Abstract
Summary: Repeated elements can be widely abundant in eukaryotic genomes, composing more than 50% of the human genome, for example. It is possible to classify repeated sequences into two large families, “tandem repeats” and “dispersed repeats.” Each of these two families can be itself divided into subfamilies. Dispersed repeats contain transposons, tRNA genes, and gene paralogues, whereas tandem repeats contain gene tandems, ribosomal DNA repeat arrays, and satellite DNA, itself subdivided into satellites, minisatellites, and microsatellites. Remarkably, the molecular mechanisms that create and propagate dispersed and tandem repeats are specific to each class and usually do not overlap. In the present review, we have chosen in the first section to describe the nature and distribution of dispersed and tandem repeats in eukaryotic genomes in the light of complete (or nearly complete) available genome sequences. In the second part, we focus on the molecular mechanisms responsible for the fast evolution of two specific classes of tandem repeats: minisatellites and microsatellites. Given that a growing number of human neurological disorders involve the expansion of a particular class of microsatellites, called trinucleotide repeats, a large part of the recent experimental work on microsatellites has focused on these particular repeats, and thus we also review the current knowledge in this area. Finally, we propose a unified definition for mini- and microsatellites that takes into account their biological properties and try to point out new directions that should be explored in a near future on our road to understanding the genetics of repeated sequences.
INTRODUCTION
At the dawn of the 21st century, the human genome was sequenced, and even though it was only the fifth eukaryotic genome to be analyzed as such, it opened a new era for geneticists. With over 150 eukaryotic genomes sequenced within the last few years, we are now provided with a wealth of DNA sequence information, an unprecedented event in the history of science. However, several years before reliable and convenient sequencing methods were published (324, 440), scientists already knew that vertebrate genomes contained a large proportion of repeated sequences. In denaturation-renaturation experiments, the rate of renaturation of genomic DNA after heat denaturation is proportional to its concentration. The C0t parameter was defined as the value at which the reassociation is half completed under controlled conditions. Each organism could then be defined by its C0t value. Using this approach, it was shown that the C0t value of the slowly reassociating fraction in calf DNA was 0.03, while the C0t value of the rapidly reassociating fraction was 3,000, proving that the concentration of DNA in the rapidly reassociating fraction was 100,000 times the concentration of the slowly reassociating fraction (52). Three different values of C0t parameters were identified for mice and in other eukaryotes. Highly repetitive sequences had the highest C0t value and accounted for approximately 10% of the mouse genome (1,000,000 copies). They corresponded to what was called satellite DNA. Moderately repetitive sequences represented 20% of the mouse genome (approximatively 1,000 to 100,000 copies), and unique sequences represented approximately 70% of the mouse genome. Although the C0t method slightly underestimated the real amount of repetitive sequences, probably due to slow renaturation of diverged or rearranged repetitive elements (a common characteristic of transposons and retrotransposons), it is remarkable that this method gave a globally accurate picture of genome composition. C0t-based DNA fractionation is still used today to produce genomic DNA libraries that are specific for highly repetitive, moderately repetitive, and single-copy sequences (390).
Early observers of chromosomes found that different species contained different amounts of DNA in their nucleus, also called the “C value.” This apparently benign observation of DNA content caused a lot of trouble when it was shown that amphibians and fishes contained 20 times more DNA per nucleus than mammals, considered to contain more genes than primitive fish due to their higher developmental complexity. Even more surprising, it was subsequently found that the DNA content of the unicellular amoeba Amoeba dubia was 200 times higher than that in humans. This was called the “C-value paradox” and was for a long time the argument of choice for the early opponents of the DNA-based theory of heredity (493). Later on, it was discovered that the increased C value in these organisms was actually due to the presence of abundant repetitive sequences and that the numbers of coding genes are of the same order of magnitude in all eukaryotes, from about 6,000 in the unicellular Saccharomyces cerevisiae to approximately 20,000 to 25,000 in the human genome (which is 200 times bigger than the genome of budding yeast).
From Biophysics to Whole-Genome Sequencing
At the present time, we know that repetitive elements can be widely abundant in some eukaryotes, composing more than 50% of the human genome, for example. It is possible to classify repeated sequences into two large families, called “tandem repeats” and “dispersed repeats.” Each of these two families can be itself divided into several subfamilies, as shown in Fig. 1. Dispersed repeats contain all transposons, tRNA genes, and gene paralogues, whereas tandem repeats contain gene tandems, ribosomal DNA (rDNA) repeat arrays, and satellite DNA, itself subdivided into satellites, minisatellites, and microsatellites. Remarkably, the molecular mechanisms that create and propagate dispersed and tandem repeats are specific to each class and usually do not overlap. In the present review, we have chosen in the first section to describe the nature and distribution of dispersed and tandem repeats in eukaryotic genomes, in the light of the complete (or nearly complete) genome sequences that are available. In the second part, we will focus on the molecular mechanisms responsible for the fast evolution of two specific classes of tandem repeats: minisatellites and microsatellites. Given that a growing number of human neurological disorders involve the expansion, sometimes massive, of a particular class of microsatellites, called trinucleotide repeats, a large part of the recent experimental work on microsatellites has focused on these particular repeats, and thus we will also review the current knowledge in this area. Finally, we will propose a unified definition for mini- and microsatellites that takes into account their biological properties and will try to point out new directions that should be explored in the near future on our road to understanding the genetics of repeated sequences.
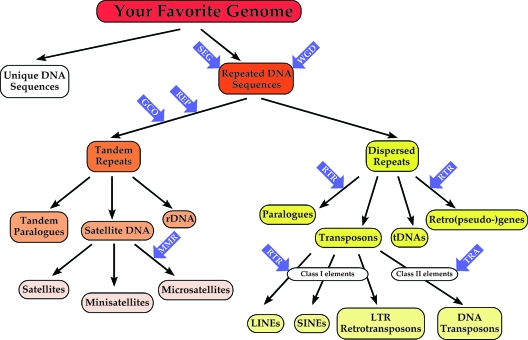
FIG. 1.
Repeated DNA sequences in eukaryotic genomes and mechanisms of evolution. The two main categories of repeated elements (tandem repeats and dispersed repeats) are shown, along with subcategories, as described in the text. Blue arrows point to molecular mechanisms that are involved in propagation and evolution of repeated sequences. REP, replication slippage; GCO, gene conversion; WGD, whole-genome duplication; SEG, segmental duplications; RTR, reverse transcription; TRA, transposition.
Centromeres, telomeres, and mitochondrial and chloroplastic DNA, which are particular kinds of repeated sequences as well, will not be reviewed here, and readers are encouraged to consult appropriate publications concerning these topics. Note also that this review focuses on tandem repeat elements in eukaryotes but such elements can also be found in prokaryotes, although generally less abundantly (510).
REPEATED DNA SEQUENCES IN EUKARYOTIC GENOMES
Whole-Genome Duplications and Segmental Duplications
Whole-genome and segmental duplications in hemiascomycetes.
Saccharomyces cerevisiae was the first eukaryotic organism whose nuclear genome was fully sequenced (167). As such, it has also been the first eukaryotic genome to be investigated for genome redundancy and gene duplications. In a seminal paper published shortly after the complete sequence was released, Wolfe and Shields (538) proposed that the modern yeast genome is derived from an ancestral tetraploid genome, followed by massive gene loss and translocations. Based on two observations, namely, the orientation of the 55 duplicated regions compared to what is seen for centromeres and the absence of any triplicated region (i.e., duplication of a duplication), the authors favored the hypothesis that an ancestral whole-genome duplication, as opposed to several successive segmental duplications, was responsible for the structure of the modern yeast genome. Later on, following the partial sequencing of 13 hemiascomycetous yeast species by the Genolevures consortium, it was found that ancestral chromosomal segments did not entirely coincide with S. cerevisiae duplicated blocks. It was therefore proposed that the hemiascomycete genomes evolved by successive segmental duplications, an alternative to the whole-genome duplication model (290). These apparently conflicting data were reconciled when other hemiascomycete genomes were completely sequenced. The ancestral whole-genome duplication was found to have occurred in the genome of the ancestor of S. cerevisiae and Candida glabrata, but no evidence of whole-genome duplication was found in the genomes of Ashbya gossypii, Kluyveromyces waltii, and Kluyveromyces lactis, three hemiascomycetes phylogenetically more distant from S. cerevisiae than C. glabrata is (108, 117, 242). Following this whole-genome duplication, genes have been lost differentially between the duplicated species (140). Other various studies showed that duplicated genes evolve or are lost at different rates during the evolution of yeast genomes (271), and that rates of large genome rearrangements—based on synteny conservation—were highly variable among hemiascomycetes (141), suggesting that the remodeling of duplicated blocks and the loss of duplicated genes were both subject to constraints specific to each organism. It must be noted that segmental duplications have been experimentally reproduced in S. cerevisiae. Using a gene dosage selection system, Koszul et al. (256) showed that large inter- and intrachromosomal duplications, covering from 41 kb up to 655 kb in size and encompassing up to several hundreds of genes, occurred with a frequency of close to 10−7 per cell/generation. Junctions of segmental duplications frequently contain either microsatellites or transposable elements. The stability of these segmental duplications during meiosis and mitosis was shown to rely both on the size of the duplication and on its structure (257). Replication-based mechanisms leading to segmental duplications rely on both homologous and nonhomologous events (384a). Similarly, by use of a positive selection screen relying on a mutated allele of the URA2 gene, segmental duplications covering from 5 kb to up to 90 kb were found to occur spontaneously in baker's yeast and to be independent of the product of the RAD52 gene, which is necessary for homologous recombination (449, 450). Whole-genome duplications, segmental duplications, and genome redundancies in hemiascomycetes have been recently reviewed (115), and possible molecular mechanisms leading to the formation of segmental duplications were reviewed by Koszul and Fischer (258).
Schizosaccharomyces pombe, an archiascomycetous yeast, is another model organism, whose complete sequence was published in 2002. Its genome does not exhibit the signature of an ancient whole-genome or large-scale duplication, but it contains many duplicated blocks at the subtelomeres of chromosomes I and II (539). These blocks contain several groups of two to four genes whose DNA sequences are 100% identical and which are predicted to be cell surface proteins, an observation reminiscent of what is observed at S. cerevisiae subtelomeres (87, 167).
Whole-genome and segmental duplications in vertebrates.
It has long been postulated that vertebrate genomes resulted from two rounds of whole-genome duplications that occurred early in their evolution (369). By comparing gene duplications between humans and two invertebrates (Drosophila melanogaster and Caenorhabditis elegans), McLysaght and colleagues (326) showed that a number of large paralogous regions detected in the human genome were significantly larger than what would be expected by chance. Molecular clock analysis of invertebrate and human orthologs revealed that a burst of gene duplications occurred in an early chordate ancestor, suggesting that at least one round of whole-genome duplication occurred in this distant ancestor. More recently, distantly related chordate genomes, namely, those of the tunicate Ciona intestinalis, the pufferfish Takifugu rubripes, the mouse, and the human, were compared. The pattern of gene duplications observed was indicative of two successive rounds of whole-genome duplications in vertebrates (98). In addition, the complete sequence of Tetraodon nigroviridis revealed that a whole-genome duplication occurred in an ancestor of this actinopterygian fish after diverging from sarcopterygians (including tetrapods and like organisms). It was shown that one region of synteny in humans was typically associated with two regions of synteny in Tetraodon, a distinctive signature of whole-genome duplications (210). It was possible to reconstruct the ancestral genome of Tetraodon, given that ancestral chromosome duplications were easy to identify due to there being few rearrangements following duplication. This is very different from what is seen for mammalian genomes, which have been extensively reshuffled compared to that of Tetraodon. This might be due to the fact that, compared to the genome of Tetraodon, they contain many transposable elements that may be directly involved in the numerous rearrangements observed. Recent segmental duplications have also been found in the human (26, 27, 270a, 463), rat (504), and mouse (25, 494) genomes. Segmental duplications show a statistical bias for pericentromeric and subtelomeric regions in these three species, although interstitial duplications are more abundant. Interestingly, although segmental duplications in humans are enriched for short interspersed elements (SINEs), no such enrichment was found for rats, except for a fourfold enrichment for centromeric satellite repeats, suggesting that these repeats could be involved in the formation of segmental duplications in rats (504). Comparisons between the human and chimpanzee (Pan troglodytes) genomes revealed that a surprisingly large fraction of duplicated DNA in humans (approximatively 32 Mb) is not duplicated in the chimpanzee. These human-specific duplications represent 515 regions, with biases for chromosomes 5 and 15. Reciprocally, 202 regions of duplicated sequences (approximatively 36 Mb) in the chimpanzee are unique in the human genome (79). When junctions of subtelomeric segmental duplications were analyzed in the chimpanzee genome, it was found that a majority of them (49 out of 53) probably resulted from nonhomologous end joining (NHEJ), whereas only 4 of the events involved nonallelic homologous recombination between repeated elements. Surprisingly, only microhomology sequences (less than 5 bp) and no microsatellites were found at the junctions (284), a situation different from experimental segmental duplications in budding yeast, in which microsatellites were found at 14% of new junctions (256). By comparing segmental duplications in humans, chimpanzees, and macaques (Macaca mulatta), Jiang et al. (222) were able to identify ancestral duplication blocks. Among those, “core duplicons” were defined as ancestral duplications that were present in more than 67% of the blocks. The 14 core duplicons identified are shared by human and chimpanzee and they have a higher gene density and match with more spliced expressed sequence tags than nonduplicated regions of the genome, suggesting that they carry some selective advantage that allows rapid expansion and fixation during great ape evolution.
Whole-genome and segmental duplications in angiosperms.
In a diploid organism, one round (or more) of whole-genome duplication leads to polyploidy, a well-described phenomenon in flowering plants (reviewed in reference 533). Among monocotyledons, maize (Zea mays) is an allotetraploid, resulting from the fusion of two diverged ancestors approximately 11.4 million years (My) ago (161). Wheat (Triticum aestivum) is an allohexaploid, containing three sets of homoeologous chromosomes (i.e., chromosomes that were completely homologous in an ancestral form), whereas rice (Oryza sativa) does not show any evidence for polyploidy (44). Among dicotyledons, soybean (Glycine subgenus soja) has probably undergone more than one round of duplication, since there is an abundance of triplicate and quadruplicate sequences in its genome (465). Using a global analysis of age distribution of paralogous pairs of genes among 11 dicotyledons, Blanc and Wolfe (44) found that seven species (namely, tomato [Lycopersicon esculentum], potato [Solanum tuberosum], soybean [Glycine max], barrel medic [Medicago truncatula], cotton [both Gossypium arboreum and Gossypium herbaceum], and Arabidopsis thaliana) exhibited large-scale gene duplications reminiscent of polyploidy or aneuploidy events. Analysis of the complete genome sequence of the model flowering plant Arabidopsis thaliana revealed 24 large duplicated regions of at least 100 kb, covering 58% of the genome (65.6 Mb) (12a). Further analyses showed that duplicated blocks fall into four different groups based on their ages. The most recent group corresponds to the duplication of approximately 9,000 genes at the same time, reminiscent of a whole-genome duplication event. The three other classes are older and may represent successive large-scale duplication events (519). The evolution of gene content has also been studied for this species, and studies have shown that all gene duplicates are not evenly lost among functional categories, i.e., signal transduction and transcription genes have been preferentially retained, whereas DNA replication and repair genes have been preferentially lost (43). This suggests that the expression of genes involved in genome maintenance and transmission is finely tuned and that any imbalance could be lethal and therefore rapidly counterselected.
Whole-genome duplications in Paramecium.
One cannot review whole-genome duplications without mentioning the unexpected and so far unique case of Paramecium tetraurelia. Sequencing of the macronuclear genome of this ciliate revealed three whole-genome duplications (and possibly a fourth, more ancient), comprising a very recent event occurring before the divergence of P. tetraurelia and P. octaurelia, an old event that occurred before the divergence of Paramecium and Tetrahymena and an intermediate event. A striking feature of the recent whole-genome duplication is the high number of genes retained in duplicate (about 68% of the proteome is composed of two-gene families). This is in contrast with whole-genome duplications discovered in yeast or fish, in which such events could not be detected without the comparison with a nonduplicated reference genome. Maintenance of such a high number of duplicated genes may be driven by gene dosage constraints, since many of the recent duplicates are functionally redundant and are under strong purifying selection, indicative of events in which deleterious mutations, affecting one of the two copies, are not complemented by the expression of the other copy (20).
Dispersed DNA Repeats
Paralogous genes and gene families.
Whole-genome duplications and segmental duplications are two active phenomena that create redundancy by duplicating a very large amount or the totality of the genes in a genome. When this happens, coding sequences (exons) and noncoding sequences (introns 5′ untranslated region [5′-UTR] and 3′-UTR) are duplicated and may undergo purifying selection or accumulate mutations and become pseudogenes. Another way of duplicating genes is to reverse transcribe mRNA and recombine the resulting cDNA. In mammalian genomes, many examples are well characterized in which genes were created by the retrotranscription of a spliced mRNA into cDNA, followed by the integration of the resulting cDNA into the genome. The 5′ sequences of these cDNAs are sometimes truncated and their upstream regulatory and promoter sequences are lacking, since they are not part of the mature transcript. They do not contain introns. These retrogenes are therefore not functional and are called retropseudogenes, but there are a few cases described in which transcription initiation started upstream of the normal promoter and reverse transcription gave rise to a functional retrogene. The process of making retropseudogenes can be dramatically efficient in mammals, since more than 200 copies of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene were found in rat and mouse (reviewed in reference 528). Analysis of the human genome sequence revealed that it contains approximately 10,000 retropseudogenes, including more than 1,700 ribosomal pseudogenes, while the C. elegans genome contains slightly more than 200 retropseudogenes and 2,000 pseudogenes (189, 555). Consistent with a reverse transcription intermediate in the formation of retropseudogenes, there seems to be a positive correlation between the number of retropseudogenes for one given gene and its level of transcription, i.e., more pseudogenes are found for highly transcribed genes (555). By establishing an experimental system in budding yeast requiring transcription, splicing, and reverse transcription of a selectable marker, Derr et al. showed that such events occurred at a frequency of about 10−7 per cell/generation and were dependent on the expression of Ty retroelement reverse transcriptase (105). However, no evidence for the presence of retrogenes in S. cerevisiae has been published thus far. Retrotransposition, like segmental and whole-genome duplications, therefore contributes to the formation of paralogues (or pseudoparalogues), thereby increasing the overall level of redundancy in eukaryotic genomes.
By definition, all the paralogues of a given genome belong to a family whose size ranges from two members to up to several hundreds for immunoglobulin genes, for example (270a, 294). Gene families represent a rather large proportion of all protein-encoding genes in almost all eukaryotes. In S. cerevisiae, 40% of predicted open reading frame (ORF) products are not unique, showing significant identity with from 1 to 22 paralogues (487). In other hemiascomycetes, the number of genes belonging to gene families ranges from 31.8% for K. lactis (the least duplicated genome) to 51.5% for Debaryomyces hansenii (117), figures that are similar to what is observed for D. melanogaster and C. elegans. The situation is very different for S. pombe, in which 93% of predicted protein-coding genes do not belong to recognizable gene families (539). In D. melanogaster, 40% of predicted genes are duplicated (433), whereas in C. elegans, figures range from 32% to 49%, depending on whether proteins (541) or protein domains (433) are considered. It must be noted that approximatively 7% of nematode duplicated genes are thought to have resulted from block duplications involving more than one gene (541). As expected from the genome of A. thaliana, which underwent several large-scale duplications, 65% of its genes belong to gene families, and a substantially higher proportion of genes (37.4%) belong to families containing more than five members, compared to what is seen for D. melanogaster (12.1%) and C. elegans (24%) (12a). In mice and humans, genes belonging to families correspond to 60 to 80% of all genes, a figure similar to what was found for other mammals (103) (Table 1).
TABLE 1.
Occurrences and distribution of repeated DNA sequences in model eukaryotic genomes, as determined from whole-genome sequencing data: dispersed DNA repeats
| Organism | Size (Mb) | No. of WGDa | % Paraloguesb | Retro-(pseudo)-genes | No. of indicated dispersed DNA repeats
|
||||
|---|---|---|---|---|---|---|---|---|---|
| tDNAsc | LINEsd | SINEsd | LTR retrotransposonsd | DNA transposonsd | |||||
| S. cerevisiae | 12.5 | 1 | 40.3 | 274 | 52 (268-280)e | ||||
| S. pombe | 13.8 | 7 | 174 | 11 (249)e | |||||
| A. thaliana | 125f | 4 | 65 | 69 | 589 (13) | 373 | 142 | 1,619 | 3,468 |
| C. elegans | 97f | 32 | 208 | 659 (29) | 648 | 25 | 3,502 | ||
| D. melanogaster | 180f | 40 | 34 | 292 | 486 (0.87) | 682 (2.65) | 404 (0.35) | ||
| T. nigroviridis | 342f | 3 | 4g | NAh | NA | 2,100 | 94 | 819 | 1,239 |
| M. musculus | 2,500f | 2 | 81 | ≈4,700 | 335 (163) | 660,000 (19) | 1,498,000 (8) | 631,000 (10) | 112,000 (0.9) |
| H. sapiens | 2,900f | 2 | 62 | 9,747 | 345 (167) | 850,000 (21) | 1,500,000 (13) | 443,000 (8) | 300,000 (3) |
Number of whole-genome duplications (WGD) or large-scale duplications.
Fraction of the proteome belonging to a gene family.
Numbers in parentheses represent numbers of pseudo-tDNAs.
Numbers in parentheses represent the genome fractions (%) covered by the elements.
Numbers in parentheses represent solo LTRs.
Estimated total genome size, including unsequenced heterochromatin regions and gaps.
Only gene pairs were considered.
NA, not available.
Gene duplications may give rise to different outcomes. One of the two copies may become nonfunctional by the accumulation of point mutations, insertions, and deletions (nonfunctionalization), or one copy may acquire a novel function, while the other retains the original function (neofunctionalization). Alternatively, the two copies may accumulate point mutations so that neither of the two copies is functional by itself and requires the presence of the other copy, or each copy loses one or more enzymatic activities and becomes specialized (subfunctionalization). Studies of substitutions in paralogues encoded by several eukaryotic genomes suggested that gene duplications are a rather frequent phenomenon, arising at the average rate of 0.01 per gene per 1 My, indicating that 50% of all genes in a genome are expected to be duplicated at least once in a 35- to 350-My time scale (301). As a corollary, since random duplications of genes occur in each genome, gene families must expand and contract compared with each other in closely related organisms. That is indeed what was observed for hemiascomycetes (179) as well as for mammals (103). Thus, even in the absence of whole-genome duplication, a large fraction of all genes in a genome are expected to become duplicated, generating a powerful source of novelty in eukaryotes.
Genes encoding tRNA: tDNA.
Transfer RNAs, the genetic link between transcription and translation, are essential for cell viability in all living organisms. In eukaryotes, there is no correlation between genome size and tDNA copy number (note that figures given hereafter only refer to nucleus-encoded tDNA and do not include organelle-encoded tDNA). A. thaliana contains 589 tDNAs and 13 tDNA pseudogenes, a number higher than that for any other eukaryotic genome sequenced so far, including the human genome. D. melanogaster contains 292 tDNAs, C. elegans has 659 tDNAs and 29 pseudogenes, and in the human genome 345 tDNAs and 167 pseudogenes were detected (5, 12a, 73a, 270a). In the mouse genome, 335 putative tDNAs were found but analysis was complicated by the presence of thousands of active B2 sequences (see the following section), which are derived from an ancient tDNA. It is therefore possible that several tDNAs detected are not functional (526). An in-depth analysis of tDNAs in nine hemiascomycetes and one archiascomycete (S. pombe) revealed 2,335 genes, which ranged in distribution from 131 in Candida albicans to 510 in Yarrowia lipolytica (313). In hemiascomycetes, tDNAs generally appear scattered throughout the genome, except in D. hansenii, in which eight identical tandem copies of a tDNA-Lys are found on chromosome B, reminiscent of the frequent occurrence of gene tandems in this organism (see “Tandem repeats of paralogues” below). Clusters of tDNAs have also been found in other eukaryotes. In S. pombe, 22 tDNAs were found in a 50-kb pericentromeric region on chromosome II and two other clusters were found around the two other centromeres (265, 484). In D. melanogaster, a genome region contains a cluster of 10 tDNAs (102), and in humans 140 tDNAs are found in a 4-Mb region on chromosome 6. This rather small region (0.1% of the whole genome) contains representatives for 36 of the 49 anticodons found in the human genome (270a).
It was experimentally shown using S. cerevisiae that when tDNAs are transcribed in the orientation opposite to replication fork progression, they promote the formation of replication pause sites (106). One may therefore wonder what the effect of large tDNAs arrays (such as those mentioned above) might be on replication and, more generally, on the stability of genomic regions encompassing them (see “Fragile sites and cancer” below).
The mechanism(s) by which tDNAs propagate in genomes is at the present time only speculative. One may imagine that reverse transcription of tRNAs followed by integration at an ectopic position in the genome is a possible mechanism. However, to the best of our knowledge, there is no experimental evidence for such a mechanism being active on tRNAs.
Transposable elements.
Transposable elements were elegantly discovered by Barbara McClintock several years before the biochemical structure of DNA itself was solved (325). Since that time, transposons have been found in prokaryotes and eukaryotes and can be classified into two large families: retrotransposons (class I elements) and DNA transposons (class II elements). Recently, a more sophisticated classification based on the mode of transposition and on insertion mechanisms was proposed. Using this classification, retrotransposons were themselves divided into five “orders,” including long terminal repeat (LTR) retrotransposons, long interspersed nuclear elements (LINEs), SINEs, DIRS (Dictyostelium intermediate repeat sequence) elements, and PLE (Penelope-like elements), the last two being less widely spread (535). In each family, autonomous elements, which are able to catalyze their own transposition, and nonautonomous elements, which rely on autonomous elements in order to transpose, are found. Their abundance in genomes is highly variable, from one complete copy of a Ty element in Candida glabrata to millions of copies in the human genome (≈50% of the total sequence) (270a). Homologous (or homeologous) recombination between transposons may induce chromosomal rearrangements such as deletions, inversions, translocations, and segmental duplications as well as mutational events when they transpose into genes (34). For these reasons, transposable elements have been considered to be an important drive in eukaryotic genome evolution (139).
(i) LINEs.
LINEs are non-LTR class I elements whose best characterized member is the LINE-1 (L1) mammalian retrotransposon. This 6- to 8-kb element contains two ORFs (ORF1 and ORF2) that are cotranscribed from the same promoter but can be processed into several distinct messenger RNAs. ORF1 encodes a trimeric nucleic acid chaperone protein that binds to L1 mRNA to form ribonucleoprotein complexes, considered to be transposition intermediates. ORF2 contains endonuclease and reverse transcriptase activities, both required for retrotransposition in wild-type cell lines (34). L1 elements transpose in three steps: (i) formation of a nick in double-stranded DNA at AT-rich sequences, mediated by the endonuclease activity of ORF2; (ii) annealing of the L1 mRNA poly(A) tail to the 5′ poly(T) tail at the nick and cDNA synthesis by the reverse transcriptase activity of ORF2; and (iii) degradation of the L1 mRNA and second-strand synthesis followed by ligation (227). It is worth noting that this mechanism is reminiscent of budding yeast group II mitochondrial intron transposition, in which the intron-encoded protein makes a double-strand break (DSB) at the exon-exon junction, which serves as a primer for reverse transcription of the intron RNA (557, 558). It is therefore possible that mitochondrial group II introns are distant ancestors of mammalian LINE elements, although it could also be a case of convergent evolution.
In the human genome, approximately 850,000 LINEs were found (270a), and 660,000 were found in the mouse genome (526); both figures represent approximately 20% of their respective genome sequences. Several instances of “exonization” of active LINE elements, leading to the creation of new genes, have been recently reviewed (62). By comparison, the genome of another vertebrate, Tetraodon nigroviridis, contains 700 times fewer transposable elements, almost half of them being LINEs (210, 426). Aside from sometimes being involved in large chromosomal rearrangements, like segmental duplications (see “Whole-genome and segmental duplications in vertebrates” above), LINEs may also play a role in evolution by regulating global genome transcription. It was shown that the presence of the human L1 retrotransposon inhibited transcriptional elongation and induced the premature polyadenylation of the transcript containing the L1 sequence. Given the high repetitive nature of L1 elements in the human genome (Table 1), it is likely that these elements play an active role in regulating gene expression genome-wide and are therefore a key component of mammalian genome evolution (182).
(ii) SINEs.
SINEs are the most abundant elements in mammalian genomes. They include Alu and MIR elements in primates and B1, B2, and ID elements in rodents as well as many other elements in mammalian and nonmammalian genomes. Alu elements are composed of two 130-bp monomers separated by a short A-rich linker region. Each monomer was ancestrally derived from the 7SL RNA, following a duplication of this gene before the time of the mammalian radiation (233, 507). They transpose through a mechanism basically similar to that seen for L1 elements, needing only the product of ORF2 to be active (34). They are classified into several families, themselves classified into subfamilies, based on sequence conservation. The S and J families are the oldest, having their origin 35 to 55 My ago, followed by the Y family, at the radiation between green monkeys and the branch leading to African apes, some 25 My ago. This family was itself expanded 4 to 6 My ago, after the divergence of humans and African apes, to give rise to “young” Alu elements (32).
Both the human and the mouse genomes contain approximately 1,500,000 SINEs, which make them the most abundant repeated elements in these genomes (270a, 526). Interestingly, Alu elements are threefold more active in humans than in chimpanzees, since approximatively 7,000 lineage-specific Alu sequences were found in the human genome, compared to 2,300 lineage-specific copies in the chimpanzee genome. Homologous recombination between more or less diverged Alu elements can produce deletions of the sequence located between the repeats. More than 600 such deletions were found in the human genome and more than 900 were found in the chimpanzee genome, underlining the dramatic role that Alu elements can play on genome rearrangements (79a).
Although occasional examples of exon capture by Alu insertion have been recorded (32), a very recent work identified a SINE family, AmnSINE1, that is conserved in all amniota (mammals, birds, and reptiles) and that may be involved in mammalian brain formation. Two of these conserved AmnSINE1 elements were found to behave as distal transcriptional enhancers of developmental genes. Out of 124 conserved AmnSINE1 elements in the human genome, one-fourth are located near genes involved in brain development, leading the authors to speculate that this conserved family could have played a central role in the development of the central nervous system in mammals (444).
(iii) LTR retroelements and retroviruses.
These class I elements transpose by a mechanism different from that seen for LINEs/SINEs. It also involves a reverse transcription step but one that is usually primed by annealing of a tRNA to the primer binding site, the 3′ end of the transposon RNA, followed by reverse synthesis of the first cDNA strand and then synthesis of the second DNA strand. This process occurs in the cytoplasm and the transposon is subsequently transferred to the nucleus, in which integration occurs by a mechanism similar to what is seen for type II DNA elements, with a nuclease making specific nicks at the integration site to catalyze the process (99). Some retroviruses share a similar method of propagation, including the formation of a cytoplasmic particle containing the viral genome, and can therefore be classified as LTR retrotransposons (364). It must be noted that homologous recombination between the two LTRs of a transposon results in “popping out” of the element, leaving as a scar a solo LTR. Most of the LTR retrotransposon copies (85%) are detected as solo LTRs both in the yeast genome (247) and in the human genome (270a). The number of such elements varies greatly among sequenced genomes, from one single full-length copy of a Ty element in the hemiascomycetous yeast Candida glabrata to 443,000 copies of retrovirus-like elements in the human genome (8% of the genome). Fifty-one Ty elements, classified in five families, have been detected in the S. cerevisiae genome, along with 268 to 280 solo LTRs (184, 247), with this number varying between budding yeast strains. Yeast Ty elements tend to be clustered around tRNA genes and genes transcribed by RNA polymerase III (Pol III), this preference most likely being mediated by interactions between the Pol III complex and the integration complex (184). Similarly to the human genome, the macaque (M. mulatta) genome contains around half a million recognizable copies of retroviruses, among which 2,750 copies are lineage specific and result from at least eight instances of horizontal transmission, a figure higher than that for the human lineage (183). LTR retroelements are particularly abundant in plants, representing up to 80% of the DNA sequence in some genomes. Massive expansions of such elements may lead to a rapid increase in genome size, as in Oryza australiensis (a wild relative of the Asian cultivated O. sativa), in which 90,000 retrotransposon copies have been accumulated in the last 3 My, leading to a doubling of genome size (393).
(iv) DNA transposons.
Class II elements are mobile DNA elements that utilize a transposase and single- or double-strand DNA breaks to transpose. They can be classified into three major subclasses: (i) elements that excise as double-stranded DNA and transpose by a classical “cut-and paste” mechanism, such as Drosophila P elements; (ii) elements that utilize a rolling-circle mechanism, such as Helitrons (398); and (iii) elements that probably utilize a self-encoded DNA polymerase but whose transposition mechanism is not well understood, such as Mavericks (399). Based on transposase sequence similarities and phylogenetic analyses, they can be classified into 10 different families (131). Similarly to LTR retrotransposons, “popping out” of the element by homologous recombination between the two LTRs results in a solo LTR. Their number and proportion, compared to those of retrotransposons, are highly variable among eukaryotic genomes. The human genome contains about 300,000 copies of DNA transposons, 100 times more than the C. elegans genome (119, 315) and 700 times more than the D. melanogaster genome (230). In the genome of the protist pathogen Trichomonas vaginalis, an estimated 3,000 Maverick copies are found, which occupy approximately 37% of the genome size (399). Given that most transposon copies in this genome show a very low level of polymorphism (2.5% on the average), and by comparison with its sister taxon Trichomonas tenax (a trichomonad of the oral cavity), it was suggested that the T. vaginalis genome was recently invaded by such elements, leading to a very substantial increase of its size (71). By comparison, some eukaryotic genomes, such as those of S. cerevisiae and S. pombe, do not contain any DNA transposons, although they do contain retroelements. It is, however, not a rule in all ascomycetes, since Y. lipolytica and C. albicans contain several DNA transposons (366). Interestingly, it was shown that 100 to 200 copies of a MITE (miniature inverted-repeat transposable element)-type transposon in the rice genome, called Micron, was flanked by (TA)n repeats, suggesting that this transposon specifically targets a microsatellite (7). This peculiar example shows how a dispersed repeated element may propagate among tandemly repeated elements such as microsatellites.
(v) Inactivation of repeated elements in fungi.
Ectopic recombination between transposable elements is detrimental to genome structure and organization. Numerous examples of large-scale chromosomal rearrangements in plants, animals, and budding yeast have been reported (32, 131, 256). Therefore, although a rapid expansion of such elements in a genome may sometimes be viable and will increase genetic diversity, it may also rapidly reduce fitness and eventually have lethal consequences. Transposable elements are not rare in the genomes of some filamentous fungi (88), and several species have developed specific mechanisms to counter their propagation. Neurospora crassa uses a process called RIP (for repeat-induced point mutation) to efficiently detect and mutate duplicated sequences. RIP recognizes duplications of at least 400 bp in length and introduces C:G-to-T:A mutations into both copies of the duplicated sequence. Since its discovery in N. crassa, evidence for a similar process operating in other filamentous fungi has been reported (155). In Ascobolus immersus, a nonmutagenic process called MIP (for methylation induced premeiotically) methylates cytosines contained in duplicated sequences with a high efficiency, reducing meiotic crossovers dramatically. By decreasing the efficiency of homologous recombination between duplicated sequences, MIP therefore reduces the chance of nonallelic translocations occurring between repeats (305).
Tandem DNA Repeats
Contrary to dispersed repeats, tandem DNA repeats are sequentially repeated. This sophism must not mask the fact that out of two possible orientations for tandem repeats (head-to-tail repeats, also called “direct repeats,” and head-to-head repeats, also called “inverted repeats”), only direct repeats are frequently found in genomes. This is demonstrated by the biased distribution of Alu tandem repeats. Alu elements are frequently found in tandem within the human genome, sometimes separated by only a few base pairs. It was found that nearly identical inverted Alu repeats are 70-fold less frequent than the same repeats in direct orientation when the two copies are separated by less than 20 bp, but this difference is abolished when the two copies are separated by more than 100 bp (292). It was postulated that such repeats are able to form hairpin or cruciform structures in vivo, and Lobachev et al. (291) showed that inverted Alu elements induce DSBs in budding yeast. These breaks require the Mre11-Rad50-Xrs2 complex (a multifunctional protein complex conserved in all eukaryotes [176]) in order to be correctly processed and repaired. Another study with the fission yeast S. pombe showed that a 160-bp palindrome induced homologous recombination and that this induction was dependent on the Rad50 orthologue, Rhp50 (126). Similarly, palindromes also induce DSBs during budding yeast meiosis (358, 362). In Escherichia coli, a very recent work showed that a 246-bp palindrome integrated into the bacterial chromosome was cleaved in vivo by the SbcCD protein complex, the prokaryotic orthologue of the Rad50-Mre11 complex, giving rise to a two-ended DSB that can be detected by Southern blotting (123). It must also be noted that large inverted repeats can be formed in yeast by a mechanism similar to rDNA palindrome formation in Tetrahymena thermophila, a highly regulated process involving the generation of a DSB near a short inverted DNA repeat (63, 64).
The effect of palindrome-induced homologous recombination can be dramatic for cells, since chromosomal rearrangements reminiscent of those found in human tumors, such as internal deletions and inverted duplications, frequently occur in yeast cells harboring such inverted repeats (361). In humans, a long AT-rich palindrome suspected to form a cruciform structure in vivo is found at the constitutive t(11;22) breakpoint, the most frequently occurring non-Robertsonian translocation (266, 267). Since inverted repeats are deleterious sequences leading to large chromosomal rearrangements, they must be counterselected for, and the vast majority of tandem repeats found in eukaryotic genomes are repeats in direct orientation (292). This is the case of all tandem repeat classes detailed in the present chapter, from the large rDNA arrays covering hundreds to thousands of kilobases to the more discrete but widely abundant microsatellites.
Tandem repeats of paralogues.
Gene tandems are not particularly frequent in hemiascomycetes (a few dozen arrays per genome), except in Debaryomyces hansenii, in which 247 tandem arrays were detected throughout its genome, including large arrays of up to nine copies that were not found in other yeast genomes, significantly contributing to global genome redundancy. Like Alu tandems in humans, most of the tandems (80 to 90%) were found to be in direct orientation (117). Given the fragmented structure of most genes in higher eukaryotes, tandem repeats of paralogues are rare, but they are not completely absent. The mouse genome draft sequence contains a high proportion of regions that could not be assembled or anchored on the chromosomes due to the repetitive nature of these regions. One striking example is a large region on chromosome 1 containing a tandem expansion of the Sp100-rs gene, repeated approximately 60 times and covering a 6-Mb region. This region is highly variable in size among mouse species and laboratory strains, ranging from 6 to 200 Mb, suggesting that an active process frequently expands and contracts this region (526). In S. cerevisiae, the CUP1 gene, encoding a copper metallothionein, can be tandemly amplified, conferring resistance to high concentrations of copper to yeast cells. Laboratory strains are polymorphic at this locus, usually exhibiting 10 to 12 tandem copies of CUP1. Losses and gains of repeat units occur mainly by meiotic homologous recombination, and both gene conversions between repeat arrays and unequal crossovers are observed (529). This is reminiscent of what is observed for minisatellites in yeast and humans (see “Rearrangements during homologous recombination” below) and suggests that homologous recombination may lead to expansions and contractions of gene tandem repeats in both budding yeast and humans.
rDNA repeated arrays.
rDNA is another essential genetic element linking transcription to translation. rRNA is at the same time the main structural and the catalytic component of the ribosome. rRNA is translated from a large tandem repeat found at one or more loci in each haploid genome. It is essential for cell viability since it is transcribed in rRNA, the central component of the whole ribosomal translational machinery. Each repeat unit contains the 28S large subunit, the 18S small subunit, and the 5.8S gene as well as two internal transcribed spacers (ITS1 and ITS2) and a large intergenic nontranscribed spacer (294). Another gene, the 5S rRNA gene, may be present within the rDNA array, as is the case in most hemiascomycetes (117), or is encoded elsewhere in the genome, as is the rule with most other eukaryotes. The number of repeat units varies greatly among eukaryotes, from 40 to 19,300 in animals and from 150 to 26,000 in plants, and is positively correlated with genome size (400). Given the repetitive nature of rDNA arrays, it is not always easy to determine whether all of the repeat units share an identical sequence. In a recent study, nonassembled rDNA sequences generated during whole-genome shotgun sequencing of five fungi have been examined in order to look for possible polymorphisms between rDNA repeat units. Few base variations were found, from 4 in S. cerevisiae to 37 in Cryptococcus neoformans, and there was no obvious bias toward their localization to spacer regions (158). These results show that rDNA tandem arrays are evolving through concerted evolution and suggest that sequence quasi-identity is maintained by homogenization of rDNA repeat arrays. This homogenization could occur by homologous recombination between tandem repeats, since Holliday junctions (a hallmark of homologous recombination) were detected in rDNA during mitotic growth of yeast cells. Their presence is dependent on Pol α, but not on Pol δ or Pol ɛ, and they are significantly reduced in a rad52 mutant in which homologous recombination is abolished (560). RAD52 is also directly involved in the formation of extrachromosomal circles (ERCs) in old yeast cells (382). ERCs are DNA minicircles whose formation is dependent on several cis- and trans-acting factors. A replication block is located 3′ of each rDNA repeat unit in budding yeast that arrests the replication fork coming from the 3′ end so that it cannot collide with the RNA polymerase complex transcribing the repeat unit in the opposite orientation. This replication fork block is dependent on the presence of the Fob1 protein and its mechanism has been extensively studied and reviewed elsewhere (268, 333, 432). Interestingly, mutations in the FOB1 gene lead to an increase in budding yeast life span and a decrease in the amount of ERCs (96). The molecular link between the amount of ERCs and aging in yeast is unclear, but both depend on the presence of the SGS1 helicase and on the SIR complex, involved in chromatin silencing (244, 468). Yeast cells mutated for the SGS1 helicase contain a higher proportion of ERCs, exhibit nucleolar fragmentation, and age prematurely compared to wild-type cells (469). SGS1 encodes an S-phase DEAH-box DNA helicase that was first identified as a suppressor of a mutation in the topoisomerase TOP3 gene (156). It was subsequently shown to play several roles during homologous recombination and probably also during replication in yeast (85, 125, 157, 207, 283, 425). SGS1 has orthologues in E. coli (RecQ), in all hemiascomycetous yeasts (419), and in mammals. In humans, five orthologues are found, namely, WRN, BLM, and RTS, involved in Werner, Bloom, and Rothmund-Thomson syndromes, respectively, and two shorter forms, RecQL and RecQ5. Interestingly, it was shown in humans that rDNA organization depends on the WRN gene. Using single-molecule analysis, Caburet and colleagues (65) have shown that rDNA tandem arrays frequently differ from the canonical organization. The size of the intergenic nontranscribed spacer varies from 9 kb to 72 kb, and palindromic structures are found in one-third of the molecules analyzed in wild-type cells. However, the proportion of palindromes increases to 50% in cells deficient for the WRN helicase, suggesting that some form of illegitimate recombination controlled by this helicase is responsible for making rearrangements within human rDNA tandem arrays. In conclusion, homologous recombination is very frequent within rDNA and is tightly linked to DNA replication of the tandem arrays.
As mentioned above, 5S rDNA genes are not often encoded within the large rDNA array but are found as dispersed elements, themselves sometimes amplified in tandem repeats, as in Drosophila species. Comparison of 5S tandem repeat sequences in several Drosophila species revealed that insertions and deletions were very frequent between species and were often flanked by conserved nucleotides, suggesting that they could occur by slippage of the newly synthesized strand during DNA replication or alternatively by gene conversion (380) (see “Molecular mechanisms involved in mini- and microsatellite expansions” below). A more recent work on four completely sequenced filamentous fungi (Aspergillus nidulans, Fusarium graminearum, Magnaporthe grisea, and Neurospora crassa) revealed an interesting property of 5S genes in these species. It was shown that 5S genes located at different loci share more identity to other 5S genes in other species than with 5S genes in the same species (5S clusters are interspecies instead of being intraspecies) (429). This suggests that 5S genes in a given species do not coevolve by a mechanism similar to large rDNA arrays and are not homogenized during evolution of a given species. This also suggests that an active mechanism of constant “birth-and-death” creates new 5S sequences, as opposed to the model of concerted evolution that seems to apply to large rDNA tandem arrays (365). Interestingly, a class of SINE (SINE3) deriving from a 5S gene was discovered in the zebrafish genome and is probably mobilized in trans by zebrafish LINE elements (234). This raises the interesting possibility that some 5S rDNA genes could also be reverse transcribed and transposed elsewhere in the genome, therefore themselves behaving like transposable elements. In plants, a retroelement called Cassandra exhibits the unique property of carrying universally conserved 5S sequences in each of its two LTRs. Transposition of Cassandra would therefore propagate 5S sequences in plant genomes, providing an explanation for the lack of concerted evolution and of rapid rearrangements of 5S loci in plants (228).
Satellite DNA.
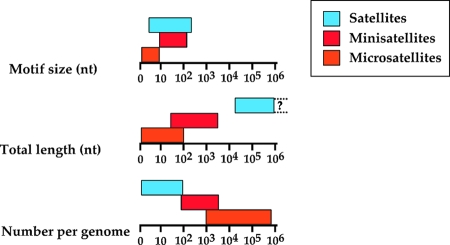
Historically, satellite DNA was identified as a DNA fraction that sedimented as a strong and localized band, above or below the main band in cesium chloride density gradients, hence its name (520). It is widespread in eukaryotic genomes, such as D. melanogaster (293), plants (461), and mammals (505) but is absent from hemiascomycetes and S. pombe, although the large fission yeast centromeres contain many repetitive elements essential for their function (539). Satellite DNA is found in heterochromatin regions, such as mammalian centromeres, the D. melanogaster Y chromosome, and plant subtelomeres and centromeres but may also be found as intercalary DNA (76). Although its unusual buoyant density was the hallmark of a strong nucleotide composition bias, molecular analyses of satellite DNA showed its highly repetitive nature. It is characterized by large tandem repeats, whose total length may reach several millions of nucleotides and whose repeat units show a great variation in size, ranging from 5 nucleotides for human satellite III up to several hundreds of base pairs. Repeat units are not strictly identical and exhibit sequence polymorphisms (505). The motif sizes, total lengths, and the numbers of satellites per genome are summarized in Fig. 2. In humans, several centromeric satellites are known, and their repeat unit lengths range from 5 to 171 nucleotides. The 5-bp satellite is an imperfect GGAAT repeat present in most if not all chromosomes, spanning up to hundreds of kilobases, and it might be a functional component of the centromere. The 171-bp satellite (generally called the α-satellite) is also found on all chromosomes and was shown to bind the centromere protein CENP-B (505). This protein is thought to be derived from transposases encoded by ancient DNA transposons. It was recently shown that S. pombe homologues of human CENP-B localize to Tf2 retrotransposons and recruit histone deacetylases to silence these retroelements. Therefore it is possible that CENP-B binding at human satellites similarly helps to recruit histone deacetylases to silence these heterochromatin regions (69). The β-satellite is normally present in tandem arrays, covering hundreds of kilobases on the short arms of acrocentric chromosomes. Remarkably, the insertion of 18 complete β-satellite repeat units (68 bp) within the TMPRSS3 gene led to both congenital and childhood onset autosomal recessive deafness in humans. It is totally unclear how this satellite was propagated and inserted into this gene, leading to its inactivation (458). In Mus musculus, the minor satellite often maps close to a telomeric (TTAGGG)n sequence, whereas the major satellite is pericentromeric (53, 406). Plant satellites were identified in centromeric and telomeric positions for dozens of species and harbor repeat unit lengths ranging from 118 to 755 bp. Some of them are also present in distinct regions on both chromosomal arms in several Triticeae genomes (461). Satellite DNA has been extensively studied and mapped on each chromosome in D. melanogaster. Repeat unit sizes range from 5 to 359 bp, with the larger units being found essentially within heterochromatin covering about half of chromosome X. Shorter repeat unit satellites are localized on all chromosomes [like the (AAGAGAG)n and the (AATAT)n satellites] or on only a subset (293). The Y chromosome, almost entirely heterochromatic, carries nine satellites whose repeat unit sizes range from 5 to 7 nucleotides, three of them mapping only to the Y chromosome and the others being present on other chromosomes (47). In Tetraodon nigroviridis, a 118-bp tandem repeat is found at all centromeres. This centromeric satellite DNA remarkably shows a high conservation of the first half of the repeat unit (approximatively 60 bp) and a more variable second half of the repeat unit, suggesting that both halves of the repeat unit are not under the same constraints (426). Remarkably, transposons, although scarce in T. nigroviridis, are preferentially found within heterochromatic regions, in proximity to satellite elements, suggesting either preferential insertions of transposons in these regions or selective elimination of transposed elements in euchromatic regions as a way to reduce the deleterious incidence of homologous recombination between them (138).
FIG. 2.
Motif sizes, lengths, and abundances of satellite sequences in eukaryotes. For each category (satellites, minisatellites, and microsatellites), the distribution of motif sizes, total lengths of repeat arrays, and numbers of occurences of each repeat category per eukaryotic genome are shown on a logarithmic scale. Satellite DNA can extend over megabases of DNA but its maximum length is unknown, due to the lack of sequence information (dotted lines and question mark).
One may wonder about the putative function of satellite DNA, given that some of these elements are conserved over large evolutionary distances, like the human α-satellite, which was detected in chicken and zebrafish (281). An old hypothesis suggested that heterochromatin satellites would help proper meiotic disjunction, increasing the chance of correctly segregating chromosomes during meiosis (520). Given the centromeric or pericentromeric locations of several mammalian satellites, and the binding of centromere-specific proteins, like CENP-B or histone CEN H3, it is reasonable to assume that they may play a role either in replicating or in correctly segregating centromeres during mitosis (and/or meiosis). Interestingly, several authors reported that satellites are transcribed in a variety of organisms, including invertebrates, vertebrates, and plants. These polyadenylated transcripts are highly regulated, being differentially expressed at particular developmental stages or in specific tissues, raising a possible role for satellites in development. Short interfering RNAs originating from satellite DNA have also been detected, and they may play a role either in control of the initial formation or subsequent maintenance of heterochromatin or in the expression of particular genes embedded in satellites (506). Due to the highly repetitive nature of satellite DNA, whole-genome sequencing studies of eukaryotic organisms have focused on sequencing euchromatic regions, and little information has been obtained about the heterochromatic nature of such genomes. Sequencing satellites is still a challenge, and the possible presence of protein-coding genes or other genetic or regulatory elements in such regions is still questionable.
Microsatellites and minisatellites.
Mini- and microsatellites are tandem repeats composed of short repeat units. The repeat unit size is used as the main feature to classify a short tandem repeat as a mini- or microsatellite. However, there is at the present time no consensus about the precise definition of both kinds of repeats. Some authors do not consider mononucleotide repeats [poly(A) tracts, for example] as microsatellites, whereas for others the threshold between micro- and minisatellites may vary between 6 and 10 repeats. There is no consensus either for the minimal number of repeat units to be considered as a micro- or minisatellite. Some authors consider that two repeat units are not enough and fix the threshold at three, four, or even five units. It has also been proposed that any distinction between the two types of repeats would be purely academic. In the present chapter, we will review experimental data showing that although molecular mechanisms involved in mini- and microsatellite size changes are basically similar (if not identical), it is possible to propose their classification as two different types of repeats based on their distributions and functions in eukaryotic genomes. The motif sizes, total lengths, and the numbers of minisatellites and microsatellites per genome are summarized in Fig. 2.
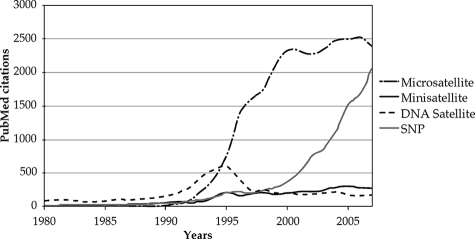
Historically, the first minisatellite (also called VNTR [for variable number of tandem repeats]) was discovered by Wyman and White (543), who identified a human locus exhibiting restriction fragment length polymorphism among individuals with various degrees of proximity. Later on, several hypervariable loci were identified in the human genome and called “minisatellites,” as a reference to megabase-large variable satellite DNA (221). One of the first minisatellites was found in an intron of the human myoglobin gene and comprised four 33-bp tandem repeats with some sequence similarities with other minisatellites discovered previously. It was flanked by a 9-bp direct repeat, a characteristic signature of transposable elements, suggesting that this minisatellite was able to transpose in some way (221). Although the first microsatellite was characterized by Weller and colleagues (531) as a polymorphic (GGAT)165 repeat in the human myoglobin gene, the term “microsatellite” (also called short sequence repeat) entered the literature a few years later, with the demonstration that a (TG)n repeat in the human genome exhibited size polymorphisms when amplified by PCR on genome samples from several individuals (286). The increasing availability of DNA amplification by PCR at the beginning of the 1990s triggered a tremendous number of studies using the amplification of microsatellites as genetic markers for forensic medicine, paternity testing, or positional cloning. Among the most prominent and original studies are the identifications by microsatellite genotyping of the skeletal remains of an 8-year old murder victim (178) and of Josef Mengele, who escaped to South America following World War II (215). DNA analysis of the descendants of the U.S. president Thomas Jefferson showed that he was the father of one of his slave's children, a long-standing debate among historians (146). Microsatellite typing started to be used in yeast studies 12 years ago to identify laboratory strains of S. cerevisiae (413) and more recently to identify industrial yeast strains or pathogenic strains of S. cerevisiae (196, 304), Candida albicans (300), and Candida glabrata (S. Brisse, C. Pannier, A. Angoulvant, T. de Meeus, O. Faure, P. Lacube, H. Muller, J. Peman, A. M. Viviani, R. Grillot, B. Dujon, C. Fairhead, and C. Hennequin, unpublished data) involved in human infections. Population geneticists also extensively used microsatellite typing to study population structures and evolution (213) and to study specific questions concerning the origin of domestic horses (516) or French wine grapes (49), to give just a couple of examples. Before the completion of whole-genome sequences, several linkage maps were built using microsatellites as genetic markers for channel catfish (Ictalurus punctatus), rainbow trout (Oncorhynchus mykiss), wheat (Triticum aestivum), Arabidopsis thaliana, pine (Pinus taeda), and Homo sapiens (107), to name just a few. Minisatellite size polymorphisms were used in a similar way for paternity testing (194) or to determine the source of saliva on a used postage stamp (201) as well as various other forensic studies (163). But the abundance of variable microsatellites, compared to minisatellites, made the former the marker of choice for similar studies. This is represented in Fig. 3, in which the numbers of citations per year in the PubMed database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed) for the words “microsatellite” and “minisatellite” have been plotted. In a few years, the number of citations for microsatellites went from 2 in 1989 to 433 in 1994 and to more than 2,000 in 1999, well above levels of citations attained by minisatellites and DNA satellites. The relatively recent development of single-nucleotide polymorphisms as genetic markers most probably led to the clear inflection in the citation curve observed for microsatellites (Fig. 3). Undoubtedly, genotyping human populations by use of variable microsatellites (or single-nucleotide polymorphisms) has become a powerful tool, not only for human geneticists who study population differentiation in modern humans (21, 30, 404) but also for governments in regulating immigration flows, as already enforced by laws in several states of the European Union.
FIG. 3.
Number of citations per year in the PubMed database for different search terms.
(i) Distribution of microsatellites in eukaryotic genomes.
Over the last 10 years, a large number of studies on microsatellite distribution in eukaryotic genomes have been published. Unfortunately, it is not always an easy task to compare published results, since different authors often use different algorithms (sometimes homemade and not necessarily published), and they do not always agree on the definition of a microsatellite and therefore use different settings and thresholds to detect what they think should be defined as a microsatellite. However, a recent study describes a comparative analysis of the main algorithms available to exhaustively search for tandem repeats in DNA sequences. Five algorithms were compared, namely, Mreps (255), Sputnik (C. Abajian, University of Washington, Seattle [http://espressosoftware.com/pages/sputnik.jsp]), TRF (35), RepeatMasker (A. Smit, R. Hubley, and P. Green [http://repeatmasker.org/]), and STAR (101). It was shown that the total number of perfect microsatellites detected varies greatly among the five algorithms, ranging from 6,228 detections per megabase, for Sputnik, to 76 detections per megabase, for RepeatMasker. STAR and RepeatMasker, which are less efficient for the detection of abundant microsatellites of two to three repeat units in length, generally detect fewer but longer microsatellites than do TRF, Mreps, and Sputnik. Most microsatellites detected by RepeatMasker and STAR are also detected by the three other algorithms, whereas the reverse is not true (273). Note that other algorithms have been developed to detect tandem repeats in DNA sequences (434), some of them, like ACMES or MREPATT, dedicated to the detection of perfect tandem repeats (410, 431), and others, like TandemSWAN, designed to specifically detect imperfect (or “fuzzy”) tandem repeats (46), but their efficiency, compared to that of other more widely used algorithms, has not been carefully evaluated. Therefore, one must be advised when undertaking a microsatellite search in a given genome to consider many parameters before selecting one software, particularly if short, imperfect, or compound microsatellites are being researched, since the efficiency of detection of such genetic objects greatly depends on the algorithm used. It must be noted that recent works tried to define the main parameters that are associated with tandem repeat polymorphism in order to predict variable (or hypervariable) micro- and minisatellites. In one approach, it was found that G+C content and a measure of redundant patterns of mutation (called HistoryR) were both strongly correlated with minisatellite polymorphism (104). In the other approach, a numeric score (called the VARscore) dependent on several parameters, including the number of units, length, and purity, was assigned to each tandem repeat. A good correlation was found between the VARscore and tandem repeat polymorphism, as determined experimentally (275).
The S. cerevisiae genome was the first eukaryotic genome to be completely sequenced and, as such, was also the first in which microsatellites could be exhaustively analyzed. Given our previous comments on the different algorithms available for such a study and the lack of a clear consensus on the definition of a microsatellite by the scientific community, it therefore is not surprising that the outcomes of such studies showed large variations in the numbers of microsatellites detected. If one compares only trinucleotide repeats (a class of microsatellites), for which several independent studies with budding yeast are available, absolute numbers of such repeats in the budding yeast genome vary from 92 to 1,769 (Table 2), depending on parameters chosen by the authors (19, 95, 133, 235, 306, 415, 548). Despite these expected discrepancies, authors agree that microsatellites are generally excluded from yeast genes, except for trinucleotide repeats and hexanucleotide repeats, which are found both in ORFs and in intergenic regions (499). Careful analysis of amino acids encoded by trinucleotide repeats showed an overrepresentation of charged residues, such as glutamine, asparagine, and glutamatic and aspartic acids. These genes often encode nuclearly located proteins, particularly transcription factors and regulators of gene expression (312a, 415, 442, 548). This is also the case for 13 other hemiascomycetous yeast genomes analyzed (306) and seems to be true for other eukaryotes, including humans (129). A recent study of 12 sequenced genomes from Drosophila species showed that the most frequent codon found in gene-encoded trinucleotide repeats was CAG, although CAA was the most frequent triplet encoded by triplet repeats detected in noncoding regions (203), reminiscent of what was observed in Saccharomyces bayanus var. uvarum (306).
TABLE 2.
Differences in trinucleotide repeat distributions in the budding yeast genome, depending on threshold and definition chosen
| Repeat typea | No. of repeat unitsb | Total no. | Density (microsatellites per Mb) | Software used (reference)c | Reference |
|---|---|---|---|---|---|
| Imperfect + perfect | 4 | 1,769 | 147 | TRF (35) | 415 |
| Imperfect + perfect | 5 | 924 | 77 | TRF (35) | 415 |
| NS | 6 | 395 | 33 | Homemade | 95 |
| Perfect | 8 | 92 | 8 | Homemade | 133 |
| Perfect | 5 | 394 | 33 | Homemade | 548 |
| NS | 9 | 141 | 12 | TRF (35) | 19 |
| Perfect | 5 | 621 | 51 | TRF (35) | 306 |
| Perfect | 3 | 396 | 28 | Homemade | 235 |
Imperfect repeats refer to trinucleotide repeats interrupted by at least one nucleotide differing from the repeat consensus and whose total score is above a given threshold. Perfect repeats are uninterrupted trinucleotide repeats. NS, not specified.
Minimal number of repeat units in the trinucleotide repeat array.
Homemade software was specifically developed for this study and is available on request to the authors.
Early studies of human DNA sequences found in public databases concluded that CAG and CGG trinucleotide repeats were overrepresented in the human genome (181, 475), but more-recent studies on the human genome public sequence revealed that this overrepresentation was limited to coding regions (478). This discrepancy is probably due to a bias in sequences cloned and sequenced in public databases at the time of the former study. In humans, as in yeast, microsatellites are generally underrepresented in exons, except for trinucleotide and hexanucleotide repeats. The densities of microsatellites are similar on all chromosomes, even though chromosomes 17, 19, and 22 show a slight increase in density (479). Analysis of five complete plant genomes showed that microsatellites are preferentially found in unique regions of the genomes and exhibit a lack of association with transposon-rich regions. The frequencies of each microsatellite class vary, but imperfect trinucleotide repeat densities range from 77 repeats/Mb in soybean (Glycine max) to 159 repeats/Mb in Arabidopsis thaliana, with an average of 105 repeats/Mb, a density significantly higher than that seen for budding yeast for the same class of repeats (imperfect trinucleotide repeats at least four units long) (Table 2) (346). Compared to plants, yeast, and humans, teleostean fishes are remarkably rich in perfect microsatellites, since 1,700/Mb and 1,176/Mb are detected in the genomes of Tetraodon nigroviridis and Fugu rubripes, respectively, compared to 281/Mb on average for plants (346), 145/Mb for budding yeast (306), and 87/Mb in the human genome (270a) (Table 3).
TABLE 3.
Occurrences and distribution of repeated DNA sequences in model eukaryotic genomes, as determined from whole-genome sequencing data: tandem DNA repeats
| Organism | Satellite size (Mb)a | No. of indicated tandem DNA repeats
|
||
|---|---|---|---|---|
| Minisatellites | Microsatellitesb | Microsatellites/Mb | ||
| S. cerevisiae | 113 | 1,818 | 145 | |
| S. pombe | NA | 879 | 64 | |
| A. thaliana | ≈10 (8) | ≈720 | ≈38,000 | 300 |
| C. elegans | ≈650 | NAc | NA | |
| D. melanogaster | ≈60 (33) | NA | ≈29,000d | 160 |
| T. nigroviridis | ≈1.2 (0.34) | NA | ≈580,000 | 1,700 |
| M. musculus | ≈250 (10) | NA | ≈960,000 | 380 |
| H. sapiens | ≈212 (7) | ≈6,000 | ≈253,000 | 90 |
Numbers in parentheses represent the genome fractions covered by the elements. Sizes are as estimated from unsequenced heterochromatin regions.
Occurrences of di-, tri-, and tetranucleotide repeats only.
NA, not available.
Occurrences of dinucleotide repeats only.
It must also be noted that microsatellites are not homogeneously distributed along budding yeast chromosomes; rather, they exhibit repeat-rich and repeat-poor regions (418). Interestingly, some of these regions also correspond to regions highly biased for G+C content (116, 462), suggesting that forces shaping chromosome structure have an influence on microsatellite formation or maintenance. A similar observation was made for dinucleotide repeats in D. melanogaster, which were frequently found in clusters (22). Similar analyses of other eukaryotic genomes will be required in order to determine if these observations underlie a more general rule.
In summary, several algorithms were designed to search for tandem repeats in DNA sequences, but one may keep in mind that some of them are more efficient at finding specific kinds of repeats (short and perfect repeats or long and imperfect ones, for example). Therefore, before selecting a given program to perform a search, it is recommended first to define the kind of repeats that are being looked for and then to select the best program suited to perform this search among those available. An alternative approach could be to select two or more programs, run them, and compare their outputs in order to get a list of repeats that would be more exhaustive than one that would be obtained with one single program.
(ii) Distribution of minisatellites in eukaryotic genomes.
Minisatellites have been less studied than microsatellites (Fig. 3) and few genome-wide analyses have been performed. Former attempts at systematic minisatellite cloning gave a rough estimate for a few hundred minisatellites in the human genome (18, 359), but other estimations were in the range of a few thousand. By analyzing the sequence from human chromosomes 21 and 22, Denoeud et al. (104) found 127 minisatellites fulfilling their criteria. Extrapolating this number to the complete human genome gives a rough estimation of 6,000 minisatellites. Among these, a majority (75%) are expected to show some degree of polymorphism in the population. In a more recent work, Vergnaud and Denoeud (513) analyzed the minisatellite content of human chromosome 22, A. thaliana chromosome 4, and C. elegans chromosome 1 by use of the TRF software (35). In this study, minisatellites were defined as tandem repeats with units longer than 16 bp and covering at least 100 bp with a high G+C content and a strong strand bias. By use of this definition, half of the 62 minisatellites detected on chromosome 22 were located within the terminal 10% of chromosome 22, confirming previous studies (104). The same analysis revealed that minisatellite densities were similar in A. thaliana and C. elegans, with the same subtelomeric bias in the nematode, whereas in A. thaliana, minisatellites tend to cluster in the pericentromeric region. In the genome of the teleostean T. nigroviridis, one tandem repeat that could qualify as a minisatellite was detected. Its repeat unit is 10 bp long and is repeated in very large variable-size arrays on 10 of the 11 short arms of subtelocentric chromosomes, with the exception being the rDNA-bearing chromosome (426).
To our knowledge, the only eukaryotic genome that was entirely analyzed for the presence of minisatellites is the budding yeast genome. By use of different algorithms, 44 minisatellite-containing genes were found by Verstrepen et al. (515) and 49 were detected by Richard and Dujon (414), with a 95% overlap between the two sets of minisatellites. In addition, 11 minisatellites were detected in intergenic regions, but they are, on the average, shorter than those included in ORFs, and 18 were found in subtelomeric Y′ elements (298). If one excludes Y′ minisatellites, there is no obvious bias for their distribution in subtelomeric or centromeric regions. They are, on the average, GC rich, a property shared by human minisatellites, and they generally exhibit a strong negative GC skew (more cytosines than guanines on the coding strand). The conservation of S. cerevisiae minisatellites in other completely sequenced hemiascomycetous yeasts reveals that a large number of them are not conserved during yeast evolution, although the genes that contain them are conserved. In Saccharomyces paradoxus, a close relative to S. cerevisiae, 73% are conserved, but in more distantly related yeast species (Candida glabrata, Kluyveromyces lactis, Debaryomyces hansenii, and Yarrowia lipolytica), only 25 to 47% of minisatellites are conserved. In addition, in each species a few pseudominisatellites were detected, testifying that in the distant past, the minisatellite was present at the same location in the same gene. Remarkably, in several instances, a different minisatellite was found at the same location in a given gene among different species. This is the case for the PRY2 gene, which contains a six-copy 18-bp repeat in S. cerevisiae, a nine-copy 15-bp repeat in S. paradoxus, a six-copy 15-bp repeat (with a different motif) in Y. lipolytica, and a pseudominisatellite in D. hansenii and does not contain any minisatellite in C. glabrata and K. lactis. This suggests that the evolution rate of minisatellites is much higher than that for their containing genes or, alternatively, that minisatellites are able to invade genes at specific locations, like transposable elements (414). Interestingly, among minisatellite-containing genes, 50 to 60% encode cell wall proteins or proteins involved in cell wall metabolism (48, 414, 515). Minisatellites found in these genes encode mostly serine and threonine residues (59% of the total), which are believed to be the sites of O mannosylations by the Pmt4 protein, these posttranslational modifications being important for maintaining the protein at the cell wall surface (121, 272). Therefore, it seems reasonable to assume that these minisatellites were positively selected for, as their presence is essential for protein function. Note that, as expected, all of the minisatellites detected in yeast genes contain a motif that is a multiple of 3 nucleotides, allowing unit additions and deletions without disrupting the reading frame. A more recent global analysis of minisatellites in the pathogenic yeast Candida glabrata revealed that this hemiascomycete contains very large minisatellites, often included in cell wall genes and genes involved in cell-to-cell adhesion, making these large minisatellites good candidates to play a role in pathogenicity (489). In support of this hypothesis, a recent work using Aspergillus fumigatus revealed that this pathogenic filamentous fungus contains several minisatellites, some of which contain large repeat units of up to 255 bp. Approximately 3% of these minisatellites are located in genes that encode potential cell surface proteins. One of these genes (Afu3g08990) was inactivated and the corresponding mutant conidia showed decreased cellular adhesion, although no effect on virulence in a mouse model was detected (279).
(iii) Alu elements and microsatellites.
We have seen that Alu elements are widely spread in the human genome, representing more than 10% of its total size (Table 1). Since Alu repeats contain a poly(A) tail and a central linker region rich in adenines, their association with A-rich microsatellites has been studied. Nadir et al. (356) showed a significant association between the 3′ end of Alu sequences not only with (A)n mononucleotide repeats but also with (AAC)n, (AAT)n, and A-rich tetra- to hexanucleotide repeats, but this association was surprisingly weaker with (AT)n dinucleotide repeats. In another study, (AC)n dinucleotide repeats were found to be preferentially associated with Alu elements, 75% of them being found at the 3′ end of the element, while the remainder were found in the central linker region (15). Interestingly, the (GAA)n trinucleotide repeat, involved in Friedreich's ataxia, may have arisen with the insertion of an Alu element. Out of 788 human genomic loci containing (GAA)n repeats, 63% (501 loci) map within 25 bp of an Alu element. Among them, 94% are associated with the poly(A) tail and the remaining are associated either with the 5′ end of the element or with the central linker region (83). Genome-wide studies using complete genome draft sequences will now be necessary to determine the real impact of Alu and other transposable elements on the spreading of microsatellites in eukaryotic genomes.
MINI- AND MICROSATELLITE SIZE CHANGES: FROM HUMAN DISORDERS TO SPECIATION
We have seen in the first chapter that both dispersed and tandem repeats are abundant in all eukaryotic genomes sequenced so far, although their relative numbers vary among organisms. Due to the repetitive nature of these elements, their presence in a genome may cause reciprocal or nonreciprocal translocations, segmental duplications, gene amplifications, and other kinds of spontaneous chromosomal rearrangements that may ultimately lead to cell death. The molecular mechanisms creating such large genome rearrangements mainly involve defects during S-phase replication and during homologous recombination. These mechanisms, along with their effect on genome stability, will be studied in the present chapter.
Fragile Sites and Cancer
Fragile sites were defined cytologically within human metaphasic chromosomes as chromatid constrictions or breaks after cells were grown in the presence of drugs involved in impairing DNA metabolism or replication. Two types of fragile sites were distinguished; common fragile sites were found in all individuals, whereas rare fragile sites were present only in a small proportion of the population (5% or less). Common fragile sites are expressed in the presence of aphidicolin (DNA polymerase inhibitor), bromodeoxyuridine, or 5-azacytidine (nucleotide analogues). Rare fragile sites are expressed in the presence of folic acid (a cofactor in nucleotide anabolism), distamycin A (an antibiotic that binds to AT-rich DNA), or bromodeoxyuridine. Fragile sites have been extensively studied for years, since they are associated with hereditary mental retardations and several types of cancers in humans. Several reviews specifically focused on fragile sites have been published (93, 164, 483), but we will more specifically discuss in the present review the link between DNA repeats and fragile sites and how the replication of regions containing repeated elements may induce the expression of a fragile site.
DNA repeats found at fragile sites.
Three types of repeated elements are found at fragile sites: transposons, microsatellites, and minisatellites. The common fragile site FRA3B was sequenced over 10 years ago (205) and was shown to contain numerous LINEs (L1 and L2), SINEs (Alu and MIR), LTR retroviruses (HERVs and MalRVs), and DNA transposons (mariner and MER) in direct and inverted repeat orientations. Forty Alu repeats, 41 MIR elements, and 39 MER repeats were recognized, covering 12.3% of the region. Altogether, repeated elements covered between 23% and 43% of the FRA3B region, with variations in subregions. Several types of cancers are associated with FRA3B (see below). The mouse orthologous region Fra14A2 is also an aphidicolin-inducible fragile site, and comparison of human and mouse sequences showed a good sequence conservation (72.6% identity). Numerous repeated elements were also detected. Overall, dispersed repeats represent 32.6% of the Fra14A2 region. Surprisingly, almost all L1 elements were inserted after the divergence of human and mouse lineages, since they disrupt the alignment, suggesting that this region was a hot spot for transposition in both lineages (464). Two fragile sites, FRA10B and FRA16B, are AT-rich regions containing tandem repeats of a 42-bp minisatellite and of a 33-bp minisatellite, respectively (198, 552). Both regions exhibit size polymorphisms and intergenerational as well as somatic instability, and fragility is associated with a large expansion of these minisatellites. Thus far, no pathology has been found to be associated with these two fragile sites. This is not the case for the rare fragile site FRAXA, which is the most common cause of hereditary mental retardation in humans. This disorder is due to an expansion of a (CGG)n microsatellite into the 5′-UTR region of the FMR1 gene (152, 514). The FMR1 gene is one of several genes involved in X-linked mental retardation (430), although most of them are not fragile sites. Nevertheless, at least three other loci containing (CCG)n microsatellites are responsible for mental retardation, namely, FRAXE (251), FRAXF (424), and FRA11B (225, 226). Note that although the fragile site FRA16A is also due to the expansion of a (CCG)n microsatellite (360), no phenotype has been associated with it thus far. Interestingly, in the filamentous fungus Candida albicans, chromosomal translocations and chromosome losses are often associated with the major repeat sequence (MRS). The MRS is a large and complex tandem repeat which is found at nine different loci in the haploid genome. The MRS is composed of a 2-kb sequence (RPS) that itself includes seven to nine copies of a 16-bp repeated sequence and six to eight copies of a 29-bp sequence. The RPS can be tandemly repeated up to 39 times in a single MRS, and size heterogeneity of the MRS is a major cause of chromosome length polymorphism in C. albicans (302).
Interestingly, fragile sites are not always associated with repeated sequences. This is the case for FRA7H, which does not exhibit any particular enrichment in repeated elements but has a sequence that is AT rich (58%). Computer analysis of sequence flexibility (443) showed several peaks of high flexibility within the region (337). The same observation was made for FRA16D (423) and FRA7E (559). In addition, it was predicted for FRA7E that these flexible peaks enriched in AT base pairs are able to form secondary structures, at least in silico. Therefore, it seems that a fragile site is determined not only by the presence of many repeated elements or large microsatellites but also by the propensity of the sequence to form some kind of secondary structure that could impede replication through its locus, thus causing fragility.
Molecular basis for fragility.
At the present time, the precise cause for chromosomal fragility is still under debate, but several hypotheses, not mutually exclusive, have been proposed. First, as mentioned above, the molecular structure of the region, particularly its ability to form secondary structures that may stall replication forks, seems to be important. However, one may wonder whether breaks result from single-strand nicks on one DNA strand that are transformed into DSBs when the replication fork encounters these nicks or from another origin, like a nuclease that would recognize and cleave those secondary structures. In Schizosaccharomyces pombe, mating-type switching is induced by the replication fork encountering a single-strand nick at the mat1 locus, transforming this nick into a DSB, thus initiating homologous recombination with one of the two homologous mat cassettes (references 13, 14, 237, and 237a). This is a highly regulated process under the control of several proteins, including the conserved fork protection Swi1/Swi3 complex (Tim/Tipin in mammals), responsible for the temporary replication fork pausing near mat1 (200, 237). The possible presence of single-strand nicks at fragile sites in vivo, during or after replication, is an open question.
The search for trans-acting factors that may regulate fragile site expression led to the finding that the downregulation of Rad51 (involved in homologous recombination [376]) or DNA-protein kinase (PK) and ligase IV, both involved in NHEJ (263), increased the expression of FRA3B and FRA16D in the presence of aphidicolin (453). Inactivation of the ATR protein also increases FRA3B and FRA16D expression, but ATM has no effect (72), suggesting that stalled replication forks but not DSBs are the signal activating the replication checkpoint (187). Similarly, the inhibition of BRCA1, a key player in DNA damage response (530), increases the expression of FRA3B and FRA16D (17). The Werner syndrome helicase, a member of the RecQ family of helicases and involved in the resolution of DNA structures during replication (55), was also shown to be involved in fragile site expression. In the presence of aphidicolin, Werner syndrome helicase-deficient cells show a significantly high level of gaps and breaks, compared to wild-type cells, at FRA7H, FRA16D, and FRA3B (396). Altogether, these data point to a role of stalled forks in promoting single-stranded damage at fragile sites, triggering checkpoints, and increasing fragility, but the precise mechanism remains elusive.
Several studies with the model budding yeast have also tried to decipher the molecular basis for chromosomal fragility. Freudenreich and colleagues designed an elegant experimental system to look at chromosomal fragility in yeast. The sequence to be tested for fragility was cloned into a yeast artificial chromosome, between the centromere and a URA3 selectable marker. If the cloned sequence induced fragility, the distal chromosome part containing the URA3 marker would be lost and yeast cells would become resistant to the drug 5-fluoroorotic acid (148). Using this system, they were able to show that (CAG)n and (CCG)n repeats exhibited length-dependent fragility in vivo (28, 148). They also showed that fragility was increased in cells mutated for MEC1 (yeast ATR homologue), RAD9, and RAD53 (269), and in a checkpoint-deficient allele of the MRC1 gene, mrc1-1, involved in replication fork stability (149). Using the same experimental system, they looked at the fragility of FRA16D and showed that a short (AT)n repeat within the fragile site sequence was able to induce chromosome breakage in a length-dependent manner. By two-dimensional gel electrophoresis, they were able to visualize the accumulation of DNA molecules during replication at the (AT)n repeat, proving that replication forks stall within or near this sequence in vivo in a length-dependent manner (554). Other experimental systems were designed in yeast, using the loss of a genetic marker on a yeast chromosome following natural or I-SceI-induced chromosome breakage (488). These studies showed that chromosomal fragility was increased in DNA damage checkpoint mutants (6, 354) and when Pol α levels were reduced (276). Interestingly, fragility sometimes involved repeated elements, like tDNAs and LTRs (6) or Ty retrotransposons (276), but spontaneous chromosome rearrangements were also observed for regions lacking such elements (354), suggesting that replication stalling and DNA breaks (either single or double stranded) may occur in nonrepetitive regions. These regions have been called replication slow zones by Cha and Kleckner (74), who determined that DSBs do not occur stochastically but rather in specific regions on yeast chromosome III during replication of strains carrying a mutant allele of the MEC1 checkpoint gene (mec1-1). In another study using the same mec1-1 mutant, it was shown that 17 early-firing regions in the yeast genome were not efficiently replicated, but they do not seem to correspond to replication slow zones (407). Hence, at the present time, it is clear that yeast fragile sites and mammalian fragile sites share some properties, such as nonrandom breakage at specific programmed sites within the genome, an increase in fragility when replication is slowed down with drugs, and the implication of the MEC1/ATR checkpoint in regulating fragility. However, the majority of yeast fragile sites do not involve repeated elements, nor do they occur in regions of nucleotide composition bias, suggesting that not all fragile sites in mammals may occur in such regions.
Fragile sites and chromosomal rearrangements in cancers.
There is a long-standing debate among cancer specialists of how to determine whether large chromosomal abnormalities detected in cancer cells are the result of uncontrolled cell proliferation or are required to transform a normal cell into a cancerous one (318). Early studies described extensive karyotype alterations (2), suggesting that spontaneous DNA damage during replication could promote formation of DSBs that are highly recombinogenic (517) and would give rise to chromosomal translocations and other genome-wide rearrangements (451). More-recent work helped to refine this model. By comparing different stages of human tumors and normal tissues, Bartkova et al. (31) showed that the DNA damage response is activated very early in tumor life and precedes the appearance of p53 mutations. At the same time, Gorgoulis et al. (170) reached an identical conclusion, showing in addition that FRA3B was frequently lost in neoplastic tissues, a signature of an unrepaired chromosomal break at this fragile site. Several lines of evidence for the involvement of FRA3B in cancer exist (421) and are strengthened by a very recent study in which human-mouse chromosome 3 somatic hybrid cells were exposed to aphidicolin-mediated replication stress. Between 13% and 23% of clones exhibited deletions of the FRA3B region, spanning 200 to 600 kb and matching deletions observed for several gastrointestinal, colon, lung, breast, and cervical cancers (120). Similarly, loss of heterozygosity was frequently observed at FRA16D for breast and prostate cancers, and a recurrent t(14;16) translocation involving FRA16D has been identified for multiple myeloma (397). Sequence analysis of one deletion showed that an (AT)n dinucleotide repeat and an AT-rich minisatellite were found at both endpoints, showing the implication of AT-rich repeats in FRA16D fragility. Most cell lines with FRA16D deletions also exhibited FRA3B deletions, showing that chromosomal breakage, following replication stress, may affect more than one common fragile site (137).
In conclusion, human fragile sites are associated with the presence of dispersed or tandem repeats, although this is not necessarily the case in yeast. Therefore, rather than envisioning the presence of a fragile site as the direct effect of the presence of such repeats, we should try to analyze replication processes in eukaryotic cells more thoroughly. It is possible that repeated elements just play the role of enhancers of “replication defects” that occur at several other places within genomes but are not detected since they do not give rise to fragile sites.
Trinucleotide Repeat Expansions
Trinucleotide repeats belong to the category of microsatellites. Since the discovery 17 years ago of the first neurological disorders involving trinucleotide repeat expansions, more than two dozen human diseases involving what are sometimes called “dynamic mutations” (422) have been brought to light. Despite extensive studies undertaken by many groups, using different prokaryotic or eukaryotic model systems, the molecular mechanism(s) underlying such dramatic expansions of triplet repeats in one single generation in humans is still unknown. This is not the place to extensively review all that is known about the disorders or about the peculiar properties of these particular microsatellites, since many review articles have been dedicated to that purpose in the last few years (84, 160, 277, 334-336, 357, 372, 385, 420, 532). Instead, we are going to give a general overview of the diverse molecular mechanisms involved in trinucleotide repeat instability and, more importantly, address what we think are crucial questions for better understanding of these mechanisms.
Researchers usually classify triplet expansions into two main categories, those that occur within genes and generate a protein containing an expansion of a given amino acid, and those that occur in noncoding regions. For the first category, only expansions of polyglutamine and polyalanine have been discovered thus far. It must be noted that the “rule of three” (308) was broken when expansions of a GC-rich dodecamer (270), of a tetranucleotide repeat (285), and of a pentanucleotide repeat (320) were discovered as being involved in human neurological disorders. Therefore, the most commonly accepted view is that expansions are unrelated to the trinucleotide repeat nature of the sequence but are related to the propensity of the sequence to form secondary structures (see below) and to interfere with DNA replication, repair, or recombination.
Trinucleotide repeat expansions in noncoding sequences.
Compared to those in coding regions, expansions in noncoding sequences include a large number and variety of tandem repeats. As already mentioned, the most common form of inherited mental retardation in humans, called fragile X syndrome, is due to expansions of a (CCG)n trinucleotide repeat responsible for the fragility (152, 514). Type 1 myotonic dystrophy (DM1) and type 8 spinocerebellar ataxia (SCA8) involve a (CTG)n repeat in the 3′-UTR of the DMPK gene (54, 153, 186), whereas DM2 involves expansion of a (CCTG)n repeat in an intron (285). Friedreich's ataxia is induced by the expansion of a (GAA)n repeat in an intron (70), whereas SCA10 is linked to the expansion of a pentanucleotide (ATTCT)n, also in an intron (320), and progressive myoclonus epilepsy involves a (C4GC4GCG)n dodecamer in the 5-UTR of the EPM1 gene (270, 518). Intuitively, one could think that noncoding regions would show a higher flexibility than coding regions to accommodate tandem repeats of various unit lengths, since any change of the unit number that is not a multiple of three would be fatal for gene function if they were in exons. This is what is observed, since several of these repeat unit lengths are not multiples of three. Even though these repeats are located in nontranslated regions, their repetitive sequences sometimes interfere with other cellular processes, such as replication, transcription, and splicing, etc. In the fragile X syndrome, the FMR1 gene is extensively methylated and inactivated in full-mutation alleles containing more than 230 CCG repeats (394). Within shorter alleles, the locus is not methylated and the mRNA levels are high (245), but protein translation is decreased, due to ribosome stalling in the expanded CCG repeats in the 5′-UTR of the FMR1 mRNA, leading to a dramatic reduction in protein levels (130, 223). Hypermethylation of CpG islands adjacent to FRAXE and FRAXF alleles, containing an expanded (CCG)n repeat, were also reported (251, 424). In Friedreich's ataxia, the expansion of the (GAA)n sequence in the first intron of the FRDA gene results in a reduction in gene expression, due to the inhibition of transcriptional elongation (70). This inhibition was recapitulated in vitro and in vivo in bacteria by Krasilnikova et al. (260). They showed that long (GAA)n repeats carried by an E. coli plasmid interfered with transcription by dramatically reducing the amount of full-size mRNA containing these repeats. A similar observation was made in an in vitro reconstituted transcription system by the same authors. Transcriptional study of expanded (CAG)n or (CTG)n repeats also showed a reduction in the amount of normally sized mRNA molecules containing these repeats, but surprisingly longer mRNA molecules were detected, suggesting that some kind of transcription slippage could occur, leading to transcripts longer than expected (124). In DM1, the expanded (CTG)n repeat in the 3′-UTR of the DMPK gene reduces the expression of the downstream gene DMAHP, probably by locally remodeling chromatin (250, 495). Two CTCF-binding sites were identified flanking the (CTG)n repeat, and methylation of the DM1 locus in congenital myotonic dystrophy disrupts their function, probably by modifying nucleosome positioning in this region (136). DMPK mRNAs, containing expanded CUG repeats, form nuclear foci in vivo and the transcripts are not properly exported (91, 486). When a URA3 reporter gene containing in its 3′-UTR an expanded CUG repeat tract was expressed into yeast cells, CUG-containing foci were detected but were not specifically clustered in the nucleus, since cytoplasmic labeling was visible. This suggests that some but not all CUG-containing RNA defects can be recapitulated in budding yeast (124). The DMPK mRNA binds to the muscleblind-like protein (MBLN) (127, 330) and the CUG-binding protein (CUG-BP) (392), deregulating splicing of several transcripts and causing defects in a muscle-specific chloride channel (77, 309). Expanded CUG repeats also form hairpins that activate the double-stranded RNA-dependent protein kinase PKR, interfering with its normal cellular function (496). Note that in type DM2, due to an expansion of a (CCTG)n tetranucleotide repeat, the CCUG-containing mRNAs also form nuclear foci and bind muscleblind-like proteins (128, 310). In conclusion, expanded repeats in noncoding regions interfere with the metabolism of several cellular pathways, such as methylation, transcription, splicing, RNA processing, nuclear export, and translation, and the resulting expanded mRNAs often acquire a dominant negative altered function that is directly involved in pathogenicity.
Trinucleotide repeat expansions in coding sequences.
In comparison to noncoding sequences, trinucleotide repeat expansions within coding sequences are homogeneous. First, they concern only triplets, since any other size change would disrupt the reading frame and lead to gene loss of function. Second, they have been found thus far to concern only two types of amino acids, namely, glutamine and alanine. Third, expansions are always of moderate size compared to noncoding expansions, which may reach several hundreds or even thousands of triplets in one generation. Although mechanisms leading to polyglutamine and polyalanine expansions are not necessarily different, they have their own specificities that will be discussed below.
(i) Polyglutamine expansions.
Expansions of (CAG)n repeats inside exons are found in Huntington disease (HD) and several SCAs. In these neurodegenerative disorders, the reading frame consists of an expanded CAG triplet that always encodes a polyglutamine tract. It is peculiar that out of two possible codons for glutamine, CAA and CAG, only CAG expansions have been discovered thus far, probably pointing to a requirement for a GC-rich triplet in order to trigger expansions. Mechanisms leading to polyglutamine pathogenesis have been reviewed elsewhere (160, 371, 372). Polyglutamine aggregates were described more than 10 years ago as intranuclear inclusions that cause a progressive neurological phenotype in mice that is similar to polyglutamine disorders in humans (90, 370, 452). Formation of these detergent-resistant aggregates (238) depends on the length of the polyglutamine tract (262, 317) and on the presence of chaperone proteins in several model organisms, including yeast (262, 350), Drosophila (75, 239), and C. elegans (445). Aggregates were initially thought to be directly involved in pathogenicity, but it was subsequently shown that neuronal death was directly correlated not to their presence (446) but rather to the nuclear presence of a soluble fraction of polyglutamine proteins (249). These proteins induce cell death in neurons by apoptosis (280, 446), although a mechanism of cellular death not involving apoptosis has also been reported (503). Protein-protein interaction domains often contain polyglutamine tracts and are therefore more prone to self-aggregation than other protein domains (236). We want to point out that the formation of polyglutamine aggregates is reminiscent of abnormal protein aggregation seen for other neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, and prion diseases (401, 452), and therefore searching for genes encoding polyglutamine tracts in fully sequenced genomes could help to predict which proteins might share the same properties (329).
It is intriguing that several trinucleotide repeat disorders affect the central nervous system. This observation has not found any satisfying explanation thus far. A possibility would be that genes involved in neural development are richer in microsatellites than other genes, increasing the chance to contain an expandable trinucleotide repeat tract. It may be the case in Drosophila, in which long amino acid repeats were preferentially found in genes involved in developmental control and in central nervous system development (236). Similar studies on completely sequenced mammalian genomes should help to clarify this point.
(ii) Polyalanine expansions.
Alanine tracts are expanded in several human developmental disorders, such as type II synpolydactyly (8), oculopharyngeal muscular dystrophy (50), cleidocranial dysplasia (351), and holoprosencephaly (57). Expansions are rather shorter than polyglutamine expansions and they involve imperfect trinucleotide repeat tracts, since polyalanine tracts are often encoded by two to four different alanine codons (56). This is very different from what is seen for other trinucleotide repeat expansions, in which repeats are stabilized by the presence of an imperfect triplet in the sequence. These observations suggest that mechanisms of poly-Gln and poly-Ala expansions could differ. It was therefore suggested that polyglutamine expansions mainly rely on a replication slippage mechanism, whereas polyalanine expansions are due to unequal crossover (525). However, careful examination of poly-Ala-expanded alleles reveals similarities between their pattern and the pattern of minisatellite rearrangements during meiotic recombination (see “Molecular mechanisms involved in mini- and microsatellite expansions” below), suggesting that gene conversion (with or without crossover) could be involved in polyalanine expansions. Interestingly, it was shown that polyglutamine tracts were often mistranslated, leading to polyalanine tracts by a ribosomal −1 frameshift (159, 500).
The timing of expansions.
One of the early questions related to trinucleotide repeats was the time of their expansion in human cells. Were expansions meiotic, prezygotic, postzygotic, or somatic? Addressing this question was a way to define the precise mechanism involved: S-phase mitotic or meiotic replication, meiotic recombination, or yet another mechanism. Since expansions are detected in every tissue (with some degree of mosaicism), it is tempting to think that they mainly occur either during parental meiosis, during zygote formation, or very shortly thereafter (348, 537). Earlier studies of the fragile X syndrome showed that (CCG)n expansions were absent from sperm cells, although they could be detected in lymphocytes, suggesting that expansions occurred in the female germ line and subsequently contracted to shorter allele sizes in the male germ line but not in somatic cells (411). Strengthening this hypothesis, further analyses of intact fetus ovaries showed that oocytes contained expanded alleles (307). Similarly, expansions were detected in sperm DNA, but not in lymphocyte DNA, in patients affected by SCA1 (321).
Single sperm analysis of (CAG)n repeats in the HD gene in humans revealed that the frequency of expansions was dependent on allele size, with longer alleles being more prone to expansions (274). It was subsequently shown by single-molecule analysis of sperm cell DNA that expansions of HD gene repeats occur before the completion of meiosis, and some of the expansions were detected before the beginning of meiosis (547). Mouse models have been particularly helpful in addressing this question. Single-cell analysis of mice transgenics for the DM1 repeats showed that both sperm cells and somatic cells exhibited a bias toward expansions, suggesting that at least some of the intergenerational expansions observed for DM1 originated from somatic expansions (343). When male mouse germ cells were sorted according to their maturation stage and DM1 allele size was analyzed by PCR, it was shown that no size change was detectable in spermatogonias, spermatocytes, or spermatids, but increases were visible in mature spermatozoa, suggesting that some mechanism(s) was generating expansions after meiotic replication and recombination took place (259). However, a more recent study using an improved method to sort germ cells in order to reduce as much as possible contamination by other cell types, revealed that expansions of the DM1 allele were detected very early, in spermatogonia, before meiotic replication took place (448).
Analysis of the sperm DNA of a Friedreich's ataxia carrier of a premutation allele (around 100 repeats) showed that an expanded allele of approximately 320 repeats was present. This carrier's son was affected by the disease and his DNA exhibited expansions up to 1,040 repeats. This suggested that a first expansion occurred in the father's germ cells (from 100 to 320 repeats) and another one occurred very early after zygote formation (from 320 to 1,040 repeats) (100).
In summary, trinucleotide repeat expansions may occur at different stages during cell life, probably reflecting the fact that different repeat sequences and different genetic locations may trigger different mechanisms leading to these expansions.
Micro- and Minisatellite Size Polymorphism: an Evolutionary Driving Force
Besides being involved in a number of fragile sites and associated cancers as well as in several neurological and developmental diseases, tandem repeats have a more positive role in eukaryotic genome evolution by allowing the rapid adaptation of a given organism to its environment, namely, the fast evolution of morphological features or modulation of sociobehavioral traits, as will be exemplified now.
Evolution of FLO genes in Saccharomyces cerevisiae.
As described above, several genes in S. cerevisiae involved in cell wall biogenesis contain minisatellites. Among them, genes belonging to the FLO family of mannoproteins also contain various minisatellites, whose repeat unit sizes are 30 bp (FLO11), 81 bp (FLO10), and 135 bp (FLO1, FLO5, and FLO10) (414). These genes are homologous to the ALS and EPA gene families in C. albicans and C. glabrata, respectively, which are involved in cell-to-cell adhesion and pathogenicity. By using several alleles of FLO1 differing only in their numbers of minisatellite repeat units, Verstrepen et al. (515) showed that the length of the minisatellite was directly correlated to cell adhesion. Yeasts with longer FLO1 minisatellites exhibit a better adhesion to polystyrene but also to other cells, as demonstrated by increased flocculation of liquid cultures. Similarly, it was shown that a yeast strain used to make cherry wine exhibits higher hydrophobicity and cell-cell adhesion, giving it the property to form a buoyant biofilm at the wine surface. This property is dependent both on the level of expression of the FLO11 gene and on the number of minisatellite repeat units in this gene (132).
Roles of microsatellites in vertebrate evolution.
Morphological variations are common among canine species. By studying 36 developmental genes that contain microsatellites, Fondon and Garner (145) found that 29 out the 36 loci exhibit fewer interruptions in the repeat tract than their human orthologues. Probably due to the greater repeat purity, some microsatellites were highly polymorphic in dogs and these size variations were correlated with morphological changes, such as digit polydactyly and skull morphology variations. Some of the genes involved, such as Hox-D13, Runx-2, and Zic-2, are also involved in the genetic abnormalities found associated with polyalanine expansions in humans (see above), suggesting that the others could also well be associated with developmental phenotypes in humans. This is a blatant example of how the rapid evolution of a gene sequence by microsatellite size changes may lead to phenotypic diversity in a modern domesticated animal species, probably much faster than would be allowed by accumulation of point mutations in the same genes.
An example of the involvement of microsatellite polymorphism in social behavior is given by voles. The prairie species of this little rodent is biparental and shows high levels of social interest, in contrast to the closely related meadow vole. Length differences in a (GA)n dinucleotide microsatellite in the 5′-UTR of the vasopressin receptor gene (V1aR) underlies this difference. In species with a long, expanded microsatellite, males show higher levels of V1aR in the brain, concomitant with higher rates of pup licking and grooming, along with higher levels of partner preference formation, compared to species with a short microsatellite (180). Remarkably, comparison of the V1aR orthologues in humans, chimpanzees (Pan troglodytes), and bonobos (Pan paniscus) shows that the microsatellite is conserved in humans and bonobos, both species sharing similar sociosexual behaviors, whereas in chimpanzees, a 360-bp sequence encompassing the microsatellite is deleted (180).
Molecular Mechanisms Involved in Mini- and Microsatellite Expansions
The frequent size variability of mini- and microsatellites has generated a large number of studies trying to understand the mechanisms involved in this size variability. In addition to the work of Alec Jeffreys and colleagues on minisatellite instability during human meiosis, numerous studies with model organisms (mainly but not exclusively E. coli, yeast, and mouse) have been helpful in dissecting such mechanisms. Given experimental data on human minisatellite size changes during meiosis (216, 220), it was initially thought that minisatellites expanded and contracted by homologous recombination, whereas microsatellites were subject to unrepaired slippage events between the newly synthesized strand and its template during S-phase DNA synthesis (also called “replication slippage”) (Fig. 4) (476). Nevertheless, subsequent experiments with both kinds of tandem repeats revealed that the differences between them were less pronounced than initially believed. Extensive studies of microsatellites, particularly of trinucleotide repeats, have shown that several mechanisms were involved in their instability, including replication, meiotic and mitotic homologous recombination, and postreplicational DNA repair. In the meantime, it was shown that minisatellites also evolved by slippage during S-phase replication. Most of what we know today about the molecular mechanisms involved in micro- and minisatellite instability comes from studies in model organisms, mainly but not exclusively E. coli, budding yeast, and mice. Although this review focuses on tandem repeats in eukaryotes, some of the most important papers deriving from studies with bacteria have been included in the present chapter. We will see that although microsatellites and minisatellites are generally rearranged in vivo by similar mechanisms, important details distinguish both types of repeats.
FIG. 4.
The “replication slippage” model of tandem repeat instability. The template strands are drawn in red, and the newly synthesized strands are drawn in blue. During replication of a repeat-containing sequence (A), the replication machinery may pause on the lagging strand, due to secondary structures or other kinds of lesions (B). (C) Partial unwinding of the lagging strand may lead to replication slippage when replication restarts, giving rise to an expansion or a contraction of the repeat tract, depending on what strand (template or newly synthesized strand) slippage occurred. (D) Alternatively, partial unwinding of the lagging strand may lead to lesion bypass by homologous recombination with the sister chromatid, also leading to contractions or expansions of the repeat tract (Fig. 7).
DNA secondary structures are involved in microsatellite instability.
Due to their repetitive nature and highly biased nucleotide composition, mini- and microsatellites were early suspected to form secondary structures that may play an important role in the mutational process. Earlier studies on homopurine-homopyrimidine (GA)-(TC) dinucleotide repeat tracts showed that they were able to form triple helices in vitro that have the property to block DNA synthesis in vitro (29). This is a general property of all homopurine-homopyrimidine tracts, even though they are not repeated in tandem (16). It was subsequently shown that (GA)n, (AT)n, and (GC)n dinucleotide repeats were very poor substrates for binding E. coli single-strand binding protein (SSB) and RecA or Rad51 recombination proteins. This was interpreted as the inability of these proteins to bind to structured DNA (41). With the discovery of the first trinucleotide repeat disorders, data on structural properties of such repeats have blossomed. Biophysical and biochemical analyses showed that (GTC)n, (CAG)n, and (CTG)n repeats form hairpin structures in vitro, in which cytosines and guanines are paired and adenines or thymines are excluded (Fig. 5) (154, 338, 340, 550, 551). The same repeats also form slipped structures on double-stranded DNA in which both DNA strands carry a hairpin (387). A more recent analysis of (CTG)n repeats shows that two nucleotides at the base of the stem are sensitive to single-strand-specific nucleases, suggesting that some sort of secondary hairpin arises from the stem base (10). In RNA, (CUG)n repeats are able to fold into a triangular tubelike structure that looks like the chocolate bar confection “Toblerone” and is more similar to a triplex than to a classical hairpin (395). Bending properties of (CCG)n, (GGC)n, and (CCA)n trinucleotide repeats were also determined and correlate well with data obtained by X-ray crystallography (58). At the same time, it was shown that (CCG)n and (CGG)n repeats form stable hairpins in vitro (154, 339, 355, 549). The same repeats are also able to fold into stable tetraplex structures similar to those found at telomeres (Fig. 5) (144, 151). (GAA)n trinucleotide repeats form several distinct types of secondary structures: at low temperature they form hairpins (193, 480); they may also form triple helices, like other homopurine-homopyrimidine sequences (314); and two triplexes may associate in a structure called “sticky DNA” (435). Both DNA triplex and sticky DNA inhibit transcription (172, 437). Secondary structure formation and transcription inhibition depend on repeat purity, since interrupted repeats lose these properties (436). (GGA)n trinucleotide repeats, which are also homopurine-homopyrimidine tracts, form intramolecular tetraplexes in vitro, similarly to (CCG)n repeats (319).
FIG. 5.
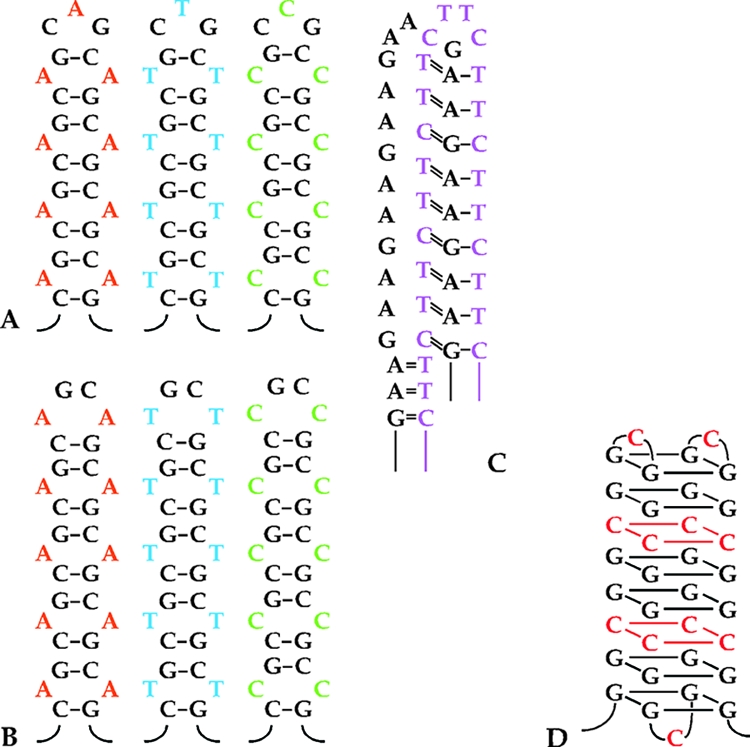
Secondary structures formed by some trinucleotide repeats. (A) CAG, CTG, and CCG hairpins formed by an odd number of repeat units. Bases making no pairing within the stem are colored. (B) CAG, CTG, and CCG hairpins formed by an even number of repeat units. Bases making no pairing within the stem are colored. (C) Triple helice formed by (GAA)n repeats. Watson-Crick pairings are shown by double lines, and Hogsteen pairings are shown by single lines (314). (D) Tetraplex structure formed by (CCG)n repeats. Cytosines and cytosine bonds are shown in red.
Interestingly, (CCG)n and (CGG)n repeats block DNA synthesis in vitro, suggesting that secondary structures make significant obstacles to replication factors (509), this defect being partially alleviated by the WRN helicase in vitro (229). Note that DNA synthesis in vitro can be efficiently arrested by a short G16CG(GGT)3 motif, which was proposed to form a tetraplex-like structure (540). Hairpin structures formed by trinucleotide repeats are resistant to cleavage by the FEN-1 nuclease, suggesting that if DNA flaps form in vivo during replication, they cannot be accurately processed by FEN-1 if they contain trinucleotide repeats that form hairpins (474).
Given that all trinucleotide repeat disorders involve sequences that are able to form secondary structures in vitro, it was postulated previously that these structures were essential to the expansion mechanism (327). However, at the present time, there is no direct proof that the same (or similar) structures form in vivo within these repeats, although several lines of evidence that will be discussed below suggest that at least some kinds of trinucleotide repeat-containing structures form in living cells.
Chromatin assembly is modified by trinucleotide repeats.
Since trinucleotide repeats form efficient secondary structures in vitro, it was a valid concern to assay the efficiency of nucleosome formation on these structures. Nucleosomes are normally formed by 146 bp of DNA wrapped around an octamer of histone proteins, and it is possible to reconstitute them in vitro using purified histones. When (CCG)n repeats were used in this assay, a strong nucleosomal exclusion was found, characterized by a reduced amount of histone-DNA complexes compared to nonrepetitive DNA (498, 522). This nucleosome exclusion was actually one of the strongest known at that time (523). Methylation of CpG dinucleotides inside the repeat tract reinforced nucleosome exclusion, suggesting that expanded (CCG)n repeats that are heavily methylated in fragile X patients may strongly inhibit nucleosome formation in vivo (524). Surprisingly, the effect is the opposite with (CTG)n trinucleotide repeats. These repeats apparently enhance nucleosome assembly in vitro in a length-dependent manner (longer repeats are more efficient at assembling nucleosomes than shorter ones) (521), while other studies demonstrated that as few as six CTG repeats facilitate nucleosome assembly (165, 439). No effect of GGA repeats on nucleosome formation was found (498).
It was very recently shown that inhibition of the SIRT1 histone deacetylase, the homologue of yeast SIR2 involved in rDNA maintenance (see “rDNA repeated arrays” above), reactivated alleles of FMR1 silenced by methylation. This reactivation was accompanied by an increase in histone H3 and H4 acetylation (40). This result shows that changing the methylation of FMR1 is possible by modulating the levels of histone acetylase/deacetylase in vivo, although this approach will most certainly lead to undesired effects on global gene expression.
DNA replication of mini- and microsatellites.
On one hand, unrepaired slippage events between the newly synthesized strand and its template, the so-called “replication slippage” model (Fig. 4), was the main pathway initially proposed to account for microsatellite instability. It was subsequently shown that trinucleotide repeats could be rearranged by gene conversion, during homologous recombination. On the other hand, meiotic gene conversion was first identified as being responsible for minisatellite size changes; therefore, studies focused mainly on homologous recombination until it was also shown that strand slippage within minisatellites could also occur during mitotic S-phase replication. At the present time, it is generally accepted that any mechanism which involves new synthesis of DNA, such as replication, recombination, and repair, may generate size changes within tandem repeats, the frequency and extent of such changes being clearly dependent on the chromosomal location, sequence, length, and purity of the repeat and on the genetic background.
(i) Effect of DNA replication on microsatellites.
Several experimental setups have been designed for model organisms in order to look at microsatellite stability in replication mutants. Since almost all of the genes involved in replication are essential for cell viability, conditional mutants of the replication fork were tested. In budding yeast, (GT)n microsatellites are destabilized in a length-dependent manner, with the rate of mutation varying by 500-fold between (GT)15 and (GT)105 repeats and with the longest being the most unstable (536). A mutation in POL30 (yeast PCNA [proliferating cell nuclear antigen] or “clamp”) (Fig. 6) increases the instability of mono-, di-, penta-, and octanucleotide repeats by several orders of magnitude, with mononucleotide repeats being the most strongly destabilized (252). Similarly, a transposon insertion into one of the subunits of the RFC complex, RFC1 (“clamp loader”) (Fig. 6), increases by 10-fold the instability of a (GT)16 tract (544). A specific allele of the POL2 gene (yeast Pol ɛ, the main polymerase of the leading strand [402]), pol2-C1089Y, was identified as having a moderate effect on mononucleotide repeat stability, whereas another allele, pol2-4, has no effect on the same tracts (248). In contrast, the pol3-01 allele of the POL3 gene (yeast Pol δ, the main polymerase of the lagging strand [367]) destabilizes the same mononucleotide tract 75-fold compared to what is seen for the wild type. This suggests that replication defects on the lagging strand are more prone to induce microsatellite size changes than replication defects on the leading strand.
FIG. 6.
Effect of mutants of the replication fork on microsatellite instability. The budding yeast replication fork is schematized. The Mcm DNA helicase opens the double helix to allow leading- and lagging-strand synthesis. The DNA Pol α-primase complex is believed to initiate replication by synthesis of a short RNA primer on both DNA strands. It is then replaced by the more processive Pol δ (on lagging stand) or Pol ɛ (on leading strand) associated with PCNA, the clamp processivity factor, loaded by the Rfc complex (“clamp loader”). Okazaki fragments on the lagging strand are subsequently processed by several enzymes, namely, the RNase H complex, Rad27, and Dna2, before ligation with the preceding Okazaki fragment is catalyzed by Cdc9. The mismatch repair complex scans newly synthesized DNA in order to check for replication errors. Single-stranded DNA is covered by the SSB complex Rpa. Note that PCNA also interacts with Cdc9 and Mlh1, in addition to proteins represented here. The trinucleotide repeat orientation represented here corresponds to the CTG orientation (or orientation II), in which CTGs are located on the lagging-strand template. Each box details the increase of microsatellite instability for each of the replication fork mutants tested over the wild-type level. Data were compiled from references given in the text.
The above-mentioned experimental setups were designed by cloning a repeat tract into a reporter gene and looking for mutations that restore (or lose) the reading frame. Mutations recovered were almost always additions or deletions of one repeat unit, suggesting that these mutations are the most prevalent in yeast and strengthening the “stepwise mutation model” of microsatellite evolution (66, 173, 556). Although elegantly designed, these experimental setups were not adapted to study trinucleotide repeat instabilities, since any change of one or more repeats would maintain the reading frame, and thus alternative systems needed to be designed. Several groups have used molecular approaches such as PCR and Southern blotting to detect trinucleotide repeat size changes, but although these methods are perfectly qualified to detect frequent size changes, they become tedious when the frequency of instability is low, and they are impossible to use in a genome-wide screen. The first genetic assay designed to overcome these limitations took advantage of the S. pombe ADH1 promoter, which exhibits specific spacing requirements in order to function in S. cerevisiae. If the distance between the TATA element and the transcription initiation start is below or above a given size, the promoter does not function and the reporter gene is turned off. This setup allows one to screen for contractions of a “long promoter” or expansions of a “short promoter”; however, the main limitation of this assay is that it does not allow screening for expansions of trinucleotide repeat tracts that are longer than 28 repeat units, since at that size or above, the promoter is turned off (109). The same genetic assay was used to look for trinucleotide repeat instability in human cells by cloning the (CAG)n-containing reporter gene into a shuttle vector that replicates both in yeast and in human cells (82). An alternative assay was designed by cloning triplet repeats in the intron of a yeast suppressor tDNA, SUP4. The tDNA is correctly transcribed and spliced as long as the repeat tract is no longer than 30 repeat units, and it suppresses a point mutation in the ADE2 reporter gene. When the repeat tract is longer than this threshold, the suppressor tDNA is no longer active and yeast cells are ade2− (416). Unfortunately, this system suffers from a similar drawback, since it is impossible to use for screening expansions of a trinucleotide repeat tract longer than 30 repeat units. Nevertheless, using either assay, as well as molecular methods, the effects of several replication mutants on trinucleotide repeat instability have been assayed. Most of the key players of the replication fork show to some extent a destabilization of triplet repeats in the corresponding mutant background (Fig. 6). Notable exceptions are mutants of the RNase H complex, with mutations of the pol3-01 allele of Pol δ and the pol2-4 allele of Pol ɛ, both deficient in proofreading function, namely, the pol2-18 and pol1-17 mutants. Knockouts of Pol ζ (REV7) and Pol η (RAD30), both involved in error-prone translesion bypass synthesis, have no effect on trinucleotide repeat stability (111). The most drastic effects on triplet repeat stability have been observed for mutants of PCNA (POL30), ligase I (CDC9), and yeast FEN-1 (RAD27), the latter showing the highest effect for all replication mutants. FEN-1/RAD27 is a structural homologue of RAD2 (209, 408), harboring both endo- and exonuclease activities, interacting with PCNA (202, 542), and is involved in Okazaki fragment processing. The RNA primer of Okazaki fragments is normally removed by RNase H, but the last ribonucleotide is cleaved by Rad27p (352, 353, 403). It was shown that Rad27p processes 5′ flaps very efficiently but at a much lower rate when these flaps are structured as hairpins (197). Mutants that separate endo- and exonuclease functions have been isolated, and several studies suggest that efficient processing of trinucleotide repeat-containing flaps necessitate all biochemical activities of Rad27p (288, 289, 470, 545). Interestingly, Dna2p, which is an essential nuclease also involved in Okazaki fragment processing, exhibits only weak phenotypes on trinucleotide repeat instability (Fig. 6). Dna2p and Rad27p interact with each other (61), and further studies suggest that Dna2p first cleaves the RNA-containing 5′ flap of Okazaki fragments, before Rad27p cleaves a second time to obtain a fully processed Okazaki fragment (24, 232). It is not completely obvious to reconcile this model with the effects of dna2-1 and rad27Δ mutants on triplet repeat instability, as the latter has a strong effect, while the former exhibits only a very moderate increase in instability. It is not completely clear either why DNA2, along with all other genes involved in replication in yeast, is essential for cell viability, while RAD27 is dispensable, although the knockout strain shows a strong mutator phenotype (497). One explanation could be that RAD27, in addition to its role at the replication fork, also plays another role in DNA metabolism, perhaps required for efficient processing of some recombination intermediates that could arise during the course of replication, as was suggested by some authors (188). In that case, in the absence of Rad27p more lesions would be made during replication of trinucleotide repeats, and these lesions could not be properly repaired, leading to the very high instability observed in this mutant background. One could also simply argue that comparing a point mutation in DNA2 to a complete deletion of RAD27 is unfair and that a point mutation in RAD27 exhibits a reduced level of trinucleotide repeat instability compared to what is seen for the complete inactivation of the gene (545). Nevertheless, it is clear that processing of Okazaki fragments during replication is an essential step leading to both contractions and expansions of triplet repeats and more generally of microsatellites. Strong destabilization of all microsatellites has also been observed in different alleles of PCNA, but this large processivity complex interacts with several replication proteins, including Pol δ, Pol ɛ, Rad27, and Cdc9, as well as the mismatch repair complex (508) (see “Defects in mismatch repair dramatically increase microsatellite instability” below). The observed increase in instability is probably the result of defects at several stages during replication. These data were collected from many publications and are summarized on Fig. 6 (68, 208, 253, 409, 416, 455, 456, 477, 534).
(ii) Effect of DNA replication on minisatellites.
Compared to the abundant literature on microsatellite instability in replication mutants, little has been done for minisatellites. Initially, the human MS32 minisatellite known to exhibit high rates of meiotic rearrangements (0.8% per molecule in sperm) was found to be also unstable in blood cells, although with a much lower frequency (<0.06%). Mutations observed include simple duplications or deletions of a given number of repeat units and can all be explained by intra-allelic events. No evidence of complex events involving interallelic recombination was found, and it was therefore proposed that minisatellite mitotic rearrangements involve replication slippage or sister-chromatid recombination (217).
Following the discovery that microsatellites—particularly trinucleotide repeats—were destabilized by inactivating RAD27, the stabilities of several minisatellites have been assayed in yeast strains deficient for this nuclease. A short minisatellite made up of 20-bp repeat units, tandemly repeated three times, shows an 11-fold increase in instability in a rad27Δ strain and a 13-fold increase in a pol3-t strain (253). Further studies of several human minisatellites integrated into the budding yeast genome showed that, in the absence of RAD27, their instability is highly increased, whereas they are only moderately increased in a dna2-1 mutant and unchanged in a rnh35Δ mutant, recapitulating the respective effects of the same mutants on trinucleotide repeat instability (295, 303). It was subsequently shown that minisatellite rearrangements in rad27Δ cells occur by homologous recombination, since they are suppressed by deletions of RAD51 or RAD52, both key players in homologous recombination, and they exhibit specific features usually associated with homology-driven mechanisms (296). A recent work identified the zinc transporter ZRT1 as also involved in minisatellite instability during mitotic divisions in budding yeast. Although the pathway involved in this process is not fully understood, it was shown that instability is reduced in a rad50Δ mutant, suggesting a role for this protein, whose functions in DSB repair are numerous (243).
(iii) cis-acting effects: repeat location, purity, and orientation.
Since the discovery of the first trinucleotide repeat disorders, it has been clear that cis-acting elements played a central role in the expansion process, as reviewed by several authors (277, 335, 385). We are now going to discuss three specific properties of trinucleotide repeats that, in our opinion, are essential to keep in mind when dealing with triplet repeats. As far as we know, other microsatellites do not exhibit the same properties, although specific experiments should certainly be designed to eventually answer this question.
First, it is noteworthy that trinucleotide repeat expansions are locus specific, meaning that a patient with an expansion in DM1 or FRAXA does not show any sign of expansion at other triplet repeat loci. This is exemplified by work published almost 10 years ago in which trinucleotide repeat polymorphisms at the DM1 locus and the SCA1 locus were compared in 29 families affected by HD. They found frequent polymorphism at the DM1 locus, whereas the SCA1 locus and other genome-wide microsatellites were stable. Moreover, they analyzed the same loci in 26 families affected by colorectal cancer, a type of cancer characterized by a high frequency of microsatellite instability, due to deficiencies in the mismatch repair system (see “Defects in mismatch repair dramatically increase microsatellite instability” below). They found that microsatellites, including triplet repeats at DM1 and SCA1, were generally highly variable, strongly suggesting that the molecular mechanisms involved in genome-wide microsatellite instability and those for trinucleotide repeat expansions were fundamentally different (166). This also explains why unstable human trinucleotide repeats integrated at different locations in the mouse genome exhibit variable levels of somatic and intergenerational instability (171, 282, 342, 460) and may even become highly stable (166) or lead to large expansions similar to those observed for humans (168). Interestingly, it was shown that (GT)16 dinucleotide repeats exhibited different rates of variation, depending on the locus at which they were integrated in the yeast genome. A 16-fold difference in the stability rate was measured between the most stable and the most unstable locus, suggesting that different regions of the genome do not replicate microsatellites with the same accuracy (190).
Another remarkable property of trinucleotide repeats is their propensity to be stabilized by the presence of interruptions in the repeat sequence, i.e., one or several repeat units that differ from the repeat consensus. This was first noticed in the fragile X locus FRAXA, in which AGG triplets are frequently found interrupting the (CGG)n repeat tract in the normal population, while patients exhibiting fragile X syndrome show pure, uninterrupted tracts (112, 199, 472). This is also true for (CAG)n repeat tracts in SCA1 alleles that are interrupted by a CAT triplet and for (CAG)n tracts in SCA2 alleles that are interrupted by CAA triplets, these interruptions being lost in the corresponding expanded alleles (80, 81). Similarly, a human (CA)n dinucleotide repeat shows a high stability when interrupted by a TA dinucleotide compared to what is seen for an uninterrupted allele (23). In budding yeast, it was shown that the introduction of an interruption within a perfect (CAG)n repeat tract stabilizes the tract by almost 2 orders of magnitude (427) and that contractions of the interrupted repeat tract almost always removed the interruption, transforming it into a perfect tract (110, 322). Similarly, Petes and colleagues (391) showed that interrupted (GT)n dinucleotide repeat instability was fivefold decreased compared to what was seen for a perfect (GT)n tract, suggesting that the property of interruptions that stabilize a repeat tract could be generalized to other microsatellites. One possible (but not exclusive) explanation would be that the presence of a different repeat unit within a microsatellite could disfavor the formation of potential secondary structures, hence reducing the chance of replication slippage due to these structures.
Last but not least of the cis-acting effects, the orientation of the repeat tract compared to the replication origin was first described for E. coli in a seminal paper showing that (CTG)n trinucleotide repeats cloned into a plasmid are more unstable when the CTG strand is the lagging-strand template than when the CAG strand is the lagging-strand template (231). This effect was confirmed in E. coli when repeats were integrated into the bacterial chromosome (553). Similar experiments were performed in yeast cells, in which (CTG)n or (CCG)n repeats were integrated in yeast chromosomes in the two possible orientations. With (CCG)n repeats, the repeat tract is more unstable when the lagging-strand template carries the CCG triplets (28). With (CTG)n repeats, the same orientation dependence seen for E. coli was found for yeast, with unstable repeats carrying CTG triplets on the lagging-strand template (150, 323, 331), although in one early report no difference was detected between the two orientations (332). The underlying model for this orientation effect relies on the propensity for CTG repeats to form secondary hairpins more stable than those formed by CAG repeats. Since it is suspected that the lagging-strand template is more prone to have single-stranded regions during replication than the leading-strand template, then when CTG repeats are exposed on such single-stranded regions, they are more prone to form hairpins than CAG repeats, and therefore they are more unstable. Note that at the present time, it is not formally proven that single-strand regions on the lagging-strand template are exposed long enough during the course of replication to allow the formation of secondary structures. It is also a possibility that some structures are left behind the replication fork and are inherited by the daughter cell, raising the chance of replication errors during the next round of replication.
It would be interesting to determine the orientation of trinucleotide repeats undergoing expansions in humans, but, unfortunately, most replication origins remain to be determined. Recently, a replication origin was identified in the promoter region of the FMR1 gene (174). The replication fork coming from this origin would replicate the (CCG)n trinucleotide repeat tract expanded in FRAXA such that the CGG sequence would be on the lagging-strand template, the orientation that was found to be more unstable in yeast, although this orientation leads more frequently to contractions than to expansions (28).
Given that authors generally use different experimental systems along with different nomenclatures to name repeat orientations, it is not always straightforward to know in which orientation repeats are replicated in a given system, sometimes adding confusion to the results. We therefore propose to adopt a general terminology to name repeat orientation. Since the lagging-strand template is supposed to be a key player in the instability process, we propose that repeats are named according to the sequence found on the lagging-strand template, i.e., CTG repeats when the CTG sequence is on the lagging-strand template (equivalent to the leading strand). This would correspond to what is generally (but not systematically) described as orientation II in the literature. The advantage of such a nomenclature is that is applicable to all kinds of microsatellites, without the need to define what is orientation I or II and what is orientation C or D, etc. Hopefully, there will be studies on other types of microsatellites that will help to determine if the orientation effect is restricted to trinucleotide repeats or is a general property of microsatellites.
(iv) Replication fork stalling and fork reversal.
One of the early questions about trinucleotide repeats was the possibility that secondary structure formation could impede the progression of the replication fork through the repeat tracts. This question was first addressed in E. coli, in which plasmids containing (CCG)n and (CAG)n repeats were cloned and transformed. By analysis of replication intermediates using two-dimensional gel electrophoresis, Samadashwily et al. (438) showed that (CCG)n repeat tracts induced strong replication blocks when located near a replication origin, and they addressed the effect of CCG or CGG triplets cloned on the lagging-strand template. The replication block was length dependent, such that longer repeats showed a stronger effect. In contrast, (CTG)n repeats induced stalling of the replication fork, only when the CTG and not when the CAG triplets were cloned in the lagging-strand template and only when chromosomal replication (but not plasmidic replication) was inhibited with chloramphenicol. Repeats shorter than (CTG)70 did not show any effect on replication. In budding yeast, by use of a similar experimental setting and the same technical procedure, (CCG)n trinucleotide repeats were also found to stall the replication fork in both orientations, even when short repeat tracts (18 repeat units) were examined. To the contrary, a very weak replication slowdown for (CAG)80 repeats was observed for both orientations (388). Similarly, no strong replication block was detected when (CAG)n repeats were integrated into a yeast chromosome (A. Kerrest, R. Anand, R. Sundararajan, R. Bermejo, G. Liberi, B. Dujon, C. H. Freudenreich, and G. F. Richard, unpublished data). When (GAA)n repeats were examined using the same approach, replication stalling was strong when GAA triplets, but not TTC triplets, were cloned on the lagging-strand template, showing that impediment of replication on the lagging strand was promoted by the homopurine but not the homopyrimidine sequence (260).
Replication fork stalling and replication restart have been intensely studied in the past few years and are the topic of several recent reviews (268, 333, 432). The fate of the replication fork after stalling or blocking was investigated in model organisms. It was first proposed by Seigneur and colleagues (459) that arrested replication forks are transiently “reversed” into Holliday junctions by the RuvAB complex, serving as a substrate for homologous recombination. Fork reversal is under the control of several proteins in E. coli, including the helicase UvrD (143, 278), a structural homologue of the Srs2 helicase that is involved in homologous recombination in budding yeast (261, 512) (see “Role of the error-free postreplication repair pathway on trinucleotide repeat expansions” below). In budding yeast, it was proposed that replication fork reversal occurs during chromosomal replication when it is slowed down with hydroxyurea and that it is controlled by Rad53, a key player in the checkpoint response (297). Using plasmid-borne (CAG)n repeat tracts, Fouché et al. (147) observed by electron microscopy “chicken foot” structures representing fork reversals and showed that these structures were dependent on the presence of the repeat tract. It would now be interesting to know if trinucleotide repeats that form strong secondary structures, like (CCG)n and (GAA)n, are also able to induce fork reversal in vivo or if this is restricted to the more labile secondary structures formed by (CAG)n repeats. It is nevertheless interesting that fork reversal cannot occur spontaneously in supercoiled regions and could occur in vivo only if topoisomerases are present to relax the supercoiling induced by the progression of the replication fork (134, 135).
(v) Effect of DNA damage checkpoints on trinucleotide repeat instability.
As presented above, mutations in DNA damage checkpoints increase fragile site expression, both in human and yeast cells (see “Molecular basis for fragility” above). The stability of triplet repeats has also been studied for such mutants. The contraction frequency of (CAG)n trinucleotide repeats is increased in yeast cell mutants for MEC1, DDC2, RAD17, RAD24, and RAD53 genes, with the effects of other checkpoint mutants being less pronounced (269). Interestingly, the expansion frequency of the same repeat tract is not increased in a comparable way, suggesting that an increase in contractions is specific to checkpoint mutants. One possibility is that checkpoint mutants increase DNA fragility (and therefore DNA breaks) within the repeat tract and that these breaks are repaired mainly by annealing of both sides of the break by single-strand annealing (SSA), a mechanism that was shown to preferentially produce contractions of the repeat tract (416). A different result was obtained with mice heterozygous for a mutation in the ATR gene (the mammalian homologue of yeast MEC1), in which more expansions of a (CGG)n repeat tract were detected (122). It is unclear if the different results obtained for yeast and for mice may reflect a difference in the intrinsic functionality of checkpoint proteins in both organisms, or whether CAG and CGG repeats behave differently in checkpoint mutants.
Defects in mismatch repair dramatically increase microsatellite instability.
The mismatch repair pathway (or MMR) is a complex of several proteins conserved from bacteria to all known eukaryotes and responsible for detecting replication errors such as transitions, transversions, insertions, and deletions, etc., and signaling them to the cell so that they can be repaired. The MMR complex includes several genes belonging to the MutS family, such as MSH2, MSH3, and MSH6 in budding yeast or to the MutL family, such as PMS1, MLH1, and MLH2 in yeast, and an endonuclease, EXO1 (254). Mismatch repair became famous when it was shown to be directly involved in sporadic colon cancer and in human nonpolyposis colon cancer (341, 389). In these two classes of colon cancer, the rate of mutation of microsatellites is several orders of magnitude higher than that in noncancerous cell types, suggesting a somatic origin for the instability in these tumors (287, 383). It was shown for these cancer patients that the MSH2 gene was mutated, leading to a high increase of microsatellite instabilities but also of point mutations (142). Several genes that could be directly involved in the tumorigenesis were subsequently found to be altered by this hypermutator phenotype. The type II transforming growth factor β receptor gene, involved in epithelial cell growth, contains an insertion in a (GT)3 dinucleotide repeat or a 1- or 2-nucleotide deletion in an (A)10 mononucleotide repeat in cancerous cells (316). IGFIIR, the insulin-like growth factor II receptor, contains a 1- or 2-bp deletion in a (G)8 mononucleotide repeat (473), and deletions and insertions in a (G)8 mononucleotide repeat in the BAX gene, involved in apoptosis, were found in tumors (405). Human nonpolyposis colon cancer-like cancer predisposition was also found in mice inactivated for the MSH3 or MSH6 genes (106a), and microsatellite instability is also elevated in C. elegans when MSH2 is inactivated (97).
Several experimental systems have been designed in budding and fission yeasts to study the effect of the MMR on microsatellite instability. Most of them rely on the insertion of a mono- or dinucleotide repeat tract within the coding sequence of a reporter gene, in such a way that a loss or gain of one repeat unit leads to gene inactivation (or activation, depending on how the construct was initially made). With such genetic tools, it was found that a deletion of any of the MutS or MutL yeast homologues led to a dramatic increase in microsatellite instability (Fig. 6) (185, 299, 312, 466, 467, 476, 501). Surprisingly, a deletion of mutS in Haemophilus influenzae has no effect on tetranucleotide repeat stability, while it has an effect on dinucleotide repeat stability, suggesting that this bacterial species has a specific way of regulating microsatellite instability. This might be linked to the fact that most of the microsatellites in H. influenzae are tetranucleotide repeats that are encoded by genes involved in virulence and adaptation to environmental changes and might have been selected to be more refractory to genome-wide microsatellite instability (33).
The stability of trinucleotide repeats was also assayed in MMR-deficient cells by use of assays similar to those used to assess the effect of replication mutants. Experimental systems initially designed in budding yeast were aimed at identifying repeat expansions or contractions of several triplets. Since most repeat size changes in MMR-deficient cells are additions or deletions of one repeat unit, mismatch repair mutants were found to have little or no effect in such experimental settings, compared to what was observed for other microsatellites (331, 332, 416). More surprisingly, when all small deletions and additions of triplets were scored using molecular approaches, it was found that trinucleotide repeats exhibited a much higher stability in MMR-deficient cells than other microsatellites (454) (Fig. 6). This suggested that secondary structures formed by trinucleotide repeats efficiently escaped recognition by the mismatch repair system or other DNA repair pathways, as suggested by some authors (345), and that trinucleotide repeat instability was largely independent of mismatch repair. However, additional studies revealed that the human MSH2 protein binds in vitro to secondary structures formed by (CAG)n and (CTG)n triplets in a length-dependent manner and with a higher efficiency for (CAG)n repeats (386). It was subsequently shown that transgenic mice deficient for Msh2 show a reduction in (CAG)n instability, a surprising result that is in contrast to what is commonly observed for other microsatellites, which are destabilized in MMR-deficient cells (311). The loss of Msh2 actually decreases the frequency of expansions, while contractions become much more frequent (447). These Msh2-dependent expansions seem to occur premeiotically, at the beginning of spermatogenesis (448). It was further shown that Msh3, but not Msh6, is also involved in (CAG)n expansions in mice, involving the heteroduplex Msh2-Msh3 in the expansion process (374). The same authors showed that purified Msh2-Msh3 complex binds efficiently to (CAG)n hairpins and it was proposed that this binding stabilizes the loop structure and inhibits correct repair by the MMR system or other DNA repair complexes. Thus, Msh2-Msh3 would have an opposite effect on trinucleotide repeats compared to other microsatellites, actually reducing the chance of correct repair of the hairpin. Strengthening the results obtained with Msh2 and Msh3, Pms2 null mice exhibit the same phenotype, a reduction in (CAG)n expansions, showing that another component of the MMR pathway is involved in making expansions (169).
Role of the error-free postreplication repair pathway on trinucleotide repeat expansions.
A new way of stabilizing trinucleotide repeats has been recently discovered, when a whole-genome screen for mutants affecting the stability of (CAG)n repeats identified the SRS2 gene of S. cerevisiae, as involved in this process. SRS2 was originally cloned as a suppressor of the radiation sensitivity phenotype of rad18 mutants, and is involved in the postreplication repair pathway of DNA damage (4). The Srs2 protein exhibits a 3′-to-5′ ATP-dependent helicase activity in vitro (428) and was shown to disrupt a Rad51p nucleoprotein filament, an intermediate of homologous recombination (261, 512). The same property was found for the bacterial functional homologue of SRS2 (UvrD) in unwinding RecA nucleoprotein filaments in E. coli (511). Further biochemical analysis of the protein substrates suggests that the Srs2 helicase could act as an antirecombinogenic protein that unwinds toxic recombination intermediates (118). Bhattacharyya and Lahue (38) showed that (CTG)13, (CTG)25, (CAG)25, and (CGG)25 trinucleotide repeats were more prone to expansions in an srs2 mutant and that these expansions were largely independent of RAD51-mediated homologous recombination. Interestingly, we recently found that longer (CTG)n repeats were highly destabilized in an srs2 mutant but that this instability was completely dependent on homologous recombination (Kerrest et al., unpublished), suggesting that two distinct pathways lead to trinucleotide repeat instability in srs2 mutants, depending on the size of repeat tracts. Mutants in the RAD18 and RAD5 genes or a mutation that abolishes the ubiquitination and sumoylation of PCNA (pol30-K164R) increase (CAG)n and (CTG)n repeat instability, confirming the role of the error-free postreplication pathway in this process (89). In addition, biochemical studies revealed that the Srs2 helicase is also able to unwind CTG hairpins or CTG-containing double-stranded DNA (39). Many years ago, it was published that a (GT)14 dinucleotide repeat tract was stabilized approximately 10-fold in a rad5 mutant (224), but this effect has been interpreted as an error-prone function of the RAD5 gene (89). RAD5 encodes a multifunctional protein which exhibits ATPase activity, is involved in DSB repair of cohesive ends in a way similar to that seen for the Mre11 protein (78), and is proposed to be a key player in replication fork reversal (45). Additional work will be required to determine whether replication fork reversal mediated by RAD5 (or SRS2) could be a way of stabilizing trinucleotide repeat tracts. It is interesting that SRS2 has a homologue in humans and in Schizosaccharomyces pombe, the FBH1 gene (F-Box DNA helicase). FBH1 in S. pombe seems to play some of the roles of SRS2 (347, 373), and it would be interesting to test the effect of FBH1 on trinucleotide repeats in human cells.
Mini- and microsatellite rearrangements during homologous recombination.
When the first massive expansions of trinucleotide repeats were discovered, it was originally thought that expansions occurred during S-phase DNA replication and most probably that the mismatch repair system was in some way involved in this process. These hypotheses were not completely wrong, although the precise mechanism responsible for these large expansions was not simple “replication slippage,” as was observed with other microsatellites in MMR mutants. After a while, some authors looked toward another mechanism, homologous recombination, as a possible cause of the large expansions, helped by the large body of evidence published on minisatellite instability during human meiosis.
(i) Expansions and contractions during meiotic recombination.
Several human minisatellites were initially found to be highly variable in the human population (218, 219, 221), and this variability was subsequently shown to arise in the germ line (60, 485). Molecular analysis of minisatellite rearrangements in sperm cells revealed complex mutation events, including both intra- and interallelic exchanges. This observation, along with the polarity of rearrangements (preferential modifications at one end of the tandem array), together with the absence of evidence for the exchange of flanking markers, suggested that rearrangements occur mainly through gene conversion not associated with crossover (59, 220). A meiotic recombination hot spot was mapped upstream of the MS32 minisatellite and was shown to be responsible for its meiotic instability (216). It was concluded that minisatellites are frequently rearranged by meiotic gene conversion when they are located near a meiotic recombination hot spot. The minisatellite mutation rate in the germ line was studied among children born in the area contaminated by radiation spills after the Chernobyl accident. It was shown that the mutation rate in the exposed population was twofold higher than the mutation rate in the control population, suggesting that ionizing radiations induce minisatellite rearrangements, probably by making low levels of DSBs that activate homologous recombination (114). In support of this hypothesis, it was shown that the mutation rate of mouse minisatellites was increased about twofold when mice were exposed to 0.5 Gy or 1 Gy of γ-irradiation (113), a dose more acute than what was estimated for the Chernobyl accident. A more recent work, looking specifically at the mouse minisatellite Ms6hm, showed that its mutation rate was also increased twofold following exposure to a 6-Gy γ-irradiation (368). Minisatellite instability was also observed in SCID (severe combined immuno deficiency) mouse cells, in which the catalytic unit of the DNA-dependent protein kinase (DNA-PKcs) was impaired, resulting in impairment in end joining. These cells exhibited a higher rate of minisatellite instability, suggesting that inactivating end joining increases minisatellite instability, possibly by increasing homologous recombination (204). It is interesting that meiotic instability is not restricted to natural minisatellites, since an artificial transgene array composed of 8 kb of human and bacterial sequences was dramatically amplified in mice. Expansions of from 5 to 8 initial repeat units of the transgene to from 200 to 300 copies were observed to happen in one generation, reminiscent of the dramatic expansions of trinucleotide repeats found to be associated with neurological disorders in humans. It was likely that these transgenic expansions occurred during gametogenesis in the male germ line (240).
S. cerevisiae has been a powerful tool to study minisatellite rearrangements during meiosis. MS1 was the first human minisatellite to be introduced in the genome of a haploid yeast cell. Frequent size changes of the minisatellite were observed and it was concluded that recombination between homologous chromosomes was therefore not a prerequisite for minisatellite rearrangements (73). The rate of instability of the human minisatellite MS32 was then compared during meiosis and mitosis in diploid yeast cells. MS32 was integrated near a well-characterized meiotic hot spot on yeast chromosome III. The frequency of size changes following meiosis was around 10%, a 40-fold increase over the frequency found during mitotic growth of yeast cells (12). Size changes occurred by gene conversion, with or without crossover (11). Similar features were found for two other human minisatellites (MS205 and MS1) integrated at the same locus in the yeast genome (36, 191, 192). A further step was made when Debrauwère and colleagues (94) showed that the meiotic instability of the human CEB1 minisatellite integrated near a yeast meiotic hot spot was dependent on the presence of the Spo11 endonuclease responsible for initiating meiotic recombination by making DSBs (37, 241). They also showed that minisatellite instability required the activity of Rad50, a protein involved in processing meiotic DSBs in order for recombination to occur properly. Subsequent analyses of human minisatellite instability during yeast meiosis showed that it did not depend on the mismatch repair system or on the Sgs1 helicase (42) but that the Rad1 protein was specifically increasing the frequency of minisatellite expansions (214). RAD1 is a gene whose product is essential to remove nonhomologous tails during gene conversion and SSA in yeast (377, 481), suggesting that such structures are formed during minisatellite recombination that need to be removed in order for expansions to occur.
All of the collective observations made for humans, along with experiments performed in budding yeast, formerly proved that meiotic gene conversion was the most important drive for minisatellite instability. This was proposed to be very different for microsatellites, however. Earlier experiments comparing the rates of instability of a (GT)16 dinucleotide repeat tract did not find significant differences between a strain mutated for the RAD52 gene and the wild-type yeast strain, suggesting that homologous recombination was not involved in microsatellite instability (195). At the same time, large expansions of (CGG)n repeats in FRAXA patients were found to occur in the absence of recombination of flanking markers (no crossover), which was too rapidly interpreted as ruling out meiotic homologous recombination as a possible cause for expansions (152), since the exchange of genetic information during meiosis is not always accompanied by crossovers (376). However, when (GT)n dinucleotide repeat tracts were introduced into a diploid yeast chromosome and these cells underwent meiosis, it was found that the presence of the microsatellite increased by severalfold the frequency of crossover and the frequency of multiple recombination events (involving more than two chromatids) in its vicinity (502). Later experiments in budding yeast showed that the presence of a (GT)39 dinucleotide repeat interfered with crossover resolution and increased the frequency of multiple recombination events. The microsatellite was found to be more unstable in recombinant spores than in parental spores (162). The instability of a (CAG)n trinucleotide repeat tract was increased during meiosis, compared to the mitotic instability, when the repeat tract was inserted near a yeast recombination hot spot (212). Most meiotic rearrangements (95%) of the repeat tract were found to be dependent on Spo11 (211). Similarly, (CAG)n trinucleotide repeats integrated in a yeast artificial chromosome were more unstable during meiosis than during mitotic cell divisions in yeast (86). Contrary to this, working with much smaller and interrupted (CAG)n trinucleotide repeats, Schweitzer et al. (457) did not find any evidence for increased instability, gene conversion, or crossover events associated with the presence of the microsatellite during yeast meiosis. The conclusion of these experiments is not qualitatively different from what was observed for minisatellites during meiosis: whenever perfect microsatellites are introduced in close proximity to a meiotic hot spot, they show a higher rate of instability during meiosis than during mitosis, this instability being dependent on Spo11 and therefore on the formation of meiotic DSBs. However, it was unclear if the microsatellite by itself was able to act as a meiotic hot spot, hence inducing meiotic recombination, although some data suggested that it could be the case (212). This question was first addressed by Moore and colleagues (345), who showed that the presence of (CAG)10 or (CGG)10 trinucleotide repeat tracts did not increase recombination between two flanking markers and were not the preferential site of meiotic DSBs. Additional experiments using much longer trinucleotide repeat tracts, (CAG)98 and (CAG)255, confirmed that even these long repeats did not act as meiotic recombination hot spots in yeast, the meiotic recombination rate being independent of the presence or absence of the microsatellite (412). Using the same experimental setting, a unique meiotic DSB was introduced by the specific endonuclease I-SceI in one of the two homologues, the other homologue carrying either a (CAG)98 or a (CAG)255 repeat tract. Contractions and expansions of the (CAG)98 repeat tract occurred in about 5% of the meiotic gene conversions, while such events were 10-fold more frequent when the (CAG)255 repeat tract was used as a template. Surprisingly, the frequency of rearrangements dropped to less than 1% when I-SceI was expressed during the mitotic growth of the cells, suggesting that meiotic recombination is more prone to make errors than mitotic recombination when (CAG)n repeat tracts are copied (412).
(ii) Expansions and contractions during mitotic recombination.
Experimental systems have also been set up in model organisms to study the fate of tandem repeats during mitotic recombination. In D. melanogaster, a tandem array of 5S ribosomal genes located within a P element was shown to be highly unstable following excision of the transposon. Contractions and expansions of the repeat array occurred in 40% of the progeny and were proposed to be the result of rearrangements within the 5S genes during DSB repair of the DSB generated by the transposon excision (375, 381). This experimental system was transposed into budding yeast and it was elegantly shown that the 5S tandem array underwent frequent expansions and contractions during repair of a single DSB induced by the HO endonuclease. Interestingly, the DSB could be repaired using two ectopic overlapping donor sequences, showing that homologous recombination was able to find and assemble overlapping sequences located at different loci (378). The same experimental system was used to study the fate of a 36-bp minisatellite during HO-induced DSB repair in yeast. Contractions and expansions of the minisatellite were found in 16% of the repair events, and it was shown that Msh2 and Rad1 proteins were required to promote contractions of the minisatellite during gene conversion (379), a result opposite to what was observed during meiotic recombination of a minisatellite (214). A similar experimental system was used to study (CAG)n size changes during HO- or I-SceI-induced DSB repair in budding yeast. When the DSB is repaired using a (CAG)39 as a template, frequent contractions but no expansions of the repeat tract were observed (416), whereas both expansions and contractions were found when a longer (CAG)98 repeat tract was used as the template (420). It was shown that the MRE11-RAD50-XRS2 complex was responsible for making large expansions, and it was proposed that these expansions occurred either through cleavage of secondary structures formed by the repeats and mediated by the endonuclease activity of Mre11 or, alternatively, by an unwinding of these secondary structures by the MRE11-RAD50-XRS2 complex (417). It is worth noting that when the endonuclease cleavage site was flanked by two short (CAG)5 repeats, a majority of the repair events occurred by annealing between the two repeats, often leading to a shorter repeat tract and therefore making SSA an attractive mechanism to generate repeat contractions (416). Note that repeat contractions and expansions of 5S genes or trinucleotide repeats during mitotic gene conversion are not associated with crossovers, ruling out the hypothesis of unequal crossover as a possible source of instability (378, 412) and supporting the hypothesis of a “DSB repair slippage” that would occur during gene conversion and would be 2 to 3 orders of magnitude more prone to errors than classical replication slippage (416). One of the possible models describing how DSB repair slippage may occur during gene conversion is shown in Fig. 7.
FIG. 7.
The “DSB repair slippage” model of tandem repeat instability. The broken molecule (recipient) is drawn in blue, the template molecule (donor) is drawn in red, and the newly synthesized strands are drawn in orange. (A) Following a DSB, gene conversion is initiated by strand invasion, forming a “D-loop.” (B) DNA synthesis within the repeat tract may be faithful or associated with slippage. After capture of the second end of the break, DNA synthesis of the second strand may be faithful (C) or associated with slippage (D). Slippage events will lead to expansions of the repeat tract (as shown in panels C and D) or to contractions if slippage occurs on the template strand. Alternatively, after capture of both ends followed by DNA synthesis, the two newly synthesized strands may unwind and anneal with each other in frame or out of frame, leading to expansions or contractions of the repeat tract. (E) This last alternative pathway is adapted from the synthesis-dependent strand annealing mechanism, proposed by several authors to explain tandem repeat rearrangements during gene conversion (363, 378, 420).
Studies with transgenic mice showed that neither Rad52 nor Rad54 had any effect on (CAG)n trinucleotide repeat instability (447). However, it was proposed that the functional homologue of yeast RAD52 in mammals might be Brca2 (349, 384, 482), and therefore the possible involvement of this gene in trinucleotide repeat instability should be assayed.
It is noteworthy that several human ataxias involve defects in DNA repair. Ataxia telangiectasia (ATM) and ATM-like disorders are characterized by early-onset cerebellar ataxia and the progressive degeneration of the cerebellum and spinocerebellar tract. These two ataxias respectively involve defects in the ATM checkpoint gene and the MRE11 gene, which are required for proper DSB repair response. These ataxias also exhibit radiosensitivity, chromosomal instability, and a high occurrence of cancers. Mutations in other genes involved in DSB repair lead to neural death by apoptosis. This is the case for NHEJ genes like those for the Ku complex, XRCC4, and ligase IV or for genes involved in homologous recombination like those for XRCC2 or BRCA1 (3). Other types of ataxias involve deficiencies in DSB repair. This is the case for SCA with axonal neuropathy (SCAN1) due to a defect in the TDP1 gene required for proper single-strand break repair and of ataxia-ocular motor apraxia (AOA1), involving the aprataxin gene, which is required for single-strand break signaling and repair (67). However, in these two last cases, no evidence for a general increase in genetic instability has been recorded and the defects are apparently restricted to the nervous system. At the present time, it is unclear why deficiencies in the above-mentioned genes seem to affect preferentially the nervous system, although one can advance the hypothesis that neurons are more sensitive than other cells to DNA breaks and enter more efficiently apoptosis when unrepaired breaks occur. It is possible that mild deficiencies in DSB repair pathways would lead to an increase in the level of endogenous single-strand breaks or DSBs and that these breaks could in turn trigger trinucleotide repeat expansions, increasing the chance of further breaks. Some of these unrepaired breaks could also trigger neuron apoptosis, increasing the severity of the clinical phenotype.
Revisiting the trinucleotide repeat expansion model.
At the end of the section on molecular mechanisms generally involved in mini- and microsatellite instability, particularly those related to trinucleotide repeat expansions, we want to propose a simple model that recapitulates data obtained both from human patients and from experiments in model organisms to explain how trinucleotide repeat instabilities occur (Fig. 8). In this model, the fate of a trinucleotide repeat depends exclusively on its propensity to form secondary structures and on the size of these structures. Short structures will be covered and protected by Msh2, preserving them and favoring replication slippage within the repeats. Slippage will eventually be corrected by the postreplicational repair pathway under the control of the Srs2 protein. Longer trinucleotide repeats will form longer and more stable hairpins that will also be substrates for Msh2. In addition, these longer hairpins may stall replication forks, leading to single-strand breaks and eventually DSBs. If DSBs are not correctly recognized by the checkpoint machinery, they will escape repair and lead to chromosomal fragility. DSB repair is also under the control of the Srs2 (and the Rad51) protein and may lead to repeat contractions and expansions by gene conversion. Other DNA damage, like oxidative damage, may also lead to fork stalling and DSBs. Given that Srs2 has a central role in this model, it would be interesting to test the role of its human homologue, FBH1, on trinucleotide repeat instability in mammalian cells.
FIG. 8.
Revisiting the trinucleotide repeat expansion model. Following this model, the fate of a given trinucleotide repeat tract depends only on its size. Due to secondary structures, short repeats are prone to replication slippage during S-phase replication, subsequently repaired by postreplication slippage under the control of the Srs2 protein. This will eventually lead to repeat expansion when Srs2 is deficient. Longer repeats are also prone to slippage but may stall forks, leading to DNA damage and DSBs. Checkpoint deficiency may lead to unrepaired DSBs and fragile site expression. DSB repair under the control of Srs2 may lead to repeat instability by gene conversion. Msh2 would bind to repeat hairpins, stabilizing them and maybe increasing the chance of slippage and/or breakage.
Finally, the link between replication and homologous recombination needs to be explored further. In model organisms like budding yeast and bacteria, several lines of evidence connect these two processes (175), but little is known on their interconnection in mammalian cells. As an alternative to DSB repair, fork reversal after DNA damage and fork stalling could supply cells with a mechanism that does not involve homologous recombination and would therefore be less prone to chromosomal rearrangements leading to segmental duplications and to microsatellite instabilities.
PERSPECTIVES: REPEATED QUESTIONS AND NEW CHALLENGES
Going from One to Two: Birth and Death of Microsatellites
As we have seen, several mechanisms relying on the formation of secondary structures, and on replication and homologous recombination, interplay with each other to generate contractions and expansions of tandem repeats. Even though the precise molecular steps remain to be clarified, the logic of going from two repeat units to hundreds or thousands is straightforward. However, it is still unclear how one goes from a single unit to two repeat units, or said in another way, how tandem repeats are born. One may easily imagine that point mutations can create mono-, di-, or even trinucleotide duplications, but what about longer microsatellites, minisatellites, and longer tandem repeats? It is unlikely that minisatellite birth occurs by successive point mutations. A seductive hypothesis was proposed by Haber and Louis (177), who noticed that the repeat units of several yeast and human minisatellites were flanked by short (5-bp) direct repeats and suggested that initial slippage between these direct repeats could duplicate the DNA sequence between them, giving birth to a minisatellite. A similar observation was made on many natural minisatellites found in budding yeast (414). Given the number of whole-genome sequences now available for eukaryotes, systematic sequence comparisons of closely related genomes will certainly shed a new light on the molecular mechanisms involved in micro- and minisatellite birth (328, 527, 556). Alternatively (or complementarily), sophisticated experimental systems in powerful model organisms like S. cerevisiae should also reveal some of these answers.
How microsatellites evolve is another intriguing question. The most recently accepted view is that short microsatellites tend to expand, while longer ones tend to contract. This was demonstrated by analyzing 122 human tetranucleotide repeat loci and showing that the rate of expansions is constant for all alleles, whereas the rate of contractions increases exponentially over repeat length. This led to a model in which microsatellites tend to expand until they reach a critical size, above which they will tend to contract (546). A corollary to this model is that the size distribution of microsatellites, in an equilibrium in a given genome, should be centered around the critical size. Another model proposes that microsatellite evolution is driven by slippage that will tend to increase repeat size and by point mutations that will tend to interrupt the tandem repeat sequence, therefore stabilizing it (66, 264, 441). Note that these two hypotheses are not mutually exclusive and that experimental results on microsatellite instability in model organisms support both models.
Toward a Unique Definition of Micro- and Minisatellites
Both microsatellites and minisatellites are frequently found within genes, at least in hemiascomycete genomes, in which they have been the most studied. Remarkably, they are not contained by the same type of genes, with microsatellites being found mainly in nuclear transcription factors, while minisatellites are contained by cell wall genes. Therefore, the distinction between both types of tandem repeats, originally based on historical grounds, finds a biological justification. We therefore propose that since the shortest minisatellite unit found in a cell wall gene was 9 nucleotides long (414), mono- to octanucleotide repeats should be called microsatellites, whereas nonanucleotide repeats and above should be called minisatellites. Hopefully, this definition will be adopted by a majority of people who work in the field, so that everyone will use the same terminology when dealing with tandem repeats. It would also be helpful if the same terminology could be used to define the orientation of a repeat tract according to replication: we therefore propose that repeats be named according to the sequence found on the lagging-strand template, i.e., CTG repeats when the CTG sequence is on the lagging-strand template (equivalent to the leading strand). This nomenclature is also applicable to all kinds of microsatellites besides trinucleotide repeats, as long as the direction of replication is determined.
Adding to the trouble coming from the lack of clear definitions for these elements, it is difficult to find two scientific reports using the same algorithm to detect tandem repeats, and whenever this occasionally happens, differences in parameters and thresholds for detection are so different that it makes any comparison between data sets quite ambiguous (Table 3). It is therefore very difficult to compare microsatellite distributions among eukaryotes, and it is still unclear for us if the density of trinucleotide repeats in the human genome (11.8 per megabase) (270a) is higher or lower than the density of trinucleotide repeats in the yeast genome, which varies from 8 to 147 repeats per megabase (Table 3). Imperfect and perfect microsatellites should at least be computed separately, which is not systematically the case.
A Final Word
As one of the pioneers of molecular biology and a Nobel prize winner, Jacques Monod was amazed by the conservation of structures in living organisms, given the relatively high frequency of some mutations in human beings: “Altogether, we may estimate that in the present-day human population of approximately three thousand million there occur, with each new generation, some hundred thousand million to a billion mutations… . Considering the scope of this gigantic lottery and the speed with which nature draws the numbers, it may well seem that the amazing and indeed paradoxical thing, hard to explain, is not evolution but rather the stability of the ‘forms’ that make up the biosphere” (344). What would Jacques Monod have thought of mutations that are even more frequent, to the order of 10−2 to 10−3 for some microsatellites, and even higher for trinucleotide repeat expansions? He would probably have been fascinated by the fact that morphological shapes in dogs rely on such unstable sequences (145). However, these mutations must still be exposed to the process of natural selection and, in the end, a dog might remain a dog or evolve into another species.
Acknowledgments
We are greatly indebted to the many colleagues who contributed over the past years to stimulating discussions on the evolution of repeated sequences, especially to all the members, past and present, of the Unité de Génétique Moléculaire des Levures; to Jim Haber, Alain Nicolas, Benoit Arcangioli, and all the members of their respective labs; and to Geneviève Gourdon, Catherine Freudenreich, and Frédéric Pâques for more-specific discussions on tandem repeat instabilities. We apologize to many colleagues working on microsatellites, since it was not possible to extensively cite all their studies. We are very grateful to Allyson Holmes, Gilles Fischer, and Cécile Neuvéglise for many suggestions that improved the overall quality of the present work and to Agnès Ullmann for lending us her personal copy of Chance and Necessity.
This work was supported by grant 3738 from the Association pour la Recherche contre le Cancer (ARC) and grant ANR-05-BLAN-0331 from the Agence Nationale de la Recherche. B.D. is a member of the Institut Universitaire de France.
REFERENCES
- 1.Reference deleted.
- 2.Abdel-Rahman, W. M., K. Katsura, W. Rens, P. A. Gorman, D. Sheer, D. Bicknell, W. F. Bodmer, M. J. Arends, A. H. Wyllie, and P. A. Edwards. 2001. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc. Natl. Acad. Sci. USA 982538-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abner, C. W., and P. J. McKinnon. 2004. The DNA double-strand break response in the nervous system. DNA Repair 31141-1147. [DOI] [PubMed] [Google Scholar]
- 4.Aboussekhra, A., R. Chanet, Z. Zgaga, C. Cassier-Chauvat, M. Heude, and F. Fabre. 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 177211-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, R. A. George, S. E. Lewis, S. Richards, M. Ashburner, S. N. Henderson, G. G. Sutton, J. R. Wortman, M. D. Yandell, Q. Zhang, L. X. Chen, R. C. Brandon, Y. H. Rogers, R. G. Blazej, M. Champe, B. D. Pfeiffer, K. H. Wan, C. Doyle, E. G. Baxter, G. Helt, C. R. Nelson, G. L. Gabor, J. F. Abril, A. Agbayani, H. J. An, C. Andrews-Pfannkoch, D. Baldwin, R. M. Ballew, A. Basu, J. Baxendale, L. Bayraktaroglu, E. M. Beasley, K. Y. Beeson, P. V. Benos, B. P. Berman, D. Bhandari, S. Bolshakov, D. Borkova, M. R. Botchan, J. Bouck, P. Brokstein, P. Brottier, K. C. Burtis, D. A. Busam, H. Butler, E. Cadieu, A. Center, I. Chandra, J. M. Cherry, S. Cawley, C. Dahlke, L. B. Davenport, P. Davies, B. de Pablos, A. Delcher, Z. Deng, A. D. Mays, I. Dew, S. M. Dietz, K. Dodson, L. E. Doup, M. Downes, S. Dugan-Rocha, B. C. Dunkov, P. Dunn, K. J. Durbin, C. C. Evangelista, C. Ferraz, S. Ferriera, W. Fleischmann, C. Fosler, A. E. Gabrielian, N. S. Garg, W. M. Gelbart, K. Glasser, A. Glodek, F. Gong, J. H. Gorrell, Z. Gu, P. Guan, M. Harris, N. L. Harris, D. Harvey, T. J. Heiman, J. R. Hernandez, J. Houck, D. Hostin, K. A. Houston, T. J. Howland, M. H. Wei, C. Ibegwam, et al. 2000. The genome sequence of Drosophila melanogaster. Science 2872185-2195. [DOI] [PubMed] [Google Scholar]
- 6.Admire, A., L. Shanks, N. Danzl, M. Wang, U. Weier, W. Stevens, E. Hunt, and T. Weinert. 2006. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev. 20159-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akagi, H., Y. Yokozeki, A. Inagaki, K. Mori, and T. Fujimura. 2001. Micron, a microsatellite-targeting transposable element in the rice genome. Mol. Genet. Genomics 266471-480. [DOI] [PubMed] [Google Scholar]
- 8.Akarsu, A. N., I. Stoilov, E. Yilmaz, B. S. Sayli, and M. Sarfarazi. 1996. Genomic structure of HOXD13 gene: a nine polyalanine duplication causes synpolydactyly in two unrelated families. Hum. Mol. Genet. 5945-952. [DOI] [PubMed] [Google Scholar]
- 9.Reference deleted.
- 10.Amrane, S., B. Sacca, M. Mills, M. Chauhan, H. H. Klump, and J. L. Mergny. 2005. Length-dependent energetics of (CTG)n and (CAG)n trinucleotide repeats. Nucleic Acids Res. 334065-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Appelgren, H., H. Cederberg, and U. Rannug. 1999. Meiotic interallelic conversion at the human minisatellite MS32 in yeast triggers recombination in several chromatids. Gene 23929-38. [DOI] [PubMed] [Google Scholar]
- 12.Appelgren, H., H. Cederberg, and U. Rannug. 1997. Mutations at the human minisatellite MS32 integrated in yeast occur with high frequency in meiosis and involve complex recombination events. Mol. Gen. Genet. 2567-17. [DOI] [PubMed] [Google Scholar]
- 12a.Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408796-815. [DOI] [PubMed] [Google Scholar]
- 13.Arcangioli, B. 1998. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 174503-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arcangioli, B. 2000. Fate of mat1 DNA strands during mating-type switching in fission yeast. EMBO Rep. 1145-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arcot, S. S., Z. Wang, J. L. Weber, P. L. Deininger, and M. A. Batzer. 1995. Alu repeats: a source for the genesis of primate microsatellites. Genomics 29136-144. [DOI] [PubMed] [Google Scholar]
- 16.Arimondo, P. B., F. Barcelo, J. S. Sun, J. C. Maurizot, T. Garestier, and C. Helene. 1998. Triple helix formation by (G,A)-containing oligonucleotides: asymmetric sequence effect. Biochemistry 3716627-16635. [DOI] [PubMed] [Google Scholar]
- 17.Arlt, M. F., B. Xu, S. G. Durkin, A. M. Casper, M. B. Kastan, and T. W. Glover. 2004. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol. Cell. Biol. 246701-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armour, J. A., S. Povey, S. Jeremiah, and A. J. Jeffreys. 1990. Systematic cloning of human minisatellites from ordered array charomid libraries. Genomics 8501-512. [DOI] [PubMed] [Google Scholar]
- 19.Astolfi, P., D. Bellizzi, and V. Sgaramella. 2003. Frequency and coverage of trinucleotide repeats in eukaryotes. Gene 317117-125. [DOI] [PubMed] [Google Scholar]
- 20.Aury, J. M., O. Jaillon, L. Duret, B. Noel, C. Jubin, B. M. Porcel, B. Segurens, V. Daubin, V. Anthouard, N. Aiach, O. Arnaiz, A. Billaut, J. Beisson, I. Blanc, K. Bouhouche, F. Camara, S. Duharcourt, R. Guigo, D. Gogendeau, M. Katinka, A. M. Keller, R. Kissmehl, C. Klotz, F. Koll, A. Le Mouel, G. Lepere, S. Malinsky, M. Nowacki, J. K. Nowak, H. Plattner, J. Poulain, F. Ruiz, V. Serrano, M. Zagulski, P. Dessen, M. Betermier, J. Weissenbach, C. Scarpelli, V. Schachter, L. Sperling, E. Meyer, J. Cohen, and P. Wincker. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444171-178. [DOI] [PubMed] [Google Scholar]
- 21.Ayub, Q., A. Mohyuddin, R. Qamar, K. Mazhar, T. Zerjal, S. Q. Mehdi, and C. Tyler-Smith. 2000. Identification and characterisation of novel human Y-chromosomal microsatellites from sequence database information. Nucleic Acids Res. 28e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachtrog, D., S. Weiss, B. Zangerl, G. Brem, and C. Schlotterer. 1999. Distribution of dinucleotide microsatellites in the Drosophila melanogaster genome. Mol. Biol. Evol. 16602-610. [DOI] [PubMed] [Google Scholar]
- 23.Bacon, A. L., S. M. Farrington, and M. G. Dunlop. 2000. Sequence interruptions confer differential stability at microsatellite alleles in mismatch repair-deficient cells. Hum. Mol. Genet. 92707-2713. [DOI] [PubMed] [Google Scholar]
- 24.Bae, S.-H., and Y.-S. Seo. 2000. Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 27538022-38031. [DOI] [PubMed] [Google Scholar]
- 25.Bailey, J. A., D. M. Church, M. Ventura, M. Rocchi, and E. E. Eichler. 2004. Analysis of segmental duplications and genome assembly in the mouse. Genome Res. 14789-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey, J. A., Z. Gu, R. A. Clark, K. Reinert, R. V. Samonte, S. Schwartz, M. D. Adams, E. W. Myers, P. W. Li, and E. E. Eichler. 2002. Recent segmental duplications in the human genome. Science 2971003-1007. [DOI] [PubMed] [Google Scholar]
- 27.Bailey, J. A., A. M. Yavor, H. F. Massa, B. J. Trask, and E. E. Eichler. 2001. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 111005-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balakumaran, B. S., C. H. Freudenreich, and V. A. Zakian. 2000. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 993-100. [DOI] [PubMed] [Google Scholar]
- 29.Baran, N., A. Lapidot, and H. Manor. 1991. Formation of DNA triplexes accounts for arrests of DNA synthesis at d(TC)n and d(GA)n tracts. Proc. Natl. Acad. Sci. USA 88507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barreiro, L. B., G. Laval, H. Quach, E. Patin, and L. Quintana-Murci. 2008. Natural selection has driven population differentiation in modern humans. Nat. Genet. 40340-345. [DOI] [PubMed] [Google Scholar]
- 31.Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434864-870. [DOI] [PubMed] [Google Scholar]
- 32.Batzer, M. A., and P. L. Deininger. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3370-379. [DOI] [PubMed] [Google Scholar]
- 33.Bayliss, C. D., T. van de Ven, and E. R. Moxon. 2002. Mutations in polI but not mutSLH destabilize Haemophilus influenzae tetranucleotide repeats. EMBO J. 211465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belancio, V. P., D. J. Hedges, and P. Deininger. 2008. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 18343-358. [DOI] [PubMed] [Google Scholar]
- 35.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berg, I., H. Cederberg, and U. Rannug. 2000. Tetrad analysis shows that gene conversion is the major mechanism involved in mutation at the human minisatellite MS1 integrated in Saccharomyces cerevisiae. Genet. Res. 751-12. [DOI] [PubMed] [Google Scholar]
- 37.Bergerat, A., B. de Massy, D. Gadelle, P. C. Varoutas, A. Nicolas, and P. Forterre. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386414-417. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharyya, S., and R. S. Lahue. 2004. Saccharomyces cerevisiae Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol. Cell. Biol. 247324-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharyya, S., and R. S. Lahue. 2005. Srs2 helicase of Saccharomyces cerevisiae selectively unwinds triplet repeat DNA. J. Biol. Chem. 28033311-33317. [DOI] [PubMed] [Google Scholar]
- 40.Biacsi, R., D. Kumari, and K. Usdin. 2008. SIRT1 inhibition alleviates gene silencing in fragile X mental retardation. PLoS Genet. 41-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biet, E., J.-S. Sun, and M. Dutreix. 1999. Conserved sequence preference in DNA binding among recombination proteins: an effect of ssDNA secondary structure. Nucleic Acids Res. 27596-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bishop, A. J. R., E. J. Louis, and R. H. Borts. 2000. Minisatellite variants generated in yeast meiosis involve DNA removal during gene conversion. Genetics 1567-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanc, G., and K. H. Wolfe. 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 161679-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanc, G., and K. H. Wolfe. 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 161667-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blastyak, A., L. Pinter, I. Unk, L. Prakash, S. Prakash, and L. Haracska. 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boeva, V., M. Regnier, D. Papatsenko, and V. Makeev. 2006. Short fuzzy tandem repeats in genomic sequences, identification, and possible role in regulation of gene expression. Bioinformatics 22676-684. [DOI] [PubMed] [Google Scholar]
- 47.Bonaccorsi, S., and A. Lohe. 1991. Fine mapping of satellite DNA sequences along the Y chromosome of Drosophila melanogaster: relationships between satellite sequences and fertility factors. Genetics 129177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowen, S., C. Roberts, and A. E. Wheals. 2005. Patterns of polymorphism and divergence in stress-related yeast proteins. Yeast 22659-668. [DOI] [PubMed] [Google Scholar]
- 49.Bowers, J., J.-M. Boursiquot, P. This, K. Chu, H. Johansson, and C. Meredith. 1999. Historical genetics: the parentage of Chardonnay, Gamay, and other wine grapes of northeastern France. Science 2851562-1565. [DOI] [PubMed] [Google Scholar]
- 50.Brais, B., J. P. Bouchard, Y. G. Xie, D. L. Rochefort, N. Chretien, F. M. Tome, R. G. Lafreniere, J. M. Rommens, E. Uyama, O. Nohira, S. Blumen, A. D. Korczyn, P. Heutink, J. Mathieu, A. Duranceau, F. Codere, M. Fardeau, and G. A. Rouleau. 1998. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 18164-167. [DOI] [PubMed] [Google Scholar]
- 51.Reference deleted.
- 52.Britten, R. J., and D. E. Kohne. 1968. Repeated sequences in DNA. Science 161529-540. [DOI] [PubMed] [Google Scholar]
- 53.Broccoli, D., O. J. Miller, and D. A. Miller. 1992. Isolation and characterization of a mouse subtelomeric sequence. Chromosoma 101442-447. [DOI] [PubMed] [Google Scholar]
- 54.Brook, J. D., M. E. McCurrach, H. G. Harley, A. J. Buckler, D. Church, H. Aburatani, K. Hunter, V. P. Stanton, J. P. Thirion, T. Hudson, R. Sohn, B. Zemelman, R. G. Snell, S. A. Rundle, S. Crow, J. Davies, P. Shelbourne, J. Buxton, C. Jones, V. Juvonen, K. Johnson, P. S. Harper, D. J. Shaw, and D. E. Housman. 1992. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68799-808. [DOI] [PubMed] [Google Scholar]
- 55.Brosh, R. M., J. Waheed, and J. A. Sommers. 2002. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 27723236-23245. [DOI] [PubMed] [Google Scholar]
- 56.Brown, L. Y., and S. A. Brown. 2004. Alanine tracts: the expanding story of human illness and trinucleotide repeats. Trends Genet. 2051-58. [DOI] [PubMed] [Google Scholar]
- 57.Brown, L. Y., S. Odent, V. David, M. Blayau, C. Dubourg, C. Apacik, M. A. Delgado, B. D. Hall, J. F. Reynolds, A. Sommer, D. Wieczorek, S. A. Brown, and M. Muenke. 2001. Holoprosencephaly due to mutations in ZIC2: alanine tract expansion mutations may be caused by parental somatic recombination. Hum. Mol. Genet. 10791-796. [DOI] [PubMed] [Google Scholar]
- 58.Brukner, I., R. Sanchez, D. Suck, and S. Pongor. 1995. Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 141812-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buard, J., and A. J. Jeffreys. 1997. Big, bad minisatellites. Nat. Genet. 15327-328. [DOI] [PubMed] [Google Scholar]
- 60.Buard, J., and G. Vergnaud. 1994. Complex recombination events at the hypermutable minisatellite CEB1 (D2S90). EMBO J. 133203-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Budd, M. E., and J. L. Campbell. 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 172136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burns, K. H., and J. D. Boeke. 2008. Great exaptations. J. Biol. 75-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butler, D. K., L. E. Yasuda, and M. C. Yao. 1995. An intramolecular recombination mechanism for the formation of the rRNA gene palindrome of Tetrahymena thermophila. Mol. Cell. Biol. 157117-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Butler, D. K., L. E. Yasuda, and M. C. Yao. 1996. Induction of large DNA palindrome formation in yeast: implications for gene amplification and genome stability in eukaryotes. Cell 871115-1122. [DOI] [PubMed] [Google Scholar]
- 65.Caburet, S., C. Conti, C. Schurra, R. Lebofsky, S. J. Edelstein, and A. Bensimon. 2005. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 151079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calabrese, P. P., R. T. Durrett, and C. F. Aquadro. 2001. Dynamics of microsatellite divergence under stepwise mutation and proportional slippage/point mutation models. Genetics 159839-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caldecott, K. W. 2004. DNA single-strand breaks and neurodegeneration. DNA Repair 3875-882. [DOI] [PubMed] [Google Scholar]
- 68.Callahan, J. L., K. J. Andrews, V. A. Zakian, and C. H. Freudenreich. 2003. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol. Cell. Biol. 237849-7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cam, H. P., K.-I. Noma, H. Ebina, H. L. Levin, and S. I. S. Grewal. 2008. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451431-437. [DOI] [PubMed] [Google Scholar]
- 70.Campuzano, V., L. Montermini, M. D. Molto, L. Pianese, M. Cossee, F. Cavalcanti, E. Monros, F. Rodius, F. Duclos, A. Monticelli, F. Zara, J. Canizares, H. Koutnikova, S. I. Bidichandani, C. Gellera, A. Brice, P. Trouillas, G. De Michele, A. Filla, R. De Frutos, F. Palau, P. I. Patel, S. Di Donato, J.-L. Mandel, S. Cocozza, M. Koenig, and M. Pandolfo. 1996. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 2711423-1427. [DOI] [PubMed] [Google Scholar]
- 71.Carlton, J. M., R. P. Hirt, J. C. Silva, A. L. Delcher, M. Schatz, Q. Zhao, J. R. Wortman, S. L. Bidwell, U. C. Alsmark, S. Besteiro, T. Sicheritz-Ponten, C. J. Noel, J. B. Dacks, P. G. Foster, C. Simillion, Y. Van de Peer, D. Miranda-Saavedra, G. J. Barton, G. D. Westrop, S. Muller, D. Dessi, P. L. Fiori, Q. Ren, I. Paulsen, H. Zhang, F. D. Bastida-Corcuera, A. Simoes-Barbosa, M. T. Brown, R. D. Hayes, M. Mukherjee, C. Y. Okumura, R. Schneider, A. J. Smith, S. Vanacova, M. Villalvazo, B. J. Haas, M. Pertea, T. V. Feldblyum, T. R. Utterback, C. L. Shu, K. Osoegawa, P. J. de Jong, I. Hrdy, L. Horvathova, Z. Zubacova, P. Dolezal, S. B. Malik, J. M. Logsdon, Jr., K. Henze, A. Gupta, C. C. Wang, R. L. Dunne, J. A. Upcroft, P. Upcroft, O. White, S. L. Salzberg, P. Tang, C. H. Chiu, Y. S. Lee, T. M. Embley, G. H. Coombs, J. C. Mottram, J. Tachezy, C. M. Fraser-Liggett, and P. J. Johnson. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Casper, A. M., P. Nghiem, M. F. Arlt, and T. W. Glover. 2002. ATR regulates fragile site stability. Cell 111779-789. [DOI] [PubMed] [Google Scholar]
- 73.Cederberg, H., E. Agurell, M. Hedenskog, and U. Rannug. 1993. Amplification and loss of repeat units of the human minisatellite MS1 integrated in chromosome III of a haploid yeast strain. Mol. Gen. Genet. 23838-42. [DOI] [PubMed] [Google Scholar]
- 73a.C. elegans sequencing consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 2822012-2018. [DOI] [PubMed] [Google Scholar]
- 74.Cha, R. S., and N. Kleckner. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297602-606. [DOI] [PubMed] [Google Scholar]
- 75.Chan, H. Y., J. M. Warrick, G. L. Gray-Board, H. L. Paulson, and N. M. Bonini. 2000. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum. Mol. Genet. 92811-2820. [DOI] [PubMed] [Google Scholar]
- 76.Charlesworth, B., P. Sniegowski, and W. Stephan. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371215-220. [DOI] [PubMed] [Google Scholar]
- 77.Charlet-B., N., R. S. Savkur, G. Singh, A. V. Philips, E. A. Grice, and T. A. Cooper. 2002. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell 1045-53. [DOI] [PubMed] [Google Scholar]
- 78.Chen, S., A. A. Davies, D. Sagan, and H. D. Ulrich. 2005. The RING finger ATPase Rad5p of Saccharomyces cerevisiae contributes to DNA double-strand break repair in a ubiquitin-independent manner. Nucleic Acids Res. 335878-5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng, Z., M. Ventura, X. She, P. Khaitovich, T. Graves, K. Osoegawa, D. Church, P. DeJong, R. K. Wilson, S. Paabo, M. Rocchi, and E. E. Eichler. 2005. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 43788-93. [DOI] [PubMed] [Google Scholar]
- 79a.Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 43769-87. [DOI] [PubMed] [Google Scholar]
- 80.Choudhry, S., M. Mukerji, A. K. Srivastava, S. Jain, and S. K. Brahmachari. 2001. CAG repeat instability at SCA2 locus: anchoring CAA interruptions and linked single nucleotide polymorphisms. Hum. Mol. Genet. 102437-2446. [DOI] [PubMed] [Google Scholar]
- 81.Chung, M. Y., L. P. Ranum, L. A. Duvick, A. Servadio, H. Y. Zoghbi, and H. T. Orr. 1993. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat. Genet. 5254-258. [DOI] [PubMed] [Google Scholar]
- 82.Claassen, D. A., and R. S. Lahue. 2007. Expansions of CAG.CTG repeats in immortalized human astrocytes. Hum. Mol. Genet. 163088-3096. [DOI] [PubMed] [Google Scholar]
- 83.Clark, R. M., G. L. Dalgliesh, D. Endres, M. Gomez, J. Taylor, and S. I. Bidichandani. 2004. Expansion of GAA triplet repeats in the human genome: unique origin of the FRDA mutation at the center of an Alu. Genomics 83373-383. [DOI] [PubMed] [Google Scholar]
- 84.Cleary, J. D., and C. E. Pearson. 2005. Replication fork dynamics and dynamic mutations: the fork-shift model of repeat instability. Trends Genet. 21272-280. [DOI] [PubMed] [Google Scholar]
- 85.Cobb, J. A., L. Bjergbaek, K. Shimada, C. Frei, and S. M. Gasser. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 224325-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cohen, H., D. D. Sears, D. Zenvirth, P. Hieter, and G. Simchen. 1999. Increased instability of human CTG repeat tracts on yeast artificial chromosomes during gametogenesis. Mol. Cell. Biol. 194153-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Coissac, E., E. Maillier, and P. Netter. 1997. A comparative study of duplications in bacteria and eukaryotes: the importance of telomeres. Mol. Biol. Evol. 141062-1074. [DOI] [PubMed] [Google Scholar]
- 88.Daboussi, M.-J., J.-M. Davière, S. Graziani, and T. Langin. 2002. Evolution of the Fot1 transposons in the genus Fusarium: discontinuous distribution and epigenetic inactivation. Mol. Biol. Evol. 19510-520. [DOI] [PubMed] [Google Scholar]
- 89.Daee, D. L., T. Mertz, and R. S. Lahue. 2007. Postreplication repair inhibits CAG-CTG repeat expansions in Saccharomyces cerevisiae. Mol. Cell. Biol. 27102-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davies, S. W., M. Turmaine, B. A. Cozens, M. DiFiglia, A. H. Sharp, C. A. Ross, E. Scherzinger, E. E. Wanker, L. Mangiarini, and G. P. Bates. 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90537-548. [DOI] [PubMed] [Google Scholar]
- 91.Davis, B. M., M. E. McCurrach, K. L. Taneja, R. H. Singer, and D. E. Housman. 1997. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. USA 947388-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reference deleted.
- 93.Debacker, K., and R. F. Kooy. 2007. Fragile sites and human disease. Hum. Mol. Genet. 16R150-R158. [DOI] [PubMed] [Google Scholar]
- 94.Debrauwère, H., J. Buard, J. Tessier, D. Aubert, G. Vergnaud, and A. Nicolas. 1999. Meiotic instability of human minisatellite CEB1 in yeast requires double-strand breaks. Nat. Genet. 23367-371. [DOI] [PubMed] [Google Scholar]
- 95.Debrauwère, H., C. G. Gendrel, S. Lechat, and M. Dutreix. 1997. Differences and similarities between various tandem repeat sequences: minisatellites and microsatellites. Biochimie 79577-586. [DOI] [PubMed] [Google Scholar]
- 96.Defossez, P. A., R. Prusty, M. Kaeberlein, S. J. Lin, P. Ferrigno, P. A. Silver, R. L. Keil, and L. Guarente. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3447-455. [DOI] [PubMed] [Google Scholar]
- 97.Degtyareva, N. P., P. Greenwell, E. R. Hofmann, M. O. Hengartner, L. Zhang, J. G. Culotti, and T. D. Petes. 2002. Caenorhabditis elegans DNA mismatch repair gene msh-2 is required for microsatellite stability and maintenance of genome integrity. Proc. Natl. Acad. Sci. USA 992158-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dehal, P., and J. L. Boore. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 31700-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deininger, P., and M. A. Batzer. 2002. Mammalian retroelements. Genome Res. 121455-1465. [DOI] [PubMed] [Google Scholar]
- 100.Delatycki, M. B., D. Paris, R. J. M. Gardner, K. Forshaw, G. A. Nicholson, N. Nassif, R. Williamson, and S. M. Forrest. 1998. Sperm DNA analysis in a Friedreich ataxia premutation carrier suggests both meiotic and mitotic expansion in the FRDA gene. J. Med. Genet. 35713-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Delgrange, O., and E. Rivals. 2004. STAR: an algorithm to search for tandem approximate repeats. Bioinformatics 202812-2820. [DOI] [PubMed] [Google Scholar]
- 102.DeLotto, R., and P. Schedl. 1984. A Drosophila melanogaster transfer RNA gene cluster at the cytogenetic locus 90BC. J. Mol. Biol. 179587-605. [DOI] [PubMed] [Google Scholar]
- 103.Demuth, J. P., T. De Bie, J. E. Stajich, N. Cristianini, and M. W. Hahn. 2006. The evolution of mammalian gene families. PLoS ONE 1e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Denoeud, F., G. Vergnaud, and G. Benson. 2003. Predicting human minisatellite polymorphism. Genome Res. 13856-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Derr, L. K., J. N. Strathern, and D. J. Garfinkel. 1991. RNA-mediated recombination in S. cerevisiae. Cell 67355-364. [DOI] [PubMed] [Google Scholar]
- 106.Deshpande, A. M., and C. S. Newlon. 1996. DNA replication fork pause sites dependent on transcription. Science 2721030-1033. [DOI] [PubMed] [Google Scholar]
- 106a.de Wind, N., M. Dekker, N. Claij, L. Jansen, Y. van Klink, M. Radman, G. Riggins, M. van der Valk, K. van't Wouk, and H. te Riele. 1999. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat. Genet. 23359-362. [DOI] [PubMed] [Google Scholar]
- 107.Dib, C., S. Faure, C. Fizames, D. Samson, N. Drouot, A. Vignal, P. Millasseau, S. Marc, J. Hazan, E. Seboun, M. Lathrop, G. Gyapay, J. Morissette, and J. Weissenbach. 1996. A comprehensive genetic map of the human genome based on 5,264 sequences. Nature 380152-154. [DOI] [PubMed] [Google Scholar]
- 108.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Philippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304304-307. [DOI] [PubMed] [Google Scholar]
- 109.Dixon, M. J., S. Bhattacharyya, and R. S. Lahue. 2004. Genetic assays for triplet repeat instability in yeast. Methods Mol. Biol. 27729-45. [DOI] [PubMed] [Google Scholar]
- 110.Dixon, M. J., and R. S. Lahue. 2004. DNA elements important for CAG*CTG repeat thresholds in Saccharomyces cerevisiae. Nucleic Acids Res. 321289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dixon, M. J., and R. S. Lahue. 2002. Examining the potential role of DNA polymerases η and ζ in triplet repeat instability in yeast. DNA Repair 1763-770. [DOI] [PubMed] [Google Scholar]
- 112.Dombrowski, C., S. Levesque, M. L. Morel, P. Rouillard, K. Morgan, and F. Rousseau. 2002. Premutation and intermediate-size FMR1 alleles in 10572 males from the general population: loss of an AGG interruption is a late event in the generation of fragile X syndrome alleles. Hum. Mol. Genet. 11371-378. [DOI] [PubMed] [Google Scholar]
- 113.Dubrova, Y. E., A. J. Jeffreys, and A. M. Malashenko. 1993. Mouse minisatellite mutations induced by ionizing radiation. Nat. Genet. 592-94. [DOI] [PubMed] [Google Scholar]
- 114.Dubrova, Y. E., V. N. Nesterov, N. G. Krouchinsky, V. A. Ostapenko, R. Neumann, D. L. Neil, and A. J. Jeffreys. 1996. Human minisatellite mutation rate after the Chernobyl accident. Nature 380683-686. [DOI] [PubMed] [Google Scholar]
- 115.Dujon, B. 2006. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 22375-387. [DOI] [PubMed] [Google Scholar]
- 116.Dujon, B., D. Alexandraki, B. André, W. Ansorge, V. Baladron, and J. P. G. Ballesta. 1994. Complete DNA sequence of yeast chromosome XI. Nature 369371-378. [DOI] [PubMed] [Google Scholar]
- 117.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 43035-44. [DOI] [PubMed] [Google Scholar]
- 118.Dupaigne, P., C. Le Breton, F. Fabre, S. Gangloff, E. Le Cam, and X. Veaute. 2008. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol. Cell 29243-254. [DOI] [PubMed] [Google Scholar]
- 119.Duret, L., G. Marais, and C. Biémont. 2000. Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics 1561661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Durkin, S. G., R. L. Ragland, M. F. Arlt, J. G. Mulle, S. T. Warren, and T. W. Glover. 2008. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc. Natl. Acad. Sci. USA 105246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ecker, M., V. Mrsa, I. Hagen, R. Deutzmann, S. Strahl, and W. Tanner. 2003. O-mannosylation precedes and potentially controls the N-glycosylation of a yeast cell wall glycoprotein. EMBO Rep. 4628-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Entezam, A., and K. Usdin. 2008. ATR protects the genome against CGG.CCG-repeat expansion in fragile X premutation mice. Nucleic Acids Res. 361050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eykelenboom, J., J. K. Blackwood, E. Okely, and D. R. F. Leach. 2008. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol. Cell 29644-651. [DOI] [PubMed] [Google Scholar]
- 124.Fabre, E., B. Dujon, and G.-F. Richard. 2002. Transcription and nuclear transport of CAG/CTG trinucleotide repeats in yeast. Nucleic Acids Res. 303540-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fabre, F., A. Chan, W.-D. Heyer, and S. Gangloff. 2002. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 9916887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Farah, J. A., E. Hartsuiker, K. Mizuno, K. Ohta, and G. R. Smith. 2002. A 160-bp palindrome is a Rad50. Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics 161461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fardaei, M., K. Larkin, J. D. Brook, and M. G. Hamshere. 2001. In vivo co-localization of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 292766-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fardaei, M., M. T. Rogers, H. M. Thorpe, K. Larkin, M. G. Hamshere, P. S. Harper, and J. D. Brook. 2002. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 11805-814. [DOI] [PubMed] [Google Scholar]
- 129.Faux, N. G., G. A. Huttley, K. Mahmood, G. I. Webb, M. G. de la Banda, and J. C. Whisstock. 2007. RCPdb: an evolutionary classification and codon usage database for repeat-containing proteins. Genome Res. 171118-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Feng, Y., F. Zhang, L. K. Lokey, J. L. Chastain, L. Lakkis, D. Eberhart, and S. T. Warren. 1995. Translational suppression by trinucleotide repeat expansion at FMR1. Science 268731-734. [DOI] [PubMed] [Google Scholar]
- 131.Feschotte, C., and E. J. Pritham. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41331-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fidalgo, M., R. R. Barrales, J. I. Ibeas, and J. Jimenez. 2006. Adaptive evolution by mutations in the FLO11 gene. Proc. Natl. Acad. Sci. USA 10311228-11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Field, D., and C. Wills. 1998. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc. Natl. Acad. Sci. USA 951647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fierro-Fernandez, M., P. Hernandez, D. B. Krimer, and J. B. Schvartzman. 2007. Replication fork reversal occurs spontaneously after digestion but is constrained in supercoiled domains. J. Biol. Chem. 28218190-18196. [DOI] [PubMed] [Google Scholar]
- 135.Fierro-Fernandez, M., P. Hernandez, D. B. Krimer, and J. B. Schvartzman. 2007. Topological locking restrains replication fork reversal. Proc. Natl. Acad. Sci. USA 1041500-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Filippova, G. N., C. P. Thienes, B. H. Penn, D. H. Cho, Y. J. Hu, J. M. Moore, T. R. Klesert, V. V. Lobanenkov, and S. J. Tapscott. 2001. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 28335-343. [DOI] [PubMed] [Google Scholar]
- 137.Finnis, M., S. Dayan, L. Hobson, G. Chenevix-Trench, K. Friend, K. Ried, D. Venter, E. Woollatt, E. Baker, and R. I. Richards. 2005. Common chromosomal fragile site FRA16D mutation in cancer cells. Hum. Mol. Genet. 141341-1349. [DOI] [PubMed] [Google Scholar]
- 138.Fischer, C., L. Bouneau, J.-P. Coutanceau, J. Weissenbach, J.-N. Volff, and C. Ozouf-Costaz. 2004. Global heterochromatic colocalization of transposable elements with minisatellites in the compact genome of the pufferfish Tetraodon nigroviridis. Gene 336175-183. [DOI] [PubMed] [Google Scholar]
- 139.Fischer, G., S. A. James, I. N. Roberts, S. G. Oliver, and E. J. Louis. 2000. Chromosomal evolution in Saccharomyces. Nature 405451-454. [DOI] [PubMed] [Google Scholar]
- 140.Fischer, G., C. Neuveglise, P. Durrens, C. Gaillardin, and B. Dujon. 2001. Evolution of gene order in the genomes of two related yeast species. Genome Res. 112009-2019. [DOI] [PubMed] [Google Scholar]
- 141.Fischer, G., E. P. Rocha, F. Brunet, M. Vergassola, and B. Dujon. 2006. Highly variable rates of genome rearrangements between hemiascomycetous yeast lineages. PLoS Genet. 2e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fishel, R., M. K. Lescoe, M. R. S. Rao, N. G. Copeland, N. A. Jenkins, J. Garber, M. Kane, and R. Kolodner. 1993. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 751027-1038. [DOI] [PubMed] [Google Scholar]
- 143.Flores, M. J., V. Bidnenko, and B. Michel. 2004. The DNA repair helicase UvrD is essential for replication fork reversal in replication mutants. EMBO Rep. 5983-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fojtik, P., and M. Vorlickova. 2001. The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH. Nucleic Acids Res. 294684-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fondon, J. W., and H. R. Garner. 2004. Molecular origins of rapid and continuous morphological evolution. Proc. Natl. Acad. Sci. USA 10118058-18063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Foster, E. A., M. A. Jobling, P. G. Taylor, P. Donnelly, P. de Knijff, R. Mieremet, T. Zerjal, and C. Tyler-Smith. 1998. Jefferson fathered slave's last child. Nature 39627-28. [DOI] [PubMed] [Google Scholar]
- 147.Fouché, N., S. Özgür, D. Roy, and J. D. Griffith. 2006. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 346044-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Freudenreich, C. H., S. M. Kantrow, and V. A. Zakian. 1998. Expansion and length-dependent fragility of CTG repeats in yeast. Science 279853-856. [DOI] [PubMed] [Google Scholar]
- 149.Freudenreich, C. H., and M. Lahiri. 2004. Structure-forming CAG/CTG repeat sequences are sensitive to breakage in the absence of Mrc1 checkpoint function and S-phase checkpoint signaling: implications for trinucleotide repeat expansion diseases. Cell Cycle 31370-1374. [DOI] [PubMed] [Google Scholar]
- 150.Freudenreich, C. H., J. B. Stavenhagen, and V. A. Zakian. 1997. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol. 172090-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fry, M., and L. A. Loeb. 1994. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. USA 914950-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fu, Y.-H., D. P. A. Kuhl, A. Pizzuti, M. Pieretti, J. S. Sutcliffe, S. Richards, A. J. M. H. Verkerk, J. J. A. Holden, R. G. Fenwick, S. T. Warren, B. A. Oostra, D. L. Nelson, and C. T. Caskey. 1991. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 671047-1058. [DOI] [PubMed] [Google Scholar]
- 153.Fu, Y. H., A. Pizzuti, R. G. Fenwick, Jr., J. King, S. Rajnarayan, P. W. Dunne, J. Dubel, G. A. Nasser, T. Ashizawa, P. de Jong, et al. 1992. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 2551256-1258. [DOI] [PubMed] [Google Scholar]
- 154.Gacy, A. M., G. Goellner, N. Juranic, S. Macura, and C. T. McMurray. 1995. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81533-540. [DOI] [PubMed] [Google Scholar]
- 155.Galagan, J. E., and E. U. Selker. 2004. RIP: the evolutionary cost of genome defense. Trends Genet. 20417-423. [DOI] [PubMed] [Google Scholar]
- 156.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 148391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25192-194. [DOI] [PubMed] [Google Scholar]
- 158.Ganley, A. R., and T. Kobayashi. 2007. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 17184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gaspar, C., M. Jannatipour, P. Dion, J. Laganière, J. Sequeiros, B. Brais, and G. A. Rouleau. 2000. CAG tract of MJD-1 may be prone to frameshifts causing polyalanine accumulation. Hum. Mol. Genet. 91957-1966. [DOI] [PubMed] [Google Scholar]
- 160.Gatchel, J. R., and H. Y. Zoghbi. 2005. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 6743-755. [DOI] [PubMed] [Google Scholar]
- 161.Gaut, B. S., and J. F. Doebley. 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 946809-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gendrel, C.-G., A. Boulet, and M. Dutreix. 2000. (CA/GT)n microsatellites affect homologous recombination during yeast meiosis. Genes Dev. 141261-1268. [PMC free article] [PubMed] [Google Scholar]
- 163.Gill, P., A. J. Jeffreys, and D. J. Werrett. 1985. Forensic application of DNA ‘fingerprints.’ Nature 318577-579. [DOI] [PubMed] [Google Scholar]
- 164.Glover, T. W., M. F. Arlt, A. M. Casper, and S. G. Durkin. 2005. Mechanisms of common fragile site instability. Hum. Mol. Genet. 142R197-R205. [DOI] [PubMed] [Google Scholar]
- 165.Godde, J. S., and A. P. Wolffe. 1996. Nucleosome assembly on CTG triplet repeats. J. Biol. Chem. 27115222-15229. [DOI] [PubMed] [Google Scholar]
- 166.Goellner, G. M., D. Tester, S. Thibodeau, E. Almqvist, Y. P. Goldberg, M. R. Hayden, and C. T. McMurray. 1997. Different mechanisms underlie DNA instability in Huntington disease and colorectal cancer. Am. J. Hum. Genet. 60879-890. [PMC free article] [PubMed] [Google Scholar]
- 167.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274546-567. [DOI] [PubMed] [Google Scholar]
- 168.Gomes-Pereira, M., L. Foiry, A. Nicole, A. Huguet, C. Junien, A. Munnich, and G. Gourdon. 2007. CTG trinucleotide repeat “big jumps”: large expansions, small mice. PLoS Genet. 3e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gomes-Pereira, M., M. T. Fortune, L. Ingram, J. P. McAbney, and D. G. Monckton. 2004. Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 131815-1825. [DOI] [PubMed] [Google Scholar]
- 170.Gorgoulis, V. G., L. V. Vassiliou, P. Karakaidos, P. Zacharatos, A. Kotsinas, T. Liloglou, M. Venere, R. A. Ditullio, Jr., N. G. Kastrinakis, B. Levy, D. Kletsas, A. Yoneta, M. Herlyn, C. Kittas, and T. D. Halazonetis. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434907-913. [DOI] [PubMed] [Google Scholar]
- 171.Gourdon, G., F. Radvanyi, A.-S. Lia, C. Duros, M. Blanche, M. Abitbol, C. Junien, and H. Hofmann-Radvanyi. 1997. Moderate intergenerational and somatic instability of a 55-CTG repeat in transgenic mice. Nat. Genet. 15190-192. [DOI] [PubMed] [Google Scholar]
- 172.Grabczyk, E., and K. Usdin. 2000. Alleviating transcript insufficiency caused by Friedreich's ataxia triplet repeats. Nucleic Acids Res. 284930-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Graham, J., J. Curran, and B. S. Weir. 2000. Conditional genotypic probabilities for microsatellite loci. Genetics 1551973-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gray, S. J., J. Gerhardt, W. Doerfler, L. E. Small, and E. Fanning. 2007. An origin of DNA replication in the promoter region of the human fragile X mental retardation (FMR1) gene. Mol. Cell. Biol. 27426-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Haber, J. E. 1999. DNA recombination: the replication connection. Trends Biochem. Sci. 24271-275. [DOI] [PubMed] [Google Scholar]
- 176.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95583-586. [DOI] [PubMed] [Google Scholar]
- 177.Haber, J. E., and E. J. Louis. 1998. Minisatellite origins in yeast and humans. Genomics 48132-135. [DOI] [PubMed] [Google Scholar]
- 178.Hagelberg, E., I. C. Gray, and A. J. Jeffreys. 1991. Identification of the skeletal remains of a murder victim by DNA analysis. Nature 352427-429. [DOI] [PubMed] [Google Scholar]
- 179.Hahn, M. W., T. De Bie, J. E. Stajich, C. Nguyen, and N. Cristianini. 2005. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 151153-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hammock, E. A. D., and L. J. Young. 2005. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science 3081630-1634. [DOI] [PubMed] [Google Scholar]
- 181.Han, J., C. Hsu, Z. Zhu, J. W. Longshore, and W. H. Finley. 1994. Over-representation of the disease associated (CAG) and (CGG) repeats in the human genome. Nucleic Acids Res. 221735-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Han, J. S., S. T. Szak, and J. D. Boeke. 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429268-274. [DOI] [PubMed] [Google Scholar]
- 183.Han, K., M. K. Konkel, J. Xing, H. Wang, J. Lee, T. J. Meyer, C. T. Huang, E. Sandifer, K. Hebert, E. W. Barnes, R. Hubley, W. Miller, A. F. Smit, B. Ullmer, and M. A. Batzer. 2007. Mobile DNA in Old World monkeys: a glimpse through the rhesus macaque genome. Science 316238-240. [DOI] [PubMed] [Google Scholar]
- 184.Hani, J., and H. Feldmann. 1998. tRNA genes and retroelements in the yeast genome. Nucleic Acids Res. 26689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Harfe, B. D., and S. Jinks-Robertson. 2000. Sequence composition and context effects on the generation and repair of frameshift intermediates in mononucleotide runs in Saccharomyces cerevisiae. Genetics 156571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Harley, H. G., J. D. Brook, S. A. Rundle, S. Crow, W. Reardon, A. J. Buckler, P. S. Harper, D. E. Housman, and D. J. Shaw. 1992. Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature 355545-546. [DOI] [PubMed] [Google Scholar]
- 187.Harper, J. W., and S. J. Elledge. 2007. The DNA damage response: ten years after. Mol. Cell 28739-745. [DOI] [PubMed] [Google Scholar]
- 188.Harrington, J. J., and M. R. Lieber. 1994. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 131235-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Harrison, P. M., N. Echols, and M. B. Gerstein. 2001. Digging for dead genes: an analysis of the characteristics of the pseudogene population in the Caenorhabditis elegans genome. Nucleic Acids Res. 29818-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hawk, J. D., L. Stefanovic, J. C. Boyer, T. D. Petes, and R. A. Farber. 2005. Variation in efficiency of DNA mismatch repair at different sites in the yeast genome. Proc. Natl. Acad. Sci. USA 1028639-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.He, Q., H. Cederberg, J. A. Armour, C. A. May, and U. Rannug. 1999. Cis-regulation of inter-allelic exchanges in mutation at human minisatellite MS205 in yeast. Gene 232143-153. [DOI] [PubMed] [Google Scholar]
- 192.He, Q., H. Cederberg, and U. Rannug. 2002. The influence of sequence divergence between alleles of the human MS205 minisatellite incorporated into the yeast genome on length-mutation rates and lethal recombination events during meiosis. J. Mol. Biol. 319315-327. [DOI] [PubMed] [Google Scholar]
- 193.Heidenfelder, B. L., A. M. Makhov, and M. D. Topal. 2003. Hairpin formation in Friedreich's ataxia triplet repeat expansion. J. Biol. Chem. 2782425-2431. [DOI] [PubMed] [Google Scholar]
- 194.Helminen, P., C. Ehnholm, M. L. Lokki, A. Jeffreys, and L. Peltonen. 1988. Application of DNA “fingerprints” to paternity determinations. Lancet i574-576. [DOI] [PubMed] [Google Scholar]
- 195.Henderson, S. T., and T. D. Petes. 1992. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 122749-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hennequin, C., A. Thierry, G.-F. Richard, G. Lecointre, H. V. Nguyen, C. Gaillardin, and B. Dujon. 2001. Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J. Clin. Microbiol. 39551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Henricksen, L. A., S. Tom, Y. Liu, and R. A. Bambara. 2000. Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J. Biol. Chem. 27516420-16427. [DOI] [PubMed] [Google Scholar]
- 198.Hewett, D. R., O. Handt, L. Hobson, M. Mangelsdorf, H. J. Eyre, E. Baker, G. R. Sutherland, S. Schuffenhauer, J. Mao, and R. I. Richards. 1998. FRA10B structure reveals common elements in repeat expansion and chromosomal fragile site genesis. Mol. Cell 1773-781. [DOI] [PubMed] [Google Scholar]
- 199.Hirst, M. C., P. K. Grewal, and K. E. Davies. 1994. Precursor arrays for triplet repeat expansion at the fragile X locus. Hum. Mol. Genet. 31553-1560. [DOI] [PubMed] [Google Scholar]
- 200.Holmes, A. M., A. Kaykov, and B. Arcangioli. 2005. Molecular and cellular dissection of mating-type switching steps in Schizosaccharomyces pombe. Mol. Cell. Biol. 25303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hopkins, B., N. J. Williams, M. B. Webb, P. G. Debenham, and A. J. Jeffreys. 1994. The use of minisatellite variant repeat-polymerase chain reaction (MVR-PCR) to determine the source of saliva on a used postage stamp. J. Forensic Sci. 39526-531. [PubMed] [Google Scholar]
- 202.Hosfield, D. J., C. D. Mol, B. Shen, and J. A. Tainer. 1998. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell 95135-146. [DOI] [PubMed] [Google Scholar]
- 203.Huntley, M. A., and A. G. Clark. 2007. Evolutionary analysis of amino acid repeats across the genomes of 12 Drosophila species. Mol. Biol. Evol. 242598-2609. [DOI] [PubMed] [Google Scholar]
- 204.Imai, H., H. Nakagama, K. Komatsu, T. Shiraishi, H. Fukuda, T. Sugimura, and M. Nagao. 1997. Minisatellite instability in severe combined immunodeficiency mouse cells. Proc. Natl. Acad. Sci. USA 9410817-10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Inoue, H., H. Ishii, H. Alder, E. Snyder, T. Druck, K. Huebner, and C. M. Croce. 1997. Sequence of the FRA3B common fragile region: implications for the mechanism of FHIT deletion. Proc. Natl. Acad. Sci. USA 9414584-14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Reference deleted.
- 207.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ireland, M. J., S. S. Reinke, and D. M. Livingston. 2000. The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics 1551657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jacquier, A., P. Legrain, and B. Dujon. 1992. Sequence of 10.7kb segment of yeast chromosome XI identifies the APN1 and the BAF1 loci and reveals one tRNA gene and several new open reading frames including homologues to RAD2 and kinases. Yeast 8121-132. [DOI] [PubMed] [Google Scholar]
- 210.Jaillon, O., J. M. Aury, F. Brunet, J. L. Petit, N. Stange-Thomann, E. Mauceli, L. Bouneau, C. Fischer, C. Ozouf-Costaz, A. Bernot, S. Nicaud, D. Jaffe, S. Fisher, G. Lutfalla, C. Dossat, B. Segurens, C. Dasilva, M. Salanoubat, M. Levy, N. Boudet, S. Castellano, V. Anthouard, C. Jubin, V. Castelli, M. Katinka, B. Vacherie, C. Biemont, Z. Skalli, L. Cattolico, J. Poulain, V. De Berardinis, C. Cruaud, S. Duprat, P. Brottier, J. P. Coutanceau, J. Gouzy, G. Parra, G. Lardier, C. Chapple, K. J. McKernan, P. McEwan, S. Bosak, M. Kellis, J. N. Volff, R. Guigo, M. C. Zody, J. Mesirov, K. Lindblad-Toh, B. Birren, C. Nusbaum, D. Kahn, M. Robinson-Rechavi, V. Laudet, V. Schachter, F. Quetier, W. Saurin, C. Scarpelli, P. Wincker, E. S. Lander, J. Weissenbach, and H. Roest Crollius. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431946-957. [DOI] [PubMed] [Google Scholar]
- 211.Jankowski, C., and D. K. Nag. 2002. Most meiotic CAG repeat tract-length alterations in yeast are SPO11 dependent. Mol. Genet. Genomics 26764-70. [DOI] [PubMed] [Google Scholar]
- 212.Jankowski, C., F. Nasar, and D. K. Nag. 2000. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl. Acad. Sci. USA 972134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jarne, P., and J. L. Lagoda. 1996. Microsatellites, from molecules to populations and back. Trends Ecol. Evol. 11424-429. [DOI] [PubMed] [Google Scholar]
- 214.Jauert, P. A., S. N. Edmiston, K. Conway, and D. T. Kirkpatrick. 2002. RAD1 controls the meiotic expansion of the human HRAS1 minisatellite in Saccharomyces cerevisiae. Mol. Cell. Biol. 22953-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jeffreys, A. J., M. J. Allen, E. Hagelberg, and A. Sonnberg. 1992. Identification of the skeletal remains of Josef Mengele by DNA analysis. Forensic Sci. Int. 5665-76. [DOI] [PubMed] [Google Scholar]
- 216.Jeffreys, A. J., J. Murray, and R. Neumann. 1998. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hot spot. Mol. Cell 2267-273. [DOI] [PubMed] [Google Scholar]
- 217.Jeffreys, A. J., and R. Neumann. 1997. Somatic mutation processes at a human minisatellite. Hum. Mol. Genet. 6129-136. [DOI] [PubMed] [Google Scholar]
- 218.Jeffreys, A. J., R. Neumann, and V. Wilson. 1990. Repeat unit sequence variation in minisatellites: a novel source of DNA polymorphism for studying variation and mutation by single molecule analysis. Cell 60473-485. [DOI] [PubMed] [Google Scholar]
- 219.Jeffreys, A. J., N. J. Royle, V. Wilson, and Z. Wong. 1988. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature 332278-281. [DOI] [PubMed] [Google Scholar]
- 220.Jeffreys, A. J., K. Tamaki, A. McLeod, D. G. Monckton, D. L. Neil, and J. A. L. Armour. 1994. Complex gene conversion events in germline mutation at human minisatellites. Nat. Genet. 6136-145. [DOI] [PubMed] [Google Scholar]
- 221.Jeffreys, A. J., V. Wilson, and S. L. Thein. 1985. Hypervariable ‘minisatellite’ regions in human DNA. Nature 31467-73. [DOI] [PubMed] [Google Scholar]
- 222.Jiang, Z., H. Tang, M. Ventura, M. F. Cardone, T. Marques-Bonet, X. She, P. A. Pevzner, and E. E. Eichler. 2007. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 391361-1368. [DOI] [PubMed] [Google Scholar]
- 223.Jin, P., and S. T. Warren. 2000. Understanding the molecular basis of fragile X syndrome. Hum. Mol. Genet. 9901-908. [DOI] [PubMed] [Google Scholar]
- 224.Johnson, R. E., S. T. Henderson, T. D. Petes, S. Prakash, M. Bankmann, and L. Prakash. 1992. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 123807-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jones, C., R. Müllenbach, P. Grossfeld, R. Auer, R. Favier, K. Chien, M. James, A. Tunnacliffe, and F. Cotter. 2000. Co-localisation of CCG repeats and chromosome deletion breakpoints in Jacobsen syndrome: evidence for a common mechanism of chromosome breakage. Hum. Mol. Genet. 91201-1208. [DOI] [PubMed] [Google Scholar]
- 226.Jones, C., L. Penny, T. Mattina, S. Yu, E. Baker, L. Voullaire, W. Y. Langdon, G. R. Sutherland, R. I. Richards, and A. Tunnacliffe. 1995. Association of a chromosome deletion syndrome with a fragile site within the proto-oncogene CBL2. Nature 376145-149. [DOI] [PubMed] [Google Scholar]
- 227.Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. USA 941872-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kalendar, R., J. Tanskanen, W. Chang, K. Antonius, H. Sela, O. Peleg, and A. H. Schulman. 2008. Cassandra retrotransposons carry independently transcribed 5S RNA. Proc. Natl. Acad. Sci. USA 1055833-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kamath-Loeb, A. S., L. A. Loeb, E. Johansson, P. M. Burgers, and M. Fry. 2001. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 27616439-16446. [DOI] [PubMed] [Google Scholar]
- 230.Kaminker, J. S., C. M. Bergman, B. Kronmiller, J. Carlson, R. Svirskas, S. Patel, E. Frise, D. A. Wheeler, S. E. Lewis, G. M. Rubin, M. Ashburner, and S. E. Celniker. 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3RESEARCH0084. [DOI] [PMC free article] [PubMed]
- 231.Kang, S., A. Jaworski, K. Ohshima, and R. D. Wells. 1995. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 10213-217. [DOI] [PubMed] [Google Scholar]
- 232.Kao, H. I., J. Veeraraghavan, P. Polaczek, J. L. Campbell, and R. A. Bambara. 2004. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 27915014-15024. [DOI] [PubMed] [Google Scholar]
- 233.Kapitonov, V., and J. Jurka. 1996. The age of Alu subfamilies. J. Mol. Evol. 4259-65. [DOI] [PubMed] [Google Scholar]
- 234.Kapitonov, V. V., and J. Jurka. 2003. A novel class of SINE elements derived from 5S rRNA. Mol. Biol. Evol. 20694-702. [DOI] [PubMed] [Google Scholar]
- 235.Karaoglu, H., C. M. Lee, and W. Meyer. 2005. Survey of simple sequence repeats in completed fungal genomes. Mol. Biol. Evol. 22639-649. [DOI] [PubMed] [Google Scholar]
- 236.Karlin, S., and C. Burge. 1996. Trinucleotide repeats and long homopeptides in genes and proteins associated with nervous system disease and development. Proc. Natl. Acad. Sci. USA 931560-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kaykov, A., A. M. Holmes, and B. Arcangioli. 2004. Formation, maintenance and consequences of the imprint at the mating-type locus in fission yeast. EMBO J. 23930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237a.Kaykov, A., and B. Arcangioli. 2004. A programmed strand-specific and modified nick in S. pombe constitutes a novel type of chromosomal imprint. Curr. Biol. 141924-1928. [DOI] [PubMed] [Google Scholar]
- 238.Kazantsev, A., E. Preisinger, A. Dranovsky, D. Goldgaber, and D. Housman. 1999. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc. Natl. Acad. Sci. USA 9611404-11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kazemi-Esfarjani, P., and S. Benzer. 2000. Genetic suppression of polyglutamine toxicity in Drosophila. Science 2871837-1840. [DOI] [PubMed] [Google Scholar]
- 240.Kearns, M., C. Morris, and E. Whitelaw. 2001. Spontaneous germline amplification and translocation of a transgene array. Mutat. Res. 486125-136. [DOI] [PubMed] [Google Scholar]
- 241.Keeney, S., C. N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88375-384. [DOI] [PubMed] [Google Scholar]
- 242.Kellis, M., B. W. Birren, and E. S. Lander. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428617-624. [DOI] [PubMed] [Google Scholar]
- 243.Kelly, M. K., P. A. Jauert, L. E. Jensen, C. L. Chan, C. S. Truong, and D. T. Kirkpatrick. 2007. Zinc regulates the stability of repetitive minisatellite DNA tracts during stationary phase. Genetics 1772469-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kennedy, B. K., M. Gotta, D. A. Sinclair, K. Mills, D. S. McNabb, M. Murthy, S. M. Pak, T. Laroche, S. M. Gasser, and L. Guarente. 1997. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 89381-391. [DOI] [PubMed] [Google Scholar]
- 245.Kenneson, A., F. Zhang, C. H. Hagedorn, and S. T. Warren. 2001. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 101449-1454. [DOI] [PubMed] [Google Scholar]
- 246.Reference deleted.
- 247.Kim, J. M., S. Vanguri, J. D. Boeke, A. Gabriel, and D. F. Voytas. 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8464-478. [DOI] [PubMed] [Google Scholar]
- 248.Kirchner, J. M., H. Tran, and M. A. Resnick. 2000. A DNA polymerase ɛ mutant that specifically causes +1 frameshift mutations within homonucleotide runs in yeast. Genetics 1551623-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Klement, I. A., P. J. Skinner, M. D. Kaytor, H. Yi, S. M. Hersch, H. B. Clark, H. Y. Zoghbi, and H. T. Orr. 1998. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 9541-53. [DOI] [PubMed] [Google Scholar]
- 250.Klesert, T. R., A. D. Otten, T. D. Bird, and S. J. Tapscott. 1997. Trinucleotide repeat expansion at the myotonic dystrophy locus reduces expression of DMAHP. Nat. Genet. 16402-406. [DOI] [PubMed] [Google Scholar]
- 251.Knight, S. J. L., A. V. Flannery, M. C. Hirst, L. Campbell, Z. Christodoulou, S. R. Phelps, J. Pointon, H. R. Middleton-Price, A. Barnicoat, M. E. Pembrey, J. Holland, B. A. Oostra, M. Bobrow, and K. E. Davies. 1993. Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell 74127-134. [DOI] [PubMed] [Google Scholar]
- 252.Kokoska, R. J., L. Stefanovic, A. B. Buermeyer, R. M. Liskay, and T. D. Petes. 1999. A mutation of the yeast gene encoding PCNA destabilizes both microsatellite and minisatellite DNA sequences. Genetics 151511-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Kokoska, R. J., L. Stefanovic, H. T. Tran, M. A. Resnick, D. A. Gordenin, and T. D. Petes. 1998. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t). Mol. Cell. Biol. 182779-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kolodner, R. 1996. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 101433-1442. [DOI] [PubMed] [Google Scholar]
- 255.Kolpakov, R., G. Bana, and G. Kucherov. 2003. mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res. 313672-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Koszul, R., S. Caburet, B. Dujon, and G. Fischer. 2004. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 23234-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Koszul, R., B. Dujon, and G. Fischer. 2006. Stability of large segmental duplications in the yeast genome. Genetics 1722211-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Koszul, R., and G. Fischer. A prominent role for segmental duplications in modeling eukaryotic genomes. C. R. Biol., in press. [DOI] [PubMed]
- 259.Kovtun, I. V., and C. T. McMurray. 2001. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 27407-411. [DOI] [PubMed] [Google Scholar]
- 260.Krasilnikova, M. M., M. L. Kireeva, V. Petrovic, N. Knijnikova, M. Kashlev, and S. M. Mirkin. 2007. Effects of Friedreich's ataxia (GAA)n*(TTC)n repeats on RNA synthesis and stability. Nucleic Acids Res. 351075-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy, H. Klein, T. Ellenberger, and P. Sung. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423305-309. [DOI] [PubMed] [Google Scholar]
- 262.Krobitsch, S., and S. Lindquist. 2000. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA 971589-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Krogh, B. O., and L. S. Symington. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38233-271. [DOI] [PubMed] [Google Scholar]
- 264.Kruglyak, S., R. Durrett, M. D. Schug, and C. F. Aquadro. 2000. Distribution and abundance of microsatellites in the yeast genome can be explained by a balance between slippage events and point mutations. Mol. Biol. Evol. 171210-1219. [DOI] [PubMed] [Google Scholar]
- 265.Kuhn, R. M., L. Clarke, and J. Carbon. 1991. Clustered tRNA genes in Schizosaccharomyces pombe centromeric DNA sequence repeats. Proc. Natl. Acad. Sci. USA 881306-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kurahashi, H., and B. S. Emanuel. 2001. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 102605-2617. [DOI] [PubMed] [Google Scholar]
- 267.Kurahashi, H., and B. S. Emanuel. 2001. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat. Genet. 29139-140. [DOI] [PubMed] [Google Scholar]
- 268.Labib, K., and B. Hodgson. 2007. Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 8346-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lahiri, M., T. L. Gustafson, E. R. Majors, and C. H. Freudenreich. 2004. Expanded CAG repeats activate the DNA damage checkpoint pathway. Mol. Cell 15287-293. [DOI] [PubMed] [Google Scholar]
- 270.Lalioti, M. D., H. S. Scott, C. Buresi, C. Rossier, A. Bottani, M. A. Morris, A. Malafosse, and S. E. Antonarakis. 1997. Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature 386847-851. [DOI] [PubMed] [Google Scholar]
- 270a.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409860-921. [DOI] [PubMed] [Google Scholar]
- 271.Langkjaer, R. B., P. F. Cliften, M. Johnston, and J. Piskur. 2003. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature 421848-852. [DOI] [PubMed] [Google Scholar]
- 272.Latgé, J.-P., and R. Calderone. 2005. The fungal cell wall. In K. Esser and R. Fischer (ed.), The mycota XIII. Springer, Berlin, Germany.
- 273.Leclercq, S., E. Rivals, and P. Jarne. 2007. Detecting microsatellites within genomes: significant variation among algorithms. BMC Bioinformatics 8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Leeflang, E. P., L. Zhang, S. Tavare, R. Hubert, J. Srinidhi, M. E. MacDonald, R. H. Myers, M. de Young, N. S. Wexler, J. F. Gusella, et al. 1995. Single sperm analysis of the trinucleotide repeats in the Huntington's disease gene: quantification of the mutation frequency spectrum. Hum. Mol. Genet. 41519-1526. [DOI] [PubMed] [Google Scholar]
- 275.Legendre, M., N. Pochet, T. Pak, and K. J. Verstrepen. 2007. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 171787-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lemoine, F. J., N. P. Degtyareva, K. Lobachev, and T. D. Petes. 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120587-598. [DOI] [PubMed] [Google Scholar]
- 277.Lenzmeier, B. A., and C. H. Freudenreich. 2003. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet. Genome Res. 1007-24. [DOI] [PubMed] [Google Scholar]
- 278.Lestini, R., and B. Michel. 2007. UvrD controls the access of recombination proteins to blocked replication forks. EMBO J. 263804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Levdansky, E., J. Romano, Y. Shadkchan, H. Sharon, K. J. Verstrepen, G. R. Fink, and N. Osherov. 2007. Coding tandem repeats generate diversity in Aspergillus fumigatus genes. Eukaryot. Cell 61380-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Li, S. H., S. Lam, A. L. Cheng, and X. J. Li. 2000. Intranuclear huntingtin increases the expression of caspase-1 and induces apoptosis. Hum. Mol. Genet. 92859-2867. [DOI] [PubMed] [Google Scholar]
- 281.Li, Y. X., and M. L. Kirby. 2003. Coordinated and conserved expression of alphoid repeat and alphoid repeat-tagged coding sequences. Dev. Dyn. 22872-81. [DOI] [PubMed] [Google Scholar]
- 282.Libby, R. T., D. G. Monckton, Y. H. Fu, R. A. Martinez, J. P. McAbney, R. Lau, D. D. Einum, K. Nichol, C. B. Ware, L. J. Ptacek, C. E. Pearson, and A. R. La Spada. 2003. Genomic context drives SCA7 CAG repeat instability, while expressed SCA7 cDNAs are intergenerationally and somatically stable in transgenic mice. Hum. Mol. Genet. 1241-50. [DOI] [PubMed] [Google Scholar]
- 283.Liberi, G., G. Maffioletti, C. Lucca, I. Chiolo, A. Baryshnikova, C. Cotta-Ramusino, M. Lopes, A. Pellicioli, J. E. Haber, and M. Foiani. 2005. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 19339-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Linardopoulou, E. V., E. M. Williams, Y. Fan, C. Friedman, J. M. Young, and B. J. Trask. 2005. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 43794-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Liquori, C. L., K. Ricker, M. L. Moseley, J. F. Jacobsen, W. Kress, S. L. Naylor, J. W. Day, and L. P. W. Ranum. 2001. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 293864-867. [DOI] [PubMed] [Google Scholar]
- 286.Litt, M., and J. A. Luty. 1989. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet. 44397-401. [PMC free article] [PubMed] [Google Scholar]
- 287.Liu, B., N. C. Nicolaides, S. Markowitz, J. K. V. Willson, R. E. Parsons, J. Jen, N. Papadopolous, P. Peltomäki, A. de la Chapelle, S. R. Hamilton, K. W. Kinzler, and B. Vogelstein. 1995. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat. Genet. 948-55. [DOI] [PubMed] [Google Scholar]
- 288.Liu, Y., and R. A. Bambara. 2003. Analysis of human flap endonuclease 1 mutants reveals a mechanism to prevent triplet repeat expansion. J. Biol. Chem. 27813728-13739. [DOI] [PubMed] [Google Scholar]
- 289.Liu, Y., H. Zhang, J. Veeraraghavan, R. A. Bambara, and C. H. Freudenreich. 2004. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol. Cell. Biol. 244049-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Llorente, B., A. Malpertuy, C. Neuveglise, J. de Montigny, M. Aigle, F. Artiguenave, G. Blandin, M. Bolotin-Fukuhara, E. Bon, P. Brottier, S. Casaregola, P. Durrens, C. Gaillardin, A. Lepingle, O. Ozier-Kalogeropoulos, S. Potier, W. Saurin, F. Tekaia, C. Toffano-Nioche, M. Wesolowski-Louvel, P. Wincker, J. Weissenbach, J. Souciet, and B. Dujon. 2000. Genomic exploration of the hemiascomycetous yeasts. 18. Comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae. FEBS Lett. 487101-112. [DOI] [PubMed] [Google Scholar]
- 291.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108183-193. [DOI] [PubMed] [Google Scholar]
- 292.Lobachev, K. S., J. E. Stenger, O. G. Kozyreva, J. Jurka, D. A. Gordenin, and M. A. Resnick. 2000. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 193822-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Lohe, A. R., A. J. Hilliker, and P. A. Roberts. 1993. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 1341149-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Long, E. O., and I. B. Dawid. 1980. Repeated genes in eukaryotes. Ann. Rev. Biochem. 49727-764. [DOI] [PubMed] [Google Scholar]
- 295.Lopes, J., H. Debrauwère, J. Buard, and A. Nicolas. 2002. Instability of the human minisatellite CEB1 in rad27Δ and dna2-1 replication-deficient yeast cells. EMBO J. 213201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Lopes, J., C. Ribeyre, and A. Nicolas. 2006. Complex minisatellite rearrangements generated in the total or partial absence of Rad27/hFEN1 activity occur in a single generation and are Rad51 and Rad52 dependent. Mol. Cell. Biol. 266675-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412557-561. [DOI] [PubMed] [Google Scholar]
- 298.Louis, E. J., E. S. Naumova, A. Lee, G. Naumov, and J. E. Haber. 1994. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics 136789-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Lühr, B., J. Scheller, P. Meyer, and W. Kramer. 1998. Analysis of in vivo correction of defined mismatches in the DNA mismatch repair mutants msh2, msh3 and msh6 of Saccharomyces cerevisiae. Mol. Gen. Genet. 257362-367. [DOI] [PubMed] [Google Scholar]
- 300.Lunel, F. V., L. Licciardello, S. Stefani, H. A. Verbrugh, W. Melchers, J. G., J. F. G. M. Meis, S. Scherer, and A. Van Belkum. 1998. Lack of consistent short sequence repeat polymorphisms in genetically homologous colonizing and invasive Candida albicans strains. J. Bacteriol. 1803771-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Lynch, M., and J. S. Conery. 2000. The evolutionary fate and consequences of duplicate genes. Science 2901151-1155. [DOI] [PubMed] [Google Scholar]
- 302.Magee, P. T. 2007. Genome structure and dynamics in Candida albicans, p. 7-26. In C. d'Enfert and B. Hube (ed.), Candida comparative and functional genomics. Caister Academic Press, Norfolk, United Kingdom.
- 303.Maleki, S., H. Cederberg, and U. Rannug. 2002. The human minisatellites MS1, MS32, MS205 and CEB1 integrated into the yeast genome exhibit different degrees of mitotic instability but are all stabilised by RAD27. Curr. Genet. 41333-341. [DOI] [PubMed] [Google Scholar]
- 304.Malgoire, J. Y., S. Bertout, F. Renaud, J. M. Bastide, and M. Mallie. 2005. Typing of Saccharomyces cerevisiae clinical strains by using microsatellite sequence polymorphism. J. Clin. Microbiol. 431133-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Maloisel, L., and J. L. Rossignol. 1998. Suppression of crossing-over by DNA methylation in Ascobolus. Genes Dev. 121381-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Malpertuy, A., B. Dujon, and G.-F. Richard. 2003. Analysis of microsatellites in 13 hemiascomycetous yeast species: mechanisms involved in genome dynamics. J. Mol. Evol. 56730-741. [DOI] [PubMed] [Google Scholar]
- 307.Malter, H. E., J. C. Iber, R. Willemsen, E. de Graaff, J. C. Tarleton, J. Leisti, S. T. Warren, and B. A. Oostra. 1997. Characterization of the full fragile X syndrome mutation in fetal gametes. Nat. Genet. 15165-169. [DOI] [PubMed] [Google Scholar]
- 308.Mandel, J.-L. 1997. Breaking the rule of three. Nature 386767-769. [DOI] [PubMed] [Google Scholar]
- 309.Mankodi, A., M. P. Takahashi, H. Jiang, C. L. Beck, W. J. Bowers, R. T. Moxley, S. C. Cannon, and C. A. Thornton. 2002. Expanded CUG repeats trigger aberrant splicing of CIC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell 1035-44. [DOI] [PubMed] [Google Scholar]
- 310.Mankodi, A., C. R. Urbinati, Q.-P. Yuan, R. T. Moxley, V. Sansone, M. Krym, D. Henderson, M. Schalling, M. S. Swanson, and C. A. Thornton. 2001. Musclebind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 102165-2170. [DOI] [PubMed] [Google Scholar]
- 311.Manley, K., T. L. Shirley, L. Flaherty, and A. Messer. 1999. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 23471-473. [DOI] [PubMed] [Google Scholar]
- 312.Mansour, A. A., C. Tornier, E. Lehmann, M. Darmon, and O. Fleck. 2001. Control of GT repeat stability in Schizosaccharomyces pombe by mismatch repair factors. Genetics 15877-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312a.Mar Albà, M. M., M. F. Santibañez-Koref, and J. M. Hancock. 1999. Amino acid reiterations in yeast are overrepresented in particular classes of proteins and show evidence of a slippage-like mutational process. J. Mol. Evol. 49789-797. [DOI] [PubMed] [Google Scholar]
- 313.Marck, C., R. Kachouri-Lafond, I. Lafontaine, E. Westhof, B. Dujon, and H. Grosjean. 2006. The RNA polymerase III-dependent family of genes in hemiascomycetes: comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res. 341816-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Mariappan, S. V., P. Catasti, L. A. Silks III, E. M. Bradbury, and G. Gupta. 1999. The high-resolution structure of the triplex formed by the GAA/TTC triplet repeat associated with Friedreich's ataxia. J. Mol. Biol. 2852035-2052. [DOI] [PubMed] [Google Scholar]
- 315.Marin, I., P. Plata-Rengifo, M. Labrador, and A. Fontdevila. 1998. Evolutionary relationships among the members of an ancient class of non-LTR retrotransposons found in the nematode Caenorhabditis elegans. Mol. Biol. Evol. 151390-1402. [DOI] [PubMed] [Google Scholar]
- 316.Markowitz, S., J. Wang, L. Myeroff, R. Parsons, L. Sun, J. Lutterbaugh, R. S. Fan, E. Zborowska, K. W. Kinzler, B. Vogelstein, M. Brattain, and J. K. V. Willson. 1995. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science 2681336-1338. [DOI] [PubMed] [Google Scholar]
- 317.Martindale, D., A. Hackam, A. Wieczorek, L. Ellerby, C. Wellington, K. McCutcheon, R. Singaraja, P. Kazemi-Esfarjani, R. Devon, S. U. Kim, D. E. Bredesen, F. Tufaro, and M. R. Hayden. 1998. Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat. Genet. 18150-154. [DOI] [PubMed] [Google Scholar]
- 318.Marx, J. 2002. Debate surges over the origins of genomic defects in cancer. Science 297544-546. [DOI] [PubMed] [Google Scholar]
- 319.Matsugami, A., T. Okuizumi, S. Uesugi, and M. Katahira. 2003. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J. Biol. Chem. 27828147-28153. [DOI] [PubMed] [Google Scholar]
- 320.Matsuura, T., P. Fang, C. E. Pearson, P. Jayakar, T. Ashizawa, B. B. Roa, and D. L. Nelson. 2006. Interruptions in the expanded ATTCT repeat of spinocerebellar ataxia type 10: repeat purity as a disease modifier? Am. J. Hum. Genet. 78125-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Maurer, D. J., K. A. Benzow, L. J. Schut, L. P. Ranum, and D. M. Livingston. 1998. Comparison of expanded CAG repeat tracts in sperm and lymphocyte DNA from Machado Joseph disease and spinocerebellar ataxia type I patients. Hum. Mutat. Suppl. 1S74-S77. [DOI] [PubMed] [Google Scholar]
- 322.Maurer, D. J., B. L. O'Callaghan, and D. M. Livingston. 1998. Mapping the polarity of changes that occur in interrupted CAG repeat tracts in yeast. Mol. Cell. Biol. 184597-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Maurer, D. J., B. L. O'Callaghan, and D. M. Livingston. 1996. Orientation dependence of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 166617-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.McClintock, B. 1950. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 36344-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.McLysaght, A., K. Hokamp, and K. H. Wolfe. 2002. Extensive genomic duplication during early chordate evolution. Nat. Genet. 31200-204. [DOI] [PubMed] [Google Scholar]
- 327.McMurray, C. T. 1999. DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl. Acad. Sci. USA 961823-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Messier, W., S.-H. Li, and C.-B. Stewart. 1996. The birth of microsatellites. Nature 381483. [DOI] [PubMed] [Google Scholar]
- 329.Michelitsch, M. D., and J. S. Weissman. 2000. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA 9711910-11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Miller, J. W., C. R. Urbinati, P. Teng-umnuay, M. G. Stenberg, B. J. Byrne, C. A. Thornton, and M. S. Swanson. 2000. Recruitment of human musclebind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 194439-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Miret, J. J., L. Pessoa-Brandão, and R. S. Lahue. 1998. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 9512438-12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Miret, J. J., L. Pessoa-Brandao, and R. S. Lahue. 1997. Instability of CAG and CTG trinucleotide repeats in Saccharomyces cerevisiae. Mol. Cell. Biol. 173382-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Mirkin, E. V., and S. M. Mirkin. 2007. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 7113-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Mirkin, S. M. 2006. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biol. 16351-358. [DOI] [PubMed] [Google Scholar]
- 335.Mirkin, S. M. 2007. Expandable DNA repeats and human disease. Nature 447932-940. [DOI] [PubMed] [Google Scholar]
- 336.Mirkin, S. M. 2005. Toward a unified theory for repeat expansions. Nat. Struct. Mol. Biol. 12635-637. [DOI] [PubMed] [Google Scholar]
- 337.Mishmar, D., A. Rahat, S. W. Scherer, G. Nyakatura, B. Hinzmann, Y. Kohwi, Y. Mandel-Gutfroind, J. R. Lee, B. Drescher, D. E. Sas, H. Margalit, M. Platzer, A. Weiss, L. C. Tsui, A. Rosenthal, and B. Kerem. 1998. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl. Acad. Sci. USA 958141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Mitas, M. 1997. Trinucleotide repeats associated with human diseases. Nucleic Acids Res. 252245-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Mitas, M., A. Yu, J. Dill, and I. S. Haworth. 1995. The trinucleotide repeat sequence d(CGG)15 forms a heat-stable hairpin containing Gsyn.Ganti base pairs. Biochemistry 3412803-12811. [DOI] [PubMed] [Google Scholar]
- 340.Mitas, M., A. Yu, J. Dill, T. J. Kamp, E. J. Chambers, and I. S. Haworth. 1995. Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucleic Acids Res. 231050-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Modrich, P., and R. Lahue. 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996101-133. [DOI] [PubMed] [Google Scholar]
- 342.Monckton, D. G., M. I. Coolbaugh, K. T. Ashizawa, M. J. Siciliano, and C. T. Caskey. 1997. Hypermutable myotonic dystrophy CTG repeats in transgenic mice. Nat. Genet. 15193-196. [DOI] [PubMed] [Google Scholar]
- 343.Monckton, D. G., L. J. Wong, T. Ashizawa, and C. T. Caskey. 1995. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum. Mol. Genet. 41-8. [DOI] [PubMed] [Google Scholar]
- 344.Monod, J. 1972. Chance and necessity. William Collins Sons & Co. Ltd., Glasgow, United Kingdom.
- 345.Moore, H., P. W. Greenwell, C.-P. Liu, N. Arnheim, and T. D. Petes. 1999. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl. Acad. Sci. USA 961504-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Morgante, M., M. Hanafey, and W. Powell. 2002. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30194-200. [DOI] [PubMed] [Google Scholar]
- 347.Morishita, T., F. Furukawa, C. Sakaguchi, T. Toda, and A. M. Carr. 2005. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol. Cell. Biol. 258074-8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Moutou, C., M.-C. Vincent, V. Biancalana, and J.-L. Mandel. 1997. Transition from premutation to full mutation in fragile X syndrome is likely to be prezygotic. Hum. Mol. Genet. 6971-979. [DOI] [PubMed] [Google Scholar]
- 349.Moynahan, M. E., A. J. Pierce, and M. Jasin. 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7263-272. [DOI] [PubMed] [Google Scholar]
- 350.Muchowski, P. J., G. Schaffar, A. Sittler, E. E. Wanker, M. K. Hayer-Hartl, and F. U. Hartl. 2000. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc. Natl. Acad. Sci. USA 977841-7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Mundlos, S., F. Otto, C. Mundlos, J. B. Mulliken, A. S. Aylsworth, S. Albright, D. Lindhout, W. G. Cole, W. Henn, J. H. Knoll, M. J. Owen, R. Mertelsmann, B. U. Zabel, and B. R. Olsen. 1997. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89773-779. [DOI] [PubMed] [Google Scholar]
- 352.Murante, R. S., L. A. Henricksen, and R. A. Bambara. 1998. Junction ribonuclease: an activity in Okazaki fragment processing. Proc. Natl. Acad. Sci. USA 952244-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Murante, R. S., J. A. Rumbaugh, C. J. Barnes, J. R. Norton, and R. A. Bambara. 1996. Calf RTH-1 nuclease can remove the initiator RNAs of Okazaki fragments by endonuclease activity. J. Biol. Chem. 27125888-25897. [DOI] [PubMed] [Google Scholar]
- 354.Myung, K., A. Datta, and R. D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104397-408. [DOI] [PubMed] [Google Scholar]
- 355.Nadel, Y., P. Weisman-Shomer, and M. Fry. 1995. The fragile X syndrome single strand d(CGG)n nucleotide repeats readily fold back to form unimolecular hairpin structures. J. Biol. Chem. 4828970-28977. [DOI] [PubMed] [Google Scholar]
- 356.Nadir, E., H. Margalit, T. Gallily, and S. A. Ben-Sasson. 1996. Microsatellite spreading in the human genome: evolutionary mechanisms and structural implications. Proc. Natl. Acad. Sci. USA 936470-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Nag, D. K. 2003. Trinucleotide repeat expansions: timing is everything. Trends Mol. Med. 9455-457. [DOI] [PubMed] [Google Scholar]
- 358.Nag, D. K., and A. Kurst. 1997. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics 146835-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Nakamura, Y., M. Leppert, P. O'Connell, R. Wolff, T. Holm, M. Culver, C. Martin, E. Fujimoto, M. Hoff, E. Kumlin, et al. 1987. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science 2351616-1622. [DOI] [PubMed] [Google Scholar]
- 360.Nancarrow, J. K., K. Holman, M. Mangelsdorf, T. Hori, M. Denton, G. R. Sutherland, and R. I. Richards. 1995. Molecular basis of p(CGG)n repeat instability at the FRA16A fragile site locus. Hum. Mol. Genet. 4367-372. [DOI] [PubMed] [Google Scholar]
- 361.Narayanan, V., P. A. Mieczkowski, H. M. Kim, T. D. Petes, and K. S. Lobachev. 2006. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 1251283-1296. [DOI] [PubMed] [Google Scholar]
- 362.Nasar, F., C. Jankowski, and D. K. Nag. 2000. Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol. Cell. Biol. 203449-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Nassif, N., J. Penney, S. Pal, W. R. Engels, and G. B. Gloor. 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 141613-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Negroni, M., and H. Buc. 2001. Retroviral recombination: what drives the switch? Nat. Rev. Mol. Cell Biol. 2151-155. [DOI] [PubMed] [Google Scholar]
- 365.Nei, M., X. Gu, and T. Sitnikova. 1997. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 947799-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Neuvéglise, C., F. Chalvet, P. Wincker, C. Gaillardin, and S. Casaregola. 2005. Mutator-like element in the yeast Yarrowia lipolytica displays multiple alternative splicings. Eukaryot. Cell 4615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Nick McElhinny, S. A., D. A. Gordenin, C. M. Stith, P. M. J. Burgers, and T. A. Kunkel. 2008. Division of labor at the eukaryotic replication fork. Mol. Cell 30137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Niwa, O., and R. Kominami. 2001. Untargeted mutation of the maternally derived mouse hypervariable minisatellite allele in F1 mice born to irradiated spermatozoa. Proc. Natl. Acad. Sci. USA 981705-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, Berlin, Germany.
- 370.Ordway, J. M., S. Tallaksen-Greene, C. A. Gutekunst, E. M. Bernstein, J. A. Cearley, H. W. Wiener, L. S. Dure IV, R. Lindsey, S. M. Hersch, R. S. Jope, R. L. Albin, and P. J. Detloff. 1997. Ectopically expressed CAG repeats cause intranuclear inclusions and a progressive late onset neurological phenotype in the mouse. Cell 91753-763. [DOI] [PubMed] [Google Scholar]
- 371.Orr, H. T. 2001. Beyond the Qs in the polyglutamine diseases. Genes Dev. 15925-932. [DOI] [PubMed] [Google Scholar]
- 372.Orr, H. T., and H. Y. Zoghbi. 2007. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30575-621. [DOI] [PubMed] [Google Scholar]
- 373.Osman, F., J. Dixon, A. R. Barr, and M. C. Whitby. 2005. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol. Cell. Biol. 258084-8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Owen, B. A., Z. Yang, M. Lai, M. Gajek, J. D. Badger II, J. J. Hayes, W. Edelmann, R. Kucherlapati, T. M. Wilson, and C. T. McMurray. 2005. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 12663-670. [DOI] [PubMed] [Google Scholar]
- 375.Pâques, F., B. Bucheton, and M. Wegnez. 1996. Rearrangements involving repeated sequences within a P element preferentially occur between units close to the transposon extremities. Genetics 142459-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Pâques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Pâques, F., and J. E. Haber. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 176765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Pâques, F., W.-Y. Leung, and J. E. Haber. 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 182045-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Pâques, F., G.-F. Richard, and J. E. Haber. 2001. Expansions and contractions in 36-bp minisatellites by gene conversion in yeast. Genetics 158155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Pâques, F., M.-L. Samson, P. Jordan, and M. Wegnez. 1995. Structural evolution of the Drosophila 5S ribosomal genes. J. Mol. Evol. 41615-621. [DOI] [PubMed] [Google Scholar]
- 381.Pâques, F., and M. Wegnez. 1993. Deletions and amplifications of tandemly arranged ribosomal 5S genes internal to a P element occur at a high rate in a dysgenic context. Genetics 135469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Park, P. U., P. A. Defossez, and L. Guarente. 1999. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 193848-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Parsons, R., G.-M. Li, M. J. Longley, W.-H. Fang, N. Papadopoulos, J. Jen, A. de la Chapelle, K. W. Kinzler, B. Vogelstein, and P. Modrich. 1993. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 751227-1236. [DOI] [PubMed] [Google Scholar]
- 384.Patel, K. J., V. P. Yu, H. Lee, A. Corcoran, F. C. Thistlethwaite, M. J. Evans, W. H. Colledge, L. S. Friedman, B. A. Ponder, and A. R. Venkitaraman. 1998. Involvement of Brca2 in DNA repair. Mol. Cell 1347-357. [DOI] [PubMed] [Google Scholar]
- 384a.Payen, C., R. Koszul, B. Dujon, and G. Fischer. 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 4e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Pearson, C. E., K. N. Edamura, and J. D. Cleary. 2005. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6729-742. [DOI] [PubMed] [Google Scholar]
- 386.Pearson, C. E., A. Ewel, S. Acharya, R. A. Fishel, and R. R. Sinden. 1997. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum. Mol. Genet. 61117-1123. [DOI] [PubMed] [Google Scholar]
- 387.Pearson, C. E., M. Tam, Y. H. Wang, S. E. Montgomery, A. C. Dar, J. D. Cleary, and K. Nichol. 2002. Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 304534-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Pelletier, R., M. M. Krasilnikova, G. M. Samadashwily, R. Lahue, and S. M. Mirkin. 2003. Replication and expansion of trinucleotide repeats in yeast. Mol. Cell. Biol. 231349-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Peltomaki, P. 2001. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum. Mol. Genet. 10735-740. [DOI] [PubMed] [Google Scholar]
- 390.Peterson, D. G., S. R. Schulze, E. B. Sciara, S. A. Lee, J. E. Bowers, A. Nagel, N. Jiang, D. C. Tibbitts, S. R. Wessler, and A. H. Paterson. 2002. Integration of Cot analysis, DNA cloning, and high-throughput sequencing facilitates genome characterization and gene discovery. Genome Res. 12795-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Petes, T. D., P. W. Greenwell, and M. Dominska. 1997. Stabilization of microsatellite sequences by variant repeats in the yeast Saccharomyces cerevisiae. Genetics 146491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Philips, A. V., L. T. Timchenko, and T. A. Cooper. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280737-740. [DOI] [PubMed] [Google Scholar]
- 393.Piegu, B., R. Guyot, N. Picault, A. Roulin, A. Saniyal, H. Kim, K. Collura, D. S. Brar, S. Jackson, R. A. Wing, and O. Panaud. 2006. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 161262-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Pieretti, M., F. Zhang, Y.-H. Fu, S. T. Warren, B. A. Oostra, C. T. Caskey, and D. L. Nelson. 1991. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66817-822. [DOI] [PubMed] [Google Scholar]
- 395.Pinheiro, P., G. Scarlett, A. Rodgers, P. M. Rodger, A. Murray, T. Brown, S. F. Newbury, and J. A. McClellan. 2002. Structures of CUG repeats in RNA. J. Biol. Chem. 27735183-35190. [DOI] [PubMed] [Google Scholar]
- 396.Pirzio, L. M., P. Pichierri, M. Bignami, and A. Franchitto. 2008. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 180305-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Popescu, N. C. 2003. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 1921-17. [DOI] [PubMed] [Google Scholar]
- 398.Pritham, E. J., and C. Feschotte. 2007. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc. Natl. Acad. Sci. USA 1041895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Pritham, E. J., T. Putliwala, and C. Feschotte. 2007. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 3903-17. [DOI] [PubMed] [Google Scholar]
- 400.Prokopowich, C. D., T. R. Gregory, and T. J. Crease. 2003. The correlation between rDNA copy number and genome size in eukaryotes. Genome 4648-50. [DOI] [PubMed] [Google Scholar]
- 401.Prusiner, S. B., M. R. Scott, S. J. DeArmond, and F. E. Cohen. 1998. Prion protein biology. Cell 93337-348. [DOI] [PubMed] [Google Scholar]
- 402.Pursell, Z. F., I. Isoz, E.-B. Lundström, E. Johansson, and T. A. Kunkel. 2007. Yeast polymerase ɛ participates in leading-strand DNA replication. Science 317127-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Qiu, J., Y. Qian, P. Frank, U. Wintersberger, and B. Shen. 1999. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol. 198361-8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Quintana-Murci, L., C. Krausz, T. Zerjal, S. H. Sayar, M. F. Hammer, S. Q. Mehdi, Q. Ayub, R. Qamar, A. Mohyuddin, U. Radhakrishna, M. A. Jobling, C. Tyler-Smith, and K. McElreavey. 2001. Y-chromosome lineages trace diffusion of people and languages in southwestern Asia. Am. J. Hum. Genet. 68537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Rampino, N., H. Yamamoto, Y. Ionov, Y. Li, H. Sawai, J. C. Reed, and M. Perucho. 1997. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science 275967-969. [DOI] [PubMed] [Google Scholar]
- 406.Rattner, J. B. 1991. The structure of the mammalian centromere. Bioessays 1351-56. [DOI] [PubMed] [Google Scholar]
- 407.Raveendranathan, M., S. Chattopadhyay, Y. T. Bolon, J. Haworth, D. J. Clarke, and A. K. Bielinsky. 2006. Genome-wide replication profiles of S-phase checkpoint mutants reveal fragile sites in yeast. EMBO J. 253627-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Reagan, M. S., C. Pittenger, W. Siede, and E. C. Friedberg. 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Refsland, E. W., and D. M. Livingston. 2005. Interactions among ligase I, the flap endonuclease and proliferating cell nuclear antigen in the expansion and contraction of CAG repeat tracts in yeast. Genetics 171923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Reneker, J., C. R. Shyu, P. Zeng, J. C. Polacco, and W. Gassmann. 2004. ACMES: fast multiple-genome searches for short repeat sequences with concurrent cross-species information retrieval. Nucleic Acids Res. 32W649-W653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Reyniers, E., L. Vits, K. De Boulle, B. Van Roy, D. Van Velzen, E. de Graaff, A. J. Verkerk, H. Z. Jorens, J. K. Darby, B. Oostra, et al. 1993. The full mutation in the FMR-1 gene of male fragile X patients is absent in their sperm. Nat. Genet. 4143-146. [DOI] [PubMed] [Google Scholar]
- 412.Richard, G.-F., C. Cyncynatus, and B. Dujon. 2003. Contractions and expansions of CAG/CTG trinucleotide repeats occur during ectopic gene conversion in yeast, by a MUS81-independent mechanism. J. Mol. Biol. 326769-782. [DOI] [PubMed] [Google Scholar]
- 413.Richard, G.-F., and B. Dujon. 1996. Distribution and variability of trinucleotide repeats in the genome of the yeast Saccharomyces cerevisiae. Gene 174165-174. [DOI] [PubMed] [Google Scholar]
- 414.Richard, G.-F., and B. Dujon. 2006. Molecular evolution of minisatellites in hemiascomycetous yeasts. Mol. Biol. Evol. 23189-202. [DOI] [PubMed] [Google Scholar]
- 415.Richard, G.-F., and B. Dujon. 1997. Trinucleotide repeats in yeast. Res. Microbiol. 148731-744. [DOI] [PubMed] [Google Scholar]
- 416.Richard, G.-F., B. Dujon, and J. E. Haber. 1999. Double-strand break repair can lead to high frequencies of deletions within short CAG/CTG trinucleotide repeats. Mol. Gen. Genet. 261871-882. [DOI] [PubMed] [Google Scholar]
- 417.Richard, G.-F., G. M. Goellner, C. T. McMurray, and J. E. Haber. 2000. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11/RAD50/XRS2 complex. EMBO J. 192381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Richard, G.-F., C. Hennequin, A. Thierry, and B. Dujon. 1999. Trinucleotide repeats and other microsatellites in yeasts. Res. Microbiol. 150589-602. [DOI] [PubMed] [Google Scholar]
- 419.Richard, G.-F., A. Kerrest, I. Lafontaine, and B. Dujon. 2005. Comparative genomics of hemiascomycete yeasts: genes involved in DNA replication, repair, and recombination. Mol. Biol. Evol. 221011-1023. [DOI] [PubMed] [Google Scholar]
- 420.Richard, G.-F., and F. Pâques. 2000. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 1122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Richards, R. I. 2001. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 17339-345. [DOI] [PubMed] [Google Scholar]
- 422.Richards, R. I., and G. R. Sutherland. 1997. Dynamic mutation: possible mechanisms and significance in human disease. Trends Biochem. Sci. 22432-436. [DOI] [PubMed] [Google Scholar]
- 423.Ried, K., M. Finnis, L. Hobson, M. Mangelsdorf, S. Dayan, J. K. Nancarrow, E. Woollatt, G. Kremmidiotis, A. Gardner, D. Venter, E. Baker, and R. I. Richards. 2000. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 91651-1663. [DOI] [PubMed] [Google Scholar]
- 424.Ritchie, R. J., S. J. L. Knight, M. C. Hirst, P. K. Grewal, M. Bobrow, G. S. Cross, and K. E. Davies. 1994. The cloning of FRAXF: trinucleotide repeat expansion and methylation at a third fragile site in distal Xqter. Hum. Mol. Genet. 32115-2121. [DOI] [PubMed] [Google Scholar]
- 425.Robert, T., D. Dervins, F. Fabre, and S. Gangloff. 2006. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 252837-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Roest Crollius, H., O. Jaillon, C. Dasilva, C. Ozouf-Costaz, C. Fizames, C. Fischer, L. Bouneau, A. Billault, F. Quetier, W. Saurin, A. Bernot, and J. Weissenbach. 2000. Characterization and repeat analysis of the compact genome of the freshwater pufferfish Tetraodon nigroviridis. Genome Res. 10939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Rolfsmeier, M. L., and R. S. Lahue. 2000. Stabilizing effects of interruptions on trinucleotide repeat expansions in Saccharomyces cerevisiae. Mol. Cell. Biol. 20173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Rong, L., and H. L. Klein. 1993. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2681252-1259. [PubMed] [Google Scholar]
- 429.Rooney, A. P., and T. J. Ward. 2005. Evolution of a large ribosomal RNA multigene family in filamentous fungi: birth and death of a concerted evolution paradigm. Proc. Natl. Acad. Sci. USA 1025084-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Ropers, H. H., and B. C. Hamel. 2005. X-linked mental retardation. Nat. Rev. Genet. 646-57. [DOI] [PubMed] [Google Scholar]
- 431.Roset, R., J. A. Subirana, and X. Messeguer. 2003. MREPATT: detection and analysis of exact consecutive repeats in genomic sequences. Bioinformatics 192475-2476. [DOI] [PubMed] [Google Scholar]
- 432.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 141-10. [PubMed] [Google Scholar]
- 433.Rubin, G. M., M. D. Yandell, J. R. Wortman, G. L. Gabor Miklos, C. R. Nelson, I. K. Hariharan, M. E. Fortini, P. W. Li, R. Apweiler, W. Fleischmann, J. M. Cherry, S. Henikoff, M. P. Skupski, S. Misra, M. Ashburner, E. Birney, M. S. Boguski, T. Brody, P. Brokstein, S. E. Celniker, S. A. Chervitz, D. Coates, A. Cravchik, A. Gabrielian, R. F. Galle, W. M. Gelbart, R. A. George, L. S. Goldstein, F. Gong, P. Guan, N. L. Harris, B. A. Hay, R. A. Hoskins, J. Li, Z. Li, R. O. Hynes, S. J. Jones, P. M. Kuehl, B. Lemaitre, J. T. Littleton, D. K. Morrison, C. Mungall, P. H. O'Farrell, O. K. Pickeral, C. Shue, L. B. Vosshall, J. Zhang, Q. Zhao, X. H. Zheng, and S. Lewis. 2000. Comparative genomics of the eukaryotes. Science 2872204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Sagot, M.-F., and E. W. Myers. 1998. Identifying satellites and periodic repetitions in biological sequences. J. Comp. Biol. 5539-554. [DOI] [PubMed] [Google Scholar]
- 435.Sakamoto, N., P. D. Chastain, P. Parniewski, K. Ohshima, M. Pandolfo, J. D. Griffith, and R. D. Wells. 1999. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich's ataxia. Mol. Cell 3465-475. [DOI] [PubMed] [Google Scholar]
- 436.Sakamoto, N., J. E. Larson, R. R. Iyer, L. Montermini, M. Pandolfo, and R. D. Wells. 2001. GGA*TCC-interrupted triplets in long GAA*TTC repeats inhibit the formation of triplex and sticky DNA structures, alleviate transcription inhibition, and reduce genetic instabilities. J. Biol. Chem. 27627178-27187. [DOI] [PubMed] [Google Scholar]
- 437.Sakamoto, N., K. Ohshima, L. Montermini, M. Pandolfo, and R. D. Wells. 2001. Sticky DNA, a self-associated complex formed at long GAA*TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J. Biol. Chem. 27627171-27177. [DOI] [PubMed] [Google Scholar]
- 438.Samadashwily, G., G. Raca, and S. M. Mirkin. 1997. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 17298-304. [DOI] [PubMed] [Google Scholar]
- 439.Sandman, K., and J. N. Reeve. 1999. Archaeal nucleosome positioning by CTG repeats. J. Bacteriol. 1811035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 745463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Santibanez-Koref, M. F., R. Gangeswaran, and J. M. Hancock. 2001. A relationship between lengths of microsatellites and nearby substitution rates in mammalian genomes. Mol. Biol. Evol. 182119-2123. [DOI] [PubMed] [Google Scholar]
- 442.Sapolsky, R. J., V. Brendel, and S. Karlin. 1993. A comparative analysis of distinctive features of yeast protein sequences. Yeast 91287-1298. [DOI] [PubMed] [Google Scholar]
- 443.Sarai, A., J. Mazur, R. Nussinov, and R. L. Jernigan. 1989. Sequence dependence of DNA conformational flexibility. Biochemistry 287842-7849. [DOI] [PubMed] [Google Scholar]
- 444.Sasaki, T., H. Nishihara, M. Hirakawa, K. Fujimura, M. Tanaka, N. Kokubo, C. Kimura-Yoshida, I. Matsuo, K. Sumiyama, N. Saitou, T. Shimogori, and N. Okada. 2008. Possible involvement of SINEs in mammalian-specific brain formation. Proc. Natl. Acad. Sci. USA 1054220-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Satyal, S. H., E. Schmidt, K. Kitagawa, N. Sondheimer, S. Lindquist, J. M. Kramer, and R. I. Morimoto. 2000. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 975750-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Saudou, F., S. Finkbeiner, D. Devys, and M. E. Greenberg. 1998. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 9555-66. [DOI] [PubMed] [Google Scholar]
- 447.Savouret, C., E. Brisson, J. Essers, R. Kanaar, A. Pastink, H. te Riele, C. Junien, and G. Gourdon. 2003. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 222264-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Savouret, C., C. Garcia-Cordier, J. Megret, H. te Riele, C. Junien, and G. Gourdon. 2004. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell. Biol. 24629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Schacherer, J., J. de Montigny, A. Welcker, J. L. Souciet, and S. Potier. 2005. Duplication processes in Saccharomyces cerevisiae haploid strains. Nucleic Acids Res. 336319-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Schacherer, J., Y. Tourrette, J. L. Souciet, S. Potier, and J. De Montigny. 2004. Recovery of a function involving gene duplication by retroposition in Saccharomyces cerevisiae. Genome Res. 141291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Schär, P. 2001. Spontaneous DNA damage, genome instability, and cancer—when DNA replication escapes control. Cell 104329-332. [DOI] [PubMed] [Google Scholar]
- 452.Scherzinger, E., R. Lurz, M. Turmaine, L. Mangiarini, B. Hollenbach, R. Hasenbank, G. P. Bates, S. W. Davies, H. Lehrach, and E. E. Wanker. 1997. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90549-558. [DOI] [PubMed] [Google Scholar]
- 453.Schwartz, M., E. Zlotorynski, M. Goldberg, E. Ozeri, A. Rahat, C. le Sage, B. P. Chen, D. J. Chen, R. Agami, and B. Kerem. 2005. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 192715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Schweitzer, J. K., and D. M. Livingston. 1997. Destabilization of CAG trinucleotide repeat tracts by mismatch repair mutations in yeast. Hum. Mol. Genet. 6349-355. [DOI] [PubMed] [Google Scholar]
- 455.Schweitzer, J. K., and D. M. Livingston. 1998. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 769-74. [DOI] [PubMed] [Google Scholar]
- 456.Schweitzer, J. K., and D. M. Livingston. 1999. The effect of DNA replication mutations on CAG tract stability in yeast. Genetics 152953-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Schweitzer, J. K., S. S. Reinke, and D. M. Livingston. 2001. Meiotic alterations in CAG repeat tracts. Genetics 1591861-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Scott, H. S., J. Kudoh, M. Wattenhofer, K. Shibuya, A. Berry, R. Chrast, M. Guipponi, J. Wang, K. Kawasaki, S. Asakawa, S. Minoshima, F. Younus, S. Q. Mehdi, U. Radhakrishna, M. P. Papasavvas, C. Gehrig, C. Rossier, M. Korostishevsky, A. Gal, N. Shimizu, B. Bonne-Tamir, and S. E. Antonarakis. 2001. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet. 2759-63. [DOI] [PubMed] [Google Scholar]
- 459.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95419-430. [DOI] [PubMed] [Google Scholar]
- 460.Seznec, H., A.-S. Lia-Baldini, C. Duros, C. Fouquet, C. Lacroix, H. Hofmann-Radvanyi, C. Junien, and G. Gourdon. 2000. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum. Mol. Genet. 91185-1194. [DOI] [PubMed] [Google Scholar]
- 461.Sharma, S., and S. N. Raina. 2005. Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet. Genome Res. 10915-26. [DOI] [PubMed] [Google Scholar]
- 462.Sharp, P., and A. Lloyd. 1993. Regional base composition variation along yeast chromosome III: evolution of chromosome primary structure. Nucleic Acids Res. 21179-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.She, X., Z. Jiang, R. A. Clark, G. Liu, Z. Cheng, E. Tuzun, D. M. Church, G. Sutton, A. L. Halpern, and E. E. Eichler. 2004. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431927-930. [DOI] [PubMed] [Google Scholar]
- 464.Shiraishi, T., T. Druck, K. Mimori, J. Flomenberg, L. Berk, H. Alder, W. Miller, K. Huebner, and C. M. Croce. 2001. Sequence conservation at human and mouse orthologous common fragile regions, FRA3B/FHIT and Fra14A2/Fhit. Proc. Natl. Acad. Sci. USA 985722-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Shoemaker, R. C., K. Polzin, J. Labate, J. Specht, E. C. Brummer, T. Olson, N. Young, V. Concibido, J. Wilcox, J. P. Tamulonis, G. Kochert, and H. R. Boerma. 1996. Genome duplication in soybean (Glycine subgenus soja). Genetics 144329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Sia, E. A., M. Dominska, L. Stefanovic, and T. D. Petes. 2001. Isolation and characterization of point mutations in mismatch repair genes that destabilize microsatellites in yeast. Mol. Cell. Biol. 218157-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Sia, E. A., R. J. Kokoska, M. Dominska, P. Greenwell, and T. D. Petes. 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 172851-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 911033-1042. [DOI] [PubMed] [Google Scholar]
- 469.Sinclair, D. A., K. Mills, and L. Guarente. 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 2771313-1316. [DOI] [PubMed] [Google Scholar]
- 470.Singh, P., L. Zheng, V. Chavez, J. Qiu, and B. Shen. 2007. Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n- and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J. Biol. Chem. 2823465-3477. [DOI] [PubMed] [Google Scholar]
- 471.Reference deleted.
- 472.Snow, K., D. J. Tester, K. E. Kruckeberg, D. J. Schaid, and S. N. Thibodeau. 1994. Sequence analysis of the fragile X trinucleotide repeat: implications for the origin of the fragile X mutation. Hum. Mol. Genet. 31543-1551. [DOI] [PubMed] [Google Scholar]
- 473.Souza, R. F., R. Appel, J. Yin, S. Wang, K. N. Smolinski, J. M. Abraham, T.-T. Zou, Y.-Q. Shi, J. Lei, J. Cottrell, K. Cymes, K. Biden, L. Simms, B. Leggett, P. M. Lynch, M. Frazier, S. M. Powell, N. Harpaz, H. Sugimura, J. Young, and S. J. Meltzer. 1996. Microsatellite instability in the insulin-like growth factor II receptor gene in gastrointestinal tumours. Nat. Genet. 14255-257. [DOI] [PubMed] [Google Scholar]
- 474.Spiro, C., R. Pelletier, M. L. Rolfsmeier, M. J. Dixon, R. S. Lahue, G. Gupta, M. S. Park, X. Chen, S. V. Mariappan, and C. T. McMurray. 1999. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell 41079-1085. [DOI] [PubMed] [Google Scholar]
- 475.Stallings, R. L. 1994. Distribution of trinucleotide microsatellites in different categories of mammalian genomic sequence: implications for human genetic diseases. Genomics 21116-121. [DOI] [PubMed] [Google Scholar]
- 476.Strand, M., T. A. Prolla, R. M. Liskay, and T. D. Petes. 1993. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365274-276. [DOI] [PubMed] [Google Scholar]
- 477.Subramanian, J., S. Vijayakumar, A. E. Tomkinson, and N. Arnheim. 2005. Genetic instability induced by overexpression of DNA ligase I in budding yeast. Genetics 171427-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Subramanian, S., V. M. Madgula, R. George, R. K. Mishra, M. W. Pandit, C. S. Kumar, and L. Singh. 2003. Triplet repeats in human genome: distribution and their association with genes and other genomic regions. Bioinformatics 19549-552. [DOI] [PubMed] [Google Scholar]
- 479.Subramanian, S., R. K. Mishra, and L. Singh. 2003. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 4R13.1-R13.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Suen, I. S., J. N. Rhodes, M. Christy, B. McEwen, D. M. Gray, and M. Mitas. 1999. Structural properties of Friedreich's ataxia d(GAA) repeats. Biochim. Biophys. Acta 144414-24. [DOI] [PubMed] [Google Scholar]
- 481.Sugawara, N., F. Pâques, M. Colaiacovo, and J. H. Haber. 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 949214-9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Sung, P., L. Krejci, S. Van Komen, and M. G. Sehorn. 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 27842729-42732. [DOI] [PubMed] [Google Scholar]
- 483.Sutherland, G. R., E. Baker, and R. I. Richards. 1998. Fragile sites still breaking. Trends Genet. 14501-506. [DOI] [PubMed] [Google Scholar]
- 484.Takahashi, K., S. Murakami, Y. Chikashige, O. Niwa, and M. Yanagida. 1991. A large number of tRNA genes are symmetrically located in fission yeast centromeres. J. Mol. Biol. 21813-17. [DOI] [PubMed] [Google Scholar]
- 485.Tamaki, K., C. A. May, Y. E. Dubrova, and A. J. Jeffreys. 1999. Extremely complex repeat shuffling during germline mutation at human minisatellite B6.7. Hum. Mol. Genet. 8879-888. [DOI] [PubMed] [Google Scholar]
- 486.Taneja, K. L., M. McCurrach, M. Schalling, D. Housman, and R. H. Singer. 1995. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 128995-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Tekaia, F., and B. Dujon. 1999. Pervasiveness of gene conversion and persistence of duplicates in cellular genomes. J. Mol. Evol. 49591-600. [DOI] [PubMed] [Google Scholar]
- 488.Tennyson, R. B., N. Ebran, A. E. Herrera, and J. E. Lindsley. 2002. A novel selection system for chromosome translocations in Saccharomyces cerevisiae. Genetics 1601363-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Thierry, A., C. Bouchier, B. Dujon, and G.-F. Richard. Megasatellites: a peculiar class of giant minisatellites in genes involved in cell adhesion and pathogenicity in Candida glabrata. Nucleic Acids Res. 365970-5982. [DOI] [PMC free article] [PubMed]
- 490.Reference deleted.
- 491.Reference deleted.
- 492.Reference deleted.
- 493.Thomas, C. A., Jr. 1971. The genetic organization of chromosomes. Annu. Rev. Genet. 5237-256. [DOI] [PubMed] [Google Scholar]
- 494.Thomas, J. W., M. G. Schueler, T. J. Summers, R. W. Blakesley, J. C. McDowell, P. J. Thomas, J. R. Idol, V. V. Maduro, S. Q. Lee-Lin, J. W. Touchman, G. G. Bouffard, S. M. Beckstrom-Sternberg, and E. D. Green. 2003. Pericentromeric duplications in the laboratory mouse. Genome Res. 1355-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Thornton, C. A., J. P. Wymer, Z. Simmons, C. McClain, and R. T. Moxley III. 1997. Expansion of the myotonic dystrophy CTG repeat reduces expression of the flanking DMAHP gene. Nat. Genet. 16407-409. [DOI] [PubMed] [Google Scholar]
- 496.Tian, B., R. J. White, T. Xia, S. Welle, D. H. Turner, M. B. Mathews, and C. A. Thornton. 2000. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA 679-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Tishkoff, D. X., N. Filosi, G. M. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88253-263. [DOI] [PubMed] [Google Scholar]
- 498.Tomita, N., R. Fujita, D. Kurihara, H. Shindo, R. D. Wells, and M. Shimizu. 2002. Effects of triplet repeat sequences on nucleosome positioning and gene expression in yeast minichromosomes. Nucleic Acids Symp. Ser. 2231-232. [DOI] [PubMed] [Google Scholar]
- 499.Toth, G., Z. Gaspari, and J. Jurka. 2000. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10967-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Toulouse, A., F. Au-Yeung, C. Gaspar, J. Roussel, P. Dion, and G. A. Rouleau. 2005. Ribosomal frameshifting on MJD-1 transcripts with long CAG tracts. Hum. Mol. Genet. 142649-2660. [DOI] [PubMed] [Google Scholar]
- 501.Tran, H. T., J. D. Keen, M. Kricker, M. A. Resnick, and D. A. Gordenin. 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 172859-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Treco, D., and N. Arnheim. 1986. The evolutionarily conserved repetitive sequence d(TG·AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol. Cell. Biol. 63934-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Turmaine, M., A. Raza, A. Mahal, L. Mangiarini, G. P. Bates, and S. W. Davies. 2000. Nonapoptotic neurodegeneration in a transgenic mouse model of Huntington's disease. Proc. Natl. Acad. Sci. USA 978093-8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Tuzun, E., J. A. Bailey, and E. E. Eichler. 2004. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res. 14493-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Tyler-Smith, C., and H. F. Willard. 1993. Mammalian chromosome structure. Curr. Opin. Genet. Dev. 3390-397. [DOI] [PubMed] [Google Scholar]
- 506.Ugarkovic, D. 2005. Functional elements residing within satellite DNA. EMBO Rep. 61035-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Ullu, E., and C. Tschudi. 1984. Alu sequences are processed 7SL RNA genes. Nature 312171-172. [DOI] [PubMed] [Google Scholar]
- 508.Umar, A., A. B. Buermeyer, J. A. Simon, D. C. Thomas, A. B. Clark, R. M. Liskay, and T. A. Kunkel. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 8765-73. [DOI] [PubMed] [Google Scholar]
- 509.Usdin, K., and K. J. Woodford. 1995. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 234202-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Veaute, X., S. Delmas, M. Selva, J. Jeusset, E. Le Cam, I. Matic, F. Fabre, and M. A. Petit. 2005. UvrD helicase, unlike Rep. helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 24180-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Veaute, X., J. Jeusset, C. Soustelle, S. C. Kowalczykowski, E. Le Cam, and F. Fabre. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423309-312. [DOI] [PubMed] [Google Scholar]
- 513.Vergnaud, G., and F. Denoeud. 2000. Minisatellites: mutability and genome architecture. Genome Res. 10899-907. [DOI] [PubMed] [Google Scholar]
- 514.Verkerk, A. J. M. H., M. Pieretti, J. S. Sutcliffe, Y.-H. Fu, D. P. A. Kuhl, A. Pizzuti, O. Reined, S. Richards, M. F. Victoria, F. Zhang, B. E. Eussen, G.-J. B. Van Ommen, L. A. J. Blonden, G. J. Riggins, J. L. Chastain, C. B. Kunst, H. Galjaard, C. T. Caskey, D. L. Nelson, B. A. Oostra, and S. T. Warren. 1991. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65905-914. [DOI] [PubMed] [Google Scholar]
- 515.Verstrepen, K. J., A. Jansen, F. Lewitter, and G. R. Fink. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37986-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Vilà, C., J. A. Leonard, A. Götherström, S. Marklund, K. Sandberg, K. Lidén, R. K. Wayne, and H. Ellegren. 2001. Widespread origin of domestic horse lineages. Science 291474-477. [DOI] [PubMed] [Google Scholar]
- 517.Vilenchik, M. M., and A. G. Knudson. 2003. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA 10012871-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Virtaneva, K., E. D'Amato, J. Miao, M. Koskiniemi, R. Norio, G. Avanzini, S. Franceschetti, R. Michelucci, C. A. Tassinari, S. Omer, L. A. Pennacchio, R. M. Myers, J. L. Dieguez-Lucena, R. Krahe, A. de la Chapelle, and A.-E. Lehesjoki. 1997. Unstable minisatellite expansion causing recessively inherited myoclonus epilepsy, EPM1. Nat. Genet. 15393-396. [DOI] [PubMed] [Google Scholar]
- 519.Vision, T. J., D. G. Brown, and S. D. Tanksley. 2000. The origins of genomic duplications in Arabidopsis. Science 2902114-2117. [DOI] [PubMed] [Google Scholar]
- 520.Walker, P. M. B. 1971. Origin of satellite DNA. Nature 229306-308. [DOI] [PubMed] [Google Scholar]
- 521.Wang, Y. H., S. Amirhaeri, S. Kang, R. D. Wells, and J. D. Griffith. 1994. Preferential nucleosome assembly at DNA triplet repeats from the myotonic dystrophy gene. Science 265669-671. [DOI] [PubMed] [Google Scholar]
- 522.Wang, Y. H., R. Gellibolian, M. Shimizu, R. D. Wells, and J. Griffith. 1996. Long CCG triplet repeat blocks exclude nucleosomes: a possible mechanism for the nature of fragile sites in chromosomes. J. Mol. Biol. 263511-516. [DOI] [PubMed] [Google Scholar]
- 523.Wang, Y. H., and J. Griffith. 1995. Expanded CTG triplet blocks from the myotonic dystrophy gene create the strongest known natural nucleosome positioning elements. Genomics 25570-573. [DOI] [PubMed] [Google Scholar]
- 524.Wang, Y. H., and J. Griffith. 1996. Methylation of expanded CCG triplet repeat DNA from fragile X syndrome patients enhances nucleosome exclusion. J. Biol. Chem. 27122937-22940. [PubMed] [Google Scholar]
- 525.Warren, S. T. 1997. Polyalanine expansion in synpolydactyly might result from unequal crossing-over of HOXD13. Science 275408-409. [DOI] [PubMed] [Google Scholar]
- 526.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, K. Barlow, S. Beck, E. Berry, B. Birren, T. Bloom, P. Bork, M. Botcherby, N. Bray, M. R. Brent, D. G. Brown, S. D. Brown, C. Bult, J. Burton, J. Butler, R. D. Campbell, P. Carninci, S. Cawley, F. Chiaromonte, A. T. Chinwalla, D. M. Church, M. Clamp, C. Clee, F. S. Collins, L. L. Cook, R. R. Copley, A. Coulson, O. Couronne, J. Cuff, V. Curwen, T. Cutts, M. Daly, R. David, J. Davies, K. D. Delehaunty, J. Deri, E. T. Dermitzakis, C. Dewey, N. J. Dickens, M. Diekhans, S. Dodge, I. Dubchak, D. M. Dunn, S. R. Eddy, L. Elnitski, R. D. Emes, P. Eswara, E. Eyras, A. Felsenfeld, G. A. Fewell, P. Flicek, K. Foley, W. N. Frankel, L. A. Fulton, R. S. Fulton, T. S. Furey, D. Gage, R. A. Gibbs, G. Glusman, S. Gnerre, N. Goldman, L. Goodstadt, D. Grafham, T. A. Graves, E. D. Green, S. Gregory, R. Guigo, M. Guyer, R. C. Hardison, D. Haussler, Y. Hayashizaki, L. W. Hillier, A. Hinrichs, W. Hlavina, T. Holzer, F. Hsu, A. Hua, T. Hubbard, A. Hunt, I. Jackson, D. B. Jaffe, L. S. Johnson, M. Jones, T. A. Jones, A. Joy, M. Kamal, E. K. Karlsson, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420520-562. [DOI] [PubMed] [Google Scholar]
- 527.Webster, M. T., N. G. Smith, and H. Ellegren. 2002. Microsatellite evolution inferred from human-chimpanzee genomic sequence alignments. Proc. Natl. Acad. Sci. USA 998748-8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Weiner, A. M., P. L. Deininger, and A. Efstratiadis. 1986. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu. Rev. Biochem. 55631-661. [DOI] [PubMed] [Google Scholar]
- 529.Welch, J. W., D. H. Maloney, and S. Fogel. 1990. Unequal crossing-over and gene conversion at the amplified CUP1 locus of yeast. Mol. Gen. Genet. 222304-310. [DOI] [PubMed] [Google Scholar]
- 530.Welcsh, P. L., and M. C. King. 2001. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 10705-713. [DOI] [PubMed] [Google Scholar]
- 531.Weller, P., A. J. Jeffreys, V. Wilson, and A. Blanchetot. 1984. Organization of the human myoglobin gene. EMBO J. 3439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Wells, R. D., R. Dere, M. L. Hebert, M. Napierala, and L. S. Son. 2005. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 333785-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Wendel, J. F. 2000. Genome evolution in polyploids. Plant Mol. Biol. 42225-249. [PubMed] [Google Scholar]
- 534.White, P. J., R. H. Borts, and M. C. Hirst. 1999. Stability of the human fragile X (CCG)n triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol. Cell. Biol. 195675-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Wicker, T., F. Sabot, A. Hua-Van, J. L. Bennetzen, P. Capy, B. Chalhoub, A. Flavell, P. Leroy, M. Morgante, O. Panaud, E. Paux, P. SanMiguel, and A. H. Schulman. 2007. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8973-982. [DOI] [PubMed] [Google Scholar]
- 536.Wierdl, M., M. Dominska, and T. D. Petes. 1997. Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics 146769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Wöhrle, D., I. Hennig, W. Vogel, and P. Steinbach. 1993. Mitotic stability of fragile X mutations in differentiated cells indicates early post-conceptional trinucleotide repeat expansion. Nat. Genet. 4140-142. [DOI] [PubMed] [Google Scholar]
- 538.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387708-713. [DOI] [PubMed] [Google Scholar]
- 539.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schafer, S. Muller-Auer, C. Gabel, M. Fuchs, A. Dusterhoft, C. Fritzc, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambutt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dreano, S. Gloux, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415871-880. [DOI] [PubMed] [Google Scholar]
- 540.Woodford, K. J., R. M. Howell, and K. Usdin. 1994. A novel K(+)-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J. Biol. Chem. 26927029-27035. [PubMed] [Google Scholar]
- 541.Woollard, A. 25 June 2005, posting date. Gene duplications and genetic redundancy in C. elegans, p. 1-6. In The C. elegans Research Community (ed.),WormBook. doi/ 10.1895/wormbook.1.2.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 542.Wu, X., J. Li, X. Li, C.-L. Hsieh, P. M. J. Burgers, and M. R. Lieber. 1996. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 242036-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Wyman, A. R., and R. White. 1980. A highly polymorphic locus in human DNA. Proc. Natl. Acad. Sci. USA 776754-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Xie, Y., C. Counter, and E. Alani. 1999. Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics 151499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Xie, Y., Y. Liu, J. L. Argueso, L. A. Henricksen, H. I. Kao, R. A. Bambara, and E. Alani. 2001. Identification of rad27 mutations that confer differential defects in mutation avoidance, repeat tract instability, and flap cleavage. Mol. Cell. Biol. 214889-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Xu, X., M. Peng, Z. Fang, and X. Xu. 2000. The direction of microsatellite mutations is dependent upon allele length. Nat. Genet. 24396-399. [DOI] [PubMed] [Google Scholar]
- 547.Yoon, S. R., L. Dubeau, M. de Young, N. S. Wexler, and N. Arnheim. 2003. Huntington disease expansion mutations in humans can occur before meiosis is completed. Proc. Natl. Acad. Sci. USA 1008834-8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Young, E. T., J. S. Sloan, and K. Van Riper. 2000. Trinucleotide repeats are clustered in regulatory genes in Saccharomyces cerevisiae. Genetics 1541053-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Yu, A., M. D. Barron, R. M. Romero, M. Christy, B. Gold, J. Dai, D. M. Gray, I. S. Haworth, and M. Mitas. 1997. At physiological pH, d(CCG)15 forms a hairpin containing protonated cytosines and a distorted helix. Biochemistry 363687-3699. [DOI] [PubMed] [Google Scholar]
- 550.Yu, A., J. Dill, S. S. Wirth, G. Huang, V. H. Lee, I. S. Haworth, and M. Mitas. 1995. The trinucleotide repeat sequence d(GTC)15 adopts a hairpin conformation. Nucleic Acids Res. 232706-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Yu, A., and M. Mitas. 1995. The purine-rich trinucleotide repeat sequences d(CAG)15 and d(GAC)15 form hairpins. Nucleic Acids Res. 234055-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Yu, S., M. Mangelsdorf, D. Hewett, L. Hobson, E. Baker, H. J. Eyre, N. Lapsys, D. Le Paslier, N. A. Doggett, G. R. Sutherland, and R. I. Richards. 1997. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell 88367-374. [DOI] [PubMed] [Google Scholar]
- 553.Zahra, R., J. K. Blackwood, J. Sales, and D. R. F. Leach. 2007. Proofreading and secondary structure processing determine the orientation dependence of CAG.CTG trinucleotide repeat instability in Escherichia coli. Genetics 17627-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Zhang, H., and C. H. Freudenreich. 2007. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol. Cell 27367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Zhang, Z., P. M. Harrison, Y. Liu, and M. Gerstein. 2003. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 132541-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Zhu, Y., D. C. Queller, and J. E. Strassmann. 2000. A phylogenetic perspective on sequence evolution in microsatellite loci. J. Mol. Evol. 50324-338. [DOI] [PubMed] [Google Scholar]
- 557.Zimmerly, S., H. Guo, R. Eskes, J. Yang, P. S. Perlman, and A. M. Lambowitz. 1995. A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell 83529-538. [DOI] [PubMed] [Google Scholar]
- 558.Zimmerly, S., H. Guo, P. S. Perlman, and A. M. Lambowitz. 1995. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell 82545-554. [DOI] [PubMed] [Google Scholar]
- 559.Zlotorynski, E., A. Rahat, J. Skaug, N. Ben-Porat, E. Ozeri, R. Hershberg, A. Levi, S. W. Scherer, H. Margalit, and B. Kerem. 2003. Molecular basis for expression of common and rare fragile sites. Mol. Cell. Biol. 237143-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 9087-96. [DOI] [PubMed] [Google Scholar]