Abstract
Three-dimensional (3D) domain swapping creates a bond between two or more protein molecules as they exchange their identical domains. Since the term `3D domain swapping' was first used to describe the dimeric structure of diphtheria toxin, the database of domain-swapped proteins has greatly expanded. Analyses of the now about 40 structurally characterized cases of domain-swapped proteins reveal that most swapped domains are at either the N or C terminus and that the swapped domains are diverse in their primary and secondary structures. In addition to tabulating domain-swapped proteins, we describe in detail several examples of 3D domain swapping which show the swapping of more than one domain in a protein, the structural evidence for 3D domain swapping in amyloid proteins, and the flexibility of hinge loops. We also discuss the physiological relevance of 3D domain swapping and a possible mechanism for 3D domain swapping. The present state of knowledge leads us to suggest that 3D domain swapping can occur under appropriate conditions in any protein with an unconstrained terminus. As domains continue to swap, this review attempts not only a summary of the known domain-swapped proteins, but also a framework for understanding future findings of 3D domain swapping.
Keywords: Functional unit, protein oligomerization, RNase A, IX/X-binding protein, glyoxalase I, T7 gene 4 ring helicase, human prion, human cystatin
Protein oligomers have evolved because of their advantages over their monomers. These advantages include the possibility of allosteric control, higher local concentration of active sites, larger binding surfaces, new active sites at subunit interfaces, and economic ways to produce large protein interaction networks and molecular machines. However, the mechanisms for the evolution of oligomeric interfaces and for the assembly of oligomers during protein synthesis or refolding remain unclear. Different mechanisms have been proposed for the evolution of protein oligomers, among which is three-dimensional (3D) domain swapping (Bennett et al. 1995; Heringa and Taylor 1997). 3D domain swapping holds additional interest because it can also serve as a mechanism for reversible oligomerization, and conceivably for pathological oligomerization, as in amyloids.
Historic background of 3D domain swapping
3D domain swapping is a mechanism for two or more protein molecules to form a dimer or higher oligomer by exchanging an identical structural element ("domain"). If both the monomer and the dimer of a molecule exist in stable forms, in which the dimer adopts a domain-swapped conformation and the monomer adopts a closed conformation, then this protein is considered to be a bona fide example of 3D domain swapping (Fig. 1 ▶). Some proteins form intertwined, apparently domain-swapped oligomers without a known closed monomer. If these proteins have homologs known to be closed monomers, these oligomers are considered to be `quasidomain swapped.' If a protein forms an oligomer by exchanging domains, but there is no monomeric form or homolog for the protein, this protein is considered a candidate for 3D domain swapping (Schlunegger et al. 1997).
Fig. 1.
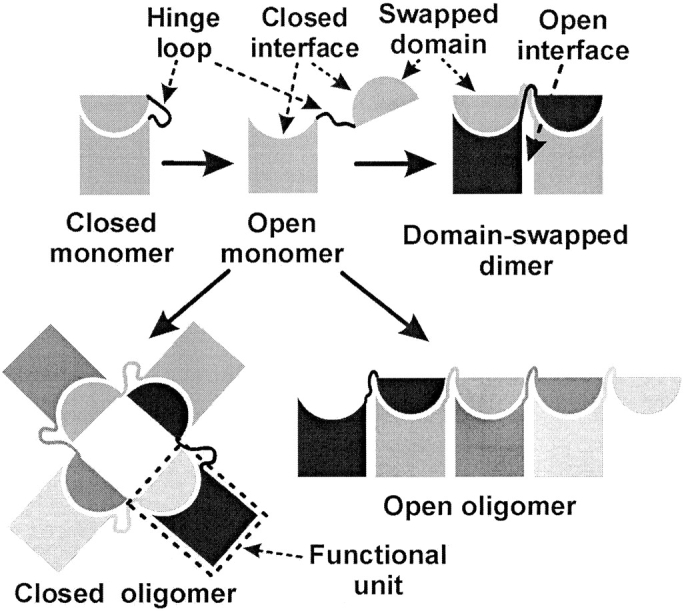
Schematic diagram illustrating terms related to 3D domain swapping. The swapped domain in an oligomer is a globular domain or a structural element of one subunit that extends into another subunit and interacts with the main domain of this subunit. This interaction is essentially identical to that of the same domain in the monomer. The hinge loop is a segment of polypeptide chain that links the swapped domain and the main domain. This loop adopts different conformations in the monomer and the domain-swapped oligomer. The closed interface is the interface between the swapped domain and the main domain that exists in both the monomer and the domain-swapped oligomer. The open interface exists only in the domain-swapped oligomer, but not in the monomer. The functional unit is shown in the dashed box. Its swapped domain and main domain are from different polypeptide chains. Under certain circumstances, a conformational change in the hinge loop converts a closed monomer to an open monomer with its closed interface exposed to the solvent. Two or more such open monomers form a domain-swapped dimer or oligomer. Domain-swapped oligomers are divided into two types: open oligomers and closed oligomers. The open oligomer is linear and has one closed interface exposed to solvent, whereas the closed oligomer is cyclic and does not expose a closed interface.
The term `3D domain swapping' was first used to describe the structure of a diphtheria toxin dimer (Bennett et al. 1994a). However, the concept of 3D domain swapping can be traced back 40 years. Bovine pancreatic ribonuclease (RNase A) forms dimers during lyophilization in acetic acid. Based on elegant chemical modification experiments, Crestfield et al. (1962) proposed that the dimer forms by exchanging the N-terminal fragments. This mechanism is essentially identical to what is now called 3D domain swapping. Similar ideas emerged from other work (Jackson and Yanofsky 1969; London et al. 1974; Miles 1991). Later, several domain-swapped structures were reported before the concept of 3D domain swapping was generalized. The first structural evidence of 3D domain swapping was shown in the cro repressor from bacteriophage λ, which forms a dimer by swapping its C-terminal strands (Anderson et al. 1981). The structure of the monomeric cro with lengthened hinge loop was reported in 1996 to show cro as an example of 3D domain swapping (Albright et al. 1996). Other possibly domain-swapped structures reported before the term 3D domain swapping include chicken citrate synthase (Remington et al. 1982), beef liver catalase (Fita and Rossmann 1985), βB2-crystallin (Bax et al. 1990), Rec A from E. coli. (Story et al. 1992), human CksHs2 (Parge et al. 1993), recombinant human interleukin-5 (IL-5, Milburn et al. 1993), and bovine seminal ribonuclease (BS-RNase, Mazzarella et al. 1993). The structure of γB-crystallin, a homolog of βB2-crystallin, was determined as a monomer in 1981 (Blundell et al. 1981). Thus, βB2-crystallin represents the first structural evidence of quasidomain swapping. Biochemical data showed that the monomeric form of BS-RNase exists (Piccoli et al. 1992), but the structure of the monomer has not been reported. The structure of CksHs1, a homolog of CksHs2, was determined as a monomer in 1995 (Arvai et al. 1995), and the structure of the GM-CSF monomer, a homolog of IL-5, was reported in 1991 (Diederichs et al. 1991). No monomer or monomeric homolog has been reported for chicken citrate synthase, beef liver catalase, or Rec A from E. coli. Diphtheria toxin offered the first structural evidence for bona fide 3D domain swapping, the structures of whose monomer and domain-swapped dimer were both reported in 1994 (Bennett et al. 1994a; Bennett and Eisenberg 1994b). These structures also led to the proposal that 3D domain swapping could be a general mechanism for switching between two protein conformers (Bennett et al. 1994c, 1995).
The definition of several terms related to 3D domain swapping and the possible mechanism for domain swapping can be found in previous reviews (Bennett et al. 1995; Schlunegger et al. 1997). The possible role of 3D domain swapping in the evolution of protein oligomers has been discussed in other reviews (Bennett et al. 1995; Heringa and Taylor 1997; Schlunegger et al. 1997). In the past few years, the number of structures of domain-swapped proteins has vastly increased. This increase elevates our understanding of 3D domain swapping to a higher level, and forms the foundation of this review.
Definition of functional unit
The definitions of several terms related to 3D domain swapping are summarized in Figure 1 ▶. Here we introduce a new term: functional unit (FU).
The FU of a 3D domain-swapped oligomer consists of the portions of two bonded polypeptide chains which form the swapped domain and its associated main domain. It is similar to a closed monomer, except that a closed monomer consists of one polypeptide chain, whereas an FU is composed of two polypeptide chains (Fig. 1 ▶).
Advances in 3D domain swapping
Swapping at the N or C terminus
To date, about 40 domain-swapped proteins have known structures. One common feature of these domain-swapped proteins is that all the swapped domains (except in one protein) are from either the N terminus or the C terminus. In several cases, half of the molecule is swapped, such as in βB2-crystallin (Bax et al. 1990), calbindin D9k (Hakansson et al. 2001), and cyanovirin-N (Yang et al. 1999). In these cases, the proteins are composed of two homologous domains, one of which is swapped. Consequently, it is difficult to define which domain is swapped.
Swapping of more than one domain in a protein
One advance in understanding 3D domain swapping is that more than one domain in a protein can swap. In previous examples of 3D domain swapping, each protein was found with only one domain swapped. It was recently shown that both the N- and C-termini swap in RNase A dimers. RNase A forms a dimer during lyophilization in acetic acid (Crestfield et al. 1962). Further studies showed that there are two types of dimers of RNase A, formed with one dimer more abundant than the other dimer (Libonati et al. 1996; Gotte et al. 1999). Structures of both dimers (Fig. 2 ▶) reveal that the N-terminal helix is swapped in the less abundant dimer (the N-terminal swapped dimer, previously named the minor dimer) (Liu et al. 1998), whereas the C-terminal strand is swapped in the other dimer (the C-terminal swapped dimer, previously named the major dimer) (Liu et al. 2001). RNase A also forms trimers (Gotte et al. 1999). Based on the structures of the N- and C-terminal swapped dimers, a trimeric model with both types of swapping (Fig. 2 ▶) was proposed (Liu et al. 2001). Further biochemical studies support this model and indicate that the model belongs to the more abundant trimer (linear N- and C-terminal swapped trimer, previously named the major trimer). Thus, structural studies of 3D domain swapping in RNase A show that a protein can have domains swapped at both the N- and C-termini, and that these two types of swapping can occur simultaneously in one oligomer. In the less abundant trimer, only the C-terminal strand is swapped, and the structure is cyclic (cyclic C-terminal swapped trimer, previously named the minor trimer, Fig. 2 ▶) (Liu et al. 2002).
Fig. 2.
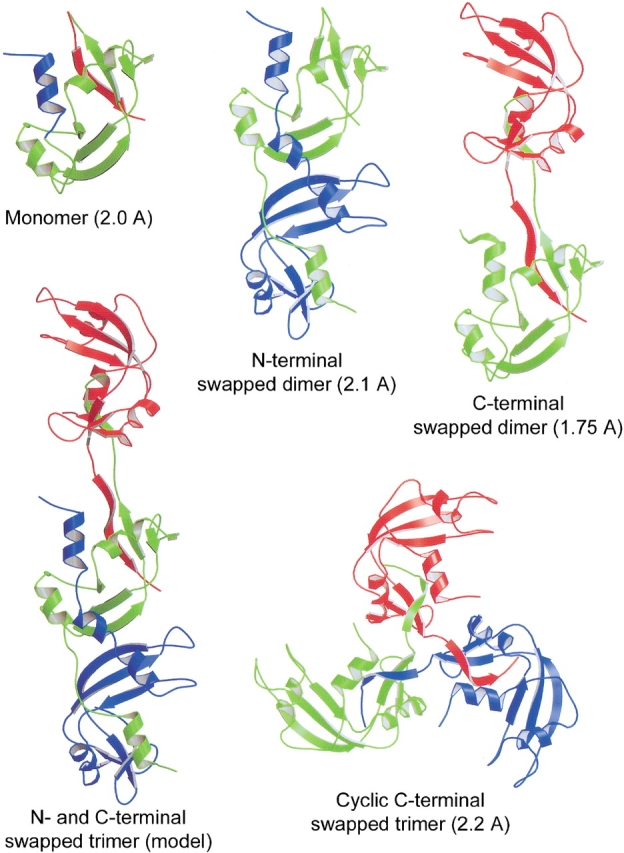
Ribbon diagrams of the structures of the RNase A monomer (2.0 Å, Wlodawer et al. 1982), the N-terminal swapped dimer (2.1 Å, Liu et al. 1998), the C-terminal swapped dimer (1.75 Å, Liu et al. 2001), the N- and C-terminal swapped trimer model (Liu et al. 2001), and the cyclic C-terminal swapped trimer (2.2 Å, Liu et al. 2002). The N- and C-termini are labeled. The N-terminal helix and the C-terminal strand that are swapped in the oligomers are colored blue and red, respectively, in the monomer. The N-terminal swapped dimer swaps its N-terminal helix, whereas the C-terminal swapped dimer swaps its C-terminal strand. Both types of swapping take place in the N- and C-terminal swapped trimer model: The green subunit swaps its C-terminal strand with the red subunit, and swaps its N-terminal helix with the blue subunit. The cyclic C-terminal swapped trimer is 3D domain-swapped at its three C-terminal strands. The three subunits of the molecule are related by a three-fold axis, giving the molecule the shape of a propeller. The figure was created using Raster 3D (Merritt and Bacon 1997).
RNase A also forms tetramers (Gotte et al. 1999). Based on the structures of the dimers and trimers, models are proposed for the tetramers of RNase A (Fig. 3 ▶). Two of these models are linear, with two types of swapping occurring in one molecule (Fig. 3A,B). One model is a combination of cyclic and linear oligomerization, also with both types of swapping occurring in one molecule (Fig. 3C ▶). The last model is a cyclic tetramer, with the swapping only of the C-terminal strand (Fig. 3D ▶).
Fig. 3.
Ribbon representations of hypothetical models of RNase A tetramers. RNase A is known to form tetramers, but their structures are unknown. These models are based on the structures of the RNase A minor dimer, major dimer, and minor trimer. (A) A tetramer model with two C-terminal strands swapped and one N-terminal helix swapped. (B) A tetramer model with one C-terminal strand swapped and two N-terminal helices swapped. (C) A tetramer model with a combination of cyclic and linear oligomerization. In this model, three subunits form a cyclic trimer by swapping the C-terminal strand. One of these three subunits swaps its N-terminal helix with the fourth subunit. This mode of domain swapping can lead to branching chains. (D) A cyclic tetramer model with the swapping of four C-terminal strands. The figure was created with Raster3D (Merritt and Bacon 1997).
Thus, RNase A oligomers display two basic types of 3D domain swapping, and show that with two types of swapping, a protein can form a variety of oligomers.
Diversity of the swapped domains
The swapped domains have diverse sizes and sequences. A swapped "domain" can be one structural element made of several residues. It can also be an entire tertiary domain consisting of hundreds of residues (Tables 1–3). Sequence comparison shows the lack of sequence similarity among these domains. No specific sequence motif can be found among these domains. Therefore, based on its sequence, a protein cannot be predicted to be domain-swapped or not. The diverse size and sequence of the swapped domains indicates that the closed interfaces of these domain-swapped proteins are different from each other. In addition, various types of interactions are formed at different closed interfaces, including hydrophobic interactions, hydrogen-bonding, electrostatic interactions, and even disulfide bridge interactions (Diederichs et al. 1991; Milburn et al. 1993; Knaus et al. 2001). The interactions at the closed interface contribute to the energy required for the disruption of the closed interface during domain swapping. This energy is called the activation energy for 3D domain swapping (Bennett et al. 1995). Therefore, the diverse sizes and sequences of the swapped domains suggest that the activation energy for 3D domain swapping varies among domain-swapped proteins.
Table 1.
Examples of bona fide 3D domain-swapped proteinsa
| Protein (Reference) | PDB code | Number of residues per subunit | Number of residues in swapped domain | Residues in hinge loop | Function | Structure of swapped domain |
| Barnase monomer (Buckle and Fersht, 1994) | 1BRN | 110 | Ribonuclease | |||
| Barnase trimer (Zegers et al. 1999) | 1YVS | 110 | 36 | 37–41 | Ribonuclease | N-terminal helices |
| Calbindin D9k wild-type monomer (Svensson et al. 1992) | 4ICB | 76 | Transcellular calcium transport and magnesium uptake in the intestine | |||
| Calbindin D9k mutant dimer (Hakansson et al. 2001) | 1HT9 | 76 | 27 | 38–47 | Unknown | C-terminal helices |
| Soluble domain CD2 monomer (Jones et al. 1992)b | 1HNG | 177 | Binding domain of lymphocyte adhesion protein | |||
| N-terminal domain of CD2 dimer (Murray et al. 1995)b | 1CDC | 99 | 43 | 44–50 | Unknown | N-terminal β-strands |
| Insertion mutant cro monomer (Albright et al. 1996) | 1ORC | 71 | 55–56d | Unknown | ||
| Wild-type cro dimer (Anderson et al. 1981)b | 1CRO | 66 | 11 | 55 | DNA repressor | C-terminal β-strand |
| Cyanovirin-N monomer (Bewley et al. 1998) | 2EZM | 101 | HIV inactivation | |||
| Cyanovirin-N dimer (Yang et al. 1999) | 3EZM | 101 | 48 | 50–53 | Unknown | N- or C-terminal globular domain |
| DT monomer (Bennett and Eisenberg 1994b)b | 1MDT | 535 | ADP-ribosylating toxin | |||
| DT dimer (Bennett et al. 1994a)b | 1DDT | 535 | 148 | 379–387 | Speculative receptor binding | C-terminal globular domain |
| Human prion monomer (Zahn et al. 2000) | 1QLX | 108 | Unknown | |||
| Human prion dimer (Knaus et al. 2001) | 1I4M | 108 | 28 | 188–198 | Unknown | C-terminal helices |
| Protein L B1 domain monomer (O'Neill et al. 2001) | 1HZ5 | 62 | Binding of Ig G | |||
| Protein L B1 domain dimer (Kuhlman et al. 2001) | 1JML | 61 | 7 | 52–55 | Unknown | C-terminal β-strand |
| RNase A monomer (Wlodawer et al. 1982) | 5RSA | 124 | Ribonuclease | |||
| RNase A N-terminal swapped dimer (Liu et al. 1998) | 1A2W | 124 | 14 | 15–22 | Ribonuclease | N-terminal α-helix |
| RNase A C-terminal swapped dimer (Liu et al. 2001) | 1F0V | 124 | 9 | 112–115 | Ribonuclease | C-terminal β-strand |
| RNase A cyclic C-terminal swapped trimer (Liu et al. 2002) | 1JS0 | 124 | 9 | 112–115 | Ribonuclease | C-terminal β-strand |
| BS-RNase dimer (Mazzarella et al. 1993)b | 1BSR | 124 | 14 | 15–22 | Ribonuclease | N-terminal α-helix |
| Single-chain Fv monomer of antibody NC10 (Malby et al. 1998) | 1NMC | 246 | 113–127 | Antigen binding | ||
| Diabody (Perisic et al. 1994) | 1LMK | 243 | 122 or 116 | 123–127 | Antigen binding | N- or C-terminal globular domain |
| Triabody (Pei et al. 1997) | 1NQB | 236 | 120 or 116 | 0 | Antigen binding | N- or C-terminal globular domain |
| Phosphorylated N-Spo0A (Lewis et al. 1999) | 1QMP | 129 | Response regulator related to sporulation | |||
| N-domain of Spo0A dimer (Lewis et al. 2000) | 1DZ3 | 129 | 21 | 103–109 | Response regulator related to sporulation | C-terminal α-helix |
| Wild-type staphylococcal nuclease monomer (Loll and Lattman 1989)b | 1SNC | 149 | 112–120 | Nuclease | ||
| Deletion mutant of staphylococcal nuclease dimer (Green et al. 1995) | 1SND | 143 | 29 | 112–114 | Unknown | C-terminal α-helix |
| suc1 monomer (Endicott et al. 1995) | 1SCE | 113 | Cell cyle regulation | |||
| suc1 dimer (Khazanovich et al. 1996)b | 1PUC | 113 | 22 | 85–91 | Cell cycle regulation | C-terminal β-stand |
| TrkA-d5 and NGF complex (Wiesmann et al. 1999) | 1WWW | 120 | Binding of nerve growth factor | |||
| TrkA-d4 dimer | 1WWA | 109 | 13 | 297–299 | Unknown | N-terminal β-strand |
| TrkC-d5 dimer | 1WWB | 103 | 14 | 299–301 | ||
| TrkC-d5 dimer (Ultsch et al. 1999) | 1WWC | 118 | 14 | 317–319 |
a The oligomers of these proteins are domain-swapped and their monomers adopt a closed conformation.
b These entries are taken from the previous review (Schlunegger et al. 1997).
Table 2.
Examples of proteins that exhibit quasi-domain swappinga
| Protein (Reference) | PDB code | Number of residues per subunit | Number of residues in swapped domain | Residues in hinge loop | Function | Structure of swapped domain |
| CksHs1 monomer (Arvai et al. 1995)b | 1DKS | 79 | Cell cycle regulation | |||
| CksHs2 dimer (Parge et al. 1993)b | 1CKS | 79 | 14 | 60–65 | Cell cycle regulation | C-terminal β-strand |
| γ-crystallin II monomer (Blundell et al. 1981) | 4GCR | 174 | Eye lens protein | |||
| βB2-crystallin dimer (Bax et al. 1990) | 1BLB | 204 | 97 | 79–87 | Eye lens protein | N- or C-terminal globular domain |
| Chicken cystatin monomer (Bode et al. 1988) | 1CEW | 108 | Cysteine protease inhibitor | |||
| Human cystatin C dimer (Janowski et al. 2001) | 1G96 | 120 | 54 | 55–59 | Unknown | N-terminal helix and strand |
| E. coli glyoxalase I dimer (He et al. 2000) | 1FA5 | 135 | Interconversion of glutathione thiohemiacetal of methylglyoxal and S-D-lactoyl-glutathione | |||
| Human glyoxalase I dimer (Cameron et al. 1997) | 1BH5 | 183 | 19 | 20–32 | Interconversion of glutathione thiohemiacetal of methylglyoxal and S-D-lactoyl-glutathione | N-terminal α-helix |
| GM-CSF monomer (Diederichs et al. 1991)b | 1GMF | 127 | 87–99 | Granulocyte macrophage growth factor | ||
| IL-5 dimer (Milburn et al. 1993)b | 1HUL | 113 | 26 | 82–89 | B and T cell growth factor | C-terminal strand and helix |
| IFN-β monomer (Senda et al. 1992)b | 1RM1 | 160 | 97–114 | Fibroblast interferon | ||
| IL-10 dimer (Zdanov et al. 1995)b | 1ILK | 160 | 46 | 108–118 | Cytokine inhibitory synthesis factor | C-terminal helices |
| Mannose binding protein (Weis et al. 1991) | 1MSB | 115 | Mannose binding | |||
| IX/X-binding protein (Mizuno et al. 1997) | 1IXX | Heterodimer 129 and 123 | 17 | Loop1: 72–75 Loop2: 93–98 | Anticoagulation | Middle loop |
| Major urinary protein monomer (Bocskei et al. 1992)b | 1MUP | 166 | 126–130 | Rodent pheromone transporter | ||
| Odorant binding protein dimer (Tegoni et al. 1996)b | 1OBP | 159 | 35 | 121–124 | Odorant binding and transport | C-terminal stand and helix |
| Human pancreatic ribonuclease chimera (Canals et al. 2001) | 1H8X | 128 | 15 | 16–23 | RNA digestion (homolog of RNase A and BS-RNase) | N-terminal α-helix |
| Grb2 adaptor (SH2 + SH3) (Maignan et al. 1995) | 1GR1 | 217 | Signal transduction | |||
| Grb2-SH2 domain dimer (Schiering et al. 2000) | 1FYR | 93 | 27 | 121–123 | Binding phosphorylated peptide | C-terminal α-helix |
| Fyn-SH3 monomer (Musacchio et al. 1994)b | 1FYN | 62 | 112–118 | Signal transduction | ||
| SH3 domain of Eps8 (Kishan et al. 1997) | 1AOJ | 65 | 26 | 34–39 | Recognition of proline-rich sequences | C-terminal β-strands |
| SigE dimer (Luo et al. 2001) | 1K3S | 113 | Molecular chaperone in type III secretion system | |||
| CesT dimer (Luo et al. 2001) | 1K3E | 156 | 32 | 33–36 | Molecular chaperone in type III secretion system | N-terminal α-helix and β-strand |
| Monomer of two repeats of α-spectrin (Grum et al. 1999) | 1CUN | 211 | Cytoskeletal protein | |||
| Dimer of one repeat of α-spectrin (Yan et al. 1993)b | 2SPC | 107 | 32 | 72–75 | Cytoskeletal protein | C-terminal α-helix |
a These proteins show oligomeric structures with 3D domain swapping, and their homologs adopt the closed monomeric conformation.
b These entries are taken from the previous review (Schlunegger et al. 1997).
Table 3.
Candidates for 3D domain swappinga
| Protein (Reference) | PDB code | Number of residues per subunit | Number of residues in swapped domain | Residues in hinge loop | Function | Structure of swapped domain |
| Bleomycin resistance protein dimer (Dumas et al. 1994) | 1BYL | 122 | 8 | 9–11 (?) | Bleomycin resistance | N-terminal β-strand |
| BTB domain of PLZF (Ahmad et al. 1998) | 1BUO | 120 | 12 | 13–23 (?) | Evolutionarily conserved protein-protein interaction motif | N-terminal β-strand |
| Cab-type β class carbonic anhydrase (Strop et al. 2001) | 1G5C | 170 | 12 | 13–23 (?) | Catalyzing reversible hydration of CO2 | N-terminal α-helix |
| Catalase dimer (beef liver) (Fita and Rossmann 1985) | 7CAT | 500 | 65 | 66–70 (?) | H2O2 hydrolysis | N-terminal helices |
| Citrate synthase dimer from chicken heart (Remington et al. 1982) | 1CTS | 433 | 10 | 417–423 (?) | Citrate synthesis | C-terminal helix |
| Designed coiled coil dimer (Ogihara et al. 2001) | 1G6U | 48 | 14 | 33–34 (?) | Unknown | C-terminal α-helix |
| dUTPase trimer (Larsson et al. 1996) | 1DUD | 136 | 10 | 125–126 (?) | dUTP hydrolysis | C-terminal strand |
| Heat shock protein 33 dimer (Vijayalakshmi et al. 2001) | 1HW7 | 255 | 75 | 178–184 (?) | Molecular chaperone | C-terminal helices and strands |
| Phosphoenolpyruvate mutase dimer (Huang et al. 1999) | 1PGM | 295 | 35 | 240–260 (?) | Converting phosphoenolpyruvate to phosphonopyruvate | C-terminal helices |
| T4 endonuclease VII dimer (Raaijmakers et al. 1999) | 1EN7 | 157 | 60 | 63–72 (?) | Mismatch repair, Resolving branchpoint before phage package | N-terminal helix and strands |
| T7 gene 4-ring helicase hexamer fragment (Singleton et al. 2000) | 1E0J | 326 | 40 | 283–305 (?) | Separation of nucleic acid duplexes into strands | N-terminal α-helix and β-strand |
| T7 gene 4-ring helicase fragment (Sawaya et al. 1999) | 1CR0 | 296 | 12 | 283–305 (?) | Unknown | N-terminal α-helix |
| RecA hexamer from E. coli (Story et al. 1992) | 2REB | 352 | 26 | 27–39 (?) | DNA recombination and repair | N-terminal α-helix |
| Simian virus 40 oligomer (Stehle et al. 1996) | 1SVA | 361 | 61 | 296–300 (?) | Virus coat protein | C-terminal helix and strands |
a These proteins show oligomeric structures with exchanging domains, but have not been shown to have monomers or monomeric homologs with a similar structure. Therefore, the residues forming hinge loops in these proteins are speculative, and are indicated here with a question mark.
The swapped domains also have diverse secondary structures. Among the domain-swapped proteins with reported structures, a swapped domain can be one α-helix (BS-RNase, RNase A N-terminal swapped dimer, staphylococcal nuclease dimer, Spo0A, etc.), one β-strand (CksHs2 dimer, cro dimer, RNase A C-terminal swapped dimer, BTB domain of PLZF, etc.), several α-helices (calbindin D9k, barnase, phosphoenolpyruvate mutase, human IL-10, etc.), several β-strands (β-B2 crystallin, diphtheria toxin dimer, SH3 domain of Eps8, N-terminal domain of CD2, etc.), or a mixture of α-helix and β-strand (T7 gp 4 ring helicase, human cystatin C, human IL-5, T4 endonuclease VII, etc.). This diversity shows that 3D domain swapping does not require or prefer certain types of secondary structure.
In summary, the diversity of the swapped domains indicates that 3D domain swapping does not depend on the protein sequence or secondary structure.
Flexibility and diversity of the hinge loops
According to the definition in Figure 1 ▶, a hinge loop has the intrinsic flexibility to adopt different conformations in the monomer and in the domain-swapped oligomer. Several recent structural studies of 3D domain swapping further support the flexibility of the hinge loops. The flexibility is evident in RNase A, BS-RNase, and human pancreatic ribonuclease (hRNase) chimera. RNase A and BS-RNase show 80% sequence identity, and BS-RNase and hRNase chimera share the common hinge loop. All three of these proteins swap the N-terminal helix; however, the relative orientations of the subunits in their dimers are different, resulting in different conformations for the three hinge loops (Mazzarella et al. 1993; Liu et al. 1998; Canals et al. 2001). Flexibility is also displayed in the C-terminal hinge loop of RNase A: the C-terminal strand of RNase A is swapped in both the C-terminal swapped dimer and the cyclic C-terminal swapped trimer of RNase A; however, the subunits are related by a two-fold axis in the C-terminal swapped dimer and by a three-fold axis in the cyclic C-terminal swapped trimer (Fig. 2 ▶). Therefore, the same hinge loop adopts different conformations in the monomer, the C-terminal swapped dimer, and the cyclic C-terminal swapped trimer of RNase A, showing the great flexibility of this hinge loop.
Hinge loops display a variety of secondary structures in domain-swapped proteins. Some hinge loops are coils, some form β-strands, and others form α-helices. In the RNase A N-terminal swapped dimer, one hinge loop forms a coil, and the other forms a helix (Liu et al. 1998). A common feature is that when the hinge loop forms a β-strand or an α-helix, the oligomeric form is favored over the monomer. These proteins usually exist as dimers in vivo or have dimeric forms more stable than the monomeric forms. Other cases of domain-swapped oligomers that are more stable than their monomers are those for which the hinge loop is not long enough for the swapped domain to fold back to the same peptide chain (Bennett et al. 1995).
New examples of 3D domain swapping in proteins
To date, about 40 domain-swapped proteins have been reported. These proteins are involved in different biological functions. The reported domain-swapped proteins are listed in Tables 1–3. Here, we discuss six domain-swapped proteins presenting aspects of 3D domain swapping that were not known at the time of previous reviews.
IX/X-binding protein
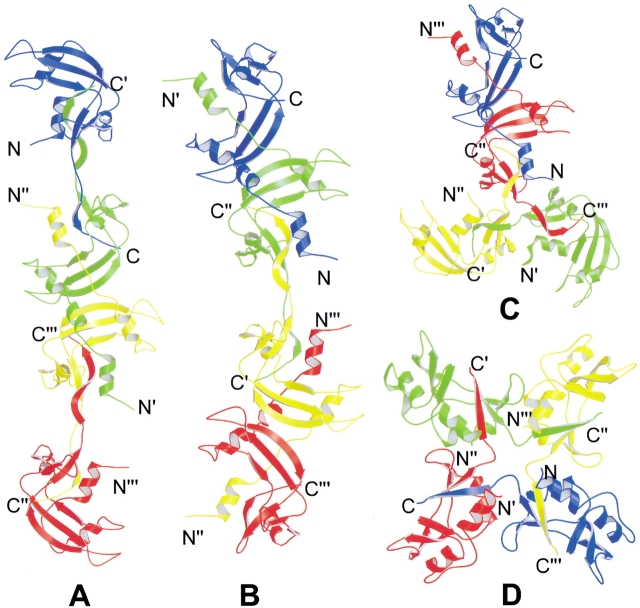
As mentioned above, most swapped domains are at either the N or C terminus. The only exception found to date is blood coagulant factors IX/X-binding protein (IX/X-bp, Mizuno et al. 1997). IX/X-bp is an anticoagulant isolated from the venom of the habu snake. It consists of two homologous subunits linked by an intermolecular disulfide bond. The two subunits form a heterodimer by exchanging a loop in the central part of the molecules (Fig. 4 ▶). Structural comparison of the two subunits with mannose binding protein (MBP) shows that they adopt the same fold, except that the exchanged loop in the IX/X-bp folds back to the same polypeptide chain in MBP. Thus, IX/X-bp is quasidomain-swapped. IX/X-bp is the only known example of 3D domain swapping taking place in the middle of the molecule, in contrast to other domain-swapped proteins, in which domain swapping takes place at either the N or C terminus. Because domain swapping takes place in the middle of IX/X-bp, there are two hinge loops in each subunit. In addition, IX/X-bp is the only known example of a domain-swapped heterodimer; all other domain-swapped proteins are homooligomers.
Fig. 4.

Ribbon diagram of the structure of blood coagulant factor IX/X-bp (Mizuno et al. 1997). IX/X-bp is a heterodimer, with subunit A in red and subunit B in green. The N- and C-termini are indicated. The middle loops of the two subunits are swapped. IX/X-bp is the first example of 3D domain swapping of a heterodimer and with the central part of the molecule swapped. The figure was created with Raster3D (Merritt and Bacon 1997).
RNase A linear N- and C-terminal swapped trimer and cyclic C-terminal swapped trimer
The structures of the RNase A trimers support the earlier proposal that a protein can form a linear or cyclic oligomer by 3D domain swapping. RNase A forms two types of trimers during lyophilization in acetic acid (Gotte et al. 1999). One trimer is slightly more abundant than the other trimer. A linear model with the swapping of both the N- and C-termini was proposed for the more abundant trimer (Fig. 2 ▶; Liu et al. 2001). Further biochemical studies support this model (Liu et al. 2002). With both types of swapping taking place, the linear model does not have an exposed closed interface, and therefore has no dangling domains. This model is thermodynamically more favorable than the previous linear model (Fig. 1 ▶; Bennett et al. 1995), which had an exposed closed interface with one type of swap. In addition, the crystal structure of the RNase A N-terminal swapped trimer shows that it is a cyclic molecule, with swapping of the C-terminal strands (Fig. 2 ▶; Liu et al. 2002). Thus, RNase A represents the first protein found to form both linear and cyclic domain-swapped oligomers.
The two different modes of domain swapping in the RNase A linear N- and C-terminal swapped trimer result in two hinge loops in the molecule. However, this is different from the dimer of IX/X-bp, which also has two hinge loops. In the RNase A major trimer, there is a swapped domain at both termini, whereas in IX/X-bp, there is only one domain swapped in the middle of the molecule.
Glyoxalase I
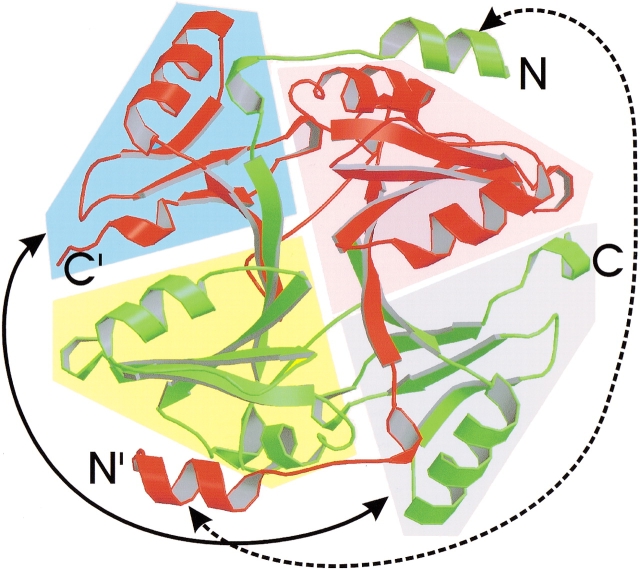
Glyoxalase I (Glx I) catalyzes the interconversion of the glutathione thiohemiacetal of methylglyoxal and S-D-lactoyl-glutathione. Human Glx I is a dimer with 183 amino acid residues per monomer. Each monomer is composed of an N-terminal helix and two homologous domains. The crystal structure of this dimer is 3D domain-swapped at the N-terminal helix (Fig. 4 ▶; Cameron et al. 1997). Sequence comparison with E. coli Glx I shows that this N-terminal helix does not exist in E. coli Glx I. The function of this N-terminal helix may be to stabilize the human Glx I dimer, although it is not required for dimerization of Glx I (He et al. 2000). The active sites exist at the dimer interface, and are therefore composite. According to Cameron et al. (1997), the dimer also swaps its C-terminal globular domain, in addition to swapping the N-terminal helix. The swapping of the C-terminal domain is ambiguous, however, because there are two ways to define the monomer. As shown in Figure 5 ▶, in one way, the monomer can be divided into two domains in the pink- and blue-shaded areas, as seen in the crystal structure. These two domains are from the same polypeptide chain, and therefore, the C-terminal domain is not swapped in the crystal structure. In such a monomer, the two domains have extensive interactions and would be stable in solution. But in this conformation, the active site is incomplete and the monomer would be inactive. The second way to define a monomer is to divide the monomer into two domains in the pink- and gray-shaded areas (Fig. 5 ▶), as proposed by Cameron et al. (1997). According to this definition, one has to move the C-terminal domain in the blue-shaded area in the crystal structure to replace the same domain in the gray-shaded area from the other subunit to obtain a monomer (shown by the arrowheads connected by a solid line in Fig. 5 ▶). In such a case, the C-terminal domain is swapped and the active site is complete, but the interactions between the two domains are limited, which may result in an unstable monomer. Biochemical data show an equilibrium of monomer and dimer in Glx I from Pseudomonas putida (55% sequence identity to human Glx I), and both the monomer and the dimer are active (Saint-Jean et al. 1998). This indicates that the active site in the monomer of P. putida Glx I should be complete, as in the second definition of monomer mentioned above. Based on these biochemical data, we regard human Glx I as a protein with two domains swapped, as proposed by Cameron et al. (1997). The crystal structure of the monomer will give a definitive answer to this question.
Fig. 5.
Ribbon diagram of the structure of human glyoxalase I (Cameron et al. 1997). Glyoxalase I is a homodimer, shown with one subunit colored red and one green. The N-and C-termini are labeled. The N-terminal helix is swapped (shown by the arrowheads on the dashed line). There are two ways to define the monomer. In one way, the monomer is composed of the domains in the pink- and blue-shaded areas. These two globular domains of the monomer are from the same polypeptide chain (red or green), and therefore, the C-terminal domain is not swapped. In the second way, the monomer is composed of the domains in the blue- and gray-shaded areas. To obtain such a monomer, the domain in the gray-shaded area must be replaced by the domain in the blue-shaded area in the crystal structure, as shown by the arrowheads on the solid line. Then the C-terminal domain is considered to be swapped, and therefore there are two swapped domains in this case. The figure was created with Raster3D (Merritt and Bacon 1997).
T7 gene 4 ring helicase
T7 gene 4 ring helicase (T7 helicase) is a 5`-3` helicase from bacteriophage T7 whose crystal structure displays a screw-axis form of open oligomeric 3D domain swapping. It contains 566 amino acid residues, and comprises separate helicase and primase domains (Bird et al. 1997; Frick et al. 1998). The active form of T7 helicase is a hexamer. The helicase loses the ability to form a hexamer, and thus loses its activity, after the truncation of its N-terminal residues 1–271. However, in crystals, T7 helicase 4E fragment (residues 272–566) forms a helical filament along a six-fold screw axis by swapping its N-terminal helix (Fig. 6 ▶; Sawaya et al. 1999). The active fragment, T7 helicase 4D (residues 241–566), forms a stable hexamer by swapping its N-terminal helix and strand (Singleton et al. 2000). Therefore, 3D domain swapping is required for the hexamerization and the activity of T7 helicase. T7 helicase provides the structural evidence for 3D domain swapping in a hexamer, supporting the proposal that 3D domain swapping is a mechanism for extended protein oligomerization (Bennett et al. 1995).
Fig. 6.
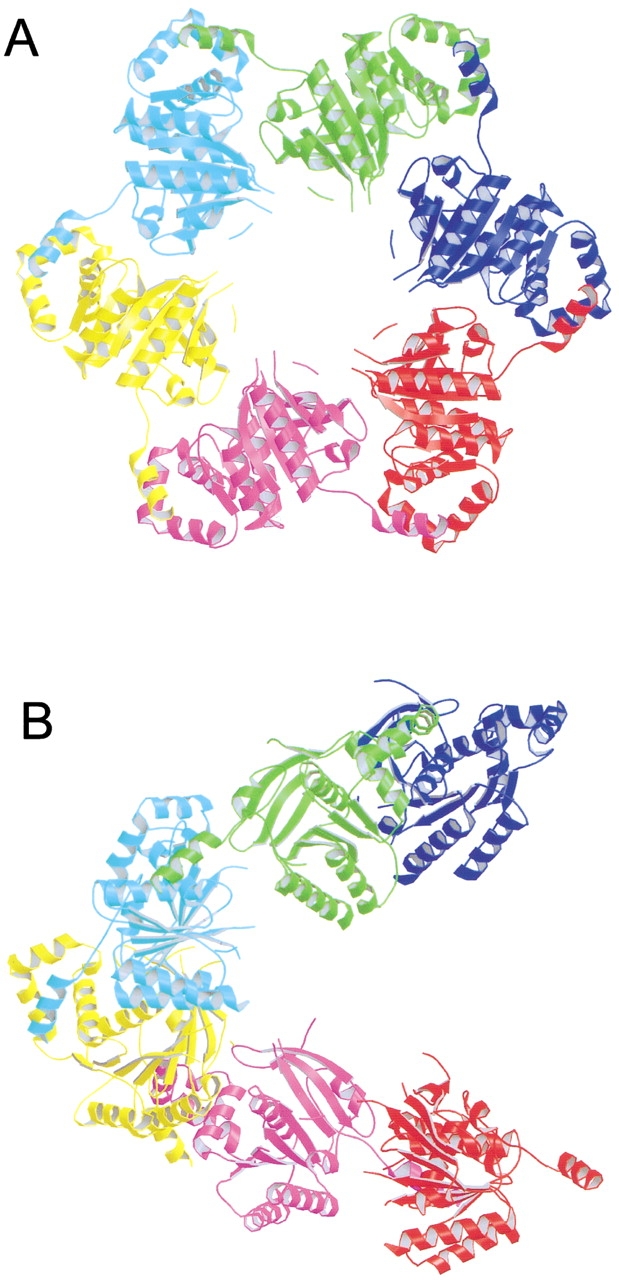
An open oligomeric domain-swapped structure, illustrated by a ribbon diagram of the structure of T7 helicase 4E fragment (Sawaya et al. 1999). The hexamer of T7 helicase 4E has a six-fold screw axis in the molecule, which is coincident with the crystallographic symmetry. 3D domain swapping is displayed along the screw axis throughout the crystal. (A) A view of the hexamer along the screw axis. (B) A view of the hexamer perpendicular to the screw axis. The figure was created with Raster3D (Merritt and Bacon 1997).
In T7 helicase 4E, each subunit involved in domain swapping is related by a six-fold screw axis. Therefore, these molecules do not form a closed ring and 3D domain swapping is extended in the helical filaments throughout the whole crystal (Sawaya et al. 1999). This type of swapping is what was proposed in the linear open oligomer (Fig.1 ▶; Bennett et al. 1995). This phenomenon is also seen in the crystal structure of the "cab"-type β class carbonic anhydrase from Archaeon Methanobacterium thermoautotrophicum, and RecA from E. coli. In RecA, domain swapping is also displayed along a six-fold screw axis (Story et al. 1992), whereas in carbonic anhydrase, domain swapping is displayed along a two-fold screw axis throughout the crystal (Strop et al. 2001).
In summary, the "cab"-type β class carbonic anhydrase, RecA, and the T7 helicase display open-oligomeric 3D domain swapping, which can be seen in crystals because the oligomer screw-axis symmetry is also a symmetry of the crystal.
Human prion and human cystatin C
3D domain swapping has been proposed as a mechanism for amyloid formation (Klafki et al. 1993; Schlunegger et al. 1997; Cohen and Prusiner 1998; Liu et al. 1998, 2001). However, there was no structural evidence for amyloidogenic proteins to be domain-swapped until human prion (Knaus et al. 2001) and human cystatin C (Janowski et al. 2001; Staniforth et al. 2001) were reported to be domain-swapped. Both human prion and cystatin C form fibers and are related to amyloid diseases. Crystal and NMR structures of these proteins show that they form domain-swapped dimers. Based on these structures, models with 3D domain swapping were proposed for amyloid formation (Janowski et al. 2001; Knaus et al. 2001; Staniforth et al. 2001). In addition, Rousseau et al. (2001) observed a qualitative correlation between domain swapping and aggregation propensity of p13suc1 mutants. Although it is unclear whether the domain-swapped dimer is the building block for these fibers, these studies show that domain swapping and amyloid formation may share common intermediates, and thus suggest that 3D domain swapping is a possible mechanism for amyloid formation.
Physiological relevance or crystallographic artifact?
As the number of domain-swapped proteins continues to increase, the question of the physiological relevance of 3D domain swapping grows in importance. If domain swapping is biologically relevant, how does domain swapping regulate biological functions of the swapped molecules? By examining domain-swapped proteins, we conclude that domain swapping is physiologically relevant in some proteins, but not in others. Here, we list several examples for both situations.
Physiological relevance
As mentioned above, RNase A forms dimers and trimers during lyophilization in acetic acid (Crestfield et al. 1962; Gotte et al. 1999). Can these oligomers form under physiological conditions? RNase A was reported to dimerize at pH 6.5 and 37°C, similar to the physiological conditions. The dissociation constant for the dimer at this condition is ∼2 mM, which is about 20-fold greater than the concentration of RNase A in the bovine pancreas, suggesting that a small amount of RNase A dimer does exist in vivo (Park and Raines 2000). In addition, RNase A oligomers display higher enzyme activity on double-strand RNA than does the monomer (Gotte et al. 1999). These results suggest that 3D domain swapping in RNase A exists in vivo and may play some physiological role.
In addition, 3D domain swapping in bovine seminal ribonuclease (BS-RNase, sharing 80% sequence identity with RNase A, Mazzarella et al. 1993) was reported to be necessary for its immunosuppression activity and allostery activity (Piccoli et al. 1988; Cafaro et al. 1995), indicating the physiological role of 3D domain swapping.
Important progress has recently been made in understanding the role of 3D domain swapping in biological functions. Single point mutations at the closed interface of the suppressor of cyclin dependent kinase 1 (p13suc1) shifted the equilibrium between monomer and dimer (Schymkowitz et al. 2001). Since the closed interface exists in both the monomer and the dimer, the mutation at the closed interface should affect both forms, and therefore should not affect the equilibrium. It was reported that there is a strain at the hinge loop of suc1 which controls the equilibrium of the monomer and the dimer through 3D domain swapping (Rousseau et al. 2001). The mutations that shift the equilibrium are distant from the hinge loop, suggesting that the strain at the hinge loop can be "sensed" by the remote mutation sites. Ligand binding to suc1, which is distant from the hinge loop, also shifted the equilibrium, further supporting the suggestion of sensing remote strain (Schymkowitz et al. 2001). These studies provide evidence for 3D domain swapping as a mechanism for allostery and signal sensing in a macromolecule, and therefore for regulating biological functions of proteins.
Macromolecular crowding supports the possible physiological relevance of 3D domain swapping. Cells are crowded with macromolecules (Goodsell 1993). The effect of other macromolecules on a specific macromolecule in cells has been studied and termed "macromolecular crowding" (Minton 2001). Macromolecular crowding increases protein local concentration and facilitates protein oligomerization (Cole and Ralston 1994; Lindner and Ralston 1995; Rivas et al. 1999). Since high concentration of a protein favors 3D domain swapping, macromolecular crowding may facilitate 3D domain swapping in vivo. Macromolecular crowding also stabilizes protein oligomers (Eggers and Valentine 2001). This effect was seen during the crystallization of the RNase A minor trimer, which is stabilized by polyethylene glycol (PEG) 10,000 even at pH 3.5 (Liu et al. 2002). Therefore, even though the dissociation constant for the dimer of RNase A is greater than the concentration of RNase A in bovine pancreas, the amount of dimers formed in vivo may be higher than the amount calculated from the dissociation constant, due to macromolecular crowding. That is, thermodynamic activity exceeds concentration. Thus, macromolecular crowding in cells increases the population of domain-swapped oligomers and thus in a general way adds support to the possible physiological relevance to 3D domain swapping.
3D domain swapping induced by receptor/ligand binding provides more evidence for the physiological relevance of domain swapping. Diphtheria toxin (DT), which enters cells by endocytosis, was first found to form a domain-swapped dimer upon lowering pH (Carroll et al. 1986; Bennett et al. 1994a). Although this low pH may mimic the environment of an endosome, more direct evidence of the physiological relevance of 3D domain swapping in DT comes from the crystal structure of the complex of DT and a domain of its receptor showing that DT forms domain-swapped dimer upon binding to its receptor at neutral pH (Louie et al. 1997). 3D domain swapping regulated by ligands was reported in glyoxalase I (Saint-Jean et al. 1998) and p13suc1 (Schymkowitz et al. 2001), where the equilibrium between the monomer and the dimer is regulated by glutathione and phosphopeptide, respectively, suggesting that ligand binding may regulate the functions of its receptor through 3D domain swapping. Similarly, 3D domain swapping was proposed as a mechanism for the oligomerization of membrane-associated guanylate kinases regulated by their ligand binding (McGee et al. 2001).
In addition, support for physiological relevance of 3D domain swapping can be found in the proteins that exist as domain-swapped oligomers in vivo. These proteins include BS-RNase (Mazzarella et al. 1993), T7 helicase (Singleton et al. 2000), cro repressor (Anderson et al. 1981), phosphoenolpyruvate mutase (Huang et al. 1999), T4 endonuclease VII (Raaijmakers et al. 1999), IX/X-binding protein (Mizuno et al. 1997), and bleomycin resistance protein (Dumas et al. 1994). The specific role of 3D domain swapping in these proteins is still unclear. However, the active forms of these proteins are domain-swapped, suggesting that their 3D domain swapping is related to their biological functions in vivo.
Artifact
Several domain-swapped oligomers are obtained under nonphysiological low pH, and the biological functions of the oligomers are unknown. Barnase is active as a monomer. At pH 4.5, it forms a domain-swapped trimer (Zegers et al. 1999). The N-terminal domain of sporulation response regulator Spo0A forms a domain-swapped dimer at pH 4.0. However, when this domain is phosphorylated, it exists as a monomer. The contradictory fact is that the phosphorylated whole Spo0A is a dimer in solution (Lewis et al. 2000). It is unclear whether domain swapping takes place in this dimer, since the structure of the whole Spo0A is not available. Cyanovirin-N (Yang et al. 1999) was also crystallized under low pH and was shown to be domain-swapped. Although low pH environments exist in some compartments of cells, there is no indication of a relationship of these domain-swapped proteins to those compartments.
In addition, several domain-swapped proteins are fragments of their complete molecules, whereas the intact molecule is a monomer. The most obvious example is Domain 5 of TrkA, TrkB, and TrkC. Domain 5 alone is domain-swapped at its N-terminal strand (Ultsch et al. 1999). However, the domain-swapped dimer is incapable of binding to the natural ligand (Urfer et al. 1995). In addition, there are four domains N-terminal to Domain 5 in the intact molecule. These four domains may block the swapping of the N-terminal strand of Domain 5 and result in a monomer of the intact molecule. Therefore, domain swapping in Domain 5 is regarded as a consequence of the truncation of the whole protein (Ultsch et al. 1999) and is not of physiological significance.
Other examples include the N-terminal domain of Spo0A (Lewis et al. 2000), the BTB domain from PLZF (Ahmad et al. 1998), the SH3 domain of Eps8 (Kishan et al. 1997), and the SH2 domain of Grb2 (Schiering et al. 2000), which are also part of their whole molecules, and show domain swapping. However, whether their entire polypeptide chains are domain-swapped remains to be seen. Although the physiological relevance of domain swapping in these domains remains controversial, these examples suggest that smaller domains form domain-swapped oligomers more easily than larger domains, and may do so under nonphysiological conditions.
The mechanism of 3D domain swapping
Although about 40 proteins have been reported to be domain-swapped, studies on the mechanism of 3D domain swapping are few (Hayes et al. 1999; Kuhlman et al. 2001; Rousseau et al. 2001; Schymkowitz et al. 2001), and to date, the mechanism of 3D domain swapping remains elusive. Based on the monomeric and dimeric structures of DT and the conditions to form its dimer, a free energy diagram was proposed for the pathways of 3D domain swapping (Bennett et al. 1995). According to this proposal, the closed interface in a closed monomer is disrupted under certain conditions to form an open monomer. There is a high energy difference between the closed and the open monomers, which is the activation energy. Two or more open monomers aggregate to form a domain-swapped dimer or oligomer. The free energy difference between the closed monomer and domain-swapped oligomer is small, because they share the same structures except at the hinge loop. Therefore, there is a high energy barrier between the closed monomer and the domain-swapped oligomer. This energy barrier can be reduced under certain circumstances, such as change of pH, change of temperature, mutation in the protein, presence of denaturants, and binding of a ligand. In short, the current energetic model for the formation of 3D domain swapping is that of a high energy barrier that can be reduced by a change in solution conditions.
Recent studies on 3D domain swapping show that there are three factors that affect the free energy difference between the monomer and the domain-swapped oligomer. First, the greater entropy of the monomer makes it more favored thermodynamically. Second, hinge loops may form new interactions in the domain-swapped dimer, which favor dimerization. Also, there may be strains introduced or relieved when a protein forms a domain-swapped dimer. Therefore, the conformational changes at the hinge loop also contribute to this free energy difference. Third, new interactions at the open interface make the domain-swapped oligomer more favorable thermodynamically (Kuhlman et al. 2001; Liu et al. 2001; Rousseau et al. 2001; Schymkowitz et al. 2001). Therefore, by changing the hinge loop or engineering the open interface, one can change the equilibrium between the monomer and the domain-swapped oligomer.
Engineering the hinge loop has been shown to affect 3D domain swapping. After the hinge loop is shortened, Domain 1 of CD2 (Murray et al. 1995), staphylococcal nuclease (Green et al. 1995), and single chain Fv (Kortt et al. 1994; Perisic et al. 1994) form domain-swapped dimers. On the other hand, lengthening the hinge loop of the domain-swapped dimer of cro repressor leads to the monomer formation (Albright et al. 1996).
Other examples of the effect of the hinge loop on domain swapping include p13suc1, in which there are two prolines at the hinge loop. These two prolines control the balance of the monomeric and the dimeric forms by the strains at the hinge loop. In the monomer, there is a strain on residue Pro90 but not Pro92, whereas in the dimer there is a strain on residue Pro92 but not Pro90. Therefore, there is an equilibrium between the monomer and the dimer in the wild-type p13suc1. By mutating the hinge loop to change the strains on the hinge loop, the authors shifted the equilibrium and obtained all monomer or all dimer (Rousseau et al. 2001). Similar examples include cystatin (Staniforth et al. 2001) and Protein L (Kuhlman et al. 2001; O'Neill et al. 2001), in whose monomers there is a strain on residues at their hinge loops. By dimerization, this strain is removed and thus the dimer is thermodynamically favored.
Structural studies of DT and RNase A suggest that 3D domain swapping occurs in these proteins through partial unfolding of the monomer, to a core whose structure remains intact and to intact terminal domains free to move and to swap (Bennett et al. 1995; Liu et al. 2001). However, studies on p13suc1, CD2, and Protein L suggest that these proteins are completely unfolded on the pathway to 3D domain swapping (Hayes et al. 1999; Kuhlman et al. 2001; Rousseau et al. 2001). Apparently different proteins have different pathways for 3D domain swapping. Nevertheless, they all require the disruption of the closed interface, which contributes to the high activation energy.
Summary
In this review, we have summarized structures showing that 3D domain swapping takes place in proteins involved in diverse biological functions. Although biochemical data indicate that 3D domain swapping may affect the regulation of protein functions, further studies are required to understand the role of domain swapping in these biological functions. In several proteins, 3D domain swapping is found in the active form of these proteins, whereas in other proteins, domain swapping seems an artifact of truncation of the whole molecules.
We also discussed several domain-swapped proteins with unique features. These examples show that: (1) one protein can swap more than one domain; (2) a protein can also swap its middle domain, in addition to the domains at the termini; (3) the swapped domains have diverse primary and secondary structures; (4) the hinge loops have high flexibility and display diverse primary and secondary structures; (5) domain-swapped open oligomers can form using a screw-axis symmetry element; and (6) two amyloid proteins have been reported to be domain-swapped, strengthening the link of 3D domain swapping to amyloid formation. These structures broaden our view of 3D domain swapping.
Studies of the mechanisms of 3D domain swapping have been reported, but much remains to be learned. The independence of 3D domain swapping from protein sequence, secondary structure and hinge loop suggests that any protein can be domain-swapped under appropriate conditions where the terminal domain of the protein is unconstrained. As domains continue to swap, new examples will raise our understanding of 3D domain swapping to a higher level, and new functions and mechanisms of 3D domain swapping will be revealed.
Acknowledgments
We thank the NSF and the NIH for support and Drs. Gary Kleiger, Massimo Libonati, and Michael Sawaya for discussions and suggestions.
Abbreviations
BS-RNase, bovine seminal ribonuclease
DT, diphtheria toxin
FU, functional unit
Glx I, glyoxalase I
hRNase, human pancreatic ribonuclease
Hsp, heat shock protein
IL, interleukin
IX/X-bp, blood coagulant factors IX/X-binding protein
MBP, mannose binding protein
p13suc1, suppressor of cyclin-dependent kinase 1
PEG, polyethylene glycol
RNase A, bovine pancreatic ribonuclease
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0201402.
References
- Ahmad, K.F., Engel, C.K., and Prive, G.G. 1998. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. 95 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albright, R.A., Mossing, M.C., and Matthews, B.W. 1996. High-resolution structure of an engineered Cro monomer shows changes in conformation relative to the native dimer. Biochemistry 35 735–742. [DOI] [PubMed] [Google Scholar]
- Anderson, W.F., Ohlendorf, D.H., Takeda, Y., and Matthews, B.W. 1981. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature 290 754–758. [DOI] [PubMed] [Google Scholar]
- Arvai, A.S., Bourne, Y., Hickey, M.J., and Tainer, J.A. 1995. Crystal structure of the human cell cycle protein CksHs1: Single domain fold with similarity to kinase N-lobe domain. J. Mol. Biol. 249 835–842. [DOI] [PubMed] [Google Scholar]
- Bax, B., Lapatto, R., Nalini, V., Driessen, H., Lindley, P.F., Mahadevan, D., Blundell, T.L., and Slingsby, C. 1990. X-ray analysis of beta B2-crystallin and evolution of oligomeric lens proteins. Nature 347 776–780. [DOI] [PubMed] [Google Scholar]
- Bennett, M.J., Choe, S., and Eisenberg, D. 1994a. Refined structure of dimeric diphtheria toxin at 2.0 A resolution. Protein Sci. 3 1444–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.J., Choe, S., and Eisenberg, D. 1994c. Domain swapping: Entangling alliances between proteins. Proc. Natl. Acad. Sci. 91 3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.J. and Eisenberg, D. 1994b. Refined structure of monomeric diphtheria toxin at 2.3 A resolution. Protein Sci. 3 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.J., Schlunegger, M.P., and Eisenberg, D. 1995. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 4 2455–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, C.A., Gustafson, K.R., Boyd, M.R., Covell, D.G., Bax, A., Clore, G.M., and Gronenborn, A.M. 1998. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat. Struct. Biol. 5 571–578. [DOI] [PubMed] [Google Scholar]
- Bird, L.E., Hakansson, K., Pan, H., and Wigley, D.B. 1997. Characterization and crystallization of the helicase domain of bacteriophage T7 gene 4 protein. Nucleic Acids Res. 25 2620–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell, T., Lindley, P., Miller, L., Moss, D., Slingsby, C., Tickle, I., Turnell, B., and Wistow, G. 1981. The molecular structure and stability of the eye lens: X-ray analysis of gamma-crystallin II. Nature 289 771–777. [DOI] [PubMed] [Google Scholar]
- Bocskei, Z., Groom, C.R., Flower, D.R., Wright, C.E., Phillips, S.E., Cavaggioni, A., Findlay, J.B., and North, A.C. 1992. Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography. Nature 360 186–188. [DOI] [PubMed] [Google Scholar]
- Bode, W., Engh, R., Musil, D., Thiele, U., Huber, R., Karshikov, A., Brzin, J., Kos, J., and Turk, V. 1988. The 2.0 A X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J. 7 2593–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckle, A.M. and Fersht, A.R. 1994. Subsite binding in an RNase: Structure of a barnase-tetranucleotide complex at 1.76-A resolution. Biochemistry 33 1644–1653. [DOI] [PubMed] [Google Scholar]
- Cafaro, V., De Lorenzo, C., Piccoli, R., Bracale, A., Mastronicola, M.R., Di Donato, A., and D'Alessio, G. 1995. The antitumor action of seminal ribonuclease and its quaternary conformations. FEBS Lett. 359 31–34. [DOI] [PubMed] [Google Scholar]
- Cameron, A.D., Olin, B., Ridderstrom, M., Mannervik, B., and Jones, T.A. 1997. Crystal structure of human glyoxalase I—Evidence for gene duplication and 3D domain swapping. EMBO J. 16 3386–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals, A., Pous, J., Guasch, A., Benito, A., Ribo, M., Vilanova, M., and Coll, M. 2001. The structure of an engineered domain-swapped ribonuclease dimer and its implications for the evolution of proteins toward oligomerization. Structure 9 967–976. [DOI] [PubMed] [Google Scholar]
- Carroll, S.F., Barbieri, J.T., and Collier, R.J. 1986. Dimeric form of diphtheria toxin: Purification and characterization. Biochemistry 25 2425–2430. [DOI] [PubMed] [Google Scholar]
- Cohen, F.E. and Prusiner, S.B. 1998. Pathologic conformations of prion proteins. Annu. Rev. Biochem. 67 793–819. [DOI] [PubMed] [Google Scholar]
- Cole, N. and Ralston, G.B. 1994. Enhancement of self-association of human spectrin by polyethylene glycol. Int. J. Biochem. 26 799–804. [DOI] [PubMed] [Google Scholar]
- Crestfield, A.M., Stein, W.H., and Moore, S. 1962. On the aggregation of bovine pancreatic ribonuclease. Arch. Biochem. Biophys. 1 217–222. [PubMed] [Google Scholar]
- Diederichs, K., Jacques, S., Boone, T., and Karplus, P.A. 1991. Low-resolution structure of recombinant human granulocyte-macrophage colony stimulating factor. J. Mol. Biol. 221 55–60. [DOI] [PubMed] [Google Scholar]
- Dumas, P., Bergdoll, M., Cagnon, C., and Masson, J.M. 1994. Crystal structure and site-directed mutagenesis of a bleomycin resistance protein and their significance for drug sequestering. EMBO J. 13 2483–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers, D.K. and Valentine, J.S. 2001. Molecular confinement influences protein structure and enhances thermal protein stability. Protein Sci. 10 250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott, J.A., Noble, M.E., Garman, E.F., Brown, N., Rasmussen, B., Nurse, P., and Johnson, L.N. 1995. The crystal structure of p13suc1, a p34cdc2-interacting cell cycle control protein. EMBO J. 14 1004–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fita, I. and Rossmann, M.G. 1985. The NADPH binding site on beef liver catalase. Proc. Natl. Acad. Sci. 82 1604–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick, D.N., Baradaran, K., and Richardson, C.C. 1998. An N-terminal fragment of the gene 4 helicase/primase of bacteriophage T7 retains primase activity in the absence of helicase activity. Proc. Natl. Acad. Sci. 95 7957–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell, D.S. 1993. The machinery of life Springer-Verlag New York Inc., New York, p. 56.
- Gotte, G., Bertoldi, M., and Libonati, M. 1999. Structural versatility of bovine ribonuclease A. Distinct conformers of trimeric and tetrameric aggregates of the enzyme. Eur. J. Biochem. 265 680–687. [DOI] [PubMed] [Google Scholar]
- Green, S.M., Gittis, A.G., Meeker, A.K., and Lattman, E.E. 1995. One-step evolution of a dimer from a monomeric protein. Nat. Struct. Biol. 2 746–751. [DOI] [PubMed] [Google Scholar]
- Grum, V.L., Li, D., MacDonald, R.I., and Mondragon, A. 1999. Structures of two repeats of spectrin suggest models of flexibility. Cell 98 523–535. [DOI] [PubMed] [Google Scholar]
- Hakansson, M., Svensson, A., Fast, J., and Linse, S. 2001. An extended hydrophobic core induces EF-hand swapping. Protein Sci. 10 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, M.V., Sessions, R.B., Brady, R.L., and Clarke, A.R. 1999. Engineered assembly of intertwined oligomers of an immunoglobulin chain. J. Mol. Biol. 285 1857–1867. [DOI] [PubMed] [Google Scholar]
- He, M.M., Clugston, S.L., Honek, J.F., and Matthews, B.W. 2000. Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation. Biochemistry 39 8719–8727. [DOI] [PubMed] [Google Scholar]
- Heringa, J. and Taylor, W.R. 1997. Three-dimensional domain duplication, swapping and stealing. Curr. Opin. Struct. Biol. 7 416–421. [DOI] [PubMed] [Google Scholar]
- Huang, K., Li, Z., Jia, Y., Dunaway-Mariano, D., and Herzberg, O. 1999. Helix swapping between two alpha/beta barrels: Crystal structure of phosphoenolpyruvate mutase with bound Mg(2+)-oxalate. Structure Fold. Des. 7 539–548. [DOI] [PubMed] [Google Scholar]
- Jackson, D.A. and Yanofsky, C. 1969. Restoration of enzymic activity by complementation in vitro between mutant alpha subunits of tryptophan synthetase and between mutant subunits and fragments of the alpha subunit. J. Biol. Chem. 244 4539–4546. [PubMed] [Google Scholar]
- Janowski, R., Kozak, M., Jankowska, E., Grzonka, Z., Grubb, A., Abrahamson, M., and Jaskolski, M. 2001. Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat. Struct. Biol. 8 316–320. [DOI] [PubMed] [Google Scholar]
- Jones, E.Y., Davis, S.J., Williams, A.F., Harlos, K., and Stuart, D.I. 1992. Crystal structure at 2.8 A resolution of a soluble form of the cell adhesion molecule CD2. Nature 360 232–239. [DOI] [PubMed] [Google Scholar]
- Khazanovich, N., Bateman, K., Chernaia, M., Michalak, M., and James, M. 1996. Crystal structure of the yeast cell-cycle control protein, p13suc1, in a strand-exchanged dimer. Structure 4 299–309. [DOI] [PubMed] [Google Scholar]
- Kishan, K.V., Scita, G., Wong, W.T., Di Fiore, P.P., and Newcomer, M.E. 1997. The SH3 domain of Eps8 exists as a novel intertwined dimer. Nat. Struct. Biol. 4 739–743. [DOI] [PubMed] [Google Scholar]
- Klafki, H.W., Pick, A.I., Pardowitz, I., Cole, T., Awni, L.A., Barnikol, H.U., Mayer, F., Kratzin, H.D., and Hilschmann, N. 1993. Reduction of disulfide bonds in an amyloidogenic Bence Jones protein leads to formation of "amyloid-like" fibrils in vitro. Biol. Chem. Hoppe Seyler 374 1117–1122. [DOI] [PubMed] [Google Scholar]
- Knaus, K.J., Morillas, M., Swietnicki, W., Malone, M., Surewicz, W.K., and Yee, V.C. 2001. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 8 770–774. [DOI] [PubMed] [Google Scholar]
- Kortt, A.A., Malby, R.L., Caldwell, J.B., Gruen, L.C., Ivancic, N., Lawrence, M.C., Howlett, G.J., Webster, R.G., Hudson, P.J., and Colman, P.M. 1994. Recombinant anti-sialidase single-chain variable fragment antibody. Characterization, formation of dimer and higher-molecular-mass multimers and the solution of the crystal structure of the single-chain variable fragment/sialidase complex. Eur. J. Biochem. 221 151–157. [DOI] [PubMed] [Google Scholar]
- Kuhlman, B., O'Neill, J.W., Kim, D.E., Zhang, K.Y., and Baker, D. 2001. Conversion of monomeric protein L to an obligate dimer by computational protein design. Proc. Natl. Acad. Sci. 98 10687–10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, G., Svensson, L.A., and Nyman, P.O. 1996. Crystal structure of the Escherichia coli dUTPase in complex with a substrate analogue (dUDP). Nat. Struct. Biol. 3 532–538. [DOI] [PubMed] [Google Scholar]
- Lewis, R.J., Brannigan, J.A., Muchova, K., Barak, I., and Wilkinson, A.J. 1999. Phosphorylated aspartate in the structure of a response regulator protein. J. Mol. Biol. 294 9–15. [DOI] [PubMed] [Google Scholar]
- Lewis, R.J., Muchova, K., Brannigan, J.A., Barak, I., Leonard, G., and Wilkinson, A.J. 2000. Domain swapping in the sporulation response regulator Spo0A. J. Mol. Biol. 297 757–770. [DOI] [PubMed] [Google Scholar]
- Libonati, M., Bertoldi, M., and Sorrentino, S. 1996. The activity on double-stranded RNA of aggregates of ribonuclease A higher than dimers increases as a function of the size of the aggregates. Biochem. J. 318 287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner, R. and Ralston, G. 1995. Effects of dextran on the self-association of human spectrin. Biophys. Chem. 57 15–25. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Gotte, G., Libonati, M., and Eisenberg, D. 2001. A domain-swapped RNase A dimer with implications for amyloid formation. Nat. Struct. Biol. 8 211–214. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Gotte, G., Libonati, M., and Eisenberg, D. 2002. Structures of the two 3D domain-swapped RNase A trimers. Protein Sci. 11 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Hart, P.J., Schlunegger, M.P., and Eisenberg, D. 1998. The crystal structure of a 3D domain-swapped dimer of RNase A at a 2.1-A resolution. Proc. Natl. Acad. Sci. 95 3437–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll, P.J. and Lattman, E.E. 1989. The crystal structure of the ternary complex of staphylococcal nuclease, Ca2+, and the inhibitor pdTp, refined at 1.65 A. Proteins 5 183–201. [DOI] [PubMed] [Google Scholar]
- London, J., Skrzynia, C., and Goldberg, M.E. 1974. Renaturation of Escherichia coli tryptophanase after exposure to 8 M urea. Evidence for the existence of nucleation centers. Eur. J. Biochem. 47 409–415. [DOI] [PubMed] [Google Scholar]
- Louie, G.V., Yang, W., Bowman, M.E., and Choe, S. 1997. Crystal structure of the complex of diphtheria toxin with an extracellular fragment of its receptor. Mol. Cell 1 67–78. [DOI] [PubMed] [Google Scholar]
- Luo, Y., Bertero, M.G., Frey, E.A., Pfuetzner, R.A., Wenk, M.R., Creagh, L., Marcus, S.L., Lim, D., Sicheri, F., Kay, C., Haynes, C., Finlay, B.B., and Strynadka, N.C. 2001. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat. Struct. Biol. 8 1031–1036. [DOI] [PubMed] [Google Scholar]
- Maignan, S., Guilloteau, J.P., Fromage, N., Arnoux, B., Becquart, J., and Ducruix, A. 1995. Crystal structure of the mammalian Grb2 adaptor. Science 268 291–293. [DOI] [PubMed] [Google Scholar]
- Malby, R.L., McCoy, A.J., Kortt, A.A., Hudson, P.J., and Colman, P.M. 1998. Three-dimensional structures of single-chain Fv-neuraminidase complexes. J. Mol. Biol. 279 901–910. [DOI] [PubMed] [Google Scholar]
- Mazzarella, L., Capasso, S., Demasi, D., G., D.L., Matia, C.A., and Zagari, A. 1993. Bovine seminal ribonuclease: Structure at 1.9 A resolution. Acta. Crystallogr. D Biol. Crystallogr. 49 389–402. [DOI] [PubMed] [Google Scholar]
- McGee, A.W., Dakoji, S.R., Olsen, O., Bredt, D.S., Lim, W.A., and Prehoda, K.E. 2001. Structure of the SH3-guanylate kinase-like module from PSD-95 suggests a mechanism for regulated assembly of MAGUK scaffolding proteins. Mol. Cell 8 1291–1301. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A. and Bacon, D.J. 1997. Raster3D: Photorealistic molecular graphics. Meth. Enzymol 277 505–524. [DOI] [PubMed] [Google Scholar]
- Milburn, M.V., Hassell, A.M., Lambert, M.H., Jordan, S.R., Proudfoot, A.E., Graber, P., and Wells, T.N. 1993. A novel dimer configuration revealed by the crystal structure at 2.4 A resolution of human interleukin-5. Nature 363 172–176. [DOI] [PubMed] [Google Scholar]
- Miles, E.W. 1991. Structural basis for catalysis by tryptophan synthase. Adv. Enzymol. Relat. Areas. Mol. Biol. 64 93–172. [DOI] [PubMed] [Google Scholar]
- Minton, A.P. 2001. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 276 10577–10580. [DOI] [PubMed] [Google Scholar]
- Mizuno, H., Fujimoto, Z., Koizumi, M., Kano, H., Atoda, H., and Morita, T. 1997. Structure of coagulation factors IX/X-binding protein, a heterodimer of C-type lectin domains [letter]. Nat. Struct. Biol. 4 438–441. [DOI] [PubMed] [Google Scholar]
- Murray, A.J., Lewis, S.J., Barclay, A.N., and Brady, R.L. 1995. One sequence, two folds: A metastable structure of CD2. Proc. Natl. Acad. Sci. 92 7337–7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio, A., Saraste, M., and Wilmanns, M. 1994. High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides. Nat. Struct. Biol. 1 546–551. [DOI] [PubMed] [Google Scholar]
- Ogihara, N.L., Ghirlanda, G., Bryson, J.W., Gingery, M., DeGrado, W.F., and Eisenberg, D. 2001. Design of three-dimensional domain-swapped dimers and fibrous oligomers. Proc. Natl. Acad. Sci. 98 1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, J.W., Kim, D.E., Baker, D., and Zhang, K.Y. 2001. Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution. Acta. Crystallogr. D Biol. Crystallogr. 57 4), 480–487. [DOI] [PubMed] [Google Scholar]
- Parge, H.E., Arvai, A.S., Murtari, D.J., Reed, S.I., and Tainer, J.A. 1993. Human CksHs2 atomic structure: A role for its hexameric assembly in cell cycle control. Science 262 387–395. [DOI] [PubMed] [Google Scholar]
- Park, C. and Raines, R.T. 2000. Dimer formation by a "monomeric" protein. Protein Sci. 9 2026–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, X.Y., Holliger, P., Murzin, A.G., and Williams, R.L. 1997. The 2.0-A resolution crystal structure of a trimeric antibody fragment with noncognate VH-VL domain pairs shows a rearrangement of VH CDR3. Proc. Natl. Acad. Sci. 94 9637–9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic, O., Webb, P.A., Holliger, P., Winter, G., and Williams, R.L. 1994. Crystal structure of a diabody, a bivalent antibody fragment. Structure 2 1217–1226. [DOI] [PubMed] [Google Scholar]
- Piccoli, R., Di Donato, A., and D'Alessio, G. 1988. Co-operativity in seminal ribonuclease function. Kinetic studies. Biochem. J. 253 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli, R., Tamburrini, M., Piccialli, G., Di Donato, A., Parente, A., and D'Alessio, G. 1992. The dual-mode quaternary structure of seminal RNase. Proc. Natl. Acad. Sci. 89 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers, H., Vix, O., Toro, I., Golz, S., Kemper, B., and Suck, D. 1999. X-ray structure of T4 endonuclease VII: A DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J. 18 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington, S., Wiegand, G., and Huber, R. 1982. Crystallographic refinement and atomic models of two different forms of citrate synthase at 2.7 and 1.7 A resolution. J. Mol. Biol. 158 111–152. [DOI] [PubMed] [Google Scholar]
- Rivas, G., Fernandez, J.A., and Minton, A.P. 1999. Direct observation of the self-association of dilute proteins in the presence of inert macromolecules at high concentration via tracer sedimentation equilibrium: Theory, experiment, and biological significance. Biochemistry 38 9379–9388. [DOI] [PubMed] [Google Scholar]
- Rousseau, F., Schymkowitz, J.W., Wilkinson, H.R., and Itzhaki, L.S. 2001. Three-dimensional domain swapping in p13suc1 occurs in the unfolded state and is controlled by conserved proline residues. Proc. Natl. Acad. Sci. 98 5596–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jean, A.P., Phillips, K.R., Creighton, D.J., and Stone, M.J. 1998. Active monomeric and dimeric forms of Pseudomonas putida glyoxalase I: Evidence for 3D domain swapping. Biochemistry 37 10345–10353. [DOI] [PubMed] [Google Scholar]
- Sawaya, M.R., Guo, S., Tabor, S., Richardson, C.C., and Ellenberger, T. 1999. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell 99 167–177. [DOI] [PubMed] [Google Scholar]
- Schiering, N., Casale, E., Caccia, P., Giordano, P., and Battistini, C. 2000. Dimer formation through domain swapping in the crystal structure of the Grb2-SH2-Ac-pYVNV complex. Biochemistry 39 13376–13382. [DOI] [PubMed] [Google Scholar]
- Schlunegger, M.P., Bennett, M.J., and Eisenberg, D. 1997. Oligomer formation by 3D domain swapping: A model for protein assembly and misassembly. Adv. Protein Chem. 50 61–122. [DOI] [PubMed] [Google Scholar]
- Schymkowitz, J.W., Rousseau, F., Wilkinson, H.R., Friedler, A., and Itzhaki, L.S. 2001. Observation of signal transduction in three-dimensional domain swapping. Nat. Struct. Biol. 8 888–892. [DOI] [PubMed] [Google Scholar]
- Senda, T., Shimazu, T., Matsuda, S., Kawano, G., Shimizu, H., Nakamura, K.T., and Mitsui, Y. 1992. Three-dimensional crystal structure of recombinant murine interferon-beta. EMBO J. 11 3193–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, M.R., Sawaya, M.R., Ellenberger, T., and Wigley, D.B. 2000. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101 589–600. [DOI] [PubMed] [Google Scholar]
- Staniforth, R.A., Giannini, S., Higgins, L.D., Conroy, M.J., Hounslow, A.M., Jerala, R., Craven, C.J., and Waltho, J.P. 2001. Three-dimensional domain swapping in the folded and molten-globule states of cystatins, an amyloid-forming structural superfamily. EMBO J. 20 4774–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle, T., Gamblin, S.J., Yan, Y., and Harrison, S.C. 1996. The structure of simian virus 40 refined at 3.1 A resolution. Structure 4 165–182. [DOI] [PubMed] [Google Scholar]
- Story, R.M., Weber, I.T., and Steitz, T.A. 1992. The structure of E. coli. recA protein monomer and polymer. Nature 355, 318–325. [DOI] [PubMed] [Google Scholar]
- Strop, P., Smith, K.S., Iverson, T.M., Ferry, J.G., and Rees, D.C. 2001. Crystal structure of the "cab"-type beta class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum. J. Biol. Chem. 276 10299– 10305. [DOI] [PubMed] [Google Scholar]
- Svensson, L.A., Thulin, E., and Forsen, S. 1992. Proline cis-trans isomers in calbindin D9k observed by X-ray crystallography. J. Mol. Biol. 223 601–606. [DOI] [PubMed] [Google Scholar]
- Tegoni, M., Ramoni, R., Bignetti, E., Spinelli, S., and Cambillau, C. 1996. Domain swapping creates a third putative combining site in bovine odorant binding protein dimer. Nat. Struct. Biol. 3 863–867. [DOI] [PubMed] [Google Scholar]
- Ultsch, M.H., Wiesmann, C., Simmons, L.C., Henrich, J., Yang, M., Reilly, D., Bass, S.H., and de Vos, A.M. 1999. Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC. J. Mol. Biol. 290 149–159. [DOI] [PubMed] [Google Scholar]
- Urfer, R., Tsoulfas, P., O'Connell, L., Shelton, D.L., Parada, L.F., and Presta, L.G. 1995. An immunoglobulin-like domain determines the specificity of neurotrophin receptors. EMBO J. 14 2795–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayalakshmi, J., Mukhergee, M.K., Graumann, J., Jakob, U., and Saper, M.A. 2001. The 2.2 A crystal structure of Hsp33: A heat shock protein with redox-regulated chaperone activity. Structure (Camb.) 9 367–375. [DOI] [PubMed] [Google Scholar]
- Weis, W.I., Kahn, R., Fourme, R., Drickamer, K., and Hendrickson, W.A. 1991. Structure of the calcium-dependent lectin domain from a rat mannose-binding protein determined by MAD phasing. Science 254 1608–1615. [DOI] [PubMed] [Google Scholar]
- Wiesmann, C., Ultsch, M.H., Bass, S.H., and de Vos, A.M. 1999. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature 401 184–188. [DOI] [PubMed] [Google Scholar]
- Wlodawer, A., Bott, R., and Sjolin, L. 1982. The refined crystal structure of ribonuclease A at 2.0 A resolution. J. Biol. Chem. 257 1325–1332. [PubMed] [Google Scholar]
- Yan, Y., Winograd, E., Viel, A., Cronin, T., Harrison, S.C., and Branton, D. 1993. Crystal structure of the repetitive segments of spectrin. Science 262 2027–2030. [DOI] [PubMed] [Google Scholar]
- Yang, F., Bewley, C.A., Louis, J.M., Gustafson, K.R., Boyd, M.R., Gronenborn, A.M., Clore, G.M., and Wlodawer, A. 1999. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J. Mol. Biol. 288 403–412. [DOI] [PubMed] [Google Scholar]
- Zahn, R., Liu, A., Luhrs, T., Riek, R., von Schroetter, C., Lopez Garcia, F., Billeter, M., Calzolai, L., Wider, G., and Wuthrich, K. 2000. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. 97 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanov, A., Schalk-Hihi, C., Gustchina, A., Tsang, M., Weatherbee, J., and Wlodawer, A. 1995. Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon gamma. Structure 3 591–601. [DOI] [PubMed] [Google Scholar]
- Zegers, I., Deswarte, J., and Wyns, L. 1999. Trimeric domain-swapped barnase. Proc. Natl. Acad. Sci. 96 818–822. [DOI] [PMC free article] [PubMed] [Google Scholar]