Abstract
In this study, the in vitro and in vivo functions of the only two identified protein phosphatases, Saci-PTP and Saci-PP2A, in the crenarchaeal model organism Sulfolobus acidocaldarius were investigated. Biochemical characterization revealed that Saci-PTP is a dual-specific phosphatase (against pSer/pThr and pTyr), whereas Saci-PP2A exhibited specific pSer/pThr activity and inhibition by okadaic acid. Deletion of saci_pp2a resulted in pronounced alterations in growth, cell shape and cell size, which could be partially complemented. Transcriptome analysis of the three strains (Δsaci_ptp, Δsaci_pp2a and the MW001 parental strain) revealed 155 genes that were differentially expressed in the deletion mutants, and showed significant changes in expression of genes encoding the archaella (archaeal motility structure), components of the respiratory chain and transcriptional regulators. Phosphoproteome studies revealed 801 unique phosphoproteins in total, with an increase in identified phosphopeptides in the deletion mutants. Proteins from most functional categories were affected by phosphorylation, including components of the motility system, the respiratory chain, and regulatory proteins. In the saci_pp2a deletion mutant the up-regulation at the transcript level, as well as the observed phosphorylation pattern, resembled starvation stress responses. Hypermotility was also observed in the saci_pp2a deletion mutant. The results highlight the importance of protein phosphorylation in regulating essential cellular processes in the crenarchaeon S. acidocaldarius.
Protein phosphorylation is an important post-translational modification (PTM)1 that has been reported in all three domains of life, Eukarya, Bacteria and Archaea (1–3). The reversible character of the modification allows for subtle and immediate regulation by modulating protein activity in different cellular processes (3–5). The addition of a phosphoryl group is catalyzed by protein kinases and dephosphorylation by their cognate phosphatases. Interestingly, in the Crenarchaeota, one of the phyla of the Archaea, only Ser/Thr/Tyr phosphorylation is predicted by bioinformatics analyses, whereas two-component systems (involving His/Asp phosphorylation) identified in the Euryarchaeota phylum are absent (6–9).
A phosphoproteome study of the thermoacidophilic crenarchaeon Sulfolobus solfataricus P2 revealed a vast amount of Ser/Thr/Tyr-phosphorylated proteins (540 detected in total), in almost all arCOGs categories, with an unexpectedly high number of Tyr phosphorylation. A differential phosphorylation pattern was observed in cells grown on glucose versus tryptone, and a role of protein phosphorylation in regulating the glycolytic flux in Sulfolobus was proposed (10). Further, a phosphoproteome study of the mesophilic euryarchaeon Halobacterium salinarum revealed 69 phosphorylated proteins in total (2).
Although a few archaeal protein kinases and phosphatases have been investigated in more detail (7, 11–19), there is still a knowledge gap regarding signal transduction pathways in Archaea and the impact of Ser/Thr/Tyr phosphorylation on cellular processes. Comparative database searches revealed only two protein phosphatases encoded in the genomes of all members of the Sulfolobales order, in comparison to eight predicted protein kinases in S. solfataricus alone (7). Similarly, in eukaryotes fewer protein phosphatases are encoded in the genome compared with protein kinases, though the phosphatases of the PPP family often act as multimeric proteins with different catalytic, regulatory and core subunits (20, 21).
Protein phosphatases can be categorized into different families. Ser/Thr-specific PPPs contain a 220 amino acid long catalytic domain that includes motif I (GDXHG), motif II (GDXXDRG), and motif III (GNHE) (22), and Mg2+ or Mn2+ dependent Ser/Thr phosphatases (PPMs) contain 11 specific motifs (23, 24). Conversely, the phosphotyrosine phosphatase family (PTP) can be subdivided into the PTPs, which are specific for Tyr dephosphorylation, the dual specific PTPs, which can dephosphorylate Ser/Thr and Tyr, and the low molecular weight PTPs. PTP phosphatase family members share a common amino acid motif, CX5R (20).
Only a few archaeal Ser/Thr- or Tyr-specific protein phosphatases have been characterized. The Ser/Thr phosphatase PP1-arch1 from S. solfataricus displays Mn2+-dependent protein phosphatase activity in vitro (16). The crystal structure of SsoPTP from S. solfataricus has been solved and the specificity toward phosphotyrosine determined in vitro (18). Only one additional archaeal PTP has been characterized, the Tk-PTP from the euryarchaeon Thermococcus kodakaraensis KOD, which exhibits phosphotyrosine as well as phosphoserine, but no phosphothreonine phosphatase activity (17). Finally, the only characterized archaeal member of the PPM family is found in Thermoplasma volcanium and has a divalent metal-ion dependent dual-specificity toward phosphorylated Ser/Thr as well as Tyr residues (25).
The creanarcheon S. acidocaldarius, which grows optimally at pH 3 and 76 °C, contains two protein phosphatases, the Saci-PTP (phospho Tyr phosphatase), encoded by the saci0545 gene (saci_ptp) and the Saci-PP2A (phospho Ser/Thr phosphatise) encoded by saci0884 (saci_pp2a), which is homologous to the PP2A catalytic subunit of eukaryotic phosphatases. In this study, Saci-PTP and Saci-PP2A were characterized with respect to substrate specificity and kinetic properties. Furthermore, in vivo analyses were performed with deletion mutants to investigate the resulting phenotypes, and additional characterization was carried out by RNA-seq and phosphoproteome analysis.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
Escherichia coli K12 DH5α (Invitrogen, Breda, The Netherlands) and Rosetta (DE3) (Stratagene, La Jolla, CA) were used for cloning and expression studies, respectively. Both strains were grown under standard conditions as reported recently (26).
The markerless in-frame deletion mutants and the uracil auxotrophic parental strain S. acidocaldarius MW001 (27) were grown at 76 °C in Brock‘s basal medium at pH 3.5 (28). The medium was supplemented with 0.1% (w/v) NZ-amine, 0.2% (w/v) sucrose, and 10 μg/ml uracil (water dissolved). The trans-complementation strains of the deletion mutants were grown without uracil because the uracil auxotrophic marker was encoded by the complementation plasmid.
Chemicals
All chemicals were purchased from Sigma-Aldrich (Taufkirchen, Germany), VWR (St. Louis, MO), Carl Roth (Karlsruhe, Germany) or Roche Diagnostics (Mannheim, Germany) in analytical grade. Para-nitrophenylphosphate (pNPP) was purchased from Sigma-Aldrich. The p-Thr-peptide RRA(pT)VA was purchased from Promega (Madison, WI). The p-peptide TEVGKRI(pY)RLVGDKN was synthesized according to the p-peptide of Saci_1938 determined in the phosphoproteome analysis and purchased from PROTEINMODS (Madison, WI). For the characterization of Saci_PTP the p-peptides NIDAIRA(pS)LNIMSR and ETTYERW(pT)TITQRER derived from the respective p-peptides of Saci_1346 and Saci_1857, respectively, determined in the phospho-proteome analysis were purchased from Thermo Fisher Scientific.
Heterologous Expression and Protein Purification
For expression of Saci-PTP (Saci0545) and Saci-PP2A (Saci0884) the genes were amplified by PCR using specific primers (Table I) and cloned into the pET vector system (pET-15b, or pETDuet-1, Novagen). The constructed plasmids were verified by sequencing of both strands (AGOWA, Berlin and MWG Operon, Ebersberg, Germany).
Table I. Kinetic properties of protein phosphatases in S. acidocaldarius at 70 °C.
| Substrate | Saci-PTP |
Saci-PP2A |
||
|---|---|---|---|---|
| Km (μm) | Vmax (U/mg) | Km (μm) | Vmax (U/mg) | |
| pNPP | 530 | 3.7 | 2500 | 4.3 |
| TEVGKRI(pY)RLVGDKN | 530 | 9.2 | No activity | No activity |
| RRA(pT)VA | - | - | 53.6 | 192.6 |
| NIDAIRA(pS)LNIMSR | 423 | 0.3 | - | - |
| ETTYERW(pT)TITQRER | 490 | 0.07 | - | - |
Heterologous expression of Saci-PTP and Saci-PP2A was performed in E. coli Rosetta (DE3)-RIL as expression host strain. For purification of the recombinant enzymes, the resulting E. coli crude extracts were diluted 1:1 with 50 mm NaH2PO4, 300 mm NaCl (pH 8.0, RT), and subjected to a heat precipitation for 20 min at 80 °C. After heat precipitation, the samples were cleared by centrifugation (60,000 × g for 30 min at 4 °C).
For Saci-PP2A and Saci-PTP the supernatant was dialyzed overnight against 50 mm NaH2PO4, 300 mm NaCl (pH 8.0, RT), subjected to immobilized metal affinity chromatography (Ni-TED 2000, Macherey-Nagel, pre-equilibrated in 50 mm NaH2PO4, 300 mm NaCl (pH 8.0, RT), and eluted in 50 mm NaH2PO4, 300 mm NaCl, 250 mm imidiazole (pH 8.0, RT). Fractions containing the recombinant enzymes (analyzed by SDS-PAGE, enzyme activity) were pooled and concentrated via centrifugal concentrators (Vivaspin6, Sartorius Stedim Biotech). From 3.8 g of recombinant cells (wet weight), 1.7 mg Saci-PTP and from 17 g of recombinant cells (wet weight) 0.4 mg Saci-PP2A were obtained.
Saci-PP2A was further purified by ion exchange chromatography and size exclusion chromatography. Briefly, Saci-PP2A was dialyzed overnight against 20 mm Tris/HCl (pH 6.5, 70 °C), subjected to ion exchange chromatography (UNO Q-17; Bio-Rad Laboratories, Hercules, CA) (pre-equilibrated in 20 mm Tris/HCl (pH 6.5, 70 °C)), and eluted with a linear salt gradient from 0 to 1 m NaCl. Afterward, the sample was dialyzed overnight against 50 mm Tris/HCl, 300 mm KCl (pH 6.5, 70 °C) and applied to size exclusion chromatography (HiLoad 26/60 Superdex 200 prep grade; Amersham Biosciences) (pre-equilibrated in 50 mm Tris/HCl, 300 mm KCl (pH 6.5, 70 °C)). The fractions containing the recombinant enzymes were analyzed via Coomassie stained SDS-PAGE and enzyme activity tests, pooled and further used for the enzymatic characterization.
Enzyme Assays
Phosphatase activity was determined in a continuous enzyme assay at 70 °C with pNPP as substrate. For Saci-PTP the standard assay was performed in 0.1 m HEPES/KOH (pH 6.5) with 7 μg protein, whereas for Saci_0884 the assay was performed in 20 mm Tris/HCl, 300 mm KCl (pH 6.5), 5 mm Mn2+, 1 mm EGTA containing 0.25 μg protein in a reaction volume of 300 μl. Enzymatic activity was determined by monitoring the change in absorbance at 403 nm because of the formation of paranitrophenyl (εp-NPP = 18 mm/cm). For each assay three independent measurements were performed.
Protein tyrosine phosphatase activity was determined by using the tyrosine phosphatase assay system (Promega) following the manufacturer's instruction. The artificial p-peptides TEVGKRI(pY)RLVGDKN, NIDAIRA(pS)LNIMSR, and ETTYERW(pT)TITQRER were used as substrate. The enzyme activity was determined in a discontinuous enzyme assay at 70 °C in 0.1 m HEPES/KOH (pH 6.5) containing 2 μg Saci-PTP. The phosphate release was measured by using the Serine and Threonine phosphatase malachite-green-based assay system from Promega following the instructions of the manufacturer. For each assay three independent measurements were performed.
Protein serine/threonine phosphatase activity was determined by using the serine/threonine phosphatase assay system (Promega) following the manufacturer's instruction. The artificial p-peptides RRA(pT)VA and TEVGKRI(pY)RLVGDKN were used as substrate. The enzyme activity was determined in a discontinuous enzyme assay at 70 °C in 20 mm Tris/HCl, 300 mm KCl (pH 6.5), 5 mm Mn2+, 1 mm EGTA containing 0.1 μg Saci-PP2A. The phosphate release was measured and visualized by using malachite green. For each assay, three independent measurements were performed.
For enzyme reactions following Michaelis-Menten kinetics the kinetic parameters (Vmax and Km) were calculated by iterative curve-fitting using the program Origin 8.6G 64 bit (Microcal Software, Northampton, MA).
Construction of In-frame Deletion Mutants in S. acidocaldarius
Plasmids for deletion mutant construction were cloned using the PCR product of the upstream- and downstream-region of the gene of interest (Primers, supplemental Table S5), overlap PCR was performed to fuse both fragments and the overlap product was ligated with the gene targeting plasmid pSVA406 (27). The constructed plasmids were methylated using E. coli ER1821 and transformed into the background strain S. acidocaldarius MW001 (uracil auxotrophic strain) as described before (27). The subsequent steps were performed as described by Lassak et al. 2012 (29). In short, transformants were first selected on gelrite plates lacking uracil and then counterselected on plates with uracil and 5-FOA. The markerless in-frame deletion mutants of both phosphatase genes (saci0545 and saci0884), designated Δsaci_ptp and Δsaci_pp2a, respectively, were identified by colony PCR and confirmed by sequencing (MWG Operon, Ebersberg, Germany).
Flow Cytometry Analysis
Aliquots from growing cultures of strains MW001, Δsaci_ptp and Δsaci_pp2ac were transferred to ice-cold ethanol (final concentration 70% (v/v)). For DNA staining, ∼0.5 ml of cell suspension in ethanol was centrifuged at 4 °C for 10 min at 16,000 × g. The pellet was resuspended in 1 ml of 10 mm Tris-buffer (pH 7.4) containing 10 mm MgCl2, centrifuged again, and then resuspended in 70 μl of the same buffer. Equal volumes (65 μl) of cell suspension and DNA-specific stain (0.2 mg/ml mithramycin A and 0.04 mg/ml ethidium bromide, also in Tris-MgCl2 buffer) were mixed. The samples were incubated on ice for 30 min before analysis in an Apogee A40 Analyzer flow cytometer equipped with a 405 nm solid-state laser.
Analysis of Cell Size
Microscopy pictures were taken using an Axio Imager.M1 microscope (Zeiss) equipped with a Zeiss Plan Apochromat × 100/1.40 Oil DIC objective and a Cascade:1K CCD camera (Photometrics). Cell size was analyzed automatically with ImageJ and ObjectJ using a modified version of the filaments-91i.ojj project. The results were evaluated with Origin 6.1 (OriginLab Corporation, Northampton, MA, USA).
RNA Isolation and Sample Preparation for RNA-seq Analysis
S. acidocaldarius MW001, Δsaci_ptp, and Δsaci_pp2a were inoculated in triplicate. Cultures were grown to reach an OD600 of 0.6 (exponential growth phase) and subsequently samples of the three independent cultures of each strain were pooled in equal amounts to generate one mixed sample per strain. Total RNA samples were isolated from 10 ml of exponentially growing shaking cultures. TRIzol reagent (Invitrogen) was used for total RNA isolation following the instructions of the manufacturer. Residual chromosomal DNA present in RNA samples was removed by RNase-free DNase I (Roche) treatment for 2 h at 37 °C. DNA-free RNA samples were confirmed by PCR amplification using saci0574 (secY) primer pairs. Before cDNA synthesis DNA-free RNA samples were fragmented to achieve a molecule size range of 50 to 500 nucleotides. Thus, 6 μg of DNA-free RNA samples were mixed with 4 μl of 5× fragmentation buffer (200 mm Tris acetate, pH 8.2; 500 mm potassium acetate; 150 mm magnesium acetate). The reaction mix was filled up with DEPC H2O up to 20 μl, incubated at 95 °C for 2.5 min and immediately transferred to ice. The reaction was then cleaned up by using sephadex columns (illustra MicroSpin™ G-25, GE Healthcare). RNA fragmentation range was checked by polyacrylamide gels. Six hundred nanograms of fragmented RNA was used for cDNA synthesis using the SuperScript Double-stranded cDNA Synthesis kit (Invitrogen) and following manufacturer's instructions. The reaction was cleaned by using phenol/chloroform/isoamyl alcohol and cDNA samples were then precipitated by using 3 m NaOAc/100% ethanol and incubated over night at −20 °C. Finally, cDNA samples were washed with 500 μl 70% ethanol. Five nanograms of cDNA were used as starting material for the generation of single-end sequencing libraries with the NEBnext DNA sample preparation kit, as described by manufacturer's protocol. The DNA was ligated to Illumina adaptors, which carried a unique four letter barcode sequence. DNA fragments of 150–500 bp were selected for sequencing. To retain strand specificity the DNA was treated with Uracil DNA-Glycosilase (UDGase) before the PCR amplification. The sequencing was performed by an Illumina Genome Analyzer IIx, multiplexed together with a total of eight samples. The reads were partitioned according to their barcode, stripped from adapter sequences and barcodes and mapped, using default settings, to the Sulfolobus acidocaldarius DSM 639 (NC_007181) reference genome, with segemehl (30).
Bioinformatic Analysis of RNA-seq Data
For each protein coding and ncRNA gene, according to the NCBI database, the associated reads were counted. An overview where the reads map to, can be found in supplemental Fig. S8A. The overall Illumina sequencing represented 26.6%, 30.2%, 26.5% of genome coverage by the mapped reads for MW001, Δsaci_ptp and Δsaci_pp2a, respectively. To identify differentially expressed genes among the sample conditions, we used DESeq (version 1.5) (31), which allows estimation of the dispersion within one condition. This estimation is based on the assumption that only a few genes are truly differentially expressed; hence variance across different conditions can be seen as a too conservative estimation of the variance within one condition. From the estimated expected variance and the observed fold change in read counts, the statistical significance of altered RNA abundance can be calculated for every gene. Genes with a p value below 0.054 were considered to be significantly altered in their expression. supplemental Figs. S8B and S8C shows the correlation among the observed read counts, the fold change in read count among the sample conditions and the significance of the alteration.
RT-qPCR
First Strand cDNA Synthesis Kit (Fermentas) was used for cDNA synthesis according to the manufacturer's instructions. The synthesis was thus performed using random hexamer primers and 1 μg of DNA-free RNA as template. Quantitative PCR (qPCR) analysis was carried out using Maxima SYBR Green/ROX qPCR Master Mix (Fermentas) based on the Sybr green detection system (Real-Time 7300 PCR machine; Applied Biosystems, Foster City, CA). The efficiency of each primer pair was calculated from the average slope of a linear regression curve, which resulted from qPCRs using a 10-fold dilution series (10 pg–10 ng) of S. acidocaldarius chromosomal DNA as template. Cq values (quantification cycle) were automatically determined by Real-Time 7300 PCR software (Applied Biosystems) after 40 cycles. Cq values of each transcript of interest were standardized to the Cq value of the reference gene saci0574 (secY) (32). qPCR reactions with DNA-free RNA samples as template were performed as control. At least three biological replicates of each assessed condition and two technical replicates per qPCR reaction were performed.
Immunoblotting Analysis After Nutrient-limited Growth Conditions
The expression levels of the archaella components FlaB and FlaX were determined as described by Reimann et al. 2012 (33). Therefore the background strain S. acidocaldarius MW001 and both phosphatase deletion mutants (Δsaci_ptp and Δsaci_pp2a) were transferred to nutrient-limited medium. The same amount of cells referring to OD600 with and without starvation stress treatment were lysed and the proteins separated by SDS-PAGE. The expression levels of FlaB and FlaX were detected by immunoblotting using specific antibodies.
Motility Assay on Semisolid Gelrite Plates
To compare the motility of the background strain S. acidocaldarius MW001 to the phosphatase deletion strains (Δsaci_ptp and Δsaci_pp2a) the two control strains ΔaapF (hypermotile strain) and ΔaapFΔflaH (nonmotile strain) were used. The strains were spotted on semisolid gelrite plates (0.15% gelrite (w/v)) containing only 0.005% (w/v) tryptone, 0.2% (w/v) dextrin and 10 μg/ml uracil as described before (29, 33). Plates were incubated for 5 days in a humid chamber at 75 °C. Swimming behavior of the different S. acidocaldarius strains was analyzed by measuring the swimming radius.
Protein Sample Preparation for Mass Spectrometry Analysis
For phosphoproteome analysis S. acidocaldarius MW001, MW010 (Δsaci_ptp), and MW025 (Δsaci_pp2a) were aerobically grown in a 10 liter fermenter with constant rotational mixing of the cultures. At the OD600 0.8 2 liter samples were taken. The cells were harvested by centrifugation at 9000 × g (Beckman Coulter Avanti J-26XP, rotor JLA 8.1) and the pellets immediately frozen in liquid nitrogen.
Cells were washed with cold water and then resuspended in 50 mm phosphate buffer (pH 7.5) and phosphatase inhibitors (5 mm each) comprising 2-glycerol phosphate, sodium fluoride, sodium vanadate, and sodium pyrophosphate (1). Cell extraction was performed using a combination of both ultrasonication and liquid nitrogen (manual grinding with a mortar and pestle). First, cells were sonicated using an ultrasonicator (Branson, Mexico) at a power of 70% for eight times (alternatively 45 s of sonication and 45 s of incubation in ice). Samples were then frozen using liquid nitrogen and ground manually five times. Furthermore, DNase I (100 μg/ml) and N-Octyl- glucoside (a final concentration of 1% (w/v)) were also added to samples to increase solubilization of membrane proteins (1). Samples were then finally sonicated in a water bath containing ice for 10 min before centrifugation at 21,000 × g for 20 min at 4 °C. Protein supernatant was collected and quantified using the RC-DC Protein Quantification Assay (Bio-Rad, UK). All chemicals were purchased from Sigma.
A total of 2 mg protein for each sample was denatured in 8 m urea, reduced with 10 mm DTT in 50 mm ammonium bicarbonate at 56 °C for 1 h, then alkylated with 55 mm idoacetamide in 50 mm ammonium bicarbonate at 37 °C for 30 min in the dark. Before trypsin digestion was performed, samples were diluted to a final urea concentration less than 1 m by 40 mm ammonium bicarbonate in 9% acetonitrile (ACN) (34). A trypsin/protein ratio of 1:25 was used for trypsin digestion at 37 °C overnight. Samples were then dried in a vacuum concentrator (Eppendorf, Germany) before resuspension in buffer A consisting of 10 mm KH2PO4, 30% acetonitrile, and 0.1% trifluoroacetic acid pH 3.0 for fractionation and cleaning of the sample. Strong cation exchange (SCX) was performed using a HPLC system with UV detector (Dionex, UK) with buffer B used for peptide elution comprising of 10 mm KH2PO4, 30% ACN, 0.1% TFA, and 500 mm KCl, pH 3.0 (see (35) for details). Fractions of the sample were then collected every 2 min and peptides were subsequently dried in a vacuum concentrator (Eppendorf, Germany).
Mass Spectrometry Analysis and Data Processing
All dried peptides were then cleaned using a C18 Discovery DSC-18 SPE column (Supelco, Sigma) as described elsewhere (36); all fractions were subsequently combined, before analysis on a HCT-Ultra ion trap (Bruker Daltonics, UK) coupled with an Ultimate 3000 nano-flow HPLC (Dionex, UK) consisting of an analytical column (3 mm C18, Dionex-LC Packings) operating at a flow rate of 300 nL/min. Buffer A contained 3% acetonitrile and 0.1% (w/v) formic acid, whereas buffer B contained 97% (w/v) acetonitrile and 0.1% (w/v) formic acid. The 90 min gradient comprised ramping from 3% B to 35% B in the first 70 min, then ramping to 90% B in 5 min, then switching back to 3% of buffer A for 15 min. Peptides eluting from HPLC column were directly submitted to the ion trap.
The PAcIFIC technique was applied for analyzing (combined) cleaned peptides. Details of this technique can be found elsewhere (10, 37). Briefly, resuspended peptides were injected multiple times on the ion trap with collision induced association at each of 10 continuous 1.0 m/z intervals across a range of 10 m/z for each LC-MS analysis using a 2.0 m/z width and fragmented ions scanned from 100 to 1600 m/z with a dynamic ion charge threshold of 160,000. Subsequently, a next (forward) new ten ions was started in the same format as the first run, then this process continued until the considered precursors covered the range from 450 to 1100 m/z.
Data from mass spectrometry were then extracted to mgf format using Bruker Data Analysis V4.0 with a MRM script, these were then were searched against the S. acidocaldarius database downloaded from NCBI in March 2010 (containing 2223 proteins) using Phenyx V 2.6 (Genebio, Geneva). The searches were performed using parameters as follows: carbamidomethylation of cysteine (fixed modification), oxidation of methionine (variation), and phosphorylation of serine, tyrosine, threonine (variation), trypsin with two missed cleavages. Furthermore, other parameters such as parent, MS/MS, tolerances were set at 2.0 and 0.8 Da, respectively, whereas minimum peptide length, z-score, p value and AC score were set at 5, 5.5, 10–15, and 5.5 respectively.
The spectra for all identified phosphopeptides were then automatically processed using Mathematica 9.0 (Wolfram Research, Inc.) in order orthogonally to confirm (1) the neutral loss of phosphoric acid (H3PO4, mass shift = −98) or metaphosphoric acid (HPO3, mass shift = −80) from the precursor or/and fragment ions and (2) the presence of at least 5 b or y sequencing ion series. For the automatic annotation, peaks of intensity lower than 10% of the maximal intensity were discarded. The annotated spectra were then manually to ensure the quality of the annotations. Moreover, an independent biological duplicate sample was generated for each strain and an identical analysis carried out. Scores of phosphorylation sites are shown in Table S7. Raw spectra have been submitted to the PRIDE database (http://www.ebi.ac.uk/pride/, with ProteomeXchange accession: PXD000289, and are waiting for an accession number) (38).
RESULTS
Characterization of saci_ptp and saci_pp2a Deletion Mutants
Marker-less gene deletion mutants of the two putative phosphatases, Saci-PTP and Saci-PP2A, were generated using the uracil auxotrophic S. acidocaldarius MW001 strain (supplemental Fig. S1). Δsaci_pp2a showed slow growth (doubling time 9.5 h) and reached the stationary phase after 130 h, whereas MW001 and Δsaci_ppt showed similar growth characteristics (doubling times 5.7 h and 6.3 h, respectively; stationary phase reached after 60 h; supplemental Fig. S2). Notably, the Δsaci_ptp cells appeared normal in cell size (1.3 μm ± 0.55 μm), whereas the Δsaci_pp2a deletion mutant showed significant size variation (1.46 μm ± 1.08 μm) in comparison to MW001 (1.27 μm ± 0.42 μm; supplemental Fig. S3) in microscopy analysis. The larger variation in the cell size distribution of the saci_pp2a deletion mutant, as compared with the parental strain and Δsaci_ptp, was confirmed by flow cytometry analysis (supplemental Fig. S4B). No changes in the DNA content distributions, reflecting the relative lengths of the cell cycle periods (39), could be detected in either deletion mutant, as compared with the parental strain (supplemental Fig. S4A), indicating no major effects on cell cycle regulation or progression through the different cell cycle phases. The aberrant phenotype of the Δsaci_pp2a mutant could be partially restored in a trans-complementation experiment, resulting in a growth curve and cell shape distribution similar to those of the parental strain (supplemental Figs. S5A and S5B).
Enzymatic Characterization of Saci-PTP and Saci-PP2A
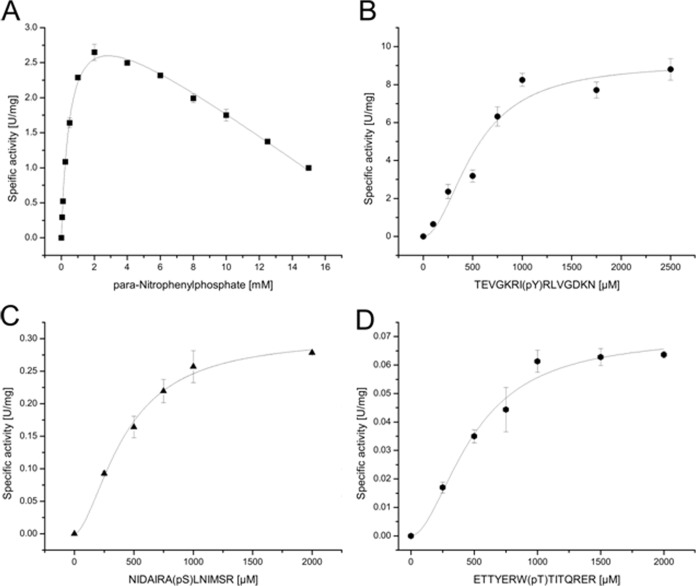
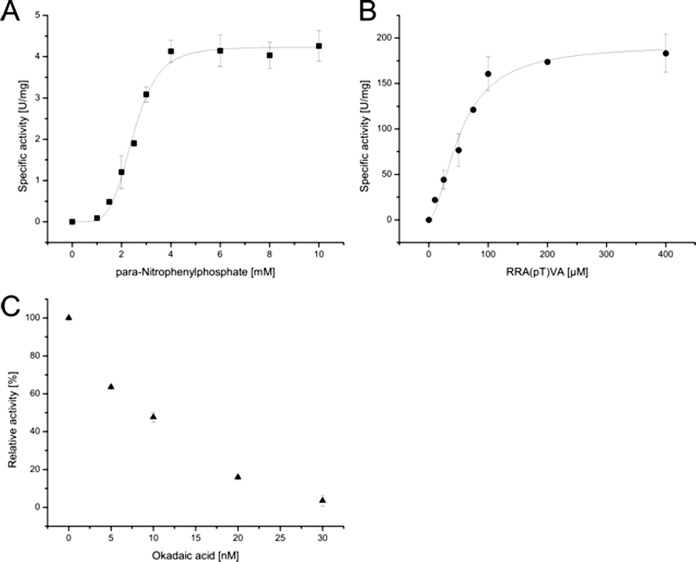
Saci-PTP and Saci-PP2A were heterologously expressed (supplemental Fig. S6) in E. coli, purified, and analyzed in vitro. Both enzymes showed similar phosphatase activity with the pNPP substrate at 70 °C (Table I). For Saci-PTP substrate inhibition was observed at pNPP concentrations above 2 mm (kirr-value of 0.17 (U/mg)mm−1) (Fig. 1A). For Saci-PP2A cooperativity was observed at pNPP concentrations below 2 mm (Hill coefficient of 4.9) (Fig. 2A).
Fig. 1.
Biochemical characterization of the protein phosphatases Saci-PTP. The activity of Saci-PTP was analyzed in the presence of pNPP (A), of the phospho-Tyr peptide TEVGKRI(pY)RLV (B), the phospho-Ser peptide NIDAIRA(pS)LNIMSR (C), and the phospho-Thr peptide ETTYERW(pT)TITQRER (D). All experiments were performed at 70 °C in triplicate. Saci_PTP showed general phosphatase activity with pNPP as substrate (A). Furthermore, Saci-PTP displayed a dual-specific phosphatase activity using p-Tyr peptides (B) as well as p-Ser (C) and p-Thr (D) peptides as substrate.
Fig. 2.
Biochemical characterization of the protein phosphatases Saci-PP2A. The activity of Saci-PP2A was determined in the presence of pNPP (A), the phospho-Thr peptide RRA(pT)VA (B) as well as in the presence of okadaic acid (p-Thr peptide RRA(pT)VA)(C). All experiments were performed at 70 °C in triplicate. Saci_PP2A revealed phosphatase activity with pNPP as well as the p-Thr peptide, whereas no activity was observed with the p-Tyr peptide. Furthermore, Saci_PP2A was significantly inhibited in presence of okadaic acid, a typical inhibitor of PPPs.
The substrate specificity of Saci-PTP was addressed with the p-Tyr peptide TEVGKRI(pY)RLVGDKN as substrate (Table I). In addition, low activities were observed with the p-Thr peptide ETTYERW(pT)TITQRER and the p-Ser peptide ETTYERW(pT)TITQRER, indicating dual protein phosphatase specificity (Table I; Fig. 1). However, the maximal velocity (Vmax-value) for the p-Tyr peptide was 30- and 131-fold higher as compared with the p-Ser and the p-Thr peptide, respectively (Table I). The affinity (Km value) toward all three phospho-peptides was similar (Table I), indicating that p-Tyr was the preferred substrate.
Phosphatase activity was only observed with the p-Thr peptide RRA(pT)VA for Saci-PP2A (Table I; Fig. 2). In addition, the analysis revealed that Saci-PP2A requires divalent metal ions for activity. The highest activity was observed with Cu2+, followed by Mn2+, Ni2+, Mg2+, Cd2+ and Co2+, in the presence of EGTA (supplemental Fig. S7). When EGTA was replaced by EDTA, no PP2A activity could be observed. In the absence of divalent metal ions the enzyme was inactive. Activity tests in the presence of okadaic acid, an inhibitor of PPPs, revealed a strong inhibitory effect (56% activity in presence of 10 nm okadaic acid; Fig. 2C).
Whole-transcriptome Profiling of Δsaci_ptp and Δsaci_pp2a by RNA-seq
Three cDNA libraries were constructed and we obtained 1,835,618, 1,472,609, and 1,560,420 qualified Illumina read pairs for MW001, Δsaci_ptp and Δsaci_pp2a, respectively. An average of 82.6% of the reads were mapped to rRNA and tRNA sequences, whereas 14.5% mapped to protein-coding sequences (CDS) and 2.8% to intergenic regions in the S. acidocaldarius genome (supplemental Fig. S8).
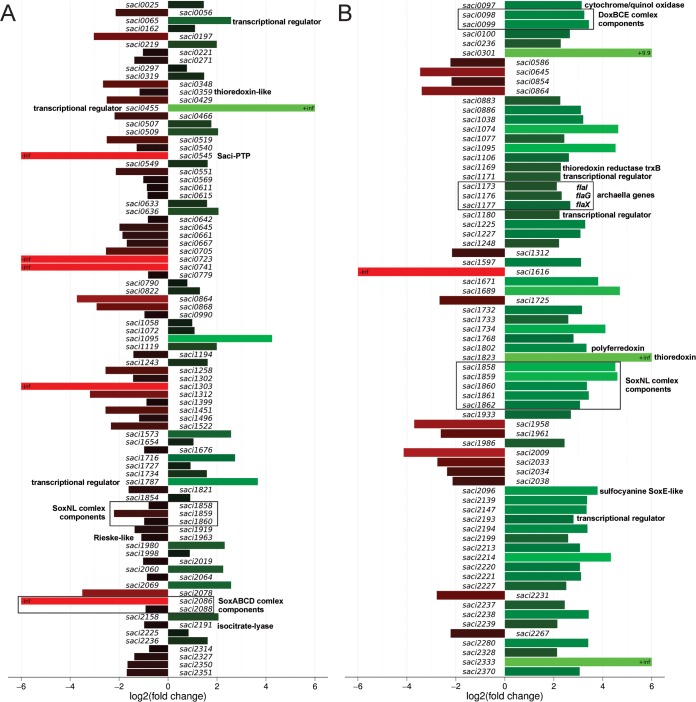
A differential gene expression analysis was performed for each mutant strain, as compared with the MW001 parent strain. The transcript levels of 84 and 71 genes were significantly (threshold of twofold change) altered in Δsaci_ptp and Δsaci_pp2a, respectively (Fig. 3A and Fig. 4; supplemental Table S3A and S3B). Although the majority of the changes corresponded to down-regulated genes in Δsaci_ptp (52 of 84 genes), the opposite was observed in Δsaci_pp2a (56 of 71 genes up-regulated; Fig. 3A and Fig. 4). To functionally categorize the regulated genes, an arCOG category analysis was performed (40, 41). A majority of the differentially affected transcripts encoded proteins belonging to the categories unknown function (arCOG S), energy production and conversion (arCOG C), and transcription (arCOG K) (supplemental Table S4).
Fig. 3.
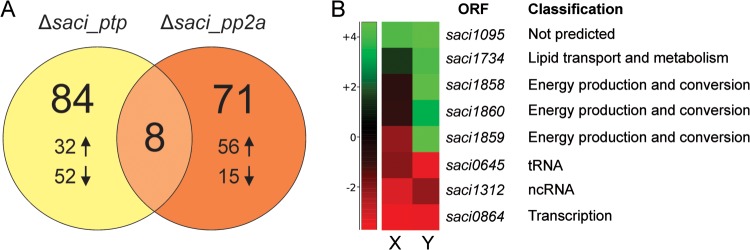
Transcriptionally regulated genes in Δsaci_ptp and Δsaci_pp2a as compared with the strain MW001. A, The number of differentially regulated gene products in Δsaci_ptp and Δsaci_pp2a are depicted in a Venn diagram. Up and down arrows represent the number of genes which were found to be either up- or down-regulated for each deletion strain. In Δsaci_ptp, most genes were down-regulated, whereas in Δsaci_pp2a most were up-regulated. Eight genes were differentially regulated in both phosphatase deletion strains. B, Heat-map representation of all eight commonly regulated genes in Δsaci_ptp (X) and Δsaci_pp2a (Y). The graph is based on the differential gene expression analysis by RNA-seq, showing all genes that are significantly altered in their expression in both deletion strains. The right side panel shows the functional classification for each gene product.
Fig. 4.
Bar plot representation of the transcriptomic changes in Δsaci_ptp and Δsaci_pp2a. A differential gene expression analysis was performed for each deletion strain, Δsaci_ptp (A) and Δsaci_pp2a (B), in comparison to the wild-type strain MW001. All genes of whose expression was significantly altered (≥twofold) are shown. Red and green bars represent down- and up-regulated genes, respectively. Genes with a p value below 0.054 were considered to be significantly differentially regulated. The annotation of selected genes is indicated to the right of each column.
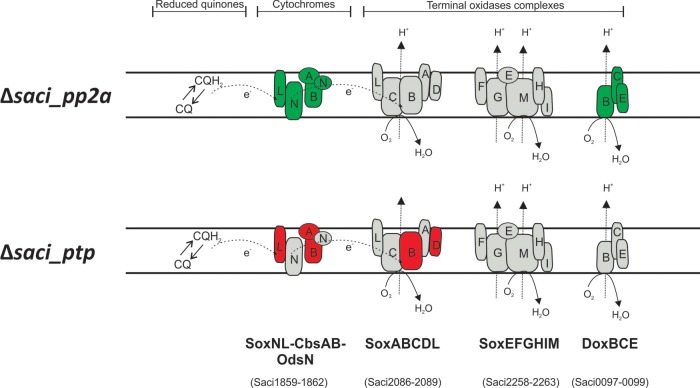
Only eight common genes were significantly affected in both deletion strains (Fig. 3B). Five of these were affected in the same direction (either up- or down-regulated), whereas three were divergently expressed (down-regulated in Δsaci_ptp whereas up-regulated in Δsaci_pp2a; Fig. 3B and Fig. 4). These three, saci1858, saci1859, and saci1860, corresponded to genes encoding SoxNL complex components (complex III of the respiratory chain; saci1858 and saci1859 encode mono-heme cytochrome b558–566 (CbsA and CbsB), whereas saci1860 encodes a Riske Fe-S protein (SoxL)) (42). The two downstream genes of the soxNL complex operon, saci1861 (soxN) and saci1862 (odsN), were also up-regulated in Δsaci_pp2a (Fig. 4). Moreover, Δsaci_pp2a displayed augmented transcript levels of components of one of the three terminal oxidases (complex IV; the DoxBCE complex (saci0098 and saci0099); (Fig. 4). Four additional genes belonging to the energy metabolism and conversion arCOG category were up-regulated in Δsaci_pp2a, encoding a cytochrome/quinol oxidase (Saci0097), a polyferredoxin (Saci1802), a thioredoxin (Saci1823), and the sulfocyanine SoxE-like protein (Saci2096) (Fig. 4).
Increased transcript levels of three transcriptional regulators (Saci1171, Saci1180, and Saci2193) were detected in Δsaci_pp2a. Interestingly, saci1180 and saci1171 are located up- and downstream of the archaellum operon (formerly archaeal flagellum (43)) (Fig. 4) and have been shown to activate archaella gene expression (44). Three additional genes of the archaellum operon, encoding FlaI, FlaG, and FlaX (saci1173, saci1176, and saci1177), were found to be up-regulated in Δsaci_pp2a cells, in agreement with a regulatory role for protein phosphorylation in archaella expression (33).
In contrast to Δsaci_pp2a, the Δsaci_ptp strain showed a predominant down-regulation of gene expression, including genes encoding components of the SoxABCD oxidase complex (saci2088 (soxB) and saci2086 (soxD)) (Fig. 4) (45). Additionally, transcript levels of ORFs encoding a thioredoxin-like (Saci0359) protein, a Rieske-like protein (Saci1963) and an isocitrate-lyase (Saci2191) were down-regulated. To illustrate the transcription changes, a model of the S. acidocaldarius branched respiratory electron transport chain is depicted in Fig. 5 with the transcriptional response of each phosphatase mutant on the oxidase complexes indicated. Among the few up-regulated genes in Δsaci_ptp, three predicted transcriptional regulators were among the highest expressed genes (saci0065, saci0455, and saci1787). The target genes for these regulators are, unknown.
Fig. 5.
Proposed model of S. acidocaldarius branched aerobic respiratory chain. The respiratory chain model is based on separate biochemical characterization studies. Electrons from NADH or succinate oxidation presumably enter electron transport chain via NADH:quinone oxidoreductases or succinate:quinone oxidoreductases (not shown). Reduced quinones (CQH2) then deliver electrons to cytochrome oxidase bc1 complex SoxLN-CbsAB-OdsN (42) and subsequently to one of the three terminal oxidase complexes. These terminal complexes are SoxABCD-SoxL (45, 54, 55), the bb3 terminal oxidase complex SoxEFGHIM (56–58), and the DoxBCE (59). Components whose transcript levels were altered in each deletion strain are depicted either in red or green for down- and up-regulation, respectively. Gene identifiers for each component are shown below.
To confirm the RNA-seq data the expression of all archaellum operon genes, together with selected respiratory chain genes, was analyzed by quantitative RT-PCR. Whereas no significant changes in expression levels could be observed in the Δsaci_ptp deletion strain, all seven genes of the archaellum operon were found to be up-regulated in the Δsaci_pp2a cells (Fig. 6), confirming the result of the analysis (Fig. 4). The induction of the genes encoding SoxNL complex components and the SoxABCDL terminal oxidase in Δsaci_pp2a was also confirmed (Fig. 6).
Fig. 6.
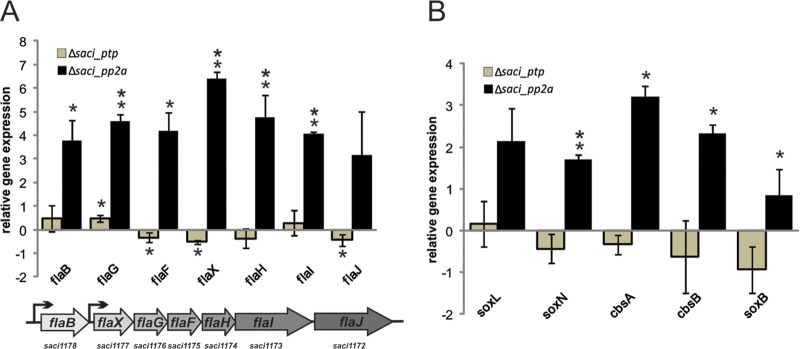
Differential expression levels of archaella operon genes and respiratory chain genes in Δsaci_pp2a and Δsaci_ptp. Total RNA isolated from S. acidocaldarius MW001, Δsaci_pp2a and Δsaci_ptp cultures were used for cDNAs synthesis. qRT-PCR analysis was performed using specific primers for each archaella component (A) and components of the terminal oxidase complexes (B). Relative transcript levels of each gene were normalized to an internal control gene secY. The values reflect the fold change compared with cDNA prepared from MW001. The means and standard deviations of three biological replicates are shown. Up-regulation of all archaella and the terminal oxidase genes in the saci_pp2a deletion strain reflect the RNA-seq results. * significant (p value ≤ 0.05), ** highly significant (p value ≤ 0.01).
Phosphoproteomic Analysis of MW001, Δsaci_ptp and Δsaci_pp2a
Phosphoproteomic analyses (including cytoplasmic and membrane proteins) were performed on the three strains (MW001, Δsaci_pp2a and Δsaci_ptp) in late exponential growth phase (OD600 nm of 0.8), as described previously for S. solfataricus P2 (10). Two biological replicates were analyzed for each strain and a false discovery rate (FDR) for each experiment was also estimated. As a result, averaged FDRs of 3.4%, 3.0%, and 3.6% were estimated for MW001, Δsaci_ptp, and Δsaci_pp2a strains, respectively. Overall, 1206 phosphorylated peptides (p-peptides) from 801 unique phosphorylated proteins (phosphoproteins) were identified, with an overall pS/pT/pY %-ratio of 35.6/28.1/36.2. In MW001, 54 phosphopeptides from 54 phosphoproteins (pS/pT/pY %-ratio of 32.9/21.3/45.7) were detected, in Δsaci_pp2a 477 phosphopeptides from 387 phosphoproteins (35.0/27.2/37.7), and in Δsaci_ptp 715 phosphopeptides from 551 phosphoproteins (36.4/29.1/34.5) (supplemental Table S1, Table II). The identified phosphoproteins were categorized according to arCOG functional code (40, 41). Unique phosphoproteins were identified in 21 out of 26 arCOG functional categories (supplemental Table S2), indicating an important role of phosphorylation in most cellular processes.
Table II. Comparison of different phosphoproteome studies.
| Organism | Genome size (Mb) | Strain | P-proteins (no.) | P-peptides (no.) | P-sites (no.) | pSer (%) | pThr (%) | pTyr (%) |
|---|---|---|---|---|---|---|---|---|
| S. acidocaldarius Total | 2.2 | 801 | 1246 | 1855 | 35.6 | 28.1 | 36.2 | |
| S. acidocaldarius | MW001 | 54 | 54 | 77 | 31.2 | 23.4 | 45.5 | |
| Δsaci_pp2a | 387 | 477 | 734 | 35.0 | 27.2 | 37.7 | ||
| Δsaci_ptp | 551 | 715 | 1044 | 36.4 | 29.1 | 34.5 | ||
| S. solfataricus Total (40) | 2.9 | P2 | 540 | 690 | 1318 | 25.8 | 20.6 | 53.6 |
| S. solfataricus (40) | 2.9 | P2 | 311Glu | 343Glu | 580Glu | 25.2Glu | 18.5Glu | 55.3Glu |
| 311Tryp | 384Tryp | 810Tryp | 25.1Tryp | 21.5Tryp | 53.4Tryp | |||
| Halobacterium salinarum (2) | 2.7 | R1 | 69 | 90 | 81 | 86 | 12 | 1 |
| Lactococcus lactis (60) | 2.4 | Il1403 | 63 | 102 | 73 | 46.5 | 50.6 | 2.7 |
| Bacillus subtilis (1) | 4.2 | 168 | 78 | 103 | 78 | 69.2 | 20.5 | 10.3 |
| Escherichia coli K12(61) | 4.6 | K12-MG1655 | 79 | 105 | 81 | 67.9 | 23.5 | 8.6 |
| Zea mays leaf (62) | 2.065 | B73 | 125 | 149 | 157 | 89.8 | 9.6 | 0.6 |
| Synechococcus sp. (63) | 3.4 | PCC 7002 | 245 | 280 | 410 | 43.9 | 42.44 | 13.66 |
| Pseudomonas putida(51) | 5.9 | PNL-MK25 | 40 | 56 | 53 | 52.8 | 39.6 | 7.5 |
| Pseudomonas aeruginosa (51) | 6.3 | PAO1 | 23 | 57 | 55 | 52.7 | 32.7 | 14.5 |
| Campylobacter jejuni (64) | 1.6 | NCTC11168 | 36 | 58 | 35 | - | - | - |
| Streptococcus pneumonia (65) | 2.1 | D39 | 84 | 102 | 163 | 47.2 | 43.8 | 9 |
| Streptomyces coelicolor (66) | 9.1 | A3 (2) | 40 | 44 | 44 | 34.1 | 52.3 | 13.6 |
| Streptomyces coelicolor (67) | 9.1 | M145 | 127 | 260 | 289 | 46.8 | 48 | 5.2 |
| Klebsiella pneumonia (68) | 5.5 | NTUH-K2044 | 81 | 117 | 93 | 31.2 | 15.1 | 25.8 |
| Mycoplasma pneumoniae (69) | 0.8 | M129 | 63 | 16 | 16 | 53.3 | 46.7 | 0 |
| Helibacter pylori (70) | 1.7 | 26695 | 67 | 80 | 126 | 42.8 | 38.7 | 18.5 |
| Mycobacterium tuberculosis (71) | 4.4 | H37Rv | 301 | 381 | 516 | 60 | 40 | 0 |
| Listeria monocytogenes (72) | 2.9 | EGDe | 112 | 155 | 143 | 65 | 30.1 | 4.9 |
| Clostridium acetobutyicum (73) | 4.1 | ATCC824 | 61 | 82 | 107 | 42 | 47.7 | 10.3 |
| Plasmodium falciparum (74) | 23.3 | 3D7 | 919 | 2418 | 2541 | 84.4 | 13.2 | 2.4 |
| Trichomonas vaginalis (75) | 176.4 | ATCC30236 | 82 | 93 | 103 | 71.8 | 21.4 | 6.8 |
| Thermus thermophilus (76) | 2.1 | HB8 | 48 | 52 | 50 | 64 | 26 | 10 |
| Rhodopseudomonas palustris (77) | 5.5 | CGA010 | 54CH | 100CH | 60CH | 63.3CH | 16.1CH | 19.4CH |
| 42PH | 74PH | 56PH | 58.9PH | 23.2PH | 17.9PH | |||
| Trypanosoma cruzi (78) | 67 | DM28c | 753 | - | 2572 | 84.1 | 14.9 | 1.0 |
| Toxoplasma gondii (79) | 80 | RH | 2793i | 11,822i | 12,793i | 87.2 | 12.6 | 0.25 |
| 3506p | 21,498p | 24,298p | 89.3 | 10.5 | 0.25 | |||
| Plasmodium falciparum (79) | 23 | 3D7 | 1673 | 7835 | 8463 | 89.1 | 10.4 | 0.51 |
| Arabidopsis thaliana (80) | 119.7 | - | 598 | 962 | 609 | 86 | 14 | 0.16 |
Target Proteins for Phosphorylation
The phosphoproteome data set was specifically screened for proteins belonging to the aerobic respiration chain and the archaellum (Table III). Six proteins of the arCOG family C (energy production and conversion) were found to be phosphorylated, including SoxC (Saci2087) and SoxI (Saci2258), which belong to different terminal oxidase complexes (Fig. 5). Furthermore, three von Willebrand domain-containing proteins (Saci0977, Saci1209, and Saci1211) involved in archaellum synthesis were phosphorylated at different residues. One of these, ArnB (Saci1211), has previously been shown to be involved in negative regulation of the archaellum in vivo (33). Also the membrane-associated FlaJ protein (Saci1172), involved in archaella assembly, contained four phosphorylation sites and was identified in both phosphatase deletion mutants. The predicted positive transcriptional regulator ArnR1 (Saci1171)(44) located directly adjacent to the archaellum operon was also phosphorylated. Seventeen additional predicted transcriptional regulators also contained phosphorylation sites, demonstrating the importance of this PTM in transcriptional regulation in Archaea. Five predicted serine/threonine kinases were also identified as phosphorylated in the deletion strains.
Table III. Phosphoproteins related to aerobic respiration, motility, transcriptional regulation and phosphorylation identified in the S. acidocaldarius phosphoproteome.
| Protein | arCOG category | Functional arCOG annotation | Strain |
|---|---|---|---|
| Saci2087 | C | Cytochrome b subunit of the bc complex (SoxC) | Δsaci_ptp |
| Saci2250 | C | Pyruvate:ferredoxin oxidoreductase or related 2-oxoacid:ferredoxin oxidoreductase, gamma subunit | Δsaci_ptp |
| Saci2258 | C | Predicted subunit of heme/copper-type cytochrome/quinol oxidase (SoxI) | Δsaci_ptp |
| Saci2263 | C | Heme/copper-type cytochrome/quinol oxidase, subunit 1 and 3 | Δsaci_pp2a |
| Saci2273 | C | Fe-S oxidoreductase | Δsaci_pp2a |
| Saci2316 | C | Fe-S oxidoreductase | Δsaci_pp2a Δsaci_ptp |
| Saci0977 | R | Uncharacterized protein containing a von Willebrand factor type A (vWA) domain | MW001 Δsaci_ptp |
| Saci1209 | R | Uncharacterized protein containing a von Willebrand factor type A (vWA) domain | Δsaci_ptp |
| Saci1211 | R | Protein containing a von Willebrand factor type A (vWA) domain ArnB | Δsaci_ptp |
| Saci1172 | NU | Archaeal flagella assembly protein J | Δsaci_ptp Δsaci_pp2a |
| Saci1171 | K | Predicted transcriptional regulator | Δsaci_ptp |
| Saci0446 | K | Transcriptional regulator, contains HTH domain | Δsaci_ptp |
| Saci0501 | K | Predicted transcriptional regulator, PadR family | Δsaci_pp2a |
| Saci0882 | K | Predicted transcriptional regulator | Δsaci_pp2a |
| Saci1068 | K | Transcriptional regulator, MarR family | Δsaci_ptp |
| Saci1161 | K | Sugar-specific transcriptional regulator TrmB | Δsaci_ptp |
| Saci1242 | K | Transcriptional regulator, contains HTH domain | Δsaci_pp2a |
| Saci1344 | K | Predicted transcriptional regulator | MW001 Δsaci_ptp |
| Saci1502 | K | Predicted transcriptional regulator, contains C-terminal CBS domains | Δsaci_ptp |
| Saci1523 | K | Predicted transcriptional regulator, contains ATP-binding and HTH domains | Δsaci_ptp Δsaci_pp2a |
| Saci1528 | K | Predicted transcriptional regulator, contains ATP-binding and HTH domains | Δsaci_pp2a |
| Saci1588 | K | Transcriptional regulator (IclR family) | Δsaci_ptp |
| Saci1658 | K | Transcriptional regulator (Lrp/AsnC family) | Δsaci_ptp |
| Saci2116 | K | Predicted transcriptional regulator, c-terminal HTH-like domain | Δsaci_pp2a |
| Saci2352 | K | Predicted transcriptional regulator, PadR family | Δsaci_ptp |
| Saci0635 | K | Transcriptional regulator, xre family | Δsaci_ptp |
| Saci1505 | K | Transcriptional regulator, xre family | Δsaci_ptp |
| Saci2005 | K | Predicted transcriptional regulator | Δsaci_pp2a |
| Saci0965 | TD | Serine/threonine protein kinase involved in cell cycle control | Δsaci_pp2a |
| Saci1193 | RTKL | Membrane associated serine/threonine protein kinase | Δsaci_pp2a Δsaci_ptp |
| Saci1694 | RTKL | Membrane associated serine/threonine protein kinase | Δsaci_pp2a Δsaci_ptp |
| Saci1869 | RTKL | Membrane associated serine/threonine protein kinase | Δsaci_ptp |
| Saci1875 | RTKL | Membrane associated serine/threonine protein kinase | Δsaci_pp2a |
Regulation of Cell Motility Via Reversible Protein Phosphorylation
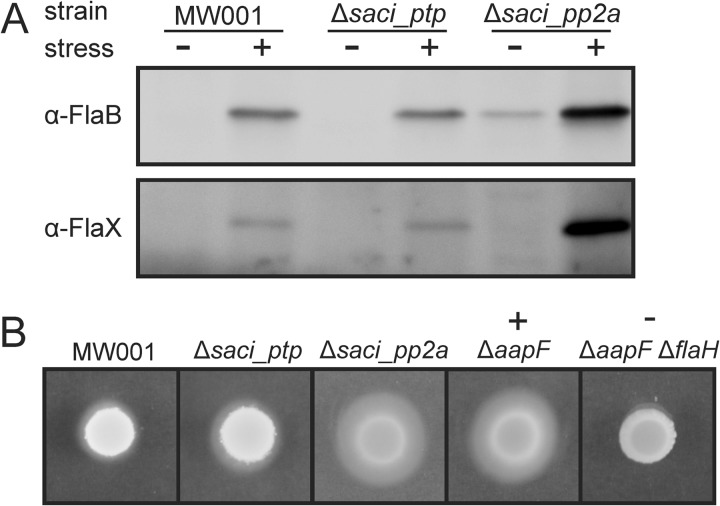
The up-regulation of the seven archaella genes in Δsaci_pp2a (Figs. 4 and 6), was investigated at the protein phosphorylation level. The parental strain and both deletion strains were cultivated in rich medium and then transferred to medium without supplemented nutrients for 5 h, a treatment that has been shown to induce archaella biosynthesis (29). Samples before and after induction were analyzed by immunoblotting with specific antibodies against the archaellin protein FlaB and the accessory protein FlaX. Both proteins were induced by nutrient limitation stress in the parent strain MW001, and to the same extent in Δsaci_ptp. In contrast, Δsaci_pp2a showed a FlaB signal already before induction, and highly increased levels of FlaB and FlaX after nutrient limitation as compared with MW001 and Δsaci_ptp (Fig. 7A).
Fig. 7.
Impact of phosphatase deletion on archaella expression and motility. A, For the nutrient limitation assay MW001, Δsaci_ptp and Δsaci_pp2a were grown to an OD600 of about 0.4 in Brock medium supplemented with 0.1% NZ-amine, 0.2% sucrose and 10 μg/ml uracil and then transferred to medium lacking the nutrient source. After 6 h samples were collected and immunoblotting was performed with specific antibodies against the archaellum proteins FlaB and FlaX. The amount of protein in Δsaci_ptp is comparable to the strain MW001, whereas for Δsaci_pp2a highly elevated expression of FlaB and FlaX was observed after the starvation stress. Interestingly, already before stress induction the FlaB levels were increased in the Saci-PP2A deletion mutant. B, The wild type strain MW001, the two phosphatase deletion strains Δsaci_ptp and Δsaci_pp2a, the hypermotile positive control strain ΔaapF and the negative nonmotile strain ΔaapFΔflaH were spotted on semi-solid Gelrite dishes and incubated for 5 days at 76 °C. Only a small halo is visible for MW001 and Δsaci_ptp, whereas Δsaci_pp2a was as hypermotile as the positive control strain.
The swimming ability of the wild-type strain and both deletion mutants was investigated using a motility assay. MW001, Δsaci_ptp, Δsaci_pp2a, a hypermotile positive control (ΔaapF) and a nonmotile negative control (ΔaapFΔflaH) were spotted on semi-solid Gelrite-plates containing low amounts of nutrients (29, 46). After 5 days of incubation at 76 °C, MW001 and Δsaci_ptp showed only slight motility. In contrast, the motility of Δsaci_pp2a was comparable to the hypermotile ΔaapF, which lacks the Aap pili (Archaeal adhesive pili) (Fig. 7B), indicating a major impact of protein phosphorylation on cell motility in S. acidocaldarius.
DISCUSSION
We report a detailed characterization of two protein phosphatases of S. acidocaldarius, Saci-PTP and Saci-PP2A. Saci-PTP displayed dual-specific Ser/Thr and Tyr phosphatase activity, whereas Saci-PP2A was specific for Ser/Thr dephosphorylation. Differences in phosphorylation and gene expression patterns were observed in deletion strains, suggesting roles for both phosphatases in signal transduction pathways. The changes observed when the gene encoding Saci-PP2A was deleted were similar to responses observed when S. acidocaldarius cultures experience severe nutrient limitation. Up-regulation of archaellar and respiratory chain components may enable the organism to evade unfavorable conditions by promoting archaellar assembly and providing energy for archaella-driven motility.
Comparison of the phosphoproteome of S. acidocaldarius with that of S. solfataricus (40) revealed an overlap of 94 proteins, although the different experimental approaches (deletion mutants compared with growth on different carbon sources) should be taken into account. Conserved phosphorylation sites were identified in 13 of the 94 overlapping proteins. A single conserved site was identified in ten of these, whereas two sites were detected in Saci0940, Saci0381, and Saci2134. None of the 15 sites had a score higher than 0.5 (required to score as a phosphorylation site) in searches using DISPHOS 1.3 software. The highest scores were obtained for the conserved pThr sites in Saci1549 (V-type ATP synthase subunit B; score 0.477) and Saci1715 (dihydroxyacid dehydratase; score 0.263). Although the 15 phosphorylation sites only represent a small fraction of all identified sites and therefore may not provide sufficient data, difficulties in prediction of archaeal phosphorylation sites were also reported in a phosphoproteome study of H. salinarium, using the Motif X tool (2).
The number of identified phosphorylation targets in S. acidocaldarius is high, with 801 phosphoproteins identified in the three strains. Only for Plasmodium falciparum (1673) and Toxoplasma gondii (intracellular samples 2793; purified samples 3506) have higher numbers been recorded. However, both are eukaryotes and possess several-fold larger proteomes as compared with S. acidocaldarius. Also, the number of identified phosphoproteins in the S. acidocaldarius parent strain MW001 alone was low (54), and similar to the number observed in the euryarchaeon H. salinarum (69).
Deletion of the phosphatases increased the number of phosphoproteins, as expected. When comparing the Δsaci_pp2a strain with the MW001 parent, the number of phosphorylation sites increased from 77 to 734, whereas the pSer/pThr/pTyr %-ratio remained similar. In the Δsaci_ptp strain the phosphorylation sites increased even more, from 77 to 1044, and the pSer/pThr/pTyr %-ratio was also influenced by the deletion (31.2/23.4/45.5 compared with 36.4/29.1/34.5). In Δsaci_pp2a the dephosphorylation of pSer and pThr is prevented, which should result in a higher amount of pSer and pThr, whereas in Δsaci_ptp strain the amount of pTyr should also be increased. A similar discrepancy was reported for H. salinarum, in which a ΔserB strain was compared with the corresponding wild type. The gene serB encodes the only phosphoserine phosphatase annotated in the genome of H. salinarium and the deletion increased the number of phosphoproteins from 26 (wild type) up to 62 (ΔserB strain). In addition, the amount of pSer was increased in a similar way from 25 (wild type) up to 64 (ΔserB strain). However, the amount of pThr as well as pTyr was also increased from 5 (wild type) to 10 (ΔserB strain) and 0 (wild type) to 1 (ΔserB strain). Usually, one would expect that the deletion of SerB only affects the number of phosphoproteins and the amount of pSer and that the amount of pThr and pTyr should be the same. Nonetheless, the enzymatic properties of SerB were not investigated until now, the data imply that the protein is not specific for pSer.
Bioinformatic analysis revealed that SerB shares no similarity with Saci-PTP or Saci-PP2A. For S. acidocaldarius, the unexpected change of the pSer/pThr/pTyr %-ratio could partly be explained by the biochemical properties of Saci-PTP and Saci-PP2a, as the Saci-PTP displays dual activity. It is also possible that deletion of the protein phosphatases induces other PTMs to compensate for the loss of regulation.
The high amount of Tyr phosphorylation in both Sulfolobus species appears to be a crenarchaeal feature, rather than a thermoadaptation property or a general archaeal feature. The phosphoproteome of the hyperthermophilic bacterium Thermus thermophilus was recently analyzed, revealing 48 phosphoproteins with 50 phosphorylation sites and a pSer/pThr/pTyr %-ratio of 64/26/10. The amount of pTyr is in accordance with amounts identified in other prokaryotic phosphoproteome studies (Table II), supporting the conclusion that Tyr phosphorylation is a crenarchaeal feature rather than a thermoadaptation. Both T. thermophilus and H. salinarum contain two-component phosphorylation systems, which are absent in crenarchaea. It is thus possible that that the extensive Tyr phosphorylation in Sulfolobus species provides a means to compensate for the absence of two-component systems in these organisms.
The expression of several transcription factors was highly affected in the deletion mutants. However, in the absence of information about target genes, the physiological consequences remain unknown. In contrast, an explanation can be provided for the effects of deletion of Saci-PP2A on archaellum and respiration chain genes. In S. acidocaldarius two regulatory systems have been shown to be involved in control of archaella biosynthesis, the ArnA (Saci1210) and ArnB (Saci1211) repressors of the flaX-flaJ gene cluster (33), and the ArnR (Saci1180) and ArnR1 (Saci1171) positive regulators of flaB expression (See model in Fig. 8) (44). ArnR1 and ArnR, were up-regulated in the Δsaci_pp2a strain and, in addition, ArnR1 was found to be phosphorylated in the Δsaci_ptp strain. Further, in the biochemical characterization, ArnA and ArnB were phosphorylated by the Saci1193 kinase and dephosphorylated by the Saci-PP2A phosphatase (for a model see Fig. 8) (33). In the phosphoproteome data, this kinase and ArnB revealed in vivo phosphorylation in the kinase domain and the C-terminal region, respectively, (see supplemental Table S1). In addition, the saci1193 gene displays strong cell-cycle-specific induction of transcription (47), with >16-fold up-regulation around the cell division stage, just before a two- to fourfold up-regulation of the archaellum-encoding genes (flab - flaJ; saci1174 - saci1178), further linking these functions together. In S. tokodaii, ArnA has been shown to bind to the FlaX promoter (48). However, no effect on transcription of FlaB and FlaX was observed in the S. acidocaldarius arnA deletion strain, suggesting that regulation instead may occur via protein-protein interaction (33). The C-terminal phosphorylation sites of ArnB shown here could thus affect docking of ArnA (33). Under nutrient starvation conditions archaella biosynthesis is highly induced and expression levels are comparable to those in the Δsaci_pp2a strain, therefore both conditions lead to a situation, at least regarding to archaella expression, where the negative regulators (ArnA/B) are inactive because of permanent phosphorylation and the positive regulators (ArnR/R1) can enhance archaella expression (see Fig. 8).
Fig. 8.
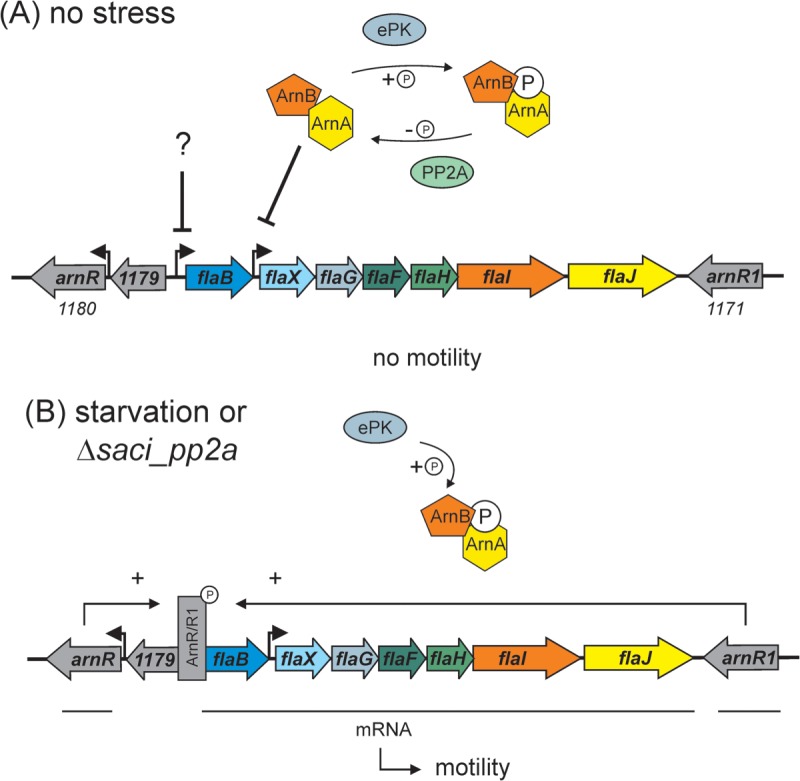
Model of the involvement of phosphorylation in the regulation of motility in S. acidocaldarius. During exponential growth (A) archaellum synthesis from the flaB-flaJ (saci1178–1172) operon is repressed. Part of the repression network are ArnA (Saci1210) and ArnB (Saci1211) that are phosphorylated and dephosphorylated by ePK (Saci1193) and PP2A, respectively. ArnA and B closely interact and repress the flaX promoter (35). Probably another, yet unknown, factor is repressing the flaB promoter. When S. acidocaldarius cells enter stationary phase and undergo starvation, or in the Δsaci_pp2a deletion mutant (B), the ArnaA/B complex exists only in its phosphorylated form and repression is released. Moreover, the expression of ArnR (Saci1180) and ArnR1 (Saci1171) is induced and these factors then drive high expression of FlaB which leads to the assembly of archaella on the cell surface and therefore to increased motility.
Thioredoxin (Saci1823), thioredoxin reductase (Saci1169) and peroxiredoxin (Saci2227) encoding genes were up-regulated in the Δsaci_pp2a strain. These were also up-regulated in a study on on oxidative stress in S. solfataricus, but no archaealla gene up-regulation was found (49). In all three domains of life, these proteins are involved in disulfide bond shuffling during oxidative stress conditions. The thioredoxin reductase gene is located immediately adjacent to the archaella genes and the ArnR1 regulator and the protein was, in addition, found to be phosphorylated. Hence, nutrient limitation leads both to archaella expression and to oxidative stress responses to avoid cellular damage. In summary, the ArnA/ArnB and ArnR/R1 regulatory systems of archaella expression constitute the first complex regulatory network involving protein phosphorylation to be investigated in crenarchaea.
Surface structure-driven motility is an energy-consuming process and archaella assembly therefore has to be tightly controlled. Several genes involved in energy conversion, including those encoding the SoxNL, SoxABCDL, and DoxBCE respiratory chain complexes, were differentially regulated in the phosphatase deletion strains as compared with the parental strain (Fig. 4). Recently, an association of the flagellar switch-motor complex to the NADH-ubiquinone oxidoreductase and the F0F1 ATP synthase was demonstrated in the bacterium Escherichia coli, and predicted to function as a local energy supply for flagellar movement (50). The up-regulation of archaella genes in S. acidocaldarius may similarly be connected to the up-regulation of genes involved in the respiratory chain. Possible interactions of energy generating complexes with the archaella basal body should therefore be investigated. Phosphorylation of respiratory chain components has also been reported in phosphoproteomic studies of Pseudomonas spp (51)., Saccharomyces cerevisiae mitochondria (52), human muscle mitochondria (53) and S. solfataricus (10).
This study provides the first insights into the in vitro and in vivo functions of the two S. acidocaldarius protein phosphatases, Saci-PTP and Saci-PP2A. The RNA-seq and phosphoproteomics studies of the deletion mutants shed light on the multitude of cellular processes affected by reversible protein phosphorylation. In particular, we demonstrate that SaciPP2A is involved in the complex signal transduction pathway regulating the expression of the S. acidocaldarius archaellum, and also plays a major role in the control of aerobic respiration and oxidative stress responses.
Supplementary Material
Footnotes
* JR was supported by a DFG grant (AL 1206/4-1) and SVA and AO were supported by intramural funds of the Max Planck Society. DE was supported by DFG grant (SI 642/10-1). FA was supported by GEN-AU project “regulatory ncRNAs”. PCW and TKP were supported by the EPSRC (EP/E036252/1 and EP/I031812/1). RB and ACL were supported by Swedish Research Council grant 621-2010-5551.
 This article contains supplemental Figs S1 to S9 and Tables S1 to S7.
This article contains supplemental Figs S1 to S9 and Tables S1 to S7.
1 The abbreviations used are:
- PTM
- post-translational modification
- arCOG
- archaeal clusters of orthologous groups
- qPCR
- quantitative PCR.
REFERENCES
- 1. Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., Mann M. (2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697–707 [DOI] [PubMed] [Google Scholar]
- 2. Aivaliotis M., Macek B., Gnad F., Reichelt P., Mann M., Oesterhelt D. (2009) Ser/Thr/Tyr protein phosphorylation in the archaeon Halobacterium salinarum–a representative of the third domain of life. PloS One 4, e4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pawson T., Scott J. D. (2005) Protein phosphorylation in signaling - 50 years and counting. Trends Biochem. Sci. 30, 286–290 [DOI] [PubMed] [Google Scholar]
- 4. Johnson L. N., Lewis R. J. (2001) Structural basis for control by phosphorylation. Chem. Rev. 101, 2209–2242 [DOI] [PubMed] [Google Scholar]
- 5. Westheimer F. H. (1987) Why nature chose phosphates. Science 235, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 6. Galperin M. Y. (2005) A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kennelly P. J. J. (2003) Archaeal protein kinases and protein phosphatases: insights from genomics and biochemistry. Biochem. J. 370, 373–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim D., Forst S. (2001) Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology 147, 1197–1212 [DOI] [PubMed] [Google Scholar]
- 9. Koretke K. K., Lupas A. N., Warren P. V., Rosenberg M., Brown J. R. (2000) Evolution of two-component signal transduction. Mol. Biol. Evolution 17, 1956–1970 [DOI] [PubMed] [Google Scholar]
- 10. Esser D., Pham K. T., Reimann J., Albers S. V., Siebers B., Wright P. C. (2012) Change of carbon source causes dramatic effects in the phospho-proteome of the Archaeon Sulfolobus solfataricus. J. Proteome Res. 11, 4823–4833 [DOI] [PubMed] [Google Scholar]
- 11. Lower B. H., Bischoff K. M., Kennelly P. J. (2000) The archaeon Sulfolobus solfataricus contains a membrane-associated protein kinase activity that preferentially phosphorylates threonine residues in vitro. J. Bacteriol. 182, 3452–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lower B. H., Kennelly P. J. (2003) Open reading frame sso2387 from the Archaeon Sulfolobus solfataricus encodes a polypeptide with protein-serine kinase activity. J. Bacteriol. 185, 3436–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lower B. H., Potters M. B., Kennelly P. J. (2004) A phosphoprotein from the archaeon Sulfolobus solfataricus with protein-serine/threonine kinase activity. J. Bacteriol. 186, 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang B., Yang S., Zhang L., He Z. G. (2010) Archaeal eukaryote-like serine/threonine protein kinase interacts with and phosphorylates a forkhead-associated-domain-containing protein. J. Bacteriol. 192, 1956–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oxenrider K. A., Kennelly P. J. (1993) A protein-serine phosphatase from the halophilic Archaeon. Haloferax volcanii. Biochem. Biophys. Res. Commun. 194, 1330–1335 [DOI] [PubMed] [Google Scholar]
- 16. Leng J., Cameron A. J., Buckel S., Kennelly P. J. (1995) Isolation and cloning of a protein-serine/threonine phosphatase from an archaeon. J. Bacteriol. 177, 6510–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeon S.-J., Fujiwara S., Takagi M., Tanaka T., Imanaka T. (2002) Tk-PTP, protein tyrosine/serine phosphatase from hyperthermophilic archaeon Thermococcus kodakaraensis KOD1: enzymatic characteristics and identification of its substrate proteins. Biochem. Biophys. Res. Commun. 295, 508–514 [DOI] [PubMed] [Google Scholar]
- 18. Chu H. M., Wang A. H. (2007) Enzyme-substrate interactions revealed by the crystal structures of the archaeal Sulfolobus PTP-fold phosphatase and its phosphopeptide complexes. Proteins 66, 996–1003 [DOI] [PubMed] [Google Scholar]
- 19. Solow B., Young J. C., Kennelly P. J. (1997) Gene cloning and expression and characterization of a toxin-sensitive protein phosphatase from the methanogenic archaeon Methanosarcina thermophila TM-1. J. Bacteriol. 179, 5072–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi L., Potts M., Kennelly P. J. (1998) The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 22, 229–253 [DOI] [PubMed] [Google Scholar]
- 21. Hubbard M. J., Cohen P. (1993) On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem. Sci. 18, 172–177 [DOI] [PubMed] [Google Scholar]
- 22. Barton G. J., Cohen P. T., Barford D. (1994) Conservation analysis and structure prediction of the protein serine/threonine phosphatases. Sequence similarity with diadenosine tetraphosphatase from Escherichia coli suggests homology to the protein phosphatases. Eur. J. Biochem. 220, 225–237 [DOI] [PubMed] [Google Scholar]
- 23. Bork P., Brown N. P., Hegyi H., Schultz J. (1996) The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 5, 1421–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barford D. (1996) Molecular mechanisms of the protein serine/threonine phosphatases. Trends Biochem. Sci. 21, 407–412 [DOI] [PubMed] [Google Scholar]
- 25. Dahche H., Abdullah A., Ben Potters M., Kennelly P. J. (2009) A PPM-family protein phosphatase from the thermoacidophile Thermoplasma volcanium hydrolyzes protein-bound phosphotyrosine. Extremophiles 13, 371–377 [DOI] [PubMed] [Google Scholar]
- 26. Zaparty M., Esser D., Gertig S., Haferkamp P., Kouril T., Manica A., Pham T. K., Reimann J., Schreiber K., Sierocinski P., Teichmann D., van, Wolferen M., von, Jan M., Wieloch P., Albers S. V., Driessen A. J., Klenk H. P., Schleper C., Schomburg D., van, der, Oost J., Wright P. C., Siebers B. (2010) “Hot standards” for the thermoacidophilic archaeon Sulfolobus solfataricus. Extremophiles 14, 119–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wagner M., van Wolferen M., Wagner A., Lassak K., Meyer B. H., Reimann J., Albers S. V. (2012) Versatile genetic tool box for the Crenarchaeote Sulfolobus acidocaldarius. Frontiers Microbiol. 3, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brock T. D., Brock K. M., Belly R. T., Weiss R. L. (1972) Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84, 54–68 [DOI] [PubMed] [Google Scholar]
- 29. Lassak K., Neiner T., Ghosh A., Klingl A., Wirth R., Albers S. V. (2012) Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83, 110–124 [DOI] [PubMed] [Google Scholar]
- 30. Hoffmann S., Otto C., Kurtz S., Sharma C. M., Khaitovich P., Vogel J., Stadler P. F., Hackermüller J. (2009) Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Computat. Biol. 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van der Sluis E. O., Nouwen N., Koch J., De Keyzer J., Van der Does C., Tampé R., Driessen A. J. (2006) Identification of two interaction sites in SecY that are important for the functional interaction with SecA. J. Mol. Biol. 361, 839–849 [DOI] [PubMed] [Google Scholar]
- 33. Reimann J., Lassak K., Khadouma S., Ettema T. J., Yang N., Driessen A. J., Klingl A., Albers S. V. (2012) Regulation of archaella expression by the FHA and von Willebrand domain-containing proteins ArnA and ArnB in Sulfolobus acidocaldarius. Mol. Microbiol. 86, 24–36 [DOI] [PubMed] [Google Scholar]
- 34. Gan C. S., Reardon K. F., Wright P. C. (2005) Comparison of protein and peptide prefractionation methods for the shotgun proteomic analysis of Synechocystis sp. PCC 6803. Proteomics 5, 2468–2478 [DOI] [PubMed] [Google Scholar]
- 35. Zaparty M., Esser D., Gertig S., Haferkamp P., Kouril T., Manica A., Pham T. K., Reimann J., Schreiber K., Sierocinski P., Teichmann D., Van Wolferen M., Von Jan M., Wieloch P., Albers S. V., Driessen A. J. M., Klenk H.-P., Schleper C., Schomburg D., Van der Oost J., Wright P. C., Siebers B. (2010) “Hot standards” for the thermoacidophilic archaeon Sulfolobus solfataricus. Extremophiles 14, 119–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chong P. K., Wright P. C. (2005) Identification and characterization of the Sulfolobus solfataricus P2 proteome. J. Proteome Res. 4, 1789–1798 [DOI] [PubMed] [Google Scholar]
- 37. Panchaud A., Scherl A., Shaffer S. A., Haller P. D., Von, Kulasekara H. D., Miller S. I., Goodlett D. R. (2009) Precursor acquisition independent from ion count : how to dive deeper into the proteomics ocean. Anal. Chem. 81, 6481–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A, Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bernander R., Poplawski A. (1997) Cell cycle characteristics of thermophilic archaea. J. Bacteriol. 179, 4963–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Esser D., Kouril T., Zaparty M., Sierocinski P., Chan P. P., Lowe T., Van der Oost J., Albers S.-V., Schomburg D., Makarova K. S., Siebers B. (2011) Functional curation of the Sulfolobus solfataricus P2 and S. acidocaldarius 98–3 complete genome sequences. Extremophiles 15, 711–712 [DOI] [PubMed] [Google Scholar]
- 41. Makarova K. S., Sorokin A. V., Novichkov P. S., Wolf Y. I., Koonin E. V. (2007) Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol. Direct 2, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hiller A., Henninger T., Schäfer G., Schmidt C. L. (2003) New genes encoding subunits of a cytochrome bc1-analogous complex in the respiratory chain of the hyperthermoacidophilic crenarchaeon Sulfolobus acidocaldarius. J. Bioenergetics Biomembranes 35, 121–131 [DOI] [PubMed] [Google Scholar]
- 43. Jarrell K. F., Albers S.-V. (2012) The archaellum: an old motility structure with a new name. Trends Microbiol. 20, 307–312 [DOI] [PubMed] [Google Scholar]
- 44. Lassak K., Peeters E., Wróbel S., Albers S. V. (2013) The one-component system ArnR: a membrane-bound activator of the crenarchaeal archaellum. Mol. Microbiol. 88, 125–139 [DOI] [PubMed] [Google Scholar]
- 45. Lübben M., Warne A., Albracht S. P., Saraste M. (1994) The purified SoxABCD quinol oxidase complex of Sulfolobus acidocaldarius contains a novel haem. Mol. Microbiol. 13, 327–335 [DOI] [PubMed] [Google Scholar]
- 46. Henche A. L., Koerdt A., Ghosh A., Albers S. V. (2012) Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environmental Microbiol. 14, 779–793 [DOI] [PubMed] [Google Scholar]
- 47. Lundgren M., Bernander R. (2007) Genome-wide transcription map of an archaeal cell cycle. Proc. Natl. Acad. Sci. U.S.A. 104, 2939–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Duan X., He Z. G. (2011) Characterization of the specific interaction between archaeal FHA domain-containing protein and the promoter of a flagellar-like gene-cluster and its regulation by phosphorylation. Biochem. Biophys. Res. Commun. 407, 242–247 [DOI] [PubMed] [Google Scholar]
- 49. Maaty W. S., Wiedenheft B., Tarlykov P., Schaff N., Heinemann J., Robison-Cox J., Valenzuela J., Dougherty A., Blum P., Lawrence C. M., Douglas T., Young M. J., Bothner B. (2009) Something old, something new, something borrowed; how the thermoacidophilic archaeon Sulfolobus solfataricus responds to oxidative stress. PloS One 4, e6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zarbiv G., Li H., Wolf A., Cecchini G., Caplan S. R., Sourjik V., Eisenbach M. (2012) Energy complexes are apparently associated with the switch-motor complex of bacterial flagella. J. Mol. Biol. 416, 192–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ravichandran A., Sugiyama N., Tomita M., Swarup S., Ishihama Y. (2009) Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics 9, 2764–2775 [DOI] [PubMed] [Google Scholar]
- 52. Ohlmeier S., Hiltunen J. K., Bergmann U. (2010) Protein phosphorylation in mitochondria–a study on fermentative and respiratory growth of Saccharomyces cerevisiae. Electrophoresis 31, 2869–2881 [DOI] [PubMed] [Google Scholar]
- 53. Zhao X., León I. R., Bak S., Mogensen M., Wrzesinski K., Højlund K., Jensen O. N. (2011) Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes. Mol. Cell. Proteomics 10, M110.000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gleissner M., Kaiser U., Antonopoulos E., Schafer G. (1997) The archaeal SoxABCD complex is a proton pump in Sulfolobus acidocaldarius. J. Biol. Chem. 272, 8417–8426 [DOI] [PubMed] [Google Scholar]
- 55. Lübben M., Kolmerer B., Saraste M. (1992) An archaebacterial terminal oxidase combines core structures of two mitochondrial respiratory complexes. EMBO J. 11, 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Castresana J., Lübben M., Saraste M. (1995) New archaebacterial genes coding for redox proteins: implications for the evolution of aerobic metabolism. J. Mol. Biol. 250, 202–210 [DOI] [PubMed] [Google Scholar]
- 57. Komorowski L., Verheyen W., Schäfer G. (2002) The archaeal respiratory supercomplex SoxM from S. acidocaldarius combines features of quinole and cytochrome c oxidases. Biol. Chem. 383, 1791–1799 [DOI] [PubMed] [Google Scholar]
- 58. Lübben M., Arnaud S., Castresana J., Warne a, Albracht S. P., Saraste M. (1994) A second terminal oxidase in Sulfolobus acidocaldarius. Eur. J. Biochem. 224, 151–159 [DOI] [PubMed] [Google Scholar]
- 59. Purschke W. G., Schmidt C. L., Petersen A., Schäfer G. (1997) The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J. Bacteriol. 179, 1344–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Soufi B., Gnad F., Jensen P. R., Petranovic D., Mann M., Mijakovic I., Macek B. (2008) The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics 8, 3486–3493 [DOI] [PubMed] [Google Scholar]
- 61. Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics : MCP 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 62. Bi Y. D., Wang H. X., Lu T. C., Li X. H., Shen Z., Chen Y. B., Wang B. C. (2011) Large-scale analysis of phosphorylated proteins in maize leaf. Planta 233, 383–392 [DOI] [PubMed] [Google Scholar]
- 63. Yang M., Qiao Z., Zhang W., Xiong Q., Zhang J., Li T., Ge F., Zhao J. (2013) Global phosphoproteomic analysis reveals diverse functions of serine/threonine/tyrosine phosphorylation in the model Cyanobacterium Synechococcus sp. strain PCC 7002. J. Proteome Res. 12, 1909–1923 [DOI] [PubMed] [Google Scholar]
- 64. Voisin S., Watson D. C., Tessier L., Ding W., Foote S., Bhatia S., Kelly J. F., Young N. M. (2007) The cytoplasmic phosphoproteome of the Gram-negative bacterium Campylobacter jejuni: Evidence for modification by unidentified protein kinases. Proteomics 7, 4338–4348 [DOI] [PubMed] [Google Scholar]
- 65. Sun X., Ge F., Xiao C. L., Yin X. F., Ge R., Zhang L. H., He Q. Y. (2009) Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. J.Proteome Res. 9, 275–282 [DOI] [PubMed] [Google Scholar]
- 66. Parker J. L., Jones A. M., Serazetdinova L., Saalbach G., Bibb M. J., Naldrett M. J. (2010) Analysis of the phosphoproteome of the multicellular bacterium Streptomyces coelicolor A3 (2) by protein/peptide fractionation, phosphopeptide enrichment and high-accuracy mass spectrometry. Proteomics 10, 2486–2497 [DOI] [PubMed] [Google Scholar]
- 67. Manteca A., Ye J., Sanchez J., Jensen O. N. (2011) Phosphoproteome analysis of Streptomyces development reveals extensive protein phosphorylation accompanying bacterial differentiation. J. Proteome Res. 10, 5481–5492 [DOI] [PubMed] [Google Scholar]
- 68. Lin M. H., Hsu T. L., Lin S. Y., Pan Y. J., Jan J. T., Wang J. T., Khoo K. H., Wu S. H. (2009) Phosphoproteomics of Klebsiella pneumoniae NTUH-K2044 reveals a tight link between tyrosine phosphorylation and virulence. Mol. Cell. Proteomics 8, 2613–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schmidl S. R., Gronau K., Pietack N., Hecker M., Becher D., Stülke J. (2010) The phosphoproteome of the minimal bacterium Mycoplasma pneumoniae analysis of the complete known ser/thr kinome suggests the existence of novel kinases. Mol. Cell. Proteomics 9, 1228–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ge R., Sun X., Xiao C., Yin X., Shan W., Chen Z., He Q. Y. (2011) Phosphoproteome analysis of the pathogenic bacterium Helicobacter pylori reveals over representation of tyrosine phosphorylation and multiply phosphorylated proteins. Proteomics 11, 1449–1461 [DOI] [PubMed] [Google Scholar]
- 71. Prisic S., Dankwa S., Schwartz D., Chou M. F., Locasale J. W., Kang C. M., Bemis G., Church G. M., Steen H., Husson R. N. (2010) Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. U. S. A. 107, 7521–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Misra S. K., Milohanic E., Aké F., Mijakovic I., Deutscher J., Monnet V., Henry C. (2011) Analysis of the serine/threonine/tyrosine phosphoproteome of the pathogenic bacterium Listeria monocytogenes reveals phosphorylated proteins related to virulence. Proteomics 11, 4155–4165 [DOI] [PubMed] [Google Scholar]
- 73. Bai X., Ji Z. (2012) Phosphoproteomic investigation of a solvent producing bacterium Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 1–11 [DOI] [PubMed] [Google Scholar]
- 74. Lasonder E., Green J. L., Camarda G., Talabani H., Holder A. A., Langsley G., Alano P. (2012) The Plasmodium falciparum schizont phosphoproteome reveals extensive phosphatidylinositol and cAMP-protein kinase A signaling. J. Proteome Res. 11, 5323–5337 [DOI] [PubMed] [Google Scholar]
- 75. Yeh Y. M., Huang K. Y., Richie Gan R. C., Huang H. D., Wang T. C. V., Tang P. (2013) Phosphoproteome profiling of the sexually transmitted pathogen Trichomonas vaginalis. J. Microbiol. Immunol. Infect. 46, 366–373 [DOI] [PubMed] [Google Scholar]
- 76. Takahata Y., Inoue M., Kim K., Iio Y., Miyamoto M., Masui R., Ishihama Y., Kuramitsu S. (2012) Close proximity of phosphorylation sites to ligand in the phosphoproteome of the extreme thermophile Thermus thermophilus HB8. Proteomics 12, 1414–1430 [DOI] [PubMed] [Google Scholar]
- 77. Hu C. W., Lin M. H., Huang H. C., Ku W. C., Yi T. H., Tsai C. F., Chen Y. J., Sugiyama N., Ishihama Y., Juan H. F., Wu S. H. (2012) Phosphoproteomic analysis of Rhodopseudomonas palustris reveals the role of pyruvate phosphate dikinase phosphorylation in lipid production. J. Proteome Res. 11, 5362–5375 [DOI] [PubMed] [Google Scholar]
- 78. Marchini F. K., De Godoy L. M., Rampazzo R. C., Pavoni D. P., Probst C. M., Gnad F., Mann M., Krieger M. A. (2011) Profiling the Trypanosoma cruzi phosphoproteome. PLoS ONE 6, e25381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Treeck M., Sanders J. L., Elias J. E., Boothroyd J. C. (2011) The Phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii Reveal Unusual Adaptations Within and Beyond the Parasites' Boundaries. Cell Host Microbe 10, 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mayank P., Grossman J., Wuest S., Boisson-Dernier A., Roschitzki B., Nanni P., Nühse T., Grossniklaus U. (2012) Characterization of the phosphoproteome of mature Arabidopsis pollen. Plant J. 72, 89–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.