Abstract
TCP proteins constitute a family of plant transcription factors with more than 20 members in angiosperms. They can be divided in two classes based on sequence homology and the presence of an insertion within the basic region of the TCP DNA binding and dimerization domain. Here, we describe binding site selection studies with the class I protein TCP16, showing that its DNA binding preferences are similar to those of class II proteins. Through sequence comparison and the analysis of mutants and chimeras of TCP16, TCP20 (class I), and TCP4 (class II), we established that the identity of residue 11 of the class I TCP domain or the equivalent residue 15 of the class II domain, whether it is Gly or Asp, determines a preference for a class I or a class II sequence, respectively. Footprinting analysis indicated that specific DNA contacts related to these preferences are established with one of the strands of DNA. The dimerization motif also influences the selectivity of the proteins toward class I and class II sequences and determines a requirement of an extended basic region in proteins with Asp-15. We postulate that differences in orientation of base-contacting residues brought about by the presence of either Gly or Asp are responsible for the binding site preferences of TCP proteins. Expression of repressor forms of TCP16 with Asp-11 or Gly-11 differently affects leaf development. TCP16-like proteins with Asp-11 in the TCP domain arose in rosids and may be related to developmental characteristics of this lineage of eudicots.
Keywords: Arabidopsis, Development, DNA-binding Protein, Plant, Transcription Factors
Introduction
TCP proteins are plant transcription factors that contain the TCP domain, a conserved domain involved in DNA binding and dimerization (1). The N-terminal portion of the TCP domain is enriched in basic amino acids and is followed by a region that is predicted to contain two amphipathic α-helices connected by a disordered loop (2). These features give the TCP domain a resemblance with the bHLH5 domain present in eukaryotic transcription factors. The basic region, however, differentiates these two structures because this region is longer and contains helix-breaking amino acids in the TCP domain. This makes theoretical predictions about the nature of its contacts with DNA rather inaccurate when bHLH domain-DNA complex structures are used as templates.
A broad separation of TCP domains can be made based on amino acid similarities. This produces two main classes of TCP domains that also differ in the number of residues of the basic region because class II proteins contain a 4-amino acid insertion in this region (1, 2). The function of most TCP proteins studied to date is associated with the regulation of different developmental processes in plants (3–11). However, other functions have also been proposed, such as the coordination of mitochondrial biogenesis (12–14), regulation of the circadian clock (15), control of jasmonic acid biosynthesis (16), and determination of the embryonic growth potential in seeds (17). The fact that there are >20 different TCP proteins in most angiosperm species raises the question of whether there is a high degree of redundancy or different proteins perform different functions and, if the latter case is correct, the additional question is what is the basis for specificity. Studies using mutants and plants overexpressing native or modified forms of TCP proteins have suggested that partial redundancy overlaps with specific functions of different TCP proteins (8, 9, 16, 18).
One of the sources of functional specificity may be the existence of different DNA binding preferences among TCP proteins. Previous studies have provided consensus DNA sequences preferentially bound by different class I and class II proteins that, with the sole exception of Arabidopsis TCP11, can be described as GTGGGNCC for class I and GTGGNCCC for class II (11, 16, 19). Because these kinds of study have only been performed with a limited number of proteins, it is not known whether these consensus sequences apply to all members of each class or not.
Studies on the molecular basis of DNA binding specificity of TCP proteins will help to understand how different TCP proteins perform their function and eventually construct a code linking the presence of certain residues to the DNA binding preferences of the respective proteins. In this work, we have studied the DNA binding properties of the class I TCP protein TCP16 from Arabidopsis and determined that it has a preference for a class II binding site. We show that the identity of residue 11 of the class I TCP domain and the equivalent residue 15 of the class II domain is an important determinant of the preference of TCP proteins for class I or class II sequences. In addition, we have uncovered an influence of the HLH domain on the selectivity of the proteins toward each sequence. Phylogenetic analyses indicated that TCP16-like proteins with Asp-11 in the TCP domain arose in rosids and may be related to developmental characteristics of this lineage of eudicots.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Recombinant Proteins
The full-length coding region of Arabidopsis thaliana TCP16 was cloned in-frame with the maltose-binding protein (MBP) in the XbaI and SalI sites of plasmid pMAL-c2 (New England Biolabs). TCP20 was cloned in the BamHI and HindIII sites of the same plasmid. A clone expressing TCP4 fused to MBP was sent to us by Drs. Carla Schommer and Javier Palatnik (IBR, Rosario, Argentina). For the analysis of mutants and chimeras, shorter forms of TCP16 (amino acids 1–80), TCP20 (amino acids 1–157), and TCP4 (amino acids 1–131) were used. These proteins showed the same DNA binding preferences as the full-length proteins. Mutants and chimeras were constructed by overlap extension mutagenesis (20) using complementary oligonucleotides with the desired mutations or chimeric sequences (supplemental Table 1). All constructs were checked by DNA sequencing.
For expression, Escherichia coli BL21 (DE3) cells were cultured and induced as described previously (21). Purification of recombinant proteins was performed as indicated by the manufacturers of the pMAL-c2 system.
DNA Binding Assays
For electrophoretic mobility shift assays (EMSAs), aliquots of purified proteins were incubated with labeled double-stranded DNA generated by hybridization of complementary synthetic oligonucleotides (supplemental Table 1) as described previously (22). DNA binding assays were performed with the proteins fused to MBP. Controls made with proteins obtained after cleavage with factor Xa indicated that the MBP moiety does not affect the behavior of the recombinant proteins. For quantitative analysis, the amount of radioactivity in gels was measured using phosphor storage technology with a Typhoon (GE Healthcare) scanner.
Binding Site Selection (SELEX)
To select DNA sequences preferentially bound by TCP16, the random oligonucleotide selection technique (SELEX) (23) was applied, using procedures described by Blackwell and Weintraub (24). A 52-mer double-stranded oligonucleotide containing a 10-bp central core with random and fixed positions (5′-NNGGNNCCNN-3′) was incubated with TCP16 as described above. Bound DNA was separated by EMSA and eluted from the gel with 0.5 ml of 0.5 m ammonium acetate, 10 mm MgCl2, 1 mm EDTA, and 0.1% (w/v) SDS. Eluted DNA molecules were amplified using oligonucleotides R1 and R2 (supplemental Table 1) during 30 cycles of 1 min at 94 °C, 1 min at 53 °C, and 1 min at 72 °C. After purification through polyacrylamide gels, the amplified molecules were subjected to new cycles of binding, elution, and amplification. Enrichment in sequences bound by TCP16 was monitored by binding and competition analysis in EMSAs. After selection, the population of DNA sequences was cloned into the pCR 2.1-TOPO vector (Invitrogen), and individual clones were sequenced.
Footprinting Analysis
For hydroxyl radical footprinting (25), double-stranded oligonucleotides with BamHI- and EcoRI-compatible cohesive ends containing the desired binding sites were cloned into pBluescript SK−. DNA fragments from the respective clones were obtained by PCR using reverse and universal primers followed by cleavage with HindIII and XbaI. The fragments were labeled in one of their 3′ ends by filling in with the Klenow fragment of DNA polymerase and [α-32P]dATP prior to cleavage with the second enzyme and subsequently purified by PAGE. Binding of proteins (3 μg) to these fragments (200,000 cpm) was performed in 15 μl of 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 10 mm β-mercaptoethanol, 0.1 mm EDTA, 22 ng/μl BSA, and 10 ng/μl poly(dI-dC). After binding, DNA was subjected to hydroxyl radical cleavage by the addition of 10.5 μl of 6.6 mm sodium ascorbate, 0.66 mm EDTA (pH 8.0), 0.33 mm (NH4)2Fe(SO4)2, and 0.2% H2O2, and bound and free forms were separated by EMSA. The corresponding fractions were excised from the gel, eluted, and analyzed on denaturing polyacrylamide gels.
One-hybrid Analysis in Yeast
To obtain yeast strains carrying class I or class II binding sequences inserted into the genome, tandem copies of oligonucleotides with the corresponding binding sites were cloned in front of the LacZ reporter gene preceded by the CYC1 minimal promoter in the pLacZi vector (Clontech). Plasmids linearized in their NcoI sites were introduced into the URA3 locus of the yeast aW303 strain (MATa ade2–1 his3–11,15 leu2–3,112 trp1–1 ura3–1). The presence of the fragment of interest in the genome of transformants was analyzed by PCR with specific oligonucleotides. To express fusions of TCP proteins to the GAL4 activation domain, fragments encoding the different proteins were cloned in plasmid pGADT7 (Clontech). DNA was introduced into yeast using the lithium acetate transformation method (26). β-Galactosidase activity was assayed as described by Reynolds et al. (27) using o-nitrophenylgalactoside as substrate.
Gene Cloning and Plant Transformation
For expression of wild-type and mutant TCP16 fused to the EAR repressor domain (28), full-length TCP16 coding sequences were amplified with specific primers (supplemental Table 1), digested with XhoI, and ligated with a double-stranded synthetic oligonucleotide with a compatible end encoding the EAR domain. The fusions were amplified with primers T16-F and EAR-XK and cloned in the binary vector pBI121 under the control of the 35SCaMV promoter.
Constructs were checked by DNA sequencing and introduced into Agrobacterium tumefaciens strain LB4404. Arabidopsis plants were transformed by the floral dip procedure (29). Transformed plants were selected on the basis of kanamycin resistance and genotyping. Expression of the transgene was analyzed by RT-PCR with specific oligonucleotides.
Phylogenetic Analysis
To identify TCP proteins, BLAST sequence searches were conducted on genomes of 22 members of Land Plants (Embryophytes) available at the Phytozome V. 6.0 data base. Additional searches were performed using the nucleotide collection, genomic survey sequence, the nonredundant protein sequence, and the EST sequence databases at NCBI. BLASTN, BLASTP, and TBLASTX searches were conducted using the consensus sequences for the class I and class II TCP domains (KDRHTKVDGRGRRIRMPALCAARVFQLTRELGHKSDGETIEWLL and KDRHSKVCTAKGPRDRRVRLSVGTAIQFYDLQDRLGFDKPSKAVDWLL, respectively). Gene names and identifiers of selected sequences are presented in supplemental Table 2. Sequences were aligned using MUSCLE (30, 31), and the alignment was corrected manually in MEGA V. 4 (32). Trees were generated by the maximum likelihood method, using RAxML 7.2.7 (33) on the CIPRES Science Gateway V. 3.1 under the GTRGAMMA model (using the GTRCAT setting) with 25 categories of rate variation. Node support was estimated using 100 bootstrap replicates (34). Maximum parsimony ancestral state reconstructions were estimated in Mesquite V. 2.74 (see Mesquite Project Web site).
To test whether there was any evidence for positive selection on sites of the TCP domain, we have calculated the ω ratio (calculated as the number of nonsynonymous substitutions versus the number of synonymous substitutions, dN/dS). The ω ratio was estimated using two different codon-based maximum likelihood methods available in the HyPhy package accessed through the Datamonkey web server (36, 37): (i) single-likelihood ancestor counting (SLAC), and (ii) fixed effects likelihood (FEL). For selection analysis, incomplete and duplicate sequences were removed from the alignment. Nucleotide sites were explored using the General Reversible Model (REV) of selection. Specified significance levels were p = 0.1.
RESULTS
TCP16 Prefers Class II Sequence
The TCP domain of TCP16 contains 11 nonconservative substitutions respective to the class I consensus sequence, three of them within the basic region (supplemental Fig. 1). We then decided to analyze the DNA binding properties of TCP16, assuming that the results may give new insights into the DNA binding specificity of TCP proteins. As a first step, we analyzed binding of a recombinant form of TCP16 to an oligonucleotide with the sequence GTGGGCCCAC, designed on the basis of the known DNA binding preferences of other class I and class II TCP proteins (GTGGGNCC and GTGGNCCC, respectively) (11, 19). Binding assays indicated that TCP16 is able to bind specifically to this oligonucleotide and that a single change at position 4 or 7 (position 7 is not shown) or double changes at positions 5 and 6 or 3 and 8 produce a marked decrease in binding (supplemental Fig. 2). Because these characteristics are shared by TCP16 and other TCP proteins, we decided to perform a SELEX experiment using a population of oligonucleotides (NNGGNNCCNN) containing fixed nucleotides at positions 3, 4, 7, and 8 and variable nucleotides around them, a strategy previously used with other TCP proteins (11).
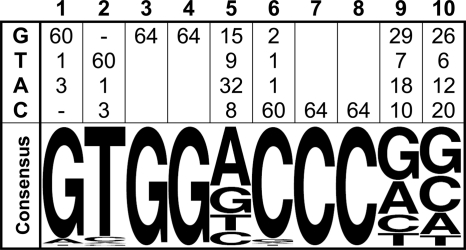
TCP16 was able to select specific sequences from the population of oligonucleotides, as indicated by the fact that increased binding was observed after progressive rounds of selection. After three rounds, when no further increases in DNA binding were observed, the oligonucleotide mixture was cloned and analyzed by sequencing (supplemental Fig. 3A). The results indicated that TCP16 selects a sequence of the type GTGGNCCCNN (selected nucleotides underlined) with almost 100% efficiency (Fig. 1). A slight preference for purines was observed at positions 5 and 9, and G or C were selected more frequently at position 10. In addition, clones with one of the flanking sequences (i.e. the one containing T at position −1 respective to the preferred binding site) were more abundant (48/64) that those in the reverse orientation. This may indicate a preference for sequences present in arm regions as also suggested by competition experiments using oligonucleotides with the same core sequence but exchanged flanking regions (supplemental Fig. 3B).
FIGURE 1.
TCP16 selects a class II sequence. Upper, nucleotide frequencies at different positions in the population of oligonucleotides selected by TCP16. Lower, consensus binding site, based on nucleotide frequencies, expressed as a sequence logo. The sequences of the 64 different clones that were analyzed are shown in supplemental Fig. 3A.
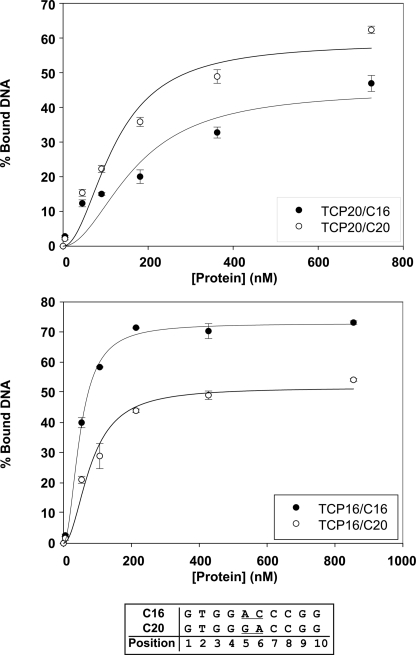
The sequence preferred by TCP16, GTGGNCCCNN, matches the consensus described for class II proteins (16, 19) and is different from GTGGGNCCNN, described for other class I proteins such as PCF2, TCP15, and TCP20 (11, 19). To confirm the SELEX results, we performed binding curves using different amounts of TCP16 and oligonucleotides containing the sequences GTGGACCCGG (named C16; based on the TCP16 consensus) and GTGGGACCGG (named C20; based on the TCP20 consensus) (11). We also performed binding curves with TCP20 for comparison. The curves confirmed that the two proteins show different sequence preferences (Fig. 2).
FIGURE 2.
TCP16 and TCP20 show different sequence preferences. Binding of different amounts of TCP20 (upper) and TCP16 (lower) to oligonucleotides carrying the sequences GTGGACCCGG (C16; black symbols) or GTGGGACCGG (C20; white symbols) was analyzed in EMSAs. The amount of bound DNA was calculated from signal intensities obtained with phosphor storage technology. The data are mean (±S.D.) of three independent experiments.
Single Residue of TCP Domain Determines Preference for Class I or Class II Binding Site
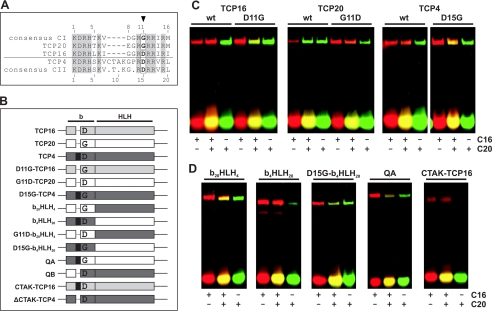
Analysis of residues that differ in TCP16 respective to other class I proteins showed the presence of Gly (instead of Asp, Asn, or Glu) at position 8 and Asp (instead of Gly) at position 11 of the TCP domain, both within the basic region that presumably establishes contacts with DNA (Fig. 3A). We then performed reciprocal mutations of these residues in TCP16 and TCP20 (Fig. 3B) to analyze their influence on the DNA binding properties of the respective proteins. The relative preference of each protein for C16 or C20 was analyzed in EMSAs using oligonucleotides labeled with different fluorophores combined in the same binding reaction (Fig. 3C). The results indicated that the introduction of Asp-11 in TCP20 produces a protein with a preference for C16 (Fig. 3C), whereas the incorporation of Gly-8 has no effect on the binding properties of the protein (data not shown). This points to residue 11 as a main determinant of the different binding preferences of TCP16 respective to other class I proteins. In agreement with this, introduction of Gly-11 in TCP16 increased the preference of this protein for C20 (Fig. 3C).
FIGURE 3.
Residue 11 of the class I TCP domain and residue 15 of the class II domain determine the binding preferences of TCP proteins. A, alignment of the basic regions of TCP16, TCP20, and TCP4, together with the consensus sequences of class I and class II TCP proteins. Conserved residues are shaded. Numbering indicates residue position within the respective TCP domain. The arrowhead indicates the residues that were exchanged in the mutant proteins under study. B, schematic structure of the mutants and chimeric proteins used in this study. Portions from TCP16, TCP20, or TCP4 are indicated in different colors. The black rectangle represents the 4-amino acid insertion of the TCP4 basic region. D or G indicates the presence of either Asp or Gly at position 11 (or 15 in proteins with the insertion). b and HLH indicate the basic region and the helix-loop-helix motif, respectively. The names of the proteins are indicated on the left. C, EMSA of TCP proteins and the respective mutants using C16, C20, or both oligonucleotides in the same binding reaction. C16 was 5′ end-labeled with Cy5, whereas C20 was 5′ end-labeled with 6-carboxyfluorescein. The images correspond to scans performed at the excitation and emission wavelengths of the corresponding fluorophores that were superimposed and colored in red and green for C16 and C20, respectively. D, EMSA similar to that in C with chimeric proteins with portions of TCP20 and TCP4. A TCP16 protein with the 4-amino acid insertion of TCP4 was also tested.
Because the binding preferences of TCP16 resemble those described for class II proteins, we also looked at the equivalent position 15 of the TCP domain of these proteins (class II TCP proteins have a 4-amino acid insertion within the basic region) and found that they contain a conserved Asp, just as TCP16 at position 11 (Fig. 3A). Change of this Asp to Gly shifted the binding preferences of the class II protein TCP4 toward C20, although the selectivity of the mutant was less pronounced than the one showed by the other proteins (Fig. 3C).
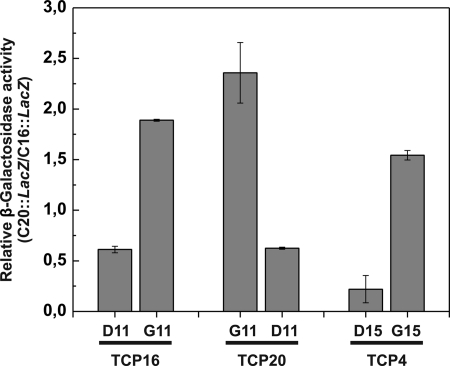
The ability of the different proteins to interact with class I and class II sequences was also evaluated in vivo using one-hybrid assays in yeast. For this purpose, tandem copies of C16 or C20 were introduced in the yeast genome in front of the β-galactosidase gene containing a minimal promoter, and the respective strains were transformed with clones expressing TCP proteins and their mutants fused to the GAL4 activation domain. For each protein, the β-galactosidase activity obtained with C20 was divided by that obtained with C16 so that values indicate a preference for C20 or C16 if they are >1 or <1, respectively. The results show that TCP20 activates more efficiently the construct that contains C20 in the promoter, whereas TCP16 and TCP4 produce higher activation levels with C16 (Fig. 4). Reciprocal mutations of residues located at position 11 originate a change in the relative preferences of both class I proteins, in agreement with the experiments performed in vitro (Fig. 4). The same is true for mutation of Asp-15 to Gly in TCP4. We conclude that the residue present at position 11 of the class I TCP domain, or the equivalent position 15 of the class II domain, is a main determinant of the preference for a class I or a class II binding sequence.
FIGURE 4.
Binding preferences of TCP proteins and the corresponding mutants in vivo in yeast. Yeast strains carrying a fusion of oligonucleotides C16 or C20 (six copies) to the LacZ gene containing a minimal promoter were transformed with constructs expressing the indicated TCP proteins fused to the GAL4 activation domain. Values indicate the specific β-galactosidase activity of the strain carrying the C20 construct divided by the activity of the one carrying the C16 construct and indicate a relative preference for C20 or C16 whether they are >1 or <1, respectively. The mean (±S.D., error bars) of three independent measurements is shown.
We also performed binding curves with different amounts of the native and mutated forms of TCP16, TCP20, and TCP4 and a fixed amount of either C20 or C16. The results were fitted to the Hill equation with a Hill number of 2, assuming that dimer formation is required for DNA binding. The model produced a good fit to experimental results, suggesting that monomers and dimers are in equilibrium in the binding assay. Examination of the respective dissociation constants (Table 1) indicates that the presence of Asp-11 (Asp-15 in TCP4) produces proteins with higher affinity for C16 than for C20. Asp-11, then, acts on the DNA binding preferences of the respective proteins mainly influencing their affinity for C16. In the case of proteins with Gly-11 or Gly-15, changes in affinity were not evident, with the sole exception of TCP20 (Table 1). It can be speculated that Gly-11 produces a more relaxed specificity and that, in the case of class I proteins like TCP20, additional factors may also influence the preference for C20.
TABLE 1.
Apparent dissociation constant (Kd) values for the interaction of TCP proteins and their respective mutants with C20 and C16
Binding curves were fitted to the Hill equation with a Hill number of 2, assuming that dimer formation is required for DNA binding. ΔKd indicates the S.D. of three independent experiments. Values indicate concentration of TCP monomers. The root mean square deviation (r.m.s.d.) errors and correlation coefficients (r2) of the fits to the experimental data are also indicated.
| Protein | C20 |
C16 |
||||||
|---|---|---|---|---|---|---|---|---|
| Kd | ΔKd | r.s.m.d. | r2 | Kd | ΔKd | r.s.m.d. | r2 | |
| nm | nm | % | nm | nm | % | |||
| TCP16 | 81.2 | 7.2 | 12.3 | 0.980 | 49.2 | 2.2 | 3.2 | 0.996 |
| D11G-TCP16 | 238.6 | 8.4 | 10.7 | 0.962 | 239.3 | 25.1 | 10.9 | 0.961 |
| TCP20 | 122.0 | 5.6 | 9.7 | 0.956 | 169.1 | 13.8 | 14.5 | 0.892 |
| G11D-TCP20 | 138.4 | 1.0 | 9.6 | 0.970 | 68.4 | 5.1 | 11.6 | 0.939 |
| TCP4 | 157.4 | 13.9 | 9.8 | 0.932 | 99.1 | 7.1 | 10.6 | 0.944 |
| D15G-TCP4 | 162.6 | 23.1 | 6.1 | 0.983 | 162.6 | 1.5 | 7.2 | 0.979 |
Helix-Loop-Helix Motif Influences Selectivity of Basic Region
To evaluate the influence of different parts of the TCP domain on the preferences and selectivity of TCP proteins, we constructed a series of chimeras exchanging modules between TCP20 and TCP4 (Fig. 3B). Two of the chimeric proteins consisted of the basic region of TCP20 fused to the HLH motif of TCP4 (b20HLH4) and vice versa (b4HLH20). Analysis of the binding behavior of these proteins showed that the basic region dictates the sequence preferences (Fig. 3D), in agreement with the results described above. In addition, it became evident that the presence of the HLH motif of TCP20 produced a protein with enhanced selectivity (i.e. compare TCP4 with b4HLH20 and TCP20 with b20HLH4 in Fig. 3). Thus, the nature of the HLH motif influences the capacity of the basic region to select among different target sequences, and the TCP20 HLH motif seems to contain features that enhance selectivity. To confirm these results further, we mutated the basic regions of the chimeric proteins, introducing Gly instead of Asp-15 in b4HLH20 and Asp instead of Gly-11 in b20HLH4 (Fig. 3B) and performed DNA binding assays with these proteins. As observed with the native proteins, introduction of Gly in b4HLH20 shifted the preference of this protein toward a class I sequence (Fig. 3D). Notably, the opposite mutation in b20HLH4 produced a protein unable to bind DNA (data not shown).
Additional chimeras consisted of the N-terminal 11 residues of the TCP4 basic region (including the 4-amino acid insertion present in class II proteins) fused to the rest of the TCP20 TCP domain and the N-terminal 7 residues of the TCP20 basic region fused to the rest of the TCP4 TCP domain without the insertion (called QA and QB respectively; Fig. 3B). The first of these chimeras, QA, showed a preference for C20 (Fig. 3D), indicating that the C-terminal portion of the basic region, which contains Gly (Gly-15 in the chimera), is responsible for this behavior. In addition, the presence of the 4-amino acid insertion had no influence on binding behavior. QB, on the other hand, was unable to bind DNA (data not shown). Analysis of the structure of QB revealed some common features with G11D-b20HLH4 because these were the only proteins analyzed that contain the HLH motif of TCP4 and Asp-11 and lack the 4-amino acid insertion (Fig. 3B). This points to the existence of an important role of the insertion, either for folding or DNA binding, in proteins that contain the TCP4 HLH motif and Asp-15. In agreement with this, deletion of the 4-amino acid insertion in TCP4 produced a protein unable to bind DNA (data not shown). Insertion of the 4 amino acids in TCP16, on the contrary, did not modify its binding preferences (Fig. 3D).
Contacts Established with Central Part of One of the DNA Strands Correlate with Binding Site Preferences
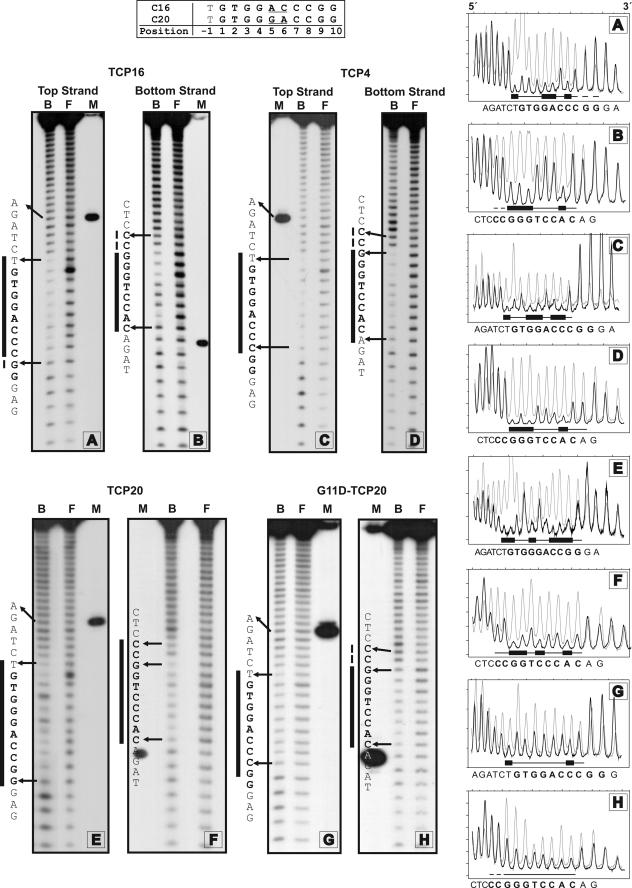
Further insight into the interaction of TCP proteins with DNA was obtained using hydroxyl radical footprinting. Upon binding to C16, TCP16 establishes interactions with a 9-bp region (Fig. 5, A and B). The sequence TGGACCC is contacted in both strands. In the top strand (the one containing GTGGACCC), the strongest protection is observed in T-1, G4, A5, and C7, whereas A2, G6, G7, and G8 are more strongly protected in the bottom strand. The bottom strand of C16 is contacted in a similar way by TCP4 and Asp-11-TCP20, but contacts with position 6 (now a T) are decreased upon interaction of TCP20 with C20 (Fig. 5, D, F, and H). Instead, TCP20 protects more strongly C5, which conforms the C:G pair selected by this protein and not by TCP16 or TCP4 (Fig. 5F). Thus, contacts established with positions 5 and 6 of the bottom strand seem to be specific and may be the basis for the different binding preferences of the respective proteins.
FIGURE 5.
Hydroxyl radical footprinting patterns of the complexes of TCP16, TCP4, TCP20, and G11D-TCP20 with their DNA target sites. C16 or C20 was labeled in the 3′ end of one strand (either top or bottom strand, named arbitrarily). After binding of the respective proteins, the DNA was subjected to hydroxyl radical attack. Free (F) and bound (B) DNA were separated and analyzed. The same fragment digested with restriction enzymes was used to calculate the position of the footprint (M). Letters beside each panel indicate the DNA sequence of the corresponding strand (5′ end in the upper part). Vertical bars indicate the region protected from the attack after protein binding. Densitometric scans of the lanes corresponding to free (gray line) and bound (black line) DNA are shown on the right.
In the top strand, protection patterns differ among the different proteins, with the sole exception of T−1 and C7, which are strongly protected by all of them (Fig. 5, A, C, E, and G). TCP4 establishes a symmetric protection pattern around A5, comprising 3 nucleotides at each side, with G3 showing equivalent protection to C7 (Fig. 5C). As opposed to TCP16, TCP4 shows almost no protection of A5. For TCP20, the protection pattern extends along 11 nucleotides, starting at T−1, and is weaker at G3 and G5 (Fig. 5E). In Asp-11-TCP20, position 5 (A5) shows protection similar to its surrounding nucleotides (Fig. 5G). The contacts with positions 5 and 6 of the top strand detected by footprinting are probably established with the phosphodiester backbone because they are not correlated with the capacity of nucleotide discrimination of the different proteins at these positions. Contacts established with this strand reveal protein-specific differences, suggesting that even proteins with similar DNA binding preferences accommodate differently on DNA, perhaps due to different orientations of residues that establish contacts with the phosphodiester backbone. This points to the existence of flexibility in the way the TCP domain interacts with its target site(s).
To evaluate the influence of the DNA binding sequence on the footprinting pattern, we also performed experiments with the sequence GTGGGCCCGG, which contains G:C pairs at positions 5 and 6 and is thus efficiently bound by all the proteins. With TCP16, the most obvious change was an increased relative protection of position 5 (now a C) in the bottom strand, respective to C3 and C4 (supplemental Fig. 4). A similar observation was made for TCP4. For TCP20, G6 from the bottom strand was relatively more protected than G7 compared with T6 in C20 (supplemental Fig. 4). Nevertheless, protection of C5, which conforms the C:G pair selected by this protein and not by TCP16 or TCP4, remained stronger, as with C20. These observations reinforce the notion that contacts established with positions 5 and 6 of the bottom strand reflect the different binding preferences of the respective proteins. In addition, they suggest that the presence of a C:G pair at the nonselected position 5 or 6 enhances the relative protection at this position, even if the proteins are not capable of selecting it. In the top strand, changes in protection patterns dependent on the target sequence were observed for all of the proteins but were more evident for TCP20 in the region G3 to G5 (supplemental Fig. 4). Because these residues are identical in both target sequences analyzed, this may be a reflection of changes that occur to accommodate the basic region for optimal binding to a different target site. Again, this suggests the existence of high flexibility in the interaction of the TCP domain with DNA. The notion of flexibility is indeed implicit in the fact that a TCP dimer is able to interact with a rather asymmetric binding site.
Identity of Residue 11 Determines TCP16 Function in Vivo
To ascertain whether changes in the identity of residue 11 influence the action of TCP proteins, we analyzed the effect of expressing in Arabidopsis repressor forms of TCP16 with either Asp-11 of Gly-11 (Fig. 6). Expression of wild-type TCP16 (with Asp-11) fused to the EAR repressor domain produced a change in leaf form, originating plants with rounder leaves respective to nontransformed plants (Fig. 6, A, B, D, and E). Transformation with Gly-11-TCP16-EAR, in turn, affected leaf form in a different way. Young leaves from these plants were wider at the base and had acute tips, contrasting with the round and elliptical shapes of TCP16-EAR and wild-type plants, respectively (Fig. 6, A–C and J). In addition, these plants had altered cotyledons with cup-shaped structure, whereas nontransformed and TCP16-EAR plants had slightly epinastic cotyledons (Fig. 6, A–C, G, and H). Upon progression of development, newly formed leaves in some Gly-11-TCP16-EAR plants showed altered lamina development, with invaginations due to uneven growth of different laminar sectors, whereas rather flat laminar surfaces were observed in wild-type and TCP16-EAR plants (Fig. 6, F and I). This is reminiscent of alterations observed in leaves that express repressor fusions to the class I TCP proteins TCP14 and TCP15 (38, 39), which possess Gly at position 11. There is increasing evidence that class I and class II TCP proteins affect leaf development and form (5, 6, 11, 18, 38–40). Even if our results do not necessarily imply that TCP16 is involved in these processes, they clearly show that the identity of residue 11 has functional implications, most likely because it influences the interaction of TCP16 with different target sites within the genome.
FIGURE 6.
TCP16 and Gly-11-TCP16 alter leaf development in different ways. A–H, phenotype of plants transformed with constructs that express fusions of TCP16 (B and E) or its Gly-11 mutant (C, F, and H) to the EAR repressor domain compared with plants transformed with vector pBI121, used as control (A, D, and G). A–C, 15-day-old plants. D and F, 22-day-old plants. E, 28-day-old TCP16-EAR plant. G and H, cotyledons from 15-day-old plants. I and J show third pair leaves from 28-day-old plants with severe (I) or mild (J) phenotype (left, wild-type; center, TCP16-EAR; right, Gly-11-TCP16-EAR). Scale bars represent 2.5 mm.
DISCUSSION
Possible Roles of Residues of Basic Region and HLH Motif in DNA Binding Preferences of TCP Proteins
Our results indicate that position 11 of the class I TCP domain is important to determine a preference for a class I or a class II target site. Rather than establishing direct contacts with DNA, it is possible that the presence of Gly-11 in most class I proteins influences the orientation of adjacent residue(s) that are in direct contact with the bases (Arg-10, Arg-12, Arg-13, and Arg-15, conserved in class I and class II proteins, are good candidates). Asp-11, present in TCP16 and a reduced group of class I proteins, may be directly involved in DNA recognition or may also influence the positioning of adjacent amino acids.
The region containing Asp-11 and adjacent arginines in TCP16 (RDRR) is similar to the sequence RERR contained in MyoD, where the Glu residue establishes specific contacts with a C of the target site (41). One possibility is that Asp-11 in TCP16 and the equivalent Asp-15 present in class II proteins interact with position 6 of C16, thus determining the preference for a C:G pair at this position. According to footprinting experiments, the presence of Asp increases protection of G6 from the bottom strand, suggesting that this may be the contacted nucleotide. The establishment of direct contacts with DNA by Asp-11, and not by Gly-11, is consistent with our results showing that the presence of Asp produces an increase in affinity for C16. Aggarwal et al. (2) also considered the possibility that Asp-15 of TCP4 may interact with DNA but argued that the side chain of Asp-15 may be too short to establish direct contacts. Another possibility is that the presence of either Asp or Gly at position 11 of class I proteins influences the orientation of another base-contacting residue. A candidate for this may be Arg-15, whose importance for the recognition of G:C pairs located at positions 5, 6, and/or 8 has been documented earlier (11). Indeed, mutation of Arg-15 to Thr in TCP20 abolishes specific recognition of a G:C pair at position 8 and establishes a preference for G:C pairs at position 6 and, with relaxed specificity, at position 5 (11). Based on this, we postulate that Arg-15 from one monomer may be located near positions 5 and 6 and, according to its orientation, interact preferentially with a G:C pair at one of these positions. Arg-15 of TCP16 would be located one helical turn C-terminal to Asp-11, suggesting that both side chains may interact. Interactions of this type, which help to fix DNA-contacting residues in the correct orientation, have been reported for Glu-345 and Arg-348 in the basic region of the E47 bHLH domain (42).
It is also noteworthy that the HLH motif influences the selectivity toward different DNA sequences. A direct role of this motif in DNA binding is conceivable from the fact that HLH residues establish nonspecific contacts with DNA in bHLH proteins (42–45) and was suggested by Aggarwal et al. (2) for TCP proteins. Additionally, the packing of the four-helix bundle putatively formed by the HLH motif may aid in fixing the basic region in a specific conformation on DNA. Specific residues located in the HLH motifs of class I and class II proteins may be responsible for the different effects of these motifs on the selectivity of the respective proteins. A combined effect of the HLH motif, Asp-15, and the 4-amino acid insertion of the basic region of class II proteins on the DNA binding behavior of TCP proteins was also uncovered in our study. Aggarwal et al. (2) postulated that Thr-9 of the TCP4 domain insertion may establish contacts with the phosphodiester backbone from the fact that its mutation severely affected binding to DNA. Another possibility is that the insertion, which most likely constitutes a loop that links two helices of the basic region, interacts with specific residues of the HLH motif and/or Asp-15, and this helps to position the helices on DNA correctly. If so, class I proteins may use a different mechanism based on the nature of the HLH motif and/or the presence of Gly instead of Asp.
Evolutionary Aspects of TCP Domain Binding Preferences
From an evolutionary point of view, the ancestor of the TCP domain is unknown. TCP proteins appeared in Streptophyta, before the divergence of the Zygnemophyta, which already contain class I and class II proteins (46). As a consequence, it is not known if the ancestral TCP gene encoded a protein with or without the 4-amino acid insertion. Our results suggest that current class II proteins depend on this insertion for DNA binding if they contain Asp-15. Interestingly, all class II proteins available in databases contain this residue (supplemental Fig. 5). For class I proteins, a vast majority of proteins with Gly-11 is observed (supplemental Fig. 5). However, the appearance of class I proteins with Asp-11, like TCP16, seems to have occurred during evolution. Ancestral character state reconstruction shows that the ancestor of class I proteins probably had Gly-11 and that proteins with Asp-11, like TCP16, seem to have arisen at least twice during class I protein evolution due to a G-to-A transition at the second position of codon 11 (supplemental Fig. 6). Interestingly, these transitions occurred only in eurosids (members of the eudicots). In fact, apart from TCP16 and a close homologue from Arabidopsis lyrata, we have identified class I proteins with Asp-11 in Populus trichocarpa, Cucumis sativus, and Carica papaya. To test whether these substitution patterns represent potentially adaptive evolution for TCP genes, we estimated the nonsynonymous (dN) to synonymous rate (dS) ratio (ω) on the codon alignment of the TCP domain using two different codon-based maximum likelihood methods. Both methods suggest that none of the sites along the TCP domain are under positive selection constrains (supplemental Table 3). These findings are in agreement with studies of adaptative evolution on the LEGCYC locus of legumes (47) that found no positive selective sites along the TCP domain. It is possible that the failure to detect positive selection on the TCP domain is given by the preponderance of purifying selection over most of the domain, indicating the importance of most positions for the correct function of TCP proteins.
The fact that class I proteins with Asp-11 are only present in eurosids suggests that these unique proteins may be involved in the control of some distinctive characters of this lineage. TCP16 has been shown to play a role during early pollen development (35), and many eurosids have pollen grains with unique complex apertures and exine ornamentations. Thus, the emergence of TCP16 and related proteins may have contributed to these aspects of pollen evolution. Another possibility is that the emergence of proteins with Asp-11 has influenced the evolution of leaf form, as suggested by the different effects of expression of TCP16 with either Asp-11 or Gly-11 shown here.
In conclusion, we have determined that the identity of the residue present at position 11 of the class I TCP domain or the equivalent position 15 of the class II domain is a main determinant of the target site preferences of TCP proteins. We have also shown that the HLH motif influences the selectivity of the basic region, allowing more or less efficient discrimination among related sequences. Selection among class I and class II sequences is probably dictated by the orientation of base contacting amino acids, most likely arginines, located around residues 11 or 15. Subtle changes in orientation of these base-contacting amino acids, brought about by interactions with other regions of the TCP domain or with other proteins, may be relevant in vivo for the recognition of specific target genes by different TCP proteins. The distribution of Gly-11 and Asp-11 or Asp-15 in TCP proteins indicates that these residues are at once important for TCP protein function and a source of evolutionary novelties.
Supplementary Material
Acknowledgments
We thank Drs. Sergio Garay and Daniel Rodrigues (Departmento de Física, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral) for helpful discussions on the structure of the TCP domain, and Drs. Carla Schommer and Javier Palatnik (IBR, Rosario) for providing a clone expressing TCP4.
This work was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) and Universidad Nacional del Litoral.

This article contains supplemental Figs. 1–6 and Tables 1–3.
- bHLH
- basic helix-loop-helix
- MBP
- maltose-binding protein.
REFERENCES
- 1. Martín-Trillo M., Cubas P. (2010) Trends Plant Sci. 15, 31–39 [DOI] [PubMed] [Google Scholar]
- 2. Aggarwal P., Das Gupta M., Joseph A. P., Chatterjee N, Srinivasan N., Nath U. (2010) Plant Cell 22, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996) Nature 383, 794–799 [DOI] [PubMed] [Google Scholar]
- 4. Doebley J., Stec A., Hubbard L. (1997) Nature 386, 485–488 [DOI] [PubMed] [Google Scholar]
- 5. Nath U., Crawford B. C., Carpenter R., Coen E. (2003) Science 299, 1404–1407 [DOI] [PubMed] [Google Scholar]
- 6. Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., Weigel D. (2003) Nature 425, 257–263 [DOI] [PubMed] [Google Scholar]
- 7. Li C., Potuschak T., Colón-Carmona A., Gutiérrez R. A., Doerner P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aguilar-Martínez J. A., Poza-Carrión C., Cubas P. (2007) Plant Cell 19, 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007) Plant Cell 19, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hervé C., Dabos P., Bardet C., Jauneau A., Auriac M. C., Ramboer A., Lacout F., Tremousaygue D. (2009) Plant Physiol. 149, 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Viola I. L., Uberti Manassero N. G., Ripoll R., Gonzalez D. H. (2011) Biochem. J. 435, 143–155 [DOI] [PubMed] [Google Scholar]
- 12. Welchen E., Gonzalez D. H. (2006) Plant Physiol. 141, 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gonzalez D. H., Welchen E., Attallah C. V., Comelli R. N., Mufarrege E. F. (2007) Plant J. 51, 105–116 [DOI] [PubMed] [Google Scholar]
- 14. Giraud E., Ng S., Carrie C., Duncan O., Low J., Lee C. P., Van Aken O., Millar A. H., Murcha M., Whelan J. (2010) Plant Cell 22, 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pruneda-Paz J. L., Breton G., Para A., Kay S. A. (2009) Science 323, 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schommer C., Palatnik J. F., Aggarwal P., Chételat A., Cubas P., Farmer E. E., Nath U., Weigel D. (2008) PLoS Biol. 6, e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tatematsu K., Nakabayashi K., Kamiya Y., Nambara E. (2008) Plant J. 53, 42–52 [DOI] [PubMed] [Google Scholar]
- 18. Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010) Plant Cell 22, 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kosugi S., Ohashi Y. (2002) Plant J. 30, 337–348 [DOI] [PubMed] [Google Scholar]
- 20. Silver J., Limjoco T., Feinstone S. (1995) in PCR Strategies (Innis M. A., Gelfand D. H., Sninsky J. J., eds) pp. 179–188, Academic Press, San Diego, CA [Google Scholar]
- 21. Viola I. L., Gonzalez D. H. (2009) Biochimie 91, 974–981 [DOI] [PubMed] [Google Scholar]
- 22. Viola I. L., Gonzalez D. H. (2007) Biochemistry 46, 7416–7425 [DOI] [PubMed] [Google Scholar]
- 23. Oliphant A. R., Brandl C. J., Struhl K. (1989) Mol. Cell. Biol. 9, 2944–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blackwell T. K., Weintraub H. (1990) Science 250, 1104–1110 [DOI] [PubMed] [Google Scholar]
- 25. Jain S. S., Tullius T. D. (2008) Nat. Protoc. 3, 1092–1100 [DOI] [PubMed] [Google Scholar]
- 26. Gietz D., St Jean A., Woods R. A., Schiestl R. H. (1992) Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reynolds A., Lundblad V., Dorris D., Keavency M. (1997) in Current Protocols in Molecular Biology (Ausubel F., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., eds) pp. 13.6.1–13.6.6, Green Publishing and Wiley-Interscience, New York [Google Scholar]
- 28. Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003) Plant J. 34, 733–739 [DOI] [PubMed] [Google Scholar]
- 29. Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 30. Edgar R. C. (2004) BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 33. Stamatakis A. (2006) Bioinformatics 22, 2688–2690 [DOI] [PubMed] [Google Scholar]
- 34. Stamatakis A., Hoover P., Rougemont J. (2008) Syst. Biol. 57, 758–771 [DOI] [PubMed] [Google Scholar]
- 35. Takeda T., Amano K., Ohto M. A., Nakamura K., Sato S., Kato T., Tabata S., Ueguchi C. (2006) Plant Mol. Biol. 61, 165–177 [DOI] [PubMed] [Google Scholar]
- 36. Kosakovsky Pond S. L., Frost S. D. W. (2005) Bioinformatics 21, 2531–2533 [DOI] [PubMed] [Google Scholar]
- 37. Kosakovsky Pond S. L., Frost S. D. W. (2005) Mol. Biol. Evol. 22, 1208–1222 [DOI] [PubMed] [Google Scholar]
- 38. Kieffer M., Master V., Waites R., Davies B. (2011) Plant J. 68, 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Uberti Manassero N. G., Lucero L. E., Viola I. L., Vegetti A. C., Gonzalez D. H. (October 20, 2011) J. Exp. Bot. 10.1093/jxb/err305 [DOI] [PubMed] [Google Scholar]
- 40. Ori N., Cohen A. R., Etzioni A., Brand A., Yanai O., Shleizer S., Menda N., Amsellem Z., Efroni I., Pekker I., Alvarez J. P., Blum E., Zamir D., Eshed Y. (2007) Nat. Genet. 39, 787–791 [DOI] [PubMed] [Google Scholar]
- 41. Ma P. C., Rould M. A., Weintraub H., Pabo C. O. (1994) Cell 77, 451–459 [DOI] [PubMed] [Google Scholar]
- 42. Ellenberger T., Fass D., Arnaud M., Harrison S. C. (1994) Genes Dev. 8, 970–980 [DOI] [PubMed] [Google Scholar]
- 43. Ferré-D'Amaré A. R., Pognonec P., Roeder R. G., Burley S. K. (1994) EMBO J. 13, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shimizu T., Toumoto A., Ihara K., Shimizu M., Kyogoku Y., Ogawa N., Oshima Y., Hakoshima T. (1997) EMBO J. 16, 4689–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Longo A., Guanga G. P., Rose R. B. (2008) Biochemistry 47, 218–229 [DOI] [PubMed] [Google Scholar]
- 46. Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C. (2007) J. Mol. Evol. 65, 23–33 [DOI] [PubMed] [Google Scholar]
- 47. Ree R. H., Citerne H. L., Lavin M., Cronk Q. C. B. (2004) Mol. Biol. Evol. 21, 321–331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.