Abstract
DNA replication and repair are critical processes for all living organisms to ensure faithful duplication and transmission of genetic information. Flap endonuclease 1 (Fen1), a structure-specific nuclease, plays an important role in multiple DNA metabolic pathways and maintenance of genome stability. Human FEN1 mutations that impair its exonuclease activity have been linked to cancer development. FEN1 interacts with multiple proteins, including proliferation cell nuclear antigen (PCNA), to form various functional complexes. Interactions with these proteins are considered to be the key molecular mechanisms mediating FEN1's key biological functions. The current challenge is to experimentally demonstrate the biological consequence of a specific interaction without compromising other functions of a desired protein. To address this issue, we established a mutant mouse model harboring a FEN1 point mutation (F343A/F344A, FFAA), which specifically abolishes the FEN1/PCNA interaction. We show that the FFAA mutation causes defects in RNA primer removal and long-patch base excision repair, even in the heterozygous state, resulting in numerous DNA breaks. These breaks activate the G2/M checkpoint protein, Chk1, and induce near-tetraploid aneuploidy, commonly observed in human cancer, consequently elevating the transformation frequency. Consistent with this, inhibition of aneuploidy formation by a Chk1 inhibitor significantly suppressed the cellular transformation. WT/FFAA FEN1 mutant mice develop aneuploidy-associated cancer at a high frequency. Thus, this study establishes an exemplary case for investigating the biological significance of protein-protein interactions by knock-in of a point mutation rather than knock-out of a whole gene.
Keywords: FEN1, PCNA, Okazaki fragment maturation, long patch base excision repair, tetraploidy, aneuploidy, cancer
Introduction
DNA replication and repair are essential processes for maintenance of the integrity of genomes. Various forms of DNA intermediates are produced during DNA replication and repair and they must be efficiently processed to avoid genome instabilities. The DNA intermediates with bifurcated nucleic acid structures are recognized and cleaved by structure-specific nucleases, typified by flap endonuclease 1 (FEN1), to generate ligatable DNA ends. FEN1 is best known for its involvement in RNA primer removal and long patch base excision repair (LP-BER) 1, 2, 3. It recognizes and cleaves the DNA 5′-flap substrate generated by Pol δ- or β-mediated strand-displacement DNA synthesis. The versatile FEN1 is implicated to be involved in other major DNA metabolic pathways, including resolution of tri-nucleotide repetitive sequence-derived secondary structures and rescue of stalled DNA replication forks, as well as apoptotic DNA fragmentation 4, 5, 6, 7. During its actions in these pathways, the concerted actions of the 5′ exonuclease and gap-dependent endonuclease activities of FEN1 remove a few nucleotides from the 5′ blunt ends or nicks and cut the template strand of a gapped DNA duplex, to cleave the double-stranded DNA hairpin structures or generate DNA double-strand breaks. Yeast cells harboring a deletion of the rad27 gene (FEN1 yeast homologue) display slow growth, hypersensitivity to DNA-damaging agents, and mutator phenotypes 8, 9, 10. Mice that are homozygous for Fen1 knockout have an embryonic lethal phenotype 11, 12. Furthermore, FEN1 mutations have been identified in humans and linked to cancer development 6. Together, these findings suggest the biological significance of FEN1 in DNA replication and repair.
It appears that FEN1 is in complex with other proteins to execute its function in different pathways. More than 30 FEN1-interacting proteins have been identified, including (1) DNA replication proteins (proliferation cell nuclear antigen (PCNA), hnRNP A1, Pol α/ɛ, replication protein A [RPA] and DNA ligase I) 13, 14, 15, 16, 17; (2) LP-BER specific proteins (APE1) and other DNA repair proteins (9-1-1 and WRN) 18, 19, 20; (3) telomere proteins (TRF2 and TERT) 21, 22; (4) chromosome cohesion proteins (Chlr1) 23; and (5) apoptotic DNA fragmentation protein endonuclease G (endo G) 4. However, the physiological relevance of these protein-protein interactions in FEN1-mediated DNA metabolic pathways remains unknown. A critical question is whether FEN1 mutations that abolish these protein-protein interactions, such as the FEN1/PCNA interaction, impair its cellular functions and cause cancer and other disease phenotypes.
Of these FEN1 interaction partners, PCNA, which was initially identified as a replication accessory protein, accompanies FEN1 in all FEN1-involved DNA metabolic pathways except for the apoptotic DNA fragmentation pathway, suggesting a critical role of the FEN1/PCNA interaction in regulating FEN1's functions in different pathways. Previous studies suggested that the interaction with PCNA was the key mechanism for recruiting FEN1 to the replication factories 24. On the other hand, PCNA stimulates FEN1 activity, facilitating its functions in RNA primer removal and LP-BER 25. PCNA has also been suggested to coordinate the sequential actions of different enzymes during Okazaki fragment maturation 26. The FEN1/PCNA interaction is mediated via the PIP box (337QGRLDDFFK345) at the C-terminal end of FEN1, and Phe343 and Phe344 in the PIP box are two key residues for mediating the FEN1/PCNA interaction 27, 28. Substitution of these two residues with two Ala residues (FFAA) specifically abolishes the physical interaction of FEN1 and PCNA 27, 28. To study the in vivo requirement of this interaction for the cellular function of FEN1, we previously generated a mouse model carrying the FFAA mutation 24. We found that disruption of the FEN1/PCNA interaction resulted in slow cell proliferation and embryonic lethality of homozygous mutant mice 24. However, the underlying molecular mechanism that causes such phenotypes remains unclear. In this study, we show that the FFAA mutation has a dominant-negative or heterozygous effect on the function of FEN1 in Okazaki fragment maturation and LP-BER. As a consequence, both homozygous and heterozygous FFAA mutant cells accumulate DNA strand breaks and spontaneously become aneuploid, as well as in response to the treatment with DNA-damaging agents. Wild-type (WT)/FFAA aneuploid cells have dramatically elevated potential to undergo malignant transformation, and heterozygous FFAA mice develop aneuploidy-associated cancers at high incidence. Taken together, these findings suggest FEN1 mutations that disrupt the FEN1/PCNA interaction cause enhanced susceptibility to endogenous or exogenous DNA insults and early onset of cancer.
Results
WT/FFAA cells display defects in Okazaki fragment maturation
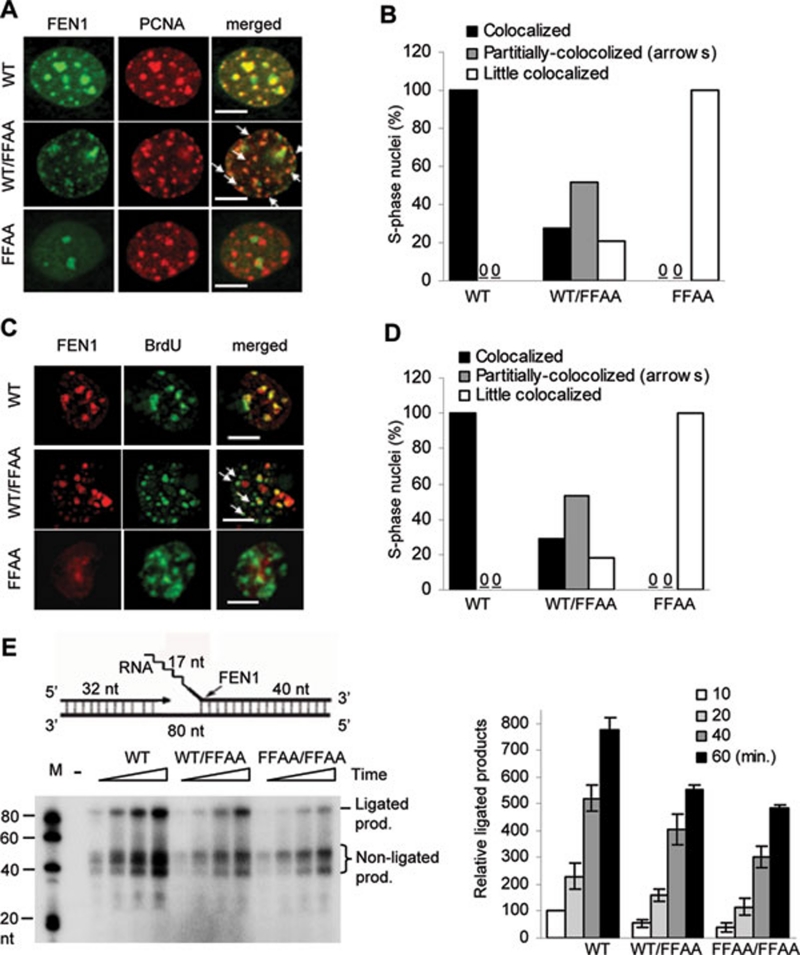
We previously showed that the FFAA mutation in the homozygous form caused defects in DNA replication, cell proliferation, and late embryonic lethality via pulmonary hypoplasia and pancytopenia 24. To test whether the FFAA mutation had a heterozygous effect, which may be more physiologically relevant, we first determined if WT/FFAA cells displayed any defect in FEN1 localization to replication foci, which is mediated via the FEN1/PCNA interaction. In all WT S phase MEF cells examined, every PCNA or BrdU focus had a corresponding FEN1 focus, indicating a complete co-localization (Figures 1A-1D). However, little FEN1/PCNA or FEN1/BrdU co-localization was observed in FFAA/FFAA MEF cells (Figures 1A-1D). Interestingly, we observed three categories of WT/FFAA nuclei, according to the degree of FEN1/PCNA or FEN1/BrdU co-localization: complete co-localization, partial co-localization, and little co-localization nuclei. Fifty-five percent of WT/FFAA MEF nuclei displayed a partial co-localization pattern, in which approximately half of the PCNA or BrdU foci had no corresponding FEN1 foci or very faint FEN1 foci, suggesting a partial defect in the recruitment of FEN1 to these replication foci (Figures 1A-1D). Of the remaining 45% of WT/FFAA nuclei, 25% had a normal FEN1/PCNA and FEN1/BrdU co-localization pattern (Figures 1B and 1D), but the other 20% had few FEN1 foci that co-localized to the PCNA and BrdU foci (Figures 1B and 1D). In addition, we found that the FFAA mutation did not change FEN1 protein levels in the cytoplasm or nucleus (Supplementary information, Figure S1). Together, these findings indicated that the WT/FFAA cells displayed a partial defect in FEN1 localization to replication foci.
Figure 1.
FFAA FEN1 mutation causes defective PCNA-coordinated Okazaki fragment maturation. (A-D) WT, WT/FFAA, and FFAA/FFAA MEF cells (Passage 1, first passage after isolation from embryos, for all three genetic backgrounds) were synchronized at the G1/S boundary by treatment with 400 μM mimosine (12 h), which has 40-50% synchronization efficiency 24. Cells were labeled with BrdU (1 h), fixed and co-stained with anti-FEN1 and anti-PCNA (A) or anti-FEN1 and anti-BrdU antibodies (C). Scale bars, 10 μm. Nuclei with different degrees of co-localization were defined as follows: co-localized nuclei, greater than 90% PCNA or BrdU foci with corresponding FEN1 foci (yellow spots in merged views); partially co-localized nuclei, approximately half of the PCNA or BrdU foci fail to correspond with equivalent FEN1 foci as indicated by arrows; little co-localized nuclei, greater than 90% of PCNA or BrdU foci have no corresponding FEN1 foci. The nuclei with varying degrees of FEN1 co-localization with PCNA or BrdU were counted and categorized, and the percentage of each category of nuclei was calculated by dividing the number of nuclei in each category by the total number of nuclei analyzed. More than 100 nuclei were analyzed per genetic background; average values of three independent assays. Data are summarized in (B) or (D). (E) A gap substrate with a 20-nt RNA-DNA flap, which consists of 17 ribonucleotides (represented by the zigzag line) and 3 deoxyribonucleotides, was prepared (upper panel). Nuclear extracts of cells with the indicated genotypes (1 μg each) were incubated with the indicated DNA substrates (1 pmol). All reactions were carried out in a buffer containing 5 μCi [α-32P] dCTP and 50 μM each of dATP, dGTP, and dTTP at 37 °C for 10, 20, 40, and 60 min and analyzed by 15% denaturing PAGE. The histogram on the right shows quantification of ligated products by ImageQuant. Values are mean ± SD of n = 3 assays.
In our recent publication, we showed that nuclear extracts (NEs) from FFAA cells displayed defects in production of fully ligated products in an in vitro Okazaki fragment maturation assay 24. Therefore, we determined if the FFAA mutation also had heterozygous effects on the chemical reactions in Okazaki fragment maturation in vitro. Incubation of WT NEs with a gapped DNA substrate with a 5′ flap produced significant amounts of non-ligated (32-50 nt) and ligated products (80 nt) (Figure 1E). However, the amount of overall product (ligated and non-ligated) produced by WT/FFAA and FFAA/FFAA NEs was considerably less than that produced by the WT NEs (Figure 1E), suggesting that the FFAA mutation affected Okazaki fragment maturation efficiency even in the heterozygous state. We then investigated whether the reduction in ligated products was due to defects in flap cleavage and/or DNA ligation. To assay the flap cleavage activity, we incubated WT, WT/FFAA, or FFAA/FFAA NEs with 5′ flap RNA-DNA flap substrates. We found that these NEs had similar flap endonuclease activity (Supplementary information, Figure S2), suggesting that the FFAA mutation did not result in obvious defects in removal of the flap structure in vitro. We then assayed the DNA end ligation activity of WT, WT/FFAA, and FFAA/FFAA NEs, by incubating the NEs with 3′ end 32P-labeled nicked DNA substrates, in which the 5′ end of the downstream oligonucleotide was phosphorylated to allow direct DNA ligation (Supplementary information, Figure S3). We observed that WT, WT/FFAA, or FFAA/FFAA NEs had little difference in production of ligated DNA on this nicked DNA substrate (Supplementary information, Figure S3). These findings raised the question of how FFAA NEs had decreased efficiency in production of fully ligated products.
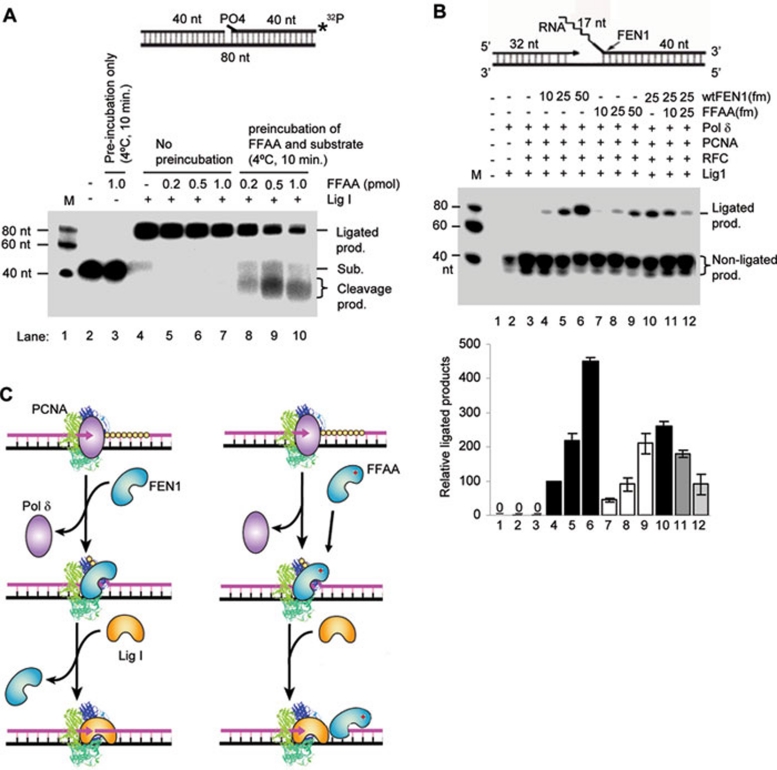
We used partially reconstituted systems to answer this question. First, we sought to confirm if the FFAA FEN1 mutant had no considerable defect in flap cleavage in the absence or presence of PCNA and replication factor C (RFC). We found that it indeed had similar flap cleavage activity to WT FEN1 in the absence or presence of PCNA/RFC (Supplementary information, Figure S4). Consistently, a previous study showed that PCNA mutations abolishing the high-affinity FEN1/PCNA interaction did not affect its stimulation of FEN1 nuclease activity 29. We then assayed whether the FFAA FEN1 mutant protein competed with Lig I for the nicked DNA substrates. There was little inhibition of production of ligated products by FFAA FEN1 when we simultaneously mixed FFAA, Lig I, and nicked DNA substrates (lanes 4-7, Figure 2A). However, when we pre-coated the nicked DNA substrates with FFAA FEN1, which mimics the situation immediately after flap cleavage by FFAA FEN1, DNA substrate-bound FFAA effectively cleaved the nicked DNA substrates with its 5′ exonuclease activity, and these reactions generated considerably less ligated products (lanes 8-9, Figure 2A) than the reactions without pre-coated substrates (lanes 4-7, Figure 2A). To further investigate if the FFAA FEN1 mutant protein causes defects in Okazaki fragment maturation, we reconstituted the chemical reactions in Okazaki fragment maturation. Reconstituted reactions with WT FEN1 effectively produced fully ligated products; however, reactions with the FFAA mutant generated considerably less fully ligated product (Figure 2B). Furthermore, reconstituted reactions with a mixture of WT and FFAA FEN1 mutant, resembling the heterozygous situation, also produced less fully ligated products (Figure 2B). Together, these data indicate the importance of prompt dissociation of FEN1 from the DNA nick site, which is thought to be mediated by PCNA 26, 30, in order to avoid futile cycles of DNA cleavage and ligation attempt. The FFAA FEN1 proteins, which lack high-affinity binding to PCNA, may fail to engage in PCNA-mediated handoff of the nuclease, resulting in unwanted cleavage by the 5′ exonuclease activity of the FEN1 mutant (Figure 2C, see Discussion for more details).
Figure 2.
Purified FFAA FEN1 mutant proteins reduces Okazaki fragment maturation in reconstituted reactions. (A) Nicked DNA substrate-bound FFAA leads to futile DNA end cleavage and inhibits Lig I-driven DNA ligation. To mimic the situation in which FFAA proteins remain bound to DNA substrates immediately after the flap cleavage, we pre-incubated the 3′ end 32P-labeled nick substrate (upper panel) with the indicated amounts of FFAA proteins at 4 °C for 10 min. Purified recombinant Lig I proteins (100 fmol) were added to the mixture, which was then incubated at 37 °C for 15 min (lanes 8-10). In one control reaction (lane 3), FFAA proteins (1 pmol) were incubated with 500 fmol DNA substrates at 4 °C for 10 min, to verify that the pre-incubation did not result in cleavage of DNA substrates by the FFAA proteins. In another set of reactions (lanes 4-7), 0, 0.2, 0.5, or 1 pmol of FFAA FEN1 proteins was mixed with the nicked substrate and Lig I simultaneously. The reactions were carried out at 37 °C for 15 min. All reactions were analyzed by 15% denaturing PAGE. (B) Chemical reactions for Okazaki fragment maturation were reconstituted with purified WT and FFAA FEN1 proteins (as specified), PCNA, Pol δ, RFC and Lig1 (100 fmol each) and gapped flap substrates (500 fmol). Reactions were carried out at 37 °C for 60 min. The bottom panel shows quantification of ligated products by ImageQuant. Values are mean ± SD of n = 3 assays. (C) A model to elucidate how FFAA may inhibit the ligation of Okazaki fragments. Left panel, PCNA mediates the sequential actions of Pol δ, FEN1, and Lig I, possibly via the hand-off mechanism 26, 30. Right panel, non-PCNA-coordinated actions of FFAA, which cause unwanted 5′ end cleavage and futile cycles of gap-filling, DNA cleavage, and ligation.
Spontaneous DNA breaks accumulate in WT/FFAA cells and activate Chk1
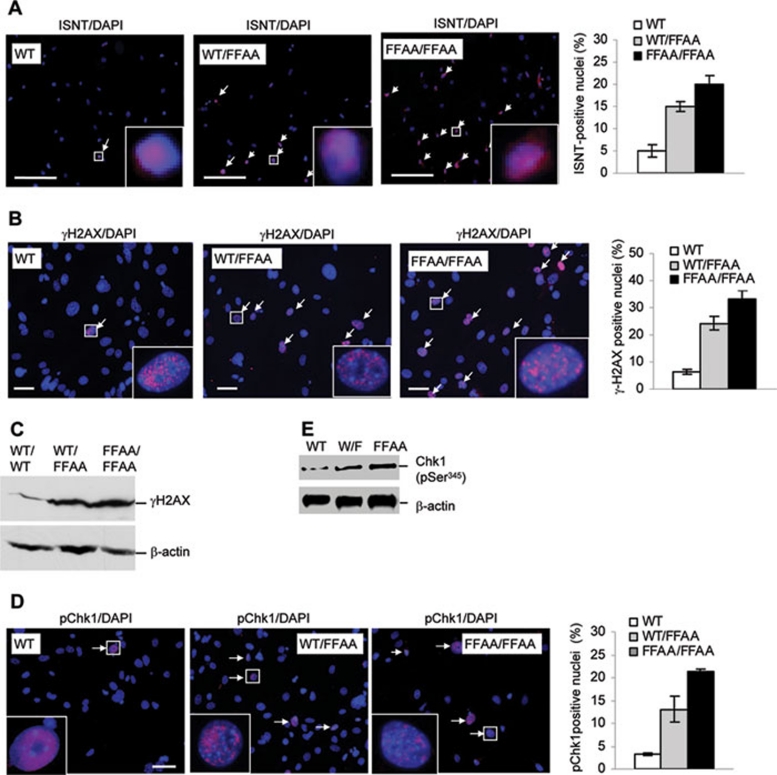
Next, we sought to determine if a delay in the ligation of Okazaki fragments, as suggested by the in vitro assay, caused spontaneous single-stranded DNA breaks (SSBs) and subsequent double-stranded breaks (DSBs). We analyzed formation of spontaneous SSBs and DSBs in primary WT, WT/FFAA, and FFAA/FFAA MEF cells using in situ nick translation (INST) labeling with Rhodamine-dCTP 31 and immunostaining with an antibody to γH2AX (Abcam) 32. The percentages of ISNT-positive nuclei for WT/FFAA and FFAA/FFAA cells were 15 and 20%, respectively, but approximately 5% for WT cells (Figure 3A). In addition, WT/FFAA and FFAA/FFAA cells had 5- and 6-fold higher percentages of γH2AX-positively stained nuclei than WT cells (Figure 3B). Western blotting analysis supported the conclusion that the FFAA mutation led to phosphorylation of H2AX (Figure 3C). These results suggest that the FFAA mutation causes DNA strand breaks, which is consistent with a previous observation that defects in the Okazaki fragment maturation process in rad27 (yeast FEN1 homologue) knockout yeast cells resulted in DNA strand breaks 10.
Figure 3.
The FFAA mutation induces spontaneous DNA damage and activates Chk1. (A, B) In situ detection of DNA strand breaks in MEF cells. Primary WT, WT/FFAA, or FFAA/FFAA cells were fixed in 4% paraformaldehyde. (A) DNA nicks or recessive double-strand DNA breaks were stained with in situ nick translation (ISNT). Briefly, fixed cells were incubated with Klenow fragment, and 1 mM each of rodamine-dCTP, dATP, dGTP, and dTTP in a reaction buffer (50 mM Tris-Cl pH 8.0, 5 mM MgCl2, and 1 mM DTT). Right-bottom image, enlarged view of the boxed area. Scale bars: 300 μm. Right panel, quantification of ISNT staining. The ISNT-positive nuclei were counted and normalized to the total number of DAPI-stained nuclei. (B) Double-strand DNA breaks were also detected with immunostaining using an antibody against γH2AX (pink). Nuclei were stained with DAPI (blue). Nuclei with at least five pink spots in the merged view were scored as positively stained nuclei. Right-bottom image, enlarged view of the boxed area. Scale bars: 50 μm. Right panel, quantification of γ-H2AX staining. The γ-H2AX-positive nuclei were counted and normalized to the total number of DAPI-stained nuclei. (C) Western analysis of γ-H2AX of NEs from WT, WT/FFAA, and FFAA/FFAA MEF cells. (D) Phosphorylated Chk1 (active form) was detected by immunostaining with an antibody against phosphorylated Chk1 (S345). Nuclei were stained with DAPI. Left-bottom image, enlarged view of the boxed area. Scale bars, 60 μm. Right panel, quantification of Chk1-positive nuclei. Ten randomly selected microscopic views were analyzed (150-250 nuclei per slide) for quantification of phosphor-Chk1 positively stained nuclei. To reduce the chance of randomly occurring red fluorescence signals, nuclei with at least 5 pink foci (indicating phosphor-Chk1) in the merged view were scored as positively stained nuclei. Values in (A, B, and D) represent mean ± SD of three distinct primary cell lines of each genetic background. (E) NEs (20 μg) were resolved by 4%-15% SDS-PAGE. Phospho-Chk1 was detected with western blot using an antibody against phosphor-Chk1 (S345). β-Actin was used as an internal control.
DNA damage during DNA replication can activate checkpoint proteins such as Chk1, arresting cells in G2/M phase and allowing cells to repair damage 33. To test whether DNA damage in WT/FFAA cells activated DNA damage checkpoints, we determined the level of phosphorylated Chk1, which is the primary player in response to DNA replication stress and spontaneous DNA damage 33. Immunostaining with an antibody against phosphorylated Chk1 (S345, active form) 34 revealed that approximately 12 and 20% of WT/FFAA and FFAA/FFAA cells had pChk1 positively stained nuclei, compared to 3% for WT cells (Figure 3D). Western blotting analysis confirmed the increase in the level of phosphorylated Chk1 in FFAA cells (Figure 3E). These results suggest that FFAA mutation-induced DNA damage results in Chk1 activation. Consistently, we also observed that the level of phosphorylated ATR, which is a major activator of Chk1 in response to DNA damage 35, was greater in WT/FFAA and FFAA/FFAA cells (Supplementary information, Figure S5).
FFAA causes defects in long-patch base excision repair
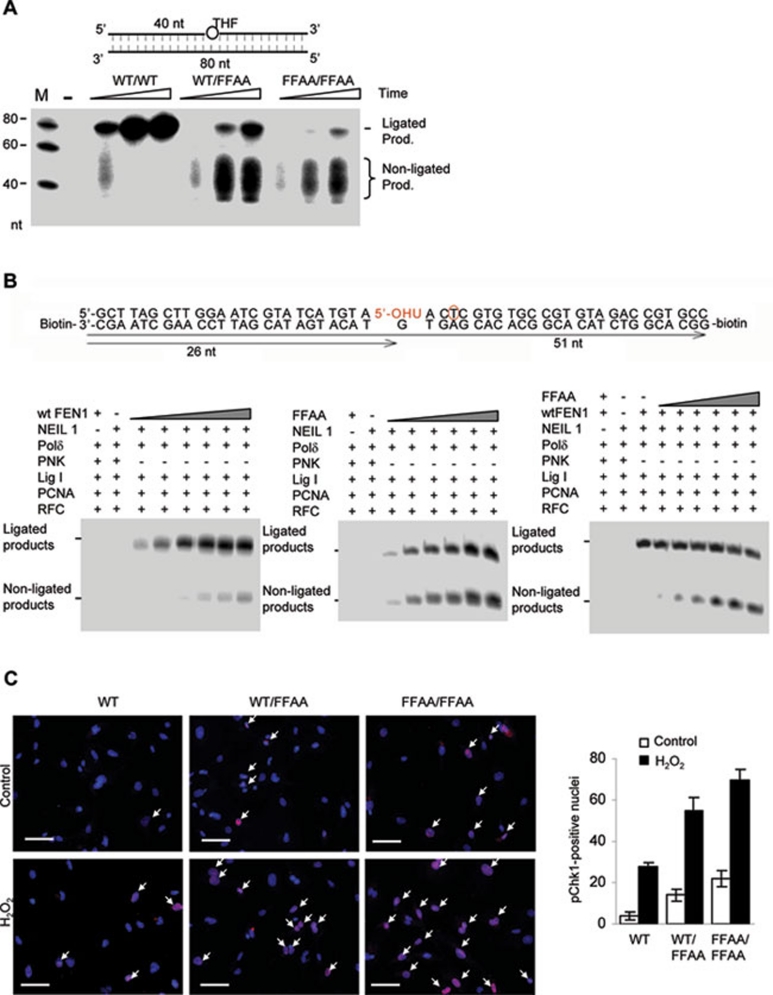
To determine if the FFAA FEN1 mutation caused any defect in LP-BER, we prepared NEs from WT, WT/FFAA, and FFAA/FFAA MEF cells and incubated the NEs with a synthetic LP-BER model substrate: a DNA duplex carrying a tetrahydrofuran (THF)/G pair in the middle. WT NE effectively removed THF, replaced it with 32P-dCTP, and generated a full-length (80 nt) product (Figure 4A). WT/FFAA or FFAA/FFAA NEs could efficiently incorporate 32P-dCTP; however, they produced significantly less ligated products than the WT NE, coincident with accumulation of non-ligated products (Figure 4A). This suggested that the FFAA FEN1 mutant suppressed DNA ligation during LP-BER. To confirm this observation, we reconstituted the reactions with purified LP-BER enzymes and a BER substrate containing a hydroxyuracil (OHU)/G pair, which resembles the real base damage in the cell. To assay the repair of the OHU/G damage by LP-BER, we analyzed the incorporation of 32P-dTTP at the position 2 nt downstream of the OHU lesion (Figure 4B). Reconstituted LP-BER with WT FEN1 efficiently produced full-length repair products (left panel, Figure 4B); however, reconstituted LP-BER with the FFAA FEN1 mutant accumulated significant amounts of non-ligated products (middle panel, Figure 4B). Furthermore, inclusion of the FFAA FEN1 mutant proteins in the reconstituted LP-BER with WT FEN1 resulted in accumulation of non-ligated products but a decrease in the ligated product (right panel, Figure 4B), suggesting a heterozygous effect of the FFAA mutation in suppressing DNA ligation, similar to what was observed on gapped flap substrates (Figure 2B).
Figure 4.
The FFAA FEN1 mutation impairs LP-BER. (A) LP-BER assays on NEs from WT, WT/FFAA, and FFAA/FFAA MEF cells with a model LP-BER substrate carrying the THF lesion (circle) (upper panel). Reactions were carried out at 37 °C for 10, 30, and 60 min and analyzed by 15% denaturing PAGE. (B) LP-BER reconstitution with purified WT and FFAA FEN1 proteins. Gapped flap substrates (500 fmol) were mixed with Neil1, PNK (polynucleotide kinase), PCNA, Pol δ, RFC, and Lig1 (100 fmol each) and 0, 10, 20, 50, 100, 200, or 500 fmol WT (left panel) or FFAA (middle panel) FEN1 proteins. To test the dominant-negative effect of FFAA, the LP-BER was reconstituted with 50 fmol WT FEN1 plus 10, 20, 50, 100, 200, or 500 fmol FFAA proteins. All reactions were carried out for 60 min. (C) Activation of Chk1 spontaneously or in response to H2O2 treatment. Primary WT, WT/FFAA, or FFAA/FFAA cells were treated with 1 mM H2O2 for 15 min or left untreated. The cells were washed and fixed in 4% paraformaldehyde. Those with the activated form of Chk1 were detected with immunostaining using an antibody against phosphorylated Chk1 (p-Ser354) (pink). Nuclei were stained with DAPI. Pink spots were scored as positive-stained nuclei. Scale bars: 100 μm.
We next tested if the FFAA-induced defects in LP-BER caused cells to be sensitive to DNA base damage. We treated WT, WT/FFAA, and FFAA/FFAA MEF cells with 1 mM H2O2 and evaluated the status of ATR and Chk1 activation, which is indicative of cell cycle arrest. Treatment with H2O2 activated ATR and Chk1 more in FFAA cells than in WT cells (Figure 4C and Supplementary information, Figure S5). In the presence of H2O2, approximately 55% and 70% of the WT/FFAA and FFAA/FFAA cells were phosphoChk1-positive, compared to 24% of WT cells (Figure 4C). Consistent with this observation, H2O2-induced growth inhibition of WT/FFAA and FFAA/FFAA cells was considerably greater than that of WT cells (data not shown).
Chk1 inhibitors suppress formation of polyploidy
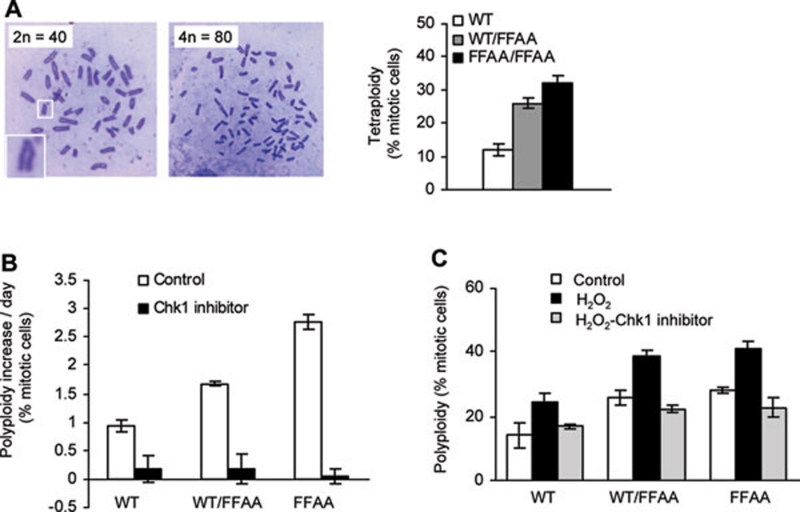
It has been hypothesized that DNA replication and repair defect-induced DNA damage may disturb chromosome segregation and cytokinesis, and cause tetraploidy and aneuploidy 36, 37. To test this hypothesis, we counted the chromosome number in primary WT and FFAA cells freshly isolated from embryos (E13.5, P1). The percentages of WT/FFAA and FFAA cells with near-tetraploid aneuploidy were approximately 25% and 30%, compared to approximately 12% for WT cells (Figure 5A). We further hypothesized that aneuploidy in FFAA cells was caused by DNA damage-activated Chk1, which arrests the cells at G2/M phase, in which chromosomes have been duplicated but chromosome segregation and/or cell division is blocked. The cells might be reprogrammed to replicate their genome, skipping mitosis and cytokinesis, leading to tetraploidy. Supporting this hypothesis, we observed that considerably greater numbers of WT/FFAA and FFAA/FFAA cells than WT cells accumulated in prophase (Supplementary information, Figure S6), and consistently fewer WT/FFAA and FFAA/FFAA cells than the WT cells were detected at anaphase (Supplementary information, Figure S6).
Figure 5.
FFAA mutant cells become polyploid in a Chk1-activation-dependent manner. (A) Chromosomes were stained with Giemsa solution and replicated chromosomes in M phase cells were counted (100-200 cells per slide). Diploid mouse cells have 40 sister chromatids and tetraploid have 80 sister chromatins for mouse cells. The square box in the lower-left corner shows the V-shape of replicated chromosomes. The percentage of near-tetraploid cells (chromosomes > 70) in WT, WT/FFAA, and FFAA/FFAA primary culture (passage 1) was quantified (right panel). (B) Inhibition of Chk1 suppresses spontaneous polyploidy. Primary MEF cells (106), in which the percentage of polyploidy was determined by chromosome counting (#P0), were seeded for approximately 18 h and treated with the Chk1 inhibitor SB218078 56 (5 h) or left untreated (day 1). Cells were washed and cultured for 2 days (day 2 and day 3). Treatment with the corresponding inhibitor was repeated once. Cells were cultured for approximately 15 h and treated with colcemid for 4 h (day 4). The metaphase spread was prepared and chromosome number was counted to determine the percentage of polyploidy (#P1) for each cell culture. The polyploidy increase per culture day was expressed as the difference of #P1 and #P0 divided by the number of cultured days (4 days). (C) Inhibition of Chk1 abolishes H2O2-induced polyploidy formation. MEF cells were left untreated or treated with 0.5 mM H2O2 (15 min). H2O2-treated cells were washed and incubated in DMEM containing 1 μg/ml SB218078 (5 h). SB 218078-treated or -untreated cells were washed and incubated for additional 15 h. The chromosome number was counted and the percentage of near polyploidy was calculated. In each panel, values are mean ± SD of n = 3 independent experiments.
We then tested whether disruption of G2/M phase arrest by Chk1 inhibitors abolished the formation of tetraploidy, while elevation of DNA damage by treatment with DNA-damaging agents promoted tetraploidization during cell proliferation. We evaluated the rate of spontaneous near-tetraploid aneuploidy formation per culture day in WT and FFAA cells in the absence or presence of the Chk1 inhibitor SB218078 (TOCRIS). In the absence of the Chk1 inhibitor, the polyploidy population in WT/FFAA or FFAA/FFAA cells increased by approximately 1.7 and 2.7% per culture day, compared to 1.0% in the WT (Figure 5B). However, the Chk1 inhibitor prevented the formation of polyploidy, regardless of the genetic background (Figure 5B). We then treated cells with H2O2, a typical DNA strand break inducer. We found that H2O2, which dramatically activates Chk1 (Figure 4C), enhanced polyploidy formation in WT, WT/FFAA, and FFAA/FFAA cells (Figure 5C). The tetraploidy level positively correlated to the number of cells with Chk1 activation (Figure 5C). However, inhibition of Chk1 by SB218078 suppressed the H2O2-induced polyploidization (Figure 5C). Taken together, these findings suggest that the FFAA mutation causes spontaneous and chemical-induced near-tetraploid aneuploidy in a Chk1 activation-dependent manner.
WT/FFAA cells display high transformation frequency
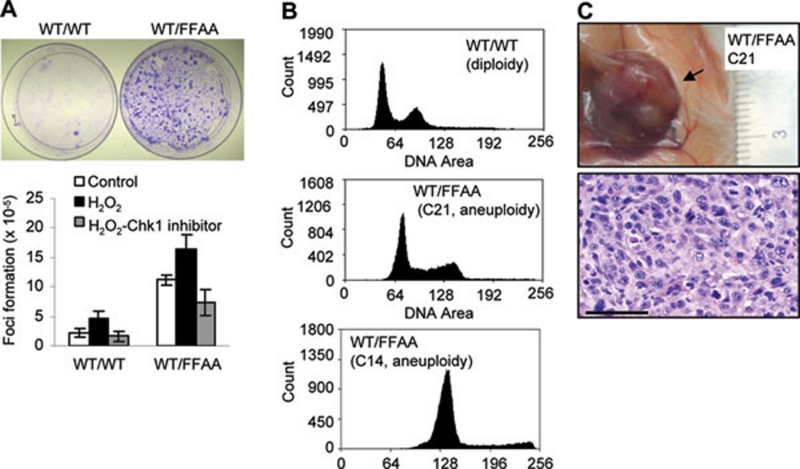
To test if the FFAA mutation promotes tumorigenesis, we evaluated the cellular transformation frequency with a focus formation assay (Figure 6A). WT/FFAA MEF cells had six-fold greater formation of colonies than the WT cells (Figure 6A). However, few visible colonies were observed with FFAA/FFAA cells. It is possible that the severe defects in DNA replication and cellular proliferation observed in FFAA homozygous mutant cells 24 could limit the expansion of FFAA/FFAA transformed cells, thus preventing colony formation.
Figure 6.
Polyploidy promotes cellular transformation and tumor formation. (A) MEF cells were left untreated or treated with 0.5 mM H2O2 (15 min). H2O2-treated cells were incubated in DMEM containing 1 μg/ml Chk1 inhibitor SB218078 (5 h), washed and replaced with fresh DMEM. All cells were incubated for an additional 24 h. MEF cells were trypsinized and seeded onto a new dish (105 cells per dish) for the colony-focus formation assay. Upper panels, colony-focus formation assays. Each focus (blue) was scored as a transformed cell, which was normalized to the total number of cells plated; mean ± SD is summarized in the lower panel, n = 3 independent experiments. (B) Representative graphs showing the DNA contents of transformed WT/FFAA cells as analyzed with flow cytometry (lower two panels) and compared to the diploid control (upper panel). (C) Anatomic and histological analysis of tumors in NOD/SCID mice injected with transformed WT/FFAA cells. Histological examination indicated that the tumor was a subcutaneous sarcoma. Scale bar: 200 μm.
Next, we determined the ploidy status of these transformed cells, which were derived from a single cell and were able to endlessly proliferate. We found that they displayed near-polyploid aneuploidy (Figure 6B). In addition, chromosome number counting on chromosome spreads of 10 independent WT/FFAA transformed cell lines confirmed that all of these lines were near-polyploid aneuploidy (data not shown). These findings suggested that WT/FFAA cells with near-polyploid aneuploidy were able to undergo cellular transformation and outgrow the diploid cells, leading to clonal expansion. To further test whether aneuploidy contributed to the cellular transformation, we treated cells with H2O2, which enhanced the aneuploidy level, and enhanced cellular transformation, particularly in the WT/FFAA cells (Figure 6A). More importantly, H2O2-induced cellular transformation in the WT/FFAA cells was completely suppressed by the Chk1 inhibitor SB218078 (Figure 6A). To test if these transformed near-polyploid cells could generate tumors in vivo, we subcutaneously grafted near-polyploid WT/FFAA cells into NOD-SCID mice. All clones of near-polyploid cells (n = 10) injected into NOD-SCID mice progressed to subcutaneous sarcoma (Figure 6C), but in the control (3 lines of WT and 3 lines of WT/FFAA with 2n) none of diploid cells advanced to cancer, indicating that the transformed polyploid cells were indeed cancerous in vivo (Figure 6C).
WT/FFAA mice develop aneuploidy-associated cancer
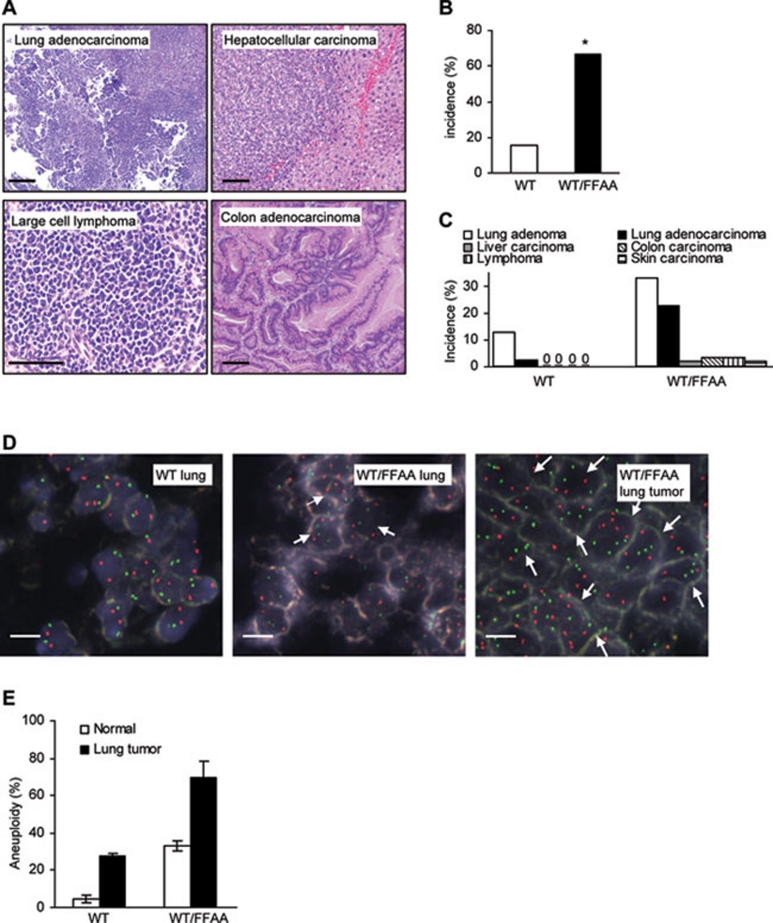
We then determined if FFAA-induced DNA damage and polyploidy/aneuploidy result in higher susceptibility to the development of spontaneous tumors. WT/FFAA mice had a near-normal life span of approximately 20 months, which was slightly less than that of their WT littermates (22 months). However, 67% of WT/FFAA mice (16-20 months, n = 57, P < 0.0001) spontaneously developed tumors, compared to their WT littermates (15%, n = 39) (Figures 7A and 7B). Lung adenoma (31%) and adenocarcinoma (22%) were the most common tumors in the WT/FFAA mice (Figure 7C). We postulate that the relatively high incidence of lung cancer is likely due to the genetic background, as WT mice also had lung adenoma but no other types of cancer. In addition, the FFAA mutant mice had an increased incidence of other cancers, suggesting the FFAA-induced cancer phenotype is not lung cancer-specific.
Figure 7.
WT/FFAA mice develop aneuploid tumors. (A) Histology (H&E) of different tumors that developed in WT/FFAA mice. Scale bar: 200 μm. (B) Overall cancer incidence of WT and WT/FFAA mice (16-20 months). *P < 0.0001 Fisher exact. (C) Cancer incidence in major organs of WT and WT/FFAA mice. (D) Micrographs of dual-color interphase FISH (probes 2qA1 (red) and 8qA1 (green)) of WT and WT/FFAA normal lung tissue and a WT/FFAA lung cancer (5-μm tissue section). Red or green spots indicate chromosome 2 or 8. Aneuploid cells are specified by arrows. (E) Quantification of aneuploidy (chromosome gain). Cells were isolated from a lung tumor and subjected to metaphase spread preparation. Chromosome number in mitotic cells was quantified and normalized to the total number of mitotic cells.
Dual-color FISH confirmed that polyploidy and aneuploidy occurred in normal WT/FFAA lung tissue (10 months, Figure 7D). The polyploid and aneuploid cells expanded and became the major population in WT/FFAA tumors (Figure 7D). To further confirm the distribution of chromosome numbers of the cells in normal and lung cancer tissues of WT and WT/FFAA mice, we isolated cells from fresh normal and lung cancer tissues and counted their chromosome number. Normal WT/FFAA lung tissues had significantly more aneuploid cells than the WT (Figure 7E). Furthermore, more than 85% of WT/FFAA lung cancer cells were aneuploid (Figure 7E), indicating that the FFAA mutant mice develop aneuploidy-associated cancer.
Discussion
We have demonstrated the biological significance of the interaction between FEN1 and PCNA in DNA replication and repair. One important role of the FEN1/PCNA interaction is that it allows FEN1 to be efficiently recruited to the replication or repair machinery. Supporting this view, the FFAA FEN1 mutation disrupting the high-affinity binding to PCNA results in fewer number of discrete FEN1 foci that co-localize to PCNA or BrdU foci. This may lead to temporary deficiency of FEN1 for the removal of the RNA-DNA flap during Okazaki fragment maturation in vivo. However, we by no means suggest that the FFAA FEN1 mutation causes a complete FEN1 deficiency at the replication site or failure to remove RNA primers in the FFAA cells. Indeed, the FFAA FEN1 mutant on its own can effectively bind to and cleave the DNA flap and nick substrates in vitro 27 (Supplementary information, Figure S4). In addition, the crystal structure of the FEN1/PCNA complex reveals other interfaces between FEN1 and PCNA, besides the high-affinity PIP box 38. Thus, the FFAA FEN1 protein may still be able to interact with PCNA, albeit at a much lower affinity. This explains why the mutant FEN1 cannot form foci or be pulled down with PCNA, but can still be stimulated by PCNA. A similar study revealed that the PCNA mutations, which disrupt the high-affinity FEN1/PCNA interaction, did not affect its stimulation of FEN1 nuclease activity 29. Unlike FEN1 knockout cells, which have a complete cell proliferation failure, the FFAA/FFAA MEF cells can proliferate, albeit at a reduced rate (60% of the rate of WT cells) 24. We suggest that disruption of the high-affinity FEN1/PCNA interaction affects the efficiency of FEN1 recruitment to the DNA replication and/or repair site but not its nuclease activity. Due to the cellular compartment effect, this may cause a temporary FEN1 deficiency at the DNA replication site, leading to a delay in the processing of the RNA-DNA flap structure in Okazaki fragment maturation or the short flap structure in LP-BER.
Our data also suggest another role of the FEN1/PCNA interaction – to ensure the timely dissociation of FEN1 from DNA nicks. Since FEN1 is both a flap endonuclease and 5′ exonuclease, after flap cleavage, the nicked DNA-bound FEN1 will continue to remove nucleotides from the 5′ end of DNA at the nick site. This may cause futile cycles of gap-filling and 5′ end cleavage, and ligation of DNA fragments 39, 40. Previous structural studies of the FEN1/PCNA or the FEN1/DNA complex suggested that PCNA mediates the hand-off of FEN1 from the nicked DNA, once flap cleavage is completed (left panel, Figure 2C) 26, 30. The FFAA FEN1 mutant protein, which loses the high-affinity site for the PCNA interaction but retains the DNA substrate-binding capacity and nuclease activities (Supplementary information, Figure S4) 27, may bind to and cleave the flap structure on its own or through a low-affinity PCNA interaction (Figure 2C). Nevertheless, PCNA fails to hand off the FFAA FEN1 mutant protein after flap cleavage. As a consequence, the FFAA mutant protein may remove a few nucleotides from the 5′-end nicked DNA ends, leading to futile cycles of gap filling, DNA cleavage and ligation (Figure 2C), regardless of whether WT FEN1 proteins are present. This also explains why the FFAA mutation has a strong heterozygous effect, which seems to be a dominant-negative effect. These findings are consistent with our recent studies showing that FEN1 methylation and phosphorylation mediate the timely binding to or dissociation from PCNA and DNA substrates, and that FEN1 mutations that abolish its methylation and phosphorylation affect the FEN1/PCNA interaction and DNA replication, even in the presence of WT FEN1 41.
Defects either in RNA-DNA flap cleavage due to failure of FEN1 recruitment or in DNA ligation because of the futile cycles may result in un-ligated single-stranded DNA breaks, which may be further converted into double-strand breaks 42. DNA strand breaks that accumulated in FFAA cells spontaneously or in response to chemical treatments activated G2/M checkpoints such as Chk1, and activation of Chk1 in the FFAA cells arrested cell cycle progression at the G2/M phase, which is critical for the avoidance of the transmission of DNA mutations to daughter cells 43. However, such mutation avoidance mechanisms have an unwanted expense, which is the formation of tetraploidy and aneuploidy. Our data indicate that activation of Chk1 is a key molecular event in the road to DNA damage-induced tetraploidization. We found that inhibition of Chk1 activation efficiently suppresses spontaneous and H2O2-induced polyploidy formation. This study supports the previous hypothesis indicating that DNA damage due to DNA replication or repair defects may act as signals, which activate checkpoints and perturb chromosome segregation and cell division 36, 37. The arrested G2 cells may be directly reprogrammed to initiate DNA replication via the endoreduplication mechanism and become tetraploid or polyploid, similar to what was observed in Emi1 depletion cells 44. If cells are aberrantly arrested during mitosis, they may become tetraploid via the mitotic slippage mechanism 45. It is commonly thought that tetraploidy is an unstable genetic state and subsequently evolves into aneuploidy, which is a hallmark of human cancer 36, 37, 46. Supporting this hypothesis, transformed or cancer cells under culture conditions or in vivo are predominantly aneuploid.
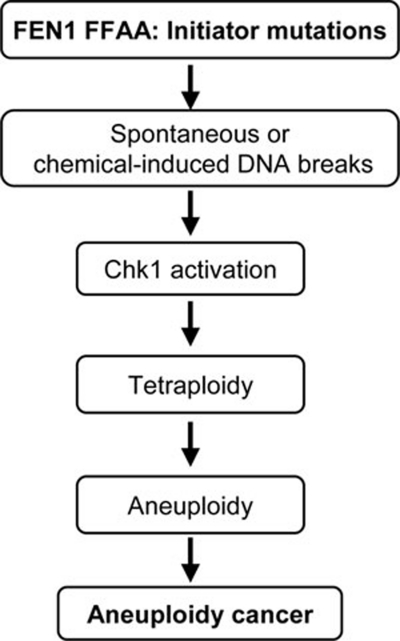
Furthermore, in vitro and in vivo characterizations of mutant cells and mice carrying the FFAA FEN1 mutation link aneuploidy and cancer development. These studies also define a new pathway for cancer development due to FEN1 mutations (Figure 8). We previously demonstrated that human FEN1 mutations, by abolishing the exonuclease activity, caused a strong mutator phenotype, which drives cancer development 6. However, mutation rate analysis of WT and FFAA cells revealed a similar mutation frequency in the WT/FFAA and FFAA/FFAA cells as for the WT cells (data not shown), suggesting that the FFAA mutation does not result in a mutator phenotype. Instead, the FFAA mutation induces formation of near-polyploid aneuploidy. The FFAA polyploid cells have great potential to undergo neoplastic transformation. Exposure of the FFAA cells to DNA-damaging agents induced aneuploidy and considerably enhanced cell transformation. Conversely, suppression of aneuploidy formation by a Chk1 inhibitor led to inhibition of cellular transformation of the FFAA cells. Moreover, the FFAA mice developed aneuploidy-associated cancer. This is consistent with many recent studies on aneuploidy and cancer development. For example, Pellman's group showed that p53-null cells with polyploid DNA contents have a considerably higher transformation frequency than diploid p53-null cells 47. Mutant mice that are deficient in mitotic checkpoints exhibit aneuploidy formation and spontaneous tumorigenesis 48, 49. These observations support the long-standing hypothesis that DNA damage may result in aneuploidy, which promotes tumorigenesis 36, 37, 50, 51.
Figure 8.
A model illustrating the path from FFAA mutant cells to aneuploidy-associated cancer cells. The FEN1 FFAA mutation-induced DNA strand breaks induce genome instabilities, spontaneously or in response to chemical treatments, which may directly contribute to cancer initiation. In addition, DNA damage activates Chk1, which arrests the cell cycle at G2/M phase. This, in turn, increases the chance of tetraploidy and aneuploidy, which have been linked to cancer development and progression.
Materials and Methods
In vitro assays of Okazaki fragment maturation
NEs from MEF cells were prepared as previously described 7. FEN1 and the FFAA FEN1 mutant, PCNA, Pol δ, and RFC were expressed and purified following published protocols 27, 52, 53. Ligase I proteins were purchased from Axxora Platform (San Diego, CA, USA). Okazaki fragment maturation reactions were reconstituted or assayed with NEs using a gapped DNA substrate with a DNA-RNA flap, as previously described 24, 54. NEs (1 μg) from each cell line or purified proteins, as indicated in the figure legend, were incubated with DNA substrates (500 fmol) in reaction buffer (50 mM HEPES-KOH (pH 7.5), 45 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 2 mM ATP, 200 unit creatine-phosphokinase, 0.5 mM NAD, and 5 mM phosphocreatine) containing 5 μCi 32P-dCTP and 50 μM each of the other three dNTPs. The reactions were carried out at 37 °C and the products analyzed by 15% denaturing PAGE and autoradiography.
In vitro LP-BER assays
LP-BER activity of NEs was assayed using a synthetic DNA duplex (80 nt) carrying a THF/G pair in the middle 54. NEs (10 μg) from each cell line were incubated with DNA substrates (500 fmol) in reaction buffer (50 mM HEPES-KOH (pH 7.5), 45 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 2 mM ATP, 200 unit creatine-phosphokinase, 0.5 mM NAD, and 5 mM phosphocreatine) containing 5 μCi 32P-dCTP and 50 μM each of the other three dNTPs. For reconstitution of LP-BER using the 5′-OHU-containing DNA substrate (51 nt), specific amounts of WT FEN1 or FFAA and other purified BER proteins, as indicated in the figure legend, were mixed with the DNA substrate and 5 μCi 32P-dTTP and 50 μM each of the other three dNTPs. In both assays, reactions were carried out at 37 °C and the incorporation of 32P-dCTP or -dTTP and formation of the full-length product were analyzed by 15% denaturing PAGE and autoradiography.
In vitro assays of Flap endonuclease and DNA ligase activities
The flap endonuclease activity was assayed with 5′-end 32P-labeled flap DNA substrates, which were prepared following a previously described protocol 27. The DNA ligase activity was assayed with 3′-end 32P-labeled nicked DNA substrates, containing a phosphate group at the 5′ end of the downstream oligonucleotide to allow immediate ligation reactions. To carry out the reactions, the NEs or a reconstituted mixture of purified FEN1, Lig I, and/or other required proteins were incubated with the flap DNA substrate (flap cleavage assay) or the nicked substrate (DNA ligation assay) in the reaction buffer (50 mM HEPES-KOH (pH 7.5), 45 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, 2 mM ATP, 200 unit creatine-phosphokinase, 0.5 mM NAD, and 5 mM phosphocreatine). All reactions were incubated at 37 °C for 15 min. The cleavage or ligation products were analyzed by 15% denaturing PAGE and visualized by autoradiography.
Metaphase spread preparation and analysis
Cells with or without colcemid treatment (0.1 μM, 2 h) were harvested, treated with hypotonic solution (75 mM KCl), fixed with Carnoy's solution, and mounted on a slide, which was then baked (65 °C, overnight), stained with a Giemsa solution, scanned under a microscope for mitotic cells and imaged using Image Pro 6.3 (Media Cybernetics, Inc, Bethesda, MD, USA). The chromosome number of each mitotic cell was analyzed and scored using Image Pro 6.3; typically 100-200 mitotic cells were analyzed.
Immunofluorescence staining
FEN1, PCNA, and BrdU foci were stained following a previously described protocol 24. To stain for phosphor-Chk1 or γH2AX, cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X100, blocked with Image iT FX signal enhancer (Invitrogen), and incubated with the indicated antibodies. Antibodies against phosphor-Chk1 (S345) and γH2AX were from Abcam (Cambridge, MA, USA). Slides were extensively washed with PBS buffer and then incubated with fluorescence-labeled secondary antibodies. Nuclei were stained with DAPI (200 ng/ml). All slides were examined by fluorescence microscopy (AX70, Olympus, Center Valley, PA, USA).
Cell transformation assay
The transformation mouse primary cells were assayed with a focus-forming assay 55. Primary MEF cells (with or without treatments as indicated in the figure legends) were plated onto the 10-cm dish, incubated (15-25 days), stained with Giemsa solution, and the number of spontaneous colonies were scored under a microscope. To culture WT or WT/FFAA colonies, cells from the colonies were picked and transferred to a 12-well plate. After 1 week cells were trypsinized, diluted and transferred to 96-well plates, approximately one cell per well. Transformed cells from a single cell were expanded and subjected to analysis.
Histopathology
Tissues were fixed in 10% formalin and tissue sections stained with hematoxylin and eosin (H&E). Slides were analyzed in a double-blind fashion. All protocols that involved animals were approved by the Research Animal Care Committee of the City of Hope in compliance with the Public Health Service Policy of the United States.
Acknowledgments
We thank V Bedell, D Bobadilla, J Collins, and M Slovak at the cytogenetic core-facility of COH for technical assistance with the karyotype analysis of mouse MEF cells and FISH detection of aneuploid cells in paraffin-embedded tissue sections of normal and lung tumor tissues. We thank QM Chen (University of Arizona) for critical review of the manuscript and SR da Costa (City of Hope) for editorial assistance. This work was supported by NIH grants R01 CA085344 and R01 CA073764 to BHS and in part by R01 CA081063 to SM and R01 CA084469 to GPP and by the Lung Cancer Program of City of Hope's Comprehensive Cancer Center.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Levels of cytoplasmic and nuclear FEN1 were similar in WT, WT/FFAA, and FFAA/FFAA MEF cells.
FFAA nuclease extracts (NEs) displayed similar flap cleavage activities to the WT NEs.
FFAA nuclease extracts (NEs) had similar ligation activities to the WT NEs on the nicked DNA substrates.
Purified recombinant FFAA FEN1 proteins have similar FEN1 activity as the WT FEN1.
ATR was activated in the FFAA cells spontaneously or in response to treatments with H2O2.
The FFAA mutation induced cell cycle arrest at the prophase.
References
- Bambara RA, Murante RS, Henricksen LA. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Yang C, Shen B, Xue D. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J. 2003;22:3451–3460. doi: 10.1093/emboj/cdg320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Zheng L, Chavez V, Qiu J, Shen B. Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n- and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J Biol Chem. 2007;282:3465–3477. doi: 10.1074/jbc.M606582200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Chai Q, et al. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 2005;6:83–89. doi: 10.1038/sj.embor.7400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Kovvali GK, Prakash L, Prakash S. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- Reagan MS, Pittenger C, Siede W, Friedberg EC. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Filosi N, Gaida GM, Kolodner RD. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- Kucherlapati M, Yang K, Kuraguchi M, et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc Natl Acad Sci USA. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen E, Gran C, Saether BE, Seeberg E, Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol Cell Biol. 2003;23:5346–5353. doi: 10.1128/MCB.23.15.5346-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas EE, Zhu FX, Biswas SB. Stimulation of RTH1 nuclease of the yeast Saccharomyces cerevisiae by replication protein A. Biochemistry. 1997;36:5955–5962. doi: 10.1021/bi962890u. [DOI] [PubMed] [Google Scholar]
- Chai Q, Zheng L, Zhou M, Turchi JJ, Shen B. Interaction and stimulation of human FEN-1 nuclease activities by heterogeneous nuclear ribonucleoprotein A1 in alpha-segment processing during Okazaki fragment maturation. Biochemistry. 2003;42:15045–15052. doi: 10.1021/bi035364t. [DOI] [PubMed] [Google Scholar]
- Li X, Li J, Harrington J, Lieber MR, Burgers PM. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- Siegal G, Turchi JJ, Myers TW, Bambara RA. A 5′ to 3′ exonuclease functionally interacts with calf DNA polymerase epsilon. Proc Natl Acad Sci USA. 1992;89:9377–9381. doi: 10.1073/pnas.89.20.9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX, Biswas EE, Biswas SB. Purification and characterization of the DNA polymerase alpha associated exonuclease: the RTH1 gene product. Biochemistry. 1997;36:5947–5954. doi: 10.1021/bi962889v. [DOI] [PubMed] [Google Scholar]
- Brosh RM Jr, von Kobbe C, Sommers JA, et al. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. doi: 10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- Wang W, Brandt P, Rossi ML, et al. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc Natl Acad Sci USA. 2004;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharia A, Guittat L, Crocker S, et al. Flap endonuclease 1 contributes to telomere stability. Curr Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathi S, Bhusari A, Shen B, Chai W. Human flap endonuclease I is in complex with telomerase and is required for telomerase-mediated telomere maintenance. J Biol Chem. 2009;284:3682–3690. doi: 10.1074/jbc.M805362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A, Shin JH, Kim DH, et al. Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1. J Biol Chem. 2008;283:20925–20936. doi: 10.1074/jbc.M802696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Dai H, Qiu J, Huang Q, Shen B. Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice. Mol Cell Biol. 2007;27:3176–3186. doi: 10.1128/MCB.01652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li J, Li X, Hsieh CL, Burgers PM, Lieber MR. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–2043. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapados BR, Hosfield DJ, Han S, et al. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- Frank G, Qiu J, Zheng L, Shen B. Stimulation of eukaryotic flap endonuclease-1 activities by proliferating cell nuclear antigen (PCNA) is independent of its in vitro interaction via a consensus PCNA binding region. J Biol Chem. 2001;276:36295–36302. doi: 10.1074/jbc.M103397200. [DOI] [PubMed] [Google Scholar]
- Gary R, Park MS, Nolan JP, et al. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol Cell Biol. 1999;19:5373–5382. doi: 10.1128/mcb.19.8.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes XV, Burgers PM. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 2000;19:3811–3821. doi: 10.1093/emboj/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore AS, Kilkenny ML, Jones SA, et al. Structure of an archaeal PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res. 2006;34:4515–4526. doi: 10.1093/nar/gkl623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara Y, Anai H, Kusumoto T, Sakaguchi Y, Sugimachi K. Nick translation detection in situ of cellular DNA strand break induced by radiation. Am J Pathol. 1989;134:7–10. [PMC free article] [PubMed] [Google Scholar]
- Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci USA. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- Capasso H, Palermo C, Wan S, et al. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J Cell Sci. 2002;115:4555–4564. doi: 10.1242/jcs.00133. [DOI] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- Sakurai S, Kitano K, Yamaguchi H, et al. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricksen LA, Veeraraghavan J, Chafin DR, Bambara RA. DNA ligase I competes with FEN1 to expand repetitive DNA sequences in vitro. J Biol Chem. 2002;277:22361–22369. doi: 10.1074/jbc.M201765200. [DOI] [PubMed] [Google Scholar]
- Ranalli TA, Tom S, Bambara RA. AP endonuclease 1 coordinates Flap endonuclease 1 and DNA ligase I activity in long patch base excision repair. J Biol Chem. 2002;277:41715–41724. doi: 10.1074/jbc.M207207200. [DOI] [PubMed] [Google Scholar]
- Guo Z, Zheng L, Xu H, et al. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat Chem Biol. 2010;6:766–773. doi: 10.1038/nchembio.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zheng L, Dai H, Zhou M, Hua Y, Shen B.Chemical-induced cancer incidence and underlying mechanisms in Fen1 mutant mice Oncogene 2010. Oct 25. doi: 10.1038/onc.2010.482 [DOI] [PMC free article] [PubMed]
- Walworth NC, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007;21:184–194. doi: 10.1101/gad.1495007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Palazzo RE. Colcemid and the mitotic cycle. J Cell Sci. 1992;102 (Part 3):387–392. doi: 10.1242/jcs.102.3.387. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. Does aneuploidy cause cancer. Curr Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Boveri T.The Origin of Malignant Tumors. Baltimore: Williams and Wilkins, 1929
- Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- Ellison V, Stillman B. Reconstitution of recombinant human replication factor C (RFC) and identification of an RFC subcomplex possessing DNA-dependent ATPase activity. J Biol Chem. 1998;273:5979–5987. doi: 10.1074/jbc.273.10.5979. [DOI] [PubMed] [Google Scholar]
- Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J Biol Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweasy JB, Lang T, Starcevic D, et al. Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. Proc Natl Acad Sci USA. 2005;102:14350–14355. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JR, Gilmartin A, Imburgia C, Winkler JD, Marshall LA, Roshak A. An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res. 2000;60:566–572. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Levels of cytoplasmic and nuclear FEN1 were similar in WT, WT/FFAA, and FFAA/FFAA MEF cells.
FFAA nuclease extracts (NEs) displayed similar flap cleavage activities to the WT NEs.
FFAA nuclease extracts (NEs) had similar ligation activities to the WT NEs on the nicked DNA substrates.
Purified recombinant FFAA FEN1 proteins have similar FEN1 activity as the WT FEN1.
ATR was activated in the FFAA cells spontaneously or in response to treatments with H2O2.
The FFAA mutation induced cell cycle arrest at the prophase.