Abstract
The eukaryotic spindle assembly checkpoint (SAC) monitors microtubule attachment to kinetochores and prevents anaphase onset until all kinetochores are aligned on the metaphase plate. In higher eukaryotes, cytoplasmic dynein is involved in silencing the SAC by removing the checkpoint proteins Mad2 and the Rod–Zw10–Zwilch complex (RZZ) from aligned kinetochores (Howell, B.J., B.F. McEwen, J.C. Canman, D.B. Hoffman, E.M. Farrar, C.L. Rieder, and E.D. Salmon. 2001. J. Cell Biol. 155:1159–1172; Wojcik, E., R. Basto, M. Serr, F. Scaerou, R. Karess, and T. Hays. 2001. Nat. Cell Biol. 3:1001–1007). Using a high throughput RNA interference screen in Drosophila melanogaster S2 cells, we have identified a new protein (Spindly) that accumulates on unattached kinetochores and is required for silencing the SAC. After the depletion of Spindly, dynein cannot target to kinetochores, and, as a result, cells arrest in metaphase with high levels of kinetochore-bound Mad2 and RZZ. We also identified a human homologue of Spindly that serves a similar function. However, dynein's nonkinetochore functions are unaffected by Spindly depletion. Our findings indicate that Spindly is a novel regulator of mitotic dynein, functioning specifically to target dynein to kinetochores.
Introduction
The spindle assembly checkpoint (SAC) is critical for preventing the onset of anaphase until all chromosomes are aligned on the metaphase plate. A single misaligned kinetochore is sufficient to generate a wait anaphase signal, thereby ensuring that all sister chromatids segregate to opposite ends of the spindle and are equally distributed to the daughter cells. Failure of the SAC can lead to premature anaphase onset and aneuploidy (Liu et al., 2003; Kops et al., 2005b; for review see Kadura and Sazer, 2005). Such defects can have consequences for a whole organism, as mice that lack a full complement of SAC genes have more frequent DNA segregation errors and are more susceptible to tumor development (Baker et al., 2005).
The presence of the SAC was initially inferred from observations that cells delay in metaphase when meiotic sex chromosomes fail to pair and align or after the spindle is perturbed by either microtubule poisons or microsurgery. Molecules responsible for the SAC were later identified in yeast genetic screens and named Mad1, -2, and -3 (Mad for mitotic arrest deficient) and Bub1, -2, and -3 (Bub for budding unperturbed by benzimidazole). Subsequent work showed that these proteins together with the MPS1 kinase form distinct complexes that target to the kinetochore (for reviews see Lew and Burke, 2003; Kadura and Sazer, 2005; Malmanche et al., 2006; Musacchio and Salmon, 2007). Two additional metazoan checkpoint proteins, Zw10 and Rough Deal (Rod), were later isolated as cell cycle mutants in Drosophila melanogaster. These two proteins, together with a third protein called Zwilch, form a complex (Rod–Zw10–Zwilch complex [RZZ]) that regulates the levels of Mad1 and Mad2 on the kinetochore (for review see Karess, 2005).
Ultimately, the SAC pathway must lead to inhibition of the anaphase-promoting complex (APC), a multisubunit ubiquitin E3 ligase that targets multiple mitotic regulators (e.g., mitotic cyclins as well as the securin protein that inhibits the cleavage of cohesin molecules) for proteosome degradation to allow mitotic exit (Acquaviva and Pines, 2006). Several studies have shown that localization of the checkpoint proteins to misaligned kinetochores is essential for establishing the SAC and keeping the APC inhibited, most likely by generating a diffusible signal that inhibits the APC (Taylor et al., 2004; Pinsky and Biggins, 2005; for review see Musacchio and Salmon, 2007). The nature of the diffusible signal is still subject to debate. However, a current model suggests that the kinetochore-bound Mad1–Mad2 complex acts as a template that coverts the free, inactive Mad2 to an active form that can diffuse away from the kinetochore and bind to and sequester Cdc20, a regulatory component of the APC (for review see Musacchio and Salmon, 2007).
The capture of microtubules by the kinetochore and the downstream activity of two different microtubule motors are required for silencing the SAC in metazoans. One of these motors is the kinesin centromere protein (CENP) E, which may act as a tension sensor that, when stretched, inactivates the BubR1-dependent inhibition of Cdc20 (Chan et al., 1999; Mao et al., 2005). The second motor is dynein, which transports Mad1, Mad2, and RZZ from the kinetochore to the spindle pole (Howell et al., 2001; Wojcik et al., 2001). Dynein-based removal of Mad1 and Mad2 from the kinetochore may disrupt the template mechanism that generates the active Mad2 that inhibits the APC (De Antoni et al., 2005; for review see Musacchio and Salmon, 2007). After inhibition or depletion of dynein or its cofactors, metazoan cells arrest in metaphase with correctly aligned chromosomes and high levels of kinetochore-bound Mad1, Mad2, and RZZ.
Resolving the mechanism of dynein recruitment to kinetochores is important for understanding how kinetochore– microtubule binding ultimately leads to inactivation of the SAC. Currently, it is thought that dynein is brought to the kinetochore by binding directly to dynactin (a multisubunit complex required for multiple dynein functions; Schroer, 2004), which, in turn, binds to the Zw10 subunit of the RZZ complex (Starr et al., 1998). Lis1, another dynein cofactor, also has been proposed to play a role in targeting dynein to kinetochores (Dzhindzhev et al., 2005). Dynactin, Lis1, and Zw10 are not kinetochore- specific factors, as they are involved in targeting dynein to multiple other locations in the cell (Cockell et al., 2004; Hirose et al., 2004). It has not been clearly established whether dynactin and Lis1 are sufficient for targeting dynein to kinetochores or whether other proteins might be involved.
To find new proteins that might participate in the SAC, we undertook an automated 7,200 gene mitotic index RNAi screen in S2 cells. This screen uncovered a novel gene, which we also identified in an independent screen of genes involved in S2 cell spreading and morphology. We show that this protein (termed Spindly) localizes to microtubule plus ends in interphase and to kinetochores during mitosis. Cells depleted of Spindly arrest in metaphase with high levels of Mad2 and Rod on aligned kinetochores, a defect caused by a failure to recruit dynein to the kinetochore. However, Spindly is not required for other dynein functions during interphase and mitosis. We also identify a human homologue of Spindly, which is similarly involved in recruiting dynein to kinetochores. Thus, our results have uncovered a novel conserved dynein regulator that is involved specifically in dynein's function in silencing the SAC.
Results
Identification of Spindly in two RNAi screens
Using a double-stranded RNA (dsRNA) library corresponding to ∼7,200 Drosophila genes (Echard et al., 2004), we performed two screens using Drosophila S2 cells (Fig. 1, b–d). The first screen measured mitotic index (the percentage of phosphohistone H3–positive cells in a population; see Materials and methods). In the second screen, the shape of S2 cells (spread on concanavalin A [Con A]–coated surfaces; Rogers et al., 2003) was evaluated by visual inspection.
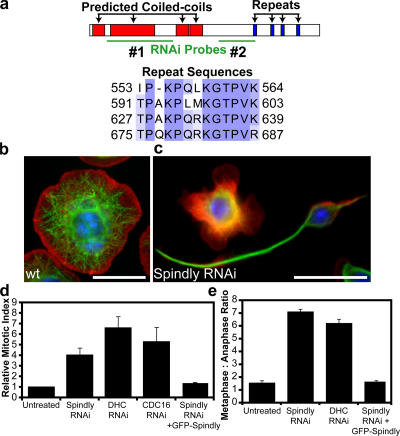
Figure 1.
RNAi of Spindly alters cell morphology and causes mitotic arrest in Drosophila S2 cells. (a) Domains of the Spindly protein showing predicted coiled-coil sequences in red and repeated motifs in blue; sequence alignment of residues in the repeat motifs is shown below. The locations of two nonoverlapping dsRNAs used to deplete Spindly are shown in green. A third dsRNA to the 3′ UTR was also used (not depicted). (b and c) Wild-type (wt) S2 cells show a uniformly spread morphology (b), whereas Spindly RNAi-treated cells (c) show marked defects in the actin lamellae as well as increased numbers of cells with long microtubule-rich projections. Actin, red; microtubules, green; DNA, blue. (d) The mitotic index of S2 cells is increased after the depletion of Spindly, dynein heavy chain (DHC), or a subunit of the APC (Cdc16; mean ± SEM [error bars]; n = 3 experiments, with 1,000–3,000 cells counted per experiment). Values are expressed as a ratio of RNAi-treated to untreated cells (untreated cells have a mitotic index of 1–3%). (e) The ratio of metaphase to anaphase cells (scored manually after staining with anti-tubulin and antiphosphohistone antibodies; see Materials and methods) reveals a selective increase in metaphase cells after Spindly and DHC RNAi (mean ± SEM; n = 2 experiments, with >200 mitotic spindles scored per experiment). (d and e) The expression of GFP-tagged Spindly can rescue the mitotic phenotype after endogenous Spindly is depleted using a dsRNA that targets the Spindly 3′ UTR. Bars, 10 μm.
RNAi of one novel gene, CG15415, produced strong phenotypes in both screens. CG15415 is a novel uncharacterized Drosophila gene encoding a 780–amino acid protein with predicted N-terminal coiled-coil sequences and four repeats with the consensus sequence TPXKPQXKGTPVK (Fig. 1 a). In the interphase screen, many of the CG15415-depleted cells showed spiky and elongated microtubule-rich projections in contrast to the rounded shape of normal spread S2 cells (Fig. 1, b and c). In the mitotic index screen, the depletion of CG15415 caused an increase in mitotic index that was comparable with that observed for RNAi of the dynein heavy chain (DHC) and the APC subunit Cdc16 (Fig. 1 d). The majority of the mitotic CG15415- depleted cells were arrested in metaphase, which is also similar to DHC depletion (Fig. 1 e). This result was confirmed in live cells expressing GFP-tubulin, in which CG15415-depleted cells failed to enter anaphase within 4 h after nuclear envelope breakdown. In contrast, untreated cells initiated anaphase within 20–85 min of nuclear envelope breakdown (unpublished data). Because the depletion of CG15415 produced spindle-shaped interphase cell morphology and arrested cells with metaphase spindles, we refer to this protein as Spindly.
The specificity of the Spindly phenotypes was confirmed using three nonoverlapping dsRNAs: two in the coding region and one dsRNA that targets the 3′ untranslated region (UTR; Fig. 1 a). Using an antibody generated against Spindly's C-terminal 357 amino acids, we confirmed that the three dsRNAs effectively depleted the protein after 5 d (Fig. S1 a, available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1). As further confirmation of the specificity of the Spindly RNAi phenotype, we found that expression of a GFP-Spindly fusion protein could rescue the metaphase block after the endogenous protein was depleted with the 3′ UTR dsRNA. This result also indicates that Spindly retains its function after fusion to GFP, enabling the localization studies described in the next section.
GFP-Spindly targets to microtubule plus ends in interphase and to kinetochores in mitosis
To learn more about Spindly's function, we examined the localization and dynamics of GFP-tagged Spindly. In live cells expressing low levels of GFP-Spindly, the protein was concentrated in punctae that continually moved to the periphery of the cell, which is behavior typical of microtubule plus end–binding proteins (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1). Fixation and staining of cells expressing low levels of GFP-Spindly with an antibody to EB1 (a well-established plus end–binding protein) confirmed this localization, although the plus end enrichment was less pronounced than that displayed by EB1 (Fig. 2 a). At higher levels of GFP-Spindly expression, the protein began to decorate along the length of the microtubule and to localize to the lamella (unpublished data).
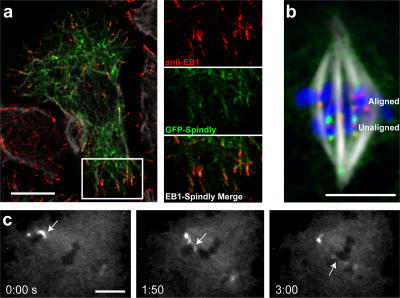
Figure 2.
Spindly binds to microtubule plus ends in interphase and to kinetochores in mitosis. (a) GFP-Spindly–expressing cells were fixed with methanol-formaldehyde and stained with anti-EB1 and anti-tubulin antibodies. The insets (magnified image of boxed area) show GFP-Spindly and anti-EB1 antibodies colocalized on the tips of microtubules. (b) Stably expressed GFP-Spindly (green) localizes to kinetochores (marked by the CENP-A homologue Cid; red) but substantially enriches on kinetochores that have not yet aligned on the metaphase plate (compare top kinetochores [aligned kinetochores] with bottom kinetochores [unaligned kinetochores]). (c) A time-lapse sequence of a GFP-Spindly–expressing cell starting in prometaphase. The arrows show an unaligned chromosome that is captured by microtubules from the opposite pole and then dragged to the metaphase plate. As this chromosome reaches the metaphase plate, the kinetochore levels of GFP-Spindly decrease until they reach the levels of the aligned chromosomes (the minutes and seconds elapsed are shown at the bottom). The video (Video 2) is available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1. Bars (a and b), 10 μm; (c) 5 μm.
After cells entered mitosis, GFP-Spindly was no longer localized to microtubule tips but instead was found on kinetochores. In prometaphase cells, GFP-Spindly was found on most kinetochores, a localization confirmed by colocalization with anti-Cid antibodies, which recognize the Drosophila homologue of CENP-A. However, in metaphase cells, the levels of GFP-Spindly were reduced considerably on the kinetochores of aligned chromosomes, and the protein was more evident on the mitotic spindle, especially at spindle poles (Fig. 2 b). During anaphase, GFP-Spindly was seen once again at high levels on kinetochores, but, after the nuclear envelope reformed in telophase, the protein was excluded from the nucleus. Time-lapse microscopy revealed that high initial levels of GFP-Spindly on misaligned chromosomes decreased as these chromosomes were pulled toward the metaphase plate (Fig. 2 c and Videos 2 and 3, available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1). A similar distribution of endogenous Spindly in mitosis was confirmed using an affinity-purified antibody in cells expressing the Drosophila homologue of the kinetochore protein Mis12 (CG18156) fused to GFP (Fig. S1). The transient targeting of Spindly to kinetochores is very similar to what has been reported for the mitotic checkpoint proteins Rod and Mad2 (Chen et al., 1996; Scaerou et al., 1999). This dynamic kinetochore localization together with the data from our mitotic index screen led us to focus our efforts on understanding Spindly's role during mitosis.
Spindly is shed from the kinetochore in a dynein-dependent manner and requires Rod to target to the kinetochore
Components of the RZZ complex as well as Mad2 accumulate on kinetochores in prometaphase and are shed from metaphase kinetochores by dynein-dependent transport along kinetochore microtubules (Howell et al., 2001; Wojcik et al., 2001). Using faster acquisition live cell imaging, we similarly observed punctae of GFP-Spindly moving processively from metaphase-aligned kinetochores toward the spindle poles (Fig. 3 a and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1). Kymograph analysis revealed that GFP-Spindly moved poleward at a mean velocity of ∼12 μm/min (Figs. 3 b and S2), which is similar to rates reported for the dynein-mediated transport of RZZ and Mad2 in Drosophila (Wojcik et al., 2001; Basto et al., 2004). However, not all GFP-Spindly particles moved uniformly; some paused or made short reversals toward the kinetochore before continuing toward the spindle pole (Video 4), which is behavior similar to that described for dynein–dynactin complexes in vitro (Ross et al., 2006).
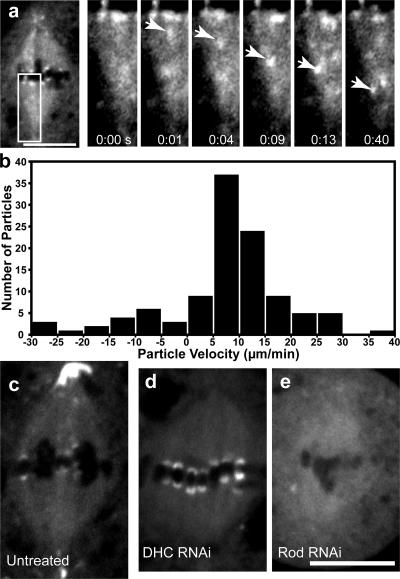
Figure 3.
GFP-Spindly is moved from kinetochores toward the spindle pole in a dynein-dependent manner. (a) By live cell microscopy, a particle of GFP-Spindly (arrows in the insets [magnified images of the boxed area]) can be seen moving from the kinetochore to the centrosome. The seconds elapsed are shown at the bottom (see Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1). (b) Kymograph analysis was performed on the GFP-Spindly particles, and a histogram of the rates of 110 GFP-Spindly particles during episodes of continuous motion was produced (data were obtained from four separate spindles). The mean speed was 11.9 ± 6.9 μm/min (±SD). (c–e) GFP-Spindly in live cells was imaged by spinning disc confocal microscopy in untreated (c), dynein (DHC) RNAi-treated (d), or Rod RNAi-treated (e) cells. Dynein depletion caused Spindly to accumulate at high levels on aligned kinetochores, whereas Rod depletion blocked the recruitment of Spindly to the kinetochore. Bars, 5 μm.
To establish whether dynein is indeed the motor responsible for the poleward transport of Spindly, we examined GFP-Spindly after RNAi-mediated depletion of the cytoplasmic DHC. Under these conditions, high levels of GFP-Spindly accumulated on metaphase-aligned kinetochores (Fig. 3 d and Video 4), which is similar to what has been described for Rod and Mad2 after the disruption of dynein (Wojcik et al., 2001; unpublished data). Immunofluorescence localization of endogenous Spindly confirmed this result (unpublished data). We also no longer observed the poleward transport of GFP-Spindly by time-lapse microscopy. RNAi-mediated depletion of the dynein regulatory proteins Lis1 and p150Glued produced similar results (Video 4 and not depicted, respectively). These results indicate that kinetochore to pole movement of Spindly depends on cytoplasmic dynein and its activators, as is true of other known components of the SAC.
We next sought to determine how Spindly is targeted to the kinetochore. It has been previously shown that recruitment of dynein–dynactin to the corona region of the kinetochore depends on the RZZ complex, which, in turn, links through Zwint-1 to the Ndc80 and Mis12 complexes of the kinetochore (Starr et al., 1998; Obuse et al., 2004; Kops et al., 2005a). The depletion of any of the three RZZ polypeptides destabilizes the whole complex and prevents the recruitment of Mad2 and dynein–dynactin (Scaerou et al., 1999, 2001; Buffin et al., 2005). When Rod was depleted by RNAi, GFP-Spindly no longer localized to kinetochores or the spindle poles (Fig. 3 e and Video 4). These results indicate that Spindly is a part of the corona region of the kinetochore and requires the RZZ complex (but not dynein or dynactin, as discussed above) for its kinetochore localization.
Spindly-depleted cells arrest in mitosis with high levels of Rod and Mad2 on aligned kinetochores
Because Spindly is required for cells to complete mitosis and localizes to kinetochores in a manner similar to known SAC proteins, we decided to investigate the role of Spindly in the kinetochore localization of Rod and Mad2. In prometaphase cells, Rod and Mad2 are more abundant on misaligned than aligned chromosomes and are also observed on the spindle and spindle poles (Fig. 4, a and d) as previously described (Chen et al., 1996; Williams et al., 1996). However, after Spindly RNAi, the levels of Rod and Mad2 were comparable on misaligned and metaphase-aligned kinetochores, which is similar to the outcome of DHC RNAi (Fig. 4, b, c, e, and f). These results indicate that both dynein and Spindly are required for the shedding of Rod and Mad2 from the kinetochore. Consistent with this interpretation, the staining of Rod and Mad2 on the spindle (likely reflecting the population of molecules undergoing transport) was severely reduced after Spindly and DHC RNAi (Fig. 4, b, c, e, and f). The retention of Rod and Mad2 on metaphase-aligned chromosomes explains the high mitotic index and increased number of metaphases seen after Spindly or DHC depletion (Fig. 2 a).
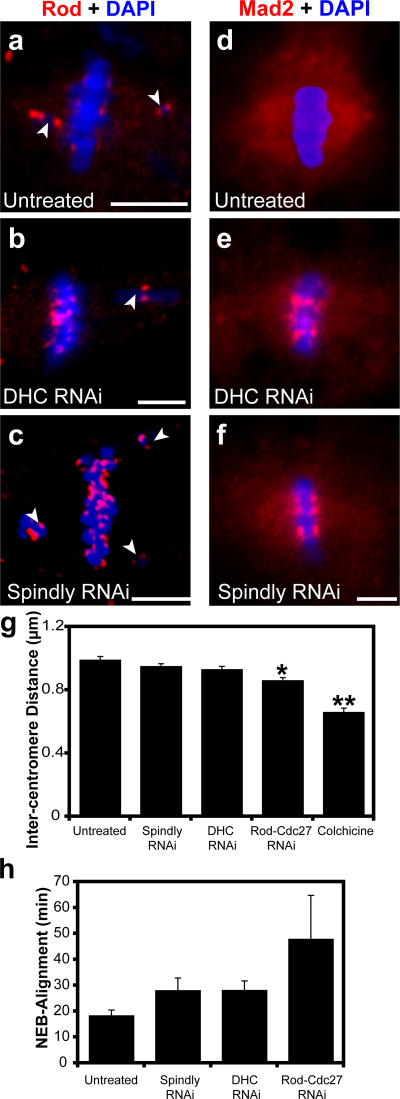
Figure 4.
Spindly is required for removing Rod and Mad2 from kinetochores. (a) Immunofluorescence of Rod (red in this overlay) enriches on chromosomes (blue) that are not aligned on the metaphase plate (arrowheads indicate misaligned chromosomes). (b and c) In DHC and Spindly-depleted cells, the levels of Rod are similar on aligned and unaligned kinetochores. (d) Similarly, Mad2 (red) is barely detectable on aligned kinetochores but is present throughout the spindle. (e and f) DHC (e) and Spindly (f) depletion causes the accumulation of Mad2 on aligned chromosomes (blue) and a decrease in Mad2 staining on the spindle. (g) Intercentromere tension, which was measured as the distance between Cid-stained centromeres, was measured in untreated cells and cells treated with the indicated dsRNAs or 6 μg/ml colchicine (4-h treatment; n ≥ 25 for each condition; error bars represent SEM; *, P < 5 × 10−5, **, P < 5 × 10−10). (h) As a second measure of kinetochore function, the time required for untreated and dsRNA-treated cells to form a metaphase spindle after nuclear envelope breakdown (NEB) was measured from time-lapse videos (n ≥ 6 cells for each condition). Bars, 5 μm.
The metaphase arrest and retention of Mad2 and Rod on aligned chromosomes seen after Spindly depletion could be the result of defects in dynein-based transport or of alterations in kinetochore–microtubule interactions, which would keep the SAC activated even on seemingly aligned kinetochores. To test the latter possibility, we examined two parameters of the spindle that probe the microtubule–kinetochore interface. First, we measured the distance between paired centromeres (as marked by anti-Cid staining); larger distances reflect higher microtubule-generated tension pulling the two sister chromatids apart. In colchicine-treated cells (no microtubule-generated tension), the distance between paired centromeres was reduced from 0.99 to 0.66 μm. Interestingly, the depletion of Rod and Cdc27 (an APC subunit; Cdc27 was codepleted with Rod to prevent premature anaphase onset) caused a statistically significant (P < 0.0001) decrease in the stretch between centromeres of 35.3 ± 6.4% (from 0.99 to 0.87 μm [±SEM]). However, the depletion of Spindly and DHC only reduced stretch between paired centromeres by 10.3 ± 5.4% and 17.9 ± 6.3% (from 0.99 to 0.95 or 0.93 μm), respectively, and neither distance was statistically different from untreated cells.
As another measure of kinetochore function, we determined the time required to align all chromosomes at the metaphase plate using a cell line expressing GFP-tagged histone H2B and mCherry-tagged α-tubulin and automated time-lapse imaging (see Materials and methods). Intriguingly, the Spindly- and DHC-depleted cells both required 50% more time to form a metaphase plate compared with untreated cells (a mean of 18.5 ± 2.3 min vs. 28.1 ± 4.9 and 28.2 ± 3.7 min [±SEM] for Spindly and dynein, respectively), which might be the result of a defect in initial kinetochore microtubule capture (Fig. 4 h and Videos 5–8, available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1). Alexander and Rieder (1991) also proposed that kinetochore-associated dynein could play an important role in making lateral attachments between chromosomes and microtubules before the final end-on attachments observed at metaphase, which could explain the delay in chromosome alignment after DHC depletion. Consistent with our results for centromere tension, cells depleted of Rod and Cdc27 took considerably longer (48 ± 16.9 min) to assemble a metaphase plate, which might reflect a requirement for the RZZ complex to incorporate multiple proteins into the outer corona of the kinetochore. In summary, these results suggest that Spindly-depleted cells do not have gross defects in kinetochores or kinetochore–microtubule interactions but rather have kinetochores that resemble those found in cells lacking dynein.
Spindly is a kinetochore-specific dynein recruitment factor
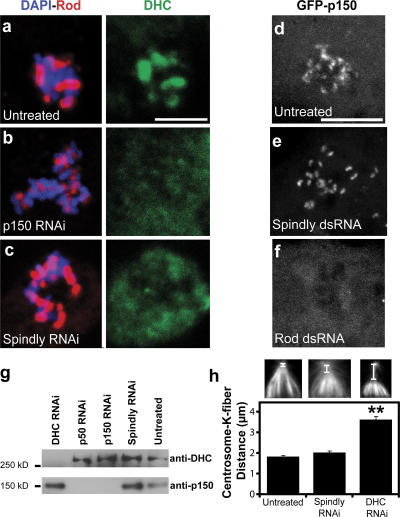
The similar Spindly and dynein RNAi phenotypes of mitotic arrest, defects in Mad2 and Rod transport, and delays in forming a metaphase plate suggested that Spindly might somehow play a role in dynein function at the kinetochore. Therefore, we next examined whether Spindly affects the kinetochore localization of dynein. To more easily assay dynein localization, microtubules were depolymerized with colchicine, which causes a substantial accumulation of dynein and dynactin on kinetochores (Fig. 5 a). Spindly RNAi resulted in a profound reduction in DHC staining at kinetochores compared with untreated cells (Fig. 5 c). Interfering with dynactin function has also been reported to abolish kinetochore staining of dynein (Vallee et al., 1995; Starr et al., 1998; Dzhindzhev et al., 2005), a finding that we repeated as well (Fig. 5 b). However, dynactin, as assayed by GFP-p150glued (Fig. 5 e) or with anti-p150glued antibodies (Fig. S3, a and c; available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1), was still recruited to kinetochores in Spindly-depleted cells (however, Rod RNAi displaces p150glued from kinetochores; Fig. 5 f). To confirm that Spindly is required for dynein kinetochore localization and not the stability of the protein, immunoblot analysis was performed, which revealed that DHC and p150glued protein levels were unaltered by Spindly RNAi (Fig. 5 g). Thus, Spindly is required for dynein but not dynactin recruitment to kinetochores.
Figure 5.
Spindly depletion blocks the recruitment of DHC but not dynactin to kinetochores. (a) In mitotic cells treated with colchicine to depolymerize the mitotic spindle, Rod (red) and DHC (green) colocalize on chromosomes (blue). (b and c) In cells depleted of p150Glued (b) or Spindly (c), Rod remains bound to kinetochores, but the DHC is displaced. (d–f) S2 cells stably expressing GFP-p150Glued (a dynactin subunit) were treated with 6 μg/ml colchicine for 4 h, and the localization of the protein was assayed after RNAi treatment. In untreated (d) and Spindly-depleted (e) cells, GFP-p150Glued still bound to the kinetochore, whereas the depletion of Rod (f) prevented the protein from associating with the kinetochore. Images are maximum intensity z projections of 2-μm-thick stacks of images taken of live cells. (g) Immunoblots of lysates from RNAi-treated S2 cells show that Spindly RNAi did not affect dynein (DHC) or dynactin (p150Glued) protein levels. (h) The distance between the minus ends of kinetochore (K) fibers and the centrosome (see insets) was measured for untreated (left inset), Spindly RNAi (middle inset), and DHC RNAi (right inset) cells (n ≥ 69 for each condition; error bars represent SEM; **, P < 5 × 10−10), revealing a defect with dynein but not Spindly depletion. Bars, 5 μm.
The aforementioned results clearly revealed an important role for Spindly in dynein function at the kinetochore. We next investigated whether Spindly participates in other dynein-mediated activities. In S2 cells, dynein is known to be important for spindle focusing, specifically in transporting kinetochore fibers along microtubules emanating from the centrosomes. After DHC RNAi, the centrosomes detach and move away from the minus ends of the K fibers (Fig. 5 h; Maiato et al., 2004; Goshima et al., 2005). However, Spindly depletion did not produce the centrosome detachment or spindle focusing defects seen in cells lacking dynein (Fig. 5 h). Additionally, after plating on Con A for 3 h, Spindly-depleted and untreated interphase cells generally cluster their endosomes (marked by GFP-Rab5) toward the cell interior, whereas endosomes in dynein- or dynactin-depleted cells tend to remain spread throughout the cell (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1; dynactin depletion data not depicted). Collectively, these experiments suggest that Spindly influences dynein function at the kinetochore but not everywhere throughout the cell.
Identification of human Spindly
We next sought to identify Spindly homologues in other species. Standard BLAST (Basic Local Alignment and Search Tool) searches identified Spindly homologues in the insects Aedes aegypti and Anopheles gambiae but not in more distant species. Multiple Em for motif elicitation was then used to identify conserved motifs present in all three insect homologues, and these motifs were used for MAST (Motif Alignment and Search Tool) searches to identify more distant homologues (Bailey and Elkan, 1994; Bailey and Gribskov, 1998). A conserved 32-amino acid motif found in a break between predicted coiled-coil domains in the N terminus of all three insect proteins also was found in the human protein RefSeq NP_060255 (Fig. S5 a, available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1). The overall primary sequence conservation between Drosophila Spindly and human NP_060255 is low (14.3% identity), and the putative human homologue is somewhat shorter (605 vs. 780 amino acids). However, the sequences in the 32–amino acid conserved motif are 56% identical (75% similar), and the first nine amino acids of this motif are 100% identical. The predicted coiled-coil organization and charge distribution of the putative human homologue also is similar to Drosophila Spindly, although the sequences of the coiled coils are not conserved.
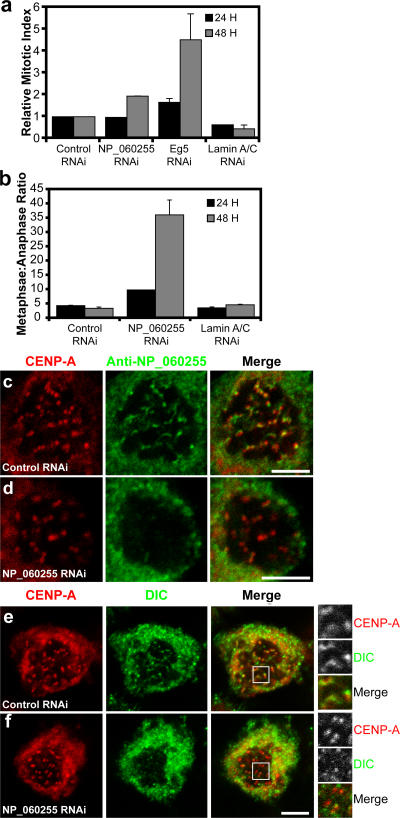
The function of the putative human homologue of Spindly had not been previously characterized. To test whether NP_060255 is a bona fide functional homologue of Drosophila Spindly, we examined whether depletion of the protein by siRNA caused mitotic defects. Transfection of a siRNA pool targeted to NP_060255 reduced NP_060255 protein levels by 86% (immunoblot analysis; not depicted) and produced a twofold increase in the mitotic index of HeLa cells after 48 h (Fig. 6 a). When these mitotic cells were examined, a dramatic increase in the ratio of metaphase versus anaphase cells was apparent (Fig. 6 b), and a substantial number of these cells had misaligned chromosomes (Fig. S5 b). A similar phenotype has been reported in HeLa cells after the depletion of either CLIP-170 or dynein, which targets CLIP-170 to the kinetochore (Tanenbaum et al., 2006). We next localized NP_060255 with a polyclonal antibody in HeLa cells treated with colchicine to depolymerize spindle microtubules. Similar to the Drosophila protein, we observed punctae of NP_060255 that were coincident with CENP-A–stained centromeres (Fig. 6 c). This staining was eliminated by treating cells with the siRNA oligonucleotides that target NP_060255 (Fig. 6 d), confirming the localization of this protein at kinetochores.
Figure 6.
Identification of a human Spindly homologue that is also required for targeting dynein to the kinetochore. (a) The mitotic index was determined (n = 3 wells per condition and >1,000 cells per well counted; error bars represent SEM) 24 or 48 h after siRNAs targeting the indicated proteins were transfected into HeLa cells. (b–d) The ratio of metaphase to anaphase cells for these treatments is shown (n = 2 experiments; at least 75 cells per condition). NP_060255 was localized using crude antisera in HeLa cells treated with colchicine to enrich for the protein on kinetochores, and we found that NP_060255 colocalizes with the centromere marker CENP-A (c). Colchicine treatment helped to accumulate NP_060255 on kinetochores; without this treatment, background spindle staining with the NP_060255 antibody made it difficult to unambiguously visualize kinetochore localization, even on prometaphase chromosome. To confirm the specificity of kinetochore localization in colchicine-treated cells, we depleted NP_060255 with siRNA oligonucleotides and found that the colocalization with CENP-A was eliminated (d). (e and f) The dynein intermediate chain (DIC) was localized in control and NP_060255 siRNA–transfected cells that had been treated with 6 μg/ml colchicine for 4 h to depolymerize all microtubules. The insets (magnified images of boxed areas) show that NP_060255 depletion eliminated the colocalization between CENP-A and DIC, demonstrating that NP_060255 is required for bringing dynein to the kinetochore. Bars, 5 μm.
To determine whether NP_060255, like Drosophila Spindly, is required to recruit dynein to the human kinetochore, we localized dynein using an antibody to its intermediate chain (dynein intermediate chain [DIC]) in colchicine-treated siRNA-transfected cells. In control siRNA-treated cells, a subset of the DIC-stained punctae colocalized with CENP-A, a marker of the centromere (Fig. 6 e). However, after siRNA against NP_060255, the colocalization of dynein with CENP-A was substantially reduced (Fig. 6 f). Similar to what was found for Drosophila Spindly, the depletion of NP_060255 also decreased the stretch between paired centromeres from 1.15 to 0.98 μm (29.6 ± 4.5% decrease; P < 0.00005), a result that is in agreement with the previously reported effect of p50dynamitin microinjection (a dominant-negative inhibitor of dynactin function) on kinetochore stretch (Howell et al., 2001). Collectively, our data show that the protein encoded by NP_060255 localizes to kinetochores and is required for localizing dynein to the kinetochore and for mitotic progression. Thus, we suggest that NP_060255 is a true homologue of Drosophila Spindly and propose to rename NP_060255 as Hs Spindly. These results also indicate that the mechanism for localizing dynein to the kinetochore to silence the SAC is conserved between humans and flies.
Discussion
Using RNAi screens in Drosophila S2 cells, we have identified Spindly, a previously uncharacterized protein, as an essential factor for docking dynein to the kinetochore. Spindly is recruited to the kinetochore in an RZZ-dependent manner, and there, together with dynactin, Spindly recruits dynein to the outermost region of the kinetochore. The dynein motor complex then transports Spindly along with Mad2 and the RZZ complex to the spindle poles to inactivate the SAC. We also identify a Spindly homologue that plays a similar role in human cells, revealing a conserved dynein kinetochore targeting mechanism in invertebrates and vertebrates. These data provide new insight into the mechanism and importance of recruiting dynein to the kinetochore to inactivate the SAC. We also find that Spindly plays a role in maintaining S2 cell morphology during interphase and localizes to the growing ends of microtubules.
Involvement of Spindly in mitotic dynein function
The depletion of Spindly creates several mitotic defects that appear to reflect a loss of dynein activity exclusively at the kinetochore. Metaphase arrest is the most evident defect observed after the RNAi-mediated depletion of Spindly in Drosophila or human cells. This metaphase arrest phenotype is most likely explained by the absence of kinetochore-bound dynein in Spindly-depleted cells, and, indeed, our data support the model of Howell et al. (2001), which proposes that kinetochore-bound dynein is required for transporting Mad2 from the kinetochore to inactivate the SAC. Nevertheless, we cannot rule out the possibility that the mitotic delay seen after dynein or Spindly depletion is caused by another kinetochore aberration that keeps the checkpoint activated. However, Spindly-depleted cells ultimately overcome metaphase arrest, as seen in our live cell imaging experiments and by the modest increases in the mitotic indices of Spindly-depleted S2 and HeLa cells (three- to sevenfold and twofold, respectively). The mechanism of slippage from this metaphase arrest (Rieder and Maiato, 2004) is not clear, but it might involve proteins (e.g., p31 comet) that silence the SAC by disrupting the interaction between Mad2 and Cdc20 (Habu et al., 2002; Xia et al., 2004).
In addition to mitotic arrest, we observed that chromosomes in Spindly- and dynein-depleted S2 cells required a longer time to align on the metaphase plate. This result may be attributable either to the displacement of CLIP-190 (a microtubule tip–binding protein) from kinetochores after Spindly or dynein depletion (Dzhindzhev et al., 2005; unpublished data) or the loss of dynein-mediated lateral attachments to microtubules in early prometaphase (Alexander and Rieder, 1991). In HeLa cells, we also have noticed a defect in chromosome alignment after Hs Spindly depletion, which also has been observed after the depletion of dynein (perhaps mediated through a loss of kinetochore-bound CLIP-170; Dujardin et al., 1998; Tanenbaum et al., 2006).
Thus, the spectrum of mitotic defects observed in Spindly-depleted cells is consistent with a loss of dynein function specifically at the kinetochore. Spindly depletion did not produce any other defects seen after dynein depletion, such as centrosome detachment and spindle defocusing. Dynactin is another protein that is required for recruiting dynein to kinetochores, but it is important for other mitotic and interphase dynein functions. Depletion of the RZZ complex inhibits the kinetochore recruitment of dynein, but this also prevents Mad1 and Mad2 recruitment and reduces kinetochore tension to a greater degree than Spindly or dynein depletion alone. Thus, Spindly depletion appears to be the most specific means identified to date for interfering with dynein function only at the kinetochore.
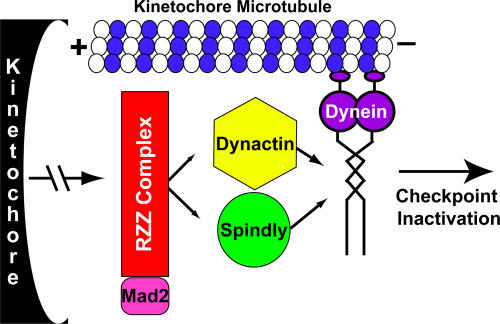
Our findings provide new insight into how dynein localizes to kinetochores. Previous studies have led to a model in which dynactin binds to the RZZ complex and then, either alone or in collaboration with Lis1, recruits dynein to the kinetochore (Vallee et al., 1995; Starr et al., 1998; Tai et al., 2002; Cockell et al., 2004; Dzhindzhev et al., 2005; Siller et al., 2005). Because we find that both dynactin and Spindly are required for dynein localization to kinetochores, we propose an updated model in which Spindly and dynactin target to the kinetochore independently and work together to recruit dynein (Fig. 7). Thus, dynein recruitment to the kinetochore may involve multiple weak interactions. Consistent with the possibility of weak interactions, endogenous dynein, dynactin, and Rod did not coprecipitate with GFP in pull-down experiments, and Spindly did not coenrich with these proteins in sucrose gradient fractions (unpublished data). Lis1 is not included in our dynein localization model, as we found that Lis1 RNAi did not block dynein recruitment to the kinetochore (using our colchicine treatment localization assay; unpublished data), although Lis1 depletion did cause a mitotic delay and substantial increase in GFP-Spindly on aligned kinetochores (Video 4). Thus, we favor a role for Lis1 in dynein activity but not in recruiting dynein to the kinetochore.
Figure 7.
A model of Spindly activity. During mitosis, the RZZ complex binds to the outer kinetochore region and recruits Mad2, Spindly, and the dynactin complex. Spindly and dynactin then cooperatively work to recruit dynein, which then transports the whole complex toward the spindle pole and silences SAC signaling on the kinetochore. See Discussion for details.
Spindly's role in regulating interphase cell morphology
Spindly's role in the spreading morphology of S2 cells makes it unusual among proteins involved in silencing the SAC (including dynein and dynactin), which did not produce phenotypes in our interphase morphology screen. The Spindly RNAi interphase phenotype of defective actin morphology and the formation of extensive microtubule projections is still not understood. However, a clue may be Spindly's dynamic localization to the growing microtubule plus end. Other plus end–binding proteins (+TIPs) interact with signaling molecules that regulate cell shape, one example being the binding and recruitment of RhoGEF2 to the microtubule plus end by EB1 (Rogers et al., 2004). Spindly may similarly interact with and carry an actin regulatory molecule to the cortex, but this hypothesis will require identifying proteins that interact with Spindly during interphase.
The mechanism of Spindly recruitment to the microtubule plus end also warrants further investigation. This interaction must be regulated by the cell cycle because GFP-Spindly no longer tracks along microtubule tips in prometaphase. Seven consensus CDK1 phosphorylation sites are present in the positively charged C-terminal repeats of Spindly, and phosphorylation of these sites could reverse the charge of these repeats and regulate the transition from microtubule tip binding to kinetochore binding at the onset of mitosis.
Spindly, an example of a cargo-specific dynein localization factor
Motor proteins must be guided to the correct subcellular site to execute their biological function. To carry out the multitude of transport activities required in eukaryotic cells, metazoans have evolved numerous kinesin motors (25 genes in Drosophila) with distinct domains that dictate their localization and regulation (Vale, 2003). In contrast, a single cytoplasmic DHC performs numerous roles in interphase and mitosis, suggesting that additional regulatory factors guide dynein to specific cargoes (e.g., organelles, mRNAs, and vesicles). The main dynein-associated proteins (the dynactin complex, Lis1, and NudEL) are involved in dynein function at many sites and, thus, do not appear to be cargo specific. Zw10 was initially thought to specifically regulate the recruitment of dynein–dynactin to the kinetochore, but it now also appears to play an essential role in targeting dynein to membrane-bound organelles (Hirose et al., 2004; Varma et al., 2006). Bicaudal D is another multifunctional adaptor molecule that has a role in the dynein-based transport of multiple cargoes such as RNA, vesicles, and nuclei (Swan et al., 1999; Bullock and Ish-Horowicz, 2001; Matanis et al., 2002). Perhaps the most site-specific dynein recruitment factor is the Saccharomyces cerevisiae Num1 protein that binds to the DIC Pac11p to target the motor to the cortex of daughter cells, where it pulls the nucleus into the bud neck (Heil-Chapdelaine et al., 2000; Farkasovsky and Kuntzel, 2001). However, dynein only serves this one function in yeast compared with its plethora of activities in metazoans, and Num1p homologues have yet to be identified in higher eukaryotes.
By our assays performed to date, Spindly appears to be a highly selective dynein-recruiting factor, and, unlike other dynein cofactors, it does not appear to be involved in the motor's nonkinetochore functions in mitosis (e.g., pole focusing) or in interphase (e.g., endosome transport). However, the mechanism by which Spindly recruits dynein to the kinetochore remains to be elucidated. Our observations that Spindly moves from kinetochores to the spindle poles as discrete punctae strongly suggests that it may incorporate into a large and somewhat stable particle that contains the RZZ complex, Mad1–Mad2, dynein, and likely additional proteins. Therefore, Spindly not only serves to recruit dynein to the kinetochore but also is part of a cargo that dynein transports. Future studies will be needed to better understand the protein composition of these transport particles and the contacts that Spindly makes within them.
Materials and methods
Cell culture, RNAi, and immunofluorescence
Drosophila Schneider cell line (S2) cells (Invitrogen) were cultured, and dsRNA incubation was performed as previously described (Goshima and Vale, 2003; Rogers et al., 2003). The 7,200 gene screens were performed with a previously described library (Echard et al., 2004). After 5 d of dsRNA treatment, cells were plated in glass-bottom 96-well plates (Whatman) coated with Con A (Sigma-Aldrich). Cell shape phenotypes were manually scored and documented on a microscope (Axioplan 200M; Carl Zeiss MicroImaging, Inc.) equipped with a 40× 1.3 NA objective and a cooled CCD camera (Sensicam HQ; The Cooke Corporation) after staining with an anti-tubulin antibody (DM1A, anti–α-tubulin; 1:500; Sigma-Aldrich) and rhodamine phalloidin. For the mitotic index screen, mitotic index was determined by dividing the number of phosphohistone H3–positive nuclei (1:1,000; Upstate Biotechnology) by the total number of nuclei (determined by DAPI staining). These cells were imaged using a 20 or 10× air objective in either an ArraySCAN HCS System (Cellomics Inc.) or an automated microscope (ImageXpressMicro; Molecular Devices).
In the follow-up experiments described in this paper, most assays were performed after 7 d of RNAi treatment as previously reported (Goshima and Vale, 2003). At the end of the RNAi treatments, cells were resuspended and seeded on Con A–coated coverglasses or dishes for 2 h before imaging or fixation. For colchicine treatment, cells were allowed to settle for 20 min, the media was removed and replaced with media containing 6 μg/ml colchicine, and imaging or fixation and staining was performed 4 h after treatment began. HeLa cells were maintained as previously described (Griffis et al., 2002). siRNA oligonucleotides were On-TARGETplus SMARTpools (Dharmacon), and transfections were performed using Dharmafect1 (Dharmacon) according to the manufacturer's instructions. Immunofluorescence was performed with affinity-purified rabbit anti-Dm Spindly (1:100), rabbit anti-Hs Spindly serum (1:100), chicken anti-Cid (1:200; provided by G. Karpen, Lawrence Berkeley National Laboratory, Berkeley, CA), rabbit anti-Rod (1:200; provided by R. Karess, Centre National de la Recherche Scientifique, Gif-sur-Yvette, France), mouse anti-DHC (1:100; provided by T. Hays, University of Minnesota, Minneapolis, MN), rabbit anti-p150Glued (1:200; provided by R. Giet, University of Rennes, Rennes, France), rat anti–α-tubulin (1:150; Serotec), mouse anti–CENP-A (1:2,000; Abcam), rabbit anti-DIC (1:500; provided by K. Vaughan, Notre Dame University, South Bend, IN), and rabbit anti-Mad2 (1:35; provided by C. Sunkel, (Instituto de Biologia Molecular e Celular, Porto, Portugal). Images were collected with either a confocal microscope (LSM510; Carl Zeiss MicroImaging, Inc.) using a 63× 1.4 NA objective or a microscope (Axioplan; Carl Zeiss MicroImaging, Inc.) outfitted with 40× 1.3 NA, 63× 1.4 NA, and 100× 1.3 NA objectives and a cooled CCD camera (Sensicam HQ; The Cooke Corporation).
Live cell imaging of GFP-Spindly and analysis
We cloned Spindly from an S2 cell cDNA pool and found that the sequenced cDNA clone lacks 27 amino acids from the predicted ORF. This ORF was cloned into the pENTR/D-TOPO vector (Invitrogen) and moved into N- or C-terminal Gateway GFP vectors under the control of the metallothionein promoter vector (N- and C-terminal fusions produced the same results). To observe the tip tracking, it was optimal to use cells without inducing GFP-Spindly protein expression with CuSO4. For observation of protein on kinetochores, GFP-Spindly expression was induced by incubating the cells with 20 μm CuSO4 for 18 h. S2 cells stably expressing GFP-tagged proteins were plated in dishes with coverslip bottoms (MatTek) that had been coated with Con A. Images were collected at 1–20-s intervals at room temperature using a cooled CCD (Orca II ERG; Hamamatsu Photonics) or iCCD (MEGA10; Stanford Photonics) camera attached to a spinning disk confocal scan head (Yokogawa Electric and Solamere Inc.) that was mounted on a microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) outfitted with a 100× 1.45 NA objective. Images were collected using either MetaMorph software (Molecular Devices), QED (Media Cybernetics), or μManager (www.micro-manager.org).
For analysis of GFP-Spindly movement from the kinetochore to poles, cells were imaged on the spinning disk confocal microscope with 300-ms exposures taken every second. Image stacks were opened in ImageJ (National Institutes of Health), and spindles were oriented horizontally. A box was drawn that was wide enough to contain all of the kinetochores on one half of the metaphase plate and long enough to contain the proximal spindle pole. A stack of kymographs (each one representing a given one-pixel–thick line within the box) was then generated. These kymograph stacks were then combined into maximum intensity z projections, and particle velocities were determined by measuring the lengths of the lines created by particles moving toward or from the spindle poles (distance traveled) and then dividing that value by the displacement in the y direction (time). To determine statistical significance, datasets were analyzed using the t test.
Antibody production and immunoblotting
A region of the Drosophila Spindly gene corresponding to amino acids 451–780 was cloned into pET28a (Novagen), and protein expression was induced in BL21 DE3 cells (Invitrogen). Full-length Hs Spindly was also cloned into pET28a, and the protein was expressed in BL21 DE3 cells. The expressed proteins were purified and used for injecting rabbits (Covance). Anti-Dm Spindly antibodies were purified on an Affi-Gel 10 column (Bio-Rad Laboratories) containing the immobilized antigen. To isolate protein from S2 and HeLa cells after RNAi treatment, 100 μl laemmli sample buffer was added per well of cells in a 96-well plate. The sample was then processed for Western blotting as previously described (Rogers et al., 2003). The blot shown in Fig. S1 was pieced together from multiple lanes of a larger gel; the blot was cut between the 100- and 150-kD markers and blotted with the indicated antibodies (rabbit anti-p150glued; 1:500; provided by E. Holzbaur, University of Pennsylvania, Philadelphia, PA). The blot shown in Fig. 5 was cut at the 250-kD marker and blotted with the indicated antibodies (mouse anti-DHC; 1:1,000; provided by T. Hays).
Online supplemental material
Fig. S1 shows that the endogenous Spindly protein also enriches on unattached, unaligned, and anaphase kinetochores. Fig. S2 shows kymograph analysis of GFP-Spindly particles. Fig. S3 shows that Spindly depletion does not alter the targeting of endogenous dynactin to the kinetochore. Fig. S4 shows that Spindly is not required for the dynein-dependent reorganization of endosomes in S2 cells. Fig. S5 shows that the depletion of NP_060255 causes defects in chromosome alignment. Video 1 shows that GFP-Spindly tracks on the plus ends of microtubules in interphase cells. Video 2 shows that GFP-Spindly concentrates on lagging chromosomes and then diminishes after alignment at the metaphase plate. Video 3 shows that GFP-Spindly returns to kinetochores during anaphase, and Video 4 shows that GFP-Spindly traffics from kinetochores to centrosomes in a dynein- and Rod-dependent manner. Videos 5–8 show that the depletion of Spindly, dynein, or Rod slows the alignment of chromosomes on the metaphase plate. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200702062/DC1.
Supplementary Material
Acknowledgments
We thank T. Hays, R. Karess, E. Holzbaur, G. Karpen, C. Sunkel, K. Vaughan, E. Salmon, and R. Giet for their gift of antibodies. We thank T. Murphy for providing us with the Drosophila Gateway Vector Collection. We are grateful to R. Wollman for use of his MatLab algorithms for automatically identifying and analyzing mitotic cells and to J. Kardon for developing the Rab5 localization assay to monitor interphase dynein activity. We thank E. Quan for assistance with quantitative immunoblotting. We also thank G. Goshima and A. Roll-Mecak for reagents, U. Wiedemann and N. Zhang for excellent technical assistance, and S. Reck-Peterson, K. Slep, J. Kardon, S. Rogers, and G. Goshima for helpful discussions. We thank K. Vaughan for sharing results in advance of publication.
E. Griffis is supported by a postdoctoral fellowship from the American Cancer Society.
Abbreviations used in this paper: APC, anaphase-promoting complex; CENP, centromere protein; Con A, concanavalin A; DHC, dynein heavy chain; DIC, dynein intermediate chain; dsRNA, double-stranded RNA; RZZ, Rod–Zw10–Zwilch complex; SAC, spindle assembly checkpoint; UTR, untranslated region.
References
- Acquaviva, C., and J. Pines. 2006. The anaphase-promoting complex/cyclosome: APC/C. J. Cell Sci. 119:2401–2404. [DOI] [PubMed] [Google Scholar]
- Alexander, S.P., and C.L. Rieder. 1991. Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J. Cell Biol. 113:805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36. [PubMed] [Google Scholar]
- Bailey, T.L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 14:48–54. [DOI] [PubMed] [Google Scholar]
- Baker, D.J., J. Chen, and J.M. van Deursen. 2005. The mitotic checkpoint in cancer and aging: what have mice taught us? Curr. Opin. Cell Biol. 17:583–589. [DOI] [PubMed] [Google Scholar]
- Basto, R., F. Scaerou, S. Mische, E. Wojcik, C. Lefebvre, R. Gomes, T. Hays, and R. Karess. 2004. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 14:56–61. [DOI] [PubMed] [Google Scholar]
- Buffin, E., C. Lefebvre, J. Huang, M.E. Gagou, and R.E. Karess. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856–861. [DOI] [PubMed] [Google Scholar]
- Bullock, S.L., and D. Ish-Horowicz. 2001. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 414:611–616. [DOI] [PubMed] [Google Scholar]
- Chan, G.K., S.A. Jablonski, V. Sudakin, J.C. Hittle, and T.J. Yen. 1999. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.H., J.C. Waters, E.D. Salmon, and A.W. Murray. 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 274:242–246. [DOI] [PubMed] [Google Scholar]
- Cockell, M.M., K. Baumer, and P. Gonczy. 2004. lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J. Cell Sci. 117:4571–4582. [DOI] [PubMed] [Google Scholar]
- De Antoni, A., C.G. Pearson, D. Cimini, J.C. Canman, V. Sala, L. Nezi, M. Mapelli, L. Sironi, M. Faretta, E.D. Salmon, and A. Musacchio. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15:214–225. [DOI] [PubMed] [Google Scholar]
- Dujardin, D., U.I. Wacker, A. Moreau, T.A. Schroer, J.E. Rickard, and J.R. De Mey. 1998. Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J. Cell Biol. 141:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev, N.S., S.L. Rogers, R.D. Vale, and H. Ohkura. 2005. Distinct mechanisms govern the localisation of Drosophila CLIP-190 to unattached kinetochores and microtubule plus-ends. J. Cell Sci. 118:3781–3790. [DOI] [PubMed] [Google Scholar]
- Echard, A., G.R. Hickson, E. Foley, and P.H. O'Farrell. 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14:1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky, M., and H. Kuntzel. 2001. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J. Cell Biol. 152:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and R.D. Vale. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., F. Nedelec, and R.D. Vale. 2005. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 171:229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis, E.R., N. Altan, J. Lippincott-Schwartz, and M.A. Powers. 2002. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell. 13:1282–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habu, T., S.H. Kim, J. Weinstein, and T. Matsumoto. 2002. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 21:6419–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine, R.A., J.R. Oberle, and J.A. Cooper. 2000. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151:1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, H., K. Arasaki, N. Dohmae, K. Takio, K. Hatsuzawa, M. Nagahama, K. Tani, A. Yamamoto, M. Tohyama, and M. Tagaya. 2004. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23:1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B.J., B.F. McEwen, J.C. Canman, D.B. Hoffman, E.M. Farrar, C.L. Rieder, and E.D. Salmon. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadura, S., and S. Sazer. 2005. SAC-ing mitotic errors: how the spindle assembly checkpoint (SAC) plays defense against chromosome mis-segregation. Cell Motil. Cytoskeleton. 61:145–160. [DOI] [PubMed] [Google Scholar]
- Karess, R. 2005. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 15:386–392. [DOI] [PubMed] [Google Scholar]
- Kops, G.J., Y. Kim, B.A. Weaver, Y. Mao, I. McLeod, J.R. Yates III, M. Tagaya, and D.W. Cleveland. 2005. a. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops, G.J., B.A. Weaver, and D.W. Cleveland. 2005. b. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 5:773–785. [DOI] [PubMed] [Google Scholar]
- Lew, D.J., and D.J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37:251–282. [DOI] [PubMed] [Google Scholar]
- Liu, S.T., J.M. van Deursen, and T.J. Yen. 2003. The role of mitotic checkpoint in maintaining genomic stability. Curr. Top. Dev. Biol. 58:27–51. [DOI] [PubMed] [Google Scholar]
- Maiato, H., C.L. Rieder, and A. Khodjakov. 2004. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167:831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmanche, N., A. Maia, and C.E. Sunkel. 2006. The spindle assembly checkpoint: preventing chromosome mis-segregation during mitosis and meiosis. FEBS Lett. 580:2888–2895. [DOI] [PubMed] [Google Scholar]
- Mao, Y., A. Desai, and D.W. Cleveland. 2005. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J. Cell Biol. 170:873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis, T., A. Akhmanova, P. Wulf, E. Del Nery, T. Weide, T. Stepanova, N. Galjart, F. Grosveld, B. Goud, C.I. De Zeeuw, et al. 2002. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 4:986–992. [DOI] [PubMed] [Google Scholar]
- Musacchio, A., and E.D. Salmon. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. [DOI] [PubMed] [Google Scholar]
- Obuse, C., O. Iwasaki, T. Kiyomitsu, G. Goshima, Y. Toyoda, and M. Yanagida. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6:1135–1141. [DOI] [PubMed] [Google Scholar]
- Pinsky, B.A., and S. Biggins. 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15:486–493. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., and H. Maiato. 2004. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell. 7:637–651. [DOI] [PubMed] [Google Scholar]
- Rogers, S.L., U. Wiedemann, N. Stuurman, and R.D. Vale. 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.L., U. Wiedemann, U. Hacker, C. Turck, and R.D. Vale. 2004. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr. Biol. 14:1827–1833. [DOI] [PubMed] [Google Scholar]
- Ross, J.L., K. Wallace, H. Shuman, Y.E. Goldman, and E.L. Holzbaur. 2006. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat. Cell Biol. 8:562–570. [DOI] [PubMed] [Google Scholar]
- Scaerou, F., I. Aguilera, R. Saunders, N. Kane, L. Blottiere, and R. Karess. 1999. The rough deal protein is a new kinetochore component required for accurate chromosome segregation in Drosophila. J. Cell Sci. 112:3757–3768. [DOI] [PubMed] [Google Scholar]
- Scaerou, F., D.A. Starr, F. Piano, O. Papoulas, R.E. Karess, and M.L. Goldberg. 2001. The ZW10 and Rough Deal checkpoint proteins function together in a large, evolutionarily conserved complex targeted to the kinetochore. J. Cell Sci. 114:3103–3114. [DOI] [PubMed] [Google Scholar]
- Schroer, T.A. 2004. Dynactin. Annu. Rev. Cell Dev. Biol. 20:759–779. [DOI] [PubMed] [Google Scholar]
- Siller, K.H., M. Serr, R. Steward, T.S. Hays, and C.Q. Doe. 2005. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol. Biol. Cell. 16:5127–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D.A., B.C. Williams, T.S. Hays, and M.L. Goldberg. 1998. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 142:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan, A., T. Nguyen, and B. Suter. 1999. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat. Cell Biol. 1:444–449. [DOI] [PubMed] [Google Scholar]
- Tai, C.Y., D.L. Dujardin, N.E. Faulkner, and R.B. Vallee. 2002. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 156:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum, M.E., N. Galjart, M.A. van Vugt, and R.H. Medema. 2006. CLIP-170 facilitates the formation of kinetochore-microtubule attachments. EMBO J. 25:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S.S., M.I. Scott, and A.J. Holland. 2004. The spindle checkpoint: a quality control mechanism which ensures accurate chromosome segregation. Chromosome Res. 12:599–616. [DOI] [PubMed] [Google Scholar]
- Vale, R.D. 2003. The molecular motor toolbox for intracellular transport. Cell. 112:467–480. [DOI] [PubMed] [Google Scholar]
- Vallee, R.B., K.T. Vaughan, and C.J. Echeverri. 1995. Targeting of cytoplasmic dynein to membranous organelles and kinetochores via dynactin. Cold Spring Harb. Symp. Quant. Biol. 60:803–811. [DOI] [PubMed] [Google Scholar]
- Varma, D., D.L. Dujardin, S.A. Stehman, and R.B. Vallee. 2006. Role of the kinetochore/cell cycle checkpoint protein ZW10 in interphase cytoplasmic dynein function. J. Cell Biol. 172:655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.C., M. Gatti, and M.L. Goldberg. 1996. Bipolar spindle attachments affect redistributions of ZW10, a Drosophila centromere/kinetochore component required for accurate chromosome segregation. J. Cell Biol. 134:1127–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik, E., R. Basto, M. Serr, F. Scaerou, R. Karess, and T. Hays. 2001. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3:1001–1007. [DOI] [PubMed] [Google Scholar]
- Xia, G., X. Luo, T. Habu, J. Rizo, T. Matsumoto, and H. Yu. 2004. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 23:3133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.