Abstract
Sprouting of grains in mature spikes before harvest is a major problem in wheat (Triticum aestivum) production worldwide. We cloned and characterized a gene underlying a wheat quantitative trait locus (QTL) on the short arm of chromosome 3A for preharvest sprouting (PHS) resistance in white wheat using comparative mapping and map-based cloning. This gene, designated TaPHS1, is a wheat homolog of a MOTHER OF FLOWERING TIME (TaMFT)-like gene. RNA interference-mediated knockdown of the gene confirmed that TaPHS1 positively regulates PHS resistance. We discovered two causal mutations in TaPHS1 that jointly altered PHS resistance in wheat. One GT-to-AT mutation generates a mis-splicing site, and the other A-to-T mutation creates a premature stop codon that results in a truncated nonfunctional transcript. Association analysis of a set of wheat cultivars validated the role of the two mutations on PHS resistance. The molecular characterization of TaPHS1 is significant for expediting breeding for PHS resistance to protect grain yield and quality in wheat production.
Keywords: DNA marker, Preharvesting sprouting, RNA interference, gene clone, wheat abiotic stress
WHEAT is a staple food crop for >40% of the world’s population and provides >20% of calories and proteins for humans (Gill et al. 2004). Preharvest sprouting (PHS) in wheat (physiologically mature grains germinating in spikes before harvest) causes significant loss in grain yield and quality, particularly in regions with prolonged wet weather during the harvest season. Direct annual losses caused by PHS approach $1 billion dollars worldwide (Black et al. 2006).
Resistance to PHS in wheat is a complex trait that is affected by both genotype and environment (Imtiaz et al. 2008). Grain color and seed dormancy have long been regarded as two major factors affecting PHS resistance (Gfeller and Svejda 1960; Bewley 1997; Groos et al. 2002). White grain wheat is usually more susceptible to PHS than red grain wheat (Gale and Lenton 1987; Groos et al. 2002; Himi et al. 2002). Demand for white grain wheat is increasing rapidly in many countries because of consumer preferences, higher flour yield, and better end-use quality; therefore, improving resistance to PHS in white wheat is imperative for successful production in environments where PHS occurs.
Seed dormancy is another important trait of PHS resistance (Bewley 1997; Mares et al. 2005; Sussman and Phillips 2009). Adequate seed dormancy can reduce or block PHS during harvest seasons, but dormancy breaks down during seed storage so seeds germinate uniformly after sowing. Several other factors have been proposed as potential contributors to overall PHS resistance in field conditions, including germination-inhibitory substances residing in chaff tissue (Derera and Bhatt 1980; Gatford et al. 2002), physical barriers to water penetration in a spike, and spike morphology such as structure and erectness of wheat spikes, openness of florets, and tenacity of glumes (King and Richards 1984). The degree to which these factors contribute to the levels of wheat PHS resistance remains unknown.
To date, PHS resistance genes have not been well characterized at the nucleotide sequence level in wheat, although several genes for seed dormancy have been reported in other species. DELAY OF GERMINATION 1 (DOG1) in Arabidopsis and seed dormancy 4 (Sdr4) in rice (Bentsink et al. 2006; Sugimotoa et al. 2010) were cloned. In wheat, quantitative trait loci (QTL) for PHS resistance have been reported on most wheat chromosomes (Groos et al. 2002; Mori et al. 2005; Imtiaz et al. 2008; Kulwal et al. 2012), and QTL on chromosomes 2B (Munkvold et al. 2009), 3A (Liu et al. 2008), and 4A (Mares et al. 2005; Liu et al. 2011) have demonstrated major effects on PHS resistance. Recently, a MOTHER OF FT AND TFL1 was identified to be involved in seed dormancy under low temperature (13°) through a microarray study (Nakamura et al. 2011). Sequencing the gene from two cultivars identified a single nucleotide polymorphism (SNP) from the promoter region as the functional SNP that regulates seed dormancy in red wheat. However, wheat PHS usually occurs before harvest in a field at much higher temperatures, so it is not known if this gene is responsible for PHS resistance in wheat under natural conditions and whether the SNP causes the change in PHS resistance.
We previously mapped a major QTL (Qphs.pseru-3AS) for PHS resistance to the distal end of the short arm of chromosome 3A of a PHS-resistant white wheat cultivar Rio Blanco (Liu et al. 2008; Liu and Bai 2010). In this study, we used comparative fine mapping and map-based cloning to (1) determine the candidate gene underlining the QTL, (2) identify the causal variations in the candidate gene responsible for the change in PHS resistance in wheat, and (3) develop a diagnostic gene assay for marker-assisted selection to improve PHS resistance in wheat.
Materials and Methods
Plant materials and PHS evaluation
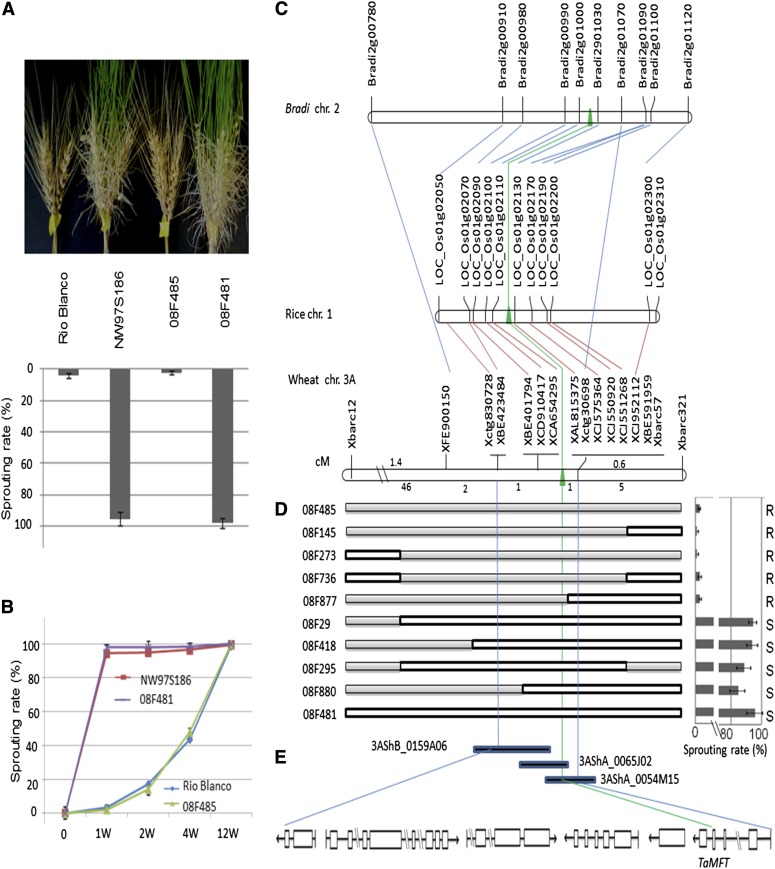
The major QTL for PHS resistance, Qphs.pseru-3AS, was previously mapped using two F6 recombinant inbred line (RIL) populations developed from the crosses of Rio Blanco/NW97S186 and Rio Blanco/NW97S078, where Rio Blanco carries Qphs.pseru-3AS, whereas NW97S186 and NW97S078 do not (Liu et al. 2008). To fine map and clone this QTL, RIL#25, a F6 RIL that segregated at Qphs.pseru-3AS locus, was selected from the Rio Blanco/NW97S186 population to develop recombinant near-isogenic lines (NILs) of Qphs.pseru-3AS using the heterogeneous inbred family method (Tuinstra et al. 1997) (Supporting Information, Figure S1A). This RIL segregated at the Qphs.pseru-3AS region represented by three closely linked markers, Xbarc321, Xbarc57, and Xbarc12, in the QTL region. A total of 1874 RIL#25-derived plants (equal to F2 plants) were screened for recombination within the QTL region using the three SSR markers. The selected heterozygous plants with a recombination event were self-pollinated to produce homozygous recombinants (equal to F3). A total of 56 homozygous NILs with recombination among the three markers were selected (Liu and Bai 2010). Two contrasting homozygous NILs, 08F485 (resistant NIL) and 08F481 (susceptible NIL) without recombination among the three markers, and the two parents, Rio Blanco and NW97S186 (Figure 1A), were used to screen polymorphic markers for further fine mapping and gene expression analysis.
Figure 1.
Map-based cloning of QTL (Qphs.pseru-3AS) for preharvest sprouting (PHS) resistance. (A) PHS phenotypes of different genotypes after 1 week in a moist chamber. The top picture shows wheat spikes of resistant (Rio Blanco) and susceptible parents (NW97S186), and resistant (08F485) and susceptible (08F481) near-isogenic lines (NILs). The bottom graph shows the difference in spike sprouting rates (%) with standard deviations for different genotypes. (B) Spike sprouting rates for both parents and NILs evaluated from harvesting date to 12 weeks after harvest. Wheat spikes were stored at room temperature for different weeks before moist treatment. (C) Comparative maps of the Qphs.pseru-3AS region across wheat chromosome 3A, Brachypodium chromosome 2, and rice chromosome 1. The green arrow indicates the location of TaPHS1. The blue line connects the other syntenic genes in Brachypodium, and the red line connects rice to wheat homologs in the Qphs.pseru-3AS region. The wheat chromosome 3A fine map was generated using a high-resolution mapping population from the analysis of 1874 F2 plants using three SSR markers Xbarc12, Xbarc57, and Xbarc321; the other markers are added to the map using the high-resolution mapping population. The numbers below the linkage map indicate the number of recombinants between the intervals of two markers on the two bars. (D) Graphical genotypes of the Qphs.pseru-3AS region in 10 near-isogenic recombinants with unique recombination among markers (left) and graphical phenotypes (10 plants for each line) showing their sprouting levels (right). Solid and open bars in the genotypic graph represent chromosomal segments from Rio Blanco and NW97S186. R, sprouting resistant; S, sprouting susceptible. (E) Six candidate genes, including TaMFT (first on the right), were identified after sequencing the three BACs covering the Qphs.pseru-3AS region.
The wheat accessions used for association analysis included 60 white and 22 red wheat accessions. Among them, 61 were from U.S. Southern and Northern Hard Winter Wheat Regional Performance Nurseries, and 21 were kindly provided by the U.S. Department of Agriculture (USDA) National Small Grain Collection, Aberdeen, ID.
Wheat PHS resistance was evaluated in the greenhouses at Kansas State University, Manhattan, KS, as described previously (Liu et al. 2008). To evaluate sprouting rates, wheat spikes were harvested from each replication at physiological maturity as characterized by loss of green color on the spike. The harvested spikes were air dried for 5 days in the greenhouse at 25° ± 5° and then stored in a freezer at –20° to maintain dormancy. Sprouting assays were conducted in a moist chamber for 7 days at 23° ± 2° with 100% humidity (Liu et al. 2008). For association mapping, phenotyping experiments were repeated twice with two replications per experiment and five spikes from different plants per accession in each replication. For fine mapping, three to five spikes per F2 plant were tested for PHS resistance using completely randomized design; the selected homozygous recombinant NILs were evaluated for PHS resistance using a randomized complete block design with two replications and five spikes per replication.
Comparative fine mapping
Qphs.pseru-3AS was initially mapped to a 2.0 cM region at the distal end of the short arm of wheat chromosome 3A (Figure S1B). Comparing the QTL map to previously published 3A maps (Song et al. 2005) revealed four common markers for the QTL (Figure S1C). Among them, Xbarc12 and Xgwm369 flanking the QTL were physically mapped in the deletion bin 3AS4-0.45-1.00 (Sourdille et al. 2004) (Figure S1D), indicating the QTL was located in the bin of 3AS4-0.45-1.00. Previously, 224 wheat expressed sequence tags (ESTs) were physically mapped in the bin (Munkvold et al. 2004) (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi). We randomly selected 114 of these ESTs to develop sequence tag site (STS) markers or sequence directly to identify polymorphisms between the two parents and the two NILs (08F485 and 08F481) for fine mapping of the QTL.
Comparative mapping allowed us to identify the syntenic region of Qphs.pseru-3AS in rice and Brachypodium genomes by BLASTN search for polymorphic wheat EST sequences in the rice and Brachypodium genome databases (http://www.jcvi.org; http://www.phytozome.org) (International Brachypodium Initiative 2010; Fu et al. 2009). The annotated gene sequences in the syntenic region of rice and Brachypodium genomes were used as queries in BLASTN (Altschul et al. 1997) to search the wheat EST database (http://www.ncbi.nlm.nih.gov/ or http://www.plantgdb.org) or Chinese Spring 5× coverage of 454 reads (http://www.cerealsdb.uk.net/CerealsDB/Documents/DOC_CerealsDB.php) to identify additional unmapped wheat ESTs or DNA sequences that may be mapped in the Qphs.pseru-3AS region to develop a fine map. In the BLASTN searches, a significant match was declared when there was at least 70% nucleotide identity for at least half of the query sequences, but no fewer than 300 bases, and with an e-value <e−20. For tBLASTX searches, significance was declared when there was at least 40% amino acid identity over at least half of the EST sequences, but no fewer than 200 amino acids at an e-value <e−11. When several significant matches were found for a single predicted rice or Brachypodium gene sequence, the best match was selected (Kuraparthy et al. 2008).
Development and analysis of STS and SNP markers
STS primers were designed using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) to target amplicons of 400–1200 bp. Polymerase chain reactions (PCRs) of 25 μl mixture contained 100 ng of template DNA; 1 mM each of reverse and M13-tailed forward primers; 1 pmole fluorescence-labeled M13 primer; and 0.2 mM each of dNTP, 1× PCR buffer, 2.5 mM MgCl2, and 1.5 units of Taq polymerase. PCR was performed in a DNA Engine Tetrad Peltier Thermal Cycler (Bio-Rad Lab, Hercules, CA) using a touch-town program (Liu et al. 2008).
Those ESTs that did not show STS polymorphism were resequenced to identify SNPs. The PCR products were sequenced using big dye-terminator chemistry after they were cleaned up by adding 2 units shrimp alkaline phosphatase and 0.4 units exonuclease I, and then incubating at 37° for 1 hr and 75° for 15 min to eliminate enzyme activity. DNA sequence data were checked for sequencing errors using Sequencer software (Gene Codes Corporation, Ann Arbor, MI). Sequences were aligned using ClustlW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/), and SNPs between the parents and between the NILs were identified manually.
After an SNP was identified, the SNP primer was designed as 18–24 bases ending right before the SNP for genotyping the fine-mapping population. SNP was analyzed using SNaPshot kit (Applied Biosystems, Foster City, CA) and following the manufacturer’s manual. All primers were checked for possible extendable primer–dimer formation using PerlPrimer v1.1.9. Fragments were scored using GeneMarker version 1.6 (SoftGenetics, State College, PA).
Physical mapping and sequencing of BACs and TaPHS1 gene in wheat accessions
Five flanking or co-segregating markers were used to screen six-dimensional BAC pools from the Chinese Spring 3AS chromosome-specific BAC library, as reported previously (Safar et al. 2007; Sehgal et al. 2012), to construct the physical map for Qphs.pseru-3AS. Each marker was PCR amplified from the BAC pools, and the positive pools were deconvoluted using in-house scripts to map the markers to individual BACs. Positive BACs were traced to fingerprint assembly, and the contig covering the QTL region was identified. The minimum tiling path BACs spanning the QTL region were identified and sequenced using the 454 GS FLX+ System (Roche, Branford, CT) as pools. The sequence was assembled by gsAssembler using default parameters. A sequence with 5951 bp was annotated with the ORF of the TaPHS1. To amplify this gene in Rio Blanco and NW97S186, a pair of TaPHS1 gene-specific primers (TaPHS1-GS) were developed using the Chinese Spring group 3 nulli-tetrasomic lines N3AT3B, N3AT3D, N3BT3A, N3BT3D, N3DT3A, and N3DT3B (Figure S2A and Table S1). Seven pairs of PCR primers (TaPHS1-P, TaPHS1-17, TaPHS1-18, TaPHS1-19, TaPHS1-20, TaPHS1-21, and TaPHS1-SNPShot) were designed to sequence the genomic DNA of the TaPHS1 in the mapping parents and the 82 accessions for association analysis (Table S1). The PCR products were purified with shrimp alkaline phosphatase and exonuclease I and then used for sequencing.
Gene expression analysis
RNA was extracted and purified using the RNeasy plant kit with on-column DNaseI treatment (Qiagen, Valencia, CA). Complementary DNA from reverse-transcription reaction using a SuperScriptII kit (Invitrogen, Grand Island, NY) was amplified by conventional reverse transcriptase–PCR (RT–PCR). Table S1 lists all primer sequences used in the gene expression analysis.
RNA interference-mediated knockdown of TaPHS1
An RNAi-based gene-silencing construct was made using the Gateway system described by Miki and Shimamoto (2004). The insert in the silencing construct consisted of a 230-bp fragment from 3′-UTR of TaPHS1. The insert DNA fragment was amplified from a cDNA pool synthesized from total RNA extracted from the embryos of Rio Blanco collected 17 days after anthesis using the primers listed in Table S1. The fragment was subcloned into pENTR/D-TOPO using a directional TOPO cloning kit (Invitrogen). The final RNAi-based silencing construct was made by recombination from an LR clonase reaction using a Gateway LR clonase enzyme mix (Invitrogen) between the entry vector carrying the TaPHS1 fragment and the pANDA-mini vector. The resulting TaPHS1 silencing construct, PALi7, was bombarded into the cultured immature embryo of two PHS-resistant cultivars, Bobwhite and Rio Blanco, as described by Altpeter et al. (1996).
Transgenic wheat plants were identified by PCR for the presence of the antisense TaPHS1 insert using the gus linker primer, Gus-F1, and the TaPHS1 specific reverse primer, RNAi-R. The sense fragment of TaPHS1 insert was confirmed using gus linker reverse primer Gus-R2 with RNAi-R (Table S1). The transformants with both sense and antisense fragments of TaPHS1 gene in RT–PCR were selected, and progenies from three T0 plants (three, two, and three T1 plants lacking or showing weak TaPHS1 expression) were evaluated for spike sprouting using nontransformed Bobwhite as control.
Bioinformatics and statistical analysis
Genomic sequences from the three BACs covering the Qphs.pseru-3AS region were annotated using FGENESH (http://linux1.softberry.com/berry.phtml). The promoter region of TaPHS1 was analyzed by PlantCare to display the cis-acting regulatory elements (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Amino acid sequence alignments were conducted using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
The TaPHS1 homologs were identified in rice, Brachypodium, maize, sorghum, Arabidopsis, and barley by BLAST at the Rice Annotation Project (RAP) Database (http://www.jcvi.org/), phytozome (http://www.phytozome.org), and Arabidopsis Information Resource (http://www.arabidopsis.org/index.jsp) using the entire deduced amino acid sequence of TaPHS1. All statistical analyses were conducted using SAS 9.0 for Windows (SAS, Cary, NC).
Thirty-three unlinked SSR markers covering 21 wheat chromosomes were selected for structure analysis using STRUCTURE 2.2 (Pritchard et al. 2000). SPAGeDi was used for kinship analysis (Hardy and Vekemans 2002). Tassel was used for association analysis (Bradbury et al. 2007) using a threshold of P < 0.001 to claim significant association between SNPs and PHS resistance. The protein structures of different TaPHS1 alleles were predicted using I-TASSER (Zhang 2008).
Results
Comparative fine mapping of Qphs.pseru-3AS
We developed a set of NILs contrasting at the Qphs.pseru-3AS region using a heterogeneous inbred family method (Tuinstra et al. 1997). PHS resistance was evaluated in 1874 F2 plants to identify recombinants in the QTL region. All homozygous NILs harboring the QTL showed a low sprouting rate (2.9%) similar to Rio Blanco (3.5%), whereas all the NILs without the QTL sprouted at a higher rate (94.5%), similar to susceptible parent NW97S186 (95.2%) (Figure S1A). The heterozygotes showed a sprouting rate of 20.5%, much lower than the middle value (49.3%) between the two parents. These results confirmed that the partially dominant Qphs.pseru-3AS allele in Rio Blanco significantly enhanced PHS resistance. We selected two NILs, 08F485 (resistant) and 08F481 (susceptible), for further gene expression and marker analysis (Figure 1, A and B) and 56 recombinant NILs with a recombination in the QTL region for further fine mapping (Table S2).
Based on the published 3A linkage and physical maps, Qphs.pseru-3AS was further located to deletion bin 3AS4-0.45-1.00 at the distal end of the short arm of chromosome 3A (Figure S1, B–D). A total of 114 ESTs in this bin were used to develop STS markers for fine mapping. One STS (XBE423484) was polymorphic between the parents and between the NILs and mapped in the QTL region (Figure 1C and Figure S2B). Comparative sequence analysis of the EST BE423484 using BLASTN against rice and Brachypodium genomes identified a rice homolog, LOC_Os01g02070, at the distal end of rice chromosome 1, but the homolog was not found in Brachypodium (Figure 1C and Table S3). Further analysis using two closely linked rice genes (LOC_Os01g02060 and LOC_Os01g02080) to LOC_Os01g02070 identified a syntenic region on Brachypodium chromosome 2. Among the 45 genes (from LOC_Os01g01800 to LOC_Os01g02510) in the rice syntenic region and 38 genes (from Bradi2g00780 to Bradi2g01150) in the Brachypodium syntenic region that were blasted in the wheat EST database, 13 wheat homologous ESTs were mapped to the Qphs.pseru-3AS region (Figure 1C, Figure S2, C and D, and Table S3). Qphs.pseru-3AS was delimited at an interval between STS markers XBE423484 and XAL815375. This interval corresponds to a 35-kb region between LOC_Os01g02070 and LOC_Os01g02130 in rice chromosome 1 and a 73-kb region between Bradi2g00910 and Bradi2g01030 in Brachypodium chromosome 2 (Figure 1D). Three wheat STS markers, XBE401794, XCD910417, and XCA654295, co-segregated with PHS resistance (Table S2).
Physical mapping and determination of the candidate gene for Qphs.pseru-3AS
The two flanking and three co-segregating markers were then used to screen a Chinese Spring 3AS chromosome-arm-specific BAC library. One 210-kb contig (Ctg619) containing 22 BACs was found to harbor the QTL (Figure S3). A high-confidence minimum tiling path of three BACs (3AShA_0054M15, 3AShA_0065J02, and 3AShB_0159A06) was identified to cover the entire Ctg619. After the three BACs were sequenced, six genes were identified and considered as candidate genes for Qphs.pseru-3AS (Figure 1E and Table S4).
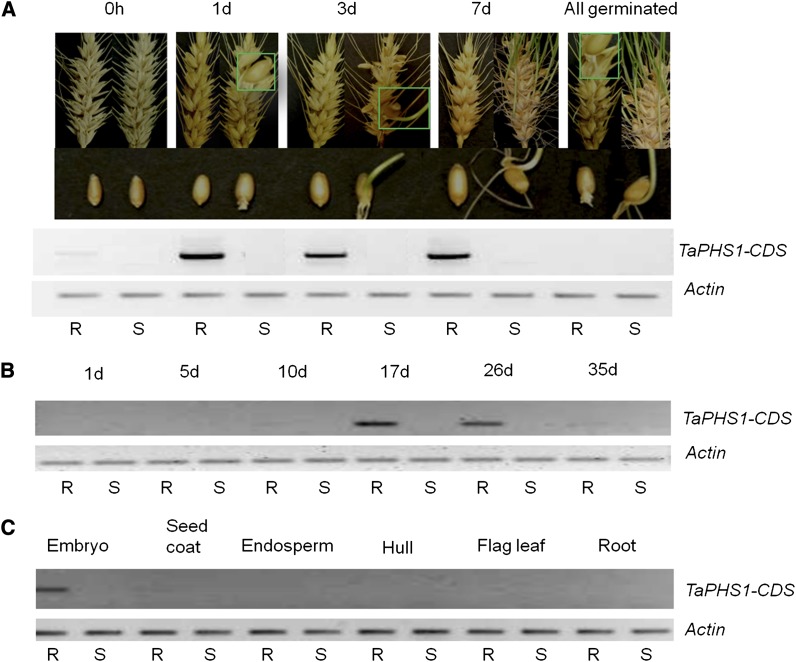
The expression of the six candidate genes was studied during seed sprouting. Only the wheat gene TaMFT, a wheat homolog to the rice gene LOC_Os01g02120, was expressed in the PHS-resistant NIL and not in the susceptible NIL (Figure 2A). MFT is involved in seed germination of Arabidopsis and encodes a phosphatidylethanolamine-binding protein (PEPB) in the embryo (Xi et al. 2010). TaMFT was highly expressed only in the embryo of the resistant wheat genotypes during seed development 17 and 26 days after anthesis and between imbibition and full seed germination during sprouting (Figure 2, B and C). Treating imbibed seeds at low temperature (4°) for 3 days terminated the gene expression in the resistant genotype (Figure 2A). Among the other five genes (Table S4), four showed no differential expression between resistant and susceptible NILs, and the fifth, a transposon gene homologous to LOC_Os01g02100 in rice, was not expressed during sprouting (data not shown). Therefore, TaMFT is the candidate gene for Qphs.pseru-3AS.
Figure 2.
Expression of all the genes in the Qphs.pseru-3AS region during spike sprouting determined the wheat MFT homolog (TaMFT) as TaPHS1. (A) TaMFT was amplified in the PHS resistant (R) NIL, but not in the susceptible (S) NIL, during sprouting. RNA was extracted from the embryos of seeds harvested from the spikes at 0 hr, 1 day, 3 days, and 7 days after imbibing. “All germinated” embryos were obtained by incubating the spikes at 4° for 3 days after imbibing and then keeping them at room temperature for 1 day. (B) TaPHS1 is expressed only in the embryo of the resistant (R) NIL after 10 days and before physical maturity during seed development, not in the susceptible (S) NIL. (C) TaPHS1 expression occurs only in the embryo, not in other tissues at 15 days after anthesis. Actin was used as control. TaPHS1-CDS refers to the primer pair P1 + P6 that were used in RT–PCR and designed from the full-length cDNA sequence of Rio Blanco (Table S1).
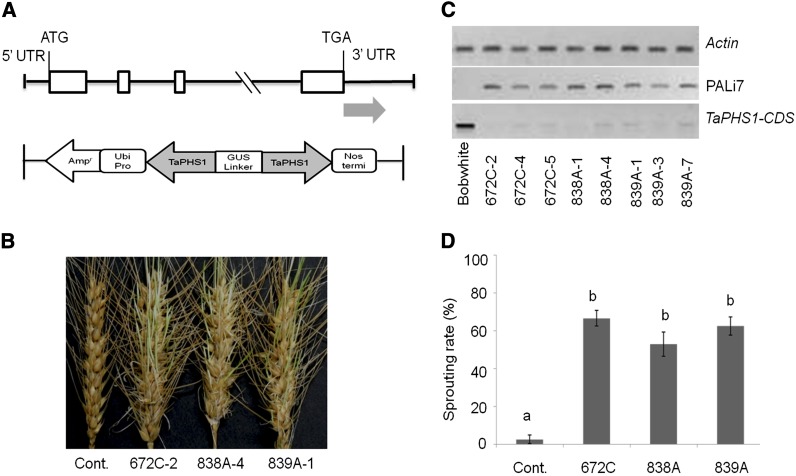
To validate the function of TaMFT for PHS resistance, an RNA interference (RNAi) construct, PALi7 (Figure 3A), was constructed and transformed into PHS-resistant wheat cultivars Rio Blanco and Bobwhite using the particle bombardment-mediated method. Bobwhite shares an identical TaMFT DNA sequence with Rio Blanco; however, regeneration of transgenic plants was not successful for Rio Blanco. Of the 10 independent T0 transformants from Bobwhite, only three (672C, 838A, and 839A) were confirmed to have both the sense and the antisense strands of TaMFT (data not shown). T1 plants from these three transformation events showed significantly reduced endogenous RNA levels of TaMFT but significantly increased sprouting rates: from 2.7% in nontransgenic control to 52.9–66.6% in transgenic Bobwhite plants (Figure 3, B–D). These results confirm that TaMFT is the gene underlying the Qphs.pseru-3AS QTL and is responsible for PHS resistance. Because of the confirmed biological function of TaMFT in wheat preharvest sprouting resistance, rather than in regulating flower time of Arabdopsis thaliana, we designated it as TaPHS1.
Figure 3.
Confirmation of TaPHS1’s function on PHS resistance by RNA interference (RNAi). (A) Structure of the RNAi construct, PALi7. The gray arrow shows that the fragment in 3′-UTR of Rio Blanco TaPHS1 was introduced into the construct as inverted repeats. (B) Representative spikes from PHS-resistant cultivar Bobwhite and three Bobwhite RNAi T1 transgenic lines at 5 days in a moist chamber. (C) Reverse transcription PCR detected PALi7 only in the transgenic T1 lines, not in the nontransgenic control; in contrast, TaPHS1 strongly expressed in nontransgenic control but not in its RNAi lines. RNA was extracted from the embryo 1 day after the spikes were put into a moist chamber. (D) Spike sprouting rates of nontransgenic Bobwhite (Cont) and its T1 lines from the three transgenic events with standard deviations.
Structure of TaPHS1
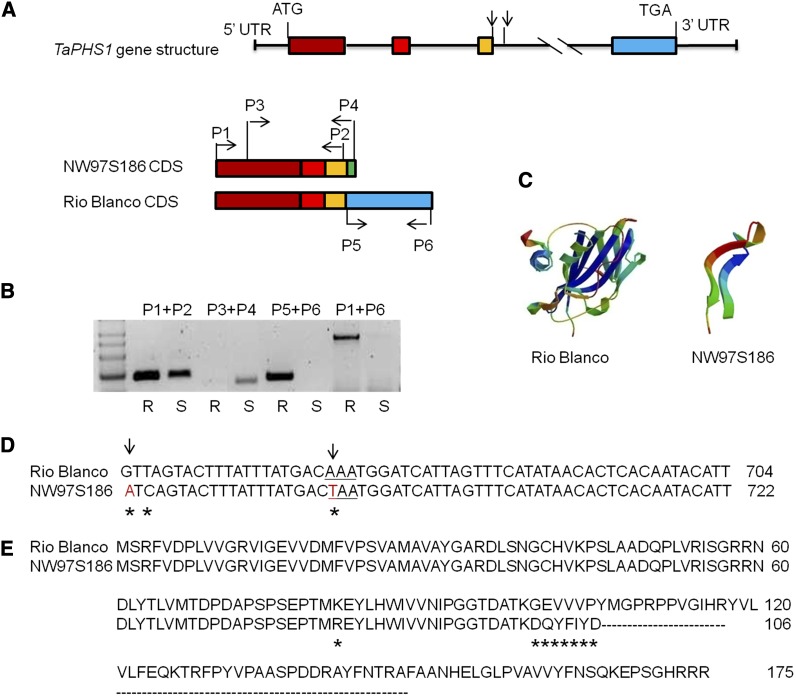
Genomic sequence of TaPHS1 (5165 bp), including a 941-bp promoter sequence, was obtained from Rio Blanco using specific primers designed from the BAC sequences and the Chinese Spring nulli-tetrasomic lines (Figure S2A and Table S1). Gene annotation predicted four exons and three introns in TaPHS1 encoding a peptide of 175 amino acids (Figure 4, A and E). To verify the coding sequence annotation, primers were designed based on a wheat full-length cDNA, AK330655, from the Triticeae Full-Length CDS database and used to amplify the full-length cDNA from Rio Blanco (Mochida et al. 2009). The resulting sequence is identical to previous coding sequence annotations, and the deduced peptide sequence is homologous to MFT proteins from several species (Figure S4).
Figure 4.
Gene and protein structures of TaPHS1 in Rio Blanco (R) and NW97S186 (S). (A) Genomic sequence and the full-length cDNA sequences of TaPHS1 from both parents. Boxes represent exons; the green box shows the extension of exon 3 caused by mis-splicing and truncation of exon 4 in NW97S186. Lines represent introns and 5′- and 3′-UTR; arrows point to two critical mutation sites (mis-splicing site and the premature stop codon) in NW97S186. P1–P6 were the primers used to confirm the truncated mRNA in NW97S186. (B) The two critical mutations in intron 3 of NW97S186 were validated by RT–PCR using four pairs of primers (see A). (C) The predicted disruption of the protein structure of the TaPHS1 in NW97S186, not in Rio Blanco. (D) Partial sequences of TaPHS1 show the two key mutations that form the mis-splicing site and a premature stop codon in intron 3 of NW97S186. Arrows point to the key mutation sites, and * indicates SNPs between two parents. (E) Deduced amino acid sequences of TaPHS1. * represents polymorphic amino acids between parents.
Mis-splicing in TaPHS1 causes PHS susceptibility
Sequence comparison of TaPHS1 alleles between Rio Blanco and NW97S186 identified 31 SNPs or InDels in the promoter region and 32 SNPs in the gene. Two important mutations in Taphs1 of NW97S186 were identified. One was a GT-to-AT transition at the 5′ donor splice site (position +646) of intron 3, which extended exon 3 into intron 3 (Figure 4, A and D); the other was an A-to-T transversion at position +666 that generated a premature stop codon and resulted in a truncated protein in NW97S186 (106 amino acids) (Figure 4, C and E). These two SNPs are most likely the candidate SNPs for wheat sprouting resistance. The primers designed from different exons of Rio Blanco and NW97S186 were used for RT–PCR. The primers designed from the identical sequence of exons 1–3 amplified one band in both Rio Blanco and NW97S186, whereas the primers designed from the fourth exon amplified one band only in Rio Blanco. In contrast, the primers designed from the truncated cDNA sequence of NW97S186 amplified one band only in NW97S186 (Figure 4, A and B), which confirmed the extension of exon 3 and truncation of the transcript in NW97S186.
Analysis of the sequences in the promoter region of Rio Blanco identified six cis-acting abscisic acid (ABA) responsive elements (ABREs) that were reported to be involved in ABA responsiveness, two RY elements involved in seed-specific regulation, one motif IIB, and an ABA responsive element (Baumlein et al. 1992; Suzuki et al. 2005) (Figure S5). Two SNPs, both transversions, between Rio Blanco and NW97S186, were detected in two ABREs at positions −314 and −222.
To further determine the SNPs for PHS resistance in TaPHS1, we sequenced both the promoter and gene-coding regions of the gene from 82 wheat cultivars and found 84 SNPs/InDels. Structure analysis using SSR markers from all 21 chromosomes stratified these cultivars into two groups (Table S5). Significant associations were found between 20 sequence polymorphisms and sprouting rates (P < 0.001), with 8 in the promoter region and 12 in the gene (Figure 5A). The two SNPs at positions +646 and +666 that resulted in the mis-splicing site and the premature stop codon, respectively, were the two most significant SNPs (P < 8.27E-5); the other 18 significant signals were most likely due to the linkage disequilibrium (LD) of the two SNPs (Figure S6). The association between sprouting rate and the two SNPs in the two ABRE cis-acting elements at positions −314 and −222 was not significant (Figure 5A).
Figure 5.
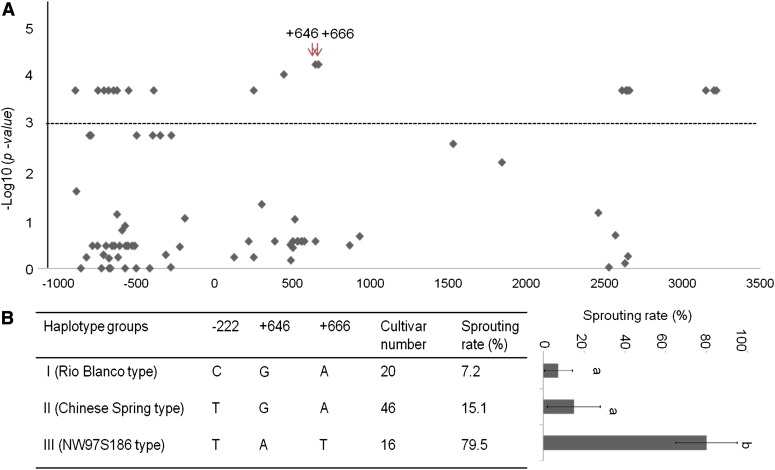
Association analysis to validate two causal SNPs in intron 3 of TaPHS1 for PHS resistance. (A) Twenty SNPs were significantly associated with PHS resistance at P < 0.001. The SNP positions are based on TaPHS1 sequence of Rio Blanco with the start codon as 0. Red arrows point to the two SNPs causing the mis-splicing and the premature stop codon in intron 3. (B) Mean sprouting rates of three contrasting haplotype groups according to the three SNPs, where haplotype I represents 20 cultivars that share the same haplotype with Rio Blanco; haplotype II represents 46 cultivars that share the same haplotype with Chinese Spring, which has one mutation at the promoter region (Nakamura et al. 2011), and haplotype III represents 16 cultivars that share the same haplotype with NW97S186, which has all mutations at three sites. Error bar denotes standard deviation. Different letters, a and b, indicate significant difference at P < 0.05.
Haplotype analysis based on the SNPs (+646 and +666) and the SNP in the promoter region (−222) allowed the 82 cultivars to be divided into three groups: Rio Blanco type (group I), Chinese Spring type (group II), and NW97S196 type (group III). Group I differed from group II in the SNP at position −222; their sprouting rates were not significantly different, but group I had a slightly lower sprouting rate than group II (Figure 5B). The SNPs in group III differed from groups I and II at both positions +646 and +666, where mis-splicing and the premature stop codon mutations occurred. Group III had significantly higher sprouting rates (79.5%, P = 2.98 × 10−5) than groups I and II (Figure 5B), demonstrating that two SNPs at positions +646 and +666 were responsible for the change in PHS resistance. Although red wheat had a slightly lower sprouting rate than white wheat in both haplotypes (Figure S7), the two SNPs at +646 and +666 were primarily responsible for the shift from PHS-resistant to susceptible. Thus, the two mutations that cause mis-splicing and the premature stop codon are the SNPs for PHS resistance. We designed two SNP markers, TaPHS1-SNP1 based on the mis-splicing SNP and TaPHS1-SNP2 based on the premature stop codon. Because the mis-splicing mutation at position +646 was required for the splicing change and both SNPs appeared together in all cultivars tested, we propose to use TaPHS1-SNP1 (Figure S8 and Table S1) as the diagnostic marker for identifying the resistance allele of TaPHS1 in breeding.
Discussion
In a recent microarray study, TaMFT was identified as one of several genes that regulate seed dormancy; it is located in the region with a previously reported QTL for seed dormancy, QPhs.ocs-3A.1, from a red wheat (Nakamura et al. 2011). However, whether the gene regulates overall PHS resistance was not determined in that study. In our study, we independently cloned the same TaMFT gene using map-based cloning of a QTL (Qphs.pseru-3AS) on chromosome 3AS for PHS resistance. Fine mapping of the QTL was achieved by taking advantage of draft reference genome sequences of rice (http://www.jcvi.org) and the Brachypodium and wheat EST database (Munkvold et al. 2004; International Brachypodium Initiative 2010). By measuring the spike-sprouting rate, we directly evaluated overall PHS resistance in the fine-mapping population. We used comparative mapping, physical mapping, BAC sequencing, and candidate gene expression to pinpoint TaMFT as the only gene underlining Qphs.pseru-3AS. The result of RNA interference-mediated knockdown of TaPHS1 confirmed that TaPHS1 positively regulates overall PHS resistance. Thus, the gene underlining the PHS resistance conditioned by Qphs.pseru-3AS is the same gene that regulates seed dormancy as reported by Nakamura et al. (2011).
An SNP in the promoter region (at position −222) of TaMFT was previously reported to be the key SNP responsible for the change in seed dormancy level (Nakamura et al. 2011). We observed a high frequency of sequence variation in both promoter and coding regions of TaPHS1 among wheat cultivars. Gene structure analysis of these SNPs revealed five SNPs (two in the promoter region and three in the gene) that could be potential candidates for the causal SNPs for changing PHS resistance to susceptibility. Two SNPs at positions −222 and −314 of the promoter region are ABREs binding sites; one SNP at position +386 in the second exon leads to one amino acid change; one SNP at the 5′ donor splicing site at +646 results in extending exon 3 into intron 3; and one SNP at +666 in intron 3 creates a premature stop codon when exon 3 is extended. The association between the two SNPs in the promoter region (−222 and −314) and PHS resistance, however, was not significant (Figure 5A). The SNP at position −222 of the promoter region might regulate differences in seed dormancy between PHS-resistant and moderately resistant cultivars at low temperatures (Nakamura et al. 2011). The discrepancy between the two studies is most likely due to different materials used in different studies and different traits evaluated under different conditions. In this study, we evaluated sprouting rate per spike for overall PHS resistance at normal wheat growing temperatures (∼21°–25°) instead of evaluating germination rate after threshing at 13° (Nakamura et al. 2011), and we used a population developed from two parents with the largest contrast in sprouting rates. The susceptible cultivar (Chinese Spring) used in the previous study (Nakamura et al. 2011) showed moderate PHS resistance in our study. These results suggested that the same gene conditioned both seed dormancy and PHS resistance on the 3AS chromosome, and other morphological factors did not play a significant role in PHS resistance expression in this study. This might be because all tested spikes were provided with sufficient moisture for sprouting in a moist chamber, and the difference in spike structures that attract water for seed germination in natural conditions did not contribute to PHS resistance; thus, PHS resistance was mainly contributed by long seed dormancy in this study. Genetic control of different levels of PHS resistance, however, may be different among different sources, so it is possible that quantitative difference in seed dormancy between highly PHS-resistant and moderately resistant genotypes is regulated by sequence variation in the promoter region of TaPHS1 (Nakamura et al. 2011), whereas qualitative difference in PHS resistance between resistant and susceptible genotypes is determined by the sequence variation in the coding region through functional change of the gene. The coding region of TaPHS1 determining qualitative PHS resistance was validated in a panel of 82 wheat cultivars in this study. The difference in germination temperatures between the two studies may also contribute to difference in the gene regulation.
Mis-splicing in Vp1 transcripts in wheat has been shown to cause reduction in Vp1 protein activity, thus compromising the gene function (McKibbin et al. 2002). The mis-splicing at position +646 of TaPHS1 together with the mutation at +666 that forms a premature stop codon results in a nonfunctional transcript, which leads to a change from a PHS-resistant genotype to a susceptible genotype. When all SNPs from a panel of 82 wheat cultivars were analyzed, the highest association occurred between the two SNPs in intron 3 and PHS resistance (Figure 5A), which validated our results. These two key SNPs were not found in the previous study (Nakamura et al. 2011, Figure 5B). Gene expression analysis using the primers designed from different exons of both resistant and susceptible genotypes confirmed the extension of exon 3 and truncation of exon 4 in the TaPHS1 transcript of the susceptible genotypes. Thus, these newly identified SNPs are the key mutations for changing PHS resistance and together cause the loss in function of TaPHS1, changing wheat from PHS resistant to susceptible. Based on these results, we designed two SNP markers, TaPHS1-SNP1 and TaPHS1-SNP2, at the two mutation sites for screening of TaPHS1 in breeding. The two mutations always come together in all wheat germplasm screened to date, so TaPHS1-SNP1 for the mutation at the mis-splicing site suffices for diagnosing the presence of TaPHS1 in wheat breeding.
Red wheat usually shows a higher level of PHS resistance than white wheat. Red color pericarp/testa has been used as a genetic marker for selecting PHS resistance (Gale and Lenton 1987; Groos et al. 2002; Himi et al. 2002). Several other DNA markers for grain color genes have been developed recently (Himi et al. 2011). In this study, both parents of the mapping populations are white wheat, suggesting that major differences in PHS resistance are independent of grain color. In the association panel, both resistant white wheat and highly susceptible red wheat were identified, so this study indicated that TaPHS1 is much more important for PHS resistance than seed color genes are. However, seed color may modify PHS resistance that is regulated by TaPHS1. In either group containing the resistance or susceptibility allele of TaPHS1, the red wheat subgroup had a slightly lower sprouting rate than the white wheat subgroup, although the differences were not significant (Figure S7). This suggested that TaPHS1 is the major gene determining PHS resistance (qualitative), although seed color genes may modify the level of PHS (quantitative) in either PHS-resistant or susceptible genotype groups as determined by TaPHS1. Thus, TaPHS1 is a highly valuable PHS resistance gene for breeding white wheat cultivars.
Global climate change with unstable temperatures and rainfall threatens wheat production. Developing PHS-resistant cultivars that can adapt to unpredictable environments is essential to reduce losses from grain sprouting in fields under wet weather conditions. The characterization and functional determination of TaPHS1 as the PHS resistance gene for QTL Qphs.pseru-3AS in white wheat significantly advances the understanding of the mechanisms underlying PHS resistance in wheat and identifying causal SNPs and developing the diagnostic markers for TaPHS1 and should facilitate effective manipulation of PHS resistance to improve grain yield and quality in wheat.
Supplementary Material
Acknowledgments
We thank Hyeonju Lee and Dehlia McAfee from the Kansas State University Department of Plant Pathology for technical assistance in wheat transformation and R. Graybosch, USDA/ARS in Lincoln, Nebraska, for providing original recombinant inbred line population. This project was partially supported by the National Research Initiative Competitive Grants (NRICG) CAP project 2011-68002-30029, and NRICG grants 2006-35604-17248 and 2008-35300-04588 from the National Institute of Food and Agriculture of the U.S. Department of Agriculture. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. Contribution number 13-277-J is from the Kansas Agricultural Experiment Station.
Footnotes
Communicating editor: S. Poethig
Literature Cited
- Altpeter F., Vasil V., Srivastava V., Stoger E., Vasil I. K., 1996. Accelerated production of transgenic wheat (Triticum aestivum L.) plants. Plant Cell Rep. 16: 12–17. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaeffer A. A., Zhang J., Zhang Z., et al. , 1997. A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumlein H., Nagy I., Villarroel R., Inze D., Wobus U., 1992. Cis-analysis of a seed protein gene promoter: The conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J. 2: 233–239. [PubMed] [Google Scholar]
- Bentsink L., Jowett J., Hanhart C. J., Koornneef M., 2006. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J. D., 1997. Seed germination and dormancy. Plant Cell 9: 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, M., J. D. Bewley, and P. Halmer, 2006, p. 528 in The Encyclopedia of Seeds Science, Technology and Uses. CABI Publishing, Oxfordshire, United Kingdom. [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., et al. , 2007. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- Derera N. F., Bhatt G. M., 1980. Germination inhibition of the bracts in relation to preharvest sprouting tolerance in wheat. Cereal Res. Commun. 8: 199–201. [Google Scholar]
- Fu D., Uauy C., Distelfeld A., Blechl A., Epstein L., et al. , 2009. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 326: 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M. D., Lenton J. R., 1987. Pre-harvest sprouting in wheat: a complex genetic and physiological problem affecting bread making quality in UK wheat. Asp. Appl. Biol. 15: 115–124. [Google Scholar]
- Gatford K. T., Eastwood R. F., Halloran G. M., 2002. Germination inhibitors in bracts surrounding the grain of Triticum tauschii. Fucnt. Plant Biol. 29: 881–890. [DOI] [PubMed] [Google Scholar]
- Gfeller F., Svejda F., 1960. Inheritance of post-harvest seed dormancy and kernel color in spring wheat lines. Can. J. Plant Sci. 40: 1–6. [Google Scholar]
- Gill B. S., Appels R., Botha-Oberholster A. M., Buell C. R., Bennetzen J. L., et al. , 2004. A workshop report on wheat genome sequencing: international genome research on wheat consortium. Genetics 168: 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groos C., Gay G., Perretant M. R., Gervais L., Bernard M., et al. , 2002. Study of the relationship between pre- harvest sprouting and grain color by quantitative trait loci analysis in a white - red grain bread wheat cross. Theor. Appl. Genet. 104: 39–47. [DOI] [PubMed] [Google Scholar]
- Hardy O. J., Vekemans X., 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2: 618–620. [Google Scholar]
- Himi E., Mars D. J., Yanagisawa A., Noda K., 2002. Effect of grain color gene (R) on grain dormancy and sensitivity of the embryo to abscisic acid (ABA) in wheat. J. Exp. Bot. 53: 1569–1574. [DOI] [PubMed] [Google Scholar]
- Himi E., Maekawa M., Miura H., Nada K., 2011. Development of PCR markers for Tamyb10 related to R-1, red grain color gene in wheat. Theor. Appl. Genet. 122: 1561–1576. [DOI] [PubMed] [Google Scholar]
- Imtiaz M., Ogbonnaya F. C., Oman J., Ginkel M., 2008. Characterization of quantitative trait loci controlling genetic variation for preharvest sprouting in synthetic backcross-derived wheat lines. Genetics 178: 1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative , 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–767. [DOI] [PubMed] [Google Scholar]
- King R. W., Richards R. A., 1984. Water uptake in relation to pre-harvest sprouting damage in wheat: Ear characteristics. Aust. J. Agric. Res. 35: 327–336. [Google Scholar]
- Kulwal P., Ishikawa G., David-Benscher D., Feng Z., Yu L. X., et al. , 2012. Association mapping for pre-harvest sprouting resistance in white winter wheat. Theor. Appl. Genet. 125: 793–805. [DOI] [PubMed] [Google Scholar]
- Kuraparthy V., Sood S., Gill B. S., 2008. Genomic targeting and mapping of tiller inhibition gene (tin3) of wheat using ESTs and synteny with rice. Funct. Integr. Genomics 8: 33–42. [DOI] [PubMed] [Google Scholar]
- Liu S., Bai G., 2010. Dissection and fine mapping of a major QTL for preharvest sprouting resistance in white wheat Rio Blanco. Theor. Appl. Genet. 121: 1395–1404. [DOI] [PubMed] [Google Scholar]
- Liu S., Cai S., Graybosch R., Chen C., Bai G., 2008. Quantitative trait loci for resistance to pre-harvest sprouting in U.S. hard white winter wheat Rio Blanco. Theor. Appl. Genet. 117: 691–699. [DOI] [PubMed] [Google Scholar]
- Liu S., Bai G., Cai S., Chen C., 2011. Dissection of genetic components of preharvest sprouting resistance in white wheat. Mol. Breed. 27: 511–523. [Google Scholar]
- Mares D., Mrva K., Cheong J., Williams K., Watson B., et al. , 2005. A QTL located on chromosome 4A associated with dormancy in white- and red-grained wheats of diverse origin. Theor. Appl. Genet. 111: 1357–1364. [DOI] [PubMed] [Google Scholar]
- McKibbin R. S., Wilkinson M. D., Bailey P. C., Flintham J. E., Andrew L. M., et al. , 2002. Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proc. Natl. Acad. Sci. USA 99: 10203–10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D., Shimamoto K., 2004. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45: 490–495. [DOI] [PubMed] [Google Scholar]
- Mochida K., Yoshida T., Sakurai T., Ogihara Y., Shinozaki K., 2009. TriFLDB: a database of clustered full-length coding sequences from Triticeae with applications to comparative grass genomics. Plant Physiol. 150: 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Uchino N., Chono M., Kato K., Miura H., 2005. Mapping QTLs for grain dormancy on wheat chromosome 3A and group 4 chromosomes, and their combined effect. Theor. Appl. Genet. 110: 1315–1323. [DOI] [PubMed] [Google Scholar]
- Munkvold J. D., Greene R. A., Bermudez-Kandianis C. E., Rota C. M. L., Edwards H., et al. , 2004. Group 3 chromosome bin maps of wheat and their relationship to rice chromosome 1. Genetics 168: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold J. D., Tanaka J., Benscher D., Sorrells M. E., 2009. Mapping quantitative trait loci for preharvest sprouting resistance in white wheat. Theor. Appl. Genet. 119: 1223–1235. [DOI] [PubMed] [Google Scholar]
- Nakamura S., Abe F., Kawahigashi H., Nakazono K., Tagiri A., et al. , 2011. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23: 3215–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar J., Simkova H., Kubaláková M., Suchankova P., Cihalikova J., et al. , 2007. Generating resources for genomics of wheat homoeologous chromosome group 3: 3AS- and 3DS-specific BAC libraries. J. Genet. Breed. 61: 151–160. [Google Scholar]
- Sehgal S. K., Li W., Rabinowicz P. D., Chan A., Šimková H., et al. , 2012. Chromosome arm-specific BAC end sequences permit comparative analysis of homoeologous chromosomes and genomes of polyploid wheat. BMC Plant Biol. 12: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q. J., Shi J. R., Singh S., Fickus E. W., Costa J. M., et al. , 2005. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 110: 550–560. [DOI] [PubMed] [Google Scholar]
- Sourdille P., Singh S., Cadalen T., Brown-Guedira G. L., Gay G., et al. , 2004. Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct. Integr. Genomics 4: 12–25. [DOI] [PubMed] [Google Scholar]
- Sugimotoa K., Takeuchib Y., Ebanaa K., Miyaoa A., Hirochikaa H., et al. , 2010. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 107: 5792–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M. R., Phillips G. N., 2009. How plant cell go to sleep for a long time. Science 326: 1356–1357. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Ketterling M. G., McCarty D. R., 2005. Quantitative statistical analysis of cis-regulatory sequences in ABA/VP1- and CBF/DREB1-regulated genes of Arabidopsis. Plant Physiol. 139: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinstra M. R., Ejeta G., Goldsbrough P. B., 1997. Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor. Appl. Genet. 95: 1005–1011. [Google Scholar]
- Xi W., Liu C., Hou X., Yu H., 2010. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22: 1733–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.