Abstract
Helicobacter hepaticus causes chronic hepatitis and liver cancer in mice. It is the prototype enterohepatic Helicobacter species and a close relative of Helicobacter pylori, also a recognized carcinogen. Here we report the complete genome sequence of H. hepaticus ATCC51449. H. hepaticus has a circular chromosome of 1,799,146 base pairs, predicted to encode 1,875 proteins. A total of 938, 953, and 821 proteins have orthologs in H. pylori, Campylobacter jejuni, and both pathogens, respectively. H. hepaticus lacks orthologs of most known H. pylori virulence factors, including adhesins, the VacA cytotoxin, and almost all cag pathogenicity island proteins, but has orthologs of the C. jejuni adhesin PEB1 and the cytolethal distending toxin (CDT). The genome contains a 71-kb genomic island (HHGI1) and several genomic islets whose G+C content differs from the rest of the genome. HHGI1 encodes three basic components of a type IV secretion system and other virulence protein homologs, suggesting a role of HHGI1 in pathogenicity. The genomic variability of H. hepaticus was assessed by comparing the genomes of 12 H. hepaticus strains with the sequenced genome by microarray hybridization. Although five strains, including all those known to have caused liver disease, were indistinguishable from ATCC51449, other strains lacked between 85 and 229 genes, including large parts of HHGI1, demonstrating extensive variation of genome content within the species.
Keywords: genomics, pathogenicity island, evolution
In 1992, an unusually high rate of liver tumors was noted in mouse colonies at the U.S. National Cancer Institute (1). An extensive search for the cause of these tumors showed that an infectious agent, Helicobacter hepaticus, infected the livers of these mice and induced chronic hepatic inflammation and subsequently liver cancer in a high percentage of animals (2). H. hepaticus infection has since been shown to be widespread in mouse colonies worldwide (3), and in addition to liver disease it has been linked to inflammatory bowel disease in immunocompromised mice (4). H. hepaticus is currently the best studied of the enterohepatic Helicobacter species, a diverse group that comprises bacteria that colonize the intestinal tracts and/or livers of susceptible hosts and that includes two human diarrhoeal pathogens, Helicobacter fennelliae and Helicobacter cinaedi (5). DNA from enterohepatic Helicobacter species has been found in patients with hepatobiliary diseases, but a causal role of the bacteria in human liver disease has not yet been established (5, 6). H. hepaticus has many features in common with Helicobacter pylori: both persistently infect their hosts, leading to chronic inflammation, and in both cases this inflammation can progress to carcinoma (7). However, H. hepaticus does not colonize the stomach, but instead shares the same lower bowel habitat with Campylobacter jejuni, the most frequent bacterial cause of diarrhea in humans. We therefore expected that a genomic comparison of H. hepaticus with H. pylori and C. jejuni would reveal new insights into host and habitat specificity of bacterial pathogens, as well as mechanisms leading to inflammation and cancer. To this end, we determined the whole genome sequence of H. hepaticus.
The sequence analysis has generated testable hypotheses about mechanisms of adaptation to the gastric vs. the enteric and hepatobiliary habitat. We also identified a putative pathogenicity island that encodes components of a type IV secretion system and other putative virulence genes. This paper provides the definitive resource for systematic functional analysis of the pathogenic and carcinogenic mechanisms of this bacterium in its natural murine host.
Materials and Methods
Genome Sequencing. H. hepaticus ATCC51449 was isolated from liver tissue in the course of the initial investigation of the outbreak of hepatitis and hepatocellular carcinoma in control mice used in carcinogenesis assays (1). The sequence was assembled from 22,034 end reads (giving 8.7× coverage) from several shotgun libraries (insert sizes, 700–3,000 bp). End and walking sequences from a cosmid library were used as a scaffold. Vectorette PCRs (8) and combinatorial PCRs were used to assemble the sequence and fill in gaps.
ORF Prediction. The identification of ORFs was performed by using multiple software packages and databases. ORFs were predicted with GLIMMER2 (9) and FLIP (N. Brossard, ftp://megasun.bch. umontreal.ca/pub/flip/flip.tar.Z), ribosome binding sites with rbs_finder (B. E. Suzek, www.tigr.org/software/), and tRNA genes with trnascan-se (10). In parallel, the fasta package (11) was used to identify orthologs of H. pylori and C. jejuni genes. Additional genes were found by comparison with the GenBank bacteria subdivision database. An automated annotation was performed with interproscan (EBI, Cambridge, U.K.). The results of these methods were evaluated and ambiguities resolved manually.
Functional Annotation. To characterize the biochemical and cellular functions of the predicted gene products, GeneData phylosopher 3.5 (GeneData, Basel) was used. Based on a large-scale comparison with 25 other complete genomes, clusters of orthologous genes were calculated. The resulting protein families (12) represented the basis for the automated functional annotation. Assignment of putative function to protein families was done by phylosopher in an automated process that takes into account existing annotations of the proteins sorted into a family. In some cases, a conserved gene order indicating operon structures provided additional information about the protein's function. Additionally, phylogenetic pattern correlations were used to assign function to uncharacterized proteins (13). The final assignment of putative functions to the H. hepaticus ORFs was done manually, by using the results of the automated annotations. A functional categorization was performed on the basis of gene ontology, a universally applicable annotation system whose three organizing principles are molecular function, biological process and cellular component (www.geneontology.org).
Microarray Hybridizations. The Massachusetts Institute of Technology H. hepaticus strain collection comprises strains from the U.S. (11 strains), The Netherlands (one strain), and Germany (one strain). All of these were used for comparison with the sequenced strain by microarray hybridizations (3). The identification of all strains as H. hepaticus was confirmed by 16S rDNA sequence analysis and phenotypical characterization. The MWG H. hepaticus array (MWG Biotech AG, Ebersberg, Germany) consists of 50-mer oligonucleotides permitting the detection of 1,863 of the 1,875 ORFs. The design strategy for MWG arrays has been described (14). Fluorescent labeling of DNAs and competitive hybridizations were essentially performed as described by Salama et al. (15) (more details are available in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org). Microarray scanning and data processing were performed as described previously (14). Categorization of genes as “present” and “missing” was done with the program GACK (16), which uses the signal-ratio distribution rather than a fixed cutoff. The total or partial absence of HHGI1 from seven H. hepaticus isolates was confirmed by PCR analyses using primers in ORFs flanking the island (empty site PCR), as well as primers targeting representative ORFs within the island. The primer sequences are available on request. The precise location of the deletion was determined by sequencing of the empty site PCR product. The absence of representative ORFs from the other strains was similarly verified.
Results and Discussion
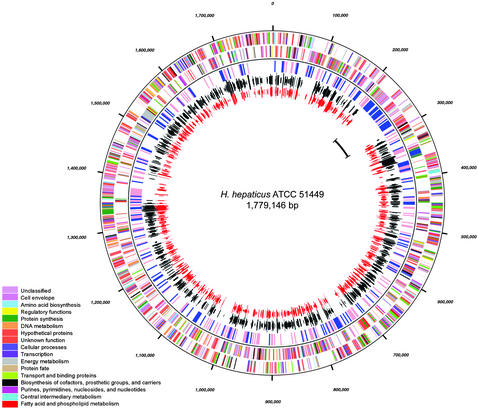
General Features of the H. hepaticus Genome. The genome of H. hepaticus strain ATCC51449 (Fig. 1), with 1,799,146 bp (G+C content 35.9%), is slightly larger than the genomes of both H. pylori (1.64 and 1.67 Mbp) and C. jejuni (1.64 Mbp) (17–19). Its general features are summarized in Table 1. Of the 1,875 predicted proteins, a function could be assigned to 713 (38.0%) with a high level of confidence, 673 (35,9%) were conserved hypothetical proteins (309 with some evidence of the function, 364 without assignment of function), and 489 proteins (26.1%) had no significant database match (Table 2, which is published as supporting information on the PNAS web site). A total of 938 and 941 (50,2%) of the H. hepaticus ORFs have orthologs in the completely sequenced H. pylori strains 26695 and J99, respectively, and 953 (50.8%) have an ortholog in C. jejuni NCTC 11168. A total of 821 H. hepaticus proteins have the same ortholog in both H. pylori and C. jejuni. A total of 109 H. hepaticus ORFs have orthologs in both H. pylori genomes, but none in C. jejuni. A total of 130 H. hepaticus ORFs have an ortholog in C. jejuni but lack one in H. pylori.
Fig. 1.
Circular representation of the H. hepaticus genome. From the outside to the inside, the first two circles represents the positions of coding sequences transcribed in clockwise and anticlockwise direction, respectively. The colors represent the functional categories of the encoded proteins, as shown in the color legend. The third circle depicts areas of the chromosome where the G+C content is >5% higher (pink) or lower (blue) than the average G+C content (window size = 2000). The fourth and fifth circles represent genes with orthologs in H. pylori (black) and C. jejuni (red). The length of the lines representing the orthologs is proportional to the percentage of amino acid identity. The position of the HHGI1 genomic island is marked in the innermost circle.
Table 1. General features of the H. hepaticus genome.
| Total size, bp | 1,799,146 |
| GC content, % | 35.9 |
| Coding sequences | 1,875 |
| Average gene length, bp | 1,082 |
| Coding density, % | 93.04 |
| Predicted secreted proteins | 347 |
| Predicted membrane proteins | 358 |
| Predicted proteins with assigned functions | 1,022 |
| Ribosomal RNA | 1 × 16S-23S-5S |
| tRNA | 37 (7 clusters, 15 single genes) |
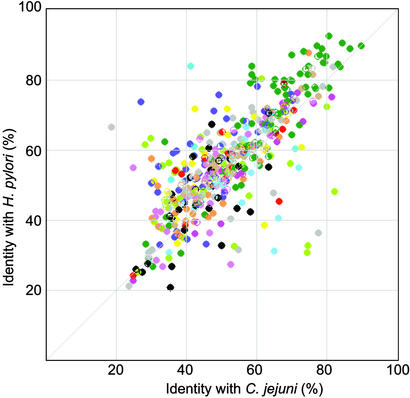
The average percentage of amino acid sequence identity was 60.0% between H. hepaticus proteins and their H. pylori orthologs, and 54.3% between H. hepaticus proteins and their C. jejuni orthologs. However, among the H. hepaticus genes with orthologs in both H. pylori and C. jejuni, there are some that encode proteins much more similar to their C. jejuni than their H. pylori orthologs or vice versa (Fig. 2). H. hepaticus proteins more similar to C. jejuni than H. pylori include HH0646 encoding ferredoxin (FrxA) and several enzymes involved in biotin metabolism (BioA, BioC, BioF). Examples for proteins much more similar to H. pylori than C. jejuni include many flagellar and chemotaxis proteins, such as FlaA, FlaB, FlgK, CheV, or FliS. Such proteins with unusually high homology to either H. pylori or C. jejuni are likely to be the result of strong selection for protein properties favourable in the gut or gastric environment, respectively, and are candidates for future functional studies aimed at identifying proteins involved in the specificity of a pathogen for its particular ecological niche.
Fig. 2.
Similarity of H. hepaticus proteins that have orthologs with known function in both H. pylori and C. jejuni with their H. pylori 26695 and C. jejuni orthologs. The color coding is as in Fig. 1. Each of the axes represents the percentages of amino acid identity with the H. pylori and C. jejuni orthologs, respectively.
Urease Production, Nickel and Iron Uptake. H. hepaticus, like H. pylori produces large amounts of urease. The function of urease during H. hepaticus infection is currently not known, but urease is essential for colonization (E. Chin, J. Sohn, V. Young, M. Villar-Prados, M. Whary, J.G.F., and D.B.S., unpublished data). One prerequisite for high level urease activity is the ability to acquire sufficient amounts of nickel from the environment. H. hepaticus has a urease gene cluster (ureABIEFGH) similar to that of H. pylori (20–22). Downstream of the urease gene cluster, transcribed in the opposite direction, is a cluster of homologs of E. coli nickel transport genes [nikAB(C+D)E, ref. 23), not present in H. pylori or C. jejuni. This predicted nickel uptake ABC transporter system is likely to complement for the absence in H. hepaticus of orthologs of the H. pylori nickel transporter NixA, the C-terminal nickel-binding domain of the HspA heat shock protein, and the histidine-rich protein Hpn, which are all involved in nickel trafficking by H. pylori (22). The urease system illustrates that the H. hepaticus and H. pylori genomes encode different combinations of subsystems that could either have been acquired from different sources, or were once jointly present in an ancestor and subsequently partly deleted during the adaptation of each species to its respective habitat. In contrast to the differences in nickel uptake, uptake systems for ferric (Fe3+) and ferrous (Fe2+) iron as well as other ions (potassium, sodium, copper, magnesium, molybdenum) are very similar between H. hepaticus, H. pylori, and C. jejuni.
Motility and Chemotaxis. The flagellar biosynthesis system of H. hepaticus is similar to that of H. pylori, with genes encoding two flagellin types FlaA and FlaB under control of respective σ28 and σ54 promoters. Remarkably, there are two identical copies of flaA, including the promoter (HH1364 and HH1653), indicating a relatively recent duplication. The duplicated flaA genes were present in all 13 H. hepaticus strains tested, as shown by PCR with one primer binding in flaA and one outside the repeat (in HH1365 for the first and HH1654 for the second copy). The significance of this duplication is currently unknown. Inactivation of one of the copies of flaA (HH1364) did not affect flagellar biosynthesis and motility (S. Ragnum and D.B.S., unpublished data). Major differences between the motility systems of H. pylori and the Salmonella paradigm have been described (24), and H. hepaticus shares these differences. H. hepaticus possesses nine (H. pylori: 4, C. jejuni: 10) predicted chemosensor proteins, suggesting that H. hepaticus can recognize a larger number of chemicals for spatial orientation than H. pylori, consistent with its more diverse habitat. Like almost all other motile bacteria with the exception of H. pylori, H. hepaticus has a pair of cheR/cheB genes encoding a protein methyl transferase and methyl esterase involved in chemotaxis adaptation. Only one of the putative chemosensors (HH1088) has a methylation motif (EQVAAS) that fully matches the consensus sequence (GlxGlxXXA-S/T) (25). However, all of the other H. hepaticus putative chemosensors have one or multiple Glx–Glx motifs followed by varying amino acid residues. Evidence from other bacteria suggests that at least some variant motifs can become methylated, albeit less efficiently than consensus motifs (25). Similar variant methylation motifs exist in C. jejuni (which has cheB/cheR) and H. pylori (which lacks cheB/cheR). Thus, the role of methylation and of the different sensor proteins in chemotaxis adaptation of the three organisms remains to be clarified.
Toxins, Adhesins, and Outer Membrane Proteins. H. hepaticus lacks orthologs of most colonization and virulence factors of H. pylori. There is no ortholog of the H. pylori vacuolating cytotoxin gene vacA (26). Like C. jejuni, H. hepaticus has a cluster of genes (cdtABC) encoding a cytolethal distending toxin (27). C. jejuni cytolethal distending toxin (CDT) causes cell cycle arrest, chromatin fragmentation, and eventually apoptotic cell death by a type I DNase-like activity (28). Although CDT may cause only limited damage in acute infections, such as those caused by C. jejuni, its genotoxic effects may contribute to carcinogenesis in persistent H. hepaticus infection.
In contrast to C. jejuni (19), H. hepaticus has 11 genes that encode proteins with homology to the large family of H. pylori outer membrane proteins (17) (Fig. 4, which is published as supporting information on the PNAS web site). This family of 33 paralogous genes has been subdivided into two subfamilies, named Hop and Hor (29). Members of the Hop subfamily, which comprises the H. pylori adhesins BabA, SabA, and AlpA/AlpB, as well as the porins HopA–E, all contain the cleavable N-terminal motif AEX[D,N]G, whereas the Hor proteins, whose function is still unknown, lack this motif. Detailed sequence comparisons of the H. hepaticus OMPs with the other outer membrane proteins do not allow clear conclusions about their functions. None of the H. hepaticus proteins contains the characteristic N-terminal motif of the Hop proteins. In a phylogenetic tree that includes the H. hepaticus OMPs, the Hop and Hor proteins and selected E. coli porins, five proteins (HH0525, HH1713, HH0661, HH1543, and HH0812) cluster with the E. coli porins (Fig. 4a), suggesting that these proteins may in fact represent porins. The other OMPs form several smaller clusters, generally most related to Hor proteins, such as HorG. Surprisingly, the H. hepaticus OMPs do not have noticeable similarity to the few characterized outer membrane proteins of C. jejuni.
With the exception of three proteins with homology to basic components of a type IV secretion system (see below), H. hepaticus has no orthologs of genes of the H. pylori cag pathogenicity island (30), which has been implicated in the pathogenesis of inflammation and carcinogenesis by H. pylori. There is also no evidence for the presence of a type III secretion system, except for the flagellar apparatus. The lack of orthologs of most well characterized genes involved in the pathogenesis of H. pylori infection plausibly explains why H. hepaticus does not colonize the stomach, and underlines the complexity of the adaptation of H. pylori to the gastric niche.
The molecular basis of C. jejuni colonization and virulence is still not well understood, permitting only more limited comparisons between C. jejuni and H. hepaticus. Three proteins, PEB1 (also called CBF1) (31), CadF (32), and JlpA (33) have been shown to be involved in adhesion of C. jejuni to epithelial cells. H. hepaticus has a protein (HH1481) with strong homology (72% amino acid identity) to PEB1, which is not present in H. pylori. HH1481 is thus a candidate adhesin that might be involved in intestinal colonization by H. hepaticus. H. hepaticus and H. pylori both lack orthologs of CadF and JlpA (Cj0983). The latter finding is consistent with a report that jlpA (together with the hippurate hydrolase gene hipO) has been acquired by C. jejuni quite recently, and was not present in the common ancestor of C. jejuni and Campylobacter coli (33).
H. hepaticus has a well-conserved prepilin peptidase gene (HH0603), in addition to a classical signal peptidase I (HH1367), as well as a type IV pilin gene (HH1285) and other pilus-related genes (HH1115–1117) indicating that H. hepaticus may be able to assemble type IV pili, which might play a role in adhesion and/or in DNA uptake. Pili have not yet been observed in H. hepaticus and the function of the type IV pilus genes remains to be elucidated.
Transcriptional Regulation and Contingency Genes. H. hepaticus possesses only a small set of genes encoding transcriptional regulators (three sigma factors, σ70, σ54, σ28, one flagellar anti-sigma factor, FlgM, and a flagellar transcriptional activator, FlgR). Similar to both H. pylori (17, 18) and C. jejuni (19), this dearth of dedicated regulators is compensated by abundant “contingency genes” predicted to be phase variable because of slipped strand mispairing-mediated length variations in homopolymeric or dinucleotide repeats. At 36 positions (17 in coding regions), length variation of poly(G) or poly(C) tracts was observed in different shotgun clones, indicating that phase variation occurs so frequently that significant heterogeneity developed in the very few passages required from single colony isolation to DNA preparation (Table 3, which is published as supporting information on the PNAS web site). In addition, there are 33 more genes likely to exhibit phase variation, even if not observed in the shotgun clones. Three of the genes with observed phase variation and five of the hypothetical phase-variable genes encode fucosyl transferases or other glycosyl transferases. Phase variable fucosyl transferases play a role in lipopolysaccharide (LPS) modification and antigenic mimicry in H. pylori (34), suggesting that LPS antigenic variation may also contribute to immune evasion by H. hepaticus.
Restriction Modification Systems and Natural Competence. In contrast to H. pylori with its very large number of restriction–modification systems, H. hepaticus has only two complete restriction-modification systems (HH238/239 and HH1050/1051). Like H. pylori, H. hepaticus is naturally transformable (T.S. and S.S., unpublished data). H. hepaticus lacks the unusual type IV secretion system (comB locus) required for natural competence in H. pylori (35). However, the presence of components of a type IV pilus biogenesis machinery suggests that H. hepaticus might be able to take up DNA via a type IV pilus-like structure.
Respiratory Chain and Citric Acid Cycle. The metabolic capabilities of H. hepaticus, H. pylori and C. jejuni as inferred from the genome sequences are generally quite similar. However, there are notable differences that offer interesting insights into the basic physiology of the three organisms. One example is the respiratory chain of the three species. Energy generation by the electron transport chain depends on the pathways for proton extrusion leading to ATP synthesis, via proton reentry through ATP synthase. The types of dehydrogenases and oxidases expressed in the respiratory chain of a bacterium determine proton export and thus control the efficiency of ATP synthesis. H. hepaticus has the possibility of expressing an NDH-1 or NDH-2 dehydrogenase as well as a cytochrome bd or a cytochrome cbb3 terminal oxidase, and thus has the most versatile respiratory chain of all three species, which may allow this bacterium to adapt to very different environments such as those of the intestinal tract and hepatobiliary tree. The respiratory chain of H. pylori with only an NDH-1 dehydrogenase and a cytochrome cbb3 terminal oxidase in contrast is less versatile, but may have evolved to provide the highest possible number of outward translocated protons per electron transferred to oxygen (36).
The three genomes encode all of the enzymes of the tricarboxylic acid branch of the citric acid cycle, suggesting that this segment operates in the oxidative direction. However, there are important differences between the three bacteria in the genes encoding putative enzymes of the dicarboxylic acid segment of the cycle. Genes encoding the five enzymes oxidizing metabolites from α-ketoglutarate to oxaloacetate have been annotated in the genome of C. jejuni, suggesting the presence of a full oxidative cycle in this bacterium. The H. pylori and H. hepaticus genomes lack four and three of these genes, respectively. Based on biochemical and genomic data, two proposals have been put forward for the H. pylori cycle, a branched pathway in which this segment functions in the reductive direction (37), or an oxidative cyclic pathway in which the functions of the four missing enzymes are substituted by α-ketoglutarate/ferredoxin oxidoreductase, succinyl-CoA acetoacetyl-CoA transferase (SCOT), fumarate reductase and malate/quinone oxidoreductase encoded by its genome (38). With the exception of SCOT, the genes coding for these other three enzymes are present in the C. jejuni and H. hepaticus genomes. The absence of gene coding for a SCOT and the presence of ORF HH1571 encoding a malate dehydrogenase suggest that the dicarboxylic acid branch of the H. hepaticus citric acid cycle functions in the reductive direction, a characteristic of the cycle of many anaerobes. This would be in agreement with the more stringent microaerobic conditions required to culture H. hepaticus than C. jejuni or H. pylori.
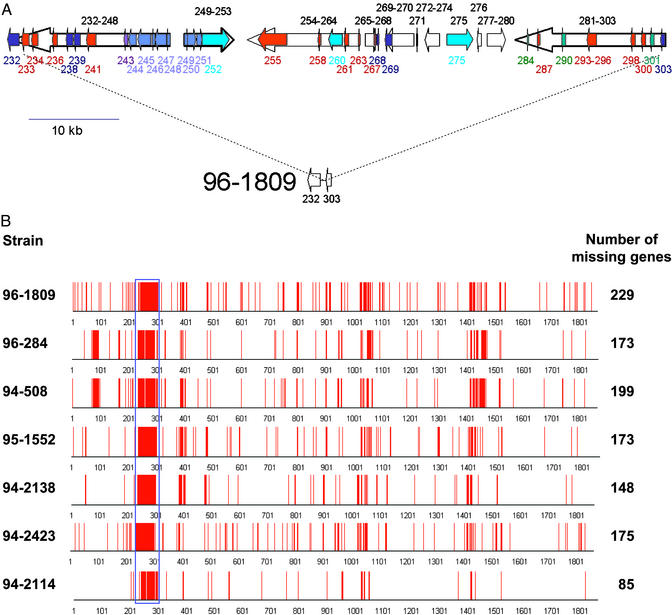
The Genomic Island HHGI1, a Candidate Pathogenicity Island. The H. hepaticus genome contains one large region as well as numerous smaller regions that differ from the rest of the chromosome by their G+C content, suggesting that they may have been acquired by horizontal gene transfer (Fig. 1). The largest region, termed H. hepaticus genomic island 1 (HHGI1, G+C content 33.2%) contains 70 ORFs (HH0233-HH0302, 71.0 kb, Fig. 3A). Most genes within HHGI1 encode hypothetical proteins. However, the island harbors three proteins with homology to structural components of type IV secretion systems. The percentages of amino acid identity and similarity were 7% and 13% for HH252 and A. tumefaciens VirB10, 11% and 24% for HH275 and VirD4, and 12% and 29% for HH260 and VirB4 (for sequence alignments, see Fig. 5, which is published as supporting information on the PNAS web site). HH252 also has high similarity to IcmF of Vibrio cholerae and Legionella pneumophila, a protein involved in macrophage killing and bacterial conjugation (39). The island also contains a gene with homology to Vibrio cholerae hcp, which encodes a secreted protein coregulated with the V. cholerae hemolysin (40). Furthermore, the island contains a gene cluster (HH244–251) with significant homology to clusters of genes of unknown function on the small chromosome of V. cholerae (VCA0107-0115) and the Yersinia pestis genome. Unlike many pathogenicity islands, HHGI1 is not associated with a tRNA gene, and not flanked by direct repeats. However, it contains a prophage P4-like integrase gene (HH269), a feature that has been found in several pathogenicity islands (41). Taken together, the presence of secretion system components and several secreted virulence proteins strongly suggests that HHGI1 is a pathogenicity island.
Fig. 3.
(A) The large genomic island of H. hepaticus (HHGI1). Red arrows represent genes encoding putative membrane-associated proteins, green arrows represent genes encoding proteins with a leader peptide, and blue arrows indicate genes coding for apparent homologs of other bacterial proteins (light blue, V. cholerae VCA0107–VCA0115; violet, V. cholerae Hcp; dark blue, proteins from other bacteria). Turquoise arrows indicate the three genes that encode proteins (HH0252, HH0259, and HH0275) with homology to VirB10, VirD4, and VirB4. Some smaller ORFs transcribed in the same orientation are not depicted individually but shown as open arrows representing a block of genes. The lower part of the figure shows the same genomic region in one strain, 96-1809, where the complete island is lacking and where only the flanking sequences, HH0232 and HH0303, are present. (B) Genomic variation in H. hepaticus. Twelve H. hepaticus strains were tested for hybridization with a whole-genome DNA microarray. Five strains (ATCC51448, ATCC51450, 95-225, 95-557, and 94-739) contained all genes detected by the array. The other seven strains did not hybridize with 85-229 probes, and the positions of these missing genes in the genome of the sequenced strain ATCC51449 are indicated by red lines. The location of the genomic island HHGI1 that is totally or partially deleted in all seven strains is indicated by the blue rectangle. The array experiments identified six more clusters of at least five genes that were not detected in at least one of the strains.
Genome Content Variation in Different H. hepaticus Strains. Because genomic islands and pathogenicity islands are frequently not present in all strains of a species, we analyzed the gene content for 12 other H. hepaticus strains by hybridization with a whole-genome DNA microarray. Five of these strains, all isolated in the U.S., contained all of the genes present in ATCC51449. However, the other seven strains did not hybridize with probes for 85–229 genes, and all of these lacked large parts or all of HHGI1 (Fig. 3B). Because all seven strains differed in the number of remaining HHGI1 genes, it seems most likely that they have arisen from a HHGI1-carrying ancestor by one or multiple steps of deletion and/or rearrangement. Although not flanked by repeats, HHGI1 contains several long tandem repeats (up to 1,007 bp of perfect identity), including two copies of a 330-bp repeat, one of which is situated at the very 5′ border (Fig. 3A). Such repeats may have played a role in these deletions, which is supported by the genomic configuration in strain 96-1809, where the deletion point is located within the 330-bp repeat (Fig. 3A).
The lack of the HHGI1 island in some strains raised the question whether strains carrying the island are more virulent than strains lacking parts of the island. Pathology records were available for all mice from whom HHGI1-carrying strains had been isolated, and four of seven mice infected with a strain lacking HHGI1. Five of the six mice infected with HHGI1-carrying strains had liver disease, whereas none of the four mice infected with HHGI1-negative strains showed evidence of liver disease. The available information about the H. hepaticus strains tested is summarized in Table 4, which is published as supporting information on the PNAS web site. These data are consistent with a higher virulence of HHGI1-carrying strains, and experiments are now in progress to systematically investigate the role of the HHGI1 island in H. hepaticus-induced liver disease.
Summary. The genome of H. hepaticus exhibits a unique combination of features from H. pylori, C. jejuni, as well as other enteric bacteria such as V. cholerae and E. coli. Together with 489 species-specific genes, they make H. hepaticus an organism with a unique habitat and pathogenic potential. Although the absence of many H. pylori colonization and virulence factors explains the inability of H. hepaticus to colonize the stomach, and extensive physiological similarities with C. jejuni are likely to be involved in enteric colonization, the reasons for its tropism for the hepatobiliary tract and, in particular, its carcinogenic potential are not immediately apparent from the genome sequence. Because both the pathogen, H. hepaticus, and its host, are amenable to genetic manipulation, the availability of the genome sequence now provides the opportunity for a systematic exploration of the mechanisms of tissue tropism and carcinogenesis induced by H. hepaticus, and, by way of comparative functional genomics, by H. pylori.
Supplementary Material
Acknowledgments
We thank Eike Niehus, Allison Stack, and Fang Ye for help with the microarray analysis, and the Phylosopher development team for support of the Phylosopher analyses. Work in S.S.'s laboratory was funded by the Bundesministerium für Bildung und Forschung PathoGenoMik network and Deutsche Forschungsgemeinschaft Grant SFB479/A5. The work was supported by National Institutes of Health Grants R01AI50952, R01CA67529, and P01CA26723 (to J.G.F.) and DK52413 (to D.B.S.). G.L.M. is thankful for the support of the Australian Research Council.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AE017125). The annotated sequence and further information are available at www.THE-MWG.com.
References
- 1.Ward, J. M., Fox, J. G., Anver, M. R., Haines, D. C., George, C. V., Collins, M. J. J., Gorelick, P. L., Nagashima, K., Gonda, M. A., Gilden, R. V., et al. (1994) J. Natl. Cancer Inst. 86, 1222-1227. [DOI] [PubMed] [Google Scholar]
- 2.Fox, J. G., Dewhirst, F. E., Tully, J. G., Paster, B. J., Yan, L., Taylor, N. S., Collins, M. J. J., Gorelick, P. L. & Ward, J. M. (1994) J. Clin. Microbiol. 32, 1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saunders, K. E., McGovern, K. J. & Fox, J. G. (1997) J. Clin Microbiol. 35, 2859-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahill, R. J., Foltz, C. J., Fox, J. G., Dangler, C. A., Powrie, F. & Schauer, D. B. (1997) Infect. Immun. 65, 3126-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solnick, J. V. & Schauer, D. B. (2001) Clin. Microbiol. Rev. 14, 59-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox, J. G., Dewhirst, F. E., Shen, Z., Feng, Y., Taylor, N. S., Paster, B. J., Ericson, R. L., Lau, C. N., Correa, P., Araya, J. C., et al. (1998) Gastroenterology 114, 755-763. [DOI] [PubMed] [Google Scholar]
- 7.Suerbaum, S. & Michetti, P. (2002) N. Engl. J. Med. 347, 1175-1186. [DOI] [PubMed] [Google Scholar]
- 8.Arnold, C. & Hodgson, I. J. (1991) PCR Methods Appl. 1, 39-42. [DOI] [PubMed] [Google Scholar]
- 9.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson, W. R. (2000) Methods Mol. Biol. 132, 185-219. [DOI] [PubMed] [Google Scholar]
- 12.Tatusov, R. L., Koonin, E. V. & Lipman, D. J. (1997) Science 278, 631-637. [DOI] [PubMed] [Google Scholar]
- 13.Freiberg, C. (2001) Drug Discov. Today 6, Suppl., S72-S80. [Google Scholar]
- 14.Josenhans, C., Niehus, E., Amersbach, S., Hörster, A., Betz, C., Drescher, B., Hughes, K. T. & Suerbaum, S. (2002) Mol. Microbiol. 43, 307-322. [DOI] [PubMed] [Google Scholar]
- 15.Salama, N., Guillemin, K., McDaniel, T. K., Sherlock, G., Tompkins, L. & Falkow, S. (2000) Proc. Natl. Acad. Sci. USA 97, 14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, C. C., Joyce, E. A., Chan, K. & Falkow, S. (2002) Genome Biol. 3, RESEARCH0065. [DOI] [PMC free article] [PubMed]
- 17.Tomb, J.-F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., Ketchum, K. A., Klenk, H. P., Gill, S., Dougherty, B. A., et al. (1997) Nature 388, 539-547. [DOI] [PubMed] [Google Scholar]
- 18.Alm, R. A., Ling L.-S. L., Moir D. T., King, B. L., Brown, E. D., Doig, P. C., Smith, D. R., Noonan B., Guild, B. C., deJonge, B. L., et al. (1999) Nature 397, 176-180. [DOI] [PubMed] [Google Scholar]
- 19.Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., Chillingworth, T., Davies, R. M., Feltwell, T., Holroyd, S., et al. (2000) Nature 403, 665-668. [DOI] [PubMed] [Google Scholar]
- 20.Labigne, A., Cussac, V. & Courcoux, P. (1991) J. Bacteriol. 173, 1920-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beckwith, C. S., McGee, D. J., Mobley, H. L. & Riley, L. K. (2001) Infect. Immun. 69, 5914-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mobley, H. L. (2001) in Helicobacter pylori: Molecular and Cellular Biology, eds. Achtman, M. & Suerbaum, S. (Horizon Scientific, Wymondham, U.K.), pp. 155-170.
- 23.Navarro, C., Wu, L. F. & Mandrand-Berthelot, M. A. (1993) Mol. Microbiol. 9, 1181-1191. [DOI] [PubMed] [Google Scholar]
- 24.Josenhans, C. & Suerbaum, S. (2001) in Helicobacter pylori: Molecular and Cellular Biology, eds. Achtman, M. & Suerbaum, S. (Horizon Scientific, Wymondham, U.K.), pp. 171-184.
- 25.Le Moual, H. & Koshland, D. E., Jr. (1996) J. Mol. Biol. 261, 568-585. [DOI] [PubMed] [Google Scholar]
- 26.Cover, T. L., Tummuru, M. K. R., Cao, P., Thompson, S. A. & Blaser, M. J. (1994) J. Biol. Chem. 269, 10566-10573. [PubMed] [Google Scholar]
- 27.Young, V. B., Knox, K. A. & Schauer, D. B. (2000) Infect. Immun. 68, 184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lara-Tejero, M. & Galan, J. E. (2000) Science 290, 354-357. [DOI] [PubMed] [Google Scholar]
- 29.Alm, R. A., Bina, J., Andrews, B. M., Doig, P., Hancock, R. E. & Trust, T. J. (2000) Infect. Immun. 68, 4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Censini, S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pei, Z., Burucoa, C., Grignon, B., Baqar, S., Huang, X. Z., Kopecko, D. J., Bourgeois, A. L., Fauchere, J. L. & Blaser, M. J. (1998) Infect. Immun. 66, 938-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konkel, M. E., Garvis, S. G., Tipton, S. L., Anderson, D. E., Jr., & Cieplak, W., Jr. (1997) Mol. Microbiol. 24, 953-963. [DOI] [PubMed] [Google Scholar]
- 33.Jin, S., Joe, A., Lynett, J., Hani, E. K., Sherman, P. & Chan, V. L. (2001) Mol. Microbiol. 39, 1225-1236. [DOI] [PubMed] [Google Scholar]
- 34.Wang, G., Ge, Z., Rasko, D. A. & Taylor, D. E. (2000) Mol. Microbiol. 36, 1187-1196. [DOI] [PubMed] [Google Scholar]
- 35.Hofreuter, D., Odenbreit, S. & Haas, R. (2001) Mol. Microbiol. 41, 379-391. [DOI] [PubMed] [Google Scholar]
- 36.Smith, M. A., Finel, M., Korolik, V. & Mendz, G. L. (2000) Arch. Microbiol. 174, 1-10. [DOI] [PubMed] [Google Scholar]
- 37.Pitson, S. M., Mendz, G. L., Srinivasan, S. & Hazell, S. L. (1999) Eur. J. Biochem. 260, 258-267. [DOI] [PubMed] [Google Scholar]
- 38.Kather, B., Stingl, K., van der Rest, M. E., Altendorf, K. & Molenaar, D. (2000) J. Bacteriol. 182, 3204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal, G., Purcell, M. & Shuman, H. A. (1998) Proc. Natl. Acad. Sci. USA 95, 1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams, S. G., Varcoe, L. T., Attridge, S. R. & Manning, P. A. (1996) Infect. Immun. 64, 283-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hacker, J., Blum-Oehler, G., Mühldorfer, I. & Tschäpe, H. (1997) Mol. Microbiol. 23, 1089-1097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.