were resected and fixed in formalin. Each xenograft tissue section was imaged by immunofluorescence for MYC protein and γH2AX, and by MYC-FISH for ecMYC content, with DAPI nuclear stain. γH2AX foci denote sites of DNA damage signaling, whereas homogenous γH2AX nuclear signals denote apoptotic nuclei. Dashed borders mark ecMYChigh cells, and solid borders mark ecMYClow cells. Scale bar = 10 μm. E, Distributions of γH2AX foci/nucleus. Treatment resulted in bimodal distributions with a clear threshold between damaged and undamaged cells (dashed line). F, Normalized MYC immunofluorescence vs. MYC-FISH area per nucleus. r = Pearson correlation coefficient, p = significance of correlation. G and H, MYC immunofluorescence (G) or MYC-FISH area (H) in nuclei with high or low γH2AX foci (with or without DNA damage), as measured by γH2AX foci/nucleus threshold in E. Created with BioRender.com.
In untreated tumors, cells with multiple γH2AX foci were rare but became abundant after EP or OT (Fig. 3B–D), and the comparison of treated and untreated xenografts revealed a clear threshold to delineate damaged cells (Fig. 3E). MYC protein levels correlated strongly with ecMYC abundance in both treated and untreated cells (Fig. 3F), consistent with the known capacity of ecDNAs to drive high-level oncogene expression (42). Strikingly, DNA damage induced by EP and OT was concentrated in the MYClow cells, and almost all cells with the highest MYC expression were spared (Fig. 3G). This difference could reflect ecMYC-driven therapy resistance, but could also result from diminished MYC protein synthesis or stability in cells responding to DNA damage. To distinguish these possibilities, we compared ecMYC abundance with γH2AX foci directly. Undamaged cells had markedly higher ecMYC levels than damaged cells, and as with MYC protein, the cells with the highest ecMYC levels were spared regardless of regimen (Fig. 3H). Variation in ecMYC copy number between tumor cells in the same xenograft results from random segregation, and the cells that inherited more copies of ecMYC incurred less DNA damage from both EP and OT.
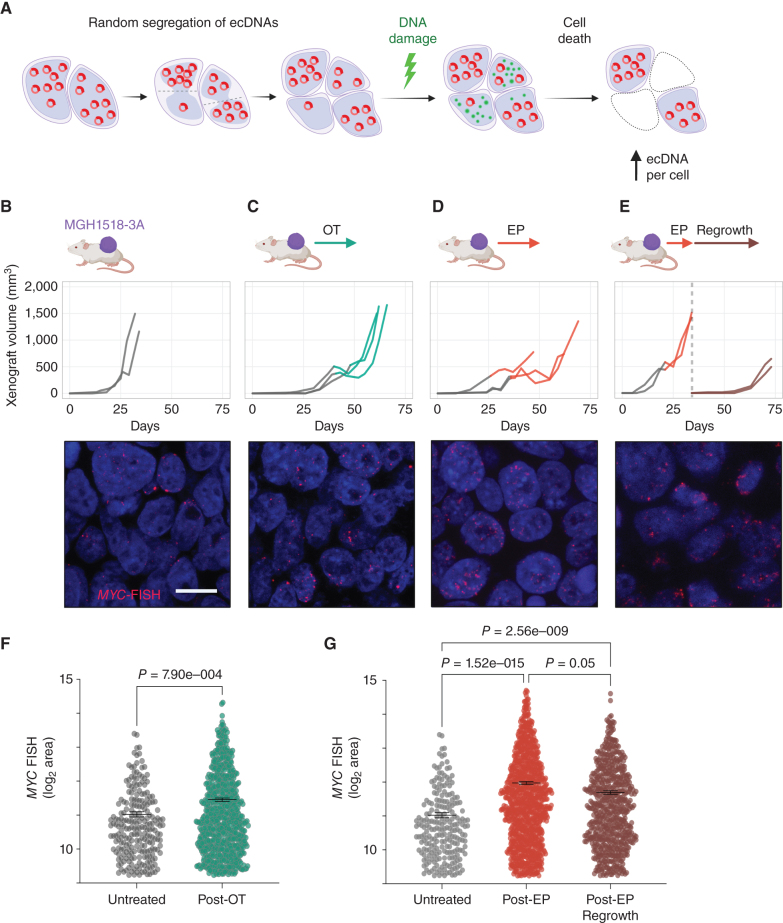
MGH1518-3A xenografts are cross-resistant compared with MGH1518-1BX, but they are still affected by chemotherapy (Fig. 1C). Tumor growth stalls transiently during treatment, with occasional brief regressions in some xenografts. Because a single day of EP or OT selectively damages ecMYClow tumor cells (Fig. 3H), we reasoned that the growth delay induced by the full regimens could represent selection against these damaged cells, which would result in higher overall copy numbers (Fig. 4A; Supplementary Fig. S5E). To test whether ecMYC confers a survival advantage, we performed MYC-FISH on MGH1518-3A xenografts resected after regrowth following full OT and EP regimens and compared them with untreated xenografts (Fig. 4B–D). To assess the persistence of any change in ecMYC copy number, EP-treated xenografts were passaged to new mice and regrown off therapy (Fig. 4E). Following treatment, the ecMYC copy-number distributions of xenografts increased significantly, and these changes in copy number were consistent between individual xenografts (Fig. 4F and G; Supplementary Fig. S6A–S6D). The increase in copy number after EP was particularly striking, and this increase persisted through at least one subsequent passage. This demonstrated a selective advantage for cells with high ecMYC in the presence of these drugs. Together with the low number of γH2AX foci in ecMYChigh cells during treatment, these results suggest that ecMYC confers dose-dependent resistance to DNA-damaging agents.
Figure 4.
Selection of tumor cells with the highest ecMYC after chemotherapy. A, Following xenograft progression on chemotherapy, the distributions of ecMYC copy numbers in treated tumors are compared with untreated tumors to determine whether ecMYC promotes DNA damage survival. B–E, MGH1518-3A xenografts tumor volume curves from implantation to resection (top) and representative MYC-FISH images (bottom). For growth curves, the transition from gray to color at the start of therapy. Xenografts were either untreated (B) or treated with full regimens of OT (C) or EP (D). E, Xenografts were treated with slightly reduced doses of EP from the standard regimen (5 mg/kg cisplatin day 1 + 8 mg/kg etoposide days 1–3 of 7 days × 2 cycles), then allowed to grow to 1,200–1,600 mm3, resected and cryopreserved. Posttreatment samples were then implanted and grown off therapy to at least 500 mm3. F and G, Measurement of MYC-FISH area per cell in untreated and treated xenografts. Comparison of distributions, and nonparametric test P values. Created with BioRender.com.
PDX Models of SCLC Recapitulate Acquired Cross-Resistance to DNA-Damaging Agents
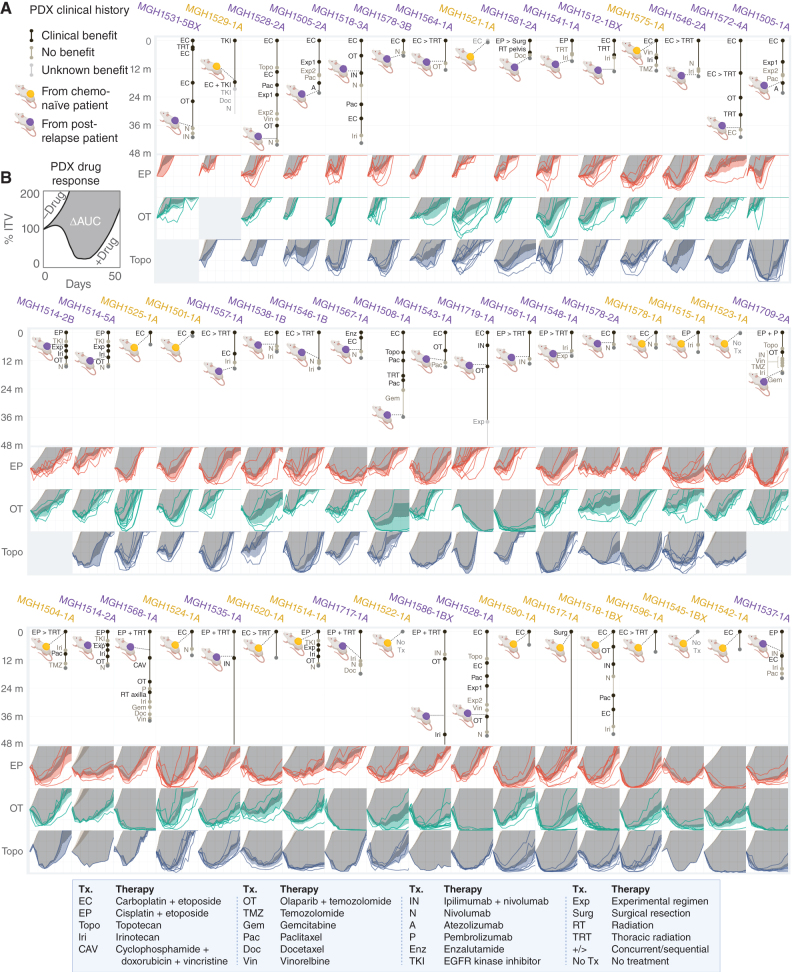
In patient MGH1518's SCLC, an ecMYC amplification was acquired during treatment and conferred resistance to both EP and OT. To understand whether ecDNA amplifications are generalized drivers of cross-resistance, we developed a PDX-based experimental system to quantify therapy responses in SCLC and to investigate their association with molecular features. A panel of 51 PDX models were derived from 42 patients with SCLC, 19 models before treatment and 32 after at least one line of therapy. For each model, the clinical history was annotated retrospectively to catalog all systemic therapies administered before and after model derivation, whether these resulted in clinical benefit, and the duration of therapies (Fig. 5A; Supplementary Table S1). For all but four patients (MGH1517, MGH1535, MGH1586, and MGH1719), a four-year window captured these clinical histories.
Figure 5.
Clinical and functional annotation of PDX panel to measure acquired cross-resistance in SCLC. A, Patient treatment histories before and after model derivation, for 48 months following the start of first-line therapy. Xenograft color denotes models derived before (yellow) or after (purple) first-line chemotherapy. Segment length = therapy duration. Segment shade = early progression (tan) vs. disease stabilization or regression (black) vs. unknown benefit (gray). Therapy abbreviation key below. B, PDX responses in vivo to EP, OT, and topotecan. TV curves for individual treated xenografts, average TV curves for models with or without treatment ± 95% CI, and ΔAUC as depicted for MGH1518 models in Fig. 1C; Supplementary Fig. S1A–S1C. PDX models are arranged by increasing chemosensitivity (ΔAUCavg, Fig. 6A) from top left to bottom right. Created with BioRender.com.
To measure cross-resistance directly, each model was treated in vivo with two to three clinical regimens that induce DNA damage through distinct mechanisms: EP, OT, and topotecan. Treatment was initiated at a large tumor volume (300–600 mm3) to measure regression precisely. Tumor-volume curves were compiled for each regimen, and an area-under-the-curve (AUC) bounded by 50 days and 200% initial tumor volume (ITV) was calculated for each xenograft (Supplementary Fig. S1B). The AUC metric was less subject to single-measurement variation than maximum regression or time-to-volume endpoints that are typically reported. Multiple xenografts were treated for each model and regimen, resulting in 694 tumor-volume curves (Fig. 5B). To control for intrinsic growth rate, the untreated AUC for each model was calculated from 1,184 control xenografts (Supplementary Fig. S7A and S7B).
Model drug effect was calculated as the difference in mean AUC between drug-treated and control xenografts (ΔAUC; Supplementary Fig. S1A–S1C). There was a strong correlation between ΔAUC values for each pair of regimens, with the correlation between EP and topotecan particularly strong (Pearson r = 0.69, P = 6.2e−8; Supplementary Fig. S8A–S8C). Furthermore, the in vivo regimens for EP, OT, and topotecan were calibrated such that the means and distributions of their ΔAUC values across the PDX panel were indistinguishable (Supplementary Fig. S8D). Therefore, we use a simple average of ΔAUC as a metric for modeling cross-resistance (ΔAUCavg; Fig. 6A; Supplementary Table S6). When models were ordered by ΔAUCavg, cross-resistance between EP, OT, and topotecan became apparent (Fig. 5B).
Figure 6.
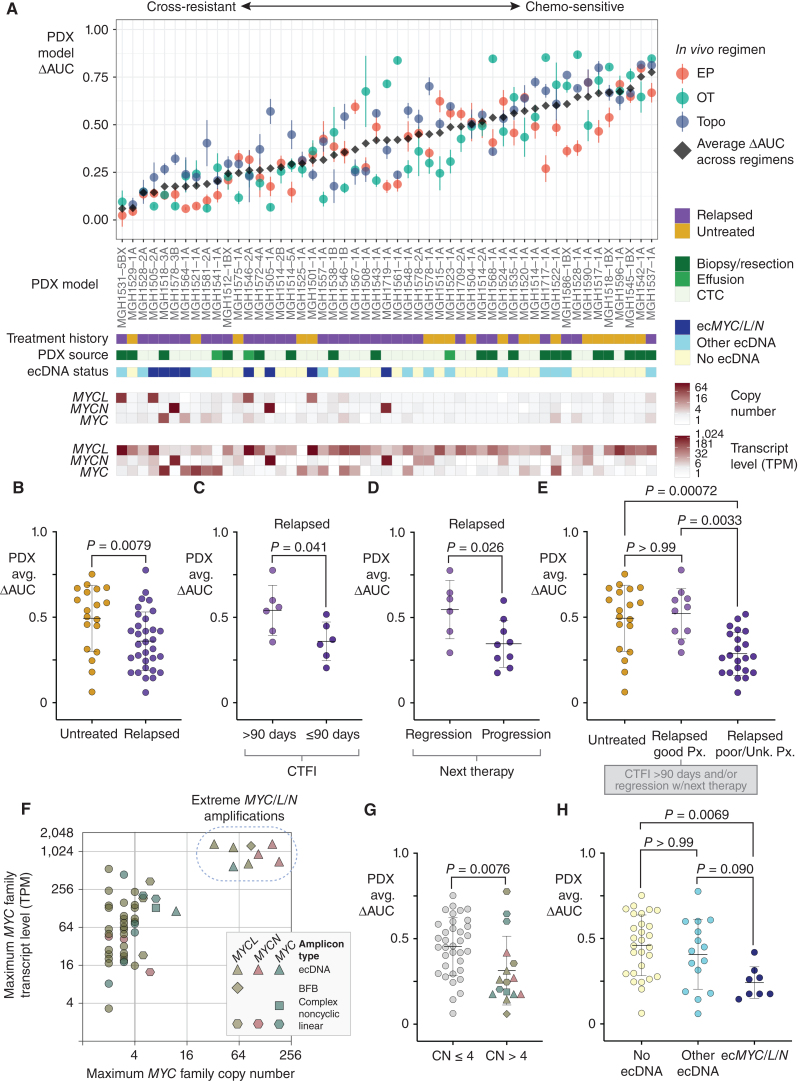
ecDNA amplifications of MYC paralogs are recurrent in cross-resistant PDX models derived from patients with relapsed SCLC. A, Integrated clinical–functional–molecular landscape of MYC paralogs across the SCLC PDX panel. Top scatter plot: In vivo cross-resistance metrics. Models are arranged left-to-right by increasing the average of ΔAUC for EP, OT, and topotecan (ΔAUCavg). Error bars = SEM of ΔAUC for each regimen. Bars below (9 total): (1) Annotated clinical treatment history (chemo-naïve vs. post-relapse), (2) PDX source (CTC vs. biopsy vs. effusion), (3) ecDNA status (ecMYC/L/N vs. other ecDNA vs. no ecDNA), (4–6) MYC paralog copy numbers, and (7–9) MYC paralog transcript levels. B–E, Performance of PDX ΔAUCavg as a metric of clinical cross-resistance by comparison with patient clinical histories. B, Comparison of ΔAUCavg of PDX models derived from patients with untreated vs. relapsed SCLC. C, Comparison of ΔAUCavg of PDX models derived from patients after first-line therapy with CTFI > 90 days (platinum-sensitive) vs. ≤ 90 days (platinum-resistant). D, Comparison of ΔAUCavg of PDX models derived from patients with relapsed SCLC who responded to the next line of chemotherapy after model derivation vs. those who received chemotherapy but did not have significant tumor regression. E, Refinement of Fig. 6B to compare PDX ΔAUCavg between models derived from untreated patients, relapsed patients with positive prognostic features described in C and D (platinum sensitivity and response to next therapy), or relapsed patients with unknown or negative prognostic features. F, Comparison of transcript level vs. copy number for the MYC paralog with the highest expression level in each model. Amplicon type is annotated. Solid vertical line at CN = 4, beyond which we classify MYC paralogs as amplified. Models with > 30-copy amplifications form a clear separate cluster (“extreme amplifications,” dashed circle). G, Comparison of ΔAUCavg between models with or without MYC paralog amplifications (CN > 4), with annotation of gene and amplification structure. H, Comparison of ΔAUCavg between models without ecDNAs and models with ecDNAs with or without MYC paralogs. Statistical tests: Mann–Whitney test P values for B–D and G, and Kruskal–Wallis test P values for E and H. Created with BioRender.com.
To determine whether PDX ΔAUCavg recapitulated clinical features of cross-resistance in SCLC, we compared this metric against known clinical determinants of treatment efficacy. Unlike established SCLC cell lines (32), PDX models derived from relapsed patients are significantly more cross-resistant than models from chemo-naïve patients (Fig. 6B). The overlap of ΔAUCavg between these cohorts reasonably approximates the proportions of chemoresistance among untreated SCLC and chemosensitivity in the relapsed population. To further test the fidelity of PDX model drug responses to clinical histories, we compared models by CTFI and by whether the patient's next regimen was effective. Models derived after the first relapse from patients with a CTFI > 90 days (“platinum-sensitive”) had significantly higher ΔAUCavg values that were comparable to models derived from untreated patients (Fig. 6C). Similarly, models derived from relapsed patients who responded to their next line of therapy were more chemosensitive than models derived just before an ineffective therapy (Fig. 6D). Clinical history distinctions were not confounded by PDX source, as there was no difference in ΔAUCavg between CTC- and biopsy- or effusion-derived models (Supplementary Fig. S8E).
These results demonstrate a remarkable degree of fidelity for PDX model drug responses to the clinical histories of their patients. They also demonstrate the value of functional testing, as the history of relapse alone is less predictive of cross-resistance. Based on the history of relapse, highly sensitive models derived from patients after a prolonged CTFI or from patients who responded to further chemotherapy would be grouped with cross-resistant SCLC, and would confound the search for candidate drivers of resistance (Fig. 6E). We conclude that the PDX panel with both clinical and functional annotation is suitable for investigating the molecular features of cross-resistant SCLC.
Relapsed Cross-Resistant SCLC Is Enriched for MYC Paralog Amplifications on ecDNA (ecMYC/L/N)
To address whether ecDNA amplifications are recurrent with acquired cross-resistance in SCLC, we generated comprehensive genomic and transcriptomic profiles of each model by WGS and paired-end RNA sequencing. AA reconstruction from WGS revealed ecDNAs in 23 of 51 PDX models (45%; Fig. 6A; Supplementary Figs. S9A–S9E; S10A–S10B), with MYC and its paralogs MYCN and MYCL as the most frequently amplified genes (ecMYC/L/N; Supplementary Fig. S11A). Only the presence of a MYC family member on these ecDNAs was recurrent, as their structures were otherwise highly divergent (Supplementary Fig. S11B). All but one ecMYC/L/N+ model was derived after relapse (7/8), whereas ecDNAs lacking MYC paralogs were evenly distributed between treated and untreated patients (Fig. 6A; Supplementary Figs. S9 and S10).
The relationship between MYC paralog expression level and copy number was compared across the PDX panel (Fig. 6F). Sixteen of 51 PDX models harbored amplifications of one MYC paralog [copy number (CN) > 4], and eight of these models clustered apart with extreme amplifications and transcript levels. ecDNAs accounted for all but one extreme amplification, a breakage–fusion–bridge (BFB) structure that resulted in 88 copies of MYCL in the highly resistant model MGH1531-5BX. PDX models with a MYC paralog amplification (CN > 4) were more resistant than unamplified models, with ecMYC/L/N marking the most resistant (Fig. 6G; Supplementary Table S6).
Comparison of model chemosensitivity by ecDNA status, regardless of CN or expression, yielded similar results (Fig. 6H; Supplementary Table S6). Models with ecMYC/L/N were significantly more cross-resistant than models that lacked ecDNAs, whereas there was no significant association between non-MYC ecDNAs and cross-resistance. The single chemo-naïve model harboring ecMYCL, MGH1501-1A, was also relatively resistant (Fig. 6A; Supplementary Fig. S9C and S9D), suggesting that de novo ecMYC/L/N amplifications may underly poor responses to first-line therapy. The presence of ecMYCL in MGH1501-1A was confirmed by MYCL FISH in metaphase tumor cells (Supplementary Fig. S9E). It is unknown how patient MGH1501 would have responded to chemotherapy, as he passed away due to critical illness shortly after the first doses were administered (Fig. 5A; Supplementary Table S1). Regression analysis of CN, transcript level, and presence on an ecDNA revealed that all three features were significantly associated with cross-resistance (low ΔAUCavg), and ecMYC/L/N was associated with resistance to each regimen (EP, OT, and topotecan; Supplementary Fig. S11C). We conclude that ecDNA formation is the most common mechanism for extreme amplification of MYC paralogs, and ecMYC/L/N amplifications are recurrent among relapsed cross-resistant SCLC tumors.
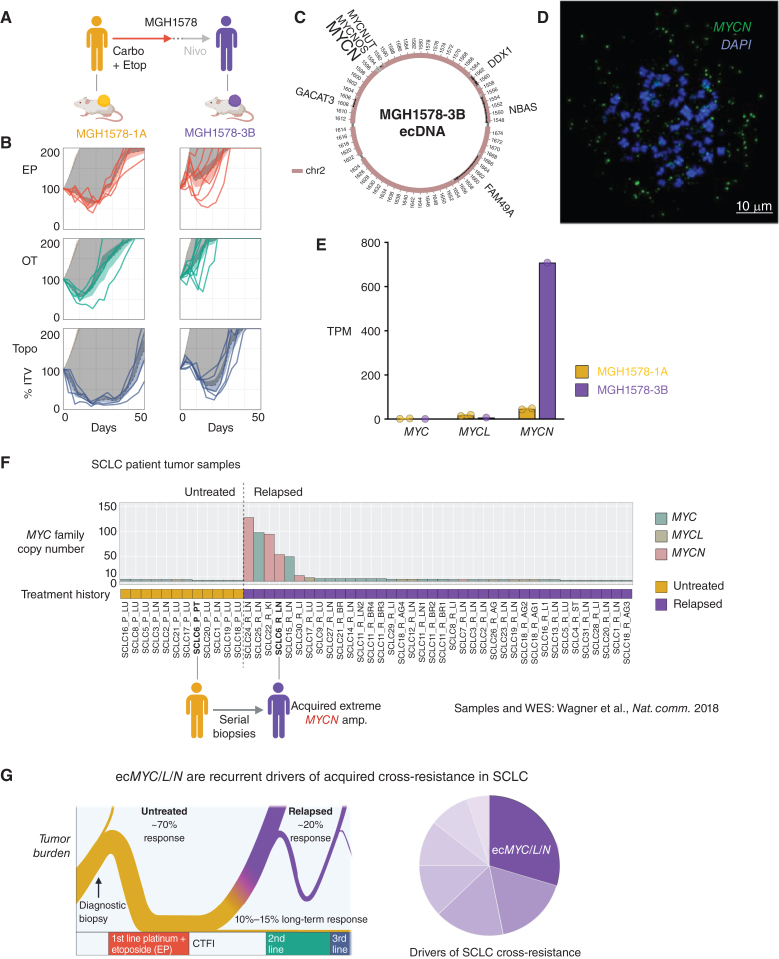
The association of ecMYC/L/N with acquired cross-resistance was bolstered by a second set of serial models from patient MGH1578 (Fig. 7A). MGH1578-1A was derived from CTCs before first-line EC. After initial tumor regression, early relapse (CTFI ≤ 90 days), and progression through second-line nivolumab, MGH1578-3B was derived from CTCs. MGH1578-3B demonstrated acquired resistance to EP as well as cross-resistance to OT and topotecan (Fig. 7B). WGS analysis revealed a massive 186-copy MYCN amplification in MGH1578-3B that was not present in the pretreatment model (Supplementary Fig. S12A), and AA reconstruction revealed an ecDNA amplification confirmed by MYCN FISH in metaphase tumor cells (Fig. 7C and D; Supplementary Fig. S12B). Transcriptome comparison between MGH1578-1A and MGH1578-3B revealed a 15.5-fold increase in MYCN expression (Fig. 7E) that was reflected in differential expression of MYC-regulated gene sets (Supplementary Fig. S12C–S12E), as was observed in the MGH1518 serial models.
Figure 7.
ecDNA amplifications of MYC paralogs are recurrent in tumor samples from patients with relapsed SCLC. A–E, Serial PDX models derived from the second patient, MGH1578, in which ecDNA amplification of an MYC paralog was detected only following relapse and acquired cross-resistance. A, Clinical treatment history of patient MGH1578. Prior to therapy, PDX model MGH1578-1A was derived from CTCs. The patient then received five cycles of EC reduced for cytopenias, followed by 14 days off therapy before progression. Second-line nivolumab was ineffective, and then MGH1578-3B was derived from CTCs. B, MGH1578 serial PDX responses to EP, OT, and Topo as in Fig. 1C, demonstrating acquired cross-resistance. C, AA reconstruction of ecMYCN detected in MGH1578-3B but not MGH1578-1A. D, Confirmation of ecMYCN in MGH1578-3B with MYCN-FISH in metaphase cells, as in Fig. 2C for ecMYC, and Supplementary Fig. S9E for ecMYCL. E, MGH1578 PDX MYC paralog transcripts per million (TPM). Circles = replicate xenografts. Error bars = mean and SEM, as in Fig. 1D. F, Maximum copy number of MYC, MYCL, or MYCN in SCLC patient tumor samples, with whole-exome sequencing and annotated treatment histories (untreated vs. relapsed) as reported in Wagner et al. (62). Post-relapse samples were reported for 30 patients, with multiple relapsed biopsies from 2 patients (SCLC11 and SCLC18). For 12 of 30 patients, paired pretreatment biopsies were also reported. Reanalysis of MYC paralog copy number by unsegmented exon counts as described in text and Methods. G, The biphasic clinical trajectory of SCLC, progressing from broad chemosensitivity to acquired cross-resistance, as presented in Fig. 1A, with our proposal that ecMYC/N/L amplifications are recurrent drivers of cross-resistance after relapse. Created with BioRender.com.
Of note, MYC paralog expression has been associated with transcriptional subtype differences in SCLC, with reported coexpression of ASCL1 and MYCL in SCLC with classic histology, versus NEUROD1 and MYC in SCLC with variant histology (37, 52–55). The serial model pairs with acquired ecMYC/L/N did not follow this trend (Supplementary Fig. S12F and S12G). In MGH1518-3A, acquired ecMYC was accompanied by increased expression of ASCL1, and in MGH1578-3B, there was no change in NEUROD1 expression with ecMYCN. Across the PDX panel, expression of ASCL1 and NEUROD1 were significantly anticorrelated, and positive correlations were detected between the expression of MYC and POU2F3, as has been reported in cell lines (56), and between MYCN and NEUROD1 (Supplementary Fig. S12H). However, there was no significant difference in the expression of ASCL1, NEUROD1, or POU2F3 in models with ecMYC/L/N (Supplementary Fig. S12I–S12K).
Patient-derived SCLC cell lines echo our results in PDX models. They harbor frequent ecMYC/L/N amplifications, and this frequency is highest among cell lines derived after relapse (Supplementary Fig. S13A; Supplementary Table S7 and refs. 57–60). However, it is possible that this reflects a competitive advantage in cell line or PDX derivation conferred by ecMYC/L/N that is not representative of SCLC in patients. Genomic profiles of relapsed SCLC tumors are rare, but we analyzed the available data. In the largest WGS study of patients with SCLC, only 5 of 96 samples were derived after relapse, with 2 additional autopsy samples with unknown treatment histories (61). MYC paralog ecDNAs were identified in 12 of 89 pretreatment samples, mostly ecMYCL with up to 33 copies, but the two highest ecDNA copy numbers were observed in a relapsed sample (ecMYCN with 104 copies) and an unknown treatment history (ecMYCL with 61 copies; Supplementary Fig. S13B and Supplementary Table S7; ref. 44). Although anecdotal, these two amplifications are comparable in magnitude with the ecMYC/L/N amplifications in relapsed PDX models.
The largest genomic study of relapsed SCLC, by Wagner and colleagues, reported whole-exome sequencing of elective research biopsies from 30 previously treated patients, with paired pretreatment biopsies from 12 patients (62). The authors did not report any high-level MYC paralog amplifications. However, in PDX models, we observed that segmented copy-number analysis (CNA) of exons failed to identify 5 of 8 ecMYC/L/N amplifications that were nonetheless detected by AA reconstruction (Supplementary Fig. S13C and Supplementary Table S7). These include ecMYC in MGH1518-3A and ecMYCL in MGH1501-1A, both of which were confirmed by FISH on metaphase cells (Fig. 2C; Supplementary Fig. S9E). In contrast, raw exon counts faithfully detected all 8 focal ecMYC/L/N amplifications, without false positives (Supplementary Fig. S13D).
We reanalyzed the Wagner and colleagues’, data set and observed the same phenomenon. Only 2 MYCN amplifications were detected with segmented CNA, both in relapsed patients (Supplementary Fig. S13E and S13F; Supplementary Fig. S14A and S14B; Supplementary Table S7). However, raw exon CNA detected 4 additional focal amplifications, including 97 copies of MYC in patient SCLC25, 94 copies of MYCN in patient SCLC22, and 49 copies of MYC in patient SCLC15 (Supplementary Fig. S14C–S14E). Of particular interest, paired biopsies from patient SCLC6 demonstrated a 53-copy MYCN amplification that was not detected in the pretreatment biopsy but acquired post-relapse (Fig. 7F; Supplementary Fig. S14F). Overall, high-level amplifications of MYC paralogs were detected in 6 of 30 patients after relapse versus 0 of 12 prior to therapy (Fig. 7F; Supplementary Fig. S13F; Supplementary Table S7). This frequency is not significantly different than in relapsed PDX models (8/32). Together with the direct detection of ecMYC in the biopsy that gave rise to MGH1518-3A (Fig. 2D), this strongly suggests that the genomic features of the PDX models are faithful to SCLC in patients.
In summary, ecMYC/L/N amplifications were enriched in cross-resistant PDX models of relapsed SCLC, and focal MYC/L/N amplifications consistent with ecDNAs were enriched in patients with relapsed SCLC. For two sets of serial PDX models, MGH1518 and MGH1578, the MYC paralog ecDNA was detected only after relapse and resistance. For MGH1518, the emergence of ecMYC after relapse could be demonstrated from mutation signature analysis, and ecMYC conferred cross-resistance to EP and OT (Figs. 2–4). These results show that ecMYC/L/N amplifications are recurrent drivers of cross-resistance in SCLC (Fig. 7G). However, most cross-resistant PDX models lack any MYC paralog amplification, and there was no difference in overall survival between patients who contributed ecMYC/L/N-positive or negative models (Fig. 6A; Supplementary Fig. S14G). Acquired cross-resistance is nearly inevitable in SCLC with successive lines of therapy, and there are likely multiple alterations beyond ecMYC/L/N amplifications that are responsible for treatment failure (Fig. 7G).
DISCUSSION
For most solid tumors, including NSCLC, chemotherapy is contraindicated if cancer progression has incapacitated a patient to the point of hospitalization. Chemotherapy acts too slowly to reverse advanced disease before the side effects exact a toll that the patient cannot endure. This rule is broken for SCLC because first-line chemotherapy works so reliably and quickly that the benefit outweighs and outpaces the side effects (63). Critically ill patients can recover to leave the hospital after one cycle of treatment. Equally reliable is that this response is transient, and after relapse, the benefit of additional lines of therapy diminishes rapidly.
One of the enduring mysteries in SCLC is that when it acquires resistance to one DNA-damaging regimen, it usually acquires cross-resistance to others. This aspect of the disease has proven stubbornly difficult to recapitulate in the laboratory, and the molecular alterations underlying this transformation remain unknown. Here, we reported 51 SCLC PDX models with comprehensive genomic, transcriptomic, functional, and clinical annotation. For individual patients, serial PDXs derived before therapy and after acquired resistance reproduced this transition. For population-level characteristics of acquired resistance, the full panel of PDX models captured the distinctions between untreated and relapsed SCLC, between relapsed platinum-sensitive and platinum-resistant cancers (CTFI ≤ 90 days), and between relapsed SCLC that continues to respond to therapy and SCLC that no longer responds. These models constitute an experimental system that captures clinical cross-resistance in SCLC with high fidelity.
We used this system to test the recurrence of candidate drivers of cross-resistance. To discover these candidates, we performed an integrated genomic comparison of serial models derived from patient MGH1518. Following second-line OT, this patient's SCLC acquired cross-resistance, and during OT, it acquired an ecDNA amplification of MYC (ecMYC) that radically altered the epigenome and transcriptome. We exploited the intrinsic random segregation of ecDNAs to demonstrate that ecMYC was a dose-dependent driver of cross-resistance. Of note, this approach can be applied broadly to assess the impact of ecDNAs on cancer cell fitness under a variety of experimental conditions. Across the PDX panel, we found additional cross-resistant PDX models with ecMYC, ecMYCL, and ecMYCN, including a second serial pair from patient MGH1578, in which ecMYCN was acquired after therapy. Overall, ecDNA amplifications of MYC paralogs were enriched in cross-resistant models from relapsed patients.
The current consensus on the molecular evolution of SCLC during disease progression is that genetic alterations drive initial tumorigenesis and possibly metastasis, but thereafter, epigenetic mechanisms drive resistance to therapy (7). Promising candidates include the upregulation of multidrug-resistance efflux pumps (64), soluble guanylate cyclase (sGC; ref. 65), the WNT signaling pathway (62), and the epithelial-to-mesenchymal transition (66, 67), as well as downregulation of the putative RNA/DNA helicase SLFN11 (67–70) and neuroendocrine differentiation factors (71). However, discovering the initiating events that induce these gene-expression changes has proven challenging.
To the best of our knowledge, ecMYC is the second genomic driver of resistance to be discovered in a patient with SCLC for any therapy, and the first acquired driver of cross-resistance. The previous driver was discovered in 1983, also on an ecDNA (72). A cell line, NCI-H249P, was derived from a patient with SCLC after one year of high-dose methotrexate (MTX). This line harbored an ecDNA amplification of dihydrofolate reductase (ecDHFR), the target of MTX. Unlike ecMYC, ecDHFR is specific for MTX resistance, and retention in NCI-H249P required continuous MTX treatment. MTX has since fallen out of the SCLC armamentarium, but the ecDHFR example raises the possibility that other ecDNAs with drug-specific resistance genes may vanish during model derivation unless drug treatment is maintained.
Both ecDNAs and MYC paralog amplifications have a 40-year history of research in SCLC. Amplification of MYC was the first genomic alteration to be identified in SCLC (also in 1983; ref. 73), followed by MYCL (74) and MYCN (75), preceding the discoveries of biallelic loss of RB1 and TP53 that have become the genomic hallmarks of this disease (76, 77). Cytogenetic analyses revealed frequent MYC paralog amplifications on ecDNAs (double-minute chromosomes; refs. 73, 75), and these were corroborated by genome sequencing of an SCLC cell line as one of the first examples of chromothripsis (78). The pathologic functions of these genes have been investigated continuously in SCLC, with roles described in tumorigenesis, metastasis, intratumoral heterogeneity, and lineage plasticity (53, 54, 79–82). However, the ecMYC amplification in MGH1518-3A played no role in initial tumorigenesis or metastasis. It was acquired during therapy and conferred drug resistance, and the roles of MYC paralogs in drug resistance are less clear.
Early suspicion that MYC amplifications might drive cross-resistance arose from cell lines established at the NCI–Navy Medical Oncology Branch from 1977 to 1992 (83). Amplifications of MYC paralogs were more frequent in cell lines derived from patients with relapsed SCLC (57–59), and two cell lines treated in vitro until cisplatin resistance demonstrated elevated MYC protein levels (84). However, further discovery was limited because cell lines derived from untreated or relapsed patients demonstrate similar responses to chemotherapy in vitro (32). Our main advance was to develop PDX models that recapitulate clinical cross-resistance and, among them, isogenic serial models in which ecMYC was acquired after relapse. Within the relapsed model, cells that inherited more ecMYC copies incurred less DNA damage and survived chemotherapy. This naturally occurring gene dose–drug response relationship captures the evolution of drug resistance and complements the seminal finding by Grunblatt and colleagues, that ectopic expression of both MYCN and MYCL in a chemosensitive PDX can confer resistance to EP (85). Together, these results show that ecMYC/L/N amplifications can drive cross-resistance in disease models that recapitulate the clinical phenomenon.
It will be important to demonstrate that ecMYC/L/N amplifications drive cross-resistance in patients with SCLC, in addition to models, but this will not be easy. Relapsed tumor samples are scarce, and not all relapsed SCLC is resistant to further therapy. The largest WGS profile of patient tumors revealed recurrent amplifications of MYC paralogs on ecDNAs (44, 61), but these were primarily surgical resection specimens, representing fewer than 5% of SCLC cases (7, 8). Within the only published genomic comparison of SCLC tumor samples obtained before and after chemotherapy (62), we found a strong enrichment of MYC paralog amplifications in relapsed samples, at a similar frequency to our relapsed PDX models. These amplifications were both focal and extreme, consistent with ecDNAs, but WGS would be required to demonstrate circular structures. A well-powered comparison of untreated and relapsed SCLC by WGS will represent a major step forward for the field.
We show that ecMYC/L/N amplifications render SCLC broadly cross-resistant to chemotherapy and inhibitors of DNA repair. This discovery opens major epidemiologic, mechanistic, and therapeutic questions. The fraction of refractory SCLC cases that can be ascribed to ecMYC/L/N, and whether each paralog contributes equally, remains to be determined. Furthermore, the inciting events that produce these amplifications are not clear. Some SCLCs may harbor preexisting ecDNAs that confer drug resistance, which would undergo positive selection in the presence of the drug. In other cases, therapy-induced DNA damage may drive the formation of ecDNAs that confer resistance to further therapy. Our results are consistent with both processes, and noninvasive CTC-based assays may enable prospective monitoring of ecDNAs to address these questions. The mechanism by which ecMYC causes resistance is largely unexplored, including whether the cross-resistance functions are distinct from those that drive tumorigenesis, whether complete elimination of ecMYC is necessary to resensitize, and whether cross-resistance extends to experimental therapies such as BH3 mimetics or DLL3-targeted agents. Pharmacologic targeting of MYC is one of the enduring challenges in cancer research, but strategies to disrupt ecDNA maintenance mechanisms could also be effective. Agents that induce replication stress, such as hydroxyurea, can reduce ecMYC in cell lines in vitro (86), but this may not translate to cancers that have already acquired resistance to DNA-damaging agents. Finally, the discovery of one category of cross-resistance will help to distinguish others and to determine whether they are overlapping or mutually exclusive. A landscape of cross-resistance drivers will be a major step toward personalized therapy for relapsed SCLC.
METHODS
PDX Model Generation
All tissue and blood samples from patients were collected per IRB-approved protocols at Massachusetts General Hospital and the University of Texas Southwestern Medical Center, with written informed consent from the patients and in accordance with the Declaration of Helsinki. Major inclusion criteria were age ≥18 and diagnosis of lung cancer, with further inclusion and exclusion criteria as previously described (34). Deidentified subject demographic data including age at diagnosis, sex as a biological variable, ethnicity, and race are reported in Supplementary Table S1. As these are tissue procurement protocols, randomization, blinding, and power analysis are not relevant. All mouse studies were conducted through Institutional Animal Care and Use Committee–approved animal protocols in accordance with Massachusetts General Hospital and University of Texas Southwestern Medical Center institutional guidelines. PDX models were generated from CTCs, biopsy/resection specimens, or malignant effusions as described previously (34). The following 9 PDX models are first reported in this study: MGH1505-1A, MGH1508-1A, MGH1517-1A, MGH1567-1A, MGH1572-4A, MGH1578-1A, MGH1709-2A, MGH1717-1A, MGH1719-1A. All other models have been reported previously (34, 35, 87). Palpable tumors were measured with electronic calipers until tumor volume exceeded 1,500 mm3. Measurement frequency was weekly unless drug treatment was initiated. Tumor volume was estimated by using the spheroid formula: tumor volume = [(tumor length) × (tumor width2)] × 0.52. Upon mouse euthanasia and xenograft resection, scalpel-dissected fragments were either implanted immediately into the right flank of an NSG mouse (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) for passaging, processed for downstream molecular analysis or short-term culture growth, or preserved in cryopreservation media for future use. For downstream molecular analyses, xenograft tissue samples were either fixed or flash-frozen within 5 minutes of mouse euthanasia. Tissue samples were fixed overnight in 10% neutral buffered formalin for histology and IHC studies, or fresh-frozen in liquid nitrogen for western blot and nucleic acid assays.
PDX Model Drug Testing
Drugging studies were initiated at xenograft volumes = 300 to 600 mm3, and tumors were measured twice weekly at least three days apart. Drugs were used as follows (unless otherwise stated): EP treatment regimen: cisplatin (7 mg/kg, intraperitoneal) once on days 1 and 8, and etoposide (10 mg/kg, intraperitoneal) once on days 1 to 3 and 8 to 10. Two exceptions to this regimen. Exception #1 EP regimen in Fig. 4E and Supplementary Fig. S6D: cisplatin (5 mg/kg, intraperitoneal) once on days 1 and 8, and etoposide (8 mg/kg, intraperitoneal) once on days 1 to 3 and 8 to 10. Exception #2 EP regimen in Fig. 3D: cisplatin (7 mg/kg, intraperitoneal) and etoposide (10 mg/kg, intraperitoneal) each once on day 1. EC treatment regimen: carboplatin (40 mg/kg, intraperitoneal) once a day 1, and day 8, and etoposide (10 mg/kg, intraperitoneal) once on days 1 to 3 and 8 to 10. OT treatment regimen: olaparib (50 mg/kg, oral gavage) twice daily for 5 days and TMZ (25 mg/kg, oral gavage) for 5 days once daily. Drugging with TMZ was performed 4 hours after olaparib treatment, and a gap of 8 to 9 hours was maintained between two olaparib administrations. Exception OT regimen for Fig. 3C: olaparib (50 mg/kg, oral gavage), followed 4 hours later by TMZ (25 mg/kg, oral gavage), followed 4 hours later by a second dose of olaparib (50 mg/kg, oral gavage), on day 1 only. Topotecan regimen: topotecan (1 mg/kg, intraperitoneal) was administered once daily on days 1 to 5 and 8 to 12. For analysis, tumor measurements were included for tumor volume > 2× ITV or 50 days after the start of treatment, whichever was attained earlier.
PDX Model Drug-Response Analysis
PDX drug responses were quantified as a function of change in tumor volume with or without treatment over time, as depicted for MGH1518-1BX treated with EP in Supplementary Fig. S1A–S1C. Tumor volume was expressed relative to ITV on the first day of treatment, within endpoints of 50 days or 200% ITV. For treated xenografts, the area under the tumor volume curve (AUC) was estimated as the sum of right trapezoids between volume measurements, bounded by endpoints. The AUC for a model–regimen combination (AUCdrug) was calculated as the mean AUCs of all xenografts treated with that regimen. In the absence of treatment, some PDX models grew too rapidly for precise measurement of tumor doubling (200% ITV) using twice-weekly caliper measurements. Therefore, the exponential growth rates of untreated xenografts were calculated by linear regression of log-transformed volume measurements during growth from a starting range of 100 to 300 mm3 to a final range of 1,200 to 1,500 mm3. The mean growth constant for each model was calculated from 6 to 94 replicate xenografts (median, 16 per model) and used to estimate the AUC without treatment to 200% ITV over 50 days (AUCno.drug). The effect of a drug regimen on model tumor volume was calculated as the difference between AUCno.drug and AUCdrug, ΔAUC. All volume calculations, tumor volume curves, and derived metrics were generated in R from compiled spreadsheets containing xenograft identities and biweekly electronic caliper measurements.
PDX Short-Term Cultures
Xenografts were resected and collected in cold PBS and the cells were manually dissociated using a scalpel. Live-cell clusters were enriched by repeated passes of gravity sedimentation in 15 mL conical tubes, with interval assessment of supernatant to identify fractions with live-cell clusters and minimal debris. ACK lysis buffer (Gibco, cat no. A1049201) was used to remove red blood cells. Cell clusters were plated and grown in HITES media with 5% FBS until use.
WGS
Genomic DNA was extracted from flash-frozen PDX tumor tissue and germline controls using the Qiagen DNeasy Blood and Tissue Kit (cat no. 69506). WGS of the 51 PDX models (41 with matched germline control) was performed by Novogene (www.novogene.com; NovaSeq 6000 PE150 with 60× coverage). Sequencing reads were aligned to the human reference genome GRCh38.v21 or the mouse reference genome GRCm38.p3 using bwa-0.7.12 (github.com/lh3/bwa). Mouse reads were filtered when the mouse alignment score (AS flag in bam files) was greater than the human alignment score. Duplicated reads were then marked using Picard MarkDuplicates (v2.27.5; https://broadinstitute.github.io/picard/).
Somatic Variants
Somatically acquired variants in the 41 germline-matched samples were determined with Mutect2 and FilterMutectCalls from GATK (gatk-4.1.2; github.com/broadinstitute/gatk). For the 10 nonmatched samples, we used HaplotypeCaller, SelectVariants, and VariantFiltration from GATK. Gene annotation was then done for both matched and unmatched samples with SnpEff (pcingola.github.io/SnpEff/). Potential germline variants in unmatched samples were removed using the NCBI dbSNP database of common germline variants: (ftp.ncbi.nih.gov/snp/organisms/human_9606_b151_GRCh38p7/VCF/common_all_20180418.vcf.gz, minus variants from COSMIC [cancer.sanger.ac.uk/cosmic]) as well as our own set of 187 germline controls. Further variant filtering was done by removing variants with allele fraction <0.01 and allele depth <2, by removing variants with LOW impact (in SnpEff output).
Copy-Number Variants
Copy-number variation (CNV) was estimated from the distribution of exon read depth in WGS using circular binary segmentation (implemented in the R package “DNAcopy”; ref. 88) with the following adjustments: Choice of diploid controls: Because of possible batch effects, diploid controls were generally selected from the same sequencing run as tumor samples (but not necessarily from the same patient DNA). Recalibration: Because tumor samples are generally not diploid overall, CNs were recalibrated by visual inspection of each DNAcopy-generated plot so that areas of the genome with 2 copies should have log2(tumor/control) = 0. Two CNV outputs were obtained, one with “raw” exon copy numbers that are highly variable and one with “segmented” copy numbers that contains large areas of constant copy number. Because the latter tends to obscure small focal amplifications, these were put back by searching the raw CNV data for areas of at least 3 highly amplified consecutive exons.
Amplicon Structural Analysis
For amplicon analysis, aligned WGS bam files were processed by AmpliconSuite-pipeline (v0.1477.1, previously named PrepareAA) to generate seeds for AmpliconArchitect (AA; v1.3.r3), which returned the architectures of amplicons. Then, outputs of AA were interpreted by AmpliconClassifier (v0.4.16) to annotate the types of amplicons (ecDNA, BFB, complex noncyclic, and linear amplification). Finally, circular plots of ecDNAs were generated by CycleViz (v0.1.3).
Mutation Signature Analysis
Mutations called with Mutect2 were filtered by a variant allele fraction of greater than or equal to 0.01 and those with a VAD greater than or equal to 2. To deconvolve mutation signatures, single-nucleotide substitutions from 39 SCLC whole-genome sequences with matched normal were tallied by their 3′ and 5′ reference base resulting in a 96-category base substitution matrix. Using nonnegative matrix factorization (k = 4), mutational signatures were calculated in MATLAB (v2017b). Mutational signatures are displayed based on the trinucleotide context and substitution type, with the frequency of mutations represented by the bar height (“Lego” plot). The four major deconvolved mutational signatures were those associated with smoking, APOBEC, aging, and TMZ. MGH1518-1BX showed a mutational pattern associated with smoking, SBS4, dominated by C:G > A:T mutations regardless of 3′ or 5′ context. MGH1518-3A showed a striking pattern of numerous C:G > T:A mutations, SBS11, associated with temozolomide methylation of guanines. To determine the variant allele fraction distribution across the identified MYC-containing ecDNA, mutations were filtered to single-nucleotide variants representing the associated TMZ substitutions C:G > T:A and restricted to those within the regions identified by AA as part of the complex ecDNA.
Allelic Imbalance Analysis
Counts of base calls (A, C, G, T) were tallied at all positions within the 15-megabase stretch of chromosome 8 affected by rearrangement and amplification in patient MGH1518, separately for the germline, treatment-naïve, and posttreatment samples. Amplified segments were annotated according to AmpliconArchitect. Heterozygous SNP sites were identified in the germline sample as follows: At each position, a major allele and minor allele were identified, by taking the first- and second-most commonly called base. Sites at which total coverage was at least 10 reads, and the major and minor alleles each contributed at least 3 reads and at least 25% of the total reads, were designated heterozygous sites. Loss of heterozygosity (LOH) or retention of heterozygosity (ROH) in each of the tumor samples (pre- and posttreatment) was determined as follows: Sites that were heterozygous in the germline sample, but in a tumor sample had zero counts for either the major or minor allele, were designated as LOH in that tumor sample. Sites that retained at least 3 reads of coverage for each allele were designated as ROH in that tumor. Supplementary Fig. S3C shows the regions affected by LOH or amplification in the posttreatment tumor. Supplementary Fig. S3D compares the pre- and posttreatment tumors, showing the minor allele fraction of each heterozygous SNP, in each tumor. Positions in segments affected by either LOH or amplification are colored red or blue, respectively. The three-copy state of chr 8 in the pretreatment tumor allows phasing of the two parental alleles. SNPs that had a minor allele fraction of ∼0.33 in the pretreatment tumor (left cloud of points) were located on parental allele “B.” They disappeared posttreatment (if located in LOH regions) or got enriched to ∼98% allele fraction (if located in an amplified segment). SNPs with a minor allele fraction of ∼0.67 pretreatment (right cloud of points) pretreatment were located on parental allele “A” and showed the opposite pattern: they became the only remaining allele posttreatment (if located in LOH regions), or else got diluted to ∼2% allele fraction (if located in an amplified segment, where the other allele got amplified).
RNA Sequencing Alignment, Mutation Calling and Transcript Abundance Measurement
RNA was extracted from flash-frozen PDX tumor tissue using the Qiagen RNeasy Mini Kit (cat no. 74104). RNA quality assessment (1% agarose gel electrophoresis, Nano-drop for RNA amount and purity, Agilent2100 for RNA integrity number), library preparation (poly-A enrichment: 250–300 bp insert cDNA library), and paired-end RNA sequencing (NovaSeq PE150 strategy) were performed by Novogene or as previously described (34, 35). Samples with an RNA integrity number of >8 were selected for library construction. Sequencing reads were aligned to the human reference genome GRCh38.v21 or the mouse reference genome GRCm38.p3 using STAR-2.7.1a (github.com/alexdobin/STAR). Mouse reads were filtered when the mouse AS (flag in bam files) was greater than the human alignment score. Duplicated reads were then removed using Picard MarkDuplicates (v2.27.5; broadinstitute.github.io/picard/). FPKM values were generated with cufflinks-2.2.1 (cole-trapnell-lab.github.io/cufflinks/) and then normalized to transcripts per million (TPM). Alternatively, mouse-filtered bam files were reverted to fastq format with samtools fastq (samtools-1.16; www.htslib.org) then used to generate TPMs with salmon-1.9.0 (github.com/COMBINE-lab/salmon) and R package tximport (ioconductor.org/packages/release/bioc/html/tximport.html).
Chromatin Immunoprecipitation Sequencing
Nuclei and cross-linked chromatin were prepared from flash-frozen xenograft tissue as previously described (89). Briefly, fragments were cut by scalpel, resuspended in a buffer containing 0.1% NP40, 0.1% Tween-20, and 0.01% Digitonin, homogenized using a TissueLyser II (Qiagen) set at 30 Hz for 1 minute and incubated on ice for 10 minutes. Lysates were filtered through a 40-μm strainer. Nuclei collected after cold centrifugation were cross-linked with 1.5% formaldehyde for 10 minutes and quenched with 0.125 mol/L glycine. Sonication was performed on a Bioruptor (Diagenode) on high power, 30 seconds on/off (50/50) for 25 minutes at 4°C, and samples were centrifuged 20,000 g × 10 minutes at 4°C. An aliquot of supernatant was frozen for chromatin input control. Chromatin was immunoprecipitated overnight with an H3K27ac antibody (Abcam, cat no. ab4729) conjugated to Dynabeads (LifeTech). Samples were eluted at 65°C in 50 mmol/L Tris, 10 mmol/L EDTA, 1% SDS, and cross-links were reversed overnight. DNA was extracted using MaXtract High Density tubes (Qiagen) and ChIP-seq libraries were prepared using the ThruPLEX DNA-Seq Kit (Takara). 75 bp single-end reads were sequenced on a Nextseq instrument (Illumina). Reads were aligned by bwa-mem2 (v2.2.1) to the hg38 reference genome. Reads were then sorted, and duplicates were removed, by sambamba (v1.0.0).
Hi-C and 4C
Hi-C libraries were generated using the Arima HiC kit (Arima Genomics) from two replicates. One million cells were cross-linked with 2% formaldehyde, followed by restriction enzyme digestion, end filling and biotin-labeled DNA incorporation, and proximal ligation. 1.5 μg of ligated DNA was then sheared by the Covaris ME220 (peak power: 75W; duty factor, 20%; cycles per burst: 200; treatment time, 3.5 minutes), size-selected around 300 bp using DNA purification beads, and enriched for biotin-labeled proximity-ligated DNA by using T1 streptavidin beads. Sequencing libraries were generated using the Arima library prep module from these biotin-enriched Hi-C DNA. The quality of the final libraries was examined with Agilent TapeStation 4150. Libraries were then pooled and sequenced on the Illumina NextSeq 2000 with the paired-end 100 mode. At least 600 million reads were sequenced for each library. Raw Hi-C sequencing reads were first processed by fastp (v0.22.0) to trim adapters and then analyzed by the HiC-Pro pipeline (v3.1.0) using hg38 as the reference genome. The Hi-C contact heat map and virtual 4C track were generated by the hicpro2juicebox.sh and make_viewpoints.py scripts, respectively, which were included in the HiC-Pro suite.
Western Blotting
Whole-cell protein was extracted from flash-frozen PDX tumor tissue. Tissue samples were lysed in RIPA buffer supplemented with protease and phosphatase inhibitors (Roche, cat no. 04693159001, 04906837001) using TissueLyser II (Qiagen) homogenizer at 4°C following the manufacturer's instructions. Samples were then centrifuged for 10 minutes at 15,000 × g and supernatant was collected for the assay. Protein concentration was measured by the Pierce BCA protein assay (Thermo Fisher, cat no.23225). Samples were separated on 4% to 20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad) after heat denaturation in Laemmli sample buffer, and proteins were transferred to nitrocellulose membranes. Membranes were blocked with EveryBlot Blocking Buffer (Bio-Rad, cat no. 12010020) for 10 minutes at room temperature. The membrane was probed overnight at 4°C with the following antibodies: Myc (1:1,000; Cell Signaling Technology, 5605S), cyclophillin A (1:1,000, Cell Signaling Technology, 2175s) followed by incubation with secondary antibody [Li-Cor, IRDye 800CW Goat anti-Rabbit IgG (1:15,000) and IRDye 680RD Goat anti-Mouse IgG (1:15,000)] in EveryBlot Blocking Buffer for 1 hour at room temperature. Following washes in TBST, membranes were imaged using the Bio-Rad ChemiDoc MP imaging system.
Tissue Fixation and Sectioning
Freshly dissected xenograft samples were fixed overnight at 4°C in 10% neutral buffered formalin, rinsed in PBS 3 × 5 minutes on ice and stored in 70% ethanol for up to 1 week. Dehydration and paraffin embedding were performed by standard method with an automated machine by the UT Southwestern Tissue Management and Pathology Core. Formalin-fixed paraffin-embedded (FFPE) tissue blocks were incubated in room-temperature water for 2 minutes followed by 2 minutes in ice-cold water before sectioning. Tissue sections were cut at 4-μm in thickness with a microtome. Sections were transferred to positively charged slides (Fisher Scientific, cat no. 1255015) and allowed to air-dry overnight before staining.
Hematoxylin and Eosin Staining and IHC
Slides with FFPE sections were warmed for 20 to 30 minutes in a 60°C oven before deparaffinization. The sections were then deparaffinized with xylene 3 × 5 minutes, rehydrated by incubation in 100% ethanol 2 × 5 minutes, 90% ethanol 1 × 3 minutes, 70% ethanol 2 × 3 minutes, 50% ethanol 1 × 3 minutes, 30% ethanol 1 × 3 minutes, and 2 × 10 minutes in deionized water and stained with hematoxylin and eosin (H&E). Post rehydration, antigen retrieval was performed in SignalStain Citrate Unmasking Solution (Cell Signaling Technology, cat no. 14746S) at 98°C for 10 to 20 minutes. Following antigen retrieval and inhibition of endogenous peroxidase activity with 3% H2O2 for 15 minutes, the slides were incubated with 10% normal goat serum for 1 hour at room temperature. Tissue sections were incubated with primary antibody: Myc (1:100; Cell Signaling Technology, 5605S) diluted in SignalStain Antibody Diluent (Cell Signaling Technology, cat no. 8112) overnight at 4°C in a humid chamber. Following 3 × 5-minute washes in TBST, tissue sections were incubated with HRP-conjugated secondary antibody (Vector Laboratories) for 1 hour at room temperature followed by development using chromogenic substrate DAB (SignalStain DAB Substrate Kit, Cell Signaling Technology, cat no. 8059s) for 10 minutes at room temperature. Slides were scanned and images were captured using a NanoZoomer (Hamamatsu).
Dual Immunofluorescence and FISH on FFPE
For dual staining of FFPE sections, slides were stained for immunofluorescence (IF) first following staining for FISH as shown in Supplementary Fig. S5D. Tissue deparaffinization, rehydration, and antigen retrieval were performed using the same protocol as described for IHC. Following antigen retrieval, sections were permeabilized for 45 minutes in 0.2% Triton X in PBS. Sections were blocked in 10% goat serum, 0.05% Triton X, and 0.05% Tween 20 in PBS for 1 hour followed by incubation in primary antibodies (MYC 1:100, Abcam, cat no. 32072; γH2AX 1:100 Abcam, cat no. 303656) for overnight at 4°C in a humid chamber. Following 3 × 10-minute washes in PBST sections were then incubated with secondary antibodies for 60 minutes at room temperature. After 3×10-minute washes in PBST, sections were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 minutes. To quench autofluorescence, slides were treated with the Vector TrueVIEW autofluorescence quenching kit (Vector Laboratories, cat no. SP-8400) as per the manufacturer's instructions and mounted using Vectashield Vibrance antifade mounting media (cat no. H-1700). Slides were cured for at least 2 hours or overnight before imaging. Images were taken at 100× using Keyence BZX microscope and the navigation setting was used to identify and save the XY coordinates for each image. To prepare the slides for FISH, coverslips were removed by incubating the slides in PBS overnight. Slides were then treated with 0.2N HCl at room temperature for 20 minutes and followed by incubation in SignalStain Citrate Unmasking Solution at 95°C for 20 minutes. This step removes any IF signal from the slides. Slides were then rinsed 2× SSC for 5 minutes and digested by 1% Proteinase K (NEB) in TE buffer at room temperature for 1 minute, and then rinsed again in 2× SSC for 5 minutes. Slides were dehydrated with 70%, 90%, and 100% ethanol for 2 minutes each and let completely dry for 10 minutes. Of note, in addition to dehydrating the slides, the alcohol series also eliminates any residual IF signal that remains after the citric acid treatment. Following the application of the MYC probe (Empire Genomics, cat no. MYC-20-RE), slides were then denatured at 75°C for 5 minutes and hybridized at 37°C overnight. Slides were washed twice in 0.4× SSC with 0.3% IGEPAL CA-630 at 40–60°C for 5 minutes each and once with 2× SSC with 0.1% IGEPAL CA-630 for 5 minutes and stained again with DAPI. Autofluorescence was quenched as described previously and slides were mounted using Vectashield Vibrance. FISH images were taken for the same microscopy fields for which IF images were taken previously using the same microscope and magnification. MYC IF intensity, the number of gH2AX foci, and MYC-FISH area within each nucleus were analyzed by CellProfiler (v4.2.4). Image registration was performed to correct any alignment issues using the DAPI images within IF and FISH data conditions. The reference image selected for this alignment process was the DAPI FISH image, and the alignment was performed using “the “imreg'form” function of MATLAB version R2023 (MathWorks, Inc.). An “affine” transformation was chosen, enabling adjustments in translation, rotation, anisotropic scaling, and shearing. We optimized the transformation metric by maximizing mutual information between the two images in multimodel mode. At least 200 cells were analyzed for each condition, from 3 to 7 fields on 2 slides (3 fields for control, 6 fields for OT, and 7 fields for EP).
FISH
For metaphase FISH, cultured cells were treated with 30 ng/mL KaryoMAX Colcemid overnight and washed once by PBS. Cells were then resuspended in 75 mmol/L KCl and swollen at 37°C for 30 minutes. Next, cell samples were fixed by 3:1 methonal:glacial acetic acid (v/v) for 4 times and dropped onto microscopic slides. Prior to FISH probe (purchased from Empire Genomics) hybridization, slides were dried overnight, equilibrated by 2× SSC buffer, and serially dehydrated by 70%, 85%, and 100% ethanol. Slides were then denatured at 75°C for 2 minutes and hybridized at 37°C overnight. Finally, slides were washed twice with 0.4× SSC with 0.3% IGEPAL CA-630 and once with 2× SSC with 0.1% IGEPAL CA-630, stained with DAPI, and mounted with anti-fade media. For FISH on interphase nuclei in FFPE samples, slides were aged at 75°C for 20 minutes and deparaffinized by xylene. After being washed with 100% and 70% ethanol, slides were treated with 0.2N HCl at room temperature for 20 minutes, 10 mmol/L citric acid at 90°C for 20 minutes, rinsed with 2× SSC, and digested briefly (< 1 minute) by 1% Proteinase K (NEB) in TE buffer at room temperature. Next, slides were serially dehydrated with ethanol and hybridized as above. To quench autofluorescence, slides were treated with Vector TrueVIEW (Vector Laboratories) following the instructions from the manufacturer. FISH images were taken by ZEISS Apotome 3 wide-field microscope with a 63× oil lens. The total area of the FISH signal within each nucleus was analyzed by CellProfiler (v4.2.4).
Statistical Analysis
Statistical methods are indicated in the corresponding figure legends. All statistical tests are two-sided. Unless otherwise indicated in figure legends or methods, all statistical tests were performed in GraphPad Prism 10. Differential gene-expression analysis was performed with R (v4.2.3) using package DESeq2. Gene set enrichment analysis was performed using the GSEA function in the clusterProfiler R package. Regression analyses were performed with R using the linear model fitting function lm(). For patients with SCLC who donated PDX models, OS rates were estimated by the Kaplan–Meier method, and log-rank (Mantel–Cox) test was used to detect any significant difference in survival curves between patients whose models lacked ecDNAs, or harbored ecDNAs with or without MYC paralogs. Survival analysis was performed using GraphPad Prism 10, and data cutoffs of 48 months after diagnosis, or August 14th, 2023, were used.
Figures
All main and supplementary figures were assembled or created with BioRender.com (www.biorender.com) and are presented under academic license.
Data Availability Statement
The data generated in this study are presented in the paper or the supplementary materials. WGS and RNA-seq data files are publicly available in the database of Genotypes and Phenotypes (dbGaP) under accession number phs003486.v1.p1, study name “Acquired Cross-Resistance in Small Cell Lung Cancer Patient-Derived Xenografts.” All other raw data files are available upon request to the corresponding authors. PDX models reported in this study will be made available to the scientific community upon reasonable request to the corresponding author B. Drapkin, contingent upon material transfer agreement.
Supplementary Material
Supplementary Table S1. Clinical Histories of PDX Models.
Supplementary Table S2. Differential Gene Expression for MGH1518 Serial Models.
Supplementary Table S3. Mutations in the ecMYC region in MGH1518-3A.
Supplementary Table S4. Mutations in the chromosome 17 ecDNA region in MGH1531-5BX.
Supplementary Table S5. Mutations at ecMYC junctions.
Supplementary Table S6. Focused PDX model clinical, molecular, and functional profiles.
Supplementary Table S7. MYC paralog amplifications in cell lines, PDX models and patient tumor samples.
Calculation of delta-AUC and comparison of EC with EP in PDX models.
Comparison gene expression and H3K27ac between MGH1518 serial models
Genomic analysis of chromosome 8 focal amplification in MGH1518-3A
Genomic analysis of chromosome 17 focal amplification in MGH1531-5BX and analysis of ecDNA junction mutations in MGH1518-3A.
Method to compare ecMYC level with resistance to drug-induced DNA damage.
Levels of ecMYC in untreated and treated MGH1518-3A xenografts
Untreated PDX growth curves.
Comparison of chemotherapy regimen delta-AUC and comparison of average delta-AUC by PDX source.
Reconstruction of ecDNAs containing MYC paralogs and MYCL FISH in MGH1501-1A metaphase chromosomes.
Reconstruction of ecDNAs that do not contain MYC paralogs.
Frequency of amplified oncogenes across PDX panel, similarity of ecMYC/L/N structures, and regression analysis of average drug response versus MYC paralog copy number, expression and amplicon type.
Acquired ecMYCN amplification in MGH1578 serial models and lineage oncogene expression compared with ecDNA status across the PDX panel.
MYC paralog copy number and ecDNA status in SCLC cell lines, PDX models and biopsy samples derived before treatment or after relapse.
MYC paralog amplifications that were detected or omitted by segmented copy number analysis, and patient survival compared with ecDNA status of PDX models.
Acknowledgments
We are deeply grateful to the patients and families who participated in these research studies, without whose selfless generosity our work would not have been possible. We thank the members of the MGH and UT Southwestern thoracic oncology groups for their assistance with this research. We are grateful to John D. Minna for his ongoing scientific guidance and critical reading of this manuscript. We thank Siddhartha Devarakonda and Obi Griffith for their constructive review of the genomic data and analysis. We thank Gaudenz Danuser for his ongoing support of this project. This work was supported by NCI grant U01CA220323 (N.J. Dyson), the Rullo Family Foundation (M.S. Lawrence), the Cancer Prevention and Research Institute of Texas (CPRIT) grant RR210034 (S. Wu), the University of Texas SPORE in Lung Cancer award number P50CA070907 (L. Girard; J. Minna P.I.), NIH grant RM1 GM145399 (H. Mazloom-Farsibaf), CPRIT grant RR20007 (B.J. Drapkin), NIH grant 1K08CA237832 (B.J. Drapkin) and a Disease-Oriented Scholar Award from UT Southwestern Medical Center (B.J. Drapkin). This work was delivered as part of the eDyNAmiC team supported by the Cancer Grand Challenges partnership funded by Cancer Research UK (P.S.M CGCATF-2021/100012, S.W. CGCATF-2021/100023) and the National Cancer Institute (P.S.M. OT2CA278688, S.W. OT2CA278683). We thank the UT Southwestern Whole Brain Microscopy Facility (RRID: SCR_017949) for assistance with whole slide scanning. Research reported in this publication was supported by the NCI of the NIH under award number P30CA142543.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
V.D. Chien reports personal fees from Texas Health Resources outside the submitted work. K. Wong reports grants from Mirati, Boehringer Ingelheim, Novartis, Pfizer, Takeda, and Bridgebio outside the submitted work. Y.P. Hung reports personal fees from Elsevier outside the submitted work. A.F. Farago reports personal fees from Novartis outside the submitted work. N.J. Dyson reports grants from NIH during the conduct of the study. S. Wu reports being supported by the Cancer Prevention and Research Institute of Texas (CPRIT) grant RR210034. This work was delivered as part of the eDyNAmiC team supported by the Cancer Grand Challenges partnership funded by Cancer Research UK (P.S.M. CGCATF-2021/100012, S.W. CGCATF-2021/100023) and the National Cancer Institute (P.S.M. OT2CA278688, S.W. OT2CA278683). S. Wu is a member of the scientific advisory board of Dimension Genomics Inc. B.J. Drapkin reports grants from NIH and Cancer Prevention Research Institute of Texas during the conduct of the study; personal fees from AstraZeneca, Sonata Therapeutics, Dialectic Therapeutics, and grants from National Foundation for Cancer Research outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
S. Pal Choudhuri: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. L. Girard: Conceptualization, resources, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J. Lim: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.F. Wise: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. B. Freitas: Data curation, supervision, investigation, methodology. D. Yang: Data curation, software, formal analysis, visualization, methodology. E. Wong: Data curation, investigation. S. Hamilton: Data curation, investigation. V.D. Chien: Data curation, investigation. Y. Kim: Investigation. C. Gilbreath: Data curation, investigation. J. Zhong: Data curation, investigation. S. Phat: Investigation. D.T. Myers: Data curation, investigation, methodology. C.L. Christensen: Data curation, supervision, investigation, methodology. H. Mazloom-Farsibaf: Resources, data curation, software, supervision. M. Stanzione: Conceptualization, resources, supervision, funding acquisition, methodology, project administration. K. Wong: Resources, data curation, supervision, validation, visualization, writing–original draft, writing–review and editing. Y.P. Hung: Conceptualization, resources, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing. A.F. Farago: conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. C.B. Meador: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. N.J. Dyson: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M.S. Lawrence: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. S. Wu: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. B.J. Drapkin: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
REFERENCES
- 1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17–48. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Colombet M, Mery L, Piñeros M, Znaor A, Zanetti R. Cancer incidence in five continents. Volume XI. IARC Scientific Publication No. 166; 2021.
- 3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 4. Wang S, Tang J, Sun T, Zheng X, Li J, Sun H, et al. Survival changes in patients with small cell lung cancer and disparities between different sexes, socioeconomic statuses and ages. Sci Rep 2017;7:1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med 2020;383:640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.H.R.733. Recalcitrant Cancer Research Act. United States of America: 112th Congress; 2012.
- 7. Rudin CM, Brambilla E, Faivre-Finn C, Sage J. Small-cell lung cancer. Nat Rev Dis Primers 2021;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blackhall F, Girard N, Livartowski A, McDonald L, Roset M, Lara N, Juarez Garcia A. Treatment patterns and outcomes among patients with small-cell lung cancer (SCLC) in Europe: a retrospective cohort study. BMJ Open 2023;13:e052556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang CF, Chan DY, Speicher PJ, Gulack BC, Wang X, Hartwig MG, et al. Role of adjuvant therapy in a population-based cohort of patients with early-stage small-cell lung cancer. J Clin Oncol 2016;34:1057–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Small Cell lung cancer (Version 2.2024). National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology. [cited 2023 December 10]. Available from: https://www.nccn.org/professionals/physician_gls/pdf/sclc.pdf.
- 11. Rossi A, Di Maio M, Chiodini P, Rudd RM, Okamoto H, Skarlos DV, et al. Carboplatin- or cisplatin-based chemotherapy in first-line treatment of small-cell lung cancer: the COCIS meta-analysis of individual patient data. J Clin Oncol 2012;30:1692–8. [DOI] [PubMed] [Google Scholar]
- 12. Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 13. Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med 2017;377:829–38. [DOI] [PubMed] [Google Scholar]
- 14. Planchard D, Kim TM, Mazieres J, Quoix E, Riely G, Barlesi F, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:642–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Noda K, Nishiwaki Y, Kawahara M, Negoro S, Sugiura T, Yokoyama A, et al. Irinotecan plus cisplatin compared with etoposide plus cisplatin for extensive small-cell lung cancer. N Engl J Med 2002;346:85–91. [DOI] [PubMed] [Google Scholar]
- 16. Schmittel A, Fischer von Weikersthal L, Sebastian M, Martus P, Schulze K, Hortig P, et al. A randomized phase II trial of irinotecan plus carboplatin versus etoposide plus carboplatin treatment in patients with extended disease small-cell lung cancer. Ann Oncol 2006;17:663–7. [DOI] [PubMed] [Google Scholar]
- 17. Livingston RB, Moore TN, Heilbrun L, Bottomley R, Lehane D, Rivkin SE, et al. Small-cell carcinoma of the lung: combined chemotherapy and radiation: a Southwest Oncology Group study. Ann Intern Med 1978;88:194–9. [DOI] [PubMed] [Google Scholar]
- 18. Hansen HH, Dombernowsky P, Hansen M, Hirsch F. Chemotherapy of advanced small-cell anaplastic carcinoma: superiority of a four-drug combination to a three-drug combination. Ann Intern Med 1978;89:177–81. [DOI] [PubMed] [Google Scholar]
- 19. Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol 2008;26:3543–51. [DOI] [PubMed] [Google Scholar]
- 20. Breathnach OS, Freidlin B, Conley B, Green MR, Johnson DH, Gandara DR, et al. Twenty-two years of phase III trials for patients with advanced non-small-cell lung cancer: sobering results. J Clin Oncol 2001;19:1734–42. [DOI] [PubMed] [Google Scholar]
- 21. Liu SV, Reck M, Mansfield AS, Mok T, Scherpereel A, Reinmuth N, et al. Updated overall survival and PD-L1 subgroup analysis of patients with extensive-stage small-cell lung cancer treated with Atezolizumab, carboplatin, and etoposide (IMpower133). J Clin Oncol 2021;39:619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paz-Ares L, Chen Y, Reinmuth N, Hotta K, Trukhin D, Statsenko G, et al. Durvalumab, with or without tremelimumab, plus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer: 3-year overall survival update from CASPIAN. ESMO Open 2022;7:100408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Turrisi AT 3rd, Kim K, Blum R, Sause WT, Livingston RB, Komaki R, et al. Twice-daily compared with once-daily thoracic radiotherapy in limited small-cell lung cancer treated concurrently with cisplatin and etoposide. N Engl J Med 1999;340:265–71. [DOI] [PubMed] [Google Scholar]
- 24. Faivre-Finn C, Snee M, Ashcroft L, Appel W, Barlesi F, Bhatnagar A, et al. Concurrent once-daily versus twice-daily chemoradiotherapy in patients with limited-stage small-cell lung cancer (CONVERT): an open-label, phase 3, randomised, superiority trial. Lancet Oncol 2017;18:1116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gronberg BH, Killingberg KT, Flotten O, Brustugun OT, Hornslien K, Madebo T, et al. High-dose versus standard-dose twice-daily thoracic radiotherapy for patients with limited stage small-cell lung cancer: an open-label, randomised, phase 2 trial. Lancet Oncol 2021;22:321–31. [DOI] [PubMed] [Google Scholar]
- 26. Trigo J, Subbiah V, Besse B, Moreno V, Lopez R, Sala MA, et al. Lurbinectedin as second-line treatment for patients with small-cell lung cancer: a single-arm, open-label, phase 2 basket trial. Lancet Oncol 2020;21:645–54. [DOI] [PubMed] [Google Scholar]
- 27. von Pawel J, Schiller JH, Shepherd FA, Fields SZ, Kleisbauer JP, Chrysson NG, et al. Topotecan versus cyclophosphamide, doxorubicin, and vincristine for the treatment of recurrent small-cell lung cancer. J Clin Oncol 1999;17:658–67. [DOI] [PubMed] [Google Scholar]
- 28. Zugazagoitia J, Paz-Ares L. Extensive-stage small-cell lung cancer: first-line and second-line treatment options. J Clin Oncol 2022;40:671–80. [DOI] [PubMed] [Google Scholar]
- 29. Owonikoko TK, Behera M, Chen Z, Bhimani C, Curran WJ, Khuri FR, et al. A systematic analysis of efficacy of second-line chemotherapy in sensitive and refractory small-cell lung cancer. J Thorac Oncol 2012;7:866–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baize N, Monnet I, Greillier L, Geier M, Lena H, Janicot H, et al. Carboplatin plus etoposide versus topotecan as second-line treatment for patients with sensitive relapsed small-cell lung cancer: an open-label, multicentre, randomised, phase 3 trial. Lancet Oncol 2020;21:1224–33. [DOI] [PubMed] [Google Scholar]
- 31. Steffens CC, Elender C, Hutzschenreuter U, Dille S, Binninger A, Spring L, et al. Treatment and outcome of 432 patients with extensive-stage small cell lung cancer in first, second and third line: results from the prospective German TLK cohort study. Lung Cancer 2019;130:216–25. [DOI] [PubMed] [Google Scholar]
- 32. Polley E, Kunkel M, Evans D, Silvers T, Delosh R, Laudeman J, et al. Small cell lung cancer screen of oncology drugs, investigational agents, and gene and microRNA expression. J Natl Cancer Inst 2016;108:djw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897–903. [DOI] [PubMed] [Google Scholar]
- 34. Drapkin BJ, George J, Christensen CL, Mino-Kenudson M, Dries R, Sundaresan T, et al. Genomic and functional fidelity of small cell lung cancer patient-derived xenografts. Cancer Discov 2018;8:600–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Farago AF, Yeap BY, Stanzione M, Hung YP, Heist RS, Marcoux JP, et al. Combination olaparib and temozolomide in relapsed small-cell lung cancer. Cancer Discov 2019;9:1372–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439:353–7. [DOI] [PubMed] [Google Scholar]
- 37. Gazdar AF, Carney DN, Nau MM, Minna JD. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 1985;45:2924–30. [PubMed] [Google Scholar]
- 38. Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012;151:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lange JT, Rose JC, Chen CY, Pichugin Y, Xie L, Tang J, et al. The evolutionary dynamics of extrachromosomal DNA in human cancers. Nat Genet 2022;54:1527–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Deshpande V, Luebeck J, Nguyen ND, Bakhtiari M, Turner KM, Schwab R, et al. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat Commun 2019;10:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 2017;543:122–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 2019;575:699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim H, Nguyen NP, Turner K, Wu S, Gujar AD, Luebeck J, et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet 2020;52:891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pongor LS, Schultz CW, Rinaldi L, Wangsa D, Redon CE, Takahashi N, et al. Extrachromosomal DNA amplification contributes to small cell lung cancer heterogeneity and is associated with worse outcomes. Cancer Discov 2023;13:928–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu S, Bafna V, Chang HY, Mischel PS. Extrachromosomal DNA: an emerging hallmark in human cancer. Annu Rev Pathol 2022;17:367–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DI, Zairis S, et al. Clonal evolution of glioblastoma under therapy. Nat Genet 2016;48:768–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stanzione MS, Zhong J, Wong E, LaSalle TJ, Wise JF, Simoneau A, et al. Translesion DNA synthesis mediates acquired resistance to olaparib plus temozolomide in small cell lung cancer. Sci Adv 2022;8:eabn1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yi E, Gujar AD, Guthrie M, Kim H, Zhao D, Johnson KC, et al. Live-cell imaging shows uneven segregation of extrachromosomal DNA elements and transcriptionally active extrachromosomal DNA hubs in cancer. Cancer Discov 2022;12:468–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. deCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet 2018;50:708-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep 2016;16:1259–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mollaoglu G, Guthrie MR, Bohm S, Bragelmann J, Can I, Ballieu PM, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell 2017;31:270–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ireland AS, Micinski AM, Kastner DW, Guo B, Wait SJ, Spainhower KB, et al. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell 2020;38:60–78 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang YH, Klingbeil O, He XY, Wu XS, Arun G, Lu B, et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev 2018;32:915–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Johnson BE, Ihde DC, Makuch RW, Gazdar AF, Carney DN, Oie H, et al. myc family oncogene amplification in tumor cell lines established from small cell lung cancer patients and its relationship to clinical status and course. J Clin Invest 1987;79:1629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brennan J, O'Connor T, Makuch RW, Simmons AM, Russell E, Linnoila RI, et al. myc family DNA amplification in 107 tumors and tumor cell lines from patients with small cell lung cancer treated with different combination chemotherapy regimens. Cancer Res 1991;51:1708–12. [PubMed] [Google Scholar]
- 59. Johnson BE, Russell E, Simmons AM, Phelps R, Steinberg SM, Ihde DC, et al. MYC family DNA amplification in 126 tumor cell lines from patients with small cell lung cancer. J Cell Biochem Suppl 1996;24:210–7. [DOI] [PubMed] [Google Scholar]
- 60. Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wagner AH, Devarakonda S, Skidmore ZL, Krysiak K, Ramu A, Trani L, et al. Recurrent WNT pathway alterations are frequent in relapsed small cell lung cancer. Nat Commun 2018;9:3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mamesaya N, Kodama H, Iida Y, Kobayashi H, Ko R, Wakuda K, et al. Efficacy and safety of carboplatin and etoposide in older extensive-stage small-cell lung cancer patients with a poor performance status. Thorac Cancer 2023;14:805–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 1992;258:1650–4. [DOI] [PubMed] [Google Scholar]
- 65. Schenk MW, Humphrey S, Hossain A, Revill M, Pearsall S, Lallo A, et al. Soluble guanylate cyclase signalling mediates etoposide resistance in progressing small cell lung cancer. Nat Commun 2021;12:6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stewart CA, Gay CM, Xi Y, Sivajothi S, Sivakamasundari V, Fujimoto J, et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nature Cancer 2020;1:423–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A 2012;109:15030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sousa FG, Matuo R, Tang SW, Rajapakse VN, Luna A, Sander C, et al. Alterations of DNA repair genes in the NCI-60 cell lines and their predictive value for anticancer drug activity. DNA Repair 2015;28:107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Reinhold WC, Varma S, Sunshine M, Rajapakse V, Luna A, Kohn KW, et al. The NCI-60 methylome and its integration into CellMiner. Cancer Res 2017;77:601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thomas A, Takahashi N, Rajapakse VN, Zhang X, Sun Y, Ceribelli M, et al. Therapeutic targeting of ATR yields durable regressions in small cell lung cancers with high replication stress. Cancer Cell 2021;39:566–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Curt GA, Carney DN, Cowan KH, Jolivet J, Bailey BD, Drake JC, et al. Unstable methotrexate resistance in human small-cell carcinoma associated with double minute chromosomes. N Engl J Med 1983;308:199–202. [DOI] [PubMed] [Google Scholar]
- 73. Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature 1983;306:194–6. [DOI] [PubMed] [Google Scholar]
- 74. Nau MM, Brooks BJ, Battey J, Sausville E, Gazdar AF, Kirsch IR, et al. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature 1985;318:69–73. [DOI] [PubMed] [Google Scholar]
- 75. Nau MM, BrooksBJ Jr, Carney DN, Gazdar AF, Battey JF, Sausville EA, et al. Human small-cell lung cancers show amplification and expression of the N-myc gene. Proc Natl Acad Sci U S A 1986;83:1092–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Harbour JW, Lai SL, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science 1988;241:353–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, et al. p53: a frequent target for genetic abnormalities in lung cancer. Science 1989;246:491–4. [DOI] [PubMed] [Google Scholar]
- 78. Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011;144:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ciampricotti M, Karakousi T, Richards AL, Quintanal-Villalonga A, Karatza A, Caeser R, et al. Rlf-Mycl gene fusion drives tumorigenesis and metastasis in a mouse model of small cell lung cancer. Cancer Discov 2021;11:3214–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Olsen RR, Ireland AS, Kastner DW, Groves SM, Spainhower KB, Pozo K, et al. ASCL1 represses a SOX9(+) neural crest stem-like state in small cell lung cancer. Genes Dev 2021;35:847–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 2014;156:1298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kim DW, Wu N, Kim YC, Cheng PF, Basom R, Kim D, et al. Genetic requirement for Mycl and efficacy of RNA Pol I inhibition in mouse models of small cell lung cancer. Genes Dev 2016;30:1289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Phelps RM, Johnson BE, Ihde DC, Gazdar AF, Carbone DP, McClintock PR, et al. NCI-Navy Medical Oncology Branch cell line database. J Cell Biochem Suppl 1996;24:32–91. [DOI] [PubMed] [Google Scholar]
- 84. Chalishazar MD, Wait SJ, Huang F, Ireland AS, Mukhopadhyay A, Lee Y, et al. MYC-driven small-cell lung cancer is metabolically distinct and vulnerable to arginine depletion. Clin Cancer Res 2019;25:5107–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Grunblatt E, Wu N, Zhang H, Liu X, Norton JP, Ohol Y, et al. MYCN drives chemoresistance in small cell lung cancer while USP7 inhibition can restore chemosensitivity. Genes Dev 2020;34:1210–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Von Hoff DD, McGill JR, Forseth BJ, Davidson KK, Bradley TP, Van Devanter DR, et al. Elimination of extrachromosomally amplified MYC genes from human tumor cells reduces their tumorigenicity. Proc Natl Acad Sci U S A 1992;89:8165–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mooradian MJ, Piotrowska Z, Drapkin BJ, Dias-Santagata D, Marcoux N, Arnaoutakis K, et al. Clonal evolution and the role of serial liquid biopsies in a case of small-cell lung cancer-transformed EGFR mutant non-small-cell lung cancer. JCO Precis Oncol 2017;1:10.1200/PO.17.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004;5:557–72. [DOI] [PubMed] [Google Scholar]
- 89. Cejas P, Xie Y, Font-Tello A, Lim K, Syamala S, Qiu X, et al. Subtype heterogeneity and epigenetic convergence in neuroendocrine prostate cancer. Nat Commun 2021;12:5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Clinical Histories of PDX Models.
Supplementary Table S2. Differential Gene Expression for MGH1518 Serial Models.
Supplementary Table S3. Mutations in the ecMYC region in MGH1518-3A.
Supplementary Table S4. Mutations in the chromosome 17 ecDNA region in MGH1531-5BX.
Supplementary Table S5. Mutations at ecMYC junctions.
Supplementary Table S6. Focused PDX model clinical, molecular, and functional profiles.
Supplementary Table S7. MYC paralog amplifications in cell lines, PDX models and patient tumor samples.
Calculation of delta-AUC and comparison of EC with EP in PDX models.
Comparison gene expression and H3K27ac between MGH1518 serial models
Genomic analysis of chromosome 8 focal amplification in MGH1518-3A
Genomic analysis of chromosome 17 focal amplification in MGH1531-5BX and analysis of ecDNA junction mutations in MGH1518-3A.
Method to compare ecMYC level with resistance to drug-induced DNA damage.
Levels of ecMYC in untreated and treated MGH1518-3A xenografts
Untreated PDX growth curves.
Comparison of chemotherapy regimen delta-AUC and comparison of average delta-AUC by PDX source.
Reconstruction of ecDNAs containing MYC paralogs and MYCL FISH in MGH1501-1A metaphase chromosomes.
Reconstruction of ecDNAs that do not contain MYC paralogs.
Frequency of amplified oncogenes across PDX panel, similarity of ecMYC/L/N structures, and regression analysis of average drug response versus MYC paralog copy number, expression and amplicon type.
Acquired ecMYCN amplification in MGH1578 serial models and lineage oncogene expression compared with ecDNA status across the PDX panel.
MYC paralog copy number and ecDNA status in SCLC cell lines, PDX models and biopsy samples derived before treatment or after relapse.
MYC paralog amplifications that were detected or omitted by segmented copy number analysis, and patient survival compared with ecDNA status of PDX models.
Data Availability Statement
The data generated in this study are presented in the paper or the supplementary materials. WGS and RNA-seq data files are publicly available in the database of Genotypes and Phenotypes (dbGaP) under accession number phs003486.v1.p1, study name “Acquired Cross-Resistance in Small Cell Lung Cancer Patient-Derived Xenografts.” All other raw data files are available upon request to the corresponding authors. PDX models reported in this study will be made available to the scientific community upon reasonable request to the corresponding author B. Drapkin, contingent upon material transfer agreement.