Abstract
The Beclin 1 gene is a haplo-insufficient tumor suppressor and plays an essential role in autophagy. However, the molecular mechanism by which Beclin 1 functions remains largely unknown. Here we report the crystal structure of the evolutionarily conserved domain (ECD) of Beclin 1 at 1.6 Å resolution. Beclin 1 ECD exhibits a previously unreported fold, with three structural repeats arranged symmetrically around a central axis. Beclin 1 ECD defines a novel class of membrane-binding domain, with a strong preference for lipid membrane enriched with cardiolipin. The tip of a surface loop in Beclin 1 ECD, comprising three aromatic amino acids, acts as a hydrophobic finger to associate with lipid membrane, consequently resulting in the deformation of membrane and liposomes. Mutation of these aromatic residues rendered Beclin 1 unable to stably associate with lipid membrane in vitro and unable to fully rescue autophagy in Beclin 1-knockdown cells in vivo. These observations form an important framework for deciphering the biological functions of Beclin 1.
Keywords: Beclin 1, autophagy, crystal structure, membrane binding, membrane deformation, omegasome
Introduction
The Beclin 1 gene, which encodes a Bcl-2-interacting protein 1, is mono-allelically deleted in a large percentage of sporadic breast and ovarian cancers 2. Compared to normal breast epithelia, Beclin 1 is expressed at much lower levels in human breast cancer cell lines 3. Consistent with these observations, Beclin 1 was found to promote autophagy and inhibit tumorigenesis 3. Subsequent genetic studies revealed that Beclin 1 null mice die early in embryogenesis and Beclin 1 heterozygous mice develop spontaneous tumors, establishing Beclin 1 as a haplo-insufficient tumor suppressor 4, 5.
Beclin 1 shares ∼30% sequence identity with its yeast ortholog ATG6/VPS30, which was first identified to play an important role in vacuolar protein sorting 6. In Saccharomyces cerevisiae, ATG6 and ATG14 form a protein complex essential for autophagy 7, later known as the yeast complex I with class III phosphatidylinositol 3-kinase (PI3KC3) activity. In mammals, Beclin 1 appears to be an obligate component of two distinct PI3K complexes, one involving the human ortholog of ATG14/Barkor and the other involving UVRAG 8, 9. The ATG14/Barkor-containing PI3K complex, corresponding to the yeast complex I, is thought to be responsible for autophagic vesicle nucleation 10. The UVRAG-containing complex functions in the vacuolar protein sorting pathway. Despite more than one decade of intense investigation on Beclin 1, its precise role in autophagosome formation and membrane trafficking remains unknown 11, 12.
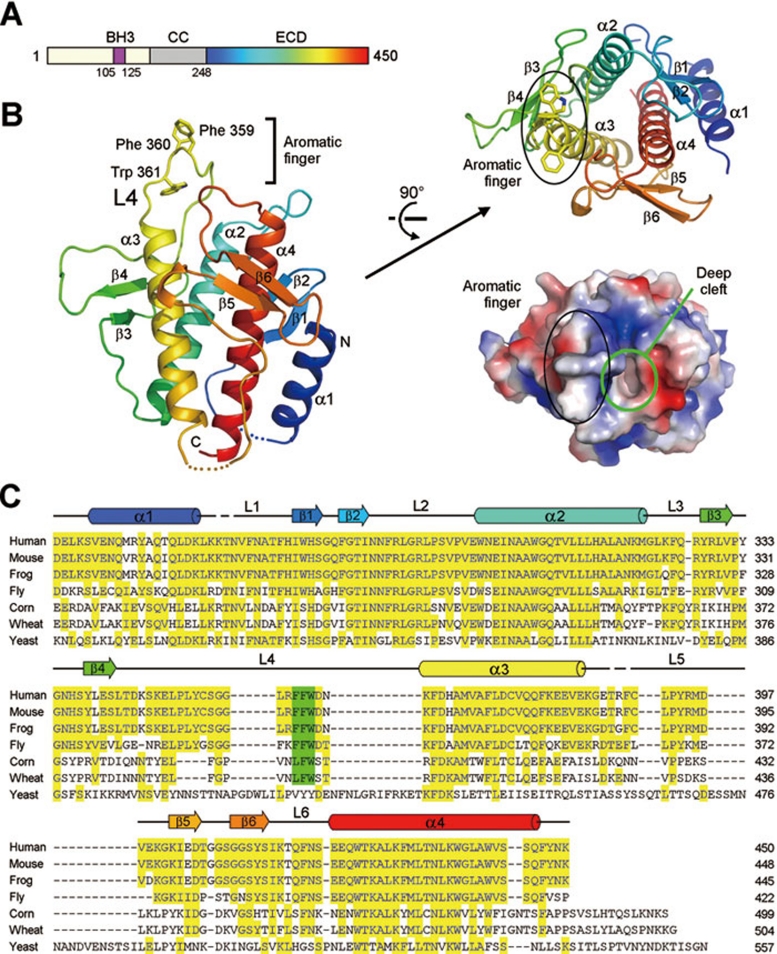
The 450-amino acid Beclin 1 protein contains three identifiable domains: a short BH3 motif (residues 105-125), a coiled-coil segment, and a C-terminal half encompassing the so-called evolutionarily conserved domain (ECD, residues 244-337 13) (Figure 1A). The BH3 domain is capable of binding to Bcl-2 family proteins such as Bcl-XL14, 15, 16, whereas the coiled-coil segment is likely involved in homo-dimerization or hetero-dimerization with other proteins such as UVRAG 17 and ATG14/Barkor 18. The ECD domain, which exhibits no sequence homology with any protein of known structure, constitutes the C-terminal half of Beclin 1 and is required for all known functions of Beclin 1. Elucidating the structure of the ECD may be central to deciphering the unresolved biochemical roles of Beclin 1.
Figure 1.
Structure of the Beclin 1 ECD. (A) Beclin 1 contains a BH3 domain, a coiled coil and an ECD. Rainbow color for the ECD domain is preserved in panel B. (B) Overall structure of the ECD (residues 248-447) of human Beclin 1. The structure is shown in rainbow color, with the N- and C-termini colored blue and red, respectively. An aromatic finger, comprising Phe359, Phe360 and Trp361, protrudes from the core structure. Beclin 1 ECD contains three structural repeats, each comprising a pair of β-strands and a long α-helix (top right). A deep pocket is located next to the aromatic finger (bottom right). (C) Sequence alignment of Beclin 1 homologs from multiple organisms. Conserved sequences are colored yellow. The three hydrophobic residues in the aromatic finger, highlighted in green, are highly conserved in all organisms except yeast. Secondary structural elements are indicated above the sequences. All structural images were generated using PyMol 45.
In this study, we report the crystal structure of Beclin 1 C-terminal half encompassing ECD and identify it as a novel lipid membrane-binding protein. Three consecutive aromatic residues have been identified to be responsible for binding to lipid membrane and mutation of these residues compromised the autophagy function of Beclin 1. These findings provide important clues for understanding the cellular function of Beclin 1.
Results
Structure of Beclin 1 ECD
To search for mechanistic clues of Beclin 1 function, we sought structural information. Unfortunately, the full-length human Beclin 1 protein was prone to aggregation and unfit for crystallization. Truncation of the N-terminal 100-120 amino acids in Beclin 1 drastically improved its solution behavior; yet the resulting protein defied repeated attempts of crystallization, likely due to the flexible nature of the BH3 motif and the coiled-coil domain. Limited proteolysis of the Beclin 1 variant (residues 118-450) led to identification of the entire C-terminal half (residues 248-450) as a stable structural core domain. For ease of description, the ECD-containing C-terminal half is hereafter referred to as Beclin 1 ECD. After numerous trials, we succeeded in generating high-quality crystals of human Beclin 1 ECD. The structure was determined at 1.6 Å resolution by multi-wavelength anomalous dispersion (MAD) (Table 1 and Figure 1B).
Table 1. Statistics of data collection and refinement.
| Data | Native | Selenium-SAD |
|---|---|---|
| Integration package | HKL2000 | HKL2000 |
| Space Group | C2 | C2 |
| Unit Cell (a,b,c, in Å, β in degrees) | a=80.75, b=33.56, c=74.77, β=113.49 | a=82.2, b=33.69, c=75.2, β=114.09 |
| No. of molecules in ASU | 1 | 1 |
| Wavelength (Å) | 1.00000 | 0.97894 |
| Resolution (Å) | 50~1.55 (1.61~1.55) | 50~1.55 (1.61~1.55) |
| Rmerge (%) | 7.9 (56.1) | 9.9 (63.4) |
| I/sigma | 16.4 (2.1) | 22.7 (1.9) |
| Completeness (%) | 97.8 (93.3) | 94.2 (69.5) |
| No. of reflections | 89,885 | 180,936 |
| No. of unique reflections | 26,572 | 26,274 |
| Redundancy | 3.4 (3.2) | 6.9 (5.3) |
| Wilson B factor (Å2) | 20.4 | 23.5 |
| R / Rfree (%) | 17.55 / 20.34 | |
| No. atoms | ||
| Overall | 1816 | |
| Main chain | 776 | |
| Side chain | 925 | |
| Water | 115 | |
| Average B value (Å2) | ||
| Overall | 37.59 | |
| Main chain | 33.80 | |
| Side chain | 40.31 | |
| Water | 41.36 | |
| Other entities | ||
| R.m.s. deviations | ||
| Bonds (Å) | 0.012 | |
| Angle (°) | 1.336 | |
| Ramachandran plot statistics (%) | ||
| Most favourable | 88.8 | |
| Additionally allowed | 8.9 | |
| Generously allowed | 1.8 | |
| Disallowed | 0.6 |
Values in parentheses are for the highest resolution shell. Rmerge=ΣhΣi|Ih,i-Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of symmetry related reflections of h. R=Σ|Fobs-Fcalc|/ΣFobs, where Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections selected).
The 24-kDa Beclin 1 ECD forms a compact ellipsoid, with four α-helices (α1-α4), three pairs of anti-parallel β-strands (β1-β6) and six intervening loops (Figure 1B and 1C). These structural elements are organized into three internal repeats, each comprising a pair of short β-strands and an ensuing α-helix. These three repeats can be superimposed with each other with a pair-wise root-mean-squared deviation (RMSD) of 1.4-3.1 Å (Supplementary information, Figure S1A). The three α-helices (α2-α4) of the structural repeats form a parallel, left-handed helical bundle at the center, surrounded by three β-sheets, six loops and the N-terminal α1 helix. The surface of Beclin 1 ECD is highly charged (Supplementary information, Figure S1B).
A prominent feature of the Beclin 1 ECD structure is protrusion of loop L4 away from the main ellipsoid. Strikingly, three consecutive aromatic amino acids, Phe359, Phe360 and Trp361, are located at the tip of the protrusion (Figure 1B). For simplicity of description, we refer to this structural element as an aromatic finger. In the crystal lattice, neighboring ECD molecules stack up against each other by placing the aromatic finger of one molecule into a hydrophobic pocket of another molecule, forming an elongated rod (Supplementary information, Figure S2). These stacking interactions, together with the highly conserved nature of the three aromatic residues in all metazoans (Figure 1C), suggest potential functional importance for the aromatic finger. Another notable feature of Beclin 1 ECD is a deep cleft next to the aromatic finger (Figure 1B).
Because structural homology frequently offers insights into the function of a previously uncharacterized protein, we subjected the atomic coordinates of Beclin 1 ECD to extensive search of the Protein Data Bank using the program DALI 19. Unfortunately, we failed to identify any entry that is structurally homologous to Beclin 1 ECD over its entire length. This analysis suggests that Beclin 1 ECD may represent a previously undocumented protein fold.
Beclin 1 ECD directly associates with lipid membrane
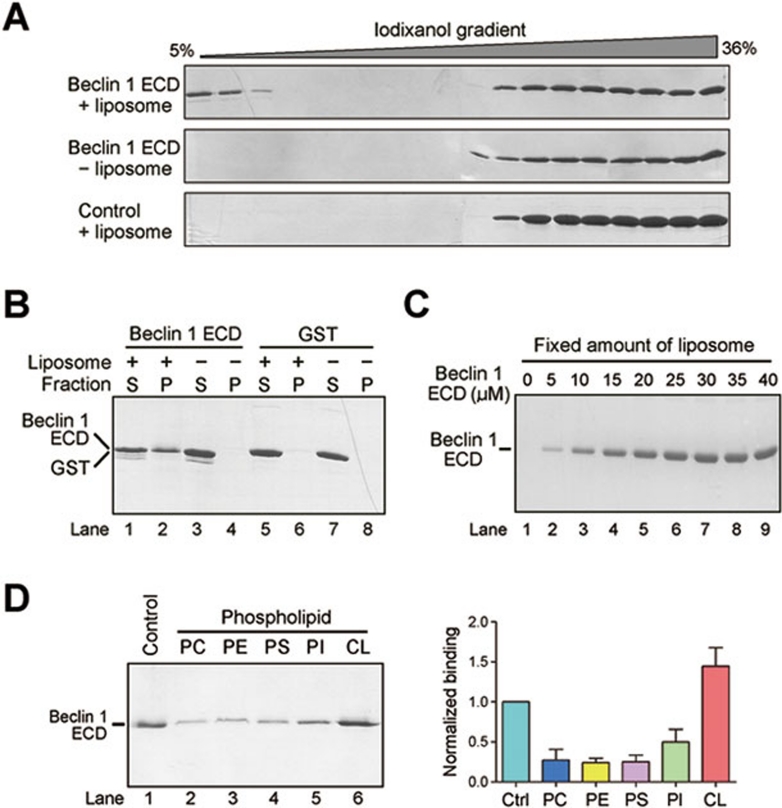
The core function of Beclin 1 is linked to lipid membrane 20, and a PI3K complex containing Beclin 1 is responsible for autophagic vesicle nucleation 10. The prominent location of the aromatic finger (Figure 1B) and the crystal lattice interactions in Beclin 1 ECD (Supplementary information, Figure S2) are all reminiscent of features of a membrane-binding domain such as the C2 domain of human coagulation factor V 21, 22 (Supplementary information, Figure S3). We hypothesized that Beclin 1 ECD may directly associate with lipid membrane. To investigate this possibility, we prepared liposomes using lipid composition similar to that of Xenopus mitochondria 23 and examined their association with Beclin 1 ECD using density gradient centrifugation analysis. The results, employing either iodixanol (Figure 2A) or sucrose (Supplementary information, Figure S4A), showed that Beclin 1 ECD, but not other cytosolic protein, directly associated with the liposomes. The nonionic reagent iodixanol is iso-osmotic over a wider range of density gradients and thus offers better separation than sucrose for most biological samples 24, 25.
Figure 2.
Beclin 1 ECD directly binds to lipid membrane. (A) Beclin 1 ECD associates with liposome by iodixanol gradient sedimentation analysis. Liposome co-sediments with low percentage of iodixanol, where Beclin 1 ECD was also detected (top panel). Free Beclin 1 ECD co-sediments with high percentage of iodixanol (top and middle panels). The control protein Nampt 46 failed to co-sediment with liposome (bottom panel). (B) Beclin 1 ECD binds to liposome by ultracentrifugation analysis. Beclin 1 ECD or GST was ultracentrifuged in the presence or absence of liposome. The pellet and supernatant fractions were analyzed by SDS-PAGE and coomassie staining. (C) Analysis of liposome binding by Beclin 1 ECD. A fixed amount of liposome was ultracentrifuged in the presence of increasing amounts of Beclin 1 ECD. The pellets were analyzed by SDS-PAGE and coomassie staining. (D) Beclin 1 ECD preferentially binds to liposome enriched with cardiolipin. Liposomes of five distinct compositions were prepared, each containing fixed components (70% PC and 20% PE) and a variable component (10% each of the five specific phospholipids). Binding to Beclin 1 ECD was analyzed for these five liposomes. The control liposome has the same phospholipid composition as that of the Xenopus mitochondria 23.
We performed two additional experiments to investigate the potential interactions between Beclin 1 ECD and the liposomes. In the first experiment, we pre-incubated liposomes with Beclin 1 ECD, pelleted the membrane fraction through ultracentrifugation, and analyzed the presence of ECD in both supernatant and pellet (Figure 2B). This experiment showed that, only in the presence of the liposomes, Beclin 1 ECD could be detected in the pellet (Figure 2B). The soluble protein glutathione S-transferase (GST) failed to be pelleted in the presence of the liposomes. In the second experiment, we incubated a fixed concentration of liposomes (0.6 mg/ml) with increasing concentrations of ECD and examined the ability of the liposomes to bind Beclin 1 ECD. Consistent with direct interactions between Beclin 1 ECD and the liposomes, the result showed saturable binding by Beclin 1 ECD (Figure 2C).
Beclin 1 ECD preferentially binds to liposomes enriched with cardiolipin
Next, we investigated whether Beclin 1 ECD exhibits any binding preference towards liposomes constituted by different phospholipids. Using 70% phosphatidylcholine (PC) and 20% phosphatidyl ethanolamine (PE) as the fixed components, we prepared five distinct liposomes, each with a variable component of 10% of the phospholipids PC, PE, phosphatidyl serine (PS), phosphatidyl inositol (PI) and cardiolipin (CL). An identical concentration of Beclin 1 ECD (10 μM) was incubated with these liposomes (1.2 mg/ml); the outcome of binding was determined by ultracentrifugation analysis. This preliminary analysis revealed that Beclin 1 ECD preferentially bound to liposomes that contained 10% cardiolipin as the variable component (Figure 2D).
To further examine lipid preferences, we performed binding experiments using liposomes composed of only two phospholipids. These liposomes contain decreasing percentages of PC as the fixed component and increasing percentages of a specific phospholipid. Confirming earlier observations, Beclin 1 ECD was found to have the highest preference for liposomes that were enriched with cardiolipin, followed by the two acidic phospholipids PI and PS (Supplementary information, Figure S4B). By sharp contrast, Beclin 1 ECD exhibited little binding for liposomes that were composed of the two neutral phospholipids PC and PE. These liposomes exhibited normal appearance as judged by electron microscopic analysis (Supplementary information, Figure S5A), ruling out potential problems associated with liposome formation. Finally, in contrast to Beclin 1 ECD, the C2 domain of MFG-E8 exhibited a strong preference for PS-enriched liposome (Supplementary information, Figure S5B). The result of this control experiment is consistent with previous reports 26, 27 and supports the validity of results derived from our liposome binding assays.
Aromatic finger is required for membrane association
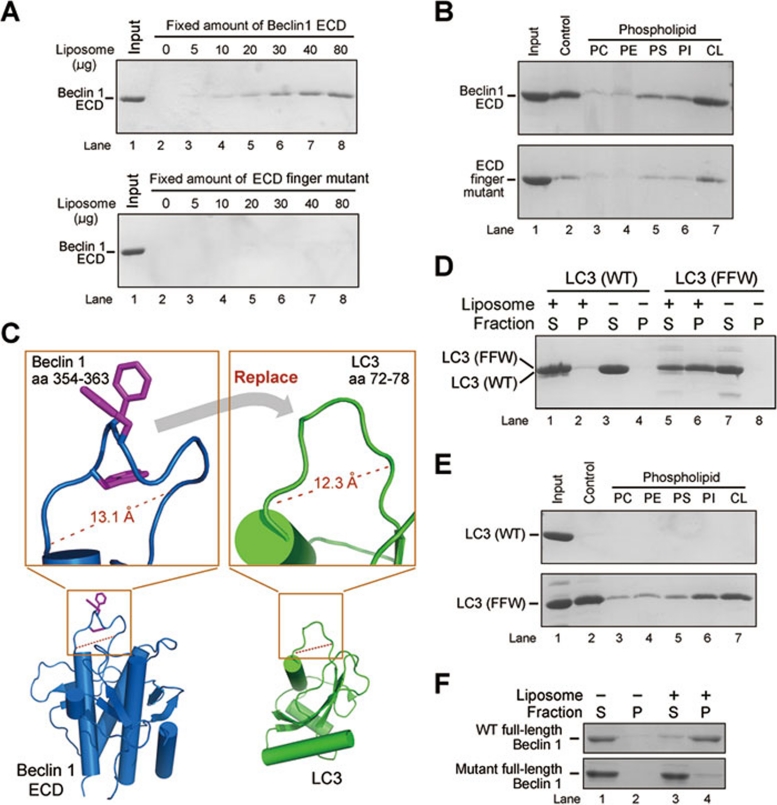
To examine the role of the aromatic finger in membrane association, we generated a variant of Beclin 1 ECD that had the three aromatic residues Phe359/Phe360/Trp361 replaced by negatively charged aspartate residues (Beclin 1 ECD, F359D, F360D and W361D). These mutations do not compromise the overall structure or thermal stability of the ECD (Supplementary information, Figure S6A and S6B), and the mutant ECD maintains its native fold in the presence of liposomes (Supplementary information, Figure S6C). Strikingly, the finger mutant displayed a sharply reduced ability to associate with liposomes (Figure 3A) and no longer exhibited the same level of preference for cardiolipin-enriched liposomes as the wild-type (WT) ECD (Figure 3B). These experiments indicate that the aromatic finger of Beclin 1 ECD is required for membrane association and may be responsible for membrane lipid preference.
Figure 3.
Mutations in the aromatic finger of Beclin 1 ECD cripple its binding to mitochondria and liposomes. (A) The WT Beclin 1 ECD, but not the finger mutant (F359D/F360D/W361D), bound to liposome by ultracentrifugation analysis. (B) The WT Beclin 1 ECD, but not the finger mutant (F359D/F360D/W361D), exhibited a strong preference for cardiolipin-enriched liposomes. (C) A close-up view of the rationale for engineering a LC3 variant with grafted aromatic finger from Beclin 1. Amino acids 72-78 of LC3 constitute a surface loop with features similar to those of residues 354-363 of Beclin 1 ECD. Notably, the main chain Cα-Cα distances are similar in both cases. (D) The LC3 variant with engrafted aromatic finger, but not the WT LC3, associated with liposomes. S: supernatant; P: pellet (i.e., liposome fraction). (E) The LC3 variant with engrafted aromatic finger, but not the WT LC3, exhibited a strong preference for cardiolipin-enriched liposomes. (F) The full-length WT Beclin 1, but not the finger mutant (F359D/F360D/W361D), bound to liposome. S: supernatant; P: pellet (i.e., liposome fraction).
The aromatic finger is positioned away from the ECD and barely interacts with other sequence elements of the ECD. This structural feature engendered an intriguing hypothesis: the aromatic finger might allow a heterologous protein to associate with membrane if an appropriate conformation of the aromatic finger can be maintained. We chose the mammalian protein LC3 to test this hypothesis, because the high-resolution crystal structure of LC3 28 (PDB code 1UGM) allowed us to determine where to engraft the aromatic finger. The surface loop residues 72-78 of LC3 are positioned away from the core LC3 structure; the Cα-Cα distance between residues 72 and 78 of LC3 is similar to that between residues 354 and 363 of the L4 loop of Beclin 1 ECD (Figure 3C). We thus generated a LC3 variant with residues 72-78 replaced by the Beclin 1 residues 354-363 and examined its ability to associate with lipid membrane. Although the WT LC3 failed to bind to liposomes under our experimental conditions, the LC3 variant with engrafted aromatic finger gained the ability to stably interact with liposomes (Figure 3D). Similar to Beclin 1 ECD, this LC3 variant also exhibited a strong binding preference for liposomes enriched with cardiolipin (Figure 3E).
Our experimental evidence identifies the ECD-containing C-terminal half of Beclin 1 as a structural core capable of binding to lipid membrane in vitro, where the aromatic finger plays an essential role. To examine whether the full-length Beclin 1 exhibits a similar property, we managed to purify sufficient amounts of the full-length WT Beclin 1 protein as well as the full-length finger mutant F359D/F360D/W361D. Consistent with the conclusion for ECD, the full-length WT Beclin 1, but not the finger mutant, associated stably with liposomes in an in vitro membrane binding assay (Figure 3F). Interestingly, the full-length Beclin 1 protein has been reported to mainly localize in the cytoplasm, associating with various membrane fractions such as mitochondria 29, 30, ER 29, trans-Golgi network 10, 18, or simply intra-cytoplasmic organelles 31. These contrasting claims are likely a reflection of the fact that cellular localization of Beclin 1 may be affected by a multitude of interacting proteins and environmental cues.
Does Beclin 1 ECD associate with membrane in cells? To explore this scenario, we transfected NRK cells with Beclin 1 ECD and examined its cellular localization. Strikingly, the WT ECD predominantly co-localized with Mitotracker (a mitochondria-specific dye), whereas the finger mutant ECD partially lost this ability as judged by its diffuse staining pattern (Supplementary information, Figure S7A). This finding is further confirmed by cellular fractionation (Supplementary information, Figure S7B). In addition, proteinase K treatment of mitochondria in the absence of the solubilizing detergent Triton X-100 led to complete degradation of Beclin 1 ECD, but not the mitochondria residential proteins AIF or TIM23 (Supplementary information, Figure S7C). This result confirmed that Beclin 1 ECD is associated with the outer membrane of mitochondria.
These results strongly suggest that Beclin 1 ECD binds to lipid membrane in cells. We caution, however, that the physiological relevance and significance of mitochondrial association by Beclin 1 ECD remain to be determined, because the isolated Beclin 1 ECD does not exist in cells. Nonetheless, we note that Beclin 1 was thought to undergo caspase-mediated cleavages between the BH3 motif and the coiled-coil domain 30, 32 and the C-terminal cleavage product (residues 150-450, encompassing the coiled-coil and ECD) appeared to mainly localize to mitochondria 30. Furthermore, mitochondria were recently reported to be a major source of membrane for autophagosome biogenesis during starvation 33.
Beclin 1 ECD deforms liposome
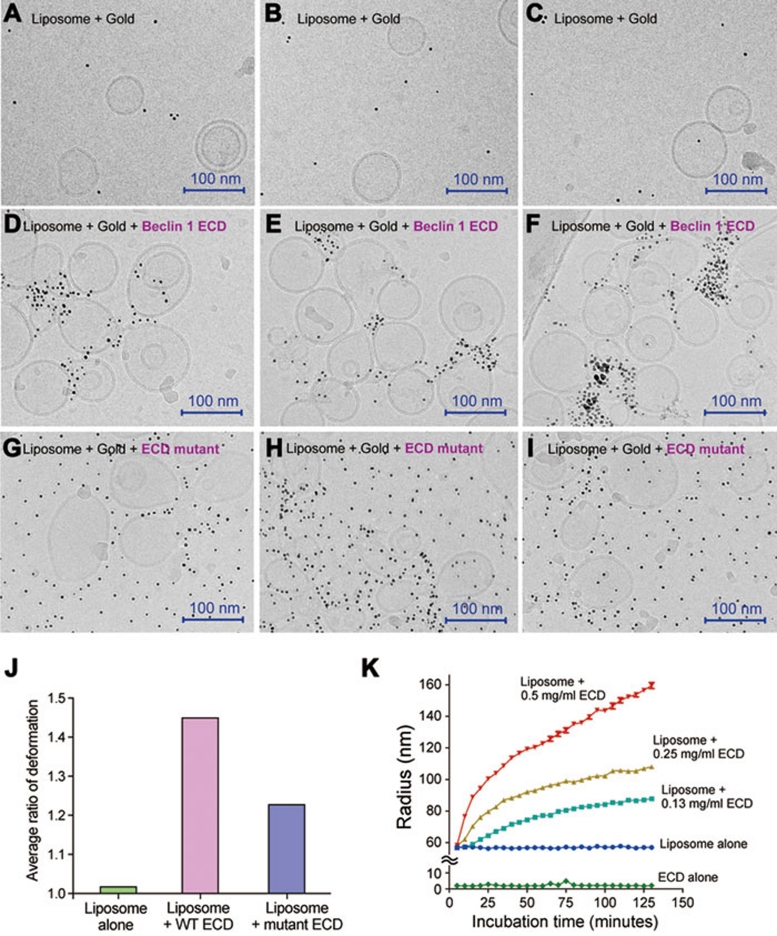
Our experimental evidence strongly suggests that the aromatic finger of Beclin 1 ECD may directly associate with the membrane by inserting into the lipid leaflet. In principle, insertion into lipid membrane by any motif will result in surface expansion of the leaflet where insertion occurs and consequently promotes membrane curvature 34, 35. We reasoned that Beclin 1 ECD may exert the same impact on liposomes. To examine this scenario, we investigated the morphology of liposomes in the absence and presence of Beclin 1 ECD by cryo-electron microscopy. In these experiments, the N-terminus of ECD contained a hexa-histidine tag, which was recognized by Ni2+-NTA-linked nanogold particles (5 nm in diameter each). In the absence of Beclin 1 ECD, the liposomes mostly adopted a normal, spherical morphology and the nanogold particles were rarely associated with the liposomes (Figure 4A-4C). In the presence of WT Beclin 1 ECD, the liposomes appeared to undergo deformation, and the nanogold particles were concentrated around the deformed surface areas, but not on the regular spherical surface (Figure 4D-4F). In particular, the nanogold particles were associated with smaller curved vesicles or areas where membrane protrusions appeared to be forming. By contrast, the presence of the mutant Beclin 1 ECD (F359D/F360D/W361D) had much less pronounced effect on the liposomes, and the nanogold particles were distributed throughout the microscopic view (Figure 4G-4I). These observations suggest that the WT, but not the mutant, Beclin 1 ECD efficiently induces membrane deformation, perhaps by modifying membrane curvature. Identical results were obtained for untagged Beclin 1 ECD (Supplementary information, Figure S8A), ruling out potential problems (such as aggregation) caused by the hexa-histidine tag.
Figure 4.
The Beclin 1 ECD deforms liposome. Free liposomes (panels A-C), liposomes pre-incubated with WT ECD (panels D-F), and liposomes pre-incubated with mutant ECD (panels G-I) were treated with nanogold particles and visualized under cryo-electron microscope. Beclin 1 ECD contains a 6×His tag, and the nanogold particle is linked to Ni2+-NTA. The WT Beclin 1 ECD, but not the mutant ECD, allowed the nanogold particles to be concentrated in the areas of liposome deformation, which might be liposome fusion, vesicle budding, and/or membrane vesiculation. (J) Quantitative analysis of liposome deformation by Beclin 1 ECD. The extent of liposome deformation is defined by the ratio of the longest dimension over the shortest dimension for each liposome. These ratios for 65 randomly chosen liposomes for each of three categories, control (no ECD), with WT ECD, and with mutant ECD, were measured and averaged. Liposomes incubated with WT ECD exhibited a larger average ratio than that for liposomes incubated with mutant ECD. (K) Incubation with Beclin 1 ECD led to increased radius for the liposomes. The increase is correlated with the concentrations of Beclin 1 ECD.
Free liposomes are usually spherical in shape due to surface tension (Figure 4A-4C). Liposome deformation of any type will result in alteration of the spherical shape. The extent of liposome deformation can be quantitatively defined by the ratio of the longest dimension over the shortest dimension for each liposome. A larger ratio indicates greater deformation. Relying on representative EM images, we measured the deformation ratios for 65 randomly chosen liposomes for each of three categories: control (no ECD), with WT ECD, and with mutant ECD. Then, an average ratio was calculated for each category. Free liposomes, which showed no signs of deformation, displayed an average deformation ratio of 1.02 (Figure 4J). By sharp contrast, liposomes incubated with WT ECD exhibited an average ratio of 1.45, considerably larger than the value of 1.23 for liposomes incubated with mutant ECD.
Liposome deformation may result in increase of its hydrodynamic radius, which can be detected by dynamic light scattering. To investigate this possibility for Beclin 1 ECD, we measured the average hydrodynamic radius of the liposomes every 5 min following incubation with Beclin 1 ECD (Figure 4K). Indeed, the radius expanded with time and the expansion was proportionally faster with increasing concentrations of Beclin1 ECD (Figure 4K). Mutation of the aromatic finger in Beclin 1 ECD markedly reduced the radius expansion (Supplementary information, Figure S8B). Because physical association between liposomes and Beclin 1 ECD is likely to reach equilibrium within a few minutes, the continued expansion of the liposome radius over a time span of 125 min suggested the possibility of continued tubulation or other forms of elongation over time.
Compromised ability of mutant Beclin 1 to rescue autophagy
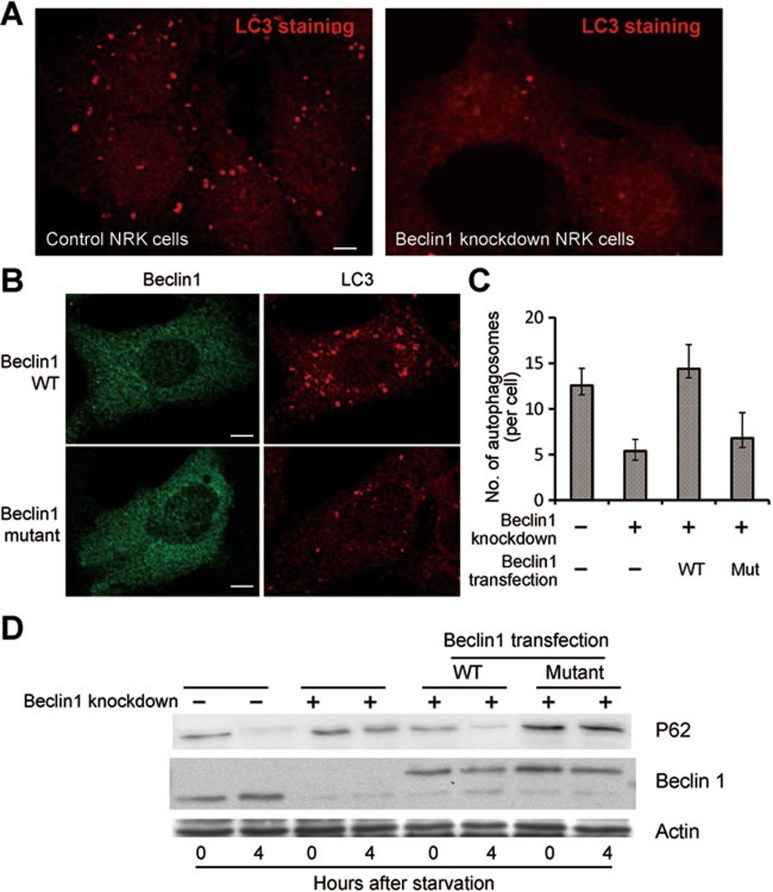
Beclin 1 is known to play an essential role in autophagy, particularly for formation of the autophagosomes 36, 37. If binding to lipid membrane is biologically important, mutation of the aromatic finger would likely compromise Beclin 1-mediated autophagy. To investigate this scenario, we generated NRK cells with stable knockdown of Beclin 1. Upon starvation, these cells were examined for signs of autophagy. In the control NRK cells, LC3 displayed a punctate staining pattern that is characteristic of normal autophagy (Figure 5A, left panel). In the Beclin 1 knockdown cells, however, the punctate staining disappeared, suggesting a defect in autophagy (Figure 5A, right panel). Next, we transfected these cells with WT or mutant Beclin 1. Only the WT Beclin 1, but not the mutant (F359D/F360D/W361D), restored LC3 staining to normal levels (Figure 5B). The number of autophagosome in cells transfected with WT Beclin 1 is considerably more than that in cells transfected with mutant Beclin 1 (Figure 5C). Consistent with these observations, only the WT Beclin 1, but not the mutant, was able to restore the autophagic degradation of p62 upon starvation (Figure 5D).
Figure 5.
The aromatic finger of Beclin 1 plays an important role in autophagy. (A) Beclin 1-knockdown cells exhibited a markedly reduced level of autophagy upon starvation. Control and Beclin 1 stable knockdown NRK cells were stained with anti-LC3 antibody 4 h after starvation. Scale bar, 5 μm. (B) The WT Beclin 1, but not the finger mutant (F359D/F360D/W361D), rescued the autophagy defect in NRK cells. Beclin 1 stable knockdown NRK cells were transfected with WT or mutant Beclin 1-YFP. 24 h after transfection, cells were starved for 4 h. Then the cells were stained with anti-GFP or anti-LC3 antibody. Scale bar, 5 μm. (C) Average numbers of autophagosome per cell are shown for panels A and B. 50 cells were counted for each sample. (D) NRK cells transfected with the WT Beclin 1, but not with the finger mutant (F359D/F360D/W361D), had significant autophagic degradation of p62, a hallmark of autophagy. Cells from panels A and B were analyzed by western blot using antibodies specific for p62, Beclin 1 and actin. The position of transfected Beclin 1 is higher because Beclin 1 was fused to YFP.
Compromised ability to rescue autophagy by mutant Beclin 1 can be explained by a number of scenarios. For example, mutation of the aromatic finger could negatively affect Beclin 1 interactions with a host of other cellular proteins, ultimately affecting autophagy. Similarly, the mutation could somehow lead to altered targeting of Beclin 1-binding proteins to the endoplasmic reticulum (ER). We experimentally scrutinized these possibilities. The results clearly demonstrate that mutation of the aromatic finger had no detectable consequence on the interactions between Beclin 1 and UVRAG, Barkor, or PI3KC3 (Supplementary information, Figure S9). In addition, the ER targeting of Barkor or PI3KC3 was unaffected by mutation of the aromatic finger (Supplementary information, Figures S10 and S11). Furthermore, mutation of the aromatic finger in Beclin 1 had no impact on the cellular distribution pattern of the early endosome antigen 1 (EEA1) (Supplementary information, Figure S12). However, in our hands, UVRAG was not significantly co-localized with EEA1 (Supplementary information, Figure S13). Nonetheless, mutation of the aromatic finger in Beclin 1 had no detectable effect on the cellular localization of UVRAG (Supplementary information, Figure S13).
A potential role of the Beclin 1 aromatic finger in omegasome formation
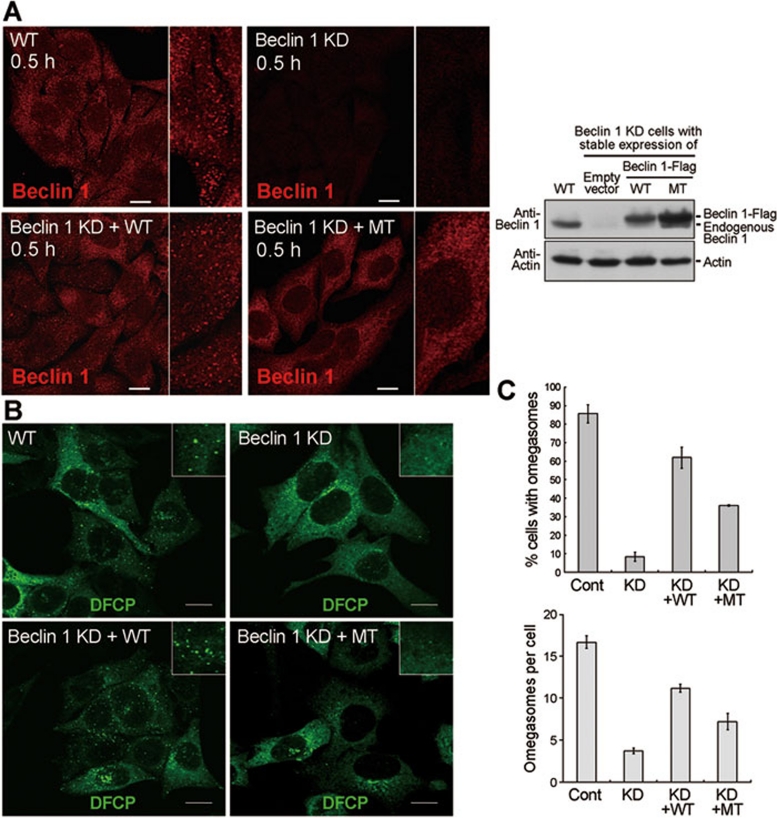
Collectively, our experimental results are consistent with the notion that the mutation in the aromatic finger compromises autophagy through crippled membrane association by Beclin 1, rather than through reduced interactions with other cellular proteins. Because Beclin 1 prefers to localize in regions of membrane deformation (Figure 4), we examined whether mutation of the aromatic finger in Beclin 1 affected its localization pattern, or whether the mutation affected formation of the omegasome, a specialized structure that is thought to be the precursor of autophagososme. The results indicate that, 30 min following starvation, only the WT Beclin 1, but not the mutant, was localized to punctate spots with the cells (Figure 6A). Importantly, the punctate staining of DFCP (a marker of omegasome) was considerably more in cells with stable expression of the WT Beclin 1, but not mutant (Figure 6B). Quantitative analysis shows that both the percentage of omegasome-positive cells and the average number of omegasomes in these cells are significantly reduced for cells with stable expression of the mutant Beclin 1 (Figure 6C). These preliminary results suggest that the aromatic finger of Beclin 1 may play an important role in formation of the omegasome and its mutation results in defective genesis of omegasome.
Figure 6.
The aromatic finger of Beclin 1 might be involved in omegasome formation. (A) The WT, full-length Beclin 1 exhibited punctate staining 30 min after starvation. Shown here are staining pattern for Beclin 1 in normal HeLa cells (WT), Beclin 1 stable knockdown HeLa cells (Beclin 1 KD), WT Beclin 1 stably expressing Beclin 1 KD cells (Beclin 1 KD+WT), mutant Beclin 1 stably expressing Beclin 1 KD cells (Beclin 1KD+MT). Scale bar, 10 μm. The right panel shows the expression levels of WT and mutant Beclin 1 in the 4 different types of cells. (B) The cellular localization of DFCP was restored by WT Beclin 1, but not mutant. Shown here are cellular staining pattern for DFCP in normal HeLa cells (WT), Beclin 1 KD cells, Beclin 1 KD+WT cells transfected with DFCP-GFP, and Beclin 1 KD+MT cells transfected with DFCP-GFP. 18 h after DFCP transfection, cells were starved for 0.5 h and stained for antibody against GFP. Scale bar, 5 μm. (C) The WT Beclin 1, but not the mutant, supported formation of a greater number of omegasomes (upper panel). Importantly, for the omegasome-containing cells, the average number of omegasomes in Beclin 1 KD+WT cells is ∼11.3, 57% more than that (7.2) in Beclin 1 KD+MT cells. Cells from panel B were quantified. 50 cells were counted for each category. Error bars represent standard deviation. For the top panel, the statistical P values are 0.0001/0.0055/0.0001 between “Cont” and “KD”/“KD+WT”/“KD+MT”, 0.0001/0.0001 between “KD” and “KD+WT”/“KD+MT”, and 0.0015 between “KD+WT” and “KD+MT”. For the bottom panel, the statistical P values are 0.0001/0.0022/0.0012 between “Cont” and “KD”/“KD+WT”/“KD+MT”, 0.0001/0.0216 between “KD” and “KD+WT”/“KD+MT”, and 0.0163 between “KD+WT” and “KD+MT”.
Discussion
The finding that Beclin 1 ECD preferentially associates with cardiolipin-enriched membrane suggests two distinct possibilities: (1) ECD directly binds cardiolipin, or (2) ECD does not directly bind cardiolipin but favors a general physical property of lipid membrane, such as curvature or charge, that can be supported by cardiolipin. These two scenarios have many precedents, exemplified by the phosphoinositide-specific PH domains and the membrane curvature-sensing BAR domains 22. Intriguingly, the bacterial protein MinD co-localized with cardiolipin to membrane regions with highly negative curvature; yet MinD was still distributed to these negatively curved membrane regions in cardiolipin-deficient bacteria 38. We examined the potential direct interactions of Beclin 1 ECD with cardiolipin by a variety of biochemical and biophysical methods, including lipid overlay studies and isothermal titration calorimetry. Despite repeated trials, we have been unable to detect specific interaction between cardiolipin and Beclin 1 ECD.
Next we assessed the possibility that Beclin 1 ECD may bind to specific membrane regions with defined topology or charge. A unique physical property of cardiolipin — a very small head group and a bulky tail — gives rise to local membrane structure with negative curvature; Beclin 1 ECD might simply favor such membrane regions. In this regard, mitochondria localization by Beclin 1 ECD could be a coincidence, because mitochondria, which is thought to contain cardiolipin in its outer and inner membrane 23, 39, 40, may exhibit such desirable membrane property for binding by Beclin 1 ECD. Each molecule of cardiolipin can be thought of as two phosphatidic acid (PA) molecules linked by an isopropanol unit; thus membrane curvature induced by one molar equivalence of cardiolipin is predicted to be similar to that induced by two molar equivalence of PA. Consistent with this analysis, Beclin 1 ECD binds almost equally well to liposomes with one molar equivalence of cardiolipin or with two molar equivalence of PA (Supplementary information, Figure S14). We caution that this analysis merely provides a clue, not an answer, to the question of why Beclin 1 ECD favors cardiolipin-enriched membrane. Conclusive answer to this question may embody a separate study. It should be noted that, at pH 7, each cardiolipin molecule carries an average of −1.3 negative charges (not −2) due to formation of intramolecular hydrogen bonds 41, whereas each molecule of PA has about −1.1 negative charges (pKa1∼3.0, pKa2∼8.0). This analysis does not favor net lipid charge as a dominant factor for Beclin 1 ECD binding to membrane.
Our experimental evidence shows that Beclin 1 ECD binds to lipid membrane through its aromatic finger. Mutation of these aromatic residues abrogated the membrane-binding ability of the ECD and compromised autophagy function of the full-length Beclin 1. These findings support the notion that the membrane-binding activity of Beclin 1 may be central to its function in autophagy. Curiously, however, the full-length Beclin 1 contrasts the ECD and mainly exhibits a diffuse staining pattern in normal cells (Supplementary information, Figure S15). Interestingly, upon starvation, the staining pattern for Beclin 1 becomes more punctate (Supplementary information, Figure S15), particularly at the 30-min time point (Figure 6A). This observation suggests that the full-length Beclin 1 might exist in an inhibitory conformation (perhaps through homo-dimerization via the coiled-coil domain), where the aromatic finger is sequestered and unable to associate with membrane. We further speculate that, at the onset of autophagy, binding to other protein(s) may allow Beclin 1 to expose the aromatic residues for association with lipid membrane, resulting in an altered cellular localization pattern. How could this be accomplished? One possibility is that hetero-dimerization with other binding proteins such as ATG14/Barkor relieves the inhibitory conformation of Beclin 1.
The aromatic finger in loop L4 of Beclin 1 appears to be highly conserved in all metazoans, but not in yeast where the three aromatic residues Phe359-Phe360-Trp361 were replaced by Val418-Tyr419-Tyr420 (Figure 1C). The L4 loop in ATG6 is also considerably longer than the corresponding loops in metazoans (Figure 1C). This sequence analysis strongly suggests that the function of aromatic finger may not be conserved in yeast. Supporting this analysis, mutation of Val418-Tyr419-Tyr420 in ATG6 had no apparent consequence on the autophagy or Cvt pathways in Saccharomyces cerevisiae (Supplementary information, Figure S16).
Beclin 1 plays an indispensable role in nucleation of the autophagic vesicles through a multi-protein complex containing ATG14/Barkor and a class III PI3K 10. This process is thought to involve formation of a PI3P-enriched ER domain named omegasome 42, 43, 44. The observations reported in this study make Beclin 1 ECD a potential candidate for initiating the omegasome formation. The finding that Beclin1 localized on punctate structure in starved cells on an aromatic finger-dependent manner and the observation that recombinant Beclin 1 ECD can induce liposome deformation, raise an interesting possibility that Beclin 1 may bind or induce membrane structures with high curvature, which is required for omegasome formation. Recently, mitochondria were reported to be a major source of membrane for autophagosome biogenesis during starvation 33; whether Beclin 1 plays a role in this process remains to be investigated.
In summary, our experimental evidence unequivocally demonstrates that Beclin 1 ECD adopts a previously unreported protein fold, and unexpectedly, associates with lipid membrane through its aromatic finger. Mutation of the aromatic finger compromises the autophagic function of Beclin 1 without affecting its interactions with a host of other cellular proteins. These findings have important ramifications for understanding Beclin 1 function in omegasome and autophagosome formation. Despite these advances, we recognize that the detailed molecular mechanisms underlying autophagy remain largely enigmatic and many questions about Beclin 1 remain to be investigated. For example, what is the precise role of Beclin 1 in the formation of omegasome? How is Beclin 1 regulated? Can the membrane-binding activity of the Beclin 1 aromatic finger be explored for the regulation of autophagy? Answers to these questions await future investigations.
Materials and Methods
Protein and plasmid preparation
The WT and mutant ECD domain of human Beclin 1 (residues 248-450) was cloned into pET15b (Novagen) with an N-terminal 6×His tag. The aromatic finger mutant (F359D/F360D/W361D) was generated by PCR-based mutagenesis. Both WT and mutant ECD constructs were overexpressed in E. coli BL21(DE3) at 15 °C overnight after induction by 0.2 mM β-D-thiogalactopyranoside (IPTG) at OD of 1.2 at 600 nm. The ECD domains were purified by Ni2+-NTA affinity column (Qiagen). The protein was eluted from the affinity resin by 350 mM imidazole, 25 mM Tris (pH 8.0), 150 mM NaCl, and concentrated to around 10 mg/ml before further purification by gel filtration (Superdex 75, GE Healthcare) in a buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl, 2 mM dithiothreitol. The peak fraction was collected and concentrated to ∼10 mg/ml for crystallization. Selenomethionine-substituted Beclin 1 ECD was similarly prepared. For all liposome-related assays, proteins were in buffer containing 50 mM Na2HPO4/NaH2PO4 (pH7.4). For in vivo localization assay, the Beclin 1 ECD (residues 272-450) was cloned into pEYFP-N1 (Clontech) with a C-terminal YFP. The C2 domain (residues 306-426) of MFG-E8 was cloned into pET21b (Novagen) with a C-terminal 8×His tag. The LC3 variant was cloned into pET15b (Novagen) with an N-terminal 6×His tag. MFG-E8 and LC3 variants were similarly prepared as Beclin 1 ECD. Beclin1-flag WT and mutant (F359D/F360D/W361D) were cloned into pcDNA4 with a Flag tag at the C-terminus. Barkor-YFP and PI3KC3-YFP were cloned into pEYFP-N1 (Clontech) with a C-terminal YFP. Flag-UVRAG, Barkor-Myc, and PI3KC3-Flag were generous gifts from Dr Qing Zhong of UC Berkeley, USA.
Crystallization
Crystals were grown at 18 °C by the hanging-drop vapor-diffusion method mixing 1.5 μl of Beclin 1 ECD domain with 1.5 μl of reservoir solution contain 30% PEG3350, 0.3 M NaCl and 100 mM Tris buffer (pH 8.5). The Se-Met protein crystals used for MAD phasing were obtained in a similar manner. Both native and Se-Met crystals were directly flash frozen in a cold nitrogen stream at 100 K.
Data collection and structural determination
All data were collected at the SPring-8 beamline BL41XU, integrated and scaled using the HKL2000 package 47. Further processing was carried out using programs from the CCP4 suite 48. Data collection statistics are summarized in Table 1. The selenium positions were determined using the program SHELXD 49. The identified sites were refined and initial phases were generated in the program PHASER 50 with the SAD experimental phasing module. Solvent flattening and histogram matching were performed using DM 51. The initial model was traced automatically using the program BUCCANEER 52 and was manually rebuilt in COOT 53. The final structure was refined with PHENIX 54.
Cell culture and transfection
NRK cells were obtained from American Type Culture Condition (ATCC) and cultured in DMEM (Hyclone) medium supplemented with 10% FBS. Beclin 1-YFP NRK cell lines were selected with 1 mg/ml G418. Beclin 1 stable knockdown HeLa cells, a generous gift from Dr Quan Chen, were maintained with 200 μg/ml G418 and further transfected with WT or mutant Beclin 1-flag under selection of 500 μg/ml Zeocin. Cells were transfected via AmaxanucleofectionTM using solution T and program X-001, using 200 pmol RNAi or 2 μg DNA.
Antibodies and dyes
Mouse anti-GFP (Roche), mouse anti-LC3 (MBL), rabbit anti-P62 (MBL), rabbit anti-Atg14 (MBL), mouse anti-UVRAG (MBL), mouse anti-Beclin 1 (BD Transduction Laboratories), rabbit anti-vps34 (Sigma), mouse anti-FLAG (Sigma), mouse anti-myc (Sigma-Aldrich), mouse anti-EEA1 (BD Transduction Laboratories), mouse anti-Tom20 (BD Transduction Laboratories), mouse anti-Tim23 (BD Transduction Laboratories), rabbit anti-Calreticulin (abcam), rabbit anti-GFP (Invitrogen). Mitotracker Red and secondary antibodies were all purchased from Invitrogen.
Live cell imaging
Transfected cells were re-plated in Lab Tek Chambered coverglass (NUNC) the night before imaging, and cells were maintained at 37 °C with 5% CO2 in a LCI chamber (LCI, Seoul, Korea). Images were acquired by an Olympus FV-1000 confocal microscope.
Immunofluorescence staining
Cells were washed with phosphate buffered saline (PBS), fixed in 4% paraformaldehyde for 10 min. After wash cells with PBS 3 times for ten min each, incubate for 10 min with PBS containing 0.1% saponin or 30 min with 100 μM digitonin on ice for permeabilization. After blocking with 10% FBS in PBS for 30 min, cells were stained with primary antibody in blocking buffer for 1 h and washed with PBS three times. Cells were then stained with fluorescein isothiocyanate-conjugated secondary antibody in PBS for 1 h and washed with PBS 3 times.
Liposome preparation
Liposomes were prepared with the lipid composition of Xenopus mitochondrial membranes. Chicken egg PA, Chicken egg PC, chicken egg PE, porcine brain PS, bovine liver PI, and bovine heart CL (Avanti Polar Lipids) all dissolved in chloroform, were then mixed in a glass vial with 47% PC:28%PE:9%PS:9%PI:7%CL. Unless specifically indicated, liposomes refer to above lipid compositions. Chloroform was evaporated under a stream of N2 for 30 min. Lipids were further dried overnight in Concentrator Plus (Eppendorf). Dry lipid films were hydrated at room temperature with constant mixing in 50 mM Na2HPO4/NaH2PO4 (pH 7.4). Following hydration, lipid vesicles were subjected to freeze-thaw cycles in liquid N2 and room temperature water bath, and then sized by extruder (Avanti). For liposome binding assay, we used large unilamellar vesicles (LUV) that were sized using 100 nm polycarbonate filters (Whatman).
Discontinuous density gradient floatation assay
Human Beclin 1 ECD and LUVs were mixed together for at least 5 min at room temperature. The protein-liposome mixtures were then mixed with stock iodixanol solution (Sigma) to generate 1 ml 36% iodixanol solution with 2.4 mg/ml LUVs and 20 μM protein, which was loaded at the bottom of a centrifuge tube. 1 ml 31% and 0.4 ml 5% iodixanol solutions were sequentially loaded on top of the sample. After centrifugation at 199 000× g for 3 h, fractions were collected from the top to the bottom and then subjected to SDS-PAGE followed by coomassie blue staining.
Sucrose flotation assay
Beclin 1 ECD (10 μM) and 1.2 mg/ml LUVs were mixed with stock sucrose (68% w/v) to generate 1 ml 50% (w/v) sucrose liposome-protein solution, which was overlaid with 1 ml 40% (w/v) sucrose and 0.25 ml buffer containing 50 mM Na2HPO4/NaH2PO4 (pH 7.4). After centrifugation at 75 000× g for 16 h at 4 °C, 0.16 ml per fraction was collected from top to bottom. Then each fraction was analyzed by SDS-PAGE followed by coomassie blue staining.
Co-immunoprecipitation
293T cells were transiently transfected using the chemical-based transfection method with indicated plasmids. All the following steps were performed at 4 °C. For co-immunoprecipitation, cells were lysed in TNE buffer (150 mM NaCl, 10 mM Tris (pH 7.4), 1% NP-40, 1 mM EDTA, 1 mM PMSF) for 30 min. Then after 3 cycles of freezing-thawing, lysates were centrifuged for 18 000× g for 30 min. Supernatants were incubated with 50 μl protein G (Thermo) for 2 h with agitation and were centrifuged at 1 000× g for another 2 min. The bead pellet was discarded and the supernatants were centrifuged at 18 000× g for 30 min to remove contaminants. Then the supernatants were subjected to co-immunoprecipitation using primary antibody and protein G. The mixture was incubated for 1 h. Then, lysates with antibody and protein G bead were centrifuged for 1 000× g. Immune complexes were washed 3 times with TNE buffer, then subjected to SDS-PAGE, and analyzed by western blot.
Mitochondrial fractionation
Cells expressing indicated variant Beclin 1 were suspended in (150 mM NaCl, 10 mM HEPES (pH 7.4), 10 mM mannitol, 50 mM sucrose, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF) and homogenized by Dounce homogenizer. The cell homogenate was centrifuged at 1 000× g for 10 min to remove the nucleus and unbroken cells. Then the supernatant fraction was centrifuged at 18 000× g for 30 min to obtain crude mitochondria. The pellet was resuspended to the same volume of supernatant by mitochondria isolation buffer as mitochondrial fraction. The supernatant was collected as the cytosol fraction.
Proteinase K protection assay
Isolated mitochondria from NRK cells stably expressing Beclin 1 ECD were resuspended in ice-cold mitochondria isolation buffer and treated with proteinase K (50 μg/ml) for 15 min on ice with or without 1% Triton X-100. The proteinase K reaction was stopped by adding 5 mM PMSF and then subjected to SDS-PAGE and analyzed by western blot.
ER isolation
HeLa cells were trypsinized and resuspended in 10 mM HEPES pH 7.8, with 1 mM EGTA and 25 mM KCl after being washed twice by PBS. After cells were fully swollen for 20 min on ice, cells were centrifuged and resuspended into 10 mM HEPES pH 7.8 with 0.25 M sucrose, 1 mM EGTA and 25 mM KCl. Cells were lysed by Dounce homogenizer (with 30%-50% breakage, determined by Typan Blue) and subjected to centrifugation at 1 000× g for 10 min at 4 °C. Supernatant was collected as total fraction and centrifuged at 12 000× g for 15 min at 4 °C. Supernatant was carefully removed and applied to ultracentrifugation at 100 000× g for 60 min at 4 °C. Collect supernatant as post-mitochondrial cytosol fraction and resuspend the pellet to the same volume of supernatant as crude ER fraction.
Liposome sedimentation assay
5 μM of human Beclin 1 ECD was incubated with 0.6 mg/ml LUVs in 50 mM Na2HPO4/NaH2PO4 (pH 7.4) at 25 °C for 1 h in a total volume of 40 μl. For lipid preference assay, 10 μM protein were incubated with 1.2 mg/ml liposomes in a total volume of 80 μl. Samples were centrifuged in a Beckman Optima L-90K ultracentrifuge using a 42.2 Ti rotor at 4 °C for 15 min at 100 000× g. The supernatant was removed for determination of free protein that is not bound to liposomes. The pellets were washed twice with the same buffer and brought up to the same volume as the supernatant. The supernatant and pellet fractions were subjected to SDS-PAGE followed by coomassie blue staining.
Circular dichroism (CD)
CD scanning spectrum were collected between 190 and 260 nm at 25 °C using a quartz cuvette with 1 mm path length on Photophysics Chirascan spectropolarimeter equipped with thermoelectric temperature control. Measurements were performed on protein samples at 5 μM and liposome at 0.6 mg/ml in 50 mM Na2HPO4/NaH2PO4 (pH 7.4). The protein-liposome samples were incubated at 25 °C for 1 h before scan. Spectrum were collected at 1.0 nm interval at a scan rate of 50 nm/min and a response time of 5 s, subtracted for buffer signal, and averaged for 3 scans. Melting temperature spectra were collected between 25 °C and 90 °C at 222 nm, averaging from 3 scans. The temperature increment was 1 °C and the equilibration time was 3 min with 0.1 °C tolerance.
Dynamic light scattering (DLS)
LUVs (0.6 mg/ml) were incubated with ECD of different concentrations at 25 °C and subjected to analysis by dynamic light scattering in a DynoPro Nanostar (Wyatt Technology). A standard 10 μl cuvette was used and the samples were incubated for 5 min before the first reading was recorded. A measurement was performed every 5 min afterwards. Ten consecutive acquisitions were averaged for each measurement. DYNAMICS V7 was used to analyze the calculated average sizes and radius.
Cryo-electron microscopy
Protein (20 μΜ) and liposome (2.4 mg/ml) were incubated for 1 h at 25 °C. The samples were then incubated with 5 nm Ni-NTA-Nanogold (Nanoprobes, NY, USA) for 30 min at room temperature. An aliquot of 5 μl of mixture was applied to a 300 mesh glow-discharged Quantifoil 2/4 grid (Quantifoil Micro Tools GmbH, Jena, Germany) blotted inside an Vitrobot (FEI, Hillsboro, OR, USA) at 22 °C and plunged into liquid ethane. The specimens were examined at liquid nitrogen temperature in an FEI Titan Krios cryo-electron microscope operated at 300 kV using a nominal magnification of 59 000× g. Images were recorded under low-dose conditions with a dose of ∼20 e-/A2 on an FEI Eagle 4k×4k CCD camera with a post-magnification ratio of 1.7.
Acknowledgments
We thank Jianhua He and Sheng Huang at Shanghai Synchrotron Radiation Source and staff at the SPring-8 beamlines for help. This work was supported by funds from the Ministry of Science and Technology (2009CB918801 to YS, 2010CB833704 and 2011CB910100 to LY, 2010CB912402 to NG), the National Natural Science Foundation of China (30888001 to YS, 30971484 and 31030043 to LY), and Tsinghua University (2010THZ0 and 2009THZ03071 to LY). The atomic coordinates of Beclin 1 ECD have been deposited in the Protein Data Bank with the accession code 4DDP.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Structural features of Beclin 1 ECD.
Beclin 1 ECD molecules form an elongated rod in the crystals, relying on the aromatic finger.
Structural comparison of Beclin 1 ECD with the C2 domain of human coagulation factor V.
Characterization of liposome binding by Beclin 1 ECD.
Validation of the membrane binding experiments by Beclin 1 ECD.
Mutation of the aromatic finger does not compromise the structure or stability of the Beclin 1 ECD.
Characterization of mitochondria binding by Beclin 1 ECD.
Mutation of the aromatic finger compromises the ability of Beclin 1 ECD to deform membrane.
Mutation of the aromatic finger does not affect interactions between Beclin 1 and UVRAG, Barkor, or PI3KC3.
Mutation of the aromatic finger does not alter targeting of Barkor or PI3KC3 to endoplasmic reticulum (ER).
Mutation of the aromatic finger does not alter targeting of Barkor or PI3KC3 to endoplasmic reticulum (ER).
Mutation of the aromatic finger does not affect the cellular distribution pattern of EEA1.
Mutation of the aromatic finger does not alter cellular localization of UVRAG or EEA1 before or after starvation.
Characterization of liposome binding by Beclin 1 ECD.
Cellular localization pattern of endogenous Beclin 1 in NRK cells following starvation.
Mutation of the residues VYY in loop L4 of ATG6 does not appear to cause detectable disruption of autophagy or Cvt pathway in Saccharomyces cerevisiae.
References
- Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aita VM, Liang XH, Murty VV, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, Marcusson EG, Cereghino JL, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5:534–536. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS) FEBS Lett. 2010;584:1280–1286. doi: 10.1016/j.febslet.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo-Ribeiro S, Bode W, Huber R, et al. Crystal structures of the membrane-binding C2 domain of human coagulation factor V. Nature. 1999;402:434–439. doi: 10.1038/46594. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Mackey MR, Perkins G, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- Ford T, Graham J Rickwood D. Iodixanol: a nonionic iso-osmotic centrifugation medium for the formation of self-generated gradients. Anal Biochem. 1994;220:360–366. doi: 10.1006/abio.1994.1350. [DOI] [PubMed] [Google Scholar]
- Gias E, Nielsen SU, Morgan LA, Toms GL. Purification of human respiratory syncytial virus by ultracentrifugation in iodixanol density gradient. J Virol Methods. 2008;147:328–332. doi: 10.1016/j.jviromet.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisenko GG, Iverson SL, Ahlberg S, Kagan VE, Fadeel B. Milk fat globule epidermal growth factor 8 (MFG-E8) binds to oxidized phosphatidylserine: implications for macrophage clearance of apoptotic cells. Cell Death Differ. 2004;11:943–945. doi: 10.1038/sj.cdd.4401421. [DOI] [PubMed] [Google Scholar]
- Nakatani H, Aoki N, Nakagawa Y, et al. Weaning-induced expression of a milk-fat globule protein, MFG-E8, in mouse mammary glands, as demonstrated by the analyses of its mRNA, protein and phosphatidylserine-binding activity. Biochem J. 2006;395:21–30. doi: 10.1042/BJ20051459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, et al. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 2004;9:611–618. doi: 10.1111/j.1356-9597.2004.00750.x. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Wirawan E, Vande Walle L, Kersse K, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–3449. [PubMed] [Google Scholar]
- Zhu Y, Zhao L, Liu L, et al. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell. 2010;1:468–477. doi: 10.1007/s13238-010-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- Graham TR, Kozlov MM. Interplay of proteins and lipids in generating membrane curvature. Curr Opin Cell Biol. 22:430–436. doi: 10.1016/j.ceb.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27 Suppl 1:S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD, Weibel DB. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc Natl Acad Sci USA. 2011;108:6264–6269. doi: 10.1073/pnas.1015757108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N, Joshi AS, Kutik S, et al. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr Biol. 2009;19:2133–2139. doi: 10.1016/j.cub.2009.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kroon AI, Dolis D, Mayer A, Lill R, de Kruijff B. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane. Biochim Biophys Acta. 1997;1325:108–116. doi: 10.1016/s0005-2736(96)00240-4. [DOI] [PubMed] [Google Scholar]
- Kates M, Syz JY, Gosser D, Haines TH. pH-dissociation characteristics of cardiolipin and its 2′-deoxy analogue. Lipids. 1993;28:877–882. doi: 10.1007/BF02537494. [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson HE, de Lartigue J, Rigden DJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- DeLano WL.The PyMOL Molecular Graphics System on World Wide Web http://www.pymol.org 2002
- Wang T, Zhang X, Bheda P, et al. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat Struct Mol Biol. 2006;13:661–662. doi: 10.1038/nsmb1114. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- McCoy A, Grosse-Kunstleve R, Adams P, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. DM: An automated procedure for phase improvement by density modification. in Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. Acta Crystallogr D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Adams P, Grosse-Kunstleve R, Huang LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural features of Beclin 1 ECD.
Beclin 1 ECD molecules form an elongated rod in the crystals, relying on the aromatic finger.
Structural comparison of Beclin 1 ECD with the C2 domain of human coagulation factor V.
Characterization of liposome binding by Beclin 1 ECD.
Validation of the membrane binding experiments by Beclin 1 ECD.
Mutation of the aromatic finger does not compromise the structure or stability of the Beclin 1 ECD.
Characterization of mitochondria binding by Beclin 1 ECD.
Mutation of the aromatic finger compromises the ability of Beclin 1 ECD to deform membrane.
Mutation of the aromatic finger does not affect interactions between Beclin 1 and UVRAG, Barkor, or PI3KC3.
Mutation of the aromatic finger does not alter targeting of Barkor or PI3KC3 to endoplasmic reticulum (ER).
Mutation of the aromatic finger does not alter targeting of Barkor or PI3KC3 to endoplasmic reticulum (ER).
Mutation of the aromatic finger does not affect the cellular distribution pattern of EEA1.
Mutation of the aromatic finger does not alter cellular localization of UVRAG or EEA1 before or after starvation.
Characterization of liposome binding by Beclin 1 ECD.
Cellular localization pattern of endogenous Beclin 1 in NRK cells following starvation.
Mutation of the residues VYY in loop L4 of ATG6 does not appear to cause detectable disruption of autophagy or Cvt pathway in Saccharomyces cerevisiae.