Abstract
Molecular expression profiling of tumors initiated by transgenic overexpression of c-myc, c-neu, c-ha-ras, polyoma middle T antigen (PyMT) or simian virus 40 T/t antigen (T-ag) targeted to the mouse mammary gland have identified both common and oncogene-specific events associated with tumor formation and progression. The tumors shared great similarities in their gene-expression profiles as compared with the normal mammary gland with an induction of cell-cycle regulators, metabolic regulators, zinc finger proteins, and protein tyrosine phosphatases, along with the suppression of some protein tyrosine kinases. Selection and hierarchical clustering of the most variant genes, however, resulted in separating the mouse models into three groups with distinct oncogene-specific patterns of gene expression. Such an identification of targets specified by particular oncogenes may facilitate development of lesion-specific therapeutics and preclinical testing. Moreover, similarities in gene expression between human breast cancers and the mouse models have been identified, thus providing an important component for the validation of transgenic mammary cancer models.
Keywords: cDNA microarray‖mammary cancer‖oncogenes‖gene-expression profiles
Gene expression profiling of human breast cancers has increased our understanding of the clinical diversity of the disease and has been instrumental in the classification of tumors into subtypes and studying their response to drug treatment (1, 2). Recently, attempts have been made to delineate pathways characteristic of hereditary breast cancer with BRCA1 and BRCA2 mutations (3). However, most breast cancers are sporadic, develop through the accumulation of more than one genetic lesion, and cannot be studied within the same patient, thereby posing difficulties in the identification of stage-specific events involved in the initiation and progression of cancer (4).
Many cancers in transgenic mice arise from the targeted overexpression of a particular oncogene in a well defined genetic background, thus offering particular advantages for studying tumor progression caused by a single initiating event. Although characteristic differences in the histopathology of the mammary cancers from many of these transgenic models have been defined (5), little is known about the gene-expression profiles that distinguish the tumor types on the basis of the initiating oncogenic event.
In this study, we have compared six well characterized mouse models of human breast cancer to determine the fundamental differences in gene expression between the normal mammary gland and mammary tumors and to define genes that are associated specifically with each oncogenic pathway. Gene-expression patterns of mouse mammary tumor virus long terminal repeat (MMTV)-c-myc, MMTV-neu, MMTV-Ha-ras, MMTV-polyoma middle T antigen (PyMT), C3 (1)/simian virus 40 (SV40) T/t antigen, and WAP-SV40 T/t antigen (T-ag; refs. 6–11) transgenic mice were established by high-density cDNA microarray analysis. Several of these models are directly relevant to molecular alterations found in human breast cancer. For example, amplification of erbb2/her2/neu, an epidermal growth factor receptor family tyrosine kinase, and the proto-oncogene, c-myc, have been associated with 30% and 17% of human breast cancers, respectively (12, 13). The SV40 T antigen functionally inactivates both tumor suppressor genes pRb and p53, which is often mutated in human breast cancer (14–15).
Analysis of over 8,600 unique genes demonstrated that despite different initiating oncogenic events, the mouse models were remarkably similar in their molecular expression profiles, and they differed greatly from normal mammary gland. Changes in gene expression observed in human breast cancer were often found in the mouse models. In addition, our analysis identified a small subset of genes capable of assigning oncogenic signatures to the tumor types, which may potentially be useful in identifying previously uncharacterized, oncogene-specific therapeutic targets for the treatment of human breast cancer. We propose that gene-expression profiles of mouse models of cancer will further identify models most suitable for preclinical testing of novel therapeutic molecules and serve as an important means of model validation.
Methods
cDNA Clones.
The mouse oncochip of National Cancer Institute 2.7K array (2,700 features) and mouse Incyte GEM1 8.7K array (8,700 features) of cDNA clones were arrayed separately at the National Cancer Institute Applied Technology Center. The gene list is available at http://nciarray.nci.nih.gov. Approximately 11,000 features comprising 8,680 unique genes were analyzed. The cDNA set spanned 4,246 named genes, 2,288 expressed sequence tags (EST), and 2,146 Riken cDNAs.
Animals.
All transgenic mice studied were of the FVB strain background. Animals were housed and cared for in accordance with National Institutes of Health guidelines and palpitated for tumors twice every week. Tumors (0.6–0.8 cm) were dissected out, fixed in 4% (wt/vol) paraformaldehyde for histology, and the remainder was snap frozen in liquid nitrogen. All mammary gland tumors from the same mouse were pooled, and tumors from four to six different mice were analyzed for each mouse model. Mammary glands from 10 randomly selected 11-week old FVB females at various stages of estrous cycle were pooled and used as the reference RNA for all arrays. Three batches of reference RNA were prepared.
RNA Isolation and cDNA Microarray Analysis.
RNA from the normal mammary gland and tumor samples was extracted by the guanidine isothiocyanate method (16). Total RNA from each sample (20 μg) was labeled and hybridized as described (17), except that the reactions were purified by using Microcon YM-30 columns. The array slides were scanned with an Axon 4000 scanner (Axon Instruments, Foster City, CA) at a resolution of 10 μm. The reference RNA was labeled by using Cyanine 3-dUTP (Cy3), and the tumor samples were labeled with Cyanine 5-dUTP (Cy5), except for the reverse-labeling experiments.
Data Filtering and Normalization.
Image analysis and the calculation of average foreground signal adjusted for local channel specific background was performed with GENEPIX software. All statistical analyses were performed with the S+ package. Spots with signal intensities in both channels less than 100 were excluded. If at least one channel had an intensity above 100, the intensity under 100 was set at 100. The average number of clones filtered from analysis was 110 for the NCI 2.7K array and 85 for the GEM1 8.7K arrays. Each array was separately globally normalized to make the median value of log2 ratio equal to zero.
Evaluation of Gene-Specific Dye Bias.
Because we consistently labeled the reference normal breast epithelium with Cy3 and the tumors with Cy5, it is possible that the down-regulation detected in tumors for some genes could reflect dye bias that was not removed by the normalization process. To evaluate this possibility, we examined pairs of arrays in which the tumor sample was labeled with Cy5 and the reference with Cy3 for one array, and the labeling was reversed in the paired array. We analyzed 17 such pairs of arrays for the NCI 2.7K array and 11 such pairs containing clones from the GEM1 8.7K array. Reverse-labeled experiments were performed by using a variety of tumor samples, including prostate tumors from a related study. We computed the average difference in log2 ratio between normalized forward- and reverse-labeled experiments for each clone as an estimate of the residual dye bias for that clone. Of the clones on the NCI 2.7K array, only 7 had an average difference in log2 ratios greater in absolute value than 1 (i.e., 2-fold difference), and for the GEM1 8.7K array, the number was 19. There was no general systematic bias favoring either dye after normalization. For comparing tumor models with each other, the use of an internal reference consistently labeled in all arrays should not result in spurious claims that genes are differentially expressed among models, but may result in missing the detection of differential expression of genes that do not stain adequately with Cy5.
Sample Clustering and Multidimensional Scaling.
Average linkage hierarchical clustering of samples was based on a Pearson correlation similarity metric using all available genes or genes selected by F test (see below). Multidimensional scaling is the process of representing the distances (1 minus similarities) of a group of objects in a low dimensional (i.e., three-dimensional) space (18). Multidimensional scaling analyses were performed with the same distance matrix as was used for hierarchical clustering for genes selected by F test.
F Tests and Gene Clustering.
To identify genes that distinguish transgenic mouse models from each other, an F test was performed separately for each gene represented on the arrays. The F test is a generalization of the t test for more than two groups. Statistical significance levels were calculated with all available log ratios for genes in GEM1 8.7K arrays and NCI2.7K arrays. Genes showing variation between models greater than expected at the 0.001 level of statistical significance were identified by using a stringent level of significance controls for the large number of genes tested. Approximately 10 genes significant at the 0.001 level would be expected by chance, but the statistical significance level is approximate and limited by the accuracy of the normal distribution approximation. Average linkage hierarchical analysis of these genes was performed by using a Pearson correlation similarity metric to group genes based on their patterns of variation across the transgenic models. Gene clusters were selected based on a cut of the dendrogram at a correlation coefficient of 0.7. The clusters and associated image-plots were displayed with TREEVIEW software (19).
Northern and Western Blot Analysis.
Total RNA (20 μg) was electrophoresed through a 1.2% agarose formaldehyde gel, and Northern blot analysis was performed by using the method of Church and Gilbert (20). Gene-specific inserts of sequence-verified cDNA clones (Incyte Pharmaceuticals, Palo Alto, CA) were labeled with radioactive [32P]dCTP by using the Ready-To-Go DNA-labeling kit (Invitrogen); blots were washed by using standard protocols and exposed to Kodak XO-MAT films. For Western blots, 30 μg of protein extracts from tumors were analyzed as described (21). Anti-proliferating cell nuclear antigen (PCNA) and anti-actin antibodies (Santa Cruz Biotechnology) were used at a 1:500 dilution. The detection of antibodies was performed by an Enhanced Chemiluminescence Kit (NEN/Du Pont).
Results
We have performed microarray analysis of mouse models of human breast cancer. The data collected was analyzed to determine “cancer-related” genes by comparing the expression profiles of tumors to that of the normal mammary gland. Secondly, individual mouse models were compared with each other to define “oncogene signatures” characteristic of the initiating oncogenic event. Results are available at the National Cancer Institute Mouse Mammary Models Collective web site: http://emice.nci.nih.gov.
Cancer Genes.
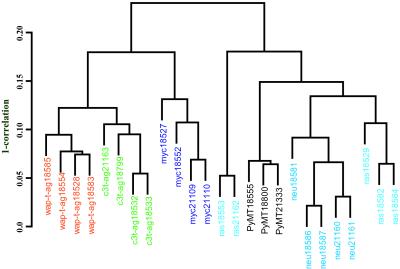
Genes that were commonly regulated in mouse models as compared with normal tissue were identified as those with an average log2 ratio of at least 1 or less than −1, when the average was computed across all arrays. Such analysis resulted in the selection of 627 features from the mouse GEM1 8.7K array and 276 features from the NCI 2.7K array. The relationship between the tumor types as determined by hierarchical clustering using these genes is shown in Fig. 1. The tumor models seem to be highly similar in their expression profiles as determined by the high correlation coefficients in the dendrogram. Interestingly, tumors from the same cohort of transgenic animals clustered together. In addition, we observed modulation in several genes previously implicated in human breast cancer (Table 1). The named genes were classified by using GENECARDS (http://bioinformatics.weizmann.ac.il) and by extensive review of the literature (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). Regardless of the transgenic model, the most striking feature of the tumors was the high induction of genes in the glycolytic pathway involved in the conversion of glucose to pyruvate, including high levels of lactate dehydrogenase. Accelerated rates of metabolism were evident by increased expression of translation elongation factors and structural RNA genes. Several cell-cycle regulators, signaling receptors and their effectors, including G proteins and downstream transcription factors, were significantly induced in all mammary gland tumors. A unique observation of our profiling data was the induction in the expression of several zinc-finger proteins in all mouse models studied. However, expression of soluble protein tyrosine kinases was diminished in most tumors with a concomitant induction in the protein tyrosine phosphatase activity. Several cytoskeletal proteins like tubulin 4 and tubulin 5 isoforms displayed identical regulatory patterns in all tumors studied. In addition, several ESTs with as yet uncharacterized function were modulated in the tumors, thus identifying a vast number of genes potentially important in the process of oncogenesis.
Figure 1.
Comparison of mouse models of cancer. A dendrogram depicting the degree of similarity between the six mouse models of human cancer is shown. Average linkage hierarchical clustering using Pearson correlation similarity was carried out for all genes studied with at least a two-fold geometric mean relative to the normal mammary gland (reference RNA). The scale on the right shows 1 minus correlation. The high correlation coefficients suggest that the tumors are highly similar in their gene-expression profiles, irrespective of the initiating oncogenic event.
Table 1.
Comparison of genes that have been implicated in human breast cancer with mouse models of cancer
| Gene | Status in human cancer | Status and tumor type in mouse |
|---|---|---|
| Cortactin | Amplification of chromosome 11q13 in 15% of breast cancers amplifies cortactin expression | Induced in myc |
| Galectin 3 | Increased grade of breast tumor has decreased expression | Induced in T-ag |
| Tieg | Not expressed in invasive lesions but highly expressed in normal breast epithelium, moderately in noninvasive tumors | Induced expression in neu-ras-PyMT group |
| Phopholipase A2, VII | Increases in breast cancer | Induced in T-ag |
| Tumor antigen CO-029 | High in breast carcinomas | Induced in neu |
| Aldolase C | Found in human DCIS lesions | Induced in neu |
| Prothymosin alpha | A Myc target used as a marker to measure proliferation index of human breast tumors | Highly induced in Tag and myc |
| STAT3 | Increased activity in human breast and prostate cancer cells | Induced in models except myc |
| CD24 antigen | Differential expression in human mammary cell lines | Induced in all models |
| Lipocalin 2 | Increased expression in normal ductal lumens surrounding cancer tissue, but low expression in normal ductal epithelium | Induced in all models |
| Acid β glucosidase | Inhibitors to this enzyme displayed decreased metastasis | Induced in all models |
| Riken cDNA | Similar to DRIM-1, down regulated in metastasis, isolated from MDA-Mb 435 cells | Induced in all models |
| Procollagen, Iα and IIIα | Induced in stromal cells of malignant lesions of human breast cancer | Suppressed in all tumors |
Several genes were commonly down-regulated in all tumor models studied, including fat-specific gene 27, enolase, and carbonic anhydrase. It is likely that a subset of these genes appeared suppressed because of the comparison of an enriched population of epithelial cells in the tumor to a mixture of adipose and epithelial cells of the normal mouse mammary gland. We compared RNA from fat pads devoid of epithelial cells to our reference RNA to identify such a subset of genes (see Fig. 5, which is published as supporting information on the PNAS web site).
Oncogene Signatures.
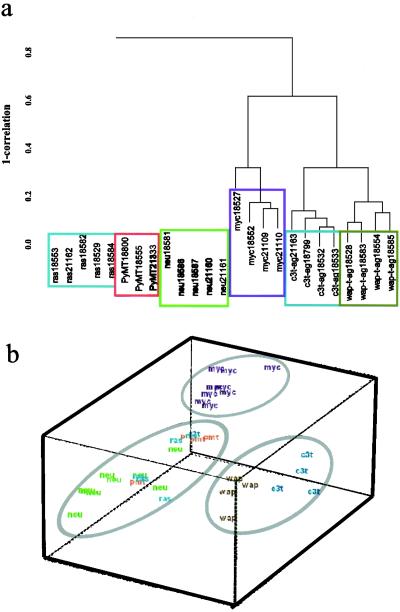
Analysis of commonly regulated genes was powerful in determining pathways that differed between a normal and cancerous state of mouse mammary gland but could not completely identify discrete oncogene-specific clusters that would aid the identification of pathway-specific targets for further characterization. Therefore, F tests were performed on the microarray data described in Methods and which resulted in the identification of a total of 930 genes. Average linked hierarchical clustering of the arrays with regard to genes identified by F tests separated the tumor types into three groups (Fig. 2a). The first group was composed of SV40 T/t antigen tumors. This group seemed to be more similar to the MMTV-myc tumors but was distantly related to the third group of tightly clustered neu-, ras-, and PyMT-induced tumors. Multidimensional scaling analysis of the data visually depicts the separation of the three groups (Fig. 2b). The image plots of the clustered genes is shown in Fig. 6 a and b (which is published as supporting information on the PNAS web site).
Figure 2.
Comparative cDNA microarray analysis of mouse models of human breast cancer. (a) A subset of 930 genes selected by F test (P < 0.001) were clustered as described in Methods to determine oncogene-specific signatures. The tumors fall into three distinct groups: (i) MMTV-neu, MMTV-ras, and MMTV-PyMT; (ii) MMTV-myc; and (iii) the T antigen group. (b) Data are represented in three dimensions by multidimensional scaling.
T-Antigen Cluster.
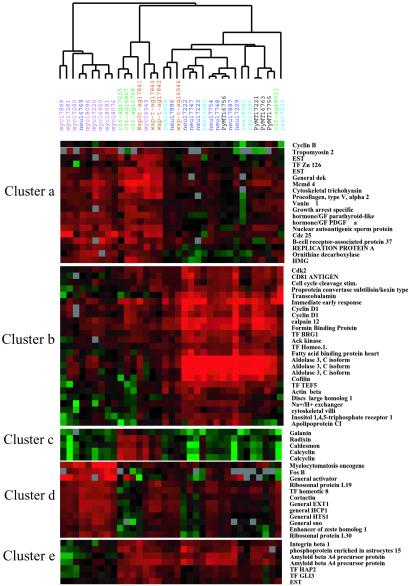
Of the 930 genes differentially expressed between the tumor models, more than 100 genes were uniquely modulated by SV40 T/t antigen (Table 3, which is published as supporting information on the PNAS web site). This group was the largest of tumor signature genes among all of the oncogenes studied. T-ag expression perturbed several important cellular pathways, including cell cycle, DNA replication, RNA metabolism, signal transduction, cell death, and genes implicated in human cancer (Table 3). The most unique observation was the induction of calcium binding and/or calcium-regulated genes like calcyclin, calcium calmodulin-dependent kinase II (CamKII), annexin A2 and 5, caldesmon, and calumenin exclusively in the Tag group (Fig. 3, cluster c). Interestingly, only a few genes involved in DNA repair, RAB4A and pRb, showed appreciable down-regulation in these tumors.
Figure 3.
Oncogene-specific clusters spanning interesting variant genes are shown. Each tumor type is color-coded; gene names are shown on the right-hand side of the figure. Complete hierarchical clustering of the 258 genes and 672 genes that varied significantly by F test (P < 0.001) in their expression profiles across the tumors studied are shown in Fig. 6 a and b, respectively.
Myc Cluster.
The myc-derived tumors cluster closely with the T-ag tumors but display only a subset of the gene-expression changes observed in the T-ag tumors, including overexpression of cyclin E, PCNA, and cdc25A (Fig. 6a). Myc overexpression seemed to suppress Rb- and E2F-related gene. In addition, the myc cluster included previously identified myc targets like c-fos, ornithine decarboxylase, and dihydrofolate reductase (22), along with up-regulation of several transcription factors like enhancer of zeste homolog, hox proteins, general transcription activators and repressors like sno and other DNA-binding proteins. As reported (23), we observed increased expression of ribosomal RNA genes in myc tumors.
Neu-ras-PyMT Cluster.
These tumors showed very similar profiles of gene expression and clustered very tightly with each other, with maximal changes observed in tumors derived from neu overexpression. GTPase activating proteins (GAPs) and related G proteins were predominantly induced in this group (Fig. 6b). In addition, neu tumors showed up-regulation of E2F, cdk-2, and cyclin D1, as has been documented earlier (24). A unique class of tetraspanin family membrane glycoproteins previously isolated as tumor antigens, including CD81 and CO-029, were up-regulated in neu tumors. In addition, neu-derived tumors displayed induction of proteases like calpains and MMP15 as well as extracellular proteinase inhibitor. Within the genes analyzed, we failed to detect changes in the PyMT tumors that may suitably explain the high incidence of metastasis observed in this mouse model.
Validation of the Observed Gene-Expression Profiles.
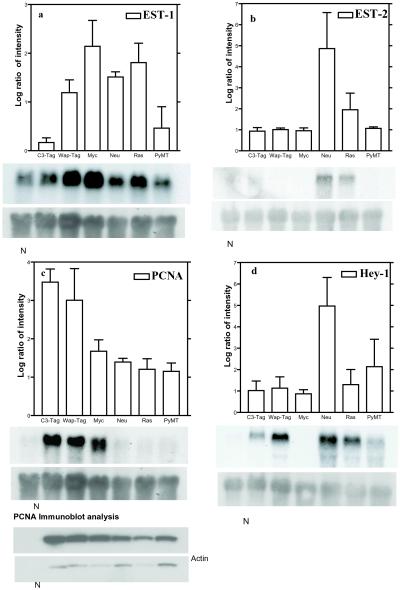
Northern blot analyses were performed for 11 differentially expressed genes identified by microarray studies. Of these, three known genes displayed results similar to the array data. Of the eight ESTs studied, five probes labeled poorly and could not give sufficient and/or specific signal, suggesting that EST data from arrays require rigorous validation before interpretation. Representative blots are shown in Fig. 4. The bar charts in each panel represent the levels of gene expression observed by microarray hybridization; Northern blots for the corresponding EST or named gene(s) are shown below. The N lane denotes total RNA from normal mammary glands. EST-1 was highly induced in all tumors and was recently annotated as a structural ribosomal RNA gene (Fig. 4a). Fig. 4b shows a previously uncharacterized EST (EST-2) that was specifically induced in the neu and ras tumors. PCNA was specifically induced in the myc-T-ag group, but this induction was absent from the neu-ras-PyMT group. This variant expression pattern was retained at the protein level (Fig. 4c). Also identified in the myc tumors was the specific down-regulation of hey-1, hairy enhancer-of-split-related gene with YRPW motif, involved in the Notch-signaling pathway (Fig. 4d). These results demonstrate that the modulations observed by cDNA microarray analyses could be validated by a second technique.
Figure 4.
Comparison of microarray and Northern blot analysis. The average log ratio of intensity (to the base 2) observed by microarray hybridization for each selected gene is depicted as a bar graph and is plotted on the y axis, with the tumor type on the x axis. (Bars = SD.) The results of Northern blot analysis performed for the gene are shown below each bar graph. Tumor samples are listed on the top of each lane of the Northern blot; the N lane denotes normal mammary gland RNA sample. (a) EST-1. (b) EST-2. PCNA and Hey-1 were modulated in an oncogene-specific manner (c and d, respectively). The differential regulation of PCNA was retained at the protein level by Western blot. 18S rRNA was used as an internal control for RNA quantification for Northern blots.
Discussion
We hypothesized that comparative gene-expression profiling of oncogene-derived tumors would broaden our understanding of oncogene-specific pathways, a study difficult to perform in the human disease owing to the potential multiplicity of genetic changes associated with human breast cancer. First, we determined differences in gene profiles of the normal mammary gland and mammary tumors, irrespective of the initiating oncogene, thus identifying pathways that change during tumor formation and disease progression. Second, by an analysis of variance, we studied the genes that differed in their expression profiles depending upon the initiating oncogenic event. We proposed that the first analysis would identify genes that display altered expression in mammary cancer irrespective of the causal genetic lesion. The second approach would identify genes and potential pathways uniquely perturbed by the specific initiating oncogenic event.
Results of the first analysis indicate that the mouse models studied display common and highly similar patterns of gene expression that differ greatly from the normal mammary gland. We found many similarities in the expression of genes previously implicated in human breast cancer, thus supporting the use of transgenic mouse models in studying aspects of human cancer. All mouse models displayed increased expression of transcription factors bearing the zinc finger motif and certain cytoskeletal proteins like tubulin 4 and tubulin 5. Paclitaxel, a cytotoxic agent and potent inducer of tubulin polymerization, has been used in combinatorial chemotherapy of advanced or metastatic breast cancer (25). Our data suggest that such compounds potentially could be tested in vivo in mouse models of cancer. Although several receptor tyrosine kinases have been targeted for breast cancer therapy, we observed suppression of several protein tyrosine kinases concomitant with the induction of protein tyrosine phosphatases, thereby identifying a unique subset of potential targets that are cancer-related but are independent of initial events in tumor formation.
We performed F tests to identify a subset of genes that constituted the oncogene-induced signatures. Hierarchical clustering with this subset of genes segregated the tumors into three major groups: the nuclear oncogene groups, (i) MMTV-myc and (ii) C3(1)-T-ag and WAP-T-ag, and the extranuclear signaling oncogene group, (iii) MMTV-neu, MMTV-ras, and MMTV-PyMT. The tight association of neu-ras-PyMT seems to be because of the convergence of the neu, PyMT, and ras pathways, as documented earlier (26). Despite the use of two different promoters to express T-ag [C3 (1) prostatein and WAP] with some distinct histopatholic differences in tumors, the T-ag tumors clustered tightly, suggesting that the oncogene was the primary determinant of gene-expression profiles.
T-ag inactivates both p53 and pRb, deregulating two important checkpoints in cell cycle. p53 seems to regulate gene expression at both the G1/S transition, by induction of cell-cycle inhibitors, and G2/M transition, by suppression of genes, including cdc2 and cdc25A (27). pRb interacts with E2F, HDAC, and Mcmd7, as well as SWI-SNF proteins that collectively result in suppression of cell-cycle genes leading to cell-cycle arrest (28). We observed induction of cdc2, cdc25A, G1 phase cyclin E, PCNA, and histone acetylases, as well as proteins involved in DNA replication, including the Mcmd family, thereby indicating that T-ag expression results in unchecked progression through both cell-cycle check points leading to increased cell duplication. The MMTV-myc tumors displayed cell-cycle gene-expression profiles that overlapped with the T-ag models. In contrast, the Neu-ras-PyMT group was characterized by the induction of cyclin D1, cdk-2, and E2F, with no apparent changes in genes involved in G2/M transition or those affecting DNA replication. This observation implies that tumors derived from Neu-ras-PyMT display events more similar to those that occur after mitogenic stimulation of cells and the activation of the ras pathway (29).
The most unique gene cluster specific to T-ag was the induction of calcium-signaling pathways and S100A4 and S100A1, which have been associated with invasive lesions in mammary models of cancer (30). Treatment of breast cancer by the antiestrogen, tamoxifen, has been clinically most successful for estrogen-dependent tumors. In addition to functionally blocking estrogen receptor (ER) signaling, tamoxifen has been shown to inhibit other enzymatic activities in the cell, like those of protein kinase C and calmodulin-dependent cAMP phosphodiesterases (31). For this reason, calmodulin inhibitors are being evaluated in vitro as potential antiproliferative and chemopreventive agents in ER-positive as well as ER-negative human breast cancer cells (32, 33). C3(1)-T-ag tumors lose ER expression during tumor progression from mammary intra-epithelial neoplasia lesions to adenocarcinomas (21). Because only the T-ag models show an increase in calcium-signaling molecules, they may be well suited for the in vivo evaluation of novel calmodulin inhibitors.
Most of the genes that were modulated in myc tumors could be classified into three major groups: (i) the cell-cycle pathway, (ii) transcription factors, and (iii) ribosomal RNA genes. Interestingly, the enhancer of zeste homolog 1 (ezh1) and hoxb8 gene have been mapped to mouse chromosomes 11 (34). Increased copy number and rearrangements of chromosome 11 have been described previously in the MMTV-myc-derived cell lines (35). Hence, the observed myc-specific induction of these genes may be a result of chromosomal aberrations in addition to increased gene expression. In contrast, the complete suppression of hey-1, a downstream effector of Notch signaling (36), was characteristic of most myc tumors analyzed, whereas it was induced in other models (Fig. 4d). It is possible that Notch signaling through hey-1 is specifically down-regulated in myc-derived tumors, because myc is known to transcriptionally repress several genes with diverse functions including CDK1, p21, H-ferritin, collagen, and fibronectin (37).
Tumors from transgenic mice overexpressing neu, ras, and PyMT clustered very tightly owing to remarkable similarities in their gene-expression profiles. However, only three changes were unique to ras-derived tumors, and nine were specific to PyMT. Many GAPs and serine-threonine kinases were up-regulated in these tumors. Increased processing of pre-pro-ligands of the epidermal growth factor super-family by induction of specific proteases belonging to the calpain and metalloprotease family has been documented previously in human cell lines and tumors bearing increased erbB2/neu expression (38). We observed the induction of calpains and MMP15 mostly in neu tumors and in some ras tumors. In addition, members of the tetraspanin family including CD9, CD81, and several ESTs similar to this family were up-regulated in MMTV-neu tumors. However, loss of CD9 expression is associated with invasive lesions in human breast carcinomas and is shown to be a marker of poor prognosis of the disease (39, 40).
Gene-based approaches have been successful in breast cancer therapy and have led to the development of several classes of drugs (41). Often, combinatorial drug therapy is more effective for the treatment of advanced cancer than single agents, where development of drug resistance is common. Microarray technology offers a high throughput screening to help identify such targets that may lead to “smart” combinatorial therapies for the treatment of clinically diverse human disease. We have profiled late-stage tumors from six mouse models of human breast cancer, thus developing a comparative platform for cancer-related genes and/or oncogene-specific pathways. However, cross-species differences may exist in gene-expression patterns, gene functions in the process of oncogenesis, and in the metabolism of the selected candidate therapeutic agent(s). Moreover, certain changes may be limited to the specific genetic background of the mouse strain used in the present study. After careful consideration of these issues, our database may be used to select a mouse model for particular purposes, including preclinical testing of potential targets depending upon the status of that pathway in the model. Moreover, comparative expression profiling of tumors at various stages of development in mouse models would profoundly enhance our knowledge of progressive genetic changes associated with oncogene-induced events.
Supplementary Material
Acknowledgments
We thank David Petersen and the staff of the Advanced Technology Center, National Cancer Institute, for printing the microarrays, Lalage Wakefield for providing MMTV-neu mice, Lisa Bailey for assistance with animal handling, and Jason Kang for useful discussions. This work was supported in part by the National Cancer Institute Mouse Models of Human Cancer Consortium.
Abbreviations
- MMTV
mouse mammary tumor virus
- EST
expressed sequence tag
- PCNA
proliferating cell nuclear antigen
References
- 1.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, et al. Nature (London) 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Perou C M, Jeffrey S S, van de Rijn M, Rees C A, Eisen M B, Ross D T, Pergamenschikov A, Williams C F, Zhu S X, Lee J C, et al. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi O P, et al. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 4.Bieche I, Lidereau R. Genes Chromosomes Cancer. 1995;4:227–251. doi: 10.1002/gcc.2870140402. [DOI] [PubMed] [Google Scholar]
- 5.Cardiff R D, Anver M R, Gusterson B A, Hennighausen L, Jensen R A, Merino M J, Rehm S, Russo J, Tavassoli F A, Wakefield L M, et al. Oncogene. 2000;19:968–988. doi: 10.1038/sj.onc.1203277. [DOI] [PubMed] [Google Scholar]
- 6.Leder A, Pattengale P K, Kuo A, Stewart T A, Leder P. Cell. 1986;45:485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 7.Bouchard L, Lamarre L, Tremblay P J, Jolicoeur P. Cell. 1989;57:931–936. doi: 10.1016/0092-8674(89)90331-0. [DOI] [PubMed] [Google Scholar]
- 8.Tremblay P J, Pothier F, Hoang T, Tremblay G, Brownstein S, Liszauer A, Jolicoeur P. Mol Cell Biol. 1989;9:854–859. doi: 10.1128/mcb.9.2.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guy C T, Cardiff R D, Muller W J. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tzeng Y J, Guhl E, Graessmann M, Graessmann A. Oncogene. 1993;8:1965–1971. [PubMed] [Google Scholar]
- 11.Maroulakou I G, Anver M, Garrett L, Green J E. Proc Natl Acad Sci USA. 1994;91:11236–11240. doi: 10.1073/pnas.91.23.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slamon D J, Clark G M, Wong S G, Levin W J, Ullrich A, McGuire W L. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 13.Wright C, Angus B, Nicholson S, Sainsbury J R, Cairns J, Gullick W J, Kelly P, Harris A L, Horne C H. Cancer Res. 1989;49:2087–2090. [PubMed] [Google Scholar]
- 14.Mietz J A, Unger T, Huibregtse J M, Howley P M. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 16.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 17.Hegde P, Qi R, Abernathy R, Gay C, Dharap S, Gaspard R, Earle-Hughes J, Snesrud E, Lee N H, Quackenbush J. BioTechniques. 2000;29:548–562. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- 18.Khan J, Simon R, Bittner M, Chen Y, Leighton S B, Pohida T, Smith P D, Jiang Y, Gooden G C, Trent J M, Meltzer P S. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- 19.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshidome K, Shibata M A, Couldrey C, Korach K S, Green J E. Cancer Res. 2000;60:6901–6910. [PubMed] [Google Scholar]
- 22.Coller H A, Grandori C, Tamayo P, Colbert T, Lander E S, Eisenman R N, Golub T R. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Q M, Malek R L, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee N H, Dang C V, Liu E T. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- 24.Lee R J, Albanese C, Fu M, D'Amico M, Lin B, Watanabe G, Haines G K, III, Siegel P M, Hung M C, Yarden Y, et al. Mol Cell Biol. 2000;20:672–683. doi: 10.1128/mcb.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esteva F J, Valero V, Pusztai L, Boehnke-Michaud L, Buzdar A U, Hortobagyi G N. Oncologist. 2001;6:133–146. doi: 10.1634/theoncologist.6-2-133. [DOI] [PubMed] [Google Scholar]
- 26.Dankort D L, Muller W J. Oncogene. 2000;19:1038–1044. doi: 10.1038/sj.onc.1203272. [DOI] [PubMed] [Google Scholar]
- 27.Taylor W R, Stark G R. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 28.Harbour J W, Dean D C. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 29.Frame S, Balmain A. Curr Opin Genet Dev. 2000;10:106–113. doi: 10.1016/s0959-437x(99)00052-0. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Rudland P S, White M R, Barraclough R. J Biol Chem. 2000;275:11141–11146. doi: 10.1074/jbc.275.15.11141. [DOI] [PubMed] [Google Scholar]
- 31.Colletta A A, Benson J R, Baum M. Breast Cancer Res Treat. 1994;31:5–9. doi: 10.1007/BF00689672. [DOI] [PubMed] [Google Scholar]
- 32.Newton C J, Eycott K, Green V, Atkin S L. J Steroid Biochem Mol Biol. 2000;73:29–38. doi: 10.1016/s0960-0760(00)00047-9. [DOI] [PubMed] [Google Scholar]
- 33.Jacobs E, Bulpitt P C, Coutts I G, Robertson J F. Anticancer Drugs. 2000;11:63–68. doi: 10.1097/00001813-200002000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Laible G, Haynes A R, Lebersorger A, O'Carroll D, Mattei M G, Denny P, Brown S D, Jenuwein T. Mamm Genome. 1999;10:311–314. doi: 10.1007/s003359900993. [DOI] [PubMed] [Google Scholar]
- 35.Weaver Z A, McCormack S J, Liyanage M, du Manoir S, Coleman A, Schrock E, Dickson R B, Ried T. Genes Chromosomes Cancer. 1999;25:251–260. [PubMed] [Google Scholar]
- 36.Leimeister C, Externbrink A, Klamt B, Gessler M. Mech Dev. 1999;85:173–177. doi: 10.1016/s0925-4773(99)00080-5. [DOI] [PubMed] [Google Scholar]
- 37.Grandori C, Cowley S M, James L P, Eisenman R N. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 38.Yarden Y, Sliwkowski M X. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 39.Shi W, Fan H, Shum L, Derynck R. J Cell Biol. 2000;148:591–602. doi: 10.1083/jcb.148.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berditchevski F, Odintsova E. J Cell Biol. 1999;146:477–492. doi: 10.1083/jcb.146.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bange J, Zwick E, Ullrich A. Nat Med. 2001;7:548–552. doi: 10.1038/87872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.